- 1Department of Physiology, Ajou University School of Medicine, Suwon, South Korea
- 2Department of Molecular Science and Technology, Ajou University, Suwon, South Korea
- 3College of Pharmacy, Sunchon National University, Suncheon, South Korea
Particulate matter (PM) in polluted air can be exposed to the human body through inhalation, ingestion, and skin contact, accumulating in various organs throughout the body. Organ accumulation of PM is a growing health concern, particularly in the cardiovascular system. PM emissions are formed in the air by solid particles, liquid droplets, and fuel – particularly diesel – combustion. PM2.5 (size < 2.5 μm particle) is a major risk factor for approximately 200,000 premature deaths annually caused by air pollution. This study assessed the deleterious effects of diesel-derived PM2.5 exposure in HL-1 mouse cardiomyocyte cell lines. The PM2.5-induced biological changes, including ultrastructure, intracellular reactive oxygen species (ROS) generation, viability, and intracellular ATP levels, were analyzed. Moreover, we analyzed changes in transcriptomics using RNA sequencing and metabolomics using gas chromatography-tandem mass spectrometry (GC-MS/MS) and liquid chromatography-tandem mass spectrometry (LC-MS/MS) in PM2.5-treated HL-1 cells. Ultrastructural analysis using transmission electron microscopy revealed disruption of mitochondrial cristae structures in a PM2.5 dose-dependent manner. The elevation of ROS levels and reduction in cell viability and ATP levels were similarly observed in a PM2.5 dose-dependently. In addition, 6,005 genes were differentially expressed (fold change cut-off ± 4) from a total of 45,777 identified genes, and 20 amino acids (AAs) were differentially expressed (fold change cut-off ± 1.2) from a total of 28 identified AAs profiles. Using bioinformatic analysis with ingenuity pathway analysis (IPA) software, we found that the changes in the transcriptome and metabolome are highly related to changes in biological functions, including homeostasis of Ca2+, depolarization of mitochondria, the function of mitochondria, synthesis of ATP, and cardiomyopathy. Moreover, an integrated single omics network was constructed by combining the transcriptome and the metabolome. In silico prediction analysis with IPA predicted that upregulation of mitochondria depolarization, ROS generation, cardiomyopathy, suppression of Ca2+ homeostasis, mitochondrial function, and ATP synthesis occurred in PM2.5-treated HL-1 cells. In particular, the cardiac movement of HL-1 was significantly reduced after PM2.5 treatment. In conclusion, our results assessed the harmful effects of PM2.5 on mitochondrial function and analyzed the biological changes related to cardiac movement, which is potentially associated with cardiovascular diseases.
Introduction
Air pollution has become a major global health problem. Through inhalation, ingestion, and skin contact, it has led to the accumulation of particulate matter (PM) in various organs throughout the body (1). Emissions of particulate matter from fossil fuels – particularly diesel – are a major source of air pollution (2). The toxicological evaluation of fine (size < 2.5 μm) particulate matter (PM2.5) has been conducted in the epidermal (2), respiratory (3), immune (4), nervous (5), and cardiovascular systems (6). The cardiovascular system in particular is affected by PM2.5. As such, PM2.5 is a crucial risk factor for approximately 200,000 premature deaths annually owing to its effect on vascular dysfunction, atherosclerosis, autonomic dysfunction, and hypertension (7). Oxidative stress has been reported to be one of the major factors in PM2.5-induced toxicity (7, 8); however, the underlying mechanisms in the cardiovascular system are still not fully understood.
Mitochondria are the major target of oxidative stress. They are highly developed in cardiac muscle cells compared to other cell types (9, 10), occupying over 40% of the spatial portion of the cells. They generate over 90% of ATP in the cardiac muscle cells through aerobic respiration (9). Calcium homeostasis and regulation are particularly important for the proper functioning of the mitochondria (11, 12) and are highly related to the cardiac contraction machinery and mitochondrial polarization (11, 13). As such, mitochondria, as well as calcium homeostasis, are the major targets for PM2.5-induced toxicity; however, the underlying biological relationship between them remains poorly understood.
Comprehensive analyses using omics technology have been applied for the toxicological assessment of nanomaterials, polystyrene particles, and urban PM (14–21). Fundamentally, the toxicities of these particles, including PM2.5, occur non-specifically and disturb various biological homeostasis simultaneously.
Several limitations in conventional toxicological evaluations have been previously documented (20, 22, 23). Integrated analysis of multi-layer molecules such as genes, proteins, and metabolites has been suggested to overcome these limitations and better understand the complex biological changes. This method is called systems toxicology (24). Moreover, integrated omics analysis can synergize the strengths of single omics analysis and overcome their weaknesses (20–22). In this study, we evaluated the toxicological mechanism of PM2.5-induced toxicity in the mitochondria of cardiomyocytes using transcriptomics, metabolomics, and integrated omics analysis.
Materials and methods
PM2.5 preparation and treatment
Diesel particulate matter (NIST® SRM® 1650b, Sigma-Aldrich, St. Louis, MO, USA) was diluted in dimethyl sulfoxide (DMSO) (10 mg/mL) to form a stock solution and was stored at -20°C. Before use, the stock solution was sonicated for 30 min to prevent particle aggregation (2). The stock solution was then diluted in a Claycomb medium (Sigma-Aldrich) for treatment. Cells were each treated with different concentrations of PM2.5 for 12 h. After treatment, each well was washed twice with PBS to reduce the effect of the remaining particles.
Cell culture
Culture flasks and plates were coated with a 0.02% gelatin (Sigma-Aldrich) and 5 μg/mL fibronectin (Sigma-Aldrich) extracellular matrix (ECM) solution before seeding the cells. HL-1, a murine cardiomyocyte cell line (Merck, Darmstadt, Germany), was cultured in a Claycomb medium supplemented with 10% fetal bovine serum (FBS; Corning, NY, USA), 0.1 mM norepinephrine (Sigma-Aldrich), 2 mM L-glutamine (Sigma-Aldrich), and 100 U/mL penicillin-streptomycin (Thermo Fisher Scientific, Waltham, MA, USA) at 37°C and 5% CO2 in a humidified atmosphere. The culture medium was replaced daily.
Cell viability
Cell viability was measured using a 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) Cell Proliferation Kit I (Roche, Basel, Switzerland). HL-1 cells were treated with 0, 10, 20, 50, 100, or 200 μg/mL PM2.5 for 12 h, followed by treatment with an MTT reagent for 4 h, and then the MTT formazan was dissolved using the solubilization solution provided in the kit and left overnight. Absorbance was measured at 590 nm using a microplate spectrophotometer (Epoch 2; BioTek, Winooski, VT, USA). The influence of DMSO on HL-1 cells was not detected in 0, 0.1, 0.5, 1.0, and 2.0% DMSO, respectively (data not shown).
Measurement of intracellular ATP level
Relative levels of intracellular ATP were measured using the CellTiter-Glo® luminescent cell viability assay as in previous study (Promega, Madison, WI, USA) (25). The 0, 10, and 100 μg/mL PM2.5-treated HL-1 cells were incubated at room temperature (RT) for 30 min, and assay reagent was added equally to the volume of the culture medium. The contents were mixed for 2 min and incubated for 10 min at RT. Luminescence signals were detected using a multi-mode microplate reader (SpectraMax® iD3; Molecular Devices, San Jose, CA, USA).
Measurement of ROS level
Intracellular ROS levels were measured using the ROS-Glo™ H2O2 assay (Promega). After PM2.5 treatment (0, 10, and 100 μg/mL) for 6 h, H2O2 substrate was added and incubated for another 6 h at 37°C. The detection solution was added to each well and incubated for 20 min at RT. Luminescence signals were detected using a multi-mode microplate reader (SpectraMax® iD3).
RNA sequencing and data processing
HL-1 cells were treated with 10 and 100 µg/mL PM2.5 for 12 h and then lysed. Total RNA was isolated using QIAzol® (Qiagen, Valencia, CA, USA). Total RNA concentration was quantified using Quant-IT RiboGreen (Invitrogen, Waltham, MA, USA). The integrity of total RNA was determined using TapeStation RNA screentape (Agilent Technologies, Atlanta, GA, USA), and samples with RNA integrity numbers higher than 7 were used for library generation. A library was generated with 1 µg of total RNA from each sample using the Illumina TruSeq Stranded mRNA Sample Prep Kit (Illumina Inc., San Diego, CA, USA). First-strand cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen). Second-strand cDNA was synthesized using DNA polymerase I, RNase H, and dUTP. The cDNA samples were processed via end repair by adding a single “A” base and ligating the adapters. The library samples were analyzed using Illumina NovaSeq (Illumina Inc.).
Raw reads from the sequencer were preprocessed to eliminate low-quality and adapter sequences before analysis, and the processed reads were aligned to the Mus musculus (mm10) genome using HISAT v2.1.0 (26). Transcript assembly and abundance estimations were performed using StringTie (27, 28). The aligned reads were assembled, and gene abundance was determined using StringTie v2.1.3b. Biological functions were investigated using bioinformatics ingenuity pathway analysis (IPA, http://www.ingenuity.com) software (Qiagen) (29). A 4-fold expression change for the transcriptome and a 1.2-fold expression change for AA levels were used as cut-off values for significant changes.
Gas chromatography-tandem mass spectrometry
Profiling analysis of AAs in HL-1 cells was performed using ethoxycarbonylation (EOC)/tert-butyldimethylsilyl (TBDMS) derivatives as previously described (30, 31). Briefly, deproteinization was performed by adding acetonitrile (100 μL) to lysed cardiomyocytes (1.0 × 106) containing norvaline as an internal standard (IS; 0.2 µg). After centrifugation, the supernatants were spiked into deionized water (1.0 mL) and added to dichloromethane (2.0 mL) containing ethyl chloroformate (ECF) adjusted to pH ≥ 12 using 5 M sodium hydroxide. A two-phase EOC reaction was performed by vortex mixing for 10 min. After the EOC reaction, the aqueous phase was adjusted to pH ≤ 2 using 10% H2SO4, saturated with sodium chloride, and sequentially extracted using diethyl ether (3.0 mL) and ethyl acetate (2.0 mL). The extracts were evaporated to dryness under a gentle stream of nitrogen (40°C). Before GC-MS/MS analysis, toluene (20 μL), N-Methyl-N-tert-butyldimethylsilyl trifluoroacetamide (MTBSTFA) (15 μL), and triethylamine (TEA) were added to the residue and then heated at 60°C for 60 min to generate a TBDMS derivative.
GC-MS/MS analysis was performed using a Shimadzu TQ 8040 triple quadruple mass spectrometer (Shimadzu, Kyoto, Japan) equipped with an Ultra-2 (5% phenyl-95% methylpolysiloxane bonded phase; 25 m × 0.20 mm I.D., 0.11 μm film thickness) cross-linked capillary column (Agilent Technologies). Samples (1.0 μL) were injected and conducted in a split-injection mode (10:1). GC oven temperature was set initially at 140°C for 3 min and increased to 300°C at a rate of 8°C/min with a holding time of 5 min. Helium (0.5 mL/min) and argon were used as carrier and collision gases, respectively. Ionization was performed in the electron impact ionization (EI) mode at 70 eV. SRM transitions and optimized mass parameters for each AA are summarized in Supplementary Table 1.
Liquid chromatography-tandem mass spectrometry
Profiling analysis of AAs and oxidized glutathione in HL-1 cells was performed without derivatization by LC-MS/MS. Briefly, deproteinization was performed by adding acetonitrile (ACN, 40 μL) and IS (13C1-phenylalanine; 25 ng) to lysed cardiomyocytes (5.0 × 104) in an Eppendorf tube, followed by mixing for 1 min. After centrifugation, the supernatant was transferred to an auto vial and injected into the LC-MS/MS system as described previously (20).
LC-MS/MS was performed using a Triple Quadrupole LCMS-8050 system (Shimadzu). An Intrada Amino Acid column (50 mm × 3.0 mm, 3 µm) was used to separate AAs. The system parameters used were as follows: ionization mode, electrospray ionization (ESI) mode; nebulizing gas flow, 3.0 L/min; heating gas flow, 10.0 L/min; interface temperature, 300°C; desolvation line (DL) temperature, 250°C. The mobile phase for AAs and oxidized glutathione was applied with a gradient elution of acetonitrile (ACN)/tetrahydrofuran (THF)/25 mM ammonium formate in water/formic acid = 9/75/16/0.3 (v/v/v/v) (A) and ACN/100 mM ammonium formate in water = 20:60 (v/v) (B). SRM transitions and optimized mass parameters for each AA are summarized in Supplementary Table 1.
Star pattern recognition analysis
AA levels were determined based on a calibration curve. The mean AA levels in the PM2.5 treated cells were normalized to the corresponding mean levels of the control. Normalized values were plotted as lines radiating from a common central point, and the far ends were joined to produce star patterns using MS Excel (32, 33).
Transmission electron microscope
Following PM2.5 treatment, cells were fixed using Karnovsky’s fixative solution (Sigma-Aldrich) for 12 h at 4°C. The fixed cells were washed with 0.1 M cacodylate buffer (pH 7.4) and treated with 1% osmium tetroxide (Polysciences, Warrington, PA, USA) in a 0.1 M cacodylate buffer for post-fixation (2 h, RT) as previously reported (20). Samples were dehydrated in a series of ethanol solutions (50 to 100%), infiltrated with propylene oxide, embedded in Epon Mixture (Polysciences), and sequentially incubated at 60°C (24 h). The sample blocks were sectioned using an ultramicrotome (Reichert-Jung, Bayreuth, Germany). For contrast staining, sections were stained with 2% uranyl acetate (Electron Microscopy Sciences, Hatfield, PA, USA) for 10 min and lead citrate (Thermo Fisher Scientific) for 5 min. Images were acquired using a transmission electron microscope (SIGMA500; Carl Zeiss, Oberkochen, Germany).
Mitochondrial membrane potential assay
To assess mitochondrial membrane potential, 0, 10, and 100 μg/mL PM2.5-treated cells were incubated with 100 nM tetramethylrhodamine ethyl ester perchlorate (TMRE; Sigma-Aldrich) for 20 min at 37°C. After rinsing each well with 0.2% BSA in PBS, the red fluorescence intensity of live cells was measured at an excitation wavelength/emission wavelength of 549/575 nm using a multi-mode microplate reader (SpectraMax® iD3), and fluorescence images were acquired using a fluorescence microscope (Axiovert 200M; Carl Zeiss) at the 3D immune system imaging core facility of Ajou University.
Calcium fluctuation detection
To analyze changes in calcium fluctuations in PM2.5-treated HL-1 cells, following PM2.5-treatment (0, 10, and 100 μg/mL) for 12 h, cells were incubated with 5 μM Fluo-4 AM (green-fluorescent calcium indicator; Invitrogen) in Hepes Buffered Tyrode’s Solution for 1 h at 37°C. After rinsing each well with Tyrode’s solution, calcium fluctuation signals were detected using a fluorescence microscope (Axiovert 200M). The calcium fluctuation videos were randomly obtained six times for each condition.
RNA extraction and quantitative real-time PCR
Total RNA was isolated from cells treated with PM2.5 (0, 10, and 100 μg/mL) for 12 h using Direct-zol RNA Miniprep (Zymo Research, Irvine, CA, USA), and a cDNA library from isolated RNA was synthesized using the iScript Advanced cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) and a Thermocycler (T3000; Biometra, Jena, Germany). The transcriptomic network-related gene expression levels were measured by qRT-PCR using the SsoAdvanced™ Universal SYBR® Green Supermix real-time PCR kit (Bio-Rad) using a Rotor Gene-Q system (Qiagen). Relative quantification of gene expression was calculated using the 2-ΔΔCt method. The primer information is summarized in Supplementary Table 2.
Statistical analysis
One-way analysis of variance (ANOVA) and Bonferroni’s post-hoc test were used for data analysis using the IBM SPSS Statistics 20 software (IBM Corporation, Armonk, NY, USA). *p-value < 0.05, vs. control; #p-value < 0.05, vs. 10 μg/mL PM2.5-treated cells were considered statistically significant.
Results
Viability and intracellular ATP and ROS levels in PM2.5-treated HL-1 cells
To assess the cytotoxicity of PM2.5, we analyzed the viability of 0–200 μg/mL PM2.5-treated HL-1 cells for 12 h. Viability was retained at approximately 90% in cells treated with up to 100 μg/mL of PM2.5; however, it was decreased to ~60% in those treated with 200 μg/mL of PM2.5 (Figure 1A). Therefore, we set a low dose of 10 μg/mL and a high dose of 100 μg/mL for further experiments. Intracellular ATP concentration and ROS levels showed a significant change in cells treated with 100 μg/mL PM2.5 compared to untreated and 10 μg/mL PM2.5-treated cells (Figures 1B, C). In cells treated with 100 μg/mL PM2.5, the ATP concentration decreased by approximately 20% (Figure 1B), and ROS accumulated over three times more than in controls (Figure 1C). These results suggest that a high dose of PM2.5, even though it might not induce cell death, may affect biological functions such as energy and redox homeostasis.
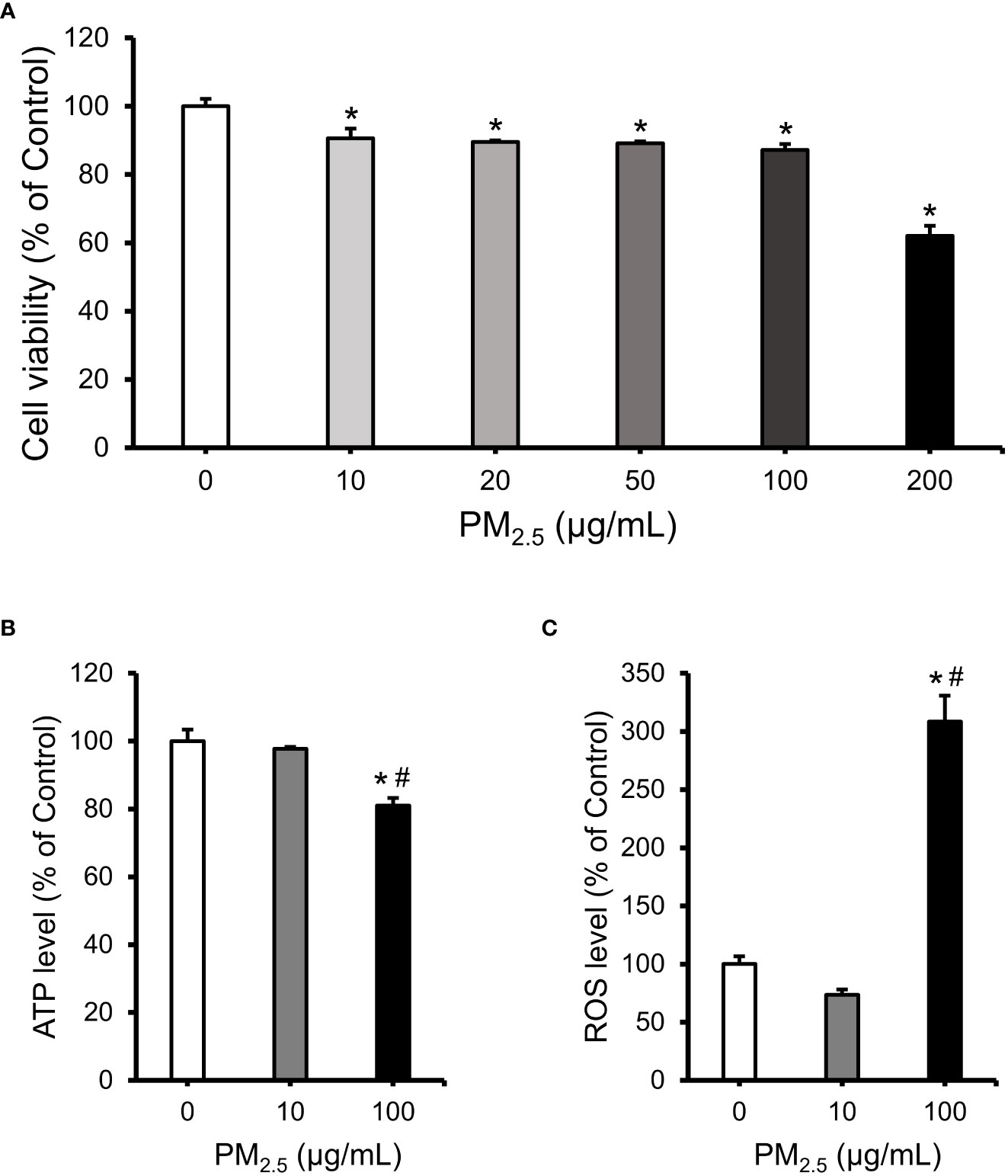
Figure 1 Evaluation of cell viability, intracellular ATP concentration, and ROS level in PM2.5-treated HL-1 cells. (A) Cell viability of HL-1 cells treated with PM2.5 (0, 10, 20, 50, 100, and 200 μg/mL) for 12 h. (B) The relative intracellular ATP levels in PM2.5-treated cells. (C) The relative intracellular ROS level in PM2.5-treated cells. Each data is shown from independently performed triplicate experiments. *p-value < 0.05 vs. control; #p-value < 0.05 vs. 10 μg/mL PM2.5-treated cells.
Transcriptomic analysis of PM2.5-treated cardiomyocytes
To investigate detailed biological changes, we analyzed the transcriptome of HL-1 cells using RNA sequencing after treatment with 0, 10, and 100 μg/mL PM2.5 for 12 h. We identified a total of 45,777 genes and found 6,005 genes to be differentially expressed (fold change cut-off of ± 4). We then analyzed the differentially expressed genes related to canonical pathways, diseases, and biological functions (Supplementary Tables 3 and 4), as well as the most significantly changed gene cluster among them (Figure 2A). When comparing the 10 μg/mL PM2.5-treated HL-1 cells to the control cells, two differentially expressed genes were found. We also identified a total of 37 differentially expressed genes in the gene cluster of the 100 μg/mL PM2.5-treated HL-1 cells. Next, we analyzed the transcriptomic network of the 37 genes using IPA in the 10 and 100 μg/mL PM2.5-treated HL-1 cells. Our analysis showed that these genes were related to six biological functions: ROS generation, homeostasis of Ca2+, depolarization of mitochondria, function of mitochondria, synthesis of ATP, and cardiomyopathy (Supplementary Table 5, Supplementary Figures 1 and 2). The transcriptomic network prediction revealed upregulation of ROS generation, depolarization of mitochondria, and cardiomyopathy with a downregulation in the homeostasis of Ca2+, function of mitochondria, and synthesis of ATP (Figure 2B). Among the genes in the network, the expression of Bcl2-interacting killer (Bik), histidine triad nucleotide binding protein 2 (Hint2), mitochondrial elongation factor 2 (Mief2), phospholipase C-like 2 (Plcl2), presenilin 2 (Psen2), and prostaglandin E receptor 4 (Ptger4) was validated by qPCR, and the expression trend was similar to the IPA network (Figure 2C).
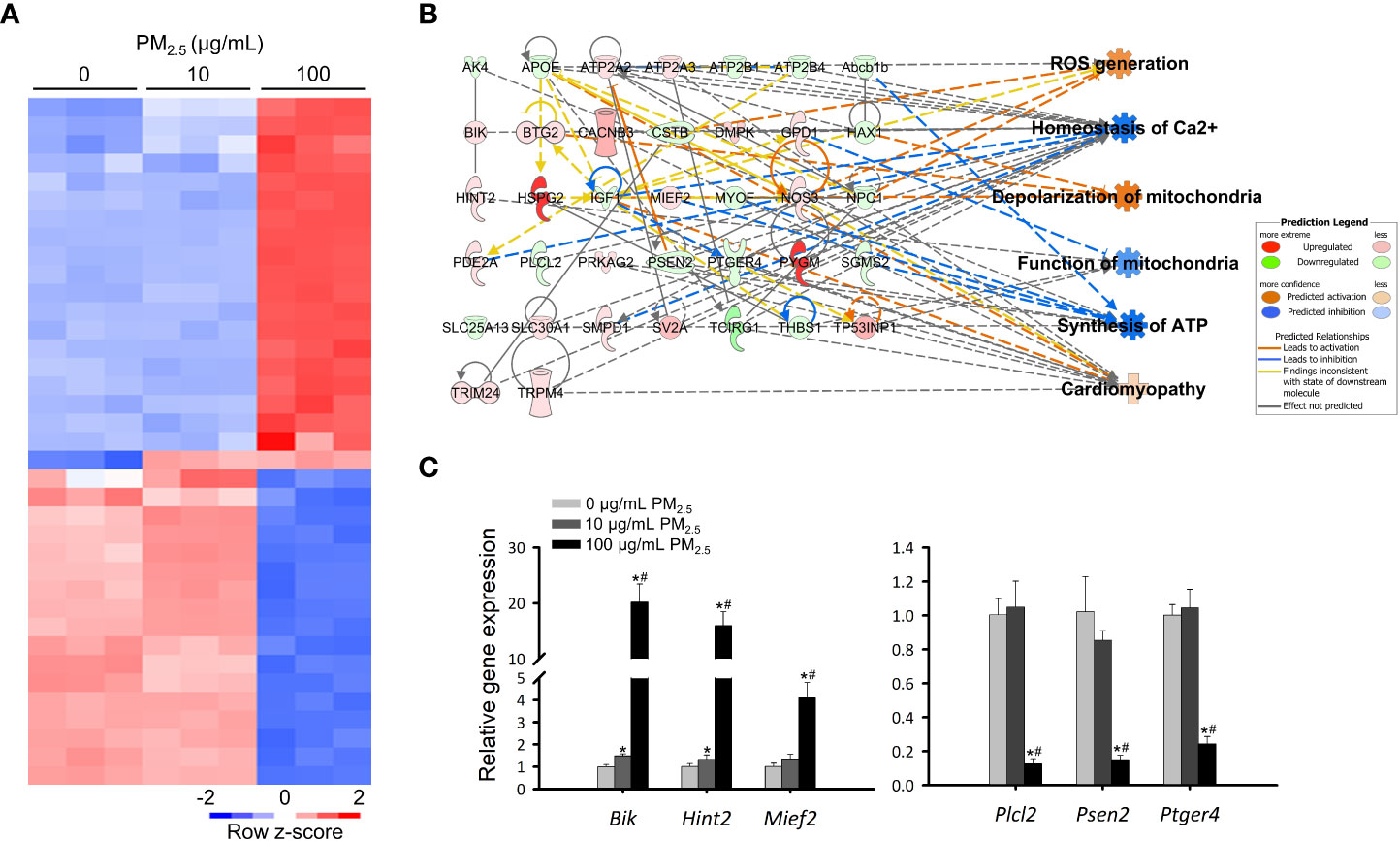
Figure 2 Transcriptomics analysis of PM2.5-treated HL-1 cells. (A) Heatmap of differentially expressed genes in 0, 10, and 100 μg/mL PM2.5-treated cells. Gene expression levels were analyzed three times per condition. (B) Transcriptome network with an in silico prediction of PM2.5-treated cells. A network of cellular functions and disease-related genes was generated using IPA (https://www.ingenuity.com). Green and red indicate downregulated and upregulated genes, respectively. Dotted and solid lines indicate indirect and direct relationships, respectively. Details for shape are provided in Supplementary Figure 1. (C) qPCR analysis for Bik, Hint2, Mief2, Plcl2, Psen2, and Ptger4 in 0, 10, and 100 μg/mL PM2.5-treated cells. Gapdh was used as a reference gene. Data represent independently performed triplicate experiments. *p-value < 0.05 vs. control; #p-value < 0.05 vs. 10 μg/mL PM2.5-treated cells.
Metabolomic analysis of PM2.5-treated cardiomyocytes
Changes in AA profiles in 10 and 100 μg/mL PM2.5-treated HL-1 cells were quantified using EOC/TBDMS derivatization followed by GC-MS/MS and LC-MS/MS analyses. A total of 28 AAs were identified, of which 20 were differentially expressed (fold change cut-off of ± 1.2). In the 10 and 100 μg/mL PM2.5-treated HL-1 cells, 16 AAs were differentially expressed (Figure 3A). We analyzed the differentially expressed AAs related to canonical pathways, diseases, and biological functions (Supplementary Tables 6 and 7). A metabolomics network was also created using IPA, which showed the relationship between AAs and those associated with cellular functions found in the transcriptomics analysis (Supplementary Figures 3 and 4, Supplementary Table 8). Like the transcriptomics analysis, the network prediction also revealed upregulation of ROS generation, depolarization of mitochondria, and cardiomyopathy in the 10 and 100 μg/mL PM2.5 treated HL-1 cells (Figure 3B). Representative selected ion monitoring (SIM) chromatography results for the four AAs in the network, including aspartic acid, glutathione disulfide, glutamine, and methionine, are presented in Figure 3C.
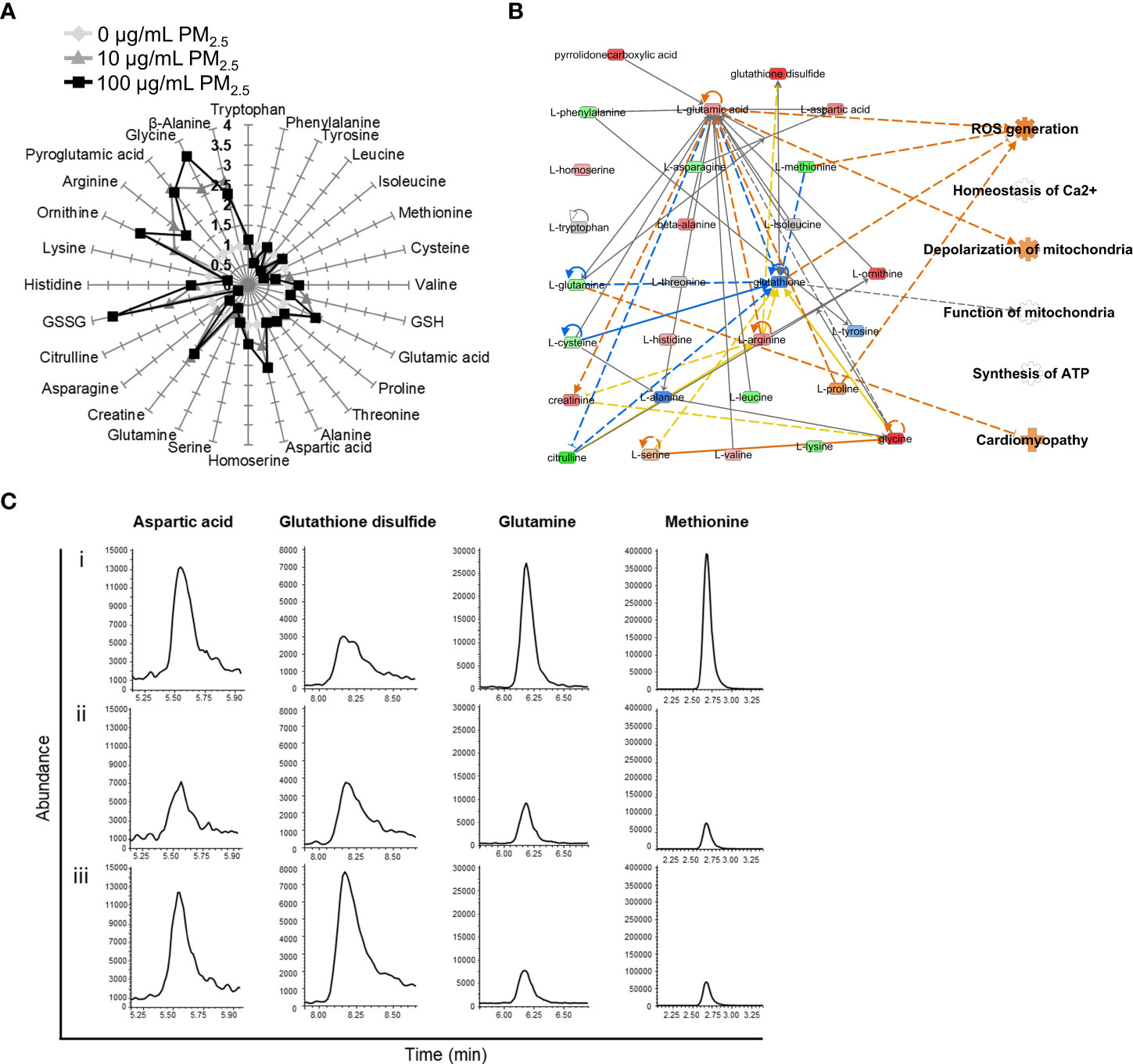
Figure 3 AA profiles of PM2.5-treated HL-1 cells. (A) The relative levels of 28 amino acids in 0 μg/mL PM2.5-treated (white), 10 μg/mL PM2.5-treated (gray), and 100 μg/mL PM2.5-treated cells (black). Each amino acid level was analyzed three times per condition. (B) The AA network with an in silico prediction of 100 μg/mL PM2.5-treated cells. The network was generated using IPA. Green and red indicate downregulated and upregulated amino acids, respectively. Dotted and solid lines indicate indirect and direct relationships, respectively. Details for shape and color are provided in Supplementary Figure 1 and Figure 2B. (C) SIM chromatograms of aspartic acid, glutathione disulfide, glutamine, and methionine in (i) 0 μg/mL PM2.5-treated, (ii) 10 μg/mL PM2.5-treated, and (iii) 100 μg/mL PM2.5-treated cells.
Metabotranscriptomic analysis of PM2.5-treated cardiomyocytes
Integrative omics analysis was used to compensate for the limitations of single omics analysis (34–37). The transcriptome and metabolome were integrated based on biological functions, including ROS generation, homeostasis of Ca2+, depolarization of mitochondria, function of mitochondria, synthesis of ATP, and cardiomyopathy. Genes and AAs profiles were found to be interconnected and tightly related to the biological functions in the 10 and 100 μg/mL PM2.5 treated HL-1 cells (Supplementary Figures 5 and 6). The integrated transcriptomics and metabolomics network, namely the metabotranscriptomic network, is predicted to increase ROS generation, depolarization of mitochondria, and cardiomyopathy, and decrease homeostasis of Ca2+, function of mitochondria, and synthesis of ATP. This trend was more pronounced than in the single omics networks (Figure 4).
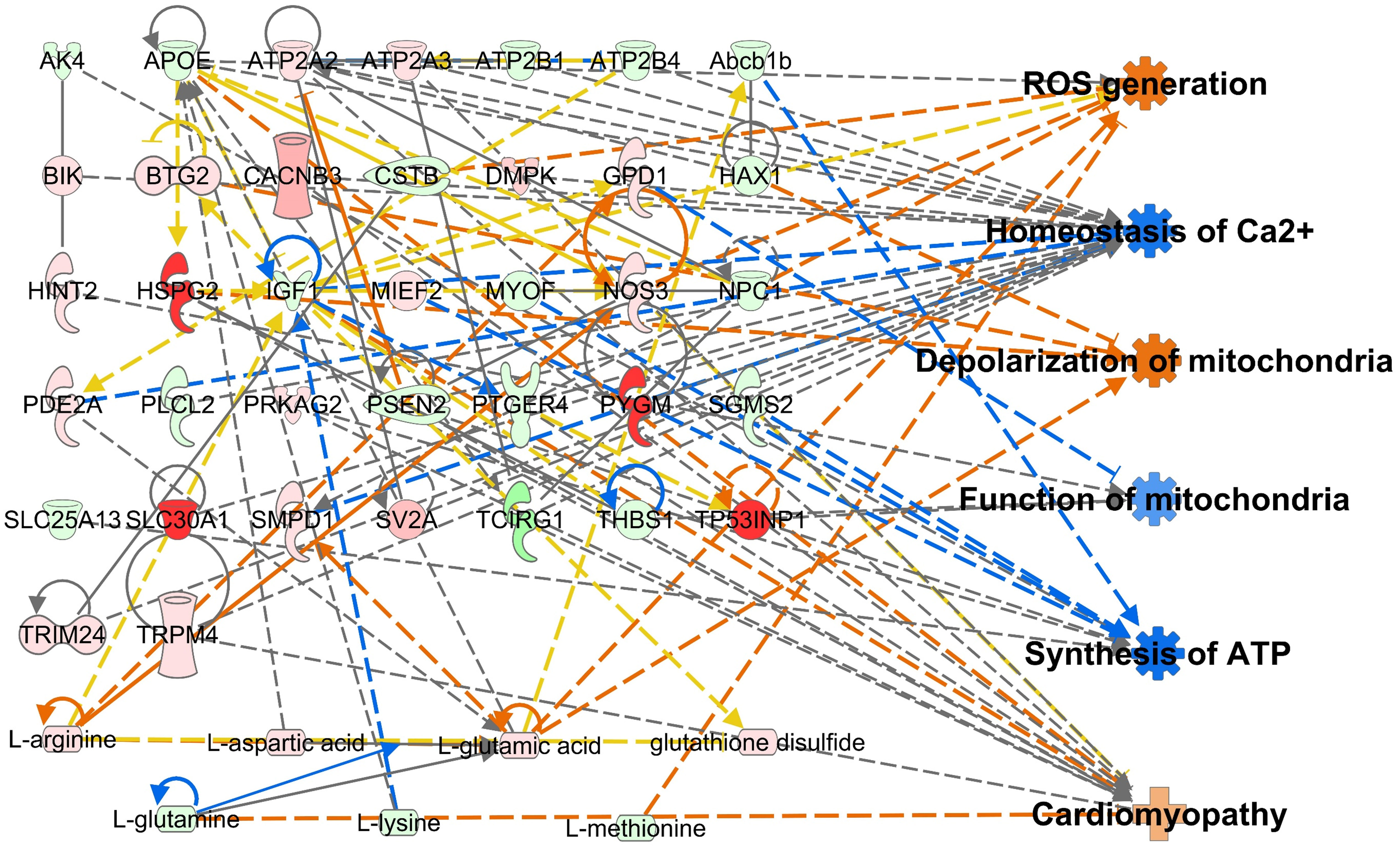
Figure 4 Metabotranscriptomic network with an in silico prediction of PM2.5-treated HL-1 cells. The network was generated using IPA. Green and red indicate downregulated and upregulated genes and amino acids, respectively. Dotted and solid lines indicate indirect and direct relationships, respectively. Details for shape and color are provided in Supplementary Figure 1 and Figure 2B.
Reduction in mitochondrial membrane potential in PM2.5-treated cardiomyocytes
To validate the predicted biological changes, we investigated the mitochondrial membrane potential, which is highly related to homeostasis of Ca2+, depolarization of mitochondria, and function of mitochondria (11) in PM2.5-treated HL-1 cells using the fluorescence of tetramethylrhodamine ethyl ester perchlorate (TMRE) (Figure 5A). The TMRE intensity decreased insignificantly in 10 μg/mL PM2.5-treated cells (~87% that of the control); however, it decreased significantly (~64% that of the control) in the cells treated with 100 μg/mL PM2.5 (Figure 5B). This result suggests that a high dose of PM2.5 could reduce the electrochemical proton gradient of mitochondria, which is a critical factor in ATP synthesis.
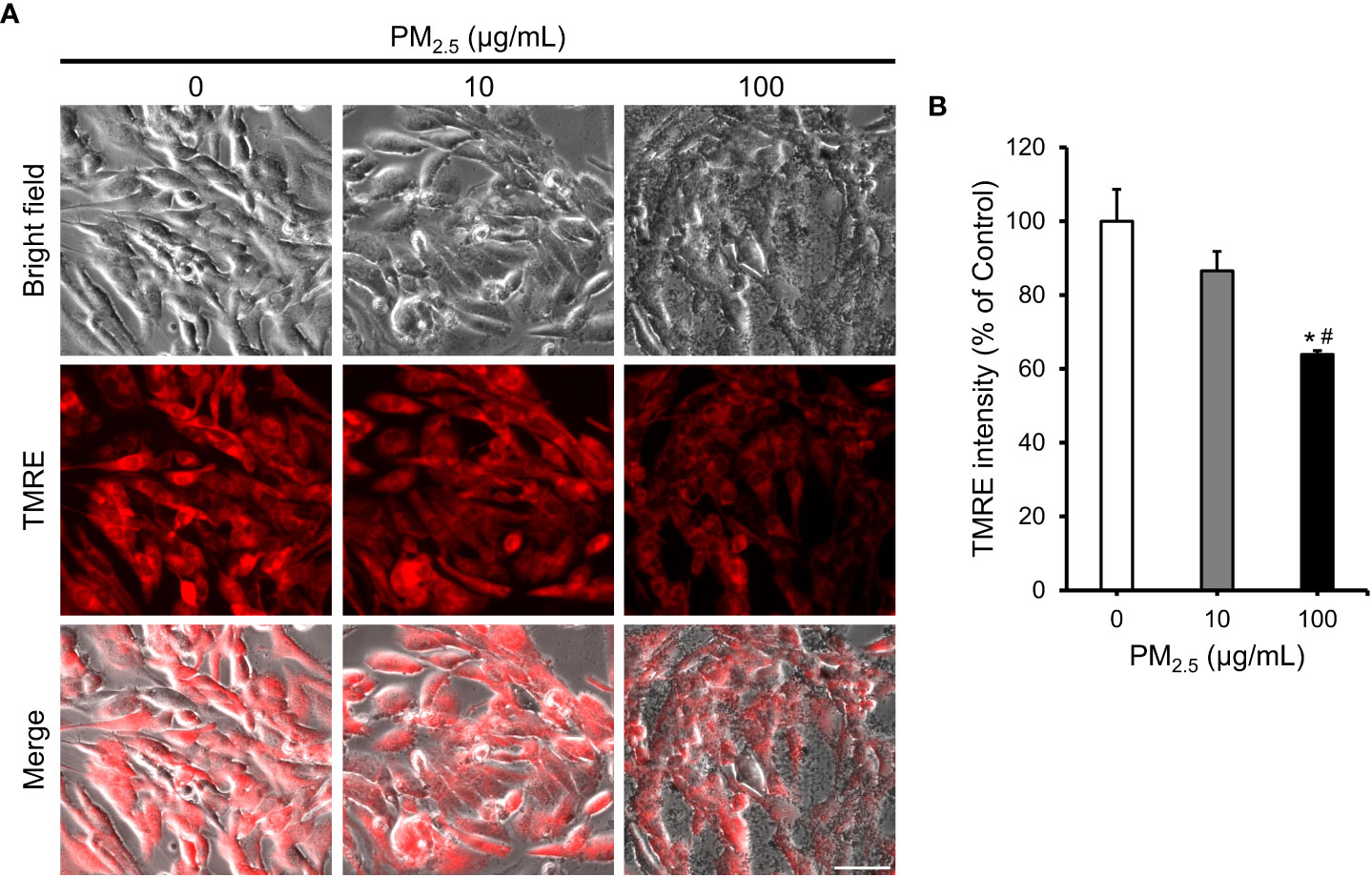
Figure 5 Reduction in mitochondrial membrane potential in PM2.5-treated HL-1 cells. (A) TMRE staining and bright-field images of PM2.5-treated cells. Scale bar = 50 μm. (B) Measurement of TMRE intensity in 0, 10, and 100 μg/mL PM2.5-treated cells. Data represent independently performed triplicate experiments. *p-value < 0.05 vs. control; #p-value < 0.05 vs. 10 μg/mL PM2.5-treated cells.
Impairment in movement of PM2.5-treated cardiomyocytes
The HL-1 cell line retained differentiated cardiac electrophysiological properties (38). Therefore, to assess the effect of PM2.5 on cardiac electrophysiological properties, we observed calcium fluctuation in PM2.5-treated HL-1 cells using a calcium indicator (Supplementary Movies 1, 2, and 3). Both 10 μg/mL and 100 μg/mL PM2.5-treated HL-1 cells showed a more rapid calcium fluctuation rate than untreated HL-1 cells (Control: 30.7 ± 1.5 signals/min, 10 μg/mL PM2.5: 38.0 ± 2.0 signals/min, 100 μg/mL PM2.5: 40.7 ± 6.0 signals/min) (Figure 6A). Interestingly, the calcium fluctuation area of 100 μg/mL PM2.5-treated HL-1 cells was significantly decreased, whereas that of 10 μg/mL PM2.5-treated HL-1 cells was unaffected (Figure 6B). Moreover, calcium fluctuation signals in some regions were rarely detected in 100 µg/mL PM2.5-treated HL-1 cells (only; ~8% of the total region). These results suggested that PM2.5 could affect the electrophysiological properties of a cardiomyocytes even in 10 μg/mL PM2.5-treated conditions, and especially 100 μg/mL of PM2.5 could impair the calcium fluctuation ability of cardiomyocytes required for heartbeat regulation.
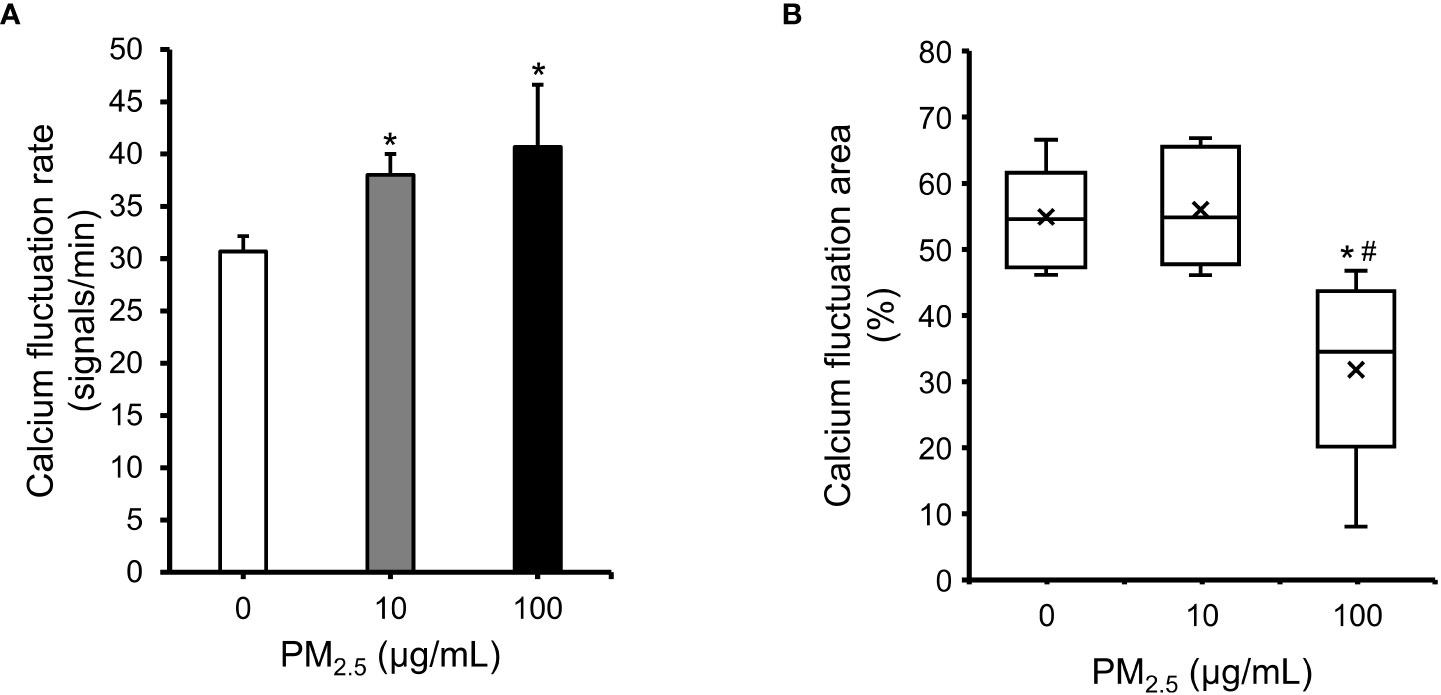
Figure 6 Impairment in cardiac calcium fluctuation in PM2.5-treated HL-1 cells. (A) Calcium fluctuation rates of 0, 10, and 100 μg/mL PM2.5-treated cells. The graph indicates the number of calcium fluctuation signals per min. (B) Calcium fluctuation area of PM2.5-treated cells. The box plot shows the cell area with calcium fluctuation signals from randomly selected microscope fields. Calcium fluctuation videos were acquired in six fields per condition. × indicates average. The internal horizontal line indicates the median value. *p-value < 0.05 vs. control; #p-value < 0.05 vs. 10 μg/mL PM2.5-treated cells.
TEM image analysis of PM2.5-treated cardiomyocytes
To observe the mitochondrial structure of PM2.5-treated HL-1 cells, we captured TEM images of cells treated with 0, 10, and 100 μg/mL PM2.5 (Figure 7). In the 100 μg/mL PM2.5-treated cell, the structure of mitochondrial cristae was disintegrated, whereas those of non-treated cells maintained the elaborate system. In addition, cells treated with 10 and 100 μg/mL PM2.5 had fewer mitochondria compared to non-treated control in a dose-dependent manner. These results suggested that PM2.5 has an adverse effect on the mitochondria of cardiomyocytes, in particular the structure of cristae which is critical for mitochondrial functions.
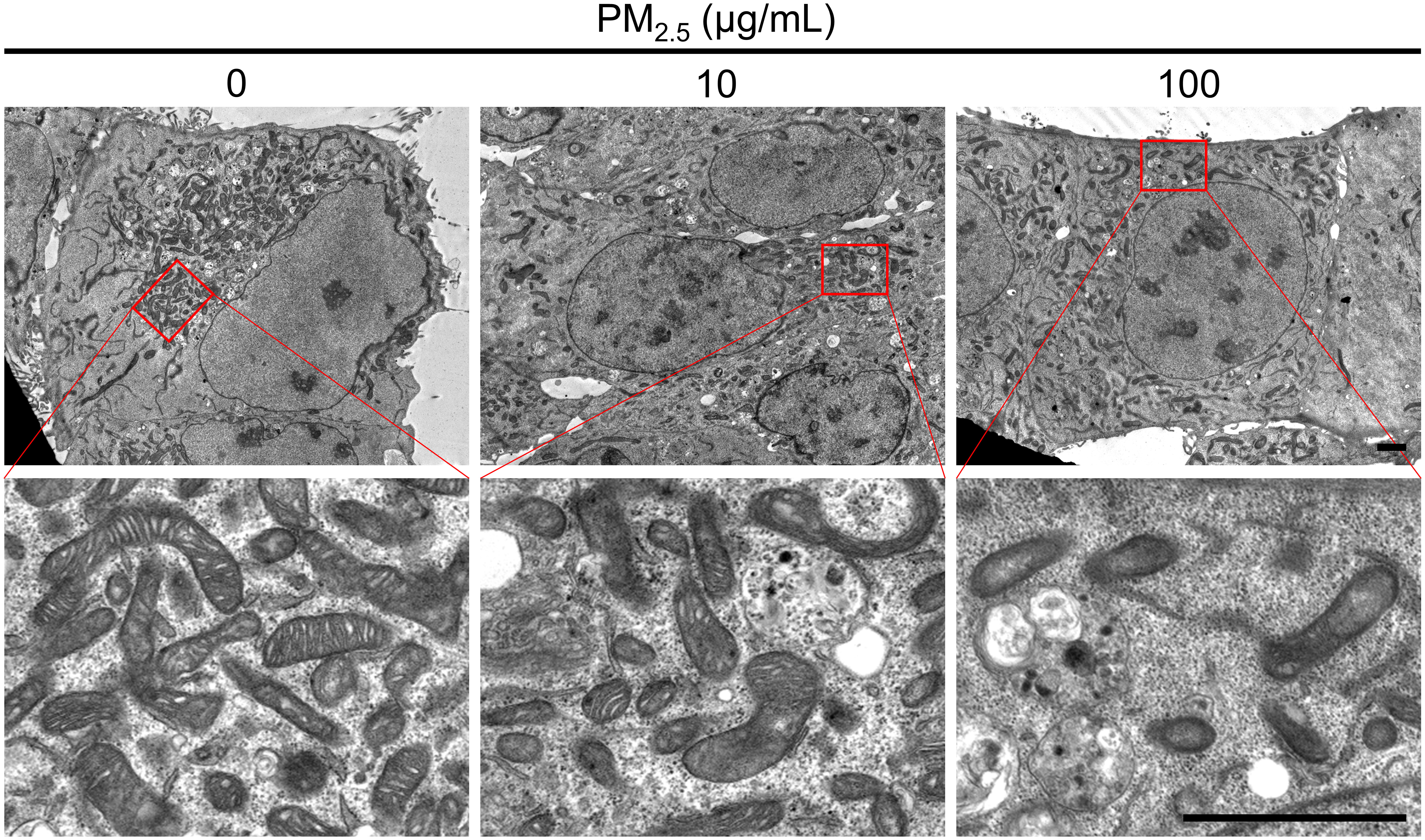
Figure 7 Ultrastructural analysis using TEM. TEM images represent the mitochondrial morphology of PM2.5-treated and non-treated HL-1 cells. Each representative image was acquired from three TEM images per condition. Magnified images are presented in each bottom panel. Scale bar = 2 μm.
Discussion
The present study analyzed the molecular and biological changes and alterations in the transcriptome and amino acid composition in PM2.5-treated cardiomyocytes. Our analysis mainly focused on mitochondrial function and calcium homeostasis. We also documented the disturbances caused by PM2.5 in cardiomyocytes, namely its effect on cardiac movement and its relation to cardiomyopathy. We carried out the PM2.5-treated in vitro experiments and found six biological functions affected by PM2.5: ROS generation, homeostasis of Ca2+, depolarization of mitochondria, function of mitochondria, synthesis of ATP, and cardiomyopathy. Furthermore, to compensate for any experimental weakness, we integrated transcriptome and metabolome and created an integrated single omics network as in previous reports (14, 15, 20). As a result, in silico prediction of metabotranscriptome for six biological functions highly corresponded with the results of in vitro experiments. Therefore, this approach provides reliable data that reveals PM2.5 inducing toxicity with deleterious effects on the cardiovascular system. The overall analysis for PM2.5-induced biological changes is summarized in Figure 8. To assess the effects of diesel-derived PM2.5 exposure in HL-1 mouse cardiomyocyte cell lines, we performed both experimental and bioinformatic analysis in PM2.5-treated HL-1 cells. The PM2.5-induced biological changes, including ROS accumulation, mitochondrial dysfunction, ATP depletion, and cardiomyopathy, were analyzed in in vitro cell line and via in silico prediction.
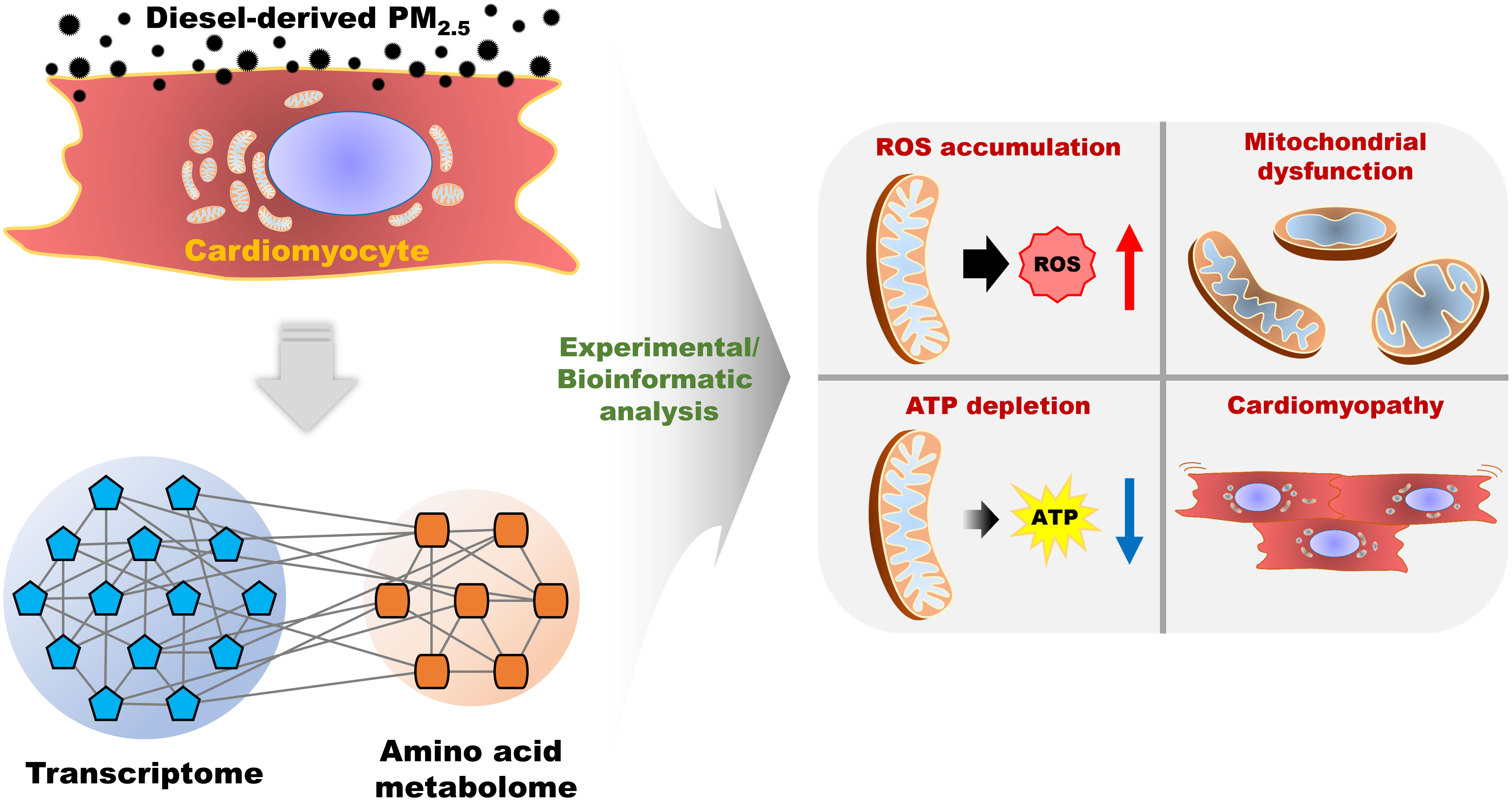
Figure 8 The summarized scheme of overall analysis for diesel-derived PM2.5 exposure in HL-1 mouse cardiomyocyte cell line.
The standard values for PM2.5 exposure set by WHO and the Chinese National Secondary Standard concentration of environmental air pollutants are 25 μg/m3 24 h mean and 75 μg/m3 24 h mean, respectively (39). To mimic the in vivo situation based on these criteria, Liu et al. reported that for the establishment of PM2.5 exposure mouse model (forty-eight male C57BL/6 mice, six weeks old), the 24 h mean concentration for eight weeks of exposure of control and treatment group were 23.71 and 225.05 μg/m3, and for 16 weeks of exposure was 21.75 and 168.00 μg/m3, respectively. For in vitro model, they treated 0, 25, 50, and 100 μg/mL of PM2.5 for 24 h in HL-1 cells, respectively. The biomarker expression pattern of in vitro and in vivo was found to be similar (39). Specifically, after PM2.5 exposure in vivo, the mRNA levels of an atrial natriuretic factor (ANF), brain natriuretic peptide (BNP), and β-myosin heavy chain (β-MHC) were statically upregulated by 1.28-, 0.99- and 1.16-fold, respectively. In the analysis of transcriptome after PM2.5 exposure in HL-1 cells, ANF, BNP, and β-MHC were significantly upregulated to 1.89-,1.65-, 1.86-fold after treatment of 50 μg/mL PM2.5 and increased to 2.28-, 1.99-, 2.16-fold after treatment of 100 μg/mL PM2.5. In addition, Sivakumar et al. reported that 100 μg/mL PM2.5-treated H9c2 cardiomyocytes cells showed mitochondrial dysfunction and inactivation of the PI3K/Akt signaling pathway (40). These results are consistent with our findings. Recently, this group also reported that myocardial changes in the animal model were significantly altered in 0.5 mg/mL concentration of PM2.5, compared to 0.05 mg/mL concentration (41). However, the pathological changes in test group with 0.05 mg/mL concentration were not statistically different from the control. Hence, they used PM2.5 at a concentration of 0.5 mg/mL, yielding 250 µg/m3 of PM2.5 in the exposure chamber. Based on previous reports and our preliminary data, treatment of 10 μg/mL and 100 μg/mL PM2.5 were established as test groups in in vitro experiments for extrapolating the findings to predict biological effects in in vivo. However, there are several limitations to analyze the clinical relevance of the concentration at which the toxic effects were observed in our study. It is difficult to assess the accurate amount of component accumulation of diesel-derived PM2.5 concentration in the heart due to the heterogeneous component of diesel-derived PM2.5 and the limitation of instrumental analysis. To validate this, there is a pressing need for large-scale in vitro experiments, clinical studies, and the development of high-sensitive instrumental analysis. In addition, further studies on the specific concentrations of PM2.5 linked with clinical relevance are warranted.
The toxicity of fine and nanoscale particles is highly dependent on the composition of the particles due to physicochemical differences of the materials and differences in toxicity of the composed chemicals (22, 42). Moreover, the composition of the PM2.5 varies according to PM2.5 sources, including region, natural circumstance, and anthropogenic factors (43). In this study, we evaluated the toxicity of diesel-derived PM2.5, which is certified as Standard Reference Material (SRM) by the National Institute of Standards & Technology (NIST), and the PM2.5 is composed with polycyclic aromatic hydrocarbons (PAHs) and nitro-substituted polycyclic aromatic hydrocarbons (Nitro-PAHs) (44, 45). PAHs and Nitro-PAHs are highly related to the toxicity of diesel-derived PM2.5 (46, 47). To analyze the adverse effects of PM2.5, it is necessary to evaluate the specific toxicity of both PAHs and Nitro-PAHs.
Zhou et al. reported that 100 or 200 μg/mL of the water-soluble fraction of PM2.5 or DMSO-soluble fraction of PM2.5 identically induced ROS and apoptosis in KGN cells, i.e., ovarian granulosa cell-like human granulosa cells (48). Regardless of the solvent, chemical components of PM2.5 were responsible for the biological effects in the cells. Even though the cell lines are different, we assume that chemical components of PM2.5 are responsible for biological effects in PM2.5-treated HL-1 cells.
The present study showed a decrease in intracellular ATP levels and an increase in intracellular ROS levels in PM2.5-treated cardiomyocytes, with no significant difference in cell viability. Moreover, disruption of the mitochondrial cristae structures in PM2.5-treated cardiomyocytes was detected (Figure 7). We reported similar findings in our previous study, where human embryonic kidney 293 (HEK293) cells were treated with 50 nm silica-coated magnetic nanoparticles containing rhodamine B isothiocyanate [MNPs@SiO2(RITC)], which have similar characteristics to ultrafine PM (14). We believe that the generation of ROS and treatment time (12 h) were insufficient to induce cell death in both studies (14). In addition, we noted an increase in glutamate levels detected in HEK293 cells treated with MNPs@SiO2(RITC) (14, 49) and in cardiomyocytes treated with PM2.5. Thus, we concluded that the biological responses were similar for both MNPs@SiO2(RITC) and PM2.5 cells and that ROS may be a major trigger for mitochondria-related biological changes in PM2.5-treated cardiomyocytes.
Reduced glutathione (GSH) is the most significant non-protein thiol in mammalian cells. It acts as a scavenger of ROS through a redox reaction that yields oxidized glutathione disulfide (GSSG) (50, 51). Therefore, the GSH/GSSG ratio can be used as an indicator of oxidative stress (51, 52). In 10 μg/mL PM2.5-treated HL-1 cells, the level of GSSG was increased ~2-fold compared to that in controls, and the GSH level was increased ~1.5-fold. In contrast, the level of GSSG in 100 μg/mL PM2.5-treated HL-1 cells was increased ~3.5-fold, while that of GSH remained constant. These altered levels of GSH and GSSG suggest that the ROS-scavenging system may be impaired in PM2.5-treated HL-1 cells.
PM2.5-induced ROS disturbs calcium regulation and thus can affect cardiac movement. Cardiac movement requires calcium cycling via the influx and efflux of calcium in the sarcoplasmic reticulum, cytosol, and mitochondria. Any impairment of this cycle is highly related to heart failure (53). In this study, we found disturbances in the expression of ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2/3 (Atp2a2 and Atp2a3), ATPase plasma membrane Ca2+ transporting 1/4 (Atp2b1 and Atp2b4), and voltage-gated calcium channel auxiliary subunit beta 3 (Cacnb3) in PM2.5-treated cardiomyocytes. Moreover, a decrease in intracellular ATP level is also highly related to the impairment of calcium cycling and, thus, cardiac movement (54). Imbalanced calcium homeostasis can also induce depolarization and mitochondrial dysfunction (55, 56). Thus, PM2.5-induced impairment of cardiac movement might be caused by disturbances in calcium regulation-related gene expression and a concomitant reduction in mitochondrial activity.
Besides the impairment of cardiac movement in 100 μg/mL PM2.5-treated HL-1 cells, our result showed the increment of calcium fluctuation rate in both 10 and 100 μg/mL PM2.5-treated cells compared to non-treated control (Figure 6A). This phenomenon supports previous epidemiological studies that PM2.5 exposure was highly related with elevated heart rate (57, 58). Since increased heart rate is considered the major cardiovascular risk factor (59), our result suggests that even relatively low concentrations of PM2.5 also have an adverse effect on cardiomyocytes.
Cell internalization of particles occurs through multiple pathways, including passive diffusion, phagocytosis, pinocytosis, micropinocytosis, receptor-mediated endocytosis, clathrin-mediated endocytosis, and caveolin-mediated endocytosis (60, 61). Previous studies have reported that the internalization of the fluorescence dye-labeled PM2.5 depends on clathrin- and caveolin-mediated endocytosis as well as other pathways according to PM2.5 component diversity (62). In contrast, PM emission from cells has not been well documented. Therefore, we analyzed exocytosis using integration omics in PM2.5-treated HL-1 cells (Supplementary Figures 7 and 8, Supplementary Table 9). We predicted that exocytosis was suppressed using the metabotranscriptomic analysis of the PM2.5-treated HL-1 cells (Supplementary Figure 9). This process is energy-dependent, and ATP levels dropped due to mitochondrial dysfunction in PM2.5-treated HL-1 cells. Thus, the suppressed exocytosis might correlate with the energy homeostasis of the HL-1; however, further studies using fluorescence dye labeling and super-resolution live-cell imaging are needed to elucidate the exact mechanism of exocytosis of PM2.5.
In addition to maintaining blood circulation, the heart also functions as an endocrine organ that produces hormones, such as atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP), growth differentiation factor (GDF)-15, and endothelin-1 (63–65). As such, abnormal hormonal regulation of the heart is closely related to diseases such as hypertension, heart failure, chronic renal failure, and septic shock (64, 65). We studied the synthesis and secretion of hormone-related differentially expressed genes, AAs, and their relationship via the metabotranscriptomic analysis of PM2.5-treated HL-1 cells (Supplementary Figures 10 and 11, Supplementary Table 10), and these functions were predicted to be suppressed in PM2.5-treated HL-1 cells (Supplementary Figure 12). However, our study could not accurately describe the mechanism by which PM2.5 affects these functions. Future studies are needed to further document the effect of PM2.5 on the hormonal homeostasis of cardiomyocytes.
In this study, we analyzed molecular biological phenomena by combining transcriptomics and metabolomics with AA profiling. Recent studies have been conducted using multi-omics integration analysis for a precise and detailed understanding of the biological changes under specific conditions using transcriptomics, miRNAs, proteomics, phosphoproteomics, and metabolomics (18, 20, 66). Moreover, data processing helps find strong relationships in data using machine learning algorithms for clustering and reducing the dimensionality of the data (20). Thus, future studies with additional omics analysis combined with machine learning algorithms will further improve our understanding of PM2.5-induced toxicity in cardiomyocytes. In conclusion, our results suggest that exposure to PM2.5 can induce deleterious effects on cardiac mitochondrial function and calcium homeostasis, leading to impaired cardiac movement. This further highlights the major role fossil fuel-induced air pollution plays in the pathogenesis of cardiovascular diseases.
Data availability statement
Data have been uploaded to the Gene Expression Omnibus https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE211949 under accession GSE211949.
Author contributions
TS, SK, MP, and GL conceived and designed experiments. MJ, DK, JH, DE, and CP collected information and analyzed the data. TS, SK, MJ, NG, MP and GL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Research Foundation (NRF) funded by the Ministry of Science and ICT (MSIT) in Korea (2020R1C1C1008366, 2020R1A4A4079722, and 2020M3E5D9080661).
Acknowledgments
The authors thank Bunsoon Choi and the 3D immune system imaging core facility of Ajou University for technical assistance and cell imaging analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.999475/full#supplementary-material
References
1. Calderón-Garcidueñas L, Stommel EW, Rajkumar RP, Mukherjee PS, Ayala A. Particulate air pollution and risk of neuropsychiatric outcomes. What we breathe, swallow, and put on our skin matters. Int J Environ Res Public Health (2021) 18(21):11568. doi: 10.3390/ijerph182111568
2. Piao MJ, Ahn MJ, Kang KA, Ryu YS, Hyun YJ, Shilnikova K, et al. Particulate matter 2.5 damages skin cells by inducing oxidative stress, subcellular organelle dysfunction, and apoptosis. Arch Toxicol (2018) 92(6):2077–91. doi: 10.1007/s00204-018-2197-9
3. Liu Q, Xu C, Ji G, Liu H, Shao W, Zhang C, et al. Effect of exposure to ambient Pm2.5 pollution on the risk of respiratory tract diseases: A meta-analysis of cohort studies. J BioMed Res (2017) 31(2):130–42. doi: 10.7555/JBR.31.20160071
4. Zhao J, Gao Z, Tian Z, Xie Y, Xin F, Jiang R, et al. The biological effects of individual-level Pm(2.5) exposure on systemic immunity and inflammatory response in traffic policemen. Occup Environ Med (2013) 70(6):426–31. doi: 10.1136/oemed-2012-100864
5. Wang Y, Xiong L, Tang M. Toxicity of inhaled particulate matter on the central nervous system: Neuroinflammation, neuropsychological effects and neurodegenerative disease. J Appl Toxicol (2017) 37(6):644–67. doi: 10.1002/jat.3451
6. Du Y, Xu X, Chu M, Guo Y, Wang J. Air particulate matter and cardiovascular disease: The epidemiological, biomedical and clinical evidence. J Thorac Dis (2016) 8(1):E8–E19. doi: 10.3978/j.issn.2072-1439.2015.11.37
7. Aryal A, Harmon AC, Dugas TR. Particulate matter air pollutants and cardiovascular disease: Strategies for intervention. Pharmacol Ther (2021) 223:107890. doi: 10.1016/j.pharmthera.2021.107890
8. Hamanaka RB, Mutlu GM. Particulate matter air pollution: Effects on the cardiovascular system. Front Endocrinol (Lausanne) (2018) 9:680. doi: 10.3389/fendo.2018.00680
9. Hussain A, Ghosh S, Kalkhoran SB, Hausenloy DJ, Hanssen E, Rajagopal V. An automated workflow for segmenting single adult cardiac cells from Large-volume serial block-face scanning electron microscopy data. J Struct Biol (2018) 202(3):275–85. doi: 10.1016/j.jsb.2018.02.005
10. Li A, Gao M, Liu B, Qin Y, Chen L, Liu H, et al. Mitochondrial autophagy: Molecular mechanisms and implications for cardiovascular disease. Cell Death Dis (2022) 13(5):444. doi: 10.1038/s41419-022-04906-6
11. Giorgi C, Marchi S, Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol (2018) 19(11):713–30. doi: 10.1038/s41580-018-0052-8
12. Park SY, Gifford JR, Andtbacka RH, Trinity JD, Hyngstrom JR, Garten RS, et al. Cardiac, skeletal, and smooth muscle mitochondrial respiration: Are all mitochondria created equal? Am J Physiol Heart Circ Physiol (2014) 307(3):H346–52. doi: 10.1152/ajpheart.00227.2014
13. No MH, Heo JW, Yoo SZ, Kim CJ, Park DH, Kang JH, et al. Effects of aging and exercise training on mitochondrial function and apoptosis in the rat heart. Pflugers Arch (2020) 472(2):179–93. doi: 10.1007/s00424-020-02357-6
14. Shim W, Paik MJ, Nguyen DT, Lee JK, Lee Y, Kim JH, et al. Analysis of changes in gene expression and metabolic profiles induced by silica-coated magnetic nanoparticles. ACS Nano (2012) 6(9):7665–80. doi: 10.1021/nn301113f
15. Shin TH, Lee DY, Lee HS, Park HJ, Jin MS, Paik MJ, et al. Integration of metabolomics and transcriptomics in nanotoxicity studies. BMB Rep (2018) 51(1):14–20. doi: 10.5483/bmbrep.2018.51.1.237
16. Phukan G, Shin TH, Shim JS, Paik MJ, Lee JK, Choi S, et al. Silica-coated magnetic nanoparticles impair proteasome activity and increase the formation of cytoplasmic inclusion bodies in vitro. Sci Rep (2016) 6:29095. doi: 10.1038/srep29095
17. Shin TH, Seo C, Lee DY, Ji M, Manavalan B, Basith S, et al. Silica-coated magnetic nanoparticles induce glucose metabolic dysfunction in vitro Via the generation of reactive oxygen species. Arch Toxicol (2019) 93(5):1201–12. doi: 10.1007/s00204-019-02402-z
18. Shin TH, Lee DY, Manavalan B, Basith S, Na Y-C, Yoon C, et al. Silica-coated magnetic nanoparticles activate microglia and induce neurotoxic d-serine secretion. Particle Fibre Toxicol (2021) 18(1):30. doi: 10.1186/s12989-021-00420-3
19. Shin TH, Nithiyanandam S, Lee DY, Kwon DH, Hwang JS, Kim SG, et al. Analysis of nanotoxicity with integrated omics and mechanobiology. Nanomaterials (2021) 11(9):2385. doi: 10.3390/nano11092385
20. Shin TH, Manavalan B, Lee DY, Basith S, Seo C, Paik MJ, et al. Silica-coated magnetic-Nanoparticle-Induced cytotoxicity is reduced in microglia by glutathione and citrate identified using integrated omics. Particle Fibre Toxicol (2021) 18(1):42. doi: 10.1186/s12989-021-00433-y
21. Peng T, Wei C, Yu F, Xu J, Zhou Q, Shi T, et al. Predicting nanotoxicity by an integrated machine learning and metabolomics approach. Environ pollut (2020) 267:115434. doi: 10.1016/j.envpol.2020.115434
22. Marcoccia M, Ronci L, De Matthaeis E, Setini A, Perrino C, Canepari S. In-vivo assesment of the genotoxic and oxidative stress effects of particulate matter on echinogammarus veneris. Chemosphere (2017) 173:124–34. doi: 10.1016/j.chemosphere.2017.01.019
23. Chakkarapani SK, Shin TH, Lee S, Park KS, Lee G, Kang SH. Quantifying intracellular trafficking of silica-coated magnetic nanoparticles in live single cells by site-specific direct stochastic optical reconstruction microscopy. J Nanobiotechnol (2021) 19(1):398. doi: 10.1186/s12951-021-01147-1
24. Sturla SJ, Boobis AR, FitzGerald RE, Hoeng J, Kavlock RJ, Schirmer K, et al. Systems toxicology: From basic research to risk assessment. Chem Res Toxicol (2014) 27(3):314–29. doi: 10.1021/tx400410s
25. Shin TH, Lee DY, Jang YE, Kwon DH, Hwang JS, Kim SG, et al. Reduction in the migration activity of microglia treated with silica-coated magnetic nanoparticles and their recovery using citrate. Cells (2022) 11(15):2393. doi: 10.3390/cells11152393
26. Kim D, Langmead B, Salzberg SL. Hisat: A fast spliced aligner with low memory requirements. Nat Methods (2015) 12(4):357–60. doi: 10.1038/nmeth.3317
27. Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. Stringtie enables improved reconstruction of a transcriptome from rna-seq reads. Nat Biotechnol (2015) 33(3):290–5. doi: 10.1038/nbt.3122
28. Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of rna-seq experiments with hisat, stringtie and ballgown. Nat Protoc (2016) 11(9):1650–67. doi: 10.1038/nprot.2016.095
29. Kramer A, Green J, Pollard J Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics (2014) 30(4):523–30. doi: 10.1093/bioinformatics/btt703
30. Seo C, Kim SH, Lee HS, Ji M, Min J, Son YJ, et al. Metabolomic study on bleomycin and polyhexamethylene guanidine phosphate-induced pulmonary fibrosis mice models. Metabolomics (2019) 15(8):111. doi: 10.1007/s11306-019-1574-6
31. Choi RY, Ji M, Lee MK, Paik MJ. Metabolomics study of serum from a chronic alcohol-fed rat model following administration of defatted tenebrio molitor larva fermentation extract. Metabolites (2020) 10(11):436. doi: 10.3390/metabo10110436
32. Lee HS, Seo C, Kim YA, Park M, Choi B, Ji M, et al. Metabolomic study of polyamines in rat urine following intraperitoneal injection of gamma-hydroxybutyric acid. Metabolomics (2019) 15(4):58. doi: 10.1007/s11306-019-1517-2
33. Seo C, Park S, Kim Y, Ji M, Lee HS, Hwang YH, et al. Metabolomic analysis of amino acids and organic acids in aging mouse eyes using gas chromatography-tandem mass spectrometry. BioMed Chromatogr (2022) 36(3):e5298. doi: 10.1002/bmc.5298
34. Baginsky S, Hennig L, Zimmermann P, Gruissem W. Gene expression analysis, proteomics, and network discovery. Plant Physiol (2010) 152(2):402–10. doi: 10.1104/pp.109.150433
35. Seyfried NT, Dammer EB, Swarup V, Nandakumar D, Duong DM, Yin L, et al. A multi-network approach identifies protein-specific Co-expression in asymptomatic and symptomatic alzheimer's disease. Cell Syst (2017) 4(1):60–72 e4. doi: 10.1016/j.cels.2016.11.006
36. Rehrauer H, Opitz L, Tan G, Sieverling L, Schlapbach R. Blind spots of quantitative rna-seq: The limits for assessing abundance, differential expression, and isoform switching. BMC Bioinf (2013) 14:370. doi: 10.1186/1471-2105-14-370
37. Evans TG. Considerations for the use of transcriptomics in identifying the 'Genes that matter' for environmental adaptation. J Exp Biol (2015) 218(Pt 12):1925–35. doi: 10.1242/jeb.114306
38. Claycomb WC, Lanson NA, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, et al. Hl-1 cells: A cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci (1998) 95(6):2979–84. doi: 10.1073/pnas.95.6.2979
39. Liu Q, Han B, Zhang Y, Jiang T, Ning J, Kang A, et al. Potential molecular mechanism of cardiac hypertrophy in mice induced by exposure to ambient Pm2.5. Ecotoxicol Environ Saf (2021) 224:112659. doi: 10.1016/j.ecoenv.2021.112659
40. Sivakumar B, Kurian GA. Pm2.5 from diesel exhaust attenuated fisetin mediated cytoprotection in H9c2 cardiomyocytes subjected to ischemia reoxygenation by inducing mitotoxicity. Drug Chem Toxicol (2021) 1–9. Advance online publication. doi: 10.1080/01480545.2021.2003698
41. Sivakumar B, Kurian GA. Pm2.5 exposure lowers mitochondrial endurance during cardiac recovery in a rat model of myocardial infarction. Cardiovasc Toxicol (2022) 22(6):545–57. doi: 10.1007/s12012-022-09737-7
42. Jiang W, Rutherford D, Vuong T, Liu H. Nanomaterials for treating cardiovascular diseases: A review. Bioact Mater (2017) 2(4):185–98. doi: 10.1016/j.bioactmat.2017.11.002
43. Basith S, Manavalan B, Shin TH, Park CB, Lee WS, Kim J, et al. The impact of fine particulate matter 2.5 on the cardiovascular system: A review of the invisible killer. Nanomater (Basel) (2022) 12(15):2656. doi: 10.3390/nano12152656
44. Poster DL, Lopez de Alda MJ, Schantz MM, Sander LC, Wise SA, Vangel MG. Development and analysis of three diesel particulate-related standard reference materials for the determination of chemical, physical, and biological characteristics. Polycyclic Aromatic Compounds (2003) 23(2):141–91. doi: 10.1080/10406630308056
45. Poster DL, Benner BA, Schantz MM, Sander LC, Wise SA, Vangel MG. Determination of methyl-substituted polycyclic aromatic hydrocarbons in diesel particulate-related standard reference materials. Polycyclic Aromatic Compounds (2003) 23(2):113–39. doi: 10.1080/10406630308059
46. Patel AB, Shaikh S, Jain KR, Desai C, Madamwar D. Polycyclic aromatic hydrocarbons: Sources, toxicity, and remediation approaches. Front Microbiol (2020) 11:562813. doi: 10.3389/fmicb.2020.562813
47. Abbas I, Badran G, Verdin A, Ledoux F, Roumié M, Courcot D, et al. Polycyclic aromatic hydrocarbon derivatives in airborne particulate matter: Sources, analysis and toxicity. Environ Chem Lett (2018) 16(2):439–75. doi: 10.1007/s10311-017-0697-0
48. Zhou S, Xi Y, Chen Y, Zhang Z, Wu C, Yan W, et al. Ovarian dysfunction induced by chronic whole-body Pm2.5 exposure. Small (2020) 16(33):e2000845. doi: 10.1002/smll.202000845
49. Udintsev NA, Ivanov VV. Antioxidant action of glutamic acid. Patol Fiziol Eksp Ter (1984) (4):60–2.
50. Franco R, Cidlowski JA. Apoptosis and glutathione: Beyond an antioxidant. Cell Death Differ (2009) 16(10):1303–14. doi: 10.1038/cdd.2009.107
51. Zitka O, Skalickova S, Gumulec J, Masarik M, Adam V, Hubalek J, et al. Redox status expressed as Gsh:Gssg ratio as a marker for oxidative stress in paediatric tumour patients. Oncol Lett (2012) 4(6):1247–53. doi: 10.3892/ol.2012.931
52. Jones DP. Redox potential of Gsh/Gssg couple: Assay and biological significance. Methods Enzymol (2002) 348:93–112. doi: 10.1016/s0076-6879(02)48630-2
53. Kho C, Lee A, Hajjar RJ. Altered sarcoplasmic reticulum calcium cycling–targets for heart failure therapy. Nat Rev Cardiol (2012) 9(12):717–33. doi: 10.1038/nrcardio.2012.145
54. Boerries M, Most P, Gledhill JR, Walker JE, Katus HA, Koch WJ, et al. Ca2+ -dependent interaction of S100a1 with F1-atpase leads to an increased atp content in cardiomyocytes. Mol Cell Biol (2007) 27(12):4365–73. doi: 10.1128/MCB.02045-06
55. Son G, Han J. Roles of mitochondria in neuronal development. BMB Rep (2018) 51(11):549–56. doi: 10.5483/BMBRep.2018.51.11.226
56. Zundorf G, Reiser G. Calcium dysregulation and homeostasis of neural calcium in the molecular mechanisms of neurodegenerative diseases provide multiple targets for neuroprotection. Antioxid Redox Signal (2011) 14(7):1275–88. doi: 10.1089/ars.2010.3359
57. Xie X, Wang Y, Yang Y, Xu J, Zhang Y, Tang W, et al. Long-term exposure to fine particulate matter and tachycardia and heart rate: Results from 10 million reproductive-age adults in China. Environ pollut (2018) 242(Pt B):1371–8. doi: 10.1016/j.envpol.2018.08.022
58. Zhu C, Fu Z, Liu L, Shi X, Li Y. Health risk assessment of Pm2.5 on walking trips. Sci Rep (2021) 11(1):19249. doi: 10.1038/s41598-021-98844-6
59. Perret-Guillaume C, Joly L, Benetos A. Heart rate as a risk factor for cardiovascular disease. Prog Cardiovasc Dis (2009) 52(1):6–10. doi: 10.1016/j.pcad.2009.05.003
60. Manzanares D, Cena V. Endocytosis: The nanoparticle and submicron nanocompounds gateway into the cell. Pharmaceutics (2020) 12(4):371. doi: 10.3390/pharmaceutics12040371
61. Lee Y, Geckeler KE. Carbon nanotubes in the biological interphase: The relevance of noncovalence. Advanced Mater (2010) 22(36):4076–83. doi: 10.1002/adma.201000746
62. Su R, Jin X, Li H, Huang L, Li Z. The mechanisms of Pm2.5 and its main components penetrate into huvec cells and effects on cell organelles. Chemosphere (2020) 241:125127. doi: 10.1016/j.chemosphere.2019.125127
63. McGrath MF, de Bold ML, de Bold AJ. The endocrine function of the heart. Trends Endocrinol Metab (2005) 16(10):469–77. doi: 10.1016/j.tem.2005.10.007
64. Ogawa T, de Bold AJ. The heart as an endocrine organ. Endocr Connect (2014) 3(2):R31–44. doi: 10.1530/EC-14-0012
65. Zhao J, Pei L. Cardiac endocrinology: Heart-derived hormones in physiology and disease. JACC Basic Transl Sci (2020) 5(9):949–60. doi: 10.1016/j.jacbts.2020.05.007
Keywords: diesel particulate matter, mitochondria, cardiomyocytes, metabolomics, transcriptomics, integrated omics
Citation: Shin TH, Kim SG, Ji M, Kwon DH, Hwang JS, George NP, Ergando DS, Park CB, Paik MJ and Lee G (2022) Diesel-derived PM2.5 induces impairment of cardiac movement followed by mitochondria dysfunction in cardiomyocytes. Front. Endocrinol. 13:999475. doi: 10.3389/fendo.2022.999475
Received: 21 July 2022; Accepted: 13 September 2022;
Published: 28 September 2022.
Edited by:
Yong Seek Park, Kyung Hee University, South KoreaReviewed by:
Hye Hyun Yoo, Hanyang University, South KoreaCuiqing Liu, Zhejiang Chinese Medical University, China
Copyright © 2022 Shin, Kim, Ji, Kwon, Hwang, George, Ergando, Park, Paik and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Man Jeong Paik, cGFpazgxNUBzdW5jaG9uLmFjLmty; Gwang Lee, Z2xlZUBham91LmFjLmty
†These authors have contributed equally to this work
 Tae Hwan Shin
Tae Hwan Shin Seok Gi Kim2†
Seok Gi Kim2† Man Jeong Paik
Man Jeong Paik Gwang Lee
Gwang Lee