
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol., 11 August 2022
Sec. Gut Endocrinology
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.958218
This article is part of the Research TopicThe Role of Bile Acid (BA) and Related Metabolites and Hormone Abnormalities in Metabolic DiseasesView all 12 articles
Type 2 diabetes mellitus (T2DM), one of the fastest growing metabolic diseases, has been characterized by metabolic disorders including hyperglycemia, hyperlipidemia and insulin resistance (IR). In recent years, T2DM has become the fastest growing metabolic disease in the world. Studies have indicated that patients with T2DM are often associated with intestinal flora disorders and dysfunction involving multiple organs. Metabolites of the intestinal flora, such as bile acids (BAs), short-chain fatty acids (SCFAs) and amino acids (AAs)may influence to some extent the decreased insulin sensitivity associated with T2DM dysfunction and regulate metabolic as well as immune homeostasis. In this paper, we review the changes in the gut flora in T2DM and the mechanisms by which the gut microbiota modulates metabolites affecting T2DM, which may provide a basis for the early identification of T2DM-susceptible individuals and guide targeted interventions. Finally, we also highlight gut microecological therapeutic strategies focused on shaping the gut flora to inform the improvement of T2DM progression.
T2DM is the most common metabolic disease and is mainly characterized by metabolic disorders, such as hyperglycemia, hyperlipidemia and IR. Diabetes can lead to various serious complications, such as coronary artery disease, lower extremity arteriopathy, retinopathy and diabetic nephropathy, which affect the life quality of a large populations worldwide (1, 2). The rapid growth of diabetes poses an alarming burden on global social and economic development (3). Recent data by the International Diabetes Federation (IDF) in 2021 have demonstrated that the global prevalence of diabetes in 20- to 79-year-olds was estimated at 10.5% (approximately 537 million people) and predicted to increase to 12.2% (783.2 million people) in 2045. The diabetes prevalence is projected to be higher in middle-income countries (21.1%) than in low-income countries (11.9%) and high-income countries (12.2%) between 2021 and 2045 (4, 5). The prevalence of T2DM is greater than 25% in older patients over 65 years of age; treatment of older patients is more difficult because complications are more likely to occur (6, 7). Today, the main treatment strategies for T2DM include surgery, pharmacotherapy, exercise therapy, diet, nutrition and multifactorial therapy (8, 9). The core of the treatment of T2DM is to solve the problem of hyperglycemia. The use of medication based on lifestyle modification is the cornerstone of the treatment of T2DM. If blood glucose remains uncontrolled, additional treatment with insulin and other injectable drugs will eventually be needed (10). Insulin injection therapy is the most effective way to control blood glucose (11). However, with increased IR, the risk of hypoglycemia is increased if insulin injections are increased, thus escalating the risk of cardiovascular-related complications. Therefore, the underlying mechanisms of T2DM need to be identified for early diagnosis and treatment.
Recognized risk factors for diabetes include obesity, poor dietary habits, positive family history and genetics (12, 13). A variety of factors play a critical part in the development of diabetes. Recently, plenty of studies have proposed a link of the intestinal microbiome and its metabolites with the metabolic health of the human body (14, 15). Genome-wide association reports show significant associations between specific gut microbes, bacterial genes and metabolic pathway variants in T2DM (16). The possible mechanisms of T2DM induced by the gut microbiota may be linked to IR, BA metabolism, disturbances of lipid metabolism and endotoxemia (17). Furthermore, many researches have shown that the relationship between BAs and microbiome plays a key role in the pathogenesis of metabolic disorders including T2DM (18, 19). The study showed that the imbalance in the interplay between BAs and the intestinal flora led to composition changes of the BA pool, as well as alterations in the intestinal flora structure and endocrine signaling pathways, which they suggest will influence the development of T2DM (20, 21). In addition, increased production of trimethylamine N-oxide (TMAO) and branched-chain amino acids (BCAAs) as well as changes in BA and SCFA metabolism may lead to changes in fat levels, thereby affecting insulin signaling (22–24).
Disorders of energy balance due to gut microbial dysbiosis have an integral role in the progression of T2DM (25). However, the mechanisms of intestinal flora in T2DM are not fully understood. Thus, the purpose of this review is to discuss the altered intestinal microbiota in T2DM and the role of metabolites in T2DM relative to a healthy human host. We provide a summary of microbiota-based therapeutic approaches to improve T2DM.
The impact of societal advances and changes in people’s lifestyles, which have led to a greater intake of high-energy foods and a lack of physical activity, has left individuals in a suboptimal state of metabolic health with a high incidence of metabolic diseases, such as obesity, T2DM, nonalcoholic liver disease and cardiometabolic disease (CMD) (26, 27). The two main pathogenic mechanisms of diabetes are the relative lack of insulin secretion and IR, with IR being the main cause of T2DM. Diabetes biomarkers, such as venous glucose levels and glycated hemoglobin (HbA1c), have limitations as predictive biomarkers, especially in elderly or hyperlipidemic patients. Therefore, the identification of other early predictors is needed.
The collection of all gut microbial genes in an individual (i.e., the microbiome) represents a gene pool that is an order of magnitude higher than the human genome (28). Therefore, the intestinal microbiome is considered an “ organ” with important roles in enhancing host immunity, facilitating the digestion of food, regulating intestinal endocrine function, regulating neural signaling, modifying drug action, modifying metabolism and eliminating toxins. This symbiotic relationship with the host ensures proper development of the human metabolic system. Gut microbial metabolites absorbed by the host act on receptors in organs such as the liver, gut, brown adipose tissue (BAT), white adipose tissue (WAT) and the central nervous system (CNS), and they are involved in micronutrient synthesis, intestinal motility, mineral absorption and electrolyte absorption (29, 30). A growing number of studies have demonstrated a strong link between the intestinal flora and human disease, particularly its role in obesity and T2DM (17, 31–35). Improvement of insulin sensitivity by altering the composition of the intestinal microbiota has gained attention from many researchers, and gut flora therapy has become a new treatment modality.
Approximately 86% of patients with T2DM are overweight or obese, and obesity is considered the greatest risk factor for T2DM (36), which is commonly used as an early warning marker for T2DM. Compared to healthy adults, obese and T2DM populations show significant changes in gut microbes and their metabolites (Table 1). Studies have shown that modern lifestyle changes and the use of antimicrobial drugs, among other factors, have led to a decrease in gut microbial diversity in many developed populations (72).
Studies have shown that individuals with low abundance of intestinal flora are predisposed to obesity, IR and dyslipidemia (43). A study of gut microbial diversity in patients with low gene count (LGC) and high gene count (HGC) has shown that Lactobacillus, Prevotella, Bacteroides, Desulfovibrio and Oxalobacter spp. in the intestinal microbiota are decreasing in abundance; this decrease in diversity may promote the development of metabolic disorders. Functional changes in the microbiota of individuals with LGC mainly include a reduction in butyrate-producing bacteria and an elevation in the ratio of Akkermansia to R. torque/gnavus, leading to enhanced mucus degradation, decreased hydrogen production potential, decreased methane production potential, increased Campylobacter/Shigella abundance and increased peroxidase activity (43). This metabolic disturbance caused by an imbalance between pro- and anti-inflammatory bacterial species in LGC individuals puts them at an increased risk for metabolic diseases, such as prediabetes and T2DM (73).
A previous study has shown that feces from obese twins transplanted into germ-free mice results in weight gain (61). Additionally, fecal transplantation studies have shown that obese subjects with metabolic syndrome have increased insulin sensitivity after the transfer of gut microbiota from a lean donor (37, 74, 75). These studies provide a theoretical basis for linking intestinal flora and whole-body energy metabolism, and they also highlight the important role of the intestinal flora in host metabolism (76, 77). Recent studies have suggested that the human intestinal microbiota is altered in obese individuals relative to lean individuals, probably primarily by an elevation in the obesity-associated phylum Ligustrum and a reduction in Bacteroidetes (78, 79). A study of 416 twin pairs has shown that Dehalobacteriaceae, Christensenellaceae and SHA-98 were found to be significantly lower in the intestinal flora of obese individuals compared to those of lighter weight. The Christensenellaceae family is known to be producer of SCFAs (80). In addition, it has been demonstrated that Christensenella minuta amended and transplanted into recipient mice results in a tendency to lose weight. Oscillospira also produces SCFAs by degrading host blood glucose. In recent studies, we observed that Oscillospira and the methanogenic archaeon Methanobrevibacter smithii are abundant in healthy weight subjects and may promote leanness (65, 66, 81). Lean individuals also exist in T2DM (82). A study on the intestinal flora of lean diabetic patients has demonstrated that lean T2DM patients show a reduced Akkermansia muciniphila abundance, which is positively associated with a decrease in their insulin secretion (37). Studies have shown that Ruminococcus, Clostridium, Bifidobacterium, Bacteroides, Eubacterium, Listeria and others are the main intestinal microorganisms that affect the formation of secondary bile acids (49, 50). It has been shown that BAs modulate the metabolism and energy homeostasis of host by activating FXR receptors and TGR5, thereby shaping the intestinal flora (19).
Microorganisms including Blastocystis spp. and Prevotella copri have been found to be indicators of favorable postprandial glucose metabolism. And studies have shown that Prevotella copri supplementation improves glucose metabolism in mice (83). However, a study has demonstrated that Prevotella copri was associated with the production of BCAA, which is correlated with IR and glucose intolerance (17). It has been suggested that the elevated richness of Bacteroides vulgatus and Prevotella copri in IR individuals relative to the normal group may lead to an increased potential for BCAA synthesis; however, Butyrivibrio crossotus and Eubacterium siraeum abundance decreases, leading to reduced BCAA catabolism (17). Increased concentrations of BCAAs in circulation (including leucine, isoleucine and valine) may be biomarkers of IR and increased risk of T2DM (54).
Patients with T2DM are often treated with multiple drugs for glycemic control and prevention of cardiovascular complications. Thus, drug therapy usually affects the gut microbiota of patients, and the role of the gut microbiota may be confounded by the effects of various drugs (84). Therefore, many recent studies on T2DM flora have focused on the early stages of T2DM (not yet taking drugs). Although plasma glucose concentrations have not yet reached the diabetic range in the early stages of diabetes, individuals are at escalated risk of developing significant T2DM and cardiovascular disease (85). Alterations in the intestinal microbiota of unmedicated prediabetic patients have been reported to be dominated by butyrate-producing bacteria, Akkermansia muciniphila, and some bacteria with pro-inflammatory potential (86).
A previous study on the effect of metformin treatment on intestinal microbes in individuals with T2DM has reported a reduction in Roseburia, Subdoligranulum and a cluster of butyrate-producing Clostridium spp. in metformin-naïve T2DM patients, consistent with previous indications (37, 38). Metformin treatment modifies the intestinal microbiota in individuals with diabetes as indicated by enrichments in mucin-degrading Akkermansia muciniphila and SCFA-producing bacteria such as Bifidobacterium bifidum and Butyrivibrio (87). An independent amplicon-based T2DM cohort analysis has similarly validated an elevation in Escherichia coli and a reduction in Enterobacteriaceae abundance in metformin-treated patients with several gut microbial genera more similar in abundance to normal control levels, particularly Subdoligranulum and Akkermansia. Therefore, we hypothesized that metformin may alleviate T2DM by affecting the microbiota and its microbial production of SCFAs. It has been shown that probiotic therapy based on flora modulation increases the richness of SCFAs while achieves a reduction in IR, and this therapy has been reported to have a similar effect under metformin treatment (41, 42, 88). Some studies have shown that probiotics act as adjuvants to metformin by increasing the production of butyrate, thus allowing enhanced glucose management (89).
All of these findings provide strong evidence that the gut microbiome can be used to improve the prediction of metabolic diseases, such as T2DM, and to discover new targets for diabetes treatment. In this review, we focus on how the intestinal microbiota and its metabolites affect the pathogenesis of T2DM. We summarize examples of microbiota-targeted interventions for T2DM and highlight the great promise of this therapeutic approach in the field of T2DM research.
In the liver, BAs are synthesized from cholesterol and act as promoters for emulsification, absorption and transportation of dietary lipids as well as fat-soluble vitamins in the intestinal lumen. BAs are mainly metabolized by the actions of intestinal bacteria, which play a critical role in glucose regulation. BAs also act as regulators of intestinal microbiota as well as signaling molecules that modulate metabolic homeostasis (Figure 1). Interactions of BAs and intestinal flora have a profound impact on IR and the progression of T2DM.
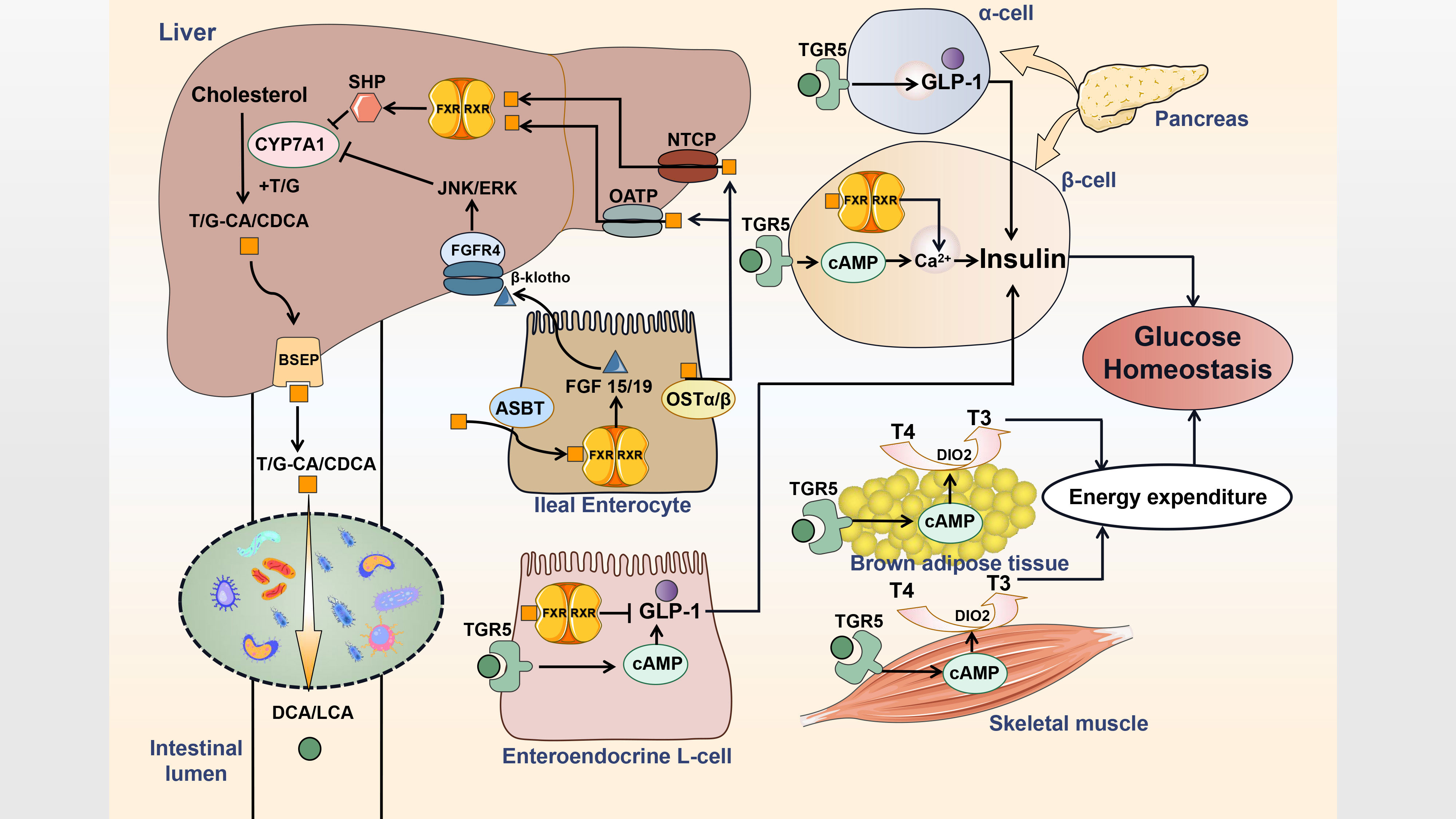
Figure 1 Interactions of BAs and gut microbe influence host metabolism. Primary bile acids are synthesized and then conjugated with taurine or glycine in hepatocytes. Conjugated bile acids are transported into the bile duct by BSEP. Most conjugated bile acids are reabsorbed via ASBT and circulate to the liver by OSTα/β, OATP and NTCP, while a small part is converted into secondary bile acids by deconjugation and dehydroxylation of gut flora. Bile acids acts as the endogenous ligands for FXR and TGR5 to generate distinct effects on metabolism regulation. T, taurine; G, glycine; CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; LCA, lithocholic acid; BSEP, bile salt export protein; FGF, fibroblast growth factor; FGFR, FGF receptor; RXR, retinoid X receptor; SHP, small heterodimer partner; JNK, c-Jun N-terminal kinase; ERK, extracellular signal-regulated kinase; OST, organic solute transporter; OATP, organic anion-transporting polypeptide; NTCP, sodium taurocholate cotransporting polypeptide; ASBT, apical sodium-dependent bile acid transporter; DIO2, type 2 iodothyronine deiodinase; T4, thyroxine; T3, thyroid hormone.
Cholesterol 7α-hydroxylase (CYP7A1) predominantly regulates the classic pathway (namely, the neutral pathway) of BA synthesis, while the alternative pathway (namely, the acidic pathway) depends mainly on CYP27A1. CYP8A1 and CYP7B1 also play distinct roles in the process (49). Glucose and insulin act as the main postprandial factors inducing CYP7A1 gene expression as well as BA synthesis (90). Mechanistically, glucose induces CYP7A1 gene transcription by increasing ATP to reduce the AMP/ATP ratio, which inhibits AMP-activated protein kinase (AMPK) activity, or by epigenetically modifying the acetylation state of the CYP7A1 chromatin structure (91). Cold exposure may also trigger hepatic conversion of cholesterol to BAs through the alternative pathway by induction of CYP7B1, leading to increased plasma levels and fecal excretion of BAs accompanied by alterations in intestinal microbiota that are characterized by higher abundance of Deferribacteraceae and Lachnospiraceae as well as lower abundance of Porphyromonadaceae and Clostridiales. Increased energy expenditure is also related to hepatic CYP7B1 induction, which is reduced in individuals with T2DM, suggesting that the alternative pathway plays a role in metabolic homeostasis (92). Another regulator of BA composition, CYP8B1 (an 12α-hydroxylase) has been reported to contribute to metabolic anomalies in T2DM as greater 12α-hydroxy/non12α-hydroxy BA ratios are correlated with decreased insulin sensitivity (93).
Hepatocytes conjugate bile acids with taurine or glycine to increase their solubility before secretion (18). In the distal ileum, roughly 95% of conjugated bile acids (CBAs) are reuptaken via the apical sodium-dependent BA transporter (IBAT or ASBT) in conjugated form, while a small portion are deconjugated by the gut microbiota prior to resorption or converted into secondary bile acids (DCA from CA and LCA from CDCA, respectively), which are either absorbed through passive diffusion or excreted in the feces (94). BAs absorbed recirculate through the portal vein to the liver, where they are conveyed into hepatocytes to be reconjugated and then resecreted with newly synthesized BAs (95).
Microbial deconjugation refers to enzymatic hydrolysis by bile salt hydrolases (BSHs), namely, the removal of taurine or glycine conjugates, and it impedes active reabsorption of BAs via ASBT in the small intestine. In the human intestinal microbiome, BSHs are distributed among various microbial species and were identified early in several microbial genera, such as Lactobacillus (96–99), Clostridium (100), Bifidobacterium (101–103), Listeria (104, 105), Enterococcus (106) and Bacteroides (107). BSH activity exerts beneficial effects on bacteria by strengthening resistance to CBAs, assisting surviving in the gastrointestinal environment without causing bacterial overgrowth or damage to the host. This may facilitate colonization and development of the gut microbiota (108). High levels of BSH have also been reported to decrease body weight gain and plasma cholesterol in conventionally raised mice (109). However, some controversial results have suggested that deconjugated bile acids (DBAs) with higher binding affinity to farnesoid X receptor (FXR) may suppress the transcription of CYP7A1 and reduce BA synthesis from cholesterol (110). Moreover, some studies have deviated from previous concepts, indicating that the intrinsic chemical features and enzymatic preferences of various BSHs change the Lactobacillus transcriptome in a BA-dependent manner and contribute to the toxicity during Lactobacillus growth (111). Therefore, the importance of BSHs classification is highlighted as distinct BSHs in the identical Bacteroides strain differ in deconjugation ability. Generally, BSHs benefit metabolism, but the higher relative abundance (RA) of BSH phylotype with high activity may exert detrimental effects. To explore the relationship between the RA of BSHs and disease, it is important to first assess activity to identify particular BSHs rather than use an estimated RA of total BSHs (112).
BAs are C-24 steroids hydroxylated at the C-3, C-7 position, and also the C-12 position in the case of CA (113). 7α/β-dehydroxylating bacteria play an important role in transformations of BAs as the pool of secondary bile acids mainly consists of 7α/β-dehydroxylated BAs (e.g., LCA and DCA) (114). Different from deconjugation, BA 7α/β-dehydroxylation is confined to a limited number of gut bacteria, which lead to the salvage of BAs that evade active reuptake in the distal ileum, as deconjugation and 7α/β-dehydroxylation increase Pka and hydrophobicity of BAs to further permit passive absorption across the colonic epithelium (115). Because 7α/β-dehydroxylation is restricted to free bile acids, deconjugation is a prerequisite. 7α/β-Hydroxysteroid dehydrogenases (7α/β-HSDHs) have been identified in gut bacteria such as Clostridium (116–118), Bacteroides (119–121), Escherichia (122–124) and Eubacterium (125–127). While 7α-dehydroxylation functions as the most quantitatively vital bacterial biotransformation of BAs, 7β-dehydroxylation appears to be not essential in the human colon (128).
In diabetic patients, the BA pool composition and the associated pathways of biosynthesis are altered, which may involve a higher conversion of primary bile acids to secondary bile acids by the gut microbiota (129). Cirrhotics show lower abundance of Ruminococcaceae, Lachonospiraceae and Blautia (7α-dehydroxylating bacteria) while increased Enterobacteriaceae (potentially pathogenic), resulting in decreased conversion of primary to secondary fecal BAs (130). Clostridium scindens with 7α-HSDHs activity strengthens resistance to Clostridium difficile infection in a secondary bile acid-dependent manner upon administration (131). In addition to 7α/β-dehydroxylation, 3β-HSDH is identified in Odoribacteraceae strains to produce isoalloLCA, which exerts antibacterial effects against Gram-positive pathogens including Enterococcus faecium and Clostridioides difficile to maintain the intestinal homeostasis (132). 12α-dehydrogenation is involved in synthetic process from CA to ursodeoxycholic acid (UDCA), which functions in the therapy of certain gastrointestinal tract diseases, and contributes to counter the effects of LCA and DCA (133). Clostridium scindens, Clostridium hylemonae and Peptacetobacter hiranonis are identified as 12α-HSDH expressing strains (134). A new fecal isolate, Eggerthella lenta strain C592, is found to be a critical microbe in BA metabolism and express 3α-, 3β-, 7α-, and 12α-HSDHs (135, 136). 12β-HSDH interconverts the 12-oxo bile acids to the 12β-configuration to completes the epimerization, forming less toxic and more hydrophilic bile acids. 12β-HSDH activity is proved in Clostridium paraputrificum (137).
The effects of acarbose are based on the gut microbiota-plasma BA axis as acarbose elevates the RA of Bifidobacterium and Lactobacillus while depleting Bacteroides, leading to altered RA of microbial genes that are responsible for BA metabolism. The results of acarbose treatment depend on microbiota compositions in the gut before treatment, and diverse abilities of the gut flora to metabolize BAs result in distinguishing therapeutic effects (138). Antibiotics modify the intestinal microbiota to change high-fat diet (HFD)-driven inflammatory signaling and BA composition, resulting in improved glucose metabolism and IR. These effects depend on interactions with the host’s systemic inflammatory response and genetic background (139).Patients undergoing bariatric surgery (especially gastric bypass procedures) show improvements in insulin sensitivity, which are correlated with increased BA concentrations in circulation and alterations in intestinal microbiome, indicating that elevations in circulating BAs and the interaction of BAs and gut flora following bariatric surgery as well as calorie restriction and body mass loss lead to T2DM remission (140).
FXR, which is mainly activated by the primary BA CDCA, plays a critical role in modulating glucose homeostasis as well as insulin sensitivity (141, 142). In ileal enterocytes, activation of FXR promotes transcription of FGF19 (FGF15 in mouse), which circulates through the portal vein to the liver and further activates JNK/ERK signaling by binding to FGFR4/β-klotho heterodimer complex to inhibit expression of CYP7A1 in hepatocytes (143). In the liver, FXR activation not only induces small heterodimer partner (SHP) expression to inhibit CYP7A1, but also negatively interferes with glycolysis by inhibiting the carbohydrate response element binding protein (ChREBP), which is responsible for the expression of hepatic glycolytic genes (144–146). In intestinal L cells, FXR interacts with ChREBP to inhibit proglucagon mRNA levels induced by glucose. The effects of FXR on ChREBP also decrease intracellular ATP levels by inhibiting the transcription of glycolytic enzymes, resulting in a reduction in ATP-dependent Glucagon-like peptide-1 (GLP-1) secretion (147). In pancreatic β cells, FXR activation has been suggested to inhibit the KATP current and increase the cytosolic Ca2+ concentration, eventually resulting in increased insulin secretion (148, 149). These observations highlight the importance of the FXR-dependent control of BAs and glucose homeostasis.
In T2DM individuals, metformin decreases the abundance and BSH activity of Bacteroides fragilis to increase intestinal levels of glycoursodeoxycholic acid (GUDCA), which has been identified as a novel endogenous FXR antagonist in the gut. This finding demonstrates that metformin improves glucose metabolic dysfunction via a Bacteroides fragilis-GUDCA-intestinal FXR axis (150). In NAFLD patients with T2DM, obeticholic acid (OCA, an FXR agonist) activates FXR to mediate an increase in FGF19, resulting in weight loss and improved insulin sensitivity, accompanied by decreased endogenous BA production and 7a-hydroxy-4-cholesten-3-one (C4, a biomarker for BA synthesis) levels (151). Although serum levels of BA and C4 vary significantly among individuals, C4 plasma levels are considerably elevated in patients with T2DM and metabolic syndrome (MetS) (152). Studies in FXR-deficient mice fed a HFD have demonstrated that intestinal flora takes part in the development of IR and obesity by modulating BA and FXR signaling, and conversely, FXR may contribute to adiposity increase by changing the composition of gut microbiome, which is characterized by elevation in Bacteroidetes and reduction in Firmicutes (153).
TGR5, a transmembrane G protein-coupled receptor, is activated mainly by the secondary bile acids, DCA and LCA (154). Previous studies have shown that TGR5 activation results in increased intracellular cyclic AMP (cAMP), leading to the maintenance of glucose homeostasis, preservation of pancreatic function and insulin sensitivity as a consequence of increased GLP-1 secretion and energy expenditure induced by enhanced mitochondrial function (155). In enteroendocrine L cells, activation triggers an increase in the synthesis and release of GLP-1. In systemic circulation, BAs escaping hepatic clearance activate TGR5 in BAT and muscle to activate type 2 iodothyronine deiodinase (DIO2), which converts inactive thyroxine (T4) into active thyroid hormone (T3) to promote energy expenditure (156). In addition, TGR5 positively regulates insulin secretion via a cAMP/Ca2+ pathway in pancreatic β cells, while it reprograms pancreatic α cells to produce GLP-1 to mediate the proliferation and mass of β cells (157, 158).
The FXR agonist, fexaramine (FEX), activates FXR to shape the gut flora inducing Bacteroides and Acetatifactor, which convert CDCA and UDCA to LCA. LCA not only activates TGR5 and further stimulates L cells to secrete GLP-1, resulting in improved insulin sensitivity, but also induces WAT browning by activating TGR5/cAMP signaling to improve energy metabolism and ultimately reduce weight (159). Wu et al. demonstrated that ablation of intestinal HIF-2a leads to an elevation in Ruminococcus torques abundance and a decrease in Bacteroides vulgatus by reducing lactate synthesis, resulting in elevated levels of taurine-conjugated cholic acid (TCA) and DCA. These changes activate adipose TGR5 to upregulate WAT thermogenesis, ultimately improving obesity and IR (160). Cholic acid-7-sulfate (CA7S), an endogenous BA sulfated metabolite, has been identified as a potent gut-restricted TGR5 agonist that induces the secretion of GLP1 from enteroendocrine L cells. CA7S is increased in the gastrointestinal tract after sleeve gastrectomy (SG), exhibiting anti-diabetic effects by enhancing glucose tolerance and reducing blood glucose levels (161). LCA has been identified to mediate the synthesis of CA7S in hepatocytes to influence host metabolism. Changes in the gut flora post-SG, particularly a decline in Clostridia, lead to a reduction in the production of LCA, resulting in increased production of CA7S (162). As CA7S is generated through microbiome-dependent signaling, its stability may differ among individual gut microbiomes (163).
GLP-1 is a proteolytic product of the proglucagon gene and regulates glucose homeostasis in the body. GLP-1 reduces appetite and slows gastric emptying in response to ingestion, and it acts on the pancreas to reduce glucagon secretion. GLP-1 also promotes glucose uptake and storage in muscle and adipose tissues while inhibiting glucose production in the liver (164, 165). Moreover, GLP-1 activates the GLP-1 receptor (GLP-1R) to promote proliferation and inhibit apoptosis of islet β cells to increase insulin biosynthesis and secretion (166). In T2DM patients, postprandial BA concentration is higher, and BA metabolism is upregulated, which may be due to the advantage of glucose-induced BA stimulation over other inhibiting factors to maintain GLP-1 levels as a compensation. Thus, the requirement of higher BA levels to stimulate enteroendocrine L cells to release GLP-1 as resistance to BA in L cells is suggested (167). Recently, studies have found that Akkermansia muciniphila secretes P9, a novel protein identified to induce the secretion of GLP-1 (168, 169).
The fermentation of undigested dietary components (e.g. fibre and resistant starch) in the large intestine mainly produces three SCFAs including acetate, propionate as well as butyrate, and various biochemical pathways are involved in the process (170). Thus far, as the most well studied metabolites of gut flora, SCFAs control immunomodulatory functions, promote the integrity of intestinal epithelium, as well as regulate insulin secretion and the proliferation of pancreatic β cell, playing a variety of roles in IR and T2DM (171). Most enteric bacteria are identified as acetate producers such as Akkermansia muciniphila, Blautia hydrogenotrophica, Clostridium spp., Ruminococcus spp., Prevotella spp., Bifidobacterium spp., Bacteroides spp., Streptococcus spp. Propionate producers include Roseburia inulinivorans, Phascolarctobacterium succinatutens, Megasphaera elsdenii, Coprococcus catus, Ruminococcus obeum, Bacteroides spp., Dialister spp., Veillonella spp., while Roseburia spp., Anaerostipes spp., Faecalibacterium prausnitzii, Coprococcus catus, Coprococcus eutactus, Coprococcus comes, Eubacterium hallii, Eubacterium rectale are proved to be burytate producers (170, 172, 173).
SCFAs exert benefits by activating G-protein coupled receptors (GPCRs) such as GPR41, GPR43 and GPR109A (174). Acetate, as well as propionate promotes GPR43 and GPR41 to release peptide YY (PYY) and GLP-1 to influence satiety and intestinal transit (175). Butyrate has also been proved to induce GLP-1 and PYY to increase insulin secretion and maintain glucose homeostasis (176, 177). In addition, butyrate not only activates GPR109A and inhibits histone deacetylases (HDACs) to exert anti-inflammatory effects, but also produces antimicrobial peptides to promote epithelial barrier function (178). Another study has shown that butyrate induces satiety to reduce food intake and activate BAT to promote fat oxidation by influencing the gut-brain neural circuit (179). A study suggested that butyrate produced in the gut mainly improves dynamic insulin response to food ingestion, instead of maintaining glucose homeostasis in the fasted state, and increased butyrate production driven by a genetically influenced change in the gut microbiome is beneficial for pancreatic β-cell function (180). Furthermore, butyrate was reported to maintain the gut microecological balance by limiting the bioavailability of respiratory electron acceptors of Enterobacteriaceae in the colonic lumen and thus preventing a dysbiotic proliferation of opportunistic pathogenic Salmonella and Escherichia (181). Dysosmobacter welbionis J115T, a new butyrate-producing bacterium belonging to the Ruminococcaceae family and isolated from human feces, is identified to be negatively correlated with fasting plasma glucose, HbA1c and BMI in overweight or obese individuals with metabolic syndrome, and influences host metabolism in beneficial and direct way (182). However, another study highlighted the importance of functional dysbiosis of gut flora in association with T2DM pathophysiology, rather than a specific microbial species. They found only a mild degree dysbiosis of gut flora in T2DM patients, while a decrease in butyrate-producing bacteria, which seemed to be metabolically beneficial (38). In T2DM patients, reduced SCFA levels caused by decreased abundance of SCFA-producing bacteria may contribute to the progression of IR and T2DM (31). Yet animal and clinical researches have demonstrated that the levels of fecal SCFA are positively correlated with IR and body weight (183), controversies over the role of SCFAs in T2DM and IR still remain, and further investigations are required.
Research has shown that elevated concentrations of AAs, particularly BCAAs, are correlated with increased IR and risks of T2DM (184). BCAAs also promote the development of IR associated with obesity in a setting of a HFD (185). The intestinal microbiota acts as an independent factor of increased serum BCAA levels in individuals with IR. Prevotella copri and Bacteroides vulgatus have been identified as the major gut microbes correlated with the biosynthesis of BCAAs and IR (17). Among BCAAs, a previous study has further identified a critical role of isoleucine in metabolic health as a diet with low isoleucine activates the FGF21-UCP1 axis to reprogram the metabolism of adipose tissue and liver, improving energy expenditure and hepatic insulin sensitivity. Reducing dietary valine induces similar but milder metabolic effects, whereas low leucine does not show these effects, thus suggesting that decreasing dietary isoleucine is important to prevent and treat diabetes (186). Of note, IR may induce the protein degradation normally suppressed by insulin and impair the oxidative metabolism of BCAA to result in aminoacidemia, suggesting that increased BCAA may be a marker of insulin action loss instead of a causation (187).
Similar to BCAAs, aromatic amino acids (AAAs) also take part in the pathogenesis of hyperglycemia as the metabolisms of BCAAs and AAAs change prior to alteration of glucose homeostasis (188). BCAAs and AAAs both act as markers of IR development in young adults with normoglycemia, suggesting that their correlation with diabetes risk partly depends on IR (189). Clostridium sporogenes has been identified to metabolize all three AAAs in the human gut (190). Ka et al. demonstrated that excessive consumption of monosodium glutamate (MSG) positively correlates with overweight and BMI among healthy Chinese adults, suggesting a correlation between glutamate and obesity (191). In obese individuals, Liu et al. observed a decrease in Bacteroides thetaiotaomicron abundance, which is negatively correlated with serum levels of glutamate. These microbial and metabolic changes are partly reversed by bariatric surgery, and the reduced glutamate levels in circulation contribute to the improvement of IR and hyperglycemia (81).
Gut flora convert choline in diet to trimethylamine (TMA), the precursor of TMAO. A high-fat diet elevates choline catabolism of Escherichia coli by changing the physiology of colonic epithelium, thus increases TMAO levels in circulation (192). Intestinal microbiota (predominantly Enterobacteriaceae) also reduces TMAO to TMA, which is untaken by the host and circulated to the liver to be further converted into TMAO by hepatic enzymes. Conversely, TMAO influences the metabolism and growth of intestinal microbiota in a taxon and source-dependent manner (193). Urocanate produced by histidine metabolism is converted to imidazole propionate by microbiota expressing urocanate reductase. T2DM patients show higher concentrations of imidazole propionate in peripheral and portal blood than healthy individuals, indicating that imidazole propionate may impair insulin signaling. Eggerthella lenta and Streptococcus mutans were identified to produce imidazole propionate, which was proved to aggravate glucose intolerance in mice (55). In addition, elevated serum levels of imidazole propionate are correlated with decreased abundance of intestinal flora (194). Gut flora-produced indolepropionic acid, a promising biomarker for T2DM progression, is suggested to be positively correlated with insulin secretion and negatively associated with low-grade inflammation as it preserves function of islet β cells (195).
In summary, the intestinal microbiota and its metabolites are involved in the occurrence and progression of T2DM, and the process can be reversed to some extent by regulating the intestinal microbiota and its metabolites, providing a new idea for clinical treatment (196) (Figure 2).
When applied in appropriate amounts, probiotics, which are described as the living microbes, provide the host with health benefits (197). Studies have shown that the administration of probiotics regulates intestinal flora, suppresses inflammatory responses, improves intestinal barrier function, antagonizes each other with pathogens and produces beneficial microbial metabolites, including SCFAs and BAs (198). Studies on the molecular mechanisms of probiotic intervention in T2DM have shown that probiotics have ameliorative effects on IR and hyperglycemia (199).
Studies have shown that Akkermansia muciniphila treatment improves liver function, reduces oxidative stress and inhibits inflammation in diabetic mice. Akkermansia muciniphila supplementation reduces chronic low-grade inflammation and increases anti-inflammatory factors, such as α-tocopherol and β-sitosterol (200). Glucose tolerance and insulin sensitivity in diabetic mice were also ameliorated by supplementation with Lactobacillus mucilaginosa. In addition, it has been shown that supplementation with Lactobacillus acidophilus improves intestinal barrier function, and a reduced inflammatory response in the liver and colon has been observed in animal models of diabetes. Studies have shown that Akkermansia muciniphila strains are not even required to colonize the intestine to exert beneficial metabolic health effects (201, 202). In addition, 14 probiotics were isolated from fermented camel milk, and the effects of these 14 probiotics encompassed an increase in SCFA, an improvement in function of intestinal barrier, an upregulation of GLP-1 secretion, all of which showed significant improvements in blood glucose in db/db mice (203). A randomized, double-blinded, placebo-controlled study has reported that the administration of probiotic improves glucose metabolism in T2DM individuals, while the consumption of fermented milk results in other metabolic alterations including an elevation in acetic acid and a reduction in inflammatory cytokines (TNF-α and resistin) (204).The role of probiotics in T2DM deserves recognition, but the FDA and EFSA do not yet have any approval statements for probiotics (205–207). To verify the effectiveness of probiotics, more clinical trials are required.
Microbiologists have isolated many probiotic bacteria in the last century, and although studies have demonstrated their efficacy in standard animal models, single microorganisms are weak in preventing and treating human diseases (208, 209). Consequently, their clinical benefits are limited (210). Therefore, it is necessary for multiple microbes to work together to remodel the intestinal microbiota. In recent years, FMT has received increasing attention in the field of biomedical and clinical medicine (211). FMT refers to the transfer of fecal microbiota from a healthy donor to an unhealthy recipient via capsules with freeze-dried stool (or other ways), it may correct dysbiosis by increasing microbial diversity and restoring microbial function (212).
Official guidelines have ratified FMT as a standard treatment for recurrent Clostridium difficile infection (CDI) (213). According to data available on clinicaltrials.gov, ongoing trials on FMT are focused on indications other than CDI, and many clinical trials have shown that it also has strong potential in the treatment of refractory ulcerative colitis, Crohn’s disease, irritable bowel syndrome and other intestinal diseases as well as many extraintestinal diseases, including diabetes, cancer, cirrhosis, entero-brain disease and other metabolic diseases (214–217).
Early studies have shown that germ-free mice receiving samples from obese donors gained more weight than those from lean suppliers (78). After transplantation of Kazaks normal glucose tolerance (KNGT) fecal bacteria to db/db mice, the richness of Clostridium coccoides and Desulfovibrio in the intestine markedly declined, while the fecal abundance of Akkermansia muciniphila are increased. Moreover, the levels of fasting blood glucose and postprandial glucose are significantly downregulated in db/db mice, while the levels of HDL-cholesterol are upregulated, suggesting that fecal bacteria from KNGT may be a promising source for treating T2DM individuals with FMT (218, 219).
Previous studies have demonstrated that BA concentrations are changed in T2DM patients (147, 220). In addition, a study on the functional identification of bacterial and viral genes before and after FMT has shown that metabolic pathways such as the degradation of fluorobenzoate and the biosynthesis of secondary bile acids were significantly altered. A recent study has shown that the composition of the intestinal flora affects metabolism of BAs (211). The formation of BAs, SCFAs, and intestinal transit time of the organism are altered after FMT treatment. A previous study showed that allogenic FMT using feces from post-Roux-en-Y gastric bypass donor (rYgB-D) exerted short-term effects on metabolism of glucose, adipose tissue inflammation and intestinal transit time in obese and treatment-naive male individuals with IR, compared with feces from a metabolic syndrome donor (MetS-D) (74, 212).
Some studies have shown high interindividual variability in strain-level outcomes following FMT (221). Changes in plasma metabolites are also reported. The improvement in glucose metabolism, and the regulation of gut microbiota and plasma metabolites by FMT from lean donors are dependent on the reduction in fecal microbial diversity at baseline. Thus, pretreatment fecal microbiota characteristics may differ in response to bacterial species of lean donors, and thus pretreatment status may be correlated with the treatment effects (44, 58, 74).
However, some contradictory results have been reported. A 12-week double-blinded randomized placebo-controlled pilot trial of oral FMT capsules has shown that weekly treatment of FMT capsules to obese adults led to intestinal microbiota transplantation in most recipients for at least 12 weeks. Despite the successful implantation of the colonies, obviously clinical metabolic effects were not observed during the study period (222). Because researchers tend to publish studies with positive data, it makes it more difficult to determine the actual results. Studies have shown that performing FMT is also risky, and that harmful microorganisms may be transferred to the recipient via FMT. A previous study showed that an individual receiving feces from an overweight but healthy donor had developed new-onset obesity. Therefore, it is essential to publish studies with negative data so that we can have an unbiased understanding of the intestinal microbiota and its role in diseases (223).
To explore the potential of FMT in metabolic diseases (T2DM), further detailed work is needed, such as determining the indications for recipients, optimal donor microbiome profile and appropriate dose frequency. Modulation of the gut microbiota by techniques including FMT may become a potential therapeutic option for T2DM management.
There is growing evidence that many herbs or their herbal compounds may have a therapeutic effect on T2DM by modulating the intestinal microbiota. Radix scutellariae can eliminate heat and dampness, cure jaundice and quench thirst. A modern pharmacological study has shown that scutellaria achieves hypoglycemic and lipid-regulating effects by reducing intrahepatic cholestasis or increasing BA excretion in the feces (224). Studies have shown that baicalin improves diabetes by modulating the interplay between BAs and intestinal flora, an effect that may be mediated by FXR (225).
A study has shown that licorice extract improves IR, endotoxemia-related colonic inflammation and serum lipids in diabetic mice. In addition, licorice extract reshapes the intestinal flora by reducing Lachnospiraceae_NK4A136_group, while increasing Akkermansia and Bacteroides.These results indicate that the modulation of intestinal microbiota and colonic TLR4/NF-κB signaling pathway in diabetic mice may be the main reason for the anti-diabetic effect of licorice extracts (226). Studies have shown that the antidiabetic effect of Scutellaria-coptis herb couple (SC), one of the famous herbal compounds in traditional herbal combinations for diabetes treatment, is attributed to its modulation of gut microbiota and anti-inflammatory effects involving TLR4 signaling pathway (227). A rich-polyphenols extract of Dendrobium loddigesii (DJP) has been used to treat diabetic db/db mice, possibly due to the effects of DJP-induced reduction of inflammation and oxidative stress and improved intestinal flora balance, resulting in improved diabetic symptoms and complications in mice (228). Gegen Qinlian Decoction (GQD), a traditional Chinese medicine formula, has already been applied to treat common metabolic diseases such as T2DM. The mechanism of GQD for T2DM is mainly through altering the structure of the entire intestinal microbiota, enriching it with many butyrate-producing bacteria, thereby reducing intestinal inflammation and lowering blood glucose (229). Berberine, a hypothetical key active pharmaceutical ingredient of GQD, is also isolated from rhizoma coptidis and acts as an active alkaloid to achieve pharmacological effects by regulating the intestinal microbiota (230). In animal models of T2DM, berberine has been shown to increase the number of beneficial bacteria, reduce potentially pathogenic bacteria, and protect the islets and protect insulin target organs by reducing the invasion of inflammatory cells and inhibiting the development of a systemic inflammatory response (231). In a mouse model with T2DM, ginsenosides have been shown to modulate the intestinal flora, thereby reducing gut mucosal damage and a range of inflammatory responses (232, 233). Purified citrus polymethoxyflavone-rich extract (PMFE) significantly increases the abundance of Bacteroides ovatus, Bacteroides uniformis and Bacteroides thetaiotaomicron. The enrichment of Bacteroides ovatus by PMFE contributes to lower BCAA levels, weight loss and MetS relief (234). Ginsenoside Rb1 (Rb1) significantly changes the composition of the intestinal microbiota as it significantly increases the abundance of the bacterium Akkermansia spp. to which circulating alanine levels are related. Modulation of alanine may be correlated with Akkermansia spp., which is elevated in abundance by Rb1 to gain glucose homeostasis (235).
The role of herbal medicines in improving the intestinal microbiota in the treatment of T2DM deserves recognition. However, from the perspective of modern science, the underlying mechanism requires more explorations. Most of the effects of herbal medicines on microbiota are associated with intestinal flora structure modulation, increased richness of beneficial bacteria and butyrate concentration in gut, inhibition of opportunistic pathogens. Most of the findings are based on animal experiments, thus indicating the need of further evidence from human studies, and the underlying principles remain to be explored.
Studies have shown that weight loss of approximately 15 kg achieved through calorie restriction resulted in T2DM remission in 80% of obese and T2DM individuals. In addition, this remission is proportional to the amount of weight loss (236). Therefore, optimizing carbohydrate intake and increasing dietary fiber supplementation is particularly important for patients with T2DM. Fiber has already been recognized to play a critical role in regulating metabolism and preventing chronic gastroenterological diseases (237). The lower fiber content in the Western diet (especially in industrialized countries) is closely related to the elevated prevalence of metabolic disease states. Thus, alteration of the microbiota through dietary fiber interventions can improve health. Associations have been found between plant-based diets and taxa, such as Roseburia, Faecalibacterium prausnitzii and Eubacterium rectale, as well as an elevation in total SCFAs (238). SCFAs are produced by microbial fermentation of dietary fiber, which have cholesterol-lowering and glucose-control effects (239). Dietary fiber has been identified to markedly increase the RA of Bifidobacterium and total SCFAs while reducing glycated hemoglobin (240). Another study has reported that Bifidobacterium pseudocatenulatum, an acetate producer, is one of the most significantly promoted SCFA-produced microbiota by dietary fibers, and inoculation with this strain results in improved IR and postprandial glycemic response (31). A previous study has also shown that an almond-based low carbohydrate diet may improve glucose metabolism in patients with T2DM by increasing SCFA-producing bacteria, including Roseburia, Ruminococcus and Eubacterium, to elevate SCFA production and activate GPR43 to sustain GLP-1 secretion (48). Consistently, another study has suggested that almond-based diets may promote SCFA-producing bacteria while decreasing hemoglobin and body mass index (BMI) in T2DM individuals (241). The Green-Mediterranean diet with a gradual increase in plant components induces specific alterations in the intestinal flora and BCAA metabolism, including an elevation in Prevotella abundance and BCAA degradation as well as a decrease in Bifidobacterium abundance and BCAA biosynthesis, leading to increased insulin sensitivity (242).
Similarly, increasing physical activity and fitness is another important factor in alleviating T2DM. Physical activity is essential for reducing blood glucose and improving insulin sensitivity (243). A previous study has shown that exercise increases the abundance of Akkermansia muciniphila in the intestinal flora of athletes and increases the diversity of the intestinal microbiota (244). Regular exercise has also been reported to influence the composition of intestinal flora, elevate the production of SCFAs and plasma SCFA concentrations to ameliorate IR in skeletal muscle (245). Furthermore, exercise-induced elevation of SCFA-producing bacteria and improvement of intestinal barrier integrity play critical roles in T2DM improvement (246). Studies have shown that taxa producing SCFAs are positively correlated with changes in locomotion and mass, indicating that SCFAs may be involved in improving locomotor performance (247, 248).
The recognized risk factors for the development of diabetes include the interaction among different elements such as genetic susceptibility, diet, physical activity, smoking, and stress. The interplay of diet and gut microbiota determines the formation and absorption of different metabolites. It has been found that T2DM individuals can be divided into different clusters depending on characteristics of patients and risk of developing complications. Differences in pathophysiology of various groups in the pathogenesis were also found in individuals with prediabetes (249). Different patient subgroups may respond to the same treatment in very different ways. Thus, early identification of patients with diverse traits of T2DM has allowed us to predict the disease progression and response of individuals to the corresponding treatment regimen (250).
Only recently has the importance of the gut microbiota been more widely recognized. Bacteria that live in symbiosis with humans play a critical part in health and disease, and this makes microbiology one of the most active frontiers in biomedicine today. The microbial “organ” of the intestinal flora has involved in the regulation of human health and metabolism (251).Numerous animal studies and clinical trials have strongly supported the role of intestinal flora in obesity, IR and T2DM, as well as the proposition that the gut microbiome may influence a range of host systems and metabolic pathways through the production of metabolites (39, 40, 77, 252, 253). As mentioned before, Bacteroidetes, Firmicutes, Ruminococcus torques, and other species regulate changes in BAs. Akkermansia muciniphila, Roseburia spp., Prevotella spp., and others play important roles in the production of SCAAs. Prevotella copri and Bacteroides vulgatus are closely related to the biosynthesis of BCAAs. The metabolites of these microorganisms are highly associated with the evolution of obesity and T2DM in humans. However, changes in other microbial metabolites, such as imidazole propionate, indole, and TMAO, also play an important role in T2DM, but the corresponding clinical evidences are not sufficient.
Microbially directed interventions for diabetes can be divided into nontargeted and targeted therapies. Nontargeted interventions include exercise, personalized nutrition, Probiotics and FMT, which act on the overall improvement of flora composition and function. Targeted interventions include engineered microorganisms and drugs that target the metabolism of specific microorganisms, which act on specific changes in the metabolism-related flora. Numerous animal experiments and clinical trials have shown that groups with distinct base states respond very differently to therapeutic measures targeting the gut microbiota (45, 46). This discrepancy may be closely related to the composition of the microbial flora present in the organism itself. Therefore, an important prerequisite for the application of microbial therapies is the clarification of the pathogenic mechanism and the baseline microbial level of the organism. It is also the basis for successfully colonizing the transplanted microorganisms in an individualized manner. Overall, by analyzing the causal relationship between T2DM and gut flora, we conclude that gut flora can be used not only as a diagnostic biomarker but also as a promising therapeutic target for T2DM. The utilization of gut microbiota may contribute to a more precise and personalized treatment of T2DM patients.
Conceptualization: YW, GR. Investigation: LL, JZ, GR, YC, MZ, and ZX. Writing—original draft preparation: LL, JZ, and YW. Writing—review and editing: LL, JZ, GR, YC, and MZ. Supervision: YW, GR. All authors contributed to the article and approved the submitted version.
This work was funded by grants from the National Natural Science Foundation of China (NSFC 82070571), Chongqing Science and Health Joint Project (2022MSXM052), and Army Medical Center Military Medical Frontier Innovation Capability Program (CX2019JS222).
We thank Servier Medical Art (http://smart.servier.com/) for the reference of Figures in this review, licensed under a Creative Common Attribution 3.0 Generic License (https://creativecommons.org/licenses/by/3.0/).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. American Diabetes A. Diagnosis and classification of diabetes mellitus. Diabetes Care (2014) 37 Suppl 1:S81–90. doi: 10.2337/dc14-S081
2. Sung KC, Lee MY, Kim YH, Huh JH, Kim JY, Wild SH, et al. Obesity and incidence of diabetes: Effect of absence of metabolic syndrome, insulin resistance, inflammation and fatty liver. Atherosclerosis (2018) 275:50–7. doi: 10.1016/j.atherosclerosis.2018.05.042
3. Jaacks LM, Siegel KR, Gujral UP, Narayan KM. Type 2 diabetes: A 21st century epidemic. Best Pract Res Clin Endocrinol Metab (2016) 30:331–43. doi: 10.1016/j.beem.2016.05.003
4. Sun H, Saeedi P, Karuranga S, Pinkepank M, Ogurtsova K, Duncan BB, et al. IDF diabetes atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res Clin Pract (2022) 183:109119. doi: 10.1016/j.diabres.2021.109119
5. Ayadurai S, Hattingh HL, Tee LB, Md Said SN. A narrative review of diabetes intervention studies to explore diabetes care opportunities for pharmacists. J Diabetes Res (2016) 2016:5897452. doi: 10.1155/2016/5897452
6. Umpierrez GE, Pasquel FJ. Management of inpatient hyperglycemia and diabetes in older adults. Diabetes Care (2017) 40:509–17. doi: 10.2337/dc16-0989
7. Munshi MN. Cognitive dysfunction in older adults with diabetes: What a clinician needs to know. Diabetes Care (2017) 40:461–7. doi: 10.2337/dc16-1229
8. Durrer Schutz D, Busetto L, Dicker D, Farpour-Lambert N, Pryke R, Toplak H, et al. European Practical and patient-centred guidelines for adult obesity management in primary care. Obes Facts (2019) 12:40–66. doi: 10.1159/000496183
9. Koch TR, Shope TR. Laparoscopic vertical sleeve gastrectomy as a treatment option for adults with diabetes mellitus. Adv Exp Med Biol (2021) 1307:299–320. doi: 10.1007/5584_2020_487
10. Silver B, Ramaiya K, Andrew SB, Fredrick O, Bajaj S, Kalra S, et al. EADSG guidelines: Insulin therapy in diabetes. Diabetes Ther (2018) 9:449–92. doi: 10.1007/s13300-018-0384-6
11. Home P, Riddle M, Cefalu WT, Bailey CJ, Bretzel RG, Del Prato S, et al. Insulin therapy in people with type 2 diabetes: opportunities and challenges? Diabetes Care (2014) 37:1499–508. doi: 10.2337/dc13-2743
12. DeFronzo RA. Pathogenesis of type 2 diabetes mellitus. Med Clin North Am (2004) 88:787–835. doi: 10.1016/j.mcna.2004.04.013
13. Forbes JM, Cooper ME. Mechanisms of diabetic complications. Physiol Rev (2013) 93:137–88. doi: 10.1152/physrev.00045.2011
14. Zhou W, Sailani MR, Contrepois K, Zhou Y, Ahadi S, Leopold SR, et al. Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature (2019) 569:663–71. doi: 10.1038/s41586-019-1236-x
15. Yu F, Han W, Zhan G, Li S, Jiang X, Wang L, et al. Abnormal gut microbiota composition contributes to the development of type 2 diabetes mellitus in db/db mice. Aging (Albany NY) (2019) 11:10454–67. doi: 10.18632/aging.102469
16. Vallianou NG, Stratigou T, Tsagarakis S. Microbiome and diabetes: Where are we now? Diabetes Res Clin Pract (2018) 146:111–8. doi: 10.1016/j.diabres.2018.10.008
17. Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature (2016) 535:376–81. doi: 10.1038/nature18646
18. Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov (2008) 7:678–93. doi: 10.1038/nrd2619
19. Wahlstrom A, Sayin SI, Marschall HU, Backhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab (2016) 24:41–50. doi: 10.1016/j.cmet.2016.05.005
20. Zhao L, Lou H, Peng Y, Chen S, Zhang Y, Li X. Comprehensive relationships between gut microbiome and faecal metabolome in individuals with type 2 diabetes and its complications. Endocrine (2019) 66:526–37. doi: 10.1007/s12020-019-02103-8
21. Zhao L, Lou H, Peng Y, Chen S, Fan L, Li X. Elevated levels of circulating short-chain fatty acids and bile acids in type 2 diabetes are linked to gut barrier disruption and disordered gut microbiota. Diabetes Res Clin Pract (2020) 169:108418. doi: 10.1016/j.diabres.2020.108418
22. Munoz-Garach A, Diaz-Perdigones C, Tinahones FJ. Gut microbiota and type 2 diabetes mellitus. Endocrinol Nutr (2016) 63:560–8. doi: 10.1016/j.endonu.2016.07.008
23. Heianza Y, Sun D, Li X, DiDonato JA, Bray GA, Sacks FM, et al. Gut microbiota metabolites, amino acid metabolites and improvements in insulin sensitivity and glucose metabolism: the POUNDS lost trial. Gut (2019) 68:263–70. doi: 10.1136/gutjnl-2018-316155
24. Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine (2020) 51:102590. doi: 10.1016/j.ebiom.2019.11.051
25. Patterson E, Ryan PM, Cryan JF, Dinan TG, Ross RP, Fitzgerald GF, et al. Gut microbiota, obesity and diabetes. Postgrad Med J (2016) 92:286–300. doi: 10.1136/postgradmedj-2015-133285
26. Jaacks LM, Vandevijvere S, Pan A, McGowan CJ, Wallace C, Imamura F, et al. The obesity transition: Stages of the global epidemic. Lancet Diabetes Endocrinol (2019) 7:231–40. doi: 10.1016/S2213-8587(19)30026-9
27. Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology (2016) 64:73–84. doi: 10.1002/hep.28431
28. Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med (2016) 375:2369–79. doi: 10.1056/NEJMra1600266
29. Olofsson LE, Backhed F. The metabolic role and therapeutic potential of the microbiome. Endocr Rev (2022). doi: 10.1210/endrev/bnac004
30. Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science (2006) 312:1355–9. doi: 10.1126/science.1124234
31. Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science (2018) 359:1151–6. doi: 10.1126/science.aao5774
32. Boulange CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med (2016) 8:42. doi: 10.1186/s13073-016-0303-2
33. Chobot A, Gorowska-Kowolik K, Sokolowska M, Jarosz-Chobot P. Obesity and diabetes-not only a simple link between two epidemics. Diabetes Metab Res Rev (2018) 34:e3042. doi: 10.1002/dmrr.3042
34. Peters BA, Shapiro JA, Church TR, Miller G, Trinh-Shevrin C, Yuen E, et al. A taxonomic signature of obesity in a large study of American adults. Sci Rep (2018) 8:9749. doi: 10.1038/s41598-018-28126-1
35. Cani PD. Human gut microbiome: hopes, threats and promises. Gut (2018) 67:1716–25. doi: 10.1136/gutjnl-2018-316723
36. Singer-Englar T, Barlow G, Mathur R. Obesity, diabetes, and the gut microbiome: An updated review. Expert Rev Gastroenterol Hepatol (2019) 13:3–15. doi: 10.1080/17474124.2019.1543023
37. Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature (2013) 498:99–103. doi: 10.1038/nature12198
38. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature (2012) 490:55–60. doi: 10.1038/nature11450
39. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature (2015) 528:262–6. doi: 10.1038/nature15766
40. Wu H, Tremaroli V, Schmidt C, Lundqvist A, Olsson LM, Kramer M, et al. The gut microbiota in prediabetes and diabetes: A population-based cross-sectional study. Cell Metab (2020) 32:379–390.e3. doi: 10.1016/j.cmet.2020.06.011
41. Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, et al. An increase in the akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut (2014) 63:727–35. doi: 10.1136/gutjnl-2012-303839
42. Lee H, Ko G. Effect of metformin on metabolic improvement and gut microbiota. Appl Environ Microbiol (2014) 80:5935–43. doi: 10.1128/AEM.01357-14
43. Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature (2013) 500:541–6. doi: 10.1038/nature12506
44. Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, et al. Dietary intervention impact on gut microbial gene richness. Nature (2013) 500:585–8. doi: 10.1038/nature12480
45. De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell (2014) 156:84–96. doi: 10.1016/j.cell.2013.12.016
46. Volzke H, Alte D, Schmidt CO, Radke D, Lorbeer R, Friedrich N, et al. Cohort profile: The study of health in pomerania. Int J Epidemiol (2011) 40:294–307. doi: 10.1093/ije/dyp394
47. Maldonado-Contreras A, Noel SE, Ward DV, Velez M, Mangano KM. Associations between diet, the gut microbiome, and short-chain fatty acid production among older Caribbean Latino adults. J Acad Nutr Diet (2020) 120:2047–60.e6. doi: 10.1016/j.jand.2020.04.018
48. Ren M, Zhang H, Qi J, Hu A, Jiang Q, Hou Y, et al. An almond-based low carbohydrate diet improves depression and glycometabolism in patients with type 2 diabetes through modulating gut microbiota and GLP-1: A randomized controlled trial. Nutrients (2020) 12:3036. doi: 10.3390/nu12103036
49. Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol (2018) 15:111–28. doi: 10.1038/nrgastro.2017.119
50. Chiang JYL, Ferrell JM. Bile acids as metabolic regulators and nutrient sensors. Annu Rev Nutr (2019) 39:175–200. doi: 10.1146/annurev-nutr-082018-124344
51. Yue SJ, Liu J, Wang AT, Meng XT, Yang ZR, Peng C, et al. Berberine alleviates insulin resistance by reducing peripheral branched-chain amino acids. Am J Physiol Endocrinol Metab (2019) 316:E73–85. doi: 10.1152/ajpendo.00256.2018
52. Tett A, Huang KD, Asnicar F, Fehlner-Peach H, Pasolli E, Karcher N, et al. The prevotella copri complex comprises four distinct clades underrepresented in westernized populations. Cell Host Microbe (2019) 26:666–679.e7. doi: 10.1016/j.chom.2019.08.018
53. Li Q, Chang Y, Zhang K, Chen H, Tao S, Zhang Z. Implication of the gut microbiome composition of type 2 diabetic patients from Northern China. Sci Rep (2020) 10:5450. doi: 10.1038/s41598-020-62224-3
54. Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med (2011) 17:448–53. doi: 10.1038/nm.2307
55. Koh A, Molinaro A, Stahlman M, Khan MT, Schmidt C, Manneras-Holm L, et al. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell (2018) 175:947–961.e17. doi: 10.1016/j.cell.2018.09.055
56. Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J (2010) 4:232–41. doi: 10.1038/ismej.2009.112
57. Hong S, Zhou W, Fang B, Lu W, Loro E, Damle M, et al. Dissociation of muscle insulin sensitivity from exercise endurance in mice by HDAC3 depletion. Nat Med (2017) 23:223–34. doi: 10.1038/nm.4245
58. Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: Relationship with gut microbiome richness and ecology. Gut (2016) 65:426–36. doi: 10.1136/gutjnl-2014-308778
59. Lukovac S, Belzer C, Pellis L, Keijser BJ, de Vos WM, Montijn RC, et al. Differential modulation by akkermansia muciniphila and faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. mBio (2014) 5:e01438–14. doi: 10.1128/mBio.01438-14
60. Schwiertz A, Taras D, Schafer K, Beijer S, Bos NA, Donus C, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obes (Silver Spring) (2010) 18:190–5. doi: 10.1038/oby.2009.167
61. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science (2013) 341:1241214. doi: 10.1126/science.1241214
62. Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One (2010) 5:e9085. doi: 10.1371/journal.pone.0009085
63. Sung MM, Kim TT, Denou E, Soltys CM, Hamza SM, Byrne NJ, et al. Improved glucose homeostasis in obese mice treated with resveratrol is associated with alterations in the gut microbiome. Diabetes (2017) 66:418–25. doi: 10.2337/db16-0680
64. Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio (2014) 5:e00889. doi: 10.1128/mBio.00889-14
65. Beaumont M, Goodrich JK, Jackson MA, Yet I, Davenport ER, Vieira-Silva S, et al. Heritable components of the human fecal microbiome are associated with visceral fat. Genome Biol (2016) 17:189. doi: 10.1186/s13059-016-1052-7
66. Konikoff T, Gophna U. Oscillospira: a central, enigmatic component of the human gut microbiota. Trends Microbiol (2016) 24:523–4. doi: 10.1016/j.tim.2016.02.015
67. Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell (2014) 159:789–99. doi: 10.1016/j.cell.2014.09.053
68. Le Roy CI, Beaumont M, Jackson MA, Steves CJ, Spector TD, Bell JT. Heritable components of the human fecal microbiome are associated with visceral fat. Gut Microbes (2018) 9:61–7. doi: 10.1080/19490976.2017.1356556
69. Tims S, Derom C, Jonkers DM, Vlietinck R, Saris WH, Kleerebezem M, et al. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J (2013) 7:707–17. doi: 10.1038/ismej.2012.146
70. Newman TM, Shively CA, Register TC, Appt SE, Yadav H, Colwell RR, et al. Diet, obesity, and the gut microbiome as determinants modulating metabolic outcomes in a non-human primate model. Microbiome (2021) 9:100. doi: 10.1186/s40168-021-01069-y
71. Orsso CE, Peng Y, Deehan EC, Tan Q, Field CJ, Madsen KL, et al. Composition and functions of the gut microbiome in pediatric obesity: Relationships with markers of insulin resistance. Microorganisms (2021) 9:1490. doi: 10.3390/microorganisms9071490
72. Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, et al. The microbiome of uncontacted amerindians. Sci Adv (2015) 1:e1500183. doi: 10.1126/sciadv.1500183
73. Ouchi N, Parker JL, Lugus JJ, Walsh K. Adipokines in inflammation and metabolic disease. Nat Rev Immunol (2011) 11:85–97. doi: 10.1038/nri2921
74. Kootte RS, Levin E, Salojarvi J, Smits LP, Hartstra AV, Udayappan SD, et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab (2017) 26:611–619.e6. doi: 10.1016/j.cmet.2017.09.008
75. Zhang X, Shen D, Fang Z, Jie Z, Qiu X, Zhang C, et al. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One (2013) 8:e71108. doi: 10.1371/journal.pone.0071108
76. Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe (2008) 3:213–23. doi: 10.1016/j.chom.2008.02.015
77. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature (2006) 444:1027–31. doi: 10.1038/nature05414
78. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature (2006) 444:1022–3. doi: 10.1038/4441022a
79. Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes (2010) 59:3049–57. doi: 10.2337/db10-0253
80. Morotomi M, Nagai F, Watanabe Y. Description of christensenella minuta gen. nov., sp. nov., isolated from human faeces, which forms a distinct branch in the order clostridiales, and proposal of christensenellaceae fam. nov. Int J Syst Evol Microbiol (2012) 62:144–9. doi: 10.1099/ijs.0.026989-0
81. Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med (2017) 23:859–68. doi: 10.1038/nm.4358
82. Daousi C, Casson IF, Gill GV, MacFarlane IA, Wilding JP, Pinkney JH. Prevalence of obesity in type 2 diabetes in secondary care: Association with cardiovascular risk factors. Postgrad Med J (2006) 82:280–4. doi: 10.1136/pmj.2005.039032
83. Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of prevotella. Cell Metab (2015) 22:971–82. doi: 10.1016/j.cmet.2015.10.001
84. Vich Vila A, Collij V, Sanna S, Sinha T, Imhann F, Bourgonje AR, et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun (2020) 11:362. doi: 10.1038/s41467-019-14177-z
85. Allin KH, Tremaroli V, Caesar R, Jensen BAH, Damgaard MTF, Bahl MI, et al. Aberrant intestinal microbiota in individuals with prediabetes. Diabetologia (2018) 61:810–20. doi: 10.1007/s00125-018-4550-1
86. Zhong H, Ren H, Lu Y, Fang C, Hou G, Yang Z, et al. Distinct gut metagenomics and metaproteomics signatures in prediabetics and treatment-naive type 2 diabetics. EBioMedicine (2019) 47:373–83. doi: 10.1016/j.ebiom.2019.08.048
87. de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velasquez-Mejia EP, Carmona JA, Abad JM, et al. Metformin is associated with higher relative abundance of mucin-degrading akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care (2017) 40:54–62. doi: 10.2337/dc16-1324
88. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA (2013) 110:9066–71. doi: 10.1073/pnas.1219451110
89. Palacios T, Vitetta L, Coulson S, Madigan CD, Lam YY, Manuel R, et al. Targeting the intestinal microbiota to prevent type 2 diabetes and enhance the effect of metformin on glycaemia: A randomised controlled pilot study. Nutrients (2020) 12:2041. doi: 10.3390/nu12072041
90. Li T, Francl JM, Boehme S, Ochoa A, Zhang Y, Klaassen CD, et al. Glucose and insulin induction of bile acid synthesis: Mechanisms and implication in diabetes and obesity. J Biol Chem (2012) 287:1861–73. doi: 10.1074/jbc.M111.305789
91. Li T, Chanda D, Zhang Y, Choi HS, Chiang JY. Glucose stimulates cholesterol 7alpha-hydroxylase gene transcription in human hepatocytes. J Lipid Res (2010) 51:832–42. doi: 10.1194/jlr.M002782
92. Worthmann A, John C, Ruhlemann MC, Baguhl M, Heinsen FA, Schaltenberg N, et al. Cold-induced conversion of cholesterol to bile acids in mice shapes the gut microbiome and promotes adaptive thermogenesis. Nat Med (2017) 23:839–49. doi: 10.1038/nm.4357
93. Haeusler RA, Astiarraga B, Camastra S, Accili D, Ferrannini E. Human insulin resistance is associated with increased plasma levels of 12alpha-hydroxylated bile acids. Diabetes (2013) 62:4184–91. doi: 10.2337/db13-0639
94. de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab (2013) 17:657–69. doi: 10.1016/j.cmet.2013.03.013
95. Chavez-Talavera O, Tailleux A, Lefebvre P, Staels B. Bile acid control of metabolism and inflammation in obesity, type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology (2017) 152:1679–94.e3. doi: 10.1053/j.gastro.2017.01.055
96. Christiaens H, Leer RJ, Pouwels PH, Verstraete W. Cloning and expression of a conjugated bile acid hydrolase gene from lactobacillus plantarum by using a direct plate assay. Appl Environ Microbiol (1992) 58:3792–8. doi: 10.1128/aem.58.12.3792-3798.1992
97. Corzo G, Gilliland SE. Bile salt hydrolase activity of three strains of lactobacillus acidophilus. J Dairy Sci (1999) 82:472–80. doi: 10.3168/jds.S0022-0302(99)75256-2
98. Wang Z, Zeng X, Mo Y, Smith K, Guo Y, Lin J. Identification and characterization of a bile salt hydrolase from lactobacillus salivarius for development of novel alternatives to antibiotic growth promoters. Appl Environ Microbiol (2012) 78:8795–802. doi: 10.1128/AEM.02519-12
99. Chae JP, Valeriano VD, Kim GB, Kang DK. Molecular cloning, characterization and comparison of bile salt hydrolases from lactobacillus johnsonii PF01. J Appl Microbiol (2013) 114:121–33. doi: 10.1111/jam.12027
100. Coleman JP, Hudson LL. Cloning and characterization of a conjugated bile acid hydrolase gene from clostridium perfringens. Appl Environ Microbiol (1995) 61:2514–20. doi: 10.1128/aem.61.7.2514-2520.1995
101. Tanaka H, Hashiba H, Kok J, Mierau I. Bile salt hydrolase of bifidobacterium longum-biochemical and genetic characterization. Appl Environ Microbiol (2000) 66:2502–12. doi: 10.1128/AEM.66.6.2502-2512.2000
102. Kim GB, Miyamoto CM, Meighen EA, Lee BH. Cloning and characterization of the bile salt hydrolase genes (bsh) from bifidobacterium bifidum strains. Appl Environ Microbiol (2004) 70:5603–12. doi: 10.1128/AEM.70.9.5603-5612.2004
103. Kim GB, Brochet M, Lee BH. Cloning and characterization of a bile salt hydrolase (bsh) from bifidobacterium adolescentis. Biotechnol Lett (2005) 27:817–22. doi: 10.1007/s10529-005-6717-3
104. Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, et al. Comparative genomics of listeria species. Science (2001) 294:849–52. doi: 10.1126/science.1063447
105. Dussurget O, Cabanes D, Dehoux P, Lecuit M, Buchrieser C, Glaser P, et al. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol (2002) 45:1095–106. doi: 10.1046/j.1365-2958.2002.03080.x
106. Wijaya A, Hermann A, Abriouel H, Specht I, Yousif NM, Holzapfel WH, et al. Cloning of the bile salt hydrolase (bsh) gene from enterococcus faecium FAIR-e 345 and chromosomal location of bsh genes in food enterococci. J Food Prot (2004) 67:2772–8. doi: 10.4315/0362-028x-67.12.2772
107. Stellwag EJ, Hylemon PB. Purification and characterization of bile salt hydrolase from bacteroides fragilis subsp. fragilis. Biochim Biophys Acta (1976) 452:165–76. doi: 10.1016/0005-2744(76)90068-1
108. Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA (2008) 105:13580–5. doi: 10.1073/pnas.0804437105
109. Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci USA (2014) 111:7421–6. doi: 10.1073/pnas.1323599111
110. Choi SB, Lew LC, Yeo SK, Nair Parvathy S, Liong MT. Probiotics and the BSH-related cholesterol lowering mechanism: a Jekyll and Hyde scenario. Crit Rev Biotechnol (2015) 35:392–401. doi: 10.3109/07388551.2014.889077
111. Foley MH, O'Flaherty S, Allen G, Rivera AJ, Stewart AK, Barrangou R, et al. Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization. Proc Natl Acad Sci USA (2021) 118:e2017709118. doi: 10.1073/pnas.2017709118
112. Song Z, Cai Y, Lao X, Wang X, Lin X, Cui Y, et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome (2019) 7:9. doi: 10.1186/s40168-019-0628-3
113. Hofmann AF, Hagey LR. Key discoveries in bile acid chemistry and biology and their clinical applications: history of the last eight decades. J Lipid Res (2014) 55:1553–95. doi: 10.1194/jlr.R049437
114. Marion S, Studer N, Desharnais L, Menin L, Escrig S, Meibom A, et al. In vitro and in vivo characterization of clostridium scindens bile acid transformations. Gut Microbes (2019) 10:481–503. doi: 10.1080/19490976.2018.1549420
115. Lee JY, Arai H, Nakamura Y, Fukiya S, Wada M, Yokota A. Contribution of the 7beta-hydroxysteroid dehydrogenase from ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J Lipid Res (2013) 54:3062–9. doi: 10.1194/jlr.M039834
116. Coleman JP, Hudson LL, Adams MJ. Characterization and regulation of the NADP-linked 7 alpha-hydroxysteroid dehydrogenase gene from clostridium sordellii. J Bacteriol (1994) 176:4865–74. doi: 10.1128/jb.176.16.4865-4874.1994
117. Bakonyi D, Hummel W. Cloning, expression, and biochemical characterization of a novel NADP(+)-dependent 7alpha-hydroxysteroid dehydrogenase from clostridium difficile and its application for the oxidation of bile acids. Enzyme Microb Technol (2017) 99:16–24. doi: 10.1016/j.enzmictec.2016.12.006
118. Lou D, Wang Y, Tan J, Zhu L, Ji S, Wang B. Functional contribution of coenzyme specificity-determining sites of 7alpha-hydroxysteroid dehydrogenase from clostridium absonum. Comput Biol Chem (2017) 70:89–95. doi: 10.1016/j.compbiolchem.2017.08.004
119. Hirano S, Masuda N. Enhancement of the 7 alpha-dehydroxylase activity of a gram-positive intestinal anaerobe by bacteroides and its significance in the 7-dehydroxylation of ursodeoxycholic acid. J Lipid Res (1982) 23:1152–8.
120. Bennett MJ, McKnight SL, Coleman JP. Cloning and characterization of the NAD-dependent 7alpha-hydroxysteroid dehydrogenase from bacteroides fragilis. Curr Microbiol (2003) 47:475–84. doi: 10.1007/s00284-003-4079-4
121. Fukiya S, Arata M, Kawashima H, Yoshida D, Kaneko M, Minamida K, et al. Conversion of cholic acid and chenodeoxycholic acid into their 7-oxo derivatives by bacteroides intestinalis AM-1 isolated from human feces. FEMS Microbiol Lett (2009) 293:263–70. doi: 10.1111/j.1574-6968.2009.01531.x
122. Zhang X, Fan D, Hua X, Zhang T. Large-Scale production of ursodeoxycholic acid from chenodeoxycholic acid by engineering 7alpha- and 7beta-hydroxysteroid dehydrogenase. Bioprocess Biosyst Eng (2019) 42:1537–45. doi: 10.1007/s00449-019-02151-4
123. Kim KH, Lee CW, Pardhe BD, Hwang J, Do H, Lee YM, et al. Crystal structure of an apo 7alpha-hydroxysteroid dehydrogenase reveals key structural changes induced by substrate and co-factor binding. J Steroid Biochem Mol Biol (2021) 212:105945. doi: 10.1016/j.jsbmb.2021.105945
124. Li D, Liu R, Wang M, Peng R, Fu S, Fu A, et al. 3beta-hydroxysteroid dehydrogenase expressed by gut microbes degrades testosterone and is linked to depression in males. Cell Host Microbe (2022) 30:329–339.e5. doi: 10.1016/j.chom.2022.01.001
125. Baron SF, Franklund CV, Hylemon PB. Cloning, sequencing, and expression of the gene coding for bile acid 7 alpha-hydroxysteroid dehydrogenase from eubacterium sp. strain VPI 12708. J Bacteriol (1991) 173:4558–69. doi: 10.1128/jb.173.15.4558-4569.1991
126. de Prada P, Setchell KD, Hylemon PB. Purification and characterization of a novel 17 alpha-hydroxysteroid dehydrogenase from an intestinal eubacterium sp. VPI 12708. J Lipid Res (1994) 35:922–9.
127. Mallonee DH, Lijewski MA, Hylemon PB. Expression in escherichia coli and characterization of a bile acid-inducible 3 alpha-hydroxysteroid dehydrogenase from eubacterium sp. strain VPI 12708. Curr Microbiol (1995) 30:259–63. doi: 10.1007/BF00295498
128. Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res (2006) 47:241–59. doi: 10.1194/jlr.R500013-JLR200
129. Suhre K, Meisinger C, Doring A, Altmaier E, Belcredi P, Gieger C, et al. Metabolic footprint of diabetes: A multiplatform metabolomics study in an epidemiological setting. PLoS One (2010) 5:e13953. doi: 10.1371/journal.pone.0013953
130. Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol (2013) 58:949–55. doi: 10.1016/j.jhep.2013.01.003
131. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to clostridium difficile. Nature (2015) 517:205–8. doi: 10.1038/nature13828
132. Sato Y, Atarashi K, Plichta DR, Arai Y, Sasajima S, Kearney SM, et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature (2021) 599:458–64. doi: 10.1038/s41586-021-03832-5
133. Sutherland JD, Macdonald IA, Forrest TP. The enzymic and chemical synthesis of ursodeoxycholic and chenodeoxycholic acid from cholic acid. Prep Biochem (1982) 12:307–21. doi: 10.1080/00327488208065679
134. Doden H, Sallam LA, Devendran S, Ly L, Doden G, Daniel SL, et al. Metabolism of oxo-bile acids and characterization of recombinant 12alpha-hydroxysteroid dehydrogenases from bile acid 7alpha-dehydroxylating human gut bacteria. Appl Environ Microbiol (2018) 84:e00235–18. doi: 10.1128/AEM.00235-18
135. Mythen SM, Devendran S, Mendez-Garcia C, Cann I, Ridlon JM. Targeted synthesis and characterization of a gene cluster encoding NAD(P)H-dependent 3alpha-, 3beta-, and 12alpha-hydroxysteroid dehydrogenases from eggerthella CAG:298, a gut metagenomic sequence. Appl Environ Microbiol (2018) 84:e02475-17. doi: 10.1128/AEM.02475-17
136. Harris SC, Devendran S, Mendez-Garcia C, Mythen SM, Wright CL, Fields CJ, et al. Bile acid oxidation by eggerthella lenta strains C592 and DSM 2243(T). Gut Microbes (2018) 9:523–39. doi: 10.1080/19490976.2018.1458180
137. Doden HL, Wolf PG, Gaskins HR, Anantharaman K, Alves JMP, Ridlon JM. Completion of the gut microbial epi-bile acid pathway. Gut Microbes (2021) 13:1–20. doi: 10.1080/19490976.2021.1907271
138. Gu Y, Wang X, Li J, Zhang Y, Zhong H, Liu R, et al. Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat Commun (2017) 8:1785. doi: 10.1038/s41467-017-01682-2
139. Fujisaka S, Ussar S, Clish C, Devkota S, Dreyfuss JM, Sakaguchi M, et al. Antibiotic effects on gut microbiota and metabolism are host dependent. J Clin Invest (2016) 126:4430–43. doi: 10.1172/JCI86674
140. Kaska L, Sledzinski T, Chomiczewska A, Dettlaff-Pokora A, Swierczynski J. Improved glucose metabolism following bariatric surgery is associated with increased circulating bile acid concentrations and remodeling of the gut microbiome. World J Gastroenterol (2016) 22:8698–719. doi: 10.3748/wjg.v22.i39.8698
141. Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. J Clin Invest (2006) 116:1102–9. doi: 10.1172/JCI25604
142. Prawitt J, Abdelkarim M, Stroeve JH, Popescu I, Duez H, Velagapudi VR, et al. Farnesoid X receptor deficiency improves glucose homeostasis in mouse models of obesity. Diabetes (2011) 60:1861–71. doi: 10.2337/db11-0030
143. Potthoff MJ, Kliewer SA, Mangelsdorf DJ. Endocrine fibroblast growth factors 15/19 and 21: From feast to famine. Genes Dev (2012) 26:312–24. doi: 10.1101/gad.184788.111
144. Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell (2000) 6:507–15. doi: 10.1016/s1097-2765(00)00050-2
145. Duran-Sandoval D, Mautino G, Martin G, Percevault F, Barbier O, Fruchart JC, et al. Glucose regulates the expression of the farnesoid X receptor in liver. Diabetes (2004) 53:890–8. doi: 10.2337/diabetes.53.4.890
146. Caron S, Huaman Samanez C, Dehondt H, Ploton M, Briand O, Lien F, et al. Farnesoid X receptor inhibits the transcriptional activity of carbohydrate response element binding protein in human hepatocytes. Mol Cell Biol (2013) 33:2202–11. doi: 10.1128/MCB.01004-12
147. Trabelsi MS, Daoudi M, Prawitt J, Ducastel S, Touche V, Sayin SI, et al. Farnesoid X receptor inhibits glucagon-like peptide-1 production by enteroendocrine l cells. Nat Commun (2015) 6:7629. doi: 10.1038/ncomms8629
148. Popescu IR, Helleboid-Chapman A, Lucas A, Vandewalle B, Dumont J, Bouchaert E, et al. The nuclear receptor FXR is expressed in pancreatic beta-cells and protects human islets from lipotoxicity. FEBS Lett (2010) 584:2845–51. doi: 10.1016/j.febslet.2010.04.068
149. Dufer M, Horth K, Wagner R, Schittenhelm B, Prowald S, Wagner TF, et al. Bile acids acutely stimulate insulin secretion of mouse beta-cells via farnesoid X receptor activation and K(ATP) channel inhibition. Diabetes (2012) 61:1479–89. doi: 10.2337/db11-0815
150. Sun L, Xie C, Wang G, Wu Y, Wu Q, Wang X, et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat Med (2018) 24:1919–29. doi: 10.1038/s41591-018-0222-4
151. Mudaliar S, Henry RR, Sanyal AJ, Morrow L, Marschall HU, Kipnes M, et al. Efficacy and safety of the farnesoid X receptor agonist obeticholic acid in patients with type 2 diabetes and nonalcoholic fatty liver disease. Gastroenterology (2013) 145:574–82.e1. doi: 10.1053/j.gastro.2013.05.042
152. Steiner C, Othman A, Saely CH, Rein P, Drexel H, von Eckardstein A, et al. Bile acid metabolites in serum: intraindividual variation and associations with coronary heart disease, metabolic syndrome and diabetes mellitus. PLoS One (2011) 6:e25006. doi: 10.1371/journal.pone.0025006
153. Parseus A, Sommer N, Sommer F, Caesar R, Molinaro A, Stahlman M, et al. Microbiota-induced obesity requires farnesoid X receptor. Gut (2017) 66:429–37. doi: 10.1136/gutjnl-2015-310283
154. Chen X, Lou G, Meng Z, Huang W. TGR5: A novel target for weight maintenance and glucose metabolism. Exp Diabetes Res (2011) 2011:853501. doi: 10.1155/2011/853501
155. Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab (2009) 10:167–77. doi: 10.1016/j.cmet.2009.08.001
156. Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature (2006) 439:484–9. doi: 10.1038/nature04330
157. Kumar DP, Rajagopal S, Mahavadi S, Mirshahi F, Grider JR, Murthy KS, et al. Activation of transmembrane bile acid receptor TGR5 stimulates insulin secretion in pancreatic beta cells. Biochem Biophys Res Commun (2012) 427:600–5. doi: 10.1016/j.bbrc.2012.09.104
158. Kumar DP, Asgharpour A, Mirshahi F, Park SH, Liu S, Imai Y, et al. Activation of transmembrane bile acid receptor TGR5 modulates pancreatic islet alpha cells to promote glucose homeostasis. J Biol Chem (2016) 291:6626–40. doi: 10.1074/jbc.M115.699504
159. Pathak P, Xie C, Nichols RG, Ferrell JM, Boehme S, Krausz KW, et al. Intestine farnesoid X receptor agonist and the gut microbiota activate G-protein bile acid receptor-1 signaling to improve metabolism. Hepatology (2018) 68:1574–88. doi: 10.1002/hep.29857
160. Wu Q, Liang X, Wang K, Lin J, Wang X, Wang P, et al. Intestinal hypoxia-inducible factor 2alpha regulates lactate levels to shape the gut microbiome and alter thermogenesis. Cell Metab (2021) 33:1988–2003.e7. doi: 10.1016/j.cmet.2021.07.007
161. Chaudhari SN, Harris DA, Aliakbarian H, Luo JN, Henke MT, Subramaniam R, et al. Bariatric surgery reveals a gut-restricted TGR5 agonist with anti-diabetic effects. Nat Chem Biol (2021) 17:20–9. doi: 10.1038/s41589-020-0604-z
162. Chaudhari SN, Luo JN, Harris DA, Aliakbarian H, Yao L, Paik D, et al. A microbial metabolite remodels the gut-liver axis following bariatric surgery. Cell Host Microbe (2021) 29:408–424.e7. doi: 10.1016/j.chom.2020.12.004
163. Ridlon JM. Bariatric surgery stirs symbionts to counteract diabesity by CA(7)Sting a liver-generated bile acid into the mix. Cell Host Microbe (2021) 29:320–2. doi: 10.1016/j.chom.2021.02.011
164. Baggio LL, Drucker DJ. Biology of incretins: GLP-1 and GIP. Gastroenterology (2007) 132:2131–57. doi: 10.1053/j.gastro.2007.03.054
165. Junker AE, Gluud LL, van Hall G, Holst JJ, Knop FK, Vilsboll T. Effects of glucagon-like peptide-1 on glucagon secretion in patients with non-alcoholic fatty liver disease. J Hepatol (2016) 64:908–15. doi: 10.1016/j.jhep.2015.11.014
166. Drucker DJ. Mechanisms of action and therapeutic application of glucagon-like peptide-1. Cell Metab (2018) 27:740–56. doi: 10.1016/j.cmet.2018.03.001
167. Vincent RP, Omar S, Ghozlan S, Taylor DR, Cross G, Sherwood RA, et al. Higher circulating bile acid concentrations in obese patients with type 2 diabetes. Ann Clin Biochem (2013) 50:360–4. doi: 10.1177/0004563212473450
168. Yoon HS, Cho CH, Yun MS, Jang SJ, You HJ, Kim JH, et al. Akkermansia muciniphila secretes a glucagon-like peptide-1-inducing protein that improves glucose homeostasis and ameliorates metabolic disease in mice. Nat Microbiol (2021) 6:563–73. doi: 10.1038/s41564-021-00880-5
169. Cani PD, Knauf C. A newly identified protein from akkermansia muciniphila stimulates GLP-1 secretion. Cell Metab (2021) 33:1073–5. doi: 10.1016/j.cmet.2021.05.004
170. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol (2014) 12:661–72. doi: 10.1038/nrmicro3344
171. Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes (2016) 7:189–200. doi: 10.1080/19490976.2015.1134082
172. Scott KP, Martin JC, Campbell G, Mayer CD, Flint HJ. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium "Roseburia inulinivorans". J Bacteriol (2006) 188:4340–9. doi: 10.1128/JB.00137-06
173. Rey FE, Faith JJ, Bain J, Muehlbauer MJ, Stevens RD, Newgard CB, et al. Dissecting the in vivo metabolic potential of two human gut acetogens. J Biol Chem (2010) 285:22082–90. doi: 10.1074/jbc.M110.117713
174. Bolognini D, Dedeo D, Milligan G. Metabolic and inflammatory functions of short-chain fatty acid receptors. Curr Opin Endocr Metab Res (2021) 16:1–9. doi: 10.1016/j.coemr.2020.06.005
175. Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell (2016) 165:1332–45. doi: 10.1016/j.cell.2016.05.041
176. Bindels LB, Dewulf EM, Delzenne NM. GPR43/FFA2: physiopathological relevance and therapeutic prospects. Trends Pharmacol Sci (2013) 34:226–32. doi: 10.1016/j.tips.2013.02.002
177. Everard A, Cani PD. Gut microbiota and GLP-1. Rev Endocr Metab Disord (2014) 15:189–96. doi: 10.1007/s11154-014-9288-6
178. Parada Venegas D, de la Fuente MK, Landskron G, Gonzalez MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol (2019) 10:277. doi: 10.3389/fimmu.2019.00277
179. Li Z, Yi CX, Katiraei S, Kooijman S, Zhou E, Chung CK, et al. Butyrate reduces appetite and activates brown adipose tissue via the gut-brain neural circuit. Gut (2018) 67:1269–79. doi: 10.1136/gutjnl-2017-314050
180. Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Vosa U, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet (2019) 51:600–5. doi: 10.1038/s41588-019-0350-x
181. Byndloss MX, Olsan EE, Rivera-Chavez F, Tiffany CR, Cevallos SA, Lokken KL, et al. Microbiota-activated PPAR-gamma signaling inhibits dysbiotic enterobacteriaceae expansion. Science (2017) 357:570–5. doi: 10.1126/science.aam9949
182. Le Roy T, Moens de Hase E, Van Hul M, Paquot A, Pelicaen R, Regnier M, et al. Dysosmobacter welbionis is a newly isolated human commensal bacterium preventing diet-induced obesity and metabolic disorders in mice. Gut (2022) 71:534–43. doi: 10.1136/gutjnl-2020-323778
183. Teixeira TF, Grzeskowiak L, Franceschini SC, Bressan J, Ferreira CL, Peluzio MC. Higher level of faecal SCFA in women correlates with metabolic syndrome risk factors. Br J Nutr (2013) 109:914–9. doi: 10.1017/S0007114512002723
184. Gaggini M, Carli F, Rosso C, Buzzigoli E, Marietti M, Della Latta V, et al. Altered amino acid concentrations in NAFLD: Impact of obesity and insulin resistance. Hepatology (2018) 67:145–58. doi: 10.1002/hep.29465
185. Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab (2009) 9:311–26. doi: 10.1016/j.cmet.2009.02.002
186. Yu D, Richardson NE, Green CL, Spicer AB, Murphy ME, Flores V, et al. The adverse metabolic effects of branched-chain amino acids are mediated by isoleucine and valine. Cell Metab (2021) 33:905–922.e6. doi: 10.1016/j.cmet.2021.03.025
187. Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol (2014) 10:723–36. doi: 10.1038/nrendo.2014.171
188. Wurtz P, Tiainen M, Makinen VP, Kangas AJ, Soininen P, Saltevo J, et al. Circulating metabolite predictors of glycemia in middle-aged men and women. Diabetes Care (2012) 35:1749–56. doi: 10.2337/dc11-1838
189. Wurtz P, Soininen P, Kangas AJ, Ronnemaa T, Lehtimaki T, Kahonen M, et al. Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care (2013) 36:648–55. doi: 10.2337/dc12-0895
190. Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature (2017) 551:648–52. doi: 10.1038/nature24661
191. He K, Du S, Xun P, Sharma S, Wang H, Zhai F, et al. Consumption of monosodium glutamate in relation to incidence of overweight in Chinese adults: China health and nutrition survey (CHNS). Am J Clin Nutr (2011) 93:1328–36. doi: 10.3945/ajcn.110.008870
192. Yoo W, Zieba JK, Foegeding NJ, Torres TP, Shelton CD, Shealy NG, et al. High-fat diet-induced colonocyte dysfunction escalates microbiota-derived trimethylamine n-oxide. Science (2021) 373:813–8. doi: 10.1126/science.aba3683
193. Hoyles L, Jimenez-Pranteda ML, Chilloux J, Brial F, Myridakis A, Aranias T, et al. Metabolic retroconversion of trimethylamine n-oxide and the gut microbiota. Microbiome (2018) 6:73. doi: 10.1186/s40168-018-0461-0
194. Molinaro A, Bel Lassen P, Henricsson M, Wu H, Adriouch S, Belda E, et al. Imidazole propionate is increased in diabetes and associated with dietary patterns and altered microbial ecology. Nat Commun (2020) 11:5881. doi: 10.1038/s41467-020-19589-w
195. de Mello VD, Paananen J, Lindstrom J, Lankinen MA, Shi L, Kuusisto J, et al. Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the Finnish diabetes prevention study. Sci Rep (2017) 7:46337. doi: 10.1038/srep46337
196. Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ (2018) 361:k2179. doi: 10.1136/bmj.k2179
197. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol (2014) 11:506–14. doi: 10.1038/nrgastro.2014.66
198. Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol (2019) 16:605–16. doi: 10.1038/s41575-019-0173-3
199. Won G, Choi SI, Kang CH, Kim GH. Lactiplantibacillus plantarum MG4296 and Lacticaseibacillus paracasei MG5012 Ameliorates Insulin Resistance in Palmitic Acid-Induced HepG2 Cells and High Fat Diet-Induced Mice. Microorganisms (2021) 9:1139. doi: 10.3390/microorganisms9061139
200. Zhao S, Liu W, Wang J, Shi J, Sun Y, Wang W, et al. Akkermansia muciniphila improves metabolic profiles by reducing inflammation in chow diet-fed mice. J Mol Endocrinol (2017) 58:1–14. doi: 10.1530/JME-16-0054
201. Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell (2018) 174:1388–405.e21. doi: 10.1016/j.cell.2018.08.041
202. Suez J, Zmora N, Zilberman-Schapira G, Mor U, Dori-Bachash M, Bashiardes S, et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell (2018) 174:1406–23.e16. doi: 10.1016/j.cell.2018.08.047
203. Wang Y, Dilidaxi D, Wu Y, Sailike J, Sun X, Nabi XH. Composite probiotics alleviate type 2 diabetes by regulating intestinal microbiota and inducing GLP-1 secretion in db/db mice. BioMed Pharmacother (2020) 125:109914. doi: 10.1016/j.biopha.2020.109914
204. Tonucci LB, Olbrich Dos Santos KM, Licursi de Oliveira L, Rocha Ribeiro SM, Duarte Martino HS. Clinical application of probiotics in type 2 diabetes mellitus: A randomized, double-blind, placebo-controlled study. Clin Nutr (2017) 36:85–92. doi: 10.1016/j.clnu.2015.11.011
205. Rittiphairoj T, Pongpirul K, Janchot K, Mueller NT, Li T. Probiotics contribute to glycemic control in patients with type 2 diabetes mellitus: A systematic review and meta-analysis. Adv Nutr (2021) 12:722–34. doi: 10.1093/advances/nmaa133
206. Tao YW, Gu YL, Mao XQ, Zhang L, Pei YF. Effects of probiotics on type II diabetes mellitus: a meta-analysis. J Transl Med (2020) 18:30. doi: 10.1186/s12967-020-02213-2
207. Kocsis T, Molnar B, Nemeth D, Hegyi P, Szakacs Z, Balint A, et al. Probiotics have beneficial metabolic effects in patients with type 2 diabetes mellitus: a meta-analysis of randomized clinical trials. Sci Rep (2020) 10:11787. doi: 10.1038/s41598-020-68440-1
208. Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, et al. Expert consensus document: The international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol (2017) 14:491–502. doi: 10.1038/nrgastro.2017.75
209. Brodmann T, Endo A, Gueimonde M, Vinderola G, Kneifel W, de Vos WM, et al. Safety of novel microbes for human consumption: Practical examples of assessment in the European union. Front Microbiol (2017) 8:1725. doi: 10.3389/fmicb.2017.01725
210. Miller LE, Zimmermann AK, Ouwehand AC. Contemporary meta-analysis of short-term probiotic consumption on gastrointestinal transit. World J Gastroenterol (2016) 22:5122–31. doi: 10.3748/wjg.v22.i21.5122
211. Zhang F, Cui B, He X, Nie Y, Wu K, Fan D, et al. Microbiota transplantation: Concept, methodology and strategy for its modernization. Protein Cell (2018) 9:462–73. doi: 10.1007/s13238-018-0541-8
212. Allegretti JR, Kassam Z, Osman M, Budree S, Fischer M, Kelly CR. The 5D framework: a clinical primer for fecal microbiota transplantation to treat clostridium difficile infection. Gastrointest Endosc (2018) 87:18–29. doi: 10.1016/j.gie.2017.05.036
213. van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent clostridium difficile. N Engl J Med (2013) 368:407–15. doi: 10.1056/NEJMoa1205037
214. Cui B, Li P, Xu L, Zhao Y, Wang H, Peng Z, et al. Step-up fecal microbiota transplantation strategy: A pilot study for steroid-dependent ulcerative colitis. J Transl Med (2015) 13:298. doi: 10.1186/s12967-015-0646-2
215. Bak SH, Choi HH, Lee J, Kim MH, Lee YH, Kim JS, et al. Fecal microbiota transplantation for refractory crohn's disease. Intest Res (2017) 15:244–8. doi: 10.5217/ir.2017.15.2.244
216. Johnsen PH, Hilpusch F, Cavanagh JP, Leikanger IS, Kolstad C, Valle PC, et al. Faecal microbiota transplantation versus placebo for moderate-to-severe irritable bowel syndrome: A double-blind, randomised, placebo-controlled, parallel-group, single-centre trial. Lancet Gastroenterol Hepatol (2018) 3:17–24. doi: 10.1016/S2468-1253(17)30338-2
217. He Z, Cui BT, Zhang T, Li P, Long CY, Ji GZ, et al. Fecal microbiota transplantation cured epilepsy in a case with crohn's disease: The first report. World J Gastroenterol (2017) 23:3565–8. doi: 10.3748/wjg.v23.i19.3565
218. Tao Y, Mao X, Xie Z, Ran X, Liu X, Wang Y, et al. The prevalence of type 2 diabetes and hypertension in uygur and kazak populations. Cardiovasc Toxicol (2008) 8:155–9. doi: 10.1007/s12012-008-9024-0
219. Zhang PP, Li LL, Han X, Li QW, Zhang XH, Liu JJ, et al. Fecal microbiota transplantation improves metabolism and gut microbiome composition in db/db mice. Acta Pharmacol Sin (2020) 41:678–85. doi: 10.1038/s41401-019-0330-9
220. Sonne DP, van Nierop FS, Kulik W, Soeters MR, Vilsboll T, Knop FK. Postprandial plasma concentrations of individual bile acids and FGF-19 in patients with type 2 diabetes. J Clin Endocrinol Metab (2016) 101:3002–9. doi: 10.1210/jc.2016-1607
221. Khanna S, Kraft CS. Fecal microbiota transplantation: Tales of caution. Clin Infect Dis (2021) 72:e881–2. doi: 10.1093/cid/ciaa1492
222. Yu EW, Gao L, Stastka P, Cheney MC, Mahabamunuge J, Torres Soto M, et al. Fecal microbiota transplantation for the improvement of metabolism in obesity: The FMT-TRIM double-blind placebo-controlled pilot trial. PLoS Med (2020) 17:e1003051. doi: 10.1371/journal.pmed.1003051
223. Alang N, Kelly CR. Weight gain after fecal microbiota transplantation. Open Forum Infect Dis (2015) 2:ofv004. doi: 10.1093/ofid/ofv004
224. Waisundara VY, Siu SY, Hsu A, Huang D, Tan BK. Baicalin upregulates the genetic expression of antioxidant enzymes in type-2 diabetic goto-kakizaki rats. Life Sci (2011) 88:1016–25. doi: 10.1016/j.lfs.2011.03.009
225. Zhao L, Ma P, Peng Y, Wang M, Peng C, Zhang Y, et al. Amelioration of hyperglycaemia and hyperlipidaemia by adjusting the interplay between gut microbiota and bile acid metabolism: Radix scutellariae as a case. Phytomedicine (2021) 83:153477. doi: 10.1016/j.phymed.2021.153477
226. Zhang Y, Xu Y, Zhang L, Chen Y, Wu T, Liu R, et al. Licorice extract ameliorates hyperglycemia through reshaping gut microbiota structure and inhibiting TLR4/NF-kappaB signaling pathway in type 2 diabetic mice. Food Res Int (2022) 153:110945. doi: 10.1016/j.foodres.2022.110945
227. Zhang CH, Sheng JQ, Sarsaiya S, Shu FX, Liu TT, Tu XY, et al. The anti-diabetic activities, gut microbiota composition, the anti-inflammatory effects of scutellaria-coptis herb couple against insulin resistance-model of diabetes involving the toll-like receptor 4 signaling pathway. J Ethnopharmacol (2019) 237:202–14. doi: 10.1016/j.jep.2019.02.040
228. Li XW, Chen HP, He YY, Chen WL, Chen JW, Gao L, et al. Effects of rich-polyphenols extract of dendrobium loddigesii on anti-diabetic, anti-inflammatory, anti-oxidant, and gut microbiota modulation in db/db mice. Molecules (2018) 23:3254. doi: 10.3390/molecules23123245
229. Xu X, Gao Z, Yang F, Yang Y, Chen L, Han L, et al. Antidiabetic effects of gegen qinlian decoction via the gut microbiota are attributable to its key ingredient berberine. Genomics Proteomics Bioinf (2020) 18:721–36. doi: 10.1016/j.gpb.2019.09.007
230. Habtemariam S. Berberine pharmacology and the gut microbiota: A hidden therapeutic link. Pharmacol Res (2020) 155:104722. doi: 10.1016/j.phrs.2020.104722
231. Huang J, Guan B, Lin L, Wang Y. Improvement of intestinal barrier function, gut microbiota, and metabolic endotoxemia in type 2 diabetes rats by curcumin. Bioengineered (2021) 12:11947–58. doi: 10.1080/21655979.2021.2009322
232. Gao Y, Li J, Wang J, Li X, Li J, Chu S, et al. Ginsenoside Rg1 prevent and treat inflammatory diseases: A review. Int Immunopharmacol (2020) 87:106805. doi: 10.1016/j.intimp.2020.106805
233. Wei Y, Yang H, Zhu C, Deng J, Fan D. Hypoglycemic effect of ginsenoside Rg5 mediated partly by modulating gut microbiota dysbiosis in diabetic db/db mice. J Agric Food Chem (2020) 68:5107–17. doi: 10.1021/acs.jafc.0c00605
234. Zeng SL, Li SZ, Xiao PT, Cai YY, Chu C, Chen BZ, et al. Citrus polymethoxyflavones attenuate metabolic syndrome by regulating gut microbiome and amino acid metabolism. Sci Adv (2020) 6:eaax6208. doi: 10.1126/sciadv.aax6208
235. Yang X, Dong B, An L, Zhang Q, Chen Y, Wang H, et al. Ginsenoside Rb1 ameliorates glycemic disorder in mice with high fat diet-induced obesity via regulating gut microbiota and amino acid metabolism. Front Pharmacol (2021) 12:756491. doi: 10.3389/fphar.2021.756491
236. Magkos F, Hjorth MF, Astrup A. Diet and exercise in the prevention and treatment of type 2 diabetes mellitus. Nat Rev Endocrinol (2020) 16:545–55. doi: 10.1038/s41574-020-0381-5
237. O'Keefe SJ. The association between dietary fibre deficiency and high-income lifestyle-associated diseases: Burkitt's hypothesis revisited. Lancet Gastroenterol Hepatol (2019) 4:984–96. doi: 10.1016/S2468-1253(19)30257-2
238. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature (2014) 505:559–63. doi: 10.1038/nature12820
239. O'Grady J, O'Connor EM, Shanahan F. Review article: dietary fibre in the era of microbiome science. Aliment Pharmacol Ther (2019) 49:506–15. doi: 10.1111/apt.15129
240. Ojo O, Feng QQ, Ojo OO, Wang XH. The role of dietary fibre in modulating gut microbiota dysbiosis in patients with type 2 diabetes: A systematic review and meta-analysis of randomised controlled trials. Nutrients (2020) 12:3239. doi: 10.3390/nu12113239
241. Ojo O, Wang XH, Ojo OO, Adegboye ARA. The effects of almonds on gut microbiota, glycometabolism, and inflammatory markers in patients with type 2 diabetes: A systematic review and meta-analysis of randomised controlled trials. Nutrients (2021) 13:3377. doi: 10.3390/nu13103377
242. Rinott E, Meir AY, Tsaban G, Zelicha H, Kaplan A, Knights D, et al. The effects of the green-Mediterranean diet on cardiometabolic health are linked to gut microbiome modifications: A randomized controlled trial. Genome Med (2022) 14:29. doi: 10.1186/s13073-022-01015-z
243. Zaharieva DP, McGaugh S, Davis EA, Riddell MC. Advances in exercise, physical activity, and diabetes. Diabetes Technol Ther (2020) 22:S109–18. doi: 10.1089/dia.2020.2508
244. Clarke SF, Murphy EF, O'Sullivan O, Lucey AJ, Humphreys M, Hogan A, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut (2014) 63:1913–20. doi: 10.1136/gutjnl-2013-306541
245. Yang L, Lin H, Lin W, Xu X. Exercise ameliorates insulin resistance of type 2 diabetes through motivating short-chain fatty acid-mediated skeletal muscle cell autophagy. Biol (Basel) (2020) 9:203. doi: 10.3390/biology9080203
246. Valder S, Brinkmann C. Exercise for the diabetic gut-potential health effects and underlying mechanisms. Nutrients (2022) 14:203. doi: 10.3390/nu14040813
247. Allen JM, Mailing LJ, Niemiro GM, Moore R, Cook MD, White BA, et al. Exercise alters gut microbiota composition and function in lean and obese humans. Med Sci Sports Exerc (2018) 50:747–57. doi: 10.1249/MSS.0000000000001495
248. O'Sullivan O, Cronin O, Clarke SF, Murphy EF, Molloy MG, Shanahan F, et al. Exercise and the microbiota. Gut Microbes (2015) 6:131–6. doi: 10.1080/19490976.2015.1011875
249. Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature (2012) 489:242–9. doi: 10.1038/nature11552
250. Musso G, Gambino R, Cassader M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu Rev Med (2011) 62:361–80. doi: 10.1146/annurev-med-012510-175505
251. de Groot PF, Belzer C, Aydin O, Levin E, Levels JH, Aalvink S, et al. Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLoS One (2017) 12:e0188475. doi: 10.1371/journal.pone.0188475
252. Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes (2012) 61:364–71. doi: 10.2337/db11-1019
Keywords: gut microbiota, type 2 diabetes mellitus, bile acids, short-chain fatty acids, amino acids, fecal microbiota transplantation, probiotics, herbal medicines
Citation: Liu L, Zhang J, Cheng Y, Zhu M, Xiao Z, Ruan G and Wei Y (2022) Gut microbiota: A new target for T2DM prevention and treatment. Front. Endocrinol. 13:958218. doi: 10.3389/fendo.2022.958218
Received: 31 May 2022; Accepted: 22 July 2022;
Published: 11 August 2022.
Edited by:
Yanli Pang, Peking University Third Hospital, ChinaCopyright © 2022 Liu, Zhang, Cheng, Zhu, Xiao, Ruan and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanling Wei, bGluZ3ppMDE2QDEyNi5jb20=; Guangcong Ruan, cnVhbmd1YW5nY29uZ0AxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.