- 1Key Laboratory of Freshwater Fisheries and Germplasm Resources Utilization, Ministry of Agriculture, Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences, Wuxi, China
- 2Wuxi Fisheries College, Nanjing Agricultural University, Wuxi, China
Pyruvate dehydrogenase E1 (PDHE1) is thought to play essential roles in energy metabolism, and a previous study suggested that it also has potential regulatory roles in male sexual development in the oriental river prawn, Macrobrachium nipponense. In this study, we used rapid amplification of cDNA ends, quantitative polymerase chain reaction (qPCR), in situ hybridization, western blotting, RNA interference (RNAi), and histological analyses to assess the potential functions of Mn-PDHE1 in the sexual development of male M. nipponense. The full cDNA sequence of Mn-PDHE1 was 1,614 base pairs long, including a 1,077 base pair open reading frame that encodes 358 amino acids. qPCR analysis revealed the regulatory functions of PDHE1 in male sexual development in M. nipponense and in the metamorphosis process. In situ hybridization and western blot results indicated that PDHE1 was involved in testis development, and RNAi analysis showed that PDHE1 positively regulated the expression of insulin-like androgenic gland factor in M. nipponense. Compared with the cell types in the testes of control prawns, histological analysis showed that the number of sperm was dramatically lower after test subjects were injected with Mn-PDHE1 dsRNA, whereas the numbers of spermatogonia and spermatocytes were higher. Sperm constituted only 1% of cells at 14 days after injection in the RNAi group. This indicated that knockdown of the expression of PDHE1 delayed testis development. Thus, PDHE1 has positive effects on male sexual development in M. nipponense. This study highlights the functions of PDHE1 in M. nipponense and its essential roles in the regulation of testis development.
Introduction
The oriental river prawn, Macrobrachium nipponense (Crustacea; Decapoda; Palaemonidae), is widely distributed in China and other Asian countries, and had an annual aquaculture production of 205,010 tons in 2016 (1–4). Male prawns show better growth performance than their female counterparts, as they grow faster and reach larger size by harvest time (2). However, the rapid development of the testis during the reproductive season restricts the sustainable development of M. nipponense aquaculture. Jin et al. (5) reported that the testis of M. nipponense can reach sexual maturity within 40 days after hatching, thus inbreeding can occur between newly hatched prawns. This will lead to small-sized prawns, degradation of germplasm resources, and decreased ability of offspring to tolerate adverse conditions. A better understanding of male sexual differentiation and development mechanisms is urgently needed to develop a technique to produce all male progeny on a commercial scale and to regulate the process of testis development in M. nipponense.
The androgenic gland and its secreted hormones play important positive regulatory roles in the process of male sexual differentiation and development in crustacean species, and it is especially true for testis development (6, 7). The testis also has significant effects on male sexual differentiation, sexual maturity, and reproductive capability in crustacean species (8–11). Jin et al. (5) previously showed that the androgenic gland regulates testis development in M. nipponense.
Many environmental factors, including temperature, illumination, and the presence of chemical pollutants, can regulate sexual differentiation and development by affecting the expression profiles of sex-related genes (12). Jin et al. (13) previously described significant morphological differences in the testis and androgenic gland of M. nipponense between the reproductive and non-reproductive seasons. Using transcriptome profiling to analyse the testis and androgenic gland during the two seasons, it was found that glycolysis/gluconeogenesis and the tricarboxylic acid (TCA) cycle may play essential roles in promoting the process of male sexual differentiation and development in M. nipponense by providing ATP. Pyruvate dehydrogenase E1 (PDHE1) is an important gene that is enriched in both the glycolysis/gluconeogenesis and TCA metabolic pathways (13). The PDH complex (PDHc) plays essential roles in the glycolysis pathway. Glucose is reduced to form pyruvate through glycolysis. PDHc can catalyse the oxidative decarboxylation of pyruvate to become acetyl-CoA, which is required by the TCA cycle, depending on the needs of cells (14, 15). PDH deficiency means that pyruvate cannot be converted into acetyl-CoA but instead is reduced to lactic acid, thus affecting the TCA cycle and resulting in decreased ATP production (16). An abnormal TCA cycle leads to metabolic disorders and tissue damage. Therefore, PDHc plays an important role in maintaining the normal metabolism of animals. PDHE1 is the key enzyme component of PDHc, as it catalyses the rate-limiting step of the oxidative decarboxylation of pyruvate (17, 18).
The goal of this study was to verify the important functions of PDHE1 in the process of male sexual development in M. nipponense using rapid amplification of cDNA ends (RACE) cloning, quantitative polymerase chain reaction (qPCR), in situ hybridization, western blot, RNA interference (RNAi), and histological analyses. We focused on the potential regulatory roles of PDHE1 on male sexual development in M. nipponense. Results of this study provide essential data that can be used to establish a technique to regulate testis development in M. nipponense.
Materials and Methods
Ethics Statement
All experiments involving M. nipponense were approved by the Institutional Animal Care and Use Ethics Committee of the Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences (Wuxi, China).
Sample Preparation
Healthy adult male and female M. nipponense with the body weight of 2.87-4.51 g for male prawns and the body weight of 2.37-3.45 g for female prawns were obtained from Tai Lake in Wuxi, China (120°13’44”E, 31°28’22”N). These specimens were transferred to a 500 L tank in the lab and maintained in aerated freshwater at room temperature (28°C) with a dissolved oxygen content ≥ 6 mg/L. The specimens were maintained under lab conditions for 72 h prior to tissue collection. The testis, ovary, hepatopancreas, muscle, eyestalk, gill, heart, and brain were collected from five different prawns (19). The ovarian sample was the mixed sample of the whole ovarian reproductive cycle, which included ovarian stage I (O I), O II, O III, O IV, O V. The five standard phases of the ovarian reproductive cycle have been well described by previous study (20). Additionally, one male prawn and one female prawn were mated and young were hatched in the lab to produce the full-sib population. Specimens for the different developmental stages were collected from this full-sib population during their maturation process (19). For males, the testis and androgenic gland were collected from five specimens during the reproductive season (at 28°C in summer) and from five specimens during the non-reproductive season (at 15°C in winter). Five prawns were also sampled from each of the five standard phases of the ovarian reproductive cycle. The samples were treated with phosphate buffered saline and immediately frozen in liquid nitrogen until used for RNA and protein extraction to prevent total RNA and protein degradation.
RACE
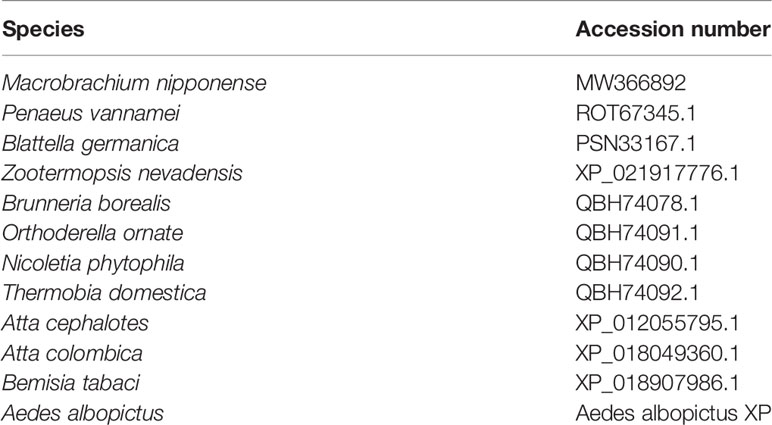
We followed previously described procedures to conduct RACE cloning (21, 22). The specific primers used for Mn-PDHE1 RACE cloning were listed in Table 1, designed by the Primer-BLAST tool in NCBI (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Table 1). Table 2 lists sequences of PDHE1 from different species. The phylogenetic tree was constructed by MEGA X using the maximum-likelihood method with 1000 bootstrap replications.
qPCR Analysis
The relative mRNA expression levels of Mn-PDHE1 in the different tissues were measured using qPCR, which was performed on the Bio-Rad iCycler iQ5 Real-Time PCR System (Bio-Rad, Hercules, CA, USA) and used to carry out the SYBR Green RT-qPCR assay (21, 22). Table 1 lists the primers used for qPCR analysis. The expression levels between different tissues were measured by using 2-ΔΔCT method (23).
In Situ Hybridization
In situ hybridization (22, 24) was performed to analyse the mRNA locations of Mn-PDHE1. Primer5 software was used to design the anti-sense and sense probes for chromogenic in-situ hybridization, with the digoxygenin signal based on the cDNA sequence of Mn-PDHE1 (Table 1). The primers for in situ hybridization analysis were synthesized by Shanghai Sangon Biotech Company (Shanghai, China). Slides were examined under light microscope for evaluation the cell types.
Western Blot Analysis
Western blot analysis was conducted on the 20 mg tissues samples following the previous study (25). The total protein concentrations were quantified using the Bradford method (26). A total of 50 mg protein from each sample was separated on a 10% sodium dodecyl-polyacrylamide gel. The gel was then transferred to a polyvinylidene fluoride membrane (Millipore, Bedford, MA, USA).
RNAi Analysis
RNAi was performed to analyse the potential regulatory roles of Mn-PDHE1 in M. nipponense. For this analysis, 300 healthy mature male M. nipponense were collected (body weight 3.34–4.47 g). These prawns were randomly divided into the RNAi group and the control group, each containing 150 male prawns. Each group was further divided into three replicates of 50 prawns, which were maintained in 500 L tanks at 28°C with dissolved oxygen content ≥ 6 mg/L. The procedures for designing specific RNAi primer and synthesizing the dsRNA have been well described in previous studies (24, 27). The concentration of Mn-PDHE1 dsRNA was measured using a spectrophotometer and diluted to 4 μg/μl. The prawns in the RNAi group were injected with 4 μg/g of Mn-PDHE1 dsRNA (24, 27) and those in the control group were injected with an equal volume of green fluorescent protein, based on their body weights. qPCR was used to investigate the PDHE1 mRNA expression in the testis after injection (N=5). To assess the regulatory relationship between Mn-PDHE1 and insulin-like androgenic gland hormone (Mn-IAG), the IAG mRNA expression levels were also measured in the same Mn-PDHE1 dsRNA-treated prawns on the same days (N=10).
Histological Observations
Morphological differences between the testis of prawns in the RNAi group and the testis of samples in the control group were observed using haematoxylin and eosin (HE) staining after Mn-PDHE1 dsRNA injection (N = 5). We followed the methods described in previous studies (28, 29). The prepared testis samples were observed using an Olympus SZX16 microscope (Olympus Corporation, Tokyo, Japan).
Statistical Analysis
SPSS Statistics 23.0 (IBM, Armonk, NY, USA) was used to conduct all statistical analyses. Statistical differences were identified by one-way analysis of variance followed by least significant difference and Duncan’s multiple range tests. The statistical difference between the control group and the RNAi group on the same day was assessed using the paired t-test. Quantitative data were expressed as mean ± standard deviation. A p-value < 0.05 was considered to be statistically significant.
Results
Genome and cDNA Sequence Analysis
The full cDNA sequence of Mn-PDHE1 was 1,614 base pairs (bp) long, with a 5′ untranslated region (UTRs) and a 3′ UTR of 264 bp and 273 bp, respectively (Figure 1). Mn-PDHE1 included a 1,077 bp open reading frame that encoded 358 amino acids. The sequence was submitted to NCBI with accession number MW366892. The molecular weight of the Mn-PDHE1 protein was 38.539 kDa, and the theoretical isoelectric point of Mn-PDHE1 protein was 6.56.

Figure 1 Nucleotide and deduced amino acid sequence of Mn-PDHE1. The nucleotide sequence is displayed in the 5′–3′ directions and numbered at the left. The deduced amino acid sequence is shown in a single capital letter amino acid code. 3′ UTR and 5′ UTR are shown with lowercase letters. Codons are numbered at the left with the methionine (ATG) initiation codon, an asterisk denotes the termination codon (TGA).
The BLASTP similarity analysis in NCBI showed that Mn-PDHE1 had the highest sequence similarity with that of Penaeus vannamei (90.72%). The similarities with other PDHE1 sequences from insect species were also > 70%, including Blattella germanica (81.66%), Zootermopsis nevadensis (81.47%), Brunneria borealis (77.31%), and Nicoletia phytophila (79.30%) (Figure 2). The amino acid sequences listed in Table 2 were used to construct the phylogenetic tree, which showed two main clusters. The amino acid sequence of Mn-PDHE1 clustered with that of P. vannamei, whereas the amino acid sequences of PDHE1 from the insect species clustered together as another group. Thus, Mn-PDHE1 has the closest evolutionary relationship with that of P. vannamei and a dramatically distant evolutionary relationship with those from insect species (Figure 3).
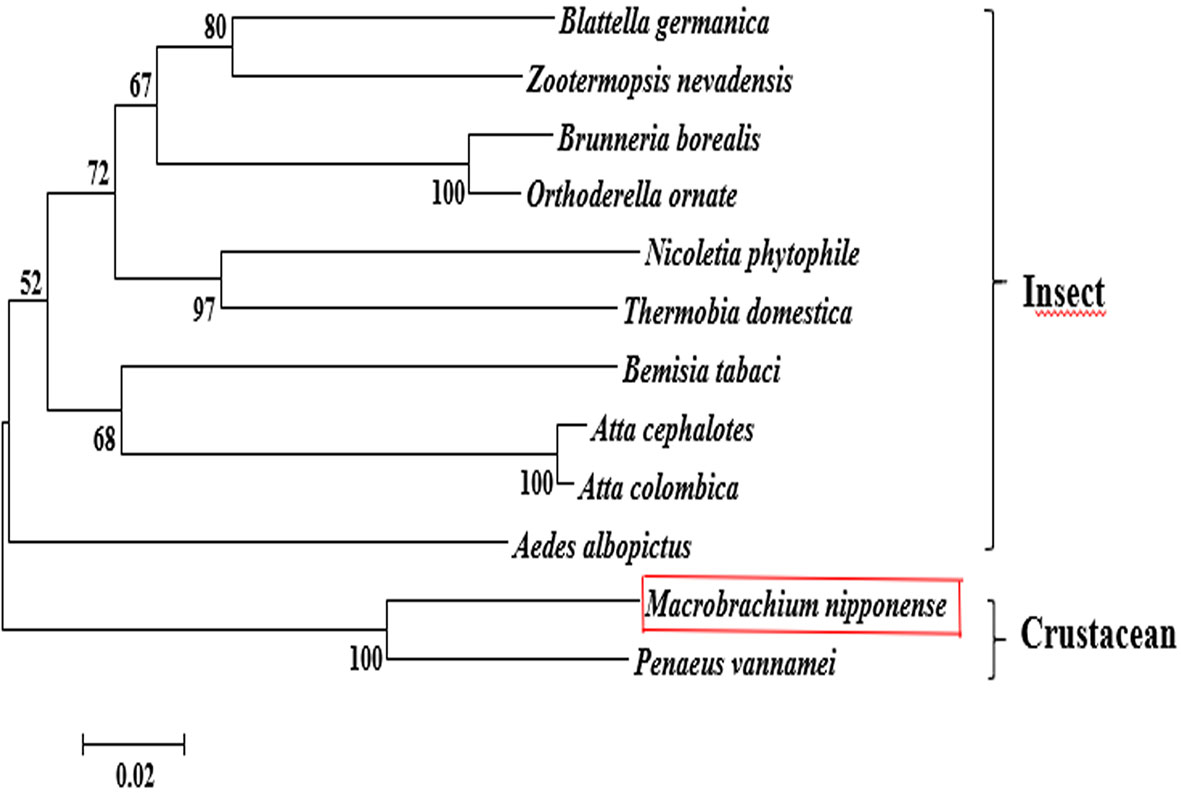
Figure 3 The phylogenetic tree of PDHE1 from different organisms based on amino acid sequence comparisons. Species names and types of PDHE1 are listed on the right of the tree. Red rectangles indicated M. nipponense.
Expression of Mn-PDHE1 in Different Tissues and Developmental Stages
Among the tissues tested, the Mn-PDHE1 mRNA expression level was highest in the testis, followed by the heart and ovary, which showed significant difference with other tested tissues (p < 0.05). The lowest expression was observed in muscle, and the expression levels in the testis and ovary were 20.37-fold and 13.18-fold higher than that in muscle, respectively (Figure 4A).
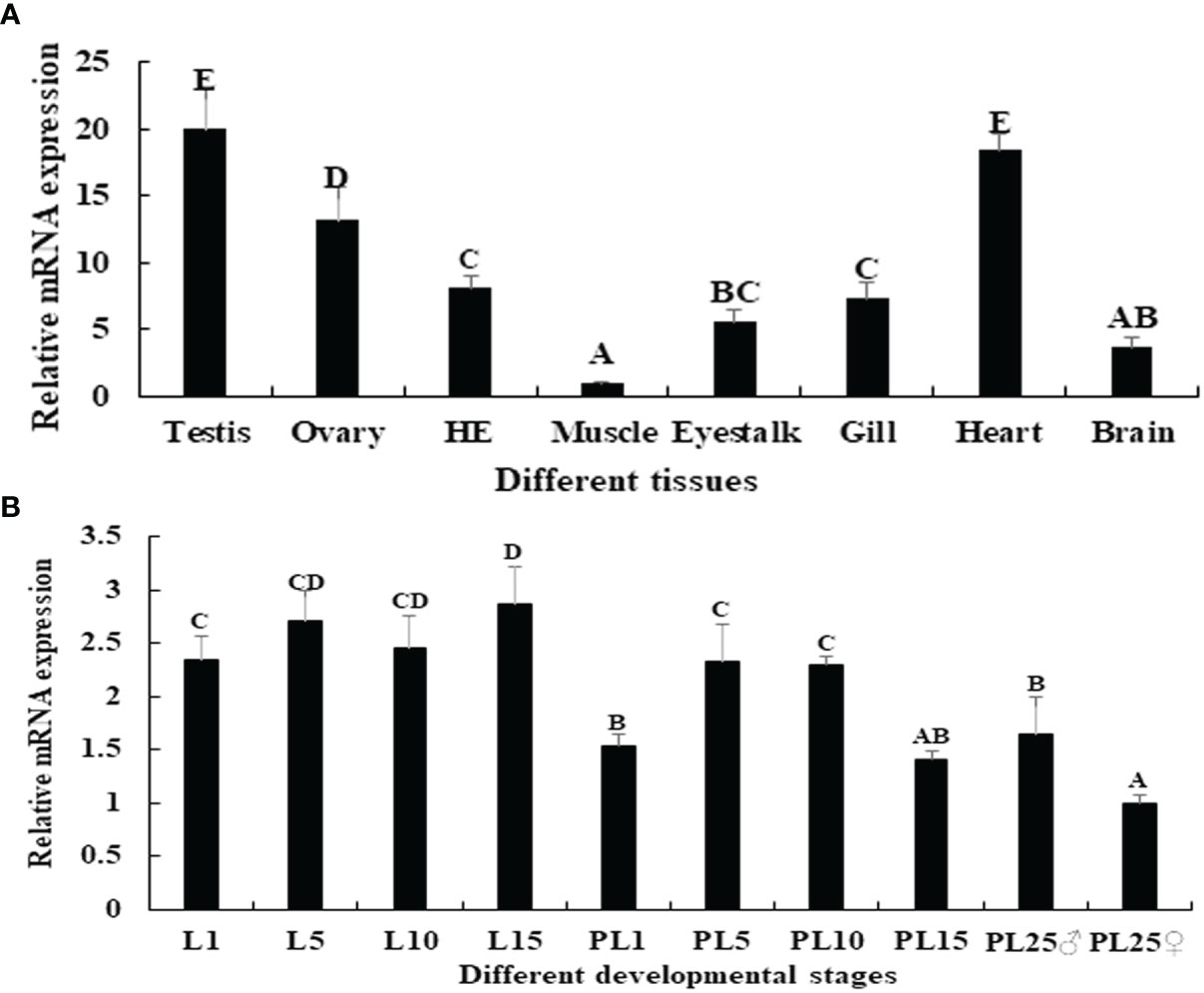
Figure 4 (A) Expression characterization of Mn-PDHE1 in different tissues. (B) Expression characterization of Mn-PDHE1 in different developmental stages. The amount of Mn-PDHE1 mRNA was normalized to the EIF transcript level. Data are shown as mean ± SD (standard deviation) of tissues from three separate individuals. Capital letters indicate expression difference between different samples.
Juvenile prawns can be visually distinguished as male or female for the first time at post-larval developmental stage 25 (PL25) by observing the presence of external gonadal features. Overall, the expression levels of Mn-PDHE1 were higher in the larval developmental stages compared with the post-larval developmental stages. During larval and post-larval development, the highest expression level of Mn-PDHE1 was observed in larval developmental stage 15 (L15), whereas the lowest expression level was observed in PL25 females (PL25♀). The expression at L15 was 2.87-fold higher than that of PL25♀. During post-larval development, expression peaked at PL5 and then gradually decreased to PL15. The expression level in PL25 males (PL25♂) was 1.64-fold higher and significantly different than that of PL25♀ (p < 0.05) (Figure 4B).
Expression of Mn-PDHE1 in Different Ovarian Developmental Stages
The expression pattern of Mn-PDHE1 was analysed in different ovarian developmental cycle of M. nipponense. The expression of Mn-PDHE1 reached the bottom at O III during the ovarian developmental cycle, and showed significant difference with other tested stages (p < 0.05). The highest expression level was observed in O V, followed by O I, which is 2.13-folder and 1.98-folder higher than that of O III, respectively (Figure 5).
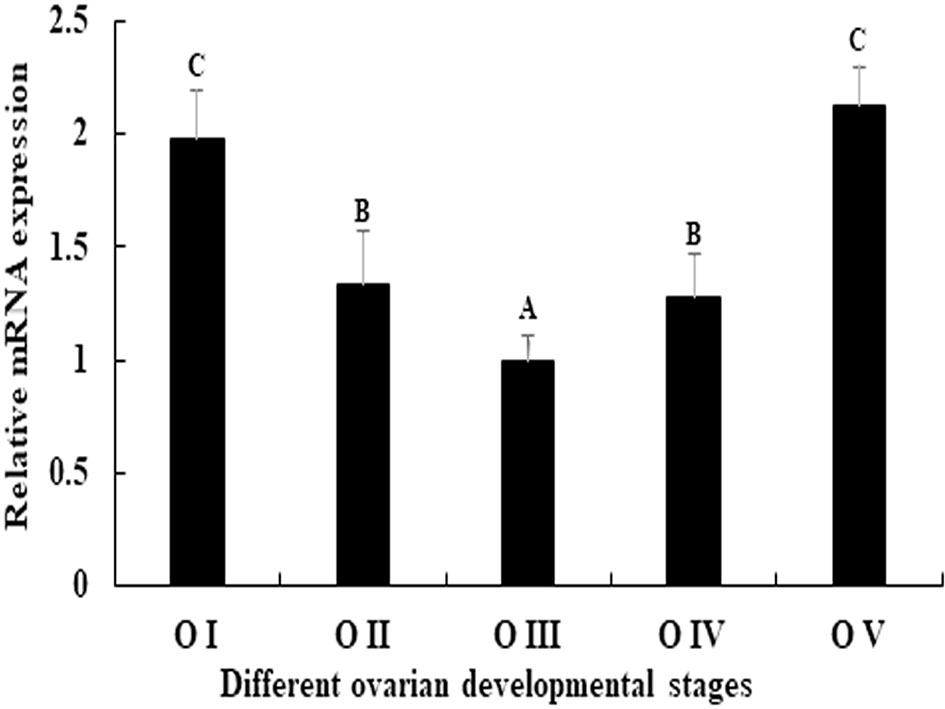
Figure 5 Expression characterization of Mn-PDHE1 in different reproductive cycles of ovary. The amount of Mn-PDHE1 mRNA was normalized to the EIF transcript level. Data are shown as mean ±SD (standard deviation) of tissues from three separate individuals. Capital letters indicate expression difference between different samples.
Western Blot Analysis
The western blot analysis showed that the molecular mass of Mn-PDHE1 was approximately 50 kDa, which was slightly larger than that of the predicted molecular weight. Jin et al. (13) previously reported that mRNA expression of Mn-PDHE1 in the testis was higher during the reproductive season than during the non-reproductive season. Clear protein bands were visible in the testis samples from the reproductive season, but the bands were blurry in the samples from the non-reproductive season. However, clear bands were observed in the androgenic gland for both of the two seasons (Figure 6A).
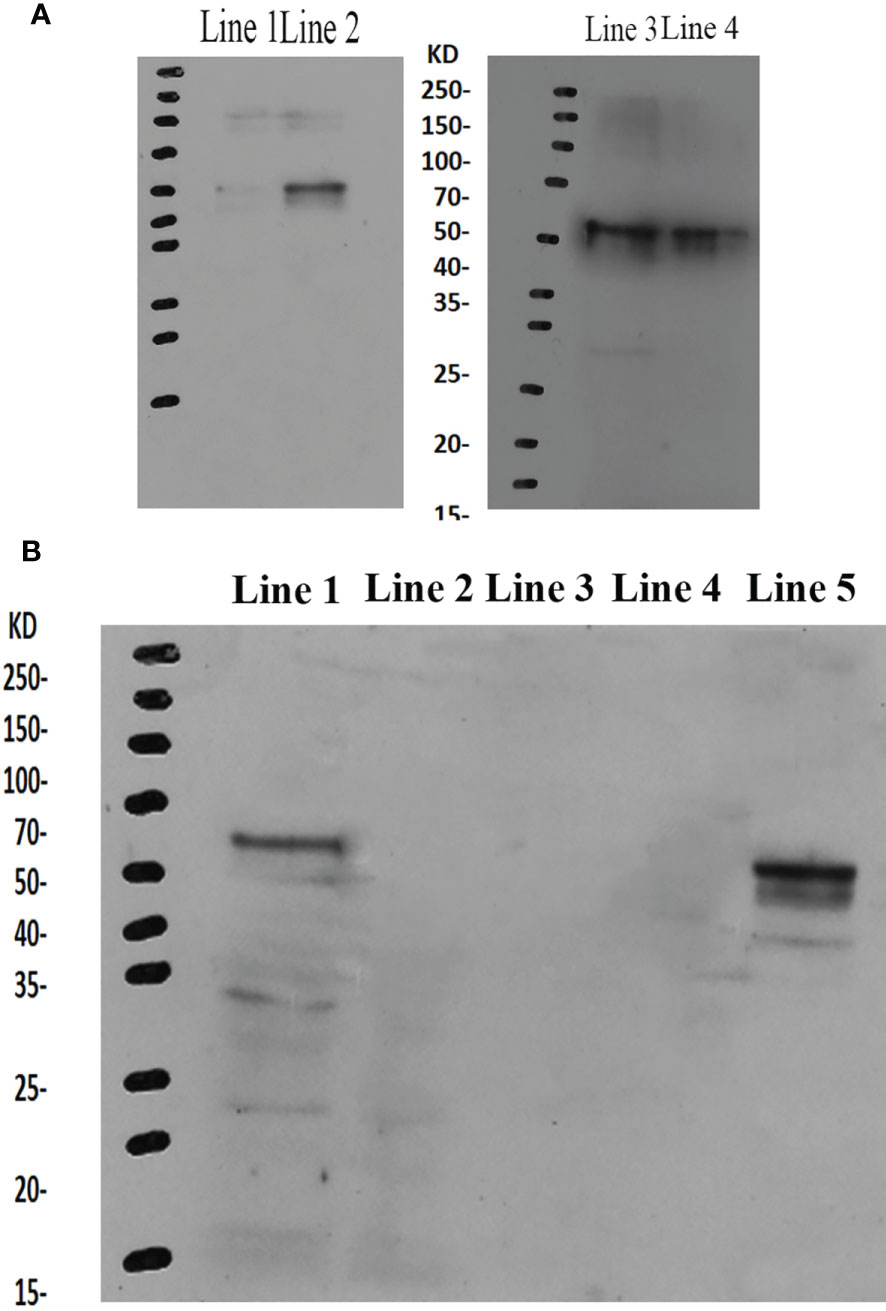
Figure 6 (A) Western-blot analysis of Mn-PDHE1 in the testis and androgenic gland from the reproductive season and non-reproductive season. Line 1: Testis from the non-reproductive season; Line 2: Testis from the reproductive season; Line 3: Androgenic gland from reproductive season; Line 4: Androgenic gland from the non-reproductive season. (B) Western-blot analysis of Mn-PDHE1 in the different ovarian developmental stages. Line 1: Ovarian developmental stage I; Line 2: Ovarian developmental stage II; Line 3: Ovarian developmental stage III; Line 4: Ovarian developmental stage IV; Line 5: Ovarian developmental stage V.
During ovarian development, clear bands were only observed for stages OI and OV, indicating that Mn-PDHE1 was transcribed during these stages. Mn-PDHE1 protein expression was more up-regulated in the OV stage than in the OI stage. No bands were observed in stages OII, OIII, and OIV, indicating that Mn-PDHE1 protein was not transcribed during these periods of development (Figure 6B).
In Situ Hybridization Analysis
HE staining showed that the androgenic gland included androgenic gland cells and the ejaculatory bulb. The cells in the testis included spermatogonia, spermatocytes, sperm, and collecting tissues. Strong mRNA signals for Mn-PDEH1 in the androgenic gland were only observed in the ejaculatory bulb surrounding the androgenic gland cells (Figure 7). mRNA signals for Mn-PDHE1 were only observed in spermatogonia in the testis (Figure 7).
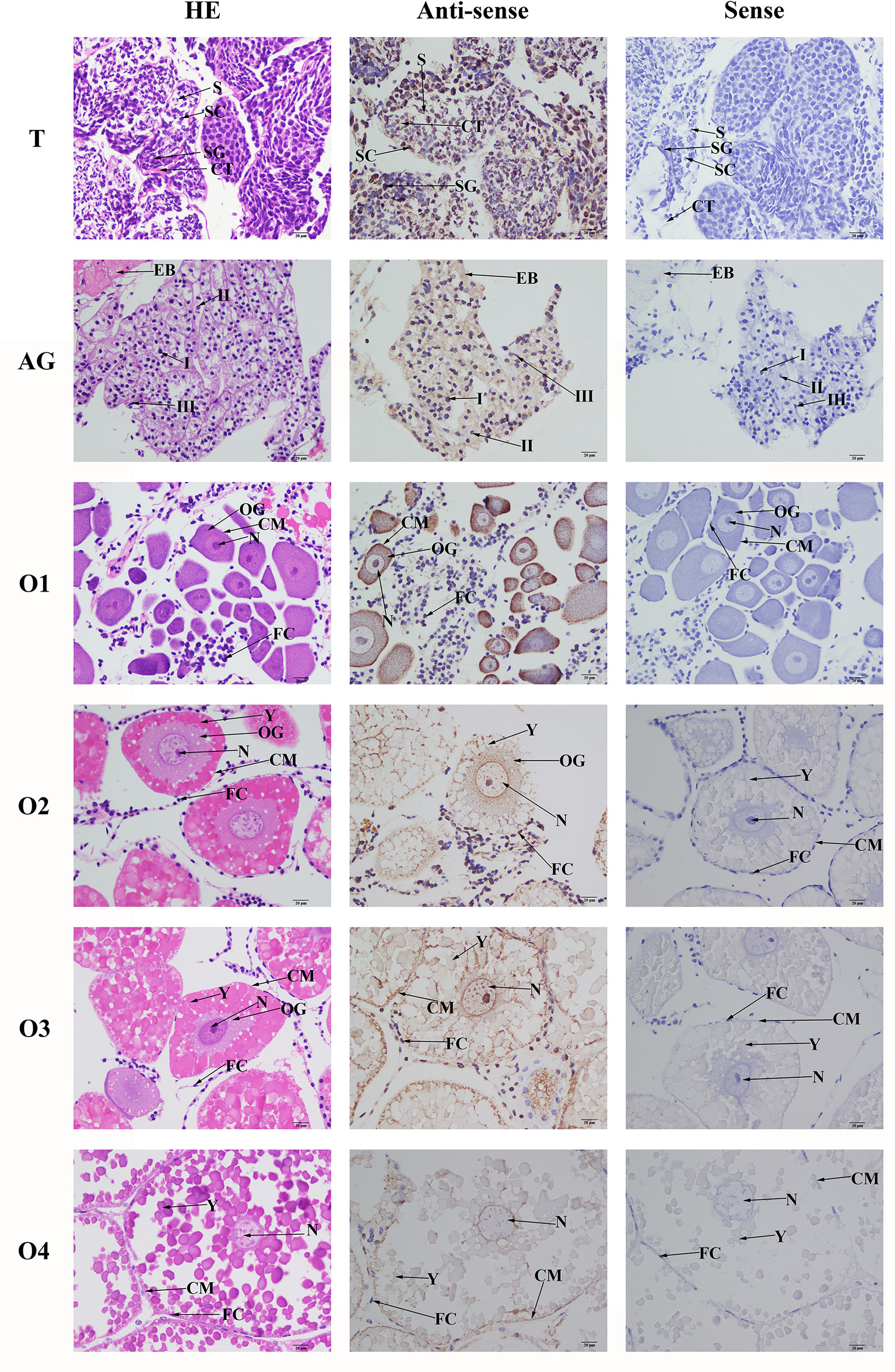
Figure 7 Location of PDHE1 gene was detected in testis, androgenic gland and ovary of M. nipponense by using in situ hybridization. ST, seminiferous tubule; SG, Spermatogonia; SC, spermatocyte; S, sperm; CT, collecting tissue; E, wall epithelium; EM, eosinophilic matrix; VD, vas deferens; EB, ejaculatory bulb; OG, oogonium; OC, oocyte; CM, cytoplasmic membrane; N, nucleus; Y, yolk granule; FC, follicle membrane. Scale bars = 20 μm.
Oogonia and follicle cells, which are derived from ovarian epithelial cells, were observed in stage OI. The follicular cavity formed in stage OII, oocyte volume gradually increased in stage OIII, and yolk granules accumulated in the oocytes in stage OIV. mRNA signals for Mn-PDHE1 were observed in all of the cell types and organelles from stages OI to OIV, including the nucleus, oogonia, oocytes, cytoplasmic membrane, yolk granules, follicle cells, and follicle membrane (Figure 7).
RNAi Analysis
The relative mRNA expression levels of Mn-PDEH1 were measured in the testis on days 1, 7, and 14 after the Mn-PDHE1 dsRNA treatment in both the RNAi and control groups. Mn-PDHE1 mRNA expression remained at a stable level in the control group and did not differ significantly over time (p > 0.05). In contrast, Mn-PDHE1 mRNA expression in the RNAi group gradually decreased from day 1 to 7, reached the lowest level on day 7, and increased slightly by day 14. The Mn-PDHE1 mRNA expression level was > 95% and 85% lower on days 7 and 14 in the RNAi group, respectively, compared to the control group on the same day (p < 0.01) (Figure 8A).
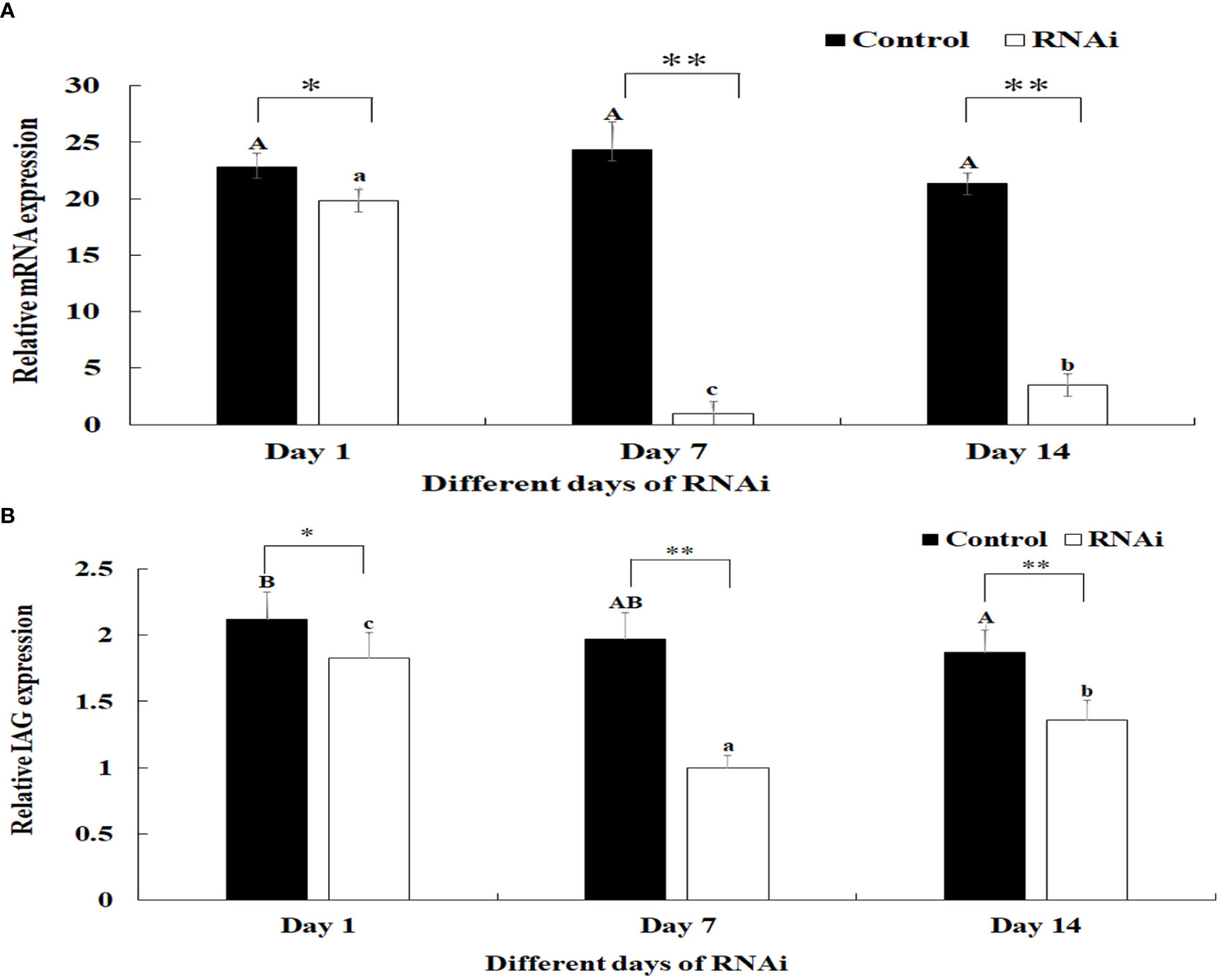
Figure 8 (A) Expression characterization of Mn-PDHE1 at different days after Mn-PDHE1 dsRNA injection. (B) Expression characterization of Mn-IAG at different days after Mn- PDHE1 dsRNA injection. The amount of Mn-PDHE1 and Mn-IAG mRNA was normalized to the EIF transcript level. Data are shown as mean ± SD (standard deviation) of tissues from three separate individuals. Capital letters indicated expression difference between different days after vehicle injection in control group. Lowercase indicated expression difference between different days after Mn-PDHE1 dsRNA injection in RNAi group. *(p < 0.05) and **(p < 0.01) indicates significant expression difference between the RNAi group and control group at the sample day.
Knockdown of the expression of Mn-PDHE1 also had a positive regulatory effect on the expression of Mn-IAG. In the RNAi group, mRNA expression of Mn-IAG was > 49% and 31% lower on days 7 and 14, respectively, compared to the control group on the same day (p < 0.01) (Figure 8B).
HE staining showed that sperm (> 60%) were the dominant cells in the testis in the control group, and spermatogonia and spermatocytes were rare. The morphology of the cells did not differ significantly over time. Compared with the control group, the number of sperm in the RNAi group was dramatically lower on day 7. On day 14, sperm accounted for only about 1% of the cells in this group, whereas the numbers of spermatogonia and spermatocytes had increased (Figure 9).
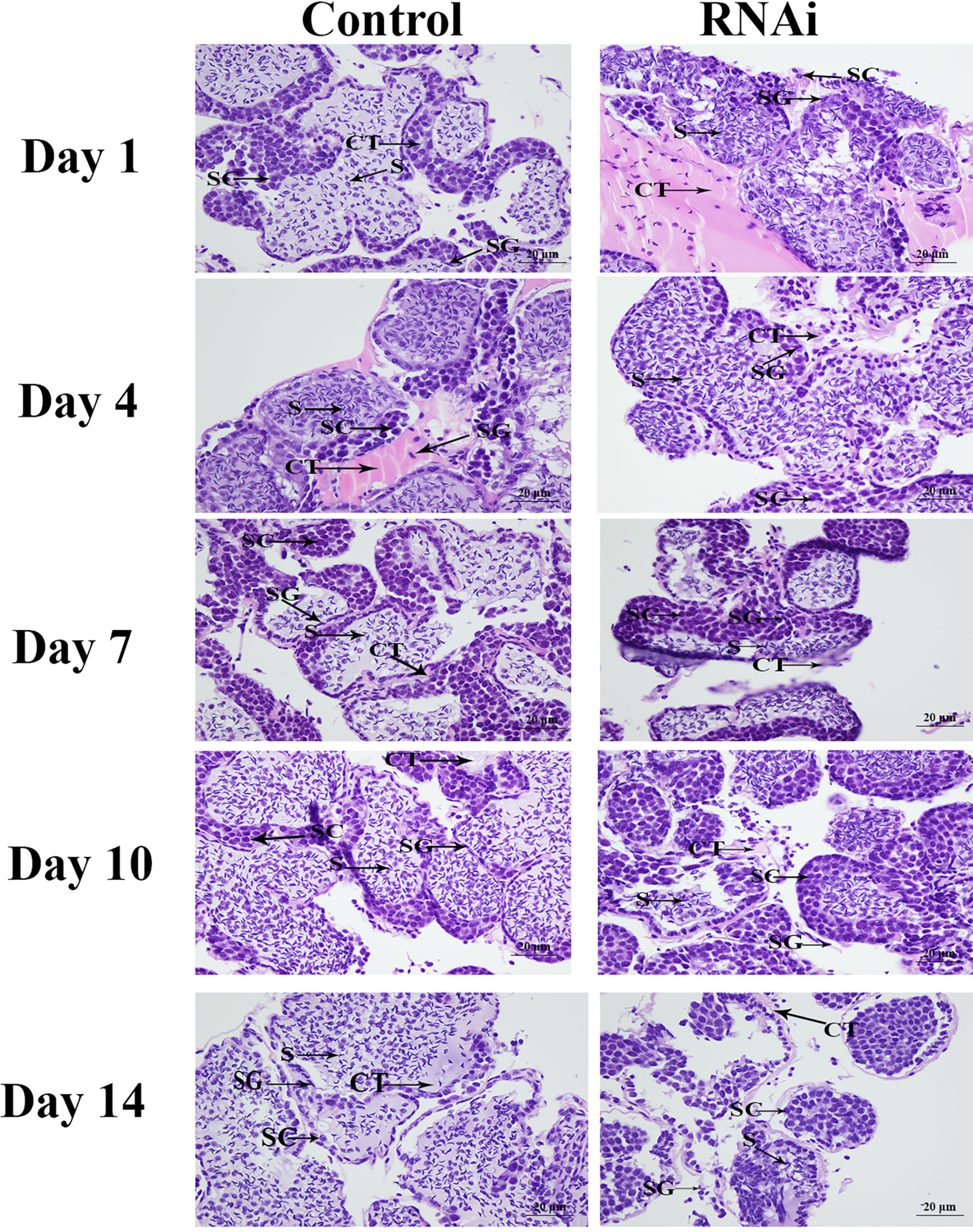
Figure 9 The histological observations of testis between RNAi and control group. SG, Spermatogonia; SC, spermatocyte; S, sperm; CT, collecting tissue. Scale bars = 20 μm.
Discussion
The PDHc is involved in the glycolysis pathway, catalysing the oxidative decarboxylation of pyruvate to become acetyl-CoA, which is required by the TCA cycle to generate ATP (14, 15). PDHE1 is the key enzyme component of the PDHc, as it promotes ATP generation in the glycolysis/gluconeogenesis and TCA metabolic pathways (16, 17). Jin et al. (13) previously predicted that PDHE1 plays important roles in generating ATP for male sexual development in M. nipponense. In this study, we analysed the functions of PDHE1 in M. nipponense, focusing especially on the regulatory effects of Mn-PDHE1 on Mn-IAG expression and on testis development. Our results may be useful for developing a technique to regulate testis development in this species. To date, the actual functions of PDHE1 have not been reported for any crustacean species.
The BLASTP analysis showed 90.72% similarity between Mn-PDHE1 and PDHE1 of P. vannamei, and Mn-PDHE1 also shared over 70% sequence identity with PDHE1 from other insect species. To the best of our knowledge, the only PDHE1 sequence available for a crustacean species prior to our study was that for P. vannamei. The neighbour-joining analysis showed that Mn-PDEH1 and the PDHE1 from P. vannamei clustered together as a group, and those from the insect species clustered together as another group. Thus, Mn-PDEH1 has the closest evolutionary relationship with the crustacean species and a much longer evolutionary relationship with insect species, which is consistent with evolutionary analysis of other genes in M. nipponense (22, 30, 31). More PDHE1 sequences from other species are needed, especially for crustacean species, in order to better analyse the evolutionary relationship of Mn-PDHE1.
The physiological functions of PDHE1 in M. nipponense can be reflected by the qPCR results of expression in various adult tissues and developmental stages. For example, qPCR analysis of PDHE1 in the roundworm Ascaris suum revealed that the Type I sequence was highly expressed in adult muscle, whereas the Type II sequence was abundant in the third-stage larvae as well as in adult muscle (32). In Streptococcus mutans, Korithoski et al. (33) found that PDHE1α expression dramatically increased during adaptation to acidic growth conditions. They also reported that PDHE1α expression increased in conditions favouring heterofermentative growth, decreased in the presence of excess glucose, and increased during the stationary phase compared with the mid-log phase of growth. In adult mice, PDHE1α showed testis-specific expression as well as somatic forms (34). They detected expression of PDHE1α in spermatogonia, Leydig cells, and Sertoli cells at a low level in somatic form. Our in situ hybridization analyses showed that PDHE1α was abundant in spermatocytes. The highest expression of Mn-PDHE1 was detected in the testis, which indicated that the activities of the TCA cycle were sufficient to generate ATP and promote male sexual development in M. nipponense. Mn-PDHE1 was more highly expressed during the larval developmental stages than during the post-larval developmental stages of juvenile M. nipponense, and the highest expression was observed in L15, indicating that PDHE1 promotes the metamorphosis process in M. nipponense (35, 36). During post-larval development, the highest expression level of Mn-PDHE1 occurred in PL5, and it gradually decreased to PL15. Previous histological studies showed that both the testis and ovary could be differentiated at PL5, and they matured at PL19 and PL22, respectively (5). Thus, the peak expression in PL5 indicated that PDHE1 plays essential roles in promoting gonad differentiation. The gender of M. nipponense can be distinguished for the first time by the naked eye at PL25, and the expression in PL25♂ was 2-fold higher than that in PL25♀. This result was consistent with results of the qPCR analysis of different tissues, which showed that PDHE1 plays more essential roles in male sexual development in M. nipponense.
Western blot analysis revealed that the molecular weight of Mn-PDHE1 protein was about 50 kDa, which was slightly larger than that of predicted molecular weight. This result indicates that the Mn-PDHE1 protein was modified after transcription. Jin et al. (13) previously reported that Mn-PDHE1 mRNA was more highly expressed in the testis and androgenic gland during the reproductive season than during the non-reproductive season. Our western blot results showed that Mn-PDHE1 proteins were transcribed in the testis and androgenic gland during the reproductive season and non-reproductive season, while the protein expression in the testis from non-reproductive season may be quite low. Together these findings indicate that PDHE1 promotes testis development during the reproductive season. qPCR analysis of the ovarian reproductive cycle revealed that the high expression levels of PDHE1 were observed at the OI and OV stages. Western-blot analysis of the ovarian reproductive cycle indicated that the Mn-PDHE1 protein was only transcribed in the OI and OV stages, and expression in stage OV was up-regulated relative to that in stage OI, which is consistent with that of qPCR analysis. Oogonia and follicle cells, which were derived from ovarian epithelial cells, were observed in stage OI. In stage OV, the mature oocytes were excreted (24).
In situ hybridization and immunostaining analysis showed that PDHE1α was abundant in spermatocytes in adult mice (34). We only observed strong mRNA signals of Mn-PDHE1 in spermatogonia in the testis in M. nipponense, indicating that PDHE1 plays essential roles in activating the testis developmental process. In the androgenic gland, strong mRNA signals were only observed in the ejaculatory bulb surrounding the androgenic gland cells. This finding indicated that PDHE1 was not directly secreted by the androgenic gland cells in M. nipponense but that it plays essential roles in maintaining the normal structure and function of the androgenic gland (22, 30, 31). In the ovary, PDHE1 was widely observed in all cell types, indicating that it was involved in the whole ovarian developmental process (24).
RNAi is an efficient method to analyse gene functions (37–39), and it has been widely used in studies of M. nipponense (24, 40). PDHE1 was shown to be involved in the glycolysis/gluconeogenesis and TCA metabolic pathways, promoting the ATP generation, which has been proven to play essential roles in testis development in adult mice (34). Jin et al. (13) predicted that PDHE1 might be involved in male sexual development in M. nipponense. Our qPCR analysis revealed that injection of Mn-PDHE1 dsRNA efficiently inhibited the mRNA expression of Mn-PDHE1 on days 7 and 14 after treatment. To analyse the regulatory roles of PDHE1 on IAG, the expression level of Mn-IAG was also measured in the same prawns. The expression of Mn-IAG was also significantly decreased on days 7 and 14 after treatment with Mn-PDHE1 dsRNA, indicating that PDHE1 positively regulated the expression of IAG in M. nipponense. Previous studies reported that IAG, which is secreted by the androgenic gland, is involved in male sexual differentiation and development in many crustacean species (41–43). Knockdown of the expression of IAG resulted in sex reversal in Macrobrachium rosenbergii (44). The positive regulatory relationship between PDHE1 and IAG highlights the important functions of PDHE1 in male sexual development in M. nipponense, as stipulated by Jin et al. (13). Morphologically, the number of sperm was significantly lower beginning on day 7 in the Mn-PDHE1 dsRNA treatment group compared to the control group. On day 14, sperm accounted for only 1% of the cells in the RNAi group, which indicated that Mn-PDHE1 positively regulates testis development in M. nipponense.
In conclusion, our results indicate that PDHE1 is involved in the metamorphosis and gonad development of M. nipponense, and it is especially important for male sexual development. Mn-PDHE1 was shown to positively regulate the expression of IAG and testis development in M. nipponense. This study provides essential data needed to establish a technique to regulate testis development in M. nipponense.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences.
Author Contributions
SJin designed and wrote the manuscript. YH performed the RNAi analysis. HF supervised the experiment. SJia and YX provided the experimental prawns. HQ performed the qPCR analysis. WZ performed the western-blot analysis. YG performed the in situ hybridization analysis. YW performed the histological observations. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by grants from the National Key R&D Program of China (2018YFD0900201); Central Public-interest Scientific Institution Basal Research Fund CAFS (2021JBFM02; 2020TD36); Jiangsu Agricultural Industry Technology System (JATS[2020]461); the China Agriculture Research System-48 (CARS-48); the New cultivar breeding Major Project of Jiangsu province (PZCZ201745).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Cai Y, Shokita S. Report on a Collection of Freshwater Shrimps (Crustacea: Decapoda: Caridea) From the Philippines, With Descriptions of Four New Species. Raffles B Zool (2006) 54:245–70.
2. Ma KY, Feng JB, Lin JY, Li JL. The Complete Mitochondrial Genome of Macrobrachium nipponense. Gene (2011) 487(2):160–5. doi: 10.1016/j.gene.2011.07.017
3. Salman SD, Page TJ, Naser MD, Yasser AG. The Invasion of Macrobrachium Nipponense (De Haan, 1849) (Caridea: Palaemonidae) Into the Southern Iraqi Marshes. Aquat Invasions (2006) 1(3):109–15. doi: 10.3391/ai.2006.1.3.2
4. Wang YB, Jin SB, Fu HT, Qiao H, Sun SM, Zhang WY, et al. Identification and Characterization of the DMRT11E Gene in the Oriental River Prawn Macrobrachium Nipponense. Int J Mol Sci (2019) 20(7):1734. doi: 10.3390/ijms20071734
5. Jin SB, Zhang Y, Guang HH, Fu HT, Jiang SF, Xiong YW, et al. Histological Observation of Gonadal Development During Post-Larva in Oriental River Prawn, Macrobrachium Nipponense. Chin J Fish (2016) 29(4):11–6. doi: 10.3969/j.issn.1005-3832.2016.04.003
6. Sagi A, Cohen D, Wax Y. Production of Macrobrachium Rosenbetgii in Momosex Population: Yield Characteristes Under Intensive Monoculture Conditions in Cages. Aquaculture (1986) 51(3-4):265–75. doi: 10.1016/0044-8486(86)90318-2
7. Sagi A, Cohen D, Milner Y. Effect of Androgenic Gland Ablation on Morphotypic Differentiation and Sexual Characteristics of Male Freshwater Prawns, Macrobrachium Rosenbergii. Gen Comp Endocr (1990) 77:15–22. doi: 10.1016/0016-6480(90)90201-V
8. Cao JX. The Molecular Mechanism in Male Reproductive Tract of the Prawn, Macrobrachium Rosenbergii. Zhejiang University. (2006).
9. Guo ZH. Study on Proliferation and Differentiation of Spermatogenic Cells From Macrobrachium Nipponense In Vitro. Hebei University. (2007).
10. Qiu GF, Du NS, Lai W. Studies on the Male Reproductive System of the Freshwater Prawn, Macrobrachium Nipponense. J Shanghai Ocean Uni (1995) 4:107–11.
11. Yang WX, Du NS, Lai W. Functional Relationship Between Spermatogenic Cells and Sertoli Cells During Spermatogenesis of Freshwater Shrimp, Macrobrachium Nipponense. Acta Zool Sinc (1999) 45(2):178–86. doi: 10.3969/j.issn.1674-5507.1999.02.009
12. Wedekind C. Demographic and Genetic Consequences of Disturbed Sex Determination. Philos T R Soc B (2017) 372:1729. doi: 10.1098/rstb.2016.0326
13. Jin SB, Hu YN, Fu HT, Sun SM, Jiang SF, Xiong YW, et al. Analysis of Testis Metabolome and Transcriptome From the Oriental River Prawn (Macrobrachium Nipponense) in Response to Different Temperatures and Illumination Times. Comp Biochem Phys D (2020) 34:100662. doi: 10.1016/j.cbd.2020.100662
14. Berg A, De Kok A. 2-Oxo Acid Dehydrogenase Multienzyme Complex. The Central Role of the Lipoyl Domain. Biol Chem (1997) 378:617–34. doi: 10.1002/bies.950190716
15. Harris RA, Bowker-kinley MM, Huang BL, Wu PF. Regulation of the Activity of the Pyruvate Dehydrogenase Complex. Adv Enzym. Regul (2015) 42:249–59.
16. Carlsson J, Kujala U, Edlund MJK. Pyruvate Dehydrogenase Activity in Streptococcus Mutans. Infect Immun (1985) 49:674–8. doi: 10.1128/iai.49.3.674-678.1985
17. Modak J, Wolf-Dieter D, Zeng AP. Metabolic Control Analysis of Eurkaryotic Pyruvate Dehydrogenase Multienzyme Complex. Biotechnol Progr (2002) 18:1157–69. doi: 10.1021/bp025613p
18. Snoep JL, de Graef MR, Westphal AH, de Kok A, de Mattos MJT, Neijssel OM. Differences in Sensitivity to NADH of Purified Pyruvate Dehydrogenase Complexes of Enterococcus Faecalis, Lactococcus Lactis, Azotobacter Vinelandii and Escherichia Coli: Implications for Their Activity In Vivo. . FEMS Microbiol Lett (1993) 114:279–84. doi: 10.1111/j.1574-6968.1993.tb06586.x
19. Jin SB, Hu YN, Fu HT, Jiang SF, Xiong YW, Qiao H, et al. Identification and Characterization of the Succinate Dehydrogenase Complex Iron Sulfur Subunit B Gene in the Oriental River Prawn, Macrobrachium Nipponense. Front Genet (2021) 12:698318. doi: 10.3389/fgene.2021.698318
20. Qiao H, Xiong YW, Zhang WY, Fu HT, Jiang SF, Sun SM, et al. Characterization, Expression, and Function Analysis of Gonad-Inhibiting Hormone in Oriental River Prawn, Macrobrachium Nipponense and its Induced Expression by Temperature. Comp Biochem Phys A (2015) 185:1–8. doi: 10.1016/j.cbpa.2015.03.005
21. Jin SB, Jiang SF, Xiong YW, Qiao H, Sun SM, Zhang WY, et al. Molecular Cloning of Two Tropomyosin Family Genes and Expression Analysis During Development in Oriental River Prawn, Macrobrachium Nipponense. Gene (2014) 546:390–7. doi: 10.1016/j.gene.2014.05.014
22. Jin SB, Fu HT, Jiang SF, Xiong YW, Sun SM, Qiao H, et al. Molecular Cloning, Expression, and In Situ Hybridization Analysis of Forkhead Box Protein L2 During Development in Macrobrachium Nipponense. J World Aquacult Soc (2018) 49(2):429–40. doi: 10.1111/jwas.12510
23. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–ΔΔCT Method. Methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
24. Li F, Qiao H, Fu HT, Sun SM, Zhang WY, Jin SB, et al. Identification and Characterization of Opsin Gene and its Role in Ovarian Maturation in the Oriental River Prawn Macrobrachium Nipponense. Comp Biochem Phys B (2018) 218:1–12. doi: 10.1016/j.cbpb.2017.12.016
25. Sun SM, Xuan FJ, Fu HT, Zhu J, Ge XP, Wu X. Molecular Cloning, mRNA Expression and Characterization of Membrane Bound Hemoglobin in Oriental River Prawn Macrobrachium Nipponense. Comp Biochem Phys A (2017) 207:36–42. doi: 10.1016/j.cbpa.2017.02.010
26. Bradford MM. A Rapid and Sensitive Method for the Quantification of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal Biochem (1976) 72:248–54. doi: 10.1016/0003-2697(76)90527-3
27. Jiang FW, Fu HT, Qiao H, Zhang WY, Jiang SF, Xiong YW, et al. The RNA Interference Regularity of Transformer-2 Gene of Oriental River Prawn Macrobrachium Nipponense. Chin Agr Sci Bull (2014) 30(32):32–7.
28. Ma XK, Liu XZ, Wen HS, Xu YJ, Zhang LJ. Histological Observation on Gonadal Sex Differentiation in Cynoglossus Semilaevis Günther. Mar Fish Res (2006) 27(2):55–61. doi: 10.3969/j.issn.1000-7075.2006.02.008
29. ShangGuan BM, Liu ZZ, Li SQ. Histological Studies on Ovarian Development in Scylla Serrata. J Fish Sci China (1991) 15(2):96–103.
30. Jin SB, Hu YN, Fu HT, Jiang SF, Xiong YW, Qiao H, et al. Potential Functions of Gem-Associated Protein 2-Like Isoform X1 in the Oriental River Prawn Macrobrachium Nipponense: Cloning, qPCR, in Situ Hybridization, and RNAi Analysis. Int J Mol Sci (2019) 20:3995. doi: 10.3390/ijms20163995
31. Jin SB, Hu YN, Fu HT, Jiang SF, Xiong YW, Qiao H, et al. Identification of Potentially Novel Functions of DNA Polymerase Zeta Catalytic Subunit in Oriental River Prawn, Macrobrachium Nipoponense: Cloning, qPCR, in Situ Hybridization and RNAi Analysis. 3 Biotech (2019) 9:330. doi: 10.1007/s13205-019-1857-7
32. Johnson KR, Komuniecki R, Sun YH, Wheelock MJ. Characterization of cDNA Clones for the Alpha Subunit of Pyruvate Dehydrogenase From Ascaris Suum. Mol Biochem Parasitol (1992) 51:37–48. doi: 10.1016/0166-6851(92)90198-S
33. Korithoski B, Levesque CM, Cvitkovitch DG. The Involvement of the Pyruvate Dehydrogenase E1asubunit, in Streptococcus Mutans Acid Tolerance. FEMS Microbiol Lett (2008) 289:13–9. doi: 10.1111/j.1574-6968.2008.01351.x
34. Takakubo F, Dahl HM. The Expression Pattern of the Pyruvate Dehydrogenase Elα Subunit Genes During Spermatogenesis in Adult Mouse. Exp Cell Res (1992) 199:39–49. doi: 10.1016/0014-4827(92)90459-L
35. Zhang YP, Fu HT, Qiao H, Jin SB, Gong YS, Jiang SF, et al. Molecular Cloning and Expression Analysis of Transformer-2 Gene During Development in Macrobrachium Nipponense (De Haan 1849). J World Aquacult Soc (2013) 44(3):338–49. doi: 10.1111/jwas.12039
36. Zhang YP, Jiang SF, Xiong YW, Sun SM, Qiao H, Jin SB, et al. Molecular Cloning and Expression Analysis of Extra Sex Combs Gene During Development in Macrobrachium Nipponense. Turk J Fish Aquat Sc (2013) 13:331–40. doi: 10.1111/jwas.12039
37. Jones HD. Wheat Transformation: Current Technology and Applications to Grain Development and Composition. J Cereal. Sci (2005) 41(2):137–47. doi: 10.1016/j.jcs.2004.08.009
38. Kusaba M. RNA Interference in Crop Plants. Curr Opin Microbiol (2004) 15:139–43. doi: 10.1016/j.copbio.2004.02.004
39. Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, et al. The Avian Z-Linked Gene DMRT1 is Required for Male Sex Determination in the Chicken. Nature (2009) 461:267–71. doi: 10.1038/nature08298
40. Li FJ, Bai HK, Xiong YW, Fu HT, Jiang FW, Jiang SF, et al. Molecular Characterization of Insulin-Like Androgenic Gland Hormone-Binding Protein Gene From the Oriental River Prawn Macrobrachium Nipponense and Investigation of its Transcriptional Relationship With the Insulin-Like Androgenic Gland Hormone Gene. Gen Comp Endocr (2015) 216(6):152–60. doi: 10.1016/j.ygcen.2014.12.007
41. Rosen O, Manor R, Weil S, Gafni O, Linial A, Aflalo ED, et al. A Sexual Shift Induced by Silencing of a Single Insulin-Like Gene in Crayfish: Ovarian Upregulation and Testicular Degeneration. PloS One (2010) 5:e15281. doi: 10.1371/journal.pone.0015281
42. Ventura T, Manor R, Aflalo ED, Weil S, Raviv S, Glazer L, et al. Temporal Silencing of an Androgenic Gland-Specific Insulin-Like Gene Affecting Phenotypical Gender Differences and Spermatogenesis. Endocrinology (2009) 150:1278–86. doi: 10.1210/en.2008-0906
43. Ventura T, Manor R, Aflalo ED, Weil S, Sagi A. Expression of an Androgenic Gland-Specific Insulin-Like Peptide During the Course of Prawn Sexual and Morphotypic Differentiation. ISRN Endocrinol (2011) 2011:476283. doi: 10.5402/2011/476283
Keywords: Macrobrachium nipponense, PDHE1, qPCR analysis, RNAi, male sexual development
Citation: Jin S, Hu Y, Fu H, Jiang S, Xiong Y, Qiao H, Zhang W, Gong Y and Wu Y (2021) Identification and Characterization of the Pyruvate Dehydrogenase E1 Gene in the Oriental River Prawn, Macrobrachium nipponense. Front. Endocrinol. 12:752501. doi: 10.3389/fendo.2021.752501
Received: 03 August 2021; Accepted: 16 September 2021;
Published: 01 November 2021.
Edited by:
Xianhu Zheng, Chinese Academy of Fishery Sciences, ChinaReviewed by:
Qingguo Meng, Nanjing Normal University, ChinaShihao Li, Institute of Oceanology (CAS), China
Copyright © 2021 Jin, Hu, Fu, Jiang, Xiong, Qiao, Zhang, Gong and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongtuo Fu, ZnVodEBmZnJjLmNu
 Shubo Jin
Shubo Jin Yuning Hu2
Yuning Hu2 Hongtuo Fu
Hongtuo Fu Sufei Jiang
Sufei Jiang Hui Qiao
Hui Qiao