
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol. , 06 June 2022
Sec. Clinical Diabetes
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.915031
This article is part of the Research Topic Advances in the Research of Diabetic Retinopathy View all 20 articles
Retinopathy is one of the most devastating complications of diabetes, which a patient fears the most. Hyperglycemic environment results in many structural, functional, molecular and biochemical abnormalities in the retina, and overproduction of mitochondrial superoxide, induced by hyperglycemic milieu, is considered to play a central role in the development of diabetic retinopathy. Expression of many genes associated with maintaining mitochondrial homeostasis is also altered. Recent research has shown that several long noncoding RNAs, RNAs with more than 200 nucleotides but without any reading frames, are aberrantly expressed in diabetes, and altered expression of these long noncoding RNAs is now being implicated in the development of diabetes and its complications including retinopathy. This review focuses the role of long noncoding RNAs in the development of diabetic retinopathy, with a special emphasis on the maintenance of mitochondrial homeostasis.
Incidence of diabetes is increasing at an alarming rate around the world; 151 million people had diabetes in 2000, this number is now 463 million adults, and is expected to pass 780 million by 2045 (1). Diabetes accounts for 6.7 million deaths in 2021-one death every 5 seconds, and this devastating disease does not know any social, economic or ethnic boundaries. Elevated blood glucose in the circulation, either due to impaired insulin production or resistance to properly use it, results in damaging small and large blood vessel in the body, resulting in many micro- and macro-vascular complications including nephropathy (kidney), neuropathy (nerve), retinopathy (retina, back of the eye) and cardiomyopathy (heart) (2). Over time, chronic high blood glucose affects almost every organ in the body, one in three adults with diabetes present chronic kidney disease, one in two peripheral nerve abnormalities and over 80% patients show some signs of retinopathy (3). Thus, diabetes is a whole-body chronic disease, and is commonly called as a ‘disease of its complications’.
Technological development in transcriptome-wide sequencing has now documented that although majority of the mammalian genome is actively transcribed into RNA, only 2-3% is further translated into protein (4). Thus, most of the DNA does not code for the proteins, and this ‘non-coding DNA’ has a heterogeneous group of noncoding RNAs including transfer RNA, circular RNA, micro RNA (miRNA, less than 20 base pairs) and long noncoding RNA (LncRNAs, more than 200 base pairs) (Figure 1). The exact number of non-coding RNAs in the human genome is still not clear, but recent transcriptomic/bioinformatic studies have put their numbers in the tens of thousands (5). These noncoding RNAs, however, are not totally unfunctional, they can be alternatively spliced and/or processed into smaller products, and may have a hidden layer of internal signals (especially those derived from introns) that could control transcription, chromatin architecture etc. Noncoding RNAs are implicated in numerous cellular processes including cell cycle and metabolism (6–8). Thus far more than 2000 miRNAs and 30,000 LncRNAs have been discovered in humans, and these noncoding RNAs are responsible for regulating one third of the genes in the genome (9, 10). Although both miRNAs and LncRNAs RNAs are transcribed by RNAs polymerase II, miRNAs are better conserved than LncRNAs, and while miRNAs regulate mRNA at transcriptional and post-transcriptional levels, LncRNAs can remodel chromatin and genomic imprinting and regulate gene expression through sequence complementarity with RNAs or DNAs (11, 12). LncRNAs have four-fold higher tissue specificity compared to miRNAs (13, 14). LncRNAs are mRNA-like transcripts, but they do not possess any stable open reading frames, and compared to mRNAs, expression levels of LncRNAs are typically lower (14). Furthermore, while mRNAs have to be translated into proteins for carrying out specific cellular functions, LncRNAs are the functional unit and can function in different subcellular compartments based on local molecular interactions, and stronger tissue-specificity (15).
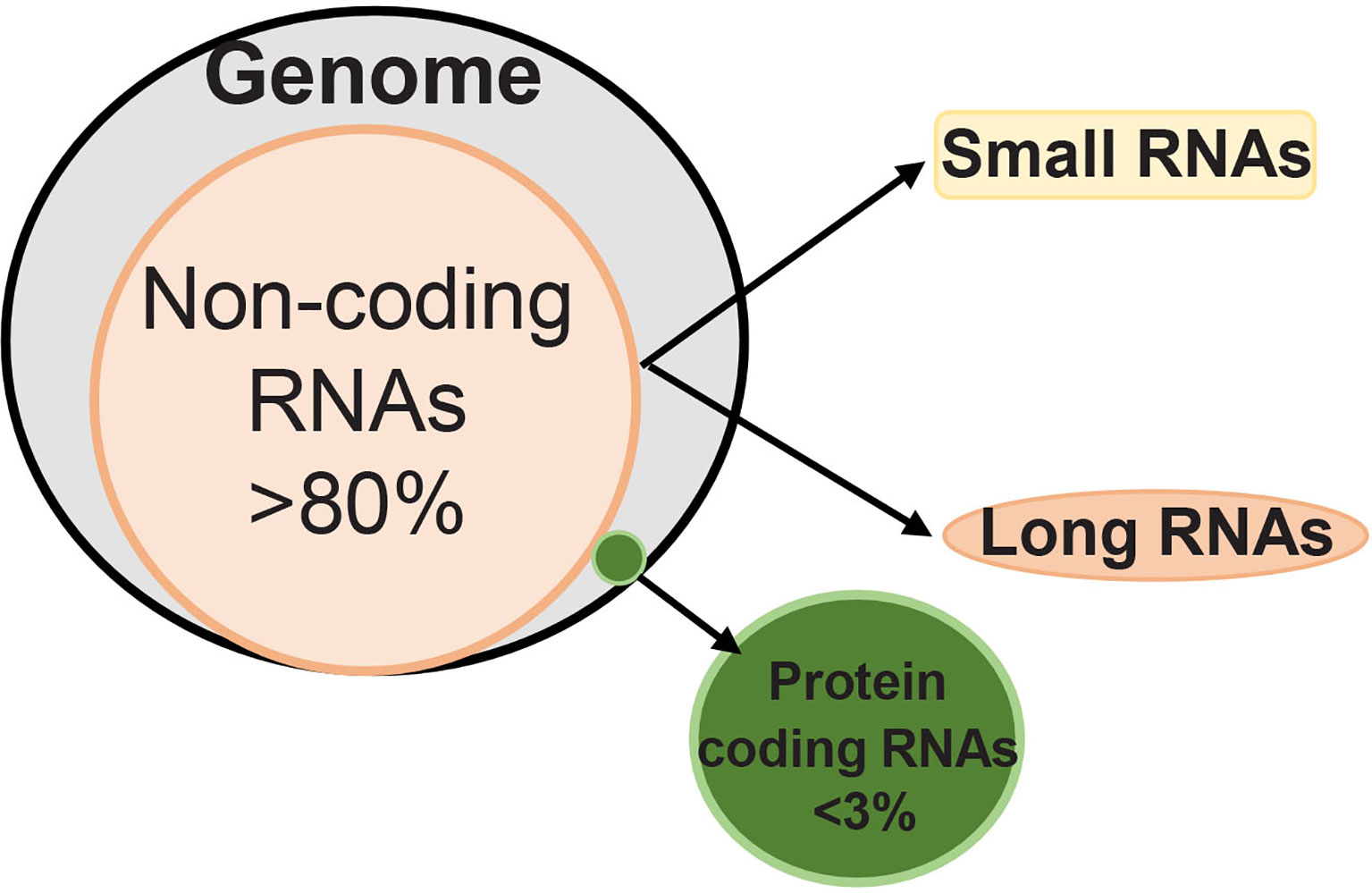
Figure 1 Human genome has less than 3% RNAs that can code for proteins, and over 80% of RNAs are noncoding. Depending on the length of their transcripts, noncoding RNAs are classified as ‘small’ and ‘long’ noncoding RNAs. While small noncoding RNAs have less than 200 nucleotides, long noncoding RNAs have over 200 nucleotides.
Based on their genomic location, LncRNAs are classified into various subclasses, ‘intergenic LncRNAs’ located between two protein coding-genes without intersecting with any protein-coding genes, ‘intronic LncRNAs’ transcribed from intronic region of a protein coding-gene and overlap with protein-coding genes, ‘antisense LncRNAs’ transcribed from complementary strand of either protein coding or non-protein coding genes, ‘bidirectional LncRNA’ initiating in a divergent fashion from a promoter of protein-coding gene, and ‘enhancer LncRNA’ synthesized at the enhancers (Figure 2). LncRNAs act as a guide via interacting with the modifying complexes or transcription factors, directing them to specific genes or loci, or they may function as a scaffold by binding its multiple effector partners at the same time with proteins. LncRNAs also act as ‘decoy’ by binding to the transcription factors or miRNAs, repressing gene expression, or as an ‘enhancer’ by changing chromatin architecture to recruit transcription factors and promote transcription (16). They bind to DNA or RNA in a sequence-specific manner, or can act as sponges for miRNAs or as scaffolds to provide stability, and facilitate transcription factor binding (7, 17, 18). Thus, LncRNAs are capable of interacting with RNA and DNA.
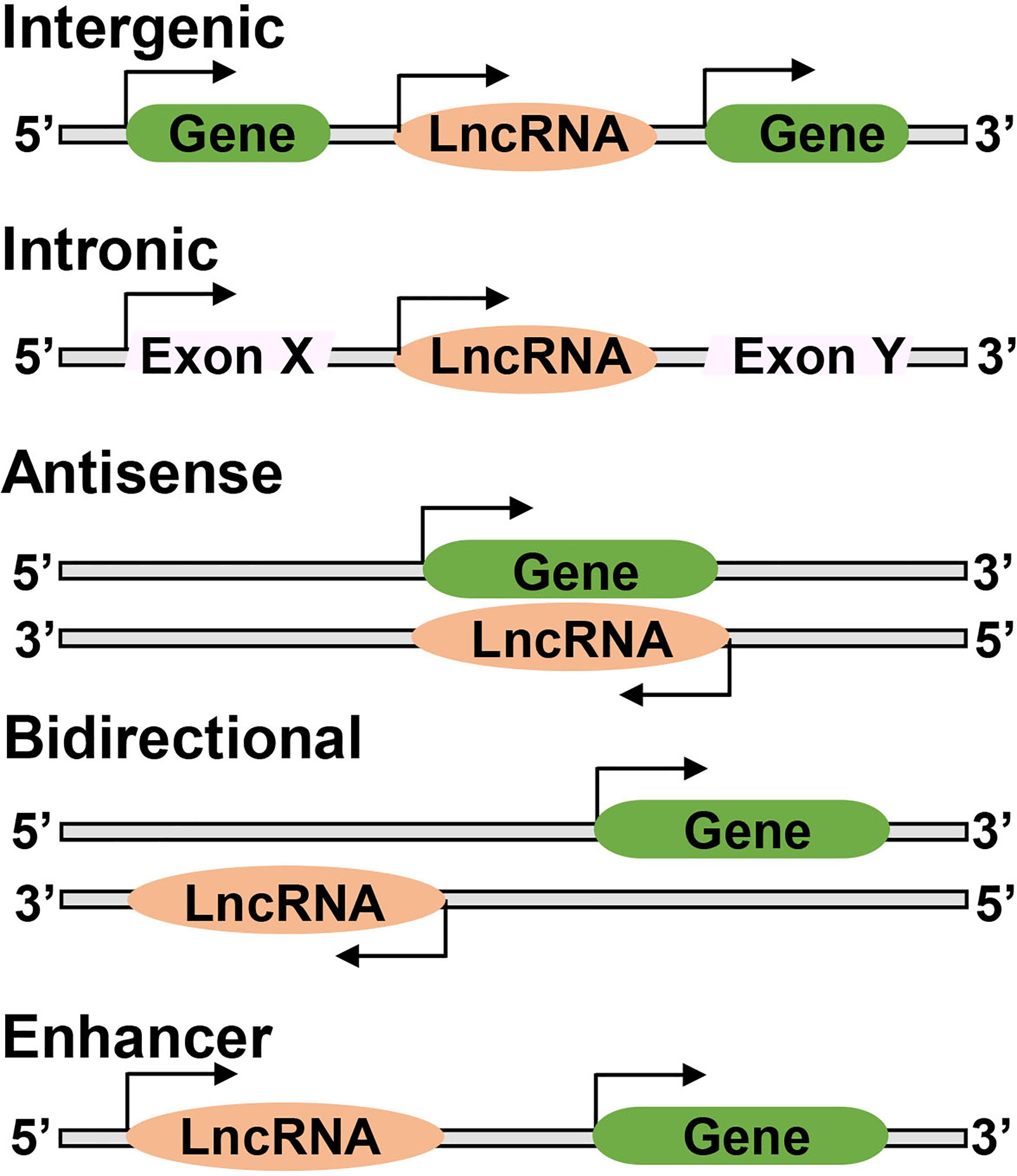
Figure 2 Long noncoding RNAs may have different genomic locations; between two protein coding genes ‘Intergenic’, or located within an intron of a coding gene ‘Intronic’, or overlap with one or more introns and exons of a different transcript on the same or opposite strand, respectively ‘Antisense’, or be within 100 kb of a coding transcript of the opposite strand ‘Bidirectional’, or present in the enhancer regions close to the promoter ‘Enhancer’.
LncRNAs are mainly encoded by nuclear genome and are overall more numerous in the nucleus, a significant number is also present in the cytoplasm, where they are more stable than their nuclear counterparts (19–21). LncRNAs modulate chromatin interactions in the nucleus, and generally help in signal transduction and posttranscriptional control of gene expression in the cytoplasm (22). In addition to nuclear genome, ~15% of the human mitochondrial transcriptome is made of noncoding RNAs, including three LncRNAs- LncRNA ND5, LncRNA ND6 and LncRNA CytB. Mitochondrial LncRNAs are located in the regions of the mtDNA complementary to the genes that encode for ND5, ND6 of complex I and cytochrome B of complex III respectively, and these mitochondrial genome encoded LncRNAs primarily form intermolecular duplexes with their respective complementary mRNAs (23, 24). However, these mitochondrial genome-encoded LncRNAs are not restricted to the mitochondria, instead, they can operate in the nucleus (24). Nuclear genome-encoded LncRNAs can also move from the nucleus to the mitochondria and the mitochondrial genome-encoded LncRNAs can be aberrantly transported to the nucleus, and this aberrant shuttling of crosstalk of LncRNAs is associated with abnormalities in energy metabolism (25). Accumulating research indicates that LncRNAs play important roles in diverse biological processes, and are identified as key regulators in pathological processes in a range of diseases including cancer, diabetes and Alzheimer (26–29), and their role in diabetic macro- and microvascular complications is an emerging area of research (30).
High circulating glucose leads to increased risk of many eye problems, including increase in intraocular pressure and damage to the lens and the retina. Damage to the retinal microvasculature, commonly known as diabetic retinopathy, is the leading cause of acquired blindness in the working age adults in developing countries. In the initial stages of this progressing disease, patients may be asymptomatic, but their retinal microvasculature begins to become fragile, and as the duration of diabetes increases, retinopathy also progresses and the damaged retinal vasculature begins to leak. With time, lack of oxygen in the microvasculature leads to neovascularization, and ultimately, to vision loss (31, 32). Early diagnosis and treatment options of diabetic retinopathy, however, are limited, and it is critical to identify effective methods of its diagnosis and therapeutic targets to retard progression of this sight-threatening disease. In the first five years of diabetes, retinopathy is rare in type 1 diabetic patients, but after 20 years of diabetes, over 80% of patients develop retinopathy. In type 2 diabetic patients, 21% of patients present early signs of retinopathy at the time of their diagnosis of diabetes, and as with type 1 diabetic patients, most type 2 diabetic patients also develop some degree of retinopathy over time. In 2020, over 100 million diabetic adults had retinopathy, and among those, 28.5 million had vision-threatening retinopathy, and with increase in the incidence of diabetes, these numbers are expected to increase to over 160 million and 45 million, respectively in 2045 (33). Although pathological evidence is mainly seen in the retinal microvasculature, research has shown that nonvascular cells of the retina are also affected; they experience functional and structural alterations including retinal ganglion cell apoptosis, loss of contrast sensitivity and alterations in the electroretinogram (34, 35). Normal cell-cell interactions are also affected leading to the blood-retinal barrier breakdown and neuronal cell dysfunction (6, 36). In addition, Optical Coherence Tomography has also shown ganglion cell loss in the retina of diabetic patients with or without retinopathy, however, diabetic patients with moderate or severe retinopathy show more thinning of their ganglion cell-inner plexiform layers compared to patients without any retinopathy (37).
Diabetic retinopathy is a multifactorial disease with a complex etiology, and increase in oxidative stress-mitochondrial damage is considered to play a major role in the development of this blinding disease (2, 38–42). Retina is rich in polyunsaturated fatty acids and has a high oxygen uptake (43), making it a good target of increased oxidative stress in diabetes. In diabetes, increased cytosolic ROS, generated by activation of Ras-related C3 botulinum toxin substrate 1 (Rac1)-NADPH oxidase 2 (Nox2)-activation of gelatinase matrix metalloproteinases in diabetes damage mitochondria in retinal capillaries (44, 45). In addition, due to high circulating glucose, electron flux through the electron transport chain system is increased, and the complex III becomes dysfunctional, resulting in increased accumulation of ROS (40, 46). Mitochondrial structural and functional stability is impaired including increased production of superoxide radicals and damage to their membrane potential. Cytochrome C leaks out from the mitochondria in the cytosol, which accelerates capillary cell apoptosis, a phenomenon which is followed by the histopathology characteristic of retinopathy (38, 41, 47, 48). Mitochondria have their own DNA (mtDNA), which encodes genes for 13 proteins that are essential for the electron transport chain functioning; diabetes damages mtDNA and reduces the gene transcripts of mitochondrial genome-encoded ND1 and ND6 of complex I, and cytochrome B of complex III are reduced. In addition, mitochondrial dynamics is imbalanced, while fusion is decreased, fission is increased, and the removal of damaged mitochondria in impaired. Impairment in mitochondrial homeostasis in diabetes is further exacerbated by the suboptimal biogenesis of mtDNA (47, 49–52). Thus, instability in mitochondrial structure, function and genome plays a central role in the development of diabetic retinopathy.
Aldose reductase, an enzyme with a high ‘Km’ for glucose, using NADPH as a cofactor, converts glucose to sorbitol. This reduces availability of NADPH for glutathione biosynthesis, increasing oxidative stress. In diabetes, polyol pathway is activated in the retina and its capillary cells (53, 54). Diabetes increases non-enzymatic glycation of amino acids in proteins, lipids and nucleic acids, and this ultimately leads to advanced glycation end products (AGEs) (55); retinal capillaries have increased AGEs and their receptors (RAGEs), and AGEs-RAGEs are implicated in increased inflammation and capillary cell loss in diabetic retinopathy (56). Diacyl glycerol-protein kinase C cascade is also activated by high glucose, and protein kinase C activation in retinal vasculature is associated with increased vascular permeability, alterations in blood flow and stimulation of neovascularization seen in diabetic retinopathy (57). Furthermore, activation of protein kinase C, polyol pathway and increased AGEs accumulation can also increase oxidative stress, and can also be initiated by hyperglycemia-induced overproduction of mitochondrial superoxide, further strengthening the role of mitochondrial damage in diabetic retinopathy (40, 58). In addition, many inflammatory markers including cytokines and leukostasis, are also elevated in diabetic retinopathy (59), and retinal glial cells, the cells that are between vasculature and neurons of the retina, also initiate the inflammatory cascade (60).
Hyperglycemia is the main instigator, but systemic factors, such as high blood pressure, cholesterol and smoking are also closely associated with the development and progression of diabetic retinopathy (61), and also in the exacerbated and accelerated mitochondrial damage (62). However, multifactorial nature and complex etiology of diabetic retinopathy makes it difficult to identify a link between any specific metabolic abnormality and the development of this progressive blinding disease.
Genetic factors appear to play some role in diabetic retinopathy, but the exact role of genetic contribution in the development/progression of diabetic retinopathy has remained unclear. Significant association between the variations in the polyol pathway gene encoding aldo-keto reductase family 1-member B1, AKR1B1gene, has been documented by a meta-analysis (63). Type 2 diabetic patients in Pakistan with Pro12Ala polymorphism are at a reduced risk of developing diabetic retinopathy (64). Jackson Heart Study has shown an association between P-selectin and diabetic retinopathy (65). Another genome-wide association study (GWAS) has shown association with a zinc finger protein associated with transcriptional regulation, ZNF600. Polymorphisms at the regulatory regions of some of the genes including VEGF and endothelial nitric oxide synthase and paraoxonase1, have been considered as risk alleles for the susceptibility or progression of diabetic retinopathy (66). Genotyping of single nucleotide polymorphism has identified new loci on chromosome 6 associated with diabetic retinopathy, but none of the variants have shown any statistical significance (67). A recent GWAS meta-analyses of type 2 diabetic patients have shown enrichment of variants in the genes involved lipid catabolism and transport, oxidative stress and cell degeneration pathways and the risk of diabetic retinopathy (68). In addition, an association for the z-2 microsatellite and rs759853 on aldo-keto reductase family 1 member B, and some evidence for polymorphisms in VEGF, intercellular adhesion molecule 1 genes have also been documented by systemic meta-analyses, but a follow up of the analysis has not reported any loci after adjustment for multiple (69). A study of cohort from China with proliferative diabetic retinopathy has identified INSR gene as a possible susceptibility candidate for severe diabetic retinopathy, but this study is underpowered (70). Siblings of affected individuals has been shown to have three-fold higher risk of severe diabetic retinopathy, but diabetic patients with similar risk factors do not show the same range in the severity of retinopathy, when evaluated by Early Treatment Diabetic Retinopathy Study Grading System (71). Thus, despite recent technological advancement in the genetic field, multifactorial pathogenesis of this blinding disease has made it difficult to identify definite genetic associations. In addition, variations in the severity of hyperglycemia and other systemic factors such as hyperlipidemia and blood pressure in the diabetic population have further compounded the problem.
Gene function is expressed in the form of protein, and changes in the DNA sequence can alter how it is transcribed into RNA, which eventually is coded for a particular protein. The expression of a gene is also affected by external factors, such as environment and lifestyle, and these ‘epigenetic’ changes can turn a gene ‘on’ and ‘off’ without altering the DNA sequence. Although epigenetic processes are essential to many organism functions, their up- or down-regulation can have major adverse health and behavioral effects; they are now implicated in many diseases including cancer, cardiovascular diseases and diabetes and its complications. Some of the major epigenetic modifications include methylation of cytosine (DNA methylation), methylation and acetylation of histones and noncoding RNAs (72). Methylation of the 5′ position of the cytosine forms 5-methyl cytosine (5mC), and this process is facilitated by DNA methyl transferases (Dnmts) and condenses the chromatin, both genomic DNA and mitochondrial DNA undergo methylation. However, dioxygenases-ten-eleven translocation (Tets) can rapidly convert 5mC to hydroxymethylated to 5-hydroxymethyl cytosine (5hmC), resulting in transcriptional activation (73–75). Lysine in the histone proteins can be acetylated, relaxing the chromatin structure to allow transcription factors binding, and this is modulated by a balance between acetyl transferring and removing enzymes, histone acetyltransferases and histone deacetylases, respectively (76–78). Lysine or arginine residues in a histone can also be modified by the addition of one, two, or three methyl groups, and methylation status of histones is also maintained by opposing enzymes, addition of methyl group by histone methyltransferases and removal of methyl group by demethylases. Unlike histone acetylation, gene repression or activation is determined by the site of methylation and the number of methyl groups (79, 80). Moreover, epigenetics modifications have potential to modify the gene expression independently, or one modification can lead to the other, to ultimately regulate gene expression (75, 81). Noncoding RNAs may not be the main epigenetic components, but they can also regulate gene expression without altering the DNA sequence.
The role of epigenetic modifications in many chronic diseases, such as cancer and Alzheimer disease, is being investigated for several years, and their role in diabetes and its complications is now emerging (82–85). In diabetes, Dnmts and Tets are activated in the retina and its vasculature, and dynamic DNA methylation-hydroxymethylation at the promoters of Rac1 and matrix metalloproteinase 9 is associated with their activation and mitochondrial damage. Furthermore, DNA at the promoter of polymerase gamma, a gene associated with mtDNA biogenesis, and at MutLH1 (associated with mtDNA damage repair) is hypermethylated, further damaging the mitochondria. Mitochondrial DNA itself is also hypermethylated in diabetes, which impairs its transcription resulting in a compromised electron transport chain system (41, 86, 87). Thus, epigenetics plays a vital role in maintaining mitochondrial homeostasis in diabetic retinopathy.
LncRNAs are now considered as a new class of epigenetic regulators because they can regulate epigenetic modification by modulating histone or DNA modifications, and can also act as a scaffold that interacts with epigenetic enzymes or methylation and acetylation complexes. LncRNAs can regulate histone methylation and acetylation simultaneously, subsequently regulating the gene expression (88). Thus, LncRNAs can epigenetically modify the expression of a gene without altering its DNA sequence.
Leading scientists in the field have reviewed how LncRNAs play a role in the development of diabetic retinopathy, focusing mainly on inflammation and angiogenesis, but the field is still in its infancy. Many laboratories are now focusing on identifying LncRNAs associated with various metabolic and functional abnormalities implicated in the development of diabetic retinopathy (89–94). LncRNA profiling performed in the retina of streptozotocin-induced diabetic mice has shown over 300 LncRNAs that are aberrantly expressed at two months of diabetes, with 214 showing downregulation and 89 upregulation (95), and profiling in the fibrovascular membranes of patients with diabetic retinopathy have identified 427 differentially expressed LncRNAs, 263 upregulated and 164 downregulated (96). Both of these profiling studies have identified differentially expressed LncRNAs that are enriched in inflammatory and angiogenic pathways including Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), Hypoxia-inducible factor 1 HIF-1 and tumor necrosis factor alpha.
In diabetic retinopathy, LncMALAT1, which is one of the most highly expressed and conserved LncRNAs, is upregulated in the in vivo and in vitro models, and also in the circulation of the patients with diabetic retinopathy (97). Aqueous humor and fibrovascular membranes of diabetic patients also have increased LncMALAT1 (95). Inhibition of LncMALAT1 protects retinal photoreceptors, and alleviate diabetic neurodegeneration (98). Although LncMALAT1 is implicated in normal physiologic functions such as vascular growth (99), its elevation in diabetes is shown to act both as pro-inflammatory and apoptotic (100–102). MALAT1 sequence has NF-kB transcription factor binding sites, and NF-kB is intimately associated with the induction of proinflammatory cytokines; in diabetic retinopathy, NF-kB is activated and many proinflammatory cytokines are upregulated (59, 103). Upregulation of cytokines is shown to play a major role in the development of diabetic retinopathy; in fact, diabetic retinopathy is also considered as a low grade chronic inflammatory disease. In experimental models of diabetic retinopathy, LncMALAT1 expression is augmented by hypoxia contributing to the proliferative response in endothelial cells (104). In retinal endothelial cells LncMALAT1 inhibition ameliorates cell migration-angiogenesis, neurodegeneration and monocyte chemotactic protein-1, and also prevents inactivation of the master transcription factor, nuclear factor erythroid 2-related factor, Nrf2 (91, 98, 105). Our recent studies have documented an antioxidant defense role of LncMALAT1 in diabetic retinopathy; via affecting the interactions of nuclear factor erythroid 2-related factor 2 (Nrf2) and its intracellular inhibitor Kelch-like ECH-associated protein 1, LncMALAT1 upregulation suppresses transcriptional activity of Nrf2, and this impairs the transcription of antioxidant defense genes, such as heme oxygenase 1 and superoxide dismutase (91). Inhibition of LncRNA MALAT1 alleviates diabetes-induced neurodegeneration (98). LncRNA, antisense RNA to INK4 locus (LncANRIL) with binding sites for NF-kB, is also upregulated in diabetes, and is implicated tube formation-proliferation in endothelial cells via regulation of VEGF expression (106). Another LncRNA Myocardial infarction associated transcript (LncMIAT), is also regulated by NF-kB; high glucose increases the NF-kB and MIAT binding, further regulating inflammatory cytokines and apoptosis (107). LncRNA maternally expressed gene 3 (LncMEG3), which increases endoplasmic reticulum stress and inhibits cell proliferation (108), is considered to negatively correlate with VEGF, and in patients with diabetic retinopathy, while the serum levels of VEGF are upregulated, LncMEG3 are downregulated (109). Another LncRNA HOX antisense intergenic RNA (LncHOTAIR) is upregulated in the vitreous of diabetic retinopathy patients and in retinal endothelial cells by high glucose, and its inhibition is shown to prevent increase in retinal vascular permeability and VEGF in diabetic rodents (110). Thus, the role of LncRNAs in inflammation, oxidative stress and angiogenesis is now at the forefront of ongoing research.
Mitochondrial dysfunction plays a critical role in many diabetic complications, and as mentioned above, in diabetic retinopathy damaged mitochondria accelerate apoptosis of capillary cells, which precedes the development of histopathology (48, 111). Retinal mitochondrial homeostasis is impaired, they become swollen and their damaged membranes leak cytochrome C in the cytosol, superoxide levels are elevated, mitochondrial dynamics are disturbed, mtDNA is damaged and mitochondrial genome-encoded genes including cytochrome B is downregulated, and the complex III of the electron transport chain is inhibited (41, 47). LncRNAs, whether encoded by mitochondria or nuclear genome, are also implicated in mitochondrial homeostasis, nuclear genome encoded LncMALAT1 is translocated in the mitochondria of hepatoma cells, where it is shown to regulate mitochondrial apoptosis and mitophagy (25), and LncMEG3, via mitochondrial pathway, is shown to induce renal cell carcinoma apoptosis (112). Mice with MALAT1 knocked out have lower levels of ROS and protein carbonylation in hepatocyte and islets (113). Furthermore, LncMALAT1 is also shown to interact with multiple loci on the mtDNA in hepatoma cells (114). In addition, LncHOTAIR induces mitochondria-mediated apoptosis in head and neck squamous cell carcinoma via Bcl-2-BAX-Caspase-3 pathway (115), and cardiac apoptosis-related LncRNA (LncCARL) suppresses mitochondrial fission (116). In diabetic nephropathy, nuclear genome encoded LncRNA taurine-upregulated gene 1 is shown to modulate mitochondrial biogenesis by directly interacting with PGC1α (117), and depletion of Nuclear Enriched Abundant Transcript 1 (LncNEAT1) is shown to alter mitochondrial dynamics (118). This clearly shows that many nuclear genome-encoded LncRNAs have potential to regulate mitochondrial function.
Focusing on diabetic retinopathy, our recent work has suggested a role of nuclear genome encoded LncMALAT1 and LncNEAT1 in regulation of mitochondrial functions, we have demonstrated that high glucose increases their mitochondrial localization in retinal endothelial cells, and siRNA of LncMALAT1 or LncNEAT1 prevents increase in ROS and alterations in mitochondrial membrane potential. As mentioned earlier, diabetes damages retinal mtDNA and impairs the transcription of mitochondrial genome-encoded genes, important for the functioning of the electron transport chain, which results in a vicious cycle of free radicals (41, 42, 119). LncMALAT1 is shown to interact with multiple loci on mtDNA in hepatoma cells (114), and regulation of LncMALAT1 or LncNEAT1 by their respective siRNAs in retinal endothelial cells, prevents glucose-induced mtDNA damage and decrease in its transcription (93). Although mtDNA does not contain histones, it has nucleoprotein complexes, the nucleoids, and these nucleoids provide a stable environment for replication and repair of mtDNA (120). In addition, nucleoids also help in controlling mitochondrial metabolism in response to cellular demands (121, 122). Regulation of LncMALAT1 or LncNEAT1 also protects glucose-induced decrease in mtDNA nucleoids in retinal endothelial cells, suggesting a close cross-talk between nuclear genome-encoded LncRNAs and mitochondrial stability via their role(s) in increasing the vulnerability of the histone-free mtDNA in hyperglycemic milieu (Figure 3).
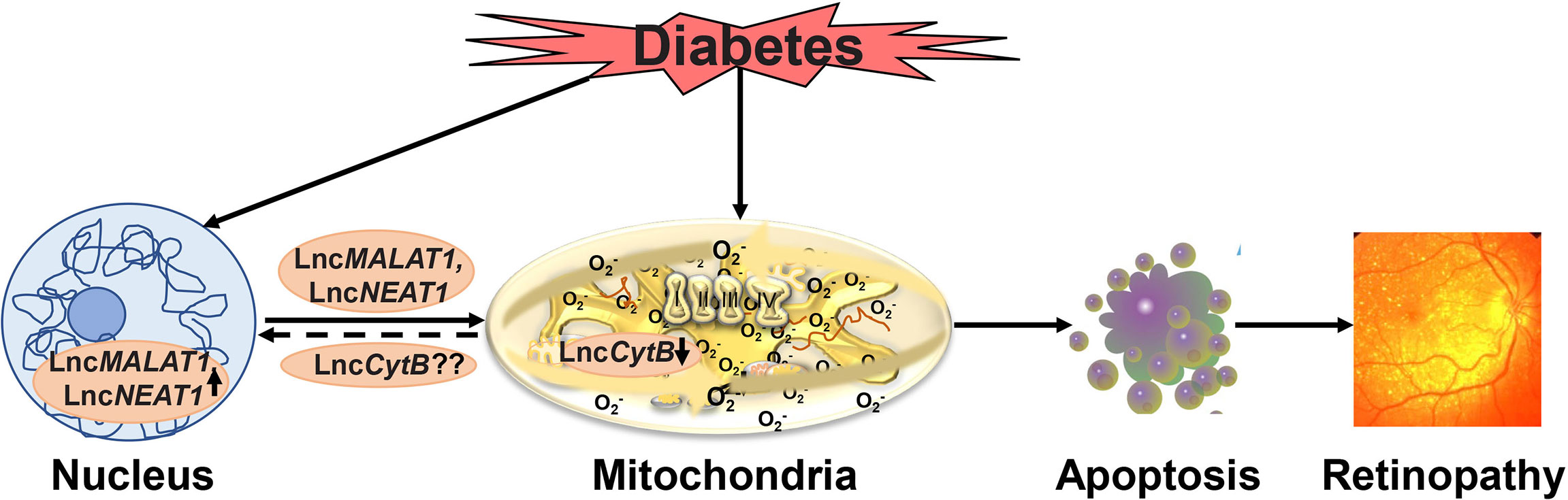
Figure 3 Hyperglycemic environment damages mitochondria and also elevates levels of various nuclear genome encoded LncRNAs including LncMALAT1 and LncNEAT1. These LncRNAs, in addition to affecting metabolic abnormalities associated with diabetic retinopathy including inflammation, also move inside the mitochondria, and damage mitochondrial structural, functional and genomic stability. Furthermore, possible decrease in the mitochondrial genome encoded LncCytB further damages the mitochondria, and the damaged mitochondria accelerates apoptosis, ultimately, resulting in the development of diabetic retinopathy.
As reported above, mtDNA itself also encodes three LncRNAs, two of them transcribed from its L strand, and one from the H strand, the regions of the mitochondrial genome that are complementary to the genes that encode ND5, cytochrome B and ND6 mRNAs (23, 24). LncND5, LncND6 and LncCytB are 58, 34 and 14% as abundant as their complementary coding ND5, ND6, and cytochrome B mRNAs, and these LncRNAs primarily form intermolecular duplexes with their respective complementary mRNAs, suggesting their functional role in either stabilizing or regulating their partner mRNAs (23, 24). Their role in metabolism/diseases, however, is still unclear. Our recent work (unpublished) has demonstrated decreased expression of LnCytB in retinal endothelial cells in high glucose conditions, and shown its role in protecting mtDNA vulnerability to nuclease damage and cytochrome B expression; overexpression of LnCytB ameliorates glucose-induced increase in mtDNA damage, decrease in mitochondrial genome-encoded nucleoids and decrease in cytochrome B expression. These studies have suggested that downregulation of LncCytB in the mitochondria in hyperglycemic milieu reduces the protective nucleoids, which makes mtDNA more susceptible to the damage. The damaged mtDNA compromises the electron transport chain and the vicious cycle of free radicals continues to self-propagate, thus implying an important role of this mitochondrial genome-encoded LncRNA in mitochondrial homeostasis in diabetic retinopathy. Although our initial results show significant decrease in LncCytB, we do not see any significant decrease in the other two mitochondrial genome-encoded LncRNAs-LncND5 or LncND6 in either in vitro or in vivo models of diabetic retinopathy. Despite significantly lower abundance of LncCytB is significantly lower compared to the other two mitochondrial genome-encoded LncRNAs, complex III is considered to play a major role in superoxide generation in diabetic retinopathy, and the activity of complex III, but not complex I, is downregulated (2, 38, 40)
As described above, nuclear genome-encoded LncRNAs can move inside the mitochondria and regulate mitochondrial homeostasis, and aberrant transport of mitochondrial genome-encoded LncCytB in the nucleus of hepatoma HepG2 cells as compared with normal hepatic HL7702 cells suggests that the aberrant shuttling of LncRNAs in mitochondria-nucleus crosstalk could be important for abnormal energy metabolism in malignant cells (25). Weather mitochondrial genome-encoded LncRNAs also translocate outside the mitochondria in diabetic retinopathy needs investigation.
The focus of this review thus far has been on how LncRNAs can directly affect mitochondrial homeostasis in diabetic retinopathy, but many epigenetic modifications including histone modifications and methylation of DNA of many genes associated with mitochondrial metabolism and mitochondrial DNA itself, also affect mitochondrial stability (47, 87). Long noncoding RNAs are considered to play a significant role in regulating histone modifications and DNA methylation, for example, LncHOTAIR is shown to interact with the histone modifying polycomb repressive complex II and histone demethylase 1 (LSD1) (18). In diabetic retinopathy, LSD1 is activated, and its recruitment at the promoter of mitochondrial damaging matrix metalloproteinase-9 is increased. Silencing of LSD1 prevents hyperglycemia-induced damage to mitochondrial structure and function in retinal endothelial cells, suggesting an important role of this enzyme in mitochondrial homeostasis in diabetic retinopathy (123). Furthermore, in hyperglycemic milieu, MALAT1 silencing directly reduces Enhancer of Zeste homolog 2 (Ezh2), an enzyme which catalyzes dimethylation/trimethylation of histone 3 lysine 27 (90), and Ezh2 activation in diabetes helps recruit DNA methylation enzymes at the promoter of matrix metalloproteinase-9, activating its transcription (124). Also, role of LncRNAs in regulating methylation of mtDNA or DNA at the promoters of other genes important genes associated with mitochondrial homeostasis needs further investigation. LncRNAs can also act as microRNA sponges, and in retinal pigment epithelium cells exposed to high glucose, upregulation of miR-34a is reversed by upregulation of MEG3 and Sirtuin 1 (125); overexpression of Sirtuin1 prevents mitochondrial damage and the development of retinopathy in diabetic mice (126). LncMALAT1, via targeting mi125b, inhibits glucose-induced angiogenesis in retinal endothelial cells (127). As the list of miRNAs implicated in the development of diabetic retinopathy grows, their interactions with LncRNAs in diabetic retinopathy is an open area that would need further attention.
Recent technical advances have allowed the LncRNA field to make great progress, the list of LncRNAs and their potential association with diseases is expanding rapidly, and their relatively stable nature in the body fluids, ease of obtaining body fluids and detecting them using commonly used molecular biology techniques makes them attractive biomarker candidates (128, 129). The results of a meta-analysis using data from over 1400 patients have shown that LncRNAs can serve as promising diagnostic biomarkers (130). For example, higher levels of LncRNA LIPCAR are associated with a higher risk of cardiovascular mortality (131), and LncRNA NONHSAT112178 can be a possible predictive biomarkers for coronary artery disease (132), LncHOTAIR is highly expressed in saliva samples of oral squamous cells from carcinoma patients, making it a strong candidate for metastatic oral cancer diagnosis (133), and LncMALAT1 is elevated in plasma and urine of prostate cancer patients (134). In diabetic retinopathy, LncHOTAIR and LncMALAT1 have shown some discrimination between patients with nonproliferative and proliferative retinopathy (135) and plasma and aqueous humor levels of LncRNA small nucleolar RNA host gene 5 are negatively correlated with the development of diabetic macular edema (136). A recent study has shown a strong correlation between the differential expressions of nine LncRNAs including LncANRIL, LncHOTAIR and LncMIAT, in the vitreous and serum samples of patients with or without diabetic retinopathy; these promising results suggest the possible use of serum LncRNAs as a potential biomarker of diabetic retinopathy, however, their validation in a larger cohort is needed (137). Thus, many LncRNA are now being implicated in regulating various pathways associated with the development of diabetic retinopathy, and future research and technical advancements may show us their role as biomarker or therapeutic target for its development, and/or its progression.
Like any new technology, LncRNA field also faces some challenges- the transcriptome studies suggest that ~80% of the genome is transcribed into RNA (138), but it is not clear how many of those LncRNAs are functional and what concentration of that LncRNA is required to achieve their biological effect. Also, LncRNAs can affect gene expression via many different mechanisms, it is difficult to predict how many different mechanisms one single LncRNA might use to control expression of a gene in a given disease. Fortunately, technical advances in genome editing and in surveying LncRNA molecular mechanisms have significantly advanced our ability to definitively characterize function of LncRNAs, making these molecules attractive targets for therapeutic intervention for diabetic retinopathy. LncRNAs in the mitochondria are also being investigated as potential blood-based biomarkers for cardiac remodeling in patients with hypertrophic cardiomyopathy (139), and with additional research in the field and technical advancements, their use in maintaining mitochondrial homeostasis in diabetic retinopathy is very optimistic, which will give diabetic patients hope of not losing their vision.
The author confirms being the sole contributor of this work and has approved it for publication.
This article based on the work supported in part by grants from the National Institutes of Health (EY014370, EY017313, and EY022230) and from The Thomas Foundation to RK, and an unrestricted grant from Research to Prevent Blindness from the Department of Ophthalmology, Wayne State University.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The author thanks her laboratory associates (past and present) for their contributions to the work cited in this review article.
1. Sun H, Saeedi P, Karuranga S, Pinkepank M, Ogurtsova K, Duncan BB, et al. IDF Diabetes Atlas: Global, Regional and Country-Level Diabetes Prevalence Estimates for 2021 and Projections for 2045. Diabetes Res Clin Pract (2022) 183:109119. doi: 10.1016/j.diabres.2021.109119
2. Brownlee M. The Pathobiology of Diabetic Complications: A Unifying Mechanism. Diabetes (2005) 54:1615–25. doi: 10.2337/diabetes.54.6.1615
3. Deshpande AD, Harris-Hayes M, Schootman M. Epidemiology of Diabetes and Diabetes-Related Complications. Phys Ther (2008) 88:1254–64. doi: 10.2522/ptj.20080020
4. Bär C, Chatterjee S, Thum T. Long Noncoding RNAs in Cardiovascular Pathology, Diagnosis, and Therapy. Circulation (2016) 134:1484–99. doi: 10.1161/circulationaha.116.023686
5. Ma L, Cao J, Liu L, Du Q, Li Z, Zou D, et al. LncBook: A Curated Knowledgebase of Human Long Non-Coding RNAs. Nucleic Acids Res (2019) 47:D128–d134. doi: 10.1093/nar/gky960
6. Pescosolido N, Barbato A, Stefanucci A, Buomprisco G. Role of Electrophysiology in the Early Diagnosis and Follow-Up of Diabetic Retinopathy. J Diabetes Res (2015) 2015:319692. doi: 10.1155/2015/319692
7. Beermann J, Piccoli MT, Viereck J, Thum T. Non-Coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev (2016) 96:1297–325. doi: 10.1152/physrev.00041.2015
8. Chan JJ, Tay Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int J Mol Sci (2018) 19:1310. doi: 10.3390/ijms19051310
9. Hammond SM. An Overview of microRNAs. Adv Drug Deliv Rev (2015) 87:3–14. doi: 10.1016/j.addr.2015.05.001
10. Beck ZT, Xing Z, Tran EJ. LncRNAs: Bridging Environmental Sensing and Gene Expression. RNA Biol (2016) 13:1189–96. doi: 10.1080/15476286.2016.1240139
11. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA Translation and Stability by microRNAs. Annu Rev Biochem (2010) 79:351–79. doi: 10.1146/annurev-biochem-060308-103103
12. Rafiee A, Riazi-Rad F, Havaskary M, Nuri F. Long Noncoding RNAs: Regulation, Function and Cancer. Biotechnol Genet Eng Rev (2018) 34:153–80. doi: 10.1080/02648725.2018.1471566
13. Washietl S, Pedersen JS, Korbel JO, Stocsits C, Gruber AR, Hackermüller J, et al. Structured RNAs in the ENCODE Selected Regions of the Human Genome. Genome Res (2007) 17:852–64. doi: 10.1101/gr.5650707
14. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative Annotation of Human Large Intergenic Noncoding RNAs Reveals Global Properties and Specific Subclasses. Genes Dev (2011) 25:1915–27. doi: 10.1101/gad.17446611
15. Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, et al. Localization and Abundance Analysis of Human lncRNAs at Single-Cell and Single-Molecule Resolution. Genome Biol (2015) 16:20. doi: 10.1186/s13059-015-0586-4
16. Bhat SA, Ahmad SM, Mumtaz PT, Malik AA, Dar MA, Urwat U, et al. Long Non-Coding RNAs: Mechanism of Action and Functional Utility. Noncoding RNA Res (2016) 1:43–50. doi: 10.1016/j.ncrna.2016.11.002
17. Fang Y, Fullwood MJ. Roles, Functions, and Mechanisms of Long Non-Coding RNAs in Cancer. Genomics Proteomics Bioinf (2016) 14:42–54. doi: 10.1016/j.gpb.2015.09.006
18. Schmitz SU, Grote P, Herrmann BG. Mechanisms of Long Noncoding RNA Function in Development and Disease. Cell Mol Life Sci (2016) 73:2491–509. doi: 10.1007/s00018-016-2174-5
19. Dietrich A, Wallet C, Iqbal RK, Gualberto JM, Lotfi F. Organellar Non-Coding RNAs: Emerging Regulation Mechanisms. Biochimie (2015) 117:48–62. doi: 10.1016/j.biochi.2015.06.027
20. Gordon AD, Biswas S, Feng B, Chakrabarti S. MALAT1: A Regulator of Inflammatory Cytokines in Diabetic Complications. Endocrinol Diabetes Metab (2018) 1:e00010–0. doi: 10.1002/edm2.10
21. Han L, Yang L. Multidimensional Mechanistic Spectrum of Long Non-Coding RNAs in Heart Development and Disease. Front Cardiovasc Med (2021) 8:728746. doi: 10.3389/fcvm.2021.728746
22. Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA Localization and Function. J Cell Biol (2021) 220:e202009045. doi: 10.1083/jcb.202009045
23. Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, et al. The Human Mitochondrial Transcriptome. Cell (2011) 146:645–58. doi: 10.1016/j.cell.2011.06.051
24. Dong Y, Yoshitomi T, Hu JF, Cui J. Long Noncoding RNAs Coordinate Functions Between Mitochondria and the Nucleus. Epigenet Chromatin (2017) 10:41. doi: 10.1186/s13072-017-0149-x
25. Zhao Y, Liu S, Zhou L, Li X, Meng Y, Li Y, et al. Aberrant Shuttling of Long Noncoding RNAs During the Mitochondria-Nuclear Crosstalk in Hepatocellular Carcinoma Cells. Am J Cancer Res (2019) 9:999–1008.
26. Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. 3rd; Kenny, P.J.; Wahlestedt, C. Expression of a Noncoding RNA Is Elevated in Alzheimer's Disease and Drives Rapid Feed-Forward Regulation of Beta-Secretase. Nat Med (2008) 14:723–30. doi: 10.1038/nm1784
27. Sun M, Kraus WL. From Discovery to Function: The Expanding Roles of Long Noncoding RNAs in Physiology and Disease. Endocr Rev (2015) 36:25–64. doi: 10.1210/er.2014-1034
28. Singer RA, Sussel L. Islet Long Noncoding RNAs: A Playbook for Discovery and Characterization. Diabetes (2018) 67:1461–70. doi: 10.2337/dbi18-0001
29. Simion V, Haemmig S, Feinberg MW. LncRNAs in Vascular Biology and Disease. Vascul Pharmacol (2019) 114:145–56. doi: 10.1016/j.vph.2018.01.003
30. Tanwar VS, Reddy MA, Natarajan R. Emerging Role of Long Non-Coding RNAs in Diabetic Vascular Complications. Front Endocrinol (Lausanne) (2021) 12:665811. doi: 10.3389/fendo.2021.665811
31. Aiello LP, Gardner TW, King GL, Blankenship G, Cavallerano JD, Ferris FL, et al. Diabetic Retinopathy. Diabetes Care (1998) 21:143–56. doi: 10.2337/diacare.21.1.143
32. Cheung N, Mitchell P, Wong TY. Diabetic Retinopathy. Lancet (2010) 376:124–36. doi: 10.1016/S0140-6736(09)62124-3
33. Teo ZL, Tham YC, Yu M, Chee ML, Rim TH, Cheung N, et al. Global Prevalence of Diabetic Retinopathy and Projection of Burden through 2045: Systematic Review and Meta-analysis. Ophthalmology (2021) 128:1580–91. doi: 10.1016/j.ophtha.2021.04.027
34. Aung MH, Kim MK, Olson DE, Thule PM, Pardue MT. Early Visual Deficits in Streptozotocin-Induced Diabetic Long Evans Rats. Invest Ophtahlmol Vis Sci (2013) 54:1370–7. doi: 10.1167/iovs.12-10927
35. Barber AJ, Baccouche B. Neurodegeneration in Diabetic Retinopathy: Potential for Novel Therapies. Vision Res (2017) 139:82–92. doi: 10.1016/j.visres.2017.06.014
36. Antonetti DA, Silva PS, Stitt AW. Current Understanding of the Molecular and Cellular Pathology of Diabetic Retinopathy. Nat Rev Endocrinol (2021) 17:195–206. doi: 10.1038/s41574-020-00451-4
37. Ng DS, Chiang PP, Tan G, Cheung CG, Cheng CY, Cheung CY, et al. Retinal Ganglion Cell Neuronal Damage in Diabetes and Diabetic Retinopathy. Clin Exp Ophthalmol (2016) 44:243–50. doi: 10.1111/ceo.12724
38. Kanwar M, Chan PS, Kern TS, Kowluru RA. Oxidative Damage in the Retinal Mitochondria of Diabetic Mice: Possible Protection by Superoxide Dismutase. Invest Ophtahlmol Vis Sci (2007) 48:3805–11. doi: 10.1167/iovs.06-1280
39. Kowluru RA. Diabetic Retinopathy, Oxidative Stress and Antioxidants. Curr Topics Nutraceutical Res (2005) 3:209–18.
40. Giacco F, Brownlee M. Oxidative Stress and Diabetic Complications. Circ Res (2010) 107:1058–70. doi: 10.1161/CIRCRESAHA.110.223545
41. Kowluru RA. Mitochondrial Stability in Diabetic Retinopathy: Lessons Learned From Epigenetics. Diabetes (2019) 68:241–7. doi: 10.2337/dbi18-0016
42. Kowluru RA, Mishra M. Oxidative Stress, Mitochondrial Damage and Diabetic Retinopathy. Biochim Biophys Acta (2015) 1852:2474–83. doi: 10.1016/j.bbadis.2015.08.001
43. Anderson RE, Rapp LM, Wiegand RD. Lipid Peroxidation and Retinal Degeneration. Curr Eye Res (1984) 3:223–7. doi: 10.3109/02713688408997203
44. Kowluru RA, Mishra M. Regulation of Matrix Metalloproteinase in the Pathogenesis of Diabetic Retinopathy. Prog Mol Biol Transl Sci (2017) 148:67–85. doi: 10.1016/bs.pmbts.2017.02.004
45. Sahajpal N, Kowluru A, Kowluru RA. The Regulatory Role of Rac1, a Small Molecular Weight GTPase, in the Development of Diabetic Retinopathy. J Clin Med (2019) 8:965. doi: 10.3390/jcm8070965
46. Madsen-Bouterse SA, Mohammad G, Kanwar M, Kowluru RA. Role of Mitochondrial DNA Damage in the Development of Diabetic Retinopathy, and the Metabolic Memory Phenomenon Associated With Its Progression. Antioxid Redox Signal (2010) 13:797–805. doi: 10.1089/ars.2009.2932
47. Kowluru RA, Kowluru A, Mishra M, Kumar B. Oxidative Stress and Epigenetic Modifications in the Pathogenesis of Diabetic Retinopathy. Prog Retin Eye Res (2015) 48:40–61. doi: 10.1016/j.preteyeres.2015.05.001
48. Mizutani M, Kern TS, Lorenzi M. Accelerated Death of Retinal Microvascular Cells in Human and Experimental Diabetic Retinopathy. J Clin Invest (1996) 97:2883–90. doi: 10.1172/JCI118746
49. Tewari S, Santos JM, Kowluru RA. Damaged Mitochondrial DNA Replication System and the Development of Diabetic Retinopathy. Antioxid Redox Signal (2012) 17:492–504. doi: 10.1089/ars.2011.4333
50. Mohammad G, Radhakrishnan R, Kowluru RA. Epigenetic Modifications Compromise Mitochondrial DNA Quality Control in the Development of Diabetic Retinopathy. Invest Ophtahlmol Vis Sci (2019) 60:3943–51. doi: 10.1167/iovs.19-27602
51. Duraisamy AJ, Mohammad G, Kowluru RA. Mitochondrial Fusion and Maintenance of Mitochondrial Homeostasis in Diabetic Retinopathy. Biochim Biophys Acta Mol Basis Dis (2019) 1865:1617–26. doi: 10.1016/j.bbadis.2019.03.013
52. Mohammad G, Kowluru RA. Mitochondrial Dynamics in the Metabolic Memory of Diabetic Retinopathy. J Diabetes Res (2022) 2022:3555889. doi: 10.1155/2022/3555889
53. Katakami N, Kaneto H, Takahara M, Matsuoka TA, Imamura K, Ishibashi F, et al. Aldose Reductase C-106T Gene Polymorphism Is Associated With Diabetic Retinopathy in Japanese Patients With Type 2 Diabetes. Diabetes Res Clin Pract (2011) 92:e57–60. doi: 10.1016/j.diabres.2011.02.017
54. Sorbinil Retinopathy Trial Research Group. A Randomized Trial of Sorbinil, an Aldose Reductase Inhibitor, in Diabetic Retinopathy. Arch Ophthalmol (1990) 108:1234–44. doi: 10.1001/archopht.1990.01070110050024
55. Ott C, Jacobs K, Haucke E, Navarrete Santos A, Grune T, Simm A. Role of Advanced Glycation End Products in Cellular Signaling. Redox Biol (2014) 2:411–29. doi: 10.1016/j.redox.2013.12.016
56. Stitt AW. AGEs and Diabetic Retinopathy. Invest Ophtahlmol Vis Sci (2010) 51:4867–74. doi: 10.1167/iovs.10-5881
57. Geraldes P, Hiraoka-Yamamoto J, Matsumoto M, Clermont A, Leitges M, Marette A, et al. Activation of PKC-Delta and SHP-1 by Hyperglycemia Causes Vascular Cell Apoptosis and Diabetic Retinopathy. Nat Med (2009) 15:1298–306. doi: 10.1038/nm.2052
58. Nishikawa T, Edelstein D, Brownlee M. THe Missing Link: A Single Unifying Mechanism for Diabetic Complications. Kidney Int Supplement (2000) 77:S26–30. doi: 10.1046/j.1523-1755.2000.07705.x
59. Kern TS. Contributions of Inflammatory Processes to the Development of the Early Stages of Diabetic Retinopathy. Exp Diabetes Res (2007) 2007:95103. doi: 10.1155/2007/95103
60. Rübsam A, Parikh S, Fort PE. Role of Inflammation in Diabetic Retinopathy. Int J Mol Sci (2018) 19:942. doi: 10.3390/ijms19040942
61. Frank RN. Diabetic Retinopathy and Systemic Factors. Middle East Afr J Ophthalmol (2015) 22:151–6. doi: 10.4103/0974-9233.154388
62. Kowluru RA. Diabetic Retinopathy and NADPH Oxidase-2: A Sweet Slippery Road. Antioxid (Basel) (2021) 10:783. doi: 10.3390/antiox10050783
63. Abhary S, Hewitt AW, Burdon KP, Craig JE. A Systematic Meta-Analysis of Genetic Association Studies for Diabetic Retinopathy. Diabetes (2009) 58:2137–47. doi: 10.2337/db09-0059
64. Tariq K, Malik SB, Ali SH, Maqsood SE, Azam A, Muslim I, et al. Association of Pro12Ala Polymorphism in Peroxisome Proliferator Activated Receptor Gamma With Proliferative Diabetic Retinopathy. Mol Vision (2013) 19:710–7.
65. Penman A, Hoadley S, Wilson JG, Taylor HA, Chen CJ, Sobrin L.. P-selectin Plasma Levels and Genetic Variant Associated With Diabetic Retinopathy in African Americans. Am J Ophthalmol (2015) 159:1152–1160.e1152. doi: 10.1016/j.ajo.2015.03.008
66. Cilensek I, Mankoc S, Globocnik Petrovic M, Petrovic D. The 4a/4a Genotype of the VNTR Polymorphism for Endothelial Nitric Oxide Synthase (eNOS) Gene Predicts Risk for Proliferative Diabetic Retinopathy in Slovenian Patients (Caucasians) With Type 2 Diabetes Mellitus. Mol Biol Rep (2012) 39:7061–7. doi: 10.1007/s11033-012-1537-8
67. Lin HJ, Huang YC, Lin JM, Wu JY, Chen LA, Tsai FJ. Association of Genes on Chromosome 6, GRIK2 , TMEM217 and TMEM63B (Linked to MRPL14 ) With Diabetic Retinopathy. Ophthalmologica (2013) 229:54–60. doi: 10.1159/000342616
68. Sobrin L, Susarla G, Stanwyck L, Rouhana JM, Li A, Pollack S, et al. Gene Set Enrichment Analsyes Identify Pathways Involved in Genetic Risk for Diabetic Retinopathy. Am J Ophthalmol (2022) 233:111–23. doi: 10.1016/j.ajo.2021.06.014
69. Dahlstrom E, Sandholm N. Progress in Defining the Genetic Basis of Diabetic Complications. Curr Diabetes Rep (2017) 17:80. doi: 10.1007/s11892-017-0906-z
70. Cheung CY, Hui EY, Lee CH, Kwok KH, Gangwani RA, Li KK, et al. Impact of Genetic Loci Identified in Genome-Wide Association Studies on Diabetic Retinopathy in Chinese Patients With Type 2 Diabetes. Invest Ophtahlmol Vis Sci (2016) 57:5518–24. doi: 10.1167/iovs.16-20094
71. Looker HC, Nelson RG, Chew E, Klein R, Klein BE, Knowler WC, et al. Genome-Wide Linkage Analyses to Identify Loci for Diabetic Retinopathy. Diabetes (2007) 56:1160–6. doi: 10.2337/db06-1299
72. Handy DE, Castro R, Loscalzo J. Epigenetic Modifications: Basic Mechanisms and Role in Cardiovascular Disease. Circulation (2011) 123:2145–56. doi: 10.1161/CIRCULATIONAHA.110.956839
73. Zhu JK. Active DNA Demethylation Mediated by DNA Glycosylases. Annu Rev Genet (2009) 43:143–66. doi: 10.1146/annurev-genet-102108-134205
74. Wu H, Zhang Y. Reversing DNA Methylation: Mechanisms, Genomics, and Biological Functions. Cell (2014) 156:45–68. doi: 10.1016/j.cell.2013.12.019
75. Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA Methylation Pathways and Their Crosstalk With Histone Methylation. Nat Rev Mol Cell Biol (2015) 16:519–32. doi: 10.1038/nrm4043
76. Strahl BD, Allis CD. The Language of Covalent Histone Modifications. Nature (2000) 403:41–5. doi: 10.1038/47412
77. Vaissiere T, Sawan C, Herceg Z. Epigenetic Interplay Between Histone Modifications and DNA Methylation in Gene Silencing. Mutat Res (2008) 659:40–8. doi: 10.1016/j.mrrev.2008.02.004
78. Xu YM, Du JY, Lau AT. Posttranslational Modifications of Human Histone H3: An Update. Proteomics (2014) 14:2047–60. doi: 10.1002/pmic.201300435
79. Martin C, Zhang Y. The Diverse Functions of Histone Lysine Methylation. Nat Rev Mol Cell Biol (2005) 6:838–49. doi: 10.1038/nrm1761
80. Black JC, Van Rechem C, Whetstine JR. Histone Lysine Methylation Dynamics: Establishment, Regulation, and Biological Impact. Mol Cell (2012) 48:491–507. doi: 10.1016/j.molcel.2012.11.006
81. Molina-Serrano D, Schiza V, Kirmizis A. Cross-Talk Among Epigenetic Modifications: Lessons From Histone Arginine Methylation. Biochem Soc Trans (2013) 41:751–9. doi: 10.1042/bst20130003
83. Cooper ME, El-Osta A. Epigenetics: Mechanisms and Implications for Diabetic Complications. Circ Res (2010) 107:1403–13. doi: 10.1161/CIRCRESAHA.110.223552
84. Shanmugam MK, Sethi G. Role of Epigenetics in Inflammation-Associated Diseases. Subcell Biochem (2012) 61:627–57. doi: 10.1007/978-94-007-4525-4_27
85. Tammen SA, Friso S, Choi SW. Epigenetics: The Link Between Nature and Nurture. Mol Aspects Med (2013) 34:753–64. doi: 10.1016/j.mam.2012.07.018
86. Kowluru RA, Mohammad G. Epigenetic Modifications in Diabetes. Metabolism (2021) 126:154920. doi: 10.1016/j.metabol.2021.154920
87. Mishra M, Kowluru RA. Epigenetic Modification of Mitochondrial DNA in the Development of Diabetic Retinopathy. Invest Ophtahlmol Vis Sci (2015) 56:5133–42. doi: 10.1167/iovs.15-16937
88. Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, et al. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int J Mol Sci (2019) 20:5573. doi: 10.3390/ijms20225573
89. Raut SK, Khullar M. The Big Entity of New RNA World: Long Non-Coding RNAs in Microvascular Complications of Diabetes. Front Endocrinol (Lausanne) (2018) 9:300. doi: 10.3389/fendo.2018.00300
90. Biswas S, Sarabusky M, Chakrabarti S. Diabetic Retinopathy, lncRNAs, and Inflammation: A Dynamic, Interconnected Network. J Clin Med (2019) 8:1033. doi: 10.3390/jcm8071033
91. Radhakrishnan R, Kowluru RA. Long Noncoding RNA MALAT1 and Regulation of the Antioxidant Defense System in Diabetic Retinopathy. Diabetes (2021) 70:227–39. doi: 10.2337/db20-0375
92. Huang Q, Li J. Research Progress of lncRNAs in Diabetic Retinopathy. Eur J Ophthalmol (2021) 31:1606–17. doi: 10.1177/1120672120970401
93. Mohammad G, Kowluru RA. Nuclear Genome-Encoded Long Noncoding RNAs and Mitochondrial Damage in Diabetic Retinopathy. Cells (2021) 10:3271. doi: 10.3390/cells10123271
94. Lu X, Tan Q, Ma J, Zhang J, Yu P. Emerging Role of LncRNA Regulation for NLRP3 Inflammasome in Diabetes Complications. Front Cell Dev Biol (2021) 9:792401. doi: 10.3389/fcell.2021.792401
95. Yan B, Tao ZF, Li XM, Zhang H, Yao J, Jiang Q. Aberrant Expression of Long Noncoding RNAs in Early Diabetic Retinopathy. Invest Ophtahlmol Vis Sci (2014) 55:941–51. doi: 10.1167/iovs.13-13221
96. Wang J, Gao X, Liu J, Wang J, Zhang Y, Zhang T, et al. Effect of Intravitreal Conbercept Treatment on the Expression of Long Noncoding RNAs and mRNAs in Proliferative Diabetic Retinopathy Patients. Acta Ophthalmol (2019) 97:e902–12. doi: 10.1111/aos.14083
97. Alfaifi M, Ali Beg MM, Alshahrani MY, Ahmad I, Alkhathami AG, Joshi PC, et al. Circulating Long Non-Coding RNAs NKILA, NEAT1, MALAT1, and MIAT Expression and Their Association in Type 2 Diabetes Mellitus. BMJ Open Diabetes Res Care (2021) 9:e001821. doi: 10.1136/bmjdrc-2020-001821
98. Zhang YL, Hu HY, You ZP, Li BY, Shi K. Targeting Long Non-Coding RNA MALAT1 Alleviates Retinal Neurodegeneration in Diabetic Mice. Int J Ophthalmol (2020) 13:213–9. doi: 10.18240/ijo.2020.02.03
99. Zhang X, Tang X, Liu K, Hamblin MH, Yin K-J. Long Noncoding RNA Malat1 Regulates Cerebrovascular Pathologies in Ischemic Stroke. J Neurosci (2017) 37:1797–806. doi: 10.1523/JNEUROSCI.3389-16.2017
100. Sathishkumar C, Prabu P, Mohan V, Balasubramanyam M. Linking a Role of lncRNAs (Long Non-Coding RNAs) With Insulin Resistance, Accelerated Senescence, and Inflammation in Patients With Type 2 Diabetes. Hum Genomics (2018) 12:41. doi: 10.1186/s40246-018-0173-3
101. Biswas S, Thomas AA, Chen S, Aref-Eshghi E, Feng B, Gonder J, et al. MALAT1: An Epigenetic Regulator of Inflammation in Diabetic Retinopathy. Sci Rep (2018) 8:6526. doi: 10.1038/s41598-018-24907-w
102. Abdulle LE, Hao JL, Pant OP, Liu XF, Zhou DD, Gao Y, et al. MALAT1 as a Diagnostic and Therapeutic Target in Diabetes-Related Complications: A Promising Long-Noncoding RNA. Int J Med Sci (2019) 16:548–55. doi: 10.7150/ijms.30097
103. Kowluru RA, Koppolu P, Chakrabarti S, Chen S. Diabetes-Induced Activation of Nuclear Transcriptional Factor in the Retina, and Its Inhibition by Antioxidants. Free Radic Res (2003) 37:1169–80. doi: 10.1080/10715760310001604189
104. Kolling M, Genschel C, Kaucsar T, Hubner A, Rong S, Schmitt R, et al. Hypoxia-Induced Long Non-Coding RNA Malat1 Is Dispensable for Renal Ischemia/reperfusion-Injury. Sci Rep (2018) 8:3438. doi: 10.1038/s41598-018-21720-3
105. Dong N, Xu B, Shi H. Long Noncoding RNA MALAT1 Acts as a Competing Endogenous RNA to Regulate Amadori-Glycated Albumin-Induced MCP-1 Expression in Retinal Microglia by a microRNA-124-Dependent Mechanism. Inflammation Res (2018) 67:913–25. doi: 10.1007/s00011-018-1184-1
106. Thomas AA, Feng B, Chakrabarti S. ANRIL: A Regulator of VEGF in Diabetic Retinopathy. Invest Ophtahlmol Vis Sci (2017) 58:470–80. doi: 10.1167/iovs.16-20569
107. Zhang J, Chen M, Chen J, Lin S, Cai D, Chen C, et al. Long Non-Coding RNA MIAT Acts as a Biomarker in Diabetic Retinopathy by Absorbing miR-29b and Regulating Cell Apoptosis. Biosci Rep (2017) 37:BSR20170036. doi: 10.1042/bsr20170036
108. Wang G, Ye Q, Ning S, Yang Z, Chen Y, Zhang L, et al. LncRNA MEG3 Promotes Endoplasmic Reticulum Stress and Suppresses Proliferation and Invasion of Colorectal Carcinoma Cells Through the MEG3/miR-103a-3p/PDHB ceRNA Pathway. Neoplasma (2021) 68:362–74. doi: 10.4149/neo_2020_200813N858
109. Zhang D, Qin H, Leng Y, Li X, Zhang L, Bai D, et al. LncRNA MEG3 Overexpression Inhibits the Development of Diabetic Retinopathy by Regulating TGF-β1 and VEGF. Exp Ther Med (2018) 16:2337–42. doi: 10.3892/etm.2018.6451
110. Biswas S, Feng B, Chen S, Liu J, Aref-Eshghi E, Gonder J, et al. The Long Non-Coding RNA HOTAIR Is a Critical Epigenetic Mediator of Angiogenesis in Diabetic Retinopathy. Invest Ophtahlmol Vis Sci (2021) 62:20. doi: 10.1167/iovs.62.3.20
111. Kowluru RA, Abbas SN. Diabetes-Induced Mitochondrial Dysfunction in the Retina. Invest Ophtahlmol Vis Sci (2003) 44:5327–34. doi: 10.1167/iovs.03-0353
112. Wang M, Huang T, Luo G, Huang C, Xiao X-Y, Wang L, et al. Long Non-Coding RNA MEG3 Induces Renal Cell Carcinoma Cells Apoptosis by Activating the Mitochondrial Pathway. J Huazhong Univ Sci Technol [Med Sci] (2015) 35:541–5. doi: 10.1007/s11596-015-1467-5
113. Gong W, Zhu G, Li J, Yang X. LncRNA MALAT1 Promotes the Apoptosis and Oxidative Stress of Human Lens Epithelial Cells via p38MAPK Pathway in Diabetic Cataract. Diabetes Res Clin Pract (2018) 144:314–21. doi: 10.1016/j.diabres.2018.06.020
114. Zhao Y, Zhou L, Li H, Sun T, Wen X, Li X, et al. Nuclear-Encoded lncRNA MALAT1 Epigenetically Controls Metabolic Reprogramming in HCC Cells Through the Mitophagy Pathway. Mol Ther Nucleic Acids (2021) 23:264–76. doi: 10.1016/j.omtn.2020.09.040
115. Kong L, Zhou X, Wu Y, Wang Y, Chen L, Li P, et al. Targeting HOTAIR Induces Mitochondria Related Apoptosis and Inhibits Tumor Growth in Head and Neck Squamous Cell Carcinoma In Vitro and In Vivo. Curr Mol Med (2015) 15:952–60. doi: 10.2174/1566524016666151123112716
116. Wang K, Long B, Zhou L-Y, Liu F, Zhou Q-Y, Liu C-Y, et al. CARL lncRNA Inhibits Anoxia-Induced Mitochondrial Fission and Apoptosis in Cardiomyocytes by Impairing miR-539-Dependent PHB2 Downregulation. Nat Commun (2014) 5:3596. doi: 10.1038/ncomms4596
117. Long J, Badal SS, Ye Z, Wang Y, Ayanga BA, Galvan DL, et al. Long Noncoding RNA Tug1 Regulates Mitochondrial Bioenergetics in Diabetic Nephropathy. J Clin Invest (2016) 126:4205–18. doi: 10.1172/jci87927
118. Wang Y, Hu SB, Wang MR, Yao RW, Wu D, Yang L, et al. Genome-Wide Screening of NEAT1 Regulators Reveals Cross-Regulation Between Paraspeckles and Mitochondria. Nat Cell Biol (2018) 20:1145–58. doi: 10.1038/s41556-018-0204-2
119. Kowluru RA, Mishra M. Therapeutic Targets for Altering Mitochondrial Dysfunction Associated With Diabetic Retinopathy. Expert Opin Ther Targets (2018) 22:233–45. doi: 10.1080/14728222.2018.1439921
120. Kolesnikov AA. The Mitochondrial Genome. The Nucleoid. Biochem (Mosc) (2016) 81:1057–65. doi: 10.1134/s0006297916100047
121. Sumitani M, Kasashima K, Ohta E, Kang D, Endo H. Association of a Novel Mitochondrial Protein M19 With Mitochondrial Nucleoids. J Biochem (2009) 146:725–32. doi: 10.1093/jb/mvp118
122. Kucej M, Kucejova B, Subramanian R, Chen XJ, Butow RA. Mitochondrial Nucleoids Undergo Remodeling in Response to Metabolic Cues. J Cell Sci (2008) 121:1861–8. doi: 10.1242/jcs.028605
123. Zhong Q, Kowluru RA. Regulation of Matrix Metalloproteinase-9 by Epigenetic Modifications and the Development of Diabetic Retinopathy. Diabetes (2013) 62:2559–68. doi: 10.2337/db12-1141
124. Duraisamy AJ, Mishra M, Kowluru RA. Crosstalk Between Histone and DNA Methylation in Regulation of Retinal Matrix Metalloproteinase-9 in Diabetes. Invest Ophtahlmol Vis Sci (2017) 58:6440–8. doi: 10.1167/iovs.17-22706
125. Tong P, Peng QH, Gu LM, Xie WW, Li WJ. LncRNA-MEG3 Alleviates High Glucose Induced Inflammation and Apoptosis of Retina Epithelial Cellsvia regulating miR-34a/SIRT1 axis. Exp Mol Pathol (2019) 107:102–9. doi: 10.1016/j.yexmp.2018.12.003
126. Mishra M, Duraisamy AJ, Kowluru RA. Sirt1- a Guardian of the Development of Diabetic Retinopathy. Diabetes (2018) 67:745–54. doi: 10.2337/db17-0996
127. Liu P, Jia SB, Shi JM, Li WJ, Tang LS, Zhu XH, et al. LncRNA-MALAT1 Promotes Neovascularization in Diabetic Retinopathy Through Regulating miR-125b/VE-Cadherin Axis. Biosci Rep (2019) 39:BSR20170036. doi: 10.1042/bsr20181469
128. Boon RA, Jaé N, Holdt L, Dimmeler S. Long Noncoding RNAs: From Clinical Genetics to Therapeutic Targets? J Am Coll Cardiol (2016) 67:1214–26. doi: 10.1016/j.jacc.2015.12.051
129. Huang CK, Kafert-Kasting S, Thum T. Preclinical and Clinical Development of Noncoding RNA Therapeutics for Cardiovascular Disease. Circ Res (2020) 126:663–78. doi: 10.1161/circresaha.119.315856
130. Chen B, Zhang RN, Fan X, Wang J, Xu C, An B, et al. Clinical Diagnostic Value of Long Non-Coding RNAs in Colorectal Cancer: A Systematic Review and Meta-Analysis. J Cancer (2020) 11:5518–26. doi: 10.7150/jca.46358
131. Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, et al. Circulating Long Noncoding RNA, LIPCAR, Predicts Survival in Patients With Heart Failure. Circ Res (2014) 114:1569–75. doi: 10.1161/circresaha.114.303915
132. Cai Y, Yang Y, Chen X, He D, Zhang X, Wen X, et al. Circulating "LncPPARδ" From Monocytes as a Novel Biomarker for Coronary Artery Diseases. Medicine (2016) 95:e2360. doi: 10.1097/md.0000000000002360
133. Tang H, Wu Z, Zhang J, Su B. Salivary lncRNA as a Potential Marker for Oral Squamous Cell Carcinoma Diagnosis. Mol Med Rep (2013) 7:761–6. doi: 10.3892/mmr.2012.1254
134. Bolha L, Ravnik-Glavač M, Glavač D. Long Noncoding RNAs as Biomarkers in Cancer. Dis Markers (2017) 2017:7243968–7243968. doi: 10.1155/2017/7243968
135. Shaker OG, Abdelaleem OO, Mahmoud RH, Abdelghaffar NK, Ahmed TI, Said OM, et al. Diagnostic and Prognostic Role of Serum miR-20b, miR-17-3p, HOTAIR, and MALAT1 in Diabetic Retinopathy. IUBMB Life (2019) 71:310–20. doi: 10.1002/iub.1970
136. He J, Rui Z, Gao J, Chen Y, Li Y, Xu T, et al. Expression of Long Non-Coding RNA (lncRNA) SNHG5 in Patients with Refractory Diabetic Macular Edema and Its Regulatory Mechanism. Med Sci Monit (2022) 28:e932996. doi: 10.12659/msm.932996
137. Biswas S, Coyle A, Chen S, Gostimir M, Gonder J, Chakrabarti S. Expressions of Serum lncRNAs in Diabetic Retinopathy – A Potential Diagnostic Tool. Front Endocrinol (Lausanne) (2022) 13:851967. doi: 10.3389/fendo.2022.851967
138. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The Transcriptional Landscape of the Mammalian Genome. Science (2005) 309:1559–63. doi: 10.1126/science.1112014
139. Kitow J, Derda AA, Beermann J, Kumarswarmy R, Pfanne A, Fendrich J, et al. Mitochondrial Long Noncoding RNAs as Blood Based Biomarkers for Cardiac Remodeling in Patients With Hypertrophic Cardiomyopathy. Am J Physiol Heart Circulatory Physiol (2016) 311:H707–712. doi: 10.1152/ajpheart.00194.2016
Keywords: Diabetic retinopathy, long noncoding RNAs, mitochondria, epigenetics, diabetes
Citation: Kowluru RA (2022) Long Noncoding RNAs and Mitochondrial Homeostasis in the Development of Diabetic Retinopathy. Front. Endocrinol. 13:915031. doi: 10.3389/fendo.2022.915031
Received: 07 April 2022; Accepted: 29 April 2022;
Published: 06 June 2022.
Edited by:
Michele Lanza, University of Campania Luigi Vanvitelli, ItalyReviewed by:
Subrata Chakrabarti, Western University, CanadaCopyright © 2022 Kowluru. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Renu A. Kowluru, cmtvd2x1cnVAbWVkLndheW5lLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.