- 1Department of Urology, The First Affiliated Hospital of Dalian Medical University, Dalian, Liaoning, China
- 2Department of Neurology, The First Affiliated Hospital of Dalian Medical University, Dalian, Liaoning, China
- 3Department of Andrology, The Second Affiliated Hospital of Dalian Medical University, Dalian, Liaoning, China
Objective: Studies have found that gut microbiota may be associated with the development of erectile dysfunction (ED); however, the exact link between the two remains unclear. This study aimed to elucidate the relationship between the gut microbiota and the risk of ED from a genetic perspective.
Methods: We investigated the relationship between the gut microflora and ED using two-sample Mendelian randomization. GWAS-pooled data for ED were obtained from 223805 participants in Europe. GWAS summary data for ED were obtained from 223805 subjects in Europe and that for the gut microbiota were obtained from 18340 participants in 24 cohorts. We used the inverse-variance weighted (IVW) estimator as the primary method for the preliminary analysis, and the MR-Egger, weighted median (WM), simple model, and weighted model as secondary methods. We used Cochrane’s Q-test, to detect heterogeneity, MREgger to detect pleiotropy, and the leave-one-out method to test the stability of the MR results. Ultimately, we genetically predicted a causal relationship between 211 gut microbiota and ED.
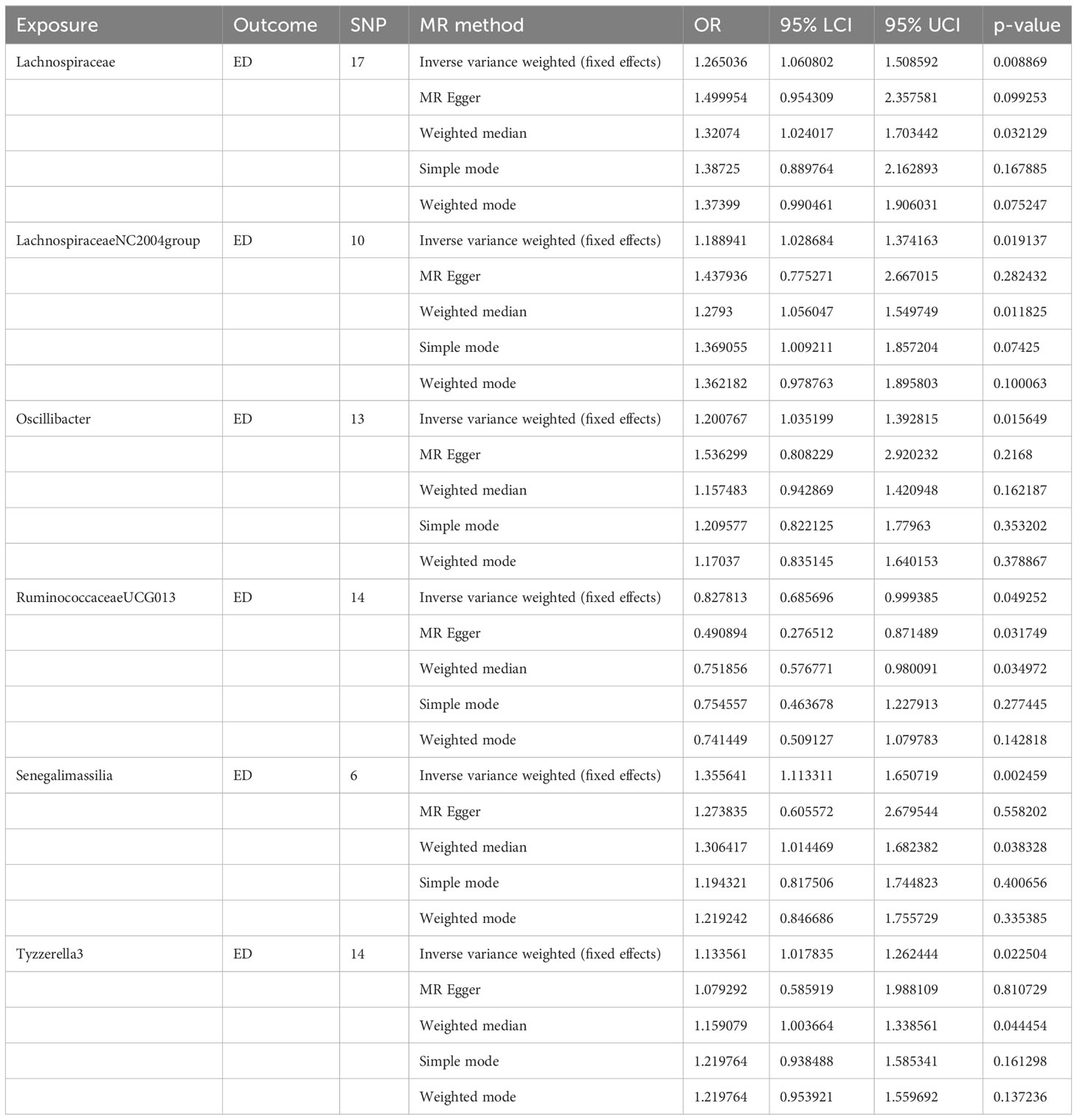
Results: A total of 2818 SNPs associated with gut microflora were screened in the ED correlation analysis based on the assumption of instrumental variables. The results of MR analysis showed a causal relationship between the six gut microbes and ED occurrence. The results of the fixed effects IVW method revealed five gut microflora, including Lachnospiraceae (OR, 1.265; P = 0.008), Lachnospiraceae NC2004 group (OR, 1.188; P = 0.019), Oscillibacter (OR, 1.200; P = 0.015), Senegalimassilia (OR, 1.355; P = 0.002), Tyzzerella3 (OR, 1.133; P = 0.022), to be negatively associated with ED. In addition, the IVW method revealed Ruminococcaceae UCG-013 (OR, 0.827; P = 0.049) to be positively associated with ED. Quality control results showed no heterogeneity or horizontal pleiotropy in the MR analysis (P > 0.05).
Conclusions: Six gut microbes were genetically associated with ED; of which, Ruminococcaceae UCG-013 was causally associated with a reduced risk of ED development. Our findings provide a new direction for research on the prevention and treatment of ED; however, the mechanisms and details require further investigation.
1 Introduction
ED is a global health problem that seriously affects the physical and mental health of patients and their sexual partners, leading to depression, anxiety, and other psychiatric disorders, as well as affecting the sexual harmony of couples and family stability. Epidemiological surveys have shown that approximately 150 million males worldwide have varying degrees of ED, and the number of people with ED is estimated to increase to 300 million by 2025 (1–3). Inflammation plays an important role in ED development. Damaged endothelial cells stimulate an inflammatory response in the vessel wall by increasing the production of inflammatory factors and cell adhesion molecules, leading to the formation of atheromatous plaques in penile blood vessels (4, 5).
The intestinal flora is involved in regulating the metabolic, immune, endocrine, neurological, and other local and systemic physiological processes of the human body through its metabolites and derivatives, which greatly expand the metabolic capacity of the human body and is called the “second brain” (6). An impaired intestinal barrier and bacterial translocation caused by intestinal flora dysbiosis can promote the production of inflammatory cytokines and lead to systemic inflammation, which may further accelerate the progression of ED and cardiovascular diseases. Previous studies that targeted the intestinal flora to investigate the pathogenesis of ED were mainly based on observational cross-sectional analyses, and such results may be invalidated by confounding factors and cannot accurately reflect the causal relationship between intestinal flora and ED (7, 8).
Mendelian randomization was used to assess causality. The purpose was to estimate the causal relationship between exposure and outcome by modeling instrumental variables (IV) with genetic variants (such as single nucleotide polymorphisms [SNPs]) that are strongly associated with exposure (9). According to Mendel’s law of inheritance “parental alleles are randomly assigned to offspring,” the process of gamete formation in MR is therefore similar to a “natural” randomized controlled trial, largely excluding the interference of unobserved confounding factors. MR is widely used as an efficient and accurate method to investigate the causal relationship between health risk factors and disease outcomes (10–12).
In this study, we performed a two-sample MR analysis to investigate the causal relationship between the gut microflora and ED risk.
2 Methods
2.1 Study design
To explore the relationship between the gut microflora and ED at the genetic level, a two-sample Mendelian randomization approach was used in this study.
2.2 Data source
GWAS summary statistics for the gut microbiota were obtained from a large GWAS study by Kurilshikov et al. (MiBioGen Consortium, www.MiBioGen.org), which analyzed 18340 individuals from 24 cohorts and recorded 211 gut microbiota and 122110 associated SNPs (13). Summary statistics for ED were downloaded from the MRC IEU OpenGWAS dataset (GWAS ID:ebi-a-GCST006956) as outcome variables. This GWAS included data from 6175 patients and 217630 controls (14).
2.3 Selection of instrumental variables
As instrumental variables, three basic requirements need to be met: first, genetic variants must be strongly associated with exposure (in this case, gut microbiota); second, these genetic variants must be independent of any confounding factors in the exposure-outcome association; and third, genetic variants should not affect the outcome (in this case, ED) unless an association through exposure is possible.
To investigate the above hypothesis, the following conditions were met for SNP screening: first, SNPs were extracted from GWAS data related to gut microbiota (that is, exposure) using P < 1 × 10−5 as a screening criterion to demonstrate a strong association with exposure. Second, SNPs containing linkage disequilibrium (LD) were removed by running the ‘TwoSampleMR’ package with r2 = 0.001 and kb = 10000. Third, we searched the ED-related database for the SNPs corresponding to the gut microflora and the non-corresponding or palindromic SNPs were deleted. After the initial screening of SNPs, we calculated the F-value of each IV to exclude bias caused by weak IVs. IVs were considered weak when the F-value was greater than 10.
2.4 Estimation of causal effects
We used the inverse variance weighted method (IVW) to estimate the causal effect between the gut microbiota and ED. When heterogeneity was present, the random-effects IVW method was adopted. When no heterogeneity was observed, the fixed effects IVW method was used. In addition, we performed additional analyses using MR-Egger, weighted median method, simple model, and weighted model. Sufficient evidence of a causal effect was consistently provided by statistically significant IVW results and the direction of the results across all five analyses.
2.5 Quality controls
Heterogeneity refers to the presence of variability among the included studies. We used IVW model estimation to calculate heterogeneity and Q test for heterogeneity and P > 0.05 was used to indicate the absence of heterogeneity among the IVs. Horizontal pleiotropy refers to the phenomenon by which IVs influence outcomes through pathways other than exposure and is a potential source of bias. We examined the pleiotropy of the IVS using the MR-Egger regression method when P value was >0.05, which represented the absence of horizontal pleiotropy between the IVs and outcomes. Confounding factors may affect the results through multiple mechanisms dominated by the inclusion of genetic variants. Therefore, we used leave-one-out sensitivity analysis for IVs to determine whether the presence of a single SNP strongly influenced the results of the two-sample Mendelian randomization analysis.
The statistical analysis in this study was conducted using RStudio software (version 4.1.2). The resources used were mainly obtained from the TwoSampleMR R package developed by Hemani et al.
3 Results
3.1 Screening and validation of IVs
We screened the instrumental variables for 211 microflora and obtained 5,548,018 exposure-related instrumental variables. After removing chain imbalance effects, excluding variables weakly associated with exposure factors (F < 10), and using the online analysis software PhenoScanner to exclude variables that might be associated with outcome confounders (15), 2818 IVs from 211 microflora were finally included in the analysis.
3.2 Calculation of causal effects
Six causal associations between gut microbiota and ED risk were identified in this study. A higher genetically predicted family Lachnospiraceae, genus Lachnospiraceae NC2004 group, genus Oscillibacter, genus Senegalimassilia, genus Tyzzerella3 were associated with a higher risk of ED. In contrast, the genus Ruminococcaceae UCG-013 was associated with a lower risk.
3.2.1 Lachnospiraceae
Through a series of quality controls, we obtained 17 SNPs as IVs for the MR analysis of Lachnospiraceae and ED, with one palindromic SNP (rs11755180) removed during quality control. Information on the instrumental variables is provided in Supplementary Table 1.
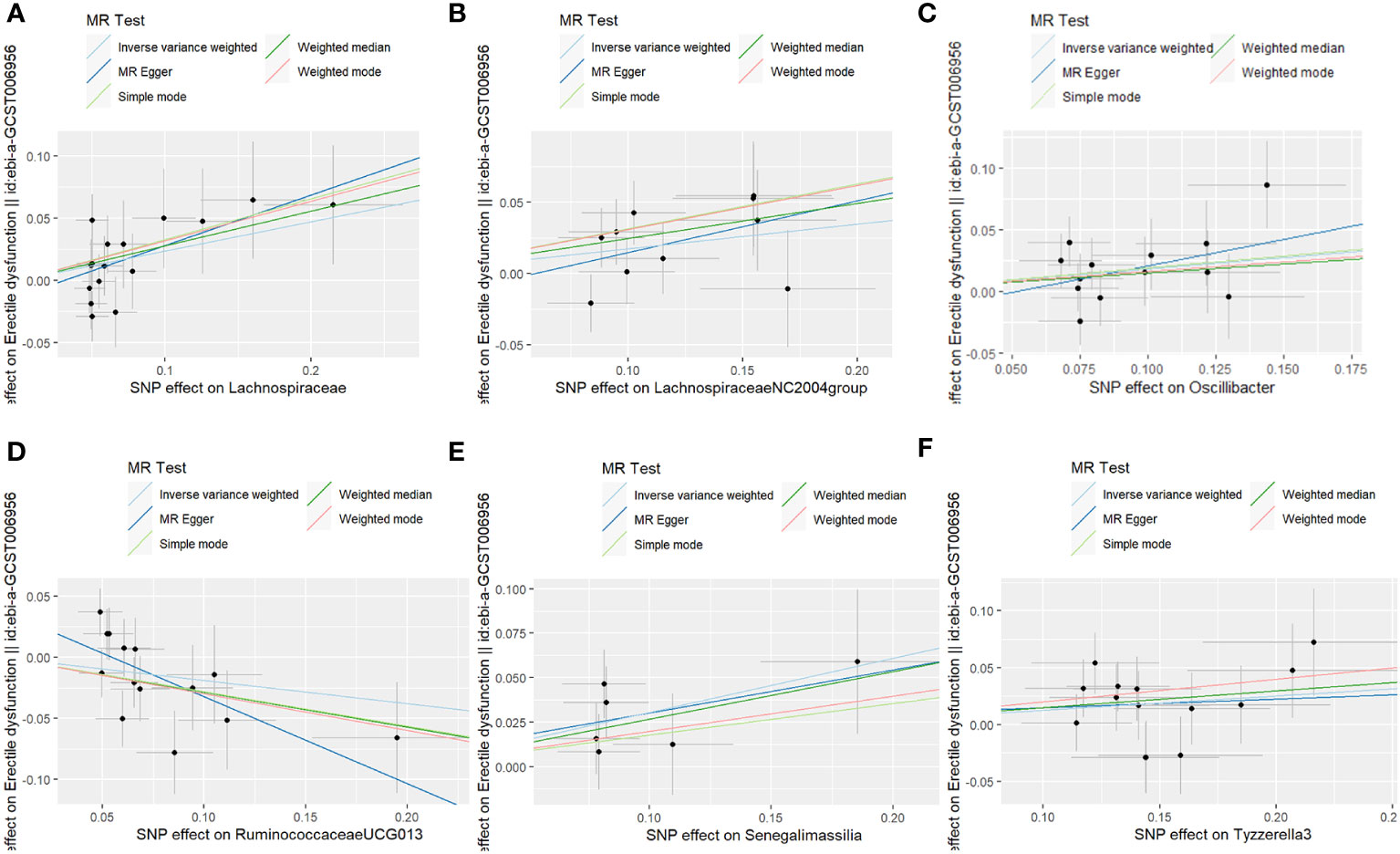
The fixed-effects IVW results indicated that Lachnospiraceae (P = 0.008; OR, 1.265) negatively correlated with ED. The weighted median analysis showed similar results (P = 0.032; OR, 1.320). MR-Egger and simple model analyses indicated that Lachnospiraceae was not genetically causally related to ED (P > 0.05). The results of this analysis are presented in Table 1 and Figure 1.
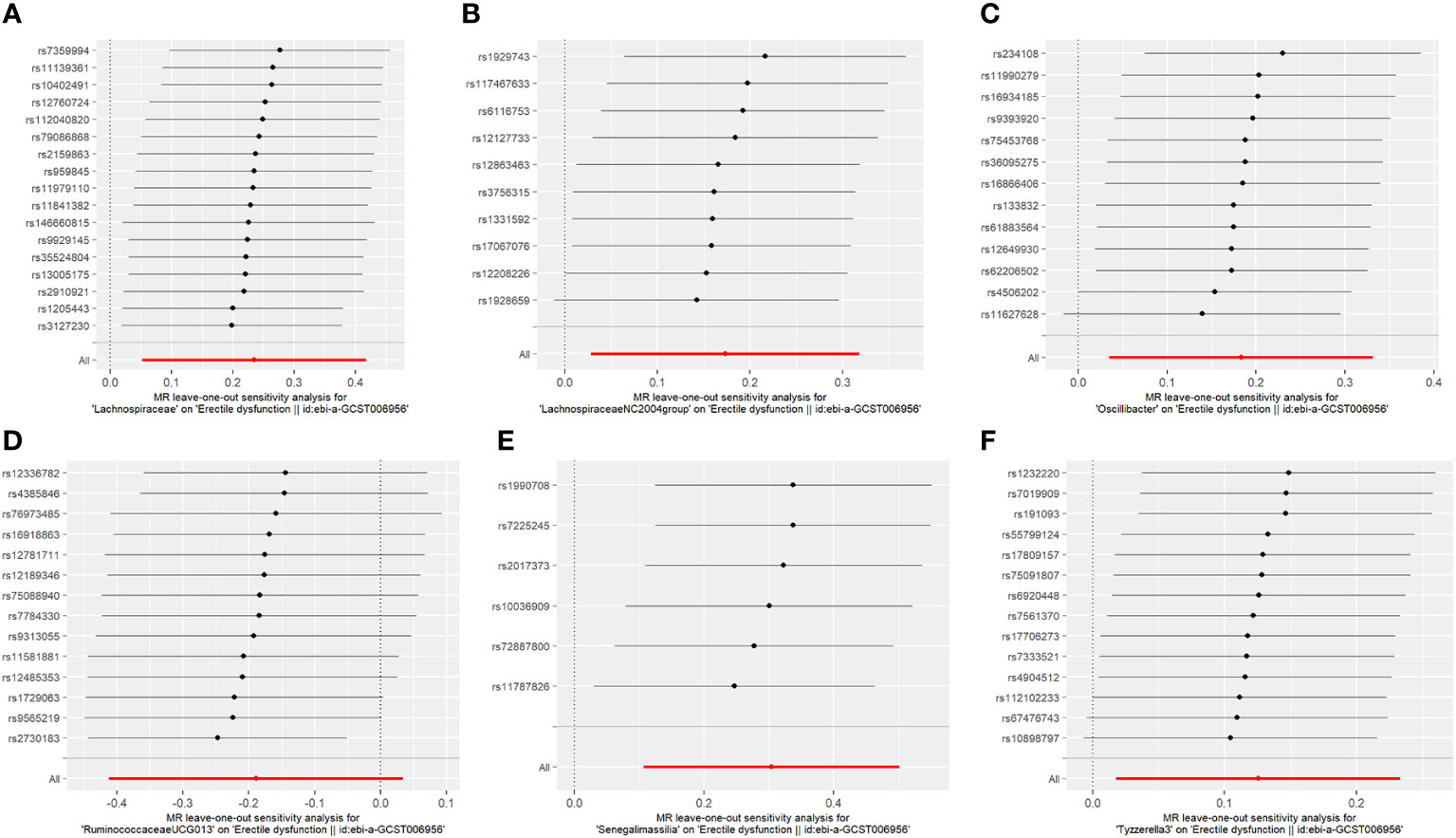
The results of the MR-IVW test showed no heterogeneity in the results of our MR analysis (P = 0.347, Q = 16.534). The results of the MR-Egger test for horizontal multiplicity indicated no horizontal multiplicity in our MR analysis (P = 0.431, SE = 0.015). The results of the sensitivity analysis are shown in Figure 2, which indicates that there was no particular SNP that would change the results, and that the final result was stable.
3.2.2 LachnospiraceaeNC2004group
A total of 10 SNPs were included in the MR analysis of the Lachnospiraceae NC2004 group and ED, with no palindromic SNPs removed (Supplementary Table 2). The fixed-effects IVW results indicated that Lachnospiraceae (P = 0.019; OR, 1.188) negatively correlated with ED. The results for w were similar to those for IVW (P = 0.011; OR, 1.279). MR-Egger and simple model analyses indicated that Lachnospiraceae was not genetically causally related to ED (P > 0.05). MR-IVW and Q tests revealed the absence of horizontal pleiotropy and heterogeneity. No single SNP outliers were identified using the leave-one-out method.
3.2.3 Oscillibacter
Fourteen SNPs were included in the analysis after quality control, including three palindromic SNPs (rs12417956, rs137917150, and rs6901560) (Supplement Table 3). The fixed-effects IVW results indicated that Oscillibacter (P = 0.015; OR, 1.200) had a negative causal relationship with ED. The other four analysis methods indicated that Oscillibacter was not causally related to ED (P > 0.05). No significant heterogeneity or horizontal pleiotropy was found according to the Cochrane’s Q and MR-Egger tests (Supplement Table 7). When the SNPs were removed individually, the results remained stable (Figure 2C).
3.2.4 Ruminococcaceae UCG-013
Fourteen SNPs were included in the analysis after quality control, with one palindromic SNP (rs2428106) removed (Supplement Table 4). Fixed effects IVW, MR-Egger, and weighted median results indicated that Ruminococcaceae UCG-013 had a positive causal relationship with ED. The simple and weighted models did not consider the relationship between Ruminococcaceae UCG-013 and ED. Quality control analysis revealed no significant heterogeneity or horizontal pleiotropy.
3.2.5 Senegalimassilia
Six SNPs were included in the analysis after quality control, including two palindromic SNPs (rs13383270 and rs57512504) (Supplementary Table 5). The results of the fixed effects IVW, MR-Egger, and weighted median analyses indicated that Senegalimassilia had a negative causal relationship with ED (P = 0.002 vs. P = 0.038). The MR-Egger, simple, and weighted models showed no association with ED. No significant heterogeneity or horizontal pleiotropy was found in quality control.
3.2.6 Tyzzerella3
After quality control, 14 SNPs were included in the analysis. No palindromic SNPs were identified (Supplementary Table 6). Using the IVW method, Tyzzerella3 was found to have a negative genetic association with ED (P = 0.022; OR=1.133). The other four analytical methods indicated that Tyzzerella3 had no causal genetic relationship with ED. In addition, the results of the MR-Egger tests confirmed that there was no horizontal pleiotropy (P = 0.875, SE = 0.045), and the results of the Cochrane Q-test showed that there was no notable heterogeneity among the selected SNPs (P = 0.584, Q = 11.312). In addition, no outlier SNPs were identified after applying the leave-one-out method.
4 Discussion
Common risk factors for ED include age, diabetes, dyslipidemia, hypertension, cardiovascular disease, obesity, metabolic syndrome, hyperhomocysteinaemia, physical inactivity, and smoking (16–18). The intestinal microflora is closely related to the development, growth, and metabolism of the host, and a stable intestinal flora environment plays an important role in regulating the immune response of the body (19). The composition and metabolites of intestinal flora can be influenced by external environmental factors. If the normal intestinal environment is disturbed, the gut is unable to function as an antimicrobial agent and the release of inflammatory factors in the gut increases intestinal permeability, leading to increased levels of inflammation and oxidative stress, and this proinflammatory state leads to endothelial cell dysfunction (20). It is common in the pathogenesis of cardiovascular diseases and type 2 diabetes, and the development of ED is closely linked to the pathology of these diseases. Therefore, we hypothesized that an association exists between the intestinal flora and its metabolites and the development of ED.
To our knowledge, this is the first large-scale, comprehensive MR study to examine the causative role of gut microbes in ED. The advantage of two-sample Mendelian randomization studies over traditional single-sample studies is that they can be analyzed using aggregated GWAS data, thus extending the range of available data (21–23). According to the core principles of Mendelian randomization, studies conducted in this manner can largely avoid the effects of reverse causality and can effectively avoid bias from confounding factors that cannot be controlled for in traditional observational studies of the ED (including smoking, alcohol consumption, and obesity associated with the ED). The quality control results showed no heterogeneity or horizontal multiplicity for any outcome. Based on the principle of MR method selection, that is, preferentially using IVW fixed-effects model estimates in the absence of heterogeneity and multiplicity, and preferentially using results calculated using the MR-Egger method in the presence of multiplicity, we finally adopted the IVW fixed-effects model estimates as the primary study outcomes (24). We identified six microbial communities associated with ED occurrence; of which, five (Lachnospiraceae, Lachnospiraceae NC2004 group, Oscillibacter, Senegalimassilia, and Tyzzerella3) were associated with an increased risk of ED, and one (Ruminococcaceae UCG-013) was associated with a decreased risk of ED.
At the family level, Lachnospiraceae, which colonizes the intestinal lumen from birth and increases in number, is part of the core gut microbiota. Several studies have shown that a high abundance of Lachnospiraceae positively correlates with glucose and/or lipid metabolism, indicating metabolic disorders (25, 26). Metabolic syndromes, including arterial hypertension, insulin resistance, and hypertriglyceridemia, significantly increase the risk of developing diabetes and cardiovascular diseases (27) and are the risk factors for ED. In a study by Zhang et al., a significant increase in Lachnospiraceae was observed in the feces of hyperlipidemic mice (28). One study reported that different OTUs of Lachnospiraceae were associated with changes in lipid metabolism and specific nutrients such as saturated and total fats, and thus obesity (29). In addition, a positive correlation exists between the different taxa of Lachnospiraceae (particularly Anaerostipes, Bhatia, Dorea, and Lachnospiraceae incertae sedis) and major depression (30–32), whereas depression and other emotional or psychiatric disorders have been shown to trigger ED (33, 34). Therefore, we speculate that Lachnospiraceae may increase the risk of ED by affecting blood lipid levels and interfering with neurotransmission in the central nervous system (35).
In addition, at the genus level, Oscillibacter, Tyzzerella 3, and Senegalimus ssilia were associated with an increased risk of ED development. Barandouzi et al. found an increased abundance of Oscillibacter in the feces of patients with depression (36). Kelly et al. found that an increased abundance of Tyzzerella 4 may be a risk factor for cardiovascular disease progression (37). Wang et al. found that Senegalimassilia spp. was associated with increased blood pressure (38). These findings suggest that gut microbes have important effects on vascular endothelial function, depression, and cognitive function in humans, thereby increasing the risk of ED development. Interestingly, gut flora may play different roles in different ethnic groups. Liu et al. analyzed the relationship between the gut microbiome and blood metabolites in an Asian population and found that Oscillibacter was causally linked to decreased triglyceride concentrations (39), suggesting that Oscillibacter may play a role in reducing ED risk in Asian populations; however, these results need to be validated in future studies.
Our genetic prediction-based study also found a strong causal relationship between Ruminococcaceae and reduced risk of ED (OR, 1.13; 95% CI, 1.02–1.25; P = 0.02). Combined with the results of previous studies, the targeted modulation of bacterial abundance appears to be a novel approach for reducing the risk of ED. Feng et al. found that Ruminococcaceae_UCG-013 was more abundant in the gut microbes of lean mice than in obese mice. Further analysis revealed that Ruminococcaceae UCG-013 was positively correlated with serum HDL-C levels and negatively correlated with serum TC, TG, and LDL-C levels. This suggests that an increased abundance of Ruminococcaceae lowers total blood cholesterol levels, thereby reducing the risk of ED (40).
Hyperhomocysteinemia is an emerging risk factor for ED. Folic acid and vitamin B12 supplements are commonly used in clinical practice to reduce HCY levels (41, 42). Studies have shown that some gut microbiota, such as red bacteria and bifidobacteria, can synthesize B vitamins and folic acid, which, to some extent, complements the human body’s requirements for folic acid and B vitamins. We speculate that gut microbiota may also influence the incidence of ED.
In addition, the current treatment of ED mainly includes oral drugs, psychotherapy and other methods. Although there are many treatment methods for ED, there are also problems such as the treatment rate is difficult to further improve. At present, probiotics and fecal bacteria transplantation and other treatment methods related to intestinal flora have been used to regulate glucose and lipid metabolism and improve vascular endothelial injury. Therefore, the treatment of ED by regulating the intestinal flora is a potential therapeutic method in the future, which is worthy of further exploration. Our results provide a potential target for the treatment of ED by regulating the intestinal flora. In the follow-up, we will further explore the specific mechanism of these six flora in ED.
This study has some limitations. First, the data in this study were all from European populations, and whether the conclusions can be generalized to other populations requires further validation. Second, this study was only a statistical result and could not further explore the biological mechanism between the gut microbiota and ED. Third, this study lacked data from subgroups stratified by age, previous surgical information and hormone levels and could not compare the causal effect differences between subgroups.
5 Conclusion
This study assessed the causal relationship between the gut microbiota and ED in 211 people using two-sample Mendelian randomization. The results showed that Ruminococcaceae UCG-013 may reduce the risk of ED. Lachnospiraceae and Tyzzerella3 may increase the risk of ED. Our study provides a new research direction for preventing and treating ED. We plan to further investigate the specific relationship between these gut microbes and ED and explore their potential value in the pathogenesis of ED.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
This study was partly based on the publicly available data. It did not include interaction with humans or use personal identifying information. Thus, the informed consent for this study was not required.
Author contributions
Conceptualization: QT. Data curation: QT, QS. Formal analysis: QS. Funding acquisition: QT, QS. Methodology: QT, LZ. Supervision: QT, TJ. Validation: KW, YYL. Writing-original draft: QS. Writing – review & editing: TJ, YXL. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
All authors would like to acknowledge MiBioGen Consortium for their selfless public sharing of GWAS summary data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1216746/full#supplementary-material
References
1. Laumann EO, Paik A, Rosen RC. Sexual dysfunction in the United States: prevalence and predictors. JAMA (1999) 281(6):537–44. doi: 10.1001/jama.281.6.537
2. Pathak RA, Broderick GA. Color doppler duplex ultrasound parameters in men without organic erectile dysfunction. Urology (2020) 135:66–70. doi: 10.1016/j.urology.2019.09.002
3. Salonia A, Bettocchi C, Boeri L, Capogrosso P, Carvalho J, Cilesiz NC, et al. European association of urology guidelines on sexual and reproductive health-2021 update: male sexual dysfunction. Eur Urol (2021) 80(3):333–57. doi: 10.1016/j.eururo.2021.06.007
4. Luboshitzky R, Aviv A, Hefetz A, Herer P, Shen-Orr Z, Lavie L, et al. Decreased pituitary-gonadal secretion in men with obstructive sleep apnea. J Clin Endocrinol Metab (2002) 87(7):3394–8. doi: 10.1210/jcem.87.7.8663
5. Matos G, Hirotsu C, Alvarenga TA, Cintra F, Bittencourt L, Tufik S, et al. The association between TNF-alpha and erectile dysfunction complaints. Andrology (2013) 1(6):872–8. doi: 10.1111/j.2047-2927.2013.00136.x
6. Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med (2018) 24(4):392–400. doi: 10.1038/nm.4517
7. Okamoto T, Hatakeyama S, Imai A, Yamamoto H, Yoneyama T, Mori K, et al. The association between gut microbiome and erectile dysfunction: a community-based cross-sectional study in Japan. Int Urol Nephrol (2020) 52(8):1421–8. doi: 10.1007/s11255-020-02443-9
8. Russo GI, Bongiorno D, Bonomo C, Musso N, Stefani S, Sokolakis I, et al. The relationship between the gut microbiota, benign prostatic hyperplasia, and erectile dysfunction. Int J Impot Res (2022) 35(4):350–5. doi: 10.1038/s41443-022-00569-1
9. Birney E. Mendelian randomization. Cold Spring Harb Perspect Med (2022) 12(4). doi: 10.1101/cshperspect.a041302
10. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet (2014) 23(R1):R89–98. doi: 10.1093/hmg/ddu328
11. Wang M, Jian Z, Gao X, Yuan C, Jin X, Li H, et al. Causal associations between educational attainment and 14 urological and reproductive health outcomes: A mendelian randomization study. Front Public Health (2021) 9:742952. doi: 10.3389/fpubh.2021.742952
12. Zhang S, Xie X, Yu L, Jiang N, Wei X, Hu Y. Investigating causal relations between genetic-related intermediate endophenotype and risk of chronic prostatitis: mendelian randomization study. Oxid Med Cell Longev (2022) 2022:4560609. doi: 10.1155/2022/4560609
13. Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet (2021) 53(2):156–65. doi: 10.1038/s41588-020-00763-1
14. Bovijn J, Jackson L, Censin J, Chen CY, Laisk T, Laber S, et al. GWAS identifies risk locus for erectile dysfunction and implicates hypothalamic neurobiology and diabetes in etiology. Am J Hum Genet (2019) 104(1):157–63. doi: 10.1016/j.ajhg.2018.11.004
15. Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics (2016) 32(20):3207–9. doi: 10.1093/bioinformatics/btw373
16. Nguyen HMT, Gabrielson AT, Hellstrom WJG. Erectile dysfunction in young men-A review of the prevalence and risk factors. Sex Med Rev (2017) 5(4):508–20. doi: 10.1016/j.sxmr.2017.05.004
17. Terentes-Printzios D, Ioakeimidis N, Rokkas K, Vlachopoulos C. Interactions between erectile dysfunction, cardiovascular disease and cardiovascular drugs. Nat Rev Cardiol (2022) 19(1):59–74. doi: 10.1038/s41569-021-00593-6
18. Defeudis G, Mazzilli R, Tenuta M, Rossini G, Zamponi V, Olana S, et al. Erectile dysfunction and diabetes: A melting pot of circumstances and treatments. Diabetes Metab Res Rev (2022) 38(2):e3494. doi: 10.1002/dmrr.3494
19. Zhang CX, Wang HY, Chen TX. Interactions between intestinal microflora/probiotics and the immune system. BioMed Res Int (2019) 2019:6764919. doi: 10.1155/2019/6764919
20. Xu Q, Ni JJ, Han BX, Yan SS, Wei XT, Feng GJ, et al. Causal relationship between gut microbiota and autoimmune diseases: A two-sample mendelian randomization study. Front Immunol (2021) 12:746998. doi: 10.3389/fimmu.2021.746998
21. Bowden J, Holmes MV. Meta-analysis and Mendelian randomization: A review. Res Synth Methods (2019) 10(4):486–96. doi: 10.1002/jrsm.1346
22. Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol (2003) 32(1):1–22. doi: 10.1093/ije/dyg070
23. Sekula P, Del Greco MF, Pattaro C, Kottgen A. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol (2016) 27(11):3253–65. doi: 10.1681/ASN.2016010098
24. Sproviero W, Winchester L, Newby D, Fernandes M, Shi L, Goodday SM, et al. High blood pressure and risk of dementia: A two-sample mendelian randomization study in the UK biobank. Biol Psychiatry (2021) 89(8):817–24. doi: 10.1016/j.biopsych.2020.12.015
25. Lippert K, Kedenko L, Antonielli L, Kedenko I, Gemeier C, Leitner M, et al. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef Microbes (2017) 8(4):545–56. doi: 10.3920/BM2016.0184
26. Salonen A, Lahti L, Salojarvi J, Holtrop G, Korpela K, Duncan SH, et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J (2014) 8(11):2218–30. doi: 10.1038/ismej.2014.63
27. Kachur S, Morera R, De Schutter A, Lavie CJ. Cardiovascular risk in patients with prehypertension and the metabolic syndrome. Curr Hypertens Rep (2018) 20(2):15. doi: 10.1007/s11906-018-0801-2
28. Zhang W, Zhang QY, Wang JJ, Zhang LL, Dong ZZ. Efficiency assessment of bacterial cellulose on lowering lipid levels in vitro and improving lipid metabolism in vivo. Molecules (2022) 27(11). doi: 10.3390/molecules27113495
29. De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut (2016) 65(11):1812–21. doi: 10.1136/gutjnl-2015-309957
30. Zheng P, Zeng B, Zhou C, Liu M, Fang Z, Xu X, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry (2016) 21(6):786–96. doi: 10.1038/mp.2016.44
31. Cheung SG, Goldenthal AR, Uhlemann AC, Mann JJ, Miller JM, Sublette ME. Systematic review of gut microbiota and major depression. Front Psychiatry (2019) 10:34. doi: 10.3389/fpsyt.2019.00034
32. Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y, et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun (2015) 48:186–94. doi: 10.1016/j.bbi.2015.03.016
33. Bozman AW, Beck JG. Covariation of sexual desire and sexual arousal: the effects of anger and anxiety. Arch Sex Behav (1991) 20(1):47–60. doi: 10.1007/BF01543007
34. Seidman SN. Exploring the relationship between depression and erectile dysfunction in aging men. J Clin Psychiatry (2002) 63 Suppl 5:5–12.
35. Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry (2013) 18(6):666–73. doi: 10.1038/mp.2012.77
36. Barandouzi ZA, Starkweather AR, Henderson WA, Gyamfi A, Cong XS. Altered composition of gut microbiota in depression: A systematic review. Front Psychiatry (2020) 11:541. doi: 10.3389/fpsyt.2020.00541
37. Kelly TN, Bazzano LA, Ajami NJ, He H, Zhao J, Petrosino JF, et al. Gut microbiome associates with lifetime cardiovascular disease risk profile among bogalusa heart study participants. Circ Res (2016) 119(8):956–64. doi: 10.1161/CIRCRESAHA.116.309219
38. Wang B, Liu J, Lei R, Xue B, Li Y, Tian X, et al. Cold exposure, gut microbiota, and hypertension: A mechanistic study. Sci Total Environ (2022) 833:155199. doi: 10.1016/j.scitotenv.2022.155199
39. Liu X, Tong X, Zou Y, Lin X, Zhao H, Tian L, et al. Mendelian randomization analyses support causal relationships between blood metabolites and the gut microbiome. Nat Genet (2022) 54(1):52–61. doi: 10.1038/s41588-021-00968-y
40. Feng J, Ma H, Huang Y, Li J, Li W. Ruminococcaceae_UCG-013 promotes obesity resistance in mice. Biomedicines (2022) 10(12). doi: 10.3390/biomedicines10123272
41. Gupta A, Priyadarshi S, Vyas N, Sharma G, Swain PK. Novel predictive risk factor for Erectile Dysfunction: Serum folic acid. Andrologia (2021) 53(1):e13890. doi: 10.1111/and.13890
Keywords: erectile dysfunction, gut microbiota, Mendelian randomization, prevention, diagnosis
Citation: Su Q, Long Y, Luo Y, Jiang T, Zheng L, Wang K and Tang Q (2023) Specific gut microbiota may increase the risk of erectile dysfunction: a two-sample Mendelian randomization study. Front. Endocrinol. 14:1216746. doi: 10.3389/fendo.2023.1216746
Received: 09 June 2023; Accepted: 28 November 2023;
Published: 18 December 2023.
Edited by:
Sara Baldassano, University of Palermo, ItalyReviewed by:
Guiting Lin, University of California, San Francisco, United StatesConrad Leitsmann, Medical University of Graz, Austria
Copyright © 2023 Su, Long, Luo, Jiang, Zheng, Wang and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kenan Wang, d2FuZ2tlbmFuODZAaG90bWFpbC5jb20=; Qizhen Tang, bG5mc3RxekAxNjMuY29t
†These authors share first authorship
 Quanxin Su
Quanxin Su Yanxi Long1†
Yanxi Long1† Tao Jiang
Tao Jiang Qizhen Tang
Qizhen Tang