
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol. , 02 November 2022
Sec. Cellular Endocrinology
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.1035159
This article is part of the Research Topic Insights in Cellular Endocrinology: 2022 View all 9 articles
 Melissa Yepmo1†
Melissa Yepmo1† Jean-Baptiste Potier1,2†
Jean-Baptiste Potier1,2† Michel Pinget1
Michel Pinget1 Anastasia Grabarz2
Anastasia Grabarz2 Karim Bouzakri1,2
Karim Bouzakri1,2 Aurore Dumond Bourie1*
Aurore Dumond Bourie1*Circular RNAs (circRNAs) are class of non-coding RNA, which are characterized by a covalently closed loop structure. Functionally they can act on cellular physiology, notably by sponging microRNAs (miR), regulating gene expression or interacting with binding protein. To date, circRNAs might represent an interesting, underexploited avenue for new target discovery for therapeutic applications, especially in the liver. The first characteristic of non-alcoholic fatty liver disease (NAFLD) is hepatic cholesterol accumulation, followed by its advanced form of the affection, nonalcoholic steatohepatitis (NASH), due to the occurrence of lobular inflammation, irreversible fibrosis, and in some cases hepatocellular carcinoma (HCC). Therefore, studies have investigated the importance of the dysregulation of circRNAs in the onset of metabolic disorders. In this review, we summarize the potential role of circRNAs in the development of metabolic diseases associated with the liver such as NAFLD or NASH, and their potential to become therapeutic strategies for these pathologies.
Metabolic associated liver diseases, such as non-alcoholic fatty liver disease (NAFLD) have been emerging these last decades all around the world due to the rising of type 2 diabetes, western diets and sedentary lifestyle (1). NAFLD is a chronic liver disease that affects 25% of adults worldwide and is associated with insulin resistance, de novo lipogenesis, dyslipidemia, obesity and a higher cardiovascular risk. The pathophysiology of NAFLD is characterized by the presence of steatosis, defined by the accumulation of lipid vacuoles in more than 5% of all hepatocytes (2). In addition to metabolic syndrome, type 2 diabetes and obesity, numerous NAFLD risk factors exist such as gender, ethnicity and genetic predispositions (3). Among these predispositions, genetic polymorphism such as those related to variants of the genes PNPLA3 and TM6SF2 have become widely studied and have proved their crucial association with the onset of NAFLD during the last few years (4).
In itself, NAFLD can remain clinically silent and undetected for most patients (5). However, in 25% of the cases, NAFLD can progress towards a more severe affection: non-alcoholic steatohepatitis (NASH) (6). NASH is a chronic liver disease whose pathophysiology includes the presence of lobular inflammation, hepatocyte ballooning, oxidative stress, and fibrosis (7) and can be related to more severe outcomes, such as cirrhosis, acute liver failure (8), liver transplantation (8) or hepatocellular carcinoma (HCC) (9, 10). In this context, the homeostasis of the hepatic microenvironment is crucial in the progression of NAFLD to NASH. Three main cell type are implied in this process: hepatocytes, hepatic stellate cells (HSC) and liver macrophages. Hepatocytes are the hub of liver metabolic functions (11) and the first liver injured cell type in NAFLD, through steatosis (12). Resident and circulatory liver macrophages are widely responsible for the onset and progression of liver inflammation during NASH (13), even if their decisive role beyond inflammation has been emerging lately (14). Finally, hepatic stellate cells are a quiescent cell type that mainly store vitamin A in healthy individuals (15). However, in the context of liver injury and NASH, they can become activated, transdifferentiate into myofibroblasts and acquire a pro-fibrotic and pro-inflammatory phenotype, leading to the development of liver fibrosis (16).
Despite the multiplication of potential new drug candidates and clinical trials, there is still no approved treatment for NASH, justifying the urge of a finding (17). The last few years have been critical for the elucidation of new pathways involved in liver diseases pathophysiology, thanks, notably, to the democratization of new breakthrough methods in the field of metabolomics, proteomics and transcriptomics, opening the way to new therapeutic hopes (18–20). Among these new emerging subjects, non-coding RNAs and their pleiotropic roles in chronic liver diseases has been widely studied (21, 22). Many types of non-coding RNAs exist including long non-coding RNAs (lncRNAs), transfer RNAs (tRNAs), micro RNAs (miR), ribosomal RNAs (rRNAs) and circular RNAs (circRNAs) and can represent promising therapeutic agents (23).
Circular RNAs were first discovered in 1976 in plant viroids, observed by electron microscopy (24). They were then identified in eukaryotes in 1979 in the cytoplasmic fraction of eukaryotic cell lines (HeLa cells) (25). Their structure is characterized by a covalently closed loop and lacks polyadenylation at the 3’ and 5’ end, protecting them from exonucleases and strengthening their stability (26). Recent studies have shown that they are generally generated by reverse splicing or exon skipping of pre-messenger RNAs (mRNA) (27). For a long time, due to technical limitations in research, circRNAs were considered as abnormal splicing products from pre-mRNA and their functions were not yet fully understood (28). The last 10 years resulted in major discoveries of the functional role of circRNAs. With the use of high-throughput sequencing and bioinformatics, increased numbers of circRNAs have been identified in eukaryotes, whether in humans (29), animals or plants (30) and the number of published studies on circRNAs has increased exponentially. Their main role as microRNA (miR) sponges has been highlighted (31), fine tuning the transcriptome’s activity. MicroRNAs (miR) are small non-coding RNAs that fine-tune gene expression at the post-transcriptional level, by binding to the 3′ untranslated regions of target mRNAs and inhibiting their expression. Cytoplasmic circRNAs can contain miR binding sites in their sequences and therefore sequester these small RNAs, preventing the interaction with specific mRNA targets (32, 33). As a result, circRNAs represent one of the most remarkable molecules in RNA biology (34), constituting an important part of the cellular transcriptome (31, 32). To date, there are three main classes of circRNAs located in different cellular compartments. Most of them have a cytoplasmic localization (29), in particular with the Exonic circRNAs (EcircRNAs), which have a sponge role of inhibiting the action of target microRNAs (31). There are also the Exon-intronic circRNAs (EIcircRNAs) and Intronic circRNAs (IcircRNAs), which are found in the cell nuclei and have a role in the regulation of gene expression (33).
Importantly, these circRNAs are related to proliferation, invasion, migration, angiogenesis, apoptosis, and metastasis of cells in liver diseases and act as oncogenic agents or suppressors and are linked to clinical manifestations. In this context, circRNAs gained importance in the discovery of new potential therapeutic targets, particularly in liver-associated metabolic diseases. Thus, we will review their role in the etiology of NAFLD and NASH.
As of today, most of the studies about circRNAs and the liver have been focusing on hepatocellular carcinoma (35) and few studies concern their contribution to NASH development, in this review, we will discuss the role of different circRNAs and their implication in NASH, as well as their potential connection with biological mechanisms linked to the onset of NASH.
Non-alcoholic fatty liver disease (NAFLD) is one of the most common liver diseases in developed countries, with the fastest growing in the USA, France and the UK. A strong increase is also observed in developing countries and worldwide in general (36, 37). In 20–30% of cases, NAFLD slowly progresses to non-alcoholic steatohepatitis (NASH), fibrosis, cirrhosis and hepatocellular carcinoma (HCC). NAFLD’s main hallmark is the accumulation of triglycerides in the liver (38).
Recent studies have reported that circRNAs are important regulators of hepatic steatosis, the main characteristic of NAFLD (39). However, the roles and mechanisms of circRNAs in NAFLD are still poorly understood (40). In different studies, RNAseq techniques as well as bioinformatics analysis has allowed to map for the first time 93 dysregulated circRNAs in NAFLD mice. As a result, 57 overexpressed and 36 underexpressed circRNAs were identified as potential cellular biomarkers in the pathogenesis of NAFLD (37). Furthermore, abnormal lipid metabolism in the liver is often accompanied by disordered expression of circRNAs (41). For example, JAK2/STAT5 pathway plays an important role in NAFLD by regulating the growth hormone pathway and their aberrant expression can lead to abnormal lipid metabolism (42). It has been identified that circSCD1 stimulates protein expression of JAK2 and STAT5 and ultimately affects the pathogenesis of NAFLD by promoting hepatic steatosis via the JAK2/STAT5 pathway (43).
Some studies also highlighted the effects of circRNA_0000660 on cellular lipid accumulation and its target gene in AML-12 hepatocytes. CircRNA_0000660 is a specific antagonist of miR_693 that directly targets Igfbp1 (44). Indeed, knockdown of circRNA_0000660 has been shown to increase lipid accumulation through the downregulation of Igfbp1 level (45).
Recent studies highlighted the crosstalk between circRNA hsa_circ_000313 with miR-6512-3p-PEG10 by identifying its role in the physiopathogenesis of NAFLD, but further studies are needed to deepen its beneficial role and to determine if this circRNA could become a potential biomarker for the disease (46).
All these results have shown that circRNAs have a huge involvement in the development of lipid disorder through different signaling pathway and can be potential blood biomarkers to detect an early stage of NAFLD.
Autophagy, also referred to as “self-eating”, is a process found across many species in the eukaryotes. It involves the delivery and degradation of cytoplasmic materials by lysosomes and plays a prominent role in cell survival, differentiation, development, and homeostasis by controlling cytoplasmic physiology through maintaining energy balance and the removal of misfolded proteins, damaged organelles, and lipid droplets (47, 48).
Alterations in the cell autophagic machinery have been implicated in several disease conditions, including non-alcoholic fatty liver disease and non-alcoholic steatohepatitis (49). Accumulating evidence suggest that maintaining intact autophagy pathways may represent a valuable target in NAFLD, because of its anti-steatogenic properties in hepatocytes (lipophagy) and its beneficial effects in inhibiting the progression to NASH through the hepatoprotective effects of mitophagy in hepatocytes and its anti-inflammatory properties in macrophages (50).
For example, circRNA_002581 has been described to play a role in hepatic autophagy by sponging mir-122. Studies already showed the involvement of this mir-RNA, thereby a decrease of mir-122 was observed with an increase of hepatic triglyceride accumulation and a total stop of fat oxidation in NASH patients through the expression of cytoplasmic polyadenylation element-binding protein 1 (CPEB1) (51). As a result, endogen CircRNA_002581 have a negative role by sponging mir-122 and thus leads to an increase of CPEB1. This dysfunction act on the autophagy by a decrease through PTEN/AMPK/mTOR signaling pathway and increase NAFLD progression (52). Thus, knockout of CircRNA_002581 leads to partial autophagy restoration. Finally, these results have shown that the circRNA_002581–miR-122–CPEB1 axis has therapeutic potential in NASH via autophagy (52).
The role of circRNAs in the regulation of autophagy in NAFLD is not yet clearly described but nevertheless represents an important key regulator and therapeutic target (Figure 1).
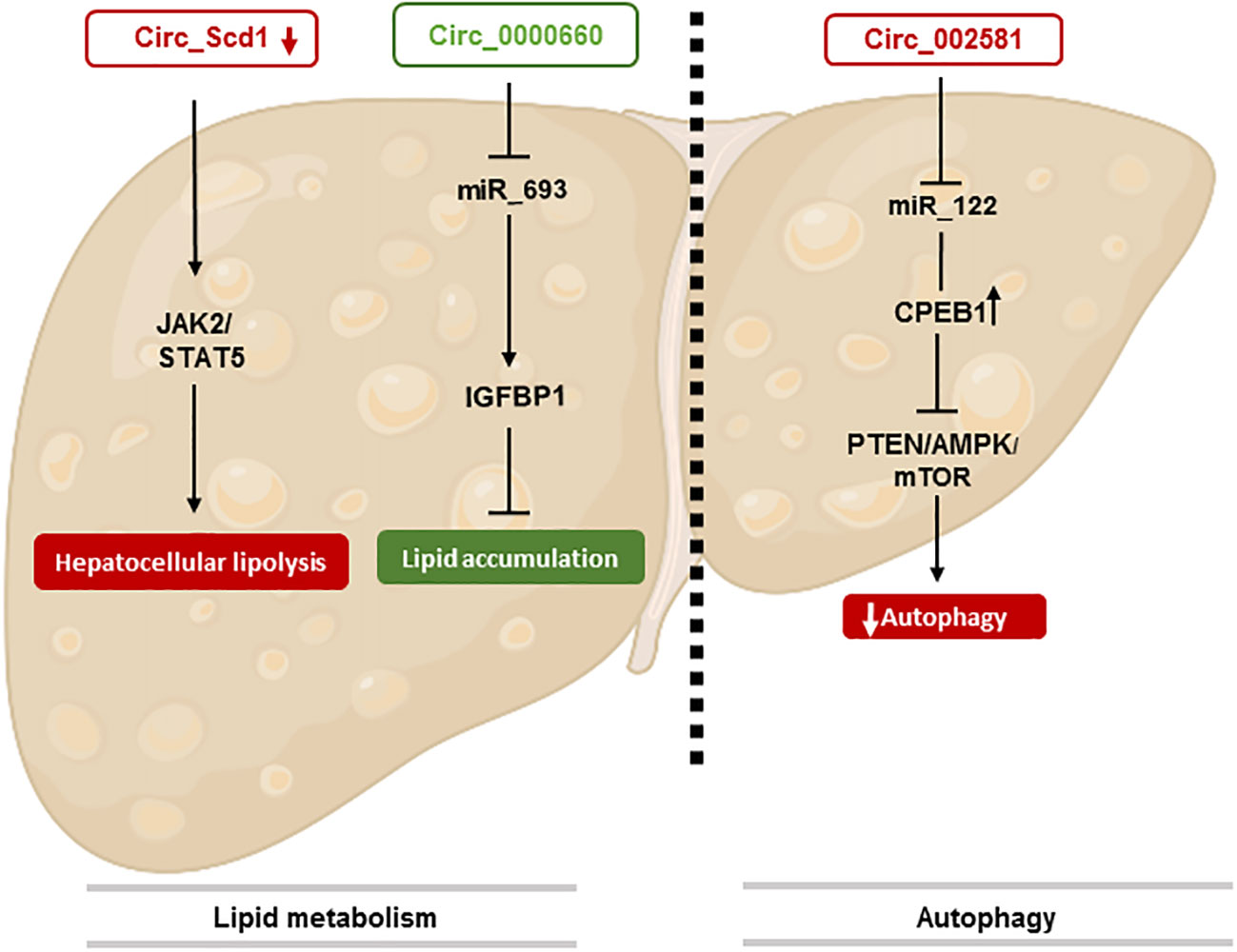
Figure 1 CircRNAs involved in NAFLD stage. This figure represents the effect of circRNAs in lipid metabolism through different pathways. The identification of circRNA involved in autophaghy is also shown.
Among all patients suffering from NAFLD, 25% to 44% of them will develop the more severe form of this disease: NASH (53). A theory aiming to explain the relationship between NAFLD and NASH has emerged during the last decade: the “two-hit” hypothesis. According to this theory, the “first hit”: NAFLD, as previously described is a hepatic metabolic disorder with increased de novo lipogenesis, unpaired β-oxidation and steatosis. Those first injuries will enhance the susceptibility of the liver to progress to the “second hit”: NASH, characterized by lobular fibrosis, inflammation and oxidative stress (54). Thus, in the next part, we will focus on the impact of circRNAs in the onset of the NASH hallmarks (Figure 2).
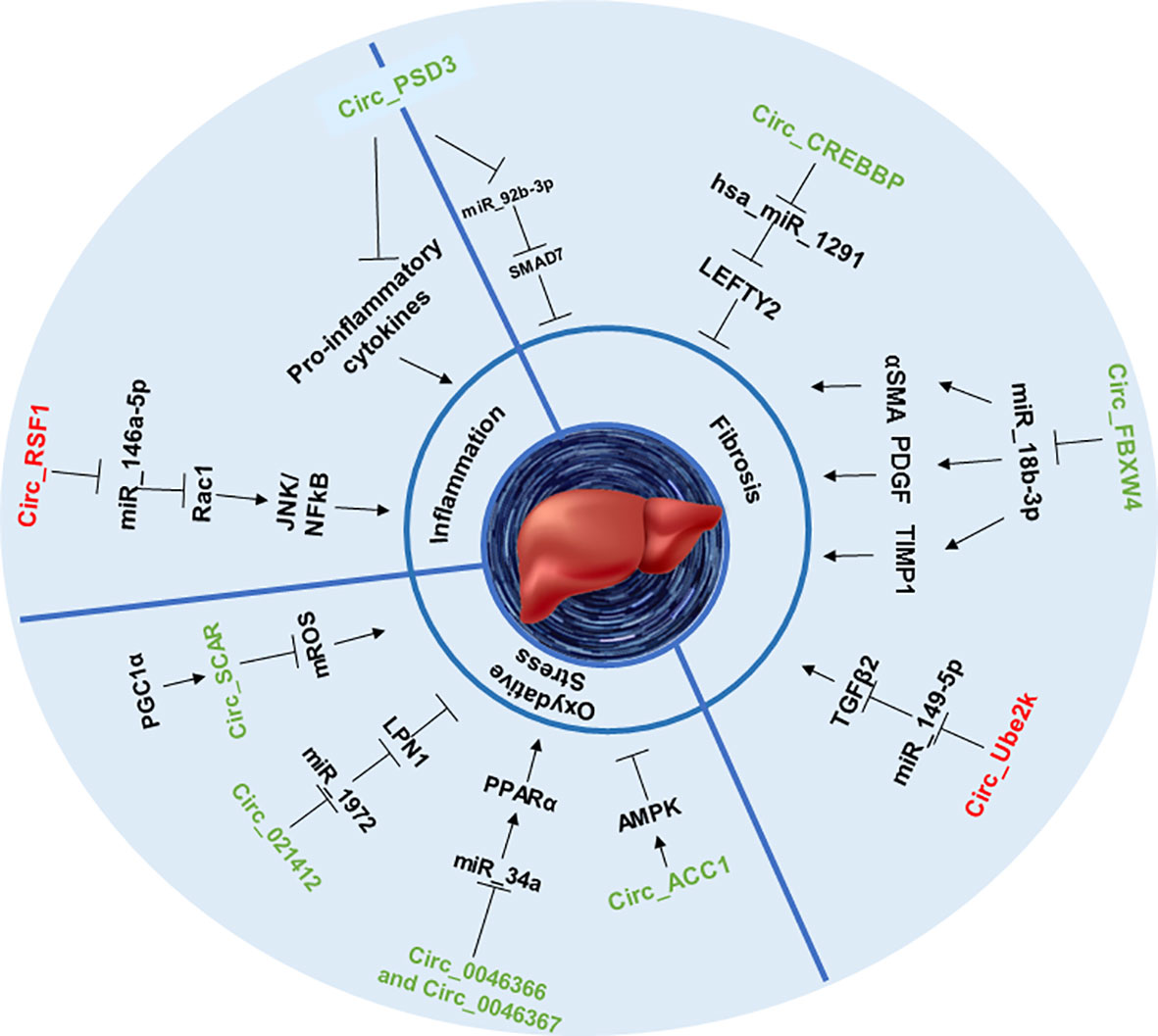
Figure 2 CircRNAs involved in NASH stage. This figure represents the effect of circRNAs in inflammation, fibrosis and oxidative stress through sponging specific miRNAs that can upregulate or downregulate specific proteins and signaling pathways.
Fibrosis remains the most important parameter defining the severity and outcome of patients with NASH (55), especially regarding the evolution of the disease toward cirrhosis and HCC (10). Liver fibrosis can be measured by staging, from F0, indicating no fibrosis to F4 being the most advanced form of fibrosis, namely cirrhosis (56). In this context, the activated HSC will secrete significant amounts of collagen and extracellular matrix in the hepatic microenvironment, inducing an abnormal level of nonfunctional scar tissue, replacing progressively healthy tissue, leading to cirrhosis and diminished liver function. Among all NASH hallmarks, liver fibrosis is undoubtedly the one for which circRNAs have been the most widely studied. Induction of NASH with repeated doses of carbon tetrachloride (CCl4) in mice is a widely used in vivo model of liver fibrosis. Using this approach, several authors highlighted the role of circRNAs in the onset of liver fibrosis.
Some circRNAS, for example, circPSD3 was shown to be downregulated in HSC and in the liver of fibrotic mice. By sponging miR-92b-3p, circPSD3 promoted the expression of Smad7, a protein known as an antagonist of the pro-fibrotic TGF-beta pathway (57, 58). Thus, circPSD3 reduced HSC proliferation and activation and alleviated the accumulation of collagen as well as the expression of pro-fibrotic genes in vitro and in vivo (59). Using similar methods, another circRNA, circCREBBP, was shown to prevent CCl4-induced fibrosis progression by sponging hsa-miR-1291, thereby activating the expression of LEFTY2, a protein from the TGF-β family with anti-fibrotic properties (60, 61). Again, circFBXW4 showed a similar anti-fibrotic capacity by targeting miR-18b-3p, an inducer of the expression of fibrogenic factors such as α-SMA (62), PDGF (63) and TIMP-1 (64) in HSCs (65).
However, the properties of circRNAs are not always beneficial, as recent studies highlighted their role in the promotion of fibrosis as well. For example, by sponging miR-149-5p, circUbe2k induced the upregulation of the TGF-β2 pathway, resulting in the exacerbation of CCl4-induced liver fibrosis (57, 66).
Taken together, it appears that circRNAs can exert bimodal effects on the onset of liver fibrosis, depending on their sponging capacities and miR targets.
Inflammation is one of the key features that characterize the progression of NAFLD to NASH. This process is often coming from change inside the hepatic microenvironment following pro-inflammatory cytokines’ circulation and macrophages’ infiltration, leading to lobular inflammation (67). Lobular inflammation is a common hallmark of NASH caused by an accumulation of immune cells inside the hepatic lobules, due to chronic inflammation, the anatomic units of the liver (68, 69). In this context, inflammatory cytokines can be secreted by various cell types, such as activated hepatic stellate cells (70) or M1 macrophages (71). CircRNAs are showing potential implications in these phenomena.
As discussed previously, circRNAs can impact HSC activation in several ways and could therefore be involved in the onset of inflammation in the context of NASH. For example, HSC activation can be obtained by challenging cells with irradiation, however, further studies are necessary to determine if irradiation challenge is physiologically relevant to mimicking the onset of NASH. In this context, it was shown that irradiation of the hepatic stellate cell line LX2 leads to the upregulation of circRSF1 and its sponging of miR‐146a‐5p. CircRSF1, by inhibiting miR‐146a‐5p-downregulation of Rac1, a protein involves in liver injury (72), increased pro-inflammatory pathways such as JNK (73) or NF-κB (74) and the secretion of pro-inflammatory cytokines such as TNFα, IL-1β or IL-6 in LX2 cell line (75). In another previously cited study (59), the authors found that the expression of circPSD3 was highly downregulated in the liver of mice treated with CCl4 and also within tissues of humans suffering from hepatic fibrosis. Infection of the animals with an adeno-associated vector carrying circPSD3 (AAV8-circPSD3) alleviated the production of pro-inflammatory cytokines by activated HSC and the infiltration of inflammatory cell infiltration (59), suggesting the positive role of this circular RNA in the prevention of liver inflammation.
In another recent study, circRNA_1639 was found to act as a sponger of the miR-122 within the liver and hepatic macrophages, triggering the inflammatory response (76). However, these findings were concerning mice liver in the context of alcoholic liver injury and could therefore be less relevant to NASH injury, thus, further studies are needed to confirm the same role of circRNA_1639 in the specific mechanism of NASH inflammation.
Since few studies have been focusing on this topic, it would be particularly interesting to study the role of circRNAs in the activation or migration of liver macrophages in the context of NASH, independently of HSC activation.
The pathophysiology of steatohepatitis and its progression are also influenced by multiple environmental and genetic factors, in which oxidative stress most likely plays a primary role as a starting point for hepatic and extra-hepatic damage (77). Moreover, numerous studies have indicated that this pathology is characterized by the presence of mitochondrial dysfunction (78). Indeed, circACC1 promotes fatty acid β-oxidation by facilitating the assembly, stability, and activity of the AMP-activated protein kinase (AMPK) holoenzyme in LO2 hepatocytes. Loss of AMPK-binding activity of circACC1 resulted in impaired mitochondrial fatty acid β-oxidation and increased steatosis in LO2 cells (78, 79).
CircRNA_021412 is increasing fatty acid β-oxidation and decreases the synthesis of triglyceride in HepG2 hepatocytes through the miR‐1972/lipin1 (LPIN1) pathway (44). As a result, downregulation of CircRNA_021412 plays an important role in the induction of steatosis-related genes via PPAR alpha activation. Indeed, after high fat stimulation there is a decrease in circRNA_021412 inhibition of mir-1972, so a reactivation of mir-1972 leading to the down-regulation of LIPIN 1 level, which will induce the activation of steatosis-related genes (80, 81).
CircRNA_0046366 and circRNA_0046367 promoted fatty acid β-oxidation in HepG2 cells (44, 82). They both prevent the binding of miR-34a to PPARα and alleviate hepatic steatosis by restoring the lipid metabolism pathways and genes (83, 84).
Recently, Zhao et al. have demonstrated that, in healthy conditions, PGC1-α activates the binding of circ_SCAR on ATP5B, which inhibits mPTP’s interaction with CypD in the mitochondria and so the activation of mROS (85). In a patient with NASH, lipid-induced ER stress reduces PGC-1α-mediated circRNA_SCAR expression and thus increases the production of mROS-induced pro-inflammatory activation of liver fibroblasts. Thus, circ_SCAR restoration in NASH may be a potential therapeutic target (85).
As previously stated, patients suffering from NASH are at higher risks of developing hepatic complications, such as liver failure, infectious diseases or hepatocellular carcinoma (55). Hepatocellular carcinoma is the most common form of liver cancer, representing up to 90% of hepatic tumors (86). HCC is often described as the final stage of NASH and occurs predominantly among patients suffering from the late stage of fibrosis: cirrhosis (10, 87). The pathophysiology of NASH-related HCC is not yet fully understood, but several approaches do exist. It is widely accepted that the perturbation of the hepatic microenvironment occurring in patients with severe NASH largely contributes to DNA mutations and abnormal metabolic homeostasis, as well as uncontrolled liver regeneration and scarring, and thus to the onset of HCC (10) (Figure 3).
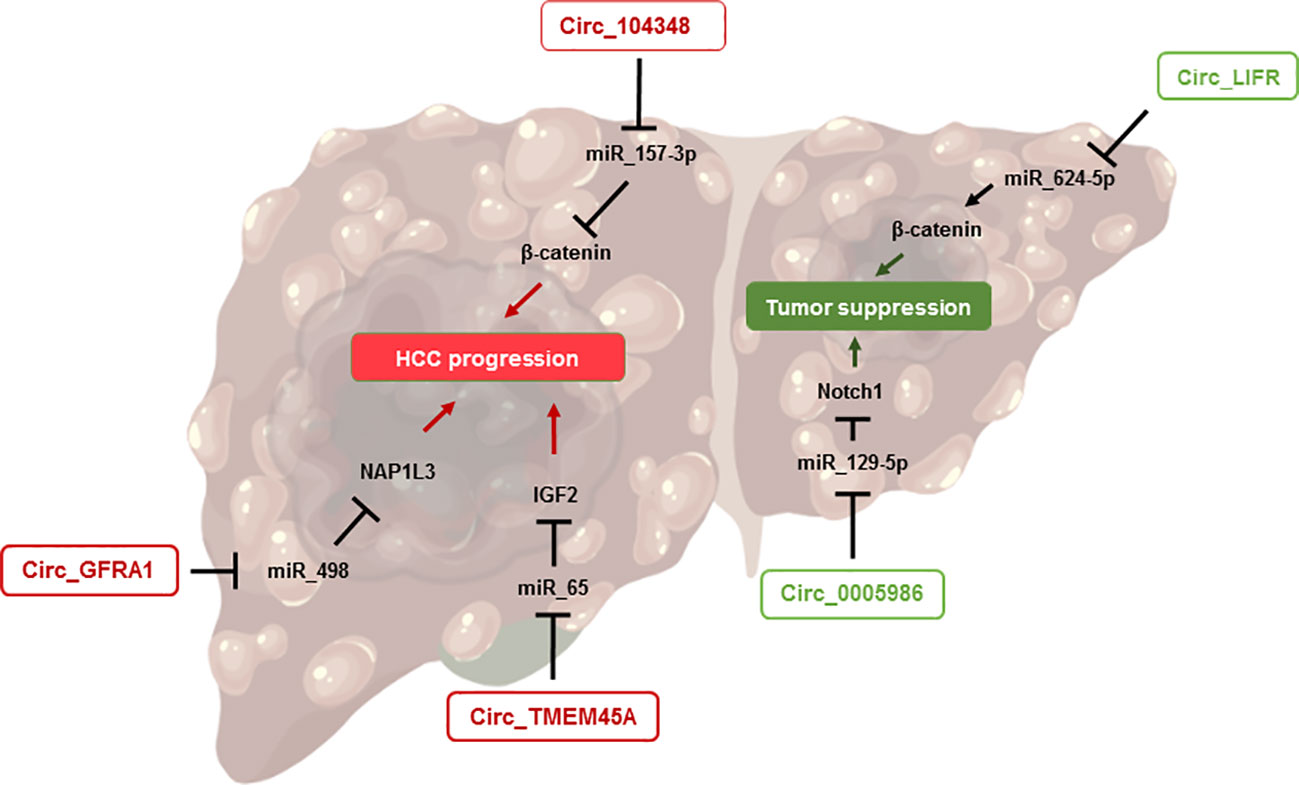
Figure 3 CircRNAs involved in HCC onset. This figure shows the implication of circRNAs in the pathophysiology of HCC. The right side represents those promoting tumor suppression while the left side highlights the circRNAs exacerbating HCC progression.
During the last decade, various biological functions of circRNAs in HCC have been studied. For example, circLIFR has been highlighted as a tumor suppressor in HCC, by sponging miR-624-5p and downregulating the β-catenin signaling pathway (88), which acts as an oncogenic signal through its ability to induce the transcription of several genes related to proliferation in hepatocytes (89). Another circRNA, circ_0005986, has been suggested as a promising candidate in the study of HCC, both as an inhibitor of carcinogenesis and as a prognostic biomarker. For the first aspect, circ_0005986 acts by inhibiting liver cell proliferation through the sponging of miR-129-5p and the regulation of Notch1, whose signaling regulates growth, apoptosis, and cell differentiation (90). Furthermore, low levels of circ_0005986 in HCC tumors were correlated with larger tumor size, while higher levels were associated with improved survival (90, 91), suggesting a promising role of this circRNA as an HCC prognostic biomarker.
Some circRNAs have been identified lately as promoters of hepatocellular carcinoma as well. For instance, circTMEM45A was upregulated in patients with HCC, and its expression was correlated with the severity of the disease. By using in vitro and in vivo approaches, the authors showed that circTMEM45A could promote the progression of HCC by sponging miR-65, resulting in the activation of the IGF2 pathway and ultimately to enhanced cell proliferation and cancer development (92). Similar results were obtained for CircRNA circ_GFRA1, which modulates the miR-498/NAP1L3 axis, subsequently promoting HCC development (93). Another circRNA, hsa_circRNA_104348, showed opposite properties in relation to circLIFR discussed earlier. Indeed, hsa_circRNA_104348 can promote HCC progression, through the upregulation of the Wnt/β-catenin pathway, by sponging miR-187-3p (94).
In this review, we discussed the involvement of circRNAs in the progression of NAFLD. Currently, this emerging class of circRNAs represents a key regulator of various biological and pathophysiological processes. Nonalcoholic fatty liver disease (NAFLD) is the most prevalent chronic liver disease and is strongly linked to the global epidemic of obesity and type 2 diabetes mellitus (T2DM), occurring in 70% of type-2 diabetes patients and in 80% of obese patients (95). Insulin resistance, leading to the development of prediabetes and its progression to T2DM, also results in a spontaneous onset of NAFLD (96). To date, there is no specific treatment for NAFLD. Current treatments paradigms, such as lifestyle improvements and weight loss, can help slow down disease progression but are not effective due to low patients’ compliance. Oftentime, anti-diabetics are used to treat NASH, but are not curative, uneffective for most patients and do not specifically target liver pathology (97). As the prevalence is rising, it becomes urgent to develop new therapies for NASH. In this review, we highlighted the important role of circRNAs affecting the different hallmarks of NAFLD, NASH and HCC: steatosis, autophagy, fibrosis, inflammation and oxidative stress (Table 1). The sponging effects of circRNAs on some miRs implicated in NASH onset and progression have shown their potential as therapeutic targets. Also, some existing treatments, such as metformin (anti-diabetic), have already shown their efficacy in the treatment of T2DM through the modification of non-coding RNAs expression (98). Thus, as we have discussed, circRNAs have an important role, although not yet fully understood, in the pathophysiology of NASH, highlighting the importance of developing specific treatments for NASH targeting circRNAs’ expression, in order to improve outcomes and halt disease progression. The emergence of technological advance, like high throughput sequencing, specific to circRNAs, will allow in the coming years a better understanding of their involvement in new biological processes.
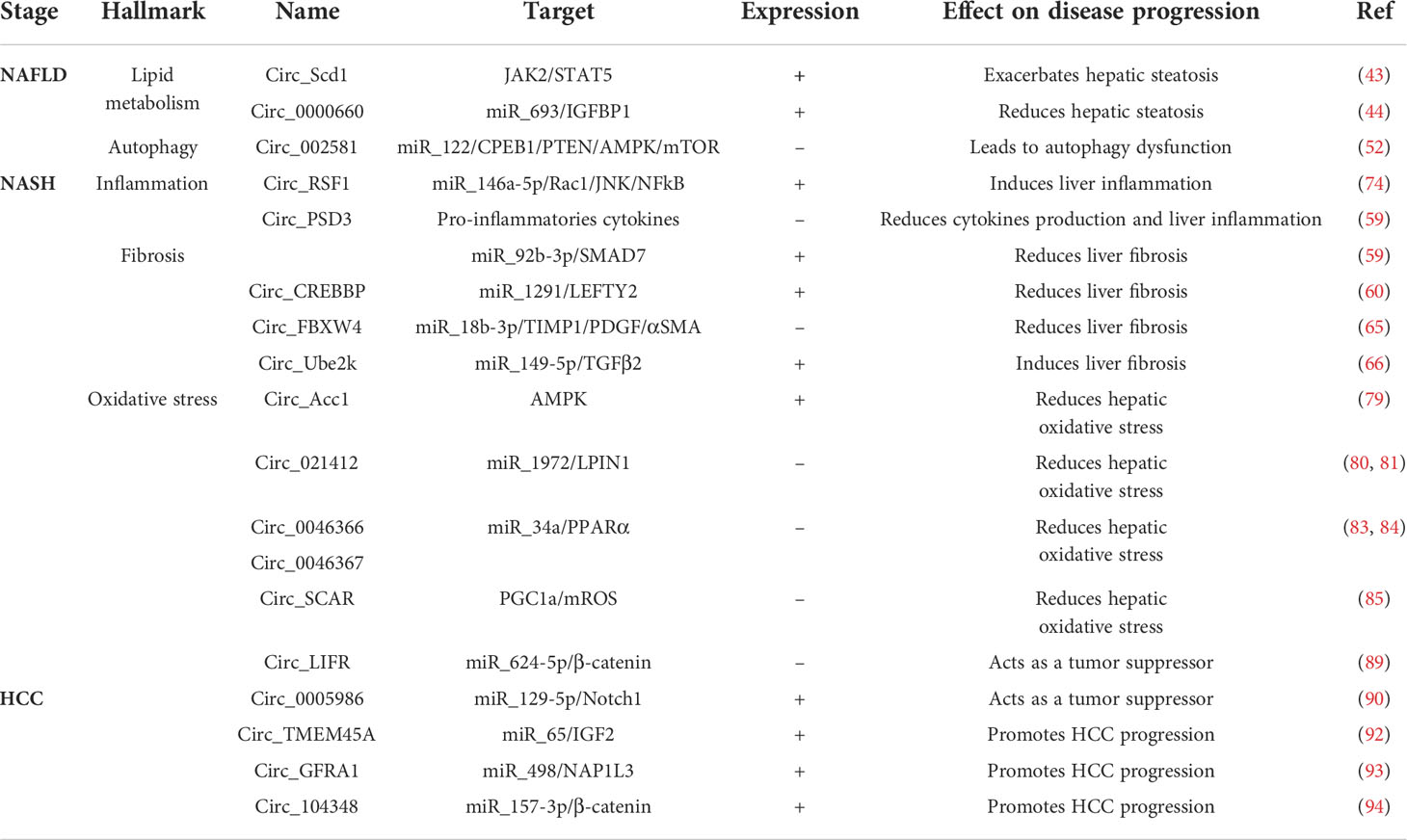
Table 1 Implication of cirRNAs in liver diseases: NAFLD, NASH and hepatocellular carcinomas, and their effects.
MY, J-BP and AD wrote the review. MP, KB, AG and AD edit the text. AD designed and edited all the review. All authors contributed to the article and approved the submitted version.
Thank you to Fanny Krumm for her help for the visual abstract.
J-BP, AG, and KB were employed by ILONOV. J-BP’s thesis is co-funded by an individual financial aid for training through research received by Ilonov from the State, through the Ministry in charge of Research.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
CCl4, Carbon tetrachloride; circRNA, circular RNA; HSC, Hepatic stellate cells; lncRNA, long non coding RNA; miR, micro RNA; mRNA, messenger RNA; mROS, Mitochondrial ROS; rRNA, ribosomal RNA; tRNA, transfer RNA.
1. Younossi ZM, Golabi P, de Avila L, Paik JM, Srishord M, Fukui N, et al. The global epidemiology of NAFLD and NASH in patients with type 2 diabetes: A systematic review and meta-analysis. J Hepatol (2019) 71(4):793–801. doi: 10.1016/j.jhep.2019.06.021
2. Parthasarathy G, Revelo X, Malhi H. Pathogenesis of nonalcoholic steatohepatitis: An overview. Hepatol Commun (2020) 4(4):478–492. doi: 10.1002/hep4.1479
3. Huh Y, Cho YJ, Nam GE. Recent epidemiology and risk factors of nonalcoholic fatty liver disease. J Obes Metab Syndr (2022) 31(1):17–27. doi: 10.7570/jomes22021
4. Krawczyk M, Rau M, Schattenberg JM, Bantel H, Pathil A, Demir M, et al. Combined effects of the PNPLA3 rs738409, TM6SF2 rs58542926, and MBOAT7 rs641738 variants on NAFLD severity: a multicenter biopsy-based study. J Lipid Res (2017) 58(1):247–255. doi: 10.1194/jlr.P067454
5. Abd El-Kader SM, El-Den Ashmawy EMS. Non-alcoholic fatty liver disease: The diagnosis and management. World J Hepatol (2015) 7(6):846–858. doi: 10.4254/wjh.v7.i6.846
6. Ong JP, Younossi ZM. Epidemiology and natural history of NAFLD and NASH. Clin Liver Dis (2007) 11(1):1–16. doi: 10.1016/j.cld.2007.02.009
7. Caligiuri A, Gentilini A, Marra F. Molecular pathogenesis of NASH. Int J Mol Sci (2016) 17(9):E1575. doi: 10.3390/ijms17091575
8. Axley P, Ahmed Z, Arora S, Haas A, Kuo Y-F, Kmath PS, et al. NASH is the most rapidly growing etiology for acute-on-Chronic liver failure-related hospitalization and disease burden in the united states: A population-based study. Liver Transplant Off Publ Am Assoc Study Liver Dis Int Liver Transplant Soc (2019) 25(5):695–705. doi: 10.1002/lt.25443
9. Noureddin M, Sanyal AJ. Pathogenesis of NASH: the impact of multiple pathways. Curr Hepatol Rep (2018) 17(4):350–360. doi: 10.1007/s11901-018-0425-7
10. Anstee QM, Reeves HL, Kotsiliti E, Govaere O, Heikenwalder M. From NASH to HCC: current concepts and future challenges. Nat Rev Gastroenterol Hepatol (2019) 16(7):411–8. doi: 10.1038/s41575-019-0145-7
11. Rui L. Energy metabolism in the liver. Compr Physiol (2014) 4(1):177–197. doi: 10.1002/cphy.c130024
12. Lu Q, et al. Metabolic changes of hepatocytes in NAFLD. Front Physiol (2021) 12:710420. doi: 10.3389/fphys.2021.710420
13. Thibaut R, Gage MC, Pineda-Torra I, Chabrier G, Venteclef N, Alzaid F, et al. Liver macrophages and inflammation in physiology and physiopathology of non-alcoholic fatty liver disease. FEBS J (2021) 289(11):3024–57. doi: 10.1111/febs.15877
14. Barreby E, Chen P. Et m. aouadi, macrophage functional diversity in NAFLD - more than inflammation. Nat Rev Endocrinol (2022) 18(8):461–472. doi: 10.1038/s41574-022-00675-6
15. Senoo H, Kojima N, Sato M. Vitamin a-storing cells (stellate cells). Vitam Horm (2007) 75:131–159. doi: 10.1016/S0083-6729(06)75006-3
16. Schwabe RF, Tabas I, Pajvani UB. Mechanisms of fibrosis development in nonalcoholic steatohepatitis. Gastroenterology (2020) 158(7):1913–1928. doi: 10.1053/j.gastro.2019.11.311
17. Ratziu V, Friedman SL. Why do so many NASH trials fail? Gastroenterology (2020) S0016-5085(20):30680–6. doi: 10.1053/j.gastro.2020.05.046
18. Masoodi M, Gastaldelli A, Hyötyläinen T, Arretxe E, Alonso C, Gaggini M, et al. Metabolomics and lipidomics in NAFLD: biomarkers and non-invasive diagnostic tests. Nat Rev Gastroenterol Hepatol (2021) 18(12):835–856. doi: 10.1038/s41575-021-00502-9
19. Boeckmans J, Gatzios A, Heymans A, Rombaut M, Rogiers V, De Kock J, et al. Transcriptomics reveals discordant lipid metabolism effects between In vitro models exposed to elafibranor and liver samples of NAFLD patients after bariatric surgery. Cells (2022) 11(5):893. doi: 10.3390/cells11050893
20. Guo M, Xiang L, Yao J, Zhang J, Zhu S, Wang D, et al. Comprehensive transcriptome profiling of NAFLD- and NASH-induced skeletal muscle dysfunction. Front Endocrinol (2022) 13:851520. doi: 10.3389/fendo.2022.851520
21. Shabgah AG, Norouzi F, Hedayati-Moghadam M, Soleimani D, Pahlavani N, Navashenaq JG, et al. A comprehensive review of long non-coding RNAs in the pathogenesis and development of non-alcoholic fatty liver disease. Nutr Metab (2021) 18(1):22. doi: 10.1186/s12986-021-00552-5
22. Sulaiman SA, Muhsin NIA, Jamal R. Regulatory non-coding RNAs network in non-alcoholic fatty liver disease. Front Physiol (2019) 10:279. doi: 10.3389/fphys.2019.00279
23. Zhang P, Wu W, Chen Q, Chen M. Non-coding RNAs and their integrated networks. J Integr Bioinform (2019) 16(3):20190027. doi: 10.1515/jib-2019-0027
24. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci (1976) 73(11):3852–3856. doi: 10.1073/pnas.73.11.3852
25. Hsu M-T, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature (1979) 280(5720):339–340. doi: 10.1038/280339a0
26. Chen L-L, Yang L. Regulation of circRNA biogenesis. RNA Biol (2015) 12(4):381–388. doi: 10.1080/15476286.2015.1020271
27. Zhang X-O, Dong R, Zhang Y, Zhang J-L, Luo Z, Zhang J, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res (2016) 26(9):1277–1287. doi: 10.1101/gr.202895.115
28. Geng X, Jia Y, Zhang Y, Shi L, Li Q, Zang A, et al. Circular RNA: biogenesis, degradation, functions and potential roles in mediating resistance to anticarcinogens. Epigenomics (2020) 12(3):267–283. doi: 10.2217/epi-2019-0295
29. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One (2012) 7(2):e30733. doi: 10.1371/journal.pone.0030733
30. Wang PL, Bao Y, Yee M-C, Barrett SP, Hogan GJ, Olsen MN, et al. Circular RNA is expressed across the eukaryotic tree of life. PloS One (2014) 9(3):e90859. doi: 10.1371/journal.pone.0090859
31. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature (2013) 495(7441):384–388. doi: 10.1038/nature11993
32. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature (2013) 495(7441):333–338. doi: 10.1038/nature11928
33. Rong Z, Xu J, Shi S, Tan Z, Meng Q, Hua J, et al. Circular RNA in pancreatic cancer: a novel avenue for the roles of diagnosis and treatment. Theranostics (2021) 11(6):2755–2769. doi: 10.7150/thno.56174
34. Yu C-Y, Kuo H-C. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci (2019) 26(1):29. doi: 10.1186/s12929-019-0523-z
35. Zhang Y, Wang Y. Circular RNAs in hepatocellular carcinoma: Emerging functions to clinical significances. Front Oncol (2021) 11:667428. doi: 10.3389/fonc.2021.667428
36. Huang DQ, El-Serag HB. Et r. loomba, global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol (2021) 18(4):223–238. doi: 10.1038/s41575-020-00381-6
37. Yuan X, Diao J, Du A, Wen S, Zhou L, Pan Y, et al. Circular RNA expression profiles and features in NAFLD mice: a study using RNA-seq data. J Transl Med (2020) 18(1):476. doi: 10.1186/s12967-020-02637-w
38. Fernando DH, Forbes JM, Angus PW, Herath CB. Development and progression of non-alcoholic fatty liver disease: The role of advanced glycation end products. Int J Mol Sci (2019) 20(20):5037. doi: 10.3390/ijms20205037
39. Wree A, Broderick L, Canbay A, Hoffman HM, Feldstein AE. From NAFLD to NASH to cirrhosis–new insights into disease mechanisms. Nat Rev Gastroenterol Hepatol (2013) 10(11):627–636. doi: 10.1038/nrgastro.2013.149
40. Ou Q, Zhao Y, Zhou J, Wu X. Comprehensive circular RNA expression profiles in a mouse model of nonalcoholic steatohepatitis. Mol Med Rep (2019) 19(4):2636–48. doi: 10.3892/mmr.2019.9935
41. Singh AK, Aryal B, Zhang X, Fan Y, Price NL, Suarez Y, et al. Posttranscriptional regulation of lipid metabolism by non-coding RNAs and RNA binding proteins. Semin Cell Dev Biol (2018) 81:129–140. doi: 10.1016/j.semcdb.2017.11.026
42. Nordstrom SM, Tran JL, Sos BC, Wagner K-U, Weiss EJ. Disruption of JAK2 in adipocytes impairs lipolysis and improves fatty liver in mice with elevated GH. Mol Endocrinol (2013) 27(8):1333–1342. doi: 10.1210/me.2013-1110
43. Li P, Shan K, Liu Y, Zhang Y, Xu L, Xu L, et al. CircScd1 promotes fatty liver disease via the janus kinase 2/Signal transducer and activator of transcription 5 pathway. Dig Dis Sci (2019) 64(1):113–122. doi: 10.1007/s10620-018-5290-2
44. Wu Y-L, Li H-F, Chen H-H, Lin H. Emergent roles of circular RNAs in metabolism and metabolic disorders. Int J Mol Sci (2022) 23(3):1032. doi: 10.3390/ijms23031032
45. Chen Q, Liu M, Luo Y, Yu H, Zhang J, Li D, et al. Maternal obesity alters circRNA expression and the potential role of mmu_circRNA_0000660 via sponging miR_693 in offspring liver at weaning age. Gene (2020) 731:144354. doi: 10.1016/j.gene.2020.144354
46. Du C, Shen L, Ma Z, Du J, Jin S. Bioinformatic analysis of crosstalk between circRNA, miRNA, and target gene network in NAFLD. Front Genet (2021) 12:671523. doi: 10.3389/fgene.2021.671523
47. Wu H, Chen S, Ammar A-B, Xu J, Wu Q, Pan K, et al. Crosstalk between macroautophagy and chaperone-mediated autophagy: Implications for the treatment of neurological diseases. Mol Neurobiol (2015) 52(3):1284–1296. doi: 10.1007/s12035-014-8933-0
48. Scrivo A, Bourdenx M, Pampliega O, Cuervo AM. Selective autophagy as a potential therapeutic target for neurodegenerative disorders. Lancet Neurol (2018) 17(9):802–815. doi: 10.1016/S1474-4422(18)30238-2
49. Udoh U-AS, Rajan PK, Nakafuku Y, Finley R, Sanabria JR. Cell autophagy in NASH and NASH-related hepatocellular carcinoma. Int J Mol Sci (2022) 23(14):7734. doi: 10.3390/ijms23147734
50. Allaire M, Rautou P-E, Codogno P, Lotersztajn S. Autophagy in liver diseases: Time for translation? J Hepatol (2019) 70(5):985–998. doi: 10.1016/j.jhep.2019.01.026
51. Cheung O, Puri P, Eicken C, Contos MJ, Mirshashi F, Maher JW, et al. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology (2008) 48(6):1810–1820. doi: 10.1002/hep.22569
52. Jin X, Gao J, Zheng R, Yu M, Ren Y, Yan T, et al. Antagonizing circRNA_002581–miR-122–CPEB1 axis alleviates NASH through restoring PTEN–AMPK–mTOR pathway regulated autophagy. Cell Death Dis (2020) 11(2):123. doi: 10.1038/s41419-020-2293-7
53. Calzadilla Bertot L, Adams LA. The natural course of non-alcoholic fatty liver disease. Int J Mol Sci (2016) 17(5):E774. doi: 10.3390/ijms17050774
54. Buzzetti E, Pinzani M, Tsochatzis EA. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism (2016) 65(8):1038–1048. doi: 10.1016/j.metabol.2015.12.012
55. Ekstedt M, Hagström H, Nasr P, Fredrikson M, Stal P, Kechagias S, et al. Fibrosis stage is the strongest predictor for disease-specific mortality in NAFLD after up to 33 years of follow-up. Hepatol Baltim Md (2015) 61(5):1547–1554. doi: 10.1002/hep.27368
56. Bataller R, Brenner DA. Liver fibrosis. J Clin Invest (2005) 115(2):209–218. doi: 10.1172/JCI24282
57. Biernacka A, Dobaczewski M, Frangogiannis NG. TGF-β signaling in fibrosis. Growth Factors Chur Switz (2011) 29(5):196–202. doi: 10.3109/08977194.2011.595714
58. Dooley S, Hamzavi J, Breitkopf K, Wiercinska E, Said HM, Lorenzen J, et al. Smad7 prevents activation of hepatic stellate cells and liver fibrosis in rats. Gastroenterology (2003) 125(1):178–191. doi: 10.1016/s0016-5085(03)00666-8
59. Bu F-T, Zhu Y, Chen X, Wang A, Zhang Y-F, You H-M, et al. Circular RNA circPSD3 alleviates hepatic fibrogenesis by regulating the miR-92b-3p/Smad7 axis. Mol Ther Nucleic Acids (2021) 23:847–862. doi: 10.1016/j.omtn.2021.01.007
60. Yang Y-R, Hu S, Bu F-T, Li H, Huang C, Meng X-M, et al. Circular RNA CREBBP suppresses hepatic fibrosis Via targeting the hsa-miR-1291/LEFTY2 axis. Front Pharmacol (2021) 12:741151. doi: 10.3389/fphar.2021.741151
61. Yang Y-R, Bu F-T, Yang Y, Li H, Huang C, Meng X-M, et al. LEFTY2 alleviates hepatic stellate cell activation and liver fibrosis by regulating the TGF-β1/Smad3 pathway. Mol Immunol (2020) 126:31–39. doi: 10.1016/j.molimm.2020.07.012
62. Carpino G, Morini S, Ginanni Corradini S, Franchitto A, Merli M, Siciliano M, et al. Alpha-SMA expression in hepatic stellate cells and quantitative analysis of hepatic fibrosis in cirrhosis and in recurrent chronic hepatitis after liver transplantation. Dig Liver Dis Off J Ital Soc Gastroenterol Ital Assoc Study Liver (2005) 37(5):349–356. doi: 10.1016/j.dld.2004.11.009
63. Ying H-Z, Chen Q, Zhang W-Y, Zhang H-H, Ma Y, Zhang S-Z, et al. PDGF signaling pathway in hepatic fibrosis pathogenesis and therapeutics. Mol Med Rep (2017) 16(6):7879–7889. doi: 10.3892/mmr.2017.7641
64. Yoshiji H, Kuriyama S, Miyamoto Y, Thorgeirsson UP, Gomez DE, Kawata M, et al. Tissue inhibitor of metalloproteinases-1 promotes liver fibrosis development in a transgenic mouse model. Hepatol Baltim Md (2000) 32(6):1248–1254. doi: 10.1053/jhep.2000.20521
65. Chen X, Li H-D, Bu F-T, Li X-F, Chen Y, Zhu S, et al. Circular RNA circFBXW4 suppresses hepatic fibrosis via targeting the miR-18b-3p/FBXW7 axis. Theranostics (2020) 10(11):4851–4870. doi: 10.7150/thno.42423
66. Zhu S, Chen X, Wang J-N, Xu J-J, Wang A, Li J-J, et al. Circular RNA circUbe2k promotes hepatic fibrosis via sponging miR-149-5p/TGF-β2 axis. FASEB J Off Publ Fed Am Soc Exp Biol (2021) 35(6):e21622. doi: 10.1096/fj.202002738R
67. Schuster S, Cabrera D, Arrese M, Feldstein AE. Triggering and resolution of inflammation in NASH. Nat Rev Gastroenterol Hepatol (2018) 15(6):349–364. doi: 10.1038/s41575-018-0009-6
68. Takahashi Y, Fukusato T. Histopathology of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol (2014) 20(42):15539–15548. doi: 10.3748/wjg.v20.i42.15539
69. Krishna M. Microscopic anatomy of the liver. Clin Liver Dis (2013) 2(Suppl 1):S4–S7. doi: 10.1002/cld.147
70. Weiskirchen R, Tacke F. Cellular and molecular functions of hepatic stellate cells in inflammatory responses and liver immunology. Hepatobil Surg Nutr (2014) 3(6):344–363. doi: 10.3978/j.issn.2304-3881.2014.11.03
71. Oates JR, McKell MC, Moreno-Fernandez ME, Damen MSMA, Deepe GS Jr, Qualls JE, et al. Macrophage function in the pathogenesis of non-alcoholic fatty liver disease: The mac attack. Front Immunol (2019) 10:2893. doi: 10.3389/fimmu.2019.02893
72. Choi SS, Sicklick JK, Ma Q, Yang L, Huang J, Qi Y, et al. Sustained activation of Rac1 in hepatic stellate cells promotes liver injury and fibrosis in mice. Hepatol Baltim Md (2006) 44(5):1267–1277. doi: 10.1002/hep.21375
73. Win S, Than TA, Zhang J, Oo C, Min RWM, Kaplowitz N, et al. New insights into the role and mechanism of c-Jun-N-terminal kinase signaling in the pathobiology of liver diseases. Hepatol Baltim Md (2018) 67(5):2013–2024. doi: 10.1002/hep.29689
74. Sunami Y, Leithäuser F, Gul S, Fiedler K, Güldiken N, Espenlaub S, et al. Hepatic activation of IKK/NFκB signaling induces liver fibrosis via macrophage-mediated chronic inflammation. Hepatol Baltim Md (2012) 56(3):1117–1128. doi: 10.1002/hep.25711
75. Chen Y, Yuan B, Chen G, Zhang L, Zhuang Y, Niu H, et al. Circular RNA RSF1 promotes inflammatory and fibrotic phenotypes of irradiated hepatic stellate cell by modulating miR-146a-5p. J Cell Physiol (2020) 235(11):8270–8282. doi: 10.1002/jcp.29483
76. Lu X, Liu Y, Xuan W, Ye J, Yao H, Huang C, et al. Circ_1639 induces cells inflammation responses by sponging miR-122 and regulating TNFRSF13C expression in alcoholic liver disease. Toxicol Lett (2019) 314:89–97. doi: 10.1016/j.toxlet.2019.07.021
77. Masarone M, Rosato V, Dallio M, Gerarda Gravina A, Aglitti A, Loguercio C, et al. Role of oxidative stress in pathophysiology of nonalcoholic fatty liver disease. Oxid Med Cell Longev (2018) 2018:1–14. doi: 10.1155/2018/9547613
78. Caldwell SH, Swerdlow RH, Khan EM, Ieozzoni JC, Hespenheide EE, Parks JK, et al. Mitochondrial abnormalities in non-alcoholic steatohepatitis. J Hepatol (1999) 31(3):430–434. doi: 10.1016/S0168-8278(99)80033-6
79. Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R, et al. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab (2019) 30(1):157–173.e7. doi: 10.1016/j.cmet.2019.05.009
80. Guo X-Y, He C-X, Wang Y-Q, Sun C, Li G-M, Su Q, et al. Circular RNA profiling and bioinformatic modeling identify its regulatory role in hepatic steatosis. BioMed Res Int (2017) 2017:1–13. doi: 10.1155/2017/5936171
81. Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, et al. Lipin 1 is an inducible amplifier of the hepatic PGC-1a/PPARa regulatory pathway. Cell Metab (2006) 4(3):199–210. doi: 10.1016/j.cmet.2006.08.005.
82. Guo X-Y, Sun F, Chen J-N, Wang Y-Q, Pan Q, Fan J-G, et al. circRNA_0046366 inhibits hepatocellular steatosis by normalization of PPAR signaling. World J Gastroenterol (2018) 24(3):323–337. doi: 10.3748/wjg.v24.i3.323
83. Guo X-Y, Chen J-N, Sun F, Wang Y-Q, Pan Q, Fan J-G, et al. circRNA_0046367 prevents hepatoxicity of lipid peroxidation: An inhibitory role against hepatic steatosis. Oxid Med Cell Longev (2017) 2017:1–16. doi: 10.1155/2017/3960197
84. Veselovskiǐ NS, Nalivaǐko ED. Isolation, identification and properties of the calcium component of the action potential in isolated neurons of the spinal ganglia of the rat. Neirofiziol Neurophysiol (1986) 18(1):126–129.
85. Zhao Q, Liu J, Deng H, Ma R, Liao J-Y, Liang H, et al. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell (2020) 183(1):76–93.e22. doi: 10.1016/j.cell.2020.08.009
86. Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular carcinoma. Nat Rev Dis Primer (2021) 7(1):6. doi: 10.1038/s41572-020-00240-3
87. Cholankeril G, Patel R, Khurana S, Satapathy SK. Hepatocellular carcinoma in non-alcoholic steatohepatitis: Current knowledge and implications for management. World J Hepatol (2017) 9(11):533–543. doi: 10.4254/wjh.v9.i11.533
88. Yang L, Tan W, Wei Y, Xie Z, Li W, Ma X, et al. CircLIFR suppresses hepatocellular carcinoma progression by sponging miR-624-5p and inactivating the GSK-3β/β-catenin signaling pathway. Cell Death Dis (2022) 13(5):464. doi: 10.1038/s41419-022-04887-6
89. Dahmani R, Just P-A, Perret C. The wnt/β-catenin pathway as a therapeutic target in human hepatocellular carcinoma. Clin Res Hepatol Gastroenterol (2011) 35(11):709–713. doi: 10.1016/j.clinre.2011.05.010
90. Fu L, Chen Q, Yao T, Li T, Ying S, Hu Y, et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget (2017) 8(27):43878–43888. doi: 10.18632/oncotarget.16709
91. Kim G, Han JR, Park SY, Tak WY, Kweon Y-O, Lee YR, et al. Circular noncoding RNA hsa_circ_0005986 as a prognostic biomarker for hepatocellular carcinoma. Sci Rep (2021) 11(1):14930. doi: 10.1038/s41598-021-94074-y
92. Zhang T, Jing B, Bai Y, Zhang Y, Yu H. Circular RNA circTMEM45A acts as the sponge of MicroRNA-665 to promote hepatocellular carcinoma progression. Mol Ther Nucleic Acids (2020) 22:285–297. doi: 10.1016/j.omtn.2020.08.011
93. Lv S, Li Y, Ning H, Zhang M, Jia Q, Wang X, et al. CircRNA GFRA1 promotes hepatocellular carcinoma progression by modulating the miR-498/NAP1L3 axis. Sci Rep (2021) 11(1):386. doi: 10.1038/s41598-020-79321-y
94. Huang G, Liang M, Liu H, Huang J, Li P, Wang C, et al. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating wnt/β-catenin pathway. Cell Death Dis (2020) 11(12):1065. doi: 10.1038/s41419-020-03276-1
95. Targher G, Bertolini L, Padovani R, Rodella S, Tessari R, Zenari L, et al. Prevalence of nonalcoholic fatty liver disease and its association with cardiovascular disease among type 2 diabetic patients. Diabetes Care (2007) 30(5):1212–1218. doi: 10.2337/dc06-2247
96. Zhang Y, Wang H, Xiao H. Metformin actions on the liver: Protection mechanisms emerging in hepatocytes and immune cells against NASH-related HCC. Int J Mol Sci (2021) 22(9):5016. doi: 10.3390/ijms22095016
97. Moseley RH. Therapy for nonalcoholic fatty liver disease. J Clin Gastroenterol (2008) 42(4):332–335. doi: 10.1097/MCG.0b013e31815e4079
Keywords: NASH, NAFLD, circRNA, liver, crosstalk, hepatocellular carcinoma, insulin resistance
Citation: Yepmo M, Potier J-B, Pinget M, Grabarz A, Bouzakri K and Dumond Bourie A (2022) Discussing the role of circular RNA in the pathogenesis of non-alcoholic fatty liver disease and its complications. Front. Endocrinol. 13:1035159. doi: 10.3389/fendo.2022.1035159
Received: 02 September 2022; Accepted: 17 October 2022;
Published: 02 November 2022.
Edited by:
Ralf Jockers, Université de Paris, FranceReviewed by:
Toshio Takahashi, Suntory Foundation for Life Sciences, JapanCopyright © 2022 Yepmo, Potier, Pinget, Grabarz, Bouzakri and Dumond Bourie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aurore Dumond Bourie, YS5kdW1vbmRAY2VlZC1kaWFiZXRlLm9yZw==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.