- 1Department of Biology, University of Alabama at Birmingham, Birmingham, AL, United States
- 2National Center for Cool and Cold Water Aquaculture, Agricultural Research Service (USDA), Kearneysville, WV, United States
Methionine restriction (MR) has been studied extensively over the last 25 years for its role in altering metabolic hallmarks of disease. Animals subjected to MR, display changes in metabolic flexibility demonstrated by increases in energy expenditure, glucose tolerance, and lifespan. These changes have been well characterized in a number of model systems and significant progress has been made in understanding how hepatic fibroblast growth factor 21 links MR to several components of its metabolic phenotype. Despite these advances, a complete understanding of mechanisms engaged by dietary MR remains elusive. In this review, we offer a brief history of MR and its known mechanisms associated with stress, metabolism, and lifespan extension. We consider the role of epigenetics in the response of animals to MR and propose a novel epigenetic pathway involving the regulation of microRNAs during MR.
Introduction
Methionine restriction (MR) is known to produce lifespan extension and has been more recently investigated for its role in improving metabolic health. While much is known about the resulting phenotype following MR, a complete picture of mechanisms involved in MRs metabolic phenotype is less clear. At a molecular level, methionine metabolism creates S-adenosylmethionine (SAM), which acts as a master methyl donor. Methylation of cytosines in promoter regions of genes can regulate gene expression by controlling binding of transcription factors (1). Changes in methionine availability through dietary MR have the potential to decrease these available methyl donors and thus change transcriptional status of certain genes. One of these potential genes is fibroblast growth factor 21 (FGF21) which has been implicated in the metabolic responses to MR (i.e., increased energy intake and expenditure as well as increased insulin sensitivity) (2, 3). While increases in circulating and hepatic FGF21 are responsible for certain aspects of MRs metabolic phenotype, FGF21−/− mice did not exhibit changes in transcriptional responses to MR in the liver (2). This FGF21 independent mechanism could potentially be linked to epigenetic changes that take place during MR in the liver when there is a reduction in methyl donors.
In this review, we explore the mechanisms of MR that could drive these epigenetic responses. One possible mechanism that has emerged as a factor in MR phenotypes is microRNAs (miRNAs). Studies have shown that MR alters miRNA expression levels in rainbow trout myosatellite cells (4, 5) and bone structures of mice (6), supporting a proposed connection between MR and miRNAs. This review will discuss the history and phenotypes associated with MR, as well as the role for miRNAs in MR phenotypes.
Methionine Restriction
Dietary interventions have been studied for many decades in fields from nutrition to healthy aging. The most investigated intervention has been caloric restriction (CR), which has been shown to increase lifespan and health span (7). While CR, defined as a decrease of 30–60% from ad libitum feeding, is a well-known and effective treatment, first demonstrated in 1935 (8), compliance is often low. Alternatives to CR such as protein restriction (de novo amino acid restriction) (9, 10) have been used in patients with renal failure and more recently in mice as a lifespan extension strategy (11). Following studies on protein restriction, Segall et al. began to explore the effects of long-term tryptophan restriction in rats (12–14). While this proved to be a lifespan extension strategy hypothesized to work by decreasing levels of serotonin in the brain, early mortality of animals was a significant problem and led to abandonment of this strategy (15, 16). Orentreich et al. were the first to propose MR as an alternative therapy to increase lifespan (17). Early on, it was established that the effects seen during MR were not due to de novo CR, but rather some other mechanism. Orentreich et al. pair-fed control diets based on amounts consumed by rats receiving MR diets to control for differences in food consumption and size to investigate whether the extension in lifespan was due to CR rather than MR. These pair-fed animals had no impairment in growth, meaning the modest reduction in feed intake by the MR animals did not account for the extension in lifespan. Further investigation revealed that MR animals weighed less but consumed more food per gram than their age matched counterparts (i.e., they were slightly hyperphagic).
To identify whether there is genetic variation in the MR response, Zimmerman et al. evaluated MR diets in three diverse strains of rat; Brown Norway, Sprague-Dawley, and Wistar Hannover (18), and demonstrated comparable increases in median lifespan in all of these strains. Miller et al. were the first to report increased lifespan in (BALB/cJ × C57BL/6J) F1 mice on MR diets. In addition, these MR mice displayed lower amounts of plasma Insulin-Like Growth Factor-1 (IGF-1), insulin, glucose, and thyroxine (T4) (19), suggesting novel markers of an endocrine response. These markers were validated in F344 rats fed an MR diet, where MR rats had lower levels of glucose and leptin with corresponding increases in adiponectin, triiodothyronine (T3), and daily energy expenditure when compared to pair-fed controls (20). In the same study, a separate cohort of MR animals were subjected to an oral glucose tolerance test to test the hypothesis that MR preserves insulin action with age. The MR animals responded in a similar fashion to the oral glucose tolerance test as rats fed a control diet at 0, 23, and 72 weeks, although MR animals showed a marked decrease in area under the curve for insulin at both 23 and 72 weeks, demonstrating a preservation of insulin action with age (20). Furthermore, these rats displayed decreased accumulation of visceral fat that, in combination with the endocrine response, supports MR as a strategy to extend lifespan in rodents.
In addition, data support the role of decreased fat deposition, preserved insulin sensitivity, and disruption of the lipogenic/lipolytic balance in adipose tissue as mediators of MRs metabolic phenotype (20–22). This balance was shown to be distrupted by MR through a cycle of increased lipolysis and increased lipogenesis in vitro hypothesized to lead to decreases in adipose tissue in older F344 rats (21). In addition, a study found that young and mature animals, as well as obesity prone Osborne–Mendel rats, on a MR diet exhibited long-term increases in energy expenditure and uncoupling protein-1 (UCP-1) in both brown and white adipose tissue. This change in UCP-1 was also accompanied by decreased leptin and increased adiponectin proposing a remodeling of the adipose tissue during MR (23). Authors suggested that MR acts to increase energy expenditure and decrease fat deposition by lowering metabolic efficiency during the night when increases in lipogenesis typically occur (23). These changes explain the extension of healthspan observed in these animals with a complete mechanism for MRs ability to decrease metabolic efficiency (i.e., hyperphagia with decreased growth) (21–24). A recent study detailed that animals introduced to the diet either postweaning or at 80% of mature size had differential effects of hyperphagia (~50 and ~20% increase in energy intake per unit body weight in juvenile and adults, respectively) indicating that the MR diet has differential effects on energy intake and expenditure depending on age (25).
The work detailed thus far has focused on the metabolic effects of MR in mice and rats, which occupies much of the current literature. However, work has also been done in the unique teleost fish model, the rainbow trout; Oncorhynchus mykiss. Methionine requirements of these fish are well-known due to their economically important aquaculture status (26–29) and MR has been studied in part due to the implementation of naturally methionine-deficient plant-based diets. In addition, rainbow trout are naturally glucose intolerant making them a potential model for studying the glucose-intolerant phenotype associated with human type-II diabetes and metabolic syndrome (30). In rainbow trout, MR, paired with carbohydrate-enriched diets, was shown to reduce genes associated with de novo fatty acid synthesis and reduce circulating plasma glucose 6 h postprandial (30); however, these results must be interpreted cautiously as the design of the diets included no methionine (i.e., methionine deprivation), but adequate cysteine levels. On the other hand, consistently, juvenile rainbow trout fed a formulated MR diet over 6 weeks displayed transcriptional markers of GCN2/eIF2α activation mirroring the effects observed in mammals (31). Conversely, brood stock females fed a similar MR diet for 6 months prior to spawning had reduced triacylglycerol levels with increased total cholesterol and LDL-cholesterol, opposing the cholesterol lowering effects observed in mammals (32). In addition, 48 h of MR in vitro in trout hepatocytes resulted in greater levels of glucose uptake due primarily to an increase in the sodium-glucose transporter 2 (33). While the literature is less extensive on the metabolic effects associated with MR in teleosts, it is clear that there is at least a partial conservation of the MR phenotype.
Physiological Mechanisms of MR
Lifespan Extension
A proposed mechanism of lifespan extension associated with CR is a decreased rate of mitochondrial reactive oxygen species generation (mitROS) (34). It has been shown that some long-lived species have markedly lower rates of mitROS production compared to their short-lived counterparts (35, 36). Similarly, animals on a MR diet display lower rates of mitROS production and less oxidative damage of mtDNA in both the heart and liver (37). Previous studies have also established that neither carbohydrate restriction nor lipid restriction can replicate the phenotypes seen in CR, PR, and MR (38, 39). This information leads to the hypothesis that changes in mitROS production during both CR and PR could be attributed in part to MR. MR-induced reductions in mitROS production in the heart occurred primarily at complex I, previously established to be the main complex targeted in CR (40). However, in liver MR reduced mitROS in complexes I/III, while CR appears to reduce mitROS only at complex I (41). It should be noted that the studies done on mitROS production during MR had similar effects at both 40 and 80% MR. Since the time of publication, it has been shown that most physiological effects of MR only occur at 80% (42) so the results must be interpreted cautiously.
An additional potential mechanism in the life span response to MR includes an enhanced capacity for a cellular autophagic response and the concomitant acidification of the vacuole (43). Studies done in Saccharomyces cerevisiae (Bakers yeast) utilized three different strains of yeast which varied in their ability to synthesize methionine (100% capability, moderate impairment, no de novo synthesis). The authors found that lifespan extension was achieved in both the moderate and devoid strains in normal media with the devoid strain demonstrating decreased survival in media with a 90% reduction in methionine. This extension in lifespan was found along with a sharp increase in autophagy followed by acidification of the vacuoles. Additionally, when key genes essential to autophagy were deleted (ATG 5,7,8) this increase in lifespan was ameliorated (43), providing some support for a relationship between autophagic capacity and lifespan, at least in yeast [full review Ref. (44)], although this mechanism has yet to be investigated in MR mammals.
Metabolic Changes
Two mechanisms related to the sensing of amino acid deficiency may partially explain the metabolic adaptations that take place during MR (Figure 1). A decrease in methionine leads to an increase in uncharged tRNAs that can activate the general control non-derepressible 2 (GCN2) kinase (45, 46), leading to metabolic adaptation during MR. However, GCN2 may not be indispensable for the response as metabolic adaptation can also take place through a noncanonical PKR-like endoplasmic reticulum kinase (PERK)/NRF2 pathway (47). Both of these mechanisms are significant for the MR-induced integrated stress response (ISR) that contributes to cellular homeostasis and regulation of the physiological response. They converge through their mutual regulation of eukaryotic initiation factor-2α (eIF2α) phosphorylation and the subsequent effects on translational capacity via eIF2 and synthesis of the active full-length activating transcription factor 4 (ATF4). Active ATF4 is defined as an amino acid sensor (48) and master regulator (49) of metabolism central for controlling expression of genes associated with the stress response, including genes related to lipid metabolism (50), autophagy (51), and maintenance of oxidative stress (52). However, recent evidence indicates that MR restriction can induce ATF4 independent of the eIF2 pathway (53), suggesting that additional mechanisms may be regulating MR-induced ATF4 response.
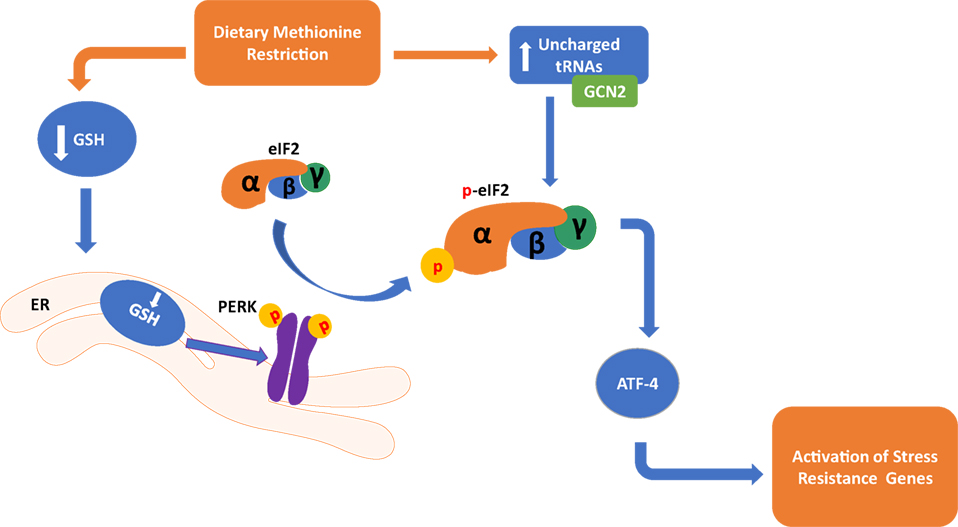
Figure 1. Dietary methionine restriction has been shown to occur through two distinct pathways in the cell. In the endoplasmic reticulum, a reduction of Glutathione (GSH) causes an activation of the PKR-like endoplasmic reticulum kinase (PERK), which activates eukaryotic translation initiation factor 2a (elF2a). In the cytoplasm, a buildup of unchanged tRNAs activates general control non-derepressible 2 (GCN2) which in turn also activates elF2a. Both of these pathways converge in the activation of activating transcription factor 4 (ATF4) and activation of genes that respond to stress.
Molecular Mechanisms of MR
Epigenetics
Nutrigenomics is a rapidly evolving area of research that identifies how nutrition and diet interact with the genome and affect gene expression (54). Epigenetic regulation includes methylation of cytosines within genes or regulatory regions and can activate, but more commonly inhibit, gene expression. Epigenetic regulation also includes histone modifications as methylation, phosphorylation, and acetylation that affect chromatin structure and transcriptional activation. Regulation of methylation status is predominantly controlled by de novo methyltransferases and demethylases that acquire their methyl groups from the downstream methionine metabolite, SAM, converting it to S-adenosylhomocysteine (SAH).
Age-related changes in the epigenome are implicated in the aging process (55) and although it is established that the epigenome responds to nutrient signals, a direct link between the aging response and nutrient restriction-induced epigenetic effects has yet to be clearly established (56). However, recent evidence indicates that age-related methylation drift, which is hypothesized to be a determinant of mammalian lifespan (57), is attenuated by CR in rhesus monkeys and mice and affects gene expression (58).
Epigenetic effects of protein or MR have largely emphasized consumption during gestation and subsequent effects on offspring development. This concept of “nutritional programming” actually supports dietary supplementation of methyl donors like folate, choline, and methionine to improve offspring health and growth potential, and the extent to which this response is regulated by differential methylation of genes critical for development remains unclear (59). There is, however, a small body of work examining the epigenetic response to MR as it relates to aging. In vitro findings indicate that physiologically relevant concentrations of methionine in cell culture media alter the SAM/SAH ratio and methylation status of H3K4trimethyl, leading to regulation of gene transcription (60). The methionine levels in the serum of humans exhibit enough variation during MR to alter histone methylation (60). Similar findings were observed in adult (but not young) mice in which short-term (12 weeks) MR (0.12%-MR, 0.84%-CD) decreased SAH concentration and increased global DNA methylation in liver, although the opposite was observed in adipose tissue (61). These studies collectively support a role for changes in epigenetic marks by MR, although additional research is needed to further characterize the gene-specific responses.
MicroRNAs
MicroRNAs are small endogenous 18–22 base pair nucleotide sequences (62) that play roles in mediating posttranscriptional gene silencing in mammals, plants, and invertebrate organisms (63, 64). They are synthesized from primary miRNAs by two enzymes Drosha in the nucleus and Dicer in the cytoplasm (65) and are bound by Argonaute subfamily of proteins before integration into the RISC complex. After integration miRNAs have the ability to post transcriptionally regulate gene expression through the formation of RNA duplexes (62). The miRNA profile changes with age and numerous miRNAs that are affected by age are also regulated by CR (66), suggesting these regulatory RNA molecules are associated with CR-induced extension of lifespan. Many of these miRNAs associated with the IGF-1 PI3K/AKT mTOR signaling pathways are important for nutrient sensing and subsequent physiological response during CR.
Recent evidence also supports a role for miRNAs in the MR phenotype in teleosts and mammals. Rainbow trout myosatellite cells exposed to methionine-deficient cell media regulate miRNAs that reduce their capacity for differentiation (miR-133a, miR-206, and miR-210) (4). Rainbow trout fed an MR (0.775%) diet for 4 weeks had lower levels of miR-133a in the skeletal muscle at 4 weeks and increased glucose tolerance following a glucose challenge at 8 weeks (5). Mice consuming an MR diet (0.12%) exhibit increased expression of miR-133a, miR-335-5p, and miR-204 in the bone marrow as well as miR-31 in the plasma and liver (5, 6, 67, 68). Although these studies did not directly investigate the aging response, they do indicate that miRNAs are regulated by methionine availability and potentially play a role in the metabolic phenotype.
While miRNAs have traditionally been known to repress transcription, evidence suggests that cells undergoing an ISR, such as those under MR, may activate miRNAs to regulate a unique suite of mRNAs to maintain homeostasis (69). As previously mentioned, a potential mechanism of MR is through the GCN2 amino acid deprivation pathway and the PERK/NRF2 (Figure 1) pathway. Both of these kinases phosphorylate eIF2α, which activates ATF4 and induces the formation of stress granules (70) containing miRNAs, mRNA targets, and Argonaute proteins (71). Similar to MR, leucine deficiency activates GCN2/ATF4 (72), which increases expression of miRNA-212-5p that subsequently reduces lipid accumulation (72) and enhances gluconeogenesis (73). Although the effect of leucine restriction on lifespan is opposite that of MR, activation of GCN2/ATF4 and subsequent regulation of specific miRNAs appears to be a common response to cellular stress induced by amino acid restriction.
While the notion of miRNA involvement in the MR phenotype is new, it is well-known that a few miRNAs regulate cellular stress response in other situations. miR-211 is known as a pro-survival miRNA during ER stress activated by the PERK pathway (74). In this circumstance, miR-211 is used to attenuate the amount of the pro-apoptotic transcription factor chop and keep the cell from initiating apoptosis during periods of short-term ER stress. miR-122 causes a repression of the cationic amino acid transporter-1 under normal conditions. Under varying conditions of stress, this miR can de-repress the mRNA transcript and cause it to be localized to the polysomes (75). For a full review of how miRNAs are regulated during stress responses, see Leung and Sharp (76). Because MR is known to activate the PERK pathway without induction of ER stress (47) the connection between MR and miRNAs must be interpreted carefully and warrants further study.
Conclusion and Future Perspectives
The evidence presented clearly shows MRs role in altering metabolic phenotypes in both mammals and teleosts. The activation of miRNAs during MR provides a potential link between changes in methylation and the ISRs in cells. Studies utilizing rainbow trout myosatellite cells in vitro and juvenile rainbow trout in vivo (4, 5) have shown that methionine can regulate the level of expression of miRNAs in teleosts (Figure 2). Studies done in mammalian systems have also shown that miRNAs are differentially regulated in the plasma, liver, and bone marrow of MR mice (6). This review explores mechanisms responsible for the MR phenotype including miRNAs, amino acid starvation, and stress response pathways.
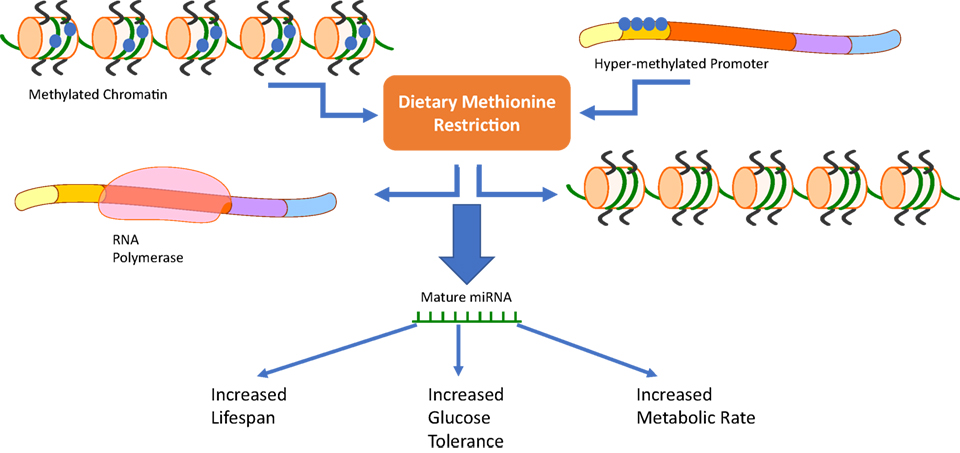
Figure 2. Our review suggests that a third pathway should be added to the already known mechanisms of methionine restriction (MR). Changes in chromatin methylation or methylation status of DNA promoters due to dietary MR has the ability to activate or repress microRNAs (miRNAs) involved in the cells response to MR. These distinct changes could then be related to MRs phenotype characterized by increased lifespan, glucose tolerance, and metabolic rate.
Future research should investigate miRNAs in circulation (77) during MR. miRNAs found to be altered during MR (78) can then be analyzed for their role in controlling muscle-specific transcription factors like MyoD and myogenin (79) and can also be explored in other tissues to observe conservation of function during stress responses in tissue (80). Indeed, the master regulator of many of these mechanisms may lie in epigenetic changes that occur during MR. These changes have been hinted at in previous literature (60, 61) but have not yet been fully explored.
Author Contributions
ML: writing, reviewing, and conception. KF: writing. BC: writing and reviewing. PB: writing, reviewing, and conception.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
The first author of this manuscript was supported by a training grant awarded to the UAB Nutrition and Obesity Research Center (NHLBI T32 HL105349). The Nutrition and Obesity Research Center is supported by Award Number P30DK056336 from the National Institute of Diabetes and Digestive and Kidney Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases or the National Institutes of Health.
References
1. Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet (2007) 39(1):61–9. doi:10.1038/ng1929
2. Wanders D, Forney LA, Stone KP, Burk DH, Pierse A, Gettys TW. FGF21 mediates the thermogenic and insulin-sensitizing effects of dietary methionine restriction but not its effects on hepatic lipid metabolism. Diabetes (2017) 66(4):858–67. doi:10.2337/db16-1212
3. Lees EK, Król E, Grant L, Shearer K, Wyse C, Moncur E, et al. Methionine restriction restores a younger metabolic phenotype in adult mice with alterations in fibroblast growth factor 21. Aging Cell (2014) 13(5):817–27. doi:10.1111/acel.12238
4. Latimer M, Sabin N, Le Cam A, Seiliez I, Biga P, Gabillard JC. miR-210 expression is associated with methionine-induced differentiation of trout satellite cells. J Exp Biol (2017) 220(Pt 16):2932–8. doi:10.1242/jeb.154484
5. Latimer MN, Cleveland BM, Biga PR. Dietary methionine restriction: effects on glucose tolerance, lipid content and micro-RNA composition in the muscle of rainbow trout. Comp Biochem Physiol C Toxicol Pharmacol (2017). doi:10.1016/j.cbpc.2017.10.012
6. Plummer J, Park M, Perodin F, Horowitz MC, Hens JR. Methionine-restricted diet increases miRNAs that can target RUNX2 expression and alters bone structure in young mice. J Cell Biochem (2017) 118(1):31–42. doi:10.1002/jcb.25604
7. Mercken EM, Carboneau BA, Krzysik-Walker SM, de Cabo R. Of mice and men: the benefits of caloric restriction, exercise, and mimetics. Ageing Res Rev (2012) 11(3):390–8. doi:10.1016/j.arr.2011.11.005
8. McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size. 1935. Nature (1989) 5:155–71.
9. Klahr S, Levey AS, Beck GJ, Caggiula AW, Hunsicker L, Kusek JW, et al. The effects of dietary protein restriction and blood-pressure control on the progression of chronic renal disease. Modification of Diet in Renal Disease Study Group. N Engl J Med (1994) 330(13):877–84. doi:10.1056/NEJM199403313301301
10. Pedrini MT, Levey AS, Lau J, Chalmers TC, Wang PH. The effect of dietary protein restriction on the progression of diabetic and nondiabetic renal diseases: a meta-analysis. Ann Intern Med (1996) 124(7):627–32. doi:10.7326/0003-4819-124-7-199604010-00002
11. JH Chen, Martin-Gronert MS, Tarry-Adkins J, Ozanne SE, et al. Maternal protein restriction affects postnatal growth and the expression of key proteins involved in lifepspan regulation in mice. PLoS One (2009) 4(3):e4950. doi:10.1371/journal.pone.0004950
12. Segall P. Long-term tryptophan restriction and aging in the rat. Aktuelle Gerontol (1977) 7(10):535–8.
13. Segall PE, Ooka H, Rose K, Timiras PS. Neural and endocrine development after chronic tryptophan deficiency in rats: I. Brain monoamine and pituitary responses. Mech Ageing Dev (1978) 7(1):1–17. doi:10.1016/0047-6374(78)90048-9
14. Segall PE, Timiras PS. Patho-physiologic findings after chronic tryptophan deficiency in rats: a model for delayed growth and aging. Mech Ageing Dev (1976) 5(2):109–24. doi:10.1016/0047-6374(76)90012-9
15. Marte MLD, Enesco HE. Influence of low tryptophan diet on survival and organ growth in mice. Mech Ageing Dev (1986) 36:161–71. doi:10.1016/0047-6374(86)90017-5
16. Ooka H, Segall PE, Timiras PS. Histology and survival in age-delayed low-tryptophan-fed rats. Mech Ageing Dev (1988) 43(1):79–98. doi:10.1016/0047-6374(88)90099-1
17. Orentreich N, Matias JR, DeFelice A, Zimmerman JA. Low methionine ingestion by rats extends life span. J Nutr (1993) 123(2):269–74.
18. Zimmerman JA, Malloy V, Krajcik R, Orentreich N, et al. Nutritional control of aging. Exp Gerontol (2003) 38(1–2):47–52. doi:10.1016/S0531-5565(02)00149-3
19. Miller RA, Buehner G, Chang Y, Harper JM, Sigler R, Smith-Wheelock M, et al. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell (2005) 4(3):119–25. doi:10.1111/j.1474-9726.2005.00152.x
20. Malloy VL, Krajcik RA, Bailey SJ, Hristopoulos G, Plummer JD, Orentreich N. Methionine restriction decreases visceral fat mass and preserves insulin action in aging male Fischer 344 rats independent of energy restriction. Aging Cell (2006) 5(4):305–14. doi:10.1111/j.1474-9726.2006.00220.x
21. Perrone CE, Mattocks DA, Hristopoulos G, Plummer JD, Krajcik RA, Orentreich N. Methionine restriction effects on 11 -HSD1 activity and lipogenic/lipolytic balance in F344 rat adipose tissue. J Lipid Res (2008) 49(1):12–23. doi:10.1194/jlr.M700194-JLR200
22. Stone KP, Wanders D, Orgeron M, Cortez CC, Gettys TW. Mechanisms of increased in vivo insulin sensitivity by dietary methionine restriction in mice. Diabetes (2014) 63(11):3721–33. doi:10.2337/db14-0464
23. Hasek BE, Stewart LK, Henagan TM, Boudreau A, Lenard NR, Black C, et al. Dietary methionine restriction enhances metabolic flexibility and increases uncoupled respiration in both fed and fasted states. Am J Physiol Regul Integr Comp Physiol (2010) 299(3):R728–39. doi:10.1152/ajpregu.00837.2009
24. Wanders D, Burk DH, Cortez CC, Van NT, Stone KP, Baker M, et al. UCP1 is an essential mediator of the effects of methionine restriction on energy balance but not insulin sensitivity. FASEB J (2015) 29(6):2603–15. doi:10.1096/fj.14-270348
25. Wanders D, Forney LA, Stone KP, Hasek BE, Johnson WD, Gettys TW, et al. The components of age-dependent effects of dietary methionine restriction on energy balance in rats. Obesity (Silver Spring) (2018) 26(4):740–6. doi:10.1002/oby.22146
26. Kim K-I, Kayes TB, Amundson CH. Requirement for sulfur amino acids and utilization of D-methionine by rainbow trout (Oncorhynchus mykiss). Aquaculture (1992) 101(1–2):95–103. doi:10.1016/0044-8486(92)90235-D
27. Walton MJ, Cowey CB, Adron JW. Methionine metabolism in rainbow trout fed diets of differing methionine and cystine content. J Nutr (1982) 112(8):1525–35. doi:10.1093/jn/112.8.1525
28. Rumsey GL, Page JW, Scott ML. Methionine and cystine requirements of rainbow trout. Prog Fish Cult (1983) 45(3):139–43. doi:10.1577/1548-8659(1983)45[139:MACROR]2.0.CO;2
29. NRC. Nutrient Requirements of Fish and Shrimp. Washington, DC: The National Academies Press (2011).
30. Craig PM, Moon TW. Methionine restriction affects the phenotypic and transcriptional response of rainbow trout (Oncorhynchus mykiss) to carbohydrate-enriched diets. Br J Nutr (2013) 109(3):402–12. doi:10.1017/S0007114512001663
31. Skiba-Cassy S, Geurden I, Panserat S, Seiliez I. Dietary methionine imbalance alters the trascnriptional regulation of genes involved in glucose, lipid and amino acid metabolism in the liver of raibow trout (Oncorhynchus mykiss). Aquaculture (2015) 454:56–65. doi:10.1016/j.aquaculture.2015.12.015
32. Fontagne-Dicharry S, Alami-Durante H, Aragão C, Kaushik SJ, Geurden I. Parental and early-feeding effects of dietary methionine in rainbow trout (Oncorhynchus mykiss). Aquaculture (2017) 469:16–27. doi:10.1016/j.aquaculture.2016.11.039
33. Craig PM, Massarsky A, Moon TW. Understanding glucose uptake during methionine deprivation in incubated rainbow trout (Oncorhynchus mykiss) hepatocytes using a non-radioactive method. Comp Biochem Physiol B Biochem Mol Biol (2013) 166(1):23–9. doi:10.1016/j.cbpb.2013.06.005
34. Gredilla R, Barja G. Minireview: the role of oxidative stress in relation to caloric restriction and longevity. Endocrinology (2005) 146(9):3713–7. doi:10.1210/en.2005-0378
35. Barja G. Aging in vertebrates, and the effect of caloric restriction: a mitochondrial free radical production-DNA damage mechanism? Biol Rev Camb Philos Soc (2004) 79(2):235–51. doi:10.1017/S1464793103006213
36. Barja G. Free radicals and aging. Trends Neurosci (2004) 27(10):595–600. doi:10.1016/j.tins.2004.07.005
37. Sanz A, Caro P, Ayala V, Portero-Otin M, Pamplona R, Barja G. Methionine restriction decreases mitochondrial oxygen radical generation and leak as well as oxidative damage to mitochondrial DNA and proteins. FASEB J (2006) 20(8):1064–73. doi:10.1096/fj.05-5568com
38. Sanz A, Caro P, Sanchez JG, Barja G. Effect of lipid restriction on mitochondrial free radical production and oxidative DNA damage. Ann N Y Acad Sci (2006) 1067:200–9. doi:10.1196/annals.1354.024
39. Sanz A, Gómez J, Caro P, Barja G. Carbohydrate restriction does not change mitochondrial free radical generation and oxidative DNA damage. J Bioenerg Biomembr (2006) 38(5–6):327–33. doi:10.1007/s10863-006-9051-0
40. Gredilla R, Sanz A, Lopez-Torres M, Barja G. Caloric restriction decreases mitochondrial free radical generation at complex I and lowers oxidative damage to mitochondrial DNA in the rat heart. FASEB J (2001) 15(9):1589–91. doi:10.1096/fj.00-0764fje
41. Gredilla R, Barja G, Lopez-Torres M. Effect of short-term caloric restriction on H2O2 production and oxidative DNA damage in rat liver mitochondria and location of the free radical source. J Bioenerg Biomembr (2001) 33(4):279–87. doi:10.1023/A:1010603206190
42. Forney LA, Wanders D, Stone KP, Pierse A, Gettys TW. Concentration-dependent linkage of dietary methionine restriction to the components of its metabolic phenotype. Obesity (Silver Spring) (2017) 25(4):730–8. doi:10.1002/oby.21806
43. Ruckenstuhl C, Netzberger C, Entfellner I, Carmona-Gutierrez D, Kickenweiz T, Stekovic S, et al. Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification. PLoS Genet (2014) 10(5):e1004347. doi:10.1371/journal.pgen.1004347
44. Tyler JK, Johnson JE. The role of autophagy in the regulation of yeast life span. Ann N Y Acad Sci (2018). doi:10.1111/nyas.13549
45. Plaisance EP, Van N, Orgeron M, McDaniel AN, Behrens PH, Gettys TW, et al. Role of general control nonderepressible 2 (GCN2) kinase in mediating responses to dietary methionine restriction. FASEB J (2012) 26(1).
46. Gallinetti J, Harputlugil E, Mitchell JR. Amino acid sensing in dietary-restriction-mediated longevity: roles of signal-transducing kinases GCN2 and TOR. Biochem J (2013) 449(1):1–10. doi:10.1042/BJ20121098
47. Wanders D, Stone KP, Forney LA, Cortez CC, Dille KN, Simon J, et al. Role of GCN2-independent signaling through a noncanonical PERK/NRF2 pathway in the physiological responses to dietary methionine restriction. Diabetes (2016) 65(6):1499–510. doi:10.2337/db15-1324
48. Maurin AC, Chaveroux C, Lambert-Langlais S, Carraro V, Jousse C, Bruhat A, et al. The amino acid sensor GCN2 biases macronutrient selection during aging. Eur J Nutr (2012) 51(1):119–26. doi:10.1007/s00394-011-0205-4
49. Hinnebusch AG, Natarajan K. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot Cell (2002) 1(1):22–32. doi:10.1128/EC.01.1.22-32.2002
50. Wilson GJ, Lennox BA, She P, Mirek ET, Al Baghdadi RJ, Fusakio ME, et al. GCN2 is required to increase fibroblast growth factor 21 and maintain hepatic triglyceride homeostasis during asparaginase treatment. Am J Physiol Endocrinol Metab (2015) 308(4):E283–93. doi:10.1152/ajpendo.00361.2014
51. B’chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, et al. The eIF2 alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res (2013) 41(16):7683–99. doi:10.1093/nar/gkt563
52. Rajesh K, Krishnamoorthy J, Kazimierczak U, Tenkerian C, Papadakis AI, Wang S, et al. Phosphorylation of the translation initiation factor eIF2alpha at serine 51 determines the cell fate decisions of Akt in response to oxidative stress. Cell Death Dis (2015) 6:e1591. doi:10.1038/cddis.2014.554
53. Pettit AP, Jonsson WO, Bargoud AR, Mirek ET, Peelor FF III, Wang Y, et al. Dietary methionine restriction regulates liver protein synthesis and gene expression independently of eukaryotic initiation factor 2 phosphorylation in mice. J Nutr (2017) 147(6):1031–40. doi:10.3945/jn.116.246710
54. Park JH, Yoo Y, Park YJ. Epigenetics: linking nutrition to molecular mechanisms in aging. Prev Nutr Food Sci (2017) 22(2):81–9. doi:10.3746/pnf.2017.22.2.81
55. McClay JL, Aberg KA, Clark SL, Nerella S, Kumar G, Xie LY, et al. A methylome-wide study of aging using massively parallel sequencing of the methyl-CpG-enriched genomic fraction from blood in over 700 subjects. Hum Mol Genet (2014) 23(5):1175–85. doi:10.1093/hmg/ddt511
56. Moreno CL, Mobbs CV. Epigenetic mechanisms underlying lifespan and age-related effects of dietary restriction and the ketogenic diet. Mol Cellular Endocrinol (2017) 455:33–40. doi:10.1016/j.mce.2016.11.013
57. Mendelsohn AR, Larrick J. Epigenetic drift is a determinant of mammalian lifespan. Rejuvenation Res (2017) 20:430–36. doi:10.1089/rej.2017.2024
58. Maegawa S, Lu Y, Tahara T, Lee JT, Madzo J, Liang S, et al. Caloric restriction delays age-related methylation drift. Nat Commun (2017) 8(1):539. doi:10.1038/s41467-017-00607-3
59. Amarger V, Giudicelli F, Pagniez A, Parnet P. Perinatal high methyl donor alters gene expression in IGF system in male offspring without altering DNA methylation. Future Science OA (2017) 3(1):FSO164. doi:10.4155/fsoa-2016-0077
60. Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, et al. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab (2015) 22(5):861–73. doi:10.1016/j.cmet.2015.08.024
61. Mattocks DA, Mentch SJ, Shneyder J, Ables GP, Sun D, Richie JP Jr, et al. Short term methionine restriction increases hepatic global DNA methylation in adult but not young male C57BL/6J mice. Exp Gerontol (2017) 88:1–8. doi:10.1016/j.exger.2016.12.003
62. Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet (2002) 30(4):363–4. doi:10.1038/ng865
63. Slack FJ. Regulatory RNAs and the demise of ‘junk’ DNA. Genome Biol (2006) 7(9):328. doi:10.1186/gb-2006-7-9-328
64. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet (2011) 12(12):861–74. doi:10.1038/nrg3074
65. Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol (2009) 10(2):126–39. doi:10.1038/nrm2632
66. Mico V, Berninches L, Tapia J, Daimiel L. NutrimiRAging: micromanaging nutrient sensing pathways through nutrition to promote healthy aging. Int J Mol Sci (2017) 18(5):1–21. doi:10.3390/ijms18050915
67. Clarke JD, Sharapova T, Lake AD, Blomme E, Maher J, Cherrington NJ. Circulating microRNA 122 in the methionine and choline-deficient mouse model of non-alcoholic steatohepatitis. J Appl Toxicol (2014) 34(6):726–32. doi:10.1002/jat.2960
68. Dolganiuc A, Petrasek J, Kodys K, Catalano D, Mandrekar P, Velayudham A, et al. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol Clin Exp Res (2009) 33(10):1704–10. doi:10.1111/j.1530-0277.2009.01007.x
69. Leung AK, Sharp PA. microRNAs: a safeguard against turmoil? Cell (2007) 130(4):581–5. doi:10.1016/j.cell.2007.08.010
70. Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci (2008) 33(3):141–50. doi:10.1016/j.tibs.2007.12.003
71. Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci U S A (2006) 103(48):18125–30. doi:10.1073/pnas.0608845103
72. Guo Y, Yu J, Wang C, Li K, Liu B, Du Y, et al. miR-212-5p suppresses lipid accumulation by targeting FAS and SCD1. J Mol Endocrinol (2017) 59(3):205–17. doi:10.1530/JME-16-0179
73. Li K, Zhang J, Yu J, Liu B, Guo Y, Deng J, et al. MicroRNA-214 suppresses gluconeogenesis by targeting activating transcriptional factor 4. J Biol Chem (2015) 290(13):8185–95. doi:10.1074/jbc.M114.633990
74. Chitnis NS, Pytel D, Bobrovnikova-Marjon E, Pant D, Zheng H, Maas NL, et al. miR-211 is a prosurvival microRNA that regulates chop expression in a PERK-dependent manner. Mol Cell (2012) 48(3):353–64. doi:10.1016/j.molcel.2012.08.025
75. Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell (2006) 125(6):1111–24. doi:10.1016/j.cell.2006.04.031
76. Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell (2010) 40(2):205–15. doi:10.1016/j.molcel.2010.09.027
77. Olivieri F, Spazzafumo L, Santini G, Lazzarini R, Albertini MC, Rippo MR, et al. Age-related differences in the expression of circulating microRNAs: miR-21 as a new circulating marker of inflammaging. Mech Ageing Dev (2012) 133(11–12):675–85. doi:10.1016/j.mad.2012.09.004
78. Sun J, Sonstegard TS, Li C, Huang Y, Li Z, Lan X, et al. Altered microRNA expression in bovine skeletal muscle with age. Anim Genet (2015) 46(3):227–38. doi:10.1111/age.12272
79. Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet (2006) 38(2):228–33. doi:10.1038/ng1725
Keywords: microRNA, methionine restriction, stress, physiological, caloric restriction, mechanisms
Citation: Latimer MN, Freij KW, Cleveland BM and Biga PR (2018) Physiological and Molecular Mechanisms of Methionine Restriction. Front. Endocrinol. 9:217. doi: 10.3389/fendo.2018.00217
Received: 20 February 2018; Accepted: 17 April 2018;
Published: 04 May 2018
Edited by:
Encarnación Capilla, Universitat de Barcelona, SpainReviewed by:
Sergio Polakof, Institut National de la Recherche Agronomique (INRA), FranceThomas W. Gettys, Pennington Biomedical Research Center, United States
Copyright: © 2018 Latimer, Freij, Cleveland and Biga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peggy R. Biga, cGVnYmlnYUB1YWIuZWR1
 Mary Neslund Latimer
Mary Neslund Latimer Khalid Walid Freij
Khalid Walid Freij Beth M. Cleveland
Beth M. Cleveland Peggy R. Biga
Peggy R. Biga