- Department of Obstetrics and Gynecology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, National Clinical Research Center for Obstetric and Gynecologic Diseases, Beijing, China
Endometriosis has a detrimental effect on oocyte quality, and ovarian endometriosis (OEM) and peritoneal endometriosis (PEM) may have different effects on female fertility. Therefore, we conducted a study to explore the circular RNA (circRNA) expression profiles of cumulus cells (CCs) in patients with OEM (n = 3), PEM (n = 3), and tubal factor infertility (TFI, n = 3) using high-throughput sequencing techniques and attempted to identify common and unique circRNAs in the OEM and PEM groups. The CIRCexplorer2 program was used to identify circRNAs. Seven candidate circRNAs were validated in 30 samples using quantitative real-time polymerase chain reaction (qRT-PCR). Finally, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed to annotate the function of circRNA-targeted genes, which were verified by sequencing results and constructed circRNA–miRNA–mRNA networks. A total of 11833 circRNAs were identified in nine samples. The numbers of differentially expressed circRNAs between the OEM and TFI groups, PEM and TFI groups, and OEM and PEM groups were 130, 71, and 191, respectively. After taking intersections, 11 circRNAs were considered common circRNAs in the OEM and PEM groups; 39 circRNAs in the OEM group and 17 circRNAs in the PEM group were identified as unique key circRNAs. During qRT-PCR validation, hsa_circ_0003638 was significantly upregulated in the PEM group compared to that in the OEM and TFI groups. Functional analysis of circRNA-targeted genes revealed that apoptosis, PI3K-AKT, and p53 signaling pathways were enriched in the PEM–TFI comparison groups, whereas the functions of target genes involved in the JAK–STAT and TGF-β signaling pathways were enriched in the PEM–OEM comparison groups. Our findings confirmed differences in circRNA expression profiles of CCs between patients with OEM and PEM infertility and provide new insights into the different effects of various endometriosis phenotypes on oocytes.
1 Introduction
Endometriosis is closely related to infertility (1), and many patients seek assisted reproductive technology (ART); however, the pregnancy outcomes of patients with endometriosis remain adverse (2). Patients with endometriosis tend to have poor quality oocytes and embryos (3, 4), causing low pregnancy rates. Thus, investigating the factors that influence oocyte quality in patients with infertility and endometriosis is critical. Cumulus cells (CCs) are somatic cells that surround oocytes and originate from undifferentiated granulosa cells (5). As a component of the cumulus–oocyte complex, CCs communicate constantly with the oocyte and influence its developmental competence (6). Therefore, gene expression profile analysis in human oocyte CCs can be used as a non-invasive approach for oocyte quality assessment (7).
Circular RNAs (circRNAs), a subset of long non-coding RNAs (lncRNAs), are characterized by a covalently closed loop structure without a 5’-3’ polarity or a poly(A) tail, which increases stability and resistance to RNase R (8, 9). In recent years, next-generation sequencing technology has accelerated the discovery of circRNAs, allowing investigation into their biological roles in gene regulation; for instance, circRNAs act as microRNA (miRNA) sponges to bind miRNAs in competitive endogenous RNA (ceRNA) networks (10, 11). Emerging evidence suggests that circRNAs may contribute to disease development, including endometriosis (8, 11). However, there are few studies on the function of circRNAs in CCs of patients with infertility along with endometriosis. To the best of our knowledge, there are no studies regarding the expression profiles of CCs in different phenotypes of endometriosis. Previous studies report that ovarian endometriosis (OEM) and peritoneal endometriosis (PEM) have different effects on female fertility; for instance, PEM is a risk factor for endometriosis-associated infertility while OEM is not, and OEM rarely induces inflammation in nearby follicles during in-vitro fertilization (12, 13). Hence, it is necessary to explore whether there are differences in circRNA expression profiles and functions between the two phenotypes.
In this study, we investigated circRNA expression profiles of CCs in patients with two different phenotypes of endometriosis (OEM and PEM) and tubal factor infertility (TFI) using high-throughput sequencing techniques. Subsequently, we constructed circRNA–miRNA–mRNA ceRNA networks and analyzed the function of circRNA-targeted genes using bioinformatic analysis. Our findings provide new evidence regarding the various effects of different endometriosis phenotypes on oocytes.
2 Materials and methods
2.1 Patient recruitment
This study was approved by the Institutional Review Board of the Peking Union Medical College Hospital (number: JS-3376). Informed consent was obtained in accordance with the institutional guidelines.
A total of 39 patients with infertility were enrolled from December 2021 to September 2022 at our hospital and divided into three groups of 13 patients each. In the OEM and PEM groups, endometriosis was surgically confirmed to ensure that the patients in each group had only one phenotype of endometriosis and not a mixture of the phenotypes. Endometriosis was classified according to the revised American Society for Reproductive Medicine criteria (14), and all lesions were removed prior to ART. In the OEM group, nine patients had stage III disease and four patients had stage IV disease, while in the PEM group, all patients had stage I disease. All patients in the control group had TFI without endometriosis lesions, as confirmed by surgery.
Other inclusion criteria included: age between 20 and 40 years, normal ovulation, baseline follicle stimulating hormone (FSH) levels< 10 IU/L and anti-Müllerian hormone > 1.1 ng/mL; no hormone therapy within three months prior to ART, and body mass index (BMI)< 30 kg/m2. The exclusion criteria included ovulatory dysfunction; untreated hydrosalpinx; cardiovascular diseases; dyslipidemia; endocrine diseases, including thyroid disorder, hyperprolactinemia or diabetes mellitus; autoimmune diseases; history of tuberculosis; smoking, drug, and/or alcohol habits; and patients with ovarian malignant tumors (15–17).
2.2 Controlled ovarian stimulation protocols
The patients underwent controlled ovarian stimulation based on their age, BMI, ovarian reserve, and previous ovarian response. Most patients in the TFI and PEM groups underwent gonadotropin-releasing hormone (GnRH) antagonist protocols, whereas most patients in the OEM group underwent long GnRH agonist protocols. Gonadotropins utilized in this study included recombinant FSH (Gonal-F, Merck Serono, Geneva, Switzerland), recombinant luteinizing hormone (Luveris, Merck Serono), and human menopausal gonadotropin (Livzon, Guangdong, China). Recombinant human chorionic gonadotropin (Ovidrel, Merck Serono) was used to trigger ovulation, when transvaginal ultrasonography detected the presence of at least three follicles larger than 16 mm. Subsequently, oocyte retrieval was performed 36 h later in the outpatient operating room.
2.3 Cumulus cell collection and purification
On the day of oocyte retrieval, CCs were collected and purified, as previously described (16, 18). Briefly, following follicular puncture, cumulus–oocyte complexes were collected in G-MOPS medium (Vitrolife, Goteborg, Sweden) under a microscope by experienced ART laboratory personnel. After mechanically stripping the oocytes under microscope, the remaining CCs were immediately transferred to a 15-mL disposable sterile centrifuge tube containing phosphate-buffered saline, and centrifuged at 500 × g for 2 min. After centrifugation and removal of the supernatant, 3–5 times the cell volume of red blood cell lysis buffer (Beyotime, Shanghai, China) was added to the pellets. The tube was maintained at 23–26°C, occasionally agitated for 1–2 min and centrifuged at 500 × g for 5 min. CCs were then washed twice with phosphate-buffered saline and transferred to a 1.5-mL Eppendorf tube. TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was immediately added to the EP tube to prevent RNA degradation, and the samples were stored at −80°C for subsequent RNA extraction.
2.4 Total RNA extraction, library construction, and sequencing
Library construction and Illumina sequencing were performed by the Shanghai Lifegenes Technology Co., Ltd. (Shanghai, China). Briefly, total RNAs were extracted using the TRIzol reagent kit, following the manufacturer’s protocol. The purity and concentration of RNA were assessed using a NanoPhotometer spectrophotometer (IMPLEN, Munich, Germany). Moreover, the integrity was measured using the Agilent fragment analyzer 5400 system (Agilent Technologies, Santa Clara, CA, USA). A total amount of 3 μg RNA per sample was used for the strand-specific library construction. First, ribosomal RNA (rRNA) was removed using the Epicentre Ribo-zero rRNA removal kit (Epicentre, Madison, WI, USA), according to the manufacturer’s instructions. Subsequently, the sequencing library was generated from the rRNA-depleted RNA using the NEBNext ultra directional RNA library prep kit for Illumina (NEB, Ipswich, MA, USA) according to the manufacturer’s instructions. When synthesizing the second strand of cDNA, dTTP was replaced by dUTP and the USER Enzyme (NEB) was then used to degrade the second strand of cDNA containing the U base, which allowed us to determine whether the transcript was from the positive-sense or antisense DNA strand. Subsequently, the products were purified using the AMPure XP system (Beckman Coulter, Brea, CA, USA), and the quality of the library was assessed using the Agilent fragment analyzer 5400 system (Agilent Technologies). The qualified library was sequenced on an Illumina NovaSeq 6000 platform following the manufacturer’s instructions and 150 bp paired-end reads were generated. Finally, we obtained raw reads, including the sequences of circRNAs, mRNAs and lncRNAs. We did not obtain the miRNAs sequences because the RNA content in each sample was not sufficient to construct another sequencing library.
2.5 Data processing and circRNA and mRNA data analyses
The obtained raw reads of Fastq format were processed using in-house PERL scripts. In this step, we obtained high-quality clean reads for further analysis by removing reads containing adapters, poly-N, and low-quality reads. HISAT2 (version 2.2.1) was used for mapping the clean reads to the reference genome with parameter “rna-strandness RF” (19). The reference genome used for alignment was the GRCh38 version from the Ensembl database.
circRNAs were identified using the CIRCexplorer2 program (version 2.3.8) with default parameters (20). The circRNAs were then searched in circBase for annotation, and those that could not be found were identified as novel circRNAs named by their source genes (21). Since circBase uses the hg19 version of the genome, CrossMap was used to transform the aligned circRNA genome coordinates from GRCh38 to hg19 version. The expression levels of circRNAs in each sample were estimated by transcript per million value using the following equation (22):
Subsequently, the limma R package (version 3.46.0) was used for differential expression analysis between the two groups. circRNAs with fold change (FC) ≥ 2 and P< 0.05 were considered as statistically differentially expressed circRNAs (DECs).
In addition, differential expression analysis of the mRNAs was performed for subsequent ceRNA network construction. After calculating the expected number of fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM) value in each sample using StringTie (version 2.1.6) (23), the DESeq2 R package (version 1.32.0) was used for differential expression analysis between each of the two groups. mRNAs with FC ≥ 2, P< 0.01 and FPKM ≥ 1 in at least one sample were considered as significantly differentially expressed mRNAs.
2.6 Finding common and respective unique circRNAs in the OEM and PEM groups
To explore the impact of different phenotypes of endometriosis on oocytes, we attempted to identify common and unique circRNAs in the OEM and PEM groups. After taking the intersection of the three clusters of DECs, the circRNAs that were shared by the OEM and PEM groups with consistent up/down-regulation were considered common circRNAs that might contribute to oocyte damage in patients with OEM and PEM. Meanwhile, the circRNAs that were significantly upregulated or downregulated in the OEM group compared with the other two groups were selected, as they might be involved in the unique effect on oocytes in patients with OEM. The unique circRNAs in the PEM group were screened in the same manner.
2.7 Quantitative real−time polymerase chain reaction validation
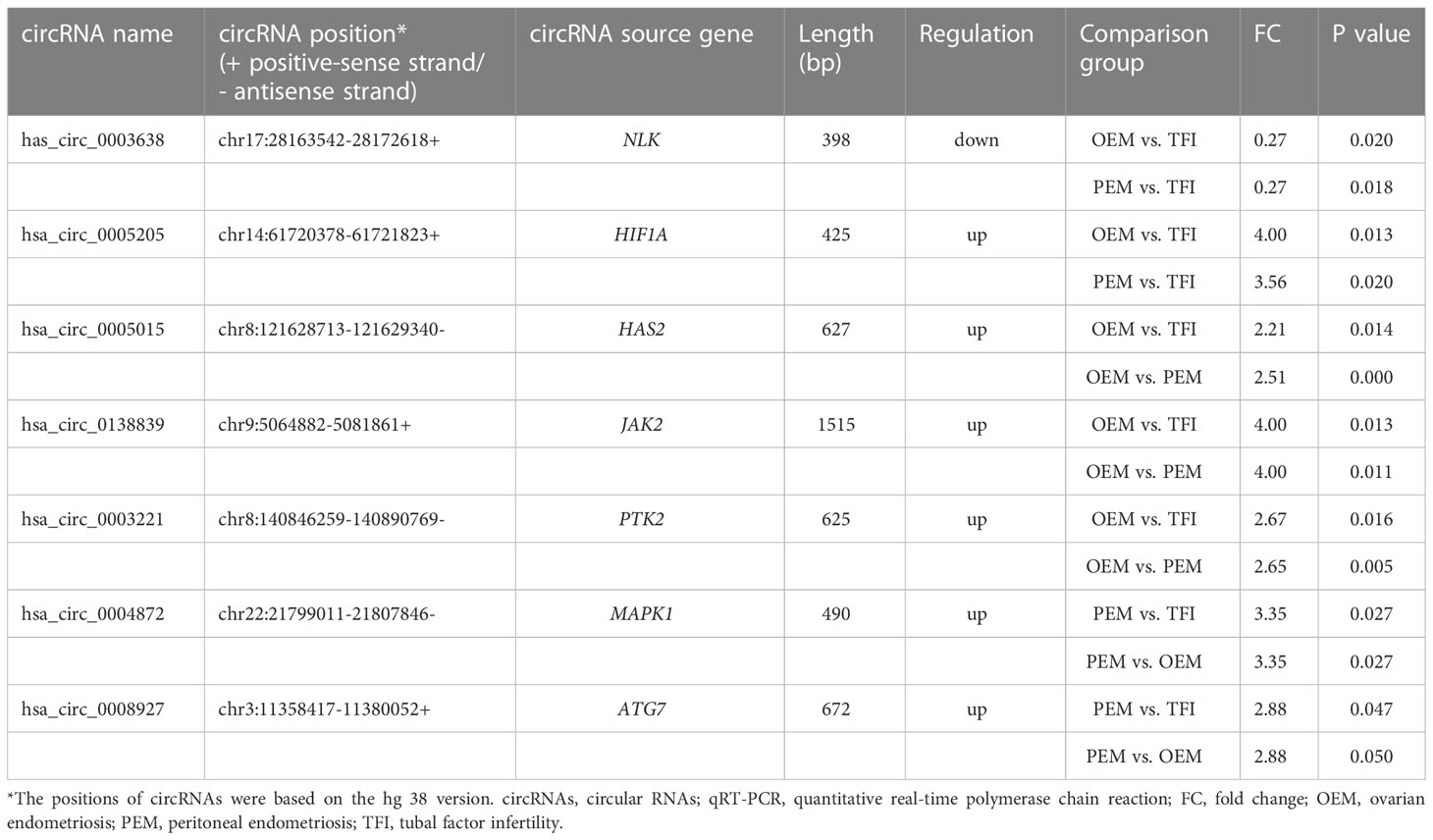
During quantitative real-time polymerase chain reaction (qRT-PCR) validation, we selected circRNAs for validation based on the following aspects: the FC and P values of circRNAs; the source genes of circRNAs (24); the length of circRNAs; and previous reports about the circRNAs. Finally, two circRNAs (hsa_circ_0003638 and hsa_circ_0005205) from the common circRNA assemblage of the OEM and PEM groups, three circRNAs (hsa_circ_0005015, hsa_circ_0138839, and hsa_circ_0003221) from the OEM unique assemblage, and two circRNAs (hsa_circ_0004872 and hsa_circ_0008927) from the unique assemblage of PEM were chose for validation. qRT-PCR was performed in 10 samples in each group.
Total RNA was isolated from the samples that had been treated with TRIzol reagent and then reverse transcribed into cDNA using Superscript III reverse transcriptase (Invitrogen). The primers of the seven candidate circRNAs were synthesized by BGI BEIJING Co. (Beijing, China) (Supplementary Table 1), and β-actin was used as a reference gene. The design of the primers was in the light of the back-spliced junction sites of circRNAs. qRT-PCR was performed using 2 × PCR mix (QIAGEN, Venlo, Netherlands) in an ABI ViiA 7 RT-PCR system (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturer’s instructions. Samples with more than 30 cycle threshold (CT) values were discarded to prevent false positives and make the results more convincing. The relative circRNA expression levels were calculated using the 2-ΔΔCt method (25). The relative expression levels of the selected circRNAs are presented as the mean ± standard error of the mean. Student’s t-test was performed if the data were normally distributed; otherwise, the Mann–Whitney U test was used for analysis. Statistical significance was set at P< 0.05. Statistical analysis was performed using the SPSS version 23.0 software (IBM, Armonk, NY, USA), and the results were displayed using GraphPad Prism version 9.4.0 (GraphPad Software, San Diego, CA, USA).
2.8 Construction of ceRNA networks
When constructing the interaction network between circRNAs and miRNAs, miRanda (version 3.3a) was used to predict the miRNAs that could bind to the verified circRNAs, with the parameter (-en -30). We used two prediction algorithms, TargetScan (version 8.0) and miRDB, to predict the mRNAs that could bind the same miRNAs. Subsequently, the overlapped mRNAs that were significantly differentially expressed according to the sequencing results were selected. The selected mRNAs also needed to follow the principle of positive correlation with circRNA expression levels. Finally, the verified circRNAs, predicted miRNAs, and overlapped mRNAs were used to construct ceRNA networks. The ceRNA networks were visualized using Cytoscape software (version 3.7.1).
2.9 Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analysis of circRNA-targeted genes
Based on the mRNAs in the ceRNA networks, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed to annotate the functions and pathways of circRNA-targeted genes, respectively. GO analysis was performed using the clusterProfiler R package (version 3.12.0), and KOBAS software (version 3.0) was used to test the statistical enrichment in KEGG pathways (http://www.genome.jp/kegg/) (26). Statistical significance regarding enrichment in the GO and KEGG analyses was set as P< 0.05. Only the most significant GO terms that contained the same genes were retained. The top 15 significantly enriched biological process (BP) terms in GO analysis and pathways in KEGG analysis were determined.
3 Results
3.1 DECs in CCs from the three groups
A total of 11833 circRNAs were identified from the nine samples, of which 3141 circRNAs were novel. The length of circRNAs was concentrated in the interval from 401 to 600 bp (Supplementary Figure 1). According to our results, 130 circRNAs were differentially expressed between the OEM and TFI groups, of which 82 were upregulated and 48 were downregulated in the OEM group. In the PEM group, 71 circRNAs (29 upregulated and 42 downregulated) were differentially expressed compared to those in the TFI group. A total of 191 circRNAs were found to be differentially expressed between the OEM and PEM groups, of which 69 were upregulated and 122 were downregulated in the PEM group. Volcano plots and heatmaps showed differences in circRNA expression levels between each of the two groups (Figure 1).
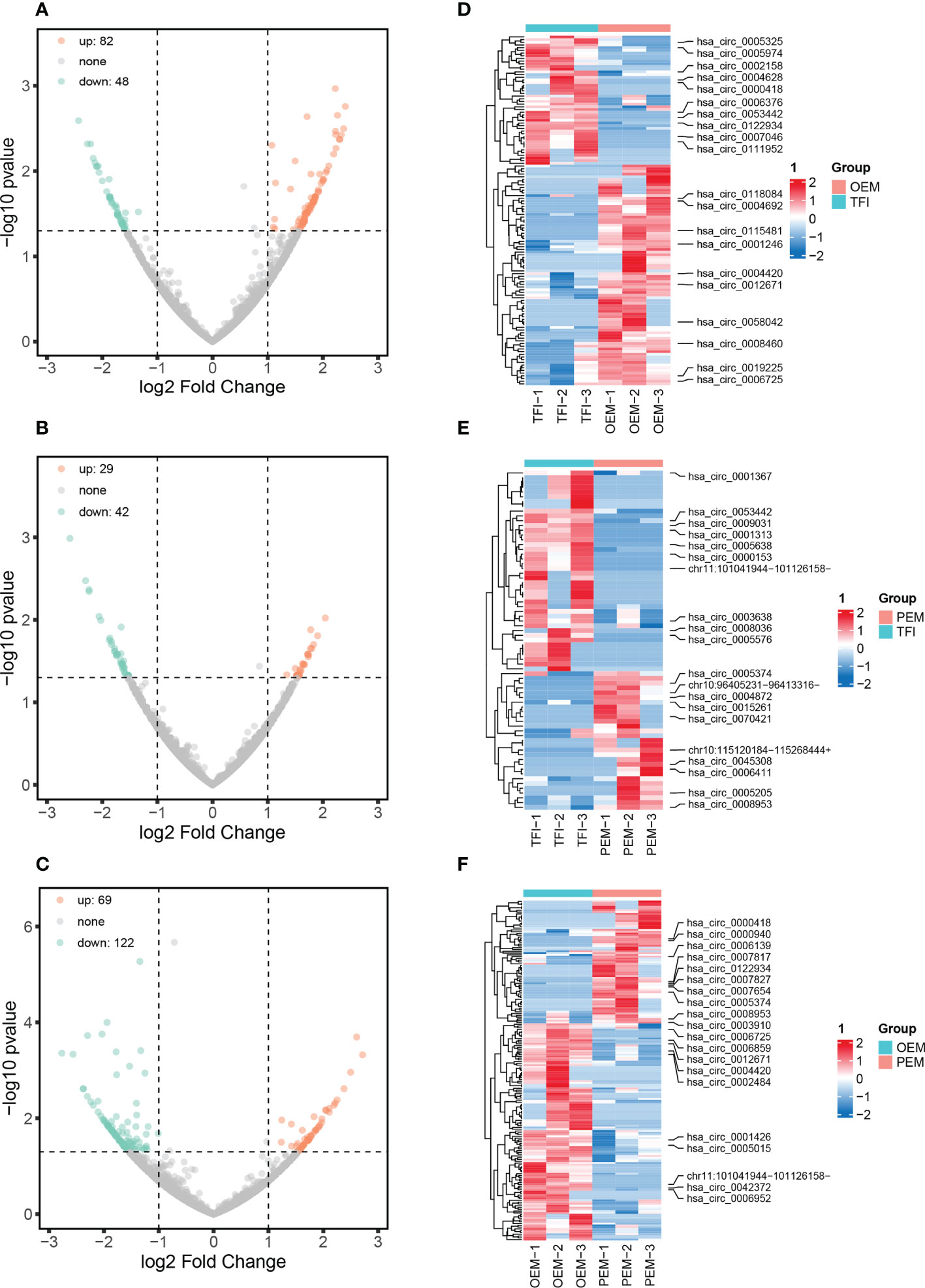
Figure 1 circRNA expression patterns of cumulus cells in patients with OEM (n = 3), PEM (n = 3), and TFI (n = 3). (A–C) Volcano plots showing all differentially expressed circRNAs between each of the two groups: (A) OEM and TFI; (B) PEM and TFI; and (C) OEM and PEM groups. (D–F) Heatmaps demonstrate the top 10 up- and downregulated circRNAs between the respective two groups. The positions of novel circRNAs are based on the hg 38 version. circRNAs, circular RNAs; OEM, ovarian endometriosis; PEM, peritoneal endometriosis; TFI, tubal factor infertility.
3.2 Finding common and respective unique circRNAs in the OEM and PEM groups
After taking the intersection of the DECs in each of the two groups (Figure 2), we found that 11 circRNAs were differentially expressed in the OEM and PEM groups compared to those in the TFI group, and the tendency of up/down-regulation was consistent. A total of 39 circRNAs were considered unique DECs in the OEM group, which were consistently upregulated or downregulated compared with that in the PEM and TFI groups. Similarly, 17 circRNAs were identified as unique differentially expressed circRNAs in the PEM group. Supplementary Table 2 presents information on all circRNAs in the three assemblages.
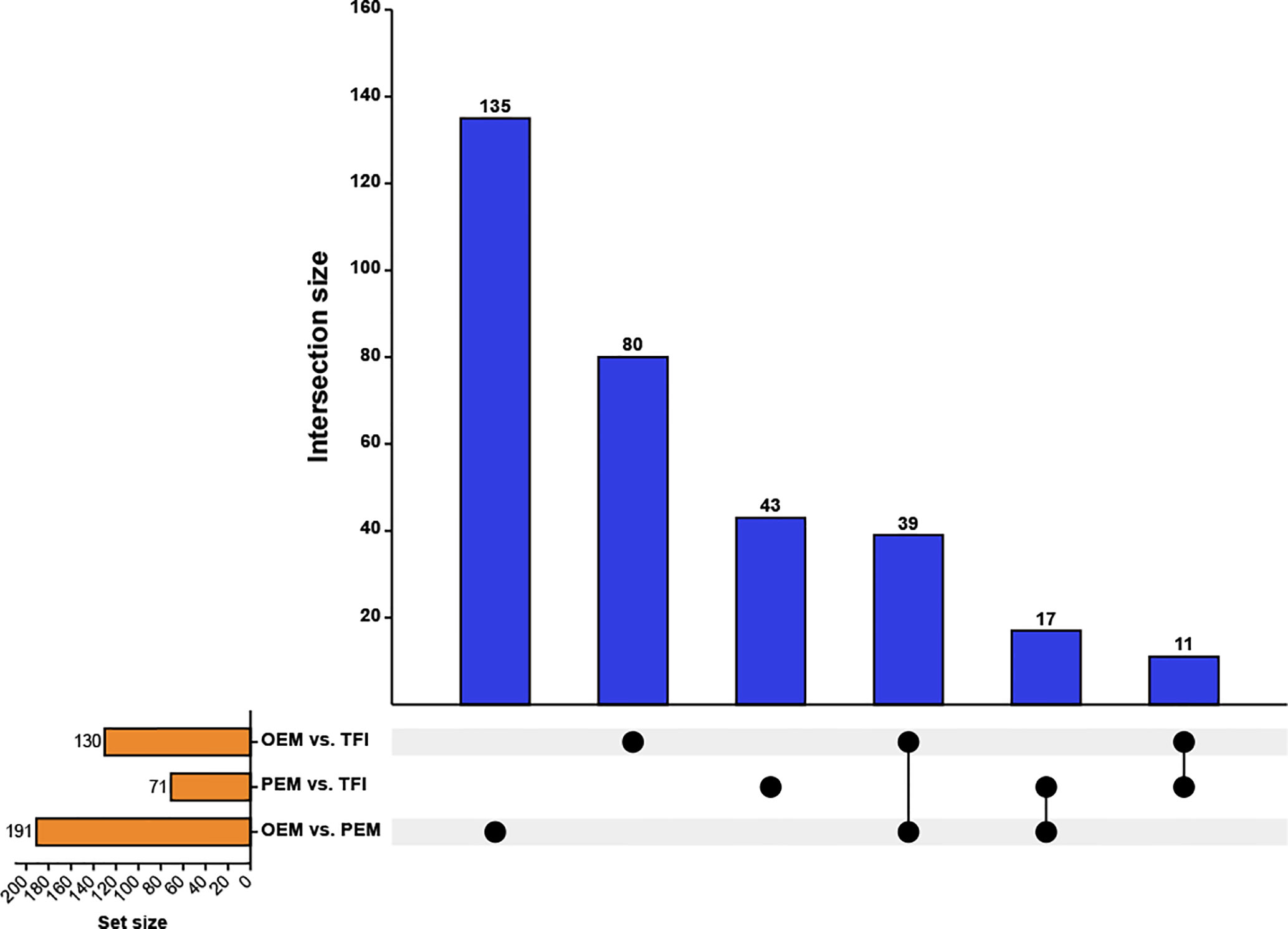
Figure 2 Upset plot depicting the intersections of the DECs. The blue columns represent the intersection size, and the yellow columns represent the size of the DECs of each comparison group. DECs, differentially expressed circular RNAs; OEM, ovarian endometriosis; PEM, peritoneal endometriosis; TFI, tubal factor infertility.
3.3 qRT-PCR validation of selected circRNAs
We selected seven circRNAs for qRT-PCR validation in 30 samples; Table 1 contains corresponding detailed information. According to our results (Figure 3), only hsa_circ_0004872 from the PEM unique cluster was significantly upregulated compared to the OEM and TFI groups, with P values of 0.038 and 0.005, respectively. hsa_circ_0003638 was found to be significantly upregulated in the OEM group compared to that in the TFI group (P = 0.035), which was opposite to the results of RNA sequencing; no difference was observed in the expression levels of hsa_circ_0003638 between the PEM and TFI groups (P = 0.412). The expression level of hsa_circ_0005205 was increased in the PEM group (P = 0.016) but was comparable in the OEM group (P = 0.413) compared with the TFI group. Another circRNA from the PEM unique cluster, hsa_circ_0008927, was marginally upregulated when compared with the TFI group (P = 0.047), but no significant difference in expression was found when compared with the OEM group (P = 0.132).
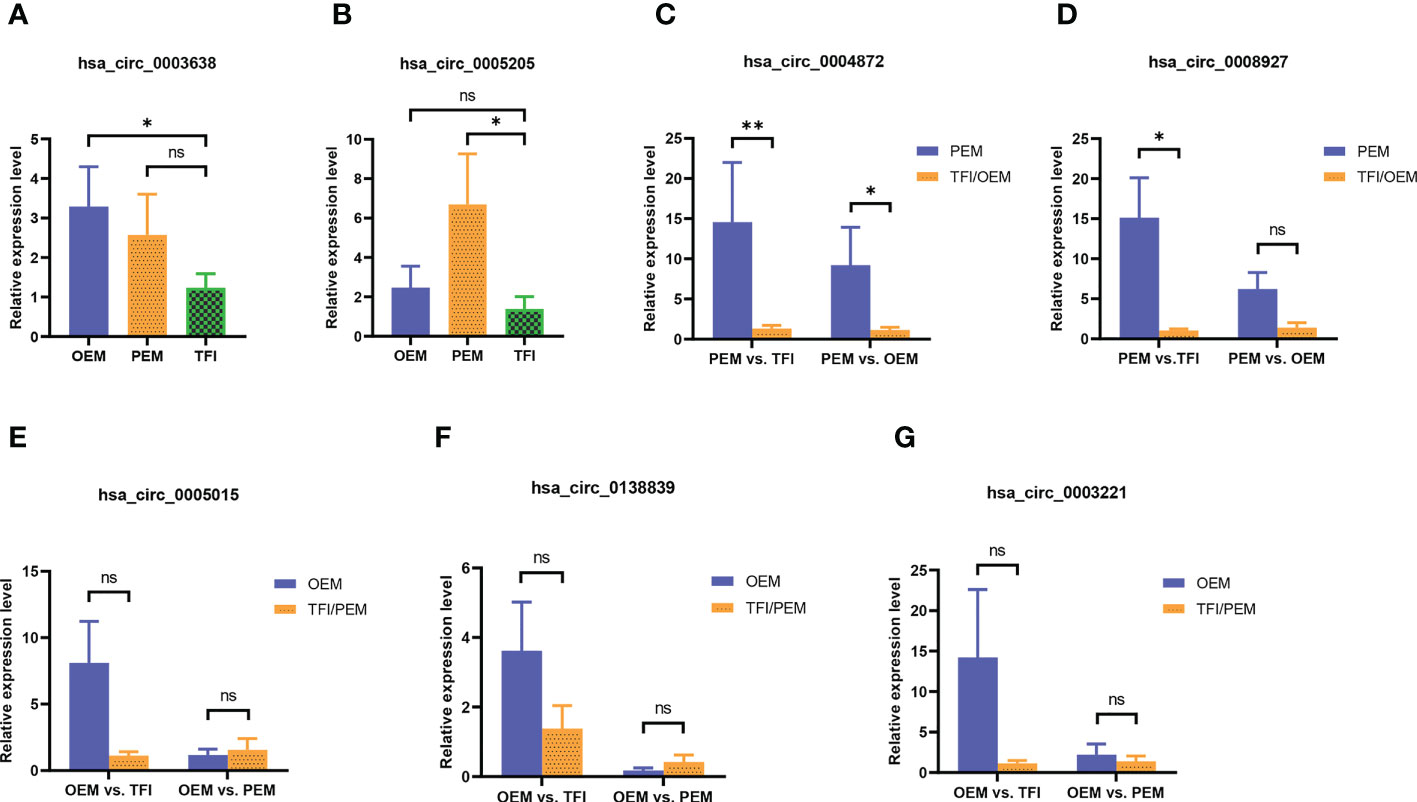
Figure 3 (A–G) The relative candidate circRNA expression levels of cumulus cells in patients with OEM (n = 10), PEM (n = 10), and TFI (n = 10) by qRT-PCR. **P < 0.01; *P < 0.05; ns, not significant. circRNAs, circular RNAs; OEM, ovarian endometriosis; PEM, peritoneal endometriosis; TFI, tubal factor infertility; qRT-PCR, quantitative real−time polymerase chain reaction.
3.4 Construction of ceRNA networks
We constructed two ceRNA networks based on hsa_circ_0004872, which were validated using qRT-PCR (Figure 4). Five miRNAs were predicted to bind to hsa_circ_0004872, namely hsa-miR-608, hsa-miR-638, hsa-miR-4469, hsa-miR-4721, and hsa-miR-6895-5p. The ceRNA network constructed by the PEM–TFI comparison groups contained 59 mRNAs, and 120 mRNAs were included in the network constructed by the PEM–OEM comparison groups.
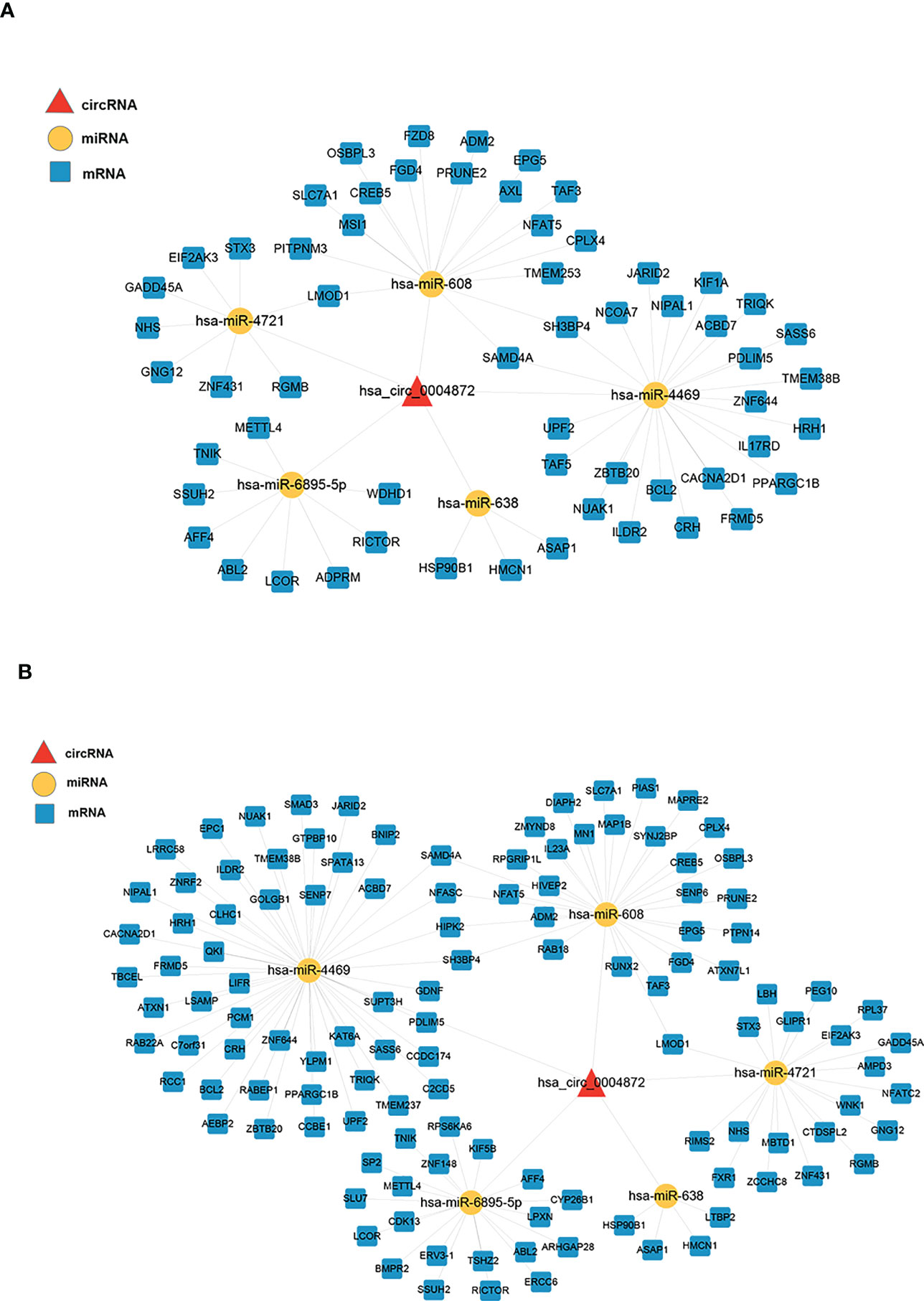
Figure 4 ceRNA networks constructed using the validated circRNAs, predicted miRNAs, and overlapped mRNAs. The circRNAs, miRNAs, and mRNAs are denoted by red triangles, yellow nodes, and blue round rectangles, respectively. (A) The PEM–TFI comparison groups and (B) PEM–OEM comparison groups. ceRNA, competitive endogenous RNA; circRNAs, circular RNAs; PEM, peritoneal endometriosis; TFI, tubal factor infertility; OEM, ovarian endometriosis.
3.5 GO and KEGG enrichment analysis of circRNA-targeted genes
GO and KEGG enrichment analyses were performed for further functional investigation of the circRNA-targeted genes. In the PEM–TFI comparison groups, the results of GO analysis showed that these target genes were mainly involved in cellular response to glucose starvation, mitochondrial transcription, and signal transduction by p53 class mediators (Figure 5A). As for the PEM–OEM comparison groups, the mainly enriched BP terms included transmembrane receptor protein serine/threonine kinase signaling pathway, BMP signaling pathway, and positive regulation of intracellular protein transport (Figure 5A). According to the results of KEGG analysis, apoptosis and PI3K–AKT and p53 signaling pathways were significantly enriched in the PEM–TFI comparison groups (Figure 5B), whereas the JAK–STAT signaling pathway and TGF-β signaling pathway were included in the PEM–OEM comparison groups (Figure 5B).
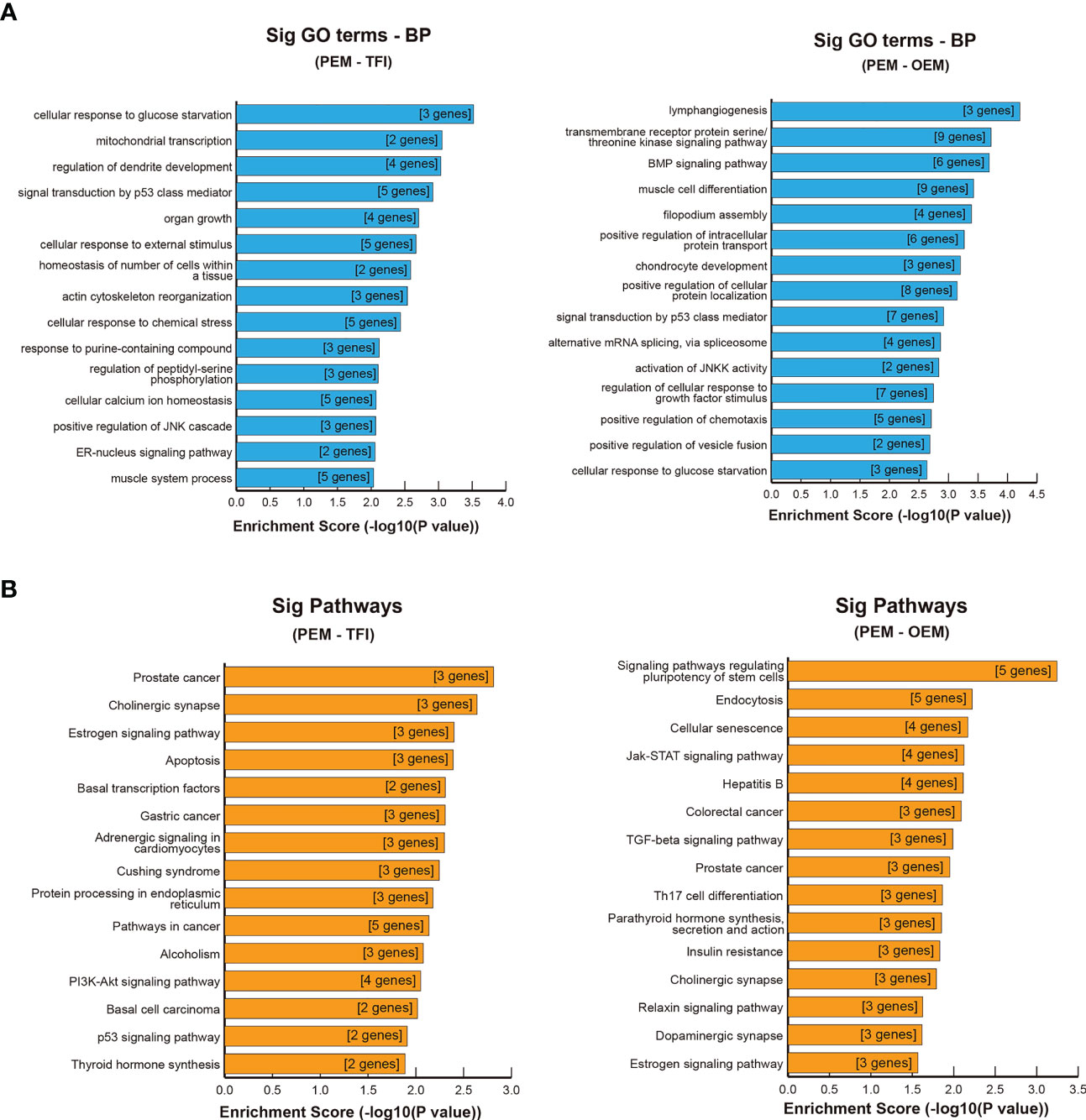
Figure 5 GO and KEGG enrichment analysis of circRNA-targeted genes. (A) Significant BP terms of GO analysis in the PEM–TFI and PEM–OEM comparison groups. (B) Significant KEGG pathways in the PEM–TFI and PEM–OEM comparison groups. The top 15 BP terms and pathways are displayed. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; circRNAs, circular RNAs; BP, biological process; PEM, peritoneal endometriosis; TFI, tubal factor infertility; OEM, ovarian endometriosis.
4 Discussion
Endometriosis is a common benign disease usually accompanied by infertility problems in women of childbearing age. Endometriosis damages the quality of oocytes, leading to lower rate of mature oocytes, higher rates of morphologically abnormal oocytes and abnormal mitochondria (27–29). Thus, it is necessary to study the mechanisms underlying endometriosis-damaging oocytes. However, owing to the precision of human oocytes, direct use of oocytes for further studies is usually not feasible. CCs and follicular fluid provide an important intrafollicular environment that supports oocyte development (6). Intercellular dialogs between CCs and oocytes are accomplished by gap junctions and paracrine signals, and these contacts are critical for follicular growth, oocyte maturation, and oocyte competence acquisition (6). Because of this close relationship, CCs are often used as an indirect approach for studying oocytes.
Traditionally, endometriosis lesions have three phenotypes: peritoneal, ovarian (endometrioma), and deep (30). Different phenotypes may have varying effects on female fertility. A large observational, cross-sectional study including 2208 patients illustrated that peritoneal superficial endometriosis was a risk factor for endometriosis-associated infertility; however, endometrioma was not found to be a significant risk factor for infertility (12). Another retrospective study observed lower mature oocyte and fertilization rates in patients with PEM than in endometrioma-affected patients, confirming the different impacts of PEM and OEM on the acquisition of oocyte competence (31). Against the background of elevated levels of various proinflammatory cytokines in peritoneal fluid in patients with PEM, Opøien et al. evaluated the relationship between OEM and local intrafollicular inflammation by testing the cytokine concentrations in the follicular fluid (13). They found that cytokine levels in the follicular fluid were comparable among patients with unilateral endometriomas, bilateral endometriomas, and without endometriosis, indicating that OEM, whether unilateral or bilateral, does not induce an inflammatory environment in nearby follicles. However, there is a lack of studies on this issue; therefore, more evidence is needed to explore the different effects of PEM and OEM on female fertility, oocyte competence, and oocyte quality.
circRNAs, as members of non-coding RNAs, have been reported to regulate gene expression in different ways, such as binding miRNAs like sponges, regulating transcription, influencing splicing, and modifying the cellular function of bound factors as protein decoys (10). In recent years, some studies have shown that the expression profile of circRNAs in CCs is altered in patients with gynecological diseases. For instance, Ma et al. identified 286 DECs in CCs between patients with and without polycystic ovary syndrome, and three circRNAs (hsa_circ_0043533, hsa_circ_0043532, and hsa_circ_0097636) were validated to be differentially expressed by qRT-PCR (32). circRNA_103827 and circRNA_104816 were upregulated in CCs in women over 38 years old, and their expression levels were negatively correlated with embryo quality (33). Two studies performed by Wu et al. and Guo et al. confirmed the differences in circRNA expression profiles in CCs between patients with and without OEM (16, 34). Nonetheless, there are no studies on the circRNA expression profiles of CCs in patients with PEM, especially comparing the differences in circRNA expression profiles between patients with PEM and OEM infertility.
In this study, we explored the circRNA expression profiles of CCs in patients with infertility related to OEM, PEM, or TFI via high-throughput sequencing and compared the differences in circRNA expression between the (1) OEM and TFI groups, (2) PEM and TFI groups, and (3) OEM and PEM groups. The results showed that abundant circRNAs were identified, and approximately 26 percent of them were novel. We detected 130, 71, and 191 significantly DECs in the OEM–TFI, PEM–TFI, and OEM–PEM comparison groups, respectively, indicating that the circRNA expression profiles differed among the three groups. To investigate the similarities and differences in the effects of different endometriosis phenotypes on oocytes, we examined the intersections of DECs with consistent expression tendencies in each of the two comparison groups. Finally, we found 11 circRNAs involved in the common mechanisms of oocyte impairment in patients with OEM or PEM; 39 and 17 circRNAs were identified as unique key circRNAs in the OEM and PEM groups, respectively. Subsequently, seven circRNAs from three assemblages were selected for qRT-PCR validation. hsa_circ_0004872 was found to be significantly upregulated in the PEM group compared to both the OEM and TFI groups, implying that it might play a critical role in influencing oocyte quality and competence in patients with PEM. hsa_circ_0004872, spliced from the mitogen-activated protein kinase 1 (MAPK1), is downregulated in gastric cancer tissues and inhibits cancer cell proliferation and invasion by binding miR-224 or coding protein MAPK1-109aa (35, 36).
To date, the most studied function of circRNAs is to act as “sponges” to bind miRNAs and affect the expression levels of downstream target mRNAs, that is, to form ceRNA networks to regulate gene expression. Thus, we constructed ceRNA networks of hsa_circ_0004872 to identify sponged miRNAs and their downstream target genes. In the ceRNA network, five miRNAs were predicted to bind to hsa_circ_0004872. Among these, hsa-miR-608 and hsa-miR-638 have been reported to be involved in the occurrence, development, and progression of cancer (37, 38). Hsa-miR-4469 can act as a tumor suppressor by regulating the infiltration of inflammatory cells in colorectal cancer, and hsa-miR-4721 has been found to promote the invasion capacity of nasopharyngeal carcinoma cells (39, 40).
To determine the function of circRNA-targeted genes, GO and KEGG enrichment analyses were performed, revealing that several important signaling pathways might be involved in the detrimental effects of PEM on oocytes. In the PEM–TFI comparison groups, some target genes were enriched in the apoptosis, PI3K–AKT signaling pathway, and p53 signaling pathway, which have been reported to affect oocyte development. A systematic review of 22 articles revealed that the PI3K/AKT/PTEN pathway can regulate the progression to MII and blastulation of mammalian oocytes during in vitro maturation (41). Wei et al. illustrated that two downregulated long non-coding RNAs (NEAT1 and NORAD) impair oocyte maturation and genome integrity by modulating PI3K–AKT pathway genes, leading to recurrent oocyte maturation arrest in women (42). The activity of the p53 tumor suppressor protein can induce cell cycle arrest or apoptosis and is related to the apoptosis of granulosa cells (43). Several experiments have confirmed the close relationship between the p53 pathway and granulosa cell death. For example, Yang et al. elucidated that the p53 pathway is involved in oxidative stress-induced apoptosis of granulosa cells using COV434, a human granulosa cell line (44). Another study reported that p53 induction in CCs impaired oocyte function and quality by disturbing maturation and fertilization in mouse models (45). In contrast, functional analysis of target genes in the PEM–OEM comparison groups demonstrated that the JAK–STAT and TGF-β signaling pathways may be involved in the different effects of PEM and OEM on oocyte quality and competence. The JAK–STAT signaling pathway is a well-established pathway involved in cell proliferation, and STAT3 signaling is extensively enriched in human primordial and primary follicles (46). Some findings have revealed the involvement of the JAK–STAT pathway in follicular development in mares and mice (47, 48). Furthermore, Frost et al. confirmed that the JAK–STAT signaling pathway contributes to the crosstalk between oocytes and CCs in humans using the COV434 cell line (49). Another enriched pathway is the TGF-β signaling pathway, which is reported to mediate the activation of primordial follicles, folliculogenesis, and oocyte–CC communication (50). In the Caenorhabditis elegans model, TGF-β signaling was found to regulate reproductive aging by modulating many aspects, such as oocyte morphology and quality, oocyte fertilization ability, chromosome segregation fidelity, and embryo viability and integrity (51). TGF-β is believed to be involved in the etiology of PEM. The “Warburg-like” effect induced by TGF-β, which refers to the metabolic reprogramming of glucose to lactate via glycolysis, may play a role in the development of PEM lesion (52). In addition, a review including 95 studies demonstrated that increased levels of TGF-β1 are related to PEM lesion development by changing ectopic endometrial and peritoneal cell metabolism and initiating neoangiogenesis, while increased levels of TGF-β ligands lead to increased attachment, invasion, and proliferation of ectopic endometrial cells (53). Thus, it is supposed that the TGF-β signaling pathway not only plays a critical role in the development of PEM lesions but may also be an important unique mechanism by which PEM damages oocyte quality and competence.
Our study had several limitations. First, the sample size for the qRT-PCR validation was small. Second, all patients in the OEM group had stage III–IV disease, while in the PEM group, all patients had stage I disease. Thus, larger samples including different stages of the OEM and PEM are warranted for further validation. Third, due to the insufficient total amount of RNA in each sample, we could not directly obtain the sequences of miRNAs. Finally, the molecular mechanisms and functions of the validated circRNAs should be investigated in the future, for example, by performing knockdown and overexpression experiments using the human granulosa cell line.
In summary, we revealed for the first time that circRNA expression patterns of CCs in patients with infertility showing different endometriosis phenotypes (OEM and PEM). We verified that the OEM and PEM groups shared 11 common circRNAs that may be involved in the common mechanisms of impaired oocyte quality in patients with OEM or PEM. circRNAs (n = 39) in the OEM group and 17 in the PEM group were identified as unique circRNAs. hsa_circ_0004872 was found to be significantly upregulated in the PEM group compared to that in the OEM and TFI groups, and downstream target genes were verified by constructing ceRNA networks and sequencing. Based on the results of functional analysis of target genes, we speculate that PEM damages oocyte competence and development via the PI3K–AKT and p53 signaling pathways, while JAK–STAT, especially the TGF-β signaling pathway, may be involved in a unique impairment mechanism different from that in the OEM. Our findings provide novel insights into the effects of different endometriosis phenotypes on oocytes. In addition, hsa_circ_0004872, as the PEM unique circRNA, may be used as a potential biomarker to help noninvasively identify whether infertility patients have concealed PEM and whether OEM affected patients have concomitant PEM lesions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://ngdc.cncb.ac.cn/gsa-human/, HRA003694.
Ethics statement
The studies involving human participants were reviewed and approved by Institutional Review Board of the Peking Union Medical College Hospital. The patients/participants provided their written informed consent to participate in this study.
Author contributions
XH performed sample collection and purification, analyzed the qRT-PCR data, visualized the results, and compiled the manuscript. QY contributed to the study design and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Key Research and Developmental Program of China (2022YFC2703800) and the National High Level Hospital Clinical Research Funding (2022-PUMCH-B-081).
Acknowledgments
We sincerely appreciate the cooperation of all the patients and reproductive laboratory staff involved in this study and for obtaining samples. We also thank Shanghai Lifegenes Technology Co., Ltd. (Shanghai, China), data acquisition, and BGI BEIJING Co. (Beijing, China).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1137235/full#supplementary-material
Abbreviations
ART, assisted reproductive technology; BMI, body mass index; CCs, cumulus cells; ceRNA, competitive endogenous RNA; circRNAs, circular RNAs; DEC, differentially expressed circRNA; FSH, follicle stimulating hormone; GnRH, gonadotropin-releasing hormone; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; OEM, ovarian endometriosis; PEM, peritoneal endometriosis; TFI, tubal factor infertility.
References
1. Meuleman C, Vandenabeele B, Fieuws S, Spiessens C, Timmerman D, D'Hooghe T. High prevalence of endometriosis in infertile women with normal ovulation and normospermic partners. Fertil Steril (2009) 92(1):68–74. doi: 10.1016/j.fertnstert.2008.04.056
2. Muteshi CM, Ohuma EO, Child T, Becker CM. The effect of endometriosis on live birth rate and other reproductive outcomes in art cycles: A cohort study. Hum Reprod Open (2018) (4):1–7. doi: 10.1093/hropen/hoy016
3. Yanushpolsky EH, Best CL, Jackson KV, Clarke RN, Barbieri RL, Hornstein MD. Effects of endometriomas on ooccyte quality, embryo quality, and pregnancy rates in in vitro fertilization cycles: A prospective, case-controlled study. J Assist Reprod Genet (1998) 15(4):193–7. doi: 10.1023/a:1023048318719
4. Borges E Jr., Braga DP, Setti AS, Vingris LS, Figueira RC, Iaconelli A Jr. Endometriosis affects oocyte morphology in intracytoplasmic sperm injection cycles? JBRA Assist Reprod (2015) 19(4):235–40. doi: 10.5935/1518-0557.20150046
5. Turathum B, Gao EM, Chian RC. The function of cumulus cells in oocyte growth and maturation and in subsequent ovulation and fertilization. Cells (2021) 10(9):2292. doi: 10.3390/cells10092292
6. Da Broi MG, Giorgi VSI, Wang F, Keefe DL, Albertini D, Navarro PA. Influence of follicular fluid and cumulus cells on oocyte quality: Clinical implications. J Assist Reprod Genet (2018) 35(5):735–51. doi: 10.1007/s10815-018-1143-3
7. Barcelos ID, Donabella FC, Ribas CP, Meola J, Ferriani RA, de Paz CC, et al. Down-regulation of the Cyp19a1 gene in cumulus cells of infertile women with endometriosis. Reprod BioMed Online (2015) 30(5):532–41. doi: 10.1016/j.rbmo.2015.01.012
8. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular rna: A new star of noncoding rnas. Cancer Lett (2015) 365(2):141–8. doi: 10.1016/j.canlet.2015.06.003
9. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular rnas are abundant, conserved, and associated with alu repeats. Rna (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
10. Panda AC, Grammatikakis I, Munk R, Gorospe M, Abdelmohsen K. Emerging roles and context of circular rnas. Wiley Interdiscip Rev RNA (2017) 8(2). doi: 10.1002/wrna.1386
11. Tu J, Yang H, Chen Y, Chen Y, Chen H, Li Z, et al. Current and future roles of circular rnas in normal and pathological endometrium. Front Endocrinol (Lausanne) (2021) 12:668073. doi: 10.3389/fendo.2021.668073
12. Santulli P, Lamau MC, Marcellin L, Gayet V, Marzouk P, Borghese B, et al. Endometriosis-related infertility: Ovarian endometrioma per Se is not associated with presentation for infertility. Hum Reprod (2016) 31(8):1765–75. doi: 10.1093/humrep/dew093
13. Opøien HK, Fedorcsak P, Polec A, Stensen MH, Åbyholm T, Tanbo T. Do endometriomas induce an inflammatory reaction in nearby follicles? Hum Reprod (2013) 28(7):1837–45. doi: 10.1093/humrep/det087
14. Medicine ASfR. Revised American society for reproductive medicine classification of endometriosis: 1996. Fertil Steril (1997) 67(5):817–21. doi: 10.1016/s0015-0282(97)81391-x
15. da Silva LFI, Da Broi MG, da Luz CM, da Silva L, Ferriani RA, Meola J, et al. Mir-532-3p: A possible altered mirna in cumulus cells of infertile women with advanced endometriosis. Reprod BioMed Online (2021) 42(3):579–88. doi: 10.1016/j.rbmo.2020.10.010
16. Wu R, Li J, Li J, Yan X, Zhou W, Ling C, et al. Circular rna expression profiling and bioinformatic analysis of cumulus cells in endometriosis infertility patients. Epigenomics (2020) 12(23):2093–108. doi: 10.2217/epi-2020-0291
17. Ferraretti AP, La Marca A, Fauser BC, Tarlatzis B, Nargund G, Gianaroli L. Eshre consensus on the definition of 'Poor response' to ovarian stimulation for in vitro fertilization: The Bologna criteria. Hum Reprod (2011) 26(7):1616–24. doi: 10.1093/humrep/der092
18. Aghadavod E, Zarghami N, Farzadi L, Zare M, Barzegari A, Movassaghpour AA, et al. Isolation of granulosa cells from follicular fluid; applications in biomedical and molecular biology experiments. Adv BioMed Res (2015) 4:250. doi: 10.4103/2277-9175.170675
19. Kim D, Langmead B, Salzberg SL. Hisat: A fast spliced aligner with low memory requirements. Nat Methods (2015) 12(4):357–60. doi: 10.1038/nmeth.3317
20. Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, et al. Diverse alternative back-splicing and alternative splicing landscape of circular rnas. Genome Res (2016) 26(9):1277–87. doi: 10.1101/gr.202895.115
21. Glažar P, Papavasileiou P, Rajewsky N. Circbase: A database for circular rnas. Rna (2014) 20(11):1666–70. doi: 10.1261/rna.043687.113
22. Zhou L, Chen J, Li Z, Li X, Hu X, Huang Y, et al. Integrated profiling of micrornas and mrnas: Micrornas located on Xq27.3 associate with clear cell renal cell carcinoma. PloS One (2010) 5(12):e15224. doi: 10.1371/journal.pone.0015224
23. Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. Stringtie enables improved reconstruction of a transcriptome from rna-seq reads. Nat Biotechnol (2015) 33(3):290–5. doi: 10.1038/nbt.3122
24. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular rnas regulate transcription in the nucleus. Nat Struct Mol Biol (2015) 22(3):256–64. doi: 10.1038/nsmb.2959
25. Schmittgen TD, Livak KJ. Analyzing real-time pcr data by the comparative C(T) method. Nat Protoc (2008) 3(6):1101–8. doi: 10.1038/nprot.2008.73
26. Wu J, Mao X, Cai T, Luo J, Wei L. Kobas server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res (2006) 34(Web Server issue):W720–4. doi: 10.1093/nar/gkl167
27. Shebl O, Sifferlinger I, Habelsberger A, Oppelt P, Mayer RB, Petek E, et al. Oocyte competence in in vitro fertilization and intracytoplasmic sperm injection patients suffering from endometriosis and its possible association with subsequent treatment outcome: A matched case-control study. Acta Obstet Gynecol Scand (2017) 96(6):736–44. doi: 10.1111/aogs.12941
28. Kasapoglu I, Kuspinar G, Saribal S, Turk P, Avci B, Uncu G. Detrimental effects of endometriosis on oocyte morphology in intracytoplasmic sperm injection cycles: A retrospective cohort study. Gynecol Endocrinol (2018) 34(3):206–11. doi: 10.1080/09513590.2017.1391203
29. Xu B, Guo N, Zhang XM, Shi W, Tong XH, Iqbal F, et al. Oocyte quality is decreased in women with minimal or mild endometriosis. Sci Rep (2015) 5:10779. doi: 10.1038/srep10779
30. International working group of AAGL E, ESHRE and WES, Vermeulen N, Horne AW, Johnson NP, Lee TTM, Missmer S. Endometriosis classification, staging and reporting systems: A review on the road to a universally accepted endometriosis classification. Hum Reprod Open (2021) 00(4):1–28. doi: 10.1093/hropen/hoab025
31. Li A, Zhang J, Kuang Y, Yu C. Analysis of Ivf/Icsi-fet outcomes in women with advanced endometriosis: Influence on ovarian response and oocyte competence. Front Endocrinol (Lausanne) (2020) 11:427. doi: 10.3389/fendo.2020.00427
32. Ma Z, Zhao H, Zhang Y, Liu X, Hao C. Novel circular rna expression in the cumulus cells of patients with polycystic ovary syndrome. Arch Gynecol Obstet (2019) 299(6):1715–25. doi: 10.1007/s00404-019-05122-y
33. Cheng J, Huang J, Yuan S, Zhou S, Yan W, Shen W, et al. Circular rna expression profiling of human granulosa cells during maternal aging reveals novel transcripts associated with assisted reproductive technology outcomes. PloS One (2017) 12(6):e0177888. doi: 10.1371/journal.pone.0177888
34. Guo J, Zeng H, Li T, Liang X, Peng J. Mrna, lncrna and circular rna expression profiles in granulosa cells of infertile women with ovarian endometriosis. Reprod Sci (2022) 29(10):2937–46. doi: 10.1007/s43032-022-00966-3
35. Ma C, Wang X, Yang F, Zang Y, Liu J, Wang X, et al. Circular rna Hsa_Circ_0004872 inhibits gastric cancer progression Via the mir-224/Smad4/Adar1 successive regulatory circuit. Mol Cancer (2020) 19(1):157. doi: 10.1186/s12943-020-01268-5
36. Jiang T, Xia Y, Lv J, Li B, Li Y, Wang S, et al. A novel protein encoded by Circmapk1 inhibits progression of gastric cancer by suppressing activation of mapk signaling. Mol Cancer (2021) 20(1):66. doi: 10.1186/s12943-021-01358-y
37. Lu J, Zhu D, Li L. Biological functions and molecular mechanisms of mir-608 in cancer. Front Oncol (2022) 12:870983. doi: 10.3389/fonc.2022.870983
38. Hasheminasabgorji E, Mishan MA, Tabari MAK, Bagheri A. Mir-638: A promising cancer biomarker with therapeutic potential. Curr Mol Med (2022) 23(5):377–89. doi: 10.2174/1566524022666220405125900
39. Qi L, Wang L, Song F, Ding Z, Zhang Y. The role of mir-4469 as a tumor suppressor regulating inflammatory cell infiltration in colorectal cancer. Comput Struct Biotechnol J (2022) 20:3755–63. doi: 10.1016/j.csbj.2022.07.021
40. Tang Z, Chen W, Xu Y, Lin X, Liu X, Li Y, et al. Mir-4721, induced by ebv-Mir-Bart22, targets Gsk3β to enhance the tumorigenic capacity of npc through the Wnt/β-catenin pathway. Mol Ther Nucleic Acids (2020) 22:557–71. doi: 10.1016/j.omtn.2020.09.021
41. Alcaráz LP, Prellwitz L, Alves G, Souza-Fabjan JMG, Dias AJB. Role of phosphoinositide 3-kinase/ protein kinase b/ phosphatase and tensin homologue (Pi3k/Akt/Pten) pathway inhibitors during In vitro maturation of mammalian oocytes on In vitro embryo production: A systematic review. Theriogenology (2022) 189:42–52. doi: 10.1016/j.theriogenology.2022.06.009
42. Wei L, Xia H, Liang Z, Yu H, Liang Z, Yang X, et al. Disrupted expression of long non-coding rnas in the human oocyte: The possible epigenetic culprits leading to recurrent oocyte maturation arrest. J Assist Reprod Genet (2022) 39(10):2215–25. doi: 10.1007/s10815-022-02596-9
43. Hosokawa K, Aharoni D, Dantes A, Shaulian E, Schere-Levy C, Atzmon R, et al. Modulation of Mdm2 expression and P53-induced apoptosis in immortalized human ovarian granulosa cells. Endocrinology (1998) 139(11):4688–700. doi: 10.1210/endo.139.11.6280
44. Yang H, Xie Y, Yang D, Ren D. Oxidative stress-induced apoptosis in granulosa cells involves jnk, P53 and puma. Oncotarget (2017) 8(15):25310–22. doi: 10.18632/oncotarget.15813
45. Haraguchi H, Hirota Y, Saito-Fujita T, Tanaka T, Shimizu-Hirota R, Harada M, et al. Mdm2-P53-Sf1 pathway in ovarian granulosa cells directs ovulation and fertilization by conditioning oocyte quality. FASEB J (2019) 33(2):2610–20. doi: 10.1096/fj.201801401R
46. Ernst EH, Franks S, Hardy K, Villesen P, Lykke-Hartmann K. Granulosa cells from human primordial and primary follicles show differential global gene expression profiles. Hum Reprod (2018) 33(4):666–79. doi: 10.1093/humrep/dey011
47. Hall SE, Upton RMO, McLaughlin EA, Sutherland JM. Phosphoinositide 3-Kinase/Protein kinase b (Pi3k/Akt) and janus Kinase/Signal transducer and activator of transcription (Jak/Stat) follicular signalling is conserved in the mare ovary. Reprod Fertil Dev (2018) 30(4):624–33. doi: 10.1071/rd17024
48. Sutherland JM, Frost ER, Ford EA, Peters AE, Reed NL, Seldon AN, et al. Janus kinase Jak1 maintains the ovarian reserve of primordial follicles in the mouse ovary. Mol Hum Reprod (2018) 24(11):533–42. doi: 10.1093/molehr/gay041
49. Frost ER, Ford EA, Peters AE, Reed NL, McLaughlin EA, Baker MA, et al. Signal transducer and activator of transcription (Stat) 1 and Stat3 are expressed in the human ovary and have janus kinase 1-independent functions in the Cov434 human granulosa cell line. Reprod Fertil Dev (2020) 32(12):1027–39. doi: 10.1071/rd20098
50. Patton BK, Madadi S, Pangas SA. Control of ovarian follicle development by tgfβ family signaling. Curr Opin Endocr Metab Res (2021) 18:102–10. doi: 10.1016/j.coemr.2021.03.001
51. Luo S, Kleemann GA, Ashraf JM, Shaw WM, Murphy CT. Tgf-β and insulin signaling regulate reproductive aging Via oocyte and germline quality maintenance. Cell (2010) 143(2):299–312. doi: 10.1016/j.cell.2010.09.013
52. Young VJ, Brown JK, Maybin J, Saunders PT, Duncan WC, Horne AW. Transforming growth factor-β induced warburg-like metabolic reprogramming may underpin the development of peritoneal endometriosis. J Clin Endocrinol Metab (2014) 99(9):3450–9. doi: 10.1210/jc.2014-1026
Keywords: circular RNAs, ovarian endometriosis, peritoneal endometriosis, expression profiling, competitive endogenous RNA, cumulus cells
Citation: Huang X and Yu Q (2023) Bioinformatic analysis confirms differences in circular RNA expression profiles of cumulus cells between patients with ovarian and peritoneal endometriosis-associated infertility. Front. Endocrinol. 14:1137235. doi: 10.3389/fendo.2023.1137235
Received: 04 January 2023; Accepted: 06 March 2023;
Published: 15 March 2023.
Edited by:
Eytan R. Barnea, BioIncept LLC, United StatesReviewed by:
Ettore Luzi, University of Florence, ItalyNicoletta Di Simone, Humanitas University, Italy
Copyright © 2023 Huang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qi Yu, eXVxaTIwMDgwMDFAc2luYS5jb20=
 Xiaodi Huang
Xiaodi Huang Qi Yu
Qi Yu