
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 12 July 2019
Sec. Clinical Diabetes
Volume 10 - 2019 | https://doi.org/10.3389/fendo.2019.00461
 Zihao Wen1†
Zihao Wen1† Xiaoqian Zou1†
Xiaoqian Zou1† Xin Xie1†
Xin Xie1† Shaoling Zheng1
Shaoling Zheng1 Xiaojing Chen1
Xiaojing Chen1 Kehui Zhu1
Kehui Zhu1 Shirui Dong1
Shirui Dong1 Jiayu Liang1
Jiayu Liang1 Xiuxia Huang1
Xiuxia Huang1 Dandan Liu1
Dandan Liu1 Yao Wang1
Yao Wang1 Yumei Liu1
Yumei Liu1 Jing Wu1
Jing Wu1 Yuting Ying1
Yuting Ying1 Kailiang Liu1
Kailiang Liu1 Congying Lu1
Congying Lu1 Baohuan Zhang1
Baohuan Zhang1 Guang Yang2,3*
Guang Yang2,3* Chunxia Jing1,3*
Chunxia Jing1,3* Lihong Nie4*
Lihong Nie4*Objective: To evaluate the potential association between the genetic variants in miRNA processing genes (RAN, XPO5, DICER1, and TARBP2) and susceptibility to type 2 diabetes mellitus (T2DM) and its vascular complications, as well as to further investigate their interaction with environmental factors in type 2 diabetes.
Methods: We conducted a case-control study in genotyping of five polymorphic loci, including RAN rs14035, XPO5 rs11077, DICER1 rs13078, DICER1 rs3742330, and TARBP2 rs784567, in miRNA processing genes to explore the risk factors for T2DM and diabetic vascular complications. Haplotype analyses, interactions of gene-gene and interactions of gene-environment were performed too.
Results: We identified a 36% decreased risk of developing T2DM in individuals with the minor A allele in DICER1 rs13078 (OR: 0.64; 95%CI: 0.42–0.95; P: 0.026). The AA haplotype in DICER1 was also associated with a protective effect on T2DM compared with the AT haplotype (OR: 0.63; 95%CI: 0.42–0.94; P-value: 0.023). T2DM patients with the TT+TC genotype at RAN rs14035 had a 1.89-fold higher risk of developing macrovascular complications than patients with the CC genotype (OR: 1.89; 95%CI: 1.04–3.45; P-value: 0.037). We also identified two three-factor interaction models. One is a three-factor [DICER1 rs13078, body mass index (BMI), and triglyceride (TG)] interaction model for T2DM (OR: 5.93; 95%CI: 1.25–28.26; P = 0.025). Another three-factor [RAN rs14035, hypertension (HP), and duration of T2DM (DOD)] interaction model was found for macrovascular complications of T2DM (OR = 41.60, 95%CI = 11.75–147.35, P < 0.001).
Conclusion: Our study provides new evidence that two single nucleotide polymorphisms (SNPs) of the miRNA processing genes, DICER1 and RAN, and their interactions with certain environmental factors might contribute to the risk of T2DM and its vascular complications in the southern Chinese population.
Diabetes mellitus (DM) is a chronic disease that occurs when there are increased levels of blood glucose because the body is unable to produce any or enough of the hormone insulin or use insulin effectively (1). According to the International Diabetes Federation (IDF) report, there were 4.25 billion people with diabetes in 2017, and the number of diabetes patients will increase to 6.29 billion globally by 2040 (2). In 2017, more than 4 million people died because of diabetes and diabetic complications, which means that worldwide, a patient died every 8 s because of diabetes (2). The total global healthcare expenditure due to DM was estimated at 727 billion dollars in 2017 (3). As the most populous and largest developing country in the world, China has the highest number of diabetes patients worldwide (114.4 million in 2017) (2). The number of diabetes patients soared from 4.8 million in 1980 to 39.8 million in 2007 (4).
T2DM as the most common type of diabetes, accounts for approximately 90% of diabetes cases and is affected by both environmental factors and genetic factors (5–7). Type 2 diabetes mellitus is a long-term metabolic disorder characterized by damage to insulin secretion and sensitivity, resulting in hyperglycemia (8, 9), which can lead to the development of diabetic vascular complications, such as diabetic nephropathy, peripheral artery disease (PAD) and coronary heart disease (CHD) (2, 10, 11). Risk factors for diabetic vascular complications include lifestyle (such as smoking and drinking), duration of diabetes, age of onset and genetic factors, etc. (12, 13).
MicroRNAs (miRNAs) are a class of short, single-strand, non-coding, and endogenous RNA molecules of 21–23 nts in length (14). Although miRNAs constitute at most 3% of the human genome, it has been reported that approximately one-third of human genes were regulated by miRNAs (15, 16). Recently, the evidences that series of miRNAs were related to the T2DM and diabetic vascular complication, have been proved by several studies (17), such as miR-200 family (18), miR-124a (19), miR-21-5P (20), and miR-125a-3P (20). RAS-related nuclear protein (RAN), exportin 5(XPO5), DICER1, and TARBP2, which are known as microRNA processing enzyme, are the key to complete the biosynthesis of mammalian miRNAs (21, 22). First, RNA II polymerase transcribed miRNAs into long precursors called pri-miRNAs, which are cleaved in the nucleus to release a stem loop intermediate about 60–70 nt, known as the miRNA precursor hairpin (pre-miRNA). Secondly, XPO5 and RAN export the pre-miRNA from the nucleus to the cytoplasm. And the DICER1 and TARBP2 further cooperatively dice the pre-miRNA into a double-stranded, short miRNA duplex (23, 24). Last, the double-stranded miRNA is preferentially incorporated into RNA-induced silencing complex, targeting endogenous mRNA silencing (25, 26). Therefore, if the expression and structure of these miRNA processing proteins have been altered in the biosynthesis of mammalian miRNAs, it would directly impact the biosynthesis of mature miRNAs and further change the function and structure of miRNAs (21, 22) (Figure 1).
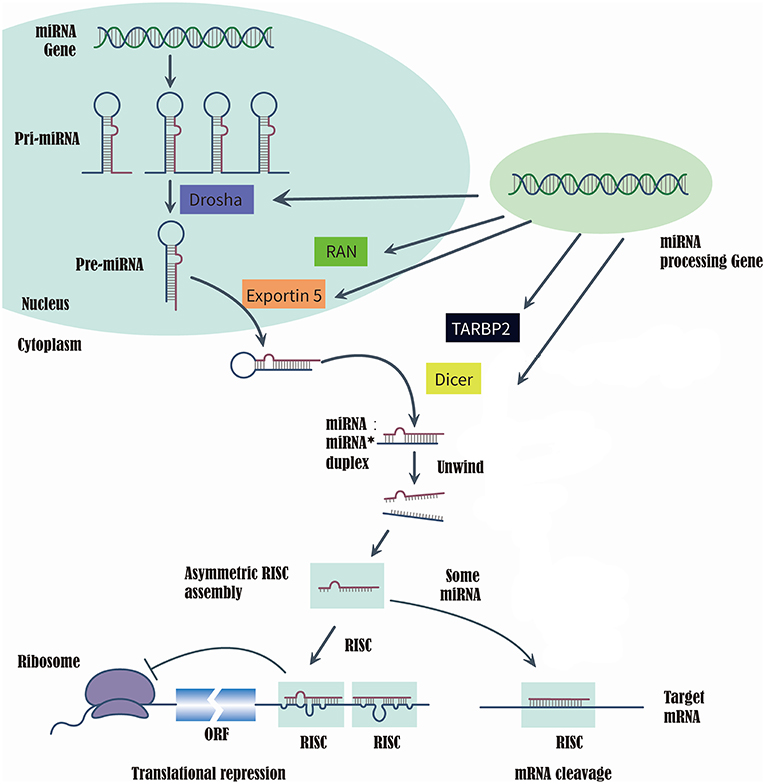
Figure 1. Summarize of the biosynthesis process of miRNA and the function of miRNA processing Gene and enzyme.
A variation in a single nucleotide is called a single nucleotide polymorphism (SNP) that occurs in polymorphisms of the genomic DNA sequence, and is the simplest form of DNA variation among individuals and might affect the expression and function of genes (27). It has been reported that SNPs are associated with the development of type 2 diabetes mellitus and its vascular complications (28–30). GWAS (genome-wide association studies) have identified over 80 SNPs were closely related to type 2 diabetes mellitus (31–34). In addition, GWAS also identified over 20 SNPs related to the various chronic T2DM complications (e.g., diabetic nephropathy, diabetic retinopathy, diabetic cardiopathy and diabetic painful neuropathy) (35). RAN rs14035, DICER1 rs3742330, and DROSHA rs10719 are located in the 3′-UTR, which may disturb the function of these miRNA processing proteins by impacting the binding site of their miRNAs, resulting in dysregulation in processing of related gene expression and impact the biosynthesis of related proteins (36–39). We hypotheses that the SNPs in miRNA processing genes may impact the susceptibility to T2DM and its vascular complications by disrupting the structure, binding sites, or processing of miRNA (40, 41). To our knowledge, few studies evaluated the potential association between the risk of T2DM and its vascular complications and specific SNPs in miRNA processing genes. Thus, the aim of our study is investigating the effects of variations in miRNA processing genes, RAN (rs14035), XPO5 (rs11077), DICER1 (rs3742330 and rs13078), and TARBP2 (rs784567), as well as their interactions with environmental factors on T2DM and its vascular complications in a Southern Han Chinese population.
This case-control study included a total of 1,275 unrelated Han Chinese Southern residents in Guangzhou. A total of 743 T2DM subjects were inpatients of the Endocrinology Departments of the First Affiliated Hospital of Jinan University in Guangzhou from September 2011 to January 2018, and all patients were diagnosed using the 2003 American Diabetes Association criteria (42). Patients with impaired glucose tolerance and type 1 diabetes mellitus were excluded from the study. During the same period, a total of 532 healthy control individuals without a family history of diabetes and with normal fasting glucose were also randomly recruited to match T2DM patients by sex and age (±5 years). Before participating in the study, all of the included individuals agreed and signed informed consent, and the study protocol was approved by the Ethics Committee of Medical School at Jinan University. Within the 743 included T2DM patients, 46, 96, 108, and 227 cases were diagnosed with T2DM with other complications, micro-macrovascular complications, macrovascular complications, and microvascular complications, respectively, while 266 cases were categorized into T2DM without any complication. According to the 8th edition IDF Diabetes Atlas (2) and Gregg et al. (43), macrovascular complications included coronary artery disease (CAD), resulting in myocardial infarction or stenocardia, and peripheral artery disease (PAD), leading to diabetic encephalopathy and cerebral infarction, whereas microvascular complications are retinopathy, neuropathy and nephropathy. Micro-macrovascular complications were defined as presenting with both macrovascular microvascular and complications.
We determined the targeted SNPs of RAN/XPO5/DICER1/TARBP2 were through an electronic search of the HapMap database based on the genotype information of Han Chinese in Beijing, China. And, the tarSNPs were determined based on the following criteria: (1) the minor allele frequency (MAF) of SNPs >5%; (2) pairwise tagging with r2 ≥ 0.8; and (3) P-value of Hardy-Weinberg > 0.001. Five SNPs were identified, RAN rs14035, XPO5 rs11077, DICER1 rs3742330 and rs13078, and TARBP2 rs784567. Finally, five target SNPs were selected for further analysis. The detailed information of the five tarSNPs, including alleles, MAFs, genes locations, the result of Hardy-Weinberg equilibrium and call rate, is shown in Table S1. However, rs14035 in the RAN gene did not meet the Hardy-Weinberg equilibrium (P < 0.05). Therefore, the further analyses of the polymorphism associations within T2DM group and health control group excluded rs14035 in the RAN gene.
A QIAmp Blood DNA Mini Kit (Qiagen, Hilden, Germany) was used to extract the genomic DNA from peripheral whole blood samples. A Sequenom MassARRAYiPLEX Gold analyzer (Sequenom, Life Technologies, Shanghai) was used to genotype the five selected SNPs. MassARRAY Assay Design 3.1 software were used to design PCR conditions and primers (Table S1).
The demographic data and clinical characteristics of the included individuals are presented as the mean ± S.D. or number (percentage). The Shapiro-Wilk test was performed to identify the normality of the included demographic data and clinical characteristics in each comparison group (Tables S2–S5). For continuous variables with normal distribution, the t-test and ANOVA were performed on them, otherwise the non-parametric test (Kruskal-Wallis test and Mann-Whitney test) was conducted. For categorical variable, the χ2-test was performed. The logistic regression with different genetic model (codominant, dominant, and recessive models) was performed to calculate the ORs and 95% confidence intervals (CI) for estimating risk (44). Hardy-Weinberg equilibrium (HWE) of each SNP in the control group was calculated by χ2 statistics. All the above statistical analyses were performed by SPSS software v.22.0 (SPSS, Inc.).
The MDR (multifactor dimensionality reduction) software (45) was used to performed to identified the best model of gene-gene interaction models and gene-environment interaction models of the five selected SNPs. The environmental factors we included in MDR were age, gender, BMI (body mass index), TG (triglyceride), TC (total cholesterol), LDL (low density lipoprotein), HDL (high density lipoprotein), FBG (fasting blood glucose), post-prandial blood glucose, HbA1c, duration of T2DM, patients with family history of diabetes, former smoking, current smoking, current smoking, current drinking, and patents with hypertension. SHEsis, an online software (http://analysis.bio-x.cn/myAnalysis.php), was used to identified the effects of haplotype frequencies on T2DM and its vascular complications (46–48).
The baseline population characteristics of all included individuals are presented in Table 1. In comparison with healthy controls, the patients in T2DM group had higher levels of body mass index (BMI), triglyceride (TG), fasting blood glucose (FBG), GPT (glutamic-pyruvic, transaminase), serum creatinine, and blood uric acid but lower levels of low-density lipoprotein (LDL) and high-density lipoprotein (HDL), when compared with healthy controls. In comparison with T2DM patients without any complications, all T2DM patients with microvascular complications, macrovascular complications and micro-macrovascular complications had higher average age, longer duration of T2DM, and high levels of serum creatinine, but they had lower estimated glomerular filtration rate (eGFR) and GPT levels. Additionally, in comparison with T2DM patients without any complications, hypertension was more prevalent in T2DM patients with vascular complications (macrovascular, microvascular, and micro-macrovascular complications). And T2DM patients with macrovascular complications included a larger number of former smokers (Table 2).
XPO5 rs11077, TARBP2 rs784567, DICER1 rs13078, and rs3742330 met the requirement of the HWE in healthy controls. However, RAN rs14035 deviated from HWE in healthy controls (Tables S2, S3).
We identified the effects of XPO5, DICER1, and TARBP2 SNPs on T2DM. As presented in Table 2, there was a significant association between rs13078 in the DICER1 gene and a decreased risk of developing T2DM under the allelic mode (A vs. T: OR: 0.64; 95CI%: 0.42–0.95; P: 0.026), suggesting that individuals carrying the A allele had a 36% lower risk of developing T2DM than those carrying the T allele. However, there were no significant differences between T2DM under the allelic and codominant, dominant, recessive models, and other SNPs.
The effects of RAN, XPO5, DICER1, and TARBP2 SNPs on diabetes progression were further analyzed. We found that rs14035 in the RAN gene was associated with T2DM with macrovascular complications (Tables 3, 4). In comparison with T2DM patients with the CC genotype, T2DM patients carrying TT+CT genotypes at rs14035 had 1.89-fold higher risk of suffering T2DM macrovascular complications (TT+CT vs. CC: OR: 1.89; 95%CI: 1.04–3.45; P: 0.037). However, we did not find that the rest of target SNPs in XPO5, DICER1, and TARBP2 genes were associated with the susceptibility of T2DM vascular complications.
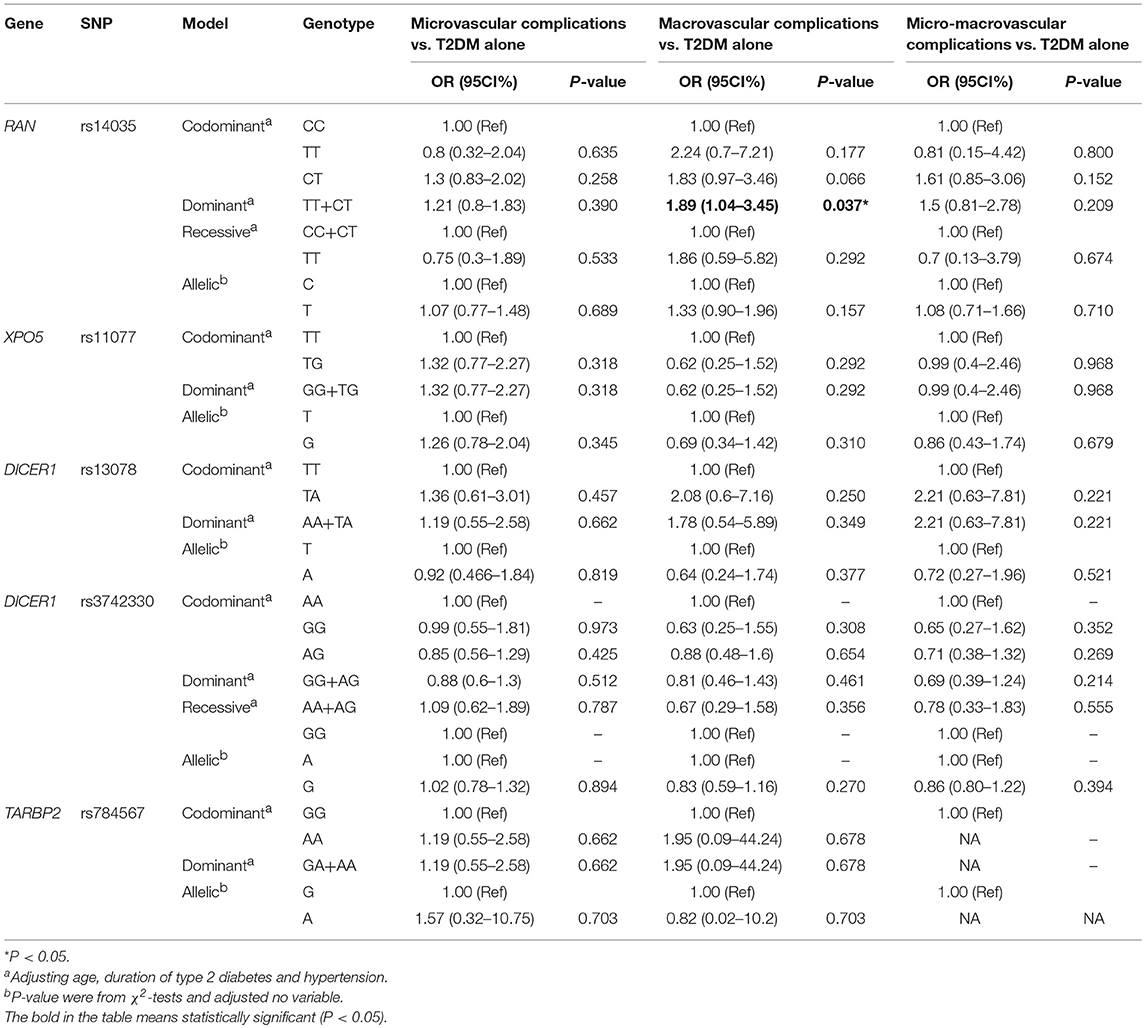
Table 4. Association of RAN, XPO5, DICER1, and TARBP2 polymorphisms with vascular complications of T2DM.
To explore the association among gene-environment interaction, T2DM and its vascular complications, MDR analysis was conducted to analysis the five SNPs in the RAN, XPO5, DICER1, and TARBP2 genes as well as environment factors (Table S4). The best gene-environmental interaction model on T2DM and its macrovascular complications were identified (DICER1 rs13078, BMI and TG; RAN rs14035, DOD and HP), with a significant TBA value and the highest CVC value (Table 5). The risk analysis of the three-way interaction in the model was also performed (Table S5). As shown in Figure 2, in comparison with the reference group (wild-type rs13078, normal BMI, and normal TG), the individuals with all three factors (mutation of rs13078, high BMI, and high TG) exhibited a 5.93-fold higher possibility of developing T2DM (OR: 5.93; 95%CI:1.25–28.26; P: 0.025). Figure 3 shows that the individuals with mutation of RAN rs14035, DOD more than 5 years and hypertension had a 40.60-fold higher possibility of suffering diabetic macrovascular complications when compared with the reference group (wild-type RAN rs14035, DOD <5 years and without hypertension) (OR: 41.60; 95%CI:11.75–147.35; P < 0.001). However, no other gene-environment interactions were identified for T2DM and T2DM vascular complications.

Table 5. The gene-environment interaction model using MDR analysis for T2DM and its vascular complications.
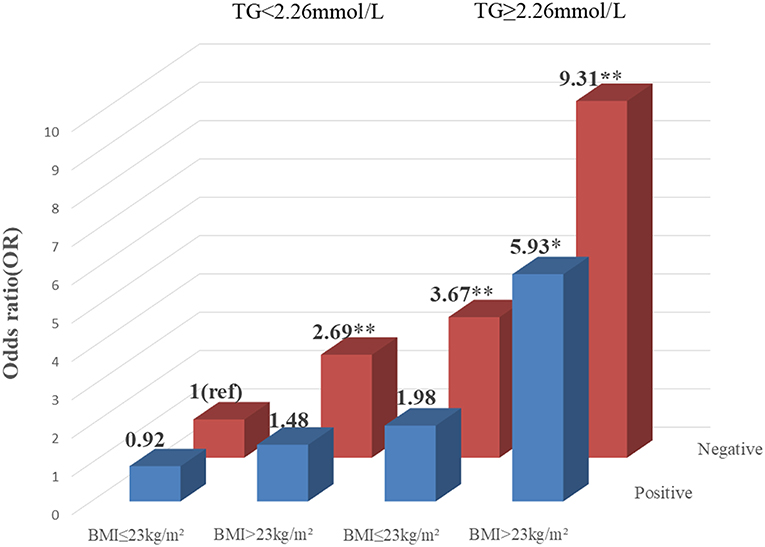
Figure 2. Risk analysis of gene-environment interaction Model: DICER1 rs13078, BMI, TG. The reference group is the interaction of wild-type for rs13078, normal BMI and normal TG. The OR value is presented in the figure. *P < 0.05 and **P < 0.001.
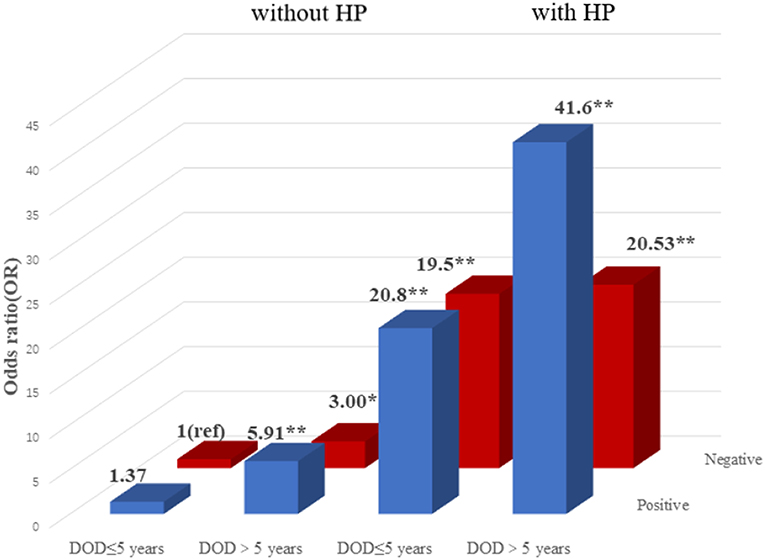
Figure 3. Risk analysis of gene-environment interaction for T2DM with macrovascular complications Model: RAN rs14035, DOD, HP. The reference group is the interaction of wild-type for rs14035, duration of T2DM <5 years and without hypertension. The OR value is shown in the figure. *P < 0.05 and **P < 0.001.
However, no gene-gene interaction was found to be associated with the increase risk of T2DM and its vascular complications using MDR analysis.
The two SNPs (rs3742330 and rs13078) constitute a haplotype block spanning 3 kb of the DICER1 gene (Figure 3). We conducted a haplotype analysis among the DICER1 gene, T2DM and T2DM vascular complications. As shown in Table S6, compared with the highest frequency haplotype AT, the haplotype AA was significantly related to a lower risk of T2DM (OR: 0.63; 95%CI: 0.42–0.94; P: 0.023).
Alterations in the miRNA machinery play important roles in the pathogenesis of a variety of disorders (49), which may account for abnormal profiles of miRNA in various diseases. Recently, more and more studies have provided evidence that alterations in miRNA machinery may result in dysregulation or upregulation of miRNAs in diabetes (40, 50, 51). In our study, a significant 0.64-fold decrease was found in the A allele frequency at DICER1 rs13078 in T2DM patients than healthy individuals in the allelic model, suggesting that individuals carring the A allele at DICER1 rs13078 had a decreased possibility of developing T2DM than those carrying the T allele. Additionally, we also identified a protective effect of the AA haplotype in DICER1.
DICER1, which situate in chromosome 14q32.13, contains 1922 amino acids in humans, encoding an approximately 218 kDa RNase III endonuclease (21, 52). The DICER1 enzyme is responsible for the processing of gene-encoded pre-miRNAs into mature miRNAs, and it plays a key role in the highly conserved cellular pathway (52). DICER1 is also well known to be an important component in the oncogenic process of several cancers, such as breast cancer (53), hepatocellular carcinoma (54), lung cancer (55), and ovarian cancer (56). In metabolic diseases, Noren Hooten et al. (57) hypothesized that alterations of the levels of DICER1 gene may play an important role in organismal aging and the upregulation of expression of DICER1 gene may provide us a new pharmacotherapeutic approaches for age-related disease, such as T2DM. Furthermore, it has been demonstrated that different levels of expression of microRNAs were identified in exosomes isolated from the serum and in the blood of healthy control individuals compared with patients with T2DM, hypercholesterolemia or metabolic syndrome (58, 59). It has been reported that the site of DICER1 rs13078 located in the 3′-UTR of the DICER1 gene and might be an important component in the expression process of the DICER1 gene by influencing the miRNAs binding site, which might impact the DICER1 enzyme's function by DICER1 gene regulation and sequentially impact the expression of microRNAs (52, 60, 61). Additionally, rs13078 genetic variants in DICER1 have been reported to be related to other diseases, such as gestational hypertension (62) and larynx cancer (63).
In the association analysis between diabetic vascular complications and SNPs, we found that T2DM patients who carried TT+TC genotype of rs14035 in RAN gene had a 1.89-fold increased risk of developing macrovascular complications compared with those with the CC wild-type genotype. RAN rs14035 is also located in the 3′-UTR of the RAN gene at chr12:130876696 (64). The RAN enzyme as the member of the Ras superfamily of GTPases, plays a key component in the translocation of pre-miRNAs from the nucleus to the cytoplasm through the nuclear pore complex (65). RAN-GTP is depleted as a result of RAN guanine nucleotide exchange factor inhibition, and pre-miRNA export is greatly reduced, indicating that miRNA transport is mediated by a RAN-GTP-binding export receptor (66). These data not only suggest that mutations in RAN might play an important role in pathology-related changes in the transport and expression of miRNA but also imply that polymorphisms of the RAN gene might also be related to the biosynthesis and translation of miRNAs. Vorpahl et al. (67) suggested that inhibition or modulation of RAN-GTPase-activating protein expression might be sufficient to attenuate the vascular proliferative response following vascular injury. Additionally, RAN-GTPase signaling might also be essential for postnatal pancreatic islet development and glucose homeostasis (68). It has been reported that a significant upregulation of circulating miR-126 has been detected in patients with angina and acute myocardial infarction, while downregulation of miR-126 has been detected in plasma from patients with cancer, diabetes or heart failure (58, 59). Moreover, it has been reported that circulating miR-126-3p, which may be a reliable biomarker of physiological endothelial senescence in elderly subjects with normoglycemia, underlies a mechanism that may be disrupted in elderly patients with DM (69). These studies give evidence the rs14035 polymorphism in RAN might be related to the occurrence and deterioration of the macrovascular complications of T2DM by regulating the expression level of RAN.
Recently, it has been reported that the susceptibility of type 2 diabetes mellitus and vascular complications of T2DM could be affected by various factors, including interactions between various environmental factors and the different polymorphisms (10). An MDR analysis was conducted to further investigate the multiple-factor, higher-order interaction of T2DM and the vascular complications of T2DM. The best three-factor interaction model among DICER1 rs13078, BMI (body mass index) and TG (triglyceride), suggested that rs13078 in DICER1 more likely interacts with overweigh and high TG to increase the possibility of developing T2DM. This outcome is consistent with the analysis of the clinical data between healthy control individuals and T2DM patients which showed that T2DM patients had significantly higher BMI and TG than healthy controls. Shai et al. (70) and Astrup et al. (71) also supported that individuals with abnormal lipid metabolism (high TG, TC, HDL, and low HDL) had a higher risk of developing T2DM. It has also been reported that BMI above normal weight level has been demonstrated to be one of the risk factors of T2DM and its complications (71–73). Moreover, our interaction results were supported by the synergistic effect and individual effect that overweight, increased triglyceride, high LDL and low HDL are associated with T2DM risk (74). In addition, another best three-factor interaction model (RAN rs14035, with hypertension and more than 5 years of T2DM duration) was also identified to be related to the increased risk of macrovascular complications of T2DM. Hypertension is known as one of the independent risk factors of T2DM (75). Moreover, the coexistence of hypertension and diabetes will significantly increase the risk of developing cardiovascular complications of DM, which means that hypertension may be an important risk factor for diabetic macrovascular complications (76, 77). According to the ADA guidelines, with the increase in the duration of diabetes, the risk of developing diabetic vascular complications will significantly increase (78). Many studies also supported that the duration of diabetes played an important role in developing diabetic vascular complications (79, 80). However, our study is the first time we found these two interaction models in T2DM and diabetic macrovascular complications, thus further studies with larger population samples are needed.
Similarly, there might be more environmental factors, such as diet behavior, passive smoke exposure and PM2.5, which might also modify the effects of genetic variants on the susceptibility of T2DM and its vascular complications (72, 73). For example, the study by Eze et al. demonstrated the interaction between long-term PM10 and gene polymorphisms at IL6-572 on T2DM (81). Therefore, more environmental factors could be included in further studies on interaction.
Our study has several limitations. First, the sample in the study was not very large, which might restrict the ability to explore weaker associations among T2DM, vascular complications of T2DM and SNPs. Second, the basic characteristics of the included individuals collected were not very comprehensive. Several clinical characteristics were not collected, such as the actual values of systolic and diastolic blood pressure. In addition, we should also include more meaningful environmental factors, such as diet, PM10 and passive smoke, in our analysis. Therefore, more large and well-designed investigation is needed to support our findings.
As far as we know, this is the first study to investigate the association of these five polymorphisms (rs14035 in RAN, rs11077 in XPO5, rs13078 in DICER1, rs3742330 in DICER1 and rs784567 in TARBP2) with the risk of T2DM and its vascular complications. Our findings offer us the first evidence that rs13078 in DICER1 is related to a reduced risk of T2DM and that RAN rs14035 is relevant to an increased risk of macrovascular complication of T2DM in southern Chinese population. Additionally, our results also demonstrate two feasible interactions, DICER1 rs13078, BMI and TG in T2DM and RAN rs14035, hypertension and duration of T2DM in diabetic macrovascular complications. Our study may provide a new clue of epidemiology about the importance of miRNA processing genes (RAN, XPO5, DICER1, and TARBP2) in type 2 diabetes mellitus and diabetic vascular complications.
Our study was conducted in conformity with the rules of the Ethics Committee of Jinan University by written informed consent to all participant subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The Ethics Committee of Jinan University approved the protocol.
ZW, XZ, and XX contributed equally to the writing of this paper. LN provided all the samples for this study. SZ, XC, KZ, SD, JL, XH, DL, YW, JW, YL, YY, KL, CL, and BZ carried out data collection and the extraction of DNA. GY and CJ carried out whole design.
Our work was supported by Project from Jinan university (grant no: JNUPHPM2016002, JNUPHPM2016001, 21615426, and 21612426), Guangdong Province Medical Research Foundation (grant no: A2014374,A2015310), the Guangdong Natural Science Foundation (grant no: S2011010002526, 10151063201000036, 2016030313089, and 2018A030313601), National Natural Science Foundation of China (grant no: 81101267, 30901249, and 81541070), and the Major Research Plan of the National Natural Science Foundation of China (91543132).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2019.00461/full#supplementary-material
1. DeCherney GS. International textbook of diabetes mellitus. JAMA. (1993) 270:1871. doi: 10.1001/jama.1993.03510150107047
3. Cho NH, Shaw JE, Karuranga S, Huang Y, da Rocha Fernandes JD, Ohlrogge AW, et al. IDF Diabetes Atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diab Res Clin Pract. (2018) 138:271–81. doi: 10.1016/j.diabres.2018.02.023
4. Reversing the rising tide of diabetes in China. Lancet. (2016) 388:2566. doi: 10.1016/S0140-6736(16)32384-4
6. Evans JM, Newton RW, Ruta DA, MacDonald TM, Morris AD. Socio-economic status, obesity and prevalence of Type 1 and Type 2 diabetes mellitus. Diab Med. (2000) 17:478–80. doi: 10.1046/j.1464-5491.2000.00309.x
7. Hou QT, Li Y, Li SY, Tian HM. The global burden of diabetes mellitus. Chin J Diabetes. (2016) 24:92–6.
8. Tang L, Tong N. Interpretation of American Diabetes Association Standards of Medical Care in Diabetes-2018. West China Med J. (2018) 33:513–19. doi: 10.7507/1002-0179.201803136
9. O'Brien MG. National Institute of Diabetes and Digestive and Kidney Diseases Bethesda: George M.O' brien (2019).
10. Lontchi-Yimagou E, Sobngwi E, Matsha TE, Kengne AP. Diabetes mellitus and inflammation. Curr Diab Rep. (2013) 13:435–44. doi: 10.1007/s11892-013-0375-y
11. Mehravar F, Mansournia MA, Holakouie-Naieni K, Nasli-Esfahani E, Mansournia N, Almasi-Hashiani A. Associations between diabetes self-management and microvascular complications in patients with type 2 diabetes. Epidemiol Health. (2016) 38:e2016004. doi: 10.4178/epih.e2016004
12. Kennedy JM, Zochodne DW. Impaired peripheral nerve regeneration in diabetes mellitus. J Peripher Nervous Syst. (2010) 10:144–57. doi: 10.1111/j.1085-9489.2005.0010205.x
13. Yasuda H, Terada M, Maeda K, Kogawa S, Sanada M, Haneda M, et al. Diabetic neuropathy and nerve regeneration. Progr Neurobiol. (2003) 69:229–85. doi: 10.1016/S0301-0082(03)00034-0
14. Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. (2009) 136:642–55. doi: 10.1016/j.cell.2009.01.035
16. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are MicroRNA targets. Cell. (2005) 120:15–20. doi: 10.1016/j.cell.2004.12.035
17. Novák J, Olejníčková V, Tkáčová N, Santulli G. Mechanistic role of MicroRNAs in coupling lipid metabolism and atherosclerosis. Adv Exp Med Biol. (2015) 887:79–100. doi: 10.1007/978-3-319-22380-3_5
18. Belgardt BF, Ahmed K, Spranger M, Latreille M, Denzler R, Kondratiuk N, et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med. (2015) 21:619–27. doi: 10.1038/nm.3862
19. Sebastiani G, Po A, Miele E, Ventriglia G, Ceccarelli E, Bugliani M, et al. MicroRNA-124a is hyperexpressed in type 2 diabetic human pancreatic islets and negatively regulates insulin secretion. Acta Diabetol. (2015) 52:523–30. doi: 10.1007/s00592-014-0675-y
20. Olivieri F, Spazzafumo L, Bonafè M, Recchioni R, Prattichizzo F, Marcheselli F, et al. MiR-21-5p and miR-126a-3p levels in plasma and circulating angiogenic cells: relationship with type 2 diabetes complications. Oncotarget. (2015) 6:35372–82. doi: 10.18632/oncotarget.6164
21. Melo SA, Manel E. A precursor microRNA in a cancer cell nucleus: get me out of here! Cell Cycle. (2011) 10:922–5. doi: 10.4161/cc.10.6.15119
22. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. (2014) 15:509–24. doi: 10.1038/nrm3838
23. Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. (2001) 409:363–6. doi: 10.1038/35053110
24. Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. (2004) 432:235–40. doi: 10.1038/nature03120
25. Eamens AL, Smith NA, Curtin SJ, Wang MB, Waterhouse PM. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA. (2009) 15:2219–35. doi: 10.1261/rna.1646909
26. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. (2009) 136:215–33. doi: 10.1016/j.cell.2009.01.002
27. Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. (2001) 409:928–33. doi: 10.1038/35057149
28. Gao K, Wang J, Li L, Zhai Y, Ren Y, You H, et al. Polymorphisms in Four Genes (KCNQ1rs151290,KLF14rs972283,GCKRrs780094 andMTNR1Brs10830963) and Their Correlation with Type 2 Diabetes Mellitus in Han Chinese in Henan Province, China. Int J Environ Res Public Health. (2016) 13:E260. doi: 10.3390/ijerph13030260
29. Imamura M, Maeda S. Genetics of type 2 diabetes: the GWAS era and future perspectives [Review]. Endocr J. (2011) 58:723–39. doi: 10.1507/endocrj.EJ11-0113
30. Ntzani EE, Kavvoura FK. Genetic risk factors for type 2 diabetes: insights from the emerging genomic evidence. Current vasc Pharmacol. (2012) 10:147–55. doi: 10.2174/157016112799305030
31. Mehta NN. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes mellitus. Circ Cardiovasc Genet. (2012) 5:708–10. doi: 10.1161/CIRCGENETICS.112.965350
32. Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. (2010) 42:579–89. doi: 10.1038/ng.609
33. DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium, Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium, South Asian Type 2 Diabetes (SAT2D) Consortium, Mexican American Type 2 Diabetes (MAT2D) Consortium, Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. (2014) 46:234–44. doi: 10.1038/ng.2897
34. Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. (2008) 40:638–45. doi: 10.1038/ng.120
35. Chang YC, Chang EY, Chuang LM. Recent progress in the genetics of diabetic microvascular complications. World J Diab. (2015) 6:715–25. doi: 10.4239/wjd.v6.i5.715
36. Yuan L, Chu H, Wang M, Gu X, Shi D, Ma L, et al. Genetic variation in DROSHA 3'UTR regulated by hsa-miR-27b is associated with bladder cancer risk. PLoS ONE. (2013) 8:e81524. doi: 10.1371/journal.pone.0081524
37. Rah H, Jeon YJ, Lee BE, Kim JO, Shim SH, Lee WS, et al. Association of polymorphisms in microRNA machinery genes (DROSHA, DICER1, RAN, and XPO5) with risk of idiopathic primary ovarian insufficiency in Korean women. Menopause J North Am Menopause Soc. (2013) 20:1067–73. doi: 10.1097/GME.0b013e3182883907
38. Qin Y, Xia Y, Wu W, Han X, Lu C, Ji G, et al. Genetic variants in microRNA biogenesis pathway genes are associated with semen quality in a Han-Chinese population. Reproduct Biomed Online. (2012) 24:454–61. doi: 10.1016/j.rbmo.2012.01.006
39. Staff TPO. Correction: genetic variants in MicroRNA machinery genes are associate with idiopathic recurrent pregnancy loss risk. PLos ONE. (2014) 9:e95803. doi: 10.1371/journal.pone.0095803
40. Gong W, Xiao D, Ming G, Yin J, Zhou H, Liu Z. Type 2 diabetes mellitus-related genetic polymorphisms in microRNAs and microRNA target sites. J Diab. (2014) 6:279–89. doi: 10.1111/1753-0407.12143
41. Ciccacci C, Di Fusco D, Cacciotti L, Morganti R, D'Amato C, Greco C, et al. MicroRNA genetic variations: association with type 2 diabetes. Acta Diabetol. (2013) 50:867–72. doi: 10.1007/s00592-013-0469-7
42. Genuth S, Alberti KG, Bennett P, Buse J, Defronzo R, Kahn R, et al. Follow-up report on the diagnosis of diabetes mellitus. Diab Care. (2003) 26:3160–7. doi: 10.2337/diacare.26.11.3160
43. Gregg EW, Sattar N, Ali MK. The changing face of diabetes complications. Lancet Diab Endocrinol. (2016) 4:537–47. doi: 10.1016/S2213-8587(16)30010-9
44. Li Y, Wu QH, Jiao ML, Fan XH, Hu Q, Hao YH, et al. Gene-environment interaction between adiponectin gene polymorphisms and environmental factors on the risk of diabetic retinopathy. J Diab Investig. (2015) 6:56–66. doi: 10.1111/jdi.12249
45. Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. (2003) 19:376–82. doi: 10.1093/bioinformatics/btf869
46. Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. (2009) 19:519–23. doi: 10.1038/cr.2009.33
47. Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. (2005) 15:97–8. doi: 10.1038/sj.cr.7290272
48. El Hajj Chehadeh SW, Jelinek HF, Al Mahmeed WA, Tay GK, Odama UO, Elghazali GE, et al. Relationship betweenMTHFRC677T and A1298C gene polymorphisms and complications of type 2 diabetes mellitus in an Emirati population. Meta Gene. (2016) 9:70–5. doi: 10.1016/j.mgene.2016.04.002
49. Jafari N, Dogaheh HP, Bohlooli S, Oyong GG, Shirzad Z, Alibeiki F, et al. Expression levels of microRNA machinery components Drosha, Dicer and DGCR8 in human (AGS, HepG2, and KEYSE-30) cancer cell lines. Int J Clin Exp Med. (2013) 6:269–74. doi: 10.3727/096368912X656144
50. Özcan S. microRNAs in pancreatic β-cell physiology. Adv Exp Med Biol. (2015) 887:101–17. doi: 10.1007/978-3-319-22380-3_6
51. Rahimi G, Jafari N, Khodabakhsh M, Shirzad Z, Dogaheh HP. Upregulation of microRNA processing enzymes Drosha and Dicer in gestational diabetes mellitus. Gynecol Endocrinol. (2015) 31:156–9. doi: 10.3109/09513590.2014.969700
52. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
53. Grelier G, Voirin N, Ay AS, Cox DG, Chabaud S, Treilleux I, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. (2010) 101:673–83. doi: 10.1038/sj.bjc.6605193
54. Wu JF, Shen W, Liu NZ, Zeng GL, Yang M, Zuo GQ, et al. Down-regulation of Dicer in hepatocellular carcinoma. Med Oncol. (2011) 28:804–9. doi: 10.1007/s12032-010-9520-5
55. Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. (2010) 96:111–5. doi: 10.1111/j.1349-7006.2005.00015.x
56. Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. New Engl J Med. (2015) 359:2641–50. doi: 10.1056/NEJMoa0803785
57. Noren Hooten N, Martin-Montalvo A, Dluzen DF, Zhang Y, Bernier M, Zonderman AB, et al. Metformin-mediated increase in DICER1 regulates microRNA expression and cellular senescence. Aging Cell. (2016) 15:572–81. doi: 10.1111/acel.12469
58. Wronska A, Kurkowska-Jastrzebska I, Santulli G. Application of microRNAs in diagnosis and treatment of cardiovascular disease. Acta Physiol. (2015) 213:60–83. doi: 10.1111/apha.12416
59. Santulli G. MicroRNAs and endothelial (Dys) function. J Cell Physiol. (2016) 231:1638–44. doi: 10.1002/jcp.25276
60. Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. (2006) 16:203–8. doi: 10.1016/j.gde.2006.02.012
61. Galehdari H, Azarshin SZ, Bijanzadeh M, Shafiei M. Polymorphism studies on microRNA targetome of thalassemia. Bioinformation. (2018) 14:252–8. doi: 10.6026/97320630014252
62. Huang X, An Y, Li X, Wang D, Tan H, Lei J. Genetic variants in DICER1, DROSHA, RAN, and XPO5 genes and risk of pregnancy-induced hypertension. Pregnancy Hypertens. (2019) 16:161–6. doi: 10.1016/j.preghy.2019.04.005
63. Osuch-Wojcikiewicz E, Bruzgielewicz A, Niemczyk K, Sieniawska-Buccella O, Nowak A, Walczak A, et al. Association of polymorphic variants of miRNA processing genes with larynx cancer risk in a polish population. Biomed Res Int. (2015) 2015:298378. doi: 10.1155/2015/298378
64. Cho SH, Ko JJ, Kim JO, Jeon YJ, Yoo JK, Oh J, et al. 3′-UTR polymorphisms in the MiRNA machinery genes DROSHA, DICER1, RAN, and XPO5 are associated with colorectal cancer risk in a korean population. PLoS ONE. (2015) 10:e0131125. doi: 10.1371/journal.pone.0131125
65. Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. (2004) 303:95–8. doi: 10.1126/science.1090599
66. Izaurralde E, Jarmolowski A, Beisel C, Mattaj IW, Dreyfuss G, Fischer U. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. (1997) 137:27–35. doi: 10.1083/jcb.137.1.27
67. Vorpahl M, Schönhofer-Merl S, Michaelis C, Flotho A, Melchior F, Wessely R. The Ran GTPase-activating protein (RanGAP1) is critically involved in smooth muscle cell differentiation, proliferation and migration following vascular injury: implications for neointima formation and restenosis. PLoS ONE. (2014) 9:e101519. doi: 10.1371/journal.pone.0101519
68. Xia F, Dohi T, Martin NM, Raskett CM, Liu Q, Altieri DC. Essential role of the small GTPase Ran in postnatal pancreatic islet development. PLoS ONE. (2011) 6:e27879. doi: 10.1371/journal.pone.0027879
69. Olivieri F, Bonafè M, Spazzafumo L, Gobbi M, Prattichizzo F, Recchioni R, et al. Age- and glycemia-related miR-126-3p levels in plasma and endothelial cells. Aging. (2014) 6:771–86. doi: 10.18632/aging.100693
70. Shai I, Jiang R, Manson JE, Stampfer MJ, Willett WC, Colditz GA, et al. Ethnicity, obesity, and risk of type 2 diabetes in women: a 20-year follow-up study. Diab Care. (2006) 29:1585–90. doi: 10.2337/dc06-0057
71. Astrup A, Finer N. Redefining type 2 diabetes: “diabesity” or “obesity dependent diabetes mellitus”? Obes Rev. (2000) 1:57–9. doi: 10.1046/j.1467-789x.2000.00013.x
72. Hu FB, Manson JE, Stampfer MJ, Colditz G, Liu S, Solomon CG, et al. Diet, lifestyle, and the risk of type 2 diabetes mellitus in women. New Engl J Med. (2001) 345:790–7. doi: 10.1056/NEJMoa010492
73. de Mutsert R, Sun Q, Willett WC, Hu FB, van Dam RM. Overweight in early adulthood, adult weight change, and risk of type 2 diabetes, cardiovascular diseases, and certain cancers in men: a cohort study. Am J Epidemiol. (2014) 179:1353–65. doi: 10.1093/aje/kwu052
74. Reusch JEB. Diabetes, microvascular complications, and cardiovascular complications: what is it about glucose? J Clin Investig. (2003) 112:986. doi: 10.1172/JCI200319902
75. Lindholm LH, Ibsen H, Borch-Johnsen K, Olsen MH, Wachtell K, Dahlöf B, et al. Risk of new-onset diabetes in the Losartan Intervention for Endpoint reduction in hypertension study. J Hypertens. (2002) 20:1879–86. doi: 10.1097/00004872-200209000-00035
76. Weinberger MH, Fineberg NS, Fineberg SE. The influence of blood pressure and carbohydrate tolerance on vascular compliance in humans. Am J Hypertens. (2002) 15:678–82. doi: 10.1016/S0895-7061(02)02955-2
77. Bell DS. Hypertension and antihypertensive therapy as risk factors for type 2 diabetes mellitus. New Engl J Med. (2000) 343:580–1. doi: 10.1056/NEJM200008243430813
78. Marathe PH, Gao HX, Close KL. American diabetes association standards of medical care in diabetes 2017. J Diab. (2017) 9:320–4. doi: 10.1111/1753-0407.12524
79. Cheng WY, Jiang YD, Chuang LM, Huang CN, Heng LT, Wu HP, et al. Quantitative sensory testing and risk factors of diabetic sensory neuropathy. J Neurol. (1999) 246:394–8. doi: 10.1007/s004150050370
80. Choe C, Edelman SV. The role of self-monitoring of blood glucose during the treatment of type 2 diabetes with medications targeting postprandial hyperglycemia. Southern Med J. (2007) 100:1123–31. doi: 10.1097/01.smj.0000286751.78656.45
Keywords: T2DM, vascular complication, miRNA processing gene, polymorphism, interaction, RAN gene, DICER1 gene
Citation: Wen Z, Zou X, Xie X, Zheng S, Chen X, Zhu K, Dong S, Liang J, Huang X, Liu D, Wang Y, Liu Y, Wu J, Ying Y, Liu K, Lu C, Zhang B, Yang G, Jing C and Nie L (2019) Association of Polymorphisms in miRNA Processing Genes With Type 2 Diabetes Mellitus and Its Vascular Complications in a Southern Chinese Population. Front. Endocrinol. 10:461. doi: 10.3389/fendo.2019.00461
Received: 26 December 2018; Accepted: 25 June 2019;
Published: 12 July 2019.
Edited by:
Jan Polák, Charles University, CzechiaReviewed by:
Shalini Mani, Jaypee Institute of Information Technology, IndiaCopyright © 2019 Wen, Zou, Xie, Zheng, Chen, Zhu, Dong, Liang, Huang, Liu, Wang, Liu, Wu, Ying, Liu, Lu, Zhang, Yang, Jing and Nie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guang Yang, dHlhbmdndWFuZ0BlbWFpbC5qbnUuZWR1LmNu; Chunxia Jing, amN4cGhkQGdtYWlsLmNvbQ==; Lihong Nie, bGlob25nbmllMDAxQG91dGxvb2suY29t
†These authors have contributed equally to the work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.