- 1First Affiliated Hospital of University of Science and Technology of China (USTC), Hefei National Laboratory for Physical Sciences at Microscale, School of Basic Medical Sciences, Division of Life Sciences and Medicine, Chinese Academy of Sciences (CAS) Center for Excellence in Molecular Cell Science, University of Science and Technology of China, Hefei, China
- 2Medical Genetics Research Laboratory, Department of Biotechnology, Quaid-i-Azam University, Islamabad, Pakistan
- 3Reproductive and Genetic Hospital, The First Affiliated Hospital of University of Science and Technology of China (USTC), Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
Male infertility is a prevalent disorder distressing an estimated 70 million people worldwide. Despite continued progress in understanding the causes of male infertility, idiopathic sperm abnormalities such as multiple morphological abnormalities of sperm flagella (MMAF) still account for about 30% of male infertility. Recurrent mutations in DNAH1 have been reported to cause MMAF in various populations, but the underlying mechanism is still poorly explored. This study investigated the MMAF phenotype of two extended consanguineous Pakistani families without manifesting primary ciliary dyskinesia symptoms. The transmission electron microscopy analysis of cross-sections of microtubule doublets revealed a missing central singlet of microtubules and a disorganized fibrous sheath. SPAG6 staining, a marker generally used to check the integration of microtubules of central pair, further confirmed the disruption of central pair in the spermatozoa of patients. Thus, whole-exome sequencing (WES) was performed, and WES analysis identified two novel mutations in the DNAH1 gene that were recessively co-segregating with MMAF phenotype in both families. To mechanistically study the impact of identified mutation, we generated Dnah1 mice models to confirm the in vivo effects of identified mutations. Though Dnah1△iso1/△iso1 mutant mice represented MMAF phenotype, no significant defects were observed in the ultrastructure of mutant mice spermatozoa. Interestingly, we found DNAH1 isoform2 in Dnah1△iso1/△iso1 mutant mice that may be mediating the formation of normal ultrastructure in the absence of full-length protein. Altogether we are first reporting the possible explanation of inconsistency between mouse and human DNAH1 mutant phenotypes, which will pave the way for further understanding of the underlying pathophysiological mechanism of MMAF.
Introduction
Male infertility is a global health concern with social and psychological impact on more than 70 million infertile men (1). Recent studies have demonstrated that sperm defects, such as reduced sperm count, decreased sperm motility, and abnormal morphology, are directly linked with male infertility (2). Sperm motility is required for normal fertilization, and around 80% of infertility is caused by compromised sperm motility (3). Subsequently, sperm morphology is also an important player necessary for sperm locomotion, and defects in sperm morphologies are associated with a major form of male infertility (4). Defects in sperm morphology are generally manifested with a wide range of phenotypic variations in the head, neck, mid-piece, and tail of spermatozoa. Among these, the most common type of abnormities is found in the form of multiple morphological abnormalities of sperm flagella (MMAF) in asthenospermia patients. The most common defects of sperm flagella observed in MMAF phenotype include coiled, bent, irregular, short, absent, or irregular-width flagella (5). Normal sperm flagella contain a typical axonemal arrangement of nine peripheral microtubules and two central microtubules (CP), which are further surrounded by dense outer fiber and fibrous sheath (6). This basic structure of flagella remains conserved through the process of evolution. Any structural aberrations in these components of sperm flagella are associated with a wide range of MMAF phenotypes which further caused a reduction of sperm motility or even sperm immobility and eventually male infertility.
DNAH1 (MIM 603332) is an essential gene that encodes inner dynein arm (IDA) heavy chain, which are believed to strengthen the link between radial spokes and outer doublet (7). IDA, an ATPase-based protein complex, is responsible for beat generation and regulation of flagellar movement (8, 9). Recurrent mutations in DNAH1 are commonly associated with MMAF in various populations (7, 10, 11). Moreover, researchers are extensively investigating the molecular mechanism of MMAF by generating knockout (KO) mice models of various genes that have been associated with MMAF phenotype in humans (6). Until now, mutations in various genes, such as CFAP43, CFAP44, CFAP47, CFAP58, CFAP65, CFAP69, CFAP70, CFAP74, CFAP91, CFAP206, CFAP251, DNAH1, DNAH2, DNAH6, DNAH8, DNAH10, DNAH17, WDR19, ARMC2, TTC21A, TTC29, FSIP2, AK7, CEP135, SPEF2, QRICH2, DZIP1, BRWD1, DRC1, STK33, etc., have been reported with MMAF phenotype (10, 12–25). However, a discrepancy exists in the Dnah1 KO mouse model and human phenotype (26, 27). A previous study demonstrated that Dnah1 KO mice are completely infertile due to defects in sperm motility, but the ultrastructure of flagella did not show any abnormalities in their components (26). On the other hand, DNAH1 mutant human sperm ultrastructure was severely compromised because of lacking CP and other defects. Thus, uncovering the inconsistency between human and mouse phenotypes of the DNAH1 gene required a deeper study to discover the unknown isoforms mediating the intact formation of normal ultrastructure of mouse sperm flagella in the absence of full-length DNAH1 protein.
In the present study, we recruited two extended consanguineous Pakistani families suffering from male infertility due to the MMAF phenotype. Subsequently, whole-exome sequencing (WES) identified two novel mutations in the DNAH1 gene that were recessively co-segregating with infertility phenotype in both families. Furthermore, we generated Dnah1 mice models to confirm the in vivo effects of identified mutations. Interestingly, we found Dnah1 isoform2 in Dnah1△iso1/△iso1 mutant mice that may be mediating the formation of normal ultrastructure in the absence of full-length protein. Altogether we are the first one to present the possible explanation of inconsistency between mouse and human DNAH1 mutant phenotype, which will pave the way for further understanding of the underlying pathophysiological mechanism of MMAF.
Materials and Methods
Recruitment of Families and Phenotype Confirmation
Two consanguineous families from Pakistan, comprising six male patients, were enrolled to investigate the underlying genetic cause of male infertility. Detailed pedigrees were constructed according to the provided information by the parents of the patients. An initial physical and andrological examination displayed that all patients have normal body mass index and no primary ciliary dyskinesia (PCD) symptoms. Other associated diseases, such as hypogonadotropic hypogonadism, cryptorchidism, varicocele, seminal ductal obstruction, testicular trauma, and andrological tumor, were also not observed. Two repeated semen analyses were performed according to the WHO manual of 2010, and all the parameters were recorded (Table 1). We also recruited the fertile siblings of patients and their parents to serve as a positive control for genetic analysis. The study was approved by a bioethical committee of Pakistan and the ethical review board of the University of Science and Technology of China. Informed consent was obtained from all participants of both families.
WES and Linkage Analysis
Genomic DNA was extracted from all available members of both families by using QIAamp Blood DNA Mini Kit (QIAGEN) as per the protocol of the manufacturer. WES was conducted on III:2, IV:1, III:3, V:1, V:2, V:3, and V:5 from family members of PK-INF-15 and PK-INF-319, respectively, previously reported in (28). Briefly, WES data from the selected members were enriched by the Agilent SureSelect XT Human All Exon Kit. The Illumina HiSeq XTEN platform accompanied next-generation sequencing. The obtained raw reads were aligned to the human genome reference assembly (GRCh37/hg19) using the Burrows-Wheeler Aligner (29). Next, the Picard software was engaged to remove polymerase chain reaction (PCR) duplicates and evaluate the quality of variants. DNA sequence variants were analyzed by the best practice genome analysis kit (30). VCF files were used for parametric linkage analysis, as described previously (31), and four linkage regions were identified with a logarithm of odds (LOD) score of more than 0.5. Further screening was opted for the variants that were located on linkage regions and following Mendelian inheritance pattern.
Filtering and Selection of Candidate Variants
Only variants that have depth >×20, genotype quality >90, and 0.5-cM intervals between each other were designated as markers. MERLIN software was employed on genotyped single-nucleotide polymorphism for linkage analysis with these parameters: due to consanguinity in the families, recessive mode of inheritance was adopted, with a disease allele frequency of 0.001 and 100% penetrance. A number of five peaks with a LOD score of more than 0.5 were considered as linkage regions. The variants that resided in the linkage regions were further annotated by ANNOVAR using the NCBI RefSeq gene annotation. We adopted the following filtration strategy to select potential candidate disease-causing variants in patients: (i) variants that were heterozygous in the father and control brothers and homozygous in patients were retained; (ii) variants with minor allele frequency of more than 0.01 in public databases such as 1000 Genome project (32), ESP6500 (33), or ExAC database (34), and variants homozygous in our in-house WES variants call set generated from 578 fertile male samples (41 Pakistanis, 254 Chinese, and 283 Europeans) were excluded; (iii) variants that affect protein sequence and predicted to be deleterious by around 10 of 13 in silico employed tools were kept; and (iv) variants which have a predominant expression in the testes and their inactivation affects male fertility were retained based on spermatogenesis online database (35) and were checked by Sanger sequencing to verify Mendel inheritance pattern. All these strategies are more clearly explained in a flow chart diagram in Supplementary Figure S2. Finally, only DNAH1 variants followed the inheritance pattern and thus were considered the potential candidates causing MMAF phenotype in subjects under investigation. The list of primers used for Sanger sequencing is shown in Supplementary Table S1.
Immunofluorescence Staining
Immunofluorescence on the spermatozoa of patients was performed, as we have previously reported (36). Briefly, sperm from patients were smeared on the clean slide and fixed with 4% paraformaldehyde followed by three times of washing in phosphate-buffered saline (PBS). The slides were permeabilized with 0.5% Triton X-100 for 30 min and blocked with 1% BSA. Next, the slides were incubated with primary antibodies, including α-tubulin (Sigma, F2168), DNAH1 (Abcam, ab122367), and SPAG6 (Proteintech, 12462-1-AP), overnight at 4°C. On the next day, the slides were washed with PBST (PBS containing Triton X-100) and incubated with secondary antibodies DAR555 (Molecular Probes, A31572) and GAM488 (Molecular Probes, A21121) for 1 h at 37°C. Finally, the slides were washed again and sealed with Hoechst and Vectashield. Images were captured by using a laser scanning confocal microscope (Olympus).
Electron Microscopy Analysis
Sperm samples from patients were processed in 0.1 M phosphate buffer (pH 7.4) containing 4% paraformaldehyde, 8% glutaraldehyde, and 0.2% picric acid at 4°C. Scanning and transmission electron microscopy (SEM and TEM) were performed as previously described, with minor modifications (36). Briefly, spermatozoa were washed in 0.1 M PBS four times, followed by fixation in 1% OsO4 and dehydration. Then, samples were infiltrated for acetone and epon resin mixture and embedded in sputter-coated (SCD 500, Bal-Tel). Thinly sectioned (70 mm) samples were stained with uranyl acetate and lead citrate. The morphology of spermatozoa was examined by Hitachi S-4800 field emission scanning electron microscope under an accelerating voltage of 15 kV. The ultrastructure of the samples was analyzed and captured by Tecnai 10 or 12Microscope (Philips) at 100 or 120 kV or by H-7650 microscope (Hitachi) at 100 kV.
Generation of Mutant Mice Models
Dnah1-/- and Dnah1△iso1/△iso1 mice were generated by using CRISPR/Cas9 genome editing technology as previously reported (37). Briefly, single guide RNAs (sgRNAs) designed to target exon 2 and 49 of Dnah1 to generate KO and mutant mice, respectively, were transcribed in vitro (Addgene, 5132). Single-strand oligodeoxynucleotides (ssODNs), with a mutation similar to family A2 patients and a synonymous mutation at the protospacer adjacent motif, were created by the Sangon Biotech system. The designed sgRNAs and ssODNs were microinjected with Cas9 mRNAs into the zygotes of B6D2F1 (C57BL/6×DBA/2J) mice, following a previously published methodology (38). The genotyping of newborn pups was determined by Sanger sequencing, and heterozygous Dnah1+/- and Dnah1+/△iso1 mice were bred to obtain the desired homozygous results of Dnah1-/- and Dnah1△iso1/△iso1 mice for future experiments. The sequence of guide RNAs and all the primers used for genotyping is shown in Supplementary Table S1.
Western Blotting
Testes from 8-week-old Dnah1-/- and Dnah1△iso1/△iso1 mice were detached and processed in radio-immunoprecipitation assay buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate) complemented with protease inhibitors cocktail using a Tissuelyzer and then cleared by centrifugation. Western blotting was performed as previously reported (39). Nitrocellulose membranes were incubated with primary antibodies against DNAH1 (this study) and GAPDH overnight at 4°C. On the next day, membranes were washed in PBST and incubated with secondary antibodies for 1 h at room temperature. The blots were developed with chemiluminescence (Image Quant LAS 4000, GE Healthcare).
RNA Extraction and Reverse Transcriptase-PCR
According to the protocol of the manufacturer, total RNA from mice and control human testes was extracted by using Trizol (Invitrogen) kit. RT-PCR was conducted as we have previously described (40). The primer used for RT-PCR is listed in Supplementary Table S1.
Phenotypic Investigation of Mutant Mice
The fertility status of Dnah1△iso1/△iso1 mice was assessed by caging each mutant mouse with two wild-type females (C57/BL) for 3 months. A total of five Dnah1△iso1/△iso1 mice were tested, and none of them produce a single pup (Table 2). Testes morphology, sperm count, H&E, sperm morphology, and sperm motility were performed as previously described (36, 39).
Results
Recruitment of Infertile Patients Manifesting MMAF Phenotype
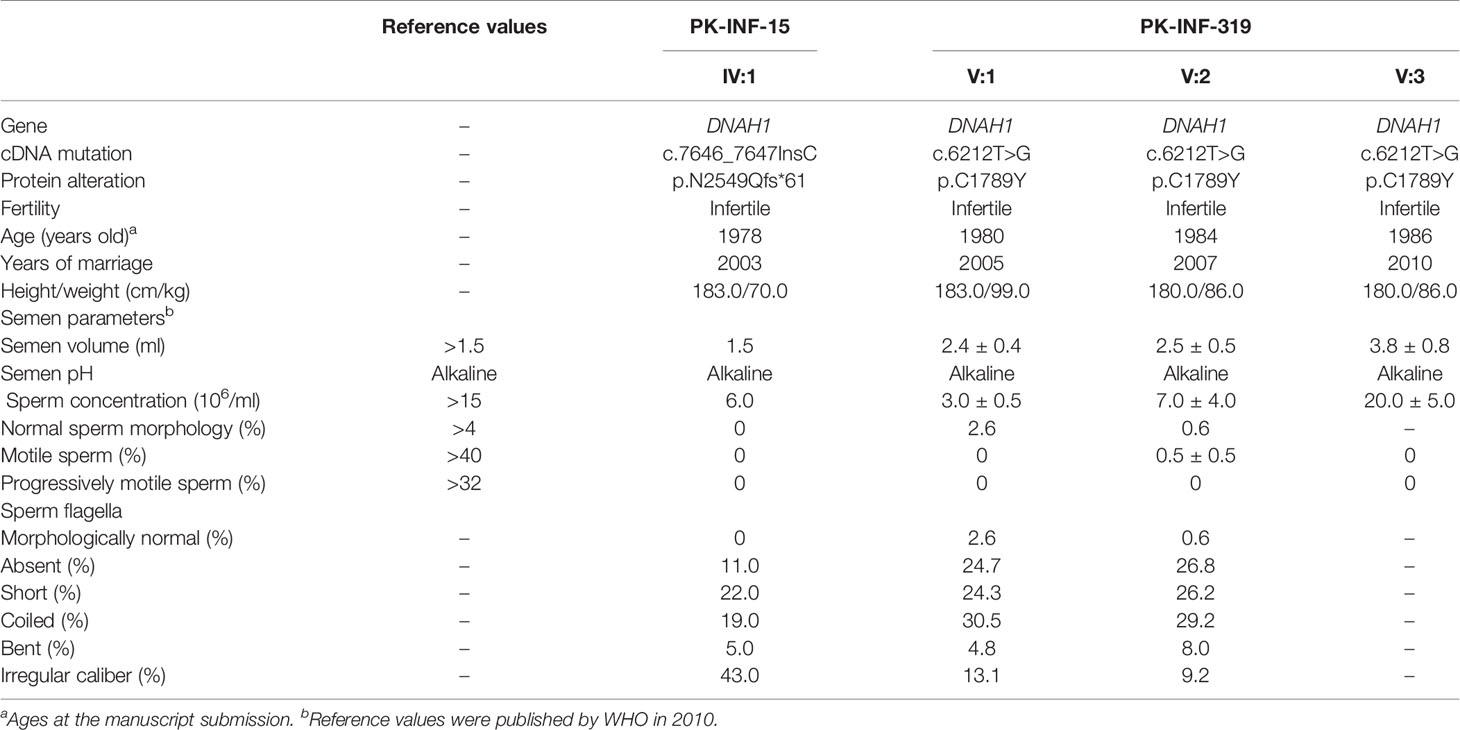
In the present study, we recruited two infertile consanguineous Pakistani families (PK-INF-15 and PK-INF-319) consisting of six patients that were suffering from primary infertility (Figure 1A). The semen analysis of four patients showed a sufficient sperm concentration, while all sperms were immotile (Table 1). SEM and light microscopy investigation revealed abnormalities in sperm tail, displaying different defects of sperm flagella, including absent, short, coiled, bent, and irregular caliber, suggesting MMAF (Supplementary Figures S1A, B). Similarly, the SEM of sperm from patients also confirmed the MMAF phenotype (Figure 1B). Altogether these results indicated that MMAF accompanies the underlying cause of infertility in these families.
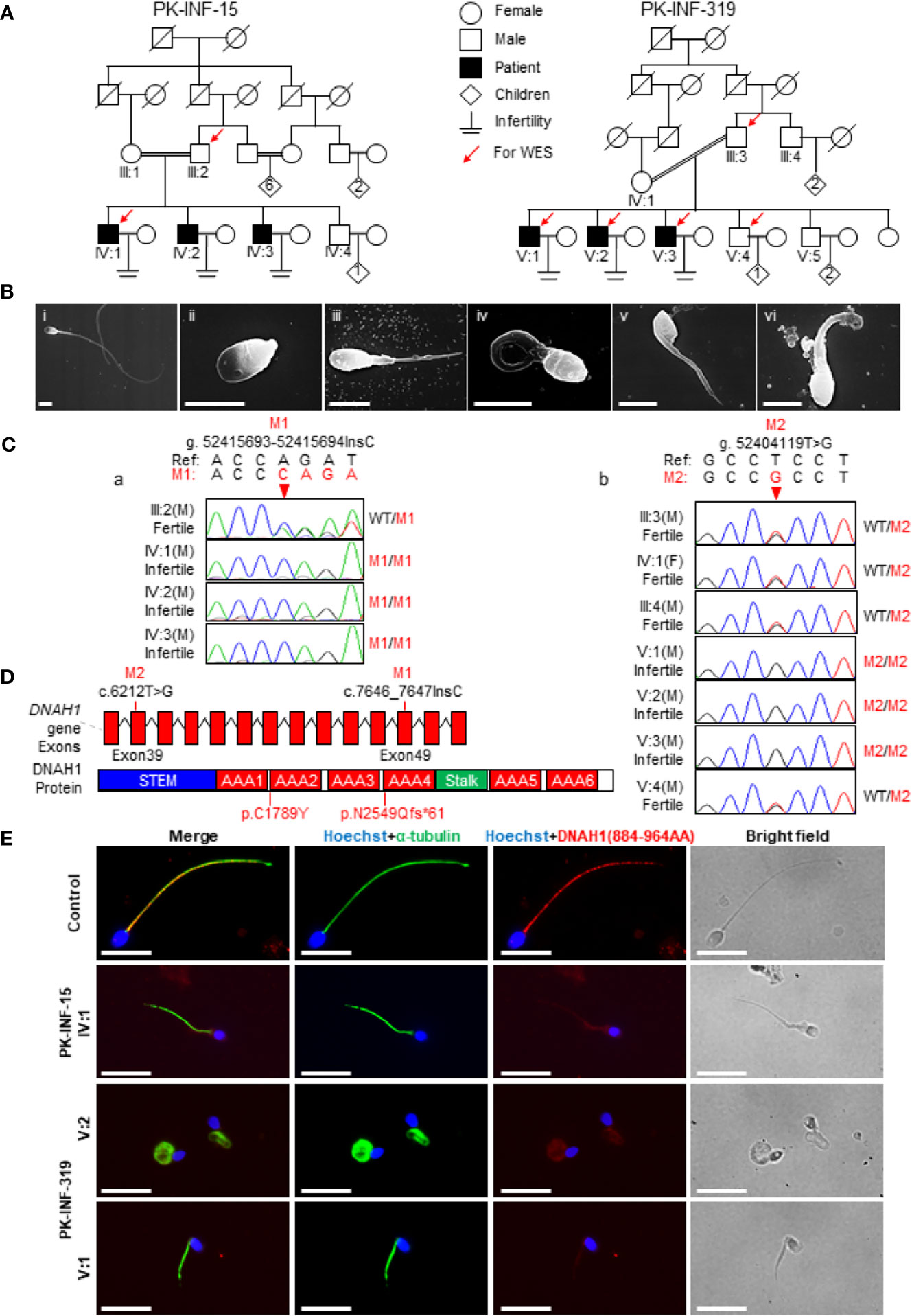
Figure 1 Recruitment of infertile families displaying multiple morphological abnormalities of sperm flagella phenotype. (A) Two consanguineous Pakistani infertile families (PK-INF-15 and PK-INF-319), comprising six male patients, were recruited. Male individuals are represented by squares, while female individuals were indicated with a circle. Diamonds indicate offspring, and the numerals inside designate the number of offspring. Solid squares are used to indicate patients, slashes represent deceased family members, and parallel slash lines specify consanguineous marriages. The red arrows indicate the members selected for whole-exome sequencing. (B) Scanning electron microscopy of sperm control (i) and from patients [PK-INF-319], V:1 (ii, ii, and iv) and V:2 (v and vi)] displayed various defects of flagella, including absent, short, coiled, and irregular calibers. Scale bar, 10 um. (C) Chromatograms representing the segregation of DNAH1 mutations in available members from families (a) PK-INF-15 and (b) PK-INF-319. The red arrows show the genomic position of DNAH1 mutations. (D) An illustrative representation of DNAH1 gene and subsequent protein structure showing the identified mutation positions. (E) No DNAH1 signals were found on paraformaldehyde-fixed sperm from a patient stained against DNAH1 and α-tubulin, while intact DNAH1 signals were observed on sperm obtained from a healthy control. Scale bar, 10 um.
Whole-Exome-Identified Novel Loss-of-Function Mutations in DNAH1
In order to explore the underlying genetic cause of infertility, WES was performed on four patients, the control brother and their parents (III:2, IV:1, III:3, V:1, V:2, V:3, and V:4). Considering the consanguinity within the families, we screened the WES data and only retained the variants that displayed recessive inherence patterns. Furthermore, we filter the variants based on minor allele frequency, testis-specific expression, and review of literature about the genes that have an association with the MMAF phenotype. In short, we identified two novel mutations in DNAH1: one was the insertion of C (c.7646_7647InsC, p.N2549Qfs*61), causing a truncation of protein in the family A1, and the other was a missense mutation (c.6212T>G, p.C1789Y) in family A2 (Figure 1D). Sanger sequencing on DNA from all available members of both families indicated that the mutations are recessively segregating in all patients with infertility phenotype (Figures 1Ca, b). To check the effect of these mutations on protein function, we performed immunofluorescence (IF) of DNAH1 and alpha-tubulin on sperm smear slides from patients IV:1, V:1, and V:2. The IF result showed the absence of DNAH1 signals on the sperm tail of patients, while positive signals of DNAH1 were observed on the control sample (Figure 1E). Thus, these results indicated that the identified mutations are loss-of-function mutations and caused transcript decay by non-sense mRNA-mediated decay.
Ultrastructural Analysis of Spermatozoa Displayed Numerous Defects in the Axonemal Structures
In order to investigate the axonemal structures, we performed TEM on patient (IV:1) spermatozoa. We statistically analyzed good-quality cross-sections of microtubule doublets from the patient and control spermatozoa samples and found that the central singlet of microtubules was missing in 66% of these sections (Figures 2A, B). Furthermore, the fibrous sheath was disorganized in 10% of the sections, and missing microtubule doublets were observed in 6% of analyzed sections. To further confirm the disruption of the central pair, we stained the sperm smear slides of the patients with SPAG6 antibody, which is a well-known marker generally used to check the integration of microtubules of the central pair. No signals of SPAG6 were observed on the sperm flagella of the patients, while sperm from the control individual displayed well-decorated signals on the entire tail of the flagella (Figure 2C). Overall, these results intimidated that the identified mutations have a deleterious effect on the axonemal structures of sperm flagella.
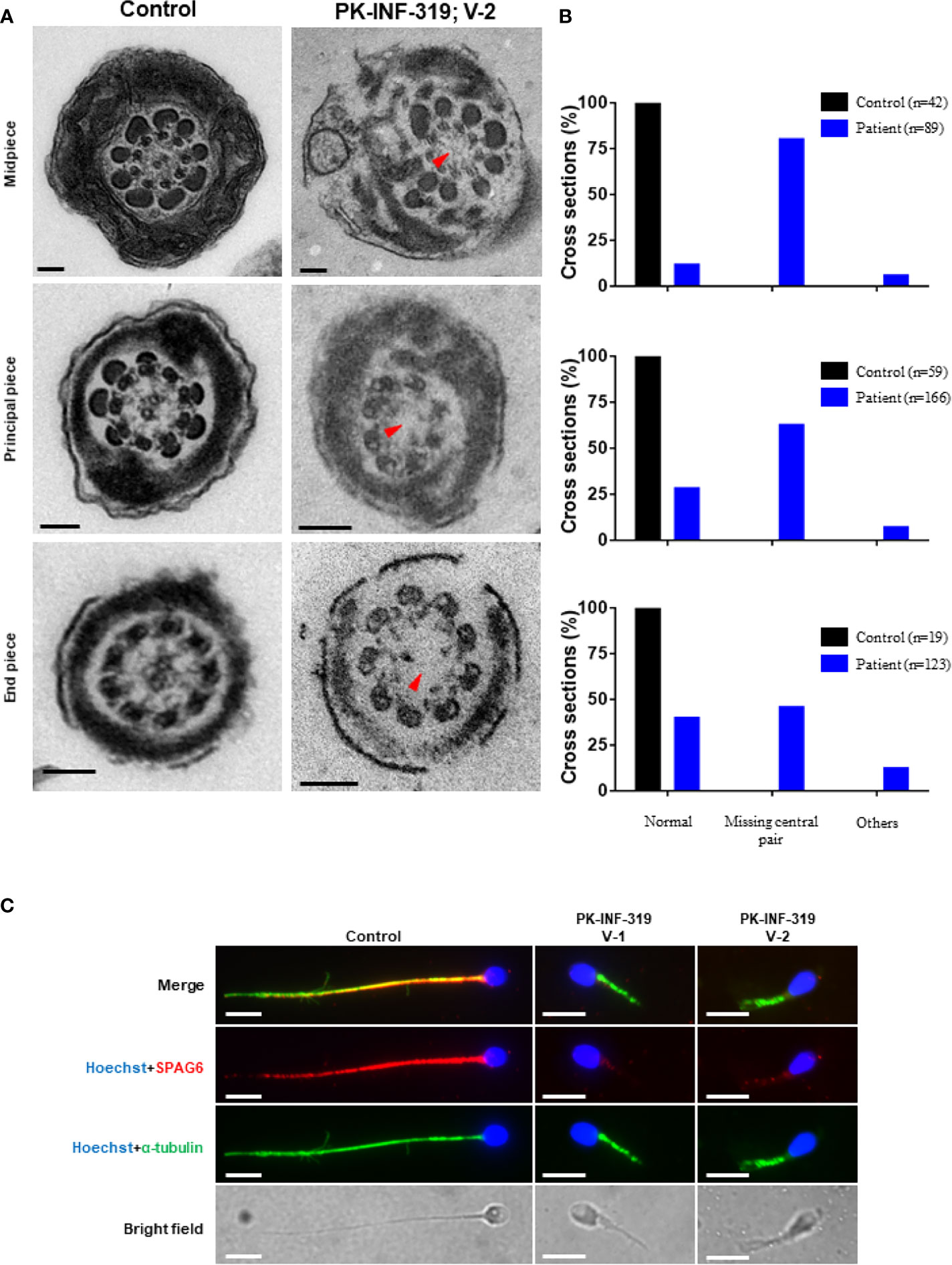
Figure 2 Transmission electron microscopic investigation of spermatozoa from patients and healthy control. (A) The cross-section of a healthy control spermatozoa represented an intact axoneme structure comprised of doublets of microtubules, circularly arranged around a central pair complex of microtubules (nine + two organization), surrounded by outer dense fibers and fibrous sheath. The cross-section of a patient spermatozoa displayed a missing central singlet of microtubules and disorganized fibrous sheath. Scale bars, 500 um. (B) Graphs representing the cross-sections of microtubule doublets corresponding to midpiece, principal piece, and end piece were statistically analyzed in control and patient. (C) The patient spermatozoa were devoid of SPAG6 signals (marker to check the integration of microtubules of the central pair), indicating disruption of inner dynein arm, while well-decorated SPAG6 signals were observed on the tail of spermatozoa obtained from a healthy control. Scale bars, 10 um.
Generation of Mutant Mice Model
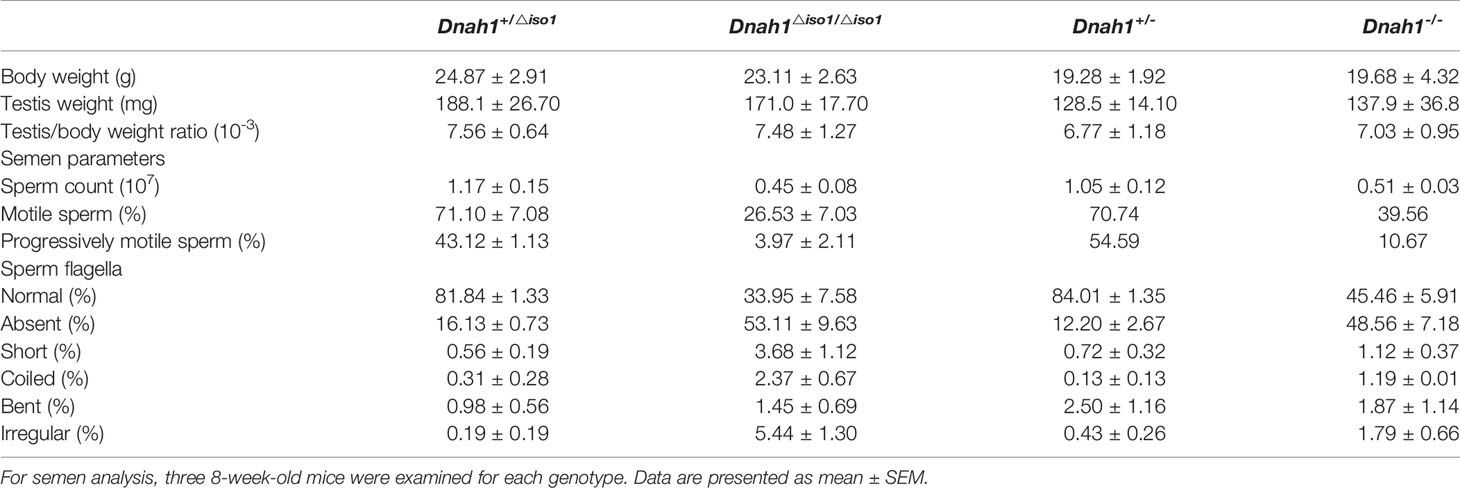
In order to study the in vivo effects of mutations, we generated Dnah1-/- mice by deletion of the Dnah1 transcript and Dnah1△iso/△iso1 mutant mice similar to a mutation in family A2 (c.7646_7647InsC) by using CRISPR/Cas9 genome editing technology (Figure 3A). Sanger sequencing of the PCR products confirmed the genotype of Dnah1-/-and Dnah1△iso/△iso1 mice (Supplementary Figure S3A). Furthermore, western blotting confirmed the absence of full-length DNAH1 protein in both mice models, while intact full-length DNAH1 (herein referred to as DNAH1 isoform1) protein was observed in DNAH1+/△iso1 and DNAH1+/- mice testes (Figure 3B). Interestingly, we observed the presence of a short isoform of DNAH1 (referred to as isoform2) in the testes of Dnah1△iso1/△iso1 mice, while no such isoform was found in Dnah1-/- mice (Figure 3C). Thus, it can be deduced that the Dnah1△iso1/△iso1 mice still harbor the short-length DNAH1 protein. Similarly, reverse transcriptase PCR with a specific primer corresponding to isoform2 of DNAH1 also verified the presence of a small-length transcript in Dnah1△iso1/△iso1 and wild-type (WT) mice (Supplementary Figures S2A, B). Moreover, we performed RT-PCR on control human spermatozoa to check the presence of isoform2. However, no isoform2 was detected in control spermatozoa, while a clear band corresponding to isoform1 was observed (Supplementary Figure S2C).
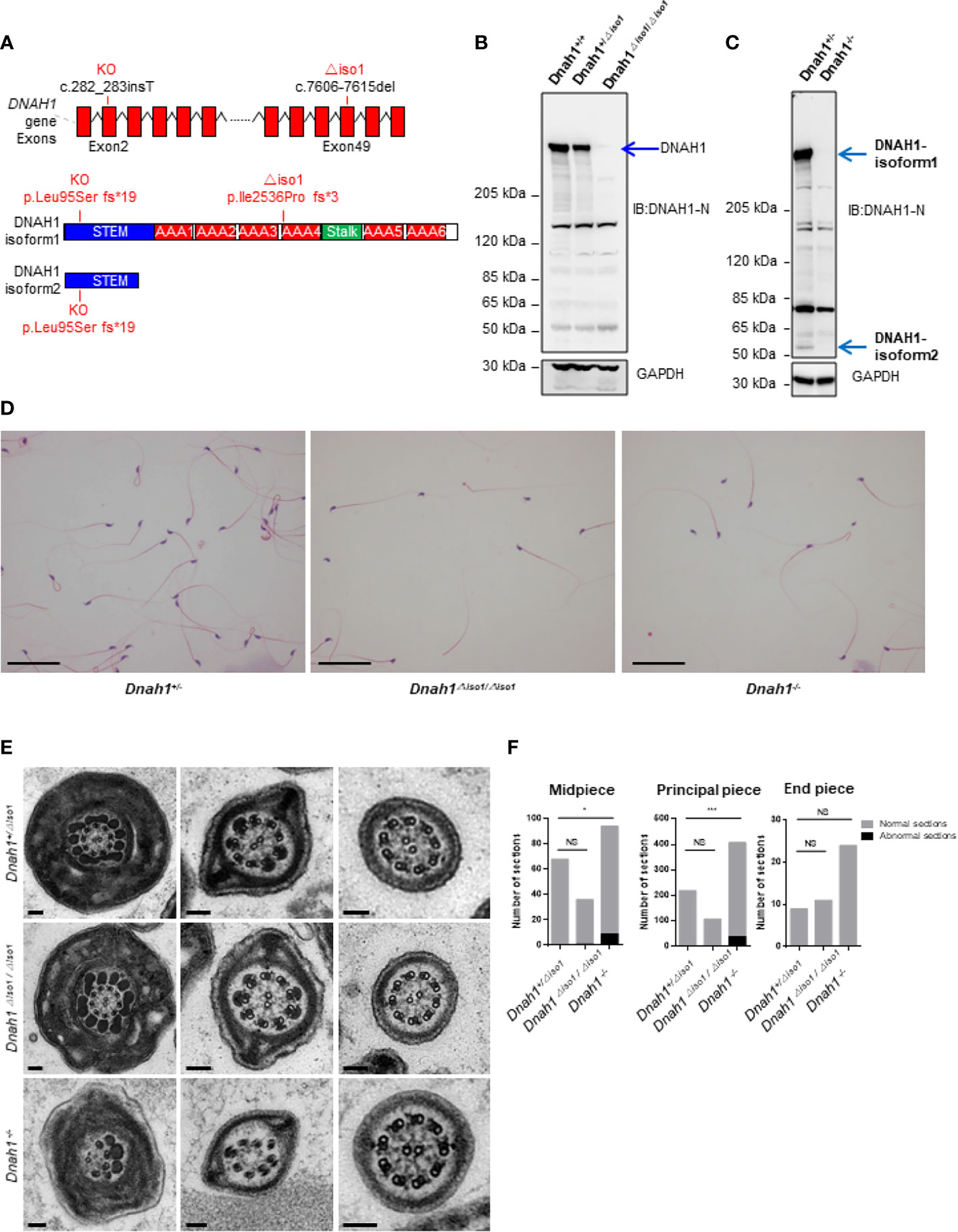
Figure 3 Generation of Dnah1-/- and Dnah1△iso1/△iso1 mice. (A) Representative Dnah1 structure and strategy used to generate Dnah1-/- and Dnah1△iso1/△iso1 mice through CRISPR/Cas9 genome editing technology by targeting exon 2 and exon 49, respectively. (B, C) Western blotting analysis confirmed the deletion of full-length DNAH1 protein in Dnah1-/- and Dnah1△iso1/△iso1 mice testes, while isoform2 (50 kDa) still existed in Dnah1△iso1/△iso1 mice testes. GAPDH was used as an internal control. (D) The sperm morphology analysis of Dnah1△iso1/△iso1 mice displayed spermatozoa without a sperm tail. Scale bars, 10 um (E) Transmission electron microscopy investigation of Dnah1△iso1/△iso1 mice spermatozoa showing an intact ultrastructure with the presence of a central pair and outer dense fibers, while abnormal sections were observed in Dnah1-/- mice spermatozoa. Scale bars, 10 um. (F) Quantification of normal and abnormal sections observed in the spermatozoa of Dnah1-/- and Dnah1△iso1/△iso1 mice. *p < 0.05, ***p < 0.001 and NS, no significant difference.
Mutant Mice With Mild Ultrastructural Defects of Sperm Flagella
After confirming the deficiency of full-length DNAH1 protein in Dnah1△iso1/△iso1 mice, we first scrutinized the fertility of mutant mice by caging one mutant mouse with two WT females for 3 months. During this duration, the female mice display vaginal plugs, demonstrating normal mating efficiency. However, no pregnancy was conceived, indicating that the Dnah1△iso1/△iso1 mice were completely infertile (Table 2). Subsequently, testes morphology (Supplementary Figure S3A) and testes-to-body-weight ratio were comparable in mutant and WT mice (Table 2). On the other hand, reduced sperm count was evident in Dnah1-/- and Dnah1△iso1/△iso1 mice (Table 2). Further examination of testis and epididymis section from Dnah1△iso1/△iso1 mouse displayed an intact seminiferous tubule structure and the presence of all types of germ cells ranging from spermatogonia to spermatozoa (Supplementary Figure S2C). Similarly, an ample number of mature spermatozoa were present in the lumen of cauda in Dnah1△iso1/△iso1 mouse, indicating that sperm production is not severely affected on the grass root level (Supplementary Figure S3D). Next, we analyzed sperm motility by CASA system and observed that progressive motility is severely compromised in Dnah1△iso1/△iso1 mouse, while no progressive motile sperm was observed in Dnah1-/- mouse (Table 2). To explore the reason of reduced motility, we performed a comprehensive inspection of sperm morphology of Dnah1△iso1/△iso1 mouse and Dnah1-/- mouse. Interestingly, we found that most of the spermatozoa (53/49%) lack full-length flagella structure, only possessing an intact head structure (Figure 3D), while few other abnormalities were observed (Table 2). Interestingly, there are still some (34/45%) sperm with normal flagella in Dnah1△iso1/△iso, and Dnah1-/- mouse, and the flagella without sperm head also have a normal morphology. On the other hand, ultrastructural investigation of sperm flagella from Dnah1△iso1/△iso1 and Dnah1-/- mice displayed no significant variation in the architecture of axoneme (Figures 3E, F). However, statistical analysis indicated more abnormal sections in Dnah1-/- mouse than Dnah1△iso1/△iso1 mouse, indicating that isoform2 may have a role in mediating the assembly and stabilization of axonemal structures. Altogether our study provided clues about the phenotype differences in Dnah1 KO and DNAH1 mutant human spermatozoa ultrastructure for the first time.
Discussion
Sperm flagellum is a highly complex structure constituted by a series of proteins that mediate its assembly, composition, and function (41). Defects in sperm flagellum can occur by exogenous and endogenous factors, including genetic mutations that can also reduce sperm motility (42). Studies on mouse models have identified that several genes are required to properly develop the sperm flagellum. However, mutations in these genes are rarely reported in humans suffering from impaired motility due to the morphological defects of flagella. To date, variants in AKAP4, CCDC39, DNAH1, CFAP43, CFAP44, and CFAP69 have been associated with MMAF phenotype in humans and mice (16, 27, 43–45). DNAH1 is one of the most important members of the inner dynein arm and is considered an essential candidate gene for male infertility. The first mutation in DNAH1 causing male infertility was reported in 2001 (26). Since then, various pathogenic mutations in DNAH1 have been reported in various populations, causing male infertility (10, 11, 46, 47). However, most of the cases of altered sperm motility leading to human male infertility remain idiopathic. Thus, it is still difficult to identify the association between genetic mutations and impaired sperm motility.
In the current study, we recruited two extended consanguineous Pakistani families suffering from male infertility due to MMAF without having any symptoms of PCD. Next-generation sequencing identified two novel loss-of-function mutations in DNAH1 (c.6212T>G, p.C1789Y, and c.7646_7647InsC, p.N2549Qfs*61) in family A1 and A2, respectively. These two variants were pathogenic in nature as no DNAH1 signals were observed on the sperm smear slides of the families of both patients, indicating that identified mutations caused the decay of mRNA transcript and the subsequent disruption of DNAH1 protein. Ultrastructural examination of patient spermatozoa revealed the absence of central singlet of microtubules, suggesting that mutations have adversely affected the axonemal structure. SPAG6 is an important marker that is widely used to check the integration microtubules of the central pair. No SPAG6 signals were observed on the patient spermatozoa, suggesting the disruption of the central pair. Subsequently, we generated mutant mice by targeting exon 49 of Dnah1, and western blotting confirmed the absence of a full-length DNAH1 protein. However, a short protein, possibly encoded by N-terminal (isoform2), was observed in the mutant and WT mice.
Interestingly, we observed mild defects in the ultrastructure of spermatozoa from mutant mice. We thought that maybe the isoform2 is mediating the assembly of microtubules in the absence of full-length DNAH1 protein in mice. Previous studies suggested that the N-terminal of the heavy chain of DNAH1 forms a stem structure and is required for the assembly dynein complex and cargo attachment (26, 48). In order to eliminate the effect of N-terminal-mediated isoform2, we generated Dnah1-/- mice by targeting exon 2 that completely disrupted the N-terminal, and western blotting confirmed the absence of full-length as well as isoform2 DNAH1 protein. Surprisingly, the TEM analysis of the ultrastructure of spermatozoa from Dnah1△iso1/△iso1 and Dnah1-/- mice still showed no significant phenotype. Though more abnormal sections were observed in Dnah1-/- mouse as compared to Dnah1△iso1/△iso1 mouse, Dnah1-/- mice spermatozoa still displayed intact central singlet of microtubules, suggesting that KO animals have some hidden factors that could ensure correct axonemal biogenesis and organization or that complete disruption of the heavy chain did not influence the assembly of the other components of an axonemal structure in mice. Alternatively, it can be hypothesized that the role of DNAH1 in axonemal structure is not as central in mice as it is in humans.
Surprisingly, most of the spermatozoa from Dnah1△iso1/△iso1 mouse and Dnah1-/- mice displayed an absence of the entire flagellum tail, a different phenotype from patients. The inner dynein arms are arranged in seven molecular complexes, viewed as globular heads arranged in three-two-two groups and corresponding to three different types of inner arms (IDA1 to IDA3). We assumed that one of the IDA has been disorganized in Dnah1 mutant mouse spermatozoa, leading to loss of connection between external doublets of the microtubules and the two central microtubules and ultimately affected the cargo system required for cytoskeleton organization. Another possibility is that Dnah1△iso1/△iso1 and Dnah1-/- spermatozoa are defective in organizing the α- and β-tubulins required for spermiogenesis (49). Thus, it can be inferred that DNAH1 also regulates the α- and β-tubulin complexes, which play crucial roles in sperm tail elongation during spermiogenesis.
It is reasonable to hypothesize that the position of mutations in DNAH1 may have a different effect on the sperm flagellum structure, which can completely reduce the immobility of spermatozoa. Furthermore, the mutations in the N-terminal and C-terminal of DNAH1 can display different effects on the axonemal structure of human spermatozoa. Altogether our study described that the role of DNAH1 is more central in regulating the axonemal assembly and dimerization in humans than in mice. The finding of this study will pave the way for finding out other intrinsic factors responsible for different phenotypes of DNAH1 mutations in humans and mice. Finally, it is suggested that screening the deleterious mutations of DNAH1could be important for the clinical molecular diagnosis of male infertility.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Bioethical Committee of Pakistan and the ethical review board of the University of Science and Technology of China (USTC). The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the ethical review board of the USTC.
Author Contributions
RK, JC, AM, and BZ performed the experiments. QZ, ManK, AA, MZ, WS, and MazK recruited the patients, performed semen analysis, and collected patient samples. MN gave insightful discussion and constructive comments on the manuscript. DZ and JZ performed the whole-exome sequencing (WES) and WES data analysis. QS, HZ, BX, YZ, and RK conceived and supervised the study, designed and analyzed the experiments, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (32070850) and National Key Research and Developmental Program of China (2018YFC1004700).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.765639/full#supplementary-material
Supplementary Figure S1 | Sperm morphology analysis. (A) Typical flagellar appearance in control subject. (B) Various defects of sperm flagella, such as absent, short, coiled, and bent, were common in the spermatozoa of patients.
Supplementary Figure S2 | Presence of DNAH1 isoform 2 in mutant and wild-type (WT) mice. (A) Gel electrophoresis of cDNA products (one, three, and five WT mice cDNA product; two, four, and six Dnah1△iso1/△iso1 cDNA mice products) confirmed the presence of isoform 2 mRNA. (B) Corresponding Sanger sequencing of WT and Dnah1△iso1/△iso1 mice cDNA further verified the presence of isoform 2 in Dnah1△iso1/△iso1 mice. (C) Gel electrophoresis of cDNA product from control human spermatozoa confirmed the absence of isoform2 in humans.
Supplementary Figure S3 | Genotyping and spermatogenesis in Dnah1-/- and Dnah1△iso1/△iso1 mice. (A) Representative chromatograms from Dnah1+/-, Dnah1+/△iso1, Dnah1-/-, and Dnah1△iso1/△iso1 mice confirming their genotype. The red letters indicate the changes in the DNA sequences. (B) Representative image of testes from adult rom Dnah1+/-, Dnah1+/△iso1, Dnah1-/-, and Dnah1△iso1/△iso1 mice. Scale bars, 1 cm. (C) H&E staining of testes and caudal epididymis from adult Dnah1+/△iso1 and Dnah1△iso1/△iso1 mice. Scale bars, 50 μm.
References
1. Krausz C, Riera-Escamilla A. Genetics of Male Infertility. Nat Rev Urol (2018) 15(6):369–84. doi: 10.1038/s41585-018-0003-3
2. Coutton C, Escoffier J, Martinez G, Arnoult C, Ray PF. Teratozoospermia: Spotlight on the Main Genetic Actors in the Human. Hum Reprod Update (2015) 21(4):455–85. doi: 10.1093/humupd/dmv020
3. Curi SM, Ariagno JI, Chenlo PH, Mendeluk GR, Pugliese MN, Sardi Segovia LM, et al. Asthenozoospermia: Analysis of a Large Population. Arch Androl (2003) 49(5):343–9. doi: 10.1080/01485010390219656
4. Beurois J, Cazin C, Kherraf ZE, Martinez G, Celse T, Touré A, et al. Genetics of Teratozoospermia: Back to the Head. Best Pract Res Clin Endocrinol Metab (2020) 2(101473):101473. doi: 10.1016/j.beem.2020.101473
5. Yang SM, Li HB, Wang JX, Shi YC, Cheng HB, Wang W, et al. Morphological Characteristics and Initial Genetic Study of Multiple Morphological Anomalies of the Flagella in China. Asian J Androl (2015) 17(3):513–5. doi: 10.4103/1008-682X.146100
6. Lehti MS, Sironen A. Formation and Function of Sperm Tail Structures in Association With Sperm Motility Defects. Biol Reprod (2017) 97(4):522–36. doi: 10.1093/biolre/iox096
7. Yang X, Zhu D, Zhang H, Jiang Y, Hu X, Geng D, et al. Associations Between DNAH1 Gene Polymorphisms and Male Infertility: A Retrospective Study. Med (Baltimore) (2018) 97(49):e13493. doi: 10.1097/MD.0000000000013493
8. Wilson KS, Gonzalez O, Dutcher SK, Bayly PV. Dynein-Deficient Flagella Respond to Increased Viscosity With Contrasting Changes in Power and Recovery Strokes. Cytoskeleton (Hoboken) (2015) 72(9):477–90. doi: 10.1002/cm.21252
9. Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K, et al. Dynein Structure and Power Stroke. Nature (2003) 421(6924):715–8. doi: 10.1038/nature01377
10. Sha Y, Yang X, Mei L, Ji Z, Wang X, Ding L, et al. DNAH1 Gene Mutations and Their Potential Association With Dysplasia of the Sperm Fibrous Sheath and Infertility in the Han Chinese Population. Fertil Steril (2017) 107(6):1312–8.e2. doi: 10.1016/j.fertnstert.2017.04.007
11. Amiri-Yekta A, Coutton C, Kherraf ZE, Karaouzène T, Le Tanno P, Sanati MH, et al. Whole-Exome Sequencing of Familial Cases of Multiple Morphological Abnormalities of the Sperm Flagella (MMAF) Reveals New DNAH1 Mutations. Hum Reprod (2016) 31(12):2872–80. doi: 10.1093/humrep/dew262
12. Tang S, Wang X, Li W, Yang X, Li Z, Liu W, et al. Biallelic Mutations in CFAP43 and CFAP44 Cause Male Infertility With Multiple Morphological Abnormalities of the Sperm Flagella. Am J Hum Genet (2017) 100(6):854–64. doi: 10.1016/j.ajhg.2017.04.012
13. Liu C, Tu C, Wang L, Wu H, Houston BJ, Mastrorosa FK, et al. Deleterious Variants in X-Linked CFAP47 Induce Asthenoteratozoospermia and Primary Male Infertility. Am J Hum Genet (2021) 108(2):309–23. doi: 10.1016/j.ajhg.2021.01.002
14. He X, Liu C, Yang X, Lv M, Ni X, Li Q, et al. Bi-Allelic Loss-Of-Function Variants in CFAP58 Cause Flagellar Axoneme and Mitochondrial Sheath Defects and Asthenoteratozoospermia in Humans and Mice. Am J Hum Genet (2020) 107(3):514–26. doi: 10.1016/j.ajhg.2020.07.010
15. Li W, Wu H, Li F, Tian S, Kherraf ZE, Zhang J, et al. Biallelic Mutations in CFAP65 Cause Male Infertility With Multiple Morphological Abnormalities of the Sperm Flagella in Humans and Mice. J Med Genet (2020) 57(2):89–95. doi: 10.1136/jmedgenet-2019-106344
16. Dong FN, Amiri-Yekta A, Martinez G, Saut A, Tek J, Stouvenel L, et al. Absence of CFAP69 Causes Male Infertility Due to Multiple Morphological Abnormalities of the Flagella in Human and Mouse. Am J Hum Genet (2018) 102(4):636–48. doi: 10.1016/j.ajhg.2018.03.007
17. Beurois J, Martinez G, Cazin C, Kherraf ZE, Amiri-Yekta A, Thierry-Mieg N, et al. CFAP70 Mutations Lead to Male Infertility Due to Severe Astheno-Teratozoospermia. A Case Report. Hum Reprod (2019) 34(10):2071–9. doi: 10.1093/humrep/dez166
18. Sha Y, Wei X, Ding L, Ji Z, Mei L, Huang X, et al. Biallelic Mutations of CFAP74 may Cause Human Primary Ciliary Dyskinesia and MMAF Phenotype. J Hum Genet (2020) 65(11):961–9. doi: 10.1038/s10038-020-0790-2
19. Martinez G, Beurois J, Dacheux D, Cazin C, Bidart M, Kherraf ZE, et al. Biallelic Variants in MAATS1 Encoding CFAP91, A Calmodulin-Associated and Spoke-Associated Complex Protein, Cause Severe Astheno-Teratozoospermia and Male Infertility. J Med Genet (2020) 57(10):708–16. doi: 10.1136/jmedgenet-2019-106775
20. Shen Q, Martinez G, Liu H, Beurois J, Wu H, Amiri-Yekta A, et al. Bi-Allelic Truncating Variants in CFAP206 Cause Male Infertility in Human and Mouse. Hum Genet (2021) 140(9):1367–77. doi: 10.1007/s00439-021-02313-z
21. Li W, He X, Yang S, Liu C, Wu H, Liu W, et al. Biallelic Mutations of CFAP251 Cause Sperm Flagellar Defects and Human Male Infertility. J Hum Genet (2019) 64(1):49–54. doi: 10.1038/s10038-018-0520-1
22. Touré A, Martinez G, Kherraf ZE, Cazin C, Beurois J, Arnoult C, et al. The Genetic Architecture of Morphological Abnormalities of the Sperm Tail. Hum Genet (2021) 140(1):21–42. doi: 10.1007/s00439-020-02113-x
23. Lv M, Liu W, Chi W, Ni X, Wang J, Cheng H, et al. Homozygous Mutations in DZIP1 can Induce Asthenoteratospermia With Severe MMAF. J Med Genet (2020) 57(7):445–53. doi: 10.1136/jmedgenet-2019-106479
24. Zhang J, He X, Wu H, Zhang X, Yang S, Liu C, et al. Loss of DRC1 Function Leads to Multiple Morphological Abnormalities of the Sperm Flagella and Male Infertility in Human and Mouse. Hum Mol Genet (2021) 30(21):1996–2011. doi: 10.1093/hmg/ddab171
25. Ma H, Zhang B, Khan A, Zhao D, Ma A, Jianteng Z, et al. Novel Frameshift Mutation in STK33 Is Associated With Asthenozoospermia and Multiple Morphological Abnormality of the Flagella. Hum Mol Genet (2021) 30(21):1977–84. doi: 10.1093/hmg/ddab165
26. Neesen J, Kirschner R, Ochs M, Schmiedl A, Habermann B, Mueller C, et al. Disruption of an Inner Arm Dynein Heavy Chain Gene Results in Asthenozoospermia and Reduced Ciliary Beat Frequency. Hum Mol Genet (2001) 10(11):1117–28. doi: 10.1093/hmg/10.11.1117
27. Ben Khelifa M, Coutton C, Zouari R, Karaouzène T, Rendu J, Bidart M, et al. Mutations in DNAH1, Which Encodes an Inner Arm Heavy Chain Dynein, Lead to Male Infertility From Multiple Morphological Abnormalities of the Sperm Flagella. Am J Hum Genet (2014) 94(1):95–104. doi: 10.1016/j.ajhg.2013.11.017
28. Yin H, Ma H, Hussain S, Zhang H, Xie X, Jiang L, et al. A Homozygous FANCM Frameshift Pathogenic Variant Causes Male Infertility. Genet Med (2019) 21(1):62–70. doi: 10.1038/s41436-018-0015-7
29. Li H, Durbin R. Fast and Accurate Short Read Alignment With Burrows-Wheeler Transform. Bioinformatics (2009) 25(14):1754–60. doi: 10.1093/bioinformatics/btp324
30. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing Next-Generation DNA Sequencing Data. Genome Res (2010) 20(9):1297–303. doi: 10.1101/gr.107524.110
31. Smith KR, Bromhead CJ, Hildebrand MS, Shearer AE, Lockhart PJ, Najmabadi H, et al. Reducing the Exome Search Space for Mendelian Diseases Using Genetic Linkage Analysis of Exome Genotypes. Genome Biol (2011) 12(9):2011–12. doi: 10.1186/gb-2011-12-9-r85
32. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A Global Reference for Human Genetic Variation. Nature (2015) 526(7571):68–74. doi: 10.1038/nature15393
33. Fu W, O'Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, et al. Analysis of 6,515 Exomes Reveals the Recent Origin of Most Human Protein-Coding Variants. Nature (2013) 493(7431):216–20. doi: 10.1038/nature11690
34. Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of Protein-Coding Genetic Variation in 60,706 Humans. Nature (2016) 536(7616):285–91. doi: 10.1038/nature19057
35. Zhang Y, Zhong L, Xu B, Yang Y, Ban R, Zhu J, et al. SpermatogenesisOnline 1.0: A Resource for Spermatogenesis Based on Manual Literature Curation and Genome-Wide Data Mining. Nucleic Acids Res (2013) 41(Database issue):D1055–62. doi: 10.1093/nar/gks1186
36. Zhang B, Ma S, Khan T, Ma A, Li T, Zhang H, et al. A DNAH17 Missense Variant Causes Flagella Destabilization and Asthenozoospermia. J Exp Med (2020) 217(2):20182365. doi: 10.1084/jem.20182365
37. Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step Generation of Mice Carrying Reporter and Conditional Alleles by CRISPR/Cas-mediated Genome Engineering. Cell (2013) 154:1370–9. doi: 10.1016/j.cell.2013.08.022
38. Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, et al. Efficient Genome Modification by CRISPR-Cas9 Nickase With Minimal Off-Target Effects. Nat Methods (2014) 11(4):399–402. doi: 10.1038/nmeth.2857
39. Gao Q, Khan R, Yu C, Alsheimer M, Jiang X, Ma H, et al. The Testis-Specific LINC Component SUN3 Is Essential for Sperm Head Shaping During Mouse Spermiogenesis. J Biol Chem (2020) 295(19):6289–98. doi: 10.1074/jbc.RA119.012375
40. Khan R, Ye J, Yousaf A, Shah W, Aftab A, Shah B, et al. Evolutionarily Conserved and Testis-Specific Gene, 4930524B15Rik, Is Not Essential for Mouse Spermatogenesis and Fertility. Mol Biol Rep (2020) 47(7):5207–13. doi: 10.1007/s11033-020-05595-0
41. Vyklicka L, Lishko PV. Dissecting the Signaling Pathways Involved in the Function of Sperm Flagellum. Curr Opin Cell Biol (2020) 63:154–61. doi: 10.1016/j.ceb.2020.01.015
42. Inaba K. Molecular Architecture of the Sperm Flagella: Molecules for Motility and Signaling. Zoolog Sci (2003) 20(9):1043–56. doi: 10.2108/zsj.20.1043
43. Turner RM, Musse MP, Mandal A, Klotz K, Jayes FC, Herr JC, et al. Molecular Genetic Analysis of Two Human Sperm Fibrous Sheath Proteins, AKAP4 and AKAP3, in Men With Dysplasia of the Fibrous Sheath. J Androl (2001) 22(2):302–15. doi: 10.1002/j.1939-4640.2001.tb02184.x
44. Merveille AC, Davis EE, Becker-Heck A, Legendre M, Amirav I, Bataill G, et al. CCDC39 Is Required for Assembly of Inner Dynein Arms and the Dynein Regulatory Complex and for Normal Ciliary Motility in Humans and Dogs. Nat Genet (2011) 43(1):72–8. doi: 10.1038/ng.726
45. Coutton C, Vargas AS, Amiri-Yekta A, Kherraf ZE, Ben Mustapha SF, Le Tanno P, et al. Mutations in CFAP43 and CFAP44 Cause Male Infertility and Flagellum Defects in Trypanosoma and Human. Nat Commun (2018) 9(1):017–02792. doi: 10.1038/s41467-017-02792-7
46. Wambergue C, Zouari R, Fourati Ben Mustapha S, Martinez G, Devillard F, Hennebicq S, et al. Patients With Multiple Morphological Abnormalities of the Sperm Flagella Due to DNAH1 Mutations Have a Good Prognosis Following Intracytoplasmic Sperm Injection. Hum Reprod (2016) 31(6):1164–72. doi: 10.1093/humrep/dew083
47. Wang X, Jin H, Han F, Cui Y, Chen J, Yang C, et al. Homozygous DNAH1 Frameshift Mutation Causes Multiple Morphological Anomalies of the Sperm Flagella in Chinese. Clin Genet (2017) 91(2):313–21. doi: 10.1111/cge.12857
48. Myster SH, Knott JA, Wysocki KM, O'Toole E, Porter ME. Domains in the 1alpha Dynein Heavy Chain Required for Inner Arm Assembly and Flagellar Motility in Chlamydomonas. J Cell Biol (1999) 146(4):801–18. doi: 10.1083/jcb.146.4.801
Keywords: MMAF, male infertility, mutant mice, DNAH1, central singlet
Citation: Khan R, Zaman Q, Chen J, Khan M, Ma A, Zhou J, Zhang B, Ali A, Naeem M, Zubair M, Zhao D, Shah W, Khan M, Zhang Y, Xu B, Zhang H and Shi Q (2021) Novel Loss-of-Function Mutations in DNAH1 Displayed Different Phenotypic Spectrum in Humans and Mice. Front. Endocrinol. 12:765639. doi: 10.3389/fendo.2021.765639
Received: 27 August 2021; Accepted: 22 October 2021;
Published: 17 November 2021.
Edited by:
Furhan Iqbal, Bahauddin Zakariya University, PakistanReviewed by:
Yanwei Sha, Xiamen University, ChinaFaiz Rasul, Peking University, China
Saima Malik, University of Alabama at Birmingham, United States
Copyright © 2021 Khan, Zaman, Chen, Khan, Ma, Zhou, Zhang, Ali, Naeem, Zubair, Zhao, Shah, Khan, Zhang, Xu, Zhang and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Xu, YmlveHVib0BtYWlsLnVzdGMuZWR1LmNu; Huan Zhang, bGlrZW1vb25yaXZlckAxMjYuY29t; Qinghua Shi, cXNoaUB1c3RjLmVkdS5jbg==
†Present address: Manan Khan, Department of Biotechnology and Genetic Engineering Hazara University, Mansehra, Pakistan
‡These authors have contributed equally to this work
 Ranjha Khan
Ranjha Khan Qumar Zaman
Qumar Zaman Jing Chen
Jing Chen Manan Khan
Manan Khan Ao Ma
Ao Ma Jianteng Zhou
Jianteng Zhou Beibei Zhang1
Beibei Zhang1 Muhammad Naeem
Muhammad Naeem Muhammad Zubair
Muhammad Zubair Daren Zhao
Daren Zhao Wasim Shah
Wasim Shah Mazhar Khan
Mazhar Khan Yuanwei Zhang
Yuanwei Zhang Huan Zhang
Huan Zhang Qinghua Shi
Qinghua Shi