- 1Department of Genetics and Endocrinology, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangdong Provincial Clinical Research Center for Child Health, Guangzhou, China
- 2Center of Big Data and Business Itelligent, South China University of Technology, Guangzhou, China
Objective: Hyperthyroidism in Chinese children is relatively high and has been increasing in recent years, which has a significant impact on their healthy development. Hyperthyroidism is a polygenic disorder that presents greater challenges in terms of prediction and treatment than monogenic diseases. This study aims to elucidate the associated functions and gene sets of mutated genes in children with hyperthyroidism in terms of the gene ontology through GO enrichment analysis and in terms of biological signaling pathways through KEGG enrichment analysis, thereby enhancing our understanding of the expected effects of multiple mutated genes on hyperthyroidism in children.
Methods: Whole-exome sequencing was performed on the DNA samples of children with hyperthyroidism. Screening for pathogenic genes related to hyperthyroidism in affected children was performed using the publicly available disease databases Malacards, MutationView, and Clinvar, and the functions and influences of the identified pathogenic genes were analyzed using statistical analysis and the gene enrichment approach.
Results: Through GO enrichment analysis, it was found that the most significant gene ontology enrichment was the function “hormone activity” in terms of gene ontology molecular function. The corresponding mutated genes set that has common effects on hyperthyroidism in children included TG, CALCA, POMC, CGA, PTH, GHRL, FBN1, TRH, PRL, LEP, ADIPOQ, INS, GH1. The second most significant gene ontology enrichment was the function “response to peptide hormone” in terms of biological process. The corresponding mutated genes set that has common effects on hyperthyroidism in children included LRP6, TSC2, KANK1, COL1A1, CDKN1B, POMC, STAT1, MEN1, APC, GHRL, TSHR, GJB2, FBN1, GPT, LEP, ADIPOQ, INS, GH1. Through KEGG enrichment analysis, it was found that the most significant biological signaling pathway enrichment was the pathway “Thyroid hormone signaling pathway” function. The corresponding mutated genes set that has common effects on hyperthyroidism in children included NOTCH3, MYH7, TSC2, STAT1, MED13L, MAP2K2, SLCO1C1, SLC16A2, and THRB. The second most significant biological signaling pathway enrichment was the pathway “Hypertrophic cardiomyopathy” in terms of biological process. The corresponding mutated genes set that has common effects on hyperthyroidism in children included IGF1, CACNA1S, MYH7, IL6, TTN, CACNB2, LAMA2, and DMD.
Conclusion: The mutated genes in children with hyperthyroidism were closely linked to function involved in “hormone activity” and “response to peptide hormone” in terms of the biological signaling pathway, and to the functional pathways involved in “Thyroid hormone signaling pathway” and “Hypertrophic cardiomyopathy” in terms of the biological signaling pathway.
1 Introduction
Hyperthyroidism is characterized by the excessive synthesis and release of thyroid hormones, leading to increased metabolism and sympathetic nervous system activity, resulting in symptoms such as palpitations, sweating, increased appetite, increased bowel movements, and weight loss. Its hallmark is an increase in the synthesis and secretion of thyroid hormones and thyroxine (T4) and triiodothyronine (T3) (1). According to an epidemiological survey report released in 2021, the overall incidence rate of hyperthyroidism in mainland China is approximately 1.78% (2). The overall incidence of hyperthyroidism in European children and adolescents is approximately 4.58 per 10,000 per year, with a higher prevalence in girls than boys (3). Children with hyperthyroidism may experience thyroid enlargement, increased appetite with weight loss, bulging eyes, irritability, excessive sweating, tachycardia, hyperactivity, and tremors (4). They may also have comorbid conditions such as attention deficit hyperactivity disorder (ADHD), regulatory disorders, anxiety, bipolar disorder, depression, and an increased risk of suicide. Children with hyperthyroidism’s physical and mental health can be severely affected (5). Therefore, early and accurate diagnosis, precise treatment, and improved prognosis of pediatric hyperthyroidism are of great clinical significance.
Currently, more than 100 genes have been found to be related to the development of hyperthyroidism (according to the Malacards database, MutationView database, and ClinVar database). Among these genes, those that have received more attention include (1): the thyroid stimulating hormone receptor (TSHR) gene. TSHR gene mutations are one of the causes of non-autoimmune hyperthyroidism (6, 7), and neonatal non-autoimmune hyperthyroidism is associated with the TSHR gene c.1856A>G (p.Asp619Gly) heterozygous mutation (8); (2) the cytotoxic T-lymphocyte associated protein 4 (CTLA4) gene. CTLA-4 gene mutations in the Chinese Han population are related to the development of hyperthyroidism (9–11); (3) the thyroid globulin (TG) gene. Specific TG SNP haplotypes are associated with hyperthyroidism (12), and hyperthyroidism patients with the TG gene E33SNP C/C genotype are more likely to relapse after discontinuing medication (13); (4) the GNAS complex locus (GNAS) gene. Gene mutations in GNAS are found in approximately 70% of patients with autonomous thyroid adenomas (14), and non-autoimmune hyperthyroidism is also associated with GNAS mutations (15); (5) the thyroid hormone receptor beta (THRB) gene. THRB gene mutations are related to a tendency towards thrombosis in patients with hyperthyroidism (16).
Research on gene mutations related to hyperthyroidism has mainly focused on individual genes, such as the study of TSHR gene mutations in congenital non-autoimmune hyperthyroidism (17–19), the impact of novel heterozygous TSHR gene mutations on hyperthyroidism (20), and special attention paid to its effects on hyperthyroidism (21). The role of BAFF gene mutations in the pathogenesis of hyperthyroidism is also studied (22). Other studies have explored the relationship between hyperthyroidism and other related diseases, such as the analysis of KCNJ18 gene mutations in hyperthyroidism associated with hypokalemic periodic paralysis (23), the study of BRAF gene mutations in hyperthyroidism associated with thyroid papillary carcinoma (24), and the study of UGT1A1 gene mutations in hyperthyroidism associated with liver failure (25).
Studies on gene mutations in children with hyperthyroidism have also mainly focused on single genes, including TSHR gene mutation analysis in children with hyperthyroidism (26, 27) and its relationship with congenital non-autoimmune hyperthyroidism in newborns (28), as well as functional gain-of-function mutations in the TSHR gene leading to increased function and thyroid growth (29). Additionally, there have been studies on gene sets, such as the age-related HLADRB1*03 allele in the Polish population (30) and gene sets significantly associated with early-onset hyperthyroidism, including BTNL2, NOTCH4, TNFAIP3, and CXCR4 (31). Other recent studies on pediatric hyperthyroidism have focused on prevention (32, 33) and treatment options (34–39), as well as research on related clinical symptoms (40, 41).
In summary, the genetic research on hyperthyroidism, especially pediatric hyperthyroidism, has mainly focused on single genes that affect the development of the disease. There are relatively few studies analyzing the expected effects of multiple mutated genes in hyperthyroidism. Therefore, more comprehensive understanding of the disease and help promote healthy development for children and adolescents, it is necessary to conduct more in-depth research and analysis on the functional implications of mutated genes in pediatric hyperthyroidism.
2 Materials and methods
2.1 WES data generation and gene processing
The cases were obtained from the Guangzhou Women and Children’s Medical Center. After approval from the Center’s Ethics Committee and obtaining informed consent from the guardians of the children with hyperthyroidism, peripheral blood samples were collected from 39 children with hyperthyroidism and DNA was extracted.
Whole-exome sequencing was performed on the DNA samples with GRCh38 as the reference genome. The main steps of Whole-exome sequencing include data filtering, data quality control, map to reference, mark duplicates, InDel realignment, base recalibration, the second data quality control, variant calling, variant filtering and variant annotation. GATK was used to detect the SNP and InDel. The reads map rate of the resulting data is 99%, indicating that the selection of the reference sequence is more accurate, and the average sequencing depth is 150X, which is enough for analysis, and 99% reaches 4X coverage.
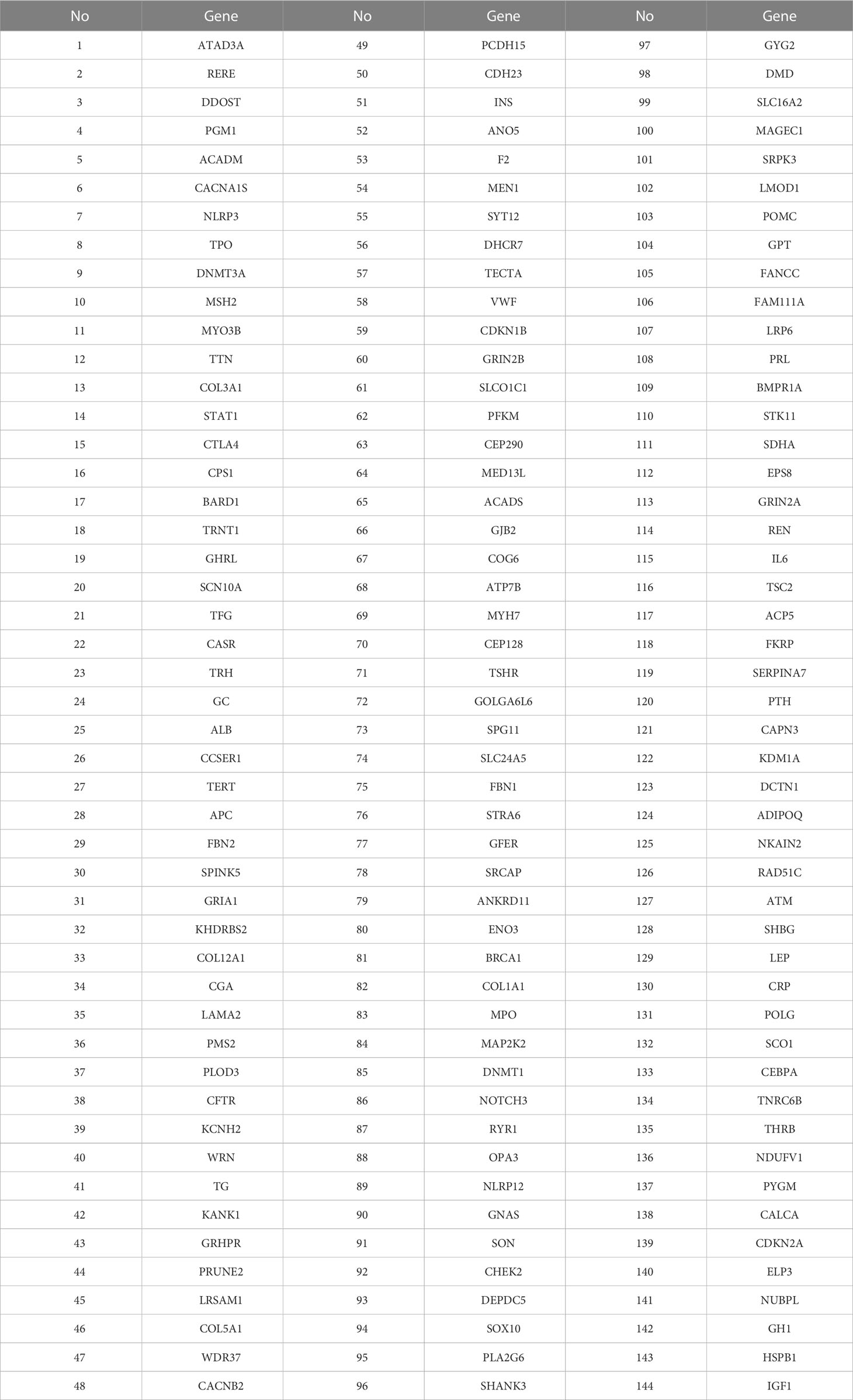
The Whole-exome sequencing data were screened. This study obtained genes related to hyperthyroidism by searching public disease databases such as Malacards, MutationView, and Clinvar, comparing them with the mutated genes of 39 patients, and a total of 144 genes related to hyperthyroidism were screened. The specific genes are shown in Table 1.
2.2 Clinical data of the children studied
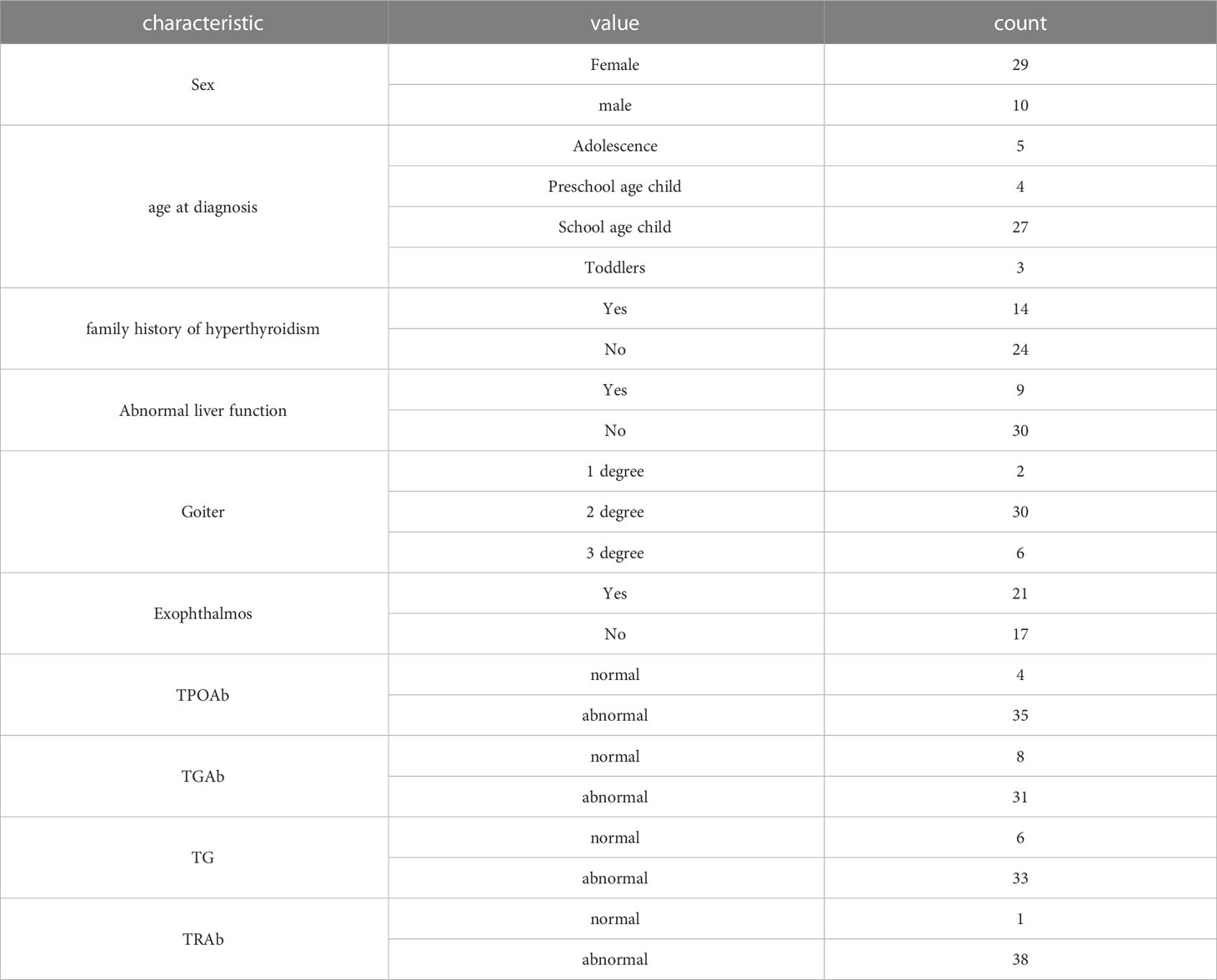
All the 39 children with hyperthyroidism in this study have corresponding clinical characteristics. The statistics of their clinical data are shown in the Table 2 below. Female patients have more than male patients, Schoolagechild accounts for the largest proportion, 14 patients with family history of hyperthyroidism, 9 patients with abnormal liver function, the largest proportion of patients with second degree goiter, most of the patients’ TPOAb, TGAb, TG and TRAb indexes are abnormal.
2.3 Functional enrichment analysis
The “clusterProfiler” R package was used for enrichment analysis of this paper. The basic principle of enrichment analysis is to compare the gene set to be analyzed from patients in section 1.1 with the reference gene set to determine which biological functions or pathways appear significantly more frequently in the gene set to be analyzed than in the reference gene set in the database, so as to find the biological processes, functions or pathways that have a significant impact on patients and the corresponding gene set.
2.4 Mutation data analysis and visualization
The “maftools” R package (42) was used to analyze a large amount of whole-exome sequencing data and visualize the data. First of all, we made a statistical analysis of the whole exon sequencing data set of screened genes: the distribution of high-frequency genes in each patient, the number of rare mutations compared with the 1000G_ALL database in the sample patients. Finally, the data distribution is displayed visually.
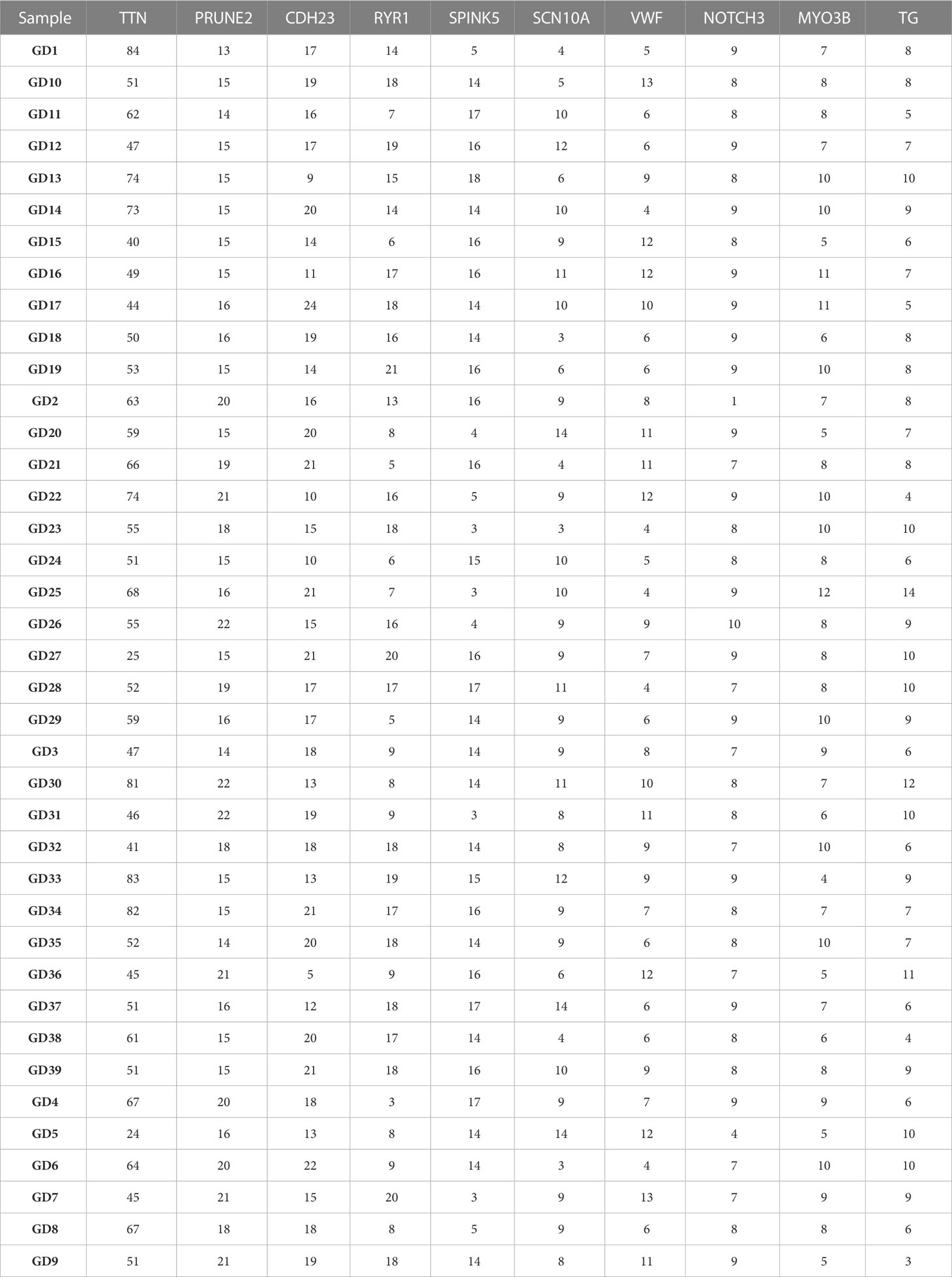
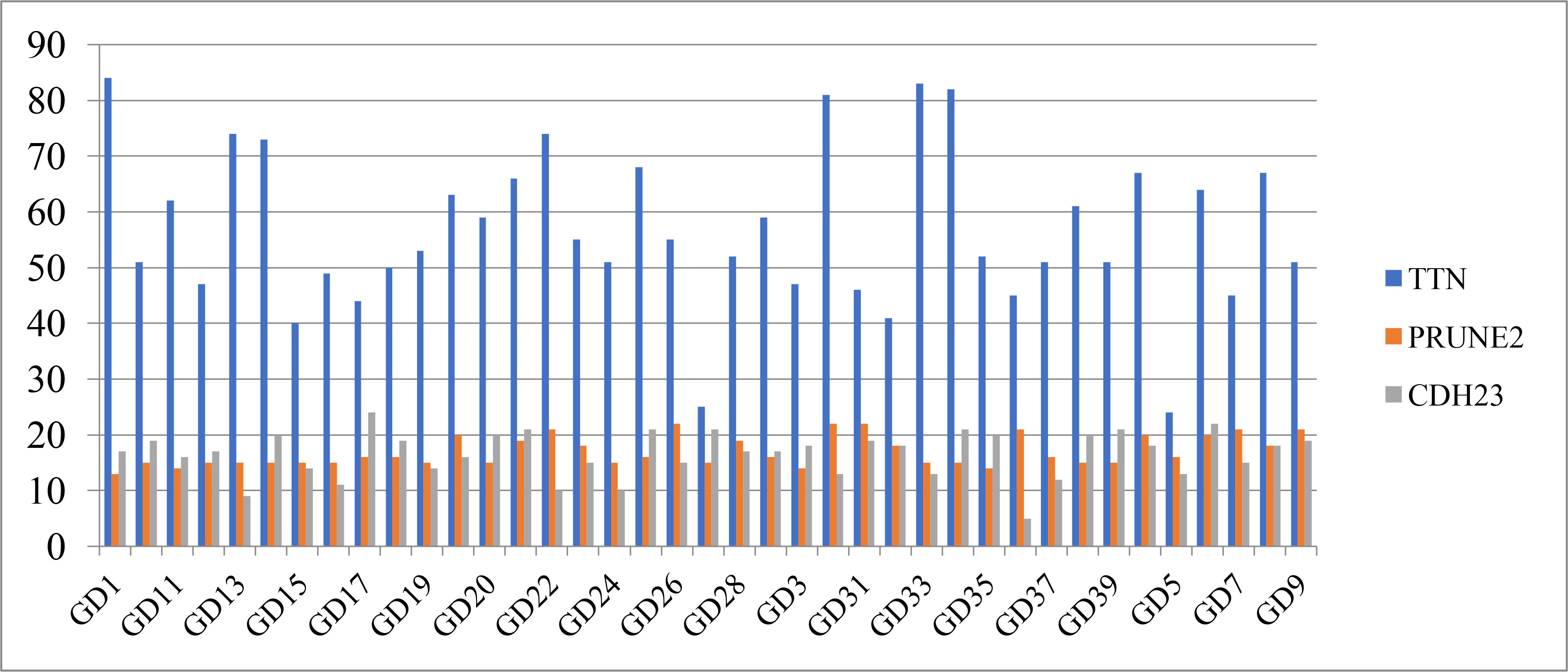
Table 3 shows the distribution of the top 10 high-frequency mutated genes on each sample. In this paper, the top 3 genes were selected for graphical display. Figure 1 shows that the TTN gene has the highest mutation frequency in patient GD1, the PRUNE2 gene has the highest mutation frequency in patients GD30 and GD31, and the CDH23 gene has the highest mutation frequency in patient GD17.
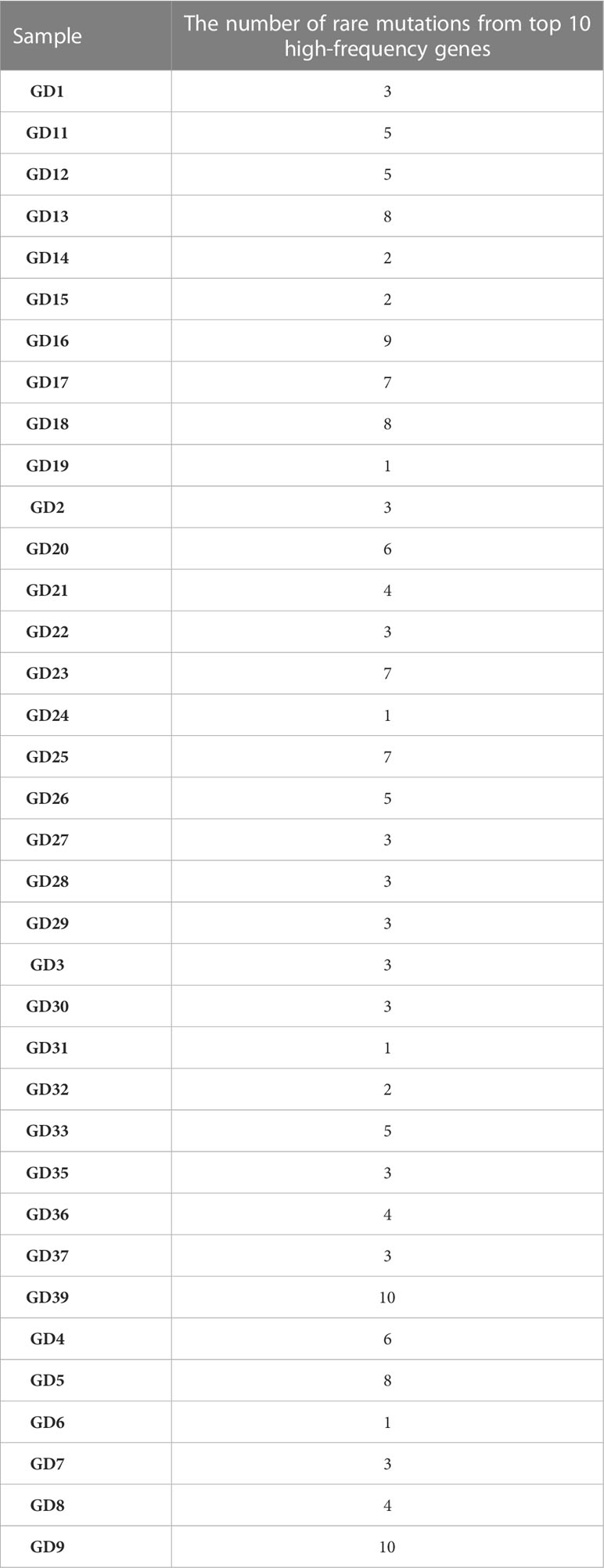
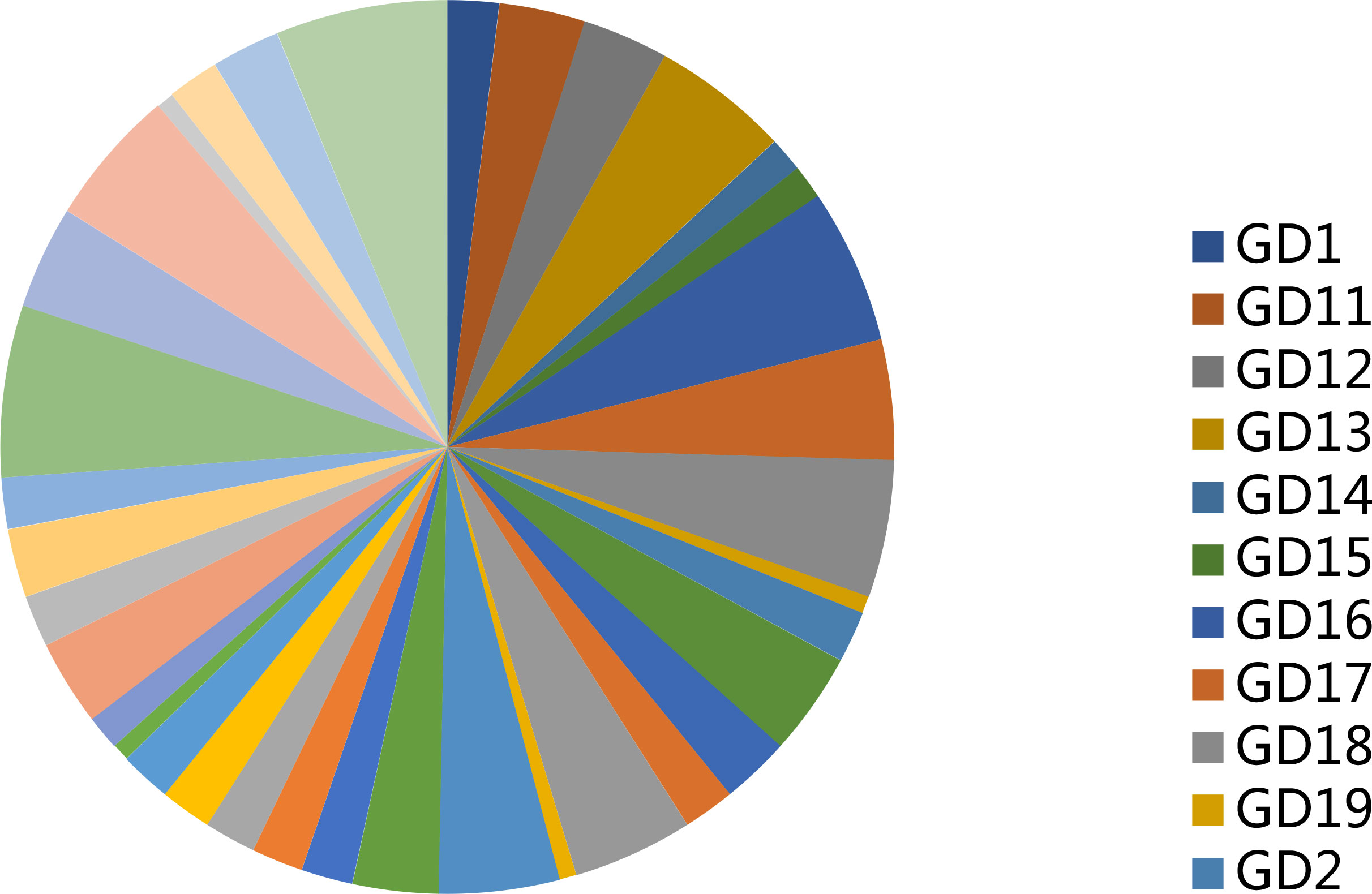
Table 4 shows the rare variance of the top 10 high-frequency variant genes in each sample compared with the 1000G_ALL database, and Figure 2 shows the proportion of rare variants corresponding to Table 4 for each patient. It can be found that the proportion of rare variants in GD9 and GD39 is the largest.
A summary of the mutation information of hyperthyroidism in children was visualizing in Figure 3. The top 15 mutated genes of hyperthyroidism in children by percentage was shown in the figure, including TTN, PRUNE2, CDH23, RYR1, SPINK5, SCN10A, VWF, NOTCH3, MYO3B, TG, LAMA2, KANK1, APC, TECTA, PCDH15. The mutation rate of the 15 genes in 39 children with hyperthyroidism is 100% (Figures 3A, B). The synonymous mutation was the most common type of mutation in 39 children with hyperthyroidism, followed by missense mutations (Figure 3A). Among the SNV classes, T>C was the predominant type of SNVs detected (Figure 3B). Different mutated genes have different tendencies for mutation types in children with hyperthyroidism. Gene WDR37 tends to have Splice site mutations, Gene COL3A1, GRHPR, OPA3 and SOX10 tend to have Silent mutations, and Gene INS and SLC24A5 tend to have missense mutations (Figure 3C).
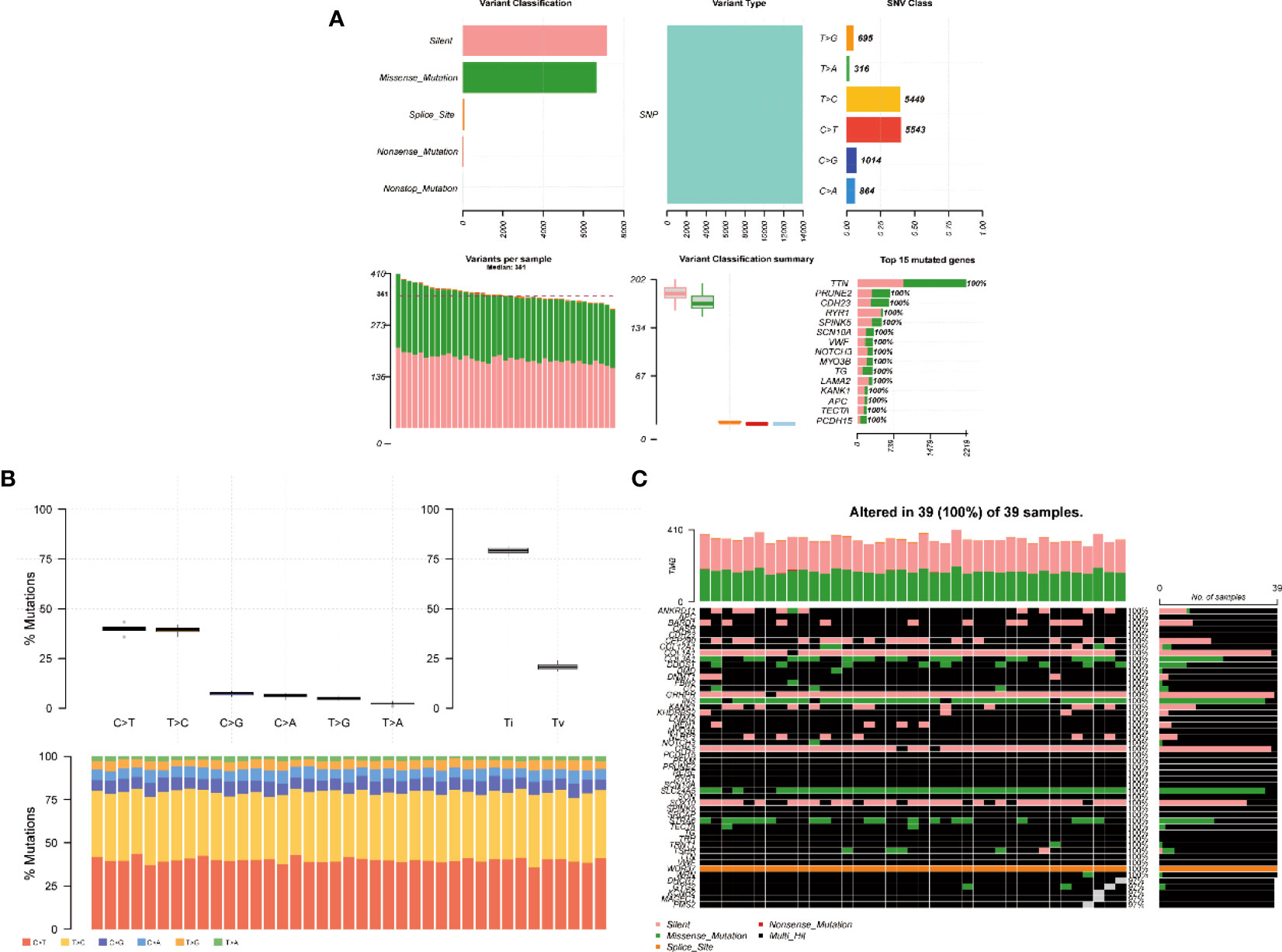
Figure 3 Somatic mutation profiles of hyperthyroidism in children. (A) Cohort summary plot of the mutation information among 39 cases of children with hyperthyroidism, the first row shows the statistical distribution of mutation classification, type, SNV class, the second row shows the distribution of mutation frequency and classification for each sample, and the last stacked statistic plot shows the top 15 most mutated genes. (B) Transition (Ti) and transversion(Tv) plot displays the distribution of SNVs in children with hyperthyroidism, and the stacked bar statistics show the distribution of SNVs in each child. (C) Landscape of mutation profiles shows mutation information of 39 cases of children with hyperthyroidism. The mutated genes are ordered by their mutation frequency.
3 Results
3.1 GO enrichment analysis
With the ORA method, the screened 144 genes were used to perform a statistical analysis of the gene set using the GO database as a reference. The returned p-values were subjected to multiple testing FDR correction, deriving a significance index denoted as p.adjust. A lower p.adjust value indicates a higher significance level in the enrichment analysis.
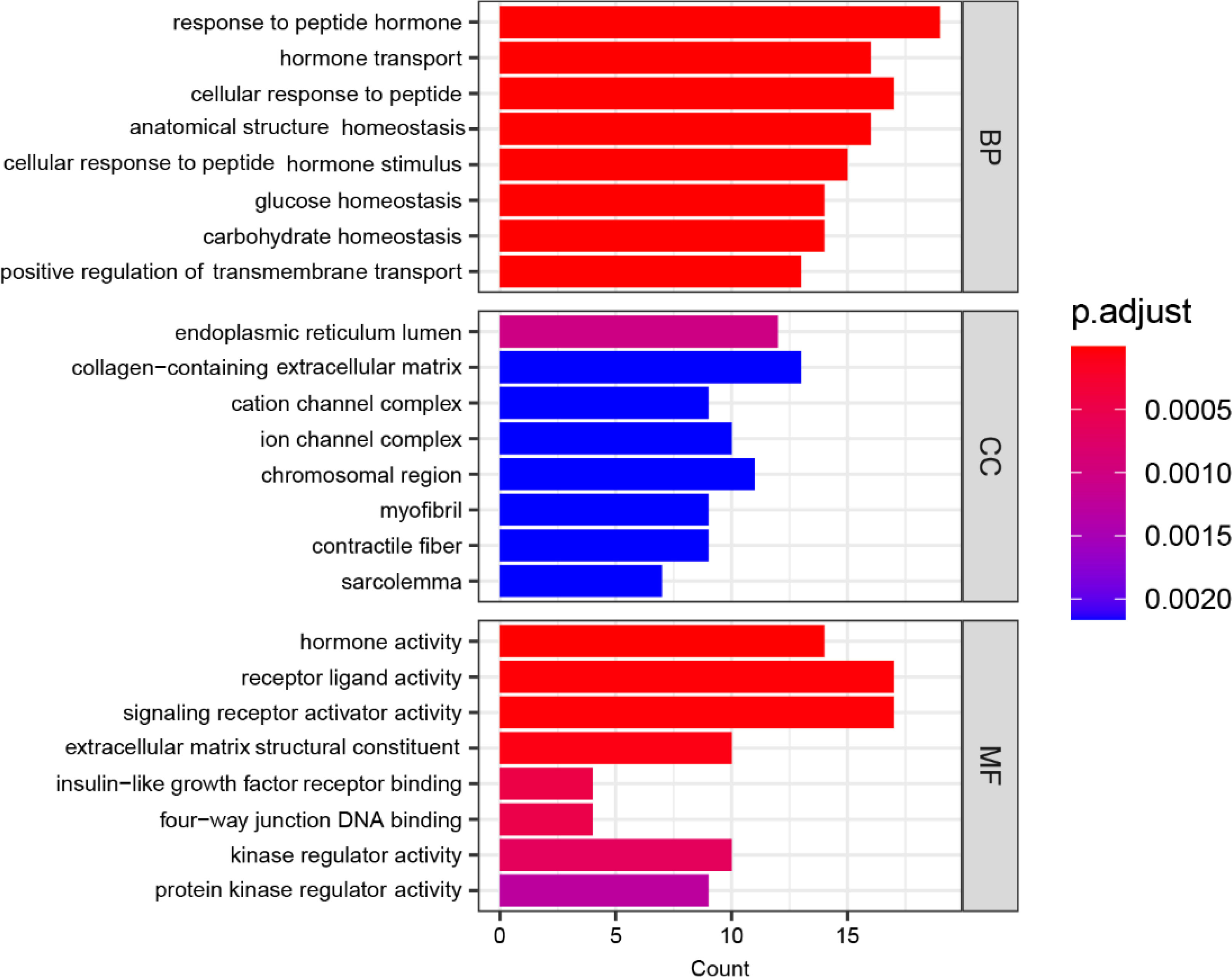
Figure 4 displays the functional enrichment of the 144 mutated genes in three dimensions of gene ontology. BP represents Biological Process: characterizing the biological processes in which genes are involved, such as cell differentiation and DNA replication; CC represents Cellular Component: describing where the gene product acts in which cellular component or gene product; MF represents Molecular Function: describing the activities of individual molecules in molecular biologies, such as protein kinase activity and insulin receptor activity. In Figure 4, the horizontal axis labeled “Count” denotes the quantity of genes in the enriched gene set, with longer bars indicating a more significant number of genes. The color coding represents the significance of the enrichment, with a redder color and a smaller p.adjust value indicating a higher significance level.
Based on the results presented in Figure 4, in terms of the Biological Process (BP) category, the most significant enrichment of mutated genes is in the “response to peptide hormone” function, followed by enrichment in “hormone transport”, “cellular response to peptide”, “anatomical structure homeostasis”, “cellular response to peptide hormone stimulus” and “glucose homeostasis”. In terms of the Cellular Component (CC) category, the most significant enrichment of mutated genes is in the “endoplasmic reticulum lumen” function, followed by enrichment in “collagen-containing extracellular matrix”, “cation channel complex”, “ion channel complex”, “chromosomal region” and “myofibril”. Similarly, within the Molecular Function (MF) category, the “hormone activity” function exhibits the most significant enrichment of mutated genes, followed by enrichments in functions “receptor ligand activity”, “signaling receptor activator activity”, “extracellular matrix structural constituent”, “insulin-like growth factor receptor binding” and “four-way junction DNA binding”.
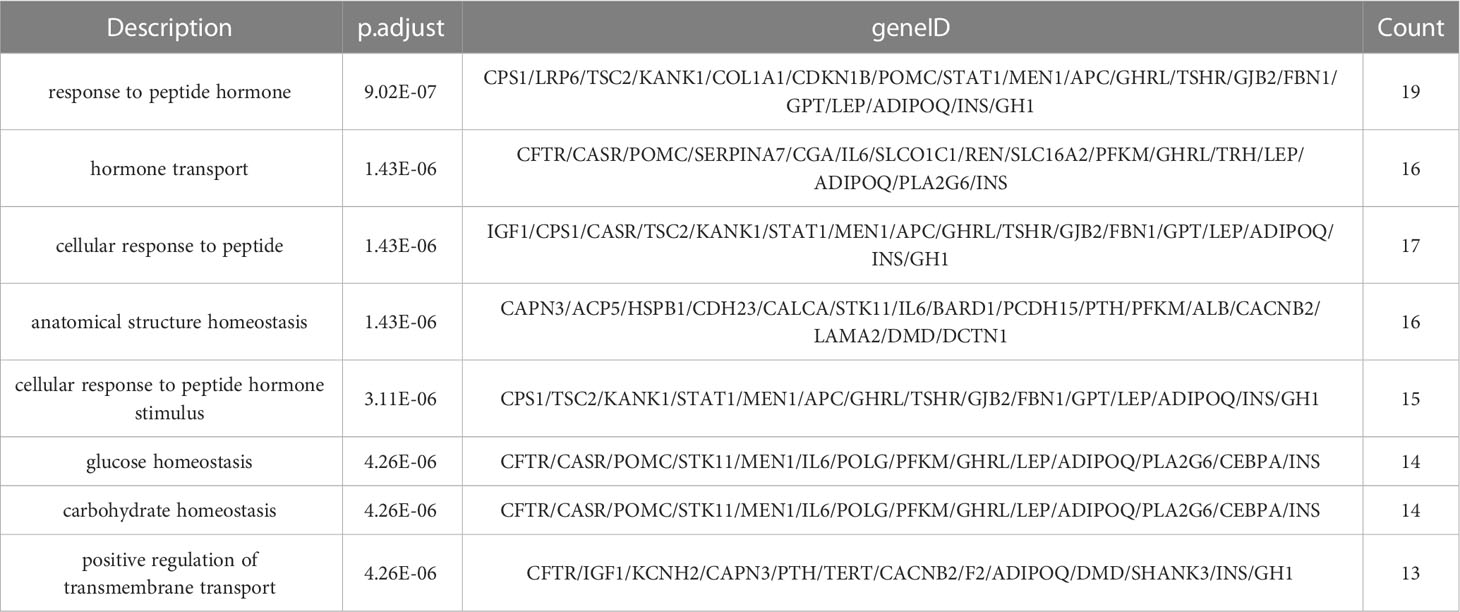
The complete information of the top 8 significantly enriched biological processes (BP) terms are shown in Table 5, ranked by their level of significance based on the p.adjust values. The “Description” column describes the biological process associated with the enriched gene set, the “geneID” column lists all the genes in the gene set that has common effects, and the “Count” column indicates the number of genes in the enriched gene set.
In the BP terms, the most significant enrichment of mutant genes was in the function of “response to peptide hormone” (p. adjust = 9.01×10-7) and the corresponding gene set that has common effects contained 19 genes: CPS1, LRP6, TSC2, KANK1, COL1A1, CDKN1B, POMC, STAT1, MEN1, APC, GHRL, TSHR, GJB2, FBN1, GPT, LEP, ADIPOQ, INS, GH1, followed by “hormone transport” (p. adjust = 1.43×10-6), with the corresponding gene set that has common effects containing 16 genes: CFTR, CASR, POMC, SERPINA7, CGA, IL6, SLCO1C1, REN, SLC16A2, PFKM, GHRL, TRH, LEP, ADIPOQ, PLA2G6, INS, “cellular response to peptide” (p. adjust = 1.43×10-6), with the corresponding gene set that has common effects containing 17 genes: IGF1, CPS1, CASR, TSC2, KANK1, STAT1, MEN1, APC, GHRL, TSHR, GJB2, FBN1, GPT, LEP, ADIPOQ, INS, GH1, “anatomical structure homeostasis” (p. adjust = 1.43×10-6), with the corresponding gene set that has common effects containing 16 genes: CAPN3, ACP5, HSPB1, CDH23, CALCA, STK11, IL6, BARD1, PCDH15, PTH, PFKM, ALB, CACNB2, LAMA2, DMD, DCTN1, “cellular response to peptide hormone stimulus” (p. adjust = 3.11×10-6), with the corresponding gene set that has common effects containing 15 genes: CPS1, TSC2, KANK1, STAT1, MEN1, APC, GHRL, TSHR, GJB2, FBN1, GPT, LEP, ADIPOQ, INS, GH1 and “glucose homeostasis” (p. adjust = 4.26×10-6), with the corresponding gene set that has common effects containing 14 genes: CFTR, CASR, POMC, STK11, MEN1, IL6, POLG, PFKM, GHRL, LEP, ADIPOQ, PLA2G6, CEBPA, INS.
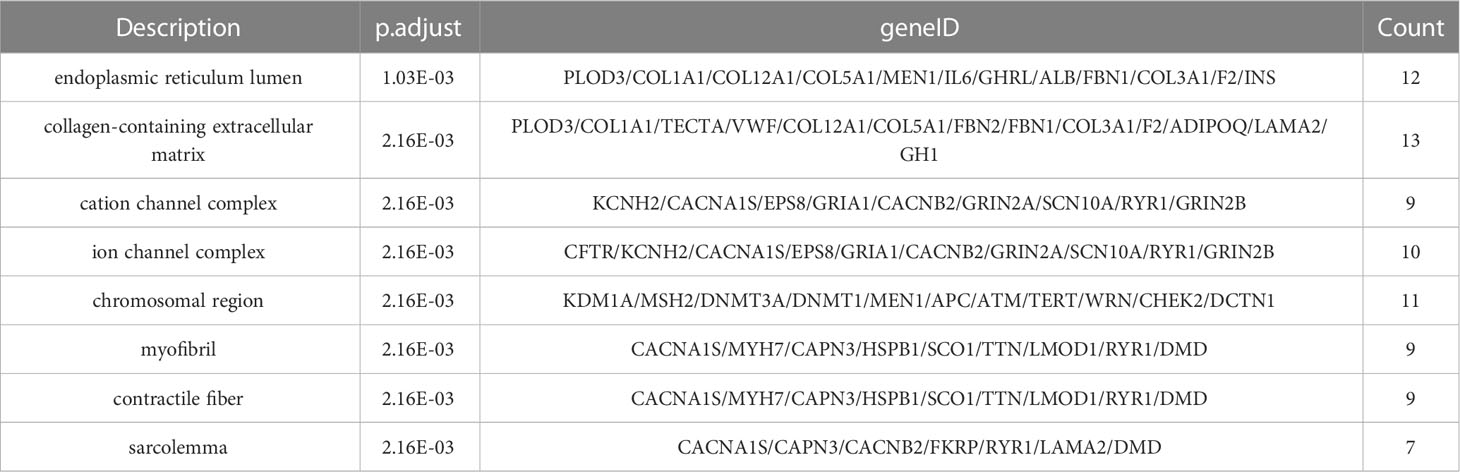
The complete information of the top 8 significantly enriched cellular component (CC) terms are shown in Table 6, ranked by their level of significance based on the p.adjust values. The “Description” column provides a description of the cellular component associated with the enriched gene set, the “geneID” column lists all the genes in the gene set that has common effects and the “Count” column indicates the number of genes in the enriched gene set.
In the CC terms, the most significant enrichment of mutant genes was in the function of “endoplasmic reticulum lumen” (p. adjust = 1.03×10-3) and the corresponding gene set that has common effects contained 12 genes: PLOD3, COL1A1, COL12A1, COL5A1, MEN1, IL6, GHRL, ALB, FBN1, COL3A1, F2, INS, followed by “collagen-containing extracellular matrix” (p. adjust = 2.16×10-3), with the corresponding gene set that has common effects containing 13 genes: PLOD3, COL1A1, TECTA, VWF, COL12A1, COL5A1, FBN2, FBN1, COL3A1, F2, ADIPOQ, LAMA2, GH1, “cation channel complex” (p. adjust = 2.16×10-3), with the corresponding gene set that has common effects containing 9 genes: KCNH2, CACNA1S, EPS8, GRIA1, CACNB2, GRIN2A, SCN10A, RYR1, GRIN2B, “ion channel complex” (p. adjust = 2.16×10-3), with the corresponding gene set that has common effects containing 10 genes: CFTR, KCNH2, CACNA1S, EPS8, GRIA1, CACNB2, GRIN2A, SCN10A, RYR1, GRIN2B, “chromosomal region” (p. adjust = 2.16×10-3), with the corresponding gene set that has common effects containing 11 genes: KDM1A, MSH2, DNMT3A, DNMT1, MEN1, APC, ATM, TERT, WRN, CHEK2, DCTN1 and “myofibril” (p. adjust = 2.16×10-3), with the corresponding gene set that has common effects containing 9 genes: CACNA1S, MYH7, CAPN3, HSPB1, SCO1, TTN, LMOD1, RYR1, DMD.
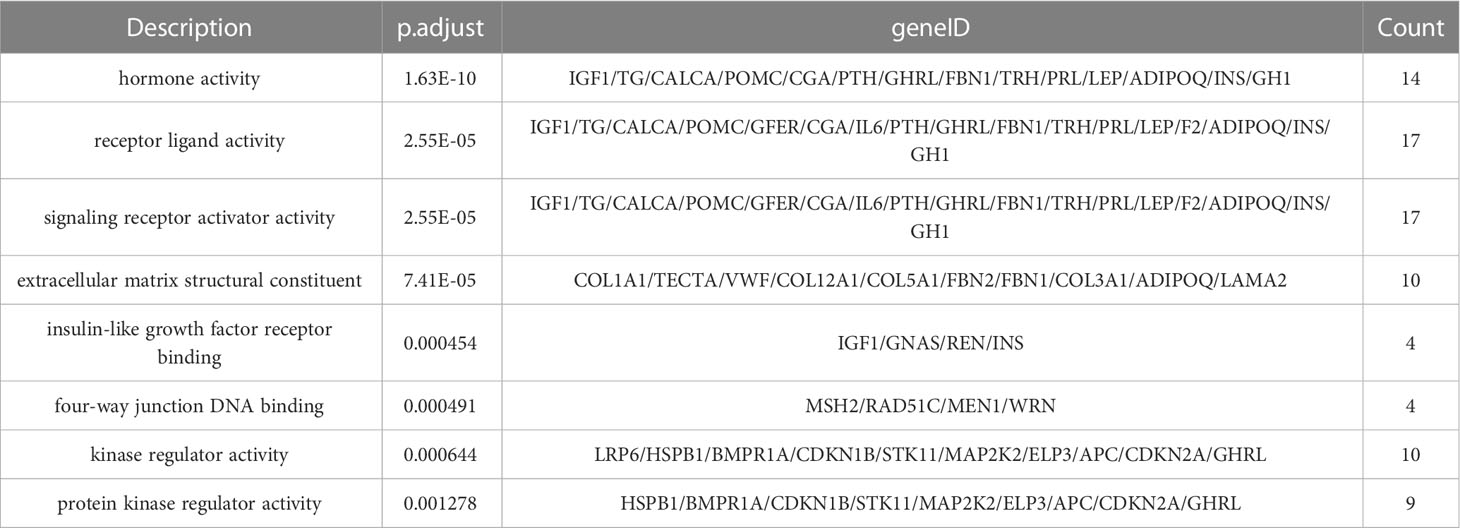
The complete information of the top 8 significantly enriched molecular function (MF) terms are shown in Table 7, ranked by significance level based on the p.adjust values. The “Description” column describes the molecular function associated with the enriched gene set, the “geneID” column lists all the genes in the gene set that has common effects and the “Count” column indicates the number of genes in the enriched gene set.
In the MF terms, the most significant enrichment of mutant genes was in the function of “hormone activity” (p. adjust = 1.63×10-10) and the corresponding gene set that has common effects contained 14 genes: IGF1, TG, CALCA, POMC, CGA, PTH, GHRL, FBN1, TRH, PRL, LEP, ADIPOQ, INS, GH1, followed by “receptor ligand activity” (p. adjust = 2.55×10-5), with the corresponding gene set that has common effects containing 17 genes: IGF1, TG, CALCA, POMC, GFER, CGA, IL6, PTH, GHRL, FBN1, TRH, PRL, LEP, F2, ADIPOQ, INS, GH1, “signaling receptor activator activity” (p. adjust = 2.55×10-5), with the corresponding gene set that has common effects containing 17 genes: IGF1, TG, CALCA, POMC, GFER, CGA, IL6, PTH, GHRL, FBN1, TRH, PRL, LEP, F2, ADIPOQ, INS, GH1, “extracellular matrix structural constituent” (p. adjust = 7.41×10-5), with the corresponding gene set that has common effects containing 10 genes: COL1A1, TECTA, VWF, COL12A1, COL5A1, FBN2, FBN1, COL3A1, ADIPOQ, LAMA2, “insulin-like growth factor receptor binding” (p. adjust = 4.54×10-4), with the corresponding gene set that has common effects containing 4 genes: IGF1, GNAS, REN, INS and “four-way junction DNA binding” (p. adjust = 4.91×10-4), with the corresponding gene set that has common effects containing 4 genes: MSH2, RAD51C, MEN1, WRN.
Overall, the functional significance of mutated genes in CC terms was relatively low in affected children and relatively high in BP terms. In all GO enrichments, the most significant functional enrichment of mutated genes in hyperthyroidism children was observed in “hormone activity” and “response to peptide hormone" functions.
3.2 KEGG enrichment analysis
With the ORA method, the 144 screened genes were used to perform a statistical analysis of the gene set using the KEGG biological signaling pathway database as a reference. The returned p-values were subjected to multiple testing FDR correction, deriving a significance index denoted as p.adjust. A lower p.adjust value indicates a higher significance level in the enrichment analysis.
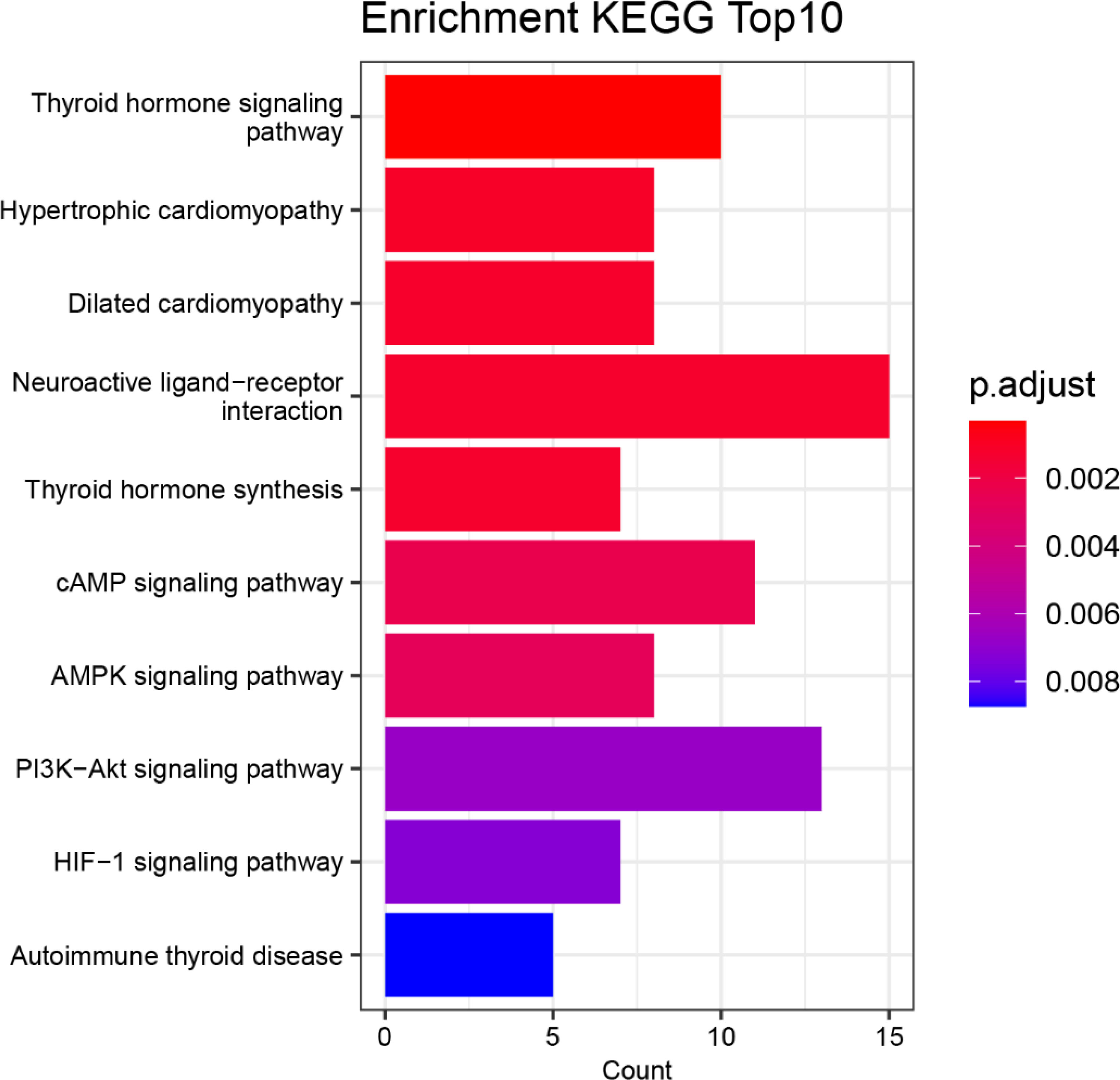
Figure 5 displays the top 10 functional biological signaling pathways of the 144 mutant genes that were most significantly enriched. The horizontal axis labeled “Count” denotes the quantity of genes in the enriched gene set, with longer bars indicating a more significant number of genes. The color coding represents the significance of the enrichment, with a redder color and a smaller p.adjust value indicating a higher significance level.
Based on the results presented in Figure 5, the most significant enrichment of mutated genes is in the biological signaling pathway “Thyroid hormone signaling pathway” function, followed by enrichment in the biological signaling pathway “Hypertrophic cardiomyopathy”, “Dilated cardiomyopathy”, “Neuroactive ligand-receptor interaction”, “Thyroid hormone synthesis” and “cAMP signaling pathway”.
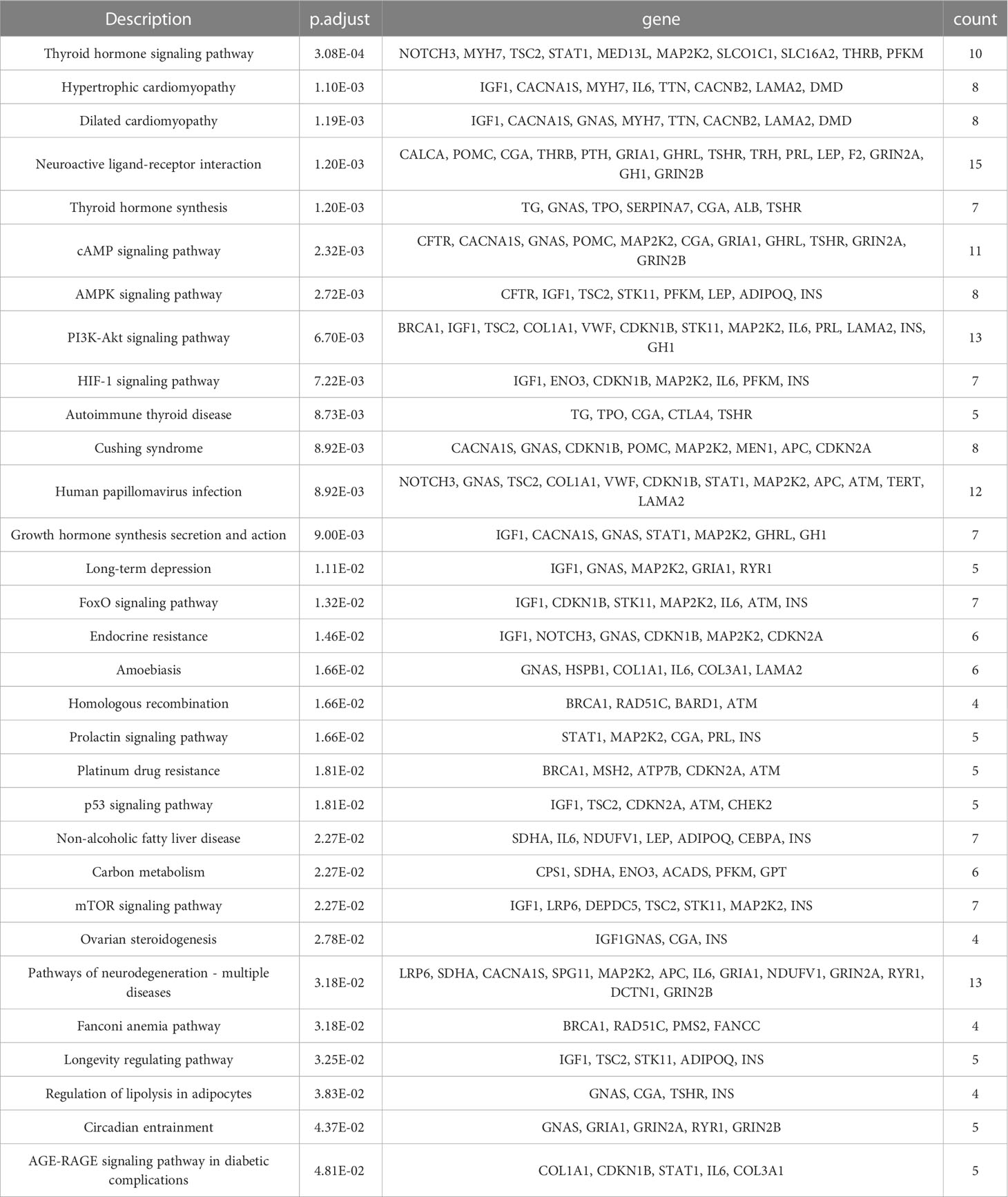
The complete information of the top 10 significantly functional biological signaling pathways are shown in Table 8, ranked by their significance level based on the p.adjust values. The “Description” column provides a description of the biological signaling pathways associated with the enriched gene set, the “geneID” column lists all the genes in the gene set that has common effects and the “Count” column indicates the number of genes in the enriched gene set.
The most significant biological signaling pathways enrichment of mutant genes was in the functional pathway of “Thyroid hormone signaling pathway” (p. adjust = 3.08×10-4) and the corresponding gene set that has common effects contained 10 genes: NOTCH3, MYH7, TSC2, STAT1, MED13L, MAP2K2, SLCO1C1, SLC16A2, THRB, PFKM, followed by the functional pathway “Hypertrophic cardiomyopathy” (p. adjust = 1.10×10-3), with the corresponding gene set that has common effects containing 8 genes: IGF1, CACNA1S, MYH7, IL6, TTN, CACNB2, LAMA2, DMD, the functional pathway “Dilated cardiomyopathy” (p. adjust = 1.19×10-3), with the corresponding gene set that has common effects containing 8 genes: IGF1, CACNA1S, GNAS, MYH7, TTN, CACNB2, LAMA2, DMD, the functional pathway “Neuroactive ligand-receptor interaction” (p. adjust = 1.20×10-3), with the corresponding gene set that has common effects containing 15 genes: CALCA,POMC,CGA,THRB,PTH, GRIA1, GHRL, TSHR, TRH, PRL, LEP, F2, GRIN2A, GH1, GRIN2B, the functional pathway “Thyroid hormone synthesis” (p. adjust = 1.20×10-3), with the corresponding gene set that has common effects containing 7 genes: TG, GNAS, TPO, SERPINA7, CGA, ALB, TSHR and the functional pathway “cAMP signaling pathway” (p. adjust = 2.32×10-3), with the corresponding gene set that has common effects containing 11 genes: CFTR, CACNA1S, GNAS, POMC, MAP2K2, CGA, GRIA1, GHRL, TSHR, GRIN2A, GRIN2B.
4 Discussion
The collective action of multiple genes and various genetic factors contribute to the development of hyperthyroidism. The improvement of bioinformatics has provided new approaches and tools for studying diseases such as hyperthyroidism. In this study, we processed and statistically analyzed the whole-exome sequencing data of 39 children with hyperthyroidism. We used bioinformatics tools to perform GO enrichment analysis and KEGG enrichment analysis to obtain the significant functional gene sets associated with hyperthyroidism in children, and to help understand the pathogenesis of hyperthyroidism in polygenic diseases and the common effects of its mutated genes on different functions.
Through the mutation data visualization, we found that among the 144 mutated genes in 39 children with hyperthyroidism, the top 15 most mutated genes were TTN, PRUNE2, CDH23, RYR1, SPINK5, SCN10A, VWF, NOTCH3, MYO3B, TG, LAMA2, KANK1, APC, TECTA, PCDH15, and the top 45 genes with high mutation frequencies had mutations in each patient. In addition, Gene WDR37 tends to have Splice site mutations, Gene COL3A1, GRHPR, OPA3 and SOX10 tend to have Silent mutations, and Gene INS and SLC24A5 tend to have missense mutations. Among the mutation classifications, Synonymous mutation was the most common mutation classification in children with hyperthyroidism, followed by missense mutations. Regarding single nucleotide variations, T>C had the highest mutation frequency.
Through GO enrichment analysis, we found that in terms of the biological process (BP), the enriched functional gene set associated with hyperthyroidism in children was most significant for function “response to peptide hormone” with the p.adjust value of 9.01×10-7 and the corresponding gene set with mutations included 19 genes: CPS1, LRP6, TSC2, KANK1, COL1A1, CDKN1B, POMC, STAT1, MEN1, APC, GHRL, TSHR, GJB2, FBN1, GPT, LEP, ADIPOQ, INS, GH1. In terms of the cellular component (CC), the enriched functional gene set associated with hyperthyroidism in children was most significant for the function “endoplasmic reticulum lumen”, its p.adjust value is 1.03×10-3. The corresponding gene set with mutations included 12 genes: PLOD3, COL1A1, COL12A1, COL5A1, MEN1, IL6, GHRL, ALB, FBN1, COL3A1, F2, INS. In terms of the molecular function (MF), the enriched functional gene set associated with hyperthyroidism in children was most significant for function “hormone activity” with the p.adjust value of 1.63×10-10 and the corresponding gene set with mutations included 14 genes: IGF1, TG, CALCA, POMC, CGA, PTH, GHRL, FBN1, TRH, PRL, LEP, ADIPOQ, INS, GH1. In all GO enrichments, the significance of function “hormone activity” and “response to peptide hormone” was the highest. From the overall view of GO enrichment, the significance of mutated genes in children with hyperthyroidism was relatively low in the function of cell sublocalization level, and relatively high in the biological process level. This indicates that the mutated gene may affect more biological processes and molecular functions of children, such as internal circulation, hormone secretion, and the cell structure is not affected. From the gene ontology level, the mutant genes in hyperthyroidism children have the highest significance in the function of “hormone activity” and “response to peptide hormones”. In the GO database, “hormone activity” includes corticotropin releasing hormone activity, thyrotropin releasing hormone activity and other series of hormone activities. Studies have shown that corticotropin releasing hormone can affect the degree of hyperthyroidism (43). Thyrotropin releasing hormone is one of the hormones that regulate the level of serum thyroid hormone in a certain range, so the mutated gene can affect the incidence of hyperthyroidism by affecting the secretion of these hormones (44). Mutated genes also affect hyperthyroidism by influencing biological processes called “response to peptide hormones.”
Through KEGG enrichment, the enriched functional gene set associated with hyperthyroidism in children was most significant for function “Thyroid hormone signaling pathway” in the signal pathway with the p.adjust value of 3.08×10-4. The corresponding gene set with mutations included 10 genes: NOTCH3, MYH7, TSC2, STAT1, MED13L, MAP2K2, SLCO1C1, SLC16A2, THRB, PFKM. “Hypertrophic cardiomyopathy” in the signal pathway was followed with the p.adjust value of 1.10×10-3 and the corresponding collective action gene set with mutations included 8 genes: IGF1, CACNA1S, MYH7, IL6, TTN, CACNB2, LAMA2, DMD. Therefore, from a pathway perspective, the mutated gene affects the thyroid hormone-related interactions of biomolecules and chemical reactions in children leading to their disease and increases the risk of hyperthyroid hypertrophic cardiomyopathy in children.
Data availability statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by Guangzhou Women and Children’s Medical Center. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author contributions
Conceptualization ideas: XM, LT. Data curation: XM, LT, HL,YH. Investigation: XM, WZ. Statistical analysis and interpretation: XM, LT, HL, LL,YH. Methodology: XM, LT, HL, WZ, LL, AH. Resources: XM, HL, WZ, LL. Supervision: XM. Project administration: XM,YH. Visualization: LT, HL, AH. Writing – original draft: XM, LT, HL,YH. Writing—review and editing: XM, LT, WZ. All authors read, contributed to the research design, and approved the final manuscript.
Funding
This research was supported by Project of Guangzhou Science and technology plan project, Grant No. 202201020609.
Acknowledgments
This study was performed with the support of Department of Genetics and Endocrinology, Guangzhou Women and Children’s Medical Center. The authors are indebted to the patients, physicians, and nurses who made this work possible.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
2. Wang C, Li Y, Teng D, Shi X, Ba J, Chen B, et al. Hyperthyroidism prevalence in China after universal salt iodization. Front Endocrinol (Lausanne). (2021) 12:651534. doi: 10.3389/fendo.2021.651534
3. Simon M, Rigou A, Le Moal J, Zeghnoun A, Le Tertre A, De Crouy-Chanel P, et al. Epidemiology of childhood hyperthyroidism in France: a nationwide population-based study. J Clin Endocrinol Metab (2018) 103(8):2980–7. doi: 10.1210/jc.2018-00273
4. Xie H, Chen D, Zhang J, Yang R, Gu W, Wang X. Characteristics of graves' disease in children and adolescents in nanjing: a retrospective investigation study. Front Public Health (2022) 10:993733. doi: 10.3389/fpubh.2022.993733
5. Schneider Aguirre R, Fuqua JS. Mental health disorders and hyperthyroidism in the pediatric population. Pediatrics (2019) 144(5):e20192140. doi: 10.1542/peds.2019-2140
6. Patel KA, Knight B, Aziz A, Babiker T, Tamar A, Findlay J, et al. Utility of systematic TSHR gene testing in adults with hyperthyroidism lacking overt autoimmunity and diffuse uptake on thyroid scintigraphy. Clin Endocrinol (Oxf). (2019) 90(2):328–33. doi: 10.1111/cen.13892
7. Jaeschke H, Eszlinger M, Lueblinghoff J, Coslovsky R, Paschke R. Prolonged inappropriate TSH suppression during hypothyroidism after thyroid ablation in a patient with nonautoimmune familial hyperthyroidism. Horm Metab Res (2011) 43(7):500–4. doi: 10.1055/s-0031-1277184
8. Taha D, Adhikari A, Flore LA. Familial neonatal nonautoimmune hyperthyroidism due to a gain-of-function (D619G) thyrotropin-receptor mutation. J Pediatr Endocrinol Metab (2020) 34(2):267–71. doi: 10.1515/jpem-2020-0291
9. Huang CJ, Jap TS. A systematic review of genetic studies of thyroid disorders in Taiwan. J Chin Med Assoc (2015) 78(3):145–53. doi: 10.1016/j.jcma.2014.09.010
10. Chen X, Hu Z, Liu M, Li H, Liang C, Li W, et al. Correlation between CTLA-4 and CD40 gene polymorphisms and their interaction in graves' disease in a Chinese han population. BMC Med Genet (2018) 19(1):1–9. doi: 10.1186/s12881-018-0665-y
11. Ting WH, Chien MN, Lo FS, Wang CH, Huang CY, Lin CL, et al. Association of cytotoxic T-Lymphocyte-Associated protein 4 (CTLA4) gene polymorphisms with autoimmune thyroid disease in children and adults: case-control study. PloS One (2016) 11(4):1–13. doi: 10.1371/journal.pone.0154394
12. Wang LQ, Wang TY, Sun QL, Qie YQ. Correlation between thyroglobulin gene polymorphisms and autoimmune thyroid disease. Mol Med Rep (2015) 12(3):4469–75. doi: 10.3892/mmr.2015.3978
13. Hsiao JY, Hsieh MC, Tien KJ, Hsu SC, Shin SJ, Lin SR. Association between a C/T polymorphism in exon 33 of the thyroglobulin gene is associated with relapse of graves’ hyperthyroidism after antithyroid withdrawal in Taiwanese[J]. J Clin Endocrinol Metab (2007) 92(8):3197–201. doi: 10.1210/jc.2007-0675
14. Calebiro D, Grassi ES, Eszlinger M, Ronchi CL, Godbole A, Bathon K, et al. Recurrent EZH1 mutations are a second hit in autonomous thyroid adenomas. J Clin Invest. (2016) 126(9):3383–8. doi: 10.1172/JCI84894
15. França MM, Levine RL, Pappa T, Ilaka-Chibuluzo S, Rothberger GD, Dumitrescu AM, et al. Nonautoimmune hyperthyroidism caused by a somatic mosaic GNAS mutation involving part of the thyroid gland. Thyroid (2020) 30(4):640–2. doi: 10.1089/thy.2019.0471
16. Elbers LP, Moran C, Gerdes VE, van Zaane B, Meijers J, Endert E, et al. The hypercoagulable state in hyperthyroidism is mediated via the thyroid hormone β receptor pathway. Eur J Endocrinol (2016) 174(6):755–62. doi: 10.1530/EJE-15-1249
17. Chen YQ, Zhu M. Clinical study of congenital thyroid dysfunction caused by TSHR gene mutation. Adv Clin Med (2022) 12(5):3861–5. doi: 10.12677/ACM.2022.125558
18. Li J, Liu YF. Research progress on the mechanism of TSHR gene in hyperthyroid diseases. Chin J Integr Med Cardio-Cerebrovascular Dis (2022) 20(08):1435–7.
19. Li CK, Liu GL. TSH receptor gene mutations and congenital nonautoimmune hyperthyroidism. J Pract Diabetol (2021) 17(01):3–5.
20. Nakamura A, Morikawa S, Aoyagi H, Ishizu K, Tajima T. A Japanese family with nonautoimmune hyperthyroidism caused by a novel heterozygous thyrotropin receptor gene mutation. Pediatr Res (2014) 75(6):749–53. doi: 10.1038/pr.2014.34
21. Pujol-Borrell R, Giménez-Barcons M, Marín-Sánchez A, Colobran R. Genetics of graves’ disease: special focus on the role of TSHR gene. Hormone Metab Res (2015) 47(10):753–66. doi: 10.1055/s-0035-1559646
22. Lane LC, Allinson KR, Campbell K, Bhatnagar I, Ingoe L, Razvi S, et al. Analysis of BAFF gene polymorphisms in UK Graves' disease patients. Clin Endocrinol (2019) 90(1):170–4. doi: 10.1111/cen.13872
23. Yang M, Luo CH, Fu LJ, Lu WH. Analysis of KCNJ18 gene mutations in patients with thyrotoxic periodic paralysis in dongguan. Shandong Med J (2019) 59(29):46–8.
24. Yuan MG, Wang XF, Guo YJ, Guo ZQ, Zhao WJ. Correlation between BRAF gene mutation and primary hyperthyroidism combined with papillary thyroid carcinoma. J Modern Oncol (2019) 27(17):3027–31.
25. Yan QY. UGT1A1 gene mutation and Graves' disease complicated with liver failure: a case report and literature review Kunming Medical University (2019).
26. Mao XJ, Ma XD, Liu L, Huang YL, Zhou ZH, Cheng J, et al. Analysis of thyroid stimulating hormone receptor gene mutation in children with hyperthyroidism. Chin J Endocrinol Metab (2019) 35(2):5. doi: 10.3760/cma.j.issn.1000-6699.2019.02.008
27. Ma XD. Clinical study of methimazole treatment and TSHR gene mutation analysis in children with hyperthyroidism. Guangzhou Medical University (2015).
28. Brown RS, Lombardi A, Hasham A, Greenberg DA, Gordon J, Concepcion E, et al. Genetic analysis in young-age-of-onset graves' disease reveals new susceptibility loci. J Clin Endocrinol Metab (2014) 99(7):E1387–91. doi: 10.1210/jc.2013-4358
29. Roberts SA, Moon JE, Dauber A, Smith JR. Novel germline mutation (Leu512Met) in the thyrotropin receptor gene (TSHR) leading to sporadic non-autoimmune hyperthyroidism. J Pediatr Endocrinol Metab (2017) 30(3):343–7. doi: 10.1515/jpem-2016-0185
30. Jurecka-Lubieniecka B, Ploski R, Kula D, Krol A, Bednarczuk T, Kolosza Z, et al. Association between age at diagnosis of graves' disease and variants in genes involved in immune response. PloS One (2013) 8(3):e59349. doi: 10.1371/journal.pone.0059349
31. Brown RS, Lombardi A, Hasham A, Greenberg DA, Gordon J, Concepcion E, et al. Genetic analysis in young-age-of-onset graves' disease reveals new susceptibility loci. J Clin Endocrinol Metab (2014) 99(7):E1387–91. doi: 10.1210/jc.2013-4358
32. Pyrżak B, Rumińska M, Witkowska-Sędek E, Kucharska A. Follow-up of thyroid function in children with neonatal hyperthyroidism. Front Endocrinol (Lausanne). (2022) 13:877119. doi: 10.3389/fendo.2022.877119
33. Niedziela M. Hyperthyroidism in adolescents. Endocr Connect. (2021) 10(11):R279–92. doi: 10.1530/EC-21-0191
34. Yuan RH, Yuanli JL, Shu YL, Jin YX, Shi ZH. Efficacy and safety of methimazole combined with thyroxine in the treatment of hyperthyroidism in children:a meta-analysis. Strait Pharma J (2022) 34(11):90–5.
35. Zhang H. Adverse reactions comparison of methimazole and propylthionracil in the treatment of hyperthy roidism. . .China Modern Med (2018) 25(4):165–7.
36. Wang J, Su Y, Gu YH, Li BQ, Li SX. The treatment progress of graves' disease in children and the timing of discontinuation of antithyroid drugs. Maternal Child Health Care China (2019) 34(19):4588–91.
37. Hazkani I, Stein E, Samis J, Josefson J, Maddalozzo J, Johnston D, et al. The effect of hyperthyroidism at thyroidectomy on complication rates in children with graves’ disease. Int J Pediatr otorhinolaryngology (2022) 163:111360. doi: 10.1016/j.ijporl
38. Wu X, Qin X, Yao Y. Methimazole plus levothyroxine for treating hyperthyroidism in children: a systematic review and meta-analysis. Transl Pediatr (2022) 11(1):41–57. doi: 10.21037/tp-21-497
39. Chung SK, Asban A, Hur J, Iyer P, Chen H. Hyperthyroidism symptoms, management, and outcomes in children and adults seeking definitive surgical treatment. Ann Surg (2021) 273(5):e181–2. doi: 10.1097/SLA.0000000000004053
40. Du CC, Gao L. Assessment of left ventricular global systolic function using real-time three-dimensional speckle tracking imaging in children with hyperthyroidism. Med Equip (2022) 35(7):9–14.
41. Gong XP, Jiang YH, Ran QF. Regulation of blood lipid and blood glucose levels between thyroid hormone and heart rate in children with hyperthyroidism. Med Innovation China (2021) 18(20):143–6.
42. Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res (2018) 28(11):1747–56. doi: 10.1101/gr.239244.118
43. Cao ZH, Chen Q. Relationship between hyperthyroidism and serum cortisol and adrenocorticotropic hormone levels. Chin J Hum Sexuality (2020) 29(06):24–7.
Keywords: hyperthyroidism, bioinformatics, gene mutation, enrichment analysis, gene ontology enrichment
Citation: Mao X, Tang L, Li H, Zhang W, Liu L, Wang H and Headar A (2023) Functional enrichment analysis of mutated genes in children with hyperthyroidism. Front. Endocrinol. 14:1213465. doi: 10.3389/fendo.2023.1213465
Received: 28 April 2023; Accepted: 27 July 2023;
Published: 09 October 2023.
Edited by:
Takayoshi Ubuka, International Cancer Laboratory Co., Ltd., JapanReviewed by:
Mirian Romitti, Université libre de Bruxelles, BelgiumRasim Mogulkoc, Selçuk University, Türkiye
Copyright © 2023 Mao, Tang, Li, Zhang, Liu, Wang and Headar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heyong Wang, d2FuZ2hleUBzY3V0LmVkdS5jbg==
 Xiaojian Mao1
Xiaojian Mao1 Heyong Wang
Heyong Wang