
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol., 23 September 2022
Sec. Neuroendocrine Science
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.973862
This article is part of the Research TopicGenomic and Transcriptomic Insights into Neuroendocrine Evolution and FunctionView all 8 articles
Bilateria have bilateral symmetry and are subdivided into Deuterostomia (animals like vertebrates) and Protostomia (animals like insects and mollusks). Neuropeptides occur in both Proto- and Deuterostomia and they are frequently structurally related across these two lineages. For example, peptides belonging to the oxytocin/vasopressin family exist in both clades. The same is true for the G protein-coupled receptors (GPCRs) of these peptides. These observations suggest that these neuropeptides and their GPCRs were already present in the common ancestor of Proto- and Deuterostomia, which lived about 700 million years ago (MYA). Furthermore, neuropeptides and their GPCRs occur in two early-branching phyla that diverged before the emergence of Bilateria: Cnidaria (animals like corals and sea anemones), and Placozoa (small disk-like animals, feeding on algae). The sequences of these neuropeptides and their GPCRs, however, are not closely related to those from Bilateria. In addition, cnidarian neuropeptides and their receptors are not closely related to those from Placozoa. We propose that the divergence times between Cnidaria, Placozoa, and Bilateria might be too long for recognizing sequence identities. Leucine-rich repeats-containing GPCRs (LGRs) are a special class of GPCRs that are characterized by a long N-terminus containing 10-20 leucine-rich domains, which are used for ligand binding. Among the ligands for LGRs are dimeric glycoprotein hormones, and insulin-like peptides, such as relaxin. LGRs have been found not only in Proto- and Deuterostomia, but also in early emerging phyla, such as Cnidaria and Placozoa. Humans have eight LGRs. In our current review, we have revisited the annotations of LGRs from the sea anemone Nematostella vectensis and the placozoan Trichoplax adhaerens. We identified 13 sea anemone LGRs and no less than 46 LGRs from T. adhaerens. All eight human LGRs appear to have orthologues in sea anemones and placozoans. LGRs and their ligands, therefore, have a long evolutionary history, going back to the common ancestor of Cnidaria and Placozoa.
Neuropeptides have a broad distribution in the animal kingdom and a long evolutionary history, occurring in both Deuterostomia (animals like mammals and other chordates) and Protostomia (animals like insects, crustaceans, mollusks, round- and flatworms) (Figure 1) (1, 2). The Proto- and Deuterostomia diverged about 700 million years ago (MYA) (3), which also marked the birth of the Bilateria (animals with a bilateral symmetry) (Figure 1). When comparing the neuropeptides from Proto- and Deuterostomia, we can occasionally observe that their sequences are conserved (1, 2, 4, 5). Examples of such conserved sequences between the two evolutionary lineages are peptides belonging to the oxytocin/vasopressin family, neuropeptide-Y family, gastrin/cholecystokinin (CCK)-like peptides, and tachykinin-like peptides (1, 2, 4–10). The existence of identical or nearly identical neuropeptide families in Proto- and Deuterostomia suggests that these peptides originated in the common ancestor of these two evolutionary lineages.
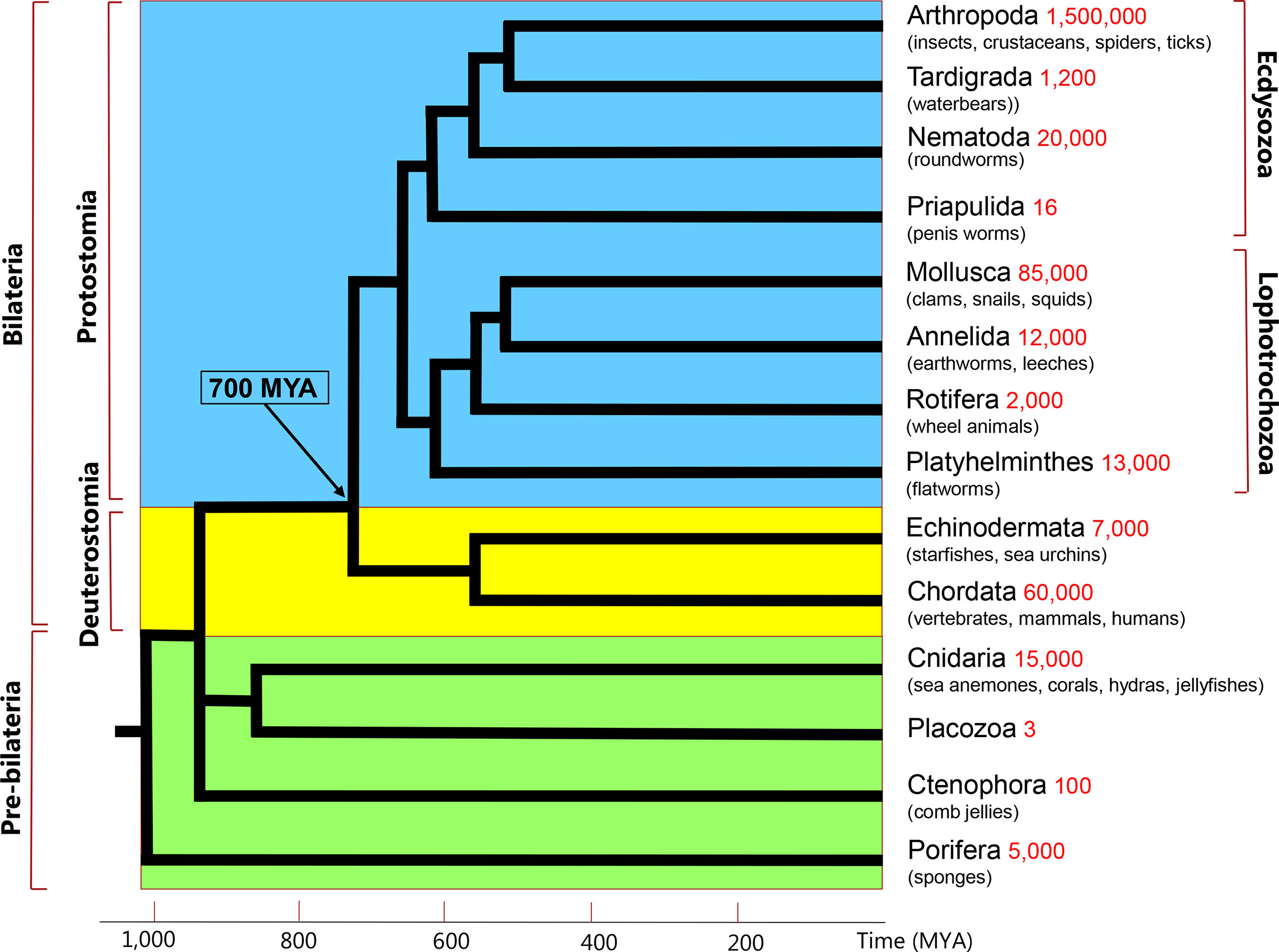
Figure 1 Simplified phylogenetic tree of metazoans. These animals can be subdivided into Protostomia (branches with blue background), and Deuterostomia (braches with yellow background). The Proto- and Deuterostomia originated 700 million years ago (MYA), see arrow and time scale on the X-axis. These two taxa together are also called Bilateria, because of their bilateral symmetry. There are four phyla (Cnidaria, Placozoa, Ctenophora, Porifera) that diverged before the emergence of Bilateria (branches with green background). Two of these early-branching phyla, the Cnidaria and Placozoa, which probably form a monophyletic clade, are discussed in the current review. The phylogenetic position of Ctenophora is still uncertain. The phyla listed in the right column are extant phyla and do not represent all extant animal phyla. Their estimated species numbers are given in red.
For some of the neuropeptide families, their members have striking sequence identities in Proto- and Deuterostomia, such as members of the oxytocin/vasopressin neuropeptide family, where six out of nine amino acid residues are identical, including an essential cystine bridge between Cys-1 and Cys-6 of the peptide sequence (7). Many chordate gastrin/CCKs have YGWMDFamide, YMGWMDFamide, or YYGWMDFamide as their C-terminal sequences, where the Y residues are sulfated (10). Sulfation is unusual in neuropeptides and is a hallmark for chordate gastrin/CCKs (10). Protostomes also have gastrin/CCK-like peptides called sulfakinins. For example, the cockroach Leucophea maderae, which was the first insect from which sulfakinins have been isolated, has a sulfakinin peptide with the C-terminal sequence YGHMRFamide. This peptide has a sulfated Y residue at position 6 counted from the C-terminus, which corresponds to the positions of the sulfated Y residues in chordate gastrin/CCKs (8, 9). The cockroach C-terminal six amino acid residue sequence has four amino acid residues in common with mammalian gastrin/CCK, including the characteristic sulfated Y residue and is, thus, a likely orthologue of the mammalian peptide. For many other protostome neuropeptides, however, the situation is not clear-cut and other independent methods are required to establish robust evolutionary relationships between neuropeptide families in Proto- and Deuterostomia. One powerful method in this context is to look at the neuropeptide receptors, because, in our experience, neuropeptide receptors are much more conserved than their neuropeptide ligands. Furthermore, receptors are much larger (about 350 amino acid residues) than their smaller neuropeptide ligands (3-20 residues) and are, thus, easier to identify, using bioinformatics tools (11, 12).
Most neuropeptide receptors are G protein-coupled receptors (GPCRs). Many of these receptors have nowadays been deorphanized (= “matched” with their neuropeptide ligands), both in Protostomia (13, 14) and Deuterostomia (15, 16). Thus, when neuropeptide orthologues are claimed between the two bilaterian evolutionary lineages (Figure 1), one should always try to test these claims by showing the presence of orthologous GPCRs. Indeed, the identifications of oxytocin/vasopressin, neuropeptide Y, gastrin/CCK, and tachykinin peptides in Protostomia, have always been followed up by the subsequent identifications of their cognate GPCRs in this animal taxon and the demonstrations that these GPCRs are the orthologues of their deuterostome counterparts (4–7, 17–24). Figure 2 shows that the protostome (in this case, insect) oxytocin/vasopressin, neuropeptide Y, gastrin/CCK, and tachykinin GPCRs are the orthologues of their deuterostome (in this case, human) counterparts. These findings confirm that the complete ligand/receptor couples have been conserved during 700 million years of evolution in a process of ligand/receptor co-evolution.
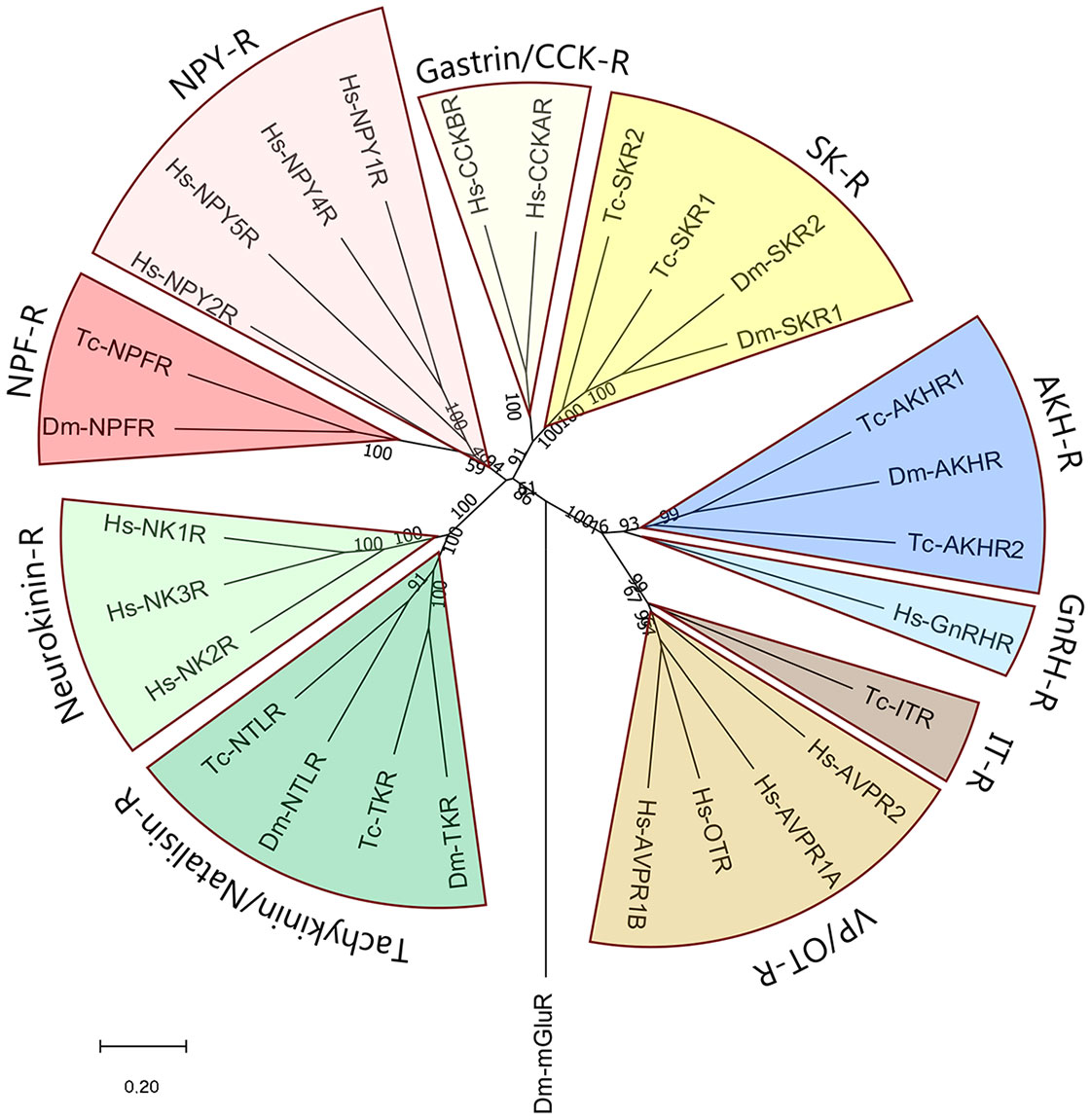
Figure 2 Phylogenetic tree of some neuropeptide GPCRs that have orthologues in both proto- and deuterostomes. As representatives of the deuterostomes we have taken humans (Homo sapiens, abbreviated Hs). As representatives of the protostomes, we have selected two model insects: The fruit fly Drosophila melanogaster (abbreviated Dm), and the red flour beetle Tribolium castaneum (abbreviated Tc). The figure shows that the two sulfakinin receptors (SK-R) found in each of these two insects are the orthologues of the two human gastrin/CCK receptors (gastrin/CCK-R); that the insect AKH receptors (AKH-R) are the orthologues of the human GnRH receptors (GnRH-R); that the insect oxytocin/vasopressin (= inotocin) receptors (IT-R) are the orthologues of the human vasopressin/oxytocin receptors (VP/OT-R); that the insect tachykinin/natalisin receptors are the orthologues of the human neurokinin receptors; and that the insect neuropeptide-F receptors (NPF-R) are the orthologues of the human neuropeptide-Y receptors (NPY-R). The numbers associated with the branches are bootstrap support values. The tree is rooted with the metabotropic glutamate receptor CG11144 from D. melanogaster (Dm-mGluR). The accession numbers of the GPCR sequences used to calculate this tree are provided in Supplementary Table S1.
In the following, we would like to give an example, where we could recognize two gonadotropin-releasing hormone (GnRH) receptor orthologs, one in Deuterostomia and the other one in Protostomia, but where we initially were unable to identify the ligand for the protostome GPCR. GnRH is a neuropeptide produced by a population of neuroendocrine cells in the vertebrate hypothalamus, where it stimulates the release of the glycoprotein hormones, luteinizing hormone (LH), and follicle stimulating hormone (FSH) from the anterior pituitary (2). GnRH and the two glycoprotein hormones are essential for vertebrate reproduction (2). To gain more insight into the reproduction of the fruitfly Drosophila melanogaster, we cloned a GPCR from this fruitfly that was a clear orthologue of the mammalian GnRH receptor (25). Next, we started to search for the D. melanogaster GnRH receptor ligand, which we expected would be GnRH or a GnRH-like peptide. However, we were unsuccessful with finding such a peptide, even after the publication of the Drosophila genome sequence, two years later (26), and carrying out bioinformatic searches. Therefore, we started a project where we purified the Drosophila GnRH receptor ligand from an aqueous extract of Drosophila larvae, using second messenger production in Chinese Hamster Ovary (CHO) cells that were expressing the Drosophila GnRH receptor, as a bioassay (27). This project resulted in the isolation and sequencing of a neuropeptide, which, to our surprise, was adipokinetic hormone (AKH), an insect neuropeptide, which had been known for more than twenty-five years (28), but which had an amino acid sequence that was completely different from mammalian GnRH (27). AKH is a metabolic neuropeptide that mobilizes lipids and carbohydrates from the insect fat body during a flight and other energy-requiring activities (28). Thus, not only are the structures of the mammalian GnRH and insect AKH completely different, but also are their actions. Yet, the presence of the two orthologous receptors in Proto- and Deuterostomia implies that these two receptors must have had a common ancestor that evolved before the split of Proto- and Deuterostomia at 700 MYA (Figure 1).
In conclusion, the following main lessons can be learned from the examples described in this chapter 1: (i) Many bilaterian neuropeptides and their GPCRs evolved in the common ancestor of Proto- and Deuterostomia, which lived around 700 MYA; (ii) Neuropeptide GPCRs are often more conserved than their ligands. Identifying (= deorphanizing) neuropeptide GPCRs may reveal evolutionary trajectories that otherwise would stay obscured (example: GnRH and AKH receptors).
Neuropeptides are synthesized as proteins (preprohormones) on the ribosomes of the rough endoplasmic reticulum (RER). After translation and during transport across the RER membrane, the pre-part of the preprohormone (also called signal peptide) is removed by a signal peptidase, after which the prohormone is directed into the secretory pathway. Here, the prohormones are packed into dense-core vesicles together with at least four prohormone processing enzymes, which catalyse the conversion of the prohormones into biologically active neuropeptides: (i) Prohormone convertase 1/3 (PC 1/3) that cleaves C-terminally of R, RR, or KR sequences. The resulting peptide products are still C-terminally elongated by R, RR, or KR residues. (ii) These C-terminal residues are removed by a carboxypeptidase specific for basic residues. (iii) About 50% of all neuropeptides carry a C-terminal amide group, which protects against C-terminal enzymatic degradation. This C-terminal amide group is formed from a C-terminal Gly residue with the help of peptidylglycine alpha-monooxygenase. (iv) Many neuropeptides carry an N-terminal pGlu residue, which protects the peptide against N-terminal enzymatic degradation. This N-terminal pGlu residue is formed by cyclization of an N-terminal Gln residue catalyzed by the enzyme glutaminyl cyclase. In a few cases, neuropeptides have N-terminal XP, or XPP sequences, which have a secondary amine, instead of a primary amine, involved in the peptide bond, which also protects against N-terminal enzymatic degradation (29–32).
Neuroendocrine systems evolved in a group of ancient animals that originated before the split of Proto- and Deuterostomia (Figure 1). These “pre-bilaterian” animals were related to four extant animal phyla: Porifera (sponges); Ctenophora (comb jellyfishes); Placozoa (1-mm small, disk-like animals, feeding on algae); and Cnidaria (animals like sea anemones, corals, and jellyfishes). Only Cnidaria and Placozoa will be discussed in this review, because they probably form a monophyletic clade (explained below) and cnidarians have well-established peptidergic nervous systems. A recent comment on neuropeptides and nervous systems in Ctenophora can be found in (33).
Cnidarians are a diverse group of animals characterized by having cnidocytes, which are stinging cells used for capturing prey. The phylum Cnidaria is sub-divided into several classes and subphyla (Figure 3). These are the subphylum Anthozoa, which includes sea anemones and corals; the subphylum Medusozoa, which consists of the classes Cubozoa (box jellyfishes), Scyphozoa (true jellyfishes), Staurozoa (stalked jellyfishes), and Hydrozoa (animals like Hydra and hydromedusae); and the subphylum/class Endocnidozoa, which are small endoparasites, belonging to two taxa, the Polypodiozoa, and Myxozoa. In the past, neuropeptides have only been isolated and sequenced from just a few model cnidarians, like Hydra magnipapillata, Clythia hemisphaerica, and the sea anemone Anthopleura elegantissima (35–55). These peptides are 4-10 amino acid residues long and are protected at their C-termini by amide groups against carboxypeptidases, and at their N-termini by either pGlu (pQ), XP, or XPP residues against aminopeptidases. Thus, the overall organization of these neuropeptides resembles the overall structures of the neuropeptides isolated from Bilateria. Yet, with respect to the actual amino acid sequences, these isolated cnidarian neuropeptides had no homologues in the Bilateria. We and other researchers raised antibodies against the various isolated cnidarian neuropeptides and used them for whole-mount immunocytochemistry. This procedure enabled us to stain the cnidarian nervous systems with unprecedented details (35–37, 56–61).
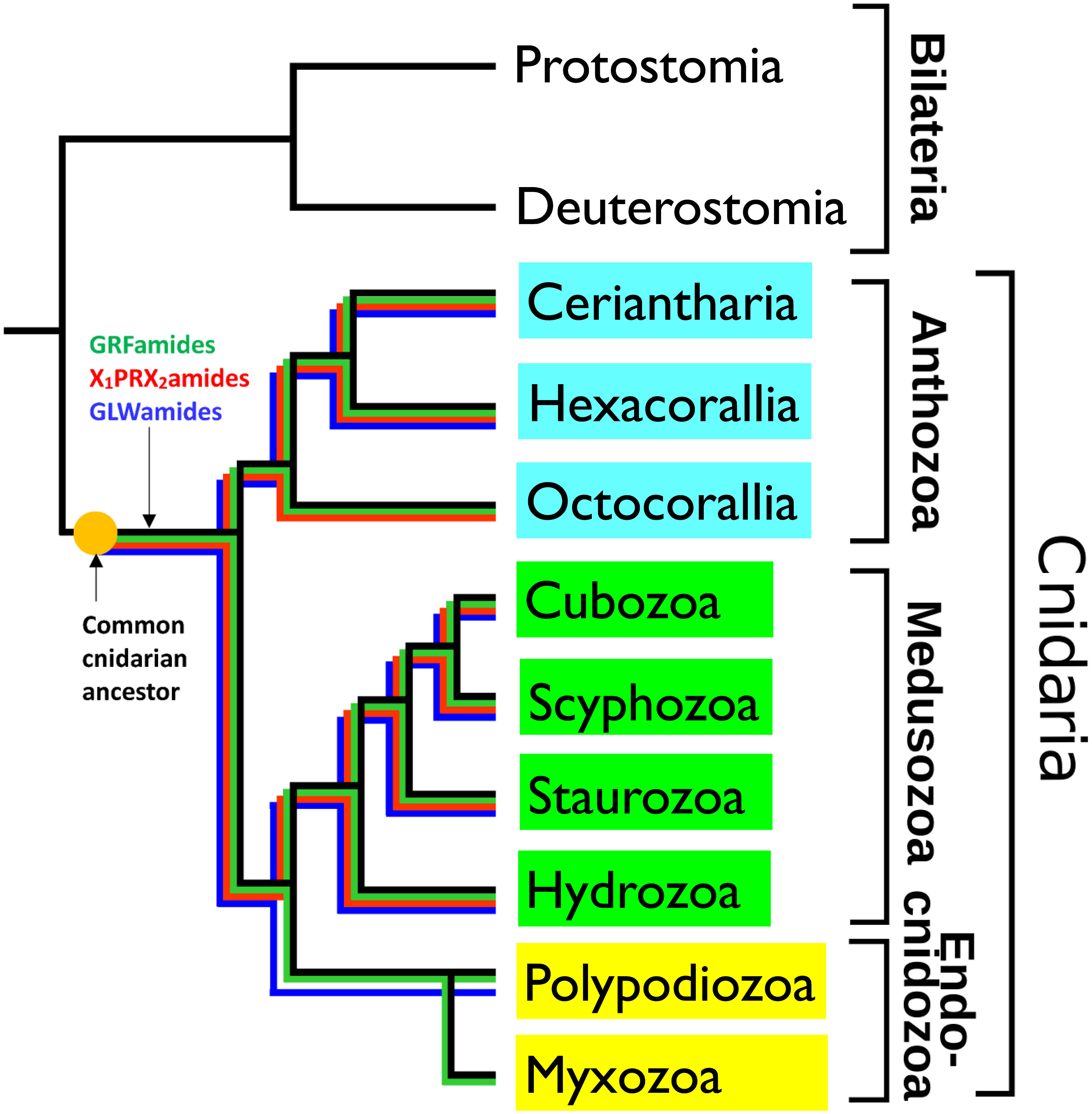
Figure 3 Schematic drawing, showing the phylogenetic positions of the cnidarian subclasses Ceriantharia, Hexacorallia and Octocorallia (belonging to the subphylum Anthozoa, highlighted in blue), the classes Cubozoa, Scyphozoa, Staurozoa, Hydrozoa (belonging to the subphylum Medusozoa, highlighted in green), and Polypodiozoa, and Myxozoa (belonging to the subphylum Endocnidozoa, highlighted in light yellow). The figure also shows the presence of three ancestral neuropeptide families in these clades by colored lines: the X1PRX2amide family (highlighted in red), the GRFamide family (highlighted in green), and the GLWamide family (highlighted in dark blue). These neuropeptide families are present in all the investigated Ceriantharia, Hexacorallia, Cubozoa, Scyphozoa, Staurozoa, and Hydrozoa species. However, the Octocorallia have apparently lost their GLWamide genes. The endoparasitic Polypodiozoa and Myxozoa are strongly reduced. Only a few species produce GRFamide and GLWamide peptides, while in all endocnidozoan species the X1PRX2amide peptides are absent. The presence of the three neuropeptide families in all the other cnidarian classes and subclasses suggests that they also were present in the common cnidarian ancestor. Adapted from (34) with permission.
Cnidarian neuropeptides control physiological processes like reproduction, development, metamorphosis, feeding, and muscle contraction (35–37, 46, 49–51, 53–55, 62–64).
We and other research groups also cloned the cnidarian neuropeptide preprohormones (35–37, 65–71). These preprohormones often have very high copy numbers of the immature neuropeptides. For example, the A. elegantissima preprohormone for Antho-RFamide has 21 copies of Antho-RFamide (pQGRFamide) and the Antho-RFamide preprohormone from the sea anemone Calliactis parasitica has 19 copies of this neuropeptide sequence (65, 66). At the C-terminal sides of each of the immature cnidarian neuropeptide sequences, we found classical processing signals for PC1/3 and, as in Bilateria, C-terminal Gly residues, which are converted into amide groups (35–37, 65–71). We also cloned the sea anemone peptidylglycine alpha-monooxygenase and determined its intron/exon organization, showing that this enzyme is evolutionarily closely related to its bilaterian counterparts (31, 72, 73). Thus, cnidarian preprohormone processing is “classical” at the C-terminal sites of their immature neuropeptide sequences. To our surprise, however, we found that processing at the N-terminal sites of the neuropeptide sequences was rather different, because no basic residues were present for cleavage by PC1/3. Instead, acidic residues (D, E) and sometimes other residues were flanking the N-termini of the immature neuropeptides. In the preprohormone for the isolated and sequenced sea anemone neuropeptide Antho-RFamide (pQGRFamide), for example, we find 14 copies of the immature sequence DQGRFGKR and 5 copies of EQGRFGKR (65). We do not know the identity of the N-terminal processing enzyme, which could either be an aminopeptidase or endoproteinase. Unorthodox N-terminal processing of neuropeptide sequences appears to be specific for ancient peptidergic nervous and endocrine systems. This type of processing is not present in Bilateria, but, interestingly, it also seems to occur in Placozoa (see Chapter 4). These findings would suggest a close phylogenetic relationship between Cnidaria and Placozoa.
Cnidaria consists of more than 15,000 species that are distributed over the nine classes or subclasses shown in Figure 3. As mentioned above, neuropeptides from only five cnidarian species have been isolated and sequenced (35–55). Therefore, we wanted to know, whether the other cnidarians also produced neuropeptides and what the structures of these neuropeptides were. We assumed that investigating this question might lead to the discovery of novel neuropeptides with novel actions, thereby promoting cnidarian biology. In addition, this work might shed light on the evolution of neuropeptides in Cnidaria and tell us more about the neuropeptides that existed in the common cnidarian ancestor. To answer these questions, we investigated the sequenced genomes and transcriptomes from 80 cnidarian species, belonging to different families of the Ceriantharia, Hexacorallia, Octocorallia, Cubozoa, Scyphozoa, Staurozoa, Hydrozoa, Polypodiozoa, and Myxozoa (Figure 3) (34, 74–77).
For screening this large number of genomes and transcriptomes, we developed a bioinformatics script that recognized proteins, containing at least three repetitive sequences, each having the C-terminal sequence GKR, GRR, or GR (34, 74–77). In addition, we applied TBLASTN screening with queries corresponding to known cnidarian, placozoan, and bilaterian neuropeptide sequences (34, 74–77). We found that most cnidarians produced three to eight neuropeptide families. Nearly all cnidarian species produced peptides with the C-terminal sequence GRFamide, GLWamide, or X1PRX2amide (Figure 3). The only two exceptions were species belonging to the Octocorallia, where apparently a gene loss has occurred of their GLWamide peptide genes, and Endocnidozoa, where only a few species produced GRFamide, and GLWamide peptides, while the X1PRX2amides were lacking (Figure 3). Due to their endoparasitic lifestyle, endocnidozoans often consist of just a handful of cells and they have one of the smallest genomes in the animal kingdom. These strongly reduced genomes probably explain the gene losses of some of their neuropeptide genes (34).
The presence of GRFamides, GLWamides, and X1PRX2amides in nearly all cnidarian species clearly shows that these three neuropeptide families must have been present in the common cnidarian ancestor (Figure 3). They belong, thus, to the most ancient neuropeptide families known, so far.
The amino acid sequences of the three primordial neuropeptide families can vary somewhat, although their hallmarks are conserved. For the GRFamide family, for example, hexacoral and octocoral species (Figure 3) all have a short peptide version: pQGRFamide (75, 76). In hydrozoans, the GRFamides are N-terminally elongated. For instance, C. hemisphaerica contains a preprohormone, with 17 copies of the neuropeptide pQWLNGRFamide (34).
Some cnidarians have only one gene coding for GRFamides, one gene for GLWamides, and one gene for X1PR2Xamide peptides, such as the hydrozoans Dynamena pumila, Porpita porpita, Vellela vellela, and Millipora alconis (34). These three neuropeptide genes are apparently sufficient for a hydrozoan animal to live its daily hydrozoan life, including reproduction, development, metamorphosis, and feeding. Other cnidarians have duplicated one or more of these primordial genes, assumedly to give them a more advanced neuropeptide repertoire, which enables them to carry out a more advanced behavior. An extreme example of such gene multiplications can be observed in the freshwater hydromedusa Craspedacusta sowerbii, which has six GRFamide preprohormone genes, three GLWamide preprohormone genes, and two X1PRX2amide preprohormone genes (34). Furthermore, C. sowerbii has created an additional novel gene for a preprohomone that contains two copies of pQFLRPamide and one copy of pQFIRPamide (34). The presence of twelve neuropeptide preprohormone genes in C. sowerbii (compared to only three) is surprising and we do not know the reason for this.
Normally, cnidarians have genes for each of the three primordial neuropeptide families and, in addition, zero to three additional neuropeptide genes. These additional neuropeptide genes are taxon-specific and we assume that the animals need them for group-specific behaviour or physiology.
Do the current cnidarian neuropeptide genes have orthologues in the Bilateria and could we perhaps follow their evolution from pre-bilaterians to bilaterians? We have investigated this question by TBLASTN screening of the genomes and transcriptomes from species belonging to the two animal groups and found little or no resemblance between cnidarian and bilaterian neuropeptides or preprohormones. The best candidates for having orthologues in cnidarians and bilaterians would be members of the RFamide peptide family. In the following, therefore, we would like to focus on the RFamide neuropeptide family. As mentioned earlier in this chapter, RFamide peptides are abundant in nearly all cnidarians, where they have the C-terminal sequence GRFamide, which is N-terminally elongated by one to four amino acid residues (34, 75, 76). RFamide peptides are also occurring in many protostomes and to a lesser degree in deuterostomes. In Supplementary Figure S1, we have aligned the amino acid sequences of RFamide peptides from some cnidarian species (N. vectensis, H. magnipapillata) with those from several protostomes (the mollusc Lymnaea stagnalis; the brachiopod Terebratalia transversa; the nematode Caenorhabditis elegans; the fruitfly Drosophila melanogaster) and deuterostomes (the lancelet Branchiostoma floridae; and Homo sapiens). The results from these alignments were that, of course, all neuropeptides had the RFamide sequence in common, but that the N-terminal extensions were different. The structural similarities between the peptides were, therefore, very few and limited to the last two amino acid residues. We also compared the RFamide peptides, using phylogenetic tree analyses, which yielded trees with low bootstrap values and unreliable phylogenetic relationships, except for the cnidarian peptides (Supplementary Figure S1). Next, we compared the preprohormones of all the selected RFamides. This showed that some of the protostome RFamide preprohormones had a similar overall structure as the cnidarian RFamide preprohormones with a high number of immature RFamide neuropeptide sequences arranged in a repetitive manner directly after each other. In deuterostomes, these highly repetitive RFamide sequences were generally absent, such as in the kisspeptin-preprohormone and structurally related vertebrate preprohormones (Supplementary Figure S1) (78). We also analysed these preprohormones using phylogenetic tree analyses, which resulted, again, in trees with low bootstrap values and unreliable phylogenetic relationships. (Supplementary Figure S1). Based on these results, therefore, we cannot conclude that bilaterian RFamide neuropeptides are phylogenetically related to the cnidarian RFamide neuropeptides.
In analogy to the GnRH/AKH receptor story in Bilateria (Chapter 1), the amino acid sequences of GPCRs may be more conserved than the amino acid sequences of their neuropeptide ligands in Cnidaria. Finding GPCR orthologues in Cnidaria and Bilateria, followed by deorphanization, could help to identify orthologous GPCR/neuropeptide couples and give us useful information about their evolution (78). GPCR conservation between cnidarians and bilaterians has been investigated by Thiel et al. (79), but they were unable to find GPCR orthologues between the two animal groups. In our current paper, we extended this approach by comparing virtually all neuropeptide GPCRs from humans, the model sea anemone N. vectensis and the placozoan T. adhaerens (see Chapter 4) in a large phylogenetic tree analysis. GPCRs can be subdivided in three major classes: Family-A, -B, and -C, where family-A (also called rhodopsin-like GPCRs) is the largest class, comprising, among others, neuropeptide receptors, glycoprotein hormone receptors, olfactory receptors, and the opsins. We, therefore, collected the sequences from 723 human family-A receptors and removed the 437 olfactory receptors known to exist in humans, resulting in 286 human family-A receptors and compared them with 843 family-A GPCRs from N. vectensis and 679 family-A GPCRs from T. adhaerens (altogether 1826 receptors) in phylogenetic tree analyses. These analyses are described in Supplementary Figures S3 and S4. They showed that we were unable to identify the existence of neuropeptide GPCR orthologues in the three species in a convincing way, except for a group of special GPCRs, the leucine-rich repeats-containing GPCRs, which we will discuss in Chapter 5.
So far, only one neuropeptide GPCR has been deorphanized in cnidarians, which is the receptor for an oocyte maturation inducing hormone (MIH) from the hydromedusa Clytia hemisphaerica (80). We found, however, that also this MIH GPCR had no convincing orthologues in bilaterians, in N. vectensis, and in T. adhaerens (as tested by TBLASTN).
Finding neuropeptide GPCR orthologues between bilaterians, cnidarians, and placozoans might be hard, due to the long divergence times, existing between these three animal clades. In addition, deorphanizing the large number of family-A GPCRs in cnidarians or placozoans would also be a huge task, as it would require the molecular cloning and expression of around 800 cnidarian or placozoan GPCRs, followed by functional expression of their cDNAs in human cell lines and screening of neuropeptide libraries. However, a recent publication shows that there are possibilities for identifying neuropeptide-GPCR couples by machine learning-assisted strategies, which potentially would shortcut the first labor-intensive steps (81, 82).
Recently, the precursor of a presumed neuropeptide, phoenixin, was reported to occur in bilatarians, cnidarians, ctenophores, and sponges (83). However, a cognate GPCR for this peptide could not be identified in any of the four animal clades (83).
In 1883, F.E. Schulze, a German zoologist at the University of Graz (Austria), discovered tiny animals, feeding on algae in a sea water aquarium and named them Trichoplax adhaerens (84, 85). T. adhaerens has remained the only representative of the phylum Placozoa during more than 130 years, but in the last few years, several other placozoan species or strains were isolated from marine waters from different parts of the globe (85). So far, the genomes from three species/strains have been sequenced: T. adhaerens (strain H1), T. adhaerens (strain H2), and Hoilungia hongkongensis (85). Nikitin identified twelve peptide preprohormones in T. adhaerens (strain H1) (86). We repeated his experiments in all three placozoan species/strains, using the script described in Chapter 3 and could confirm the existence of all twelve preprohormones (77).
In the following, we want to discuss the PRWamide preprohormones as an example of a placozoan peptide preprohormone (Figure 4). The PRWamide preprohormone from T. adhaerens (strain H1) contains 11 copies of DQPPRWGR and three copies of NQPPRWGR (Figure 4). We propose that these immature peptide sequences are likely to be processed into 14 copies of mature pQPPRWamide. It is intriguing that N-terminal peptide processing in Placozoa apparently follows the same principles as N-terminal neuropeptide processing in Cnidaria, as shown by the presence of acidic and other residues (Asp and Asn), but not basic residues, preceding the immature neuropeptides sequences. We find the same prohormone processing in T. adhaerens (strain H2), where the PRWamide preprohormone contains 11 copies of DQPPRWamide, which we propose are processed into 11 copies of mature pQPPRWamide. Furthermore, the PRWamide preprohormone from H. hongkongensis contains 12 copies of DQPPRWGR and three copies of NQPPRWGR. We propose that these immature peptide sequences will be converted into 15 copies of mature pQPPRWamide. In addition to these pQPPRWamides, the preprohormones from each of the three placozoan species/strains contain a few PRWamide sequences that are slightly different from pQPPRWamide.

Figure 4 Amino acid residue sequences of three RWamide preprohormones from three placozoan species or lineages: T. adhaerens, T. adhaerens sp. H2, and Hoilungia hongkongensis. The signal sequences are underlined, the immature neuropeptide sequences are highlighted in yellow, the cleavage sites for prohormone convertase 1/3 are highlighted in green and the C-terminal G residues that are converted into C-terminal amide residues are highlighted in red. Furthermore, each immature neuropeptide sequence contains an N-terminal Q residue that is converted into an N-terminal pQ residue by the enzyme pyroglutaminyl cyclase. Note that each preprohormone is processed into a large number of identical or similar mature neuropeptides, which are protected at their C-termini by amide groups and at their N-termini by pQ groups or pQPP sequences.
The placozoan pQPPRWamide peptide structure reminds us of the LRWamide peptides from Hexacorallia and Ceriantharia (Supplementary Figure S5) (76) and of some other peptides with the Wamide C-terminus from other animals (87), but are they really orthologues? We do not believe that this is the case, because: (i) LRWamides have been isolated and sequenced from the hexacoral A. elegantissima (41, 42) and have subsequently been annotated from the genomes and transcriptomes of 17 species belonging to the Hexacorallia and Ceriantharia (76). Each species expresses one to four genes, coding for an LRWamide preprohormone and each preprohormone contains one LRWamide peptide. This yields knowledge from altogether 39 LRWamide peptides (Supplementary Figure S5). These 39 peptides nearly always have the C-terminal sequence LRWamide and in a few cases IRWamide, or MRWamide (L, I, and M residues are amino acid residues with similar physico-chemical properties) and never PRWamide, or PPRWamide. L and P residues do not have similar properties and P residues disturb the secondary structure of the peptides, making them more rigid. This means that the two peptide families have different secondary structures. (ii) In 38 out of 39 sea anemone LRWamide preprohormones, the LRWamide neuropeptide sequence directly follows the signal sequence (Supplementary Figure S5) (76). For the placozoan PRWamide preprohormones, in contrast, there are always long stretches of amino acid residues following the signal sequence, before the first PRWamide neuropeptide sequence appears (Figure 4). Besides, there are multiple copies (about 15) of these PRWamide peptides and not only one, as in sea anemones. Thus, both the peptide and the overall preprohormone structures of the LRWamide and PRWamide peptides are essentially different, arguing strongly against the possibility that the two peptide families are orthologous.
For the other placozoan peptide families, we also cannot find orthologues in Cnidaria, or Bilateria (as tested with TBLASTN). In addition, we were unable to discover orthologues, when comparing placozoan GPCRs with GPCRs from Cnidaria (N. vectensis), or Bilateria (H. sapiens), using extensive phylogenetic tree analyses (Supplementary Figures S3, S4), except for the LGRs, which we will discuss in Chapter 5. These results suggest a low evolutionary pressure to conserve amino acid sequences in neuropeptides or neuropeptide GPCRs between the three animal groups, which is surprising, because there has been clear neuropeptide conservation, and a clear neuropeptide GPCR conservation in Proto- and Deuterostomia over a period of 700 million years (Chapter 1; Figures 1, 2).
Leucine rich repeat-containing GPCRs (LGRs) belong to the family-A GPCRs, but distinguish themselves from the other members of this family by the presence of a long N-terminus, containing multiple leucine-rich repeats (Figure 5). These leucine-rich repeats are an important part of the ligand binding domains of the receptors (88–90). The ligands of the LGRs are various protein hormones, such as the glycoprotein hormones: Luteinizing hormone (LH), choriogonadotropin (CG), follicle stimulating hormone (FSH), and thyroid stimulated hormone (TSH). LH, CG, FSH, and TSH have one common subunit (the glycoprotein alpha subunit, also called GPA1), while the second subunit of these dimeric hormones is a beta subunit specific for each of the four mentioned hormones (LHbeta, CGbeta, FSHbeta, TSHbeta). These beta subunits are also called GPB1 to GPB4) (91). LH and CG bind to the same receptor, while FSH and TSH have their own specific receptors. There also exist other glycoprotein hormone subunits in humans: Glycoprotein alpha subunit-2 (GPA2), and glycoprotein beta subunit-5 (GPB5) (92). GPA2/GPB5 (also called thyrostimulin) is a dimeric glycoprotein hormone stimulating the TSH receptor, thus acting similarly to TSH (93). Altogether, there are eight LGRs in humans: LGR1 to LGR3 are the FSH, LH/CG, and TSH receptors (in this order); LGR4 to LGR6 are activated by various R-spondin proteins, which signal through the canonical Wnt/beta-catenin pathway (94–97); LGR7 and LGR8 are receptors for relaxins and other insulin-like peptides (88, 98).
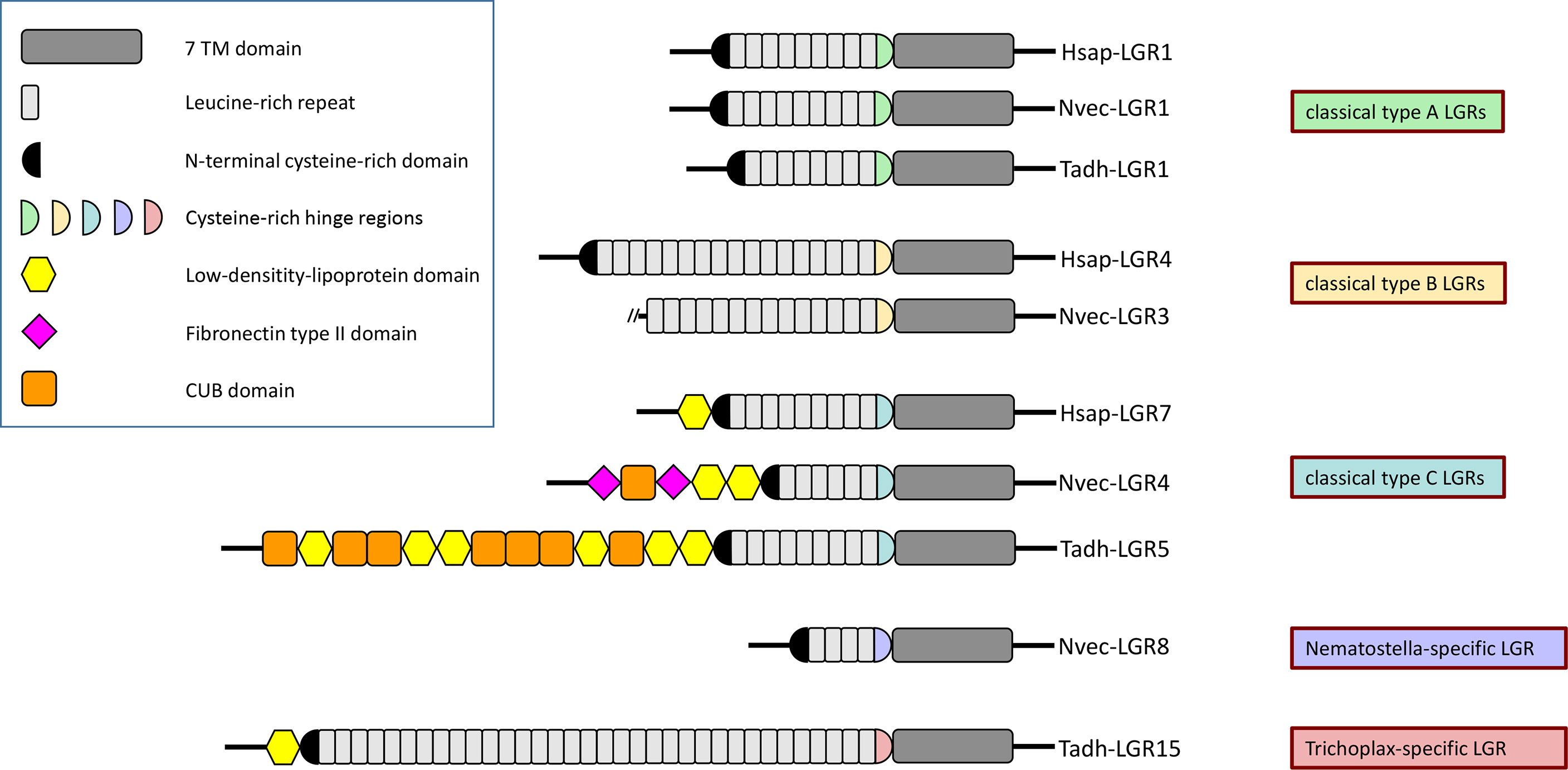
Figure 5 Protein domain composition of the extracellular N-termini of human (Hsap-) LGRs, N. vectensis (Nvec-) LGRs, and T. adhaerens (Tadh-) LGRs. Humans have three type-A, three type-B, and two type-C LGRs, depending on the structure of the hinge domains connecting the extracellular N-termini to their seven-transmembrane (7TM) regions. These human LGRs are also called LGR-1 to -3, LGR-4 to -6, and LGR-7 to -8, respectively (see main text). The top of this figure shows Hsap-LGR-1 (human FSH receptor), a typical human type-A LGR, containing a type-A-like cysteine-rich hinge region (highlighted in green) that connects the 7TM domain (dark gray) to the 9 leucine-rich repeats (highlighted in light gray) of its N-terminus. The N-terminus starts with another cysteine-rich domain (highlighted in black), preceding the leucine-rich domains. Hsap-LGR-2 and -3 (= human TSH and LH/CG receptors), Nvec-LGR-1 and -2, and Tadh-LGR-1 and -2 have similar overall structures and domains. This figure shows also Hsap-LGR-4, a typical human type-B LGR, containing a type-B-like cysteine-rich hinge region (drawn as a yellow half circle) that connects the 7TM domain to the 16-18 leucine-rich repeats of its N-terminus. Nvec-LGR-3 has a similar type-B-like cysteine-rich hinge region, but its N-terminus is incomplete. Surprisingly, a type-B LGR appears to be lacking in T. adhaerens (see Figure 6). Furthermore, the figure shows Hsap-LGR-7 (= human relaxin receptor), which is a typical type-C LGR. This receptor has a C-type cysteine-rich hinge region (drawn as a light blue half circle) that connects the 7TM domain with the 9 leucine-rich repeats of the N-terminus. This N-terminus starts with a low-density-lipoprotein (LDL) domain (yellow hexagon) followed by the usual cysteine-rich domains preceding the N-terminal leucine-rich domains. Nvec-LGR4 and Tadh-LGR-5 have similar overall structures including the type C-like hinge regions and the LDL domains at the start of their N-termini. These receptors, however, have also additional N-terminal domains, such as CUB domains (a structural motif of about 110 residues found almost exclusively in extracellular and plasma membrane-associated proteins) (orange boxes), and fibronectin type II domains (pink squares). Finally, N. vectensis and T. adhaerens have a large number of novel LGRs that do not belong to one of the above-mentioned canonical LGR families. We give here two examples: Nvec-LGR-8, which only contains 4 leucine-rich repeats and that has its own, specific hinge region; and Tadh-LGR-15, which has as much as 34 leucine-rich repeats and, again, its own specific cysteine-rich hinge region (see also Figure 6). The accession numbers of the LGR sequences used in this figure can be found in Supplementary Table S2.
LGRs can be classified as type-A, -B or -C, based on the structures of their N-terminal ectodomains and the hinge regions connecting these ectodomains to the transmembrane regions of the LGRs (99) (see also Figure 5 for details). Type-A and -C LGRs contain about 7-9 leucine-rich repeats (LRRs), whereas type-B LGRs contain about 14-18 LRRs (Figure 5). The hinge regions are cysteine-rich and have a subtype-specific consensus sequence with a well-defined number of cysteine residues forming disulfide bridges that stabilize the structure (99). The classification of the LGRs in type-A, -B, and -C, categorizes the human LGRs in the same way as described in the first paragraph of this Chapter (Figure 5): Type-A LGRs are the TSH, LH/CG, and FSH receptors; Type-B are LGR4 to LGR6; Type-C are LGR7 and LGR8 (99). This implies (at least in mammals) that each group uses one category of ligands (Type-A: Glycoprotein hormones; Type-B: R-spondins; Type-C: Relaxins and related insulin-like peptides) (99).
In 1997, three years before the sequencing of the Drosophila genome, we cloned an LGR from Drosophila, which we named Drosophila LGR-1 (DLGR1), the first arthropod (and even protostome) LGR to be identified (100). Three years later followed the cloning of a second Drosophila LGR, DLGR2 (101). Subsequently the ligands for both LGRs were identified, being Drosophila GPA2/GPB5 for DLGR1 (102) and the Drosophila glycoprotein hormone bursicon for DLGR2 (103, 104). Bursicon is a heterodimeric glycoprotein and its structure resembles that of human LH, CG, FSH, and TSH. Bursicon promotes darkening (hardening) of the insect exoskeleton after a moult and has several other important roles in insects among them glucose homeostasis (105, 106). Drosophila has two other LGRs, Lgr3 and Lgr4, of which Drosophila Lgr3 is a receptor for a Drosophila insulin-like peptide, Dilp8 (107). These examples from LGRs in Bilateria, therefore, can be added to the impressive list of GPCRs and their ligands that have orthologues in both Proto- and Deuterostomia, showing that these receptor/neurohormone couples must have originated in the common ancestor of these two lineages (Figure 1).
In 1993, when the whole genome sequences from the sea anemone N. vectensis or other cnidarians had not been established yet, we were able to clone the first pre-bilaterian LGR from the sea anemone A. elegantissima (108–111). Our results from A. elegantissima showed, for the first time, that we could follow the evolution of a pre-bilaterian LGR to a bilaterian LGR and that this pre-bilaterian LGR was strongly conserved. We recently created a transcriptome from the cubomedusa Tripedalia cystophora and found that this cnidarian contained two LGRs (74). In our current paper, we analysed the genome from the sea anemone N. vectensis (110) and discovered that it had thirteen LGRs. After a phylogenetic tree analysis of these sea anemone LGRs and comparing them with the eight human LGRs, we could see that two sea anemone LGRs belonged to type-A, one sea anemone LGR belonged to type-B, and one sea anemone LGR belonged to type-C (Figure 6; Supplementary Table S2). In addition, there were nine sea anemone LGRs that were neither type-A, -B, or -C. The two sea anemone LGRs from type-A were closely associated with the human TSH, LH/CG, and FSH receptors (Figure 6), suggesting that also the sea anemone ligands are dimeric glycoprotein hormones. Similarly, the ligand for the sea anemone LGR from type-B might be an R-spondin; and the ligand for the sea anemone LGR from type-C might be a relaxin or related insulin-like peptide.
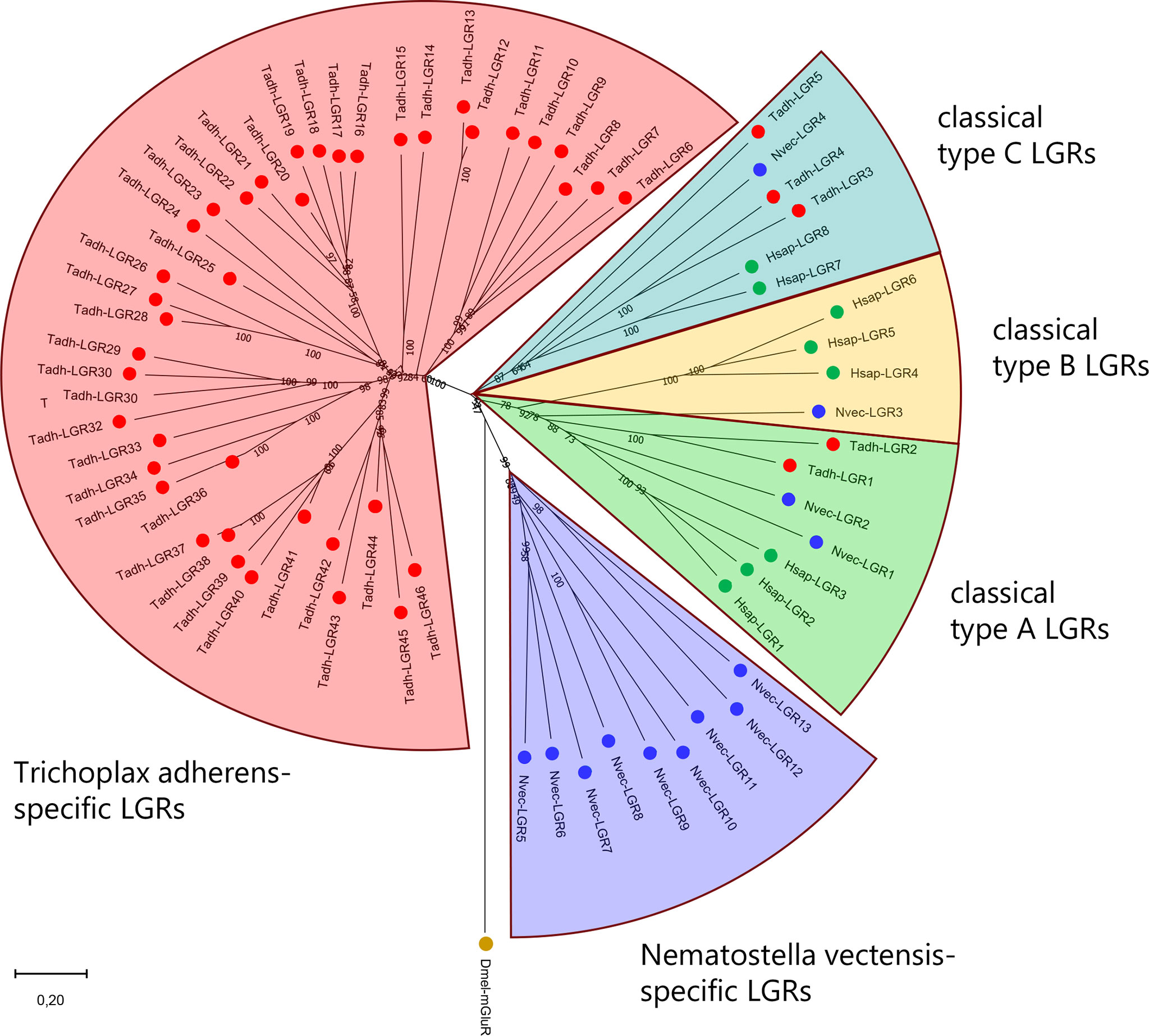
Figure 6 Phylogenetic tree analysis of LGRs from H. sapiens (Hsap), the sea anemone N. vectensis (Nvec), and the placozoan T. adherens (Tadh). Green dots indicate human LGRs, blue dots indicate N. vectensis LGRs. Red dots indicate T. adhaerens LGRs. Numbers associated with branches of the tree are bootstrap support values. The tree is rooted with the D. melanogaster metabotropic glutamate receptor CG11144 (Dm-mGluR). LGRs belong to three families: Type-A (located in the green field of this tree), type-B (yellow field), and type-C (light blue field). Humans have three type-A, three type-B, and two type-C LGRs. N. vectensis has two type-A, one type-B, and one type-C LGR. T. adhaerens has two type-A, no type-B, and three type-C LGRs. These assignments are independently supported by the presence of specific protein domains in the N-termini of the LGRs (see Figure 5). In addition to these three canonical LGR types, N. vectensis (purple field) and T. adhaerens (red field) have large numbers of novel LGRs that do not belong to the three canonical families: nine in N. vectensis and forty-one in T. adhaerens. The accession numbers of the LGRs used to calculate this tree are provided in Supplementary Table S2.
Using TBLASTN, we screened the genome from N. vectensis with queries corresponding to the alpha and beta subunits of bilaterian glycoprotein hormones and found several candidate ligands for the two type-A sea anemone LGRs (Supplementary Figure S7). The next step would be to produce these candidate ligands and test them on mammalian cells transfected with cDNA coding for the sea anemone type-A LGRs. This procedure might be the roadmap for deorphanizing LGRs in pre-bilaterian animals.
There are nine N. vectensis-specific LGRs (located in the purple field of Figure 6) that in a phylogenetic tree cannot be associated with bilaterian LGRs. Therefore, we cannot predict the ligand classes for them. These LGRs have a lower number of leucine rich repeats in their N-termini and different hinge regions compared the LGRs from group-A, -B, and -C (Figure 5). We do not understand why sea anemones would need these nine additional LGRs, but LGRs in sea anemones have apparently a broad range of actions.
We also analysed the LGRs from Placozoa. Nikitin and Roch et al. were the first to discover two LGRs in the genome from T. adhaerens (86, 112). We revisited these annotations, using updated genomic and transcriptomic databases from T. adhaerens (85), and found no less than forty-six placozoan LGRs (Figure 6) (77). Among them were two type-A LGRs, no type-B, and three type-C LGRs. In addition to these five LGRs, there were forty-one LGRs that were specific for T. adhaerens (located in the red field of Figure 6) (77). These T. adhaerens-specific LGRs had hinge regions that were different from the placozoan type-A and type-C LGRs and also had an unusually high number of leucine-rich repeats in their N-termini (Figure 5). The presence of forty-six placozoan LGRs is surprising for such a primitive animal like T. adhaerens, which only has a simple behavioral repertoire (113).
In conclusion, LGRs have a long evolutionary history, going all the way back to the common ancestor of Cnidaria and Placozoa, which is far beyond 700 MYA (Figure 1). Furthermore, due to high amino acid sequence conservations, it is possible to follow the evolutionary “jump” of LGRs from pre-bilaterian animals to bilaterians, something that has not been possible, so far, with the neuropeptide GPCRs or their ligands. During evolution, both the LGRs (Figure 6), and their potential ligands (Supplementary Figure S7) have been well conserved.
Roch and Sherwood (112) described one or two genes for an LGR and a potential glycoprotein hormone candidate in genomic databases from Ctenophora and Porifera, which are the other two early-branching animal phyla that diverged before the emergence of Bilateria. Thus, the LGRs and their ligands were apparently broadly present in the early metazoans that lived on Earth before the emergence of the Bilateria.
Peptidergic signaling through GPCRs is slow, due to the use of many steps involved in second messenger cascades. For a few neuropeptides, however, peptidergic signalling can be fast, namely for those peptides that directly gate ion channels, which leads to fast postsynaptic depolarizations. The ion channels involved belong to a small subfamily of the degenerin/epithelial Na+ channel (DEG/ENaC) protein family that are channels for Na+ ions. To this DEG/ENaC family also belong other physiologically important channel proteins, such as the acid-sensitive ion channels (ASICs). DEG/ENaC proteins consist of three subunits. Each subunit crosses the cell membrane twice and has a large extracellular loop (114, 115). Neuropeptide-gated Na+ channels were first discovered by Cottrell and co-workers in 1990 during patch-clamp experiments, where molluscan FMRFamide was applied to neuronal outside-out cell membrane patches from the snail Helix aspersa (116). This FMRFamide-gated Na+ channel was subsequently cloned by Lingueglia and colleagues in 1995, showing that it was a member of the DEG/ENaC family, and afterwards functionally expressed in frog oocytes, showing that it was a homotrimer (117).
In the following years, FMRFamide-gated ion channels were discovered in a number of molluscs, belonging to different molluscan classes, among them the gastropods Lymnaea stagnalis (118), Aplysia kurodai (119), and Lottia gigantea (120), the bivalve Crassostrea gigas (120), and the cephalopod Octopus bimaculoides (120). Already in the beginning of these studies, these FMRFamide-gated DEG/ENaC proteins were named FaNaCs (118).
In 2007 Gründer and colleagues found that the cnidarian Hydra magnipapillata also contained ion channels acting like the molluscan FMRFamide-gated channels and that these cnidarian channels were gated by two Hydra neuropeptides: Hydra-RFamide-1 (pQWLGGRFamide) and Hydra-RFamide-2 (pQWFNGRFamide) (121). These channels were, therefore, called HyNaCs (121). Subsequent studies showed that the HyNaCs were heterotrimers, consisting of combinations of three out of ten different HyNaC subunits that were expressed in Hydra and that some of these trimer combinations not only had Na+ permeability, but also high Ca2+ permeability. The HyNaCs were, in fact, the first members of the DEG/ENaC family to be discovered with high Ca2+ permeability (121–124).
Several phylogenetic trees of the HyNaCs, FaNaCs and other members of the DEG/ENaC family from molluscs, nematodes, arthropods and chordates can be found in (125). These cladograms, using both Bayesian and maximum likelihood analyses, show that the HyNaC subunits are not closely related to the FaNaCs, because they are sorted into two clearly different branches of the DEG/ENaC family tree with the HyNaCs grouped together with the ASICs, and the FaNaCs grouped together with the DEGs and ENaCs (125). This is an important finding, as it reveals that the Hydra and the snail neuropeptide-gated channels are not close orthologues. In Chapter 3, we have explained that we cannot conclude that cnidarian neuropeptides (for example the Hydra-RFamides-1 and -2) are evolutionary related to RFamide neuropeptides from bilaterians (for example FMRFamide). The presence of neuropeptide-gated ion channels in cnidarians and molluscs, therefore, cannot necessarily be regarded as the evolutionary conservation of a neuropeptide/receptor pair from pre-bilaterians to bilaterians. Of course, several members of the DEG/ENaC family can be gated by neuropeptides and this property has been conserved from pre-bilaterians to bilaterians. The peptide-gated family members in cnidarians, however, might not be directly related to one of the peptide-gated family members that we find in bilaterians today.
Although members of the DEG/ENaC family are probably present in all bilaterians, neuropeptide-gated DEG/ENaC family members have not been found in deuterostomes. In protostomes, however, in addition to the earlier-mentioned FaNaCs from molluscs, also the annelid Platynereis dumerilii expresses peptide gated ion channels. These channels, however, are not gated by FMRFamide or RFamide peptides, but by annelid-specific peptides having the C-terminal Wamide residue, such as GWKQGASYSWamide (= myoinhibitory peptide-2). They were, therefore, named myoinhibitory peptide-gated ion channels (MGICs) (126) and recently re-named as Wamide-gated Na+ channels (WaNaCs) (120). In various phylogenetic tree analyses, the molluscan FaNaCs and the annelid WaNaCs turned out to be orthologues, while the cnidarian HyNaC subunits were not orthologous to the molluscan/annelid subunits (120, 126). These results confirm our earlier suggestion that the cnidarian HyNaCs might not have direct evolutionary relationships to the peptide-gated ion channels from bilaterians.
In a recent paper, Dandamudi and co-workers tested a large number of molluscs, annelids, platyhelminths and other lophotrochozoans (Figure 1) for the presence of FaNaCs and WaNaCs (120). The results from that work showed that FaNaCs were present in the majority of lophotrochozoans, but also that a small group of annelids, among them P. dumerilii, had WaNaCs instead of FaNaCs.
No reports have been published about the presence of peptide-gated DEG/ENaC channels in Ecdysozoa (Figure 1) and, as mentioned earlier, in Deuterostomia. It seems, therefore, that these peptide-gated DEG/ENaCs are limited to two animal clades: To the larger clade of lophotrochozoans and the smaller clade of cnidarians. It is possible, however, that peptide-gated ion channels also occur in placozoans, because references (77, 125) independently describe the presence of around ten DEG/ENaC subunits in the placozoans T. adhaerens, and H. hongkongensis. In phylogenetic tree analyses, the placozoan subunits lie on the same branch as HyNaCs and ASICs, and far away from the FaNaCs, suggesting that they might have properties similar to HyNaCs (77, 125). However, both publications describe the presence of one placozoan subunit that lies on the same branch as the FaNaCs, suggesting that placozoans might have two types of peptide-gated DEG/ENaCs. These possibilities based on phylogenetic tree analyses, however, need to be validated by laboratory experiments.
For long, it had been uncertain whether placozoans had a nervous system. We answered this question by raising antibodies against the various predicted placozoan neuropeptide sequences and applying these antibodies to fixed placozoans during whole mount immunocytochemistry (77). However, already in the beginning of our work, we were “surpassed” by a publication from Varoqueaux and colleagues (113), who showed that all known neuropeptides in placozoans were produced by endocrine cells. We reached the same conclusions when we stained T. adhaerens with our own neuropeptide antibodies (77). Many neuropeptides in T. adhaerens have an N-terminal pyroglutamate residue. Therefore, we also raised antibodies against the processing enzyme pyroglutaminyl cyclase from T. adhaerens, which we annotated from its genome sequence (77). Figure 7 shows the staining of T. adhaerens by one such antibody. Again, only endocrine cells could be stained (77).
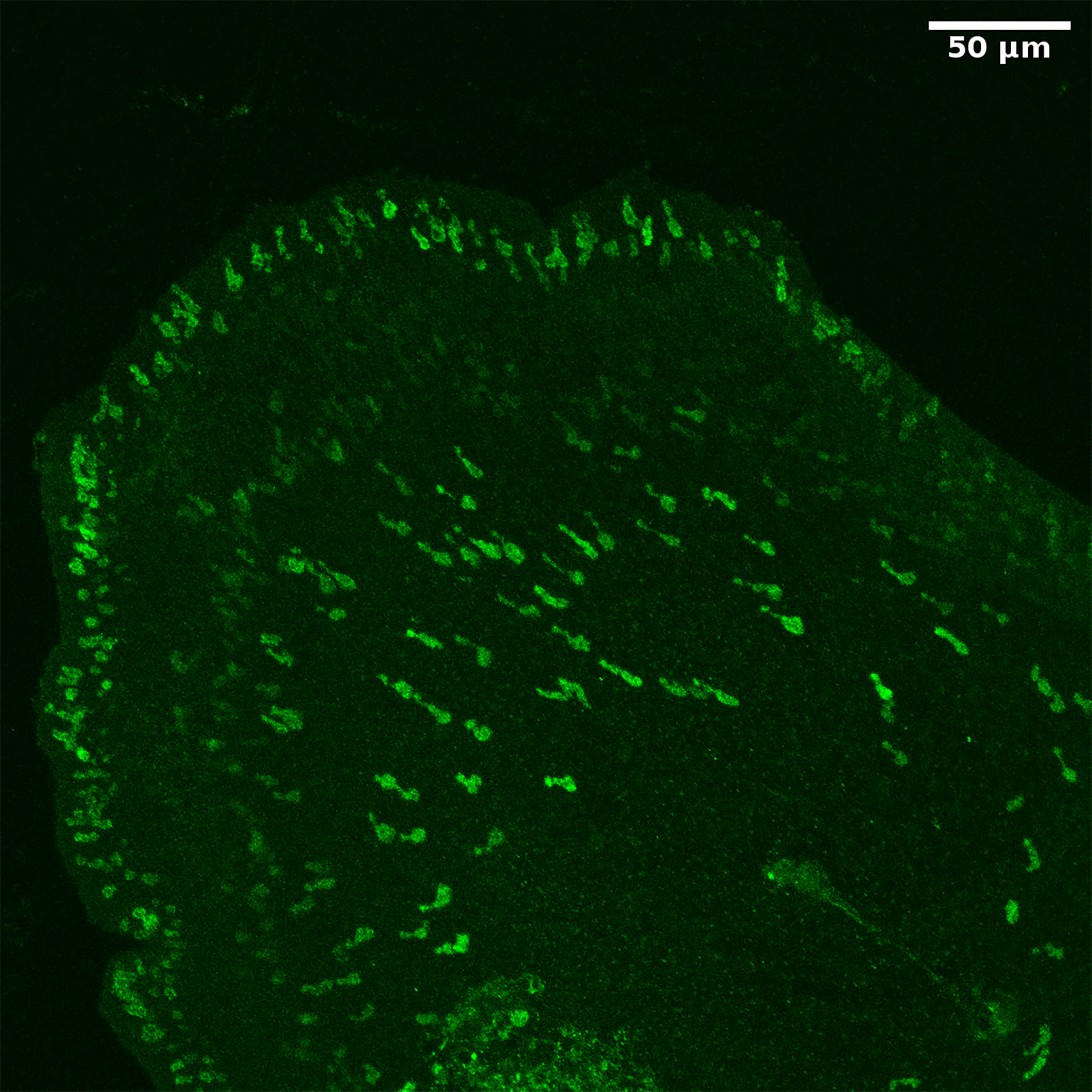
Figure 7 Whole mount staining of T. adhaerens, using an antiserum against the neuropeptide processing enzyme glutaminyl cyclase from T. adhaerens. We can observe staining of a large number of endocrine cells. These cells are flask-shaped and have protrusions towards the surface of the animal (please, see its margin), where they probably receive sensory input.
In mammals, many neuropeptides are produced by both nerve cells and endocrine cells. A classical example of this phenomenon is somatostatin, which was one of the first neuropeptides to be isolated from mammalian hypothalami in the 1960s and 1970s (127, 128). Ten years later, when immunocytochemistry had been introduced as an extremely versatile cell staining technique, Dubois found that somatostatin was not only produced by neurons from the hypothalamus, but also by endocrine cells from the stomach, the so-called D-cells (129). Somatostatin released from the D-cells inhibits the release of gastrointestinal hormones by other endocrine cells in the stomach, thereby blocking gastric acid secretion (130).
Also in Protostomia, like D. melanogaster, there are several neuropeptides that are produced in enteroendocrine cells of the gut, such as diuretic hormone-31, the allatostatins-A, -B, and -C, CCHamides, neuropeptide-F, and tachykinins (131–134). These peptides, however, like the other neuropeptides from Drosophila, are also expressed in neurons of the central nervous system (135). Both in protostomes and deuterostomes, therefore, neuropeptide genes may be expressed in neurons and endocrine cells.
How is the situation in Cnidaria? Yearlong ultrastructural work by Jane Westfall on cnidarians has shown that two types of neurons exist in cnidarians: “sensory cells” and “ganglion cells” (136). Cnidarians have two cell layers, ectoderm and endoderm, which are separated by a layer of extracellular matrix, called the mesoglea. Sensory cells are part of either the ectoderm or endoderm, and are orientated perpendicularly to the mesoglea. These cells have protrusions towards the apical parts of their cell layer and are typically bearing a sensory cilium that reaches out into the gastric cavity of the animal, or into the surrounding seawater (136). The ganglion cells are lying in the basal portions of the two cell layers and are a part of in the nerve net of the animal. Ganglion cells also connect to the sensory cells via synapses. Both sensory cells and ganglion cells have long processes and form multiple synapses, so they have the properties of genuine neurons (136).
As mentioned in Chapter 3, cnidarian nervous systems are strongly peptidergic (34–71, 74–77). By using antibodies against the different cnidarian neuropeptides, therefore, it is possible to stain the various populations of nerve cells in high detail. Such staining shows both sensory cells and ganglion cells connected to densely or loosely woven nerve nets or giant nerve fibres (35–37, 56–61). We have never realized that among the stained cells also might be endocrine cells, simply because their cell shapes make them look like sensory cells. Yet, we were sometimes wondering about scattered single immunostained sensory cells with no processes that were located in the endoderm of anthozoans, for example in the endodermal ovaries of octocorals (60). An example of these cells can be seen in Supplementary Figure S8. Therefore, it did not come as a complete surprise to us, when a publication appeared this year, showing that the anthozoan N. vectensis had genuine endocrine cells (137). These cells became visible in transgenic N. vectensis embryos, expressing the transcription factor N. vectensis insulinoma-associated protein 1 (NvInsml-1) coupled to green fluorescent protein (GFP). In vertebrates, insml-1 expression is required for the development of central and peripheral neurons and for stem cells that are committed to differentiate into endocrine cells (138). The orthologous gene in N. vectensis, NvInsml-1, appears to have similar properties. The gastrula stage was the first stage in which N. vectensis embryos, transgenically expressing the NvInsml-1-GFP gene, showed labelled cells, which anatomically looked like endocrine cells. These presumed endocrine cells had the appearance of sensory cells with protrusions towards the apical part of the ectoderm and an apical cilium, but lacking their basal nerve processes (dendrites) (137). In their planula stages, the transgenic embryos expressed GFP in typical sensory cells, and in their primary polyp stages, these transgenic lines expressed GFP in ganglion cells. Cnidarians, therefore, appear to have (i) endocrine cells (ii) sensory cells, and (iii) ganglion cells, while placozoans only have endocrine cells (113). The situation in cnidarians resembles that of mammals and other bilaterians, while placozoans are unique for only having endocrine cells that are responsible for peptidergic signaling.
Cnidaria and Placozoa probably form a monophyletic clade based on their common ways of prohormone processing, which is somewhat different from that in Bilateria (Chapter 3, Chapter 4). Also, molecular phylogenetic work from other laboratories supports a single clade of Cnidaria and Placozoa (139). This brings up the question of whether the common ancestor of cnidarians and placozoans had (i) endocrine cells (like in placozoans); or (ii) endocrine cells, sensory cells, and ganglion cells (like in cnidarians). Of course, this question is hard to answer in an experimental way. The above-mentioned experiments with transgenic N. vectensis embryo’s (137), however, very neatly show a sequential appearance, during sea anemone embryonic development, of first endocrine cells (in gastrula stages), then sensory cells (in planula stages), and finally ganglion cells (in primary polyps). If one could accept that essential parts of Ernst Haeckel’s “Biogenetic Law” (“ontogeny recapitulates phylogeny”) is still valid in modern biology (140, 141), this consecutive appearance of the three cell types would support the idea that endocrine cells evolved first.
Neuropeptides and their GPCRs are present in bilaterians, cnidarians, placozoans, and possibly other animal taxa. In placozoans, it would be more appropriate to call these peptides “endocrine peptides”, because nerve cells are apparently lacking in this early-branching animal phylum. It is hard to see evolutionary relationships between the neuropeptide/GPCR couples across bilaterians, cnidarians, and placozoans, although within bilaterians, these relationships between proto- and deuterostomes are obvious. Yet, a special group of neuropeptide-related hormones, the glycoprotein hormones and their GPCRs, the LGRs, are considerably conserved over a long evolutionary period. Especially in the early-branching phyla, the LGRs have strongly radiated, which culminated in placozoans having forty-six LGRs compared to eight in humans.
Genomics, transcriptomics, and bioinformatics have been invaluable tools for our understanding of cnidarian and placozoan neuro-endocrine evolution (34, 74–76). These techniques have already transformed large areas of biology today and will certainly continue to do so in the near future.
For Supplementary Figure S3, the human family-A GPCRs, excluding the olfactory receptors, were downloaded from GPCR-PEn (http://gpcr.utep.edu). Cnidarian family-A GPCRs, predicted from the sequenced genome of Nematostella vectensis, were downloaded from uniprot (http://uniprot.org). Placozoan family-A GPCRs, annotated from the re-annotated genome and the predicted proteome of T.adhaerens, were downloaded from genomeevolution.org (142). The receptors were extracted using the HMMER hmmsearch method using the EMBL-EBI Pfam rhodopsin GPCR model (pfam.xfam.org/). Sequences with at least 5 predicted transmembrane domains, as predicted by TMHMM, were kept for analysis.
The receptors were aligned using MAFFT v7.487, using the FFT-NS-2 alignment strategy. The alignments were trimmed using trimAl v1.4 with the -gt 0.1 settings. The trees were constructed using IQ-Tree v1.6.12 with the JTT model on a single thread. Bootstrap values were calculated using IQ-Tree’s UFboot method with 1000 replicates.
For the phylogenetic tree analyses of Figures 2, 6, the sequences were aligned by ClustalW using MEGA v11.0.11, but their N-termini were not trimmed. The accession numbers of the proteins used for Figure 2 are shown in Supplementary Table S1. The accession numbers of the proteins used for Figure 6 are shown in Supplementary Table S2. The trees were constructed with the neighbor-joining method. Bootstrap values were calculated with 1000 replicates.
This paper is part of the research topic “Genomic and Transcriptomic Insights into Neuroendocrine Evolution and Function”, hosted by Drs Dan Larhammar, Isabel Beets, Maurice Richard Elphick, and Elizabeth Amy Williams.
FH, TK, and CG conceived and designed the project and analyzed the data. CG wrote the paper with continuous input from the other authors. All authors approved the final manuscript.
We thank the Danish Council for Independent Research (grant number 7014-000088 to CG) for financial support. This funding body played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.973862/full#supplementary-material
1. Grimmelikhuijzen CJP, Hauser F. Mini-review: the evolution of neuropeptide signaling. Regul Pept (2012) 177:S6–9. doi: 10.1016/j.regpep.2012.05.001
2. Dufour S, Quérat B, Tostivint H, Pasqualini C, Vaudry H, Rousseau K. Origin and evolution of the neuroendocrine control of reproduction in vertebrates, with special focus on genome and gene duplications. Physiol Rev (2020) 100:869–943. doi: 10.1152/physrev.00009.2019
3. Douzery EJ, Snell EA, Bapteste E, Delsuc F, Philippe H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc Natl Acad Sci USA (2004) 101:15386–91. doi: 10.1073/pnas.0403984101
4. Nässel DR, Wegener C. A comparative review of short and long neuropeptide F signaling in invertebrates: Any similarities to vertebrate neuropeptide Y signaling? Peptides (2011) 32:1335–55. doi: 10.1016/j.peptides.2011.03.013
5. Odekunle EA, Elphick MR. Comparative and evolutionary physiology of vasopressin/oxytocin-type neuropeptide signaling in invertebrates. Front Endocrinol (2020) 11:225. doi: 10.3389/fendo.2020.00225
6. Nässel DR, Zandawala M, Kawada T, Satake H. Tachykinins: Neuropeptides that are ancient, diverse, widespread, and functionally pleiotropic. Front Neurosci (2019) 13:1262. doi: 10.3389/fnins.2019.01262
7. Stafflinger E, Hansen KK, Hauser F, Schneider M, Cazzamali G, Williamson M, et al. Cloning and identification of an oxytocin/vasopressin-like receptor and its ligand from insects. Proc Natl Acad Sci USA (2008) 105:3262–7. doi: 10.1073/pnas.0710897105
8. Nässel DR, Wu SF. Cholecystokinin/sulfakinin peptide signaling: conserved roles at the intersection between feeding, mating and aggression. Cell Mol Life Sci (2022) 79:188. doi: 10.1007/s00018-022-04214-4
9. Nachmann RJ, Holman GM, Haddon WF, Ling N. Leucosulfakinin, a sulfated insect neuropeptide with homology to gastrin and cholecystokinin. Science (1986) 234:71–3. doi: 10.1126/science.3749893
10. Rehfeld JF. Cholecystokinin and the hormone concept. Endocrine Connect (2021) 10:R139–50. doi: 10.1530/EC-21-0025
11. Hauser F, Grimmelikhuijzen CJP. Evolution of the AKH/corazonin/ACP/GnRH receptor superfamily and their ligands in the Protostomia. Gen Comp Endocrinol (2014) 209:35–49. doi: 10.1016/j.ygcen.2014.07.009
12. Li S, Hauser F, Skadborg SK, Nielsen SV, Kirketerp-Møller N, Grimmelikhuijzen CJP. Adipokinetic hormones and their G protein-coupled receptors emerged in Lophotrochozoa. Scient Rep (2016) 6:32789. doi: 10.1038/srep32789
13. Hauser F, Cazzamali G, Williamson M, Blenau W, Grimmelikhuijzen CJP. A review of neurohormone GPCRs present in the fruitfly Drosophila melanogaster and the honey bee Apis mellifera. Prog Neurobiol (2006) 80:1–19. doi: 10.1016/j.pneurobio.2006.07.005
14. Hauser F, Cazzamali G, Williamson M, Park Y, Li B, Tanaka Y, et al. A genome-wide inventory of neurohormone GPCRs in the red flour beetle Tribolium castaneum. Front Neuroendocrinol (2008) 29:142–65. doi: 10.1016/j.yfrne.2007.10.003
15. Civelli O, Nothacker H-P, Saito Y, Wang Z, Lin SH, Reinscheid RK. Novel neurotransmitters as natural ligands of orphan G-protein-coupled receptors. Trends Neurosci (2001) 24:230–7. doi: 10.1016/S0166-2236(00)01763-X
16. Civelli O. GPCR deorphanizations: the novel, the known and the unexpected transmitters. Trends Pharm Sci (2005) 26:15–9. doi: 10.1016/j.tips.2004.11.005
17. Aikins MJ, Schooley DA, Begum K, Detheux M, Beeman RW, Park Y. Vasopressin-like peptide and its receptor function in an indirect diuretic signaling pathway in the red flour beetle. Insect Biochem Mol Biol (2008) 38:740–8. doi: 10.1016/j.ibmb.2008.04.006
18. van Kesteren RE, Tensen CP, Smit AB, van Minnen J, van Soest PF, Kits KS, et al. A novel G protein-coupled receptor mediating both vasopressin- and oxytocin-like functions of Lys-conopressin in Lymnaea stagnalis. Neuron (1995) 15:897–908. doi: 10.1016/0896-6273(95)90180-9
19. Kanda A, Takuwa-Kuroda K, Iwakoshi-Ukena E, Furukawa Y, Matsushima O, Minakata H. Cloning of Octopus cephalotocin receptor, a member of the oxytocin/vasopressin superfamily. J Endocrinol (2003) 179:281–91. doi: 10.1677/joe.0.1790281
20. Kanda A, Satake H, Kawada T, Minakata H. Novel evolutionary lineages of the invertebrate oxytocin/vasopressin superfamily peptides and their receptors in the common octopus (Octopus vulgaris). Biochem J (2005) 387:85–91. doi: 10.1042/BJ20041230
21. Levoye A, Mouillac B, Rivière G, Vieau D, Salzet M, Breton C. Cloning, expression and pharmacological characterization of a vasopressin-related receptor in an annelid, the leech Theromyzon tessulatum. J Endocrinol (2005) 184:277–89. doi: 10.1677/joe.1.05833
22. Brown MR, Crim JW, Arata RC, Cai HN, Chun C, Shen P. Identification of a Drosophila brain-gut peptide related to the neuropeptide Y family. Peptides (1999) 20:1035–42. doi: 10.1016/S0196-9781(99)00097-2
23. Garczynski SF, Brown MR, Shen P, Murray TF, Crim JW. Characterization of a functional neuropeptide F receptor from Drosophila melanogaster. Peptides (2002) 23:773–80. doi: 10.1016/S0196-9781(01)00647-7
24. Kubiak TM, Larsen MJ, Burton KJ, Bannow CA, Martin RA, Zantello MR, et al. Cloning and functional expression of the first Drosophila melanogaster sulfakinin receptor DSK-R1. Biochem Biophys Res Commun (2002) 291:313–20. doi: 10.1006/bbrc.2002.6459
25. Hauser F, Søndergaard L, Grimmelikhuijzen CJP. Molecular cloning, genomic organization and developmental regulation of a novel receptor from Drosophila melanogaster structurally related to gonadotropin-releasing hormone receptors from vertebrates. Biochem Biophys Res Commun (1998) 249:822–8. doi: 10.1006/bbrc.1998.9230
26. Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, et al. The genomic sequence of Drosophila melanogaster. Science (2000) 287:2185–95. doi: 10.1126/science.287.5461.2185
27. Staubli F, Jørgensen TJD, Cazzamali G, Williamson M, Lenz C, Søndergaard L, et al. Molecular identification of the insect adipokinetic hormone receptors. Proc Natl Acad Sci USA (2002) 99:3446–51. doi: 10.1073/pnas.052556499
28. Stone JV, Mordue W, Batley KE, Morris HR. Structure of locust adipokinetic hormone, a neurohormone that regulates lipid utilisation during flight. Nature (1976) 263:207–21. doi: 10.1038/263207a0
29. Hook V, Funkelstein L, Lu D, Bark S, Wegrzyn J, Hwang S-Z. Proteases for processing proneuropeptide into peptide neurotransmitters and hormones. Ann Rev Pharmacol Toxicol (2008) 48:393–423. doi: 10.1146/annurev.pharmtox.48.113006.094812
30. Seidah NG, Prat A The biology and therapeutic targeting of the proprotein convertases. Nature Rev Drug Discover (2012) 11:367–83. doi: 10.1038/nrd3699
31. Eipper BA, Stoffers DA, Mains RE. The biosynthesis of neuropeptides: Peptide alpha-amidation. Ann Rev Neurosci (1992) 15:57–85. doi: 10.1146/annurev.ne.15.030192.000421
32. Schilling S, Hoffmann T, Manhart S, Hoffmann M, Demuth HU. Glutaminyl cyclases unfold glutaminyl cyclase activity under mild acid conditions. FEBS Lett (2004) 563:191–6. doi: 10.1016/S0014-5793(04)00300-X
33. Williams EA, Jékely G. Nervous systems: Neuropeptides define enigmatic comb-jelly neurons. Curr Biol (2021) 31:R1515–7. doi: 10.1016/j.cub.2021.10.054
34. Koch TL, Hauser F, Grimmelikhuijzen CJP. An evolutionary genomics view on neuropeptide genes in Hydrozoa and Endocnidozoa (Myxozoa). BMC Genomics (2021) 22:862. doi: 10.1186/s12864-021-08091-2
35. Grimmelikhuijzen CJP, Carstensen K, Darmer D, Moosler A, Nothacker HP, Reinscheid RK, et al. Coelenterate neuropeptides: structure, action and biosynthesis. Amer Zool (1992) 32:1–12. doi: 10.1093/icb/32.1.1
36. Grimmelikhuijzen CJP, Leviev I, Carstensen K. Peptides in the nervous systems of cnidarians: Structure, function and biosynthesis. Int Rev Cytol (1996) 167:37–89. doi: 10.1016/S0074-7696(08)61345-5
37. Grimmelikhuijzen CJP, Williamson M, Hansen GN. Neuropeptides in cnidarians. Can J Zool (2002) 80:1690–702. doi: 10.1139/z02-137
38. Grimmelikhuijzen CJP, Graff D. Isolation of <Glu-Gly-Arg-Phe-NH2 (Antho-RFamide), a neuropeptide from sea anemones. Proc Natl Acad Sci USA (1986) 83:9817–21. doi: 10.1073/pnas.83.24.9817
39. Grimmelikhuijzen CJP, Groeger A. Isolation of the neuropeptide pGlu-Gly-Arg-Phe-amide from the pennatulid Renilla köllikeri. FEBS Lett (1987) 211:105–8. doi: 10.1016/0014-5793(87)81283-8
40. Grimmelikhuijzen CJP, Hahn M, Rinehart KL, Spencer AN. Isolation of <Glu-Leu-Leu-Gly-Gly-Arg-Phe-NH2 (Pol-RFamide), a novel neuropeptide from hydromedusae. Brain Res (1988) 475:198–203. doi: 10.1016/0006-8993(88)90219-3
41. Graff D, Grimmelikhuijzen CJP. Isolation of <Glu-Ser-Leu-Arg-Trp-NH2, a novel neuropeptide from sea anemones. Brain Res (1988) 442:354–8. doi: 10.1016/0006-8993(88)91525-9
42. Graff D, Grimmelikhuijzen CJP. Isolation of <Glu-Gly-Leu-Arg-Trp-NH2 (Antho-RWamide II), a novel neuropeptide from sea anemones. FEBS Lett (1988) 239:137–40. doi: 10.1016/0014-5793(88)80560-X
43. Grimmelikhuijzen CJP, Rinehart KL, Spencer AN. Isolation of the neuropeptide <Glu-Trp-Leu-Lys-Gly-Arg-Phe-NH2 (Pol-RFamide II) from the hydromedusa polyorchis penicillatus. Biochem Biophys Res Commun (1992) 183:375382. doi: 10.1016/0006-291X(92)90491-3
44. Carstensen K, Rinehart KL, McFarlane ID, Grimmelikhuijzen CJP. Isolation of leu-Pro-Pro-Gly-Pro-Leu-Pro-Arg-Pro-NH2 (Antho-RPamide), an N-terminally protected, biologically active neuropeptide from sea anemones. Peptides (1992) 13:851–7. doi: 10.1016/0196-9781(92)90040-A
45. Carstensen K, McFarlane ID, Rinehard KL, Hudman D, Sun F, Grimmelikhuijzen CJP. Isolation of <Glu-Asn-Phe-His-Leu-Arg-Pro-NH2 (Antho-RPamide II), a novel, biologically active neuropeptide from sea anemones. Peptides (1993) 14:131–5. doi: 10.1016/0196-9781(93)90020-H
46. Leitz T, Morand K, Mann M. Metamorphosin a: A novel peptide controlling development of the lower metazoan Hydractinia echinata (Coelenterata, Hydrozoa). Dev Biol (1994) 163:440–6. doi: 10.1006/dbio.1994.1160
47. Moosler A, Rinehart KL, Grimmelikhuijzen CJP. Isolation of four novel neuropeptides, the Hydra-RFamides I-IV, from Hydra magnipapillata. Biochem Biophys Res Commun (1996) 229:596–602. doi: 10.1006/bbrc.1996.1849
48. Moosler A, Rinehardt KL, Grimmelikhuijzen CJP. Isolation of three novel neuropeptides, the Cyanea-RFamides I-III, from scyphomedusae. Biochem Biophys Res Commun (1997) 236:743–9. doi: 10.1006/bbrc.1997.7022
49. Takahashi T, Muneoka Y, Lohmann J, Lopez de Haro MS, Solleder G, Bosch TCG, et al. Systematic isolation of peptide signal molecules regulating development in Hydra; LWamide and PW families. Proc Natl Acad Sci USA (1997) 94:1241–6. doi: 10.1073/pnas.94.4.1241
50. Yum S, Takahashi T, Koizumi O, Ariura Y, Kobayakawa Y, Mohri S, et al. A novel neuropeptide, Hym-176, induces contraction of the ectodermal muscle in Hydra. Biochem Biophys Res Commun (1998) 248:584–90. doi: 10.1006/bbrc.1998.8831
51. Takahashi T, Koizumi O, Ariura Y, Romanovitch A, Bosch TC, Kobayakawa Y, et al. A novel neuropeptide, Hym-355, positively regulates neuron differentiation in Hydra. Development (2000) 127:997–1005. doi: 10.1242/dev.127.5.997
52. Hayakawa E, Takahashi T, Nishimiya-Fujisawa C, Fujisawa T. A novel neuropeptide (FRamide) family identified by a peptidomic approach in Hydra magnipapillata. FEBS J (2007) 274:5438–48. doi: 10.1111/j.1742-4658.2007.06071.x
53. Takahashi T, Takeda N. Insight into the molecular and functional diversity of cnidarian neuropeptides. Int J Mol Sci (2015) 16:2610–25. doi: 10.3390/ijms16022610
54. Takeda N, Kon Y, Quiroga Artigas G, Lapébie P, Barreau C, Koizumi O, et al. Identification of jellyfish neuropeptides that act directly as oocyte maturation-inducing hormones. Development (2018) 145:dev156786. doi: 10.1242/dev.156786
55. Takahashi T. Comparative aspects of structure and function of cnidarian neuropeptides. Front Endocrinol (2020) 11:339. doi: 10.3389/fendo.2020.00339
56. Grimmelikhuijzen CJP, Spencer AN. FMRFamide immunoreactivity in the nervous system of the medusa Polyorchis penicillatus. J Comp Neurol (1984) 230:361–71. doi: 10.1002/cne.902300305
57. Grimmelikhuijzen CJP. Antisera to the sequence Arg-Phe-amide visualize neuronal centralization in hydroid polyps. Cell Tissue Res (1985) 241:171–82. doi: 10.1007/BF00214639
58. Grimmelikhuijzen CJP, Spencer AN, Carré D. Organization of the nervous system of physonectid siphonophores. Cell Tissue Res (1986) 246:463–79. doi: 10.1007/BF00215186
59. Mackie GO. The elementary nervous system revisited. Amer Zool (1990) 30:907–20. doi: 10.1093/icb/30.4.907
60. Pernet V, Anctil M, Grimmelikhuijzen CJP. Antho-RFamide-containing neurons in the primitive nervous system of the anthozoan Renilla koellikeri. J Comp Neurol (2004) 472:208–20. doi: 10.1002/cne.20108
61. Nielsen SKD, Koch TL, Wiisbye SH, Grimmelikhuijzen CJP, Garm A. Neuropeptide expression in the box jellyfish Tripedalia cystophora – New insights into the complexity of a “simple” nervous system. J Comp Neurol (2021) 529:2865–82. doi: 10.1002/cne.25133
62. McFarlane ID, Anderson PAV, Grimmelikhuijzen CJP. Effects of three anthozoan neuropeptides, Antho-RWamide I, Antho-RWamide II and Antho-RFamide, on slow muscles from sea anemones. J Exp Biol (1991) 156:419–31. doi: 10.1242/jeb.156.1.419
63. Katsukura Y, David CN, Grimmelikhuijzen CJP, Sugiyama T. Inhibition of metamorphosis by RFamide neuropeptides in planula larvae of Hydractinia echinata. Dev Genes Evol (2003) 213:579–86. doi: 10.1007/s00427-003-0361-5
64. Katsukura Y, Ando H, David CN, Grimmelikhuijzen CJP, Sugiyama T. Control of planula migration by LWamide and RFamide neuropeptides in Hydractinia echinata. J Exp Biol (2004) 207:1803–10. doi: 10.1242/jeb.00974
65. Darmer D, Schmutzler C, Diekhoff D, Grimmelikhuijzen CJP. Primary structure of the precursor for the sea anemone neuropeptide Antho-RFamide (<Glu-Gly-Arg-Phe-NH2). Proc Natl Acad Sci USA (1991) 88:2555–59. doi: 10.1073/pnas.88.6.2555
66. Schmutzler C, Darmer D, Diekhoff D, Grimmelikhuijzen CJP. Identification of a novel type of processing sites in the precursor for the sea anemone neuropeptide Antho-RFamide (<Glu-Gly-Arg-Phe-NH2) from Anthopleura elegantissima. J Biol Chem (1992) 267:22534–41. doi: 10.1016/S0021-9258(18)41705-X
67. Schmutzler C, Diekhoff D, Grimmelikhuijzen CJP. The primary structure of the Pol-RFamide neuropeptide precursor protein from the hydromedusa Polyorchis penicillatus indicates a novel processing proteinase activity. Biochem J (1994) 299:431–6. doi: 10.1042/bj2990431
68. Leviev I, Grimmelikhuijzen CJP. Molecular cloning of a preprohormone from sea anemones containing numerous copies of a metamorphosis inducing neuropeptide: A likely role for dipeptidyl aminopeptidase in neuropeptide precursor processing. Proc Natl Acad Sci USA (1995) 92:11647–51. doi: 10.1073/pnas.92.25.11647
69. Leviev I, Williamson M, Grimmelikhuijzen CJP. Molecular cloning of a preprohormone from Hydra magnipapillata containing multiple copies of Hydra-LWamide (Leu-Trp-NH2) neuropeptides: Evidence for processing at Ser and Asn residues. J Neurochem (1997) 68:1319–25. doi: 10.1046/j.1471-4159.1997.68031319.x
70. Darmer D, Hauser F, Nothacker H-P, Bosch TCG, Williamson M, Grimmelikhuijzen CJP. Three different prohormones yield a variety of Hydra-RFamide (Arg-Phe-NH2) neuropeptides in Hydra magnipapillata. Biochem J (1998) 332:403–12. doi: 10.1042/bj3320403
71. Yum S, Takahashi T, Hatta M, Fujisawa T. The structure and expression of a preprohormone of a neuropeptide, Hym-176 in Hydra magnipapillata. FEBS Lett (1998) 439:31–4. doi: 10.1016/S0014-5793(98)01314-3
72. Hauser F, Williamson M, Grimmelikhuijzen CJP. Molecular cloning of a peptidylglycine α-hydroxylating monooxygenase from sea anemones. Biochem Biophys Res Commun (1997) 241:509–12. doi: 10.1006/bbrc.1997.7854
73. Williamson M, Hauser F, Grimmelikhuijzen CJP. Genomic organization and splicing variants of a peptidylglycine α-hydroxylating monooxygenase from sea anemones. Biochem Biophys Res Commun (2000) 277:7–12. doi: 10.1006/bbrc.2000.3629
74. Nielsen SKD, Koch TL, Hauser F, Garm A, Grimmelikhuijzen CJP. De novo transcriptome assembly of the cubomedusa Tripedalia cystophora, including the analysis of a set of genes involved in peptidergic neurotransmission. BMC Genomics (2019) 20:175. doi: 10.1186/s12864-019-5514-7
75. Koch TL, Grimmelikhuijzen CJP. Global neuropeptide annotations from the genomes and transcriptomes of Cubozoa, Scyphozoa, Staurozoa (Cnidaria: Medusozoa), and Octocorallia (Cnidaria: Anthozoa). Front Endocrinol (2019) 10:831. doi: 10.3389/fendo.2019.00831
76. Koch KL, Grimmelikhuijzen CJP. A comparative genomics study of neuropeptide genes in the cnidarian subclasses Hexacorallia and Ceriantharia. BMC Genomics (2020) 21:666. doi: 10.1186/s12864-020-06945-9
77. Koch TL. Molecular evolution of peptide signaling in Placozoa and Cnidaria. Ph.D. thesis. University of Copenhagen (2020). Available at: http://www2.bio.ku.dk/docs/Thomas%20Lund%20Koch.pdf.
78. Yun S, Kim D-K, Furlong M, Hwang J-I, Vaudry H, Seong JY. Does kisspeptin belong to the proposed RFamide peptide family? Front Endocrinol (2014) 5:134. doi: 10.3389/fendo.2014.00134
79. Thiel D, Franz-Wachtel M, Aguilera F, Hejnol A. Xenacoelomorph neuropeptidomes reveal a major expansion of neuropeptide systems during early bilaterian evolution. Mol Biol Evol (2018) 35:2528–43. doi: 10.1093/molbev/msy160
80. Quiroga Artigas G, Lapébie P, Leclère L, Bauknecht P, Uveira J, Chevalier S, et al. A G protein-coupled receptor mediates neuropeptide-induced oocyte maturation in the jellyfish Clytia. PloS Biol (2020) 18:e3000614. doi: 10.1371/journal.pbio.3000614
81. Shiraishi A, Okuda T, Myasaka N, Osugi T, Okuno Y, Inoue J, et al. Repertoires of G protein-coupled receptors for Ciona-specific neuropeptides. Proc Natl Acad Sci USA (2019) 116:7847–56. doi: 10.1073/pnas.1816640116
82. Kawada T, Osugi T, Matsubara S, Sakai T, Shiraishi A, Yamamoto T, et al. Omics studies for the identification of ascidian peptides, cognate receptors, and their relevant roles in ovarian follicular development. Front Endocrinol (2022) 13:858885. doi: 10.3389/fendo.2022.858885
83. Yañez-Guerra LA, Thiel D, Jékely G. Premetazoan origin of neuropeptide signaling. Mol Biol Evol (2022) 39:msac051. doi: 10.1093/molbev/msac051
85. Schierwater B, Osigus H-J, Bergmann T, Blackstone NW, Hadrys H, Hauslage J, et al. The enigmatic Placozoa part 1: Exploring evolutionary controversies and poor ecological knowledge. Bioessays (2021) 43:e2100080. doi: 10.1002/bies.202100080
86. Nikitin M. Bioinformatic prediction of Trichoplax adhaerens regulatory peptides. Gen Comp Endocrinol (2015) 212:145–55. doi: 10.1016/j.ygcen.2014.03.049
87. Williams EA. Function and distribution of the Wamide neuropeptide superfamily in metazoans. Front Endocrinol (2020) 11:344. doi: 10.3389/fendo.2020.00344
88. Sudo S, Kumagai J, Nishi S, Layfield S, Ferraro T, Bathgate RAD, et al. H3 relaxin is a specific ligand for LGR7 and activates the receptor by interacting with both the ectodomain and the exoloop 2. J Biol Chem (2003) 278:7855–62. doi: 10.1074/jbc.M212457200
89. Zebisch M, Jones EY. Crystal structure of r-spondin 2 in complex with the ectodomains of its receptors LGR5 and ZNRF3. J Struct Biol (2015) 191:149–55. doi: 10.1016/j.jsb.2015.05.008
90. Xu J-G, Huang C, Yang Z, Jin M, Fu P, Zhang N, et al. Crystal structure of LGR4-Rspo1 complex. insights into the divergent mechanisms of ligand recognition by leucine-rich repeat G protein-coupled receptors (LGRs). J Biol Chem (2015) 290:2455–65. doi: 10.1074/jbc.M114.599134
91. Hsu SY, Nakabayashi K, Nishi S, Kumagai J, Kudo M, Sherwood OD, et al. Activation of orphan receptors by the hormone relaxin. Science (2002) 295:671–4. doi: 10.1126/science.1065654
92. Hsu SY, Nakabayashi K, Bhalla A. Evolution of glycoprotein hormone subunit genes in bilateral metazoa: identification of two novel human glycoprotein hormone subunit family genes, GPA2 and GPB5. Mol Endocrinol (2002) 16:1538–51. doi: 10.1210/mend.16.7.0871
93. Nakabayashi K, Matsumi H, Bhalla A, Bae J, Mosselman S, Hsu SY, et al. Thyrostimulin, a heterodimer of two new human glycoprotein hormone subunits, activates the thyroid-stimulating hormone receptor. J Clin Invest (2002) 109:1445–52. doi: 10.1172/JCI0214340
94. Ordaz-Ramos A, Rossales-Gallegos VH, Melendez-Zajgla J, Maldonado V, Vazquez-Santillan K. The role of LGR4 (GPR48) in normal and cancer processes. Int J Mol Sci (2021) 22:4690. doi: 10.3390/ijms22094690
95. Hsu SY, Liang S-G, Hsueh AJW. Characterization of two LGR genes homologous to gonadotropin and thyrotropin receptors with extracellular leucine-rich repeats and a G protein coupled, seven transmembrane region. Mol Endocrinol (1998) 12:1830–45. doi: 10.1210/mend.12.12.0211
96. Xu L, Lin W, Wen L, Li G. Lgr5 in cancer biology: functional identification of Lgr5 in cancer progression and potential opportunities for novel therapy. Stem Cell Res Ther (2019) 10:219. doi: 10.1186/s13287-019-1288-8
97. Huang PY, Kandyba E, Jabouille A, Sjolund J, Kumar A, Halliwill K, et al. Lgr6 is a stem cell marker in mouse skin squamous cell carcinoma. Nat Genet (2017) 49:1624–32. doi: 10.1038/ng.3957
98. Hsu SY, Kudo M, Chen T, Nakabayashi K, Bhalla A, van der Spek PJ, et al. The three subfamilies of leucine-rich repeat-containing G protein-coupled receptors (LGR): Identification of LGR6 and LGR7 and the signaling mechanism for LGR7. Mol Endocrinol (2000) 14:1257–71. doi: 10.1210/mend.14.8.0510
99. Van Hiel MB, Vandersmissen HP, Van Loy T, Vanden Broeck J. An evolutionary comparison of leucine-rich repeat containing G protein-coupled receptors reveals a novel LGR subtype. Peptides (2012) 34:193–200. doi: 10.1016/j.peptides.2011.11.004
100. Hauser F, Nothacker H-P, Grimmelikhuijzen CJP. Molecular cloning, genomic organization and developmental regulation of a novel receptor from Drosophila melanogaster structurally related to members of the thyroid-stimulating hormone, follicle-stimulating hormone, luteinizing hormone/choriogonadotropin receptor family from mammals. J Biol Chem (1997) 272:1002–10. doi: 10.1074/jbc.272.2.1002
101. Eriksen KK, Hauser F, Schiøtt M, Pedersen K-M, Søndergaard L, Grimmelikhuijzen CJP. Molecular cloning, genomic organization, developmental regulation, and a knock out mutant of a novel leu-rich repeats-containing G protein-coupled receptor (DLGR-2) from Drosophila melanogaster. Genome Res (2000) 10:924–38. doi: 10.1101/gr.10.7.924
102. Sudo S, Kuwabara Y, Park JI, Hsu SY, Hsueh AJW. Heterodimeric fly glycoprotein hormone-alpha2 (GPA2) and glycoprotein hormone-beta5 (GPB5) activate fly leucine-rich repeat-containing G protein-coupled receptor-1 (DLGR1) and stimulation of human thyrotropin receptors by chimeric fly GPA2 and human GPB5. Endocrinol (2005) 146:3596–3604. doi: 10.1210/en.2005-0317
103. Mendive FM, Van Loy T, Claeysen S, Poels J, Williamson M, Hauser F, et al. Drosophila molting neurohormone bursicon is a heterodimer and the natural agonist of the orphan receptor DLGR2. FEBS Lett (2005) 579:2171–6. doi: 10.1016/j.febslet.2005.03.006
104. Luo CW, Dewey EM, Sudo S, Ewer J, Hsu SY, Honegger HW, et al. Bursicon, the insect cuticle-hardening hormone, is a heterodimeric cystine knot protein that activates G protein-coupled receptor LGR2. Proc Natl Acad Sci USA (2005) 102:2820–5. doi: 10.1073/pnas.0409916102
105. Honegger HW, Dewey EM, Ewer J. Bursicon, the tanning hormone of insects: recent advances following the discovery of its molecular identity. J Comp Physiol A (2008) 194:989–1005. doi: 10.1007/s00359-008-0386-3
106. Scopelliti A, Bauer C, Yu Y, Zhang T, Kruspig B, Murphy DJ, et al. A neuronal relay mediates a nutrient responsive gut/fat body axis regulating energy homeostasis in adult Drosophila. Cell Metab (2019) 29:269–284.e10. doi: 10.1016/j.cmet.2018.09.021
107. Vallejo DM, Juarez-Carreño S, Bolivar J, Morante J, Dominguez M. A brain circuit that synchronizes growth and maturation revealed through Dilp8 binding to Lgr3. Science (2015) 350:aac6767. doi: 10.1126/science.aac6767
108. Nothacker H-P, Grimmelikhuijzen CJP. Molecular cloning of a novel, putative G protein-coupled receptor from sea anemones structurally related to members of the FSH, TSH, LH/CG receptor family from mammals. Biochem Biophys Res Commun (1993) 197:1062–9. doi: 10.1006/bbrc.1993.2586
109. Vibede N, Hauser F, Williamson M, Grimmelikhuijzen CJP. Genomic organization of a receptor from sea anemones, structurally and evolutionarily related to glycoprotein hormone receptors from mammals. Biochem Biophys Res Commun (1998) 252:497–501. doi: 10.1006/bbrc.1998.9661
110. Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, et al. Sea Anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science (2007) 317:86–94. doi: 10.1126/science.1139158
111. Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T. The dynamic genome of Hydra. Nature (2010) 464:592–6. doi: 10.1038/nature08830
112. Roch GJ, Sherwood NM. Glycoprotein hormones and their receptors emerged at the origin of metazoans. Genome Biol Evol (2014) 6:1466–79. doi: 10.1093/gbe/evu118
113. Varoqueaux F, Williams EA, Grandemange S, Truscello L, Kamm K, Schierwater B, et al. High cell diversity and complex peptidergic signaling underlie placozoan behavior. Curr Biol (2018) 28:3495–501. doi: 10.1016/j.cub.2018.08.067
114. Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 a resolution and low pH. Nature (2007) 449:316–23. doi: 10.1038/nature06163
115. Baconguis I, Bohlen CJ, Goehring A, Julius D, Gouaux E. X-Ray structure of acid-sensing ion channel 1-snake toxin complex reveals open state of a Na(+)-selective channel. Cell (2014) 156:717–29. doi: 10.1016/j.cell.2014.01.011
116. Cottrell GA, Green KA, Davies NW. The neuropeptide Phe-Met-Arg-Phe-NH2 (FMRFamide) can activate a ligand-gated ion channel in Helix neurones. Pflügers Arch (1990) 416:612–4. doi: 10.1007/BF00382698
117. Lingueglia E, Champigny G, Lazdunski M, Barbry P. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature (1995) 378:730–3. doi: 10.1038/378730a0
118. Perry SJ, Straub VA, Schofield MG, Burke JF, Benjamin PR. Neuronal expression of an FMRFamide-gated Na+ channel and its modulation by acid pH. J Neurosci (2001) 21:5559–67. doi: 10.1523/JNEUROSCI.21-15-05559.2001
119. Furukawa Y, Myawaki Y, Abe G. Molecular cloning and functional characterization of the Aplysia FMRFamide-gated Na+ channel. Pfügers Arch - Eur J Physiol (2006) 451:646–56. doi: 10.1007/s00424-005-1498-z
120. Dandamudi M, Hausen H, Lynagh T. Comparative analysis defines a broader FMRFamide-gated sodium channel family and determinants of neuropeptide sensitivity. J Biol Chem (2022) 298:102086. doi: 10.1016/j.jbc.2022.102086
121. Golubovic A, Kuhn A, Williamson M, Kalbacher H, Holstein TW, Grimmelikhuijzen CJP, et al. A peptide-gated ion channel from the freshwater polyp Hydra. J Biol Chem (2007) 282:35098–103. doi: 10.1074/jbc.M706849200
122. Dürrnagel S, Kuhn A, Tsiairis CD, Williamson M, Kalbacher H, Grimmelikhuijzen CJP, et al. Three homologous subunits form a high affinity peptide-gated ion channel in Hydra. J Biol Chem (2010) 285:11958–65. doi: 10.1074/jbc.M109.059998
123. Dürrnagel S, Falkenburger BH, Gründer S. High Ca (2+) permeability of a peptide-gated DEG/ENaC from Hydra. J Gen Physiol (2012) 140:391–402. doi: 10.1085/jgp.201210798
124. Assman M, Kuhn A, Dürrnagel S, Holstein TW, Gründer S. The comprehensive analysis of DEG/ENaC subunits in Hydra reveals a large variety of peptide-gated channels, potentially involved in neuromuscular transmission. BMC Biol (2014) 12:84. doi: 10.1186/s12915-014-0084-2
125. Gründer S, Assmann M. Peptide-gated ion channels and the simple nervous system of Hydra. J Exp Biol (2015) 218:551–61. doi: 10.1242/jeb.111666
126. Schmidt A, Bauknecht P, Williams E Å, Augustinowski K, Gründer S, Jékely G. Dual signaling of Wamide myoinhibitory peptides through a peptide-gated channel and a GPCR in Platynereis. FASEB J (2018) 32:5338–49. doi: 10.1096/fj.201800274R
127. Brazeau P, Vale W, Burgus R, Ling N, Butcher M, Rivier J, et al. Hypothalamic polypeptide that inhibits the secretion of immunoreactive pituitary growth hormone. Science (1973) 179:77–9. doi: 10.1126/science.179.4068.77
128. Arimura A, Sato H, Dupont A, Nishi N, Schally AV. Somatostatin: Abundance of immunoreactive hormone in rat stomach and pancreas. Science (1975) 189:1007–9. doi: 10.1126/science.56779
129. Dubois MP. Immunoreactive somatostatin is present in discrete cells of the endocrine pancreas. Proc Natl Acad Sci USA (1975) 72:1340–3. doi: 10.1073/pnas.72.4.1340
130. Alumets J, Ekelund M, El Munshid HA, Håkanson R, Lorén I, Sundler F. Topography of somatostatin cells in the stomach of the rat: possible functional significance. Cell Tissue Res (1979) 202:177–88. doi: 10.1007/BF00232233
131. Veenstra JA, Agricola HJ, Sellami A. Regulatory peptides in fruit fly midgut. Cell Tissue Res (2008) 334:499–516. doi: 10.1007/s00441-008-0708-3
132. Li S, Torre-Muruzabal T, Søgaard KC, Ren GR, Hauser F, Engelsen SM, et al. Expression patterns of the Drosophila neuropeptide CCHamide-2 and its receptor may suggest hormonal signaling from the gut to the brain. PloS One (2013) 8:e76131. doi: 10.1371/journal.pone.0076131
133. Williamson M, Lenz C, Winther ÅME, Nässel DR, Grimmelikhuijzen CJP. Molecular cloning, genomic organization, and expression of a C-type (Manduca sexta-type) allatostatin preprohormone from Drosophila melanogaster. Biochem Biophys Res Comm (2001) 282:124–30. doi: 10.1006/bbrc.2001.4565
134. Williamson M, Lenz C, Winther ÅME, Nässel DR, Grimmelikhuijzen CJP. Molecular cloning, genomic organization, and expression of a B-type (cricket-type) allatostatin preprohormone from Drosophila melanogaster. Biochem Biophys Res Comm (2001) 281:544–50. doi: 10.1006/bbrc.2001.4402
135. Nässel DR, Winther ÅM. Drosophila neuropeptides in regulation of physiology and behavior. Prog Neurobiol (2010) 92:42–104. doi: 10.1016/j.pneurobio.2010.04.010
136. Westfall JA, Elliott CF, Carlin RW. Ultrastructural evidence for two-cell and three-cell neuronal pathways in the tentacle epidermis of the sea anemone Aiptasia pallida. J Morphol (2002) 251:83–92. doi: 10.1002/jmor.1075
137. Tournière O, Gahan JM, Busengdal H, Bartsch N, Rentzsch F. Insm1-expressing neurons and secretory cells develop from a common pool of progenitors in the sea anemone Nematostella vectensis. Sci Adv (2022) 8:eabi7109. doi: 10.1126/sciadv.abi7109
138. Farkas LM, Haffner C, Giger T, Khaitovich P, Nowick K, Birchmeier C, et al. Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron (2008) 60:40–55. doi: 10.1016/j.neuron.2008.09.020
139. Laumer CE, Gruber-Vodicka H, Hadfield MG, Pearse VB, Riesgo A, Marioni JC, et al. Support for a clade of Placozoa and Cnidaria in genes with minimal compositional bias. eLife (2018) 7:e36278. doi: 10.7554/eLife.36278
140. Olsson L, Levit GS, Hossfeld U. The “Biogenetic Law” in zoology: from Ernst Haeckel’s formulation to current approaches. Theory Biosci (2017) 136:19–29. doi: 10.1007/s12064-017-0243-4
141. Cárdenas A, Borell V. Molecular and cellular evolution of corticogenesis in amniotes. Cell Mol Life Sci (2020) 77:1435–60. doi: 10.1007/s00018-019-03315-x
Keywords: peptide, receptor, evolution, G protein-coupled receptor (GPCR), leucine-rich repeat-containing GPCR (LGR), DEG/ENaC, nervous system, endocrine system
Citation: Hauser F, Koch TL and Grimmelikhuijzen CJP (2022) Review: The evolution of peptidergic signaling in Cnidaria and Placozoa, including a comparison with Bilateria. Front. Endocrinol. 13:973862. doi: 10.3389/fendo.2022.973862
Received: 20 June 2022; Accepted: 26 August 2022;
Published: 23 September 2022.
Edited by:
Dan Larhammar, Uppsala University, SwedenReviewed by:
Tsuyoshi Kawada, Suntory Foundation for Life Sciences, JapanCopyright © 2022 Hauser, Koch and Grimmelikhuijzen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cornelis J. P. Grimmelikhuijzen, Y2dyaW1tZWxpa2h1aWp6ZW5AYmlvLmt1LmRr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.