
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 26 November 2020
Sec. Experimental Endocrinology
Volume 11 - 2020 | https://doi.org/10.3389/fendo.2020.589994
Nonalcoholic fatty liver disease (NAFLD) is the common disease in the liver, which is associated with metabolic syndrome and hepatocellular carcinoma. Accumulated evidence establishes that small non-coding microRNAs (miRNAs) contribute to the initiation and progression of NAFLD. However, the molecular repertoire of miRNA in NAFLD is still largely unknown. Here, using an integrative approach spanning bioinformatic analysis and functional approaches, we demonstrate that miR-124-3p participates in the development of NAFLD by directly targeting preadipocyte factor-1 (Pref-1). In response to high-fat diet (HFD), expression of miR-124-3p was increased in the liver. Inhibition of miR-124-3p expression led to a dramatic reduction of triglyceride contents in hepatocytes, in parallel with decreased inflammatory factors. Mechanistically, miR-124-3p directly controls the transcription of Pref-1, a secretory factor that has been proved to resist metabolic syndrome. Our work identifies a novel molecular axis in hepatosteatosis, and highlights miR-124-3p/Pref-1 as potential targets for clinical interventions of NAFLD.
Non-alcoholic fatty liver disease (NAFLD) is the most common liver disease, which is characterized by excessive hepatic fat deposition and a wide range of pathologies encompassing steatohepatitis, fibrosis, cirrhosis, liver failure, and hepatocellular carcinoma (1, 2). A number of molecular abnormalities that occur in hepatosteatosis confer to the initiation and progression of NAFLD. Evidence from experimental and clinical studies suggests a complicated interplay of multiple biological processes in the disease development, including obesity, dysbiosis of the intestinal microbiome, heightened intestinal barrier permeability, metabolic endotoxemia and inflammations (3–5). Despite the high prevalence and clinical importance, factors leading to NAFLD were still poorly understood and few efficacious therapies exist (6). Therefore, molecular understandings of the initiation and progression of NAFLD are urgently needed to provide a basis for therapeutic design.
MicroRNAs (miRNAs) are small non-coding RNAs with 18~24 nucleotides, which regulates gene expression by binding to mRNAs and impairs the process of translation (7). MiRNAs are emerging as new regulators of glucose and lipid metabolism during liver development and disease progression, including NAFLD (8, 9). For example, hepatic miR-122 affects gene expressions in cholesterol and lipid metabolism, and thereby maintains liver homeostasis (10, 11). miR-27a attenuated hepatic de novo lipogenesis and alleviated obesity-initiated NAFLD by inhibiting Fasn and Scd1 in liver (12). miR-375 is up-regulated in NASH patients, and its inhibition ameliorates lipid accumulation and decreases inflammatory cytokines (13, 14). Despite these studies, however, the molecular repertoire of miRNA in NAFLD is still largely unknown.
In this study, we reveal a novel function of miR-124-3p in hepatic lipid metabolism. MiR-124-3p expression in the liver is increased under high-fat diet (HFD). Inhibition of MiR-124-3p reduces triglyceride contents as well as inflammatory factors in hepatocytes, and vice versa. Mechanistically, miR-124-3p directly controls the transcription of Pref-1, a secretory factor that resists metabolic syndrome. Our work establishes the novel function of miR-124-3p in maintaining liver homeostasis, and defines miR-124-3p as a potential target for clinical interventions of NAFLD.
C57BL/6J mice were obtained and raised in the animal center of Sichuan Academy of Medical Science. Experimental mice were kept in SPF conditions with standard housing conditions in a temperature-controlled environment with 12-h light/dark cycles and received chow diet (CD) or high-fat diet (HFD, from 8-week age) and water ad libitum for 12 weeks (15). Animals studies were conducted in accordance with institutional guidelines. For primary hepatocyte culture, we conducted a two-step collagenase perfusion method as described (16). Isolated hepatocytes were cultured in Williams E medium (Sigma) including HepExtend™ Supplement (Gibco), plus penicillin/streptomycin and gentamycin in 37°C under 5% CO2.
For oil red O staining to validate the hepatosteatosis in mice with HFD, fresh liver samples were fixed in 4% paraformaldehyde (PFA) solution, and embedded with OCT for subsequent staining with Oil Red O, according to standard protocols (17).
To detect the profile of miRNAs in mouse livers under HFD and CD, a total of miRNAs were extracted using miRNeasy mini kit (QIAGEN). The microarray hybridization of miRNAs was performed using GeneChip miRNA 3.0 Array (Affymetrix). Microarray datasets were presented in volcano plot and GO biological process classification, and miRNAs with 2-fold or greater fold change with P-value < 0.05 were considered differentially expressed.
To confirm that miR-124-3p directly targets Pref-1, a luciferase reporter assay was performed. Briefly, the pmirGL-control luciferase vector, pmirGLO dual-Luciferase miRNA target expression vector and luciferase reporter assay system were purchased from Youbia (China). The pmirGLO-control vector was used to construct pGL-pref-1, which contained the 3’UTR of mouse pref-1 (Genbank accession NM003836). DNA fragment of Pref-1 was constructed into vector (18). For luciferase reporter assay, control vector or miR-124-3p plasmid and reporter plasmids were co-transfected into cells. Luciferase activities were measured at 48 h post-transfection by using the dual-luciferase assay system (Turner BioSystem, USA).
For western blots, cultured cells were lysed in 2% SDS buffer plus protease and phosphatase inhibitors (Thermo Scientific, USA). The lysates of equivalent total proteins were separated on SDS-PAGE. Then proteins were transferred into PVDF membrane (Millipore) for incubation with primary/secondary antibodies. The primary antibodies used were as follows, Pref-1 (Cell Signaling Technology, Cat#2069) and GAPDH (Cell Signaling Technology, Cat#2118).
Total RNA was extracted by Animal miRNA Isolation Kit (Foregene, China) and Cell Total RNA Isolation Kit (Foregene, China) followed by standard procedure. The PrimeScriptTM RT reagent Kit (Takara) was used to reverse transcript to cDNA, and SYBR Premix Ex TaqTM II (Takara) was used to qPCR assay in Bio-rad system. The qPCR primers were list as follows: miR-124-3p, F: 5’-TAAGGCACGCGGTGAATGCC-3’, R: 5’-GATTGAATCGAGCACCAGTTAC-3’; TNFα, F: 5’-CAGGCGGTGCCTATGTCTC-3’, R: 5’-CGATCACCCCGAAGTTCAGTAG-3’; Pref-1, F: 5’-TTCGGCCACAGCACCTATG-3’, R: 5’-GGGGCAGTTACACACTTGTCA-3’.
For Pref-1 knockdown, targeted siRNA were transfected into cells using Lipofectamine RNAiMAX reagent (Invitrogen, USA) in Opti-MEM (Invitrogen, USA) over 72 h. The siRNA sequences were as follows, siRNA negative-control: sense-UCCGGAACUGUUACGUGAA; antisense-UUCACGUAACAGUUCCGGA. siRNA Pref-1: sense: UCCUGAAGGUGUCCAUGAA; antisense: UUCAUGGACACCUUCAGGA.
Statistical analysis was performed in GraphPad Prism. Data were expressed as mean ± SD from at least three independent experiments. Statistical differences between two groups were analyzed by Student’s t test, and multi-group comparisons by one-way ANOVA followed by Tukey post hoc tests. P<0.05 was considered statistically significant; **indicates p<0.01, and *** p<0.001.
To investigate the miRNAs involved in hepatosteatosis, we assessed the hepatic miRNA profile in C57BL/6J mice fed with high fat diet (HFD) for 12 weeks, in comparison to littermate controls fed with chow diet (CD). Then we extracted the total miRNA from mouse livers and performed microarray hybridization using GeneChip miRNA 3.0 Array (Affymetrix). Histochemical results confirmed the lipid accumulation by Oil Red O staining in the liver of HFD mice (Figure 1A). By miRNA screening, we found that altered miRNAs between HFD and CD mice were enriched in metabolic process by GO (Gene Ontology) biological process classification (Figure 1B). As shown in the volcano plot, we differentiated the increased and decreased miRNAs. Particularly, it’s found that miR-124-3p was dramatically increased in the liver of HFD mice (Figure 1C). And this increasing was further validated by quantitative evaluation from more paired HFD/CD mice (Figure 1D). All these data suggests that increased miR-124-3p may play a vital role in HFD-induced hepatosteatosis.
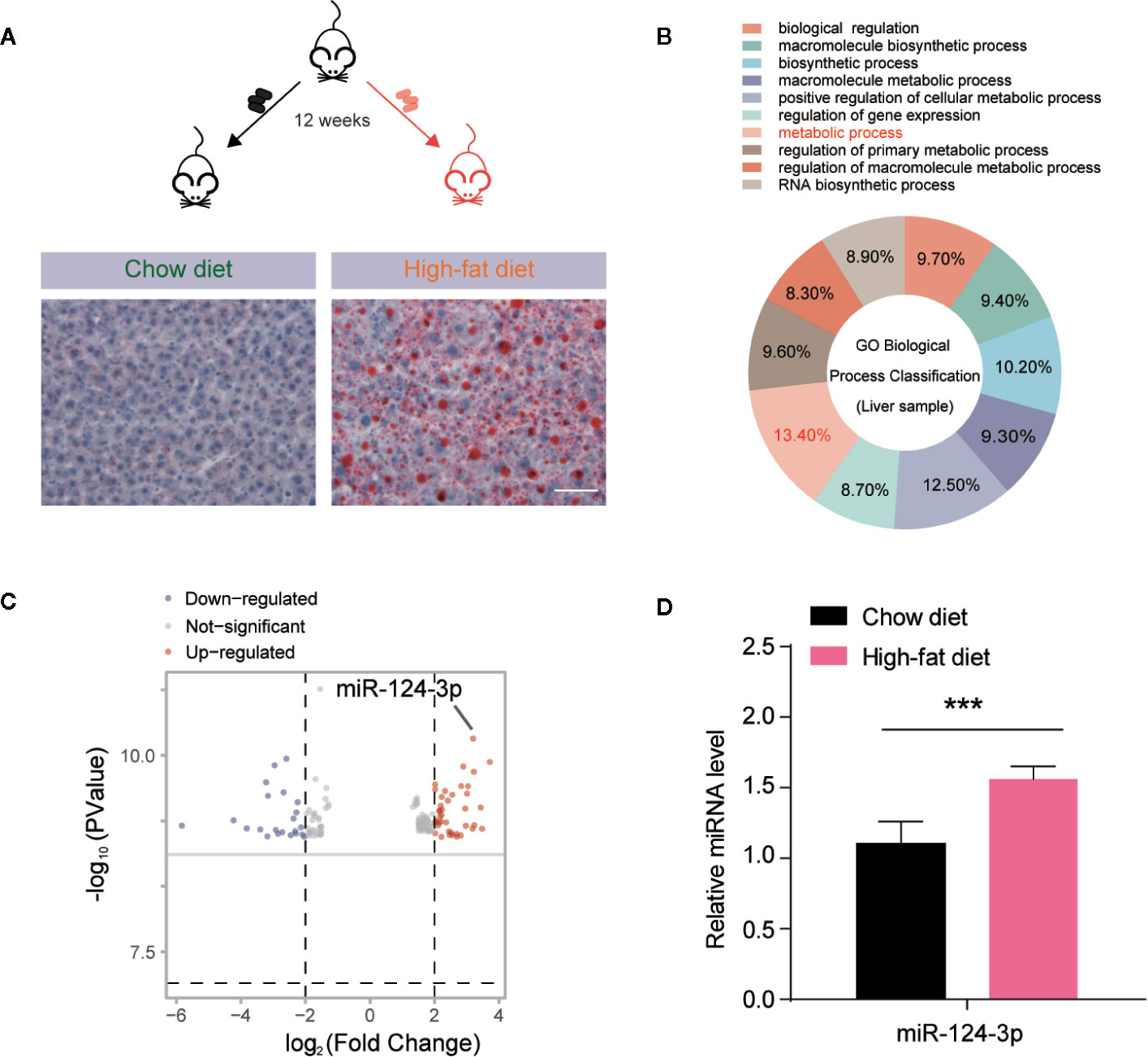
Figure 1 MiR-124-3p expression is increased by high-fat diet (HFD)-induced hepatosteatosis. (A) A schematic model illustrating that mice fed with chow diet (CD) and high fat diet (HFD) for 12 weeks, and hepatosteatosis were examined by Oil Red O staining. Scale bar 100 μm. (B) A diagram showing that metabolic process are relevant to miRNA expression analyzed by GO biological classification in the liver of HFD mice compared to CD controls. (C) Volcano Plots showing the different miRNA expression in the liver of HFD mice compared to CD controls. The arrow indicates miR-124-3p. (D) qPCR results showing that miR-124-3p is upregulated in the liver of HFD mice compared to CD controls. Results were averages of three independent experiments. Data represent mean ± SD. ***P < 0.001.
Based on the finding that miR-124-3p was induced by HFD in mouse liver, we asked whether induction of miR-124-3p has a causal effect on lipid accumulation. Therefore, we examined the lipid contents in hepatocytes after miR-124-3p inhibition. As a first step, we knocked down miR-124-3p expression by transfection of its inhibitors in primary hepatocytes, and the transfection efficiency was confirmed by RT-qPCR (Figure 2A). Next, we inactivated miR-124-3p expression in palmetric acid (PA) treated hepatocytes (Figure 2B). Although PA could increase triglycerides in control hepatocytes (19), however, miR-124-3p inhibitors dramatically reduced triglyceride contents by PA treatment (Figure 2C). Excessive lipid accumulation often coincides with increased inflammation (20). We found that miR-124-3p inhibitors could also decrease the level of TNFα by PA treatment, indicating that miR-124-3p inactivation ameliorates liver inflammation (Figure 2D). Reciprocally, we examined whether overexpression of miR-124-3p by its mimics is sufficient to increase lipid contents and inflammations in hepatocytes. Results showed that miR-124-3p overexpression increased triglyceride levels and TNFα expression, suggesting that miR-124-3p may have a causal effect on steatosis in the liver (Figures 2E–H). Taken together, these results suggest that miR-124-3p positively regulates lipid contents and inflammation in hepatocytes.
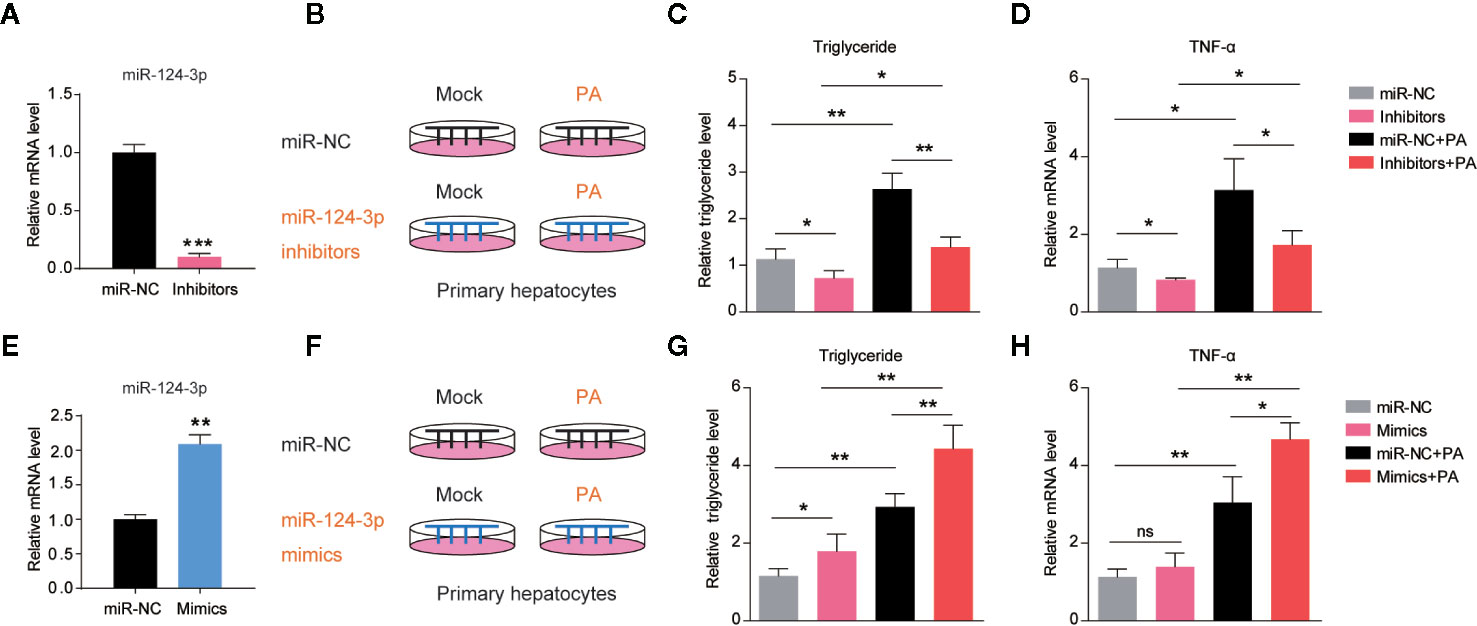
Figure 2 MiR-124-3p positively regulates lipid contents in hepatocytes. (A) qPCR results showing that miR-124-3p inhibitors decreased endogenous miR-124-3p level in primary hepatocytes. (B–D) A schematic model (B) illustrating that primary hepatocytes were treated with palmetric acid (PA) with or without miR-124-3p inhibitors. Biochemical results (C) showing that miR-124-3p inhibition decreased PA-induced triglyceride in hepatocytes. qPCR results (D) showing that miR-124-3p inhibition decreased PA-induced TNF-α in hepatocytes. (E) qPCR results showing that miR-124-3p mimics increased miR-124-3p level in primary hepatocytes. (F–H) A schematic model (F) illustrating that primary hepatocytes were treated with palmetric acid (PA) with or without miR-124-3p mimics. Biochemical results (G) showing that miR-124-3p overexpression increased PA-induced triglyceride in hepatocytes. qPCR results (H) showing that miR-124-3p overexpression increased PA-induced TNF-α in hepatocytes. Results were averages of three independent experiments. Data represent mean ± SD. *P < 0.05, **P < 0.01 and ***P < 0.001.
To clarify how miR-124-3p participates in the regulation of lipid metabolism, we next examined the potential targets of miR-124-3p. By sequence analysis, we found that Preadipocyte factor 1 (Pref-1) was a potential candidate of miR-124-3p in the liver (Figure 3A). Pref-1 is a transmembrane protein that could be cleaved at the extracellular domain to generate a soluble form that reduces hepatosteatosis and hyperglycemia (21, 22). To confirm that miR-124-3p directly targets Pref-1, we performed a luciferase reporter assay. Data showed that the activity of the reporter plasmid with miR-124-3p mimic was decreased, indicating that miR-124-3p binds to the 3’-UTR of Pref-1 and thus regulates Pref-1 expression (Figure 3B). Results of qPCR showed that the mRNA level of Pref-1 was decreased by miR-124-3p overexpression and increased by its knockdown (Figure 3C). Consistently, the protein levels of both full and cleaved Pref-1 were both negatively regulated by miR-124-3p (Figures 3D–F). Therefore, we propose that miR-124-3p directly targets Pref-1 to regulate its expression.
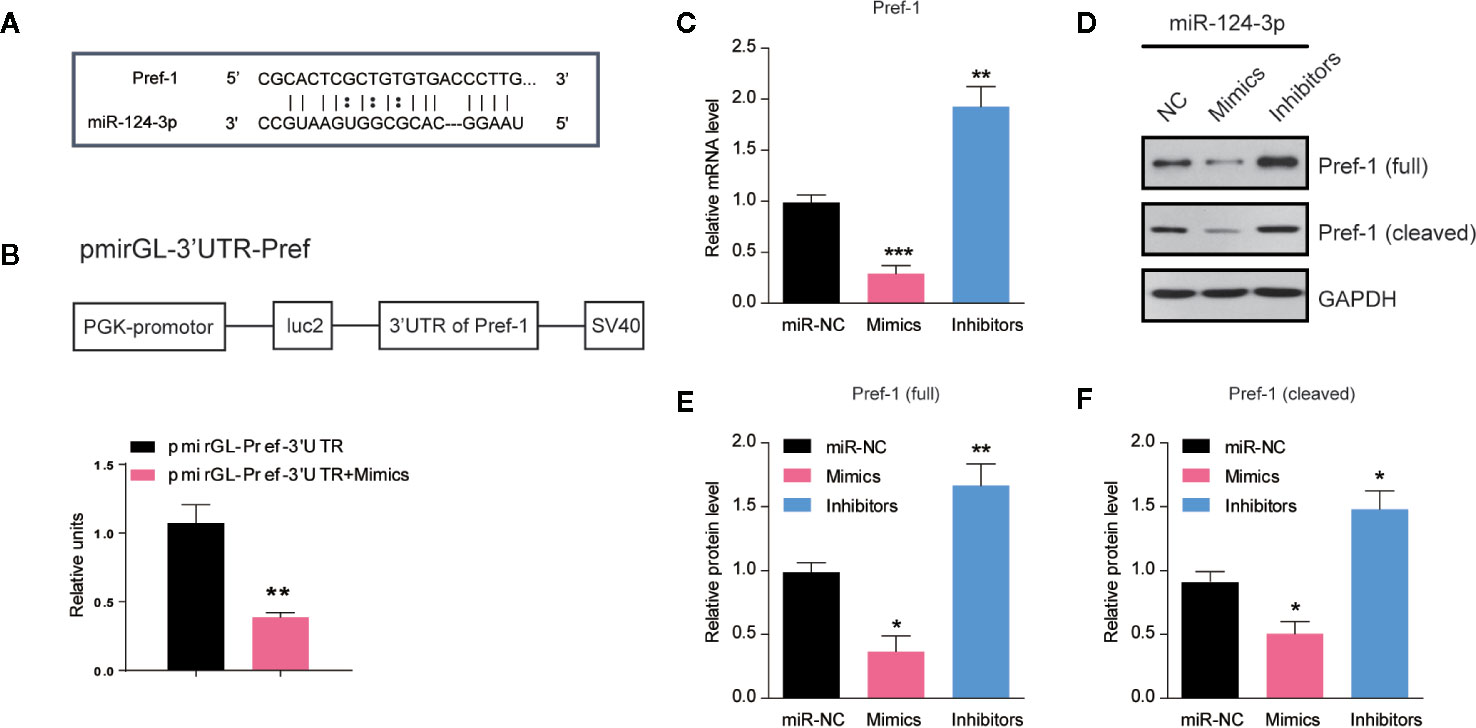
Figure 3 MiR-124-3p directly targets Pref-1 in hepatocytes. (A) Sequence analysis of miR-124-3p binding with the 3’-UTR of Pref-1. (B) A schematic diagram showing the pmirGL-3’UTR Pref-1 vector, and luciferase activities were measured by the luciferase reporter assay. (C) qPCR results showing the mRNA levels of Pref-1 in hepatocytes transfected with miR-124-3p mimics, inhibitors or scramble sequence. (D, E) Western blots (D) and quantifications (E, F) showing the protein levels of full-length and cleaved Pref-1 in hepatocytes transfected with miR-124-3p mimics, inhibitors or scramble sequence. Results were averages of three independent experiments. Data represent mean ± SD. *P < 0.05, **P < 0.01 and ***P < 0.001.
Pref-1 has been proved to resist HFD–induced hepatosteatosis and obesity (23, 24). To study whether miR-124-3p regulates hepatic lipid metabolism through Pref-1, we assessed the changes of Pref-1 after PA treatment in miR-124-3p inactivated hepatocytes. Western blot results showed that miR-124-3p inhibitors could induce expression of full and cleaved Pref-1 with or without PA (Figures 4A–C). It’s noted that Pref-1 expression was slightly increased by PA, however, dramatically increased by miR-124-3p inhibitors (Figures 4A–C), suggesting that miR-124-3p inactivation was necessary for Pref-1 expression under lipid stress conditions.
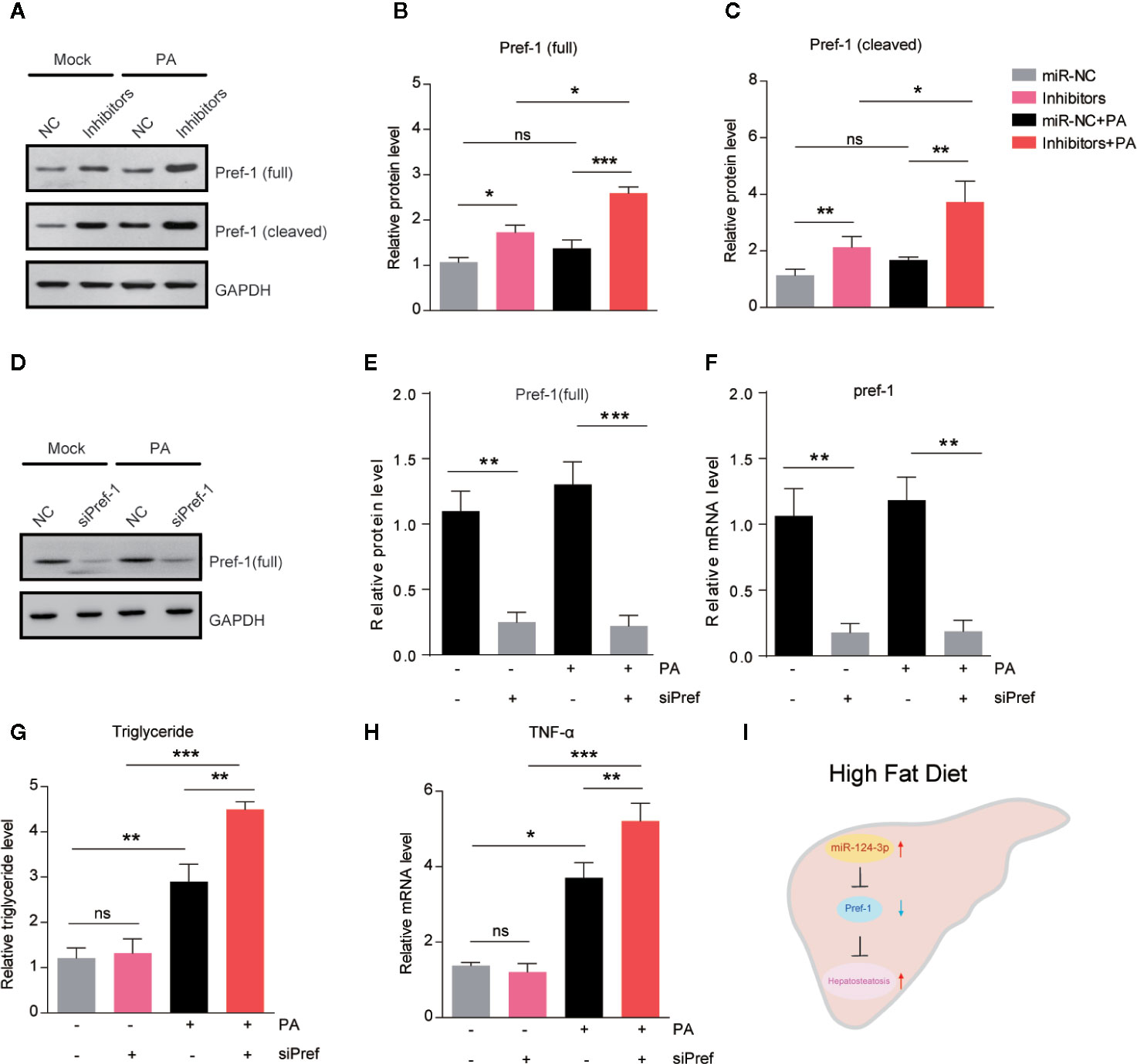
Figure 4 MiR-124-3p regulates lipid metabolism via Pref-1 in hepatocytes. (A–C) Western blots (A) and quantifications (B, C) showing the protein levels of full-length and cleaved Pref-1 in hepatocytes transfected with miR-124-3p inhibitors or scramble sequence and treated with palmetric acid (PA). Results were averages of three independent experiments. (D–F) Western blots and qPCR showing the decreased protein and mRNA level of Pref-1 in cells transfected with siPref-1. (G) Biochemical results showing that siPref-1 increased PA-induced triglyceride. (H) qPCR results showing that siPref-1 increased PA-induced TNF-α expression. (I) A schematic model highlighting the role of miR-124-3p alleviates hepatosteatosis by directly targeting Pref-1 in hepatocytes under lipid stress. Data represent mean ± SD. *P < 0.05, **P < 0.01 and ***P < 0.001.
To strengthen the point that miR-124-3p regulates hepatosteatosis through Pref-1, we examined the effect of Pref-1 knockdown on the lipid metabolism and inflammation in hepatocytes (Figures 4D–F). Results showed that Pref-1 knockdown did not dramatically alter triglyceride levels or TNFα expression under basal condition. However, in the presence of PA, Pref-1 knockdown caused a more dramatic increasing of triglyceride levels or TNFα expression, indicating that Pref-1 is required for the resistance of triglyceride accumulation under lipid stress (Figures 4G–H). Thus, we conclude that miR-124-3p regulated Pref-1 expression is critical for the maintenance of lipid homeostasis in hepatocytes.
Altered miRNA expression in response to genetic/epigenetic factors or environmental conditions confers to the onset of NAFLD (25). However, the molecular repertoire of miRNA in the initiation and progression of NAFLD is largely unknown. The present study demonstrates that miR-124-3p participates in hepatosteatosis in response to lipid stress. Inhibition of miR-124-3p in hepatocytes promotes Pref-1 expression and thus ameliorates lipid accumulation (Figure 4I).
In decades, numerous miRNAs were identified to regulate a wide spectrum of metabolic processes, including lipid homeostasis, glucose catabolism, and inflammation, which were known to be epigenetically deregulated in NAFLD (26). Altered hepatic miRNA profile has been described in both in humans and animal models of NAFLD (27). A recent study assessed circulating miRNAs in NASH patients and found that among 84 circulating miRNAs (13). For example, miR-122 and miR-192 were significantly upregulated and others were downregulated. Comparing to these studies, we revealed that miR-124-3p was particularly increased in the liver of HFD mice. This increasing was consistent with our finding that PA treatment could induce miR-124-3p expression in primary hepatocytes. Therefore, we conclude that miR-124-3p expression is dynamically regulated by lipid stress in vitro and in vivo. Moreover, we revealed that increased miR-124-3p expression may have a causal effect on the lipid accumulation in hepatocytes. Our data showed that inhibition of miR-124-3p decreased PA-induced triglyceride levels, and particularly, overexpression of miR-124-3p is sufficient to increase triglyceride level (Figure 2G), which is more pronounced after PA treatment. Thus, miR-124-3p is a critical regulator in lipid homeostasis of the liver. The impact of miR-124-3p on lipid metabolism indicates its potential therapeutic implication of NAFLD.
MiR-124-3p is a widely expressed miRNA in mammalian cells, and previous studies reveal its biological function in tumorigenesis and neural diseases. For example, miR-124-3p regulates aerobic glycolysis and induces chemoresistance in glioma cells through AMPK pathway (28). miR-124-3p also represses the migration and invasion of bladder cancer cells via ROCK1 (29). In the liver, miR-124-3p modulates autophagy through Beclin and LC3 and thus participates in hepatic impact injury (30). However, the role of miR-124-3p in metabolic diseases is not yet well understood. A recent study showed that miR-124 represses genes associated with fatty acid and triglyceride breakdown, and thus promotes triglyceride accumulation in hepatoma cells (31). Consistent with this finding, our study demonstrate that miR-124-3p has a direct effect in the liver under lipid stress conditions. We further revealed that miR-124-3p directly targets Pref-1 that resists hepatic lipid accumulation. Pref-1 belongs to the NOTCH family of epidermal growth factor-like repeat-containing proteins, which can inhibit adipogenesis and resist to high fat diet-induced obesity in mice (21, 32). In this study, we identified that miR-124-3p negatively regulates the expression of Pref-1 and its soluble fragment. It’s noticed that miR-124-3p overexpression could induce a slight but reproducible triglyceride increasing in hepatocytes under basal condition (Figure 2G). However, Pref-1 knockdown alone failed to produce a similar effect like miR-124-3p overexpression (Figure 4G), suggesting that miR-124-3p may regulate lipid metabolism partly through Pref-1 independent mechanisms. Interestingly, after PA treatment, Pref-1 knockdown phenocopies the effect of miR-124-3p overexpression, indicating that miR-124-3p-controlled lipid accumulation is largely dependent on Pref-1 under lipid stress condition. Nevertheless, our study at least reveals that miR-124-3p/Pref-1 is a novel molecular axis in hepatosteatosis. Future studies would be directed at the assessment of miR-124-3p/Pref-1 axis in resistance to hepatosteatosis in animal models and clinical trials.
In summary, our study identifies a novel function of miR-124-3p in the liver. Hepatocytes with miR-124-3p knockdown have decreased triglyceride contents and inflammatory cytokines. This work provides a therapeutic target for potential interventions.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by Sichuan Provincial People’s Hospital.
GW, YY, and GX designed the study and wrote the manuscript. GW, HZ, CL, XH, YY, and GX performed the experiments and analyzed the data. All authors contributed to the article and approved the submitted version.
This work was supported by grants from Specialized Fund for Basic Scientific Research, Sichuan Provincial Science and Technology Department (2018YSKY0017).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Huang TD, Behary J, Zekry A. Non-alcoholic fatty liver disease (NAFLD): a review of epidemiology, risk factors, diagnosis and management. Internal Med J (2020) 50(9):1038–47. doi: 10.1111/imj.14709
2. Samuel VT, Shulman GI. Nonalcoholic Fatty Liver Disease as a Nexus of Metabolic and Hepatic Diseases. Cell Metab (2018) 27(1):22–41. doi: 10.1016/j.cmet.2017.08.002
3. Byrne CD, Targher G. NAFLD: a multisystem disease. J Hepatol (2015) 62(1 Suppl):S47–64. doi: 10.1016/j.jhep.2014.12.012
4. Arab JP, Arrese M, Trauner M. Recent Insights into the Pathogenesis of Nonalcoholic Fatty Liver Disease. Annu Rev Pathol (2018) 13:321–50. doi: 10.1146/annurev-pathol-020117-043617
5. Bedi O, Aggarwal S, Trehanpati N, Ramakrishna G, Krishan P. Molecular and Pathological Events Involved in the Pathogenesis of Diabetes-Associated Nonalcoholic Fatty Liver Disease. J Clin Exp Hepatol (2019) 9(5):607–18. doi: 10.1016/j.jceh.2018.10.004
6. Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med (2018) 24(7):908–22. doi: 10.1038/s41591-018-0104-9
7. Bjorkman S, Taylor HS. MicroRNAs in endometriosis: biological function and emerging biomarker candidatesdagger. Biol Reprod (2019) 100(5):1135–46. doi: 10.1093/biolre/ioz014
8. Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol (2012) 13(4):239–50. doi: 10.1038/nrm3313
9. Sud N, Taher J, Su Q. MicroRNAs and Noncoding RNAs in Hepatic Lipid and Lipoprotein Metabolism: Potential Therapeutic Targets of Metabolic Disorders. Drug Dev Res (2015) 76(6):318–27. doi: 10.1002/ddr.21269
10. Csak T, Bala S, Lippai D, Satishchandran A, Catalano D, Kodys K, et al. microRNA-122 regulates hypoxia-inducible factor-1 and vimentin in hepatocytes and correlates with fibrosis in diet-induced steatohepatitis. Liver Int (2015) 35(2):532–41. doi: 10.1111/liv.12633
11. Yamada H, Suzuki K, Ichino N, Ando Y, Sawada A, Osakabe K, et al. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin Chim Acta (2013) 424:99–103. doi: 10.1016/j.cca.2013.05.021
12. Zhang M, Sun W, Zhou M, Tang Y. MicroRNA-27a regulates hepatic lipid metabolism and alleviates NAFLD via repressing FAS and SCD1. Sci Rep (2017) 7(1):14493. doi: 10.1038/s41598-017-15141-x
13. Pirola CJ, Fernandez Gianotti T, Castano GO, Mallardi P, San Martino J, Mora Gonzalez Lopez Ledesma M, et al. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut (2015) 64(5):800–12. doi: 10.1136/gutjnl-2014-306996
14. Lei L, Zhou C, Yang X, Li L. Down-regulation of microRNA-375 regulates adipokines and inhibits inflammatory cytokines by targeting AdipoR2 in non-alcoholic fatty liver disease. Clin Exp Pharmacol Physiol (2018) 45(8):819–31. doi: 10.1111/1440-1681.12940
15. Liu Z, Patil IY, Jiang T, Sancheti H, Walsh JP, Stiles BL, et al. High-fat diet induces hepatic insulin resistance and impairment of synaptic plasticity. PLoS One (2015) 10(5):e0128274. doi: 10.1371/journal.pone.0128274
16. Li WC, Ralphs KL, Tosh D. Isolation and culture of adult mouse hepatocytes. Methods Mol Biol (2010) 633:185–96. doi: 10.1007/978-1-59745-019-5_13
17. Lee J-S, Zheng Z, Mendez R, Ha S-W, Xie Y, Zhang K. Pharmacologic ER stress induces non-alcoholic steatohepatitis in an animal model. Toxicol Lett (2012) 211(1):29–38. doi: 10.1016/j.toxlet.2012.02.017
18. Kim YJ, Min TS, Seo KS, Kim SH. Expression of pref-1/dlk-1 is regulated by microRNA-143 in 3T3-L1 cells. Mol Biol Rep (2015) 42(3):617–24. doi: 10.1007/s11033-014-3807-0
19. Moravcova A, Cervinkova Z, Kucera O, Mezera V, Rychtrmoc D, Lotkova H. The effect of oleic and palmitic acid on induction of steatosis and cytotoxicity on rat hepatocytes in primary culture. Physiol Res (2015) 64(Suppl 5):S627–36. doi: 10.33549/physiolres.933224
20. Joshi-Barve S, Barve SS, Amancherla K, Gobejishvili L, Hill D, Cave M, et al. Palmitic acid induces production of proinflammatory cytokine interleukin-8 from hepatocytes. Hepatology (2007) 46(3):823–30. doi: 10.1002/hep.21752
21. Lee YH, Yun MR, Kim HM, Jeon BH, Park BC, Lee BW, et al. Exogenous administration of DLK1 ameliorates hepatic steatosis and regulates gluconeogenesis via activation of AMPK. Int J Obes (2016) 40(2):356–65. doi: 10.1038/ijo.2015.173
22. Lee K, Villena JA, Moon YS, Kim KH, Lee S, Kang C, et al. Inhibition of adipogenesis and development of glucose intolerance by soluble preadipocyte factor-1 (Pref-1). J Clin Invest (2003) 111(4):453–61. doi: 10.1172/JCI15924
23. Villena JA, Choi CS, Wang Y, Kim S, Hwang YJ, Kim YB, et al. Resistance to high-fat diet-induced obesity but exacerbated insulin resistance in mice overexpressing preadipocyte factor-1 (Pref-1): a new model of partial lipodystrophy. Diabetes (2008) 57(12):3258–66. doi: 10.2337/db07-1739
24. O’Connell J, Lynch L, Hogan A, Cawood TJ, O’Shea D. Preadipocyte factor-1 is associated with metabolic profile in severe obesity. J Clin Endocrinol Metab (2011) 96(4):E680–4. doi: 10.1210/jc.2010-2026
25. Panera N, Gnani D, Crudele A, Ceccarelli S, Nobili V, Alisi A. MicroRNAs as controlled systems and controllers in non-alcoholic fatty liver disease. World J Gastroenterol (2014) 20(41):15079–86. doi: 10.3748/wjg.v20.i41.15079
26. Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol (2013) 10(9):542–52. doi: 10.1038/nrgastro.2013.87
27. Enache LS, Enache EL, Ramiere C, Diaz O, Bancu L, Sin A, et al. Circulating RNA molecules as biomarkers in liver disease. Int J Mol Sci (2014) 15(10):17644–66. doi: 10.3390/ijms151017644
28. Zhaohui W, Yingli N, Hongli L, Haijing W, Xiaohua Z, Chao F, et al. Amentoflavone induces apoptosis and suppresses glycolysis in glioma cells by targeting miR-124-3p. Neurosci Lett (2018) 686:1–9. doi: 10.1016/j.neulet.2018.08.032
29. Xu X, Li S, Lin Y, Chen H, Hu Z, Mao Y, et al. MicroRNA-124-3p inhibits cell migration and invasion in bladder cancer cells by targeting ROCK1. J Trans Med (2013) 11:276. doi: 10.1186/1479-5876-11-276
30. Niu Y, Zhang Y, Li K, Sun C, Xing C. miR-124-3p and miR-140-3p.2 act as negative regulators of Beclin1 and LC3 expression in the liver of rat model with hepatic impact injury. Biomed Res (Tokyo) (2018) 29(1):50–6. doi: 10.4066/biomedicalresearch.29JRG-17-1393
31. Shaw TA, Singaravelu R, Powdrill MH, Nhan J, Ahmed N, Ozcelik D, et al. MicroRNA-124 Regulates Fatty Acid and Triglyceride Homeostasis. iScience (2018) 10:149–57. doi: 10.1016/j.isci.2018.11.028
Keywords: nonalcoholic fatty liver disease, high-fat diet, miR-124-3p, Pref-1, miRNA
Citation: Wang G, Zou H, Lai C, Huang X, Yao Y and Xiang G (2020) Repression of MicroRNA-124-3p Alleviates High-Fat Diet–Induced Hepatosteatosis by Targeting Pref-1. Front. Endocrinol. 11:589994. doi: 10.3389/fendo.2020.589994
Received: 31 July 2020; Accepted: 28 October 2020;
Published: 26 November 2020.
Edited by:
Oliana Carnevali, Marche Polytechnic University, ItalyReviewed by:
Claudia P. Oliveira, University of São Paulo, BrazilCopyright © 2020 Wang, Zou, Lai, Huang, Yao and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yutong Yao, eGlhbnlpeWFvQDEyNi5jb20=; Guangming Xiang, NTMxOTA2NzgzQHFxLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.