- 1Department of Human Genetics, Amsterdam Movement Sciences, Tissue Function & Regeneration and Musculoskeletal Health, Amsterdam University Medical Centers (UMC) location Vrije Universiteit Amsterdam, Amsterdam, Netherlands
- 2Department of Traumatology and Orthopeadics, Institute of Clinical Medicine, The University of Tartu, Tartu, Estonia
- 3Department of Clinical Chemistry, Amsterdam Movement Sciences, Tissue Function & Regeneration and Ageing & Vitality, Amsterdam University Medical Centers (UMC) location Vrije Universiteit Amsterdam, Amsterdam, Netherlands
- 4Department of Endocrinology and Metabolism, Amsterdam Rare Bone Disease Center, Amsterdam University Medical Centers (UMC), Amsterdam, Netherlands
Introduction: Osteogenesis Imperfecta is a rare genetic connective tissue disorder, characterized by skeletal dysplasia and fragile bones. Currently only two mouse models have been reported for haploinsufficient (HI) mild Osteogenesis Imperfecta (OI); the Col1a1+/Mov13 (Mov13) and the Col1a1+/-365 mouse model. The Mov13 mice were created by random insertion of the Mouse Moloney leukemia virus in the first intron of the Col1a1 gene, preventing the initiation of transcription. Since the development of the Mov13 mice almost four decades ago and its basic phenotypic characterization in the 90s, there have not been many further studies. We aimed to extensively characterize the Mov13 mouse model in order to critically evaluate its possible use for preclinical studies of HI OI.
Methods: Bone tissue from ten heterozygous Mov13 and ten wild-type littermates (WT) C57BL/6J mice (50% males per group) was analyzed at eight weeks of age with bone histomorphometry, micro computed tomography (microCT), 3-point bending, gene expression of different collagens, as well as serum markers of bone turnover
Results: The Mov13 mouse presented a lower bone strength and impaired material properties based on our results of 3-point bending and microCT analysis respectively. In contrast, no significant differences were found for all histomorphometric parameters. In addition, no significant differences in Col1a1 bone expression were present, but there was a significant lower P1NP concentration, a bone formation marker, measured in serum. Furthermore, bone tissue of Mov13 mice presented significantly higher expression of collagens (Col1a2, Col5a1 and Col5a2), and bone metabolism markers (Bglap, Fgf23, Smad7, Edn1 and Eln) compared to WT. Finally, we measured a significantly lower Col1a1 expression in heart and skin tissue and also determined a higher expression of other collagens in the heart tissue.
Conclusion: Although we did not detect a significant reduction in Col1a1 expression in the bone tissue, a change in bone structure and reduction in bone strength was noted. Regrettably, the variability of the bone phenotype and the appearance of severe lymphoma in adult Mov13 mice, does not favor their use for the testing of new long-term drug studies. As such, a new HI OI type 1 mouse model is urgently needed.
1 Introduction
Osteogenesis Imperfecta (OI), also known as brittle bone disease, is a rare connective tissue disorder affecting 1 in 15-20,000 births (1). It is mainly characterized by skeletal fragility and dysplasia, substantial growth deficiency, blue sclerae and early-onset hearing loss. OI can present multiple genetic causes, most of which are directly related to collagen type I synthesis and regulation. Approximately 85 to 90% of the patients have autosomal dominant OI caused by pathogenic variants α1(I) collagen gene (COL1A1) and α1(II) collagen gene (COL1A2) (2). These genes code for the α1(I) and α2(I) chains of collagen type I, which is the most abundant protein of bone, skin and tendon extracellular matrices. Other genetic causes include pathogenic variants in genes coding for proteins that facilitate collagen folding or post-translational modification and other aspects of bone metabolism (3).
Collagen type I defects can be divided into two groups, one showing less collagen type I production (haploinsufficiency (HI)) and the other showing abnormal collagen type I structure (dominant negative (DN)). HI mainly causes mild OI whereas the DN defects mostly cause severe OI forms, which highlights the necessity for distinct mouse models for their study. At the moment, multiple mouse models are in existence for OI, but only two are available for HI in mild OI (Sillence type 1), which represents the majority of the patients (4–6). The Mov13 model was developed by Jaenish et al. in 1983 and characterized by Bonadio et al. and Jepsen et al. (7–13) It was created by microinjection of the Moloney murine leukemia virus (MMuLV) at midgestation of C57BL/6 mice (7). The virus integrated in the first intron, 19 bp away from the splice donor site of the Col1a1 (9). The insertion prevents initiation of Col1a1 transcription by causing a change in the chromatin conformation and de novo methylation of the gene (12). In the case of homozygous embryos this leads to a lethal developmental arrest between day 11 and 12 of gestation, while heterozygous embryos serve as a model for mild HI OI (9, 10). Heterozygous Mov13 mice are reported to show around 50% reduction in type I collagen in non-mineralized connective tissue and display reduced mechanical and material properties in the long bones (10, 14). In addition, the mice also present progressive hearing loss, resembling the human phenotype (10, 11).
As the bone phenotype is the most pronounced feature in OI, Mov13 mouse studies have been mostly focused around long bone mechanical and histological properties (10, 11, 15). The brittleness of the bones has been explained by altered microstructure, reduced ductility and increase in porosity (11). Studies in skin, revealed approximately 50% reduction in collagen type I and thin collagen fibers (10, 16). Aside from the bone and skin, limited research has been performed on other collagen type I-rich tissues presenting the OI phenotype. Otopathology analyses determined hearing loss which appears to be connected with the cochlear hair cell loss in relation to bony changes (10, 17). In addition, Stankovic et al. confirmed a 50% reduction in Col1a1 expression in the otic capsule (18). Derwin et al. studied the mouse tail tendon fascicle, which revealed reduced fibril size but no change in tissue mechanics (19, 20). A histological study of homozygous Mov13 embryos revealed that the rupture of major blood vessels between day 12 and 14 during the gestational was the cause of their lethality, due to the lack of collagen type I in combination with increased blood pressure at that age (21). Though observed in patients with OI type 1, dentinogenesis imperfecta and impaired stability of the aorta and the cardiac valves have not been well characterized in Mov13 mice. Concerning dentinogenesis, tooth germ cells taken from homozygous Mov13 embryos were capable of differentiating into all dental tissues and odontoblasts produced a normal layer of dentin (22). Moreover, Kratochwil et al. failed to identify a transcriptional block in the odontoblasts of Mov13 mice which were determined to produce normal amount of collagen type I (22).
It has been estimated that Sillence type 1 OI represents ~ 46–71% of the whole OI population; in approximately 40% of type 1 patients OI is caused by COL1A1 HI (23, 24). Even though the disease in HI patients is milder regarding skeletal deformities, these patients can still experience a vast number of fractures. A cohort of 86 young HI OI patients showed a 95-fold increase in long-bone fracture risk (24). Another study of 364 Italian OI patients showed that regarding the HI OI type 1 patients ~48% showed osteopenia and ~38% had osteoporosis. Also, 28% presented deafness and in 24% cardiac defects were recorded (25). Thus, considering the high number of these patients, the impact of OI on their quality of life and lack of therapy, the existence of a reliable mouse model is paramount to adequately study the underlying pathology of HI OI. Based on the above, it is evident that phenotyping studies of Mov13 mice are mostly limited to bone and skin tissue. Replicability of these results and further investigation of the phenotype is required with extended methodological approaches to be able to use this model in future studies on new therapies for HI OI type 1. In an effort to address this need, we performed an in-depth characterization of the bone phenotype of Mov13 mice regarding their histomorphological and gene expression properties in order to provide a reference for the suitability of this model for testing of new therapies and long-term drug studies.
2 Materials and methods
2.1 Animal & ethics claim
Male and female mice, heterozygous for the insertion of MMuLV in the Col1a1 locus, were purchased from Jackson Laboratories (USA) and mated to obtain ten heterozygous Mov13 mice (50% males) and ten wild-type (WT) (C57BL/6J) littermates (50% males). All animal experiments were approved by the Central Committee of Animal experiments (CCD) of the Netherlands and in agreement with the Animal Welfare Body of the Amsterdam UMC in full compliance with the directive 2010/63/EU.
2.2 Tissue preparation for histochemistry, microCT and 3-point bending
Four-week-old mice were weighted twice a week for four weeks and sacrificed at eight weeks of age by CO2-induced hypoxia. Blood serum, humeri, femora, tibiae, the ventricles of the heart and abdominal skin tissue were obtained immediately after.
Blood was collected by heart puncture. Serum was acquired by centrifuging the blood at 4°C twice for 10min at 1000g and stored at -20°C. Both humeri and the right femur were centrifuged, after being cleaned of soft tissue, to remove bone marrow, before storing in RNAlater™-ICE solution (Thermo Fisher Scientific, Waltham, MA, US) at -20°C for gene expression measurements. Immediately after extraction, both heart and skin tissue were also stored in RNAlater™-ICE solution at -20°C.
The cleaned left femora were stored at 4°C in a PBS-soaked gauze before micro computed tomography (microCT) analysis and consecutive 3-point bending test. The cleaned, undecalcified left tibiae were stored in 70% ethanol and processed for methylmethacrylate (MMA) embedding. MMA (BDH Chemicals, Poole, UK) was supplemented with 20% dibutyl phtalate (Merck, Darmstadt, Germany), 8.0 g/l dibenzoyl peroxide (AKZO Nobel, Deventer, The Netherlands), and 22 ml/10 ml N,N-dimethyl-p-toluidine (Merck). Sections of 5µm thickness were cut with a Leica Polycut SM 2500S microtome (Leica Microsystems, Wetzlar, Germany) and mounted onto gelatin-coated glass slides (VWR, Super premium microscope Slides).
2.3 Histomorphometry analysis
Left tibia MMA-embedded sections were stained following standard protocols for Masson-Goldner trichrome, Von Kossa and tartrate-resistant acid phosphatase (TRAP) staining for analysis of bone microarchitecture and turnover, and Safranin O staining for analysis of cartilage. Bright field images were captured using a microscope Nikon eclipse E800 (Nikon Instruments Inc., Melville, NY, US) and camera Nikon digital sight DS-5Mc (Nikon Instruments, Inc., Melville, NY, US) for Safranin O staining and Olympus SLIDEVIEWTM VS200 Slide Scanner (Olympus Corp., Tokyo, Japan) for Masson-Goldner trichrome, Von Kossa and TRAP staining. Images were analyzed using the NIS-Elements Imaging Software, version AR 4.10.01 (Nikon Instruments Inc., Melville, NY, US).
Sections stained for Safranin O (n=2 sections per mouse (eight WT and ten Mov13)) were measured for the length, thickness, column index and percentage of cells in columns. Images were captured at 10x10 magnification. The length of the growth plate was measured by following the middle of the growth plate (light green line on Supplemental Figure 1). The thickness was measured in five different places perpendicular to the top of the growth plate. The three different zones (resting zone (RZ, dark green, Supplemental Figure 1), proliferative zone (PZ, dark blue, Supplemental Figure 1) and hypertrophic zone (HZ, light blue, Supplemental Figure 1) were similarly measured at the same location within the growth plate. The ratio of the different regions was presented as a percentage of the total thickness of the growth plate. A column was defined as: 1) Being composed of a minimum of 3 cells, 2) the angle between each cell in the column must range from -155 to -179° or 155 to 180° (blue and green arrow on Supplemental Figure 1) and 3) the cells cannot be more than 20 pixels apart. The length of the column was measured by using the distance between each cell starting from the center of the cell (yellow line on Supplemental Figure 1). The percentage of cells in columns was counted manually by dividing the number of cells counted in the columns (green triangles on Supplemental Figure 1) by the total amount of cells counted (cells in column + cells outside column (yellow dots) in Supplemental Figure 1). Afterwards the column index was calculated by multiplying the percentage of cells in columns with the average column length measured (26).
Masson-Goldner trichrome, TRAP and Von Kossa analyses were performed (n=2 sections per mouse (ten WT and ten Mov13 mice)). For Goldner trichrome and TRAP-stained slides 10x objective magnification was used, and for Von Kossa-stained slides 20x objective magnification was used. Primary measured parameters for Goldner trichrome included cortical width (Ct.Wi), tissue area (T.Ar), bone area (B.Ar), bone perimeter (B.Pm), osteocyte number (N.Ot); osteoclast number (N.Oc), osteoclast perimeter (Oc.Pm) for TRAP; for Von Kossa this included osteoid perimeter (O.Pm) and osteoid width (O.Wi). Cortical width (Ct.Wi) were measured four times for both sites of each section following a spacing grid of 200µm, at least 200-400µm distance from the growth plate. The trabecular bone was assessed in a defined area between 200-400µm from the growth plate, and half of Ct.Wi from the cortical bone, in order to exclude primary spongiosa and endocortical bone. All measurements and analysis were performed according to the American Society for Bone and Mineral Research (ASBMR) nomenclature committee (27).
2.4 Micro computed tomography
The left femora were scanned using the Scanco Medical AG microCT scanner (MicroCT42, version 6.1). Scans were automatically reconstructed in 2D slices and were analyzed using the Skyscan CT-analyzer (version 6.6). A trabecular section of 2.37mm (395 slices) of the distal left femur and a cortical section of 1.11mm (185 slices) of the midshaft were scanned. Femora were placed vertically in a 11.5mm scanner holder and scanned with the following settings: 10µm voxel size, 55kVp and 145μA, with an integration time of 200ms. Three-dimensional reconstruction and analysis were performed with CTan software (Bruker, Billerica, MA, USA, version 1.13.11.0). The trabecular bone parameters measured include trabecular bone volume fraction (Tb. BV/TV), trabecular bone surface density (Tb.BS/TV), trabecular thickness (Tb.Th), trabecular number (Tb.N), trabecular separation (Tb.S) and connectivity density (Conn.D). For cortical bone, the total cross-sectional area inside the periosteal envelope (Tt.Ar), cortical bone area (Ct.Ar), cortical area fraction (Ct.Ar/Tt.Ar), average cortical thickness (Ct.Th), cortical porosity (Ct.Po), total pore volume (Po.V) and pore density (Po.Dn) were measured. The cortical thickness was measured as the average measurement between the outer edge of the periosteum and the endosteum with the pores filled in. Both the trabecular and cortical parameters were corrected for body weight using the linear regression method as described in Jepsen et al. (28)
2.5 Mechanical testing
After microCT the mechanical properties of the left femora were measured by three-point bending analysis with the Instron E1000 (Electrodynamic Testing System V1.4 Upgrade, United Kingdom). Each bone was placed on two supports, that have a span of 7mm, with the anterior surface facing down and a transverse force was applied at the mid-diaphysis at 5mm/s until failure occurred. Force (F) and displacement (d) data were collected with the WaveMatrix 2 (Instron, version 2.0) software and used for the calculation of the ultimate load (N), breaking load (N), stiffness (N/mm), yield load (N), post-yield displacement (mm) and work-to-fracture (N/mm). These mechanical properties were calculated using standard definition and corrected for midshaft total area (Tt.Ar) measured with microCT using the linear regression method as mentioned before in Jepsen et al. (Supplemental Figure 2) (28).
2.6 ELISA
Blood was collected from eight-week-old heterozygous Mov13 (n=10) and WT littermates (n=10). After isolation, the serum was analyzed for the levels of Bone Turnover Markers (BTM), procollagen type I N-terminal propeptide (P1NP) (P1NP EIA kit, immunodiagnostic systems, AC-33F1) and Tartrate-resistant acid phosphatase 5b (TRAcP 5b) (TRAcP 5b, immunodiagnostic systems, SB-TR103), by enzyme-linked immunosorbent assay (ELISA) according to manufacturer’s protocol.
2.7 Quantitative real-time PCR
Humeri and right femora of nine Mov13 (60% males) and ten WT littermates (50% males) C57BL/6J mice were combined and pulverized in liquid nitrogen with the 6775 Freezer/Mill® Cryogenic Grinder (SPEX Sample Prep, Metuchen, NJ, US). Total bone RNA was extracted first with TRIzol® Reagent (Thermo Fisher Scientific, Waltham, MA, US) and then isolated using a RNeasy kit (Qiagen, Hilden, Germany). Heart ventricle RNA and abdominal total skin RNA (ten Mov13 and ten WT) was isolated using the RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. RNA concentrations were measured with Nanodrop spectrophotometer (NanoDrop Technologies, Wilmington, NC, US) and cDNA was synthesized from 140 ng of RNA with a SuperScript VILO kit (Thermo Fisher Scientific, Waltham, MA, US) in 20 µl final volume. The statistical expression of Col1a1, Col1a2, other collagens and tissue-specific markers was tested by quantitative real-time PCR (qPCR). qPCR analysis was performed in 10µl volume in duplicates with LightCycler® 480 SYBR Green I Master (Roche, Basel, Switzerland) and performed with the LightCycler® 480 System (Roche, Basel, Switzerland) according to the following program: 10 min at 95°C; 45 cycles consisting of 10s at 95°C, 5s at 60°C, 10s at 72°C, 5s at 78°C; 1 cycle of 1s at 95°C, 1s at 60°C and 1s at 95°C; and 30s at 40°C. List of used primers is presented in Table 1. Raw data was analyzed with the LightCycler® 480 Software, version 1.5 (Roche, Basel, Switzerland). Expressions of target genes were normalized towards the housekeeping TATA-box binding protein (Tbp) gene The relative gene expression was calculated with the ΔΔCt method.
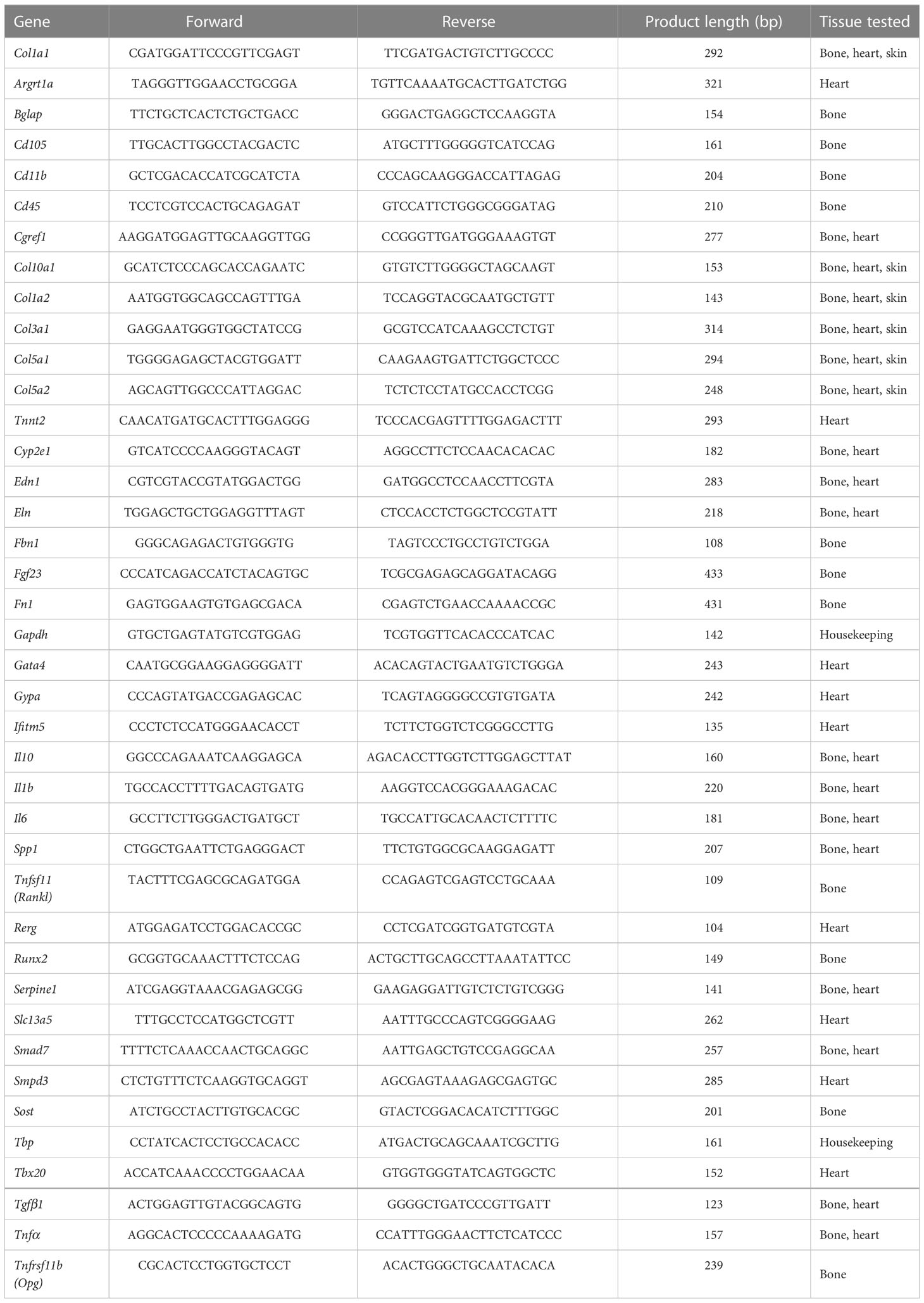
Table 1 Primers used in qPCR analysis and tissue, in which corresponding gene expression was tested.
2.8 Statistics
All data were tested for a normal distribution with the Kolmogorov-Smirnov test and analyzed using the unpaired T-test in GraphPad Prism version 9 (GraphPad Software Inc., San Diego, CA, US). For analysis of the body weight on the different timepoints a two-way ANOVA corrected for time and with multiple comparisons was used. A P-value of ≤ 0.05 indicated the statistically significant difference. Graphs, including Heatmaps were generated using GraphPad Prism version 9 (GraphPad Software Inc., San Diego, CA, US). For Heatmaps values of ΔΔCt were used.
3 Results
3.1 Reduced body weight in male Mov13 mice
The initial average mean weight of the Mov13 mice and of their WT littermates at four weeks of age were 16.32 ± 1.28g and 17.69 ± 1.41g respectively. The final mean weight at eight weeks before sacrifice of the Mov13 mice was 20.27 ± 2.36g and of the wild-type littermates 21.8 ± 3.02g. Weights were not significantly different between Mov13 and WT mice (Figure 1A). However, we found a considerably lower weight in the male Mov13 mice compared to male WT mice (p<0.001) from the second week onwards (Figure 1B). In addition, mice at the age of 15 weeks started presenting symptoms of lymphoma (fluffy coat, jerky motor skills, reduced appetite, increased size of liver and spleen) which was confirmed by the analysis of the liver and spleen by pathology examination.
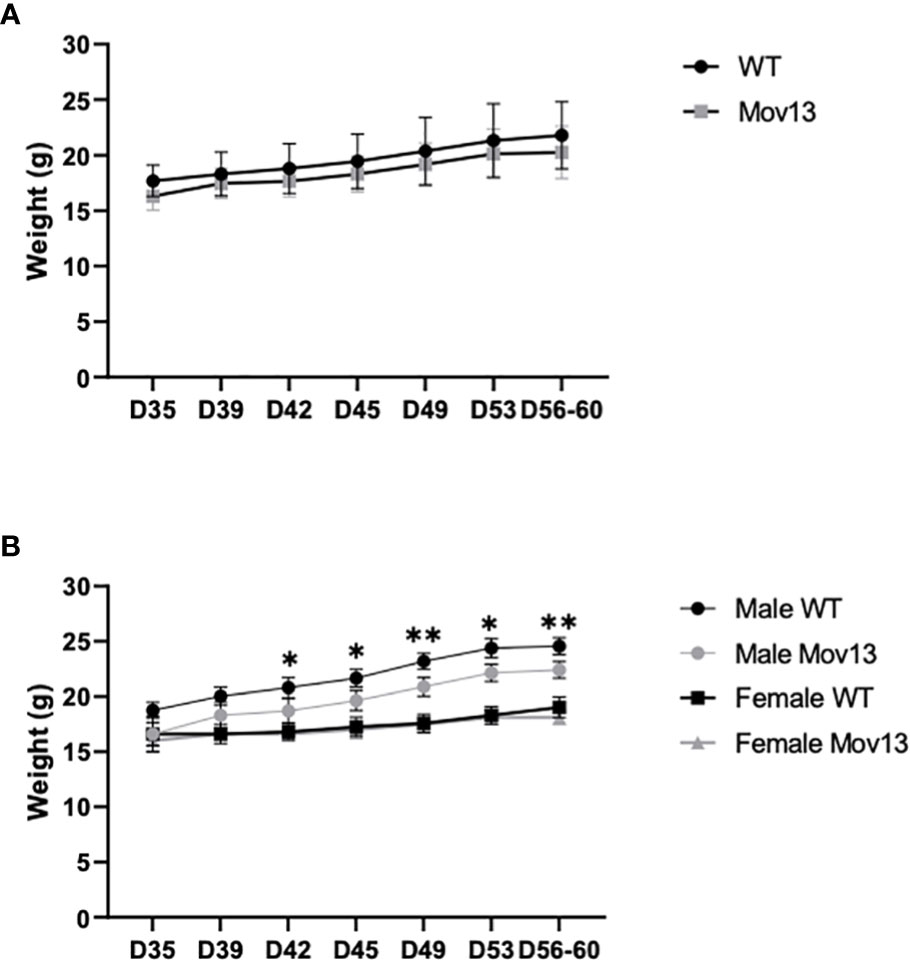
Figure 1 Weight of four-week-old till eight-week-old mice. (A) The weight of four-week-old Mov13 and WT littermates at different time points during four weeks. The age of the mice is presented on the x-axis in days. (B) Weight of four-week-old Mov13 and WT littermates divided in males and females during four weeks. Values of standard deviations (SD) are shown for each group: WT (n=10, 50%male) and Mov13 (n=10, 50%male). Statistical significance is indicated on the graphs (P-values ≤0.05 (*), ≤ 0.01 (**)).
3.2 Increased cortical microCT measurements in Mov13 mice
MicroCT scanning of the distal trabecular femoral bone and midshaft cortical femoral bone indicated significant differences in many structural parameters between Mov13 and WT littermates (Figure 2A). Significantly lower trabecular bone surface density (Tb. BS/TV) (p<0.05) and a significantly higher trabecular thickness (Tb.Th) (p<0.05) were detected in Mov13 mice (Figure 2B). Furthermore, Mov13 mice revealed a pattern of less trabecular number (Tb.N) and trabecular connectivity density (Conn.D) (Figure 2B). In addition, a trend of higher trabecular separation (Tb.Sp) was documented (Figure 2B). No difference was observed in trabecular bone volume fraction (Tb. BV/TV) (Figure 2B). In addition, Mov13 mice presented a significantly higher cortical midshaft total area (Tt.Ar) (p<0.001), cortical bone area (Ct.Ar) (p<0.001), cortical area fraction (Ct.Ar/Tt.Ar) (p<0.05), cortical thickness (Ct.Th) (p<0.01), periosteal perimeter (Ps.Pm) (p<0.0001), cortical porosity (Ct.Po) (p<0.01) and total pore volume (Po.V) (p<0.01) (Figure 2B). Moreover, a trend of a higher cortical pore density (Po.Dn) was also noted in Mov13 mice (Figure 2B). This data indicates a reduction in trabecular bone in contrast to increased formation of cortical bone with higher porosity in Mov13 mice.
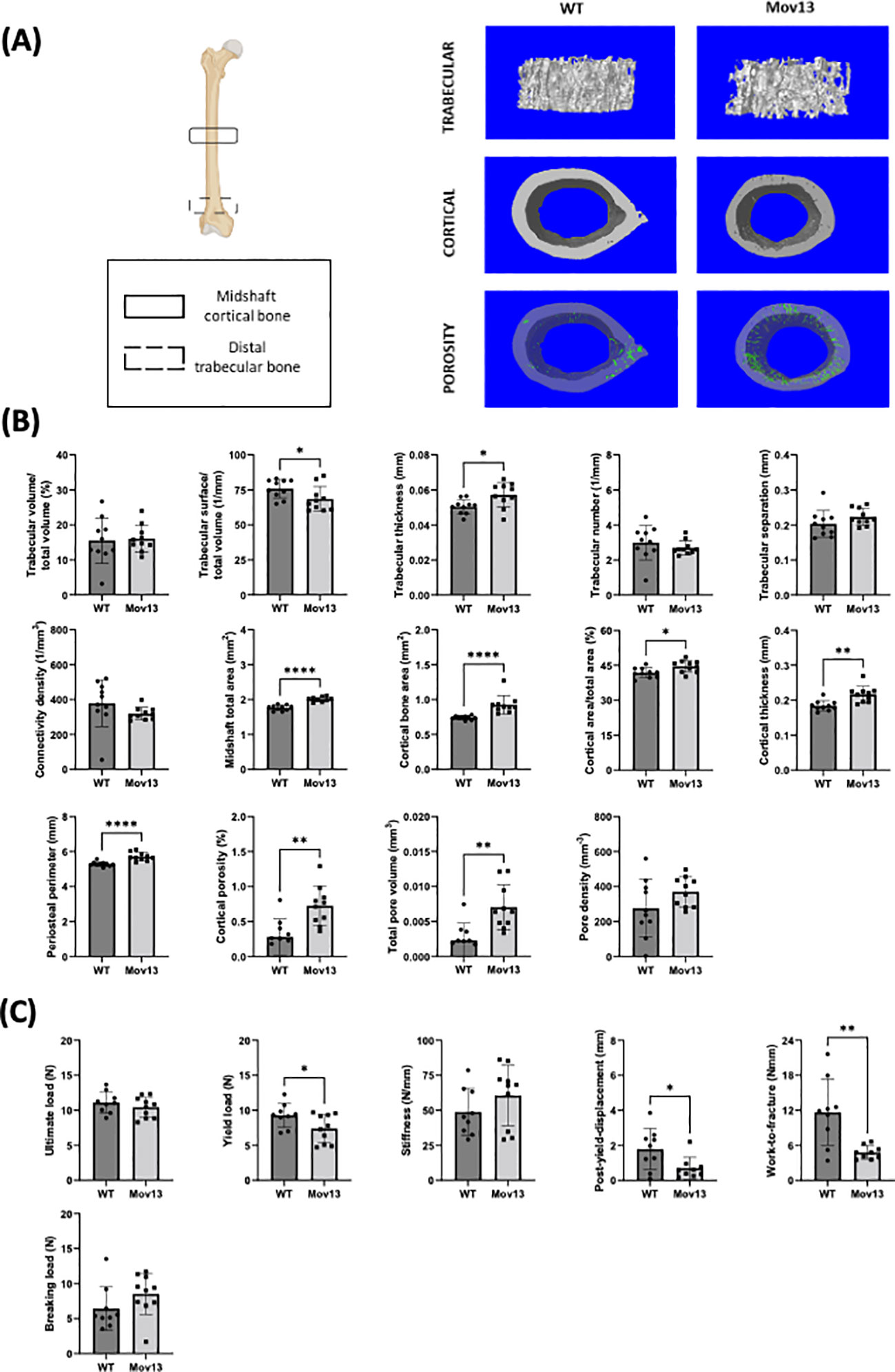
Figure 2 Biomechanical and structural measurements of the Mov13 mouse. (A) Schematic overview of femur showing the regions used to measure the midshaft cortical bone and distal trabecular bone and the 3D reconstructions of the microCT scans of the trabecular bone, cortical bone and cortical porosity of a wild-type and Mov13 mouse. (B) Trabecular and cortical parameters were measured from microCT scans, corrected for body weight, of Mov13 mice and wild-type littermates. Values of standard deviation (SD) are shown for each group: WT (n=10, 50%male), Mov13 (n=10, 50%male). (C) The biomechanical properties (Ultimate load, yield load, breaking load, stiffness, post-yield displacement and work-to-fracture) were measured using the load-displacement curve after 3-point bending and corrected for midshaft total area (Tt.Ar). Values of standard deviation (SD) are shown for each group: WT (n=9, 45%male), Mov13 (n=10, 50%male). Statistical significance is indicated on the graphs (P-values ≤0.05 (*), ≤ 0.01 (**), ≤0.0001 (****)). Tb.N, Ct.Ar and the breaking load were measured with a non-parametric unpaired T-test.
3.3 Reduced bone strength in Mov13 femur
The biomechanical properties of the femur were measured by 3-point bending and calculated from the resulting load-displacement curve and corrected for the cortical midshaft total area (Tt.Ar). Mov13 mice presented a significantly lower yield load (p<0.05), post-yield displacement (p<0.05) and Work-to-fracture (p<0.001) compared to their WT littermates (Figure 2C). In addition, no significant difference in the ultimate load, stiffness and breaking load was detected in Mov13 mice a trend of higher stiffness and breaking load (Figure 2C). A reduced PYD and WTF in Mov13 mice was expected considering the lower yield load. This suggests that Mov13 femora are more brittle due to a reduced amount of load needed before the bone suffers permanent damage and reduced ductility measured by PYD.
3.4 Histomorphometric parameters of Mov13 tibia
None of the histomorphometric parameters presented significant differences between Mov13 and WT mice (Figure 4). However, there was a trend of a lower cortical width (Ct.Wi), higher trabecular width (Tb.Wi) and higher osteoid perimeter (O.Pm/B.Pm) in Mov13 compared to WT mice (Figure 4B).
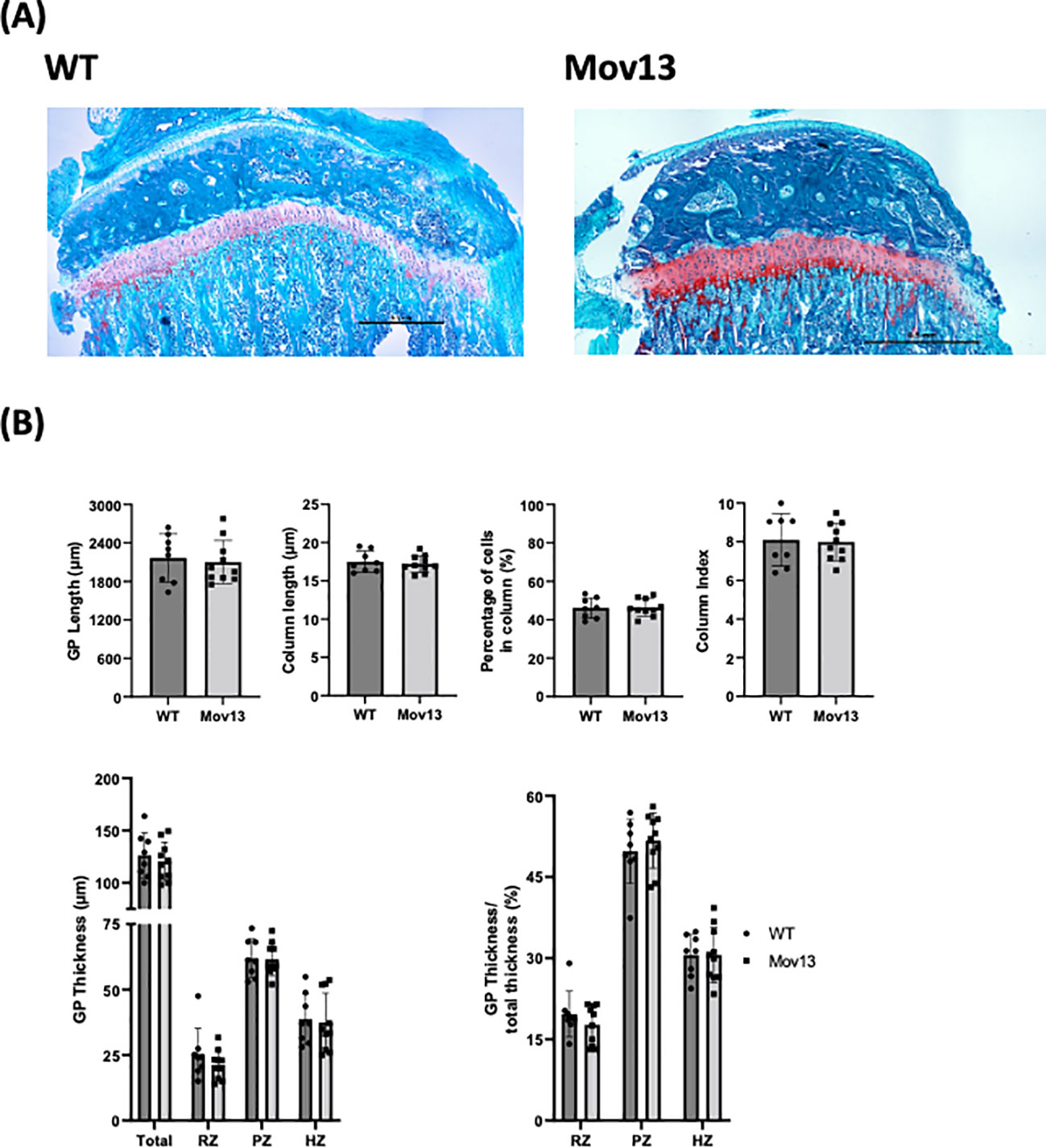
Figure 3 Histomorphometrical analysis of mouse growth plate. (A) Representative images of Safranin O staining of left tibia from eight-week-old Mov13 mice and WT littermates (10x10 magnification). (B) GP length, column length, column index (CI), percentage of cells in column, thickness (total and the different zones; resting zone (RZ), proliferative zone (PZ) and hypertrophic zone (HZ)) and ratio of thickness. Values of standard deviation (SD) are shown for each group: WT (n=8, 40%male) and Mov13 (n=10, 50%male). Statistical significance is indicated on the graphs (P-values ≤0.05 (*)). GP thickness RZ and PZ were measured with a non-parametric unpaired T-test.
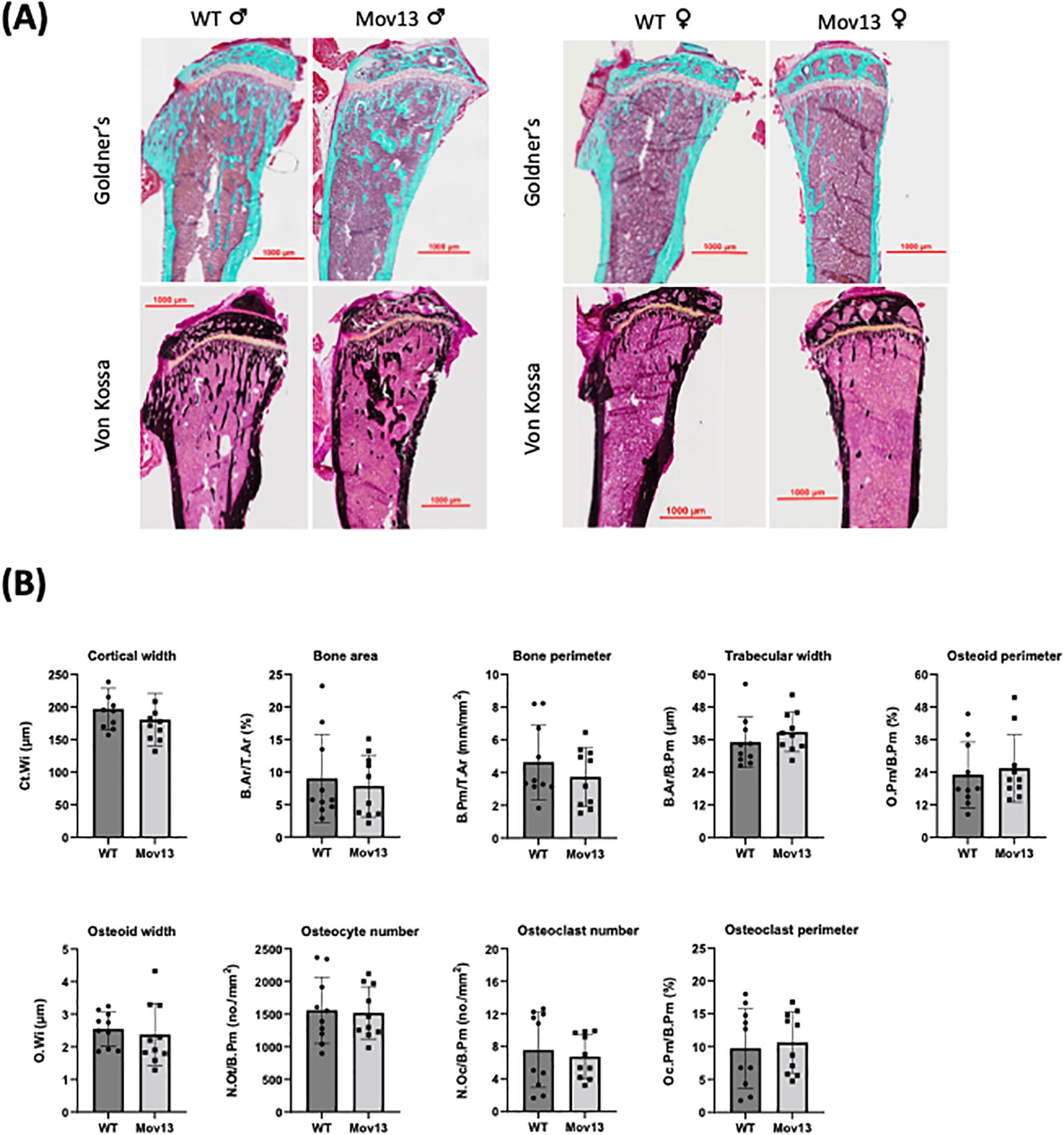
Figure 4 Histomorphometrical analysis of Mov13 and WT tibias. (A) Representative images of Mov13 and WT mice tibia bone histology. Masson Goldner’s trichrome (10x objective) and Von Kossa (20x objective) staining of male and female mice tibiae. (B) Cortical and trabecular measurements of Mov13 and WT tibias: Cortical width (Ct.Wi), Bone area (B.Ar/T.Ar), Bone perimeter (B.Pm/T.Ar), Trabecular width (B.Ar/B.Pm), Osteoid perimeter (O.Pm/B.Pm), Osteoid width (O.Wi), Number of osteocytes (N.Ot/B.Pm), Number of osteoclasts (N.Oc/B.Pm), Osteoclast perimeter (Oc.Pm/B.Pm). Values of standard deviation (SD) are shown for each group: WT (n=10, 50%male), Mov13 (n=10, 50%male). Statistical significance is indicated on the graphs (P-values ≤0.05 (*)). Bv/Tv, B.Pr/T.Ar and O.Wi were measured with a non-parametric unpaired T-test.
We analyzed the growth plate (GP), since morphological GP changes were previously found in OI patients (29). However, our GP analysis revealed no significant differences between the Mov13 and WT mice for any of the measurements (Figure 3). However, a discernible lower total growth plate thickness was detected in Mov13 mice (Figure 3B). We also perceived a slightly lower ratio for the resting zone of Mov13 mice, no difference was discovered for the hypertrophic zone. Though, we found a marginally higher ratio of the proliferative zone in the Mov13 mice compared to the WT mice (Figure 3B).
3.5 Bone formation marker is reduced in Mov13 mice
Serum P1NP levels, a bone formation marker, were 36.13% lower in the 8-week-old Mov13 than in WT mice, 74.55 ± 28.93ng/ml and 107.43 ± 25.33ng/ml respectively (p<0.05) (Figure 5). We did not observe a difference in the bone resorption marker, TRAcP 5b, between Mov13 and WT mice (Figure 5). This substantiates the expected reduced Col1a1 production in Mov13 mice and might indicate an overall reduced bone formation.
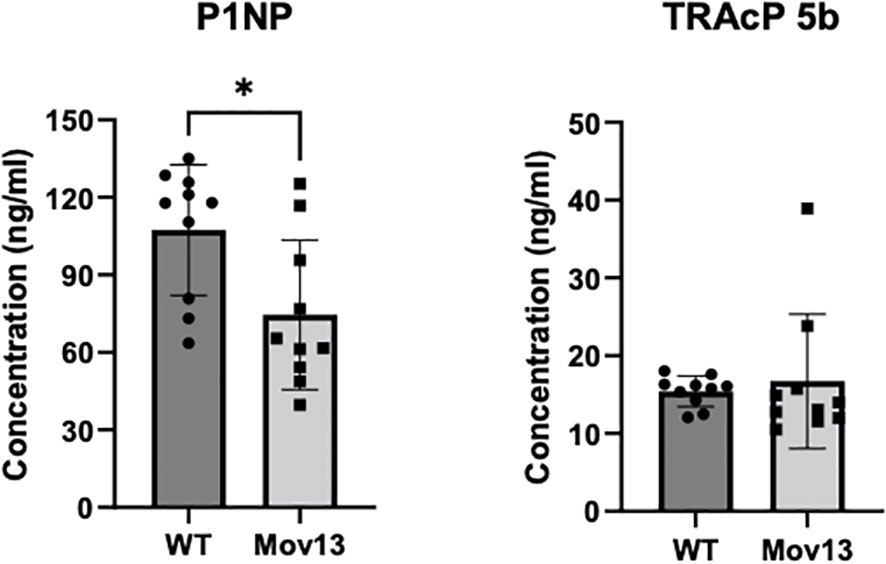
Figure 5 ELISA of bone turnover markers in serum. Markers for bone formation (P1NP) and bone resorption (TRAcP 5b) were measured in mice blood serum with ELISA. P1NP and TRAcP 5b measurement of eight-week-old Mov13 and WT littermates. Standard deviations (SD) are shown for each group: WT (n=10, 50%male) and Mov13 (n=10, 50%male). Statistical significance is indicated on the graphs (P-values ≤0.05 (*)). TRAcP 5b was measured with a non-parametric unpaired T-test.
3.6 Expression of Col1a1, osteogenic, collagen and immune marker genes in bone tissue of Mov13 mice
No difference in transcriptional expression of Col1a1 was measured between Mov13 and WT mice. However, the Col1a1/Col1a2 ratio was significantly lower (p<0.001) in the overall Mov13 group, due to higher Col1a2 expression in Mov13 (p<0.05) (Figure 6A). Surprisingly, Mov13 mice also showed upregulation of collagen type V: Col5a1, Col5a2 (p<0.05 and p<0.01 respectively); osteogenic and related markers: Bglap (p<0.05), Edn1 (p<0.01), Eln (p<0.05), Fgf23 (p<0.001) and immune marker Smad7 (p<0.01) (Figure 6A, Supplemental Figures 3, 5A). Several common differentially expressed genes in osteogenesis imperfecta mouse (oim) and Col1a1Jrt/+ mice were also tested (30). None of these genes (Cgref, Cyp2e1, Ifitm5, Rerg, Slc13a5, Smpd3, Gypa) presented expression differences in Mov13 compared to WT, highlighting differences in bone fragility mechanisms of DN and HI OI mouse models (Supplemental Figures 3, 5A). In addition, many other osteogenic and related markers (Fn1, Opg, Rankl, Runx2, Sost, Spp1, Tgfβ, Agtr1a, Fbn1) and immune markers (Il10, Il1b, Il6, Serpine1, Tnfα) did not present any difference between Mov13 and WT mice (Supplemental Figure 3).
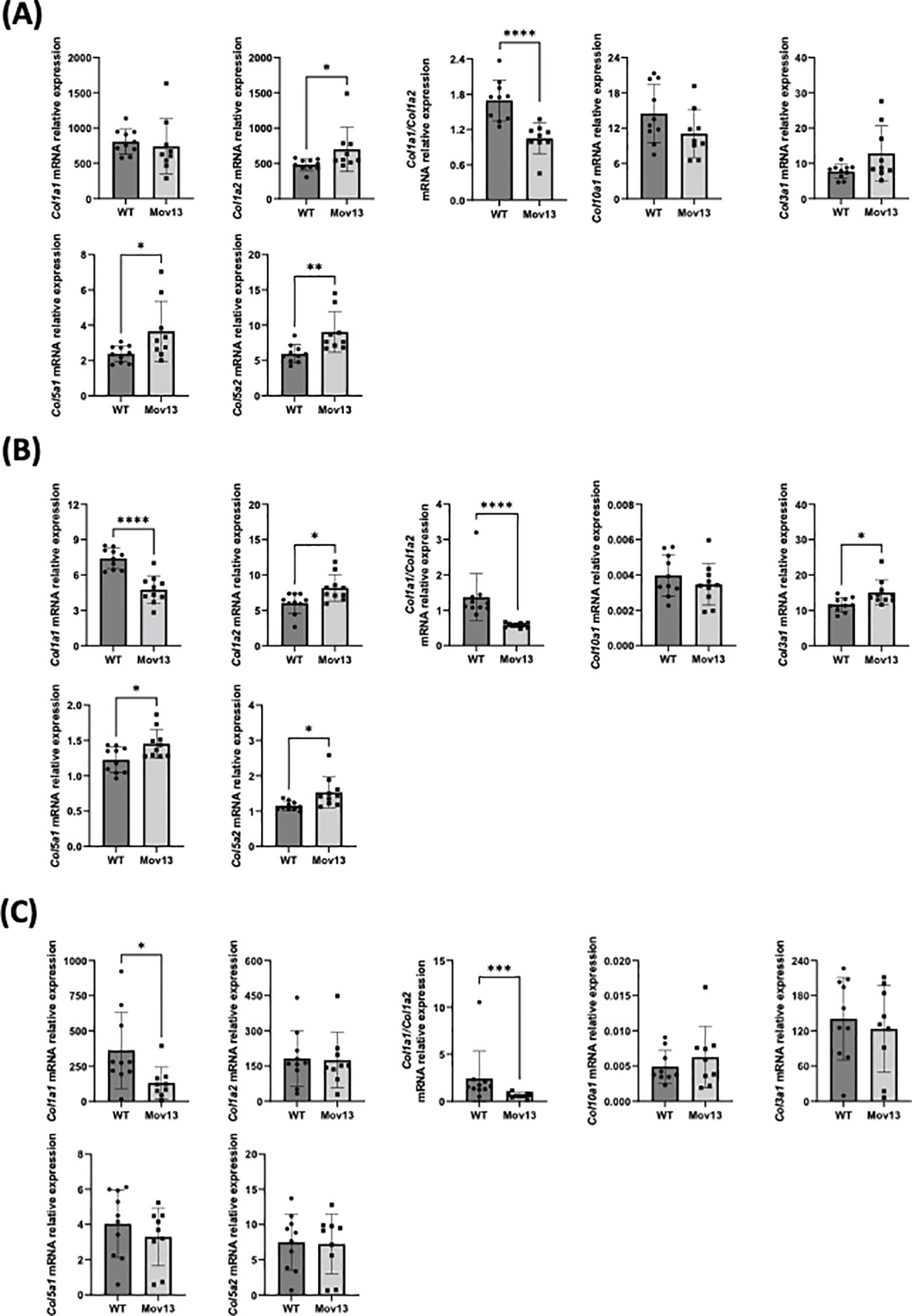
Figure 6 Relative mRNA expression of collagens in bone, heart and skin tissues. Relative mRNA expression of different collagen types in (A) bone, (B) heart and (C) skin tissue of Mov13 and WT mice. Relative mRNA expression is represented as ΔΔCt values. P-values are indicated as follows: (P-values ≤0.05 (*), ≤ 0.01 (**), ≤ 0.001 (***), ≤0.0001 (****)). Bone Col1a2, Col5a2, Col1a1/Col1a2, heart Col1a2, Col3a1, Col1a1/Col1a2, skin Col1a1, Col10a1 and Col1a1/Col1a2 were measured with a non-parametric unpaired T-test.
3.7 Expression of Col1a1 and target genes in heart and skin tissue of Mov13 mice
Heart tissue presented a 35.76% lower expression of Col1a1 (p<0.0001) and a lower Col1a1/Col1a2 ratio (p<0.01) in Mov13 mice. Mov13 mice appeared to have a significantly higher expression of other collagens (Col1a2, Col3a1, Col5a1, Col5a2) (p<0.05), cardiological markers (Tbx20 (p<0.05), Spp1 (p<0.05)) and immune markers (Tnfα (p<0.05), Il10 (p<0.05)). As in the bone tissue, several common differentially expressed genes (Cgref, Cyp2e1, Ifitm5, Slc13a5, Smpd3, Gypa) of the oim and Col1a1Jrt/+ mice were measured in the heart and presented no difference between Mov13 and WT mice (Supplemental Figures 4, 5B). However, the Rerg gene expression was significantly lower in Mov13 mice compared to WT (p<0.05) (Figure 6B, Supplemental Figures 4, 5B). No difference was seen in the other cardiological and related markers (Agtr1a, Tnnt2, Gata4, Edn1, Eln), immune markers (Il1b, Il6, Smad7, Serpine1) and osteogenic markers (Fn1, Tgfβ) (Supplemental Figures 4, 5B). Similarly to heart tissue, Col1a1 in the skin of Mov13 mice was reduced by 63.58% (p<0.05), (Figure 6C). No difference was observed in the expression of other collagen genes in the skin (Figure 6C, Supplemental Figure 5C).
4 Discussion
In this study, a thorough characterization of heterozygous Mov13 mice with a null mutation in the alpha1 collagen type I locus is provided. The reduced mechanical and material properties in the Mov13 mice compared to healthy litter-mate controls were demonstrated with microCT and 3-point bending. No significantly lower expression of Col1a1 mRNA level was detected in Mov13 mineralized connective tissue, although this was the case in the heart and skin tissue.
The most important measurement in support of Mov13 as a suitable mouse model for HI OI type 1, is the expected reduced mechanical properties of its long bones. Based on previous loading test outcomes on Mov13 femora, a distinctly lower work-to-fracture in Mov13 were expected (10, 11, 15, 31). It has to be stated that all previous loading tests in Mov13 mice were performed with 4-point bending; as such, our findings with 3-point bending are not entirely directly comparable. However, in addition to a significantly lower work-to-fracture, post-yield displacement and yield load, a trend of higher breaking load and stiffness in Mov13 mice were also detected, corroborating previous studies about the causative role of reduced collagen type I in bone fragility (10, 15, 31). All together, we can hypothesize that the bone brittleness is caused by a reduced tissue bending strength and ductility resulting in a a lower yield load, post-yield displacement and work-to-fracture (15).
A lower trabecular bone surface density (Tb. BS/TV) and significantly higher trabecular width, were measured in Mov13 mice with microCT. The difference in trabecular BS/TV could suggest a possible alteration in the tendency of the trabecular tissue for microstructure remodeling, caused by a reduced available surface for osteoclasts and osteoblasts for replacing old and damaged bone matrix (32). It can be hypothesized that this reflects a mechanism by which bone tissue may compensate for this with an increase in trabecular thickness. Even though no other significant differences were found in the trabecular bone, we did observe a different trabecular bone structure with few but large trabeculae based on a lower trabeculae number, connectivity density and a higher trabecular separation and the previously mentioned lower Tb. BS/TV and higher Tb.Th. However, a difference in the trabecular bone volume fraction (Tb. BV/TV) was not observed. In agreement with previous studies, both the cortical bone area and cortical porosity were higher in the Mov13 mice (11, 15, 31). In addition, higher midshaft total area, cortical area fraction, cortical thickness, total pore volume and periosteal perimeter in Mov13 mice were noted. Hence, the hypothesis of Bonadio et al. and Jepsen et al, that the bone tissue is adapting via the periosteal surface to compensate for the loss of load-bearing capacity due to the altered microstructure associated with a higher porosity in Mov13 mice was substantiated by our data (11, 15, 31).
No significant differences were found in both cortical and trabecular histomorphometrical measurements. However, this did not correlate to our significantly higher cortical measurements determined by microCT. Yet, a trend of decreased trabecular bone was determined by both histomorphometry and microCT. This was in contrast to the reduced mechanical properties noted in the Col1a1 ± 365 mouse model, in which the underlying cause was attributed to a lower and sparser amount of trabecular bone and a higher number of osteoclasts instead of changes in the cortical bone (6). In addition, no difference in any of the osteoid parameters was detected; this is in contrast with findings in OI patients, including HI OI type 1, in which a difference in osteoid surface has been reported (33, 34). This clearly emphasizes the importance of measuring the bone phenotype not only histomorphometrically but also based on structural parameters by microCT. In addition, this might also demonstrate that more tissue sections are necessary to analyze in the future. Sanguinetti et al. found a disruption in the growth plate in OI type 1 and 3 patients due to an increased thickness of the hypertrophic zone and shorter columns in the proliferative zone (26, 29). However, we failed to find any significant differences in any growth plate measurement between Mov13 and WT mice.
Our results did not reveal any difference in weight between Mov13 and WT mice. However, significantly lower weight was observed in Mov13 males in agreement with male Col1a1 ± 365 mice (6). This warrants further research to further explain the lack of weight difference between female WT and Mov13 mice.
Significantly lower P1NP concentration was noted in Mov13 mice. Since almost 90% of the organic portion of the bone matrix consists of type I collagen and P1NP is cleaved off when collagen type I is secreted, P1NP can be perceived as a good marker for collagen type I production and bone formation (35). As such, a decreased bone formation was clearly indicated by the lower P1NP concentration in Mov13 mice which corresponds with the lower concentration found in HI OI type 1 patients and Col1a1 ± 365 mice (6, 36). No difference in TRAcP 5b concentrations was found in Mov13 mice. TRAcP 5b measurement informs about osteoclast number rather than osteoclasts activity and as such corroborates our histomorphological findings in the tibia (37, 38). In comparison, an increased number of TRAP-positive osteoclasts was observed in the trabecular femur of eight-week-old Col1a1 ± 365 mice (6). In addition, equal levels in the gene expression of osteoclast related markers, Opg and Rankl, were also noted between Mov13 and WT mice. Conclusively, the lower level of P1NP is in line with the expected reduction in collagen type I synthesis in Mov13 mice but more investigation is needed to fully comprehend the catabolic mechanism involved in the bone turnover of HI OI.
Despite transcription inactivation of one Col1a1 allele by MMuLV, we did not detect a lower Col1a1 mRNA expression by qPCR in the bone tissue of heterozygous Mov13 mice. In agreement with our findings a previous study reported a 30% increase in Col1a1 gene expression by northern blotting in the femur of eight-week-old Mov13 mice and returned to a comparable level at 15 weeks old (31). This is in contrast with Col1a1 ± 365 mice where a decreased pro-α1(I) expression in femur of eight-week-old mice was measured (6). Additionally, in the tibia of 17-week-old Mov13 mice a nine-fold Col1a1 reduction was observed by qPCR (18). Similarly, lower expression of Col1a1 was observed in otic capsule, while parietal bone Col1a1 expression was almost 2-fold higher compared to WT littermates (18). Though, Mov13 embryos and embryonic fibroblast cultures also did not reveal a convincing decrease in Col1a1 expression by northern blotting (39). In comparison with Iruela-Arispe et al., a significantly lower Col1a1 expression was measured in total skin of our Mov13 mice. Regardless of mRNA expression findings, Mov13 skin fibroblasts from 15-week-old mice have been shown to produce half the normal amount of radiolabeled type I collagen (6, 10). In addition, decreased collagen is recorded in the supernatant and extracellular matrix of the embryonic Mov13 fibroblast cultures (39). Finally, although no reduced Col1a1 expression was measured in the bone tissue of Mov13 mice, as mentioned above, a decreased P1NP concentration was detected. As such, this indicates a divergence between Col1a1 gene expression and the eventual collagen type I secretion in the bone tissue. Furthermore, the high variability in Col1a1 gene expression in the different studies and different bone tissues (femora, tibiae and otic capsule) also hamper the validation of this mouse model for HI OI type 1.
Regarding the expression of other collagen types in fibroblast-like cells of homozygous Mov13 mice, no compensation was seen on protein level (40). Unfortunately, our study could not include protein measurement of collagen types and the correlation between Col1a1 mRNA and protein expression has previously only been examined in Mov13 fibroblasts (39). Consequently, it remains unclear what the Col1a1 mRNA expression changes mean on protein level in the different Mov13 tissues. Equally lacking is the biological significance of the increased expression of Col1a2, Col5a1, Col5a2 and Col3a1 in the bone and heart tissues of Mov13 mice. We can hypothesize that the reduction of Col1a1 could influence the expression of genes encoding the fibril-forming collagens type III and V with which it forms fibrils in the bone and myocardium (41, 42). However, this has not been shown before as no difference in Col1a1 expression levels were found in a Ehlers-Danlos syndrome HI Col5a1+/- mouse model (43).
No change was found in Runx2 expression whereas Bglap expression was higher in Mov13 mice. In contrast, lower expression of Runx2 and Bglap were found in the femur of eight-week-old Col1a1 ± 365 mice. However collagen type I regulates osteoblast differentiation, which may potentially justify this downregulation in the Col1a1 ± 365 mice (6, 44). Notably, a higher expression of Smad7, which is an inhibitor of TGFβ signaling, was also found in the Mov13 mice (45). Excessive TGFβ signaling and bioactivity have previously been found in OI mouse models (Crtap-/- and Col1a2tm1.1Mcbr) and serum of OI patients respectively (46, 47). It can be speculated that Smad7 upregulation may oppose potential TGFβ activity. Recent studies have hinted towards the presence of inflammation in OI patients (48, 49). No upregulation of inflammation factors was identified in bone tissue, whereas TNFα, Il10, Tbx20 and Spp1, were upregulated in heart tissue. All four genes play a crucial role in cardiac remodeling, revealing signs of possible cardiac dysfunction caused by significant reduction of Col1a1 expression in the hearts of Mov13 mice (50–53). Dysregulated expression of estrogen signaling regulator Rerg was previously reported in bone of oim and Col1a1Jrt/+ mice. However, in contrast to mice with DN defect, Mov13 have significantly downregulated Rerg expression in heart.
A concerning factor interfering with gene expression may be the presentation of severe lymphoma in adult Mov13 mice. Even though the mice only visibly started deteriorating starting from 15 weeks old on, it cannot be excluded that the gene expression was affected by a precancerous state. Notably, in addition to a high variability in bone tissue, which was shown in the literature and our results, a significant decrease in Col1a1 gene expression was detected in the heart and skin tissue of our Mov13 mice. Even though the heart and skin may be less likely affected by lymphoma, given the severe nature of the disease, it is not possible to safely conclude if these gene expression changes can be attributed to cancer or tissue-dependent collagen regulation as reported in Mov13 odontoblasts (18, 54, 55). The Col1a1 expression variability present in Mov13 mice can also be explained with the presence of cell-specific, orientation-dependent transcriptional elements in the first intron of the Col1a1, known to exist in both mice and human genomes (4, 56–60). It is also worth to note that no obvious evidence of lymphoma was detected by the analysis of our mice, given that there was no severe trabecular and cortical bone loss, no decreased osteoclast number, serum TRAcP 5b concentration or no downregulation of Runx2 and Bglap detected (61, 62). However, elevated levels of Fgf23 expression, found in our Mov13 mice and which is known to suppress mineralization of osteoblasts and negatively regulate bone homeostasis, are associated with acute myeloid leukemia (63–66). The higher expression of Edn1 and Eln could also underline metastatic processes in the bone (67–69). On the other hand, it could be plausible that the upregulation of both genes is a consequence of the bone defect in Mov13 mice. Similarly to Mov13, oim mice have shown upregulation of Edn1, important for angiogenesis, osteoblastic proliferation and bone development, whereas Eln overexpression might be an attempt to compensate for the loss of bone extracellular matrix elasticity (30). High content of elastin was shown in biopsies of human OI type 4 and Ehlers-Danlos syndrome patients (70, 71). Due to the mixed effect of the transgenesis and possible development of lymphoma, the biological interpretation of the gene expression pattern in Mov13 mice remains unclear.
In conclusion, our findings corroborate that the Mov13 mice present a reduced mechanical strength and altered material properties in accordance to the expected reduced Col1a1 in the bone tissue. Unfortunately, the presentation of severe lymphoma in adult mice and the challenging variability of the Mov13 bone phenotype prevents us from recommending this mouse model for therapeutic studies for of HI OI type 1, especially when older mice are required. A new reliable mouse model, which recapitulates HI OI type 1, is clearly urgently needed to facilitate mild OI research and the testing of new therapies.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by the Central Committee of Animal experiments (CCD) of the of the Netherlands and in agreement with the Animal Welfare Body of the Amsterdam UMC in full compliance with the directive 2010/63/EU.
Author contributions
All authors have contributed substantially to the conception of the article and critical revision. LC, together with the help of HE, performed all the experimental work for Figures 1, 2 and supplemental Figures 1 and 2. LZ and LC together performed all the experiments for Figures 3, 4. LZ, together with the help of LW, performed all the experimental work and analysis for Figure 6 and supplemental Figures 3–5. LC performed the analysis for Figures 1–3, 5 while LZ also performed the analysis for Figure 4. DM, NB, GP and EE supervised the work of LC, LZ, LW and HE and contributed to the experimental design and writing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The work of LC, DM and this project was funded by the kind donation of Hans Horsting and Mary Horsting-stuit of the Horstingstuit foundation. The work of LZ is supported by the Estonian Research Council, grant PUTJD1009.
Acknowledgments
Figure 2A was created with Biorender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1145125/full#supplementary-material
Glossary
References
1. Forlino A, Cabral WA, Barnes AM, Marini JC. New perspectives on osteogenesis imperfecta. Nat Rev Endocrinol (2011) 7(9):540–57. doi: 10.1038/nrendo.2011.81
2. Forlino A, Marini JC. Osteogenesis imperfecta. Lancet (2016) 387:1657–71. doi: 10.1016/S0140-6736(15)00728-X
3. Claeys L, Storoni S, Eekhoff M, Elting M, Wisse L, Pals G, et al. Collagen transport and related pathways in osteogenesis imperfecta. Hum Genet (2021) 140(8):1121–41. doi: 10.1007/s00439-021-02302-2
4. Hormuzdi SG, Penttinen R, Jaenisch R, Bornstein P. A gene-targeting approach identifies a function for the first intron in expression of the α1(I) collagen gene. Mol Cell Biol (1998) 18(6):3368–75. doi: 10.1128/MCB.18.6.3368
5. Enderli TA, Burtch SR, Templet JN, Carriero A. Animal models of osteogenesis imperfecta: Applications in clinical research. Orthop Res Rev (2016) 8:41–55. doi: 10.2147/orr.s85198
6. Liu Y, Wang J, Liu S, Kuang M, Jing Y, Zhao Y, et al. A novel transgenic murine model with persistently brittle bones simulating osteogenesis imperfecta type I. Bone (2019) 127:646–55. doi: 10.1016/j.bone.2019.07.021
7. Jaenisch R, Jähner D, Nobis P, Simon I, Löhler J, Harbers K, et al. Chromosomal position and activation of retroviral genomes inserted into the germ line of mice. Cell (1981) 24(2):519–29. doi: 10.1016/0092-8674(81)90343-3
8. Jaenisch R. Retroviruses and mouse embryos: A model system in which to study gene expression in development and differentiation. Ciba Found Symp (1983) 98:44–62. doi: 10.1002/9780470720790.ch4
9. Harbers K, Kuehn M, Delius H, Jaenisch R. Insertion of retrovirus into the first intron of alpha 1(I) collagen gene to embryonic lethal mutation in mice. Proc Natl Acad Sci USA (1984) 81(5):1504–8. doi: 10.1073/pnas.81.5.1504
10. Bonadio J, Saunders TL, Tsai E, Goldstein SA, Morris-Wiman J, Brinkley L, et al. Transgenic mouse model of the mild dominant form of osteogenesis imperfecta. Proc Natl Acad Sci United States America (1990) 87(18):7145–9. doi: 10.1073/pnas.87.18.7145
11. Jepsen KJ, Goldstein SA, Kuhn JL, Schaffler MB, Bonadio J. Type-I collagen mutation compromises the post-yield behavior of Mov13 long bone. J Orthop Res (1996) 14(3):493–9. doi: 10.1002/jor.1100140320
12. Hartung S, Jaenisch R, Breindl M. Retrovirus insertion inactivates mouse alpha 1(I) collagen gene by blocking initiation of transcription. Nature (1986) 320(6060):365–7. doi: 10.1038/320365a0
13. Schnieke A, Harbers K, Jaenisch R. Embryonic lethal mutation in mice induced by retrovirus insertion into the α1(I) collagen gene. Nature (1983) 304(5924):315–20. doi: 10.1038/304315a0
14. Jacobsen CM, Schwartz MA, Roberts HJ, Lim KE, Spevak L, Boskey AL, et al. Enhanced wnt signaling improves bone mass and strength, but not brittleness, in the Col1a1(+/mov13) mouse model of type I osteogenesis imperfecta. Bone (2016) 90:127–32. doi: 10.1016/j.bone.2016.06.005
15. Jepsen KJ, Schaffler MB, Kuhn JL, Goulet RW, Bonadio J, Goldstein SA. Type I collagen mutation alters the strength and fatigue behavior of Mov13 cortical tissue. J Biomech (1997) 30(11-12):1141–7. doi: 10.1016/S0021-9290(97)00088-2
16. Del Prete Z, Antoniucci S, Hoffman AH, Grigg P. Viscoelastic properties of skin in mov-13 and tsk mice. J Biomech (2004) 37(10):1491–7. doi: 10.1016/j.jbiomech.2004.01.015
17. Altschuler RA, Dolan DF, Ptok M, Gholizadeh G, Bonadio J, Hawkins JE. An evaluation of otopathology in the MOV-13 transgenic mutant mouse. Ann N Y Acad Sci (1991) 630:249–52. doi: 10.1111/j.1749-6632.1991.tb19595.x
18. Stankovic KM, Kristiansen AG, Bizaki A, Lister M, Adams JC, McKenna MJ. Studies of otic capsule morphology and gene expression in the Mov13 mouse – an animal model of type I osteogenesis imperfecta. Audiol Neurotol (2007) 12(5):334–43. doi: 10.1159/000104789
19. Derwin KA, Soslowsky LJ, Kimura JH, Plaas AH. Proteoglycans and glycosaminoglycan fine structure in the mouse tail tendon fascicle. J Orthop Res (2001) 19(2):269–77. doi: 10.1016/S0736-0266(00)00032-2
20. Derwin KA, Soslowsky LJ. A quantitative investigation of structure-function relationships in a tendon fascicle model. J Biomech Eng (1999) 121(6):598–604. doi: 10.1115/1.2800859
21. Löhler J, Timpl R, Jaenisch R. Embryonic lethal mutation in mouse collagen I gene causes rupture of blood vessels and is associated with erythropoietic and mesenchymal cell death. Cell (1984) 38(2):597–607. doi: 10.1016/0092-8674(84)90514-2
22. Kratochwil K, von der Mark K, Kollar EJ, Jaenisch R, Mooslehner K, Schwarz M, et al. Retrovirus-induced insertional mutation in Mov13 mice affects collagen I expression in a tissue-specific manner. Cell (1989) 57(5):807–16. doi: 10.1016/0092-8674(89)90795-2
23. Zhytnik L, Simm K, Salumets A, Peters M, Märtson A, Maasalu K. Reproductive options for families at risk of osteogenesis imperfecta: A review. Orphanet J Rare Dis (2020) 15(1):128. doi: 10.1186/s13023-020-01404-w
24. Ben Amor IM, Roughley P, Glorieux FH, Rauch F. Skeletal clinical characteristics of osteogenesis imperfecta caused by haploinsufficiency mutations in COL1A1. J Bone Miner Res (2013) 28(9):2001–7. doi: 10.1002/jbmr.1942
25. Maioli M, Gnoli M, Boarini M, Tremosini M, Zambrano A, Pedrini E, et al. Genotype–phenotype correlation study in 364 osteogenesis imperfecta Italian patients. Eur J Hum Genet (2019) 27(7):1090–100. doi: 10.1038/s41431-019-0373-x
26. Killion CH, Mitchell EH, Duke CG, Serra R. Mechanical loading regulates organization of the actin cytoskeleton and column formation in postnatal growth plate. Mol Biol Cell (2017) 28(14):1862–70. doi: 10.1091/mbc.e17-02-0084
27. Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H, et al. Standardized nomenclature, symbols, and units for bone histomorphometry: A 2012 update of the report of the ASBMR histomorphometry nomenclature committee. J Bone Miner Res (2013) 28(1):2–17. doi: 10.1002/jbmr.1805
28. Jepsen KJ, Silva MJ, Vashishth D, Guo XE, van der Meulen MC. Establishing biomechanical mechanisms in mouse models: Practical guidelines for systematically evaluating phenotypic changes in the diaphyses of long bones. J Bone Miner Res (2015) 30(6):951–66. doi: 10.1002/jbmr.2539
29. Sanguinetti C, Greco F, De Palma L, Specchia N, Falciglia F. Morphological changes in growth-plate cartilage in osteogenesis imperfecta. J Bone Joint Surg Br (1990) 72(3):475–9. doi: 10.1302/0301-620X.72B3.2187879
30. Moffatt P, Boraschi-Diaz I, Marulanda J, Bardai G, Rauch F. Calvaria bone transcriptome in mouse models of osteogenesis imperfecta. Int J Mol Sci (2021) 22(10):5290. doi: 10.3390/ijms22105290
31. Bonadio J, Jepsen KJ, Mansoura MK, Jaenisch R, Kuhn JL, Goldstein SA. A murine skeletal adaptation that significantly increases cortical bone mechanical properties. implications for human skeletal fragility. J Clin Invest. (1993) 92(4):1697–705. doi: 10.1172/jci116756
32. Lerebours C, Thomas CDL, Clement JG, Buenzli PR, Pivonka P. The relationship between porosity and specific surface in human cortical bone is subject specific. Bone (2015) 72:109–17. doi: 10.1016/j.bone.2014.11.016
33. Roschger P, Fratzl-Zelman N, Misof BM, Glorieux FH, Klaushofer K, Rauch F. Evidence that abnormal high bone mineralization in growing children with osteogenesis imperfecta is not associated with specific collagen mutations. Calcif Tissue Int (2008) 82(4):263–70. doi: 10.1007/s00223-008-9113-x
34. Rauch F, Travers R, Parfitt AM, Glorieux FH. Static and dynamic bone histomorphometry in children with osteogenesis imperfecta. Bone (2000) 26(6):581–9. doi: 10.1016/S8756-3282(00)00269-6
35. Seo W-Y, Kim J-H, Baek D-S, Kim S-J, Kang S, Yang WS, et al. Production of recombinant human procollagen type I c-terminal propeptide and establishment of a sandwich ELISA for quantification. Sci Rep (2017) 7(1):15946. doi: 10.1038/s41598-017-16290-9
36. Garnero P, Schott AM, Prockop D, Chevrel G. Bone turnover and type I collagen c-telopeptide isomerization in adult osteogenesis imperfecta: Associations with collagen gene mutations. Bone (2009) 44(3):461–6. doi: 10.1016/j.bone.2008.11.006
37. Halleen JM, Alatalo SL, Suominen H, Cheng S, Janckila AJ, Väänänen HK. Tartrate-resistant acid phosphatase 5b: A novel serum marker of bone resorption. J Bone Mineral Res (2000) 15(7):1337–45. doi: 10.1359/jbmr.2000.15.7.1337
38. Alatalo SL, Halleen JM, Hentunen TA, Mönkkönen J, Väänänen HK. Rapid screening method for osteoclast differentiation in vitro that measures tartrate-resistant acid phosphatase 5b activity secreted into the culture medium. Clin Chem (2000) 46(11):1751–4. doi: 10.1093/clinchem/46.11.1751
39. Iruela-Arispe ML, Vernon RB, Wu H, Jaenisch R, Sage EH. Type I collagen-deficient mov-13 mice do not retain SPARC in the extracellular matrix: Implications for fibroblast function. Dev Dyn (1996) 207(2):171–83. doi: 10.1002/(SICI)1097-0177(199610)207:2<171::AID-AJA5>3.0.CO;2-E
40. Dziadek M, Timpl R, Jaenisch R. Collagen synthesis by cell lines derived from mov-13 mouse embryos which have a lethal mutation in the collagen alpha 1(I) gene. Biochem J (1987) 244(2):375–9. doi: 10.1042/bj2440375
41. Sofia C, Georgiadou P, Sbarouni E, Voudris V. Chronic heart failure and serum collagen. In: Preedy VR, Patel VB, editors. General methods in biomarker research and their applications. Dordrecht: Springer Netherlands (2015). p. 689–707.
42. Wenstrup RJ, Florer JB, Brunskill EW, Bell SM, Chervoneva I, Birk DE. Type V collagen controls the initiation of collagen fibril assembly *. J Biol Chem (2004) 279(51):53331–7. doi: 10.1074/jbc.M409622200
43. Wenstrup RJ, Florer JB, Davidson JM, Phillips CL, Pfeiffer BJ, Menezes DW, et al. Murine model of the ehlers-danlos syndrome: col5a1 HAPLOINSUFFICIENCY DISRUPTS COLLAGEN FIBRIL ASSEMBLY AT MULTIPLE STAGES*. J Biol Chem (2006) 281(18):12888–95. doi: 10.1074/jbc.M511528200
44. Vater C, Kasten P, Stiehler M. Culture media for the differentiation of mesenchymal stromal cells. Acta Biomater (2011) 7(2):463–77. doi: 10.1016/j.actbio.2010.07.037
45. Yan X, Liu Z, Chen Y. Regulation of TGF-beta signaling by Smad7. Acta Biochim Biophys Sin (Shanghai) (2009) 41(4):263–72. doi: 10.1093/abbs/gmp018
46. Grafe I, Yang T, Alexander S, Homan EP, Lietman C, Jiang MM, et al. Excessive transforming growth factor-β signaling is a common mechanism in osteogenesis imperfecta. Nat Med (2014) 20(6):670–5. doi: 10.1038/nm.3544
47. Infante A, Cabodevilla L, Gener B, Rodríguez CI. Circulating TGF-β pathway in osteogenesis imperfecta pediatric patients subjected to MSCs-based cell therapy. Front Cell Dev Biol (2022) 10:830928. doi: 10.3389/fcell.2022.830928
48. Cao YJ, Wei Z, Zhang H, Zhang ZL. Expanding the clinical spectrum of osteogenesis imperfecta type V: 13 additional patients and review. Front Endocrinol (Lausanne) (2019) 10:375. doi: 10.3389/fendo.2019.00375
49. Salter L, Offiah AC, Bishop N. Elevated platelet counts in a cohort of children with moderate-severe osteogenesis imperfecta suggest that inflammation is present. Arch Dis Child. (2018) 103(8):767–71. doi: 10.1136/archdischild-2017-313859
50. Rolski F, Błyszczuk P. Complexity of TNF-α signaling in heart disease. J Clin Med (2020) 9(10):3267. doi: 10.3390/jcm9103267
51. Stafford N, Assrafally F, Prehar S, Zi M, De Morais AM, Maqsood A, et al. Signaling via the interleukin-10 receptor attenuates cardiac hypertrophy in mice during pressure overload, but not isoproterenol infusion. Front Pharmacol (2020) 11:559220. doi: 10.3389/fphar.2020.559220
52. Takeuchi JK, Mileikovskaia M, Koshiba-Takeuchi K, Heidt AB, Mori AD, Arruda EP, et al. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development (2005) 132(10):2463–74. doi: 10.1242/dev.01827
53. Renault MA, Robbesyn F, Réant P, Douin V, Daret D, Allières C, et al. Osteopontin expression in cardiomyocytes induces dilated cardiomyopathy. Circ Heart Fail (2010) 3(3):431–9. doi: 10.1161/CIRCHEARTFAILURE.109.898114
54. Kratochwil K, Ghaffari-Tabrizi N, Holzinger I, Harbers K. Restricted expression of Mov13 mutant alpha 1(I) collagen gene in osteoblasts and its consequences for bone development. Dev Dyn (1993) 198(4):273–83. doi: 10.1002/aja.1001980405
55. Xu S, Xu H, Wang W, Li S, Li H, Li T, et al. The role of collagen in cancer: From bench to bedside. J Trans Med (2019) 17(1):309. doi: 10.1186/s12967-019-2058-1
56. Chan H, Hartung S, Breindl M. Retrovirus-induced interference with collagen I gene expression in Mov13 fibroblasts is maintained in the absence of DNA methylation. Mol Cell Biol (1991) 11(1):47–54. doi: 10.1128/mcb.11.1.47-54.1991
57. Liska DJ, Slack JL, Bornstein P. A highly conserved intronic sequence is involved in transcriptional regulation of the alpha 1(I) collagen gene. Cell Regul (1990) 1(6):487–98. doi: 10.1091/mbc.1.6.487
58. Bornstein P, McKay J, Liska DJ, Apone S, Devarayalu S. Interactions between the promoter and first intron are involved in transcriptional control of alpha 1(I) collagen gene expression. Mol Cell Biol (1988) 8(11):4851–7. doi: 10.1128/mcb.8.11.4851-4857.1988
59. Bornstein P. Regulation of expression of the alpha 1 (I) collagen gene: A critical appraisal of the role of the first intron. Matrix Biol (1996) 15(1):3–10. doi: 10.1016/S0945-053X(96)90121-3
60. Rahkonen O, Su M, Hakovirta H, Koskivirta I, Hormuzdi SG, Vuorio E, et al. Mice with a deletion in the first intron of the Col1a1 gene develop age-dependent aortic dissection and rupture. Circ Res (2004) 94(1):83–90. doi: 10.1161/01.RES.0000108263.74520.15
61. Frisch BJ, Ashton JM, Xing L, Becker MW, Jordan CT, Calvi LM. Functional inhibition of osteoblastic cells in an in vivo mouse model of myeloid leukemia. Blood (2012) 119(2):540–50. doi: 10.1182/blood-2011-04-348151
62. Giannoni P, Marini C, Cutrona G, Matis S, Capra MC, Puglisi F, et al. Chronic lymphocytic leukemia cells impair osteoblastogenesis and promote osteoclastogenesis: Role of TNFα, IL-6 and IL-11 cytokines. Haematologica (2021) 106(10):2598–612. doi: 10.3324/haematol.2019.231456
63. Beck-Nielsen SS, Mughal Z, Haffner D, Nilsson O, Levtchenko E, Ariceta G, et al. FGF23 and its role in X-linked hypophosphatemia-related morbidity. Orphanet J Rare Dis (2019), 14, 58. doi: 10.1186/s13023-019-1014-8
64. Larsson T, Marsell R, Schipani E, Ohlsson C, Ljunggren O, Tenenhouse HS, et al. Transgenic mice expressing fibroblast growth factor 23 under the control of the α1(I) collagen promoter exhibit growth retardation, osteomalacia, and disturbed phosphate homeostasis. Endocrinology (2004) 145(7):3087–94. doi: 10.1210/en.2003-1768
65. Reinert RB, Bixby D, Koenig RJ. Fibroblast growth factor 23-induced hypophosphatemia in acute leukemia. J Endocr Soc (2018) 2:437–43. doi: 10.1210/js.2018-00010
66. Weidner H, Baschant U, Lademann F, Ledesma Colunga MG, Balaian E, Hofbauer C, et al. Increased FGF-23 levels are linked to ineffective erythropoiesis and impaired bone mineralization in myelodysplastic syndromes. JCI Insight (2020) 5(15). doi: 10.1172/jci.insight.137062
67. Kristianto J, Johnson MG, Afzal R, Blank RD. Endothelin signaling in bone. Endocrinol Metab Clin North Am (2017) 46(1):51–62. doi: 10.1016/j.ecl.2016.09.014
68. Li J, Xu X, Jiang Y, Hansbro NG, Hansbro PM, Xu J, et al. Elastin is a key factor of tumor development in colorectal cancer. BMC Cancer (2020) 20(1):217. doi: 10.1186/s12885-020-6686-x
69. Kasperk CH, Börcsök I, Schairer HU, Schneider U, Nawroth PP, Niethard FU, et al. Endothelin-1 is a potent regulator of human bone cell metabolism in vitro. Calcif Tissue Int (1997) 60(4):368–74. doi: 10.1007/s002239900245
70. Sarathchandra P, Pope FM, Kayser MV, Ali SY. A light and electron microscopic study of osteogenesis imperfecta bone samples, with reference to collagen chemistry and clinical phenotype. J Pathol (2000) 192(3):385–95. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH704>3.0.CO;2-U
Keywords: Mov13, osteogenesis imperfecta, Col1a1, haploinsufficient, bone, collagen, mouse model, skeletal dysplasia
Citation: Claeys L, Zhytnik L, Wisse LE, van Essen HW, Eekhoff EMW, Pals G, Bravenboer N and Micha D (2023) Exploration of the skeletal phenotype of the Col1a1+/Mov13 mouse model for haploinsufficient osteogenesis imperfecta type 1. Front. Endocrinol. 14:1145125. doi: 10.3389/fendo.2023.1145125
Received: 15 January 2023; Accepted: 17 February 2023;
Published: 08 March 2023.
Edited by:
Daniela Merlotti, University of Siena, ItalyReviewed by:
Arantza Infante, Biocruces Bizkaia Health Research Institute, SpainRoy Morello, University of Arkansas for Medical Sciences, United States
Copyright © 2023 Claeys, Zhytnik, Wisse, van Essen, Eekhoff, Pals, Bravenboer and Micha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dimitra Micha, ZC5taWNoYUBhbXN0ZXJkYW11bWMubmw=
†These authors contributed equally to this work and share last authorship
 Lauria Claeys
Lauria Claeys Lidiia Zhytnik
Lidiia Zhytnik Lisanne E. Wisse1
Lisanne E. Wisse1 E. Marelise W. Eekhoff
E. Marelise W. Eekhoff Gerard Pals
Gerard Pals Nathalie Bravenboer
Nathalie Bravenboer Dimitra Micha
Dimitra Micha