
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol. , 26 February 2020
Sec. Molecular and Structural Endocrinology
Volume 11 - 2020 | https://doi.org/10.3389/fendo.2020.00078
This article is part of the Research Topic The Brain–Gut–Microbiome Network in Metabolic Regulation and Dysregulation View all 5 articles
 Sandra Dedrick1
Sandra Dedrick1 Bharathi Sundaresh1
Bharathi Sundaresh1 Qian Huang1
Qian Huang1 Claudia Brady1
Claudia Brady1 Tessa Yoo1
Tessa Yoo1 Catherine Cronin1
Catherine Cronin1 Caitlin Rudnicki1
Caitlin Rudnicki1 Michael Flood1
Michael Flood1 Babak Momeni1
Babak Momeni1 Johnny Ludvigsson2
Johnny Ludvigsson2 Emrah Altindis1*
Emrah Altindis1*Type 1 Diabetes (T1D) is regarded as an autoimmune disease characterized by insulin deficiency resulting from destruction of pancreatic β-cells. The incidence rates of T1D have increased worldwide. Over the past decades, progress has been made in understanding the complexity of the immune response and its role in T1D pathogenesis, however, the trigger of T1D autoimmunity remains unclear. The increasing incidence rates, immigrant studies, and twin studies suggest that environmental factors play an important role and the trigger cannot simply be explained by genetic predisposition. Several research initiatives have identified environmental factors that potentially contribute to the onset of T1D autoimmunity and the progression of disease in children/young adults. More recently, the interplay between gut microbiota and the immune system has been implicated as an important factor in T1D pathogenesis. Although results often vary between studies, broad compositional and diversity patterns have emerged from both longitudinal and cross-sectional human studies. T1D patients have a less diverse gut microbiota, an increased prevalence of Bacteriodetes taxa and an aberrant metabolomic profile compared to healthy controls. In this comprehensive review, we present the data obtained from both animal and human studies focusing on the large longitudinal human studies. These studies are particularly valuable in elucidating the environmental factors that lead to aberrant gut microbiota composition and potentially contribute to T1D. We also discuss how environmental factors, such as birth mode, diet, and antibiotic use modulate gut microbiota and how this potentially contributes to T1D. In the final section, we focus on existing recent literature on microbiota-produced metabolites, proteins, and gut virome function as potential protectants or triggers of T1D onset. Overall, current results indicate that higher levels of diversity along with the presence of beneficial microbes and the resulting microbial-produced metabolites can act as protectors against T1D onset. However, the specifics of the interplay between host and microbes are yet to be discovered.
The human microbiome consists of trillions of bacterial, viral, and fungal microorganisms (1). They have symbiotically co-evolved with humans for hundred thousands of years (2). The extensive diversity of these microorganisms results from host lifestyle, geographical location, infections, diet, sex, age, and genetic background (3–6). It is shown that the human microbiome can affect many aspects of host physiology, such as metabolism, immunity, and behavior (7–9). Therefore, the disruption of human microbiota could result in disease. A plethora of studies have demonstrated that the human microbiome has the potential to affect the pathogenesis of immune diseases, particularly autoimmune diseases in which the immune system fails to distinguish self from non-self proteins and attacks self-tissues. Examples of such diseases include multiple sclerosis (8), rheumatoid arthritis, systemic lupus erythematosus, anti-phospholipid syndrome (10), Crohn's disease (11–13), ulcerative colitis (14), inflammatory bowel diseases (15, 16), coeliac disease (17), and Type 1 Diabetes (T1D) (18, 19).
The presence of specific gut microbiota, along with overall diversity, are critical in the development of the nascent immune system. Studies conducted in germ-free (GF) and gnotobiotic mouse models reveal their important role in the modulation and differentiation of innate immune cell-types, most notably IL-17 producing CD4+ T cells (Th17) and Foxp3+ T regulatory cells (Tregs) (20, 21). Specifically, segmented filamentous bacteria (SFB) are known to induce the expression of pro-inflammatory Th17 cells, important for maintaining the mucosal barrier, and protect NOD mice from developing type 1 diabetes (20, 22, 23). However, in other mouse models of autoimmune disease (e.g., K/BxN mouse model of autoimmune arthritis), SFB are shown to promote disease progression via increased Th17 accumulation, suggesting their role in autoimmunity is etiologically-specific (24). Other protective bacteria, such as Lactobacilli, Bifidobacteria and Clostridium species, are implicated in the induction of anti-inflammatory Treg cells while Bacteroides fragilis polysaccharide A (PSA) induces IL-10 production and is known to suppress Th17 cell responses (25, 26).
While the pathogenesis of autoimmune disease remains largely unknown, genetic predisposition (27, 28) and environmental factors (29, 30) have been suspected as major causes. Among environmental factors, the microbiome is correlated with autoimmune diseases via direct and indirect interactions with innate and adaptive immune cells. This results in loss of immune tolerance, chronic inflammation, and immune response against host tissues (31–33). The proposed mechanisms of the microbiome's role in autoimmunity include molecular mimicry, epitope spreading, bystander activation, and prolonged infection (32, 34, 35). Therefore, the loss of immune tolerance might be induced by alteration of microbiome composition. Several studies have, indeed, shown that the gut microbiome composition of individuals with autoimmune diseases is significantly different from those of healthy controls (36–41).
With a few exceptions (e.g., Sardinia, Italy), autoimmune disease incidence follows a north-south trajectory, with higher incidences found in Nordic countries, such as Finland, Sweden and Norway (42, 43). Gut microbiota studies on the effect of extreme climate conditions (i.e., polar expeditions) (44) and birth month/place (45, 46) illustrate how climate, particularly sunlight exposure, affects microbiota composition and immune dysregulation. Factors prominent in northern populations—circadian rhythm disruption and vitamin D deficiencies—have also been shown to result in immune dysregulation via shifts in gut microbiota, leading to autoimmune diseases like T1D (47–49).
Type 1 diabetes (T1D) is regarded as a disease characterized by insulin deficiency resulting from autoimmune destruction or loss of function of pancreatic β-cells. It is important to note that although autoimmunity is seen as the main factor causing T1D by a majority of scientists, some scientists think that autoimmunity is secondary to other factors including endoplasmic reticulum stress leading to beta cell loss (50). Historically, the diagnosis of T1D has considered elevated blood glucose levels (hyperglycemia), and the presence of one or more autoantibodies, all of which occur/are present prior to β-cell ablation (51). Autoimmunity can be mounted against insulin (IAA), glutamic acid decarboxylase (GADA), insulinoma-associated autoantigen 2 (IA2A) and/or zinc transporter 8 (ZnT8A) and may occur many years prior to symptom onset (51). In addition to these main autoantibodies, a newly discovered family of neoepitopes formed with post-translational modification were identified (52, 53). Hybrid insulin peptides (HIPs) are one of the most interesting neopeptides that are formed with the fusion of an insulin peptide with a secretory granule peptide in the granules (54). The most common autoantibody observed prior to disease onset is against IAA, with IAA concentrations correlating strongly with the rate of progression to overt T1D in children (51). This process is commonly referred to as seroconversion, an important term found throughout this review.
While T1D is one of the most commonly represented chronic diseases in childhood, around 25% of people diagnosed with the disease are adults (55, 56). Globally, the incidence of T1D has been increasing for several decades; specifically, after World War II in the western world (57–59). Despite the significant genetic influence, the rise of T1D prevalence and variable incidence rates across different countries, and even between some neighboring areas in Europe (e.g., Russian Karelia and Finnish Karelia), implies an interplay between predisposition genes and environmental influences (60, 61). Discordant results for T1D in identical twin studies (30–60% concordant rate), along with migrant studies demonstrating that second generation immigrants to Sweden have an increased risk of developing T1D, all suggest that environmental affects likely play a significant role in disease pathogenesis (62–64).
The non-obese diabetic (NOD) mouse model is the most widely used animal model for the study of T1D due to significant similarities with human T1D in terms of autoantigen recognition, immunopathology, and gene susceptibility (65). Similar to humans, the NOD mouse model displays T1D-associated variance in MHC class II which effects the presentation of islet-derived antigen to T cells. Additionally, multiple non-MHC susceptible genes common to both NOD mice and humans are associated with T1D risk including PTPN22, CTLA-4, IL2RA (66). It was initially observed that colony hygiene status impacts T1D incidence in NOD mice (67). Other NOD mice studies have also demonstrated that the gut microbiome can interact with the immune system to regulate diabetes pathogenesis in mice (68). However, unlike humans, female NOD mice have significantly higher T1D incidence than male NOD mice. A study by Markle et al. (69) found that male NOD mice under specific-pathogen-free (SPF) conditions were more protected against T1D than female SPF mice. However, in germ-free conditions, both male and female mice had equal incidence rates. Certain commensal bacteria known to elevate testosterone levels are also found to be capable of protecting male NOD mice against T1D onset (69). Interestingly, transferring of the gut microbiota from male NOD mice to female mice altered the composition of gut microbiome in recipient mice and, consequently, protected them from T1D (69). This implies that alterations of the gut microbiome can influence immune system and disease pathogenesis.
Despite the apparent differences in NOD mouse model T1D etiology, in vivo studies offer a mechanistic approach not possible in human studies and have proven vital in understanding disease onset and progression. In this review, we will discuss the findings from both human longitudinal studies and NOD mice studies and the role of gut microbiota as it relates to environmental factors in type 1 diabetes pathogenesis.
Genome-wide association studies have identified ~50 genetic regions that affect the risk of developing T1D (70). One's risk for developing T1D is initially determined by identifying genetic factors, such as T1D-associated single nucleotide polymorphisms (SNPs) in the human leukocyte antigen (HLA) gene; more specifically, the HLA-DQ and HLA-DR protein-coding genes DQA1 and DQB1 (71, 72). However, as previously described, genetic predisposition alone is not sufficient to explain the onset of T1D. Cross-sectional studies have successfully identified several environmental factors that might be potential triggers for disease onset (73). Thus far, results from such studies have been broad and primarily correlative, alluding to the multi-faceted nature of the disease.
To identify the causative environmental triggers of disease onset, longitudinal studies of large at-risk cohorts are required across a wide geographical range. Such efforts have been initiated and include: TEDDY, DIABIMMUNE, BABYDIET (a substudy of the larger BABYDIAB), ABIS, TRIGR, and FINDIA (a sub-study with in the Type 1 Diabetes Prediction and Prevention-DIPP Study). In the following section, we will discuss the main findings of TEDDY, DIABIMMUNE, and ABIS, focusing on the role of gut microbiome in the development of autoimmunity. Due to the diet intervention used in BABYDIET, TRIGR, and FINDIA, these longitudinal studies will be discussed in the Diet section of this review.
The Environmental Determinants of Diabetes in the Young (TEDDY) study was designed to monitor children identified as at-risk for the development of T1D. The study was conducted at six different clinical centers in Colorado, Washington State, Georgie/Florida in the United States and Finland, Germany and Sweden. Between 2004 and 2010, general population (GP) and first-degree relative (FDR) newborns were screened for human leukocyte antigen (HLA) types (72). For all six geographical regions, the same high-risk haplogenotypes were identified; four of these haplogenotypes were considered for the GP inclusion criteria (DR3/4, DR4/4, DR4/8, DR3/3) while nine haplogenotypes were considered for FDR (DR3/4, DR4/4, DR4/8, DR3/3, DR4/4b, DR4/1, DR4/13, DR4/9, and DR3/9) (https://teddy.epi.usf.edu/research).
Two additional studies were recently conducted using TEDDY samples. In Vatanen et al. (19), stool sample metagenomes were collected and sequenced from 783 children monthly from age 3 months until the clinical end-point (seroconversion, detection of autoantibodies). In islet autoimmunity (IA) case-control cohorts, IA had a higher prevalence of Streptococcus group mitis/oralis/pneumoniae while controls had higher abundances of Lactobacillus rhamnosus and Bifidobacterium dentium; two commonly used species in probiotic cocktails (Supplementary Table 1). In T1D case-control cohorts, T1D cases had a higher abundance of Bifidobacterium pseudocatenulatum, Roseburia hominis, and Alistepes shahii while healthy controls had a higher prevalence of Streptococcus thermophilus and Lactococcus lactis (Table 1, Supplementary Table 1c). In another study, Stewart et al. (74) analyzed stool samples from 903 children between 3 months and 46 months of age using 16s rRNA (V4) and metagenomic sequencing data. A nested case-control analysis showed that alpha diversity, microbiota maturation and microbiota-by-age Z-scores (MAZ) were similar between cases and controls for both IA and T1D cohorts. The relative abundance of the most abundant genera showed subtle compositional alterations including a higher prevalence of an unclassified Erysipelotrichaceae in IA cases (Supplementary Table 1a). Compared to healthy controls, T1D cases had a higher abundance of Parabacteroides (P < 0.001) and a decreased prevalence of 11 genera including four unclassified Ruminococcaceae, Lactococcus, Streptococcus, and Akkermansia (Table 1). Overall, TEDDY identified several bacteria weakly correlate with T1D onset, however, further studies are needed in order to dissect the mechanisms at play.
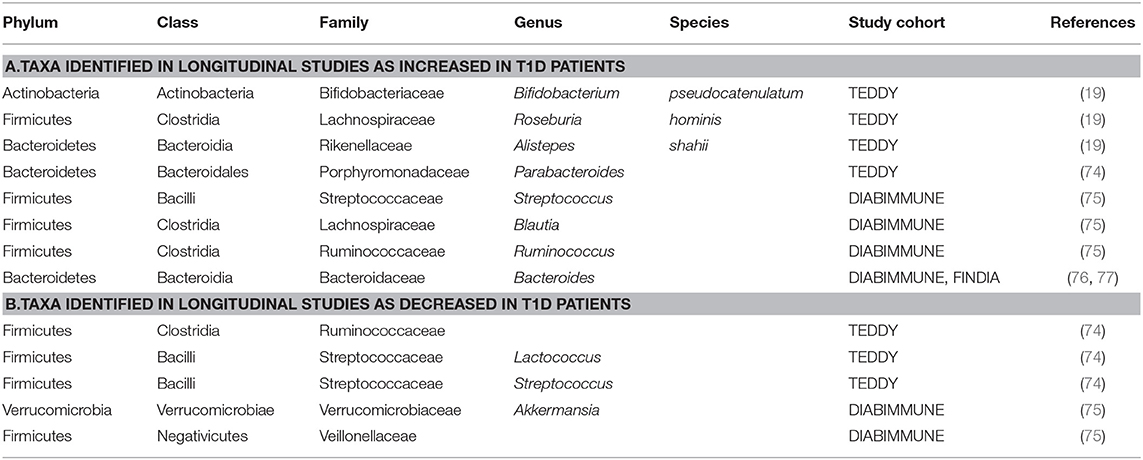
Table 1. Taxa with increased/decreased abundance in T1D subjects identified in longitudinal human studies. Summary of findings from all longitudinal human studies discussed in this review.
In northern Europe, highly disparate incidence rates for autoimmune disease have been documented between neighboring countries. For example, although the frequency of HLA genotypes are very similar between Finland and Russian Karelia, the incidence rates of T1D are 6-fold higher in Finland (78). These observed trends are thought to be dependent upon national public health standards and personal hygiene practices; with “developed” nations carrying a higher incidence rate for autoimmune diseases compared to their “less developed” counterparts. Hygiene and autoimmunity (i.e., hygiene hypothesis) are purportedly linked by the observed decrease in gut microbiota diversity in more hygienic conditions (79–81). The loss of alpha diversity is thought to increase the risk of pathogenic invasion (e.g., antibiotic-induced Clostridium difficile infections) (82) and has been shown to exacerbate autoimmunity in predisposed individuals. The DIABIMMUNE longitudinal study sought to identify environmental factors that could be attributed to the higher incidence of autoimmune and allergenic diseases.
Starting in 2008, DIABIMMUNE recruited ~1,000 newborn infants with high-risk HLA haplotypes from Finland, Estonia, and Russia. Blood and stool samples, along with clinical metadata, were collected from 1 month to 3 years of age. An initial analysis published in 2015 focused on the Finnish and Estonian subjects (75). This study examined a cohort of 33 infants genetically predisposed to T1D. Of this cohort, 11 children developed autoantibodies (seroconverted), and out of the 11 seroconverted subjects, four subjects developed T1D. Microbiome analyses were performed using 16S rRNA and metagenomic shotgun sequencing data. Overall, this study found a decrease in microbial diversity and a reduction in bacterial gene content in autoantibody-positive children during progression toward T1D. Specifically, they found a decrease in Lachnospiraceae and Veillonellaceae in children who developed diabetes, and an increase in Streptococcus, Blautia, and Ruminococcus genera (Table 1, Figure 1). A functional analysis found that bacterial metabolism in autoantibody-positive subjects is characterized by a higher prevalence of genes involved in sugar transport and a lower prevalence of genes associated with amino acid biosynthesis compared to subjects that did not seroconvert.
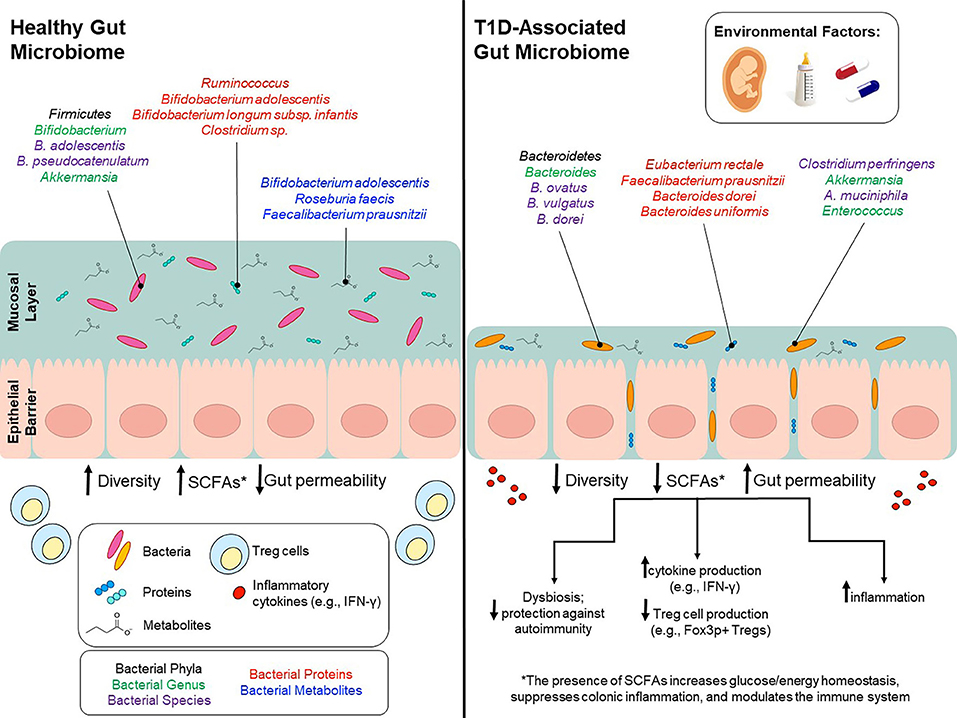
Figure 1. Environmental factors modulate gut microbiota and potentially contribute to T1D onset. Environmental factors, such as birth mode, diet early in life, and use of antibiotics can influence gut microbiota composition and can to lead to lower bacterial diversity, decreased SCFA production and increased gut permeability. Bacterial phyla/genus/species that are affected by environmental factors and differ between T1D patients and healthy controls are depicted in the colors black/green/purple, respectively. Bacterial genus/species that have been identified in proteomic analyses and are increased in either T1D patients or healthy controls are shown in red. Bacterial genus/species that have been identified in metabolomics analyses and are increased in either T1D patients or healthy controls are shown in blue.
In a subsequent DIABIMMUNE study (76), a metagenomic analysis of 222 Finnish, Estonian, and Russian children (sub-cohorts of 74 children from each country), matched for HLA risk and gender, found no correlation between islet autoantibodies or T1D status and microbiome composition. However, the authors reported differential abundance of Bacteroides species. Bacteroides species were lowly abundant in Russian children compared to neighboring Finland and Estonia. The authors postulate that the higher incidence rates of T1D in Finland and Estonia are associated with early life lipopolysaccharide (LPS) exposure. LPS exposure would potentially arise from Escherichia coli for Russian children whereas children from Finland and Estonia would more likely have early-life exposure to Bacteroides species' LPS.
As with TEDDY, DIABIMMUNE identified differences in gut microbiota composition and an overall decrease in microbial diversity in T1D patients. This study also demonstrated a potential mechanism using in vitro experiments which elucidated the structural and functional characteristics of Bacteroides dorei LPS compared to E. coli LPS. The results of the latter study and the effects of early life LPS exposure is further discussed in the Breastfeeding section (Breast Milk and Its Role in Gut Microbiota Modulation).
All Babies in Southeast Sweden (ABIS) is a prospective birth cohort to address the etiology of immune-mediated diseases, specifically T1D (83). All mothers in the Southeast of Sweden that gave birth between October 1997 and October 1999 were asked to participate. In total, ~17,000 babies participated in the study (78.6% of those born in the region), and to date, 147 of them went on to develop T1D. Blood, fecal and other biological samples were collected from newborns at 1, 2.5–3, 5–6, 8, and 11–12 years of age. In a recent study using the ABIS cohort, authors investigated the correlation between HLA haplotype and its effect on gut microbiota composition (23). Previous work in mice has shown that the inability to present class II antigens and/or different MHC class II haplotypes results in differences in gut microbiota composition (23, 84–86). In one study, the lack of class II antigen presentation led to a decrease in Lactobacillus species and an increase in segmented filamentous bacteria (85). In the ABIS study, Russell et al. (23) found similar results in humans in which the genetic risk for developing T1D autoimmunity was associated with alterations in gut microbiota composition. They discovered that the core gut microbiota composition and the beta diversity differed depending on the HLA risk group and overall genotype. Moreover, certain protective HLA haplotypes were correlated with the genera Intestinbacter and Romboutsia (23). Another interesting study examined the relationship between exposure to pets and T1D. They found that 45.5% of pregnant women recruited for the study owned pets. Most common were cats and dogs, and neither exposure to dogs nor cats were associated to T1D risk (87). However, they found exposure to hamster's significantly increased T1D risk. This is a curious finding since a recent study based on the Canadian Healthy Infant Longitudinal Development Study (CHILD) showed that early exposure to household pets alter the gut microbiota composition of the infants which may reduce the risk of some of the diseases including obesity and allergic diseases (88).
Between birth and age 3, the infant microbiota changes drastically due to its constant exposure to novel environmental/maternal microbes and food/animal-born antigens. By age 3, these fluctuations diminish as the microbiota composition settles into its adult-like state (74). Early life factors, such as birth method, breastfeeding, exposure to cow's milk, and introduction to solid foods has been shown to influence early-life microbiota composition, thereby dictating its successional composition. Thus, early life factors have consequences beyond infancy and most likely influence health and disease development later on in life. In the following section we will discuss (i) how environmental factors influence microbiota composition, (ii) how environmental factors influence T1D risk, and (iii) the interplay between environmental factors, gut microbiota, and T1D onset and progression.
Previous research has implicated that the uterus is a sterile environment, completely devoid of bacteria. However, current research efforts have called this into question. A number of studies have found that during the intrauterine period of development, the fetus is exposed to maternal microbiota via transplacental passage into the amniotic fluid (89). Pre-partum, unidirectional shifts in the maternal gut microbiota have also been observed between the first and third trimester, presumably influenced by hormonal and immunologic changes during this period of time (90). Interestingly, this shift enriches for butyrate-producing taxa that promote increased levels of immunomodulatory T regulatory cells (90). This is thought to reduce the chances of maternal rejection of the fetus (90). A second shift in gut microbiota is then observed just prior to giving birth. This shift leads to increased heterogeneity, reduced alpha diversity, and an overrepresentation of taxa that are known to induce inflammation (90, 91). Studies in humanized germ-free mice have suggested that these changes are adaptive and promote maximum energy transfer between mother and fetus (91). These taxa are also thought to promote the successful colonization of anaerobic species that dominate the neonate's early life microbiota. The mother's vaginal microbiota also undergoes alterations just prior to delivery. During pregnancy, four major Lactobacillus species increase in abundance and contribute to the stability of the community while also lowering the alpha diversity (90). These dominant taxa do not play a significant role in maintaining the vaginal pH or in preventing infection, but instead are thought to increase in abundance due to their importance in initial neonate colonization (90).
A number of studies have correlated birth method with distinct variations in neonatal microbiota (92–96). Links between birth mode and an increased risk of obesity, asthma, allergies and autoimmune diseases have also been reported (96–105). Taken together, the evidence strongly suggests that delivery mode has a profound impact on microbiota composition and succession. Vaginally-delivered infants have a microbiome most similar to the vaginal microbiome of the mother while C-section-delivered babies are typically colonized with species found on the mother's skin (96). Differences in taxa are most distinct during the first 3 months of life. The microbiome of infants delivered vaginally are initially dominated by commensal bacteria, such as Lactobacillus and Bifidobacterium while infants born by C-section are predominantly colonized by Clostridium species and skin bacterial communities, most notably, Staphylococcus species (96). TEDDY also found that children delivered vaginally had a higher level of Bacteroides which, in turn, was linked to higher levels of diversity and faster maturation of the gut microbiota (74).
The first stages of microbial exposure/colonization ultimately determines the composition and succession of the infant's microbiota and influences overall metabolism and immunomodulation. Thus, early colonization by aberrant microbiota can have long-term effects on immune function and can increase a child's risk for developing an autoimmune-related disease, such as T1D. Several studies have found that the colonization of Lactobacillus, Bifidobacterium, and Bacteroides is delayed or completely absent in infants born via C-section (92, 96, 106, 107) and the lack of exposure to vaginally-derived Lactobacillus species can lead to differences in microbial succession. Subsequent diversity for the major phyla of gut microbiota—Actinobacteria (dominated by Bifidobacterium), Bacteroidetes, and Proteobacteria—is also significantly lower in C-section-delivered infants (92, 107–112).
A 2008 meta-analysis by Cardwell and colleagues compared the results of 20 different studies to determine if T1D risk is correlated with birth mode. After adjusting for covariates, including birth weight, gestation time, maternal age, maternal T1D incidence, and breastfeeding, there was still a 20% increased risk of T1D onset in infants born via C-section (113). Yet, taking individual research results into consideration reveals a number of discrepancies. Several studies have observed a significant correlation between birth mode and T1D risk (113–116) while others have revealed little to no association (103, 116–118).
Research into the direct link between birth mode, microbiota and T1D incidence is scarce; nevertheless, evidence to date suggests that C-section delivery and aberrant microbiota has an influence on T1D onset and progression. In an NOD mouse study, researchers identified distinct differences in microbiota composition between C-section and vaginally delivered pups although they did not find a significant correlation between birth method and T1D incidence, (119). They also observed lower levels of Foxp3+ T-regulatory cells, anti-inflammatory IL-10, and tolerogenic CD103+ dendritic cells, suggesting that C-section delivery has long-term effects that potentially lead to dysfunctional immunosuppression, thereby, increasing the risk of developing anti-islet autoimmunity (119).
Further analyses into the specific genera implicated in disease onset reveals striking inconsistencies in the role that Bacteroidetes plays. As previously mentioned, neonates delivered via C-section have a decreased abundance of Bacteroides species. Bacteroides play an important role in overall immune system development and function by stimulating the plasmocyte production of secretory IgA (116, 120). More specifically, B. thetaiotaomicron is involved in maintaining the gut barrier while B. fragilis and B. subtilis have been shown to promote gut-associated lymphoid tissue maturation and aid in pre-immunity antibody production (116, 121–123). B. fragilis also inhibits the pro-inflammatory cytokine IL-17 in the intestine (116, 124).
Conversely, one of the most notable distinctions in microbiota for seroconverted high-risk children is the increased abundance of Bacteroides. One example is the longitudinal T1D Prediction and Prevention study (DIPP); a Finnish research effort that started collecting stool and blood samples from children with the high-risk HLA genotype in 1994 (125, 126). A more recent study used four matched case-control pairs from DIPP to identify microbiota that differed between cases and controls (127). Based on their analyses, one major distinction was the abundance of Bacteroidetes and Firmicutes. Cases had a significantly higher abundance of Bacteroidetes, which increased over time, compared to controls. In support of these results, amongst the taxa identified in the DIABIMMUNE study, Bacteroides species were found to be most abundant in the seroconverted cohorts from Finland and Estonia (76). Similar studies have found that Bacteroides species were overrepresented in case subjects; specifically, the species Bacteroides ovatus, which was responsible for 24% of the observed increase (127). A more recent Finnish study (128) corroborates these findings using a cohort of 76 at-risk children which they followed from birth to 2 years of age. Metagenomic analyses identified two Bacteroides species, B. dorei and B. vulgatus, that were significantly higher in cases compared to controls prior to seroconversion (128). However, a number of studies have come to different conclusions or have found no correlation at all between Bacteroides abundance and T1D onset (129). Table 1 and Figure 1 summarize the findings of these studies and the differences in the microbial composition of T1D patients and the controls.
Extensive research has elucidated the many benefits breast milk provides for an infant's growth and development (130–133). Bioactive agents contained in breast milk, including antimicrobial and immunomodulatory components, have been shown to modulate gastrointestinal and immune function along with gut microbiota composition through a multitude of mechanisms (130, 131, 133, 134).
Alongside bioactive compounds, the maternal microbiota is transferred in breast milk and changes in the infant gut microbiota are correlated with breastfeeding frequency in a dose-dependent manner (135). Specifically, lactic acid bacteria in the genera Bifidobacterium and Lactobacillus, capable of breaking down human milk oligosaccharides (HMOs), are transferred from mother to child (132, 136, 137). These genera are known to maintain the intestinal barrier, stimulate the production of IgA antibodies and are involved in the production of short chain fatty acids (SCFAs) (132, 138–144). Of note, the species Bifidobacterium longum subsp. infantis is enriched in infants that are strictly breastfed the first 6 months of life (74). B. infantis is particularly efficient at metabolizing HMOs into SCFAs (indirectly) and has also been shown to promote mucus production, alleviate diet-induced colonic mucus deterioration, and play an important in immuno-regulation (145, 146) (Figure 1).
Successful immunologic development consists of interactions between the host and its microbes at the mucosal surface of the intestine (145, 147). Murine model studies have identified critical developmental windows in which successful microbe-driven immuno-regulation can occur. The same is shown to be true in humans as discussed in the Diet section Gluten-Based Foods. These data suggest that the microbes present and the timing of introduction are key to proper immune system development. For children genetically predisposed to T1D, breast milk is thought to contain protective properties against T1D development (148, 149). Moreover, infants that are strictly breastfed the first 6 months of life have a distinct microbiota composition. It is postulated that the protective properties of breast milk work by modulating gut microbiota composition (94, 132, 150–155).
In a 2015 Norwegian study (MIDIA) investigating the association between breastfeeding duration, age of introduction of solid foods, and the risk of islet autoimmunity/T1D in genetically susceptible children found that any frequency of breastfeeding for 12 months or longer was correlated with a slower progression toward, and an overall decrease in, T1D incidence (156). However, neither the age of solid food introduction nor whether infants were being breastfed at the time of introduction had any effect on islet autoimmunity and/or T1D progression (156). These results indicate that breastfeeding alone has a significant impact on T1D progression/onset while the introduction of other food sources, such as solid foods, may not play as significant of a role (157–159).
Microbiota studies have often looked to the Firmicutes:Bacteroidetes ratio as a potential harbinger for disease development. Although the correlation between disease and shifts in higher-order ratios is currently speculative, similar shifts between Bifidobacterium and Bacteroides have been observed in children at-risk for T1D. Since both genera metabolize HMOs, this inverse correlation is thought to stem from interspecific competition for a common resource (160, 161). Interestingly, these shifts are paralleled in comparisons between breastfed (more Bifidobacterium) and formula-fed (more Bacteroides) infants. Perhaps unsurprisingly, an inverse correlation between Bifidobacterium colonization and T1D disease development has been observed in a number of cross-sectional and longitudinal human studies (145, 161). Moreover, Mejía-Leon et al. discovered an enriched population of Bacteroides in recently diagnosed type 1 diabetics (162). Similarly, a higher incidence of T1D in Finnish and Estonian populations has been documented alongside a higher prevalence of Bacteroides in at-risk children when compared to at-risk children in nearby Russia (76).
In the second DIABIMMUNE study discussed above, authors showed that Bacteroides species, such as B. dorei, inhibit immune stimulation and inflammatory cytokine responses to E. coli which results in suppressed innate immune signaling and a lowered endotoxin tolerance (161). This immune suppression is modulated by high levels of Bacteroides-derived lipopolysaccharides (LPS) that are structurally and functionally distinct from those produced by E. coli; the dominant form of LPS present in Russian infants (161). Moreover, in vitro experiments show that LPS produced by various taxa can either inhibit or trigger TLR4, NF-κB activation, and endotoxin tolerance (163–167). Specifically, when NOD mice were injected with an immunogenic LPS derived from E. coli, endotoxin tolerance was elicited in conjunction with a decreased incidence of T1D (161). B. dorei LPS did not provide the same protection against disease onset (161). These results support previous NOD mouse studies in which LPS were found to have a direct impact on T1D progression (161, 168). The mechanisms involved in LPS-modulated immunity and its link to T1D development are not fully understood, but previous work by Gulden et al. (169), and Wen et al. (68) has revealed the importance of toll like receptor-3 (TLR3) and Innate Immune Signal Transduction Adaptor (MyD88) in T1D onset in NOD mice; two components of the LPS/TLR4 signal transduction pathway (68, 161, 169).
Taken together, results derived from both in vivo and human studies have elucidated the importance of certain breast milk taxa in immune system development and modulation. Some genera, such as Bifidobacterium, play an important role in the overall health and development of an infant, and have also been shown to play a specific role in protecting at-risk children against T1D onset (Figure 1). However, in recent studies, even strictly breastfed infants were void of Bifidobacterium species, suggesting that mothers are not being colonized by these genera to the same extent as their predecessors. In one U.S. study, 30% of strictly breastfed infants had no detectable Bifidobacterium, and for those infants with Bifidobacterium colonization, only 30% had a microbiota in which Bifidobacterium represented over 50% of the population (145, 170).
The overall incidence of T1D has increased considerably over the past half of the twentieth century (171, 172). Current evidence has pointed toward dietary factors as important contributors to T1D development. Some factors have been suggested to trigger or accelerate disease progression, while others protect against the development of T1D-related autoantibodies and overall disease progression (171, 173, 173–183). Both animal and human studies have indicated that early life exposure to foreign food antigens, such as gluten and bovine insulin can influence β-cell autoimmunity (184–186). However, how and to what extent these dietary factors affect disease outcome remains to be discovered.
A common physiological feature of T1D pathogenesis is a weakened intestinal barrier (i.e., leaky gut syndrome) which contributes to increased inflammation in T1D patients (38, 187). Gluten, which consists primarily of monomeric gliadin and polymeric glutenins, can also contribute to gut permeability and induce inflammation by triggering the release of cytokines (188–191). Thus, intestinal microflora that help regulate intestinal barrier function are crucial for dampening the pro-inflammatory response. Gluten-based diets are thought to contribute to gut permeability through the modulation of gut microbiota (192–194). In support of this, taxa associated with a gluten-free diet, such as Akkermansia species, have been shown to regulate human epithelial tight junction proteins, protect against pathogen invasion, and play a protective role against T1D onset (195, 196).
Thus far, the data on the effect of gluten-based foods in humans has shown a wide range of variance. A number of studies have found that placing patients on a gluten-free diet does not markedly improve their autoantibody titers (i.e., reduce). However, these same studies have also shown that a gluten-free diet can improve an individual's insulin response in a glucose tolerance test (197, 198). A few studies focused on patients with both coeliac disease and T1D observed a significant improvement in overall health and diabetic control in subjects provided with a gluten-free diet (193, 199). Nonetheless, other studies have not observed such affects (200, 201).
Despite these apparent inconsistencies, one factor has been ubiquitously identified as playing a crucial role in disease development; the timing and mode of gluten introduction. Human studies have found that early exposure (<3 months of age) to gluten increases the risk of islet autoimmunity and that this risk is mitigated if gluten is introduced while the child continues to breastfeed (51, 202–204). This trend is mirrored in animal studies (205) and has been found to affect T1D onset in a dose-dependent manner (206, 207).
The diet of the mother during pregnancy has also been investigated in relation to a child's T1D risk. When NOD mice mothers were fed a gluten-free diet, the incidence of T1D in their offspring was significantly reduced (208, 209). One such study observed an astounding 7-fold+ decrease in T1D incidence in NOD pups (62.5–8.3%) (209). However, the results from human studies have not been so consistent. A recent study by Antvorskov et al. (210) found that in a cohort of Danish women, the risk of developing T1D was directly proportional to the maternal gluten intake during pregnancy (210). Women that consumed the highest amount of gluten (>20 g/day) had double the risk of T1D onset in their children. In contrast, two previous studies found no strong correlation between a mother's gluten intake and T1D incidence in the child (211, 212).
Early life exposure to diabetogenic dietary factors has a major influence on T1D onset. Even so, eliminating gluten from the diet of older children diagnosed with T1D has been shown to improve diabetic metrics including prolonged remission periods and reduced HBA1c (213, 214). A similar study reported improved glucose tolerance and insulin sensitivities, but no improved titers in islet autoantibodies in older children placed on a gluten-free diet for 6 months (median age = 16 years old) (197).
Overall, these research efforts have provided vital information on the role that gluten plays in T1D onset. Nonetheless, human-subject longitudinal studies are needed to make the connection between diet, its influence on the gut microbiome, and how this effects T1D onset and progression. One such study is the German longitudinal cohort study, BABYDIET study.
BABYDIET, a substudy of the larger BABYDIAB study, was initiated to determine if early-life gluten exposure plays a role in T1D onset. This study followed 22 case children displaying autoimmunity along with 22 matched controls for a little over 3 years. Stool samples were collected monthly between ages 3 and 36 months, and at 6-month intervals thereafter. All subjects had at least one first-degree relative with T1D. Overall, no significant differences were observed between cases and control for microbial diversity (i.e., richness, evenness) and composition once data was corrected for multiple comparisons (215, 216). However, when comparing the overall microbiota network [i.e., Eigenvector Centrality (EC), Node Degree] of children displaying autoimmunity vs. autoantibody-negative children, significant variations emerged. Autoantibody-positive children showed markedly different centrality distributions compared to healthy controls at both 6 months and 2 years of age. There were also more network nodes that had intermediate levels of connectivity in the autoantibody-positive cohort. Furthermore, specific genera had variations between cases and controls in an age-dependent manner. For example, at 6 months of age, the bacterial genera Enterococcus, Sarcina, Prevotella, and Corynebacterium showed high EC in networks of children who became autoantibody positive. In contrast, genera that displayed high EC at 2 years of age in autoantibody-positive children include Barnsiella and Candidatus Nardonella, whereas Staphylococcus and Nocardioides had high EC in the autoantibody-negative group. It is worth noting that these observations were in the genera level as a consequence of 16S sequencing.
Animal model studies and human studies alike have found associations between T1D incidence and the consumption of cow's milk at both weaning and later in development. Retrospective meta-analyses on national dietary consumption records have been used to determine if national and/or global patterns of cow's milk consumption correlates with regional T1D incidence rates (184, 217, 218). Several analyses performed by Muntoni and colleagues found a positive association between T1D and a region's intake of animal-based energy along with a region's overall milk supply (217). A meta-analysis specifically comparing a nation's T1D incidence rate with its annual cow milk protein consumption also found a positive correlation between a nation's T1D incidence rate and its annual milk protein consumption (184, 218). A negative association between breastfeeding (at 3 months) and T1D risk was also observed (184).
The Finnish Diabetes Prediction and Prevention Project (DIPP) studied general food consumption in children with advanced β-cell autoimmunity and found that the general intake of cow's milk products was one of the few factors directly associated with β-cell autoimmunity. This was not true of other dairy products, such as sour milk products and cheese which suggests that the increased risk of β-cell autoimmunity comes specifically from cow's milk-associated proteins (219). Looking more closely at the immune response to these proteins, several studies have observed elevated levels of cow's milk-specific IgG and IgA antibodies in early onset T1D (220–222). Interestingly, Elliott and colleagues found that the total protein consumption was not correlated with T1D incidence, but the specific consumption of cow's milk-derived β-casein (A1 variant) was (218). These results indicate that an elevated humoral response is mounted against cow's milk proteins in T1D patients, making them a potential contributor to the autoimmune process leading up to T1D onset (222).
While evidence supports a role for cow's milk proteins in T1D onset, other studies have found no such correlation (223). Conversely, other studies have suggested that cow's milk-based formula plays a protective role against T1D incidence (224). Human studies investigating the relationship between cow's milk proteins, gut microbiota composition and T1D derive from two major longitudinal studies. These dietary studies include: (1) the Trial to Reduce IDDM in the Genetically at Risk (TRIGR) and (2) the Finnish Dietary Intervention Trial for the Prevention of T1D (FINDIA). While these studies do not specifically address the association between the gut microbiome and T1D incidence, a cross-sectional study derived from these studies was subsequently conducted to investigate this affect.
In the TRIGR study, exposure to foreign dietary proteins was postponed until 6–8 months of age and the development of autoantibodies (insulin, GAD, IA-2, and ZnT8) was monitored until the age of 6 years (172). The aim of this study was to test whether supplementing breast milk with highly hydrolyzed milk formula is able to inhibit T1D onset. Infants (n = 230) with confirmed HLA-associated susceptibility and at least one family member with T1D were enrolled in the study. Subjects received either casein hydrolysate formula or conventional formula whenever breast milk was not available and were subsequently followed up to age ten. Over the course of the study, 17 children developed at least one autoantibody in the casein hydrolysate group (17%) compared to the 33 children in the control group (30%). Eight children in the casein hydrolysate group (8%) and 17 (16%) in the control group developed two or more autoantibodies. Insulin-autoantibodies were the most frequently observed first-autoantibodies with anti-islet autoantibodies being a close second. Overall, this study found weak associations between formula intervention and T1D development. A more recent update on this study published in 2018 found that weaning to hydrolyzed formula did not reduce the risk of T1D in children with an increased disease risk up to the median age of 11.5 years (225).
FINDIA is a multisite double-blind clinical trial in which newborn infants were randomized to receive a bovine insulin-free cow's milk formula (CMF), a whey-based hydrolyzed formula (WHF), or a whey-based FINDIA formula (FINDIA group) from which bovine insulin was removed. Between 2002 and 2005, subjects were recruited from three pediatric hospitals in Finland. One hundred thirteen infants with HLA-conferred susceptibility to T1D were randomly assigned to receive one of the three formulas. Results from the FINDIA study (77) suggest that administering bovine insulin-free formula during the first 6 months of life reduces a child's risk of developing β-cell autoantibodies by age three. When the microbiota of seroconverted subjects were compared to non-seroconverted subjects, researchers found an increased abundance of Bacteroides, and a decreased abundance of Bifidobacterium. In the intention-to-treat analysis, 6.3% of children in the CMF group, 4.9% in the WHF group, and 2.6% of children in the FINDIA formula group were positive for at least one autoantibody by age three. The researchers found that compare to ordinary formula, weaning to an insulin-free formula (FINDIA group) reduced the cumulative incidence of autoantibodies by age 3 years in children at genetic risk of T1D.
In a study by de Gaffau et al. (36), gut microbiota composition was analyzed and compared between autoantibody-positive (n = 18; tested positive for at least two autoantibodies) and matched autoantibody-negative (n = 18) children. Subjects were recruited from TRIGR and FINDIA. Compared to autoantibody-negative children, autoantibody-positive children had higher relative abundance of Bacteroidetes (Bacteroides genus). However, at the species level, there were only a few Bacteroides species associated with autoimmunity, alongside a higher abundance of various Firmicutes, such as Clostridium perfringens; both of which are known to play a role in increased gut permeability. This analysis also suggests that low abundances of Bifidobacterium adolescentis and Bifidobacterium pseudocatenulatum (<12% combined) are significantly correlated with increased levels of beta-cell autoimmunity (Figure 1).
Antibiotics are used to control bacterial infections and have a range of effects on the gut microbiota. Antibiotic exposure has been found to significantly decrease bacterial diversity; however, several studies have shown that adult gut microbiota often recovers to resemble microbiota prior to treatment (226). Nevertheless, a 4-day intervention with a cocktail of three antibiotics (meropenem, gentamicin and vancomycin) caused the loss of nine bacterial species in adult men 180 days after antibiotic use (226). This same study also observed an initial bloom of pathobionts including Enterococcus faecalis and Fusobacterium nucleatum and a depletion of Bifidobacterium (226).
On average, a child in the U.S. receives three antibiotic courses before they are two, and 10 courses before they are 10 (227). Despite their common administration to infants and children, their effects on gut microbiota and how it relates to human diseases, such as T1D, is still not fully understood. In a study on infants, antibiotic exposure was associated with diminished Clostridiales and Ruminococcus in the first 3–9 months of life, and delayed microbiota maturation (228). In contrast to this, a large Norwegian Mother and Daughter Cohort study did not find an association with acetaminophen use and T1D risk. In this study, the use of antibiotics by the mothers and the child in the first 6–9 months was also not found to be correlated with T1D risk (229). Likewise, TEDDY found that frequency and use of antibiotics during the first 4 years of life did not influence the risk of developing autoimmunity for T1D or Coeliac Disease (230). A population-based case-control study that included all T1D cases of children born between 1997 did not find a correlation between antibiotic use and T1D. However, a positive association was seen between children who took broad-spectrum antibiotics in the first 2 years and T1D (231). In a population-based mother-child cohort that included all children born in Finland between 1996 and 2000 who were diagnosed with T1D, they found an increased risk of T1D in children of mothers who took phenoxymethyl penicillins or quinolone before pregnancy. They proposed that use of antibiotics by mothers may diminish the transfer of healthy microflora to the baby. A higher risk of T1D was found when the mother took macrolides before pregnancy and the child took macrolides compared to mother-child pairs where neither took macrolides. A high risk of antibiotics, defined as seven or more purchases of antibiotics, was also found to have an increased risk of T1D. Bifidobacterium and Lactobacillus species are associated with a healthy microflora and are particularly sensitive to macrolides, quinolines, and penicillins. The study proposes that macrolides and quinolones might increase the risk of T1D in children by preventing the synthesis of DNA and enzymes in β-cells resulting in beta cell death (232). Another case-control study used THIN, a UK population medical record database that includes complete medical records of about ten million patients, to look at the effect of antibiotic exposure on both type 1 and type 2 diabetes. The study found that exposure to a single antibiotic was not associated with higher diabetes risk. However, taking two to five antibiotic courses of penicillins, cephalosporins, macrolides, or quinolines was associated with an increase in diabetes risk, and taking more than five courses of quinoline was associated with an increased T1D risk. Exposure to more than five courses of penicillin was also associated with an increased diabetes risk (233).
Several studies have been performed on Bio-breeding Diabetes-Prone (BB-DP) rats, LEW1.WR1 rats, and NOD mice to determine the impact of antibiotic treatment on T1D incidence and the gut microbiome composition. In a study performed by Brugman et al. (192) male and female BB-DP rats on either a conventional plant based (CON) or hydrolyzed casein (HC) diet continuously received antibiotic treatment with broad spectrum antibiotics Bactrimel (sulfamethoxazole and trimethoprim) and colistine sulfate. Antibiotic treatment while on the CON diet was found to reduce T1D incidence and delay onset to 30 days whereas antibiotic treatment while on the HC diet was found to protect against T1D onset (192). Additionally, BB-DP rats that later developed T1D were shown to have higher abundance of Bacteroides when compared to controls and rats that did not develop T1D (192). Similarly, a 2012 Hara et al. study investigating LEW.WR1 rats infected with kilham rat virus (KRV) in order to induce T1D found that virus-induced T1D was suppressed by treatment with the antibiotic sulfatrim. Treatment with sulfatrim was also found to suppress the virus-induced anti-islet responses in the LEW1.WR1 rat via the down-modulation of the immune system. Furthermore, infection with KRV caused an increase in abundance of Bifidobacterium and Clostridium (234). Contrary to these studies, Livanos et al. found that pulses of therapeutic doses of the macrolide antibiotic tylosin tartrate (pulsed therapeutic antibiotics, PAT) accelerated T1D and insulitis development in male NOD mice. The study also found that the group receiving PAT had larger abundance of Akkermansia and Enterococcus when compared to control groups which were more abundant in Bifidobacterium (235). A 2012 study performed by Hansen et al. investigating the effects of early life treatment with the antibiotic vancomycin on NOD mice found that mice treated with vancomycin from birth until 28 days of age had a significantly lower incidence of T1D and higher clusters of CD4+ T cells producing pro-inflammatory cytokines. Conversely, NOD mice that received vancomycin from 8 weeks of age until onset of T1D had higher diabetes incidence, higher insulitis scores, and increased blood glucose levels, indicating that early life treatment significantly impacts T1D progression. Treatment with vancomycin was also seen to affect microbial diversity as Akkermansia muciniphila dominated the gut microbiome of all groups treated with vancomycin (236). In a similar study, the effects of antibiotic treatment on T1D progression in the offspring of NOD mice treated with antibiotics while pregnant further found the importance of antibiotic treatment early in life (237). The authors treated pregnant NOD mice with a combination of three antibiotics; neomycin, polymyxin B, and streptomycin, and found the offspring of treated mice to be significantly more protected than mice that received the antibiotic immediately after birth and mice that received antibiotic treatment at 3 weeks of age. The study illustrates the importance of early life or prenatal antibiotic treatment in order to combat T1D as the age the mice received treatment negatively correlated with protection from T1D. Additionally, the antibiotic treatment was seen to deplete gram negative proteobacteria and lead to dominance by gram positive bacteria of the Firmicutes family. According to the studies mentioned, it is unclear whether antibiotic treatment consistently increases or decreases T1D risk in animals, but it is always seen to have an effect on both microbial diversity and T1D incidence and there is evidence to suggest that early intervention is more effective.
The birth of sequencing altered the way by which scientists asked questions pertaining to both environmental and human associated microbiota and has enabled the use of big data in a way that was not previously possible. As sequencing technologies advance, more researchers are turning to such methodologies to investigate the association between the human microbiome and autoimmune disease. Currently, in T1D research, scientists have investigated differences present in the gut proteome, virome and metabolome of T1D patients. In this section, we will discuss the results and advancements made in these ‘omics related fields.
16S and shotgun sequencing studies inform us regarding the microbial composition of the gut microbiome. On the other hand, proteomics studies are needed to understand which proteins are actively produced by these gut microbes. Few studies to date have been conducted to analyze the intestinal microbial proteome of T1D patients. However, in one such study Pinto and colleagues (238) investigated the differences in the intestinal microbial proteome between children with established T1D (3 children) compared with the proteome of healthy children (3 children). In control samples, bacterial proteins from Bifidobacterium adolescentis, Bifidobacterium longum subsp. infantis, Ruminococcus, Collinsella aerofaciens, Coprococcus comes, and Clostridium sp. were found to be the most abundant while proteins originating from Eubacterium rectale, Faecalibacterium prausnitzii, Bacteroides dorei and Bacteroides uniformis varied between control and case samples (Figure 1). More recently, one study (239) compared stool samples from newly-onset type 1 diabetes patients, islet autoantibody-positive, and low-risk subjects. Metaproteomic analysis was utilized to differentiate these stool samples into these three categories based on the presence of human (host-derived) and microbial-derived proteins. In patients with new-onset T1D, there was a significant reduction in microbial-associated host proteins that are responsible for maintaining the mucous barrier, microvilli adhesion, and exocrine pancreas. Thus, T1D patients had a higher prevalence of intestinal inflammation and decreased barrier function. A previous study, published in 2016, sets the stage for larger-scale metagenomics studies by demonstrating, on a smaller-scale, how specific gastrointestinal microbial communities and host-microbe phenotypic relationships are associated with onset of human disorders (18). Although the role of exocrine pancreas function in T1D onset remain unclear, they showed that exocrine pancreas enzymes, including amylase, carboxypeptidase CPA1 and CUZD1are less abundant in T1D patients (18). These initial results are promising and provides the field with an initial insight into how the T1D metaproteome differs in functionality compared to controls. Further research into this area will provide more information on disease progression and prevention methods.
Viruses have long been considered potential triggers of T1D onset and progression (240). However, much of the research to date has focused on the role that viral infections play in this process and the data have been inconclusive. More recently, researchers have begun to investigate the role of the gut virome. The gut virome is highly understudied as it relates to both states of health and disease, however, characterization of the intestinal virome from birth to the development of autoimmunity has potential to be an important component to understanding the etiology of T1D. Using the Finnish Diabetes Prediction and Prevention (DIPP) cohort, stool viromes were collected for 19 cases (who turned autoantibody positive before the age of 2 and later went on to develop T1D) and 19 matched controls at months 0, 3, and 6 before the onset of islet autoimmunity (241). Viruses made up to 2% of all identified sequences, with a major predominance of bacteriophages which were identified in 52 of the 96 samples (54.2%). Surprisingly, only 10.4% of samples contained one or more human viruses including parechovirus, bocavirus, annelovirus, enterovirus, and/or sapovirus. Kramna and colleagues also found no association between the gut virome and islet autoimmunity (241).
A similar study conducted in 2017 (242) examined stool samples from 11 children who developed serum autoantibodies associated with T1D (of whom, five developed T1D). In contrast with Kramna et al. (241), this study detected a significantly lower abundance of Circoviridae-related sequences (the group of viruses includes circoviruses) and lower virome diversity in T1D cases. Most importantly, the researchers found that multiple lines of evidence suggest that over time, the developmental progression of viromes significantly differed between children who developed autoimmunity and those who did not. The dynamic change in Shannon diversity over time was different between controls and T1D cases for Microviridae, Myoviridae, and Podoviridae. Additionally, the changes in richness over time differed between controls and cases for Microviridae, Myoviridae, and Podoviridae. The Random Forests analysis of age-discriminative contigs also suggested differences between cases and controls (242). This study reports virome changes occurred before seroconversion which suggests possible causality. Using virome capture sequencing methods, a more recent study characterized the gut virome of pregnant women with (n = 35) and without (n = 26) T1D throughout the Environmental Determinants of Islet Autoimmunity longitudinal study. Two viruses, picobirnavirus and tobamovirus, were more frequently observed in pregnant women with T1D, and the abundance of 77 viruses differed between the two maternal groups including 88 enterovirus B types (243).
Taken together, these data suggest that there are significant differences in the gut virome of T1D patients and progressors. However, caution should be taken in interpreting these results as very few studies with very small sample size have been conducted on this subject. As with any new avenue of interrogation, differences in methodology between studies may contribute to variation observed in the conclusions.
In vivo studies have demonstrated the importance of gut microbiota in the production of host metabolites (244–246). Of the metabolites known to be modulated and/or produced by gut microbiota, short chain fatty acids in particular have been shown to play a significant role in many aspects of general homeostasis. When dietary fiber is consumed, it is digested and subsequently fermented in the colon by gut microbiota into short chains fatty acids (SCFAs), such as acetate, propionate, and butyrate (246). Diets high in fiber alter gut microbiota composition by facilitating the growth of SCFA-producing species. Furthermore, a 4-fold increase in SCFAs has been observed in breast-fed infants compared to their formula-fed counterparts (247).
Diets high in fiber have also been shown to reduce systemic inflammation. A study by Trompette et al. (248) demonstrated that mice fed a diet high in fiber had higher levels of SCFAs and were protected against allergic inflammation of the lung (248). In humans, a decrease in dietary fiber has been directly connected to a decrease in the SCFA butyrate (249). Of the SCFAs, butyrate is thought to be particularly important. Amongst its multiple roles, it improves glucose and energy homeostasis, is an energy substrate for colonic epithelium and suppresses colonic inflammation and carcinogenesis. Butyrate also plays an essential role in modulating the immune response by inducing differentiation of regulatory T cells (Tregs), particularly Fox3p+ Tregs, and inhibiting inflammatory cytokines, such as IFN-γ (26, 246, 250–253). Interestingly, when NOD mice were fed a diet designed to increase acetate and butyrate levels they experienced a decrease in T1D incidence when compared to controls (254).
In the longitudinal studies TRIGR and FINDIA, SCFA-producing species including Bifidobacterium adolescentis, Roseburia faecis, and Faecalibacterium prausnitzii were negatively correlated with the number of autoantibodies detected (36). Thus, children positive for three or more antibodies had the lowest levels of SCFA-producing species. In agreement with this, the TEDDY study observed a higher prevalence of microbial genes associated with fermentation and SCFA biosynthesis in healthy controls. Interestingly, the taxon associated with these functions were not consistent across geographic regions suggesting that the function, rather than the taxa, is conserved amongst healthy children (19). Together, these results indicate that species capable of producing butyrate are potentially protective against autoantibody formation in at-risk children; more specifically, various Clostridium clusters capable of forming butyrate from acetate alongside Bifidobacterium species which can form butyrate via lactate metabolism (Figure 1).
Mirroring the results derived from animal models, human studies have demonstrated a metabolic shift in patients preceding autoantibody development, insulitis, and/or T1D. Sardinia has the second highest T1D incidence rate just below Finland, making it a good candidate for T1D studies (255). In one Sardinian-based study, Culeddu et al. (256) discovered that gut microbial metabolic products, such as p-cresol sulfate and phenylacetylglycine were involved in the classification of T1D patients294. Using data acquired through the Finland-based DIPP longitudinal study, serum metabolomic profiles of children who progressed to T1D were compared to controls that did not have autoantibodies and/or had not developed T1D. For individuals who progressed to T1D, reduced levels of succinic acid and phosphatidylcholine (PC) were observed at birth while reduced levels of triglycerides and antioxidant ether phospholipids were observed in follow-up serum samples. Increased levels of pro-inflammatory lysoPCs, reduced levels of ketoleucine and increased levels of glutamic acid were also amongst the alterations detected months before the documented seroconversion to autoantibody positivity (257). Interestingly, metabolic profiles partially normalized for T1D patients after seroconversion. This implies that autoimmunity is a later manifestation of an earlier alteration which may include disturbances in metabolic function (257).
As demonstrated, extensive research has been done to gain a complete understanding of T1D etiology, onset and progression. What's largely apparent in this body of work is that T1D is a multi-faceted disease with no one “smoking gun”; no one simple answer. Furthermore, the methods used to investigate this complex disease may influence the outcomes observed. One major distinction are the conclusions gained from human studies compared to animal model studies. Although animal model studies have provided in-depth knowledge on the specific mechanisms at play, significant differences intrinsic to mouse/rat models force one to take caution when attempting to interpret findings to human health applications. For one, histological work has shown distinct differences in both the physiology of the pancreas in mouse models and the pathophysiological progression of T1D within the β-cell islets (258, 259). Moreover, epidemiological data on T1D demonstrates a slight male predominance of incidence rate before puberty, however, the incidence is twice as high in males compared to females between the ages of 15–39 (260–262). In mice, female NOD mice develop T1D at a much higher frequency than their male counterparts. One study showed that a number of Bacteroides genus correlations were sex-dependent. One specific example is B. ovatis, which was found to be positively correlated with autoantibody-positive male children while this same species was negatively correlated with female autoantibody-positive subjects (36). Thus, further study into these seemingly elusive differences is needed in order to determine if sex differences are indeed driven by intrinsic differences in gut microbiota. It is important to note that researchers have identified around 500 different therapeutics to prevent or reverse T1D in NOD mice model, none of these treatments have successfully translated to an effective tool for human T1D (263, 264). The differences between NOD mice model and human data indicate that it is time to think about alternative animal models for the T1D research and focus on human studies.
The advancement of sequencing technologies has opened up entirely novel avenues for microbiota exploration; an area of research that is just gaining its foothold in biomedical research. Thanks to these advancements, researchers have been able to explore the role of microbiota as it pertains to a number of chronic life-threatening diseases, including T1D. On the other hand, we are living in a bacteria-centric microbiota approach. As summarized in section T1D and the Gut Microbiota: Looking Beyond Bacterial Species, there are very few virome, proteome and metabolomics studies. More strikingly, we are not aware of any T1D study focusing on the phageome (265), fungal microbiome (266) and archeome (267), and metatranscriptome (268). Gut microbiota and its products are still a “dark matter” and our knowledge is quite limited.
Another limitation is being gut-centric in the microbiota studies. We are not aware of any nasal (269), skin (270) or vaginal microbiota studies (271) in the T1D field. These are other important mucosal surfaces for microbiota-host interaction and might have a role in the autoimmune diseases including T1D.
It is important to note that there is no causal relation identified yet with any human disease and microbiota. Instead of large screening studies, a parallel approach might be to focus on hypotheses based on specific mechanisms. We and others have demonstrated the importance of microbial interactions in shaping microbial communities through both contact-dependent and chemically-mediated mechanisms (272). Recent work also suggests that community function, rather than specific taxonomic composition, may play a more integral role in disease development (19). Taken together, perhaps future efforts should look further into specific community interactions and overall function to gain a better understanding of the varying disease etiologies. For instance, we recently hypothesized that the presence of microbes carrying an insulin-like peptide could instigate a molecular mimicry mechanism in T1D and have since discovered viral insulins (273, 274). We also showed the presence of the sequences of these viruses in human fecal and plasma samples.
To conclude, the available data suggest that there is a correlation between T1D onset and gut microbiota but we are far away to claim a causal link. Going forward, mechanism-based investigations beyond taxonomic characterization has potential to be the key in elucidating the mechanisms at play between gut microbiota, the immune system and T1D pathogenesis.
SD contributed intellectually to the conceptualization, literature review, writing and editing of the review, contributed to all sections of the review including tables and the figure. BS contributed intellectually to the conceptualization, literature review, writing and editing of the longitudinal studies along with the tables, figure, and proteomics section. QH contributed to the introduction. CB, TY, CC, CR, and MF contributed to the environmental factors sections. JL contributed intellectually to the review, providing feedback and edits along with insight into the field of T1D research. BM contributed intellectually to the review, providing feedback and edits. EA provided supervision and contributed intellectually to the conceptualization, writing and editing of the review.
This work was supported by NIH NIDDK 1K01DK117967-01 and G. Harold & Leila Y. Mathers Foundation grants to EA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2020.00078/full#supplementary-material
2. Gordo I. Evolutionary change in the human gut microbiome: From a static to a dynamic view. PLoS Biol. (2019) 17:e3000126. doi: 10.1371/journal.pbio.3000126
3. Miller FW, Pollard KM, Parks CG, Germolec DR, Leung PS, Selmi C, et al. Criteria for environmentally associated autoimmune diseases. J Autoimmun. (2012) 39:253–8. doi: 10.1016/j.jaut.2012.05.001
4. Ramos-Casals M, Brito-Zeron P, Kostov B, Siso-Almirall A, Bosch X, Buss D, et al. Google-driven search for big data in autoimmune geoepidemiology: analysis of 394,827 patients with systemic autoimmune diseases. Autoimmun Rev. (2015) 14:670–9. doi: 10.1016/j.autrev.2015.03.008
5. De Luca F, Shoenfeld Y. The microbiome in autoimmune diseases. Clin Exp Immunol. (2019) 195:74–85. doi: 10.1111/cei.13158
6. Marietta E, Horwath I, Balakrishnan B, Taneja V. Role of the intestinal microbiome in autoimmune diseases and its use in treatments. Cell Immunol. (2019) 339:50–8. doi: 10.1016/j.cellimm.2018.10.005
7. Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. (2017) 550:61–6. doi: 10.1038/nature23889
8. Maruvada P, Leone V, Kaplan LM, Chang EB. The human microbiome and obesity: moving beyond associations. Cell Host Microbe. (2017) 22:589–99. doi: 10.1016/j.chom.2017.10.005
9. Young VB. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ. (2017) 356:j831. doi: 10.1136/bmj.j831
10. Rinaldi M, Perricone R, Blank M, Perricone C, Shoenfeld Y. Anti-Saccharomyces cerevisiae autoantibodies in autoimmune diseases: from bread baking to autoimmunity. Clin Rev Allergy Immunol. (2013) 45:152–61. doi: 10.1007/s12016-012-8344-9
11. Lamps LW, Madhusudhan KT, Havens JM, Greenson JK, Bronner MP, Chiles MC, et al. Pathogenic Yersinia DNA is detected in bowel and mesenteric lymph nodes from patients with Crohn's disease. Am J Surg Pathol. (2003) 27:220–7. doi: 10.1097/00000478-200302000-00011
12. Chassaing B, Rolhion N, De Vallee A, Salim SY, Prorok-Hamon M, Neut C, et al. Crohn disease–associated adherent-invasive E. coli bacteria target mouse and human Peyer's patches via long polar fimbriae. J Clin Invest. (2011) 121:966–75. doi: 10.1172/JCI44632
13. Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. (2014) 15:382–92. doi: 10.1016/j.chom.2014.02.005
14. Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. (2015) 26:26191. doi: 10.3402/mehd.v26.26191
15. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. (2007) 104:13780–5. doi: 10.1073/pnas.0706625104
16. Navaneethan U, Venkatesh PG, Shen B. Clostridium difficile infection and inflammatory bowel disease: understanding the evolving relationship. World J Gastroenterol. (2010) 16:4892–904. doi: 10.3748/wjg.v16.i39.4892
17. Quagliariello A, Aloisio I, Bozzi Cionci N, Luiselli D, D'auria G, Martinez-Priego L, et al. Effect of bifidobacterium breve on the intestinal microbiota of coeliac children on a gluten free diet: a pilot study. Nutrients. (2016) 8:660. doi: 10.3390/nu8100660
18. Heintz-Buschart A, May P, Laczny CC, Lebrun LA, Bellora C, Krishna A, et al. Erratum: integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes. Nat Microbiol. (2016) 2:16227. doi: 10.1038/nmicrobiol.2016.227
19. Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. (2018) 562:589–94. doi: 10.1038/s41586-018-0620-2
20. Atarashi K, Honda K. Microbiota in autoimmunity and tolerance. Curr Opin Immunol. (2011) 23:761–8. doi: 10.1016/j.coi.2011.11.002
21. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. (2016) 352:539–44. doi: 10.1126/science.aad9378
22. Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci USA. (2011) 108:11548–53. doi: 10.1073/pnas.1108924108
23. Russell JT, Roesch LFW, Ordberg M, Ilonen J, Atkinson MA, Schatz DA, et al. Genetic risk for autoimmunity is associated with distinct changes in the human gut microbiome. Nat Commun. (2019) 10:3621. doi: 10.1038/s41467-019-11460-x
24. Wu HJ, Ivanov Ii, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. (2010) 32:815–27. doi: 10.1016/j.immuni.2010.06.001
25. Di Giacinto C, Marinaro M, Sanchez M, Strober W, Boirivant M. Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGF-beta-bearing regulatory cells. J Immunol. (2005) 174:3237–46. doi: 10.4049/jimmunol.174.6.3237
26. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. (2011) 331:337–41. doi: 10.1126/science.1198469
27. Rai E, Wakeland EK. Genetic predisposition to autoimmunity–what have we learned? Semin Immunol. (2011) 23:67–83. doi: 10.1016/j.smim.2011.01.015
28. Ceccarelli F, Agmon-Levin N, Perricone C. Genetic factors of autoimmune diseases 2017. J Immunol Res. (2017) 2017:2789242. doi: 10.1155/2017/2789242
29. Vojdani A, Pollard KM, Campbell AW. Environmental triggers and autoimmunity. Autoimmune Dis. (2014) 2014:798029. doi: 10.1155/2014/798029
30. Jorg S, Grohme DA, Erzler M, Binsfeld M, Haghikia A, Muller DN, et al. Environmental factors in autoimmune diseases and their role in multiple sclerosis. Cell Mol Life Sci. (2016) 73:4611–22. doi: 10.1007/s00018-016-2311-1
31. Agmon-Levin N, Ram M, Barzilai O, Porat-Katz BS, Parikman R, Selmi C, et al. Prevalence of hepatitis C serum antibody in autoimmune diseases. J Autoimmun. (2009) 32:261–6. doi: 10.1016/j.jaut.2009.02.017
32. Getts DR, Chastain EM, Terry RL, Miller SD. Virus infection, antiviral immunity, and autoimmunity. Immunol Rev. (2013) 255:197–209. doi: 10.1111/imr.12091
33. Vojdani A. A potential link between environmental triggers and autoimmunity. Autoimmune Dis. (2014) 2014:437231. doi: 10.1155/2014/437231
34. Vanderlugt CJ, Miller SD. Epitope spreading. Curr Opin Immunol. (1996) 8:831–6. doi: 10.1016/S0952-7915(96)80012-4
35. Guilherme L, Kalil J, Cunningham M. Molecular mimicry in the autoimmune pathogenesis of rheumatic heart disease. Autoimmunity. (2006) 39:31–9. doi: 10.1080/08916930500484674
36. De Goffau MC, Luopajarvi K, Knip M, Ilonen J, Ruohtula T, Harkonen T, et al. Fecal microbiota composition differs between children with beta-cell autoimmunity and those without. Diabetes. (2013) 62:1238–44. doi: 10.2337/db12-0526
37. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. (2014) 157:121–41. doi: 10.1016/j.cell.2014.03.011
38. De Goffau MC, Fuentes S, Van Den Bogert B, Honkanen H, De Vos WM, Welling GW, et al. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia. (2014) 57:1569–77. doi: 10.1007/s00125-014-3274-0
39. Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. (2016) 8:43. doi: 10.1186/s13073-016-0299-7
40. Jangi S, Gandhi R, Cox LM, Li N, Von Glehn F, Yan R, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. (2016) 7:12015. doi: 10.1038/ncomms12015
41. De Groot PF, Belzer C, Aydin O, Levin E, Levels JH, Aalvink S, et al. Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLoS ONE. (2017) 12:e0188475. doi: 10.1371/journal.pone.0188475
42. Maahs DM, West NA, Lawrence JM, Mayer-Davis EJ. Epidemiology of type 1 diabetes. Endocrinol Metab Clin North Am. (2010) 39:481–97. doi: 10.1016/j.ecl.2010.05.011
43. Patterson CC, Dahlquist GG, Gyurus E, Green A, Soltesz G, Group ES. Incidence trends for childhood type 1 diabetes in Europe during 1989-2003 and predicted new cases 2005-20: a multicentre prospective registration study. Lancet. (2009) 373:2027–33. doi: 10.1016/S0140-6736(09)60568-7
44. Jin JS, Touyama M, Yamada S, Yamazaki T, Benno Y. Alteration of a human intestinal microbiota under extreme life environment in the Antarctica. Biol Pharm Bull. (2014) 37:1899–906. doi: 10.1248/bpb.b14-00397
45. Weets I, Kaufman L, Van Der Auwera B, Crenier L, Rooman RP, De Block C, et al. Seasonality in clinical onset of type 1 diabetes in belgian patients above the age of 10 is restricted to HLA-DQ2/DQ8-negative males, which explains the male to female excess in incidence. Diabetologia. (2004) 47:614–21. doi: 10.1007/s00125-004-1369-8
46. Soderstrom U, Aman J, Hjern A. Being born in Sweden increases the risk for type 1 diabetes - a study of migration of children to Sweden as a natural experiment. Acta Paediatr. (2012) 101:73–7. doi: 10.1111/j.1651-2227.2011.02410.x
47. Voigt RM, Forsyth CB, Green SJ, Mutlu E, Engen P, Vitaterna MH, et al. Circadian disorganization alters intestinal microbiota. PLoS ONE. (2014) 9:e97500. doi: 10.1371/journal.pone.0097500
48. Liang X, Bushman FD, Fitzgerald GA. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci USA. (2015) 112:10479–84. doi: 10.1073/pnas.1501305112
49. Voigt RM, Forsyth CB, Green SJ, Engen PA, Keshavarzian A. Circadian rhythm and the gut microbiome. Int Rev Neurobiol. (2016) 131:193–205. doi: 10.1016/bs.irn.2016.07.002
50. Marre ML, James EA, Piganelli JD. beta cell ER stress and the implications for immunogenicity in type 1 diabetes. Front Cell Dev Biol. (2015) 3:67. doi: 10.3389/fcell.2015.00067
51. Atkinson M, Gale EA. Infant diets and type 1 diabetes: too early, too late, or just too complicated? JAMA. (2003) 290:1771–2. doi: 10.1001/jama.290.13.1771
52. Mannering SI, Di Carluccio AR, Elso CM. Neoepitopes: a new take on beta cell autoimmunity in type 1 diabetes. Diabetologia. (2019) 62:351–6. doi: 10.1007/s00125-018-4760-6
53. Purcell AW, Sechi S, Dilorenzo TP. The evolving landscape of autoantigen discovery and characterization in type 1 diabetes. Diabetes. (2019) 68:879–86. doi: 10.2337/dbi18-0066
54. Delong T, Wiles TA, Baker RL, Bradley B, Barbour G, Reisdorph R, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. (2016) 351:711–4. doi: 10.1126/science.aad2791
55. Duncan GE. Prevalence of diabetes and impaired fasting glucose levels among US adolescents: National Health and Nutrition Examination Survey, 1999-2002. Arch Pediatr Adolesc Med. (2006) 160:523–8. doi: 10.1001/archpedi.160.5.523
56. Group SFDIYS, Liese AD, D'agostino RB Jr., Hamman RF, Kilgo PD, Lawrence JM, et al. The burden of diabetes mellitus among US youth: prevalence estimates from the SEARCH for Diabetes in Youth Study. Pediatrics. (2006) 118:1510–8. doi: 10.1542/peds.2006-0690
57. Bingley PJ, Gale EA. Rising incidence of IDDM in Europe. Diabetes Care. (1989) 12:289–95. doi: 10.2337/diacare.12.4.289
58. Gale EA. The rise of childhood type 1 diabetes in the 20th century. Diabetes. (2002) 51:3353–61. doi: 10.2337/diabetes.51.12.3353
59. Ludvigsson J. Increasing incidence but decreasing awareness of Type 1 diabetes in Sweden. Diabetes Care. (2017) 40:e143–4. doi: 10.2337/dc17-1175
60. Podar T, Solntsev A, Karvonen M, Padaiga Z, Brigis G, Urbonaite B, et al. Increasing incidence of childhood-onset type I diabetes in 3 Baltic countries and Finland 1983-1998. Diabetologia. (2001) 44 (Suppl. 3):B17–20. doi: 10.1007/PL00002947
61. Dabelea D. The accelerating epidemic of childhood diabetes. Lancet. (2009) 373:1999–2000. doi: 10.1016/S0140-6736(09)60874-6
62. Barnett AH, Eff C, Leslie RD, and Pyke DA. Diabetes in identical twins. A study of 200 pairs. Diabetologia. (1981) 20:87–93. doi: 10.1007/BF00262007
63. Kaprio J, Tuomilehto J, Koskenvuo M, Romanov K, Reunanen A, Eriksson J, et al. Concordance for type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in Finland. Diabetologia. (1992) 35:1060–7. doi: 10.1007/BF02221682
64. Redondo MJ, Jeffrey J, Fain PR, Eisenbarth GS, Orban T. Concordance for islet autoimmunity among monozygotic twins. N Engl J Med. (2008) 359:2849–50. doi: 10.1056/NEJMc0805398
65. Thayer TC, Wilson SB, Mathews CE. Use of nonobese diabetic mice to understand human type 1 diabetes. Endocrinol Metab Clin North Am. (2010) 39:541–61. doi: 10.1016/j.ecl.2010.05.001
66. Wicker LS, Clark J, Fraser HI, Garner VE, Gonzalez-Munoz A, Healy B, et al. Type 1 diabetes genes and pathways shared by humans and NOD mice. J Autoimmun. (2005) 25 (Suppl):29–33. doi: 10.1016/j.jaut.2005.09.009
67. Pozzilli P, Signore A, Williams AJ, Beales PE. NOD mouse colonies around the world–recent facts and figures. Immunol Today. (1993) 14:193–6. doi: 10.1016/0167-5699(93)90160-M
68. Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. (2008) 455:1109–13. doi: 10.1038/nature07336
69. Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. (2013) 339:1084–8. doi: 10.1126/science.1233521
70. Onengut-Gumuscu S, Chen WM, Burren O, Cooper NJ, Quinlan AR, Mychaleckyj JC, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. (2015) 47:381–6. doi: 10.1038/ng.3245
71. She JX. Susceptibility to type I diabetes: HLA-DQ and DR revisited. Immunol Today. (1996) 17:323–9. doi: 10.1016/0167-5699(96)10014-1
72. Hagopian WA, Erlich H, Lernmark A, Rewers M, Ziegler AG, Simell O, et al. The Environmental Determinants of Diabetes in the Young (TEDDY): genetic criteria and international diabetes risk screening of 421 000 infants. Pediatr Diabetes. (2011) 12:733–43. doi: 10.1111/j.1399-5448.2011.00774.x
73. Rewers M, Ludvigsson J. Environmental risk factors for type 1 diabetes. Lancet. (2016) 387:2340–8. doi: 10.1016/S0140-6736(16)30507-4
74. Stewart CJ, Ajami NJ, O'brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. (2018) 562:583–8. doi: 10.1038/s41586-018-0617-x
75. Kostic AD, Gevers D, Siljander H, Vatanen T, Hyotylainen T, Hamalainen AM, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. (2015) 17:260–73. doi: 10.1016/j.chom.2015.01.001
76. Vatanen T, Kostic AD, D'hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. (2016) 165:842–53. doi: 10.1016/j.cell.2016.04.007
77. Vaarala O, Ilonen J, Ruohtula T, Pesola J, Virtanen SM, Harkonen T, et al. Removal of bovine insulin from cow's milk formula and early initiation of beta-cell autoimmunity in the FINDIA pilot study. Arch Pediatr Adolesc Med. (2012) 166:608–14. doi: 10.1001/archpediatrics.2011.1559
78. Kondrashova A, Reunanen A, Romanov A, Karvonen A, Viskari H, Vesikari T, et al. A six-fold gradient in the incidence of type 1 diabetes at the eastern border of Finland. Ann Med. (2005) 37:67–72. doi: 10.1080/07853890410018952
79. Gale EA. A missing link in the hygiene hypothesis? Diabetologia. (2002) 45:588–94. doi: 10.1007/s00125-002-0801-1
80. Bach JF. Six questions about the hygiene hypothesis. Cell Immunol. (2005) 233:158–61. doi: 10.1016/j.cellimm.2005.04.006
81. D'angeli MA, Merzon E, Valbuena LF, Tirschwell D, Paris CA, Mueller BA. Environmental factors associated with childhood-onset type 1 diabetes mellitus: an exploration of the hygiene and overload hypotheses. Arch Pediatr Adolesc Med. (2010) 164:732–8. doi: 10.1001/archpediatrics.2010.115
82. Britton RA, Young VB. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology. (2014) 146:1547–53. doi: 10.1053/j.gastro.2014.01.059
83. Akerman L, Ludvigsson J, Swartling U, Casas R. Characteristics of the pre-diabetic period in children with high risk of type 1 diabetes recruited from the general Swedish population-The ABIS study. Diabetes Metab Res Rev. (2017) 33. doi: 10.1002/dmrr.2900
84. Silverman M, Kua L, Tanca A, Pala M, Palomba A, Tanes C, et al. Protective major histocompatibility complex allele prevents type 1 diabetes by shaping the intestinal microbiota early in ontogeny. Proc Natl Acad Sci USA. (2017) 114:9671–6. doi: 10.1073/pnas.1712280114
85. Kubinak JL, Stephens WZ, Soto R, Petersen C, Chiaro T, Gogokhia L, et al. MHC variation sculpts individualized microbial communities that control susceptibility to enteric infection. Nat Commun. (2015) 6:8642. doi: 10.1038/ncomms9642
86. Toivanen P, Vaahtovuo J, Eerola E. Influence of major histocompatibility complex on bacterial composition of fecal flora. Infect Immun. (2001) 69:2372–7. doi: 10.1128/IAI.69.4.2372-2377.2001
87. Faresjo AO, Ludvigsson J. Pet exposure in the family during pregnancy and risk for type 1 diabetes-The prospective ABIS study. Pediatr Diabetes. (2018) 19:1206–10. doi: 10.1111/pedi.12721
88. Tun HM, Konya T, Takaro TK, Brook JR, Chari R, Field CJ, et al. Exposure to household furry pets influences the gut microbiota of infant at 3-4 months following various birth scenarios. Microbiome. (2017) 5:40. doi: 10.1186/s40168-017-0254-x
89. Walker WA. The importance of appropriate initial bacterial colonization of the intestine in newborn, child, and adult health. Pediatr Res. (2017) 82:387–95. doi: 10.1038/pr.2017.111
90. Blaser MJ, Dominguez-Bello MG. The human microbiome before birth. Cell Host Microbe. (2016) 20:558–60. doi: 10.1016/j.chom.2016.10.014
91. Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Backhed HK, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. (2012) 150:470–80. doi: 10.1016/j.cell.2012.07.008
92. Gronlund MM, Lehtonen OP, Eerola E, Kero P. Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J Pediatr Gastroenterol Nutr. (1999) 28:19–25. doi: 10.1097/00005176-199901000-00007
93. Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. (1999) 69:1035S−45S. doi: 10.1093/ajcn/69.5.1035s
94. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. (2006) 118:511–21. doi: 10.1542/peds.2005-2824
95. Biasucci G, Benenati B, Morelli L, Bessi E, Boehm G. Cesarean delivery may affect the early biodiversity of intestinal bacteria. J Nutr. (2008) 138:1796S−1800S. doi: 10.1093/jn/138.9.1796S
96. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. (2010) 107:11971–5. doi: 10.1073/pnas.1002601107
97. Pistiner M, Gold DR, Abdulkerim H, Hoffman E, Celedon JC. Birth by cesarean section, allergic rhinitis, and allergic sensitization among children with a parental history of atopy. J Allergy Clin Immunol. (2008) 122:274–9. doi: 10.1016/j.jaci.2008.05.007
98. Thavagnanam S, Fleming J, Bromley A, Shields MD, Cardwell CR. A meta-analysis of the association between Caesarean section and childhood asthma. Clin Exp Allergy. (2008) 38:629–33. doi: 10.1111/j.1365-2222.2007.02780.x
99. Almqvist C, Cnattingius S, Lichtenstein P, Lundholm C. The impact of birth mode of delivery on childhood asthma and allergic diseases–a sibling study. Clin Exp Allergy. (2012) 42:1369–76. doi: 10.1111/j.1365-2222.2012.04021.x
100. Huh SY, Rifas-Shiman SL, Zera CA, Edwards JW, Oken E, Weiss ST, et al. Delivery by caesarean section and risk of obesity in preschool age children: a prospective cohort study. Arch Dis Child. (2012) 97:610–6. doi: 10.1136/archdischild-2011-301141
101. Penders J, Gerhold K, Thijs C, Zimmermann K, Wahn U, Lau S, et al. New insights into the hygiene hypothesis in allergic diseases: mediation of sibling and birth mode effects by the gut microbiota. Gut Microbes. (2014) 5:239–44. doi: 10.4161/gmic.27905
102. Kuhle S, Tong OS, Woolcott CG. Association between caesarean section and childhood obesity: a systematic review and meta-analysis. Obes Rev. (2015) 16:295–303. doi: 10.1111/obr.12267
103. Sevelsted A, Stokholm J, Bonnelykke K, Bisgaard H. Cesarean section and chronic immune disorders. Pediatrics. (2015) 135:e92–8. doi: 10.1542/peds.2014-0596
104. Yuan C, Gaskins AJ, Blaine AI, Zhang C, Gillman MW, Missmer SA, et al. Association between cesarean birth and risk of obesity in offspring in childhood, adolescence, and early adulthood. JAMA Pediatr. (2016) 170:e162385. doi: 10.1001/jamapediatrics.2016.2385
105. Tun HM, Bridgman SL, Chari R, Field CJ, Guttman DS, Becker AB, et al. Roles of birth mode and infant gut microbiota in intergenerational transmission of overweight and obesity from mother to offspring. JAMA Pediatr. (2018) 172:368–77. doi: 10.1001/jamapediatrics.2017.5535
106. Adlerberth I, Lindberg E, Aberg N, Hesselmar B, Saalman R, Strannegard IL, et al. Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res. (2006) 59:96–101. doi: 10.1203/01.pdr.0000191137.12774.b2
107. Rutayisire E, Huang K, Liu Y, Tao F. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants' life: a systematic review. BMC Gastroenterol. (2016) 16:86. doi: 10.1186/s12876-016-0498-0
108. Mitsou EK, Kirtzalidou E, Oikonomou I, Liosis G, Kyriacou A. Fecal microflora of Greek healthy neonates. Anaerobe. (2008) 14:94–101. doi: 10.1016/j.anaerobe.2007.11.002
109. Kabeerdoss J, Ferdous S, Balamurugan R, Mechenro J, Vidya R, Santhanam S, et al. Development of the gut microbiota in southern Indian infants from birth to 6 months: a molecular analysis. J Nutr Sci. (2013) 2:e18. doi: 10.1017/jns.2013.6
110. Hesla HM, Stenius F, Jaderlund L, Nelson R, Engstrand L, Alm J, et al. Impact of lifestyle on the gut microbiota of healthy infants and their mothers-the ALADDIN birth cohort. FEMS Microbiol Ecol. (2014) 90:791–801. doi: 10.1111/1574-6941.12434
111. Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. (2014) 63:559–66. doi: 10.1136/gutjnl-2012-303249
112. Dogra S, Sakwinska O, Soh SE, Ngom-Bru C, Bruck WM, Berger B, et al. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. MBio. (2015) 6:e02419-14. doi: 10.1128/mBio.02419-14
113. Cardwell CR, Stene LC, Joner G, Cinek O, Svensson J, Goldacre MJ, et al. Caesarean section is associated with an increased risk of childhood-onset type 1 diabetes mellitus: a meta-analysis of observational studies. Diabetologia. (2008) 51:726–35. doi: 10.1007/s00125-008-0941-z
114. Algert CS, Mcelduff A, Morris JM, Roberts CL. Perinatal risk factors for early onset of Type 1 diabetes in a 2000-2005 birth cohort. Diabet Med. (2009) 26:1193–7. doi: 10.1111/j.1464-5491.2009.02878.x
115. Black M, Bhattacharya S, Philip S, Norman JE, Mclernon DJ. Planned cesarean delivery at term and adverse outcomes in childhood health. JAMA. (2015) 314:2271–9. doi: 10.1001/jama.2015.16176
116. Magne F, Puchi Silva A, Carvajal B, Gotteland M. The elevated rate of cesarean section and its contribution to non-communicable chronic diseases in Latin America: the growing involvement of the microbiota. Front Pediatr. (2017) 5:192. doi: 10.3389/fped.2017.00192
117. Samuelsson U, Lindell N, Bladh M, Akesson K, Carlsson A, Josefsson A. Caesarean section per se does not increase the risk of offspring developing type 1 diabetes: a Swedish population-based study. Diabetologia. (2015) 58:2517–24. doi: 10.1007/s00125-015-3716-3
118. Clausen TD, Bergholt T, Eriksson F, Rasmussen S, Keiding N, Lokkegaard EC. prelabor cesarean section and risk of childhood type 1 diabetes: a nationwide register-based cohort study. Epidemiology. (2016) 27:547–55. doi: 10.1097/EDE.0000000000000488
119. Hansen CH, Andersen LS, Krych L, Metzdorff SB, Hasselby JP, Skov S, et al. Mode of delivery shapes gut colonization pattern and modulates regulatory immunity in mice. J Immunol. (2014) 193:1213–22. doi: 10.4049/jimmunol.1400085
120. He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. (2007) 26:812–26. doi: 10.1016/j.immuni.2007.04.014
121. Lopez-Boado YS, Wilson CL, Hooper LV, Gordon JI, Hultgren SJ, Parks WC. Bacterial exposure induces and activates matrilysin in mucosal epithelial cells. J Cell Biol. (2000) 148:1305–15. doi: 10.1083/jcb.148.6.1305
122. Rhee KJ, Sethupathi P, Driks A, Lanning DK, Knight KL. Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire. J Immunol. (2004) 172:1118–24. doi: 10.4049/jimmunol.172.2.1118
123. Lutgendorff F, Akkermans LM, Soderholm JD. The role of microbiota and probiotics in stress-induced gastro-intestinal damage. Curr Mol Med. (2008) 8:282–98. doi: 10.2174/156652408784533779
124. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. (2008) 453:620–5. doi: 10.1038/nature07008
125. Nejentsev S, Sjoroos M, Soukka T, Knip M, Simell O, Lovgren T, et al. Population-based genetic screening for the estimation of Type 1 diabetes mellitus risk in Finland: selective genotyping of markers in the HLA-DQB1, HLA-DQA1 and HLA-DRB1 loci. Diabet Med. (1999) 16:985–92. doi: 10.1046/j.1464-5491.1999.00186.x
126. Kupila A, Muona P, Simell T, Arvilommi P, Savolainen H, Hamalainen AM, et al. Feasibility of genetic and immunological prediction of type I diabetes in a population-based birth cohort. Diabetologia. (2001) 44:290–7. doi: 10.1007/s001250051616
127. Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. (2011) 5:82–91. doi: 10.1038/ismej.2010.92
128. Davis-Richardson AG, Ardissone AN, Dias R, Simell V, Leonard MT, Kemppainen KM, et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front Microbiol. (2014) 5:678. doi: 10.3389/fmicb.2014.00678
129. Stene LC, Magnus P, Lie RT, Sovik O, Joner G, Norwegian Childhood Diabetes Study G. No association between preeclampsia or cesarean section and incidence of type 1 diabetes among children: a large, population-based cohort study. Pediatr Res. (2003) 54:487–90. doi: 10.1203/01.PDR.0000081301.25600.5D
130. Knol J, Scholtens P, Kafka C, Steenbakkers J, Gro S, Helm K, et al. Colon microflora in infants fed formula with galacto- and fructo-oligosaccharides: more like breast-fed infants. J Pediatr Gastroenterol Nutr. (2005) 40:36–42. doi: 10.1097/00005176-200501000-00007
131. Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. (2007) 5:e177. doi: 10.1371/journal.pbio.0050177
132. Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG. The infant microbiome development: mom matters. Trends Mol Med. (2015) 21:109–17. doi: 10.1016/j.molmed.2014.12.002
133. Lopez NM, Ponce S, Piccinini D, Perez E, Roberti M. From hospital to home care: creating a domotic environment for elderly and disabled people. IEEE Pulse. (2016) 7:38–41. doi: 10.1109/MPUL.2016.2539105
134. Gale C, Logan KM, Santhakumaran S, Parkinson JR, Hyde MJ, Modi N. Effect of breastfeeding compared with formula feeding on infant body composition: a systematic review and meta-analysis. Am J Clin Nutr. (2012) 95:656–69. doi: 10.3945/ajcn.111.027284
135. Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. (2017) 171:647–54. doi: 10.1001/jamapediatrics.2017.0378
136. Martin R, Heilig GH, Zoetendal EG, Smidt H, Rodriguez JM. Diversity of the Lactobacillus group in breast milk and vagina of healthy women and potential role in the colonization of the infant gut. J Appl Microbiol. (2007) 103:2638–44. doi: 10.1111/j.1365-2672.2007.03497.x
137. Solis G, De Los Reyes-Gavilan CG, Fernandez N, Margolles A, Gueimonde M. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe. (2010) 16:307–10. doi: 10.1016/j.anaerobe.2010.02.004
138. Duffy LC, Zielezny MA, Riepenhoff-Talty M, Dryja D, Sayahtaheri-Altaie S, Griffiths E, et al. Effectiveness of Bifidobacterium bifidum in mediating the clinical course of murine rotavirus diarrhea. Pediatr Res. (1994) 35:690–5. doi: 10.1203/00006450-199406000-00014
139. Gibson GR, Wang, X. Regulatory effects of bifidobacteria on the growth of other colonic bacteria. J Appl Bacteriol. (1994) 77:412–20. doi: 10.1111/j.1365-2672.1994.tb03443.x
140. Perdigon G, Alvarez S, Rachid M, Aguero G, Gobbato N. Immune system stimulation by probiotics. J Dairy Sci. (1995) 78:1597–606. doi: 10.3168/jds.S0022-0302(95)76784-4
141. Fujiwara S, Hashiba H, Hirota T, Forstner JF. Proteinaceous factor(s) in culture supernatant fluids of bifidobacteria which prevents the binding of enterotoxigenic Escherichia coli to gangliotetraosylceramide. Appl Environ Microbiol. (1997) 63:506–12. doi: 10.1128/AEM.63.2.506-512.1997
142. Sudo N, Sawamura S, Tanaka K, Aiba Y, Kubo C, Koga Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol. (1997) 159:1739–45.
143. Lievin V, Peiffer I, Hudault S, Rochat F, Brassart D, Neeser JR, et al. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut. (2000) 47:646–52. doi: 10.1136/gut.47.5.646
144. Picard C, Fioramonti J, Francois A, Robinson T, Neant F, Matuchansky C. Review article: bifidobacteria as probiotic agents – physiological effects and clinical benefits. Aliment Pharmacol Ther. (2005) 22:495–512. doi: 10.1111/j.1365-2036.2005.02615.x
145. Insel R, Knip M. Prospects for primary prevention of type 1 diabetes by restoring a disappearing microbe. Pediatr Diabetes. (2018) 19:1400–6. doi: 10.1111/pedi.12756
146. Schroeder BO, Birchenough GMH, Stahlman M, Arike L, Johansson MEV, Hansson GC, et al. Bifidobacteria or fiber protects against diet-induced microbiota-mediated colonic mucus deterioration. Cell Host Microbe. (2018) 23:27–40.e27. doi: 10.1016/j.chom.2017.11.004
147. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. (2012) 336:1268–73. doi: 10.1126/science.1223490
148. Gerstein HC. Cow's milk exposure and type I diabetes mellitus. A critical overview of the clinical literature. Diabetes Care. (1994) 17:13–9. doi: 10.2337/diacare.17.1.13
149. Alves JG, Figueiroa JN, Meneses J, Alves GV. Breastfeeding protects against type 1 diabetes mellitus: a case-sibling study. Breastfeed Med. (2012) 7:25–8. doi: 10.1089/bfm.2011.0009
150. Lundequist B, Nord CE, Winberg J. The composition of the faecal microflora in breastfed and bottle fed infants from birth to eight weeks. Acta Paediatr Scand. (1985) 74:45–51. doi: 10.1111/j.1651-2227.1985.tb10919.x
151. Fanaro S, Chierici R, Guerrini P, Vigi V. Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl. (2003) 91:48–55. doi: 10.1111/j.1651-2227.2003.tb00646.x
152. Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. (2013) 185:385–94. doi: 10.1503/cmaj.121189
153. Albenberg LG, Wu GD. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology. (2014) 146:1564–72. doi: 10.1053/j.gastro.2014.01.058
154. O'sullivan A, Farver M, Smilowitz JT. The influence of early infant-feeding practices on the intestinal microbiome and body composition in infants. Nutr Metab Insights. (2015) 8:1–9. doi: 10.4137/NMI.S41125
155. Madan JC, Hoen AG, Lundgren SN, Farzan SF, Cottingham KL, Morrison HG, et al. Association of cesarean delivery and formula supplementation with the intestinal microbiome of 6-week-old infants. JAMA Pediatr. (2016) 170:212–9. doi: 10.1001/jamapediatrics.2015.3732
156. Lund-Blix NA, Stene LC, Rasmussen T, Torjesen PA, Andersen LF, Ronningen KS. Infant feeding in relation to islet autoimmunity and type 1 diabetes in genetically susceptible children: the MIDIA Study. Diabetes Care. (2015) 38:257–63. doi: 10.2337/dc14-1130
157. Sadauskaite-Kuehne V, Ludvigsson J, Padaiga Z, Jasinskiene E, Samuelsson U. Longer breastfeeding is an independent protective factor against development of type 1 diabetes mellitus in childhood. Diabetes Metab Res Rev. (2004) 20:150–7. doi: 10.1002/dmrr.425
158. Wahlberg J, Vaarala O, Ludvigsson J, Group AB-S. Dietary risk factors for the emergence of type 1 diabetes-related autoantibodies in 21/2 year-old Swedish children. Br J Nutr. (2006) 95:603–8. doi: 10.1079/BJN20051676
159. Holmberg H, Wahlberg J, Vaarala O, Ludvigsson J, Group AS. Short duration of breast-feeding as a risk-factor for beta-cell autoantibodies in 5-year-old children from the general population. Br J Nutr. (2007) 97:111–6. doi: 10.1017/S0007114507210189
160. Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. (2011) 10:507–14. doi: 10.1016/j.chom.2011.10.007
161. Vatanen T, Kostic AD, D'hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. (2016) 165:1551. doi: 10.1016/j.cell.2016.05.056
162. Mejía-Leon ME, Petrosino JF, Ajami NJ, Dominguez-Bello MG, De La Barca AM. Fecal microbiota imbalance in Mexican children with type 1 diabetes. Sci Rep. (2014) 4:3814. doi: 10.1038/srep03814
163. Qureshi ST, Lariviere L, Leveque G, Clermont S, Moore KJ, Gros P, et al. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4). J Exp Med. (1999) 189:615–25. doi: 10.1084/jem.189.4.615
164. Nomura F, Akashi S, Sakao Y, Sato S, Kawai T, Matsumoto M, et al. Cutting edge: endotoxin tolerance in mouse peritoneal macrophages correlates with down-regulation of surface toll-like receptor 4 expression. J Immunol. (2000) 164:3476–9. doi: 10.4049/jimmunol.164.7.3476
165. Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. (2008) 42:145–51. doi: 10.1016/j.cyto.2008.01.006
166. Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. (2009) 30:475–87. doi: 10.1016/j.it.2009.07.009
167. Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, et al. Pillars article: cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. (1999) 162:3749–52.
168. Sai P, Rivereau AS. Prevention of diabetes in the nonobese diabetic mouse by oral immunological treatments. Comparative efficiency of human insulin and two bacterial antigens, lipopolysacharide from Escherichia coli and glycoprotein extract from Klebsiella pneumoniae. Diabetes Metab. (1996) 22:341–8.
169. Gulden E, Ihira M, Ohashi A, Reinbeck AL, Freudenberg MA, Kolb H, et al. Toll-like receptor 4 deficiency accelerates the development of insulin-deficient diabetes in non-obese diabetic mice. PLoS ONE. (2013) 8:e75385. doi: 10.1371/journal.pone.0075385
170. Frese SA, Hutton AA, Contreras LN, Shaw CA, Palumbo MC, Casaburi G, et al. Persistence of supplemented bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere. (2017) 2:e00501-17. doi: 10.1128/mSphere.00501-17
171. Virtanen SM, Knip M. Nutritional risk predictors of beta cell autoimmunity and type 1 diabetes at a young age. Am J Clin Nutr. (2003) 78:1053–67. doi: 10.1093/ajcn/78.6.1053
172. Knip M, Virtanen SM, Seppa K, Ilonen J, Savilahti E, Vaarala O, et al. Dietary intervention in infancy and later signs of beta-cell autoimmunity. N Engl J Med. (2010) 363:1900–8. doi: 10.1056/NEJMoa1004809
173. Lampeter EF, Klinghammer A, Scherbaum WA, Heinze E, Haastert B, Giani G, et al. The Deutsche Nicotinamide Intervention Study: an attempt to prevent type 1 diabetes. DENIS Group Diabetes. (1998) 47:980–4. doi: 10.2337/diabetes.47.6.980
174. Kolb H, Burkart V. Nicotinamide in type 1 diabetes. Mechanism of action revisited. Diabetes Care. (1999) 22 (Suppl. 2):B16–20.
175. Ohly P, Dohle C, Abel J, Seissler J, Gleichmann H. Zinc sulphate induces metallothionein in pancreatic islets of mice and protects against diabetes induced by multiple low doses of streptozotocin. Diabetologia. (2000) 43:1020–30. doi: 10.1007/s001250050009
176. Ho E, Quan N, Tsai YH, Lai W, Bray TM. Dietary zinc supplementation inhibits NFkappaB activation and protects against chemically induced diabetes in CD1 mice. Exp Biol Med. (2001) 226:103–11. doi: 10.1177/153537020122600207
177. Hypponen E, Laara E, Reunanen A, Jarvelin MR, Virtanen SM. Intake of vitamin D and risk of type 1 diabetes: a birth-cohort study. Lancet. (2001) 358:1500–3. doi: 10.1016/S0140-6736(01)06580-1
178. Gale EA, Bingley PJ, Emmett CL, Collier T, European Nicotinamide Diabetes Intervention Trial G. European Nicotinamide Diabetes Intervention Trial (ENDIT): a randomised controlled trial of intervention before the onset of type 1 diabetes. Lancet. (2004) 363:925–31. doi: 10.1016/S0140-6736(04)15786-3
179. Kawasaki E, Abiru N, Eguchi K. Prevention of type 1 diabetes: from the view point of beta cell damage. Diabetes Res Clin Pract. (2004) 66 (Suppl. 1):S27–32. doi: 10.1016/j.diabres.2003.09.015
180. Luong K, Nguyen LT, Nguyen DN. The role of vitamin D in protecting type 1 diabetes mellitus. Diabetes Metab Res Rev. (2005) 21:338–46. doi: 10.1002/dmrr.557
181. Zipitis CS, Akobeng AK. Vitamin D supplementation in early childhood and risk of type 1 diabetes: a systematic review and meta-analysis. Arch Dis Child. (2008) 93:512–7. doi: 10.1136/adc.2007.128579
182. Cantorna MT. Vitamin D and autoimmunity: is vitamin D status an environmental factor affecting autoimmune disease prevalence? Proc Soc Exp Biol Med. (2000) 223:230–3. doi: 10.1046/j.1525-1373.2000.22333.x
183. Gregori S, Giarratana N, Smiroldo S, Uskokovic M, Adorini L. A 1alpha,25-dihydroxyvitamin D(3) analog enhances regulatory T-cells and arrests autoimmune diabetes in NOD mice. Diabetes. (2002) 51:1367–74. doi: 10.2337/diabetes.51.5.1367
184. Scott FW. Cow milk and insulin-dependent diabetes mellitus: is there a relationship? Am J Clin Nutr. (1990) 51:489–91. doi: 10.1093/ajcn/51.3.489
185. Verge CF, Howard NJ, Irwig L, Simpson JM, Mackerras D, Silink M. Environmental factors in childhood IDDM. A population-based, case-control study. Diabetes Care. (1994) 17:1381–9. doi: 10.2337/diacare.17.12.1381
186. Scott FW, Cloutier HE, Kleemann R, Woerz-Pagenstert U, Rowsell P, Modler HW, et al. Potential mechanisms by which certain foods promote or inhibit the development of spontaneous diabetes in BB rats: dose, timing, early effect on islet area, and switch in infiltrate from Th1 to Th2 cells. Diabetes. (1997) 46:589–98. doi: 10.2337/diab.46.4.589
187. Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. (2013) 11:46. doi: 10.1186/1741-7015-11-46
188. Maiuri L, Troncone R, Mayer M, Coletta S, Picarelli A, De Vincenzi M, et al. In vitro activities of A-gliadin-related synthetic peptides: damaging effect on the atrophic coeliac mucosa and activation of mucosal immune response in the treated coeliac mucosa. Scand J Gastroenterol. (1996) 31:247–53. doi: 10.3109/00365529609004874
189. Lammers KM, Lu R, Brownley J, Lu B, Gerard C, Thomas K, et al. Gliadin induces an increase in intestinal permeability and zonulin release by binding to the chemokine receptor CXCR3. Gastroenterology. (2008) 135:194–204.e193. doi: 10.1053/j.gastro.2008.03.023
190. Fasano A. Zonulin and its regulation of intestinal barrier function: the biological door to inflammation, autoimmunity, and cancer. Physiol Rev. (2011) 91:151–75. doi: 10.1152/physrev.00003.2008
191. Lammers KM, Khandelwal S, Chaudhry F, Kryszak D, Puppa EL, Casolaro V, et al. Identification of a novel immunomodulatory gliadin peptide that causes interleukin-8 release in a chemokine receptor CXCR3-dependent manner only in patients with coeliac disease. Immunology. (2011) 132:432–40. doi: 10.1111/j.1365-2567.2010.03378.x
192. Brugman S, Klatter FA, Visser JT, Wildeboer-Veloo AC, Harmsen HJ, Rozing J, et al. Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia. (2006) 49:2105–8. doi: 10.1007/s00125-006-0334-0
193. Hansen AK, Ling F, Kaas A, Funda DP, Farlov H, Buschard K. Diabetes preventive gluten-free diet decreases the number of caecal bacteria in non-obese diabetic mice. Diabetes Metab Res Rev. (2006) 22:220–5. doi: 10.1002/dmrr.609
194. Marietta EV, Gomez AM, Yeoman C, Tilahun AY, Clark CR, Luckey DH, et al. Low incidence of spontaneous type 1 diabetes in non-obese diabetic mice raised on gluten-free diets is associated with changes in the intestinal microbiome. PLoS One. (2013) 8:e78687. doi: 10.1371/journal.pone.0078687
195. Qin H, Zhang Z, Hang X, Jiang Y. L. plantarum prevents enteroinvasive Escherichia coli-induced tight junction proteins changes in intestinal epithelial cells. BMC Microbiol. (2009) 9:63. doi: 10.1186/1471-2180-9-63
196. Karczewski J, Troost FJ, Konings I, Dekker J, Kleerebezem M, Brummer RJ, et al. Regulation of human epithelial tight junction proteins by Lactobacillus plantarum in vivo and protective effects on the epithelial barrier. Am J Physiol Gastrointest Liver Physiol. (2010) 298:G851–9. doi: 10.1152/ajpgi.00327.2009
197. Pastore MR, Bazzigaluppi E, Belloni C, Arcovio C, Bonifacio E, Bosi E. Six months of gluten-free diet do not influence autoantibody titers, but improve insulin secretion in subjects at high risk for type 1 diabetes. J Clin Endocrinol Metab. (2003) 88:162–5. doi: 10.1210/jc.2002-021177
198. Fuchtenbusch M, Ziegler AG, Hummel M. Elimination of dietary gluten and development of type 1 diabetes in high risk subjects. Rev Diabet Stud. (2004) 1:39–41. doi: 10.1900/RDS.2004.1.39
199. Saadah OI, Zacharin M, O'callaghan A, Oliver MR, and Catto-Smith AG. Effect of gluten-free diet and adherence on growth and diabetic control in diabetics with coeliac disease. Arch Dis Child. (2004) 89:871–6. doi: 10.1136/adc.2002.012799
200. Westman E, Ambler GR, Royle M, Peat J, Chan A. Children with coeliac disease and insulin dependent diabetes mellitus–growth, diabetes control and dietary intake. J Pediatr Endocrinol Metab. (1999) 12:433–42. doi: 10.1515/JPEM.1999.12.3.433
201. Rami B, Sumnik Z, Schober E, Waldhor T, Battelino T, Bratanic N, et al. Screening detected celiac disease in children with type 1 diabetes mellitus: effect on the clinical course (a case control study). J Pediatr Gastroenterol Nutr. (2005) 41:317–21. doi: 10.1097/01.mpg.0000174846.67797.87
202. Strobel S, Mowat AM. Immune responses to dietary antigens: oral tolerance. Immunol Today. (1998) 19:173–81. doi: 10.1016/S0167-5699(97)01239-5
203. Norris JM, Barriga K, Klingensmith G, Hoffman M, Eisenbarth GS, Erlich HA, et al. Timing of initial cereal exposure in infancy and risk of islet autoimmunity. JAMA. (2003) 290:1713–20. doi: 10.1001/jama.290.13.1713
204. Ziegler AG, Schmid S, Huber D, Hummel M, Bonifacio E. Early infant feeding and risk of developing type 1 diabetes-associated autoantibodies. JAMA. (2003) 290:1721–8. doi: 10.1001/jama.290.13.1721
205. Schmid S, Koczwara K, Schwinghammer S, Lampasona V, Ziegler AG, Bonifacio E. Delayed exposure to wheat and barley proteins reduces diabetes incidence in non-obese diabetic mice. Clin Immunol. (2004) 111:108–18. doi: 10.1016/j.clim.2003.09.012
206. Funda DP, Kaas A, Tlaskalova-Hogenova H, Buschard K. Gluten-free but also gluten-enriched (gluten+) diet prevent diabetes in NOD mice; the gluten enigma in type 1 diabetes. Diabetes Metab Res Rev. (2008) 24:59–63. doi: 10.1002/dmrr.748
207. Mueller DB, Koczwara K, Mueller AS, Pallauf J, Ziegler AG, Bonifacio E. Influence of early nutritional components on the development of murine autoimmune diabetes. Ann Nutr Metab. (2009) 54:208–17. doi: 10.1159/000220416
208. Hansen CH, Krych L, Buschard K, Metzdorff SB, Nellemann C, Hansen LH, et al. A maternal gluten-free diet reduces inflammation and diabetes incidence in the offspring of NOD mice. Diabetes. (2014) 63:2821–32. doi: 10.2337/db13-1612
209. Antvorskov JC, Josefsen K, Haupt-Jorgensen M, Fundova P, Funda DP, Buschard K. Gluten-free diet only during pregnancy efficiently prevents diabetes in NOD mouse offspring. J Diabetes Res. (2016) 2016:3047574. doi: 10.1155/2016/3047574
210. Antvorskov JC, Halldorsson TI, Josefsen K, Svensson J, Granstrom C, Roep BO, et al. Association between maternal gluten intake and type 1 diabetes in offspring: national prospective cohort study in Denmark. BMJ. (2018) 362:k3547. doi: 10.1136/bmj.k3547
211. Lamb MM, Myers MA, Barriga K, Zimmet PZ, Rewers M, Norris JM. Maternal diet during pregnancy and islet autoimmunity in offspring. Pediatr Diabetes. (2008) 9:135–41. doi: 10.1111/j.1399-5448.2007.00311.x
212. Virtanen SM, Uusitalo L, Kenward MG, Nevalainen J, Uusitalo U, Kronberg-Kippila C, et al. Maternal food consumption during pregnancy and risk of advanced beta-cell autoimmunity in the offspring. Pediatr Diabetes. (2011) 12:95–9. doi: 10.1111/j.1399-5448.2010.00668.x
213. Sildorf SM, Fredheim S, Svensson J, Buschard K. Remission without insulin therapy on gluten-free diet in a 6-year old boy with type 1 diabetes mellitus. BMJ Case Rep. (2012) 2012:bcr0220125878. doi: 10.1136/bcr.02.2012.5878
214. Svensson J, Sildorf SM, Pipper CB, Kyvsgaard JN, Bojstrup J, Pociot FM, et al. Potential beneficial effects of a gluten-free diet in newly diagnosed children with type 1 diabetes: a pilot study. Springerplus. (2016) 5:994. doi: 10.1186/s40064-016-2641-3
215. Endesfelder D, Zu Castell W, Ardissone A, Davis-Richardson AG, Achenbach P, Hagen M, et al. Compromised gut microbiota networks in children with anti-islet cell autoimmunity. Diabetes. (2014) 63:2006–14. doi: 10.2337/db13-1676
216. Paun A, Yau C, Danska JS. The influence of the microbiome on Type 1 diabetes. J Immunol. (2017) 198:590–5. doi: 10.4049/jimmunol.1601519
217. Muntoni S, Cocco P, Aru G, Cucca F. Nutritional factors and worldwide incidence of childhood type 1 diabetes. Am J Clin Nutr. (2000) 71:1525–9. doi: 10.1093/ajcn/71.6.1525
218. Elliott RB, Harris DP, Hill JP, Bibby NJ, Wasmuth HE. Type I (insulin-dependent) diabetes mellitus and cow milk: casein variant consumption. Diabetologia. (1999) 42:292–6. doi: 10.1007/s001250051153
219. Virtanen SM, Nevalainen J, Kronberg-Kippila C, Ahonen S, Tapanainen H, Uusitalo L, et al. Food consumption and advanced beta cell autoimmunity in young children with HLA-conferred susceptibility to type 1 diabetes: a nested case-control design. Am J Clin Nutr. (2012) 95:471–8. doi: 10.3945/ajcn.111.018879
220. Savilahti E, Akerblom HK, Tainio VM, Koskimies S. Children with newly diagnosed insulin dependent diabetes mellitus have increased levels of cow's milk antibodies. Diabetes Res. (1988) 7:137–40.
221. Dahlquist G, Savilahti E, Landin-Olsson M. An increased level of antibodies to beta-lactoglobulin is a risk determinant for early-onset type 1 (insulin-dependent) diabetes mellitus independent of islet cell antibodies and early introduction of cow's milk. Diabetologia. (1992) 35:980–4. doi: 10.1007/BF00401429
222. Savilahti E, Saukkonen TT, Virtala ET, Tuomilehto J, Akerblom HK. Increased levels of cow's milk and beta-lactoglobulin antibodies in young children with newly diagnosed IDDM. The Childhood Diabetes in Finland Study Group. Diabetes Care. (1993) 16:984–9. doi: 10.2337/diacare.16.7.984
223. Robertson L, Harrild K. Maternal and neonatal risk factors for childhood type 1 diabetes: a matched case-control study. BMC Public Health. (2010) 10:281. doi: 10.1186/1471-2458-10-281
224. Savilahti E, Saarinen KM. Early infant feeding and type 1 diabetes. Eur J Nutr. (2009) 48:243–9. doi: 10.1007/s00394-009-0008-z
225. Writing Group for The TSG, Knip M, Akerblom HK, Al Taji E, Becker D, Bruining J, et al. Effect of hydrolyzed infant formula vs conventional formula on risk of type 1 diabetes: the TRIGR randomized clinical trial. JAMA. (2018) 319:38–48. doi: 10.1001/jama.2017.19826
226. Palleja A, Mikkelsen KH, Forslund SK, Kashani A, Allin KH, Nielsen T, et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat Microbiol. (2018) 3:1255–65. doi: 10.1038/s41564-018-0257-9
227. Hicks LA, Taylor TH Jr., Hunkler RJ. U.S. outpatient antibiotic prescribing, 2010. N Engl J Med. (2013) 368:1461–2. doi: 10.1056/NEJMc1212055
228. Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. (2016) 8:343ra382. doi: 10.1126/scitranslmed.aad7121
229. Tapia G, Stordal K, Marild K, Kahrs CR, Skrivarhaug T, Njolstad PR, et al. Antibiotics, acetaminophen and infections during prenatal and early life in relation to type 1 diabetes. Int J Epidemiol. (2018) 47:1538–48. doi: 10.1093/ije/dyy092
230. Kemppainen KM, Vehik K, Lynch KF, Larsson HE, Canepa RJ, Simell V, et al. association between early-life antibiotic use and the risk of islet or celiac disease autoimmunity. JAMA Pediatr. (2017) 171:1217–25. doi: 10.1001/jamapediatrics.2017.2905
231. Mikkelsen KH, Knop FK, Vilsboll T, Frost M, Hallas J, Pottegard A. Use of antibiotics in childhood and risk of Type 1 diabetes: a population-based case-control study. Diabet Med. (2017) 34:272–7. doi: 10.1111/dme.13262
232. Kilkkinen A, Virtanen SM, Klaukka T, Kenward MG, Salkinoja-Salonen M, Gissler M, et al. Use of antimicrobials and risk of type 1 diabetes in a population-based mother-child cohort. Diabetologia. (2006) 49:66–70. doi: 10.1007/s00125-005-0078-2
233. Boursi B, Mamtani R, Haynes K, Yang YX. The effect of past antibiotic exposure on diabetes risk. Eur J Endocrinol. (2015) 172:639–48. doi: 10.1530/EJE-14-1163
234. Hara N, Alkanani AK, Ir D, Robertson CE, Wagner BD, Frank DN, et al. Prevention of virus-induced type 1 diabetes with antibiotic therapy. J Immunol. (2012) 189:3805–14. doi: 10.4049/jimmunol.1201257
235. Livanos AE, Greiner TU, Vangay P, Pathmasiri W, Stewart D, Mcritchie S, et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat Microbiol. (2016) 1:16140. doi: 10.1038/nmicrobiol.2016.140
236. Hansen CH, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sorensen SJ, et al. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia. (2012) 55:2285–94. doi: 10.1007/s00125-012-2564-7
237. Hu Y, Peng J, Tai N, Hu C, Zhang X, Wong FS, et al. Maternal antibiotic treatment protects offspring from diabetes development in nonobese diabetic mice by generation of tolerogenic APCs. J Immunol. (2015) 195:4176–84. doi: 10.4049/jimmunol.1500884
238. Pinto E, Anselmo M, Calha M, Bottrill A, Duarte I, Andrew PW, et al. The intestinal proteome of diabetic and control children is enriched with different microbial and host proteins. Microbiology. (2017) 163:161–74. doi: 10.1099/mic.0.000412
239. Gavin PG, Mullaney JA, Loo D, Cao KL, Gottlieb PA, Hill MM, et al. Intestinal metaproteomics reveals host-microbiota interactions in subjects at risk for type 1 diabetes. Diabetes Care. (2018) 41:2178–86. doi: 10.2337/dc18-0777
240. Coppieters KT, Wiberg A, Von Herrath MG. Viral infections and molecular mimicry in type 1 diabetes. APMIS. (2012) 120:941–9. doi: 10.1111/apm.12011
241. Kramna L, Kolarova K, Oikarinen S, Pursiheimo JP, Ilonen J, Simell O, et al. Gut virome sequencing in children with early islet autoimmunity. Diabetes Care. (2015) 38:930–3. doi: 10.2337/dc14-2490
242. Zhao G, Vatanen T, Droit L, Park A, Kostic AD, Poon TW, et al. Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc Natl Acad Sci USA. (2017) 114:E6166–75. doi: 10.1073/pnas.1706359114
243. Kim KW, Horton JL, Pang CNI, Jain K, Leung P, Isaacs SR, et al. Higher abundance of enterovirus A species in the gut of children with islet autoimmunity. Sci Rep. (2019) 9:1749. doi: 10.1038/s41598-018-38368-8
244. Martin FP, Wang Y, Sprenger N, Yap IK, Lundstedt T, Lek P, et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol. (2008) 4:157. doi: 10.1038/msb4100190
245. Wikoff WR, Anfora AT, Liu J, Schultz, PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. (2009) 106:3698–703. doi: 10.1073/pnas.0812874106
246. Aw W, Fukuda S. Toward the comprehensive understanding of the gut ecosystem via metabolomics-based integrated omics approach. Semin Immunopathol. (2015) 37:5–16. doi: 10.1007/s00281-014-0456-2
247. Bridgman SL, Azad MB, Field CJ, Haqq AM, Becker AB, Mandhane PJ, et al. Fecal short-chain fatty acid variations by breastfeeding status in infants at 4 months: differences in relative versus absolute concentrations. Front Nutr. (2017) 4:11. doi: 10.3389/fnut.2017.00011
248. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. (2014) 20:159–66. doi: 10.1038/nm.3444
249. Benus RF, Van Der Werf TS, Welling GW, Judd PA, Taylor MA, Harmsen HJ, et al. Association between Faecalibacterium prausnitzii and dietary fibre in colonic fermentation in healthy human subjects. Br J Nutr. (2010) 104:693–700. doi: 10.1017/S0007114510001030
250. Donohoe DR, Garge N, Zhang X, Sun W, O'connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. (2011) 13:517–26.
251. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. (2013) 504:446–50. doi: 10.1038/nature12721
252. De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. (2014) 156:84–96. doi: 10.1016/j.cell.2013.12.016
253. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. (2014) 40:128–39. doi: 10.1016/j.immuni.2013.12.007
254. Marino E, Richards JL, Mcleod KH, Stanley D, Yap YA, Knight J, et al. Erratum: gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol. (2017) 18:1271. doi: 10.1038/ni1117-1271c
255. Songini M, Bernardinelli L, Clayton D, Montomoli C, Pascutto C, Ghislandi M, et al. The Sardinian IDDM study: 1. Epidemiology and geographical distribution of IDDM in Sardinia during 1989 to 1994. Diabetologia. (1998) 41:221–7. doi: 10.1007/s001250050893
256. Culeddu N, Chessa M, Porcu MC, Fresu P, Tonolo G, Virgilio G, et al. NMR-based metabolomic study of type 1 diabetes. Metabolomics. (2012) 8:1162–9. doi: 10.1007/s11306-012-0420-x
257. Oresic M, Simell S, Sysi-Aho M, Nanto-Salonen K, Seppanen-Laakso T, Parikka V, et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med. (2008) 205:2975–84. doi: 10.1084/jem.20081800
258. Atkinson MA, Leiter EH. The NOD mouse model of type 1 diabetes: as good as it gets? Nat Med. (1999) 5:601–4. doi: 10.1038/9442
259. Dolensek J, Rupnik MS, Stozer A. Structural similarities and differences between the human and the mouse pancreas. Islets. (2015) 7:e1024405. doi: 10.1080/19382014.2015.1024405
260. Blohme G, Nystrom L, Arnqvist HJ, Lithner F, Littorin B, Olsson PO, et al. Male predominance of type 1 (insulin-dependent) diabetes mellitus in young adults: results from a 5-year prospective nationwide study of the 15-34-year age group in Sweden. Diabetologia. (1992) 35:56–62. doi: 10.1007/BF00400852
261. Williams AJ, Bingley PJ, Moore WP, Gale EA, Trial ESGENDI. Islet autoantibodies, nationality and gender: a multinational screening study in first-degree relatives of patients with Type I diabetes. Diabetologia. (2002) 45:217–23. doi: 10.1007/s00125-001-0749-6
262. Wandell PE, Carlsson AC. Time trends and gender differences in incidence and prevalence of type 1 diabetes in Sweden. Curr Diabetes Rev. (2013) 9:342–9. doi: 10.2174/15733998113099990064
263. Roep BO, Atkinson M, Von Herrath M. Satisfaction (not) guaranteed: re-evaluating the use of animal models of type 1 diabetes. Nat Rev Immunol. (2004) 4:989–97. doi: 10.1038/nri1502
264. Atkinson MA, Roep BO, Posgai A, Wheeler DCS, Peakman M. The challenge of modulating beta-cell autoimmunity in type 1 diabetes. Lancet Diabetes Endocrinol. (2019) 7:52–64. doi: 10.1016/S2213-8587(18)30112-8
265. Manrique P, Bolduc B, Walk ST, Van Der Oost J, De Vos WM, Young MJ. Healthy human gut phageome. Proc Natl Acad Sci USA. (2016) 113:10400–5. doi: 10.1073/pnas.1601060113
266. Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, et al. Fungal microbiota dysbiosis in IBD. Gut. (2017) 66:1039–48. doi: 10.1136/gutjnl-2015-310746
267. Koskinen K, Pausan MR, Perras AK, Beck M, Bang C, Mora M, et al. First insights into the diverse human archaeome: specific detection of archaea in the gastrointestinal tract, lung, and nose and on skin. MBio. (2017) 8:e00824-17. doi: 10.1128/mBio.00824-17
268. Abu-Ali GS, Mehta RS, Lloyd-Price J, Mallick H, Branck T, Ivey KL, et al. Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nat Microbiol. (2018) 3:356–66. doi: 10.1038/s41564-017-0084-4
269. Wilson MT, Hamilos DL. The nasal and sinus microbiome in health and disease. Curr Allergy Asthma Rep. (2014) 14:485. doi: 10.1007/s11882-014-0485-x
270. Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. (2018) 16:143–55. doi: 10.1038/nrmicro.2017.157
271. Ma B, Forney LJ, Ravel J. Vaginal microbiome: rethinking health and disease. Annu Rev Microbiol. (2012) 66:371–89. doi: 10.1146/annurev-micro-092611-150157
272. Niehaus L, Boland I, Liu M, Chen K, Fu D, Henckel C, et al. Microbial coexistence through chemical-mediated interactions. Nat Commun. (2019) 10:2052. doi: 10.1038/s41467-019-10062-x
273. Altindis E, Cai W, Sakaguchi M, Zhang F, Guoxiao W, Liu F, et al. Viral insulin-like peptides activate human insulin and IGF-1 receptor signaling: a paradigm shift for host-microbe interactions. Proc Natl Acad Sci USA. (2018) 115:2461–6. doi: 10.1073/pnas.1721117115
Keywords: microbiome, longitudinal studies children, type 1 diabetes, environmental factors, autoimmunity
Citation: Dedrick S, Sundaresh B, Huang Q, Brady C, Yoo T, Cronin C, Rudnicki C, Flood M, Momeni B, Ludvigsson J and Altindis E (2020) The Role of Gut Microbiota and Environmental Factors in Type 1 Diabetes Pathogenesis. Front. Endocrinol. 11:78. doi: 10.3389/fendo.2020.00078
Received: 21 November 2019; Accepted: 06 February 2020;
Published: 26 February 2020.
Edited by:
Nathalie Delzenne, Catholic University of Louvain, BelgiumReviewed by:
Edgar Zenteno, National Autonomous University of Mexico, MexicoCopyright © 2020 Dedrick, Sundaresh, Huang, Brady, Yoo, Cronin, Rudnicki, Flood, Momeni, Ludvigsson and Altindis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emrah Altindis, YWx0aW5kaXNAYmMuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.