- 1State Key Laboratory of Developmental Biology of Freshwater Fish, College of Life Sciences, Hunan Normal University, Changsha, China
- 2Department of Anatomy, Physiology and Pharmacology, College of Veterinary Medicine, Auburn University, Auburn, AL, United States
Melanocortin-4 receptor (MC4R) plays important roles in regulation of multiple physiological processes, and interaction of MC4R and melanocortin receptor accessory protein 2 (MRAP2) is suggested to play pivotal role in energy balance of vertebrates. Topmouth culter (Culter alburnus) is an economically important freshwater fish in China. Herein we cloned culter mc4r, mrap2a, and mrap2b. Culter mc4r consisted of a 981 bp open reading frame encoding a protein of 326 amino acids. qRT-PCR revealed that mc4r, mrap2a, and mrap2b were primarily expressed in the central nervous system. In the periphery, mc4r and mrap2b were expressed more widely in the male, while mrap2a was expressed more widely in the female. Culter MC4R could bind to four peptide agonists and increase intracellular cAMP production dose dependently. Culter MC4R was constitutively active in both cAMP and ERK1/2 pathways, which was differentially regulated by culter MRAP2a and MRAP2b. Culter MRAP2a significantly increased Bmax and decreased agonist-stimulated cAMP, while MRAP2b increased cell surface and total expression but did not affect Bmax and agonist-stimulated cAMP. These results will aid the investigation of the potential physiological processes that MC4R might be involved in topmouth culter.
Introduction
The melanocortins are derived from tissue-specific post-translational processing of proopiomelanocortin (POMC), including α-, β-, γ-melanocyte-stimulating hormones (α-, β-, γ-MSH) and adrenocorticotropic hormone (ACTH) (1, 2). The physiological functions of these peptides are exerted by five melanocortin receptors (MC1R-MC5R), members of rhodopsin-like Family A G-protein-coupled receptors (GPCRs), including regulation of pigmentation, adrenal steroidogenesis, energy homeostasis, lipolysis, stress, cardiovascular, and sexual function (3–5). MC4R is highly expressed in the central nervous system and involved in regulating energy homeostasis via modulating both food intake and energy expenditure; it is also involved in regulating sexual function and reproduction by affecting the secretion of reproductive hormones [reviewed in (6, 7)]. Targeted deletion of Mc4r increases food intake and decreases energy expenditure, resulting in obesity in mice (8, 9). MC4R mutation is the leading cause of human monogenic obesity (10) [reviewed in (11, 12)].
The MC4R is primarily coupled to the stimulatory heterotrimeric G protein (Gs). MC4R activation leads to stimulation of adenylyl cyclase activity, which will increase the intracellular level of the second messenger cyclic adenosine monophosphate (cAMP) to trigger downstream signaling (13). MC4R activation also phosphorylates extracellular signal-regulated kinase 1 and 2 (ERK1/2) (14–17).
Melanocortin receptor accessory proteins (MRAPs, including MRAP1 and MRAP2), small single transmembrane-domain proteins, are identified as receptor-specific molecular chaperones, regulating MC2R trafficking, ligand binding and cAMP generation (18–21). MRAP2 is involved in modulating energy homeostasis due to its high expression in central nervous system. MRAP2 deceases agonist-stimulated signaling but does not affect basal signaling of human (h) MC4R (20), while MRAP2 in mice decreases the basal activity and increases maximal response (Rmax) (22). MRAP2 mutations were shown to be potentially pathogenic for early-onset obesity (22, 23). Mice with whole body and brain-specific deletion of Mrap2 also have early-onset severe obesity (22). These results suggest that MRAP2 is involved in MC4R signaling and regulating body weight in mammals (21, 22).
MC4R has also been characterized in teleosts (21, 24–32), and shown to act as a regulator in energy balance, sexual behavior, and reproduction (33–37). Non-mammalian MRAPs are different from those of mammals. Some fishes only have MRAP2, lacking MRAP1 (38, 39), and some have two MRAP2s (MRAP2a and MRAP2b) (21, 40). In zebrafish, two MRAP2s have different expression patterns at different stages, with MRAP2a expressed from embryos to adults, stimulating growth by blocking MC4R action in larvae, and MRAP2b mainly expressed in adults, enhancing MC4R response (21). In vitro, MRAP2a decreases the MC4R affinity for α-MSH, but MRAP2b increases ligand sensitivity of zebrafish MC4R (21). Hence, a better understanding of the vital functions of MC4R in modulating energy homeostasis and reproductive function and the interaction between MRAP2 and MC4R is important for feeding and artificial breeding of economical species.
The topmouth culter (Culter alburnus), which belongs to Cyprinidae, Cultrinae, Erythroculter, is an economically important freshwater fish widely distributed in large rivers, reservoirs, and lakes in China (41–44). With excellent taste, rapid growth, and strong performance in culture, this fish has been extensively cultured over the past few decades due to high market demand (45). In this study, we cloned culter mc4r, mrap2a, and mrap2b, and investigated the tissue distribution of these genes. We then studied the pharmacological properties of caMC4R and the effects of MRAP2a and MRAP2b on caMC4R.
Materials and Methods
Ligands and Plasmids
[Nle4, D-Phe7]-α-MSH (NDP-MSH) was purchased from Peptides International (Louisville, KY, USA), human α- and β-MSHs from Pi Proteomics (Huntsville, AL, USA), ACTH (1-24) from Phoenix Pharmaceuticals (Burlingame, CA, USA), and THIQ (N-[(3R)-1,2,3,4-Tetrahydroisoquinolinium-3-ylcarbonyl]-(1R)-1-(4-chlorobenzyl)-2-[4-cyclohexyl-4-(1H-1,2,4-triazol-1-ylmethyl)piperidin-1-yl]-2-oxoethylamine) (46) from Tocris Bioscience (Ellisville, MO, USA). We analyzed culter pomc from culter genome, and found that homology of culter α-MSH, ACTH (1-24) and β-MSH with human counterparts were 100, 87.5, and 57.1%, respectively (Supplementary Figure 1). [125I]-NDP-MSH and [125I]-cAMP were iodinated using chloramine T method (17, 47). The N-terminal c-myc-tagged wild-type hMC4R was subcloned into pcDNA3.1 vector as previously described (48). N-terminal c-myc-tagged caMC4R, N-terminal FLAG-tagged caMRAP2a, and N-terminal FLAG-tagged caMRAP2b were synthesized and subcloned into pcDNA3.1 vector by GenScript (Piscataway, NJ, USA) to obtain the expression plasmids.
Gene Cloning and Sequence Alignment
Adult culter were collected from Engineering Research Center of Polyploid Fish Reproduction and Breeding of the Ministry of Education at Hunan Normal University. Fish were anesthetized before decapitation and tissues were excised and stored at −80°C. All experiments were approved by Animal Care Committee of Hunan Normal University and followed guidelines of the Administration of Affairs Concerning Experimental Animals of China.
Total RNA was purified using Trizol™ Reagent (Invitrogen, Carlsbad, CA, USA). PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, Tokyo, Japan) was used for first-strand cDNA synthesis. Primer Premier 5.0 was used to design specific primers (Supplementary Table 1) to obtain partial cDNA fragment of the coding region and untranslated region (UTR) by PCR and touch-down PCR with TaKaRa LA Taq® (TaKaRa), respectively. PCR products were detected with 1.2% agarose gels. Target fragments were purified by SanPrep Column DNA Gel (Sangon Biotech, Shanghai, China), and then subcloned into the pMD18-T vector (TaKaRa), and sequenced (Sangon Biotech).
Tissue Distribution of mc4r, mrap2a, and mrap2b
To analyze tissue distribution of these genes, the olfactory bulb, telencephalon, mesencephalon, medulla, cerebellum, pituitary, hypothalamus, liver, heart, head kidney, gonads, skin, kidney, muscle, spleen, and gill, were taken from three males and three females, respectively. qRT-PCR was carried out by a Prism 7,500 Sequence Detection System (ABI, Foster City, CA, USA) according to manufacturer's instructions. Primers were designed by AlleleID 6, and β-actin was used as the internal control (Supplementary Table 1). The reaction consisted of 5 μL SYBR green PCR Master Mix, 3 μL water, 1 μL cDNA sample, 0.5 μL QPCR-x-F, and 0.5 μL QPCR-x-R. The condition was: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 61°C for 45 s. To ensure the accuracy, experimental samples were added to a 96-well plate repeated thrice. The 2−ΔΔCT method was used for analyzing the relative expression of the genes (49).
Cell Culture and Transfection
Human embryonic kidney (HEK) 293T cells (ATCC, Manassas, VA, USA) were cultured at 37°C in a 5% CO2-humidified atmosphere. The medium contained Dulbecco's Modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA, USA), 10% newborn calf serum, 10 mM HEPES, 50 μg/mL of gentamicin, 0.25 μg/mL of amphotericin B, 100 μg/mL of streptomycin and 100 IU/mL of penicillin. Cells were plated into 24-well or 6-well plates (Corning, NY, USA) pre-coated with 0.1% gelatin. At ~60–70% confluency, cells were transfected with plasmids by calcium phosphate precipitation method (50). Total DNA was normalized using empty vector pcDNA3.1.
Flow Cytometry Assay
The influence of caMRAP2a or caMRAP2b on the cell surface and total expression levels of caMC4R was studied using flow cytometry as described earlier (31). Cells were plated into 6-well plates and transfected with caMC4R (N-terminal c-myc tag) and caMRAP2a or caMRAP2b plasmids in four ratios (1:0, 1:1, 1:3, and 1:5). The C6 Accuri Cytometer (Accuri Cytometers, Ann Arbor, MI, USA) was used for analysis. The empty vector (pcDNA3.1) fluorescence was used for background staining. The expression of the caMC4R was calculated as the percentage of 1:0 (caMC4R/caMRAP2a or caMC4R/caMRAP2b) group (51).
Ligand Binding Assays
Binding assay was used to study the binding properties of caMC4R to different ligands as described previously (48, 52). Culter MC4R or hMC4R plasmid (0.25 μg/μL) were transfected into cells (6-well plate). The ligands and final concentrations were: NDP-MSH (from 10−11 to 10−6 M), β-MSH (from 10−10 to 10−5 M), α-MSH (from 10−10 to 10−5 M), ACTH(1-24) (from 10−10 to 10−5 M) or THIQ (from 10−11 to 10−6 M). To study the effects of caMRAP2a or caMRAP2b on the binding property of caMC4R, caMC4R (0.25 μg/μL) with caMRAP2a or caMRAP2b in two ratios (1:0 and 1:5) were transfected into cells (6-well plate), and two ligands, α-MSH (from 10−10 to 10−5 M) and ACTH(1-24) (from 10−10 to 10−5 M) were used.
cAMP Assays
Radioimmunoassay for intracellular cAMP was performed as described previously (47, 48). The final concentration of ligands used for signaling assays were NDP-MSH (from 10−11 to 10−6 M), α-MSH (from 10−10 to 10−5 M), β-MSH (from 10−10 to 10−5 M), ACTH(1-24) (from 10−10 to 10−5 M), or THIQ (from 10−10 to 10−5 M).
To study the potential effect of caMRAP2a and caMRAP2b on caMC4R signaling, caMC4R (0.25 μg/μL) and caMRAP2a or caMRAP2b plasmids in two ratios (1:0 and 1:5) were co-transfected into cells (24-well plate), and two ligands, α-MSH (from 10−10 to 10−5 M) and ACTH(1-24) (from 10−10 to 10−5 M) were used. To study the dose-dependent effect of caMRAP2a or caMRAP2b on the Rmax of cAMP levels to α-MSH stimulation, caMC4R (0.25 μg/μL) and caMRAP2a or caMRAP2b plasmids in four ratios (1:0, 1:1, 1:3 and 1:5) were co-transfected into cells (24-well plate). To explore the constitutive activity of Gs-cAMP, cells were transfected with caMC4R plasmid in different concentrations (0, 0.007, 0.015, 0.030, 0.060, 0.125, and 0.250 μg/μL) (6-well plate).
ERK1/2 Phosphorylation Assay
To explore the constitutive pERK1/2 level, cells were transfected with caMC4R plasmid in different concentrations (0, 0.007, 0.015, 0.030, 0.060, 0.125, and 0.250 μg/μL). The phosphorylated ERK1/2 levels were detected as described previously (16, 17). α-MSH (10−6 M) was used for stimulation. Rabbit anti-pERK1/2 antibody (Cell Signaling Technology, Danvers, MA) and mouse anti-β-tubulin antibody (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) were used in this study. ImageJ 1.44 software (National Institute of Health, Bethesda, MD) were used to quantify the films. The pERK1/2 levels were normalized as a ratio of pERK1/2 over β-tubulin in the same gel.
Statistical Analysis
All data were shown as mean ± SEM. Prism 8.3 software (GraphPad, San Diego, CA, USA) was used to calculate parameters including maximal binding (Bmax), IC50, maximal response (Rmax), basal activity, and EC50. The significance of differences in ligand binding and signaling between caMC4R and hMC4R were determined by Student's t-test. Ligand binding, cAMP, flow cytometry parameters of caMC4R regulated by MRAP2s and ERK1/2 signaling were analyzed for significance of differences by one-way ANOVA. Statistical analysis was also performed with GraphPad Prism 8.3 software.
Results
Nucleotide and Deduced Amino Acid Sequences of caMC4R, caMRAP2a and caMRAP2b
The cloned topmouth culter mc4r had 981 bp open reading frame (ORF) that encoded a putative protein of 326 amino acids with 36.57 kDa molecular mass (Figure 1A). The culter MC4R had seven putative hydrophobic transmembrane domains (TMDs) with an extracellular N-terminus, three extracellular loops (ECLs), three intracellular loops (ICLs), and an intracellular C-terminus (Figure 1A and Supplementary Figure 2). The deduced amino acid sequence in the TMDs of caMC4R was significantly conserved with those of other species. The PMY, DRY, and DPxxY motifs were predicted at homologous positions with MC4Rs of other species (Supplementary Figure 2). Two potential N-linked glycosylation site (Asn2 and Asn15) in N-terminus, 15 cysteine residues and consensus sequence for protein kinase C phosphorylation (Thr310Phe311Lys312) in C-terminus were observed in the caMC4R primary structure (Supplementary Figure 2). By multiple sequence alignment analysis, we found that caMC4R shared high identities with other piscine MC4Rs, with 99.1% homology to grass carp, 98.5% to zebrafish, 86.9% to flounder, 87.7% to fugu, and 87.5% to sea bass, as well as to mammalian MC4Rs with 81.4% to human, 81.2% to mouse, and 82.0% to pig. Phylogenetic tree between caMC4R and other MC4Rs revealed that caMC4R was localized in a clade of grass carp and zebrafish (Figure 1B).
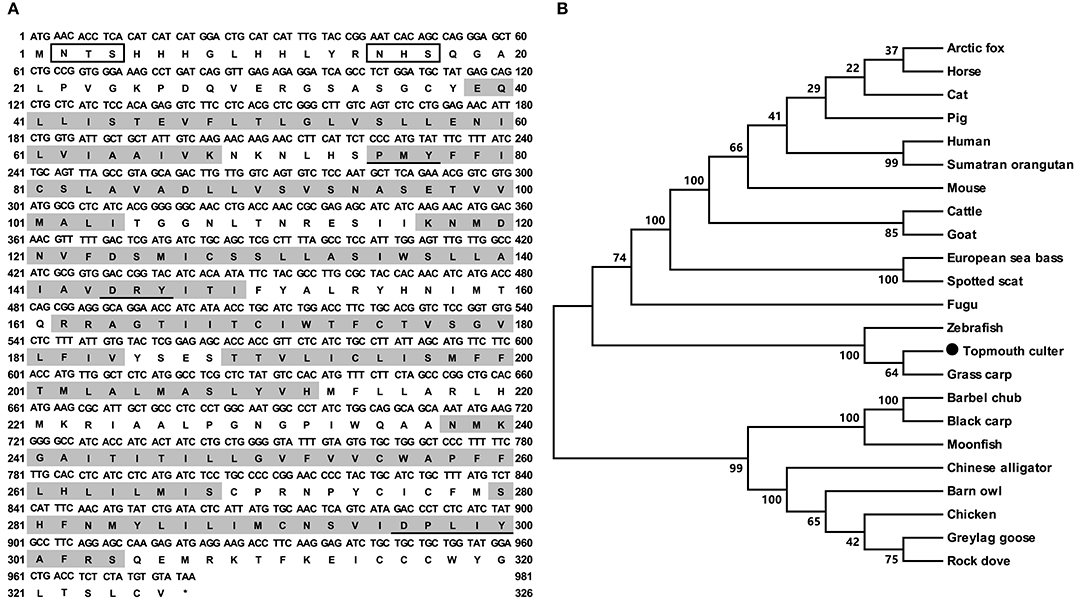
Figure 1. Nucleotide and deduced amino acid sequences and phylogenetic tree of caMC4R. (A) Nucleotide and deduced amino acid sequences of caMC4R. Positions of nucleotide and amino acid sequences are indicated on both sides. Shaded boxes denote putative TMD1-7. Potential phosphorylation sites are present in oval frame. Underline show PMY, DRY, DPxxY motifs. Open boxes denote the consensus sequence for N-linked glycosylation sites. Asterisk (*) indicates stop codon. (B) Phylogenetic tree of MC4R proteins. The tree was constructed by the neighbor-joining (NJ) method. Numbers at nodes indicate the bootstrap value, as percentages, obtained for 1,000 replicates. Black dot shows caMC4R. MC4Rs: Culter alburnus (topmouth culter, MT163518), Scatophagus argus (spotted scat, KU760724.1), Dicentrarchus labrax (European sea bass, CBN82190.1), Lepisosteus oculatus (spotted gar, XP_006634516.1), Danio rerio (zebrafish, NP_775385.1), Vulpes lagopus (arctic fox, ACN55093.1), Ctenopharyngodon idella (grass carp, AOZ60534.1), Paralichthys olivaceus (Japanese flounder, ADP09415.1), Squaliobarbus curriculus (barbel chub, ADV40875.1), Takifugu rubripes (fugu, NP_001027732.1), Mylopharyngodon piceus (black carp, ADV40871.1), Tyto alba (barn owl, ATN96237.1), Columba livia (rock dove, XP_021153678.1), Bos taurus (cattle, NP_776535.1), Alligator sinensis (Chinese alligator, XP_006025279.1), Anser anser (greylag goose, ABF19809.1), Pongo abelii (Sumatran orangutan, XP_002828309.1), Equus caballus (horse, XP_001489706.1), Felis catus (cat, XP_019670932.2), Gallus gallus (chicken, AEP17334.10), Sus scrofa (pig, ABD28176.1), Mus musculus (mouse, NP_058673.2), Capra hircus (goat, NP_001272520.1), and Homo sapiens (human, NP_005903.2).
The cloned culter mrap2a had 654 bp ORF that encoded a putative protein of 217 amino acids with 24.37 kDa molecular mass (Figure 2A). The cloned culter mrap2b had 594 bp ORF that encoded a putative protein of 197 amino acids with 22.20 kDa molecular mass (Figure 2B). The culter MRAP2a and MRAP2b had similar features to other MRAP2s that contained a potential N-linked glycosylation site (Asn8 of caMRAP2a and Asn6 of caMRAP2b) in N-terminus, a single TMD, and a long C-terminal tail with many conserved residues (Supplementary Figure 3). In addition, a putative motif (LKAHKYS) was also found in caMRAP2a and caMRAP2b (Supplementary Figure 3), which is vital in the formation of antiparallel homodimers (53). Multiple sequence alignment showed that caMRAP2a shared high identity (93.6%) with zebrafish MRAP2a, and low identities (61.0%) with caMRAP2b and other piscine MRAP2, with 64.4% to rainbow trout MRAP2, 60.9% to tilapia MRAP2, as well as to mammalian MRAP2s with 61.8% to human MRAP2, 62.2% to mouse MRAP2 and 60.3% to goat MRAP2. Similar to caMRAP2a, caMRAP2b shared high identity with zebrafish MRAP2b with 80.9%, and low identities with other piscine MRAPs, with 61.1% to rainbow trout MRAP2, 59.0% to tilapia MRAP2, as well as to mammalian MRAP2s with 58.3% to human, 60.1% to mouse and 57.7% to goat. Phylogenetic tree analysis showed that caMRAP2a was clustered with teleost MRAP2a, nested with zebrafish MRAP2a, and MRAP2b was clustered with teleost MRAP2b, nested with kanglang fish MRAP2b (Figure 2C).
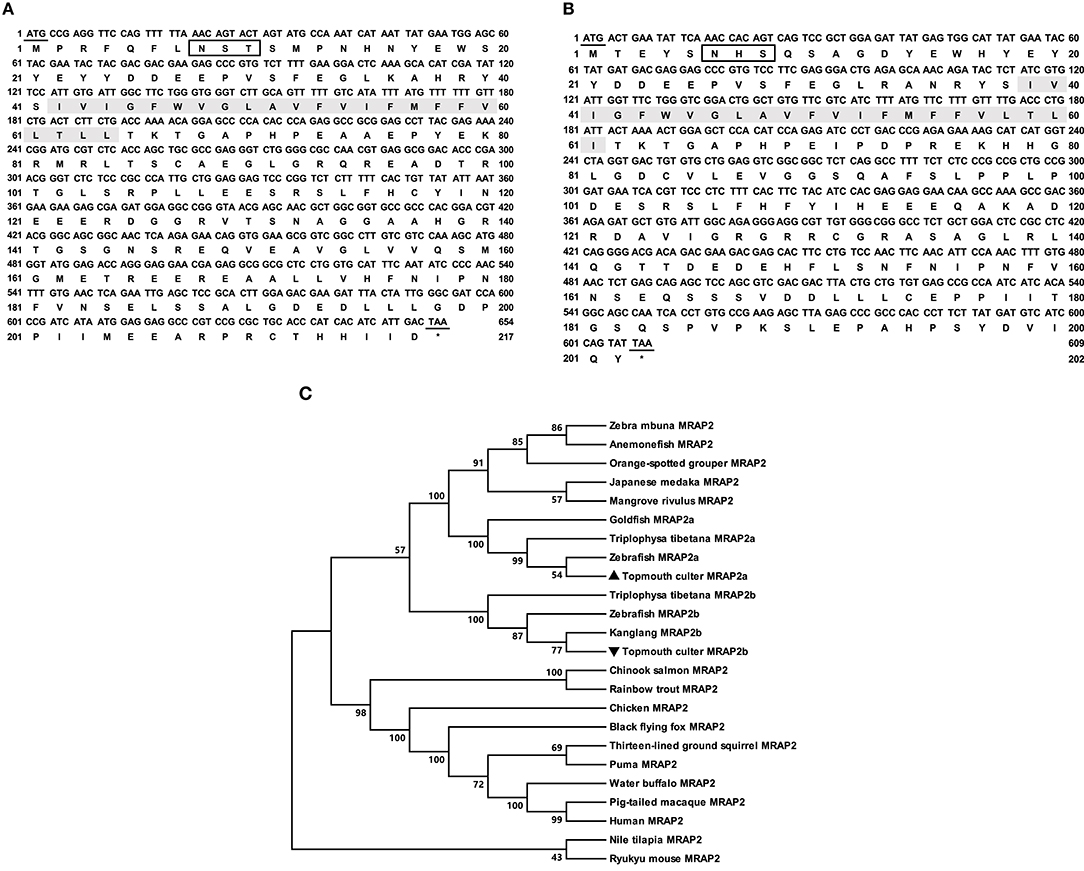
Figure 2. Nucleotide and deduced amino acid sequences and phylogenetic tree of caMRAP2a and caMRAP2b. (A) Nucleotide and deduced amino acid sequences of caMRAP2a. (B) Nucleotide and deduced amino acid sequences of caMRAP2b. The positions of nucleotide and amino acid sequences are indicated on both sides. Shaded box shows putative TMD. Open boxes denote putative N-linked glycosylation sites. Underline show initiation and stop codons. Asterisk (*) indicates stop codon. (C) Phylogenetic tree of MRAP2s. The tree was constructed by the NJ method. Numbers at nodes indicate the bootstrap value, as percentages, obtained for 1,000 replicates. Black triangle dot show caMRAP2a and caMRAP2b. MRAP2s: Culter alburnus (topmouth culter, MRAP2a: MT163516, and MRAP2b: MT163517), Danio rerio (zebrafish, MRAP2a: F8W4H9.1, and MRAP2b: F8W4H9.1), Epinephelus coioides (orange-spotted grouper, QED39647.1), Oryzias latipes (Japanese medaka, XP_023809099.1), Maylandia zebra (zebra mbuna, XP_004568825.1), Amphiprion ocellaris (anemonefish, XP_023122806.1), Kryptolebias marmoratus (mangrove rivulus, XP_017267334.1), Oncorhynchus mykiss (rainbow trout, XP_021467183.1), Oreochromis niloticus (Nile tilapia, XP_003458293.2), Gallus gallus (chicken, ALO81626.1), Ictidomys tridecemlineatus (thirteen-lined ground squirrel, XP_021581743.1), Oncorhynchus tshawytscha (Chinook salmon, XP_024278413.1), Mus caroli (Ryukyu mouse, XP_021029091.1), Pteropus alecto (black flying fox, XP_006926405.1), Puma concolor (puma, XP_025781535.1), Homo sapiens (human, AAH10003.2), Bubalus bubalis (water buffalo, XP_006054803.2), Macaca nemestrina (pig-tailed macaque, XP_011764298.1), Triplophysa tibetana (MRAP2a: KAA0703529.1 and MRAP2b: KAA0720858.1), Anabarilius grahami (Kanglang fish, MRAP2b ROJ29330.1), Carassius auratus (goldfish, MRAP2a: XP_026139519).
Tissue Expression of Culter mc4r, mrap2a, and mrap2b
The relative mRNA expression of culter mc4r, mrap2a, and mrap2b was analyzed by qRT-PCR (Figure 3). Sexual dimorphism was observed in mc4r expression (Figures 3A,B). Our results showed that in the male culter, mc4r was expressed more widely, in brain (telencephalon, mesencephalon, hypothalamus, medulla), pituitary gland, and the periphery (liver, testis, and head kidney) (Figure 3A). In the female culter, expression of mc4r was higher in mesencephalon, olfactory bulb, telencephalon, hypothalamus, medulla, and pituitary gland, but expressed at low levels in other peripheral tissues (Figure 3B).
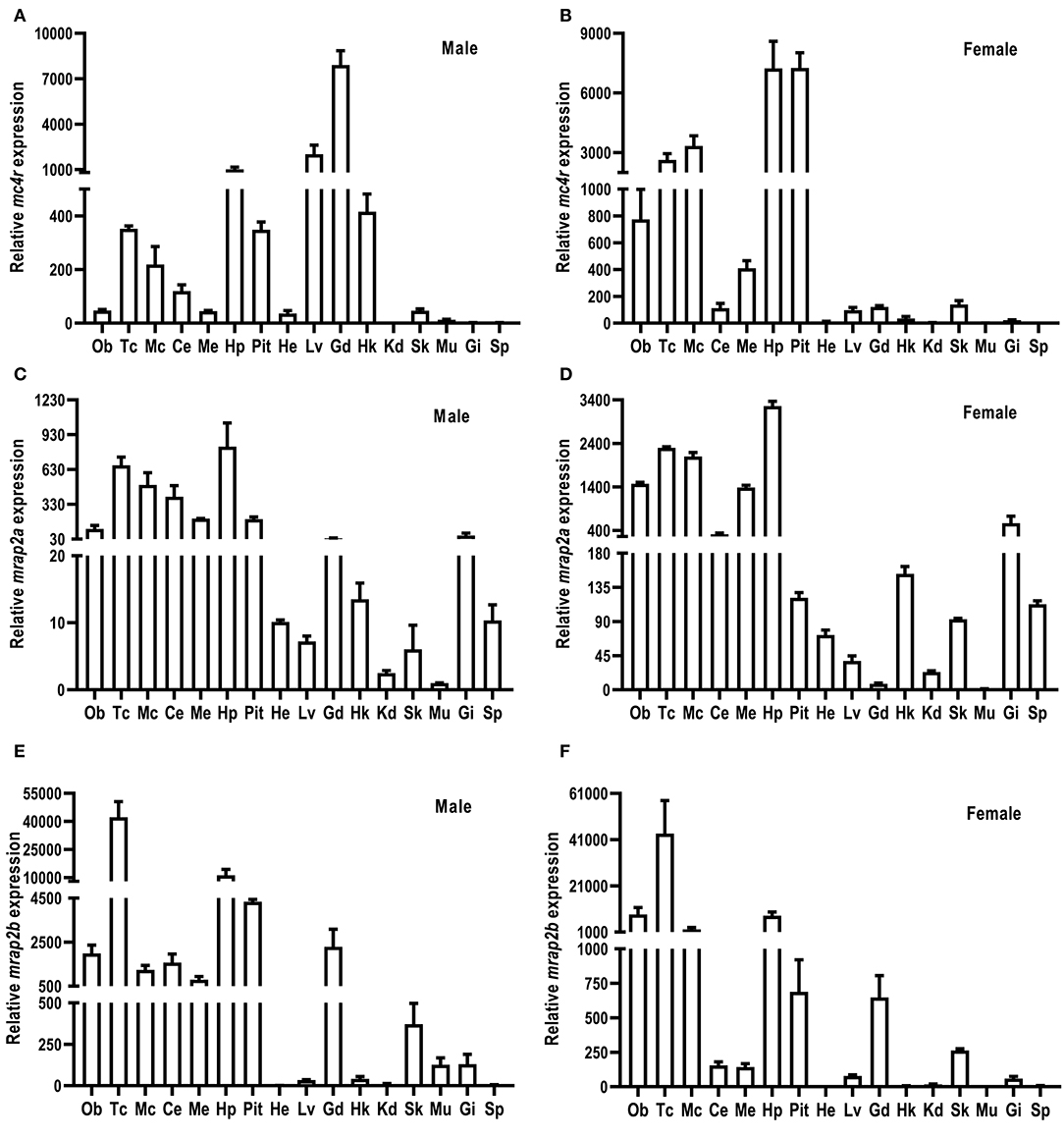
Figure 3. Expression of culter mc4r (A,B), mrap2a (C,D) and mrap2b (E,F). The mRNA levels of mc4r, mrap2a, and mrap2b were measured by qRT-PCR. The results were calculated as the ratio of β-actin. Each data point represented the mean ± SEM (n = 3). Ce, cerebellum; Ob, olfactory bulb; Mc, mesencephalon; Me, medulla; Tc, telencephalon; Pit, pituitary gland; Hp, hypothalamus; Lv, liver; He, heart; St, stomach; Int, intestine; Kd, kidney; Gd, gonad; Sk, skin; Sp, spleen; Hk, head kidney; Gi, gill; Mu, muscle.
Culter mrap2a was highly expressed in brain, and moderately expressed in testis and ovary. Culter mrap2a was expressed more widely in peripheral tissues in female than in male (Figures 3C,D). In the male, mrap2a was mainly expressed in testis and gill, and had a lower expression in other peripheral tissues studied (Figure 3C). In the female, mrap2a was present in liver, skin, kidney, gill, ovary, head kidney, and spleen (Figure 3D).
Similar to mrap2a distribution in the central nervous system, mrap2b was highly expressed in the olfactory bulb, telencephalon, cerebellum, mesencephalon, medulla, hypothalamus, and pituitary gland (Figures 3E,F). However, different from marp2a expression in peripheral tissues, mrap2b was present more widely in peripheral tissues of the male, including liver, head kidney, muscle, skin, testis, and gill (Figure 3E). In the female, mrap2b was primarily expressed in ovary and gill (Figure 3F).
Ligand Binding Properties of caMC4R
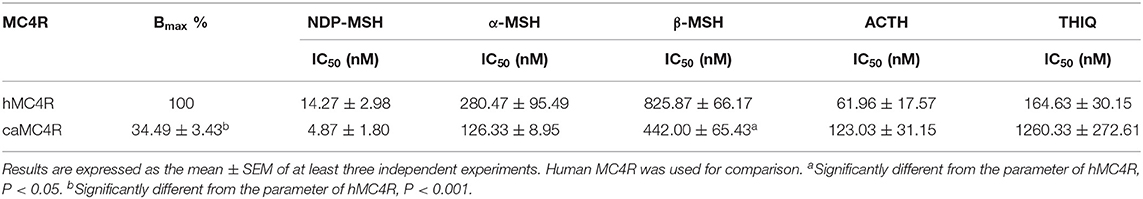
Competitive ligand binding assays were performed to investigate the binding properties of caMC4R using hMC4R for comparison. Different concentrations of five unlabeled agonists (NDP-MSH, α-MSH, β-MSH, ACTH (1-24), and THIQ) were used as competitors with a fixed amount of 125I-NDP-MSH. The maximal binding values (Bmax) of caMC4R was 34.49 ± 3.43% of that of the hMC4R (Figure 4 and Table 1). Similar as the binding affinity order of hMC4R, caMC4R bound to superpotent agonist NDP-MSH with the highest affinity (IC50, 4.87 ± 1.80 nM), followed by ACTH (1–24) (123.03 ± 31.15 nM), α-MSH (126.33 ± 8.95 nM), and β-MSH (442.00 ± 65.43 nM) (Table 1). CaMC4R had a significantly higher affinity to β-MSH than hMC4R. THIQ was able to displace the 125I-NDP-MSH bound with caMC4R, although it had a lower affinity (1260.33 ± 272.61 nM) compared with that for hMC4R (164.63 ± 30.15 nM) (Figure 4 and Table 1).
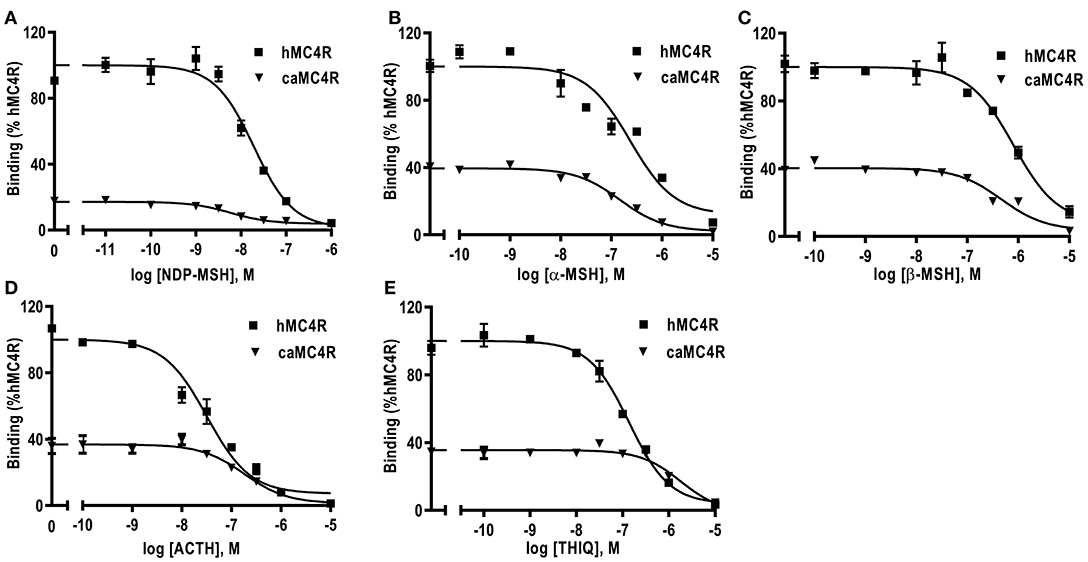
Figure 4. Ligand binding properties of caMC4R. HEK293T cells were transiently transfected with caMC4R plasmids (hMC4R was used as a control). Different concentrations of unlabeled NDP-MSH (A), α-MSH (B), β-MSH (C), ACTH (1–24) (D), and THIQ (E) was used to displace the binding of 125I-NDP-MSH. Results are expressed as % of hMC4R binding (in the absence of competitor) ± range from duplicate determinations within one experiment. All experiment was repeated at least three times independently.
Signaling Properties of caMC4R
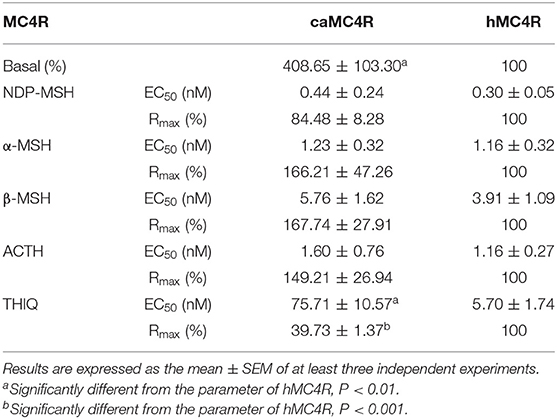
All agonists (NDP-MSH, α-MSH, β-MSH, ACTH (1-24) and THIQ) were able to stimulate caMC4R and dose-dependently increased intracellular cAMP levels (Figure 5 and Table 2). Similar maximal responses (Rmax) and EC50s to NDP-MSH, α-MSH, β-MSH, and ACTH (1-24) stimulation were observed between caMC4R and hMC4R. However, EC50 was significantly increased and Rmax was significantly decreased when stimulated by THIQ (Figure 5 and Table 2).
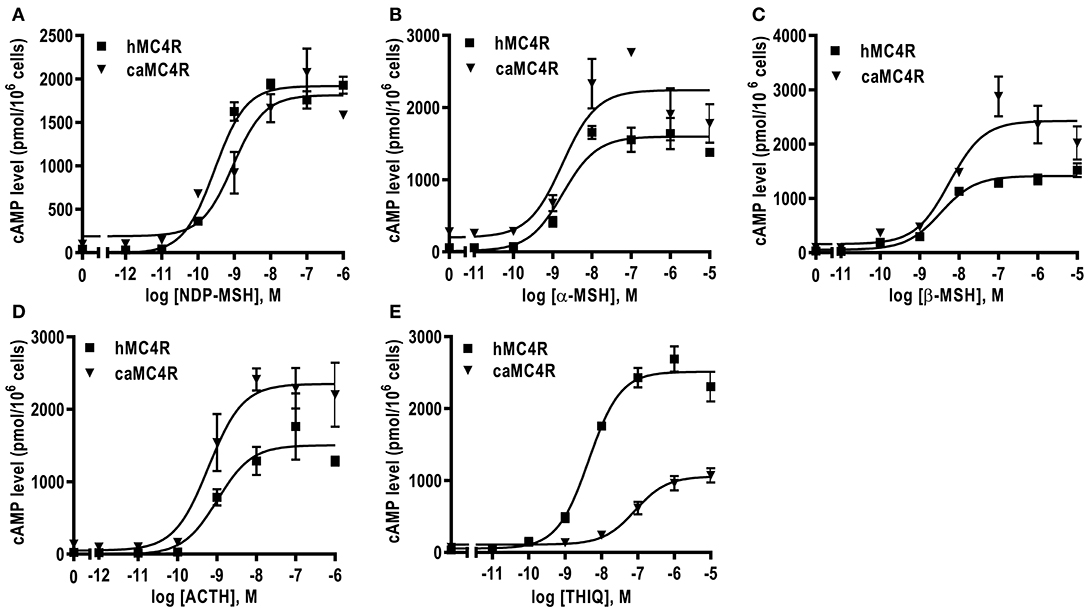
Figure 5. Signaling properties of caMC4R. HEK293T cells were transiently transfected with MC4R plasmids. Forty-eight hours after transfection, cells were stimulated with different concentrations of NDP-MSH (A), α-MSH (B), β-MSH (C), ACTH (1–24) (D), and THIQ (E). Data are mean ± SEM from triplicate measurements within one experiment. All experiments were performed at least three times independently.
In this study, we found that the basal signaling of caMC4R was 4.08 times that of hMC4R (Table 2), indicating that caMC4R might be constitutively active. Different concentrations of caMC4R plasmid were transfected into cells and basal intracellular cAMP levels measured. We found that a low amount of caMC4R plasmid (0.007 μg/μL) transfected resulted in high-level cAMP production (Figure 6A). Similar results were found in basal ERK1/2 phosphorylation, starting with a transfection of 0.015 μg/μL caMC4R that significantly increased basal pERK1/2 level (Figures 6B,C). Therefore, our data indicated that caMC4R could be constitutively active in both cAMP and ERK1/2 pathways.
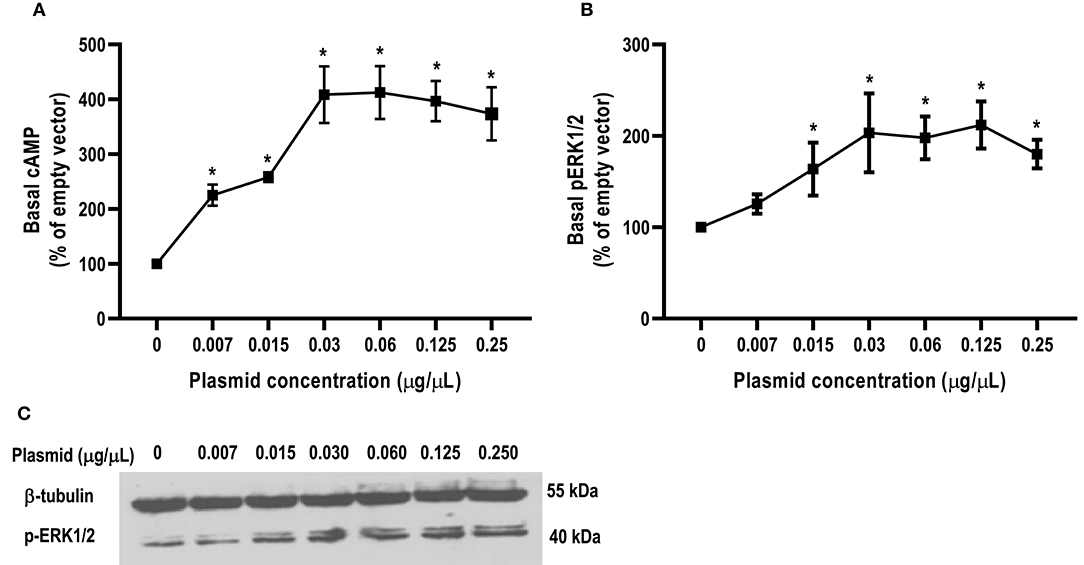
Figure 6. Constitutive activities of caMC4R in cAMP and ERK1/2 pathways. HEK293T cells were transfected with increasing concentrations of caMC4R plasmids. Cells transfected with empty pcDNA3.1 vector was considered as control group. (A) cAMP levels were were measured by RIA. The curve was made with data from three independent experiments. (B,C) The ERK1/2 phosphorylation levels were detected by western blot. Values are expressed as mean ± SEM (n = 3). Asterisk (*) showed significant difference of cAMP and pERK1/2 levels compared with control group (P < 0.05) (One-way ANOVA followed by Tukey-test).
Modulation of caMC4R Expression and Pharmacology by caMRAP2s
The influence of caMRAP2a and caMRAP2b on the cell surface and total expression levels of caMC4R was performed by flow cytometry (Figure 7). The results demonstrated that caMRAP2a increased the cell surface and total expression of caMC4R, and there were no significant differences among groups (Figures 7A,B). Culter MRAP2b significantly increased cell surface and total expression of caMC4R in 1:3 group compared with the 1:0 group (Figures 7C,D).
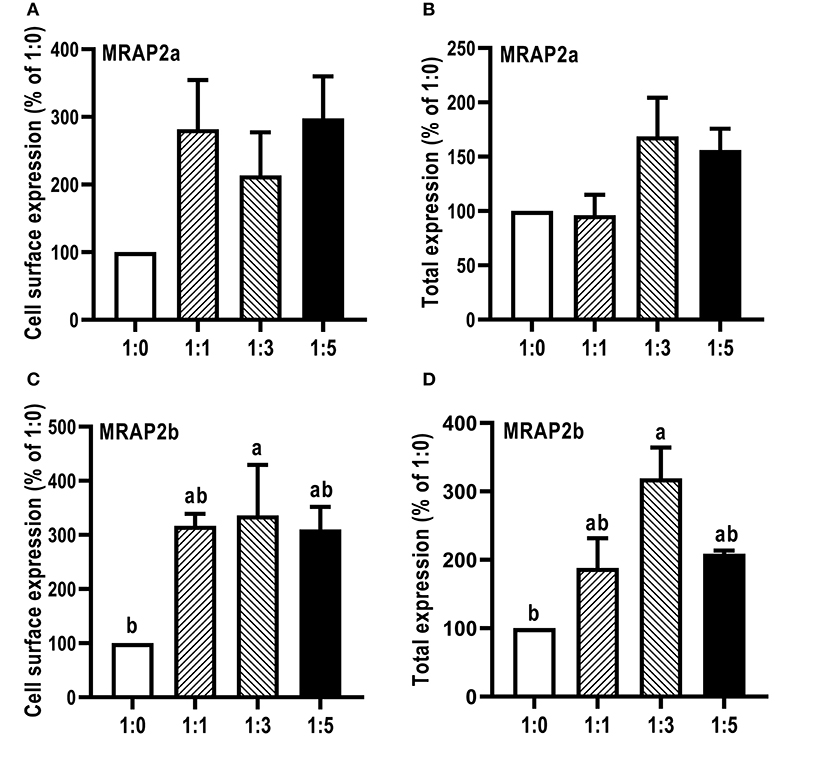
Figure 7. Modulation of caMC4R expression by caMRAP2a or caMRAP2b. Cell surface (A,C) and total expression (B,D) of caMC4R was measured by flow cytometry. HEK293T cells were cotransfected with different ratios of caMC4R/caMRAP2a or caMC4R/caMRAP2b (1:0, 1:1, 1:3, and 1:5). The results were calculated as % of 1:0 group after correction of the background staining (fluorescence in cells transfected with empty vector, pcDNA3.1). Each data point represented the mean ± SEM (n = 3–4). Different letters indicated significant difference (P < 0.05) (One-way ANOVA followed by Tukey-test).
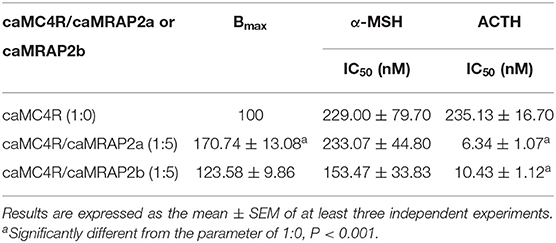
Competitive ligand binding assays with ACTH(1-24) and α-MSH showed that caMRAP2a significantly increased the Bmax of caMC4R in the 1:5 group, while caMRAP2b did not affect the Bmax (Figures 8A,B and Table 3). CaMRAP2a and caMRAP2b significantly increased affinity of caMC4R to ACTH(1-24) but did not affect the IC50s of caMC4R to α-MSH (Table 3).
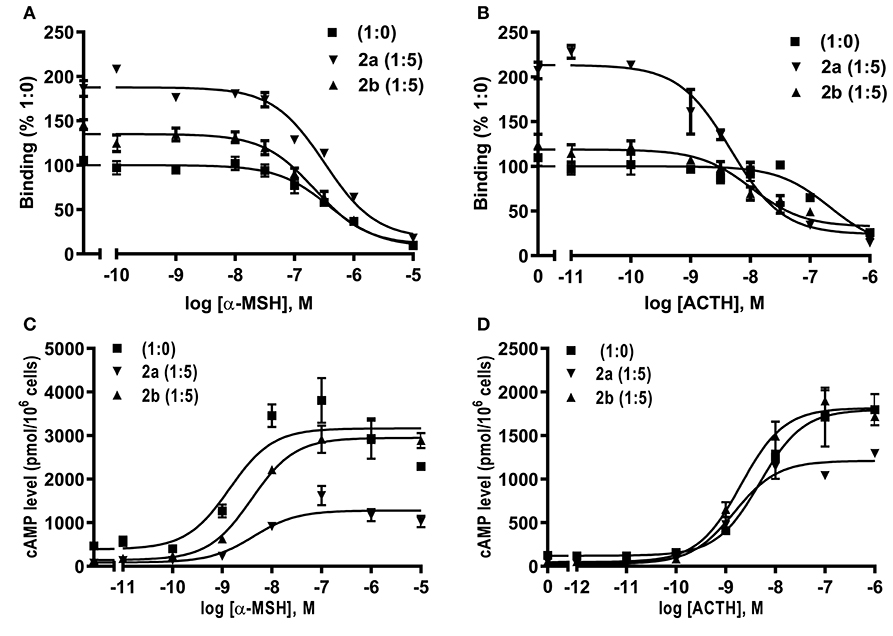
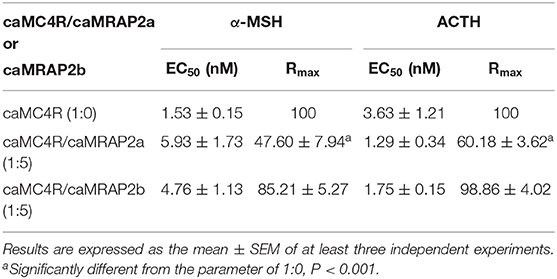
Figure 8. Modulation of caMC4R pharmacology by caMRAP2a or caMRAP2b. Binding (A,B) and signaling properties (C,D) of caMC4R to α-MSH or ACTH (1–24) upon co-expression of caMRAP2a or caMRAP2b were measured. HEK293T cells were co-transfected with caMC4R/caMRAP2a or caMC4R/caMRAP2b in two different ratios (1:0, and 1:5). Binding and signaling properties were calculated as described in detail in Figures 4, 5, respectively.
To study the effect of caMRAP2a or caMRAP2b on the cAMP signaling of caMC4R, cells were co-transfected with caMC4R/caMRAP2a or caMC4R/caMRAP2b in two different ratios (1:0 and 1:5) and ACTH (1-24) and α-MSH were used as agonists. The results showed that caMRAP2a or caMRAP2b had no effect on EC50s; caMRAP2a significantly decreased the Rmax, but caMRAP2b did not affect the Rmax (Figures 8C,D and Table 4).
We further investigated the dose-dependent effects of caMRAP2a or caMRAP2b on basal and maximal signaling to α-MSH stimulation (Figure 9). Cells were co-transfected with four ratios of caMC4R/caMRAP2a or caMC4R/caMRAP2b (1:0, 1:1, 1:3, and 1:5). We found that the basal cAMP production of caMC4R were dose-dependently decreased by both caMRAP2a and caMRAP2b (Figures 9A,B). In addition, maximal cAMP levels of caMC4R activated by 10−6 M α-MSH were also dose-dependently decreased by caMRAP2a in 1:3 and 1:5 groups but not caMRAP2b (Figures 9C,D).
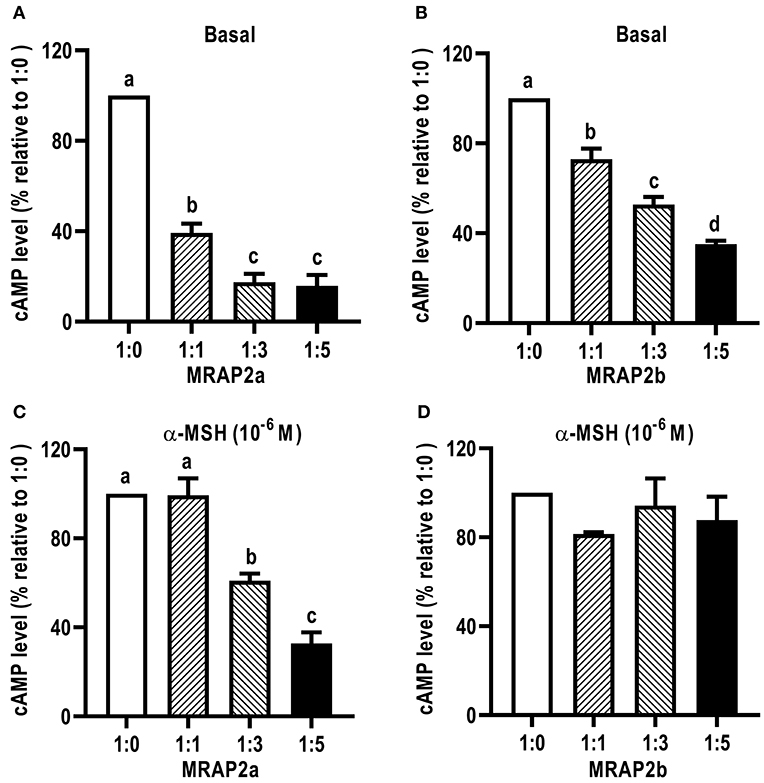
Figure 9. The effects of caMRAP2a or caMRAP2b on caMC4R signaling. HEK293T cells were co-transfected with different ratios of caMC4R/caMRAP2a or caMC4R/caMRAP2b (1:0, 1:1, 1:3, and 1:5) and cAMP levels under basal (A,B) or stimulated [with 10−6 M α-MSH, (C,D)] conditions were measured. Data are expressed as % of 1:0 group. Values are expressed as mean ± SEM (n = 3). Different letters indicate significant difference (one-way ANOVA followed by Tukey-test).
Discussion
In the present study, we demonstrated that cloned culter mc4r was predicted to encode a protein of 326 amino acids with similar structural characteristics as MC4Rs of other species (Figure 1A and Supplementary Figure 2), including other teleost MC4Rs (27–31, 37). Phylogenetic analysis revealed that caMC4R clustered with teleost MC4Rs (Figure 1B).
The distribution of MC4R in lower vertebrates is much wider than in mammals. We observed that culter mc4r was highly expressed in the central nervous system (Figure 3), consistent with it being a critical regulator of energy homeostasis (7, 24, 28, 30). In addition to brain, culter mc4r was present more widely in the periphery in the male, especially in the testis (but not in the ovary) (Figures 3A,B). These data suggest that teleost MC4Rs might play an important role in regulating reproductive function (28, 30, 37). Several studies have investigated the roles of teleost MC4R in regulating reproductive function (33, 35–37, 54, 55).
In the present study, we also cloned culter mrap2a and mrap2b, and showed that similar to MRAP2s of other species, culter MRAP2a and MRAP2b had one potential N-linked glycosylation site in the N-terminal domain and a single highly conserved TMD (Figure 2 and Supplementary Figure 3). Based on culter genomic data (43), we did not identify MRAP1 in topmouth culter, consistent with the hypothesis that MRAP1 is lost in lobe-finned fish, amphibians, and reptiles (39, 56). The tissue expression data showed that mrap2a and mrap2b were highly expressed in the central nervous system (Figures 3C–F), similar to the expression of mc4r, indicates that caMRAP2a or caMRAP2b might modulate MC4R signaling in the central nervous system. In the periphery, mrap2a was expressed more widely in the female than in the male, while mrap2b was expressed more widely in the male (Figures 3C–F). Especially, mrap2b was highly expressed in the testis and ovary, while mrap2a had lower expression in the testis and ovary (Figures 3C–F). These data suggested that caMRAP2a and caMRAP2b might have differential roles in modulating MC4R signaling in the periphery, especially in regulating gonadal function.
We also explored the pharmacology of the cloned caMC4R with binding and signaling assays. Our results showed that NDP-MSH bound to caMC4R with the highest affinity (IC50 of 4.87 nM) and activated caMC4R with the highest potency (EC50 was 0.44 nM) (Tables 1, 2). Lower binding capacity was observed in caMC4R (about 35% of that of hMC4R) (Figure 4 and Table 1), consistent with previous studies in spotted scat (27), grass carp (28), swamp eel (29), sea bass (31), and orange-spotted grouper (30). In culter, ACTH had higher affinity and was more efficacious than α-MSH for caMC4R, consistent with the suggestion that ACTH may be the original ligand for the MCRs (57).
In cAMP signaling assay, α-MSH, β-MSH and ACTH (1-24) stimulated caMC4R and hMC4R with similar potencies (Figure 5 and Table 2). THIQ, a small molecule agonist, displaced 125I-NDP-MSH from caMC4R with a lower affinity than hMC4R (Table 1), activated caMC4R and initiated cAMP accumulation with an EC50 of 75.71 nM (~15-fold higher than that of hMC4R) (Table 2). These data suggested that THIQ was not an allosteric agonist at caMC4R, different from our previous studies in grass carp and swamp eel (28, 29).
The hMC4R has been shown to have constitutive activity in Gs-cAMP signaling (58), and N-terminus is an important modulator in regulating constitutive activities in hMC4R (59). Mutations leading to decreased constitutive activity are associated with obesity pathogenesis (6, 59, 60). Compared to hMC4R, teleost MC4Rs showed much higher constitutive activity in cAMP signaling (27–31). Our present study also showed that caMC4R significantly increased basal activities in Gs-cAMP and ERK1/2 signaling (Figure 6). The potential relevance of constitutive activity in teleost MC4Rs remains to be studied more extensively.
We further investigated whether MRAP2a and MRAP2b could modulate the trafficking, ligand binding and signaling of caMC4R. CaMRAP2a and caMRAP2b both increased the cell surface expression of caMC4R (Figure 7). In zebrafish, MRAP2b dose-dependently increases the cell surface expression of MC4R, while MRAP2a has no effect on the cell surface expression of MC4R (21). Mouse MRAP1 and MRAP2 decrease the cell surface expression of MC4R (20). In chicken, MRAP and MRAP2 have no significant effect the cell surface expression of MC4R (61). In tilapia, MRAP2 dose-dependently decreases the cell surface expression of MC4R (32). MRAP2 also decreases the cell surface expression of MCRa and MCRb in sea lamprey, which has only two MCRs (62). Therefore, the effect of MRAP2 on cell surface and total expression of the MC4R varies in different species. CaMRAP2a increased the Bmax of caMC4R but caMRAP2b did not affect the Bmax (Table 3). caMRAP2a and caMRAP2b significantly increased affinity of caMC4R to ACTH(1-24) but did not affect the IC50 of caMC4R to α-MSH (Table 3).
Remarkably, culter MRAP2a and MRAP2b also inhibited the constitutive activity of caMC4R (Figure 9). Furthermore, caMRAP2a significantly decreased the Rmax but caMRAP2b had no effect on the Rmax (Table 4). MRAP2 decreases the constitutive activity of MC4R and the Rmax of MC4R in tilapia and grouper (30, 32). In zebrafish, MRAP2a suppresses the constitutive activity of MC4R, reduces the Rmax, while increases α-MSH potency and MRAP2b suppresses the constitutive activity of MC4R and increases the Rmax (21). In sea lamprey, MRAP2 increases agonist-stimulated signaling of MCRa and MCRb (62). In chicken, MRAP and MRAP2 decreases the basal activity and increases sensitivity to ACTH (61). In mouse, MRAP1 and MRAP2 decreases agonist-stimulated cAMP production (20). Therefore, different effects of MRAPs on MC4R basal and agonist-stimulated signaling are observed in different species.
One shortcoming of this study is that we used human ACTH and β-MSH for the experiments. α-MSH has been shown to be fully conserved in all species with POMC gene studied so far, including culter investigated here. For ACTH(1-24), there were 3 amino acids different between human and culter sequences, but two of these changes were very conservative (Supplementary Figure 1). Therefore, we deduce that culter ACTH(1-24) would likely behave similarly as human ACTH(1-24) that we used in the experiments. There was only 57.1% homology between human and culter β-MSHs (Supplementary Figure 1). Therefore, we need to interpret the data obtained for β-MSH with caution. In future studies, we need to identify the molecular forms of the endogenous MSHs produced and the modifications (for example, acetylated or des-acetylated) and synthesize these peptides for functional characterization experiments.
In summary, we cloned and analyzed the expression patterns of mc4r, mrap2a, and mrap2b from topmouth culter. All three genes were mainly present in the central nervous system, but differential expression was observed in the periphery. Culter MC4R had high constitutive activities and similar potencies to several agonists as hMC4R. Culter MRAP2a significantly increased the Bmax and decreased agonist-stimulated cAMP, whereas culter MRAP2b increased the cell surface and total expression but did not affect Bmax and agonist-stimulated cAMP. Therefore, these data suggested that caMRAP2a and caMRAP2b had differential effects on the expression, binding, and signaling of caMC4R. These findings lay the foundation for future physiological studies on the functions of culter MC4R that might provide new strategies to improve growth and reproduction in culter culture.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by Animal Care Committee of Hunan Normal University.
Author Contributions
MT: conceptualization, investigation, writing original draft, and funding acquisition. R-LJ: conceptualization, investigation, and writing original draft. LH, S-YF, and TL: investigation. S-JL and Y-XT: conceptualization, writing–review and editing, supervision, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (Grant Nos. 31872551, U19A2040, and 31730098), the earmarked fund for China Agriculture Research System (Grant No. CARS-45), and 111 Project (D20007). This study was also partially supported by Animal Health and Disease Research Program of College of Veterinary Medicine at Auburn University. MT, R-LJ, and TL received fellowships from China Scholarship Council, People's Republic of China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2020.00538/full#supplementary-material
References
1. Smith AI, Funder JW. Proopiomelanocortin processing in the pituitary, central nervous system, and peripheral tissues. Endocr Rev. (1988) 9:159–79. doi: 10.1210/edrv-9-1-159
2. Dores RM, Lecaude S. Trends in the evolution of the proopiomelanocortin gene. Gen Comp Endocrinol. (2005) 142:81–93. doi: 10.1016/j.ygcen.2005.02.003
3. Gantz I, Fong TM. The melanocortin system. Am J Physiol Endocrinol Metab. (2003) 284:E468–74. doi: 10.1152/ajpendo.00434.2002
4. Cone RD. Studies on the physiological functions of the melanocortin system. Endocr Rev. (2006) 27:736–49. doi: 10.1210/er.2006-0034
5. Tao YX. Melanocortin receptors. Biochim Biophys Acta. (2017) 1863:2411–3. doi: 10.1016/j.bbadis.2017.08.001
6. Tao YX. Molecular mechanisms of the neural melanocortin receptor dysfunction in severe early onset obesity. Mol Cell Endocrinol. (2005) 239:1–14. doi: 10.1016/j.mce.2005.04.012
7. Tao YX. The melanocortin-4 receptor: physiology, pharmacology, and pathophysiology. Endocr Rev. (2010) 31:506–43. doi: 10.1210/er.2009-0037
8. Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. (1997) 88:131–41. doi: 10.1016/S0092-8674(00)81865-6
9. Balthasar N, Dalgaard LT, Lee CE, Yu J, Funahashi H, Williams T, et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell. (2005) 123:493–505. doi: 10.1016/j.cell.2005.08.035
10. Farooqi IS, Keogh JM, Yeo GS, Lank EJ, Cheetham T, O'rahilly S. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med. (2003) 348:1085–95. doi: 10.1056/NEJMoa022050
11. Tao YX. Mutations in melanocortin-4 receptor and human obesity. Prog Mol Biol Transl Sci. (2009) 88:173–204. doi: 10.1016/S1877-1173(09)88006-X
12. Hinney A, Volckmar AL, Knoll N. Melanocortin-4 receptor in energy homeostasis and obesity pathogenesis. Prog Mol Biol Transl Sci. (2013) 114:147–11. doi: 10.1016/B978-0-12-386933-3.00005-4
13. Gantz I, Konda Y, Tashiro T, Shimoto Y, Miwa H, Munzert G, et al. Molecular cloning of a novel melanocortin receptor. J Biol Chem. (1993) 268:8246–50.
14. Daniels D, Patten CS, Roth JD, Yee DK, Fluharty SJ. Melanocortin receptor signaling through mitogen-activated protein kinase in vitro and in rat hypothalamus. Brain Res. (2003) 986:1–11. doi: 10.1016/S0006-8993(03)03162-7
15. Vongs A, Lynn NM, Rosenblum CI. Activation of MAP kinase by MC4-R through PI3 kinase. Regul Pept. (2004) 120:113–8. doi: 10.1016/j.regpep.2004.02.018
16. Huang H, Tao YX. Pleiotropic functions of the transmembrane domain 6 of human melanocortin-4 receptor. J Mol Endocr. (2012) 49:237–48. doi: 10.1530/JME-12-0161
17. Mo XL, Yang R, Tao YX. Functions of transmembrane domain 3 of human melanocortin-4 receptor. J Mol Endocrinol. (2012) 49:221–35. doi: 10.1530/JME-12-0162
18. Metherell LA, Chapple JP, Cooray S, David A, Becker C, Ruschendorf F, et al. Mutations in MRAP, encoding a new interacting partner of the ACTH receptor, cause familial glucocorticoid deficiency type 2. Nat Genet. (2005) 37:166–70. doi: 10.1038/ng1501
19. Sebag JA, Hinkle PM. Melanocortin-2 receptor accessory protein MRAP forms antiparallel homodimers. Proc Natl Acad Sci USA. (2007) 104:20244–9. doi: 10.1073/pnas.0708916105
20. Chan LF, Webb TR, Chung TT, Meimaridou E, Cooray SN, Guasti L, et al. MRAP and MRAP2 are bidirectional regulators of the melanocortin receptor family. Proc Natl Acad Sci USA. (2009) 106:6146−51. doi: 10.1073/pnas.0809918106
21. Sebag JA, Zhang C, Hinkle PM, Bradshaw AM, Cone RD. Developmental control of the melanocortin-4 receptor by MRAP2 proteins in zebrafish. Science. (2013) 341:278–81. doi: 10.1126/science.1232995
22. Asai M, Ramachandrappa S, Joachim M, Shen Y, Zhang R, Nuthalapati N, et al. Loss of function of the melanocortin 2 receptor accessory protein 2 is associated with mammalian obesity. Science. (2013) 341:275–8. doi: 10.1126/science.1233000
23. Schonnop L, Kleinau G, Herrfurth N, Volckmar AL, Cetindag C, Müller A, et al. Decreased melanocortin-4 receptor function conferred by an infrequent variant at the human melanocortin receptor accessory protein 2 gene. Obesity. (2016) 24:1976–82. doi: 10.1002/oby.21576
24. Cerdá-Reverter JM, Ringholm A, Schioth HB, Peter RE. Molecular cloning, pharmacological characterization, and brain mapping of the melanocortin 4 receptor in the goldfish: involvement in the control of food intake. Endocrinology. (2003) 144:2336–49. doi: 10.1210/en.2002-0213
25. Song Y, Cone RD. Creation of a genetic model of obesity in a teleost. FASEB J. (2007) 21:2042–9. doi: 10.1096/fj.06-7503com
26. Wei R, Yuan D, Zhou C, Wang T, Lin F, Chen H, et al. Cloning, distribution and effects of fasting status of melanocortin 4 receptor (MC4R) in Schizothorax prenanti. Gene. (2013) 532:10017. doi: 10.1016/j.gene.2013.09.068
27. Li JT, Yang Z, Chen HP, Zhu CH, Deng SP, Li GL, et al. Molecular cloning, tissue distribution, and pharmacological characterization of melanocortin-4 receptor in spotted scat, Scatophagus argus. Gen Comp Endocrinol. (2016) 230-231:143−52. doi: 10.1016/j.ygcen.2016.04.010
28. Li L, Yang Z, Zhang YP, He S, Liang XF, Tao YX. Molecular cloning, tissue distribution, and pharmacological characterization of melanocortin-4 receptor in grass carp (Ctenopharyngodon idella). Domest Anim Endocrinol. (2017) 59:140–51. doi: 10.1016/j.domaniend.2016.11.004
29. Yi TL, Yang LK, Ruan GL, Yang DQ, Tao YX. Melanocortin-4 receptor in swamp eel (Monopterus albus): cloning, tissue distribution, and pharmacology. Gene. (2018) 678:79–89. doi: 10.1016/j.gene.2018.07.056
30. Rao YZ, Chen R, Zhang Y, Tao YX. Orange-spotted grouper melanocortin-4 receptor: modulation of signaling by MRAP2. Gen Comp Endocrinol. (2019) 284:113234. doi: 10.1016/j.ygcen.2019.113234
31. Zhang KQ, Hou ZS, Wen HS, Li Y, Qi X, Li WJ, et al. Melanocortin-4 receptor in spotted sea bass, Lateolabrax maculatus: Cloning, tissue distribution, physiology, and pharmacology. Front Endocrinol. (2019) 10:705. doi: 10.3389/fendo.2019.00705
32. Wang M, Chen YJ, Zhu M, Xu BX, Guo WX, Lyu YS, et al. Pharmacological modulation of melanocortin-4 receptor by melanocortin receptor accessory protein 2 in Nile tilapia. Gen Comp Endocrinol. (2019) 282:113219. doi: 10.1016/j.ygcen.2019.113219
33. Lampert KP, Schmidt C, Fischer P, Volff JN, Hoffmann C, Muck J, et al. Determination of onset of sexual maturation and mating behavior by melanocortin receptor 4 polymorphisms. Curr Biol. (2010) 20:1729–34. doi: 10.1016/j.cub.2010.08.029
34. Aspiras AC, Rohner N, Martineau B, Borowsky RL, Tabin CJ. Melanocortin 4 receptor mutations contribute to the adaptation of cavefish to nutrient-poor conditions. Proc Natl Acad Sci USA. (2015) 112:9668–73. doi: 10.1073/pnas.1510802112
35. Smith CC, Harris RM, Lampert KP, Schartl M, Hofmann HA, Ryan MJ. Copy number variation in the melanocortin 4 receptor gene and alternative reproductive tactics the swordtail Xiphophorus multilineatus. Environ Biol Fishes. (2015) 98:23–33. doi: 10.1007/s10641-014-0234-y
36. Jiang DN, Li JT, Tao YX, Chen HP, Deng SP, Zhu CH, et al. Effects of melanocortin-4 receptor agonists and antagonists on expression of genes related to reproduction in spotted scat, Scatophagus argus. J Comp Physiol B. (2017) 187:603–12. doi: 10.1007/s00360-017-1062-0
37. Zhang Y, Wen HS, Li Y, Lyu LK, Zhang ZX, Wang XJ, et al. Melanocortin-4 receptor regulation of reproductive function in black rockfish (Sebastes schlegelii). Gene. (2020) 741:144541. doi: 10.1016/j.gene.2020.144541
38. Västermark Å, Schiöth HB. The early origin of melanocortin receptors, agouti-related peptide, agouti signalling peptide, and melanocortin receptor-accessory proteins, with emphasis on pufferfishes, elephant shark, lampreys, and amphioxus. Eur J Pharmacol. (2011) 660:61–9. doi: 10.1016/j.ejphar.2010.10.106
39. Valsalan R, Krishnan A, Almen MS, Fredriksson R, Schioth HB. Early vertebrate origin of melanocortin 2 receptor accessory proteins (MRAPs). Gen Comp Endocrinol. (2013) 188:123–32. doi: 10.1016/j.ygcen.2013.01.004
40. Josep Agulleiro M, Cortes R, Fernandez-Duran B, Navarro S, Guillot R, Meimaridou E, et al. Melanocortin 4 receptor becomes an ACTH receptor by coexpression of melanocortin receptor accessory protein 2. Mol Endocrinol. (2013) 27:1934–45. doi: 10.1210/me.2013-1099
42. Cao XJ, Wang WM. Haematological and biochemical characteristics of two aquacultured carnivorous cyprinids, topmouth culter Culter alburnus (Basilewsky) and yellowcheek carp Elopichthys bambusa (Richardson). Aquaculture Res. (2010) 41:1331–8. doi: 10.1111/j.1365-2109.2009.02421.x
43. Ren L, Li WH, Qin QB, Dai H, Han FM, Xiao J, et al. The subgenomes show asymmetric expression of alleles in hybrid lineages of Megalobrama amblycephala x Culter alburnus. Genome Res. (2019) 29:1805–15. doi: 10.1101/gr.249805.119
44. Ren L, Tan XJ, Xiong YF, Xu K, Zhou Y, Zhong H, et al. Transcriptome analysis reveals positive selection on the divergent between topmouth culter and zebrafish. Gene. (2014) 552:265–71. doi: 10.1016/j.gene.2014.09.053
45. Wang W, Chen LQ, Yang P, Hou L, He CB, Gu ZM, et al. Assessing genetic diversity of populations of topmouth culter (Culter alburnus) in China using AFLP markers. Biochem System Ecol. (2007) 35:662–9. doi: 10.1016/j.bse.2007.04.008
46. Sebhat IK, Martin WJ, Ye Z, Barakat K, Mosley RT, Johnston DB, et al. Design and pharmacology of N-[(3R)-1,2,3,4-tetrahydroisoquinolinium- 3-ylcarbonyl]-(1R)-1-(4-chlorobenzyl)- 2-[4-cyclohexyl-4-(1H-1,2,4-triazol- 1-ylmethyl)piperidin-1-yl]-2-oxoethylamine (1), a potent, selective, melanocortin subtype-4 receptor agonist. J Med Chem. (2002) 45:4589–93. doi: 10.1021/jm025539h
47. Steiner AL, Kipnis DM, Utiger R, Parker C. Radioimmunoassay for the measurement of adenosine 3',5'-cyclic phosphate. Proc Natl Acad Sci USA. (1969) 64:367–73. doi: 10.1073/pnas.64.1.367
48. Tao YX, Segaloff DL. Functional characterization of melanocortin-4 receptor mutations associated with childhood obesity. Endocrinology. (2003) 144:4544–51. doi: 10.1210/en.2003-0524
49. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
50. Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. (1987) 7:2745–52. doi: 10.1128/MCB.7.8.2745
51. Wang SX, Fan ZC, Tao YX. Functions of acidic transmembrane residues in human melanocortin-3 receptor binding and activation. Biochem Pharmacol. (2008) 76:520–30. doi: 10.1016/j.bcp.2008.05.026
52. Yang LK, Zhang ZR, Wen HS, Tao YX. Characterization of channel catfish (Ictalurus punctatus) melanocortin-3 receptor reveals a potential network in regulation of energy homeostasis. Gen Comp Endocrinol. (2019) 277:90–103. doi: 10.1016/j.ygcen.2019.03.011
53. Sebag JA, Hinkle PM. Opposite effects of the melanocortin-2 (MC2) receptor accessory protein MRAP on MC2 and MC5 receptor dimerization and trafficking. J Biol Chem. (2009) 284:22641–8. doi: 10.1074/jbc.M109.022400
54. Volff JN, Selz Y, Hoffmann C, Froschauer A, Schultheis C, Schmidt C, et al. Gene amplification and functional diversification of melanocortin 4 receptor at an extremely polymorphic locus controlling sexual maturation in the platyfish. Genetics. (2013) 195:1337–52. doi: 10.1534/genetics.113.155952
55. Liu RQ, Du K, Ormanns J, Adolfi MC, Schartl M. Melanocortin 4 receptor signaling and puberty onset regulation in Xiphophorus swordtails. Gen Comp Endocrinol. (2020) 295:113521. doi: 10.1016/j.ygcen.2020.113521
56. Tao YX. Molecular chaperones and G protein-coupled receptor maturation and pharmacology. Mol Cell Endocrinol. (2020) 511:110862. doi: 10.1016/j.mce.2020.110862
57. Metz JR, Peters JJ, Flik G. Molecular biology and physiology of the melanocortin system in fish: a review. Gen Comp Endocrinol. (2006) 148:150–62. doi: 10.1016/j.ygcen.2006.03.001
58. Tao YX. Constitutive activity in melanocortin-4 receptor: biased signaling of inverse agonists. Adv Pharmacol. (2014) 70:135–54. doi: 10.1016/B978-0-12-417197-8.00005-5
59. Srinivasan S, Lubrano-Berthelier C, Govaerts C, Picard F, Santiago P, Conklin BR, et al. Constitutive activity of the melanocortin-4 receptor is maintained by its N-terminal domain and plays a role in energy homeostasis in humans. J Clin Invest. (2004) 114:1158–64. doi: 10.1172/JCI200421927
60. Tao YX. Constitutive activation of G protein-coupled receptors and diseases: insights into mechanism of activation and therapeutics. Pharmacol Ther. (2008) 120:129–48. doi: 10.1016/j.pharmthera.2008.07.005
61. Zhang J, Li X, Zhou Y, Cui L, Li J, Wu C, et al. The interaction of MC3R and MC4R with MRAP2, ACTH, α-MSH and AgRP in chickens. J Endocrinol. (2017) 234:155–74. doi: 10.1530/JOE-17-0131
Keywords: topmouth culter, melanocortin-4 receptor, melanocortin receptor accessory protein 2, signaling, constitutive activity
Citation: Tao M, Ji R-L, Huang L, Fan S-Y, Liu T, Liu S-J and Tao Y-X (2020) Regulation of Melanocortin-4 Receptor Pharmacology by Two Isoforms of Melanocortin Receptor Accessory Protein 2 in Topmouth Culter (Culter alburnus). Front. Endocrinol. 11:538. doi: 10.3389/fendo.2020.00538
Received: 14 May 2020; Accepted: 02 July 2020;
Published: 14 August 2020.
Edited by:
Honoo Satake, Suntory Foundation for Life Sciences, JapanReviewed by:
Robert M. Dores, University of Denver, United StatesYajun Wang, Sichuan University, China
Copyright © 2020 Tao, Ji, Huang, Fan, Liu, Liu and Tao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shao-Jun Liu, bHNqQGh1bm51LmVkdS5jbg==; Ya-Xiong Tao, dGFveWF4aUBhdWJ1cm4uZWR1
†These authors have contributed equally to this work
 Min Tao1,2†
Min Tao1,2† Ya-Xiong Tao
Ya-Xiong Tao