
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol., 11 July 2022
Sec. Endocrinology of Aging
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.887486
This article is part of the Research TopicMetabolic Factors in Erectile DysfunctionView all 14 articles
 Xuan Zhou1†
Xuan Zhou1† Rong Cong1†
Rong Cong1† Liangyu Yao1†
Liangyu Yao1† Xiang Zhou1†
Xiang Zhou1† Jiaochen Luan1
Jiaochen Luan1 Qijie Zhang1
Qijie Zhang1 Xu Zhang1
Xu Zhang1 Xiaohan Ren1
Xiaohan Ren1 Tongtong Zhang1
Tongtong Zhang1 Xianghu Meng1*
Xianghu Meng1* Ninghong Song1,2*
Ninghong Song1,2*Background: The key regulatory roles of long non-coding RNAs (lncRNAs) in age-related erectile dysfunction (A-ED) are unknown.
Aim: This study aimed to identify putative lncRNAs that regulate age-related erectile dysfunction via transcriptome analyses, and to predict their specific regulatory routes via bioinformatics methods.
Methods: 22 geriatric male SD rats were divided into age-related erectile dysfunction (A-ED) and negative control (NC) groups after evaluations of intracavernous pressure (ICP). By comparative analysis of transcriptomes of cavernosal tissues from both groups, we identified differentially expressed lncRNAs, miRNAs, and mRNAs. Seven differentially expressed lncRNAs were selected and further verified by quantitative real-time polymerase chain reactions (RT-qPCR). The construction of the lncRNA-miRNA-mRNA network, the Gene Ontology (GO) term enrichment, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed in Cytoscape.
Results: From comparative transcriptome analyses of A-ED and NC groups, 69, 29, and 364 differentially expressed lncRNAs, miRNAs, and mRNAs were identified respectively. Differentially expressed lncRNAs were culled to seven, which were all verified by qPCR. Three of these lncRNAs (ENSRNOT00000090050, ENSRNOT00000076482, and ENSRNOT00000029245) were used to build regulatory networks, of which only ENSRNOT00000029245 was successful. Moreover, GO and KEGG analyses demonstrated that these lncRNAs possibly regulated muscle myosin complex, muscle cell cellular homeostasis, and ultimately erectile function in rats through PI3K-Akt, fluid shear stress, and atherosclerosis pathways.
Conclusion: Our study identified differentially expressed lncRNAs, miRNAs, and mRNAs through comparisons of transcriptomes of geriatric rats. An identified lncRNA verified by qPCR, was used to construct a lncRNA-miRNA-mRNA regulatory network. LncRNA ENSRNOT00000029245 possibly regulated downstream mRNAs through this regulatory network, leading to apoptosis in the cavernous tissue, fibrosis, and endothelial dysfunction, which ultimately caused ED. These findings provide seminal insights into the molecular biology of aging-related ED, which could spur the development of effective therapeutics.
Erectile dysfunction (ED) is an inability to either achieve or maintain a satisfactory penile erection for sexual activity. It is more common in middle-aged and elderly men, especially those over 40 years of age (1). Moreover, most ED patients have an organic etiology (2). ED is not only associated with aging, but also with cardiovascular and endocrine diseases, and neurogenic illnesses (3).
By 2050, 22% of the global population is predicted to have become over 60 years of age (4), and this will result in more ED patients, whose current prevalence rate is 2% and 86% for men under 40 and over 80 years of age respectively (5). Yet by 2025, 332 million people are predicted to have suffered from ED (6). Oral selective phosphodiesterase type 5 inhibitors (PDE5-Is) are common first-line therapies. They inhibit the conversion of cyclic guanosine monophosphate (cGMP) to cyclic guanosine monophosphate(GMP), leading to relaxation of vascular and cavernosal smooth cells, and this ultimately maintains an erection. Vacuum erection devices (VED), intracavernosal injections (ICI), and penile implants are used as second-line therapies. Nevertheless, they have a variety of shortcomings, resulting in the current unsatisfactory clinical treatment of ED (2). Hence, further investigations on the molecular mechanisms of ED are not only needed but could also lead to the development of effective therapies.
Senescence is a major risk factor for various chronic diseases, including type 2 diabetes mellitus (T2DM), cardiovascular disease, and cancer (7). Senescence is a programmed progression caused by accumulated DNA, protein, and lipid damage (8). Relatedly, endothelial dysfunction, which is closely related to erectile dysfunction, occurs due to aging (9–11). Specifically, accumulated damage due to aging leads to dysfunction of the vascular endothelium, which in turn leads to vasculogenic erectile dysfunction (12).
Long non-coding ribonucleic acids (lncRNAs) are ribonucleic acids (RNA) that are both longer than 200 base pairs and not translated into proteins (13, 14). They play important roles in transcriptional regulation, the formation of subcellular structures, epigenetic gene regulation, and programmed cellular development (15, 16). In cardiovascular diseases, lncRNAs regulate downstream biological functions by positive and negative modulatory effects, playing roles as miRNA sponges and co-expression with miRNAs (17). LncRNAs have been associated with ED (18, 19). For example, in diabetes mellitus-induced erectile dysfunction, lncRNA MALAT1 functioned as a sponge for miR-206 to upregulate the expression of VEGFA, thus boosting bone marrow-derived mesenchymal stem cells (BM-MSCs) differentiation into endothelial cells for rectification of ED (20). However, there exists a paucity of studies on this topic. To fill this research gap, we aimed to elucidate possible key roles of lncRNA in age-related ED (A-ED) by constructing a lncRNA-miRNA-mRNA regulatory network via comparative transcriptome analyses.
We used 22 geriatric male SD rats (20 months) purchased from the Experimental Animal Center of Nanjing Medical University, and our research was approved by the Animal Care and Use Committee of the First Affiliated Hospital of Nanjing Medical University. The animals were anesthetized with pentobarbital natrium (40 mg/kg) by intraperitoneal injection.
After anesthesia, dissections were made along the penis midline to expose the carotid artery. A neck artery was cannulated and a heparinized 25-gauge needle was inserted into the corpora cavernosum of the penis to measure both mean arterial pressure (MAP) and ICP. Electrical stimulation was set at 5 V, 15 Hz, pulse width 0.2ms, and the stimulation lasted for 1 min. The BL-420S Biological Functional System (Chengdu Taimeng Technology Co, LTD, Chengdu, China) was used to measure pressure and generate electrical stimulation. Each experiment was done in triplicate. The ratio of the maximal ICP to the corresponding MAP (ICP/MAP) was calculated to estimate the erectile function. The corpus cavernosum tissue was isolated and stored -80°C for further experiments.
Experiments were carried out as previously described (21). Penile tissue was fixed with 4% paraformaldehyde and embedded in paraffin. Masson trichrome staining was used to observe, under 200x magnification, then evaluate the ratio of smooth muscle to collagen area in penile tissue.
The rats were divided into geriatric A-ED (erectile dysfunction) and NC (negative control) groups based on the erectile evaluation results. ICP/MAP > 35% was defined as the geriatric NC group, and ICP/MAP < 35% was defined as the geriatric ED group. After stripping the urethra and other connective tissue from the penis, the cavernous tissue was retained. Cavernous tissue was minced into small pieces with a low-temperature grinder (Servicebio, KZ-III-F) under grinder parameters of 60Hz & -10°C. Then, total RNA was extracted from pieces of cavernous tissues or cells using a TRIzol (Invitrogen, Carlsbad, CA, USA) based protocol. RNA concentration and purity were determined via a NanoDrop 2000 spectrophotometer (Madison, WI, USA). Ribosomal RNA (rRNA) was separated from total RNA and RNA integrity was analyzed via an Agilent RNA 6000 Nano Kiton an Agilent 2100 Bioanalyzer. Residual total RNA was broken into short fragments for 2 min, then reverse transcribed into cDNA with random hexamers as primers via a SuperScript III First-Strand Synthesis protocol (Invitrogen). Synthetized products were purified using a QIAquick PCR Purification Kit (QIAGEN). DNA fragment ends were repaired and ligated with poly (A) tails to connect them to Illumina sequencing adapters. Then, the second strand was digested and the appropriate fragment was identified for PCR amplification. The cDNA sequencing library was made and subsequently sequenced using an Illumina HiSeq TM 4000 platform (Gene Denovo Biotechnology Co., Guangzhou, China). Only reads that met the following criteria were used as raw data (1) reads did not contain adapters; (2) reads did not contain more than 10% of unknown nucleotides (N); (3) reads did not contain more than 50% of low quality (Q-value ≤ 20) bases. These reads were aligned to Ribosome RNA (rRNA) and mapped to a rat reference genome using HISAT2 (22).
We extracted total RNA from previously isolated corpus cavernosum tissues using a Trizol (Invitrogen, USA) based protocol. Then, cDNA was synthesized using HiScript® III All-in-one RT SuperMix Perfect for qPCR (Vazyme, China). Next, we utilized the StepOne Plus Real-Time PCR system (Applied Biosystems, USA) to perform qPCR. Fold changes in mRNA expression were calculated using the 2−ΔΔCt method and normalized against β-actin with ABI Step One software version 2.1. PCR primer sequences were synthesized by TSINGKE Biological Technology (Nanjing, China) and are listed in Supplementary Table S1.
Transcriptome data were normalized and analyzed in R using the edgR package. Then three validated lncRNAs were selected (ENSRNOT00000090050, ENSRNOT00000076482, and ENSRNOT00000029245). For prediction analysis, we defined differently expressed miRNAs and mRNAs as logFC >2 and p <0.05. MiRNAs target prediction tools TargetScan, miRcode, and miRDB were used to predict miRNAs targets. All relevant annotations of miRNAs were derived from miRbase.
The network was constructed based on ceRNA theory as previously described (21). (1) Spearman rank correlation coefficient (SCC) was used to evaluate expression correlation between miRNAs-mRNAs or miRNA-lncRNAs. RNA interaction pairs that had SCC<0 were defined as either negatively co-expressed lncRNA-miRNA or mRNA-miRNA pairs. All RNAs were differentially expressed. The mRNA and lncRNA were predicted as miRNA target genes. (2) Pearson correlation coefficient (PCC) was used to assess expression correlations between lncRNA and mRNA. ceRNA interaction pairs that had PCC > 0.5 were defined as co-expressed lncRNA-mRNA pairs. RNA interaction pairs of mRNA and lncRNA were termed as negatively or postively co-expressed with a common miRNA. Pairs of mRNA and lncRNA were targeted negatively or positively co-expressed with a common miRNA. (3) The significance of common miRNA sponges differences between the two genes was tested by a hypergeometric cumulative distribution function test. The filtering condition was p< 0.05. Finally, all eligible RNA interaction pairs were collected and combined into a network, which was visualized in Cytoscape 3.8.2 (http://www.cytoscape.org/).
Gene ontology (GO, http://www.geneontology.org/) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg) pathway analysis of differentially expressed mRNAs were performed using the R package clusterProfiler. An alpha level of p<0.05 was used to test statistical significance.
The rat vascular endothelium cell line (RAOEC) was purchased from Shanghai Jingkang Biological Co., LTD (Shanghai, China) and cultured at 37 °CC in 5% CO2. The endothelial cell lines were characterized by double-immunofluorescent staining for biomarkers CD31 and vWF (Proteintech, China). DMEM medium, supplemented with 10% FBS, and 100 units/mL penicillin, were used in cell culture (Gibco).
ROAEC cells were treated with hydrogen peroxide (H2O2) to induce senescence (23, 24). Cells were seeded onto 6-well plates (106/well) in a full growth medium. We choose the optimized concentration (100 µM) to build a cellular aging model according to the references (25). After 24 hours, the cells were treated with H2O2 (100μM) and DMSO (0.01%) respectively for 24 hours. Senescence-associated β-galactosidase (sa-β-Gal) staining was performed using a β-galactosidase enzyme assay kit (Beyotime, China) between the H2O2-treated group and the DMSO group.
All data analyses were done using SPSS software (SPSS V 24.0). T-tests were used to determine the statistical significance of differences between two groups of parametric data whereas Mann-Whitney tests were used for nonparametric data. Statistical significances of GO and KEGG analyses were determined using Fisher’s exact tests. An alpha level of p<0.05 was used for all statistical tests.
Among 22 rats, we selected ICP/MAP less than 0.35 as the low-pressure group and ICP/MAP greater than 0.35 as the high-pressure group, and when combined with the results of the Masson staining, 22 rats were divided into A-ED group and NC group (n=7, n=15). There was no significant difference in MAP between the A-ED and NC groups (p=0.3777). However, the ratio of ICP/MAP was lower in the A-ED than the NC group (Figures 1A, B). Masson trichromatic staining results showed that the smooth muscle/collagen ratio was significantly lower in the A-ED than the NC group (Figures 1C, D). Thus, this A-ED rat model was valid for studies on age-related erectile dysfunction.
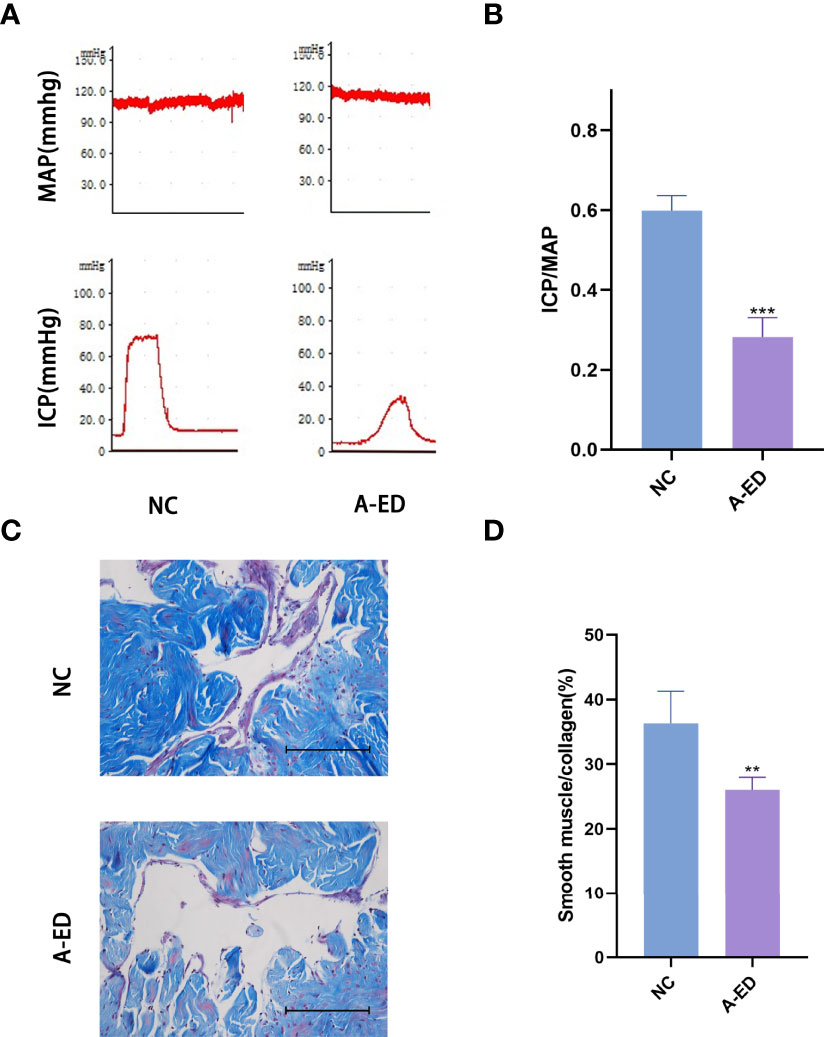
Figure 1 Evaluation of Erectile Dysfunction (A): Mean arterial pressure (MAP) and intracavernous pressure (ICP) of rats in the A-ED group and NC group. (B): Bar plot of the ICP/MAP of A-ED group and NC group. ***p < 0.001. (C): Masson-trichrome (Masson) staining showing the fibrosis in the A-ED and NC groups. (D) Bar plot of the ratio of smooth muscle to collagen in the A-ED and NC groups. **p < 0.01.
A total of 5,307 lncRNAs, 1,322 miRNAs, and 22,050 mRNAs were identified by sequencing. The expression levels of lncRNAs in six samples were assessed by FPKM, and did not show a deviated distribution of mRNAs nor lncRNAs (Figures 2A, B). Scatterplots (Figures 2C, D) demonstrated high reproducibility of samples from each group.
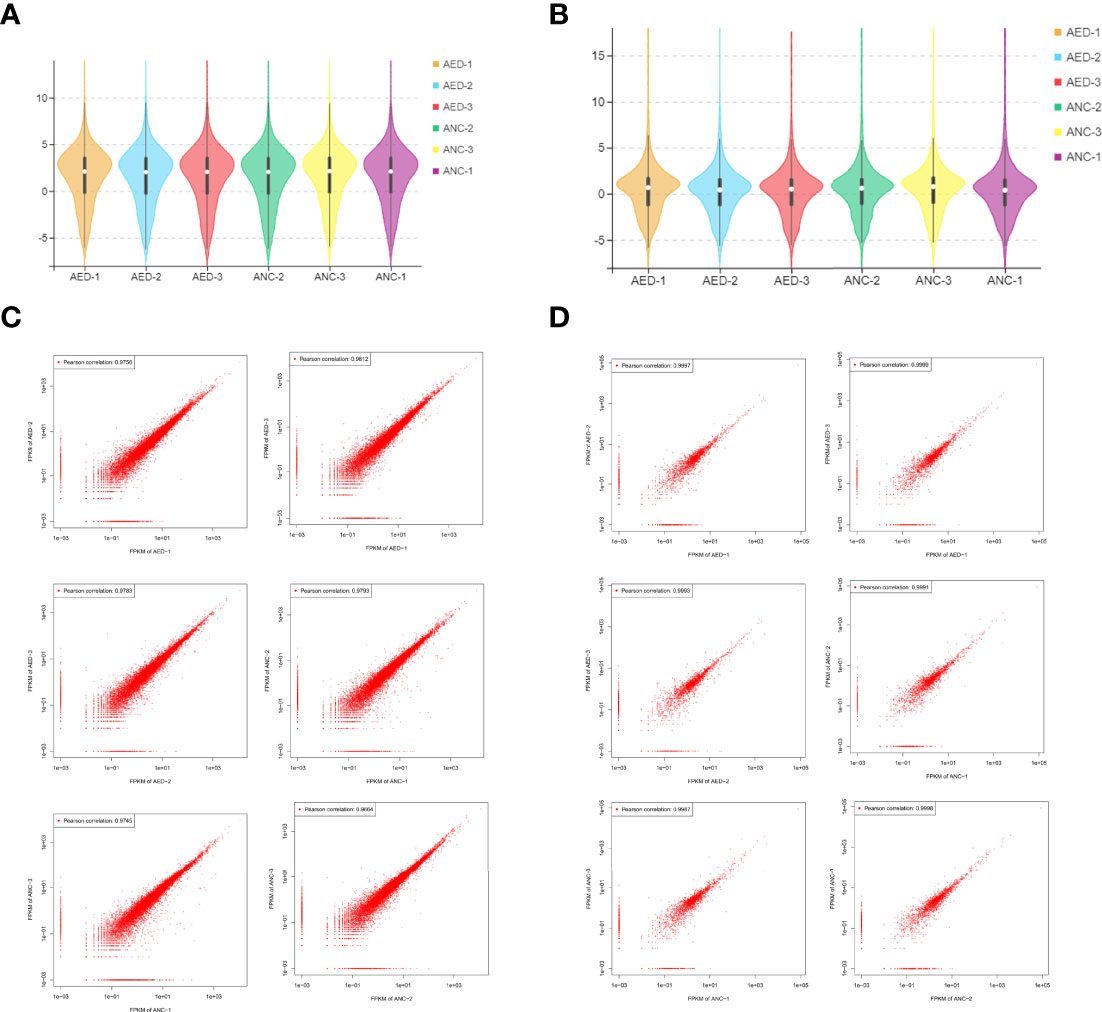
Figure 2 Data distribution and quality assessment. (A) A violin diagram of the FPKM value of all mRNAs within each sample. (B) A violin diagram of the FPKM value of all lncRNAs within each sample. (C) Scatter plot of correlation between mRNAs expression levels in two different samples. (D) Scatter plot of correlation between lncRNA expression levels of two different samples.
Differentially expressed mRNAs and lncRNAs were analyzed using volcano plots (Figures 3A, B). Upregulated and downregulated mRNAs and lncRNAs, when compared to the geriatric NC group, were shown using orange and blue dots respectively. Ultimately, a total of 69, 364 and 29 differential expressed lncRNAs (49 up-regulated, 20 down-regulated), mRNAs (143 up-regulated, 221 down-regulated), and miRNAs (18 down-regulated, 11 up-regulated) respectively, were identified (Figures 3C–E).
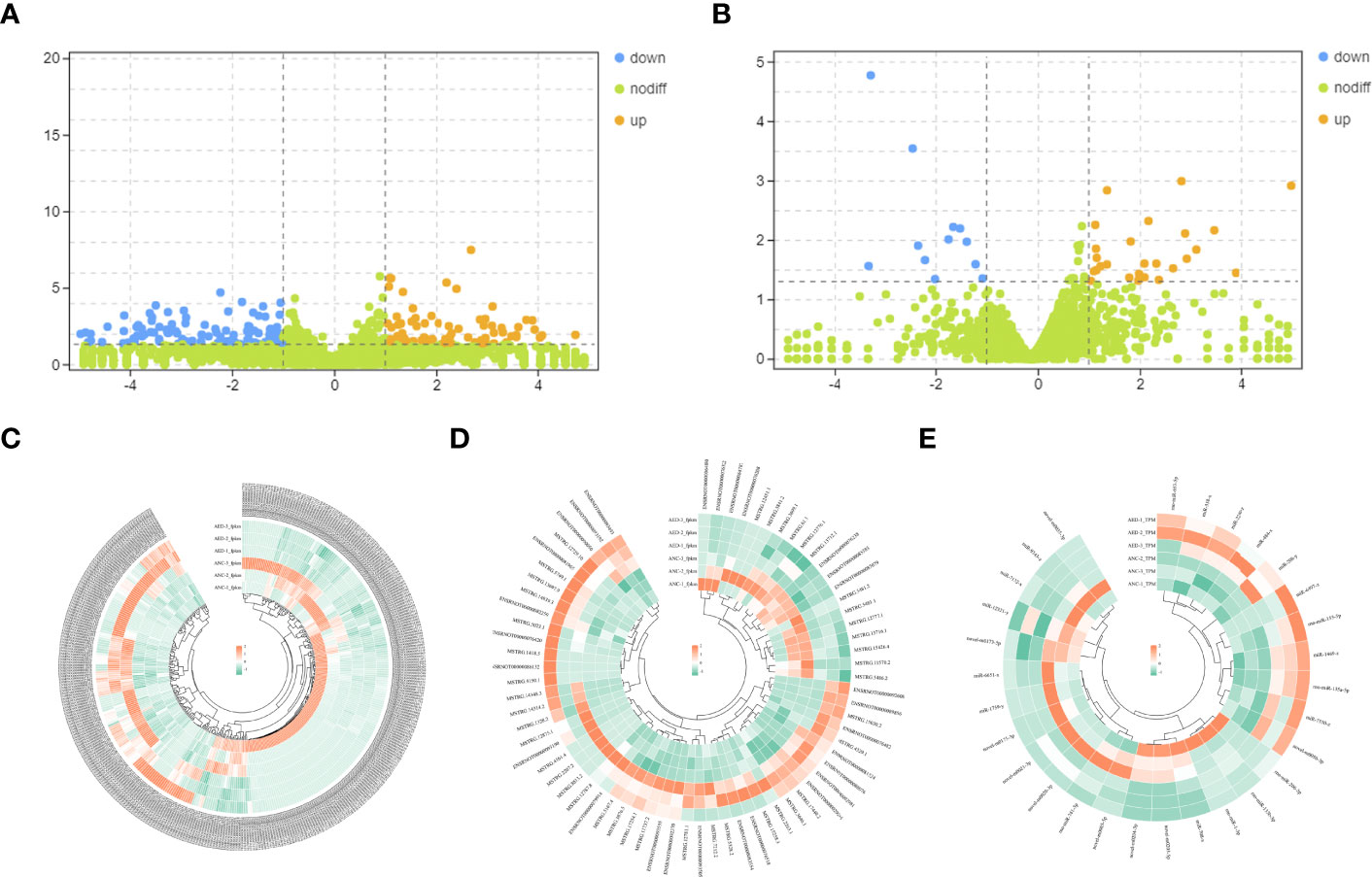
Figure 3 (A) Volcano plots of differentially expressed mRNAs. The abscissa represents the log of the multiple of the difference between the two groups, and the ordinate represents the negative Log10 value of the FDR of the difference between the two groups. (B) Volcano plots of differentially expressed lncRNAs. The abscissa represents the log of the multiple of the difference between the two groups, and the ordinate represents the negative Log10 value of the FDR of the difference between the two groups. (C–E) Heatmaps demonstrate the distribution of differentially expressed lncRNAs, miRNAs, and mRNAs.
Using an inclusion criteria for lncRNAs of 1<count number<120, fold change > 2, p< 0.05, seven lncRNAs were identified and used to validate RNA-seq results in seven pairs of geriatric ED and NC samples (MSTRG.3646.1, ENSRNOT00000085383, ENSRNOT00000093493, ENSRNOT00000081965, ENSRNOT00000029245, ENSRNOT00000090050, ENSRNOT00000076482, ENSRNOT00000085383). According to qPCR results, ENSRNOT00000076482(p=0.0175), ENSRNOT00000090050(p=0.007) and ENSRNOT00000029245(p=0.0006) were highly expressed in the ED group, corroborating sequencing results (Figure 4). However, expression levels of the other four lncRNAs did not statistical differ between groups.
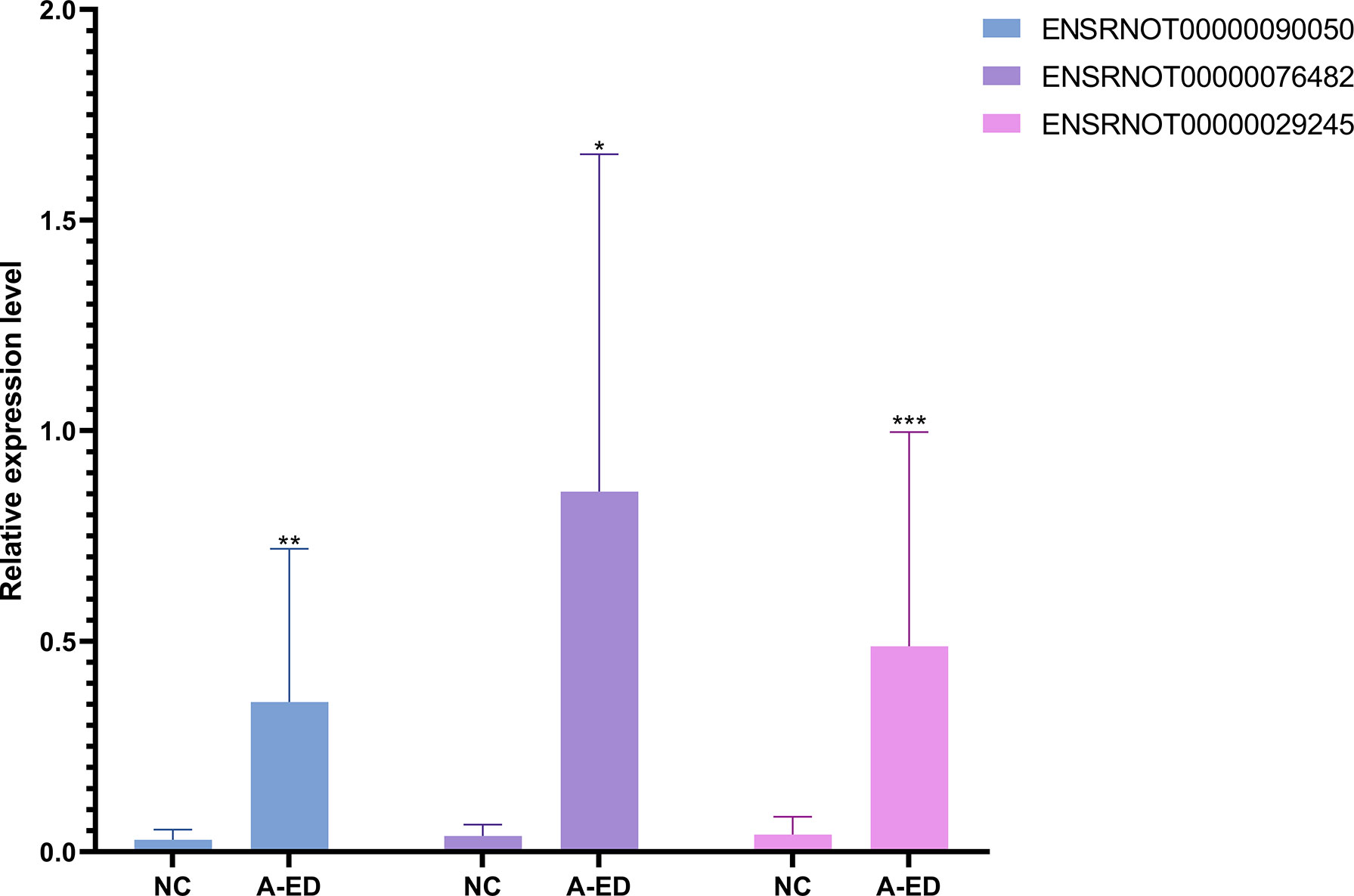
Figure 4 Bar cartoon of qPCR shows the expression of 3 lncRNAs between the A-ED and NC groups. *P < 0.05, **p < 0.01, ***p < 0.001.
To further explore the specific function of these lncRNAs, related mRNAs were used to carry out GO and KEGG enrichment analyses and the top 20 results of enrichment analysis (including KEGG and GO) according to p values, are shown in Figure 5. In doing KEGG analyses, we identified pathways for PI3K-AKT & Jak-STAT signaling, terpenoid backbone biosynthesis, synthesis and degradation of ketone bodies, EGFR tyrosine kinase inhibitor resistance, fluid shear stress and atherosclerosis.
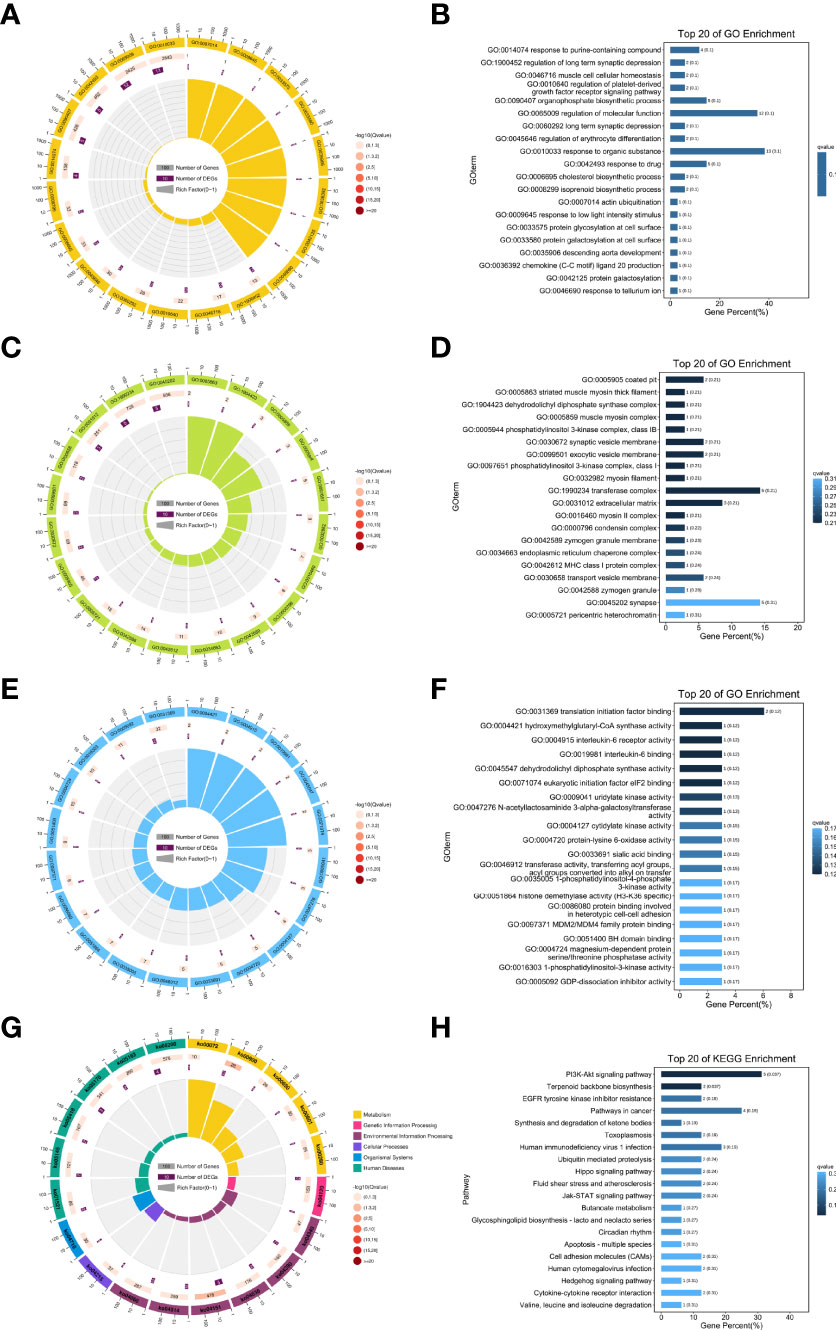
Figure 5 Results of the GO and KEGG enrichment. Top 20 enrichment results (A, B) biological processes; (C, D) cellular component; (E, F) molecular function; (G, H) KEGG pathways enrichment.
From GO enrichment analyses, cellular component (CC) annotations showed that the following components played key roles: coated pit (GO:0005905), striated muscle myosin thick filament (GO:0005863), dehydrodolichyl diphosphate synthase complex (GO:1904423), muscle myosin complex (GO:0005859), and phosphatidylinositol 3-kinase complex, class IB (GO:0005944). Corresponding annotations of molecular functions included translation initiation factor binding (GO:0031369), hydroxymethylglutaryl-CoA synthase activity (GO:0004421), and interleukin-6 receptor activity (GO:0004915).
Moreover, from the annotations of biological processes, the following were enriched (Figure 5): response to purine-containing compound (GO:0014074), regulation of long term synaptic depression (GO:1900452), muscle cell cellular homeostasis (GO:0046716), regulation of platelet-derived growth factor receptor signaling pathway (GO:0010640), and organophosphate biosynthetic process (GO:0090407).
From the results of RT-qPCR and RNA-seq, three verified lncRNAs, 29 differentially expressed miRNAs, and 364 differentially expressed mRNAs were selected, and the regulatory network was constructed according to its hypothesis. Figure 6 illustrates the lncRNA-miRNA-mRNA regulatory network of geriatric ED rats. In our network, the up-regulated lncRNA ENSRNOT00000029245 upregulated the expression of 23 mRNAs (Tspan18, Mtss1, Szrd1, Pik3cg, NEWGENE_619861, Fgf1, Btrc, B2m, Ggta1, kdm4a, Fxyd7, Naf1, Vps9d1, Ppm1l, Bcl2l1, RGD1560281, Scamp1, Sele, Nrcam, Hsp90b1, Cmpk1, Fam20b, Hiatl3) by miR484x axis. Similarly, the lncRNA ENSRNOT00000029245 regulated the 14 mRNAs (Sorcs3, LOC100910882, Morf4l2, Ptprd, Hmgcs1, Lox, Gpm6a, Trim32, Tnfrsf11b, Phf14, Ncapg2, Cyyr1, Reep3, Nus1) by rno-miR-653-5p axis. Interestingly, according to our network, the mRNA Il6r was upregulated by both the miR484x axis and the rno-miR-653-5p axis.
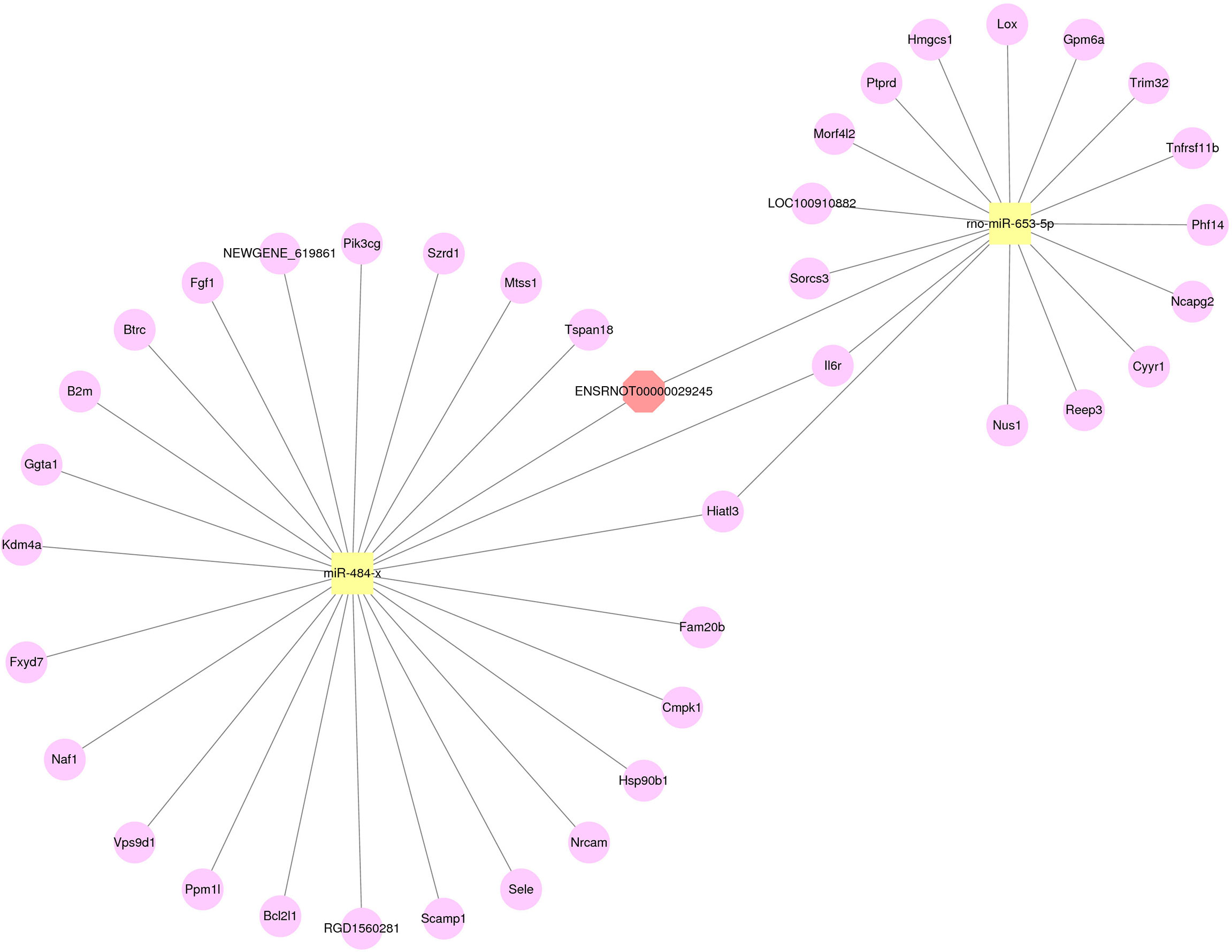
Figure 6 The network of lncRNA-miRNA-mRNA. Red octagons represent up-regulated lncRNAs, yellow squares represent up-regulated miRNAs, and pink circles represent up-regulated mRNAs.
The RAOEC cells were identified by staining with anti-CD31 antibody (red), anti-vWF antibody (green), and cell nuclei were identified using DAPI (blue) (Figure 7A). The sa-β-Gal staining demonstrated that the RAOEC cells were induced senescence at 100μM H2O2 (Figure 7B). Interestingly, the expression of the three lncRNAs (ENSRNOT00000029245, ENSRNOT00000076482, and ENSRNOT00000090050) between cell lines showed that only ENSRNOT00000029245 had a statistical significance (Figure 7C).
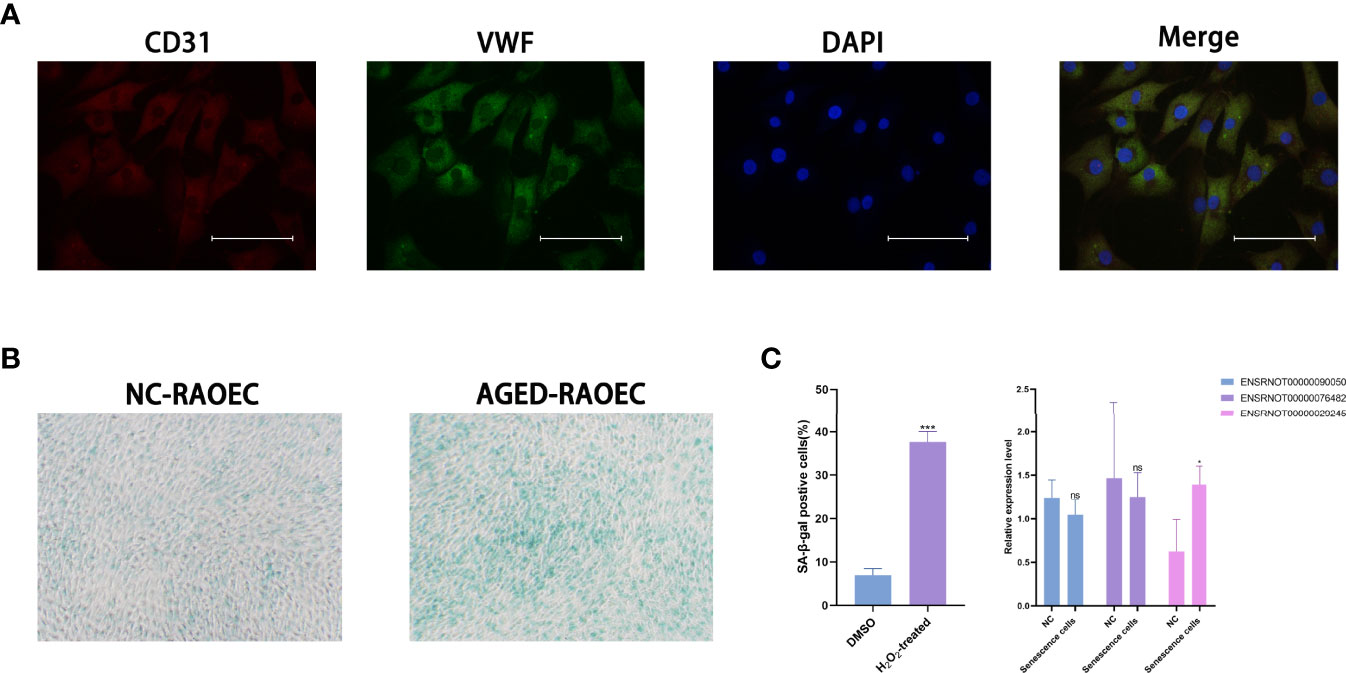
Figure 7 Cell lines validations. (A) Double-immunofluorescent staining for biomarkers CD31 and vWF. (B) β-Galactosidase staining for RAOEC cell in the DMSO and the H2O2 treated group. (C) Relative expression of three lncRNAs (ENSRNOT00000090050, ENSRNOT00000076482, and ENSRNOT00000029245) in Senescence cells and NC cells. *P < 0.05. Scale bars = 25 µm.
As the global population of the elderly rises, so does erectile dysfunction, a disease closely related to aging. Physiologically, an erection occurs when, NO, released from the endothelium and parasympathetic nerve terminals, enhances cGMP, thus promoting the loss of intracellular calcium and resulting in the relaxation of smooth muscles. The venous return is then blocked, triggering an erection.
Although PDE5Is are currently recommended as first-line treatments, they only relieve symptoms without curing, and the effective PDE5 dose increases with age (26). This, therefore, necessitates new insights into the molecular biology of age-related erectile dysfunction for the discovery of novel drug targets and therapies.
In this study, we comparatively analyzed transcriptomes based on a p< 0.05, logFC>2 criteria, to identify differentially expressed RNAs. A total of 69, 364, and 29 differentially expressed lncRNAs, mRNAs, and miRNAs respectively, were identified from comparisons of transcriptomes of geriatric ED and NC rats. Using the criteria of 1<count number<120, seven lncRNAs were identified and validated by qPCR. Furthermore, three lncRNAs were differentially expressed between the two groups. However, only lncRNA ENSRNOT00000029245 was used to successfully construct a lncRNA-miRNA-mRNA regulatory network.
Moreover, we conducted GO and KEGG enrichment analysis based on lncRNA ENSRNOT00000029245 related mRNAs. The results of these enrichment analyses showed that in terms of GO enrichment, coated pit, striated muscle myosin thick filament, dehydrodolichyl diphosphate synthase complex, muscle myosin complex, phosphatidylinositol 3-kinase complex, and translation initiation factor binding were enriched. The results of the KEGG pathway analysis indicated that PI3K-AKT and Jak-STAT signaling, terpenoid backbone biosynthesis, synthesis and degradation of ketone bodies, EGFR tyrosine kinase inhibitor resistance, fluid shear stress, and atherosclerosis pathways were enriched. The PI3K-AKT signaling pathway plays a critical role in maintaining vascular endothelial function (27). It also regulates the opening of the endothelial tight junction, as well as the permeability and mobility of endothelial cells (28). This suggests that aging leads to mutations in the PI3K pathway, which in turn leads to erectile dysfunction by affecting endothelial cell function.
ceRNA crosstalk has possible roles in numerous diseases, as lncRNAs regulate diseases through ceRNA (29, 30). Using both an exclusion criteria and qPCR results, we elected the differentially expressed lncRNA ENSRNOT00000029245 to build a lncRNA-miRNA-mRNA regulate network that could identify microregulatory processes in age-related ED.
Phosphatidylinositol 3-kinase (PI3K), an important component in the PI3K-AKT pathway, is a vascular protective factor with anti-apoptotic functions (31). Yu et al. found that PI3Kγ in vascular smooth muscle cells(VSMC) was upregulated after vascular injury, and PI3Kγ could promote proliferation and migration of VSMC (32). Thus, PI3Ks possibly contribute in large part to ED. In our study, ENSRNOT00000029245 was validated by qPCR as highly expressed in ED rats, and related to Pik3cg. In our regulatory network, ENSRNOT00000029245 regulates Pik3cg by modulating miR484-x. High expression of miR-484 downregulates eNOS transcripts, thus affecting homeostasis of endothelial function (33).
ED is accompanied by irreversible death of smooth muscle and endothelial cells (2). BCL2 family proteins are important regulators of apoptosis. Our network indicates that LncRNA ENSRNOT00000029245 may be associated with apoptosis in either cavernosal endothelium or smooth muscle cells via regulating miR-484-x-mediated Bcl2l1 expression. Moreover, tripartite motif-containing 32 (trim32), is up-regulated when cells undergo oxidative stress, promote ROS generation, and induce apoptosis (34). From our findings, trim32 was upregulated via the ENSRNOT00000029245-rno-miR-653-5p axis. This supported the hypothesis that upregulated ENSRNOT00000029245 promotes apoptosis of corporeal smooth muscle and endothelial cells, thus inducing ED in geriatric rats.
Our network findings suggest that ENSRNOT00000029245 targets miRNAs and mRNAs that affect either fibrosis or apoptosis in the corpus cavernosum and this ultimately results in erectile function.
However, our study still has some weaknesses. First, our model was based on rats and, despite lncRNAs being conserved between humans and rats, it thus does not provide direct evidence for the feasibility of our ceRNA network for humans. Ideally, this network should have been constructed from comparisons of cavernous tissues of elderly-ED patients to that of elderly healthy volunteers. Second, we constructed the ceRNA network of lncRNA without considering crosstalk from pseudogenes and circular RNAs. Lastly, we did not validate the proposed regulatory effect of this lncRNA on erectile function in vivo.
In this study, we identified 69, 29, and 364 differentially expressed lncRNAs, miRNAs, and mRNAs respectively, by comparing transcriptomes from A-ED rats to those from NC rats. Three lncRNAs (ENSRNOT00000029245, ENSRNOT00000076482, and ENSRNOT00000090050) were validated by qPCR, as highly expressed in the A-ED group. The results of GO enrichment and KEGG pathway analyses showed that A-ED may be associated with PI3K-AKT signaling, fluid shear stress, and atherosclerosis pathways. Moreover, the constructed network showed that ENSRNOT00000029245 possibly contributes to either fibrosis or apoptosis in the corpus cavernosum via either a rno-miR-653-5p-trim32 axis or by regulating miR-484-x-mediated Bcl2l1 expression. Therefore, it is worthwhile to investigate molecular mechanisms through which lncRNA ENSRNOT00000029245 regulates age-related ED.
The RNA-Seq data presented in the study are deposited in the SRA databank (www.ncbi.nlm.nih.gov/sra) and is accessible via the PRJNA848782 SRA (Bioproject) accession number.
The animal study was reviewed and approved by The Animal Care and Use Committee of the First Affiliated Hospital of Nanjing Medical University.
NS and XM designed this work. XM, XZ (1st author), LY, and RC wrote the manuscript. QZ, XiZ, and JL performed the bioinformatics analysis. XZ (7th author), XR, and TZ performed the data review. All authors have read and approved the manuscript.
This article was funded by the National Natural Science Foundation of China [grant number: 81801438; 81871151].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.887486/full#supplementary-material
Supplementary Figure 1 | The bar plot of qPCR of these 7 lncRNAs between A-ED and NC group (*p < 0.05, **p < 0.01, ***p < 0.001).
ED, Erectile Dysfunction; A-ED, Age-related Erectile dysfunction; PDE5-Is, phosphodiesterase type 5 inhibitors; GO, Gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; qPCR, Quantitative real-time PCR; MAP, Mean arterial pressure; ICP, Intracavernous pressure; lncRNAs, Long non-coding ribonucleic acids.
1. Echeverri Tirado LC, Ferrer JE, Herrera AM. Aging and Erectile Dysfunction. Sex Med Rev (2016) 4:63–73. doi: 10.1016/j.sxmr.2015.10.011
2. Yafi FA, Jenkins L, Albersen M, Corona G, Isidori A.M, Goldfarb S, et al. Erectile Dysfunction. Nat Rev Dis Primers (2016) 2:16003. doi: 10.1038/nrdp.2016.3
3. Shamloul R, Ghanem H. Erectile Dysfunction. Lancet (2013) 381:153–65. doi: 10.1016/S0140-6736(12)60520-0
4. Kanasi E, Ayilavarapu S, Jones J. The Aging Population: Demographics and the Biology of Aging. Periodontol (2000) 2016:72:13–8.doi: 10.1111/prd.12126
5. Prins J, Blanker MH, Bohnen AM, Thomas S, Bosch JL. Prevalence of Erectile Dysfunction: A Systematic Review of Population-Based Studies. Int J Impot Res (2002) 14:422–32. doi: 10.1038/sj.ijir.3900905
6. Ayta IA, McKinlay JB, Krane RJ. The Likely Worldwide Increase in Erectile Dysfunction Between 1995 and 2025 and Some Possible Policy Consequences. BJU Int (1999) 84:50–6. doi: 10.1046/j.1464-410x.1999.00142.x
7. Khosla S, Farr JN, Tchkonia T, Kirkland JL. The Role of Cellular Senescence in Ageing and Endocrine Disease. Nat Rev Endocrinol (2020) 16:263–75. doi: 10.1038/s41574-020-0335-y
8. da Costa JP, Vitorino R, Silva GM, Vogel C, Duarte A.C, Rocha-Santos T. A Synopsis on Aging-Theories, Mechanisms and Future Prospects. Ageing Res Rev (2016) 29:90–112. doi: 10.1016/j.arr.2016.06.005
9. Asai K, Kudej RK, Shen YT, Yang GP, Takagi G, Kudej AB, et al. Peripheral Vascular Endothelial Dysfunction and Apoptosis in Old Monkeys. Arterioscler Thromb Vasc Biol (2000) 20:1493–9. doi: 10.1161/01.ATV.20.6.1493
10. Donato AJ, Magerko KA, Lawson BR, Durrant JR, Lesniewski LA, Seals DR. SIRT-1 and Vascular Endothelial Dysfunction With Ageing in Mice and Humans. J Physiol (2011) 589:4545–54. doi: 10.1113/jphysiol.2011.211219
11. Donato AJ, Gano LB, Eskurza I, Silver AE, Gates PE, Jablonski K, et al. Vascular Endothelial Dysfunction With Aging: Endothelin-1 and Endothelial Nitric Oxide Synthase. Am J Physiol Heart Circ Physiol (2009) 297:H425–32. doi: 10.1152/ajpheart.00689.2008
12. Aversa A, Bruzziches R, Francomano D, Natali M, Gareri P, Spera G. Endothelial Dysfunction and Erectile Dysfunction in the Aging Man. Int J Urol (2010) 17:38–47. doi: 10.1111/j.1442-2042.2009.02426.x
13. Mercer TR, Dinger ME, Mattick JS. Long non-Coding RNAs: Insights Into Functions. Nat Rev Genet (2009) 10:155–9. doi: 10.1038/nrg2521
14. Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science (2007) 316:1484–8. doi: 10.1126/science.1138341
15. Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell (2009) 136:629–41. doi: 10.1016/j.cell.2009.02.006
16. Quinn JJ, Chang HY. Unique Features of Long non-Coding RNA Biogenesis and Function. Nat Rev Genet (2016) 17:47–62. doi: 10.1038/nrg.2015.10
17. Huang Y. The Novel Regulatory Role of lncRNA-miRNA-mRNA Axis in Cardiovascular Diseases. J Cell Mol Med (2018) 22:5768–75. doi: 10.1111/jcmm.13866
18. Huo W, Hou Y, Li Y, Li H. Downregulated lncRNA-MIAT Confers Protection Against Erectile Dysfunction by Downregulating Lipoprotein Lipase via Activation of miR-328a-5p in Diabetic Rats. Biochim Biophys Acta Mol Basis Dis (2019) 1865:1226–40. doi: 10.1016/j.bbadis.2019.01.018
19. Liao K, Chen J, Fan L, Wang Z. Long Noncoding RNA H19 Promotes the Apoptosis of Corpus Cavernsum Smooth Muscle Cells After Cavernosal Nerve Injury via JNK Signalling Pathway. Andrologia (2021) 53:e14089. doi: 10.1111/and.14089
20. Sun X, Luo L, Li J. LncRNA MALAT1 Facilitates BM-MSCs Differentiation Into Endothelial Cells via Targeting miR-206/VEGFA Axis. Cell Cycle (2020) 19:3018–28. doi: 10.1080/15384101.2020.1829799
21. Cong R, Wang Y, Wang Y, Zhang Q, Zhou X, Ji C, et al. Comprehensive Analysis of lncRNA Expression Pattern and lncRNA-miRNA-mRNA Network in a Rat Model With Cavernous Nerve Injury Erectile Dysfunction. J Sex Med (2020) 17:1603–17. doi: 10.1016/j.jsxm.2020.05.008
22. Kim D, Langmead B, Salzberg SL. HISAT: A Fast Spliced Aligner With Low Memory Requirements. Nat Methods (2015) 12:357–60. doi: 10.1038/nmeth.3317
23. Testai L, Piragine E, Piano I, Flori L, Da Pozzo E, Miragliotta V, et al. The Citrus Flavonoid Naringenin Protects the Myocardium From Ageing-Dependent Dysfunction: Potential Role of SIRT1. Oxid Med Cell Longev (2020) 2020:4650207. doi: 10.1155/2020/4650207
24. Wang B, Yang Q, Sun YY, Xing YF, Wang YB, Lu XT, et al. Resveratrol-Enhanced Autophagic Flux Ameliorates Myocardial Oxidative Stress Injury in Diabetic Mice. J Cell Mol Med (2014) 18:1599–611. doi: 10.1111/jcmm.12312
25. Du L, Chen E, Wu T, Ruan Y, Wu S. Resveratrol Attenuates Hydrogen Peroxide-Induced Aging Through Upregulation of Autophagy in Human Umbilical Vein Endothelial Cells. Drug Des Devel Ther (2019) 13:747–55. doi: 10.2147/DDDT.S179894
26. Meldrum DR, Burnett AL, Dorey G, Esposito K, Ignarro LJ. Erectile Hydraulics: Maximizing Inflow While Minimizing Outflow. J Sex Med (2014) 11:1208–20. doi: 10.1111/jsm.12457
27. Yu J, deMuinck ED, Zhuang Z, Drinane M, Kauser K, Rubanyi GM, et al. Endothelial Nitric Oxide Synthase is Critical for Ischemic Remodeling, Mural Cell Recruitment, and Blood Flow Reserve. Proc Natl Acad Sci U S A (2005) 102:10999–1004. doi: 10.1073/pnas.0501444102
28. Cong X, Kong W. Endothelial Tight Junctions and Their Regulatory Signaling Pathways in Vascular Homeostasis and Disease. Cell Signal (2020) 66:109485. doi: 10.1016/j.cellsig.2019.109485
29. Braga EA, Fridman MV, Moscovtsev AA, Filippova EA, Dmitriev AA, Kushlinskii NE. LncRNAs in Ovarian Cancer Progression, Metastasis, and Main Pathways: ceRNA and Alternative Mechanisms. Int J Mol Sci (2020) 21(22):8855. doi: 10.3390/ijms21228855
30. Su K, Wang N, Shao Q, Liu H, Zhao B, Ma S. The Role of a ceRNA Regulatory Network Based on lncRNA MALAT1 Site in Cancer Progression. BioMed Pharmacother (2021) 137:111389. doi: 10.1016/j.biopha.2021.111389
31. Gong G, Yang XX, Li YY, Geng HY, Wang H, Wang LS, et al. Protective Effects of PI3KCG Gene on Acute Myocardial Infarction. J Thorac Dis (2018) 10:941–53. doi: 10.21037/jtd.2018.01.59
32. Yu Q, Li W, Jin R, Yu S, Xie D, Zheng X, et al. Pi3kγ (Phosphoinositide 3-Kinase γ) Regulates Vascular Smooth Muscle Cell Phenotypic Modulation and Neointimal Formation Through CREB (Cyclic AMP-Response Element Binding Protein)/YAP (Yes-Associated Protein) Signaling. Arterioscler Thromb Vasc Biol (2019) 39:e91–e105. doi: 10.1161/ATVBAHA.118.312212
33. Gongol B, Marin T, Zhang J, Wang SC, Sun W, He M, et al. Shear Stress Regulation of miR-93 and miR-484 Maturation Through Nucleolin. Proc Natl Acad Sci U S A (2019) 116:12974–9. doi: 10.1073/pnas.1902844116
Keywords: lncRNA (long non-coding RNA), erectile dysfunction, aging, bioinformatics, miRNA, RNA sequence analyses
Citation: Zhou X, Cong R, Yao L, Zhou X, Luan J, Zhang Q, Zhang X, Ren X, Zhang T, Meng X and Song N (2022) Comparative Transcriptome Analyses of Geriatric Rats Associate Age-Related Erectile Dysfunction With a lncRNA-miRNA-mRNA Regulatory Network. Front. Endocrinol. 13:887486. doi: 10.3389/fendo.2022.887486
Received: 01 March 2022; Accepted: 09 June 2022;
Published: 11 July 2022.
Edited by:
James Harper, Sam Houston State University, United StatesReviewed by:
Jun Chen, Third Affiliated Hospital of Sun Yat-sen University, ChinaCopyright © 2022 Zhou, Cong, Yao, Zhou, Luan, Zhang, Zhang, Ren, Zhang, Meng and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianghu Meng, eGhtZW5nODg4QDE2My5jb20=; Ninghong Song, c29uZ25pbmdob25nQGFsaXl1bi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.