- 1Department of Endocrinology and Metabolism, Peking University People’s Hospital, Peking University Diabetes Center, Beijing, China
- 2Department of Endocrinology, Beijing Pinggu District Hospital, Beijing, China
- 3Department of Endocrinology, Beijing Liangxiang Hospital, Beijing, China
Objectives: PPARγ variants cause lipodystrophy, insulin resistance, and diabetes. This study aimed to determine the relationship between PPARγ genotypes and phenotypes and to explore the pathogenesis of diabetes beyond this relationship.
Methods: PPARγ2 exons in 1,002 Chinese patients with early-onset type 2 diabetes (diagnosed before 40 years of age) were sequenced. The functions of variants were evaluated by in vitro assays. Additionally, a review of the literature was performed to obtain all reported cases with rare PPARγ2 variants to evaluate the characteristics of variants in different functional domains.
Results: Six (0.6%) patients had PPARγ2 variant-induced diabetes (PPARG-DM) in the early-onset type 2 diabetes group, including three with the p.Tyr95Cys variant in activation function 1 domain (AF1), of which five patients (83%) had diabetic kidney disease (DKD). Functional experiments showed that p.Tyr95Cys suppresses 3T3-L1 preadipocyte differentiation. A total of 64 cases with damaging rare variants were reported previously. Patients with rare PPARγ2 variants in AF1 of PPARγ2 had a lower risk of lipodystrophy and a higher rate of obesity than those with variants in other domains, as confirmed in patients identified in this study.
Conclusion: The prevalence of PPARG-DM is similar in Caucasian and Chinese populations, and DKD was often observed in these patients. Patients with variants in the AF1 of PPARγ2 had milder clinical phenotypes and lack typical lipodystrophy features than those with variants in other domains. Our findings emphasize the importance of screening such patients via genetic testing and suggest that thiazolidinediones might be a good choice for these patients.
Introduction
Peroxisome proliferator-activated receptor γ (PPARγ) is a ligand-dependent transcription factor with key roles in adipocyte differentiation, adipogenesis, glucose homeostasis, and inflammation (1). Several rare PPARγ variants have been reported as causes of familial partial lipodystrophy (FPLD3, OMIM 604367), insulin resistance, diabetes, and hypertriglyceridemia. In some patients with FPLD3, thiazolidinediones effectively ameliorate hyperglycemia and insulin resistance (2). However, cases with rare PPARγ variants have not been reported in the Chinese population. Since thiazolidinedione-based precision medicine may be beneficial for patients with PPARγ variant-induced diabetes (PPARG-DM), it is necessary to determine the prevalence of PPARG-DM and develop clinical strategies for screening this condition.
There are four functional domains in PPARγ, including the activation function 1 (AF1) domain, DNA-binding domain (DBD), hinge domain (HD), and ligand-binding domain (LBD). Two isoforms of PPARγ (PPARγ1 and PPARγ2) differ with regard to the N-terminal region. An additional 30 amino acids in N-terminal ligand-independent AF1 of PPARγ2 results in a 5–10-fold increase in the activation function in vitro (3). PPARγ2 plays a more dominant role in glucose and lipid metabolism than PPARγ1 (4). The AF1 of PPARγ is involved in PPAR signaling via posttranslational modifications, such as phosphorylation or SUMOylation, and interactions with co-factors (5). A study has shown that a polymorphism in the AF1 domain of PPARγ2 (NP_056953.2: p.Pro12Ala) is associated with type 2 diabetes (T2DM) (6). Patients carrying a rare variant, such as p.Pro113Gln, in the AF1 of PPARγ2 show marked obesity and/or hyperglycemia (7). In contrast, patients with rare variants in domains other than AF1 (non-AF1) consistently show a reduced fat mass and severe insulin resistance. Thus, we hypothesize that the effects of PPARγ2 variants depend on the location of the variants.
Studies have shown that the p.Pro12Ala variant in PPARγ2 is associated with diabetic kidney disease (DKD) (8, 9), and this was supported by an animal study showing that mice with the PPARγ knockout develop DKD (10). However, the association between DKD and p.Pro12Ala has not been validated in Asian populations (11, 12). Furthermore, renal phenotypes for different rare PPARγ2 variants have not been fully characterized.
In this study, we estimated the prevalence of PPARG-DM in the Chinese population, confirmed the relationship between disease phenotypes and variants in different domains, and explored the pathogenesis of PPARG-DM beyond this relationship. Furthermore, we reviewed all reported cases with rare PPARγ2 genotypes in the literature and compared the clinical phenotypes of individuals with PPARγ2 variants in different domains.
Materials and Methods
Study Overview
This study was conducted in compliance with the Declaration of Helsinki. The study protocol was approved by the Ethics Committee of Peking University People’s Hospital (2014-06, 2017PHB035-01). Written informed consent was obtained from all the study participants. A synopsis of the study is shown in Figure 1.
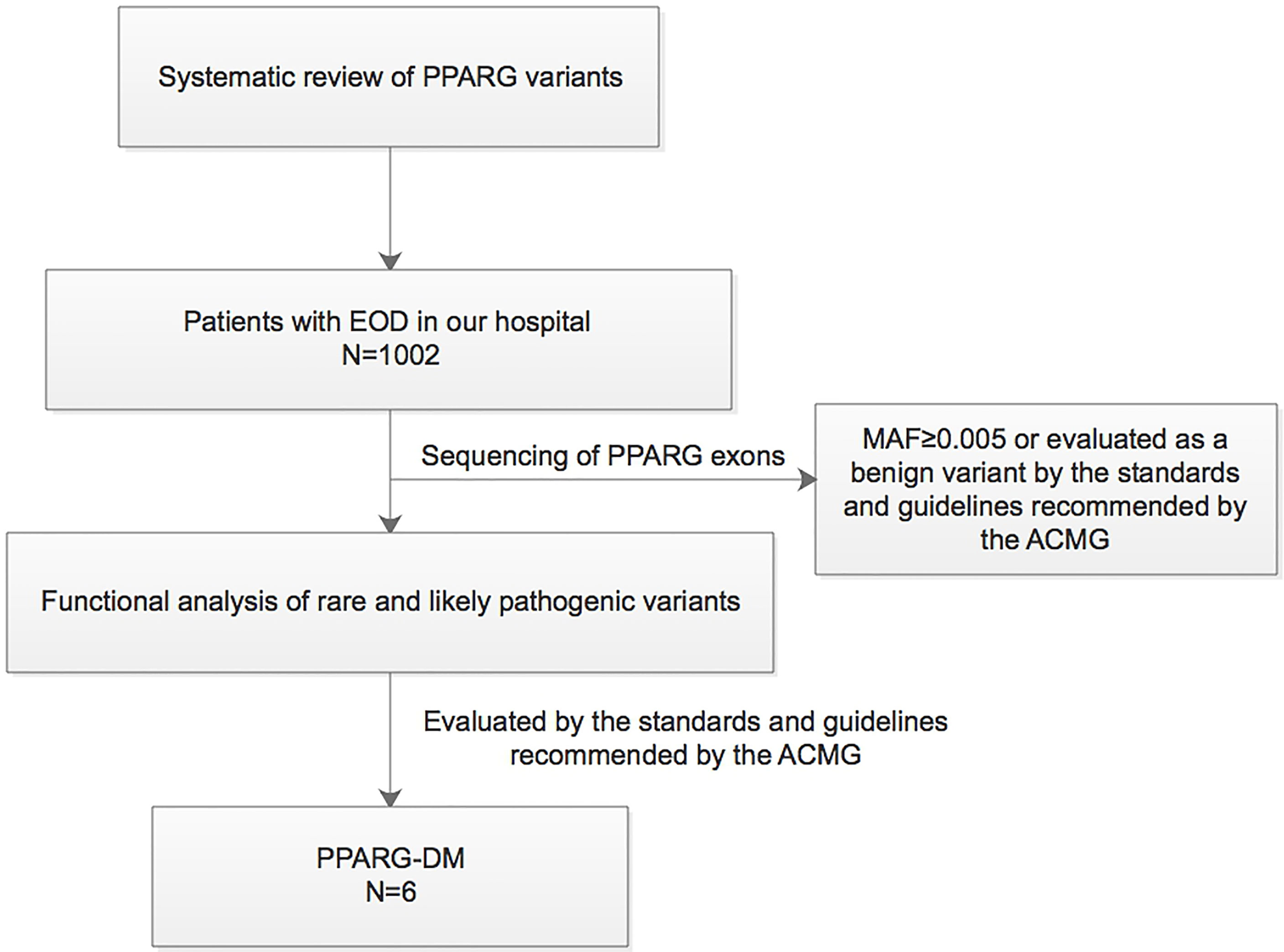
Figure 1 Synopsis of this study. EOD, early-onset type 2 diabetes; MAF, minor allele frequency; PPARG-DM, PPARγ variant-induced diabetes; ACMG, American College of Medical Genetics.
PPARγ2 Gene Sequencing of Patients With Early-Onset Diabetes (Cohort 1)
A total of 1,002 unrelated Chinese patients (age ≥ 18 years) with early-onset diabetes (EOD), who had been diagnosed with T2DM before the age of 40 years in the Endocrinology and Metabolism Department of the Peking University People’s Hospital between June 2014 and October 2018, were included. Diabetes was diagnosed according to the 1999 WHO criteria (13). Patients with typical clinical features of type 1 diabetes or other specific forms of diabetes (e.g., chronic pancreatitis) and those who were positive for anti-glutamic acid decarboxylase antibodies, anti-islet cell antibodies, or anti-insulin antibodies (among individuals not receiving insulin) were excluded from this study.
Biochemical Measurements and Clinical Information
The clinical characteristics of patients were collected at the time of enrollment. Laboratory tests and comorbidities were defined as described previously (14). The parameters were defined as follows: HbA1c National Glycohemoglobin Standardization Program/International Federation of Clinical Chemistry and laboratory medicine (NGSP/IFCC) conversion calculated the following: NGSP (%) = IFCC (mmol/mol) × 0.0915 + 2.152. Homeostasis model assessment (HOMA) was used to evaluate β-cell function and insulin sensitivity. Insulin resistance index (HOMA-IR) = [Fasting insulin (Fins, mIU/L) × fasting plasma glucose (FPG, mmol/L)]/22.5, and β-cell function (HOMA-β) = [Fins (mIU/L) × 20]/[FPG (mmol/L) − 3.5 (%)] (15). Estimated glomerular filtration rate (eGFR, mL/min/1.73 m2) = 175 × Serum creatine (Scr, mg/dl)−1.234 × age (years)−0.179 × 0.79 (if female) (16). DKD was defined as a urinary albumin/creatinine ratio (UACR) ≥30 mg/g and/or eGFR <60 ml/min/1.73 m2. Serum adiponectin (EHC120; NeoBioscience, Shanghai, China) and serum leptin (EZHL-80SK; EMD Millipore, Burlington, MA, USA) levels were determined by ELISA. Serum leptin levels were measured in 85 subjects and the reference range was determined. These subjects had normal glucose tolerance (NGT) and HbA1c <6% and normal BMI, lipid levels, and kidney and liver function, as described previously (17). The 90% interval was 0.49–5.41 ng/ml for men and 2.57–15.75 ng/ml for women.
Sequencing of Exons in PPARγ2
All DNA samples were extracted from peripheral blood. Next-generation sequencing (n = 541) and Sanger sequencing (n = 461) were used for the identification of variants in PPARγ2 exons. For Sanger sequencing, an Applied Biosystems 2720 Thermal Cycler (Waltham, MA, USA) was used [the primers used are shown in the Electronic Supplementary Material (ESM) Table 1]. For next-generation sequencing, the Roche NimbleGen Human Exon V2 capture chip was used, and sequencing was performed on the Illumina HiSeq2500 system. Sequencing data were obtained with a target gene coverage (~100×) of >99% and a depth of >200 bp. The quality scores for all FASTQ data were Q20 >90% and Q30 >85%. All rare variants of PPARγ2 were validated by Sanger sequencing.
Functional Analysis of Rare Variants of PPARγ2 Identified in This Study
The PPARγ2 wild-type construct was prepared by subcloning the full-length human PPARγ2 cDNA (NM_015869) in frame into the XhoI and KpnI sites of the GV141 vector (GeneChem, Shanghai, China) (ESM Figure 1). The p.Glu217Lys (E217K), p.Ser186Gly (S186G), p.Tyr95Cys (Y95C), and p.Ile264Thr (I264T) variants in PPARγ2 were generated by PCR, digested with XhoI and KpnI, and ligated into the GV141 plasmid. Both the wild-type and mutant clones were fully sequenced. All constructs were verified by direct sequencing.
293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM) (11995065; Invitrogen, Waltham, MA, USA) containing 10% fetal bovine serum (Gibco, Invitrogen) at 37°C on 24-well plates. Cells were grown to 60% confluence for transient transfection using X-tremeGENE HP (06366236001; Roche, Basel, Switzerland). Transfection efficiency had been evaluated by a fluorescent microscope. Luciferase activities of E217K, Y95C, S186G, and I264T, relative to those of wild-type PPARγ2, were evaluated using the Dual-Luciferase Reporter Assay System (E1910; Promega, Madison, WI, USA). The media did not contain any PPARγ agonist. The 293T cells were co-transfected with a REPO PPARγ reporter (Genomeditech, Shanghai, China, ESM Figure 2) and the constructed vectors to assay the PPARγ activity. Data are presented as the mean (± SD) values from three assays and are expressed relative to the luciferase activity of the empty vector transfectant, which was set to 1.0.
E217K, Y95C, and wild-type PPARγ2 vectors and the empty vector were transfected into the 3T3-L1 preadipocyte cell line (ATCC, Manassas, VA, USA) using Lipofectamine 3000 (L3000008; Invitrogen); the cells were cultured in DMEM containing 25 mM glucose and 10% calf serum. The transfected 3T3-L1 preadipocytes were verified by RT-PCR and Western blotting (ESM Figure 3). Total RNA was extracted from cells using the Universal RNA Extraction Kit (9767; Takara, Kusatsu, Japan) and cDNA was synthesized using the PrimeScript RT Reagent Kit with gDNA Eraser (RR047A; Takara). Gene-specific primer sequences were used for RT-PCR (human PPARγ primer, 5′-AGCAAACCCCTATTCCATGCT-3′ and 5′-CACGGAGCTGATCCCAAAGT-3′; mouse β-actin primer, 5′-CACTGTCGAGTCGCGTCCA-3′ and 5′-GACCCATTCCCACCATCACA-3′). Proteins were collected from cells treated with RIPA Lysis Buffer (Applygen, C1053, Beijing, China) and were separated by SDS-PAGE and blotted on a nitrocellulose membrane. Differentiation and lipid droplet staining in transfected 3T3-L1 preadipocytes were evaluated on 24-well plates using the Adipogenesis Assay Kit (10006908; Cayman, Ann Arbor, MI, USA) according to the protocol of the manufacturer. The proteins of interest were detected by using antibodies against PPARγ (2435; Cell Signaling, Danvers, MA, USA) and β-actin (4970; Cell Signaling).
Analysis of PPARG-DM
The pathogenicity of rare variants of PPARγ2 was evaluated in accordance with the standards and guidelines recommended by the American College of Medical Genetics and Genomics (ACMG) (18) based on five lines of computational evidence [PROVEAN and SIFT (http://provean.jcvi.org), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml), MutationTaster (http://www.mutationtaster.org), and CADD (http://cadd.gs.washington.edu)].
Screening for Hotspot Variants in the Expanded Samples (Cohort 2)
To estimate the frequency of a newly identified hotspot variant in PPARγ2 in the general Chinese population, all patients (n = 545) with clinically diagnosed T2DM from a population-based cohort (Pinggu cohort) consisting of 3,345 individuals were screened. This cohort has been described in detail previously (17). To estimate the prevalence of diabetes induced by the hotspot variant in a hospital-based population, 1,200 more patients with diabetes diagnosed at ages older than 40 years (LOD) at the Peking University People’s Hospital from 2008 to 2017 were included. Additionally, 1,243 individuals older than 40 years with NGT and HbA1c <6% from the Pinggu cohort were screened for the hotspot variant as controls. Cohort 2 includes three parts of subjects: all patients with T2DM and all subjects older than 40 years with NGT from the Pinggu cohort and patients with LOD from a hospital-based population.
Review of the Characteristics of Patients With Rare PPARγ2 Variants
A literature search was performed using PubMed, ClinVar, and the Human Gene Mutation Database for relevant PPARγ variants and their phenotypes (as of November 2019). The search terms were “PPARG,” “PPARgamma,” “PPARγ,” “Peroxisome proliferator-activated receptor γ,” “mutation,” “variant,” “lipoatrophy,” “lipodystrophy,” and “diabetes.” All articles published in English reporting patients with rare PPARγ alleles (frequency < 0.005) were included. The clinical features of patients with rare PPARγ alleles were evaluated.
Statistical Analysis
All statistical tests were performed using SPSS version 22.0 for Mac (Chicago, IL, USA). Continuous variables are presented as the means and standard deviations (± SD) for normally distributed data or medians (25% and 75% quartiles) for the non-normally distributed data. Categorical variables are presented as numbers and percentages. When fewer than five data points were obtained, ranges are presented. Chi-square tests, Fisher’s exact tests, Student’s t-tests, and Mann–Whitney tests were used to compare the clinical characteristics between cases and controls.
The results of all functional experiments are expressed as the mean ± SD from three independent assays performed in duplicate. Differences were assessed by the Student’s t-test; differences with P-values <0.05 were considered statistically significant.
Results
Screening and Functional Analysis of Rare PPARγ2 Variants in Patients With EOD
In 1,002 patients with EOD, five rare variants [p.Val48Met(AF1), p.Glu217Lys(HD), p.Ser186Gly(DBD), p.Ile264Thr(HD), and p.Tyr95Cys(AF1)] were identified (Table 1). p.Glu217Lys was probably deleterious (19) and p.Val48Met was likely benign, according to a previous in vitro study (19), and the predictions were based on three or more lines of computational tools used in this study. The functions of the other three variants predicted as deleterious have not been reported previously.
In two pedigrees, the co-segregation of rare alleles and diabetes was evaluated. We observed that the homozygotic twin brother of the proband carrying p.Glu217Lys and a sister of the proband with p.Tyr95Cys all had diabetes and carried the same variants detected in the probands (ESM Table 2 and ESM Figure 4).
In the in vitro assay, reporter gene activity levels in 293T cells expressing p.Glu217Lys (P = 0.015), p.Ile264Thr (P = 0.001), and p.Ser186Gly (P = 0.012) PPARγ2 were significantly lower than those in cells expressing wild-type PPARγ2 (Figure 2A). However, in cells transfected with the p.Tyr95Cys vector, reporter gene activity did not differ significantly (P = 0.334) from that of cells transfected with the wild-type vector.
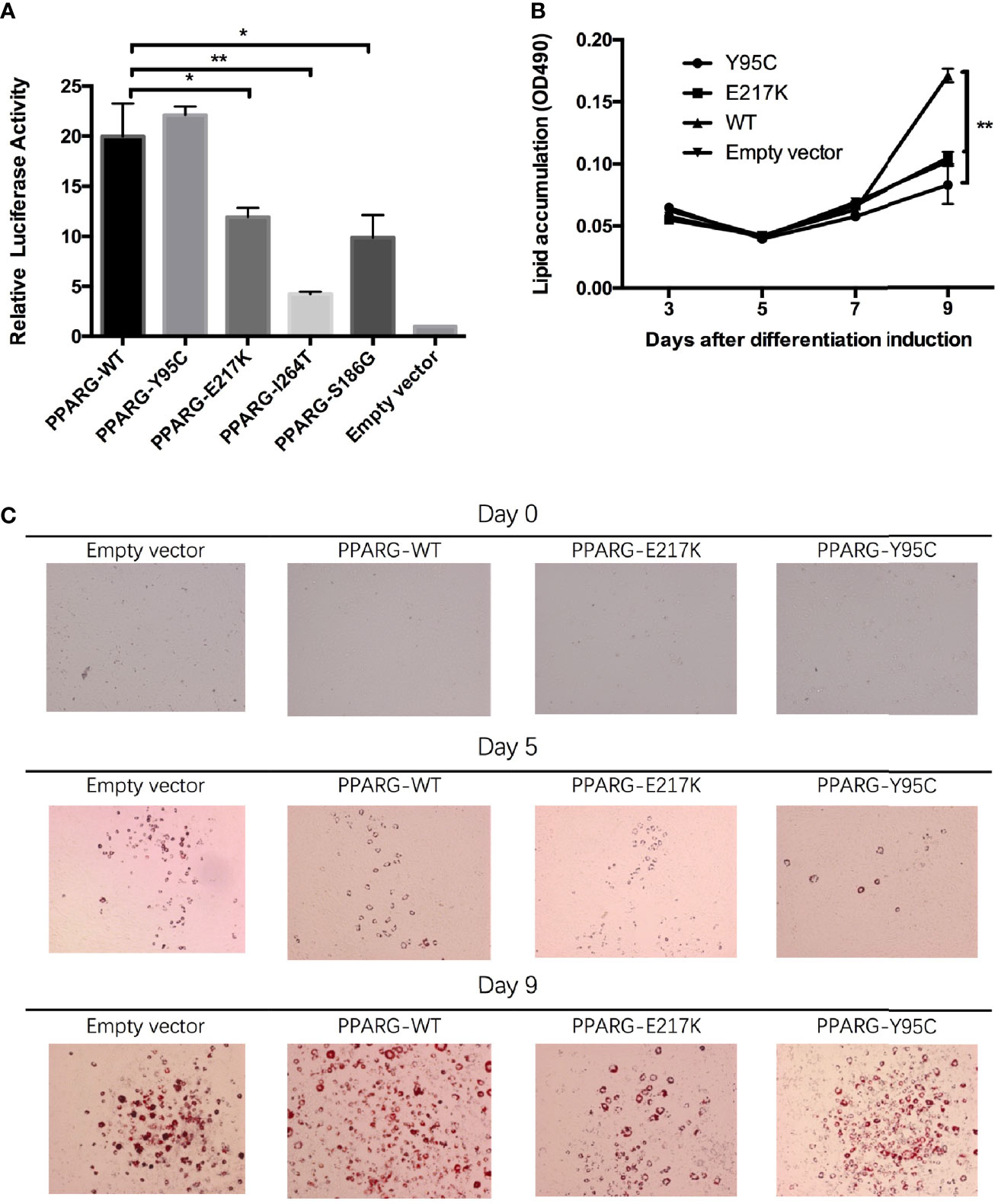
Figure 2 Functional analyses of constitutive mutant PPARγ2. Panel (A), results of reporter gene assays, 293T cells were transfected with the reporter gene and wild-type PPARγ2, Y95C, I264T, S186G, and E217K expression vectors or the empty vector as described in the Materials and Methods. Compared with wild-type PPARγ2 vector, both mutant vectors except Y95C-mutant vector show decreased relative transcription activity. Panels (B, C), NIH 3T3-L1 preadipocyte transfected with the wild-type PPARγ2 and two mutant PPARγ2 (E217K and Y95C) genes were differentiated as described in the Materials and Methods section. The cells stained with Oil Red-O at indicated time points (C) and Oil Red-O were extracted and measured by spectrometry to identify lipid accumulation (B). Images at ×100 magnification. As compared with the wild-type PPARγ2, Y95C and E217K mutants decelerated the accumulation of lipid during differentiation. *P < 0.05, **P < 0.01.
As AF1 variants may influence PPARγ2 expression via interactions with co-factors in vivo (5), we performed an adipocyte differentiation assay of the p.Tyr95Cys mutant. RT-PCR (ESM Figure 3A) and Western blotting (ESM Figure 3B) showed that the vectors containing p.Tyr95Cys, p.Glu217Lys, and wild-type PPARγ2 were successfully transfected into 3T3 cells. As determined by Oil Red-O staining, less lipid accumulation was detected in the 3T3-L1 cells with the p.Tyr95Cys variant and in the positive control samples (p.Glu217Lys) than in cells with wild-type PPARγ2 on day 9 of differentiation (Figures 2B, C, ESM Table 3).
Based on the ACMG classification system, experimental assays, or co-segregation analyses, we concluded that p.Ile264Thr, p.Tyr95Cys, and p.Ser186Gly are also likely pathogenic (ESM Table 4). Thus, six patients with PPARG-DM (0.6%) (three with p.Ile264Thr or p.Glu217Lys or p.Ser186Gly and three with Tyr95Cys; ESM Figure 5) were identified among 1,002 patients with EOD; 0.3% of these patients carried p.Tyr95Cys.
Further Screening for p.Tyr95Cys in Expanded Samples (Cohort 2) to Estimate the Allele Frequency
Of 545 patients in the Pinggu cohort, one individual with diabetes carried p.Tyr95Cys. Thus, the estimated prevalence of p.Tyr95Cys-induced diabetes was 0.18% (1/545) in the population of diabetes patients, with an allele frequency of 0.03% (1/3,345) in the whole population.
Three subjects with p.Tyr95Cys were identified in the LOD cohort (n = 1,200), and the p.Tyr95Cys variant was not found among 1,243 individuals with NGT. Thus, the prevalence rates of p.Tyr95Cys-induced diabetes were about 0.3% (3/1002) and 0.25% (3/1200) in patients with EOD and LOD in a hospital-based population, respectively.
Clinical Characteristics of Patients With PPARG-DM
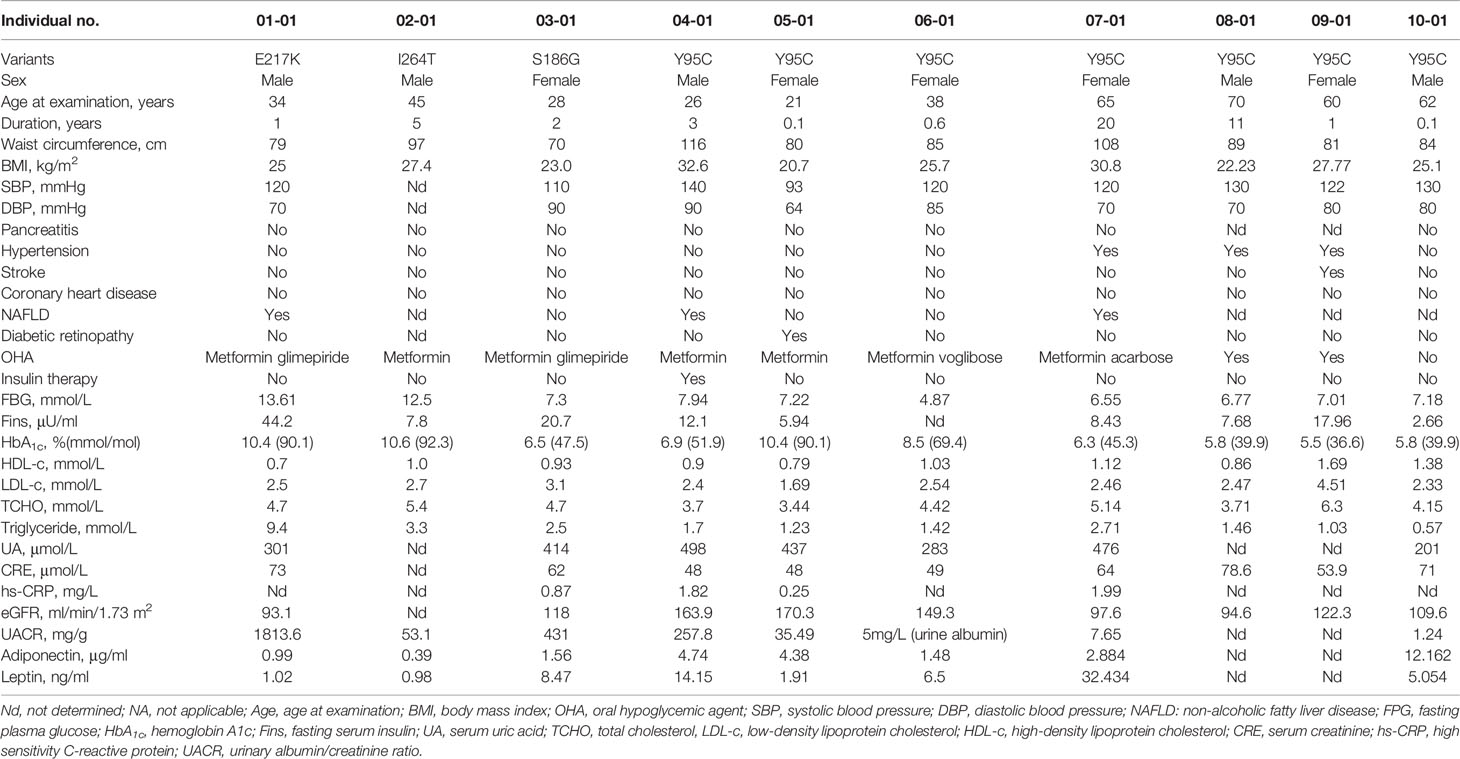
The clinical features of 10 probands with PPARG-DM identified in this study (namely, seven with AF1 variants and three with non-AF1 variants) are shown in Table 2. Since lipodystrophy is associated with low leptin and adiponectin levels, serum leptin and adiponectin levels were also measured. In total, 802 of 1,002 patients with EOD for whom leptin and adiponectin levels were available were included in the analysis of the clinical features (Table 3). Only one patient with a variant in AF1 had a low leptin level, as assessed based on our normal reference range. The clinical phenotypes of patients included hyperinsulinemia, hypertriglyceridemia, low HDL-c levels, obesity, central obesity, DKD, and hypertension without a clear clustering pattern. Compared with patients with non-PPARG-DM in EOD, patients with PPARG-DM had higher UACR levels and there was a higher proportion of individuals with DKD (Table 3). As determined by Fisher’s test, PPARG-DM was associated with DKD (odds ratio of 12.5, 95% confidence interval 1.45–107.68, P = 0.009). There were no significant differences in the leptin and adiponectin levels between these two groups.
We compared the clinical features of patients with non-AF1 and AF1 variants. All patients with non-AF1 variants had hypertriglyceridemia, DKD, and a BMI of less than 28 kg/m2. For patients with p.Tyr95Cys in AF1, the BMI values ranged from 20.7 to 32.6 kg/m2, and two of seven patients had hypertriglyceridemia. Compared with patients carrying non-AF1 variants (n = 4, including one relative with PPARG-DM), patients carrying AF1 variants (n = 8, including one relative with PPARG-DM) had higher adiponectin levels (median: 0.69 vs. 4.38 µg/ml, P = 0.03, data not shown).
To determine the effect of thiazolidinediones on patients with PPARG-DM, we followed the proband with p.Glu217Lys for 1 year and observed the response to pioglitazone. Before the patient was diagnosed with PPARG-DM, he had been taking glimepiride (4 mg/day) and metformin (1,500 mg/day) and had occasional hypoglycemia. After glimepiride was replaced with pioglitazone (30 mg/day) for 3 months, his glucose control did not change but his triglyceride, insulin, and inflammatory marker levels decreased (ESM Table 5), with no hypoglycemia or reduced eGFR.
Summary of Clinical Features of Patients With Rare PPARγ2 Alleles Reported in Previous Studies
A total of 68 probands with 50 rare PPARγ2 variants have been reported in the literature, namely, 6 with nonsense variants, 4 with frameshift variants, 1 with a compound heterozygous variant, and 57 with heterozygous variants (ESM Table 6). In total, 46 rare variants (3 in AF1, 16 in DBD, 5 in HD, and 22 in LBD) from 64 probands were classified as pathogenic or likely pathogenic according to the ACMG (ESM Table 7 and Figure 3).
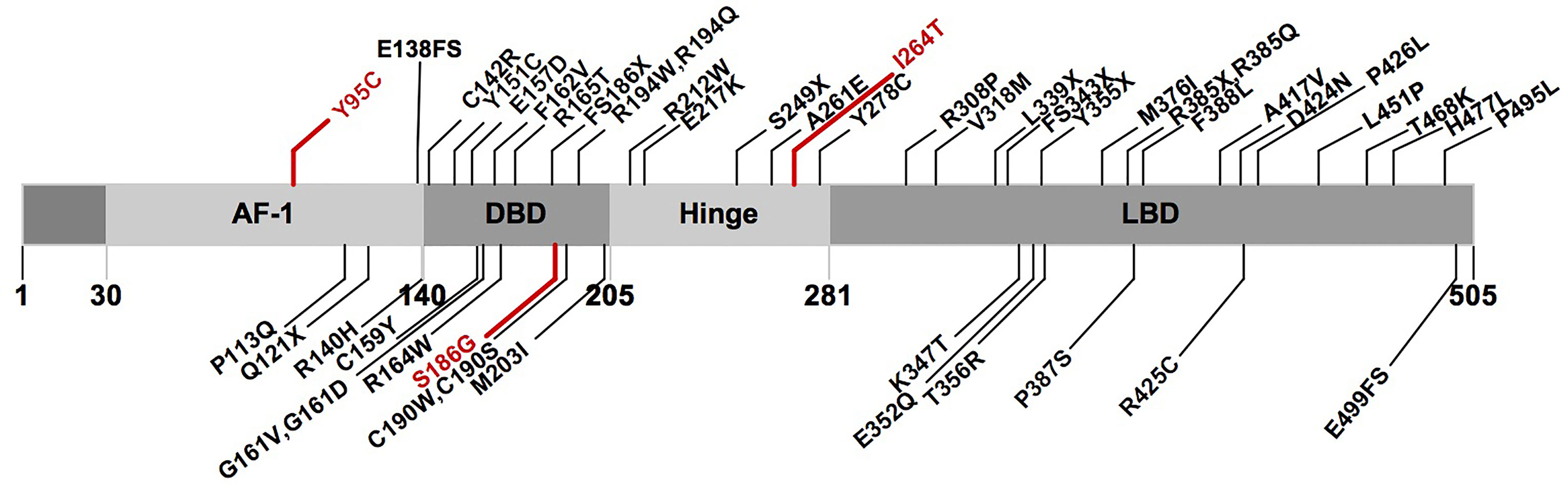
Figure 3 Schematic representation of the PPARγ2 domain structure and position of the rare variants identified in previous studies and this study. Marked in red: novel variants described in this study. AF1, activation function 1 domain; DBD, DNA-binding domain; LBD, ligand-binding domain.
Per the available clinical data for probands with likely pathogenic PPARγ2 variants, 89% (57/64) were diagnosed with diabetes or impaired glucose tolerance, 86% (32/37) were diagnosed before 40 years of age, and 85% patients (11/13) showed an improvement in glucose control after thiazolidinedione therapy. Among six probands for whom information regarding diabetic complications was available, five (83%) had DKD. Additionally, 93% (52/56) of the probands had hypertriglyceridemia, 36% (4/11) had recurrent pancreatitis, and 96% (27/28) had hepatic steatosis. Among 14 patients for whom relevant data were available, four (29%) had low leptin levels (ESM Table 6).
Compared with the probands (n = 57) with rare pathogenic variants in the non-AF1 domains of PPARγ2, the probands (n = 7) with rare pathogenic variants in AF1 had a lower prevalence of lipodystrophy (1/7), higher frequency of obesity (5/7), and relatively normal serum triglyceride levels; the triglyceride levels were only evaluated in two patients, one of whom showed an average value (over 30 years) of 0.14 mmol/L and the other showed a value of 1.7 mmol/L (Table 4).
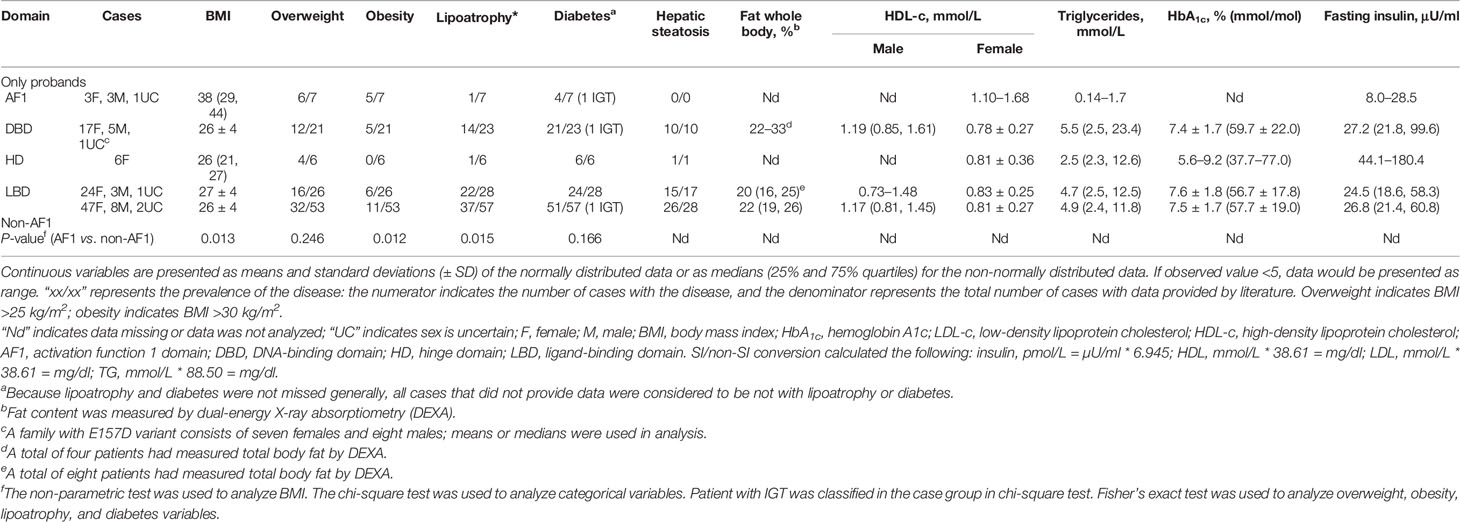
Table 4 The summary of clinical features of subjects with rare PPARγ variants from previous studies.
Discussion
In this study, we obtained the following novel findings: 1) we obtained the first estimate of the prevalence of PPARG-DM in a Chinese population with EOD, i.e., 0.6%. 2) A hotspot diabetes-related variant (p.Tyr95Cys) was identified in AF1; this variant suppressed the differentiation of 3T3-L1 preadipocytes in vitro. 3) By a review of all reported cases with rare damaging PPARγ2 variants, we found distinct phenotype differences between patients with AF1 and non-AF1 variants in PPARγ2, as confirmed in the patients evaluated in this study. 4) Most patients with PPARG-DM had DKD, supporting previous findings that PPARγ is involved in the development of DKD, based on animal studies and meta-analyses of studies on patients with diabetes.
Therefore, patients with EOD and metabolic syndrome should be screened for PPARγ variants, even if they lack typical lipodystrophy features. These patients with pathogenic PPARγ variants have a higher risk of DKD, and early genetic diagnosis is helpful to improve the clinical outcomes of these patients.
Almost all previously reported cases with rare variants were from Caucasian or Latino populations (ESM Table 6). Only one study has reported the prevalence of rare variants of PPARγ2 in East Asians (19) by the exon sequencing of nearly 20,000 individuals (9,070 patients with T2DM and 10,682 controls) from multiethnic populations; in this previous study, 0.1% of individuals with T2DM carried functional rare variants of PPARγ2. Of note, only 998 patients with T2DM and 1,146 controls from East Asian populations (Korea and Singapore) were included in this study, and it is not clear whether the rare pathogenic variants of PPARγ2 existed in East Asian subpopulations. The prevalence of T2DM in China is as high as 11.6% (20); accordingly, a larger sample size is needed to estimate the prevalence of PPARG-DM in East Asians to enable precision medicine. Because we found that most reported patients with PPARG-DM were diagnosed before the age of 40, we estimated the prevalence of PPARG-DM in this population to ensure that our results are clinically meaningful. We sequenced PPARγ2 in 1,002 Chinese patients with EOD and identified 6 patients (0.6%) with PPARG-DM, confirming the observations that PPARG-DM is more common in patients with EOD than in those with LOD. Because patients with EOD account for approximately 16.2% of all Chinese patients with T2DM (20), the estimated prevalence of PPARG-DM in patients with T2DM in China was approximately 0.1%. Thus, we concluded that the prevalence of PPARG-DM is similar in Caucasian and Chinese populations, and the smaller number of reported cases of PPARG-DM in China may be explained by the relatively low awareness of PPARG-DM among physicians in China and a lack of cost-effective genetic tests. In this study, maturity-onset diabetes of the young (MODY) was not ruled out; however, monogenic diabetes accounts for about 1%–5% of all cases of diabetes in young people. Moreover, PPARG-DM is characterized by insulin resistance, which is not consistent with the phenotype of MODY. Accordingly, we expect MODY to have a negligible impact on the results of the present study.
We identified the p.Tyr95Cys variant in AF1 in PPARγ2 as a novel rare variant. This was the most frequent variant in patients with PPARG-DM (3/6) in the EOD cohort; in a hospital-based population, about 0.3% of patients with EOD and 0.25% patients with LOD carried this variant. The frequency of p.Tyr95Cys in the Chinese population is 0.009% (1/10,588) (http://www.mbiobank.com/), which is significantly lower than that in patients with EOD (P = 0.002, data not shown). We subsequently screened for p.Tyr95Cys in a randomly sampled cohort (Pinggu cohort) and found a p.Tyr95Cys frequency of 0.18% in patients with diabetes. As reported, several population-specific rare or low-frequency variants (p.Gly319Ser in HNF1A in the Oji-Cree population, p.Glu508Lys in HNF1A in a Latino population, and p.Arg1420His in ABCC8 in a Southwest American Indian population) (21–23) are associated with an increased risk for diabetes with a large effect size. No individual was found to carry the p.Tyr95Cys variant in online databases, such as ExAC and 1000 Genomes. Thus, p.Tyr95Cys may be a Chinese-specific causative variant for diabetes.
The clinical characteristics and pathogenesis of variants in the PPARγ AF1 domain are still unclear. As summarized in Table 4, patients with rare variants in AF1 had milder insulin resistance, greater fat mass, and lower triglyceride levels than patients with rare variants in non-AF1 domains. These clinical features were similar to those of patients with p.Tyr95Cys (Table 2) in this study. The AF1 domain of PPARγ2 is involved in the transactivation and recognition of the protein by transcriptional coactivators and affects the ligand-binding affinity of the LBD (24). Previous studies have demonstrated that posttranslational modifications in AF1, including phosphorylation (25, 26), acetylation (27), O-GlcNAcylation (28), and SUMOylation (1, 29), regulate the functions of PPARγ in vitro. A variant in an important phosphorylation site, Ser112, of PPARγ2 (p.Ser112Ala) has been found to protect mice from obesity-induced insulin resistance (30). Subjects with the rare Pro113Gln variant near Ser112 show increased adipocyte differentiation via Ser112 phosphorylation. p.Pro113Gln can cause a range of metabolic symptoms, including obesity, T2DM, and high fasting insulin levels (7, 31). These phenotypic differences among patients might be partially explained by the interaction of PPARγ with co-factors or environmental factors. Further research is necessary to unravel the complex relationship between genotypes, posttranslational modification status, environmental factors, and disease risk. Differences between the effects of variants in AF1 and other domains may provide alternative therapeutic strategies to thiazolidinedione.
There is a relationship between the SUMOylation and phosphorylation of PPARγ2. The Ser112Ala variant reduces the SUMOylation of PPARγ2, while the phospho-mimicking Ser112Asp variant increases the SUMOylation of Lys107 (32). The FGF21–PPARγ pathway is a self-reinforcing regulatory loop (33). The SUMOylation of Lys107 is regulated by FGF21 in vivo (33). PPARγ2 is expressed in both adipocytes and hepatocytes. The development of fatty livers in many mouse models of obesity and diabetes is associated with the increased expression of PPARγ2 (4). FGF21 is more abundant in the liver than in adipose tissues, explaining why some patients with p.Tyr95Cys have hyperglycemia but mild hypertriglyceridemia and lipoatrophy. In addition, as reported, Lys94 is potentially modified by SUMOylation (1). Accordingly, the p.Tyr95Cys variant, in addition to affecting phosphorylation, may suppress the differentiation of adipocytes by influencing SUMOylation. Further studies are needed to verify whether p.Tyr95Cys alters phosphorylation; in particular, future studies should include a negative control transfected with a Ser112Ala mutant. Additionally, immunoprecipitation assays are needed to determine whether p.Tyr95Cys affects the SUMOylation of PPARγ.
Patients with lipodystrophy usually show low leptin levels; in our review, only 14 reported patients harboring PPARγ2 variants were evaluated for leptin levels, 4 of which had low leptin levels. In our study, leptin levels were measured in 10 patients with PPARG-DM (3 with variants in non-AF1 domains and 7 with the p.Tyr95Cys variant); no differences were observed between PPARG-DM and T2DM, and only 1 patient with p.Tyr95Cys had low leptin levels (Table 2). These observations indicate that the serum leptin cutoff value derived from the normal reference range was not helpful for distinguishing PPARG-DM from T2DM. In fact, the pathogenesis of PPARG-DM may be highly complex because more organs (e.g., the liver, adipocyte, brain, and islet) are involved in its development (34, 35). Thus, leptin levels have limited value for PPARG-DM screening in adults.
About two decades ago, p.Pro12Ala in PPARγ2 was found to be associated with T2DM in a candidate gene study, and this was confirmed in a subsequent genome-wide association study (GWAS) (36). However, this association was not replicated in a later GWAS (37) and meta-analysis (38) of the Chinese population. Similarly, this variant was recently reported to be associated with DKD in Caucasians; however, a similar association was not detected in Asians (8); this may be explained by the low frequency of Ala12 and the low effect size of this variant in Asians. Recently, an exome sequencing study has shown that the risk of diabetes was 7.22-fold higher in carriers of rare variants with reduced PPARγ function (19). However, renal phenotypes have rarely been described in subjects with PPARG-DM. Our results showed that the risk of DKD was 12.5-fold higher in patients with PPARG-DM in the EOD cohort. However, the mechanism underlying the effects of these mutations in PPARγ on DKD is not clear. Bonofiglio et al. (39) have found that PPARγ may be involved in the regulation of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) expression by binding to the peroxisomal proliferator response element (PPRE) upstream of the PTEN promoter. PTEN plays a significant role in DKD. Previous studies have found that PTEN is involved in the regulation of the epithelial-to-mesenchymal transition (EMT) in renal tubular cells, tubulointerstitial fibrosis (40), and cytoskeletal rearrangement in podocytes (41). Accordingly, PPARγ may participate in the development of DKD by regulating PTEN expression. Further studies of these variants and the expression of PTEN are needed.
Among the reported PPARG-DM patients taking thiazolidinediones, most showed improved glycemic control (ESM Table 6). The lack of response to thiazolidinediones may be due to the nearly complete loss-of-function mutations in PPARγ (19). In the present study, we also found that a patient with a partial inactivation of PPARγ had a better response to thiazolidinedione, with improvements in glucose levels, lipid metabolism, and inflammatory indicators (ESM Table 5). Thiazolidinediones could inhibit the proliferation of cultured mesangial cells (42) and downregulate the expression of vascular endothelial growth factor (VEGF) by inhibiting the expression of hypoxia-inducible factor-1α (HIF-1α) in the kidneys of diabetic rats, thereby protecting the kidneys (43). Moreover, thiazolidinediones are associated with the attenuation of DKD in patients with T2DM (44, 45); the activation of PPARγ slows the progression of DKD, and several mechanisms, both systemic and renal, have been implicated in the renoprotective effects of PPARγ (46). Thus, thiazolidinediones may serve as a good therapeutic choice for PPARG-DM. It is possible that patients carrying the rare variants in AF1 have an incomplete loss of function of PPARγ and, therefore, show a response to thiazolidinediones.
This study had a few limitations. First, fat mass was not measured. Second, cell-based studies of the pathogenicity of the variants are limited; thus, more detailed functional studies are needed to validate this aspect. Third, the association of PPARγ with DKD is still not clear and should be evaluated further. Fourth, the prevalence of PPARG-DM in a population-based sample should be estimated by screening all exons of PPARγ2. Some patients with PPARG-DM were only screened for the PPARγ gene, whereas the possibility of other diabetes-inducing genetic variants cannot be excluded. Fifth, the sample size of patients with PPARG-DM was small and potential confounders could not be adjusted in comparative analyses; hence, further studies with a larger sample size are needed.
Conclusions
The prevalence of PPARG-DM in the Chinese population is similar to that reported in other populations; PPARG-DM accounts for 0.6% of Chinese patients with EOD. The p.Tyr95Cys variant in AF1 is a novel hotspot variant in the Chinese population. Patients with PPARG-DM caused by AF1 variants often lack typical lipodystrophy features. Additionally, patients with PPARG-DM have an increased risk for DKD. The individualized application of thiazolidinedione may greatly benefit patients with PPARG-DM, especially those with DKD.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/ and rs141797536, rs1477623791, rs1421126930, and rs766913119.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Peking University People’s Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
SG conducted the research, performed the literature review, collected the data, analyzed the data, and wrote the manuscript. ML, XC, WL, YYL, S-mZ, LZ, YM, XTH, YFL, XZ, YZ, QW, LC, and QR helped to collect the data and reviewed the manuscript. PZ helped to perform the experiments. LJ and XYH designed the study, contributed to the discussion, and reviewed and edited the manuscript. SG, XYH and LJ are the guarantors of this work and, as such, have full access to all the data reported in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from the National Key Research and Development Program of China (2016YFC1304901), Beijing Municipal Science and Technology Commission Funding (Z141100007414002 and D131100005313008), National High-Technology Research and Development Program of China (2012AA02A509), and National Natural Science Foundation of China (82000759).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Xinjuan Wang, Jianhua Zhou, Na Li, Na Wu, Mei Li, and Huiting Su for their assistance in the experiments. We also wish to thank the nursing staff of the Department of Endocrinology and Metabolism for obtaining the urine and blood samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.677130/full#supplementary-material
Abbreviations
AF1, activation function 1 domain; ACMG, American College of Medical Genetics and Genomics; DKD, diabetic kidney disease; DBD, DNA-binding domain; EOD, early-onset type 2 diabetes; FPG, fasting plasma glucose; Fins, fasting insulin; GWAS, genome-wide association study; HD, hinge domain; LOD, late-onset type 2 diabetes; LBD, ligand-binding domain; NGT, normal glucose tolerance; PPARG-DM, PPARγ variant-induced diabetes; T2DM, type 2 diabetes.
References
1. Diezko R, Suske G. Ligand Binding Reduces SUMOylation of the Peroxisome Proliferator-Activated Receptor γ (Pparγ) Activation Function 1 (AF1) Domain. PloS One (2013) 8:e66947. doi: 10.1371/journal.pone.0066947
2. Agostini M, Schoenmakers E, Beig J, Fairall L, Szatmari I, Rajanayagam O, et al. Savage DB. A Pharmacogenetic Approach to the Treatment of Patients With PPARG Mutations. Diabetes (2018) 67:1086–92. doi: 10.2337/db17-1236
3. Werman A, Hollenberg A, Solanes G, Bjorbaek C, Vidal-Puig AJ, Flier JS. Ligand-Independent Activation Domain in the N Terminus of Peroxisome Proliferator-Activated Receptor Gamma (PPARgamma). Differential Activity of PPARgamma1 and -2 Isoforms and Influence of Insulin. J Biol Chem (1997) 272:20230–5. doi: 10.1074/jbc.272.32.20230
4. Lee YK, Park JE, Lee M, Hardwick JP. Hepatic Lipid Homeostasis by Peroxisome Proliferator-Activated Receptor Gamma 2. Liver Res (2018) 2:209–15. doi: 10.1016/j.livres.2018.12.001
5. van Beekum O, Brenkman AB, Grøntved L, Hamers N, van den Broek NJF, Berger R, et al. The Adipogenic Acetyltransferase Tip60 Targets Activation Function 1 of Peroxisome Proliferator-Activated Receptor γ. Endocrinology (2008) 149:1840–9. doi: 10.1210/en.2007-0977
6. Knouff C, Auwerx J. Peroxisome Proliferator-Activated Receptor-γ Calls for Activation in Moderation: Lessons From Genetics and Pharmacology. Endocrine Rev (2004) 25:899–918. doi: 10.1210/er.2003-0036
7. Ristow M, Müller-Wieland D, Pfeiffer A, Krone W, Kahn CR. Obesity Associated With a Mutation in a Genetic Regulator of Adipocyte Differentiation. N Engl J Med (1998) 339:953–9. doi: 10.1056/NEJM199810013391403
8. Zhang H, Zhu S, Chen J, Tang Y, Hu H, Mohan V, et al. Peroxisome Proliferator-Activated Receptor γ Polymorphism Pro12Ala Is Associated With Nephropathy in Type 2 Diabetes: Evidence From Meta-Analysis of 18 Studies. Diabetes Care (2012) 35:1388–93. doi: 10.2337/dc11-2142
9. Liu L, Zheng T, Wang F, Wang N, Song Y, Li M, et al. Pro12Ala Polymorphism in the PPARG Gene Contributes to the Development of Diabetic Nephropathy in Chinese Type 2 Diabetic Patients. Diabetes Care (2010) 33:144–9. doi: 10.2337/dc09-1258
10. Toffoli B, Gilardi F, Winkler C, Soderberg M, Kowalczuk L, Arsenijevic Y, et al. Nephropathy in Pparg-Null Mice Highlights Pparγ Systemic Activities in Metabolism and in the Immune System. PloS One (2017) 12:e0171474. doi: 10.1371/journal.pone.0171474
11. Liu G, Zhou T-B, Jiang Z, Zheng D, Yuan F, Li Y, et al. Relationship Between Pparγ Pro12Ala Gene Polymorphism and Type 2 Diabetic Nephropathy Risk in Asian Population: Results From a Meta-Analysis. J Recept Signal Transduct Res (2014) 34:131–6. doi: 10.3109/10799893.2013.864678
12. Wang L, Teng Z, Cai S, Wang D, Zhao X, Yu K. The Association Between the Pparγ2 Pro12Ala Polymorphism and Nephropathy Susceptibility in Type 2 Diabetes: A Meta-Analysis Based on 9,176 Subjects. Diagn Pathol (2013) 8:118. doi: 10.1186/1746-1596-8-118
13. Alberti KG, Zimmet PZ. Definition, Diagnosis and Classification of Diabetes Mellitus and Its Complications. Part 1: Diagnosis and Classification of Diabetes Mellitus Provisional Report of a WHO Consultation. Diabetes Med (1998) 15:539–53. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S
14. Gong S, Guo J, Han X, Li M, Zhou L, Cai X, et al. Clinical and Genetic Features of Patients With Type 2 Diabetes and Renal Glycosuria. J Clin Endocrinol Metab (2017) 102:1548–56. doi: 10.1210/jc.2016-2332
15. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis Model Assessment: Insulin Resistance and Beta-Cell Function From Fasting Plasma Glucose and Insulin Concentrations in Man. Diabetologia (1985) 28:412–9. doi: 10.1007/BF00280883
16. Ma Y-C, Zuo L, Chen J-H, Luo Q, Yu X-Q, Li Y, et al. Modified Glomerular Filtration Rate Estimating Equation for Chinese Patients With Chronic Kidney Disease. J Am Soc Nephrol (2006) 17:2937–44. doi: 10.1681/ASN.2006040368
17. Liu W, Han X, Zhou X, Zhang S, Cai X, Zhang L, et al. Brain Derived Neurotrophic Factor in Newly Diagnosed Diabetes and Prediabetes. Mol Cell Endocrinol (2016) 429:106–13. doi: 10.1016/j.mce.2016.04.002
18. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. ACMG Laboratory Quality Assurance Committee. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med (2015) 17:405–24. doi: 10.1038/gim.2015.30
19. Majithia AR, Flannick J, Shahinian P, Guo M, Bray M-A, Fontanillas P, et al. GoT2D Consortium, NHGRI JHS/FHS Allelic Spectrum Project, SIGMA T2D Consortium, T2D-GENES Consortium, Rosen ED, Altshuler D, Flannick J, Manning AK, Hartl C, Agarwala V, Fontanillas P, Green T, Banks E, DePristo M, Poplin R, Shakir K, Fennell T, Njolstad PR, Altshuler D, Burtt N, Gabriel S, Fuchsberger C, Kang HM, Sim X, Ma C, Locke A, Blackwell T, Jackson A, Teslovich TM, Stringham H, Chines P, Kwan P, Huyghe J, Tan A, Jun G, Stitzel M, Bergman RN, Bonnycastle L, Tuomilehto J, Collins FS, Scott L, Mohlke K, Abecasis G, Boehnke M, Strom T, Gieger C, Nurasyid MM, Grallert H, Kriebel J, Ried J, Hrabe De Angelis M, Huth C, Meisinger C, Peters A, Rathmann W, Strauch K, Meitinger T, Kravic J, Algren P, Ladenvall C, Toumi T, Isomaa B, Groop L, Gaulton K, Moutsianas L, Rivas M, Pearson R, Mahajan A, Prokopenko I, Kumar A, Perry J, Howie B, Van De Bunt M, Small K, Lindgren C, Lunter G, Robertson N, Rayner W, Morris A, Buck D, Hattersley A, Spector T, McVean G, Frayling T, Donnelly P, McCarthy M, Gupta N, Taylor H, Fox E, Cheh CN, Wilson JG, O’donnell CJ, Kathiresan S, Hirschhorn J, Seidman JG, Gabriel S, Seidman C, Altshuler D, Williams AL, Jacobs SBR, Macias HM, Chagoya AH, Churchhouse C, Luna CM, Ortiz HG, Vazquez MJG, Burtt NP, Estrada K, Mercader JM, Ripke S, Manning AK, Neale B, Stram DO, Lopez JCF, Hidalgo SR, Delfin IA, Hernandez AM, Cruz FC, Caamal EM, Monsalve CR, Andrade SI, Cordova E, Arellano ER, Soberon X, Villalpando MEG, Monroe K, Wilkens L, Kolonel LN, Le Marchand L, Riba L, Sanchez MLO, Guillen RR, Bautista IC, Torres MR, Hernandez LLM, Saenz T, Gomez D, Alvirde U, Onofrio RC, Brodeur WM, Gage D, Murphy J, Franklin J, Mahan S, Ardlie K, Crenshaw AT, Winckler W, Fennell T, MacArthur DG, Altshuler D, Florez JC, Haiman CA, Henderson BE, Salinas CAA, Villalpando CG, Orozco L, Luna TT, Abecasis G, Almeida M, Altshuler D, Asimit JL, Atzmon G, Barber M, Beer NL, Bell GI, Below J, Blackwell T, Blangero J, Boehnke M, Bowden DW, Burtt N, Chambers J, Chen H, Chen P, Chines PS, Choi S, Churchhouse C, Cingolani P, Cornes BK, Cox N, Williams AGD, Duggirala R, Dupuis J, Dyer T, Feng S, Tajes JF, Ferreira T, Fingerlin TE, Flannick J, Florez J, Fontanillas P, Frayling TM, Fuchsberger C, Gamazon ER, Gaulton K, Ghosh S, Gloyn A, Grossman RL, Grundstad J, Hanis C, Heath A, Highland H, Hirokoshi M, Huh I-S, Huyghe JR, Ikram K, Jablonski KA, Kim YJ, Jun G, Kato N, Kim J, King CR, Kooner J, Kwon M-S, Im HK, Laakso M, Lam KK-Y, Lee J, Lee S, Lee S, Lehman DM, Li H, Lindgren CM, Liu X, Livne OE, Locke AE, Mahajan A, Maller JB, Manning AK, Maxwell TJ, Mazoure A, McCarthy MI, Meigs JB, Min B, Mohlke KL, Morris A, Musani S, Nagai Y, Ng MCY, Nicolae D, Oh S, Palmer N, Park T, Pollin TI, Prokopenko I, Reich D, Rivas MA, Scott LJ, Seielstad M, Cho YS, Tai E-S, Sim X, Sladek R, Smith P, Tachmazidou I, Teslovich TM, Torres J, Trubetskoy V, Willems SM, Williams AL, Wilson JG, Wiltshire S, Won S, Wood AR, Xu W, Teo YY, Yoon J, Lee J-Y, Zawistowski M, Zeggini E, Zhang W, Zollner S. Rare Variants in PPARG With Decreased Activity in Adipocyte Differentiation are Associated With Increased Risk of Type 2 Diabetes. Proc Natl Acad Sci (2014) 111:13127–32. doi: 10.1073/pnas.1410428111
20. Xu Y, Wang L, He J, Bi Y, Li M, Wang T, et al. 2010 China Noncommunicable Disease Surveillance Group. Prevalence and Control of Diabetes in Chinese Adults. JAMA (2013) 310:948–59. doi: 10.1001/jama.2013.168118
21. Hegele RA, Cao H, Harris SB, Hanley AJ, Zinman B. The Hepatic Nuclear Factor-1alpha G319S Variant is Associated With Early-Onset Type 2 Diabetes in Canadian Oji-Cree. J Clin Endocrinol Metab (1999) 84:1077–82. doi: 10.1210/jcem.84.3.5528
22. SIGMA Type 2 Diabetes Consortium, Estrada K, Aukrust I, Bjørkhaug L, Burtt NP, Mercader JM, et al. Association of a Low-Frequency Variant in HNF1A With Type 2 Diabetes in a Latino Population. JAMA (2014) 311:2305–14. doi: 10.1001/jama.2014.6511
23. Baier LJ, Muller YL, Remedi MS, Traurig M, Piaggi P, Wiessner G, et al. ABCC8 R1420H Loss-Of-Function Variant in a Southwest American Indian Community: Association With Increased Birth Weight and Doubled Risk of Type 2 Diabetes. Diabetes (2015) 64:4322–32. doi: 10.2337/db15-0459
24. Keshet R, Bryansker Kraitshtein Z, Shanzer M, Adler J, Reuven N, Shaul Y. C-Abl Tyrosine Kinase Promotes Adipocyte Differentiation by Targeting PPAR-Gamma 2. Proc Natl Acad Sci (2014) 111:16365–70. doi: 10.1073/pnas.1411086111
25. Hu E, Kim JB, Sarraf P, Spiegelman BM. Inhibition of Adipogenesis Through MAP Kinase-Mediated Phosphorylation of PPAR. Science (1996) 274:2100–3. doi: 10.1126/science.274.5295.2100
26. Camp HS, Tafuri SR. Regulation of Peroxisome Proliferator-Activated Receptor γ Activity by Mitogen-Activated Protein Kinase. J Biol Chem (1997) 272:10811–6. doi: 10.1074/jbc.272.16.10811
27. Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, et al. Brown Remodeling of White Adipose Tissue by SirT1-Dependent Deacetylation of Pparγ. Cell (2012) 150:620–32. doi: 10.1016/j.cell.2012.06.027
28. Ji S, Park SY, Roth J, Kim HS, Cho JW. O-GlcNAc Modification of Pparγ Reduces Its Transcriptional Activity. Biochem Biophys Res Commun (2012) 417:1158–63. doi: 10.1016/j.bbrc.2011.12.086
29. Katafuchi T, Holland WL, Kollipara RK, Kittler R, Mangelsdorf DJ, Kliewer SA. Pparγ-K107 SUMOylation Regulates Insulin Sensitivity But Not Adiposity in Mice. Proc Natl Acad Sci USA (2018) 115:12102–11. doi: 10.1073/pnas.1814522115
30. Rangwala SM, Rhoades B, Shapiro JS, Rich AS, Kim JK, Shulman GI, et al. Genetic Modulation of Pparγ Phosphorylation Regulates Insulin Sensitivity. Dev Cell (2003) 5:657–63. doi: 10.1016/S1534-5807(03)00274-0
31. Blüher M, Lübben G, Paschke R. Analysis of the Relationship Between the Pro12Ala Variant in the PPAR-Gamma2 Gene and the Response Rate to Therapy With Pioglitazone in Patients With Type 2 Diabetes. Diabetes Care (2003) 26:825–31. doi: 10.2337/diacare.26.3.825
32. Yamashita D, Yamaguchi T, Shimizu M, Nakata N, Hirose F, Osumi T. The Transactivating Function of Peroxisome Proliferator-Activated Receptor γ Is Negatively Regulated by SUMO Conjugation in the Amino-Terminal Domain. Genes Cells (2004) 9:1017–29. doi: 10.1111/j.1365-2443.2004.00786.x
33. Dutchak PA, Katafuchi T, Bookout AL, Choi JH, Yu RT, Mangelsdorf DJ, et al. Fibroblast Growth Factor-21 Regulates Pparγ Activity and the Antidiabetic Actions of Thiazolidinediones. Cell (2012) 148:556–67. doi: 10.1016/j.cell.2011.11.062
34. Lu M, Sarruf DA, Talukdar S, Sharma S, Li P, Bandyopadhyay G, et al. Brain PPAR-γ Promotes Obesity and Is Required for the Insulin–Sensitizing Effect of Thiazolidinediones. Nat Med (2011) 17:618–22. doi: 10.1038/nm.2332
35. Gupta D, Kono T, Evans-Molina C. The Role of Peroxisome Proliferator-Activated Receptor γ in Pancreatic β Cell Function and Survival: Therapeutic Implications for the Treatment of Type 2 Diabetes Mellitus. Diabetes Obes Metab (2010) 12:1036–47. doi: 10.1111/j.1463-1326.2010.01299.x
36. Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. Boehnke M. A Genome-Wide Association Study of Type 2 Diabetes in Finns Detects Multiple Susceptibility Variants. Science (2007) 316:1341–5. doi: 10.1126/science.1142382
37. Tsai F-J, Yang C-F, Chen C-C, Chuang L-M, Lu C-H, Chang C-T, et al. Wu J-Y. A Genome-Wide Association Study Identifies Susceptibility Variants for Type 2 Diabetes in Han Chinese. PloS Genet (2010) 6:e1000847. doi: 10.1371/journal.pgen.1000847
38. Guo W-L, Tang Y, Han X-Y, Ji L-N. Meta-Analysis of the Association of Pro12Ala Polymorphism of Peroxisome Proliferator Activated Receptor Gamma Gene With Type 2 Diabetes in Chinese Han Population. Zhongguo Yi Xue Ke Xue Yuan Xue Bao (2011) 33:593–9.
39. Bonofiglio D, Gabriele S, Aquila S, Catalano S, Gentile M, Middea E, et al. Estrogen Receptor Alpha Binds to Peroxisome Proliferator-Activated Receptor Response Element and Negatively Interferes With Peroxisome Proliferator-Activated Receptor Gamma Signaling in Breast Cancer Cells. Clin Cancer Res (2005) 11:6139–47. doi: 10.1158/1078-0432.CCR-04-2453
40. Yan R, Wang Y, Shi M, Xiao Y, Liu L, Liu L, et al. Regulation of PTEN/AKT/FAK Pathways by Pparγ Impacts on Fibrosis in Diabetic Nephropathy. J Cell Biochem (2019) 120:6998–7014. doi: 10.1002/jcb.27937
41. Lin J, Shi Y, Peng H, Shen X, Thomas S, Wang Y, et al. Loss of PTEN Promotes Podocyte Cytoskeletal Rearrangement, Aggravating Diabetic Nephropathy. J Pathol (2015) 236:30–40. doi: 10.1002/path.4508
42. Guan Y, Zhang Y, Schneider A, Davis L, Breyer RM, Breyer MD. Peroxisome Proliferator-Activated Receptor-Gamma Activity Is Associated With Renal Microvasculature. Am J Physiol Renal Physiol (2001) 281:F1036–1046. doi: 10.1152/ajprenal.0025.2001
43. Xu X, Chen P, Zheng Q, Wang Y, Chen W. Effect of Pioglitazone on Diabetic Nephropathy and Expression of HIF-1α and VEGF in the Renal Tissues of Type 2 Diabetic Rats. Diabetes Res Clin Pract (2011) 93:63–9. doi: 10.1016/j.diabres.2011.03.019
44. Sarafidis PA, Stafylas PC, Georgianos PI, Saratzis AN, Lasaridis AN. Effect of Thiazolidinediones on Albuminuria and Proteinuria in Diabetes: A Meta-Analysis. Am J Kidney Dis (2010) 55:835–47
45. Zhou Y, Huang Y, Ji X, Wang X, Shen L, Wang Y. Pioglitazone for the Primary and Secondary Prevention of Cardiovascular and Renal Outcomes in Patients With or at High Risk of Type 2 Diabetes Mellitus: A Meta-Analysis. J Clin Endocrinol Metab (2020) 105:1670–81. doi: 10.1210/clinem/dgz252
Keywords: type 2 diabetes, activation function 1 domain, lipodystrophy, Tyr95Cys, PPARG, prevalence
Citation: Gong S, Han X, Li M, Cai X, Liu W, Luo Y, Zhang S-m, Zhou L, Ma Y, Huang X, Li Y, Zhou X, Zhu Y, Wang Q, Chen L, Ren Q, Zhang P and Ji L (2021) Genetics and Clinical Characteristics of PPARγ Variant-Induced Diabetes in a Chinese Han Population. Front. Endocrinol. 12:677130. doi: 10.3389/fendo.2021.677130
Received: 07 March 2021; Accepted: 06 October 2021;
Published: 26 October 2021.
Edited by:
Ruth Andrew, University of Edinburgh, United KingdomReviewed by:
Mitra Nourbakhsh, Iran University of Medical Sciences, IranJun Tu, Jiangxi University of Traditional Chinese Medicine, China
Copyright © 2021 Gong, Han, Li, Cai, Liu, Luo, Zhang, Zhou, Ma, Huang, Li, Zhou, Zhu, Wang, Chen, Ren, Zhang and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linong Ji, amlsbkBiam11LmVkdS5jbg==; Xueyao Han, eHVleWFvaGFuQHNpbmEuY29t
†These authors have contributed equally to this work
 Siqian Gong
Siqian Gong Xueyao Han1†*
Xueyao Han1†* Yufeng Li
Yufeng Li Xianghai Zhou
Xianghai Zhou Linong Ji
Linong Ji