- 1Department of Biochemistry and Molecular Biology, Dasman Diabetes Institute, Kuwait, Kuwait
- 2Department of Pharmacology and Toxicology, Faculty of Medicine, Kuwait University, Kuwait, Kuwait
- 3Department of Genetics and Bioinformatics, Dasman Diabetes Institute, Kuwait, Kuwait
- 4Special Service Facility Department, Dasman Diabetes Institute, Kuwait, Kuwait
The Wolfram syndrome 1 gene (WFS1) is the main causative locus for Wolfram syndrome, an inherited condition characterized by childhood-onset diabetes mellitus, optic atrophy, and deafness. Global genome-wide association studies have listed at least 19 WFS1 variants that are associated with type 2 diabetes (T2D) and metabolic traits. It has been suggested that miRNA binding sites on WFS1 play a critical role in the regulation of the wolframin protein, and loss of WFS1 function may lead to the pathogenesis of diabetes. In the Hungarian population, it was observed that a 3’ UTR variant from WFS1, namely rs1046322, influenced the affinity of miR-668 to WFS1 mRNA, and showed a strong association with T2D. In this study, we genotyped a large cohort of 2067 individuals of different ethnicities residing in Kuwait for the WFS1 rs1046322 polymorphism. The cohort included 362 Southeast Asians (SEA), 1045 Arabs, and 660 South Asians (SA). Upon performing genetic association tests, we observed significant associations between the rs1046322 SNP and obesity traits in the SEA population, but not in the Arab or SA populations. The associated traits in SEA cohort were body mass index, BMI (β=1.562, P-value=0.0035, Pemp=0.0072), waist circumference, WC (β=3.163, P-value=0.0197, Pemp=0.0388) and triglyceride, TGL (β=0.224, P-value=0.0340). The association with BMI remained statistically significant even after multiple testing correction. Among the SEA individuals, carriers of the effect allele at the SNP had significantly higher BMI [mean of 27.63 (3.6) Kg/m2], WC [mean of 89.9 (8.1) cm], and TGL levels [mean of 1.672 (0.8) mmol/l] than non-carriers of the effect allele. Our findings suggest a role for WFS1 in obesity, which is a risk factor for diabetes. The study also emphasizes the significant role the ethnic background may play in determining the effect of genetic variants on susceptibility to metabolic diseases.
1 Introduction
Obesity has now become a global epidemic with an alarmingly increasing rate of incidence (1). The World Health Organization (WHO) reports that the worldwide prevalence of obesity nearly tripled between 1975 and 2016 and is expected to double in the next 25 years (2, 3). Different ethnicities exhibit different rates of obesity. This is evident in the different rates of obesity in ethnicities such as those of Southeast Asians, Arabs, and South Asians. The Southeast Asian countries have some of the lowest rates of overweight and obesity globally ranging from 2.2 to 15.5%. A recent review of all national surveys for some of the major South Asian countries (including Afghanistan, Bangladesh, India and Sri Lanka) reported that the prevalence of being overweight or obese in adults ranged from 22.4 to 52.4% (4). As for Arabian countries, WHO reported that the prevalence of obesity has a wide range between 4 and 55% (5).
Obesity is a complex multifactorial disorder with several risk factors that contribute to its development. People with obesity not only suffer from a poor quality of life, but are also at risk of developing serious complications, such as diabetes, cardiovascular diseases, sleep disorders, or hypertension (1). Therefore, it is important to understand the underlying multifactorial causes of obesity. Although environmental factors and lifestyle practices are the main causes of obesity, genetic susceptibility also plays a very significant role. Several reports confirm the association of certain genes with obesity, fat distribution, energy expenditure, and appetite regulation. Studies have shown that 40–70% of the variation in body mass index (BMI) among individuals can be attributed to genetic factors (6, 7). Furthermore, at least 200 genetic variants have been reported to be associated with obesity in several populations; however, such studies have focused on Caucasians from Europe (8, 9). Ethnicity can play an important role in determining the genetic susceptibility of an individual to obesity (10, 11).
Wolfram syndrome 1 gene (WFS1) was identified in the year of 1998 on chromosome 4p16 as a novel gene that causes a rare autosomal recessive neurodegenerative disorder, namely Wolfram syndrome (WFS) (12) alias DIDMOAD syndrome (diabetes insipidus, diabetes mellitus, optic atrophy, and deafness). It is clinically characterized by the juvenile-onset of diabetes mellitus as the main symptom in the early stage of the disease, and by bilateral progressive optic atrophy in later stages (13–15). The WFS1 gene encodes wolframin, a protein present in the membrane of the endoplasmic reticulum (ER), and is mainly detected in certain brain regions as well as in pancreatic β-cells and the heart (16, 17). Several WFS1 polymorphic variants have been associated with the risk of developing diabetes mellitus (18, 19). Among these variants, two microRNA-single-nucleotide polymorphisms (miR-SNPs), rs1046322 and rs9457, were strongly associated with both type 1 and type 2 diabetes, and this association remained statistically significant after applying multiple corrections (19). Considering that obesity is a risk factor for diabetes associated with insulin resistance, we aimed to evaluate, in the present study, the association of these two WFS1 variants with obesity in individuals of different ethnicities.
2 Materials and methods
2.1 Study population
This study included a cohort of 2067 participants residing in Kuwait. Upon enrolment, we recorded the following information: age, sex, baseline characteristics (height, weight, waist circumference (WC)), and underlying diagnosed disorders (such as diabetes). The study protocol was reviewed and approved by the Ethical Review Committee of Dasman Diabetes Institute and was conducted in accordance with the guidelines of the Declaration of Helsinki and the US Federal Policy for the Protection of Human Subjects. All participants signed an informed consent form before participating in the study. The ethnicity of each subject was defined via self-reporting and was confirmed through detailed questioning on parental lineage up to three generations.
2.2 Sample processing
We collected blood samples in accordance with established institutional guidelines. After confirming that the participant was under an overnight fast, we collected blood samples in the morning, between 8 and 11 am. We performed DNA extraction using a Gentra Puregene kit (Qiagen, Valencia, CA, USA) and assessed quantification using Quant-iT PicoGreen dsDNA Assay Kits (Life Technologies, Grand Island, NY, USA) and an Epoch Microplate Spectrophotometer (BioTek Instruments). We checked absorbance values at 260–280 nm for adherence to an optical density range of 1.8–2.1.
2.3 Anthropometric measurements and blood biochemistry
The BMI of each participant was calculated as the ratio of their weight (Kg) to height (m) squared. We assessed lipid profiles, including triglyceride (TGL), low density lipoprotein (LDL), high density lipoprotein (HDL), and total cholesterol (TC) levels, using a Siemens Dimension RXL integrated chemistry analyzer (Diamond Diagnostics, Holliston, MA, USA).
2.4 Genotyping
We performed candidate SNP genotyping using the TaqMan Genotyping Assay on an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). We set the polymerase chain reaction (PCR) sample with 10 ng of DNA, 5 × FIREPol Master Mix (Solis BioDyne, Estonia), and 1 μl of 20 × TaqMan SNP Genotyping Assay. We set thermal cycling conditions at 60°C for 1 min and 95°C for 15 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. We used Sanger sequencing to validate certain selected cases of homozygous and heterozygous genotypes using a BigDye Terminator v3.1 Cycle Sequencing Kit on a 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA).
2.5 Quality assessment of the rs1046322 and rs9457 SNPs
We assessed the quality and statistical association of the rs1046322 and rs9457 SNPs using the PLINK genome association analysis toolset (version 1.9). Next, for quality assessments, we determined minor allele frequency (MAF) and consistency with the Hardy–Weinberg equilibrium for the WFS1 variants.
2.6 Statistical analysis
Data are presented as mean± standard deviation (SD). We determined statistical significance using Student’s t-test for quantitative variables and Fisher’s exact test for categorical variables, and P values ≤ 0.05 were considered significant. We assessed allele-based associations between the rs1046322 variant and the quantitative traits (BMI, TGL, and WC) using genetic models based on additive mode of inheritance (GG versus GA versus AA) adjusted for the confounders of age, sex and diabetes status. We assessed changes in the mean of phenotype measurement using regression coefficient (Beta), where a positive regression coefficient indicated that the minor allele increases the risk effect. Multiple comparisons were corrected by generating empirical P values (Pemp) using the max(T) permutation procedure available in PLINK, based on 10,000 permutations. A threshold of < 0.05 was set for both the P value and Pemp value to assess the statistical significance of the association signal. Any quantitative trait value lesser than Q1–1.5 × the interquartile range (IQR) or higher than Q3 + 1.5 × IQR was considered to be an outlier and was excluded from the statistical analyses. Statistical analyses were performed using PLINK, version 1.9, and R software, version 4.0.2.
3 Results
3.1 Characteristics of the study participants and genotyping data
The average rate of successful genotyping of the two SNPs rs1046322 and rs9457 in each of the three subpopulations, namely Arab, South Asians, and Southeast Asians was > 99%, and the SNP was within the Hardy–Weinberg equilibrium. Of the two SNPs, the rs9457 did not show significant associations with any of the examined obesity traits in any of the three ethnic cohorts, though an association is seen with HDL in the Arab cohort with a P value of 0.0334 albeit with an insignificant empirical P (Pemp) value (Supplementary Table S1; the allele and genotype frequencies for the variant are listed in the notes to the table). Thus, rs9457 was not included in further analyses.
The frequencies of the minor allele (A) at the WFS1 rs1046322 SNP in the Arab, South Asian, and Southeast Asian populations were 17.27%, 11.67%, and 6.77%, respectively. Of the 362 genotyped samples from the Southeast Asian population, 314 (86.7%) were homozygous for G, only 1 (0.3%) was homozygous for A, and 47 (13%) were heterozygous (GA). Of the 1045 genotyped samples from the Arab population, 715 (68.4%) were homozygous for G, 31 (3%) were homozygous for A, and 299 (28.6%) were heterozygous (GA). Of the 660 genotyped samples from the South Asian population, 515 (78%) were homozygous for G, 9 (1.4%) were homozygous for A, and 136 (20.6%) were heterozygous (GA).
The mean (SD) age of participants in the Southeast Asian cohort was 41.1 (9.4) years, in the Arab cohort was 47.6 (11.6) years and in the South Asian cohort was 42.9 (9.7) years. Thus, the participants in each of the three ethnic cohorts are uniformly largely middle-aged. Table 1 presents a genotype-wide distribution (GG versus (GA+AA)) at rs1046322 of the characteristics of the three ethnic cohorts. After examining the genotype-specific differences in the phenotypic traits, we observed statistically significant differences in the Southeast Asian population (Table 1). The phenotypic traits that showed statistically significant genotypic differences in the Southeast Asian population were as follows: (i) Mean (SD) of BMI was significantly higher in the participants that harbored the A allele as compared to non-carriers [27.63 (3.6) Kg/m2 vs. 26.02 (3.6) Kg/m2; P = 0.004] (Figure 1A and Table 1); (ii) WC of the SNP carriers was significantly higher than that of the non-carriers [89.9 (8.1) cm vs. 85.9 (9.6) cm; P = 0.006] (Figure 1B and Table 1); and (iii) Participants that harbored the A allele had higher TGL as compared to non-carriers [1.672 (0.8) mmol/l vs. 1.43 (0.7) mmol/l; P = 0.037] (Figure 1C and Table 1).
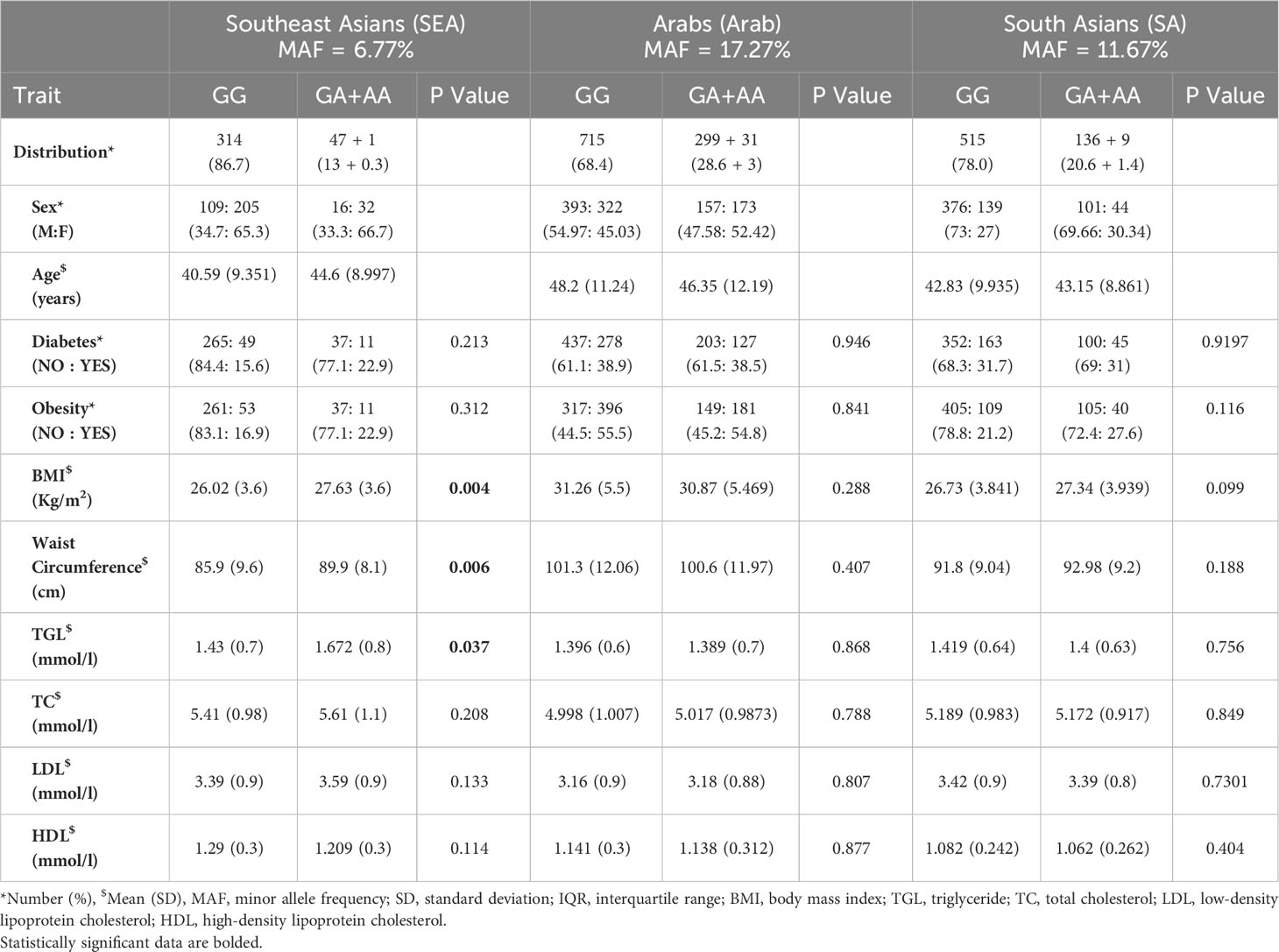
Table 1 Overview of the Southeast Asian, Arab, and South Asian populations as per genotype distribution of the WFS1 rs1046322 variant.
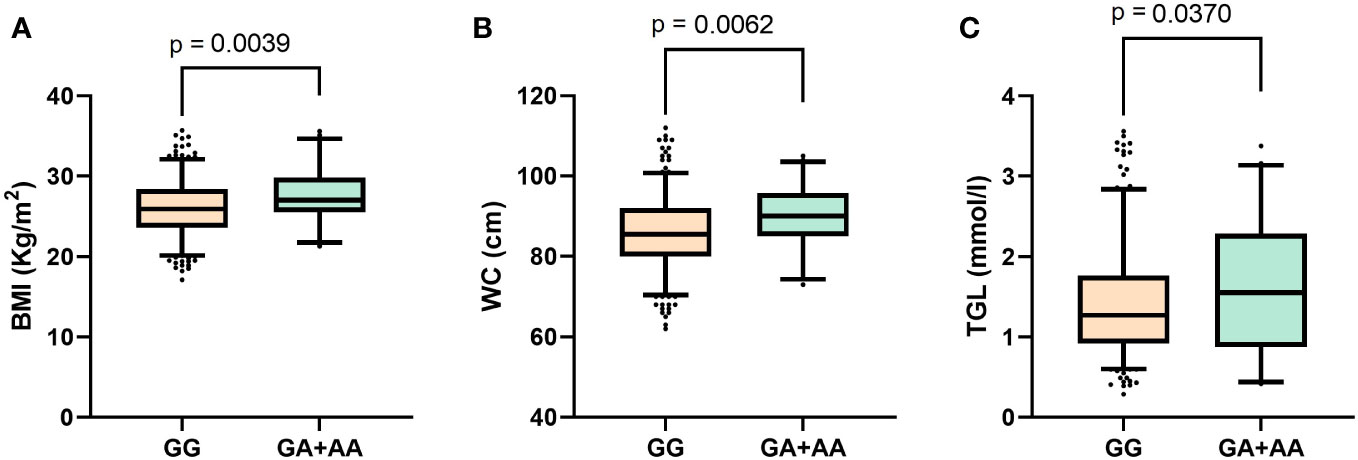
Figure 1 Boxplots displaying data distribution for the phenotype traits (A) Body mass index, (B) Waist circumference, (C) Triglyceride levels in Southeast Asian individuals with genotypes (GA+AA) containing the effect allele or homozygous (GG) genotypes for reference alleles of the WFS1 rs1046322 variant.
3.2 Association between WFS1 rs1046322 and obesity-related markers
The association tests for the variant and obesity markers showed that obesity traits were significantly associated with the WFS1 rs1046322 variant only in the SEA subpopulation, and not in the Arab or South Asian populations (Table 2). The traits showing statistically significant differences included BMI (β:1.562, P = 0.0035), WC (β:3.163, P = 0.0197), and TGL (β:0.224, P = 0.034). Further, the associations with BMI and WC also exhibited significant empirical Pempvalues of 0.0072 and 0.0388, respectively establishing the BMI and WC as strong contenders for association with the SNP.
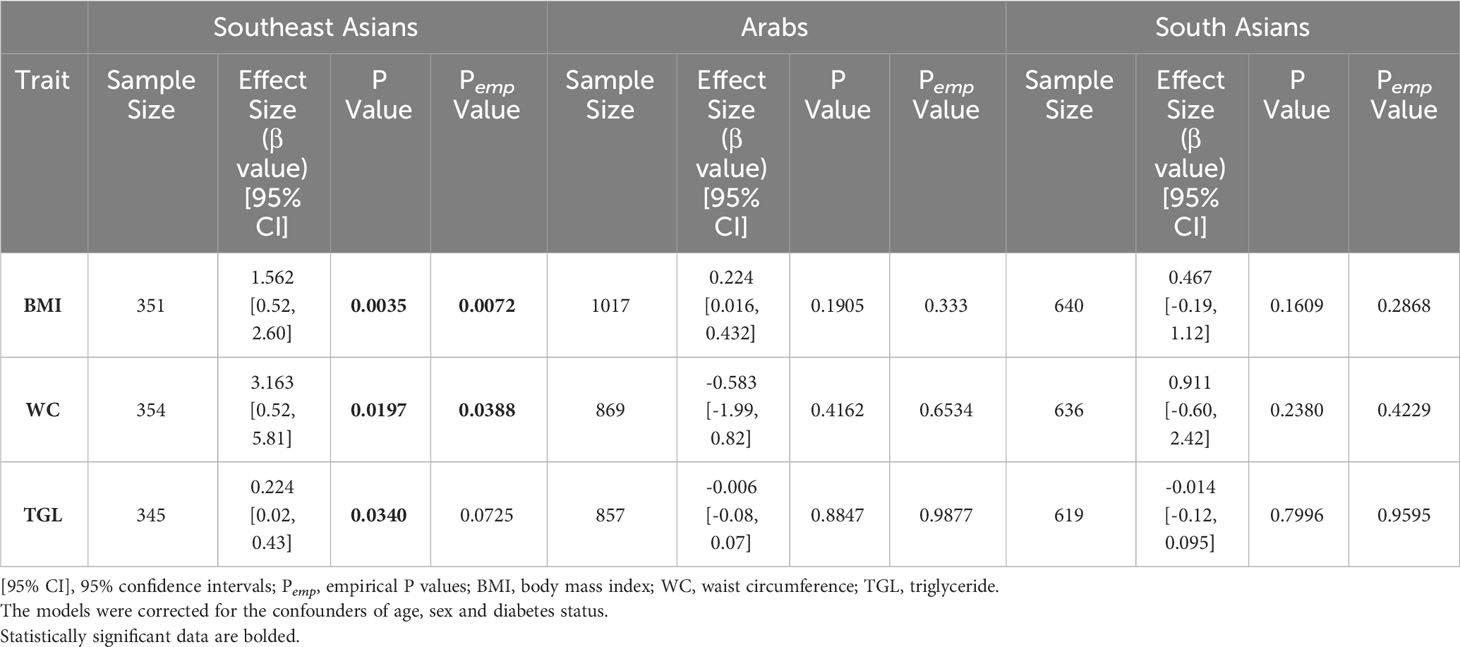
Table 2 Association tests for the WFS1 rs1046322 variant (A as the effect allele) with the phenotypic traits of BMI, WC and TGL using genetic models based on additive mode of inheritance (GG versus GA versus AA).
3.3 P value threshold after correction for multiple testing
Having found association of the SNP with triglyceride at a P value of 0.034, we investigated the associations of the SNP with the cholesterol traits of HDL, LDL and total cholesterol. We found that the associations for these cholesterol traits with the SNP were insignificant (Supplementary Table S2). As regards multiple testing for considering the cholesterol traits along with triglycerides and BMI and WC, it is to be noted that not all the cholesterol traits are independent of each other. In our earlier work (20), we queried independent variables among the four lipid traits by way of performing Pearson correlation analysis between the traits followed by matSpD analysis (21) (http://neurogenetics.qimrberghofer.edu.au/matSpD/), and found that the estimated effective number of independent traits among the lipid traits as 3. Upon including the WC and BMI as independent traits, we have a total of 5 independent traits tested for associations with the SNP. The P value threshold after multiple testing correction for significant associations turns out to be 0.01 (=0.05/5). Thus, after multiple testing correction, only the BMI association with the rs1046322 SNP (at a P value of 0.0035) remains significant.
4 Discussion
In this study, the WFS1 rs1046322 and rs9457 were assessed for association with obesity in the ethnic populations of Arabs, South Asians and Southeast Asians. The rs9457 SNP did not exhibit any significant associations with obesity traits. However, the results demonstrated that WFS1 rs1046322 is significantly associated with BMI and WC in the Southeast Asian population, but not in the Arab or South Asian populations. The SNP is also associated with TGL levels only in the Southeast Asian population, and not the other two subpopulations. Furthermore, carriers of the SNP had significantly higher BMI, WC, and TGL levels, as compared to non-carriers in the Southeast Asian subpopulation alone.
The 1000 Genome Project reported a MAF of 8% for the WFS1 rs1046322 variant. However, this SNP varies in its frequency across different populations, ranging from 7.2% in Chileans to 50% in Siberians. Although MAFs in the subpopulations of the present study fall within the lower range, we observed some differences among the three groups, with the Southeast Asian population showing the lowest MAF (6.8%), followed by the South Asian (11.7%) and Arab (17.3%) populations.
Upper body, or truncal, obesity is strongly associated with obesity-related complications, such as diabetes and cardiovascular diseases. Considering that WC is a measure of truncal obesity, it is interesting to note the effect sizes of the associations between the variant and WC and BMI (Table 2). The observed effect sizes in the SEA population indicate that the effect allele in WFS1 rs1046322 resulted in an increase by 3.163 cm in WC and by 1.562 Kg/m2 in BMI. Interestingly, a recent in vitro study by Ivask et al. (22) examined WFS1 heterozygous mouse model for response to high fat diet (HFD) in terms of body weight and metabolic characteristics. The authors found that the impaired body weight gain found in WFS1 mutant mice is prevented by HFD. They further observed that in WFS1 heterozygous mutant mice, HFD impaired the normalized insulin secretion and the expression of endoplasmic reticulum (ER) stress genes in isolated pancreatic islets. HFD increased the expression of Ire1α and Chop in pancreas and decreased the expression of Ire1α and Atf4 in liver from these mutant mice. The authors concluded that quantitative WFS1 gene deficiency predisposes carriers of single functional WFS1 copy to diabetes and metabolic syndrome and makes them susceptible to environmental factors such as HFD.
There is a lack of literature reports on associations between WFS1 and obesity. However, previous studies have linked several WFS1 SNPs with type 2 diabetes and biomarkers related to diabetes across various ethnicities, including the United Kingdom population, Swedish population and Ashkenazi population (23–26). One of the recently published large studies that confirmed the association between WFS1 and type 2 diabetes included 81,412 type 2 diabetes patients and 370,832 healthy individuals of diverse ancestries (27). Furthermore, the DESIR (Data from Epidemiological Study on the Insulin Resistance Syndrome) prospective study demonstrated in French cohorts that allelic variations at three SNPs in the WFS1 gene were associated with incident type 2 diabetes (28).
Considering the established role of wolframin as an ER stress regulator that negatively regulates ER stress signaling, discovering a link between the gene and obesity does not come as a surprise. Additional studies are required to further investigate the mechanism for this regulation. Given that wolframin plays a key role in mediating the ER export of vesicular cargo proteins, it could be speculated that it regulates the processing and release of different gut hormones or melanocortin hormones in the brain, similar to its role in regulating proinsulin cleavage and insulin secretion (29). In addition, a recent study demonstrated that WFS1 regulates anti-inflammatory responses in pancreatic β-cells. Specifically, the study reported that the pancreatic islets of WFS1 whole-body knockout mice display M1-macrophage infiltration and hypervascularization (30).
WFS1 rs1046322 is a 3’ UTR variant and is a putative miRNA (miR-668) binding site polymorphism. This variant was previously shown to influence the affinity of miR-668 to WFS1 mRNA (31). Though there exist no previous studies investigating the role of miR-668 in obesity, it has been recently shown that miR-668-3p can suppress mediators of inflammation and oxidative stress (32). Therefore, it would be interesting to examine the effect of this miRNA in obesity and its expression level in participants who suffer from obesity or metabolic syndrome.
In conclusion, the findings of the present study suggest an ethnic-specific role for WFS1 in obesity. While the current study included a large cohort with three different ethnic populations, further studies would benefit by examining the observed associations in more diverse ethnic populations. The study also highlights the importance of including ethnic groups that are under-represented in current global genetic studies of genotype–phenotype associations.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The study protocol was reviewed and approved by the Ethical Review Committee of Dasman Diabetes Institute. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
MH, MA-F, TT and JA contributed to conception and design of the study. PH and AC organized the database and performed the statistical analysis. MH and PH wrote the first draft of the manuscript. MA-F and TT wrote sections of the manuscript. EA, BC and MM performed the assays. TAT, JA and FA-M revised and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by Kuwait Foundation for the Advancement of Sciences (KFAS; research project RA HM-2018-039).
Acknowledgments
We would like to thank the National Dasman Diabetes Biobank Core Facility at DDI for their contribution in sample processing. We are also indebted to Kuwait Foundation for the Advancement of Sciences (KFAS) for providing financial support for this research project (RA HM-2018-039). The funding agency was not involved in data collection, analysis, or interpretation; trial design; patient recruitment; or any aspect pertinent to the study. Lastly, we would like to thank Ms. Lubaina Koti for editing the manuscript for language, structure, and accuracy.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1185956/full#supplementary-material
Supplementary Table 1 | Association tests for the WFS1 rs9457 variant (G as the effect allele) with phenotypic traits using genetic models based on additive mode of inheritance (CC versus GC versus GG). The model was adjusted for the confounders of age, sex and diabetes status.
Supplementary Table 2 | Association tests for the WFS1 rs1046322 variant (A as the effect allele) with phenotypic traits (including cholesterol traits) using genetic models based on additive mode of inheritance (GG versus GA versus AA). The model was adjusted for the confounders of age, sex, and diabetes status.
References
1. Hruby A, Hu FB. The epidemiology of obesity: A big picture. PharmacoEconomics (2015) 33(7):673–89. doi: 10.1007/s40273-014-0243-x
2. Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet (2014) 384(9945):766–81. doi: 10.1016/S0140-6736(14)60460-8
3. Kelly T, Yang W, Chen CS, Reynolds K, He J. Global burden of obesity in 2005 and projections to 2030. Int J Obes (2008) 32(9):1431–7. doi: 10.1038/ijo.2008.102
4. Awasthi A, Panduranga AB, Deshpande A. Prevalence of overweight/obesity in South Asia: A narrative review. Clin Epidemiol Global Health (2023) 22:101316. doi: 10.1016/j.cegh.2023.101316
5. Badran M, Laher I. Obesity in arabic-speaking countries. J Obes (2011) 2011:686430. doi: 10.1155/2011/686430
6. Loos RJF, Yeo GSH. The genetics of obesity: from discovery to biology. Nat Rev Genet (2022) 23(2):120–33. doi: 10.1038/s41576-021-00414-z
7. Loos RJ. Recent progress in the genetics of common obesity. Br J Clin Pharmacol (2009) 68(6):811–29. doi: 10.1111/j.1365-2125.2009.03523.x
8. El-Sayed Moustafa JS, Froguel P. From obesity genetics to the future of personalized obesity therapy. Nat Rev Endocrinol (2013) 9(7):402–13. doi: 10.1038/nrendo.2013.57
9. Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet (2010) 42(11):937–48. doi: 10.1038/ng.686
10. Albuquerque D, Nóbrega C, Manco L, Padez C. The contribution of genetics and environment to obesity. Br Med Bull (2017) 123(1):159–73. doi: 10.1093/bmb/ldx022
11. Byrd AS, Toth AT, Stanford FC. Racial disparities in obesity treatment. Curr Obes Rep (2018) 7(2):130–8. doi: 10.1007/s13679-018-0301-3
12. Inoue H, Tanizawa Y, Wasson J, Behn P, Kalidas K, Bernal-Mizrachi E, et al. A gene encoding a transmembrane protein is mutated in patients with diabetes mellitus and optic atrophy (Wolfram syndrome). NatGenet (1998) 20(2):143–8. doi: 10.1038/2441
13. Fraser FC, Gunn T. Diabetes mellitus, diabetes insipidus, and optic atrophy. An autosomal recessive syndrome? J Med Genet (1977) 14(3):190–3. doi: 10.1136/jmg.14.3.190
14. Richardson JE, Hamilton W. Diabetes insipidus, diabetes mellitus, optic atrophy, and deafness. 3 cases of (DIDMOAD) syndrome. Arch Dis Childhood (1977) 52(10):796. doi: 10.1136/adc.52.10.796
15. Philbrook C, Fritz E, Weiher H. Expressional and functional studies of Wolframin, the gene function deficient in Wolfram syndrome, in mice and patient cells. Exp Gerontol (2005) 40(8):671–8. doi: 10.1016/j.exger.2005.06.008
16. Rigoli L, Bramanti P, Di Bella C, De Luca F. Genetic and clinical aspects of Wolfram syndrome 1, a severe neurodegenerative disease. Pediatr Res (2018) 83(5):921–9. doi: 10.1038/pr.2018.17
17. Fonseca SG, Ishigaki S, Oslowski CM, Lu S, Lipson KL, Ghosh R, et al. Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J Clin Invest (2010) 120(3):744–55. doi: 10.1172/JCI39678
18. Fawcett KA, Wheeler E, Morris AP, Ricketts SL, Hallmans G, Rolandsson O, et al. Detailed investigation of the role of common and low-frequency WFS1 variants in type 2 diabetes risk. Diabetes (2010) 59(3):741–6. doi: 10.2337/db09-0920
19. Elek Z, Nemeth N, Nagy G, Nemeth H, Somogyi A, Hosszufalusi N, et al. Micro-RNA binding site polymorphisms in the WFS1 gene are risk factors of diabetes mellitus. PloS One (2015) 10(10):e0139519. doi: 10.1371/journal.pone.0139519
20. Hebbar P, Nizam R, Melhem M, Alkayal F, Elkum N, John SE, et al. Genome-wide association study identifies novel recessive genetic variants for high TGs in an Arab population. J Lipid Res (2018) 59(10):1951–66. doi: 10.1194/jlr.P080218
21. Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (2005) 95(3):221–7. doi: 10.1038/sj.hdy.6800717
22. Ivask M, Volke V, Raasmaja A, Koks S. High-fat diet associated sensitization to metabolic stress in Wfs1 heterozygous mice. Mol Genet Metab (2021) 134(1-2):203–11. doi: 10.1016/j.ymgme.2021.07.002
23. Minton JA, Hattersley AT, Owen K, McCarthy MI, Walker M, Latif F, et al. Association studies of genetic variation in the WFS1 gene and type 2 diabetes in U.K. populations. Diabetes (2002) 51(4):1287–90. doi: 10.2337/diabetes.51.4.1287
24. Sandhu MS, Weedon MN, Fawcett KA, Wasson J, Debenham SL, Daly A, et al. Common variants in WFS1 confer risk of type 2 diabetes. Nat Genet (2007) 39(8):951–3. doi: 10.1038/ng2067
25. Franks PW, Rolandsson O, Debenham SL, Fawcett KA, Payne F, Dina C, et al. Replication of the association between variants in WFS1 and risk of type 2 diabetes in European populations. Diabetologia (2008) 51(3):458–63. doi: 10.1007/s00125-007-0887-6
26. Wasson J, Permutt MA. Candidate gene studies reveal that the WFS1 gene joins the expanding list of novel type 2 diabetes genes. Diabetologia (2008) 51(3):391–3. doi: 10.1007/s00125-007-0920-9
27. Mahajan A, Wessel J, Willems SM, Zhao W, Robertson NR, Chu AY, et al. Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat Genet (2018) 50(4):559–71. doi: 10.1038/s41588-018-0084-1
28. Cheurfa N, Brenner GM, Reis AF, Dubois-Laforgue D, Roussel R, Tichet J, et al. Decreased insulin secretion and increased risk of type 2 diabetes associated with allelic variations of the WFS1 gene: the Data from Epidemiological Study on the Insulin Resistance Syndrome (DESIR) prospective study. Diabetologia (2011) 54(3):554–62. doi: 10.1007/s00125-010-1989-0
29. Wang L, Liu H, Zhang X, Song E, Wang Y, Xu T, et al. WFS1 functions in ER export of vesicular cargo proteins in pancreatic beta-cells. Nat Commun (2021) 12(1):6996. doi: 10.1038/s41467-021-27344-y
30. Morikawa S, Blacher L, Onwumere C, Urano F. Loss of function of WFS1 causes ER stress-mediated inflammation in pancreatic beta-cells. Front Endocrinol (Lausanne) (2022) 13:849204. doi: 10.3389/fendo.2022.849204
31. Kovacs-Nagy R, Elek Z, Szekely A, Nanasi T, Sasvari-Szekely M, Ronai Z. Association of aggression with a novel microRNA binding site polymorphism in the wolframin gene. Am J Med Genet Part B: Neuropsychiatr Genet (2013) 162(4):404–12. doi: 10.1002/ajmg.b.32157
32. Gao Z, Gao Q, Lv X. MicroRNA-668-3p protects against oxygen-glucose deprivation in a rat H9c2 cardiomyocyte model of ischemia-reperfusion injury by targeting the stromal cell-derived factor-1 (SDF-1)/CXCR4 signaling pathway. Med Sci Monitor Int Med J Exp Clin Res (2020) 26:e919601. doi: 10.12659/MSM.919601
Keywords: WFS1, ethnicity, obesity, triglycerides, polymorphism, waist circumference
Citation: Hammad MM, Abu-Farha M, Hebbar P, Anoop E, Chandy B, Melhem M, Channanath A, Al-Mulla F, Thanaraj TA and Abubaker J (2023) The miR-668 binding site variant rs1046322 on WFS1 is associated with obesity in Southeast Asians. Front. Endocrinol. 14:1185956. doi: 10.3389/fendo.2023.1185956
Received: 14 March 2023; Accepted: 11 September 2023;
Published: 04 October 2023.
Edited by:
Tarunveer Singh Ahluwalia, Steno Diabetes Center Copenhagen (SDCC), DenmarkReviewed by:
Anton Terasmaa, National Institute of Chemical Physics and Biophysics, EstoniaTajudeen Yahaya, Federal University, Birnin Kebbi, Nigeria
Edith Hofer, Medical University of Graz, Austria
Sulev Kõks, Murdoch University, Australia
Copyright © 2023 Hammad, Abu-Farha, Hebbar, Anoop, Chandy, Melhem, Channanath, Al-Mulla, Thanaraj and Abubaker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jehad Abubaker, amVoYWQuYWJ1YmFrckBkYXNtYW5pbnN0aXR1dGUub3Jn; Thangavel Alphonse Thanaraj, YWxwaG9uc2UudGhhbmdhdmVsQGRhc21hbmluc3RpdHV0ZS5vcmc=
†These authors have contributed equally to this work
 Maha M. Hammad
Maha M. Hammad Mohamed Abu-Farha
Mohamed Abu-Farha Prashantha Hebbar
Prashantha Hebbar Emil Anoop4
Emil Anoop4 Motasem Melhem
Motasem Melhem Arshad Channanath
Arshad Channanath Fahd Al-Mulla
Fahd Al-Mulla Thangavel Alphonse Thanaraj
Thangavel Alphonse Thanaraj Jehad Abubaker
Jehad Abubaker