
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 11 November 2021
Sec. Cellular Endocrinology
Volume 12 - 2021 | https://doi.org/10.3389/fendo.2021.756530
This article is part of the Research Topic Sex Determination and Developmental Mechanism of Crustaceans and Shellfish View all 8 articles
Sea urchin (Strongylocentrotus intermedius) is an economically important mariculture species in Asia, and its gonads are the only edible part. The efficiency of genetic breeding in sea urchins is hampered due to the inability to distinguish gender by appearance. In this study, we first identified a sex-associated single nucleotide polymorphism (SNP) by combining type IIB endonuclease restriction site-associated DNA sequencing (2b-RAD-seq) and genome survey. Importantly, this SNP is located within spata4, a gene specifically expressed in male. Knocking down of spata4 by RNA interference (RNAi) in male individuals led to the downregulation of other conserved testis differentiation-related genes and germ cell marker genes. We also revealed that sex ratio in this validated culture population of S. intermedius is not 1:1. Moreover, after a 58-day feeding experiment with estradiol, the expression levels of several conserved genes that are related to testis differentiation, ovary differentiation, and estrogen metabolism were dynamically changed. Taken together, our results will contribute toward improving breeding efficiency, developing sex-controlled breeding, and providing a solid base for understanding sex determination mechanisms in sea urchins.
Sea urchin (Strongylocentrotus intermedius) belongs to Echinodermata and is an important commercial species mainly distributed in several Asian countries, such as China, Korea, and Japan (1). It is well known that gonads of sea urchins are rich in unsaturated fatty acids and are the only edible parts for people (2). It has been reported that gender is one of the strongest influencing factors on the commercial value of sea urchin; for example, increasing rates of body size and gonadal growth in male sea urchins have advantages than females (3). In addition, the immunocompetence of female Paracentrotus lividus urchins might be superior to that of males (4), and the contents of free amino acids and lipid concentrations are different based on gender. As previously described, the gonads of sea urchins are classified into four stages, namely, intergametogenesis and nutritive phagocyte (NP) phagocytosis (stage 1), pregametogenesis and NP renewal (stage 2), gametogenesis and NP utilization (stage 3), and the end of gametogenesis, NP exhaustion, and spawning (stage 4) (5). Recently, three female-specific markers were identified and could be used to identify the genetic sex of Mesocentrotus nudus (6). In addition, the number of genes and micro-RNAs associated with sex determination and sex differentiation have been identified by RNA sequencing (7, 8). However, no literature about sex control was reported, and a large part about sex determination and sex differentiation in sea urchin is still unknown.
With the development of next-generation sequencing (NGS) technologies, restriction site-associated DNA sequencing (RAD-seq) was developed to identify sex-specific markers and discover the mechanisms of sex determination in various species (9–11). In Mastacembelus armatus, two male-specific DNA fragments and two single nucleotide polymorphisms (SNPs) were identified by RAD-seq, and the Tcl gene was regarded as a candidate sex determination gene (12). In crab species, sex-specific SNPs were identified via RAD-seq, and a WZ/ZZ sex determination system was suggested based on male homogamety and female heterogamety in those sex-specific SNP loci (13). Sea cucumber (Apostichopus japonicus) also belongs to Echinodermata, a male-specific DNA fragment was verified, and a rapid genetic identifying method was developed by sex-specific primer design coupled with PCR amplification (14). Therefore, sex-specific molecular markers obtained by RAD-seq will be helpful in revealing the genetic basis of sex determination and improving breeding efficiency because genetic sex could be identified at early developmental stages.
Vertebrate steroid hormones play important roles in regulating sex differentiation and maintaining secondary sexual characteristics, especially sex steroid hormones including estradiol-17β (E2), testosterone, and progesterone (15, 16). In several aquatic vertebrates, the hormonal-induced sex reversal technique had been widely used to achieve monosex culture; thus, the quality, output, and economic value had been dramatically increased (17, 18). For example, XY physiological female yellow catfish was obtained by E2 treatment, then YY super-males will be obtained by gynogenesis of XY physiological female fish, and finally all-male yellow catfish will be obtained by crossing YY super-males with normal XX males (19). What is more, the YY super-males, XY males, and XX females can be identified by Y chromosome-linked and X chromosome-linked markers (20). It has been reported that vertebrate-type steroids can be measured in some invertebrates, such as in Scylla serrata (21) and Chlamys farreri (22). However, it is still unclear whether those detected hormones are endogenously synthesized or picked up from the environment (23, 24). For example, mollusks can readily absorb vertebrate steroids from the environment and retain them for several weeks (25); thus, whether mollusks contain endogenously synthesized steroids is highly controversial (26). Moreover, steroid androgen exposure has a significant influence on vitellogenin production, ovarian development, and gene expression in some invertebrates, such as Portunus trituberculatus (27) and Saccostrea glomerata (28). In sea urchins, treatment with estradiol dipropionate will change the rate of protein synthesis in oocytes, increase gonad development, accelerate vitellogenesis, and induce maturation of the germ cells (29). Besides, feeding E2 will increase the rate of ovarian growth but inhibited oocyte growth in Lytechinus variegatus (30). However, it is still a mystery whether vertebrate-type steroids can be measured in S. intermedius and whether they have potential effects on gonadal development and gene expression patterns related to sex differentiation.
Spermatogenesis associated 4 gene (spata4) was first identified in human testes and involved in cryptorchidism development (31). Subsequently, the spata4 gene had been cloned from diverse organisms. In Mammalia, SPATA4 is specifically expressed in the testis and may play a significant role in maintaining spermatogenesis ability during adolescence (31, 32). In chicken, the spata4 gene is also specifically expressed in the testis, and along with development progress of the testis, its expression level is gradually upregulated (33). In zebrafish and rainbow trout (Oncorhynchus mykiss), the spata4 gene is expressed specifically in the testis and slightly in the ovary (34, 35). Moreover, the potential function of spata4 had been investigated by in vitro cell experiment, and it was revealed that regulatory factor X1 (RFX1) could bind the spata4 promoter, regulating its transcriptional activity and endogenous expression, and further regulate the proliferation of Sertoli cells (36). To date, no literature about spata4 was reported in sea urchin.
In the present study, 2b-RAD-seq technology was employed to identify sex‐specific tags and SNP loci of sea urchin (S. intermedius). Subsequently, we investigated the 2b-RAD markers by PCR in 100 individuals, and a sex-related SNP locus had been identified. Based on the annotation of the sex-related SNP, a male specifically expressed gene spata4 had been characterized. Then, the function implication of spata4 in testis development had been investigated by means of RNAi. Furthermore, we measured vertebrate-type steroids in S. intermedius and detected the effect of dietary administration of estradiol on testis development and sex-related gene expression in male individuals. Besides the practical implications for improving breeding efficiency, these findings also provide a molecular basis for understanding sex determination mechanisms in sea urchin.
Twenty sea urchin individuals (10 males and 10 females), randomly collected from the Key Laboratory of Mariculture & Stock Enhancement in North China Sea, Ministry of Agriculture and Rural Affairs, Dalian Ocean University, were used to identify the potential sex-specific markers by 2b-RAD-seq. In addition, 100 aquaculture individuals of S. intermedius were purchased from markets in Dalian Haibao Fishery Limited Company and used to confirm the sex-specific markers. The tube feet tissues of each sea urchin were removed and stored in absolute ethanol at −20°C for genomic DNA extraction. Genomic DNA was extracted using the protocols of a marine animal tissue genomic DNA extraction kit (Tiangen, DP324).
To determine the phenotypic sex of individuals, the corresponding gonadal tissues were removed and fixed in 4% paraformaldehyde (PFA) at 4°C overnight, and then the gonadal tissues were washed three times with phosphate buffered solution (PBS) at room temperature. Subsequently, the gonadal tissues were embedded in optimal cutting temperature (O.C.T) compound and frozen sections (5 µm) were cut. After stained with hematoxylin/eosin, the gonadal structure was observed in Leica DM4B microscope to determine the phenotypic sex of each sea urchin.
The 2b-RAD libraries were constructed at Qingdao OE Biotech Co., Ltd. (Qingdao, China) according to a previously report (37). In brief, 1 U BsaXI restriction enzyme was used to digest 200 ng genomic DNA from each individual (10 females and 10 males). The digestion products were ligated to adapter 1 and adapter 2. Then, a set of four primers that introduce individual specific barcodes and sequencing primers was used to amplify the ligation products by PCR. The PCR products were separated in 8% polyacrylamide gels by electrophoresis. After purification, each product was digested with SapI restriction enzyme and purified using streptavidin-coated magnetic beads, and 200 U T4 DNA ligase was added to the supernatant. After purification of the ligation products, barcodes were introduced by PCR with barcode-bearing primers. Then, the PCR products were purified using a MinElute PCR Purification Kit and pooled for sequencing using the Illumina NovaSeq 6000 PE150PE platform.
The raw reads were merged by Pear software (version 0.9.6) (38), and then the adaptor sequences of the merged reads were trimmed. The terminal 3-bp positions were also removed from each read. Then, low-quality reads that contain ambiguous bases (N) exceeding 8% or without restriction sites were excluded from the raw data and removed from each output read, and clean reads were obtained. The genomic information of S. intermedius is currently unavailable. Also, the 2b-RAD tags we obtained were too short to design PCR primers. One male and one female individual were thus selected to perform short-read de novo sequencing. For the genome survey, DNA was extracted from one female and one male S. intermedius for library construction by using TruSeq DNA LT Sample Prep kit. Briefly, the genomic DNA was sheared into fragments with length ~350 bp using S220 Focused-ultrasonicators (Covaris, USA). Adapters were ligated onto the 3′ end of the sheared fragments. After PCR amplification and purification, the final libraries were sequenced on the Illumina NovaSeq platform (Illumina Inc., USA) and 150 bp paired-end reads were generated. The above 150-bp paired-end reads were clustered to build reference sequences for further locus genotyping using ustacks software version 1.34 (39). To identify candidate sex-specific markers, high-quality reads were aligned to the genome reference using SOAP2 (version 2.21) with the following parameters: r = 0, M = 4, v = 2 (40). SNP loci were developed by RAD typing program with default parameter. Finally, candidate sex-specific markers for each individual were generated for further analyses.
PCR amplification was used to validate each candidate sex-specific tag and SNP locus. Firstly, specific primers of candidate sex-specific markers were designed using online software (http://biotools.nubic.northwestern.edu/OligoCalc.html) according to the flank sequences (Supplementary Table S1). Then, the PCR reactions were carried out with a total volume of 20 μl, containing 50 ng template DNA, 10 µl of 2× Taq buffer (Kang Wei, China), and 0.5 μl of each primer (10 μM), and sterile water was added to the final volume. The PCR programs were as follows: predenaturation at 94°C for 2 min, then 30 cycles of 94°C for 30 s, annealing temperature of primer pair for 30 s, followed by 72°C for 60 s, and a final extension of 10 min at 72°C. The PCR products were separated via 1% agarose gel electrophoresis, and the expected PCR fragments were directly sequenced in both directions by Tsingke Biotechnology (Tianjin, China).
Total RNA was extracted from different tissues, including the ovary, testis, tube feet, and intestines, using the SV Total RNA Isolation System (Promega Z3100). Total RNA (1 µg) of the testis was used to establish a cDNA library using the SMARTer™ Rapid Amplification of cDNA End (RACE) cDNA Amplification Kit (Clontech 634923) according to the recommendations and protocols of the manufacturer. Meanwhile, the first-strand cDNAs of each sample were synthesized according to the protocols of RevertAid First Strand cDNA Synthesis Kit (Fermentas). Real-time quantitative PCR (RT-qPCR) was performed on the LightCycler®96 Instrument (Roche) according to a previous description (41), and 18S ribosomal RNA (18S rRNA) was used as the internal control. The relative expression of the target genes was calculated with the 2−ΔΔCt method. For statistical analysis, one-way ANOVA was calculated with SPSS software and a probability (p) of <0.05 was considered statistically significant. All primers used were designed by online software http://biotools.nubic.northwestern.edu/OligoCalc.html (Supplementary Table S1).
The gene-specific dsRNA of spata4 was designed by https://www.dkfz.de/signaling/e-rnai3/ and synthesized by the T7 RiboMAX™ Express RNAi System (Promega) according to the protocol. Firstly, the genetic sex of sea urchins was identified by PCR reaction and Sanger sequencing depended on the sex-specific SNP marker. Then, a total of nine male S. intermedius with a mean shell diameter of 52.5 ± 2.10 mm was selected to inject dsRNA. Meanwhile, the testis development of selected S. intermedius was during gametogenesis and NP utilization (stage 3), and this is a critical stage in spermatogenesis. Before injection, three sea urchins were randomly removed and their gonads were surgically sampled as control samples. Then, spata4-specific dsRNA was heated to 95°C for 3min and cooled to room temperature (25–30°C). Subsequently, sea urchin was injected with 100 µl PBS containing 100 µg dsRNA according to a previous report (5). On day 3 after the first injection, three sea urchins were randomly removed and their gonads were surgically sampled. Then, a second injection was performed on the remaining sea urchins. On day 7 after the first injection, the gonadal tissues of remaining three sea urchins were surgically sampled. Histological examination of the gonads was carried out according to the method above. RT-qPCR was performed to investigate the knockdown effect and the dynamic expression change of other conserved sex-related genes.
Strongylocentrotus intermedius used for this experiment were purchased from markets in Dalian Haibao Fishery Limited Company, and after being acclimatized to laboratory conditions for 2 weeks, E2 concentration in the gonadal tissues was detected via a commercial enzyme-linked immunosorbent assay (ELISA) kit (Fish estradiol ELISA kit, Mlbio) according to the protocol of the manufacturer. A total of 54 sea urchins individuals with a mean diameter of 31.3 ± 0.43 mm were randomly divided into three groups and cultured in separate seawater tanks at 15°C–17°C. In addition, we made little plastic frames and put them in the seawater tanks, and each sea urchin was cultured and fed on the little plastic frame independently. Among the three groups, two experimental groups had diets with different estradiol contents, one group was fed with food mixed with 50 mg/kg E2 and another group was fed with food mixed with 100 mg/kg E2, while the control group was fed without E2. E2 was purchased from Sigma-Aldrich (E8875-250). Feeding experiments lasted 58 days, and sea urchins were fed with dietary supplementation of E2 every day. On days 3, the excrement of sea urchins was collected to detect E2 level. In addition, on days 15, 30, and 58, six individuals were randomly removed from each group, and their gonads were sampled to analyze the E2 level and for histological examination.
On day 58 after the feeding experiments, 12 gonadal samples from the control group and those fed with 100 mg/kg E2 were selected for RNA extraction and transcriptome sequencing. For transcriptome sequencing, every three gonadal samples from the same group were mixed as one sequencing sample and each group had two sequencing samples. The transcriptome sequencing was performed on the Illumina sequencing platform (HiSeqTM 2500 or Illumina HiSeq X Ten) by Qingdao OE Biotech Co., Ltd. (Qingdao, China) according to a previous report (42). The clean reads were obtained after being trimmed off of reads with ploy-N and of low quality. To assemble the clean reads, de novo assemblies were carried out with Trinity (version: trinityrnaseq_r20131110) software. Subsequently, the assembled unigenes were used to perform the BLAST program on the databases of NCBI RefSeq (NR), Swiss-Prot, and KOG protein databases. In order to identify the differentially expressed genes (DEGs), FPKM and read count value of each annotation unigene was calculated using bowtie2 and eXpress, and DEGs were identified using the DESeq functions estimateSizeFactors and nbinomTest with the following parameters: p < 0.05 and fold change >2 or fold change <0.5. Then, DEGs were assigned to different Gene Ontology (GO) terms using the Blast2GO software. Meanwhile, DEGs were blasted against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database by using the automated assignment server (KAAS, http://www.genome.jp/kaas-bin/kaas_main) to identify the biological pathways of these unigenes.
The amino-acid sequence was predicted by using the DNAMAN software. Multiple sequence alignment was performed using ClustalW. The phylogenetic trees were constructed using the neighbor-joining method in MEGA version 7.0, with 1,000 replicates of bootstrap analysis. The sequences used in this study were downloaded from the NCBI database (Supplementary Table S2).
In total, the 2b-RAD-seq produced 140,265,655 raw reads (GenBank: PRJNA761225), and we obtained 131,644,844 clean reads after filtering low-quality reads. In order to cluster and assemble these clean reads, two female and two male samples were selected as references based on read quality and quantity. Following alignment with reference samples, we obtained 979,411 putative tags with an average depth of 33.75× from all 10 female and 10 male samples (Table 1). In addition, 43,922 putative SNP loci were screened by alignment analysis.
A total of 102.37G clean reads with a GC content of 37.16% and 99.94G clean reads with a GC content of 36.75% were acquired from the female and male samples, respectively (GenBank: PRJNA762987). The corresponding draft genomes were then assembled for both female and male. In order to identify sex-specific tag candidates, the 2b-RAD tags of each sample were aligned with both the female and male genome survey. We obtained 38 male-specific candidates that present exclusively in 10 male samples and 13 female-specific tags that only exist in 10 female samples (Supplementary Table S3, Additional Fasta File 1). Since a smaller p-value of SNP means a higher correlation with sex, the top 10 sex‐specific SNP loci were screened with the lowest p-values for further analysis. Notably, the females appeared to be heterozygous, whereas the males seemed to be homozygous for all 10 sex-specific SNP loci candidates in all samples we analyzed (Supplementary Table S4, Additional Fasta File 2), indicating that S. intermedius may have an ZW/ZZ sex determination system.
Among the 51 candidate sex-specific tags identified initially, six were too short to be analyzed and were thus excluded. These were female-tag 2, female-tag 5, male-tag 6, male-tag 12, male-tag 13, and male-tag 16. To confirm the authenticity of the remaining 45 tags, we designed primers based on the flanking sequences of these tags and performed PCR verification in 10 female and 10 male sea urchins. Unfortunately, we obtained the same PCR products from all 45 tags in both male and female sea urchins. Sanger sequencing analysis of the PCR products suggested that no difference was observed between male and female. Therefore, all these candidate sex-specific tag candidates were excluded from further study. Next, the same method was used to assess the authenticity of the 10 candidate SNP loci. PCR amplification suggested that nine of them did not show any difference between male and female. Hence, these nine sex-related SNP loci candidates were also excluded for any further analysis. Nonetheless, Sanger sequencing of the PCR products indeed confirmed one candidate sex-specific SNP locus (ref128486). Interestingly, all male individuals we analyzed displayed an apparently homozygous type (G/G), whereas the female individuals showed a G/A heterozygous type (Figure 1). Together, these data suggest that this sex-specific SNP may be used to identify the genetic sex of S. intermedius.
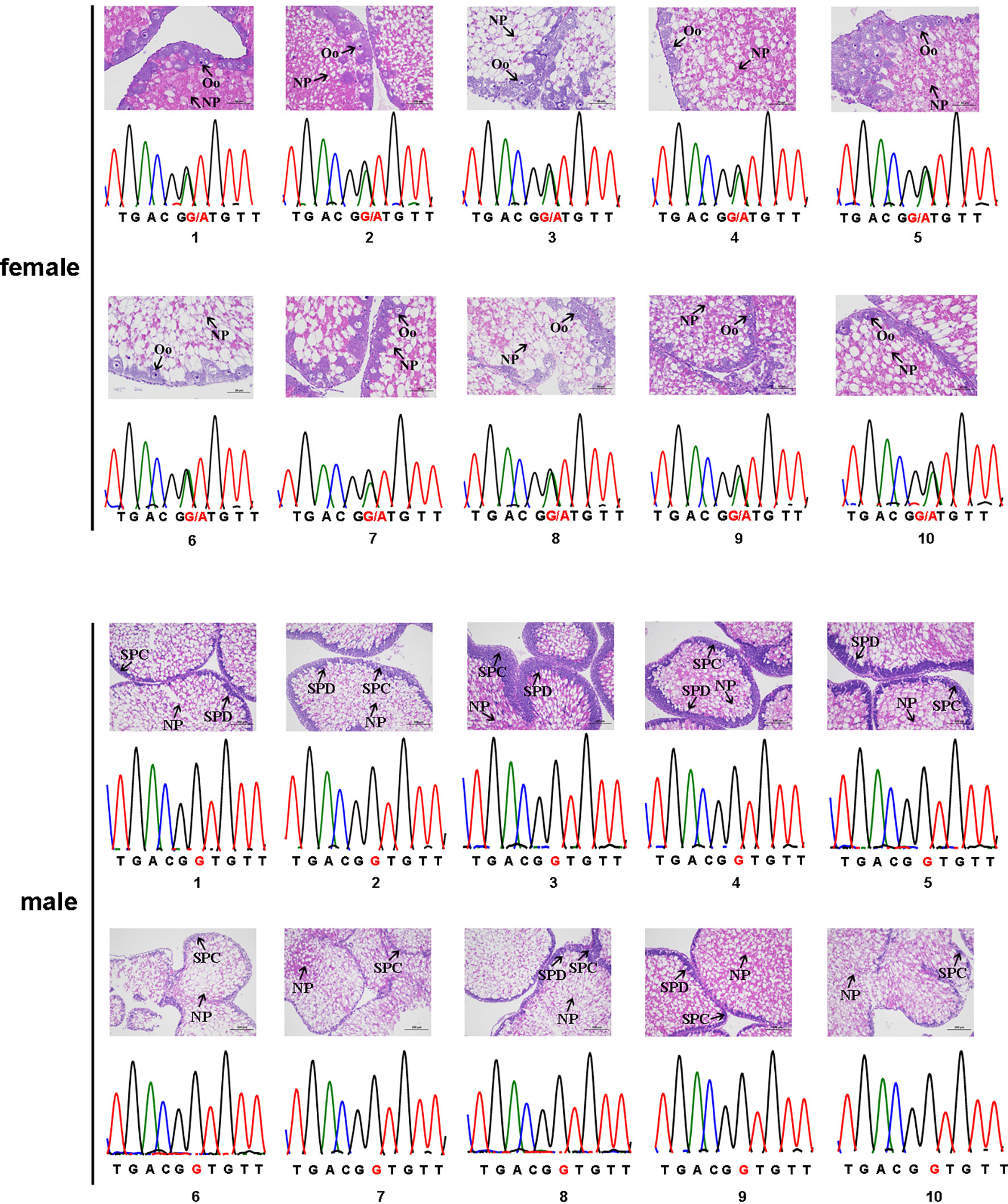
Figure 1 The sex-associated SNP of the Strongylocentrotus intermedius. The female genotype is G/A heterozygous and the male genotype is G/G homozygous. NP, nutritive phagocytes; SPC, spermatocyte; Oo, oocyte; SPD, spermatid.
To assess whether the above sex-specific SNP locus can be used to characterize the phenotypic sex of sea urchin, we ran PCR reactions with the same primers in 100 sea urchins. Prior to PCR amplification, gonadal histology analyses were performed to determine the phenotypic sex of each sea urchin and 65 females and 35 males were identified (Supplementary Figure S1). Among all 65 phenotypic female individuals, 56 (86.15%) exhibited the G/A heterozygous genotype (the dominant female genotype) and 9 (16.07%) exhibited the A/A homozygous genotype at the SNP locus. For the rest of the phenotypic male individuals, 33 (94.29%) exhibited the G/G homozygous genotype (the dominant male genotype) and 2 (5.71%) exhibited the G/A heterozygous genotype at the SNP locus (Table 2). Despite the fact that high consistency (89%) was obtained rapidly from the PCR amplification, the PCR products from 11 individuals (11%) were inconsistent with their phonotypic sex. Importantly, the segregation ratio of female to male was 65:35 (1.86:1) in this culture population. The above data indicate that sex reversal is possible in S. intermedius.
The sex-associated SNP locus is located on scaffold ref128486. Ref128486 is 6,149 bp in length. To search for homologous genes on the scaffold, we performed a BLAST program on NCBI databases. Interestingly, a spermatogenesis-associated (SPATA) family gene (spata4) was identified, and it has been reported that SPATA family genes play important roles in spermatogenesis, sperm maturation, and fertilization (43). The spata4 cDNA is 1,808 bp in length and contains an 825-bp open reading frame (ORF) that encodes a protein of 274 amino acids, a 489-bp 5′UTR, and a 494-bp 3′UTR. Because of the presence of an SNP locus, the 175th codon of the spata4 gene is CAC in males but CAU in females. However, codons of CAC and CAU are both coding for histidine (Figure 2A). Multiple protein sequence alignment analysis revealed that the Spata4 protein exhibited a high level of identity with other homologs, especially with the echinoderm, including Strongylocentrotus purpuratus (99.27%), L. variegatus (86.18%), and Acanthaster planci (63.16%; Supplementary Figure S2). Phylogenetic analysis showed that the topology of clades is basically consistent with the known taxonomic relationship among these analyzed species (Figure 2B). In addition, the mRNA expression pattern of spata4 in S. intermedius was detected by RT-qPCR in different adult tissues, including the testis, ovary, tube feet, and intestines. The results suggested that spata4 is expressed predominantly in the testis. By contrast, only a few of the spata4 transcripts can be detected in the ovary and intestine (Figure 3).
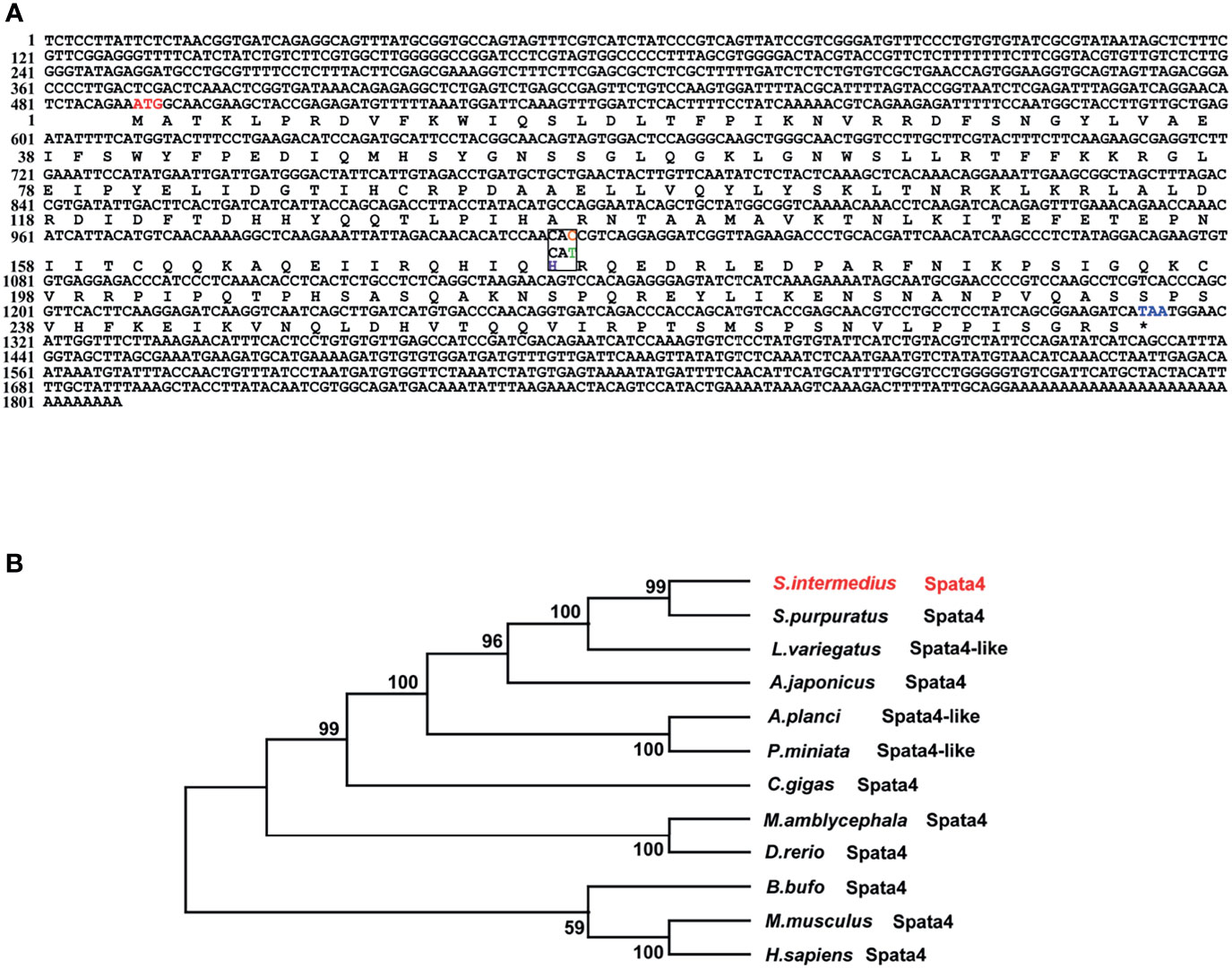
Figure 2 Sequence analysis of spata4 in S. intermedius. (A) Coding sequence and deduced amino acid sequences of spata4 in S. intermedius. (B) Phylogenetic tree of Spata4 proteins.
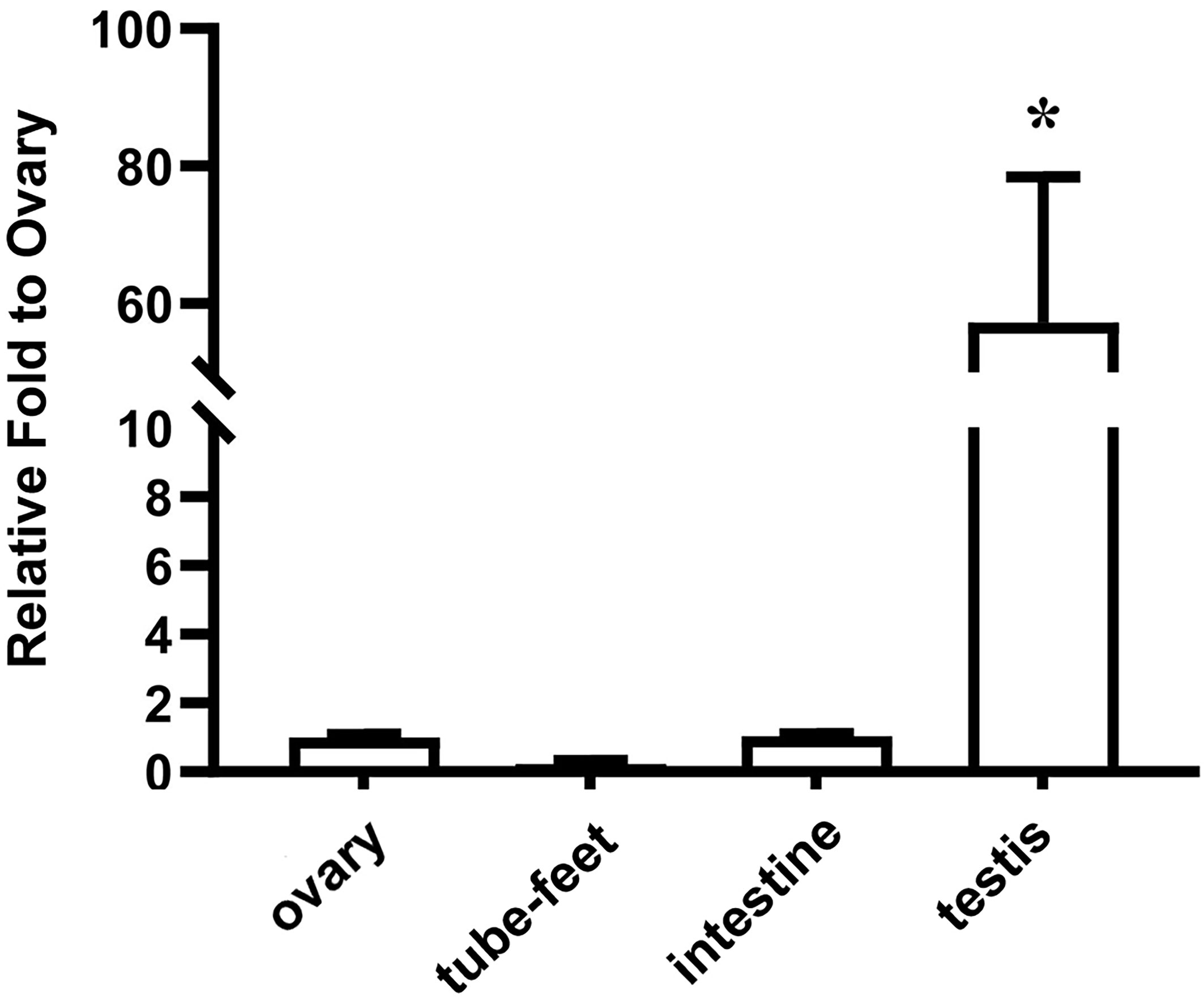
Figure 3 Expression pattern of spata4 mRNA in S. intermedius. 18S was used as the control. Each bar represents mean ± standard deviation (SD) (n = 3). One-way ANOVA was used to determine statistical analysis. Asterisks (*) indicate significant differences (p ≤ 0.05).
Considering that spata4 is expressed predominantly in the testis (Figure 3), we further explored its function in spermatogenesis by RNAi, a strategy that has been successfully utilized to study sex-related gene function in sea urchin (5). The genetic sex of nine sea urchins were first identified by PCR amplification and the corresponding products were sequenced. Gonadal tissues from three male sea urchins were removed and sampled as a control group before injection. The specific spata4-targeting double-stranded RNA (dsRNA) was then injected into adult S. intermedius. At 3 and 7 days post injection (dpi), gonadal samples were collected to perform histological analysis and to detect the spata4 mRNA expression with RT-qPCR. We observed that, compared with the control groups, the expression levels of spata4 in the testis were significantly decreased (about 60%) in knockdown samples (Figure 4A). Moreover, the results of histological examination showed that no obvious apoptotic cells were observed in the dsRNA-treated sea urchins (Supplementary Figure S3).
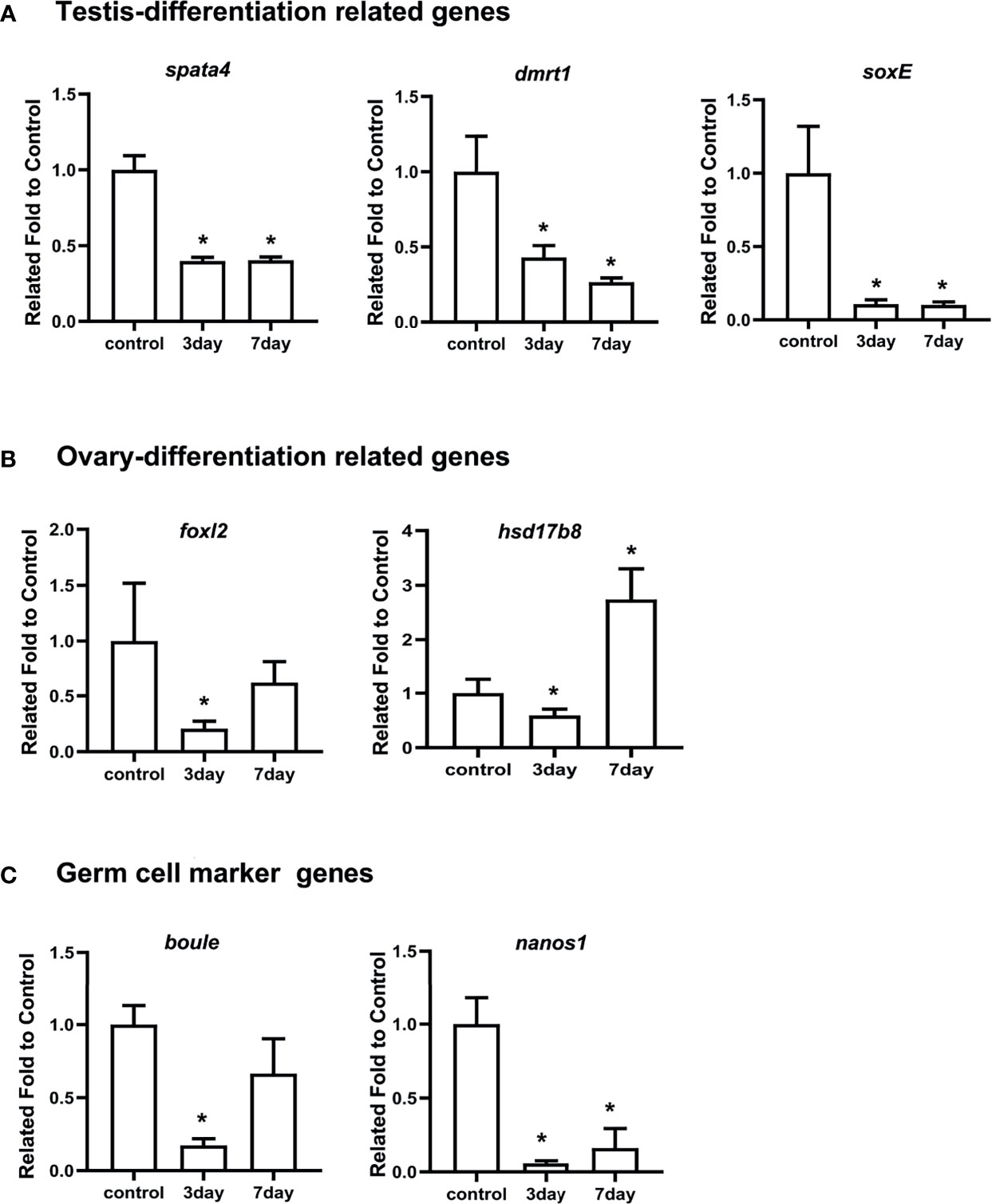
Figure 4 RT-qPCR measured the mRNA levels of spata4 and other sex-related genes after RNAi. (A) The expression of testis differentiation genes, including spata4, dmrt1, and soxE. (B) The expression of ovary differentiation genes, including foxl2 and hsd17b8. (C) The expression of germ cell marker genes, including boule and nanos1. 18S was used as the control. The gene expression in the control group was set to 1. Each bar represents mean ± standard deviation (SD) from three individuals. Independent-sample t-test was used to perform statistical analysis. Asterisks (*) indicate significant differences (p ≤ 0.05) between knockdown and control.
We also measured the expression levels of other well-studied genes involved in testis differentiation, ovary differentiation, and germ cell development. As shown in Figure 4A, the expression of dmrt1 and soxE in the testis differentiation signaling pathway significantly decreased in spata4-knockdown samples compared with the control groups at 3 and 7 dpi. Interestingly, the transcripts of foxl2 and hsd17b8 were similarly suppressed significantly in spata4-knockdown samples at 3 dpi, but the hsd17b8 expression increased about 2.74-fold in the knockdown samples at 7 dpi (Figure 4B). In addition, the germ cell marker genes, including boule and nanos1 expression, were also significantly decreased after injection in knockdown samples (Figure 4C).
Invertebrates show a high degree of variability in sex, and phenotypic sex may be affected by environmental factors, such as sex hormone, temperature, and stocking density (44). Because the genetic sex of 11 individuals identified by PCR amplification was inconsistent with their phonotypic sex and the sex ratio of females to males was not 1:1, we speculated that sex reversal is possible in S. intermedius. To test this hypothesis, we first determined the level of E2 in the gonads of S. intermedius by ELISA and found that the concentration of E2 was 17.4 and 13.75 pmol/L in the ovary and testis, respectively. We then characterized male sea urchins with the above SNP and feed them with formulated diet supplemented with 50 or 100 mg/kg E2 (defined as group 1 and group 2, respectively) for 58 days. At 3 days post feeding (dpf), the excrement was collected to detect E2 level. As shown in Figure 5A, the E2 concentration increased along with the dietary supplementation of E2. Gonadal tissues were also collected at 15, 30, and 58 dpf to examine the concentration of E2. As shown in Figure 5B, the E2 concentrations from sea urchins fed with E2 were comparable with those of the control group during a period of 15 days. On day 30, the concentrations of E2 in group 2 were 1.16 times higher than those of the control group. On day 58, the E2 concentrations from group 1 and group 2 were 1.20- and 1.20-fold against the control group, respectively (Figure 5B). These data suggested that S. intermedius can absorb estradiol from food and accumulate in the gonads.
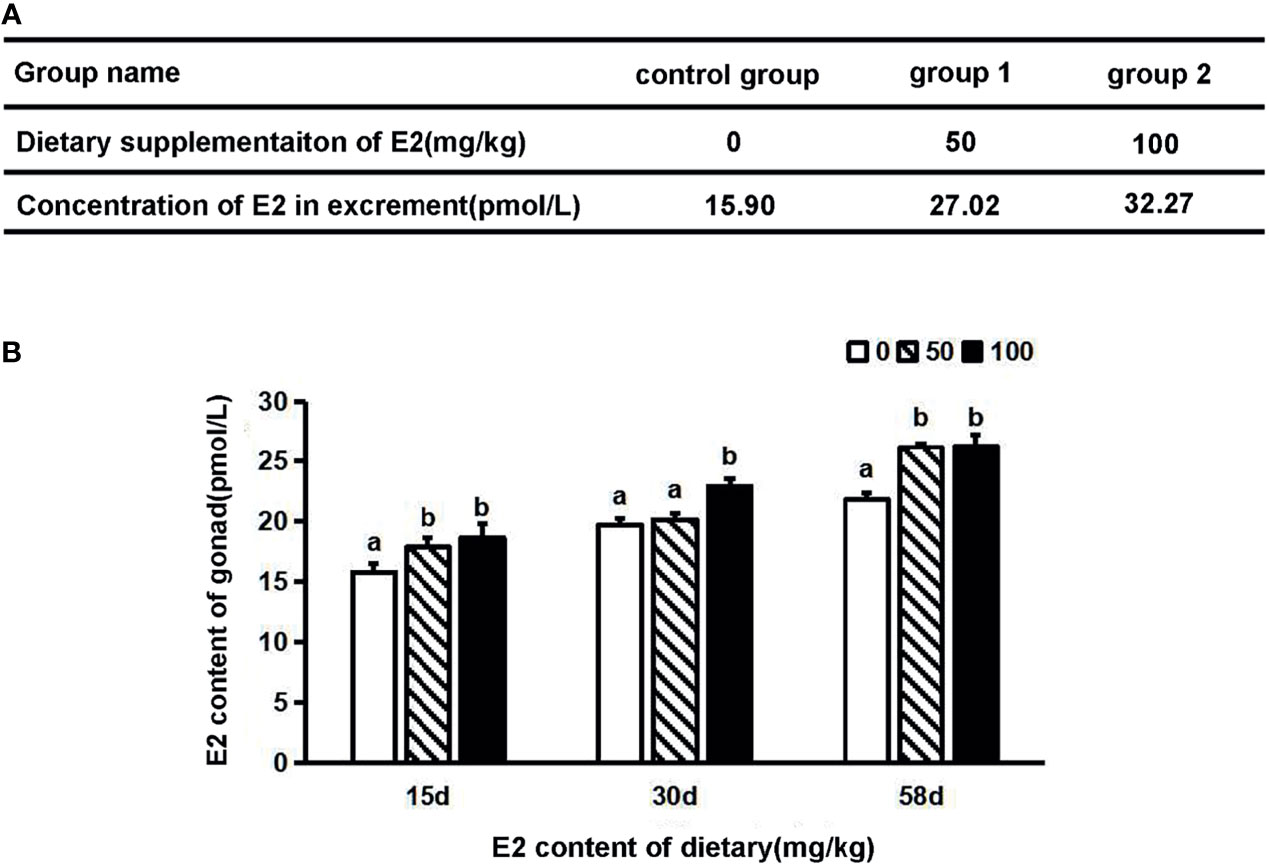
Figure 5 The concentration of estradiol levels in S. intermedius. (A) Concentration of estradiol in excrement after 3 days of dietary supplementation of E2. (B) Concentration of estradiol in testis on different days after dietary supplementation of E2 (n = 6). E2, estradiol; d, day. One-way ANOVA was used to determine statistical analysis. Different letters indicate significant differences (p ≤ 0.05).
To explore the dynamic expression changes of sex-related genes after E2 treatment, we performed comparative transcriptome analysis to investigate the expression characteristics and differentially expressed genes (DEGs) in the control group and in the 100-mg/kg E2-treated sea urchins (i.e., group 2). In total, 96.13M and 94.57M clean reads were acquired from group 2 and the control group libraries, respectively (GenBank: PRJNA761244). After assembly with de novo methods and functional annotation, 67,492 unigenes were identified. The expression levels of unigenes were then compared, and 2,355 DEGs with at least two-fold changes (p < 0.05) were identified. Of these DEGs, 1,050 were downregulated and 1,305 were upregulated in the E2 treatment group. In order to categorize the transcripts according to putative function, GO assignment was performed on all DEGs. Not surprisingly, several upregulated genes were enriched significantly in GO terms that are related to estrogen metabolism, spermatogenesis, and oogenesis (Figure 6A). These include development of primary sexual characteristics (GO:0045137), sperm-egg recognition (GO:0035036), ER-nucleus signaling pathway (GO:0006984), and sexual reproduction (GO:0019953). By contrast, many downregulated genes were significantly enriched in terms of germ plasm (GO:0060293) and positive regulation of fibroblast apoptotic process (GO:2000271) (Figure 6B). Searching the downregulated unigenes in the KOG database showed significant enrichment of unigenes in pathways previously implicated in ovarian steroidogenesis (ko04913), drug metabolism (ko00982), and cytochrome P450 estrogen signaling pathway (ko04915). Besides, the pathway of oocyte meiosis (ko04114) was recognized as a significantly upregulated term. Together, these data suggested that E2 treatment leads to downregulation of testis-biased genes but upregulation of ovary-biased genes and genes associated with estrogen metabolism pathway in S. intermedius.
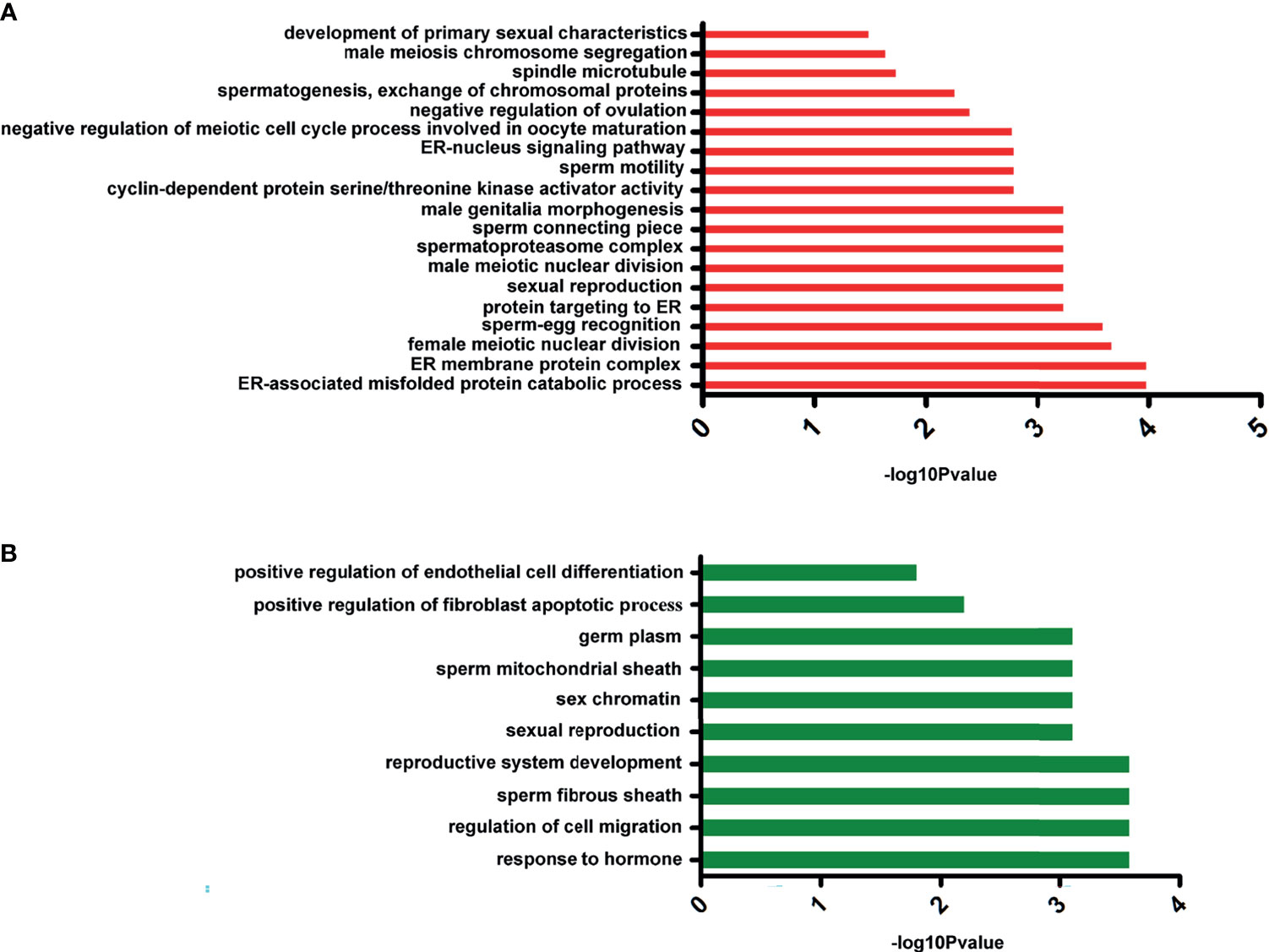
Figure 6 The significant Gene Ontology (GO) annotation of differentially expressed genes. (A) The GO cluster of upregulated genes. (B) The GO cluster of downregulated genes.
Lastly, we analyzed the dynamic expression patterns of several sex-related markers that were reported to have been involved in gonad development in the testis of E2-treated S. intermedius by RT-qPCR. As shown in Figure 7A, testis differentiation-associated genes, including dmrt1, soxE, and sp17, were significantly downregulated in the testis samples of E2-treated S. intermedius. Curiously, genes needed for ovary differentiation including hsd17b8, foxl2, and had17b14 were also downregulated (Figure 7B). Notably, E2 treatment led to a significant decrease in germ cell marker genes including nanos2, boule, seali, sycp3, and vasa in sea urchins (Figure 7C). In addition, steroidgenic enzyme genes related to active androgen biosynthesis such as cyp17a, cyp450, and nek1 were also downregulated in the testis of E2-treated sea urchins (Figure 7D). As expected, genes associated with estrogen metabolism including ER membrane protein complex subunit 4 (Emc4) were significantly upregulated (Figure 7E). However, no phenome of sex reversal was observed in the testis of E2-treated S. intermedius according to histological analysis (Supplementary Figure S4).
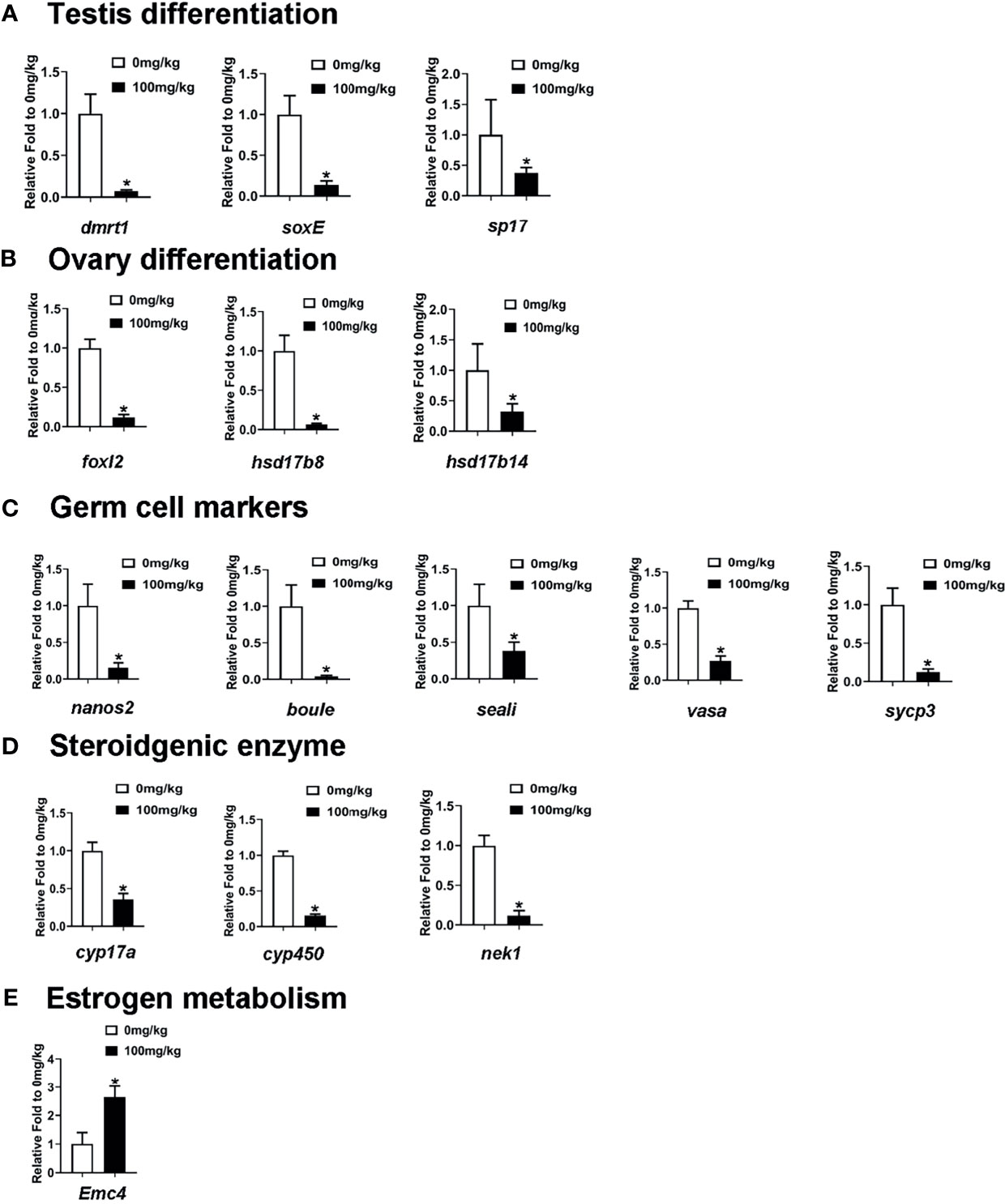
Figure 7 RT-qPCR measured the mRNA levels of sex-related genes and estrogen metabolism-related genes in the testis of E2-treated S. intermedius. (A) The expression of testis differentiation genes. (B) The expression of ovary differentiation genes. (C) The expression of germ cell marker gene. (D) The expression of steroidgenic enzyme gene. (E) The expression of estrogen metabolism gene (Emc4). 18S was used as the control. Each bar represents mean ± standard deviation (SD) (n = 3). One-way ANOVA was used to determine statistical analysis. Asterisks (*) indicate significant differences (p ≤ 0.05).
Identification of sex-specific markers is fundamental to understand the sex determination mechanism and is crucial for genetic sex identification and sex-controlled breeding (45, 46). Recently, 2b-RAD-seq has been successfully applied to identify sex-specific markers in a great diversity of aquatic organisms (9, 10). RNA interference is an effective genetic tool for studying gene function in vivo, especially in animals with low survival rates and long reproductive cycles (5). Currently, a green fluorescent protein (GFP)-derived double-stranded RNA (dsRNA-GFP) is usually used as control in RNAi studies, and no RNAi effect was detected using dsRNA-GFP suggesting target sequence specificity of the RNAi effect (47). However, according to a previous report in Apis mellifera, the expression of nearly 1,400 genes was dynamically changed in response to dsRNA-GFP (48). Hence, it is indeed a big challenge to evaluate target sequence specificity of interference effect.
In pufferfish, a female homozygous (C/C) type and a male heterozygous (C/G) SNP in anti-Mullerian hormone receptor type 2 (amhr2) were identified, and this SNP site can be used to identify the genotypic sex of pufferfish with high accuracy (49). Notably, the missense SNP that resulted in an amino acid mutation (His/Asp) in amhr2 affects its kinase activity, and the combination of the two alleles of amhr2 may determine the sex of Takifugu rubripes (50, 51). In the current study, a sex-linked SNP locus was identified in S. intermedius spata4 gene. The SNP locus is located at the 175th codon of the spata4 gene, but the CAC➔CAU change only causes a synonymous mutation in the predicted Spata4 protein. Synonymous mutations are thought to be silent for a long time. Recently, more and more studies reveal that synonymous sites do not evolve neutrally, and synonymous mutations can regulate protein conformation and function by affecting mRNA stability (52, 53). However, the effect of synonymous mutations on the expression and function of spata4 gene remains unclear and should be investigated in the future. In most of the species, spata4 is specifically expressed in the testis and may play important roles in maintaining spermatogenesis ability and infertility process (31, 43, 54). Herein, we revealed that spata4 is expressed predominantly in the testis, and only a few of spata4 transcripts can be detected in the ovary (Figure 3). As previously reported, dmrt1 and soxE were demonstrated to play critical roles in testis differentiation regulatory pathway (55, 56). Meanwhile, boule and nanos1 were previously described as germ cell markers and played conserved roles in regulating germ cell development (57, 58). Knocking down spata4 expression in sea urchins led to the downregulation of soxE, dmrt1, boule, and nanos1, indicating a close correlation between spata4 expression and testis development.
Although estradiol can be detected in the gonadal tissues of S. intermedius, we are uncertain whether it is absorbed from the environment or is endogenously synthesized. It has been reported that vertebrate steroids can be absorbed and utilized by invertebrates, especially those living in water, and all reproductively mature fish can release steroids into the environment (23, 25, 59). Sea urchins (L. variegatus, S. purpuratus, and S. intermedius) are also capable of absorbing vertebrate steroids. A notable example is that estradiol-17β exposure can induce protein synthesis in oocytes of S. intermedius (29).
It has been suggested that sex determination and sex differentiation of invertebrates may be influenced by environment factors such as temperature, nutrition level, and photoperiod (44). Because the ratio between female and male was not around 1:1 in the current study, we speculated that sex reversal might be the case in S. intermedius. After 58 days of dietary administration of estradiol, we did not observe significant sex reversal in male individuals by gonadal histology analysis. However, it should be noted that the expression levels of multiple genes have changed dynamically after estradiol treatment. In particular, many of these genes are known to be involved in testis differentiation, ovary differentiation, and germ cell development. Thus, it is possible that E2 treatment may affect gonadal development in sea urchin.
A vertebrate homologous estrogen receptor (ER) (ADL71443.1) is annotated in transcription database. Nonetheless, its expression level is relatively stable following E2 treatment, implying that the interacting mechanisms between exogenous estrogens and sea urchins may be different from that of vertebrates. In Marisa cornuarietis, estradiol-17β treatment has no obvious impact on the expression levels of ER-like or estrogen-related receptor (ERR) gene. This is not surprising because estradiol-17β does not bind to ER-like or ERR (60, 61). Hence, it is attempted to reveal the interactions between estrogen and estrogen receptor in sea urchin.
In conclusion, the current studies identified a sex-related SNP site in S. intermedius and revealed that this SNP is located within the spata4 gene. We confirmed that spata4 is expressed predominantly in the testis, and spata4 may be involved in testis development. In addition, S. intermedius can absorb vertebrate estradiol from dietary food, which caused dynamic changes of the sex-related genes. Taken together, our results provide a powerful tool for understanding the sex-determination mechanisms in sea urchins and have important implications for improving the breeding efficiency of sea urchins.
The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA761225, https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA762987, https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA761244. You can also get raw datas by searching the accession numbers (PRJNA761225, PRJNA761244, PRJNA762987) in NCBI. The accession number of spata4 sequence is OK078897. This sequence will be accessible after the indicated release date or the article was published.
Conceptualization: Z-HS. Software: Y-LH. Validation: Y-LH, SC, and BW. Investigation, Y-LH. Resources: JS and R-TZ. Data curation: Y-LH. Writing—original draft preparation: Z-HS. Writing—review and editing: Y-QC and Z-HS. Funding acquisition: Y-QC and Z-HS. All authors have read and agreed to the published version of the manuscript.
This work was funded by the National Key Research and Development Program of China (2018YFD0901603) and the National Natural Science Foundation of China (31802276).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.756530/full#supplementary-material
1. Agatsuma Y. Mesocentrotus Nudus. In: Lawrence JM, editor. Developments in Aquaculture and Fisheries Science, vol. 43. Amsterdam, FL: Elsevier Press (2020). p. 627–41. doi: 10.1016/B978-0-12-819570-3.00034-2
2. Chang YQ, Ding J, Song J, Yang W. Biology Research and Breeding of Sea Cucumber and Sea Urchin. Beijing: Ocean Press (2004).
3. Gil DG, Zaixso HE, Tolosano JA. Sex-Specific Differences in Gonopore and Gonadal Growth Trajectories in the Brooding Sea Urchin, Abatus Cavernosus (Spatangoida). Invertebr Biol (2020) 139(1):e12278. doi: 10.1111/ivb.12278
4. Arizza V, Vazzana M, Schillaci D, Russo D, Giaramita FT, Parrinello N. Gender Differences in the Immune System Activities of Sea Urchin Paracentrotus Lividus. Comp Biochem Physiol A: Mol Integr Physiol (2013) 164:447–55. doi: 10.1016/j.cbpa.2012.11.021
5. Zhang J, Han X, Wang J, Liu BZ, Wei JL, Zhang WJ, et al. Molecular Cloning and Sexually Dimorphic Expression Analysis of Nanos2 in the Sea Urchin, Mesocentrotus Nudus. Int J Mol Sci (2019) 20(11):2705. doi: 10.3390/ijms20112705
6. Cui ZP, Zhang J, Sun ZH, Liu BZ, Zhao C, Chang YQ. Identification of Sex-Specific Markers Through 2b-RAD Sequencing in the Sea Urchin (Mesocentrotus Nudus). Fron Genet (2021) 12:1433. doi: 10.3389/fgene.2021.717538
7. Wang H, Ding J, Ding S, Chang YQ. Transcriptome Analysis to Characterize the Genes Related to Gonad Growth and Fatty Acid Metabolism in the Sea Urchin Strongylocentrotus Intermedius. Genes Genomics (2019) 41:1397–415. doi: 10.1007/s13258-019-00864-0
8. Mi X, Wei Z, Zhou Z, Liu X. Identification and Profiling of Sex-Biased microRNAs From Sea Urchin Strongylocentrotus Nudus Gonad by Solexa Deep Sequencing. Comp Biochem Physiol Part D: Genomics Proteomics (2014) 10:1–8. doi: 10.1016/j.cbd.2014.01.001
9. Zhou Y, Wu J, Wang Z, Li G, Mei J, Zhou L, et al. Identification of Sex-Specific Markers and Heterogametic XX/XY Sex Determination System by 2b-RAD Sequencing in Redtail Catfish (Mystus Wyckioides). Aquac Res (2019) 50:2251–66. doi: 10.1111/are.14106
10. Zhu C, Liu H, Cheng L, Pan Z, Chang G, Wu N, et al. Identification of Sex-Specific Sequences Through 2b-RAD Sequencing in Pseudobagrus Ussuriensis. Aquaculture (2021) 539:736639. doi: 10.1016/j.aquaculture.2021.736639
11. Liu H, Pang M, Yu X, Zhou Y, Tong J, Fu B. Sex-Specific Markers Developed by Next-Generation Sequencing Confirmed an XX/XY Sex Determination System in Bighead Carp (Hypophthalmichehys Nobilis) and Silver Carp (Hypophthalmichthys Molitrix). DNA Res (2018) 25(3):257–64. doi: 10.1093/dnares/dsy009
12. Xue LZ, Guo XF, Zhou YL, Wang ZW, Fan HP, Li DP, et al. Screening and Characterization of Sex-Specific Markers by 2b-RAD Sequencing in Zig-Zag Eel (Mastacembelus Armatus) With Implication of XY Sex Determination System. Aquaculture (2020) 528:735550. doi: 10.1016/j.aquaculture.2020.735550
13. Shi X, Waiho K, Li X, Ikhwanuddin M, Miao G, Lin F, et al. Female-Specific SNP Markers Provide Insights Into a WZ/ZZ Sex Determination System for Mud Crabs Scylla Paramamosain, S. Tranquebarica and S. Serrata With a Rapid Method for Genetic Sex Identification. BMC Genomics (2018) 19:981. doi: 10.1186/s12864-018-5380-8
14. Wei JL, Cong JJ, Sun ZH, Song J, Zhao C, Chang YQ. A Rapid and Reliable Method for Genetic Sex Identification in Sea Cucumber, Apostichopus Japonicus. Aquaculture (2021) 543:737021. doi: 10.1016/J.AQUACULTURE.2021.737021
15. Mei J, Gui JF. Genetic Basis and Biotechnological Manipulation of Sexual Dimorphism and Sex Determination in Fish. Sci China Life Sci (2015) 58:124–36. doi: 10.1007/s11427-014-4797-9
16. Li XY, Gui JF. Diverse and Variable Sex Determination Mechanisms in Vertebrates. Sci China Life Sci (2018) 61(12):1503–14. doi: 10.1007/s11427-018-9415-7
17. Liu H, Guan B, Xu J, Hou C, Tian H, Chen HJ. Genetic Manipulation of Sex Ratio for the Large-Scale Breeding of YY Super-Male and XY All-Male Yellow Catfish (Pelteobagrus Fulvidraco (Richardson)). Mar Biotechnol (2013) 15:321–8. doi: 10.1007/s10126-012-9487-7
18. Ezaz MT, Myers JM, Powell SF, McAndrew BJ, Penman DJ. Sex Ratios in the Progeny of Androgenetic and Gynogenetic YY Male Nile Tilapia, Oreochromis Niloticus. Aquaculture (2004) 232(1-4):205–14. doi: 10.1016/j.aquaculture.2003.08.001
19. Liu H, Cui S, Hou C, Xu J, Hong XC. YY Supermale Generated Gynogenetically From XY Female in Pelteobagrus Fulvidraco (Richardson). Acta Hydrobiol Sinica (2007) 31(5):725. doi: 10.3321/j.issn:1000-3207.2007.05.018
20. Dan C, Mei J, Wang D, Gui JF. Genetic Differentiation and Efficient Sex-Specific Marker Development of a Pair of Y-And X-Linked Markers in Yellow Catfish. Int J Biol Sci (2013) 9(10):1043. doi: 10.7150/ijbs.7203
21. Warrier SR, Tirumalai R, Subramoniam T. Occurrence of Vertebrate Steroids, Estradiol 17β and Progesterone in the Reproducing Females of the Mud Crab Scylla Serrata. Comp Biochem Phys A: Mol Integr Physiol (2001) 130(2):283–94. doi: 10.1016/S1095-6433(01)00385-3
22. Zheng BH, An LH, Chang H, Liu Y, Jiang ZQ. Evidence for the Presence of Sex Steroid Hormones in Zhikong Scallop, Chlamys Farreri. J Steroid Biochem Mol Biol (2014) 143:199–206. doi: 10.1016/j.jsbmb.2014.03.002
23. Scott AP. Do Mollusks Use Vertebrate Sex Steroids as Reproductive Hormones? Part I: Critical Appraisal of the Evidence for the Presence, Biosynthesis and Uptake of Steroids. Steroids (2012) 77:1450–68. doi: 10.1016/j.steroids.2012.08.009
24. Scott AP. Is There Any Value in Measuring Vertebrate Steroids in Invertebrates? Gen Comp Endocr (2018) 265:77–82. doi: 10.1016/j.ygcen.2018.04.005
25. Schwarz TI, Katsiadaki I, Maskrey BH, Scott AP. Mussels (Mytilus Spp.) Display an Ability for Rapid and High Capacity Uptake of the Vertebrate Steroid, Estradiol-17β From Water. J Steroid Biochem Mol Biol (2017) 165:407–20. doi: 10.1016/j.jsbmb.2016.08.007
26. Horiguchi T, Ohta Y. Advances in Invertebrate (Neuro)Endocrinology. Oakville: Apple Academic Press (2020).
27. Andrew MN, O’Connor WA, Dunstan RH, MacFarlane GR. Exposure to 17α-Ethynylestradiol Causes Dose and Temporally Dependent Changes in Intersex, Females and Vitellogenin Production in the Sydney Rock Oyster. Ecotoxicology (2010) 19(8):1440–51. doi: 10.1007/s10646-010-0529-5
28. Lu Y, Liu M, Gong J, Cheng Y, Wu X. Effect of Exogenous Estrogen on the Ovarian Development and Gene Expression in the Female Swimming Crab Portunus Trituberculatus (Miers, 1876) (Decapoda: Brachyura: Portunidae). J Crustacean Biol (2018) 38:367–73. doi: 10.1093/jcbiol/ruy013
29. Varaksina GS, Varaksin AA. Effect of Estradiol Dipropionate on Protein Synthesis in Oocytes and Ovary of the Sea Urchin Strongylocentrotus Intermedius at Different Stages of the Reproductive Cycle. Biol Bull Russian Acad Sci (2002) 29:96–500. doi: 10.1023/A:1020422028333
30. Wasson K, Gower B, Watts SA. Responses of Ovaries and Testes of Lytechinus Variegatus (Echinodermata: Echinoidea) to Dietary Administration of Estradiol, Progesterone and Testosterone. Mar Biol (2000) 137:245–55. doi: 10.1007/s002270000360
31. Liu SF, He S, Liu BW, Zhao Y, Wang Z. Cloning and Characterization of Testis-Specific Spermatogenesis Associated Gene Homologous to Human SPATA4 in Rat. Biol Pharm Bull (2004) 27:1867–70. doi: 10.1248/bpb.27.1867
32. Liu SF, Ai C, Ge ZQ, Liu HL, Liu BW, He S, et al. Molecular Cloning and Bioinformatic Analysis of SPATA4 Gene. J Biochem Mol Biol (2005) 38:739–47. doi: 10.5483/BMBRep.2005.38.6.739
33. Xie MC, Ai C, Jin XM, Liu SF, Tao SX, Li ZD, et al. Cloning and Characterization of Chicken SPATA4 Gene and Analysis of Its Specific Expression. Mol Cell Biochem (2007) 306(1):79–85. doi: 10.1007/s11010-007-9556-9
34. Liu SF, Liu BW, He S, Zhao Y, Wang Z. Cloning and Characterization of Zebra Fish SPATA4 Gene and Analysis of Its Gonad Specific Expression. Biochem (Moscow) (2005) 70:638–44. doi: 10.1007/s10541-005-0163-7
35. Liu BW, Liu SF, He S, Zhao Y, Hu H, Wang Z. Cloning and Expression Analysis of Gonadogenesis-Associated Gene SPATA4 From Rainbow Trout (Oncorhynchus Mykiss). BMB Rep (2005) 38(2):206–10. doi: 10.5483/bmbrep.2005.38.2.206
36. Jiang J, Zhang N, Shiba H, Li L, Wang Z. Spermatogenesis Associated 4 Promotes Sertoli Cell Proliferation Modulated Negatively by Regulatory Factor X1. PloS One (2013) 8(10):e75933. doi: 10.1371/journal.pone.0075933
37. Wang S, Meyer E, McKay JK, Matz MV. 2b-RAD: A Simple and Flexible Method for Genome-Wide Genotyping. Nat Methods (2012) 9(8):808–8109. doi: 10.1038/nmeth.2023
38. Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: A Fast and Accurate Illumina Paired-End Read Merger. Bioinformatics (2014) 30(5):614–20. doi: 10.1093/bioinformatics/btt593
39. Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait J.H.J.G.G. Stacks: Building and Genotyping Loci De Novo From Short-Read Sequences. G3: Genes Genomes Genet (2011) 1:171–82. doi: 10.1534/g3.111.000240
40. Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, et al. Soap2: An Improved Ultrafast Tool for Short Read Alignment. Bioinformatics (2009) 25:1966–7. doi: 10.1093/bioinformatics/btp336
41. Sun ZH, Wei JL, Cui ZP, Han YL, Zhang J, Song J, et al. Identification and Functional Characterization of Piwi1 Gene in Sea Cucumber, Apostichopus Japonicas. Comp Biochem Physiol Part B: Biochem Mol Biol (2021) 252:110536. doi: 10.1016/j.cbpb.2020.110536
42. Sun ZH, Zhang J, Zhang WJ, Chang YQ. Gonadal Transcriptomic Analysis and Identification of Candidate Sex-Related Genes in Mesocentrotus Nudus. Gene (2019) 698:72–81. doi: 10.1016/j.gene.2019.02.054
43. Sujit KM, Singh V, Trivedi S, Singh K, Gupta G, Rajender SJA. Increased DNA Methylation in the Spermatogenesis-Associated (SPATA) Genes Correlates With Infertility. Andrology (2020) 8(3):602–9. doi: 10.1111/andr.12742
44. Cook JM. Sex Ratios: Concepts and Research Methods. Cambridge: Cambridge University Press (2002)
45. Arslan T, Phelps RPJA. Production of Monosex Male Black Crappie, Pomoxis Nigromaculatus, Populations by Multiple Androgen Immersion. Aquaculture (2004) 234(1-4):561–73. doi: 10.1016/j.aquaculture.2003.12.007
46. Chen SL, Li J, Deng SP, Tian YS, Wang QY, Zhuang ZM, et al. Isolation of Female-Specific AFLP Markers and Molecular Identification of Genetic Sex in Half-Smooth Tongue Sole (Cynoglossus Semilaevis). Mar Biotechnol (2007) 9(2):273–80. doi: 10.1007/s10126-006-6081-x
47. Ma KY, Chen J, Liu ZQ, Qiu GF. Inhibitory Effects of RNAi-Mediated Knockdown of EsDmrt-Like Gene on Testicular Development in the Chinese Mitten Crab Eriocheir Sinensis. Aquaculture (2016) 463:217–23. doi: 10.1016/j.aquaculture.2016.06.003
48. Nunes FMF, Aleixo AC, Barchuk AR, Bomtorin AD, Grozinger CM, Simões ZLP. Non-Target Effects of Green Fluorescent Protein (GFP)-Derived Double-Stranded Rna (dsRNA-GFP) Used in Honey Bee RNA Interference (RNAi) Assays. Insects (2013) 4:90–103. doi: 10.3390/insects401009
49. Gao FX, Shi Y, Duan W, Lu WJ, Huang W, Zhang XJ, et al. A Rapid and Reliable Method for Identifying Genetic Sex in Obscure Pufferfish (Takifugu Obscurus). Aquaculture (2020) 519:734749. doi: 10.1016/j.aquaculture.2019.734749
50. Kamiya T, Kai W, Tasumi S, Oka A, Matsunaga T, Mizuno N, et al. A Trans-Species Missense SNP in Amhr2 Is Associated With Sex Determination in the Tiger Pufferfish, Takifugu Rubripes (Fugu). PloS Genet (2012) 8(7):e1002798. doi: 10.1371/journal.pgen.1002798
51. Kikuchi K, Kai W, Hosokawa A, Mizuno N, Suetake H, Asahina K, et al. The Sex-Determining Locus in the Tiger Pufferfish, Takifugu Rubripes. Genetics (2007) 175(4):2039–42. doi: 10.1534/GENETICS.106.069278
52. Chamary JV, Hurst LD. Evidence for Selection on Synonymous Mutations Affecting Stability of mRNA Secondary Structure in Mammals. Genome Biol (2005) 6(9):1–12. doi: 10.1186/gb-2005-6-9-r75
53. Sauna ZE, Kimchi-Sarfaty C. Understanding the Contribution of Synonymous Mutations to Human Disease. Nat Rev Genet (2011) 12(10):683–91. doi: 10.1038/nrg3051
54. Liu SF, Li LY, Fu JJ, Liu G, Xing XW, Lu GX. Molecular Cloning of SRG2, A Mouse Testis Spermatocyte Apoptosis-Related Gene. Acta Biochim Biophys Sin (2002) 34(6):796–9. doi: 10.1007/BF02943277
55. Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D. DMRT1 Prevents Female Reprogramming in the Postnatal Mammalian Testis. Nature (2011) 476(7358):101–4. doi: 10.1038/nature10239
56. Barrionuevo F, Scherer G. SOX E Genes: SOX9 and SOX8 in Mammalian Testis Development. Int J Biochem Cell B (2010) 42.3:433–6. doi: 10.1016/j.biocel.2009.07.015
57. Han YL, Wei JL, Cui ZP, Zhang J, Zhang WJ, Wen B, et al. Expression and Cellular Localization of Maternal Factor Boule in Gonad of Sea Urchin Mesocentrotus Nudus. J Dalian Fish Univ (2021) 36(2):214–21. doi: 10.16535/j.cnki.dlhyxb.2020-074
58. Lai F, Singh A, King ML. Xenopus Nanos1 Is Required to Prevent Endoderm Gene Expression and Apoptosis in Primordial Germ Cells. Development (2012) 139(8):1476–86. doi: 10.1242/dev.079608
59. Scott AP, Ellis T. Measurement of Fish Steroids in Water—a Review. Gen Comp Endocr (2007) 153:392–400. doi: 10.1016/j.ygcen.2006.11.006
60. Bannister R, Beresford N, Granger DW, Pounds NA, Rand-Weaver M, White R, et al. No Substantial Changes in Estrogen Receptor and Estrogen-Related Receptor Orthologue Gene Transcription in Marisa Cornuarietis Exposed to Estrogenic Chemicals. Aquat Toxicol (2013) 140:19–26. doi: 10.1016/j.aquatox.2013.05.002
Keywords: SNP, sea urchin, estradiol, 2b-RAD, sex identification
Citation: Han Y-L, Sun Z-H, Chang S, Wen B, Song J, Zuo R-T and Chang Y-Q (2021) Application of SNP in Genetic Sex Identification and Effect of Estradiol on Gene Expression of Sex-Related Genes in Strongylocentrotus intermedius. Front. Endocrinol. 12:756530. doi: 10.3389/fendo.2021.756530
Received: 10 August 2021; Accepted: 22 September 2021;
Published: 11 November 2021.
Edited by:
Shubo Jin, Chinese Academy of Fishery Sciences, ChinaReviewed by:
Lijuan Wang, Institute of Oceanology (CAS), ChinaCopyright © 2021 Han, Sun, Chang, Wen, Song, Zuo and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi-Hui Sun, c3VuemhpaHVpQGRsb3UuZWR1LmNu; Ya-Qing Chang, Y2hhbmdsYWJAaG90bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.