
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 17 March 2021
Sec. Reproduction
Volume 12 - 2021 | https://doi.org/10.3389/fendo.2021.599149
 Xiaoheng Li1,2†
Xiaoheng Li1,2† Qiqi Zhu1,2†
Qiqi Zhu1,2† Zina Wen1,3†
Zina Wen1,3† Kaimin Yuan2
Kaimin Yuan2 Zhijian Su4
Zhijian Su4 Yiyan Wang1
Yiyan Wang1 Ying Zhong3*
Ying Zhong3* Ren-Shan Ge1,2*
Ren-Shan Ge1,2*The function of immature Leydig cells is regulated by hormones, such as androgen and luteinizing hormone (LH). However, the regulation of this process is still unclear. The objective of this study was to determine whether luteinizing hormone (LH) or androgens contribute to this process. Immature Leydig cells were purified from 35-day-old male Sprague Dawley rats and cultured with LH (1 ng/ml) or androgen (7α-methyl-19- nortestosterone, MENT, 100 nM) for 2 days. LH or MENT treatment significantly increased the androgens produced by immature Leydig cells in rats. Microarray and qPCR and enzymatic tests showed that LH up-regulated the expression of Scarb1, Cyp11a1, Cyp17a1, and Srd5a1 while down-regulated the expression of Sult2a1 and Akr1c14. On the contrary, the expression of Cyp17a1 was up-regulated by MENT. LH and MENT regulate Leydig cell function through different sets of transcription factors. We conclude that LH and androgens participate in the regulation of rat immature Leydig cell function through different transcriptional pathways.
Testosterone secreted by adult Leydig cells in the testis is the main androgen in the male circulation. In mature men, testosterone plays a key role in regulating spermatogenesis and maintaining secondary sexual characteristics (such as increased bone mass, muscle strength, and growth of body hair) (1).
Adult Leydig cells are developed from stem cells during puberty (1, 2). In the rat model, the postnatal development of adult Leydig cells can be conceptually divided into four different stages: stem cells, progenitor cells, immature cells, and adult Leydig cells (1, 3). Stem Leydig cells are the spindle-shaped cells that can differentiate into the Leydig cell lineage (4–6). The first committed cell in this lineage is the progenitor Leydig cell (1). It is also spindle-shaped and expresses the Leydig cell lineage biomarker cytochrome P450 cholesterol side chain cleavage enzyme (CYP11A1, encoded by Cyp11a1), 3β-hydroxysteroid dehydrogenase 1 (HSD3B1, encoded by Hsd3b1), and cytochrome P450 17α-hydroxylase/17,20-lyase (CYP17A1, encoded by Cyp17a1) and appears in interstitial compartment of the testes about 11 to 14 days after birth (1, 3). Progenitor Leydig cells do not have the last step of androgen synthetic enzyme 17β-hydroxysteroid dehydrogenase 3 (HSD17B3, encoded by Hsd17b3), which can catalyze the conversion of androstenedione to testosterone (7), but have higher androgen-metabolizing enzymes, including steroid 5α-reductase isoform 1 (SRD5A1, encoded Srd5a1) and 3α-hydroxysteroid dehydrogenase (AKR1C14, encoded by Akr1c14), which metabolize androstenedione to androstanedione, and further into androsterone (7). In the first week after the appearance of progenitor Leydig cells, they rapidly divide to increase the number of Leydig cells (8), and then differentiate into oval immature Leydig cells around 28 days postpartum (9, 10). Immature Leydig cells express the last-step androgen synthetic enzyme HSD17B3 that catalyzes androstenedione into testosterone, which is further converted into 5α-androstane-3α,17β-diol (androstanediol, DIOL), because these cells still have high capacity of androgen-metabolic enzymes SRD5A1 and AKR1C14 (7). The development of adult Leydig cells from progenitor cells includes an increase of androgen synthetic enzymes (CYP11A1, HSD3B1, CYP17A1, and HSD17B3) and a decrease of androgen metabolic enzymes (SRD5A1 and AKR1C14) (7).
In immature Leydig cells, which hormone controls the proliferation and differentiation of Leydig cells, it is still unclear, but many factors are involved. These factors include luteinizing hormone (LH), insulin-like growth factor-1, transforming growth factor α, transforming growth factor β, interleukin 1, thyroid hormone, anti-Müllerian hormone, and androgen (1). LH has been shown to be an important regulator of Leydig cell function (5, 11). LH works by binding to the LH receptor (LHCGR, encoded by Lhcgr) on the Leydig cell membrane, activating adenyyl cyclase to increase intracellular cAMP levels, thereby activating protein kinase A transcription factors (CREB and CREM) signals to regulate Leydig cell function (12). Indeed, knocking out Lhcgr in mice resulted in functional defects in Leydig cells, and there are almost no mature adult Leydig cells, only some progenitor Leydig cells exist (13, 14). Besides LH, other factors have yet to be determined. Rat immature Leydig cells contain the highest level of androgen receptor (NR3C4) (9). The role of NR3C4 in the development and function of Leydig cells is more complex, depending on the maturity of Leydig cells. Studies using feminized testis mice (Tfm) with Nr3c4 null mutations have demonstrated the role of NR3C4 in Leydig cells (15). Despite the high circulating levels of LH, the Tfm had a significant reduction in testosterone production (15–17). In fact, the activity of CYP17A1 and HSD17B3 enzymes is significantly reduced in Tfm testes (15–17). Conditional knockout of Nr3c4 in mouse Leydig cells also demonstrated that autocrine NR3C4 signal is necessary for the maturation of adult Leydig cells and the regulation of steroidogenic enzymes (18). Although there is a lot of evidence that NR3C4 is important in the early stages of Leydig cell function, such as the progenitor Leydig cell stage (19, 20), NR3C4 signaling in the maturation of immature Leydig cells to adult Leydig cells has not been determined and the detailed signaling for NR3C4 is unclear. The objective of this study was to investigate and compare the effects of LH and androgen signals on the function of rat immature Leydig cells.
Ovine LH was a gift from the National Institute of Diabetes and Digestive and Kidney (USA). Since the immature Leydig cells in the 35-day-old rat testis have higher SRD5A1 and AKR1C14 (7), they easily metabolize testosterone to the weak androgen androstanediol, so a synthetic SRD5A1-resistant androgen 7α-methyl-19-nortestosterone (MENT) was used (7, 21). MENT is more potent than testosterone in specifically binding NR3C4 (7, 21). MENT was obtained from Upjohn (Kalamazoo, MI). Sprague-Dawley rats were purchased from Shanghai Laboratory Animal Center (Shanghai, China). The animal experiment protocol was approved by the Institutional Animal Care and Use Committee of Wenzhou Medical University and carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
The method of isolating immature Leydig cells was previously described (19). Briefly, testes from eighteen 35-day-old rats were taken out to isolate immature Leydig cells. The decapsulated testes were dispersed in M199 medium with 0.25 mg/ml collagenase D (Sigma, St. Louis, MO) at 34°C with shaking (75 rpm) for 10 min. The separated cells were filtered through two layers of 100-μm nylon mesh, centrifuged at 250g, and resuspended in 55% isotonic Percoll. Subsequently, a density gradient centrifugation was performed in Percoll at 25,000g at 4°C for 45 min. The density of the immature Leydig cell fraction collected was between 1.070 and 1.080 g/ml. The cells were washed with Hank’s buffered saline, centrifuged at 250g, and then resuspended in phenol red-free Dulbecco’s modified Eagle medium:Nutrient Mixture F-12 (DMEM-F12) medium (Sigma, St. Louis, MO) supplemented with 1 mg/ml bovine serum albumin. According to the published method (22), the purity of immature Leydig cells was determined by histochemical staining of HSD3B1 using 0.4 mM etiocholanolone as a substrate and NAD+ as a cofactor. The enrichment of immature Leydig cells was typically as high as 95%. In total, eight isolations were performed.
To test the effects of MENT and LH on androgen synthesis and metabolism, we isolated immature Leydig cells. 1 × 106 Leydig cells were seeded in a 12-well plate and cultured with different concentrations of MENT (10, 100, and 500 nM) or LH (0.1, 1, and 5 ng/ml) or MENT (100 nM) + LH (1 ng/ml) for 48 h. Testosterone and androstanediol from the culture medium were measured and then compared with the control (untreated, basal). Immature Leydig cells were washed with phosphate-buffered saline (PBS) twice and warmed 1× trypsin–EDTA solution (ThermoFisher Scientific Waltham, MA) was added, and the cells were harvested. An aliquot of cell solution was added to a hemocytometer for counting Leydig cell number. Cell viability was checked by the exclusion of 0.4% trypan blue using a kit (ThermoFisher Scientific Waltham, MA) as previously described (23).
According to the manufacturer’s instructions (Invitrogen, Carlsbad, CA), total RNA was extracted from Leydig cells by homogenization in TRIzol. Briefly, rat immature Leydig cells were harvested and homogenized in TRIzol reagent and then extracted with chloroform. The aqueous supernatant containing RNA was retained and the RNA was precipitated with isopropanol. The RNA pellet was eluted with 70% ethanol, air dried, and resuspended in RNase-free water. The RNA was further purified using the RNeasy Kit according to the manufacturer’s instructions (Qiagen, Valencia, CA). A NanoDrop 2000 spectrophotometer (Fisher Scientific, NJ) was used to determine the RNA concentration, and an Agilent 2100 bioanalyzer (Santa Clara, CA) was used to determine the integrity in each individual RNA sample.
Three independently isolated batches of immature Leydig cells treated with LH and/or MENT in each group were used for the microarray array. The whole genome-expressed RatRef-12 Expression BeadChip from Illumina Inc (San Diego, CA) was used. Each BeadChip contains 21,910 genes, which are mainly selected from the NCBI RefSeq database covering the entire rat transcriptome. As previously reported (24), probe labeling, hybridization, washing, and scanning were performed according to the manufacturer’s instructions using the Illumina Total Prep kit (Applied Biosystems, Foster City, CA). First strand cDNA was synthesized in a total volume of 20 μl with the supplied reagents. The complete first-strand product was used for second-strand synthesis and then subjected to column purification. The purified product was then used for in vitro transcription using T7 polymerase. Biotin-16-dUTP was incorporated in this step, resulting in a biotinylated complementary RNA (cRNA) probe. An Agilent 2100 bioanalyzer was used to verify the integrity of the probe. The labeled cRNA (750 ng) was hybridized with the array overnight at 58°C in a total volume of 30 μl of hybridization buffer, followed by rigorous washing and scanning after hybridization.
Scanned microarray expression data was imported into BeadStudio (Illumina, San Diego, CA) for normalization, preliminary analysis, and filtering. Average normalization without background subtraction was used, and the Illumina custom error model was used to generate present/absent calls for each probe (“present” defined as p < 0.01 for signal detection) on each array and to call differentially expressed genes at each of samples (defined as p < 0.05 after false discovery rate correction). Normalized data from BeadStudio was filtered to exclude genes that were not expressed in the testis (i.e., data from probes that were classed as “absent” in all samples). The net count of target mRNA was calculated by subtracting the background count from the mRNA count. Of the 21,910 genes present in the data, further analysis was conducted. In BeadStudio, linear plot comparison between groups was made, and Heatmap profiling was generated. The data were further imported into Microsoft Access 2010, and queries were generated to find Leydig cell specific genes.
In order to characterize the biological process, Gene MicroArray Pathway Profiler 2.1 (GenMAPP2.1, San Francisco, CA) software was used to generate a list of significant (P < 0.05) regulatory pathways, and GenMAPP2.1 was used to create a map of signaling pathways. We imported the statistical results into the program and illustrated biological pathways containing differentially expressed genes. The results of the differential gene expression profile were validated by qPCR.
After reverse transcription of the isolated RNA, the level of specific mRNA species was measured by quantitative real-time PCR (qPCR) using the SYBR method. Briefly, first strand synthesis and qPCR were performed as previously described (25). QPCR was conducted in a volume of 20 µl using a 96-well plate format by SYBR Green PCR Core Reagents (Applied Biosystems, Foster City, CA). Primer titration was performed at a concentration of 300 nM. ABI 7700 system (PE Applied Biosystems) was used to detect fluorescence. Each sample was run in duplicate and in parallel with no template controls. The relative mRNA level of the targeted gene was normalized to ribosomal protein S16 (Rps16, the house-keeping gene as an internal control) by standard curve method. Ct was read, a standard curve was generated, and the RNA expression in each sample was calculated. All primers in this study were designed by Primer 3 software (Whitehead Institute for Biomedical Research, Cambridge, MA). Forward and reverse primers were placed in different exons to minimize the effects of possible DNA contamination. These genes measured in this study are: luteinizing hormone receptor (Lhcgr), scavenger receptor class B member 1 (Scarb1), steroidogenic acute regulatory protein (Star), Cyp11a1, Hsd3b1, Cyp17a1, Hsd17b3, Srd5a1, Akr1c14, Insl3, and Ccnd1. We also examined the biomarkers of mature Leydig cells: Cdk1na, Svs5, and Ptgds. Some primers were used in these papers (25, 26) and were listed in the Supplementary Table S1.
Cells were homogenized and boiled in an equal volume of sample loading buffer, a Tris-Cl buffer (pH 6.8), containing 20% glycerol, 5% sodium dodecyl sulfate, 3.1% dithiothreitol, and 0.001% bromophenol blue. A homogenized sample (50 µg protein) was electrophoresed on a 10% polyacrylamide gel containing sodium dodecyl sulfate. Proteins were transferred onto a nitrocellulose membrane, and after 1-h exposure to 5% skim milk to block nonspecific binding, the membrane was incubated with CYP17A1 (Abcam Trading Company Ltd. Shanghai, China; dilution, 1:1,000) and actin β (ACTB, Cell signaling technology, Danvers, MA; dilution, 1:1,000) antibody. The membrane was then washed and incubated with a goat anti-rabbit antiserum conjugated to horseradish peroxidase diluted 1:5,000. The washing steps were repeated, and the immunoreactive bands were visualized by chemiluminescence using Amersham ECL kit (Arlington Heights, IL). The density was calculated by ImageJ software (National Institutes of Health, USA) and the density of CYP17A1 relative to ACTB was calculated.
Sephadex LH-20 (Pharmacia Biotech, Uppsala, Sweden) column chromatography was used to separate testosterone and androstanediol as previously described (7). The elution system is chloroform: butane: ethanol (50:50:1, v/v/v) saturated with distilled water. Clear separation of testosterone and androstanediol in this system was confirmed using radiolabeled steroids (7). Separated testosterone and androstanediol were collected and their amounts were measured with a tritium-based RIA as previously described (27). In brief, samples were incubated with a mixture of rabbit testosterone antiserum and 3H-testosterone (for testosterone assay) or a mixture of rabbit DIOL antiserum and 3H-androstanediol (for androstanediol assay) at 4°C overnight. Testosterone or androstanediol standards (10–2,000 pg/100 µl) were measured in parallel. Bound steroid was separated from the free steroid by mixing with dextran-coated activated charcoal followed by centrifugation. The bound supernatant was put in a bottle with a scintillation cocktail and the relative amount of radioactivity was counted in β-counter (Packard, Meriden, CT). The coefficient of inter-assay and intraassay was 7–8%.
CYP11A1, HSD3B1, CYP17A1, HSD17B3, SRD5A1, and AKR1C14 activities of intact immature Leydig cells isolated in vitro were measured according to the method previously described (7). In brief, an unlabeled steroid (final concentration being 20 μM) spiked with its respective radiolabeled substrate (1 m Ci) for CYP11A1 (3H-hydroxychloesterol), HSD3B1 (3H-pregnenolone), CYP17A1 (3H-progesterone), and HSD17B3 (3H-androstenedione), SRD5A1 (3H-testosterone), and AKR1C14 (3H-dihydrotestosterone) was added. The reaction was started by adding an aliquot of 0.1 × 106 Leydig cells to the reaction mixture. Since testosterone can be metabolized by 5α-reduction in immature Leydig cells, 4-methyl-aza-3-oxo-5 -pregnan-20(S)-carboxylate (2 μM) was used to inhibit SRD5A1 when the activity of CYP17A1 and HSD17B3 measured. The reaction was maintained at pH 7.2. The reaction mixtures, conducted in triplicate, were maintained at 34°C in a shaking water bath (75 rpm) for 10 min. The radioactive products were produced. Reactions were terminated by adding ice-cold ether, and steroids were rapidly extracted. The ether layer was dried under nitrogen. The extracted steroid was dissolved in 100 μl ether and plotted on a thin-layer chromatography plate. The activity of HSD3B1 was determined by measuring the conversion of pregnenolone to progesterone. The activity of CYP17A1 was determined by measuring the conversion of progesterone to 17α-hydroxyprogesterone and androstenedione. The activity of HSD17B3 was determined by measuring the conversion of androstenedione to testosterone. The activity of SRD5A1 was determined by measuring the conversion of testosterone to dihydrotestosterone. The activity of AKR1C14 was determined by measuring the conversion of dihydrotestosterone to androstanediol. Steroids were separated on the thin layer chromatography plate in chloroform-methanol (97:3, v/v) for HSD3B1, HSD17B3, and SRD5A1 assays, in diethyl ether-acetone (98:2, v/v) for AKR1C14, as well as chloroform-ether (7:1, v/v) for CYP17A1 assay. Radioactivity was measured by System 200/AC3000 radioactive scanner (Bioscan, Washington DC).
The activity of CYP11A1 was determined by measuring the conversion of side-chain labeled 3H-hydroxycholesterol to radioactive 4-hydroxyl-4-methyl-pentanoic acid as previously described (7). In brief, incubation was performed at 34°C for 30 min, and after incubation, 0.5 ml NaOH (0.5 M) was added. The mixture was extracted twice with 2 ml of chloroform and mixed with neutral alumna to remove non-metabolized substrate, and an aliquot was removed for measurement by liquid scintillation counting on a scintillation counter (Packard, Meriden, CT).
The data were analyzed by Student’s t-test to find significant differences when comparing the two groups (if there were two treatments). The data were analyzed by ANOVA, and then post doc by Tukey’s multiple comparison test to determine significant differences between both groups when comparing three or more groups (including microarray and qPCR data). All data are expressed as means ± SE. The difference is considered significant at p < 0.05.
First, we established culture conditions for immature Leydig cells treated with MENT and LH. After 2 days of culture, LH and MENT did not alter the number of immature Leydig cells and cell viability. On days 1 and 2 after culture, immature Leydig cells can maintain the production of androstanediol and testosterone. However, compared with the value on day 1 (73.600 ± 4.77 ng/106 cells.24 h, Mean ± SD, n = 6), androstanediol production of immature Leydig cells in the control was significantly reduced on day 3 (43.395 ± 9.41 ng/106 cells 24 h), while maintaining testosterone production. As shown in Figure 1A (androstanediol) and Figure 1B (testosterone), MENT showed a stimulating effect on androgen production on days 1 and 2 at 100–500 nM. On the third day, MENT lost its stimulation. As shown in Figure 1C (androstanediol) and Figure 1D (testosterone), the stimulating effect of LH on androgens on days 1 and 2 was 0.1–5 ng/ml. On the third day, LH lost stimulation. LH (1 ng/ml) showed the highest increase in androstanediol and/or testosterone production on day 2.
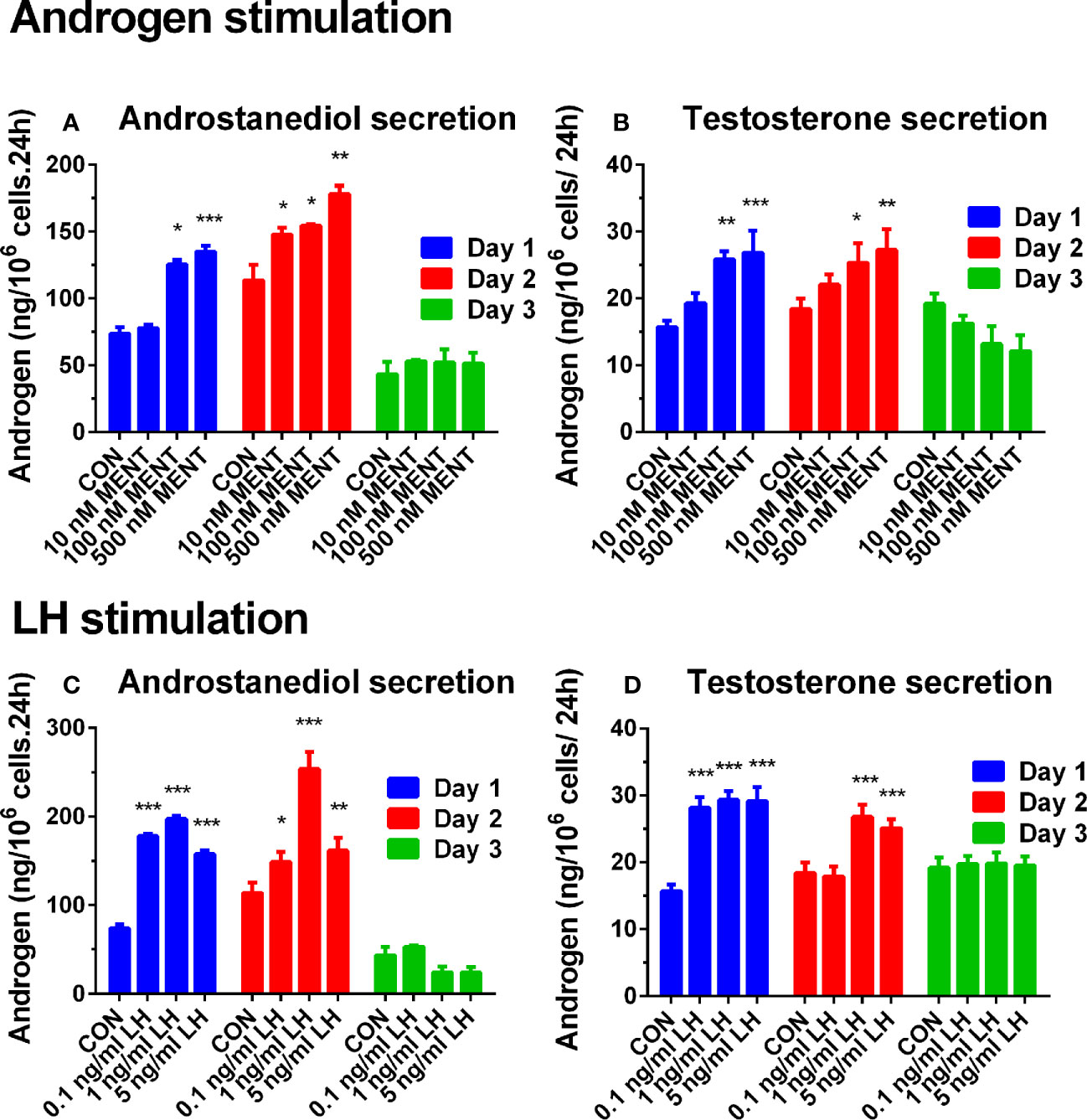
Figure 1 Effect of hormone treatment on androgen production in rat immature Leydig cells. Immature rat Leydig cells were cultured with MENT or LH for 3 days. The levels of medium testosterone and androstanediol were measured. (A, B) MENT treatment. (C, D) LH treatment. (A, C) Androstanediol level. (B, D) Testosterone level. Mean ± SEM, n = 6. *, **, and *** represent significant differences compared with the control group each day at P < 0.05, 0.01, and 0.001, respectively.
Then, we used MENT (100 nM) and LH (1 ng/ml) alone or in combination to treat rat immature Leydig cells for 2 days as described in Figure 2A. As shown in Figure 2, the combination of LH and MENT further increased the levels of androstanediol (Figure 2A) without affecting testosterone (Figure 2B), indicating that LH and MENT synergistically act on androstanediol production in rat immature Leydig cells.
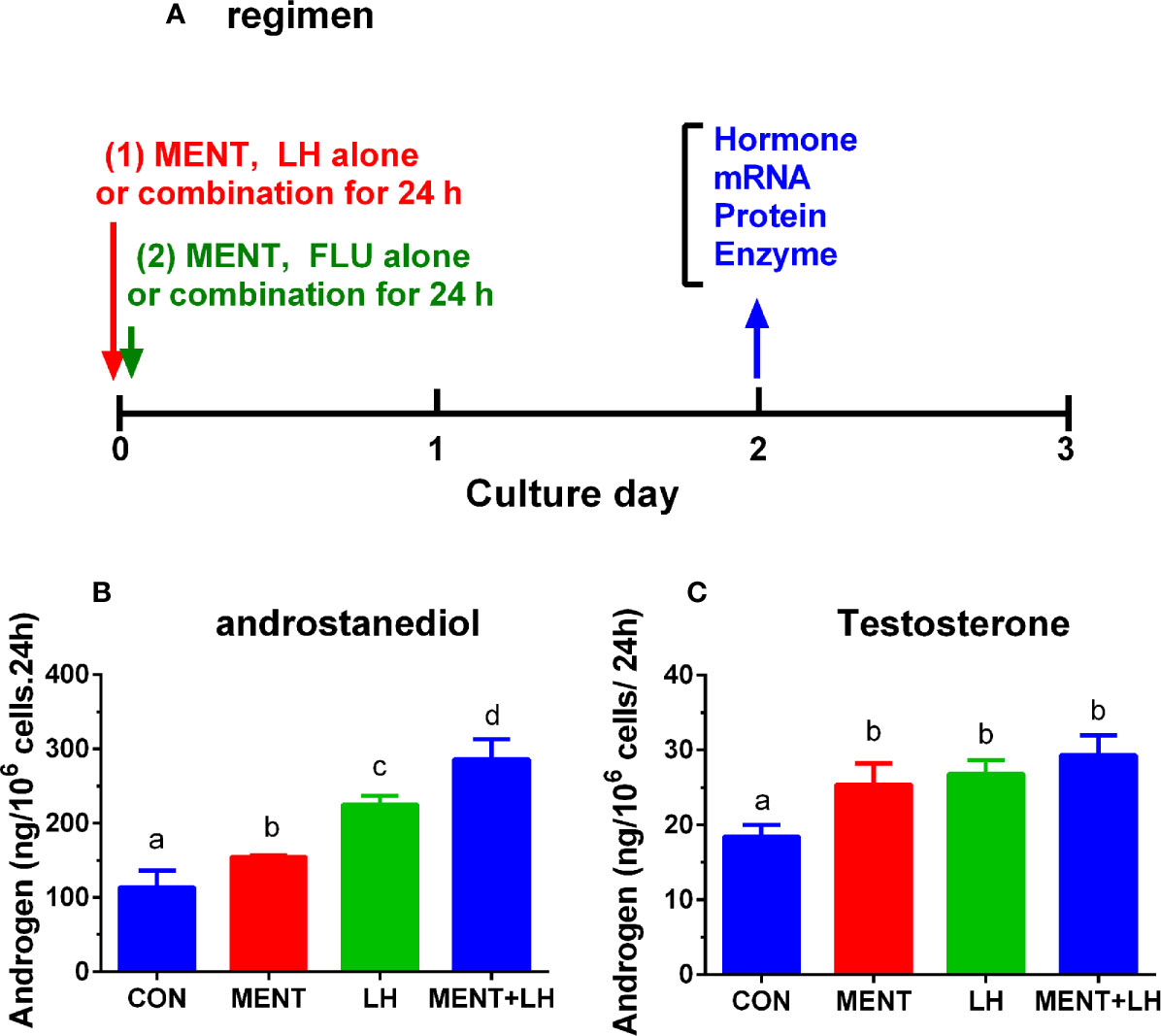
Figure 2 Regimen of treatment and effect of MENT and LH alone or in combination on androgen production in immature Leydig cells in rats. Immature rat Leydig cells were cultured with MENT or LH alone or in combination for 2 days or with FLU or MENT or in combination for another study (A). The levels of medium androstanediol and testosterone were measured. (A) Regimen for this experiment and for experiment in Figures 7 and 8. (B) Androstanediol. (C) Testosterone level. Mean ± SEM, n = 4. The same letter means that there is no significant difference between the two groups at P < 0.05.
Rat immature Leydig cells (three different replicates) were treated for 2 days without treatment (control) or 100 nM MENT, 1 ng/ml LH, or 100 nM MENT + 1 ng/ml LH (MENT + LH). The Illumina RatRef-12 expression chip was used to determine changes in mRNA levels. The whole genome expression containing 21,910 probes was analyzed. The counts of house-keeping gene Rps16 were 150.00 ± 4.00, 152.97 ± 9.37, 160.94 ± 8.88, and 145.56 ± 8.39, for control, MENT, LH, and MENT + LH, respectively and there was no significant difference between two groups. There was no statistical difference between two groups for Actb, another housing gene (data not shown). Among these probes, 8,047 probes were detected in the control group. When compared with the control group, MENT up-regulated 98 genes by approximately three times (Table 1 lists the genes whose expression has increased five times). Of these genes, 74 were also up-regulated by two folds and higher by LH, and only one (Aoc3) was down-regulated by LH (Figure 3A). Compared with controls, MENT down-regulated the expression of 67 genes by approximately three times (Table 2 lists the genes whose expression has decreased five times). Of these 67 genes, 18 genes were also down-regulated ≥2 folds, and only five were up-regulated by LH (Figure 3B). This suggests that the up-regulated genes between MENT and LH are mostly overlapping while the down-regulated genes are very different.
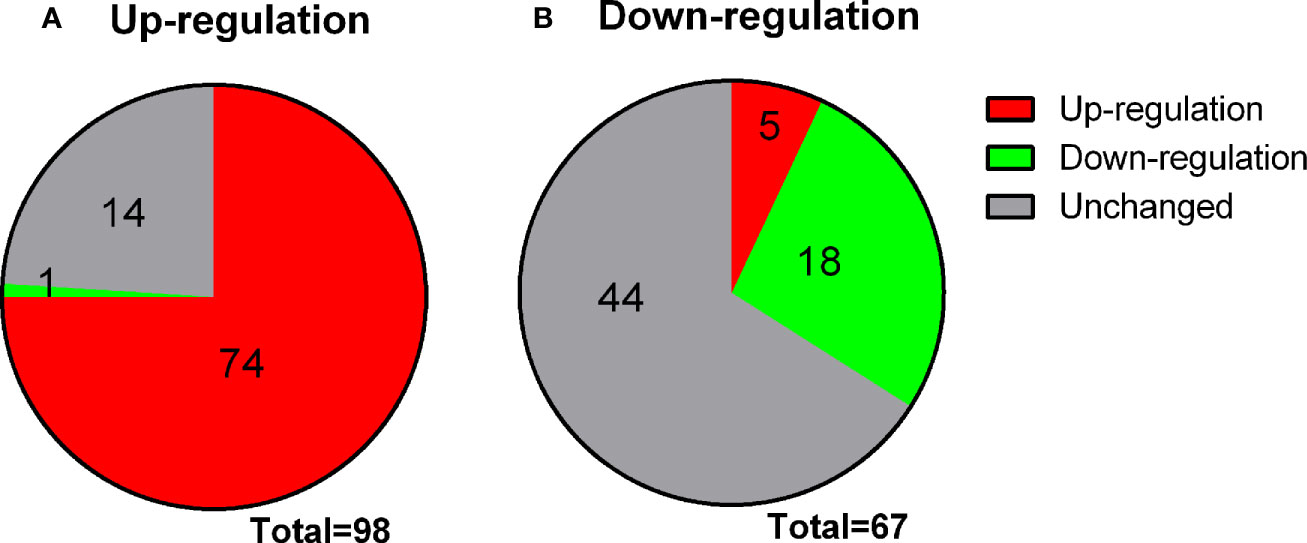
Figure 3 Changes in genes overlap between LH and MENT. (A) 98 genes up-regulated by MENT ≥3 times, 74 genes up-regulated by LH ≥3 times, 1 gene down-regulated by LH and 14 unaltered genes; (B) 67 genes down-regulated by MENT were 18 genes down-regulated, 4 genes up-regulated and 44 unaltered genes after LH treatment.
Using GenMAPP2, we discovered several pathways specific to MENT or LH regulation. As shown in Figure 4 and Supplementary Table S2, steroidogenesis-related genes (Scarb1, Cyp11a1, and Cyp17a1) were significantly increased by ≥2 folds, while the two steroid metabolic enzymes (Sult1a1 and Akr1c14) were significantly decreased by ≥2 folds after LH treatment. However, after MENT treatment, only Cyp17a1 increased significantly by ≥2 folds. This suggests that MENT and LH stimulate the steroidogenesis of immature Leydig cells by targeting different genes. Interestingly, the combination of LH and MENT led to the increases of two additional genes (Hsd3b1 and Srd5a1) besides Scarb1, Cyp11a1, and Cyp17a1, indicating that the combination of LH and MENT is conducive to optimized steroidogenesis.
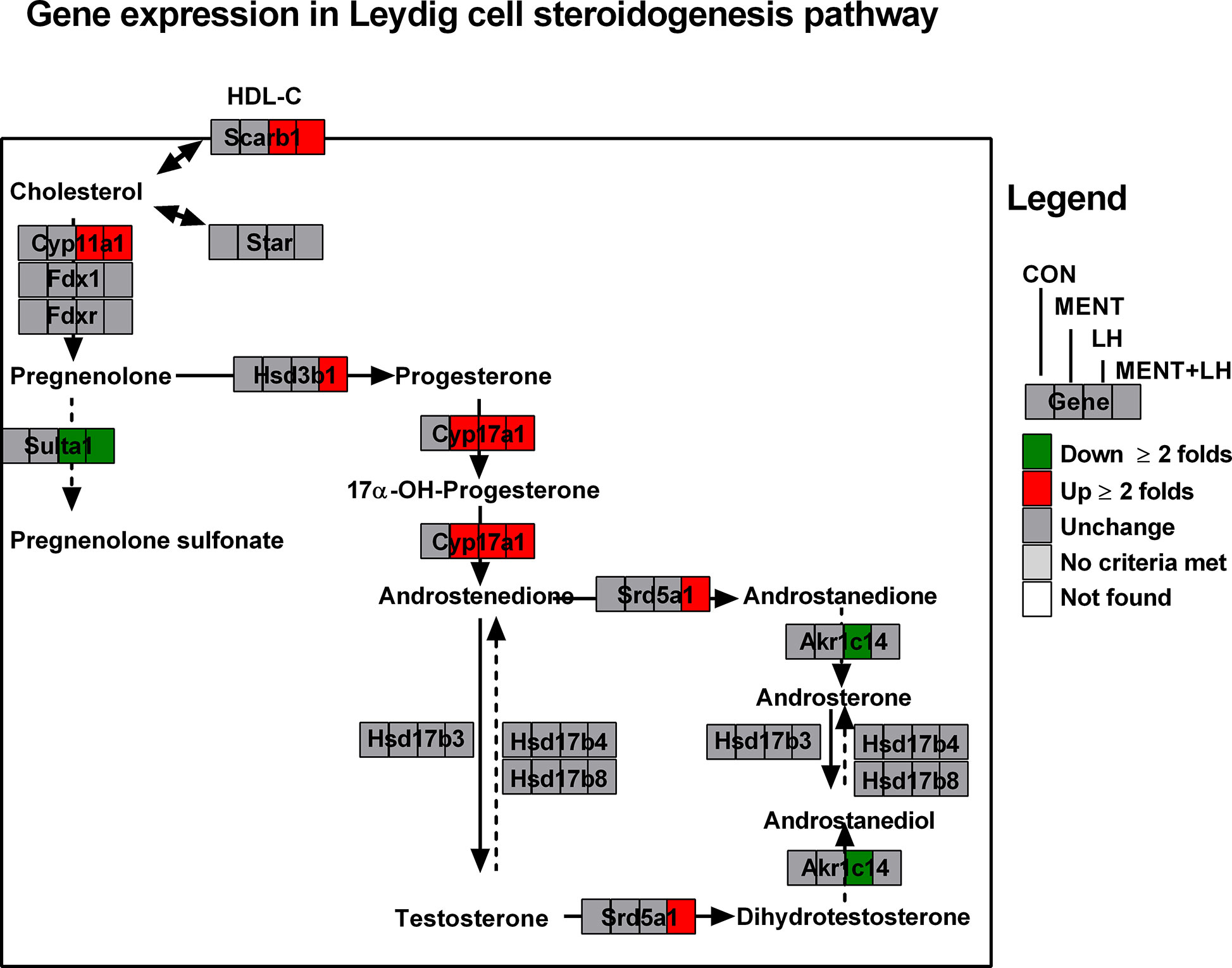
Figure 4 Analysis of gene pathways for MENT, LH alone or in combination (MENT+LH) to regulate steroidogenesis. Red indicates up-regulated genes; green indicates down-regulated genes. Solid arrow indicates androgen synthesis pathway; Dash arrow indicates steroid metabolism; Double arrows indicates transport of cholesterol.
When we examined cell cycle pathway (Supplementary Figure S1), we found that the expression of Cdkn1a, Cdkn1b, Gadd45a, Ccng2, and RGD735212 increased by ≥2 folds after MENT treatment. MENT treatment also resulted in down-regulation of Ccnd1 and Pola2. The expression of Cdkn1a, Gadd45a, and Ccng2 was also significantly up-regulated by ≥2 folds after LH treatment. These results indicate that both LH and MENT cause cell cycle arrest by inducing similar cell cycle arrest regulators.
When we examined the transcription pathway, we found that MENT and LH regulated quite different sets of transcription factors. We divided the patterns of gene expression in four categories. As shown in Supplementary Table S2 and Figure 5A (for up-regulated genes), category 1 lists the gene expression only affected by MENT, which contains Atf5, Gfi1, Mecp2, Pawr, Ppard, Raf2, and Vps36; category 2 lists the gene expression affected by MENT and MENT+LH, which contains Atf4, Ccnl1, Dit3, Per1, Stat5b, and Trib3); category 3 lists the gene expression only affected by LH, which contains Egr1, Crem1, and Runx1; category 4 lists the gene expression affected by LH, MENT, and MENT + LH, which contains Atf3, Btg2, and Notch4. As shown in Figure 5B (for down-regulated genes), category 1 lists the gene expression only affected by MENT, which contains Smarcd2; category 2 lists the gene expression affected by MENT and MENT + LH, which contains Zbtb7a; category 3 lists the gene expression only affected by LH, which contains Aes and Tceal1; category 4 lists the gene expression affected by LH, MENT, and MENT + LH, which contains Irf7 and Nr1h3. This suggests that MENT function through divergent transcriptional regulation.
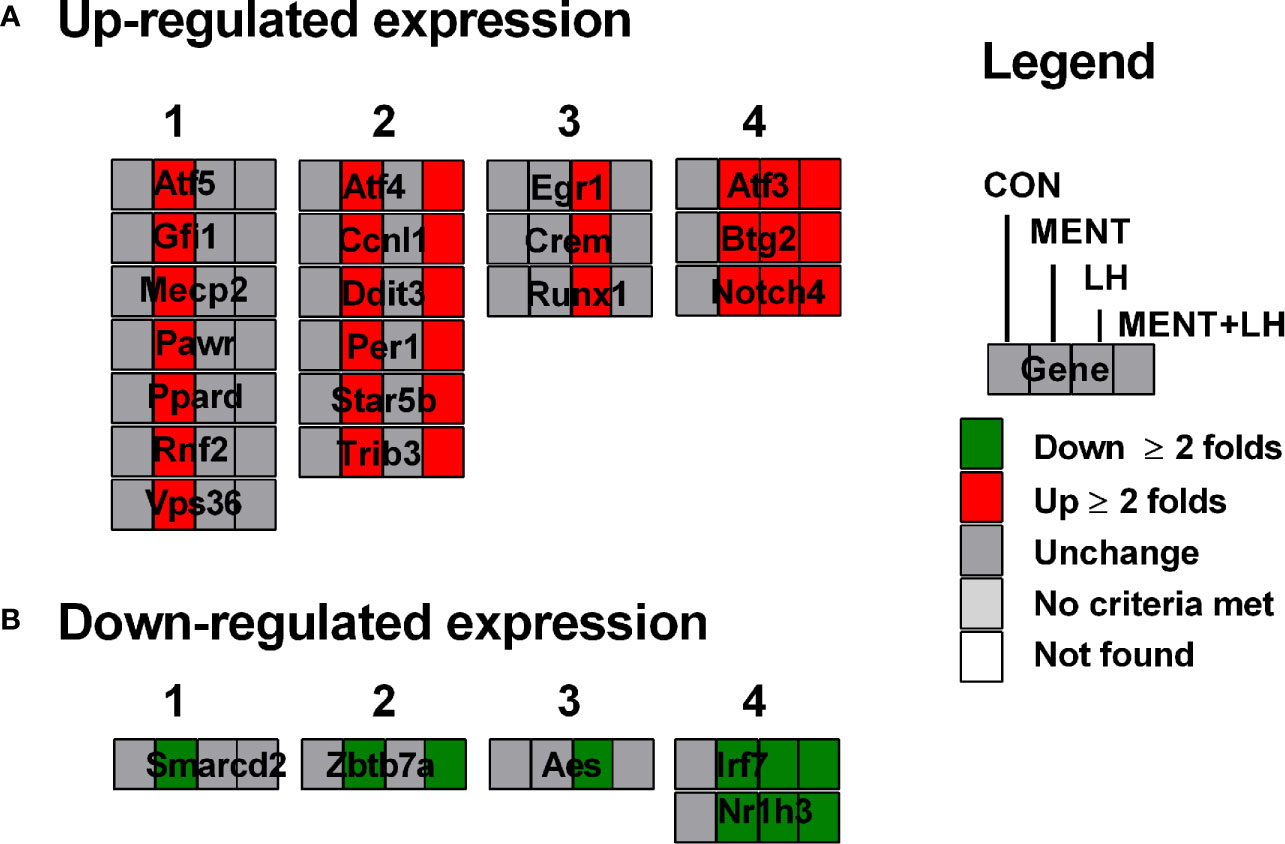
Figure 5 Transcription factors are regulated by MENT, LH alone or in combination (MENT+LH). Red indicates that the gene is up-regulated; green indicates that the gene is down-regulated. (A) Up-regulated expression of genes. (B) Down-regulated expression of genes; Four categories: (1) Only up-regulated/down-regulated in MENT-treated groups; (2) Up-regulated/down-regulated in MENT alone or MENT+LH groups; (3) Only up-regulated/down-regulated in LH-treated group; (4) Up-regulated/down-regulated in MENT, LH, and MENT + LH groups.
Rps16 was used as a house-keeping gene. Its levels by qPCR were not significantly changed between groups (control, MENT, LH, and MENT + LH, Figure 6). We selected three sets of genes: unchanged after treatment with MENT and LH (Lhcgr, Star, and Hsd17b3), up-regulated by LH (Scarb1, Cyp11a1, and Srd5a1), up-regulated by LH and MENT (Cyp17a1, Cdkn1a, and Insl3), up-regulated only by LH plus MENT (Hsd3b1), and down-regulated by LH and MENT (Ccnd1, Akr1c14, and Ccx12) and subjected to qPCR analysis. We found similar trends in these gene changes in the microarray (Figure 6), which indicates that the microarray analysis is reliable.
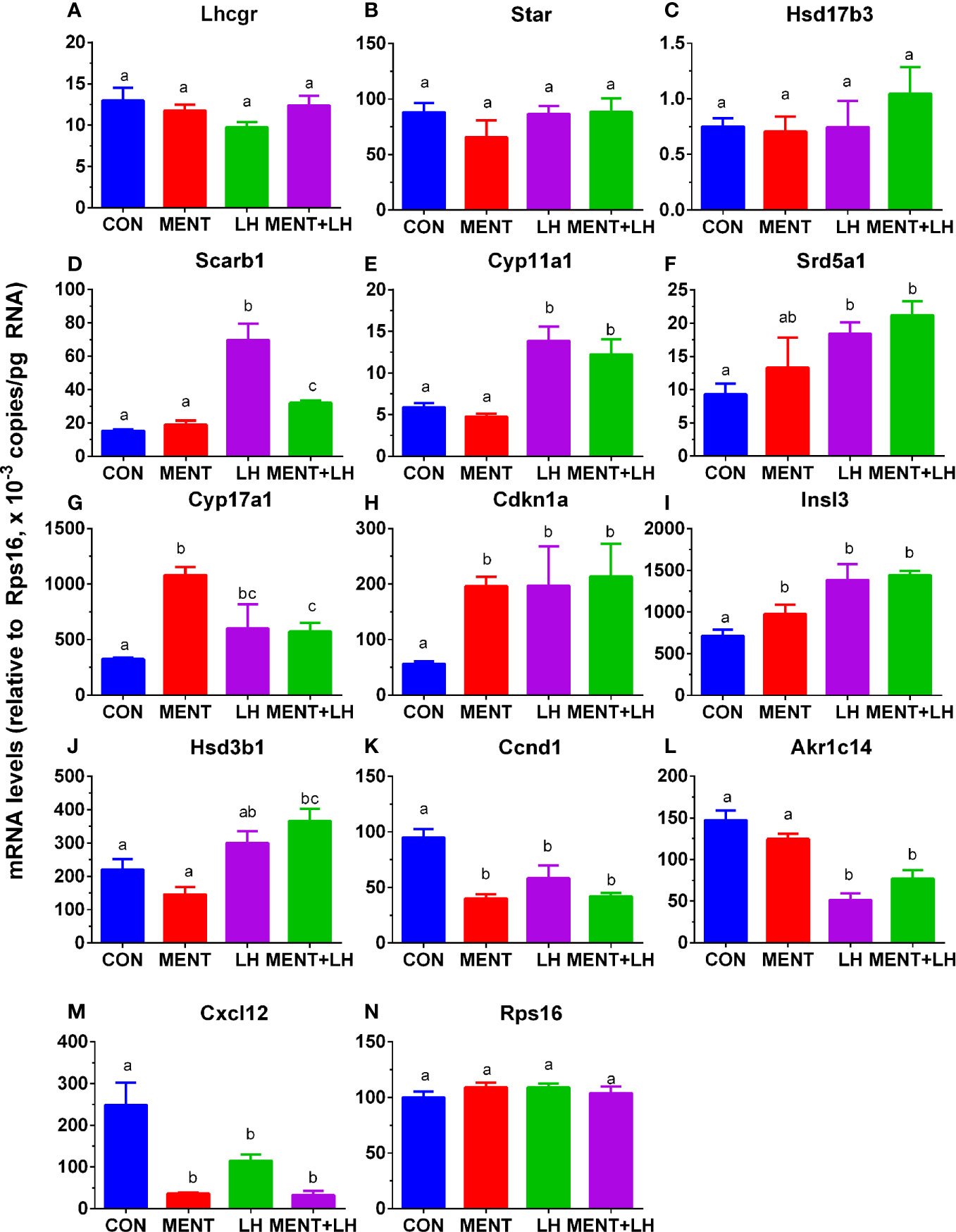
Figure 6 Messenger RNA levels are regulated by MENT or LH alone or in combination (MENT+LH) after qPCR analysis. In the presence of MENT or LH alone or in combination, immature Leydig cells were cultured for 2 days. The following genes were analyzed (A–N): Lhcgr, Star, Hsd17b3, Scarb1, Cyp11a1, Srd5a1, Cyp17a1, Cdkn1a, Insl3, Hsd3b1, Ccnd1, Akr1c14, Cxcl12, and Rps16. The data are shown as mean ± SEM, n = 4–8 preparations. The same letter indicates that the difference between the two groups is not statistically significant (P < 0.05).
We treated rat immature Leydig cells for 2 days without (control), 1 µM flutamide (FLU), 100 nM MENT, and 1 µM FLU and 100 nM MENT (FLU + MENT), and selected four genes (Cyp17a1, Cdkn1a, Svs5, and Ptgds) are biomarkers of mature Leydig cells and can be analyzed by qPCR. We found that androgen receptor antagonist flutamide reversed the regulation of these genes by MENT (Figure 7). This suggests that MENT up-regulates these genes though androgen receptors. We chose a protein CYP17A1 and found that the level of CYP17A1 changed with its mRNA (Figure 8).
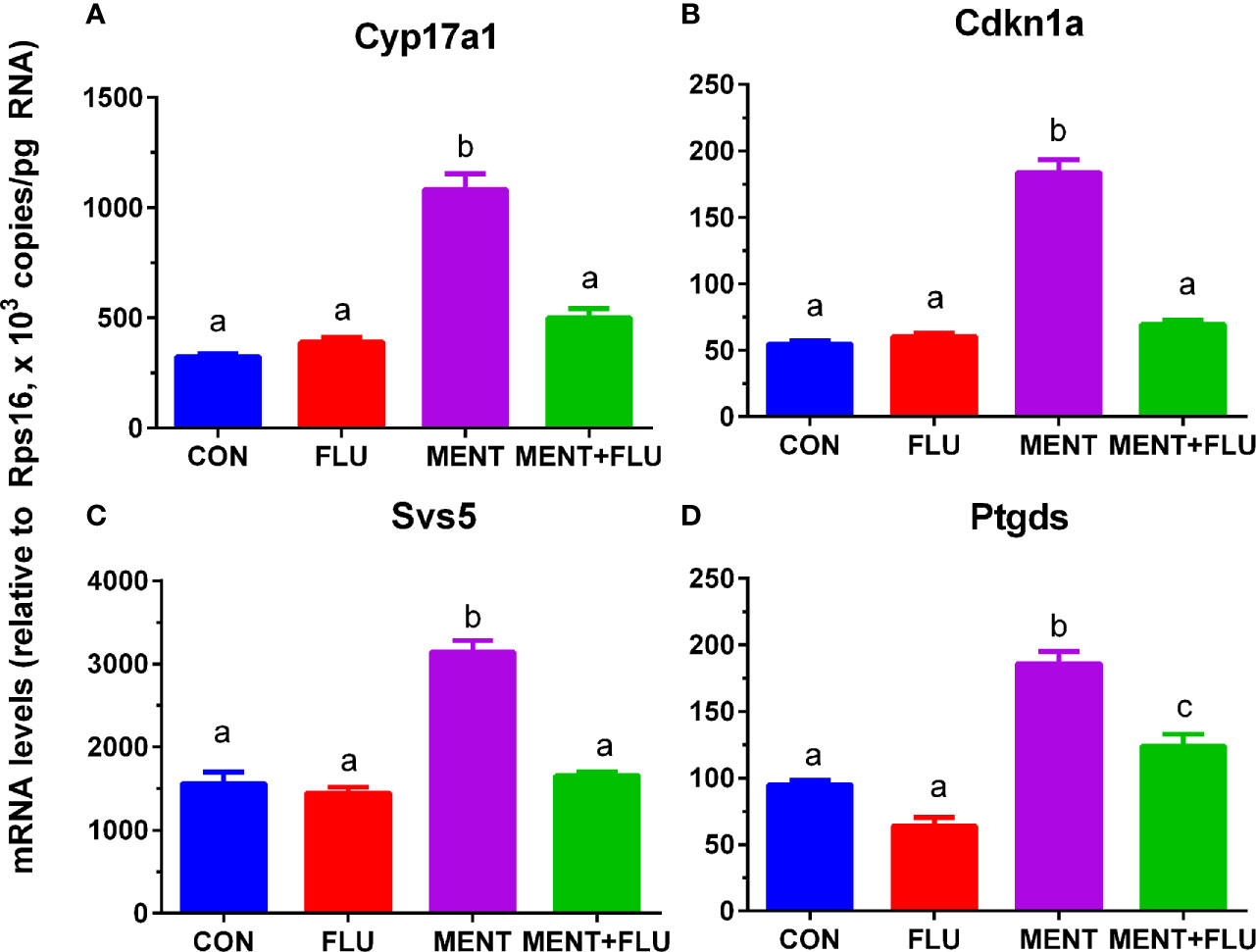
Figure 7 Antagonistic effect of flutamide (FLU) on MENT-induced gene up-regulation. In the presence of MENT, FLU alone or in combination (MENT +FLU), immature Leydig cels were cultured for 2 days. The following genes were analyzed (A–D): Cyp17a1, Cdkn1a, Svs5, and Ptgds. The data are shown as mean ± SEM, n = 4–8 preparations. The same letter indicates that the difference between the two groups is not statistically significant (P < 0.05).
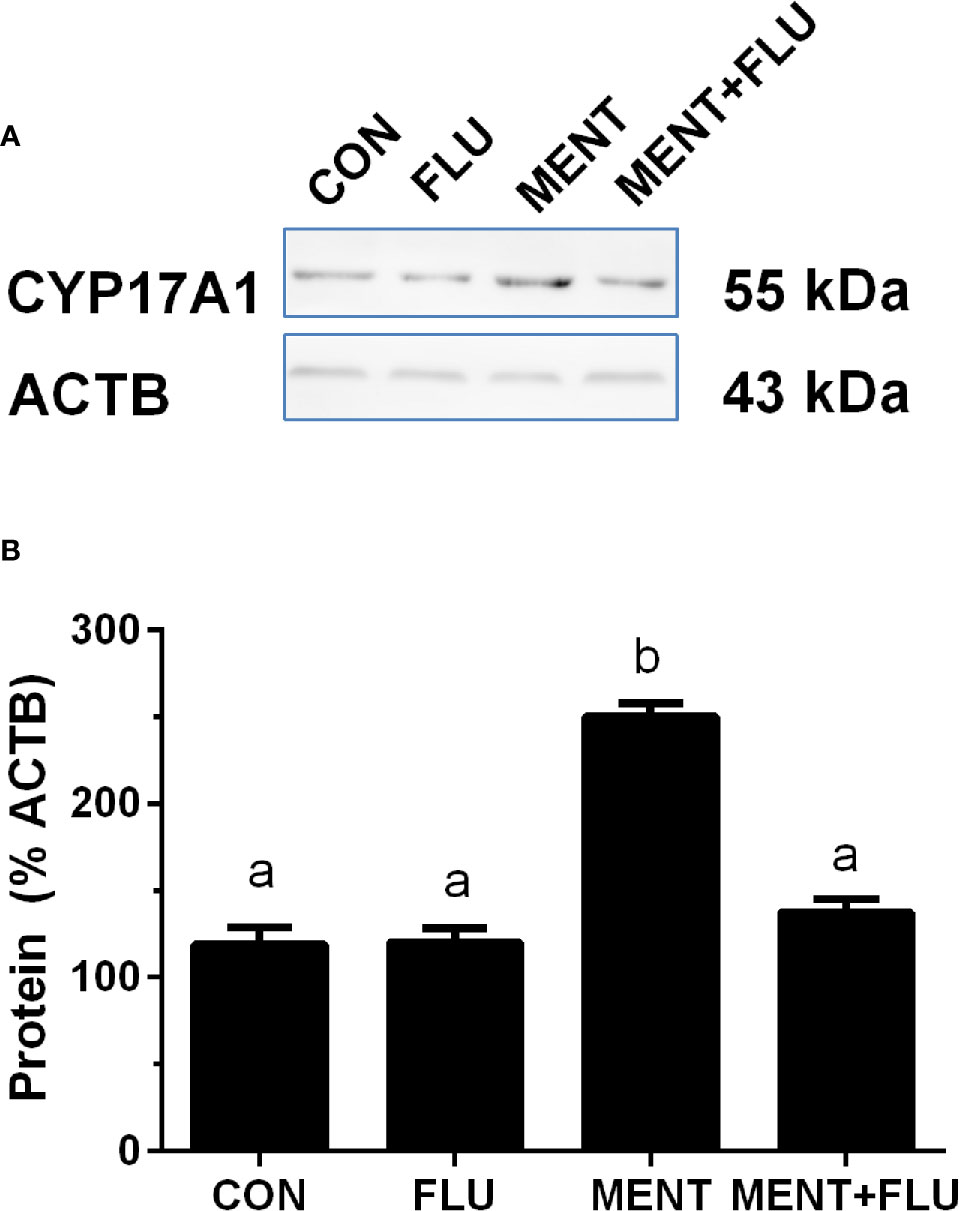
Figure 8 Androgen receptor antagonist flutamide (FLU) antagonizes MENT-induced CYP17A1 level. In the presence of MENT, FLU alone or in combination (MENT +FLU), immature Leydig cells were cultured for 2 days. Analysis of CYP17A1 and actin β (ACTB) levels. The CYP17A1 level was adjusted to ACTB. (A) Western blot; (B) Quantitative data. The data is shown as mean ± SEM, n = 3 preparations. The same letter indicates that the difference between the two groups is not statistically significant (P < 0.05).
We performed enzyme activity assays to identify protein changes in CYP11A1, HSD3B1, CYP17A1, HSD17B3, SRD5A1, and AKR1C14 in rat immature Leydig cells using radioactive substrates. As shown in Figure 9, the activity of all enzymes paralleled changes in their mRNA levels.
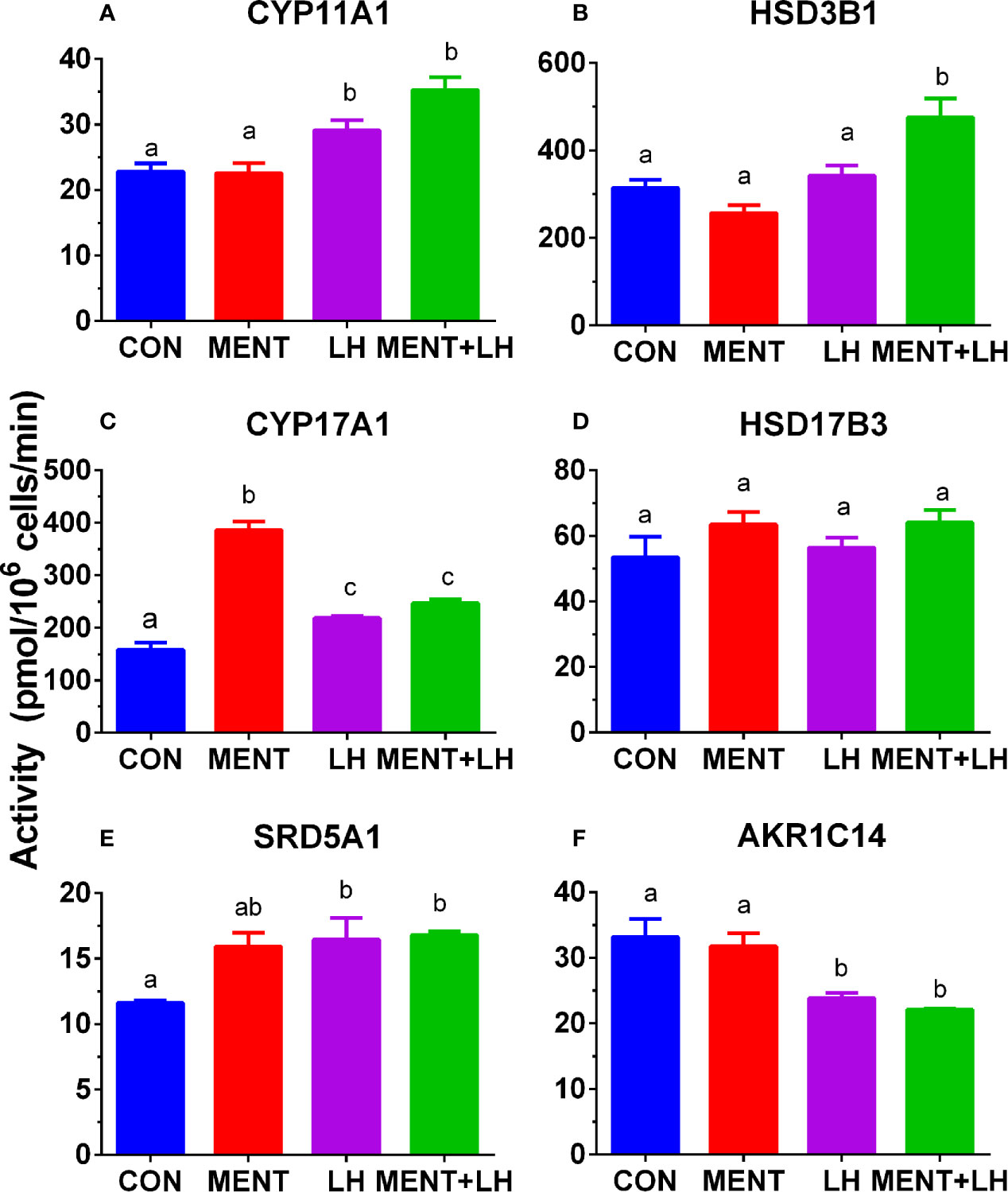
Figure 9 Effect of MENT, LH alone or in combination (MENT+LH) on the activity of steroidogenic enzymes in immature Leydig cells. In the presence of MENT, LH alone or in combination (MENT+LH), immature Leydig cells were cultured for 2 days. (A–F): CYP11A1, HSD3B1, CYP17A1, HSD17B3, SRD5A1, AKR1C14, respectively. The data is shown as mean ± SEM, n = 4 preparations. The same letter indicates that the difference between the two groups is not statistically significant (P < 0.05).
This study shows that androgen (MENT) and LH stimulate the function of immature Leydig cells in rats. androgen (MENT) and LH regulate different sets of transcription factors to control steroidogenesis. MENT mainly binds to NR3C4 to up-regulate the expression of Cyp17a1. LH mainly up-regulates the expression of Cyp11a1, Cyp17a1, and Srd5a1 through Crem1 and Egr1 transcription factors. However, LH down-regulates the expression of steroid metabolic enzyme genes, Akr1c14 and Sult1a1, thereby increasing the androgen formation of rat immature Leydig cells.
LH is essential for Leydig cell function because it is the principle stimulating hormone of androgen production in adult Leydig cells (1). Previous studies have shown that LH stimulated the development of rat progenitor Leydig cells into immature Leydig cells (5, 11, 20). In this study, we used 1 ng/ml LH instead of 100 ng/ml because high concentration of LH can cause LHCGR internalization (28, 29), and we also used a 2-day culture in which immature Leydig cells still maintained strong androgen production. Obviously, LH (1 ng/ml) can increase androgen production by two to three times, which is reasonable because these cells have lower LHCGR number compared to adult Leydig cells (9). In this study, we also demonstrated that LH stimulated the function of immature Leydig cells. In the steroidogenic pathway, LH up-regulated the expression of Scarb1, Cyp11a1, and Cyp17a1, and down-regulated the expression of Sult1a1 and Akr1c14 (Fig.4), thereby increasing androgen synthesis. Our previous studies have demonstrated that immature Leydig cells matured into adult Leydig cells by increase in the expression of Scarb1, Cyp11a1, and Cyp17a1 and down-regulation of Akr1c14 (7, 30). Therefore, LH is an important factor for function and differentiation of immature Leydig cells. The combination of MENT and LH also significantly down-regulated the expression of Akr1c14 (by 1.9 folds, Supplementary Table S2). The up-regulation of other biomarker of mature Leydig cells such as Insl3 also suggests that LH is a factor for inducing the differentiation of immature Leydig cells.
LH has been demonstrated to regulate Leydig cell function by binding to LHCGR, activating adenylyl cyclase to increase cAMP levels, thereby activating PKA/CREB/CREM signals (12). Knocking out Lhcgr in mice led to defects in the function of Leydig cells with no adult Leydig cells in the adult testis (13, 14). Indeed, in the current study, we found that CREM was significantly up-regulated (Figure 5). Although individual studies identified that several pathways besides cAMP/PKA signal are also involved, including MEK/ERK1/2 signal, the present study provided the transcription profiles of LH-mediated action in immature Leydig cells via several other pathways, including LHCGR/EGR1, Runx1 et al. (Figure 5).
In the steroidogenic pathway, MENT is different from LH, and only up-regulates the expression of Cyp17a1. Earlier evidence from the Tfm model supports the role of NR3C4 signaling in Leydig cell function. The expression levels of Leydig cell maturation markers Insl3 and Cyp17a1 in mice lacking functional NR3C4 were significantly reduced (16, 31). Although the role of NR3C4 in other cell types in Tfm cannot be ruled out, Leydig cell condition knockout mice for Nr3c4 also resulted in a significant reduction in mature Leydig cell biomarkers (Cyp17a1, Hsd17b3, and Insl3) (18). Obviously, NR3C4 acts through the androgen response element of the target gene. In this regard, in the Insl3 gene, the region that mediates androgen action has been mapped to the sequence of 2132 to 285 in the promoter (32). According to reports (16, 18, 31), in Tfm or Leydig cell conditionally knocked out NR3C4 mice, Cyp17a1 expression levels were significantly down-regulated. However, there is little information about the NR3C4 stimulation site in Cyp17a1. In contrast, in mature Leydig cells, androgens actually inhibit the expression level of Cyp17a1 through the binding sequence in the cAMP response region of the Cyp17a1 promoter (33). This difference in the androgen effect on the expression of Cyp17a1 may depend on the maturity of Leydig cells. Our data also points to androgen stimulation of immature Leydig cell maturation, as indicated by increased expression of Cyp17a1, Cdkn1a, Svs5, and Ptgds. Cyp17a1 (7), Cdkn1a (34), Svs5 (35), and Ptgds (36) have been identified as biomarkers for Leydig cell maturation. The increase in the expression of these genes (Cyp17a1, Cdkn1a, Svs5, and Ptgds) mediated by MENT was antagonized by flutamide (Figure 7), which is an inhibitor of NR3C4, indicating that MENT up-regulates the expression of these genes through androgen receptor.
Interestingly, among the up-regulated genes, MENT and LH showed higher similarity. Of the 98 genes that were up-regulated by MENT three times, 74 were also up-regulated by LH, and only 1 was down-regulated (Figure 3). Although some effects of LH on the gene expression may be mediated by testosterone, immature Leydig cells secrete a very low amount of testosterone and the major androgen is androstanediol, which almost has no binding to the androgen receptor. Furthermore, we found that LH and androgen regulate different sets of transcriptional factors related to Leydig cell differentiation. LH significantly up-regulated the expression of Crem1, Atf3, Egr1, and Runx1 (Figure 5), and down-regulated transcription factor Aes. The combination of MENT and LH also down-regulated Aes expression by 1.9-folds although the significant difference did not reach P < 0.05 (Supplementary Table S2). CREM, CREB (cAMP responsive element binding protein), and ATF (activating transcript factor) belong to the CREB transcriptional factor family, they respond to cAMP signaling and bind to cAMP response element (CRE) site in the target gene promoter (37–39). LH-LHCGR-protein kinase A signaling may lead to up-regulation of Crem1 and Atf3. It has been reported that LH up-regulated Egr1 in gonadal cells (40), which is critical for steroidogenesis in Leydig cells (41). There are also reports that Runx1 is up-regulated by LH and is involved in steroidogenesis in gonadal cells (42). Thirteen genes (Atf5, Gfi1, Mecp2, Pawr, Ppard, Raf2, Vps36, Atf4, Ccnl1, Dit3, Per1, Stat5b, and Trib3) were significantly up-regulated while two genes (Smarcd2 and Zbtb7a) were down-regulated only by MENT by 2-fold (Figure 5). These data suggest that LH and MENT function through different transcriptional regulation mechanisms.
In conclusion, this study shows that androgen and MENT can induce immature Leydig cells to differentiate into adult Leydig cells. Androgen and androgen act on their respective receptors and activated different sets of transcriptional factors to regulate the function of immature cells into adult Leydig cells (Supplementary Figure S2).
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Wenzhou Medical University.
XL, QZ, ZW, YW, KY, and ZS performed experiments. KY performed microarray analysis. XL, YZ, and R-SG designed the study. XL drafted the manuscript. R-SG edited the manuscript. All authors contributed to the article and approved the submitted version.
The work is supported by the National Natural Science Foundation of China (81730042).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.599149/full#supplementary-material
Supplementary Figure 1 | Analysis of G1 to S cell cycle-controlled gene pathway regulated by MENT, LH alone or in combination (MENT + LH). Red indicates that the gene is up-regulated; green indicates that the gene is down-regulated.
Supplementary Figure 2 | Illustration of androgen (MENT) and luteinizing hormone (LH) regulating steroid synthesis and metabolism in Leydig cells through different pathways and targets. MENT binds to androgen receptor (NR3C4) to up-regulate the expression of Cyp17a1. LH binds to its receptor (LHCGR) to trigger the cAMP/PKA pathway, thereby up-regulating the expression of Scarb1, Cyp11a1, and Cyp17a1 and down-regulating the expression of Sult1a1 and Akr1c14.
MENT, 7α-Methyl-19- nortestosterone; CYP11A1, Cytochrome P450 cholesterol side chain cleavage enzyme; HSD3B1, 3β-Hydroxysteroid dehydrogenase 1; CYP17A1, Cytochrome P450 17α-hydroxylase/17, 20-lyase; HSD17B3, 17β-Hydroxysteroid dehydrogenase 3; SRD5A1, Steroid 5α-reductase isoform 1; AKR1C14, 3α-Hydroxysteroid dehydrogenase; DIOL, 5α-Androstane-3α,17β-diol; LH, Luteinizing hormone; NR3C4, Androgen receptor; Tfm, feminized testis mice.
1. Chen P, Zirkin BR, Chen H. Stem Leydig Cells in the Adult Testis: Characterization, Regulation and Potential Applications. Endocr Rev (2020) 41:22–32. doi: 10.1210/endrev/bnz013
2. Haider SG. Cell biology of Leydig cells in the testis. Int Rev Cytol (2004) 233:181–241. doi: 10.1016/S0074-7696(04)33005-6
3. Ye L, Li X, Li L, Chen H, Ge RS. Insights into the development of the adult Leydig cell lineage from stem Leydig cells. Front Physiol (2017) 8:430. doi: 10.3389/fphys.2017.00430
4. Ge RS, Dong Q, Sottas CM, Papadopoulos V, Zirkin BR, Hardy MP. In search of rat stem Leydig cells: identification, isolation, and lineage-specific development. Proc Natl Acad Sci U.S.A. (2006) 103:2719–24. doi: 10.1073/pnas.0507692103
5. Stanley E, Lin CY, Jin S, Liu J, Sottas CM, Ge R, et al. Identification, proliferation, and differentiation of adult Leydig stem cells. Endocrinology (2012) 153:5002–10. doi: 10.1210/en.2012-1417
6. Stojkov NJ, Janjic MM, Baburski AZ, Mihajlovic AI, Drljaca DM, Sokanovic SJ, et al. Sustained in vivo blockade of alpha(1)-adrenergic receptors prevented some of stress-triggered effects on steroidogenic machinery in Leydig cells. Am J Physiol Endocrinol Metab (2013) 305:E194–204. doi: 10.1152/ajpendo.00100.2013
7. Ge RS, Hardy MP. Variation in the end products of androgen biosynthesis and metabolism during postnatal differentiation of rat Leydig cells. Endocrinology (1998) 139:3787–95. doi: 10.1210/endo.139.9.6183
8. Hardy MP, Zirkin BR, Ewing LL. Kinetic studies on the development of the adult population of Leydig cells in testes of the pubertal rat. Endocrinology (1989) 124:762–70. doi: 10.1210/endo-124-2-762
9. Shan LX, Hardy MP. Developmental changes in levels of luteinizing hormone receptor and androgen receptor in rat Leydig cells. Endocrinology (1992) 131:1107–14. doi: 10.1210/endo.131.3.1505454
10. Shan LX, Phillips DM, Bardin CW, Hardy MP. Differential regulation of steroidogenic enzymes during differentiation optimizes testosterone production by adult rat Leydig cells. Endocrinology (1993) 133:2277–83. doi: 10.1210/endo.133.5.8404681
11. Odeh HM, Kleinguetl C, Ge R, Zirkin BR, Chen H. Regulation of the proliferation and differentiation of leydig stem cells in the adult testis. Biol Reprod (2014) 90:123. doi: 10.1095/biolreprod.114.117473
12. Casarini L, Santi D, Simoni M, Poti F. ‘Spare’ Luteinizing Hormone Receptors: Facts and Fiction. Trends Endocrinol Metab (2018) 29:208–17. doi: 10.1016/j.tem.2018.01.007
13. Lei ZM, Mishra S, Zou W, Xu B, Foltz M, Li X, et al. Targeted disruption of luteinizing hormone/human chorionic gonadotropin receptor gene. Mol Endocrinol (2001) 15:184–200. doi: 10.1210/mend.15.1.0586
14. Zhang F-P, Poutanen M, Wilbertz J, Huhtaniemi I. Normal prenatal but arrested postnatal sexual development of luteinizing hormone receptor knockout (LHRKO) mice. Mol Endocrinol (2001) 15:172?183. doi: 10.1210/mend.15.1.0582
15. Murphy L, O’shaughnessy PJ. Testicular steroidogenesis in the testicular feminized (Tfm) mouse: loss of 17 alpha-hydroxylase activity. J Endocrinol (1991) 131:443–9. doi: 10.1677/joe.0.1310443
16. O’shaughnessy PJ, Murphy L. Cytochrome P-450 17 alpha-hydroxylase protein and mRNA in the testis of the testicular feminized (Tfm) mouse. J Mol Endocrinol (1993) 11:77–82. doi: 10.1677/jme.0.0110077
17. O’shaughnessy PJ, Johnston H, Willerton L, Baker PJ. Failure of normal adult Leydig cell development in androgen-receptor-deficient mice. J Cell Sci (2002) 115:3491–6.
18. O’hara L, Mcinnes K, Simitsidellis I, Morgan S, Atanassova N, Slowikowska-Hilczer J, et al. Autocrine androgen action is essential for Leydig cell maturation and function, and protects against late-onset Leydig cell apoptosis in both mice and men. FASEB J (2015) 29:894–910. doi: 10.1096/fj.14-255729
19. Hardy MP, Kelce WR, Klinefelter GR, Ewing LL. Differentiation of Leydig cell precursors in vitro: a role for androgen. Endocrinology (1990) 127:488–90. doi: 10.1210/endo-127-1-488
20. Guo JJ, Ma X, Wang CQ, Ge YF, Lian QQ, Hardy DO, et al. Effects of luteinizing hormone and androgen on the development of rat progenitor Leydig cells in vitro and in vivo. Asian J Androl (2013) 15:685–91. doi: 10.1038/aja.2013.55
21. Shan L, Hardy DO, Catterall JF, Hardy MP. Effects of luteinizing hormone (LH) and androgen on steady state levels of messenger ribonucleic acid for LH receptors, androgen receptors, and steroidogenic enzymes in rat Leydig cell progenitors in vivo. Endocrinology (1995) 136:1686–93. doi: 10.1210/endo.136.4.7895679
22. Payne AH, Downing JR, Wong KL. Luteinizing hormone receptors and testosterone synthesis in two distinct populations of Leydig cells. Endocrinology (1980) 106:1424–9. doi: 10.1210/endo-106-5-1424
23. Strober W. Trypan Blue Exclusion Test of Cell Viability. Curr Protoc Immunol (2015) 111:A3 B 1–3. doi: 10.1002/0471142735.ima03bs111
24. Griffin DK, Ellis PJ, Dunmore B, Bauer J, Abel MH, Affara NA. Transcriptional profiling of luteinizing hormone receptor-deficient mice before and after testosterone treatment provides insight into the hormonal control of postnatal testicular development and Leydig cell differentiation. Biol Reprod (2010) 82:1139–50. doi: 10.1095/biolreprod.109.082099
25. Lin H, Lian QQ, Hu GX, Jin Y, Zhang Y, Hardy DO, et al. In utero and lactational exposures to diethylhexyl-phthalate affect two populations of leydig cells in male long-evans rats. Biol Reprod (2009) 80:882–8. doi: 10.1095/biolreprod.108.072975
26. Lin H, Ge RS, Chen GR, Hu GX, Dong L, Lian QQ, et al. Involvement of testicular growth factors in fetal Leydig cell aggregation after exposure to phthalate in utero. Proc Natl Acad Sci U.S.A. (2008) 105:7218–22. doi: 10.1073/pnas.0709260105
27. Cochran RC, Ewing LL, Niswender GD. Serum levels of follicle-stimulating hormone, prolactin, testosterone, 5a-dihydrotestosterone, 5a-androstane-3a,17b-diol, 5a-androstane-3b,17b-diol and 17b-estradiol from male beagles with spontaneous or induced benign prostatic hyperplasia. Invest Urol (1981) 19:142–7.
28. Habberfield AD, Dix CJ, Cooke BA. Effects of lutropin (LH) receptor internalization and recycling on the apparent numbers of LH receptors in rat testis Leydig cells at different temperatures. Mol Cell Endocrinol (1987b) 51:153–61. doi: 10.1016/0303-7207(87)90129-8
29. Habberfield AD, Dix CJ, Cooke BA. The dynamics of LH-induced desensitization of adenylate cyclase and LH receptor internalization in rat Leydig cells at physiological levels of LH. J Endocrinol (1987a) 114:415–22. doi: 10.1677/joe.0.1140415
30. Ge RS, Dong Q, Sottas CM, Chen H, Zirkin BR, Hardy MP. Gene expression in rat leydig cells during development from the progenitor to adult stage: a cluster analysis. Biol Reprod (2005) 72:1405–15. doi: 10.1095/biolreprod.104.037499
31. Murphy L, Jeffcoate IA, O’shaughnessy PJ. Abnormal Leydig cell development at puberty in the androgen-resistant Tfm mouse. Endocrinology (1994) 135:1372–7. doi: 10.1210/endo.135.4.7925099
32. Lague E, Tremblay JJ. Antagonistic effects of testosterone and the endocrine disruptor mono-(2-ethylhexyl) phthalate on INSL3 transcription in Leydig cells. Endocrinology (2008) 149:4688–94. doi: 10.1210/en.2008-0310
33. Burgos-Trinidad M, Youngblood GL, Maroto MR, Scheller A, Robins DM, Payne AH. Repression of cAMP-induced expression of the mouse P450 17 alpha-hydroxylase/C17-20 lyase gene (Cyp17) by androgens. Mol Endocrinol (1997) 11:87–96. doi: 10.1210/mend.11.1.9871
34. Lin H, Huang Y, Su Z, Zhu Q, Ge Y, Wang G, et al. Deficiency of CDKN1A or both CDKN1A and CDKN1B affects the pubertal development of mouse Leydig cells. Biol Reprod (2015) 92:77. doi: 10.1095/biolreprod.114.118463
35. Zhang YF, Yuan KM, Liang Y, Chu YH, Lian QQ, Ge YF, et al. Alterations of gene profiles in Leydig-cell-regenerating adult rat testis after ethane dimethane sulfonate-treatment. Asian J Androl (2015) 17:253–60. doi: 10.4103/1008-682X.136447
36. Strauss L, Kallio J, Desai N, Pakarinen P, Miettinen T, Gylling H, et al. Increased exposure to estrogens disturbs maturation, steroidogenesis, and cholesterol homeostasis via estrogen receptor alpha in adult mouse Leydig cells. Endocrinology (2009) 150:2865–72. doi: 10.1210/en.2008-1311
37. Hoeffler JP, Meyer TE, Yun Y, Jameson JL, Habener JF. Cyclic AMP-responsive DNA-binding protein: structure based on a cloned placental cDNA. Science (1988) 242:1430–3. doi: 10.1126/science.2974179
38. Rehfuss RP, Walton KM, Loriaux MM, Goodman RH. The cAMP-regulated enhancer-binding protein ATF-1 activates transcription in response to cAMP-dependent protein kinase A. J Biol Chem (1991) 266:18431–4. doi: 10.1016/S0021-9258(18)55078-X
39. Foulkes NS, Mellstrom B, Benusiglio E, Sassone-Corsi P. Developmental switch of CREM function during spermatogenesis: from antagonist to activator. Nature (1992) 355:80–4. doi: 10.1038/355080a0
40. Yoshino M, Mizutani T, Yamada K, Tsuchiya M, Minegishi T, Yazawa T, et al. Early growth response gene-1 regulates the expression of the rat luteinizing hormone receptor gene. Biol Reprod (2002) 66:1813–9. doi: 10.1095/biolreprod66.6.1813
41. Tourtellotte WG, Nagarajan R, Bartke A, Milbrandt J. Functional compensation by Egr4 in Egr1-dependent luteinizing hormone regulation and Leydig cell steroidogenesis. Mol Cell Biol (2000) 20:5261–8. doi: 10.1128/MCB.20.14.5261-5268.2000
Keywords: luteinizing hormone, testosterone, development, immature Leydig cell, regulation, steroidogenesis
Citation: Li X, Zhu Q, Wen Z, Yuan K, Su Z, Wang Y, Zhong Y and Ge R-S (2021) Androgen and Luteinizing Hormone Stimulate the Function of Rat Immature Leydig Cells Through Different Transcription Signals. Front. Endocrinol. 12:599149. doi: 10.3389/fendo.2021.599149
Received: 26 August 2020; Accepted: 15 February 2021;
Published: 17 March 2021.
Edited by:
Katja Teerds, Wageningen University, NetherlandsReviewed by:
Richard Ivell, University of Nottingham, United KingdomCopyright © 2021 Li, Zhu, Wen, Yuan, Su, Wang, Zhong and Ge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ren-Shan Ge, cl9nZUB5YWhvby5jb20=; Ying Zhong, eXpob25nOEB5YWhvby5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.