
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol., 01 August 2022
Sec. Molecular and Structural Endocrinology
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.885650
This article is part of the Research TopicNon-coding Genome and Endocrinology: From Bench to BedsideView all 5 articles
Diabetes mellitus (DM) is an endocrine disorder characterized by a relative or absolute lack of insulin due to the dysfunction or destruction of β-cells. DM is one of the fastest growing challenges to global health in the 21st century and places a tremendous burden on affected individuals and their families and countries. Although insulin and antidiabetic drugs have been used to treat DM, a radical cure for the disease is unavailable. The pathogenesis of DM remains unclear. Emerging roles of circular RNAs (circRNAs) in DM have become a subject of global research. CircRNAs have been verified to participate in the onset and progression of DM, implying their potential roles as novel biomarkers and treatment tools. In the present review, we briefly introduce the characteristics of circRNAs. Next, we focus on specific roles of circRNAs in type 1 diabetes mellitus, type 2 diabetes mellitus, gestational diabetes mellitus and diabetes-associated complications.
Diabetes mellitus (DM) encompasses several heterogeneous disorders that occur when a lack of insulin or insulin resistance evokes a high level of blood glucose. Long-term hyperglycemia undermines the organs of the body, contributing to a group of DM-associated complications. The common types of DM include type 1 diabetes mellitus (T1DM), type 2 diabetes mellitus (T2DM), and gestational diabetes mellitus (GDM). Additionally, specific types of diabetes have been identified, such as monogenic diabetes syndromes, diseases of the exocrine pancreas, and drug- or chemical-induced diabetes (1). However, these specific types of diabetes are not included in this review because they are rare. Prevalence of DM is alarming. According to the latest International Diabetes Federation (IDF) Diabetes Atlas, the number of people aged 20-79 years with DM was estimated to be 536.6 million (10.5% of the global population) in 2021, and this number is predicted to reach 783.2 million (12.2%) by 2045 (2). Globally, almost one in two (44.7%; 239.7 million) adults (20–79 years of age) with DM are living with undiagnosed DM (3). Late diagnosis of DM corresponds to the greater risk of developing complications. In summary, increasing number of patients with DM and its serious complications, such as diabetic retinopathy (DR), diabetic nephropathy (DN), and diabetic cardiomyopathy (DC) (4), exerts an undue pressure on global health. The etiology of DM remains obscure despite extensive research efforts. However, as long as a patient is diagnosed early and appropriate therapy is applied, DM and its complications can be adequately controlled. Thus, a biomarker with high sensitivity and high specificity and an effective method of therapy are imperative.
In recent years, the associations between circular RNAs (circRNAs) and DM have been demonstrated (5–8). CircRNAs are endogenous biomolecules with a structure of covalently closed transcript loops. CircRNAs are expressed in various organisms, from fruit flies and worms to mice and humans (7). CircRNAs can serve as the sponges for specific miRNAs to further regulate the expression levels of the target genes. Advancement of RNA sequencing technology and bioinformatics-based prediction resulted in identification of an increasing number of circRNAs and their biological functions (6, 8–10).
Current studies have reported the effects of circRNAs on DM (Figure 1). For example, the levels of circHIPK3 and ciRS-7/CDR1as are decreased in the islets in a DM mouse model, and this decrease may disrupt β-cell function, including insulin secretion and proliferation (5). Additionally, a study demonstrated an enriched circRNA profile in human pancreatic islets from the donors with T2DM. CircCIRBP was shown to be associated with the insulin secretory index in isolated human islets, and the expression of circCIRBP and circRPH3AL is altered after the contents of fatty acids are elevated in EndoC-βH1 cells. This evidence revealed that understanding of the actions of circRNAs may elucidate the etiology and progression of DM. Additionally, circCAMSAP1 is abundant in human pancreatic islets and is expressed in peripheral blood (6), uncovering the potential use of circRNAs as biomarkers because of their easy availability. Furthermore, circ-TGFBR3, a highly β-cell-selective circRNA, encodes the transforming growth factor (TGF)-β type III receptor and acts mainly by binding with miR-29a-5p and miR-874-3p (11). A recent study used small molecule inhibitors of the TGF-β pathway to demonstrate that the suppression of TGF-β signaling facilitates human pancreatic β-cell replication in human islets transplanted into nonobese diabetic (NOD) mice (12). The destruction of β-cells is the key factor in the progression of DM; in contrast, regeneration of β-cells via self-replication has high potential to restore pancreatic islet function, offering new avenues for DM therapy.
In conclusion, these findings revealed that circRNAs are not only involved in the etiology and progression of DM but also may be used as biomarkers or even play a crucial role in regenerative medicine. Therefore, many very recent studies have focused on various circRNAs and their relevant functions under diabetic conditions with the hope of using these circRNAs to predict and cure DM. However, additional studies are warranted before clinical applications.
In the past, noncoding RNAs (ncRNAs) were regarded as transcriptional noise; however, increasing evidence identified ncRNAs as regulators of diverse biological processes (13). Based on their functions, ncRNAs can be divided into structural or housekeeping ncRNAs (e.g., transfer RNAs, ribosomal RNAs, and small nucleolar RNAs) and regulatory RNAs (14). Regulatory RNAs mainly comprise small interfering RNAs (siRNAs), piwi-RNAs (piRNAs), microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circRNAs. Previous studies were predominantly focused on miRNAs and lncRNAs. Recently, circRNAs have gained increasing attention of the researchers because of their potential as novel biomarkers for early detection of pathological states and as therapeutic targets to ameliorate the clinical burden of the diseases.
Although the circular nature of circRNAs renders their detection and functional annotation challenging, progress in high-throughput RNA sequencing technology and computational algorithms has enabled identification of many endogenous circRNAs in various tissues. These RNA species are covalently closed single-stranded transcripts derived by back-splicing of precursor messenger RNA (pre-mRNA) in eukaryotes (Figure 2) (7). According to their structures, circRNAs can be classified as exon–intron circRNAs (EIciRNAs), intronic circRNAs (ciRNAs), and exonic circRNAs (ecircRNAs) (15). Compared with their linear counterparts (4.0−7.4 h), circRNAs have longer half-lives (18.8–23.7 h) because they lack a 5’ cap and 3’ tail, protecting the molecules against RNase degradation (16). In addition to stability, age-based accumulation trends are another notable feature of circRNAs (17). Consistent with these trends, the number of circRNAs at a specific stage reflects the characteristics of host cells, and this property may be helpful for disease prediction.
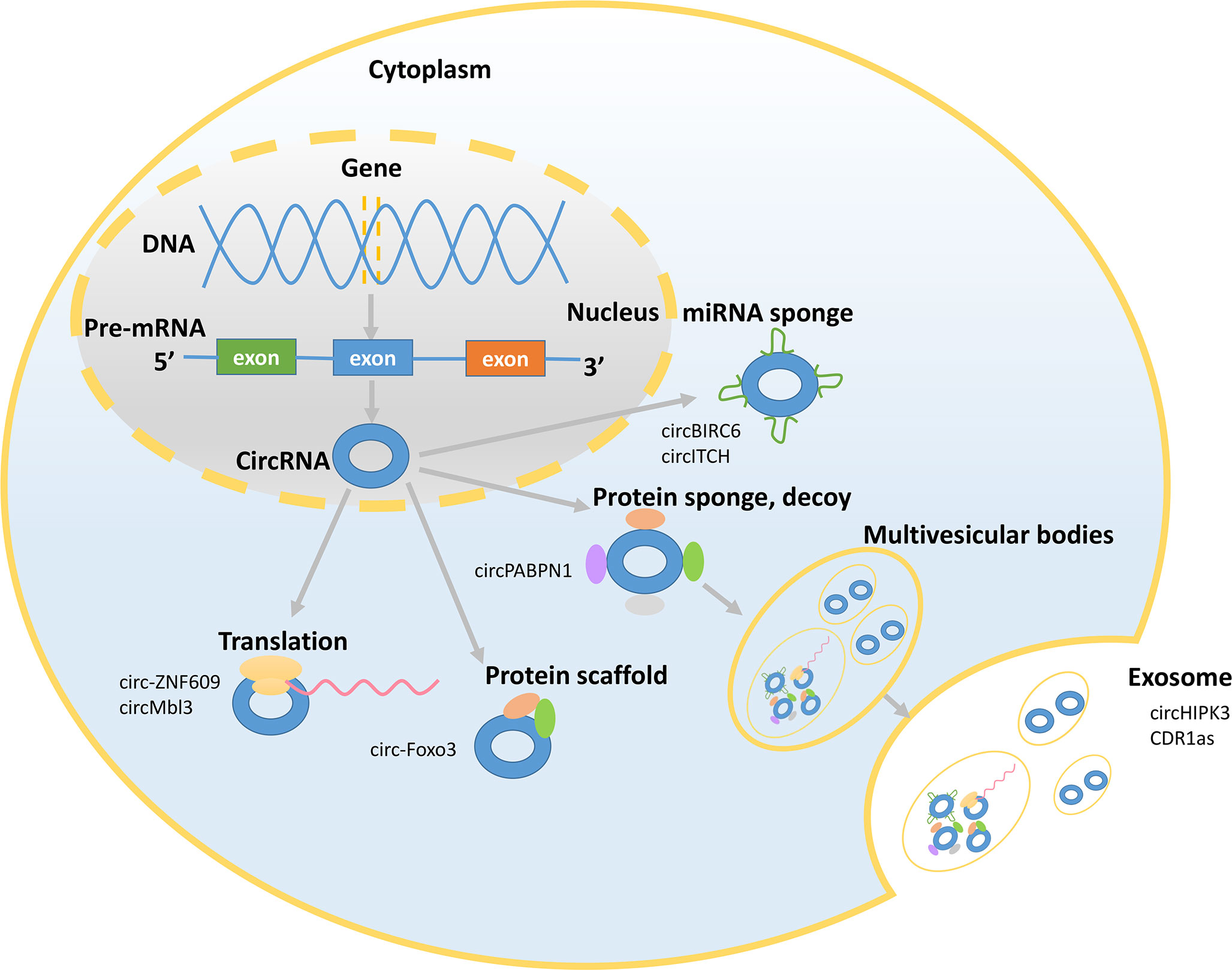
Figure 2 Mechanisms underlying circRNA functions. The picture shows that circRNAs exert their functions in the nucleus and cytoplasm and presents some representative circRNAs. CircRNAs can act as miRNA sponges and bind to RNA-binding proteins (RBPs) and can be translated and sorted to the exosomes.
CircRNAs exert their functions via various mechanisms (Figure 2). First, circRNAs can act as miRNA sponges to influence the expression of the target genes by suppressing miRNA activity (18). CircRNAs compete with parental genes for the miRNA-binding sites given that circRNAs have the reverse sequence of the parental genes. A single circRNA that functions as a competitive endogenous RNA is able to regulate one or multiple miRNAs via its miRNA-binding sites. Moreover, the miRNA-binding sites of circRNAs are located in the 3’ untranslated regions or in the noncoding transcript of a specific gene (19). For example, circBIRC6 acts as a sponge for miR-34a and miR-145 to abate the downregulation of these target genes and restrain the differentiation of human embryonic stem cells (20). CircITCH promotes the expression of ITCH and CBL by sponging miR-214 and miR-22-3p (18). Second, circRNAs can serve as decoys, transporters, or scaffolds to regulate cellular function by binding to proteins (21). CircRNAs have binding sites on RNA-binding proteins (RBPs) and can recruit RBPs. For example, circPABPN1 binds to HuR to suppress the translation of the linear form of PABPN1 mRNA (22). Additionally, circ-Foxo3 acts as a protein scaffold to suppress cell cycle progression by binding to the cell cycle proteins cyclin-dependent kinase 2 and cyclin-dependent kinase inhibitor 1 (23). Third, several studies have provided interesting evidence that circRNAs can be translated (24–26). Translated circRNAs use the start codon of a hosting mRNA and have evolutionary conserved termination codons, which are unique to the circular open reading frame. Because of their closed structure, circRNAs have been demonstrated to undergo cap-independent translation given the existence of the internal ribosome entry sites (25). For example, circ-ZNF609 contains an open reading frame, and heat shock induces intense and meaningful activation of the translation of the flagged circ-ZNF609 (26). Proteins produced by circMbl3 have also been detected. Circ-ZNF609 and circMbl3 confirm the protein-coding potential of circRNAs and may change the view that circRNAs have previously considered ncRNAs. Fourth, circRNAs are signaling molecules related to the intercellular communication. The exosomes, derived from multivesicular bodies, are secreted by most cells and are the vesicles with a diameter of 30-200 nm (27). CircRNAs are more abundant in the exosomes than their linear counterparts and are sorted to the exosomes by changing the corresponding miRNA levels in the host cells. For example, the level of circHIPK3 is higher in the exosomes derived from U-2 OS cells than the level of linear HIPK3. In addition, circRNAs in the exosomes remain circular and are stable in the serum. Some circRNAs that retain their biological activity in the exosomes can be transferred to the recipient cells to regulate their function (28, 29). For example, CDR1as circRNA in the exosomes relieves miR-7-induced growth suppression in the recipient cells. The emergence of circRNAs in the exosomes may indicate their pivotal role in cellular and organ crosstalk considering the functions of the exosomes in mediating the communications between the cells.
Current progress in the studies of circRNAs revealed several functions of circRNAs; however, additional studies are needed to detect the relationships between circRNAs and human diseases.
T1DM, also known as autoimmune diabetes mellitus, is characterized by irreversible destruction of insulin-producing β-cells in pancreatic islets (30) and an absolute paucity of endogenous insulin with subsequent hyperglycemia (31). Genetics and environmental factors are two critical pathogenic elements that contribute to the onset and development of T1DM (32, 33). Screening for islet-specific autoantibodies is the main tool to confirm and predict T1DM (34). However, this approach cannot fully meet the needs for early prediction and prevention of T1DM. CircRNAs may identify individuals at high risk of developing T1DM and help to elucidate the pathogenesis of T1DM (Table 1).
Increasing evidence has shown that circRNAs participate in the onset and development of T1DM by various mechanisms. First, circRNAs are involved in inflammation. A study in vivo identified 1,020 upregulated and 902 downregulated circRNAs in mouse pancreatic β-cells stimulated by the cytokines interferon (IFN)-γ, interleukin (IL)-1β, and tumor necrosis factor (TNF)-α. These differentially expressed circRNAs include circRNAs 000286 and 017277 whose downregulation reduces insulin biosynthesis and secretion and facilitates apoptosis of pancreatic β-cells (35), indicating that circRNAs may participate in proinflammatory cytokine-mediated β-cell dysfunction. Second, circRNAs may be associated with T1DM by influencing macrophages, which contribute to the loss of β-cells and subsequent development of hyperglycemia and are involved in the pathogenesis of T1DM (121, 122). For example, hsa_circ_0060450, circPPM1F, and hsa_circ_0002202 are upregulated in peripheral blood mononuclear cells (PBMCs) from T1DM patients and sponge miR-199a-5p to inhibit the JAK-STAT signaling pathway induced by type I IFN (IFN-I) to further inhibit macrophage-mediated inflammation (36), regulate M1 macrophage activation via the circPPM1F−HuR−PPM1F−NF-κB axis (37), and mediate IFN-I-induced macrophage inflammation (38), respectively. However, additional in vivo experiments are needed to draw stronger conclusions concerning hsa_circ_0060450 and hsa_circ_0002202.
Given that autoantigens are generated as the consequence of β-cell death (123), islet autoantibodies cannot identify high-risk individuals before β-cell dysfunction is manifested. Thus, other early biomarkers are necessary. CircARHGAP12 is decreased in the islets of NOD mice, which is a mouse model of spontaneously developing T1DM (5). Altered expression level of circARHGAP12 under diabetic conditions suggested that this circRNA may be a potential biomarker for T1DM. Another study detected 8,214 circRNAs in human plasma by microarray analysis of the samples of three subjects with new-onset T1DM and identified 61 upregulated and 7 downregulated circRNAs. Notably, the results of gene analysis revealed that these differentially expressed circRNAs participate in inflammation and the pathogenesis of T1DM (9), supporting the future applications of circRNAs as novel biomarkers of T1DM. Additionally, hsa_circ_0072697 is significantly downregulated in patients with T1DM (40). This circRNA regulates the expression of UBASH3A via the circ_0072697−miR-15a−UBASH3A network. Two T1DM-associated variants of UBASH3A, including the risk alleles at rs11203203 and rs80054410, can increase BASH3A expression in human primary CD4+ T cells (41) that mediate the destruction of β-cells (33), indicating that circ_0072697 may mediate the effects of a risk factor for T1DM via UBASH3A and may serve as a biomarker of T1DM.
Presently, insulin injection is required daily for most T1DM patients. Insulin can maintain blood glucose at an appropriate level but is not a radical treatment. Hence, a new and effective therapy is essential, and circRNAs may offer these possibilities. For example, hsa_circ_0002473, which originates from DNAJ homolog subfamily C member 3 (DNAJC3), is upregulated in T1DM patients. DNAJC3 promotes the translocation of the 78-kDa glucose-regulated protein (GRP78) to the plasma membrane to trigger a proapoptotic feedback loop in β-cells (39). This axis may provide novel therapeutic ideas for T1DM because blocking GRP78 is expected to decrease apoptosis to preserve β-cells (40).
In summary, circRNAs may be involved in T1DM via various pathways, such as inflammation and macrophages, can be considered biomarkers, and may provide new targets to treat T1DM.
T2DM is characterized by insulin resistance and relative insulin deficiency and accounts for 90 to 95% of adult DM cases (124); T2DM prevalence in children and adolescents has increased globally in the past several decades (125). The expansion of T2DM prevalence from the adult population to people of all ages has propelled this disease to become one of the greatest public health burdens in the world. Therefore, an in-depth understanding of T2DM is valuable.
CircRNAs are correlated with T2DM (Table 1) and may participate in T2DM pathogenesis. For example, rno_circRNA_008565 is downregulated in a T2DM rat model and binds rno-miR-504 to influence autophagy in rat islet β-cells (43). Additionally, the level of circ-Tulp4 is decreased in pancreatic islets of T2DM mice, and a subsequent study demonstrated that circ-Tulp4 regulates β-cell proliferation via miR-7222-3p−soat1−cyclin D1 signaling (44). Another study using a T2DM rat model induced by streptozotocin (STZ) showed that circANKRD36 expression is higher in T2DM rats, and silencing circANKRD36 decreases blood glucose and suppresses insulin resistance and inflammation by targeting miR-145−XBP1 (52). In addition to the results of the studies in rats, some circRNAs have been identified to be related to T2DM in patients. Recently, a study identified 21 differentially expressed circRNAs, including 5 upregulated and 16 downregulated circRNAs, in venous blood of fasting T2DM patients. Further analysis demonstrated that downregulated circRNAs (novel_circ_0016196, novel_circ_0016198, and novel_circ_0005686) interact with miR-877-3p to modulate mitochondrial apoptosis, and upregulated circRNAs (hsa_circ_0002590 and novel_circ_000372) interact with miR-149-5p to inhibit insulin secretion (45). Another study detected a significant downregulation of hsa-circRNA11783-2 in peripheral blood in the T2DM group, and this circRNA has a notable correlation with T2DM. Hsa-circRNA11783-2 sponges miR-608 and miR-3907; however, the relationships between these two miRNAs and T2DM need further studies (46). Additionally, circRNAs may be involved in T2DM due to an association with IL-6 and TNF-α, which are strongly correlated with T2DM (126). For example, the level of circANKRD36 in peripheral blood leukocytes is elevated in T2DM patients and positively correlates with the level of IL-6 (51). Another study highlighted that circ_CCNB1 is clearly upregulated and positively associated with IL-6 and TNF-α, whereas circ_0009024 is downregulated and negatively associated with IL-6 and TNF-α in the serum samples of T2DM patients (47). Overall, circRNAs may be related to T2DM via the inflammation-associated pathways.
The exact time of the onset of T2DM is usually challenging to determine. Delayed diagnosis may increase the risk of the development of DM-associated complications. Therefore, precise early identification of T2DM is valuable. Evidence has shown that circRNAs may function as potential biomarkers of T2DM. A study using RNA sequencing detected 683 differentially expressed circRNAs in the blood samples of T2DM patients, including 354 upregulated and 329 downregulated circRNAs. The results of functional analysis suggested that these differentially expressed circRNAs may be involved in the apoptosis pathway (10), indicating possible use of circRNAs as biomarkers in the near future. Additionally, receiver operating characteristic (ROC) curves are usually used to estimate the ability of circRNAs to distinguish T2DM patients from healthy controls. Hsa_circ_0071106 is upregulated in the T2DM group and may increase the risk of T2DM by sponging hsa-miR-29a-5p. The area under the ROC curve for this circRNA was 0.690 (48). Additionally, the areas under the curve (AUCs) for hsa_circ_CCNB1 and circ_0009024 were 0.9255 and 0.9592, respectively (47). Recently, circHIPK3 and ciRS-7 were shown to be significantly increased in the peripheral blood samples of T2DM patients and prediabetic individuals, respectively. CircHIPK3 may be able to distinguish T2DM patients from control subjects (AUC=0.665), and ciRS-7 may distinguish prediabetic patients from healthy control subjects (AUC=0.605) (42). Furthermore, the results of a retrospective case−control study in enrolled T2DM patients and control subjects indicated that hsa_circ_0054633 is gradually increased from the baseline condition to the prediabetic state and to actual T2DM in the corresponding groups. The AUCs for this circRNA for the diagnosis of prediabetes and T2DM were 0.747 and 0.72, respectively (53). Additionally, impaired fasting glucose (IFG) indicates a prediabetic status, and evaluation of IFG is vital to relieve the DM burden. Recent evidence has shown that a panel of 2 circRNAs, hsa_circ_0063425 and hsa_circ_0056891, may be able to distinguish T2DM and IFG patients from healthy controls given that the corresponding AUCs were 0.873 and 0.719 for T2DM and IFG, respectively. Moreover, hsa_circ_0063425, which sponges miR-19a-3p, and hsa_circ_0056891, which sponges miR-1-3p, are positively related to AKT and PI3K, respectively, suggesting that these circRNAs may influence PI3K/AKT signaling to modulate the pathogenesis of T2DM (49). These results suggest that circRNAs may function as potential biomarkers for the diagnosis of T2DM. Additionally, lipotoxicity contributes to insulin resistance and β-cell apoptosis (50). The expression of β-cell-derived exosomal circGlis3 is increased under lipotoxic conditions; circGlis3 may be transferred to islet endothelial cells to reduce the viability, migration, and angiogenesis of these cells. Moreover, an increase in exosomal circGlis3 was detected in the serum in the mouse models of DM and in individuals with T2DM. Since the serum is an easily accessible biological fluid, exosomal circGlis3 may be a potential biomarker for T2DM (50). Moreover, insulin treatment may relieve inflammation in T2DM patients. Serum hsa_circ_0054633, which is positively correlated with TNF-α and IL-17, is downregulated in the T2DM with insulin treatment group compared with that in the groups of healthy adults and T2DM patients without insulin treatment, suggesting that hsa_circ_0054633 may be a potential early indicator of the anti-inflammatory effects of insulin treatment (54).
CircRNAs may have potential therapeutic functions in T2DM. A decrease in the expression level of ciRS-7 in the islets of mice causes a reduction in insulin secretion. Furthermore, silencing ciRS-7 diminishes prolactin-stimulated proliferation of primary rat β-cells (5). Silencing circHIPK3 causes decreased insulin secretion and production in response to glucose and decreases β-cell proliferation. Moreover, circHIPK3 regulates the expression level of pivotal β-cell genes, such as Slc2a2, Akt1, and Mtpn, by sponging miR-124-3p and miR-338-3p (Figure 3) (5). These results indicate that a drug targeting circHIPK3 and/or ciRS-7 to maintain the appropriate levels of these circRNAs may help to avoid β-cell dysfunction and abnormal insulin secretion.
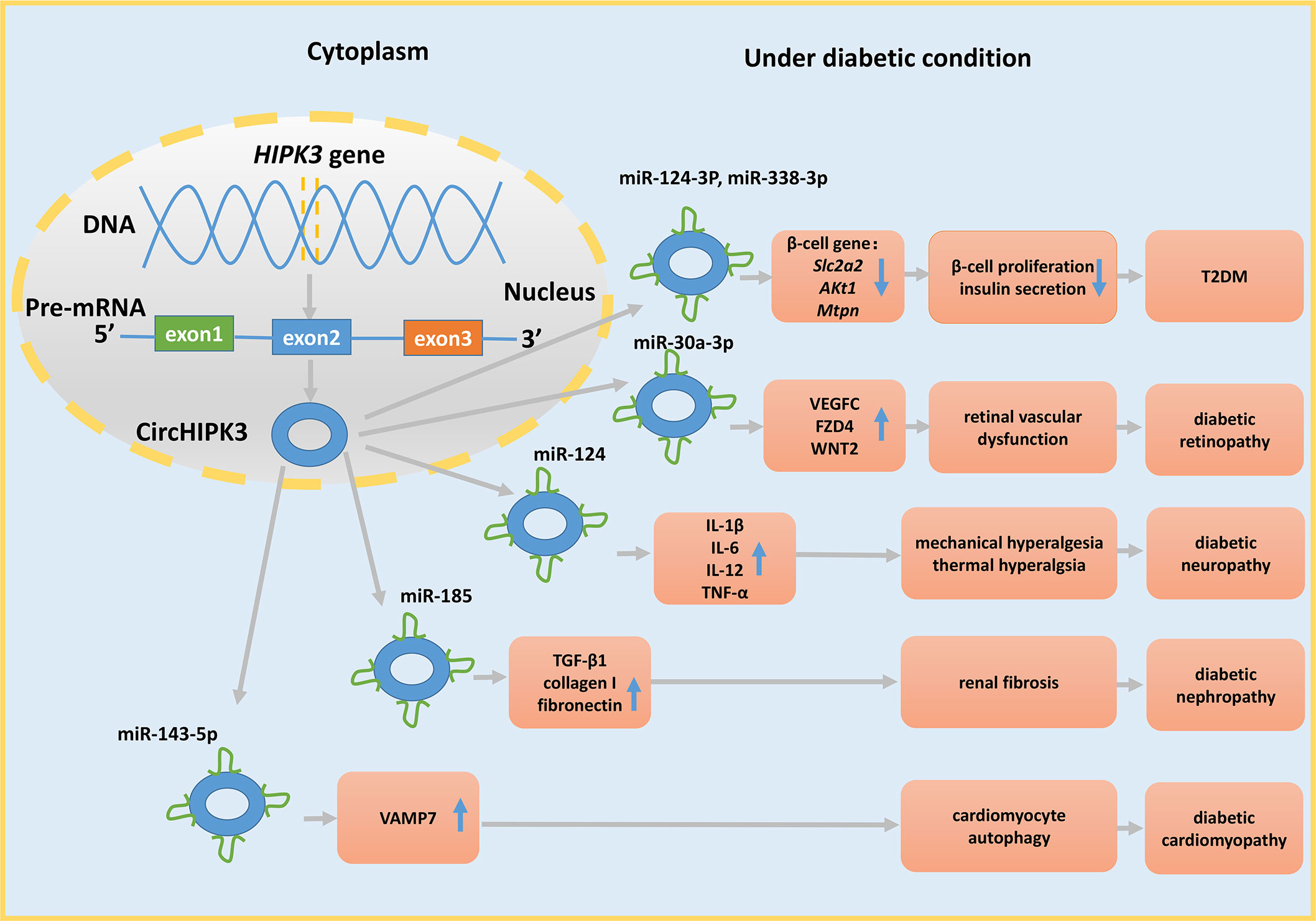
Figure 3 The miRNA sponge role of circHIPK3 in DM and its complications. CircHIPK3 is derived from exon 2 of the HIPK3 gene and acts as a sponge for various miRNAs to regulate cell function, further contributing to the development of DM and its complications.
These studies provide evidence that circRNAs may be involved in the pathogenesis, may be biomarkers for T2DM, and may even represent a therapy for T2DM; however, considerable work is needed to evaluate their safety and efficacy before these circRNAs can be widely applied in clinical practice.
GDM is manifested as impaired glucose tolerance and elevated fasting glucose levels during pregnancy and normal blood glucose levels before pregnancy. The incidence rate of GDM is increasing with prevalence of obesity (127). Unless appropriate treatment is provided, GDM will cause adverse pregnancy outcomes for the offspring and mother (128). The emerging role of circRNAs in GDM has drawn public attention (Table 1).
CircRNAs may participate in GDM progression considering associations between these circRNAs and clinical indicators in GDM patients. For example, hsa_circ_0054633 expression is positively associated with 2-h glucose, glycated hemoglobin (GHBA1), and GHBA1c (63) and was speculated to play a role in the regulation of β-cell proliferation and glucose metabolism during pregnancy because this circRNA was reported to be involved in the progression of the cell cycle and molecular catabolism (64). Hsa_circRNA_102893 is downregulated in the plasma of pregnant women with GDM and acts as a sponge for miR-33a-5p and miR-197-3p (65). Hsa_circRNA_102893 is likely to be related to GDM because miR-33a is positively associated with total serum cholesterol and low-density lipoprotein cholesterol in DM patients (66), and miR-197-3p is correlated with impaired glycemic control (129). Similar to this study, another recent study indicated that circ_0008285 expression is upregulated and circ_0001173 is downregulated in the plasma specimens of GDM patients; the former circRNA is notably correlated with total cholesterol, and the latter circRNA is correlated with glycated hemoglobin (55). A retrospective case–control study revealed that hsa_circ_102682 is downregulated in the case group and significantly correlates with triglycerides, apolipoprotein A1 (APOA1), APOB, and 1-h blood glucose in the serum of GDM patients (56). In addition to the correlations with clinical indicators, circRNAs are also related to multiple adverse events of GDM. Recently, circular RNA vascular endothelial growth factor C (circVEGFC) was identified as a novel regulator of glucose metabolism, and its level was shown to be increased in GDM patients on the day of admission and 1 month before and after delivery. This circRNA is also positively associated with fetal malformation and hypertension (57). Similarly, circRNA actin-related protein 2 homolog (circACTR2) is increased in GDM patients and is associated with higher rates of premature delivery, miscarriage, intrauterine death, fetal malformation, intrauterine infection, and hypertension (58). However, these results were derived from the association studies; therefore, additional in vivo and in vitro experiments will be indispensable to verify the relationships between circRNAs and GDM, leading to more definitive conclusions.
Fetal exposure to maternal diabetes is related to a higher lifetime risk of obesity and development of T2DM in the offspring, possibly contributing to the worldwide diabetes epidemic (130). Thus, it is essential to distinguish GDM patients from healthy individuals early. A study demonstrated that 229 circRNAs are significantly upregulated and 278 circRNAs are significantly downregulated in the umbilical cord blood exosomes of GDM patients, indicating that circRNAs may be the sentinels of GDM (131). Additionally, hsa_circRNA_0054633 expression is elevated in the blood during the second and third trimesters of pregnancy and may be a potential biomarker for GDM (AUC=0.793) (63). Hsa_circRNA_0039480 encapsulated in the exosomes is upregulated in the GDM group and has a diagnostic value in the first, second, and third trimesters of pregnancy (AUC = 0.704, AUC = 0.898, and AUC = 0.698, respectively) (59). Hsa_circRNA_102893 and hsa_circ_102682 are also valuable for the diagnosis of GDM with AUCs of 0.806 and 0.684, respectively (56, 65). The AUC of circVEGFC on the day of admission was 0.8927, indicating a higher sensitivity and specificity of this circRNA for early diagnosis of GDM (57).
CircRNAs may have a possible for GDM treatment based on their influence on the cell functions. For example, circ-PNPT1 expression is higher in placental tissues from individuals with GDM and trophoblasts treated with high glucose (HG). A further study demonstrated that circ-PNPT1 facilitates a biological dysfunction of HG-induced trophoblasts via the miR-889-3p−PAK1 axis (60). Hence, interference with this signaling pathway may maintain normal trophoblast function, implying a promising targeted therapy for GDM. Hsa_circ_0005243 is decreased in both the placenta and plasma of GDM patients. Cell proliferation and migration are inhibited after hsa_circ_0005243 depletion, which causes a trophoblast dysfunction and inflammation via the β-catenin and nuclear factor (NF)-κB pathways (61). Additionally, exosomal circ_0074673 is increased in the exosomes from GDM patients and in human umbilical vein endothelial cells (HUVECs) cocultured with the exosomes. Silencing circ_0074673 promotes the proliferation, migration, and angiogenesis of HG-HUVECs via the miR-1200−MEOX2 axis (62). In summary, a drug that maintains normal function of the cells by regulating circRNA expression levels may open new avenues for GDM therapy.
Overall, abundant evidence suggests a crucial role of circRNAs in the pathogenesis of GDM and indicates that circRNAs may be considered biomarkers and may provide potential therapies for GDM.
DM-related complications can be classified as acute and chronic. The IDF estimated that more than four million adults aged 20–79 years died because of DM and its associated complications in 2019 (132). Therefore, a thorough understanding of DM-related complications is required because of their enormous social burden (Table 1).
DR includes proliferative DR (PDR) and nonproliferative DR (NPDR); DR remains a primary cause of blindness globally, along with vascular leakage, inflammation, and angiogenesis (133). CircRNAs may be related to the pathogenesis of DR in various ways. First, some circRNAs influence DR by serving as stimulators or inhibitors of neovascularization and may thus have a certain pathogenic importance for the progression of DR (134). For example, circ-ITCH is downregulated in retinal pigment epithelial cells isolated from diabetic rats. This circRNA reduces angiogenesis by sponging miR-22 because its overexpression inhibits an increase in matrix metalloproteinase (MMP)-2 and MMP-9, which are positive regulators of neovascularization (67). Conversely, a recent study demonstrated that hsa_circ_0002570 is dramatically increased in the retinal proliferative fibrovascular membranes from DR patients and in human retinal microvascular endothelial cells (hRMECs), and knockdown of this circRNA inhibits HG-induced angiogenesis and inflammation in hRMECs via the circ_0002570−miR-1243−angiomotin axis (68). Circ_0005015 and hsa_circ_0081108 are upregulated in the plasma of DR patients and HG-induced hRMECs, respectively. The results of functional analysis showed that both these circRNAs facilitate angiogenesis by regulating the proliferation and migration of endothelial cell via the circ_0005015−miR-519d-3p−MMP-2/XIAP/STAT3 pathway (69) and the hsa_circ_0081108−miR-29b−VEGF axis (70), respectively. Circular RNA-ZNF609 expression is elevated in DM mice, and silencing this circRNA reduces retinal vessel loss and inhibits pathological angiogenesis in DM mice via the miR-615-5p−MEF2A pathway (71).
Second, circRNAs regulate DR by influencing pericyte loss, microaneurysms, and acellular microvasculature, which are the crucial characteristics of diabetic retinas (135). For example, mmu_circ_0000254 is upregulated and sponges miR-579. Silencing of this circRNA increases retinal vascular leakage and inflammation and the levels of acellular vasculature, pericyte loss, and microaneurysms in diabetic retinas (72). In contrast, silencing circHIPK3 decreases the number of retinal acellular capillaries, vascular leakage, and inflammation via the circHIPK3−miR-30a-3p−VEGFC/WNT2/FZD4 axis (Figure 3). CircHIPK3 derived from exon 2 of the HIPK3 gene (136) influences retinal endothelial cells (82). The level of this circRNA is markedly decreased in diabetic retinas and retinal endothelial cells. In vitro studies demonstrated that the changes in circHIPK3 expression affect the viability, proliferation, migration, and tube formation in retinal endothelial cells (83). Additionally, circDNMT3B is downregulated in hRMECs, and circDNMT3B overexpression reduces the number of acellular capillaries in diabetic rats via the circDNMT3B−miR-20b-5p−BAMBI axis (73). A very recent study reported that circular RNA-ZNF532 is upregulated in the retinal vessels in pericytes under diabetic stress, and silencing cZNF532 reduces the viability, proliferation, and differentiation of pericytes and aggravates retinal pericyte degeneration by sponging miR-29a-3p (74).
Considering the severity of DR, improved biomarkers are urgently needed for early identification of DR patients. The levels of seven circRNAs, including hsa_circRNA_063981, hsa_circRNA_404457, hsa_circRNA_100750, hsa_circRNA_406918, hsa_circRNA_104387, hsa_circRNA_103410, and hsa_circRNA_100192, are distinctly increased in the serum of T2DM patients with PDR (75), implying a potential role of circRNAs as biomarkers for DR. Additionally, hsa_circ_0001953 was reported to be increased in the blood of PDR patients and is positively correlated with the duration of DM and hemoglobin A1c levels. This study indicated a potential value of hsa_circ_0001953 in distinguishing PDR patients from NPDR patients and healthy controls (76).
CircRNAs have been shown to be the potential targets for treatment of DR due to their involvement in the regulation of cell death. For example, exosomal circEhmt1 released from hypoxia-pretreated pericytes reduces HG-induced apoptosis and inflammation in cultured endotheliocytes and protects endotheliocytes against HG-induced injury in vitro via the NFIA/NLRP3 pathway (77). Hsa_circ_0041795 and circRNA_0084043 are clearly upregulated in HG-treated retinal pigment epithelial cells, and circ_001209 is upregulated in HG-treated human retinal vascular endothelial cells. All these circRNAs are able to promote HG-triggered apoptosis of the cells by the miR-646−VEGFC pathway (78), circRNA_0084043−miR-140-3p−TGFA axis (79), and miR-15b-5p−COL12A1 axis (80), respectively. Additionally, ferroptosis is a form of programmed cell death involved in DR. For example, circ-PSEN1 is upregulated in HG-treated ARPE19 cells and its inhibition mitigates ferroptosis of HG-treated retinal pigment epithelial cells via the miR-200b-3p−cofilin-2 axis (81). Thus, a new treatment for DR needs to maintain an appropriate expression level of circRNAs to normalize the properties of the cells.
DN is a severe complication in DM patients. Dysfunction of mesangial cells and podocytes accelerates the development of DN, causing end-stage renal disease and representing a massive healthcare burden (137). Accumulating evidence has revealed that DN is significantly associated with circRNAs.
Fibrosis and inflammation are essential characteristics of DN (138). Circ_0123996 expression is increased in the mouse models of DN, and the expression of fibrosis-related proteins is alleviated when the level of circ_0123996 is decreased. Additionally, circ_0123996 acts as a sponge of miR-149-5p to participate in DN (84). The levels of circEIF4G2, circRNA_15698, and circ_0000491 are dramatically elevated in the kidney of db/db mice in vivo and in a DN cellular model in vitro. Knockdown of these circRNAs restrains the expression of collagen type I and fibronectin, and these circRNAs are able to promote fibrosis via the circEIF4G2−miR-218−SERBP1 pathway (85), circRNA_15698−miR‐185−TGF‐β1 axis (86), and circRNA_0000491−miR-101b−TGFβRI axis (87), respectively. The level of circLRP6 is increased in HG-treated mesangial cells, and this circRNA acts as a sponge for miR-205 to target high mobility group box 1 (HMGB1), which facilitates HG-induced proliferation, oxidative stress, extracellular matrix (ECM) accumulation, and inflammation in mesangial cells via the toll-like receptor 4 (TLR4)/NF-κB pathway. Further studies demonstrated that circLRP6 knockdown increases the expression of miR‐205 and reduces the expression of HMGB1 mRNA and protein, protecting mesangial cells from dysfunction via the circLRP6−miR‐205−HMGB1−TLR4/NF‐κB regulatory network (88). Recent studies showed that the levels of circ_0037128, circ_0000064, and circHIPK3 (Figure 3) are notably elevated in mesangial cells treated with HG, and circ_DLGAP4 and circ_0125310 are also elevated in the exosomes isolated from HG-treated mesangial cells. All these circRNAs promote mesangial cell proliferation and aggravate fibrosis via the circ_0037128−miR-17-3p−AKT3 axis (107), sponging miR-143 (109), sponging miR-185 (89), the circ_DLGAP4−miR-143−ERBB3/NF-κB/MMP-2 axis (90), and the circ_0125310−miR‐422a−IGF1R-p38 axis (91), respectively. Similarly, two in vitro studies using HG-induced HK-2 cells demonstrated that circACTR2 (92) and circ_0037128 (108) are increased and that silencing these circRNAs decreases inflammation and fibrosis in HK-2 cells. Circ_0037128 regulates DN via the circ_0037128−microRNA-497-5p−nuclear factor of activated T cells 5 (NFAT5) axis. Additionally, circ-FBXW12 and hsa_circ_0004442 are upregulated in the sera of DN patients and HG-induced human mesangial cells, and their silencing inhibits HG-induced inflammation and ECM accumulation in human mesangial cells via the miR-31-5p−LIN28B axis (93) and hsa_circ_0004442−miR-126-5p/miR-204-5p−AKT/NF-κB pathway (94), respectively.
The functions of certain circRNAs associated with DN are opposite to the functions of circRNAs listed above. Circ_0080425 is upregulated in DN mice, and circ_LARP4 is downregulated in a DN cellular model; both these circRNAs inhibit cell proliferation and fibrosis via the circ_0080425−miR-24-3p−FGF11 axis (95) and sponging miR-424 (96), respectively. Circ-AKT3 and circSMAD4 are clearly decreased in DN mice and in mouse mesangial cells treated with HG. These circRNAs inhibit fibrosis via the circ-AKT3−miR-296-3p−E-cadherin (97) and circSMAD4−miR-377-3p−BMP7 pathways (98), respectively. The expression of circ-ITCH is clearly decreased in HG-treated rat mesangial cells. Further experiments demonstrated that circ-ITCH alleviates renal inflammation and fibrosis in STZ-induced diabetic mice by regulating the miR-33a-5p−SIRT6 axis (99). Additionally, circRNA_010383 is dramatically reduced in diabetic kidneys, mesangial cells, and tubular epithelial cells stimulated under the HG conditions, and knockdown of this circRNA facilitates proteinuria and renal fibrosis in DN by serving as a sponge for miRNA-135a (100). Additionally, the expression of hsa_circ_0003928 is upregulated in the blood samples of DN patients, and an increase in hsa_circ_0003928 expression alleviates inflammation in HK-2 cells via the miR-151-3p−Anxa2 axis (101).
CircRNAs may be biomarkers. For example, hsa_circRNA_102682 is markedly downregulated in the serum of diabetic patients with high levels of homocysteine, which is an independent risk factor for DN (102), indicating that circRNAs may be associated with the pathogenesis of hyperhomocysteinemia in DN and may be potential biomarkers for DN.
CircRNAs may play a role in DN treatment considering relationships between circRNAs and cell apoptosis. For example, circ_0000712, circ_WBSCR17, and circ_0000285 are increased in DN mice and the cellular models of DN and promote HG-induced apoptosis in DN via the circ_0000712−miR-879-5p−SOX6 axis (103), circ_WBSCR17−miR-185-5p−SOX6 regulatory axis (104), and circ_0000285−miR-654-3p−mitogen-activated protein kinase 6 (MAPK6) axis (105), respectively. These results imply a possible circRNA-targeted therapy for DN by altering circRNA levels. Furthermore, circRNAs regulate the effect of other drugs. For example, germacrone alleviates kidney damage and suppresses podocyte apoptosis in a DN model in mice. The expression of mmu_circRNA_0000309 is decreased in DN mice, and its knockdown mediates drug resistance to germacrone by abolishing the antiapoptotic and anti-injury effects of germacrone via the mmu_circRNA_0000309−miR-188-3p−GPX4 axis (106). Additionally, emodin exerts a protective effect in DN via the circ_0000064−miR-30c-5p−Lmp7 axis (110).
DC is characterized by cardiac dysfunction. The pathological mechanism of DC includes myocardial fibrosis, dysfunctional remodeling, and associated diastolic dysfunction, leading to heart failure (139).
Recent studies demonstrated the associations between DC and circRNAs. For example, the expression of hsa-circRNA11783-2 is reduced in both the T2DM and coronary artery disease groups (46), indirectly implying that hsa-circRNA11783-2 may be correlated with DC. However, the specific mechanism of participation of circRNA11783-2 in DC is poorly understood and requires further investigation. Additionally, circRNA_000203, circRNA_010567, and circHIPK3 are increased in diabetic mice. These circRNAs promote myocardial fibrosis via the circRNA_000203−miR-26b-5p−Col1a2/CTGF axis (111), circRNA_010567−miR-141−TGF-β1 axis (112), and circHIPK3−miR-29b-3p−Col1a1−Col3a1 axis (Figure 3) (113), respectively, and these data elucidate the pathogenesis of DC.
Suppression of cell death and inflammation is considered an important therapy for DC (140). Pyroptosis is a type of programmed cell death associated with inflammation (141). CircRNAs may be used as a novel treatment of DC. Hsa_circ_0076631 is increased in HG-treated cardiomyocytes and in the serum of patients with DM, and circ_0071269 is upregulated in HG-treated H9c2 cells. Both these circRNAs promote the inflammatory response and pyroptosis via the hsa_circ_0076631−miR-214-3p−caspase-1 axis (114) and microRNA-145−gasdermin A axis (115), respectively. Knockdown of these circRNAs protects against diabetic cardiomyopathy injury, implying an accurate therapy for DC.
Diabetic foot (DF) is a severe diabetic complication, and diabetic foot ulceration (DFU) contributes to high mortality of DM. Therefore, an understanding of the molecular mechanism of DFU is urgently needed.
A study demonstrated that hsa_circ_0084443 located in the cytoplasm of human epidermal keratinocytes is increased in the DFU samples and decreases the migration capacity of keratinocytes via the PI3K, EGFR, and ERK signaling pathways (116), indicating that circRNAs are involved in DFU.
Another study identified 10 upregulated and 23 downregulated circRNAs significantly different between patients with DF and DM and patients with DM alone (142), indicating that circRNAs may be the biomarkers for DF. Additionally, the expression levels of hsa_circ_0000907 and hsa_circ_0057362 are high in the serum of patients with DF, and the AUCs for serum hsa_circ_0000907 and hsa_circ_0057362 in diagnosis of early DFU are 0.9389 and 0.8792, respectively, implying that these circRNAs may be used to identify DFU (117).
Recent evidence has shown that the level of circRNA_072697, which acts as a sponge of miR-3150a-3p, is increased in DFU. The level of circRNA_072697 is related to miR-3150a-3p via the MAPK signaling pathway (118). Given that p38 MAPK is a member of the MAPK family and that its inhibition relieves chronic wounds associated with DM (119), circRNA_072697 may be a new target for treatment of DFU.
DN-related pain is manifested in approximately 30% of T2DM patients and increases with the duration of DM. CircHIPK3 is not only involved in DR, DN, DC, and T2DM but is also upregulated in the serum of DM patients with neuropathic pain and in the dorsal root ganglia of STZ-induced DM rats. Furthermore, a higher grade of neuropathic pain in T2DM patients is associated with greater concentrations of circHIPK3 in the serum. Further experiments revealed that circHIPK3 acts as a sponge for miR-124, and silencing this circRNA decreases the expression levels of IL-1β, IL-6, IL-12, and TNF-α to ease mechanical and thermal hyperalgesia (Figure 3). Moreover, a study demonstrated that the treatment with intrathecal circHIPK3 shRNA alleviates neuropathic pain in diabetic rats (120). However, additional extensive research is necessary to apply this therapy to patients in an early stage of the disease.
In summary, regardless of human sample analysis or experiments in the cells or even animals, evidence indicates that circRNAs mediate DM-associated complications via various regulatory networks and may be the biomarkers and potential therapeutic targets for DM-associated complications.
DM is a chronic serious metabolic disease. T1DM, T2DM, and GDM are three common types of DM. Impaired glucose tolerance and impaired fasting glucose indicate a prediabetic state. Prediabetes is a condition in which individuals have impaired glucose metabolism but do not meet the diagnostic criteria of DM; prediabetes is positively associated with the risk of all-cause mortality and incidence of cardiovascular outcomes, stroke, chronic kidney disease, cancer, and dementia (143). Intermediate states of hyperglycemia are becoming an increasing problem, bringing major global challenges. The etiology of DM remains unclear. Therefore, individuals at high risk of DM must be identified early. Currently, despite the advancements in medical technology, many individuals develop DM and its complications, resulting in not only physical harm but also mental effects and even huge economic burden for individuals and countries. Therefore, novel biomarkers and novel treatments are required to overcome DM.
CircRNAs are abundant and stable transcripts, emerging as the key regulators of gene expression. Steady advances in RNA sequencing technology resulted in identification of many circRNAs. Accumulating evidence has demonstrated that circRNAs are related to the onset and progression of DM and its complications. Under pathological conditions, the expression levels of circRNAs are upregulated or downregulated. Differential expression of circRNAs between the diabetic and normal physiological conditions indicates a link between circRNAs and DM. CircRNAs play three roles under diabetic conditions: participation in DM pathogenesis, application as potential biomarkers, and the use as potential therapeutic targets. First, circRNAs contribute to the progression of DM by influencing cellular phenotypes, such as fibrosis and proliferation, and cellular functions, such as insulin secretion, via various signaling pathways. Second, circRNAs are able to distinguish DM patients from healthy control subjects, and diagnostic abilities of circRNAs have been demonstrated by ROC analysis. However, some circRNAs alone do not have high sensitivity and specificity, and a combination of two or more circRNAs may enhance their diagnostic value. Third, the influence of altered expression of circRNAs on disease conditions and the role of circRNAs in regenerative medicine imply that circRNAs may be used as novel agents for treatment of DM. CircRNAs have particular benefits for diagnosis and therapy, such as (1) stability (2), relatively long half-lives in the cells, and (3) availability of circRNAs in the body fluids, such as blood.
However, many hurdles for clinical application of circRNAs exist. First, evidence of the roles of circRNAs in DM is inadequate and needs to be verified by many experiments in vivo and in vitro. Second, the dynamics, pharmacokinetics, and toxic effects of circRNAs need to be further evaluated.
In conclusion, circRNAs are involved in DM and help us gain a better and deeper understanding of the mechanism of DM and its complications, although many challenges remain in the field.
WF searched references, wrote the first draft of the paper, and revised the text. HP, ZX, and ZZ critically revised the text and provided substantial scientific contributions. GH proposed the project and revised the manuscript. All authors approved the final version of the manuscript.
This study was supported by the National Key R&D Program of China (2018YFC1315600, 2018YFC1315603), National Science and Technology Major Project (2020ZX09201-028) and the National Natural Science Foundation of China (81820108007, 81800745)
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. American Diabetes Association2. classification and diagnosis of diabetes: Standards of medical care in diabetes-2021. Diabetes Care (2021) 44(Suppl 1):S15–s33. doi: 10.2337/dc21-S002
2. Sun H, Saeedi P, Karuranga S, Pinkepank M, Ogurtsova K, Duncan BB, et al. IDF diabetes atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res Clin Pract (2021) 109119. doi: 10.1016/j.diabres.2021.109119
3. Ogurtsova K, Guariguata L, Barengo NC, Ruiz PL, Sacre JW, Karuranga S, et al. IDF diabetes atlas: Global estimates of undiagnosed diabetes in adults for 2021. Diabetes Res Clin Pract (2021) 109118. doi: 10.1016/j.diabres.2021.109118
4. Harding JL, Pavkov ME, Magliano DJ, Shaw JE, Gregg EW. Global trends in diabetes complications: a review of current evidence. Diabetologia (2019) 62(1):3–16. doi: 10.1007/s00125-018-4711-2
5. Stoll L, Sobel J, Rodriguez-Trejo A, Guay C, Lee K, Veno MT, et al. Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol Metab (2018) 9:69–83. doi: 10.1016/j.molmet.2018.01.010
6. Haque S, Ames RM, Moore K, Lee BP, Jeffery N, Harries LW. Islet-expressed circular RNAs are associated with type 2 diabetes status in human primary islets and in peripheral blood. BMC Med Genomics (2020) 13(1):64. doi: 10.1186/s12920-020-0713-2
7. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol (2016) 17(4):205–11. doi: 10.1038/nrm.2015.32
8. Zaiou M. circRNAs signature as potential diagnostic and prognostic biomarker for diabetes mellitus and related cardiovascular complications. Cells (2020) 9(3). doi: 10.3390/cells9030659
9. Li Y, Zhou Y, Zhao M, Zou J, Zhu Y, Yuan X, et al. Differential profile of plasma circular RNAs in type 1 diabetes mellitus. Diabetes Metab J (2020) 44(6):854–65. doi: 10.4093/dmj.2019.0151
10. Yang F, Chen Y, Xue Z, Lv Y, Shen L, Li K, et al. High-throughput sequencing and exploration of the lncRNA-circRNA-miRNA-mRNA network in type 2 diabetes mellitus. BioMed Res Int (2020) 2020:8162524. doi: 10.1155/2020/8162524
11. Kaur S, Mirza AH, Pociot F. Cell type-selective expression of circular RNAs in human pancreatic islets. Noncoding RNA (2018) 4(4). doi: 10.3390/ncrna4040038
12. Dhawan S, Dirice E, Kulkarni RN, Bhushan A. Inhibition of TGF-β signaling promotes human pancreatic β-cell replication. Diabetes (2016) 65(5):1208–18. doi: 10.2337/db15-1331
13. Qu Z, Adelson DL. Evolutionary conservation and functional roles of ncRNA. Front Genet (2012) 3:205. doi: 10.3389/fgene.2012.00205
14. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet (2006) 15 Spec No 1:R17–29. doi: 10.1093/hmg/ddl046
15. Lodde V, Murgia G, Simula ER, Steri M, Floris M, Idda ML. Long noncoding RNAs and circular RNAs in autoimmune diseases. Biomolecules (2020) 10(7). doi: 10.3390/biom10071044
16. Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res (2016) 44(3):1370–83. doi: 10.1093/nar/gkv1367
17. Gruner H, Cortés-López M, Cooper DA, Bauer M, Miura P. CircRNA accumulation in the aging mouse brain. Sci Rep (2016) 6(1). doi: 10.1038/srep38907
18. Yu C-Y, Kuo H-C. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci (2019) 26(1). doi: 10.1186/s12929-019-0523-z
19. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol (2016) 238:42–51. doi: 10.1016/j.jbiotec.2016.09.011
20. Yu CY, Li TC, Wu YY, Yeh CH, Chiang W, Chuang CY, et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun (2017) 8(1):1149. doi: 10.1038/s41467-017-01216-w
21. Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J (2019) 38(16):e100836. doi: 10.15252/embj.2018100836
22. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol (2017) 14(3):361–9. doi: 10.1080/15476286.2017.1279788
23. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res (2016) 44(6):2846–58. doi: 10.1093/nar/gkw027
24. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res (2017) 27(5):626–41. doi: 10.1038/cr.2017.31
25. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell (2017) 66(1):9–21.e7. doi: 10.1016/j.molcel.2017.02.021
26. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell (2017) 66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017
27. Pang H, Luo S, Xiao Y, Xia Y, Li X, Huang G, et al. Emerging roles of exosomes in T1DM. Front Immunol (2020) 11:593348. doi: 10.3389/fimmu.2020.593348
28. Preusser C, Hung LH, Schneider T, Schreiner S, Hardt M, Moebus A, et al. Selective release of circRNAs in platelet-derived extracellular vesicles. J Extracell Vesicles (2018) 7(1):1424473. doi: 10.1080/20013078.2018.1424473
29. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res (2015) 25(8):981–4. doi: 10.1038/cr.2015.82
30. Kahaly GJ, Hansen MP. Type 1 diabetes associated autoimmunity. Autoimmun Rev (2016) 15(7):644–8. doi: 10.1016/j.autrev.2016.02.017
31. Atkinson MA, Eisenbarth GS, Michels AW. Type 1 diabetes. Lancet (2014) 383(9911):69–82. doi: 10.1016/S0140-6736(13)60591-7
32. Wang Z, Xie Z, Lu Q, Chang C, Zhou Z. Beyond genetics: What causes type 1 diabetes. Clin Rev Allergy Immunol (2017) 52(2):273–86. doi: 10.1007/s12016-016-8592-1
33. Katsarou A, Gudbjörnsdottir S, Rawshani A, Dabelea D, Bonifacio E, Anderson BJ, et al. Type 1 diabetes mellitus. Nat Rev Dis Primers (2017) 3:17016. doi: 10.1038/nrdp.2017.16
34. Winter WE, Schatz DA. Autoimmune markers in diabetes. Clin Chem (2011) 57(2):168–75. doi: 10.1373/clinchem.2010.148205
35. Wang Z, Deng C, Zheng Y. Involvement of circRNAs in proinflammatory cytokines-mediated β-cell dysfunction. Mediators Inflamm (2021) 2021:5566453. doi: 10.1155/2021/5566453
36. Yang L, Han X, Zhang C, Sun C, Huang S, Xiao W, et al. Hsa_circ_0060450 negatively regulates type I interferon-induced inflammation by serving as miR-199a-5p sponge in type 1 diabetes mellitus. Front Immunol (2020) 11:576903. doi: 10.3389/fimmu.2020.576903
37. Zhang C, Han X, Yang L, Fu J, Sun C, Huang S, et al. Circular RNA circPPM1F modulates M1 macrophage activation and pancreatic islet inflammation in type 1 diabetes mellitus. Theranostics (2020) 10(24):10908–24. doi: 10.7150/thno.48264
38. Yi X, Cheng X. Understanding competitive endogenous RNA network mechanism in type 1 diabetes mellitus using computational and bioinformatics approaches. Diabetes Metab Syndr Obes (2021) 14:3865–945. doi: 10.2147/DMSO.S315488
39. Vig S, Buitinga M, Rondas D, Crevecoeur I, van Zandvoort M, Waelkens E, et al. Cytokine-induced translocation of GRP78 to the plasma membrane triggers a pro-apoptotic feedback loop in pancreatic beta cells. Cell Death Dis (2019) 10(4):309. doi: 10.1038/s41419-019-1518-0
40. Luo S, Deng M, Xie Z, Li X, Huang G, Zhou Z. Circulating circular RNAs profiles associated with type 1 diabetes. Diabetes Metab Res Rev (2021) 37(3):e3394. doi: 10.1002/dmrr.3394
41. Ge Y, Paisie TK, Newman JRB, McIntyre LM, Concannon P. UBASH3A mediates risk for type 1 diabetes through inhibition of T-cell receptor-induced NF-κB signaling. Diabetes (2017) 66(7):2033–43. doi: 10.2337/db16-1023
42. Rezaeinejad F, Mirzaei A, Khalvati B, Sabz G, Alipoor B. Circulating expression levels of CircHIPK3 and CDR1as circular-RNAs in type 2 diabetes patients. Mol Biol Rep (2022) 49(1):131–8. doi: 10.1007/s11033-021-06850-8
43. Bai C, Yang W, Lu Y, Wei W, Li Z, Zhang L. Identification of circular RNAs regulating islet beta-cell autophagy in type 2 diabetes mellitus. BioMed Res Int (2019) 2019:4128315. doi: 10.1155/2019/4128315
44. Wu L, Xiong L, Li J, Peng Z, Zhang L, Shi P, et al. Circ-Tulp4 promotes beta-cell adaptation to lipotoxicity by regulating soat1 expression. J Mol Endocrinol (2020) 65(4):149–61. doi: 10.1530/JME-20-0079
45. Lv B, Bao X, Li P, Lian J, Wu Y, An T, et al. Transcriptome sequencing analysis of peripheral blood of type 2 diabetes mellitus patients with thirst and fatigue. Front Endocrinol (Lausanne) (2020) 11:558344. doi: 10.3389/fendo.2020.558344
46. Li X, Zhao Z, Jian D, Li W, Tang H, Li M. Hsa-circRNA11783-2 in peripheral blood is correlated with coronary artery disease and type 2 diabetes mellitus. Diabetes Vasc Dis Res (2017) 14(6):510–5. doi: 10.1177/1479164117722714
47. Chen X, Yin J, Zhang F, Xiao T, Zhao M. has_circ_CCNB1 and has_circ_0009024 function as potential biomarkers for the diagnosis of type 2 diabetes mellitus. J Clin Lab Anal (2020) 34(10):e23439. doi: 10.1002/jcla.23439
48. Yingying Z, Yongji Y, Qiuting C, Rifang L, Zhuanping Z. has_circ_0071106 can be used as a diagnostic marker for type 2 diabetes. Int J Med Sci (2021) 18(11):2312–20. doi: 10.7150/ijms.52575
49. Lu YK, Chu X, Wang S, Sun Y, Zhang J, Dong J, et al. Identification of circulating hsa_circ_0063425 and hsa_circ_0056891 as novel biomarkers for detection of type 2 diabetes. J Clin Endocrinol Metab (2021) 106(7):e2688–e99. doi: 10.1210/clinem/dgab101
50. Xiong L, Chen L, Wu L, He W, Chen D, Peng Z, et al. Lipotoxicity-induced circGlis3 impairs beta cell function and is transmitted by exosomes to promote islet endothelial cell dysfunction. Diabetologia (2022) 65(1):188–205. doi: 10.1007/s00125-021-05591-4
51. Fang Y, Wang X, Li W, Han J, Jin J, Su F, et al. Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus. Int J Mol Med (2018) 42(4):1865–74. doi: 10.3892/ijmm.2018.3783
52. Lu J, Pang L, Zhang B, Gong Z, Song C. Silencing circANKRD36 inhibits streptozotocin-induced insulin resistance and inflammation in diabetic rats by targeting miR-145 via XBP1. Inflammation Res (2021) 70(6):695–704. doi: 10.1007/s00011-021-01467-w
53. Liang H, Hou L, Wang Q, Zhou X, Sha L, Xu L, et al. Serum hsa_circ_0054633 is elevated and correlated with clinical features in type 2 diabetes mellitus. Ann Clin Lab Sci (2021) 51(1):90–6.
54. Dai H, Hu F, Yang X, Hu P, Chu Y, Bu S. Hsa_circ_0054633 association of c peptide is related to IL-17 and TNF-α in patients with diabetes mellitus receiving insulin treatment. J Clin Lab Anal (2021) 35(8):e23856. doi: 10.1002/jcla.23856
55. Chen H, Zhang S, Wu Y, Li Z, Wang D, Cai S, et al. The role of circular RNA circ_0008285 in gestational diabetes mellitus by regulating the biological functions of trophoblasts. Biol Res (2021) 54(1):14. doi: 10.1186/s40659-021-00337-3
56. Wu H, Zheng X, Liu Y, Shen J, Ye M, Zhang Y. Hsa_circRNA_102682 is closely related to lipid metabolism in gestational diabetes mellitus. Gynecol Endocrinol (2022) 38(1):50–4. doi: 10.1080/09513590.2021.1991911
57. She W, Li T, Liu Y, Liu X. CircRNA circVEGFC is highly expressed in gestational diabetes mellitus (GDM) and it is correlated with multiple adverse events. Diabetes Metab Syndr Obes (2021) 14:4409–14. doi: 10.2147/DMSO.S334728
58. Zhu C, Liu Y, Wu H. Overexpression of circACTR2 in gestational diabetes mellitus predicts intrauterine death, fetal malformation, and intrauterine infection. Diabetes Metab Syndr Obes (2021) 14:4655–60. doi: 10.2147/DMSO.S316043
59. Jiang B, Zhang J, Sun X, Yang C, Cheng G, Xu M, et al. Circulating exosomal hsa_circRNA_0039480 is highly expressed in gestational diabetes mellitus and may be served as a biomarker for early diagnosis of GDM. J Transl Med (2022) 20(1):5. doi: 10.1186/s12967-021-03195-5
60. Zhang L, Zeng M, Tang F, Chen J, Cao D, Tang ZN. Circ-PNPT1 contributes to gestational diabetes mellitus (GDM) by regulating the function of trophoblast cells through miR-889-3p/PAK1 axis. Diabetol Metab Syndr (2021) 13(1):58. doi: 10.1186/s13098-021-00678-9
61. Wang H, Zhou W, She G, Yu B, Sun L. Downregulation of hsa_circ_0005243 induces trophoblast cell dysfunction and inflammation via the β-catenin and NF-κB pathways. Reprod Biol Endocrinol (2020) 18(1):51. doi: 10.1186/s12958-020-00612-0
62. Huang Y, Liang B, Chen X. Exosomal circular RNA circ_0074673 regulates the proliferation, migration, and angiogenesis of human umbilical vein endothelial cells via the microRNA-1200/MEOX2 axis. Bioengineered (2021) 12(1):6782–92. doi: 10.1080/21655979.2021.1967077
63. Wu H, Wu S, Zhu Y, Ye M, Shen J, Liu Y, et al. Hsa_circRNA_0054633 is highly expressed in gestational diabetes mellitus and closely related to glycosylation index. Clin Epigenet (2019) 11(1):22. doi: 10.1186/s13148-019-0610-8
64. Zhao Z, Li X, Jian D, Hao P, Rao L, Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol (2017) 54(3):237–45. doi: 10.1007/s00592-016-0943-0
65. Yang H, Ye W, Chen R, Zeng F, Long Y, Zhang X, et al. Circulating expression of Hsa_circRNA_102893 contributes to early gestational diabetes mellitus detection. Sci Rep (2020) 10(1):19046. doi: 10.1038/s41598-020-76013-5
66. Deng X, Qin S, Chen Y, Liu HY, Yuan E, Deng H, et al. B-RCA revealed circulating miR-33a/b associates with serum cholesterol in type 2 diabetes patients at high risk of ASCVD. Diabetes Res Clin Pract (2018) 140:191–9. doi: 10.1016/j.diabres.2018.03.024
67. Zhou L, Li FF, Wang SM. Circ-ITCH restrains the expression of MMP-2, MMP-9 and TNF-α in diabetic retinopathy by inhibiting miR-22. Exp Mol Pathol (2021) 118:104594. doi: 10.1016/j.yexmp.2020.104594
68. Liu G, Zhou S, Li X, Ding X, Tian M. Inhibition of hsa_circ_0002570 suppresses high-glucose-induced angiogenesis and inflammation in retinal microvascular endothelial cells through miR-1243/angiomotin axis. Cell Stress Chaperones (2020) 25(5):767–77. doi: 10.1007/s12192-020-01111-2
69. Zhang SJ, Chen X, Li CP, Li XM, Liu C, Liu BH, et al. Identification and characterization of circular RNAs as a new class of putative biomarkers in diabetes retinopathy. Invest Ophthalmol Vis Sci (2017) 58(14):6500–9. doi: 10.1167/iovs.17-22698
70. Zou J, Liu KC, Wang WP, Xu Y. Circular RNA COL1A2 promotes angiogenesis via regulating miR-29b/VEGF axis in diabetic retinopathy. Life Sci (2020) 256:117888. doi: 10.1016/j.lfs.2020.117888
71. Liu C, Yao M-D, Li C-P, Shan K, Yang H, Wang J-J, et al. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics (2017) 7(11):2863–77. doi: 10.7150/thno.19353
72. Liu C, Ge HM, Liu BH, Dong R, Shan K, Chen X, et al. Targeting pericyte-endothelial cell crosstalk by circular RNA-cPWWP2A inhibition aggravates diabetes-induced microvascular dysfunction. Proc Natl Acad Sci U S A (2019) 116(15):7455–64. doi: 10.1073/pnas.1814874116
73. Zhu K, Hu X, Chen H, Li F, Yin N, Liu AL, et al. Downregulation of circRNA DMNT3B contributes to diabetic retinal vascular dysfunction through targeting miR-20b-5p and BAMBI. EBioMedicine (2019) 49:341–53. doi: 10.1016/j.ebiom.2019.10.004
74. Jiang Q, Liu C, Li CP, Xu SS, Yao MD, Ge HM, et al. Circular RNA-ZNF532 regulates diabetes-induced retinal pericyte degeneration and vascular dysfunction. J Clin Invest (2020) 130(7):3833–47. doi: 10.1172/JCI123353
75. Gu Y, Ke G, Wang L, Zhou E, Zhu K, Wei Y. Altered expression profile of circular RNAs in the serum of patients with diabetic retinopathy revealed by microarray. Ophthalmic Res (2017) 58(3):176–84. doi: 10.1159/000479156
76. Wu Z, Liu B, Ma Y, Chen H, Wu J, Wang J. Discovery and validation of hsa_circ_0001953 as a potential biomarker for proliferative diabetic retinopathy in human blood. Acta Ophthalmol (2021) 99(3):306–13. doi: 10.1111/aos.14585
77. Ye L, Guo H, Wang Y, Peng Y, Zhang Y, Li S, et al. Exosomal circEhmt1 released from hypoxia-pretreated pericytes regulates high glucose-induced microvascular dysfunction via the NFIA/NLRP3 pathway. Oxid Med Cell Longev (2021) 2021:8833098. doi: 10.1155/2021/8833098
78. Sun H, Kang X. hsa_circ_0041795 contributes to human retinal pigment epithelial cells (ARPE 19) injury induced by high glucose via sponging miR-646 and activating VEGFC. Gene (2020) 747:144654. doi: 10.1016/j.gene.2020.144654
79. Li Y, Cheng T, Wan C, Cang Y. circRNA_0084043 contributes to the progression of diabetic retinopathy via sponging miR-140-3p and inducing TGFA gene expression in retinal pigment epithelial cells. Gene (2020) 747:144653. doi: 10.1016/j.gene.2020.144653
80. Wang F, Zhang M. Circ_001209 aggravates diabetic retinal vascular dysfunction through regulating miR-15b-5p/COL12A1. J Transl Med (2021) 19(1):294. doi: 10.1186/s12967-021-02949-5
81. Zhu Z, Duan P, Song H, Zhou R, Chen T. Downregulation of circular RNA PSEN1 ameliorates ferroptosis of the high glucose treated retinal pigment epithelial cells via miR-200b-3p/cofilin-2 axis. Bioengineered (2021) 12(2):12555–67. doi: 10.1080/21655979.2021.2010369
82. Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep (2017) 18(9):1646–59. doi: 10.15252/embr.201643581
83. Shan K, Liu C, Liu BH, Chen X, Dong R, Liu X, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation (2017) 136(17):1629–42. doi: 10.1161/CIRCULATIONAHA.117.029004
84. Wang W, Feng J, Zhou H, Li Q. Circ_0123996 promotes cell proliferation and fibrosisin mouse mesangial cells through sponging miR-149-5p and inducing Bach1 expression. Gene (2020) 761:144971. doi: 10.1016/j.gene.2020.144971
85. Xu B, Wang Q, Li W, Xia L, Ge X, Shen L, et al. Circular RNA circEIF4G2 aggravates renal fibrosis in diabetic nephropathy by sponging miR-218. J Cell Mol Med (2020) 26(6). doi: 10.1111/jcmm.16129
86. Hu W, Han Q, Zhao L, Wang L. Circular RNA circRNA_15698 aggravates the extracellular matrix of diabetic nephropathy mesangial cells via miR-185/TGF-beta1. J Cell Physiol (2019) 234(2):1469–76. doi: 10.1002/jcp.26959
87. Mou X, Chenv JW, Zhou DY, Liu K, Chen LJ, Zhou D, et al. A novel identified circular RNA, circ_0000491, aggravates the extracellular matrix of diabetic nephropathy glomerular mesangial cells through suppressing miR−101b by targeting TGFβRI. Mol Med Rep (2020) 22(5):3785–94. doi: 10.3892/mmr.2020.11486
88. Chen B, Li Y, Liu Y, Xu Z. circLRP6 regulates high glucose-induced proliferation, oxidative stress, ECM accumulation, and inflammation in mesangial cells. J Cell Physiol (2019) 234(11):21249–59. doi: 10.1002/jcp.28730
89. Liu R, Zhang M, Ge Y. Circular RNA HIPK3 exacerbates diabetic nephropathy and promotes proliferation by sponging miR-185. Gene (2021) 765:145065. doi: 10.1016/j.gene.2020.145065
90. Bai S, Xiong X, Tang B, Ji T, Li X, Qu X, et al. Exosomal circ_DLGAP4 promotes diabetic kidney disease progression by sponging miR-143 and targeting ERBB3/NF-κB/MMP-2 axis. Cell Death Dis (2020) 11(11):1008. doi: 10.1038/s41419-020-03169-3
91. Zhu Y, Zha F, Tang B, Ji TT, Li XY, Feng L, et al. Exosomal hsa_circ_0125310 promotes cell proliferation and fibrosis in diabetic nephropathy via sponging miR-422a and targeting the IGF1R/p38 axis. J Cell Mol Med (2022) 26(1):151–62. doi: 10.1111/jcmm.17065
92. Wen S, Li S, Li L, Fan Q. circACTR2: A novel mechanism regulating high glucose-induced fibrosis in renal tubular cells via pyroptosis. Biol Pharm Bull (2020) 43(3):558–64. doi: 10.1248/bpb.b19-00901
93. Sun A, Sun N, Liang X, Hou Z. Circ-FBXW12 aggravates the development of diabetic nephropathy by binding to miR-31-5p to induce LIN28B. Diabetol Metab Syndr (2021) 13(1):141. doi: 10.1186/s13098-021-00757-x
94. Qiu B, Qi X, Wang J. CircTLK1 downregulation attenuates high glucose-induced human mesangial cell injury by blocking the AKT/NF-κB pathway through sponging miR-126-5p/miR-204-5p. Biochem Genet (2021). doi: 10.1007/s10528-021-10146-8
95. Liu H, Wang X, Wang ZY, Li L. Circ_0080425 inhibits cell proliferation and fibrosis in diabetic nephropathy via sponging miR-24-3p and targeting fibroblast growth factor 11. J Cell Physiol (2020) 235(5):4520–9. doi: 10.1002/jcp.29329
96. Wang Y, Qi Y, Ji T, Tang B, Li X, Zheng P, et al. Circ_LARP4 regulates high glucose-induced cell proliferation, apoptosis, and fibrosis in mouse mesangial cells. Gene (2021) 765:145114. doi: 10.1016/j.gene.2020.145114
97. Tang B, Li W, Ji TT, Li XY, Qu X, Feng L, et al. Circ-AKT3 inhibits the accumulation of extracellular matrix of mesangial cells in diabetic nephropathy via modulating miR-296-3p/E-cadherin signals. J Cell Mol Med (2020) 24(15):8779–88. doi: 10.1111/jcmm.15513
98. Wu R, Niu Z, Ren G, Ruan L, Sun L. CircSMAD4 alleviates high glucose-induced inflammation, extracellular matrix deposition and apoptosis in mouse glomerulus mesangial cells by relieving miR-377-3p-mediated BMP7 inhibition. Diabetol Metab Syndr (2021) 13(1):137. doi: 10.1186/s13098-021-00753-1
99. Liu J, Duan P, Xu C, Xu D, Liu Y, Jiang J. CircRNA circ-ITCH improves renal inflammation and fibrosis in streptozotocin-induced diabetic mice by regulating the miR-33a-5p/SIRT6 axis. Inflammation Res (2021) 70(7):835–46. doi: 10.1007/s00011-021-01485-8
100. Peng F, Gong W, Li S, Yin B, Zhao C, Liu W, et al. circRNA_010383 acts as a sponge for miR-135a and its downregulated expression contributes to renal fibrosis in diabetic nephropathy. Diabetes (2020). doi: 10.2337/db200203
101. An L, Ji D, Hu W, Wang J, Jin X, Qu Y, et al. Interference of Hsa_circ_0003928 alleviates high glucose-induced cell apoptosis and inflammation in HK-2 cells via miR-151-3p/Anxa2. Diabetes Metab Syndr Obes (2020) 13:3157–68. doi: 10.2147/DMSO.S265543
102. Hu F, Sha W, Dai H, Yang X, Hu P, Chu Y, et al. Lower expression of Hsa_circRNA_102682 in diabetic hyperhomocysteinemia negatively related to creatinemia is associated with TGF-β and CTGF. J Clin Lab Anal (2021) 35(8):e23860. doi: 10.1002/jcla.23860
103. Zhao L, Chen H, Zeng Y, Yang K, Zhang R, Li Z, et al. Circular RNA circ_0000712 regulates high glucose-induced apoptosis, inflammation, oxidative stress, and fibrosis in (DN) by targeting the miR-879-5p/SOX6 axis. Endocr J (2021). doi: 10.1507/endocrj.EJ20-0739
104. Li G, Qin Y, Qin S, Zhou X, Zhao W, Zhang D. Circ_WBSCR17 aggravates inflammatory responses and fibrosis by targeting miR-185-5p/SOX6 regulatory axis in high glucose-induced human kidney tubular cells. Life Sci (2020) 259:118269. doi: 10.1016/j.lfs.2020.118269
105. Yao T, Zha D, Hu C, Wu X. Circ_0000285 promotes podocyte injury through sponging miR-654-3p and activating MAPK6 in diabetic nephropathy. Gene (2020) 747:144661. doi: 10.1016/j.gene.2020.144661
106. Jin J, Wang Y, Zheng D, Liang M, He Q. A novel identified circular RNA, mmu_mmu_circRNA_0000309 involves in germacrone-mediated the improvement of diabetic nephropathy through regulating ferroptosis by targeting miR-188-3p/GPX4 signaling axis. Antioxid Redox Signal (2021). doi: 10.1089/ars.2021.0063
107. Wang Q, Cang Z, Shen L, Peng W, Xi L, Jiang X, et al. circ_0037128/miR-17-3p/AKT3 axis promotes the development of diabetic nephropathy. Gene (2021) 765:145076. doi: 10.1016/j.gene.2020.145076
108. Feng T, Li W, Li T, Jiao W, Chen S. Circular RNA_0037128 aggravates high glucose-induced damage in HK-2 cells via regulation of microRNA-497-5p/nuclear factor of activated T cells 5 axis. Bioengineered (2021) 12(2):10959–70. doi: 10.1080/21655979.2021.2001912
109. Ge X, Xi L, Wang Q, Li H, Xia L, Cang Z, et al. Circular RNA Circ_0000064 promotes the proliferation and fibrosis of mesangial cells via miR-143 in diabetic nephropathy. Gene (2020) 758:144952. doi: 10.1016/j.gene.2020.144952
110. Sun L, Han Y, Shen C, Luo H, Wang Z. Emodin alleviates high glucose-induced oxidative stress, inflammation and extracellular matrix accumulation of mesangial cells by the circ_0000064/miR-30c-5p/Lmp7 axis. J Recept Signal Transduct Res (2021) 1-11. doi: 10.1080/10799893.2021.1933028
111. Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep (2017) 7:40342. doi: 10.1038/srep40342
112. Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem Biophys Res Commun (2017) 487(4):769–75. doi: 10.1016/j.bbrc.2017.04.044
113. Wang W, Zhang S, Xu L, Feng Y, Wu X, Zhang M, et al. Involvement of circHIPK3 in the pathogenesis of diabetic cardiomyopathy in mice. Diabetologia (2021) 64(3):681–92. doi: 10.1007/s00125-020-05353-8
114. Yang F, Li A, Qin Y, Che H, Wang Y, Lv J, et al. A novel circular RNA mediates pyroptosis of diabetic cardiomyopathy by functioning as a competing endogenous RNA. Mol Ther - Nucleic Acids (2019) 17:636–43. doi: 10.1016/j.omtn.2019.06.026
115. Fu L, Zhang J, Lin Z, Li Y, Qin G. CircularRNA circ_0071269 knockdown protects against from diabetic cardiomyopathy injury by microRNA-145/gasdermin a axis. Bioengineered (2022) 13(2):2398–411. doi: 10.1080/21655979.2021.2024688
116. Wang A, Toma MA, Ma J, Li D, Vij M, Chu T, et al. Circular RNA hsa_circ_0084443 is upregulated in diabetic foot ulcer and modulates keratinocyte migration and proliferation. Adv Wound Care (New Rochelle) (2020) 9(4):145–60. doi: 10.1089/wound.2019.0956
117. Chen ZJ, Shi XJ, Fu LJ, Liu J, Shi K, Zhang WB, et al. Serum and exosomal hsa_circ_0000907 and hsa_circ_0057362 as novel biomarkers in the early diagnosis of diabetic foot ulcer. Eur Rev Med Pharmacol Sci (2020) 24(15):8117–26. doi: 10.26355/eurrev_202008_22498
118. Tian M, Dong J, Yuan B, Jia H. Identification of potential circRNAs and circRNA-miRNA-mRNA regulatory network in the development of diabetic foot ulcers by integrated bioinformatics analysis. Int Wound J (2021) 18(3):323–31. doi: 10.1111/iwj.13535
119. Medicherla S, Wadsworth S, Cullen B, Silcock D, Ma JY, Mangadu R, et al. p38 MAPK inhibition reduces diabetes-induced impairment of wound healing. Diabetes Metab Syndr Obes (2009) 2:91–100. doi: 10.2147/DMSO.S5859
120. Wang L, Luo T, Bao Z, Li Y, Bu W. Intrathecal circHIPK3 shRNA alleviates neuropathic pain in diabetic rats. Biochem Biophys Res Commun (2018) 505(3):644–50. doi: 10.1016/j.bbrc.2018.09.158
121. Kulkarni A, Pineros AR, Walsh MA, Casimiro I, Ibrahim S, Hernandez-Perez M, et al. 12-lipoxygenase governs the innate immune pathogenesis of islet inflammation and autoimmune diabetes. JCI Insight (2021) 6(14). doi: 10.1172/jci.insight.147812
122. Zirpel H, Roep BO. Islet-resident dendritic cells and macrophages in type 1 diabetes: In search of bigfoot's print. Front Endocrinol (Lausanne) (2021) 12:666795. doi: 10.3389/fendo.2021.666795
123. Ziegler AG, Rewers M, Simell O, Simell T, Lempainen J, Steck A, et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. Jama (2013) 309(23):2473–9. doi: 10.1001/jama.2013.6285
124. Henning RJ. Type-2 diabetes mellitus and cardiovascular disease. Future Cardiol (2018) 14(6):491–509. doi: 10.2217/fca-2018-0045
125. Valaiyapathi B, Gower B, Ashraf AP. Pathophysiology of type 2 diabetes in children and adolescents. Curr Diabetes Rev (2020) 16(3):220–9. doi: 10.2174/1573399814666180608074510
126. Mirza S, Hossain M, Mathews C, Martinez P, Pino P, Gay JL, et al. Type 2-diabetes is associated with elevated levels of TNF-alpha, IL-6 and adiponectin and low levels of leptin in a population of Mexican americans: a cross-sectional study. Cytokine (2012) 57(1):136–42. doi: 10.1016/j.cyto.2011.09.029
127. Casagrande SS, Linder B, Cowie CC. Prevalence of gestational diabetes and subsequent type 2 diabetes among U.S. women. Diabetes Res Clin Pract (2018) 141:200–8. doi: 10.1016/j.diabres.2018.05.010
128. Lowe WL Jr., Scholtens DM, Kuang A, Linder B, Lawrence JM, Lebenthal Y, et al. Hyperglycemia and adverse pregnancy outcome follow-up study (HAPO FUS): Maternal gestational diabetes mellitus and childhood glucose metabolism. Diabetes Care (2019) 42(3):372–80. doi: 10.2337/dc18-1646
129. Samandari N, Mirza AH, Nielsen LB, Kaur S, Hougaard P, Fredheim S, et al. Circulating microRNA levels predict residual beta cell function and glycaemic control in children with type 1 diabetes mellitus. Diabetologia (2017) 60(2):354–63. doi: 10.1007/s00125-016-4156-4
130. Fetita LS, Sobngwi E, Serradas P, Calvo F, Gautier JF. Consequences of fetal exposure to maternal diabetes in offspring. J Clin Endocrinol Metab (2006) 91(10):3718–24. doi: 10.1210/jc.2006-0624
131. Cao M, Zhang L, Lin Y, Li Z, Xu J, Shi Z, et al. Circular RNA expression profiles in umbilical cord blood exosomes from normal and gestational diabetes mellitus patients. Biosci Rep (2020) 40(11). doi: 10.1042/BSR20201946
132. Saeedi P, Salpea P, Karuranga S, Petersohn I, Malanda B, Gregg EW, et al. Mortality attributable to diabetes in 20-79 years old adults, 2019 estimates: Results from the international diabetes federation diabetes atlas, 9(th) edition. Diabetes Res Clin Pract (2020) 162:108086. doi: 10.1016/j.diabres.2020.108086
133. Stitt AW, Curtis TM, Chen M, Medina RJ, McKay GJ, Jenkins A, et al. The progress in understanding and treatment of diabetic retinopathy. Prog Retin Eye Res (2016) 51:156–86. doi: 10.1016/j.preteyeres.2015.08.001
134. Crawford TN, Alfaro DV 3rd, Kerrison JB, Jablon EP. Diabetic retinopathy and angiogenesis. Curr Diabetes Rev (2009) 5(1):8–13. doi: 10.2174/157339909787314149
135. Hammes HP, Lin J, Renner O, Shani M, Lundqvist A, Betsholtz C, et al. Pericytes and the pathogenesis of diabetic retinopathy. Diabetes (2002) 51(10):3107–12. doi: 10.2337/diabetes.51.10.3107
136. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun (2016) 7:11215. doi: 10.1038/ncomms11215
137. Tung CW, Hsu YC, Shih YH, Chang PJ, Lin CL. Glomerular mesangial cell and podocyte injuries in diabetic nephropathy. Nephrol (Carlton) (2018) 23 Suppl 4:32–7. doi: 10.1111/nep.13451
138. Thomas MC. Targeting the pathobiology of diabetic kidney disease. Adv Chronic Kidney Dis (2021) 28(4):282–9. doi: 10.1053/j.ackd.2021.07.001
139. Jia G, Hill MA, Sowers JR. Diabetic cardiomyopathy: An update of mechanisms contributing to this clinical entity. Circ Res (2018) 122(4):624–38. doi: 10.1161/CIRCRESAHA.117.311586
140. Chen D, Ruan X, Liu Y, He Y. MGCS2 silencing attenuates high glucose-induced in vitro diabetic cardiomyopathy by increasing cell viability, and inhibiting apoptosis, inflammation, and oxidative stress. Bioengineered (2022) 13(5):11417–29. doi: 10.1080/21655979.2022.2063222
141. Broz P. Immunology: Caspase target drives pyroptosis. Nature (2015) 526(7575):642–3. doi: 10.1038/nature15632
142. Zhao W, Liang J, Chen Z, Diao Y, Miao G. Combined analysis of circRNA and mRNA profiles and interactions in patients with diabetic foot and diabetes mellitus. Int Wound J (2020) 17(5):1183–93. doi: 10.1111/iwj.13420
Keywords: CircRNAs, type 1 diabetes mellitus, type 2 diabetes mellitus, gestational diabetes mellitus, diabetes-associated complications
Citation: Fan W, Pang H, Xie Z, Huang G and Zhou Z (2022) Circular RNAs in diabetes mellitus and its complications. Front. Endocrinol. 13:885650. doi: 10.3389/fendo.2022.885650
Received: 14 April 2022; Accepted: 12 July 2022;
Published: 01 August 2022.
Edited by:
George Calin, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Cong-Yi Wang, Tongji Medical College, ChinaCopyright © 2022 Fan, Pang, Xie, Huang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gan Huang, aHVhbmdnYW4wMDdAY3N1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.