
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 10 March 2023
Sec. Reproduction
Volume 14 - 2023 | https://doi.org/10.3389/fendo.2023.1138096
This article is part of the Research Topic Male Reproduction and Oxidative Stress View all 10 articles
 Lin Wang1†
Lin Wang1† Yan-Ling Yong1†
Yan-Ling Yong1† Kun-Kun Wang1†
Kun-Kun Wang1† Yun-Xia Xie1
Yun-Xia Xie1 Ying-Chen Qian2
Ying-Chen Qian2 Feng-Mei Zhou1
Feng-Mei Zhou1 Jian-Ge Qiu1*
Jian-Ge Qiu1* Bing-Hua Jiang1*
Bing-Hua Jiang1*Makorin-2 (Mkrn2) is an evolutionarily conserved gene whose biological functions are not fully known. Although recent studies have shed insights on the potential causes of male infertility, its underlining mechanisms still remain to be elucidated. We developed a Mrkn2 knockout mice model to study this gene and found that deletion of Mkrn2 in mice led to male infertility. Interestingly, the expression level of signal transducer and activator of the transcription (STAT)1 was significantly decreased in MKRN2 knockout testis and MEF cells. Co-IP assay showed an interaction between MKRN2 and STAT1. Moreover, our results further indicated that MKRN2 regulated the expression level of SIX4 and tenascin C (TNC) via the EBF transcription factor 2 (EBF2) in mice. The results of our study will provide insights into a new mechanism of male infertility.
Makorin-2 (MKRN2; HSPC070) belongs to the MKRN gene family with the ribonucleoproteins characterized by a variety of zinc-finger motifs, which contains four C3H zinc fingers and a C3HC4 really interesting new gene (RING) zinc finger domain (1, 2). MKRN2 was first identified in human CD34+stem/progenitor cells, as well as in some leukemic cell lines (1, 3, 4). Previous studies reported that mkrn2 in Xenopus laevis acted upstream of glycogen synthase kinase-3b in the phosphatidylinositol 3-kinase/Akt pathway. The third C3H zinc finger and the RING motif are required for the antineurogenesis activity (5, 6). Recent studies also investigated the role of MKRN2 in tumorigenesis, such as lung cancer and melanoma (7, 8). Though MKRN2 is a highly conserved gene, however, its function remains largely unknown.
The signal transducer and activator of the transcription (STAT) protein family mediate the transcription of several genes, such as cytokine-inducible genes and growth factors (9–12). So far, seven STAT proteins have been identified: STAT1–6 and STAT5b (13, 14). When the receptor bounds to STATs, a conserved tyrosine residue in the C-terminal domain will be phosphorylated by the Janus kinase (JAK), then two STATs assemble through reciprocal phosphotyrosine/SH2 domain interactions leading to dimerization. Once dimerized, STATs are translocated into the nucleus and regulate the transcription of several different genes. Truchet et al. reported that JAK/STAT pathway is functional during early embryonic development, and STAT1 is present in mouse oocytes and in preimplantation embryos (15). STAT1 was reported to be phosphorylated in response to capacitation and the acrosomal reaction (16). Moreover, human samples with varicocele conveyed a significant negative correlation between the phosphorylated levels of STAT1 and sperm head morphological defects (17). Moreover, The Six4 genes belong to the mammalian homolog of the Drosophila sine oculis homeobox (Six) family, and studies have reported that SIX4 is required for genital primordium formation and testicular differentiation of male gonads (18). SIX4 and tenascin C (TNC) have been reported to be functioning in a productive system. Thus, we wonder how to evaluate the regulatory mechanism between MKRN2 and STAT1/SIX4/TNC to figure out the novel mechanism of MKRN2 in male infertility.
In this study, we plan to address: (a) the role of MKRN2 in male infertility; (b) whether MKRN2 regulates the expression level of STAT1; and (c) how MKRN2 induces the expression levels of SIX4 and TNC by the transcription factor EBF transcription factor 2 (EBF2). The results of our study will provide insights into a new mechanism of MKRN2 in regulating male infertility.
To generate Mkrn2-heterozygous and Mkrn2-knockout mice, Mkrn2-floxed mice have been crossed with transgenic EIIa-cre mice with a C57BL/6 J background as we previously described (19) to obtain mosaic mice with the Mkrn2+/flox(−)·EIIa-cre genotype. The resulting mice were crossed with C57BL/6 J to obtain Mkrn2 heterozygotes. Mkrn2 knockout mice were generated by sibmating Mkrn2 heterozygotes. Genomic DNAs isolated from the tails were genotyped with indicated primers by PCR as described (19). All the animals were housed in specific pathogen-free conditions, and all the experiments were approved by the Committee of Laboratory Animal Experimentation of Zhengzhou University.
Mkrn2-WT and Mkrn2-KO primary mouse embryonic fibroblasts (MEFs) were derived from 13.5-day embryos and cultured as described (19), and 293T cells were cultured in the DMEM medium supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 μg/ml), and were maintained at 37°C.
The H&E staining procedure used here is as follows: (a) Deparaffinize all sections in xylene two times for 10 min each. (b) Gradually rehydrate sections with graded alcohol: wash in absolute alcohol two times for 5 min each, then 95% alcohol for 2 min, and 70% alcohol for 2 min. (c) Briefly wash in distilled water. (d) Stain in Harris hematoxylin solution for 8 min, then wash for 5 min under running water. (e) Differentiate in 1% acid alcohol for 30 s, then wash under running water for 1 min. (f) Blue in 0.2% ammonia water for 30 s, then wash under running water for 5 min. (g) Rinse in 95% alcohol at 10 dips. (h) Counterstain in an eosin–phloxine solution for 1 min. (i) Dehydrate with 95% alcohol and wash with absolute alcohol two times at 5 min each. (j) Clear in xylene two times at 5 min each. (k) Lastly, mount with a xylene-based mounting medium.
Cells were harvested, washed with cold PBS twice, and then suspended in 200 μl of cold cell lysed buffer with protease inhibitor. Tissues were homogenized and suspended in 500 μl of cold cell lysed buffer with protease inhibitor. All of the lysates were incubated on ice for 30 min and centrifuged for 10 min at 4°C; the supernatants were collected, and the protein concentration was quantified using the Bradford method (20). The cell lysates were resolved by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes. After being incubated in the blocking buffer (5% BSA in TBST) for 2 h at room temperature, the membranes were incubated with primer antibodies overnight at 4°C, washed three times with TBST, and incubated with HRP-conjugated secondary antibodies for 1 h at room temperature, then washed three times more with TBST. The protein–antibody complex was detected by a chemiluminescence detection system via an ECL chemiluminescence detection kit.
Quantitative real-time RT-PCR was used to determine the gene expression levels. RNAs were extracted using a Trizol reagent, reverse transcribed using SYBR Premix Dimer Eraser, and real-time PCR was performed using QuautStudio-5 Real-Time Thermal Cycler (ABI, Los Angeles, CA, USA). The following primer sequences were used for Q-PCR: Six4 forward primer: 5′-CTCCTGTCTCAGTAGCAGCTTC-3′; reverse primer: 5′-GGAACGGTGTATACCACTGCAC-3′; Ebf2 forward primer: 5′-GAGCAAGAAGGCTTGACCCATC-3′; reverse primer: 5′-CCAAACACAACCTGGAGACCATC-3′; Tnc forward primer: 5′-GAGACCTGACACGGAGTATGAG-3′; reverse primer: 5′-CTCCAAGGTGATGCTGTTGTCTG-3′; and GAPDH forward primer: 5′-ATGGGTGTGAACCATGAGAAGTATG-3′; reverse primer: 5′-GGTGCAGGAGGCATTGCT-3′. The gene expression levels were normalized to the value of GAPDH, and fold changes were calculated by relative quantification (2−ΔΔCt).
HEK293T cells were cultured in a 10-cm plate and transfected with the indicated plasmids for 48–72 h. Cells were then collected and lysed in immunoprecipitation (IP) buffer (1 M Tris, 0.5 M EDTA, 5 M NaCl, 100% glycerol, 100% Np40) and treated with antibodies at 4°C overnight. The lysis was then incubated with agarose beads (Roche, Indianapolis, IN, USA) for 4–6 h at 4°C. IP samples were separated by 8%–10% SDS-PAGE gels, and protein bands were detected by the chemiluminescent substrate.
Total proteins were extracted from Mkrn2-WT and Mkrn2-KO MEFs, with four biological repetitions in each group. The proteins of different samples were respectively subjected to trypsin enzymatic hydrolysis, and peptide fractionation as described (21). The samples were further analyzed on a Thermo HFX MS (Thermo Fisher Scientific, Waltham, MA, USA) interfaced with an EASY-nLC1200 LC system (Thermo Fisher Scientific). Raw files were searched against the mouse refseq protein database (27,414 proteins, version 04/07/2013) with Proteome Discoverer (Thermo Fisher Scientific, version 1.4). The levels of proteins were estimated with a label-free, intensity-based absolute quantification (iBAQ) approach (22). The iBAQ of each protein was normalized by dividing the sum of all identified proteins. We replaced the extremely small values with 10−8. The matrix file integrated with all samples was subjected to different protein expression between Mkrn2-WT MEFs and Mkrn2-KO MEFs by using the limma package in R 3.6.1 software (23). Different proteins were filtered by |Log2(Fold Change)|>1 and p-value < 0.05. Heatmap shows 977 differently expressed proteins.
The pathway enrichment analysis of differently expressed proteins was performed with the Reactome database, using the ReactomePA package in R software. Default parameters (OrgDb, org.Mm.eg.db; p-value cutoff, 0.05; q-value cutoff, 0.2; p-value adjust method, BH; readable, T) were set for pathway enrichment analysis. Enriched pathways with FDR < 0.2 were considered statistically significant.
The matrix data from GEO (GSE6872) is an expression profiling array, and the analysis of differently expressed genes was performed by using the limma package in R software (24). The threshold for differently expressed genes is |Log2(Fold Change)| > 1 and p-value < 0.05. The volcano plot shows analysis results including upregulated genes (red), downregulated genes (blue), and nonsignificant genes (black).
All results have been obtained from at least three independent experiments, and data were analyzed via GraphPad Prism 8 software. The statistical evaluation for data analysis was determined by a t-test. The differences were considered to be statistically significant at p < 0.05.
In order to investigate the effects of MKRN2 on male spermatogenesis, we detected the macroscopic appearance of the testis of Mkrn2 knockout and wild-type (WT) mice via HE staining. We selected the testis at phase VIII of testicular development and epididymis, and the results showed that compared to the wild-type group, there were no sperm in the testis of Mkrn2 knockout mice (Figures 1A, B). In our previous study, we showed that Mkrn2 knockout induced spermiation failure, but we also found that a few Mkrn2 knockout mice had another new phenotype in which there is no sperm generated. We then analyzed the MKRN2 expression level of four normal spermatogenesis individuals (normal) and 27 nonobstructive azoospermia samples (patients) from the GEO dataset GSE45885. The results showed that the expression level of MKRN2 was significantly lower than the normal individuals (Figure 1C). These results suggested that MKRN2 played a key role in testicular development and male spermatogenesis.
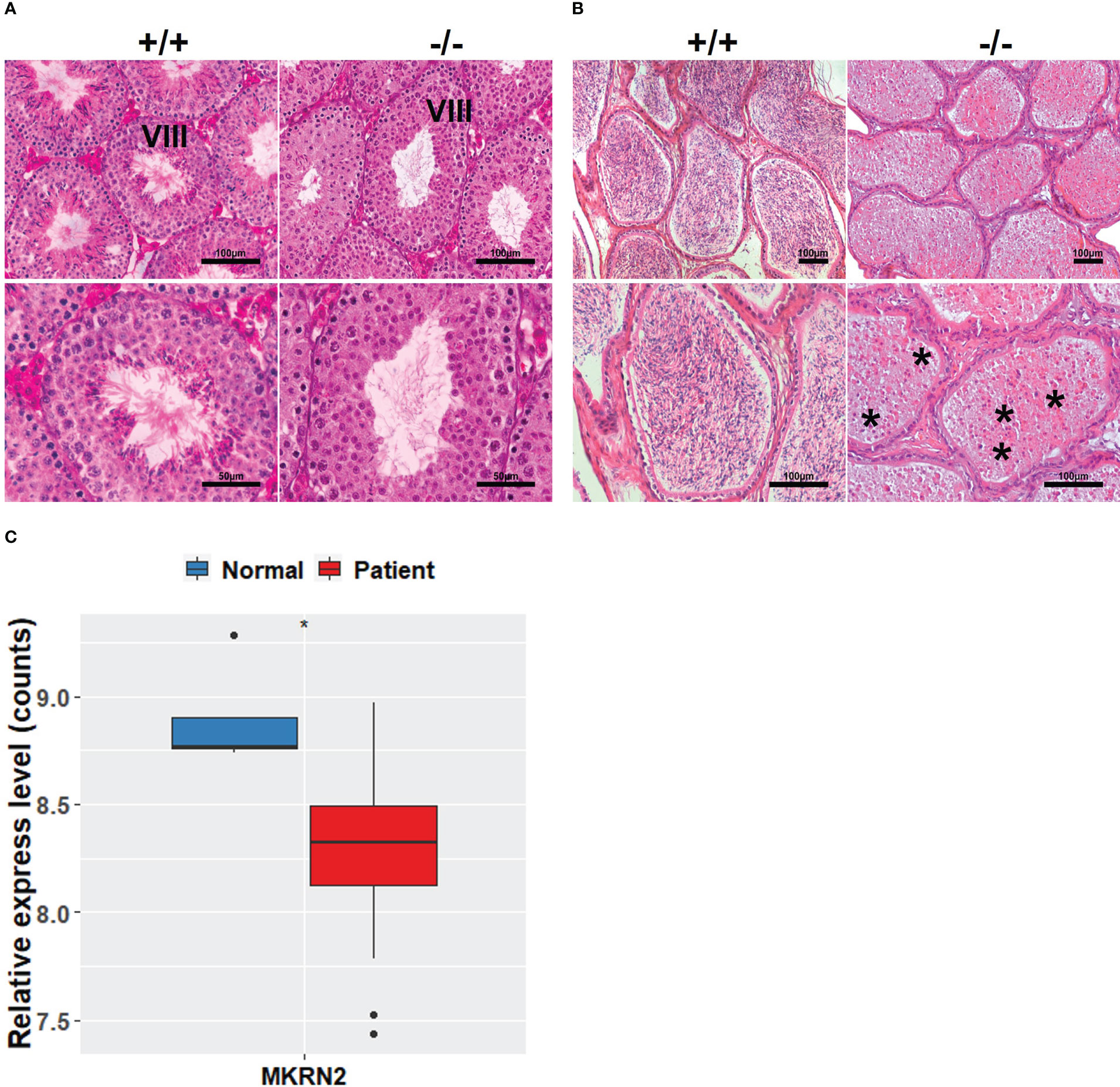
Figure 1 MKRN2 is related to male spermatogenesis. (A, B) Hematoxylin and eosin (H&E) staining sections of Mkrn2-WT (+/+) and Mkrn2-KO (−/−) mice testes in the phase VIII of sperm development and epididymis. Representative pictures are shown, Asterisk represents no sperm. (C) Expression levels of MKRN2 in four normal spermatogenesis individuals (normal) and 27 nonobstructive azoospermia samples (patients) from GEO dataset GSE45885. Data are analyzed by two-tailed, unpaired Student’s t-test. *p < 0.05, a significant difference.
To investigate the role of MKRN2 in testicular development and male spermatogenesis further, we performed a proteomics analysis of Mkrn2-WT MEFs and Mkrn2-KO MEFs (Figure 2A), as well as an analysis of differential expression genes (DEGs) in normospermic and teratozoospermic groups from the GEO dataset GSE6872 (Figure 2B). The signaling pathways represented among overlapping genes and ranked according to the number of enriched genes were screened for overlapping genes between differentially expressed proteins from proteomics and DEGs from GSE6872. The results showed that the primary signaling pathway (Figures 2C, D) was receptor tyrosine kinase signaling. We performed a transcription factor assay to compare the different transcription factors between Mkrn2-WT and Mkrn2-KO MEFS, and the overlapping genes of the transcription factor array and genes of the receptor tyrosine kinase signaling pathway were analyzed. STAT1 was the only gene that overlapped (Figure 2E). We then analyzed the expression levels of STAT family members in Mkrn2-WT MEFs and Mkrn2-KO MEFs according to the transcription factor array results, and STAT1 was the most downregulated gene in the Mkrn2-KO MEFs (Figure 2F). All of these results indicated that MKRN2-regulated STAT1 is involved in the process of male infertility.
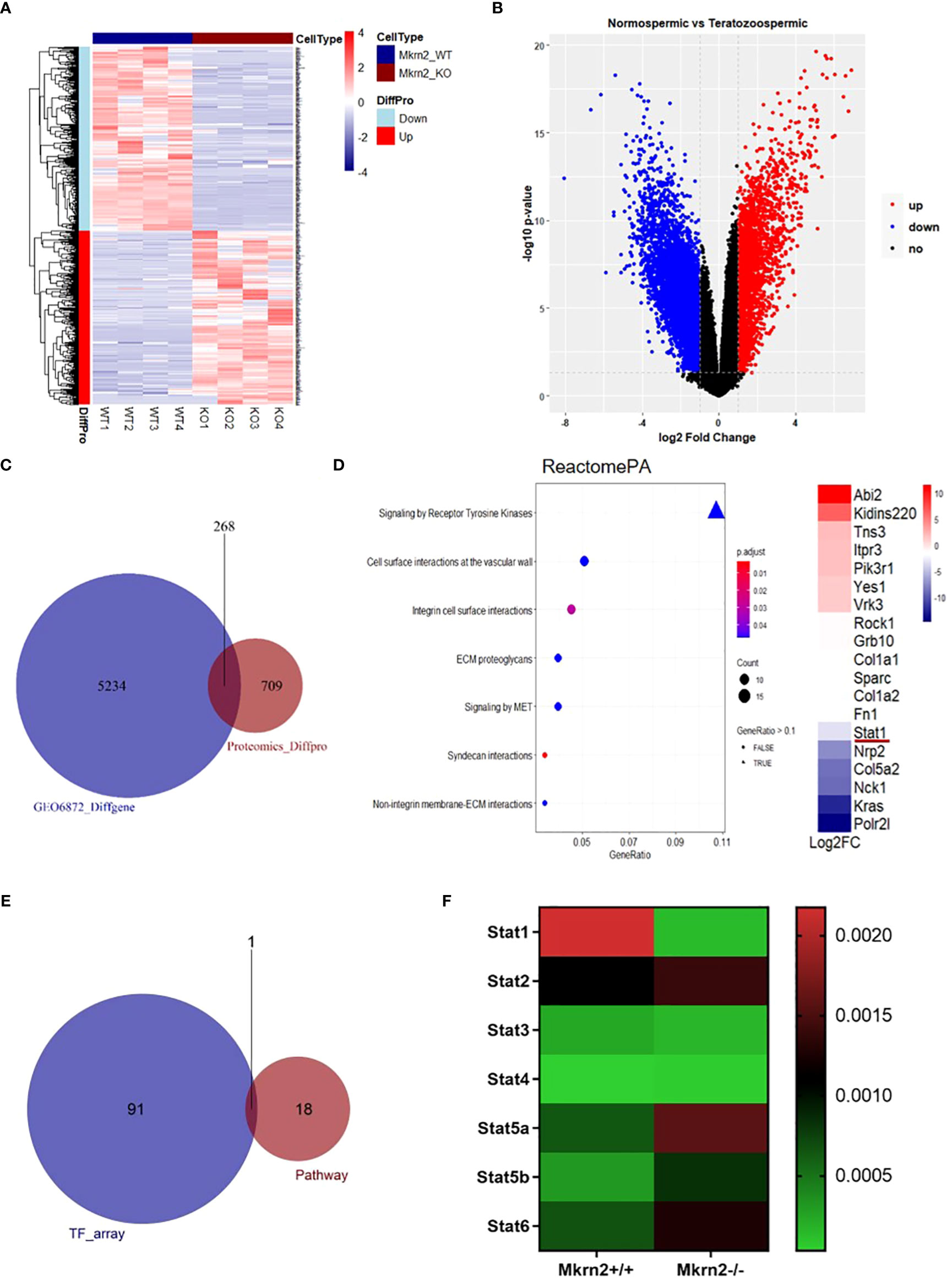
Figure 2 MKRN2 expression levels are highly correlated with the receptor tyrosine kinase signaling pathway. (A) Heatmap plot of all differentially expressed proteins in Mkrn2-WT MEFs and Mkrn2-KO MEFs. The proteins and samples are respectively clustered by Euclidean distance, and the color block shows the classification. Upregulated proteins are coded in red color and downregulated proteins are coded in blue. (B) Volcano plot of DEGs in normospermic group and teratozoospermic group from GEO dataset GSE6872. According to the values of logFC and p-value (logFC ≥ 1 or ≤ −1, p < 0.05), all genes were classified into upregulated genes (red), downregulated genes (blue), and no changed genes (black). (C) The overlapping between differentially expressed proteins from proteomics (Proteomics_Diffpro) and DEGs from GSE6872 (GSE6872_Diffgene). (D) The signaling pathways are represented among overlapping genes and ranked according to the number of enriched genes. The color of the dots represents the p-value; the size of the dot represents the gene count, and the shape of the dots represents the gene ratio. The primary signaling pathway is signaling by receptor tyrosine kinases, including 18 genes, ranked by log2FC. (E) The overlapping of the results of the transcription factors array (TF_array) and genes of the signaling by receptor tyrosine kinase pathway. (F) Expression levels of the STAT family members in Mkrn2-WT MEFs and Mkrn2-KO MEFs, according to TF array results.
To further investigate the regulatory effect of MKRN2 on STAT1, we measured the protein and mRNA expression levels of MKRN2 and STAT1 in Mkrn2-WT MEFs and Mkrn2-KO MEFs, and the results showed that the expression level of STAT1 was significantly downregulated in Mkrn2-KO MEFs (Figure 3A). We then overexpressed MKRN2 in the 293T cells and measured the protein and mRNA expression levels of STAT1. The results showed that the expression level of STAT1 was significantly upregulated in MKRN2 overexpression cells (Figure 3B). We further found that MKRN2 could interact with STAT1 using a co-immunoprecipitation assay (Figure 3C). All of these results indicated that MKRN2 could interact with STAT1 and regulate its expression level.
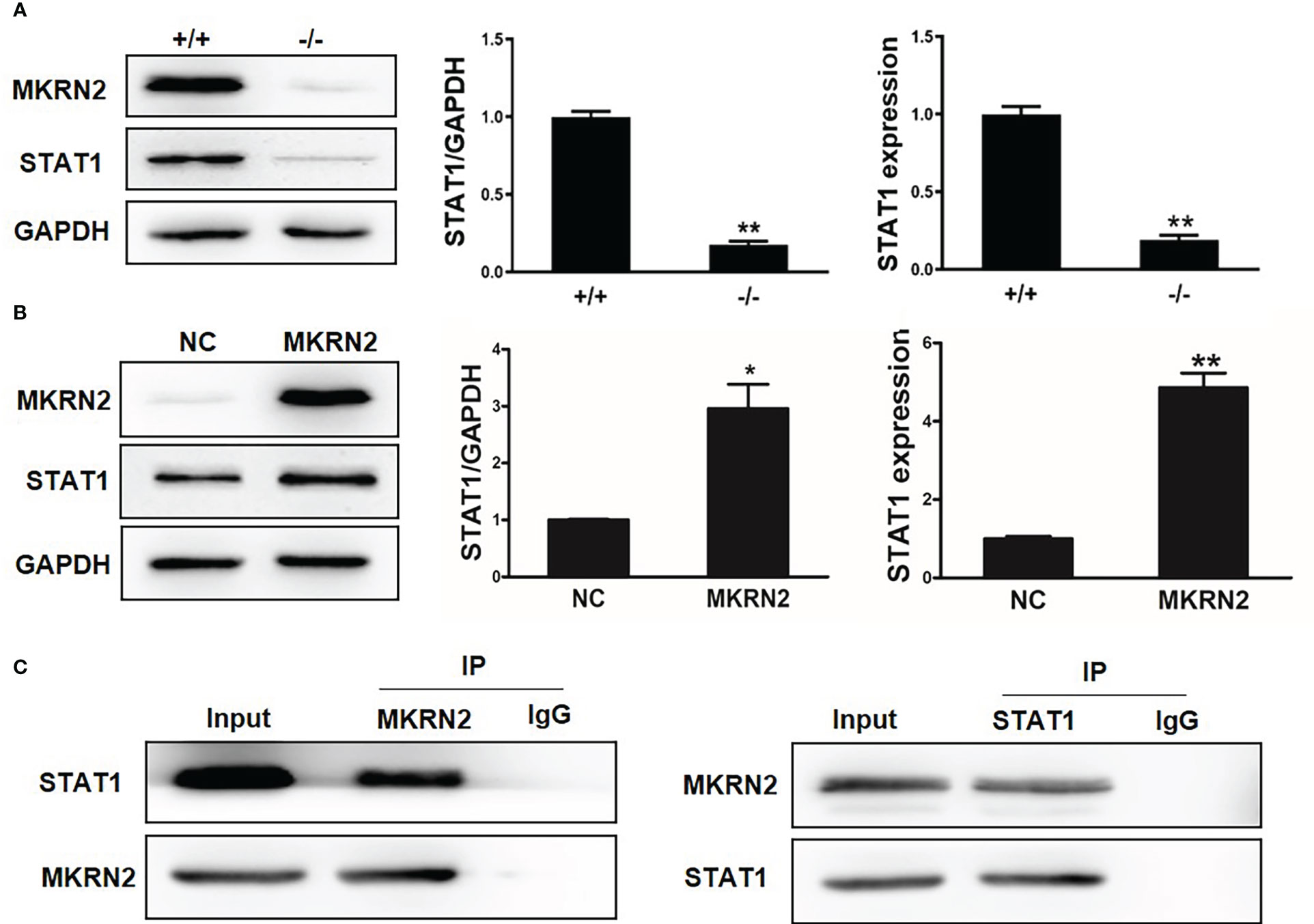
Figure 3 MKRN2 regulates the expression level of STAT1. (A) The protein and mRNA expression levels of MKRN2 and STAT1 in Mkrn2-WT MEFs and Mkrn2-KO MEFs. (B) Overexpressed MKRN2 in the 293T cells and the protein and mRNA expression levels of MKRN2 and STAT1 are shown. (C) Overexpressed MKRN2 or STAT1 in the 293T cells; the co-immunoprecipitation assay was preformed to detect the interaction of MKRN2 and STAT1, and the representative pictures are shown. *p < 0.05 and **p < 0.01—significant difference.
To verify the role of MKRN2 in the male teratozoospermia, we analyzed the expression levels of MKRN2 and STAT1 in 13 normally fertile male patients and eight infertile individuals with a severe and consistent heterogeneous teratozoospermia based on the GEO dataset GSE6872. The results showed that the expression levels of MKRN2 and STAT1 were both significantly downregulated in the infertile patients (Figure 4A). We then detected the protein and mRNA expression levels of MKRN2 and STAT1 in the testis of Mkrn2 knockout and wild-type (WT) mice and found that the expression levels of MKRN2 and STAT1were both significantly downregulated in the testis tissues of Mkrn2 knockout mice (Figures 4B–D). These results indicated that MKRN2 mediated male teratozoospermia by regulating the expression level of STAT1.
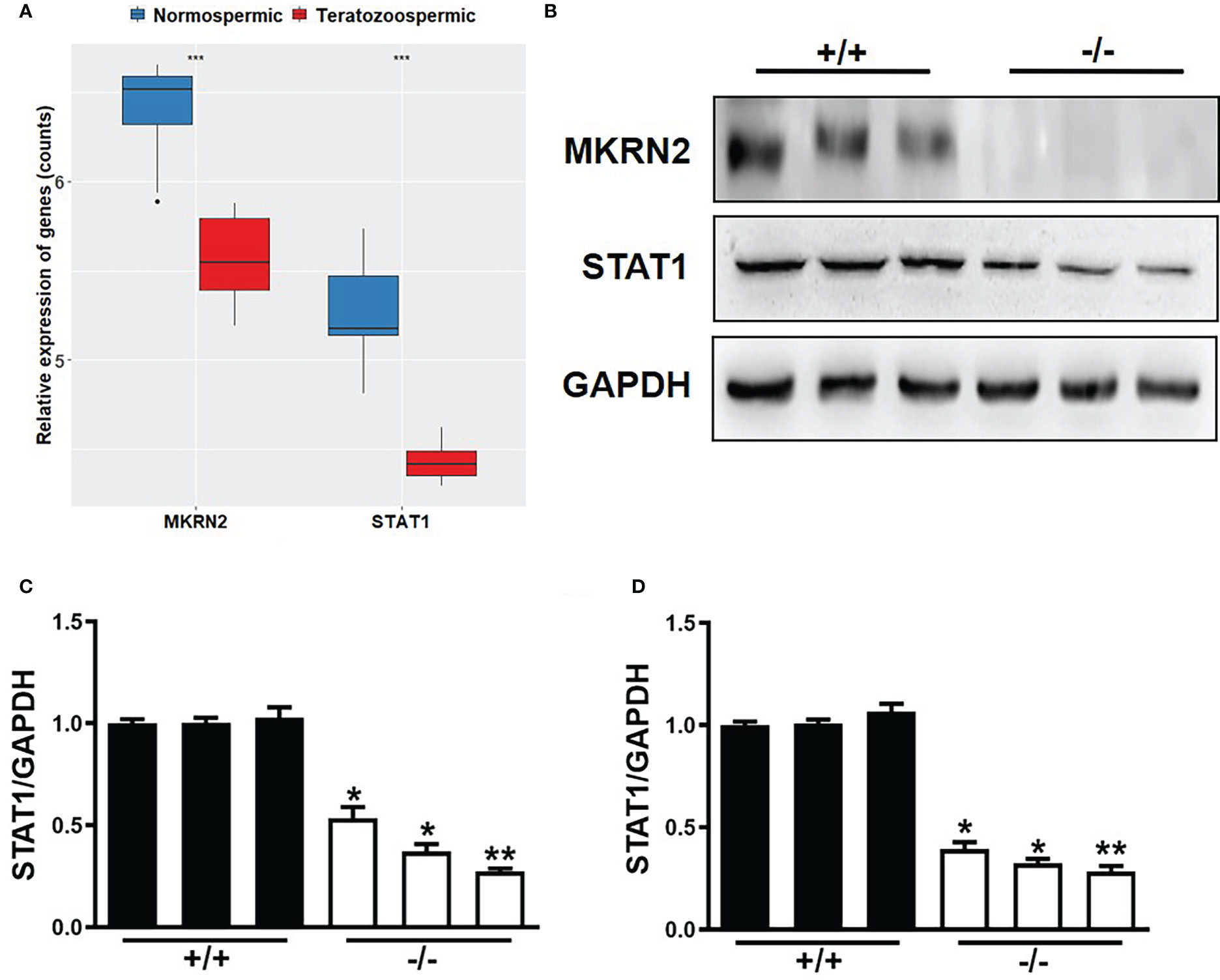
Figure 4 MKRN2 mediates male teratozoospermia by affecting the expression level of STAT1. (A) The expression levels of MKRN2 and STAT1 in the sperms of 13 normally fertile male patients and eight infertile individuals with severe and consistent heterogeneous teratozoospermia. Data are from the GEO dataset GSE6872. (B–D) The protein and mRNA expression levels of MKRN2 and STAT1 in the testis of Mkrn2-WT and Mkrn2-KO mice. *p < 0.05, **p < 0.01, and ***p < 0.0001—significant difference.
To further investigate the effects of Mkrn2 on other reproductive pathways, we obtained a dataset of all genes related to reproduction from the Mouse Genome Informatics (MGI) website (http://www.informatics.jax.org/), overlapped the differentially expressed proteins of Mkrn2-WT MEFs and Mkrn2-KO MEFs and the reproduction-related genes, and then obtained 68 overlapped genes (Figure 5A). We then looked into the overlap of differentially expressed proteins from proteomics, genes from reproduction pathway, and genes from related molecular function sets of male infertility. There were nine overlapping genes with male gonad development (Figure 5B), 18 with spermatogenesis (Figure 5C), 18 with male gamete generation (Figure 5D), and two with prostate gland development (Figure 5E). All of the above findings suggested that MKRN2 knockout cause a disruption in the reproductive pathway.
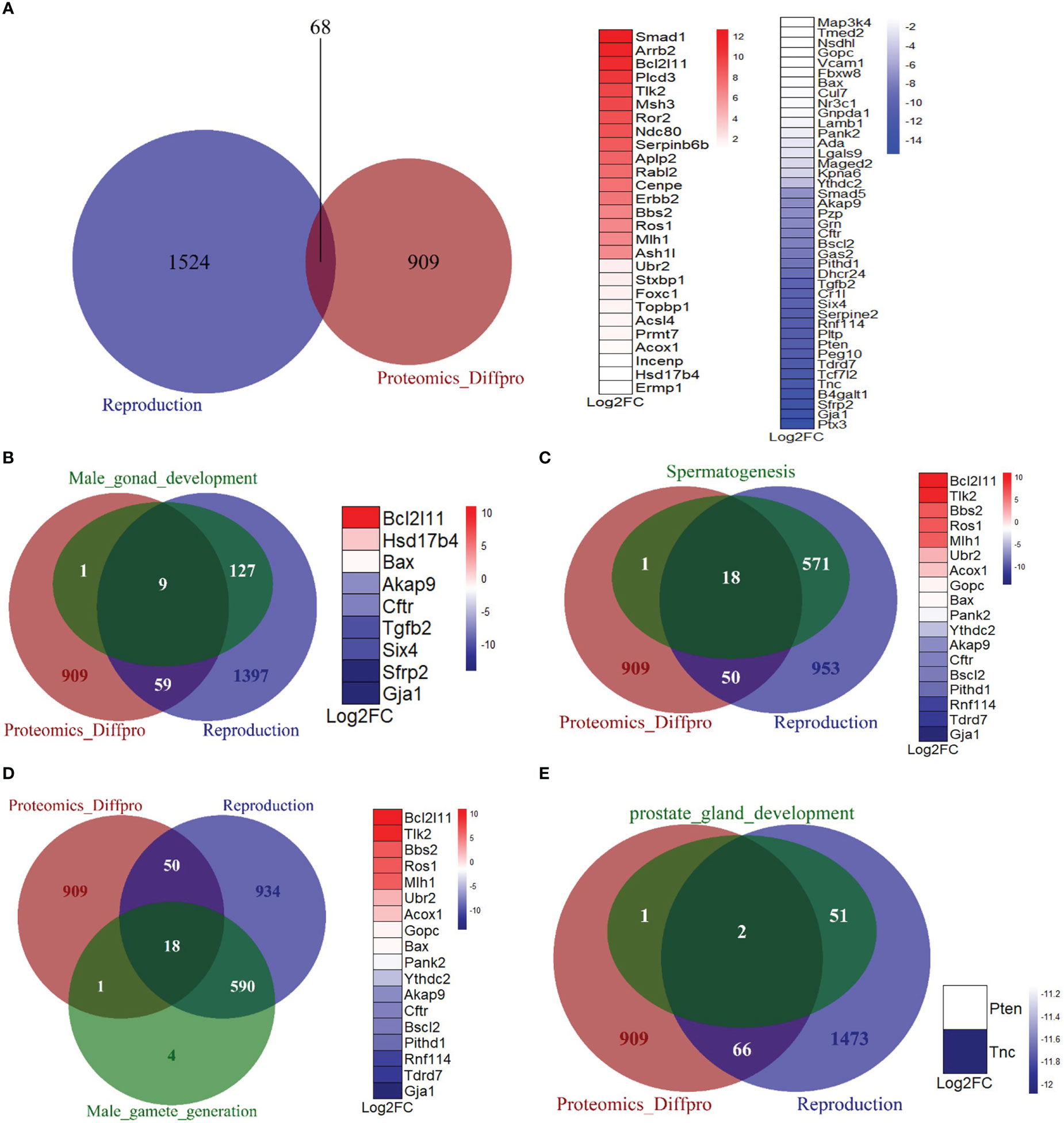
Figure 5 MKRN2 results in a disorder of the reproductive pathway. (A) The overlapping between differentially expressed proteins from proteomics (Proteomics_Diffpro) and genes from the reproduction pathway (reproduction). Upregulated proteins (blue) and downregulated proteins (red) from proteomics analysis were ranked by log2FC. (B–E) The overlapping of differentially expressed proteins from proteomics (Proteomics_Diffpro), genes from reproduction pathway (reproduction), and genes from related molecular function sets of male infertility, including male gonad development, spermatogenesis, male gamete generation, and prostate gland development. The overlapped genes are ranked by log2 fold change (Log2FC).
To further investigate the molecular mechanism of the above reproductive-related genes in male teratozoospermia, we verified expression levels of SIX4 and TNC in Mkrn2-WT MEFs and Mkrn2-KO MEFs using the proteomics data. The results showed that the expression levels of SIX4 and TNC were significantly downregulated in the Mkrn2-KO MEFs (Figure 6A). We then overlapped the putative upstream transcription factors of SIX4 or TNC according to the Jaspar website and differentially expressed proteins from proteomics and found only one transcription factor, EBF2 (Figure 6B). The mRNA expression levels of SIX4, TNC, and EBF2 were detected in Mkrn2-WT MEFs and Mkrn2-KO MEFs, and we found that the expression levels of SIX4, TNC, and transcription factor EBF2 were significantly downregulated in Mkrn2-KO MEFs (Figure 6C). We also used Western blot to examine the testis tissues of Mkrn2-WT and Mkrn2-KO mice, and the results showed that the protein expression levels of EBF2, SIX4, and TNC were decreased in Mkrn2-KO mice (Figure 6D). These findings indicated that MKRN2 regulated the expression level of SIX4 and TNC in Mkrn2 knockout mice via the transcription factor EBF2.
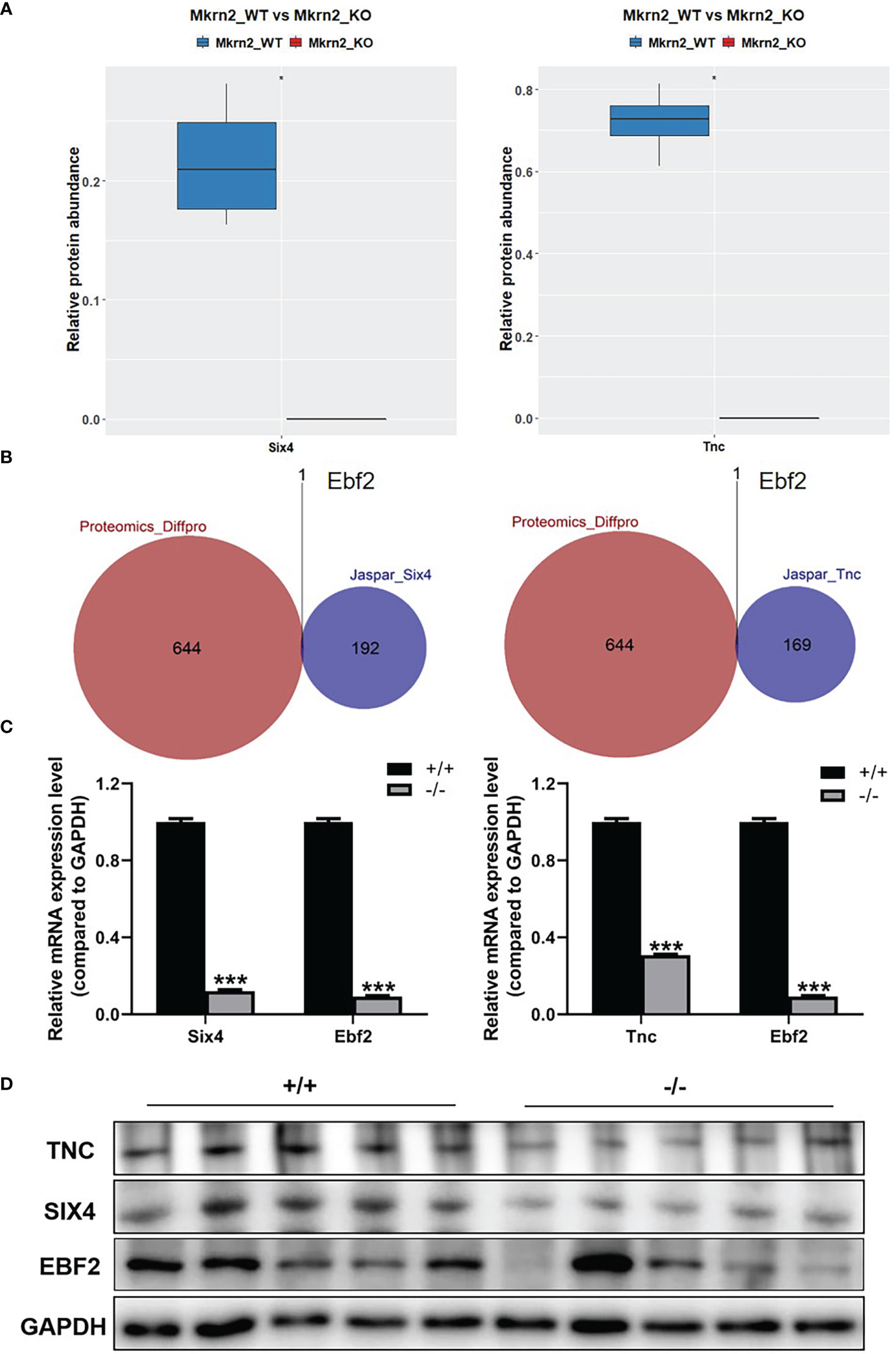
Figure 6 MKRN2 regulates the expression of SIX4 and TNC. (A) Proteomics data show SIX4 and TNC expression levels in Mkrn2-WT and Mkrn2-KO MEFs. (B) The overlapping between the putative upstream transcription factors of Six4 or Tnc according to the Jaspar website and differentially expressed proteins from proteomics. Ebf2 is the only differentially expressed transcription factor. (C) The mRNA expression levels of Six4 and Ebf2 were detected via Q-PCR, and the mRNA expression levels of Tnc and Ebf2 in Mkrn2-WT MEFs and Mkrn2-KO MEFs were also detected via Q-PCR. (D) The protein expression levels of EBF2, SIX4, and TNC were detected via Western blot in the testis tissues of Mkrn2-WT and Mkrn2-KO mice. *p < 0.05 and ***p < 0.0001—significant difference.
The MKRN2 gene is a member of the makorin gene family, which also includes MKRN1, MKRN2, and MKRN3. MKRN2 has been found to be highly conserved throughout evolution, and its ancestral origin can be traced back to 450 million years ago, possibly as a result of MKRN1 gene duplication (1). We have illustrated the functional role of Mkrn2 in Xenopus embryos to negatively regulate neurogenesis through PI3K/Akt signaling. However, the potential functions and molecular mechanisms of MKRN2 in mammals remain to be studied. This Mkrn2 knockout mouse model and an independent GEO dataset of human sperm samples were used in this study to demonstrate the important role of MKRN2 in male fertility.
Previous studies have reported that MKRN2 inhibits the cell migration and invasion of non-small cell lung cancer by downregulating the PI3K/Akt pathway and is associated with lymph node metastasis, TNM stage, and cell differentiation (7). MKRN2 also acts as an E3 ligase for the NF-κB p65 subunit and has a negative regulatory effect on the inflammatory response (25).
Few reports have focused on the regulation of MKRN2 in male infertility. Our previous results demonstrated that Mkrn2 deletion in somatic Sertoli cells disrupted ectoplasmic specialization (ES), resulting in abnormalities in sperm heads and spermiation failure, both of which are essential to spermiogenesis. Moreover, Mkrn2 is crucial for protecting germ cells from apoptosis in spermatogenesis and male fertility via the p53/PERP signaling pathway. In this study, we focus on another phenotype of Mkrn2 knockout mice: no sperm generated, which may indicate the essential role of MKRN2 in male fertility. In this study, we found a new regulatory mechanism between MKRN2 and STAT1/SIX4/TNC, which is a novel mechanism of MKRN2 in regulating male infertility.
Recent studies have shown that the testis shares many similarities with cancerous tissues, including cell division, immigration, and immortalization. Cancer–testis (CT) antigens are usually expressed only in testis tissues, with the exception of early-developing embryos. In addition, CT antigens are expressed in various tumor types (26). The CT genes, OIP5, TAF7L, and AURKC have been identified as biomarkers for breast cancer and may be promising and potent candidates for therapeutic cancer vaccines (27, 28). MKRN2 may function as a cancer–testis antigen, with dual roles in tumorigenesis and spermiogenesis that need to be further investigated.
In this study, MKRN2 could interact with STAT1 and regulate its expression level, as well as the expression levels of SIX4 and TNC via the transcription factor EBF2. These results suggested that MKRN2 played a key role in testicular development and male spermatogenesis.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by Committee of Laboratory Animal Experimentation of Zhengzhou University.
J-GQ and B-HJ designed experiments and prepared the manuscript. LW, Y-LY, and K-KW performed experiments and analyzed the data. Y-XX, Y-CQ, and F-MZ analyzed clinical samples and provided technical support. B-HJ provided advice. All authors contributed to the article and approved the submitted version.
This work was supported in part by the National Natural Science Foundation of China (No. 81903174).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Gray TA, Azama K, Whitmore K, Min A, Abe S, Nicholls RD. Phylogenetic conservation of the makorin-2 gene, encoding a multiple zinc-finger protein, antisense to the RAF1 proto-oncogene. Genomics (2001) 77(3):119–26. doi: 10.1006/geno.2001.6627
2. Gray TA, Hernandez L, Carey AH, Schaldach MA, Smithwick MJ, Rus K, et al. The ancient source of a distinct gene family encoding proteins featuring RING and C(3)H zinc-finger motifs with abundant expression in developing brain and nervous system. Genomics (2000) 66(1):76–86. doi: 10.1006/geno.2000.6199
3. Mao M, Fu G, Wu JS, Zhang QH, Zhou J, Kan LX, et al. Identification of genes expressed in human CD34(+) hematopoietic stem/progenitor cells by expressed sequence tags and efficient full-length cDNA cloning. Proc Natl Acad Sci U S A (1998) 95(14):8175–80. doi: 10.1073/pnas.95.14.8175
4. Zhang QH, Ye M, Wu XY, Ren SX, Zhao M, Zhao CJ, et al. Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells. Genome Res (2000) 10(10):1546–60. doi: 10.1101/gr.140200
5. Cheung WK, Yang PH, Huang QH, Chen Z, Chen SJ, Lin MC, et al. Identification of protein domains required for makorin-2-mediated neurogenesis inhibition in xenopus embryos. Biochem Biophys Res Commun (2010) 394(1):18–23. doi: 10.1016/j.bbrc.2010.02.041
6. Yang PH, Cheung WK, Peng Y, He ML, Wu GQ, Xie D, et al. Makorin-2 is a neurogenesis inhibitor downstream of phosphatidylinositol 3-kinase/Akt (PI3K/Akt) signal. J Biol Chem (2008) 283(13):8486–95. doi: 10.1074/jbc.M704768200
7. Jiang J, Xu Y, Ren H, Wudu M, Wang Q, Song X, et al. MKRN2 inhibits migration and invasion of non-small-cell lung cancer by negatively regulating the PI3K/Akt pathway. J Exp Clin Cancer Res (2018) 37(1):189. doi: 10.1186/s13046-018-0855-7
8. Zhang Y, Cui N, Zheng G. Ubiquitination of P53 by E3 ligase MKRN2 promotes melanoma cell proliferation. Oncol Lett (2020) 19(3):1975–84. doi: 10.3892/ol.2020.11261
9. Darnell JE Jr., Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science (1994) 264(5164):1415–21. doi: 10.1126/science.8197455
10. Ihle JN. The stat family in cytokine signaling. Curr Opin Cell Biol (2001) 13(2):211–7. doi: 10.1016/s0955-0674(00)00199-x
11. Leaman DW, Leung S, Li X, Stark GR. Regulation of STAT-dependent pathways by growth factors and cytokines. FASEB J (1996) 10(14):1578–88.
12. Schindler C. Cytokines and JAK-STAT signaling. Exp Cell Res (1999) 253(1):7–14. doi: 10.1006/excr.1999.4670
13. Darnell JE Jr. STATs and gene regulation. Science (1997) 277(5332):1630–5. doi: 10.1126/science.277.5332.1630
14. O'Shea JJ, Gadina M, Schreiber RD. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell (2002) 109 Suppl:S121–31. doi: 10.1016/s0092-8674(02)00701-8
15. Truchet S, Chebrout M, Djediat C, Wietzerbin J, Debey P. Presence of permanently activated signal transducers and activators of transcription in nuclear interchromatin granules of unstimulated mouse oocytes and preimplantation embryos. Biol Reprod (2004) 71(4):1330–9. doi: 10.1095/biolreprod.104.029405
16. Bastian Y, Zepeda-Bastida A, Uribe S, Mujica A. In spermatozoa, Stat1 is activated during capacitation and the acrosomal reaction. Reproduction (2007) 134(3):425–33. doi: 10.1530/REP-06-0264
17. Silva JV, Freitas MJ, Correia BR, Korrodi-Gregorio L, Patricio A, Pelech S, et al. Profiling signaling proteins in human spermatozoa: biomarker identification for sperm quality evaluation. Fertil Steril (2015) 104(4):845–56.e8. doi: 10.1016/j.fertnstert.2015.06.039
18. Fujimoto Y, Tanaka SS, Yamaguchi YL, Kobayashi H, Kuroki S, Tachibana M, et al. Homeoproteins Six1 and Six4 regulate male sex determination and mouse gonadal development. Dev Cell (2013) 26(4):416–30. doi: 10.1016/j.devcel.2013.06.018
19. Qian X, Wang L, Zheng B, Shi ZM, Ge X, Jiang CF, et al. Deficiency of Mkrn2 causes abnormal spermiogenesis and spermiation, and impairs male fertility. Sci Rep (2016) 6:39318. doi: 10.1038/srep39318
20. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem (1976) 72:248–54. doi: 10.1006/abio.1976.9999
21. Zhou Q, Liu M, Xia X, Gong T, Feng J, Liu W, et al. A mouse tissue transcription factor atlas. Nat Commun (2017) 8:15089. doi: 10.1038/ncomms15089
22. Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature (2011) 473(7347):337–42. doi: 10.1038/nature10098
23. van Ooijen MP, Jong VL, Eijkemans MJC, Heck AJR, Andeweg AC, Binai NA, et al. Identification of differentially expressed peptides in high-throughput proteomics data. Brief Bioinform (2018) 19(5):971–81. doi: 10.1093/bib/bbx031
24. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res (2015) 43(7):e47. doi: 10.1093/nar/gkv007
25. Shin C, Ito Y, Ichikawa S, Tokunaga M, Sakata-Sogawa K, Tanaka T. MKRN2 is a novel ubiquitin E3 ligase for the p65 subunit of NF-kappaB and negatively regulates inflammatory responses. Sci Rep (2017) 7:46097. doi: 10.1038/srep46097
26. Whitehurst AW. Cause and consequence of cancer/testis antigen activation in cancer. Annu Rev Pharmacol Toxicol (2014) 54:251–72. doi: 10.1146/annurev-pharmtox-011112-140326
27. Mobasheri MB, Shirkoohi R, Modarressi MH. Cancer/Testis OIP5 and TAF7L genes are up-regulated in breast cancer. Asian Pac J Cancer Prev (2015) 16(11):4623–8. doi: 10.7314/apjcp.2015.16.11.4623
Keywords: MKRN2, STAT1, SIX4, TNC, male infertility
Citation: Wang L, Yong Y-L, Wang K-K, Xie Y-X, Qian Y-C, Zhou F-M, Qiu J-G and Jiang B-H (2023) MKRN2 knockout causes male infertility through decreasing STAT1, SIX4, and TNC expression. Front. Endocrinol. 14:1138096. doi: 10.3389/fendo.2023.1138096
Received: 05 January 2023; Accepted: 24 February 2023;
Published: 10 March 2023.
Edited by:
Shun Bai, University of Science and Technology of China, ChinaReviewed by:
Kaiqiang Fu, Qingdao Agricultural University, ChinaCopyright © 2023 Wang, Yong, Wang, Xie, Qian, Zhou, Qiu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing-Hua Jiang, YmluZ2hqaWFuZ0B6enUuZWR1LmNu; Jian-Ge Qiu, amlhbmdlcWl1QHp6dS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.