- 1Department of Gynecology, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, China
- 2Department of Dermatology, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, China
- 3Department of Cardiology, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, China
- 4Research Center for Translational Medicine, East Hospital, Tongji University School of Medicine, Shanghai, China
Background: The incidence of endometriosis (EMs), a common disease in gynecology, has increased over the years. Women suffer from the symptoms caused by EMs, such as chronic pelvic pain, dysmenorrhea, and infertility. However, the etiology and pathophysiology of EMs remain unclear. This study aimed to identify candidate genes of endometriosis through integrated analysis of genome-wide gene expression and DNA methylation profiles.
Results: Eutopic and ectopic endometrial tissues were collected from patients who were diagnosed as ovarian EMs. Genome-wide methylation profiling identified 17551 differentially methylated loci, with 9777 hypermethylated and 7774 hypomethylated loci. Differentially methylated loci were mainly concentrated in the gene body and intergenic regions. Genome-wide gene expression profiling identified 1837 differentially expressed genes (DEGs), with 1079 genes upregulated and 758 downregulated in ectopic groups. Integrated analysis revealed that DNA methylation was negatively correlated to gene expression in most genomic regions, such as exon, 3’UTR, 5’UTR, and promoter. We also identified promoter-related (53 downregulated and 113 upregulated) and enhancer-related DMGs (212 downregulated and 232 upregulated), which were significantly correlated to the gene expression. Further validation of the top-ranked genes belonging to differentially methylated genes (DMGs) and DEGs revealed that TMEM184A, GREM2, SFN, KIR3DX1, HPGD, ESR1, BST2, PIK3CG and RNASE1 were significant candidate genes in ovarian endometriosis.
Conclusion: Our study revealed the significance of DNA methylation in the gene expression in ovary endometriosis, which provides new insights and a molecular foundation for understanding the underlying mechanisms of endometriosis.
Introduction
Endometrial tissue that exhibits growing activity outside the uterine cavity and grows abnormally is defined as endometriosis (EMs) (1). Lesions typically occur in the peritoneal cavity, including the ovaries, uterorectal depressions, and uterosacral ligaments (2). EMs affected 5% to 15% of women of reproductive age (3). Patients with EMs suffer a series of symptoms, such as pelvic pain, dysmenorrhea, and infertility (4). EMs exhibit biological behaviors of malignant tumors as a benign disease, such as cell proliferation, implant growth, infiltration, distant metastasis, and recurrence (2). The current mainstream etiological theories cannot fully explain the pathogenesis of EMs, including ‘menstrual blood reflux theory’, ‘lymphatic and venous dissemination theory’ (5), and ‘body cavity epithelial metaplasia theory’ (6), immunological mechanism, angiogenesis mechanism, cell proliferation, and apoptosis imbalance mechanism, and in-place endometrial determinism (7). In recent years, the incidence of EMs has significantly increased, and the patient’s onset age tends toward younger, calling for a deeper understanding of the pathogenesis of EMs.
DNA methylation is important in the regulation of gene expression (8). Several studies reported that DNA methylation might play an essential role in the pathogenesis of EMs (9). With the development of sequencing techniques, genome-wide methylation differences between eutopic and ectopic endometrial tissues have been investigated in recent years (10–14). However, few studies linked genome-wide DNA methylation with the changes in gene expression in EMs (10, 15), suggesting DNA methylation changes were associated with altered gene expression in endometrial function or dysfunction. Herein, we integrated the genome-wide DNA methylation with the gene expression of EMs to find candidate genes in EMs.
Materials and methods
Patient enrollment and sample collection
Thirteen female patients who were diagnosed as ovarian EMs were enrolled. DNA methylation profiles were performed on 6 patient specimens, RNA sequencing was performed on 3 patient specimens, and quantitative reverse transcription PCR (qRT-PCR) verifications were performed on 7 patient specimens. Patients (Hans, 27-42 years old) in Shanghai East Hospital undergoing laparoscopic and postoperative pathological diagnosis of ovarian EMs were selected. The typical ectopic endometrial tissue and the matched eutopic endometrium from each patient were collected in the proliferation phase. All the participants had regular menstrual cycles and had not received hormonal medication 3 months before surgery. The participants had no history of other organic diseases, such as hypertension, diabetes, tumors, chronic infections, etc. Patients with benign gynecologic diseases (leiomyoma, ovarian serous cyst, PCOS) were excluded. All patients provided written informed consent in this study. This study was approved by the Ethics Committee of the Shanghai East Hospital of Tongji University.
DNA and RNA extraction
All the samples were kept in RNAlater (Ambion, AM7020) when obtained during laparoscopic surgery. Ectopic and eutopic tissues were extracted for genomic DNA detection according to the manufacturer’s instructions (QIAamp DNA Mini Kit, 51304). Total RNA was extracted from ectopic and eutopic tissues using RNAiso plus reagent (Takara, 9109) following the manufacturer’s instructions. Briefly, tissues were grinded and crushed in liquid nitrogen, and then added 300 µL chloroform/isoamyl alcohol (24:1) and mixed thoroughly. After centrifuging, the supernatant was moved to a 1.5 mL centrifuge tube, mixed with isopropyl alcohol, and then purified to RNA for the subsequent experiments.
DNA methylation array
At least 500 ng genomic DNA was extracted and bisulfate converted (Zymo EZ-96 DNA Methylation-Direct Kit, D5023) before being processed to the methylation array. Illumina Human Methylation 450 K BeadChips were applied to get the whole genome methylation states of the samples. The experimental data were sorted out and encoded, and the data was double-converted and double-calibrated by EpidData. Background adjustment and quantile normalization pretreatment were performed according to the raw data of each probe methylation site of the Illumina methylated HM450K BeadChip chip. Then the degree of methylation at the probe level was analyzed. β value was calculated and normalized to quantify the degree of methylation. In this study, the average β value ≥ 0.2 and P-value < 0.05 was considered as significant.
Differential methylation analysis
After standardizing the original methylation data, the Pearson Correlation Coefficiency (PCC) was used to evaluate the methylation profile correlation between the ectopic and eutopic groups. The correlation between ectopic and eutopic samples was high, indicating the overall reproducibility of the samples. Since not all the samples were in accordance with normal distribution, the Wilcox test was applied to do differential methylation analysis. The criteria for screening the difference site need to meet both (1) the original P < 0.05 and (2) the difference of ectopic and eutopic group beta value (delta beta value) greater than or equal to 0.2. The results were also calculated with FDR (false discovery rate) corrected by multiple hypothesis testing to assess differential methylation levels. The differential DMR biological function analysis was performed using the hypergeometric distribution algorithm or Fisher test. Biological pathway analysis was enriched in KEGG (Kyoto Encyclopedia of Genes and Genomes, www.genome.jp/kegg/) database.
RNA-sequencing and analysis
RNA libraries were constructed with a VAHTS Universal V6 RNA-seq Library Prep Kit for MGI (Vazyme, NRM604). DNBSEQ-T7 high-throughput sequencing platform of BGI was used for sequencing. The raw data was filtered to clean data after trimming low quality reads and adapters. Clean data were mapped to mm10 mouse reference genome using Hisat2 software (version 2.2.1). The reads were quantified by featureCounts (version 2.0.1). The differential gene analysis was performed by Deseq2 (version 1.32.0). Genes with adjusted P value < 0.05 and the abs (Log2(Fold Change)) ≥ 1 was considered as differentially expressed genes (DEGs). The enrichment of Gene Ontology (GO) was conducted by clusterProfiler package (v4.0.5).
Quantitative reverse transcription PCR
Total RNA was extracted from ectopic and eutopic samples. cDNA was obtained after reverse transcription reversal by Evo M-MLV RT Kit (Accurate Biology Co. Ltd, AG11601). The qRT-PCR was performed by Evo M-MLV One Step RT-PCR Kit (Accurate Biology Co. Ltd, AG11607). Each target gene was compared to β-actin. The expression of target mRNA was calculated based on 2-ΔΔCt method. The primers used in this study were listed in the Supplementary Table S2.
Statistical analysis
All the statistical analyses were performed using R software (version 4.0.1). P-values less than 0.05 were considered statistically significant. The Kolmogorov-Smirnov test was applied to evaluate the normality of the distribution of the variables. For qRT-PCR, statistical analyses were conducted using a student t-test (data with normal distribution) or Mann-Whitney test (data with skewed distribution) as appropriate by GraphPad Prism software 8.0 (GraphPad Software Inc).
Results
Genome-wide DNA methylation changes between eutopic and ectopic endometrial tissues
We first obtained the genome-wide DNA methylation changes of eutopic and ectopic endometrial tissues (Figure 1A). The clinical characteristics of the patients were summarized in Supplementary Table S1. A total of 17551 differentially methylated loci were identified, of which 9777 were hypermethylated and 7774 were hypomethylated (Figure 1B). To get view of the distribution of differentially methylation loci, we annotated the locations to different functional elements in the genome. Hypermethylated and hypomethylated loci in ectopic groups were mainly located in gene body (39.13% of hypermethylated loci, and 37.97% of hypomethylated loci) and intergenic regions (36.22% of hypermethylated loci, and 30.89% of hypomethylated loci) (Figure 1C). Furthermore, the CpG island probe distribution of both hypermethylated and hypomethylated loci occurred in the OpenSea region (Figure 1D). Next, we performed KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis of differentially methylated loci. Both hypermethylated and hypomethylated genes in ectopic were enriched in ECM-receptor interaction, human papillomavirus (HPV) infection, focal adhesion, PI3K-Akt signaling pathway and Wnt signaling pathway. Moreover, axon guidance, estrogen signaling pathway, glutamatergic synapse, protein digestion and absorption, and relaxin signaling pathway were only enriched in hypermethylated genes. Besides, apelin signaling pathway, cholinergic synapse and Hippo signaling pathway were only enriched in hypomethylated genes in the ectopic group (Figure 1E). The above results indicated that DNA methylation was associated with various pathways in the eutopic and ectopic endometrial tissues.
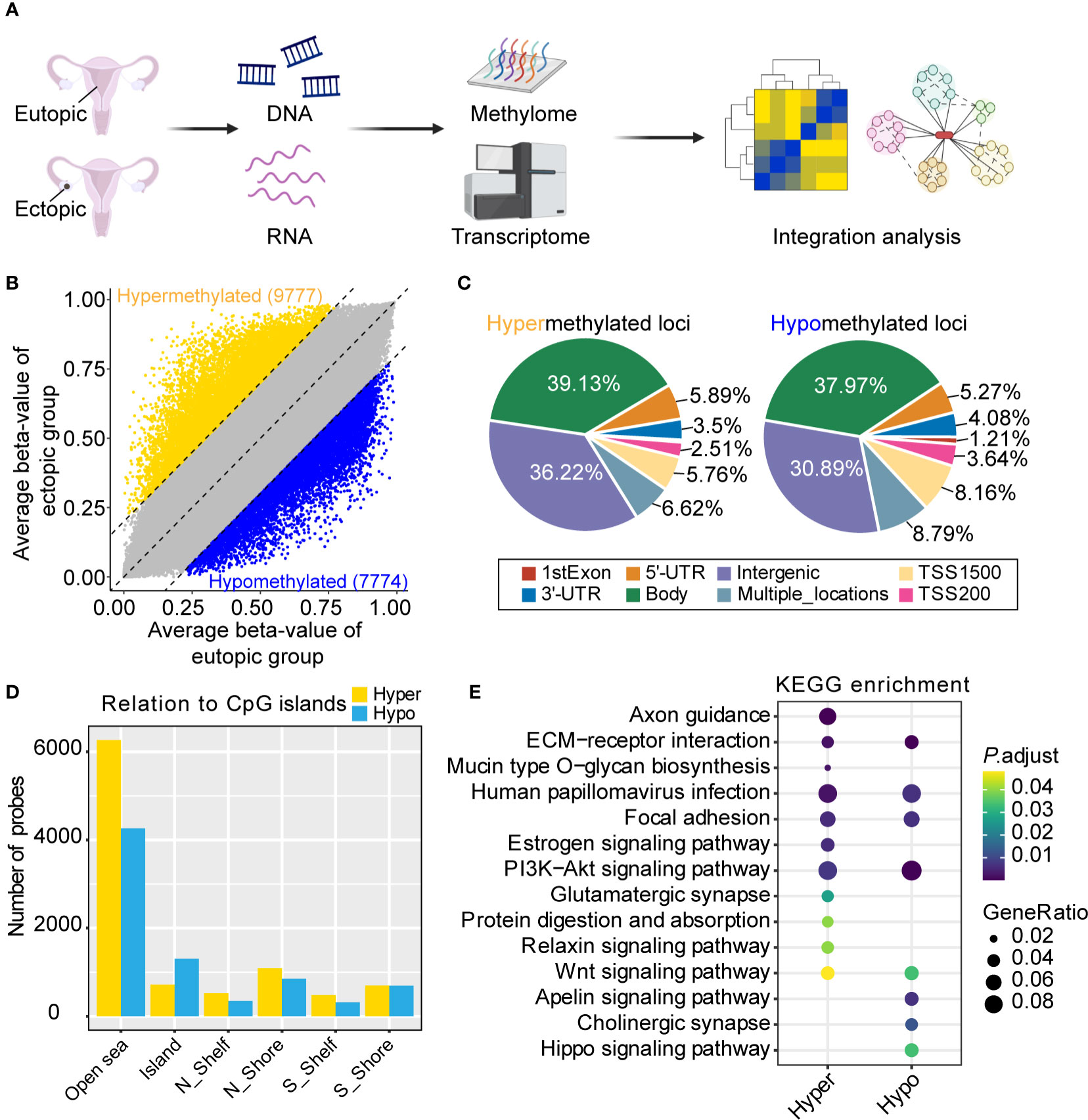
Figure 1 Genome-wide DNA methylation analysis of eutopic and ectopic endometrial tissues. (A) Schematic diagram of the study. Created with BioRender.com. (B) Differential methylated loci in eutopic and ectopic endometrial tissues. (C) The distribution of hypermethylated (left) and hypomethylated (right) loci in ectopic endometrial tissues. (D) CpG island probe distribution of hypermethylated and hypomethylated loci in ectopic endometrial tissues. (E) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the hypermethylated and hypomethylated loci in the ectopic group.
Transcriptome analysis of ectopic and eutopic samples
To view the transcriptional difference between ectopic and eutopic endometrial tissues. We performed RNA sequencing of ectopic and eutopic endometrial tissues (3 ectopic vs 3 eutopic, Supplementary Table S1). As indicated in Principal Component Analysis (PCA), ectopic and eutopic endometrial tissues were significantly separated and the samples in each group were closely related (Figure 2A), suggesting that ectopic and eutopic endometrial tissues were transcriptionally different. Differential gene expression analysis showed that there were 1837 differentially expressed genes (DEGs) (Figure 2B) in the ectopic compared to eutopic samples, with 1079 genes upregulated and 758 downregulated. Next, we compared our RNA-seq data with another RNA-seq dataset comparing the ectopic with eutopic endometria in eight patients with ovarian endometriosis (GSE105764) (16). Venn plot showed that 70.4% upregulated and 60.95% downregulated DEGs in our RNA-seq data were overlapped with DEGs in GSE105764 dataset (Figure 2C), validating the reliability of our RNA-seq data. The top 10 upregulated and downregulated genes in ectopic endometrial tissues were shown in Figure 2D. The upregulated DEGs in ectopic samples were enriched in leukocyte migration, muscle system process, humoral immune in response (Figure 2E), and downregulated DEGs are enriched in functions such as nuclear division, chromosome segregation, and mitotic nuclear division (Figure 2F). In summary, ectopic and eutopic endometrial tissues were transcriptionally different from each other.
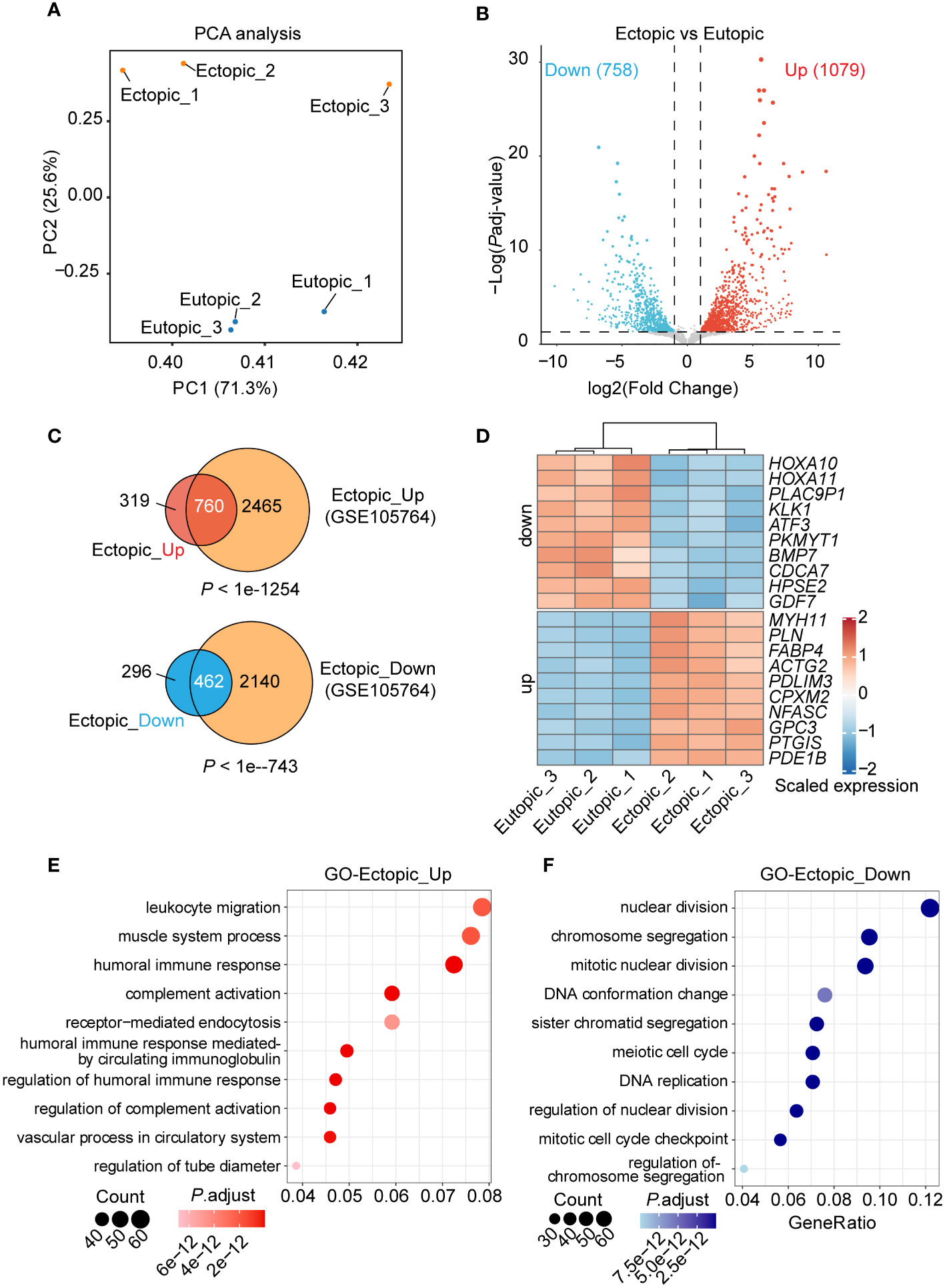
Figure 2 Genome-wide gene expression analysis of eutopic and ectopic endometrial tissues. (A) Principal components analysis (PCA) analysis of the eutopic and ectopic samples. (B) Differentially expressed genes (DEGs) analysis of eutopic and ectopic samples. (C) Venn plots of the DEGs and DEGs in the GSE105764 dataset. (D) Heatmap of the top 10 upregulated and downregulated DEGs in RNA-seq. (E) GO analysis of the upregulated genes in ectopic samples. (F) GO analysis of the downregulated genes in ectopic samples.
Integrated analysis of DNA methylation and gene expression reveals candidate genes in endometriosis
DNA methylation was important in the gene expression regulation (8). To connect DNA methylation with gene expression in endometriosis, we further integrated our DNA methylation and RNA-seq data. First, we divided the DNA methylation states to six groups (1st exon, 3’UTR, 5’UTR, gene body, multiple locations, and promoter) according to the position of methylated loci. As expected, the level of DNA methylation (delta β-value) was negatively correlated with gene expression in most of the groups (1st exon, 5’UTR, multiple locations, and promoter) (Figure 3A). We further defined genes with differentially methylated loci in promoters as ‘promoter-related DMGs (differentially methylated genes)’ and genes with differentially methylated loci not in promoters as ‘enhancer-related DMGs’. According to the relation of level of methylation (delta β-value) and gene expression, genes were separated to four quadrants (part 1 to part 4, Figure 3A). Almost half of promoter-related DMGs (48.0%, 113/235) were in part 4 (Figure 3B). Promoter-related DMGs in part 4 were enriched in sialic acid binding and lipase activity (Figure 3C). Furthermore, promoter-related DMGs in part 2 were specifically enriched in channel activity. For enhancer-related DMGs, 70.25% were in part 1 and 4 (Figure 3D). DMGs in part 1 were enriched in transcriptional activity (nuclear receptor activity, ligand-activated transcription factor activity, and DNA-binding transcription activator), steroid hormone receptor activity and channel activity (Figure 3E). These results indicated that most DMGs in promoter and enhancer were negatively correlated with gene expression.
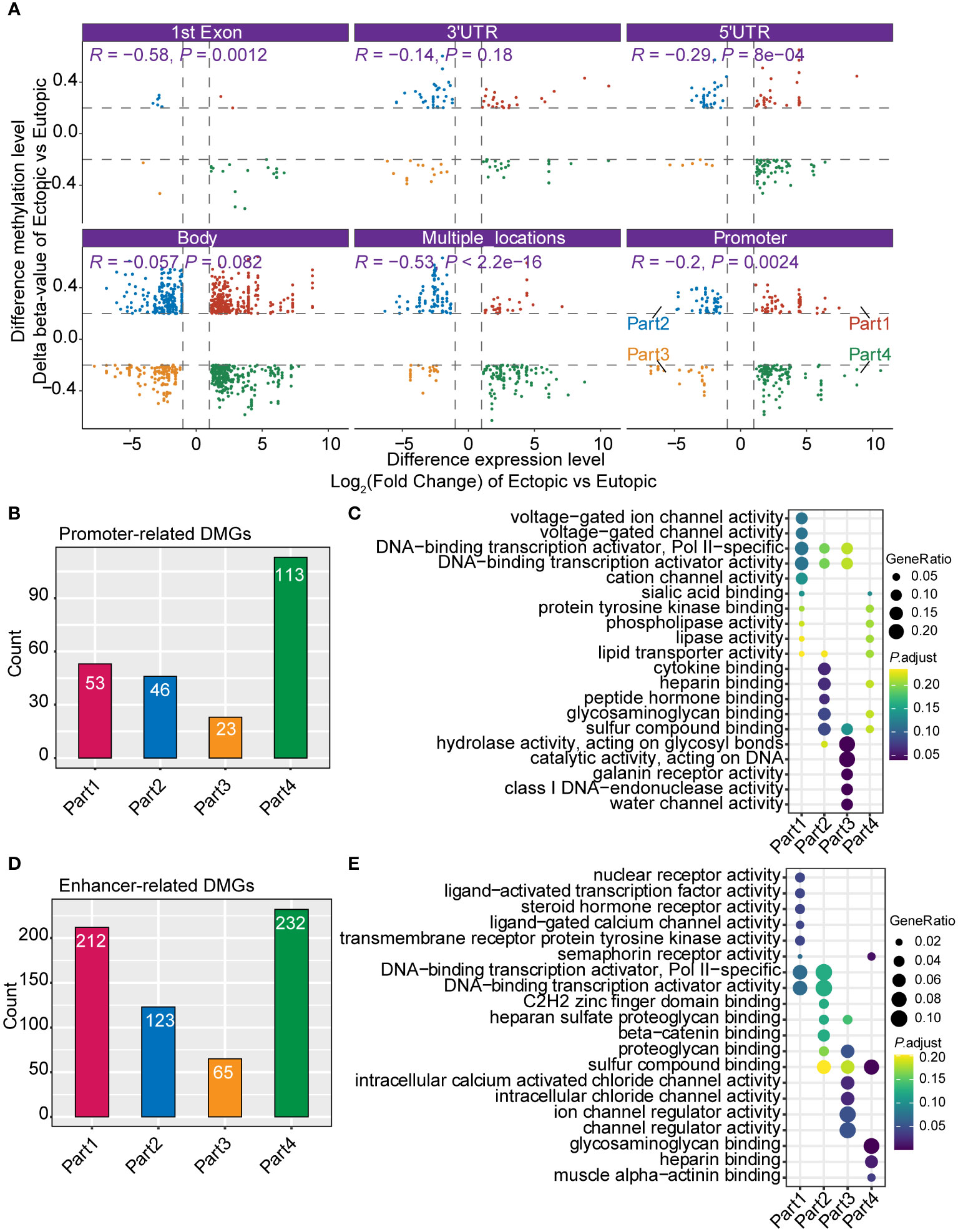
Figure 3 Integrated analysis of DNA methylation and gene expression profiles. (A) Correlation of the DNA methylation and gene expression grouped by different genomic positions (1st Exon, 3’UTR, 5’UTR, gene body, multiple locations, and promoter). (B) Bar plot shows the count of promoter-related differentially methylated genes (DMGs). (C) Gene Ontology (GO) of different groups of promoter-related DMGs (part1-part4 in Figure 3A). (D) Bar plot shows the count of enhancer-related DMGs. (E) GO analysis of different groups of enhancer-related DMGs.
Validation of candidate genes in endometriosis
To validate the candidate genes in endometriosis, we listed the top 15 DMGs (ranked by delta β-value) whose expression was upregulated (Table 1) and downregulated (Table 2). After filtering the past reported genes, 10 unreported DMGs (TMEM184A, GREM2, SFN, KIR3DX1, HPGD, ESR1, CASS4, BST2, PIK3CG and RNASE1) were kept. We further detected the expression of these 10 DMGs in ectopic and eutopic endometrial tissues (7 ectopic vs 7 eutopic, Supplementary Table S1). TMEM184A, GREM2, SFN, KIR3DX1, HPGD, and ESR1 were significantly downregulated in ectopic tissues, and BST2, PIK3CG and RNASE1 were significantly upregulated in ectopic tissues (Figure 4). In summary, we validated the top candidate genes in endometriosis which might be regulated by DNA methylation.
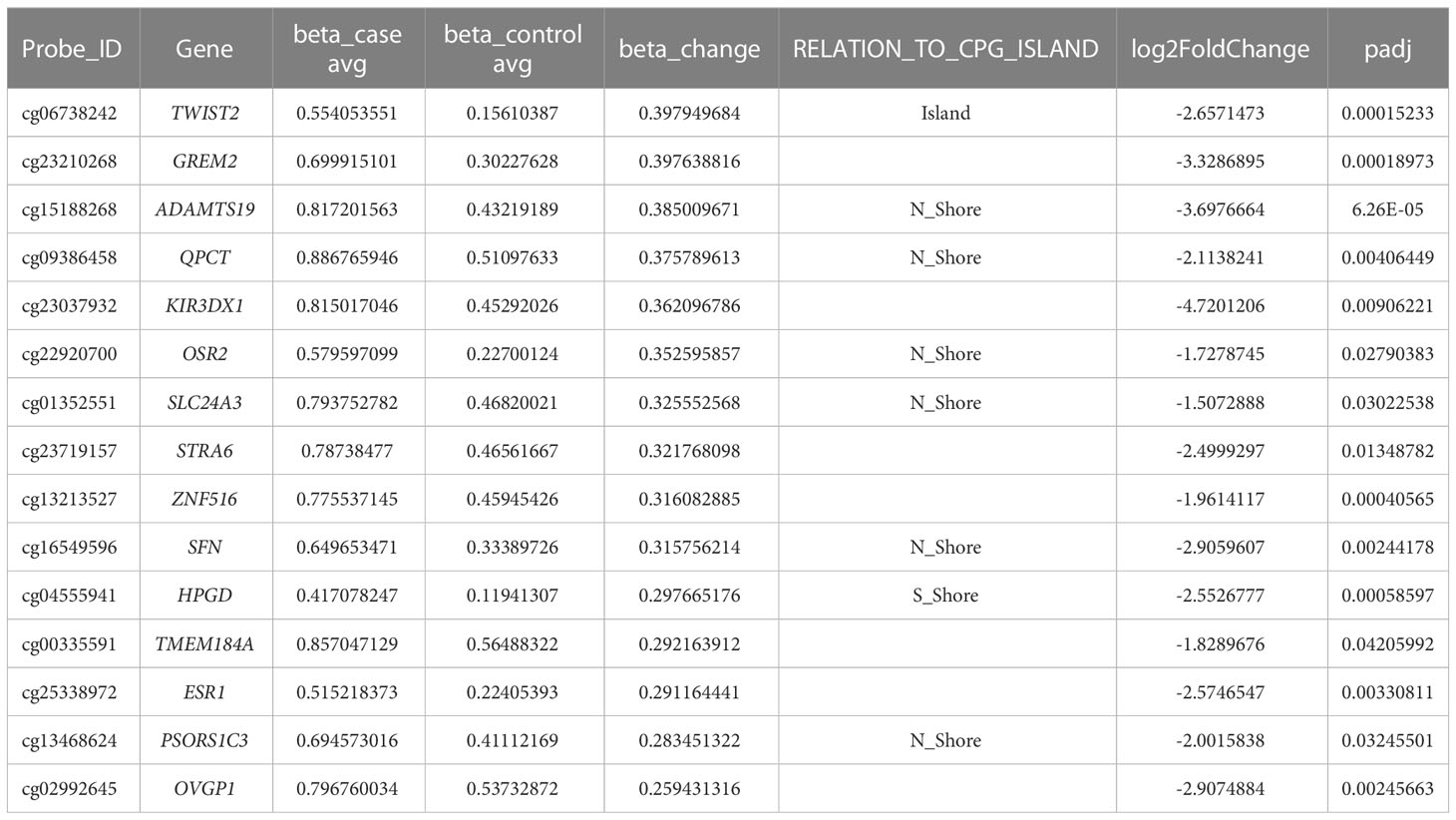
Table 1 Top candidate genes which were hypermethylated and downregulated in ectopic endometrial tissues.
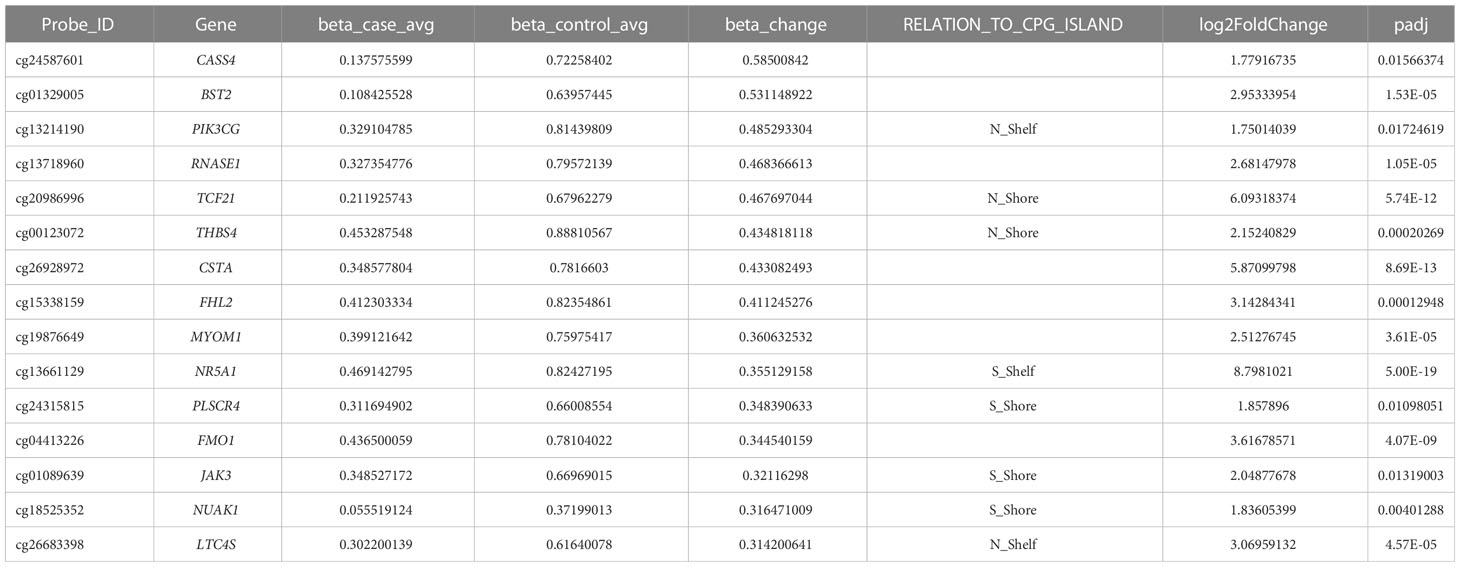
Table 2 Top candidate genes which were hypomethylated and upregulated in ectopic endometrial tissues.
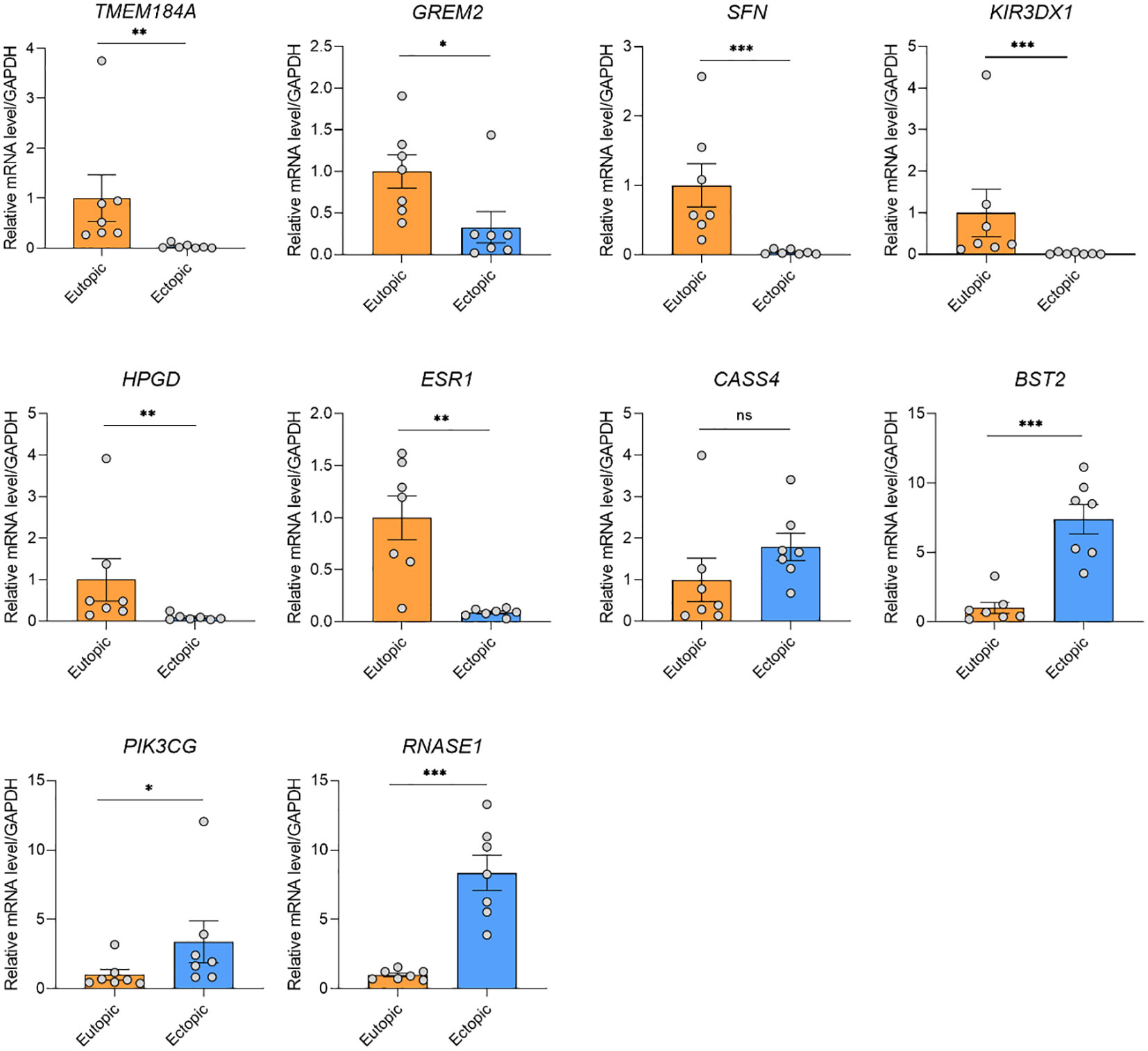
Figure 4 qRT-PCR validation of the gene expression of the candidate genes. TMEM184A, GREM2, SFN, KIR3DX1, HPGD, and ESR1 were significantly downregulated in ectopic tissues, and BST2, PIK3CG and RNASE1 were significantly upregulated in ectopic tissues. ***P < 0.001, **P < 0.01, *P < 0.05, ns, not significant.
Discussions
EMs, vividly known as ‘a kind of benign cancer’ and women’s pelvic ‘sandstorm’, has established itself as one of the most common diseases in gynecology by its high incidence and its early onset age, Symptoms like chronic pelvic pain, dysmenorrhea, and infertility caused by EMs have seriously threatened women’s health and reduced their life quality. EMs is one of the focuses of global reproductive health issues. According to the available literature reports, it is estimated that about 15% to 30% of women in the childbearing period would be affected by the disease, involving about 270 million people worldwide (17).
Although the theory of menstrual blood reflux proposed by Sampson J.J., who was the father of endometriosis in the world, has been accepted for a century and a half (5), the related factors and pathogenesis of EMs are still incomplete. Due to the wide range of lesions and diverse morphologies, malignant biological behaviors such as metastasis, invasion, and recurrence, and the possibility of malignant transformation, EMs is vividly called ‘benign cancer’ (1). More and more studies believe that its similar biological behavior to malignant tumors may be one of the important pathogenesis of EMs (18–21). The occurrence and development of malignant lesions are marked by the accumulation of tumor suppressor genes and oncogene genetic variations, especially by the abnormal methylation of tumor suppressor genes. Studies have confirmed that abnormal methylation of CpG islands regulated by non-methylated tumor suppressor genes is closely related to the occurrence and metastasis of human malignant tumors (22). A large number of studies have confirmed that EMs is a genetic disease caused by the interaction of multiple locus genes and environmental factors (23–26). HOXA10, hMLH1, PTEN promoter region 5’ CpG island methylation pattern is currently known to be related to the occurrence and development of EMs. HOXA10 gene can inhibit tumor cell proliferation, reduce invasion ability, and inhibit tumor cell metastasis. Recent studies abroad have found that the reduced expression of the HOXA10 gene in EMs is similar to malignant lesions. EMs animal model studies have further supported that methylation of the promoter region of the HOXA10 gene may be involved in the pathogenesis of EMs (27). PTEN is currently recognized as a tumor suppressor gene, and PTEN methylation is closely related to the occurrence and development of various tumors. Abnormalities in this gene have been demonstrated in many human cancers, especially in ovarian endometrioidal carcinomas, endometrial cancers, and gliomas (28). Salvesen et al. confirmed that PTEN gene methylation was prevalent in endometrial cancer, and tumor suppressor gene methylation was associated with advanced tumor metastasis in endometrial cancer and plays an important role in tumor progression (28). KF. Tam et al. (29) explored the relationship between DNA methylation and ovarian tumors, including PTEN genes. They concluded that the methylation rate of borderline ovarian tumors and ovarian cancer was significantly higher than that of benign ovarian tumors and normal ovarian tissue.
Our study identified a series of candidate genes and pathways in EMs. Of note, in KEGG pathway analysis, both hypermethylated and hypomethylated genes in ectopic were enriched in human papillomavirus (HPV) infection, suggesting a potential link between endometriosis and HPV infection. The prevalence of high-risk HPV was significantly higher in patients with EMs than in those without EMs (30). Besides, HPV-infected endometrial cells were reported to be spread through retrograde menstruation (31). The persistent HPV infection of endometriosis lesions was proposed to contribute to malignant progression. Since EMs is considered a common gynecologic problem with multifactorial origins, HPV infection is a highly possible pathophysiologic causes of EMs. We also validated the differential expression of top-ranked genes (TMEM184A, GREM2, SFN, KIR3DX1, HPGD, ESR1, BST2, PIK3CG and RNASE1) in EMs and normal tissues. TMEM184A, GREM2, SFN, KIR3DX1, HPGD, and ESR1 were hypermethylated and significantly downregulated in ectopic endometrial tissues. Conversely, BST2, PIK3CG and RNASE1 were hypomethylated and upregulated in ectopic endometrial tissues.
SFN is highly expressed in the esophagus-mucous membrane, skin, small salivary glands, and vagina (32–34), indicating its essential role in maintaining the homeostasis of a series of tissues and organs. The higher expression of SFN was significantly associated with a better prognosis of endometrial cancer (35). Accordingly, SFN was downregulated in ectopic endometrial tissues in our study, suggesting the loss of function of SFN might be involved in the pathophysiology of EMs. HPGD was reported to be involved in the pathophysiology of endometriosis (36). Decreased expression of HPGD is associated with abnormal prostaglandin metabolism in endometriosis (35, 37). Besides, the decreased expression of HPGD might be regulated by the miR-218-5p in endometrial adenocarcinoma tissue (35). ESR1 encodes estrogen receptors and ligand-activated transcription factors that can form homodimers or heterodimers with estrogen receptors. ERβ is widely expressed during endometrial hyperplasia in infertile women while decreasing or absent in women of normal childbearing age, suggesting that overexpression of ERβ may be related to infertility. Mutated ER in exon 8 with high activity in vivo may cause precocious puberty in girls (38). As reported, genetic variants in ESR1 were reported to change the susceptibility to endometriosis and might influence the fertility status in endometriosis patients (39). Moreover, ESR1 amplification might be one mechanism for ER over-expression in endometrial carcinoma (40). BST2 was one of the proteins that were found to be related to tumor metastasis (41). The anti-BST2 antibody had a potent antitumor effect against endometrial cancer both in vitro and in vivo, indicating that anti-BST2 antibody might be a potential therapeutic strategy for endometrial cancer. Interestingly, There are few studies of TMEM184A, GREM2, KIR3DX1, BST2 and RNASE1 in EMs, which needs more investigations to figure out their function in EMs in the future.
There are some limitations in this study. The low number of included patients might limit the statistical power of the findings. Further large-scale studies and investigations are needed to validate the findings. Besides, all the patients enrolled were Asian, which might introduce confounding when interpreting the results. With the increasing sequencing data of EMs being reported, meta-analyses of these sequencing data might give a more comprehensive view of the pathophysiology of EMs.
Conclusion
In conclusion, we screened the differentially methylated and expressed genes through genome-wide DNA methylation and transcriptome sequencing of ectopic and eutopic endometrial tissues. Further integrated analysis identified a series of potential candidate genes in EMs. We also verified TMEM184A, GREM2, SFN, KIR3DX1, HPGD, ESR1, BST2, PIK3CG and RNASE1 as key candidate genes in EMs.
Data availability statement
The data presented in the study are deposited in the CNGB Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb) repository, accession number CNP0003813.
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Committee of the Shanghai East Hospital of Tongji University. The patients/participants provided their written informed consent to participate in this study.
Author contributions
FL and BL conceived and designed the project. LL and FL performed all operations. BL analyzed the data and drew the figures. LL and BL wrote the manuscript. XX, CG, BL and FL revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (2021YFC2701200) to FL, and fund program of Shanghai Municipal Commission of Health and Family Planning (No. 202040132) to LL.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1093683/full#supplementary-material
References
1. Olive DL, Pritts EA. Treatment of endometriosis. N Engl J Med (2001) 345(4):266–75. doi: 10.1056/NEJM200107263450407
2. Macer ML, Taylor HS. Endometriosis and infertility: a review of the pathogenesis and treatment of endometriosis-associated infertility. Obstet Gynecol Clin North Am (2012) 39(4):535–49. doi: 10.1016/j.ogc.2012.10.002
3. Giudice LC, Kao LC. Endometriosis. Lancet (2004) 364(9447):1789–99. doi: 10.1016/S0140-6736(04)17403-5
4. Giudice LC. Clinical practice. Endometriosis. N Engl J Med (2010) 362(25):2389–98. doi: 10.1056/NEJMcp1000274
5. Nisolle M, Donnez J. Peritoneal endometriosis, ovarian endometriosis, and adenomyotic nodules of the rectovaginal septum are three different entities. Fertil Steril (1997) 68(4):585–96. doi: 10.1016/s0015-0282(97)00191-x
6. Eskenazi B, Warner ML. Epidemiology of endometriosis. Obstet Gynecol Clin North Am (1997) 24(2):235–58. doi: 10.1016/s0889-8545(05)70302-8
7. Santamaria X, Massasa EE, Taylor HS. Migration of cells from experimental endometriosis to the uterine endometrium. Endocrinology (2012) 153(11):5566–74. doi: 10.1210/en.2012-1202
8. Greenberg MVC, Bourc'his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol (2019) 20(10):590–607. doi: 10.1038/s41580-019-0159-6
9. Koukoura O, Sifakis S, Spandidos DA. DNA Methylation in endometriosis (Review). Mol Med Rep (2016) 13(4):2939–48. doi: 10.3892/mmr.2016.4925
10. Houshdaran S, Nezhat CR, Vo KC, Zelenko Z, Irwin JC, Giudice LC. Aberrant endometrial DNA methylome and associated gene expression in women with endometriosis. Biol Reprod (2016) 95(5):93. doi: 10.1095/biolreprod.116.140434
11. Naqvi H, Ilagan Y, Krikun G, Taylor HS. Altered genome-wide methylation in endometriosis. Reprod Sci (2014) 21(10):1237–43. doi: 10.1177/1933719114532841
12. Nasu K, Kawano Y, Tsukamoto Y, Takano M, Takai N, Li H, et al. Aberrant DNA methylation status of endometriosis: epigenetics as the pathogenesis, biomarker and therapeutic target. J Obstet Gynaecol Res (2011) 37(7):683–95. doi: 10.1111/j.1447-0756.2011.01663.x
13. Saare M, Modhukur V, Suhorutshenko M, Rajashekar B, Rekker K, Soritsa D, et al. The influence of menstrual cycle and endometriosis on endometrial methylome. Clin Epigenet (2016) 8:2. doi: 10.1186/s13148-015-0168-z
14. Wang L, Zhao J, Li Y, Wang Z, Kang S. Genome-wide analysis of DNA methylation in endometriosis using illumina human methylation 450 K BeadChips. Mol Reprod Dev (2019) 86(5):491–501. doi: 10.1002/mrd.23127
15. Houshdaran S, Zelenko Z, Irwin JC, Giudice LC. Human endometrial DNA methylome is cycle-dependent and is associated with gene expression regulation. Mol Endocrinol (2014) 28(7):1118–35. doi: 10.1210/me.2013-1340
16. Zhao L, Gu C, Ye M, Zhang Z, Li L, Fan W, et al. Integration analysis of microRNA and mRNA paired expression profiling identifies deregulated microRNA-transcription factor-gene regulatory networks in ovarian endometriosis. Reprod Biol Endocrinol (2018) 16(1):4. doi: 10.1186/s12958-017-0319-5
17. Nezhat C, Lewis M, Kotikela S, Veeraswamy A, Saadat L, Hajhosseini B, et al. Robotic versus standard laparoscopy for the treatment of endometriosis. Fertil Steril (2010) 94(7):2758–60. doi: 10.1016/j.fertnstert.2010.04.031
18. Choudhuri S. From waddington's epigenetic landscape to small noncoding RNA: some important milestones in the history of epigenetics research. Toxicol Mech Methods (2011) 21(4):252–74. doi: 10.3109/15376516.2011.559695
19. Wu Y, Strawn E, Basir Z, Halverson G, Guo SW. Promoter hypermethylation of progesterone receptor isoform b (PR-b) in endometriosis. Epigenetics (2006) 1(2):106–11. doi: 10.4161/epi.1.2.2766
20. Xue Q, Lin Z, Cheng YH, Huang CC, Marsh E, Yin P, et al. Promoter methylation regulates estrogen receptor 2 in human endometrium and endometriosis. Biol Reprod (2007) 77(4):681–7. doi: 10.1095/biolreprod.107.061804
21. Xue Q, Lin Z, Yin P, Milad MP, Cheng YH, Confino E, et al. Transcriptional activation of steroidogenic factor-1 by hypomethylation of the 5' CpG island in endometriosis. J Clin Endocrinol Metab (2007) 92(8):3261–7. doi: 10.1210/jc.2007-0494
22. van Kaam KJ, Delvoux B, Romano A, D'Hooghe T, Dunselman GA, Groothuis PG. Deoxyribonucleic acid methyltransferases and methyl-CpG-binding domain proteins in human endometrium and endometriosis. Fertil Steril (2011) 95(4):1421–7. doi: 10.1016/j.fertnstert.2011.01.031
23. Matsuzaki S, Canis M, Pouly JL, Wattiez A, Okamura K, Mage G. Cyclooxygenase-2 expression in deep endometriosis and matched eutopic endometrium. Fertil Steril (2004) 82(5):1309–15. doi: 10.1016/j.fertnstert.2004.03.059
24. Dyson MT, Roqueiro D, Monsivais D, Ercan CM, Pavone ME, Brooks DC, et al. Genome-wide DNA methylation analysis predicts an epigenetic switch for GATA factor expression in endometriosis. PloS Genet (2014) 10(3):e1004158. doi: 10.1371/journal.pgen.1004158
25. Ren F, Wang DB, Li T, Chen YH, Li Y. Identification of differentially methylated genes in the malignant transformation of ovarian endometriosis. J Ovarian Res (2014) 7:73. doi: 10.1186/1757-2215-7-73
26. Senthong A, Kitkumthorn N, Rattanatanyong P, Khemapech N, Triratanachart S, Mutirangura A. Differences in LINE-1 methylation between endometriotic ovarian cyst and endometriosis-associated ovarian cancer. Int J Gynecol Cancer (2014) 24(1):36–42. doi: 10.1097/IGC.0000000000000021
27. Penna I, Du H, Ferriani R, Taylor HS. Calpain5 expression is decreased in endometriosis and regulated by HOXA10 in human endometrial cells. Mol Hum Reprod (2008) 14(10):613–8. doi: 10.1093/molehr/gan055
28. Salvesen HB, MacDonald N, Ryan A, Jacobs IJ, Lynch ED, Akslen LA, et al. PTEN methylation is associated with advanced stage and microsatellite instability in endometrial carcinoma. Int J Cancer (2001) 91(1):22–6. doi: 10.1002/1097-0215(20010101)91:1<22::aid-ijc1002>3.0.co;2-s
29. Tam KF, Liu VW, Liu SS, Tsang PC, Cheung AN, Yip AM, et al. Methylation profile in benign, borderline and malignant ovarian tumors. J Cancer Res Clin Oncol (2007) 133(5):331–41. doi: 10.1007/s00432-006-0178-5
30. Heidarpour M, Derakhshan M, Derakhshan-Horeh M, Kheirollahi M, Dashti S. Prevalence of high-risk human papillomavirus infection in women with ovarian endometriosis. J Obstet Gynaecol Res (2017) 43(1):135–9. doi: 10.1111/jog.13188
31. Oppelt P, Renner SP, Strick R, Valletta D, Mehlhorn G, Fasching PA, et al. Correlation of high-risk human papilloma viruses but not of herpes viruses or chlamydia trachomatis with endometriosis lesions. Fertil Steril (2010) 93(6):1778–86. doi: 10.1016/j.fertnstert.2008.12.061
32. Esfandyari S, Aleyasin A, Noroozi Z, Taheri M, Khodarahmian M, Eslami M, et al. The protective effect of sulforaphane against oxidative stress through activation of NRF2/ARE pathway in human granulosa cells. Cell J (2021) 23(6):692–700. doi: 10.22074/cellj.2021.7393
33. Zhang H, Wu J, Li Y, Jin G, Tian Y, Kang S. Identification of key differentially Methylated/Expressed genes and pathways for ovarian endometriosis by bioinformatics analysis. Reprod Sci (2022) 29(5):1630–43. doi: 10.1007/s43032-021-00751-8
34. Coudyzer P, Lemoine P, Po C, Jordan BF, van der Smissen P, Courtoy PJ, et al. Induction of post-menstrual regeneration by ovarian steroid withdrawal in the functionalis of xenografted human endometrium. Hum Reprod (2015) 30(5):1156–68. doi: 10.1093/humrep/dev043
35. Tang FH, Chang WA, Tsai EM, Tsai MJ, Kuo PL. Investigating novel genes potentially involved in endometrial adenocarcinoma using next-generation sequencing and bioinformatic approaches. Int J Med Sci (2019) 16(10):1338–48. doi: 10.7150/ijms.38219
36. Sinreih M, Anko M, Kene NH, Kocbek V, Rizner TL. Expression of AKR1B1, AKR1C3 and other genes of prostaglandin F2alpha biosynthesis and action in ovarian endometriosis tissue and in model cell lines. Chemico-biological Interact (2015) 234:320–31. doi: 10.1016/j.cbi.2014.11.009
37. Cobellis L, Razzi S, De Simone S, Sartini A, Fava A, Danero S, et al. The treatment with a COX-2 specific inhibitor is effective in the management of pain related to endometriosis. Eur J Obstet Gynecol Reprod Biol (2004) 116(1):100–2. doi: 10.1016/j.ejogrb.2004.02.007
38. Li B, Liu L, Fu X, Zhou WQ, Zou DT, Zhao XY, et al. A novel mutation of estrogen receptor gene detected in girls with precocious puberty. Yi Chuan Xue Bao (2005) 32(10):1011–7.
39. Lamp M, Peters M, Reinmaa E, Haller-Kikkatalo K, Kaart T, Kadastik U, et al. Polymorphisms in ESR1, ESR2 and HSD17B1 genes are associated with fertility status in endometriosis. Gynecol Endocrinol (2011) 27(6):425–33. doi: 10.3109/09513590.2010.495434
40. Lebeau A, Grob T, Holst F, Seyedi-Fazlollahi N, Moch H, Terracciano L, et al. Oestrogen receptor gene (ESR1) amplification is frequent in endometrial carcinoma and its precursor lesions. J Pathol (2008) 216(2):151–7. doi: 10.1002/path.2405
Keywords: DNA methylation, gene expression, epigenetics, endometriosis, multi-omic analysis
Citation: Lei L, Xu X, Gong C, Lin B and Li F (2023) Integrated analysis of genome-wide gene expression and DNA methylation profiles reveals candidate genes in ovary endometriosis. Front. Endocrinol. 14:1093683. doi: 10.3389/fendo.2023.1093683
Received: 09 November 2022; Accepted: 10 March 2023;
Published: 23 March 2023.
Edited by:
Mainak Dutta, Birla Institute of Technology and Science, United Arab EmiratesReviewed by:
Takashi Kajitani, Sakura no Seibo Junior College, JapanMartin Stimpfel, University Medical Centre Ljubljana, Slovenia
Copyright © 2023 Lei, Xu, Gong, Lin and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Li, ZmFuZ19saUB0b25namkuZWR1LmNu; Bowen Lin, Ym93ZW5saW4wNTEyQDE2My5jb20=
 Lei Lei1
Lei Lei1 Xinxin Xu
Xinxin Xu Bowen Lin
Bowen Lin Fang Li
Fang Li