- 1Department of Obstetrics and Gynaecology, West China Second University Hospital, Sichuan University, Chengdu, China
- 2Key Laboratory of Birth Defects and Related Diseases of Women and Children, Sichuan University, Ministry of Education, Chengdu, China
- 3Reproductive Endocrinology and Regulation Laboratory, West China Second University Hospital, Sichuan University, Chengdu, China
- 4Department of Evidence-based Medicine and Clinical Epidemiology, West China Hospital, Sichuan University, Chengdu, China
- 5Center of Biostatistics, Design, Measurement and Evaluation (CBDME), West China Hospital, Sichuan University, Chengdu, China
Purpose: Controversial results existed in amounts of studies investigating the authentic association of estrogen receptor genes (ESR1 and ESR2) polymorphisms with the occurrence and progression of polycystic ovary syndrome (PCOS). The inconsistency might result from different loci, sample sizes, and ethnicities. To find the potential correlations between ESR1/ESR2 polymorphisms and PCOS risk, we conducted the first systematic review and meta-analysis to comprehensively summarize current studies in a large combined population.
Methods: Eligible studies were retrieved from PubMed, MEDLINE, EMBASE, Cochrane Library, CBM, CNKI, WANFANG, and VIP up to February 28, 2021. The quality of studies was assessed using the Newcastle–Ottawa Scale (NOS) scoring system. Odds ratios (ORs) and 95% confidence intervals (95%CIs) were calculated to synthesize data in five genetic models. Subgroup analyses were conducted by ethnicity. Heterogeneity and publication bias were also assessed. The protocol was registered in PROSPERO under the number CRD42021239200.
Results: A total of 8 studies involving 1,522 PCOS patients and 4,198 controls were included. No evidence demonstrated the association of ESR1 rs2234693 (OR=1.07 95%CI 0.98–1.18), ESR1 rs9340799 (OR=0.99 95%CI 0.69–1.43), or ESR2 rs4986938 (OR=1.06 95%CI 0.81–1.38) polymorphisms and PCOS risk in five genetic models. According to stratified subgroup analyses, ethnicity was considered the major source of heterogeneity. No publication bias was found in eligible studies.
Conclusion: The present meta-analysis found no significant associations between the variants of ESR1 rs2234693, ESR1 rs9340799, ESR2 rs4936938, and individual PCOS susceptibility, even if ethnicity was taken into account.
Systematic Review Registration: The protocol was registered in PROSPERO (available from https://www.crd.york.ac.uk/PROSPERO) with the ID number CRD42021239200.
Introduction
Although the pathogenesis and etiology of polycystic ovary syndrome (PCOS) remain hitherto unelucidated, it is widely considered multifactorial, that is, both environmental exposure and genetic susceptibility interact with each other and jointly influence PCOS incidence (1). To date, numerous studies suggested that genetic polymorphisms contribute substantially to the development of PCOS; over 100 genes are candidates, which functioned mainly in steroid biosynthesis, steroid action, glucose metabolism, and lipid metabolism (2–7). Rising interests were drawn in abnormal function of female sex hormone and hormonal receptors of PCOS women, and the impact of the mutation of some critical genes in the hormone metabolism pathways such as androgen receptor (AR) (8), luteinizing hormone receptor (LHR) (9), and follicle-stimulating hormone receptor (FSHR) (10) has been investigated in some meta-analyses. It was found that they were associated with some characteristics of PCOS such as clinical and/or biochemical hyperandrogenism, ovulatory dysfunction, and polycystic ovaries. However, the studies on estrogen receptor genes (ESR) were still controversial.
Estrogen mediates ovarian folliculogenesis and ovulation by binding to estrogen receptor α (ERα) and estrogen receptor β (ERβ), class 1 members of the superfamily of nuclear hormone receptor protein (11), which plays a prominent role in regulating cellular proliferation, differentiation, and apoptosis (12). Furthermore, two subtypes of ESR are nowadays identified, designated as ESR1 and ESR2, encoding ERα and ERβ separately. The ESR1 gene is located on chromosome 6q25.1, encompassing 140 kDa of DNA composed of 8 exons, and its introns are highly conserved (13). The ESR2 gene is located on chromosome 14q23.1, which contains 9 exons. Previous genetically engineered researches of knockout mice have revealed an interesting phenomenon that the absence of ESR1 can be associated with PCOS symptoms of irregular estrous, infertile, higher androgen level, and the formation of hemorrhagic follicles (14). Besides, the absence of ESR2 can lead to more early atretic follicles and fewer corpora lutea, suggesting partially follicular developmental and mature obstruction (15). Therefore, polymorphisms in the steroid action genes ESR1 and ESR2 may contribute to individual susceptibility to PCOS.
In recent years, an increasing number of investigations have focused on the potential association of ESR1 and ESR2 polymorphisms with PCOS susceptibility. Nonetheless, some studies held the view that these gene polymorphisms of ESR1 and ESR2 were positively related to PCOS (16, 17), whereas other studies reported no relation (18–22) or negative relation (23). The results remain inconsistent presumably due to loci and insufficient sample sizes in single study (16–23). Besides, different allelic frequencies between ethnic groups were the most crucial reason leading to long-existed controversy; thus, scientific and rigorous meta-analysis was required to summarize current studies. In this systematic review and meta-analysis, we aim to better clarify the relationship between single nucleotide polymorphisms (SNPs) in ESR1/ESR2 and the risk of PCOS in a large combined population by comprehensively collecting the existing data.
Materials and Methods
Identification of Eligible Studies
A search of literature was conducted in the PubMed, MEDLINE, EMBASE, Cochrane Library, CBM, CNKI, WANFANG, and VIP. The search period covered the start of the databases to February 28, 2021. The search strategy used to identify eligible studies included (polycystic ovary syndrome OR polycystic ovary OR PCOS OR PCO) and (Single Nucleotide Polymorphism OR SNP OR gene variant OR mutation OR genotype) and (estrogen receptor OR ESR1 OR ERα OR ESR2 OR ERβ), without language restriction. All references cited by these studies were manually screened in order to identify relevant studies.
Inclusion and Exclusion Criteria
Studies were included if they met all of the following criteria: (1) case-control studies, cross-sectional studies, case series, and cohort studies that were published in peer-reviewed journals; (2) PCOS patients diagnosed with one of the following criteria: the National Institutes of Health (NIH) (24), Rotterdam criteria (25), or the Androgen Excess and PCOS Society (AE-PCOS Society) criteria (26); and (3) the sample size and genotype frequency of case and control groups were directly provided or original data were sufficient to calculate the odds ratios (ORs) and 95% confidence intervals (95% CIs). Studies were excluded if they (1) were conference abstracts or reviews with non-extractable data; (2) deviated from Hardy–Weinberg equilibrium (HWE); or (3) recruited participants from one family. If data based on the same population were reported by different authors or research center, only the most comprehensive publication was included for meta-analysis.
Data Extraction
The titles and abstracts were scrutinized, and the data of included studies were extracted from fully retrieved texts by two investigators independently (SZ and SW). The characteristics of eligible studies were extracted according to Cochrane guidelines. The following contents were extracted: first author, year of publication, country/ethnicity, genotyping method, PCOS diagnostic criteria, the characteristics of cases and controls, HWE, and genotype frequency in cases and controls for ESR1 and ESR2 polymorphisms, respectively. Dissents were noted and resolved by discussing with the third investigator (LX).
Methodological Quality Assessment
The Newcastle–Ottawa scoring system (NOS), recommended by the Cochrane Working Group, was conducted to assess the quality of these nonrandomized studies (27). Evaluating concerns of NOS consists of the following three aspects: selection of subjects, comparability of enrolments, and outcome of interest ascertainment. The total score ranged from 0 to 9. Studies with a score from 0 to 3, 4 to 6, and 7 to 9 were considered to be of low, moderate, and high quality, respectively. Evaluation of evidence quality was determined by two reviewers (SZ and SW) independently, with any disagreement resolved by the discussion (SZ, SW, and LX).
Statistics Analysis
The HWE was evaluated for each study with a chi-square (χ²) test in the control group by the IBM SPSS statistics 25 software. P> 0.05 indicated that the study population was in HWE. The 95%CI and OR were utilized to evaluate the association strengths of the SNPs of ESR1/ESR2 with susceptibility to PCOS. And five genetic models (allele model, dominant model, recessive model, heterozygote model, and homozygote model) were conducted to evaluate the influence of the genotype. The Q-test was used for heterogeneity testing, and the inconsistency index (I²), a test statistic, was used for quantification. In the Q test, when I² ≤ 50%, the fixed-effects model was established to analyze. Otherwise, the random-effects model was performed. Next, a Z-test was performed to evaluate the statistical significance of combined ORs in the quantitative synthesis. When P value < 0.05, it was considered a statistically significant association between SNP mutation and PCOS. Subgroup analyses were stratified by ethnicity, which might be the potential source of inconsistencies. Funnel plots of the outcomes were used to detect publication bias; an asymmetric graph suggested possible publication bias. Egger’s regression test was also applied to assess the publication bias of selected studies, and as proposed by Egger, P value < 0.10 indicated significant publication bias. Sensitivity analysis was performed where appropriate to determine the robustness of the results. All analyses were carried out in the Review Manager software (Version 5.3; Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2014) and the STATA software (Version 13.0; Stata Corporation, College Station, TX).
Results
Study Selection
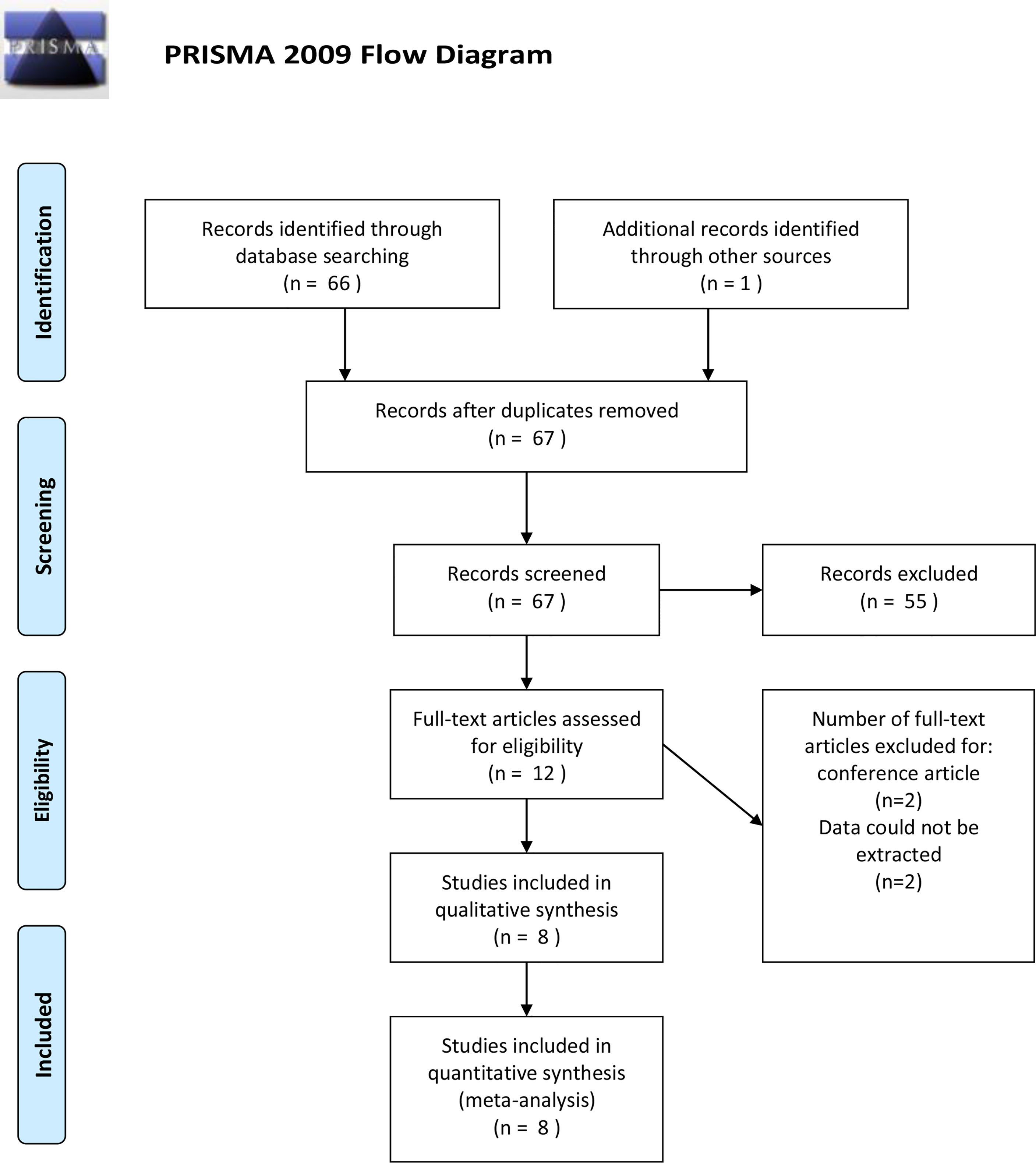
A total of 67 potential citations were acquired from electronic databases after exclusion of duplicate articles. Of these, by scanning titles and abstracts carefully, 55 records were removed for irrelevant themes. The remained 12 articles were full-text screened, and another 4 records were excluded subsequently, among which 2 were conference abstracts from the same population, 1 was retracted, and 1 reported non-extractable data. We contacted the corresponding authors for further data; nonetheless, no reply was received. Finally, eight eligible case-control studies that met our criteria were included in the qualitative and quantitative synthesis (16–23). The selection process was documented with a flowchart of Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) (Figure 1).
Characteristics and Methodological Quality of Included Studies
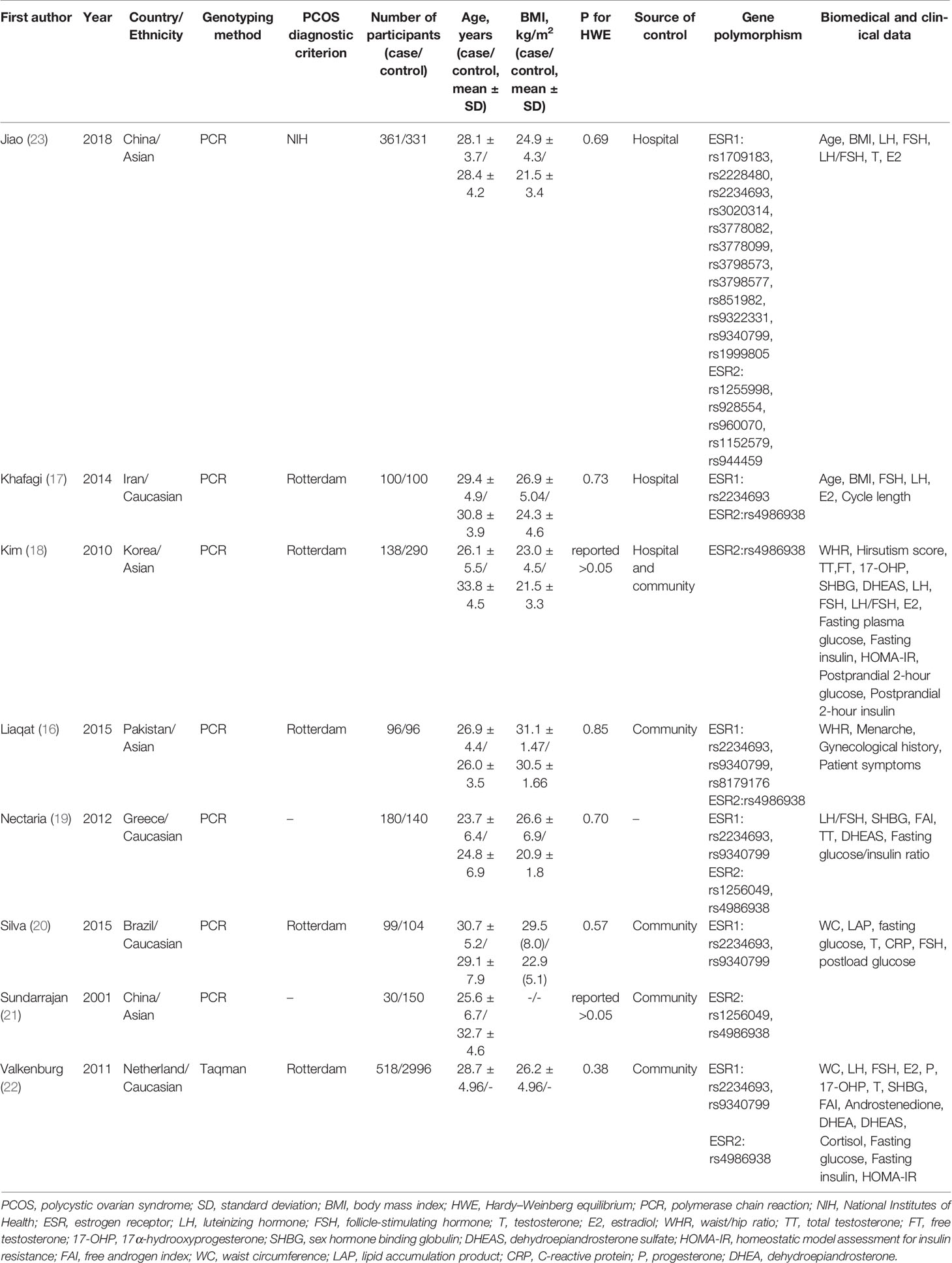
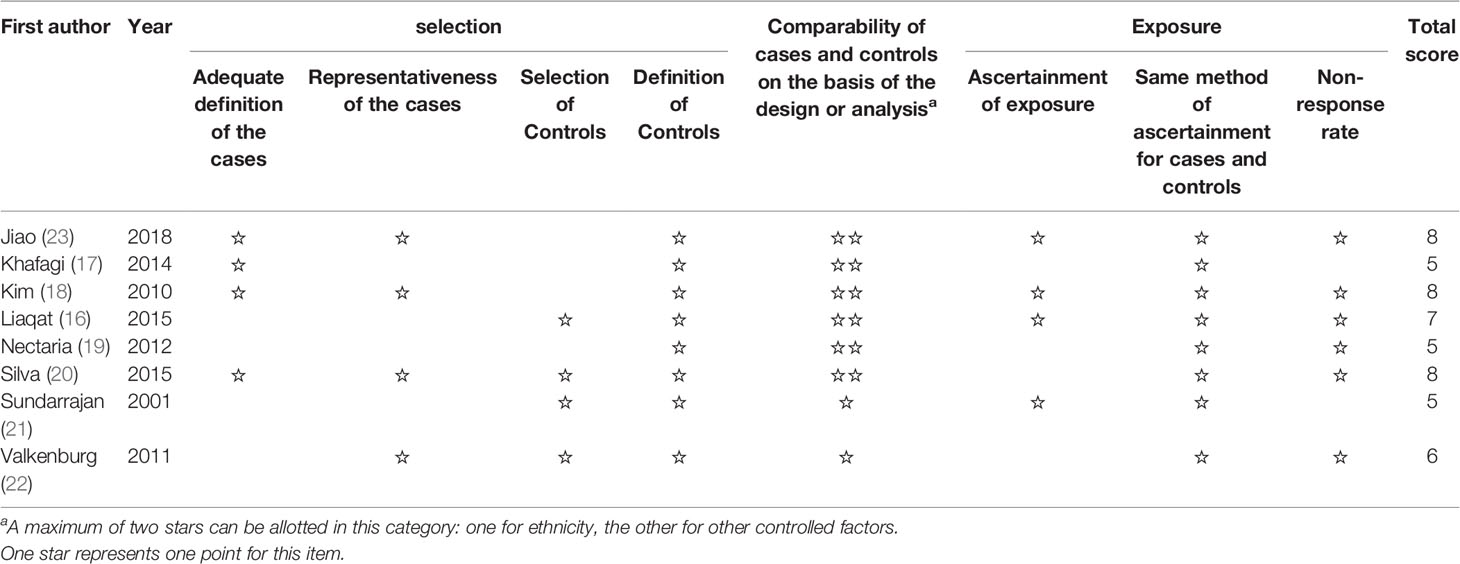
The characteristics of each eligible study are listed in Table 1. A total of 5,720 subjects were reported in 8 studies, including 1,522 PCOS patients and 4,198 controls. These studies, which ranged from 2001 to 2018, spanned seven countries and involved two ethnicities: Asian and Caucasian. Despite the fact that these studies explored the association between multiple SNPs in ESR1/ESR2 and PCOS, most SNPs were reported only once. Therefore, the SNPs that were examined in more than three populations were selected for ultimate pooled analysis. The PCOS patients and control people were genotyped for ESR1 rs2234693 in six studies (16, 17, 19, 20, 22, 23), for ESR1 rs9340799 in five studies (16, 19, 20, 22, 23), and for ESR2 rs4986938 in six studies (16–19, 21, 22). The genotype distributions were in agreement with HWE; the age and body mass index (BMI) were comparable between patients and controls in most studies as reported except that two studies (21, 22) did not record the BMI of the participants. The scores of included studies assessed by NOS were ranged from 5 to 8 (Table 2), which means all included articles were regarded as moderate or high quality.
Associations Between ESR1 rs2234693 Polymorphism and PCOS Susceptibility
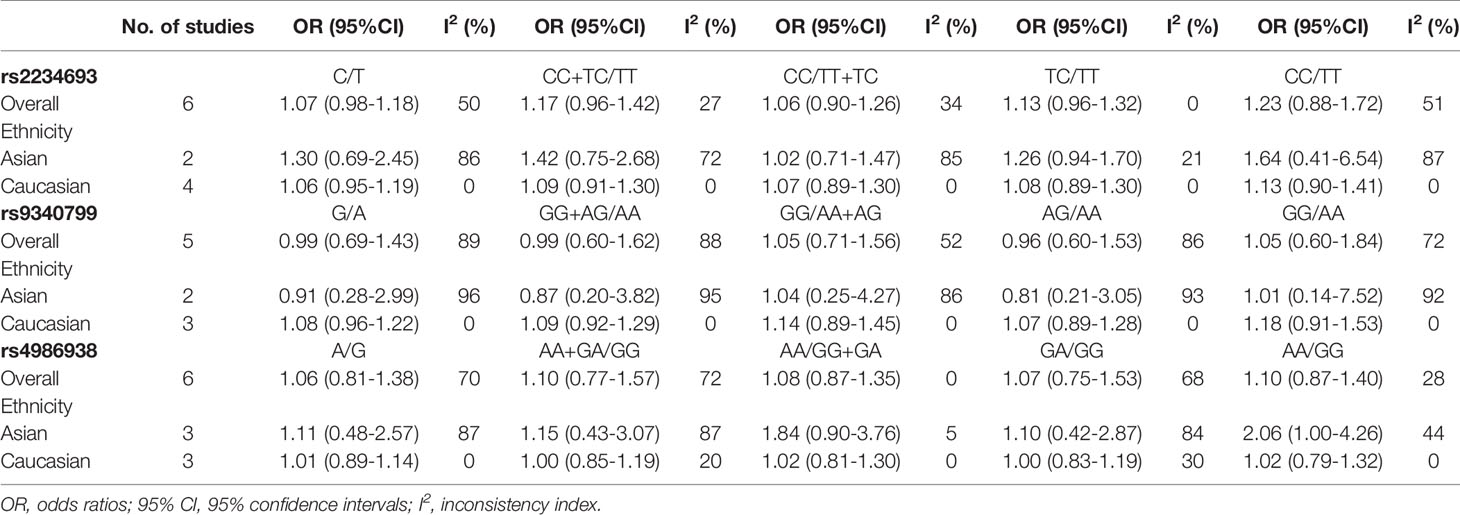
Six studies that recruited a total of 1,354 PCOS patients and 3,767 controls were measured for rs2234693. The analyses were conducted in five genetic models, and the results are presented in Figure 2. The Q-test and I2 statistic indicated that mild heterogeneity was revealed across these studies. Ultimately, no significant association was identified in five genetic models: Allele model (C versus T): OR=1.07 95%CI 0.98–1.18, I2 = 50%; Dominant model (CC+TC versus TT): OR=1.17 95%CI 0.96–1.42, I2 = 27%; Recessive model (CC versus TT+TC): OR=1.06 95%CI 0.90–1.26, I2 = 34%; Heterozygote model (TC versus TT): OR=1.13 95%CI 0.96–1.32, I2 = 0%; Homozygote model (CC versus TT): OR=1.23 95%CI 0.88–1.72, I2 = 51%. Besides, subgroup analyses were performed based on a predefined factor: ethnicity. In Asians, to reduce the influence of severe heterogeneity, the random-effects model was conducted in most genetic models except for the heterozygote model, which showed no significant statistical heterogeneity. In Caucasians, there was no between-study heterogeneity, due to which the fixed-effects model was applied. Likewise, there was no significant association between rs2234693 and PCOS susceptibility in Asians or Caucasians (Table 3).
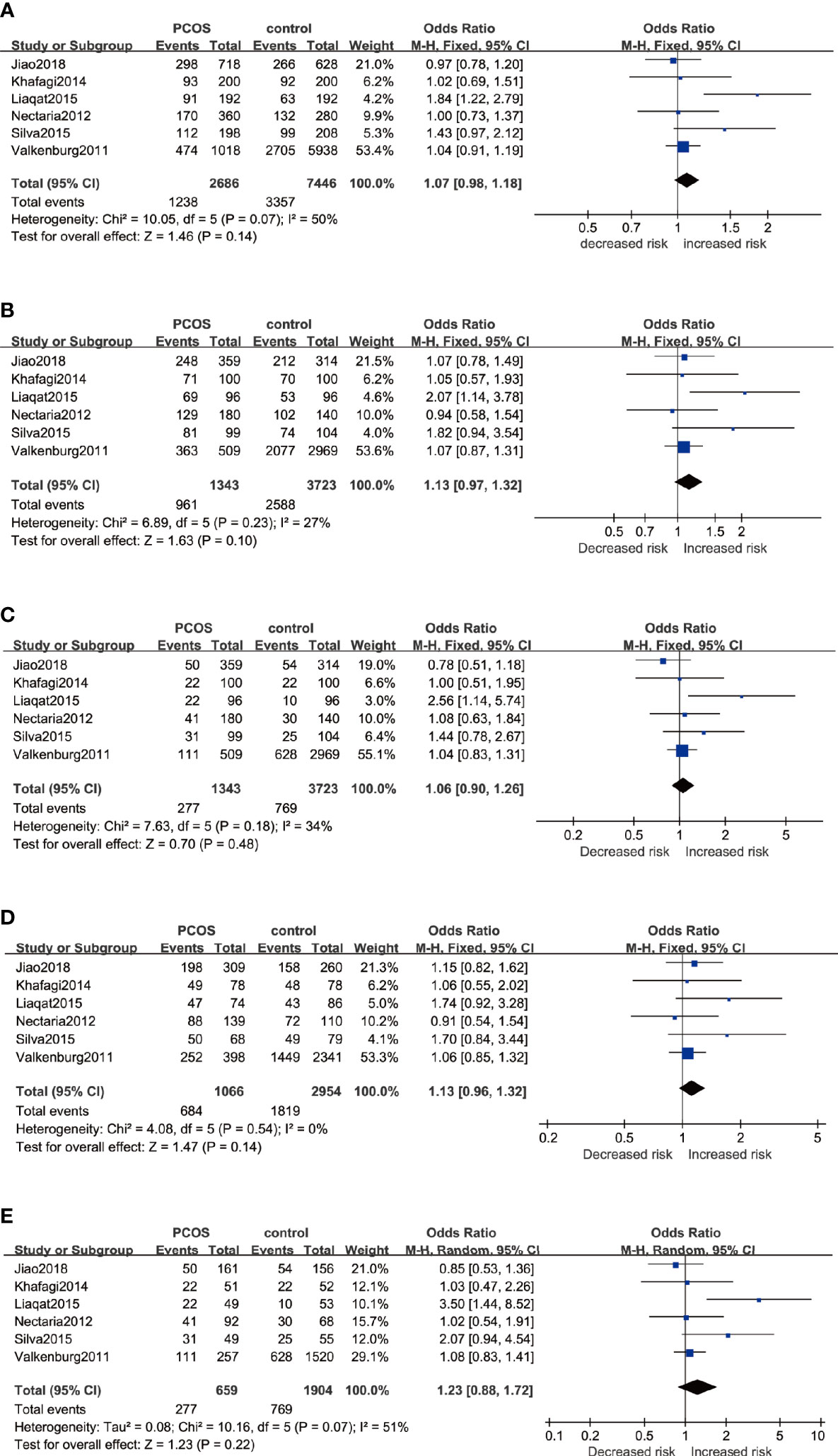
Figure 2 Forest plots of the association between the ESR1 rs2234693 gene and risk of PCOS using different genetic models in overall analysis: (A) Allele model (C versus T), (B) Dominant model (CC+TC versus TT), (C) Recessive model (CC versus TT+TC), (D) Heterozygote model (TC versus TT), and (E) Homozygote model (CC versus TT). In each model, solid squares represent the OR, horizontal lines represent 95%CI, and diamond represents the pooled OR and 95%CI. OR, odds ratios; 95% CI, 95% confidence intervals; I2, inconsistency index.
Associations Between ESR1 rs9340799 Polymorphism and PCOS Susceptibility
By pooling five eligible studies that consisted of 1,254 cases and 3,667 controls, no significant association was observed in five genetic models, respectively as depicted in Figure 3: Allele model (G versus A): OR=0.99 95%CI 0.69–1.43, I2 = 89%; Dominant model (GG+AG versus AA): OR=0.99 95%CI 0.60–1.62, I2 = 88%; Recessive model (GG versus AA+AG): OR=1.05 95%CI 0.71–1.56, I2 = 52%; Heterozygote model (AG versus AA): OR=0.96 95%CI 0.60–1.53, I2 = 86%; Homozygote model (GG versus AA): OR=1.05 95%CI 0.60–1.84, I2 = 72%. With severe heterogeneity presented, the random-effects model was applied in every genetic model to synthesize the OR and 95% CI. In subgroup analyses, noticeable heterogeneity was revealed in the Asian population; thus, the random-effects model was chosen for two studies based on Asians. No heterogeneity was found in studies based on Caucasians in which the fixed-effects model was selected. Nonetheless, no statistical association was found in the subgroups of Asians or Caucasians irrespective of whether allele, dominant, recessive, heterozygote, or homozygote comparison of inheritance was used (Table 3).
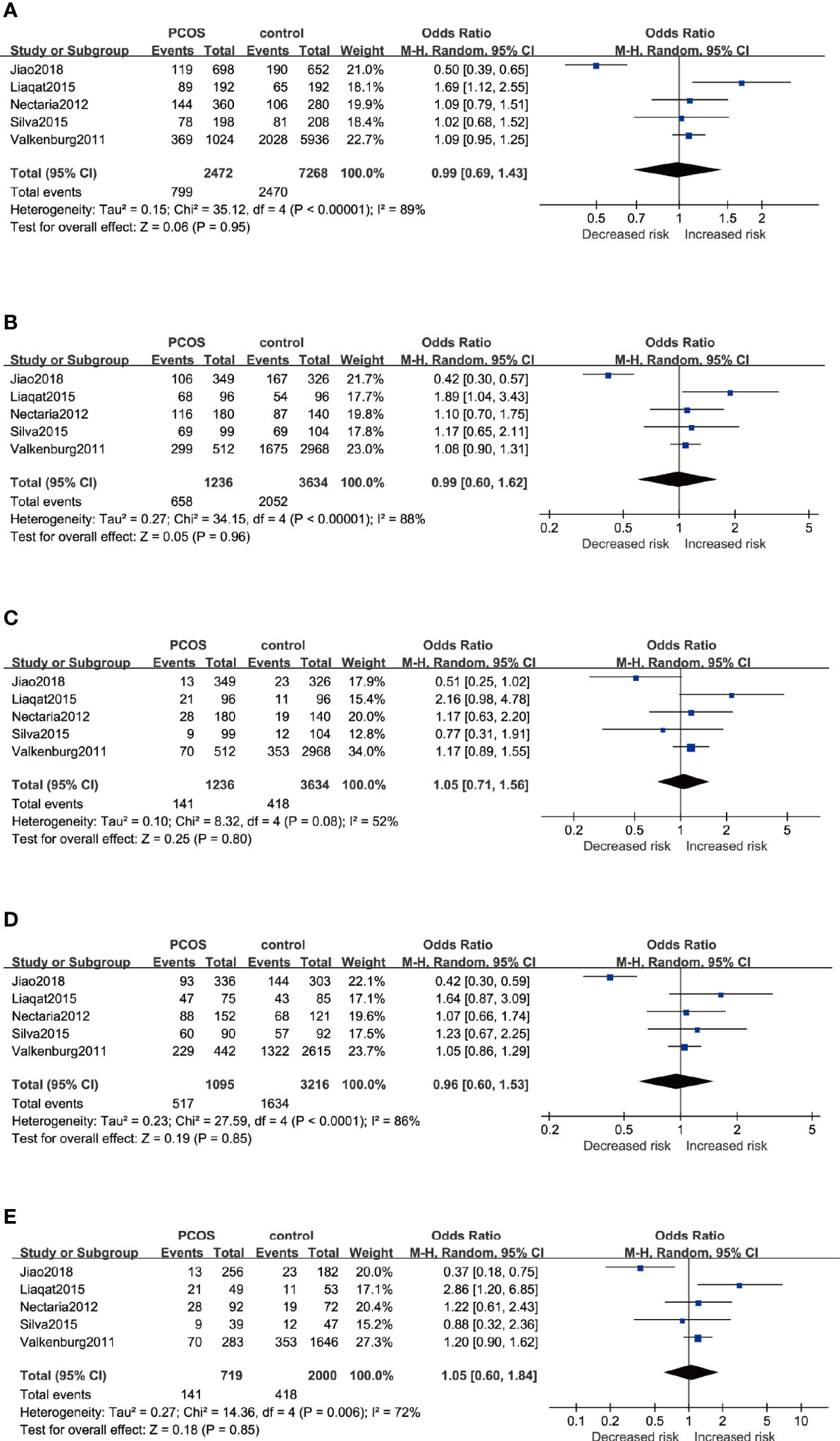
Figure 3 Forest plots of the association between the ESR1 rs9340799 gene and risk of PCOS using different genetic models in overall analysis: (A) Allele model (G versus A), (B) Dominant model (GG+AG versus AA), (C) Recessive model (GG versus AA+AG), (D) Heterozygote model (AG versus AA), and (E) Homozygote model (GG versus AA). In each model, solid squares represent the OR, horizontal lines represent 95%CI, and diamond represents the pooled OR and 95%CI. OR, odds ratios; 95% CI, 95% confidence intervals; I2, inconsistency index.
Associations Between ESR2 rs4986938 Polymorphism and PCOS Susceptibility
The association between ESR2 rs4986938 and PCOS risk was assessed in six pieces of research performed in different countries, involving 1,062 PCOS women and 3,772 healthy women. Prominent heterogeneity was revealed in the allele model, the dominant model, and the heterozygote model in which meta-analysis was performed with a random-effects model, while the fixed-effects model was adopted in the other two genetic models. The results of meta-analysis suggested that ESR2 rs4986938 was not associated with increased or decreased risk of PCOS (Figure 4): Allele model (A versus G): OR=1.06 95%CI 0.81–1.38, I2 = 70%; Dominant model (AA+GA versus GG): OR=1.10 95%CI 0.77–1.57, I2 = 72%; Recessive model (AA versus GG+GA): OR=1.08 95%CI 0.87–1.35, I2 = 0%; Heterozygote model (GA versus GG): OR=1.07 95%CI 0.75–1.53, I2 = 68%; Homozygote model (AA versus GG): OR=1.10 95%CI 0.87–1.40, I2 = 28%. Subsequently, subgroup analyses were conducted in Asians with high heterogeneity (random-effects model) and Caucasians with low heterogeneity (fixed-effects model). No association was found to be significant in different ethnicities (Table 3).
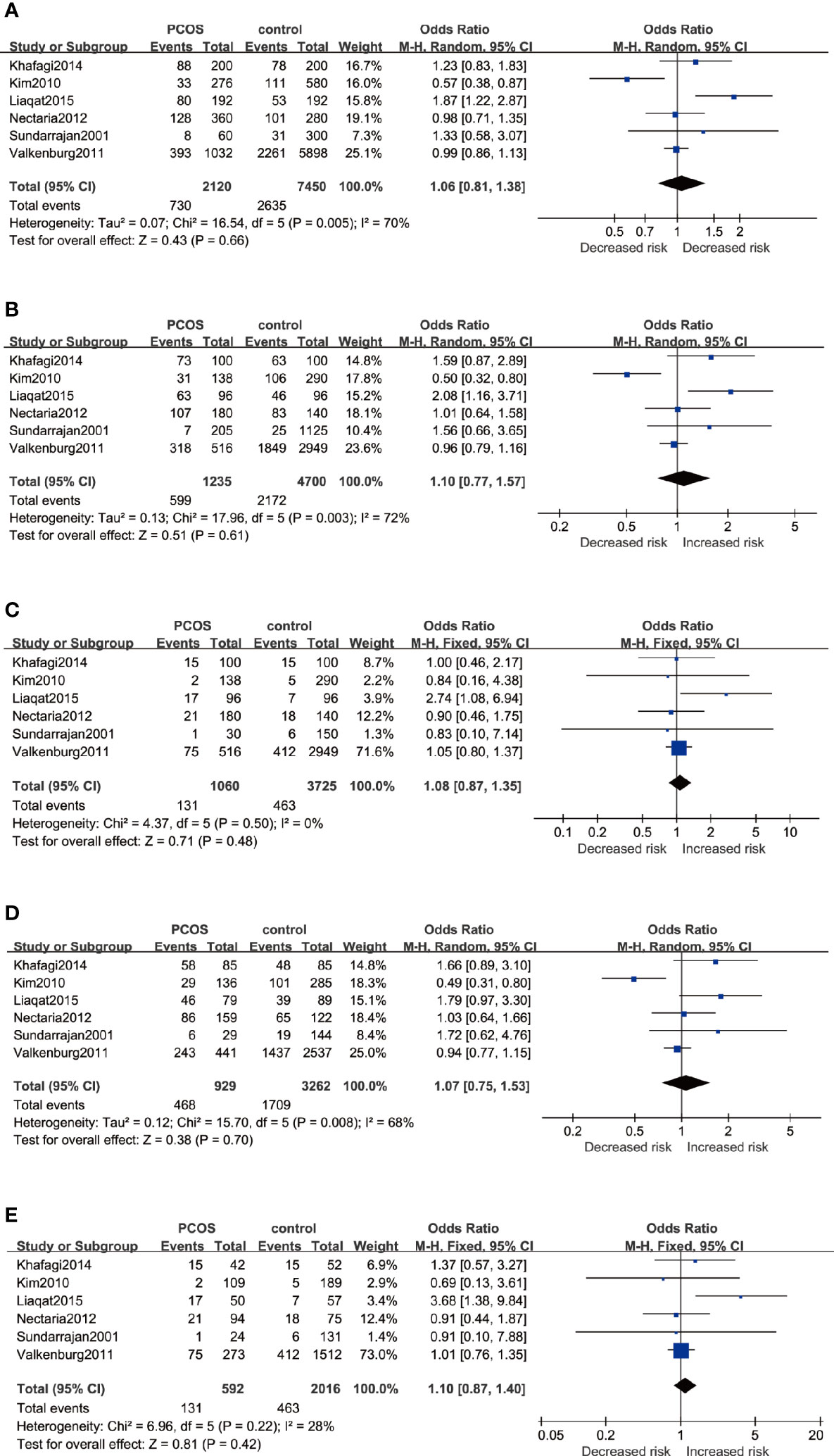
Figure 4 Forest plots of the association between the ESR2 rs4986938 gene and risk of PCOS using different genetic models in overall analysis: (A) Allele model (A versus G), (B) Dominant model (AA+GA versus GG), (C) Recessive model (AA versus GG+GA), (D) Heterozygote model (GA versus GG), and (E) Homozygote model (AA versus GG). In each model, solid squares represent the OR, horizontal lines represent 95%CI, and diamond represents the pooled OR and 95%CI. OR, odds ratios; 95% CI, 95% confidence intervals; I2, inconsistency index.
Sensitivity Analysis
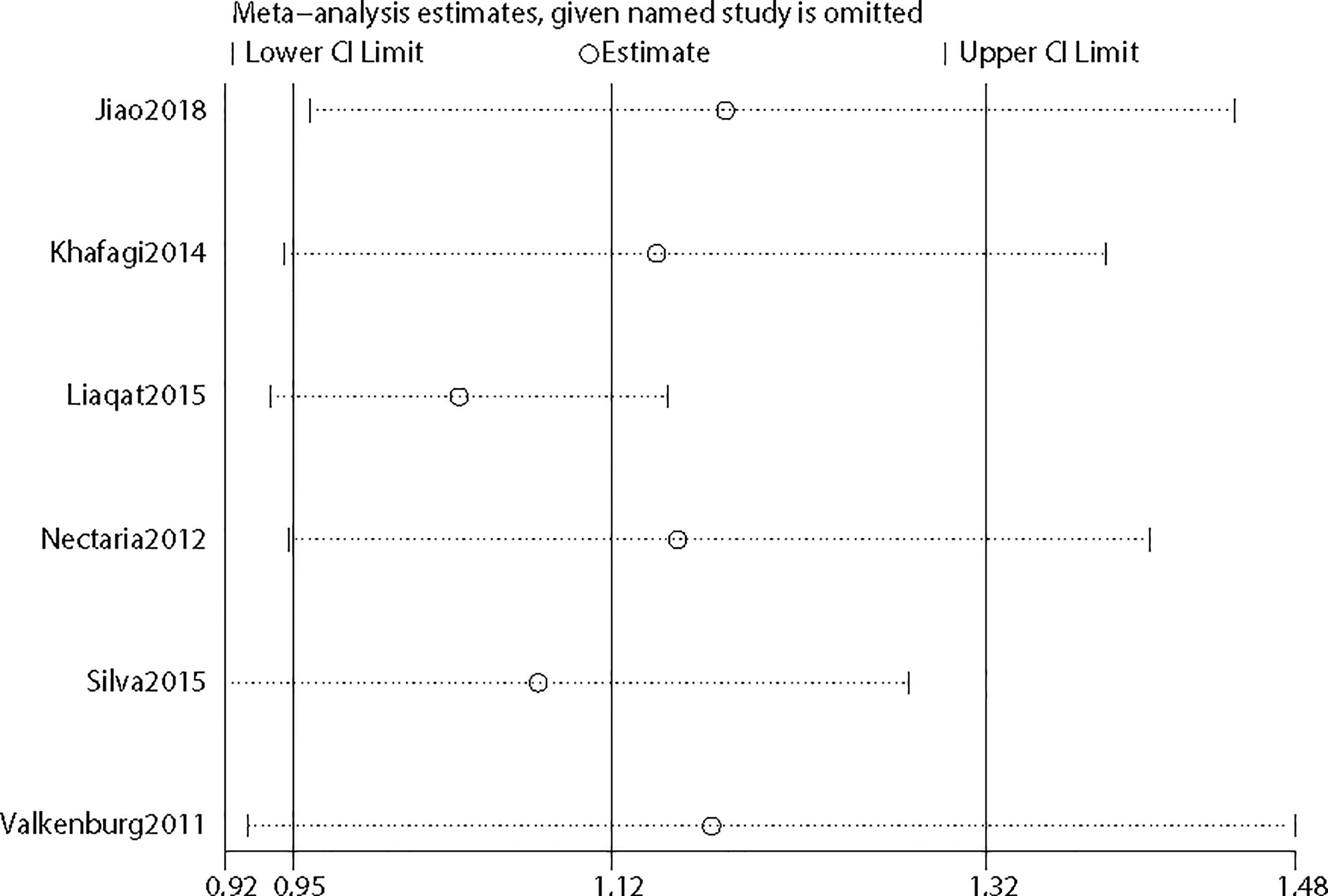
To further confirm the robustness of synthesized results, sensitivity analysis was conducted by removing each study once in five genetic models, respectively, and Figure 5 depicts the pooled ORs and 95%CIs after deleting a single study in the allele model for rs2234693. For rs2234693, one study (16) in Asians contributed greatly to the high heterogeneity, as I2 decreased from 50% to 0% with a consistent result (OR=1.07 95%CI 0.98–1.18) after excluding this study. For rs9340799, the research of Jiao et al. (23) in Asians seemed to be the source of noticeable heterogeneity. With the removal of this study, the heterogeneity decreased from high (88%) to low (2%) in the Q test, and consistently, the result demonstrated no association between rs9340799 and PCOS susceptibility (OR=1.14 95%CI 0.97–1.34). By comprehensively scrutinizing these two pieces of research, ethnicity contributed remarkably to the inconsistency, and obviously, the results of subgroup analyses also supported that ethnicity led to the potential discrepancy.
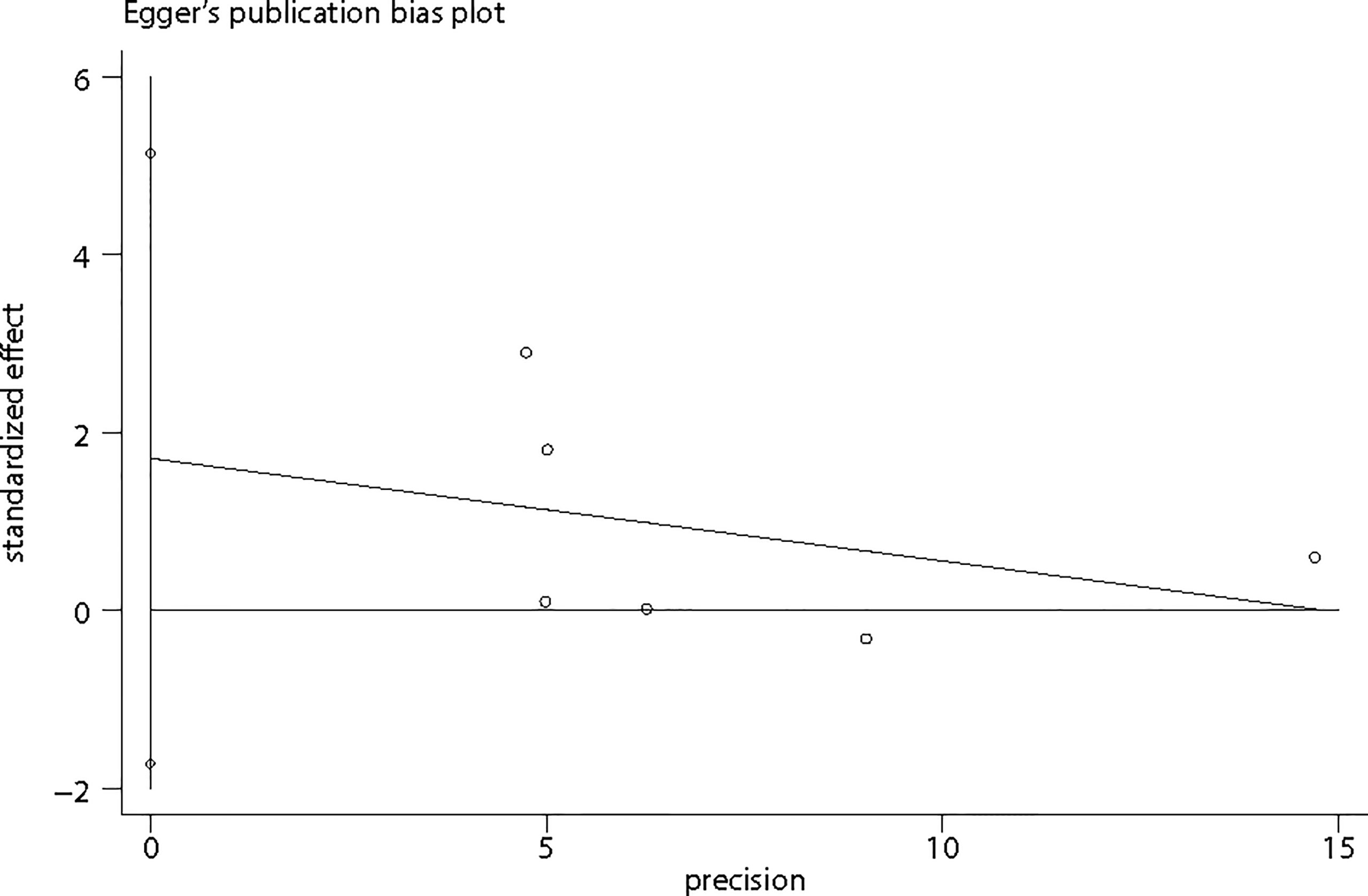
Publication Bias
Considering that the number of included studies was limited, it was inappropriate to apply Funnel plots to assess publication bias by visual inspection. Hence, Egger’s regression test was applied (Figure 6) to assess publication bias by formal statistic test. The P value for the Egger’s test was 0.238, indicating no potential publication bias among included studies. However, the results should be interpreted with caution due to the limited studies.
Discussion
PCOS was generally considered implicated with genetic and hormonal factors. Accumulating shreds of evidence brought forward over 70 candidate genes associated with different genotypes of PCOS, especially in dysfunction of reproduction and endocrine metabolism (28). To our knowledge, this is the first and the most comprehensive meta-analysis concerning the correlation between ESR1/ESR2 polymorphisms and PCOS susceptibility. Our study found that rs2234693, rs9340799, and rs4986938 polymorphisms, the most extensively investigated in ESR1 and ESR2, might not play an essential role in PCOS susceptibility in the allele model, the dominant model, the recessive model, the heterozygote model, or the homozygote model from pooled analysis. Subgroup analyses according to the predefined factor of ethnicity indicated that these gene mutations were not associated with increased or decreased risk of PCOS in Asians and Caucasians separately. Furthermore, ethnicity might be the source of inconsistency due to low heterogeneity in subgroups. After sensitivity analysis by removing individual study once sequentially, the results remained consistent, which enhanced the robustness and liability of our conclusion.
Until now, at least 2,200 SNPs and 700 SNPs in ESR1 and ESR2 were identified (www.snpper.chip.org). Two best-characterized SNPs of rs2234693 and rs9340799 in the ESR1 are located in the first intron, which are 391 and 351 bp upstream of exon 2 and named as PvuII and XbaI, respectively (29). Also, the widely investigated rs4936938 polymorphism in the ESR2 is at position 1859 in the 3’-untranslated region of exon 8 and named as AluI (16). Although the transcriptional regulation attributed to these SNPs was not clarified adequately, feasible functional mechanisms were raised that they changed the expression of ESR1 and ESR2 by altering the binding of transcription factors (30). One previous human observational research manifested that there was markedly increased ESR1 expression in the endometrium tissue from PCOS women accompanied with a higher ESR1/ESR2 ratio than the comparable healthy group (31). Another study found similar results by isolating follicles and granulosa cells from PCOS women versus regular cycling women: the expression of ESR1 was higher in follicles, while the expression of ESR2 was lower in granulosa cells compared with the control group (32). Furthermore, these three SNPs were found to be associated with various estrogen-dependent diseases such as venous miscarriage, onset of menopause, and fracture risk after menopause (33–35).
Folliculogenesis and ovulation are complicated processes that involved a variety of hormone interactions. It is a typical feature of PCOS that dominant follicles fail to develop consistently, and previous research also observed that the concentration of estrogen was low in PCOS follicular fluid compared with that in dominant follicles (36). A biological function was clarified that estrogen can directly stimulate the proliferation of granulosa cells, attenuate apoptosis and follicular atresia, and increase the expression of hormone receptors (37, 38). Estrogen performed function of proliferation by binding to ERα expressed in the theca cells, while it also mediated the differentiation action and stimulated late follicle growth by attaching with ERβ localized in granulosa cells of developing follicles at all stages. Further studies also suggested that ERα and ERβ, which were both high-affinity ligand-dependent transcription factors (39, 40), were expressed not only in the ovaries but also in the hypothalamus. They played important roles in inducing gonadotropin-releasing hormone (GnRH) release as well as upregulating the expression of GnRH receptors in the anterior pituitary, thus promoting positive regulation of estrogen on the hypothalamus–pituitary axis and maintaining regular ovulation (41–43). This estrogen-ER complex recruited coregulatory proteins (coactivators or corepressors) to the promoter and regulated gene expression by binding to estrogen response element sequences in the promoter region of estrogen-responsive genes (44). Considering direct and indirect effects of estrogen on follicle development, maturation, and ovulation mediated by ERα and ERβ, polymorphisms of ESR1 and/or ESR2 would be expected to be associated with persistent anovulation in PCOS and an ocean of studies focused on genetic variations in ESR1 and ESR2 with pathological dysfunction of ovulation. However, there was a persistent inconsistency among studies concerning SNPs in the estrogen signaling pathway and susceptibility of PCOS. Our studies synthesized all of the current studies and solved the controversy by reporting no significant associations between the variants of ESR1 rs2234693, rs9340799, and ESR2 rs4936938 with PCOS.
One thing worth being called attention to is that in spite of the fact that no statistical significance was found, PCOS patients tended to have a higher ratio of rs2234693 polymorphism in ESR1 than control women, while rs9340799 and rs4986938 polymorphisms distributed equally in two groups according to our results. Furthermore, it was found in subgroup analysis that when the population was divided into different ethnicities, the range of 95%CI was wider than the ultimate meta-result of rs2234693. It showed that accumulating sample sizes led to a more obvious result tilting to the right, indicating that sample size might contribute greatly to the distribution of SNPs. Rs2234693 could still be a promising candidate SNP involved in the occurrence of PCOS, which needed more large sample-size studies to verify. Nevertheless, the conclusion we drew from this meta-analysis might be influenced by underpowered effect. It was well known that PCOS was a multifactorial syndrome, which meant single gene mutation might contribute little, while the interaction of multiple genes might play a decisive role in the susceptibility of the diseases. In the future, genetic association studies should focus on comprehensive genotyping on candidate gene loci, analyzing the majority of variations across the entire gene region by linkage disequilibrium. Meanwhile, the gene–gene and gene–environment interactions will also be the priorities in future studies on the etiology of PCOS.
Some superiorities need to be addressed in this meta-analysis. First, the included studies in pooled analysis were all of moderate or high quality according to the most widely accepted assessment tool for observational studies, the NOS system, which guaranteed the reliability of raw data. Second, although the number of studies was limited, Egger’s linear regression test suggested that no obvious publication bias was detected. Third, we searched all mainstream databases by comprehensive search strategies and references in eligible studies irrespective of language; thus, selection bias was controlled. Finally, the results of sensitivity confirmed the robustness of our conclusion after strict inclusion and exclusion criteria.
However, several limitations should be noticed when interpreting the results of our meta-analysis. First, the ESR1 and ESR2 genes were generally considered highly polymorphic, while only 20 SNPs were under detection for correlation with PCOS and only 3 widely investigated SNPs (rs2234693, rs9340799, and rs4936938) were detected in more than three studies. Due to the fact that numerous SNPs were reported merely once or twice, for which the results cannot be meta-analyzed, there might be other potential PCOS susceptibility loci. Furthermore, the pathogenesis of PCOS was complicated, which meant a single gene mutation was unlikely to remarkably influence the risk of the disease, while disequilibrium between SNPs in ESR genes might be a risk factor. Second, although eligible studies were conducted in different countries, raw data from Africans were unavailable. We tried to contact the corresponding author, but no response was received. As the distribution of SNPs in different ethnicities was conspicuously distinct, the results should be interpreted with caution. Third, PCOS was defined as a heterogeneous disease for its complicated phenotypes. Due to the absence of biochemical parameters of PCOS patients in eligible studies, subgroup stratification analyses based on phenotypes were unable to be performed. Hence, more well-designed studies with a large sample size and elaborate clinical data conducted in different ethnicities were strongly required.
Conclusions
In conclusion, our systematic review and meta-analysis found that the rs2234693 and rs9340799 polymorphisms in the ESR1 gene and rs4936938 polymorphism in the ESR2 gene were not likely to be associated with individual PCOS susceptibility, even if ethnicity was taken into account. The conclusion we draw from the meta-analysis may guide and promote the study of pathogenesis and etiology of PCOS in gene–gene and gene–environment interactions.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author Contributions
SZ contributed to the conception of the study. SZ and SW contributed to the selection of articles, evaluation of evidence quality, and extraction of data. SZ performed the data analyses and wrote the manuscript. SW, YS, MY, XS, YC, DK, and LX helped perform the analyses with constructive discussions. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by Scientific Research Projects of The National Natural Science Fund (81671421 and 81971354).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Shengrong Zhou from Swiss Federal Institute of Technology Zurich for language support.
References
1. Diamanti-Kandarakis E, Kandarakis H, Legro RS. The Role of Genes and Environment in the Etiology of PCOS. Endocrine (2006) 30(1):19–26. doi: 10.1385/endo:30:1:19
2. Sharp L, Cardy AH, Cotton SC, Little J. CYP17 Gene Polymorphisms: Prevalence and Associations With Hormone Levels and Related Factors. A HuGE Review. Am J Epidemiol (2004) 160(8):729–40. doi: 10.1093/aje/kwh287
3. Shen W, Li T, Hu Y, Liu H, Song M. CYP1A1 Gene Polymorphisms and Polycystic Ovary Syndrome Risk: A Meta-Analysis and Meta-Regression. Genet Test Mol Biomarkers (2013) 17(10):727–35. doi: 10.1089/gtmb.2013.0209
4. Lin LH, Baracat MC, Maciel GA, Soares JM Jr., Baracat EC. Androgen Receptor Gene Polymorphism and Polycystic Ovary Syndrome. Int J Gynaecol Obstet: Off Organ Int Fed Gynaecol Obstet (2013) 120(2):115–8. doi: 10.1016/j.ijgo.2012.08.016
5. Wang X, Wang K, Yan J, Wu M. A Meta-Analysis on Associations of FTO, MTHFR and TCF7L2 Polymorphisms With Polycystic Ovary Syndrome. Genomics (2020) 112(2):1516–21. doi: 10.1016/j.ygeno.2019.08.023
6. Anagnostou E, Drakakis P, Marinopoulos S, Mavrogianni D, Loutradis D. The Impact of Genetics Profile (Gene Polymorphisms) in Obese Non-PCOS Women Entering an IVF/ICSI Program. Curr Drug Targets (2013) 14(8):850–5. doi: 10.2174/1389450111314080004
7. Feng C, Lv PP, Yu TT, Jin M, Shen JM, Wang X, et al. The Association Between Polymorphism of INSR and Polycystic Ovary Syndrome: A Meta-Analysis. Int J Mol Sci (2015) 16(2):2403–25. doi: 10.3390/ijms16022403
8. Ketefian A, Jones MR, Krauss RM, Chen YD, Legro RS, Azziz R, et al. Association Study of Androgen Signaling Pathway Genes in Polycystic Ovary Syndrome. Fertil Steril (2016) 105(2):467–73.e4. doi: 10.1016/j.fertnstert.2015.09.043
9. Deswal R, Nanda S, Dang AS. Association of Luteinizing Hormone and LH Receptor Gene Polymorphism With Susceptibility of Polycystic Ovary Syndrome. Syst Biol Reprod Med (2019) 65(5):400–8. doi: 10.1080/19396368.2019.1595217
10. Chen DJ, Ding R, Cao JY, Zhai JX, Zhang JX, Ye DQ. Two Follicle-Stimulating Hormone Receptor Polymorphisms and Polycystic Ovary Syndrome Risk: A Meta-Analysis. Eur J Obstet Gynecol Reprod Biol (2014) 182:27–32. doi: 10.1016/j.ejogrb.2014.08.014
11. Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, et al. The Nuclear Receptor Superfamily: The Second Decade. Cell (1995) 83(6):835–9. doi: 10.1016/0092-8674(95)90199-x
12. Xu XL, Deng SL, Lian ZX, Yu K. Estrogen Receptors in Polycystic Ovary Syndrome. Cells (2021) 10(2):459. doi: 10.3390/cells10020459
13. Gennari L, Merlotti D, De Paola V, Calabrò A, Becherini L, Martini G, et al. Estrogen Receptor Gene Polymorphisms and the Genetics of Osteoporosis: A HuGE Review. Am J Epidemiol (2005) 161(4):307–20. doi: 10.1093/aje/kwi055
14. Lee S, Kang DW, Hudgins-Spivey S, Krust A, Lee EY, Koo Y, et al. Theca-Specific Estrogen Receptor-Alpha Knockout Mice Lose Fertility Prematurely. Endocrinology (2009) 150(8):3855–62. doi: 10.1210/en.2008-1774
15. Krege JH, Hodgin JB, Couse JF, Enmark E, Warner M, Mahler JF, et al. Generation and Reproductive Phenotypes of Mice Lacking Estrogen Receptor Beta. Proc Natl Acad Sci USA (1998) 95(26):15677–82. doi: 10.1073/pnas.95.26.15677
16. Liaqat I, Jahan N, Krikun G, Taylor HS. Genetic Polymorphisms in Pakistani Women With Polycystic Ovary Syndrome. Reprod Sci (Thousand Oaks Calif) (2015) 22(3):347–57. doi: 10.1177/1933719114542015
17. Khafagi Z, Mozdarani H, Behmanesh M, Distribution A, Of F. ESR1 and ESR2 SNPs Among Women With Polycystic Ovary Syndrome Undergoing In Vitro Fertilization. Bull Environ Pharmacol Life Sci (2014) 3(8):128–34.
18. Kim JJ, Choi YM, Choung SH, Yoon SH, Lee GH, Moon SY. Estrogen Receptor Beta Gene +1730 G/A Polymorphism in Women With Polycystic Ovary Syndrome. Fertil Steril (2010) 93(6):1942–7. doi: 10.1016/j.fertnstert.2008.12.040
19. Nectaria X, Leandros L, Ioannis G, Agathocles T. The Importance of Erα and Erβ Gene Polymorphisms in PCOS. Gynecol Endocrinol Off J Int Soc Gynecol Endocrinol (2012) 28(7):505–8. doi: 10.3109/09513590.2011.649811
20. Silva FS, Sóter MO, Sales MF, Candido AL, Reis FM, Silva IF, et al. Estrogen Receptor αlpha Gene (ESR1) PvuII and XbaI Polymorphisms Are Associated to Metabolic and Proinflammatory Factors in Polycystic Ovary Syndrome. Gene (2015) 560(1):44–9. doi: 10.1016/j.gene.2015.01.037
21. Sundarrajan C, Liao WX, Roy AC, Ng SC. Association Between Estrogen Receptor-Beta Gene Polymorphisms and Ovulatory Dysfunctions in Patients With Menstrual Disorders. J Clin Endocrinol Metab (2001) 86(1):135–9. doi: 10.1210/jcem.86.1.7098
22. Valkenburg O, Uitterlinden AG, Themmen AP, de Jong FH, Hofman A, Fauser BC, et al. Genetic Polymorphisms of the Glucocorticoid Receptor May Affect the Phenotype of Women With Anovulatory Polycystic Ovary Syndrome. Hum Reprod (Oxford England) (2011) 26(10):2902–11. doi: 10.1093/humrep/der222
23. Jiao X, Chen W, Zhang J, Wang W, Song J, Chen D, et al. Variant Alleles of the ESR1, PPARG, HMGA2, and MTHFR Genes Are Associated With Polycystic Ovary Syndrome Risk in a Chinese Population: A Case-Control Study. Front Endocrinol (2018) 9:504. doi: 10.3389/fendo.2018.00504
24. Zawadzki J, Dunaif A. Current Issues in Endocrinology and Metabolism: Polycystic Ovary Syndrome. MA: Blackwell Scientific Publications Cambridge (1992).
25. Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 Consensus on Diagnostic Criteria and Long-Term Health Risks Related to Polycystic Ovary Syndrome (PCOS). Hum Reprod (Oxford England) (2004) 19(1):41–7. doi: 10.1093/humrep/deh098
26. Azziz R, Carmina E, Dewailly D, Diamanti-Kandarakis E, Escobar-Morreale HF, Futterweit W, et al. Positions Statement: Criteria for Defining Polycystic Ovary Syndrome as a Predominantly Hyperandrogenic Syndrome: An Androgen Excess Society Guideline. J Clin Endocrinol Metab (2006) 91(11):4237–45. doi: 10.1210/jc.2006-0178
27. Stang A. Critical Evaluation of the Newcastle-Ottawa Scale for the Assessment of the Quality of Nonrandomized Studies in Meta-Analyses. Eur J Epidemiol (2010) 25(9):603–5. doi: 10.1007/s10654-010-9491-z
28. Jones MR, Goodarzi MO. Genetic Determinants of Polycystic Ovary Syndrome: Progress and Future Directions. Fertil Steril (2016) 106(1):25–32. doi: 10.1016/j.fertnstert.2016.04.040
29. Motawi TM, El-Rehany MA, Rizk SM, Ramzy MM, El-Roby DM. Genetic Polymorphism of Estrogen Receptor Alpha Gene in Egyptian Women With Type II Diabetes Mellitus. Meta Gene (2015) 6:36–41. doi: 10.1016/j.mgene.2015.08.001
30. Araújo KL, Rezende L, Souza LS, Daltoé RD, Madeira KP, Paes MF, et al. Prevalence of Estrogen Receptor Alpha PvuII (C454-397T>C) and XbaI (C454a>G) Polymorphisms in a Population of Brazilian Women. Braz Arch Biol Technol (2011) 54:1151–8. doi: 10.1590/S1516-89132011000600010
31. Quezada S, Avellaira C, Johnson MC, Gabler F, Fuentes A, Vega M. Evaluation of Steroid Receptors, Coregulators, and Molecules Associated With Uterine Receptivity in Secretory Endometria From Untreated Women With Polycystic Ovary Syndrome. Fertil Steril (2006) 85(4):1017–26. doi: 10.1016/j.fertnstert.2005.09.053
32. Jakimiuk AJ, Weitsman SR, Yen HW, Bogusiewicz M, Magoffin DA. Estrogen Receptor Alpha and Beta Expression in Theca and Granulosa Cells From Women With Polycystic Ovary Syndrome. J Clin Endocrinol Metab (2002) 87(12):5532–8. doi: 10.1210/jc.2002-020323
33. Silva IV, Rezende LC, Lanes SP, Souza LS, Madeira KP, Cerri MF, et al. Evaluation of PvuII and XbaI Polymorphisms in the Estrogen Receptor Alpha Gene (ESR1) in Relation to Menstrual Cycle Timing and Reproductive Parameters in Post-Menopausal Women. Maturitas (2010) 67(4):363–7. doi: 10.1016/j.maturitas.2010.08.006
34. Dvornyk V, Long JR, Liu PY, Zhao LJ, Shen H, Recker RR, et al. Predictive Factors for Age at Menopause in Caucasian Females. Maturitas (2006) 54(1):19–26. doi: 10.1016/j.maturitas.2005.08.005
35. van Meurs JB, Schuit SC, Weel AE, van der Klift M, Bergink AP, Arp PP, et al. Association of 5' Estrogen Receptor Alpha Gene Polymorphisms With Bone Mineral Density, Vertebral Bone Area and Fracture Risk. Hum Mol Genet (2003) 12(14):1745–54. doi: 10.1093/hmg/ddg176
36. Jakimiuk AJ, Weitsman SR, Brzechffa PR, Magoffin DA. Aromatase mRNA Expression in Individual Follicles From Polycystic Ovaries. Mol Hum Reprod (1998) 4(1):1–8. doi: 10.1093/molehr/4.1.1
37. Kaipia A, Hsueh AJ. Regulation of Ovarian Follicle Atresia. Annu Rev Physiol (1997) 59:349–63. doi: 10.1146/annurev.physiol.59.1.349
38. Ireland JJ, Richards JS. Acute Effects of Estradiol and Follicle-Stimulating Hormone on Specific Binding of Human [125I]Iodofollicle-Stimulating Hormone to Rat Ovarian Granulosa Cells In Vivo and In Vitro. Endocrinology (1978) 102(3):876–83. doi: 10.1210/endo-102-3-876
39. Altmäe S, Haller K, Peters M, Hovatta O, Stavreus-Evers A, Karro H, et al. Allelic Estrogen Receptor 1 (ESR1) Gene Variants Predict the Outcome of Ovarian Stimulation in In Vitro Fertilization. Mol Hum Reprod (2007) 13(8):521–6. doi: 10.1093/molehr/gam035
40. Hegele-Hartung C, Siebel P, Peters O, Kosemund D, Müller G, Hillisch A, et al. Impact of Isotype-Selective Estrogen Receptor Agonists on Ovarian Function. Proc Natl Acad Sci USA (2004) 101(14):5129–34. doi: 10.1073/pnas.0306720101
41. Mahesh VB, Brann DW. Regulation of the Preovulatory Gonadotropin Surge by Endogenous Steroids. Steroids (1998) 63(12):616–29. doi: 10.1016/s0039-128x(98)00075-0
42. Shughrue PJ, Scrimo PJ, Merchenthaler I. Evidence for the Colocalization of Estrogen Receptor-Beta mRNA and Estrogen Receptor-Alpha Immunoreactivity in Neurons of the Rat Forebrain. Endocrinology (1998) 139(12):5267–70. doi: 10.1210/endo.139.12.6525
43. Scully KM, Gleiberman AS, Lindzey J, Lubahn DB, Korach KS, Rosenfeld MG. Role of Estrogen Receptor-Alpha in the Anterior Pituitary Gland. Mol Endocrinol (Baltimore Md) (1997) 11(6):674–81. doi: 10.1210/mend.11.6.0019
Keywords: polycystic ovary syndrome, polymorphisms, estrogen receptor, meta-analysis, gene variants
Citation: Zhou S, Wen S, Sheng Y, Yang M, Shen X, Chen Y, Kang D and Xu L (2021) Association of Estrogen Receptor Genes Polymorphisms With Polycystic Ovary Syndrome: A Systematic Review and Meta-Analysis Based on Observational Studies. Front. Endocrinol. 12:726184. doi: 10.3389/fendo.2021.726184
Received: 17 June 2021; Accepted: 13 September 2021;
Published: 04 October 2021.
Edited by:
Artur Mayerhofer, Ludwig Maximilian University of Munich, GermanyReviewed by:
Giovanna Motta, University Hospital of the City of Health and Science of Turin, ItalySurasak Saokaew, University of Phayao, Thailand
Copyright © 2021 Zhou, Wen, Sheng, Yang, Shen, Chen, Kang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangzhi Xu, eHVsaWFuZ3poaV9hcnRAMTI2LmNvbQ==
 Siyu Zhou
Siyu Zhou Shu Wen1,2,3
Shu Wen1,2,3 Liangzhi Xu
Liangzhi Xu