- 1Shanghai Engineering Research Center of Advanced Dental Technology and Materials, Shanghai Key Laboratory of Stomatology & Shanghai Research Institute of Stomatology, National Clinical Research Center for Oral Diseases, Shanghai Ninth People’s Hospital, College of Stomatology, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Department of Prosthodontics, Ninth People’s Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 3Department of Preventive Dentistry, Ninth People’s Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
- 4Department of Radiation Oncology, David Geffen School of Medicine at University of California, Los Angeles (UCLA), Los Angeles, CA, United States
- 5Department of Endodontics, Ninth People’ Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
Objectives: Periodontium regeneration remains a significant challenge in clinics and research, and it is essential to understand the stage-specific biological process in situ. However, differing findings have been reported, and the mechanism has yet to be elucidated. The periodontium of adult mice molars is considered to be stable remodeling tissue. At the same time, the continuously growing incisors and the developing dental follicle (DF) of postnatal mice highly represent fast remodeling tissue. In this study, we attempted to explore different clues of temporal and spatial comparisons to provide improved references for periodontal regeneration.
Methods: Periodontal tissues from the developing periodontium (DeP) of postnatal mice, and continuously growing periodontium (CgP) and stable remodeling periodontium (ReP) of adult mice were isolated and compared using RNA sequencing. Based on the Dep and CgP separately compared with the ReP, differentially expressed genes and signaling pathways were analyzed using GO, KEGG databases, and Ingenuity Pathway Analysis (IPA). The results and validation were obtained by immunofluorescence staining and RT-PCR assays. Data were expressed as means ± standard deviation (SD) and analyzed by GraphPad Prism 8 software package, and one-way ANOVA was used to test multiple groups.
Results: Principal component analysis showed that the three groups of periodontal tissue were successfully isolated and had distinct expression profiles. A total of 792 and 612 DEGs were identified in the DeP and CgP groups compared with the ReP. Upregulated DEGs in the DeP were closely related to developmental processes, while the CgP showed significantly enhanced cellular energy metabolism. The DeP and CgP showed a common downregulation of the immune response, with activation, migration, and recruitment of immune cells. IPA and further validation jointly suggested that the MyD88/p38 MAPK pathway played an essential regulatory role in periodontium remodeling.
Conclusion: Tissue development, energy metabolism, and immune response were critical regulatory processes during periodontal remodeling. Developmental and adult stages of periodontal remodeling showed different expression patterns. These results contribute to a deeper understanding of periodontal development and remodeling and may provide references for periodontal regeneration.
1 Introduction
The periodontium, composed of the gingiva, cementum, periodontal ligament, and alveolar bone, plays an essential role in supporting and maintaining tooth function. Although studies have shown that the periodontium contains a population of stem cells that can further differentiate into fibroblasts, cementoblasts, osteoblasts, and osteoclasts, with the ability to self-renew and remodel (1), it is still highly challenging to repair periodontal defects caused by trauma and congenital malformation. A unique characteristic of the periodontal complex (cement–periodontal ligament–bone) is the ordered hierarchical structure within its tiny space, posing spatiotemporal compartmentalization as a critical requirement for micron-scale regeneration and functional restoration of multiple tissues (2). However, current clinical methods, such as guided tissue regeneration, still cannot effectively prevent disorganized or spontaneous tissue formation that can lead to anomalies, such as ankyloses (3). It has gradually become a consensus that periodontal regeneration based on the biological rationales of in situ remodeling to promote orderly tissue patterns and morphogenesis could be more effective.
Recently, researchers have employed new techniques and methods to help obtain more information on tissues and cells. Biomedical “omics” approaches rely on collecting large amounts of omics data, combined with bioinformatics and biostatistics, to drive new and deeper insights into complex biology (4). In previous studies, single-cell RNA-seq analysis was successfully applied to analyze the cellular heterogeneity of cells in the periodontium and their developmental relationship with early precursor cells, such as identifying PTHrP+ cells in the dental follicle as important precursor cells that may give birth to osteoblasts, periodontal ligament cells, and cementoblasts to provide a cell source for later periodontal tissue formation (5). Some transcriptome studies have investigated the gene expression patterns of periodontal ligament cells under specific culture conditions, such as mechanical loading and osteogenic induction, to clarify the mechanism of different environmental factors stimulating the differentiation process (6, 7). Furthermore, deepening the understanding of oral proteomics and the microbiome provides meaningful clues for detecting the onset of periodontal diseases, improving treatment effects, and promoting personalized treatment (8, 9). However, the remodeling and repair of periodontal tissues result from complex cell–environment and cell-cell interactions. Detecting in vitro periodontal cells or specific cell populations is insufficient for systematically understanding periodontal remodeling. The process of how periodontal tissue characteristics regulate periodontal remodeling in situ in a healthy state has not been elucidated. Therefore, sample sources for omics exploration should be further considered to fully understand the biological process of dynamic periodontal tissue remodeling and the cellular and molecular mechanisms controlling tissue patterns and morphogenesis that contribute to the development of periodontal regeneration and self-repair methods.
Regeneration shares many mechanisms with embryonic development. However, renewal to restore original tissue structure and biological function does not appear equivalent to developmental events. Adult tissues and organs do not necessarily show the same gene expression profiles as their embryonic, fetal, or juvenile counterparts (10). For example, Driskell’s team compared genes and cells in developing and adult skin and found that a transcription factor, Lef1, which controls hair follicle formation during development, acts as a molecular switch in the skin of young mice. It is essentially turned off after skin formation, but when specific cells in adult mice were activated, their skin healed a wound without leaving a scar (11). From another aspect, although the regenerative capacity of most tissues and organs declines with age, some organs continue to grow and remodel in adulthood, providing promising clues for treating adult patients. For example, studies have shown that the continuous activation of Wnt signaling may play a role in the growth and self-renewal of the kidney, providing a therapeutic target for promoting or restoring the regenerative capacity of the kidney (12). Thus, research on tissue regeneration should be conducted throughout the life cycle. Since the developmental stages in youth and adult regeneration processes are not identical, exploring periodontal remodeling in youths and adults is equally essential for periodontium regeneration. Thus, studying the different stages of active remodeling of the periodontium is necessary.
Periodontium develops from the dental follicle. Accompanied by root development and tooth eruption, dental follicles differentiate to form periodontal tissues (13). At this stage, the follicle coordinates the extensive formation and remodeling of the extracellular matrix. It is worth noting that rodent incisors continue to grow throughout life. Compared with molars, the incisors have sites of more active remodeling of the extracellular matrix (14), which provides us with an ideal model for studying periodontal renewal and remodeling in adult individuals.
In this study, mouse dental follicles and incisor periodontium were used as representative tissues representing actively remodeling periodontal tissue in the developmental and adult stages. They were compared with the stable remodeling periodontium of molar teeth. Genome-wide mRNA expression profiling and in-depth bioinformatics analysis were performed to characterize the regulation of gene expression in the actively remodeling periodontal tissues. Attention was paid to the shared or unique signaling regulatory mechanisms during periodontal remodeling in the developmental and maturation stages. More importantly, multi-directional regulatory targets in various processes of periodontal tissue remodeling were proposed to provide a basis for a more comprehensive understanding of the underlying mechanism and the promotion of periodontal defect repair.
2 Methods
2.1 Tissue acquisition and preparation
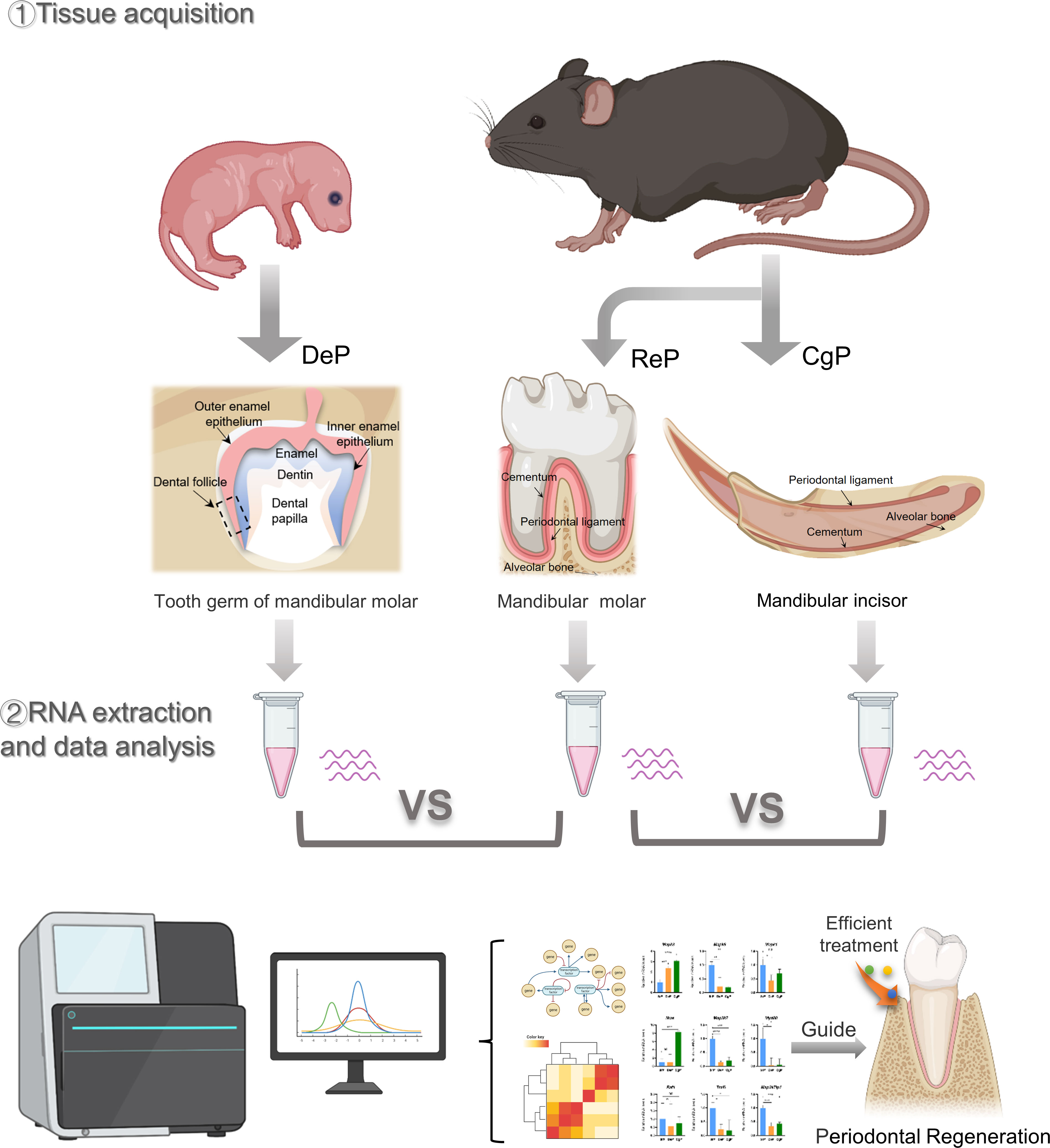
All of the experimental animal procedures were approved by the Laboratory Animal Ethics Committee at the Ninth People’s Hospital affiliated to Shanghai Jiao Tong University School of Medicine. C57 mice were used in this study. The molar tooth germs were excised from postnatal day 3 mice under a zoom stereo microscope (Olympus SZ51, Tokyo, Japan) by cutting the tooth capsule open and carefully removing the tooth papilla. Then, 1/3 of the dental follicles adjacent to the root were peeled off. The periodontal ligament of the mandibular molar and the lingual periodontal ligament of the mandibular incisor were dissected from eight-week-old adult mice. Each dental follicle sample contained tissue from eight to ten mice. Each molar and incisor periodontium sample contained tissue from three mice. There were three samples of each group, and the tissues were stored in a tissue storage solution for RNA extraction. In addition, the tissue of three mice in each group was obtained for histological experiments. Figure 1 illustrates the flow diagram of this study.
2.2 Histological analysis
The mandibles of postnatal and adult mice (n = 3) were dissected and fixed in 4% paraformaldehyde. Mandibular bones were subsequently demineralized in 0.5 M ethylenediaminetetraacetic acid. The tissue was embedded in paraffin and cut into 5-μm tissue sections.
For hematoxylin and eosin (H&E) staining, nuclei were stained with Gill’s hematoxylin, then stained blue with diluted ammonium hydroxide, and the cytoplasm was counterstained with acidified eosin. Immunofluorescent staining was performed according to a standard procedure, and 4,6-diamidino-2-phenylindole (DAPI) was used to visualize the nuclei. Mean fluorescence intensity was calculated for three fields of tissue for each group to indicate cell density using ImageJ software.
2.3 RNA preparation and sequencing
Total RNA was isolated from each sample using an RNA mini kit (Qiagen, Germany). RNA quality was examined using gel electrophoresis and a Qubit fluorometer (Thermo, Waltham, MA, USA). For RNA sequencing, strand-specific libraries were constructed using a TruSeq RNA sample preparation kit (Illumina, San Diego, CA, USA), and sequencing was carried out using an Illumina Novaseq 6000 instrument.
2.4 Real-time PCR analysis of gene expression
Total cellular RNA of periodontal tissues was extracted with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and reverse transcription was performed using a PrimeScript RT reagent kit (Takara Bio, Inc., Otsu, Japan). Gene-specific primers were synthesized commercially (Shenggong Co., Ltd., Shanghai, China), and their sequences are listed in Table 1. In one reaction, a 10 μL SYBR Premix Ex Taq kit (Takara Bio, Inc.) was used to amplify 1 μL of cDNA (mixed with 8 μL of distilled water and 0.5 μL of each primer). A Bio-Rad iQ5 real-time PCR system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was then used to detect gene expression. All of the relative gene expression values were normalized to Actb (encoding β-actin) based on the 2ΔΔCq method.
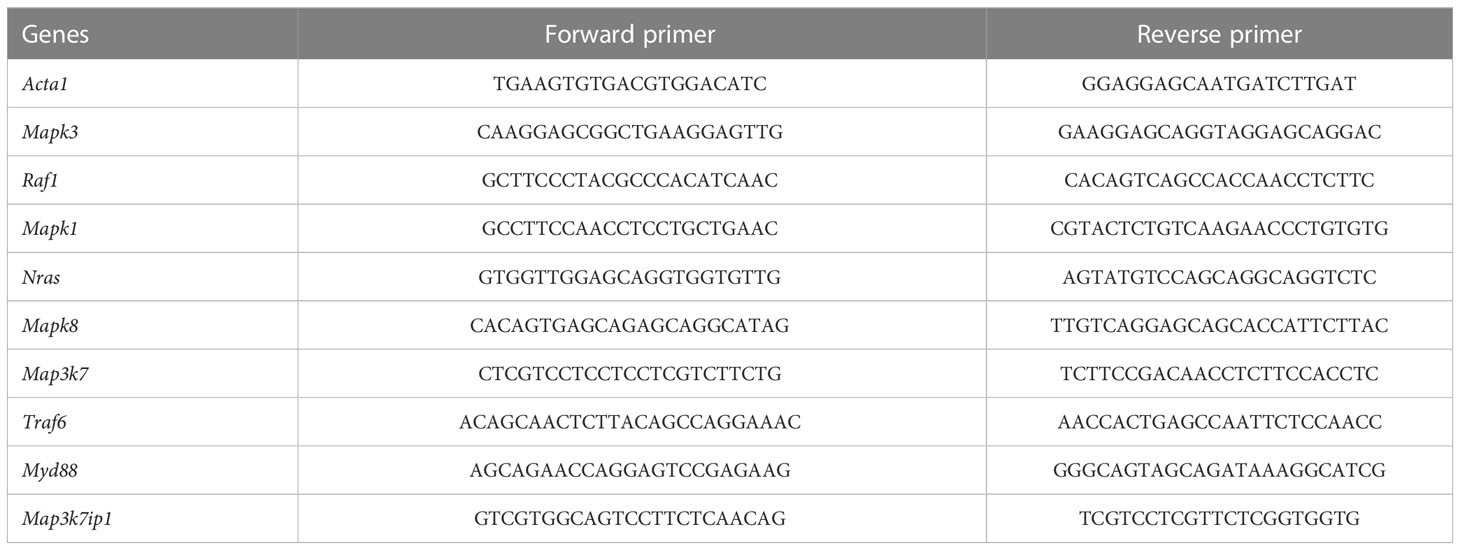
Table 1 Sequences of the primers used for a reverse transcription-quantitative polymerase chain reaction.
2.5 RNA-seq and identification of differentially expressed genes
The sequencing data have been deposited in the NCBI Sequence Read Archive (SRA) database under the accession code PRJNA928506. The raw data were processed by Skewer, and data quality was checked by FastQC v0.11.2 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The expression of the genes was calculated by fragments per kilobase of exon model per million mapped reads (FPKM) using Perl. Differentially expressed genes (DEGs) between different tissues were determined using the MA-plot-based method with random sampling (MARS) model in the DEGseq package. The thresholds for determining DEGs were q value (adjusted p-value) < 0.05 and absolute fold change ≥ 1.5.
2.6 Bioinformatics analysis
Venn diagram and principal component analyses were performed using R software. DEGs were chosen for function and signaling pathway enrichment analysis using GO and KEGG databases to discover potential regulatory networks. The significantly enriched pathways were determined when p < 0.05.
The Ingenuity Pathway Analysis system (IPA system; Qiagen China Co., Ltd.), which includes canonical pathway analysis, disease and function, regulator effects, upstream regulators, and molecular networks, was also used for subsequent bioinformatics analysis. For each analysis, a p-value < 0.05 was set as the threshold.
2.7 Statistical analysis
GraphPad Prism 8 software package was used for statistical analysis. Data were expressed as means ± standard deviation (SD), and one-way ANOVA was used to test multiple groups. A statistically significant difference was considered at *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
3 Results
3.1 Acquisition of mice periodontal tissue
Three days after birth, the mouse molars developed to the late bell stage and had not yet erupted. The periodontal part of the dental follicles would develop into the periodontium. In 8-week-old mice, the teeth had erupted completely and participated in chewing and other maxillofacial activities. The molar dental follicle (DeP), the molar periodontium (ReP), and the lingual periodontium of the mandibular incisors (CgP) were collected, as shown in Figure 2A, and were subjected to high-throughput sequencing using the Illumina Hiseq platform.
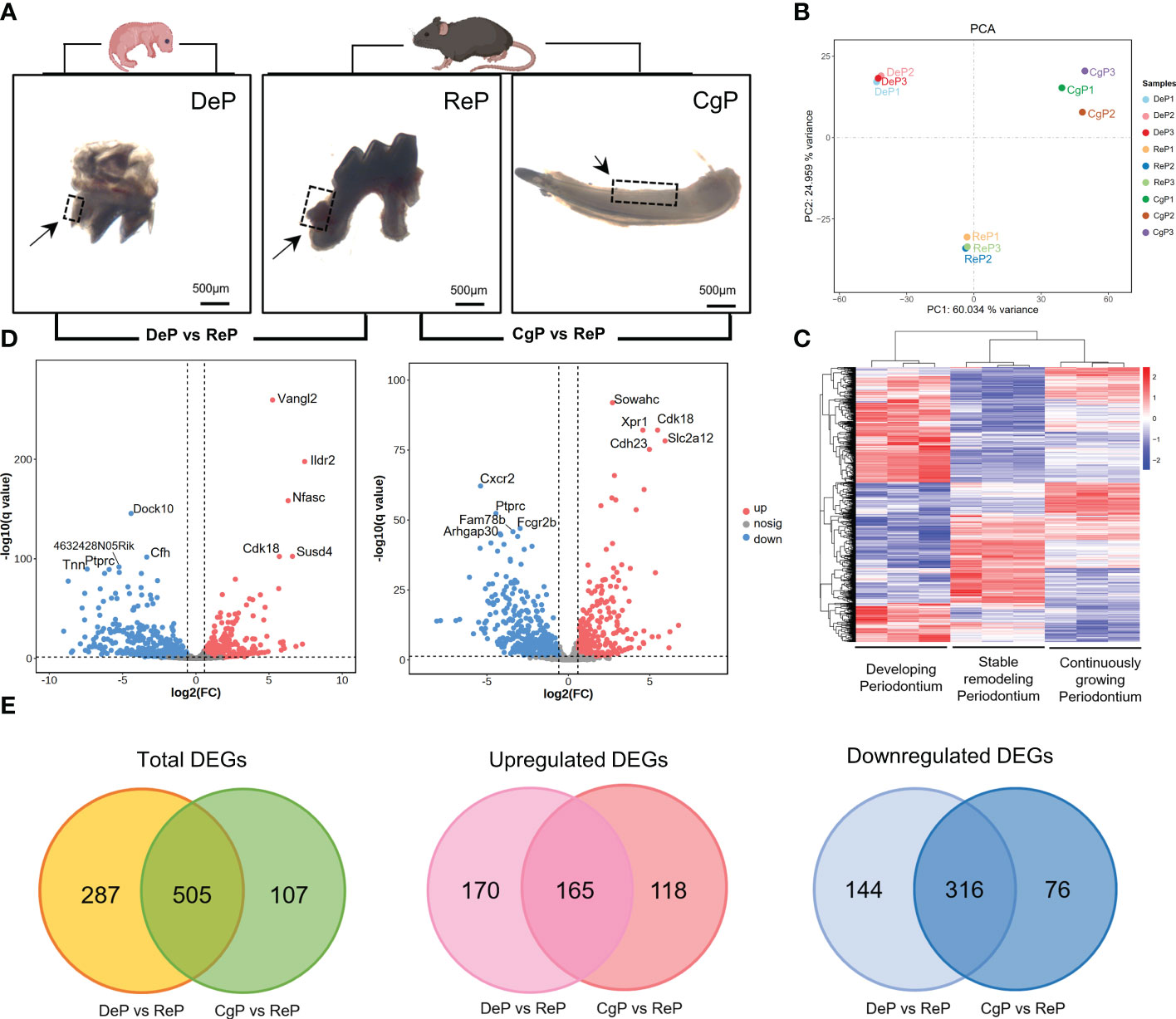
Figure 2 Comprehensive analysis of DEGs. (A) Postnatal mice dental follicles, adult mice incisors, and molars periodontal tissues were obtained. Tissues were collected around the tooth germ/tooth(inside the black dotted line). (B) Principal component analysis of the three groups. The higher the variation in gene expression patterns across samples was, the greater the distance among samples. (C) Heat map of differential gene expression levels in different periodontal tissues. The color saturation in the heat map indicates the expression of genes. Blue shows the lowest expression of genes, and red indicates the highest expression of genes. (D) Comparison of DEGs in DeP/CgP versus ReP using the Volcano plot. Red dots indicate significantly upregulated genes, blue dots indicate downregulated genes considerably, and gray dots represent nonsignificantly differentially expressed genes. (E) Venn diagram. DEGs were identified as having a q value (adjusted p-value) of < 0.05 and |fold change| of ≥ 1.5.
The transcript abundance of each gene was estimated by FPKM. Principal component analysis was performed to determine whether the expression profiles obtained for each group represented unique molecular features suggestive of developing or actively remodeling periodontal tissues. The results showed that principal component 1 (PC1) accounted for 60.034% of the variance. Principal component 2 (PC2), accounting for 24.959% of the variance, clearly separated the three groups in this study (Figure 2B). Figure 2C shows the heat map after the bidirectional hierarchical clustering of the genes and samples, which sufficiently demonstrated the intra-group consistency of the groups, and the inter-group difference was apparent.
3.2 Significant changes in gene expression were observed in DeP and CgP compared with the ReP
Significant DEGs were identified with a significance threshold of |fold change| ≥ 1.5 and q value (adjusted p-value) < 0.05. Identification of DEGs revealed different gene expression regulations of the remodeling process in DePs and CgPs. When the DeP was compared with the ReP (DeP versus ReP comparison), 335 genes were found to be upregulated, and 460 were downregulated. The top five upregulated genes were Vangl2, Ildr2, Nfasc, Susd4, and Cdk18, while the top five downregulated genes were Dock10, Cfh, 4632428N05Rik, Tnn, and Ptprc, as shown in Figure 2D (left). A comparison of the CgP and ReP (CgP versus ReP comparison) revealed 283 upregulated genes and 392 downregulated genes. The top five upregulated genes were Sowahc, Cdk18, Xpr1, Slc2a12, and Cdh23, while the top five downregulated genes were Cxcr2, Ptprc, Fcgr2b, Fam78b, and Arhgap30. A volcano plot based on DEGs is shown in Figure 2D (right). Detailed information on the DEGs is provided in Supplementary Tables 1, 2.
To further compare the guiding significance of the DeP and CgP samples for the regeneration and repair of the periodontium, we evaluated the DEGs of these two comparisons. A Venn diagram (Figure 2E) revealed the two overlapping and unique DEGs compared with the ReP. The DeP and CgP shared 165 upregulated DEGs and 316 downregulated DEGs. At the same time, 170 and 118 upregulated DEGs and 144 and 76 downregulated DEGs were unique for the DeP and CgP, respectively. The results suggested that the gene regulation of periodontal tissue remodeling processes in developing and adult individuals was generally similar but still had different properties.
3.3 DeP and CgP were characterized by enhanced tissue development and cellular energy metabolic activity, respectively
To identify the critical physiological mechanisms involved in periodontium remodeling, enrichment analyses of DEGs in the DeP and CgP versus the ReP were performed. In GO analysis, upregulated genes in the DeP versus ReP comparison were mainly enriched in pathways involved in cell development (odontogenesis of dentin-containing tooth, odontogenesis, neuron development, and neuron projection development). Among the processes related to cell metabolism, it was noticed that the metabolism of phosphate-containing compounds metabolic process and phosphorus metabolic process were particularly prominent. Figure 3A illustrates the top 20 biological processes enriched by upregulated DEGs of DeP versus ReP. The upregulated DEGs in the CgP versus ReP comparison showed significant enrichment in the cellular energy metabolism process. In particular, aerobic metabolism accounted for a high proportion of energy metabolism (aerobic respiration, oxidative phosphorylation, and energy derivation by oxidation of organic compounds).
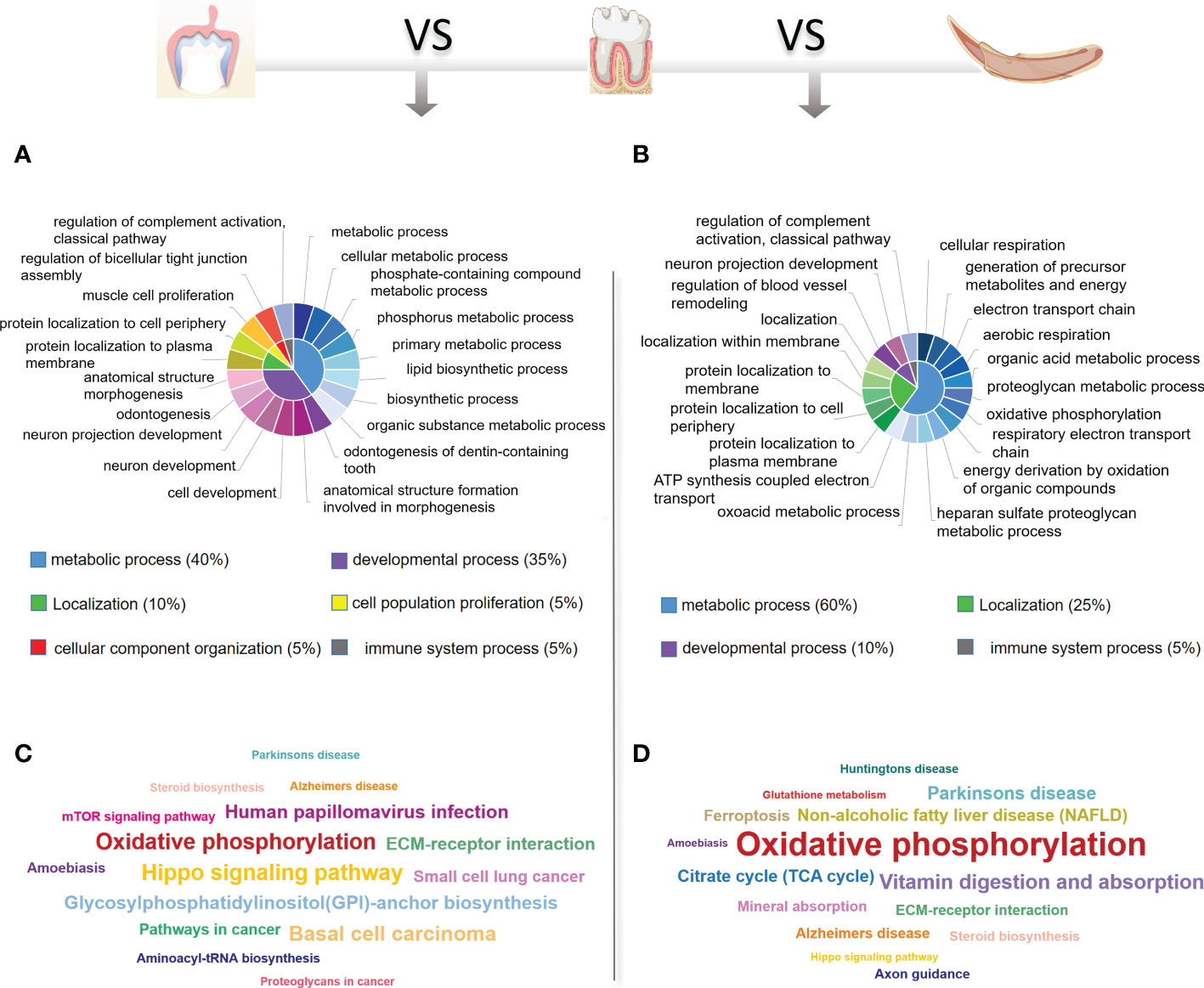
Figure 3 GO and KEGG database enrichment analyses were used to determine the critical pathways of the upregulated DEGs in DeP and CgP. (A, B) The top 20 biological processes in the GO enrichment analyses of the upregulated DEGs in DeP (A) and CgP (B). The same color in the inner circle represents the same category of pathways in the outer ring, and the area size of the inner circle represents the proportion of the pathway category. (C, D) The top 15 pathways in the KEGG enrichment analyses of the upregulated DEGs in DeP (C) and CgP (D). The size of the word in the word cloud is proportional to the -log10 (p-value).
Moreover, the significant enrichment of extracellular matrix metabolic processes suggested that they may be involved in active tissue remodeling processes, such as the proteoglycan metabolic process and the heparan sulfate proteoglycan metabolic process (Figure 3B). As shown by the KEGG analysis (Figures 3C, D), although the upregulated DEGs in DeP and CgP tissues were both enriched in the oxidative phosphorylation pathway, the Hippo signaling pathway, which is closely related to dental development, was more significantly enriched in the DeP. Correspondingly, the citrate cycle (TCA cycle) and other energy metabolism pathways in the CgP were enhanced.
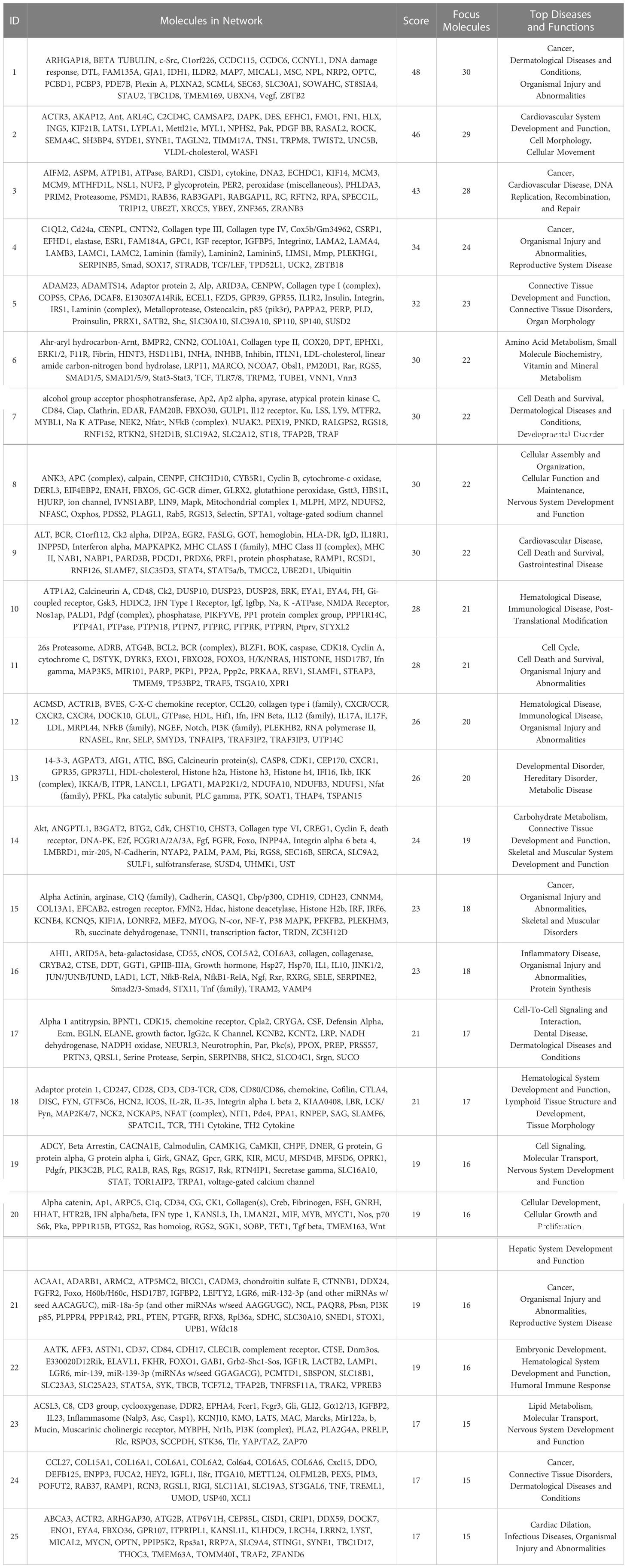
The IPA system was used to analyze the critical molecular networks involved in periodontium remodeling. The interaction network analysis was used to identify the interactions between molecules in the dataset and their related diseases and functions. Using the score value to rank all of the networks, it was found that the highest-ranked network in the DeP (score = 51) was associated with “connective tissue disorders” and involved in the disease regulation of “developmental disorders”. The results suggest that the gene expression changes in the DeP were closely related to the developmental process of the tissue. This network identified 32 molecules in the DEGs, which, together with the other three predicted molecules, constituted the corresponding molecular network, as shown in Table 2. The DEGs network affecting “connective tissue development and function” in the CgP also affected “carbohydrate metabolism”. These results suggested that the energy supply of cells plays a vital role in regulating the state of adult periodontal tissue. This network ranked 14th (score=24) and involved 19 molecules in the DEGs, which, together with 16 other predicted molecules, constituted the corresponding molecular network, as shown in Table 3. These results were consistent with GO and KEGG analyses, suggesting that the enhanced biological functions related to tissue development and energy metabolism were closely related to the formation and remodeling of periodontal tissues and may represent the characteristic cellular behavior of developing and mature periodontal tissues.
3.4 DeP and CgP both showed suppression of immune system processes
Enrichment analysis of the downregulated DEGs in the DeP and CgP compared with the ReP showed that the top 20 biological processes, according to p-value, accounted for the most significant proportion in the biological processes related to immune response regulation (7/20). Figure 4A shows the immune system processes enriched in the top 20 biological processes, including adaptive immune response, immune effector process, immune response, immune system process, leukocyte activation, positive regulation of immune system process, regulation of immune effector process, regulation of immune response, and regulation of immune system process. Consistent with this, in the KEGG analysis, both showed prominent downregulation in the pathways classified in the immune system, as shown in Figure 4B, involving the differentiation of immune cells such as Th17 cells, T cell receptor signaling pathway, and immune cell-mediated cytotoxicity. These results showed that there are immune-mediated tissue characteristics in both the DeP and CgP.
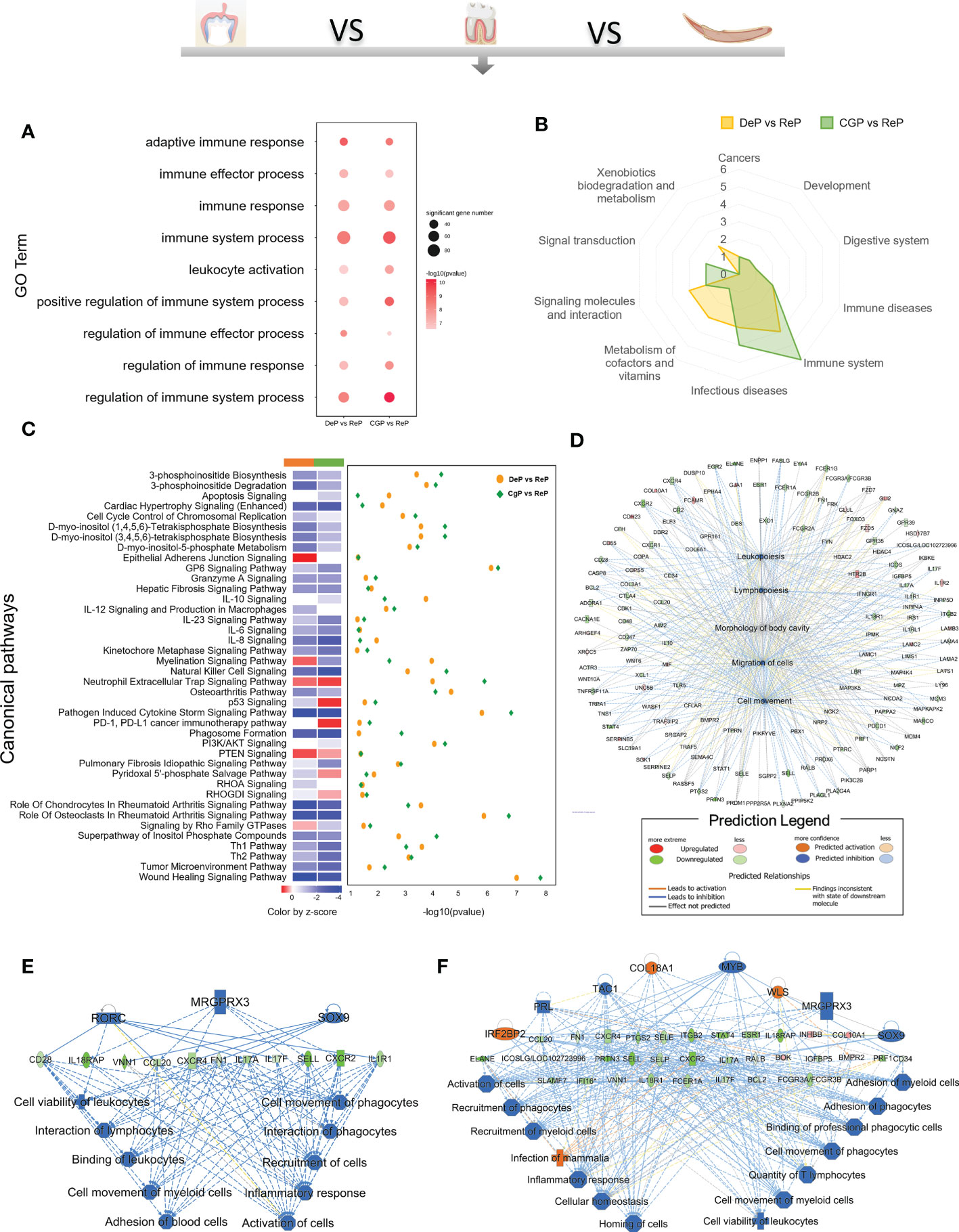
Figure 4 GO, KEGG database enrichment analyses, and IPA analysis were used to determine the critical function of the downregulated DEGs in DeP and CgP. (A) In the GO enrichment analysis of downregulated DEGs, immune system-related pathways in the top 20 pathways of DeP and CgP. (B) Radar map of the classification of the top 20 pathways in KEGG enrichment analysis of DeP and CgP. (C) The common canonical pathways enriched by DeP and CgP DEGs in IPA analysis. The z-score of the pathway is shown in the heat map, and the -log10 (p-value) of the pathway is shown in the scatter map. (D) “disease and function” predictions were made based on DEGs involved in common canonical pathways. The figure shows the top five functions. (E, F) Network diagram representing the regulatory effects with top consistency scores in DeP (E)/CgP (F) versus ReP.
The IPA system showed that the DEGs in the DeP versus the ReP and CgP versus ReP were enriched in multiple canonical pathways. Overall, 93 and 101 canonical pathways were identified by applying p < 0.05 as the threshold. Figure 4C shows the p-value and z-score of the 40 common classical pathways that were predicted. Functional predictions of DEGs involved in these common pathways showed that the functions of the top five involved leukopoiesis, lymphopoiesis, morphology of body cavity, migration of cells, and cell movement (Figure 4D).
Furthermore, the upstream regulatory network and downstream functions involved in DEGs were predicted by regulatory effect analysis. The consistency score evaluates the causal consistency of upstream regulators in the network, DEG datasets, and dense connectivity measures between disease and function. Fifty-four regulatory effects were identified in the DEGs of the DeP versus the ReP. Among them, the highest-ranking regulatory effect had a consistency score of 31.056, which strongly predicted that Mrgprx3, Rorc, and Sox9 in the DeP might participate in the downregulation of the movement and interaction of lymphocytes, myeloid cells, and phagocytes by mediating their targets (Figure 4E). Meanwhile, 47 regulatory effects were identified in the DEGs of CgP versus ReP. The highest ranked regulatory effect with a consistency score of 35.355 strongly suggested that the regulators Col18a1, Irf2bp2, Mrgprx3, Myb, Prl, Sox9, Tac1, and Wls may participate in extensive regulation of cellular behavior in immune processes (Figure 4F).
These results suggested that the altered gene expression was closely related to the immune response in the DeP and CgP. There were apparent differences in the status of leukocytes and myeloid cells in the tissues, which may represent their common tissue characteristics during active tissue remodeling.
3.5 There were differences in the regulatory genes involved in tissue formation between the DeP and CgP
To further explore tissue characteristics, tissues from the DeP, CgP, and ReP were prepared as tissue sections and subjected to H&E staining, as shown in Figure 5A. The mean DAPI fluorescence intensity was used to represent the live cell density in the tissue. The cell density in the DeP was significantly higher than that in the other two groups, which may be related to the threshold characteristics in early development (Figure 5B). During remodeling, periodontal cells can differentiate and participate in the formation of different tissues. Promoting the formation of bone, blood vessels, and nerves is an essential basis for functional periodontium regeneration (Figure 5C).
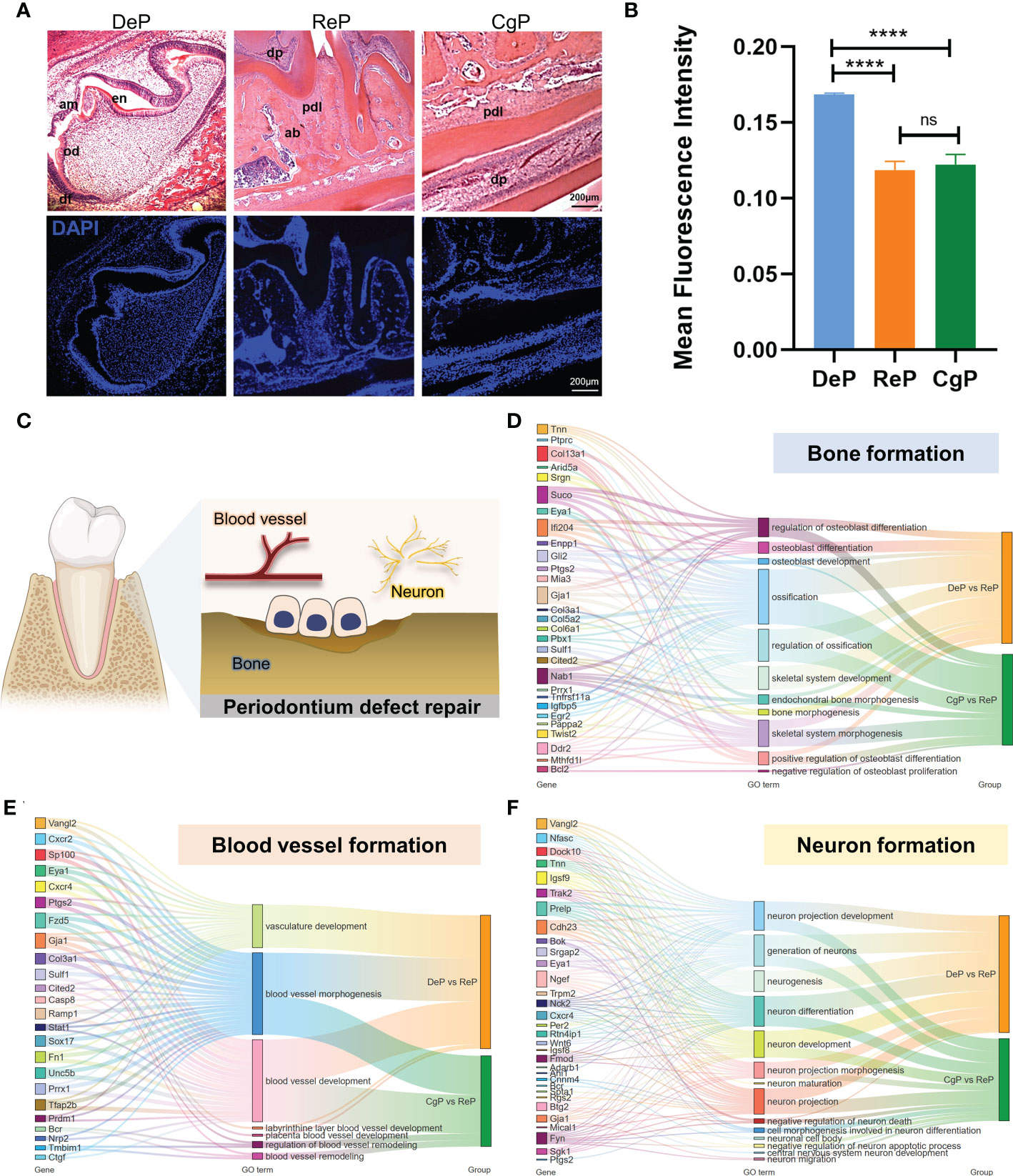
Figure 5 Comparison of the characteristics of tissue formation processes. (A) H&E staining and immunofluorescence staining of dental follicles and periodontium in mouse mandibular in sagittal sections. (B) The mean fluorescence intensity of DAPI was used to represent the cell density in the tissue. ****p < 0.0001. (C) Diagram of the formation of several vital tissues during periodontal remodeling. (D–F) Sankey maps are used to show critical genes and pathways involved in bone, blood vessel, and neuron formation. ns, no significance.
Since DeP and CgP have different characteristics in the tissue microenvironment, we summarized the significant genes (top 20 based on q value) involved in cell differentiation in these two tissues to better understand and apply their regulatory targets. As shown in Figure 5D, Suco, Ifi204, Gja1, Col13a1, and Nab1 were involved in the most extensive bone formation pathways compared with other genes and may be the core genes in the process of osteoblast differentiation and ossification in periodontal tissue. As for blood vessel formation, most genes showed enrichment in blood vessel development and morphogenesis. We noticed that only the DEGs of DeP versus ReP were enriched in the vasculature development pathway, while the blood vessel remodeling process was explicitly enriched in CgP versus ReP. Tmbim1 was a specific gene enriched during blood vessel remodeling (Figure 5E). In the formation of nerve tissue, the significant genes of the two differed. Only nine genes, Bok, Cdh23, Eya1, Igsf9, Ngef, Prelp, Trpm2, and Vangl2, were significantly different (Figure 5F). It can be seen that there were differences in the regulatory genes involved in tissue formation between the DeP and CgP.
3.6 Key regulatory signals of DeP and CgP tissues
To further explore the possible key regulatory signals during periodontal remodeling, the upstream regulators and potential associations of 795 and 675 DEGs in the two comparisons were analyzed. By applying the p-value of overlap < 0.05 threshold, 1637 upstream regulators were enriched in the DeP versus ReP, of which 65 regulators with a z-score > 2 were predicted to be activated and 173 with a z-score < −2 were predicted to be inhibited. Under the same threshold, a total of 1986 upstream regulators were enriched in the CgP versus ReP, of which 59 regulators were expected to be activated, and 135 regulators were predicted to be inhibited. The chemical drug SB203580 was the most potent activator common to both groups (Z-score = 3.798 in DeP versus ReP, and Z-score=3.405 in DeP versus ReP). Figure 6A shows its 29 target molecules enriched in the two groups of DEGs. The expression trends of other genes showed consistency between the two groups except for Skg1, Aim2, and Cd24a. In addition, poly I:rC-RNA (Z-score = −4.537, Figure 6B) and MYD88 (Z-score = −3.897, Figure 6C) were the most potent inhibitors in the DeP versus ReP and the DeP versus ReP, respectively, regulating the expression of 34 and 21 target molecules in periodontium remodeling.
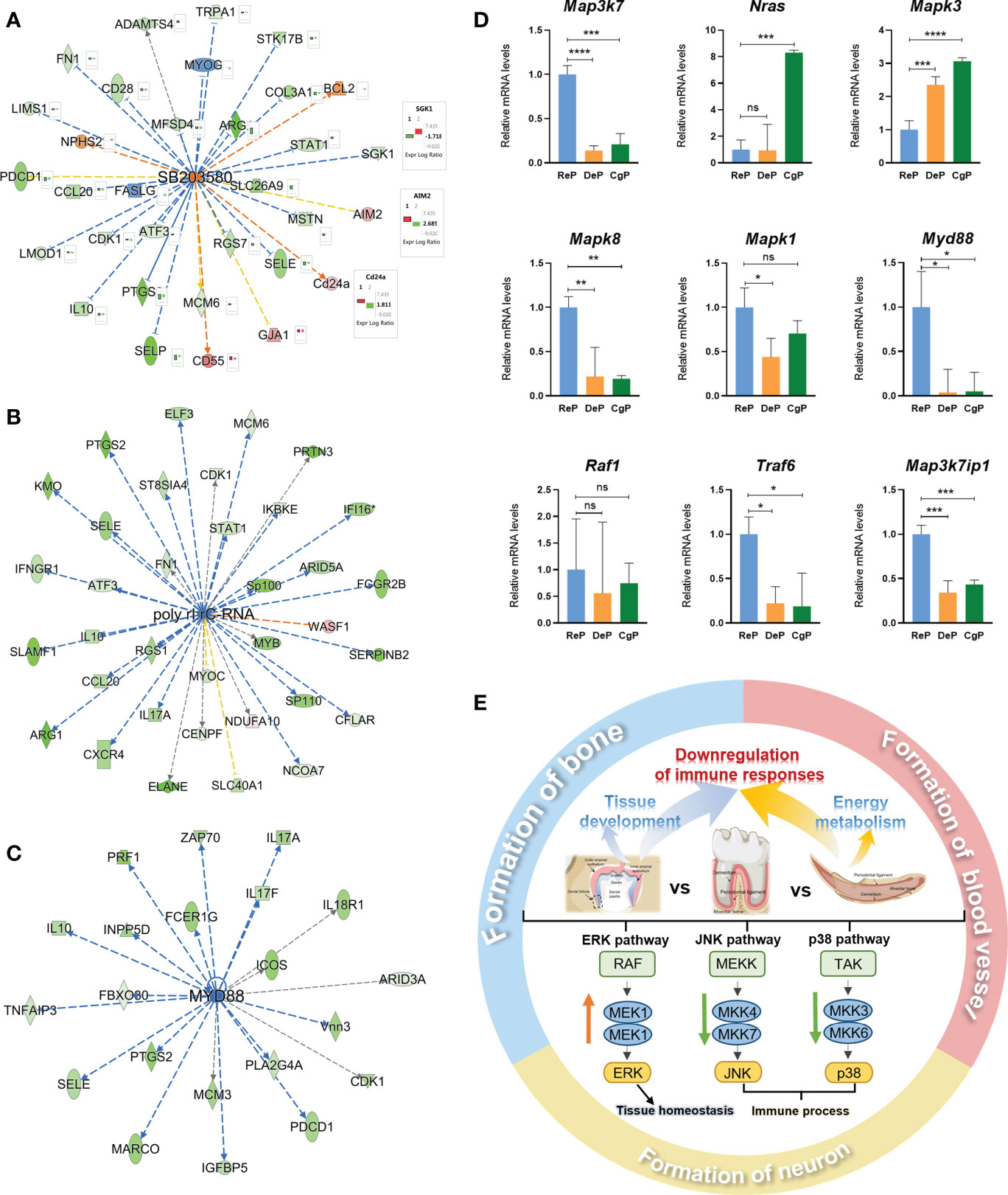
Figure 6 Prediction of upstream regulators and validation of target genes. (A–C) Network diagram representing the upstream regulators with highest and lowest z-scores. (D) mRNA expression of 9 representative genes was validated using RT-qPCR. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (E) Schematic diagram of active remodeling periodontal tissue characteristics. ns, no significance.
SB203580 is an inhibitor of the p38 MAPK pathway (15). Poly rI:rC-RNA effectively promotes the phosphorylation and activation of MAPK pathway molecules (16), while the adaptor protein encoded by Myd88 transduces Toll-like receptor signals and activates NF-κB and MAPK pathways (17). The activation and inhibition status of these molecules as upstream regulators strongly suggested that the MAPK pathway was inhibited in the DeP and ReP and may be closely related to the periodontium remodeling process. In total, nine genes involved in the MAPK pathway were selected for qPCR validation. The genes that were upregulated in the actively remodeling periodontium were Mapk3 and Nras, and the downregulated genes were Mapk8, Mapk1, Map3k7, Myd88, Raf1, Traf6, and Map3k7ip1 (Figure 6D).
4 Discussion
Exploring the mechanism of postnatal tissue development and adult regeneration is significant for understanding tissue regeneration. To improve clinical therapeutic outcomes of periodontium defects, it is necessary to understand the physiological process of periodontal remodeling in both the developmental and adult stages. In postnatal mice, the dental follicle structure forms the periodontal tissue, which regulates bone remodeling and tooth eruption. Unlike molars, mice incisors grow continuously throughout life, completing a renewal every 35–45 days (18). The mature mandibular incisor is attached to the alveolar bone only on the lingual side through the periodontal ligament tissue. Studies showed that collagen remodeling was more active on this site, making mice an ideal model for periodontal tissue formation and continuous remodeling. Here, we isolated tooth follicles and lingual periodontal tissues of the incisor, and compared them with periodontal tissue of the fully erupted molar to explore the tissue characteristics and critical mechanisms involved in the active remodeling process of periodontal tissues with different temporal and spatial distributions.
4.1 DEGs in the DeP/CgP versus ReP
Our study identified 792 and 612 DEGs in the DeP and CgP compared with the ReP. The Venn diagram results shown in Figure 2E suggested that gene regulation in the DeP and CgP was generally similar but had certain distinct features. These different molecular features emphasized that the remodeling activities occurring in developing and adult periodontium can inspire the repair and regeneration of periodontal tissues from different perspectives.
4.2 DeP and CgP showed different upregulated biological processes during periodontal remodeling
The pathway analysis suggested that upregulated DEGs in the DeP/CgP versus the ReP were enriched in various biological processes in developmental and adult stages during the active remodeling of the periodontium. The DeP showed enhanced activities related to tissue development. These results are highly consistent with previous studies showing that the periodontal tissue undergoes a developmental process from the beginning at the dental follicle stage (19). The dental follicle is a loose ectoderm-derived connective tissue surrounding the tooth germ that can differentiate into cementoblasts and secrete substances to form root cementum. At the same time, dental follicle cells secrete collagen fibers in the newly generated cementum to fix the tooth root in the alveolar fossa to form the root–bone interface and coordinate tooth eruption (20). At this stage, tooth morphogenesis is active.
Interestingly, unlike classical signals in tooth development, upregulated dental follicle DEGs in the KEGG analysis showed significant enrichment in the Hippo signaling pathway, as shown in Figure 3C. It has been confirmed that the critical effectors of the Hippo pathway, Yes-associated protein (YAP) and its homolog PDZ-binding motif transcriptional coactivator (TAZ), enhance the stability and nuclear translocation of β-catenin and participate in the regulation of osteogenic and adipogenic differentiation of oral stem cells (21). Multiple studies have shown that Hippo signaling regulates the proliferation, differentiation, apoptosis, and senescence of periodontal ligament stem cells and the differentiation and mineralization of cementoblasts through crosstalk with the ERK1/2 pathway, ROCK pathway, TGF-β/BMP pathway, Wnt/β-catenin pathway, and BCL-2 (22). Our results confirmed that the dental follicle provides the reference for the orderly generation of periodontal tissue from scratch, and Hippo signaling is an essential regulator of periodontium development.
Just as important, the processes involved in energy metabolism were the most prominent in the enrichment analysis of the upregulated DEGs in the CgP versus ReP, especially aerobic metabolic processes, such as oxidative phosphorylation and the TCA cycle, indicating that significantly different metabolic characteristics accompanied the active remodeling state in adult periodontal tissues. Indeed, in addition to growth factors and extracellular matrix signaling, there is increasing evidence that various metabolic pathways provide essential signals for the self-renewal and differentiation capacity of cells, thereby regulating the epigenome and influencing cell fate (23). The critical link of periodontal tissue remodeling is the synthesis stage of osteoblast differentiation, which requires a large amount of energy (24). The process of mitochondrial phosphate oxidation is an efficient way to generate energy. A dynamic proteomic analysis of human periodontal ligament stem cells during osteogenic differentiation showed that oxidative phosphorylation was the most critical regulatory pathway during differentiation (25), and oxygen tension significantly regulated the activity of alkaline phosphatase in periodontal ligament fibroblasts (26). The inhibition of osteoblast differentiation in the aged periodontal ligament may be directly related to the difficulty in completing cellular respiration (27). Our results suggested that metabolic status may represent the critical mechanism during the remodeling process of the adult periodontium, and promoting periodontal remodeling and osteogenic repair processes based on metabolism regulation may be a promising approach for periodontium regeneration.
4.3 Comparison of the tissue formation processes in developmental and adult stages
The aim of periodontal regeneration therapy includes the regeneration of hard and soft tissues. Through the gene analysis of bone, blood vessels, and neurons, we summarized the gene regulation paradigm, which can be introduced into animal models for more in-depth research to provide more clues to the biological events of periodontal reconstruction. Among the most significant DEGs, Suco, Ifi204, Gja1, Col13a1, and Nab1 most extensively played roles in bone formation-related biological processes. Among them, only Ifi204, the murine homolog of human IFI16, showed consistent inhibition of expression in the DeP and CgP, which regulates the release of IL-1β in tissues and plays a vital role in bone loss in experimental periodontitis. Suco, Gja1, Col13a1, and Nab1 were significantly upregulated. These genes have been widely studied in the regulation of osteoblast differentiation and bone remodeling (28–30), but their roles in regulating periodontal bone health are rarely reported. The concordant results between the DeP and CgP suggested the expression paradigm of these critical genes during alveolar bone remodeling. The combinatorial regulation of these essential genes to promote alveolar bone regeneration is worth exploring.
During the biological process of blood vessel formation and remodeling, although active blood vessel development occurs in both tissues, we noticed CgP-specific enrichment in the process of blood vessel remodeling. This process is different from vasculogenesis, in which angiogenic progenitor cells migrate to sites of vascularization, differentiate into endothelial cells, and coalesce to form the initial vascular plexus, and instead is a remodeling process of existing blood vessels (31). Blood vessel remodeling in the CgP may be related to adapting to the characteristics of the tissue microenvironment, such as precisely regulating O2 delivery to maintain oxygen homeostasis and meeting the energy metabolism demand in the periodontal tissue (32). Therefore, it is suggested that attention to the characteristics of the microenvironment of the erupted tooth tissue, such as the microstructural changes of blood vessels caused by chewing force, is of great significance to understanding the biological mechanism of blood vessel formation and remodeling to promote good blood circulation during periodontal regeneration. As there is a complex neural network in periodontal tissue, we also analyzed the nerve formation process. The local sensory and sympathetic nerves play an essential role in developing the dental follicle, maintaining periodontal health, and promoting local disease recovery (33). The abundant enrichment of related genes, shown in Figure 5F, suggests that different molecular guidance cues may exist for innervation in developing and mature periodontal tissues.
4.4 Characteristics of the immune microenvironment in the DeP and CgP
In terms of commonalities, the tissue characteristics of the DeP and CgP were surprisingly highly correlated with the immune system. In both of them, the activation, migration, and recruitment of immune cells (such as lymphocytes), the production of inflammatory factors (such as TNF and IL17), and the signaling of immune cells (such as the T cell receptor signaling pathway) were inhibited relative to the ReP. This result suggested that unique immune cell activities and cytokine levels may contribute to the characteristics of actively remodeling periodontium. There may be different reasons for this characteristic. First, there is consideration of the unique environment of periodontal tissue and its proximity to oral plaque biofilms. The oral microbiota induces many immune cells to generate periodontal immune responses due to the highly permeable junctional epithelial attachments on the tooth surface (34). Therefore, microbial biofilms in healthy individuals can significantly mobilize the periodontal immune microenvironment and mediate the maintenance of periodontal homeostasis (35). However, the dental follicle structure of the unerupted tooth and the periodontal tissue of the middle part of the incisor is relatively distant from the oral microbial community and show relatively silent immune activities. Second, there may have been an immunosuppressed population of cells in both tissues. Studies have shown that cytokines secreted by dental follicle cells inhibited lymphocyte proliferation (36), and osteoprotegerin secreted by dental follicle cells inhibited monocyte recruitment and osteoclast formation (37). Despite the need to form an osteoclastic resorption channel during tooth germ development and eruption, this seems consistent given the simultaneous alveolar bone formation.
At the same time, stem cell niches exist in the root of the mouse incisor as a cell population that inhibits inflammation and immune regulation, which may regulate the immune activity of the periodontal tissue of the incisor (38). Our results suggest that the actively remodeling periodontium has unique immune system characteristics in both developmental and adult stages. The tissue microenvironment characteristics may regulate and, in turn, coordinate the tissue remodeling process.
4.5 Possible regulatory role of MAPK pathway in periodontal remodeling
Further analysis of upstream regulators predicted that SB203580, poly rI:rC-RNA, and Myd88 were identified as potent regulators that might have significant regulatory effects on gene expression in actively remodeling periodontium. Unexpectedly, all three molecules individually suggested an inhibitory state of the mitogen-activated protein kinase (MAPK) pathway. MAPK is a highly conserved serine/threonine protein kinase in eukaryotes. It starts from the activation of upstream MAP kinase kinase kinase kinase (MKKK) (39). Eventually, it leads to the cascade activation of the pathway by targeting a series of downstream substrate proteins for phosphorylation. Three MAPK families have been characterized: the classical MAPK (also known as ERK), C-Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK), and p38 MAPK. As the MAPK signal transduction cascade plays a critical regulatory role in cell proliferation, differentiation, development, inflammatory response, and apoptosis (40), here we speculated that regulation of the MAPK pathway might be an essential factor in the microenvironment characteristics of the DeP and CgP and further modulates the cellular behavior of the remodeling process. We next selected genes of interest in the pathway for qPCR validation. The genes (Mapk3 and Nras) involved in the ERK pathway showed upregulated levels in DeP and CgP, while the genes involved in the JNK and p38 MAPK pathways (Mapk8, Mapk1, Map3k7, Myd88, Raf1, Traf6, and Map3k7ip1) were downregulated.
The expression trends of these genes showed consistency with previous studies. It has been proven that different extracellular stimuli can preferentially activate different MAP kinases, thus producing corresponding effects. Growth factors (41) and G protein-coupled receptors (42) can activate the ERK pathway and regulate cell proliferation and differentiation. Active tissue formation occurs in DeP and CgP, which may be related to the activation of the ERK pathway in response to growth factors. Meanwhile, inflammatory cytokines (such as IL-1 and TNF-α) and many cellular stress-inducing factors preferentially activate JNK/SAPK and p38 MAPK (43). Pathogen stimulation in oral biofilms is essential in activating the p38 MAPK pathway in periodontal tissue (44). The unique anatomical features of DeP and CgP reduce the activation of this pathway by inflammatory cytokines and may further regulate the characteristics of the immune microenvironment of these two groups. However, the activation of pathways by stimuli is not one or the other, and studies have demonstrated that the MAPK signaling pathway plays a complex regulatory role in periodontal regeneration. The activation of the ERK/p38 MAPK pathway is involved in the process of osteogenic differentiation of periodontal ligament cells stimulated by asarylaldehyde, but the JNK pathway is not activated (45).
On the other hand, the same pathway may play opposite roles during cell differentiation under different microenvironment signals. Pharmacological antagonism of the p38 MAPK pathway can effectively inhibit the tissue destruction process of periodontitis and promote bone regeneration (46, 47). In a word, as the MAPK pathway can regulate the regeneration of periodontium, the exploration of its key targets and corresponding stimulating factors is of great significance. Combining upstream regulators with gene expression levels, the MyD88/p38 MAPK pathway may be inhibited in the DeP and CgP, and the levels of other MAPK-related pathways in actively remodeling periodontium were altered varied according to the functional differences of each pathway.
Figure 6E shows the schematic diagram of actively remodeling periodontal tissue characteristics. The developmental periodontal tissue was enhanced in processes related to tissue development, and the enhanced energy metabolism is characteristic of the adult actively remodeling periodontium. Both stages showed a decrease in the immune response. The MAPK pathway may play an essential regulatory role in microenvironment features and be involved in tissue formation.
5 Conclusion
The developmental periodontium and continuously growing periodontium, which are actively remodeled, have shared and unique differences in microenvironmental tissue characteristics and molecular regulatory mechanisms, such as the upregulation of tissue development and metabolic state and common inhibition of the immune system. The MAPK pathway (especially MyD88/p38 MAPK) may be a promising therapeutic target to promote periodontal remodeling. This study provides a deep physiological understanding for promoting the orderly regeneration of the complex structure of periodontal tissue by considering the characteristics of tissue and microenvironment and providing a basis for selecting targets for regenerative therapy.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA928506.
Ethics statement
The animal study was reviewed and approved by the Laboratory Animal Ethics Committee in Ninth People’s Hospital Affiliated to Shanghai Jiao Tong University School of Medicine.
Author contributions
YD and NL analyzed the data and drafted the manuscript. MX performed the dissection and processing of the tissue samples. LH helped the conception of this study. RJ and NH reviewed and revised the manuscript. JW and XJ contributed to the concept and design of the study. All authors contributed to the article and approved the submitted version.
Funding
The work is supported by The National Natural Science Foundation of China (No. 81900970, No. 82130027), Clinical Research Plan of SHDC (SHDC2020CR2042B), the Young Physician Innovation Team Project (No. QC202003) of Ninth People’s Hospital affiliated to Shanghai Jiao Tong University School of Medicine, and Shanghai "Rising Stars of Medical Talent" Youth Development Program.
Acknowledgments
We thank Wenjing Ding of the Library, Shanghai Jiao Tong University School of Medicine, China, for assistance with the IPA system in preparing this manuscript.
Conflict of interest
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1154931/full#supplementary-material
References
1. Xu XY, Li X, Wang J, He XT, Sun HH, Chen FM. Concise review: Periodontal tissue regeneration using stem cells: Strategies and translational considerations. Stem Cells Transl Med (2019) 8(4):392–403. doi: 10.1002/sctm.18-0181
2. Park CH, Kim KH, Lee YM, Seol YJ. Advanced engineering strategies for periodontal complex regeneration. Materials (Basel) (2016) 9(1):57. doi: 10.3390/ma9010057
3. Bosshardt DD, Stadlinger B, Terheyden H. Cell-to-Cell communication–periodontal regeneration. Clin Oral Implants Res (2015) 26(3):229–39. doi: 10.1111/clr.12543
4. Bostanci N. Revisiting "-omics" in oral health and disease. Proteomics Clin Appl (2020) 14(3):e1900022. doi: 10.1002/prca.201900022
5. Nagata M, Chu AKY, Ono N, Welch JD, Ono W. Single-cell transcriptomic analysis reveals developmental relationships and specific markers of mouse periodontium cellular subsets. Front Dent Med (2021) 2:679937. doi: 10.3389/fdmed.2021.679937
6. Spitz A, Christovam IO, Maranon-Vasquez GA, Masterson DF, Adesse D, Maia LC, et al. Global gene expression profile of periodontal ligament cells submitted to mechanical loading: A systematic review. Arch Oral Biol (2020) 118:104884. doi: 10.1016/j.archoralbio.2020.104884
7. Saito MT, Mofatto LS, Albiero ML, Casati MZ, Sallum EA, Nociti Junior FH, et al. Transcriptome profile of highly Osteoblastic/Cementoblastic periodontal ligament cell clones. J Appl Oral Sci (2020) 28:e20200242. doi: 10.1590/1678-7757-2020-0242
8. Grover HS, Kapoor S, Saksena N. Periodontal proteomics: Wonders never cease! Int J Proteomics (2013) 2013:850235. doi: 10.1155/2013/850235
9. Di Stefano M, Polizzi A, Santonocito S, Romano A, Lombardi T, Isola G. Impact of oral microbiome in periodontal health and periodontitis: A critical review on prevention and treatment. Int J Mol Sci (2022) 23(9):5142. doi: 10.3390/ijms23095142
10. Brockes JP, Kumar A. Comparative aspects of animal regeneration. Annu Rev Cell Dev Biol (2008) 24:525–49. doi: 10.1146/annurev.cellbio.24.110707.175336
11. Phan QM, Fine GM, Salz L, Herrera GG, Wildman B, Driskell IM, et al. Lef1 expression in fibroblasts maintains developmental potential in adult skin to regenerate wounds. Elife (2020) 9:e60066. doi: 10.7554/eLife.60066
12. Rinkevich Y, Montoro DT, Contreras-Trujillo H, Harari-Steinberg O, Newman AM, Tsai JM, et al. In vivo clonal analysis reveals lineage-restricted progenitor characteristics in mammalian kidney development, maintenance, and regeneration. Cell Rep (2014) 7(4):1270–83. doi: 10.1016/j.celrep.2014.04.018
13. Zhou T, Pan J, Wu P, Huang R, Du W, Zhou Y, et al. Dental follicle cells: Roles in development and beyond. Stem Cells Int (2019) 2019:9159605. doi: 10.1155/2019/9159605
14. Suzuki H, Amizuka N, Kii I, Kawano Y, Nozawa-Inoue K, Suzuki A, et al. Immunohistochemical localization of periostin in tooth and its surrounding tissues in mouse mandibles during development. Anat Rec A Discov Mol Cell Evol Biol (2004) 281(2):1264–75. doi: 10.1002/ar.a.20080
15. Cuenda A, Rouse J, Doza YN, Meier R, Cohen P, Gallagher TF, et al. Sb 203580 is a specific inhibitor of a map kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett (1995) 364(2):229–33. doi: 10.1016/0014-5793(95)00357-f
16. De Waele J, Verhezen T, van der Heijden S, Berneman ZN, Peeters M, Lardon F, et al. A systematic review on Poly(I:C) and poly-iclc in glioblastoma: Adjuvants coordinating the unlocking of immunotherapy. J Exp Clin Cancer Res (2021) 40(1):213. doi: 10.1186/s13046-021-02017-2
17. Zhou M, Xu W, Wang J, Yan J, Shi Y, Zhang C, et al. Boosting mtor-dependent autophagy Via upstream Tlr4-Myd88-Mapk signalling and downstream nf-kappab pathway quenches intestinal inflammation and oxidative stress injury. EBioMedicine (2018) 35:345–60. doi: 10.1016/j.ebiom.2018.08.035
19. Bastos VC, Gomez RS, Gomes CC. Revisiting the human dental follicle: From tooth development to its association with unerupted or impacted teeth and pathological changes. Dev Dyn (2022) 251(3):408–23. doi: 10.1002/dvdy.406
20. Li J, Parada C, Chai Y. Cellular and molecular mechanisms of tooth root development. Development (2017) 144(3):374–84. doi: 10.1242/dev.137216
21. Pandya M, Gopinathan G, Tillberg C, Wang J, Luan X, Diekwisch TGH. The hippo pathway effectors Yap/Taz are essential for mineralized tissue homeostasis in the alveolar Bone/Periodontal complex. J Dev Biol (2022) 10(1):14. doi: 10.3390/jdb10010014
22. Wang T, Li K, Liu H, Luo E. Focusing on hippo pathway in stem cells of oral origin, enamel formation and periodontium regeneration. Organogenesis (2022) 18(1):2082236. doi: 10.1080/15476278.2022.2082236
23. Hu C, Fan L, Cen P, Chen E, Jiang Z, Li L. Energy metabolism plays a critical role in stem cell maintenance and differentiation. Int J Mol Sci (2016) 17(2):253. doi: 10.3390/ijms17020253
24. Dirckx N, Moorer MC, Clemens TL, Riddle RC. The role of osteoblasts in energy homeostasis. Nat Rev Endocrinol (2019) 15(11):651–65. doi: 10.1038/s41574-019-0246-y
25. Li J, Wang Z, Huang X, Wang Z, Chen Z, Wang R, et al. Dynamic proteomic profiling of human periodontal ligament stem cells during osteogenic differentiation. Stem Cell Res Ther (2021) 12(1):98. doi: 10.1186/s13287-020-02123-6
26. Amemiya H, Matsuzaka K, Kokubu E, Ohta S, Inoue T. Cellular responses of rat periodontal ligament cells under hypoxia and re-oxygenation conditions in vitro. J Periodontal Res (2008) 43(3):322–7. doi: 10.1111/j.1600-0765.2007.01032.x
27. Huttner EA, Machado DC, de Oliveira RB, Antunes AGF, Hebling E. Effects of human aging on periodontal tissues. Spec Care Dentist (2009) 29(4):149–55. doi: 10.1111/j.1754-4505.2009.00082.x
28. Moorer MC, Hebert C, Tomlinson RE, Iyer SR, Chason M, Stains JP. Defective signaling, osteoblastogenesis and bone remodeling in a mouse model of connexin 43 c-terminal truncation. J Cell Sci (2017) 130(3):531–40. doi: 10.1242/jcs.197285
29. Kemppainen AV, Finnila MA, Heikkinen A, Haronen H, Izzi V, Kauppinen S, et al. The Cms19 disease model specifies a pivotal role for collagen xiii in bone homeostasis. Sci Rep (2022) 12(1):5866. doi: 10.1038/s41598-022-09653-4
30. Ali M, Yang F, Plachokova AS, Jansen JA, Walboomers XF. Application of specialized pro-resolving mediators in periodontitis and peri-implantitis: A review. Eur J Oral Sci (2021) 129(1):e12759. doi: 10.1111/eos.12759
31. Semenza GL. Vasculogenesis, angiogenesis, and arteriogenesis: Mechanisms of blood vessel formation and remodeling. J Cell Biochem (2007) 102(4):840–7. doi: 10.1002/jcb.21523
32. Rademakers T, Horvath JM, van Blitterswijk CA, LaPointe VLS. Oxygen and nutrient delivery in tissue engineering: Approaches to graft vascularization. J Tissue Eng Regener Med (2019) 13(10):1815–29. doi: 10.1002/term.2932
33. Duan Y, Liang Y, Yang F, Ma Y. Neural regulations in tooth development and tooth-periodontium complex homeostasis: A literature review. Int J Mol Sci (2022) 23(22):14150. doi: 10.3390/ijms232214150
34. Hathaway-Schrader JD, Novince CM. Maintaining homeostatic control of periodontal bone tissue. Periodontol 2000 (2021) 86(1):157–87. doi: 10.1111/prd.12368
35. Hathaway-Schrader JD, Aartun JD, Poulides NA, Kuhn MB, McCormick BE, Chew ME, et al. Commensal oral microbiota induces osteoimmunomodulatory effects separate from systemic microbiome in mice. JCI Insight (2022) 7(4):e140738. doi: 10.1172/jci.insight.140738
36. Ulusoy C, Zibandeh N, Yildirim S, Trakas N, Zisimopoulou P, Kucukerden M, et al. Dental follicle mesenchymal stem cell administration ameliorates muscle weakness in musk-immunized mice. J Neuroinflamm (2015) 12:231. doi: 10.1186/s12974-015-0451-0
37. Zeng L, He H, Sun M, Gong X, Zhou M, Hong Y, et al. Runx2 and Nell-1 in dental follicle progenitor cells regulate bone remodeling and tooth eruption. Stem Cell Res Ther (2022) 13(1):486. doi: 10.1186/s13287-022-03140-3
38. Krivanek J, Soldatov RA, Kastriti ME, Chontorotzea T, Herdina AN, Petersen J, et al. Dental cell type atlas reveals stem and differentiated cell types in mouse and human teeth. Nat Commun (2020) 11(1):4816. doi: 10.1038/s41467-020-18512-7
39. Yuan J, Dong X, Yap J, Hu J. The mapk and ampk signalings: Interplay and implication in targeted cancer therapy. J Hematol Oncol (2020) 13(1):113. doi: 10.1186/s13045-020-00949-4
40. Zhang W, Liu HT. Mapk signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res (2002) 12(1):9–18. doi: 10.1038/sj.cr.7290105
41. Santos SD, Verveer PJ, Bastiaens PI. Growth factor-induced mapk network topology shapes erk response determining pc-12 cell fate. Nat Cell Biol (2007) 9(3):324–30. doi: 10.1038/ncb1543
42. Xiang YK. Arrestin orchestrates crosstalk between G protein-coupled receptors to modulate the spatiotemporal activation of erk mapk. FASEB J (2010) 24:79–88. doi: 10.1096/fasebj.24.1_supplement.lb478
43. Pua LJW, Mai CW, Chung FF, Khoo AS, Leong CO, Lim WM, et al. Functional roles of jnk and P38 mapk signaling in nasopharyngeal carcinoma. Int J Mol Sci (2022) 23(3):1108. doi: 10.3390/ijms23031108
44. Li Q, Valerio MS, Kirkwood KL. Mapk usage in periodontal disease progression. J Signal Transduct (2012) 2012:308943. doi: 10.1155/2012/308943
45. Hwang JW, Park WJ, Han Y. Asarylaldehyde enhances osteogenic differentiation of human periodontal ligament stem cells through the Erk/P38 mapk signaling pathway. Biochem Bioph Res Co (2021) 545:27–32. doi: 10.1016/j.bbrc.2021.01.053
46. Kirkwood KL, Rossa C Jr. The potential of P38 mapk inhibitors to modulate periodontal infections. Curr Drug Metab (2009) 10(1):55–67. doi: 10.2174/138920009787048347
Keywords: periodontium, bioinformatics, regeneration, signal, tissue microenvironment
Citation: Deng Y, Luo N, Xie M, He L, Jiang R, Hu N, Wen J and Jiang X (2023) Transcriptome landscape comparison of periodontium in developmental and renewal stages. Front. Endocrinol. 14:1154931. doi: 10.3389/fendo.2023.1154931
Received: 31 January 2023; Accepted: 22 February 2023;
Published: 15 March 2023.
Edited by:
Bingdong Sui, Air Force Medical University, ChinaReviewed by:
Xiaoxing Kou, Sun Yat-sen University, ChinaRan Zhang, Peking University Hospital of Stomatology, China
Copyright © 2023 Deng, Luo, Xie, He, Jiang, Hu, Wen and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinquan Jiang, eGlucXVhbmppYW5nQGFsaXl1bi5jb20=; Jin Wen, ZWNob21ldEAxMjYuY29t
†These authors have contributed equally to this work
 Yuwei Deng
Yuwei Deng Nan Luo1,3†
Nan Luo1,3† Ruixue Jiang
Ruixue Jiang Jin Wen
Jin Wen Xinquan Jiang
Xinquan Jiang