
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Endocrinol. , 26 May 2021
Sec. Reproduction
Volume 12 - 2021 | https://doi.org/10.3389/fendo.2021.668073
 Jiajie Tu1,2*†
Jiajie Tu1,2*† Huan Yang1†
Huan Yang1† Yu Chen3
Yu Chen3 Yu Chen1
Yu Chen1 He Chen1
He Chen1 Zhe Li4
Zhe Li4 Lei Li1
Lei Li1 Yuanyuan Zhang1
Yuanyuan Zhang1 Xiaochun Chen1
Xiaochun Chen1 Zhiying Yu1*
Zhiying Yu1*The uterine endometrium, which lines the mammalian uterus, is essential for embryo implantation. This lining undergoes significant changes during sexual and menstrual cycles. The endometrium is also associated with hormone-related diseases such as endometriosis and endometrial cancer. Circular RNAs (circRNAs) play a role in various biological processes. Recent studies have determined that circRNAs function in both normal and pathological endometrial environments. Here, we review high-throughput studies pertaining to circRNAs as well as individual circRNAs active in the endometrium, in order to explore the myriad functions of circRNAs in the endometrium and mechanisms underlying these functions, from panoramic and individual perspectives. Owing to their abundant expression, stability, and small size, circRNAs have displayed potential usefulness as diagnostic markers and treatment targets for endometrial-related diseases. Therefore, the specific role of circRNAs in the endometrium warrants systematic investigation in the future.
The endometrium is a layer of cells that forms the lining of the mammalian uterus. It responds to changes in the levels of both estrogen and progesterone (1). The endometrium consists of two layers: the functional layer and the basal layer. The functional layer, which comprises a dense layer and a sponge layer, changes and is shed during the ovarian cycle. The basal layer is not affected by ovarian hormones and does not undergo periodic changes.
High-throughput sequencing has enabled the discovery of a wide range of non-coding RNAs, including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), that function in various biological processes (2). In some cases, these non-coding RNAs exhibit abnormal expression patterns that may lead to multiple diseases (3). Unlike conventional linear RNAs, circRNAs are a unique class of RNAs whose 3′ and 5′ ends are covalently bonded to form a closed continuous loop that is resistant to exonuclease digestion (4). The high abundance, stability, and evolutionary conservation of circRNAs suggest that they play important regulatory roles (5). For example, as competing endogenous RNA (ceRNA), circRNAs may use miRNA response elements (MREs) to sponge miRNAs and strongly inhibit miRNA activity, resulting in the upregulation of miRNA targets, which ultimately affects cellular processes such as cell differentiation, proliferation, and apoptosis as well as other cellular functions (6). These properties make circRNAs important bioregulators of molecular mechanisms underlying various diseases, indicating that circRNAs may be useful as potential diagnostic biomarkers and therapeutic targets (5). However, studies related to the activity of circRNAs in the endometrium, including those involving endometrial development and endometrium-related diseases, such as endometrial cancer (EC) and endometriosis, are currently in the formative stages.
Investigating circRNA function in the endometrium as well as mechanisms underlying such functions may enhance our understanding of the molecular processes associated with the physiological development of the endometrium and provide new opportunities to develop more effective diagnostics and treatments for endometrium-related diseases. The current study attempted to review the properties and functions of circRNAs in the endometrium under both physiological and pathological conditions with a view toward the future of circRNA research.
EC is a type of epithelial malignancy that occurs in endometrial tissue, often in perimenopausal and postmenopausal women (7). With nearly 200,000 new cases being diagnosed each year, EC has come to be considered as one of the most common female reproductive system tumors and the third most common gynecological malignant tumor (second only to ovarian and cervical cancer) (7).
RNA sequencing (RNA-seq) analysis has indicated that the total abundance of circRNAs in 1-2 grade type I EC is lower than that in the normal endometrium. In addition, many circRNAs embedded in the “hotspot” genes of EC were transcribed, explaining the differences in circRNA expression between normal and malignant endometria (8). Although the functional significance of these circRNAs in EC remains to be determined, it is surmised that these may serve as potential biomarkers of EC.
Study has focused on the role of circRNAs in extracellular vesicles (EVs) isolated from the sera of three grade 3 EC patients (9). EVs isolated from serum samples obtained from 3 EC patients and three age-matched healthy controls were subjected to RNA-seq, which indicated that the expression levels of 209 circRNAs were upregulated while those of 66 were downregulated. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses showed that differentially expressed circRNAs were associated with several signaling pathways, such as focal adhesion, extracellular matrix-receptor interaction, amebiasis, and regulation of the actin cytoskeleton. The expression levels of circ_0109046 and circ_0002577 were confirmed using a quantitative real-time polymerase chain reaction (qRT-PCR). CircRNAs enriched in circulating EVs may be useful as potential biomarkers of EC.
Another study, which focused on the role of circRNAs in grade 3 ECs, used circRNA sequencing (circRNA-seq) to detect circRNA expression characteristics in two grade 3 ECs and adjacent non-cancerous endometrial tissues (10). In total, the expression of 75,928 circRNAs was significantly changed (p<0.05). Upregulation of hsa_circ_0000437, hsa_circ_0001776, and hsa_circ_0009043 and downregulation of hsa_circ_0039569 and hsa_circ_0001610 was verified using qPCR. Bioinformatic methods (TargetScan and miRanda) indicated that hsa_circ_0039569 has MREs for hsa-miR-542-3p and hsa-let-let-7c-5p. Downregulation of hsa-miR-542-3p and hsa-let-let-7c-5p in grade 3 EC tissues was validated using qRT-PCR. Clinical-pathological parameters indicated that the expression level of hsa_circ_0039569 was significantly correlated with tumor differentiation. These results demonstrated the presence of many differentially expressed circRNAs between level 3 EC and adjacent non-cancerous endometrial tissues, thus providing new molecular candidates for diagnosis and clinical treatment of grade 3 EC.
Dou et al., identified 234 recurrent circRNAs in EC (11). In order to determine possible regulators, they examined the correlation between RNA-binding proteins (obtained from protein sequencing), and circRNAs. The level of the kH domain RNA binding (QKI) protein was positively correlated with 35 circRNAs. QKI induces epithelial-mesenchymal transition (EMT). Since miRNAs also play a key role in EMT, circRNAs may be used as miRNA sponges to regulate miRNA activity. Therefore, researchers explored miRNA binding sites in the 35 circRNAs that were correlated with QKI levels and identified potential binding sites for 36 miRNAs. The activity of these miRNAs was negatively correlated with QKI expression, indicating that the activity of these miRNAs may be inhibited by QKI, possibly via QKI-mediated expression of circRNAs. A set of known or predicted QKI regulators (miR-200c, miR-221, miR-130a, miR-130b, and miR-183) was among the miRNAs showing the strongest negative correlations with QKI. Such positive correlation between QKI and circRNAs, and negative correlation between QKI and the activity of specific miRNAs suggested that a novel mechanism may underlie EMT in EC.
Research studies about the role of circRNA in EC are still in the early phase. Thus, circRNA-seq enables a better understanding of the features associated with the overall expression of circRNAs in EC. Firstly, circRNAs are a special class of small non-coding RNAs, and a panoramic view of circRNAs in EC should be compared with that of other types of small non-coding RNAs such as miRNAs and PIWI-interacting RNAs (piRNAs). Secondly, circRNAs-seq indicated that circRNAs may play an important role in EC. A major limitation of the aforementioned studies was that the sample size of clinical EC patients was relatively small. Future studies should include larger EC samples to assess the role of circRNAs at different stages and subtypes of EC. In addition, it would be interesting to compare circRNA expression in endometrial hyperplasia with or without cytological atypia. Endometrial hyperplasia with cytological atypia indicates a tendency toward cancer (12–14). Thus, differentially expressed circRNAs may be useful for obtaining a better understanding of EC.
Regarding the role of individual circRNAs in EC, the expression level of circ_PUM1 was significantly higher in EC tissues than in normal tissues (15). Circ_PUM1 overexpression promoted the proliferation, metastasis, and invasion of EC cells. In contrast, the ability of EC cells to grow into tumors was reduced following circ_PUM1 knockdown. Mechanistically, circ_PUM1 bound with miR-136, which targeted Notch homolog 3 (NOTCH3, an identified oncogene of EC). Sponging of miR-136 by Circ_PUM1 lead to activation of NOTCH3, thereby promoting EC development.
EC patients displaying high expression levels of circ_0002577 showed poor overall survival and advanced tumor stages (16). Overexpression of circ_002577 in EC cells promoted their proliferation, migration and invasion, whereas circ_002577 knockdown in EC cells demonstrated the opposite effect. Furthermore, circ_002577 accelerated EC progression by sponging miR-625-5p and upregulating insulin like growth factor 1 receptor (IGF1R) (17). Another recent study validated the oncogenic role of circ_0002577 in EC via miR-197/catenin delta 1(CTNND1)/Wnt axis, suggesting that circ_002577 is a potentially therapeutic factor in EC.
CircWHSC1 overexpression promoted the proliferation, migration, and invasion of EC cells while decreasing the apoptosis of these cells (18). EC-derived tumors overexpressing CircWHSC1 showed increased tumorigenicity as indicated by a study investigating nude mouse xenograft tumor models. Reportedly, circWHSC1 binds to miR-646 and induces the expression of nucleophosmin 1 (NPM1), a downstream target of miR-646, in EC cells. This suggests that CircWHSC1 promotes EC development by sponging miR-646 and targeting NPM1.
The expression of circTNFRSF21 was up-regulated in EC tumor tissues (19). circTNFRSF21 promoted EC cell growth in vitro and tumor formation in vivo. Mechanically, circTNFRSF21 acted as a sponge of miR-1227 to restore mitogen-activated protein kinase 13 (MAPK13)/activating transcription factor 2(ATF2) signaling pathway activity in EC cells. Hsa_circ_0061140 is another oncognetic circRNA in EC cells (20). Hsa_circ_0061140 acted as a sponge for miR-149-5p, which was proven as a tumor suppressor in tumor growth and metastasis (21). Signal transducer and activator of transcription 3 (STAT3) was proved as another target of miR-149-5p. Moreover, hsa_circ_0061140 exhibited its oncogenic role due to the modulation of miR-149-5p/STAT3 axis.
The number of studies of individual circRNA in EC is limited. There are only six related reports up to now. Among them, there are two independent studies that focus on a common circRNA, circ_0002577. Moreover, all these reported circRNAs involved in two basic functional processes of EC cells, proliferation and migration. Interestingly, Wnt/β-Catenin pathway has been proven as a central signaling pathway to connect the abovementioned individual circRNAs (Figure 1A). The interaction of circRNAs and Wnt pathway in EC needs further investigation.
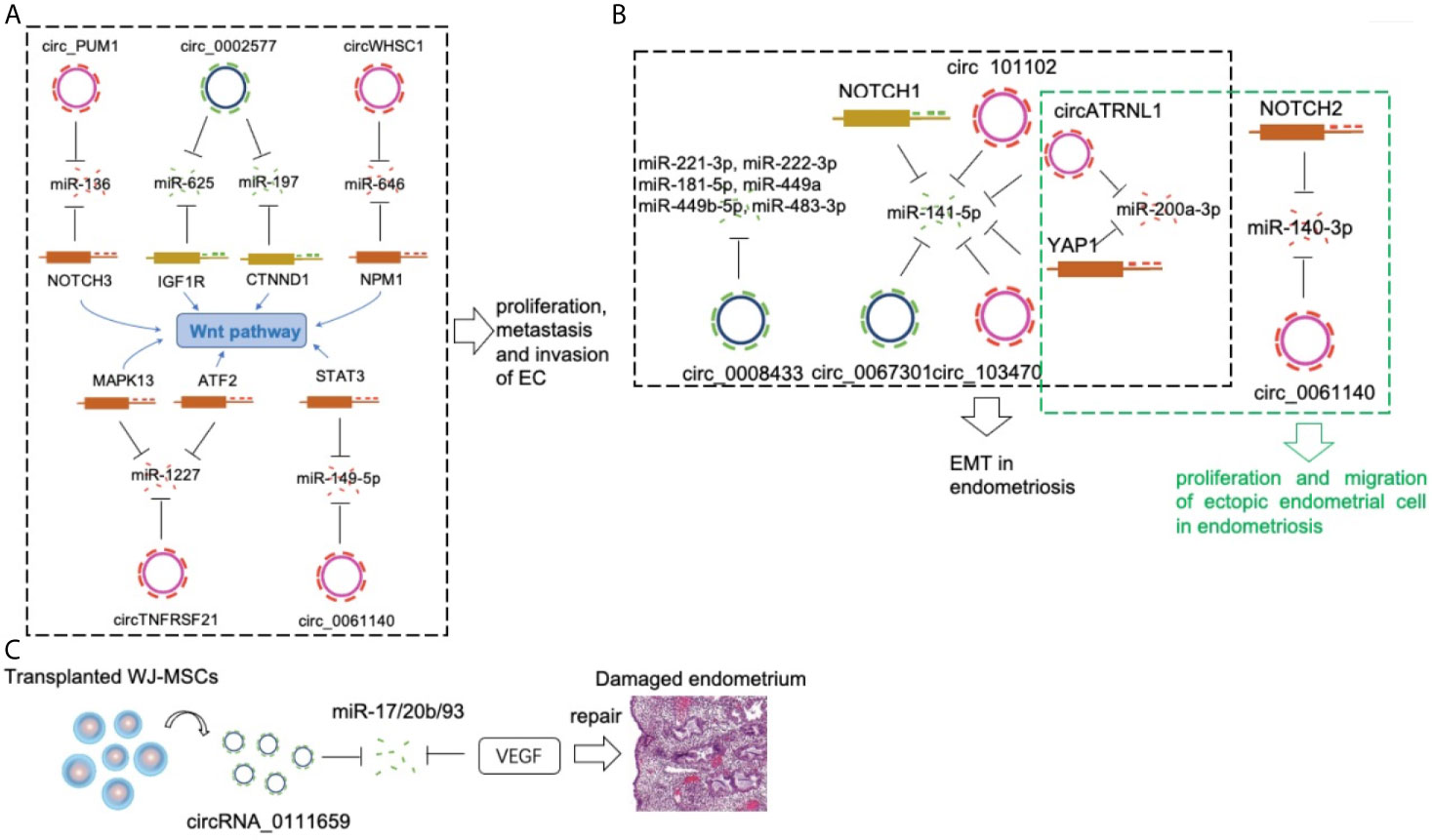
Figure 1 CircRNAs in a pathological endometrium. (A) Circ_PUM1, circ_0002577, circWHSC1, circTNFRSF21 and circ_0061140 compete with NOTCH3, IGF1R, NPM, MAPK13/ATF2 and STAT3 via sponging miR-136, miR-625/miR-197, miR-646, miR-1227 and miR-149-5p respectively, to promote EC development. (B) The circ_0067301/miR-141e-5p/NOTCH1, circ_103470/miR-141-5p, circ_101102/miR-141-5p, circATRNL1/miR-141-5p/miR-200a-3p/YAP1, circ_0061140/miR-140-3p/NOTCH2 axis played an important regulatory role in the EMT and proliferation/migration in endometriosis. (C) The circRNA_0111659-miR-17/20b/93-VEGF association is involved in regulating endometrial damage repair.
Endometriosis is a common gynecological disease in women, which is caused by active endometrial cells that are present outside the endometrium. Endometrial cells are naturally intended to grow in the uterine cavity. The uterine cavity is connected to the pelvic cavity via the fallopian tube. However, endometrial cells may grow ectopically by moving through the fallopian tube. Many theories have been proposed to explain the pathological mechanism underlying this disease (22). Emerging evidence suggests that many factors, including immune cells, adhesion molecules, extracellular matrix metalloproteinases and pro-inflammatory cytokines shape the environment for survival, adhesion and differentiation of ectopic endometrial cells (23, 24). The disease mostly occurs in women of childbearing age. Ectopic lesions may gradually shrink and degenerate during the postmenopausal period (25).
Six pairs of ectopic and eutopic endometria were subjected to high-throughput RNA-seq to analyze the expression patterns of circRNAs in endometriosis, which was followed by qRT-PCR analysis of 30 pairs of samples (26). The analyses identified 146 upregulated and 148 downregulated circRNAs. Based on 2,495 MREs, a ceRNA network consisting of 48 miRNAs and 296 mRNAs was identified. GO functional analysis and KEGG pathway analysis revealed several circRNA-associated signaling pathways, such as MAPK, PI3K-AKT and FOXO, which may be associated with the pathogenesis of endometriosis. Abnormal activation of MAPK pathway in primary eutopic endometrial stromal cells of patients with endometriosis induced cell proliferation and migration (27). The communications between these pathways and circRNAs may provide a new direction for the prevention, diagnosis and treatment of endometriosis (28).
Microarray assays were performed on endometrial tissues from patients with endometriosis and normal endometrial tissues, followed by qRT-PCR validation, which identified 262 upregulated and 291 downregulated circRNAs, binding to 1,225 MREs (29). The ceRNA networks included 122 miRNAs and 137 mRNAs, which were closely related to nine signaling pathways. Some of them, such as Rap1 pathway (30) and leukocyte transendothelial migration pathway(including Intercellular Adhesion Molecule 1[ICAM-1) and Integrin Subunit Alpha L (ITGAL)] (31, 32), have been proven as involved in endometriosis. The data suggest that these pathways might play a role in the pathogenesis and development of endometriosis via interacting with circRNAs.
Researchers analyzed the expression patterns of circRNAs in ectopic and paired eutopic endometria and constructed a circRNA-miRNA-mRNA network (33). Expression-related characteristics of circRNA and mRNA in four patients were evaluated using microarray analysis. Eight circRNAs and mRNAs in 37 patients were validated using qPCR. In addition, 1,258 upregulated and 1,061 downregulated circRNAs, as well as 1,900 upregulated and 2,535 downregulated mRNAs, were detected between ectopic and control endometria. Functional analysis showed that most of the differentially expressed mRNAs were involved in immune-inflammatory responses and cell-cycle regulation. Five circRNAs (circ_0004712, circ_0002198, circ_0003570, circ_0008951, and circ_0017248) and eight mRNAs (SCN3B, ENTPD1, IL16, BACH2, C3, CKS2, G0S2, and PGRMC1) identified using qPCR matched those shown in microarray results. In addition, a circRNA-miRNA-mRNA network was constructed, revealing the participation of cancer-related purine metabolism, glyceride metabolism, and thyroid hormone signaling pathways in the pathogenesis of endometriosis. The results of this study indicated that circRNAs have potential as promising diagnostic biomarkers of ovarian endometriosis as well as therapeutic targets for this disease.
Another study included 63 clinical samples (control endometrium, n = 22; and ectopic endometrium, n = 41) (34). Of these, four samples in each group were used for circRNA microarray assays. Of the 88 significantly changed circRNAs, 11 and 77 circRNAs were upregulated and downregulated, respectively, in the ectopic endometrium of endometriosis patients. The qRT-PCR validation results of two induced circRNAs (circ_0004712 and circ_0002198) matched microarray assay results. These two circRNAs may represent potential new biomarkers useful for diagnosing endometriosis.
Similar to EC, research of circRNA in endometriosis also focused on the illumination of a “circRNAome” via sequencing techniques. Mechanisms underlying the alteration of physiological functions caused by circRNAs in endometriosis need to be further elucidated. This will broaden application of the results, as these may function not only as potential targets for treatment of endometriosis but also as diagnostic biomarkers of the disease. In addition, studies based on in vitro experiments as well as in vivo animal models, such as primate models and patient-derived xenograft models (PDX) (35, 36), should be performed to further clarify the pathophysiological relevance of circRNAs in endometriosis.
Epithelial-mesenchymal transition (EMT) is a core pathological mechanism associated with endometriosis (37, 38). A study was designed to investigate the role of circRNAs and miRNAs during the EMT process in endometriosis (39). Compared with those in control endometria, the expression levels of hsa_circ_0067301 and miR-141e-5p in the ectopic endometria were significantly reduced. Hsa_circ_0067301 knockdown promoted the proliferation and migration of Ishikawa cells and End1/E6E7 cells. Some EMT-related markers, such as Notch-1, Hes-1, N-cadherin, and vimentin, were upregulated, whereas E-cadherin was downregulated. EMT was partially relieved following co-infection with miR-141e-5p inhibitors. These results showed that the hsa_circ_0067301/miR-141e-5p/Notch-1 pathway plays an important regulatory role in EMT during endometriosis.
Another study included four patients with endometriosis and four with general endometritis (without endometrial disease) (40). Patients with endometriosis provided ectopic endometrial tissues and paired normal endometrial tissues. This study identified 2,233 differentially expressed circRNAs among these three groups. In addition, eight circRNAs were closely associated with EMT, which is a prerequisite for the establishment of endometriotic lesions. The results also showed that two circRNAs (circ_103470 and circ_101102) regulated EMT in endometriosis by sponging miR-141-5p, which may represent a promising target for the treatment of endometriosis (41). Interestingly, miR-141-5p is a common EMT-related miRNA in both studies. These studies highlighted the importance of circRNAs in the process of EMT in endometriosis and provides unique insights into the molecular basis of the pathogenesis of endometriosis (Figure 1B).
Circ_0061140 only acts as oncogenic regulator in EC (20), it also plays a role in endometriosis (42). Knockdown of circ_0061140 inhibited proliferation and migration of ectopic endometrial cell. MiR-140-3p decreased in ectopic endometrial cells and it acted as a direct target of circ_0061140 by repressing the effects of circ_0061140 on ectopic endometrial cells. A positive correlation between circ_0061140 and Notch homolog 2 (Notch2) was demonstrated in endometriosis. Moreover, circ_0061140 could induce endometriosis progression via miR-140-3p-Notch2 axis.
Knockdown of Circ_0008433 repressed proliferation, migration and angiogenesis and induced apoptosis of endometrial stromal cells (43). Six potential target miRNAs (miR-221-3p, miR-222-3p, miR-181-5p, miR-449a, miR-449b-5p, miR-483-3p) were significantly changed in circ_0008433-overexpressing endometrial stromal cells. Furthermore, Protein-protein interaction (PPI) analysis showed that up-regulation of circ_0008433 modulated EMT in endometriosis through the circRNA–miRNA–mRNA axis.
CircATRNL1 and Yes-associated protein 1 (YAP1) increased and miR-141-3p and miR-200a-3p decreased in ectopic tissues (44). CircATRNL1 and YAP1 induced the proliferation and migration and induce EMT by targeting miR-141-3p and miR-200a-3p in endometrial cells. Rescue assays further validated the role of circATRNL1–miR-141-3p/miR-200a-3p–YAP1 axis in endometrial cells, which could contribute to the progression of endometriosis.
There are five studies of individual circRNA in endometriosis until now. Interestingly, four circRNAs (circ_0067301, circ_103470, circ_101102, and circATRNL1) promoted EMT in endometriosis by targeting a common miRNA, miR-141. The essential inhibitory effects of miR-141 on proliferation/migration (45) and EMT (46) also have been proven in endometrial cells. In addition, two circRNAs (circ_0067301 and circ_0061140) induced the expression of different members of the Notch family, Notch1 and Notch2 in endometriosis. Given that the hyperactivation of Notch family (47) and the effects of Notch pathway on EMT (48) have been reported in endometriosis, the role of circRNA-miRNA-Notch axis in EMT of endometriosis definitely should be deeply investigated in the future.
Other endometrial-related disorders, such as endometrial damage and implantation failure, also greatly affect the quality of life of women (49). Wharton’s jelly derived mesenchymal stem cells (WJ-MSCs) have shown great potential for repairing different diseases. Human endometrial stromal cells (ESCs) damaged by culturing with mifepristone were repaired by WJ-MSCs (50). The repairing of damaged ESCs in the co-culture group by WJ-MSCs, improved cell morphology, increased proliferation and decreased apoptosis. The circRNA microarray analyses indicated that 7,757 circRNAs showed differential expression in ESCs co-cultured with WJ-MSCs. In addition, researchers focused on hsa_circRNA_0111659 and predicted it to be related to miR-17/20b/93 and the target mRNA, VEGF. The circRNA-miRNA-mRNA combination may be involved in regulating endometrial damage repair. The results showed that abundant circRNAs were expressed during repair of damaged endometria by WJ-MSC, providing a new perspective in regard to the mechanism of endometrial repair by WJ-MSCs (Figure 1C).
The specific role of circRNAs in the pathogenesis of repeated implantation failure remains unclear (51). CircRNA expression in the endometrial biopsies of six women showing repeated implantation failure and a control group (six healthy women) was screened using microarrays (52). Data from this circRNA microarray assay showed that 856 unique circRNAs were significantly altered. Subsequently, qRT-PCR verified the upregulation of hsa_circRNA_070616, hsa_circRNA_103716, hsa_circRNA_104001, and hsa_circRNA_104854 and the downregulation of hsa_circRNA_ 004183, hsa_circRNA_044353, and hsa_circRNA_404686. Differentially expressed circRNAs provided new target molecular candidates for the diagnosis and clinical treatment of patients with repeated implantation failures.
Endometrial receptivity is defined as the ability of the endometrium to accept an embryo implantation (53, 54). Studies directed at the potential molecular mechanisms that may be involved, centered on protein-coding genes in the field of assisted reproduction (55). In addition, circRNA microarray assays, used to compare circRNA expression in the early secretory phase and mid-secretory phase endometrium, identified a number of circRNAs (hsa_circRNA_101280, hsa_circRNA_102293, hsa_circRNA_104789, hsa_circRNA_104791, hsa_circRNA_101263, hsa_circRNA_103493, hsa_circRNA_104625, has_circRNA_400019 and hsa_circRNA_104700), which may be useful as potential biomarkers of endometrial receptivity (56). Adenomyosis is a benign uterine disorder characterized by presence of endometrium in the myometrium (57). The down-regulation of hsa_circRNA_101280 was validated in adenomyosis samples (n=11) compared with that in the control samples (n=11), suggesting a potential mechanism underlying decreased implantation rates observed in women with adenomyosis (56). This study not only expands the knowledge of circRNAs in human endometrium but also provides useful clues for understanding the role of circRNAs in endometrial receptivity.
Researchers used Illumina Solexa techniques to analyze circRNAs in the endometria of three goats at gestational day 5 (pre-receptive endometrium, PE) and three goats at gestational day 15 (receptive endometrium, RE) (58). Overall, 21,813 circRNAs were identified, of which 5,925 circRNAs were RE- and 9,078 circRNAs were PE-specific, indicating the high stage-specificity of circRNAs. Further analyses indicated that there were 334 differentially expressed circRNAs in the RE stage compared with those in the PE stage. Analyses of the cyclic RNA-miRNA interaction network further supported the contention that circRNAs may be used as miRNA sponges to regulate gene expression. In addition, estrogen/progesterone regulated some circRNAs in the endometrial epithelial cells (EECs) and ESCs. These data were used to compile a circRNA atlas of the goat endometrium during embryo implantation.
CircRNA-miRNA-mRNA networks were constructed to explore the involvement of ceRNA in the development of RE in goats (59). Cyclic RNA8073 (ciR-8073) reduced miR-181a levels in a manner similar to that of a miRNA sponge. This effect indirectly increased the expression of neurotensin in EECs. Neurotensin promotes BCL-2 expression via the MAPK pathway and induces expression of leukemia inhibitor factor, cyclooxygenase 2 (COX-2), vascular endothelial growth factor A (VEGFA), and homeobox A10 (HOXA10). This indicated the presence of a ciR-8073-miR181a-neurotensin pathway in the endometrium of goats. CiR-8073 functions as a ceRNA sequestering miR-181a, thereby protecting neurotensin from miR-181a-mediated suppression in EECs.
Circ-8073 directly binds miR-449a and inhibits its activity (60). Centrosomal protein 55 (CEP55) is a direct target of miR-449a. Circ-8073 improves the expression of CEP55 by absorbing miR-449a in EECs in vitro. Circ-8073/miR-449a/CEP55 promotes EC proliferation via the PI3K/AKT/mTOR pathway. In addition, CEP55 regulates the expression of VEGF and FOXM1 in EECs, thereby contributing to the formation of endometrial receptors. These findings in goats suggest that circ-8073 regulates endometrial receptivity via miR-449a/CEP55 and PI3K/AKT/mTOR pathways. Another study also validated the promotive effects of circ-8073 on EEC proliferation via competitive sponging of miR-34a/c via CEP55 (61) (Figure 2A).
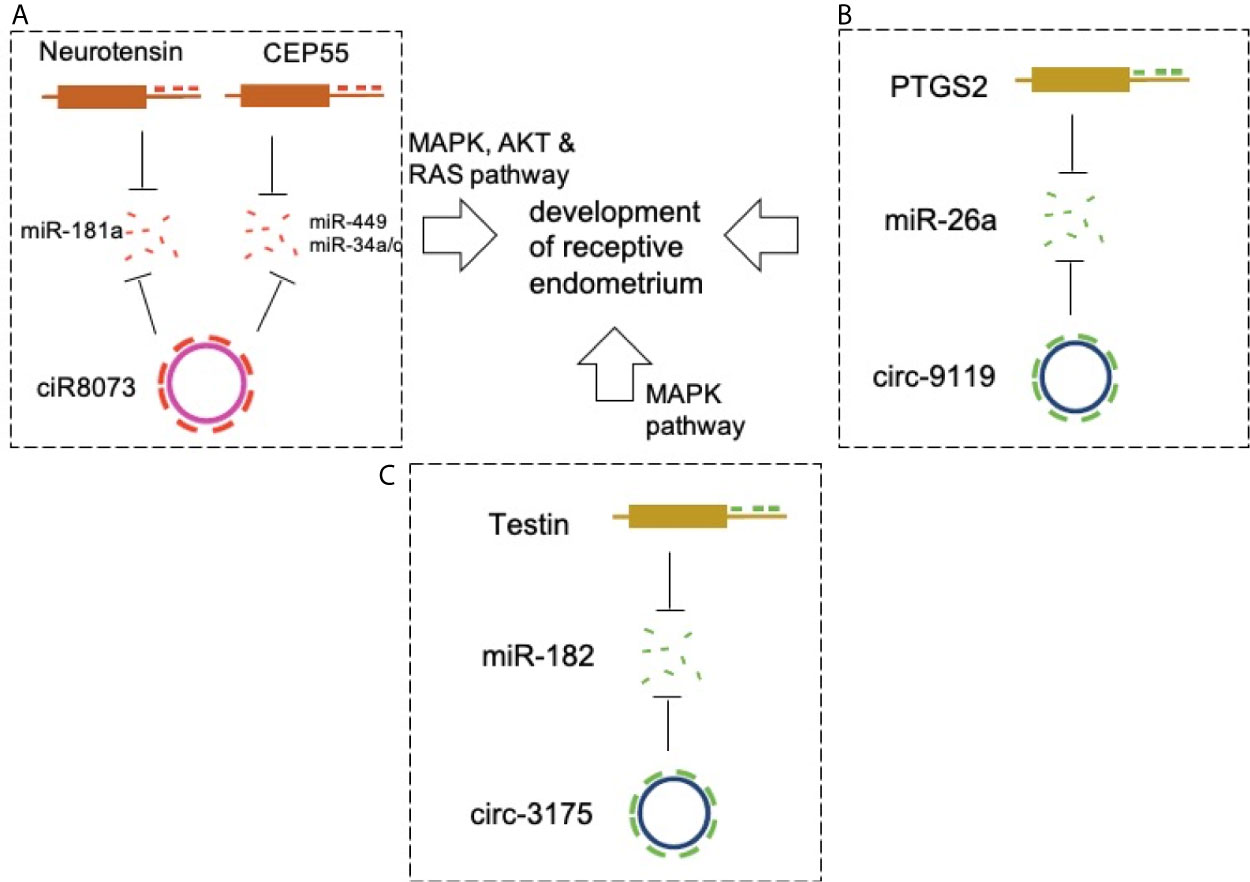
Figure 2 CircRNAs in endometrial receptivity. (A) CiR-8073 functions as a ceRNA to sequester miR-181a, thereby protecting neurotensin from miR-181a-mediated suppression in EECs. Circ-8073 regulates endometrial receptivity via miR-181a/neurotensin and miR-449a/CEP55 and miR-34/CEP55 axes. (B) Modulation of the circRNA-9119-miR-26a-PTGS2 axis in EECs may be a potential target for regulating RE development. (C) The ciR3175-miR182-testin axis is essential for development of pre-receptive endometrium in goats.
A similar study detected high expression of circRNA-9119 and prostaglandin-endoperoxide synthase 2 (PTGS2) and low levels of miR-26a at the RE stage of goats (62). Further studies showed that circRNA-9119 reduces miR-26a by acting as a miRNA sponge, and it is known that miR-26a reduces the expression of PTGS2 in goat EECs. In addition, PTGS2 is involved in the regulation of certain protein markers of endometrial receptivity in goat EECs. Therefore, modulation of the circRNA-9119-miR-26a-PTGS2 pathway in EECs may be a potential target in the regulation of RE development (Figure 2B).
The levels of circRNA3175 (ciR3175) and testin in the goat pre-receptive endometrium were high, whereas the expression level of miR-182 was low (63). Further studies showed that ciR3175 and testin functioned as ceRNAs by competitively sponging miR-182 in EECs. In addition, testin inhibited EEC apoptosis by reducing the expression levels of BCL-2/BAX via the MAPK pathway. Therefore, the ciR3175-miR182-testin may be essential for the development of pre-receptive endometrium in goats. High-quality circRNA expression profiles were obtained from endometrial tissue in these studies (Figure 2C).
A better understanding of the conditions associated with endometrial receptivity may help in improving the rate of embryonic bedding, which positively affects the treatment of female infertility (64). Mechanistic research has concentrated on ceRNA regulatory networks, such as the ciR-8073-miR181a-neurotensin, circ-8073/miR-449a/miR-34/CEP55, circRNA-9119-miR-26a-PTGS2, and ciR3175-miR182-testin networks. Many circRNAs are differentially expressed between PE and RE. Following GO and KEGG pathway analysis, interaction network analysis of circRNA-miRNA-mRNA and analysis of circRNAs and their host genes may improve our understanding of how circRNAs mediate the regulation of target genes in the development of endometrial receptivity. The above studies have demonstrated that some circRNAs exert their biological effects on endometrial receptivity by competitively sponging miRNA, thus inducing the expression of miRNA’s targets. In the studies of ciR8073, this circRNA was validated as target for three miRNAs (miR-181a, miR-449 and miR-34a/c), suggesting that a circRNA was regulated by multiple miRNAs, and further analysis should investigate whether a miRNA could be targeted for multiple circRNAs in endometrial receptivity. However, in addition to ceRNA regulatory mechanisms, other underlying mechanisms, such as circRNA-protein interactions that also lead to significant changes in circRNA expression as well as the function of individual circRNAs in endometria, need further investigation. These findings will expectedly increase the diversity of the endometrial transcriptome for circRNAs and yield new insights into the development of endometrial receptivity.
Thousands of circRNAs are encoded by the human genome in a context-dependent manner (65). Due to their abundant expression, stability, and multiple MREs, a large number of circRNAs function via ceRNA regulatory mechanisms (66). circRNAs interact with RNA-binding proteins via direct binding, which constitutes another major regulatory mechanism of circRNAs (67). In addition, inflammation of endometrium is a complex condition, which is almost involved in all endometrial disorders (68–70). Further research is needed for a better understanding of circRNAs in inflammation of reproductive tract.
Certain properties of circRNAs, such as small size and stability, are ideally suited for a role as biomarkers. Furthermore, tissue-specific expression patterns of circRNAs are closely associated with clinical phenotypes. In addition, circRNAs, which are not affected by endonuclease degradation and are stable in formalin-fixed paraffin-embedded tissues (71), can be detected in vaginal secretions (72), and therefore should be investigated further for the purpose of diagnosing related diseases.
Due to being considered as a “hotspot” of RNA research, an increasing number of circRNAs are identified via effective high-throughput sequencing techniques and bioinformatics. However, studies pertaining to the emerging role of circRNAs in the endometrium are yet in the formative stages. Advanced databases, testing tools, and research techniques have enabled circRNAs to be recognized as potential non-invasive biomarkers of reproductive as well as gynecological diseases. In addition, there is also a lack of studies evaluating the panoramic view of circRNAome and individual circRNA along the entire menstrual cycle in human endometrium. Considered together, circRNA research allows researchers to enter a new level in epigenetic regulatory networks, whereby future research may lead to a better understanding of the mechanisms regulating circRNAs in the endometrium.
JT and YC (3rd author) drafted the manuscript. HY, YC (4th author), HC, ZL, LL, YZ, XC, and ZY revised the article. All authors contributed to the article and approved the submitted version.
This study was supported by grants from the Sanming Project of Medicine in Shenzhen (SZSM201812041) and Clinical Research Funding from Shenzhen Second People’s Hospital (4001023).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Marquardt RM, Kim TH, Shin JH, Jeong JW. Progesterone and Estrogen Signaling in the Endometrium: What Goes Wrong in Endometriosis? Int J Mol Sci (2019) 20(15):3822. doi: 10.3390/ijms20153822
2. Fu XD. Non-Coding RNA: A New Frontier in Regulatory Biology. Natl Sci Rev (2014) 1:190–204. doi: 10.1093/nsr/nwu008
3. Esteller M. Non-Coding RNAs in Human Disease. Nat Rev Genet (2011) 12:861–74. doi: 10.1038/nrg3074
4. Xu S, Zhou LY, Ponnusamy M, Zhang LX, Dong YH, Zhang YH, et al. A Comprehensive Review of circRNA: From Purification and Identification to Disease Marker Potential. PeerJ (2018) 2018:1–28. doi: 10.7717/peerj.5503
5. Zhang Z, Yang T, Xiao J. Circular RNAs: Promising Biomarkers for Human Diseases. EBioMedicine (2018) 34:267–74. doi: 10.1016/j.ebiom.2018.07.036
6. Verduci L, Strano S, Yarden Y, Blandino G. The circRNA–microRNA Code: Emerging Implications for Cancer Diagnosis and Treatment. Mol Oncol (2019) 13:669–80. doi: 10.1002/1878-0261.12468
7. Burke WM, Orr J, Leitao M, Salom E, Gehrig P, Olawaiye AB, et al. Endometrial Cancer: A Review and Current Management Strategies: Part I. Gynecol Oncol (2014) 135:385–92. doi: 10.1016/j.ygyno.2014.08.035
8. Chen BJ, Byrne FL, Takenaka K, Modesitt SC, Olzomer EM, Mills JD, et al. Analysis of the Circular RNA Transcriptome in Endometrial Cancer. Oncotarget (2018) 9:5786–96. doi: 10.2147/CMAR.S197343
9. Xu H, Gong Z, Shen Y, Fang Y, Zhong S. Circular RNA Expression in Extracellular Vesicles Isolated From Serum of Patients With Endometrial Cancer. Epigenomics (2018) 10:187–97. doi: 10.2217/epi-2017-0109
10. Ye F, Tang QL, Ma F, Cai L, Chen M, Ran XX, et al. Analysis of the Circular RNA Transcriptome in the Grade 3 Endometrial Cancer. Cancer Manag Res (2019) 11:6215–27. doi: 10.2147/CMAR.S197343
11. Dou Y, Kawaler EA, Zhou DC, Liu T, Fenyö D, Gritsenko MA, et al. Proteogenomic Characterization of Endometrial Carcinoma. Cell (2020) 180:1–20. doi: 10.1016/j.cell.2020.01.026
12. Fabjani G, Kucera E, Schuster E, Minai-Pour M, Czerwenka K, Sliutz G, et al. Genetic Alterations in Endometrial Hyperplasia and Cancer. Cancer Lett (2002) 175:205–11. doi: 10.1016/S0304-3835(01)00714-5
13. Mills AM, Longacre TA. Endometrial Hyperplasia. Semin Diagn Pathol (2010) 27:199–214. doi: 10.1053/j.semdp.2010.09.002
14. Bajwa P, Nielsen S, Lombard JM, Rassam L, Nahar P, Rueda BR, et al. Overactive mTOR Signaling Leads to Endometrial Hyperplasia in Aged Women and Mice. Oncotarget (2017) 8:7265–75. doi: 10.18632/oncotarget.13919
15. Zong ZH, Liu Y, Chen S, Zhao Y. Circ_PUM1 Promotes the Development of Endometrial Cancer by Targeting the miR-136/NOTCH3 Pathway. J Cell Mol Med (2020) 24(7):4127–35. doi: 10.1111/jcmm.15069
16. Shen Q, He T, Yuan H. Hsa_Circ_0002577 Promotes Endometrial Carcinoma Progression Via Regulating miR-197/CTNND1 Axis and Activating Wnt/β-Catenin Pathway. Cell Cycle (2019) 18:1229–40. doi: 10.1080/15384101.2019.1617004
17. Wang Y, Yin L, Sun X. CircRNA Hsa_Circ_0002577 Accelerates Endometrial Cancer Progression Through Activating IGF1R/PI3K/Akt Pathway. J Exp Clin Cancer Res (2020) 39:1–16. doi: 10.1186/s13046-020-01679-8
18. Liu Y, Chen S, Zong ZH, Guan X, Zhao Y. Circrna WHSC1 Targets the miR-646/NPM1 Pathway to Promote the Development of Endometrial Cancer. J Cell Mol Med (2020) 24:6898–907. doi: 10.1111/jcmm.15346
19. Liu Y, Chang Y, Cai Y. circTNFRSF21, a Newly Identified Circular RNA Promotes Endometrial Carcinoma Pathogenesis Through Regulating miR-1227-MAPK13/ATF2 Axis. Aging (Albany NY) (2020) 12:6774–92. doi: 10.18632/aging.103037
20. Liu Y, Chang Y, Cai Y. Hsa_Circ_0061140 Promotes Endometrial Carcinoma Progression Via Regulating Mir-149-5p/STAT3. Gene (2020) 745:144625. doi: 10.1016/j.gene.2020.144625
21. Xu R, Feng F, Yu XS, Liu Z, Lao LF. miR-149-5p Inhibits Cell Growth by Regulating TWEAK/Fn14/PI3K/AKT Pathway and Predicts Favorable Survival in Human Osteosarcoma. Int J Immunopathol Pharmacol (2018) 32:1–12. doi: 10.1177/2058738418786656
22. Sourial S, Tempest N, Hapangama DK. Theories on the Pathogenesis of Endometriosis. Int J Reprod Med (2014) 2014:179515. doi: 10.1002/9781444398519.ch5
23. Filipchiuk C, Laganà AS, Beteli R, Ponce TG, Christofolini DM, Trevisan CM, et al. BIRC5/Survivin Expression as a Non-Invasive Biomarker of Endometriosis. Diagnostics (2020) 10:1–10. doi: 10.3390/diagnostics10080533
24. Laganà AS, Garzon S, Götte M, Viganò P, Franchi M, Ghezzi F, et al. The Pathogenesis of Endometriosis: Molecular and Cell Biology Insights. Int J Mol Sci (2019) 20:1–42. doi: 10.3390/ijms20225615
25. Bendon CL, Becker CM. Potential Mechanisms of Postmenopausal Endometriosis. Maturitas (2012) 72:214–9. doi: 10.1016/j.maturitas.2012.04.010
26. Wang D, Luo Y, Wang G, Yang Q. Circular RNA Expression Profiles and Bioinformatics Analysis in Ovarian Endometriosis. Mol Genet Genomic Med (2019) 7:1–13. doi: 10.1002/mgg3.756
27. Yotova IY, Quan P, Leditznig N, Beer U, Wenzl R, Tschugguel W. Abnormal Activation of Ras/Raf/MAPK and RhoA/ROCKII Signalling Pathways in Eutopic Endometrial Stromal Cells of Patients With Endometriosis. Hum Reprod (2011) 26:885–97. doi: 10.1093/humrep/der010
28. Xing Y, Li A, Yang Y. Li X Xia, Zhang L Nan, Guo H Cai. The Regulation of FOXO1 and Its Role in Disease Progression. Life Sci (2018) 193:124–31. doi: 10.1016/j.lfs.2017.11.030
29. Shen L, Zhang Y, Zhou W, Peng Z, Hong X, Zhang Y. Circular RNA Expression in Ovarian Endometriosis. Epigenomics (2018) 10:559–72. doi: 10.2217/epi-2017-0079
30. Prašnikar E, Kunej T, Knez J, Repnik K, Potočnik U, Kovačič B. Determining the Molecular Background of Endometrial Receptivity in Adenomyosis. Biomolecules (2020) 10:1–28. doi: 10.3390/biom10091311
31. Li R, Qiu Y. Diagnostic Value of Serum ICAM-1 for Endometriosis A Meta-Analysis. Med (United States) (2018) 97(31):e11760. doi: 10.1097/MD.0000000000011760
32. Smolinska N, Szeszko K, Dobrzyn K, Kiezun M, Rytelewska E, Kisielewska K, et al. Transcriptomic Analysis of Porcine Endometrium During Implantation After In Vitro Stimulation by Adiponectin. Int J Mol Sci (2019) 20(6):1335. doi: 10.3390/ijms20061335
33. Xu X, Jia SZ, Dai Y, Zhang JJ, Li X, Shi J, et al. The Relationship of Circular RNAs With Ovarian Endometriosis. Reprod Sci (2018) 25:1292–300. doi: 10.1177/1933719118759439
34. Xu XX, Jia SZ, Dai Y, Zhang JJ, Li XY, Shi JH, et al. Identification of Circular RNAs as a Novel Biomarker for Ovarian Endometriosis. Chin Med J (Engl) (2018) 131:559–66. doi: 10.4103/0366-6999.226070
35. Grümmer R. Animal Models in Endometriosis Research. Hum Reprod Update (2006) 12:641–9. doi: 10.1093/humupd/dml026
36. Bruner-Tran KL, Mokshagundam S, Herington JL, Ding T, Osteen KG. Rodent Models of Experimental Endometriosis: Identifying Mechanisms of Disease and Therapeutic Targets. Curr Women s Heal Rev (2018) 14:173–88. doi: 10.2174/1573404813666170921162041
37. He J, Chang W, Feng C, Cui M, Xu T. Endometriosis Malignant Transformation: Epigenetics as a Probable Mechanism in Ovarian Tumorigenesis. Int J Genomics (2018) 2018:1465348. doi: 10.1155/2018/1465348
38. Du Y, Zhang Z, Xiong W, Li N, Liu H, He H, et al. Estradiol Promotes EMT in Endometriosis Via MALAT1/ miR200s Sponge Function. Reproduction (2019) 157:179–88. doi: 10.1530/REP-18-0424
39. Zhang M, Wang S, Tang L, Wang X, Zhang T, Xia X, et al. Downregulated Circular RNA Hsa_Circ_0067301 Regulates Epithelial-Mesenchymal Transition in Endometriosis Via the miR-141/Notch Signaling Pathway. Biochem Biophys Res Commun (2019) 514:71–7. doi: 10.1016/j.bbrc.2019.04.109
40. Xiong Y, Liu Y, Xiong W, Zhang L, Liu H, Du Y, et al. Hypoxia-Inducible Factor 1α-Induced Epithelial-Mesenchymal Transition of Endometrial Epithelial Cells may Contribute to the Development of Endometriosis. Hum Reprod (2016) 31:1327–38. doi: 10.1093/humrep/dew081
41. Zhang M, Ren C, Xiao Y, Xia X, Fang X. Expression Profile Analysis of Circular RNAs in Ovarian Endometriosis by Microarray and Bioinformatics. Med Sci Monit (2018) 24:9240–50. doi: 10.12659/MSM.913885
42. Xu A, Jiang M, Li S, Fei Q. Down-Regulation of Circ_0061140 Attenuates Ectopic Endometrial Cell Proliferation, Migration and Invasion in Endometriosis Via Inactivating Notch2. Gene (2020) 757:144926. doi: 10.1016/j.gene.2020.144926
43. Jiang N, Pan W, Li J, Cao T, Shen H. Upregulated Circular RNA Hsa_Circ_0008433 Regulates Pathogenesis in Endometriosis Via MiRNA. Reprod Sci (2020) 27:2002–17. doi: 10.1007/s43032-020-00219-1
44. Wang D, Luo Y, Wang G, Yang Q. CircATRNL1 Promotes Epithelial–Mesenchymal Transition in Endometriosis by Upregulating Yes-Associated Protein 1 In Vitro. Cell Death Dis (2020) 11(7):594. doi: 10.1038/s41419-020-02784-4
45. Zhang Y, Yan J, Pan X. miR-141-3p Affects Apoptosis and Migration of Endometrial Stromal Cells by Targeting KLF-12. Pflugers Arch Eur J Physiol (2019) 471:1055–63. doi: 10.1007/s00424-019-02283-2
46. Wang S, Zhang M, Zhang T, Deng J, Xia X, Fang X. microRNA-141 Inhibits TGF-β1-Induced Epithelial-to-Mesenchymal Transition Through Inhibition of the TGF-β1/SMAD2 Signalling Pathway in Endometriosis. Arch Gynecol Obstet (2020) 301:707–14. doi: 10.1007/s00404-019-05429-w
47. González-Foruria I, Santulli P, Chouzenoux S, Carmona F, Chapron C, Batteux F. Dysregulation of the ADAM17/Notch Signalling Pathways in Endometriosis: From Oxidative Stress to Fibrosis. Mol Hum Reprod (2017) 23:488–99. doi: 10.1093/molehr/gax028
48. Luo Y, Wang D, Chen S, Yang Q. The Role of miR-34c-5p/Notch in Epithelial-Mesenchymal Transition (EMT) in Endometriosis. Cell Signal (2020) 72:109666. doi: 10.1016/j.gfj.2019.100488
49. Cakmak H, Taylor HS. Implantation Failure: Molecular Mechanisms and Clinical Treatment. Hum Reprod Update (2011) 17:242–53. doi: 10.1093/humupd/dmq037
50. Sun B, Shi L, Shi Q, Jiang Y, Su Z, Yang X, et al. Circular RNAs Are Abundantly Expressed and Upregulated During Repair of the Damaged Endometrium by Wharton’s Jelly-Derived Mesenchymal Stem Cells. Stem Cell Res Ther (2018) 9:1–13. doi: 10.1186/s13287-018-1046-3
51. Ma Y, Xu Y, Zhang J, Zheng L. Biogenesis and Functions of Circular RNAs and Their Role in Diseases of the Female Reproductive System. Reprod Biol Endocrinol (2020) 18:1–11. doi: 10.1186/s12958-020-00653-5
52. Liu L, Li L, Ma X, Yue F, Wang Y, Wang L, et al. Altered Circular Rna Expression in Patients With Repeated Implantation Failure. Cell Physiol Biochem (2017) 44:303–13. doi: 10.1159/000484887
53. Neykova K, Tosto V, Giardina I, Tsibizova V, Vakrilov G. Endometrial Receptivity and Pregnancy Outcome. J Matern Neonatal Med (2020) 1–15. doi: 10.1080/14767058.2020.1787977
54. Lessey BA, Young SL. What Exactly Is Endometrial Receptivity? Fertil Steril (2019) 111:611–7. doi: 10.1016/j.fertnstert.2019.02.009
55. Vannuccini S, Clifton VL, Fraser IS, Taylor HS, Critchley H, Giudice LC, et al. Infertility and Reproductive Disorders: Impact of Hormonal and Inflammatory Mechanisms on Pregnancy Outcome. Hum Reprod Update (2016) 22:104–15. doi: 10.1093/humupd/dmv044
56. Hu S, Wang Y, Yao G, Sun Y. The Expression Changes of Circular RNAs Between LH + 2 and LH + 7 Human Endometrium. Acta Biochim Biophys Sin (Shanghai) (2019) 51:1296–9. doi: 10.1093/abbs/gmz116
57. Guo. The Pathogenesis of Adenomyosis Vis-À-Vis Endometriosis. J Clin Med (2020) 9:485. doi: 10.3390/jcm9020485
58. Song Y, Zhang L, Liu X, Niu M, Cui J, Che S, et al. Analyses of circRNA Profiling During the Development From Pre-Receptive to Receptive Phases in the Goat Endometrium. J Anim Sci Biotechnol (2019) 10:1–16. doi: 10.1186/s40104-019-0339-4
59. Zhang L, Liu X, Che S, Cui J, Ma X, An X, et al. Endometrial Epithelial Cell Apoptosis Is Inhibited by a Cir8073-Mir181a-Neurotensis Pathway During Embryo Implantation. Mol Ther - Nucleic Acids (2019) 14:262–73. doi: 10.1016/j.omtn.2018.12.005
60. Liu X, Zhang L, Liu Y, Cui J, Che S, An X, et al. Circ-8073 Regulates CEP55 by Sponging miR-449a to Promote Caprine Endometrial Epithelial Cells Proliferation Via the PI3K/AKT/mTOR Pathway. Biochim Biophys Acta - Mol Cell Res (2018) 1865:1130–47. doi: 10.1016/j.bbamcr.2018.05.011
61. Liu X, Zhang L, Yang L, Cui J, Che S, Liu Y, et al. miR-34a/c Induce Caprine Endometrial Epithelial Cell Apoptosis by Regulating circ-8073/CEP55 Via the RAS/RAF/MEK/ERK and PI3K/AKT/mTOR Pathways. J Cell Physiol (2020) 235(12):10051–67. doi: 10.1002/jcp.29821
62. Zhang L, Liu X, Che S, Cui J, Liu Y, An X, et al. CircRNA-9119 Regulates the Expression of Prostaglandin-Endoperoxide Synthase 2 (PTGS2) by Sponging miR-26a in the Endometrial Epithelial Cells of Dairy Goat. Reprod Fertil Dev (2018) 30:1759–69. doi: 10.1071/RD18074
63. Zhang L, Liu X, Ma X, Liu Y, Che S, Cui J, et al. Testin Was Regulated by circRNA3175-miR182 and Inhibited Endometrial Epithelial Cell Apoptosis in Pre-Receptive Endometrium of Dairy Goats. J Cell Physiol (2018) 233:6965–74. doi: 10.1002/jcp.26614
64. Miravet-Valenciano JA, Rincon-Bertolin A, Vilella F, Simon C. Understanding and Improving Endometrial Receptivity. Curr Opin Obstet Gynecol (2015) 27:187–92. doi: 10.1097/GCO.0000000000000173
65. Holdt LM, Kohlmaier A, Teupser D. Molecular Roles and Function of Circular RNAs in Eukaryotic Cells. Cell Mol Life Sci (2018) 75:1071–98. doi: 10.1007/s00018-017-2688-5
66. Zhao X, Cai Y, Xu J. Circular RNAs: Biogenesis, Mechanism, and Function in Human Cancers. Int J Mol Sci (2019) 20(16):3926. doi: 10.3390/ijms20163926
67. Zang J, Lu D, Xu A. The Interaction of circRNAs and RNA Binding Proteins: An Important Part of circRNA Maintenance and Function. J Neurosci Res (2020) 98:87–97. doi: 10.1002/jnr.24356
68. Drizi A, Djokovic D, Lagana AS, Van Herendael B. Impaired Inflammatory State of the Endometrium: A Multifaceted Approach to Endometrial Inflammation. Current Insights and Future Directions. Prz Menopauzalny (2020) 19:90–100. doi: 10.5114/pm.2020.97863
69. Laganà AS, Scioscia M. Endometrial Cancer in Women With Adenomyosis: An Underestimated Risk? Int J Fertil Steril (2020) 14:260–1. doi: 10.22074/ijfs.2020.44413
70. Scioscia M, Noventa M, Laganà AS. Abnormal Uterine Bleeding and the Risk of Endometrial Cancer: Can Subendometrial Vascular Ultrasound Be of Help to Discriminate Cancer From Adenomyosis? Am J Obstet Gynecol (2020) 223:605–6. doi: 10.1016/j.ajog.2020.05.049
71. Zhang F, Zhao X, Dong H, Xu J. circRNA Expression Analysis in Lung Adenocarcinoma: Comparison of Paired Fresh Frozen and Formalin-Fixed Paraffin-Embedded Specimens. Biochem Biophys Res Commun (2018) 500:738–43. doi: 10.1016/j.bbrc.2018.04.145
Keywords: circRNAs, endometrium, endometriosis, endometrial cancer, high-throughput studies
Citation: Tu J, Yang H, Chen Y, Chen Y, Chen H, Li Z, Li L, Zhang Y, Chen X and Yu Z (2021) Current and Future Roles of Circular RNAs in Normal and Pathological Endometrium. Front. Endocrinol. 12:668073. doi: 10.3389/fendo.2021.668073
Received: 15 February 2021; Accepted: 11 May 2021;
Published: 26 May 2021.
Edited by:
Signe Altmäe, University of Granada, SpainCopyright © 2021 Tu, Yang, Chen, Chen, Chen, Li, Li, Zhang, Chen and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiajie Tu, dHVqaWFqaWVAYWhtdS5lZHUuY24=; Zhiying Yu, bGl6aGV5enlAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.