
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 15 March 2023
Sec. Clinical Diabetes
Volume 14 - 2023 | https://doi.org/10.3389/fendo.2023.1139805
This article is part of the Research Topic Clinical and Genetic Determinants of Diabetes and Complications View all 31 articles
 Qing Zhang1,2,3
Qing Zhang1,2,3 Yuying Qi4,5
Yuying Qi4,5 Shuanghu Wang6
Shuanghu Wang6 Fangling Zhao4,5
Fangling Zhao4,5 Lili Zou1,2,4
Lili Zou1,2,4 Quan Zhou6
Quan Zhou6 Peiwu Geng6
Peiwu Geng6 Yun Hong2,7
Yun Hong2,7 Hang Yang4,5
Hang Yang4,5 Qingfeng Luo2,7
Qingfeng Luo2,7 Jianping Cai4
Jianping Cai4 Hualan Wu1,2
Hualan Wu1,2 Dongxu Wang1,2
Dongxu Wang1,2 Hao Chen1,2*
Hao Chen1,2* Jiefu Yang1,2,3*
Jiefu Yang1,2,3* Dapeng Dai4*
Dapeng Dai4*Cytochrome P450 2C9 (CYP2C9) participates in about 15% of clinical drug metabolism, and its polymorphism is associated with individual drug metabolism differences, which may lead to the adverse drug reactions (ADRs). In this study, 1163 Chinese Han individuals were recruited to investigate their distribution pattern of CYP2C9 gene and find out the variants that may affect their drug metabolic activities. We successfully developed a multiplex PCR amplicon sequencing method and used it for the genetic screening of CYP2C9 in a large scale. Besides the wild type CYP2C9*1, totally 26 allelic variants of CYP2C9 were detected, which included 16 previously reported alleles and 10 new non-synonymous variants that had not been listed on the PharmVar website. The characteristics of these newly detected CYP2C9 variants were then evaluated after co-expressing them with CYPOR in S. cerevisiae microsomes. Immunoblot analysis revealed that except for Pro163Ser, Glu326Lys, Gly431Arg and Ile488Phe, most of newly detected variants showed comparable protein expression levels to wild type in yeast cells. Two typical CYP2C9 probe drugs, losartan and glimepiride, were then used for the evaluation of metabolic activities of variants. As a result, 3 variants Thr301Met, Glu326Lys, and Gly431Arg almost lost their catalytic activities and most of other variants exhibited significantly elevated activities for drug metabolism. Our data not only enriches the knowledge of naturally occurring CYP2C9 variants in the Chinese Han population, but also provides the fundamental evidence for its potential clinical usage for personalized medicine in the clinic.
Cytochrome P450 (CYP) is one of the critical enzymes involved in the drug metabolism in human. It is responsible for the biotransformation of most foreign substances, including 70-80% of clinically used drugs (1). Variation in clinical response to drug treatment is very common among individuals, and this variation can be affected by many factors, including age, gender, hormone, disease status, genetic polymorphism, and so forth (2–4). It is reported that most CYP enzymes exhibit marked genetic polymorphism, including copy number variation, missense mutation, insertion, deletion, and most of these genetic variations can affect the protein expression level or drug metabolic activity of enzyme. Clinical evidence has confirmed the apparent correlation between genetic polymorphisms of CYP and adverse drug reactions (ADRs), especially for drugs with narrow therapeutic windows (5–7).
The cytochrome P450 2C (CYP2C) subfamily is one of the most important members of the P450 family, with strong correlations with DNA and protein sequences (>82%) (1). Among them, CYP2C9 is the most abundantly expressed in human body, accounting for about 20% of the total liver P450 protein (8). CYP2C9 enzyme is responsible for the metabolism of approximately 15% of drugs, such as the hypoglycemic agent glimepiride and tolbutamide, the anticoagulant warfarin, the antihypertensive drug losartan, the anticonvulsant phenytoin, as well as the non-steroidal anti-inflammatory drugs flurbiprofen and diclofenac (9). Similar to other CYP2C members, the distribution pattern of CYP2C9 polymorphic alleles varies greatly among different populations, and most of allelic variants exhibited significantly changed drug metabolic activities compared with that of the wild type CYP2C9 protein (10–12). In order to carry out the individualized treatment for patients with different pharmacogenetic phenotypes and reduce the occurrence rate of related ADRs, the Pharmacological Clinical Pharmacogenetics Implementation Consortium (CPIC) recently issued three CYP2C9-related guidelines for warfarin (13), phenytoin (14), and non-steroidal anti-inflammatory drugs, respectively (15).
To date, 85 allelic variants of CYP2C9 gene have been discovered and nominated by the Pharmacogene Variation (PharmVar) Consortium (https://www.pharmvar.org/gene/CYP2C9, accessed on Dec 2022). Like other CYP2C members, CYP2C9 gene is highly polymorphic and exhibits different distribution patterns in different races and geographical regions. According to the previous reports, CYP2C9*2 is the most prevalent defective allele in the Caucasian population (11.7%), while it is rarely identified in the Asian population (<0.1%) and African population (2.4%). In contrast, the main allelic variant of CYP2C9 in the Asian population is *3 (3.4% in East Asian and 11.3% in South Asian). CYP2C9*5, *6, *8, *9, and *11 are nearly only restricted to African populations, and CYP2C9*14 is almost uniquely found in South Asian individuals (16). Thus, clinical treatment decision on CYP2C9 meditated drugs in Caucasian populations may not have good generality and adaptability for other national populations. To better understand the specific polymorphic pattern of CYP2C9 gene in the Chinese Han population, we previously conducted a large-scale genetic screening of CYP2C9 in 2124 Chinese Han individuals and reported 21 new allelic variants in healthy subjects. Since then, four additional CYP2C9 alleles CYP2C9*58-*60 and *62 were also identified in the warfarin-sensitive Chinese patients (17–20). Both in vitro and in vivo studies on these newly uncovered CYP2C9 variants revealed that almost all of them exhibited significantly changed metabolic activities, although their allele frequencies are below 1% (21–23). These data indicated that some other rare CYP2C9 alleles may still be undiscovered and need further investigation, considering that more than 1.4 billion Chinese Han populations lived in mainland China.
In this study, 1163 healthy Chinese individuals were used for the genetic polymorphism investigation on CYP2C9 gene by a time- and labor-saving sequencing method. As a consequence, 10 new allelic variants were identified and functional evaluation experiments were also conducted to characterize their impacts on the enzyme’s drug metabolic activity.
The FinePure Universal DNA Purification Kit was purchased from GENFINE Biotech (Beijing, China). The Taq plus master mix was obtained from Vazyme (Nanjing, China). PrimeSTAR Max DNA polymerase, restriction enzymes and DO Supplement-Ura were obtained from Takara Bio, Inc. (Otsu, Shiga, Japan). Saccharomyces cerevisiae strain YPH499 was obtained from ATCC (VA, USA). Yeast nitrogen base without amino acids, dextrose, galactose and losartan were purchased from Sigma-Aldrich (MO, USA). Baculosomes co-expressing human CYP2C9 and NADPH-cytochrome P450 oxidoreductase (OR) were purchased from BD Gentest (Woburn, MA, USA). The rabbit polyclonal anti-CYP2C9 antibody was obtained from Abcam (Cambridge, UK). The mouse monoclonal anti-OR antibody was from Santa Cruz Biotechnology (Dallas, Texas, USA). The Super Signal West Pico Trial Kit was obtained from Thermo Scientific (Rockford, IL, USA). Losartan was purchased from Sigma-Aldrich (St. Louis, MO, USA). Losartan carboxylic acid (E-3174), glimepiride and cyclohexyl hydroxymethyl glimepiride (M1) were obtained from Toronto Research Chemicals, Inc. (Toronto, Ontario, Canada). The NADPH-regenerating system was purchased from Promega (Madison, WI, USA). High-pressure liquid chromatography-grade solvents were purchased from Fisher Scientific Co. (Fair Lawn, NJ, USA). Other chemicals and solvents used were of analytical grade or the highest grade that was commercially available.
All participants in this experiment were healthy Chinese Han individuals recruited in the Physical Examination Center of Beijing Hospital. The written informed consent form was signed when blood collection and this study was approved by the Ethics Committee of Beijing Hospital. FinePure Universal DNA Purification Kit was used to extract DNA from white blood cells following manufacturer’s recommend protocol, and genomic DNAs were diluted to the final concentration of approximately 40 ng/μL for PCR amplification.
To get a time-saving and cost-effective method for the genotyping of CYP2C9 gene, a multiplex PCR amplicon sequencing method was developed in this study (Figure 1). The first round PCR reaction is used for the multiplex PCR amplification of all 9 exons of CYP2C9 plus the exon-intron junction regions, and the second round of PCR reaction is aimed to obtain the amplicon library for the second-generation sequencing. Primers in the first round PCR reaction were designed by MFEPrimer (version 3.1) at the website of iGeneTech(https://mfeprimer3.igenetech.com/muld). Detailed primer information was listed in Table 1. A total amount of 40 ng genomic DNA was used as the input material for two rounds of PCR amplification. After purification with AMPure XP beads (Beckman, USA), barcoded library was quantified with Qubit 3.0 Fluorometer (Thermo Fisher Scientific, USA) and Agilent 2100 Bioanalyzer system (Agilent, USA) was used to measure the concentration and length of library fragments (from 270 to 420 bp). Qualified libraries were then sequenced on NovaSeq 6000 (Illumina, USA) with pair-end 150 sequencing strategy by iGeneTech Co (Beijing, China).
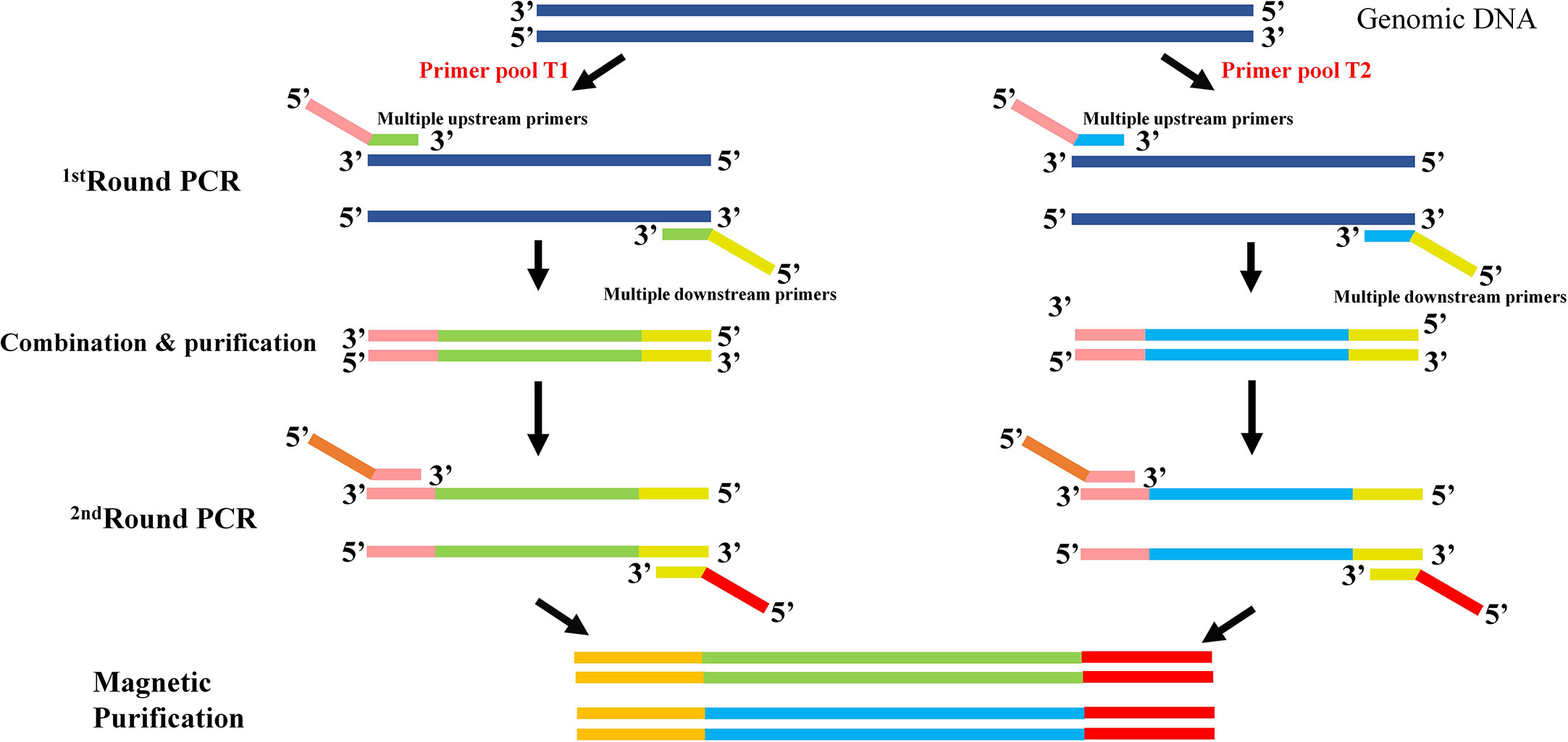
Figure 1 Schematic diagram of the multiplex PCR amplicon sequencing method for CYP2C9 genotyping. Two rounds of PCR amplification were included in this method. The first round of PCR is for the multiplex amplification of all 9 exons of CYP2C9 gene with pooled primers T1 or T2. After the combination and purification with magnetic beads, products were used as the template for the second round of PCR reaction and sequencing library construction. Universal primers in round 1 are illustrated as pink and yellow bars, and sequencing primers in round 2 are shown in brown and red bars.
Raw reads were filtered to remove low quality reads using FastQC (Version 0.11.9) and clean data were mapped to the reference genome GRCh38 and annotated using Annovar software (24). The high-quality annotated data were then obtained after filtering with these parameters: Sequencing depth >50 and detected frequency is within the range of 0.4-0.6 (heterozygote) or 0.9-1.0 (homozygote). Then, detected mutation sites were aligned with PharmVar listed CYP2C9 allele table to identify the allelic variants. For novel variants not included in the allele table, bi-directional sanger sequencing were used for sequence verification with our recently published primers (25).
Full-length cDNA of the typical defective CYP2C9 allele (CYP2C9*3) was constructed using previously described overlap extension PCR amplification method (21). Similarly, cDNA of newly discovered variants were obtained with primer pairs listed in Table 2 using wild type CYP2C9*1 cDNA as the PCR template. The resulting full-length cDNA fragments were double digested with EcoRI and XhoI, and ligated to EcoRI/XhoI digested pESC-OR vector to get the dual expression yeast vector pESC-OR-CYP2C9. Using previously described method, all newly detected CYP2C9 variants were highly expressed with co-expressed CYPOR enzyme in yeast cell microsomes (18). The quantification of expressed CYP2C9 proteins was performed according to our previously reported method (20).
Based on our previously described methods (18, 22, 26), the drug metabolic activities of the wild type, typical defective variant CYP2C9.3, and seven allelic CYP2C9 variants found in this study were assessed with 2 typical CYP2C9 substrates: losartan and glimepiride. Briefly, reaction mixture contained 2-3 pmol of P450 from yeast microsomes, 5 μL purified cytochrome b5, and 2 μL substrates stock solution (dissolved in methanol) in 0.1M K3PO4 buffer (pH 7.5). The ultimate concentrations of losartan and glimepiride were 0.5-50 μM and 0.1-20 μM per reaction, respectively. After a 5 min pre-incubation, an NADPH-regenerating system (1.3 mM NADP+, 3.3 mM glucose-6-phosphate, 3.3 mM MgCl2, and 0.4 unit/mL glucose-6-phosphate dehydrogenase) was added to start the reaction at 37°C in a final volume of 200 μL and proceeded for 30 min (losartan) or 50 min (glimepiride). The incubation was terminated by adding an equal volume of the stop solution containing 150 μL acetonitrile and 50 μL internal standard midazolam (500 ng/mL). After vortexing, the incubated mixture was centrifuged at 12,000 × g for 5 min, and 200 μL aliquots were then removed and used for the following measurements. The incubations were performed in triplicate, and the mean values and S.D. from three experiments were provided for analysis.
Detection and quantification of the metabolites after incubation were performed on the ACQUITY UPLC I-Class/Xevo TQD IVD System (Waters, Milford, MA, USA). Aliquots of samples were placed into an ACQUITY UPLC BEH C18 column (2.1 mm × 50 mm; 1.7 μM; Waters), and the column temperature was maintained at 40°C. The initial mobile phase comprised A (pure acetonitrile, >98%) and B (ultrapure water), and the flow rate was 0.4 mL/min. The detection was performed on a triple quadrupole tandem mass spectrometer equipped with positive electrospray ionization (ESI) by multiple reactions monitoring (MRM) of the transitions. The linearity gradient elution condition for losartan was as following: 0–0.5 min (30%A), 0.5–1.0 min (30-95%A), 1.0–2.0 min (95%A), 2.0–2.3 min (95–30%A); The linearity gradient elution condition for glimepiride was set as 0–0.3 min (10-25%A), 0.3–2.0 min (25-95%A), 2.0–2.5 min (95%A), 2.5–2.6 min (95–10%A). The running time for all detections was 3.0 min. MRM transitions were m/z 437.20 → 235.00, m/z 507.30 → 126.10, and m/z 325.98 → 291.07 for E-3174, hydroxyglimepiride and midazolam, respectively. Nitrogen was used as the desolvation gas (1000 L/h) and cone gas (50 L/h). The dwell time was 0.063 s for E-3174 and 0.108 s for hydroxyglimepiride, the capillary voltage was set as 3.00 kV and the desolvation temperature was maintained at 500°C.
The enzymatic kinetic parameters Km, Vmax, and clearance rate Clint (Vmax/Km) were calculated by GraphPad Prism (version 9; GraphPad Software, Inc., CA, USA). IBM SPSS software (version 25.0, Magneto, New York, USA) was then used to evaluate the catalytic activity difference between the wild type and expressed variants by independent-samples T test.
As illustrated in Figure 1, two rounds of PCR amplification were included in the newly developed multiplex PCR amplicon sequencing method. All 9 exons and exon-intron regions of CYP2C9 gene could be efficiently and specifically amplified after the first round PCR with multiplex PCR primers listed in Table 1. The products were pooled and purified with magnetic kit. Then, the purified amplicons were used for the second round of PCR amplification with universal primers to obtain the library for the second-generation sequencing. Using this system, we efficiently identified 39 allelic variants of CYP2C9 in 1163 individuals, which include 16 previously reported nonsynonymous variations, 13 synonymous variations and 10 new nonsynonymous variations (Table 3). Similar to other studies, the most common defective allele in Chinese Han population is CYP2C9*3 with a allele frequency of 3.998% and 7.57% of studied subjects are heterozygote carrying *1/*3. In addition, 16 previously reported alleles (CYP2C9*2, *8, *13, *16, *29, *31, *34, *36, *37, *39, *45, *48, *53, *56, *60, and *75) were also detected in this study, and most of these allelic variants are heterozygous with the wild type with a total genotype frequency less than 4% which indicates that these alleles are rare in the Han Chinese populations (Table 4).
In this study, 10 non-synonymous CYP2C9 variations (L71R, P163S, T301M, E326K, C372R, I389V, H396Y, N398H, G431R, and I488F) were newly identified, which have not yet been nominated by the Pharmacogene Variation (PharmVar) Consortium (https://www.pharmvar.org/gene/CYP2C9). Their sequencing electropherogram pictures are shown in Figure 2. As illustrated in Table 3, these newly detected variants are located at almost all exons which include exon 2, exon 4, exon 6 - exon 9. Individuals carrying these variants were all heterozygous with wild type CYP2C9*1 and most of the variants could be detected in only one person, except for Asn398His which was found to be carried by two subjects. Specially, 7 of these 10 variants were reported for the first time and could be regarded as novel CYP2C9 variants because they have not been registered by the dbSNP database or any other public databases currently.

Figure 2 Variation verification of newly detected CYP2C9 variants. Sanger sequencing electropherogram pictures of newly detected CYP2C9 variants. The red arrow indicates that the variation sites detected in carriers and the amino acid substitutions are illustrated at the bottom of captured pictures.
In order to characterize the biological effects of newly detected CYP2C9 variants, the yeast expression system was used to efficiently co-express CYP2C9 enzyme and NADPH-cytochrome P450 oxidoreductase (CYPOR) according to the methods described previously (18). Immunoblot results indicated that most of newly detected CYP2C9 variants exhibited comparable protein expression level to that of wild type enzyme CYP2C9.1, except for variants Pro163Ser, Glu326Lys, Gly431Arg and Ile488Phe which showed obviously lower protein expression levels than the wild type (Figure 3).
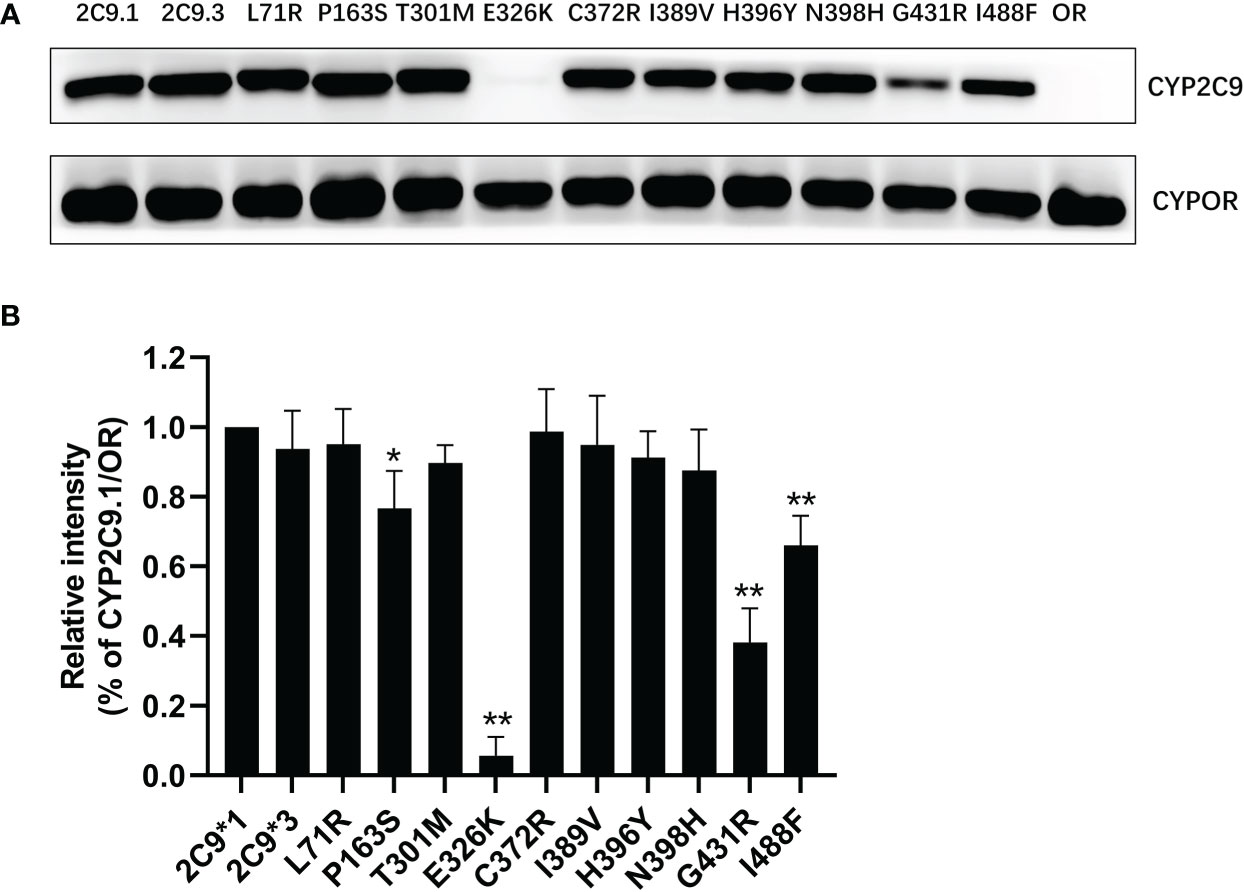
Figure 3 The immunoblotting results for expressed CYP2C9 variants in yeast microsomes. (A) Expressed CYP2C9 variants and CYPOR enzyme were detected by their corresponding antibodies after the SDS-PAGE gel separation. OR: microsome from yeast cells only expressing CYPOR enzyme. (B) Relative CYP2C9/OR intensities. Each bar represents the mean ± SD of three independently experiments. *P < 0.05, **P < 0.01 vs CYP2C9.1/OR.
To better understand the impacts of newly detected CYP2C9 variants on drug metabolic activity of enzyme, two typical CYP2C9 mediated drugs losartan and glimepiride were included in this study. As a result, three variants (Thr301Met, Glu326Lys and Gly431Arg) showed no catalytic activities towards both drugs. Whereas 4 variants exhibited elevated activities for the metabolism of losartan (Table 5) and 6 variants exhibited increased intrinsic clearance rate for glimepiride, as compared with the wild type enzyme CYP2C9.1 (Table 6). These data indicated that most of newly detected CYP2C9 variants could significantly change the metabolic ability of enzyme (Figures 4, 5).
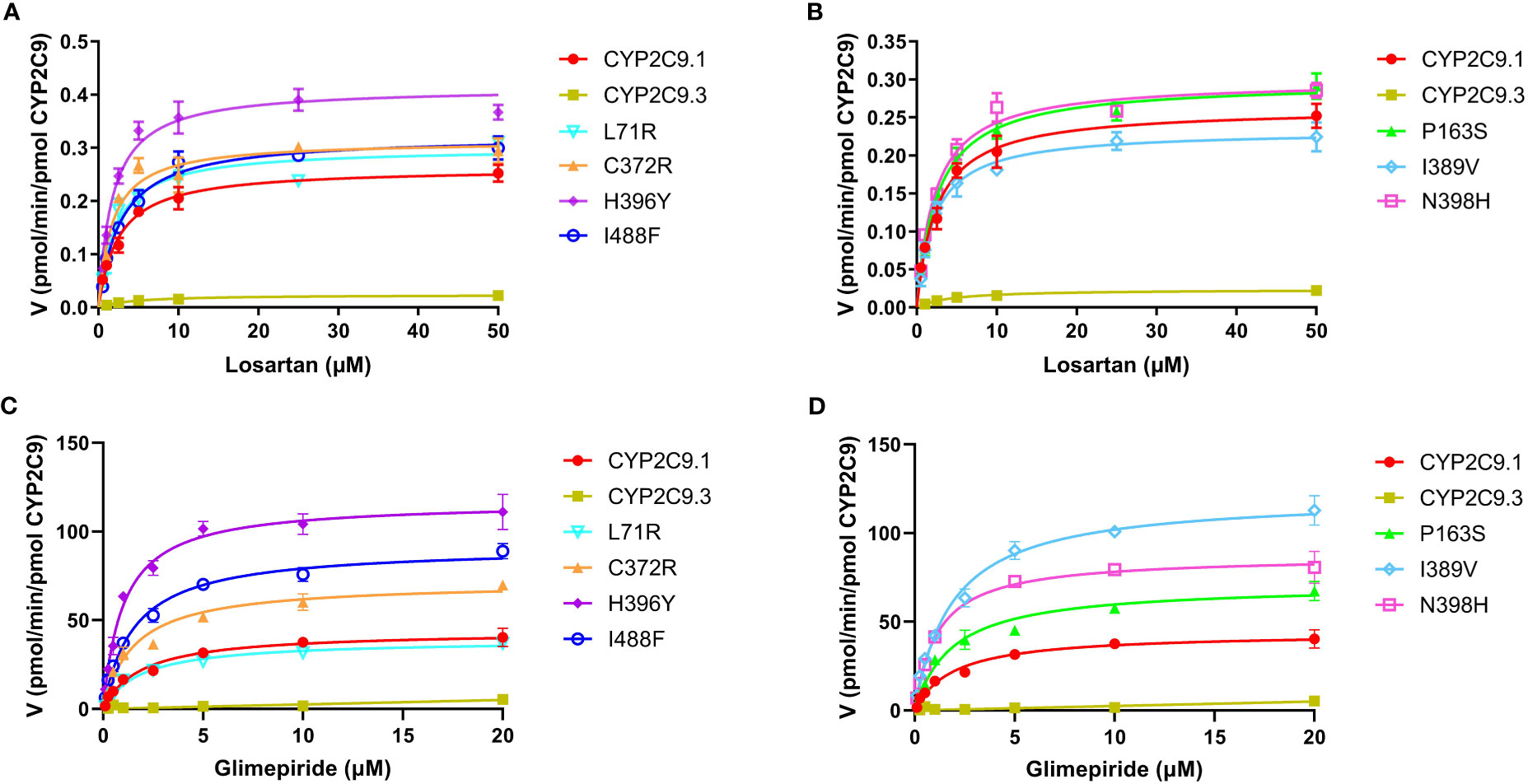
Figure 4 Michaelis-Menten curves of the enzymatic activities of expressed CYP2C9 variants toward losartan (A, B) and glimepiride (C, D). Each point represents the mean ± S.D. of 3 separate experiments.
In this study, we developed a timesaving and cost-effective genotyping method for CYP2C9 gene. This method is based on the combination of MPCR (Multiplex polymerase chain reaction) and NGS (Next generation sequencing) techniques and has many advantages over traditional Sanger sequencing method. Firstly, this method is easy to be operated in a large scale with automatic protocol, leading to the lower cost, reduced man-made errors and improved accuracy than Sanger method; Secondly, newly developed method can present overall genetic information of CYP2C9 gene in the target region, that can be used for the genotyping of previously reported alleles and for the discovery of novel variants with unreported mutations, simultaneously; Finally, short time, typically only 2-3 weeks, is needed for the genotyping of hundreds of the samples. In contrast, for traditional Sanger sequencing method, several months maybe needed for the sequencing and analyzing of all 9 exons of CYP2C9 in a large scale. In brief, our method not only reduces the costs for genotyping, but also greatly improves the efficiency and accuracy of sequencing, favoring its application in large sample scale, multi-center or multi-targets genotyping projects.
Like other CYP2C members, CYP2C9 gene shows marked differences in the allelic frequency in different biogeographic groups and races. These genetic polymorphisms are highly related to the adverse drug reactions (ADRs), especially for the drugs with narrow therapeutic window (27), such as the hypoglycemia caused by hypoglycemic drugs (28), the gastrointestinal bleeding caused by non-steroidal anti-inflammatory drugs (29, 30), and severe bleeding caused by anticoagulation therapy (31, 32), etc. Therefore, digging out the “special subgroups” with abnormal drug metabolism in the population is one of the key factors for reducing the occurrence of ADRs in the clinic. For instance, warfarin is the most commonly used oral anticoagulant, but its therapeutic index is narrow and wildly variable among different patients. Genetic polymorphisms of CYP2C9 and Vitamin K epoxide reductase complex subunit 1 (VKORC1) are one of the most concerned factors for the optimal warfarin dose determination in clinic (33). S-warfarin is mainly metabolized via CYP2C9 to 7-hydroxy warfarin. Typical missense variant CYP2C9*3 caused a remarkable decrease in the S-warfarin clearance rate, leading to the increased risk of venous thromboembolism and bleeding in patients (13). Our recent studies revealed that a lot of rare CYP2C9 alleles are carried by Chinese individuals and most of missense mutations in CYP2C9 gene are highly related to the low dose of warfarin in Chinese population (18, 20, 34). In this study, we developed one time-saving and cost-effective genotyping method for CYP2C9 and performed a genetic screening in 1163 Chinese individuals. Similar to our previous study, CYP2C9*3 is the most prevalent defective alleles in Chinese population although the allele frequency detected in this study is slightly higher than previous report (17). Additionally, CYP2C9*13 and *29 exhibited relatively higher frequencies than other allelic variants which is in agreement to our previous reports (17, 25). For the first time, we reported one Chinese individual carrying allele CYP2C9*8 which was previously regarded as only limited to individuals of African ancestry (35). Specially, we detected 10 new allelic variants that have not been listed on the PharmVar consortium website (Table 3; Figure 2). These data indicated that CYP2C9 was highly polymorphic in Chinese population and more attention should be paid to the distribution pattern and its potential clinical application in clinic, considering that more than 1.4 billion people lived in the mainland of China.
Glimepiride is one of the most used oral sulfonylureas (SU) drugs in the clinical treatment of type 2 diabetes mellitus (T2DM). Hypoglycemia is the most common adverse effect related to SU therapy and severe hypoglycemia might significantly increase the cost of medication and decrease the quality of life for T2DM patients (36, 37). Since CYP2C9 is the major enzyme involved in SUs metabolism, the risk of hypoglycemia induced by SUs would be elevated in deleterious CYP2C9 variant allele carriers (38). According to a recent meta-analysis, CYP2C9 variant alleles have increased risk of hypoglycemia than wild-type CYP2C9*1/*1 after the SUs treatment. The incidence of hypoglycemia would be increased by 80% in CYP2C9*2 carrier (39). Previous studies have also reported that the AUC of tolbutamide was increased by 150% and 190% in CYP2C9*1/*2 and *1/*3 carriers, respectively (40); Similarly, for glimepiride, the AUC was increased by 167% in CYP2C9*3 carriers in comparison to CYP2C9*1/*1 individuals (41). In this study, the in vitro metabolic activity analysis results revealed that 3 newly detected CYP2C9 variants had no catalytic activity for glimepiride metabolizing and carriers for these variants might exhibit significantly reduced drug metabolizing activity for SU. However, most of other newly detected allelic variants exhibited significantly increased enzyme activity for glimepiride metabolism which indicated that carriers with these variants might possess the higher drug metabolizing activity for SU than individuals with wild type CYP2C9*1/*1. (Figures 4, 5 and Table 6). These data indicated that different amino acid substitution at different sites of CYP2C9 protein had different effects on the drug metabolizing activity of enzyme.
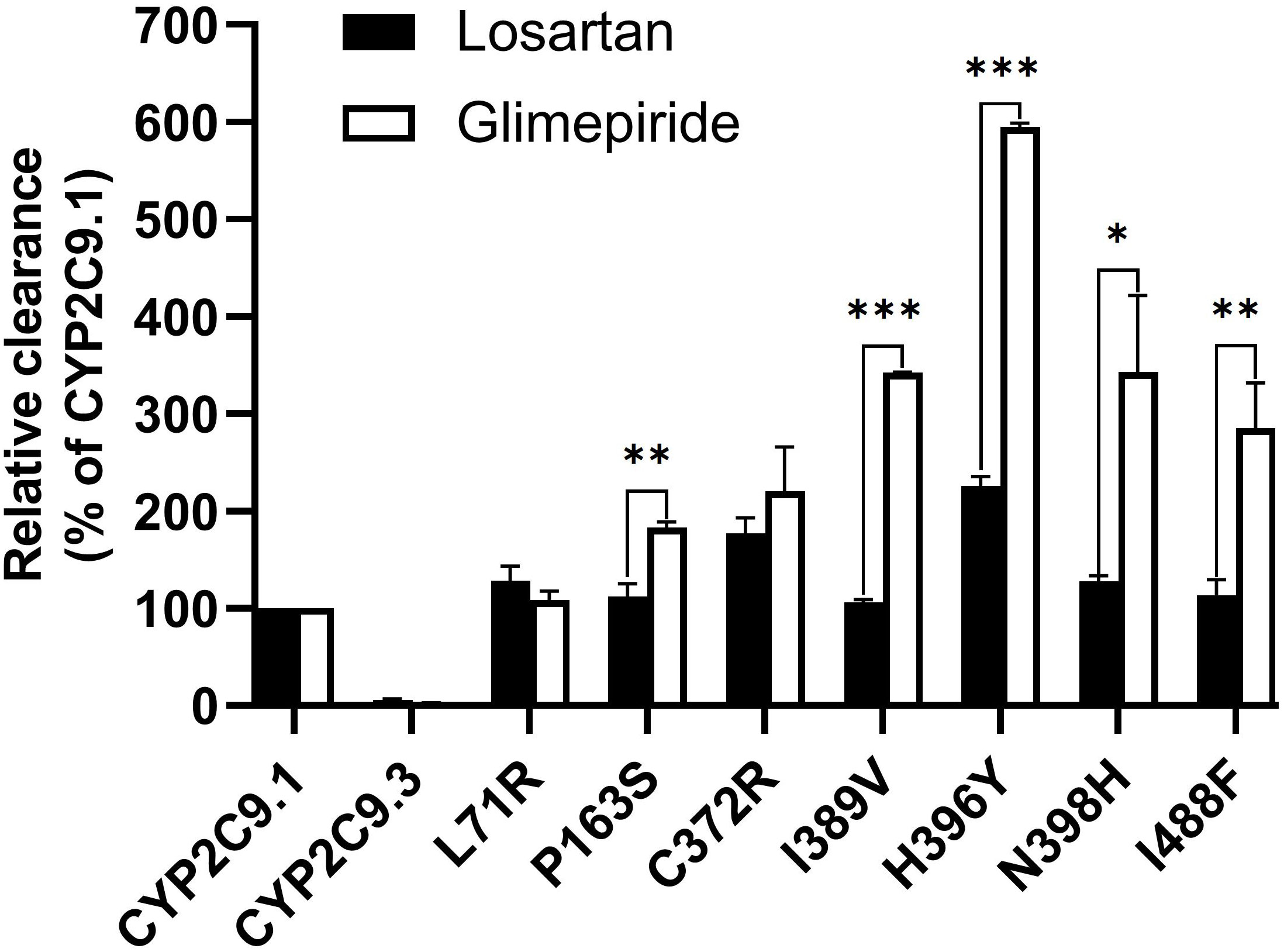
Figure 5 The relative clearance rates of losartan and glimepiride among wild type CYP2C9.1, typical defective variant CYP2C9.3 and 7 newly detected variants. *P < 0.05, **P < 0.01, and ***P < 0.005.
Typical tertiary structure of a cytochrome P450 enzyme mainly consists of twelve α-helices (A-L) and four β-sheets (1–4) with the heme locating between the helices I and L. There are six substrate recognition sites (SRSs) in CYP2C9 enzyme which locate at the amino acids 96-117 (between the helices B and C), 198-205 (between the helices F and G), 233-240 (between the helices F and G), 286-304 (in the center of the helix I), 359-369 (at the N-terminus of β strand 1-4), and 470-477 (at the turn at the end of β sheet 4), respectively (42). In this study, 3 allelic variants, Thr301Met, Glu326Lys and Gly431Arg showed activity deficiency for both losartan and glimepiride. In the crystal structure of CYP2C9, Thr301 is involved in the SRS4, and Gly431 belongs to heme-binding motif residues. Therefore, the amino acid substitution at position 301 or 431 is estimated to affect the substrate recognition or heme propionate binding capacity for CYP2C9. Similar to our results, another allelic variant at position 301, Thr301Lys, also showed no enzymatic activity (43). These data indicated that Thr301 might be crucial for the drug metabolic activity of enzyme. Glu326 is located at the helix J of CYP2C9 and it has a strong binding strength with 5 amino acid residues within 5Å distance. Previous study revealed that variant Glu326Asp (CYP2C9*65) had deleterious effect in SIFT and Polyphen prediction (12). Combined with the data in this study, it is estimated that the replacement of Glu326 might influence the enzyme activity significantly. Different from these 3 defective variants, most of other newly detected variants exhibited significantly increased metabolizing activities towards both losartan and glimepiride in vitro (Figure 5; Tables 5, 6). These data indicated that carriers with these allelic variants might have higher metabolizing activity for CYP2C9 mediated drugs.
In summary, we developed a time-saving next generation sequencing based method for CYP2C9 genotyping and performed a large-scale polymorphic screening of CYP2C9 gene in Chinese Han population. Totally 16 previously reported allelic variants and 10 new non-synonymous variations were detected in this study. When expressed in yeast microsomes, most of newly detected variations showed similar protein expression level to wild type. Further drug metabolic activity analysis revealed that 3 variants were loss of function isoforms and most of other newly detected variants exhibited significantly increased metabolizing activities for both losartan and glimepiride. Our study greatly enriched the knowledge of genetic polymorphism of CYP2C9 in Chinese Han population, and the clinical significance of newly detected CYP2C9 alleles still needs further investigation by enlarging the sample size and deep correlation analysis between genetic information and clinical features.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Genbank-BankIt2667312 Seq_C487T OQ376733, BankIt2667312 Seq_G976A OQ376734, BankIt2667312 Seq_T1114C OQ376735, BankIt2667312 Seq_A1165G OQ376736, BankIt2667312 Seq_C1186T OQ376737, BankIt2667312 Seq_A1192C OQ376738, BankIt2667312 Seq_G1291A OQ376739.
The studies involving human participants were reviewed and approved by the ethics committee of Beijing Hospital. The patients/participants provided their written informed consent to participate in this study.
DD, JY, and HC contributed to conception and design of the study. QiZ, YQ, SW, FZ, LZ, QuZ, PG, YH, and HY performed the experiments. QiZ, YQ, SW, FZ, QuZ, QL, JC, HW, and DW performed the statistical analysis. QiZ wrote the first draft of the manuscript. YQ and DD wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by grants from the National High Level Hospital Clinical Research Funding (BJ-2022-121), CAMS Innovation Fund for Medical Sciences (2021-I2M-1-050) and the National Key R&D Program of China (2020YFC2008301).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther (2013) 138(1):103–41. doi: 10.1016/j.pharmthera.2012.12.007
2. Zanger UM, Klein K, Richter T, Toscano C, Zukunft J. Impact of genetic polymorphism in relation to other factors on expression and function of human drug-metabolizing P450s. Toxicol Mech Methods (2005) 15(2):121–4. doi: 10.1080/15376520590918847
3. Parkinson A, Mudra DR, Johnson C, Dwyer A, Carroll KM. The effects of gender, age, ethnicity, and liver cirrhosis on cytochrome P450 enzyme activity in human liver microsomes and inducibility in cultured human hepatocytes. Toxicol Appl Pharmacol (2004) 199(3):193–209. doi: 10.1016/j.taap.2004.01.010
4. Harvey RD, Morgan ET. Cancer, inflammation, and therapy: Effects on cytochrome P450-mediated drug metabolism and implications for novel immunotherapeutic agents. Clin Pharmacol Ther (2014) 96(4):449–57. doi: 10.1038/clpt.2014.143
5. Saiz-Rodriguez M, Belmonte C, Roman M, Ochoa D, Koller D, Talegon M, et al. Effect of polymorphisms on the pharmacokinetics, pharmacodynamics and safety of sertraline in healthy volunteers. Basic Clin Pharmacol Toxicol (2018) 122(5):501–11. doi: 10.1111/bcpt.12938
6. Park JW, Kim KA, Park JY. Effects of ketoconazole, a Cyp4f2 inhibitor, and Cyp4f2*3 genetic polymorphism on pharmacokinetics of vitamin K1. J Clin Pharmacol (2019) 59(11):1453–61. doi: 10.1002/jcph.1444
7. Pereira NL, Rihal CS, So DYF, Rosenberg Y, Lennon RJ, Mathew V, et al. Clopidogrel pharmacogenetics. Circ Cardiovasc Interventions (2019) 12(4):e007811. doi: 10.1161/circinterventions.119.007811
8. Zhang HF, Wang HH, Gao N, Wei JY, Tian X, Zhao Y, et al. Physiological content and intrinsic activities of 10 cytochrome P450 isoforms in human normal liver microsomes. J Pharmacol Exp Ther (2016) 358(1):83–93. doi: 10.1124/jpet.116.233635
9. Rendic S. Summary of information on human cyp enzymes: Human P450 metabolism data. Drug Metab Rev (2002) 34(1-2):83–448. doi: 10.1081/dmr-120001392
10. McGraw J, Waller D. Cytochrome P450 variations in different ethnic populations. Expert Opin Drug Metab Toxicol (2012) 8(3):371–82. doi: 10.1517/17425255.2012.657626
11. Waring RH. Cytochrome P450: Genotype to phenotype. Xenobiotica fate foreign compounds Biol Syst (2020) 50(1):9–18. doi: 10.1080/00498254.2019.1648911
12. Nizamuddin S, Dubey S, Singh S, Sharma S, Machha P, Thangaraj K. Cyp2c9 variations and their pharmacogenetic implications among diverse south Asian populations. Pharmacogenomics personalized Med (2021) 14:135–47. doi: 10.2147/pgpm.s272015
13. Johnson JA, Caudle KE, Gong L, Whirl-Carrillo M, Stein CM, Scott SA, et al. Clinical pharmacogenetics implementation consortium (Cpic) guideline for pharmacogenetics-guided warfarin dosing: 2017 update. Clin Pharmacol Ther (2017) 102(3):397–404. doi: 10.1002/cpt.668
14. Karnes JH, Rettie AE, Somogyi AA, Huddart R, Fohner AE, Formea CM, et al. Clinical pharmacogenetics implementation consortium (Cpic) guideline for Cyp2c9 and hla-b genotypes and phenytoin dosing: 2020 update. Clin Pharmacol Ther (2021) 109(2):302–9. doi: 10.1002/cpt.2008
15. Theken KN, Lee CR, Gong L, Caudle KE, Formea CM, Gaedigk A, et al. Clinical pharmacogenetics implementation consortium guideline (Cpic) for Cyp2c9 and nonsteroidal anti-inflammatory drugs. Clin Pharmacol Ther (2020) 108(2):191–200. doi: 10.1002/cpt.1830
16. Zhou Y, Ingelman-Sundberg M, Lauschke VM. Worldwide distribution of cytochrome P450 alleles: A meta-analysis of population-scale sequencing projects. Clin Pharmacol Ther (2017) 102(4):688–700. doi: 10.1002/cpt.690
17. Dai DP, Xu RA, Hu LM, Wang SH, Geng PW, Yang JF, et al. Cyp2c9 polymorphism analysis in han Chinese populations: Building the largest allele frequency database. Pharmacogenomics J (2014) 14(1):85–92. doi: 10.1038/tpj.2013.2
18. Dai DP, Li CB, Wang SH, Cai J, Geng PW, Zhou YF, et al. Identification and characterization of a novel Cyp2c9 allelic variant in a warfarin-sensitive patient. Pharmacogenomics (2015) 16(13):1475–86. doi: 10.2217/pgs.15.89
19. Dai DP, Wang SH, Li CB, Geng PW, Cai J, Wang H, et al. Identification and functional assessment of a new Cyp2c9 allelic variant Cyp2c9*59. Drug Metab Dispos (2015) 43(8):1246–9. doi: 10.1124/dmd.115.063412
20. Chen H, Dai DP, Zhou S, Liu J, Wang SH, Wu HL, et al. An identification and functional evaluation of a novel Cyp2c9 variant Cyp2c9*62. Chem Biol Interact (2020) 327:109168. doi: 10.1016/j.cbi.2020.109168
21. Dai DP, Wang YH, Wang SH, Geng PW, Hu LM, Hu GX, et al. In vitro functional characterization of 37 Cyp2c9 allelic isoforms found in Chinese han population. Acta Pharmacol Sin (2013) 34(11):1449–56. doi: 10.1038/aps.2013.123
22. Dai DP, Wang SH, Geng PW, Hu GX, Cai JP. In vitro assessment of 36 Cyp2c9 allelic isoforms found in the Chinese population on the metabolism of glimepiride. Basic Clin Pharmacol Toxicol (2014) 114(4):305–10. doi: 10.1111/bcpt.12159
23. Hu GX, Pan PP, Wang ZS, Yang LP, Dai DP, Wang SH, et al. In vitro and in vivo characterization of 13 Cyp2c9 allelic variants found in Chinese han population. Drug Metab Dispos (2015) 43(4):561–9. doi: 10.1124/dmd.114.061200
24. Wang K, Li M, Hakonarson H. Annovar: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res (2010) 38(16):e164. doi: 10.1093/nar/gkq603
25. Zhao FL, Zhang Q, Wang SH, Hong Y, Zhou S, Zhou Q, et al. Identification and drug metabolic characterization of four new Cyp2c9 variants Cyp2c9*72-*75 in the Chinese han population. Front Pharmacol (2022) 13:1007268. doi: 10.3389/fphar.2022.1007268
26. Wang YH, Pan PP, Dai DP, Wang SH, Geng PW, Cai JP, et al. Effect of 36 Cyp2c9 variants found in the Chinese population on losartan metabolism in vitro. Xenobiotica (2014) 44(3):270–5. doi: 10.3109/00498254.2013.820007
27. Wang B, Wang J, Huang SQ, Su HH, Zhou SF. Genetic polymorphism of the human cytochrome P450 2c9 gene and its clinical significance. Curr Drug Metab (2009) 10(7):781–834. doi: 10.2174/138920009789895480
28. Zeng W, Guo Y, Chen P, Liu Z, Chen D, Han C. Cyp2c93 variant is associated with antidiabetes efficacy of gliclazide in Chinese type 2 diabetes patients. J Diabetes Investig (2016) 7(5):764–8. doi: 10.1111/jdi.12486
29. Pilotto A, Seripa D, Franceschi M, Scarcelli C, Colaizzo D, Grandone E, et al. Genetic susceptibility to nonsteroidal anti-inflammatory drug-related gastroduodenal bleeding: Role of cytochrome P450 2c9 polymorphisms. Gastroenterology (2007) 133(2):465–71. doi: 10.1053/j.gastro.2007.05.025
30. Macias Y, Gomez Tabales J, Garcia-Martin E, Agundez JAG. An update on the pharmacogenomics of nsaid metabolism and the risk of gastrointestinal bleeding. Expert Opin Drug Metab Toxicol (2020) 16(4):319–32. doi: 10.1080/17425255.2020.1744563
31. Kamali F, Wynne H. Pharmacogenetics of warfarin. Annu Rev Med (2010) 61:63–75. doi: 10.1146/annurev.med.070808.170037
32. Su Q, Li J, Tang Z, Yang S, Xing G, Liu T, et al. Association of Cyp2c19 polymorphism with clopidogrel resistance in patients with acute coronary syndrome in China. Med Sci Monit (2019) 25:7138–48. doi: 10.12659/MSM.915971
33. Cini M, Legnani C, Cosmi B, Guazzaloca G, Valdre L, Frascaro M, et al. A new warfarin dosing algorithm including Vkorc1 3730 G > a polymorphism: Comparison with results obtained by other published algorithms. Eur J Clin Pharmacol (2012) 68(8):1167–74. doi: 10.1007/s00228-012-1226-5
34. Wang D, Wu H, Dong M, Zhang Q, Zhao A, Zhao X, et al. Clinical significance of the series of Cyp2c9*Non3 variants, an unignorable predictor of warfarin sensitivity in Chinese population. Front Cardiovasc Med (2022) 9:1052521. doi: 10.3389/fcvm.2022.1052521
35. Parikh SJ, Kamat S, Phillips M, Boyson SP, Yarbrough T, Davie D, et al. Insights into the genetic variations of human cytochrome P450 2c9: Structural analysis, characterization and comparison. Int J Mol Sci (2021) 22(19):10206. doi: 10.3390/ijms221910206
36. Anna-Maria O, Martin Hrabě de A, Hans-Ulrich H, Staiger H. Pharmacogenetics of oral antidiabetic therapy. Pharmacogenomics (2018) 19(6):577—87. doi: 10.2217/pgs-2017-0195
37. Fokoun C, Serrier H, Rabier H, Goutelle S, Tod M, Bourguignon L. Pharmacogenetic-guided glimepiride therapy in type-2 diabetes mellitus: A cost-effectiveness study. pharmacogenomics J (2021) 21(5):559–65. doi: 10.1038/s41397-021-00232-w
38. Aquilante CL. Sulfonylurea pharmacogenomics in type 2 diabetes: The influence of drug target and diabetes risk polymorphisms. Expert Rev Cardiovasc Ther (2010) 8(3):359–72. doi: 10.1586/erc.09.154
39. Yee J, Heo Y, Kim H, Yoon HY, Song G, Gwak HS. Association between the Cyp2c9 genotype and hypoglycemia among patients with type 2 diabetes receiving sulfonylurea treatment: A meta-analysis. Clin Ther (2021) 43(5):836–43.e4. doi: 10.1016/j.clinthera.2021.03.008
40. Lee CR, Pieper JA, Hinderliter AL, Blaisdell JA, Goldstein JA. Evaluation of cytochrome P4502c9 metabolic activity with tolbutamide in Cyp2c91 heterozygotes. Clin Pharmacol Ther (2002) 72(5):562–71. doi: 10.1067/mcp.2002.127913
41. Niemi M, Cascorbi I, Timm R, Kroemer HK, Neuvonen PJ, Kivistö KT. Glyburide and glimepiride pharmacokinetics in subjects with different Cyp2c9 genotypes. Clin Pharmacol Ther (2002) 72(3):326–32. doi: 10.1067/mcp.2002.127495
42. Nair PC, McKinnon RA, Miners JO. Cytochrome P450 structure-function: Insights from molecular dynamics simulations. Drug Metab Rev (2016) 48(3):434–52. doi: 10.1080/03602532.2016.1178771
Keywords: CYP2C9, allelic variant, genetic polymorphism, drug metabolism, Chinese Han population
Citation: Zhang Q, Qi Y, Wang S, Zhao F, Zou L, Zhou Q, Geng P, Hong Y, Yang H, Luo Q, Cai J, Wu H, Wang D, Chen H, Yang J and Dai D (2023) Identification and in vitro functional assessment of 10 CYP2C9 variants found in Chinese Han subjects. Front. Endocrinol. 14:1139805. doi: 10.3389/fendo.2023.1139805
Received: 07 January 2023; Accepted: 22 February 2023;
Published: 15 March 2023.
Edited by:
Sen Li, Beijing University of Chinese Medicine, ChinaReviewed by:
Xiaoqiang Xiang, Fudan University, ChinaCopyright © 2023 Zhang, Qi, Wang, Zhao, Zou, Zhou, Geng, Hong, Yang, Luo, Cai, Wu, Wang, Chen, Yang and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dapeng Dai, ZGFpZGFwZW5nQDE2My5jb20=; Jiefu Yang, eWFuZ2pmYmpoMjAyMkAxNjMuY29t; Hao Chen, ZHIuY2hlbmhhb21kX2JqaEB2aXAuMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.