
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 14 November 2022
Sec. Gut Endocrinology
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.1053103
This article is part of the Research Topic Can Traditional Chinese Medicines affect Endocrine Diseases via Effects on the Intestinal Flora? View all 21 articles
 Xiuwen Xia1†
Xiuwen Xia1† Ya Xie1†
Ya Xie1† Qiaoqiao Chen1,2
Qiaoqiao Chen1,2 Dou Ding1,3
Dou Ding1,3 Zongqin Wang4
Zongqin Wang4 Yaji Xu1,5
Yaji Xu1,5 Yili Wang6
Yili Wang6 Xiumin Wang1,7
Xiumin Wang1,7 Weijun Ding1*
Weijun Ding1*Objective: Diarrhea-predominant irritable bowel syndrome (IBS-D) is a recurrent and common disease featuring dysbiotic intestinal microbiota, with limited treatments. Si-Jun-Zi Decoction (SJZD), a classic Chinese prescription, has been extensively used for IBS-D. This work aimed to explore the ex vivo interactions of SJZD and IBS-D’s intestinal microbiota.
Methods: Five samples of intestinal microbiota collected from IBS-D volunteers and five age-matched healthy controls were recruited from the Affiliated Hospital, Chengdu University of Traditional Chinese Medicine (TCM). A representative mixture of intestinal microbiota was composed of an equal proportion of these fecal samples. To simulate the clinical interaction, this microbiota was cocultivated with SJZD at clinical dosage in an anaerobic incubator at 37°C for 35 h. Microbiota and metabolic alterations were assessed by 16S rRNA gene sequencing in the V3/V4 regions and a nontargeted metabolome platform, respectively.
Results: After being cocultivated with SJZD, the dysbiotic intestine microbiota from IBS-D subjects was largely restored to those of the healthy controls. A total of 624 differentially expressed metabolites were detected by nontargeted metabolomics, of which 16 biomarkers were identified. These metabolites were then enriched into 11 pathways by KEGG, particularly those involved in neurotransmitter metabolism responses for the major symptom of IBS-D. Correlation analysis of bacterial metabolites demonstrated a synergistic pattern of neurotransmitter metabolism between Streptococcus and E. Shigella.
Conclusion: SJZD rescued the dysbiotic intestinal microbiota and ameliorated the dysfunctional neurotransmitter metabolism involved in IBS-D’s major symptoms.
Irritable bowel syndrome (IBS) is a common and recurrent disease, with an internationally pooled prevalence of 12.41% and limited treatments (1, 2). Diarrhea-predominant irritable bowel syndrome (IBS-D), as the major type of IBS, is characterized by perennial abdominal pain and diarrhea (3), with complex and diverse pathogenesis (4–6). Although its etiology remains unclear, microbial factors play key roles in IBS pathophysiology (7). Both the structure and function of the intestinal microbiota of IBS-D patients are intensively disturbed (8, 9). The multifactorial pathophysiology of IBS-D suggests multiple therapeutic approaches, such as altering intestine microbiota, visceral hypersensitivity, intestinal permeability, gut-brain interaction and psychological strategies (10). Therefore, complementary and alternative medicines such as traditional Chinese medicine (TCM) featuring synergistic effects may show special activities for IBS-D (11).
Few investigations have shown direct interactions between TCM formulas and gut microbiota. It is well known that oral administration is the major routine of TCM that inevitably interacts with the intestinal microbiota. The slight alterations of the construction and function in gut microbiota can significantly change the decomposition, transformation, and absorption of complex ingredients in Chinese herbs. Hence, directly detecting the potential molecular interactions between the intestinal microbiota and TCM herbs is pivotal for probing their interactive mechanisms. For instance, Si-Jun-Zi Decoction (SJZD) has been extensively used for IBS-D and other intestinal complaints for hundreds of years (11, 12). Unfortunately, the underlying mechanisms of SJZD via intestinal microbiota against IBS-D have not been fully described.
The ex vivo fermentation system is a promising platform for exploring the direct interactions between TCM formulas and intestinal microbiota. Accumulating publications have demonstrated that gut microbiota can transform ingredients in TCM herbs into diverse metabolites that show different bioavailability, bioactivity and/or toxicity (13), which can intensively impact the health and disease of mammalian hosts (14). A considerable amount of research has been performed to explore the interactions between intestinal microbiota (always from healthy volunteers) and the ingredients of certain TCM herbs (15, 16). However, few studies have explored the precise interactions of intestinal microbiota derived from particular patients and relevant TCM formulas (17, 18). Originally from long-term successful clinical practice, TCM formulas consisting of several herbs create a much more complicated mechanism because different active ingredients can work together to produce a more desired health effect or can cancel out the negative effects associated with a single herb, thereby minimizing side effects and retaining only the desired effect.
In the present work, an ex vivo cocultivation system will be established to determine the direct interactions of SJZD with the intestinal microbiota derived from IBS-D patients (19). To simulate the interactive regulation process between IBS-D intestine flora and effective TCM formulas, this anaerobic fermentation platform is of scientific significance for elucidating the interactive molecular mechanisms of a large number of TCM prescriptions and sparks a revolution in drug discovery based on Chinese formulas (20).
This work was approved by the Ethics Committee of the Chengdu University of TCM. Each participant signed an informed consent form before the experiment. Five representative IBS-D patients (group IBS-D) were diagnosed and recruited according to TCM standards (ZY/T001.9~001.9.94 & GB/T16751.2~1997) for the screen of Spleen-deficiency syndrome and Rome III diagnostic criteria for IBS-D identification. Five age-matched healthy subjects were simultaneously enrolled as normal controls (NC group). All participants had a routine diet before sample collection. None of the volunteers had taken antibiotics for the last three months. All fecal samples were collected under sterile rule. A solution for sample preservation and transportation, composed of KH2PO4 (0.45%), Na2HPO4 (0.6%), Tween-80 (0.05%), and agar (0.1%), was prepared and applied for temporary storage and transit of collected samples.
All herbs of SJZD formula were purchased from the Affiliated Hospital, Chengdu University of TCM. It consists of six herbs: Codonopsis Tangshan Oliv., Atractylodes macrocephala Koidz., Poria cocos (Schw. Wolf, Glycyrrhiza uralensis Fisch, Citrus reticulata Blanco, Zingiber oj-jicinale Rosc., 30 g for each herb. This formula was prepared under the manufacturing rule of TCM decoction (21). The applied solution of SJZD was adjusted to 1 g/ml (W/V).
Tween-80 (Kemiou Chemical reagent, Tianjin, China), vitamin K1 (Solarbio Biotechnology, Beijing, China), and protohemin (Meilun Biotech, Dalian, China) were used in anaerobic culture. The culture medium, named General Anaerobic Medium (GAM) (Hopebiol Biotechnology, Qingdao, China), was composed of peptone 5.0 g, proteose peptone 5.0 g, enzymatically digested soybean meal 3.0 g, serum powder 10.0 g, beef extract 2.2 g, yeast extract 2.5 g, liver infusion 1.2 g, soluble starch 5.0 g, dextrose 0.5 g, sodium chloride 3.0 g, monopotassium phosphate 2.5 g, L-tryptophan 0.2 g, L-arginine 1.0 g, sodium thioglycollate 0.3 g, and L-cysteine monohydrochloride 0.3 g, and water was added up to 1000 mL. pH 7.3 was adjusted, sterilized at 121 °C for 20 min, and stored at 4°C for culture. The culture solution (KH2PO4 4.5 g, Na2HPO4 6 g, Tween-80 0.5 g, agar 1 g, added distilled water to 1000 ml) was mixed with GAM 3:7 (V/V) to obtain the transfer solution.
Approximately 1 g of fecal sample was collected from each volunteer. The fecal sample was immediately put into a tube with 2 ml of the solution for sample preservation and transportation as described above. After slight mixing at 4°C for 5 min, 0.2 ml of the mixture was removed from each sample. The representative intestinal microbiota was then obtained by putting together five mixtures of groups IBS-D or NC and stored at -80°C.
The representative intestinal microbiota derived from the IBS-D and NC groups were resuscitated in GAM (1:9, V/V) at 37°C. A total of 10 ml of gut microbiota was mixed with 90 ml of drug-containing GAM (1:4, V/V) and cultured in a 250 ml flask located in an anaerobic airbag at 37°C for 35 h. NC0, NC6, NC12, NC24 and NC35 represent the coculture mixtures from group NC and sampled at 0, 6, 12, 24, and 35 h postincubation, respectively, while IBSD0, IBSD6, IBSD12, IBSD24, and IBSD35 represent those of group IBS-D. Three duplicated samples of each group were performed and centrifuged at 4000 g and 4°C for 10 min. The supernatant was collected, and three samples at each time point were equally mixed together and stored in a -80°C refrigerator.
Cetyltrimethylammonium bromide (CTAB) and sodium dodecyl sulfate (SDS) were used to extract the total DNA from each sample of fermentation solution. The V3 and V4 regions of the 16S rRNA gene were amplified with a barcode (22) by a high-fidelity PCR master mix (New England Biolabs). The PCR products were then purified with a GeneJET gel extraction kit (Thermo Scientific). Sequencing libraries were generated using the NEB Next® Ultra™ DNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations, and index codes were added. Sequencing data were analyzed by a quantitative kit (Kapa Biosystems, KK4824) using an Illumina MiSeq platform for paired-end sequencing. The operational taxonomic units (OTUs) were selected through open reference OTU selection. Using LEfSe software, the LDA score was set to 4, and the community structure differences of the samples were analyzed. Metastatic analysis was carried out by R (Version 2.15.3) to analyze the differences between groups at each classification level, and the p value was obtained. Referring to Benjamin and Hochberg’s false discovery rate, the Q value was obtained by correction of the p value (23). R (Version 2.15.3) was used to perform the t test between groups to determine the species with significant differences between groups and draw the map.
Then, 400 μL of 80% methanol solution was added to 100 μL of fermentation, vortexed well, and centrifuged at 14000 g and 4°C for 20 min. Next, 300 μL of the supernatant was placed in a 1.5-ml centrifuge tube for LC–MS analysis. The supernatant was diluted to a final concentration of 53% methanol with LC–MS grade water. The samples were subsequently transferred to a fresh tube and centrifuged at 14000 g and 4°C for 20 min. Finally, the supernatant was injected into the LC–MS/MS system for further analysis. A Conquer UHPLC system (ThermoFisher) and a track rap Q extraction series mass spectrometer (ThermoFisher) were used in positive and negative modes. The original data generated by UHPLC–MS/MS were processed by peak pairing, peak selection, and quantification of each metabolite using compound finder 3.1 (CD3.1, Thermo Fisher). Principal component analysis (PCA) and partial least squares discrimination analysis (PLS-DA) were used to analyze the significant differences in metabolites between the IBS-D and NC groups. Hierarchical clustering (HCA) and metabolite correlation analysis were used to reveal the relationship between metabolites and samples. Finally, the differentially expressed metabolites (DEMs) were matched with relevant biological processes based on the KEGG database.
Network pharmacology forecasting was performed by the “BATMAN-TCM” (Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine) (24) to predict the compounds and corresponding targets in SJZD (set score = 20, P <= 0.05). Compared with the Therapeutic Target Database (TTD) and Online Mendelian Inheritance in Man (OMIM), targets and compounds related to IBS(-D) were obtained. The pathways that contact related targets were retrieved through the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Finally, the network diagram was drawn using Cytoscape 3.8.2 software.
SPSS Software (V16.0; SPSS Inc., Chicago, IL) was used for statistical analyses. All data are expressed as the mean ± standard deviation (SD). Data were analyzed with one-way ANOVA. Differences between the groups were evaluated using Student’s t test. Data were considered to be statistically significant with each value *P < 0.05, **P < 0.01. The correlation analysis was based on the cor() function in R (v3.1.3), and the Pearson correlation coefficient R (1 ≥ R ≥ - 1) between all metabolites was calculated to analyze the correlation among the metabolites. The cor.test () function (FDR < 0.05) was used to test the significance of the correlation analysis.
From May 15 to August 04, 2018, ten stool samples, including five IBS-D subjects (IBS-D group) and five healthy subjects as normal controls (NC group), were collected from patients aged 18~35 years in the outpatient clinic of the Department of Gastroenterology, Sichuan Provincial Hospital of TCM.
The results of 16S rRNA gene sequencing showed that the dominant phyla in the cocultivations were Firmicutes, Proteobacteria, Actinobacteria and Bacteroidetes (Figure 1A), while Streptococcus, E. Shigella, Lactobacillus, Clostridium, Paraclostridium and Bifidobacterium were the main genera (Figure 1B). A total of 578 OTUs were identified from both groups (Figure 1C), and 103 were identified as core bacteria (Figure 1D). The T tests between groups showed that Proteobacteria and Firmicutes were significantly different at the phylum level (Figure 1E), and 6 microbiota were significantly different at the genus level (Figure 1F). LEfSe analysis of biomarkers (gate level to genus level) in each group revealed 10 biomarkers in the IBS-D group and 6 biomarkers in the NC group (Figure 1G).
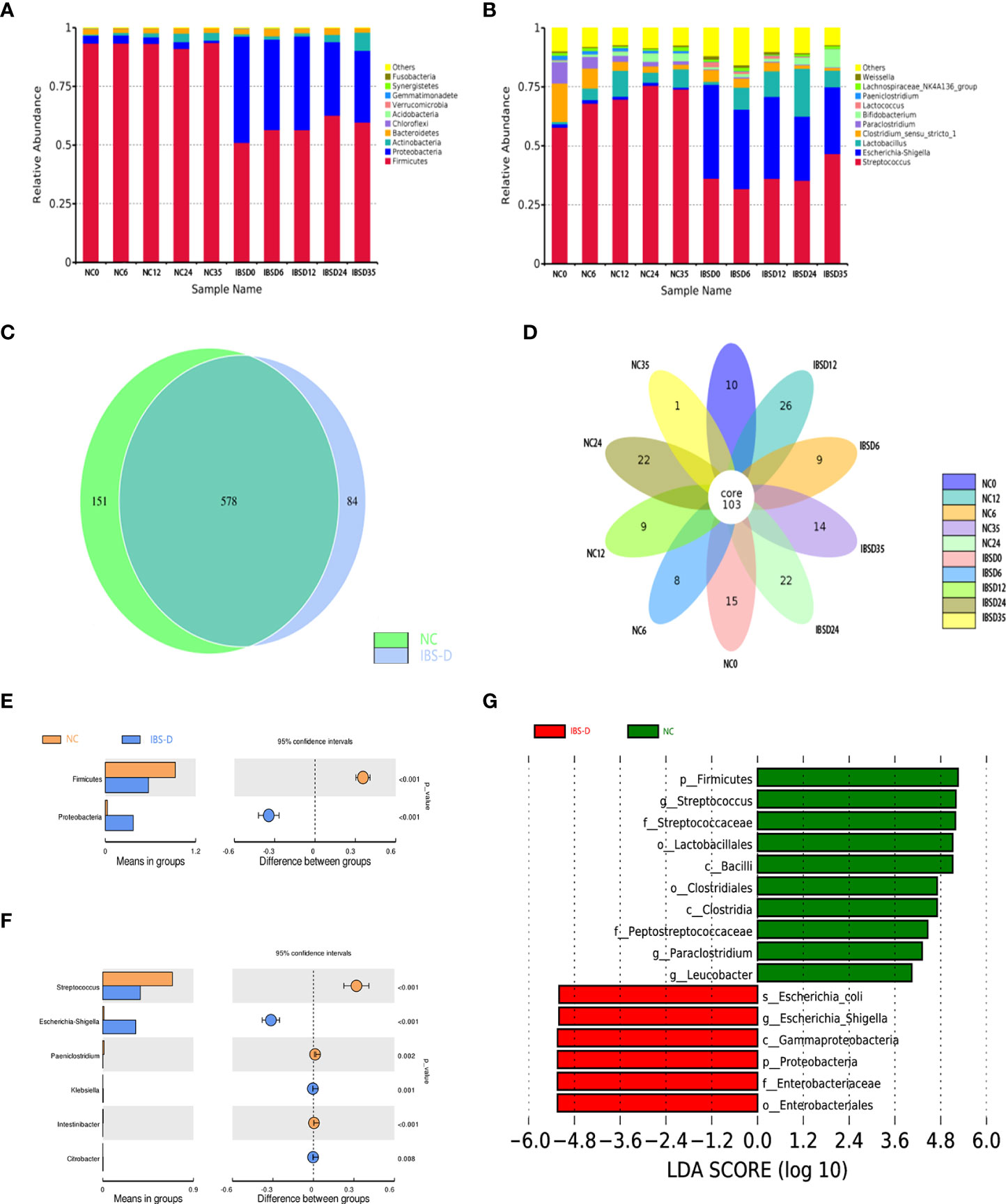
Figure 1 Basic characteristics of the cocultivated microbiota. (A, B) Histogram of relative abundance at the phylum (A) and genus (B) levels. The Arabic numerals within the sample name (0–35) indicate the culture time (hours). (C, D) Differentially expressed species between groups NC and IBS-D (C) and time series cocultivations (D). (E, F) Differentially expressed taxa at the phylum (E) and genus (F) levels. (G) LEfSe analysis. LEfSe: LDA effect size analysis. LDA score: Linear discriminant analysis (LDA) affects the influence of species with significant differences in data. IBS-D: IBS-D group. NC: Normal control group. NC0-NC35: time series cocultivations of SJZD and intestine microbiota derived from the NC group. IBSD0-IBSD35: time series cocultivations of SJZD and intestine microbiota derived from the IBS-D group. SJZD: Si-Jun-Zi Decoction. IBS-D: diarrhea-predominant irritable bowel syndrome.
The alpha diversity counted by the Shannon and Simpson indexes between the IBS-D and NC groups was significantly different (*P < 0.05, **P < 0.01), while the Ace and Chao1 indexes were not (P > 0.05) (Table 1 and Figures S1A–D). In addition, the beta diversities between the two groups also reached significant differences (Figure S1E).
SJZD restores the gut microbiota of IBS-D in vitro. The UPGMA clustering tree shows that with the development of time, the clustering distance between IBS-D samples cultured for 35 h and the NC group becomes shorter, and the difference between the IBS-D group and the NC group decreases (Figure 2A). SJZD mainly reduced the relative abundance of the phylum Proteobacteria and increased the relative abundance of Firmicutes (Figure 2A). Based on the screened differential microbiota from T test and LEfSe analysis and observing their dynamics from 0 through 35 h coculture time. We homogenized the differential microbiota using the Zscore to assess changes in microbiota abundance at different taxonomic levels in the samples before and after culture and found that most microbiota in the IBS-D group were more discrete than those in the NC group (Figure 2B). Among the top 10 genera, Escherichia-Shigella (Figure 2C), Streptococcus (Figure 2D), and Paeniclostridium (Figure 2E) were upregulated by SJZD. In addition, SJZD-upregulated Bifidobacterium was also the dominant group in the Top10 at the genus level (Figure 2F). The change in the ratio of Escherichia-Shigella to Bifidobacterium in the IBS-D group was less than that in the NC group (Figure 2G).
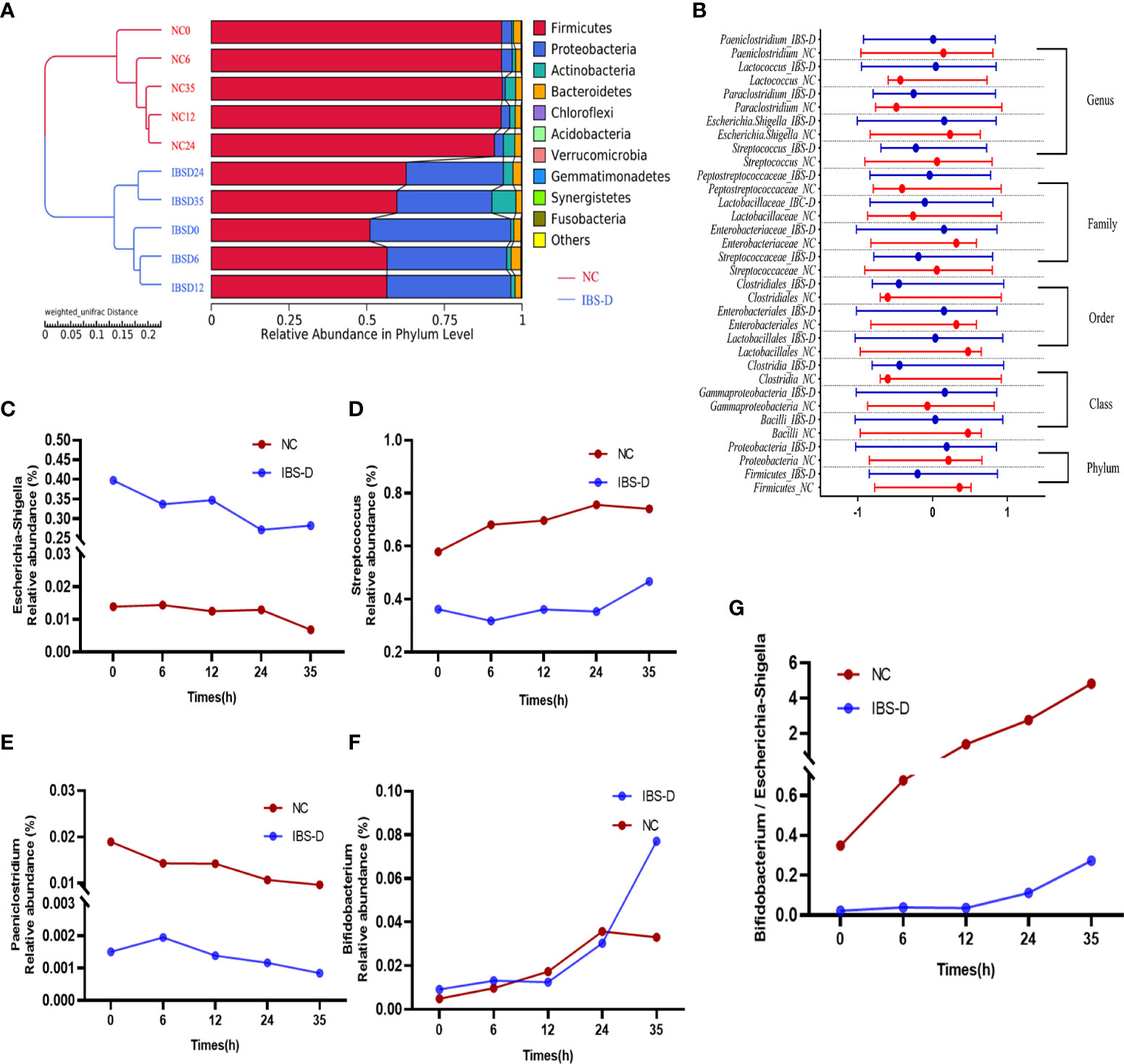
Figure 2 SJZD restored the dysbiotic intestinal microbiota of IBS-D subjects. (A) The unweighted pair-group method with arithmetic mean (UPGMA) clustering tree. (B) The Zscores of biomarkers at different taxa. The nodes in the graph show the median and interquartile ranges. Zscore = (x–μ)/σ (x: relative abundance of known bacteria, σ: standard difference, μ: average value). (C-F) Time series of relative abundances of Escherichia-Shigella (C), Streptococcus (D), Paeniclostridium (E), and Bifidobacterium (F). (G) Bifidobacterium to Escherichia-Shigella ratio. IBS-D: IBS-D group. NC: Normal controls. The Arabic numbers 0~35 indicate the coculture times (hours) of SJZD with the microbiota. SJZD: Si-Jun-Zi Decoction. IBS-D: diarrhea-predominant irritable bowel syndrome.
The potential targets of 450 compounds of SJZD were predicted by the BATMAN-TCM platform (Table 2). We then compared the TTD database with the OMIM database to select compounds and targets associated with IBS (Table 3). Third, five target genes (HTR3A, HTR4, HTR6, CRHR1, HTR3B) of SJZD acting on IBS-D were screened after comparing the above results. Finally, these target genes were enriched in four pathways (i.e., serotonergic synapse, calcium signaling pathway, neuroactive ligand–receptor interaction, long-term depression.) based on the KEGG database.
Associated with the above virtual experiments, we analyzed the metabolites in the coculture system and found that the differentially expressed metabolites (DEM) were also enriched in neuroactive ligand–receptor interactions. A total of 458 DEMs (Figure 3A, C) were detected in positive ion mode, and 166 DEMs (Figure 3B, D) were detected in negative ion mode (Table 4). Eleven KEGG pathways were statistically enriched based on the above DEMs (p<0.05), including 14 metabolites such as histamine, morphine, cytocine, tryptamine, and pyridoxamine (Table 5). It is worth mentioning that histamine, morphine and tryptamine are enriched in the neuroactive ligand–receptor interaction pathway.
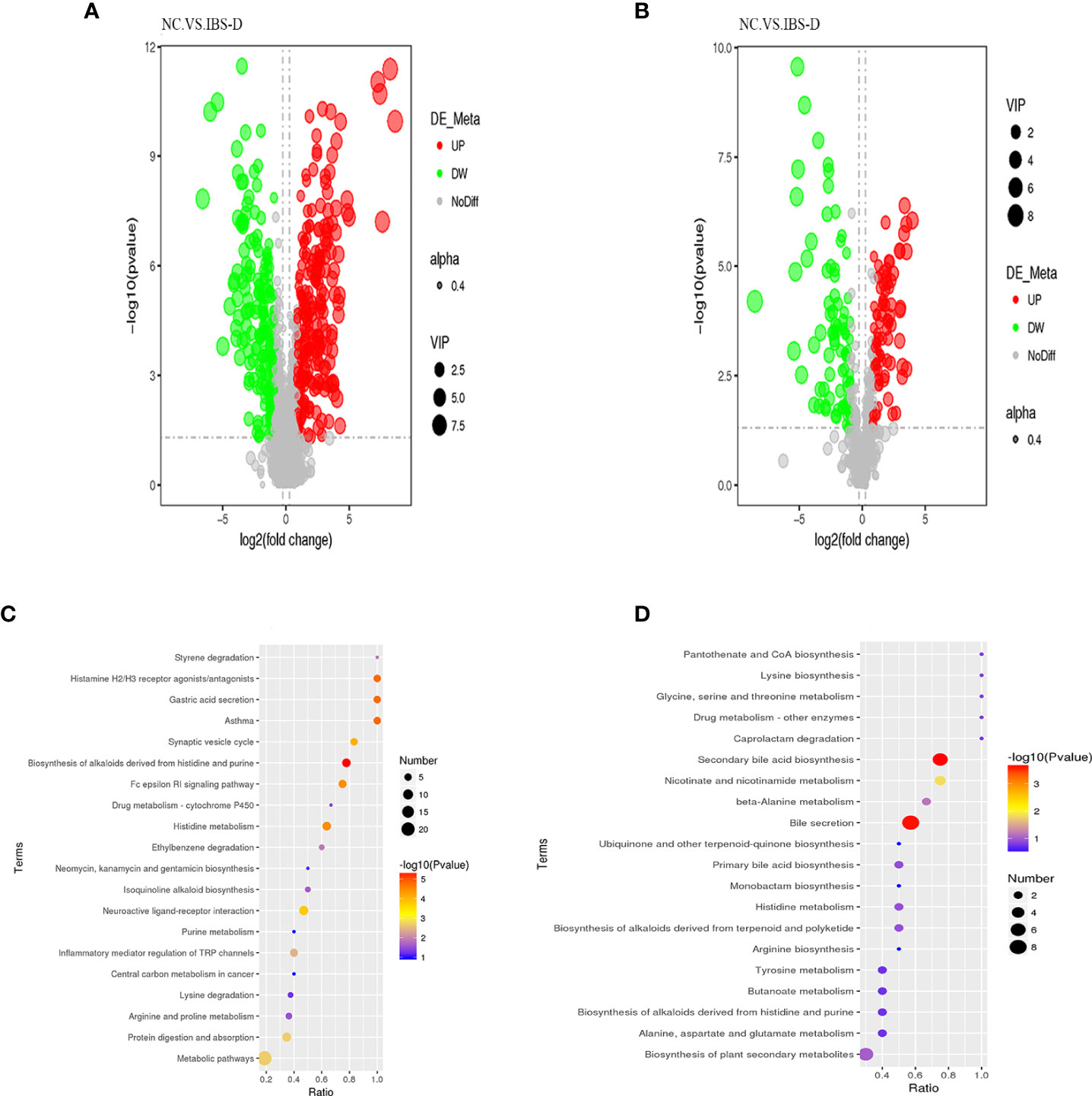
Figure 3 Nontargeted metabolites of the cocultured SJZD and intestine microbiota. (A, B) Volcano map of differentially expressed metabolites (DEM) from positive ion mode (A) and negative ion mode (B). VIP: Variable importance in the projection. alpha: Transparency of dots. Black: the metabolite with no significant difference (nodiff), red: upregulated metabolite (up), green: downregulated metabolite (DW). (C, D) Bubble chart of enriched KEGG pathways, positive ion mode (C) and negative ion mode (D). SJZD: Si-Jun-Zi Decoction. IBS-D: diarrhea-predominant irritable bowel syndrome.
The gut microbiota showed a close correlation with intestinal DEMs (S2). Correlation analysis showed that five genera, Proteobacteria and Paeniclostridium, were significantly correlated with three DEMs (histamine, morphine and tryptamine) (Figure 4A). At the phylum level, SJZD mainly affected the relative abundance of Firmicutes (Figure 4B) and Proteobacteria (Figure 4C) in IBS-D subjects; Proteobacteria was negatively correlated with Firmicutes and positively correlated with histamine, morphine and tryptamine. At the genus level, Escherichia-Shigella was negatively correlated with Streptococcus and Paeniclostridium and positively correlated with histamine, morphine and tryptamine. Using Cytoscape 3.8.2 software, the crucial intestinal microbe-metabolite-SJZD-target network diagram was drawn (Figure 4D), particularly associated with the neuroactive ligand–receptor interaction pathway (hsa04080) and key intestinal microbiota.
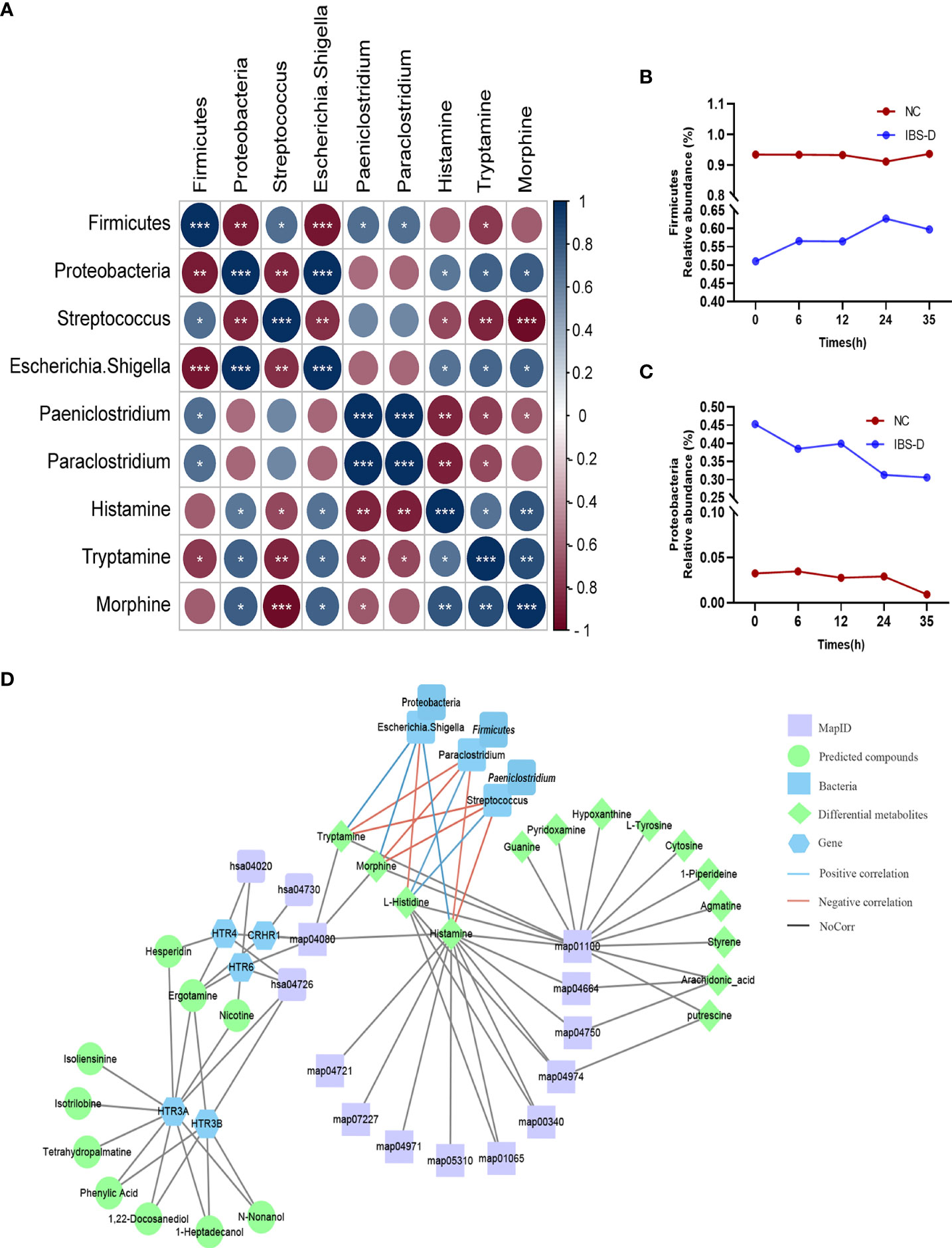
Figure 4 Significant correlations between microbiota and metabolites. (A) Correlation analysis of intestinal microbiota and differentially expressed metabolites. (B, C). Time series of relative abundances of the phyla Proteobacteria (B) and Firmicutes (C) in a cocultivation system. (D) Compound-target network based on IBS-D microbiota-metabolome-SJZD compound analysis. SJZD: Si-Jun-Zi Decoction. IBS-D: diarrhea-predominant irritable bowel syndrome.
Given that oral administration is the main approach in TCM, observation of the direct interactions between TCM formulas and gut microbiota is an extremely important and urgent research field. The intestinal microbiome of human beings is a diverse and dynamic collection of microorganisms that have much more metabolic potential than those of their mammalian host. Ever-increasing evidence indicates that the intestinal microbiota plays a pivotal role in TCM therapy by complicated interplay with Chinese components (25). This interplay includes activities such as intestinal microbiota biotransforming TCM components into metabolites with different bioavailability and bioactivity/toxicity from their precursors (15), improving the dysbiotic microbiota and consequently ameliorating associated pathological conditions (16) and mediating the synergistic and antagonistic interactions between the multiple chemicals in certain TCM formulas (26). However, the interactive mechanisms between TCM herbs/formula and relevant intestinal microbiota have always been distorted by gut ecology, such as mucosal immunity, the enteric nervous system and the hormonal environment.
This work aims to observe the direct interaction mechanism of a typical dysbiotic intestine microbiota and a TCM formula based on a cocultivation or fermentation system. IBS is a common but complex disease characterized by dysbiotic intestinal microbiota. Compared with healthy controls, the family Enterobacteriaceae (phylum Proteobacteria), family Lactobacillaceae, and genus Bacteroides were increased in IBS subjects, whereas uncultured Clostridiales I, genus Faecalibacterium and genus Bifidobacterium were decreased (27). On the other hand, the TCM formula SJZD has a long history of clinical application for functional dyspepsia and IBS-D. Therefore, we collected five intestinal microbiota samples derived from representative IBS-D subjects and five controls and established an ex vivo concultivation system to reveal the interaction pattern of SJZD compounds and the intestinal microbiota of IBS-D patients.
The results of 16S rRNA gene sequencing showed that SJZD effectively rescued intestinal dysbiosis in patients with IBS-D. Both alpha and beta diversities between the IBS-D and NC groups reached significant differences (Table 1 and Figure S1E) (28–31). Core taxa were observed in both groups (Figure 1B), consistent with other publications (32). The genera Bifidobacteria (33), Lactobacillus and Streptococcus (34) can ameliorate the symptoms of IBS; we observed that the relative abundances of these genera increased with cocultivation time. Interestingly, the abundance of the genus Streptococcus was significantly higher than that in the NC group at every time point. Previous studies have reported that the abundance of Streptococcus in constipated IBS (IBS-C) is relatively high (35), but the underlying mechanism is not clear. IBS-D is associated with increased abundances of Escherichia-Shigella (36–38). Paeniclostridium is related to intestinal injury and inflammation (39), whereas Proteobacteria is a negative factor for intestinal homeostasis (30–32). In summary, SJZD effectively rescued key abnormal bacteria in the intestinal microbiota of IBS-D patients.
Metabolome analysis revealed that SJZD beneficially tuned the altered metabolite profile of intestinal microbiota in IBS-D subjects. Abdominal pain, as one of the predominant manifestations of IBS-D, is associated with the abnormal metabolism of enteric neurotransmitters such as tryptophan. Tryptamine, as the bacterial metabolite of tryptophan, plays a pivotal role in balancing intestinal immune tolerance and maintaining intestinal microbiota (40–44) and promoting intestinal functions (45). In this work, we observed significantly higher levels of tryptamine, morphine, arachidonic acid, and histamine in the IBS-D group (S3). The concentrations of these neurotransmitters and metabolites were positively correlated with Escherichia-Shigella but negatively correlated with Streptococcus (Figures 4B, D). Although morphine has the effect of slowing down movement in the large intestine (46) and arachidonic acid may improve gastrointestinal movement (47), our present work showed that the content of morphine is not statistically high in the IBS-D group. The histamine level was negatively correlated with L-histidine (Figures 4B, C), suggesting that histidine was converted to histamine in the IBS-D group (S3). In addition, our results indicated that the genera Escherichia-Shigella and Streptococcus may synergistically regulate histidine metabolism and suggest that foods rich in L-histidine may worsen the symptoms of IBS-D. In a sentence, histamine, morphine and tryptamine, as crucial metabolites enriched in the neuroactive ligand–receptor interaction pathway, were the essential neurotransmitters derived from the fermentation of SJZD and the intestinal microbiota of IBS-D subjects. These metabolites respond to abdominal pain, the crucial symptom of IBS-D patients. Therefore, our results indicated that SJZD facilitated the ex vivo modulation of the metabolite profiles, particularly those involved in the pathway of neuroactive ligand–receptor interaction.
Correlation analysis further revealed the network outline of intestine microbiota, intestine metabolites, SJZD compounds and targets for IBS-D patients (Figure 4D), particularly associated with the neuroactive ligand–receptor interaction pathway (hsa04080) and core taxa in the intestinal microbiota. The BATMAN-TCM network pharmacology and other correlation analysis platforms revealed that, in addition to metabolic modulation of the intestinal microbiota that leads to symptom amelioration, SJZD compounds showed regulatory targets for IBS-D therapy. Specifically, the correlation analysis demonstrated that five genera were significantly correlated with IBS-D-relevant neurotransmitters (i.e., histamine, morphine and tryptamine) (Figure 4A). These results could be used to determine the correlation between SJZD and the restoration of abnormal abundances of certain intestinal microbes. This finding indicates that the cocultivation of SJZD and gut microbiota originating from IBS-D volunteers is a useful platform to explore the direct interactions of TCM formula and complex intestine microbiota.
Some shortcomings exist in this work. First, our concultivation platform does not fully mimic the intestine microecology of human beings. For instance, apart from the majority of anaerobic bacteria, there are facultative and even aerobic bacteria living within our intestine. The dynamic growth of anaerobic, facultative and aerobic bacteria is one of the key mechanisms underlying the robust intestinal air environment. Hence, our anaerobic culture environment cannot fully simulate clinical gas conditions. Second, although we used an ex vivo fermentation system and observed a direct interaction between SJZD and the intestinal microbiota of IBS-D patients, the underlying molecular regulation mechanisms need to be further explored. Third, to detect more complete mechanisms of SJZD against IBS-D subjects, comparative studies between in vivo and ex vivo models should also be executed. Finally, the representative samples of intestine microbiota were composed of only ten donors in this study. Given the marked individual diversities of the intestinal microbiota in IBS-D patients, a larger sample size might be required for further studies.
Our work demonstrated that SJZD rescued the dysbiotic intestinal microbiota and ameliorated the dysfunctional neurotransmitter metabolism involved in the major symptoms of IBS-D. The ex vivo coculture system could be extensively used to reveal the direct interactions between TCM formulas and complex gut microbiota.
The original contributions presented in the study are publicly available. This data can be found here: NCBI database; PRJNA898776; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA898776.
The studies involving human participants were reviewed and approved by The Ethics Committee of the Chengdu University of TCM. The patients/participants provided their written informed consent to participate in this study.
XWX: Writing - Original Draft, Data Curation. YX: Investigation, Data Curation, Visualization. QQC: Visualization. DD: Writing-Review and Editing. ZQW: Investigation. YJX: Investigation. YLW: Resources. XMW: Resources. WJD: Writing - Review and Editing, Project administration, Funding acquisition. All authors contributed to the article and approved the submitted version.
The author would like to express their sincere gratitude to the experimental technical assistance provided by the Institute of Chinese Medicine Innovation, Chengdu University of Traditional Chinese Medicine.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.1053103/full#supplementary-material
1. Devanarayana NM, Rajindrajith S, Pathmeswaran A, Abegunasekara C, Gunawardena NK, Benninga MA. Epidemiology of irritable bowel syndrome in children and adolescents in Asia. J Pediatr Gastroenterol Nutr (2015) 60:792–8. doi: 10.1097/MPG.0000000000000714
2. Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology (2006) 130:1480–91. doi: 10.1053/j.gastro.2005.11.061
3. Guilera M, Balboa A, Mearin F. Bowel habit subtypes and temporal patterns in irritable bowel syndrome: Systematic review. Am J Gastroenterol (2005) 100:1174–84. doi: 10.1111/j.1572-0241.2005.40674.x
4. Crowell MD, Harris L, Jones MP, Chang L. New insights into the pathophysiology of irritable bowel syndrome: Implications for future treatments. Curr Gastroenterol Rep (2005) 7:272–9. doi: 10.1007/s11894-005-0019-8
5. Hasler WL. Traditional thoughts on the pathophysiology of irritable bowel syndrome. Gastroenterol Clin North Am (2011) 40:21–43. doi: 10.1016/j.gtc.2010.12.004
6. Pimentel M, Chang C. Inflammation and microflora. Gastroenterol Clin North Am (2011) 40:69–85. doi: 10.1016/j.gtc.2010.12.010
7. Pimentel M, Lembo A. Microbiome and its role in irritable bowel syndrome. Dig Dis Sci (2020) 65:829–39. doi: 10.1007/s10620-020-06109-5
8. Distrutti E, Monaldi L, Ricci P, Fiorucci S. Gut microbiota role in irritable bowel syndrome: New therapeutic strategies. World J Gastroenterol (2016) 22:2219–41. doi: 10.3748/wjg.v22.i7.2219
9. Mohajeri MH, La Fata G, Steinert RE, Weber P. Relationship between the gut microbiome and brain function. Nutr Rev (2018) 76:481–96. doi: 10.1093/nutrit/nuy009
10. Nee J, Lembo A. Review article: Current and future treatment approaches for IBS with diarrhoea (IBS-d) and IBS mixed pattern (IBS-m). Aliment Pharmacol Ther (2021) 54 Suppl 1:S63–74. doi: 10.1111/apt.16625
11. Li L, Cui H, Li T, Qi J, Chen H, Gao F, et al. Synergistic effect of berberine-based Chinese medicine assembled nanostructures on diarrhea-predominant irritable bowel syndrome In vivo. Front Pharmacol (2020) 11:1210. doi: 10.3389/fphar.2020.01210
12. Wu Z-C, Zhao Z-L, Deng J-P, Huang J-T, Wang Y-F, Wang Z-P. Sanhuang shu’ai decoction alleviates DSS-induced ulcerative colitis via regulation of gut microbiota, inflammatory mediators and cytokines. BioMed Pharmacother (2020) 125:109934. doi: 10.1016/j.biopha.2020.109934
13. An X, Bao Q, Di S, Zhao Y, Zhao S, Zhang H, et al. The interaction between the gut microbiota and herbal medicines. BioMed Pharmacother (2019) 118:109252. doi: 10.1016/j.biopha.2019.109252
14. Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev (2010) 90:859–904. doi: 10.1152/physrev.00045.2009
15. Chen F, Wen Q, Jiang J, Li H-L, Tan Y-F, Li Y-H, et al. Could the gut microbiota reconcile the oral bioavailability conundrum of traditional herbs? J Ethnopharmacol (2016) 179:253–64. doi: 10.1016/j.jep.2015.12.031
16. Li H, Zhou M, Zhao A, Jia W. Traditional Chinese medicine: Balancing the gut ecosystem. Phytother Res (2009) 23:1332–5. doi: 10.1002/ptr.2590
17. Peterson CT, Sharma V, Iablokov SN, Albayrak L, Khanipov K, Uchitel S, et al. 16S rRNA gene profiling and genome reconstruction reveal community metabolic interactions and prebiotic potential of medicinal herbs used in neurodegenerative disease and as nootropics. PloS One (2019) 14(3):e0213869. doi: 10.1371/journal.pone.0213869
18. Su L, Su Y, An Z, Zhang P, Yue Q, Zhao C, et al. Fermentation products of danshen relieved dextran sulfate sodium-induced experimental ulcerative colitis in mice. Sci Rep (2021) 11:16210. doi: 10.1038/s41598-021-94594-7
19. Wu R, Zhao D, An R, Wang Z, Li Y, Shi B, et al. Linggui zhugan formula improves glucose and lipid levels and alters gut microbiota in high-fat diet-induced diabetic mice. Front Physiol (2019) 10:918. doi: 10.3389/fphys.2019.00918
20. Jia W, Li H, Zhao L, Nicholson JK. Gut microbiota: A potential new territory for drug targeting. Nat Rev Drug Discovery (2008) 7:123–9. doi: 10.1038/nrd2505
21. Tian S, Song X, Wang Y, Wang X, Mou Y, Chen Q, et al. Chinese Herbal medicine baoyuan jiedu decoction inhibits the accumulation of myeloid derived suppressor cells in pre-metastatic niche of lung via TGF-β/CCL9 pathway. Biomed Pharmacother (2020) 129:110380. doi: 10.1016/j.biopha.2020.110380
22. Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A, et al. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut (2012) 61:1124–31. doi: 10.1136/gutjnl-2011-301104
23. White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PloS Comput Biol (2009) 5:e1000352. doi: 10.1371/journal.pcbi.1000352
24. Liu Z, Guo F, Wang Y, Li C, Zhang X, Li H, et al. BATMAN-TCM: A bioinformatics analysis tool for molecular mechANism of traditional Chinese medicine. Sci Rep (2016) 6:21146. doi: 10.1038/srep21146
25. Xu J, Chen H-B, Li S-L. Understanding the molecular mechanisms of the interplay between herbal medicines and gut microbiota. Med Res Rev (2017) 37:1140–85. doi: 10.1002/med.21431
26. Zhang M, Long Y, Sun Y, Wang Y, Li Q, Wu H, et al. Evidence for the complementary and synergistic effects of the three-alkaloid combination regimen containing berberine, hypaconitine and skimmianine on the ulcerative colitis rats induced by trinitrobenzene-sulfonic acid. Eur J Pharmacol (2011) 651:187–96. doi: 10.1016/j.ejphar.2010.10.030
27. Pittayanon R, Lau JT, Yuan Y, Leontiadis GI, Tse F, Surette M, et al. Gut microbiota in patients with irritable bowel syndrome-a systematic review. Gastroenterology (2019) 157:97–108. doi: 10.1053/j.gastro.2019.03.049
28. Si J-M, Yu Y-C, Fan Y-J, Chen S-J. Intestinal microecology and quality of life in irritable bowel syndrome patients. World J Gastroenterol (2004) 10:1802–5. doi: 10.3748/wjg.v10.i12.1802
29. Chey WD, Kurlander J, Eswaran S. Irritable bowel syndrome: A clinical review. JAMA (2015) 313:949–58. doi: 10.1001/jama.2015.0954
30. Wang Y, Zheng F, Liu S, Luo H. Research progress in fecal microbiota transplantation as treatment for irritable bowel syndrome. Gastroenterol Res Pract (2019) 2019:9759138. doi: 10.1155/2019/9759138
31. Kassinen A, Krogius-Kurikka L, Mäkivuokko H, Rinttilä T, Paulin L, Corander J, et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology (2007) 133:24–33. doi: 10.1053/j.gastro.2007.04.005
32. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature (2010) 464:59–65. doi: 10.1038/nature08821
33. Parkes GC, Rayment NB, Hudspith BN, Petrovska L, Lomer MC, Brostoff J, et al. Distinct microbial populations exist in the mucosa-associated microbiota of sub-groups of irritable bowel syndrome. Neurogastroenterol Motil (2012) 24:31–9. doi: 10.1111/j.1365-2982.2011.01803.x
34. Lewis ED, Antony JM, Crowley DC, Piano A, Bhardwaj R, Tompkins TA, et al. Efficacy of lactobacillus paracasei HA-196 and bifidobacterium longum R0175 in alleviating symptoms of irritable bowel syndrome (IBS): A randomized, placebo-controlled study. Nutrients (2020) 12:E1159. doi: 10.3390/nu12041159
35. Matsumoto H, Shiotani A, Katsumata R, Fukushima S, Handa Y, Osawa M, et al. Mucosa-associated microbiota in patients with irritable bowel syndrome: A comparison of subtypes. Digestion (2021) 102:49–56. doi: 10.1159/000512167
36. Liu Y, Yuan X, Li L, Lin L, Zuo X, Cong Y, et al. Increased ileal immunoglobulin a production and immunoglobulin a-coated bacteria in diarrhea-predominant irritable bowel syndrome. Clin Transl Gastroenterol (2020) 11:e00146. doi: 10.14309/ctg.0000000000000146
37. Li J, Cui H, Cai Y, Lin J, Song X, Zhou Z, et al. Tong-Xie-Yao-Fang regulates 5-HT level in diarrhea predominant irritable bowel syndrome through gut microbiota modulation. Front Pharmacol (2018) 9:1110. doi: 10.3389/fphar.2018.01110
38. Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil (2012) 24:521–530, e248. doi: 10.1111/j.1365-2982.2012.01891.x
39. Tian S, Liu Y, Wu H, Liu H, Zeng J, Choi MY, et al. Genome-wide CRISPR screen identifies semaphorin 6A and 6B as receptors for paeniclostridium sordellii toxin TcsL. Cell Host Microbe (2020) 27:782–792.e7. doi: 10.1016/j.chom.2020.03.007
40. Leprun PMB, Clarke G. The gut microbiome and pharmacology: A prescription for therapeutic targeting of the gut-brain axis. Curr Opin Pharmacol (2019) 49:17–23. doi: 10.1016/j.coph.2019.04.007
41. Gao J, Xu K, Liu H, Liu G, Bai M, Peng C, et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol (2018) 8:13. doi: 10.3389/fcimb.2018.00013
42. Jennis M, Cavanaugh CR, Leo GC, Mabus JR, Lenhard J, Hornby PJ. Microbiota-derived tryptophan indoles increase after gastric bypass surgery and reduce intestinal permeability in vitro and in vivo. Neurogastroenterol Motil (2018) 30(2). doi: 10.1111/nmo.13178
43. Zucchi R, Chiellini G, Scanlan TS, Grandy DK. Trace amine-associated receptors and their ligands. Br J Pharmacol (2006) 149:967–78. doi: 10.1038/sj.bjp.0706948
44. Williams BB, Van Benschoten AH, Cimermancic P, Donia MS, Zimmermann M, Taketani M, et al. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe (2014) 16:495–503. doi: 10.1016/j.chom.2014.09.001
45. Bhattarai Y, Williams BB, Battaglioli EJ, Whitaker WR, Till L, Grover M, et al. Gut microbiota-produced tryptamine activates an epithelial G-Protein-Coupled receptor to increase colonic secretion. Cell Host Microbe (2018) 23:775–785.e5. doi: 10.1016/j.chom.2018.05.004
46. Corazziari E. Role of opioid ligands in the irritable bowel syndrome. Can J Gastroenterol (1999) 13 Suppl:A:71A–75A. doi: 10.1155/1999/598659
Keywords: diarrhea-predominant irritable bowel syndrome (IBS-D), Si-Jun-Zi decoction (SJZD), nontargeted metabolomics, 16S rRNA gene sequencing, microbiota-herbal cocultivation
Citation: Xia X, Xie Y, Chen Q, Ding D, Wang Z, Xu Y, Wang Y, Wang X and Ding W (2022) Cocultivation of Chinese prescription and intestine microbiota: SJZD alleviated the major symptoms of IBS-D subjects by tuning neurotransmitter metabolism. Front. Endocrinol. 13:1053103. doi: 10.3389/fendo.2022.1053103
Received: 25 September 2022; Accepted: 17 October 2022;
Published: 14 November 2022.
Edited by:
Zhoujin Tan, Hunan University of Chinese Medicine, ChinaReviewed by:
Xiaoliang Li, Heilongjiang University of Chinese Medicine, ChinaCopyright © 2022 Xia, Xie, Chen, Ding, Wang, Xu, Wang, Wang and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weijun Ding, ZGluZ3dlaWp1bkBjZHV0Y20uZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.