
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol. , 14 November 2022
Sec. Bone Research
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.1054827
This article is part of the Research Topic Role of Long non-Coding RNA and Circular RNA in Bone Metabolism and their Role as Circulating Biomarkers for Bone Diseases View all 6 articles
Long non-coding RNAs (lncRNAs) have been comprehensively implicated in various cellular functions by mediating transcriptional or post-transcriptional activities. MALAT1 is involved in the differentiation, proliferation, and apoptosis of multiple cell lines, including BMSCs, osteoblasts, osteoclasts, and chondrocytes. Interestingly, MALAT1 may interact with RNAs or proteins, regulating cellular processes. Recently, MALAT1 has been reported to be associated with the development of bone and cartilage diseases by orchestrating the signaling network. The involvement of MALAT1 in the pathological development of bone and cartilage diseases makes it available to be a potential biomarker for clinical diagnosis or prognosis. Although the potential mechanisms of MALAT1 in mediating the cellular processes of bone and cartilage diseases are still needed for further elucidation, MALAT1 shows great promise for drug development.
Non-coding RNAs (ncRNAs) are commonly divided into four groups, including long ncRNAs (lncRNAs), microRNAs (miRNAs), circular RNAs, and pseudogenes. Biologically, ncRNAs have been implicated in the gene expression regulation at the levels of transcription and post-transcription. LncRNAs, usually more than 200 nucleotides in length, have been demonstrated to be associated with the pathogenesis and progression of various diseases. Due to alternative cleavage and/or pre-mRNA splicing and polyadenylation, many different isoforms can be produced from the same locus by lncRNAs (1). This may lead to generation of multiple and diverse lncRNAs. However, most of lncRNAs are expressed at a low level and restricted to specific sub-cell localizations/cell lines/developmental stages/physiological conditions (2). It can be postulated that lncRNAs are remarkably involved in the physiological and pathological processes.
Metastasis‐associated lung adenocarcinoma transcript‐1 (MALAT1, also known as NEAT2 or HCN), encoded on the chromosome 11q13.1 in human and 19qA in mouse, is a highly conserved lncRNA that is retained in the nucleus and abundantly expressed in various cells (3). Evolutionally, MALAT1 shows about 50% conservation in overall sequence and more than 80% conservation in the triple-helix transcript at 3’ end (4). The expression of MALAT1 is high, and this might be related to the strong activity of the promoter and the stability of the transcribed RNA (5). MALAT1, a well-studied lncRNA in human disease, has been implicated in cell differentiation, proliferation, and death. For example, co-culture of prostate cancer PC3 cells and osteoblasts may up regulate the expression of MALAT1 in PC3 cells, and co-culture with SOSTKO osteoblasts may further enhance MALAT1 expression, promoting bone metastasis (6).
Bone undergoes constant modeling and remodeling to maintain homeostasis, which requires multiple regulations in cell differentiation, proliferation, and apoptosis. Disturbance of bone homeostasis may lead to the pathogenesis and progression of bone diseases, such as osteoporosis (OP), osteoarthritis (OA), and rheumatoid arthritis (RA) (7, 8). LncRNAs have been reported to epigenetically regulate the differentiation of mesenchymal stromal cells (MSCs) and the development of diseases (9). It has been reported that the expression of MALAT1 in patients with OP is significantly lower than that in healthy people (10). Bone mineral density (BMD) is a diagnostic factor for OP, and genome-wide association study have identified that the intronic variants at MALAT1 gene locus are associated with low BMD in the Qatari population (11). MALAT1 promotes ossification of the posterior longitudinal ligament (OPLL) by enhancing the transcriptional expression of connexin 43 (Cx43) indirectly and sponging miR-1. Deletion of MALAT1 expression may lead to inhibition of Cx43 expression, osteogenesis, proliferation, and inflammation in OPLL fibroblasts (12). The critical roles of MALAT1 in bone diseases have been demonstrated, and this article provides a review on this field.
Nuclear speckles, consisting of RNAs and proteins, are the organelles with self-assembly, governing some specific steps involved in gene expression, such as transcription, splicing, mRNA export. Most of the splicing and transcription are performed at the periphery sites of the speckles, and RNA maturation is modulated at the central sites (13). It has been reported that MALAT1 is abundantly nuclear distribution and located at the periphery sites of the speckles. A study shows that MALAT1 is not essential for the formation of nuclear speckles, and MALAT1 deficiency does not produce any effects on the number, size, distribution, assembly, and structural maintenance of nuclear speckles (14). However, it is demonstrated that depletion of MALAT1 may induce an appropriate 50% reduction of SON expression, which is a scaffold protein in the nuclear speckle. This may, at least partially, result in a decrease in nuclear speckle size in MALAT1-knockout cells (15). Interestingly, MALAT1 tends to interact with many pre-mRNA splicing factors, such as SON1, SRSF1, hnRNPH1, and hnRNPC, which are accumulated in the nuclear speckles. Thus, pre-mRNA splicing can be alternatively regulated by the expression of MALAT1 (16). In addition, MALAT1 may regulate the expression of COL1A1 at the RNA level. Specifically, depletion of MALAT1 can lead to aberrant splicing and decreased distributing sites of COL1A1 RNA (15). Genome-wide transcriptome study shows that MALAT promotes cell proliferation by regulating the pre-mRNA splicing of cell cycle-mediated transcription factor B-MYB in human diploid fibroblasts (17). Dissociation of MALAT1 with the nuclear speckles may also produce negative effects on the transcriptional regulation of target genes (18) (Figure 1).
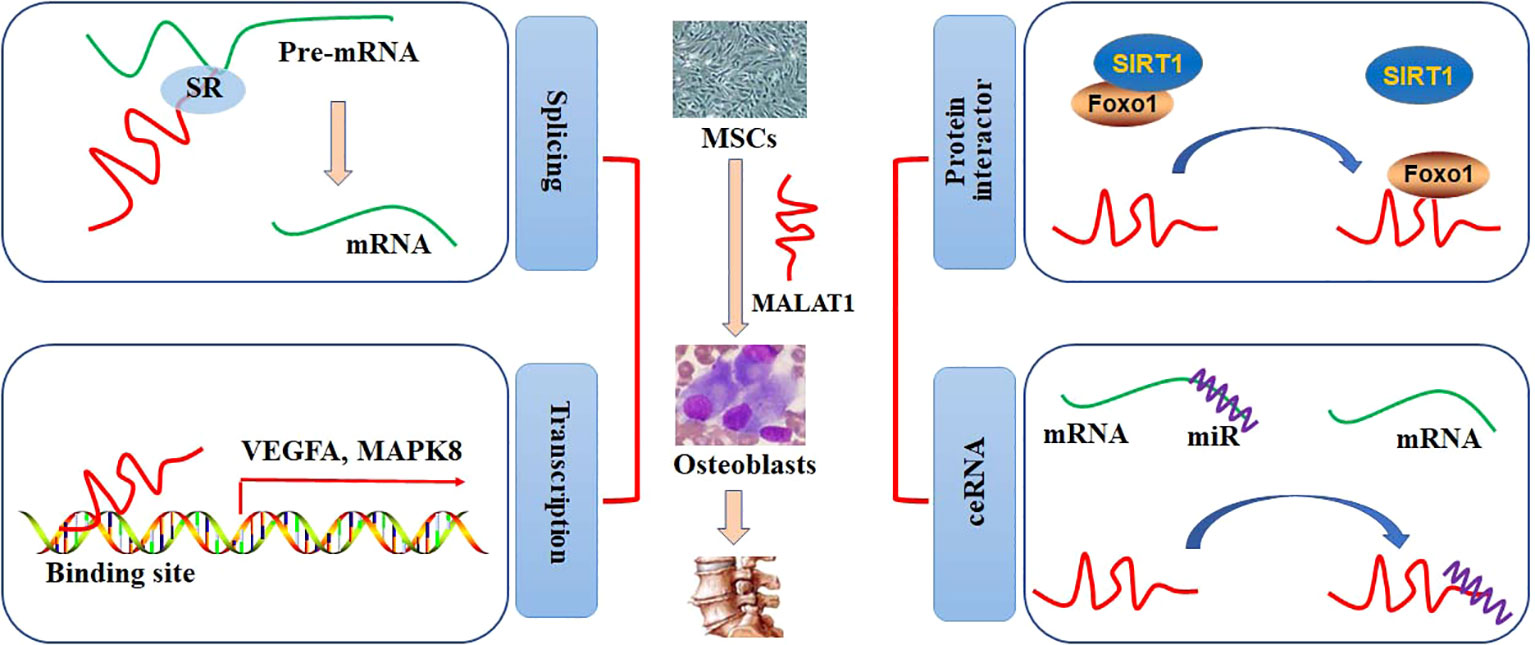
Figure 1 The roles of MALAT1 in bone and cartilage diseases. The regulatory activities of MALAT1 in osteogenic differentiation can be performed by mediating splicing and transcription and acting as a protein interactor or ceRNA.
MALAT1 has been shown to account for a high percentage of the chromatin fraction and be actively associated with transcriptional gene expression. In MCF-7 cells, MALAT1 can preferably interact with the active genomic sites. For example, MALAT1 physically binds to CTCF sites and increases the activity of the promoters (19). Spatially proximal to the target binding sites may increase the binding affinity to lncRNAs than distant binding sites. MALAT1, locates at the upstream (60 Kb approximately) of LTPB3 promoter, can be recruited to the promoter of LTPB3 with higher scores, indicating either RNA-DNA or RNA-protein-DNA interaction (19). Chromatin immunoprecipitation (ChIP) has been employed to identify that MALAT1 can regulate the gene expression of EEF1A1 in MDA-MB231 and SKRB3 cells by binding to the regulatory elements in the promoter. Knockdown of MALAT1 expression may abolish cell proliferation and invasion (20). Clinically, highly expression of MALAT1 is positively correlated with poor overall survival in patients with phase III and IV GC. However, no statistical difference is observed in patients with phase I and II GC (21). The association between MALAT1 and EZH2 may provide a novel therapeutic strategy for management of many cancers, such as GC and prostate cancer (22). MALAT1, acts as a chromatin associated RNA, binds to the target genomic sequences by proximity ligation. By employing new technologies, such as mapping RNA-genome interactions (MARGI) and global RNA interaction with DNA by deep sequencing (GRID-seq), a large body of genomic loci for MALAT1 binding has been explored (23, 24). The diagnostic and prognostic effects of MALAT1 in patients with pancreatic cancer have been evaluated by Gene Expression Omnibus, Oncomine, and The Cancer Genome Atlas databases. CCND1, MAPK8, VEGFA, FOS, CDH1, and HSP90AA1 have been identified as the target genes of MALAT1 in patients with pancreatic cancer (25) (Figure 1).
The regulatory activity of MALAT1 at the transcriptional level has been demonstrated by binding to the specific sites in the chromatin/genome or interacting with the transcriptional factors or co-activators. In mice diabetic kidney, high glucose-increased expression of MALAT1 is positively related to the levels of serum creatinine and urinary albumin. Further study shows that MALAT1 down regulates the expression of SIRT1 by competitively binding to Foxo1 and disrupting the interaction between SIRT1 and Foxo1 in HK-2 cells (26). In lung transplant ischemia-reperfusion (LTIR) rats, enhanced expression of MALAT1 may up regulate IL-8 expression by recruiting p300, ameliorating inflammatory responses. Silencing of MALAT1 expression can reversely inhibit inflammatory injury in LTIR rats (27). MALAT1 may associate with EZH2, a subunit catalyzing the trimethylation modification in histone 3 Lys27 (H3K27me3), to inhibit the expression of tumor suppressor PCH10 and increase migration and invasion in gastric cancer (GC) cells (21). MALAT1/EZH2 can also negatively inhibits KEAP1 transcriptional expression, leading to up regulation of NRF2 expression in multiple myeloma cells (28). In a high throughput experiment, 127 potential proteins are identified as the interacting targets of MALAT1. Further study shows that MALAT1 may competitively interact with DBC1, resulting in SIRT1 release, p53 deacetylation, cell proliferation, and inhibition of cell apoptosis (29). The roles of MALAT1 in cancer cells have been reviewed (30). Knockdown or overexpression of MALAT1 under different pathophysiological conditions exhibits substantial transcriptional changes (Figure 1).
A large body of studies report MALAT1 as a miRNA sponge or a ceRNA in various cell lines. In high glucose-treated HK-2 cells, up regulated expression of MALAT1 significantly inhibits cell viability, increases apoptosis, and promotes the productions of IL-1β, IL-6, and TNFα by sponging miR-16b-5p, which targets to degrade TLR4. Knockdown of MALAT1 expression alleviates high glucose-induced injury in HK-2 cells, protecting against diabetic nephropathy (31). In human liver tissues, the up regulation of MALAT1 expression is associated with the progression of nonalcoholic fatty liver disease (NAFLD). In fatty acid-treated hepatocytes in vitro, knockdown expression of MALAT1 ameliorates the inhibitory activity against miR-206, which degrades ARNT. As a transcriptional factor, ARNT can bind to and inhibit the promoter activity of PPARγ. Thus, MALAT1/miR-206/PPARγ can be a therapeutic target against NAFLD (32). Similarly, MALAT1 acts as a sponge to inhibit miR-181a, which is up regulated in angiotensin II-treated cardiomyocytes. Decreased expression of MALAT1 promotes miR-181a-induced cardiac hypertrophy by decreasing the expression of HMGB2 (33). By sequestering miRNAs, MALAT1 may directly/indirectly regulate the pathophysiological processes of many diseases, providing a potential target for clinically therapeutic management (Figure 1).
Osteogenic differentiation of bone marrow-derived MSCs (BMSCs) is critical for the formation of bone. Both osteoblasts and osteoclasts are differentiated from BMSCs. The complex regulatory mechanisms orchestrate the processes of osteogenic differentiation (Figure 1). It has been reviewed that many signaling pathways, such as Wnt/β-catenin, TGF-β1/Smad2/3, BMP, and PI3K/AKT, have been involved in the regulation of osteogenic differentiation (34). The importance of lncRNAs in gene regulation at the epigenetic, transcriptional, and post-transcriptional levels has been identified in the stages of osteogenic differentiation. These regulations include histone modification, transcriptional interference, nuclear and cytoplasmic trafficking, genomic imprinting, and X-chromosome inactivation (35). One study has been shown that 89 lncRNAs from the constructed lncRNA-miRNA-mRNA network are differentially expressed after osteogenic differentiation, mediating the expression of Wnt/β-catenin, TGF-β1/Smad2/3, and PI3K/AKT signaling pathways in dental pulp stem cells (36). Furthermore, the lncRNAs-transcription factor (TF) feedback loops are identified. Specifically, lncRNAs and TFs may compose feedback loops to mediate osteoblast differentiation, due to that TFs can directly mediate the differentiation activity by binding to the DNA-regulatory elements of lncRNAs (37).
MALAT1 has been considered as a biomarker in OP and is related to osteogenic differentiation (38). During the culture period of human MSCs, the osteogenic differentiation is developed from day 1 to 14. MALAT1 promotes the expression of osterix (Osx) and increase osteogenic differentiation by sponging miR-143 in human BMSCs (39). MALAT1 has been reported to increase RUNX2-regulated osteogenic differentiation of adipose-derived MSC (ADSCs) by specifically sponging miR-30 (40). In osteogenic hBMSCs, the expression of MALAT1 and Osx is up regulated, and the expression of miR-96 is down regulated (41). Co-culture of BMSCs-derived exosome and human osteoblasts increases SATB2-mediated osteoblast differentiation and proliferation by enhancing MALAT1 expression and attenuating miR-34c expression. Up regulation of MALAT1 expression can ameliorate the pathological changes of OP in ovariectomized (OVX) mice (42).
Simultaneously, the expression of MALAT1 is significantly increased. Knockdown of MALAT1 expression can attenuate osteogenic differentiation by mediating has-miR-214-3p/BMP2 axis (43) (Table 1). In microgravity-treated MC3T3-E1 cells, the expression of MALAT1 is down regulated, accompanied by decreased expression of alkaline phosphatase (ALP), Col1a1, and BMP4. In human aortic valve interstitial cells (hAVICs), MALAT1 functions as a positive mediator in osteogenic differentiation through Smad4 by suppressing the expression of miR-204 (Table 1), as indicated by increased expression of ALP and osteocalcin (OCN) and mineralization (44). Overexpression of MALAT1 can promote osteogenic differentiation by regulating the activity of miR-127/AKT3 axis, alleviating OP (38). Consistently, the expression of MALAT1 is up regulated after osteogenic differentiation of BMSCs and down regulated after osteoclast differentiation of mononuclear macrophages. Knockdown of MALAT1 expression suppresses osteogenic differentiation and facilitates osteoclast differentiation by mediating miR-124-3p/IGF2BP1 axis (Table 1). Overexpressed MALAT1 stimulates the activity of Wnt/β-catenin signaling pathway, ameliorating bone injury in mice (10).
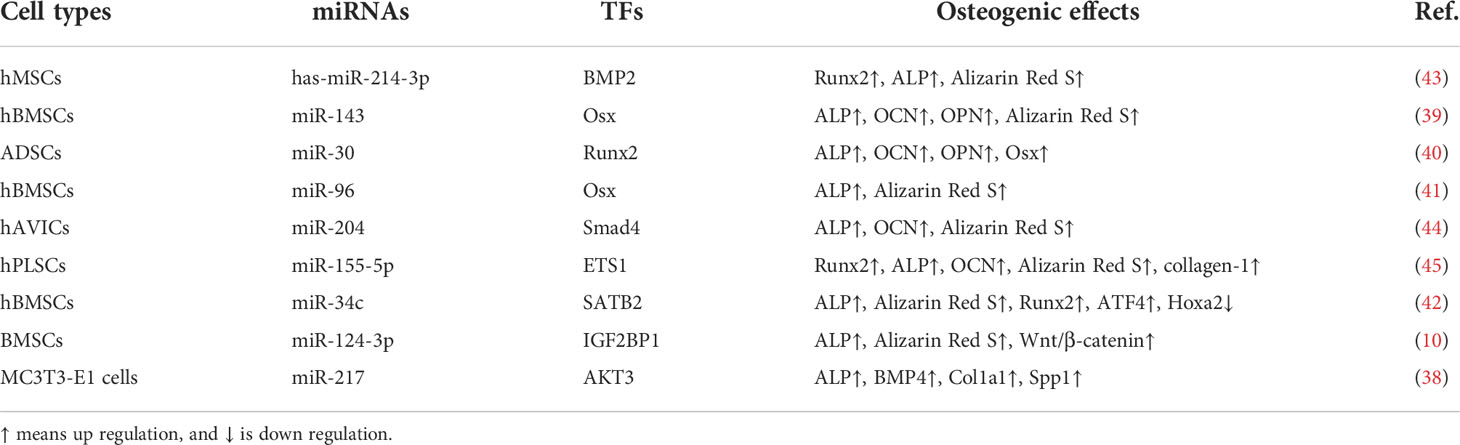
Table 1 MALAT1 promotes osteogenic differentiation by activating transcriptional factors (TFs) through interacting with miRNAs.
In a study, the expression of MALAT1 in rat OP models is down regulated. However, it is also shown that MALAT1 can induce inhibition of osteogenic differentiation by increasing the activity of p38 MAPK and ERK1/2 signaling pathways in BMSCs (46). This conclusion seems to be contradictory, due to the confusion by the different cell lines and experimental conditions. It has been reported that p38 MAPK is essential for bone formation and homeostasis in stem cells (47). Activation of p38 MAPK and ERK1/2 pathways increases the expression of RUNX2 and Osx, leading to enhancement of osteogenic differentiation (48, 49). MALAT1 deletion may ameliorate RANKL-induced inhibition of cell growth and arrest of cell cycle in hFOB 1.19 osteoblast cells (50). Consistently, MALAT1 overexpression is associated with OPG down regulation and RANKL up regulation, respectively, promoting osteoclast processes in ultra-high molecular weight polyethylene (UHMWPE)-treated hFOB 1.19 cells. Potentially, MALAT1 may increase the expression of VEGF by sponging miR-22-5p, promoting the onset of osteolysis (51).
Osteogenic differentiation of MSCs may promote bone regeneration, which involves multiple-step- and multiple-gene-mediated processes that are implicated in the recovery of bone diseases, such as OP and bone fracture. By using RNA-seq technology, 1524 differential expressed lncRNAs have been identified in osteo-inductive groups compared with the control group. The potential lncRNA-miRNA-mRNA network in rat BMMSCs has been explored (52). Many lncRNAs, such as MALAT1, H19, and HOTAIR, have been summarized to be associated with bone regeneration and balance of bone formation and resorption (53). Several reviews focusing on the contribution of MALAT1 to osteogenesis and endochondral ossification have been explored (54). In the exosomes from BMSCs in patients with postmenopausal OP, 148 lncRNAs are up regulated, and 138 are down regulated. Analysis of constructed lncRNA-miRNA-mRNA network indicates that these differential expression of lncRNAs may potentially associate with Wnt/β-catenin, PI3K/AKT, and MAPK signaling pathways (55). A total of 1878 differential expressed lncRNAs from patients with steroid-induced osteonecrosis of the femoral head have been detected using microarray and bioinformatics. MALAT1 is one of the ceRNA relating to abnormal osteogenic differentiation of BMSCs (56).
In steroid-induced avascular necrosis of the femoral head, the expression of MALAT1 is decreased. MALAT1 overexpression or miR-214 inhibition can ameliorate dexamethasone (DEX)-induced inhibition of osteogenic differentiation of BMSCs (57). The expression of MALAT1 in pre-osteoblast MC3T3-E1 cells can be down regulated by DEX, which has been demonstrated to inhibit osteoblast proliferation and promote osteoblast apoptosis (58). More specifically, forced expression of MALAT1 can abrogate DEX-induced viability repression and cell apoptosis by suppressing Ppm1e expression and activating AMPK signaling in OB-6 and hFOB1.19 osteoblast cells (59). Promoted osteoblast differentiation, inhibited osteoclast function, and balanced metabolism are important for the treatment of osteoporosis (OP). However, in rats with OP tibial fracture, the expression of MALAT1 is increased. Knockdown of MALAT1 expression can enhance osteogenic differentiation of BMSCs by increasing the expression of miR-144-3p (60). This discrepancy might be associated with different cell lines and culture conditions. Bone repair processes include bone resorption by activating osteoclastogenesis and bone formation by activating osteogenesis and neovascularization. Endothelial progenitor cells (EPCs)-derived exosomes can promote the migration and osteoclasts differentiation by increasing the expression of MALAT1, providing a therapeutic strategy for bone repair. Specifically, MALAT1 can increase the expression of ITGB1 by sponging miR-124 in EPC-derived exosomes (61).
In RAW 264.7 cells, co-culture of MCF-7 cells may activate osteoclastogenesis. Denosumab, an inhibitor of receptor activator of NF-κB ligand (RANKL), has been reported to inhibit MCF-7 cell-induced osteoclastogenesis by decreasing the expression of MALAT1 (62). In human periodontal ligament stem cells (hPLSCs), MALAT1 increases ALP activity and mineralization, enhances RUNX2, collagen I, and OCN expression, and promotes osteogenic differentiation by mediating miR-155-5p/ETS1 signaling, providing a strategy for treatment of periodontitis (45) (Table 1). Bisphosphonates (BPs), have been used for treating bone lesions, including those associated with multiple myeloma (MM). However, it is reported that BPs may potentially produce adverse effects, such as osteonecrosis of the jaw (63). The altered profile of lncRNAs in MM patients with osteonecrosis induced by BPs indicates that the down regulated expression of MALAT1 is assumed to be related to increased osteoclastogenesis in bone lesions (64). Angiogenesis contributes to bone regeneration by connecting with osteocytes. The expression of MALAT1, VEGF, and SP1 are increased in osteogenic medium-stimulated MC3T3-E1 cells. Knockdown of MALAT1 expression inhibits angiogenesis and bone regeneration, as shown by decreased proliferation, migration, and capillary tube formation of human umbilical vein endothelial cells (65).
Collectively, the expression of MALAT1 in OP is decreased, and enhanced expression of MALAT1 may promote osteogenic differentiation and osteoblast proliferation and inhibit osteoclast differentiation and osteoblast cell death.
Osteoarthritis (OA) is marked by chronic inflammation, chondrocyte apoptosis, extracellular matrix (ECM) degradation, and articular cartilage destruction, resulting in joint pain and disability. Chondrocytes, the unique cell type in cartilage, maintain the micro-environmental homeostasis. In addition, the synthesis and degradation of ECM are balanced by chondrocytes, which are orchestrated by network of signaling pathways. To provide effective prevention and treatment of OA, it is essential to understand the complex pathogenesis and pathophysiological processes. It has been demonstrated that lncRNAs are closely associated with the pathogenesis and progression of OA (66). Bioinformatics analysis reveals that 3007 lncRNAs are up regulated and 1707 lncRNAs are down regulated in OA chondrocytes. Additionally, 530 lncRNAs have been found to regulate their target genes expression by interacting with TF SP1 (67). Under IL-1β stimulation, 125 differentially expressed lncRNAs are identified and act as important regulator in the inflammatory responses in OA cartilage (68). MSCs with the capability of self-renew may promote tissue repairment. Injection of MSCs into the articular cavity can significantly impede the degradation of cartilage and improve joint biological functions (69). Extracellular vesicles (EVs) secreted from MSCs have been reported to ameliorate the pathological alternations and inflammation in cartilage. Specifically, EVs with MALAT1 overexpression inhibit the expression of MMP-13, IL-6, and caspase-3 in IL-1β-treated C28/I2 cells, promoting chondrocyte proliferation and migration and compromising inflammation (70).
In patients with OA, the expression of MALAT1 is increased. In LPS-treated rat chondrocytes, silenced expression of MALAT1 increases Collagen II expression and decreases MMP-13, IL-6, and COX-2 expression, protecting ECM against LPS-induced degradation. Mechanically, MALAT1 plays a critical role in LPS-induced pathological changes in chondrocytes by activation PI3K/AKT/mTOR signaling pathway via sponging miR-146a (71). Similarly, MALAT1 also promotes ECM degradation by activating the expression of a disintegrin and metalloproteinase with thrombospondin motifs-5 (ADAMTS-5) by binding to miR-145 in human primary chondrocytes in vivo and in vitro. Knockdown of MALAT1 expression may contribute to cell viability and the stability of ECM in IL-1β-treated chondrocytes (72). MALAT1 may activate the expression of NF-κB signaling by interacting with miR-9, increasing the productions of IL-6, MMP-13, and caspase-3 in mice chondrocytes. Resveratrol has been demonstrated to bind to the promoter of MALAT1 and inhibit its transcriptional expression, therefore inhibiting NF-κB signaling-mediated inflammation, ECM degradation, and chondrocyte apoptosis (73).
Conversely, excessive proliferation may contribute to the pathological alternations of OA. A study shows that MALAT1 may enhance cell proliferation, inhibit cell apoptosis, and ameliorate the degradation of ECM in IL-1β-treated human chondrocytes by stimulating AKT3 expression via sponging miR-150-5p (74). Consistently, MALAT1 knockdown can suppress the proliferation of human OA chondrocytes by binding to miR-127-5p, accompanied by decreased expression of PI3K/AKT signaling-related factors and OPN (75). In IL-1β-treated rat chondrocytes, the expression of MALAT1 is down regulated. PcDNA3.1-MALAT1 transfection may increase the viability of chondrocyte and the expression of collagen II and decrease the apoptotic ratio of chondrocytes and the expression of MMP-13 by inhibiting the activity of p-JNK signaling (76).
Pathological changes in OA subchondral bone tissues are also associated with the early alternations of OA. Trabecular thickening can be developed before the initiation of cartilage degeneration (77). MALAT1 expression in subchondral bone of early OA patients is enhanced, and it is induced by inflammatory stress. Depletion of MALAT1 may result in increased production of PGE2 without affecting OPG synthesis (78). In early stage of OA, synovitis is active. OA patients with synovitis have been found the radiographic signs in cartilage degradation, indicating a potential role of synovitis in accelerating the pathological changes of OA (79). In obese patients with OA, higher expression levels of inflammatory cytokines IL- 6 and CXCL8 in synovial fibroblasts are observed. RNA-seq assays identify that MALAT1 expression is significantly up regulated. Knockdown of MALAT1 expression may compromise the increased expression of inflammatory cytokines and inhibit the proliferation of synovial fibroblasts (80).
Collectively, BMSCs-derived MALAT1 expression contributes to inhibition of inflammation. In OA chondrocytes, the expression of MALAT1 is up regulated and associated with increased inflammation and cell excessive proliferation. Knockdown of MALAT1 in OA chondrocytes may become a potentially therapeutic strategy for clinic management of OA.
Intervertebral disc degeneration (IDD), an aging disease with degenerative characteristics, is often associated with low back pain in clinic. Intervertebral disc (IVD) consists of the nucleus pulposus (NP), annulus fibrosus (AF), and cartilage endplates (CEPs). The pathological changes in NP cells contribute a key role to the development of IDD (81). Differentially expressed lncRNAs from the NP cells of IDD patients have been shown the importance for the pathogenesis and progression of IDD (82). It has been demonstrated that the expression of MALAT1 in NP cells from degenerative IVD is down regulated. Overexpressed MALAT1 may decrease the production of IL-1 and IL6 and inhibit the expression of caspase-3, inhibiting inflammatory responses and cell apoptosis (83). Consistently, down regulated expression of MALAT1 is statistically correlated with decreased production of collagen II and aggrecan in rats with IDD. MALAT1 overexpression exhibits the protective activity against the biological actions produced by IL-1β in NP cells via inhibiting the expression of miR-503 and the activation of MAPK signaling pathways (84).
CEPs control the transport and distribution of nutrients in IVDs. Injury in CEPs has been reported to promote IDD development. Diabetes mellitus (DM) contributes the degeneration of CEPs, due to the detrimental effects of high glucose on CEPs by inducing oxidative stress, mitochondrial dysfunctions, and cell apoptosis (85). Controversially, the expression of MALAT1 in high glucose-treated CEP cells is up regulated. Deletion of MALAT1 may decrease CEP cells apoptosis by attenuating the phosphorylation levels of p38 MAPK (86). More efforts are still needed to clearly elucidate the relationship between MALAT1 and diabetic patients with IDD.
Rheumatoid arthritis (RA), an autoimmune disease, is marked by chronic inflammation and joint destruction. Till now, no effective and reliable biomarkers for prognosis prediction or therapeutic evaluation are available for RA. The etiological studies suggest that many factors, such as genetic susceptibility, aberrant metabolism, and dysregulated immune actions, are included in the pathogenesis and progression of RA (87). Differentially expressed lncRNAs have been demonstrated to contribute to the development of some auto-immune diseases, such as RA (88). Activation of inflammatory and immune responses and increased angiogenesis in synovial cells have been reported to promote the pathogenesis of RA (89, 90). These biological effects are also involved in the regulation of MALAT1 as discussed above. It is reasonable to postulate that MALAT1 is implicated in RA development.
MALAT1, MEG3, and NEAT1 have been identified as the key potential lncRNAs involved in certain physiological and pathological activity in RA patients. Specifically, the expression of MALAT1 in the peripheral blood mono-nuclear cells (PBMCs), plasma, and synovial fluid of RA patients is significantly up regulated (91). In the fibroblast-like synoviocytes (FLS) derived from RA patients, deletion of MALAT1 can decrease cell apoptosis and activate PI3K/AKT signaling pathway. Quercetin may exhibit the protective activity against MALAT1-induced pathological changes in RA FLS (92). It has been reported that MALAT1 can be considered as a potential biomarker for RA diagnosis, due to being statistically correlating with the expression of HSPA5 (GRP78) and MMD (PAQR1) in patients with RA (93). However, the single nucleotide polymorphism of MALAT1 gene is reported to be not correlated with the susceptibility of RA (94).
Juvenile idiopathic arthritis (JIA), a chronic rheumatic disease in children associating with chronic inflammation and inappropriate immune activation, starts before 18 years of age and lasts at least 6 months, according to the updated classification criteria (95). Drugs available for children with JIA are rather limited (96). LncRNAs have been shown to regulate immune abnormalities and the development of JIA. For example, lncRNA RP11-340f14.6 can bind to the neighbor P2X7R and increase its expression in patients with JIA. In addition, RP11-340f14.6 up regulates the differentiation of Th17 and down regulates Treg distribution in a P2X7R-independent manner (97). In patients with systemic JIA, the expression of MALAT1 is significantly up regulated, and the productions of IL-6/-17/-1β, TNFα, and MMP-8/-9 in the plasma are also increased. Knockdown of MALAT1 expression may reverse the abnormal levels of these cytokines. MALAT1 ameliorates the progression of systemic JIA by activating JAK/STAT signaling pathway through sponging miR-150-5p (98).
Ankylosing spondylitis (AS), a chronic immune-mediated inflammatory disease featured as bone erosion and syndesmophyte formation, mainly occurs in the axial skeleton and the sacroiliac joints. Bone destruction may be related to activation of RANKL system and inactivation of Wnt/β-catenin pathway (99). In addition, many immunocytes, such as Th1/Th2/Th22 cells, dendritic cells, and T/B lymphocytes, have been reported to be aberrantly activated (100). However, the molecular mechanisms underlying the pathogenesis and progression of AS remain unclear, and the clinical therapeutic management of AS is dissatisfying. Evidence shows that lncRNAs play a pivotal role in AS development (101). The expression of MALAT1 is up regulated in cartilage tissue and chondrocytes isolated from AS patients. Decreased expression of MALAT1 enhances cell viability and inhibits pyroptosis by suppressing GSDMD expression and increasing miR-558 expression in AS chondrocytes (102).
Hyperuricemia-induced gouty arthritis (GA) is marked by the deposition of monosodium urate (MSU) crystal in joints and activation of immune inflammation. MSU can be recognized by macrophages, and the system of NF-κB/RANKL is activated. Increased tophus in the joint cavity constantly destroys the bone surface and induces osteoclast differentiation by activating RANKL system (103). Urate-lowering is the critical strategy to therapeutic management of hyperuricemia and gouty. However, adverse effects of current available drugs greatly limit their applications (104). LncRNA has been implicated in the pathogenesis of inflammatory arthritis, such as GA (105). Differentially expressed lncRNAs in patients with GA have been reported, and the predicted regulatory pathways are mainly focusing on inflammation and osteoclast differentiation (106). In MSU-treated THP-1 macrophages, MALAT1 expression is up regulated, and the productions of IL-1β, TNFα, caspase-1, and NLRP3 are also enhanced. The total glucosides extracted from paeony (TGP) may significantly ameliorate the abnormal expression of these factors. Overexpression of MALAT1 can abolish the suppressive activity of TGP on MSU-induced inflammation by mediating miR-876-5p/NLRP3 axis in THP-1 macrophages (107). Targeting MALAT1 may become a potential strategy for therapeutically treating GA.
Extensive evidence shows that lncRNAs have been implicated in the various genomic processes, affecting cell physiological and pathological functions. MALAT1, a well-studied lncRNA, plays a key role in bone and cartilage diseases by regulating the biological processes of osteogenic differentiation, proliferation, and apoptosis. MALAT1 is abundantly located at nuclear speckles, which mediate the transcription, splicing, and mRNA export. Specifically, there is a facilitatory effect for MALAT1 to interact with the targets, such as mRNA or TFs. The interactions between MALAT1/protein and MALAT1/RNA are the essential ways for MALAT1 to regulate the activity in bone and cartilage diseases, including OP, OA, IDD, RA, JIA, AS, and GA. Particularly, MALAT1 may promote osteogenic differentiation in MSCs. It is interesting that decreased expression of MALAT1 is associated with the pathological changes in OA chondrocytes. Conversely, MALAT1 exhibits a stimulating effect on the pathological development of RA, JIA, AS, and GA. However, the molecular mechanisms of MALAT1 in regulating the pathogenesis and progression of bone and cartilage diseases remain largely unclear. More efforts are still needed.
FP: Conceptualization, Methodology. DZ, JX and FP: Data curation, Writing-original draft preparation, Data curation, Validation, Writing-reviewing and Editing. All authors contributed to the article and approved the submitted version.
This study was financially supported by the Science and Technology research project of the Education Department of Jiangxi Province (Grant NO. GJJ211520).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature (2016) 539(7629):452–5. doi: 10.1038/nature20149
2. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell (2013) 154(1):26–46. doi: 10.1016/j.cell.2013.06.020
3. Arun G, Aggarwal D, Spector DL. MALAT1 long non-coding RNA: Functional implications. Noncoding RNA (2020) 6(2):22. doi: 10.3390/ncrna6020022
4. Johnsson P, Lipovich L, Grandér D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta (2014) 1840(3):1063–71. doi: 10.1016/j.bbagen.2013.10.035
5. Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J (2010) 29(18):3082–93. doi: 10.1038/emboj.2010.199
6. Sebastian A, Hum NR, Hudson BD, Loots GG. Cancer-osteoblast interaction reduces sost expression in osteoblasts and up-regulates lncRNA MALAT1 in prostate cancer. Microarrays (Basel) (2015) 4(4):503–19. doi: 10.3390/microarrays4040503
7. Su P, Tian Y, Yang C, Ma X, Wang X, Pei J, et al. Mesenchymal stem cell migration during bone formation and bone diseases therapy. Int J Mol Sci (2018) 19(8):2343. doi: 10.3390/ijms19082343
8. Xia K, Yu LY, Huang XQ, Zhao ZH, Liu J. Epigenetic regulation by long noncoding RNAs in osteo-/adipogenic differentiation of mesenchymal stromal cells and degenerative bone diseases. World J Stem Cells (2022) 14(1):92–103. doi: 10.4252/wjsc.v14.i1.92
9. Chen J, Wang Y, Wang C, Hu JF, Li W. LncRNA functions as a new emerging epigenetic factor in determining the fate of stem cells. Front Genet (2020) 11:277. doi: 10.3389/fgene.2020.00277
10. Li X. LncRNA metastasis-associated lung adenocarcinoma transcript-1 promotes osteogenic differentiation of bone marrow stem cells and inhibits osteoclastic differentiation of mø in osteoporosis via the miR-124-3p/IGF2BP1/Wnt/β-catenin axis. J Tissue Eng Regener Med (2022) 16(3):311–29. doi: 10.1002/term.3279
11. Younes N, Syed N, Yadav SK, Haris M, Abdallah AM, Abu-Madi M. A whole-genome sequencing association study of low bone mineral density identifies new susceptibility loci in the phase I Qatar biobank cohort. J Pers Med (2021) 11(1):34. doi: 10.3390/jpm11010034
12. Yuan X, Guo Y, Chen D, Luo Y, Chen D, Miao J, et al. Long non-coding RNA MALAT1 functions as miR-1 sponge to regulate connexin 43-mediated ossification of the posterior longitudinal ligament. Bone (2019) 127:305–14. doi: 10.1016/j.bone.2019.06.019
13. Hall LL, Smith KP, Byron M, Lawrence JB. Molecular anatomy of a speckle. Anat Rec A Discovery Mol Cell Evol Biol (2006) 288(7):664–75. doi: 10.1002/ar.a.20336
14. Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep (2012) 2(1):111–23. doi: 10.1016/j.celrep.2012.06.003
15. Fei J, Jadaliha M, Harmon TS, Li ITS, Hua B, Hao Q, et al. Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J Cell Sci (2017) 130(24):4180–92. doi: 10.1242/jcs.206854
16. Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet (2011) 27(8):295–306. doi: 10.1016/j.tig.2011.05.006
17. Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor b-MYB. PloS Genet (2013) 9(3):e1003368. doi: 10.1371/journal.pgen.1003368
18. Wang C, Lu T, Emanuel G, Babcock HP, Zhuang X. Imaging-based pooled CRISPR screening reveals regulators of lncRNA localization. Proc Natl Acad Sci U.S.A. (2019) 116(22):10842–51. doi: 10.1073/pnas.1903808116
19. Cai L, Chang H, Fang Y, Li G. A comprehensive characterization of the function of LincRNAs in transcriptional regulation through long-range chromatin interactions. Sci Rep (2016) 6:36572. doi: 10.1038/srep36572
20. Li X, Chen N, Zhou L, Wang C, Wen X, Jia L, et al. Genome-wide target interactome profiling reveals a novel EEF1A1 epigenetic pathway for oncogenic lncRNA MALAT1 in breast cancer. Am J Cancer Res (2019) 9(4):714–29.
21. Qi Y, Ooi HS, Wu J, Chen J, Zhang X, Tan S, et al. MALAT1 long ncRNA promotes gastric cancer metastasis by suppressing PCDH10. Oncotarget (2016) 7(11):12693–703. doi: 10.18632/oncotarget.7281
22. Wang D, Ding L, Wang L, Zhao Y, Sun Z, Karnes RJ, et al. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget (2015) 6(38):41045–55. doi: 10.18632/oncotarget.5728
23. Li X, Zhou B, Chen L, Gou LT, Li H, Fu XD. GRID-seq reveals the global RNA-chromatin interactome. Nat Biotechnol (2017) 35(10):940–50. doi: 10.1038/nbt.3968
24. Sridhar B, Rivas-Astroza M, Nguyen TC, Chen W, Yan Z, Cao X, et al. Systematic mapping of RNA-chromatin interactions In vivo. Curr Biol (2017) 27(4):602–9. doi: 10.1016/j.cub.2017.01.011
25. Xie ZC, Dang YW, Wei DM, Chen P, Tang RX, Huang Q, et al. Clinical significance and prospective molecular mechanism of MALAT1 in pancreatic cancer exploration: a comprehensive study based on the GeneChip, GEO, oncomine, and TCGA databases. Onco Targets Ther (2017) 10:3991–4005. doi: 10.2147/ott.S136878
26. Zhou L, Xu DY, Sha WG, Shen L, Lu GY. Long non-coding RNA MALAT1 interacts with transcription factor Foxo1 to regulate SIRT1 transcription in high glucose-induced HK-2 cells injury. Biochem Biophys Res Commun (2018) 503(2):849–55. doi: 10.1016/j.bbrc.2018.06.086
27. Wei L, Li J, Han Z, Chen Z, Zhang Q. Silencing of lncRNA MALAT1 prevents inflammatory injury after lung transplant ischemia-reperfusion by downregulation of IL-8 via p300. Mol Ther Nucleic Acids (2019) 18:285–97. doi: 10.1016/j.omtn.2019.05.009
28. Amodio N, Stamato MA, Juli G, Morelli E, Fulciniti M, Manzoni M, et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia (2018) 32(9):1948–57. doi: 10.1038/s41375-018-0067-3
29. Chen R, Liu Y, Zhuang H, Yang B, Hei K, Xiao M, et al. Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res (2017) 45(17):9947–59. doi: 10.1093/nar/gkx600
30. Malakoti F, Targhazeh N, Karimzadeh H, Mohammadi E, Asadi M, Asemi Z, et al. Multiple function of lncRNA MALAT1 in cancer occurrence and progression. Chem Biol Drug Des (2021). doi: 10.1111/cbdd.14006
31. Yang Z, Song D, Wang Y, Tang L. lncRNA MALAT1 promotes diabetic nephropathy progression via miR-15b-5p/TLR4 signaling axis. J Immunol Res (2022) 2022:8098001. doi: 10.1155/2022/8098001
32. Xiang J, Deng YY, Liu HX, Pu Y. LncRNA MALAT1 promotes PPARα/CD36-mediated hepatic lipogenesis in nonalcoholic fatty liver disease by modulating miR-206/ARNT axis. Front Bioeng Biotechnol (2022) 10:858558. doi: 10.3389/fbioe.2022.858558
33. Chen F, Li W, Zhang D, Fu Y, Yuan W, Luo G, et al. MALAT1 regulates hypertrophy of cardiomyocytes by modulating the miR-181a/HMGB2 pathway. Eur J Histochem (2022) 66(3):3426. doi: 10.4081/ejh.2022.3426
34. Zhou Z, Chen J, Huang Y, Liu D, Chen S, Qin S. Long noncoding RNA GAS5: A new factor involved in bone diseases. Front Cell Dev Biol (2021) 9:807419. doi: 10.3389/fcell.2021.807419
35. Skvortsova K, Iovino N, Bogdanović O. Functions and mechanisms of epigenetic inheritance in animals. Nat Rev Mol Cell Biol (2018) 19(12):774–90. doi: 10.1038/s41580-018-0074-2
36. Liu Z, Xu S, Dao J, Gan Z, Zeng X. Differential expression of lncRNA/miRNA/mRNA and their related functional networks during the osteogenic/odontogenic differentiation of dental pulp stem cells. J Cell Physiol (2020) 235(4):3350–61. doi: 10.1002/jcp.29223
37. Hong S, Hu S, Kang Z, Liu Z, Yang W, Zhang Y, et al. Identification of functional lncRNAs based on competing endogenous RNA network in osteoblast differentiation. J Cell Physiol (2020) 235(3):2232–44. doi: 10.1002/jcp.29132
38. Song C, Guo Y, Chen F, Liu W. lncRNA MALAT1 promotes osteogenic differentiation through the miR-217/AKT3 axis: A possible strategy to alleviate osteoporosis. J Gene Med (2022) 24(6):e3409. doi: 10.1002/jgm.3409
39. Gao Y, Xiao F, Wang C, Wang C, Cui P, Zhang X, et al. Long noncoding RNA MALAT1 promotes osterix expression to regulate osteogenic differentiation by targeting miRNA-143 in human bone marrow-derived mesenchymal stem cells. J Cell Biochem (2018) 119(8):6986–96. doi: 10.1002/jcb.26907
40. Yi J, Liu D, Xiao J. LncRNA MALAT1 sponges miR-30 to promote osteoblast differentiation of adipose-derived mesenchymal stem cells by promotion of Runx2 expression. Cell Tissue Res (2019) 376(1):113–21. doi: 10.1007/s00441-018-2963-2
41. Zang LY, Yang XL, Li WJ, Liu GL. Long noncoding RNA metastasis-associated lung adenocarcinoma transcript 1 promotes the osteoblast differentiation of human bone marrow-derived mesenchymal stem cells by targeting the microRNA-96/Osterix axis. J Craniofac Surg (2022) 33(3):956–61. doi: 10.1097/scs.0000000000008092
42. Yang X, Yang J, Lei P, Wen T. LncRNA MALAT1 shuttled by bone marrow-derived mesenchymal stem cells-secreted exosomes alleviates osteoporosis through mediating microRNA-34c/SATB2 axis. Aging (Albany NY) (2019) 11(20):8777–91. doi: 10.18632/aging.102264
43. Li J, Zhuang H, Wang Z, Cai J, Ma X, Chen W, et al. lncRNAs MALAT1 and LINC00657 upstream to miR-214-3p/BMP2 regulate osteogenic differentiation of human mesenchymal stem cells. Mol Biol Rep (2022) 49(7):6847–57. doi: 10.1007/s11033-022-07136-3
44. Xiao X, Zhou T, Guo S, Guo C, Zhang Q, Dong N, et al. LncRNA MALAT1 sponges miR-204 to promote osteoblast differentiation of human aortic valve interstitial cells through up-regulating Smad4. Int J Cardiol (2017) 243:404–12. doi: 10.1016/j.ijcard.2017.05.037
45. Hua L, Zhang X. MALAT1 regulates osteogenic differentiation of human periodontal ligament stem cells through mediating miR-155-5p/ETS1 axis. Tissue Cell (2021) 73:101619. doi: 10.1016/j.tice.2021.101619
46. Zheng S, Wang YB, Yang YL, Chen BP, Wang CX, Li RH, et al. LncRNA MALAT1 inhibits osteogenic differentiation of mesenchymal stem cells in osteoporosis rats through MAPK signaling pathway. Eur Rev Med Pharmacol Sci (2019) 23(11):4609–17. doi: 10.26355/eurrev_201906_18038
47. Rodríguez-Carballo E, Gámez B, Sedó-Cabezón L, Sánchez-Feutrie M, Zorzano A, Manzanares-Céspedes C, et al. The p38α MAPK function in osteoprecursors is required for bone formation and bone homeostasis in adult mice. PloS One (2014) 9(7):e102032. doi: 10.1371/journal.pone.0102032
48. Ge C, Yang Q, Zhao G, Yu H, Kirkwood KL, Franceschi RT. Interactions between extracellular signal-regulated kinase 1/2 and p38 MAP kinase pathways in the control of RUNX2 phosphorylation and transcriptional activity. J Bone Miner Res (2012) 27(3):538–51. doi: 10.1002/jbmr.561
49. Lu Y, Ma ZX, Deng R, Jiang HT, Chu L, Deng ZL. The SIRT1 activator SRT2104 promotes BMP9-induced osteogenic and angiogenic differentiation in mesenchymal stem cells. Mech Ageing Dev (2022) 207:111724. doi: 10.1016/j.mad.2022.111724
50. Che W, Dong Y, Quan HB. RANKL inhibits cell proliferation by regulating MALAT1 expression in a human osteoblastic cell line hFOB 1.19. Cell Mol Biol (Noisy-le-grand) (2015) 61(1):7–14.
51. Yang X, Zhang Y, Li Y, Wen T. MALAT1 enhanced the proliferation of human osteoblasts treated with ultra−high molecular weight polyethylene by targeting VEGF via miR−22−5p. Int J Mol Med (2018) 41(3):1536–46. doi: 10.3892/ijmm.2018.3363
52. Liu J, Yao Y, Huang J, Sun H, Pu Y, Tian M, et al. Comprehensive analysis of lncRNA-miRNA-mRNA networks during osteogenic differentiation of bone marrow mesenchymal stem cells. BMC Genomics (2022) 23(1):425. doi: 10.1186/s12864-022-08646-x
53. Peng S, Cao L, He S, Zhong Y, Ma H, Zhang Y, et al. An overview of long noncoding RNAs involved in bone regeneration from mesenchymal stem cells. Stem Cells Int (2018) 2018:8273648. doi: 10.1155/2018/8273648
54. Ghafouri-Fard S, Abak A, Tavakkoli Avval S, Rahmani S, Shoorei H, Taheri M, et al. Contribution of miRNAs and lncRNAs in osteogenesis and related disorders. BioMed Pharmacother (2021) 142:111942. doi: 10.1016/j.biopha.2021.111942
55. Wang J, Fu M, He S, Cai P, Xiang X, Fang L. Expression profile analysis of lncRNA in bone marrow mesenchymal stem cells exosomes of postmenopausal osteoporosis patients through microarray and bioinformatics analyses. Pathol Res Pract (2022) 236:153985. doi: 10.1016/j.prp.2022.153985
56. Wang Q, Yang Q, Chen G, Du Z, Ren M, Wang A, et al. LncRNA expression profiling of BMSCs in osteonecrosis of the femoral head associated with increased adipogenic and decreased osteogenic differentiation. Sci Rep (2018) 8(1):9127. doi: 10.1038/s41598-018-27501-2
57. Huang XZ, Huang J, Li WZ, Wang JJ, Song DY, Ni JD. LncRNA-MALAT1 promotes osteogenic differentiation through regulating ATF4 by sponging miR-214: Implication of steroid-induced avascular necrosis of the femoral head. Steroids (2020) 154:108533. doi: 10.1016/j.steroids.2019.108533
58. Zhu Z, Wang X, Wang Z, Zhao Z, Zhou P, Gao X. Neobavaisoflavone protects osteoblasts from dexamethasone-induced oxidative stress by upregulating the CRNDE-mediated Nrf2/HO-1 signaling pathway. Drug Dev Res (2021) 82(7):1044–54. doi: 10.1002/ddr.21811
59. Fan JB, Zhang Y, Liu W, Zhu XH, Xu DW, Zhao JN, et al. Long non-coding RNA MALAT1 protects human osteoblasts from dexamethasone-induced injury via activation of PPM1E-AMPK signaling. Cell Physiol Biochem (2018) 51(1):31–45. doi: 10.1159/000495159
60. Ling S, Xu T, Sun J, Yan C, Lv B, Wang H, et al. Expression of lncRNA MALAT1 through miR-144-3p in osteoporotic tibial fracture rats and its effect on osteogenic differentiation of BMSC under traction. Evid Based Complement Alternat Med (2022) 2022:2590055. doi: 10.1155/2022/2590055
61. Cui Y, Fu S, Sun D, Xing J, Hou T, Wu X. EPC-derived exosomes promote osteoclastogenesis through LncRNA-MALAT1. J Cell Mol Med (2019) 23(6):3843–54. doi: 10.1111/jcmm.14228
62. Feng Q, Wang D, Feng J, Guo P, Geng C. Denosumab inhibits MCF-7 cell line-induced spontaneous osteoclastogenesis via the RANKL/MALAT1/miR-124 axis. Transl Cancer Res (2020) 9(4):2482–91. doi: 10.21037/tcr.2020.03.17
63. Allegra A, Innao V, Pulvirenti N, Musolino C. Antiresorptive agents and anti-angiogenesis drugs in the development of osteonecrosis of the jaw. Tohoku J Exp Med (2019) 248(1):27–9. doi: 10.1620/tjem.248.27
64. Allegra A, Mania M, D’Ascola A, Oteri G, Siniscalchi EN, Avenoso A, et al. Altered long noncoding RNA expression profile in multiple myeloma patients with bisphosphonate-induced osteonecrosis of the jaw. BioMed Res Int (2020) 2020:9879876. doi: 10.1155/2020/9879876
65. Ding A, Li CH, Yu CY, Zhou HT, Zhang ZH. Long non-coding RNA MALAT1 enhances angiogenesis during bone regeneration by regulating the miR-494/SP1 axis. Lab Invest (2021) 101(11):1458–66. doi: 10.1038/s41374-021-00649-8
66. Zhang X, Hong R, Chen W, Xu M, Wang L. The role of long noncoding RNA in major human disease. Bioorg Chem (2019) 92:103214. doi: 10.1016/j.bioorg.2019.103214
67. Fu M, Huang G, Zhang Z, Liu J, Zhang Z, Huang Z, et al. Expression profile of long noncoding RNAs in cartilage from knee osteoarthritis patients. Osteoarthr Cartilage (2015) 23(3):423–32. doi: 10.1016/j.joca.2014.12.001
68. Pearson MJ, Philp AM, Heward JA, Roux BT, Walsh DA, Davis ET, et al. Long intergenic noncoding RNAs mediate the human chondrocyte inflammatory response and are differentially expressed in osteoarthritis cartilage. Arthritis Rheumatol (2016) 68(4):845–56. doi: 10.1002/art.39520
69. Matas J, Orrego M, Amenabar D, Infante C, Tapia-Limonchi R, Cadiz MI, et al. Umbilical cord-derived mesenchymal stromal cells (MSCs) for knee osteoarthritis: Repeated MSC dosing is superior to a single MSC dose and to hyaluronic acid in a controlled randomized phase I/II trial. Stem Cells Transl Med (2019) 8(3):215–24. doi: 10.1002/sctm.18-0053
70. Pan C, Huang W, Chen Q, Xu J, Yao G, Li B, et al. LncRNA malat-1 from MSCs-derived extracellular vesicles suppresses inflammation and cartilage degradation in osteoarthritis. Front Bioeng Biotechnol (2021) 9:772002. doi: 10.3389/fbioe.2021.772002
71. Li H, Xie S, Li H, Zhang R, Zhang H. LncRNA MALAT1 mediates proliferation of LPS treated-articular chondrocytes by targeting the miR-146a-PI3K/Akt/mTOR axis. Life Sci (2020) 254:116801. doi: 10.1016/j.lfs.2019.116801
72. Liu C, Ren S, Zhao S, Wang Y. LncRNA MALAT1/MiR-145 adjusts IL-1β-Induced chondrocytes viability and cartilage matrix degradation by regulating ADAMTS5 in human osteoarthritis. Yonsei Med J (2019) 60(11):1081–92. doi: 10.3349/ymj.2019.60.11.1081
73. Zhang G, Zhang H, You W, Tang X, Li X, Gong Z. Therapeutic effect of resveratrol in the treatment of osteoarthritis via the MALAT1/miR-9/NF-κB signaling pathway. Exp Ther Med (2020) 19(3):2343–52. doi: 10.3892/etm.2020.8471
74. Zhang Y, Wang F, Chen G, He R, Yang L. LncRNA MALAT1 promotes osteoarthritis by modulating miR-150-5p/AKT3 axis. Cell Biosci (2019) 9:54. doi: 10.1186/s13578-019-0302-2
75. Liang J, Xu L, Zhou F, Liu AM, Ge HX, Chen YY, et al. MALAT1/miR-127-5p regulates osteopontin (OPN)-mediated proliferation of human chondrocytes through PI3K/Akt pathway. J Cell Biochem (2018) 119(1):431–9. doi: 10.1002/jcb.26200
76. Gao GC, Cheng XG, Wei QQ, Chen WC, Huang WZ. Long noncoding RNA MALAT-1 inhibits apoptosis and matrix metabolism disorder in interleukin-1β-induced inflammation in articular chondrocytes via the JNK signaling pathway. J Cell Biochem (2019) 120(10):17167–79. doi: 10.1002/jcb.28977
77. Quasnichka HL, Anderson-MacKenzie JM, Bailey AJ. Subchondral bone and ligament changes precede cartilage degradation in guinea pig osteoarthritis. Biorheology (2006) 43(3,4):389–97. doi: 10.1016/j.bbmt.2005.09.011
78. Alnajjar FA, Sharma-Oates A, Wijesinghe SN, Farah H, Nanus DE, Nicholson T, et al. The expression and function of metastases associated lung adenocarcinoma transcript-1 long non-coding RNA in subchondral bone and osteoblasts from patients with osteoarthritis. Cells (2021) 10(4):786. doi: 10.3390/cells10040786
79. Myers SL, Brandt KD, Ehlich JW, Braunstein EM, Shelbourne KD, Heck DA, et al. Synovial inflammation in patients with early osteoarthritis of the knee. J Rheumatol (1990) 17(12):1662–9. doi: 10.1002/art.1780370327
80. Nanus DE, Wijesinghe SN, Pearson MJ, Hadjicharalambous MR, Rosser A, Davis ET, et al. Regulation of the inflammatory synovial fibroblast phenotype by metastasis-associated lung adenocarcinoma transcript 1 long noncoding RNA in obese patients with osteoarthritis. Arthritis Rheumatol (2020) 72(4):609–19. doi: 10.1002/art.41158
81. Eisenstein SM, Balain B, Roberts S. Current treatment options for intervertebral disc pathologies. Cartilage (2020) 11(2):143–51. doi: 10.1177/1947603520907665
82. Wan ZY, Song F, Sun Z, Chen YF, Zhang WL, Samartzis D, et al. Aberrantly expressed long noncoding RNAs in human intervertebral disc degeneration: a microarray related study. Arthritis Res Ther (2014) 16(5):465. doi: 10.1186/s13075-014-0465-5
83. Zhang H, Li J, Duan D, She W, Wang L, Zhang F. The role of lncRNA MALAT1 in intervertebral degenerative disc disease. Int J Clin Exp Pathol (2017) 10(10):10611–7.
84. Zheng H, Wang T, Li X, He W, Gong Z, Lou Z, et al. LncRNA MALAT1 exhibits positive effects on nucleus pulposus cell biology in vivo and in vitro by sponging miR-503. BMC Mol Cell Biol (2020) 21(1):23. doi: 10.1186/s12860-020-00265-2
85. Jiang Z, Lu W, Zeng Q, Li D, Ding L, Wu J. High glucose-induced excessive reactive oxygen species promote apoptosis through mitochondrial damage in rat cartilage endplate cells. J Orthop Res (2018) 36(9):2476–83. doi: 10.1002/jor.24016
86. Jiang Z, Zeng Q, Li D, Ding L, Lu W, Bian M, et al. Long non−coding RNA MALAT1 promotes high glucose−induced rat cartilage endplate cell apoptosis via the p38/MAPK signalling pathway. Mol Med Rep (2020) 21(5):2220–6. doi: 10.3892/mmr.2020.11009
87. Zhang TP, Lv TT, Xu SZ, Pan HF, Ye DQ. Association of interleukin-10 gene single nucleotide polymorphisms with rheumatoid arthritis in a Chinese population. Postgrad Med J (2018) 94(1111):284–8. doi: 10.1136/postgradmedj-2017-135441
88. Liang J, Chen W, Lin J. LncRNA: An all-rounder in rheumatoid arthritis. J Transl Int Med (2019) 7(1):3–9. doi: 10.2478/jtim-2019-0002
89. Elshabrawy HA, Chen Z, Volin MV, Ravella S, Virupannavar S, Shahrara S. The pathogenic role of angiogenesis in rheumatoid arthritis. Angiogenesis (2015) 18(4):433–48. doi: 10.1007/s10456-015-9477-2
90. Malemud CJ. Defective T-cell apoptosis and T-regulatory cell dysfunction in rheumatoid arthritis. Cells (2018) 7(12):223. doi: 10.3390/cells7120223
91. Chatterjee S, Bhattcharjee D, Misra S, Saha A, Bhattacharyya NP, Ghosh A. Increase in MEG3, MALAT1, NEAT1 significantly predicts the clinical parameters in patients with rheumatoid arthritis. Per Med (2020) 17(6):445–57. doi: 10.2217/pme-2020-0009
92. Pan F, Zhu L, Lv H, Pei C. Quercetin promotes the apoptosis of fibroblast-like synoviocytes in rheumatoid arthritis by upregulating lncRNA MALAT1. Int J Mol Med (2016) 38(5):1507–14. doi: 10.3892/ijmm.2016.2755
93. Mahmoudi Z, Karamali N, Roghani SA, Assar S, Pournazari M, Soufivand P, et al. Efficacy of DMARDs and methylprednisolone treatment on the gene expression levels of HSPA5, MMD, and non-coding RNAs MALAT1, H19, miR-199a-5p, and miR-1-3p, in patients with rheumatoid arthritis. Int Immunopharmacol (2022) 108:108878. doi: 10.1016/j.intimp.2022.108878
94. Zhang TP, Zhu BQ, Tao SS, Fan YG, Li XM, Pan HF, et al. Long non-coding RNAs genes polymorphisms and their expression levels in patients with rheumatoid arthritis. Front Immunol (2019) 10:2529. doi: 10.3389/fimmu.2019.02529
95. Martini A, Ravelli A, Avcin T, Beresford MW, Burgos-Vargas R, Cuttica R, et al. Toward new classification criteria for juvenile idiopathic arthritis: First steps, pediatric rheumatology international trials organization international consensus. J Rheumatol (2019) 46(2):190–7. doi: 10.3899/jrheum.180168
96. Golhen K, Winskill C, Yeh C, Zhang N, Welzel T, Pfister M. Value of literature review to inform development and use of biologics in juvenile idiopathic arthritis. Front Pediatr (2022) 10:909118. doi: 10.3389/fped.2022.909118
97. Huang N, Fan Z, Ma L, Ma H, Huang H, Yu H, et al. Long non−coding RNA RP11−340F14.6 promotes a shift in the Th17/Treg ratio by binding with P2X7R in juvenile idiopathic arthritis. Int J Mol Med (2020) 46(2):859–68. doi: 10.3892/ijmm.2020.4618
98. Jiang H, Zhu M, Wang H, Liu H. Suppression of lncRNA MALAT1 reduces pro-inflammatory cytokines production by regulating miR-150-5p/ZBTB4 axis through JAK/STAT signal pathway in systemic juvenile idiopathic arthritis. Cytokine (2021) 138:155397. doi: 10.1016/j.cyto.2020.155397
99. Garcia-Montoya L, Gul H, Emery P. Recent advances in ankylosing spondylitis: understanding the disease and management. F1000Res (2018) 7:F1000 Faculty Rev-1512. doi: 10.12688/f1000research.14956.1
100. Zhu W, He X, Cheng K, Zhang L, Chen D, Wang X, et al. Ankylosing spondylitis: etiology, pathogenesis, and treatments. Bone Res (2019) 7:22. doi: 10.1038/s41413-019-0057-8
101. Sun R, Wang X, Sun X, Zhao B, Zhang X, Gong X, et al. Emerging roles of long non-coding RNAs in ankylosing spondylitis. Front Immunol (2022) 13:790924. doi: 10.3389/fimmu.2022.790924
102. Chen W, Wang F, Wang J, Chen F, Chen T. The molecular mechanism of long non-coding RNA MALAT1-mediated regulation of chondrocyte pyroptosis in ankylosing spondylitis. Mol Cells (2022) 45(6):365–75. doi: 10.14348/molcells.2022.2081
103. Wu M, Liu FJ, Chen J, Chen L, Wei C, Hu ZM, et al. Prevalence and factors associated with bone erosion in patients with gout. Arthritis Care Res (Hoboken) (2019) 71(12):1653–9. doi: 10.1002/acr.23816
104. Nutmakul T. A review on benefits of quercetin in hyperuricemia and gouty arthritis. Saudi Pharm J (2022) 30(7):918–26. doi: 10.1016/j.jsps.2022.04.013
105. Li X, Pan Y, Li W, Guan P, You C. The role of noncoding RNAs in gout. Endocrinology (2020) 161(11):bqaa165. doi: 10.1210/endocr/bqaa165
106. Zhong X, Peng Y, Liao H, Yao C, Li J, Yang Q, et al. Aberrant expression of long non-coding RNAs in peripheral blood mononuclear cells isolated from patients with gouty arthritis. Exp Ther Med (2019) 18(3):1967–76. doi: 10.3892/etm.2019.7816
Keywords: MALAT1, bone, cartilage, ECM degradation, osteoblasts, chondrocytes
Citation: Zhang D, Xue J and Peng F (2022) The regulatory activities of MALAT1 in the development of bone and cartilage diseases. Front. Endocrinol. 13:1054827. doi: 10.3389/fendo.2022.1054827
Received: 27 September 2022; Accepted: 01 November 2022;
Published: 14 November 2022.
Edited by:
Chong Yin, Affiliated Hospital of North Sichuan Medical College, ChinaReviewed by:
Qin Yang, Chengdu University of TCM, ChinaCopyright © 2022 Zhang, Xue and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Peng, amlhbmd4aWdhbnpob3VwZkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.