
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Endocrinol. , 17 June 2022
Sec. Cancer Endocrinology
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.895729
This article is part of the Research Topic Underlying Molecular Interconnections of the Estrogen Receptor alpha and Associated Factors involved in Breast Cancer Development: the Way to New Therapeutic Approaches View all 7 articles
 Sarah A. Jeffreys1,2*
Sarah A. Jeffreys1,2* Therese M. Becker1,3
Therese M. Becker1,3 Sarah Khan4
Sarah Khan4 Patsy Soon1,3,5
Patsy Soon1,3,5 Hans Neubauer6
Hans Neubauer6 Paul de Souza1,2,3
Paul de Souza1,2,3 Branka Powter1
Branka Powter1Background: Up to 80% of breast cancers (BCa) are estrogen receptor positive and current treatments target the estrogen receptor (endocrine therapies) and/or CDK4/6 (CDK4/6 inhibitors). CCND1 encodes the protein cyclin D1, responsible for regulation of G1 to S phase transition in the cell cycle. CCND1 amplification is common in BCa and contributes to increased cyclin D1 expression. As there are signalling interactions between cyclin D1 and the estrogen receptor, understanding the impact of CCND1 amplification on estrogen receptor positive patients’ disease outcomes, is vital. This review aims to evaluate CCND1 amplification as a prognostic and predictive biomarker in BCa.
Materials and Methods: Publications were retrieved from the databases: PubMed, MEDLINE, Embase and Cochrane library. Exclusion criteria were duplication, publication type, non-English language, in vitro and animal studies, not BCa, male BCa, premenopausal BCa, cohort size <35, CCND1 amplification not reported. Publications with cohort duplication, and inadequate recurrence free survival (RFS) and overall survival (OS) data, were also excluded. Included publications were assessed for Risk of Bias (RoB) using the Quality In Prognosis Studies tool. Statistical analyses (Inverse Variance and Mantel-Haenszel) were performed in Review Manager. The PROSPERO registration number is [CRD42020208179].
Results: CCND1 amplification was significantly associated with positive estrogen receptor status (OR:1.70, 95% CI:1.19-2.43, p = 0.004) and cyclin D1 overexpression (OR: 5.64, 95% CI: 2.32-13.74, p=0.0001). CCND1 amplification was significantly associated with shorter RFS (OR: 1.64, 95% CI: 1.13-2.38, p = 0.009), and OS (OR: 1.51, 95% CI: 1.19-1.92, p = 0.0008) after removal of studies with a high RoB. In endocrine therapy treated patients specifically, CCND1 amplification predicted shorter RFS (HR: 2.59, 95% CI: 1.96-3.41, p < 0.00001) and OS (HR: 1.59, 95% CI: 1.00-2.49, p = 0.05) also after removal of studies with a high RoB.
Conclusion: While a lack of standardised approach for the detection of CCND1 amplification is to be considered as a limitation, CCND1 amplification was found to be prognostic of shorter RFS and OS in BCa. CCND1 amplification is also predictive of reduced RFS and OS in endocrine therapy treated patients specifically. With standardised methods and cut offs for the detection of CCND1 amplification, CCND1 amplification would have potential as a predictive biomarker in breast cancer patients.
Systematic Review Registration: https://www.crd.york.ac.uk/prospero/, identifier CRD42020208179.
Chronic sustained cell proliferation is one of the hallmarks of cancer, which is achieved through signalling changes resulting in progression through the cell cycle (1). Cyclin D1, along with its binding partners cyclin dependent kinases (CDK4/6), is a key regulator of the cell cycle, mediating transition from G1 to S phase (Figure 1). The gene encoding cyclin D1, CCND1, located on chromosome 11q13.3, has been reported to be amplified in 10-35% of breast cancers (BCa) (2–4) and its amplification has been associated with increased cyclin D1 expression (3). CCND1 amplification may be an effective prognostic and predictive biomarker in BCa.
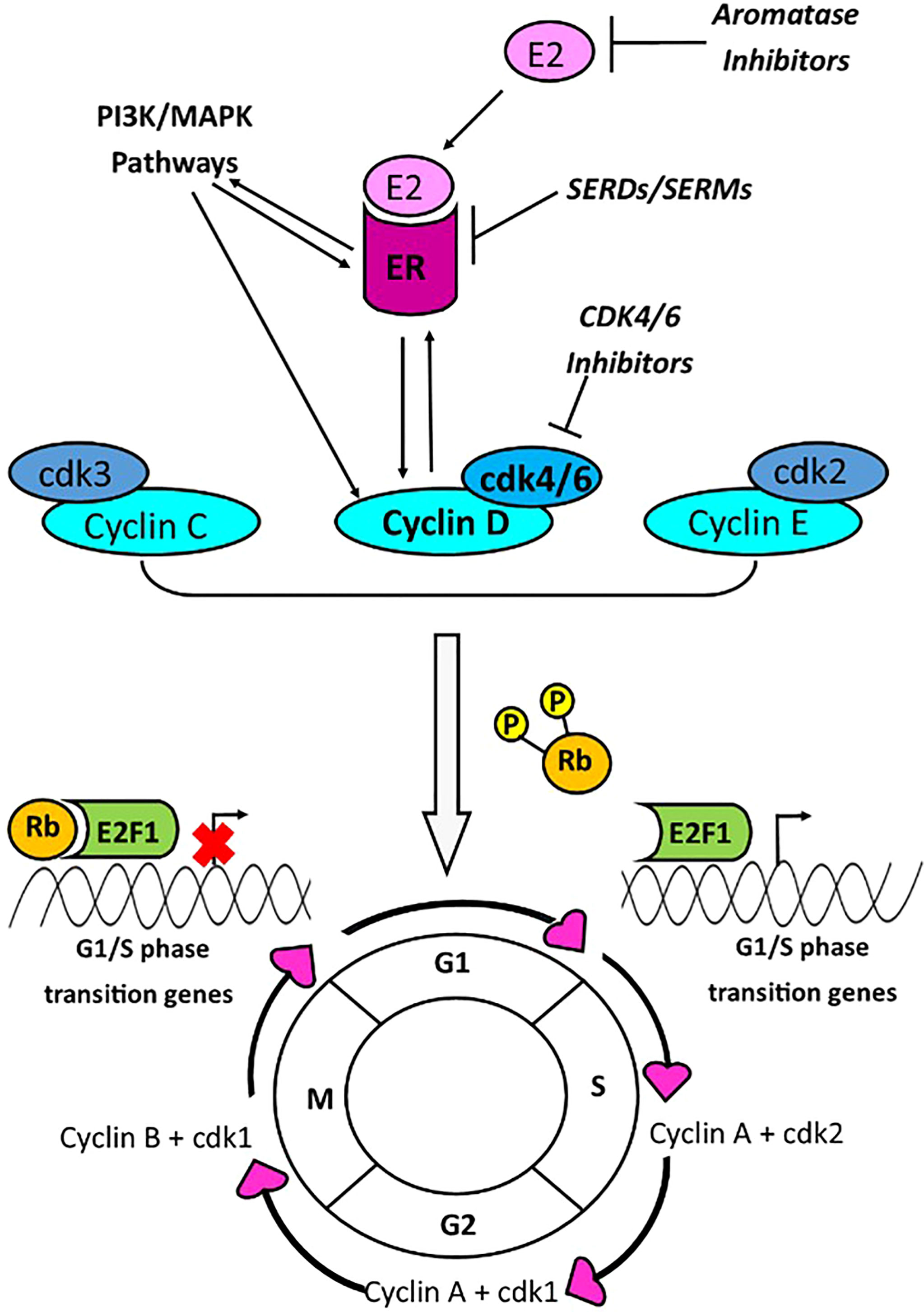
Figure 1 Cyclin D1 promotes G1/S phase cell cycle progression via interaction with CDK4/6. Transition between phases of the cell cycle is mediated by cyclins A-E, and cyclin dependent kinases (cdk) 1-6. Cyclins C-E are responsible for the transition from G1 to S phase of the cell cycle. The cyclin D and cdk4/6 complex, activated by PI3K/MAPK pathways or the estrogen receptor (ER), is a key mediator of G1-S phase transition, and this occurs through phosphorylation of retinoblastoma protein (Rb). Phosphorylation of Rb, results in its dissociation with E2F1, enabling transcription of G1/S phase genes. Transcriptional activity of the ER is stimulated by cyclin D1, and the ER may activate the CCND1 promoter. Current breast cancer drugs target estrogen production (aromatase inhibitors); the estrogen receptor [Selective Estrogen Receptor Modulators (SERMs) or Selective Estrogen Receptor Degraders (SERDs)]; or cdk4/6 (cdk4/6 inhibitors).
Progression through the cell cycle is regulated by several cyclins and cyclin dependent kinases, at each stage of cycle. During the G0 phase, retinoblastoma protein (Rb) inhibits the E2F transcription factor 1 (E2F1), thereby preventing transcription of G1/S phase genes (5). The Cyclin D1-CDK4 or 6 complex phosphorylates the Rb, leading to disassociation of Rb from E2F1, thus activating G1/S phase gene transcription and cell cycle progression (5) (Figure 1). Growth factor signalling pathways, PI3K-AKT-mTOR and MAPK, are linked with both cyclin D1 and Estrogen Receptor (ER) activity (6, 7). There is interplay between the ER, CCND1 gene and cyclin D1 protein whereby the ER promotes transcription of the CCND1 gene, and the cyclin D1 protein interacts with the ER to promote ER mediated transcription (8–10).
The ER is expressed in approximately 75% of all BCa tumours (11). CCND1 amplification is particularly common in ER positive tumours and is associated with reduced survival in these patients (3, 12–15). Many ER positive patients are treated with endocrine therapies (ET), that inhibit the ER pathway, however up to 50% of patients develop resistance (16, 17). CCND1 amplification is a proposed mechanism of ET resistance, however this remains controversial due to studies with conflicting results. Some studies have shown significant association between CCND1 amplification and poor aromatase inhibitor (AI) (18, 19) and tamoxifen response (19), whilst another study has shown no association with tamoxifen response (20). Yet, another study suggested CCND1 was predictive of resistance to aromatase inhibitors but not to tamoxifen (21).
Added to the complexity of these biological relationships, is the role of cyclin D1 overexpression. Whilst the CCND1 gene amplification is detected in 10-35% of patients (2–4), 50-70% overexpress the cyclin D1 protein, suggesting additional mechanisms of cyclin D1 regulation (14). In a study that separated patients according to CCND1 amplification status and cyclin D1 overexpression; they found that patients with CCND1 amplification had reduced Recurrence Free Survival (RFS) compared to those with normal CCDN1, but patients with cyclin D1 overexpression had longer RFS than those without (22). Some studies have reported correlation between cyclin D1 expression and poor prognosis in ER positive BCa (23–25) but not all (26, 27). A meta-analysis concluded that cyclin D1 overexpression was a significant predictor of poor prognosis, in ER positive BCa but this was not observed in unselected BCa patients (25). The effects of cyclin D1 expression may differ due to differences in active signalling mechanisms between patients (26, 28).
The role of CCND1 amplification in BCa, in relation to ET response, RFS and overall survival (OS) remains unclear. This systematic review and meta-analysis evaluates the prognostic and predictive value of CCDN1 amplification in BCa patients across studies.
This review was registered with PROSPERO: International Prospective Register of Systematic Reviews, registration number [CRD42020208179].
Publications for screening were obtained from PubMed, MEDLINE, Embase, and Cochrane library databases. These databases were searched on 31st of August 2020 with the search: (“breast cancer” OR “breast carcinoma” OR “breast tumour” OR “breast tumor” OR “breast neoplas*” OR “mammary cancer” OR “mammary carcinoma”) AND (“Cyclin D1” OR CCND1 OR PRAD1 OR BCL1 OR U21B31 OR D11S287E) AND (“Hormone receptor” OR “estrogen receptor” OR “oestrogen receptor” OR ER OR “progesterone receptor” OR PR) AND (Amplification OR “copy number”). There were no restrictions on year of publication. The results from these searches were uploaded to the Rayyan Qatar Computing Research Institute (QCRI) systematic review application (29).
Publications were screened within the Rayyan QCRI (29) platform, by two blinded investigators. Publication duplicates, non-English language publications, reviews, comments, conference abstracts and letters were excluded. In one case, the publication could not be accessed and was excluded under publication type. Other exclusions were studies reporting only in vitro or animal findings, not breast cancer, less than 35 participants, premenopausal patients only, male participants only, those that did not report CCND1 amplification findings. Studies focused on premenopausal and male BCa patients were thus excluded to reduce intra study heterogeneity resulting from biological and treatment differences. For example, premenopausal patients typically present at later stages, have worse long-term outcomes, and receive different therapeutic regimes (particularly in terms of aromatase inhibitor treatment) than postmenopausal patients (30, 31). Following exclusion, investigators were unblinded, and any discrepancies were resolved by consensus. There were 83 publications remaining, and these were assessed first for cohort duplication. Studies that were deemed as having cohorts of the same patients were grouped and the study with the most patients was selected for inclusion; where they had the same number of patients, the most recent study was selected. There were two studies which each had two cohorts of patients, and one of these cohorts was the same in both studies; in this case, data was extracted from both cohorts of one study (32) and only from the non-duplicated cohort from the second study (14). Data extraction was performed on the remaining 69 studies, and these studies were then screened for availability of survival data for analysis of hazard ratios (HR). A final number of 18 studies were included, since the remainder did not provide sufficient survival data for analysis. Of these 18 studies, only those providing the necessary data for each analysis were included, and therefore the total number of studies included in each analysis differs, as reported in the results section.
Publications were uploaded to Covidence (33), which enabled data extraction using a customisable data extraction form, by one reviewer. Collected data included: general information (title, study type, cohort size, recruitment dates and place, cohort size, inclusion and exclusion criteria), patient characteristics (type of breast cancer, ER status, menopausal status, treatment type), CCND1 detection (method of detection and cut off), cyclin D1 expression (method, cut-off, correlation with CCND1 amplification) and CCND1 amplification correlation with other factors (human epidermal growth factor receptor 2 (HER2), ER, and progesterone receptor (PR) status, histological grade, clinical stage and treatment) and finally, CCND1 and outcomes: OS, breast cancer specific survival (BCSS), disease free survival (DFS) and recurrence free survival, relapse free survival, time to progression, and recurrence event numbers.
A Risk of Bias (RoB) assessment was performed on all 18 studies included in the meta-analysis, by two blinded investigators, using a customisable quality assessment form in Covidence (33). The quality assessment form was customised to align with the Quality in Prognosis Studies (QUIPs) tool (34). This tool assesses bias across six domains, namely: study participation, study attrition, prognostic factor measurement, outcome measurement, study confounding, and statistical analysis and reporting (34). Study participation was assessed with focus on inclusion and exclusion criteria, number of patients in cohort, and reported characteristics of the cohort. Study attrition focussed on the proportion of samples not assessed for CCND1 amplification, and if reasons were provided for sample loss. For prognostic factor measurement, whether appropriate methods and controls were used for detection of CCND1 amplification, and if cut-offs were reported, were considered. Outcome measurement was assessed based on definitions of survival data reported, whether these were considered standard, and reporting of follow up times. Study confounding considered other clinicopathological features, statistical comparisons made between these and CCND1 amplification, and how stage and grade were assessed. For assessment of statistical analysis and reporting, reporting and type of statistical test were considered, as well as the proportion of CCND1 amplification and survival data reported. Each domain was rated as “low”, “moderate” or “high” RoB, and where insufficient information was provided for judgement, these were rated as “unclear”. Discrepancies between ratings of the investigators were resolved by consensus. Each study was then given an overall rating based on a method reported by Jermy et al, whereby low bias studies have ≤2 domains rated as moderate with the remainder rated low; moderate bias studies have either 3 moderate ratings or one moderate and one high with the remainder rated low; and high bias studies having either ≥2 moderate plus one high rating, or ≥2 high ratings, or ≥4 of moderate ratings for each domain (35).
Statistical analyses were performed using Review Manager (36). For statistical analyses, studies rated as low and moderate overall RoB are grouped together, whilst studies with high RoB are grouped separately, as indicated in the relevant results sections. The Mantel-Haenszel method was used for statistical analysis of CCND1 amplification and clinicopathological features (ER, PR, HER2, stage, grade, and cyclin D1 expression). Statistics for CCND1 amplification and clinicopathological features are expressed as OR as these are categorical variables. Some analyses required grouping of data for analysis. For example, for grade, grades I-II were combined and compared with grade III; for stage T1-T2 were combined and compared with T3-T4, and for cyclin D1 overexpression low was compared to moderate and high combined; these categories were as reported by individual studies. The definitions used for grade and stage were considered as part of the RoB outcome measurement assessment. For the analysis of OS and RFS, the inverse-variance method was used. In these analyses, statistics were reported as HR as these are continuous variables. A fixed effects approach was taken for analyses with I2 <50%, whilst a random effect approach was used when I2 was >50%. OS analysis included both OS and BCSS. RFS analysis included recurrence free survival, relapse free survival, disease free survival and recurrence events raw data. Where HR and standard error (SE) were not provided, these were calculated based on provided summary statistics, in accordance with previously described methods (37). In each analysis, all studies reporting the necessary data for that specific analysis were included.
The process of exclusion and inclusion of studies is summarised in Figure 2. Across the four databases, 625 results were retrieved, of which 268 were duplicates. During screening, studies were excluded for the following reasons: publication type (181 studies), language other than English (3 studies), in vitro study (25 studies), animal study (9 studies), male breast cancer (6 studies), premenopausal (7 studies), cohort size <35 patients (28 studies), CCND1 amplification not reported (13 studies) and cohort duplication (14 studies). After screening the remaining studies, 69 studies were deemed eligible for further evaluation. Of these 69 studies, 42 were excluded as they did not evaluate RFS or OS, and nine were excluded due to insufficient summary data to derive HR. The remaining 18 studies formed the basis for this meta-analysis.
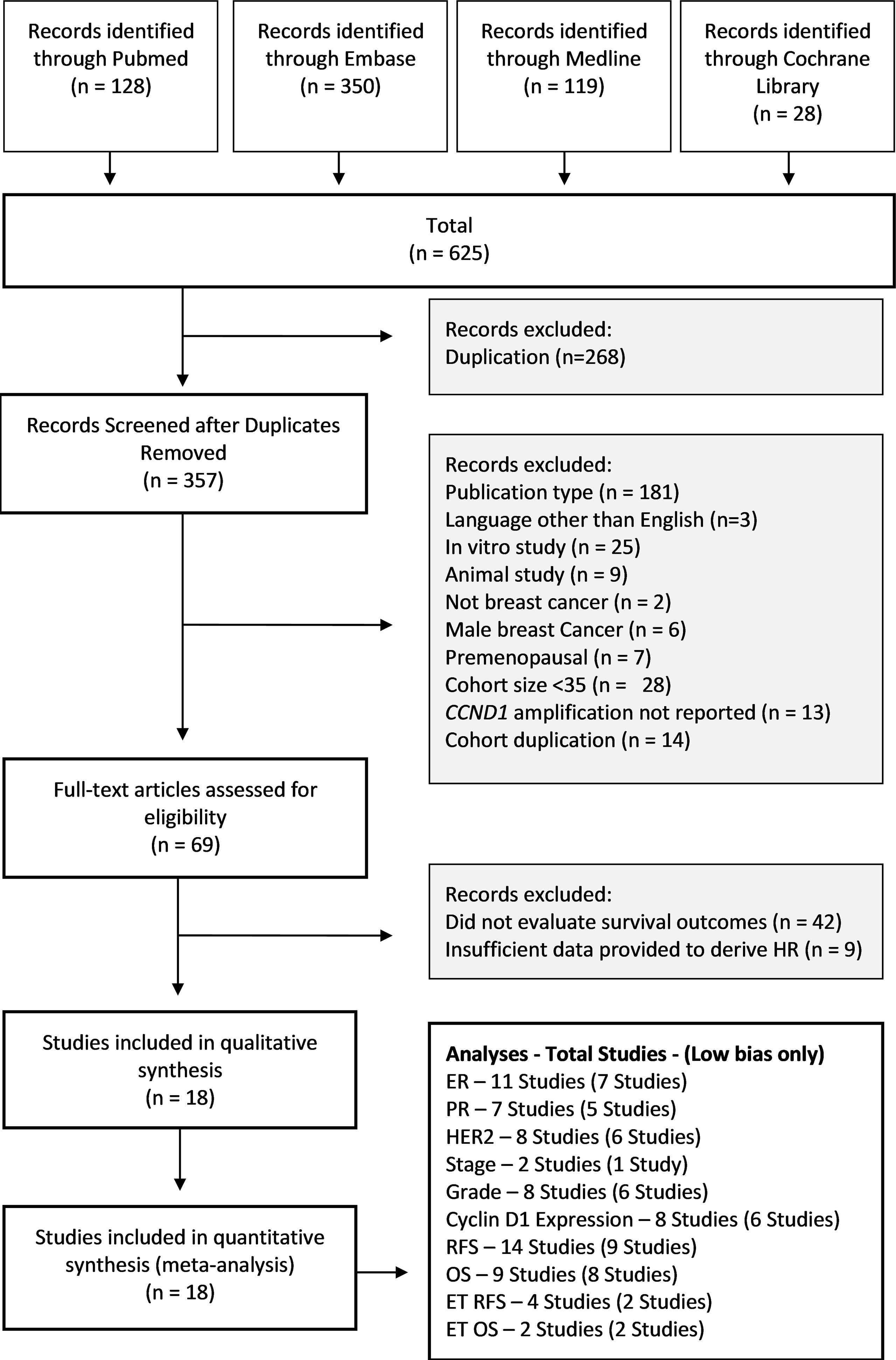
Figure 2 Study Selection PRISMA Diagram. Flow diagram shows studies retrieved from databases, and the number of studies excluded and the basis on which they were excluded. OS, Overall Survival; RFS, Relapse Free Survival; HR, Hazard Ratio.
Results of quality assessment, using the QUIPs tool, are shown in Table 1. There were three studies (22, 42, 49) that were deemed to have a low RoB across all six domains. All 18 studies were rated as low RoB for study participation, with all of them describing inclusion or exclusion criteria and all reporting key study characteristics. For study attrition, four studies (3, 19, 41, 45) had >20% of the cohort samples not assessed for CCND1 amplification, and four (26, 38, 43, 45) did not provide adequate reasoning for sample loss. For prognostic factor measurement, two studies (19, 44) inadequately reported methods and controls used for CCND1 amplification detection assays, however, all studies reported CCND1 amplification copy number cut-offs. For outcome measurement, definitions of OS and RFS were considered as well as whether follow up time was reported. Definitions of OS or RFS were not reported in six studies (20, 41, 44, 46–48), as such, for these six studies, plus an additional study (45), it was unclear whether the method of RFS and OS measurement were standard (outcome measurement). Also, for outcome measurement, four studies (20, 32, 46, 48) failed to report follow up time. For the study confounding domain, two studies (39, 46) did not adequately measure important confounders, six studies (19, 20, 32, 39, 45, 46) did not define grade or stage measurements, and five (39, 41, 43, 45, 46) made no statistical comparisons between grade and stage and either prognostic or outcome measurements. For the statistical analysis and reporting domain, two studies did not perform statistical analysis of raw data for CCND1 amplification and RFS or OS (39, 46), six studies did not adequately report statistical methods (14, 19, 20, 39, 44, 46) and three studies (20, 39, 46) selectively reported CCND1 amplification and survival data. There were four studies that were assessed as having a high overall RoB (19, 20, 45, 46); in each analysis these are represented in a separate subgroup.
Characteristics of studies are summarised in Table 2. CCND1 amplification was detected by various methods including: Florescence In Situ Hybridisation (FISH) (26, 41–45, 49, 50), Chromogenic In Situ Hybridisation (CISH) (3, 19), RT-PCR (20, 22, 46), Multiplex Ligation-dependent Probe Amplification (MLPA) (14, 38), Targeted Sequencing (32), Nanostring copy number variation assay (39), Southern Blot (22), Slot Blot Hybridisation (47) (Table 2). Amplification cut-offs varied significantly between studies, even amongst those with the same detection method. Some studies counted number of copies, some a copy number ratio relative to a control gene, some considered the number of signals and the proportion of cells with signals, whilst others had unique measures based on their method and corresponding controls. The frequency of CCND1 amplification ranged from 9%-57%. The studies collectively comprised 6400 patient samples and of these, 1135 (18%) were considered CCND1 amplified.
Several studies provided sufficient data for analysis of CCND1 amplification status and clinicopathological features, including ER (11 studies), PR (7 studies), HER2 status (8 studies), tumour stage (2 studies), histologic grade (8 studies) as well as cyclin D1 expression (8 studies). CCND1 amplification was significantly associated with ER status (OR: 1.70, 95% CI: 1.19-2.43, p = 0.004) and cyclin D1 overexpression (OR: 5.64, 95% CI: 2.32-13.74, p = 0.0001) (Table 3 and Supplementary Figures 1, 2). There was low heterogeneity (I2 = 35%, p = 0.13) for ER analysis, but high (I2 = 88%, p < 0.00001) for cyclin D1 overexpression analysis. CCND1 amplification was not significantly associated with: PR (p = 0.71) or HER2 (p = 0.39) status, tumour stage (p = 0.20) or histologic grade (p = 0.28) (Table 3 and Supplementary Figures 3–6). Analysis of CCND1 amplification and clinicopathological features excluding studies that had a high RoB did not differ substantially from the analysis of all eligible studies (Supplementary Figures 1–6).
A total of 14 studies consisting of 5083 patients had sufficient data for inclusion in the RFS analysis. CCND1 amplification was found be associated with significantly worse RFS (HR: 1.64, 95% CI: 1.07-2.52, p = 0.02), with high heterogeneity (I2 = 97%, p < 0.00001) (Figure 3A). Of the 14 studies, four studies, consisting of 2503 patients, were assessed as having a high RoB; exclusion of these studies did not substantially alter association between CCND1 amplification and RFS (HR: 1.64, 95% CI: 1.13-2.38, p = 0.008) with high heterogeneity (I2 = 93%, p < 0.00001) (Figure 3A).
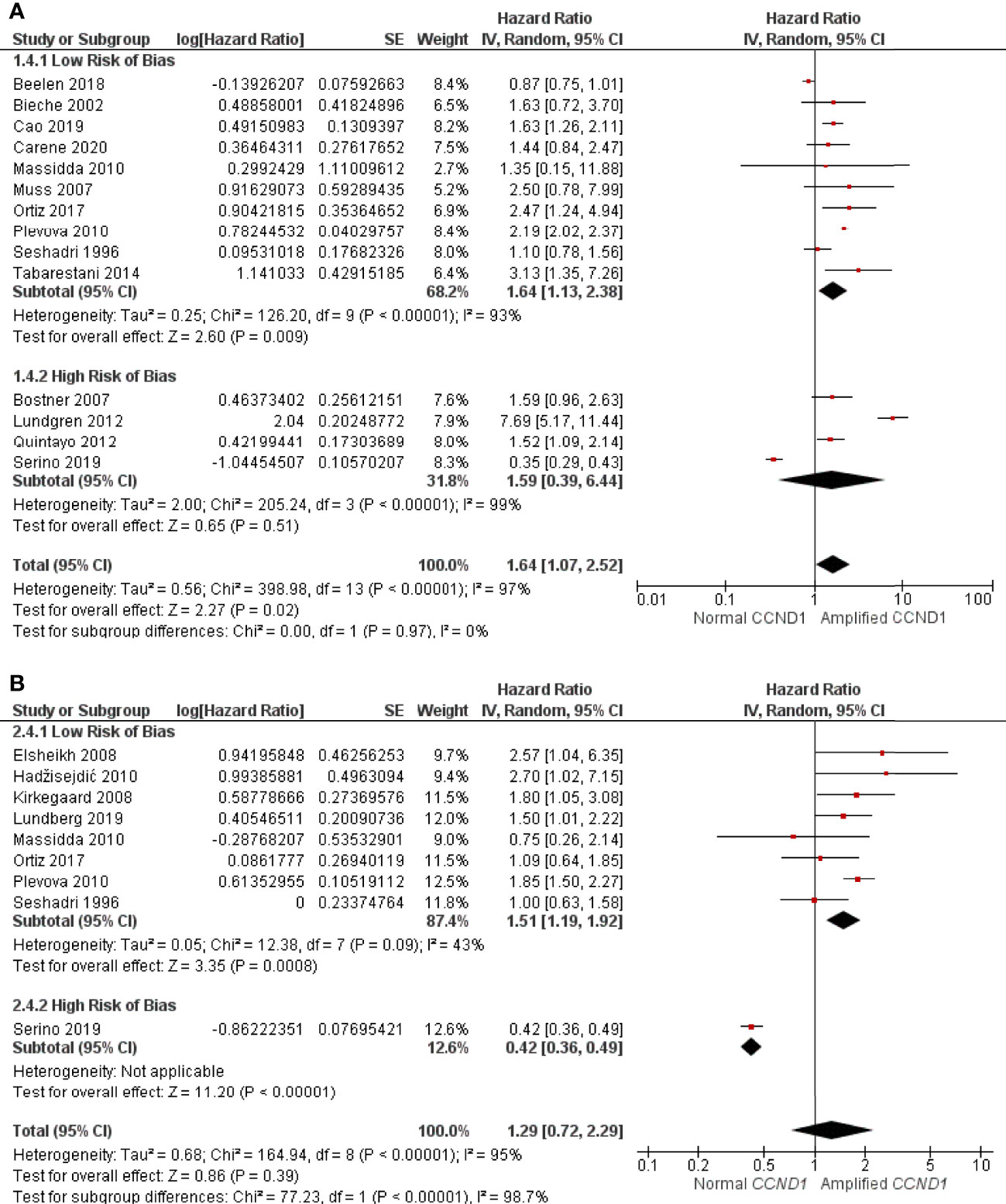
Figure 3 Forrest plot of hazard ratios for CCND1 amplification and worse relapse free survival and overall survival of breast cancer patients. (A) CCND1 Amplification and relapse free survival (B) CCND1 Amplification and overall survival. Z values indicate the magnitude of association, with p-values <0.05 indicating statistically significant association. Red squares indicate hazard ratio, with values >1 indicative of association of the outcome measure (RFS and OS) with CCND1 amplification, with strongest association towards the right of the plot. Black lines either side of squares indicate 95% confidence interval (CI). Size of red boxes is relative to specific study weight with greatest weight given to studies with minimal variance (calculated based on inverse of the variance). Large black diamond represents pooled hazard ratio estimate of the above studies. A random effects approach was taken. SE, Standard Error. Plots were generated in Review Manger.
Nine studies consisting of 2697 patients reported sufficient OS data for statistical analysis. There was no statistically significant association between CCND1 amplification and worse OS (HR: 1.29, 95% CI: 0.72-2.29, p = 0.39) with the high heterogeneity (I2 = 95%, p < 0.00001) (Figure 3B). Of these nine studies, one consisting of 56 patients, was considered as having a high RoB. After excluding this study, eight studies of 2641 patients remained for reanalysis. This reanalysis showed a statistically significant association between CCND1 amplification and worse OS (HR: 1.51, 95% CI: 1.19-1.92, p = 0.0008) with low heterogeneity (I2 = 43%, p = 0.09) (Figure 3B).
There were four studies, comprised of 1083 patients, that reported CCND1 amplification and RFS in patients on endocrine therapy. Patients with CCND1 amplification had significantly shorter RFS whilst on endocrine therapy than those without amplification (HR: 2.00, 95% CI: 1.12-3.58, p=0.02) with high heterogeneity (I2 = 72%, p=0.01) (Figure 4A). One of the studies of 226 patients was deemed as having a high RoB, and their exclusion resulted in a greater effect (HR: 2.59, 95% CI: 1.96-3.41, p<0.00001) with low heterogeneity (I2 = 0%, p = 0.90) (Figure 4A). There were two studies, comprised of 694 patients, that reported CCND1 amplification and OS in patients on endocrine therapy, and these were assessed as having a low RoB. Patients with CCND1 amplification had significantly shorter OS whilst on endocrine therapy than those without (HR: 1.59, 95% CI: 1.00-2.49, p = 0.05) with low heterogeneity (I2 = 0%, p = 0.36) (Figure 4B).
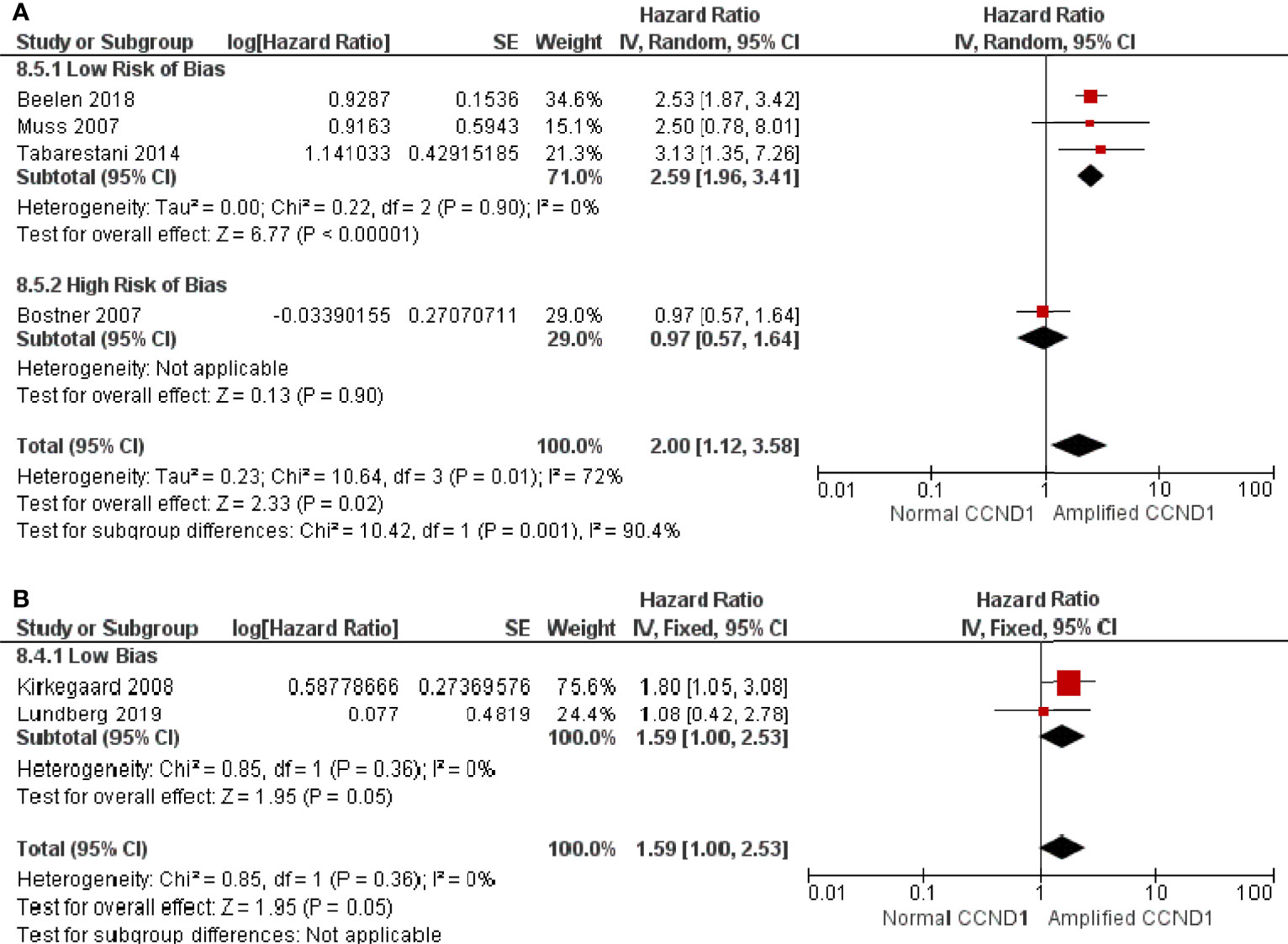
Figure 4 Forrest plot of hazard ratios for CCND1 amplification and worse relapse free survival and overall survival of endocrine therapy breast cancer patients.(A) CCND1 Amplification and relapse free survival (B) CCND1 Amplification and overall survival. Z values indicate the magnitude of association, with p-values <0.05 indicating statistically significant association. Red squares indicate hazard ratio, with values >1 indicative of association of the outcome measure (RFS and OS) with CCND1 amplification, with strongest association towards the right of the plot. Black lines either side of squares indicate 95% confidence interval (CI). Size of red boxes is relative to specific study weight with greatest weight given to studies with minimal variance (calculated based on inverse of the variance). Large black diamond represents pooled hazard ratio estimate of the above studies. A random effects approach was taken. SE, Standard Error. Plots were generated in Review Manger.
CCND1 is an oncogene that encodes the protein cyclin D1. Cyclin D1, in conjunction with CDK4/6, promotes progression through the cell cycle from G1 to S phase. Amplification of CCND1 is a proposed marker of poor prognosis, in BCa, and in some studies is associated with ET resistance. Despite several studies investigating the predictive and prognostic value of CCND1 amplification, much ambiguity remains.
This meta-analysis included 18 studies comprising 6400 patients. Of these, 1136 (18%) had CCND1 amplified tumours, which is within the 10-35% range generally reported (2–4). Previous evidence suggested an association between CCND1 amplification and ER positive status (4, 13, 15, 51). Our analysis supports these findings, demonstrating a strong relationship between CCND1 amplification and positive ER status, with low heterogeneity. However, regarding other clinicopathological features, there were no significant relationships between CCND1 amplification and PR or HER2 status, nor with histological grade or tumour stage. Other studies have yielded conflicting results with respect to CCND1 amplification and PR status, HER2 status and histological grade (13, 15, 51–54). Many have reported no association between CCND1 amplification and tumour stage (4, 52–54). A previous meta-analysis found significant association between CCND1 amplification and ER and PR status as well as histologic grade, but no association with HER2 status or stage (4). Whilst this meta-analysis yielded similar results to one by He et al. (4), the included studies differed considerably; this is in part due to the search terms and exclusion criteria used, as well as several studies being published since their review. Our systematic review and metanalysis targeted studies of hormone receptor postmenopausal patients specifically with cohorts consisting of >35 patients and included analysis of CCND1 amplification and cyclin D1 overexpression.
There are several factors that may contribute to differences observed between studies, these may include variations in detection methods, cut off definitions for clinicopathological features and of CCND1 amplification (47), as well as the composition of cohorts in terms of molecular and histological subtypes (54, 55). In this study, analysis of CCND1 amplification in terms of molecular subtypes was not possible as it was not reported for many of the analysed studies, however others have shown CCND1 amplification is more common in the luminal subtype and associated with worse breast cancer specific OS compared to other subtypes (14, 56).
Our analysis also demonstrated that CCND1 amplification is strongly associated with high level of cyclin D1 expression. However, there was high heterogeneity between the studies, which may stem from the different methods of detection and scoring of immunohistochemistry results of cyclin D1 expression, as these were not standardised. Nevertheless, previous studies have noted the association between CCND1 amplification and high levels of cyclin D1 expression (13, 15, 54). Interestingly, one study showed that cyclin D1 immunostaining with an Allred score of >6.5 (57) was predictive of CCND1 amplification in ER positive BCa patients, with high sensitivity (94.2%) and specificity (87.8%) (13). This is somewhat surprising, given that CCND1 amplification occurs in 10-35% of BCa tumours (Table 1), and cyclin D1 overexpression occurs in 50-70% of BCa tumours (2–4).
Another study found that CCND1 amplification and cyclin D1 overexpression correlated in ER positive but not ER negative BCa (58). This may be indicative of a positive feedback loop between cyclin D1 and ER, initiated by CCND1 amplification; cyclin D1 is known to stimulate ER transcriptional activity, whilst the ER forms a complex with Nuclear Factor Kappa B (NF-κB) and the cofactor, Rac Family Small GTPase 3 (RAC3) to promote CCND1 transcription (59). It has been found that high levels of Cyclin D1 mRNA was associated with positive ER status (24). Under this model, ER likely promotes expression of cyclin D1, which is further augmented by CCND1 amplification, accounting for the higher prevalence of cyclin D1 overexpression than amplification (22).
Regardless, CCND1 amplification retains its value as the preferred prognostic marker over cyclin D1 overexpression, due to several contradictory findings regarding the prognostic value of cyclin D1 mRNA and protein overexpression. There are several possible explanations for these discrepancies, i) differences between molecular subtypes, ii) treatment regime iii) mechanisms underlying cyclin D1 overexpression, iv) methodological differences. Because there is greater evidence for the CCND1 amplification as a prognostic maker, it has been suggested that studies are needed that consider patients with CCND1 amplification separately to those with cyclin D1 overexpression in the absence of amplification, in a well-defined cohort (60). Indeed, one study segregated patients into three groups; i) unamplified CCND1, cyclin D1 overexpression, ii) normal CCND1 and iii) CCND1 amplified and cyclin D overexpressed; they showed that patients with CCND1 amplification and cyclin D1 overexpression had worse RFS, whilst those with cyclin D1 overexpression but unamplified had good RFS, compared to those with normal CCND1 (22). This study demonstrated that there are important differences between these groups, that remain to be fully elucidated. Thus, the current evidence favours CCND1 amplification as the preferred prognostic marker over cyclin D1 transcript and protein expression. Additionally, our focus on CCDN1 amplification rather than transcript or protein is also based on practical considerations: with improving molecular technologies CCDN1 amplification testing could easily be moved into diagnostic settings as economic fast turnaround assay.
CCDN1 amplification is a proposed mechanism of resistance to ET. To examine this further, we analysed the effect of CCND1 amplification status on RFS and OS in ET treated patients. In these patients, CCND1 amplification was significantly associated with shorter RFS and OS. Removal of high RoB studies resulted in a stronger association between CCND1 amplification and RFS in ET treated patients. One of the limitations for these analyses is the number of studies which reported type of treatment with RFS and OS. The overall analysis for ET and RFS contained four studies, and this was further reduced to just two after exclusion of those assessed as having high RoB. Additionally, majority of studies in the ET RFS analysis reported results for treatment with tamoxifen alone, and hence may be biased towards tamoxifen treatment specifically. Comparison of different types of ET in terms of CCND1 amplification and RFS and OS, was not possible as majority of studies reported on tamoxifen only, and studies of other ETs were lacking. In fact, just one study (14) reported on ET generally, the others all focused on patients treated with tamoxifen. However, our findings agree with previous studies that reported CCND1 amplification as predictive of poor prognosis in patients treated with ET, including tamoxifen (15, 61) and AIs (12, 14, 19). However, these are not altogether unanimous; one study found that co-amplification of CCND1 and EMSY predicted tamoxifen resistance (62) whereas another found that CCND1 amplification was predictive of poor response to AIs but not tamoxifen (21). As mechanisms of ET action differ, it is possible that CCND1 amplification may contribute to resistance in some treatments but not others (63). However, differences may also be due to biological and chance differences between the cohorts, and further investigation is required to fully elucidate differences between subgroups of ET.
To explore the prognostic potential of CCND1 amplification, we compared duration of RFS and OS between patients with CCND1 amplified tumours and those without. In the analysis, including all eligible studies, we found that CCND1 amplification was significantly associated with shorter RFS but not OS. In both analyses there was a high degree of heterogeneity, and this could potentially be attributed to a range of variables. For example, RoB assessment indicated that multiple studies did not always report RFS and OS definitions or follow up times. Additionally, several studies failed to compare histological grade and tumour stage with CCND1 amplification. In support of this, removal of studies deemed as having a high RoB, yielded statistically significant association between CCND1 amplification and both RFS and OS, and reduced heterogeneity particularly in the case of OS.
There are some important limitations of the present study that should be considered. One of these is the variation amongst methods used to define CCND1 amplification and the cut off values. For example, many studies set cut offs based on a reference probe, with reference probes differing between studies, whilst others may select arbitrarily based on number of signals or the proportion of cells with positive signals. This has the potential to influence results in either direction depending on sensitivity of the assay, and on how cut offs were determined. Due to the wide variety of methods, and the overall number of studies included in this meta-analysis, it was not possible to conduct subgroup analyses between the different methods employed. Secondly, treatment regimens were generally poorly reported, making it difficult to compare amplification status with treatment responses. Thirdly, definitions of survival statistics differed in some studies, or were not reported. Fourthly, some studies only provided HR and 95% CI for subgroups of patients in which there was statistical significance; such underreporting of non-significant results, may have led to a bias in favour of associations between CCND1 and survival outcomes. Lastly, of the 18 studies included in our analysis, just three were rated as having low RoB across all six domains for quality assessment.
A major challenge with interpretation of data is due to the variation of methods used to detect CCND1 amplification, and variations in the cut-off values. Amplification detection methods include: southern blot, FISH, CISH, silver in situ hybridisation (SISH) PCR based (qPCR, quantitative fluorescence PCR, multiplex ligation-dependent probe amplification and droplet digital PCR), targeted sequencing, array comparative genomic hybridisation, and next generation sequencing (64). FISH was the most common detection method used in the literature, but there are potentially better methods. Increasingly, bright field ISH, CISH and SISH, are becoming the preferred method of amplification detection, owing to their increased resolution, and ability to simultaneously view gene amplification and tissue morphology (65). Currently, in the clinical setting, CISH and SISH are the preferred methods of HER2 amplification detection, with FISH used only in challenging cases (66). Current methods to define CCND1 amplification have not been taken up into routine diagnostic settings and development of better assays for rapid, reliable, economic, standardised CCND1 amplification testing are needed to harness the value of this prognostic and predictive biomarker.
In conclusion, our meta-analysis demonstrated that CCND1 amplification is significantly associated with positive ER status and cyclin D1 overexpression. CCND1 amplification was also predictive of both shorter RFS, and OS, in ET treated patients. As a prognostic biomarker, our meta-analysis indicated that CCND1 amplification may be effective in predicting shorter RFS and OS, after quality assessment. The lack of a standardised method of CCND1 amplification detection remains a considerable limitation, warranting future investigations aimed at establishing a standardised approach.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Conceptualisation, SJ; literature search and screening, SJ and BP; data collection SJ; risk of bias assessment, SJ and BP; Figures and tables, SJ; statistical analyses, SJ; drafting, SJ, BP, and TB; critical review of work, BP, SK, PS, HN, PdS, and TB. All authors contributed to the article and approved the submitted version.
This work was supported by a grant (13/TRC/1-01) from the Cancer Institute NSW through the CONCERT Translational Cancer Research Centre, SJ is a recipient of an Ingham Institute PhD Scholarship, generated by the Liverpool Catholic Club. BP is funded through a Clinical Academic Group Seed Grant from the Sydney Partnership for Health, Education, Research and Enterprise (SPHERE).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to acknowledge the librarians at Western Sydney University who assisted with document delivery requests.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.895729/full#supplementary-material
1. Hanahan D, Weinberg Robert A. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
2. Azarnezhad A, Tabrizi M, Javan F, Mehdipour P. Detection of CCND1 , C-MYC , and FGFR1 Amplification Using Modified SYBR Green qPCR and FISH in Breast Cancer. Turk J Med Sci (2018) 48(4):759–67. doi: 10.3906/sag-1710-93
3. Elsheikh S, Green AR, Aleskandarany MA, Grainge M, Paish CE, Lambros MB, et al. CCND1 Amplification and Cyclin D1 Expression in Breast Cancer and Their Relation With Proteomic Subgroups and Patient Outcome. Breast Cancer Res Treat (2008) 109(2):325–35. doi: 10.1007/s10549-007-9659-8
4. He Q, Wu J, Liu XL, Ma YH, Wu XT, Wang WY, et al. Clinicopathological and Prognostic Significance of Cyclin D1 Amplification in Patients With Breast Cancer: A Meta-Analysis. J BUON (2017) 22(5):1209–16.
5. Topacio BR, Zatulovskiy E, Cristea S, Xie S, Tambo CS, Rubin SM, et al. Cyclin D-Cdk4,6 Drives Cell-Cycle Progression via the Retinoblastoma Protein's C-Terminal Helix. Mol Cell (2019) 74(4):758–70.e4. doi: 10.1016/j.molcel.2019.03.020
6. Jeffreys SA, Powter B, Balakrishnar B, Mok K, Soon P, Franken A, et al. Endocrine Resistance in Breast Cancer: The Role of Estrogen Receptor Stability. Cells (2020) 9(9):2077. doi: 10.3390/cells9092077
7. Sobhani N, D’Angelo A, Pittacolo M, Roviello G, Miccoli A, Corona SP, et al. Updates on the CDK4/6 Inhibitory Strategy and Combinations in Breast Cancer. Cells (2019) 8(4):321. doi: 10.3390/cells8040321
8. Neuman E, Ladha MH, Lin N, Upton TM, Miller SJ, DiRenzo J, et al. Cyclin D1 Stimulation of Estrogen Receptor Transcriptional Activity Independent of Cdk4. Mol Cell Biol (1997) 17(9):5338–47. doi: 10.1128/MCB.17.9.5338
9. Yang C, Chen L, Li C, Lynch MC, Brisken C, Schmidt EV. Cyclin D1 Enhances the Response to Estrogen and Progesterone by Regulating Progesterone Receptor Expression. Mol Cell Biol (2010) 30(12):3111–25. doi: 10.1128/MCB.01398-09
10. Cicatiello L, Addeo R, Sasso A, Altucci L, Petrizzi VB, Borgo R, et al. Estrogens and Progesterone Promote Persistent CCND1 Gene Activation During G1 by Inducing Transcriptional Derepression via C-Jun/c-Fos/estrogen Receptor (Progesterone Receptor) Complex Assembly to a Distal Regulatory Element and Recruitment of Cyclin D1 to Its Own Gene Promoter. Mol Cell Biol (2004) 24(16):7260–74. doi: 10.1128/MCB.24.16.7260-7274.2004
11. Nadji M, Gomez-Fernandez C, Ganjei-Azar P, Morales AR. Immunohistochemistry of Estrogen and Progesterone Receptors Reconsidered: Experience With 5,993 Breast Cancers. Am J Clin Pathol (2005) 123(1):21–7. doi: 10.1309/4WV79N2GHJ3X1841
12. Giltnane JM, Hutchinson KE, Stricker TP, Formisano L, Young CD, Estrada MV, et al. Genomic Profiling of ER(+) Breast Cancers After Short-Term Estrogen Suppression Reveals Alterations Associated With Endocrine Resistance. Sci Trans Med (2017) 9(402). doi: 10.1126/scitranslmed.aai7993
13. Li Z, Cui J, Yu Q, Wu X, Pan A, Li L. Evaluation of CCND1 Amplification and CyclinD1 Expression: Diffuse and Strong Staining of CyclinD1 Could Have Same Predictive Roles as CCND1 Amplification in ER Positive Breast Cancers. Am J Trans Res (2016) 8(1):142–53.
14. Lundberg A, Lindström LS, Li J, Harrell JC, Darai-Ramqvist E, Sifakis EG, et al. The Long-Term Prognostic and Predictive Capacity of Cyclin D1 Gene Amplification in 2305 Breast Tumours. Breast Cancer Res (2019) 21(1):34. doi: 10.1186/s13058-019-1121-4
15. Roy PG, Pratt N, Purdie CA, Baker L, Ashfield A, Quinlan P, et al. High CCND1 Amplification Identifies a Group of Poor Prognosis Women With Estrogen Receptor Positive Breast Cancer. Int J Cancer (2010) 127(2):355–60. doi: 10.1002/ijc.25034
16. Haque MM, Desai KV. Pathways to Endocrine Therapy Resistance in Breast Cancer. Front Endocrinol (Lausanne) (2019) 10:573. doi: 10.3389/fendo.2019.00573
17. Ma CX, Sanchez CG, Ellis MJ. Predicting Endocrine Therapy Responsiveness in Breast Cancer. Oncology (2009) 23(2):133.
18. Giltnane JM, Hutchinson KE, Stricker TP, Formisano L, Young CD, Estrada MV, et al. Genomic Profiling of ER+ Breast Cancers After Short-Term Estrogen Suppression Reveals Alterations Associated With Endocrine Resistance. Sci Trans Med (2017) 9(402):eaai7993. doi: 10.1126/scitranslmed.aai7993
19. Lundgren K, Brown M, Pineda S, Cuzick J, Salter J, Zabaglo L, et al. Effects of Cyclin D1 Gene Amplification and Protein Expression on Time to Recurrence in Postmenopausal Breast Cancer Patients Treated With Anastrozole or Tamoxifen: A TransATAC Study. Breast Cancer Res (2012) 14(2):R57. doi: 10.1186/bcr3161
20. Bostner J, Ahnström Waltersson M, Fornander T, Skoog L, Nordenskjöld B, Stål O. Amplification of CCND1 and PAK1 as Predictors of Recurrence and Tamoxifen Resistance in Postmenopausal Breast Cancer. Oncogene (2007) 26(49):6997–7005. doi: 10.1038/sj.onc.1210506
21. Aleksakhina SN, Kramchaninov MM, Mikushina AD, Kubrina SE, Petkau VV, Ivantsov AO, et al. CCND1 and FGFR1 Gene Amplifications Are Associated With Reduced Benefit From Aromatase Inhibitors in Metastatic Breast Cancer. Clin Trans Oncol (2021) 23(4):874–81. doi: 10.1007/s12094-020-02481-w
22. Bieche I, Olivi M, Nogues C, Vidaud M, Lidereau R. Prognostic Value of CCND1 Gene Status in Sporadic Breast Tumours, as Determined by Real-Time Quantitative PCR Assays. Br J Cancer (2002) 86(4):580–6. doi: 10.1038/sj.bjc.6600109
23. Aaltonen K, Amini R-M, Landberg G, Eerola H, Aittomäki K, Heikkilä P, et al. Cyclin D1 Expression Is Associated With Poor Prognostic Features in Estrogen Receptor Positive Breast Cancer. Breast Cancer Res Treat (2009) 113(1):75–82. doi: 10.1007/s10549-008-9908-5
24. Kenny FS, Hui R, Musgrove EA, Gee JMW, Blamey RW, Nicholson RI, et al. Overexpression of Cyclin D1 Messenger RNA Predicts for Poor Prognosis in Estrogen Receptor-Positive Breast Cancer. Clin Cancer Res (1999) 5(8):2069–76.
25. Xu X-L, Chen S-Z, Chen W, Zheng W-H, Xia X-H, Yang H-J, et al. The Impact of Cyclin D1 Overexpression on the Prognosis of ER-Positive Breast Cancers: A Meta-Analysis. Breast Cancer Res Treat (2013) 139(2):329–39. doi: 10.1007/s10549-013-2563-5
26. Ortiz AB, Garcia D, Vicente Y, Palka M, Bellas C, Martin P. Prognostic Significance of Cyclin D1 Protein Expression and Gene Amplification in Invasive Breast Carcinoma. PLoS One (2017) 12(11):e0188068. doi: 10.1371/journal.pone.0188068
27. van Diest PJ, Michalides RJ, Jannink L, van der Valk P, Peterse HL, de Jong JS, et al. Cyclin D1 Expression in Invasive Breast Cancer. Correlations and Prognostic Value. Am J Pathol (1997) 150(2):705–11.
28. Montalto FI, De Amicis F. Cyclin D1 in Cancer: A Molecular Connection for Cell Cycle Control, Adhesion and Invasion in Tumor and Stroma. Cells (2020) 9(12):2648. doi: 10.3390/cells9122648
29. Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan—a Web and Mobile App for Systematic Reviews. Systematic Rev (2016) 5(1):210. doi: 10.1186/s13643-016-0384-4
30. Sella T, Ruddy KJ, Carey LA, Partridge AH. Optimal Endocrine Therapy in Premenopausal Women: A Pragmatic Approach to Unanswered Questions. JCO Oncol Practice (2021) 18(3):211–6. doi: 10.1200/OP.21.00482
31. Heer E, Harper A, Escandor N, Sung H, McCormack V, Fidler-Benaoudia MM. Global Burden and Trends in Premenopausal and Postmenopausal Breast Cancer: A Population-Based Study. Lancet Global Health (2020) 8(8):e1027–37. doi: 10.1016/S2214-109X(20)30215-1
32. Carene D, Tran-Dien A, Lemonnier J, Dalenc F, Levy C, Pierga JY, et al. Association Between FGFR1 Copy Numbers, MAP3K1 Mutations, and Survival in Axillary Node-Positive, Hormone Receptor-Positive, and HER2-Negative Early Breast Cancer in the PACS04 and METABRIC Studies. Breast Cancer Res Treat (2020) 179(2):387–401. doi: 10.1007/s10549-019-05462-y
33. Covidence Systematic Review Software, Veritas Health Innovation. Melbourne, Australia. Available at: www.covidence.org.
34. Hayden JA, van der Windt DA, Cartwright JL, Côté P, Bombardier C. Assessing Bias in Studies of Prognostic Factors. Ann Internal Med (2013) 158(4):280–6. doi: 10.7326/0003-4819-158-4-201302190-00009
35. Jermy JE, Copley PC, Poon MT, Demetriades AK. Does Pre-Operative Multifidus Morphology on MRI Predict Clinical Outcomes in Adults Following Surgical Treatment for Degenerative Lumbar Spine Disease? A Systematic Review. Eur Spine J (2020) 29(6):1318–27. doi: 10.1007/s00586-020-06423-6
37. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical Methods for Incorporating Summary Time-to-Event Data Into Meta-Analysis. Trials (2007) 8(1):16. doi: 10.1186/1745-6215-8-16
38. Beelen K, Opdam M, Severson T, Koornstra R, Vincent A, Wesseling J, et al. Mitotic Count can Predict Tamoxifen Benefit in Postmenopausal Breast Cancer Patients While Ki67 Score Cannot. BMC Cancer (2018) 18(1):761. doi: 10.1186/s12885-018-4516-1
39. Cao L, Basudan A, Sikora MJ, Bahreini A, Tasdemir N, Levine KM, et al. Frequent Amplifications of ESR1, ERBB2 and MDM4 in Primary Invasive Lobular Breast Carcinoma. Cancer Lett (2019) 461(7600053):21–30. doi: 10.1016/j.canlet.2019.06.011
40. Hadzisejdić I, Mustać E, Jonjić N, Petković M, Grahovac B. Nuclear EGFR in Ductal Invasive Breast Cancer: Correlation With Cyclin-D1 and Prognosis. Modern Pathol (2010) 23(3):392–403. doi: 10.1038/modpathol.2009.166
41. Kirkegaard T, Nielsen KV, Jensen LB, Campbell FM, Müller S, Tovey SM, et al. Genetic Alterations of CCND1 and EMSY in Breast Cancers. Histopathology (2008) 52(6):698–705. doi: 10.1111/j.1365-2559.2008.03007.x
42. Massidda B, Sini M, Budroni M, Atzori F, Deidda M, Pusceddu V, et al. Molecular Alterations in Key-Regulator Genes Among Patients With T4 Breast Carcinoma. BMC Cancer (2010) 10(1):458. doi: 10.1186/1471-2407-10-458
43. Muss HB, Bunn JY, Crocker A, Plaut K, Koh J, Heintz N, et al. Cyclin D-1, Interleukin-6, HER-2/Neu, Transforming Growth Factor Receptor-II and Prediction of Relapse in Women With Early Stage, Hormone Receptor-Positive Breast Cancer Treated With Tamoxifen. Breast J (2007) 13(4):337–45. doi: 10.1111/j.1524-4741.2007.00440.x
44. Plevova P, Cerna D, Balcar A, Foretova L, Zapletalova J, Silhanova E, et al. CCND1 and ZNF217 Gene Amplification is Equally Frequent in BRCA1 and BRCA2 Associated and non-BRCA Breast Cancer. Neoplasma (2010) 57(4):325–32. doi: 10.4149/neo_2010_04_325
45. Quintayo MA, Munro AF, Thomas J, Kunkler IH, Jack W, Kerr GR, et al. Gsk3β and Cyclin D1 Expression Predicts Outcome in Early Breast Cancer Patients. Breast Cancer Res Treat (2012) 136(1):161–8. doi: 10.1007/s10549-012-2229-8
46. Serino LTR, Jucoski TS, Morais SB, Fernandes CCC, Lima RS, Urban CA, et al. Association of FOSL1 Copy Number Alteration and Triple Negative Breast Tumors. Genet Mol Biol (2019) 42(1):26–31. doi: 10.1590/1678-4685-gmb-2017-0267
47. Seshadri R, Lee CS, Hui R, McCaul K, Horsfall DJ, Sutherland RL. Cyclin DI Amplification is Not Associated With Reduced Overall Survival in Primary Breast Cancer But may Predict Early Relapse in Patients With Features of Good Prognosis. Clin Cancer Res (1996) 2(7):1177–84.
48. Tabarestani S, Ghaderian SM, Rezvani H, Mirfakhraie R, Ebrahimi A, Attarian H, et al. Prognostic and Predictive Value of Copy Number Alterations in Invasive Breast Cancer as Determined by Multiplex Ligation-Dependent Probe Amplification. Cell Oncol (Dordrecht) (2014) 37(2):107–18. doi: 10.1007/s13402-013-0165-1
49. Hadzisejdi I, Mustac E, Jonjic N, Petkovic M, Grahovac B. Nuclear EGFR in Ductal Invasive Breast Cancer: Correlation With Cyclin-D1 and Prognosis. Modern Pathol (2010) 23(3):392–403. doi: 10.1038/modpathol.2009.166
50. Tabarestani S, Ghaderian SM, Rezvani H. Detection of Gene Amplification by Multiplex Ligation-Dependent Probe Amplification in Comparison With In Situ Hybridization and Immunohistochemistry. Asian Pacific J Cancer Prev APJCP (2015) 17:7997–8002. doi: 10.7314/APJCP.2015.16.17.7997
51. Courjal F, Louason G, Speiser P, Katsaros D, Zeillinger R, Theillet C. Cyclin Gene Amplification and Overexpression in Breast and Ovarian Cancers: Evidence for the Selection of Cyclin D1 in Breast and Cyclin E in Ovarian Tumors. Int J Cancer (1996) 69(4):247–53. doi: 10.1002/(SICI)1097-0215(19960822)69:4<247::AID-IJC1>3.0.CO;2-X
52. Al-Kuraya K, Schraml P, Torhorst J, Tapia C, Zaharieva B, Novotny H, et al. Prognostic Relevance of Gene Amplifications and Coamplifications in Breast Cancer. Cancer Res (2004) 64(23):8534–40. doi: 10.1158/0008-5472.CAN-04-1945
53. Burandt E, Grünert M, Lebeau A, Choschzick M, Quaas A, Jänicke F, et al. Cyclin D1 Gene Amplification is Highly Homogeneous in Breast Cancer. Breast Cancer (Tokyo Japan) (2016) 23(1):111–9. doi: 10.1007/s12282-014-0538-y
54. Reis-Filho JS, Savage K, Lambros MB, James M, Steele D, Jones RL, et al. Cyclin D1 Protein Overexpression and CCND1 Amplification in Breast Carcinomas: An Immunohistochemical and Chromogenic in Situ Hybridisation Analysis. Modern Pathol (2006) 19(7):999–1009. doi: 10.1038/modpathol.3800621
55. Freitag CE, Mei P, Wei L, Parwani AV, Li Z. Genetic Alterations and Their Association With Clinicopathologic Characteristics in Advanced Breast Carcinomas: Focusing on Clinically Actionable Genetic Alterations. Hum Pathol (2020) 102:94–103. doi: 10.1016/j.humpath.2020.05.005
56. Natrajan R, Weigelt B, Mackay A, Geyer FC, Grigoriadis A, Tan DSP, et al. An Integrative Genomic and Transcriptomic Analysis Reveals Molecular Pathways and Networks Regulated by Copy Number Aberrations in Basal-Like, HER2 and Luminal Cancers. Breast Cancer Res Treat (2010) 121(3):575–89. doi: 10.1007/s10549-009-0501-3
57. Allred D, Harvey JM, Berardo M, Clark GM. Prognostic and Predictive Factors in Breast Cancer by Immunohistochemical Analysis. Modern Pathol (1998) 11(2):155–68.
58. Ahlin C, Lundgren C, Embretsén-Varro E, Jirström K, Blomqvist C, Fjällskog M. High Expression of Cyclin D1 Is Associated to High Proliferation Rate and Increased Risk of Mortality in Women With ER-Positive But Not in ER-Negative Breast Cancers. Breast Cancer Res Treat (2017) 164(3):667–78. doi: 10.1007/s10549-017-4294-5
59. Rubio MF, Werbajh S, Cafferata EGA, Quaglino A, Coló GP, Nojek IM, et al. TNF-α Enhances Estrogen-Induced Cell Proliferation of Estrogen-Dependent Breast Tumor Cells Through a Complex Containing Nuclear Factor-Kappa B. Oncogene (2006) 25(9):1367–77. doi: 10.1038/sj.onc.1209176
60. Tobin NP, Bergh J. Analysis of Cyclin D1 in Breast Cancer: A Call to Arms. Curr Breast Cancer Rep (2012) 4(3):171–3. doi: 10.1007/s12609-012-0083-7
61. Jirström K, Stendahl M, Rydén L, Kronblad A, Bendahl PO, Stål O, et al. Adverse Effect of Adjuvant Tamoxifen in Premenopausal Breast Cancer With Cyclin D1 Gene Amplification. Cancer Research (2005) 65(17):8009–16.
62. Brown LA, Johnson K, Leung S, Bismar TA, Benítez J, Foulkes WD, et al. Co-Amplification of CCND1 and EMSY is Associated With an Adverse Outcome in ER-Positive Tamoxifen-Treated Breast Cancers. Breast Cancer Res Treat (2010) 121(2):347–54. doi: 10.1007/s10549-009-0479-x
63. Todorović-Raković N, Milovanović J, Durosaro SO, Radulovic M. The Prognostic Value of Cyclin D1 in Breast Cancer Patients Treated With Hormonal Therapy: A Pilot Study. Pathol Res Practice (2021) 222:153430. doi: 10.1016/j.prp.2021.153430
64. Pös O, Radvanszky J, Styk J, Pös Z, Buglyó G, Kajsik M, et al. Copy Number Variation: Methods and Clinical Applications. Appl Sci (2021) 11(2):819. doi: 10.3390/app11020819
65. Volpi CC, Gualeni AV, Pietrantonio F, Vaccher E, Carbone A, Gloghini A. Bright-Field in Situ Hybridization Detects Gene Alterations and Viral Infections Useful for Personalized Management of Cancer Patients. Expert Rev Mol Diagn (2018) 18(3):259–77. doi: 10.1080/14737159.2018.1440210
66. Farshid G, Armes JE, Bell R, Cummings M, Fox S, Francis G, et al. Establishment of the Australian In Situ Hybridization Program for the Assessment of HER2 Amplification in Breast Cancer: A Model for the Introduction of New Biomarkers Into Clinical Practice. Diagn Mol Pathol (2010) 19(4):187–93. doi: 10.1097/PDM.0b013e3181e1cc9d
Keywords: breast cancer, CCND1, cyclin D1, biomarker, meta-analysis, systematic review, amplification
Citation: Jeffreys SA, Becker TM, Khan S, Soon P, Neubauer H, de Souza P and Powter B (2022) Prognostic and Predictive Value of CCND1/Cyclin D1 Amplification in Breast Cancer With a Focus on Postmenopausal Patients: A Systematic Review and Meta-Analysis. Front. Endocrinol. 13:895729. doi: 10.3389/fendo.2022.895729
Received: 14 March 2022; Accepted: 10 May 2022;
Published: 17 June 2022.
Edited by:
Leclercq Guy, Université libre de Bruxelles, BelgiumReviewed by:
Rosemary O’Connor, University College Cork, IrelandCopyright © 2022 Jeffreys, Becker, Khan, Soon, Neubauer, de Souza and Powter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah A. Jeffreys, MTkwNTg1MTlAc3R1ZGVudC53ZXN0ZXJuc3lkbmV5LmVkdS5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.