- 1Department of Pharmacology, School of Medicine, The University of Jordan, Amman, Jordan
- 2Department of Biological Sciences, School of Science, The University of Jordan, Amman, Jordan
Background: Hypoxia plays a critical role in the tumor microenvironment by affecting cellular proliferation, metabolism, apoptosis, DNA repair, and chemoresistance. Since hypoxia provokes a distinct shift of microRNA, it is important to illustrate the relative contribution of each hypoxamiR to cancer progression.
Aims: The present study aims to shed light on the hypoxamiRs that are involved in pancreatic and breast cancer progression to highlight novel targets for the development of new therapies.
Methods: For 20 cycles, MCF7 breast cancer cells and PANC-1 pancreatic cancer cells were subjected to chronic cyclic hypoxia, which consisted of 72 hours of hypoxia followed by 24 hours of reoxygenation. After 10 and 20 cycles of hypoxia, miRNA expression alterations were profiled using RT-PCR array and further analyzed using a visual analytics platform. The MTT cell proliferation assay was used to determine hypoxic cells’ chemoresistance to doxorubicin.
Results: Under chronic cyclic hypoxia, hypoxic PANC-1 cells have a comparable doubling time with their normoxic counterparts, whereas hypoxic MCF7 cells show a massive increase in doubling time when compared to their normoxic counterparts. Both hypoxic cell lines developed EMT-like phenotypes as well as doxorubicin resistance. According to the findings of miRNet, 6 and 10 miRNAs were shown to play an important role in enriching six hallmarks of pancreatic cancer in the 10th and 20th cycles of hypoxia, respectively, while 7 and 11 miRNAs were shown to play an important role in enriching the four hallmarks of breast cancer in the 10th and 20th cycles of hypoxia, respectively.
Conclusions: miR-221, miR-21, miR-155, and miR-34 were found to be involved in the potentiation of hypoxic PANC-1 hallmarks at both the 10th and 20th cycles, while miR-93, miR-20a, miR-15, and miR-17 were found to be involved in the potentiation of hypoxic MCF7 hallmarks at both the 10th and 20th cycles. This variation in miRNA expression was also connected to the emergence of an EMT-like phenotype, alterations in proliferation rates, and doxorubicin resistance. The chemosensitivity results revealed that chronic cyclic hypoxia is critical in the formation of chemoresistant phenotypes in pancreatic and breast cancer cells. miR-181a and let-7e expression disparities in PANC1, as well as miR-93, miR-34, and miR-27 expression disparities in MCF7, may be associated with the formation of chemoresistant MCF7 and PANC-1 cells following 20 cycles of chronic cyclic hypoxia. Indeed, further research is needed since the particular mechanisms that govern these processes are unknown.
1 Introduction
Cancer is the devastating growth of cells along with inhibition of apoptotic pathways as well as the acquisition of metastatic characteristics via triggering oncogenes or frustrating tumor suppressor genes. Breast cancer (BC) ranks as the most common cancer in women. Many cases of BC have shown dissimilar clinical results despite having similar characteristics. In addition to drug resistance cases that arise as a result of complexity, heterogeneity, and metastasis (1), the level of hypoxia (oxygen deprivation) within the tumor microenvironment is also a factor that complicates tumor progression, metastasis, and the survival of breast cancer patients (2).
On the other hand, pancreatic ductal adenocarcinoma (PDAC) is one of the most fatal cancers worldwide, with a mortality rate almost equal to the incidence rate (3). The absence of early worrisome symptoms, rapid metastasis, highly malignant patterns, and an innate resistance to conventional chemotherapy treatments are the primary causes of PDAC prognosis (4). Furthermore, PDAC possesses therapeutic-obstructive features such as heterogeneity, a desmoplastic and hypoxic tumor microenvironment (TME), intrinsic resistance to chemotherapy, and immunotherapy resistance (5). The limitations of current treatment strategies for breast cancer and PDAC emphasize the pressing need to explore key molecular pathways related to early diagnosis, prognosis, and therapy improvement.
Cancer patients rarely die from primary tumors; cancer cells’ metastatic capacity is responsible for the severity, aggressiveness, and majority of cancer-related deaths. Hypoxia performs a critical function in triggering metastasis through manipulation of the tumor microenvironment (TME). It also promotes drug resistance, angiogenesis, metabolic reprogramming, and proliferation by interfering with different regulatory networks of transcription factors and signaling proteins. Accordingly, hypoxia can boost several hallmarks of cancer (6). Recent studies demonstrated that microRNA plays an important role in the adaptive response to hypoxia in tumors. Modification of specific miRNAs, collectively called hypoxamiRs, has become a remarkable feature of hypoxia (7). HypoxamiRs are microRNAs that are not only tightly regulated by hypoxia but also influence cell responses to reduced oxygen tension (8). A growing body of research suggests that hypoxia controls several aspects of hypoxamiR transcription and function (9). Thus, it is essential to define the distinct expression of miRNAs in different types of cancers to suggest specific treatment strategies and avoid any off-target effects.
Our goal was to figure out how miRNAs interact with the hypoxic microenvironment and what function this interaction plays in carcinogenesis. The hypoxic microenvironment of cancer cells is simulated, as closely as feasible, by exposing breast and pancreatic cancer cells to chronic cyclic hypoxia for 20 cycles. This enables the detection of changes in miRNA expression patterns and their consequences for cellular regulatory processes, possibly leading to the identification of novel therapeutic targets.
2 Materials and methods
2.1 Cell culture
Pancreatic cancer cell line (PANC-1) and breast adenocarcinoma cell line (MCF7) were purchased from the American Type Culture Collection (ATCC, USA). PANC-1 cells were cultured in Dulbecco’s Modified Medium High Glucose (Euro clone, Italy) while MCF7 cells were cultured in RPMI 1640 medium (Euro clone, Italy), each of which was supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Biowest, South America), 1% of 2mM L-glutamine (Euro Clone, Italy), and 100U/ml and 100µg/ml penicillin-streptomycin (Euro Clone, Italy). Cells were routinely propagated and cultured in a cell culture incubator (Nuaire, USA) at 37°C and (5% CO2) 95% atmospheric air (normoxic conditions); then each cell line was divided into non-hypoxic and hypoxic cells as a control and experimental group, respectively.
2.2 Cells passaging
When the confluence of adherent cells reached 80%, the cells were passaged using Trypsin-EDTA (0.05%) in DPBS (1x) (Capricon Scientific, Germany) to allow them to survive, grow and divide. PANC-1 and MCF7 cancer cells were counted and evaluated in terms of viability via trypan blue 0.5% solution (Biowest, South America); after 1:1 dilution of homogenized trypan blue and cells, the solution was loaded onto a hemacytometer (Neubauer Double, Zuzi, Spain) and thus could be counted. The doubling time of both the control and hypoxic cells was calculated and compared throughout the experiment.
2.3 Exposure to hypoxia
AnaeroGen Compact (Oxoid; Hampshire, UK) was used for hypoxic episode application. The system encompasses a tightly sealed bag with a gas-generating sachet. Each sachet contains activated carbon and ascorbic acid, which combine with the air when opened, consuming oxygen and reducing its concentration within the bag to less than 1%. Culture flasks and opened sachets were placed inside plastic pouches, which were then sealed. PANC-1 and MCF7 cells were exposed to a 72-hour hypoxic cycle using the abovementioned system followed by 24-48 hours of reoxygenation. Cells were subjected to 20 cycles of hypoxia. Apart from these signified cycles, control cells were split beside their hypoxic counterparts and were subjected to similar conditions except hypoxic cycles (10, 11).
2.4 Cell proliferation assay
PANC-1 and MCF7 viability, for both hypoxic and normoxic cells, was evaluated after the10th and 20th hypoxic cycles using a Cell Titer 96® Non-Radioactive Cell Proliferation Assay (MTT) (Promega, USA). Doxorubicin 50mg/25 ml (Ebewe, Austria) was used in descending concentrations, from 25µM to 0.1 µM, to assess the chemo-sensitivity of both groups. Thereafter, optical density was determined using a Synergy multi-mode reader (Biotek, USA) at a wavelength of 570 nm; consequently, the cells’ viability (%) could be compared. Statistical analysis numerical data were expressed as mean ± SD and the statistical significance was determined by using the Student’s t-test.
2.5 Total ribonucleic acid extraction
After the 10th and 20th hypoxic cycles, 3×103 cells of each cell line (PANC-1 and MCF7) were subjected to total RNA extraction according to the miRNeasy Mini Kit (Qiagen, Germany) quick start protocol; RNA samples were preserved at -80°C for the next step. RNA purity and concentrations of RNA samples were assessed using a NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, USA). Each concentration was tested in duplicate.
2.6 miRNA expression profiling
The extracted total RNA was converted to complementary DNA (cDNA) using the miScript ll RT kit (Qiagen, SABIO). The miScript SYBR® Green PCR ready master mix was used to prepare samples, and levels of expression of human breast cancer and pancreatic cancer miRNAs were investigated using a Human Hypoxia Signaling miScript miRNA PCR Array (Qiagen, SABIO). The panels of miRNA that were used in this research were ready-to-use PCR panels (Qiagen, SABIO), with a 96-well format that contained 84 miRNA assays targeting human hypoxia signaling, reference miRNAs, and control assays, according to the manufacturer’s instructions, via a Bio-Rad CFX96 instrument (Qiagen, Germany). Variation in the cDNA content was normalized using different small nuclear and small nucleolar RNAs (snRNAs and snoRNAs) like SNORD61, SNORD68, SNORD72, SNORD95, SNORD96A, and RNU6-6P (Appendix D). Reverse transcription control and positive PCR control were also performed. The 2-ΔΔCt method was used to calculate the relative expression levels of the miRNA. miRNAs expression analysis was performed using the GeneGlobe Data Analysis Center from Qiagen. The arrays’ reproducibility were determined by analyzing two samples from each group in duplicate.
2.7 miRNA pathway-enriched analysis
miRNAs with more than twofold changes in expression levels were further analyzed and classified using a miRNA-centric network visual analytics platform (miRNet). Based on the IMEx interactome, literature-curated comprehensive data from InnateDB (12), we highlighted the miRNAs that were exclusively related to our investigation (miRNAs related to breast cancer and miRNAs related to pancreatic cancer), and then the miRNAs were filtered to distinguish miRNAs that are highly related to different cancer pathways with a p-value less than 0.05.
The p-value is primarily dependent on network connectivity and indicates how significant the connections within a specified miRNA function are. The edges of a certain function were considered “internal,” and the edges connecting the nodes of that function to the rest of the graph were considered “external.” The test’s null hypothesis was that there is no difference in the number of “internal” and “external” connections to a given node in the miRNA function. A Wilcoxon rank-sum test of the “internal” and “external” degrees was used to compute the p-value.
The degree of a node in a graph network is the number of connections it has to other nodes. In a network, nodes having a higher node degree function as hubs. Nodes having a degree of 100 or higher were selected in this study.
3 Results
3.1 Total RNA quality and quantity
At cycles 10 and 20, RNA was extracted from hypoxic PANC-1 and MCF7 cells, as well as their normoxic counterparts (see Table 1 in the Supplementary Material).
3.2 Resistance to doxorubicin
Resistance to the commonly used chemotherapeutic agent, doxorubicin, was examined to ensure the development of the hypoxic phenotype and to understand the effect of hypoxia on doxorubicin resistance after 10 and 20 cycles of exposure to hypoxic conditions. For both hypoxic and normoxic cancer cells, each doxorubicin concentration was tested in triplicate.
3.2.1 PANC-1 doxorubicin resistance
When investigating the chemosensitivity of PANC-1 after cycles 10 and 20, the findings in Figures 1A, B show that the PANC-1 viability percentage was significantly higher for hypoxic cells than their normoxic counterparts per concentration of doxorubicin. The results are illustrated as mean ± SEM, and the p-values were determined using the unpaired t-test.
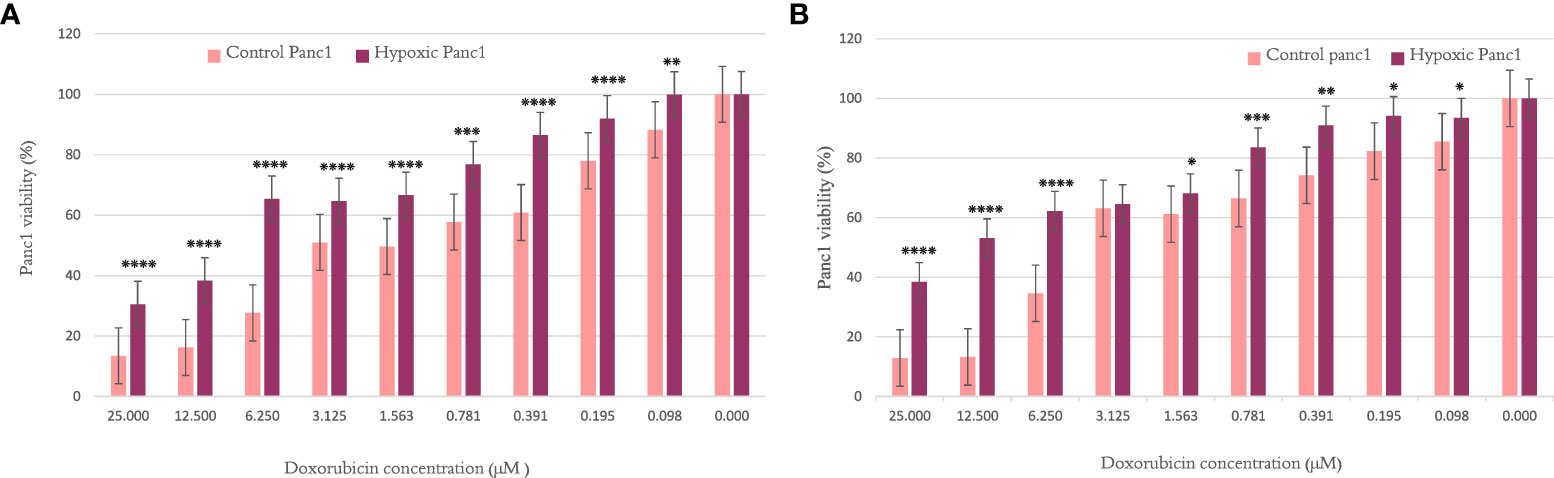
Figure 1 The effect of 10 and 20 cycles of chronic cyclic hypoxia on doxorubicin-induced cytotoxicity of PANC-1. (A) Represents the effect after 10 cycles, (B) represents the effect after 20 cycles. Statistical significance was determined by using the Student’s t-test.*p-value <0.05, **p-value <0.01, ***p-value <0.001, ****p-value <0.0001.
3.2.2 MCF7 doxorubicin resistance
The MCF7 viability percentage was significantly higher for hypoxic cells than normal counterparts per concentration of doxorubicin, as shown in Figures 2A, B, when examined after cycles 10 and 20, respectively. The results are illustrated as mean ± SEM, and the p-values were determined using the unpaired t-test.
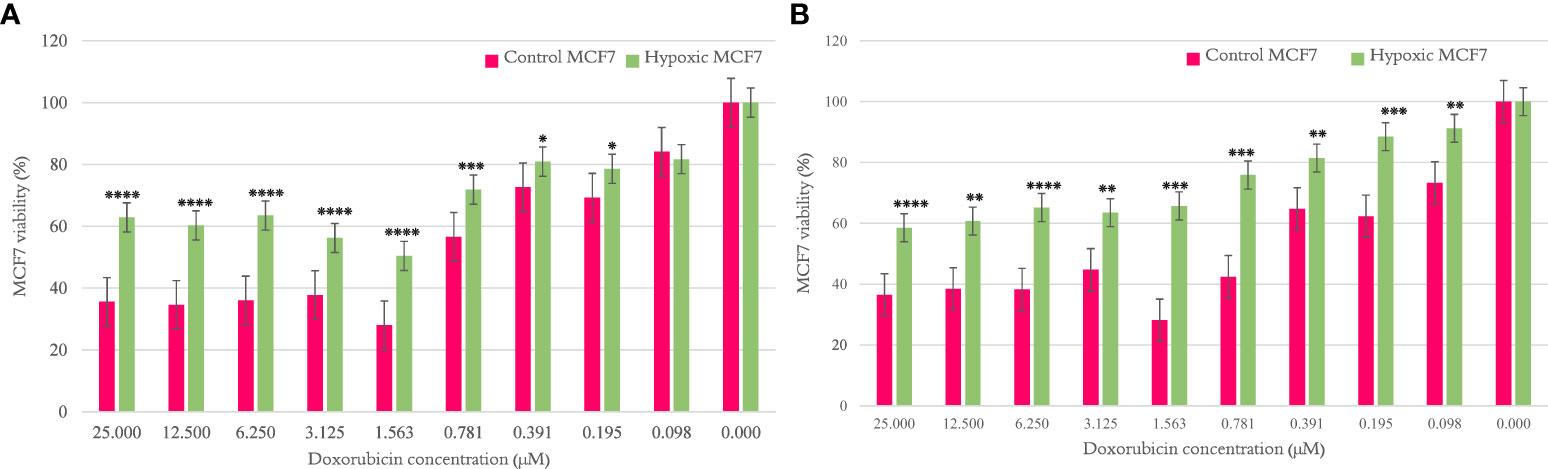
Figure 2 The effect of 10 and 20 cycles of chronic cyclic hypoxia on doxorubicin-induced cytotoxicity of MCF7. (A) Represents the effect after 10 cycles, (B) represents the effect after 20 cycles. Statistical significance was determined by using the Student’s t-test.*p-value <0.05, **p-value <0.01, ***p-value <0.001, ****p-value <0.0001.
3.3 Differential miRNA expression
In the present study, we examined miRNA expression changes after 10 and 20 cycles of chronic cyclic hypoxia, 1% O2, 72 hours of hypoxia, and 24 hours of reoxygenation (H-R: 72-24), in pancreatic and breast cancer cell lines. A standard twofold expression change was used as an arbitrary cutoff. The human hypoxia signaling miRNA PCR array revealed several deregulated miRNAs in hypoxic cells when compared with their normoxic counterparts.
3.3.1 Changes in PANC-1 miRNA expression pattern
When PANC-1 cells exposed to 10 and 20 cycles of hypoxia were compared with their normoxic counterparts, expression analysis revealed significant changes in 25 miRNA at the 10th cycle. The expression of five of these miRNAs were significantly upregulated, while those of 20 miRNAs were significantly downregulated. At the 20th cycle, significant changes in 24 miRNAs were revealed. The expressions of six of these miRNAs were significantly upregulated, while those of 18 miRNAs were significantly downregulated. The PCR array-based miRNA transcriptomic profiling of PANC-1 is presented in more detail in Supplementary Tables 2, 3.
3.3.2 Changes in MCF7 miRNA expression pattern
When MCF7 cells exposed to 10 and 20 cycles of hypoxia were compared with their normoxic counterparts, miRNA expression analysis revealed significant changes in 19 miRNA at the 10th cycle. The expressions of six of these miRNAs were significantly upregulated, while those of 13 miRNAs were significantly downregulated. At the 20th cycle, significant changes in 47 miRNAs were revealed. The expression of two of these miRNAs were significantly upregulated, while those of 45 miRNAs were significantly downregulated. The PCR array-based miRNA transcriptomic profiling of MCF7 is presented in more detail in Supplementary Tables 4, 5.
3.4 Interaction network analysis (miRNet)
We constructed the miRNA gene network based on specific miRNA–disease relationships to classify the functional units and putative enriched pathways in which the miRNAs are involved.
3.4.1 Enriched pathway of hypoxic PANC-1
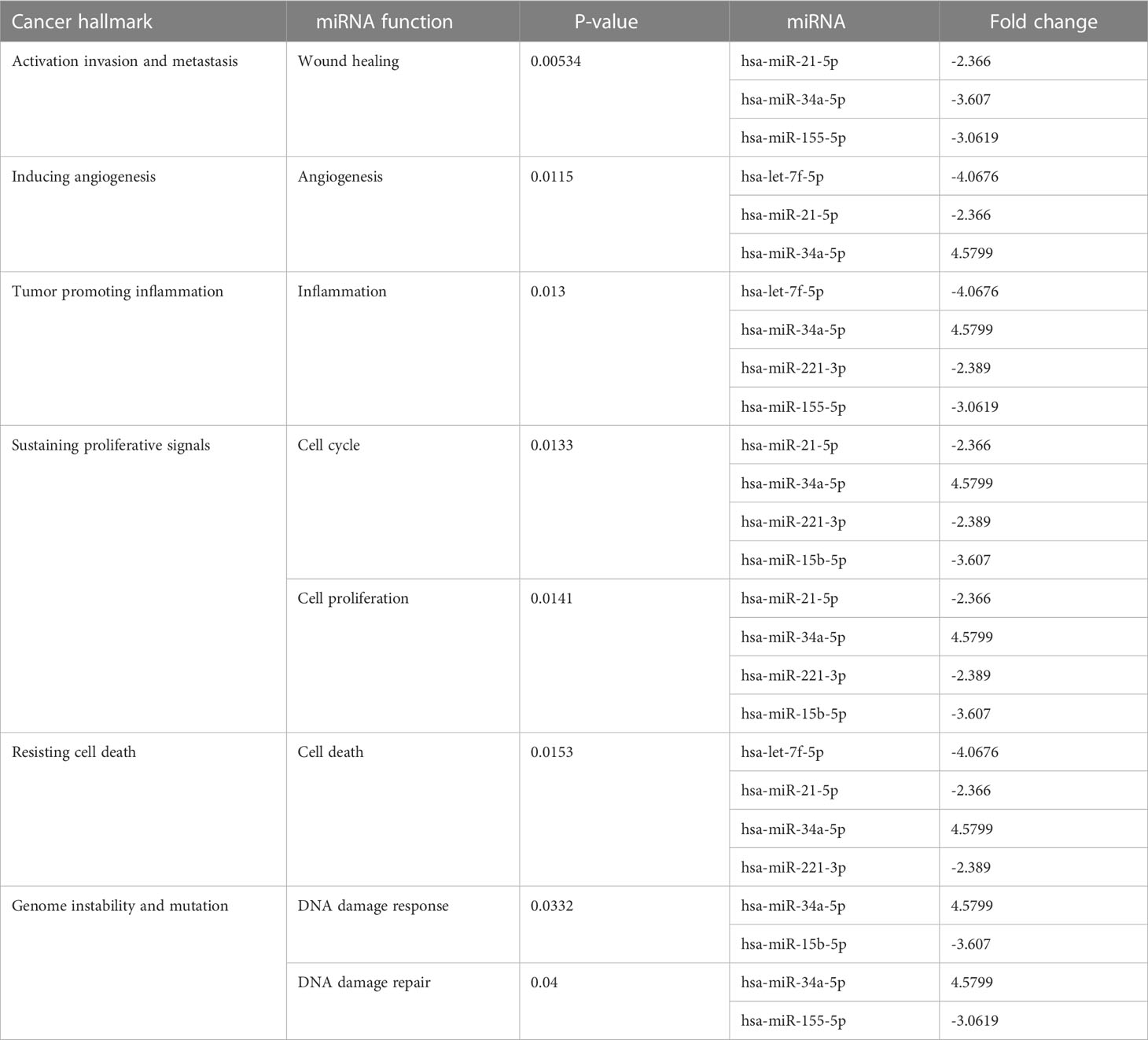
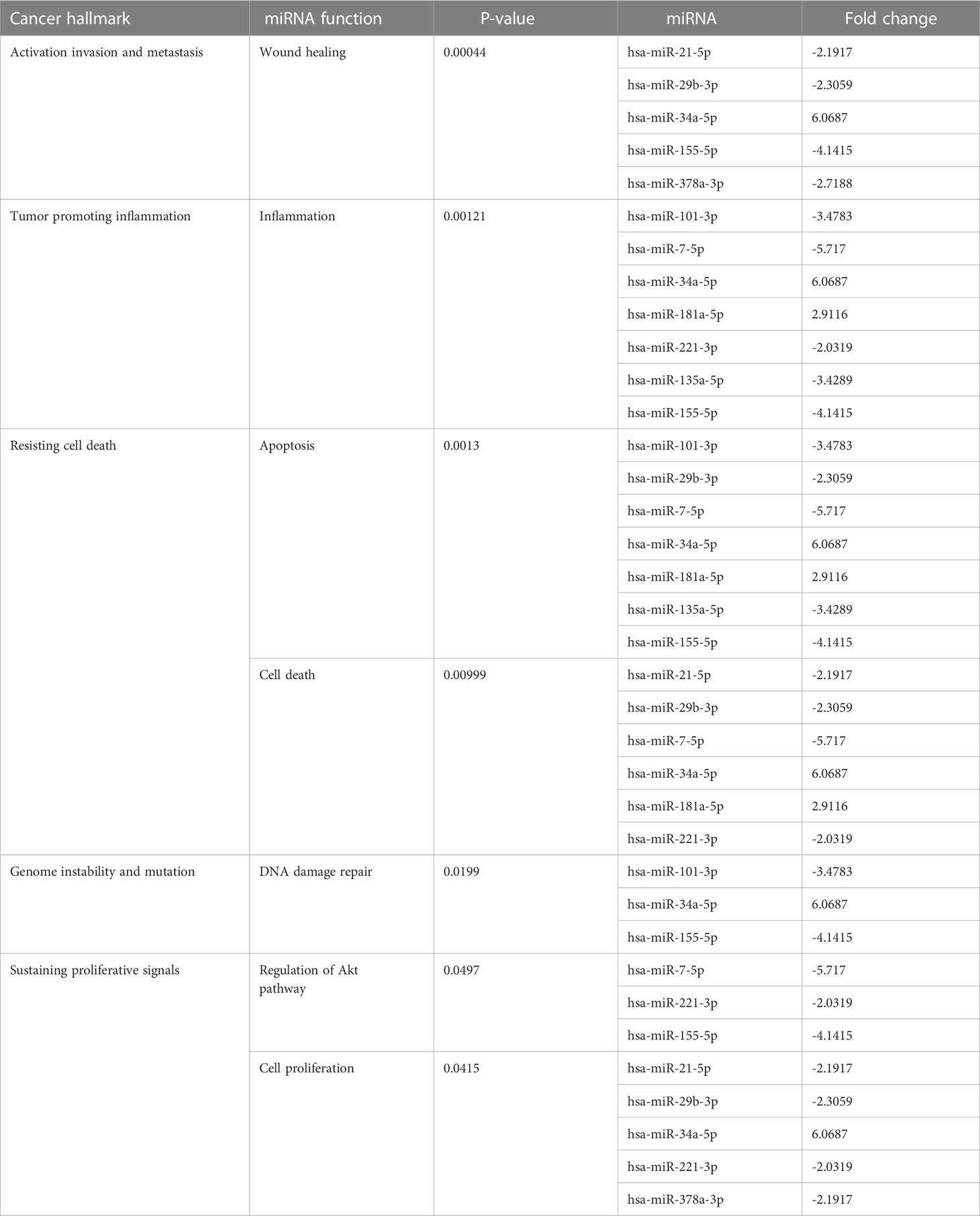
Many miRNAs with high levels of cancer pathways enrichment in pancreatic cancer were detected by the network, including let-7f and miR-15b at the 10th cycle. The 20th cycle included miR-135, miR-101, miR-7, miR-378, miR-181a, and miR-29, as well as miR-155, miR-21, miR-221, and miR-34a, which were involved in both cycles. The results are given in Tables 1, 2 for the 10th and 20th cycles, respectively. The p-values were primarily dependent on network connectivity and indicate how significant the connections within a specified miRNA function are.
3.4.2 Enriched pathway of hypoxic MCF7
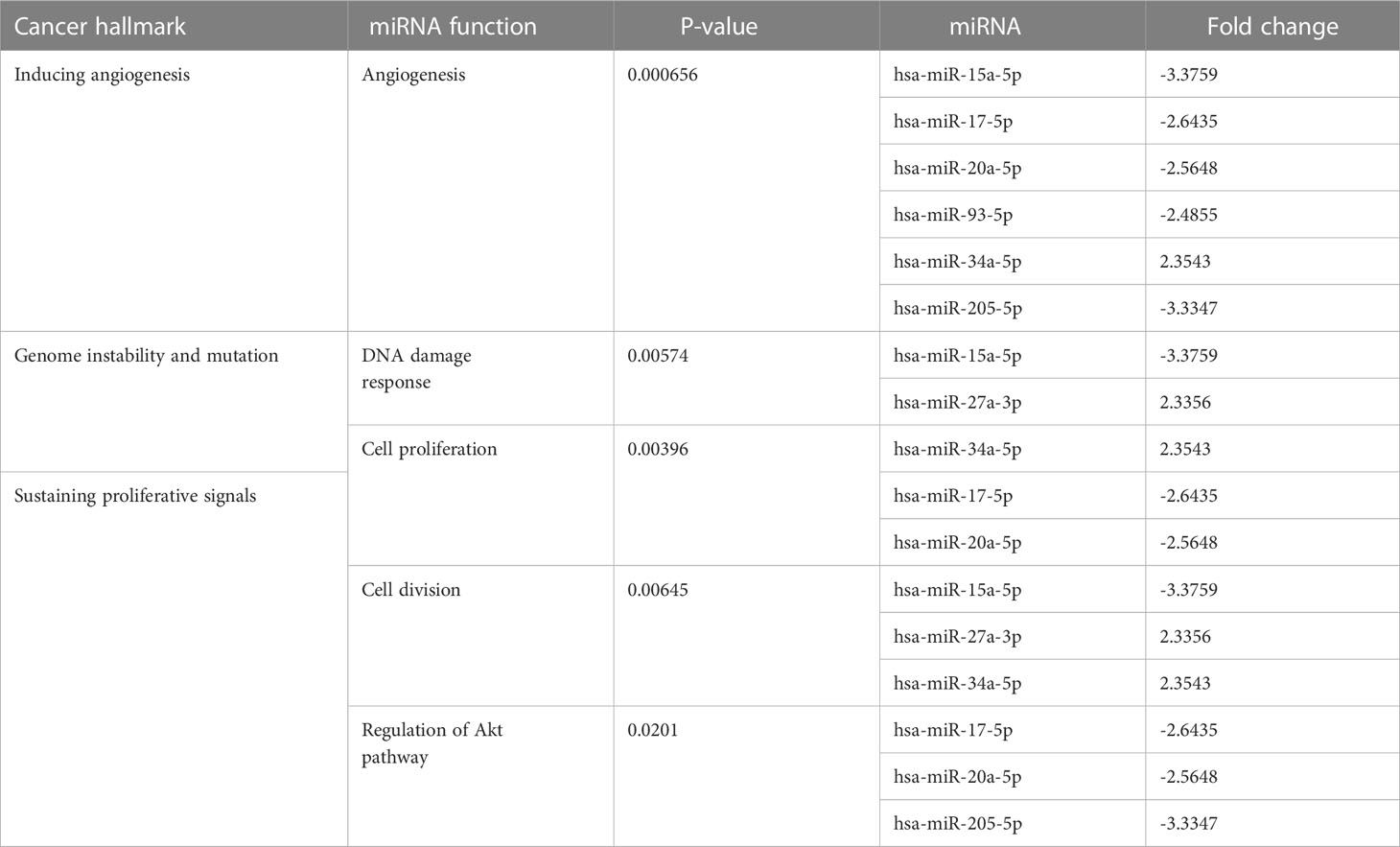
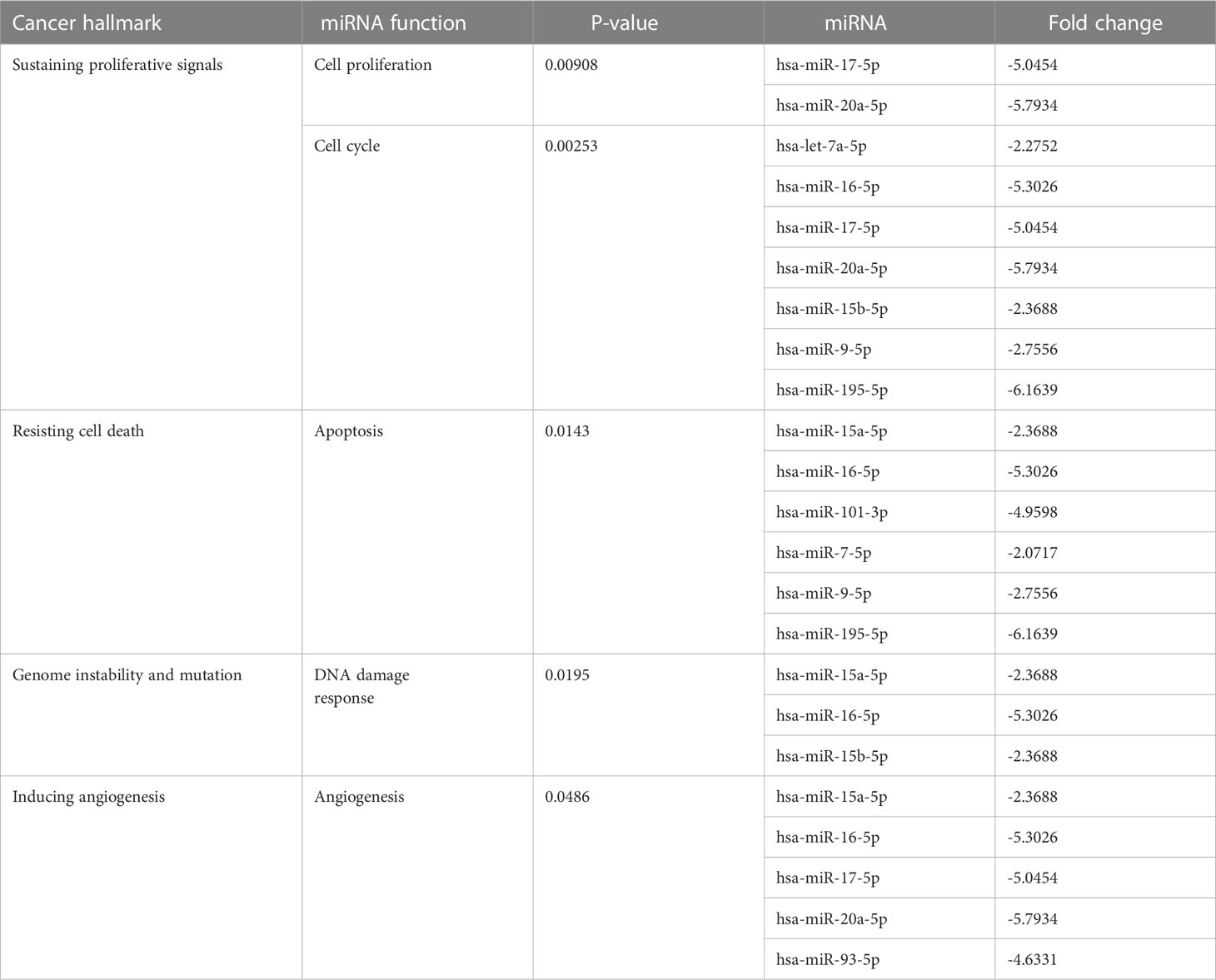
At the 10th cycle, the network identified several miRNA molecules with a high degree of hallmarks enrichment in breast cancer, including miR-27, miR-34, and miR-205. In the 20th cycle, miR-15b, miR-16, miR-7, miR-101, miR-9, and let-7a all played important roles. The two cycles both identified miR-93, miR-15a, miR-17, and miR-20a. Tables 3, 4 encompass the results for the 10th and 20th cycles, respectively. The p-values were primarily dependent on network connectivity and indicate how significant the connections within a specified miRNA function are.
3.4.3 The co-enrichment pathways between hypoxic MCF7 and hypoxic PANC1
Sustaining proliferative signals and resisting cell death were both enriched in PANC1 and MCF7 hypoxic cells, as indicated in Table 5.
3.5 Phenotypic alteration under chronic cyclic hypoxia
We merely describe a change in the morphology of cells that acquire an elongated shape and lose their polarity when observed under the microscope, but further investigation into epithelial-to-mesenchymal transition (EMT) biomarkers is necessary to confirm this change.
3.5.1 PANC-1 phenotypic switch
The morphological switch from epithelial to mesenchymal was observed throughout the hypoxia cycles, starting obviously with the fifth cycle. PANC-1 maintained a comparable proliferation rate throughout the experiment for hypoxic and normal cells, beginning with a 43-hour doubling time for both, a 28-hour doubling time for both at cycle 10, and a 22-hour doubling time for normoxic PANC-1 at cycle 20, while the doubling time for hypoxic cells remained unchanged from cycle 10. PANC-1 cells underwent morphological changes after 5, 10, 15, and 20 cycles of hypoxia, as seen in Figure 3.
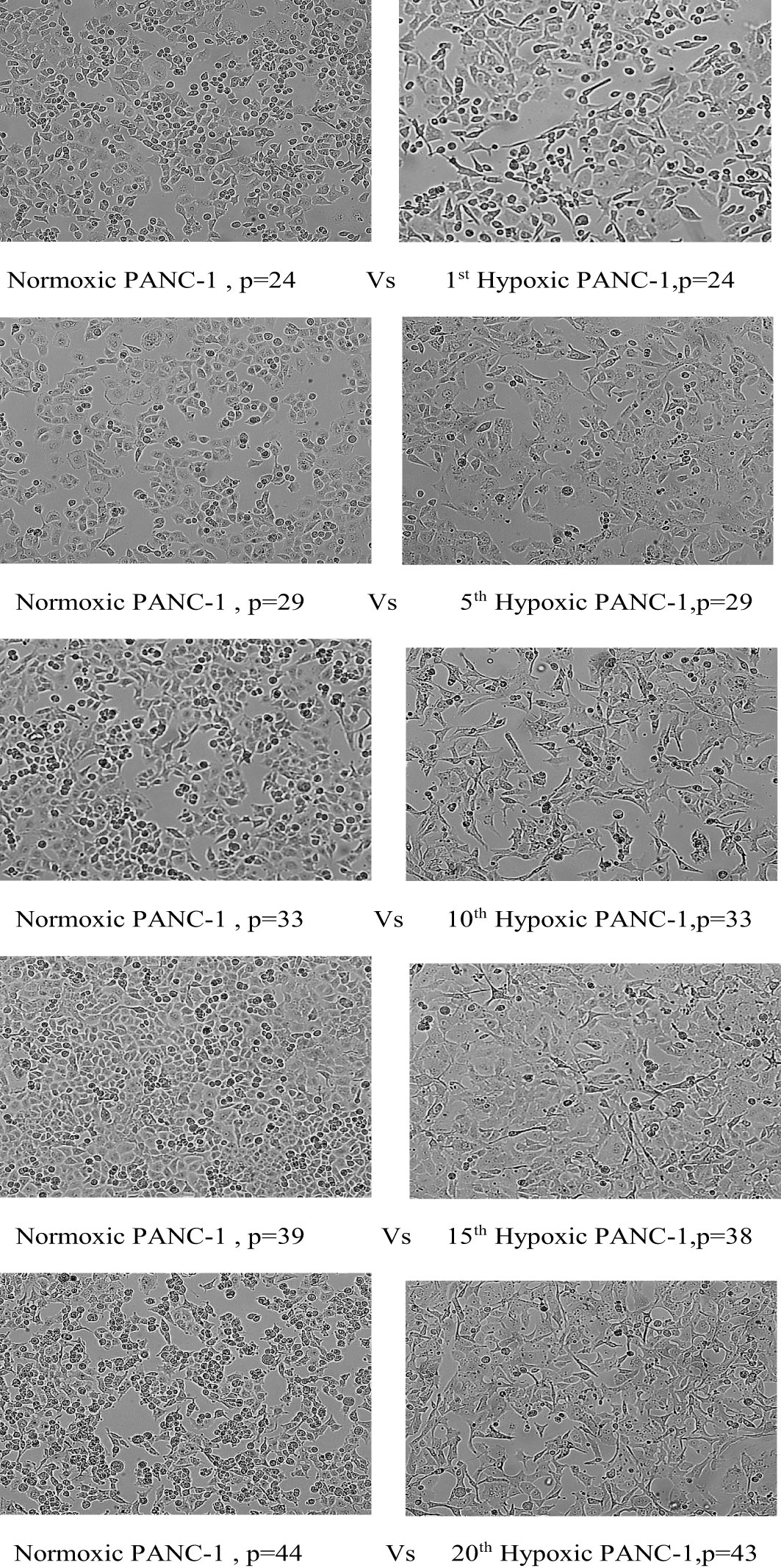
Figure 3 Morphological change of PANC-1 cell lines under chronic cyclic hypoxia. Normoxic PANC-1 vs. hypoxic PANC-1, (10x magnification). P: passage number.
3.5.2 MCF7 phenotypic switch
Hypoxic MCF7 cells proliferated at a slower rate than their normal counterparts; the doubling time of hypoxic cells was greatly increased to 84 hours in the 10th cycle, compared with 42 hours for normoxia, and in cycle 20, normal cells rapidly proliferated with a doubling time of 33 hours while the doubling time for hypoxic cells was further increased to 89 hours. However, hypoxic MCF7 cell proliferation was significantly reduced at cycle 12, allowing us to expand the reoxygenation period to 5 days between cycles 12 and 15. In terms of EMT development, MCF7 lost cell–cell adhesion during all hypoxic cycles, gaining an elongated shape and losing its polarization, as seen in Figure 4.
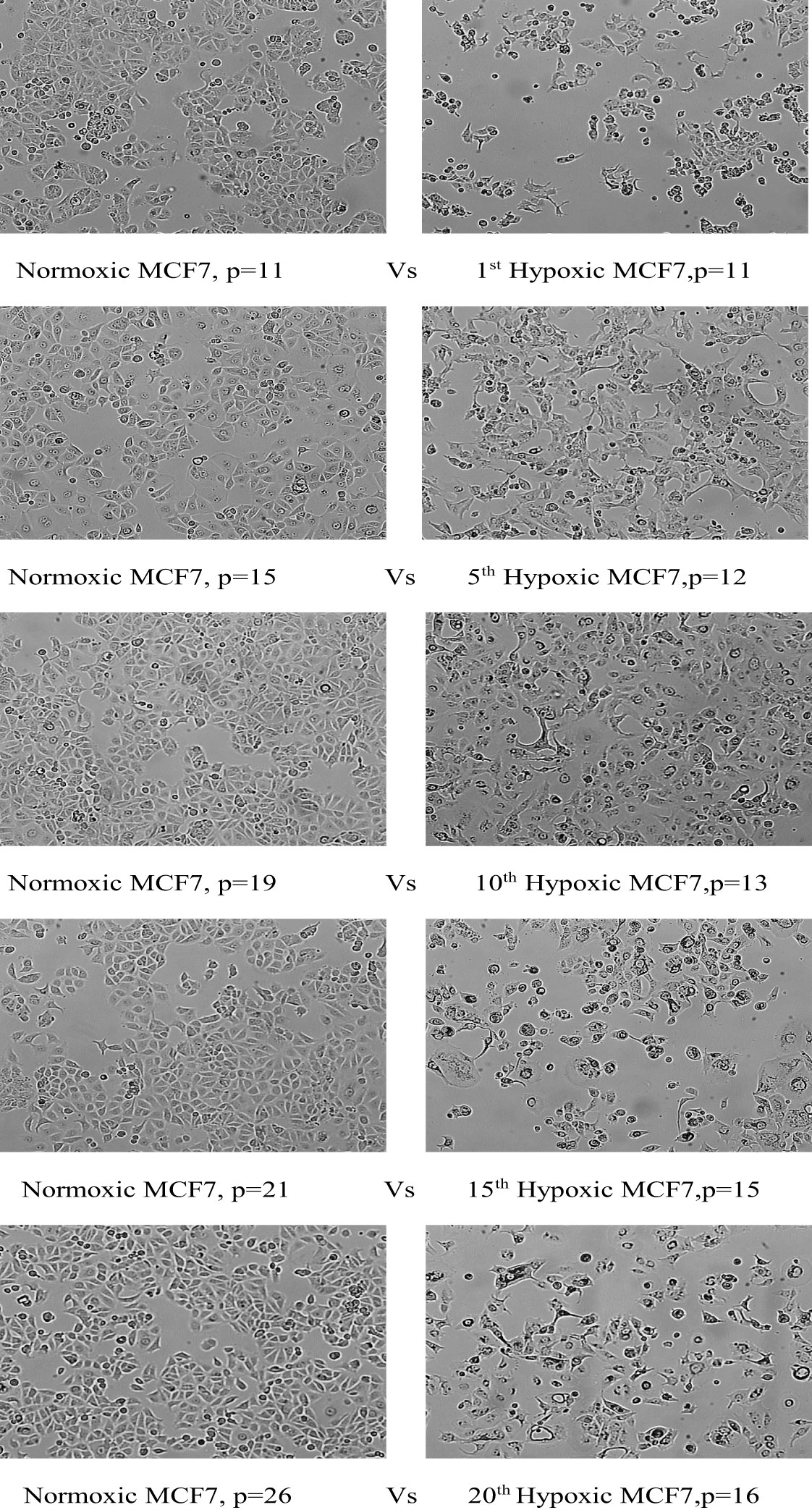
Figure 4 Morphological change of MCF7 cell lines under chronic cyclic hypoxia. Normoxic MCF7 vs. hypoxic MCF7, (10x magnification). P: passage number.
4 Discussion
To uncover novel hypoxamiR targets in cancer, we exposed MCF7 breast cancer cells and PANC-1 pancreatic cancer cells to 20 cycles of chronic cyclic hypoxia (72 h H, 24 h R). The MTT proliferation assay was used to determine resistance to doxorubicin after 10 and 20 cycles. In addition, miRNA expression profiling was performed mid-experiment (after 10 cycles of hypoxia) and at completion (after 20 cycles of hypoxia). According to the findings, chronic cyclic hypoxia had a key role in the formation of a chemoresistant phenotype in the MCF7 breast cancer cell line and the PANC-1 pancreatic cancer cell line. Furthermore, cells subjected to 20 cycles of chronic cyclic hypoxia showed substantial changes in miRNA expression profile when compared with their normoxic counterparts. Likewise, significant alterations were seen in cells subjected to 10 cycles, sharing several characteristics with those exposed to 20 cycles. We were able to induce doxorubicin resistance in MCF7 and PANC-1 cells after exposing them to 10 and 20 cycles of chronic cyclic hypoxia. This was verified by an unpaired t-test with a significant p-value in the MTT assay when administering increasing doses of doxorubicin. The obtained chemosensitivity results indicated that chronic cyclic hypoxia plays an imperative role in the development of chemoresistant phenotypes in pancreatic and breast cancer cells; thus, understanding the differential expression of miRNA between hypoxic and normal cells may assist in the introduction of new markers to target.
It has been previously reported that combining oxaliplatin with a miR-181a-5p inhibitor enhances the apoptotic pathway in pancreatic cancer cells by targeting ATMs (13). In addition, the downregulation of let-7e enhances DNA repair by targeting PARP1 and thus induces cisplatin resistance in ovarian cancer (14). Based on the finding that miR-181 is consistently overexpressed in both 10 and 20 cycles of hypoxia, whereas let-7e is consistently downregulated, we can speculate that miRNA-181 and let-7e may be involved in pancreatic cancer cells’ resistance to doxorubicin under hypoxic conditions (15). MiR-17 and miR-93 were consistently downregulated in MCF7 miRNA profiling at the 10th and 20th hypoxia cycles, with a twofold decline in the fold change for each. According to Xia et al., the loss of miR-17 and miR-20b boosted breast cancer resistance to taxol by upregulating nuclear receptor coactivator 3 (NCOA3) levels (16). Furthermore, downregulated miR-93 induced chemoresistance to paclitaxel by targeting CCND1 and E2F1 (17). Moreover, miR-34a and miR-27 were both upregulated in cycle 10 and may participate in doxorubicin chemoresistance via targeting of the delta-like ligand 1 (DLL1) receptor, a Notch pathway ligand (18), and XIAP axis (19), respectively. Based on previous studies and their positive correlation with our miRNA expression profile, we may infer that these miRNAs have an important role in hypoxic MCF7 doxorubicin resistance.
The results of this study point to several miRNAs that may play a role in modulating chemotherapy response in pancreatic and breast cancer patients. Before these associations can be suggested as biomarkers of response to chemotherapeutic treatments in pancreatic and breast cancer, more research is required to uncover the molecular mechanisms underlying them.
4.1 An illustration of how differential miRNA expression affects cancer hallmarks
4.1.1 PANC-1 enriched pathways
According to the hypoxic PANC-1 miRNA expression profile analysis, cell proliferation pathways were some of the most enriched pathways. Hypoxia reduces overall cell proliferation in most cell types, because an increased cell number, with a corresponding rise in oxygen demand, will only exacerbate hypoxia-induced stress (20). However, as the proliferation rate of hypoxic PANC-1 cells was comparable to that of normoxic cells, this assumption is irrelevant to our investigation. The deregulation of specific miRNAs could dramatically orchestrate cell proliferation after PANC-1 cells were exposed to chronic cyclic hypoxia. Previous studies have shown that miR-15b and miR-221 downregulation enhances cell proliferation through targeting FGF7, WNT3A (21), and eukaryotic translation initiation factor 5A2 (EIF5A2) (22), respectively. On the other hand, cell proliferation may be halted by the downregulation of miR-21 which targets the cell proliferation inhibitor, sprouty RTK signaling antagonist 2 (Spry2) (23), and the upregulation of miR-34 which targets CDK4 and c-Myc (24). Thus, the effect of these differentially expressed miRNAs on cell proliferation at the 10th cycle of hypoxia is debatable. At the 20th cycle of hypoxia, additional miRNAs that inhibit cell proliferation were downregulated, including miR-29b, miR-378, miR-155, and miR-7, which target CDK14 (25), BRAF (26), IGF2 (27), and SOX18 (28), respectively. Collectively these studies explained why, even after 20 cycles of hypoxia, the hypoxic PANC-1 proliferation rate was still comparable to its normoxic counterpart.
Another pathway that enriched significantly in the hypoxic PANC-1 miRNA expression profile was evading apoptosis. By targeting histone deacetylase-6 (HDAC6), a key player in the cellular control of protein aggregates, the downregulation of miR-221 suppresses autophagy (29). Moreover, downregulation of miR-101, miR-29, miR-7, miR-135, and miR-155, as well as the overexpression of miR-181a, may all have played a role in modulating PANC-1 cell death at cycle 20. MiR-181a has been shown to promote the development of pancreatic cancer by suppressing the tumor suppressor genes PTEN and MAP2K4 (30), whereas downregulated miR-155, miR-135a, miR-7, miR-29b, and miR-101 aid in apoptosis evasion by the targeting of X-linked inhibitor of apoptosis (XIAP) (31), Bmi1 (32), MAP3K9 (33), DNMT3b (3), and RLIP76 (34), respectively.
Our results contrast with some studies indicating that the induction of apoptosis is clearly linked to upregulated miR-34, which targets Bcl-2 and Notch (35), as well as downregulated let-7f, which targets caspase 3 and caspase 9 (36), and the intermittent hypoxia/re-oxygenation (IHR) downregulated miR-21, which targets TNF and EIF2 (37). This evidence suggests that hypoxic signaling does not exclusively contribute to oncogenic effects and could also be associated with tumor-suppressive and pro-apoptotic cascades.
Metastasis is a multi-step, dynamic biological process in which the EMT plays a critical role. Growing evidence shows that miRNAs have an important role in EMT and cancer metastasis (38). Regardless of miR-34 tumor suppressor activity via targeting Snail1 and Notch1 (39), other deregulated miRNAs suggest PANC-1 metastatic stimulation under 10 cycles of hypoxia. miR-21 downregulation increases Tiam1 expression which is attributed to cancer metastatic potential (15). Moreover, miR-155-null macrophages were susceptible to M2 polarization due to increased expression of C/EBPβ, a recognized miR-155 target, thus modulating solid tumor metastasis (40).
The downregulation of miR-29 and miR-378 could have played important roles in fostering metastasis, at cycle 20, by targeting matrix metalloproteinase 2 (MMP2) (41) and vimentin (42), respectively. Depending on previous studies and our findings, which indicate an evident morphological transformation in hypoxic PANC-1 cells from epithelial to mesenchymal phenotype with consistent metastasis related-miRNA deregulations, we may deduce that chronic cyclic hypoxia promotes pancreatic cancer metastasis. Angiogenesis is a well-coordinated tumor growth and metastasis process that entails the creation of new blood vessels from pre-existing ones to satisfy the cell’s needs (38). The pathways that related to angiogenesis were exclusively enriched in the 10th cycle miRNA expression profile. Intriguingly, the effect of miR-21-5p on its target, SMAD7, expression was markedly different between normoxia and hypoxia; in normoxia, the transfection of miR-21-5p mimicked enhanced tube-forming ability, but in hypoxia, it diminished. In addition, inhibiting miR-21 improved blood flow in a skin flap, indicating that miR-21 inhibits angiogenesis in human endothelial cells and rat skin flaps under hypoxic conditions (43). miR-221-3p downregulation, moreover, induce angiogenesis via targeting HIF-1 (44). In contrast, Gnanamony et al. reported that the circular dumbbell miR34a -3p and -5p suppresses angiogenesis in pancreatic cancer cells (45).
The DNA damage response (DDR) pathway was another enriched pathway. Surprisingly, the DDR mechanism is involved in both the development and treatment of cancer. A rising number of miRNAs have been found to intervene in processes regulating tumorigenesis and responses to radiation or chemotherapies by modifying the DDR (46). A study showed that the exogenous addition of miR-34 affects breast cancer cell survival after radiotherapy and suggests that anti-miR-34 may be used as a radiosensitizing agent in p53-mutant breast cancer (47). In addition, DNA repair factors are targeted by several miRNAs, including miR-155 which targets RAD51, MLH1, and MSH6 (48), miR-15b which targets the inhibitor of nuclear factor kappa B kinase subunit beta (IKBKB) and WEE1 (49), and miR-101 which targets RRM1 (50). These studies support what was reported in the miRNA expression profile that the upregulation of miR-34a, as well as the downregulation of miR-15b, miR-155, and miR-101, might improve DNA repair and hence contribute to cancer cell survival and chemoresistance.
At the 10th cycle, the downregulation of miR-155, miR-221, and let-7f, and the upregulation of miR-34a could boost hypoxic PANC-1 inflammation and promote its chemoresistance. miR-34 pro-inflammatory functioning is mediated by LGR4 (51). The downregulation of the inflammation-associated miRNA, miR-155, promotes M2 macrophages, which play a key role in the immunosuppressive microenvironment. Furthermore, let-7f and miR-221 downregulation promote inflammation by targeting NLRP3 (52), p38/NF-Kβ, and ICAM-1 (53), respectively. The downregulation of miR-101 which targets COX2 (54), miR-135a which targets toll-like receptor 4 (TLR4), and miR-7 (55), as well as the upregulation of miR-181a which is linked to IL-1, IL-6, and TNFα expression (56), all may contribute to inflammation potentiation at cycle 20. The majority of previous studies support the development of the PANC-1 inflammatory response by chronic cyclic hypoxia.
4.1.2 MCF7 enriched pathways
In the case of MCF7, chronic cyclic hypoxia had a significant impact on four cancer pathways: cell proliferation, apoptosis, angiogenesis, and DDR. The miRNA expression profile of hypoxic MCF7 cells was enriched for cell proliferation pathways, and several tumor suppressor miRNAs that target cell proliferation regulators were downregulated at the 10th cycle, including miR-17-5p that targets AIB1 and insulin-like growth factor 1 (57), miR-20a that targets MAPK1 (58), miR-205 that targets E2F1 (59), and miR-15a that targets cyclin E1 (CCNE1) (60). Moreover, the upregulation of miR-27a induces cell proliferation via the upregulation of Wnt-β-catenin and SFRP1 targeting (61). In contrast, miR-34a inhibits BC proliferation, invasion, and migration by deactivating the Wnt-β-catenin signaling pathway (62), raising the question of whether hypoxia has a negative or positive effect on the Wnt-β-catenin signaling pathway.
Other downregulated tumor suppressor miRNAs may have potentiated MCF7 proliferation at cycle 20. Downregulated let-7a-5p induces glycolysis via PKM2 and GLUT12 targeting (63, 64). Furthermore, reduced miR-16-5p, miR-9-5p, and miR-195 enhanced breast cancer cell proliferation, and accelerated the cell cycle by targeting AKT3 (65), androgen receptor signaling (66), and cyclin E1 (CCNE1) (67), respectively. The emergence of evading apoptosis hallmarks was expected to become evident at cycle 20, in line with the behavior of breast cancer cells from the 12th to the 15th cycle of hypoxia, where the proliferation rate decreased and the apoptosis rate increased significantly, and then the cells returned to grow and multiply, defeating apoptosis. Several tumor suppressor miRNAs, whose upregulation has been linked to apoptosis in previous studies, were downregulated in this study. For instance, ectopic expression of miR-15a and miR-16 induced mitochondrial reactive oxygen species, followed by cytochrome-C release into the cytosol, which triggered caspase-3 and caspase-6/9, resulting in intrinsic apoptosis. Additionally, BMI1 and BCL2 were significantly downregulated by miR-15a and miR-16 (68). miR-9-5p, miR-195, miR-7-5p, and miR-101 also exhibit pro-apoptotic activity via the targeting of MTHFD2 (69), Bcl-2 (70), proteasome activator subunit 3 (REGγ) (71), and JAK2 (72), respectively.
Increased tumor angiogenesis and upregulated expression of the proangiogenic factor VEGF-A contribute to the tumorigenicity of breast cancer cells as a result of EMT (73). Hypoxic MCF7 exhibited the downregulation of many anti-angiogenic miRNAs at cycle 10, including miR-15a/16 which targets VEGF-A (74), miR-17 which targets several tumor angiogenesis-inducing genes, including TGFBR2, HIF1, and VEGFA (75), miR-93 which targets WNK lysine deficient protein kinase 1 (WNK1) (76), and miR-20a which targets VEGF (77). The miR-205/YAP1 signaling pathway, in particular, converts normal breast fibroblasts to CAFs. Angiogenesis and therefore breast cancer cell invasion and metastasis are inhibited when miR-205 is restored in CAFs (78). ErbB3 and VEGF-A are considered direct targets for miR-205 (79). miR-34a, on the other hand, inhibits epithelial cell-mediated angiogenesis by inducing senescence through Sirt1 (80). Additionally, in terms of miRNA expression at cycle 20, downregulation of miR-16-5p targets VEGFA (81). All of this evidence is consistent with our differential miRNA expression findings, which affirm angiogenesis as a reasonable response to chronic cyclic hypoxia that breast cancer cells are exposed to, as well as reinforce the angiogenesis-EMT relationship, which fuels breast cancer metastasis.
The DDR pathway was significantly enriched in the hypoxic MCF7 miRNA expression profile. Low levels of miR-15a and miR-16 desensitize breast cancer cells to doxorubicin, resulting in less apoptotic cell death by targeting the DNA repair factor BMI1 (82). As previously mentioned, miRNA-34a facilitates DNA damage repair, too. Furthermore, miR-27a targets ATM and facilitates cell proliferation even after irradiation (83). At cycle 20, miR-15b and miR-16, in addition to miR-15a, played a role in DDR. miR-15b targets Wip1 and induces the DNA damage response (84). Collectively, these studies show an increase in DNA repair by chronic cyclic hypoxia and thus the development of resistant and aggressive cancer cells.
As expected, the miRNA expression profiles of PANC-1 and MCF7, after chronic cyclic hypoxia, deregulated differently; this can be explained by the fact that the two cell lines tolerate hypoxia in distinct manners. The pancreatic cancer cell line studied in the present study may be particularly tolerant of hypoxia since it survives oxygen levels 25 times lower than its normal counterpart. Whereas, the breast cancer cell line studied is considered less hypoxic since it survives oxygen levels 5 times less than normal breast cancer tissue (85).
5 Conclusions
According to this study, distinct patterns of dysregulated miRNA expression in PANC-1 and MCF7 cell lines under chronic cyclic hypoxia could aid in highlighting novel hypoxamiRs to target. During the 20 cycles of pancreatic and breast cancer hypoxia, four different miRNAs appeared to be key mediators of various cancer hallmarks and may have great potential as novel targets. miR-221, miR-21, miR-155, and miR-34 could orchestrate proliferation, apoptosis, DDR, metastasis, angiogenesis, and inflammation of hypoxic PANC-1, whereas miR-93, miR-20a, miR-17, and miR-15 could regulate proliferation, apoptosis, angiogenesis, and DDR of hypoxic MCF-7 cells. Furthermore, differential expression of miR-181a and let-7e in PANC-1, as well as miR-93, miR-34, and miR-27 in MCF7 may be linked to the development of chemoresistant MCF7 and PANC-1 cells after 20 cycles of chronic cyclic hypoxia. The particular mechanisms that govern the aforementioned processes are yet unknown; thus, the interpretation of miRNA profiling data and the regulatory functions of miRNA in cancer development and progression should be further investigated.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
SA-S conceptualized and conducted the experiments, carried them out, analyzed the data, constructed figures and/or tables, and wrote the manuscript. MZ conceptualized and conducted the experiments, analyzed the data, constructed figures and/or tables, and reviewed versions of the manuscript. HH conceptualized the experiments and undertook funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
The present study was funded by the Deanship of Scientific Research at the University of Jordan in Amman, Jordan (grant no: 2467).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1110743/full#supplementary-material
Glossary

References
1. Yang Z, Liu Z. The emerging role of microRNAs in breast cancer. J Oncol (2020) 2020:e9160905. doi: 10.1155/2020/9160905
2. Gervin E, Shin B, Opperman R, Cullen M, Feser R, Maiti S, et al. Chemically induced hypoxia enhances miRNA functions in breast cancer. Cancers (2020) 12(8):2008. doi: 10.3390/cancers12082008
3. Wang W, Lou W, Ding B, Yang B, Lu H, Kong Q, et al. A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging (Albany NY) (2019) 11(9):2610–27. doi: 10.18632/aging.101933
4. Sun T, Kong X, Du Y, Li Z. Aberrant microRNAs in pancreatic cancer: researches and clinical implications [Review article]. Gastroenterol Res Practice; Hindawi (2014). doi: 10.1155/2014/386561
5. Weidle UH, Birzele F, Nopora A. Pancreatic ductal adenocarcinoma: microRNAs affecting tumor growth and metastasis in preclinical in vivo models. Cancer Genomics Proteomics (2019) 16(6):451–64. doi: 10.21873/cgp.20149
6. Saxena K, Jolly MK. Acute vs. Chronic vs. Cyclic Hypoxia: Their Differential Dynamics, Molecular Mechanisms, and Effects on Tumor Progression. Biomolecules (2019) 9(8). doi: 10.3390/biom9080339
7. Gee HE, Ivan C, Calin GA, Ivan M. HypoxamiRs and cancer: from biology to targeted therapy. Antioxid Redox Signaling (2013) 21(8):1220–38. doi: 10.1089/ars.2013.5639
8. Greco S, Martelli F. MicroRNAs in hypoxia response. Antioxid Redox Signaling (2014) 21(8):1164–6. doi: 10.1089/ars.2014.6083
9. Nallamshetty S, Chan SY, Loscalzo J. Hypoxia: A master regulator of microRNA biogenesis and activity. Free Radical Biol Med (2013) 64:20–30. doi: 10.1089/ars.2013.5639
10. Alhawarat FM, Hammad HM, Hijjawi MS, Sharab AS, Abuarqoub DA, Al Shhab MA, et al. The effect of cycling hypoxia on MCF-7 cancer stem cells and the impact of their microenvironment on angiogenesis using human umbilical vein endothelial cells (HUVECs) as a model. PeerJ (2019) 7:e5990. doi: 10.7717/peerj.5990
11. Obeidat NM, Zihlif MA, Alqudah DA, Alshaer W, Alqaraleh M, Abdalla SS. Effects of cyclic acute and chronic hypoxia on the expression levels of metabolism related genes in a pancreatic cancer cell line. Biomed Rep (2022) 17(4):81. doi: 10.3892/br.2022.1564
12. Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, et al. InnateDB: Systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic Acids Res (2013) 41(Database issue):D1228–1233. doi: 10.1093/nar/gks1147
13. Meijer LL, Garajová I, Caparello C, Le Large TYS, Frampton AE, Vasile E, et al. Plasma miR-181a-5p downregulation predicts response and improved survival after FOLFIRINOX in pancreatic ductal adenocarcinoma. Ann Surg (2020) 271(6):1137–47. doi: 10.1097/SLA.0000000000003084
14. Xiao M, Guo J, Xie L, Yang C, Gong L, Wang Z, et al. Let-7e suppresses DNA damage repair and sensitizes ovarian cancer to cisplatin through targeting PARP1. Mol Cancer Res (2020) 18(3):436–47. doi: 10.1158/1541-7786.MCR-18-1369
15. He J-H, Li Y-G, Han Z-P, Zhou J-B, Chen W-M, Lv Y-B, et al. The circRNA-ACAP2/hsa-miR-21-5p/tiam1 regulatory feedback circuit affects the proliferation, migration, and invasion of colon cancer SW480 cells. Cell Physiol Biochem (2018) 49(4):1539–50. doi: 10.1159/000493457
16. Xia L, Li F, Qiu J, Feng Z, Xu Z, Chen Z, et al. Oncogenic miR-20b-5p contributes to malignant behaviors of breast cancer stem cells by bidirectionally regulating CCND1 and E2F1. BMC Cancer (2020) 20(1):949. doi: 10.1186/s12885-020-07395-y
17. Bao C, Chen J, Chen D, Lu Y, Lou W, Ding B, et al. MiR-93 suppresses tumorigenesis and enhances chemosensitivity of breast cancer via dual targeting E2F1 and CCND1. Cell Death Dis (2020) 11(8):1–14. doi: 10.1038/s41419-020-02855-6
18. Pu Y, Zhao F, Wang H, Cai S. MiR-34a-5p promotes multi-chemoresistance of osteosarcoma through down-regulation of the DLL1 gene. Sci Rep (2017) 7(1):44218. doi: 10.1038/srep44218
19. Zhou S, Huang Q, Zheng S, Lin K, You J, Zhang X. MiR-27a regulates the sensitivity of breast cancer cells to cisplatin treatment via BAK-SMAC/DIABLO-XIAP axis. Tumour Biol: J Int Soc Oncodevelopmental Biol Med (2016) 37(5):6837–45. doi: 10.1007/s13277-015-4500-1
20. Hubbi ME, Semenza GL. Regulation of cell proliferation by hypoxia-inducible factors. Am J Physiol - Cell Physiol (2015) 309(12):C775–82. doi: 10.1152/ajpcell.00279.2015
21. Zhang XJ, Ye H, Zeng CW, He B, Zhang H, Chen YQ. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J Hematol Oncol (2010) 3:46. doi: 10.1186/1756-8722-3-46
22. Yang Y, Cui H, Wang X. Downregulation of EIF5A2 by miR-221-3p inhibits cell proliferation, promotes cell cycle arrest and apoptosis in medulloblastoma cells. Biosci Biotechnol Biochem (2019) 83(3):400–8. doi: 10.1080/09168451.2018.1553604
23. Zhao Q, Chen S, Zhu Z, Yu L, Ren Y, Jiang M, et al. MiR-21 promotes EGF-induced pancreatic cancer cell proliferation by targeting Spry2. Cell Death Dis (2018) 9(12):1–16. doi: 10.1038/s41419-018-1182-9
24. Wan Y, Shen A, Qi F, Chu J, Cai Q, Sferra TJ, et al. Pien Tze Huang inhibits the proliferation of colorectal cancer cells by increasing the expression of miR−34c−5p. Exp Ther Med (2017) 14(4):3901–7. doi: 10.3892/etm.2017.4972
25. Sun Y, Wang P, Yang W, Shan Y, Zhang Q, Wu H. The role of lncRNA MSC-AS1/miR-29b-3p axis-mediated CDK14 modulation in pancreatic cancer proliferation and Gemcitabine-induced apoptosis. Cancer Biol Ther (2019) 20(6):729–39. doi: 10.1080/15384047.2018.1529121
26. Wang Z, Ma B, Ji X, Deng Y, Zhang T, Zhang X, et al. MicroRNA-378-5p suppresses cell proliferation and induces apoptosis in colorectal cancer cells by targeting BRAF. Cancer Cell Int (2015) 15(1):40. doi: 10.1186/s12935-015-0192-2
27. Luo X, Dong J, He X, Shen L, Long C, Liu F, et al. MiR-155-5p exerts tumor-suppressing functions in Wilms tumor by targeting IGF2 via the PI3K signaling pathway. Biomed Pharmacother (2020) 125:109880. doi: 10.1016/j.biopha.2020.109880
28. Shen Y, Pu K, Zheng K, Ma X, Qin J, Jiang L, et al. Differentially expressed microRNAs in MIA paCa-2 and PANC-1 pancreas ductal adenocarcinoma cell lines are involved in cancer stem cell regulation. Int J Mol Sci (2019) 20(18). doi: 10.3390/ijms20184473
29. Yang Y, Sun Y, Wang H, Li H, Zhang M, Zhou L, et al. MicroRNA-221 induces autophagy through suppressing HDAC6 expression and promoting apoptosis in pancreatic cancer. Oncol Lett (2018) 16(6):7295–301. doi: 10.3892/ol.2018.9513
30. Liu J, Xu D, Wang Q, Zheng D, Jiang X, Xu L. LPS induced miR-181a promotes pancreatic cancer cell migration via targeting PTEN and MAP2K4. Digest Dis Sci (2014) 59(7):1452–60. doi: 10.1007/s10620-014-3049-y
31. Chen W, Huang L, Hao C, Zeng W, Luo X, Li X, et al. MicroRNA-155 promotes apoptosis in SKOV3, A2780, and primary cultured ovarian cancer cells. Tumor Biol (2016) 37(7):9289–99. doi: 10.1007/s13277-016-4804-9
32. Dang Z, Xu W-H, Lu P, Wu N, Liu J, Ruan B, et al. MicroRNA-135a inhibits cell proliferation by targeting bmi1 in pancreatic ductal adenocarcinoma. Int J Biol Sci (2014) 10(7):733–45. doi: 10.7150/ijbs.8097
33. Xia J, Cao T, Ma C, Shi Y, Sun Y, Wang ZP, et al. MiR-7 suppresses tumor progression by directly targeting MAP3K9 in pancreatic cancer. Mol Ther - Nucleic Acids (2018) 13:121–32. doi: 10.1016/j.omtn.2018.08.012
34. Yang J, Song Q, Cai Y, Wang P, Wang M, Zhang D. RLIP76-dependent suppression of PI3K/AKT/Bcl-2 pathway by miR-101 induces apoptosis in prostate cancer. Biochem Biophys Res Commun (2015) 463(4):900–6. doi: 10.1016/j.bbrc.2015.06.032
35. Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PloS One (2009) 4(8):e6816. doi: 10.1371/journal.pone.0006816
36. Tie Y, Chen C, Yang Y, Qian Z, Yuan H, Wang H, et al. Upregulation of let−7f−5p promotes chemotherapeutic resistance in colorectal cancer by directly repressing several pro−apoptotic proteins. Oncol Lett (2018) 15(6):8695–702. doi: 10.3892/ol.2018.8410
37. Chen Y-C, Hsu P-Y, Su M-C, Chin C-H, Liou C-W, Wang T-Y, et al. MiR-21-5p under-expression in patients with obstructive sleep apnea modulates intermittent hypoxia with re-oxygenation-induced-cell apoptosis and cytotoxicity by targeting pro-inflammatory TNF-α-TLR4 signaling. Int J Mol Sci (2020) 21(3):999. doi: 10.3390/ijms21030999
38. Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Targeted Ther (2016) 1:15004. doi: 10.1038/sigtrans.2015.4
39. Tang Y, Tang Y, Cheng Y. MiR-34a inhibits pancreatic cancer progression through Snail1-mediated epithelial–mesenchymal transition and the Notch signaling pathway. Sci Rep (2017) 7. doi: 10.1038/srep38232
40. Yu F, Jia X, Du F, Wang J, Wang Y, Ai W, et al. MiR-155–deficient bone marrow promotes tumor metastasis. Mol Cancer Res (2013) 11(8):923–36. doi: 10.1158/1541-7786.MCR-12-0686
41. Fang J-H, Zhou H-C, Zeng C, Yang J, Liu Y, Huang X, et al. MicroRNA-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology (2011) 54(5):1729–40. doi: 10.1002/hep.24577
42. Zhang G, Zhou H, Xiao H, Li Y, Zhou T. MiR-378 is an independent prognostic factor and inhibits cell growth and invasion in colorectal cancer. BMC Cancer (2014) 14(1):109. doi: 10.1186/1471-2407-14-109
43. Chang C-H, Yen M-C, Liao S-H, Hsu Y-L, Lai C-S, Kuo Y-R, et al. Dual role of miR-21-mediated signaling in HUVECs and rat surgical flap under normoxia and hypoxia condition. Int J Mol Sci (2017) 18(9):1917. doi: 10.3390/ijms18091917
44. Li Y, Yan C, Fan J, Hou Z, Han Y. MiR-221-3p targets Hif-1α to inhibit angiogenesis in heart failure. Lab Invest (2021) 101(1):104–15. doi: 10.1038/s41374-020-0450-3
45. Gnanamony M, Demirkhanyan L, Ge L, Sojitra P, Bapana S, Norton JA, et al. Circular dumbbell miR−34a−3p and −5p suppresses pancreatic tumor cell−induced angiogenesis and activates macrophages. Oncol Lett (2021) 21(1):1–1. doi: 10.3892/ol.2020.12336
46. He M, Zhou W, Li C, Guo M. MicroRNAs, DNA damage response, and cancer treatment. Int J Mol Sci (2016) 17(12):2087. doi: 10.3390/ijms17122087
47. Kato M, Paranjape T, Müller RU, Ullrich R, Nallur S, Gillespie E, et al. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene (2009) 28(25):2419–24. doi: 10.1038/onc.2009.106
48. Czochor JR, Sulkowski P, Glazer PM. MiR-155 over-expression promotes genomic instability by reducing high-fidelity polymerase delta expression and activating error-prone DSB repair. Mol Cancer Research : MCR (2016) 14(4):363–73. doi: 10.1158/1541-7786.MCR-15-0399
49. Ramirez HA, Pastar I, Jozic I, Stojadinovic O, Stone RC, Ojeh N, et al. Staphylococcus aureus Triggers Induction of miR-15B-5P to Diminish DNA Repair and Deregulate Inflammatory Response in Diabetic Foot Ulcers. J Invest Dermatol (2018) 138(5):1187–96. doi: 10.1016/j.jid.2017.11.038
50. Fan P, Liu L, Yin Y, Zhao Z, Zhang Y, Amponsah PS, et al. MicroRNA-101-3p reverses gemcitabine resistance by inhibition of ribonucleotide reductase M1 in pancreatic cancer. Cancer Lett (2016) 373(1):130–7. doi: 10.1016/j.canlet.2016.01.038
51. Wu J, Li X, Li D, Ren X, Li Y, Herter EK, et al. MicroRNA-34 family enhances wound inflammation by targeting LGR4. J Invest Dermatol (2020) 140(2):465–476.e11. doi: 10.1016/j.jid.2019.07.694
52. Tan W, Gu Z, Leng J, Zou X, Chen H, Min F, et al. Let-7f-5p ameliorates inflammation by targeting NLRP3 in bone marrow-derived mesenchymal stem cells in patients with systemic lupus erythematosus. Biomed Pharmacother (2019) 118:109313. doi: 10.1016/j.biopha.2019.109313
53. Liu C-W, Sung H-C, Lin S-R, Wu C-W, Lee C-W, Lee I-T, et al. Resveratrol attenuates ICAM-1 expression and monocyte adhesiveness to TNF-α-treated endothelial cells: Evidence for an anti-inflammatory cascade mediated by the miR-221/222/AMPK/p38/NF-κB pathway. Sci Rep (2017) 7(1):44689. doi: 10.1038/srep44689
54. He X-P, Shao Y, Li X-L, Xu W, Chen G-S, Sun H-H, et al. Downregulation of miR-101 in gastric cancer correlates with cyclooxygenase-2 overexpression and tumor growth. FEBS J (2012) 279(22):4201–12. doi: 10.1111/febs.12013
55. Kong D, Piao Y-S, Yamashita S, Oshima H, Oguma K, Fushida S, et al. Inflammation-induced repression of tumor suppressor miR-7 in gastric tumor cells. Oncogene (2012) 31(35):3949–60. doi: 10.1038/onc.2011.558
56. Xie W, Li Z, Li M, Xu N, Zhang Y. miR-181a and inflammation: MiRNA homeostasis response to inflammatory stimuli in vivo. Biochem Biophys Res Commun (2013) 430(2):647–52. doi: 10.1016/j.bbrc.2012.11.097
57. Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol (2006) 26(21):8191–201. doi: 10.1128/MCB.00242-06
58. Si W, Shen J, Du C, Chen D, Gu X, Li C, et al. A miR-20a/MAPK1/c-Myc regulatory feedback loop regulates breast carcinogenesis and chemoresistance. Cell Death Different (2018) 25(2):406–20. doi: 10.1038/cdd.2017.176
59. Piovan C, Palmieri D, Di Leva G, Braccioli L, Casalini P, Nuovo G, et al. Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol Oncol (2012) 6(4):458–72. doi: 10.1016/j.molonc.2012.03.003
60. Shinden Y, Akiyoshi S, Ueo H, Nambara S, Saito T, Komatsu H, et al. Diminished expression of miR-15a is an independent prognostic marker for breast cancer cases. Anticancer Res (2015) 35(1):123–7.
61. Kong L-Y, Xue M, Zhang Q-C, Su C-F. In vivo and in vitro effects of microRNA-27a on proliferation, migration and invasion of breast cancer cells through targeting of SFRP1 gene via Wnt/β-catenin signaling pathway. Oncotarget (2017) 8(9):15507–19. doi: 10.18632/oncotarget.14662
62. Si W, Li Y, Shao H, Hu R, Wang W, Zhang K, et al. MiR-34a inhibits breast cancer proliferation and progression by targeting wnt1 in wnt/β-catenin signaling pathway. Am J Med Sci (2016) 352(2):191–9. doi: 10.1016/j.amjms.2016.05.002
63. Yao A, Xiang Y, Si Y-R, Fan L-J, Li J-P, Li H, et al. PKM2 promotes glucose metabolism through a let-7a-5p/Stat3/hnRNP-A1 regulatory feedback loop in breast cancer cells. J Cell Biochem (2019) 120(4):6542–54. doi: 10.1002/jcb.27947
64. Shi Y, Zhang Y, Ran F, Liu J, Lin J, Hao X, et al. Let-7a-5p inhibits triple-negative breast tumor growth and metastasis through GLUT12-mediated warburg effect. Cancer Lett (2020) 495:53–65. doi: 10.1016/j.canlet.2020.09.012
65. Ruan L, Qian X. MiR-16-5p inhibits breast cancer by reducing AKT3 to restrain NF-κB pathway. Biosci Rep (2019) 39(8). doi: 10.1042/BSR20191611
66. Bandini E, Fanini F, Vannini I, Rossi T, Plousiou M, Tumedei MM, et al. MiR-9-5p as a regulator of the androgen receptor pathway in breast cancer cell lines. Front Cell Dev Biol (2020) 8. doi: 10.3389/fcell.2020.579160
67. Luo Q, Wei C, Li X, Li J, Chen L, Huang Y, et al. MicroRNA-195-5p is a potential diagnostic and therapeutic target for breast cancer. Oncol Rep (2014) 31(3):1096–102. doi: 10.3892/or.2014.2971
68. Patel N, Garikapati KR, Ramaiah MJ, Polavarapu KK, Bhadra U, Bhadra MP. MiR-15a/miR-16 induces mitochondrial dependent apoptosis in breast cancer cells by suppressing oncogene BMI1. Life Sci (2016) 164:60–70. doi: 10.1016/j.lfs.2016.08.028
69. Selcuklu SD, Donoghue MTA, Rehmet K, de Souza Gomes M, Fort A, Kovvuru P, et al. MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells*. J Biol Chem (2012) 287(35):29516–28. doi: 10.1074/jbc.M111.335943
70. Yang G, Wu D, Zhu J, Jiang O, Shi Q, Tian J, et al. Upregulation of miR-195 increases the sensitivity of breast cancer cells to Adriamycin treatment through inhibition of Raf-1. Oncol Rep (2013) 30(2):877–89. doi: 10.3892/or.2013.2532
71. Shi Y, Luo X, Li P, Tan J, Wang X, Xiang T, et al. MiR-7-5p suppresses cell proliferation and induces apoptosis of breast cancer cells mainly by targeting REGγ. Cancer Lett (2015) 358(1):27–36. doi: 10.1016/j.canlet.2014.12.014
72. Li C, Xiong D, Huang C, He R, Liang H, Pan D, et al. Clinical Value of miR-101-3p and Biological Analysis of its Prospective Targets in Breast Cancer: A Study Based on The Cancer Genome Atlas (TCGA) and Bioinformatics. Med Sci Monitor : Int Med J Exp Clin Res (2017) 23:1857–71. doi: 10.12659/MSM.900030
73. Fantozzi A, Gruber DC, Pisarsky L, Heck C, Kunita A, Yilmaz M, et al. VEGF-mediated angiogenesis links EMT-induced cancer stemness to tumor initiation. Cancer Res (2014) 74(5):1566–75. doi: 10.1158/0008-5472.CAN-13-1641
74. Sun C-Y, She X-M, Qin Y, Chu Z-B, Chen L, Ai L-S, et al. MiR-15a and miR-16 affect the angiogenesis of multiple myeloma by targeting VEGF. Carcinogenesis (2013) 34(2):426–35. doi: 10.1093/carcin/bgs333
75. Ma H, Pan J-S, Jin L-X, Wu J, Ren Y-D, Chen P, et al. MicroRNA-17~92 inhibits colorectal cancer progression by targeting angiogenesis. Cancer Lett (2016) 376(2):293–302. doi: 10.1016/j.canlet.2016.04.011
76. Shyamasundar S, Lim JP, Bay BH. MiR-93 inhibits the invasive potential of triple-negative breast cancer cells in vitro via protein kinase WNK1. Int J Oncol (2016) 49(6):2629–36. doi: 10.3892/ijo.2016.3761
77. Pin A-L, Houle F, Guillonneau M, Paquet ER, Simard MJ, Huot J. MiR-20a represses endothelial cell migration by targeting MKK3 and inhibiting p38 MAP kinase activation in response to VEGF. Angiogenesis (2012) 15(4):593–608. doi: 10.1007/s10456-012-9283-z
78. Du Y-E, Tu G, Yang G, Li G, Yang D, Lang L, et al. MiR-205/YAP1 in activated fibroblasts of breast tumor promotes VEGF-independent angiogenesis through STAT3 signaling. Theranostics (2017) 7(16):3972–88. doi: 10.7150/thno.18990
79. Wu H, Zhu S, Mo Y-Y. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res (2009) 19(4):439–48. doi: 10.1038/cr.2009.18
80. Zhao T, Li J, Chen AF. MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. Am J Physiology-Endocrinol Metab (2010) 299(1):E110–6. doi: 10.1152/ajpendo.00192.2010
81. Qu Y, Liu H, Lv X, Liu Y, Wang X, Zhang M, et al. MicroRNA-16-5p overexpression suppresses proliferation and invasion as well as triggers apoptosis by targeting VEGFA expression in breast carcinoma. Oncotarget (2017) 8(42):72400–10. doi: 10.18632/oncotarget.20398
82. Patel N, Garikapati KR, Pandita RK, Singh DK, Pandita TK, Bhadra U, et al. MiR-15a/miR-16 down-regulates BMI1, impacting Ub-H2A mediated DNA repair and breast cancer cell sensitivity to doxorubicin. Sci Rep (2017) 7(1):4263. doi: 10.1038/s41598-017-02800-2
83. Di Francesco A, De Pittà C, Moret F, Barbieri V, Celotti L, Mognato M. The DNA-damage response to γ-radiation is affected by miR-27a in A549 cells. Int J Mol Sci (2013) 14(9):17881–96. doi: 10.3390/ijms140917881
84. Rahman M, Lovat F, Romano G, Calore F, Acunzo M, Bell EH, et al. MiR-15b/16-2 Regulates Factors That Promote p53 Phosphorylation and Augments the DNA Damage Response following Radiation in the Lung. J Biol Chem (2014) 289(38):26406–16. doi: 10.1074/jbc.M114.573592
Keywords: breast cancer, cyclin dependent kinase, epithelial-to-mesenchymal transition, miRNA, pancreatic ductal adenocarcinoma, chronic cyclic hypoxia, hypoxamirs, doxorubicin chemoresistance
Citation: Al-Sisan SM, Zihlif MA and Hammad HM (2023) Differential miRNA expression of hypoxic MCF7 and PANC-1 cells. Front. Endocrinol. 14:1110743. doi: 10.3389/fendo.2023.1110743
Received: 29 November 2022; Accepted: 21 June 2023;
Published: 31 July 2023.
Edited by:
Gianluca Baldanzi, Università degli Studi del Piemonte Orientale, ItalyReviewed by:
Ashraf M. Omar, University of Toyama, JapanPouya Goleij, Sana Institute of Higher Education, Iran
Copyright © 2023 Al-Sisan, Zihlif and Hammad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hana M. Hammad, aGFuYS5oYW1tYWRAZ21haWwuY29t
 Sandy M. Al-Sisan
Sandy M. Al-Sisan Malek A. Zihlif
Malek A. Zihlif Hana M. Hammad
Hana M. Hammad