- 1College of Biological and Environmental Sciences, Zhejiang Wanli University, Ningbo, China
- 2College of Life Sciences, Zhejiang University, Hangzhou, China
The freshwater prawn Macrobrachium rosenbergii is one kind of important economic aquaculture species and displays remarkable sexual dimorphism. The molecular mechanism of sexual differentiation in M. rosenbergii has been primarily unraveled through the research efforts of the androgenic gland and its related genes. However, the understanding of conserved genes involved in the molecular mechanism underpinning sex determination and sexual differentiation of M. rosenbergii is still fragmentary. MroDmrt11E is a member of the doublesex and mab-3-related transcription factor (Dmrt) gene family and is prominently expressed in the testis. In the present study, in vivo knockdown of MroDmrt11E at the postlarva stage in male prawn induced a complete and functional sex reversal and achieved the production of an all-male monosex population. Furthermore, a great deal of new information of upregulated and downregulated transcriptions involved in sexual differentiation of MroDmrt11E knockdown was enriched by comparative transcriptomic analysis. The effects of RNAi-mediated gene knockdown of MroDmrt11E on the differentially expressed and sex-related candidate genes, such as transformer, fruitless, feminization, insulin-like androgenic gland gene, Dmrt gene family, were primarily focused on, and their possible molecular regulatory relationships in sexual differentiation were analyzed. Meanwhile, the response of primary Kyoto Encyclopedia of Genes and Genomes (KEGG) biological pathways was investigated to expound the potential roles of MroDmrt11E in male sexual differentiation, which provided a deeper understanding of the molecular regulatory network underlying sexual differentiation of M. rosenbergii. The finding provided a novel sexual manipulation technique through silencing of Dmrt gene family for achieving a complete and functional sex reversal and offered a new insight regarding the mechanism of the Dmrt gene family in the sexual differentiation of crustaceans.
Highlights
1. MroDmrt11E silencing induced a complete and functional sex reversal in male juveniles and was applied in the production of an all-male monosex population
2. The effect of RNAi-mediated gene knockdown of MroDmrt11E on sex-related candidate genes and the changes of primary KEGG biological pathways were investigated by comparative transcriptomic analysis
Introduction
The freshwater prawn Macrobrachium rosenbergii is an important economic aquaculture species that is widely distributed in tropical and subtropical regions (1). It displays remarkable sexual dimorphism, which means the male prawn grow faster than females and show larger body sizes in adults (2). Thus, to achieve monosex production of this species, considerable research efforts have been devoted to unraveling the molecular mechanism of sex determination and sexual differentiation of M. rosenbergii (2–4).
In crustaceans, the androgenic gland (AG) provided much more information in the history of sexual regulation research. AG is known as a male-specific gland and primarily regulates male sexual differentiation and maintains male characteristics in crustaceans. Simultaneously, several insulin-like androgenic gland peptides (IAGs) have been identified in decapods (3, 5–8). Previous studies have reported that IAG from M. rosengergii (Mr-IAG) is proven to participate in male differentiation (3, 9). Mr-IAG silencing at postlarva (PL) induced a full and functional sex reversal and achieved the production of an all-male monosex population in M. rosenbergii (2, 4, 10). Moreover, long-term knockdown of Mr-IR, the receptor for Mr-IAG, also induced sex reversal and yielded neo-females in M. rosenbergii (11). For another instance, silencing Cq-IAG in Cherax quadricarinatus feminized male-related phenotypes (6). Until now, in crustaceans, the central role of IAG in orchestrating male sexual differentiation has been well characterized (10, 12), but the understanding of other conserved genes involved in sex determination and sexual differentiation is still fragmentary. On the other hand, identifying and studying the regulation mechanism of sex-related genes and noncoding RNAs also provide valuable sexual development information of decapods (13). Thus, the first question is whether the crustacean develops a novel sexual differentiation pathway, which connects the AG-specific genes and other sex-related regulatory factors.
Dmrt is a kind of well-conserved protein and is characterized by a DNA-binding region called the DM domain (14, 15). All of the arthropod doublesex (dsx) (16–18), nematode mab-3 (19), and vertebrate dmrt1 (20) are members of Dmrt gene family and involved in sex determination and/or sexual differentiation in bilaterian animals. In Drosophila, primary sex determination occurs very early in embryonic development when zygotically transcribed genes located on the X chromosome and the autosomes and activate Sxl in females but fail to activate it in males (21). Dsx is the connecting element and the central nexus of insect sex determination (18). Dmrt1 has appeared in the common vertebrate ancestor and is a regulator of male determination and testicular formation in gonadal cells (20). In mammals, Dmrt1 activates the male sexual differentiation signaling pathway and promotes testicular growth and development (20). Accumulating evidence from these studies suggested that the sexual differentiation functions of Dmrt gene family were evolutionarily conserved (14, 15).
Recently, numbers of Dmrt gene family have been identified from several crustaceans, such as the Chinese mitten crab Eriocheir sinensis (22), the water flea Daphnia (17), the eastern rock lobster Sagmariasus verreauxi (23), the Chinese shrimp Fenneropenaeus chinensis (24), the oriental river prawn Macrobrachium nipponense (25), and the giant freshwater prawn M. rosenbergii (26–28). However, one of the limitations of these studies is that these studies have not explained how the Dmrt participates in sex determination and/or sexual differentiation pathway.
In M. rosenbergii, several Dmrt gene families have been identified, including MroDmrt11E, MroDmrt99B, MroiDmrt1a, MroiDmrt1b, MroiDmrt1c, MroiDmrt1d, MroDSX, and Mr-Dsx (26–28). These Dmrts show various time-dependent expression patterns and tissue-specific distributions, which suggested that these Dmrts were possibly involved in both somitogenesis and sexual differentiation. Among them, MroDmrt11E presents a sexually dimorphic expression pattern, and the transcription is prominent in the testis but lower in the ovary (26, 27). Significantly, MroDmrt11E knockdown induced a significant decrease of the Mr-IAG transcript (26), suggesting that MroDmrt11E possibly plays an upstream role in the “IAG-switch” regulatory signaling and participates in the sexual differentiation by directly or indirectly influencing the expression of Mr-IAG (26). However, one question that needs to be addressed is: What is the role of MroDmrt11E in male sexual differentiation? There is no direct evidence of animal experiments to explore MroDmrt11E’s involvement in sex determination and/or sexual differentiation. In M. rosenbergii, the position of Dmrt gene family in the sexual differentiation signaling pathways remains largely elusive.
The present study was designed to explore the potential role of MroDmrt11E in the male sexual differentiation pathway. In this study, in vivo knockdown of MroDmrt11E in male PL by RNA interference (RNAi) was implemented to evaluate whether MroDmrt11E silencing would induce full functional sex reversal in M. rosenbergii. Meanwhile, the response of various related gene transcriptions to MroDmrt11E silencing and the effect of RNAi-mediated gene knockdown of MroDmrt11E on sex-related gene expression were investigated. The main understanding of this study would offer a novel insight regarding the biological function and mechanism of Dmrt gene family in regulating sexual differentiation of crustaceans.
2 Materials and Methods
2.1 Animals
M. rosenbergii PL of the same brood was collected from Yonggang commercial farm in Ningbo, Zhejiang, China, and acclimated in laboratory tanks 1 week before injection. The prawn at the PL15–30 stage with 1.0–1.5-cm body length (BL) were selected for the treatment. The BL of M. rosenbergii was measured as a straight line from the base of the eyestalk to the end of the telson. The injected shrimps were reared in separated tanks (100 L) under a flow-through system. The temperature was maintained at 27°C ± 2°C, and the photoperiod was 14:10 (light:dark per day). Prawn were fed with artificial food twice daily.
2.2 In Vivo Knockdown of MroDmrt11E by RNAi in Postlarvae
For RNAi experiments, the dsRNAs of MroDmrt11E and Green fluorescent protein (GFP) were produced and purified as described (26). Here, 5 μg dsRNA of MroDmrt11E in 0.9% (w/v) physiological saline was injected into each shrimp (N = 200) through the arthrodial membrane at the base of the fifth pereiopods using a microinjection needle. The control group (N = 200) received an equal amount of GFP dsRNA injection. The injection was performed monthly during the animal experiment, and three injections for 90 days were performed for each prawn.
To evaluate the RNAi efficiency, the male reproductive system of the RNAi group (N = 5) was dissected at the end of the animal experiment. The interference efficiency of MroDmrt11E silencing was detected by both real-time fluorescence quantitative PCR (qPCR) and semiquantitative-PCR. On one hand, SYBR Green RT PCR assay was carried out in a CFX384 quantitative PCR Detection System (Bio-Rad, USA) for qPCR analysis. Here, 18S (GenBank accession no. DQ642856.1) was used as an internal reference to adjust the number of cDNA templates. Mro18S-qF 5′-GAGAAACGGCTACCACATCCAA-3′ and Mro18S-qR 5′-GTGCTCATTCCAATTACGCAGACT-3′ were designed to generate a 125-bp fragment of Mro18S. And MroDmrt11E-qF 5′-CGCATCCCACCCTACTTGA-3′ and MroDmrt11E-qR 5′-GGCTTCCCTCTGCATCATGA-3′ were designed to generate an 89-bp fragment of MroDmrt11E. A total volume of 20 µl mixture [10 µl of 2× SYBR Master Mix (Applied Biosystems, USA), 1 µl of cDNA mix, 0.5 µl of each primer (10 µM), and 8 µl of sterile distilled H2O] was used for qPCR analysis. And the program of qPCR was 95°C for 1 min, followed by 40 cycles of 95°C for 15 s and 63°C for 25 s. Three replicates for each sample were performed. The relative expression level was calculated using the 2-ΔΔCt method. The data obtained from qPCR analysis were analyzed for statistical significance using GraphPad Instat (GraphPad Software Inc.).
On the other hand, one pair of 18S primers (Mro18S-F 5′-GGTAGTGACGAAAAATAACAAT-3′ and Mro18S-R 5′-CCCACCCCAGTCCGGAACTGA-3′) and another pair of MroDmrt11E primers, MroDmrt11E (MroDmrt11E-F 5′-CACTCCTCCAGTTGGTTGT-3′ and MroDmrt11E-R 5′-GCTGATGGGTGTCCTTGT-3′), were used for semiquantitative PCR analysis. The PCR products were assessed by electrophoresis on 1.2% agarose gel. Three replicates for each sample were also performed. Relative abundances were expressed as the ratio of MroDmrt11E transcript levels to those of 18S rRNA. The peak value of the GFP group was set to 100, and the rest of the value was normalized. The data obtained from PCR analysis were analyzed for statistical significance using GraphPad Instat (GraphPad Software Inc.).
2.3 The Effects of MroDmrt11E Silencing on the Induction of Sex Reversal
2.3.1 Identification of Sex Reversal by MroDmrt11E Silencing
In the freshwater prawn M. rosenbergii, the manipulation of the insulin-like androgenic gland hormone (Mr-IAG) silencing at the PL period obtained a full and functional sex reversal, leading to the production of an all-male monosex population (2). Thus, similar gene silencing is attempted to expound whether MroDmrt11E participates in male sexual differentiation in M. rosenbergii.
In the knockdown of MroDmrt11E, the male/female sex of the silenced individual was inspected and identified by both external appearance observation (1) and genetic molecular marker methods (1, 29). Since gross external signs of first sexual characteristic, such as genital pores and male appendages, are not evident at the PL stage, the gene silencing experiment lasted 3 months until clear sexual differentiation with 4.0–5.0 cm BL of the prawn (1). At the end of the RNAi experiment, male prawn were distinguished from females with the structure of male genital pores located at the coxopodite of the fifth pereiopods. Then, the muscle genomic DNA of the individuals was extracted respectively and used as the template for PCR amplification to evaluate the genetic sex as described (29). Subsequently, the sex reversal genetic male prawn induced by MroDmrt11E silencing were discriminated then kept for further analysis.
2.3.2 Confirmation of a Complete and Functional Sex Reversal and the Breeding of Monosex Progeny
The sex reversal male prawn, known as neo-females (N = 14), were then selected and cultured in the prawn hatchery in Ningbo, Zhejiang, China. All of the external appearance, the development and maturation of the ovary, the reproductive behavior, and the copulation process of the neo-females were observed and compared with those of normal females during the whole period of culture. These neo-females were then mated with normal males and spawned, and the embryos were incubated in their brood chambers. Furthermore, the various characteristic parameters of embryonic development, hatching, and metamorphosis of the progeny of neo-females were surveyed in parallel with that of the control progeny.
In addition, 10 embryos of egg-nauplius or egg-zoea period from each progeny were sampled, and the genomic DNA of each embryo was used for genetic sexual identification by amplifying the genetic sex marker. In general, the progeny of the neo-females was expected to be males, known as an all-male monosex population, whereas the progeny of normal females was composed of both males and females. Moreover, with regard to the growth processes of all-male monosex prawn, all of the embryonic development, zoea larvae metamorphosis, juvenile growth, and adult culture were monitored through the whole culture period of 3–6 months in M. rosenbergii.
2.4 Comparative Transcriptomic Analysis of MroDmrt11E Knockdown
2.4.1 RNA Isolation for Transcriptomic Sequencing
To evaluate the potential role of MroDmrt11E in the sexual differentiation pathway, the response of various related gene transcriptions and the expression of sex-related genes were investigated by comparative transcriptomic after MroDmrt11E gene knockdown in M. rosenbergii.
The individual of MroDmrt11E silencing showed clear sexual differentiation with 4.0–5.0-cm BL of the prawn at the end of the 3-month experiment. The male reproductive system (testis, sperm duct, and terminal ampullae) and the coxopodite between the third and fifth pereiopods from males were dissected from MroDmrt11E-silenced individuals (N = 10), and the organs were mixed together to provide sufficient RNA for the transcriptomic sequencing. Total RNA was extracted using the column TRIzol total RNA isolation kit (Sangon) following the manufacturer’s protocol. The OD260/280 should range from 1.8 to 2.0 to ensure the purity of the RNA sample.
2.4.2 Identification and Classification of Differentially Expressed Unigenes
The transcriptome was sequenced using the Illumina HiSeq. The raw reads were cleaned by removing adaptor sequences, empty reads, and low-quality sequences. The clean reads were assembled into non-redundant (Nr) transcripts. The resulting unigene sequences were then annotated using homology search (BLASTX) with an E-value cutoff of 10-5 against an NCBI Nr database, Swissprot, Cluster of Orthologous Groups database (COG), and Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The coding sequence and the direction of the annotated unigenes were determined based on the BLAST results from the four above mentioned databases. For the differential expression analysis, the transcript expression level of the unigenes was measured using the FPKM method (fragments per kilobase per million fragments). Genes were considered differentially expressed in the given library when the p-value was less than 0.05 and a greater than 2-fold change (with the absolute value of log2 fold change more than 1).
Furthermore, the effect of RNAi-mediated gene knockdown of MroDmrt11E on the expression of sex-related genes was also investigated. The differentially expressed sexual candidate genes were considered when the p-value was less than 0.05 and a greater than 1-fold change (with the absolute value of log2 fold change more than 0.5) in comparative transcriptomic analysis. According to these differentially expressed transcripts involved in sexual differentiation or cell proliferation signal, the putative intuitive cascade regulation axis or networks of pathways were illustrated.
3 Results
3.1 In Vivo Knockdown of MroDmrt11E by RNAi in M. rosenbergii
The brief strategy of the present study was illustrated in Figure 1A. The neo-female prawn was expected to be induced by MroDmrt11E silencing and applied for the breeding of the all-male population. The comparative transcriptomic analysis of the MroDmrt11E knockdown male reproductive system was implemented to enrich differently expressed genes.
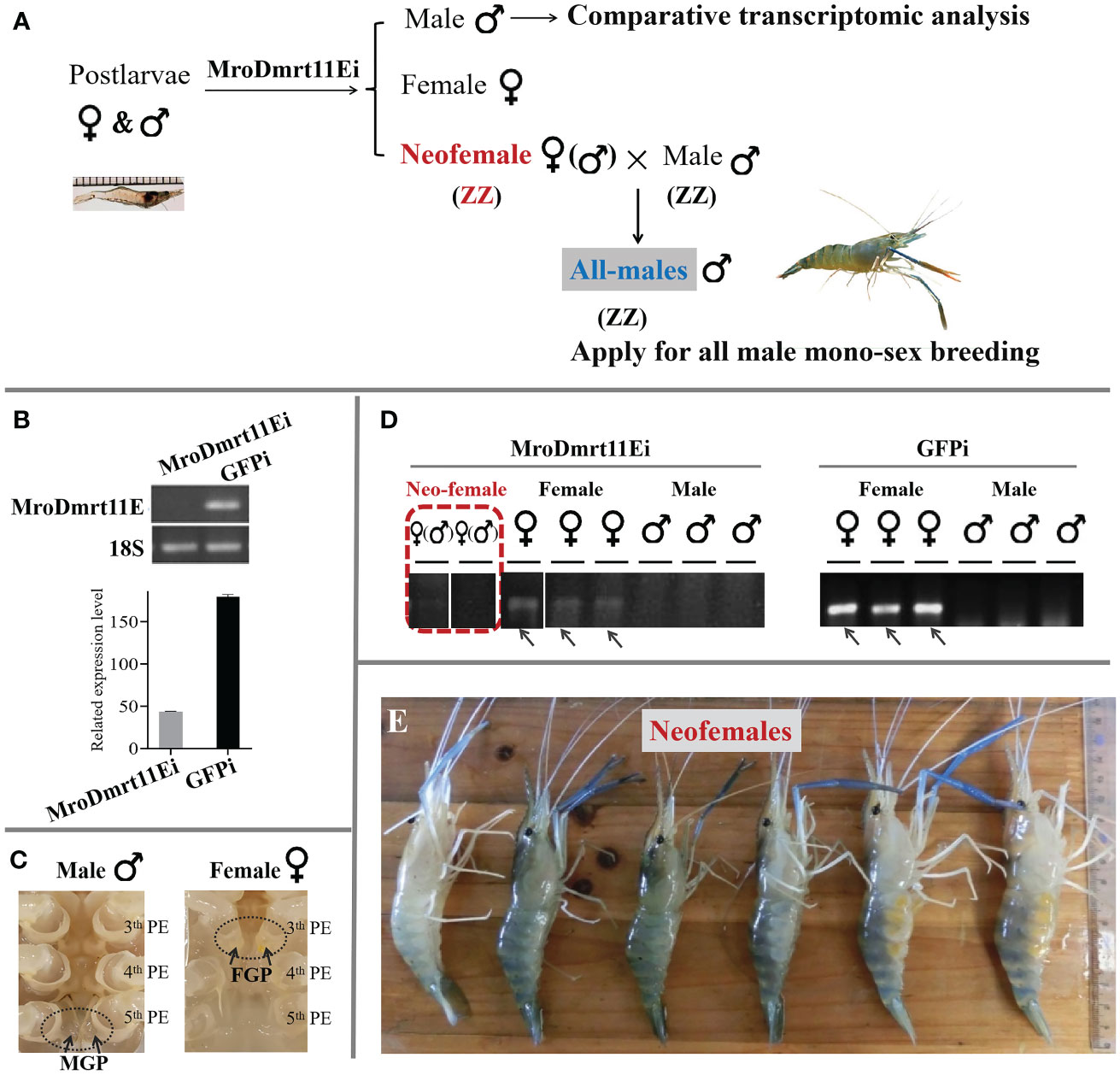
Figure 1 In vivo knockdown of MroDmrt11E by RNAi in postlarvae. (A) The diagrammatic sketch of in vivo knockdown of MroDmrt11E by RNAi in postlarvae. MroDmrt11E dsRNA injection was performed in postlarvae, and the gene silencing lasted for 3 months. (B) Detection of the relative MroDmrt11E mRNA level in RNAi induced and the control group. (C) The sex identification of differentiated male and female prawn by the appearance of sexual characteristics in M. rosenbergii. There was a pair of male genital pores located at the coxopodite of the fifth pereiopods in males, but a pair of female genital pores located at the coxopodite of the third pereiopods in females as well as neo-females. MGP, male genital pores; PE, pereiopods. (D) The gender of individuals was identified using sex molecular markers in both MroDmrt11E RNAi and the control group. The female-specific sex band indicates female genotype. The female-specific sex bands are indicated by the gray arrows in females. The representative neo-female prawn are shown in a red broken line frame. (E) The neo-female prawn with female appearance features. The MroDmrt11E-silenced genetic males (neo-females) were raised to maturity and induced appearance features of females. Some neo-females spawned and incubated the eggs/embryos in their brood chambers, located ventrally on their abdomens.
After in vivo knockdown of MroDmrt11E gene performed in M. rosenbergii PL, during the whole RNAi experiment, the mRNA expression level of MroDmrt11E was dramatically decreased to a low level compared with control samples (Figure 1B). This result indicated that a long-term efficiency of MroDmrt11E silencing was successfully maintained. Secondly, the male/female sexes of the MroDmrt11E RNAi and control group were identified by both external appearance features (a pair of genital pores) (Figure 1C) and the genetic sex molecular marker method (Figure 1D). In detail, all female controls in the GFP-silenced group and confirmations in the MroDmrt11E-silenced group were as expected, revealing the female-specific sex band (Figure 1D). Comparatively, although some MroDmrt11E-silenced individuals morphologically resembled the control females, without male genital pores at the fifth pereiopods (as evidence of appearance feature in the control male), they lacked female-specific sex bands (as evidence of genetic sex in the control female), therefore proving genetic male (Figure 1D). It was clear evidence for sex reversal that MroDmrt11E silencing in the PL males successfully induced the full functional sex reversal individuals, also known as neo-females. Moreover, these neo-females were carefully raised to maturity, with induced female appearance features and reproductive system (Figure 1E).
3.2 Confirmation of a Complete and Functional Sex Reversal and the Breeding of Monosex Progeny
To investigate the potential role of MroDmrt11E in gonad development, the MroDmrt11E-silenced genetic males (neo-females) were raised into adults. The result showed that these neo-females were characterized by the disappearance of male characteristics (such as male genital pore and male reproductive system) and the formation and development of female features (female reproductive system). Moreover, the mature neo-females developed normal-appearing ovaries as compared with normal females. As to the ovarian development of M. rosenbergii neo-females, it was closely related to the individual development process by significant changes in color and ovarian volume. With the gradual development and maturation of the ovary, the full and inflated orange-yellow ovary was displayed throughout the carapace or extended to the first ventral segment (Figure 2A).
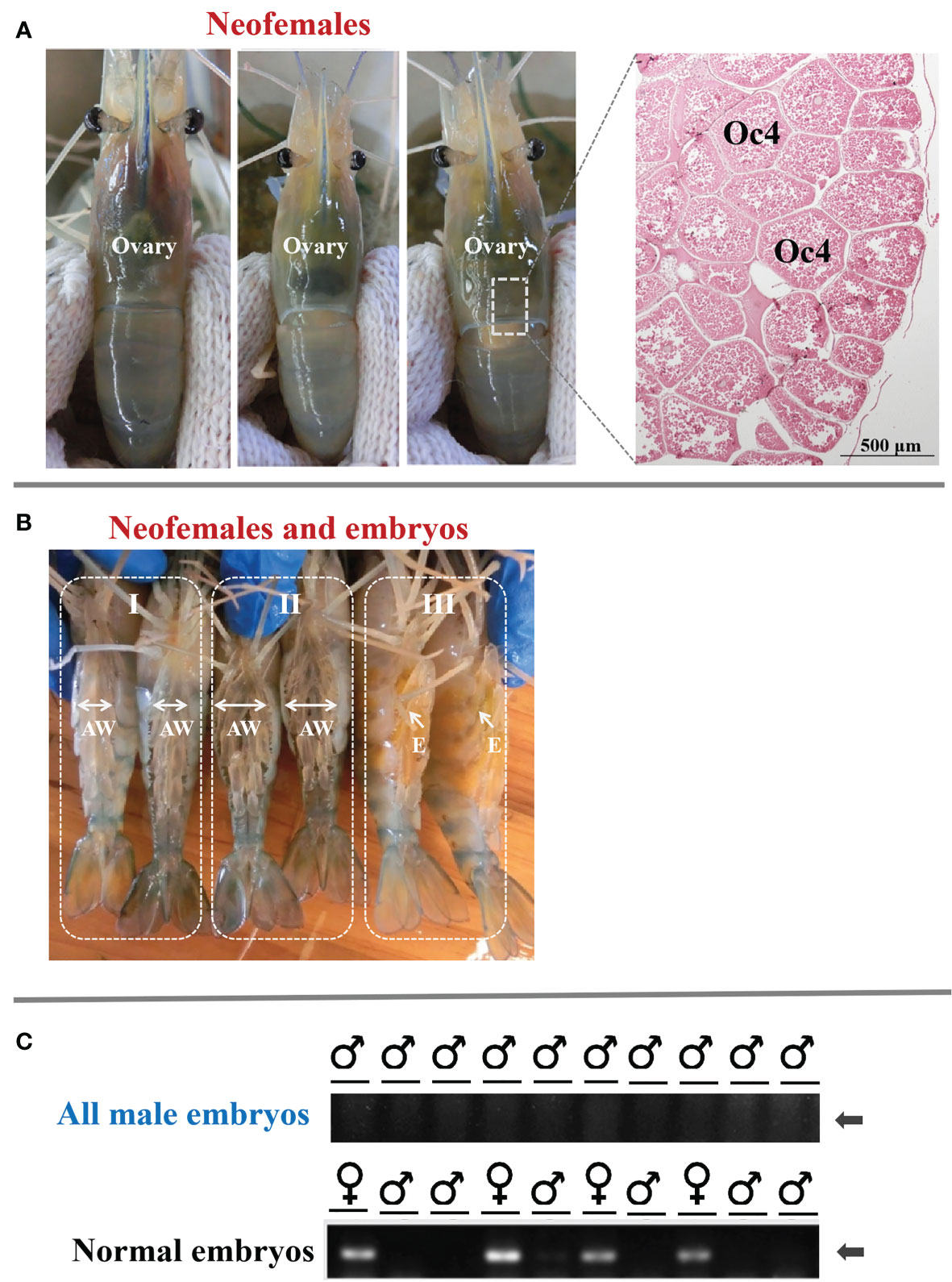
Figure 2 The ovarian development of full-functional sex reversal prawn and the breeding of all-male progeny by MroDmrt11E-silenced prawn (neo-females). (A) The ovarian development and maturation of neo-females. The inset photograph indicated the histology of the mature ovary of neo-females. The ovary was full of mature oocytes (Oc4) that were intensely eosinophilic (reddish orange). (B) Ventral view of MroDmrt11E-silenced prawn (neo-females) incubating embryos in their brood chamber. The mature neo-females (B-I) spawned and incubated the embryos in their brood chambers, located ventrally on their enlarged abdomens (B-II, B-III). The color of embryos presented gradual changes from yellow to orange (B-III) then to light-gray color (B-II) during embryonic development. AW, abdomen; E, embryos. (C) Genomic validation of representative embryos from each progeny of neo-females. The first row of PCR products was generated from neo-female embryos, and the second row was from normal females. The presented specific band in gel electrophoresis implied the female embryo.
On the other hand, the breeding of neo-females and their offspring culture was also implemented. The neo-females had successful mating and showed normal mating behavior when they mated with normal males (data are not presented). These neo-females then spawned and incubated the embryos in their brood chambers, located ventrally on the abdomens (Figure 2B). The appearance color of embryos presented gradual changes from yellow to orange, then to light-gray color during embryonic development in female (enlarged) abdomen (Figure 2B).
Furthermore, to confirm the sex ratio or composition of offspring, the sex of the embryo was also detected by the genetic sex marker method. The results showed that the progeny of the control female was composed of male and female as expected, whereas the progenies of the neo-female were only males (Figure 2C). It was proven that the offspring of these neo-females was an all-male population. Therefore, that was to say, MroDmrt11E gene silencing in male PL period caused complete and functional sex reversal into neo-female, which could be applied to produce an all-male population.
Furthermore, with regard to the sex reversal induction and neo-female production in M. rosenbergii, both the manipulations of the single gene knockdown and AG ablation have crucial effects on male sexual differentiation. The detailed results of various gene silencing, such as MroDmrt11E, Mr-IAG, and Mr-IR, and AG ablation were summarized and compared in Table 1.
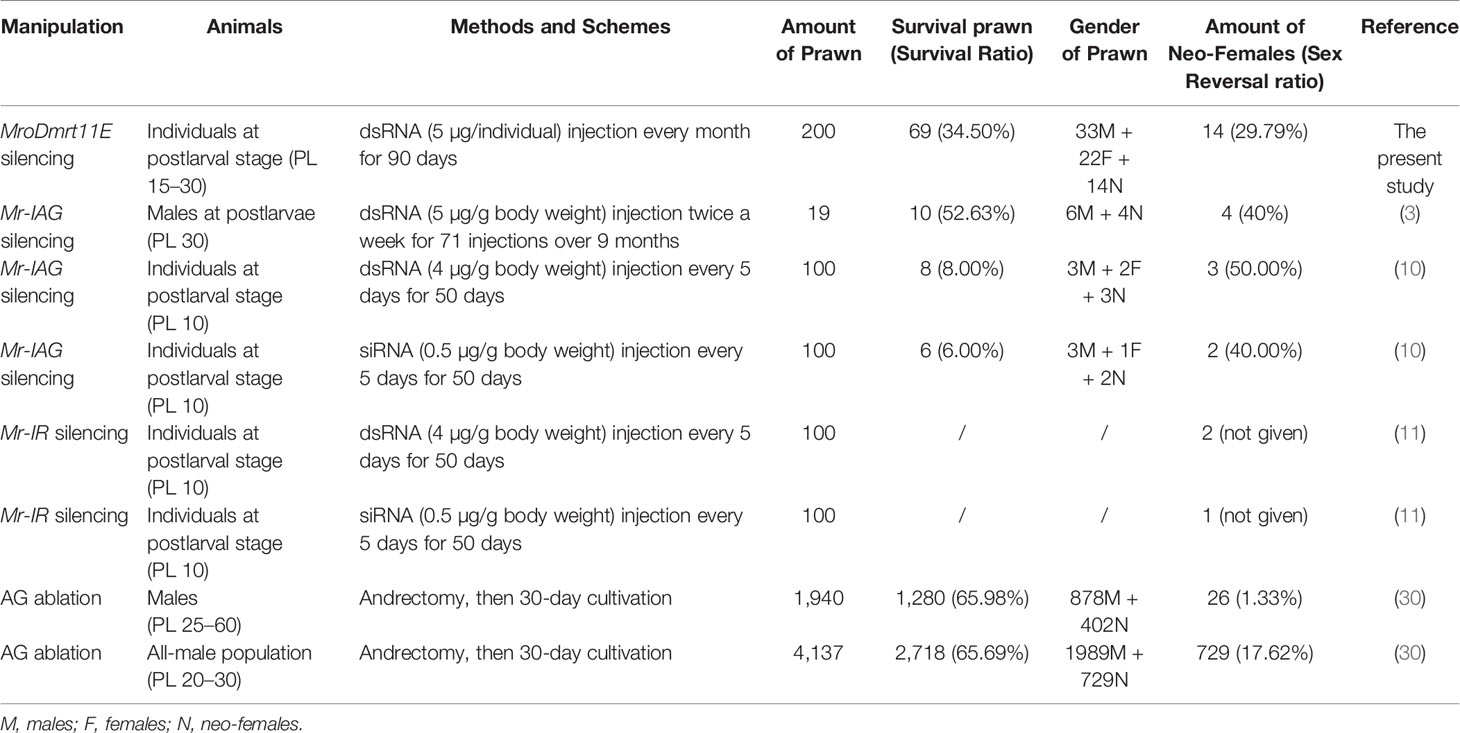
Table 1 The summary of sex reversal induction and neo-female production in Macrobrachium rosenbergii.
3.3 The Comparative Transcriptomic Analysis of MroDmrt11E Knockdown
3.3.1 The Differential Gene Expression Analysis of MroDmrt11E Silencing
To ascertain the effect of MroDmrt11E knockdown on the expression level of related genes or factors in sexual differentiation, comparative transcriptomic profiling from juvenile males of M. rosenbergii after MroDmrt11E silencing was enriched in this study. The transcriptomic libraries obtained about 50.74 million clean reads and 7.61G clean bases in MroDmrt11E RNAi group and 55.68 million clean reads and 8.35G clean bases in GFP RNAi group. The values of Q20 and Q30 were more than 97% and 93% in both groups, respectively.
Firstly, differential gene expression (DGE) analysis between MroDmrt11E silencing and control group generated 1,060 genes, of which 424 unigenes were upregulated and 636 were downregulated. Secondly, to assess how MroDmrt11E participates in the sex determination and/or sexual differentiation cascades in M. rosenbergii, the upregulated and downregulated unigenes with high percentages, which belong to the categories of response to various pathways, were identified. The top 10 representative groups of pathways with a higher percentage of differentially regulated genes were clustered into the phagosome, Phosphatidylinositide 3-kinases and protein kinase B (PKB also termed Akt) (PI3K-Akt) signaling pathway, viral carcinogenesis, cell cycle, pathogenic Escherichia coli infection, focal adhesion, apoptosis, RNA transport, estrogen signaling pathway, gap junction, and Rap1 signaling pathway. Meanwhile, the highest numbers of upregulated and downregulated unigenes were focused on zinc finger/zinc knuckle, and the higher amounts of differentially regulated genes were enriched on WD domain/G-beta repeat, tubulin/FtsZ gene family, RNA recognition motif, ribosomal protein, mitochondrial proteolipid, integrase core domain, and ATP synthase.
Furthermore, to gain insight into the biological processes being operative during the sexual differentiation in M. rosenbergii including other differentially expressed genes, we have subjected transcripts (with significant differential expression) to DEG-enriched KEGG pathway analysis. A brief illustration of these significant differentially expressed genes, which come from the four categories of the enriched KEGG pathway analysis, was detailedly summarized (Figure 3). These four kinds of categories with a higher percentage of upregulated and downregulated unigenes were primarily focused on the pathways of apoptosis (Figure 3A), regulation of actin cytoskeleton (Figure 3A), oocyte meiosis (Figure 3B), and protein processing in the endoplasmic reticulum (Figures 3A, C), respectively. Of these genes differentially upregulated and downregulated, the expression pattern of important genes playing crucial roles in these pathways was listed in Table 1S.
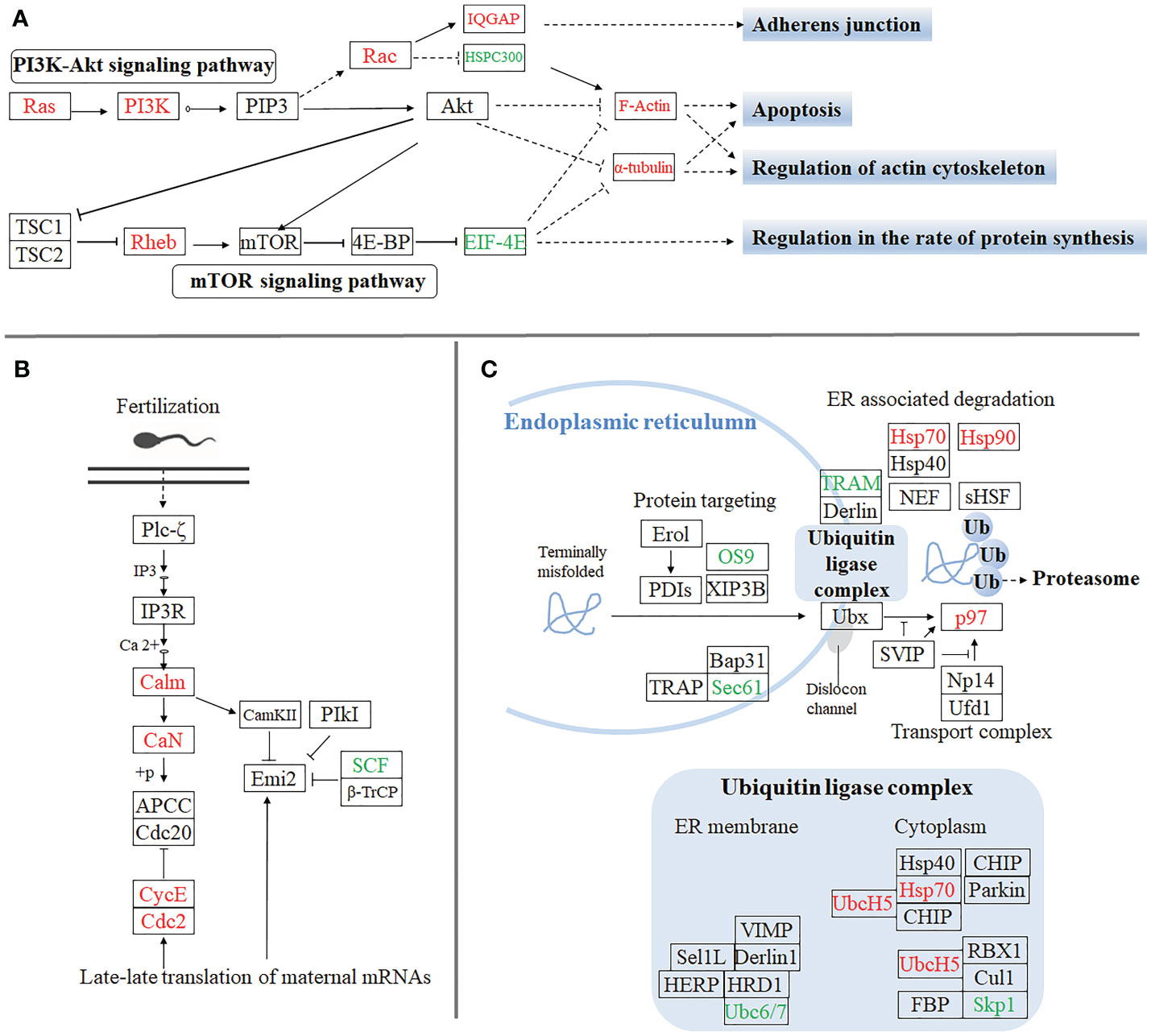
Figure 3 The significant differentially expressed unigenes enriched KEGG pathway analysis by the comparative transcriptomic analysis of MroDmrt11E knockdown. Four kinds of categories with a higher percentage of upregulated and downregulated transcripts were primarily focused on the pathways of apoptosis and regulation of actin cytoskeleton (A), oocyte meiosis (B), and protein processing in endoplasmic reticulum (C), respectively. The primary upregulated transcripts (red color) and the downregulated transcripts (green color) were briefly profiled in the various signaling pathways.
3.3.2 The Response of Sex-Related Candidate Genes to MroDmrt11E Silencing
To explore the effect of MroDmrt11E on the expression level of sex-related genes or factors, some significant differentially expressed unigenes were identified and enriched in the comparative transcriptomic profiling of M. rosenbergii. The influences or changes of these upregulated and downregulated transcripts were presented in Table 2. Furthermore, according to these differentially expressed transcripts involved in sexual differentiation or cell proliferation signal, the putative intuitive cascade regulation axis or networks of pathways were illustrated (Figure 4A).
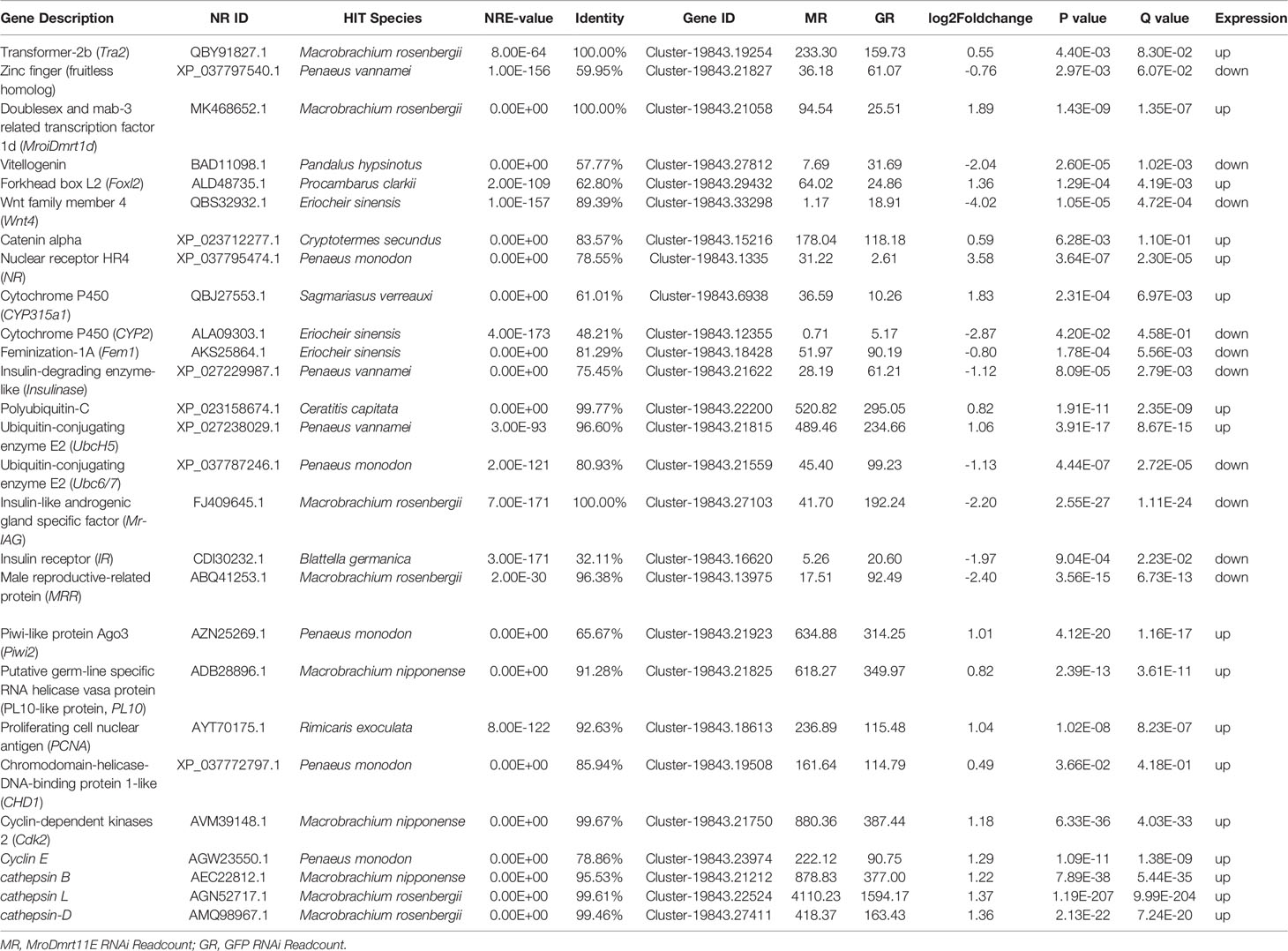
Table 2 Significantly differential expression of sex-related candidate genes in sexual differentiation and gonad development.
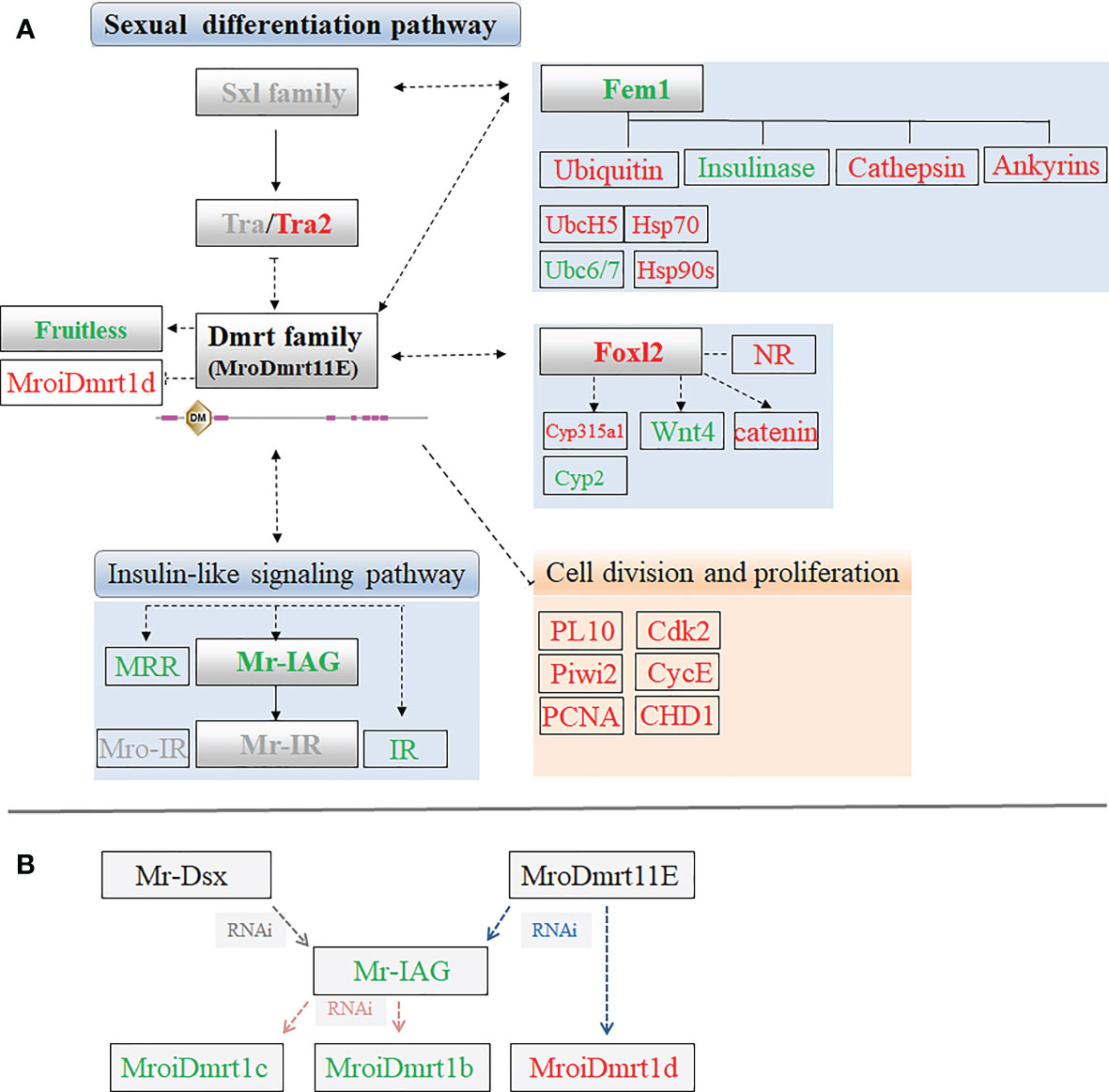
Figure 4 (A) The sex-related candidate unigenes enriched in putative sexual differentiation pathway by the comparative transcriptomic analysis of MroDmrt11E knockdown. The primary upregulated transcripts (red color) and the downregulated transcripts (green color) were briefly profiled in the signaling pathway. The genes with gray color possibly participated in the sexual differentiation but did not show significant differential expression by MroDmrt11E knockdown. (B) The brief relationship among Dmrt gene family and Mr-IAG was summarized to evaluate their potential influences involved in the sexual differentiation of M. rosenbergii.
In addition, the proven switch gene of sexual differentiation, Mr-IAG, was found in a downregulated transcript, but another member of Dmrt gene family, MroiDmrt1d, was identified as an upregulated transcript in the transcriptomic of MroDmrt11E knockdown. The possible relationships among Mr-Dsx, MroDmrt11E, Mr-IAG, and other Dmrt gene families were summarized based on the known information of the various RNAi results of these genes. Dmrt gene family probably participated in the transcriptional activation of Mr-IAG and possibly played great roles in the switch of Mr-IAG-related regulatory signaling of male differentiation. Moreover, Mr-IAG directly or indirectly influenced the expression of partial genes of Dmrt family (Figure 4B).
4 Discussion
4.1 The Regulation Relationship Between Dmrt Gene Family and Insulin Signaling Pathway in Sexual Differentiation
MroDmrt11E is a testis-prominent expressed gene and is speculated to play an important role in the developmental processes of sexual differentiation (26). The present study set out with the aim of supplying direct evidence from animal experiments and exploring the biological function of MroDmrt11E involving sexual differentiation. In addition, Dmrts were shown relative to IAG and involved in sexual differentiation (25, 28). But the molecular mechanism of Dmrts regulating sexual differentiation is little known. The position of the Dmrt gene family in the signaling pathways orchestrating sexual differentiation was also expounded by comparative transcriptomic analysis of MroDmrt11E knockdown in M. rosenbergii.
Firstly, the important highlight of this study has given remarkable evidence that MroDmrt11E played a crucial role in male sexual differentiation in M. rosenbergii and provided a novel sexual regulation illustration of the conserved Dmrt gene family in crustaceans. Although the crucial role of IAG in orchestrating male sexual differentiation has been characterized and the production of all monosex progenies on IAG or IAG’s receptor manipulation has been achieved, limited information on conserved genes involved in sexual differentiation has been published to date (13). In the present study, a gene silencing strategy was implemented to expound whether MroDmrt11E participated in male sexual differentiation of M. rosenbergii. Significantly, MroDmrt11E silencing in juvenile males successfully caused the disappearance of male characteristics (such as male genital pores and male reproductive system) while promoting the formation and development of female features and inducing full functional sex reversal individuals. Moreover, the neo-female individuals gave rise to ovarian maturity, induced appearance features of females, and were utilized to produce all-male populations.
Secondly, apart from the characterization of Dmrt itself, little information is available regarding key factors upstream or downstream of the Dmrt gene family and other elements of the signal transduction pathway involving sexual differentiation (27). Thus, we focused on the relationship between the Dmrt gene family and Mr-IAG. On one hand, it is reported that in vivo silencing of the Mr-IAG gene significantly decreased the expression of two MroDmrt genes, MroiDmrt1b and MroiDmrt1c, thereby suggesting the possible role of these two genes in the IAG switch and sexual differentiation processes (27). On the other hand, gene knockdown of Mr-Dsx resulted in a pronounced suppression of the insulin-like androgenic hormone (Mr-IAG) gene, which indicated that Mr-Dsx may participate in the transcriptional activation of IAG (28). In the present study, knockdown of MroDmrt11E was proven continuously related to the decrease of the Mr-IAG transcript. However, MroiDmrt1d was contrarily found upregulated in the male of MroDmrt11E knockdown, which meant MroDmrt11E caused pronounced suppression of MroiDmrt1d. Thus, a complex regulation network among the Dmrt gene family and Mr-IAG was summarized. In brief, both Mr-Dsx and MroDmrt11E participate in the transcriptional activation of Mr-IAG, but Mr-IAG is involved in the activation of MroiDmrt1b and MroiDmrt1c. These results implied that a feedback loop of the Dmrt gene family may exist. And, the various members of the Dmrt gene family had different effects on the expression of Mr-IAG gene by directly or indirectly influencing the switch of male sexual differentiation in M. rosenbergii (25, 28).
Differently, MniDMRT11E knockdown significantly increased insulin-like androgenic gland factor expression in the oriental river prawn M. nipponense (25), which was contrary to the result of the MroDmrt11E knockdown sustainedly repressing the expression of Mr-IAG. It indicated that the evolutionarily conserved Dmrt gene family plays widely divergent roles in the sexual differentiation of crustaceans (25, 28).
Moreover, an insulin-like receptor homolog and a male reproductive-related protein (MRR) were found significantly downregulated from the transcriptomic analysis of MroDmrt11E knockdown. Insulin-like receptor (IR) functions as the pivotal member of the insulin family signaling pathway and directs the sexual differentiation in mammals (31) and some aquatic livestock (32, 33). The binding of the insulin-like peptide ligand initiates a cascade of phosphorylation events, stimulating the downstream signal transduction and resulting cellular effect (31). Earlier evidence has shown that IR interacts with IAG to regulate sexual differentiation and spermatogenesis in crustacean (5, 11, 33, 34). In more detail, the insulin-like receptor, termed Mr-IR (33), functioned as a receptor for Mr-IAG, and its knockdown induced sex reversal and retarded the process of spermatogenesis in M. rosenbergii (11). In this study, not Mr-IR but a novel insulin-like receptor homolog was found as a significantly downregulated transcript, which indicated that more than one IR are involved in the insulin family signaling pathway and participate in the regulation of sexual differentiation. Meanwhile, a male reproductive-related protein expressed in the terminal ampullae of the male reproductive system (35) was also identified as a significantly downregulated transcript. It means that MroDmrt11E was required for the transcriptional activation of Mr-IAG or IR or MRR, but it was not clear whether the MroDmrt11E gene can directly stimulate their expressions in the prawn.
In brief, this study revealed the potential and complex regulatory relationships among various Dmrt gene families and several insulin signaling pathway-related factors. It also implied that the insulin signal pathway is crucial for female/male reproductive development or maintenance in crustaceans.
4.2 Sex-Related Candidate Genes Involved in the MroDmrt11E Knockdown
MroDmrt11E has been validated to affect the development of male gonads and played its potential function in sexual differentiation and reproductive development (26). Compared with differently expressed genes of the transcriptomic library, a great deal of sex-related candidate genes were focused on. However, due to the limitation of the molecular mechanism of sexual differentiation in crustaceans, the position of the MroDmrt11E or Dmrt gene family in the signaling pathways orchestrating sexual differentiation of M. rosenbergii is hard to be clarified (28). Recently, with the analogous transcripts addressed in many crustaceans, it suggested that crustacean species may adopt a similar sex determination and/or sexual differentiation pathway to that reported in insects (36). Therefore, the well-characterized molecular mechanism of sex determination of insects is used as an important reference clue. For instance, the primary sex determination and somatic sexual differentiation are controlled by a genetic hierarchy X:A > Sex-lethal (Sxl) > Transformer/Transformer 2 (Tra/Tra2) > Dsx and Fruitless (Fru) in Drosophila (16, 21, 37, 38). Therefore, in the present study, some sex-related homologs, including Tra2, Foxl2, Fru, Feminization-1 (Fem-1), were identified and screened as significant differential genes from the comparative transcriptomic library. In the following sections, we tried to provide more valuable information and infer the potential roles of these candidate sex-related genes in sex determination and/or sexual differentiation pathways.
4.2.1 Transformer and Transformer-2
Tra and Tra-2 are mRNA splicing factors and act as a downstream splicing complex, regulating specific splicing of target RNAs (21, 37–39). In Drosophila species, it is known that the action of Sxl results in the active splice variant of Tra in females (37). Then, Tra2 acts in concert with Tra to regulate female-specific splicing of target RNAs (21, 37, 38). In the present study, upregulated Tra2 homolog was identified in the transcriptomic library of MroDmrt11E silencing, indicating that MroDmrt11E possibly participated in the regulation of sex or gonad-specific splicing of target RNAs through suppression of the expression of Tra2. On the other hand, four Sxl isoforms, named MroSxl1, MroSxl2, MroSxl3, and MroSxl4, have been identified in both males and females in M. rosenbergii (40). However, none of these Sxl genes were screened as significantly differential transcripts from the comparative transcriptomic library until now. In addition, several Sxl and Tra-2 isoform-encoding transcripts show broad tissue expression but not sex-biased expression patterns in crustaceans, suggesting that Sxl-Tra-Dsx may be unique to Drosophilidae and not conserved among decapods (41).
4.2.2 Fruitless
Fru, defined by its zinc fingers and forkhead box L2 (Foxl2) defined by a unique DNA-binding domain, is known as a transcription factor (20, 21, 37, 38). Fru is a male-promoting gene, regulating the development of the male central nervous system and male sexual behavior (39). It is spliced in the absence of the female Tra/Tra-2 complex, carried out by non-sex-specific splicing machinery (21, 37, 38). Nevertheless, Fru showed sexual dimorphism in ovaries and testis, which displayed upregulated expression in ovaries of M. rosenbergii in the comparative transcriptomic database of gonads (42).
On the other hand, Foxl2 expression ensues and acts to inhibit the male pathway while promoting the female pathway through the action of Rspo1, Wnt family member 4 (Wnt4), and catenin in the absence of the Sry-driven expression of Sox9 (43). Interestingly, Foxl2 displayed upregulated expression in testis from the transcriptomic database of gonads of M. rosenbergii (42). Meanwhile, Sp-Wnt4, a member of Wnt4 gene in mud crab, Scylla Paramamosain, was expressed at a higher level in the ovary compared to other tissues, but the expression level of Sp-Wnt4 was significantly increased in testis after unilateral eyestalk ablation (44). In the present study, significantly upregulated Foxl2, downregulated Fru, and Wnt4 homolog were identified in MroDmrt11E knockdown. It suggested that MroDmrt11E expression might act to inhibit the expression of Foxl2 gene while promoting the action of Fru and Wnt4 in the sexual differentiation process.
In addition, it is reported that Foxl2 was able alone or with nuclear receptor subfamily 5 group A member 2 (Nr5a2) jointly to upregulate the expression of cyp19a and repress the expression of Dmrt1 in the olive flounder Paralichthys olivaceus (45). It meant that Foxl2 may play an important role in ovarian differentiation by maintaining cyp19a expression and antagonizing the expression of Dmrt1 (45). The cytochrome P450 CYP315a1, a member of the CYP superfamily gene, was implicated in the ecdysteroidogenic pathway (46). Meanwhile, CYP315a1 was also identified as a sex-related gene. From the comparative transcriptome analysis of M. rosenbergii, CYP315a1 was suggested involved in testicular development (42). Nuclear receptors are a class of proteins found within cells that are responsible for sensing steroid and thyroid hormones and certain other molecules (47, 48). Steroid hormones are well known to be responsible for controlling reproduction and development in higher organisms like arthropods (46, 47). In the present study, upregulated Foxl2, nuclear receptor (NR), and CYP315a1 homologs in MroDmrt11E knockdown were all identified, and the expression of their transcripts was by the above studies, which indicated that MroDmrt11E inhibited the expression of CYP315a1 through repressing the expression of Foxl2 and/or NR. Moreover, MroDmrt11E perhaps participated in the signaling pathway of various hormones and played great roles in reproduction and gonad differentiation.
4.2.3 Feminization-1
Fem-1 is characterized by one of the most common protein–protein interaction motifs and ankyrin repeat motifs and known as a regulatory factor of signal transduction in the sex determination signaling pathway (21, 37, 38, 49). In M. rosenbergii, Mrfem-1 was exclusively expressed in the ovary, suggesting that Mrfem-1 might be associated with ovarian maturation in prawn (50). An ovary-specific gene Fem-1 homolog, Mnfem-1, has been identified in the oriental river prawn, M. nipponense (51). The Mnfem-1 protein can be potentially interactive with cathepsin L and proteins containing the domains of insulinase (also called an insulin-degrading enzyme), ankyrin, or ubiquitin in yeast two hybridization (51). Accordantly, in the present study, downregulated Fem-1A and insulinase homolog, three upregulated cathepsin homologs, and one polyubiquitin-C homolog were identified in the comparative transcriptomic analysis of MroDmrt11E knockdown (Table 2). Meanwhile, several significant differentially expressed genes encoding the domains of ankyrin were found as upregulated transcripts (data not shown). It suggested that MroDmrt11E probably played significant roles in activating or inhibiting the expression of these genes or elements (such as cathepsin, insulinase, and ubiquitin) through suppression of the Fem-1 homolog.
Cathepsins, ubiquitously present in most organisms, belong to a family of proteases that cleaves other proteins, and some of these genes were shown to be involved in the ovarian maturation and embryo development of Penaeus japonicus (52) and M. nipponense (53). Furthermore, the gonadal transcriptomic database of Oratosquilla oratoria included eight cathepsin genes (54). Two cathepsins, cathepsin I and cathepsin D-like, were predominantly expressed in the testis of O. oratoria, suggesting that cathepsins may be involved in testicular development (54). In the present study, three upregulated cathepsin homologs were identified in the comparative transcriptomic analysis of MroDmrt11E knockdown, which indicated that MroDmrt11E may play an important role in sexual and/or gonad differentiation by antagonizing the expression of cathepsins.
4.2.4 Heat Shock Protein
Heat shock protein (Hsp) contributes to the interaction with steroid hormone receptors, temperature, estrogen signaling, etc., and several Hsps are critical for successful embryogenesis and reproduction (55, 56). It is reported that three Hsps (heat shock protein 27, heat shock protein 70, and heat shock protein 70 cognate3) exhibited upregulated expression patterns in the transcriptomic database of testis from M. rosenbergii (42). In the present study, four heat shock protein 90 homologs (Hsp90s) and one Hsp70 homolog were identified as upregulated transcripts, respectively. It was indicated that MroDmrt11E probably had a suppression effect on the expressions of Hsp90s and Hsp70, which may play a regulatory role in spermatogenesis of testis and gonad development.
In conclusion, many highly conserved sex-related candidate homologs were identified as upregulated or downregulated transcripts in the comparative transcriptomic library of M. rosenbergii testis after MroDmrt11E knockdown. Although these data were screened from the transcriptomic library and need to be further examined, this study provided new information on the sex-related candidate genes and their possible regulation mechanism in sexual differentiation. Notwithstanding its limitation, the putative position of MroDmrt11E involved in the signaling pathways orchestrating sexual differentiation of M. rosenbergii is worth further clarifying.
4.3 MroDmrt11E Participated in Gonad Development in M. rosenbergii
The MroDmrt11E presented testis-prominent distribution and were localized in spermatogonia and spermatozoa during spermatogenesis, suggesting that the MroDmrt11E participated in the development of the male reproductive system (26). In the present study, several cell division and proliferation-related candidate genes, including Piwi2, PL-10, PCNA, Cdk2, CycE, and CHD1 homologs, were found as upregulated genes that are previously implicated in regulating germ cells.
1. De novo transcriptomic analysis of the Japanese mantis shrimp O. oratoria showed that Piwi2 and vasa-like protein genes exhibited a higher expression level in ovaries than that in testis, suggesting that they may play an important role in germ cell differentiation (54).
2. Germline-specific vasa protein, also known as PL10-like protein, encodes an ATP-dependent RNA helicase belonging to the DEAD and is essential not only for germ cell specification during embryogenesis but also for the completion of meiosis of the germ cells in adults (57). A vasa-like gene, Mrvlg, is specifically expressed in the germ cells and differentially expressed during germ cell differentiation in both ovary and testis of M. rosenbergii (57).
3. Proliferating cell nuclear antigen (PCNA) is an evolutionarily well-conserved protein found in all eukaryotic species and plays an important role in cell cycle regulation and checkpoint control, DNA repair, translesion DNA synthesis, DNA methylation, chromatin remodeling, and gonadenogenesis in various species (58). The PCNA of Chinese shrimp F. chinensis, termed FcPCNA, was highly expressed in proliferating tissues, such as hematopoietic and ovary (59). In O. oratoria, PCNA was expressed at a higher level in the ovaries than in the testis, suggesting it may be related to oogenesis (36).
In conclusion, the upregulation of all of these cell division, proliferation, and differentiation-related candidate homologs suggested that MroDmrt11E probably participates in regulating both spermatogenesis and oogenesis. Despite this preliminary characteristic, this study indicated the important clues for further functional research of MroDmrt11E in gonad development.
4.4 The Changes of Primary Kyoto Encyclopedia of Genes and Genomes Pathways in MroDmrt11E Silencing
MroDmrt11E silencing induced dramatic sex-related alterations, including male feature feminization, extensive male reproductive system inhibition, oocyte meiosis promotion, and the ovarian development of neo-females. The phenomenon of complete and functional sex reversal individuals, induced by MroDmrt11E knockdown, showed a close relationship with various biological pathways. The pathway enrichment was used to capture the functional specialization of MroDmrt11E silencing and, based on known functional roles, validated various biological aspects in prawn and other closely related crustaceans.
Firstly, the pathway of protein processing, which happens in the endoplasmic reticulum (ER), was thought to be significantly involved in energy and substance metabolism during sexual differentiation and sex reversal. Abundant transcripts were exposed to protein processing in ER, including upregulated genes, such as Sec62/63, Sec61, Hsp70, Hsp90, P97, and UbcH5, and downregulated genes, such as Ubc6/7, GlcII, OS9, TRAM, and Skp1. Meanwhile, the mammalian target of rapamycin (mTOR) signaling pathway is also involved in the regulation of the rate of protein synthesis. The upregulated Rheb gene and downregulated EIF-4E gene were enriched. In addition, a series of upregulated factors such as PI3K, Rac, F-Actin, and α-tubulin, involved in PI3K/Akt signaling, were intensively activated and participated in the regulation of actin cytoskeleton and apoptosis. It was reported that the transcription of MroDmrt11E was prominent in testis and localized in spermatogonia and spermatozoa during spermatogenesis (26). The degeneration of the male reproductive system, induced by MroDmrt11E silencing in PL males, indicated that MroDmrt11E was a crucial regulator of testicular formation. The absence of male characteristics (male genital pores and reproductive system) in juvenile males probably had close relationships with the pathway of apoptosis and regulation of actin cytoskeleton, which is possibly involved in the development and reconstruction of the female reproductive system and ovary.
Moreover, the enrichment of KEGG’s oocyte meiosis pathway indicates its involvement in the ovarian development of neo-females. Several factors, upregulated genes, namely, Calm, CaN, CycE, and Cdk2, and one downregulated gene, SFC, were involved in the oocyte meiosis. Most genes downstream of the fertilization and translation of maternal mRNAs were upregulated, indicating MroDmrt11E plays an important role in the regulation of oocyte meiosis. Furthermore, many upregulated factors, such as Piwi2 (36), PL-10 (57), PCNA (58), Cdk2 (60), CycE, and CHD1 homologs, were all intensively activated to promote the germ cell proliferation and cell differentiation of the ovarian development by MroDmrt11E knockdown. These significant responses to primary KEGG pathways provided clues to understand the mechanism of sexual differentiation.
In conclusion, dsRNA-mediated gene knockdown of MroDmrt11E led to a complete and functional sexual reversal, which possibly established a crucial basis for the new development of manipulating monosex progeny of prawn. The data of comparative transcriptomic analysis, after MroDmrt11E silencing mediated, provided a deeper understanding of the molecular regulatory network underlying sexual differentiation in M. rosenbergii and provided a novel insight into the roles of Dmrt homologs in the sexual differentiation of crustaceans.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. RNA-sequencing data are available through the NCBI Sequence Read Archive (SRA) Database (https://www.ncbi.nlm.nih.gov/Traces/sra/), under BioProject Number: PRJNA796284. Further inquiries can be directed to the corresponding author.
Ethics Statement
The authors followed all applicable international, national, and/or institutional guidelines for the care and use of animals.
Author Contributions
W-MM conceived and designed the study and analyzed most of the data. H-JX, Y-LC, Y-MW, J-YL, J-WL, and S-QS performed the experiments. J-SY developed the idea for the study. All authors were involved in writing and approving the article.
Funding
This work was supported by the National Key Research and Development Program of China (grant number 2018YFD0900200), the National Natural Science Foundation of China (grant number 31872545), and the Top Disciplines of Zhejiang Province, Biology engineering (grant numbers ZS2021005 and ZS2021021).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to Wei-Jun Yang from the College of Life Sciences, Zhejiang University, for use of his lab facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.772498/full#supplementary-material
References
1. Shen SQ, Li JW, Xu HJ, Yang JS, Ma WM, Qian GY. Sexual Characteristic Development and Sex Identification of Juvenile Prawns, Macrobrachium Rosenbergii. Aquac Res (2020) 51:3718–28. doi: 10.1111/are.14721
2. Ventura T, Manor R, Aflalo ED, Weil S, Rosen O, Sagi A. Timing Sexual Differentiation: Full Functional Sex Reversal Achieved Through Silencing of a Single Insulin-Like Gene in the Prawn, Macrobrachium Rosenbergii. Biol Reprod (2012) 86:1–13. doi: 10.1095/biolreprod.111.097261
3. Ventura T, Manor R, Aflalo ED, Weil S, Khalaila I, Rosen O, et al. Expression of an Androgenic Gland-Specific Insulin-Like Peptide During the Course of Prawn Sexual and Morphotypic Differentiation. ISRN Endocrinol (2011) 2011:1–11. doi: 10.5402/2011/476283
4. Aflalo ED, Dandu RVSN, Verghese JT, Rao N, Samraj TYC, Ovadia O, et al. Neo-Females Production and All-Male Progeny of a Cross Between Two Indian Strains of Prawn (Macrobrachium Rosenbergii): Population Structure and Growth Performance Under Different Harvest Strategies. Aquaculture (2014) 428–9:7–15. doi: 10.1016/j.aquaculture.2014.02.021
5. Manor R, Weil S, Oren S, Glazer L, Aflalo ED, Ventura T, et al. Insulin and Gender: An Insulin-Like Gene Expressed Exclusively in the Androgenic Gland of the Male Crayfish. Gen Comp Endocrinol (2007) 150:326–36. doi: 10.1016/j.ygcen.2006.09.006
6. Rosen O, Manor R, Weil S, Gafni O, Linial A, Aflalo ED, et al. A Sexual Shift Induced by Silencing of a Single Insulin-Like Gene in Crayfish: Ovarian Upregulation and Testicular Degeneration. PloS One (2010) 5:1–10. doi: 10.1371/journal.pone.0015281
7. Mareddy VR, Rosen O, Thaggard HB, Manor R, Kuballa AV, Aflalo ED, et al. Isolation and Characterization of the Complete cDNA Sequence Encoding a Putative Insulin-Like Peptide From the Androgenic Gland of Penaeus Monodon. Aquaculture (2011) 318:364–70. doi: 10.1016/j.aquaculture.2011.05.027
8. Ventura T, Rosen O, Sagi A. From the Discovery of the Crustacean Androgenic Gland to the Insulin-Like Hormone in Six Decades. Gen Comp Endocrinol (2011) 173:381–8. doi: 10.1016/j.ygcen.2011.05.018
9. Ventura T, Manor R, Aflalo ED, Weil S, Raviv S, Glazer L, et al. Temporal Silencing of an Androgenic Gland-Specific Insulin-Like Gene Affecting Phenotypical Gender Differences and Spermatogenesis. Endocrinology (2009) 150:1278–86. doi: 10.1210/en.2008-0906
10. Tan K, Zhou M, Jiang H, Jiang D, Li Y, Wang W. siRNA-Mediated MrIAG Silencing Induces Sex Reversal in Macrobrachium Rosenbergii. Mar Biotechnol (2020) 22. doi: 10.1007/s10126-020-09965-4
11. Tan K, Li Y, Zhou M, Wang W. siRNA Knockdown of MrIR Induces Sex Reversal in Macrobrachium Rosenbergii. Aquaculture (2020) 523:735172. doi: 10.1016/j.scitotenv.2020.140326
12. Levy T, Sagi A. The “IAG-Switch”—A Key Controlling Element in Decapod Crustacean Sex Differentiation. Front Endocrinol (Lausanne) (2020) 11:651. doi: 10.3389/fendo.2020.00651
13. Farhadi A, Cui W, Zheng H, Li S, Zhang Y, Ikhwanuddin M, et al. The Regulatory Mechanism of Sexual Development in Decapod Crustaceans. Front Mar Sci (2021) 8:679687. doi: 10.3389/fmars.2021.679687
14. Kopp A. Dmrt Genes in the Development and Evolution of Sexual Dimorphism. Trends Genet (2012) 28:175–84. doi: 10.1016/j.tig.2012.02.002.Dmrt
15. Picard MA-L, Cosseau C, Mouahid G, Duval D, Grunau C, Toulza È., et al. The Roles of Dmrt (Double Sex/Male-Abnormal-3 Related Transcription Factor) Genes in Sex Determination and Differentiation Mechanisms: Ubiquity and Diversity Across the Animal Kingdom. C R Biol (2015) 338:451–62. doi: 10.1016/j.crvi.2015.04.010
16. Fujii T, Shimada T. Sex Determination in the Silkworm, Bombyx Mori: A Female Determinant on the W Chromosome and the Sex-Determining Gene Cascade. Semin Cell Dev Biol (2007) 18:379–88. doi: 10.1016/j.semcdb.2007.02.008
17. Toyota K, Kato Y, Sato M, Sugiura N, Miyagawa S, Miyakawa H, et al. Molecular Cloning of Doublesex Genes of Four Cladocera (Water Flea) Species. BMC Genomics (2013) 14:239. doi: 10.1186/1471-2164-14-239
18. Verhulst EC, Van de zande L. Double Nexus-Doublesex Is the Connecting Element in Sex Determination. Brief Funct Genomics (2015) 14:396–406. doi: 10.1093/bfgp/elv005
19. Yi W, Ross JM, Zarkower D. Mab-3 Is a Direct Tra-1 Target Gene Regulating Diverse Aspects of C. Elegans Male Sexual Development and Behavior. Development (2000) 127:4469–80. doi: 10.1242/dev.127.20.4469
20. Huang S, Ye L, Chen H. Sex Determination and Maintenance: The Role of DMRT1 and FOXL2. Asian J Androl (2017) 19:619–24. doi: 10.4103/1008-682X.194420
21. Millington JW, Rideout EJ. Sex Differences in Drosophila Development and Physiology. Curr Opin Physiol (2018) 6:46–56. doi: 10.1016/j.cophys.2018.04.002
22. Zhang EF, Qiu GF. A Novel Dmrt Gene Is Specifically Expressed in the Testis of Chinese Mitten Crab, Eriocheir Sinensis. Dev Genes Evol (2010) 220:151–9. doi: 10.1007/s00427-010-0336-2
23. Chandler JC, Fitzgibbon QP, Smith G, Elizur A, Ventura T. ). Y-Linked Idmrt1 Paralogue (iDMY) in the Eastern Spiny Lobster, Sagmariasus Verreauxi: The First Invertebrate Sex-Linked Dmrt. Dev Biol (2017) 430:337–45. doi: 10.1016/j.ydbio.2017.08.031
24. Li S, Li F, Yu K, Xiang J. Identification and Characterization of a Doublesex Gene Which Regulates the Expression of Insulin-Like Androgenic Gland Hormone in. Fenneropenaeus chinensis Gene (2018) 649:1–7. doi: 10.1016/j.gene.2018.01.043
25. Wang Y, Jin S, Fu H, Qiao H, Sun S, Zhang W, et al. Identification and Characterization of the DMRT11E Gene in the Oriental River Prawn Macrobrachium Nipponense. Int J Mol Sci (2019) 20:1–17. doi: 10.3390/ijms20071734
26. Yu YQ, Ma WM, Zeng QG, Qian YQ, Yang JS, Yang WJ. Molecular Cloning and Sexually Dimorphic Expression of Two Dmrt Genes in the Giant Freshwater Prawn, Macrobrachium Rosenbergii. Agric Res (2014) 3:181–91. doi: 10.1007/s40003-014-0106-x
27. Amterat Abu Abayed F, Manor R, Aflalo ED, Sagi A. Screening for Dmrt Genes From Embryo to Mature Macrobrachium Rosenbergii Prawns. Gen Comp Endocrinol (2019) 282:113205. doi: 10.1016/j.ygcen.2019.06.009
28. Zhong P, Zhou T, Zhang Y, Chen Y, Yi J, Lin W, et al. Potential Involvement of a DMRT Family Member (Mr-Dsx) in the Regulation of Sexual Differentiation and Moulting in the Giant River Prawn Macrobrachium Rosenbergii. Aquac Res (2019) 00:1–13. doi: 10.1111/are.14262
29. Jiang XH, Qiu GF. Female-Only Sex-Linked Amplified Fragment Length Polymorphism Markers Support ZW/ZZ Sex Determination in the Giant Freshwater Prawn Macrobrachium Rosenbergii. Anim Genet (2013) 44:782–5. doi: 10.1111/age.12067
30. Aflalo ED, Hoang TTT, Nguyen VH, Lam Q, Nguyen DM, Trinh Q, et al. A Novel Two-Step Procedure for Mass Production of All-Male Populations of the Giant Freshwater Prawn Macrobrachium rosenbergii. Aquaculture (2006) 256:468–78. doi: 10.1016/j.aquaculture.2006.01.03
31. Hubbard SR, Till JH. Protein Tyrosine Kinase Structure and Function. Annu Rev Biochem (2000) 69:373–93. doi: 10.1146/annurev.biochem.69.1.373
32. Gricourt L, Mathieu M, Kellner K. An Insulin-Like System Involved in the Control of Pacific Oyster Crassostrea Gigas Reproduction: hrIGF-1 Effect on Germinal Cell Proliferation and Maturation Associated With Expression of an Homologous Insulin Receptor-Related Receptor. Aquaculture (2006) 251:85–98. doi: 10.1016/j.aquaculture.2005.05.015
33. Sharabi O, Manor R, Weil S, Aflalo ED, Lezer Y, Levy T, et al. Identification and Characterization of an Insulin-Like Receptor Involved in Crustacean Reproduction. Endocrinology (2016) 157:928–41. doi: 10.1210/en.2015-1391
34. Guo Q, Li S, Lv X, Xiang J, Sagi A, Manor R, et al. A Putative Insulin-Like Androgenic Gland Hormone Receptor Gene Specifically Expressed in Male Chinese Shrimp. Endocrinology (2018) 159:2173–85. doi: 10.1210/en.2017-03253
35. Dai ZM, Zhu XJ, Yang WJ. Full-Length Normalization Subtractive Hybridization: A Novel Method for Generating Differentially Expressed cDNAs. Mol Biotechnol (2009) 43:257–63. doi: 10.1007/s12033-009-9198-0
36. Yan H, Cui X, Shen X, Wang L, Jiang L, Liu H, et al. De Novo Transcriptome Analysis and Differentially Expressed Genes in the Ovary and Testis of the Japanese Mantis Shrimp Oratosquilla Oratoria by RNA-Seq. Comp Biochem Physiol - Part D Genomics Proteomics (2018) 26:69–78. doi: 10.1016/j.cbd.2018.04.001
37. Pomiankowski A, Nöthiger R, Wilkins A. The Evolution of the Drosophila Sex-Determination Pathway. Genetics (2004) 166:1761–73. doi: 10.1534/genetics.166.4.1761
38. Sánchez L. Sex-Determining Mechanisms in Insects. Int J Dev Biol (2008) 52:837–56. doi: 10.1387/ijdb.072396ls
39. Sun X, Yang H, Sturgill D, Oliver B, Rabinow L, Samson ML. Sxl-Dependent, Tra/Tra2-Independent Alternative Splicing of the Drosophila Melanogaster X-Linked Gene Found in Neurons. G3 Genes Genomes Genet (2015) 5:2865–74. doi: 10.1534/g3.115.023721
40. Yu Y. The Molecular Characterization and Functional Analysis of Sexual Development Related Genes Sxl and Dmrt in the Prawn, Macrobrachium Rosenbergii. Hangzhou, Zhejiang: Zhejiang University. (2013).
41. Ventura T, Chandler JC, Nguyen TV, Hyde CJ, Elizur A, Fitzgibbon QP, et al. Multi-Tissue Transcriptome Analysis Identifies Key Sexual Development-Related Genes of the Ornate Spiny Lobster (Panulirus Ornatus). Genes (Basel) (2020) 11:1–17. doi: 10.3390/genes11101150
42. Jiang J, Yuan X, Qiu Q, Huang G, Jiang Q, Fu P, et al. Comparative Transcriptome Analysis of Gonads for the Identification of Sex-Related Genes in Giant Freshwater Prawns (Macrobrachium Rosenbergii) Using RNA Sequencing. Genes (Basel) (2019) 10:1–18. doi: 10.3390/genes10121035
43. Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D. DMRT1 Prevents Female Reprogramming in the Postnatal Mammalian Testis. Nature (2011) 476:101–5. doi: 10.1038/nature10239
44. Farhadi A, Fang S, Zhang Y, Cui W, Fang H, Ikhwanuddin M, et al. The Significant Sex-Biased Expression Pattern of Sp-Wnt4 Provides Novel Insights Into the Ovarian Development of Mud Crab (Scylla Paramamosain). Int J Biol Macromol (2021) 183:490–501. doi: 10.1016/j.ijbiomac.2021.04.186
45. Fan Z, Zou Y, Liang D, Tan X, Jiao S, Wu Z, et al. Roles of Forkhead Box Protein L2 (Foxl2) During Gonad Differentiation and Maintenance in a Fish, the Olive Flounder (Paralichthys Olivaceus). Reprod Fertil Dev (2019) 31:1742–52. doi: 10.1071/RD18233
46. Gilbert LI. Halloween Genes Encode P450 Enzymes That Mediate Steroid Hormone Biosynthesis in Drosophila Melanogaster. Mol Cell Endocrinol (2004) 215:1–10. doi: 10.1016/j.mce.2003.11.003
47. Lazar MA. Maturing of the Nuclear Receptor Family. J Clin Invest (2017) 127:1123–5. doi: 10.1172/JCI92949
48. Finger DS, Whitehead KM, Phipps DN, Ables ET. Nuclear Receptors Linking Physiology and Germline Stem Cells in Drosophila. Vitam Horm (2021) 116:327–62. doi: 10.1016/bs.vh.2020.12.008.Nuclear
49. Kiuchi T, Koga H, Kawamoto M, Shoji K, Sakai H, Arai Y, et al. A Single Female-Specific piRNA Is the Primary Determiner of Sex in the Silkworm. Nature (2014) 509:633–6. doi: 10.1038/nature13315
50. Zhou LX, Liu X, Ye BQ, Liu Y, Tan SP, Ma KY, et al. Molecular Characterization of Ovary-Specific Gene Mrfem-1 and siRNA-Mediated Regulation on Targeting Mrfem-1 in the Giant Freshwater Prawn, Macrobrachium Rosenbergii. Gene (2020) 754:144891. doi: 10.1016/j.gene.2020.144891
51. Ma KY, Liu ZQ, Lin JY, Li JL, Qiu GF. Molecular Characterization of a Novel Ovary-Specific Gene Fem-1 Homolog From the Oriental River Prawn, Macrobrachium Nipponense. Gene (2016) 575:244–52. doi: 10.1016/j.gene.2015.08.070
52. Qiu GF, Yamano K, Unuma T. ). Cathepsin C Transcripts Are Differentially Expressed in the Final Stages of Oocyte Maturation in Kuruma Prawn Marsupenaeus Japonicus. Comp Biochem Physiol - B Biochem Mol Biol (2005) 140:171–81. doi: 10.1016/j.cbpc.2004.09.027
53. Zhao W, Chen L, Zhang F, Wu P, Li E, Qin J. Molecular Characterization of Cathepsin L cDNA and Its Expression During Oogenesis and Embryogenesis in the Oriental River Prawn Macrobrachium Nipponense (Palaemonidae). Genet Mol Res (2013) 12:5215–25. doi: 10.4238/2013.October.30.6
54. Yang X, Ikhwanuddin M, Li X, Lin F, Wu Q, Zhang Y, et al. Comparative Transcriptome Analysis Providesinsights Into Differentially Expressed Genes and Long Non-Coding RNAs Between Ovary and Testis of the Mud Crab (Scylla Paramamosain). Mar Biotechnol (2018) 20:20–34. doi: 10.1007/s10126-017-9784-2
55. Christians ES. Heat Shock Proteins and Maternal Contribution to Oogenesis and Early Embryogenesis. Adv Anat Embryol Cell Biol (2017) 222:1–27. doi: 10.1007/978-3-319-51409-3_1
56. Jee B, Dhar R, Singh S, Karmakar S. Heat Shock Proteins and Their Role in Pregnancy: Redefining the Function of “Old Rum in a New Bottle”. Front Cell Dev Biol (2021) 9:648463. doi: 10.3389/fcell.2021.648463
57. Nakkrasae L-I, Damrongphol P. A Vasa-Like Gene in the Giant Freshwater Prawn, Macrobrachium rosenbergii. Mol Reprod Dev (2007) 1385:8–9. doi: 10.1002/mrd
58. Park SY, Jeong MS, Han CW, Yu HS, Jang SB. Structural and Functional Insight Into Proliferating Cell Nuclear Antigen. J Microbiol Biotechnol (2016) 26:637–47. doi: 10.4014/jmb.1509.09051
59. Xie Y, Wang B, Li F, Jiang H, Xiang J. Molecular Cloning and Characterization of Proliferating Cell Nuclear Antigen (PCNA) From Chinese Shrimp Fenneropenaeus Chinensis. Comp Biochem Physiol - B Biochem Mol Biol (2008) 151:225–9. doi: 10.1016/j.cbpb.2008.07.006
Keywords: MroDmrt11E gene knockdown, Macrobrachium rosenbergii, sex reversal, sexual differentiation, all-male monosex population
Citation: Xu H-J, Chen Y-L, Wang Y-M, Luo J-Y, Li J-W, Shen S-Q, Yang J-S and Ma W-M (2022) Full Functional Sex Reversal Achieved Through Silencing of MroDmrt11E Gene in Macrobrachium rosenbergii: Production of All-Male Monosex Freshwater Prawn. Front. Endocrinol. 12:772498. doi: 10.3389/fendo.2021.772498
Received: 08 September 2021; Accepted: 15 December 2021;
Published: 17 March 2022.
Edited by:
Shubo Jin, Chinese Academy of Fishery Sciences, ChinaCopyright © 2022 Xu, Chen, Wang, Luo, Li, Shen, Yang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Ming Ma, bS13bTAyQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Hai-Jing Xu1†
Hai-Jing Xu1† Wen-Ming Ma
Wen-Ming Ma