- 1Department of Nuclear Medicine, University Hospital Münster, Münster, Germany
- 2European Institute for Molecular Imaging (EIMI), University of Münster, Münster, Germany
- 3Department of General, Visceral, Tumor and Transplant Surgery, University Hospital Cologne, Cologne, Germany
- 4Institute of Zoology, University of Cologne Germany, Cologne, Germany
- 5Policlinic for Prevention, Diabetes and Endocrinology, University of Cologne, Cologne, Germany
Background: A limited number of targeted therapy options exist for papillary thyroid cancer (PTC) to date. Based on genetic alterations reported by the “The Cancer Genome Atlas (TCGA)”, we explored whether PTC shows alterations that may be targetable by drugs approved by the FDA for other solid cancers.
Methods: Databases of the National Cancer Institute and MyCancerGenome were screened to identify FDA-approved drugs for targeted therapy. Target genes were identified using Drugbank. Genetic alterations were classified into conferring drug sensitivity or resistance using MyCancerGenome, CiViC, TARGET, and OncoKB. Genomic data for PTC were extracted from TCGA and mined for alterations predicting drug response.
Results: A total of 129 FDA-approved drugs with 128 targetable genes were identified. One hundred ninety-six (70%) of 282 classic, 21 (25%) of 84 follicular, and all 30 tall-cell variant PTCs harbored druggable alterations: 259 occurred in 29, 39 in 19, and 31 in 2 targetable genes, respectively. The BRAF V600 mutation was seen in 68% of classic, 16% of follicular variant, and 93% of tall-cell variant PTCs. The RET gene fusion was seen in 8% of classic PTCs, NTRK1 and 3 gene fusions in 3%, and other alterations in <2% of classic variant PTCs. Ninety-nine of 128 (77%) FDA-approved targetable genes did not show any genetic alteration in PTC. Beside selective and non-selective BRAF-inhibitors, no other FDA-approved drug showed any frequent predicted drug sensitivity (<10%).
Conclusion: Treatment strategies need to focus on resistance mechanisms to BRAF inhibition and on genetic alteration–independent alternatives rather than on current targeted drugs.
Introduction
Papillary thyroid cancer (PTC) is quite rare and has generally a good prognosis: approximately 80% of all cases are cured after radical surgery and radioiodine ablative treatment (RAI). Up to 18% of patients are diagnosed with locoregional recurrence within the first 5 years after initial treatment (1), which is usually cured by a second RAI. Thirty-three to 55% of cases diagnosed with recurrence (2) and approximately 5% of all cases are diagnosed with RAI-refractory (RAI-R) disease (3), which is associated with a significantly poorer outcome (5-year survival <50%).
The European Thyroid Association (ETA) Guidelines state that the choice to continue with RAI therapy in a patient without RAI uptake, with a mixed pattern of uptake or progression despite RAI uptake “can only be addressed in a multidisciplinary team (MDT), weighing the benefits and disadvantages of continuing RAI eventually combined with local treatments” (3). If RAI-R disease is limited to a single lesion or more than one lesion within the same organ, the possibility of performing a local treatment should be considered, according to the ETA (3). Since so far, no systemic therapy option for RAI-R disease is curative, treatment should be started only in the setting of rapid metastatic progress according to recist criteria, taking into account patients’ age and comorbidities.
Chemotherapy plays no significant role in the systemic treatment of advanced differentiated thyroid cancer (DTC). Medical treatment is currently based on tyrosine kinase inhibitors (TKIs) as targeted molecular treatment based on studies showing that cellular dedifferentiation resulting in unresponsiveness to RAI therapy correlate with the degree of MAPK activation (4). The kinase inhibitors Sorafenib and Lenvatinib have been shown to significantly improve progression-free survival (PFS) rates in advanced RAI-R DTCs (4) and have consequently been approved by the Food and Drug Administration (FDA) and the European Medical Agency (EMA) for treatment of advanced RAI-R DTCs (5). An increase in overall patient survival, however, has been described only for Lenvatinib selectively for the subgroup of cases >65 years old (5–7).
There are, however, several drawbacks to PTC treatment with TKIs. The medication needs to be taken for the rest of the patient’s life and has a wide spectrum of adverse side effects with a significant impact on quality of life. Further, some patients develop a resistance to treatment (8). For this reason, it is generally recommended to carefully choose the starting point of treatment, with particular emphasis on the patient’s general condition and quality of life.
Based on in silico data mining, we have previously reported that drugs already approved by the FDA for different cancer types show potential to be used in other cancer entities (9). Here, we investigate whether any of the 129 currently FDA-approved drugs act on targets also commonly found in papillary thyroid cancer based on genetic mutations reported by the “The Cancer Genome Atlas (TCGA)”.
Methods
All data were obtained from open access databases and referenced accordingly. The study was conducted in accordance with the provisions of the Declaration of Helsinki and local laws.
FDA-Approved Targeted Therapy and Their Biological Targets
We identified all FDA-approved drugs for cancer therapy by searching the databases of the National Cancer Institute (10) and MyCancerGenome (11). Unspecific drugs such as Tretinoin or Cabazitaxel were excluded (Supplementary Table 1). One hundred twenty-nine FDA/EMA-approved drugs targeting genetic alterations were included. These drugs were linked to 128 genes by querying the databases of the MD Anderson Cancer Center (12) and Drugbank (13), which include information on the potential sites of binding and action (Supplementary Table 1). We further gathered information about specific genetic alterations that confer either drug sensitivity or drug resistance to targeted therapy from the MyCancerGenome (11), CiViC (14), TARGET (15), and OncoKB (16) databases (Supplementary Table 2).
Genetic Alterations in Thyroid Cancer
One of the landmarks of tumor genomics data is “The Cancer Genome Atlas Program”, shortly TCGA. The TCGA data are available through the cBioPortal platform (17, 18), which allows the user to mine the TCGA data on recent insight. The papillary thyroid cancer study (19), listed in cBioPortal namely as “Thyroid Carcinoma (TCGA, Firehose Legacy)”, was chosen as the basis for this study. Mutation data from whole exome sequencing generated by MutSig, putative copy number alterations from GISTIC 2.0, as well as fusion data in papillary thyroid cancer were taken from the supplementary material of the thyroid cancer TCGA study. Only cases with complete datasets of mutation data, putative copy number data, and clinical information about tumor histology were included in this study. The following cancer subtypes were present: 282 patients with classic type PTC, 83 patients with follicular variant PTC, and 30 patients with tall-cell variant PTC (overall n=395). Mutation variants and CNVs directly or indirectly affecting genes that have been targeted by FDA-approved drugs were identified.
Drug Response Prediction
The genetic datasets of these 395 patients were mined for (a) Gain of function, (b) CNV-Amplification, (c) gene fusion, or (d) specific genetic alterations that could potentially confer drug response to a given targeted therapy.
Gain of Function
Gene alterations resulting in gain of function were annotated according to OncoKB (16) and Civic (14). Activating gene alterations were annotated with OncoKB’s (like) gain of functions, Civic clinical significance score of “(Likely) pathogenetic” or positive, as well as whether the alteration was in a hotspot as defined by Chang et al. (20).
CNV Amplification
The data from cBioPortal (17, 18) is annotated with a copy number analysis algorithm [GISTIC 2.0 (21)], which indicates the copy number level per gene: “− 2” deep loose, “− 1” shallow loose, “0” diploid, “1” low-level gain, and “2” high-level amplification. The threshold of high-level amplification “2” was chosen, to signify an occurrence of a copy number increase in each tissue sample.
Specific Gene Alterations
Each drug shows a literature-based effectiveness upon a specific alteration found in TCGA tissue as well as an indirect gene alteration affecting resistance or sensitivity to a drug. For this we utilized MyCancerGenome (11), CiViC (14), TARGET (15) and OncoKB (16).
Mutation Hotspot Analysis
Mutation variants known to be responsive to FDA-approved drugs were searched according to the database DOCM (22).
Pathway Analysis
A pathway analysis was performed using PathwayMapper through cBioPortal.
BRAF in PanCancer Cohorts
The 395 cases of PTC as well as all TCGA-PanCancer cohorts (33 cancer cohorts, total 11,084 cases) were screened for BRAF alterations in cBioportal.
Results
Genetic Alterations in Targeted Genes
One hundred twenty-eight targetable genes of 129 FDA-approved targeted therapies were identified. There were 259 alterations in 29 drug-targetable genes in classic PTC, 39 in 19 druggable genes in follicular variant of PTC, and 31 in 2 druggable genes of tall-cell variant PTC. One hundred ninety-six (70%) of 282 classic variant, 21 (25%) of 84 follicular variant, and all 30 (100%) tall-cell variant PTC tumors harbored druggable mutations.
Of all targetable alterations, only the BRAF V600 variant shows a high frequency in the classic type of PTC (68%, 192/282 cases), 93% (28/30) of cases in tall-cell variant PTC, and 16% (13/83) of cases with follicular variant PTC (see Figure 1A). The specific BRAF mutation is nearly exclusively BRAF V600E (192/193 “BRAF-mutated cases” in classic types of PTC). All cases of tall-cell variant PTC showed at least one BRAF alteration [93% (28/30) cases with a BRAF-mutation and 7% (2/30) cases with a BRAF gene fusion]. This is followed by RET gene fusion, which occurs in 8% of cases with classic type PTC, and NTRK1 and 3 gene fusions, which occur in 3% of cases with classic type PTC. All other genetic alterations are rare (<2%, see Figure 1B). We found that 99 potentially targetable genes did not show alterations such as CNV amplification, gene fusion, or gain of function mutations in the PTC cases we analyzed.
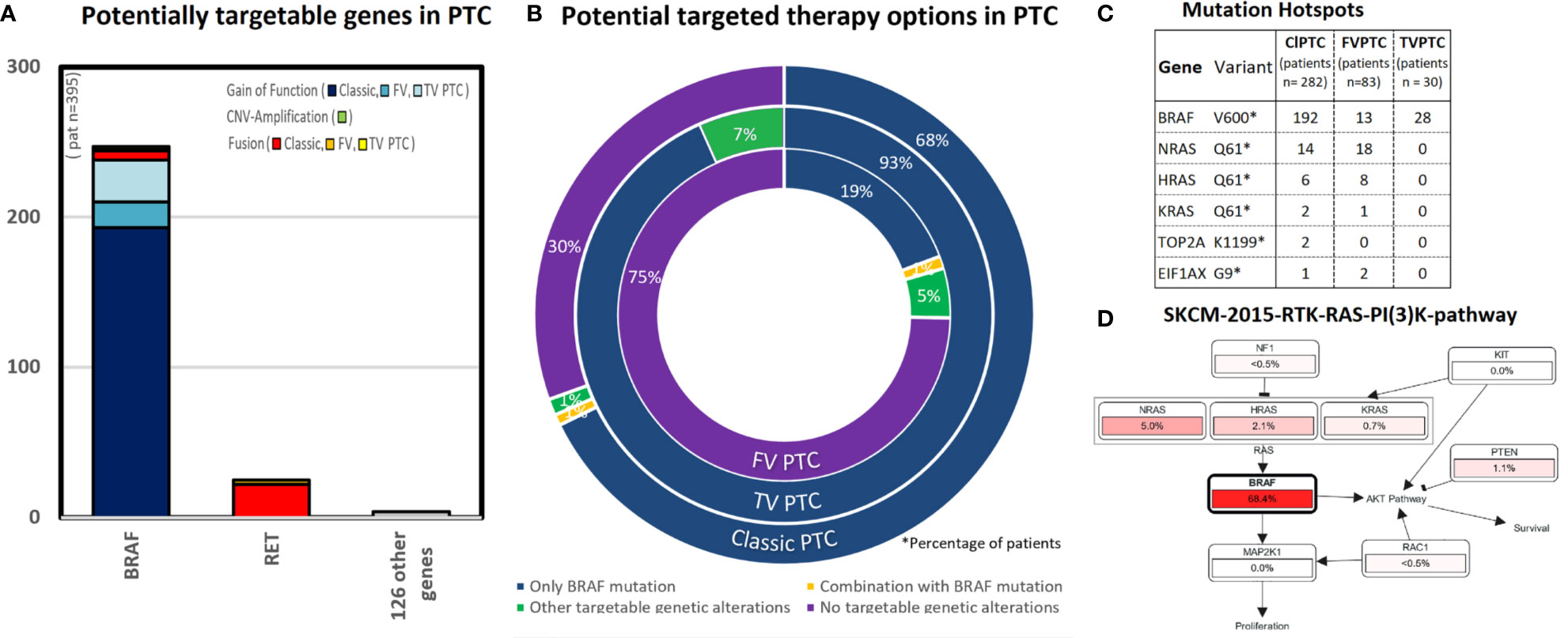
Figure 1 This figure shows the results of potential targetable genetic alterations in papillary thyroid cancer [total 395 cases PTC, respectively subtypes of papillary thyroid cancer as follows: classic type PTC (282 cases), follicular variant PTC (83 cases), and tall-cell variant PTC (30 cases)]. (A) This bar chart lists the number of cases (y-axis) with either gain of function mutation (blue colors), CNV amplification (green), and others (red colors). Almost all targetable genetic alterations occur in the gene BRAF. (B) The triple doughnut chart shows the potential targeted therapy options for classic type PTC (outer circle), tall-cell variant PTC (middle circle), and follicular thyroid cancer (inner circle). BRAF inhibitors are potential treatable in 68% cases of classic type PTC. In comparison, 75% cases of the follicular variant PTC had no targetable alterations. (C) The point mutation BRAF V600 occurs in 68% cases in classic type PTC, 16% cases in follicular variant PTC, 93% cases in tall-cell variant PTC. Interestingly, other mutation hotspots in classic type PTC occurred, as well in the RAS-pathway as follows: NRAS (5%, 14/282 cases) and HRAS (2%, 6/282 cases). (D) Pathway analysis [created by Pathway Mapper (61) through cBioportal (18)]: In the pathway analysis for classic-type papillary thyroid cancer, we identified a high match in the SKCM-2015-RTK-RAS-PI(3)K-pathway, especially in the RAS-signaling pathway (BRAF,HRAS,NRAS).
Mutation Hotspot Analysis
The highest frequency mutation hotspot in classic type PTC occurs in the point mutation BRAF V600E (68% cases). The second and third highest frequency of mutation hotspots occurred also in the RAS-Pathway: NRAS Q61 (5%, 14/282 cases) and HRAS Q61 (2%, 6/282 cases) (see Figure 1C). Moreover NRAS Q61K is a predictive biomarker in three National Comprehensive Cancer Network (NCCN) guidelines in at least one clinical setting for binimetinib, cetuximab, and panitumumab (24).
Pathway Analysis
We identified a match in the SKCM-2015-RTK-RAS-PI (3)K-pathway for the RAS-Signaling pathway (BRAF, HRAS, NRAS) as shown in the Figure 1D. Pathways other than RAS- and RET are rare in PTC (<1%).
Potential Drug Option
In our in silico analysis, BRAF inhibitors (selective BRAF inhibitors or multikinase BRAF inhibitors) were identified as potential drugs that would lead to a drug response in classic type PTC and tall-cell variant PTC based on the alterations we found. The multikinase BRAF inhibitors Regorafenib and Sorafenib showed a response prediction in all tall-cell variant, in >75% classic variant, and in >25% follicular variant PTC. The selective BRAF inhibitors Vemurafenib, Cobimetinib, and Dabrafenib displayed a response prediction in 93% of tall-cell variant, in 68% of classic variant, and in 15% of follicular variant PTC. The 122 remaining “non-BRAF inhibitors” show no or low (under 10% cases) genetically predicted drug response in PTC (see Figure 2).
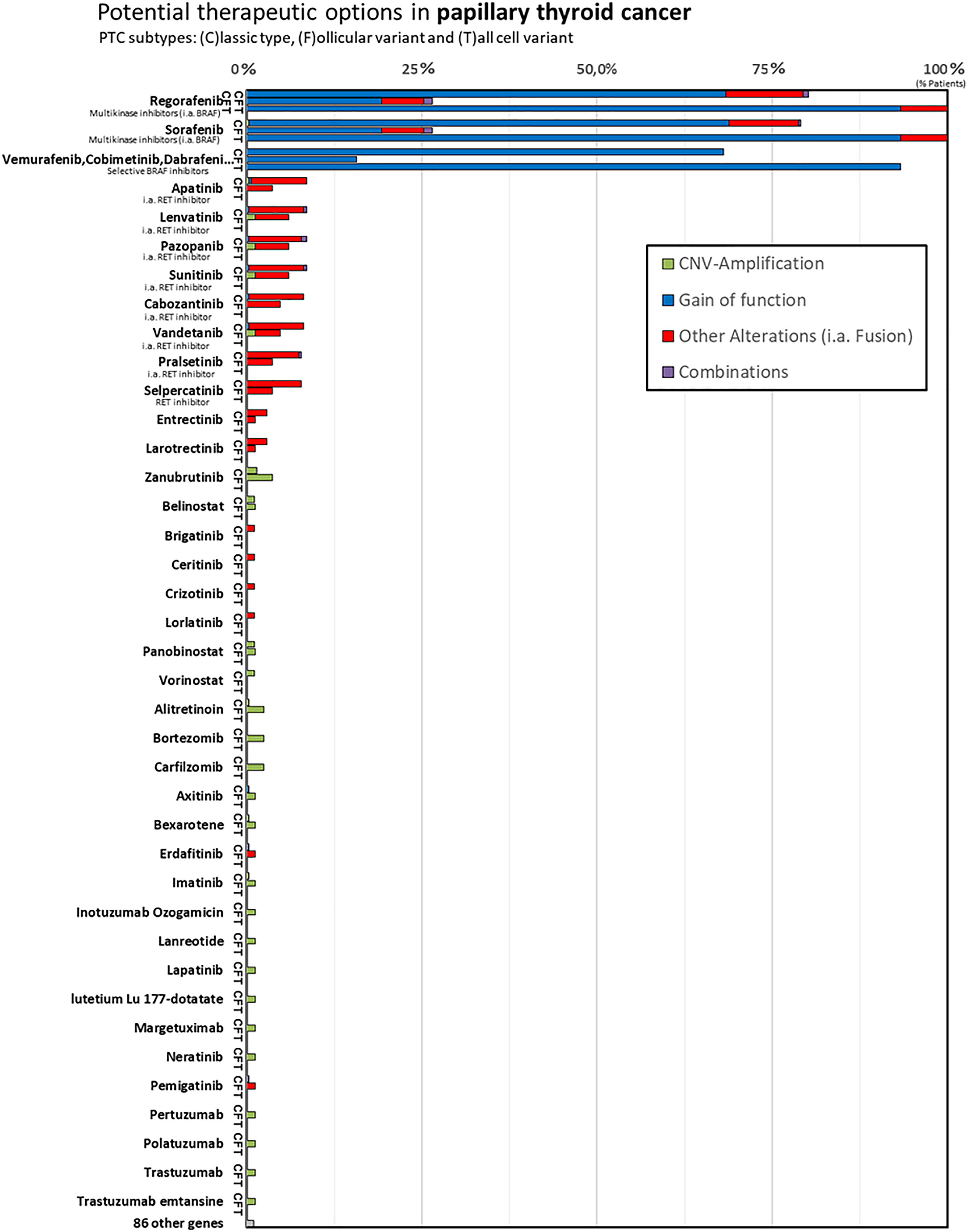
Figure 2 In the column chart y-axis lists the FDA-approved cancer drugs and the x-axis shows the number of cases with (c)lassic type PTC (shortly C), follicular variant PTC (shortly F), tall-cell variant PTC (shortly T). In our in silico analysis we identified BRAF inhibitors (selective BRAF inhibitors or multikinase, i.a., BRAF inhibitors) to be genetically predicted for drug response in papillary thyroid cancer. In comparison, the “non-BRAF inhibitors” show no or low genetically predicted drug response in differentiated thyroid cancer, although Lenvatinib, for example, represents first-line treatment for radioiodine refractory DTC currently.
Focus on BRAF
The data above suggest that the point mutation BRAF V600E possibly plays a pathognomonic role in papillary thyroid cancer. Consequently, we analyzed BRAF alterations in the TCGA-panCancer cohorts. A high frequency of BRAF alterations besides the thyroid cancers occurs in the “Melanoma TCGA-cancer” cohort (54% of 444 cases) (see Figures 3A, B).
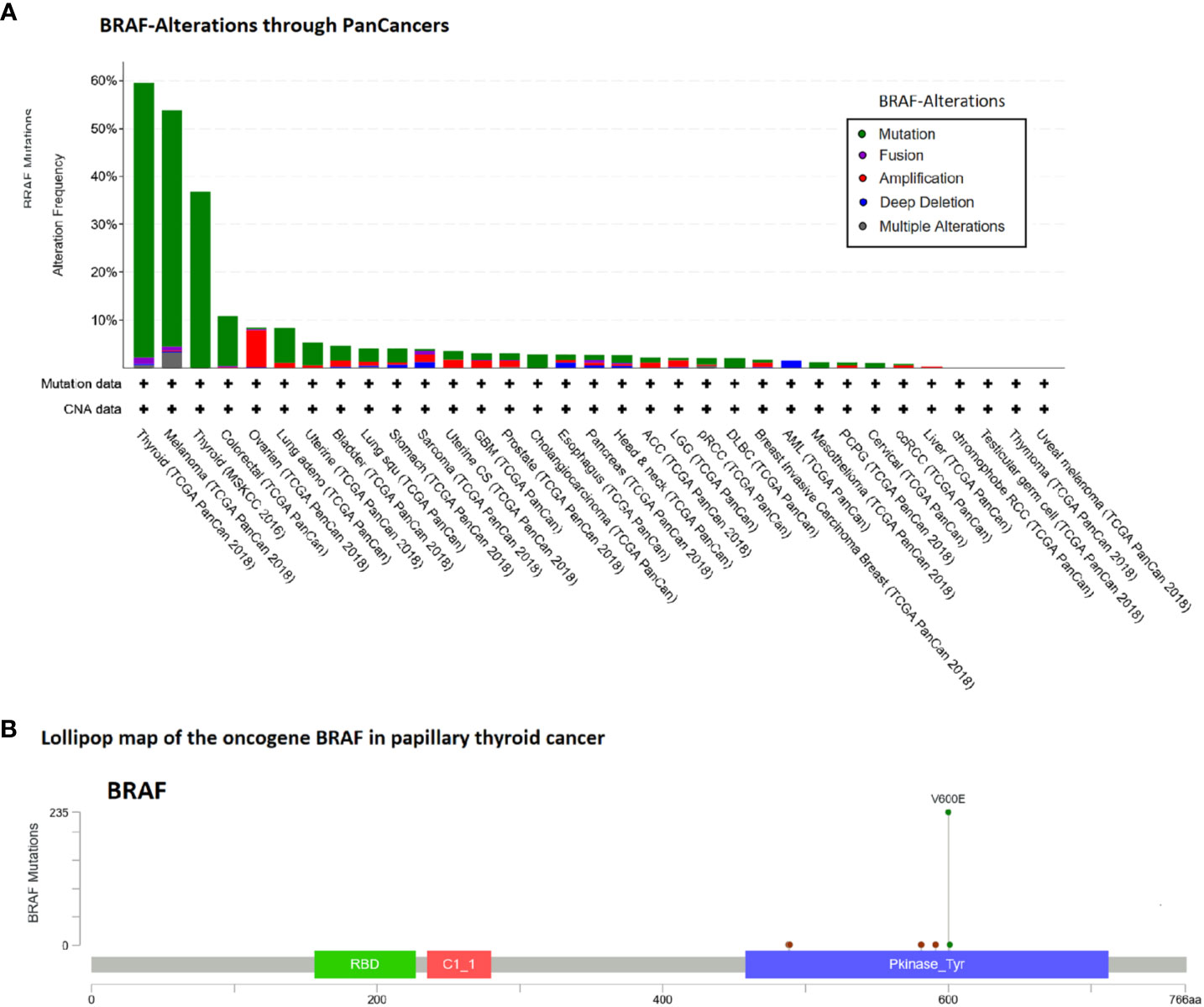
Figure 3 Alteration frequency analysis of BRAF gene in human cancers and lollipop map of BRAF mutation distribution using cBioPortal (18). (A) In the TCGA-panCancer cohorts besides the thyroid cancers, the high frequency of BRAF alterations occurs in the Melanoma TCGA-cancer cohort (53.8% of 444 cases). (B) Lollipop map of the oncogene BRAF shown are the case number of all variant types of papillary thyroid cancer (60%, 236/395 cases).
Discussion
Current targeted therapy options for radioiodine-refractory PTC are based on data from placebo-controlled randomized trials in the absence of sufficient clinical trial data comparing single agents or combinations (25). The current options encompass either mutation-specific or antiangiogenic multitargeted kinase inhibitors (aaMKIs). They frequently produce objective response, but no general prolongation of overall survival, and they have significant toxicities (4–7).
In this study we attempted to find new targetable options based on genes commonly altered in PTC. We analyzed the TCGA thyroid cancer study for commonly affected genes and found a high mutation rate in only one single point mutation: the BRAF V600 mutation. This mutation is seen in 68% of classic papillary thyroid cancer and in 93% of tall-cell variant patients. Given the predominance of BRAF mutations in RAI-R DTC (26, 27), it is possible that classic papillary thyroid cancer and tall-cell variant more often develop to refractory disease. The second highest frequency of mutation hotspots occurred in the RAS-Pathway. NRAS, HRAS, and KRAS mutations, which are seen in 30–50% of follicular thyroid cancer (FTC) (28), were detected in 8% of classic and in 33% of follicular variant PTCs. BRAF and RAS mutation are mutually exclusive in DTC (19). An association between the type of mutation (BRAF or RAS) and the metastasis pathway typical for papillary and follicular thyroid cancer (lymphogenic or hematogenic) has also been described (29). Although the RAS-Pathway displayed the second highest frequency mutation hotspots, there are no FDA-approved drugs targeting the specific mutations found in NRAS, HRAS, and KRAS for PTC. At this time, it is unclear if drugs targeting NRAS, HRAS, or KRAS (BI1701963 targeting KRAS and Tipifarnib targeting HRAS) will have any relevance for PTC in the future. Ho et al. (30) reported stable disease in 9 of 11 HRAS-mutated thyroid cancer patients treated with Tipifarnib; however, they focus on head and neck squamous cell carcinoma in their report and the specific histology of the thyroid cancer patients is not included. Given the predominance of RAS mutations in RAI avid as opposed to RAI-refractory DTC reported in the literature (31), this pathway might be less relevant for the treatment of RAI-R disease. Other potential targetable genetic alterations were found in less than 10% cases: the RET gene fusion was found in 8%, NTRK1 and 3 gene fusion in 3%, and other mutations in less than 2% of classic papillary thyroid cancers.
Accordingly, our in silico analysis predicted a high drug response for both the multikinase BRAF inhibitors (Regorafenib and Sorafenib) and the selective BRAF inhibitors (Dabrafenib, Vemurafenib, Cobimetinib), particularly in classic and tall-cell PTC (Figure 2). These results may not be surprising. Sorafenib has been shown to significantly improve median progression free survival (PFS) compared to placebo in patients with RAI-refractory DTC (10.8 vs. 5.8 months, p < 0.0001) in the phase 3 DECISION trial (32), and it is one of the two currently recommended first-line treatment agents. Regorafenib is very similar to Sorafenib in its molecular structure, except for the addition of a fluorine to the center phenyl ring. Regorafenib shows increased anti-angiogenic activity over Sorafenib (33). Studies on medullary thyroid cancer are currently recruiting (NCT02657551). However, there is currently no data or studies on Regorafenib in radioiodine-refractory papillary thyroid cancer (33).
Interestingly, our study only predicted a low drug response for the other first-line agent Lenvatinib. Our in silico analysis predicted a drug response of Lenvatinib similar to that predicted for Apatinib, Pazopanib, Sunitinib, Cabozatinib, Vandetanib, Pralsetinib, and Selpercatinib. However, Lenvatinib has not only been shown to significantly improve progression-free survival (PFS) rates in advanced RAI-R DTCs similar to Sorafenib (4), but it also represents the only current targeted therapy, for which an increase in overall survival has been shown in the >65 year old subgroup (5–7). In the SELECT study, the benefit of Lenvatinib was independent of the BRAF and RAS mutational status (34). A possible reason is the wide range of tumor cell targets affected (VEGF, VEGFR1–3, FGFR1–4, PDGFRα, KIT, and RET) (35). Studies on Apatinib (NCT03048877), Pazopanib (NCT01813136), Sunitinib (NCT00519896), and Cabozantinib (NCT03690388) on radioiodine-refractory thyroid cancer are ongoing (Table 1).
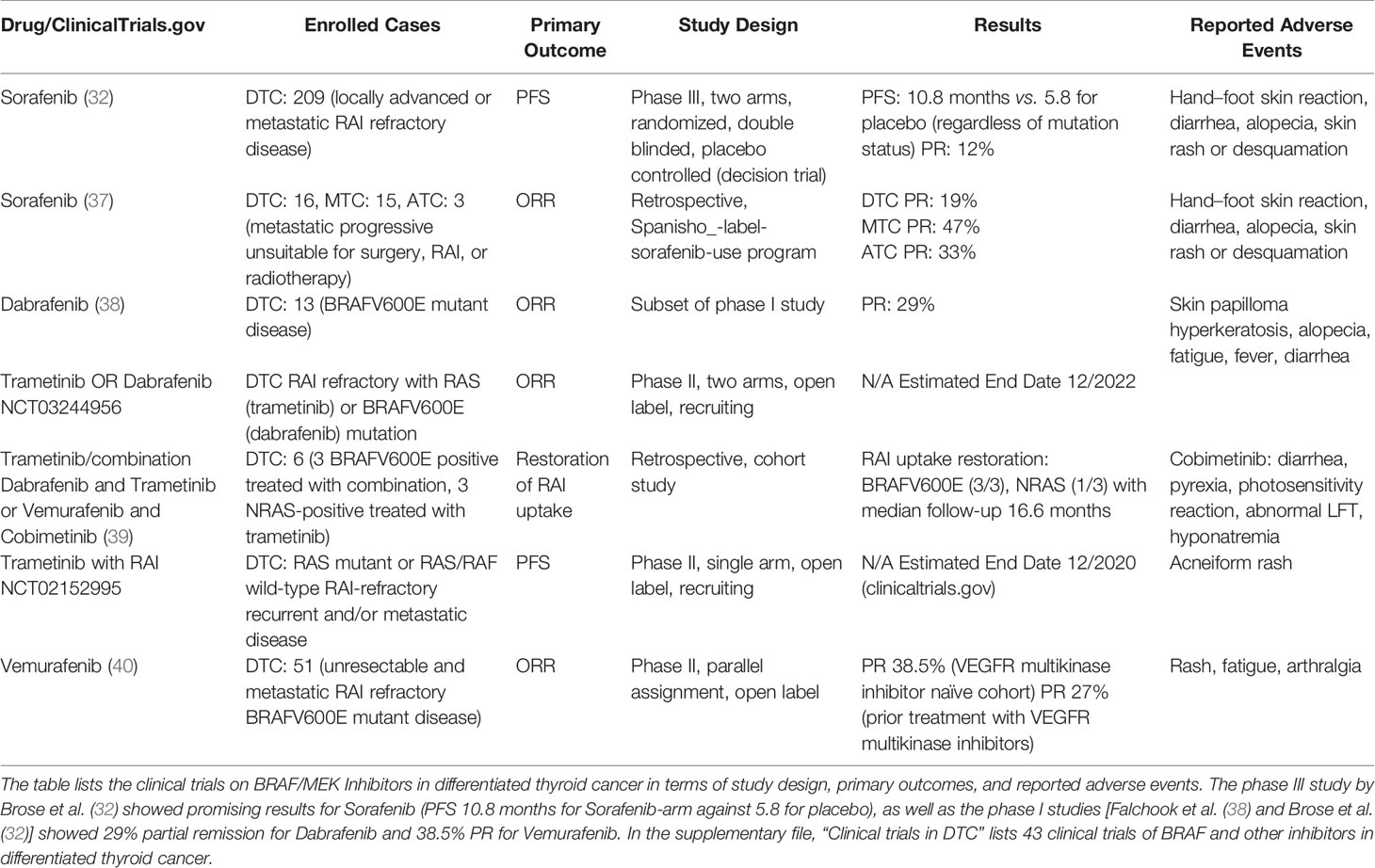
Table 1 Overview of BRAF/MEK Inhibitors in clinical trials in differentiated thyroid cancer [modified from Pottier et al. (36)].
The V600 mutant specific inhibitors Vemurafenib (PLX4032) and Dabrafenib (GSK2118436) for which the in silico analysis predicts a high drug response and which have produced excellent response rates in melanoma studies and in hairy cell leukemia (41) are typically reserved for patients who show a contraindication for first-line therapy with multikinase inhibitors and are administered in off-label use. Although objective responses and a prolongation of progression-free survival in small phase 2 trials were seen, these results have not yet been demonstrated in larger randomized trials (40, 42–51) (Table 1). Consequently, the inhibitors haven’t been approved by the FDA for thyroid cancer because toxicity seems to exceed the oncologic benefit. However, like Cobimetinib (GDC0973) (39), Vemurafenib and Dabrafenib have been reported to allow re-expression of genes responsible for iodine metabolism, restoring radioiodine uptake and permitting subsequent radioiodine treatment. In a trial with 10 BRAF V600 mutated PTC cases, 60% revealed new radioiodine uptake after 4 weeks of Dabrafenib therapy. Three months after radioiodine therapy in two of the six cases, a partial response could be observed (52). A similar partial response was also described for combined treatment with Cobimetinib and Vemurafenib in BRAF V600 mutated PTC cases by Iravani et al. (39). More data on the use of these inhibitors in PTC are critically needed, and “redifferentiation” is so far no clinical standard.
Given the fact that clinical trials with BRAF inhibitors produced various outcomes in different carcinomas, we performed a pan-cancer analysis for BRAF mutations (Figures 3A, B). A detailed knowledge and understanding of these results may also offer insights in the biology of PTC. The contrasting results of V600 mutant specific inhibitors in thyroid cancer compared to melanoma and hairy cell leukemia (53) could be due to different response and resistance mechanisms to BRAF inhibition dependent on cell types. A similar contrast was observed in clinical trials for colorectal and non-small-cell lung cancer compared to malignant melanoma (54). The mechanism responsible for a poor response could, for example, include the inhibition of the BCL2 pathway, a concurrent PI3KCA mutation and secondary alterations in the MAPK pathway (55). In colon carcinoma, for example, BRAF V600E inhibition has been shown to cause a rapid feedback activation of EGFR, which supports continued proliferation in the presence of BRAF V600E inhibition (56). Melanoma cells instead express low levels of EGFR and are therefore not subject to this feedback activation. Similarly co-treatment of thyroid cancer cell lines with an EGFR inhibitor increased antitumor efficacy and suppressed resistance to the BRAF V600E inhibition (57, 58).
Because the currently recommended first-line agents Lenvatinib and Sorafenib lack specificity for BRAF and the toxicity profile still represents a clinical hurdle, other treatment options would be useful in adjuvant setting after surgical management of radioiodine-refractory relapse. Since radioiodine-refractory papillary thyroid cancer is rare (3% of patients) and targeted therapy is currently initiated only after disease progression in a palliative setting, the present study was performed to select the most promising candidates among FDA drugs approved for solid cancer. The major limitation in this study is the fact that this is an in silico analysis. Clinical data would be critical to verify the results presented. Secondly, all data presented are based on the TCGA dataset and, thus, on primary papillary instead of radioiodine-refractory thyroid cancer. However, Sabra et al. (31) described BRAF mutations as predominant in RAI-R DTC and RAS mutations in radioiodine-avid DTC (19). For this reason, the results presented can possibly be extended to radioiodine-refractory thyroid cancer. It also needs to be mentioned that beside the histological subtype, other parameters such as pTNM stage, the patient’s age or co-morbidities must be considered for treatment stratification. In addition, spatial and temporal tumor heterogeneity, which are particularly relevant for the treatment of advanced thyroid cancer (59–61), are not sufficiently represented by the TCGA data. In the era of personalized medicine, the answer will likely require an ultra-deep sequence of the patient’s primary tumor in an attempt to identify relevant mutated genes. Appropriate drug combinations will help target the spatial and prevent the temporal heterogeneity of tumors, with the eradication of these clones (62). In addition, it is unclear how the data presented relates to pediatric patients, as the TCGA dataset does not cover patients <18 years. It is likely that the majority of pediatric PTC patients with high-risk tumors have a high genetic component and therefore druggable targets (63, 64).
Finally, several studies have demonstrated that TERT promoter mutations, particularly in association with BRAF V600E, correlate with negative clinical characteristics such as RAI- refractory disease (27, 65). TERT promoter mutations were reported in 36 (9.4%) of 384 informative tumors in the TCGA data (19). Further, for distant metastases, an enrichment in TERT mutations has been described (66). There are currently studies on telomerase inhibitors (INO5401, Telomelysin, and Imeltestat) on myeloid malignancy. These drugs might also play a role in the future for the treatment of RAI-R disease.
In summary our in silico analysis for drug prediction response based on alterations described in the TCGA data suggested BRAF inhibitors as the most promising target for most classic and tall-cell variants of thyroid cancer. The multikinase BRAF inhibitor Sorafenib is currently the first-line treatment for radioiodine-refractory thyroid cancer; however, for Regorafenib, clinical data for differentiated thyroid cancer are still lacking; and for the selective BRAF inhibitors (Dabrafenib, Vemurafenib, Cobimetinib), insufficient evidence has been collected for FDA approval in PTC treatment. Similar to colorectal and small lung cancer, resistance mechanisms might be responsible for the decreased efficacy of these medications. This needs to be considered in future studies. For Lenvatinib, which plays a major role in first-line targeted therapy of radioiodine-refractory thyroid cancer, our in silico analysis predicts a low response. Since targeted therapy does not lead to a significant improvement of overall survival in younger patients and is associated with high toxicity, alternative treatment options are urgently needed. These data suggest that resistance mechanisms to BRAF inhibition should be further investigated in order to ameliorate drug responses in the future.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
DH and HA designed the computational model and framework. DH, HA, and CC carried out the implementation. DH, BR, MH, MF, CB, HA, and CC contributed to the interpretation of the results. DH, BR, MH, MM, MF, CB, HA, and CC contributed critical feedback and helped shape the research, analysis, and manuscript. CC and DH wrote the first draft of the manuscript, and all authors critically revised the manuscript. All authors contributed to the article and approved the submitted version. All authors decided to submit this study and agreed to be accountable for all aspects of the work as recommended by the “International Committee of Medical Journal Editors” (ICMJE) authorship criteria.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.748941/full#supplementary-material
References
1. Chen W, Wei T, Li Z, Gong R, Lei J, Zhu J, et al. Association of the Preoperative Inflammation-Based Scores With TNM Stage and Recurrence in Patients With Papillary Thyroid Carcinoma: A Retrospective, Multicenter Analysis. Cancer Manag Res (2020) 12:1809–18. doi: 10.2147/CMAR.S239296
2. Kang SY, Bang J-I, Kang KW, Lee H, Chung J-K. FDG PET/CT for the Early Prediction of RAI Therapy Response in Patients With Metastatic Differentiated Thyroid Carcinoma. PloS One (2019) 14(6):e0218416. doi: 10.1371/journal.pone.0218416
3. Fugazzola L, Elisei R, Fuhrer D, Jarzab B, Leboulleux S, Newbold K, et al. 2019 European Thyroid Association Guidelines for the Treatment and Follow-Up of Advanced Radioiodine-Refractory Thyroid Cancer. Eur Thyroid J (2019) 8(5):227–45. doi: 10.1159/000502229
4. Rodon J, Soria J-C, Berger R, Miller WH, Rubin E, Kugel A, et al. Genomic and Transcriptomic Profiling Expands Precision Cancer Medicine: The WINTHER Trial. Nat Med (2019) 25(5):751–8. doi: 10.1038/s41591-019-0424-4
5. Laetitia G, Sven S, Fabrice J. Combinatorial Therapies in Thyroid Cancer: An Overview of Preclinical and Clinical Progresses. Cells (2020) 9(4):830. doi: 10.3390/cells9040830
6. Naoum GE, Morkos M, Kim B, Arafat W. Novel Targeted Therapies and Immunotherapy for Advanced Thyroid Cancers. Mol Cancer (2018) 17(1):51. doi: 10.1186/s12943-018-0786-0
7. Arcolia V, Journe F, Renaud F, Leteurtre E, Gabius H, Remmelink M, et al. Combination of Galectin-3, CK19 and HBME-1 Immunostaining Improves the Diagnosis of Thyroid Cancer. Oncol Lett (2017) 14(4):4183–9. doi: 10.3892/ol.2017.6719
8. Al-Jundi M, Thakur S, Gubbi S, Klubo-Gwiezdzinska J. Novel Targeted Therapies for Metastatic Thyroid Cancer—A Comprehensive Review. Cancers (2020) 12:1–37. doi: 10.3390/cancers12082104
9. Hescheler DA, Plum PS, Zander T, Quaas A, Korenkov M, Gassa A, et al. Identification of Targeted Therapy Options for Gastric Adenocarcinoma by Comprehensive Analysis of Genomic Data. Gastric Cancer Off J Int Gastric Cancer Assoc Jpn Gastric Cancer Assoc (2020) 23(4):627–38. doi: 10.1007/s10120-020-01045-9
10. National Cancer Institute. Copanlisib Approved for Follicular Lymphoma. Available at: https://www.cancer.gov/news-events/cancer-currents-blog/2017/aliqopa-fda-follicular-lymphoma.
11. My Cancer Genome. What Is My Cancer Genome? Available at: https://www.mycancergenome.org/about/what-is-my-cancer-genome/.
12. The University of Texas MD Anderson Cancer Center. Personalized Cancer Therapy. Available at: https://pct.mdanderson.org/.
13. Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res (2018) 46(D1):D1074–82. doi: 10.1093/nar/gkx1037
14. Griffith M, Spies NC, Krysiak K, McMichael JF, Coffman AC, Danos AM, et al. CIViC Is a Community Knowledgebase for Expert Crowdsourcing the Clinical Interpretation of Variants in Cancer. Nat Genet (2017) 49(2):170–4. doi: 10.1038/ng.3774
15. Broad Institute. TARGET (2021). Available at: https://software.broadinstitute.org/cancer/cga/target.
16. Chakravarty D, Gao J, Phillips S, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol (2017) 1):1–16. doi: 10.1200/PO.17.00011
17. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the Cbioportal. Sci Signal (2013) 6(269):pl1–1. doi: 10.1126/scisignal.2004088
18. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The Cbio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery (2012) 2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095
19. Agrawal N, Akbani R, Aksoy BA, Ally A, Arachchi H, Asa SL, et al. Integrated Genomic Characterization of Papillary Thyroid Carcinoma. Cell (2014) 159(3):676–90. doi: 10.1016/j.cell.2014.09.050
20. Chang MT, Shrestha Bhattarai T, Schram AM, Bielski CM, Donoghue MT, Jonsson P, et al. Accelerating Discovery of Functional Mutant Alleles in Cancer (2018). Available at: www.aacrjournals.org.
21. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 Facilitates Sensitive and Confident Localization of the Targets of Focal Somatic Copy-Number Alteration in Human Cancers. Genome Biol (2011) 12(4):R41. doi: 10.1186/gb-2011-12-4-r41
22. Ainscough BJ, Griffith M, Coffman AC, Wagner AH, Kunisaki J, Choudhary MNK, et al. DoCM: A Database of Curated Mutations in Cancer. Nat Methods (2016) 13:806–7. doi: 10.1038/nmeth.4000
23. Bahceci I, Dogrusoz U, La KC, Babur Ö, Gao J, Schultz N. PathwayMapper: A Collaborative Visual Web Editor for Cancer Pathways and Genomic Data. Bioinformatics (2017) 33(14):2238–40. doi: 10.1093/BIOINFORMATICS/BTX149
24. My Cancer Genome. NRAS Q61K (2021). Available at: https://www.mycancergenome.org/content/alteration/nras-q61k/)/#ref-5.
25. Cabanillas ME, Ryder M, Jimenez C. Targeted Therapy for Advanced Thyroid Cancer: Kinase Inhibitors and Beyond. Endocrine Rev (2019) 40:1573–604. doi: 10.1210/er.2019-00007
26. Shobab L, Gomes-Lima C, Zeymo A, Feldman R, Jonklaas J, Wartofsky L, et al. Clinical, Pathological, and Molecular Profiling of Radioactive Iodine Refractory Differentiated Thyroid Cancer. Thyroid (2019) 29(9):1262–8. doi: 10.1089/thy.2019.0075
27. Luo Y, Jiang H, Xu W, Wang X, Ma B, Liao T, et al. Clinical, Pathological, and Molecular Characteristics Correlating to the Occurrence of Radioiodine Refractory Differentiated Thyroid Carcinoma: A Systematic Review and Meta-Analysis. Front Oncol (2020) 10:549882. doi: 10.3389/fonc.2020.549882
28. Theurer S, Rawitzer J, Ting S, Schmid KW. Morphologic Diagnostic Criteria of Noninvasive Follicular Neoplasia With Papillary-Like Nuclear Features (NIFTP): A Diagnostic Challenge for the Patient’s Benefit. Pathologe (2021) 42(1):125–39. doi: 10.1007/s00292-020-00908-3
29. Theurer S, Rawitzer J, Ting S, Schmid KW. Diagnostic Principles of Thyroid Tumors in Pathology: Relevant Changes Due to the Current WHO Classification. Pathologe (2021) 42(1):125–39. doi: 10.1007/s00292-020-00908-3
30. Ho AL, Brana I, Haddad R, Baumann J, Bible K, Oosting S, et al. Tipifarnib in Head and Neck Squamous Cell Carcinoma With HRAS Mutations. J Clin Oncol (2021) 39(17):1856–64. doi: 10.1200/JCO.20.02903
31. Sabra MM, Dominguez JM, Grewal RK, Larson SM, Ghossein RA, Tuttle RM, et al. Clinical Outcomes and Molecular Profile of Differentiated Thyroid Cancers With Radioiodine-Avid Distant Metastases. J Clin Endocrinol Metab (2013) 98(5):E829–36. doi: 10.1210/jc.2012-3933
32. Brose MS, Nutting CM, Jarzab B, Elisei R, Siena S, Bastholt L, et al. Sorafenib in Radioactive Iodine-Refractory, Locally Advanced or Metastatic Diff Erentiated Thyroid Cancer: A Randomised, Double-Blind, Phase 3 Trial. Lancet (2014) 384(9940):319–28. doi: 10.1016/S0140-6736(14)60421-9
33. Wong SK, Chu QSC, Spratlin JL, Sangha R, McEwan AJB, Morrish DW, et al. Prolonged Response to Regorafenib in a Patient With Iodine Refractory Thyroid Cancer. Case Rep Oncol (2019) 12(3):791–5. doi: 10.1159/000503419
34. Schlumberger M, Tahara M, Wirth LJ, Robinson B, Brose MS, Elisei R, et al. Lenvatinib Versus Placebo in Radioiodine-Refractory Thyroid Cancer. N Engl J Med (2015) 372(7):621–30. doi: 10.1056/NEJMoa1406470
35. Costa R, Carneiro BA, Chandra S, Pai SG, Chae YK, Kaplan JB, et al. Spotlight on Lenvatinib in the Treatment of Thyroid Cancer: Patient Selection and Perspectives. Drug Design Dev Ther (2016) 10:873–84. doi: 10.2147/DDDT.S93459
36. Pottier C, Fresnais M, Gilon M, Jérusalem G, Longuespée R, Sounni NE. Tyrosine Kinase Inhibitors in Cancer: Breakthrough and Challenges of Targeted Therapy. Cancers (2020) 12(3):731. doi: 10.3390/cancers12030731
37. Capdevila J, Iglesias L, Halperin I, Segura Á, Martínez-Trufero J, Vaz MÁ, et al. Sorafenib in Metastatic Thyroid Cancer. Endocr Relat Cancer (2012) 19(2):209–16. doi: 10.1530/ERC-11-0351
38. Falchook GS, Millward M, Hong D, Naing A, Piha-Paul S, Waguespack SG, et al. BRAF Inhibitor Dabrafenib in Patients With Metastatic BRAF-Mutant Thyroid Cancer. Thyroid (2015) 25(1):71–7. doi: 10.1089/thy.2014.0123
39. Iravani A, Solomon B, Pattison DA, Jackson P, Ravi Kumar A, Kong G, et al. Mitogen-Activated Protein Kinase Pathway Inhibition for Redifferentiation of Radioiodine Refractory Differentiated Thyroid Cancer: An Evolving Protocol. Thyroid (2019) 29(11):1634–45. doi: 10.1089/thy.2019.0143
40. Brose MS, Cabanillas ME, Cohen EEW, Wirth LJ, Riehl T, Yue H, et al. Vemurafenib in Patients With BRAFV600E-Positive Metastatic or Unresectable Papillary Thyroid Cancer Refractory to Radioactive Iodine: A non-Randomised, Multicentre, Open-Label, Phase 2 Trial. Lancet Oncol (2016) 17(9):1272–82. doi: 10.1016/S1470-2045(16)30166-8
41. Tiacci E, Park JH, De Carolis L, Chung SS, Broccoli A, Scott S, et al. Targeting Mutant BRAF in Relapsed or Refractory Hairy-Cell Leukemia. N Engl J Med (2015) 373(18):1733–47. doi: 10.1056/NEJMoa1506583
42. Sherman SI, Ross DS, Mulder JE. Differentiated Thyroid Cancer Refractory to Standard Treatment: Systemic Therapy - UpToDate. Available at: https://www.uptodate.com/contents/differentiated-thyroid-cancer-refractory-to-standard-treatment-systemic-therapy.
43. Shah MH, Wei L, Wirth LJ, Daniels GA, De Souza JA, Timmers CD, et al. Results of Randomized Phase II Trial of Dabrafenib Versus Dabrafenib Plus Trametinib in BRAF-Mutated Papillary Thyroid Carcinoma. J Clin Oncol (2017) 35(15_suppl):6022–2. doi: 10.1200/JCO.2017.35.15_suppl.6022
44. Wirth LJ, Sherman E, Robinson B, Solomon B, Kang H, Lorch J, et al. Efficacy of Selpercatinib in RET -Altered Thyroid Cancers. N Engl J Med (2020) 383(9):825–35. doi: 10.1056/NEJMoa2005651
45. The American Association for Cancer Research. BLU-667 Controls RET-Altered Thyroid Cancers. Cancer Discov (2019) 9:OF5. doi: 10.1158/2159-8290.CD-NB2019-084
46. Hu M, Taylor M, Wirth L, Zhu V, Doebele R, Lee D, et al. Clinical Activity of Selective RET Inhibitor, BLU-667, in Advanced RET-Altered Thyroid Cancers: Updated Results From the Phase I ARROW Study. Thyroid (2018) 28(S1):A–169-A-198. doi: 10.1089/thy.2018.29071.sc.abstracts
47. Prasad ML, Vyas M, Horne MJ, Virk RK, Morotti R, Liu Z, et al. NTRK Fusion Oncogenes in Pediatric Papillary Thyroid Carcinoma in Northeast United States. Cancer (2016) 122(7):1097–107. doi: 10.1002/cncr.29887
48. Hong DS, Dowlati A, Burris HA, Lee JJ, Brose MS, Farago AF, et al. Clinical Safety and Activity From a Phase 1 Study of LOXO-101, a Selective TRKA/B/C Inhibitor, in Solid-Tumor Patients With NTRK Gene Fusions. Eur J Cancer (2017) 72:S148. doi: 10.1016/S0959-8049(17)30561-0
49. Tan DSW, Lassen UN, Albert CM, Kummar S, van Tilburg C, Dubois SG, et al. Larotrectinib Efficacy and Safety in TRK Fusion Cancer: An Expanded Clinical Dataset Showing Consistency in an Age and Tumor Agnostic Approach. Ann Oncol (2018) 29:ix23. doi: 10.1093/annonc/mdy430
50. Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of Larotrectinib in TRK Fusion–Positive Cancers in Adults and Children. N Engl J Med (2018) 378(8):731–9. doi: 10.1056/NEJMoa1714448
51. Laetsch TW, DuBois SG, Mascarenhas L, Turpin B, Federman N, Albert CM, et al. Larotrectinib for Paediatric Solid Tumours Harbouring NTRK Gene Fusions: Phase 1 Results From a Multicentre, Open-Label, Phase 1/2 Study. Lancet Oncol (2018) 19(5):705–14. doi: 10.1016/S1470-2045(18)30119-0
52. Rothenberg SM, McFadden DG, Palmer EL, Daniels GH, Wirth LJ. Redifferentiation of Iodine-Refractory BRAF V600E-Mutant Metastatic Papillary Thyroid Cancer With Dabrafenib. Clin Cancer Res (2015) 21(5):1028–35. doi: 10.1158/1078-0432.CCR-14-2915
53. Falini B, Tiacci E. New Treatment Options in Hairy Cell Leukemia With Focus on BRAF Inhibitors. Hematol Oncol (2019) 37(S1):30–7. doi: 10.1002/hon.2594
54. Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E–Mutated Colorectal Cancer. N Engl J Med (2019) 381(17):1632–43. doi: 10.1056/NEJMoa1908075
55. Porter A, Wong DJ. Perspectives on the Treatment of Advanced Thyroid Cancer: Approved Therapies, Resistance Mechanisms, and Future Directions. Front Oncol (2021) 10:592202/full. doi: 10.3389/fonc.2020.592202/full
56. Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of Colon Cancer to BRAF(V600E) Inhibition Through Feedback Activation of EGFR. Nature (2012) 483(7387):100–4. doi: 10.1038/nature10868
57. Jia Y, Zhang C, Hu C, Yu Y, Zheng X, Li Y, et al. EGFR Inhibition Enhances the Antitumor Efficacy of a Selective BRAF V600E Inhibitor in Thyroid Cancer Cell Lines. Oncol Lett (2018) 15(5):6763–9. doi: 10.3892/ol.2018.8093
58. Notarangelo T, Sisinni L, Condelli V, Landriscina M. Dual EGFR and BRAF Blockade Overcomes Resistance to Vemurafenib in BRAF Mutated Thyroid Carcinoma Cells. Cancer Cell Int (2017) 17(1):86. doi: 10.1186/s12935-017-0457-z
59. Gomes-Lima C, Shobab L, Wu D, Ylli D, Bikas A, McCoy M, et al. Do Molecular Profiles of Primary Versus Metastatic Radioiodine Refractory Differentiated Thyroid Cancer Differ? Front Endocrinol (Lausanne) (2021) 12:623182. doi: 10.3389/fendo.2021.623182
60. Sohn S, Park W, Shin H, Bae J, Ki C, Oh Y, et al. Highly Concordant Key Genetic Alterations in Primary Tumors and Matched Distant Metastases in Differentiated Thyroid Cancer. Thyroid (2016) 26(5):672–82. doi: 10.1089/thy.2015.0527
61. Masoodi T, Siraj A, Siraj S, Azam S, Qadri Z, Albalawy W, et al. Whole-Exome Sequencing of Matched Primary and Metastatic Papillary Thyroid Cancer. Thyroid (2020) 30(1):42–56. doi: 10.1089/thy.2019.0052
62. Fugazzola L MM, Pogliaghi G, Vitale M. Intratumoral Genetic Heterogeneity in Papillary Thyroid Cancer: Occurrence and Clinical Significance. Cancers (Basel) (2020) 12(2):383. doi: 10.3390/cancers12020383
63. Stenman A, Backman S, Johansson K, Paulsson J, Stålberg P, Zedenius J, et al. Pan-Genomic Characterization of High-Risk Pediatric Papillary Thyroid Carcinoma. Endocr Relat Cancer (2021) 28(5):337–51. doi: 10.1530/ERC-20-0464
64. Lee Y, Lee H, Im S, Song Y, Oh D, Kang H, et al. NTRK- and RET-Fusion-Directed Therapy in Pediatric Thyroid Cancer Yields a Tumor Response and Radioiodine Uptake. J Clin Invest (2021). doi: 10.1172/JCI144847
65. Xue J, Li S, Shi P, Chen M, Yu M, Hong S, et al. The ETS Inhibitor YK-4-279 Suppresses Thyroid Cancer Progression Independent of TERT Promoter Mutations. Front Oncol (2021) 11:649323. doi: 10.3389/fonc.2021.649323
Keywords: targeted molecular therapy, new treatment advances, human genome project, thyroid cancer, genomic analysis
Citation: Hescheler DA, Riemann B, Hartmann MJM, Michel M, Faust M, Bruns CJ, Alakus H and Chiapponi C (2021) Targeted Therapy of Papillary Thyroid Cancer: A Comprehensive Genomic Analysis. Front. Endocrinol. 12:748941. doi: 10.3389/fendo.2021.748941
Received: 28 July 2021; Accepted: 27 August 2021;
Published: 24 September 2021.
Edited by:
Yuji Nagayama, Nagasaki University, JapanReviewed by:
Marina Muzza, Fondazione Cà Granda Ospedale Maggiore Policlinico, ItalyCarl Christofer Juhlin, Karolinska Institutet (KI), Sweden
Copyright © 2021 Hescheler, Riemann, Hartmann, Michel, Faust, Bruns, Alakus and Chiapponi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hakan Alakus, aGFrYW4uYWxha3VzQHVrLWtvZWxuLmRl
†These authors have contributed equally to this work
 Daniel A. Hescheler1,2
Daniel A. Hescheler1,2 Milan J. M. Hartmann
Milan J. M. Hartmann Christiane J. Bruns
Christiane J. Bruns Costanza Chiapponi
Costanza Chiapponi