
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 28 September 2022
Sec. Cancer Endocrinology
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.989030
This article is part of the Research Topic Functional Epigenetic Regulation in Metabolic Diseases View all 9 articles
Background: Bladder cancer is the most common leading cause of mortality around the world. Previous studies have indicated that genetic factors are significantly associated with bladder cancer progression—for instance, the CYP2C8 gene is involved in bladder cancer progression. However, little is known about the impact of CYP2C8 genetic polymorphisms on bladder cancer risk. We aimed to detect the association between CYP2C8 variations and bladder cancer susceptibility.
Methods: This study included 550 healthy subjects and 217 bladder cancer patients. The odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to determine the correlation of CYP2C8 polymorphisms with bladder cancer risk. Multifactor dimensionality reduction (MDR) was carried out to investigate the influence of single-nucleotide polymorphism (SNP)–SNP interactions on bladder cancer.
Results: Our study showed that two SNPs were significantly associated with an increased risk of bladder cancer (rs1934951: OR 1.96, 95% CI 1.37–2.82, p = 2.67E-04; rs17110453: OR 1.89, 95% CI 1.35–2.67, p = 2.53E-04). On the contrary, two SNPs identified in the study had protective effects on bladder cancer (rs1934953: OR 0.26, 95% CI 0.14–0.47, p = 1.20E-05; rs2275620: OR 0.40, 95% CI 0.21–0.76, p = 0.005). The MDR analysis suggested that the combination of rs1934953, rs1934951, rs2275620, and rs17110453 was the best model to predict bladder cancer (CVC 10/10, testing accuracy 0.6720, p < 0.0001).
Conclusion: There was a significant association between CYP2C8 polymorphisms (rs1934953, rs1934951, rs2275620, and rs17110453) and susceptibility to bladder cancer.
Bladder cancer is one of the most common tumors in the urinary system all over the world (1, 2), ranking as the seventh and sixth leading cause of mortality and morbidity among women and men, respectively, with approximately 199,922 deaths and 549,393 new diagnoses in 2018 worldwide (2). The incidence of bladder cancer in men is three to four times higher than that in women, and the incidence in both men and women increases with age (3). Bladder cancer is a complex and multifactorial disease affected by some risk factors such as sex, age, tobacco smoking, environmental pollution, chemical carcinogen exposure, and lifestyle (4–8). However, not all individuals exposed to risk factors develop bladder cancer, indicating that individual genetic diversity plays a crucial role in bladder cancer occurrence. Moreover, an increasing number of studies have revealed that genetic factors have become one of the most important factors in the pathogenesis of bladder cancer (9, 10). The molecular mechanism of bladder cancer is mainly due to exogenous metabolic changes and mutations in genes related to DNA repair, cell proliferation, and tumor inhibition (11–13). Current evidence have suggested that genetic polymorphisms are significantly correlated with the development of bladder cancer (14–17). The study of genetic polymorphisms has enhanced our understanding of the pathogenesis of bladder cancer, so it is of great significance to find more genetic risk factors.
Single-nucleotide polymorphism (SNP) is not only the most common genetic diversity but also a new genetic biomarker, which can affect the gene regulation function by changing gene sequences, ultimately resulting in the alteration of its functional properties. A growing number of SNPs are observed to be related to bladder cancer (18). Cytochrome P450 2C8 (CYP2C8) is a member of the human CYP2C enzyme family. It has been certified that CYP2C8 is involved in the metabolism of many exogenous compounds (19). CYP2C8 is highly expressed in human liver, and it also can be detected in the duodenum, ovary, heart, kidney, and mammary gland (20, 21). The abnormal expression of the CYP2C8 gene is involved in the progression of many human cancers, such as hepatocellular carcinoma, breast cancer, prostate cancer, and endometrial tumor (22, 23). We noticed that CYP2C8 showed a significantly higher expression in bladder urothelial carcinoma compared with that in normal tissue (http://ualcan.path.uab.edu/cgi-bin/TCGAExResultNew2.pl?genenam=CYP2C8&ctype=BLCA). Taken together, we speculated that the polymorphisms of the CYP2C8 gene play a potential role in bladder cancer development.
To our knowledge, there is no study focusing on the association between CYP2C8 polymorphisms and bladder cancer risk. Therefore, our present study was performed to investigate whether the genetic polymorphisms (rs1934953, rs1934951, rs2275620, and rs17110453) in the CYP2C8 gene can affect the bladder cancer susceptibility in the Chinese population.
This case–control study included 217 bladder cancer patients and 550 unrelated healthy subjects admitted to the Shaanxi Provincial Cancer Hospital. We informed each subject about the purpose of the study and obtained informed consent from all participants before conducting our research. This study was approved by the Ethics Committees of Shaanxi Provincial Cancer Hospital (no. 2017SF-152). All procedures performed in the study were in accordance with the Helsinki Declaration. The case group must meet the following inclusion criteria: (1) patients with newly diagnosed, histologically confirmed bladder cancer; (2) patients with age from 18 to 80 years; (3) no preoperative chemoradiotherapy was performed; and (4) no other tumors. The exclusion criteria for all patients were as follows: (1) previous diagnosis of any cancer, metastasized cancer, and serum prostate-specific antigen (>2.5 ng/ml); (2) a family history of cancers, including bladder cancer; (3) previous chemotherapy, radiotherapy, or radical cystectomy; and (4) those with bladder tumors secondary to other malignancies. The control subjects were healthy people who have physical examinations at the same hospital with cases. The inclusion criteria for the controls were as follows: (1) healthy controls were genetically unrelated subjects and were matched to cases on age and gender; (2) there was no gross or microscopic hematuria; and (3) the ultrasonography of the bladder was normal. The controls with a previous malignancy, metastasized cancer from other or unknown origin, and family history of cancers and familial or genetic diseases were excluded. Subjects with any degree of hematuria, benign prostate hyperplasia, urinary symptoms, history of prostatitis, and pre-cancerous lesions were excluded from the study. Demographic and pathological data including gender, age, and clinical stage were obtained from the participants’ medical records. A family history of bladder cancer was considered positive when a first- or second-degree relative of the participants was diagnosed with bladder cancer. None of the individuals included in this study was under occupational exposure to hazardous carcinogens related to bladder cancer.
The detailed steps of SNP selection are as follows: (1) We obtained the physical position of the CYP2C8 gene on chromosome 10: 95,036,772–95,069,497 through the human e!GRCh37 database (http://asia.ensembl.org/Homo_sapiens/Info/Index). In the VCF to PED Converter window (http://grch37.ensembl.org/Homo_sapiens/Tools/VcftoPed), we entered the gene location, selected the Chinese Han population in Beijing as population, and downloaded the ped and info file for the SNPs of CYP2C8. We obtained 103 SNPs within CYP2C8 from the database; (2) Then, we used Haploview software for quality control [minor allele frequency >5%, minor genotype >75%, r2 < 0.8, and Hardy–Weinberg equilibrium (HWE) >0.05] to select tag-SNP. Finally, four SNPs (including rs1934953, rs1934951, rs2275620, and rs17110453) were selected for investigation. A DNA extraction kit was used for extracting the genomic DNA from peripheral blood samples. Agena Design software was used to design the PCR amplification primers. The detailed information of the primers in this study is listed in Table 1. SNP genotyping was determined using Agena MassARRAY iPLEX platform. Besides this, the data of genotypes was organized and analyzed by Agena Bioscience TYPER version 4.0 software.
All statistical tests in this study were two-sided and carried out with SPSS 22.0 software. The two-tailed p-value <0.05 was considered to have a statistical difference. χ2 test and Student’s t-test were used to detect the statistical differences in age and sex between cases and controls, respectively. HWE in controls was determined by Fisher’s exact test. The impact of CYP2C8 polymorphisms on the risk of bladder cancer was tested by logistic regression analysis under five genetic models (allele, dominant, codominant, log-additive, and recessive). We also investigated the association of stratification analyses. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to check the associations. Finally, we explored the influence of SNP–SNP interactions on bladder cancer via multifaceted dimensionality reduction (MDR) analysis.
The distributions of demographic variables of bladder cancer and healthy individuals are shown in Table 2. The average age of the cases was 64.40 ± 10.99 years, and the average age of the controls was 63.92 ± 6.62 years. There was no statistical difference in age between the patients and controls (p = 0.549), while a significant difference in sex was observed between the two groups (p = 0.001).
Four SNPs in the CYP2C8 gene were successfully genotyped in the study. As presented in Table 3, all SNPs in the control group were in line with the HWE (all p >0.05). The effect of CYP2C8 variants on bladder cancer was then evaluated with logistic regression analysis, as shown in Table 4. Our study demonstrated that rs1934951 (codominant model: OR 1.96, p = 2.67E-04; dominant model: OR 1.74, p = 0.002) and rs17110453 (codominant model: OR 1.89, p = 2.53E-04; dominant model: OR 1.63, p = 0.004; recessive model: OR 1.46, p = 0.013) were significantly associated with an increased susceptibility to bladder cancer. Conversely, rs1934953 (allele: OR 0.61, p = 2.70E-05; codominant model: OR 0.26, p = 1.20E-05; dominant model: OR 0.62, p = 0.005; recessive model: OR 0.31, p = 5.38E-05; log-additive: OR 0.58, p = 2.17E-05) and rs2275620 (codominant model: OR 0.40, p = 0.005; recessive model: OR 0.32, p = 1.41E-04) showed a protective effect on the risk of bladder cancer.
The stratification of individuals according to their age (Table 5) indicated that rs1934951 (>65 years: CT vs. CC, OR 2.17, p = 0.009; TC-TT vs. CC, OR 1.82, p = 0.037; ≤65 years: CT vs. CC, OR 1.87, p = 0.016; TC-TT vs. CC, OR 1.74, p = 0.030, respectively) and rs17110453 (>65 years: AC vs. AA, OR 1.96, p = 0.016; ≤65 years: AC vs. AA, OR 1.93, p = 0.007; AC-CC vs AA, OR 1.64, p = 0.038) significantly increased the susceptibility to bladder cancer. rs1934953 was related to decreased susceptibility to bladder cancer in people aged >65 years (T vs. C, OR 0.62, p = 0.007; TT vs. CC, OR 0.29, p = 0.005; TT vs. CC-TC, OR 0.34, p = 0.006) and aged ≤65 years (T vs. C, OR 0.58, p = 6.24E-04; TT vs. CC, OR 0.25, p = 0.003; TT vs. CC-TC, OR 0.30, p = 0.008). Besides this, rs2275620 also had a protective effect on bladder cancer risk in individuals aged >65 years (TT vs. AA-TA, OR 0.44, p = 0.034) and aged ≤65 years (TT vs. AA-TA, OR 0.21, p = 0.004).
MDR analysis was carried out to explore the correlation between SNP–SNP interactions and bladder cancer. The MDR method selects variables with attribute interactions on the basis of entropy measures for evaluating the information gain associated with attribute interactions. The patterns of entropy recapitulated the main and/or interaction effect of each pairwise combination of attributes. As shown in Figure 1, the interaction map with negative percent entropy represented the independence or redundancy of each pairwise combination of attributes (-8.61, -5.72, -5.04, -4.75, -3.27, and -3.23%, respectively, shown in blue and green), and there was a strong independence or redundancy between rs1934953 and rs2275620, with the information gain values of -8.61%. Table 6 shows that the combination of rs1934953, rs1934951, rs2275620, and rs17110453 was the best model to predict bladder cancer (CVC = 10/10, testing accuracy = 0.6720, p < 0.0001). The best single-locus model was rs17110453 (CVC = 9/10, testing accuracy = 0.6083, p < 0.0001). The best two-locus model consisted of rs1934953 and rs1934951 (CVC = 6/10, testing accuracy = 0.6242, p < 0.0001). rs1934953, rs2275620, and rs17110453 formed the best three-locus model (CVC = 7/10, testing accuracy = 0.6561, p < 0.0001).
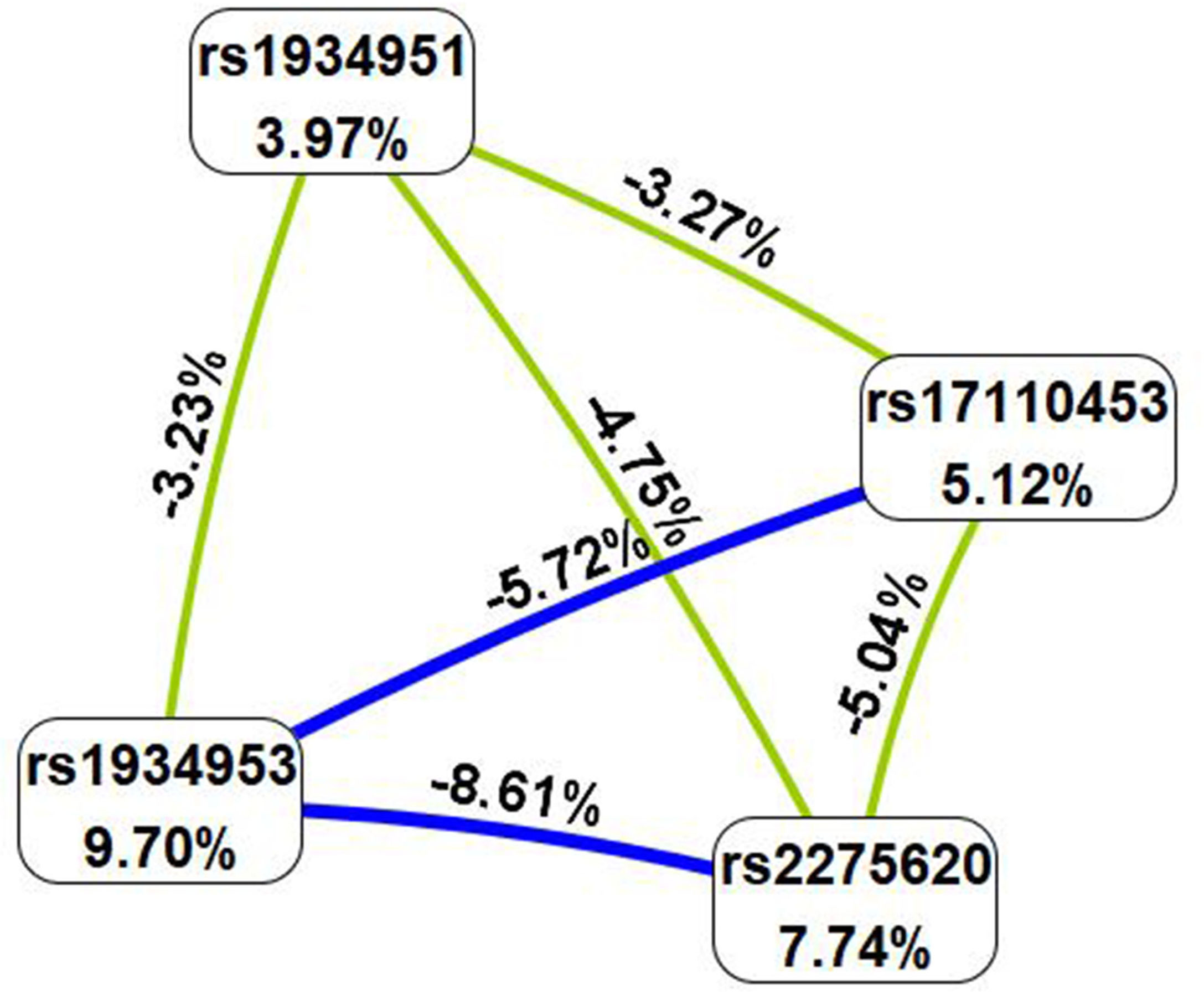
Figure 1 SNP–SNP interaction map. Values in nodes represent the information gains of individual attribute (main effects). Values between nodes are information gains of each pair of attributes (interaction effects). Blue and green with negative percent entropy indicate redundancy or independence.
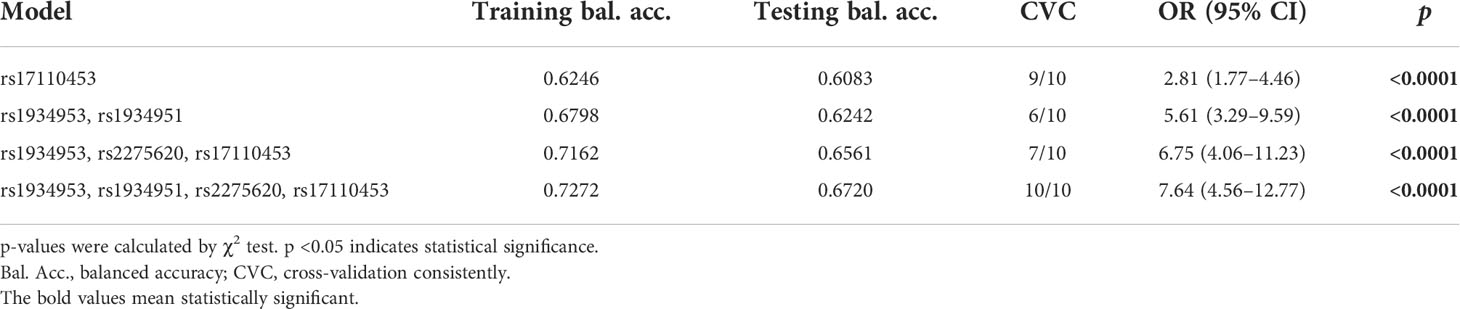
Table 6 Analysis of the SNP–SNP interaction models with the multifactor dimensionality reduction method.
Genetic factors play a significant role in the development and progression of bladder cancer. In this study, we firstly examined the correlation of CYP2C8 genetic variants with the risk of bladder cancer in the Chinese population. Our findings indicated that the SNPs in the CYP2C8 gene were significantly related to bladder cancer susceptibility. Our research provides a new perspective for understanding the molecular mechanism of the correlation between genetic background and carcinogenesis in bladder cancer.
CYP2C8 is located on chromosome 10q24. One study has shown that CYP2C8 polymorphisms have a certain functional significance (24). Some studies have shown that functional polymorphisms that affect the expression or activity of the CYP2C8 gene can significantly increase the susceptibility to bladder cancer (25, 26). Various studies have been conducted on the association of CYP2C8 polymorphisms with human cancers—for example, Golpar et al. have reported that rs1058930 of CYP2C8 could significantly increase breast cancer risk (27). It has been reported that CYP2C8 polymorphisms can significantly change the imatinib metabolism in patients with leukemia through both gain- and loss-of-function mechanism (28). Another study has indicated that CYP2C8 variations can influence ovarian cancer risk (29). However, the relationship between CYP2C8 polymorphisms and bladder cancer risk has not been reported. In our study, we found that rs1934951 and rs17110453 in CYP2C8 significantly increased the risk of bladder cancer. rs1934953 and rs2275620 were related to a reduced risk of bladder cancer. The stratification analysis suggested that the impact of CYP2C8 polymorphisms on bladder cancer susceptibility may be independent of age. This result is contrary to the fact that age is a risk factor for bladder cancer, which may be caused by the small sample size.
The study of SNP–SNP interactions is helpful to find more risk factors for bladder cancer. Interestingly, we observed that there was a strong independence or redundancy between rs1934953 and rs2275620. In addition, the combination of rs1934953, rs1934951, rs2275620, and rs17110453 was the best model to predict bladder cancer.
Some limitations in our present study should be noted. First, the sample size is relatively small, and we will further verify our conclusions by expanding the sample size in the future. Second, the associations stratified by smoking status and clinical stage were not detected on account of the limited information obtained from the medical records. Third, although we determined the impact of CYP2C8 polymorphisms on bladder cancer risk, the molecular mechanism of CYP2C8 polymorphisms affecting bladder cancer has not been investigated in this work. In spite of the abovementioned shortage, our study is the first to examine the association of CYP2C8 polymorphisms with bladder cancer risk, which may give a new biomarker for the diagnosis or prevention of bladder cancer in the Chinese population.
The original contributions presented in the study are publicly available. This data can be found here: https://doi.org/10.5281/zenodo.7074453.
The studies involving human participants were reviewed and approved by the Shaanxi Provincial Cancer Hospital and the 1964 Helsinki Declaration. The patients/participants provided their written informed consent to participate in this study.
WQ and JZ were responsible for the study design. WQ processed the data and wrote the manuscript. YC and JL recruited the study participants. FZ contributed to the primer design and data analysis. JZ revised the paper. All authors contributed to the article and approved the submitted version.
This study was supported by the Key Research & Development Projects in Shaanxi Province (grant no. 2022SF-465 to W. Qu), the Science Foundation of Shaanxi Provincial People’s Hospital (grant no. 2021YJY-30 to J. Zhou) and the Shaanxi Provincial Natural Science Foundation (grant no. 2020JM-657 to J. Zhou).
We thank all participants in this study. We also thank the Shaanxi Provincial Cancer Hospital for their helping with sample collections.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. "Bladder cancer incidence and mortality: A global overview and recent trends". Eur Urol. (2017) 71(1):96–108.
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. "Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries". CA Cancer J Clin (2018) 68(6):394–424.
3. Ferlay J, Shin H, Bray F, Forman D, Mathers C, Parkin D. "Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008". Int J Cancer (2010) 127(12):2893–917.
4. Chen C, Chiou H, Hsueh Y, Chen C, Yu H, Pu Y. "Clinicopathological characteristics and survival outcome of arsenic related bladder cancer in taiwan". J Urol (2009) 181(2):547–52.
5. Burger M, Catto J, Dalbagni G, Grossman H, Herr H, Karakiewicz P, et al. "Epidemiology and risk factors of urothelial bladder cancer". Eur Urol (2013) 63(2):234–41.
6. Polat F, Diler SB, Azazi İ, Öden A. "T-786C, G894T, and intron 4 VNTR (4a/b) polymorphisms of the endothelial nitric oxide synthase gene in bladder cancer cases". Asian Pac. J Cancer Prev (2015) 16(6):2199–202.
7. Kamat A, Hahn N, Efstathiou J, Lerner S, Malmström P, Choi W, et al. "Bladder cancer". Lancet (London England) (2016) 388(10061):2796–810.
8. Koutros S, Silverman D, Alavanja M, Andreotti G, Lerro C, Heltshe S, et al. "Occupational exposure to pesticides and bladder cancer risk". Int J Epidemiol (2016) 45(3):792–805.
9. Wu X, Hildebrandt M, Chang D. "Genome-wide association studies of bladder cancer risk: a field synopsis of progress and potential applications". Cancer metastasis Rev (2009) 28:269–80.
10. Benhamou S, Bonastre J, Groussard K, Radvanyi F, Allory Y, Lebret T. "A prospective multicenter study on bladder cancer: the COBLAnCE cohort". BMC Cancer (2016) 16(1):837.
11. Jalali Nadoushan MR, Taheri T, Jouian N, Zaeri F. "Overexpression of HER-2/neu oncogene and transitional cell carcinoma of bladder". Urol. J (2007) 4(3):151–4.
12. Karimianpour N, Mousavi-Shafaei P, Ziaee AA, Akbari MT, Pourmand G, Abedi A, et al. "Mutations of RAS gene family in specimens of bladder cancer". Urol. J (2008) 5(4):237–42.
13. Nanda MS, Sameer AS, Syeed N, Shah ZA, Murtaza I, Siddiqi MA, et al. "Genetic aberrations of the K-ras proto-oncogene in bladder cancer in kashmiri population". Urol. J (2010) 7(3):168–73.
14. Selinski S. "Discovering urinary bladder cancer risk variants: Status quo after almost ten years of genome-wide association studies". EXCLI. J (2017) 16:1288–96.
15. Selinski S, Blaszkewicz M, Ickstadt K, Gerullis H, Otto T, Roth E, et al. "Identification and replication of the interplay of four genetic high-risk variants for urinary bladder cancer". Carcinogenesis (2017) 38(12):1167–79.
16. Mao F, Niu X, Gu S, Ji L, Wei B, Wang H. "The association between matrix metalloproteinase-7 genetic variant and bladder cancer risk in a Chinese han population". Clin Exp Med (2019) 19(4):565–70.
17. Huang Y, Wang H, Fu S, Luan T, Zuo Y, Li N, et al. "Association of rs8444 polymorphism in the LASS2 3'-UTR and bladder cancer risk in Chinese population". Eur J Cancer Prev Off J Eur Cancer Prev Organisation (ECP) (2020) 29(4):329–37.
18. Fu YP, Kohaar I, Rothman N, Earl J, Figueroa JD, Ye Y, et al. "Common genetic variants in the PSCA gene influence gene expression and bladder cancer risk". Proc Natl Acad Sci U.S.A. (2012) 109(13):4974–9.
19. Totah RA, Rettie AE. "Cytochrome P450 2C8: substrates, inhibitors, pharmacogenetics, and clinical relevance". Clin Pharmacol Ther (2005) 77(5):341–52.
20. Klose TS, Blaisdell JA, Goldstein JA. "Gene structure of CYP2C8 and extrahepatic distribution of the human CYP2Cs". J Biochem Mol Toxicol (1999) 13(6):289–95.
21. Delozier TC, Kissling GE, Coulter SJ, Dai D, Foley JF, Bradbury JA, et al. "Detection of human CYP2C8, CYP2C9, and CYP2J2 in cardiovascular tissues". Drug Metab Dispos. (2007) 35(4):682–8.
22. Ren X, Ji Y, Jiang X, Qi X. "Downregulation of CYP2A6 and CYP2C8 in tumor tissues is linked to worse overall survival and recurrence-free survival from hepatocellular carcinoma". Biomed Research International (2018) 2018:5859415.
23. van Eijk M, Boosman RJ, Schinkel AH, Huitema ADR, Beijnen JH. "Cytochrome P450 3A4, 3A5, and 2C8 expression in breast, prostate, lung, endometrial, and ovarian tumors: relevance for resistance to taxanes". Cancer Chemother Pharmacol (2019) 84(3):487–99.
24. Daily EB, Aquilante CL. "Cytochrome P450 2C8 pharmacogenetics: A review of clinical studies". Pharmacogenomics (2009) 10(9):1489–510.
25. Rachakonda PS, Hosen I, de Verdier PJ, Fallah M, Heidenreich B, Ryk C, et al. "TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism". Proc Natl Acad Sci U.S.A. (2013) 110(43):17426–31.
26. Giedl J, Rogler A, Wild A, Riener MO, Filbeck T, Burger M, et al. "TERT core promotor mutations in early-onset bladder cancer". J Cancer (2016) 7(8):915–20.
27. Golmohammadzadeh G, Mohammadpour A, Ahangar N, Shokrzadeh M. "Polymorphisms in phase I (CYP450) genes CYP1A1 (rs4646421), CYP1B1 (rs1056836), CYP19A1 (rs749292) and CYP2C8 (rs1058930) and their relation to risk of breast cancer: A case-control study in mazandaran province in north of iran". Open Access Maced. J Med Sci (2019) 7(15):2488–96.
28. Barratt DT, Cox HK, Menelaou A, Yeung DT, White DL, Hughes TP, et al. "CYP2C8 genotype significantly alters imatinib metabolism in chronic myeloid leukaemia patients". Clin Pharmacokinet (2017) 56(8):977–85.
Keywords: CYP2C8, genetic variants, susceptibility, bladder cancer, case–control study
Citation: Qu W, Zhang F, Cheng Y, Li J and Zhou J (2022) The impact of genetic variants in the CYP2C8 gene on bladder cancer susceptibility. Front. Endocrinol. 13:989030. doi: 10.3389/fendo.2022.989030
Received: 08 July 2022; Accepted: 01 September 2022;
Published: 28 September 2022.
Edited by:
Wei Wu, Nanjing Medical University, ChinaReviewed by:
Octavian Sabin Tataru, George Emil Palade University of Medicine, Pharmacy, Sciences and Technology of Târgu Mureş, RomaniaCopyright © 2022 Qu, Zhang, Cheng, Li and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiancheng Zhou, MjY2NzUwODVAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.