- 1Elton Laboratory for Molecular Neuroendocrinology, Department of Human Molecular Genetics and Biochemistry, Sackler Faculty of Medicine, Adams Super Center for Brain Studies and Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel
- 2Department of Mathematics and Computer Science, The Open University of Israel, Raanana, Israel
Introduction
Activity-dependent neuroprotective protein (ADNP) (1, 2) was originally discovered at the Gozes laboratory as a glial secreted protein, in the presence of the G-protein-coupled receptors (GPCR)-neuropeptide activator, vasoactive intestinal peptide (VIP) (1). With pituitary adenylate cyclase (PACAP) exhibiting extensive homology to VIP, later studies identified PACAP regulation of ADNP (3–7). Essential for brain development and function (8, 9), ADNP is identified as one of the leading de novo mutated gene causal for an autism/intellectual disability syndrome, the ADNP syndrome (also known as Helsmoortel Van Der Aa) (10–12). Furthermore, recent studies in the Gozes laboratory identified somatic mutations in ADNP in Alzheimer’s disease brains correlating with the progression of Tau pathology (13), and paralleled by Tau depositions in the ADNP syndrome young postmortem brain (14). ADNP functions as a microtubule regulator, enhancing Tau-microtubule binding and protecting against Tauopathy (15). ADNP also functions as a chromatin remodeler (16), further involved in alternative splicing (17) and DNA damage repair (18), regulating thousands of proteins (19). As such, it is our opinion that ADNP is central to key cellular processes. Thus, paralleling disease inflicting truncating mutations in ADNP to natural protein cleavage sites will identify basic disease – related cellular mechanisms, leading to better disease management.
Similarity in ADNP Length After Cleavage by Proteases or Truncation by ADNP Mutations
Computational analysis by the eukaryotic linear motif (ELM) prediction tool (20) identified ADNP cleavage sites including the following classes (Figure 1).
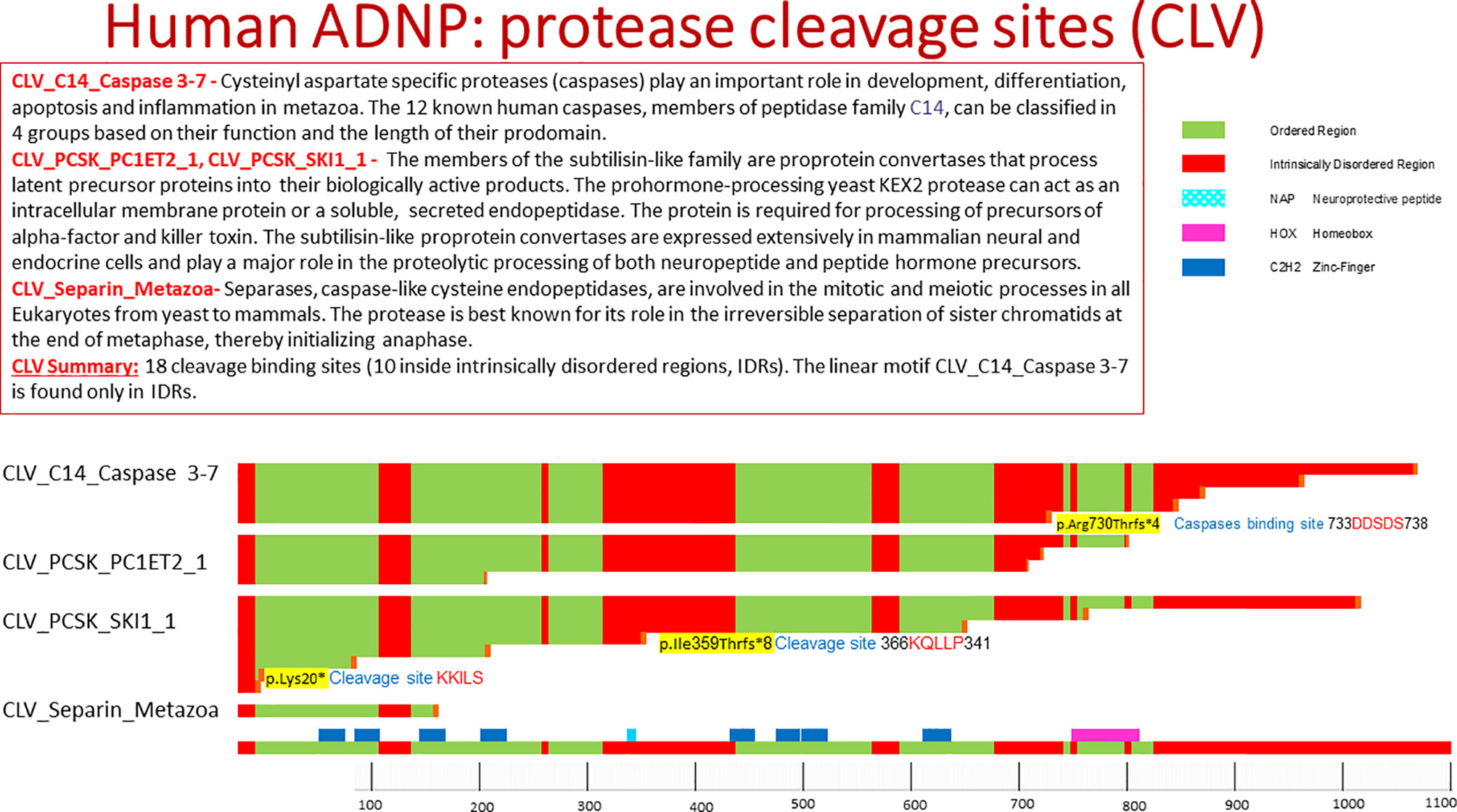
Figure 1 Human ADNP protease cleavage site (CLV). The picture shows protein cleavage sites in ADNP as obtained from the ELM data base. Highlighted in the text are cleavage sites associated with somatic ADNP mutations in postmortem Alzheimer’s disease brains. The ELM site shows ordered and disordered structures within protein, with ordered meaning a three dimensional protein structure.
Cysteinyl Aspartate Specific Proteases (Caspases)
Caspases (21) represent key players in apoptosis, development and differentiation. Caspases recognize the respective substrates by specific cleavage motifs. There are five amino acids of the substrate around the caspase cleavage site, named (N- to C-terminal): P4, P3, P2, P1, P-1. The scissile bond between the essential aspartate at P1 and P-1, usually a small amino acid, is cleaved by caspase-3 and -7, whereas positions P4 to P-1 are important for substrate specificity and recognition. ADNP residues 734-738 contain the motif DDSDS which is a recognition motif for caspase-3 and caspase-7. As indicated above, cleavage of the caspase substrates results in characteristic morphological features of apoptotic cell death, including membrane blebbing, pyknotic nuclei, cell rounding, and formation of apoptotic vesicles. Thus, activated caspase-3, a major enzyme in the apoptotic pathway, is often used as a marker for apoptotic cells. The length of the ADNP protein after the caspase cleavage is 737aa. Interestingly, one of the most prevalent autism/intellectual disability causing de novo mutations in ADNP in p.Arg730* (10), closely located near the caspase cleavage site. Furthermore the recurrent somatic ADNP frameshift mutation p.Arg730Thrfs*4, which is one of the pathogenic mutations in ADNP that is correlated to aging/Alzheimer’s disease, truncates ADNP length to a protein of 734aa (13) (Figure 1).
CLV_PCSK_SKI1_1 (http://elm.eu.org/elms/CLV_PCSK_SKI1_1.html)
The subtilisin-like proprotein convertases (PCSKs) mammalian subtilisin/kexin isozymes (SKIs) are expressed extensively in mammalian neural and endocrine cells and play major roles in the proteolytic processing of both neuropeptides and hormone precursors. The members of the subtilisin-like family are proprotein convertases that process latent precursor proteins into biologically active products. PCSK1 (proprotein convertase 1, NEC1) and PCSK2 (proprotein convertase 2, NEC2) are type I proinsulin-processing enzymes important in regulating insulin biosynthesis. These enzymes also cleave (for example) proopiomelanocortin, prorenin, proenkephalin, prodynorphin, prosomatostatin and progastrin (22–24).
ADNP residues 367-371 KQLLP include the cleavage motif of recognized by the members of the subtilisin-like family. The length of ADNP after cleavage in this site is 367aa. Interestingly, the length of ADNP after the truncating mutation p.Ile359Thrfs*8 is 367aa, found in the post mortem Alzheimer’s brain (13).
PACE4 (paired basic amino acid cleaving system 4, SPC4) is a calcium-dependent serine endoprotease that can cleave precursor protein at paired basic amino acid processing sites [e.g. p.Lys20*, Figure 1, found in the postmortem Alzheimer’s brain (13)]. Its substrates include transforming growth factor beta-related proteins, proalbumin, and von Willebrand factor and assorted neuropeptides. Several paired basic amino acids are found in ADNP (Figure 1).
Discussion
The discovery of ADNP included the identification of a short active motif within ADNP, termed NAP (NAPVSIPQ, Figure 1) (1). NAP enhances ADNP binding to microtubule end binding protein (EB1 and EB3) (25), in turn augmenting Tau microtubule interactions, protecting against tauopathy, even in the face of ADNP mutations, such as the prevalent mutation, p.Arg730* (13–15). Further studies have shown that NAP protects against activated caspase 3 associated apoptosis (26–28). Caspase 3 activation is mediated by cytochrome C (29), protected by NAP (30), whereas cytochrome C is released from mitochondria is enhanced by p53 (31) and ADNP/NAP reduce p53 (2, 32, 33). In turn, DNA damage results in posttranslational modifications of p53 (34), activating the release of cytochrome C (29).
Regarding DNA damage, R-loops are three-stranded nucleic acid structures that accumulate on chromatin in neurological diseases and cancers and contribute to genome instability. ADNP resolves/suppresses R-loops. Importantly, deletion of the ADNP homeodomain severely diminishes R-loop resolution activity, compromising neuronal differentiation. Additionally, patient-derived human induced pluripotent stem cells that contain the prevalent ADNP syndrome-causing mutation p719* exhibit R-loop and CTCF accumulation at ADNP targets (18, 35). These findings, together with our current bioinformatics observations suggest that ADNP cleavage by caspase 3, may be deleterious at two levels: 1] enhancing DNA damage, and 2] reducing ADNP-Tau-microtubule interactions, resulting in tauopathy and followed or paralleled by apoptosis. These findings implicate ADNP as part of the apoptotic pathways in neuronal cells.
Furthermore, Bend et al., identified two distinct and partially opposing genomic DNA methylation episignatures in the peripheral blood samples from 22 patients with ADNP syndrome. The “epi-ADNP-1” episignature included ~ 6000 mostly hypomethylated CpGs, and the “epi-ADNP-2” episignature included ~ 1000 predominantly hypermethylated CpGs. The two signatures correlated with the locations of the ADNP mutations. Epi-ADNP-1 mutations occupy the N- and C-terminus, and epi-ADNP-2 mutations are aggregated on the nuclear localization signal (36). These findings suggest epigenetic activities to the different ADNP cleaved fragments.
Also interesting and related are the findings of caspase 3 - dependent proteolytic cleavage of Tau causes neurofibrillary tangles and results in cognitive impairment during normal aging (37). This is coupled with the finding of plasma P-tau217 levels increasing during the early preclinical stages of Alzheimer’s disease when insoluble tau aggregates are not yet detectable by tau-positron emission tomography (PET), presenting an early biomarker (38, 39). However, an even earlier biomarker is suggested in an unfolded conformational variant of p53, apparent at least 6 years prior to disease onset (40). Phosphorylated Tau and modified p53 in prodromal Alzheimer’s disease are also associated with ADNP found to be the only protein decreasing in Alzheimer’s disease patients’ serum samples (41) and with ADNP serum levels correlating with intelligence, in cognitively intact healthy elderly (42). Importantly, ADNP indirectly interacts with sirtuin 1 (SIRT1) at the chromatin and microtubule/Tau levels (43) as well as regulates Forkhead box O3 (FOXO3) (11), two important genes associated with healthy aging.
Taken together, our studies suggest ADNP directed therapy in susceptible individuals exhibiting the modified p53 biomarker. An ideal therapy would be nasal NAP (davunetide) administration with previous human experience and cognitive score protection/enhancement in amnestic mild cognitive impairment patients (44, 45).
Author Contributions
IG orchestrated the project and wrote the paper. SS identified the cleavage sites and performed the bioinformatics. Both authors contributed to the article and approved the submitted version.
Funding
IG is supported by ERA-NET neuron ADNPinMED, as well as Drs. Ronith and Armand Stemmer (French Friends of Tel Aviv University), Holly and Jonathan Strelzik (American Friends of Tel Aviv University) and - Anne and Alex Cohen (Canadian Friends of Tel Aviv University).
Conflict of Interest
NAP (davunetide) is under patent protection for clinical use. ADNP is under patent protection for Alzheimer’s disease diagnosis (IG).
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Professor Victor OK Li and Associate Professor Jacqueline Lam, University of Hong Kong for their support and for the support of the Catalyst Award.
References
1. Bassan M, Zamostiano R, Davidson A, Pinhasov A, Giladi E, Perl O, et al. Complete Sequence of a Novel Protein Containing a Femtomolar-Activity-Dependent Neuroprotective Peptide. J Neurochem (1999) 72:1283–93. doi: 10.1046/j.1471-4159.1999.0721283.x
2. Zamostiano R, Pinhasov A, Gelber E, Steingart RA, Seroussi E, Giladi E, et al. Cloning and Characterization of the Human Activity-Dependent Neuroprotective Protein. J Biol Chem (2001) 276:708–14. doi: 10.1074/jbc.M007416200
3. Zusev M, Gozes I. Differential Regulation of Activity-Dependent Neuroprotective Protein in Rat Astrocytes by VIP and PACAP. Regul Peptides (2004) 123:33–41. doi: 10.1016/j.regpep.2004.05.021
4. Nakamachi T, Li M, Shioda S, Arimura A. Signaling Involved in Pituitary Adenylate Cyclase-Activating Polypeptide-Stimulated ADNP Expression. Peptides (2006) 27:1859–64. doi: 10.1016/j.peptides.2006.01.007
5. Nakamachi T, Ohtaki H, Yofu S, Dohi K, Watanabe J, Hayashi D, et al. Pituitary Adenylate Cyclase-Activating Polypeptide (PACAP) Type 1 Receptor (PAC1R) Co-Localizes With Activity-Dependent Neuroprotective Protein (ADNP) in the Mouse Brains. Regul Peptides (2008) 145:88–95. doi: 10.1016/j.regpep.2007.09.025
6. Sragovich S, Ziv Y, Vaisvaser S, Shomron N, Hendler T, Gozes I. The Autism-Mutated ADNP Plays a Key Role in Stress Response. Trans Psychiatry (2019) 9:235. doi: 10.1038/s41398-019-0569-4
7. Castorina A, Giunta S, Scuderi S, D’Agata V. Involvement of PACAP/ADNP Signaling in the Resistance to Cell Death in Malignant Peripheral Nerve Sheath Tumor (MPNST) Cells. J Mol Neurosci: MN (2012) 48:674–83. doi: 10.1007/s12031-012-9755-z
8. Pinhasov A, Mandel S, Torchinsky A, Giladi E, Pittel Z, Goldsweig AM, et al. Activity-Dependent Neuroprotective Protein: A Novel Gene Essential for Brain Formation. Brain Res Dev Brain Res (2003) 144:83–90. doi: 10.1016/S0165-3806(03)00162-7
9. Mandel S, Rechavi G, Gozes I. Activity-Dependent Neuroprotective Protein (ADNP) Differentially Interacts With Chromatin to Regulate Genes Essential for Embryogenesis. Dev Biol (2007) 303:814–24. doi: 10.1016/j.ydbio.2006.11.039
10. Van Dijck A, Vulto-van Silfhout AT, Cappuyns E, van der Werf IM, Mancini GM, Tzschach A, et al. Clinical Presentation of a Complex Neurodevelopmental Disorder Caused by Mutations in ADNP. Biol Psychiatry (2019) 85:287–97. doi: 10.1016/j.biopsych.2018.02.1173
11. Karmon G, Sragovich S, Hacohen-Kleiman G, Ben-Horin-Hazak I, Kasparek P, Schuster B, et al. Novel ADNP Syndrome Mice Reveal Dramatic Sex-Specific Peripheral Gene Expression With Brain Synaptic and Tau Pathologies. Biol Psychiatry (2021). doi: 10.1016/j.biopsych.2021.09.018
12. Larsen E, Menashe I, Ziats MN, Pereanu W, Packer A, Banerjee-Basu S. A Systematic Variant Annotation Approach for Ranking Genes Associated With Autism Spectrum Disorders. Mol Autism (2016) 7:44. doi: 10.1186/s13229-016-0103-y
13. Ivashko-Pachima Y, Hadar A, Grigg I, Korenkova V, Kapitansky O, Karmon G, et al. Discovery of Autism/Intellectual Disability Somatic Mutations in Alzheimer’s Brains: Mutated ADNP Cytoskeletal Impairments and Repair as a Case Study. Mol Psychiatry (2021) 26:1619–33. doi: 10.1038/s41380-019-0563-5
14. Grigg I, Ivashko-Pachima Y, Hait TA, Korenková V, Touloumi O, Lagoudaki R, et al. Tauopathy in the Young Autistic Brain: Novel Biomarker and Therapeutic Target. Trans Psychiatry (2020) 10:228. doi: 10.1038/s41398-020-00904-4
15. Ivashko-Pachima Y, Sayas CL, Malishkevich A, Gozes I. ADNP/NAP Dramatically Increase Microtubule End-Binding Protein-Tau Interaction: A Novel Avenue for Protection Against Tauopathy. Mol Psychiatry (2017) 22:1335–44. doi: 10.1038/mp.2016.255
16. Mandel S, Gozes I. Activity-Dependent Neuroprotective Protein Constitutes a Novel Element in the SWI/SNF Chromatin Remodeling Complex. J Biol Chem (2007) 282:34448–56. doi: 10.1074/jbc.M704756200
17. Schirer Y, Malishkevich A, Ophir Y, Lewis J, Giladi E, Gozes I. Novel Marker for the Onset of Frontotemporal Dementia: Early Increase in Activity-Dependent Neuroprotective Protein (ADNP) in the Face of Tau Mutation. PloS One (2014) 9:e87383. doi: 10.1371/journal.pone.0087383
18. Yan Q, Wulfridge P, Doherty J, Fernandez-Luna JL, Real PJ, Tang HY, et al. Proximity Labeling Identifies a Repertoire of Site-Specific R-Loop Modulators. Nat Commun (2022) 13:53. doi: 10.1038/s41467-021-27722-6
19. Amram N, Hacohen-Kleiman G, Sragovich S, Malishkevich A, Katz J, Touloumi O, et al. Sexual Divergence in Microtubule Function: The Novel Intranasal Microtubule Targeting SKIP Normalizes Axonal Transport and Enhances Memory. Mol Psychiatry (2016) 21:1467–76. doi: 10.1038/mp.2015.208
20. Dinkel H, Van Roey K, Michael S, Kumar M, Uyar B, Altenberg B, et al. ELM 2016–Data Update and New Functionality of the Eukaryotic Linear Motif Resource. Nucleic Acids Res (2016) 44:D294–300. doi: 10.1093/nar/gkv1291
21. Stennicke HR, Renatus M, Meldal M, Salvesen GS. Internally Quenched Fluorescent Peptide Substrates Disclose the Subsite Preferences of Human Caspases 1, 3, 6, 7 and 8. Biochem J (2000) 350 Pt 2:563–8.
22. Seidah NG, Mowla SJ, Hamelin J, Mamarbachi AM, Benjannet S, Toure BB, et al. Mammalian Subtilisin/Kexin Isozyme SKI-1: A Widely Expressed Proprotein Convertase With a Unique Cleavage Specificity and Cellular Localization. Proc Natl Acad Sci USA (1999) 96:1321–6. doi: 10.1073/pnas.96.4.1321
23. Jonsson A, Isomaa B, Tuomi T, Eriksson JG, Groop L, Lyssenko V. Effect of a Common Variant of the PCSK2 Gene on Reduced Insulin Secretion. Diabetologia (2012) 55:3245–51. doi: 10.1007/s00125-012-2728-5
24. Ramos-Molina B, Martin MG, Lindberg I. PCSK1 Variants and Human Obesity. Prog Mol Biol Trans Sci (2016) 140:47–74. doi: 10.1016/bs.pmbts.2015.12.001
25. Oz S, Kapitansky O, Ivashco-Pachima Y, Malishkevich A, Giladi E, Skalka N, et al. The NAP Motif of Activity-Dependent Neuroprotective Protein (ADNP) Regulates Dendritic Spines Through Microtubule End Binding Proteins. Mol Psychiatry (2014) 19:1115–24. doi: 10.1038/mp.2014.97
26. Leker RR, Teichner A, Grigoriadis N, Ovadia H, Brenneman DE, Fridkin M, et al. NAP, a Femtomolar-Acting Peptide, Protects the Brain Against Ischemic Injury by Reducing Apoptotic Death. Stroke (2002) 33:1085–92. doi: 10.1161/01.STR.0000014207.05597.D7
27. Idan-Feldman A, Ostritsky R, Gozes I. Tau and Caspase 3 as Targets for Neuroprotection. Int J Alzheimer’s Dis (2012) 2012:493670. doi: 10.1155/2012/493670
28. Idan-Feldman A, Schirer Y, Polyzoidou E, Touloumi O, Lagoudaki R, Grigoriadis NC, et al. Davunetide (NAP) as a Preventative Treatment for Central Nervous System Complications in a Diabetes Rat Model. Neurobiol Dis (2011) 44:327–39. doi: 10.1016/j.nbd.2011.06.020
29. Xing F, Zhan Q, He Y, Cui J, He S, Wang G. 1800mhz Microwave Induces P53 and P53-Mediated Caspase-3 Activation Leading to Cell Apoptosis In Vitro. PloS One (2016) 11:e0163935. doi: 10.1371/journal.pone.0163935
30. Zemlyak I, Sapolsky R, Gozes I. NAP Protects Against Cytochrome C Release: Inhibition of the Initiation of Apoptosis. Eur J Pharmacol (2009) 618:9–14. doi: 10.1016/j.ejphar.2009.07.013
31. Bowman KR, Kim JH, Lim CS. Narrowing the Field: Cancer-Specific Promoters for Mitochondrially-Targeted P53-BH3 Fusion Gene Therapy in Ovarian Cancer. J Ovarian Res (2019) 12:38. doi: 10.1186/s13048-019-0514-4
32. Steingart RA, Gozes I. Recombinant Activity-Dependent Neuroprotective Protein Protects Cells Against Oxidative Stress. Mol Cell Endocrinol (2006) 252:148–53. doi: 10.1016/j.mce.2006.03.029
33. Gozes I, Steingart RA, Spier AD. NAP Mechanisms of Neuroprotection. J Mol Neurosci: MN (2004) 24:67–72. doi: 10.1385/JMN:24:1:067
34. Liu X, Rong F, Tang J, Zhu C, Chen X, Jia S, et al. Repression of P53 Function by SIRT5-Mediated Desuccinylation at Lysine 120 in Response to DNA Damage. Cell Death Differ (2021). doi: 10.1038/s41418-021-00886-w
35. Kaaij LJT, Mohn F, van der Weide RH, de Wit E, Buhler M. The Chahp Complex Counteracts Chromatin Looping at CTCF Sites That Emerged From SINE Expansions in Mouse. Cell (2019) 178:1437–51.e14. doi: 10.1016/j.cell.2019.08.007
36. Bend EG, Aref-Eshghi E, Everman DB, Rogers RC, Cathey SS, Prijoles EJ, et al. Gene Domain-Specific DNA Methylation Episignatures Highlight Distinct Molecular Entities of ADNP Syndrome. Clin Epigenet (2019) 11:64. doi: 10.1186/s13148-019-0658-5
37. Means JC, Gerdes BC, Kaja S, Sumien N, Payne AJ, Stark DA, et al. Caspase-3-Dependent Proteolytic Cleavage of Tau Causes Neurofibrillary Tangles and Results in Cognitive Impairment During Normal Aging. Neurochem Res (2016) 41:2278–88. doi: 10.1007/s11064-016-1942-9
38. Janelidze S, Berron D, Smith R, Strandberg O, Proctor NK, Dage JL, et al. Associations of Plasma Phospho-Tau217 Levels With Tau Positron Emission Tomography in Early Alzheimer Disease. JAMA Neurol (2021) 78:149–56. doi: 10.1001/jamaneurol.2020.4201
39. Mattsson-Carlgren N, Janelidze S, Bateman RJ, Smith R, Stomrud E, Serrano GE, et al. Soluble P-Tau217 Reflects Amyloid and Tau Pathology and Mediates the Association of Amyloid With Tau. EMBO Mol Med (2021) 13:e14022. doi: 10.15252/emmm.202114022
40. Piccirella S, Van Neste L, Fowler C, Masters CL, Fripp J, Doecke JD, et al. A Conformational Variant of P53 (U-P53az) as Blood-Based Biomarker for the Prediction of the Onset of Symptomatic Alzheimer’s Disease. Cell Death Dis (2022). doi: 10.1101/2021.08.23.21261848
41. Yang MH, Yang YH, Lu CY, Jong SB, Chen LJ, Lin YF, et al. Activity-Dependent Neuroprotector Homeobox Protein: A Candidate Protein Identified in Serum as Diagnostic Biomarker for Alzheimer’s Disease. J Proteomics (2012) 75:3617–29. doi: 10.1016/j.jprot.2012.04.017
42. Malishkevich A, Marshall GA, Schultz AP, Sperling RA, Aharon-Peretz J, Gozes I. Blood-Borne Activity-Dependent Neuroprotective Protein (ADNP) Is Correlated With Premorbid Intelligence, Clinical Stage, and Alzheimer’s Disease Biomarkers. J Alzheimer’s Dis: JAD (2016) 50:249–60. doi: 10.3233/JAD-150799
43. Hadar A, Kapitansky O, Ganaiem M, Sragovich S, Lobyntseva A, Giladi E, et al. Introducing ADNP and SIRT1 as New Partners Regulating Microtubules and Histone Methylation. Mol Psychiatry (2021) 26:6550–61. doi: 10.1038/s41380-021-01143-9
44. Gozes I, Stewart A, Morimoto B, Fox A, Sutherland K, Schmeche D. Addressing Alzheimer’s Disease Tangles: From NAP to AL-108. Curr Alzheimer Res (2009) 6:455–60. doi: 10.2174/156720509789207895
45. Morimoto BH, Schmechel D, Hirman J, Blackwell A, Keith J, Gold M, et al. A Double-Blind, Placebo-Controlled, Ascending-Dose, Randomized Study to Evaluate the Safety, Tolerability and Effects on Cognition of AL-108 After 12 Weeks of Intranasal Administration in Subjects With Mild Cognitive Impairment. Dementia Geriatric Cogn Disord (2013) 35:325–36. doi: 10.1159/000348347
Keywords: apoptosis, activity dependent neuroprotective protein (ADNP), caspase, mutations, autism spectrum disorder, Alzheimer’s disease
Citation: Gozes I and Shazman S (2022) STOP Codon Mutations at Sites of Natural Caspase Cleavage Are Implicated in Autism and Alzheimer’s Disease: The Case of ADNP. Front. Endocrinol. 13:867442. doi: 10.3389/fendo.2022.867442
Received: 01 February 2022; Accepted: 17 February 2022;
Published: 23 March 2022.
Edited by:
Liliane Schoofs, KU Leuven, BelgiumReviewed by:
Yukio Ago, Hiroshima University, JapanCopyright © 2022 Gozes and Shazman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Illana Gozes, aWdvemVzQHRhdWV4LnRhdS5hYy5pbA==
 Illana Gozes
Illana Gozes Shula Shazman
Shula Shazman