
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol., 21 June 2021
Sec. Experimental Endocrinology
Volume 12 - 2021 | https://doi.org/10.3389/fendo.2021.674711
This article is part of the Research TopicRecent Advances in Crustacean EndocrinologyView all 16 articles
 Donald L. Mykles1,2*
Donald L. Mykles1,2*A pair of Y-organs (YOs) are the molting glands of decapod crustaceans. They synthesize and secrete steroid molting hormones (ecdysteroids) and their activity is controlled by external and internal signals. The YO transitions through four physiological states over the molt cycle, which are mediated by molt-inhibiting hormone (MIH; basal state), mechanistic Target of Rapamycin Complex 1 (mTORC1; activated state), Transforming Growth Factor-β (TGFβ)/Activin (committed state), and ecdysteroid (repressed state) signaling pathways. MIH, produced in the eyestalk X-organ/sinus gland complex, inhibits the synthesis of ecdysteroids. A model for MIH signaling is organized into a cAMP/Ca2+-dependent triggering phase and a nitric oxide/cGMP-dependent summation phase, which maintains the YO in the basal state during intermolt. A reduction in MIH release triggers YO activation, which requires mTORC1-dependent protein synthesis, followed by mTORC1-dependent gene expression. TGFβ/Activin signaling is required for YO commitment in mid-premolt. The YO transcriptome has 878 unique contigs assigned to 23 KEGG signaling pathways, 478 of which are differentially expressed over the molt cycle. Ninety-nine contigs encode G protein-coupled receptors (GPCRs), 65 of which bind a variety of neuropeptides and biogenic amines. Among these are putative receptors for MIH/crustacean hyperglycemic hormone neuropeptides, corazonin, relaxin, serotonin, octopamine, dopamine, allatostatins, Bursicon, ecdysis-triggering hormone (ETH), CCHamide, FMRFamide, and proctolin. Contigs encoding receptor tyrosine kinase insulin-like receptor, epidermal growth factor (EGF) receptor, and fibroblast growth factor (FGF) receptor and ligands EGF and FGF suggest that the YO is positively regulated by insulin-like peptides and growth factors. Future research should focus on the interactions of signaling pathways that integrate physiological status with environmental cues for molt control.
The progression of decapod crustaceans through the molt cycle depends on ecdysteroids synthesized by the Y-organ [YO; reviewed in (1)]. The molt cycle is unidirectional, progressing from the intermolt stage through premolt, ecdysis, and postmolt stages to the next intermolt stage [reviewed in (2, 3)]. Molting encompasses the preparatory processes during the premolt stage, culminating with the actual shedding of the exoskeleton (ecdysis), followed by restorative processes during the postmolt stage. Rising titers of ecdysteroids in the hemolymph initiate and coordinate premolt processes, such as synthesis of the new exoskeleton, degradation and resorption of the old exoskeleton, claw muscle atrophy, and limb regenerate growth (2, 4, 5). A precipitous drop in hemolymph ecdysteroids at the end of premolt triggers ecdysis (1). The low ecdysteroid titer during postmolt allows claw muscle growth and completion of exoskeleton synthesis and its calcification. Intermolt can last from weeks to years in adult decapods.
Molt stage transitions are determined by phenotypic changes in the activity and properties of the YO. In the intermolt stage (stage C4), inhibitory neuropeptides produced in the X-organ/sinus gland complex, such as molt-inhibiting hormone (MIH) and crustacean hyperglycemic hormone (CHH), maintain the YO in the basal state (Figure 1). A proposed model for MIH signaling couples a cAMP/Ca2+-dependent triggering phase with a NO/cGMP-dependent summation phase [reviewed in (2)]. The prolonged activation of a calmodulin-dependent NO synthase and NO-dependent guanylyl cyclase (GC-I) represses ecdysteroidogenesis between MIH pulses (2, 6–9). The decision to molt, or enter premolt, is determined by integration of environmental and physiological cues by the central nervous system that are not completely understood (10). A decrease in MIH release by the X-organ/sinus gland complex, which can be experimentally induced by eyestalk ablation (ESA), triggers YO activation and entry into early premolt (stage D0) (2, 3, 11). Multiple limb autotomy (MLA) also induces molting, as limb regenerates only become functional appendages when extended at ecdysis (4, 12, 13). It is hypothesized that MLA-induced molting is mediated by a stimulatory factor, designated limb autotomy factor – anecdysis (LAFan), produced by the developing limb buds (3). YO activation requires mechanistic Target of Rapamycin Complex 1 (mTORC1) activity, as rapamycin inhibits YO ecdysteroidogenesis in vitro and prevents YO activation in vivo (14, 15). mTORC1-dependent protein synthesis drives the initial increase in ecdysteroid synthesis by the YO. The activated YO remains sensitive to MIH, CHH, and other factors, giving the animal the flexibility to suspend or delay molting when conditions turn unfavorable (2).
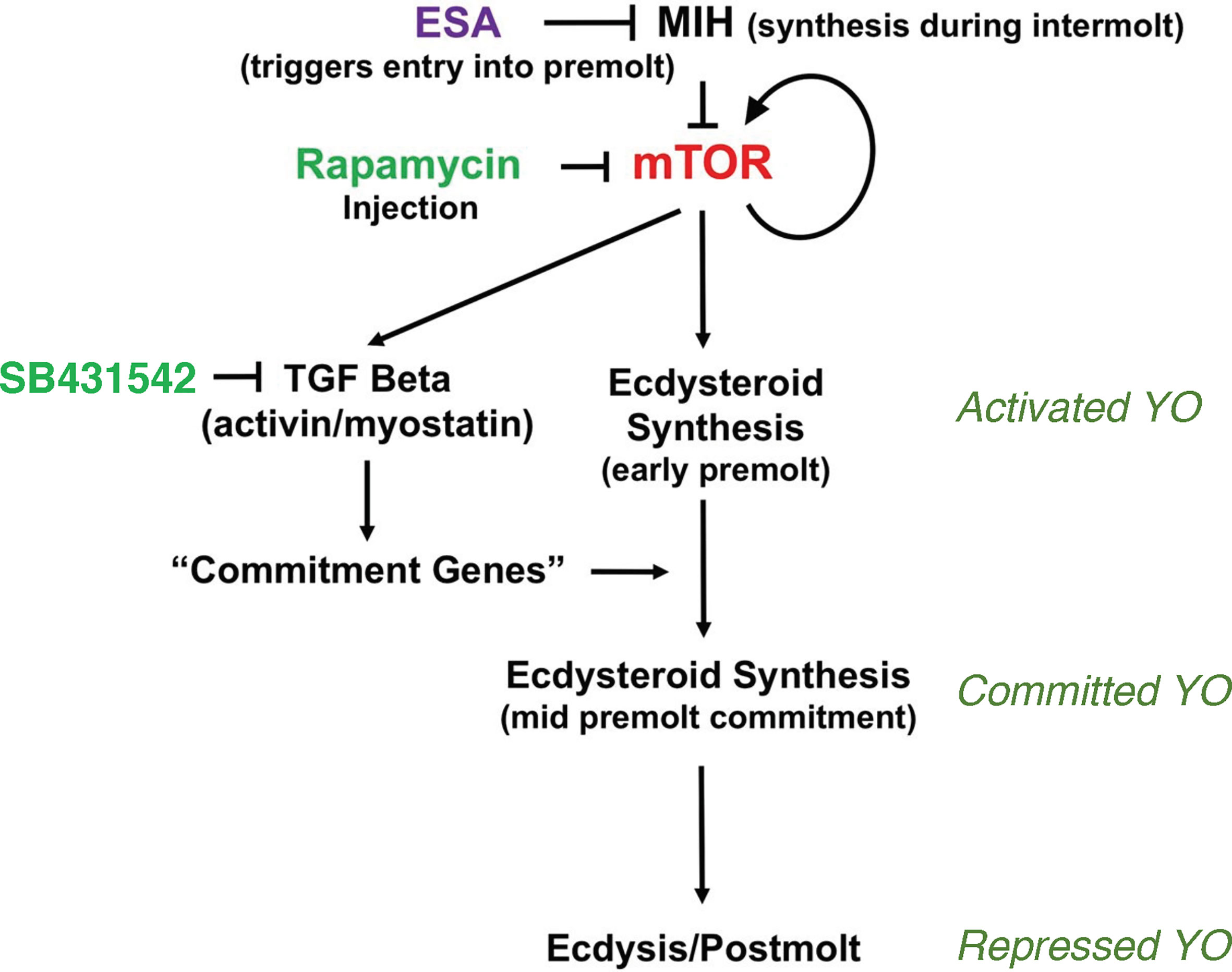
Figure 1 Organization of the signaling pathways mediating YO phenotype transitions over the molt cycle. Cyclic nucleotide-mediated MIH signaling maintains the YO in the basal state by inhibiting mTOR signaling. Reduction in MIH, such as by eyestalk ablation (ESA), stimulates mTOR activity, which is inhibited by rapamycin. mTOR stimulates ecdysteroid synthesis and up-regulates mTOR and TGFβ/Activin signaling genes, and down-regulates MIH signaling genes. Activin/myostatin signaling, which is inhibited by SB431542, up-regulates mTOR signaling genes and controls expression of commitment genes that determine the committed phenotype. High ecdysteroid titers in late premolt may trigger the repressed phenotype in postmolt. From (2).
A critical decision point occurs at the end of early premolt, when the animal becomes committed to molt. The transition of the YO from the activated to the committed state is mediated by transforming growth factor beta (TGFβ)/Activin signaling, as SB431542, an inhibitor of Activin receptor signal transduction, prevents progression of animals from early premolt to mid-premolt (stage D1; Figure 1) (15). mTORC1 activity affects the mRNA levels of thousands of genes, including those in the mTORC1 and TGFβ/Activin signaling pathways (Figure 1) (16). An invertebrate Myostatin (Mstn)-like factor, first described in scallop and crustacean muscles (17–19), appears to be the ligand for the Activin receptor. It is highly expressed in the YO and its mRNA levels are highest in the activated YO (15, 20, 21). The committed YO increases ecdysteroid synthesis, resulting in increasing ecdysteroid titers in the hemolymph during mid-premolt and reaching a peak in ecdysteroid titer at the end of late premolt (1). The committed YO also becomes insensitive to MIH and CHH (2). Limb bud autotomy, which suspends molting processes in early premolt, is no longer effective in mid- and late premolt animals (2, 12, 13).
The signaling mechanisms controlling the transition of the committed YO to the repressed YO at the end of late premolt and the transition of the repressed YO to the basal YO at the end of postmolt are not well understood. It is hypothesized that the large peak in hemolymph ecdysteroid titer triggers the transition to the repressed phenotype (Figure 1), as the YO expresses the ecdysteroid receptor (EcR/RXR) and ecdysteroid-responsive genes (2). The repressed YO has low ecdysteroid synthetic activity, which results in low hemolymph ecdysteroid titers during postmolt (1). Most of the 478 differentially-expressed genes assigned to signal transduction pathways are down-regulated to their lowest levels during the postmolt stage (21). Among these are critical components of the MIH, mTORC1, and TGFβ/Activin signaling pathways (21). These data suggest that the YO is not inhibited by MIH during postmolt and that repression of the YO involves transcriptional regulation that prevents premature reactivation of the YO until exoskeleton synthesis and calcification are completed (2). The model assumes that normal MIH control is not restored until the YO returns to the basal state in intermolt.
Transcriptomics and proteomics have revolutionized crustacean physiology (22, 23). These approaches have shown that the YO undergoes molt stage-specific changes in phenotype that differ quantitatively and qualitatively in mRNA and protein levels (2, 16, 21, 24). mTORC1 activity plays a critical role in controlling ecdysteroid synthesis at the transcriptional and translational levels (2, 14, 16). Transcriptomics and proteomics can also be tools for discovery. Analysis of the MLA Gecarcinus lateralis YO transcriptome identified 878 unique contigs assigned to 23 KEGG signaling pathways, including those for MIH/CHH, mTOR, and TGFβ/Activin [Table 1; (21)]. The YO also expresses MAP kinase, AMP kinase, ErbB, Hedgehog, HIF-1, Jak-STAT, Hippo, NF-kappa B, Notch, TNF, and Wnt signaling pathway genes among others, raising the possibility that ecdysteroidogenesis is regulated by a great many factors (20, 21). Proteomic analysis has revealed that anti-radical oxygen species, cytoskeletal, vesicular secretion, immune response, protein homeostasis proteins contribute to G. lateralis YO function (24). This review presents the current knowledge of the signaling pathways that control ecdysteroid synthesis by the YO and identifies areas for future research. It includes relevant research on signaling mechanisms that control the insect prothoracic gland.
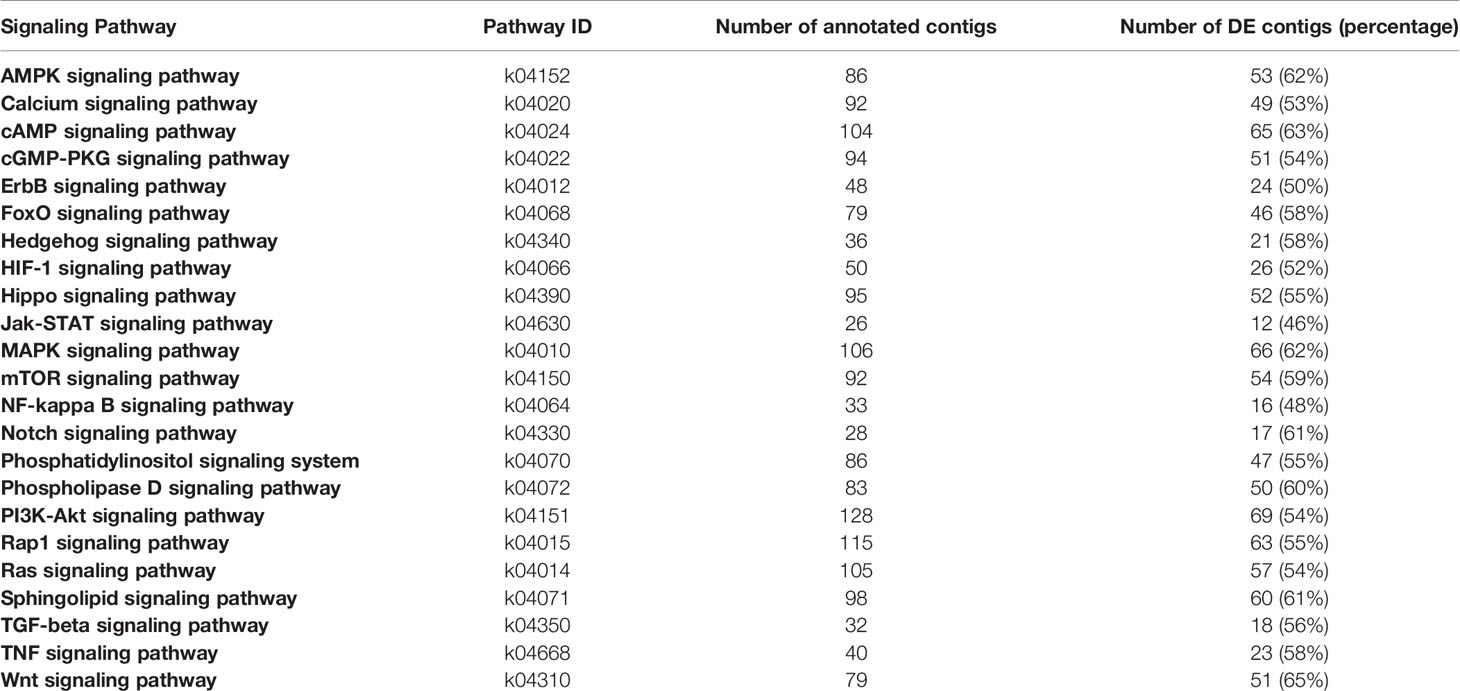
Table 1 Number of total and differentially expressed annotated contigs in the G. lateralis YO transciptome assigned to KEGG signal transduction pathways. From (21).
Transcriptomic analysis has revealed that a large number of G protein-coupled receptors (GPCRs) are expressed in decapod crustacean tissues. GPCRs are characterized by seven transmembrane domains, an external N-terminal domain, and a C-terminal cytosolic domain (25). These are divided among three large classes: rhodopsin-like (Class A), which represents the largest number of GPCRs; secretin-like (Class B); and metabotropic glutamate (Class C) (25–44).
All three GPCR classes are expressed in the YO, but represent a subset of those cataloged in decapod tissues (25, 29). In green shore crab Carcinus maenas, 62 contigs encoding GPCRs were identified in the central nervous system (29). The YO expresses 37 GPCRs annotated to 17 ligand clusters (Table 2). Thirty-two contigs are Class A and 5 contigs are in Class B; no Class C contigs were identified in the C. maenas YO (Table 2) (29). Eleven GPCRs are enriched in the YO compared to the epidermis; these were identified as gonadotropin-releasing hormone receptor, tachykinin-like R86C, relaxin R1, two rhodopsin G0-coupled receptors, two methuselah-like R1, dopamine D2-like receptor, opsin UV-sensitive receptor, serotonin R4, and GPCR161 (29). In addition, seven GPCRs are differentially expressed over the molt cycle; these include short neuropeptide F, Bursicon R2, CHHa R1, relaxin R3, ITPR-like, Moody-like, and Ast-B/MIP-R1 (29). Deep high throughput RNA sequencing and de novo assembly of the intermolt G. lateralis YO identified 99 putative GPCRs, 65 of which were annotated to 32 ligand clusters (Table 2) (25). The ligands are mostly neuropeptides, but also include biogenic amines, such as dopamine, octopamine, and serotonin (Table 2). These data suggest that the YO can potentially respond to a wide variety of ligands. The possible roles of some of these GPCRs are discussed in the sections below.
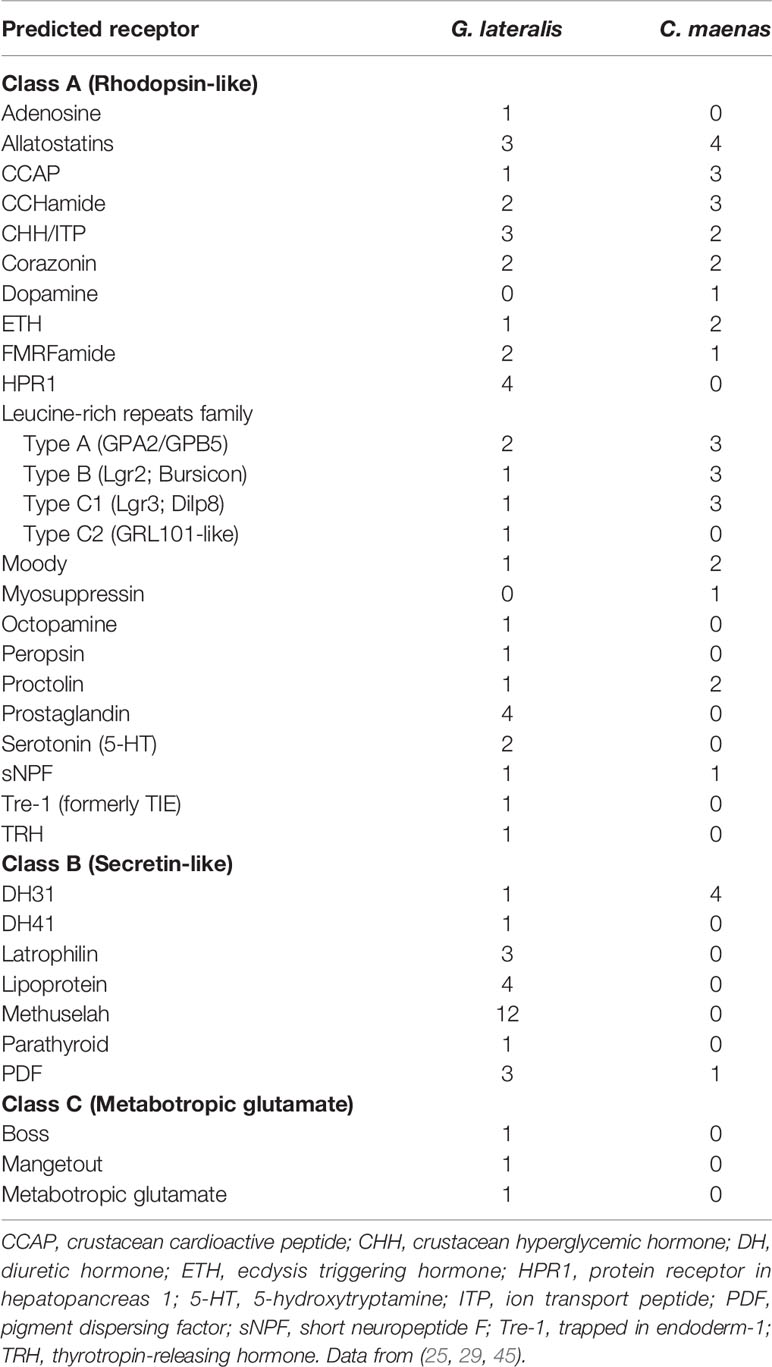
Table 2 Classification and number of contigs encoding G protein-coupled receptors in the Gecarcinus lateralis and Carcinus maenas Y-organ transcriptomes.
Surprisingly, the YO expresses a variety of peptide hormones. In C. maenas, contigs encoding 19 full-length peptides were identified (29). The six peptides that are expressed at the highest levels are Neuroparsin-1, -3, and -4; CHH-1, inotocin/vasopressin, and Eclosion Hormone-2 (29). Further research should determine if the transcripts are translated into peptides and the peptides are secreted into the hemolymph. If so, it would provide compelling evidence that the YO has endocrine functions beyond that of ecdysteroid production.
Peptides in the CHH family are divided into two types that differ in the amino acid sequences of the precursor proteins. Members of this family, which includes insect ion transport peptide (ITP), have a 66-amino acid “CHH family motif” in the mature peptide with six conserved cysteines that form three intramolecular disulfide bridges to stabilize the structure of the native protein (46–50). Type I peptides, which include CHH and ITP, are characterized by a signal peptide sequence followed by CHH/ITP precursor-related peptide (CPRP) and mature peptide sequences. Type II peptides, which include MIH, gonad-inhibiting hormone (GIH), and mandibular organ-inhibiting hormone (MOIH), lack a precursor-related sequence and have a glycine inserted between residues #11 and #12 and an invariant valine at position #20 in the mature peptide (46). Recently, a comprehensive phylogenetic analysis of crustacean ITPs proposed that ITPs be assigned to a third group (Type III) distinct from Type I CHHs (48, 50). The solution structures of MIH and CHH are similar, except that a short alpha-1 helix at positions #10 through #13 in MIH is lacking in CHH (49, 51). It is thought that the Gly12 contributes to the formation of the alpha-1 helix in Type II peptides (51). The surface structures of the N- and C-terminal regions confer specificity of binding to distinct receptors in the YO membrane (47, 49, 51–54).
The identification and characterization of the MIH receptor has remained elusive for more than three decades (46, 55). It is hypothesized that the receptors for the CHH family are GPCRs, given the similar native structures of Type I and Type II peptides (49, 51) and signal transduction mediated by cyclic nucleotide second messengers (56). In insects, studies of silk moth GPCRs (Bombyx neuropeptide G-protein coupled receptors, or BNGRs) identified BNGR-A2 and -A34 as ITP receptors, and BNGR-A24 is an ITP-like receptor (57). Based on this discovery, two full-length contigs (Pc-GPCRA52 and A53) and one partial contig (Pc-GPCRA63) from the transcriptome of adult Procambarus clarkii were identified as putative CHH-like receptors (CHHRs) (58). Subsequently, CHHR orthologs from more than ten other decapod crustacean species have been identified (25, 27, 29).
Three putative CHHRs, designated Gl-GPCR-A9, -A10, and -A12, are expressed in the G. lateralis YO transcriptome (25). Two CHH/ITP-like receptors were identified in the C. maenas YO transcriptome (29). Further phylogenetic analysis showed that the arthropod CHH/ITP GPCRs formed three clusters, designated CHHR1, CHHR2, and CHHR3/ITP-like R/tachykinin R (Figure 2). As Gl-GPCR-A9 and -A10 were grouped in the CHHR1 cluster, they are renamed Gl-CHHR-1A and -1B, respectively; Gl-GPCR-A12 is renamed Gl-CHHR-2 (Figure 2). Modeling the 3-dimensional structures of the G. lateralis CHHRs gave a highly conserved outcome of a predicted cleft at the N-terminus, suggesting a common role in binding CHH family hormones (Figure 2). End-point PCR showed that neither Gl-CHHR-1A or -2 are exclusively expressed in the YO, as would be expected for the MIH receptor; Gl-CHHR-1B was not examined (25). Interestingly, the YO is the only tissue to express both CHHRs. Eyestalk ganglia, thoracic ganglion, gill, heart, and midgut only express Gl-CHHR-1A; testis, hindgut, and hepatopancreas only express Gl-CHHR-2; and claw muscle does not express either CHHR (25). The three G. lateralis CHHRs are differentially expressed in the YO over the molt cycle, which suggests altered sensitivities to CHH neuropeptides associated with YO phenotypic changes. Gl-CHHR-1A has higher expression in intermolt and decreases during premolt stages; Gl-CHHR-1B is expressed during intermolt with higher expression during premolt stages; and Gl-CHHR-2 is expressed at high levels at late premolt (25). None of the Gl-CHHRs are expressed during postmolt (25). One of the CHH/ITP-like receptors in the C. maenas YO is differentially expressed over the molt cycle, with highest expression in late premolt (29). Future research must use a functional assay to establish which, if any, of the GPCR candidates is the MIH receptor.
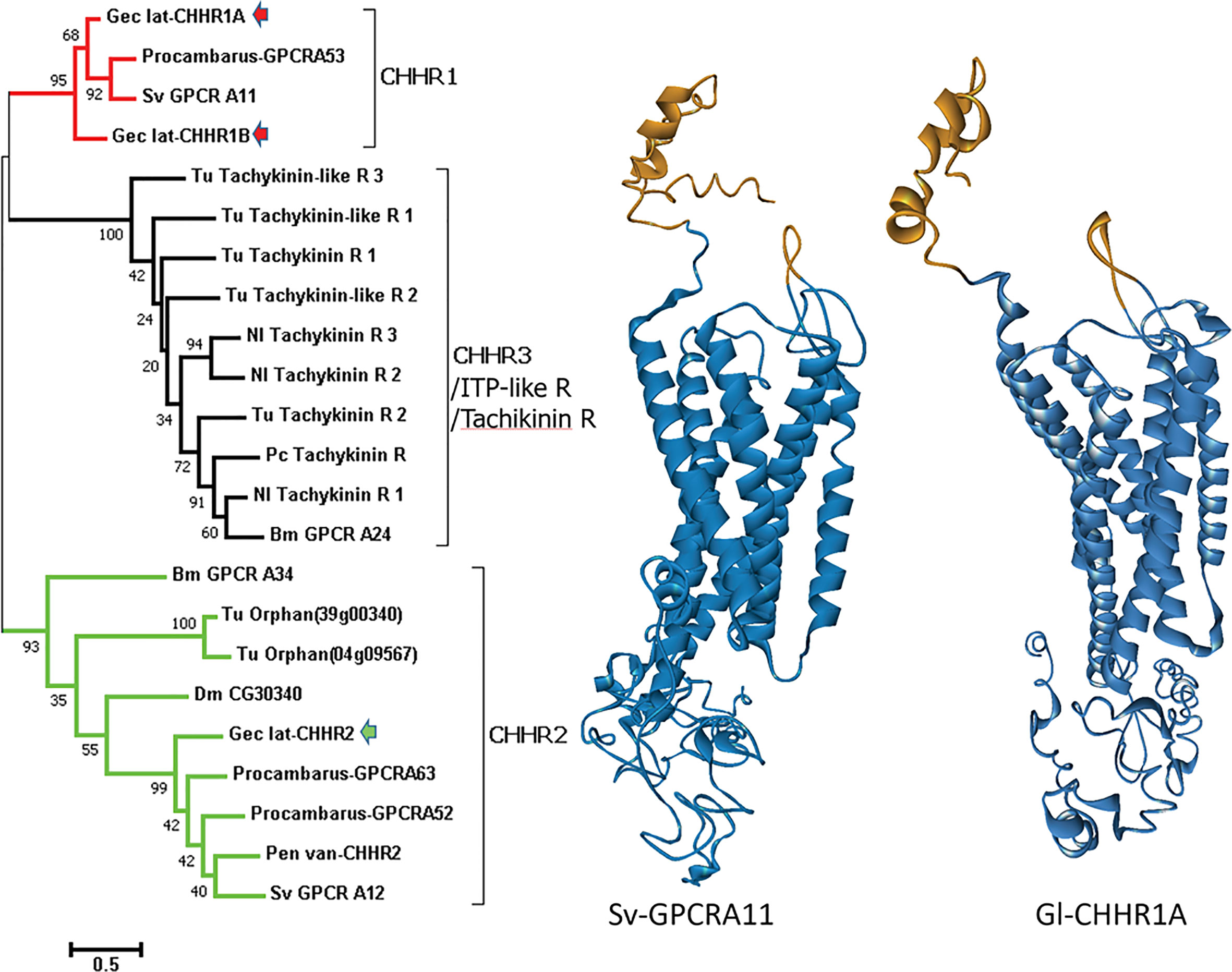
Figure 2 Comparison of decapod putative CHH and insect and mite ITP-like/tachykinin GPCRs. Left: A phylogram (left) of arthropod neuropeptide GPCRs showing three clusters designated CHHR1 (red), CHHR2 (green), and CHHR3/ITP-like R/tachykinin R (black). G. lateralis (Gec_lat) CHHR1A and 1B, originally designated Gl-GPCR-A9 and -A10, respectively (25), are within the CHHR1 cluster (red arrows) and CHHR2, originally designated Gl-GPCR-A12 (25), is within the CHHR2 cluster (green arrow). Species: Bm, Bombyx mori; Dm, Drosophila melanogaster; Nl, Nilaparvata lugens; Pen van, Penaeus (Litopenaeus) vannamei; Procambarus, P. clarkii; Sv, S. verreauxi; and Tu, Tetranychus urticae (red spider mite). Right: Structural models comparing the putative CHHR1 proteins from two decapod species predicted by GPCR-I-TASSER (27): S. verreauxi (Sv-GPCRA11) and G. lateralis Gl-CHHR1A. Gold indicates the N-terminal neuropeptide binding cleft in the extracellular domain. Blue indicates the transmembrane domain containing the 7 α-helices characteristic of GPCRs and the C-terminal intracellular domain. Figure provided by Dr. Tomer Ventura.
Corazonin (CRZ) is a conserved 11- amino acid neuropeptide with an amidated C-terminus and pGlu at the N-terminus. The sequence (pQTFQYSRGWTNa) is highly conserved among insect and crustacean species, although variants with single amino acid substitutions occur in some insects (59, 60). In Drosophila melanogaster, CRZ neurons modulate prothoracicotropic hormone (PTTH) action on basal ecdysteroidogenesis by the prothoracic gland (PG), thus controlling larval growth without affecting metamorphosis (61). C. maenas corazonin receptor (Cm-CRZR) is primarily expressed in the YO, suggesting that CRZ plays a role in regulating ecdysteroidogenesis (45). However, CRZ peptide (50 nM), which is produced in the eyestalk ganglia and other areas of the central nervous system, has only a small stimulatory effect on YO ecdysteroidogenesis in postmolt (stages A-B) C. maenas (45).
Tissue loss or injury delays molting, allowing time for regeneration or regrowth of tissues or organs prior to the next ecdysis. Molting delay by limb bud autotomy (LBA) in crustaceans and injury to imaginal discs in insects allows time for tissue regeneration, while growth of remaining or undamaged tissues slows or stops (12, 13, 62–66). In crustaceans, LBA during early premolt (stage D0) suspends premolt two to three weeks by lowering hemolymph ecdysteroid titers, so that animals can regain a full set of functional claws and legs at ecdysis (13, 67). In G. lateralis, secondary LBs produce a factor, designated Limb Autotomy Factor – proedysis (LAFpro), that lowers hemolymph ecdysteroid (3, 12, 13). In insects, regenerating imaginal discs produce a factor, identified as Drosophila insulin-like peptide 8 (Dilp8) in D. melanogaster, that delays metamorphosis by lowering ecdysteroid synthesis by the prothoracic gland (PG) (64, 68–70). Dilp8 also delays molting by activating Lgr3 neurons in the brain, which inhibit PTTH synthesis in PTTH neurons (71–74).
The LAFpro signaling pathway has not been fully characterized, but parallels with the action of Dilp8 on the insect PG suggest a common mechanism. Dilp8 is one of eight insulin-like peptides (ILPs) in D. melanogaster (74–78). The ILP superfamily consists of insulin, insulin-like growth factors (IGFs), and relaxin-like peptides (38, 71). Dilp 1 to 6 are in the insulin/IGF clade and bind to receptor tyrosine kinase receptors; Dilp 7 and 8 are in the relaxin-like clade and bind to leucine-rich repeat GPCRs (71, 74, 75). LAFpro is a peptide that is distinct from MIH. MIH is resistant to boiling in deionized water or weak acids (see (13) for references). LAFpro is stable when boiled 15 min in deionized water, but is inactivated by boiling in 0.1 M acetic acid (pH 2.9) and by proteinase K digestion (13). Dilp8 action is mediated through relaxin receptor Lgr3 and activation of NOS (73, 74, 79). NO donors inhibit ecdysteroidogenesis in both insect and crustacean molting glands (80–82). Dilp8 binding to Lgr3 stimulates production of cAMP in Drosophila cells (71). Up-regulation of Dilp8 delays expression of Halloween genes disembodied (Dib) and phantom (Phm) in the PG (68, 69). Moreover, targeted tissue damage or NOS overexpression in the PG lowers the expression of Halloween genes spookier (Spok) and Dib (79). These data suggest that Dilp8-induced NO inhibition of ecdysteroid synthesis is mediated by the down-regulation of cytochrome P450 enzymes, but it is unclear how NO represses gene transcription. A Lgr3-like GPCR is expressed in the G. lateralis and C. maenas YO transcriptomes (Table 2) (25, 29). An ILP2 is an ortholog of Dilp8 that is primarily expressed in nervous tissue, such as brain and eyestalk ganglia of Eastern spiny lobster (Sagmariasus verreauxi); brain and thoracic ganglion of red-claw crayfish (Cherax quadricarinatus); and in eyestalk ganglia, brain (males), thoracic ganglion (males), and sperm duct in ornate spiny lobster (Panulirus ornatus) (42, 83). It is not known if ILP2 is expressed in limb regenerates, and if there is higher expression in secondary than primary regenerates, as only 2˚ regenerates have LAFpro activity (13). Taken together, these data suggest that LAFpro is an Dilp8-like peptide that binds to Lgr3, activating the MIH signaling pathway to inhibit YO ecdysteroidogenesis (Figure 3).
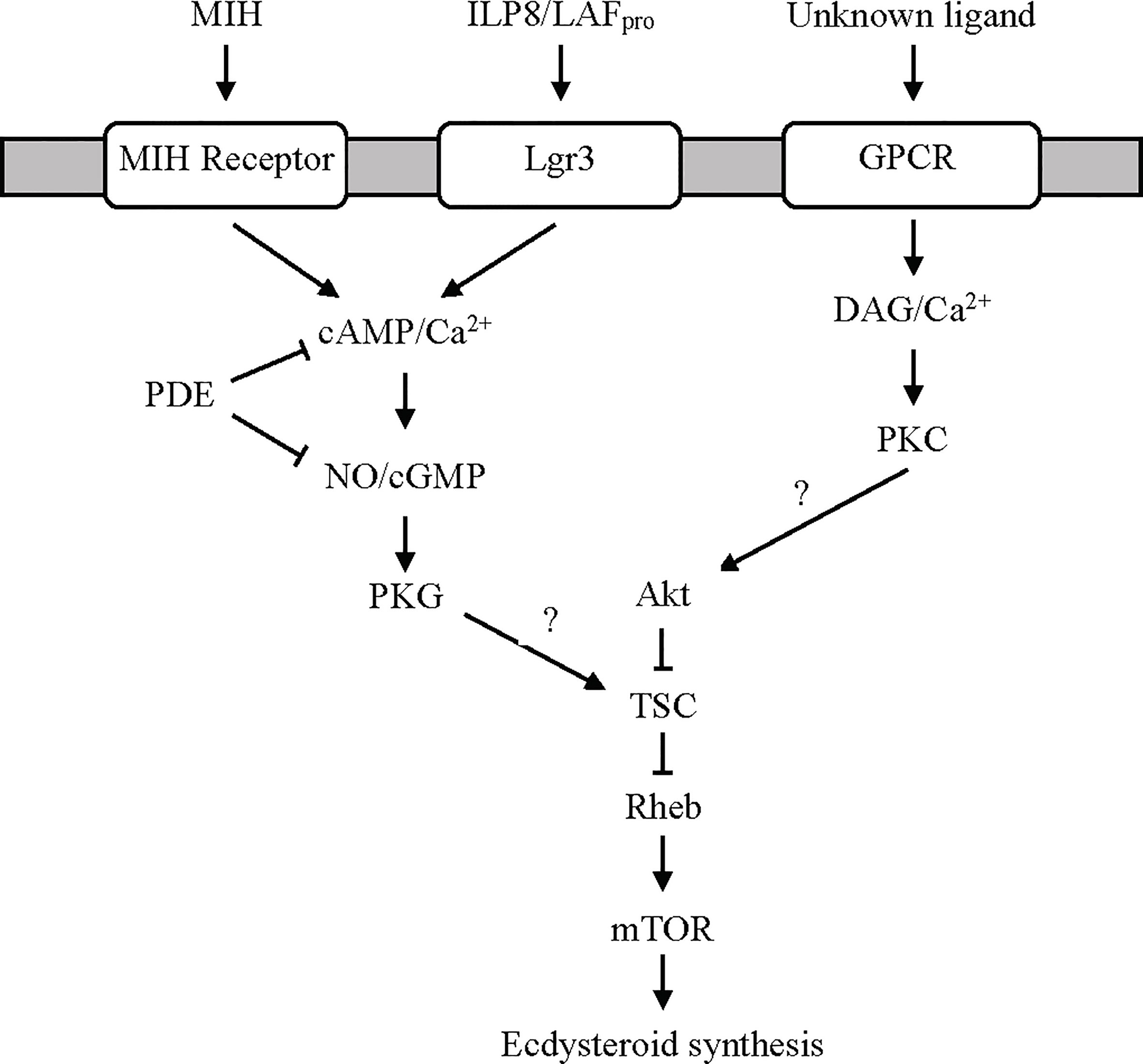
Figure 3 Proposed G protein-coupled receptor-mediated signaling pathways regulating ecdysteroidogenesis in the YO. MIH and limb autotomy factor - proecdysis (LAFpro) activate cyclic nucleotide/NO-dependent signaling via distinct receptors. It is hypothesized that LAFpro, secreted by secondary limb regenerates during early premolt, is an insulin-like peptide similar to dILP8 in Drosophila that binds to Lgr3. Cyclic phosphodiesterase (PDE) activity inhibits MIH and LAFpro signaling by hydrolyzing cAMP and cGMP to AMP and GMP, respectively. An unknown ligand, possibly serotonin or other biogenic amines (see Ca2+/Diacylglycerol/Protein Kinase C Signaling Section), binds a GPCR to activate the Ca2+/diacylglycerol (DAG)/protein kinase C (PKC) pathway. Both pathways converge on mTOR signaling, possibly by phosphorylation of tuberous sclerosis complex (TSC) by protein kinase G (PKG) to inhibit ecdysteroid synthesis or by phosphorylation of Akt by PKC to stimulate ecdysteroid synthesis (2, 12, 50, 84).
In the canonical pathway, ligand binding to a GPCR activates phospholipase C (PLC) via a Gq protein anchored in the cell membrane. PLC converts phosphatidylinositol to diacylglycerol (DAG) and inositol trisphosphate (IP3). DAG and IP3-initiated release of Ca2+ from smooth endoplasmic reticulum activate protein kinase C (PKC), which phosphorylates downstream targets to effect metabolic changes. All these components are represented in the KEGG calcium and phosphatidylinositol signaling pathways (Table 1) (21). Moreover, the YO has PKC activity (85).
Activation of PKC stimulates ecdysteroid synthesis and secretion by the YO. Studies using pharmacological reagents show that the pathway activating PKC is distinct from the MIH signaling pathway (Figure 3). A DAG analog or phorbol 12-myristate 13-acetate (PMA) stimulate PKC activity and ecdysteroid secretion by the Cancer antennarius YO in vitro (85). PMA counters the inhibitory effects of reagents that stimulate MIH signaling, such as forskolin, dibutyryl cAMP, and cyclic nucleotide phosphodiesterase inhibitor IBMX, and has no effect on cAMP levels (85). By contrast, PMA has the opposite effect on crayfish YO by inhibiting ecdysteroid secretion (86). PLC inhibitor U-73122 has no effect on crab and crayfish YO ecdysteroid production in vitro, and there are no changes in IP3 and DAG contents of YOs from intact and eyestalk-ablated crabs (86), suggesting that PKC is not involved in YO activation in early premolt. The downstream targets of PKC are unknown. mTORC1 signaling is likely involved, as PMA stimulates protein synthesis in the YO (84, 85, 87). A potential target of PKC is Akt in the mTOR signaling pathway (Figure 3).
The ligand and GPCR for the PKC pathway have not been identified. An intriguing possibility is that PKC is activated by serotonin (5-hydroxytryptamine) and other biogenic amines. The YO expresses serotonin, dopamine, and octopamine GPCRs (Table 2) (25, 29). Serotonin, dopamine, and octopamine function as neurotransmitters and neuromodulators in the crustacean central nervous system, but they may also act as neurohormones (46, 60, 88–91). Interestingly, there is evidence that the YO can synthesize serotonin (92). Much of the research of biogenic amines functioning as neurohormones has been focused on their roles in regulating decapod reproduction. For example, serotonin stimulates ovarian maturation, whereas octopamine delays gonadal development and inhibits ovarian maturation (93–95). Serotonin stimulates YO ecdysteroid production in vitro in mud crab (Scylla serrata) (96). In insects, serotonergic neurons directly innervate the PG and stimulate ecdysteroidogenesis (97–99). Octopamine acts as an autocrine factor that enhances PG ecdysteroidogenesis (100), but its effect on YO ecdysteroidogenesis is unknown. In C. maenas, dopamine D2-like and 5-hydroxytryptamine receptor 4 are up-regulated in the YO relative to their levels in the epidermis, although the receptors are not differentially expressed in the YO over the molt cycle (29). In G. lateralis YO, two serotonin receptors, designated Gl-GPCR-A30 and -A32, and an octopamine receptor, designated Gl-GPCR-A34, show different patterns of expression over the molt cycle (dopamine receptor was not identified in the G. lateralis YO transcriptome) (25). Gl-GPCR-A30 shows higher expression in early premolt and no expression in postmolt animals, while Gl-GPCR-32 is expressed in all five molt stages with higher expression in postmolt (25). Gl-GPCR-A34 is expressed in all molt stages, with higher expression during premolt (25). These data suggest that serotonin and octopamine are tropic factors in the YO. However, the signaling pathways activated by biogenic amines differ between insects and crustaceans. In the PG, serotonin and octopamine increase cAMP (97, 98), while in the YO, serotonin and octopamine action may be mediated by Ca2+/DAG (Figure 3). Future research should be directed to establishing the mode of action of biogenic amines on the YO.
The receptor tyrosine kinase (RTK) superfamily regulates animal development and homeostasis (101). There are about 58 RTKs in 20 subfamilies in mammals, with fewer in arthropods. D. melanogaster, for example, has 20 RTKs distributed among 14 subfamilies (76). RTKs have an extracellular ligand-binding domain, a single-pass transmembrane domain, and an intracellular tyrosine kinase domain (TKD). Most RTKs are heterodimers with each subunit consisting of a single polypeptide. By contrast, the insulin receptor is a heterotetramer, consisting of heterodimers of alpha and beta chains linked by disulfide bonds; the α-chain is completely extracellular and, together with the extracellular domain of the β-chain, binds ligand (101). Ligand binding activates RTK activity; autophosphorylation of the TKD activates MAPK, PI3K/Akt, PLCγ-PKC, JAK/STAT, or Rac-Rho signaling cascades (76, 101). The YO expresses both ILP receptors and growth factor receptors (Table 3).
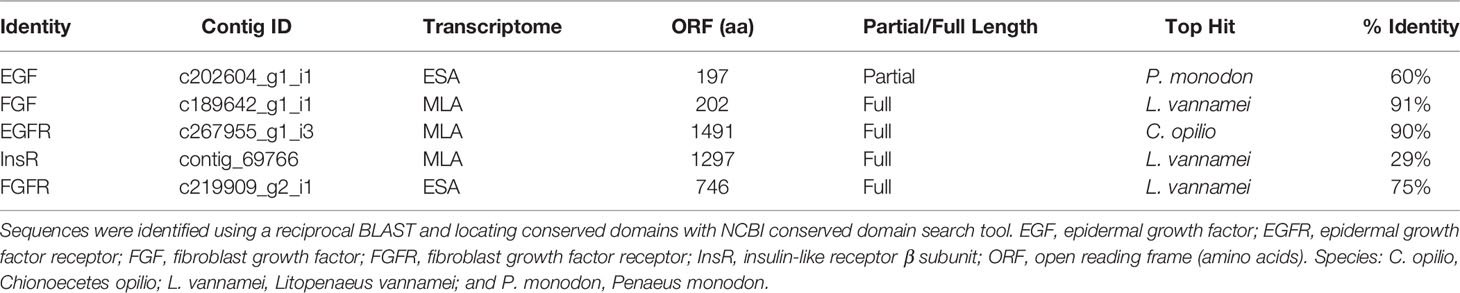
Table 3 Tyrosine receptor kinases and ligands expressed in G. lateralis MLA or ESA Y-organ transcriptomes (16, 20, 21).
In insects, ILPs are among the many factors that coordinate organismal growth and organ size and determine the timing of molting and metamorphosis (74, 78, 98, 102–105). A target of ILPs is the PG. Insulin-producing neurons in the brain secrete ILPs, in particular Dilp2, 3 and 5, that stimulate ecdysteroid production by the PG (74, 76, 102, 106). ILP/insulin-like protein receptor (InsR) signaling in the PG is mediated by PI3K/Akt/mTOR (74, 97, 106, 107). The role of ILP/InsR signaling in development and growth of crustaceans is not well understood, but it is likely that it has similar actions. Much of the research on crustacean ILP/InsR signaling has focused on reproduction (108, 109).
Insulin/ILPs are synthesized as a single polypeptide with an N-terminal signal peptide sequence, followed by B, C, and A chains (75, 77, 109). Proteolytic processing removes the signal peptide and excises the C chain, producing a B chain/A chain heterodimer stabilized by inter- and intra-chain disulfide bonds between conserved cysteines (75, 77, 109). In some ILPs, the C chain is not completely removed, producing a polypeptide, in which part of the C chain is retained (77). ILPs have been identified in decapod crustacean transcriptomes (28, 32, 33, 38, 83, 110–114). One of the best characterized ILP is the insulin-like androgenic gland hormone (IAG); it is expressed primarily in the androgenic gland and determines adult male characteristics (42, 83, 108, 109, 115, 116). In Portunus trituberculatus, Pt-IAG is expressed at very low levels in the YO (116). Other crustacean ILPs are expressed in most tissues, but at differing levels. In the oriental river prawn, Macrobrachium nipponense, Mn-ILP is expressed in brain, eyestalk ganglia, nerve cord, gonads, hepatopancreas, and muscle in adults (117). Mn-ILP expression is highest during the rapid growth stage in younger individuals and during the intermolt stage in older individuals (117). Sv-ILP1 and Cq-ILP1 are relaxin-like ILPs that are expressed in brain, antennal gland, gonads, and hepatopancreas (females only) (83, 113).
It is generally accepted that decapod crustaceans, like most invertebrates, express a single functional insulin receptor (InsR) (118). InsR has been biochemically characterized in gill, muscle, and hepatopancreas (119–121). InsR β-chain is expressed in many tissues, including the androgenic gland of male Macrobrachium rosenbergii (Mr-IR) (122), Fenneropenaeus chinensis (Fc-IAGR) (123), and S. verreauxi (Sv-TKIR) (124). Orthologs of Mr-IR have been identified in the neuropeptidomes of six other decapod species (33). Interestingly, silencing of Mr-IR led to androgenic gland hypertrophy and increased Mr-IAG production, but had no effect on somatic growth or sex determination, suggesting that molting and sexual differentiation are not solely dependent on the insulin receptor (122). S. verreauxi tyrosine kinase insulin receptor (Sv-TKIR), when expressed in a COS-7 cell reporting system, is activated by recombinant Sv-IAG and, to a lesser extent, recombinant human insulin, followed by recombinant Mr-IAG and Cq-IAG (124). In the YO, ESA results in a down-regulation of G. lateralis-InsR, which is blocked by rapamycin, suggesting that mTORC1 activity represses Gl-InsR expression (16).
Remarkably, there are no studies on the effects of insulin or ILPs on YO ecdysteroidogenesis. However, studies of other crustacean tissues indicate that insulin/ILP action is mediated through PI3K/Akt or MAPK/ERK signaling. Bovine insulin increases Sp-vitellogenin (Sp-Vtg) mRNA levels in hepatopanceas explants from Scylla paramamosain (125). Relatively high concentrations of bovine insulin were needed to elicit a response (>200 ng/ml). The insulin-induced increase in Sp-Vtg is blocked by PI3 kinase inhibitor (LY294002) and mTORC1 inhibitor rapamycin (125). Their respective recombinant IAGs increase MAPK/ERK phosphorylation in M. rosenbergii, S. verreauxi, and Cherax quadricarinatus testis explants in vitro (124). In insects, ILPs (e.g., Dilps1-6 in D. melanogaster and Bombyxin in Bombyx mori) stimulate PI3K/Akt signaling and mTORC1-dependent ecdysteroidogenesis in the insect PG (78, 97, 106, 107, 126–129). Based on these data, a model for ILP signaling in the YO is proposed (Figure 4). Binding of ILPs, such as ILP2, to InsR activates PI3K/Akt signaling, leading to mTOR activation and increased ecdysteroid synthesis. Alternatively, or perhaps in conjunction with PI3K/Akt signaling, ILP activates MAPK/ERK signaling (Figure 4). Both pathways converge on mTOR in animal cells (130, 131).
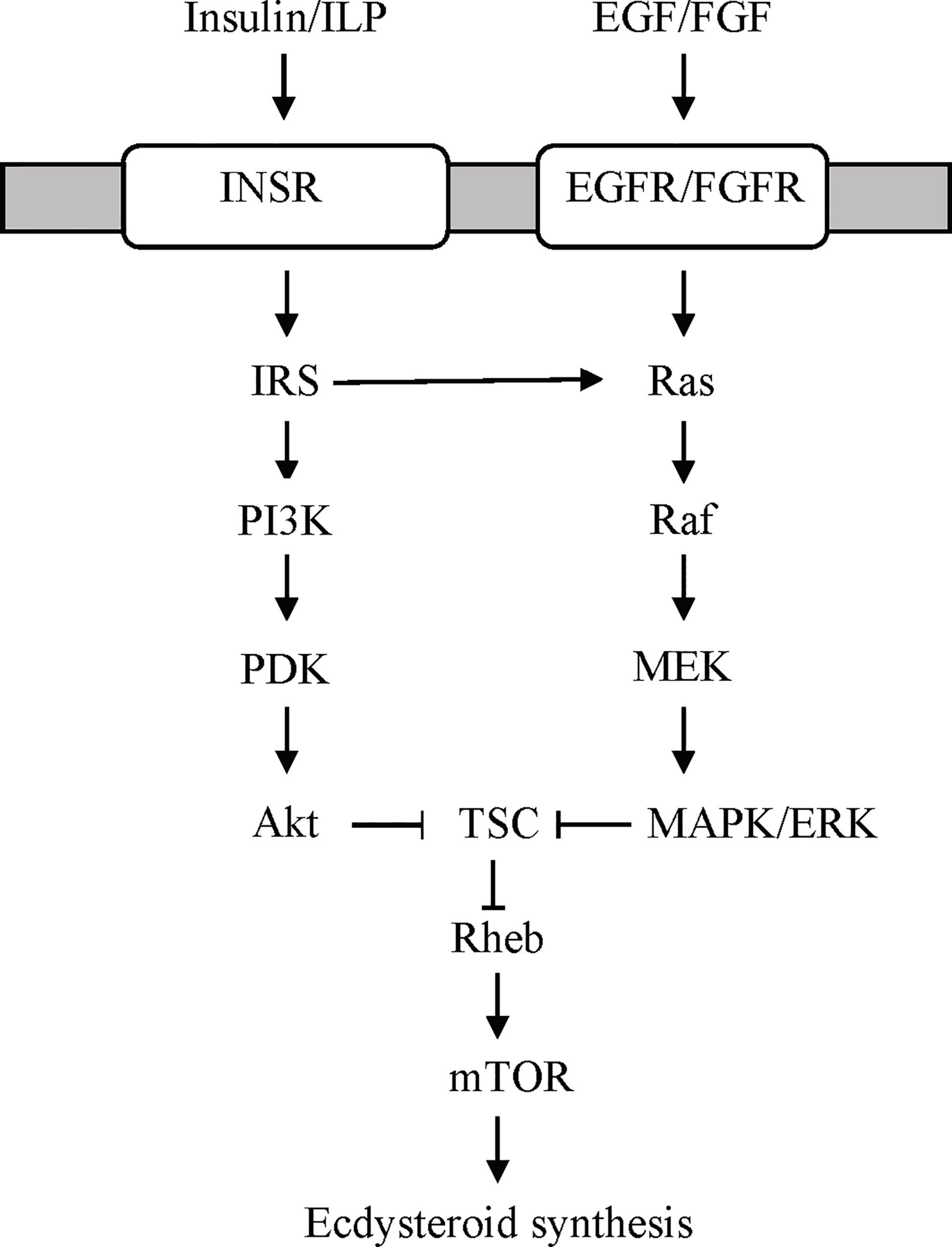
Figure 4 Proposed receptor tyrosine kinase (RTK)-mediated signaling pathways stimulating ecdysteroidogenesis in the YO based on data from the insect prothoracic gland (97, 98). YOs express both types of RTKs (Table 3). Growth factors, such as epidermal growth factor (EGF) or fibroblast growth factor (FGF), bind to EGF or FGF receptors, respectively, to activate the Ras/Raf/MEK/ERK signaling pathway. Insulin/insulin-like peptide (ILP) binds to insulin receptor (INSR) to activate PI3K/PDK/Akt and/or Ras/Raf/MEK/ERK signaling. Both pathways converge on mTOR signaling by phosphorylation of tuberous sclerosis complex (TSC) by Akt or ERK, respectively. ERK, extracellular signal-regulated kinase; IRS, insulin receptor substrate; MEK, MAPK/ERK kinase; PDK, protein 3-phosphoinositide-dependent protein kinase; PI3K, phosphoinositide 3-kinase (PI3K).
Growth factor signaling is mediated by the Ras/Raf/MAPK and PI3K/PDK1/Akt pathways (Figure 4) (132). In insects, growth factors, such as epidermal growth factor (EGF), fibroblast growth factor (FGF), platelet-derived growth factor, (PDGF) and vascular endothelial growth factor (VEGF), serve many functions critical for embryogenesis, molting, and metamorphosis (76). Activation of the Ras/Raf/MAPK pathway stimulates ecdysteroidogenesis in the insect PG (97, 106). Although not a growth factor, prothoracicotropic hormone (PTTH) activates this pathway by binding to an RTK encoded by Torso (97, 98, 102, 106, 133). PTTH is the primary factor that initiates larval molts in most insects. However, a recent study has shown that EGF receptor (EGFR) signaling supports PG ecdysteroidogenesis during the 3rd larva to pupa transition in D. melanogaster (134). The function of growth factors in YO ecdysteroidogenesis is unknown. Growth factor signaling pathways are well represented in the YO. These include the ErbB, MAPK, PI3K/Akt, and Ras KEGG signaling pathways (Table 1). Contigs can be assigned to two or more of the KEGG pathways (Figure 4) (21). The G. lateralis YO transcriptome has contigs encoding 106 MAPK signaling components, 66 of which are differentially expressed over the molt cycle (Table 1) (21). Forty-eight contigs assigned to the ErbB pathway, which includes EGFR signaling, have been identified (Table 1).
Knowledge of growth factors and their functions in crustaceans is limited. Unfortunately, there are no studies determining the effects of EGF, FGF, or VEGF on YO ecdysteroidogenesis. Immunohistochemical analysis indicated that eyestalk ganglia express a VEGF-like protein and VEGF receptor (135). The broad scale application of transcriptomics has aided the identification of growth factors and their receptors in decapod crustacean tissues. VEGFR is expressed in the embryos of Macrobrachium olfersi (136). The VEGF signaling pathway is enriched in hemocytes from pathogen-infected Eriochier sinensis, suggesting that VEGF is involved in mounting an immune response (137). Lv-EGF, Lv-EGFR, and Lv-VEGFR are expressed in embryos and larvae of Litopenaeus vannamei (138). In M. rosenbergii, Mr-EGFR is expressed in most tissues, with higher expression in thoracic ganglion, ovary, and testis (139). Knockdown of Mr-EGFR slows accumulation of mass in juvenile male M. rosenbergii, but has no effect on ecdysis frequency (139). Using transcriptomic data, a cDNA encoding the complete EGFR sequence was cloned from S. parmamosain ovary; Sp-EGFR is expressed in most tissues, with higher expression in YO, ovary, stomach, heart, and gill (140). Human EGF (1 nM and 10 nM) caused a transient increase in Sp-Vtg receptor and Sp-Cyclin B mRNA levels in ovary explants; the increases were blocked by pretreatment with EGFR tyrosine kinase inhibitors AG1478 and PD153035 (140). The G. lateralis YO expresses Gl-EGF, Gl-FGF, Gl-EGFR, and Gl-FGFR (Table 3) (16, 20). These data suggest that EGFR functions in a wide variety of tissues, including the YO (Table 3). The expression of Gl-EGF and Gl-FGF suggests that EGF, and perhaps FGF, act as autocrine factors in the YO as EGF does in the PG (134).
TGFβ signaling plays essential roles in animal cell differentiation and homeostasis, and dysregulation of TGFβ signaling contributes to many diseases including cancer (141–143). In the canonical pathway, a ligand binds to TGF receptor 2 (R2) homodimer, which associates with TGFR1 homodimer to form the active heterotetramer receptor complex by TGFR2 autophosphorylation and phosphorylation of TGFR1 and regulatory (R)-Smad (144, 145). Two phospho-R-Smads bind to one Co-Smad and the R-Smad/Co-Smad complex translocates to the nucleus to regulate gene transcription (141, 144, 145). Several proteins inhibit TGFβ signaling. Inhibitory (I)-Smads block TGFβ signaling by either preventing R-Smad phosphorylation by TGFR2 or preventing binding of Co-Smad to phospho-R-Smad. FK-506 binding protein 1A (FKBP12) binds to TGFR1 and prevents phosphorylation of TGFR1 by TGFR2 (141, 146). BMP and activin membrane bound inhibitor (BAMBI) acts as a TGFR1 pseudo receptor, as it binds ligand, but lacks the protein kinase domain for signal transduction. TGFβ signaling cross-talks with many other signaling pathways, such as MAPK, Akt, PKC, CAMKII, GSK3, JAK, JNK, Wnt, Notch, and Hedgehog (145).
TGFβ ligands include bone morphogenic proteins (BMPs), growth and differentiation factors (GDFs), Activin, anti-Müllerian hormone, nodal, and TGFβs (143, 147). They often function as autocrine factors, acting on the same tissue in which they are synthesized and secreted. TGFβ/Activin signaling, in particular, determines the competency of insect and crustacean molting glands to respond to neuropeptides (2, 97, 103). Insects express three Activins: Activin-β (Actβ), Myoglianin (Myo)/Myo-like, and Dawdle (Daw) (148, 149). In D. melanogaster, knocking out Activin signaling by targeting R-Smad dSmad2, Type I receptor Baboon (Babo), Type II receptor Punt, or Co-Smad Medea in the PG leads to 3rd instar arrest and failure of larvae to metamorphose and down-regulation of signaling genes Torso and InR and Halloween genes Dib and Spok in the PG (150). In the German cockroach Blatella germanica, an increase in Bg-Myo mRNA levels in the PG is associated with increased ecdysteroid synthesis at the end of the 5th instar (151). These data indicate that Activin signaling is necessary for the stimulation of ecdysteroidogenesis by PTTH and ILP. Interestingly, knocking out any one of the three Activin ligands Actβ, Myo, or Daw has no effect on D. melanogaster molting and metamorphosis (148). All three ligands must be knocked out in the PG in order to manifest the developmental arrest phenotype, suggesting some degree of redundancy between the three ligands and their Babo receptors (148). Adult decapod crustaceans express a single myostatin (Mstn)-like/GDF11 that is related to mammalian Mstn/GDF8 and GDF11 (2, 18). TGFβ/Mstn signaling drives the transition of the YO to the committed state, resulting in the YO becoming less sensitive to MIH and CHH in mid-premolt and late premolt (2, 15, 152). Recently, a cDNA encoding a Dawdle-like factor was characterized in S. paramansosain. Sp-Daw is expressed primarily in embryos and larvae, suggesting that it plays a role in developmental processes (153). It may also be involved in the innate immune response in adults (153).
Mstn is expressed in most crustacean tissues, with generally higher levels in the YO, heart, and muscle (15, 18, 154–161). Most studies have focused on the role of Mstn as a negative regulator of muscle growth, which contributes to organismal growth (5). In Chinese shrimp F. chinensis, Fc-Mstn mRNA levels are inversely correlated with growth traits among individuals from different genetic lineages (162). It appears that Mstn slows muscle growth by inhibiting mTORC1-dependent protein synthesis. In G. lateralis claw muscle, increased protein synthesis during premolt is correlated with decreased expression of Gl-Mstn and increased expression of Gl-Rheb, the activator of mTORC1 (17, 163). Several studies have attempted to knock down Mstn expression as a means to promote growth in aquacultural species. Surprisingly, in several cases, Mstn ds-RNA injection has just the opposite effect: an increase in molt interval and/or decrease in growth rate in Penaeus monodon, L. vannamei, and Fenneropenaeus merguiensis (155, 159, 164). These studies did not consider off-target effects. Reduced expression of Mstn in the YO could have blocked or delayed the progression from early premolt to mid-premolt, thus lengthening the interval between ecdyses. The effects on YO Mstn mRNA levels were not examined in these studies. However, injection of Es-Mstn-dsRNA or Es-Activin receptor IIB (Es-ActRIIB) dsRNA into juvenile E. sinensis and injection of Fc-Mstn-dsRNA into juvenile F. chinensis accelerated molting and growth when compared to a control group (154, 165, 166). However, control animals were injected with RNase-free water or phosphate-buffered saline, rather than an unrelated dsRNA construct (154, 164–166). Thus, one cannot rule out a nonspecific response to dsRNA injection. Only two of the studies used dsRNA products of unrelated sequences as controls, and both those showed molt inhibition (155, 159).
Molting alters TGFβ/Activin signaling gene expression in the G. lateralis YO. Analysis of RNAseq data of MLA-induced animals shows increases of Gl-Activin RI, Gl-Smad2/3 (Gl-R-Smad), and Gl-Smad4 (Gl-Co-Smad) in early premolt to mid-premolt, while TGFβ signaling inhibitors, Gl-Smad6 (Gl-I-Smad) and Gl-BAMBI, are down-regulated during premolt (21). By contrast, ESA decreases Gl-Activin RI, Gl-Smad2/3, Gl-Smad4, Gl-Smad6, and Gl-BAMBI expression (16). The differences between the MLA and ESA transcriptome results are attributed to the ESA study focusing on initial YO activation as the animals did not transition to mid-premolt (16). Neither MLA nor ESA had a significant effect on the relative expression of Gl-Mstn in the G. lateralis YO transcriptomes (16, 21), although qPCR showed that Gl-Mstn mRNA level increases by three days post-ESA (15). Future research should use qPCR to establish the precise timing of the effects of ESA ± SB431542 on the expression of TGFβ/Mstn signaling genes.
mTOR is a serine/threonine PI3-related protein kinase that allocates energy in response to nutrients, growth factors, and stress in eukaryotic cells at transcriptional and post-translational levels (167–169). mTOR associates with other proteins to form two complexes: mTOR Complex 1 (mTORC1) and Complex 2 (mTORC2) (169). mTORC1 contains Raptor and controls protein translation, lipid and nucleotide synthesis, and autophagy (169, 170). mTORC2 contains Rictor and controls cytoskeletal remodeling, ion transport, and cell survival and proliferation (169, 170). mTORC1 is inhibited by rapamycin, mediated by FKBP12, while mTORC2 is insensitive to rapamycin (167, 169). mTORC1 is activated by GTP/Ras homolog enriched in brain (Rheb). Rheb, in turn, is controlled by the tuberous sclerosis complex (TSC), a heterotrimeric protein composed of Hamartin (TSC1), Tuberin (TSC2), and TBC1 domain family member 7 (TBC1D7) (169). TSC is a GTPase activating protein (GAP) that inactivates GTP/Rheb by promoting the hydrolysis of GTP to GDP (170). Growth factor signaling pathways converge on the TSC to promote cellular growth (Figure 4) (170). Phosphorylation of TSC by Akt, ERK, p90 ribosomal S6 kinase 1 (RSK1), and other protein kinases inhibit the GAP activity to prevent inactivation of GTP/Rheb, which leads to mTORC1 activation (169). Phosphorylation of ribosome subunit 6 kinase (S6K) and 4E-binding protein (4E-BP) by mTORC1 increases translation of mRNA to protein.
mTORC1 is required for increased ecdysteroidogenesis in the arthropod molting gland. PTTH stimulates mTORC1-dependent protein synthesis, which increases ecdysteroid synthesis of the insect PG (126, 128, 171–178); see (179) for earlier references). In D. melanogaster, Rheb overexpression or TSC2 knockdown in the PG reduces the developmental delay in food-limited 3rd-stage larvae, resulting in lower adult weights (180). Additionally, Rheb overexpression under food-limiting conditions increases transcription of Halloween genes Phm and Dib (180). Comparable studies on decapod crustaceans, such as knockdown of Rheb and TSC2 with dsRNA constructs, have not been done. Incubation of hepatopancreas explants with Sp-Akt-dsRNA reduced the insulin-induced increase in Sp-Vtg mRNA level, but had no effect on Sp-Rheb mRNA level (125). Rapamycin inhibits YO ecdysteroid synthesis and secretion in vivo and in vitro and blocks entry of intermolt animals into premolt and delays the transition from early premolt to mid-premolt in eyestalk-ablated G. lateralis (14, 15).
Molting up-regulates many of the mTOR signaling components in the YO. In G. lateralis, Gl-mTOR and Gl-Akt mRNA levels increase in mid-premolt and late premolt stages in MLA animals and Gl-mTOR, Gl-Akt, and Gl-S6K mRNA levels increase by three days post-ESA in eyestalk-ablated animals (14, 15). Gl-elongation factor 2 (Gl-EF2) mRNA level is also increased by MLA and ESA, which is consistent with increased protein synthesis in the YO (14, 15). By contrast, molt stage (intermolt, early premolt, and late premolt) has no effect on Cm-EF2, Cm-mTOR, Cm-Rheb, Cm-Akt, and Cm-S6K mRNA levels in C. maenas (14). SB431542 injection blocks the ESA-induced increases in Gl-EF2, Gl-mTOR, and Gl-Akt mRNA levels and decreases Gl-Rheb mRNA level, which suggest that TGFβ/Activin signaling is required for sustained up-regulation of mTOR signaling during premolt (15). RNAseq data expands on the results from qPCR analysis. Gl-mTOR, Gl-Raptor, Gl-Rictor, Gl-S6K, and Gl-Akt are expressed at high levels during intermolt and early premolt and at their lowest levels during postmolt (21). Gl-Rheb expression increases during early premolt and mid-premolt stages (21). ESA increases expression of Gl-mTOR, Gl-Raptor, Gl-mLST8, Gl-Rheb, Gl-Akt, Gl-S6, Gl-S6K, Gl-EIF4E, and Gl-EF2 (16). These increases are inhibited by rapamycin, suggesting a positive feedback mechanism in which mTORC1 activity up-regulates expression of mTOR signaling genes (Figure 1) (16).
Temperature affects metabolic processes in crustaceans, including molting, and it is likely that mTORC1 activity contributes to the response of the YO and other tissues to temperature. Within normal physiological ranges, increasing temperature stimulates molting and growth of decapod crustaceans (3, 181–183). However, when an animal reaches its upper thermal limit, molting is inhibited, either directly on the YO or indirectly by inhibitory neuropeptide, such as CHH, secreted by the X-organ/sinus gland (183–186). The effects of temperature on survival, molting, and mTOR signaling gene expression in YO, eyestalk ganglia, and heart were determined in juvenile Dungeness crab, Metacarcinus magister. Animals at three different molt stages (12, 19, or 26 days post-ecdysis; these intervals spanned stages C to D2-3) were transferred from ambient temperature (~15°C) to 5, 10, 15, 20, 25, or 30°C for 14 days (187). None of the animals transferred to 25 and 30°C survived (187). M. magister molt successfully at 21°C, but the growth increment is less than that at 14°C (188); see (187) for additional references). These results indicate that the upper temperature limit for M. magister molting success is between 21°C and 25°C. Low temperature (5°C) inhibits molting (187). Between 10°C and 20°C, molt stage progression increases with temperature (187). Gene expression in YO, eyestalk ganglia, and heart is affected by temperature and molt stage, but there is little or no interaction in gene expression between temperature and molt stage (187). In eyestalk ganglia, Mm-MIH, Mm-CHH, Mm-Rheb, Mm-AMP kinase α subunit (AMPkα), and Mm-Akt mRNA levels decrease with increasing temperature, particularly at 20°C; Mm-S6K mRNA level is not affected by temperature (187). In heart, mRNA levels of Mm-Rheb, Mm-S6K, Mm-AMPKα, Mm-Akt, and Mm-mTOR are higher at 10°C than at 15°C and 20°C. Of the six genes quantified in the YO, only Mm-Rheb expression is affected by both molt stage and temperature (Figure 5). Mm-Rheb mRNA level is higher in premolt stages (Figure 5) and is positively correlated with hemolymph ecdysteroid titers at all three temperatures (187), which suggests that Rheb stimulates mTORC1-dependent ecdysteroid synthesis. Mm-Rheb mRNA level decreases with increasing temperature at most molt stages (Figure 5). It is noteworthy that only the mRNA level of Mm-Rheb is negatively correlated with temperature in all three tissues (Figure 6). It appears that the down-regulation of mTOR signaling serves as a compensatory mechanism for higher metabolic rates at higher temperatures, so that energy allocation to protein synthesis is maintained at relatively constant levels (Figure 6). Taken together, the data suggest that Rheb expression can be used as a proxy to assess the effects of molting and temperature on mTORC1 activity in crustacean tissues.
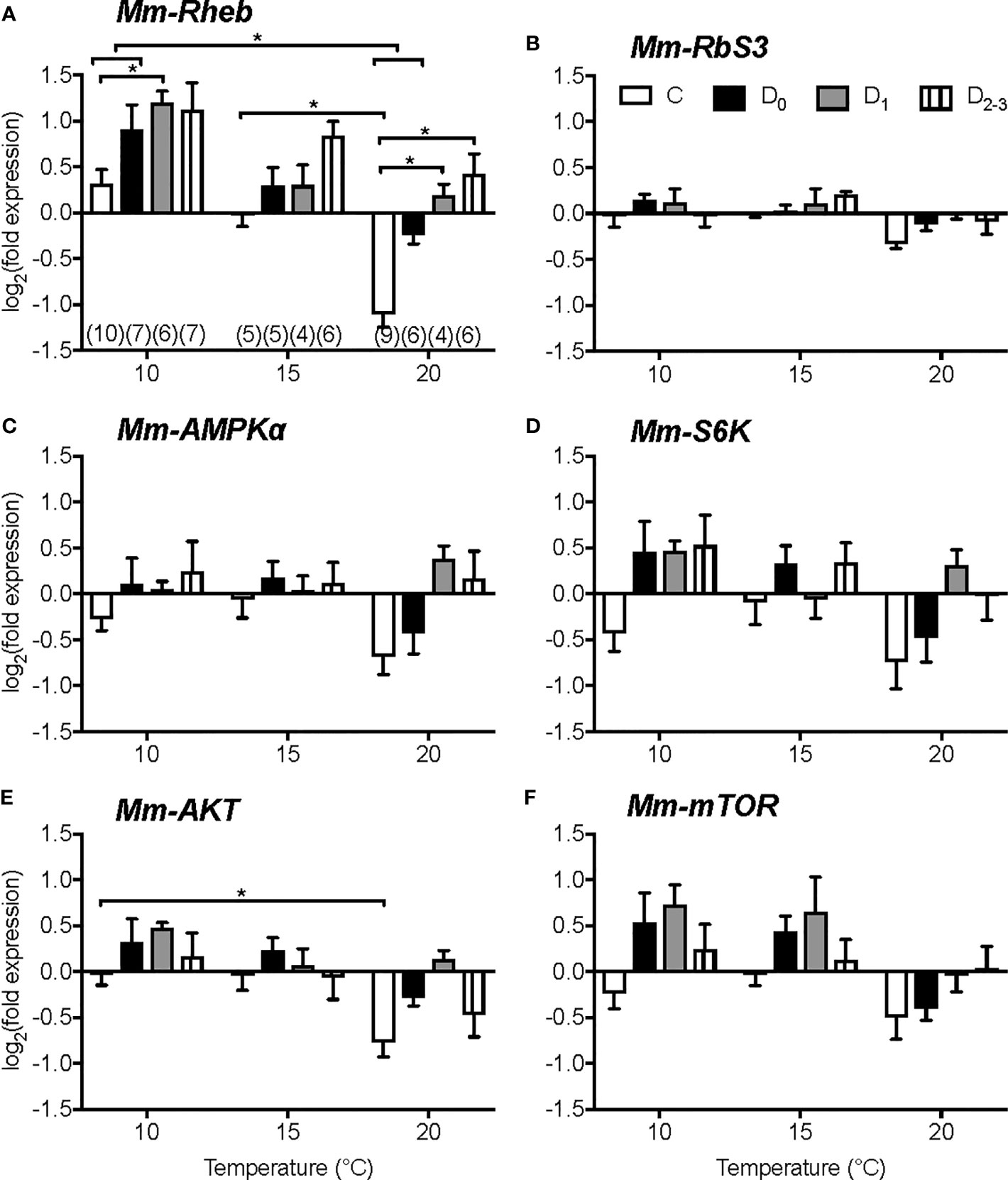
Figure 5 Effects of temperature and molt stage on gene expression in Y-organs of juvenile Dungeness crab, Metacarcinus magister. mRNA levels of Mm-Rheb (A), Mm-ribosome subunit 3 (RbS3) (B), Mm-AMPKa (C), Mm-S6K (D), Mm-AKT (E), and Mm- mTOR (F) after 14 days at 10°C, 15°C or 20°C of juveniles in intermolt (C, white), early premolt (D0, black), mid-premolt (D1, grey), or late premolt (D2-3, lines). Data are normalized to the mean absolute mRNA copy numbers in stage C at 15°C. Asterisks denote significant differences at P < 0.05. Sample size (n) given in brackets below columns in A also apply to the other genes. Data are presented as mean ± 1 S.E.M. Mm-Rheb expression is affected by temperature and molt stage. Mm-Rheb mRNA level increases during premolt stages and decrease with increasing temperature. From (187).
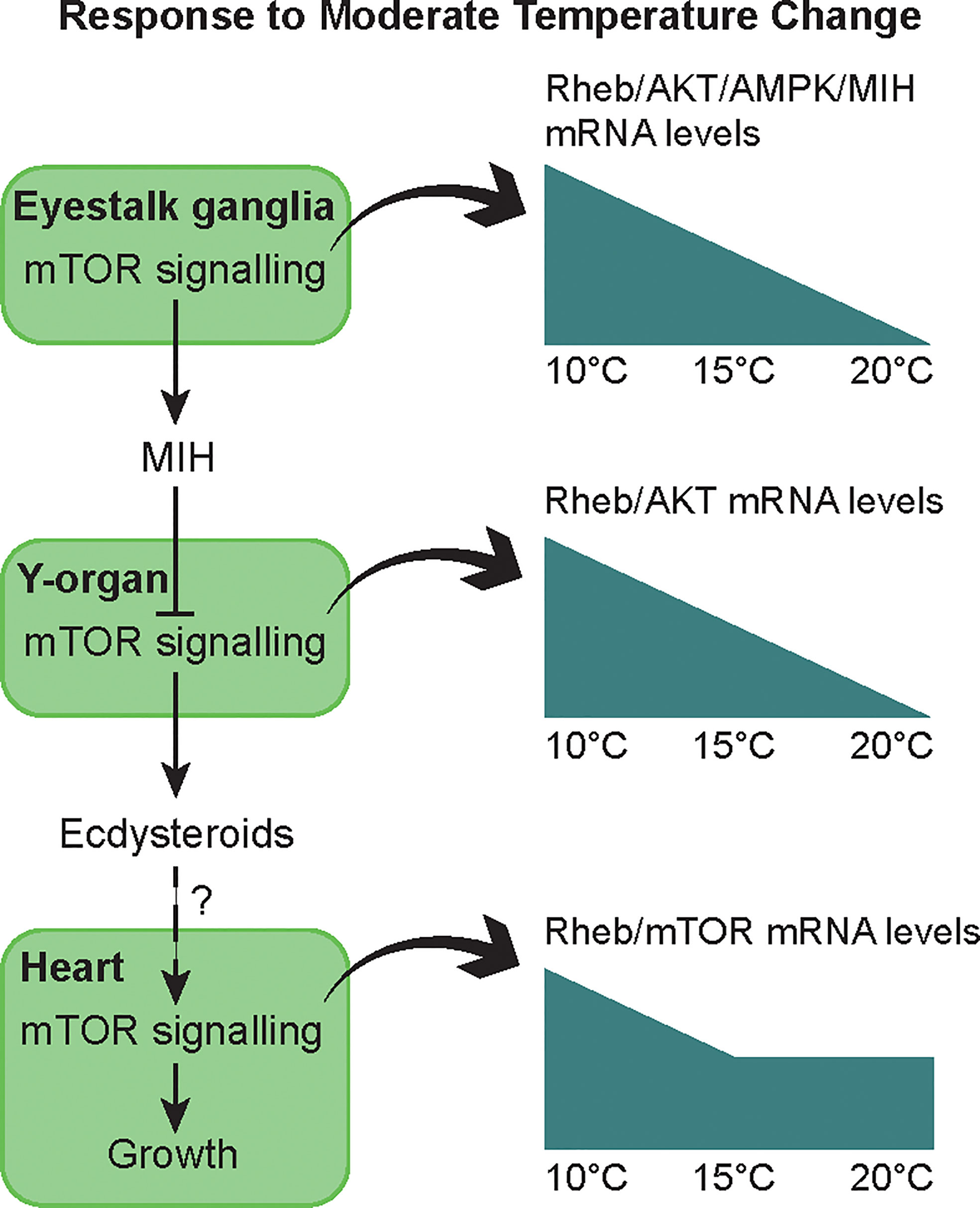
Figure 6 Summary of the effects of moderate thermal stress on molting and growth through the mTOR signaling pathway in juvenile Dungeness crabs. In intermolt, MIH keeps the YO in a basal state with low ecdysteroid secretion by inhibiting mTOR. Low levels of ecdysteroid may stimulate heart muscle growth via mTOR signaling. Moderate temperature change (10 to 20°C range) allows acclimation of the animals, at least with respect to some physiological functions. After 14 days, thermal compensation is observed in molt control, i.e. similar ecdysteroid titer across temperatures throughout the molt cycle (187). Mechanisms include up-regulation or down-regulation of Mm-MIH and mTOR signaling genes (Mm-Rheb, Mm-Akt, Mm-AMPK) during cold (10°C) or warm (20°C) exposure in the eyestalk ganglia and YO. In the heart, thermal compensation of metabolism is incomplete, as oxygen demand and heart activity increase with temperature. A sustained mRNA level of Mm-Rheb and Mm-mTOR indicates a greater allocation of energy to maintaining cardiac capacity during warm exposure. From (187).
The control of molting involves the integration of a variety of signals that affects the ecdysteroidogenic capacity and activity of the arthropod molting gland (2, 62, 78, 106). This is reflected by the diversity and actions of the factors involved. In the insect PG, tropic factors, such as PTTH, ILPs (e.g., dILP1-6, Bombyxin), growth factors (e.g., EGF and VEGF), biogenic amines (e.g., serotonin and octopamine), and FXPRLamide peptide, stimulate ecdysteroidogenesis (97, 98, 105). Static factors, such as dILP8, prothoracicostatic peptides, Bommo-myosuppressin, and FMRFamide-related peptide, inhibit ecdysteroidogenesis (97, 98, 105). By contrast, the neuropeptides MIH and CHH are the only known ligands identified for YOs in crustaceans (2, 50, 55, 189, 190). Transcriptomic analysis has revealed that the YO expresses receptors for ILPs, growth factors, biogenic amines, and neuropeptides (Tables 2, 3) (25, 29). The large number of GPCRs in particular indicates that the YO resembles the insect PG in being able to integrate a large number of signals to coordinate organ growth, development, and molting. MIH, CHH, LAFpro, and perhaps FMRFamide act as static factors on the YO. Drawing on comparisons with the insect PG, ILPs, EGF, VEGF, corazonin, and LAFan may act as tropic factors. The effects of these and other ligands (e.g., serotonin, octopamine, FGF, dopamine, pigment dispersing factor, allatostatins, ecdysis-triggering hormone, crustacean cardioactive peptide (CCAP), CCHamide, diuretic hormones DH31 and DH44, and Bursicon) on YO ecdysteroidogenesis remain to be determined.
PTTH and MIH are the primary neuropeptides controlling molting in insects and crustaceans, respectively (2, 50, 105, 106). It is remarkable that the control of a process as critical as ecdysis is to organismal growth would have evolved diametrically opposite mechanisms in these two major arthropod groups. PTTH activates the PG, while MIH inhibits the YO. Thus, molting in insects is initiated by the release of PTTH from neurosecretory neurons in the brain, while molting in crustaceans is initiated by reduced MIH release from neurosecretory neurons in the eyestalk X-organ/sinus gland complex (2, 11, 105). In D. melanogaster, PTTH stimulates PG ecdysteroidogenesis by binding to the Torso RTK and activating the Ras/Raf/MAPK signaling pathway (106). In lepidopterans (Manduca sexta and Bombyx mori), a PTTH-induced Ca2+ influx activates both Ras/Raf/MAPK signaling and cAMP-dependent signaling and both contribute to a large increase in ecdysteroid synthesis (97, 105, 179). MIH inhibits YO ecdysteroidogenesis by binding to a putative GPCR and activating a cyclic nucleotide-dependent signaling pathway (2, 50). A cAMP/Ca2+-dependent triggering phase is linked to a NO/cGMP-dependent summation phase to inhibit the YO between MIH pulses (2, 50, 55). RTKs in the YO most likely function to stimulate ecdysteroidogenesis, as they do in the PG (76, 97, 98).
The YO undergoes phenotypic changes over the molt cycle. The molt cycle is unidirectional, with YO transitions occurring at critical checkpoints that determine progression to the next molt stage. Figure 7 presents a working model for the signaling pathways that control YO basal (intermolt stage), activated (early premolt stage), and committed (mid- and late premolt stages) phenotypes. The most important decision is to initiate molting, which is determined by the integration of external cues, most likely mediated by the brain and eyestalk ganglia, which control the release of MIH from the X-organ/sinus gland complex, and internal cues, such as nutritional status, organ size, and limb regeneration, that act directly on the YO (2, 3, 11). The signaling pathways converge on Rheb/mTORC1 to regulate ecdysteroidogenesis (Figure 7). Activation of GPCR signaling by static factors (e.g., MIH and CHH) inhibits Rheb/mTORC1, maintaining the YO in the basal state. Activation of RTK signaling by tropic factors (e.g., ILPs and growth factors) stimulates Rheb/mTORC1. Although RTK signaling can potentially activate the YO, MIH release prevents molt initiation until environmental conditions are met. YO activation is mediated post-translationally by mTORC1-dependent increased global translation of mRNA to protein, resulting in increased ecdysteroid synthesis and secretion (Figure 7, dashed lines). The rising ecdysteroid titers in the hemolymph mark the entry into early premolt (1). The activated YO remains sensitive to static factors, as ecdysteroidogenesis is inhibited by MIH, CHH, and LAFpro (2). During early premolt, the YO synthesizes and secretes Mstn-like factor, which binds to Activin receptors to activate Smad transcription factors, leading to down-regulation of MIH signaling genes and up-regulation of Rheb/mTORC1 and Halloween genes in the committed YO (Figure 7, solid lines).
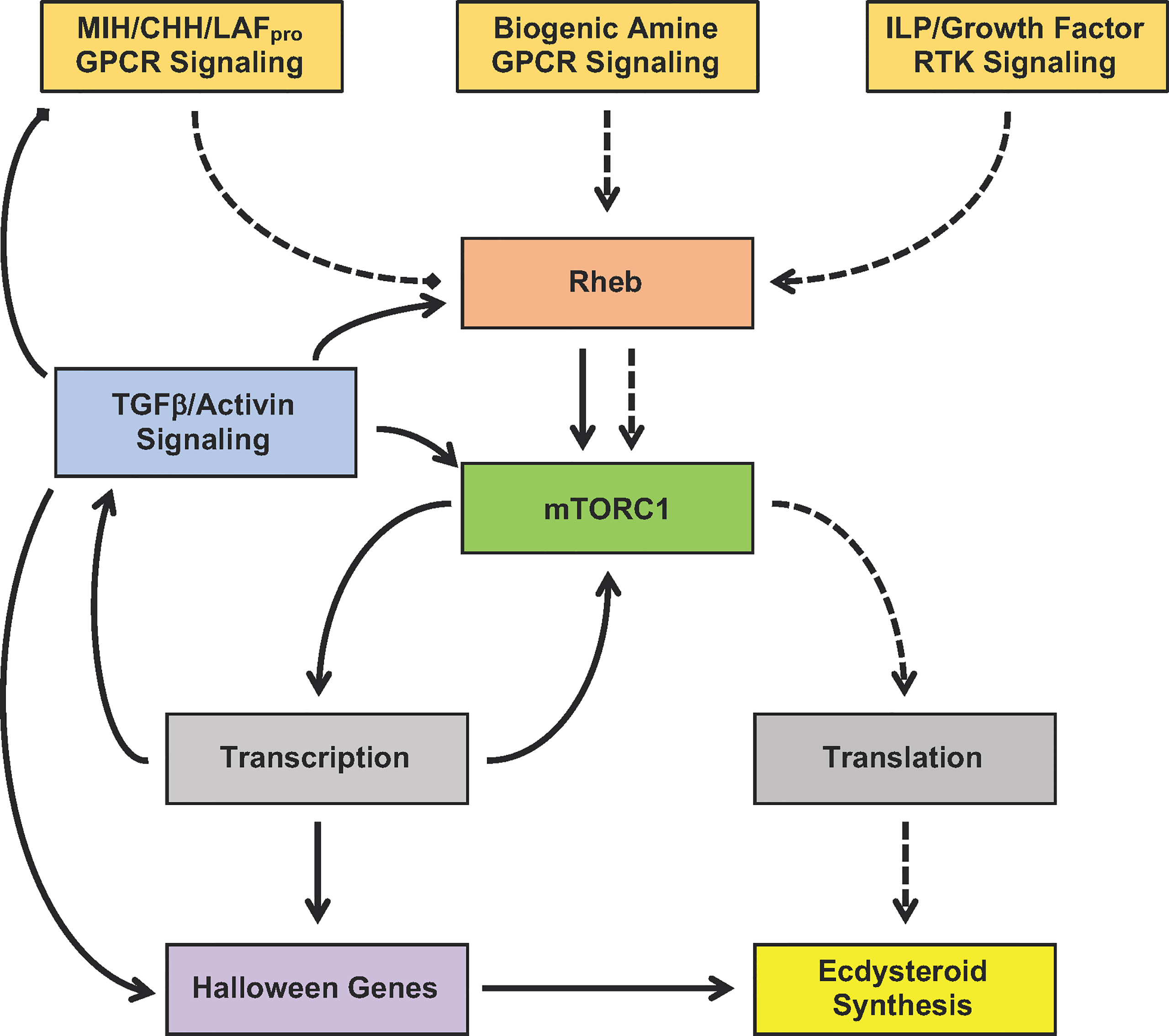
Figure 7 Proposed model for the signaling pathways mediating YO activation (dashed lines) and commitment (solid lines). Signaling pathways converge on mTORC1 by controlling Rheb activity. MIH/CHH/LAFpro GPCR signaling inhibits mTORC1 by inactivating Rheb, while biogenic amine GPCR signaling and ILP/growth factor RTK signaling stimulates mTORC1 by activating Rheb. YO activation during early premolt requires mTORC1-dependent global translation of mRNA to protein, which leads to increased ecdysteroid synthesis. YO commitment involves mTORC1-dependent changes in gene transcription, resulting in up-regulation of TGFβ/Activin, Rheb/mTORC1, and Halloween genes and down-regulation of MIH/CHH/LAFpro GPCR signaling genes.
Although much progress has been made over the last ten years, many questions remain and research efforts should be directed at answering them:
1. What is the identity of the MIH receptor? The evidence indicates that the MIH receptor is a GPCR and several potential candidates have been identified from in silico analysis of YO transcriptomes (Figure 2) (25). Moreover, there is evidence from studies of lobster muscle that the CHH receptor is a membrane receptor guanylyl cyclase (GC-II) (8, 56, 179, 191). A heterologous reporting system in COS-7 cells holds promise as a functional assay for quantifying the specificities of candidate GPCRs and GC-II to recombinant MIH and CHH (124).
2. How does MIH signaling inhibit mTORC1? The current thinking is that MIH inhibition of ecdysteroidogenesis is mediated by PKG (2, 56). The downstream substrates of PKG are unknown. A possible target is TSC (Figure 3), in which phosphorylation stimulates GAP activity, although it is not known if TSC is phosphorylated by PKG (192). Proteomic analysis using liquid chromatography-tandem mass spectrometry now provides the technology to identify and quantify phosphoproteins in the YO in response to rMIH and PKG inhibitors. A similar approach was used to show that NO synthase is phosphorylated in the activated YO (7).
3. What is the identity of LAFpro? LAFpro is a peptide factor produced by secondary limb buds to delay molting (13). As discussed in Section 2.3, the discovery of an Lgr3 GPCR in the YO transcriptome suggests that LAFpro is a Dilp8-like peptide that inhibits ecdysteroidogenesis via the MIH signaling pathway (Figure 3). Like the YO, inhibition of PG ecdysteroid synthesis by Dilp8 is through the activation of NO synthase (73, 79). This suggests that the inhibition of ecdysteroidogenesis by NO/cGMP/PKG is conserved in arthropod molting glands.
4. How does mTORC1 control gene expression? mTORC1 activity alters the mRNA levels of thousands of genes, including those for mTOR, TGFβ, and MIH signaling and ecdysteroidogenesis (16), presumably by altering the activities of transcription factors and co-factors. In D. melanogaster, the transcription factors Krüppel homolog 1 (Kr-h1), seance, ouija board, molting defective, ventral veins lacking, and Knirps are linked to Halloween gene expression (193–195). Kr-h1 is a critical component of the juvenile hormone (JH)/methyl farnesoate (MF) signaling pathway in insects (196–198), and recent studies indicate that Kr-h1 plays a role in crustacean development and reproduction (199–203). The YO expresses Kr-h1 and other MF signaling components, suggesting that it also has a role in molt regulation (204). One potential function is in the down-regulation of Halloween gene expression when the YO transitions to the repressed state in late premolt.
5. What are the gene targets of TGFβ/Activin signaling? TGFβ/Activin drives the transition of the YO from the activated to the committed state. It is hypothesized that Smad transcription factors, activated by Mstn-like factor, up-regulate the expression of commitment genes that determine the phenotypic properties of the committed YO (Figure 1), such as low sensitivity to MIH, CHH, and LAFpro and high ecdysteroid production. Possible targets are Rheb/mTORC1, MIH signaling genes, and Halloween genes (Figure 7). These and other gene targets can be identified by determining the effects of ESA ± SB431542 on the YO transcriptome and proteome.
6. What are the mechanisms mediating the transitions of the YO from the committed to repressed state and from the repressed to basal state? The repressed YO is transcriptionally inactive and has very low ecdysteroid synthesis, leading to low hemolymph ecdysteroid titers during the postmolt stage. It is hypothesized that the ecdysteroid peak at the end of premolt triggers the repressed state, mediated by ecdysteroid receptor (EcR/RXR) and ecdysone-response proteins. Even less is known about what causes the YO to return to basal state at the end of the postmolt stage. Perhaps a signal from the integument, upon completion of exoskeleton synthesis marked by the deposition of the membranous layer, is involved.
7. What is the role of RTKs in regulating ecdysteroidogenesis? Our understanding of RTKs and their ligands in the YO is largely based on inferences from research on the insect PG. It is hypothesized that ILPs and growth factors stimulate ecdysteroidogenesis (Figure 4), but their effects are dampened or nullified by MIH. The YO expresses EGF and FGF, suggesting that both have an autocrine function. In vitro assays can determine the effects of recombinant insulin, EGF, and FGF on YO ecdysteroid synthesis and secretion.
8. Is ecdysteroidogenesis regulated by biogenic amines and neuropeptides other than MIH and CHH? The YO expresses a large number and diversity of GPRCs. Of the 99 GPCRs in the G. lateralis YO, 65 are assigned to known receptors (25). Of particular interest are GPCRs for corazonin, serotonin, and octopamine, which stimulate ecdysteroidogenesis (Figure 3; Sections Corazonin Receptor and Ca2+/Diacylglycerol/Protein Kinase C Signaling). However, other ligands involved in molting, such as ecdysis triggering hormone and Bursicon, should also be investigated.
9. What are the roles of Wnt, Hedgehog, Notch, and Hippo signaling pathways in the YO? These pathways are implicated in controlling ecdysteroidogenesis in the insect PG (74, 97, 98, 205) and are well represented in the YO transcriptome (Table 1). Decapods express a large number of Wnt ligands (206). Wnt4 is implicated in having roles in limb regeneration and the immune response in decapods (207, 208). Gl-Wnt5 and Gl-Wnt7 are expressed at their highest levels in late premolt, suggesting that these ligands are involved with the ecdysteroid peak and transition of the YO to the repressed state (21). This constitutes an entirely new area of research in the coming years.
Transcriptomics and proteomics have revealed the complexities of the regulation of the arthropod molting gland. These approaches have been successfully applied to insect PG and complement functional genetic studies on D. melanogaster (176, 205, 209–215). Transcriptomic and proteomic analysis of the YO has revealed that the PG and YO are more similar than they are different. The PG and YO express the same KEGG signaling pathways. Many of these signaling pathways converge on mTORC1, which plays a central role in regulating ecdysteroid synthesis in both endocrine organs. A second shared property is that TGFβ/Activin signaling alters the ligand sensitivity of the molting gland. In insects, Activins increase the sensitivity of the PG to PTTH in preparation for the large ecdysteroid peak prior to the metamorphic molt, whereas Mstn/Activin decreases the sensitivity of the YO to MIH and CHH in mid- and late premolt. The great diversity in GPCRs indicates that the YO, like the PG, can respond to a variety of ligands, some of which are inhibitory and others are stimulatory. As RTKs stimulate ecdysteroidogenesis in the PG, it seems reasonable to postulate that RTKs have the same function in the YO. The study of insect molting and metamorphosis has informed research on crustaceans, but this does not mean that one can fully understand the control of molting in crustaceans by studying Drosophila. There are fundamental differences in evolutionary history and life history between insects and decapod crustaceans and even between insect orders. The lineages that gave rise to insects and crustaceans have been separated for more than 500 million years, allowing time for the evolution of divergent ligands and signaling pathways to become dominant (216, 217). Unlike insects, most decapod species continue to molt as adults, enabling them to grow to larger sizes. The larger size is a distinct advantage, as one can obtain the amount of YO tissue needed for transcriptomics from two or three individuals (16, 20, 21, 29) and proteomics (24, 218). This allows for increased sample sizes for statistical analysis and potentially more experimental treatments and time points for the study of molting gland function. Thus, crustacean models complement insect models for achieving a broader understanding of how arthropods integrate growth control with external and internal cues.
The author confirms being the sole contributor of this work and has approved it for publication.
This research was supported by grants from the National Science Foundation (IOS-1257732 and IOS-1922701).
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author acknowledges the collaborators, postdoctoral fellows, and students who contributed to the signaling mechanisms in the crustacean molting gland project. The collaborators are: Dr. Ernest S. Chang and Sharon A. Chang, UC Davis Bodega Marine Laboratory; Dr. David Durica, University of Oklahoma; Dr. Wen R. Zhou, Colorado State University; Dr. Lars Tomanek, California Polytechnic University, San Luis Obispo; and Dr. Tomer Ventura, University of the Sunshine Coast. The author thanks Dr. Ventura for comments on the manuscript and preparing Figure 2 and Vanessa Bentley and Sydney McAndrews, Colorado State University, for preparing Table 3. The author thanks Hector C. Horta and Rafael Polanco for collecting G. lateralis and the Ministry of Environment and Natural Resources of the Dominican Republic under Contract for Access to Genetic Resources for Research Purposes DJC-1-2019-01310 and Collection and Export Permit No. VAPS-07979.
1. Mykles DL. Ecdysteroid Metabolism in Crustaceans. J Steroid Biochem Molec Biol (2011) 127:196–203. doi: 10.1016/j.jsbmb.2010.09.001
2. Mykles DL, Chang ES. Hormonal Control of the Crustacean Molting Gland: Insights From Transcriptomics and Proteomics. Gen Comp Endocrinol (2020) 294:113493. doi: 10.1016/j.ygcen.2020.113493
3. Skinner DM. Molting and Regeneration. In: Bliss DE, Mantel LH, editors. The Biology of Crustacea. New York: Academic Press (1985). p. 43–146.
4. Hopkins PM, Das S. Regeneration in Crustaceans. In: Chang ES, Thiel M, editors. The Natural History of the Crustacea: Physiology. Oxford, U.K.: Oxford University Press (2015). p. 168–98.
5. Mykles DL, Medler S. Skeletal Muscle Differentiation, Growth, and Plasticity. In: Chang ES, Thiel M, editors. The Natural History of the Crustacea: Physiology. Oxford, U.K.: Oxford University Press (2015). p. 134–67.
6. Kim HW, Batista LA, Hoppes JL, Lee KJ, Mykles DL. A Crustacean Nitric Oxide Synthase Expressed in Nerve Ganglia, Y-Organ, Gill and Gonad of the Tropical Land Crab, Gecarcinus lateralis. J Exp Biol (2004) 207:2845–57. doi: 10.1242/jeb.01117
7. Lee SG, Mykles DL. Proteomics and Signal Transduction in the Crustacean Molting Gland. Integr Comp Biol (2006) 46:965–77. doi: 10.1093/icb/icl047
8. Lee SG, Kim HW, Mykles DL. Guanylyl Cyclases in the Tropical Land Crab, Gecarcinus lateralis: Cloning of Soluble (NO-Sensitive and -Insensitive) and Membrane Receptor Forms. Comp Biochem Physiol (2007) 2D:332–44. doi: 10.1016/j.cbd.2007.08.001
9. Lee SG, Bader BD, Chang ES, Mykles DL. Effects of Elevated Ecdysteroid on Tissue Expression of Three Guanylyl Cyclases in the Tropical Land Crab Gecarcinus lateralis: Possible Roles of Neuropeptide Signaling in the Molting Gland. J Exp Biol (2007) 210:3245–54. doi: 10.1242/jeb.007740
10. Pitts NL, Mykles DL. Localization and Expression of Molt-Inhibiting Hormone and Nitric Oxide Synthase in the Central Nervous System of the Green Shore Crab, Carcinus maenas, and the Blackback Land Crab, Gecarcinus lateralis. Comp Biochem Physiol (2017) 203A:328–40. doi: 10.1016/j.cbpa.2016.10.012
11. Hopkins PM. The Eyes Have it: A Brief History of Crustacean Neuroendocrinology. Gen Comp Endocrinol (2012) 175:357–66. doi: 10.1016/j.ygcen.2011.12.002
12. Mykles DL. Interactions Between Limb Regeneration and Molting in Decapod Crustaceans. Am Zool (2001) 41:399–406. doi: 10.1093/icb/41.3.399
13. Yu XL, Chang ES, Mykles DL. Characterization of Limb Autotomy Factor-Proecdysis (LAFpro), Isolated From Limb Regenerates, That Suspends Molting in the Land Crab Gecarcinus lateralis. Biol Bull (2002) 202:204–12. doi: 10.2307/1543470
14. Abuhagr AM, MacLea KS, Chang ES, Mykles DL. Mechanistic Target of Rapamycin (mTOR) Signaling Genes in Decapod Crustaceans: Cloning and Tissue Expression of mTOR, Akt, Rheb, and P70 S6 Kinase in the Green Crab, Carcinus maenas, and Blackback Land Crab, Gecarcinus lateralis. Comp Biochem Physiol (2014) 168A:25–39. doi: 10.1016/j.cbpa.2013.11.008
15. Abuhagr AM, MacLea KS, Mudron MR, Chang SA, Chang ES, Mykles DL. Roles of Mechanistic Target of Rapamycin and Transforming Growth Factor-Beta Signaling in the Molting Gland (Y-Organ) of the Blackback Land Crab, Gecarcinus lateralis. Comp Biochem Physiol (2016) 198A:15–21. doi: 10.1016/j.cbpa.2016.03.018
16. Shyamal S, Das S, Guruacharya A, Mykles DL, Durica DS. Transcriptomic Analysis of Crustacean Molting Gland (Y-Organ) Regulation Via the mTOR Signaling Pathway. Sci Rep (2018) 8:7307. doi: 10.1038/s41598-018-25368-x
17. Covi JA, Bader BD, Chang ES, Mykles DL. Molt Cycle Regulation of Protein Synthesis in Skeletal Muscle of the Blackback Land Crab, Gecarcinus lateralis, and the Differential Expression of a Myostatin-Like Factor During Atrophy Induced by Molting or Unweighting. J Exp Biol (2010) 213:172–83. doi: 10.1242/jeb.034389
18. Covi JA, Kim HW, Mykles DL. Expression of Alternatively Spliced Transcripts for a Myostatin-Like Protein in the Blackback Land Crab, Gecarcinus lateralis. Comp Biochem Physiol (2008) 150A:423–30. doi: 10.1016/j.cbpa.2008.04.608
19. Kim HW, Mykles DL, Goetz FW, Roberts SB. Characterization of a Myostatin-Like Gene From the Bay Scallop, Argopecten irradians. Biochim Biophys Acta (2004) 1679:174–9. doi: 10.1016/j.bbaexp.2004.06.005
20. Das S, Pitts NL, Mudron MR, Durica DS, Mykles DL. Transcriptome Analysis of the Molting Gland (Y-Organ) From the Blackback Land Crab, Gecarcinus lateralis. Comp Biochem Physiol (2016) 17D:26–40. doi: 10.1016/j.cbd.2015.11.003
21. Das S, Vraspir L, Zhou W, Durica DS, Mykles DL. Transcriptomic Analysis of Differentially Expressed Genes in the Molting Gland (Y-Organ) of the Blackback Land Crab, Gecarcinus lateralis, During Molt-Cycle Stage Transitions. Comp Biochem Physiol (2018) 28D:37–53. doi: 10.1016/j.cbd.2018.06.001
22. Mykles DL, Burnett KG, Durica DS, Joyce BL, McCarthy FM, Schmidt CJ, et al. Resources and Recommendations for Using Transcriptomics to Address Grand Challenges in Comparative Biology. Integr Comp Biol (2016) 56:1183–91. doi: 10.1093/icb/icw083
23. Das S, Mykles DL. A Comparison of Resources for the Annotation of a De Novo Assembled Transcriptome in the Molting Gland (Y-Organ) of the Blackback Land Crab, Gecarcinus lateralis. Integr Comp Biol (2016) 56:1103–12. doi: 10.1093/icb/icw107
24. Head TB, Mykles DL, Tomanek L. Proteomic Analysis of the Crustacean Molting Gland (Y-Organ) Over the Course of the Molt Cycle. Comp Biochem Physiol (2019) 29:D193–210. doi: 10.1016/j.cbd.2018.11.011
25. Tran NM, Mykles DL, Elizur A, Ventura T. Characterization of G-protein Coupled Receptors From the Blackback Land Crab Gecarcinus lateralis Y Organ Transcriptome Over the Molt Cycle. BMC Genomics (2019) 20:74. doi: 10.1186/s12864-018-5363-9
26. Ventura T, Fitzgibbon QP, Battaglene SC, Elizur A. Redefining Metamorphosis in Spiny Lobsters: Molecular Analysis of the Phyllosoma to Puerulus Transition in Sagmariasus verreauxi. Sci Rep (2015) 5:13537. doi: 10.1038/srep13537.
27. Buckley SJ, Fitzgibbon QP, Smith GG, Ventura T. In Silico Prediction of the G-protein Coupled Receptors Expressed During the Metamorphic Molt of Sagmariasus verreauxi (Crustacea: Decapoda) by Mining Transcriptomic Data: RNA-seq to Repertoire. Gen Comp Endocrinol (2016) 228:111–27. doi: 10.1016/j.ygcen.2016.02.001
28. Christie AE, Yu A. Identification of Peptide Hormones and Their Cognate Receptors in Jasus edwardsii - A Potential Resource for the Development of New Aquaculture Management Strategies for Rock/Spiny Lobsters. Aquaculture (2019) 503:636–62. doi: 10.1016/j.aquaculture.2018.11.059
29. Oliphant A, Alexander JL, Swain MT, Webster SG, Wilcockson DC. Transcriptomic Analysis of Crustacean Neuropeptide Signaling During the Moult Cycle in the Green Shore Crab, Carcinus maenas. BMC Genomics (2018) 19:711. doi: 10.1186/s12864-018-5057-3
30. Nguyen TV, Rotllant GE, Cummins SF, Elizur A, Ventura T. Insights Into Sexual Maturation and Reproduction in the Norway Lobster (Nephrops norvegicus) Via in Silico Prediction and Characterization of Neuropeptides and G Protein-Coupled Receptors. Front Endocrinol (2018) 9:430. doi: 10.3389/fendo.2018.00430
31. Christie AE, Hull JJ, Dickinson PS. Assessment and Comparison of Putative Amine Receptor Complement/Diversity in the Brain and Eyestalk Ganglia of the Lobster, Homarus americanus. Invert Neurosci (2020) 20:7. doi: 10.1007/s10158-020-0239-5
32. Bao CC, Liu F, Yang YA, Lin Q, Ye HH. Identification of Peptides and Their GPCRs in the Peppermint Shrimp Lysmata vittata, a Protandric Simultaneous Hermaphrodite Species. Front Endocrinol (2020) 11:226. doi: 10.3389/fendo.2020.00226
33. Veenstra JA. Similarities Between Decapod and Insect Neuropeptidomes. PeerJ (2016) 4:e2043. doi: 10.7717/peerj.2043
34. Manfrin C, Tom M, De Moro G, Gerdol M, Giulianini PG, Pallavicini A. The Eyestalk Transcriptome of Red Swamp Crayfish Procambarus clarkii. Gene (2015) 557:28–34. doi: 10.1016/j.gene.2014.12.001
35. Wang ZK, Luan S, Meng XH, Cao BX, Luo K, Kong J. Comparative Transcriptomic Characterization of the Eyestalk in Pacific White Shrimp (Litopenaeus vannamei) During Ovarian Maturation. Gen Comp Endocrinol (2019) 274:60–72. doi: 10.1016/j.ygcen.2019.01.002
36. Christie AE, Hull JJ. What Can Transcriptomics Reveal About the Phylogenetic/Structural Conservation, Tissue Localization, and Possible Functions of CNMamide Peptides in Decapod Crustaceans? Gen Comp Endocrinol (2019) 282:113217. doi: 10.1016/j.ygcen.2019.113217
37. Alexander J, Oliphant A, Wilcockson DC, Webster SG. Functional Identification and Characterization of the Diuretic Hormone 31 (DH31) Signaling System in the Green Shore Crab, Carcinus maenas. Front Neurosci (2018) 12:454. doi: 10.3389/fnins.2018.00454
38. Tsutsui N, Kobayashi Y, Izumikawa K, Sakamoto T. Transcriptomic Analysis of the Kuruma Prawn Marsupenaeus japonicus Reveals Possible Peripheral Regulation of the Ovary. Front Endocrinol (2020) 11:541. doi: 10.3389/fendo.2020.00541
39. Kozma MT, Ngo-Vu H, Rump MT, Bobkov YV, Ache BW, Derby CD. Single Cell Transcriptomes Reveal Expression Patterns of Chemoreceptor Genes in Olfactory Sensory Neurons of the Caribbean Spiny Lobster, Panulirus argus. BMC Genomics (2020) 21:649. doi: 10.1186/s12864-020-07034-7
40. Sun SM, Zhu MR, Pan FY, Feng JB, Li JL. Identifying Neuropeptide and G Protein-Coupled Receptors of Juvenile Oriental River Prawn (Macrobrachium nipponense) in Response to Salinity Acclimation. Front Endocrinol (2020) 11:623. doi: 10.3389/fendo.2020.00623
41. Liu JH, Zhou TT, Wang CG, Wang W, Chan SM. Comparative Transcriptomics Reveals Eyestalk Ablation Induced Responses of the Neuroendocrine-Immune System in the Pacific White Shrimp Litopenaeus vannamei. Fish Shellfish Immunol (2020) 106:823–32. doi: 10.1016/j.fsi.2020.08.029
42. Ventura T, Chandler JC, Nguyen TV, Hyde CMJ, Elizur A, Fitzgibbon QP, et al. Multi-Tissue Transcriptome Analysis Identifies Key Sexual Development-Related Genes of the Ornate Spiny Lobster (Panulirus onatus). Genes (2020) 11:1150. doi: 10.3390/genes11101150
43. Alexander JL, Oliphant A, Wilcockson DC, Brendler-Spaeth T, Dircksen H, Webster SG. Pigment Dispersing Factors and Their Cognate Receptors in a Crustacean Model, With New Insights Into Distinct Neurons and Their Functions. Front Neurosci (2020) 14:595648. doi: 10.3389/fnins.2020.595648
44. Nguyen TV, Ryan LW, Nocillado J, Le Groumellec M, Elizur A, Ventura T. Transcriptomic Changes Across Vitellogenesis in the Black Tiger Prawn (Penaeus monodon), Neuropeptides and G Protein-Coupled Receptors Repertoire Curation. Gen Comp Endocrinol (2020) 298:113585. doi: 10.1016/j.ygcen.2020.113585
45. Alexander JL, Oliphant A, Wilcockson DC, Audsley N, Down RE, Lafont R, et al. Functional Characterization and Signaling Systems of Corazonin and Red Pigment Concentrating Hormone in the Green Shore Crab, Carcinus maenas. Front Neurosci (2018) 11:752. doi: 10.3389/fnins.2017.00752
46. Webster SG, Keller R, Dircksen H. The CHH-superfamily of Multifunctional Peptide Hormones Controlling Crustacean Metabolism, Osmoregulation, Moulting, and Reproduction. Gen Comp Endocrinol (2012) 175:217–33. doi: 10.1016/j.ygcen.2011.11.035
47. Katayama H, Ohira T, Nagasawa H. Crustacean Peptide Hormones: Structure, Gene Expression and Function. Aqua-BioSci Monogr (2013) 6:49–90. doi: 10.5047/absm.2013.00602.0049
48. Toullec JY, Corre E, Mandon P, Gonzalez-Aravena M, Ollivaux C, Lee CY. Characterization of the Neuropeptidome of a Southern Ocean Decapod, the Antarctic Shrimp Chorismus antarcticus: Focusing on a New Decapod ITP-like Peptide Belonging to the CHH Peptide Family. Gen Comp Endocrinol (2017) 252:60–78. doi: 10.1016/j.ygcen.2017.07.015
49. Katayama H. Structure-Activity Relationship of Crustacean Peptide Hormones. Biosci Biotechnol Biochem (2016) 80:633–41. doi: 10.1080/09168451.2015.1116932
50. Chen HY, Toullec JY, Lee CY. The Crustacean Hyperglycemic Hormone Superfamily: Progress Made in the Past Decade. Front Endocrinol (2020) 11:578958. doi: 10.3389/fendo.2020.578958
51. Katayama H, Nagata K, Ohira T, Yumoto F, Tanokura M, Nagasawa H. The Solution Structure of Molt-Inhibiting Hormone From the Kuruma Prawn Marsupenaeus japonicus. J Biol Chem (2003) 278:9620–3. doi: 10.1074/jbc.M212962200
52. Katayama H, Ohira T, Nagata S, Nagasawa H. Structure-Activity Relationship of Crustacean Molt-Inhibiting Hormone From the Kuruma Prawn Marsupenaeus japonicus. Biochemistry (2004) 43:9629–35. doi: 10.1021/bi049433v
53. Webster SG. High-Affinity Binding of Putative Moult-Inhibiting Hormone (MIH) and Crustacean Hyperglycaemic Hormone (CHH) to Membrane-Bound Receptors on the Y-Organ of the Shore Crab Carcinus maenas. Proc R Soc Lond [Biol] (1993) 251:53–9. doi: 10.1098/rspb.1993.0008
54. Chung JS, Webster SG. Moult Cycle-Related Changes in Biological Activity of Moult-Inhibiting Hormone (MIH) and Crustacean Hyperglycaemic Hormone (CHH) in the Crab, Carcinus maenas. From Target to Transcript. Eur J Biochem (2003) 270:3280–8. doi: 10.1046/j.1432-1033.2003.03720.x
55. Webster SG. Endocrinology of Molting. In: Chang ES, Thiel M, editors. The Natural History of the Crustacea: Physiology. Oxford: Oxford Press (2015). p. 1–35.
56. Covi JA, Chang ES, Mykles DL. Conserved Role of Cyclic Nucleotides in the Regulation of Ecdysteroidogenesis by the Crustacean Molting Gland. Comp Biochem Physiol (2009) 152A:470–7. doi: 10.1016/j.cbpa.2008.12.005
57. Nagai C, Mabashi-Asazuma H, Nagasawa H, Nagata S. Identification and Characterization of Receptors for Ion Transport Peptide (ITP) and ITP-like (ITPL) in the Silkworm Bombyx mori. J Biol Chem (2014) 289:32166–77. doi: 10.1074/jbc.M114.590646
58. Veenstra JA. The Power of Next-Generation Sequencing as Illustrated by the Neuropeptidome of the Crayfish Procambarus clarkii. Gen Comp Endocrinol (2015) 224:84–95. doi: 10.1016/j.ygcen.2015.06.013
59. Nassel DR, Zandawala M. Recent Advances in Neuropeptide Signaling in Drosophila, From Genes to Physiology and Behavior. Progr Neurobiol (2019) 179:101607. doi: 10.1016/j.pneurobio.2019.02.003
60. Christie AE. Crustacean Neuroendocrine Systems and Their Signaling Agents. Cell Tiss Res (2011) 345:41–67. doi: 10.1007/s00441-011-1183-9
61. Imura E, Shimada-Niwa Y, Nishimura T, Huckesfeld S, Schlegel P, Ohhara Y, et al. The Corazonin-PTTH Neuronal Axis Controls Systemic Body Growth by Regulating Basal Ecdysteroid Biosynthesis in Drosophila melanogaster. Curr Biol (2020) 30:2156–65. doi: 10.1016/j.cub.2020.03.050
62. Mirth CK, Shingleton AW. Integrating Body and Organ Size in Drosophila: Recent Advances and Outstanding Problems. Front Endocrinol (2012) 3:49. doi: 10.3389/fendo.2012.00049
63. Holland CA, Skinner DM. Interactions Between Molting and Regeneration in the Land Crab. Biol Bull (1976) 150:222–40. doi: 10.2307/1540470
64. Andersen DS, Colombani J, Leopold P. Coordination of Organ Growth: Principles and Outstanding Questions From the World of Insects. Trends Cell Biol (2013) 23:336–44. doi: 10.1016/j.tcb.2013.03.005
65. Hackney JF, Cherbas P. Injury Response Checkpoint and Developmental Timing in Insects. Fly (2014) 8:226–31. doi: 10.1080/19336934.2015.1034913
66. Rosero MA, Abdon B, Silva NJ, Larios BC, Zavaleta JA, Makunts T, et al. Divergent Mechanisms for Regulating Growth and Development After Imaginal Disc Damage in the Tobacco Hornworm, Manduca sexta. J Exp Biol (2019) 222:200352. doi: 10.1242/jeb.200352
67. McCarthy JF, Skinner DM. Interactions Between Molting and Regeneration in Land Crab Gecarcinus lateralis. 3. Interruption of Proecdysis by Autotomy of Partially Regenerated Limbs in Land Crab, Gecarcinus lateralis. Dev Biol (1977) 61:299–310. doi: 10.1016/0012-1606(77)90300-1
68. Colombani J, Andersen DS, Leopold P. Secreted Peptide Dilp8 Coordinates Drosophila Tissue Growth With Developmental Timing. Science (2012) 336:582–5. doi: 10.1126/science.1216689
69. Garelli A, Gontijo AM, Miguela V, Caparros E, Dominguez M. Imaginal Discs Secrete Insulin-Like Peptide 8 to Mediate Plasticity of Growth and Maturation. Science (2012) 336:579–82. doi: 10.1126/science.1216735
70. Hariharan IK. How Growth Abnormalities Delay “Puberty” in Drosophila. Sci Signal (2012) 5:pe27. doi: 10.1126/scisignal.2003238
71. Gontijo AM, Garelli A. The Biology and Evolution of the Dilp8-Lgr3 Pathway: A Relaxin-Like Pathway Coupling Tissue Growth and Developmental Timing Control. Mech Dev (2018) 154:44–50. doi: 10.1016/j.mod.2018.04.005
72. Vallejo DM, Juarez-Carreno S, Bolivar J, Morante J, Dominguez M. A Brain Circuit That Synchronizes Growth and Maturation Revealed Through Dilp8 Binding to Lgr3. Science (2015) 350:aac6767. doi: 10.1126/science.aac6767
73. Jaszczak JS, Wolpe JB, Bhandari R, Jaszczak RG, Halme A. Growth Coordination During Drosophila melanogaster Imaginal Disc Regeneration Is Mediated by Signaling Through the Relaxin Receptor Lgr3 in the Prothoracic Gland. Genetics (2016) 204:703–9. doi: 10.1534/genetics.116.193706
74. Delanoue R, Romero NM. Growth and Maturation in Development: A Fly’s Perspective. Int J Molec Sci (2020) 21:1260. doi: 10.3390/ijms21041260
75. Semaniuk U, Piskovatska V, Strilbytska O, Strutynska T, Burdyliuk N, Vaiserman A, et al. Drosophila Insulin-Like Peptides: From Expression to Functions - a Review. Entomol Exp Applicata (2020), eea12981. doi: 10.1111/eea.12981
76. Mele S, Johnson TK. Receptor Tyrosine Kinases in Development: Insights From Drosophila. Int J Molec Sci (2020) 21:188. doi: 10.3390/ijms21010188
77. Li D, Chen X, Zhu F, Chen K. Insulin-Like Peptides in Model Insects. Invert Survival J (2020) 17:186–95.
78. Lin XY, Smagghe G. Roles of the Insulin Signaling Pathway in Insect Development and Organ Growth. Peptides (2019) 122:169923. doi: 10.1016/j.peptides.2018.02.001
79. Jaszczak JS, Wolpe JB, Dao AQ, Halme A. Nitric Oxide Synthase Regulates Growth Coordination During Drosophila melanogaster Imaginal Disc Regeneration. Genetics (2015) 200:1219–28. doi: 10.1534/genetics.115.178053
80. DeLalio LJ, Dion SM, Bootes AM, Smith WA. Direct Effects of Hypoxia and Nitric Oxide on Ecdysone Secretion by Insect Prothoracic Glands. J Insect Physiol (2015) 76:56–66. doi: 10.1016/j.jinsphys.2015.02.009
81. Covi J, Gomez A, S. Chang K, Chang E, Mykles D. Repression of Y-Organ Ecdysteroidogenesis by Cyclic Nucleotides and Agonists of NO-Sensitive Guanylyl Cyclase. In: Morris S, Vosloo A, editors. 4th Meeting of Comparative Physiologists & Biochemists in Africa - Mara 2008 - “Molecules to Migration: The Pressures of Life”. Bologna, Italy: Monduzzi Editore International (2008). p. 37–46.
82. Mykles DL, Adams ME, Gade G, Lange AB, Marco HG, Orchard I. Neuropeptide Action in Insects and Crustaceans. Physiol Biochem Zool (2010) 83:836–46. doi: 10.1086/648470
83. Chandler JC, Gandhi NS, Mancera RL, Smith G, Elizur A, Ventura T. Understanding Insulin Endocrinology in Decapod Crustacea: Molecular Modelling Characterization of an Insulin-Binding Protein and Insulin-Like Peptides in the Eastern Spiny Lobster, Sagmariasus verreauxi. Int J Molec Sci (2017) 18:1832. doi: 10.3390/ijms18091832
84. Spaziani E, Mattson MP, Wang WNL, McDougall HE. Signaling Pathways for Ecdysteroid Hormone Synthesis in Crustacean Y-Organs. Am Zool (1999) 39:496–512. doi: 10.1093/icb/39.3.496
85. Mattson MP, Spaziani E. Demonstration of Protein Kinase C Activity in Crustacean Y-Organs. and Partial Definition of Its Role in Regulation of Ecdysteroidogenesis. Mol Cell Endocrinol (1987) 49:159–71. doi: 10.1016/0303-7207(87)90209-7
86. Spaziani E, Jegla TC, Wang WL, Booth JA, Connolly SM, Conrad CC, et al. Further Studies on Signaling Pathways for Ecdysteroidogenesis in Crustacean Y-Organs. Am Zool (2001) 41:418–29. doi: 10.1093/icb/41.3.418
87. Mattson MP. New Insights Into Neuroendocrine Regulation of the Crustacean Molt Cycle. Zool Sci (1986) 3:733–44.
88. Manfrin C, Pallavicini A, Battistella S, Lorenzon S, Giulianini PG. Crustacean Immunity: The Modulation of Stress Responses. In: Ballarin L, Cammarata M, editors. Lessons in Immunity: From Single-Cell Organisms to Mammals. (2016). Elsevier: Amsterdam. doi: 10.1016/B978-0-12-803252-7.00008-4
89. Jung H, Lyons RE, Hurwood DA, Mather PB. Genes and Growth Performance in Crustacean Species: A Review of Relevant Genomic Studies in Crustaceans and Other Taxa. Rev Aquacult (2013) 5:77–110. doi: 10.1111/raq.12005
90. Huber R, Orzeszyna M, Pokorny N, Kravitz EA. Biogenic Amines and Aggression: Experimental Approaches in Crustaceans. Brain Behav Evol (1997) 50:60–8. doi: 10.1159/000113355
91. Nakeim J, Kornthong N, Saetan J, Duangprom S, Sobhon P, Sretarugsa P. Presence of Serotonin and Its Receptor in the Central Nervous System and Ovary and Molecular Cloning of the Novel Crab Serotonin Receptor of the Blue Swimming Crab, Portunus pelagicus. Acta Histochem (2020) 122:151457. doi: 10.1016/j.acthis.2019.151457
92. Wang B, Yang JR, Gao CC, Hao T, Li JJ, Sun JS. Reconstruction of Eriocheir sinensis Y-Organ Genome-Scale Metabolic Network and Differential Analysis After Eyestalk Ablation. Front Genet (2020) 11:532492. doi: 10.3389/fgene.2020.532492
93. Jayasankar V, Tomy S, Wilder MN. Insights on Molecular Mechanisms of Ovarian Development in Decapod Crustacea: Focus on Vitellogenesis-Stimulating Factors and Pathways. Front Endocrinol (2020) 11:577925. doi: 10.3389/fendo.2020.577925
94. Fingerman M. Crustacean Endocrinology: A Retrospective, Prospective, and Introspective Analysis. Physiol Zool (1997) 70:257–69. doi: 10.1086/639593
95. Harlioglu MM, Farhadi A, Harlioglu AG. Roles of Neurotransmitters in Decapod Reproduction. Thalassas (2020) 36:633–9. doi: 10.1007/s41208-020-00202-2
96. Girish BP, Swetha C, Reddy PS. Serotonin Induces Ecdysteroidogenesis and Methyl Farnesoate Synthesis in the Mud Crab, Scylla serrata. Biochem Biophys Res Commun (2017) 490:1340–5. doi: 10.1016/j.bbrc.2017.07.025
97. Pan X, Connacher RP, O’Connor MB. Control of the Insect Metamorphic Transition by Ecdysteroid Production and Secretion. Curr Opin Insect Sci (2021) 43:11–20. doi: 10.1016/j.cois.2020.09.004
98. Texada MJ, Koyama T, Rewitz K. Regulation of Body Size and Growth Control. Genetics (2020) 216:269–313. doi: 10.1534/genetics.120.303095
99. Shimada-Niwa Y, Niwa R. Serotonergic Neurons Respond to Nutrients and Regulate the Timing of Steroid Hormone Biosynthesis in Drosophila. Nat Commun (2014) 5:5778. doi: 10.1038/ncomms6778
100. Ohhara Y, Shimada-Niwa Y, Niwa R, Kayashima Y, Hayashi Y, Akagi K, et al. Autocrine Regulation of Ecdysone Synthesis by Beta 3-Octopamine Receptor in the Prothoracic Gland is Essential for Drosophila Metamorphosis. Proc Natl Acad Sci USA (2015) 112:1452–7. doi: 10.1073/pnas.1414966112
101. De Meyts P. The Insulin Receptor and Its Signal Transduction Network. In: Feingold KR, Anawalt B, Boyce A, et al. editors. Endotext [Internet]. South Dartmouth, MA USA: MDText.com, Inc. (2016). p. 1–35.
102. Koyama T, Texada MJ, Halberg KA, Rewitz K. Metabolism and Growth Adaptation to Environmental Conditions in Drosophila. Cell Molec Life Sci (2020) 77:4523–51. doi: 10.1007/s00018-020-03547-2
103. Malita A, Rewitz K. Interorgan Communication in the Control of Metamorphosis. Curr Opin Insect Sci (2021) 43:54–62. doi: 10.1016/j.cois.2020.10.005
104. Cobham AE, Mirth CK. The Development of Body and Organ Shape. BMC Zool (2020) 5:14. doi: 10.1186/s40850-020-00063-5
105. Marchal E, Vandersmissen HP, Badisco L, Van de Velde S, Verlinden H, Iga M, et al. Control of Ecdysteroidogenesis in Prothoracic Glands of Insects: A Review. Peptides (2010) 31:506–19. doi: 10.1016/j.peptides.2009.08.020
106. Kannangara JR, Mirth CK, Warr CG. Regulation of Ecdysone Production in Drosophila by Neuropeptides and Peptide Hormones. Open Biol (2021) 11:200373. doi: 10.1098/rsob.200373
107. Teleman AA. Molecular Mechanisms of Metabolic Regulation by Insulin in Drosophila. Biochem J (2010) 425:13–26. doi: 10.1042/BJ20091181
108. Levy T, Sagi A. The “IAG-Switch”-a Key Controlling Element in Decapod Crustacean Sex Differentiation. Front Endocrinol (2020) 11:651. doi: 10.3389/fendo.2020.00651
109. Sun R, Li YH. A Sex-Reversing Factor: Insulin-like Androgenic Gland Hormone in Decapods. Rev Aquacult (2021), raq12525. doi: 10.1111/raq.12525
110. Nguyen TV, Cummins SF, Elizur A, Ventura T. Transcriptomic Characterization and Curation of Candidate Neuropeptides Regulating Reproduction in the Eyestalk Ganglia of the Australian Crayfish, Cherax quadricarinatus. Sci Rep (2016) 6:38658. doi: 10.1038/srep38658
111. Christie AE, Pascual MG. Peptidergic Signaling in the Crab Cancer borealis: Tapping the Power of Transcriptomics for Neuropeptidome Expansion. Gen Comp Endocrinol (2016) 237:53–67. doi: 10.1016/j.ygcen.2016.08.002
112. Liu X, Ma KY, Liu ZQ, Feng JB, Ye BQ, Qiu GF. Transcriptome Analysis of the Brain of the Chinese Mitten Crab, Eriocheir sinensis, for Neuropeptide Abundance Profiles During Ovarian Development. Anim Reprod Sci (2019) 201:63–70. doi: 10.1016/j.anireprosci.2018.12.010
113. Chandler JC, Aizen J, Elizur A, Hollander-Cohen L, Battaglene SC, Ventura T. Discovery of a Novel Insulin-Like Peptide and Insulin Binding Proteins in the Eastern Rock Lobster Sagmariasus verreauxi. Gen Comp Endocrinol (2015) 215:76–87. doi: 10.1016/j.ygcen.2014.08.018
114. Christie AE. Identification of Putative Neuropeptidergic Signaling Systems in the Spiny Lobster, Panulirus argus. Invert Neurosci (2020) 20:2. doi: 10.1007/s10158-020-0235-9
115. Chandler JC, Elizur A, Ventura T. The Decapod Researcher’s Guide to the Galaxy of Sex Determination. Hydrobiologia (2018) 825:61–80. doi: 10.1007/s10750-017-3452-4
116. Jiang QH, Zheng HK, Zheng L, Wang YJ, Wang MG, Xie X, et al. Molecular Characterization of the Insulin-Like Androgenic Gland Hormone in the Swimming Crab, Portunus trituberculatus, and Its Involvement in the Insulin Signaling System. Front Endocrinol (2020) 11:585. doi: 10.3389/fendo.2020.00585
117. Li FJ, Zhang SY, Fu CP, Li TT, Cui XY. Molecular and Functional Analysis of the Insulin-Like Peptides Gene in the Oriental River Prawn Macrobrachium nipponense. Gen Comp Endocrinol (2019) 280:209–14. doi: 10.1016/j.ygcen.2019.05.006
118. Benjamin H, Joanne B, Pierre G. Divergent Evolution and Glade-Specific Duplications of the Insulin-Like Receptor in Malacostracan Crustaceans. Gen Comp Endocrinol (2018) 268:34–9. doi: 10.1016/j.ygcen.2018.07.013
119. Chuang N-N, Wang P-C. Characterization of Insulin Receptor From the Muscle of the Shrimp Penaeus japonicus (Crustacea: Decapoda). Comp Biochem Physiol (1994) 108C:289–97. doi: 10.1016/0742-8413(94)00031-5
120. Kucharski LC, Capp E, Chittó ALF, Trapp M, Da Silva RSM, Marques M. Insulin Signaling: Tyrosine Kinase Activity in the Crab Chasmagnathus granulata Gills. J Exp Zool (1999) 283:91–4. doi: 10.1002/(SICI)1097-010X(19990101)283:1<91::AID-JEZ10>3.0.CO;2-F
121. Lin CL, Wang PC, Chuang NN. Specific Phosphorylation of Membrane Proteins of Mr 44,000 and Mr 32,000 by the Autophosphorylated Insulin Receptor From the Hepatopancreas of the Shrimp Penaeus monodon (Crustacea, Decapoda). J Exp Zool (1993) 267:113–9. doi: 10.1002/jez.1402670204
122. Sharabi O, Manor R, Weil S, Aflalo ED, Lezer Y, Levy T, et al. Identification and Characterization of an Insulin-Like Receptor Involved in Crustacean Reproduction. Endocrinology (2016) 157:928–41. doi: 10.1210/en.2015-1391
123. Guo Q, Li SH, Lv XJ, Xiang JH, Sagi A, Manor R, et al. A Putative Insulin-Like Androgenic Gland Hormone Receptor Gene Specifically Expressed in Male Chinese Shrimp. Endocrinology (2018) 159:2173–85. doi: 10.1210/en.2017-03253
124. Aizen J, Chandler JC, Fitzgibbon QP, Sagi A, Battaglene SC, Elizur A, et al. Production of Recombinant Insulin-Like Androgenic Gland Hormones From Three Decapod Species: In Vitro Testicular Phosphorylation and Activation of a Newly Identified Tyrosine Kinase Receptor From the Eastern Spiny Lobster, Sagmariasus verreauxi. Gen Comp Endocrinol (2016) 229:8–18. doi: 10.1016/j.ygcen.2016.02.013
125. Huang XS, Feng BY, Huang HY, Ye HH. In Vitro Stimulation of Vitellogenin Expression by Insulin in the Mud Crab, Scylla paramamosain, Mediated Through PI3K/Akt/TOR Pathway. Gen Comp Endocrinol (2017) 250:175–80. doi: 10.1016/j.ygcen.2017.06.013
126. Gu SH, Chen CH, Hsieh YC, Lin PL, Young SC. Modulatory Effects of Bombyxin on Ecdysteroidogenesis in Bombyx mori Prothoracic Glands. J Insect Physiol (2015) 72:61–9. doi: 10.1016/j.jinsphys.2014.11.007
127. Gu SH, Young SC, Lin JL, Lin PL. Involvement of PI3K/Akt Signaling in PTTH-stimulated Ecdysteroidogenesis by Prothoracic Glands of the Silkworm, Bombyx mori. Insect Biochem Molec Biol (2011) 41:197–202. doi: 10.1016/j.ibmb.2010.12.004
128. Gu SH, Yeh WL, Young SC, Lin PL, Li S. TOR Signaling Is Involved in PTTH-stimulated Ecdysteroidogenesis by Prothoracic Glands in the Silkworm, Bombyx mori. Insect Biochem Molec Biol (2012) 42:296–303. doi: 10.1016/j.ibmb.2011.12.010
129. Gu SH, Hsieh YC, Lin PL. Signaling Involved in PTTH-stimulated 4e-BP Phosphorylation in Prothoracic Gland Cells of Bombyx mori. J Insect Physiol (2017) 96:1–8. doi: 10.1016/j.jinsphys.2016.10.007
130. Torres AS, Holz MK. Unraveling the Multifaceted Nature of the Nuclear Function of mTOR. Biochim Biophys Acta Molec Cell Res (2021) 1868:118907. doi: 10.1016/j.bbamcr.2020.118907
131. Liu GY, Sabatini DM. mTOR at the Nexus of Nutrition, Growth, Ageing and Disease. Nat Rev Molec Cell Biol (2019) 29:s41580–019-0199-y.doi: 10.1038/s41580-019-0199-y
132. Wee P, Wang ZX. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers (2017) 9:52. doi: 10.3390/cancers9050052
133. Skelly J, Pushparajan C, Duncan EJ, Dearden PK. Evolution of the Torso Activation Cassette, a Pathway Required for Terminal Patterning and Moulting. Insect Molec Biol (2019) 28:392–408. doi: 10.1111/imb.12560
134. Cruz J, Martin D, Franch-Marro X. EGFR Signaling Is a Major Regulator of Ecdysone Biosynthesis in the Drosophila Prothoracic Gland. Curr Biol (2020) 30:1547–154. doi: 10.1016/j.cub.2020.01.092
135. Fusco MA, Wajsenzon IJR, de Carvalho SL, da Silva RT, Einicker-Lamas M, Cavalcante LA, et al. Vascular Endothelial Growth Factor-Like and Its Receptor in a Crustacean Optic Ganglia: A Role in Neuronal Differentiation? Biochem Biophys Res Commun (2014) 447:299–303. doi: 10.1016/j.bbrc.2014.03.137
136. Jaramillo ML, Guzman F, Paese CLB, Margis R, Nazari EM, Ammar D, et al. Exploring Developmental Gene Toolkit and Associated Pathways in a Potential New Model Crustacean Using Transcriptomic Analysis. Dev Genes Evol (2016) 226:325–37. doi: 10.1007/s00427-016-0551-6
137. Wang YH, Xiu YJ, Bi KR, Ou JT, Gu W, Wang W, et al. Integrated Analysis of mRNA-seq in the Haemocytes of Eriocheir sinensis in Response to Spiroplasma eriocheiris Infection. Fish Shellfish Immunol (2017) 68:289–98. doi: 10.1016/j.fsi.2017.07.036
138. Wei JK, Zhang XJ, Yu Y, Huang H, Li FH, Xiang JH. Comparative Transcriptomic Characterization of the Early Development in Pacific White Shrimp Litopenaeus vannamei. PloS One (2014) 9:e106201. doi: 10.1371/journal.pone.0106201
139. Sharabi O, Ventura T, Manor R, Aflalo ED, Sagi A. Epidermal Growth Factor Receptor in the Prawn Macrobrachium rosenbergii: Function and Putative Signaling Cascade. Endocrinology (2013) 154:3188–96. doi: 10.1210/en.2013-1259
140. Lu B, Jiang QL, Liu A, Huang HY, Ye HH. Stimulatory Roles of Epidermal Growth Factor Receptor (EGFR) in Ovarian Development of Mud Crab Scylla paramamosain. Gen Comp Endocrinol (2020) 299:113616. doi: 10.1016/j.ygcen.2020.113616
141. Ark AV, Cao JC, Li XH. TGF-Beta Receptors: In and Beyond TGF-Beta Signaling. Cell Signal (2018) 52:112–20. doi: 10.1016/j.cellsig.2018.09.002
142. Teicher BA. TGF Beta-Directed Therapeutics: 2020. Pharmacol Ther (2021) 217:107666. doi: 10.1016/j.pharmthera.2020.107666
143. Tzavlaki K, Moustakas A. TGF-Beta Signaling. Biomolecules (2020) 10:487. doi: 10.3390/biom10030487
144. Budi EH, Duan D, Derynck R. Transforming Growth Factor-Beta Receptors and Smads: Regulatory Complexity and Functional Versatility. Trends Cell Biol (2017) 27:658–72. doi: 10.1016/j.tcb.2017.04.005
145. Derynck R, Budi EH. Specificity, Versatility, and Control of TGF-beta Family Signaling. Sci Signaling (2019) 12:eaav5183. doi: 10.1126/scisignal.aav5183
146. Gipson GR, Goebel EJ, Hart KN, Kappes EC, Kattamuri C, McCoy JC, et al. Structural Perspective of BMP Ligands and Signaling. Bone (2020) 140:115549. doi: 10.1016/j.bone.2020.115549
147. Nickel J, ten Dijke P, Mueller TD. TGF-Beta Family Co-Receptor Function and Signaling. Acta Biochim Biophys Sin (2018) 50:12–36. doi: 10.1093/abbs/gmx126
148. Upadhyay A, Moss-Taylor L, Kim MJ, Ghosh AC, O’Connor MB. TGF-Beta Family Signaling in Drosophila. Cold Spr Harb Perspect Biol (2017) 9. doi: 10.1101/cshperspect.a031963
149. Suh J, Lee YS. Similar Sequences But Dissimilar Biological Functions of GDF11 and Myostatin. Exp Molec Med (2020) 52:1673–93. doi: 10.1038/s12276-020-00516-4
150. Gibbens YY, Warren JT, Gilbert LI, O’Connor MB. Neuroendocrine Regulation of Drosophila Metamorphosis Requires TGF Beta/Activin Signaling. Development (2011) 138:2693–703. doi: 10.1242/dev.063412
151. Kamsoi O, Belles X. Myoglianin Triggers the Premetamorphosis Stage in Hemimetabolan Insects. FASEB J (2019) 33:3659–69. doi: 10.1096/fj.201801511R
152. Nakatsuji T, Lee CY, Watson RD. Crustacean Molt-Inhibiting Hormone: Structure, Function, and Cellular Mode of Action. Comp Biochem Physiol (2009) 152A:139–48. doi: 10.1016/j.cbpa.2008.10.012
153. Zhou YL, Li B, Xu YP, Wang LZ, Gu WB, Liu ZP, et al. The Activin-like Ligand Dawdle Regulates Innate Immune Responses Through Modulating NF-Kappa B Signaling in Mud Crab Scylla paramamosain. Dev Comp Immunol (2019) 101:103450. doi: 10.1016/j.dci.2019.103450
154. Yan YJ, Lu X, Kong J, Meng XH, Luan S, Dai P, et al. Molecular Characterization of Myostatin and Its Inhibitory Function on Myogenesis and Muscle Growth in Chinese Shrimp, Fenneropenaeus chinensis. Gene (2020) 758:144986. doi: 10.1016/j.gene.2020.144986
155. Zhuo RQ, Zhou TT, Yang SP, Chan SMF. Characterization of a Molt-Related Myostatin Gene (FmMstn) From the Banana Shrimp Fenneropenaeus merguiensis. Gen Comp Endocrinol (2017) 248:55–68. doi: 10.1016/j.ygcen.2017.03.010
156. Sarasvathi PE, Bhassu S, Maningas MBB, Othman RY. Myostatin: A Potential Growth-Regulating Gene in Giant River Prawn, Macrobrachium rosenbergii. J World Aquacult Soc (2015) 46:624–34. doi: 10.1111/jwas.12238
157. MacLea KS, Covi JA, Kim HW, Chao E, Medler S, Chang ES, et al. Myostatin From the American Lobster, Homarus Americanus: Cloning and Effects of Molting on Expression in Skeletal Muscles. Comp Biochem Physiol (2010) 157A:328–37. doi: 10.1016/j.cbpa.2010.07.024
158. Qian ZY, Mi X, Wang XZ, He SL, Liu YJ, Hou FJ, et al. cDNA Cloning and Expression Analysis of Myostatin/GDF11 in Shrimp, Litopenaeus vannamei. Comp Biochem Physiol (2013) 165A:30–9. doi: 10.1016/j.cbpa.2013.02.001
159. De Santis C, Wade NM, Jerry DR, Preston NP, Glencross BD, Sellars MJ. Growing Backwards: An Inverted Role for the Shrimp Ortholog of Vertebrate Myostatin and GDF11. J Exp Biol (2011) 214:2671–7. doi: 10.1242/jeb.056374
160. Kim K-S, Jeon J-M, Kim H-W. A Myostatin-Like Gene Expressed Highly in the Muscle Tissue of Chinese Mitten Crab, Eriocheir sinensis. Fish Aqua Sci (2009) 12:185–93. doi: 10.5657/fas.2009.12.3.185
161. Shen WY, Ren G, Zhu YR, Zhang XD. Characterization of MSTN/GDF11 Gene From Shrimp Macrobrachium nipponense and Its Expression Profiles During Molt Cycle and After Eyestalk Ablation. Genes Genomics (2015) 37:441–9. doi: 10.1007/s13258-015-0273-6
162. Kong J, Yan YJ, Lu X, Luan S, Meng XH, Dai P, et al. Integrative Phenotypic and Gene Expression Data Identify Myostatin as a Muscle Growth Inhibitor in Chinese Shrimp Fenneropenaeus Chinensis. Sci Rep (2020) 10:5985. doi: 10.1038/s41598-020-61382-8
163. MacLea KS, Abuhagr AM, Pitts NL, Covi JA, Bader BD, Chang ES, et al. Rheb, an Activator of Target of Rapamycin, in the Blackback Land Crab, Gecarcinus lateralis: Cloning and Effects of Molting and Unweighting on Expression in Skeletal Muscle. J Exp Biol (2012) 215:590–604. doi: 10.1242/jeb.062869
164. Lee JH, Momani J, Kim YM, Kang CK, Choi JH, Baek HJ, et al. Effective RNA-silencing Strategy of Lv-MSTN/GDF11 Gene and Its Effects on the Growth in Shrimp, Litopenaeus vannamei. Comp Biochem Physiol (2015) 179B:9–16. doi: 10.1016/j.cbpb.2014.09.005
165. Yue WC, Yang H, Chen YP, Wang J, Chen XW, Hou X, et al. RNA Interference Provides Insights Into the Multi-Functional Profiles of Es-MSTN Gene in Eriocheir sinensis. Aquacult Rep (2020) 17:100310. doi: 10.1016/j.aqrep.2020.100310
166. Wang JG, Zhang KJ, Hou X, Yue WC, Yang H, Chen XW, et al. Molecular Characteristic of Activin Receptor IIB and Its Functions in Growth and Nutrient Regulation in Eriocheir sinensis. PeerJ (2020) 8:e9673. doi: 10.7717/peerj.9673
167. Kim J, Guan KL. mTOR as a Central Hub of Nutrient Signalling and Cell Growth. Nat Cell Biol (2019) 21:63–71. doi: 10.1038/s41556-018-0205-1
168. Sabatini DM. Twenty-Five Years of mTOR: Uncovering the Link from Nutrients to Growth. Proc Natl Acad Sci USA (2017) 114:11818–25. doi: 10.1073/pnas.1716173114
169. Melick CH, Jewell JL. Regulation of mTORC1 by Upstream Stimuli. Genes (2020) 11:989. doi: 10.3390/genes11090989
170. Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell (2017) 168:960–76. doi: 10.1016/j.cell.2017.02.004
171. Kemirembe K, Liebmann K, Bootes A, Smith WA, Suzuki Y. Amino Acids and TOR Signaling Promote Prothoracic Gland Growth and the Initiation of Larval Molts in the Tobacco Hornworm Manduca sexta. PloS One (2012) 7:e44429. doi: 10.1371/journal.pone.0044429
172. Gu SH, Hsieh YC, Young SC, Lin PL. Involvement of Phosphorylation of Adenosine 5 ‘-Monophosphate-Activated Protein Kinase in PTTH-stimulated Ecdysteroidogenesis in Prothoracic Glands of the Silkworm, Bombyx mori. PloS One (2013) 8:e63102. doi: 10.1371/journal.pone.0063102
173. Rewitz KF, Yamanaka N, O’Connor MB. Developmental Checkpoints and Feedback Circuits Time Insect Maturation. In: Shi YB, editor. Animal Metamorphosis (2013). Elsevier: Amsterdam p. 1–33. doi: 10.1016/B978-0-12-385979-2.00001-0
174. Hatem NE, Wang Z, Nave KB, Koyama T, Suzuki Y. The Role of Juvenile Hormone and Insulin/TOR Signaling in the Growth of Manduca sexta. BMC Biol (2015) 13:14. doi: 10.1186/s12915-015-0155-z
175. Danielsen ET, Moeller ME, Yamanaka N, Ou QX, Laursen JM, Soenderholm C, et al. A Drosophila Genome-Wide Screen Identifies Regulators of Steroid Hormone Production and Developmental Timing. Dev Cell (2016) 37:558–70. doi: 10.1016/j.devcel.2016.05.015
176. Scieuzo C, Nardiello M, Salvia R, Pezzi M, Chicca M, Leis M, et al. Ecdysteroidogenesis and Development in Heliothis virescens (Lepidoptera: Noctuidae): Focus on PTTH-stimulated Pathways. J Insect Physiol (2018) 107:57–67. doi: 10.1016/j.jinsphys.2018.02.008
177. Koyama T, Rodrigues MA, Athanasiadis A, Shingleton AW, Mirth CK. Nutritional Control of Body Size Through FoxO-Ultraspiracle Mediated Ecdysone Biosynthesis. eLIFE (2014) 3:e03091. doi: 10.7554/eLife.03091
178. Iga M, Nakaoka T, Suzuki Y, Kataoka H. Pigment Dispersing Factor Regulates Ecdysone Biosynthesis Via Bombyx Neuropeptide G Protein Coupled Receptor-B2 in the Prothoracic Glands of Bombyx mori. PloS One (2014) 9:e103239. doi: 10.1371/journal.pone.0103239
179. Covi JA, Chang ES, Mykles DL. Neuropeptide Signaling Mechanisms in Crustacean and Insect Molting Glands. Invert Reprod Devel (2012) 56:33–49. doi: 10.1080/07924259.2011.588009
180. Layalle S, Arquier N, Leopold P. The TOR Pathway Couples Nutrition and Developmental Timing in Drosophila. Dev Cell (2008) 15:568–77. doi: 10.1016/j.devcel.2008.08.003
181. Hartnoll RG. Growth in Crustacea - Twenty Years on. Hydrobiologia (2001) 449:111–22. doi: 10.1007/978-94-017-0645-2_11
182. Green BS, Gardner C, Hochmuth JD, Linnane A. Environmental Effects on Fished Lobsters and Crabs. Rev Fish Biol Fish (2014) 24:613–38. doi: 10.1007/s11160-014-9350-1
183. Azra MN, Aaqillah-Amr MA, Ikhwanuddin M, Ma HY, Waiho K, Ostrensky A, et al. Effects of Climate-Induced Water Temperature Changes on the Life History of Brachyuran Crabs. Rev Aquacult (2019), raq12380. doi: 10.1111/raq.12380
185. Chang ES. Stressed-Out Lobsters: Crustacean Hyperglycemic Hormone and Stress Proteins. Integr Comp Biol (2005) 45:43–50. doi: 10.1093/icb/45.1.43
186. Chung JS, Webster SG. Dynamics of In Vivo Release of Molt-Inhibiting Hormone and Crustacean Hyperglycemic Hormone in the Shore Crab, Carcinus maenas. Endocrinology (2005) 146:5545–51. doi: 10.1210/en.2005-0859
187. Wittmann AC, Benrabaa SAM, Lopez-Ceron DA, Chang ES, Mykles DL. Effects of Temperature on Survival, Moulting, and Expression of Neuropeptide and mTOR Signalling Genes in Juvenile Dungeness Crab (Metacarcinus magister). J Exp Biol (2018) 221:jeb187492. doi: 10.1242/jeb.187492
188. Terwilliger NB, Dumler K. Ontogeny of Decapod Crustacean Hemocyanin: Effects of Temperature and Nutrition. J Exp Biol (2001) 204:1013–20. doi: 10.1242/jeb.204.5.1013
189. Webster SG. Endocrinology of Metabolism and Water Balance: Crustacean Hyperglycemic Hormone. In: Chang ES, Thiel M, editors. The Natural History of the Crustacea: Physiology. Oxford: Oxford Press (2015). p. 36–67.
190. Chung JS, Zmora N, Katayama H, Tsutsui N. Crustacean Hyperglycemic Hormone (CHH) Neuropeptides Family: Functions, Titer, and Binding to Target Tissues. Gen Comp Endocrinol (2010) 166:447–54. doi: 10.1016/j.ygcen.2009.12.011
191. Goy MF. Activation of Membrane Guanylate Cyclase by an Invertebrate Peptide Hormone. J Biol Chem (1990) 265:20220–7. doi: 10.1016/S0021-9258(17)30492-1
192. Dibble CC, Cantley LC. Regulation of mTORC1 by PI3K Signaling. Trends Cell Biol (2015) 25:545–55. doi: 10.1016/j.tcb.2015.06.002
193. Zhang TL, Song W, Li Z, Qian WL, Wei L, Yang Y, et al. Kruppel Homolog 1 Represses Insect Ecdysone Biosynthesis by Directly Inhibiting the Transcription of Steroidogenic Enzymes. Proc Natl Acad Sci USA (2018) 115:3960–5. doi: 10.1073/pnas.1800435115
194. Uryu O, Ou QX, Komura-Kawa T, Kamiyama T, Iga M, Syrzycka M, et al. Cooperative Control of Ecdysone Biosynthesis in Drosophila by Transcription Factors Seance, Ouija Board, and Molting Defective. Genetics (2018) 208:605–22. doi: 10.1534/genetics.117.300268
195. Danielsen ET, Moeller ME, Dorry E, Komura-Kawa T, Fujimoto Y, Troelsen JT, et al. Transcriptional Control of Steroid Biosynthesis Genes in the Drosophila Prothoracic Gland by Ventral Veins Lacking and Knirps. PloS Genet (2014) 10:e1004343. doi: 10.1371/journal.pgen.1004343
196. Truman JW, Riddiford LM. The Evolution of Insect Metamorphosis: A Developmental and Endocrine View. Phil Trans R Soc B Biol Sci (2019) 374:20190070. doi: 10.1098/rstb.2019.0070
197. Tsang SSK, Law STS, Li CD, Qu Z, Bendena WG, Tobe SS, et al. Diversity of Insect Sesquiterpenoid Regulation. Front Genet (2020) 11:1027. doi: 10.3389/fgene.2020.01027
198. Wu ZX, Yang LB, He QJ, Zhou ST. Regulatory Mechanisms of Vitellogenesis in Insects. Front Cell Dev Biol (2021) 8:593613. doi: 10.3389/fcell.2020.593613
199. Miyakawa H, Watanabe M, Araki M, Ogino Y, Miyagawa S, Iguchi T. Juvenile Hormone-Independent Function of Kruppel Homolog 1 in Early Development of Water Flea Daphnia pulex. Insect Biochem Molec Biol (2018) 93:12–8. doi: 10.1016/j.ibmb.2017.12.007
200. Tanaka T, Iguchi T, Miyakawa H. Establishment of a High-Sensitivity Reporter System in Mammalian Cells for Detecting Juvenoids Using Juvenile Hormone Receptors of Daphnia pulex. J Appl Toxicol (2019) 39:241–6. doi: 10.1002/jat.3713
201. Xie X, Liu MX, Jiang QH, Zheng HK, Zheng L, Zhu DF. Role of Kruppel Homolog 1 (Kr-h1) in Methyl Farnesoate-Mediated Vitellogenesis in the Swimming Crab Portunus trituberculatus. Gene (2018) 679:260–5. doi: 10.1016/j.gene.2018.08.046
202. Li XL, Chen TT, Jiang HC, Huang JW, Huang MT, Xu RH, et al. Effects of Methyl Farnesoate on Kruppel Homolog 1 (Kr-h1) During Vitellogenesis in the Chinese Mitten Crab (Eriocheir sinensis). Anim Reprod Sci (2021) 224:106653. doi: 10.1016/j.anireprosci.2020.106653
203. Liu JH, Zhou TT, Wang CG, Chan SM, Wang W. Deciphering the Molecular Regulatory Mechanism Orchestrating Ovary Development of the Pacific Whiteleg Shrimp Litopenaeus vannamei Through Integrated Transcriptomic Analysis of Reproduction-Related Organs. Aquaculture (2021) 533:736160. doi: 10.1016/j.aquaculture.2020.736160
204. Bentley VL, Mykles DL. Investigating the Function of Kruppel Homolog 1 (Kr-h1) on Molt Regulation in Gecarcinus lateralis. Integr Comp Biol (2020) 60:E282.
205. Alexandratos A, Moulos P, Nellas I, Mavridis K, Dedos SG. Reassessing Ecdysteroidogenic Cells From the Cell Membrane Receptors’ Perspective. Sci Rep (2016) 6:20229. doi: 10.1038/srep20229
206. Du JL, Zhang XJ, Yuan JB, Zhang XO, Li FH, Xiang JH. Wnt Gene Family Members and Their Expression Profiling in Litopenaeus vannamei. Fish Shellfish Immunol (2018) 77:233–43. doi: 10.1016/j.fsi.2018.03.034
207. Liu L, Fu YY, Zhu F, Mu CK, Li RH, Song WW, et al. Transcriptomic Analysis of Portunus trituberculatus Reveals a Critical Role for WNT4 and WNT Signalling in Limb Regeneration. Gene (2018) 658:113–22. doi: 10.1016/j.gene.2018.03.015
208. Wang KQ, Dai XL, Zhang C, Cao XY, Zhang RD, Zhang ZX, et al. Two Wnt Genes Regulate the Expression Levels of Antimicrobial Peptides During Vibrio Infection in Macrobrachium nipponense. Fish Shellfish Immunol (2020) 101:225–33. doi: 10.1016/j.fsi.2020.03.063
209. Ou QX, Zeng J, Yamanaka N, Brakken-Thai C, O’Connor MB, King-Jones K. The Insect Prothoracic Gland as a Model for Steroid Hormone Biosynthesis and Regulation. Cell Rep (2016) 16:1–16. doi: 10.1016/j.celrep.2016.05.053
210. Moulos P, Alexandratos A, Nellas L, Dedos SC. Refining a Steroidogenic Model: An Analysis of RNA-seq Datasets From Insect Prothoracic Glands. BMC Genomics (2018) 19:537. doi: 10.1186/s12864-018-4896-2
211. Christesen D, Yang YT, Somers J, Robin C, Sztal T, Batterham P, et al. Transcriptome Analysis of Drosophila melanogaster Third Instar Larval Ring Glands Points to Novel Functions and Uncovers a Cytochrome P450 Required for Development. Genes Genomes Genet (2017) 7:467–79. doi: 10.1534/g3.116.037333
212. Nakaoka T, Iga M, Yamada T, Koujima I, Takeshima M, Zhou XY, et al. Deep Sequencing of the Prothoracic Gland Transcriptome Reveals New Players in Insect Ecdysterodogenesis. PloS One (2017) 12:e0172951. doi: 10.1371/journal.pone.0172951
213. Li JY, Chen X, Fan W, Moghaddam SHH, Chen M, Zhou ZH, et al. Proteomic and Bioinformatic Analysis on Endocrine Organs of Domesticated Silkworm, Bombyx mori L. for a Comprehensive Understanding of Their Roles and Relations. J Proteome Res (2009) 8:2620–32. doi: 10.1021/pr8006123
214. Boerjan B, Vandingenen K, De Loof A, Schoofs L. In Search for non-Steroidogenic Functions of the Prothoracic Glands of the Desert Locust, Schistocerca Gregaria: A Peptidomic and Proteomic Approach. Peptides (2012) 34:57–64. doi: 10.1016/j.peptides.2011.07.022
215. Rewitz KF, Larsen MR, Lobner-Olesen A, Rybczynski R, O’Connor MB, Gilbert LI. A Phosphoproteomics Approach to Elucidate Neuropeptide Signal Transduction Controlling Insect Metamorphosis. Insect Biochem Molec Biol (2009) 39:475–83. doi: 10.1016/j.ibmb.2009.04.005
216. Giribet G, Edgecombe GD. The Phylogeny and Evolutionary History of Arthropods. Curr Biol (2019) 29:R592–602. doi: 10.1016/j.cub.2019.04.057
217. de Oliveira APL, Calcino A, Wanninger A. Ancient Origins of Arthropod Moulting Pathway Components. eLife (2019) 8:e46113. doi: 10.7554/eLife.46113
Keywords: Y-organ, molting (control of), mTOR - mammalian Target of Rapamycin, ecdysteroid, neuropeptide, insulin, growth factor, G protein coupled receptor (GPCR)
Citation: Mykles DL (2021) Signaling Pathways That Regulate the Crustacean Molting Gland. Front. Endocrinol. 12:674711. doi: 10.3389/fendo.2021.674711
Received: 01 March 2021; Accepted: 28 April 2021;
Published: 21 June 2021.
Edited by:
Heinrich Dircksen, Stockholm University, SwedenReviewed by:
Taisen Iguchi, Graduate University for Advanced Studies (Sokendai), JapanCopyright © 2021 Mykles. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donald L. Mykles, ZG9uYWxkLm15a2xlc0Bjb2xvc3RhdGUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.