
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol. , 30 August 2023
Sec. Developmental Endocrinology
Volume 14 - 2023 | https://doi.org/10.3389/fendo.2023.1205408
The maternal-fetal interface is defined as the interface between maternal tissue and sections of the fetus in close contact. RNA methylation modifications are the most frequent kind of RNA alterations. It is effective throughout both normal and pathological implantation and placentation during pregnancy. By influencing early embryo development, embryo implantation, endometrium receptivity, immune microenvironment, as well as some implantation and placentation-related disorders like miscarriage and preeclampsia, it is essential for the establishment of the maternal-fetal interface. Our review focuses on the role of dynamic RNA methylation at the maternal-fetal interface, which has received little attention thus far. It has given the mechanistic underpinnings for both normal and abnormal implantation and placentation and could eventually provide an entirely novel approach to treating related complications.
The maternal-fetal interface is the interface between maternal tissue and fetal components in direct contact. It is primarily made up of the placenta and maternal decidua (1). At the cellular level, these structures are predominantly composed of trophoblast, decidual, and immune cells (2). Additionally, the normal early embryonic growth is necessary for the development of the maternal-fetal interface. Poor maternal-fetal interface development predisposes to a variety of unfavorable pregnancy outcomes, like miscarriage and preeclampsia (3, 4).
RNAs play a crucial role in controlling biological functions. They have emerged as key regulators of RNA function and metabolism (5). In eukaryotes, RNA methylation modifications are the most common type of RNA alteration in eukaryotes, accounting for 60% of all RNA modifications (6). N6-methyladenosine (m6A), N1-methyladenosine (m1A), 5-methylcytidine (m5C), and others are prevalent types of RNA methylation. The most common type of RNA methylation on messenger RNAs (mRNAs) in complex organisms is m6A RNA methylation (7). Normal and abnormal implantation and placentation are both affected by RNA methylation. By influencing the early stages of embryo development, trophoblast invasion, endometrial receptivity, and immunological microenvironment, it is essential for the establishment of the maternal-fetal interface (8). Furthermore, preeclampsia and other disorders associated with implantation and placentation are closely correlated with RNA methylation (9).
Despite significant breakthroughs in the study of pregnancy, it is still exceedingly challenging to comprehend how RNA methylation works in human implantation. Our review covers the most recent RNA methylation studies and discusses the several forms of RNA methylation in humans, as well as the role of regulatory factors. Furthermore, we discuss the significance of dynamic RNA methylation in implantation and placental development. In addition, our review highlights the previously unknown involvement of RNA methylation in the formation of the maternal-fetal interface. In brief, this paper aims to review recent findings on developmental changes at the maternal-fetal interface associated with RNA methylation and its implications for implantation and placenta-associated diseases. This review is crucial because it provides a comprehensive overview of the current state of research on RNA methylation at the maternal-fetal interface, points to future research directions, and offers the prospect of a new strategy for the future treatment of related disorders (Figure 1).
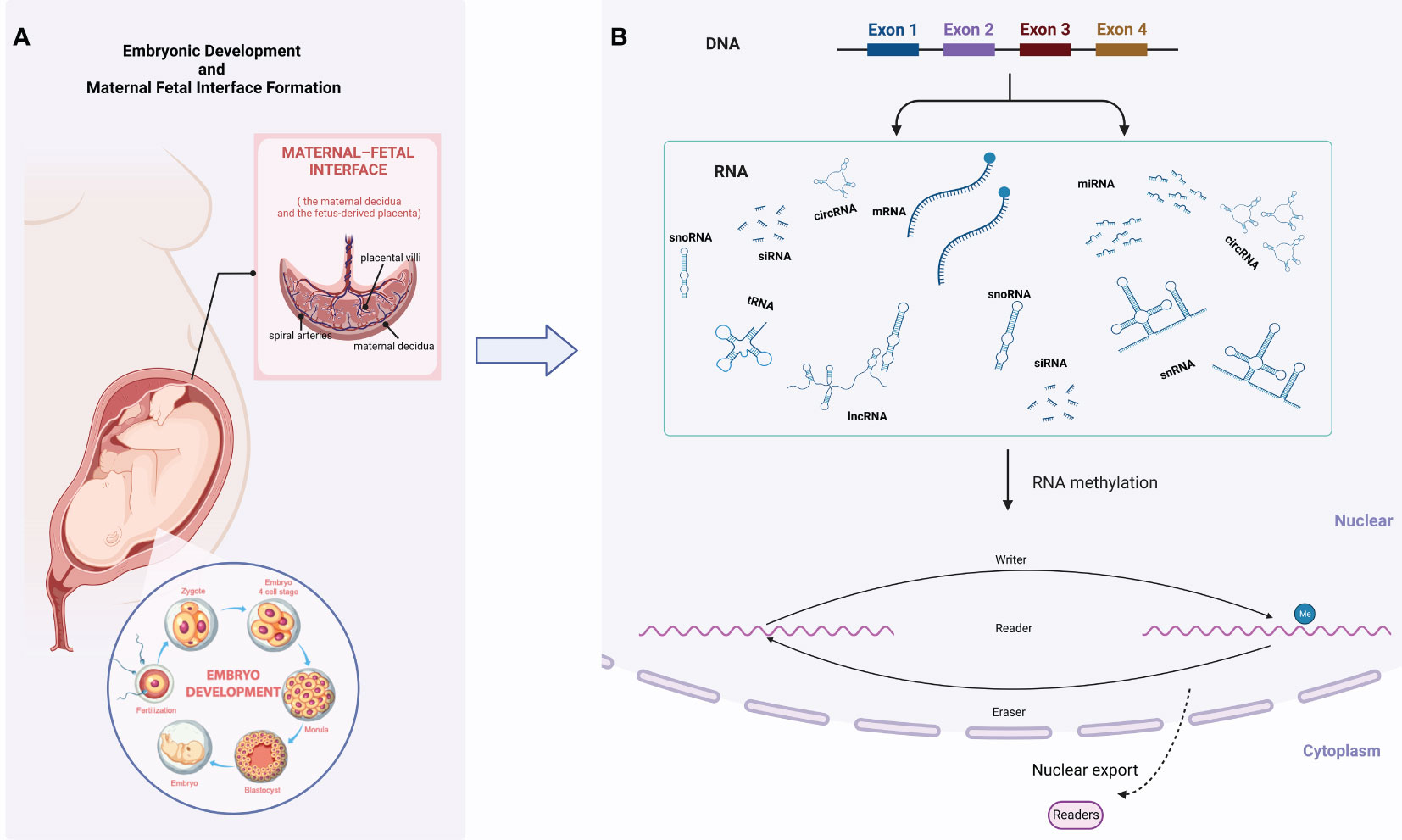
Figure 1 Graphical abstract. (A) Embryonic development and maternal-fetal interface formation; (B) RNA methylation.
In both prokaryotes and eukaryotes, over 100 different types of post-transcriptional chemical modifications of RNA have been discovered in messenger RNA (mRNA), ribosomal RNA (rRNA), transfer RNA (tRNA), and small RNA (10, 11). RNA methylation is one of the most major epigenetic modifications in post-transcriptional RNA modification (12–14). It is estimated that methylation is responsible for more than 60% of RNA alterations (15). They all play critical roles in metabolism, structural stability, and RNA synthesis (Table 1) (85, 86).
As early as 1977, Karen Beemon et al. found residues of m6A in Rous sarcoma virus RNA (87). Since two groundbreaking papers in 2012 demonstrated the distribution of m6A residues in the mammalian species, there has been an increase in interest in RNA-methylation-based research (88, 89). Previous studies reveal that m6A modification occurs preferentially at the m6A consensus motif RRACH, but not all RRACH sequences will have m6A modification (90). The dynamic network of m6A is regulated by methyltransferase complex (“writer”) and demethylase (“eraser”), with “writer” adding m6A modification and “eraser” exerting demethylation activity (91). Moreover, the downstream functions of m6A are mediated by RNA-binding proteins (“reader”) that recognize m6A and regulate RNA processing (35, 92).
METTL3 is the first methyltransferase discovered and it is one of the most fundamentally important “writers” (16). METTL14 and METTL3 share regions of sequence similarity (>35% nucleotide sequence identities) (93). They can form a heterodimeric core complex, with METTL3 acting primarily as a catalytic core and METTL14 acting as an RNA binding platform (94–96). The solution structure of the zinc finger structural domain (ZFD) of METTL3 fulfills the methyltransferase activity of the complex (97). The METTL3- METTL14 dimer mediates the deposition of m6A on nuclear RNA in mammalian cells (17). WTAP is another regulatory subunit of m6A methyltransferase in RNA (18). WTAP itself has no catalytic activity for m6A modification because it lacks a conserved catalytic methylation domain. However, it acts as a bridging protein for the interaction with METTL3 and METTL14, thus significantly affecting RNA m6A loading (17). The modification can be reversed by FTO and ALKBH5 since they are chief erasers that mediate the m6A demethylation process (28, 29). In addition, the m6A “readers” mainly belong to the YT521-B homology (YTH) family and contain the YTH structural domain (98). IGF2BPs are a recently identified class of m6A readers that preferentially recognize m6A-modified mRNAs and help to maintain the stability of thousands of possible mRNA targets, including MYC (44).
A growing body of evidence suggests that m6A is involved in RNA structural stability, translation, and degradation at the mRNA level (99–103). It can modulate protein binding by inducing RNA structural changes that alter the accessibility of the protein binding site (43). Additionally, m6A modification alteration serves several functions in linking pre-mRNA maturation to mRNA fate. Recent studies have shown that m6A in mRNA appears to be involved in the regulation of selective splicing, translation efficiency, and stability. m6A also controls the lifetime and degradation of mRNA. There is sufficient evidence that the YTH structural domain family promotes mRNA degradation. YTHDF1 interacts with AGO2 via the YTH structural domain and then undergoes liquid-liquid phase separation (LLPS) leading to mRNA degradation by P-body formation (104).
Similar to mRNA, m6A-related modifications have also been found to be involved in non-coding RNA synthesis and metabolism (60, 105). METTL3 can methylate chromosome-associated regulatory RNAs (carRNAs), such as promoter-associated RNAs (paRNAs), enhancer RNAs (eRNAs), and RNAs produced from transposable elements (repeat RNAs). These carRNAs are destabilized by m6A methylation, which affects adjacent chromatin states and downstream transcription (106).
In addition, the RNA writers, erasers, or readers mentioned above are thought to be involved in DNA methylation processes. FTO and ALKBH5, two m6A erasers, may also be related to DNA methylation. Evidence suggests that ALKBH5 demethylates DNA’s 3-methylcytosine and has limited DNA repair activity (107). FTO, however, has a negligible impact on the demethylation of 5-methyl-2’-deoxycytidine (5mdC) in DNA (108). However, this might lead to intriguing new directions for an interaction between DNA methylation and RNA methylation regulators. Recent studies have also shown that RNA m6A methylation can reverse chromatin remodeling and histone alterations. Shuang Deng et al. discovered that RNA m6A modifier reader FXR1 can recruit the DNA 5-methylcytosine dioxygenase TET1 to genomic areas and remove DNA methylation (109, 110).
Aside from m6A, m5C is another common modification found in RNAs. During m5C modification, “writers”, “erasers”, and “readers” may act through similar mechanisms compared to m6A (111). Interestingly, recent studies have revealed that the m6A and m5C modifications of the same RNA may interact and synergistically affect protein expression patterns (112, 113). m5C can be installed by any of the seven proteins of the Nol1/Nop2/SUN domain (NSUN) family as well as DNMT2 (114).
As an important epigenetic modifier in RNA, m5C plays a central role in governing transcript abundance. NOP2/NSUN1, a m5C RNA methyltransferase, inhibits transcription and promotes viral latency by competing with TAT for TAR binding and methylation (115). m5C RNA methyltransferase can bind to and stabilize mRNA. As a typical tRNA methyltransferase, methylation of p16 3’UTR by NSUN2 may stabilize p16 mRNA (116). m5c modification has also been shown to regulate the nucleo-plasmic shuttling mechanism of RNA. NSUN2 can regulate the nucleo-plasmic shuttle and RNA binding of ALYREF. After NSUN2 knockdown, nuclear retention of ALYREF is enhanced and, subsequently, cytoplasmic localization is reduced (117). In mammals, ALYREF recognizes and exports YBX2 and SMO mRNAs with m5C modifications from the nucleus to the cytoplasm (118). These findings strongly suggest that m5C is specifically recognized by ALYREF and thus promotes selective mRNA export. In addition, cytosine methylation level and the function of RNA translation are interdependent (119). m5C methylation increases translation levels in NSUN6-targeted mRNAs. Further evidence from ribosome analysis shows that NSUN6-specific methylation is connected to translation termination (120).
Additionally, these RNA m5C methylation-related writers, readers, and erasers might also be involved in DNA methylation. For example, DNA methylation is carried out by members of the DNA methyltransferase (DNMT) family (121). However, mammalian DNMT2 is also a tRNA methyltransferase with a conserved tRNA recognition mechanism (122). DNMT2-mediated m5C contributes to the secondary structure and biological properties of small non-coding RNAs (sncRNAs) and promotes the ability of sperm RNA to induce metabolic changes in offspring (123).
m5C alterations are also involved in the maturation and structural changes of non-coding RNAs. Based on miCLIP-Seq data, Han Liao et al. discovered that NOP2/NSUN1 catalyzes the deposition of m5C on 28S rRNA and regulates the processing of pre-rRNA (124). The NSUN4-mTERF4 complex is involved in the maturation of 16S rRNA (125), while NSUN5 is associated with overall protein synthesis and normal growth (126).
m5C modification in RNA is considered to be reversible. Demethylation is carried out by ten-eleven translocation (TET) enzymes. Although the catalytic activity of TET proteins was thought to be limited to DNA for several years, current research has begun to uncover their potential significance in altering RNA bases. TET2 has been shown to promote the conversion of 5-methylcytosine on tRNA to hm5C as well as to regulate the processing and stabilization of different classes of tRNA fragments (51). This may be related to intracellularly mediated m5C oxidation by TET2 (127, 128).
Furthermore, readers are responsible for reading the information on RNA m5C modification. ALYREF was characterized as a nucleoplasmic mRNA m5C reader (117). Further studies indicated that CDKN1A is an ALYREF target and a reader of m5C-modified mRNA (129). In humans, FMRP has been identified as a novel “reader” of m5C modification, promoting mRNA-dependent repair and cell survival in cancer (130). Moreover, YTHDF2 can also be considered as a type of m5C reader. It can bind directly to m5C in rRNA based on Trp432, a conserved residue located in the hydrophobic pocket of YTHDF2 (59).
Overall, the functional roles of m5C modification involve pre-RNA processing and splicing, mRNA stability, and other processes (131–135). However, the detailed mechanism is not clear. In addition, current research on m5C demethylases is restricted, and its role in RNA has to be investigated further.
As a novel form of RNA modification, m1A has received considerable attention in recent years. m1A is mainly present in the T-loop structure of tRNA and is introduced by the TRMT6/TRMT61A complex. It appears less commonly in mRNA. Modi Safra et al. discovered a m1A site catalyzed by TRMT10C in mitochondrial ND5 mRNA with high tissue-specific and tight developmental control of methylation levels (27).
Erasers act as m1A demethylases, removing the methyl group from m1A and making it functionally reversible. ALKBH1 and ALKBH3 are two common demethylases (52, 54).
Using a photocrosslinking approach, Seo KW et al. discovered that YTHDF1/2 is the reader of m1A modification on RNA and that the binding of YTHDF2 to m1A promotes the degradation of m1A RNA (136).
Nevertheless, little is known about m1A development pathways, and their function in RNA remains to be elucidated.
Notably, over the last few years, more than 100 alterations inside RNAs have been discovered, including m1G, m6G, and others. Further inquiry is required to determine their role in molecular function.
During the establishment of pregnancy, early embryonic development and implantation are important components (137). Normal preimplantation embryo development is a crucial step for a successful pregnancy. The human blastocyst begins to implant in the uterus seven days after fertilization by adhering to the receptive endometrium via its trophectoderm (138).
During the first trimester of human pregnancy, an interface is formed between the maternal decidua and the placenta of the fetus, which is also known as the maternal-fetal interface (139). It is a unique microenvironment composed of the maternal decidua and fetal trophoblast. Immune crosstalk at the maternal-fetal interface is also required for a successful pregnancy (1). Trophoblast, decidual stromal cells, and immune cells are important in metaphase vascularization, maternal immune tolerance, and maternal metabolic changes that facilitate fetal nutrient delivery (4).
RNA methylation may have a major role in the development of the maternal-fetal interaction. In this section, we summarized the function of RNA methylation in this section, focusing on four aspects in particular: early embryo development, embryo implantation, endometrial receptivity, and immunological microenvironment (Figure 2).
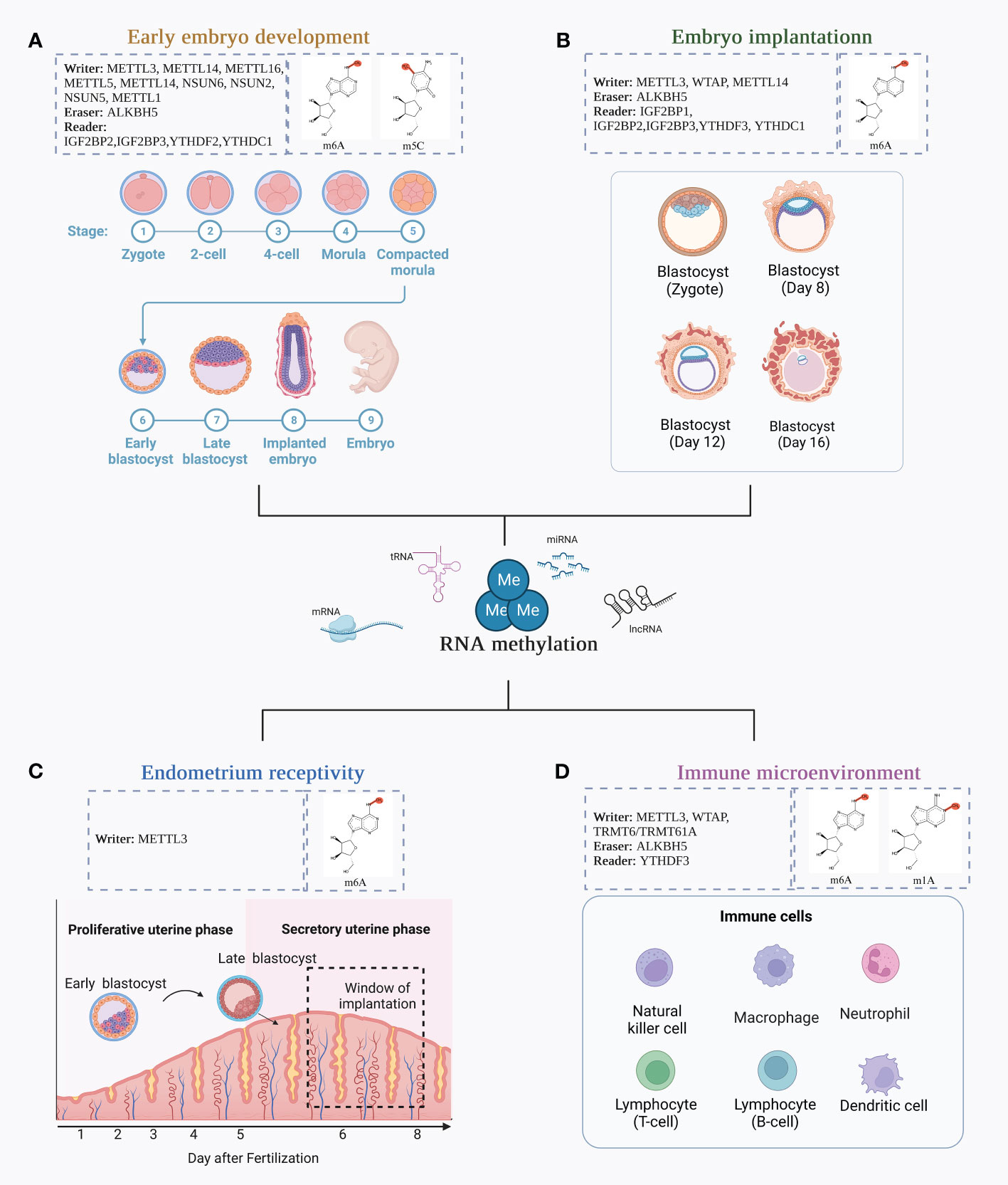
Figure 2 The function of RNA methylation in the formation of the maternal-fetal interface. (A) Early embryo development. Early embryonic development undergoes a transition from the zygote, 2-cell stage, 4-cell stage, morula, compacted morula, early blastocyst, late blastocyst, implanted embryo, and embryo; (B) The changes in blastocyst during embryo implantation; (C) The establishment of endometrium receptivity; (D) Different immune cells involved in the immune microenvironment.
Semen RNA epigenetic modifications have potential effects on early embryogenesis and paternal epigenetic inheritance (140). METTL3-mediated m6A regulation is critical for male fertility and spermatogonia differentiation during spermatogenesis (141). It also modulates protein synthesis in spermatogonial stem cells and spermatids. Dual deletion of METTL3 and METTL14 in advanced germ cells can result in poor spermatogenesis due to altered translational efficiency of m6A-containing transcripts (142). Furthermore, ALKBH5-mediated m6A erasure is required for appropriate splicing and production of longer 3’-UTR mRNAs, and reversible m6A modification is a key mechanism for post-transcriptional control of mRNA fate in late meiotic and haploid germ cells (143). These studies hint to the impact of RNA alteration on sperm development, which is responsible for the normal development of the embryo after fertilization.
Maternal RNA degradation, comprising maternal (M) and zygotic (Z) decay pathways, is also essential for oocyte maturation and embryogenesis. YAP and TUT4/7 may regulate maternal mRNA Z-decay during the human maternal-to-zygote transition (MZT) and play an important role in the early development of mouse and human embryos (144, 145). RNA methylation is believed to have a major role in maternal RNA degradation. In Drosophila, METTL3-METTL4 complex-mediated m6A modification contributes to normal embryogenesis and may be involved in the partial degradation of maternal RNA. Additionally, FMR1 preferentially binds RNAs containing the m6A-modified “AGACU” and controls the decay of its target parent mRNA in a m6A-dependent way (146). During the MZT, transcriptionally silenced embryos are dependent on post-transcriptional regulation of maternal mRNA until zygote genome activation (ZGA) (147). There is evidence that the recently identified m6A reader IGF2BP recruits the mRNA stabilizer HuR to prevent the degradation of m6A-containing mRNAs and to promote their translation (44). Maternal mRNA stability in zebrafish has been found to be significantly regulated by IGF2BP3. In zebrafish, cytoplasmic division and cytoskeletal processes are reportedly mediated by AURKB (aurora B). As a target of IGF2BP3, AURKB showed significantly reduced mRNA expression in embryos with IGF2BP3 knockdown, resulting in quick degradation of maternal mRNA (148, 149). The m6A reader YTHDF2 are also proved to primarily affect the methylated maternal mRNA at 4 h.p.f, regulating zebrafish maternal mRNA clearance and thereby controlling development through MZT (149). Further research indicated that the methyltransferase METTL16 is essential for the viability of early mouse embryos by modulating MAT2A mRNA. The loss of METTL16 causes a dramatic change in the E3.5 blastocyst transcriptome, preventing the further development of mouse embryos (150). Most recently, single-cell m6A sequencing (scm6A-seq) methods were used to analyze the m6A methylome and transcriptome in individual oocytes as well as organelles of cleavage-stage embryos. It has been demonstrated that METTL3-catalyzed m6A primarily affects RNA stability and can preferentially promotes degradation in germinal vesicle oocytes. In the field of single-cell research, the findings indicated dynamic m6A regulation in RNA metabolism during oocyte development and syncytial genome activation (151). In addition, the m6A reader YT521-B homology domain protein 1 (YTHDC1) and its target m6A RNAs function upstream of SETDB1 to maintain mouse embryonic stem cells and repress retrotransposons and bicellular phase (152).
After implantation of the embryo in the uterus, RNA post-transcriptional modifications also regulate germ layer growth. The rRNA m6A methyltransferase METTL5 is engaged in the developmental process (153). METTL5 must form a heterodimeric complex with TRMT112 to establish metabolic stability in cells (56). It methylates 18S rRNA both in vivo and in vitro, and METTL5-deposited m6 A is involved in regulating efficient translation of the F-box and WD repeat domain-containing 7 (FBXW7) to enhance mouse embryonic stem cell differentiation (154, 155). In addition, evidence suggests that m6A deposition is particularly necessary for the induction of ectoderm, but not for the differentiation of the stereotyped endoderm. METTL14 promotes ectodermal maturation, which is necessary for post-implantation development in mice. The absence of METTL14 and the associated drop in m6A levels impede the ability of ESC initiation and further differentiation, resulting in the inability of mouse ESC to transition from the naive to the initiating state (156). However, various researchers believe that m6A deposition is equally crucial in the induction of endodermal differentiation. Arginine methylation of METTL14 is a novel molecular mechanism that regulates m6A deposition. m6A deposition is regulated by PRMT1-mediated arginine methylation of METTL14 at its disordered C-terminal region (157). The R255 location of METTL14 was discovered to impact the endodermal differentiation of mouse embryonic stem cells, implying a direct link between protein methylation and RNA methylation (158). Of note, the exact effect of m6A methylation modification of RNA on which germ layer differentiation takes place still needs further study.
Other methylation changes, such as m5C methylation, have been demonstrated to have similar consequences to m6A methylation in early embryonic development.
The majority of m5C sites in protein-coding RNAs are catalyzed by NSUN6. Evidence suggests that embryonic development in mice is largely unaffected by the complete loss of NSUN6. In vitro cellular investigations and in vivo animal assays were carried out for verification. Reduced levels of NSUN6-targeted mRNAs are demonstrated in embryonic-like bodies lacking NSUN6 (120). However, there are currently conflicting perspectives on whether m5c promotes embryonic development. Jianheng Liu emphasized the functional importance of the maternal mRNA m5C by quantitative mRNA m5C maps of six vertebrate and invertebrate species at different developmental stages. They discovered that NSUN2 deletion embryos incur cell cycle stoppage or delay and fail to commence maternal-to-zygotic transition in time using D. mel as a model (159). Moreover, NSUN5 deficiency leads to decreased m5C in the exon and 3’UTR regions as well as a reduction in the translation efficiency of mitotic arrest deficient 2 like 2 (MAD2L2) and growth differentiation factor 9 (GDF9) in the ovary, inhibiting folliculogenesis and development during ovarian aging. This suggested that NSUN5/m5C-regulated maternal mRNA stability is essential for MZT transition as well (160). In zebrafish, Ybx1 and Pabpc1a coordinate to regulate the stability of m5C-modified maternal mRNA. Y-box binding protein 1 (Ybx1) recognizes m5C via π-π interaction with the key residue Trp45 in the Ybx1 cold shock domain (CSD) preferentially C-modified mRNA, which plays a role in maternal mRNA stability and early embryogenesis (161). Additionally, the RNA cytosine methyltransferase NSUN3 is an important factor in ESC differentiation, especially in neuroectodermal differentiation. Inactivation of NSUN3 skews ESC differentiation towards the mesendoderm and affects Wnt signaling and mitochondrial ROS production in embryonic stem cells (162).
Based on m7G methylated tRNA immunoprecipitation sequencing (MeRIP-Seq) and tRNA reduction and cleavage sequencing (TRAC-Seq), increased ribosome occupancy of the corresponding codon in METTL1 knockout mESCs was observed. This suggests an important role for the mammalian m7G tRNA methylome in ESCs (163).
However, the specific effects of RNA methylation on different phases of embryonic development, whether in m5C modification or m7G modification, have yet to be studied. Their different roles and mechanisms in sperm development, egg maturation, MZT, and embryo layer differentiation after embryo implantation remain the focus of future research.
Overall, RNA methylation is involved in spermatogenesis, egg maturation and MZT, and embryo layer differentiation in early embryo development (Figure 3). Further research is still required for the possible different mechanisms of RNA methylation and demethylation in the development of embryos before and after implantation. Moreover, it also needs further exploration of the function of m5C and m7G RNA modification during early embryo development.
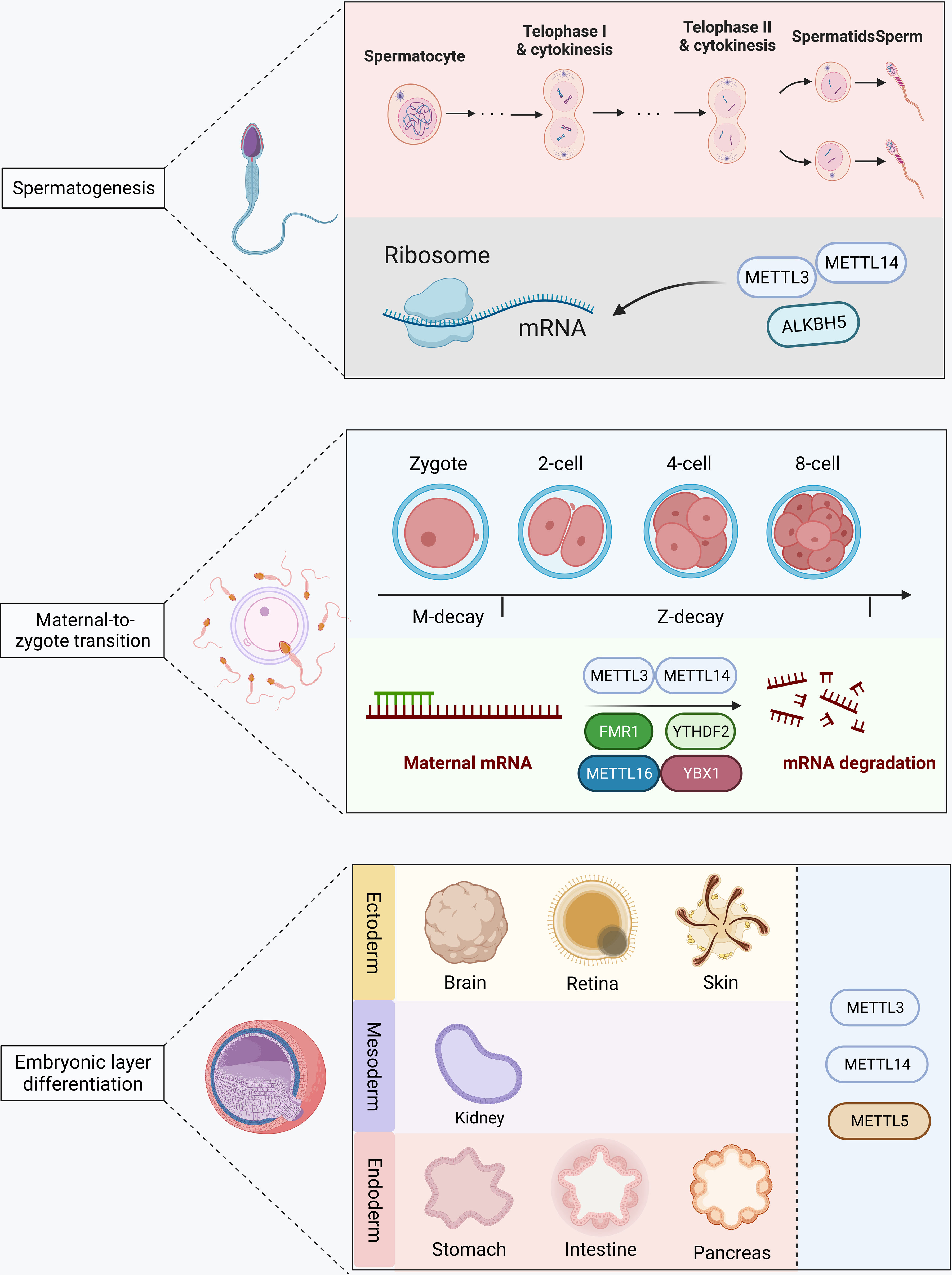
Figure 3 RNA methylation in the early embryo development. METTL3, METTL14, and ALKBH5 modify mRNA and participate in spermatogenesis. METTL3, METTL14, METTL16, FBX1, FMR1, and YTHDF2 are involved in maternal mRNA degradation during the maternal-to-zygote transition. METTL3, METTL14, and METTL15 are involved in germ layer differentiation after embryo implantation.
Embryo implantation is one of the important aspects of maternal-fetal interface formation. The embryo implantation requires the degradation of the extracellular matrix (ECM) and the establishment of uterine receptivity (164). Embryonic trophoblasts are also important for embryo implantation and placental development. The proliferation, differentiation, and invasive activity of trophoblast cells are all linked to successful embryo implantation, placenta formation, and growth and development. During embryo implantation, trophoblast cells adhere to and invade the maternal endometrium and eventually form the placenta. After implantation, trophoblasts invade and remodel the decidual spiral arteries, providing the basis for material exchange and oxygen transport between the fetus and the mother. Angiogenesis occurs at the maternal-fetal interface and plays a key role in fetal development as the embryo grows after implantation. Thus, trophoblast invasion is a hallmark of placental development in early pregnancy and plays an important role in the implantation and formation of the maternal-fetal interface during pregnancy (165). Trophoblast invasion behavior and its regulatory factors have become a hot topic of current research.
Trophoblast invasion can be regulated by dynamic methylation modifications of mRNA. By modulating MYLK expression, METTL3 enhances trophoblast cell invasion (166). Further analysis revealed that RNA methylation may affect trophoblast invasion by influencing mRNA stability. Based on stromal gel invasion assay and transwell migration assays with METTL3 over-expressing and knockdown in HTR-8 cell lines, the reduction of METTL3-mediated m6A modification attenuated the attenuation of ZBTB4 and increased the expression of ZBTB4, thereby impairing the invasive ability of trophoblast cells. m6A modification in RNA is crucial for controlling trophoblast invasion (167). Another m6A methyltransferase, WTAP, promotes trophoblast invasion and metastasis by regulating the stability of HMGN3 mRNA in a m6A-dependent way (168). Reduced m6A modification may result in inadequate early trophoblast implantation and produce poor pregnancy outcomes. By comparing ALKBH5 and m6A mRNA methylation levels in patients with recurrent miscarriage and normal population, Xiao-Cui Li et al. found that ALKBH5 controls CYR61 mRNA stability by regulating m6A methylation and regulates early gestational trophoblast invasion through its effect on mRNA m6A methylation. CYR61 mRNA levels in villi were inversely linked with ALKBH5 mRNA levels, and ALKBH5 knockdown significantly promoted cell invasion (169). DNA microarray analysis of endometrial cells revealed that IGP2BP family molecules (m6A readers), such as IGF2BP1 and IGF2BP3, were upregulated in human trophoblast ectoderm cells and receptive endometrium cells during the implantation window (170). The relationship between embryo implantation and the IGF2BP growth factor properties or the methylation effect, however, has not yet been thoroughly investigated. In addition to m6A methylation, other modifications in RNA also work in the biological behaviors of trophoblast cells. 5-hmC RNA methylation is also associated with cell invasion and early placental formation. In early pregnancy, hypoxia is the typical extrinsic factor regulating trophoblast migration invasion and placental formation. TET1 is an enzyme that converts 5-mC to 5-hydroxymethylcytosine (5-hmC). By simulating the physiological hypoxic conditions of early pregnancy, TET1 was found to be activated in cells exposed to 3% O2, and TET1 knockdown inhibited cell migration and invasion (171). It has also been proposed that m1A methylation influences trophoblast invasion. YTHDF3 selectively interacts to m1A-containing RNA, and hypoxia downregulates its expression in trophoblast HTR8/SVneo. YTHDF3 suppresses IGF1R expression by binding to m1A modifications and enhances IGF1R mRNA attenuation, inhibiting MMP9 expression and, as a result, trophoblast invasion (172).
These studies demonstrated that trophoblast cell invasion is mediated by revisable RNA methylation. Additionally, RNA methylation alterations encourage trophoblast invasion, while demethylation modifications prevent it. Nonetheless, a few considerations challenge the hypothesis. Evidence shows that hypoxia treatment can significantly upregulate ALKBH5 expression and induced ALKBH5 translocation from the nucleus to the cytoplasm, followed by ALKBH5 demethylation of m6A modification on SMAD1/5 mRNA, which promoted its expression and activated the TGF-β signaling pathway to induce MMP9 expression, thereby increasing trophoblast migration and invasion (173). ALKBH5 has also been demonstrated to reduce the m6 A level of PPARG mRNA and increase the stability of PPARG mRNA, hence increasing PPARG translation. PPARG overexpression promotes trophoblast cell proliferation and migration through the upregulation of ALCAM expression (174). Another study revealed that overexpression of METTL14 in HTR8/SVneo cells inhibited trophoblast proliferation and invasion by increasing FOXO3a expression (175). These results reveal the complexity of the regulation of trophoblast invasion by RNA methylation.
Non-coding RNA methylation also affects trophoblast cell proliferation and invasion. YTHDC1, for example, degrades circMPP1 via mA6 alteration, enhancing placental villi function (176). The m6A modification of circSETD2 represses miR-181a-5p and increases MCL1 transcription, thereby regulating trophoblast cells (177).
Mounting evidence implies that B(a)P/benzo(a) pyrene-7,8-dihydro diol-9,10-epoxide (BPDE) inhibits the proliferation of human chorionic trophoblast cells and is related to RNA m6A modification. Mengyuan Dai e al. discovered a novel lncRNA, lnc-HZ09, and its putative function in recurrent miscarriage villous tissue and human trophoblast cells exposed to BPDE. Cells over-expressing METTL3 have higher levels of m6A RNA modification on lnc-HZ09 and result in reduced stability of lnc-HZ09. lnc-HZ09 regulates trophoblast migration and invasion through the PLD1/RAC1/CDC42 pathway, and lnc-HZ09-silenced cells exhibit significantly greater migration and invasion (178). In addition, lnc-HZ01 and METTL14 were also shown to be upregulated in the recurrent miscarriage group compared to the normal group in villi tissue. METTL14 levels were positively correlated with m6A methylation levels on lnc-HZ01 levels. The downstream MXD1/EIF4E pathway is activated, which reduces trophoblast growth and causes miscarriage (179).
These findings highlight the significance of RNA methylation in controlling the biological behaviors of trophoblast cells. However, whether RNA methylation changes on trophoblast cells promote or impede invasion is debatable. Furthermore, the majority of the aforementioned pathways were discovered during the aberrant trophoblast implantation experiments. It is unclear whether these mechanisms have a role in normal trophoblast invasion and maternal-fetal interface establishment during early pregnancy.
Decidualization refers to the functional and morphological changes occurring in the endometrium. The main changes that occur during metaplasia are changes in the endometrial stromal cells, increased uterine gland secretion, increased uterine natural killer cells, and vascular remodeling. Accumulating experimental and clinical evidence demonstrated that defective decidualization or blocked decidualization may lead to abnormal maternal-fetal interface (180).
Endometrial stromal cells are the most common type of cell involved in decidualization. During embryo implantation, METTL3 is necessary for the decidualization of human endometrial stromal cells in vitro. Progesterone receptor (PGR) mRNA is a direct target of METTL3-mediated m6A modifications. In METTL3f/f mice, the m6A-modified PGR mRNA in 5 ‘- UTR is recognized by YTHDF1, thereby promoting PGR protein translation. However, in Mettl3d/d mice, the m6A modification in PGR mRNA is missing, and the PGR protein cannot be effectively translated. The low level of PGR protein ultimately leads to implantation and decidualization failure. The METTL3-PGR axis may be conserved between mice and humans (181). Of note, overexpression of METTL3 increases m6A methylation and destroys embryo attachment by inhibiting the expression of endometrial receptive biomarker HOXA10 (182).
Additionally, a major hallmark of decidualization is the mesenchymal-epithelial transformation (MET) of endometrial mesenchymal cells (183). The transformation between mesenchymal and epithelial was strongly correlated with RNA methylation. METTL3 enhances ZMYM1 mRNA expression through an m6A-HuR-dependent pathway, and epigenetic activation of ZMYM1 can lead to epithelial-mesenchymal transformation (EMT) (45). m6A-seq data revealed that during EMT, m6A modification occurs on a few genes related to cell attachment, adhesion, and migration (184). However, the effect of RNA methylation on the transition between mesenchyme and epithelium has not been demonstrated in the endometrium. Whether RNA methylation promotes decidualization through MET remains a mystery.
Collectively, the above findings imply that m6A methylation is linked to the decidualization of human endometrial stromal cells and may potentially be linked to MET. More research, however, was required to confirm the rationale behind these findings.
The immune microenvironment at the maternal-fetal interface is essential for maintaining normal embryonic development. Decidual natural killer (dNK) cells are the most prevalent in the maternal-fetal interface, followed by decidual macrophages (dM Φ) and then decidual T cells (185).
There is evidence that m6A methylation and NK cell effector function are linked to METTL3 expression levels. METTL3-mediated m6A methylation protects the signaling pathway downstream of IL-15R, thereby regulating the responsiveness of NK cells to IL-15 (186). NPM1 is associated with m6A methylation and immune infiltration. Its expression is linked to B-cell and NK-cell marker genes (187). However, there is currently no direct evidence to prove the influence of RNA m6A modification on dNKs.
RNA methylation can affect macrophage polarization. The m1A “reader” YTHDF3 may be involved in regulating macrophage polarization that promotes aortic inflammation (188). According to the findings of the ssGSEA analysis, the maternal-fetal interface comprises a substantial number of immune-related cells, with ALKBH5 serving as the key m6A regulator of macrophages. Overexpression of ALKBH5 in ESC affects macrophage differentiation through VEGF secretion (189). Mechanically, METTL3 can drive M1 macrophage polarization through direct methylation of STAT1 mRNA and may serve as an anti-inflammatory target (190).
RNA methylation also affects TCR signaling and determines T cell activation and survival. WTAP and m6A methyltransferase functions are required for thymocyte differentiation and control of activation-induced peripheral T-cell death (191). By targeting the IL-7/STAT5/SOCS pathways, m6A mRNA methylation controls T cell homeostasis (192). It also maintains Treg inhibition through the IL2-STAT5 signaling axis (193). In addition, TRMT6/TRMT61A complex-mediated m1A modification of transfer RNA (tRNA) promotes the synthesis of MYC proteins essential for T cell proliferation (194, 195). This demonstrates that m1A methylation also has important effects on T cells.
Based on the above findings, we propose that RNA methylation may have important effects on the immune microenvironment. Changes in NK cell function, macrophage polarization, and T cell activation are all linked to RNA methylation. Although the majority of the current studies on the impact of RNA methylation in the immune microenvironment focus on the tumor immune microenvironment, these studies can provide some insight into the role of RNA methylation in the maternal-fetal immune microenvironment to a certain extent.
Taken together, due to its ability to promote embryo development, regulate embryo implantation, extend uterine receptivity, and provide a stable immune microenvironment, RNA methylation is important in the formation of the maternal-fetal interface.
Common implantation and placentation-related diseases like miscarriage, preeclampsia, and preterm birth have been proposed to be associated with aberrant RNA methylation in the formation of the maternal-fetal interface. Abnormalities in RNA methylation may impact the establishment of the maternal-fetal contact, ultimately leading to illness.
Miscarriage, especially recurrent miscarriage, may be multifactorial. Problems in any part of the maternal-fetal interface formation can lead to miscarriage. In this process, RNA methylation probably plays an important role. It was discovered that m6A levels in the uterus rise throughout pregnancy. The mRNA levels of the methyltransferases METTL16 and WTAP were lower in the endometrium of patients with miscarriage and infertility, whereas the mRNA levels of ALKBH5 and IGF2BP2 were higher (196).
In the process of early embryonic development, abnormal methylation in epigenetic factors of both the father and mother can affect embryonic development (197). Abnormal RNA methylation plays a role in abnormal embryo implantation and may also lead to spontaneous abortion. High-throughput sequencing of villous tissue from the spontaneous abortion group and the normal group during early pregnancy, based on combined analysis of meRIP-seq and RNA-seq data, revealed that genes with differential m6A methylation were primarily related to the Wnt signaling pathway, phosphatase activity regulation, protein phosphatase inhibition activity, and transcriptional inhibition activity (198). The expression levels of ALKBH5 and CYR61 were negatively correlated in the villous tissue of the recurrent abortion group, which is related to the pathogenesis of recurrent abortion (169).
Moreover, FTO deficiency in the chorionic villi affects immunological tolerance, leading to aberrant methylation and oxidative stress and, eventually, spontaneous abortion. Compared with normal pregnant women, the levels of FTO-binding HLA-G, VEGFR, and MMP9 mRNA in spontaneous abortion patients were significantly reduced, and the concentration of FTO-binding MMP7 was significantly increased. This is due to oxidative stress and abnormal m6A accumulation at the maternal-fetal interface (199).
These all prove that abnormal methylation of RNA might be one of the reasons for spontaneous abortion.
Preeclampsia is classified as early-onset or late-onset based on when it appears, which is usually at 34 weeks of gestation. Early-onset preeclampsia often results in a high perinatal mortality rate, significant placental dysfunction, and poor maternal and fetal prognosis. Our review focuses on early-onset preeclampsia because it is the most severe and is a major cause of maternal and fetal death. It is strongly associated with RNA methylation. Trophoblast dysfunction is thought to be an important cause of early-onset preeclampsia, and RNA methylation has a major impact on trophoblast function in two ways, according to the existing studies.
On the one hand, RNA methylation influences trophoblast invasion in patients with preeclampsia. We discovered that DNMT3B and TET3 were significantly elevated in PE samples compared to the normal group by analyzing the mRNA expression levels of m5C RNA methylation regulators. Surprisingly, these two methylation regulators have a negative correlation. A comprehensive analysis of UMI-MeRIP Seq data revealed a decrease in the overall peak value of m5C methylation in placental tissue from patients with preeclampsia. However, compared with the normal blood pressure group, the m5C peak value before the coding sequence was stopped in the preeclampsia group significantly increased (200). In addition, preeclampsia placental samples exhibited consistently increased SMPD1 protein levels and increased 5’-UTR of m6A (201). Evidence suggests that m6A RNA methylation expression is significantly increased in trophoblast cells of preeclamptic placentas compared with normotensive placentas and that abnormal m6A modification may contribute to trophoblast cell invasion dysfunction in preeclampsia (202). An increase in m6A-modified circRNA was found to be prevalent in the PE placenta. The m6A-modified circPAPPA2 has a role in trophoblast invasion (203). Several evidences have shown that the downregulation of m6A reader IGF2BP2 is associated with trophoblast invasion disorder and PE. For example, there is a correlation between reduced IGF2BP2 expression and downregulation of MNSF in the placenta of MNSF-deficient mice and patients with severe (204). However, it is yet unknown whether methylation has a role in how IGF2BP affects PE.
RNA methylation, on the other hand, induces trophoblast apoptosis. YTHDF2 can target TMBIM6 mRNA via METTL3, resulting in ROS production and trophoblast apoptosis, exacerbating endoplasmic reticulum stress in preeclampsia (205). Similarly, m6A modification promotes circSETD2 expression, suppresses miR-181a-5p, and increases MCL1 transcription to affect the apoptosis of chorionic trophoblast cells (177).
In short, in patients with preeclampsia, a decrease in the overall peak of m5C methylation or an increase in m6A modification may lead to trophoblast invasion dysfunction. In addition, increased RNA methylation can promote trophoblast cell apoptosis, further exacerbating preeclampsia. Interestingly, the methylation trends of m5C and m6A in preeclampsia appear to be different. In addition, in the process of m5c methylation regulation, DNMT3B and TET3 seem to have opposite effects, but they are highly expressed in preeclampsia patients. In this process, the molecular mechanism of regulators is still unknown.
Preterm birth is defined by the World Health Organization (WHO) as delivery before 37 completed weeks of gestation. Between 25% and 40% of all preterm births are associated with intrauterine infection, including viral and bacterial infections (206).
m6A factors can regulate the virus life cycle. By adding m6A to the genome RNA and mRNA of a virus, changing gene expression, or the fate of corresponding RNA involved in virus or replication, the effect of m6A modification can directly or indirectly regulate virus replication. The strength and duration of innate immune response activation can also be determined by m6A methylation (207). m6A-modified HBV RNA can be coupled to YTHDF2, which can protect viral RNA from being recognized by RIG-I and prevent HBV RNA degradation (208). During the infection cycle, HSV-1 controls the redistribution of the nuclear m6A mechanism in primary fibroblasts. WTAP stays in the nucleus, while METTL3 and METTL14 are distributed throughout the cytoplasm (209). Based on the role of m6A modification in viral infection, m6A modification can be used to develop anti-viral methods. For instance, interferon-stimulating gene 20 (ISG20) can treat HBV infection by selectively recognizing and processing m6A-modified HBV transcripts, and regulated by YTHDF2 (210).
Bacterial infections, in addition to viral infections, can result in premature delivery. The P/Q/N rich domain in m6A reader YTHDF1 interacts with the DEAD domain in host factor DDX60, thereby regulating the immune response to bacterial infection by recognizing the target Traf6 transcript (211).
Moreover, in patients with spontaneous preterm delivery, abnormalities in immune function at the maternal-fetal interface may be a major cause of their preterm delivery (212). The association between RNA methylation and the immune microenvironment has primarily been examined in malignancies, but whether there is a similar relationship at the maternal-fetal interface still needs to be investigated.
Currently, research in the field of RNA modification in preterm birth is mainly focused on infection. The m6A methylation is the most common type of RNA alteration involved. However, whether or not RNA methylation alteration is one of the major biochemical pathways causing premature delivery is still being contested.
Above all, an imbalance in RNA methylation is critical in the development of implantation and placental disorders. Interestingly, the newly discovered programmed cell death pathways, including autophagy, ferroptosis, and pyroptosis are also associated with implantation and placenta disorders. Recently, autophagy has shown to have a role in miscarriage. In women who have had an early abortion, dysregulated autophagy promotes oxidative stress and aberrant production of ABC transporter proteins (213). Many individuals with unexplained spontaneous abortion have insufficient autophagy (214). Rapamycin, a known inducer of autophagy, was found to dramatically boost endometrial autophagy and NK cell residency, as well as improve spontaneous abortion in mice (215). Ferroptosis and pyroptosis have also been linked to preeclampsia in recent studies. Preeclampsia is characterized by hypoxia, which causes iron death of trophoblast cells via apoptosis, autophagy, and necrosis. Iron death-related proteins SRXN1 and NQO1 may be important in the etiology of preeclampsia (216). In early-onset pre-eclamptic placentas and human trophoblast cells exposed to hypoxia and endoplasmic reticulum stresses, pyroptosis is suggested to represent a significant inflammatory mechanism (217). Current evidence suggests that impaired autophagy and increased apoptosis in a placental microenvironment with Th1/Th2 imbalance are associated with spontaneous preterm birth (218).
There is currently a growing body of evidence linking RNA methylation to programmed cell death. m6A methylation is associated with autophagy and plays a role in apoptosis induced diseases. Silencing METTL3 enhances autophagic flux and inhibits apoptosis in H/R-treated cardiomyocytes (219). Moreover, the role of m6A modification and autophagy is of great importance in inflammatory diseases and cancer progression. METTL3-mediated m6A modification of ATG7 regulates the autophagy-GATA4 axis and promotes cellular senescence and osteoarthritis (220).
However, it is unclear whether RNA methylation affects implantation and placenta-associated diseases by influencing programmed cell death. As a result, more evidence is needed to demonstrate whether RNA methylation-mediated programmed death is directly involved in these diseases.
This is the first description of the dynamic RNA methylation function during the maternal-fetal interface development. We performed in-depth investigations of RNA methylation for this review. The three most frequent RNA modifications, m6A, m1A, and m5C, are all essential for the biogenesis, metabolism, structural stability, and function of RNA.
RNA methylation may have a considerable impact on the creation of the maternal-fetal interface by influencing early embryonic development, embryo implantation, endometrial receptivity, and the immunological microenvironment. Furthermore, abnormal RNA methylation during the formation of the maternal-fetal interface is thought to be associated to implantation and placenta-related disorders such as miscarriage, preeclampsia, and preterm birth.
Given the significance of RNA methylation in the formation of the maternal-fetal interface during pregnancy, further investigation is warranted to elucidate the underlying mechanism. The potential various mechanisms of RNA methylation and demethylation during the development of embryos before and after implantation still require further research. Furthermore, there are differing views on whether trophoblast cell RNA methylation changes encourage or impede invasion. The majority of RNA methylation mechanisms in trophoblast cells have been discovered in studies of abnormal trophoblast implantation, but it is still unknown whether these mechanisms contribute to normal trophoblast invasion and the establishment of the maternal-fetal interface in the early stages of pregnancy. There is also no direct evidence for the effects of RNA methylation on the immunological milieu at the maternal-fetal interface. It is noteworthy that whereas m6A methylation has been linked to EMT and metaphase, the underlying regulatory mechanism is yet unknown.
Additionally, it is unclear whether RNA methylation affects implantation and placenta-associated diseases by influencing programmed cell death. From a broader perspective, the role of RNA methylation and programmed cell death in implantation- and placentation-related illnesses is another significant area of challenge and promise for future research.
Together, our study highlights the importance of dynamic RNA methylation at the maternal-fetal interface. It has given the molecular underpinnings for both normal and abnormal implantation and placentation and could eventually offer a fresh approach to treating related disorders.
HY and YJ designed the work and made major revisions to the article. SW and HX wrote the manuscript. YS, XJ, and YM drafted the manuscript and revised the article. All authors contributed to the article and approved the submitted version.
This study was supported in part by the National Key Research and Development Program (2022YFC2704600 and 2022YFC2704604), the National Natural Science Foundation of China (Nos. 82271719, 82071678, and 82171683), the Shanghai Municipal Science and Technology Commission Research Fund (No. 21140903800), the Clinical Research Plan of SHDC (SHDC2020CR2059B), and the Key Research Project of Pudong New Area Population and Family Planning Commission (No. PW2020E-1).
We would like to thank BioRender (https://app.biorender.com/gallery) for its help in creating figures for the article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
mRNA, messenger RNA; rRNA, ribosomal RNA; tRNA, transfer RNA; m6A, N6-methyladenosine; m5C, 5-methylcytidine; m1A, N1-methyladenosine; ZFD, zinc finger structural domain; YTH, YT521-B homology; RNA BS-seq, RNA bisulfite sequencing; ESCs, embryonic stem cells; LLPS, liquid-liquid phase separation; eRNAs, enhancer RNAs; ECM, extracellular matrix; MZT, maternal-to-zygote transition; ZGA, zygote genome activation; scm6A-seq, single-cell m6A sequencing; PGR, progesterone receptor; MET, mesenchymal-epithelial transformation; EMT, epithelial-mesenchymal transformation.
1. Erlebacher A. Immunology of the maternal-fetal interface. Annu Rev Immunol (2013) 31:387–411. doi: 10.1146/annurev-immunol-032712-100003
2. Shi JW, Lai ZZ, Yang HL, Yang SL, Wang CJ, Ao D, et al. Collagen at the maternal-fetal interface in human pregnancy. Int J Biol Sci (2020) 16(12):2220–34. doi: 10.7150/ijbs.45586
3. Zhu D, Zou H, Liu J, Wang J, Ma C, Yin J, et al. Inhibition of hmgb1 ameliorates the maternal-fetal interface destruction in unexplained recurrent spontaneous abortion by suppressing pyroptosis activation. Front Immunol (2021) 12:782792. doi: 10.3389/fimmu.2021.782792
4. Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature (2018) 563(7731):347–53. doi: 10.1038/s41586-018-0698-6
5. Frye M, Harada BT, Behm M, He C. Rna modifications modulate gene expression during development. Science (2018) 361(6409):1346–9. doi: 10.1126/science.aau1646
6. Boulias K, Greer EL. Biological roles of adenine methylation in Rna. Nat Rev Genet (2023) 24(3):143–60. doi: 10.1038/s41576-022-00534-0
7. Zhang H, Shi X, Huang T, Zhao X, Chen W, Gu N, et al. Dynamic landscape and evolution of M6a methylation in human. Nucleic Acids Res (2020) 48(11):6251–64. doi: 10.1093/nar/gkaa347
8. Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by Mrna modifications. Nat Rev Mol Cell Biol (2017) 18(1):31–42. doi: 10.1038/nrm.2016.132
9. Apicella C, Ruano CSM, Méhats C, Miralles F, Vaiman D. The role of epigenetics in placental development and the etiology of preeclampsia. Int J Mol Sci (2019) 20(11):2837. doi: 10.3390/ijms20112837
10. Roundtree IA, Evans ME, Pan T, He C. Dynamic Rna modifications in gene expression regulation. Cell (2017) 169(7):1187–200. doi: 10.1016/j.cell.2017.05.045
11. Motorin Y, Helm M. Rna nucleotide methylation. Wiley Interdiscip Rev RNA (2011) 2(5):611–31. doi: 10.1002/wrna.79
12. Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, et al. Modomics: A database of Rna modification pathways. 2008 update. Nucleic Acids Res (2009) 37(Database issue):D118–21. doi: 10.1093/nar/gkn710
13. Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, et al. The Rna modification database, Rnamdb: 2011 update. Nucleic Acids Res (2011) 39(Database issue):D195–201. doi: 10.1093/nar/gkq1028
14. Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. Modomics: A database of Rna modification pathways. 2017 update. Nucleic Acids Res (2018) 46(D1):D303–d7. doi: 10.1093/nar/gkx1030
15. Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. The Rna modification landscape in human disease. RNA (New York NY) (2017) 23(12):1754–69. doi: 10.1261/rna.063503.117
16. Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and Cdna cloning of the adomet-binding subunit of the human Mrna (N6-adenosine)-methyltransferase. RNA (New York NY) (1997) 3(11):1233–47. Available at:https://rnajournal.cshlp.org/content/3/11/1233.long
17. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A Mettl3-Mettl14 complex mediates mamMalian nuclear Rna N6-adenosine methylation. Nat Chem Biol (2014) 10(2):93–5. doi: 10.1038/nchembio.1432
18. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. MamMalian Wtap is a regulatory subunit of the Rna N6-methyladenosine methyltransferase. Cell Res (2014) 24(2):177–89. doi: 10.1038/cr.2014.3
19. Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, et al. Perturbation of M6a writers reveals two distinct classes of mrna methylation at internal and 5’ Sites. Cell Rep (2014) 8(1):284–96. doi: 10.1016/j.celrep.2014.05.048
20. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. M(6)a Rna methylation promotes xist-mediated transcriptional repression. Nature (2016) 537(7620):369–73. doi: 10.1038/nature19342
21. Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snrna M(6)a methyltransferase Mettl16 regulates sam synthetase intron retention. Cell (2017) 169(5):824–35.e14. doi: 10.1016/j.cell.2017.05.003
22. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. Virma mediates preferential M(6)a mrna methylation in 3’utr and near stop codon and associates with alternative polyadenylation. Cell Discovery (2018) 4:10. doi: 10.1038/s41421-018-0019-0
23. Akichika S, HIrano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R, et al. (6)-methylation of Rna by an Rna polymerase Ii-associated methyltransferase. Science (2019) 363(6423):eaav0080. doi: 10.1126/science.aav0080
24. Bawankar P, Lence T, Paolantoni C, Haussmann IU, Kazlauskiene M, Jacob D, et al. Hakai is required for stabilization of core components of the M(6)a mrna methylation machinery. Nat Commun (2021) 12(1):3778. doi: 10.1038/s41467-021-23892-5
25. Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding Rna. Nucleic Acids Res (2012) 40(11):5023–33. doi: 10.1093/nar/gks144
26. Liu J, Huang T, Zhang Y, Zhao T, Zhao X, Chen W, et al. Sequence- and structure-selective mrna M(5)C methylation by Nsun6 in animals. Natl Sci Rev (2021) 8(6):nwaa273. doi: 10.1093/nsr/nwaa273
27. Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, et al. The M1a landscape on cytosolic and mitochondrial mrna at single-base resolution. Nature (2017) 551(7679):251–5. doi: 10.1038/nature24456
28. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear Rna is a major substrate of the obesity-associated Fto. Nat Chem Biol (2011) 7(12):885–7. doi: 10.1038/nchembio.687
29. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. Alkbh5 is a mamMalian Rna demethylase that impacts Rna metabolism and mouse fertility. Mol Cell (2013) 49(1):18–29. doi: 10.1016/j.molcel.2012.10.015
30. Huang W, Lan MD, Qi CB, Zheng SJ, Wei SZ, Yuan BF, et al. Formation and determination of the oxidation products of 5-methylcytosine in Rna. Chem Sci (2016) 7(8):5495–502. doi: 10.1039/c6sc01589a
31. Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, et al. Tet-mediated formation of 5-hydroxymethylcytosine in Rna. J Am Chem Soc (2014) 136(33):11582–5. doi: 10.1021/ja505305z
32. Arguello AE, Li A, Sun X, Eggert TW, Mairhofer E, Kleiner RE. Reactivity-dependent profiling of Rna 5-methylcytidine dioxygenases. Nat Commun (2022) 13(1):4176. doi: 10.1038/s41467-022-31876-2
33. Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The Rna-binding protein Sam68 modulates the alternative splicing of Bcl-X. J Cell Biol (2007) 176(7):929–39. doi: 10.1083/jcb.200701005
34. Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. Hnrnpa2b1 is a mediator of M(6)a-dependent nuclear Rna processing events. Cell (2015) 162(6):1299–308. doi: 10.1016/j.cell.2015.08.011
35. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent Rna structural switches regulate Rna-protein interactions. Nature (2015) 518(7540):560–4. doi: 10.1038/nature14234
36. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger Rna translation efficiency. Cell (2015) 161(6):1388–99. doi: 10.1016/j.cell.2015.05.014
37. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. Ythdf2 destabilizes M(6)a-containing Rna through direct recruitment of the Ccr4-not deadenylase complex. Nat Commun (2016) 7:12626. doi: 10.1038/ncomms12626
38. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear M(6)a reader Ythdc1 regulates mrna splicing. Mol Cell (2016) 61(4):507–19. doi: 10.1016/j.molcel.2016.01.012
39. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. Ythdf3 facilitates translation and decay of N(6)-methyladenosine-modified Rna. Cell Res (2017) 27(3):315–28. doi: 10.1038/cr.2017.15
40. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mamMalian spermatogenesis. Cell Res (2017) 27(9):1115–27. doi: 10.1038/cr.2017.99
41. Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, et al. N(6)-methyladenosine (M(6)a) recruits and repels proteins to regulate Mrna Homeostasis. Nat Struct Mol Biol (2017) 24(10):870–8. doi: 10.1038/nsmb.3462
42. Arguello AE, DeLiberto AN, Kleiner RE. Rna chemical proteomics reveals the N(6)-methyladenosine (M(6)a)-regulated protein-rna interactome. J Am Chem Soc (2017) 139(48):17249–52. doi: 10.1021/jacs.7b09213
43. Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters Rna structure to regulate binding of a low-complexity protein. Nucleic Acids Res (2017) 45(10):6051–63. doi: 10.1093/nar/gkx141
44. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of Rna N(6)-methyladenosine by Igf2bp proteins enhances Mrna stability and translation. Nat Cell Biol (2018) 20(3):285–95. doi: 10.1038/s41556-018-0045-z
45. Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, et al. Mettl3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer (2019) 18(1):142. doi: 10.1186/s12943-019-1065-4
46. Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mrna export machinery recruited to the 5’ End of mrna. Cell (2006) 127(7):1389–400. doi: 10.1016/j.cell.2006.10.044
47. Lyabin DN, Eliseeva IA, Ovchinnikov LP. Yb-1 protein: functions and regulation. Wiley Interdiscip Rev RNA (2014) 5(1):95–110. doi: 10.1002/wrna.1200
48. Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, et al. Human mettl16 is a N(6)-methyladenosine (M(6)a) methyltransferase that targets pre-mrnas and various non-coding rnas. EMBO Rep (2017) 18(11):2004–14. doi: 10.15252/embr.201744940
49. Brūmele B, Mutso M, Telanne L, Õunap K, Spunde K, Abroi A, et al. Human Trmt112-methyltransferase network consists of seven partners interacting with a common co-factor. Int J Mol Sci (2021) 22(24):13593. doi: 10.3390/ijms222413593
50. Okamoto M, Hirata S, Sato S, Koga S, Fujii M, Qi G, et al. Frequent increased gene copy number and high protein expression of trna (Cytosine-5-)-methyltransferase (Nsun2) in human cancers. DNA Cell Biol (2012) 31(5):660–71. doi: 10.1089/dna.2011.1446
51. He C, Bozler J, Janssen KA, Wilusz JE, Garcia BA, Schorn AJ, et al. Tet2 chemically modifies trnas and regulates Trna fragment levels. Nat Struct Mol Biol (2021) 28(1):62–70. doi: 10.1038/s41594-020-00526-w
52. Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, et al. Alkbh1-mediated Trna demethylation regulates translation. Cell (2016) 167(3):816–28.e16. doi: 10.1016/j.cell.2016.09.038
53. Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, et al. Differential M(6)a, M(6)a(M), and M(1)a demethylation mediated by Fto in the cell nucleus and cytoplasm. Mol Cell (2018) 71(6):973–85.e5. doi: 10.1016/j.molcel.2018.08.011
54. Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J, et al. Transfer Rna demethylase Alkbh3 promotes cancer progression via induction of Trna-derived small Rnas. Nucleic Acids Res (2019) 47(5):2533–45. doi: 10.1093/nar/gky1250
55. Jády BE, Ketele A, Kiss T. Dynamic association of human mrnp proteins with mitochondrial Trnas in the cytosol. RNA (New York NY) (2018) 24(12):1706–20. doi: 10.1261/rna.066738.118
56. van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, et al. The human 18s Rrna M6a methyltransferase Mettl5 is stabilized by Trmt112. Nucleic Acids Res (2019) 47(15):7719–33. doi: 10.1093/nar/gkz619
57. Pinto R, Vågbø CB, Jakobsson ME, Kim Y, Baltissen MP, O’Donohue MF, et al. The human methyltransferase Zcchc4 catalyses N6-methyladenosine modification of 28s ribosomal Rna. Nucleic Acids Res (2020) 48(2):830–46. doi: 10.1093/nar/gkz1147
58. Motorin Y, Lyko F, Helm M. 5-methylcytosine in Rna: detection, enzymatic formation and biological functions. Nucleic Acids Res (2010) 38(5):1415–30. doi: 10.1093/nar/gkp1117
59. Dai X, Gonzalez G, Li L, Li J, You C, Miao W, et al. Ythdf2 binds to 5-methylcytosine in Rna and modulates the maturation of ribosomal Rna. Analytical Chem (2020) 92(1):1346–54. doi: 10.1021/acs.analchem.9b04505
60. Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary micrornas for processing. Nature (2015) 519(7544):482–5. doi: 10.1038/nature14281
61. van den Homberg DAL, van der Kwast R, Quax PHA, Nossent AY. N-6-methyladenosine in vasoactive micrornas during hypoxia; a novel role for mettl4. Int J Mol Sci (2022) 23(3):1057. doi: 10.3390/ijms23031057
62. Chen P, Li S, Zhang K, Zhao R, Cui J, Zhou W, et al. N(6)-methyladenosine demethylase Alkbh5 suppresses Malignancy of esophageal cancer by regulating microrna biogenesis and rai1 expression. Oncogene (2021) 40(37):5600–12. doi: 10.1038/s41388-021-01966-4
63. Zhou G, Yan K, Liu J, Gao L, Jiang X, Fan Y. Fto promotes tumour proliferation in bladder cancer via the Fto/Mir-576/Cdk6 axis in an M6a-dependent manner. Cell Death Discovery (2021) 7(1):329. doi: 10.1038/s41420-021-00724-5
64. Guil S, Cáceres JF. The multifunctional Rna-binding protein hnrnp A1 is required for processing of Mir-18a. Nat Struct Mol Biol (2007) 14(7):591–6. doi: 10.1038/nsmb1250
65. Li Y, Estep JA, Karginov FV. Transcriptome-wide identification and validation of interactions between the mirna machinery and hur on mrna targets. J Mol Biol (2018) 430(3):285–96. doi: 10.1016/j.jmb.2017.12.006
66. Müller S, Bley N, Glaß M, Busch B, Rousseau V, Misiak D, et al. Igf2bp1 enhances an aggressive tumor cell phenotype by impairing mirna-directed downregulation of oncogenic factors. Nucleic Acids Res (2018) 46(12):6285–303. doi: 10.1093/nar/gky229
67. Plante I, Davidovic L, Ouellet DL, Gobeil LA, Tremblay S, Khandjian EW, et al. Dicer-derived micrornas are utilized by the fragile X mental retardation protein for assembly on target Rnas. J biomedicine Biotechnol (2006) 2006(4):64347. doi: 10.1155/jbb/2006/64347
68. Hou Y, Zhang Q, Pang W, Hou L, Liang Y, Han X, et al. Ythdc1-mediated augmentation of mir-30d in repressing pancreatic tumorigenesis via attenuation of Runx1-induced transcriptional activation of warburg effect. Cell Death differentiation (2021) 28(11):3105–24. doi: 10.1038/s41418-021-00804-0
69. Park S, Yang HD, Seo JW, Nam JW, Nam SW. Hnrnpc induces isoform shifts in Mir-21-5p leading to cancer development. Exp Mol Med (2022) 54(6):812–24. doi: 10.1038/s12276-022-00792-2
70. Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. Mettl14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding Rna xist. Mol Cancer (2020) 19(1):46. doi: 10.1186/s12943-020-1146-4
71. Yang D, Chang S, Li F, Ma M, Yang J, Lv X, et al. M(6) a transferase Kiaa1429-stabilized Linc00958 accelerates gastric cancer aerobic glycolysis through targeting Glut1. IUBMB Life (2021) 73(11):1325–33. doi: 10.1002/iub.2545
72. Li ZX, Zheng ZQ, Yang PY, Lin L, Zhou GQ, Lv JW, et al. Wtap-mediated M(6)a modification of Lncrna diaph1-As1 enhances its stability to facilitate nasopharyngeal carcinoma growth and metastasis. Cell Death differentiation (2022) 29(6):1137–51. doi: 10.1038/s41418-021-00905-w
73. Sun Z, Xue S, Zhang M, Xu H, Hu X, Chen S, et al. Aberrant Nsun2-mediated M(5)C modification of H19 lncrna is associated with poor differentiation of hepatocellular carcinoma. Oncogene (2020) 39(45):6906–19. doi: 10.1038/s41388-020-01475-w
74. Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY, et al. Alkbh5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the Lncrna neat1. J Physiol Biochem (2019) 75(3):379–89. doi: 10.1007/s13105-019-00690-8
75. Li X, Li Y, Wang Y, He X. The M(6)a demethylase Fto promotes renal epithelial-mesenchymal transition by reducing the M(6)a modification of lncrna gas5. Cytokine (2022) 159:156000. doi: 10.1016/j.cyto.2022.156000
76. Hämmerle M, Gutschner T, Uckelmann H, Ozgur S, Fiskin E, Gross M, et al. Posttranscriptional destabilization of the liver-specific long noncoding Rna Hulc by the Igf2 mrna-binding protein 1 (Igf2bp1). Hepatol (Baltimore Md) (2013) 58(5):1703–12. doi: 10.1002/hep.26537
77. Hu YP, Jin YP, Wu XS, Yang Y, Li YS, Li HF, et al. Lncrna-hgbc stabilized by hur promotes gallbladder cancer progression by regulating Mir-502-3p/set/Akt axis. Mol Cancer (2019) 18(1):167. doi: 10.1186/s12943-019-1097-9
78. Hu YP, Jin YP, Wu XS, Yang Y, Li YS, Li HF, et al. Correction to: lncrna-hgbc stabilized by hur promotes gallbladder cancer progression by regulating Mir-502-3p/Set/Akt axis. Mol Cancer (2021) 20(1):110. doi: 10.1186/s12943-021-01386-8
79. Zhang Y, Huang W, Yuan Y, Li J, Wu J, Yu J, et al. Long non-coding Rna H19 promotes colorectal cancer metastasis via binding to Hnrnpa2b1. J Exp Clin Cancer research: CR (2020) 39(1):141. doi: 10.1186/s13046-020-01619-6
80. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular Rnas driven by N(6)-methyladenosine. Cell Res (2017) 27(5):626–41. doi: 10.1038/cr.2017.31
81. Fan HN, Chen ZY, Chen XY, Chen M, Yi YC, Zhu JS, et al. Mettl14-mediated M(6)a modification of circorc5 suppresses gastric cancer progression by regulating Mir-30c-2-3p/Akt1s1 axis. Mol Cancer (2022) 21(1):51. doi: 10.1186/s12943-022-01521-z
82. Jiang Y, Wang Z, Ying C, Hu J, Zeng T, Gao L. Fmr1/Circchaf1a/Mir-211-5p/Hoxc8 feedback loop regulates proliferation and tumorigenesis via Mdm2-dependent P53 signaling in Gscs. Oncogene (2021) 40(24):4094–110. doi: 10.1038/s41388-021-01833-2
83. Pan Z, Zhao R, Li B, Qi Y, Qiu W, Guo Q, et al. Ewsr1-induced circneil3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing Igf2bp3. Mol Cancer (2022) 21(1):16. doi: 10.1186/s12943-021-01485-6
84. Di Timoteo G, Dattilo D, Centrón-Broco A, Colantoni A, Guarnacci M, Rossi F, et al. Modulation of circrna metabolism by M(6)a modification. Cell Rep (2020) 31(6):107641. doi: 10.1016/j.celrep.2020.107641
85. Rozenski J, Crain PF, McCloskey JA. The Rna modification database: 1999 update. Nucleic Acids Res (1999) 27(1):196–7. doi: 10.1093/nar/27.1.196
86. McCloskey JA, Rozenski J. The small subunit Rrna modification database. Nucleic Acids Res (2005) 33(Database issue):D135–8. doi: 10.1093/nar/gki015
87. Beemon K, Keith J. Localization of N6-methyladenosine in the rous sarcoma virus genome. J Mol Biol (1977) 113(1):165–79. doi: 10.1016/0022-2836(77)90047-x
88. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse M6a Rna methylomes revealed by M6a-seq. Nature (2012) 485(7397):201–6. doi: 10.1038/nature11112
89. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of Mrna methylation reveals enrichment in 3’ Utrs and near stop codons. Cell (2012) 149(7):1635–46. doi: 10.1016/j.cell.2012.05.003
90. Zheng H, Li S, Zhang X, Sui N. Functional implications of active N(6)-methyladenosine in plants. Front Cell Dev Biol (2020) 8:291. doi: 10.3389/fcell.2020.00291
91. Bao X, Zhang Y, Li H, Teng Y, Ma L, Chen Z, et al. Rm2target: A comprehensive database for targets of writers, erasers and readers of Rna modifications. Nucleic Acids Res (2023) 51(D1):D269–d79. doi: 10.1093/nar/gkac945
92. Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing Mrna methylation. Nat Rev Mol Cell Biol (2019) 20(10):608–24. doi: 10.1038/s41580-019-0168-5
93. Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol (2014) 16(2):191–8. doi: 10.1038/ncb2902
94. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural basis of N(6)-adenosine methylation by the Mettl3-Mettl14 complex. Nature (2016) 534(7608):575–8. doi: 10.1038/nature18298
95. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Corrigendum: structural basis of N(6)-adenosine methylation by the Mettl3-Mettl14 complex. Nature (2017) 542(7640):260. doi: 10.1038/nature21073
96. Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell (2016) 63(2):306–17. doi: 10.1016/j.molcel.2016.05.041
97. Huang J, Dong X, Gong Z, Qin LY, Yang S, Zhu YL, et al. Solution structure of the Rna recognition domain of Mettl3-Mettl14 N(6)-methyladenosine methyltransferase. Protein Cell (2019) 10(4):272–84. doi: 10.1007/s13238-018-0518-7
98. Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, et al. The Rna M6a reader ythdf2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discovery (2021) 11(2):480–99. doi: 10.1158/2159-8290.Cd-20-0331
99. Martinez de la Cruz B, Darsinou M, Riccio A. From form to function: M(6)a methylation links Mrna structure to metabolism. Adv Biol Regul (2023) 87:100926. doi: 10.1016/j.jbior.2022.100926
100. Rana AP, Tuck MT. Analysis and in vitro localization of internal methylated adenine residues in dihydrofolate reductase Mrna. Nucleic Acids Res (1990) 18(16):4803–8. doi: 10.1093/nar/18.16.4803
101. Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic M(6)a decoration: writers, erasers, readers and functions in Rna metabolism. Cell Res (2018) 28(6):616–24. doi: 10.1038/s41422-018-0040-8
102. Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol (2014) 15(5):313–26. doi: 10.1038/nrm3785
103. Nilsen TW. Molecular biology. Internal Mrna methylation finally finds functions. Science (2014) 343(6176):1207–8. doi: 10.1126/science.1249340
104. Li J, Chen K, Dong X, Xu Y, Sun Q, Wang H, et al. Ythdf1 promotes Mrna degradation via ythdf1-ago2 interaction and phase separation. Cell proliferation (2022) 55(1):e13157. doi: 10.1111/cpr.13157
105. Zhang X, Xie K, Zhou H, Wu Y, Li C, Liu Y, et al. Role of non-coding Rnas and Rna modifiers in cancer therapy resistance. Mol Cancer (2020) 19(1):47. doi: 10.1186/s12943-020-01171-z
106. Liu J, Dou X, Chen C, Chen C, Liu C, Xu MM, et al. N (6)-methyladenosine of chromosome-associated regulatory Rna regulates chromatin state and transcription. Science (2020) 367(6477):580–6. doi: 10.1126/science.aay6018
107. Akula D, O’Connor TR, Anindya R. Oxidative demethylase Alkbh5 repairs DNA alkylation damage and protects against alkylation-induced toxicity. Biochem Biophys Res Commun (2021) 534:114–20. doi: 10.1016/j.bbrc.2020.12.017
108. Zhu Y, Zhou G, Yu X, Xu Q, Wang K, Xie D, et al. Lc-Ms-Ms quantitative analysis reveals the association between Fto and DNA methylation. PloS One (2017) 12(4):e0175849. doi: 10.1371/journal.pone.0175849
109. Deng S, Zhang J, Su J, Zuo Z, Zeng L, Liu K, et al. Rna M(6)a regulates transcription via DNA demethylation and chromatin accessibility. Nat Genet (2022) 54(9):1427–37. doi: 10.1038/s41588-022-01173-1
110. Xu W, Shen H. When Rna methylation meets DNA methylation. Nat Genet (2022) 54(9):1261–2. doi: 10.1038/s41588-022-01166-0
111. Hussain S, Aleksic J, Blanco S, Dietmann S, Frye M. Characterizing 5-methylcytosine in the mamMalian epitranscriptome. Genome Biol (2013) 14(11):215. doi: 10.1186/gb4143
112. Li Q, Li X, Tang H, Jiang B, Dou Y, Gorospe M, et al. Nsun2-mediated M5c methylation and mettl3/mettl14-mediated M6a methylation cooperatively enhance P21 translation. J Cell Biochem (2017) 118(9):2587–98. doi: 10.1002/jcb.25957
113. Zheng H, Zhu M, Li W, Zhou Z, Wan X. M(5) C and M(6) a modification of long noncoding nkila accelerates cholangiocarcinoma progression via the Mir-582-3p-Yap1 axis. Liver Int (2022) 42(5):1144–57. doi: 10.1111/liv.15240
114. Aguilo F, Li S, Balasubramaniyan N, Sancho A, Benko S, Zhang F, et al. Deposition of 5-methylcytosine on enhancer rnas enables the coactivator function of Pgc-1α. Cell Rep (2016) 14(3):479–92. doi: 10.1016/j.celrep.2015.12.043
115. Kong W, Biswas A, Zhou D, Fiches G, Fujinaga K, Santoso N, et al. Nucleolar protein Nop2/Nsun1 suppresses Hiv-1 transcription and promotes viral latency by competing with tat for tar binding and methylation. PloS Pathog (2020) 16(3):e1008430. doi: 10.1371/journal.ppat.1008430
116. Zhang X, Liu Z, Yi J, Tang H, Xing J, Yu M, et al. The Trna methyltransferase nsun2 stabilizes P16ink4 Mrna by methylating the 3’-untranslated region of P16. Nat Commun (2012) 3:712. doi: 10.1038/ncomms1692
117. Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, et al. 5-methylcytosine promotes Mrna export - Nsun2 as the methyltransferase and alyref as an M(5)C reader. Cell Res (2017) 27(5):606–25. doi: 10.1038/cr.2017.55
118. Liu Y, Yang Y, Wu R, Gao CC, Liao X, Han X, et al. Mrna M(5)C inhibits adipogenesis and promotes myogenesis by respectively facilitating Ybx2 and Smo Mrna export in alyref-M(5)C manner. Cell Mol Life sciences: CMLS (2022) 79(9):481. doi: 10.1007/s00018-022-04474-0
119. Schumann U, Zhang HN, Sibbritt T, Pan A, Horvath A, Gross S, et al. Multiple links between 5-methylcytosine content of Mrna and translation. BMC Biol (2020) 18(1):40. doi: 10.1186/s12915-020-00769-5
120. Selmi T, Hussain S, Dietmann S, Heiß M, Borland K, Flad S, et al. Sequence- and structure-specific cytosine-5 Mrna methylation by nsun6. Nucleic Acids Res (2021) 49(2):1006–22. doi: 10.1093/nar/gkaa1193
121. Lyko F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat Rev Genet (2018) 19(2):81–92. doi: 10.1038/nrg.2017.80
122. Huang ZX, Li J, Xiong QP, Li H, Wang ED, Liu RJ. Position 34 of Trna is a discriminative element for M5c38 modification by human Dnmt2. Nucleic Acids Res (2021) 49(22):13045–61. doi: 10.1093/nar/gkab1148
123. Zhang Y, Zhang X, Shi J, Tuorto F, Li X, Liu Y, et al. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding Rnas. Nat Cell Biol (2018) 20(5):535–40. doi: 10.1038/s41556-018-0087-2
124. Liao H, Gaur A, McConie H, Shekar A, Wang K, Chang JT, et al. Human Nop2/Nsun1 regulates ribosome biogenesis through non-catalytic complex formation with box C/D snornps. Nucleic Acids Res (2022) 50(18):10695–716. doi: 10.1093/nar/gkac817
125. Cheng J, Berninghausen O, Beckmann R. A distinct assembly pathway of the human 39s late pre-mitoribosome. Nat Commun (2021) 12(1):4544. doi: 10.1038/s41467-021-24818-x
126. Heissenberger C, Liendl L, Nagelreiter F, Gonskikh Y, Yang G, Stelzer EM, et al. Loss of the ribosomal Rna methyltransferase nsun5 impairs global protein synthesis and normal growth. Nucleic Acids Res (2019) 47(22):11807–25. doi: 10.1093/nar/gkz1043
127. Shen H, Ontiveros RJ, Owens MC, Liu MY, Ghanty U, Kohli RM, et al. Tet-mediated 5-methylcytosine oxidation in Trna promotes translation. J Biol Chem (2021) 296:100087. doi: 10.1074/jbc.RA120.014226
128. Shen Q, Zhang Q, Shi Y, Shi Q, Jiang Y, Gu Y, et al. Tet2 promotes pathogen infection-induced myelopoiesis through Mrna oxidation. Nature (2018) 554(7690):123–7. doi: 10.1038/nature25434
129. Liu Y, Zhao Y, Wu R, Chen Y, Chen W, Liu Y, et al. Mrna M5c controls adipogenesis by promoting Cdkn1a Mrna export and translation. RNA Biol (2021) 18(sup2):711–21. doi: 10.1080/15476286.2021.1980694
130. Yang H, Wang Y, Xiang Y, Yadav T, Ouyang J, Phoon L, et al. Fmrp promotes transcription-coupled homologous recombination via facilitating Tet1-mediated M5c rna modification demethylation. Proc Natl Acad Sci United States America (2022) 119(12):e2116251119. doi: 10.1073/pnas.2116251119
131. Hussain S. The emerging roles of cytosine-5 methylation in mrnas. Trends genetics: TIG (2021) 37(6):498–500. doi: 10.1016/j.tig.2021.02.001
132. Sajini AA, Choudhury NR, Wagner RE, Bornelöv S, Selmi T, Spanos C, et al. Loss of 5-methylcytosine alters the biogenesis of vault-derived small Rnas to coordinate epidermal differentiation. Nat Commun (2019) 10(1):2550. doi: 10.1038/s41467-019-10020-7
133. Winans S, Beemon K. M(5)C goes viral. Cell Host Microbe (2019) 26(2):154–5. doi: 10.1016/j.chom.2019.07.019
134. Chen H, Yang H, Zhu X, Yadav T, Ouyang J, Truesdell SS, et al. M(5)C modification of mrna serves a DNA damage code to promote homologous recombination. Nat Commun (2020) 11(1):2834. doi: 10.1038/s41467-020-16722-7
135. Niu X, Peng L, Liu W, Miao C, Chen X, Chu J, et al. A Cis-Eqtl in nsun2 promotes esophageal squamous-cell carcinoma progression and radiochemotherapy resistance by mrna-M(5)C methylation. Signal transduction targeted Ther (2022) 7(1):267. doi: 10.1038/s41392-022-01063-2
136. Seo KW, Kleiner RE. Ythdf2 recognition of N(1)-methyladenosine (M(1)a)-modified Rna is associated with transcript destabilization. ACS Chem Biol (2020) 15(1):132–9. doi: 10.1021/acschembio.9b00655
137. Gardner DK, Lane M. Culture and selection of viable blastocysts: A feasible proposition for human Ivf? Hum Reprod Update (1997) 3(4):367–82. doi: 10.1093/humupd/3.4.367
138. Kagawa H, Javali A, Khoei HH, Sommer TM, Sestini G, Novatchkova M, et al. Human blastoids model blastocyst development and implantation. Nature (2022) 601(7894):600–5. doi: 10.1038/s41586-021-04267-8
139. Rajagopalan S, Long EO. Cell atlas reveals the landscape of early pregnancy. Nature (2018) 563(7731):337–8. doi: 10.1038/d41586-018-07317-w
140. Castillo J, Jodar M, Oliva R. The contribution of human sperm proteins to the development and epigenome of the preimplantation embryo. Hum Reprod Update (2018) 24(5):535–55. doi: 10.1093/humupd/dmy017
141. Xu K, Yang Y, Feng GH, Sun BF, Chen JQ, Li YF, et al. Mettl3-mediated M(6)a regulates spermaTogonial differentiation and meiosis initiation. Cell Res (2017) 27(9):1100–14. doi: 10.1038/cr.2017.100
142. Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou Q, et al. Mettl3-/mettl14-mediated mrna N(6)-methyladenosine modulates murine spermatogenesis. Cell Res (2017) 27(10):1216–30. doi: 10.1038/cr.2017.117
143. Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y, et al. Alkbh5-dependent M6a demethylation controls splicing and stability of long 3’-Utr mrnas in male germ cells. Proc Natl Acad Sci United States America (2018) 115(2):E325–e33. doi: 10.1073/pnas.1717794115
144. Sha QQ, Zheng W, Wu YW, Li S, Guo L, Zhang S, et al. Dynamics and clinical relevance of maternal mrna clearance during the oocyte-to-embryo transition in humans. Nat Commun (2020) 11(1):4917. doi: 10.1038/s41467-020-18680-6
145. Sha QQ, Zhu YZ, Li S, Jiang Y, Chen L, Sun XH, et al. Characterization of zygotic genome activation-dependent maternal mrna clearance in mouse. Nucleic Acids Res (2020) 48(2):879–94. doi: 10.1093/nar/gkz1111
146. Zhang G, Xu Y, Wang X, Zhu Y, Wang L, Zhang W, et al. Dynamic Fmr1 granule phase switch instructed by M6a modification contributes to maternal Rna decay. Nat Commun (2022) 13(1):859. doi: 10.1038/s41467-022-28547-7
147. Despic V, Dejung M, Gu M, Krishnan J, Zhang J, Herzel L, et al. Dynamic Rna-protein interactions underlie the zebrafish maternal-to-zygotic transition. Genome Res (2017) 27(7):1184–94. doi: 10.1101/gr.215954.116
148. Ren F, Lin Q, Gong G, Du X, Dan H, Qin W, et al. Igf2bp3 maintains maternal Rna stability and ensures early embryo development in zebrafish. Commun Biol (2020) 3(1):94. doi: 10.1038/s42003-020-0827-2
149. Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, et al. M(6)a-dependent maternal mrna clearance facilitates zebrafish maternal-to-zygotic transition. Nature (2017) 542(7642):475–8. doi: 10.1038/nature21355
150. Mendel M, Chen KM, Homolka D, Gos P, Pandey RR, McCarthy AA, et al. Methylation of structured Rna by the M(6)a writer mettl16 is essential for mouse embryonic development. Mol Cell (2018) 71(6):986–1000.e11. doi: 10.1016/j.molcel.2018.08.004
151. Yao H, Gao CC, Zhang D, Xu J, Song G, Fan X, et al. Scm(6)a-seq reveals single-cell landscapes of the dynamic M(6)a during oocyte maturation and early embryonic development. Nat Commun (2023) 14(1):315. doi: 10.1038/s41467-023-35958-7
152. Liu J, Gao M, He J, Wu K, Lin S, Jin L, et al. The Rna M6a reader Ythdc1 silences retrotransposons and guards es cell identity. Nature (2021) 591(7849):322–6. doi: 10.1038/s41586-021-03313-9
153. Ignatova VV, Stolz P, Kaiser S, Gustafsson TH, Lastres PR, Sanz-Moreno A, et al. The Rrna M(6)a methyltransferase mettl5 is involved in pluripotency and developmental programs. Genes Dev (2020) 34(9-10):715–29. doi: 10.1101/gad.333369.119
154. Xing M, Liu Q, Mao C, Zeng H, Zhang X, Zhao S, et al. The 18s Rrna M(6) a methyltransferase mettl5 promotes mouse embryonic stem cell differentiation. EMBO Rep (2020) 21(10):e49863. doi: 10.15252/embr.201949863
155. Zhang L, Yuan M, Huang X, Cao Q, Huang S, Sun R, et al. Inhibition of Mettl5 improves preimplantation development of mouse somatic cell nuclear transfer embryos. Reprod (Cambridge England) (2022) 164(5):221–30. doi: 10.1530/rep-22-0169
156. Meng TG, Lu X, Guo L, Hou GM, Ma XS, Li QN, et al. Mettl14 is required for mouse postimplantation development by facilitating epiblast maturation. FASEB J (2019) 33(1):1179–87. doi: 10.1096/fj.201800719R
157. Wang Z, Pan Z, Adhikari S, Harada BT, Shen L, Yuan W, et al. M(6) a deposition is regulated by Prmt1-mediated arginine methylation of mettl14 in its disordered C-terminal region. EMBO J (2021) 40(5):e106309. doi: 10.15252/embj.2020106309
158. Liu X, Wang H, Zhao X, Luo Q, Wang Q, Tan K, et al. Arginine methylation of Mettl14 promotes Rna N(6)-methyladenosine modification and endoderm differentiation of mouse embryonic stem cells. Nat Commun (2021) 12(1):3780. doi: 10.1038/s41467-021-24035-6
159. Liu J, Huang T, Chen W, Ding C, Zhao T, Zhao X, et al. Developmental mrna M(5)C landscape and regulatory innovations of massive M(5)C modification of maternal mrnas in animals. Nat Commun (2022) 13(1):2484. doi: 10.1038/s41467-022-30210-0
160. Ding C, Lu J, Li J, Hu X, Liu Z, Su H, et al. Rna-methyltransferase nsun5 controls the maternal-to-zygotic transition by regulating maternal mrna stability. Clin Trans Med (2022) 12(12):e1137. doi: 10.1002/ctm2.1137
161. Yang Y, Wang L, Han X, Yang WL, Zhang M, Ma HL, et al. Rna 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mrna decay. Mol Cell (2019) 75(6):1188–202.e11. doi: 10.1016/j.molcel.2019.06.033
162. Trixl L, Amort T, Wille A, Zinni M, Ebner S, Hechenberger C, et al. Rna cytosine methyltransferase Nsun3 regulates embryonic stem cell differentiation by promoting mitochondrial activity. Cell Mol Life sciences: CMLS (2018) 75(8):1483–97. doi: 10.1007/s00018-017-2700-0
163. Lin S, Liu Q, Lelyveld VS, Choe J, Szostak JW, Gregory RI. Mettl1/wdr4-mediated M(7)G trna methylome is required for normal mrna translation and embryonic stem cell self-renewal and differentiation. Mol Cell (2018) 71(2):244–55.e5. doi: 10.1016/j.molcel.2018.06.001
164. Aguilera-Castrejon A, Oldak B, Shani T, Ghanem N, Itzkovich C, Slomovich S, et al. Ex utero mouse embryogenesis from pre-gastrulation to late organogenesis. Nature (2021) 593(7857):119–24. doi: 10.1038/s41586-021-03416-3
165. Kadyrov M, Kingdom JC, Huppertz B. Divergent trophoblast invasion and apoptosis in placental bed spiral arteries from pregnancies complicated by maternal anemia and early-onset preeclampsia/intrauterine growth restriction. Am J obstetrics gynecology (2006) 194(2):557–63. doi: 10.1016/j.ajog.2005.07.035
166. Zhao J, Ding H, Ding J, Shi X, He Y, Zhu H, et al. The M(6)a methyltransferase Mettl3 promotes trophoblast cell invasion by regulating mylk expression. Placenta (2022) 129:1–6. doi: 10.1016/j.placenta.2022.09.002
167. Huang N, Gao Y, Zhang M, Guo L, Qin L, Liao S, et al. Mettl3-mediated M(6)a Rna methylation of Zbtb4 interferes with trophoblast invasion and maybe involved in Rsa. Front Cell Dev Biol (2022) 10:894810. doi: 10.3389/fcell.2022.894810
168. Bian Y, Li J, Shen H, Li Y, Hou Y, Huang L, et al. Wtap dysregulation-mediated Hmgn3-M6a modification inhibited trophoblast invasion in early-onset preeclampsia. FASEB J (2022) 36(12):e22617. doi: 10.1096/fj.202200700RR
169. Li XC, Jin F, Wang BY, Yin XJ, Hong W, Tian FJ. The M6a demethylase Alkbh5 controls trophoblast invasion at the maternal-fetal interface by regulating the stability of Cyr61 mrna. Theranostics (2019) 9(13):3853–65. doi: 10.7150/thno.31868
170. Haouzi D, Dechaud H, Assou S, Monzo C, de Vos J, Hamamah S. Transcriptome analysis reveals dialogues between human trophectoderm and endometrial cells during the implantation period. Hum Reprod (Oxford England) (2011) 26(6):1440–9. doi: 10.1093/humrep/der075
171. Zhu J, Wang K, Li T, Chen J, Xie D, Chang X, et al. Hypoxia-induced Tet1 facilitates trophoblast cell migration and invasion through Hif1α Signaling pathway. Sci Rep (2017) 7(1):8077. doi: 10.1038/s41598-017-07560-7
172. Zheng Q, Gan H, Yang F, Yao Y, Hao F, Hong L, et al. Cytoplasmic M(1)a reader Ythdf3 inhibits trophoblast invasion by downregulation of M(1)a-methylated Igf1r. Cell Discovery (2020) 6:12. doi: 10.1038/s41421-020-0144-4
173. Zheng Q, Yang F, Gan H, Jin L. Hypoxia induced Alkbh5 prevents spontaneous abortion by mediating M(6)a-demethylation of Smad1/5 mrnas. Biochim Biophys Acta Mol Cell Res (2022) 1869(10):119316. doi: 10.1016/j.bbamcr.2022.119316
174. Guo Y, Song W, Yang Y. Inhibition of Alkbh5-mediated M(6) a modification of Pparg mrna alleviates H/R-induced oxidative stress and apoptosis in placenta trophoblast. Environ Toxicol (2022) 37(4):910–24. doi: 10.1002/tox.23454
175. Fan W, Zhou W, Yan Q, Peng Y, Wang H, Kong C, et al. Upregulation of Mettl14 contributes to trophoblast dysfunction by elevating Foxo3a expression in an M(6)a-dependent manner. Placenta (2022) 124:18–27. doi: 10.1016/j.placenta.2022.05.008
176. Wang D, Guan H, Xia Y. Ythdc1 maintains trophoblasts function by promoting degradation of Ma6-modified circmpp1. Biochem Pharmacol (2023) 210:115456. doi: 10.1016/j.bcp.2023.115456
177. Wang D, Guan H, Wang Y, Song G, Xia Y. N6-methyladenosine modification in trophoblasts promotes circsetd2 expression, inhibits Mir-181a-5p, and elevates Mcl1 transcription to reduce apoptosis of trophoblasts. Environ Toxicol (2023) 38(2):422–35. doi: 10.1002/tox.23683
178. Dai M, Huang W, Huang X, Ma C, Wang R, Tian P, et al. Bpde, the migration and invasion of human trophoblast cells, and occurrence of miscarriage in humans: roles of a novel Lncrna-hz09. Environ Health Perspect (2023) 131(1):17009. doi: 10.1289/EHP10477
179. Xu Z, Tian P, Guo J, Mi C, Liang T, Xie J, et al. Lnc-hz01 with M6a Rna methylation inhibits human trophoblast cell proliferation and induces miscarriage by up-regulating Bpde-activated Lnc-hz01/mxd1 positive feedback loop. Sci total Environ (2021) 776:145950. doi: 10.1016/j.scitotenv.2021.145950
180. Ng SW, Norwitz GA, Pavlicev M, Tilburgs T, Simón C, Norwitz ER. Endometrial decidualization: the primary driver of pregnancy health. Int J Mol Sci (2020) 21(11):4092. doi: 10.3390/ijms21114092
181. Zheng ZH, Zhang GL, Jiang RF, Hong YQ, Zhang QY, He JP, et al. Mettl3 is essential for normal progesterone signaling during embryo implantation via M(6)a-mediated translation control of progesterone receptor. Proc Natl Acad Sci United States America (2023) 120(5):e2214684120. doi: 10.1073/pnas.2214684120
182. Xue P, Zhou W, Fan W, Jiang J, Kong C, Zhou W, et al. Increased mettl3-mediated M(6)a methylation inhibits embryo implantation by repressing hoxa10 expression in recurrent implantation failure. Reprod Biol endocrinology: RB&E (2021) 19(1):187. doi: 10.1186/s12958-021-00872-4
183. Owusu-Akyaw A, Krishnamoorthy K, Goldsmith LT, Morelli SS. The role of mesenchymal-epithelial transition in endometrial function. Hum Reprod Update (2019) 25(1):114–33. doi: 10.1093/humupd/dmy035
184. Lin X, Chai G, Wu Y, Li J, Chen F, Liu J, et al. Rna M(6)a methylation regulates the epithelial mesenchymal transition of cancer cells and translation of snail. Nat Commun (2019) 10(1):2065. doi: 10.1038/s41467-019-09865-9
185. Xu L, Li Y, Sang Y, Li DJ, Du M. Crosstalk between trophoblasts and decidual immune cells: the cornerstone of maternal-fetal immunotolerance. Front Immunol (2021) 12:642392. doi: 10.3389/fimmu.2021.642392
186. Song H, Song J, Cheng M, Zheng M, Wang T, Tian S, et al. Mettl3-mediated M(6)a Rna methylation promotes the anti-tumour immunity of natural killer cells. Nat Commun (2021) 12(1):5522. doi: 10.1038/s41467-021-25803-0
187. Liu XS, Zhou LM, Yuan LL, Gao Y, Kui XY, Liu XY, et al. Npm1 is a prognostic biomarker involved in immune infiltration of lung adenocarcinoma and associated with M6a modification and glycolysis. Front Immunol (2021) 12:724741. doi: 10.3389/fimmu.2021.724741
188. Wu Y, Jiang D, Zhang H, Yin F, Guo P, Zhang X, et al. N1-methyladenosine (M1a) regulation associated with the pathogenesis of abdominal aortic aneurysm through Ythdf3 modulating macrophage polarization. Front Cardiovasc Med (2022) 9:883155. doi: 10.3389/fcvm.2022.883155
189. Zhao Y, Sun J, Jin L. The N6-methyladenosine regulator Alkbh5 mediated stromal cell-macrophage interaction via Vegf signaling to promote recurrent spontaneous abortion: A bioinformatic and in vitro study. Int J Mol Sci (2022) 23(24):15819. doi: 10.3390/ijms232415819
190. Liu Y, Liu Z, Tang H, Shen Y, Gong Z, Xie N, et al. The N(6)-methyladenosine (M(6)a)-forming enzyme Mettl3 Facilitates M1 macrophage polarization through the methylation of Stat1 mrna. Am J Physiol Cell Physiol (2019) 317(4):C762–c75. doi: 10.1152/ajpcell.00212.2019
191. Yin H, Zhang X, Yang P, Zhang X, Peng Y, Li D, et al. Rna M6a methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat Commun (2021) 12(1):1394. doi: 10.1038/s41467-021-21514-8
192. Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. M(6)a mrna methylation controls T cell homeostasis by targeting the Il-7/Stat5/Socs pathways. Nature (2017) 548(7667):338–42. doi: 10.1038/nature23450
193. Tong J, Cao G, Zhang T, Sefik E, Amezcua Vesely MC, Broughton JP, et al. M(6)a Mrna methylation sustains treg suppressive functions. Cell Res (2018) 28(2):253–6. doi: 10.1038/cr.2018.7
194. Lin P, Li G, Wu M. Rna Methylation into M(1)a Era: A New Regulation over T-Cell Function. Signal transduction targeted Ther (2023) 8(1):78. doi: 10.1038/s41392-023-01360-4
195. Liu Y, Zhou J, Li X, Zhang X, Shi J, Wang X, et al. Trna-M(1)a modification promotes T cell expansion via efficient myc protein synthesis. Nat Immunol (2022) 23(10):1433–44. doi: 10.1038/s41590-022-01301-3
196. Zhao S, Lu J, Chen Y, Wang Z, Cao J, Dong Y. Exploration of the potential roles of M6a regulators in the uterus in pregnancy and infertility. J Reprod Immunol (2021) 146:103341. doi: 10.1016/j.jri.2021.103341
197. Vallet-Buisan M, Mecca R, Jones C, Coward K, Yeste M. Contribution of semen to early embryo development: fertilization and beyond. Hum Reprod Update (2023) 29(4):395–433. doi: 10.1093/humupd/dmad006
198. She J, Tan K, Liu J, Cao S, Li Z, Peng Y, et al. The alteration of M(6)a modification at the transcriptome-wide level in human villi during spontaneous abortion in the first trimester. Front Genet (2022) 13:861853. doi: 10.3389/fgene.2022.861853
199. Qiu W, Zhou Y, Wu H, Lv X, Yang L, Ren Z, et al. Rna demethylase Fto mediated Rna M(6)a modification is involved in maintaining maternal-fetal interface in spontaneous abortion. Front Cell Dev Biol (2021) 9:617172. doi: 10.3389/fcell.2021.617172
200. Wei X, Zhou S, Liao L, Liu M, Gao Y, Yin Y, et al. Comprehensive analysis of transcriptomic profiling of 5-methylcytosin modification in placentas from preeclampsia and normotensive pregnancies. FASEB J (2023) 37(2):e22751. doi: 10.1096/fj.202201248R
201. Taniguchi K, Kawai T, Kitawaki J, Tomikawa J, Nakabayashi K, Okamura K, et al. Epitranscriptomic profiling in human placenta: N6-methyladenosine modification at the 5’-untranslated region is related to fetal growth and preeclampsia. FASEB J (2020) 34(1):494–512. doi: 10.1096/fj.201900619RR
202. Gu Y, Chu X, Morgan JA, Lewis DF, Wang Y. Upregulation of Mettl3 expression and M6a Rna methylation in placental trophoblasts in preeclampsia. Placenta (2021) 103:43–9. doi: 10.1016/j.placenta.2020.10.016
203. Zhang Y, Yang H, Long Y, Zhang Y, Chen R, Shi J, et al. Circrna N6-methyladenosine methylation in preeclampsia and the potential role of N6-methyladenosine-modified circpappa2 in trophoblast invasion. Sci Rep (2021) 11(1):24357. doi: 10.1038/s41598-021-03662-5
204. Yang Q, Ma Y, Liu Y, Shao X, Jia W, Yu X, et al. Mnsfβ Regulates placental development by conjugating Igf2bp2 to enhance trophoblast cell invasiveness. Cell proliferation (2021) 54(12):e13145. doi: 10.1111/cpr.13145
205. Chen Y, Liu X, Li L, He X, Zheng F, Zhang Y, et al. Methyltransferase-like 3 aggravates endoplasmic reticulum stress in preeclampsia by targeting Tmbim6 in Ythdf2-dependent manner. Mol Med (Cambridge Mass) (2023) 29(1):19. doi: 10.1186/s10020-023-00604-x
206. Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet (London England) (2008) 371(9606):75–84. doi: 10.1016/s0140-6736(08)60074-4
207. Kostyusheva A, Brezgin S, Glebe D, Kostyushev D, Chulanov V. Host-cell interactions in Hbv infection and pathogenesis: the emerging role of M6a modification. Emerging Microbes infections (2021) 10(1):2264–75. doi: 10.1080/22221751.2021.2006580
208. Sato S, Li K, Kameyama T, Hayashi T, Ishida Y, Murakami S, et al. The Rna sensor rig-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity (2015) 42(1):123–32. doi: 10.1016/j.immuni.2014.12.016
209. Srinivas KP, Depledge DP, Abebe JS, Rice SA, Mohr I, Wilson AC. Widespread remodeling of the M(6)a Rna-modification landscape by a viral regulator of Rna processing and export. Proc Natl Acad Sci United States America (2021) 118(30):e2104805118. doi: 10.1073/pnas.2104805118
210. Imam H, Kim GW, Mir SA, Khan M, Siddiqui A. Interferon-stimulated gene 20 (Isg20) selectively degrades N6-methyladenosine modified hepatitis B virus transcripts. PloS Pathog (2020) 16(2):e1008338. doi: 10.1371/journal.ppat.1008338
211. Zong X, Xiao X, Shen B, Jiang Q, Wang H, Lu Z, et al. The N6-methyladenosine Rna-binding protein ythdf1 modulates the translation of Traf6 to mediate the intestinal immune response. Nucleic Acids Res (2021) 49(10):5537–52. doi: 10.1093/nar/gkab343
212. Miller D, Gershater M, Slutsky R, Romero R, Gomez-Lopez N. Maternal and fetal T cells in term pregnancy and preterm labor. Cell Mol Immunol (2020) 17(7):693–704. doi: 10.1038/s41423-020-0471-2
213. Shahnawaz S, Nawaz US, Zaugg J, Hussain G, Malik N, Dogar MZ, et al. Dysregulated autophagy leads to oxidative stress and aberrant expression of Abc transporters in women with early miscarriage. Antioxidants (Basel Switzerland) (2021) 10(11):1742. doi: 10.3390/antiox10111742
214. Yang HL, Lai ZZ, Shi JW, Zhou WJ, Mei J, Ye JF, et al. A defective lysophosphatidic acid-autophagy axis increases miscarriage risk by restricting decidual macrophage residence. Autophagy (2022) 18(10):2459–80. doi: 10.1080/15548627.2022.2039000
215. Lu H, Yang HL, Zhou WJ, Lai ZZ, Qiu XM, Fu Q, et al. Rapamycin prevents spontaneous abortion by triggering decidual stromal cell autophagy-mediated Nk cell residence. Autophagy (2021) 17(9):2511–27. doi: 10.1080/15548627.2020.1833515
216. Ding Y, Yang X, Han X, Shi M, Sun L, Liu M, et al. Ferroptosis-related gene expression in the pathogenesis of preeclampsia. Front Genet (2022) 13:927869. doi: 10.3389/fgene.2022.927869
217. Cheng SB, Nakashima A, Huber WJ, Davis S, Banerjee S, Huang Z, et al. Pyroptosis is a critical inflammatory pathway in the placenta from early onset preeclampsia and in human trophoblasts exposed to hypoxia and endoplasmic reticulum stressors. Cell Death Dis (2019) 10(12):927. doi: 10.1038/s41419-019-2162-4
218. Akram KM, Frost LI, Anumba DO. Impaired autophagy with augmented apoptosis in a Th1/Th2-imbalanced placental micromilieu is associated with spontaneous preterm birth. Front Mol Biosci (2022) 9:897228. doi: 10.3389/fmolb.2022.897228
219. Song H, Feng X, Zhang H, Luo Y, Huang J, Lin M, et al. Mettl3 and Alkbh5 oppositely regulate M(6)a modification of Tfeb Mrna, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy (2019) 15(8):1419–37. doi: 10.1080/15548627.2019.1586246
Keywords: implantation, placentation, epigenetic modification, post-transcriptional regulation, RNA methylation, maternal-fetal interface
Citation: Wu S, Xie H, Su Y, Jia X, Mi Y, Jia Y and Ying H (2023) The landscape of implantation and placentation: deciphering the function of dynamic RNA methylation at the maternal-fetal interface. Front. Endocrinol. 14:1205408. doi: 10.3389/fendo.2023.1205408
Received: 13 April 2023; Accepted: 15 August 2023;
Published: 30 August 2023.
Edited by:
Lu Gao, Second Military Medical University, ChinaReviewed by:
Tamas Zakar, The University of Newcastle, AustraliaCopyright © 2023 Wu, Xie, Su, Jia, Mi, Jia and Ying. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanhui Jia, eXVhbmh1aS5qaWFAMTYzLmNvbQ==; Hao Ying, c3RlcGhlbnlpbmdfMjAxMUAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.