
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol., 14 November 2022
Sec. Diabetes: Molecular Mechanisms
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.991803
This article is part of the Research TopicNovel Insights into the Pathophysiology of Diabetes-related Complications: Implications for Improved Therapeutic StrategiesView all 11 articles
Background: Diabetic retinopathy (DR) is a common and serious microvascular complication of diabetes mellitus (DM), but its pathological mechanism, especially the formation mechanism of new blood vessels remains unclear. Thrombospondin-1 (THBS1) is a potent endogenous inhibitor of angiogenesis and it was found over expressed in DR in our previous study. Our study aimed to determine whether overexpression of THBS1 is associated with its promoter methylation level, and whether methylation of THBS1 is regulated by genetic variants in DR.
Methods: Patients diagnosed with DR and DM patients without retinal problems were included in the case-control study. DNA methylation detection of THBS1 by bisulfite sequencing and genotyping of specific SNPs by MassARRAY analysis were performed in the patients recruited from 2019-2020. Real time quantitative PCR was performed to obtain mRNA expression of THBS1 in the patients recruited from August to October 2022. The differentially methylated CpG loci of THBS1 were identified by logistic regression, and associations between 13 SNPs and methylation levels of CpG loci were tested by methylation quantitative trait loci (meQTLs) analysis. Mediation analysis was applied to determine whether CpG loci were intermediate factors between meQTLs and DR.
Results: 150 patients diagnosed with DR and 150 DM patients without retinal complications were enrolled in the first recruitment, seven DR patients and seven DM patients were enrolled in the second recruitment. The patients with DR showed promoter hypomethylation of THBS1 (P value = 0.002), and six out of thirty-nine CpG sites within two CpG islands (CGIs) showed hypomethylation(P value < 0.05). THBS1 mRNA expression in peripheral blood was significantly higher in DR patients than in DM patients. Five out of thirteen cis-meQTLs were identified to be associated with CpG sites: rs13329154, rs34973764 and rs5812091 were associated with cis-meQTLs of CpG-4 (P value=0.0145, 0.0095, 0.0158), rs11070177 and rs1847663 were associated with cis-meQTLs of CpG-2 and CpG-3 respectively (P value=0.0201, 0.0275). CpG-4 methylation significantly mediated the effect of the polymorphism rs34973764 on DR (B=0.0535, Boot 95%CI: 0.004~0.1336).
Conclusion: THBS1 overexpression is related to THBS1 hypomethylation in patients with DR. DNA methylation may be genetically controlled in DR.
Diabetic retinopathy (DR) is a common and serious microvascular complication of diabetes mellitus (DM), and has long been regarded as one of the major public health problems in the world (1, 2). An up-to-date evaluation report showed that one in five patients with DM worldwide had DR, and the number of adults with DR worldwide was estimated to rise from 103.12 million in 2020 to 160.5 million in 2045 (3). Clinically, DR not only leads to visual impairment and blindness, but also signifies an enhanced risk of cardiovascular disease, atherosclerosis and diabetic peripheral neuropathy (4–7). Glycemic control is routinely recommended to prevent the complications of DM. However, multiple basic studies have found that retinopathy induced by long-term exposure to hyperglycemia persists after glycemic control (8). Despite advances in the treatment with anti-angiogenic drugs (9), patients with DR might still experience adverse effects on their quality of life and financial circumstances. Accordingly, in order to reduce the prevalence of vision impairment and blindness caused by DR, it is imperative to discover more specific and sensitive biomarkers and explore its detailed etiology.
Early clinical characteristics of DR, such as microaneurysms and intraretinal microvascular abnormalities, are caused by changes in the cellular composition of the capillary wall (10). With the occurrence of capillary blockage and retinal ischemia in DR, neovascularization arises on the retina, resulting in proliferative DR (11). However, the pathological mechanism of DR, especially the formation mechanism of new blood vessels remains unclear. Thrombospondin-1 (THBS1) is a commonly matrix-cellular glycoprotein that plays a significant role in retinal vascular homeostasis (12, 13). Importantly, THBS1 is a potent endogenous inhibitor of angiogenesis. THBS1 was found to inhibit endothelial cell migration and proliferation and stimulate apoptosis by regulating vascular endothelial growth factor activity (14). THBS1 may be involved in the development and progression of DR (15). Overexpression of THBS1 impairs retinal vascular development and neointima formation in mice (16). In addition, deficiency of THBS1 expression in retinal endothelial cells resulted in the accelerated proliferation and increased angiogenesis (16, 17). Moreover, we analyzed two gene expression datasets of DR patients (GSE94019 and GSE60436) (18, 19), which were downloaded from the Gene Expression Omnibus (GEO) (20), and found that the expression of THBS1 was significantly up-regulated in the fibrovascular membrane of DR compared to normal retinal tissue in both datasets (adjusted P value = 3.58×10-3; 3.56×10-8) (data not published). Thus, we hypothesized that THBS1 is a critical molecular in the development of DR, but the detailed mechanism underlying THBS1 over-expression in DR is still unknown.
DNA methylation is a critical epigenetic alteration with the potential to illuminate the differential expression of THBS1 in DR. DNA methylation status is closely correlated with gene expression (21). For example, hypermethylation of gene promoter regions can decreases DNA accessibility and inhibit transcription factors from binding, causing gene inactivation (i.e., inhibition of gene expression). Furthermore, since Maghbooli et al. first found higher global DNA methylation level in patients with DR (22), subsequent studies have confirmed that DNA methylation of some genes was indeed different in patients with DR (23–25). DNA methylation is therefore a critical factor related to gene expression, and cannot be ignored in the development of DR. According to our previous analysis of two datasets, differential expression of anti-angiogenic factor-THBS1 was identified in patients with DR. However, whether THBS1 methylation is involved in DR remains unknown. Moreover, the association between genetic variation and DNA methylation in humans has been extensively studied, and methylation quantitative trait loci (meQTL) may exert a regulatory effect on methylation at related cytosine-phosphate-guanine (CpG) sites (including cis and trans effect) (26–28). Particularly, meQTL pairs formed by SNPs and CpG loci tend to be enriched for functionally relevant features, such as gene expression, metabolic functions and clinical manifestations (29). Therefore, identifying the genetic regulation of DNA methylation can help us figure out the mechanisms by which genetic variation influences complex phenotypes. In light of these findings, exploring whether methylation of THBS1 gene is genetically regulated may provide insight into the role of THBS1 in DR and shed new light on the molecular networks and biological mechanisms of DR.
Our study aimed to determine whether overexpression of THBS1 is associated with its promoter methylation level, and whether methylation of THBS1 is regulated by genetic variants in DR.
Patients diagnosed with DR and DM patients without retinal complications were recruited in the case-control study from communities in Shenzhen City, Guangdong Province, China. The patients recruited from 2019 to 2020 were for the study of THBS1 methylation detection and specific SNPs genotyping, and the patients recruited from August to September 2022 were for THBS1 mRNA testing. Patients in the case group were adult males or females with a history of T2DM, impaired vision, and previously diagnosed with DR by Fluorescence fundus angiography or optical coherence tomography. Patients in the control group were previously diagnosed with T2DM {according to the Chinese Guidelines for the Prevention and Treatment of Type 2 Diabetes [2017 Edition (30)]} without retinopathy. The case and control subjects were matched for age, sex, and duration of disease. Patients with diabetic foot, diabetic nephropathy, macular edema, history of other eye diseases or eye surgery, malignant tumors, severe liver and kidney dysfunction, mental illness, and alcohol or drug abuse were excluded. This study was approved by the Ethics Committee of Shenzhen Center for Chronic Disease Control. Written informed consent was signed by all participants.
Questionnaire survey was conducted among the patients recruited from 2019 to 2020 for demographic information (gender and age), anthropometric data [height, weight and body mass index (BMI)], diabetic duration, history of hypertension and information on lifestyle (smoking and drinking frequency) of patients enrolled. BMI = weight/height2 (kg/m2). Hypertension was defined as systolic blood pressure (SBP) ≥140 mmHg and/or diastolic blood pressure (DBP) ≥ 90 mmHg, or a prior history of hypertension or use of antihypertensive medications. Smokers are defined as those who smoke one or more cigarettes a day for more than one year. Drinkers were defined as those who consumed more than 50 ml alcohol per week on average and drank for more than six months. The collected questionnaire data was sorted out by using EPIDATA software and recorded by two people.
5 ml fasting peripheral blood of all participants were collected by vacuum collection tubes after fasting for 12 hours. The blood samples of the first recruited patients were centrifuged, divided and stored in a refrigerator at -80℃. DNA was extracted from their peripheral blood by Blood Genomic DNA Extraction Kit (Omega, American). The blood samples of the secondly recruited patients were centrifuged, divided and used for subsequent mRNA level testing.
Glycosylated hemoglobin (HbA1c), fasting blood glucose (FPG), urea (UREA), creatinine (Cr), triglycerides (TG), total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C) and high-density lipoprotein cholesterol (HDL-C) were tested among the patients recruited from 2019-2020 with Beckman-LX20 automatic biochemical analyzer.
The detailed information on CpG islands and CpG sites of THBS1 gene is provided in Table S1. Bisulfite conversion of DNA was performed by EZ DNA methylation-Gold ™ Kit (ZYMO, CA, USA). The modified DNA was amplified by multiple PCR (HotStart Taq polymerase, TaKaRa, Dalian, China). Primers with Index sequences were used to introduce specific tag sequences to the end of target and region by PCR amplification (Herculase® II Fusion DNA Polymerase, Agilent Technologies, CA, USA). The information on Primer sequences of THBS1 is provided in Table S2. The PCR products were electrophoresed with 2% agarose gel and purified with TIANGEN Gel Extraction kit (TIANGEN, Beijing, China). Finally, data on THBS1 methylation was obtained using Illumina Hiseq or Nova SEQ platform for high-throughput sequencing.
Genomic DNA extracted from peripheral blood was analyzed by mass spectrometry using MassARRAY (Agena Bioscience), and then genotyping of those thirteen SNPs was performed. GTEx portal (https://www.gtexportal.org/home/index.html) was used to search for expression quantitative trait Loci (eQTL) of THBS1 gene, and thirteen eQTLs existing both in whole blood and fibroblasts were selected as SNPs to be detected. Detailed information on SNPs is listed in Table 1.
The genotyping process was completed by Shanghai Tianhao Biotechnology Company. The quality evaluation of a SNP included minor allele frequency (MAF>5%), call rate(>95%).
Peripheral blood samples were treated with Red Blood Cell Lysis Buffer (Beyotime Biotechnology, Shanghai) and total RNA was extracted from cells using the Trizol method (TRIzol, Reagent life, USA). cDNA was synthesized from 1μg of total RNA using PrimeScriptTM RT reagent Kit with gDNA Eraser (Perfect real time) (TAKARA, Japan). The concentration and purity of RNA were determined by spectrophotometry (NanoVue Plus, GE healthcare UK limited, the United Kingdom). THBS1 mRNA expression was measured by real time quantitative PCR(RT-PCR) with TB Green® Premix Ex Taq™ II (TAKARA, Japan). The optimal number of PCR cycles and the mixing ratio of primers were determined according to the instruction of TB green reagent (Code No. RR820A). PCR products were quantified using TB Green. Beta-actin (ACTB) was used as an endogenous control to normalize expression levels. The ranges of linear amplification for the target gene and for the ACTB genes were studied. Data were normalized relative to the expression level of ACTB for each sample. Primers used for RT-PCR were human THBS1-1, 5’-TTGTCTTTGGAACCACACCA-3’(sense) and 5’-TGGACAGCTCATCACAGGAG-3’ (antisense); and ACTB 5’-GATGAGATTGGCATGGCTTT-3’ (Sense) and 5’-CACCTTCACCGGTCAGTTT -3’ (antisense). The expression levels of mRNA were represented as 2-ΔΔCT.
In terms of basic characteristics, continuous variables with normal distribution were presented as mean ± standard deviation, and independent two-sample t-tests were used to compare differences between the case and control group. Continuous variables without normal distribution were presented as medians (percentile 25, percentile 75), Wilcoxon rank sum test was performed to identify differences between groups. Categorical variables were presented using percentages and chi-square tests were used to compare differences between groups. All those tests were conducted two-tailed in IBM SPSS 25 software, and P<0.05 was considered significant.
The differentially methylated CpG loci of THBS1 were identified using univariate logistic regression in SPSS software. Associations between each SNP and methylation levels of differentially methylated CpG loci were tested to identify meQTLs using linear regression in the software PLINK (31). The independent variable was the genotype of the SNP, and the additive model was adopted. Given that we had previously tried to keep the distribution of most critical confounding factors- age, gender and diabetic duration to be the same between the case group and the control group, so we did not adjust for other factors. Cis-meQTL was defined as being less than 500kb upstream or downstream of CpG sites from the associated CpG loci, otherwise as trans-meQTLs. To examine the independence of these meQTLs, linkage disequilibrium (LD) analysis was performed by Haploview 4.2 programme (32). r2 was obtained as a measure of LD based on our study population. To explore the relationship between meQTLs and DR, binary logistic regressions were performed in dominant, recessive and additive models in SPSS software. HbA1c and TC were adjusted in three models. P<0.05 was considered significant.
To determine whether methylation at CpG loci was an intermediate factor between genetic variation and DR, we performed a mediation analysis by using macro PROCESS (v3.4 by Andrew F. Hayes) in SPSS. Model 4 was used for our analysis, which represented a simple mediation model (the type occurring when only one variable regulated the effect of a cause on an outcome) (33). 5000 bias-corrected Bootstrap sample was used for the significance test. It is sufficient to support a claim of mediation effect when the 95% bootstrap confidence intervals of the indirect effect (as quantified with ab) does not include zero (34). We can determine whether there is an intermediate effect of methylation, by calculating whether the indirect effect of genetic variation on disease is significant.
From 2019 to 2020, 150 patients diagnosed with DR and 150 DM patients without retinal complications were enrolled in our study. Their clinical characteristics are summarized in Table 2. Since we matched the age, sex and duration of disease between the case group and the control group, the mean age of both groups was about 56 years, 55% were female, and the median duration of disease was 8 and 9 years, respectively. As illustrated in the table, BMI and the proportion of hypertension patients were comparable between the case and control groups. HbA1c, FPG, UREA, TC, LDL-C in the case group were significantly higher than those in the control group (P value<0.05).
From August to September 2022, seven DR patients and seven DM patients without retinal complications were enrolled in our study. Since we had matched age and gender of the participants in the case group and control group, the average ages of the case group and the control group are 60.14 ± 8.17 and 59.86 ± 7.51, with four females in each group.
There are two CpG islands (CGIs) in THBS1 gene, which contains 23 and 16 CpG sites respectively, and five CpG sites of CGI-2 are in the gene body of THBS1 (Figure 1). Methylation levels of 39 CpG loci on THBS1 gene were tested in all subjects (Table S1). The patients with DR showed a lower methylation level of THBS1 than the patients with DM (P value = 0.002) (Table S1). To be specific, the methylation levels of six CpG loci in the patients with DR were significantly different from those with DM, and all the 6 CpG loci in the patients with DR showed hypomethylation (P value<0.05) (Table 3). All these six sites are located in the promoter region of THBS1, CpG-5 locates downstream of the transcription start site (TSS), and other sites locates upstream of the TSS. The box plots of methylation levels of THBS1 and six CpG loci in the two groups are shown in Figure 2.
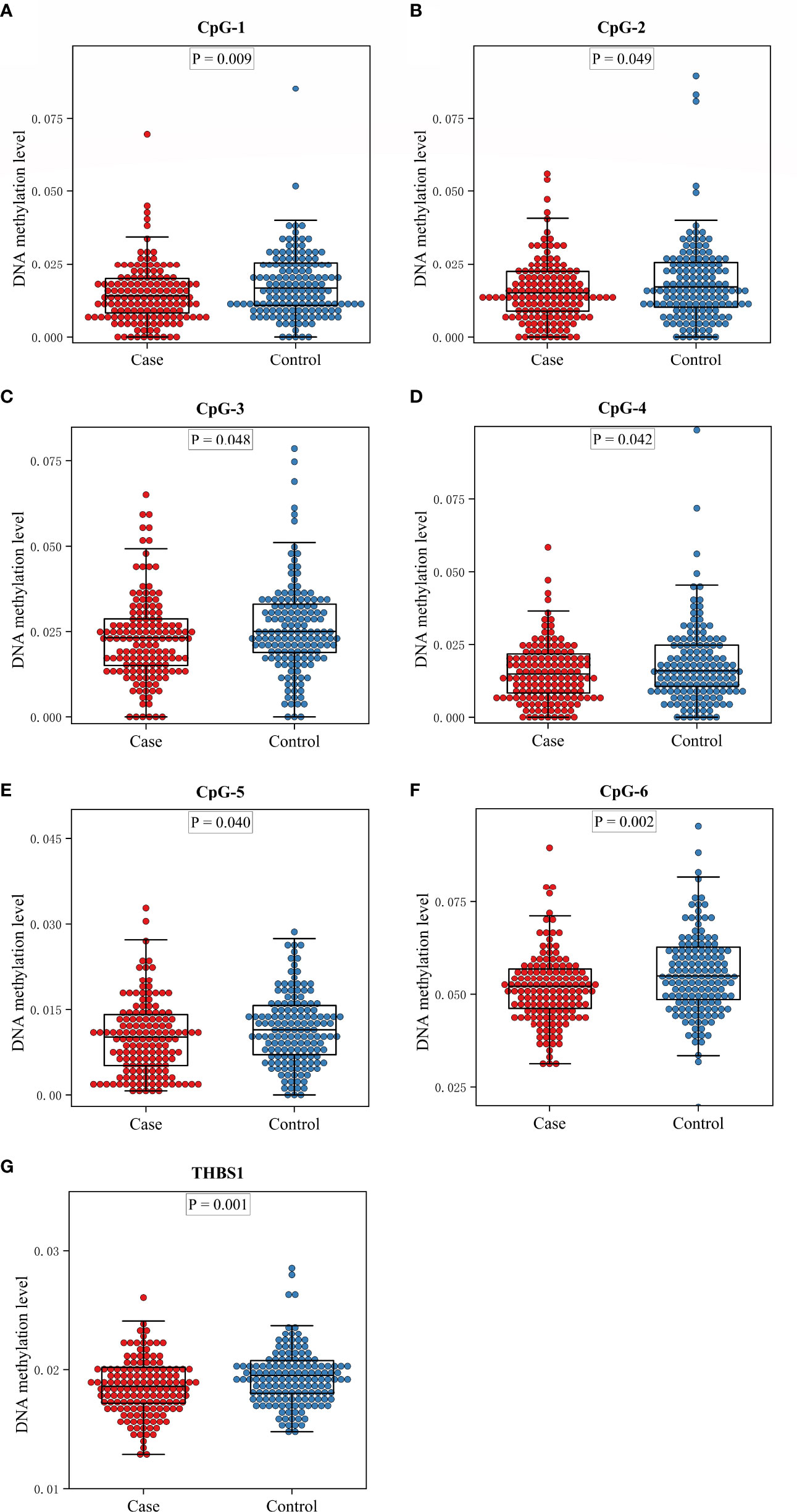
Figure 2 The box plots of methylation levels of THBS1 in the case and control groups. (A–F) Methylation levels of six CpG loci with significant difference in the case and control group. (G) Methylation levels of THBS1 in the case and control group.
The meQTL association analyses between 6 differentially methylated CpG sites and 13 SNPS were performed by the software PLINK, and the results are shown in Table S3. Five SNPs were associated with three differentially methylated CpG loci, all of which were cis-meQTLs upstream of the associated CpG loci. Box plots of methylation level plotted against genotypes for five meQTLs are shown in Figure 3. rs13329154(C>T), rs34973764(insC), rs5812091(dupC) were cis-meQTLs of CpG-4 (P value=0.0145, 0.0095, 0.0158; distance between SNPs and associated CpG site: 319kb, 297kb, 269kb respectively). Variations in these SNPs were associated with reduced methylation level. In addition, rs11070177(C>T) and rs1847663(A>G) were cis-meQTLs of CpG-2 and CpG-3 (P value=0.0201, 0.0275; distance between SNPs and associated CpG site: 323kb, 319kb), and methylation levels of CpG-2 and CpG-3 increased with the number of allelic mutations. LD analysis between these five meQTLs showed a low degree of LD (r 2<30%) (Figure 4). Association analysis between these five meQTLs and DR showed that none of these meQTLs were associated with DR in dominant, recessive and additive models (Table S4).
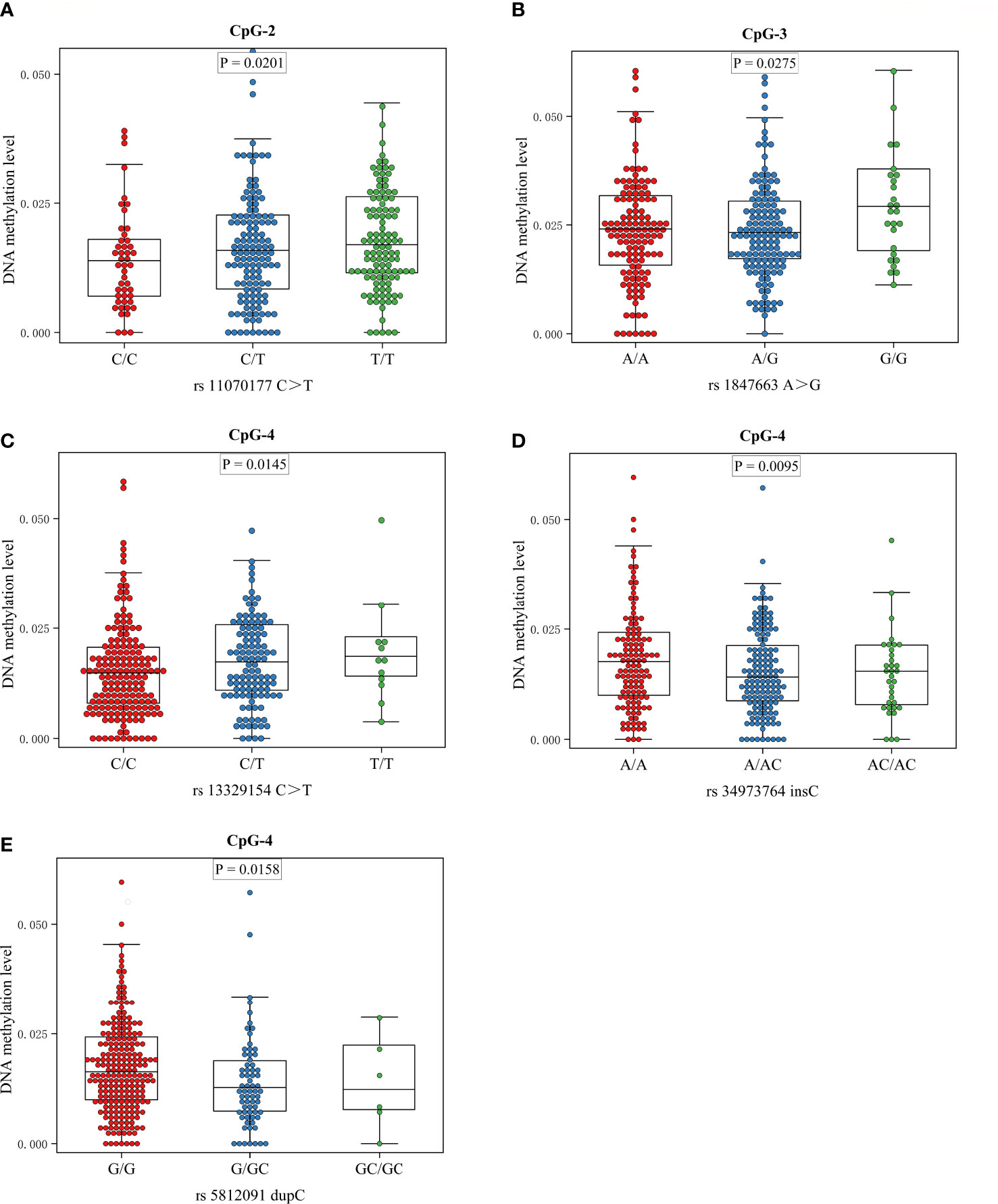
Figure 3 Boxplots showing methylation level plotted against genotypes for five meQTLs: Center lines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the 5th and 95th percentiles. (A) The association between genotype at rs11070177 and methylation level at CpG-2; (B) the association between genotype at rs1847663 and methylation level at CpG-3; (C–E) the association between genotypes at rs13329154, rs34973764, rs5812091 and methylation level at CpG-4.
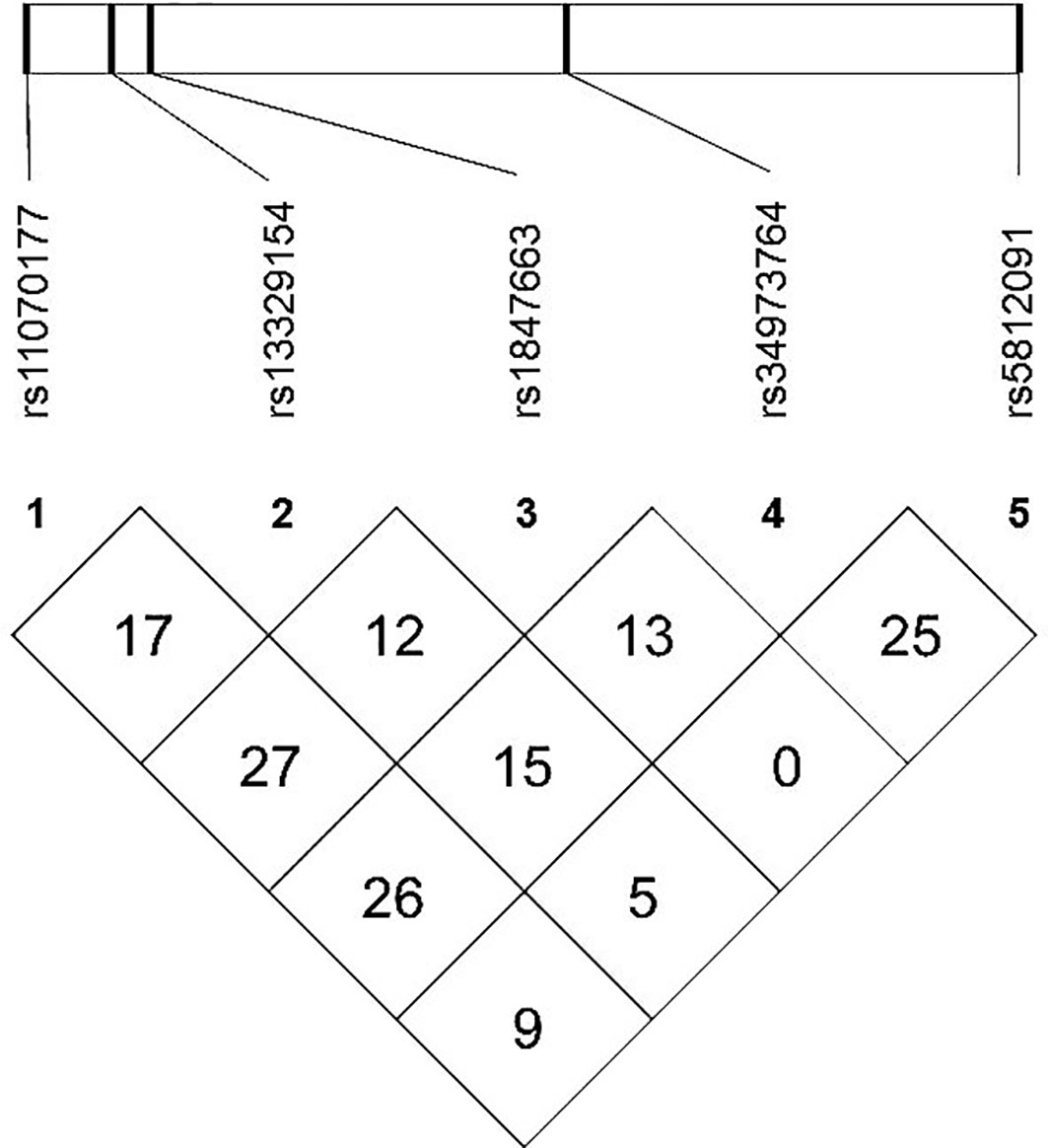
Figure 4 Linkage disequilibrium (LD) map among five meQTLs: LD r2(%) based on our study population are shown in the boxes.
The indirect mediation routes of five meQTLs on DR through methylation of three CpG loci are shown in Table 4. In the five established mediation models, CpG-4 methylation significantly mediated the effect of the polymorphism rs34973764 on DR (B=0.0535, Boot 95%CI: 0.004~0.1336) (Table 4). Analysis of indirect effects showed that the variant allele of rs34973764 predicted hypomethylation levels at CpG-4 (a=-0.272, p value=0.009), and the incidence of DR increased with hypomethylation level at CpG-4 (b=-0.197, p value=0.0596) (Figure 5). Although the total and direct effect of rs34973764 on DR were not significant (total effect: c=-0.006, p value=0.973; direct effect: c’=-.0575, p value=0.7464), the indirect effect showed significant. Therefore, rs34973764 may act as a risk SNP for DR through CpG-4.
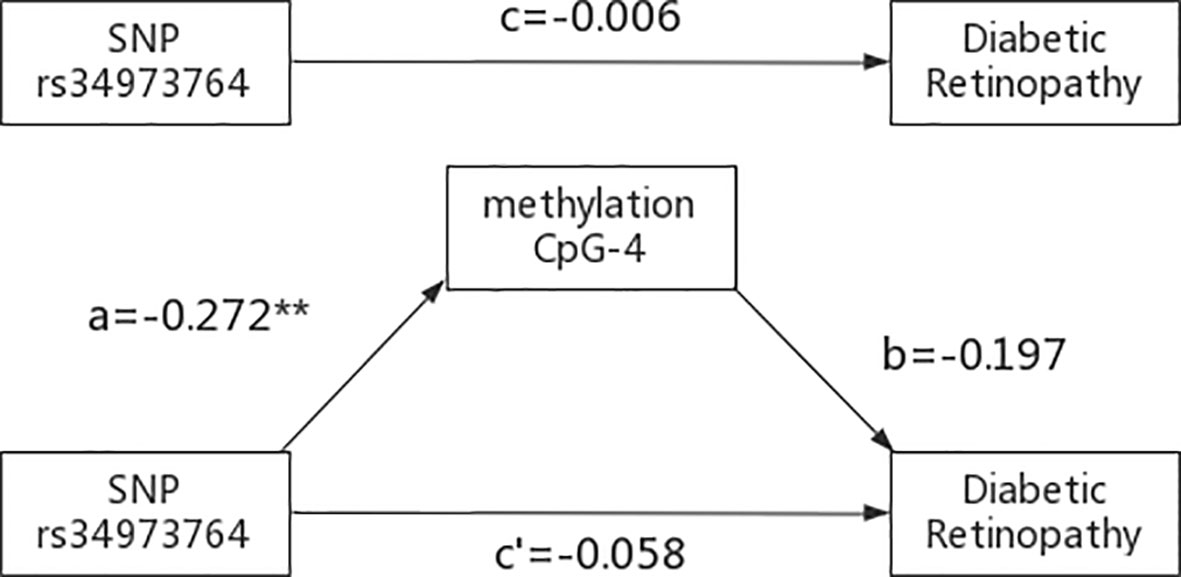
Figure 5 Results of the mediation analysis: Indirect effect of rs34973764 on diabetic retinopathy through methylation of CpG-4 and total effect (regression coefficients). **p ≤.001.
THBS1 mRNA expression in peripheral blood was significantly higher in DR patients than in DM patients (2-ΔΔCT median (percentile 25, 75): 3.67(1.41-8.03) vs. 1.00(0.53-1.33), p value=0.025). Comparison of THBS1 mRNA expression levels in the two groups is shown in Figure S1.
In our study, the patients with DR showed significant hypomethylation of THBS1 compared to those with T2DM, and 6 out of 39 CpG sites showed hypomethylation. Moreover, 5 out of 13 SNPs were identified to be associated with three differentially methylated CpG loci of THBS1. CpG-4 methylation mediated the effect of rs34973764 on DR in the mediation analysis, providing data evidence that THBS1 methylation may be regulated by genetic variation in DR.
THBS1 is an endogenous molecule functioning as anti-angiogenesis and has been proven to regulate ocular vascular homeostasis (35). Numerous studies revealed the critical role of THBS1 in the onset and progression of DR, especially in its neovascularization and vascular abnormalities. In the differential expression analysis performed prior to this study, results showed that the mRNA expression of THBS1 in the fibrovascular membrane of DR patients was significantly up-regulated in both datasets (GSE94019 and GSE60436) (18, 19). This result has been confirmed in other studies. Bian et al. found that patients with DR exhibited significantly elevated serum THBS1 compared to patients with T2DM, patients with proliferative DR in particular showed the highest THBS1 (36). Wang et al. observed high expression of THBS1 in the retina of DR rats (37). Wu et al. identified that mice with THBS1 over-expressing in the lens showed attenuated retinal vascular development and impaired neovascularization (16). Hence, the expression of THBS1 is highly associated with the formation of new blood vessels, which may have a significant impact on the development of DR.
It is widely accepted that the expression of genes is affected by genetic regulation or epigenetic modification, which is adjustable and susceptible to environmental factors. So, it is imperative to figure out the association between the expression and epigenetic modification of THBS1 in DR. Given that DNA methylation is an important epigenetic modification that has been studied the most, our study focused on how DNA methylation affects expression in DR. In our research, the methylation level of THBS1 was significantly lower in the patients with DR compared to with DM. Besides, methylation levels of six CpG sites of THBS1 showed a significant difference between those two groups, and all of these sites showed decreased DNA methylation in cases with DR. Then we examined THBS1 mRNA expression levels in another recruited patients, and the results showed that the mRNA expression level of THBS1 in peripheral blood was significantly higher in DR patients than in DM patients. In addition, patients with DR are typically poor in glycemic control, as supported by our data. Studies confirmed that hyperglycemia can result in irreversible alterations in the activity of DNA methyltransferases and hydroxymethylase (38). Moreover, THBS1 expression was significantly increased in the high-glucose environment in keratinocytes and diabetic rat model, which was induced through DNA hypomethylation (39). Thus, we speculate that high glucose may have an impact on the THBS1 methylation via enzyme activity, resulting in alterations of THBS1 expression in DR. Since the existing research is restricted, additional research is necessary to clarify the mechanism of methylation and expression of THBS1 in DR.DNA methylation has been previously described to have a genetic basis, and numerous studies have investigated the relationship between DNA methylation and genetic variation throughout the genome to identify meQTLs (40). In our meQTL analysis, there were five cis-CpG-SNPs associations identified, including three THBS1 methylation loci and five SNPs. The rare allele of rs13329154, rs34973764, rs5812091 exhibited significantly decreased levels of CpG-4 methylation, while the rare allele of rs11070177 and rs1847663 exhibited significantly increased levels of CpG-2 and CpG-3 methylation respectively. Rs13329154, rs11070177 and rs1847663 are located in the noncoding region of C15orf54 gene, while rs34973764, rs5812091 have not been characterized. C15orf54 gene has not been well understood, and up-regulated expression of C15orf54 was first found to be associated with the risk of gastric cancer in a recent bioinformatics analysis (41). In addition, there is no association between these five meQTLs and DR in dominant, recessive and additive models in our research, and these five meQTLs have not been reported to be associated to any phenotypes or diseases in prior studies. Hence, more research is needed to determine how C15orf54 gene polymorphism affects THBS1 methylation in DR.
meQTLs are highly enriched in GWAS signals (42), implying that meQTLs are associated with an elevated risk of disease. Studies have shown that meQTLs may exert an influence on disease risk via altering DNA methylation (40). In the mediation study, CpG-4 methylation mediated the effect of rs34973764 on DR, which may be supporting evidence for genetic regulation of DNA methylation in DR. Although the total effect of rs34973764 on DR was not significant, rs34973764 had a significant indirect effect on the DR via the mediator of CpG-4 methylation. One possible explanation is that rs34973764 functions in DR via two or more additional mediation pathways, and those additional pathways operate oppositely from the CpG-4 methylation pathway. However, the function of rs34973764 and its impact on CPG-4 are not yet fully understood, so the underlying molecular processes of meQTLs are unknown. The most common explanation for the cis-meQTL effect is that SNPs at protein binding sites affect the function of sequence-specific binding proteins, such as transcription factors, and thus change the methylation pattern of adjacent CpGs (40). Future research combining SNPs, DNA methylation, and gene expression are needed to better understand the pathology of DR.
There are several limitations in our study. Firstly, DR is classified as either proliferative or non-proliferative (11), but in our study, we did not divide patients into those two categories. Therefore, we could not obtain dynamic changes in THBS1 methylation level as the disease progressed. Secondly, the SNPs we detected were not from genome-wide SNPs, but eQTLs related to THBS1, which represented only a small fraction of meQTLs of THBS1. Thirdly, the sample size was relatively insufficient, especially in the detection of THBS1 mRNA expression levels, hence case-control and cohort studies with larger sample size are needed to confirm these findings.
In conclusion, THBS1 overexpression is related to THBS1 hypomethylation in patients with DR. DNA methylation may be genetically controlled in DR.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the Ethics Committee of Shenzhen Center for Chronic Disease Control. The patients/participants provided their written informed consent to participate in this study.
YL and CG contributed equally to this work. CG and WH contributed to the design of the work. YL contributed to manuscript written and statistical analyses. Data acquisition and analysis were carried out by YX and XL. JY and CG reviewed the paper. All authors contributed to the article and approved the submitted version.
This work was supported by the Sanming Project of Medicine of Shenzhen (SZSM201811057), National Natural Science of Guang dong province (2019A1515010358) and Guangdong Medical Research Fund Project (2017196).
We thank all participants for providing samples and those involved in the sample collection.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.991803/full#supplementary-material
1. Hashemi H, Rezvan F, Pakzad R, Ansaripour A, Heydarian S, Yekta A, et al. Global and regional prevalence of diabetic retinopathy; a comprehensive systematic review and meta-analysis. Semin Ophthalmol (2021) 157:1–16. doi: 10.1080/08820538.2021.1962920
2. Cheung N, Mitchell P, Wong TY. Diabetic retinopathy. Lancet (London England) (2010) 376:124–36. doi: 10.1016/S0140-6736(09)62124-3
3. Teo ZL, Tham YC, Yu M, Chee ML, Rim TH, Cheung N, et al. Global prevalence of diabetic retinopathy and projection of burden through 2045: Systematic review and meta-analysis. Ophthalmology (2021) 128:1580–91. doi: 10.1016/j.ophtha.2021.04.027
4. Shoeibi N, Bonakdaran S. Is there any correlation between diabetic retinopathy and risk of cardiovascular disease? Curr Diabetes Rev (2017) 13:81–6. doi: 10.2174/1573399812666151012115355
5. Gao L, Zhao W, Yang JK, Qin MZ. Proliferative diabetic retinopathy in patients with type 2 diabetes correlates with the presence of atherosclerosis cardiovascular disease. Diabetol Metab Syndr (2021) 13:48. doi: 10.1186/s13098-021-00666-z
6. Rasheed R, Pillai GS, Kumar H, Shajan AT, Radhakrishnan N, Ravindran GC. Relationship between diabetic retinopathy and diabetic peripheral neuropathy - neurodegenerative and microvascular changes. Indian J Ophthalmol (2021) 69:3370–5. doi: 10.4103/ijo.IJO_1279_21
7. Ellis TP, Choudhury RH, Kaul K, Chopra M, Kohner EM, Tarr JM, et al. Diabetic retinopathy and atherosclerosis: is there a link? Curr Diabetes Rev (2013) 9:146–60. doi: 10.2174/1573399811309020006
8. Zhang L, Chen B, Tang L. Metabolic memory: mechanisms and implications for diabetic retinopathy. Diabetes Res Clin Pract (2012) 96:286–93. doi: 10.1016/j.diabres.2011.12.006
9. Wang W, Lo ACY. Diabetic retinopathy: Pathophysiology and treatments. Int J Mol Sci (2018) 19:1–14. doi: 10.3390/ijms19061816
10. Lechner J, O'Leary OE, Stitt AW. The pathology associated with diabetic retinopathy. Vision Res (2017) 139:7–14. doi: 10.1016/j.visres.2017.04.003
11. Kollias AN, Ulbig MW. Diabetic retinopathy: Early diagnosis and effective treatment. Deutsches Arzteblatt Int (2010) 107:75–83. doi: 10.3238/arztebl.2010.0075
12. Hiscott P, Paraoan L, Choudhary A, Ordonez JL, Al-Khaier A, Armstrong DJ. Thrombospondin 1, thrombospondin 2 and the eye. Prog Retinal Eye Res (2006) 25:1–18. doi: 10.1016/j.preteyeres.2005.05.001
13. Sims JN, Lawler J. Thrombospondin-1-Based antiangiogenic therapy. J Ocular Pharmacol Ther (2015) 31:366–70. doi: 10.1089/jop.2015.0018
14. Huang T, Sun L, Yuan X, Qiu H. Thrombospondin-1 is a multifaceted player in tumor progression. Oncotarget (2017) 8:84546–58. doi: 10.18632/oncotarget.19165
15. Wang S, Gottlieb JL, Sorenson CM, Sheibani N. Modulation of thrombospondin 1 and pigment epithelium-derived factor levels in vitreous fluid of patients with diabetes. Arch Ophthalmol (2009) 127:507–13. doi: 10.1001/archophthalmol.2009.53
16. Wu ZF, Wang SJ, Sorenson CM, Sheibani N. Attenuation of retinal vascular development and neovascularization in transgenic mice over-expressing thrombospondin-1 in the lens. Dev Dynamics (2006) 235:1908–20. doi: 10.1002/dvdy.20837
17. Wang Y, Wang S, Sheibani N. Enhanced proangiogenic signaling in thrombospondin-1-deficient retinal endothelial cells. Microvascular Res (2006) 71:143–51. doi: 10.1016/j.mvr.2006.02.004
18. Ishikawa K, Yoshida S, Kobayashi Y, Zhou Y, Nakama T, Nakao S, et al. Microarray analysis of gene expression in fibrovascular membranes excised from patients with proliferative diabetic retinopathy. Invest Ophthalmol Visual Sci (2015) 56:932–46. doi: 10.1167/iovs.14-15589
19. Lam JD, Oh DJ, Wong LL, Amarnani D, Park-Windhol C, Sanchez AV, et al. Identification of RUNX1 as a mediator of aberrant retinal angiogenesis. Diabetes (2017) 66:1950–6. doi: 10.2337/db16-1035
20. Wang Z, Monteiro CD, Jagodnik KM, Fernandez NF, Gundersen GW, Rouillard AD, et al. Extraction and analysis of signatures from the gene expression omnibus by the crowd. Nat Commun (2016) 7:1–11. doi: 10.1038/ncomms12846
21. Bansal A, Pinney SE. DNA Methylation and its role in the pathogenesis of diabetes. Pediatr Diabetes (2017) 18:167–77. doi: 10.1111/pedi.12521
22. Maghbooli Z, Hossein-nezhad A, Larijani B, Amini M, Keshtkar A. Global DNA methylation as a possible biomarker for diabetic retinopathy. Diabetes/metabolism Res Rev (2015) 31:183–9. doi: 10.1002/dmrr.2584
23. Kowluru RA, Santos JM, Mishra M. Epigenetic modifications and diabetic retinopathy. BioMed Res Int (2013) 2013:635284. doi: 10.1155/2013/635284
24. Wu J, Liu LL, Cao M, Hu A, Hu D, Luo Y, et al. DNA Methylation plays important roles in retinal development and diseases. Exp Eye Res (2021) 211:108733. doi: 10.1016/j.exer.2021.108733
25. Kumari N, Karmakar A, Ganesan SK. Targeting epigenetic modifications as a potential therapeutic option for diabetic retinopathy. J Cell Physiol (2020) 235:1933–47. doi: 10.1002/jcp.29180
26. Zhi D, Aslibekyan S, Irvin MR, Claas SA, Borecki IB, Ordovas JM, et al. SNPs located at CpG sites modulate genome-epigenome interaction. Epigenetics (2013) 8:802–6. doi: 10.4161/epi.25501
27. Hellman A, Chess A. Extensive sequence-influenced DNA methylation polymorphism in the human genome. Epigenet Chromatin (2010) 3:11. doi: 10.1186/1756-8935-3-11
28. Smith AK, Kilaru V, Kocak M, Almli LM, Mercer KB, Ressler KJ, et al. Methylation quantitative trait loci (meQTLs) are consistently detected across ancestry, developmental stage, and tissue type. BMC Genomics (2014) 15:1–11. doi: 10.1186/1471-2164-15-145
29. Hawe JS, Wilson R, Schmid KT, Zhou L, Lakshmanan LN, Lehne BC, et al. Genetic variation influencing DNA methylation provides insights into molecular mechanisms regulating genomic function. Nat Genet (2022) 54:18–+. doi: 10.1038/s41588-021-00969-x
30. Chinese Medical Association Diabetic Society. Guidelines for the prevention and control of type 2 diabetes in China (2017 edition). Chin J Pract Internal Med (2018) 38:292–344. doi: 10.19538/j.nk2018040108
31. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet (2007) 81:559–75. doi: 10.1086/519795
32. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (2005) 21:263–5. doi: 10.1093/bioinformatics/bth457
33. Preacher KJ, Hayes AF. SPSS And SAS procedures for estimating indirect effects in simple mediation models. Behav Res Methods Instruments Comput (2004) 36:717–31. doi: 10.3758/bf03206553
34. Hayes AF, Rockwood NJ. Regression-based statistical mediation and moderation analysis in clinical research: Observations, recommendations, and implementation. Behav Res Ther (2017) 98:39–57. doi: 10.1016/j.brat.2016.11.001
35. Farnoodian M, Sorenson CM, Sheibani N. Negative regulators of angiogenesis, ocular vascular homeostasis, and pathogenesis and treatment of exudative AMD. J Ophthalmic Vision Res (2018) 13:470–86. doi: 10.4103/jovr.jovr_67_18
36. Bian Z, Sun Q, Wang J, Hu Y, Qie H, Zhao X, et al. Clinical value of thrombospondin-1 in patients with type 2 diabetic retinopathy. Department Endocrinol (2017) 25:703–6. doi: 10.3969/j.issn.1006-6187.2017.08.006
37. Wang J, Zhang X, Zhang H. Expression of thrombospondin-1 in STZ-induced diabetic retinopathy in rats. J Mol Cell Biol (2006) 39:431–7. doi: 10.3321/j.issn:1673-520X.2006.05.006
38. Mishra M, Kowluru RA. The role of DNA methylation in the metabolic memory phenomenon associated with the continued progression of diabetic retinopathy. Invest Ophthalmol Visual Sci (2016) 57:5748–57. doi: 10.1167/iovs.16-19759
39. Lan CC, Huang SM, Wu CS, Wu CH, Chen GS. High-glucose environment increased thrombospondin-1 expression in keratinocytes via DNA hypomethylation. Trans Res (2016) 169:91–101 e1-3. doi: 10.1016/j.trsl.2015.11.002
40. Villicana S, Bell JT. Genetic impacts on DNA methylation: research findings and future perspectives. Genome Biol (2021) 22:1–35. doi: 10.1186/s13059-021-02347-6
41. Liu M, Li J, Huang Z, Li Y. Gastric cancer risk-scoring system based on analysis of a competing endogenous RNA network. Transl Cancer Res (2020) 9:3889–902. doi: 10.21037/tcr-19-2977
Keywords: meQTL, DNA methylation, THBS1, diabetic retinopathy, genetic regulation
Citation: Li Y, Gong C, Xu Y, Liang X, Chen X, Hong W and Yan J (2022) Genetic regulation of THBS1 methylation in diabetic retinopathy. Front. Endocrinol. 13:991803. doi: 10.3389/fendo.2022.991803
Received: 12 July 2022; Accepted: 25 October 2022;
Published: 14 November 2022.
Edited by:
Jian Ma, Harbin Medical University, ChinaReviewed by:
Sunbin Deng, Harvard Medical School, United StatesCopyright © 2022 Li, Gong, Xu, Liang, Chen, Hong and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junxia Yan, MjA0NTc0NTZAcXEuY29t; Wenxu Hong, c3pibG9vZGNlbnRlckBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.