- 1College of Fisheries, Guangdong Ocean University, Zhanjiang, China
- 2Guangxi Key Laboratory of Beibu Gulf Marine Biodiversity Conservation, Beibu Gulf University, Qinzhou, China
Ecdysis triggering hormone (ETH) plays an important role in molting, reproduction, and courtship behavior in insects. To investigate the potential downstream pathways and genes of ETH in Scylla paramamosain, RNA interference (RNAi) was conducted on crabs at early (D0) and late (D2) premolt substages, and the transcriptome profiles of each group were compared by RNA sequencing. Real-time quantitative polymerase chain reaction (RT-qPCR) and semiquantitative polymerase chain reaction (RT-PCR) results showed a significant knockdown of ETH at D0 stage, whereas a significant increase was shown conversely in crabs at D2 substage after the injection of dsETH. A total of 242,979 transcripts were assembled, and 44,012 unigenes were identified. Transcriptomic comparison between crabs at D2 and D0 substages showed 2,683 differentially expressed genes (DEGs); these genes were enriched in ribosome and pathways related to transcription factor complex and cell part. Twenty DEGs were identified between dsETH-injected and dsGFP-injected crabs at D0 substage; these DEGs were involved in carbohydrate metabolism, one carbon pool by folate, and chitin binding. Twenty-six DEGs were identified between dsETH-injected and dsGFP-injected crabs at D2 substage; these DEGs were involved in calcium channel inhibitor activity, fat digestion and absorption, and cardiac muscle contraction. RT-qPCR verified the differential expression of the selected genes. In conclusion, crabs at D0 substage are more active in preparing the macromolecular complex that is needed for molting. Moreover, ETH has potential roles in carbohydrate metabolism, one carbon pool by folate, and chitin binding for crabs at D0 substage, while the role of ETH turns to be involved in calcium channel inhibitor activity, fat digestion and absorption, and cardiac muscle contraction at D2 substage to facilitate the occurrence of molting. The selected DEGs provide valuable insight into the role of ETH in the regulation of crustacean molting.
Introduction
Molting is a natural and biological process for survival, development, and reproduction in arthropods. The molting cycle can be divided into four stages: intermolt stage (C), premolt stage (D), ecdysis (E), and postmolt stage (A, B) (1). In crustaceans, the premolt stage can be divided into five substages: early (stage D0), mid (stage D1), and late (stages D2, D3, D4) substages (2). During the premolt stage, the old cuticle detaches and the inner layers of the old cuticle are degraded and reabsorbed, the new exoskeleton is synthesized, the claw closer muscle atrophies, and autotomized appendages regenerate (3–5). The molting process is achieved by the interaction of ecdysteroids and a complex suite of interacting neuropeptides (6). As an essential hormone in crustaceans, 20-hydroxyecdysone (20E) fluctuates with the process of molting (7) and is controlled by neuropeptides secreted from the central nervous system and then regulates a series of target genes through ecdysone receptor signaling (8, 9).
As one of the target genes, ecdysis triggering hormone (ETH) has an ecdysone response element in its promoter region. Also, Gauthier etal. (10) showed that the Drosophila homolog of ATF4 (cryptocephal) is an ecdysone receptor coactivator. The ecdysone receptor can bind to the promoter region of ETH with cryptocephal and initiates the transcription of ETH. Researchers have found that ETH has additional roles besides initiating ecdysis. In insects, ETH plays dual roles, initiation of ecdysis and trachea clearance (11, 12). In Drosophila, the knockdown of ecdysis triggering hormone receptor (ETHR) revealed differential expression of genes involved in axon guidance, courtship behavior, and chemosensory functions, which indicated a role in the regulation of courtship behavior (13). Researchers also found that ETH acts as a regulatory peptide to ensure the synthesis of juvenile hormone at the proper time (14). As a dual regulator of octopamine and juvenile hormone release, ETH also provides a link between the stress-induced elevation of ecdysone levels and consequent reduction in fecundity in Drosophila (15). ETH in Bactrocera dorsalis also plays an essential role in its reproduction, via the regulation of JH and vitellogenin, and is controlled by a pulse of 20E (16). ETH also regulates molting in crustaceans (17). In Scylla paramamosain, we found that the knockdown of ETH leads to failure of molting at the early premolt substage. Interestingly, the transcript level of ETH had a significant increase conversely when crabs at the late premolt substage (D2) were injected with the same amount of dsETH as at the early premolt stage (D0), which suggests different means of regulation of ETH at the D0 and D2 substages in S. paramamosain (18).
In crustaceans, the premolt stage is essential for adequate preparation for molting. Many biological processes are occurring at the premolt stage, amounts of them such as the regulation of apolysis have not been well-studied. Transcriptome analyses have mostly focused on the regulatory changes among A/B, C, and D molt stages. However, analyses between the different substages of the premolt stage in crustaceans are rare despite their potential to reveal the dynamic changes in transcriptome in the preparation for molting. Although the roles of ETH are well-studied in insects, related work is still limited in crustaceans, and whether ETH has different roles in crustaceans is still unknown. Notably, ETH can regulate the initiation of ecdysis in crustaceans (17); however, its downstream transcriptional pathways remain largely elusive. In this study, S. paramamosain was selected. To reveal the dynamic change of biological processes at the premolt stage and determine the roles of ETH at the D0 and D2 substages in S. paramamosain, RNAi was used. This study aimed to identify candidate genes and pathways related to ETH and determine the role of ETH at the D0 and D2 substages.
Materials and methods
Animals
Crabs were collected from the coastal area of Zhanjiang, Guangdong Province, China. Individuals (i.e., 2.4–2.9 g) were selected and cultured in 1-m3 concrete ponds with seawater at 24°C–27°C and 20‰ salinity and fed daily with oysters. After a week, healthy crabs at the D0 and D2 substages (19) were selected for the RNAi experiment.
RNAi and injection
Primers flanked by the T7 promoter sequence were designed and used to synthesize the double-stranded RNA (dsRNA) for ETH and control gene (GFP) (18). The dsRNA templates were amplified with the procedure: 95°C for 5 min; followed by 30 cycles of 95°C for 30 s, 57°C for 30 s, and 72°C for 30 s; then 72°C for 10 min. Then, PCR products were purified using FastPure Gel DNA Extraction Mini Kit (Vazyme, Nanjing, China). dsRNA was produced with the above DNA templates under the method of T7 RiboMAX™ Express RNAi Synthesis kit (Promega, Beijing, China). The final dsRNA was diluted to 1 μg/μl. Crabs (n = 20) at the D0 and D2 substages were collected and separately injected with dsETH and dsGFP. Each crab was injected in dsRNA at a dose of 5 μg/g. After 48 h, thoracic ganglia were sampled and analyzed, each sample was collected from five crabs, and each group has three samples.
RT-PCR
According to the target genetic sequence, specific primers were designed using Primer 5.0 and 18s rRNA was used as the internal reference gene (Table S1). For sequence amplification, the following program was used: 94°C for 5 min followed by 30 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 30 s, and 72°C for 10 min. RT-PCR reactions of 20 μl contained 1 μl of cDNA and each primer, 10 μl of 2× PCR Master Mix (GenStar, Beijing, China), and 7 μl ddH2O. Amplified products were electrophoresed on 1% agarose gel and imaged using ImageJ 1.51j8 software performs data conversion.
Tissue collection, RNA isolation, and cDNA library preparation
Crabs were chilled on ice and dissected for thoracic ganglia. For every group, samples were collected in three different pools; each pool contained five biological replicates. Total RNA was prepared using a column-based TransZol Up Plus RNA kit (TransGen Biotech, China). The concentration of the extractive RNA was determined using the Thermo Fisher Scientific Nanodrop 2000 (Waltham, MA, USA), and the quality and integrity were determined with an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA) and a Qubit 2.0 Fluorometer (Life Technologies, CA, USA). The qualified RNA samples were used for cDNA library construction. Sequencing was done with the NEBNext R Ultra™ RNA Library Prep Kit for Illumina R (NEB, Ipswich, MA, USA), and index codes were added to tag these sequences. The libraries were sequenced using the Illumina HiSeq platform.
Cleaning, de novo assembly, and annotation of sequencing reads
Raw reads were cleaned by removing adapter, primer sequences, reads with ambiguous nucleotides larger than 5%, and low-quality reads. De novo assembly was done using Trinity (20). For annotation, BLASTX searches were performed with e-value <1e-5 and 1e-3 against the NR (NCBI nonredundant protein sequences), NT (NCBI nonredundant nucleotide sequences), KO [Kyoto Encyclopedia of Genes and Genomes (KEGG) ortholog database], Swiss-Prot (a manually annotated and reviewed protein sequence database), Pfam (protein family), GO (Gene Ontology) databases, and KOG/COG (clusters of orthologous groups of proteins). The read count of every gene with FPKM based on the length and read count was calculated. The raw sequencing reads are archived at the NCBI Sequence Read Archive: PRJNA813292.
Differentially expressed gene analysis
Differential expression analyses of D2dsGFP vs. D0dsGFP, D0dsETH vs. D0dsGFP, and D2dsETH vs. D2dsGFP were performed using DESeq2 software (21). Unigene with false discovery rate (padj) values <0.05 and |log2(FoldChange)| >1 were set as the threshold for differentially expressed genes (DEGs). GO and KEGG database annotations were further analyzed (corrected P-value <0.05) to explore the functions of DEGs in S. paramamosain.
Validation with RT-qPCR
To analyze the knockdown efficiency of ETH and validate the DEGs from the transcriptome data, RT-qPCR reactions were performed with the RT-qPCR machines (Bio-Rad CFX Connect PCR, Bio-Rad, USA). Primers were designed using the Software Primer 5.0, and 18s rRNA was used as internal control genes (Table S1). RT-qPCR reaction system: 10 μl of SYBR qPCR Master Mix (Vazyme, Nanjing, China), 2 μl of cDNA, 7 μl of ultrapure water, and 0.5 μl of each Forward and Reverse primer (10 μM). RT-qPCR reaction condition: 95°C for 2 min, followed by 39 cycles of 95°C for 5 s and 55°C for 30 s, and then a melt curve analysis was performed. The relative mRNA abundance of each gene was calculated by the ΔΔCt method, then normalized by that of 18S for each sample. The reaction was performed in triplicate for each sample, and five mud crabs were analyzed in each group. The relative expression of the target genes between groups was statistically analyzed with GraphPad Prism 8.0.1 (GraphPad Software Inc., San Diego, CA, USA) by one-way ANOVA, followed by Tukey’s honestly significant difference test and Pearson correlation coefficients. All data were expressed as means ± standard deviation (SD).
Results
Knockdown of ETH before transcriptomic analysis
Both RT-qPCR and RT-PCR results showed that the transcript level of ETH was decreased significantly (P < 0.05) after the injection of dsETH at the D0 substage, whereas the transcript level of ETH had a significant increase conversely when crabs at the D2 substage were injected with the same amount of dsETH (Figure 1).
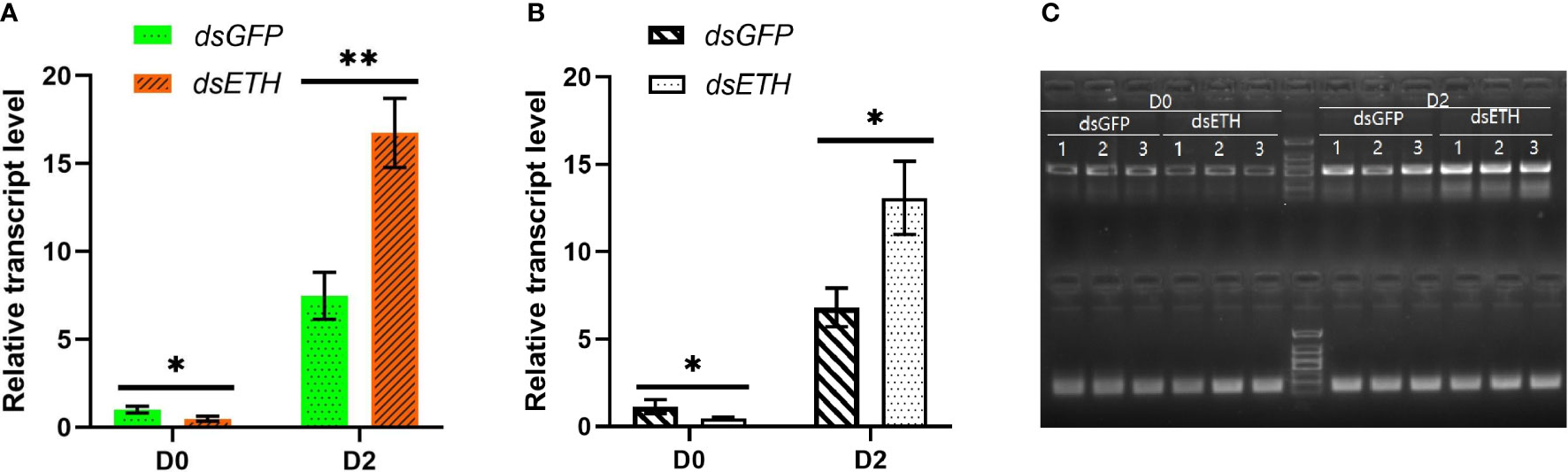
Figure 1 Relative transcript levels of ETH in thoracic ganglia of dsGFP- and dsETH-injected crabs at the D0 and D2 substages were measured respectively [by Student’s t-test, values are means ± SD (n = 3), *P < 0.05; **P < 0.01]. (A) RT-qPCR result; (B) RT-PCR result; (C) Images of the gels for RT-PCR.
Sequencing and de novo transcriptome assembly and annotation
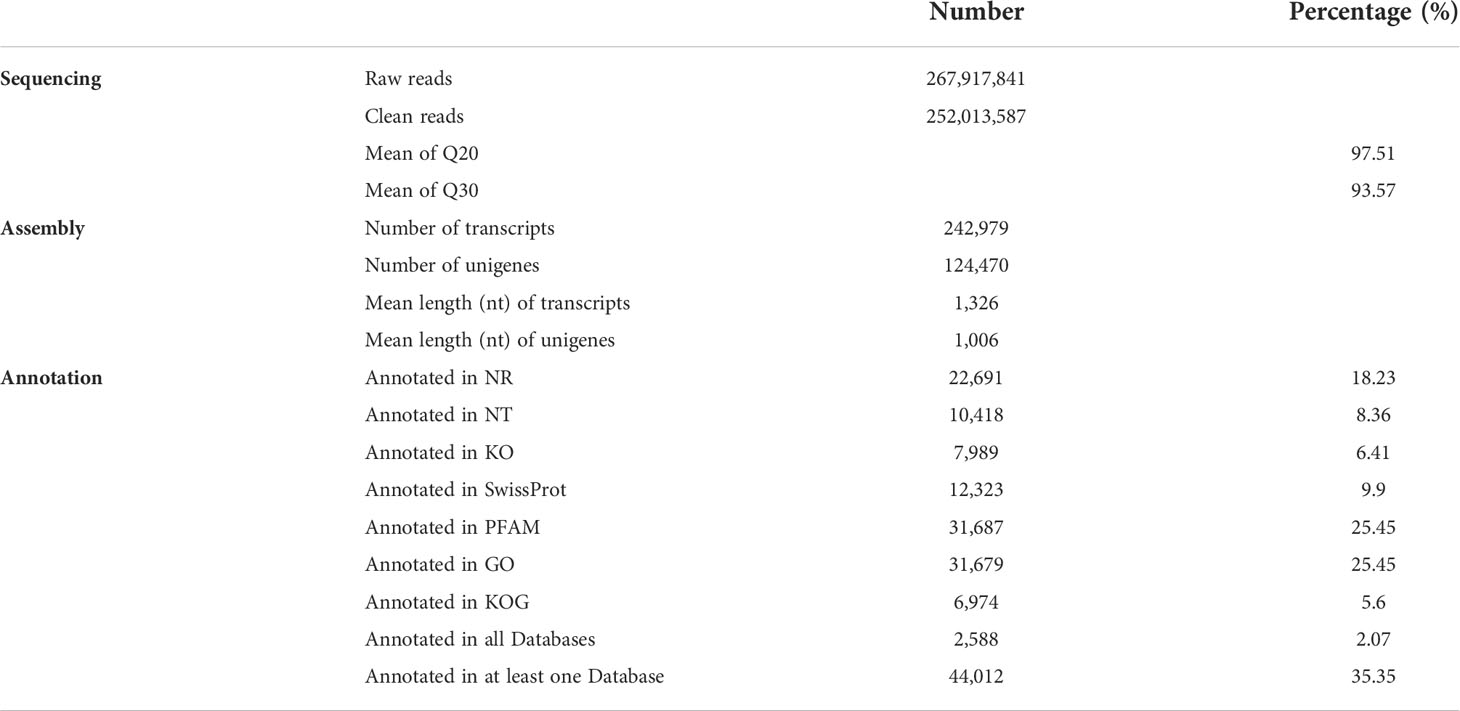
From 12 thoracic ganglia samples of the above four groups, a total of 252,013,587 clean reads from 267,917,841 raw reads were obtained after sequencing, quality trimming, and adapter clipping. None of the error rates of the 12 libraries exceeded 0.03%, and the mean Q20 and Q30 were 97.51% and 93.57%. After de novo assembly, 242,979 transcripts and 124,470 unigenes were obtained, which had mean lengths of 1,326 bp and 1,006 bp (Table 1). The length distributions of all the transcripts and unigenes are shown in Figure S1. The annotation results are shown in Table 1.
Differentially expressed gene identification
We found in total 2,708 DEGs in the transcriptomes using the corrected P-value <0.05 and |log2(fold change)| >1 (Figure 2, Table S2). The DEGs between D2dsGFP and D0dsGFP were 1,717 upregulated genes and 966 downregulated genes (Figure 2A; Figure S2). The DEGs were few in D0dsETH vs. D0dsGFP, with only 3 upregulated genes and 17 downregulated genes (Figure 2B; Figure S3); 15 upregulated genes and 11 downregulated genes were found in D2dsETH vs. D2dsGFP (Figure 2C; Figure S4).
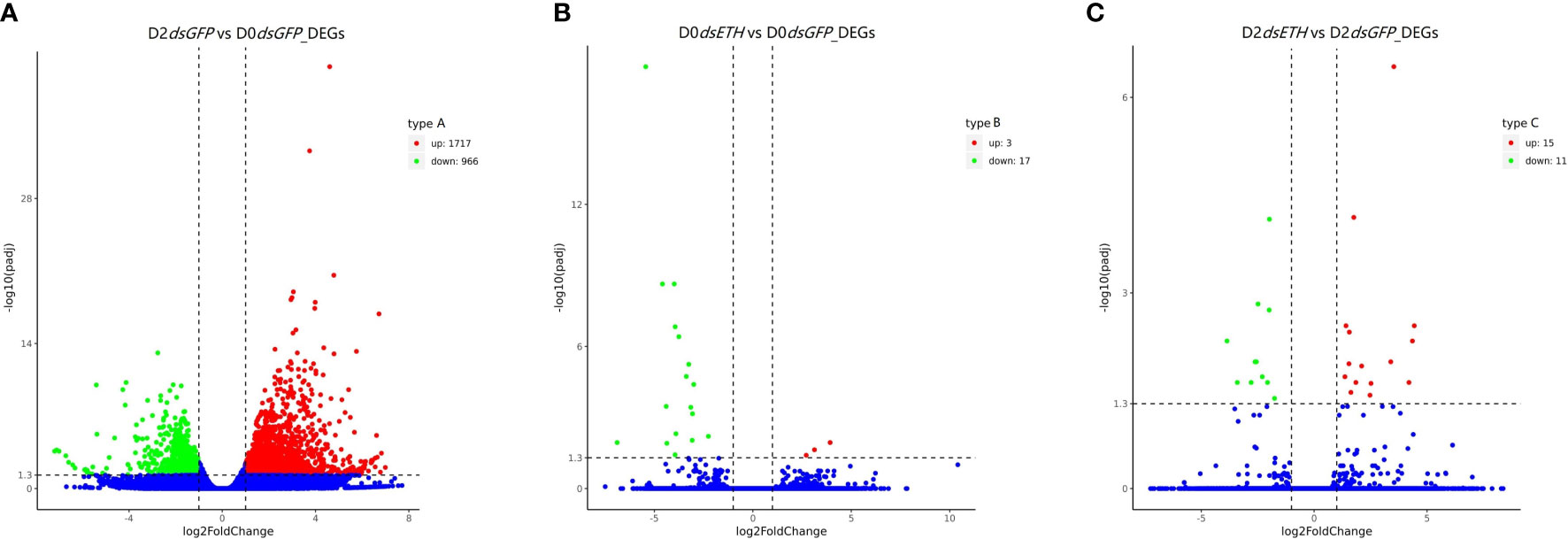
Figure 2 Differentially expressed genes (DEGs) in three comparisons of S. paramamosain [corrected P-value <0.05 and |log2(fold change)| >1]. (A) Volcano plot of DEGs in D2dsGFP (Crabs post dsGFP injection at D2 substage) vs. D0dsGFP (Crabs post dsGFP injection at D0 substage). (B) Volcano plot of DEGs in D0dsETH vs. D0dsGFP. (C) Volcano plot of DEGs in D2dsETH vs. D2dsGFP. Red spots represent upregulated genes and green spots indicate downregulated genes. Blue spots represent genes showing no obvious change in the comparison.
Enrichment analysis and functional classification of differentially expressed genes
GO and KEGG pathway analyses of DEGs were performed (corrected P-value <0.05) to determine their functions in S. paramamosain. DEGs between D2dsGFP and D0dsGFP were annotated (padj <0.05). Enrichment was not observed in either GO terms or KEGG pathways for upregulated DEGs. Compared to the D0dsGFP group, for downregulated DEGs, GO enrichment revealed that transcription factor complexes such as macromolecular complex, transcription factor complex, and RNA polymerase II transcription factor complex were the most enriched subcategories in the cellular component. In addition, cell part (means any constituent part of a cell, the basic structural and functional unit of all organisms), bind, and intracellular organelle part were enriched (Figure 3A; Table S3). KEGG results of downregulated DEGs were mainly enriched in ribosome (Figures 4A, 5; Table S4).
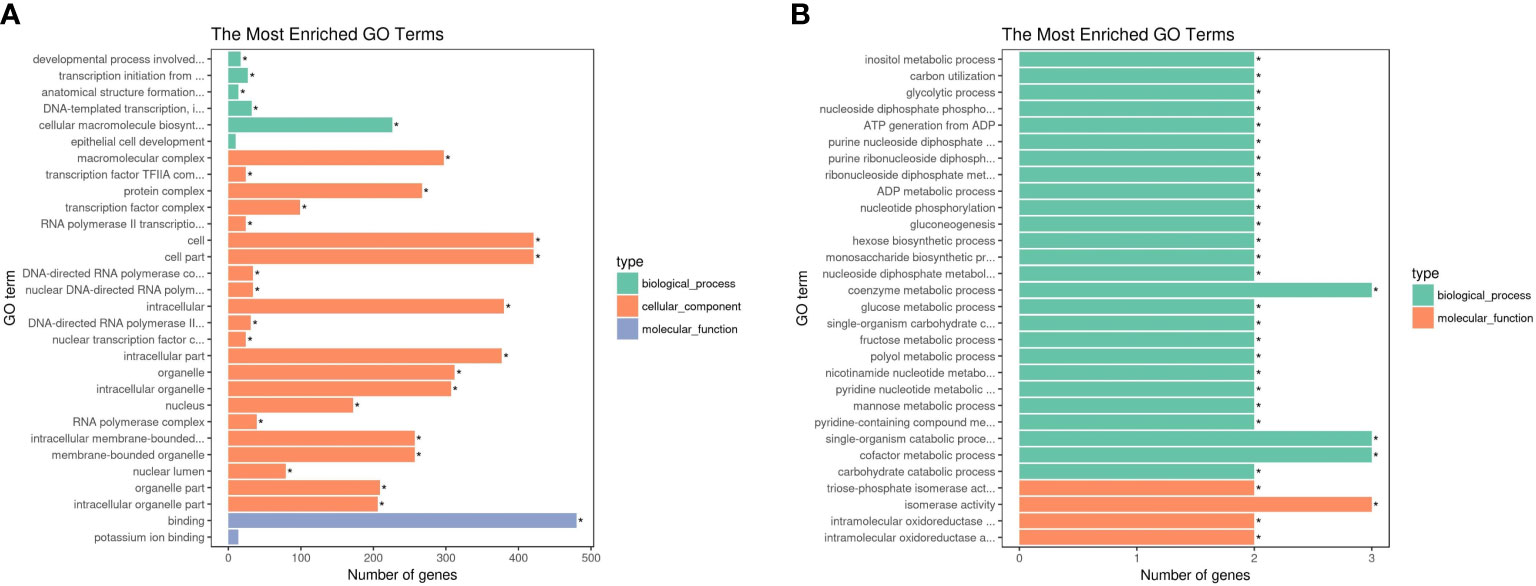
Figure 3 Enrichment of differentially expressed genes (DEGs) in GO (corrected P-value <0.05). (A) Downregulated DEGs that enriched GO terms in D2dsGFP vs. D0dsGFP. (B) Downregulated DEGs that enriched GO terms in D0dsETH vs. D0dsGFP. Significant difference compared with control indicated by an asterisk (P-value <0.05).
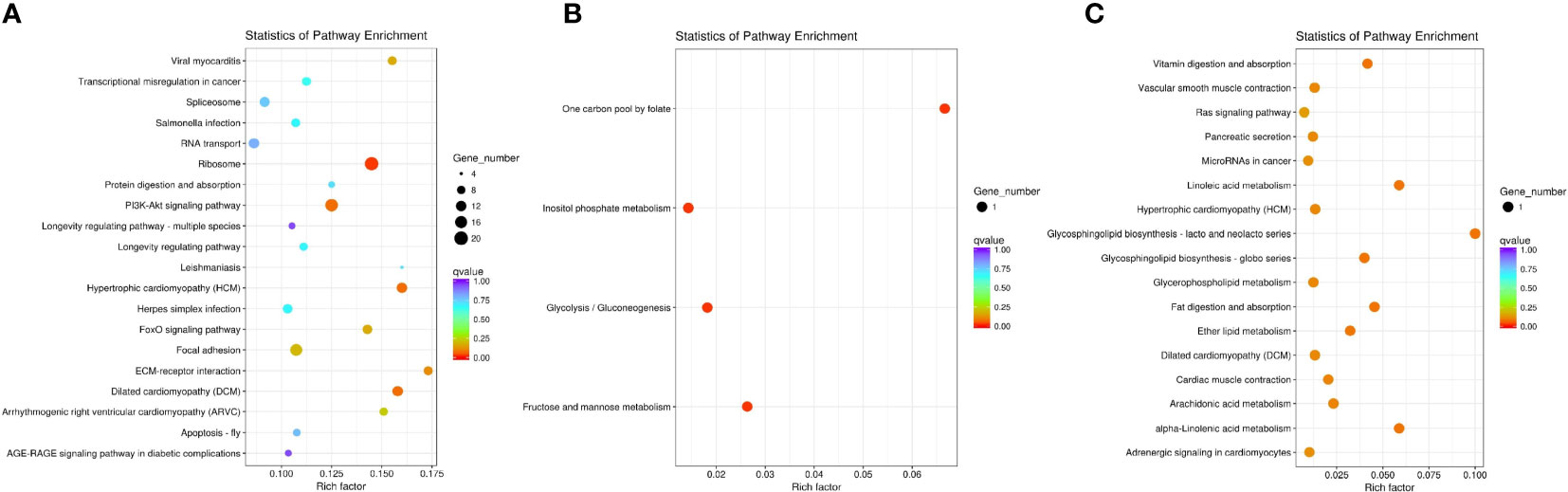
Figure 4 Enrichment of differentially expressed genes (DEGs) in KEGG (corrected P-value <0.05). (A) Downregulated DEGs that enriched KEGG terms in D2dsGFP vs. D0dsGFP. (B) Downregulated DEGs that enriched KEGG terms in D0dsETH vs. D0dsGFP. (C) Upregulated DEGs that enriched KEGG terms in D2dsETH vs. D2dsGFP.
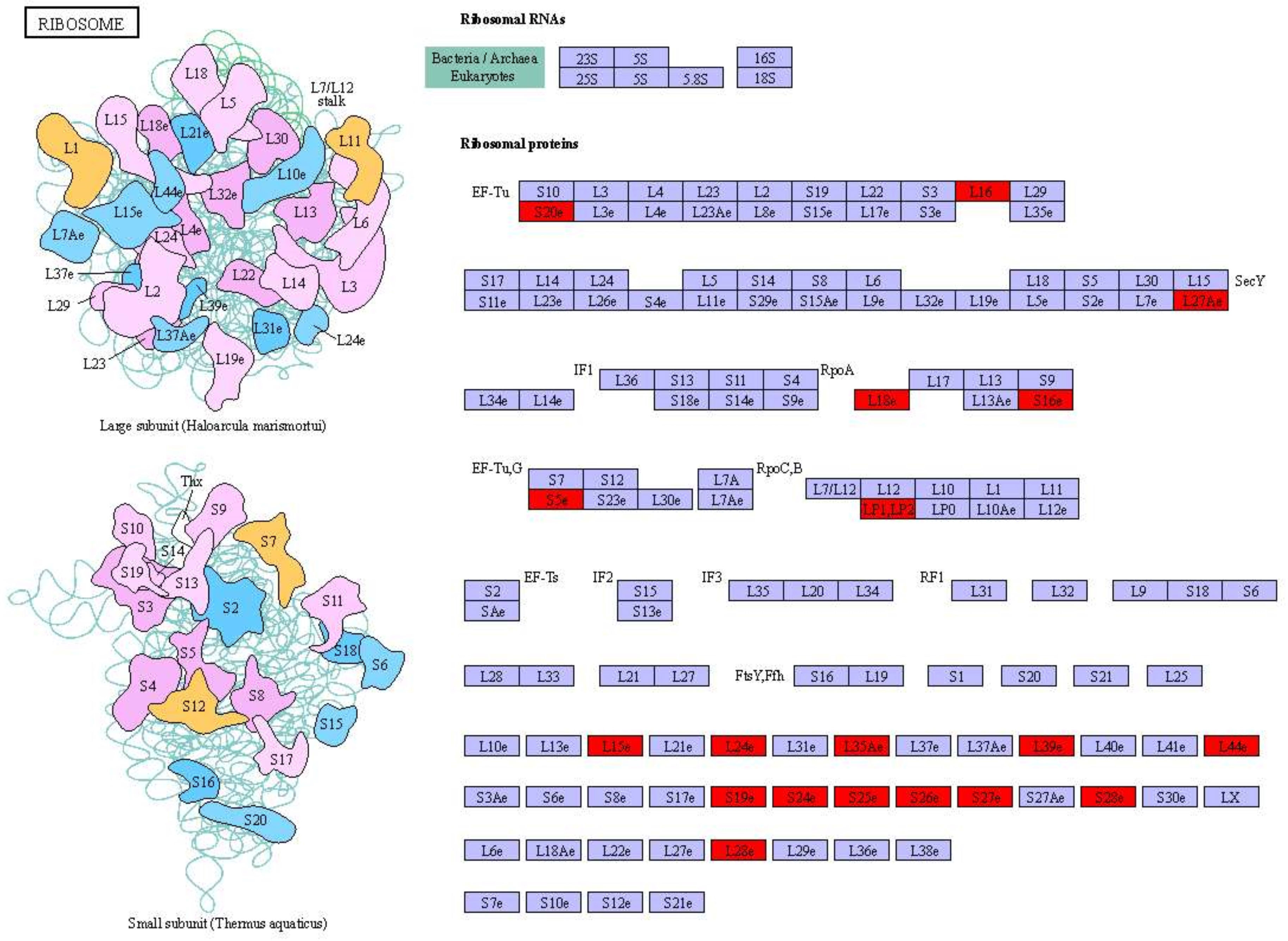
Figure 5 Significantly differentially expressed genes of ribosome-related pathways in D2dsGFP vs. D0dsGFP of S. paramamosain. Red indicates significantly upregulated genes. (For interpretation of the short letters’ meaning in this figure legend, the reader is referred to the web version of KEGG: ko03010.).
GO enrichment revealed that the downregulated DEGs of the D0dsETH group involved in the biological process were mainly enriched in energy metabolic processes such as inositol metabolic, glycolytic process, mannose and fructose metabolic, and adenosine triphosphate (ATP) generation from adenosine diphosphate (ADP). In the molecular function category, downregulated DEGs were mostly enriched in catalytic activity and binding such as triosephosphate isomerase activity and intramolecular oxidoreductase activity (Figure 3B; Table S3). Upregulated DEGs of the D0dsETH group were enriched in chitin binding (Table S3). For KEGG pathways, downregulated DEGs of the D0dsETH group were enriched in one carbon pool by folate, fructose and mannose metabolism, glycolysis/gluconeogenesis, and inositol phosphate metabolism, which were related to the energy metabolic pathway (Figure 4B; Table S4).
Enrichment was not observed in either GO terms or KEGG pathways for DEGs between the D2dsETH and D2dsGFP groups. To be as inclusive as possible, less strict screening criteria (P-value <0.05 instead of corrected P-value <0.05) were applied, and upregulated DEGs of the D2dsETH group were mainly enriched in the regulation of cell shape, regulation of Wnt signaling pathway, globoside metabolic process, alpha-(1,2)-fucosyltransferase activity, and regulation of cell morphogenesis under GO enrichment; downregulated DEGs were mainly enriched in calcium channel inhibitor activity, endopeptidase inhibitor activity, and defense response (Table S3). KEGG enrichment results show that upregulated DEGs of the D2dsETH group are involved in glycosphingolipid biosynthesis, linoleic acid metabolism, fat digestion and absorption, vitamin digestion and absorption, ether lipid metabolism, and arachidonic acid metabolism; downregulated DEGs of the D2dsETH group were not enriched in the KEGG pathway (Figure 4C; Table S4).
Differentially expressed genes involved in carbohydrate metabolism, lipid metabolism, and biological process of molting
Besides the analysis of the entire gene set, we specifically checked for genes that were significantly regulated and involved in carbohydrate metabolism, one carbon pool by folate, chitin binding, calcium channel inhibitor activity, fat digestion and absorption, and cardiac muscle contraction (Table 2). At the D0 substage, the knockdown of ETH caused the downregulation of triosephosphate isomerase-like (TIM-like) and triosephosphate isomerase A-like (TIM-A-like), genes related to glycolytic enzyme that are essential to carbohydrate metabolism. Also, a gene of methylenetetrahydrofolate reductase-like (MTHFR-like) was downregulated, which was a critical enzyme of the folate pathway and required for DNA synthesis, methylation, and amino acid metabolism. Whereas a unigene related to chitin binding (peritrophin-like1) was upregulated significantly. At the D2 substage, the knockdown of ETH showed the downregulation of fibrocystin-L that was related to calcium channel inhibitor activity and essential to cellular control of adhesion and epithelial morphogenesis (22), whereas amine oxidase (AO) related to fat digestion and absorption and the unigene of tropomyosin 1 (FBC-L) that participates in cardiac muscle contraction were upregulated. To confirm the reliability of these transcriptome data, these selected DEGs were determined with RT-qPCR (Figure 6). The correlation coefficient R was 0.926 and 0.869.
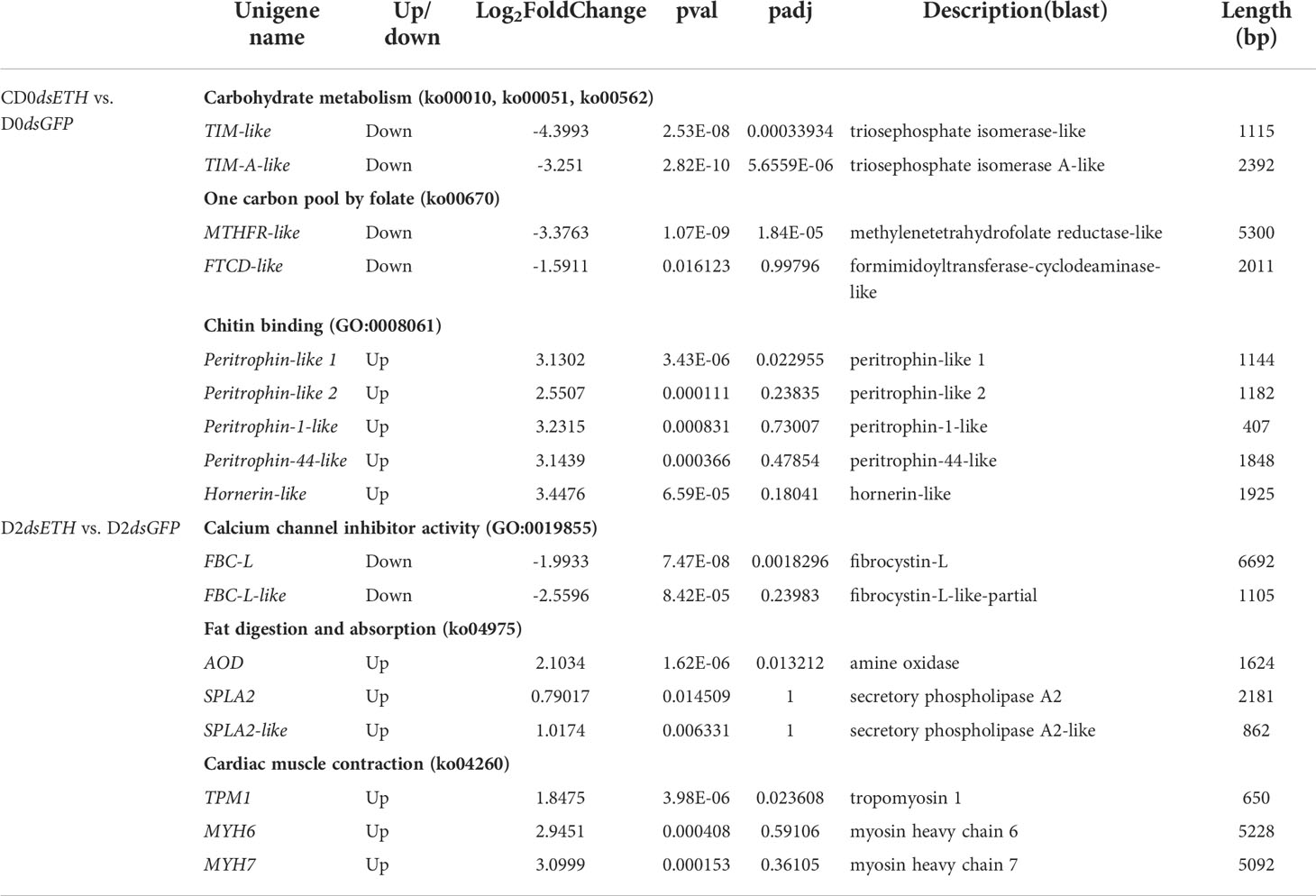
Table 2 DEGs related to metabolism and molting process from S. paramamosain responding to dsETH treatment at different premolt substages.
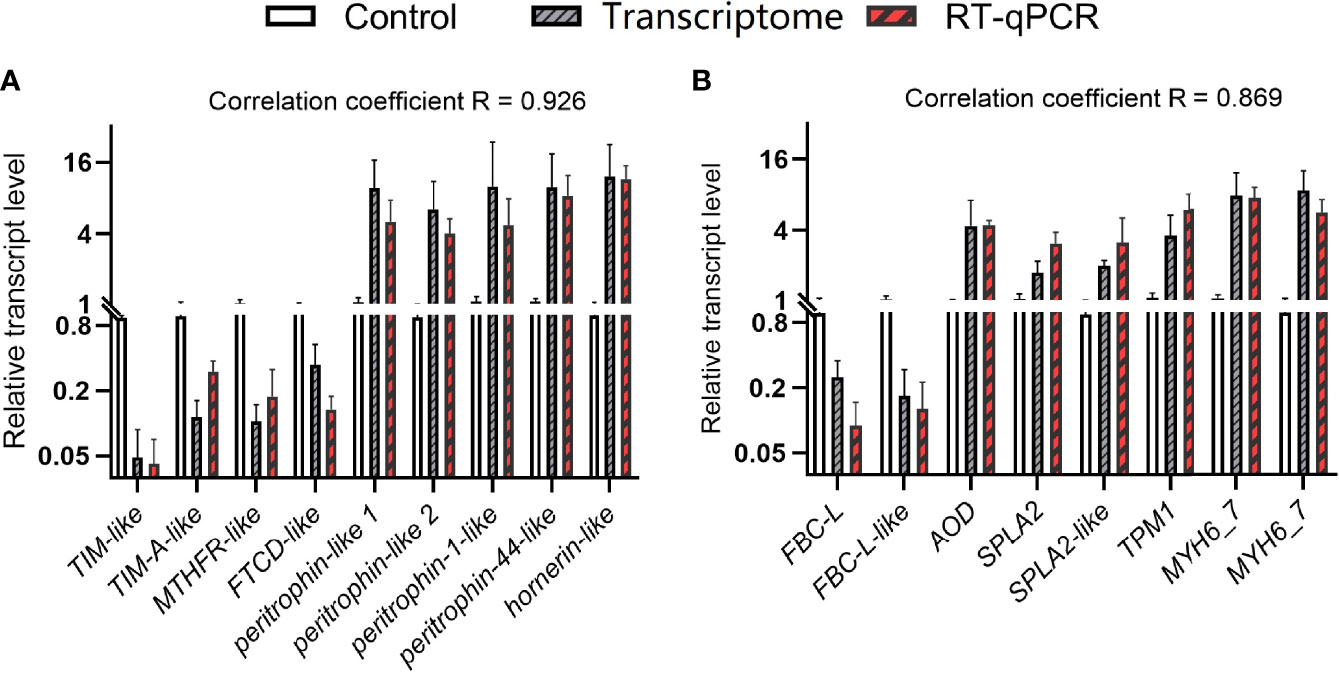
Figure 6 Comparison of gene expression patterns obtained by RNA-Seq and RT-qPCR. Values are means ± SD (n = 3). (A) D0dsETH vs. D0dsGFP. (B) D2dsETH vs. D2dsGFP.
To be as inclusive as possible, less strict screening criteria of DEGs (pval <0.05 instead of padj <0.05) were applied. From these DEGs, formimidoyltransferase-cyclodeaminase-like (FTCD-like), a folate-dependent enzyme that displays both transferase and deaminase activity (23), was selected and downregulated with the knockdown of ETH at the D0 substage. Also, another four genes (peritrophin-like 2, peritrophin-1-like, peritrophin-44-like, and hornerin-like) involved in chitin binding were upregulated. At the D2 substage, fibrocystin-L-like-partial, involved in calcium channel inhibitor activity, was downregulated with the knockdown of ETH, two genes (secretory phospholipase A2, secretory phospholipase A2-like) related to fat digestion and absorption and two genes (myosin heavy chain 6/7) related to cardiac muscle contraction were upregulated with the knockdown of ETH. These DEGs were also verified using RT-qPCR, and the result showed high consistency with transcriptome sequencing.
Discussion
In the previous study, the expression level of ETH showed a periodic fluctuation with molting, and ETH exists not only at the E stage but also at the other stages (18). Functional diversity of ETH has been proven in insects (not just triggering ecdysis and working at the E stage) (14–16). The existence of ETH at other stages meant that it may focus on triggering not only ecdysis but also other aspects. When we injected dsETH to crabs at the D0 substage, the RT-sqPCR result showed a trend: decline. It meant that we knocked down the ETH successfully. Meanwhile, RT-sqPCR results showed the increased trend at the D2 substage. It might be due to the later retaliatory rise of the transcriptive level offsetting the knockdown at an early time after the injection of dsETH. To balance, the injection of dsETH at the D2 substage led to an increase of ETH when we did sampling for transcriptome analysis. Our previous study showed that ETH was regulated by different models in S. paramamosain, and eclosion hormone was important to the regulation of ETH at the D2 substage (18).
In arthropods, apolysis is considered as an initial sign of molting and triggered by rising 20E titer (24, 25). Immediately after apolysis, the molting fluid is secreted into the new apolysial space and initiates a series of biological processes such as new cuticle secretion, old cuticle degradation, and accumulation of neuropeptides related to molting (1, 26–29). These biological processes reveal the existence of a complex and precise regulatory network at the premolt stage. To investigate the dynamic changes between the D0 and D2 substages, the transcriptomes of dsGFP-injected crabs at the D0 and D2 substages were compared. Global gene expression profiles of crabs at the D2 substage were distinct from crabs at the D0 substage, with 1,717 upregulated DEGs and 966 downregulated DEGs. Meanwhile, these downregulated DEGs related to transcription factor complex and RNA polymerase indicate that amounts of macromolecular complex are synthesized at the D0 substage to make sure the full preparation of proteins related to transcription factor that are needed for molting. Ribosomes are the important molecular machines that translate genetic information into proteins. The consistent trend of DEGs related to ribosome and transcription factor further supports the hypothesis that crab at the D0 substage is more active in preparing macromolecular complexes that are needed for molting. In S. paramamosain, the injection of dsETH with a dose of 5 µg/g could knock down ETH of crabs at the D0 substage but failed with crabs at the D2 substage (18). The expression level of ETH has a significant increase from the D0 substage to the D2 substage and then comes to a dramatic increase from the D2 substage to the E stage (18). It might be because the D0 substage is the preparatory phase of ETH mass synthesis, in which the synthesis of macromolecular complex that regulates the retaliatory increase of ETH is underway and has not been synthesized until the D2 substage. As an essential neuropeptide in the molting of S. paramamosain, the retaliatory regulatory mechanism may be a key for crab to live through the fragile stage under the changeable environment.
To investigate the correlative transcriptional pathways of ETH in S. paramamosain at the D0 substage, transcriptomes of the dsETH-injected group and the dsGFP-injected group were compared. It is interesting that knockdown of ETH leads to downregulation of genes enriched in carbohydrate metabolism, such as TPI-like and TPI-A-like, which are essential in glycolysis. Downregulation of TPI can lead to a decrease in the synthesis of ATP. Also, TPI deficiency is the most severe and lethal glycolytic enzymopathy (30, 31). We suggested that ETH may regulate the molting process by influencing glycolysis and energy supply at the D0 substage. Moreover, downregulated genes involved in one carbon pool by folate were screened, such as MTHFR-like and FTCD-like, of which the function of both is catalyzing NADH-dependent reduction of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate. It is worth mentioning that 5-methyltetrahydrofolate is essential to neural development, cell division, and embryogenesis (32, 33). As the whole molt cycle of crustacean is regulated by numerous neuropeptides released by nervous tissue, 5-methyltetrahydrofolate may affect the synthesis of neuropeptides by neural development. In contrast, transcriptional levels of peritrophins and hornerin-like involved in chitin binding were dramatically upregulated. In Eriocheir sinensis, chitinases involved in chitin binding showed a high expression level during the postmolt and ecdysis stages, and a very low level at the intermolt and premolt stages were detected (34), which indicated a potential role of chitin binding to molting. Meanwhile, researchers found that cuticular proteins analogous to peritrophins were abundantly distributed in the cuticle (35–37), and the knockdown of the genes can cause a delay of pupariation or mortality (38, 39). The relationship between ETH and peritrophins in S. paramamosain still needs more research.
At the D2 substage, transcriptomes of dsETH-injected crabs and dsGFP-injected crabs were compared. With the increase of ETH in crab, downregulated genes involved in calcium channel inhibitor activity were enriched, such as FBC-L and FBC-L-like. In mammals, FBC is localized to the primary cilium/basal body and plasma membrane in the renal epithelium and is thought to be a transcriptional target of hepatocyte nuclear factor 1β (40–42). Researchers found that a decline of FBC could result in abnormalities in ciliary morphology and possibly biliary cystogenesis in rat (43). The hepatopancreas is an important organ for the crab to store energy, substance, calcium, and so on. ETH may regulate molting by influencing the hepatopancreas through FBC. Moreover, upregulated genes were enriched in fat digestion and absorption, such as AO, SPLA2, and SPLA2-like, which are involved in the metabolism of linoleic acid, ether lipid, glycerophospholipid, and other lipids. AO is regarded as a biological regulator, especially for cell growth and differentiation (44). AO is also an important agent of pathological processes of rhabdomyolysis in rats (45). In mammals, the SPLA2 families are involved in a number of biological processes, such as modification of eicosanoid generation, inflammation, and host defense (46). In the study, ETH has a potential relationship with AO and SPLA2s in S. paramamosain at the D2 substage. We also observed the upregulation of several cardiac muscle contraction-related genes, such as TPM1, MYH6, and MYH7. TPM1 was first identified in skeletal muscles (Bailey) and plays a critical role in conjunction with troponin and the regulation of muscle contraction (47–49). Considering that muscle contraction is indispensable to shed the old cuticle for crab, ETH may trigger ecdysis through regulating TPM1.
Conclusion
ETH has been determined to function in molting in several crustaceans; how ETH affects molting is still unclear in crustaceans. In the study, it is the first time to explore the potential downstream genes of ETH that are related to molting in crustaceans. Our results show that ETH has potential roles in carbohydrate metabolism, one carbon pool by folate, and chitin binding for crabs at the D0 substage, while with the process of molt cycle, the role of ETH turns to be involved in calcium channel inhibitor activity, fat digestion and absorption, and cardiac muscle contraction to facilitate the occurrence of molting. The findings of DEGs involved in the above pathways provide valuable insight into the role of ETH signaling in the regulation of crustacean molting.
Data availability statement
The data presented in the study are deposited in the NCBI Sequence Read Archive (SRA) repository, and the sample information was registered in BioProject with accession number PRJNA813292.
Author contributions
Conceptualization, S-FC and Y-FZ; methodology, S-FC, Y-FZ, C-MA, and C-GW; software, Y-FZ and Q-QW; validation, C-MA and WW; formal analysis, Y-FZ and WW; investigation, S-FC, Y-FZ, and WW; resources, C-GW; data curation, Y-FZ and Q-QW; writing—original draft preparation, S-FC, Q-QW, and Y-FZ; writing—review and editing, Q-QW and S-FC; visualization, Q-QW and C-MA; supervision, S-FC; project administration, S-FC; funding acquisition, S-FC. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Guangdong Provisional Research Grant (#2014B020202014) and Guangxi Key Laboratory of Beibu Gulf Marine Biodiversity Conservation Fund (2019KB06).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.917088/full#supplementary-material
References
1. Stuart ER. The cuticle, growth and moulting in insects: The essential background to the action of acylurea insecticides. Pesticide Sci (1987) 20:131–46. doi: 10.1002/ps.2780200207
2. Pratoomchat B, Sawangwong P, Guedes R, Reis MD, Machado J. Cuticle ultrastructure changes in the crab Scylla serrata over the molt cycle. J Exp Zool (2002) 293:414–26. doi: 10.1002/jez.90002
3. McCarthy JF, Skinner DM. Proecdysial changes in serum ecdysone titers, gastrolith formation, and limb regeneration following molt induction by limb autotomy and/or eyestalk removal in the land crab, Gecarcinus lateralis. Gen Comp Endocrinol (1977) 33:278–92. doi: 10.1016/0016-6480(77)90252-0
4. Mykles DL. Interactions between limb regeneration and molting in decapod crustaceans 1. Am Zool (2001) 41:399–406. doi: 10.1093/icb/41.3.399
6. Zitnan D, Kim YJ, Zitnanová I, Roller L, Adams ME. Complex steroid-peptide-receptor cascade controls insect ecdysis. Gen Comp Endocrinol (2007) 153:88–96. doi: 10.1016/j.ygcen.2007.04.002
7. Chang ES, Mykles DL. Regulation of crustacean molting: a review and our perspectives. Gen Comp Endocrinol (2011) 172:323–30. doi: 10.1016/j.ygcen.2011.04.003
8. Yao TP, Forman BM, Jiang Z, Cherbas L, Chen JD, McKeown M, et al. Functional ecdysone receptor is the product of EcR and ultraspiracle genes. Nature (1993) 366:476–9. doi: 10.1038/366476a0
9. Hill RJ, Billas IML, Bonneton FO, Graham LD, Lawrence MC. Ecdysone receptors: from the ashburner model to structural biology. Annu Rev Entomol (2013) 58:251–71. doi: 10.1146/annurev-ento-120811-153610
10. Gauthier SA, VanHaaften E, Cherbas L, Cherbas P, Hewes RS. Cryptocephal, the Drosophila melanogaster ATF4, is a specific coactivator for ecdysone receptor isoform B2. PloS Genet (2012) 8:e1002883. doi: 10.1371/journal.pgen.1002883
11. Shi Y, Jiang HB, Gui SH, Liu XQ, Pei YX, Xu L, et al. Ecdysis triggering hormone signaling (ETH/ETHR-a) is required for the larva-larva ecdysis in Bactrocera dorsalis (Diptera: Tephritidae). Front Physiol (2017) 8:587. doi: 10.3389/fphys.2017.00587
12. Shen CH, Xu QY, Fu KY, Guo WC, Jin L, Li GQ. Ecdysis triggering hormone is essential for larva-pupa-adult transformation in Leptinotarsa decemlineata. Insect Mol Biol (2021) 30:241–52. doi: 10.1111/imb.12691
13. Deshpande SA, Meiselman M, Hice RH, Arensburger P, Rivera-Perez C, Kim D-H, et al. Ecdysis triggering hormone receptors regulate male courtship behavior via antennal lobe interneurons in Drosophila. Gen Comp Endocrinol (2019) 278:79–88. doi: 10.1016/j.ygcen.2018.12.003
14. Areiza M, Nouzova M, Rivera-Perez C, Noriega FG. Ecdysis triggering hormone ensures proper timing of juvenile hormone biosynthesis in pharate adult mosquitoes. Insect Biochem Mol Biol (2014) 54:98–105. doi: 10.1016/j.ibmb.2014.09.006
15. Meiselman MR, Kingan TG, Adams ME. Stress-induced reproductive arrest in Drosophila occurs through ETH defificiency-mediated suppression of oogenesis and ovulation. Bmc Biol (2018) 16:18. doi: 10.1186/s12915-018-0484-9
16. Shi Y, Liu TY, Jiang HB, Liu XQ, Dou W, Park Y, et al. The ecdysis triggering hormone system, via ETH/ETHR-b, is essential for successful reproduction of a major pest insect, Bactrocera dorsalis (Hendel). Front Physiol (2019) 10:151. doi: 10.3389/fphys.2019.00151
17. Liang GX, Fu HT, Qiao H, Sun SM, Zhang WY, Jin SB, et al. Molecular cloning and characterization of a putative ecdysis-triggering hormone receptor (ETHR) gene from macrobrachium nipponense. J World Aquacult Soc (2018) 49:1081–94. doi: 10.1111/jwas.12451
18. Zhao YF, Wen QQ, Ao CM, Wang W, Shi LL, Wang CG, et al. Ecdysis triggering hormone, eclosion hormone, and crustacean cardioactive peptide play essential but different roles in the molting process of mud crab, Scylla paramamosain. Front Mar Sci (2022) 9:855391. doi: 10.3389/fmars.2022.855391
19. Ong KS. Observations on the post larval life history of Scylla serrata forskål, reared in the laboratory. Malay Agric J (1966) 45:429–43.
20. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol (2011) 29:644–52. doi: 10.1038/nbt.1883
21. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol (2014) 15:550. doi: 10.1186/s13059-014-0550-8
22. Ziegler WH, Soetje B, Marten LP, Wiese J, Burute M, Haffner D. Fibrocystin is essential to cellular control of adhesion and epithelial morphogenesis. Int J Mol Sci (2020) 21:5140. doi: 10.3390/ijms21145140
23. Solans A, Estivill X, de la Luna S. Cloning and characterization of human FTCD on 21q22.3, a candidate gene for glutamate formiminotransferase deficiency. Cytogenet Cell Genet (2000) 88:43–9. doi: 10.1159/000015483
24. Huffman JA, Applebaum SW. Sequential action of ecdysone and 20-hydroxyecdysone on locust epidermis. Dev Responsiveness Steroid Hormones (1980), 273–84. doi: 10.1016/B978-0-08-024940-7.50021-4
25. Soares MPM, Barchuk AR, Simões ACQ, dos Santos Cristino A, de Paula Freitas FC, Canhos LL, et al. Genes involved in thoracic exoskeleton formation during the pupal-to-adult molt in a social insect model, Apis mellifera. BMC Genomics (2013) 14:576. doi: 10.1186/1471-2164-14-576
26. Hale NA. Peptidases in Drosophila melanogaster-II. the variation of peptidase activities during development. Insect Biochem (1988) 18:145–&. doi: 10.1016/0020-1790(88)90018-2
27. Reynolds SE, Samuels RI. Physiology and biochemistry of insect moulting fluid. Adv Insect Physiol (1996) 26:157–232. doi: 10.1016/S0065-2806(08)60031-4
28. Dong DJ, He HJ, Chai LQ, Jiang XJ, Wang JX, Zhao XF. Identification of genes differentially expressed during larval molting and metamorphosis of Helicoverpa armigera. BMC. Dev Biol (2007) 7:73. doi: 10.1186/1471-213X-7-73
29. Hiromichi N. The crustacean cuticle: structure, composition and mineralization. Front Biosci (2012) 4:711–20. doi: 10.2741/412
30. Orosz F, Oláh J, Ovádi J. Triosephosphate isomerase deficiency: facts and doubts. IUBMB. Life (2006) 58:703–15. doi: 10.1080/15216540601115960
31. Orosz F, Oláh J, Ovádi J. Triosephosphate isomerase deficiency: New insights into an enigmatic disease. Biochim Biophys Acta (2009) 1792:1168–74. doi: 10.1016/j.bbadis.2009.09.012
32. Ramaekers VT, Häusler M, Opladen T, Heimann G, Blau N. Psychomotor retardation, spastic paraplegia, cerebellar ataxia and dyskinesia associated with low 5-methyltetrahydrofolate in cerebrospinal fluid: A novel neurometabolic condition responding to folinic acid substitution. Neuropediatrics (2002) 33:301–8. doi: 10.1055/s-2002-37082
33. Moretti P, Sahoo T, Hyland K, Bottiglieri T, Peters S, Gaudio D, et al. Cerebral folate deficiency with developmental delay, autism, and response to folinic acid. Neurology (2005) 64:1088–90. doi: 10.1212/01.WNL.0000154641.08211.B7
34. Li JJ, Sun JS, Dong XW, Geng XY, Qiu GF. Transcriptomic analysis of gills provides insights into the molecular basis of molting in Chinese mitten crab (Eriocheir sinensis). Peerj (2019) 7:e7182. doi: 10.7717/peerj.7182
35. Behr M, Hoch M. Identification of the novel evolutionary conserved obstructor multigene family in invertebrates. FEBS Lett (2006) 579:6827–33. doi: 10.1016/j.febslet.2005.11.021
36. Dittmer NT, Hiromasa Y, Tomich JM, Lu N, Kanost MR. Proteomic and transcriptomic analyses of rigid and membranous cuticles and epidermis from the elytra and hindwings of the red flour beetle, Tribolium castaneum. J Proteome Res (2012) 11:269. doi: 10.1021/pr2009803
37. Dong Z, Zhang W, Zhang Y, Zhang X, Zhao P, Xia Q. Identification and characterization of novel chitin-binding proteins from the larval cuticle of silkworm, Bombyx mori. J Proteome Res (2016) 15:1435–45. doi: 10.1021/acs.jproteome.5b00943
38. Hou Q, Chen E, Dou W, Wang J. Knockdown of specific cuticular proteins analogous to peritrophin 3 genes disrupt larval and ovarian development in Bactrocera dorsalis (Diptera: Tephritidae). Insect Sci (2021) 28:1326–37. doi: 10.1111/1744-7917.12869
39. Jasrapuria S, Specht CA, Kramer KJ, Beeman RW, Muthukrishnan S. Gene families of cuticular proteins analogous to peritrophins (CPAPs) in Tribolium castaneum have diverse functions. PloS One (2012) 7:e49844. doi: 10.1371/journal.pone.0049844
40. Sui G, Soohoo C, Affar EB, Gay F, Shi YJ, Forrester WC, et al. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci USA (2002) 99:5515–20. doi: 10.1073/pnas.082117599
41. Wang S, Luo Y, Wilson PD, Witman GB, Zhou J. The autosomal recessive polycystic kidney disease protein is localized to primary cilia, with concentration in the basal body area. J Am Soc Nephrol (2004) 15:592–602. doi: 10.1097/01.asn.0000113793.12558.1d
42. Hiesberger T, Bai Y, Shao X, McNally BT, Sinclair AM, Tian X, et al. Mutation of hepatocyte nuclear factor-1beta inhibits Pkhd1 gene expression and produces renal cysts in mice. J Clin Invest (2004) 113:814–25. doi: 10.1172/JCI20083
43. Masyuk TV, Huang BQ, Ward CJ, Masyuk AI, Yuan D, Splinter PL, et al. Defects in cholangiocyte fibrocystin expression and ciliary structure in the PCK rat. Gastroenterology (2003) 125:1303–10. doi: 10.1016/j.gastro.2003.09.001
44. Toninello A, Pietrangeli P, De Marchi U, Salvi M, Mondovì B. Amine oxidases in apoptosis and cancer. Biochim Biophys Acta (2006) 1765:1–13. doi: 10.1016/j.bbcan.2005.09.001
45. Gudkova OO, Latyshko NV, Shandrenko SG. Amine oxidases as important agents of pathological processes of rhabdomyolysis in rats. Ukr Biochem J (2016) 88:79–87. doi: 10.15407/ubj88.01.079
46. Murakami M, Nakatani Y, Atsumi GI, Inoue K, Kudo I. Regulatory functions of phospholipase A2. Crit Rev Immunol (2017) 37:127–95. doi: 10.1615/CritRevImmunol.v37.i2-6.20
47. Cohen I, Cohen C. A tropomyosin-like protein from human platelets. J Mol Biol (1972) 68:383–7. doi: 10.1016/0022-2836(72)90220-3
48. Lazarides E. Tropomyosin antibody: the specific localization of tropomyosin in nonmuscle cells. J Cell Biol (1975) 65:549–61. doi: 10.1083/jcb.65.3.549
Keywords: ecdysis triggering hormone, RNA interference, transcriptome, molt, downstream, Scylla paramamosain
Citation: Chan S-F, Wen Q-Q, Ao C-M, Wang W, Wang C-G and Zhao Y-F (2022) Transcriptome responses of RNAi-mediated ETH knockdown in Scylla paramamosain at different premolt substages. Front. Endocrinol. 13:917088. doi: 10.3389/fendo.2022.917088
Received: 10 April 2022; Accepted: 04 July 2022;
Published: 28 July 2022.
Edited by:
Alex Ford, University of Portsmouth, United KingdomReviewed by:
Yuichiro Suzuki, Wellesley College, United StatesMatthew Meiselman, Cornell University, United States
Dusan Zitnan, Slovak Academy of Sciences, Slovakia
Copyright © 2022 Chan, Wen, Ao, Wang, Wang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan-Fei Zhao, eWZlaS5nb29kQDE2My5jb20=
 Siuming-Francis Chan
Siuming-Francis Chan Qi-Qiao Wen1
Qi-Qiao Wen1 Chun-Mei Ao
Chun-Mei Ao Wei Wang
Wei Wang Cheng-Gui Wang
Cheng-Gui Wang Yan-Fei Zhao
Yan-Fei Zhao