
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 23 May 2022
Sec. Reproduction
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.893854
This article is part of the Research Topic Endocrine and Paracrine Regulation of Spermatogenesis - A Collection of Up to Date Research Contributions on Testis Formation and Function View all 25 articles
 Zhuqing Wang1,2
Zhuqing Wang1,2 Nan Meng3,4
Nan Meng3,4 Yue Wang1,2
Yue Wang1,2 Tong Zhou1
Tong Zhou1 Musheng Li1
Musheng Li1 Shawn Wang1
Shawn Wang1 Sheng Chen1,2
Sheng Chen1,2 Huili Zheng1,2
Huili Zheng1,2 Shuangbo Kong3,4
Shuangbo Kong3,4 Haibin Wang3,4
Haibin Wang3,4 Wei Yan1,2,5*
Wei Yan1,2,5*The X-linked miR-465 cluster is highly expressed in the testis, sperm, newborn ovary, and blastocysts as well as in 8-16 cell embryos. However, the physiological role of the miR-465 cluster is still largely unknown. This study aims to dissect the role of the miR-465 cluster in murine development. Despite abundant expression in the testis, ablation of the miR-465 miRNA cluster using CRISPR-Cas9 did not cause infertility. Instead, a skewed sex ratio biased toward males (60% males) was observed among miR-465 KO mice. Further analyses revealed that the female conceptuses selectively degenerated as early as embryonic day 8.5 (E8.5). Small RNA deep sequencing, qPCR, and in situ hybridization analyses revealed that the miRNAs encoded by the miR-465 cluster were mainly localized to the extraembryonic tissue/developing placenta. RNA-seq analyses identified altered mRNA transcriptome characterized by the dysregulation of numerous critical placental genes, e.g., Alkbh1, in the KO conceptuses at E7.5. Taken together, this study showed that the miR-465 cluster is required for normal female placental development, and ablation of the miR-465 cluster leads to a skewed sex ratio with more males (~60%) due to selective degeneration and resorption of the female conceptuses.
Sexual dimorphism refers to different characteristics beyond the sex organs between the two sexes within the same species, e.g., appearance, structure, behavior, etc. (1). Data from a recent study of 14,250 wild-type (WT) and 40,192 mutant mice suggest that 9.9% of qualitative and 56.6% of quantitative traits display sexual dimorphism (2). Sexual dimorphism commences as early as embryonic development, e.g., X chromosome inactivation in the female embryo. Sexual dimorphism is also reflected by differential gene expression profiles in placental, fetal, and adult tissues (3–5). To date, a role of miRNAs in sexual dimorphism has not been reported although miRNAs are well known to be critical for early development (6–9). miRNAs are ~22 nucleotide small non-coding RNAs that regulate gene expression at post-transcriptional levels (10). Inactivation of either DICER or DROSHA, the two enzymes required for miRNA biogenesis, leads to early embryonic lethality in mice, indicating an essential role of miRNAs in early development (6–9, 11–13). Our previous studies have shown that the X-linked miR-465 cluster, which encodes 6 pre-miRNAs and 12 mature miRNAs, belongs to a large X-linked miR-506 family (14). Their high abundance in the testis, sperm, newborn ovary, blastocysts, and 8-16-cell embryos (14–17) suggests a potential role in gametogenesis and early embryonic development in mice. However, their physiological role has not been investigated in vivo. Here, we report that the miR-465 cluster miRNAs are also abundantly expressed in the developing placenta, and ablation of the miR-465 cluster does not affect fertility but causes a skewed sex ratio favoring males due to selective degradation of the female placenta during early embryonic development.
The animal use protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Nevada, Reno (Protocol# 00494). Generation of global miR-465 KO mice and mouse genotyping were performed as described (14, 18, 19). gRNA and genotyping primers are listed in Table S1.
DNA and RNA were extracted from WT and KO embryos using the mirVana™ miRNA Isolation Kit as previously described (19). The sexes of the conceptuses were determined based on PCR amplification of DYzEms3 (a Y chromosome-specific repetitive sequence) and Rn18s (a housekeeping transcript as the internal control). Males display two bands (DYzEms3 and Rn18s), while females show only one band (Rn18s). Large RNA libraries were constructed using KAPA Stranded RNA-Seq Kits with RiboErase (Cat. # 07962282001, Roche) according to the manufacturer’s instructions. Small RNA libraries were constructed using NEBNext® Small RNA Library Prep Set for Illumina® (Cat. # E7330L, NEB) according to the manufacturer’s instructions. miRNA qPCR was performed as described (14). All oligos for sex determination and qPCR are listed in Table S1.
Cryosections (10 μm) were adhered to poly-L-lysine-coated slides and fixed in 4% paraformaldehyde (Cat. # P6148, Sigma-Aldrich) solution in PBS for 1 h at room temperature. The sections were then washed 3 times in PBS for 5 min each, acetylated for 10 minutes (0.25% acetic anhydride), washed 2 times in PBS for 5 min each, and hybridized with DIG-labeled probes overnight at 50°C. Hybridization buffer contained 1X salts (200 mM NaCl, 13 mM Tris, 5 mM sodium phosphate monobasic, 5mM sodium phosphate dibasic, 5 mM EDTA), 50% formamide, 10% (w/v) dextran sulfate, 1 mg/ml yeast tRNA (Cat. # 10109509001, Roche), 1×Denhardt’s [1% (w/v) bovine serum albumin, 1% (w/v) Ficoll, 1% (w/v) polyvinylpyrrolidone], and RNA probe (final concentration: 1 μM). Post-hybridization washes were followed by an RNase treatment (20 μg/ml RNase A). After blocking in 20% heat-inactivated sheep serum (Cat. # ZLI-9021, Beijing Zhongshan Jinqiao Biotechnology Company) and 2% blocking reagent (Cat. # 12039672910, Roche) for 1 h, sections were incubated overnight in blocking solution containing anti-DIG antibody (1:2500 dilution; Cat. # 11093274910, Roche) at room temperature. After washing, the color was developed using NBT/BCIP according to the manufacturer’s instructions (NBT: Cat. # N1332, Gentihold; BCIP: Cat. # B1360, Gentihold). Sections were counterstained in Nuclear Fast Red (Cat. # G1321, Solarbio), dehydrated in gradient alcohol, cleared in xylene, and mounted in neutral resins. All oligos used for RNA ISH were listed in Table S1.
The Sailfish (20) and SPORTS1.0 (21) pipelines were used to quantify the large RNA expression and small RNA expression, respectively. Transcript per million reads (TPM) was used as the unit of gene expression level. Groupwise differential expression was estimated by the likelihood ratio test and the RNAs with a false discovery rate < 5% were deemed differentially expressed.
Luciferase assays were performed as described (22). cel-mir-67 was used as a negative control. Renilla luciferase signals were normalized to Firefly luciferase signals to correct the transfection efficiency. All oligos for constructing 3’UTR luciferase vectors are listed in Table S4.
Data are presented as mean ± SEM, and statistical differences between datasets were assessed by two samples t-test unless stated otherwise. p < 0.05, 0.01, 0.001, and 0.0001 are considered statistically significant and indicated with *, **, ***, and ****, respectively.
The miR-465 cluster consists of 6 miRNA genes encompassing a ~16.4 kb region on the X chromosome in mice (Figure 1A). Although 6 pre-miRNAs and 12 mature miRNAs are produced in mice, only 6 mature miRNAs can be distinguished based on their sequences, including miR-465a-5p, miR-465b-5p, miR-465c-5p, miR-465d-5p, miR-465a/b/c-3p and miR-465d-3p (Figure 1A). The miR-465 cluster has orthologs in humans, monkeys, and chimpanzees, which have been annotated as miR-892b in the miRBase and contain some U-to-C or A-to-G substitutions (Figure 1B). Like the miR-465 cluster, miR-892b is also flanked by Slitrk2 and Fmr1 on the X chromosome (14). To define their physiological roles, we deleted the entire miR-465 cluster in the mouse genome using CRISPR-Cas9 (Figures 1A; S1A), as previously described (14, 18, 19). PCR genotyping and Sanger sequencing confirmed that the genomic loci of these miRNAs were successfully deleted (Figures S1B, C). The miR-465 KO mice were fertile with normal testis size (Figure 1C). Both the litter size (8.4 ± 0.85, n=35) and litter interval (25.4 ± 0.86, n=34) of the KO mice were comparable to those of WT controls (Litter size: 8.6 ± 1.59, n=23; litter interval: 26.6 ± 1.42, n=22) (Figure 1D), suggesting that these miRNAs are dispensable for both spermatogenesis and folliculogenesis. Interestingly, unlike the equal distribution of the two sexes (~50%) among pups from the WT breeding pairs (+/Y × +/+), the sex ratio is significantly skewed toward the male (61%, p<0.05) among the miR-465 KO pups derived from the homozygous breeding pairs (-/Y × -/-) (Figure 1D). Of interest, ~60% appears to be the most common skewed sex ratio observed in previous reports (Table S2) (23–28).
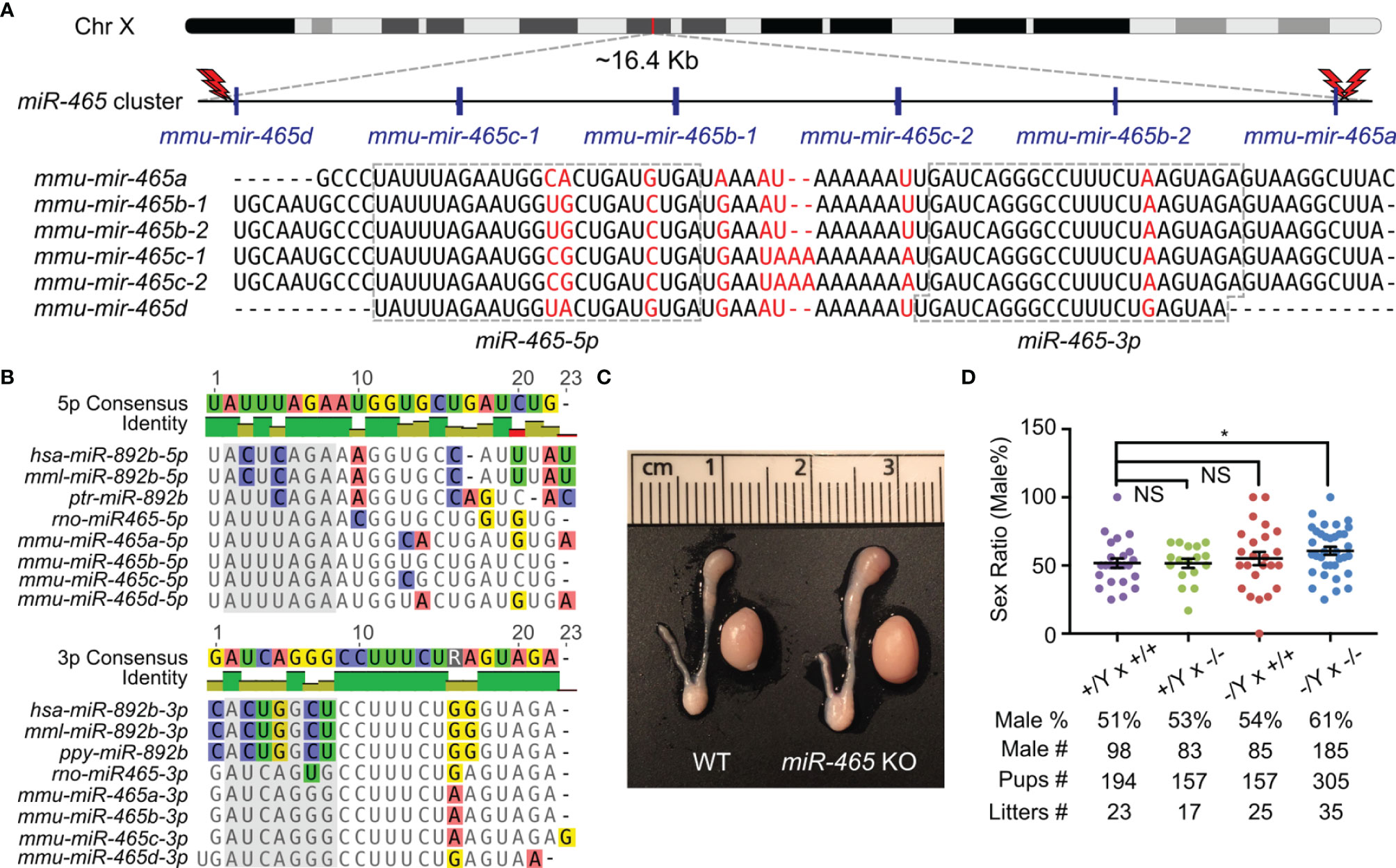
Figure 1 Generation of miR-465 KO mice. (A) The genomic location and sequences of the miR-465 cluster on the X chromosome of mice. The red lightning bolts represent the gRNAs used, and their right and left orientations indicate the reverse and forward strands targeted by the gRNAs, respectively. (B) The orthologs of the miR-465 cluster in primates and rodents. Bases highlighted in the grey background are the potential seed regions. (C) A representative image of the testis and epididymis of WT and miR-465 cluster KO mice. One unit on the ruler is 1 mm. (D) The sex ratios among pups from different breeding schemes. *, p<0.05; NS, statistically not significant.
The skewed sex ratio could result from either a distorted X/Y sperm ratio or a loss of female embryos/fetuses during development. If the sex ratio is already skewed in X/Y sperm, the bias should be observed among pups from the breeding pairs of KO males (-/Y) and WT females (+/+), but not in those from the breeding pairs of WT males (+/Y) and homozygous KO females (-/-). However, the sex ratio among the pups from the -/Y × +/+ breeding pairs was slightly, but not significantly, skewed toward males (54%) (Figure 1D), suggesting that the significantly skewed sex ratio likely occurs during development. To identify when the skewed sex ratio occurs, we collected early embryos at E3.5, E7.5, and E10.5 in the homozygous breeding pairs (-/Y × -/-). Males accounted for ~50% among all of the KO embryos at E3.5 and E7.5, whereas the ratio of the males increased to ~61% at E10.5 (Figure 2A), suggesting that some female embryos are lost between E7.5 and E10.5. Indeed, we observed that on average 1-2 conceptuses per uterus were either being resorbed or had already been resorbed between E8.5 and E10.5. More intriguingly, 6 out of 7 conceptuses that appeared to degenerate were all female KOs (Figures 2B, C). Together, these data suggest that inactivation of the miR-465 cluster leads to selective degeneration and absorption of female conceptuses between E7.5 and E10.5.
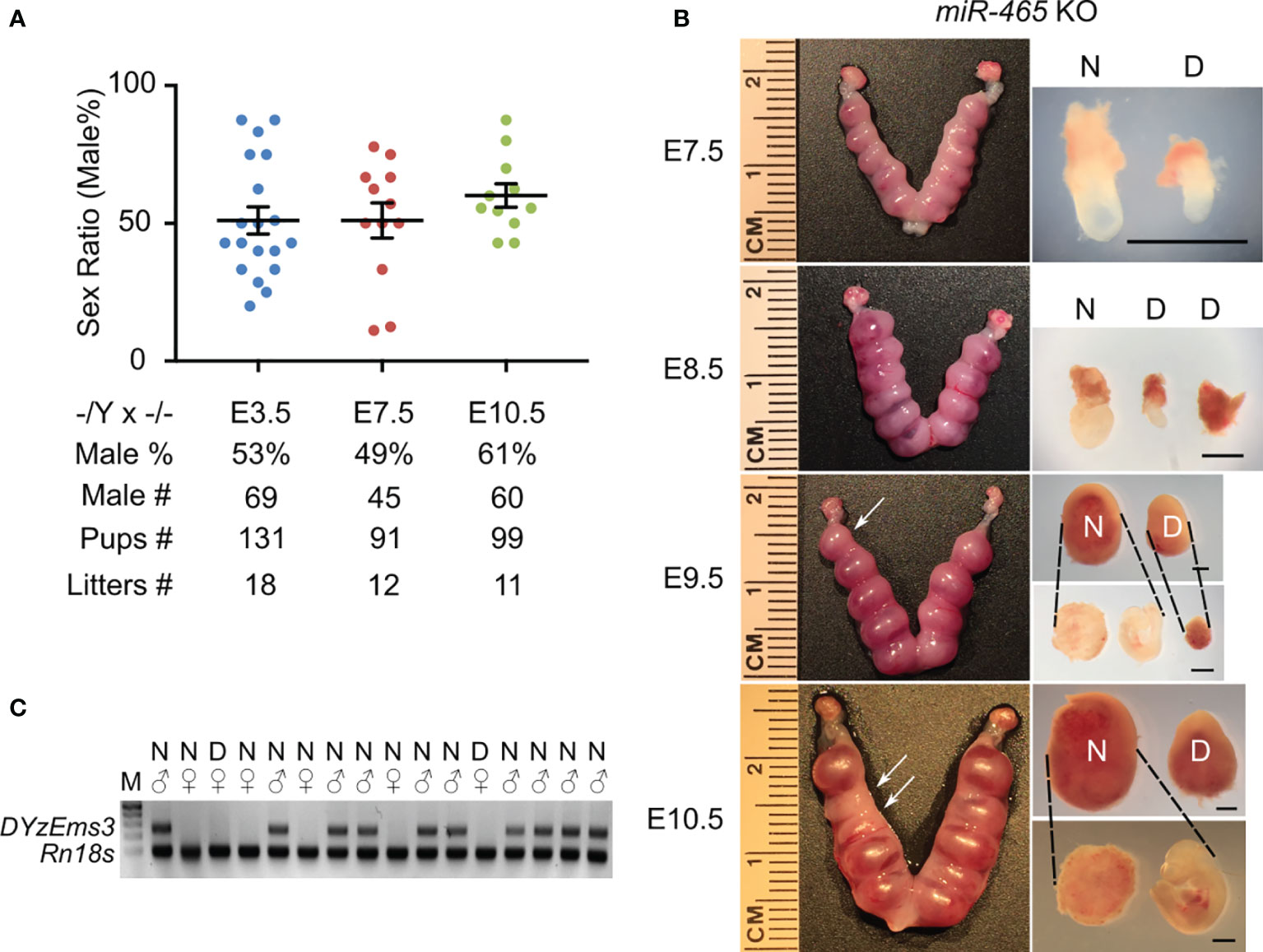
Figure 2 The phenotype of miR-465 KO mice. (A) The sex ratios of pups from homozygous inbreeding (-/Y × -/-) at E3.5, E7.5, and E10.5. (B) Representative images of the KO uteri and conceptuses collected between E7.5 and E10.5. Arrows point to the degenerating/degenerated conceptuses (D) among the normal-looking (N) ones. Scale bars = 1mm. (C) A representative gel image of genotyping results. DYzEms3, a Y-linked genomic fragment, was amplified to identify male conceptuses, and Rn18s, which encodes 18s ribosomal RNA, was used as a loading control in the PCR-based genotyping analyses.
Although the loss of the miR-465 cluster leads to female-biased lethality, it remains unknown whether the primary defects lie in the embryos or the extraembryonic/placental tissue. To address this question, we collected both WT and KO embryos and extraembryonic/placental tissues from both sexes at E7.5 and E10.5 and performed small RNA sequencing (sRNA-seq) (Figures 3A–C; S2). sRNA-seq data confirmed that the miR-465 cluster miRNAs were indeed absent in the KO embryos and extraembryonic/placental tissues (Figures 3A; S2A–C). While no significant sex differences in miRNA levels were observed in WT embryos and extraembryonic tissues at E7.5 (Figures S2D, E), the miR-465 cluster miRNAs were predominantly expressed in extraembryonic tissues, as compared to embryos of both sexes at E7.5 (Figures 3B; S2F), and these miRNAs were significantly downregulated from E7.5 to E10.5 (Figures 3C; S2G–I). Indeed, the TaqMan real-time PCR analyses further confirmed the sRNA-seq results (Figure 3D). We next further performed miRNA in situ hybridization (ISH) assays (Figure 3E) to corroborate the cellular localization of the miR-465 cluster. Consistent with the sRNA-seq and qPCR data, miRNA ISH results showed that the miR-465 cluster miRNAs were predominantly expressed in extraembryonic tissues, especially in the ectoplacental core and chorion (Figure 3E). Although the miR-465 cluster miRNAs were also detected in maternal decidua (Figure 3E), potential decidual defects are highly unlikely based on our breeding data showing normal sex ratio among offspring of the +/Y × -/- breeding pairs (Figure 1D). Given the predominant expression of the miR-465 cluster in the extraembryonic tissues, it is highly likely that the loss of some female embryos was secondary to placental defects.
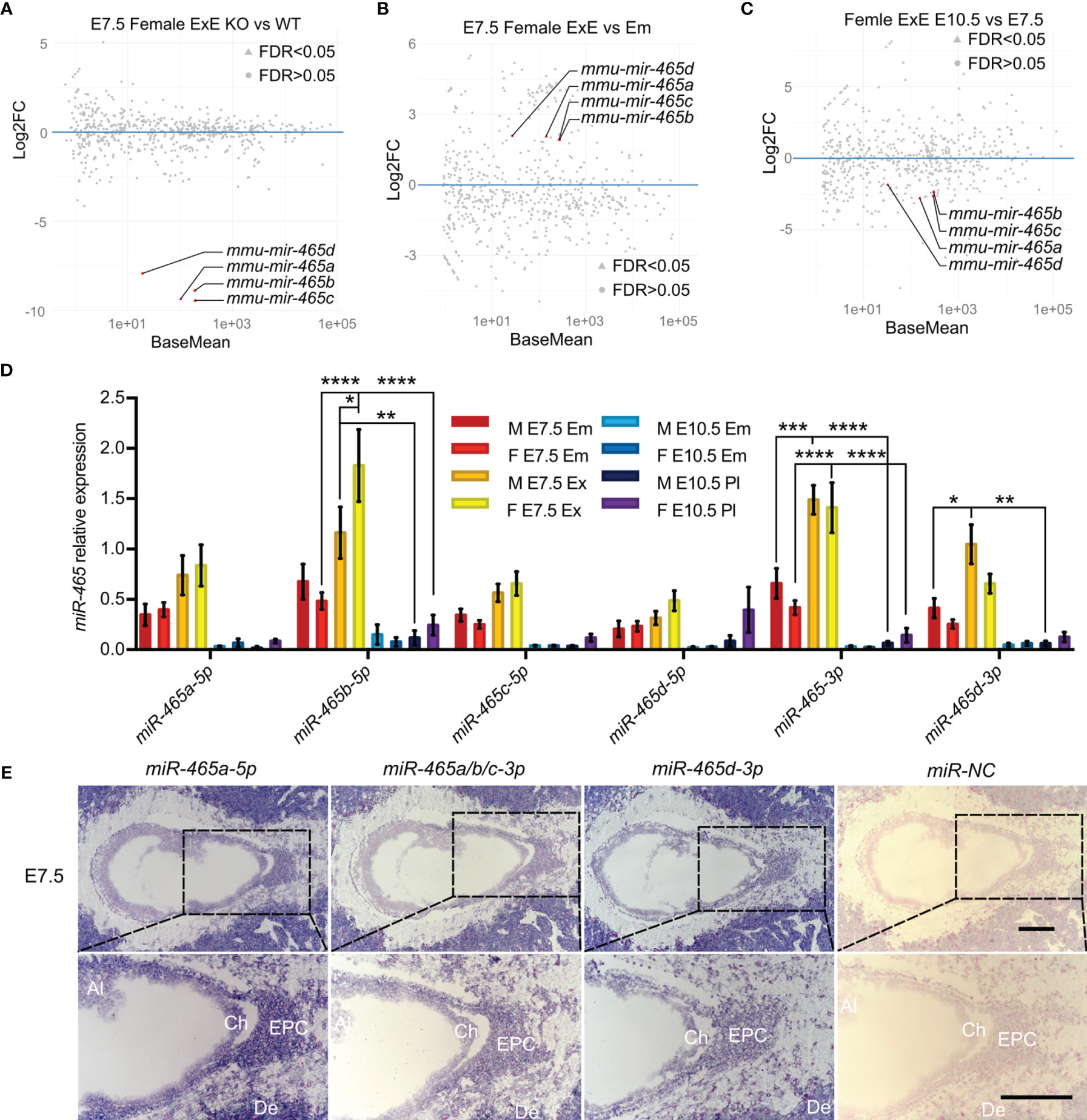
Figure 3 Expression profiles of the miR-465 cluster. (A) Differentially expressed miRNAs between WT and KO female extraembryonic tissues at E7.5. (B) Differentially expressed miRNAs between WT female extraembryonic tissues and embryos at E7.5. (C) Differentially expressed miRNAs between WT female extraembryonic tissues/placentas at E7.5 and E10.5. Data points representing the miR-465 cluster miRNAs are marked in red. sRNA-seq analyses were conducted in biological triplicates (n=3). (D) TaqMan qPCR analyses of expression levels of the miR-465 cluster miRNAs in extraembryonic tissues/placenta and embryos at E7.5 and E10.5. M, male; F, female; Em, embryo; ExE, extraembryonic tissue; Pl, placenta. *, p<0.05; **, p<0.01; *** p<0.001, **** p<0.0001. (E) Representative miRNA-ISH results showing localization of the miR-465 cluster miRNAs in female conceptuses at E7.5. Ch, chorion; EPC, ectoplacental core; Al, allantois; De, decidua. Scale bars = 200 µm.
To identify the targets of the miR-465 cluster miRNAs, we then conducted RNA-seq assays on WT and KO embryos and extraembryonic tissues of both sexes at E7.5. We chose E7.5 because, at this point, despite no obvious degeneration and resorption, the transcriptomic alterations should have accumulated in the implicated female KO conceptuses (Figure 2B). Principal component analyses (PCA) identified two major clusters, each containing either embryos or extraembryonic tissues of both WT and most of the KO of both sexes except for two outliers (Figure 4A). The two outliers turned out to be one female KO embryo and its extraembryonic tissue, suggesting that this conceptus most likely represents a “to-be-degenerating” KO female. While WT and non-degenerating KO embryos and extraembryonic tissues of both sexes displayed similar mRNA transcriptomes (Figure 4B; Table S3), numerous differentially expressed genes (DEGs) were identified between the extraembryonic tissues from the “to-be-degenerating” KO female and those from non-degenerating KO females (Figure 4B; Table S3). Gene ontology (GO) term analyses identified that the DEGs were primarily involved in extraembryonic/placental development (Figure 4C). Among the dysregulated genes responsible for placental development, 8 out of 44 were either imprinted genes or sex-biased genes (Table S4). Luciferase assays further confirmed that some of the dysregulated genes were indeed the targets of the miR-465 cluster miRNAs (Figure S3). Given the similar expression levels of the miR-465 cluster miRNAs in the extraembryonic tissues of both sexes (Figures S2D, E), it is likely that the sexually dimorphic role of the miR-465 cluster is achieved through miRNA-mediated post-transcriptional regulation of the sex-biased target genes. For example, Alkbh1, a target of miR-465 (Figure S3), is a tRNA demethylation enzyme (29) highly expressed in chorion and the ectoplacental cone at E8.5 (30); its ablation also induces female-biased lethality (27).
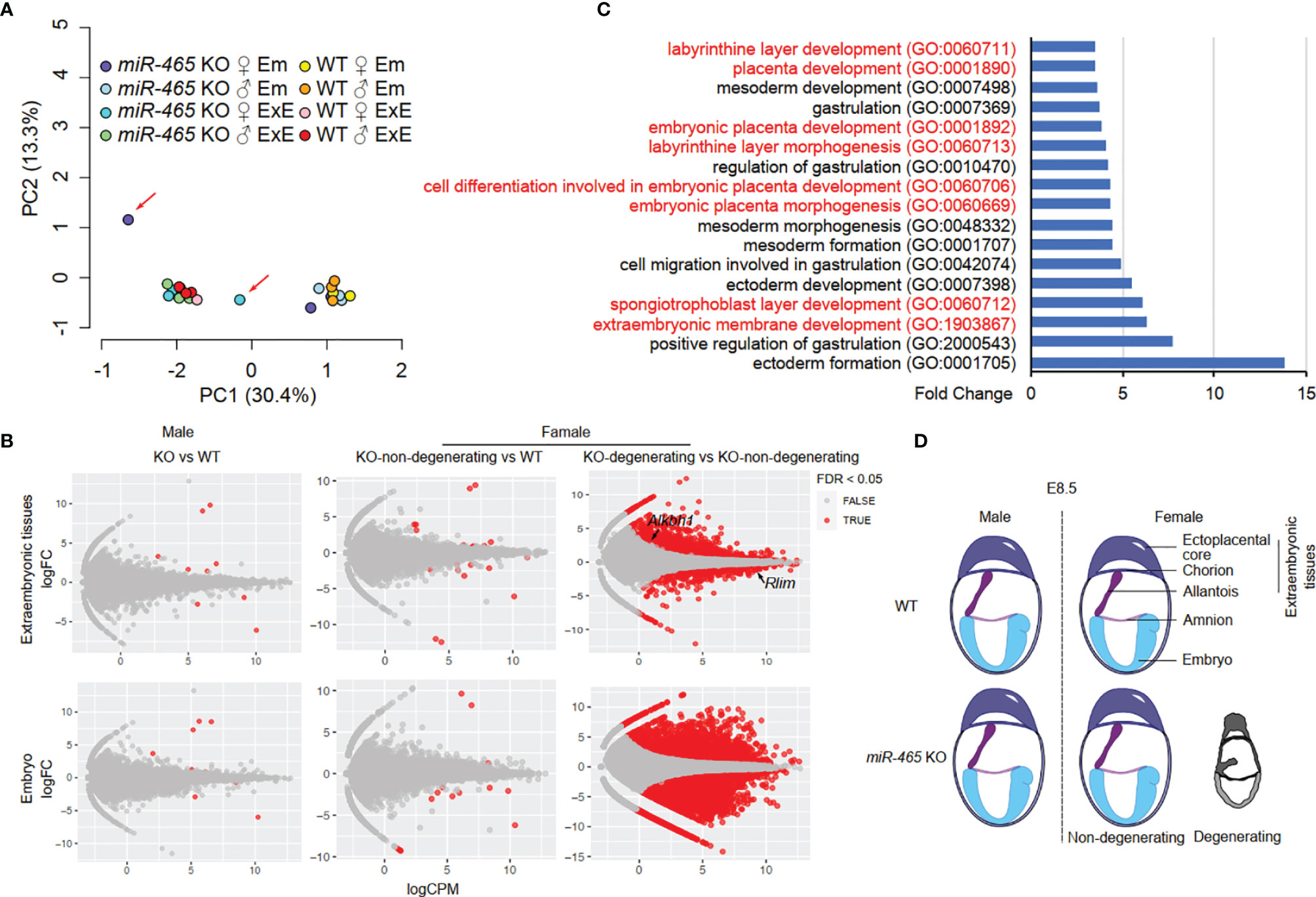
Figure 4 RNA-seq analyses of miR-465 KO and WT conceptuses. (A) Principal component (PC) analyses of RNA-seq data from embryonic (Em) and extraembryonic (ExE) tissues of miR-465 KO and WT mice. The red arrows indicate the degenerating embryo and extraembryonic tissues from a miR-465 KO female conceptus. (B) Differentially expressed genes (DEGs) identified in the following three comparisons: between the miR-465 KO and WT males (left), between the miR-465 KO non-degenerating and WT females (middle), and between miR-465 KO degenerating and non-degenerating females (right). (C) GO term enrichment analyses of DEGs between the degenerating and non-degenerating miR-465 KO female extraembryonic tissues. GO terms related to extraembryonic development are highlighted in red. (D) Schematics showing a critical role of the miR-465 cluster in supporting the full developmental potential of the female placenta and embryos.
The X chromosome is known to be enriched in protein-coding genes critical for reproduction and fertility (31, 32). Our earlier work has also shown that several large miRNA clusters, including the miR-465 cluster, are either exclusively or preferentially expressed in the testis, suggesting a role in controlling spermatogenesis and male fertility (14, 33). Although ablation of the miR-506 cluster compromises the male fertility (14), inactivation of the miR-465 miRNA cluster does not affect either gametogenesis or fertility. Surprisingly, a lack of the miR-465 miRNAs leads to a skewed sex ratio biased toward males due to selective degeneration of the female conceptuses between E7.5 and E10.5. Given its predominant expression in the extraembryonic tissue, the selective degeneration and absorption of female conceptuses in the absence of miR-465 miRNAs likely reflect the compromised development of the extraembryonic/placental tissue rather than the embryos/fetuses. Therefore, the miR-465 miRNAs appear to be required for proper development of the female, rather than the male, extraembryonic/placental tissue, supporting a role in sexual dimorphism in placental development. While sexual dimorphism is believed to mainly result from the differential gene expression between the male and female embryos (5), our study provides evidence that the placental development also displays sexual dimorphism, which can lead to a screwed sex ratio in offspring.
The 60% sex ratio seems subtle, but it is quite common in all the previous studies involving biased sex ratios (Table S2) (23–28). The X-linked miR-465 cluster belongs to the SpermiRs/miR-506 family (14, 34), and these X-linked miRNAs have no homologs on the Y chromosome. Member miRNAs of the miR-506 family share numerous targets despite their different seed sequences (14, 34). One previous study has shown that miR-465a-5p is upregulated when the miR-741 is inactivated in the cultured mouse spermatogonial stem cells (SSCs) (34), suggesting genetic compensation between these two miRNAs during spermatogenesis. Indeed, a similar phenomenon was observed in the miR-465 KO testes. Other miR-506 family members, including miR-201, miR-463, miR-471, miR-741, miR-871, miR-883a, and miR-883b, were upregulated in the miR-465 KO testes when compared to the WT testes (Figure S4A; Table S5A). Comparisons between WT male and female extraembryonic tissues at E7.5 yielded no differentially expressed miRNAs. However, comparisons between the KO counterparts, the miR-10a, miR-10b, and miR-196b were upregulated in the KO male extraembryonic tissues (Figure S4B; Table S5B). Although miR-10a, miR-10b, and miR-196b do not belong to the miR-506 family, they share a large number of target genes with the miR-465 cluster (Figure S4C), indicating that these miRNAs may compensate for the loss of the miR-465 cluster in the male extraembryonic tissues, and that the miR-465 cluster plays a sexual dimorphic role during extraembryonic tissues development. Comparisons between the miR-465 KO male and WT male or between the KO non-degenerating female and WT female extraembryonic tissues at E7.5 found no upregulated miRNAs, whereas 74 miRNAs were found dysregulated in between the KO degenerating and KO non-degenerating females (Figures S4D, E; Tables S5C–S5E), suggesting that the degenerating females are more “sensitive” to the miR-465 KO. Of interest, 26 miRNAs of the 74 dysregulated miRNAs all target Alkbh1, one of the validified targets of the miR-465 cluster that has a sex dimorphic role during extraembryonic development (27). No significant changes in mRNA transcriptome were detected between either WT and KO males, or between WT and the KO non-degenerating females; however, drastic changes were observed between the KO degenerating and KO non-degenerating females. Among these dysregulated genes, some of them are either sex-specific (e.g., Alkbh1 and Rlim) or imprinted genes, further confirming that the miR-465 cluster influences the extraembryonic development in a sex-specific manner through mediating sex differential genes.
Spontaneous embryonic resorption during early pregnancy is common in most mammalian species, including mice, rats, rabbits, voles, ewes, red pandas, swine, and humans (35–44). Moreover, spontaneous embryonic resorption during early pregnancy does not necessarily lead to reduced litter size (35, 36). Given that the embryonic resorption occurs randomly without obvious sex ratio bias, it is highly likely that both male and female embryos are resorbed at a similar rate to maintain a balanced sex ratio. Some studies have correlated embryonic loss with aberrant placental development (44). A recent in-depth survey of 103 knockout mice lines that display embryonic lethality has revealed that ~68% of these embryonic resorption cases are caused by placental dysfunction (45). Our data that almost all of the resorbed miR-465 KO embryos are females suggest a sexual dimorphic role of the miR-465 cluster in extraembryonic/placental development. Like the other X-linked miRNA clusters (14), the miR-465 has its orthologue in humans, which was named miR-892b, suggesting that the findings in mice may apply to humans. Supporting this hypothesis, a recent study in humans showed that the miR-892b was downregulated in the plasma collected from preeclampsia pregnancies (46), which is often accompanied by fetal growth restriction and placental abruption (47).
Taken together, our study uncovered an essential role of the miR-465 cluster in supporting the full developmental potential of the female, but not the male, extraembryonic tissues/placentae (Figure 4D). The male-biased sex ratio among miR-465 KO mice results from selective degeneration of the female placenta and resorption of the female embryos in the absence of the miR-465 cluster.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA669325/.
The animal study was reviewed and approved by Institutional Animal Care and Use Committee (IACUC) of the University of Nevada, Reno (Protocol# 00494).
ZW and WY designed the research. Z W, NM, YW, SW, SC, and HZ performed bench experiments. ZW, TZ, and ML performed bioinformatic analyses. SK and HW contributed reagents and protocols. ZW and WY wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the NIH (HD098593, HD0085506, HD099924 to WY) and the Templeton Foundation (PID: 61174 to WY).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.893854/full#supplementary-material
1. Williams TM, Carroll SB. Genetic and Molecular Insights Into the Development and Evolution of Sexual Dimorphism. Nat Rev Genet (2009) 10:797–804. doi: 10.1038/nrg2687
2. Karp NA, Mason J, Beaudet AL, Benjamini Y, Bower L, Braun RE, et al. Prevalence of Sexual Dimorphism in Mammalian Phenotypic Traits. Nat Commun (2017) 8:15475. doi: 10.1038/ncomms15475
3. Buckberry S, Bianco-Miotto T, Bent SJ, Dekker GA, Roberts CT. Integrative Transcriptome Meta-Analysis Reveals Widespread Sex-Biased Gene Expression at the Human Fetal-Maternal Interface. Mol Hum Reprod (2014) 20:810–9. doi: 10.1093/molehr/gau035
4. Warnefors M, Mossinger K, Halbert J, Studer T, VandeBerg JL, Lindgren I, et al. Sex-Biased Microrna Expression in Mammals and Birds Reveals Underlying Regulatory Mechanisms and a Role in Dosage Compensation. Genome Res (2017) 27:1961–73. doi: 10.1101/gr.225391.117
5. Capel B. Vertebrate Sex Determination: Evolutionary Plasticity of a Fundamental Switch. Nat Rev Genet (2017) 18:675–89. doi: 10.1038/nrg.2017.60
6. Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer Is Essential for Mouse Development. Nat Genet (2003) 35:215–7. doi: 10.1038/ng1253
7. Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. Dgcr8 Is Essential for Microrna Biogenesis and Silencing of Embryonic Stem Cell Self-Renewal. Nat Genet (2007) 39:380–5. doi: 10.1038/ng1969
8. Alisch RS, Jin P, Epstein M, Caspary T, Warren ST. Argonaute2 Is Essential for Mammalian Gastrulation and Proper Mesoderm Formation. PloS Genet (2007) 3:e227. doi: 10.1371/journal.pgen.0030227
9. Morita S, Horii T, Kimura M, Goto Y, Ochiya T, Hatada I. One Argonaute Family Member, Eif2c2 (Ago2), Is Essential for Development and Appears Not to Be Involved in DNA Methylation. Genomics (2007) 89:687–96. doi: 10.1016/j.ygeno.2007.01.004
10. Ha M, Kim VN. Regulation of Microrna Biogenesis. Nat Rev Mol Cell Biol (2014) 15:509–24. doi: 10.1038/nrm3838
11. Lin S, Gregory RI. Microrna Biogenesis Pathways in Cancer. Nat Rev Cancer (2015) 15:321–33. doi: 10.1038/nrc3932
12. Medeiros LA, Dennis LM, Gill ME, Houbaviy H, Markoulaki S, Fu D, et al. Mir-290-295 Deficiency in Mice Results in Partially Penetrant Embryonic Lethality and Germ Cell Defects. Proc Natl Acad Sci USA (2011) 108:14163–8. doi: 10.1073/pnas.1111241108
13. Parchem RJ, Ye J, Judson RL, LaRussa MF, Krishnakumar R, Blelloch A, et al. Two Mirna Clusters Reveal Alternative Paths in Late-Stage Reprogramming. Cell Stem Cell (2014) 14:617–31. doi: 10.1016/j.stem.2014.01.021
14. Wang Z, Xie Y, Wang Y, Morris D, Wang S, Oliver D, et al. X-Linked Mir-506 Family Mirnas Promote Fmrp Expression in Mouse Spermatogonia. EMBO Rep (2020) 21:e49024. doi: 10.15252/embr.201949024
15. Ohnishi Y, Totoki Y, Toyoda A, Watanabe T, Yamamoto Y, Tokunaga K, et al. Small Rna Class Transition From Sirna/Pirna to Mirna During Pre-Implantation Mouse Development. Nucleic Acids Res (2010) 38:5141–51. doi: 10.1093/nar/gkq229
16. Yuan S, Schuster A, Tang C, Yu T, Ortogero N, Bao J, et al. Sperm-Borne Mirnas and Endo-Sirnas Are Important for Fertilization and Preimplantation Embryonic Development. Development (2016) 143:635–47. doi: 10.1242/dev.131755
17. Ahn HW, Morin RD, Zhao H, Harris RA, Coarfa C, Chen ZJ, et al. Microrna Transcriptome in the Newborn Mouse Ovaries Determined by Massive Parallel Sequencing. Mol Hum Reprod (2010) 16:463–71. doi: 10.1093/molehr/gaq017
18. Wang Z, Wang Y, Wang S, Gorzalski AJ, McSwiggin H, Yu T, et al. Efficient Genome Editing by Crispr-Mb3cas12a in Mice. J Cell Sci (2020) 133:jcs240705. doi: 10.1242/jcs.240705
19. Wang Z, McSwiggin H, Newkirk SJ, Wang Y, Oliver D, Tang C, et al. Insertion of a Chimeric Retrotransposon Sequence in Mouse Axin1 Locus Causes Metastable Kinky Tail Phenotype. Mob DNA (2019) 10:17. doi: 10.1186/s13100-019-0162-7
20. Patro R, Mount SM, Kingsford C. Sailfish Enables Alignment-Free Isoform Quantification From RNA-Seq Reads Using Lightweight Algorithms. Nat Biotechnol (2014) 32:462–4. doi: 10.1038/nbt.2862
21. Shi J, Ko EA, Sanders KM, Chen Q, Zhou T. Sports1.0: A Tool for Annotating and Profiling Non-Coding RNAs Optimized for rRNA- and tRNA-Derived Small Rnas. Genom Proteom Bioinform (2018) 16:144–51. doi: 10.1016/j.gpb.2018.04.004
22. Yuan S, Liu Y, Peng H, Tang C, Hennig GW, Wang Z, et al. Motile Cilia of the Male Reproductive System Require Mir-34/Mir-449 for Development and Function to Generate Luminal Turbulence. Proc Natl Acad Sci USA (2019) 116:3584–93. doi: 10.1073/pnas.1817018116
23. Tan K, An L, Miao K, Ren L, Hou Z, Tao L, et al. Impaired Imprinted X Chromosome Inactivation Is Responsible for the Skewed Sex Ratio Following in Vitro Fertilization. Proc Natl Acad Sci USA (2016) 113:3197–202. doi: 10.1073/pnas.1523538113
24. Rosenfeld CS, Grimm KM, Livingston KA, Brokman AM, Lamberson WE, Roberts RM. Striking Variation in the Sex Ratio of Pups Born to Mice According to Whether Maternal Diet Is High in Fat or Carbohydrate. Proc Natl Acad Sci USA (2003) 100:4628–32. doi: 10.1073/pnas.0330808100
25. McNairn AJ, Chuang CH, Bloom JC, Wallace MD, Schimenti JC. Female-Biased Embryonic Death From Inflammation Induced by Genomic Instability. Nature (2019) 567:105–8. doi: 10.1038/s41586-019-0936-6
26. Wang Z, Yan W. Inflammation Induced by Faulty Replication During Embryonic Development Causes Skewed Sex Ratio. Biol Reprod (2019) 101:259–61. doi: 10.1093/biolre/ioz104
27. Nordstrand LM, Svard J, Larsen E, Nilsen A, Ougland R, Furu K, et al. Mice Lacking Alkbh1 Display Sex-Ratio Distortion and Unilateral Eye Defects. PloS One (2010) 5:e13827. doi: 10.1371/journal.pone.0013827
28. Gray C, Long S, Green C, Gardiner SM, Craigon J, Gardner DS. Maternal Fructose and/or Salt Intake and Reproductive Outcome in the Rat: Effects on Growth, Fertility, Sex Ratio, and Birth Order. Biol Reprod (2013) 89:51. doi: 10.1095/biolreprod.113.109595
29. Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, et al. Alkbh1-Mediated tRNA Demethylation Regulates Translation. Cell (2016) 167:816–28.e16. doi: 10.1016/j.cell.2016.09.038
30. Pan Z, Sikandar S, Witherspoon M, Dizon D, Nguyen T, Benirschke K, et al. Impaired Placental Trophoblast Lineage Differentiation in Alkbh1(-/-) Mice. Dev Dyn (2008) 237:316–27. doi: 10.1002/dvdy.21418
31. Wang PJ. X Chromosomes, Retrogenes and Their Role in Male Reproduction. Trends Endocrinol Metab (2004) 15:79–83. doi: 10.1016/j.tem.2004.01.007
32. Vockel M, Riera-Escamilla A, Tuttelmann F, Krausz C. The X Chromosome and Male Infertility. Hum Genet (2021) 140:203–15. doi: 10.1007/s00439-019-02101-w
33. Song R, Ro S, Michaels JD, Park C, McCarrey JR, Yan W. Many X-Linked Micrornas Escape Meiotic Sex Chromosome Inactivation. Nat Genet (2009) 41:488–93. doi: 10.1038/ng.338
34. Zhang F, Zhang Y, Lv X, Xu B, Zhang H, Yan J, et al. Evolution of an X-Linked Mirna Family Predominantly Expressed in Mammalian Male Germ Cells. Mol Biol Evol (2019) 36:663–78. doi: 10.1093/molbev/msz001
35. Flores LE, Hildebrandt TB, Kuhl AA, Drews B. Early Detection and Staging of Spontaneous Embryo Resorption by Ultrasound Biomicroscopy in Murine Pregnancy. Reprod Biol Endocrinol (2014) 12:38. doi: 10.1186/1477-7827-12-38
36. Drews B, Landaverde LF, Kuhl A, Drews U. Spontaneous Embryo Resorption in the Mouse Is Triggered by Embryonic Apoptosis Followed by Rapid Removal Via Maternal Sterile Purulent Inflammation. BMC Dev Biol (2020) 20:1. doi: 10.1186/s12861-019-0201-0
37. Conaway CH. Embryo Resorption and Placental Scar Formation in the Rat. J Mammal (1955) 36:516–32. doi: 10.2307/1375806
38. Conaway CH, Baskett TS, Toll JE. Embryo Resorption in the Swamp Rabbit. J Wildl Manage (1960) 24:197–202. doi: 10.2307/3796746
39. Martin KH, Stehn RA, Richmond ME. Reliability of Placental Scar Counts in the Prairie Vole. J Wildl Manage (1976) 40:264–71. doi: 10.2307/3800424
40. Thwaites CJ. The Time Course of Embryonic Resorption in the Ewe. Aust J Biol Sci (1972) 25:597–603. doi: 10.1071/bi9720597
41. Wang XH, Xu S, Zhou XY, Zhao R, Lin Y, Cao J, et al. Low Chorionic Villous Succinate Accumulation Associates With Recurrent Spontaneous Abortion Risk. Nat Commun (2021) 12:3428. doi: 10.1038/s41467-021-23827-0
42. Wimsatt WA. Some Comparative Aspects of Implantation. Biol Reprod (1975) 12:1–40. doi: 10.1095/biolreprod12.1.1
43. Lowe J, Curry E. Incidence of Pregnancy Loss and Characterization of Fetal Development in Red Pandas. Reprod Fertil (2021) 2:292–9. doi: 10.1530/RAF-21-0079
44. Vonnahme KA, Wilson ME, Foxcroft GR, Ford SP. Impacts on Conceptus Survival in a Commercial Swine Herd. J Anim Sci (2002) 80:553–9. doi: 10.2527/2002.803553x
45. Perez-Garcia V, Fineberg E, Wilson R, Murray A, Mazzeo CI, Tudor C, et al. Placentation Defects Are Highly Prevalent in Embryonic Lethal Mouse Mutants. Nature (2018) 555:463–8. doi: 10.1038/nature26002
46. Zhong Y, Zhu F, Ding Y. Differential Microrna Expression Profile in the Plasma of Preeclampsia and Normal Pregnancies. Exp Ther Med (2019) 18:826–32. doi: 10.3892/etm.2019.7637
Keywords: miR-465, microRNA, sexual dimorphism, sex ratio, extraembryonic tissues, placenta, CRISPR-Cas9
Citation: Wang Z, Meng N, Wang Y, Zhou T, Li M, Wang S, Chen S, Zheng H, Kong S, Wang H and Yan W (2022) Ablation of the miR-465 Cluster Causes a Skewed Sex Ratio in Mice. Front. Endocrinol. 13:893854. doi: 10.3389/fendo.2022.893854
Received: 10 March 2022; Accepted: 11 April 2022;
Published: 23 May 2022.
Edited by:
Erwin Goldberg, Northwestern University, United StatesReviewed by:
Renee Reijo Pera, McLaughlin Research Institute, United StatesCopyright © 2022 Wang, Meng, Wang, Zhou, Li, Wang, Chen, Zheng, Kong, Wang and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Yan, d2VpLnlhbkBsdW5kcXVpc3Qub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.