- 1Arthritis Research UK Biomechanics and Bioengineering Centre, School of Biosciences, Cardiff University, Cardiff, UK
- 2Institute of Molecular and Experimental Medicine, School of Medicine, Cardiff University, Cardiff, UK
- 3Division of Pathophysiology and Repair, School of Biosciences, Cardiff University, Cardiff, UK
- 4Institute of Mechanical and Manufacturing Engineering, School of Engineering, Cardiff University, Cardiff, UK
- 5School of Psychology, Cardiff University, Cardiff, UK
Mechanical loading, a potent stimulator of bone formation, is governed by osteocyte regulation of osteoblasts. We developed a three-dimensional (3D) in vitro co-culture system to investigate the effect of loading on osteocyte–osteoblast interactions. MLO-Y4 cells were embedded in type I collagen gels and MC3T3-E1(14) or MG63 cells layered on top. Ethidium homodimer staining of 3D co-cultures showed 100% osteoblasts and 86% osteocytes were viable after 7 days. Microscopy revealed osteoblasts and osteocytes maintain their respective ovoid/pyriform and dendritic morphologies in 3D co-cultures. Reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR) of messenger ribonucleic acid (mRNA) extracted separately from osteoblasts and osteocytes, showed that podoplanin (E11), osteocalcin, and runt-related transcription factor 2 mRNAs were expressed in both cell types. Type I collagen (Col1a1) mRNA expression was higher in osteoblasts (P < 0.001), whereas, alkaline phosphatase mRNA was higher in osteocytes (P = 0.001). Immunohistochemistry revealed osteoblasts and osteocytes express E11, type I pro-collagen, and connexin 43 proteins. In preliminary experiments to assess osteogenic responses, co-cultures were treated with human recombinant bone morphogenetic protein 2 (BMP-2) or mechanical loading using a custom built loading device. BMP-2 treatment significantly increased osteoblast Col1a1 mRNA synthesis (P = 0.031) in MLO-Y4/MG63 co-cultures after 5 days treatment. A 16-well silicone plate, loaded (5 min, 10 Hz, 2.5 N) to induce 4000–4500 με cyclic compression within gels increased prostaglandin E2 (PGE2) release 0.5 h post-load in MLO-Y4 cells pre-cultured in 3D collagen gels for 48, 72 h, or 7 days. Mechanical loading of 3D co-cultures increased type I pro-collagen release 1 and 5 days later. These methods reveal a new osteocyte–osteoblast co-culture model that may be useful for investigating mechanically induced osteocyte control of osteoblast bone formation.
Introduction
Osteocytes are by far the most abundant bone cell type (90–95% of all bone cells (1), forming a network of cells, connected by long cell processes that extend along canaliculi within the mineralized bone matrix. The adult human skeleton is continually being remodeled by bone forming osteoblasts and bone resorbing osteoclasts, whose activities are balanced in healthy individuals. Osteocytes are thought to integrate hormonal, growth factor, and mechanical stimuli to influence control of bone remodeling.
In vivo, osteocytes increase transcriptional and metabolic activities in response to short loading periods (2, 3), increase their dentin matrix protein 1 (DMP1) and matrix extracellular phosphoglycoprotein (MEPE) expression, controlling bone matrix mineral quality (4, 5), increase insulin growth factor 1 (IGF-1), and related proteins involved in mechanically induced bone formation (6, 7), and stimulate nitric oxide (NO) production (8), an early mediator of mechanically induced bone formation (9). Osteocyte abundance, morphology, position within bone, and ability to form an extensive network are ideally suited to this mechanoresponsive role (10–14). Osteocytic processes and primary cilia detect mechanical stimuli (15–19) whereas, proteins involved in the connection of osteocytes to surrounding cells and/or the extracellular matrix (ECM), like focal adhesions, connexin 43 (CX43), and integrins, are involved in osteocyte response to mechanical stimuli (20, 21). Mechanically loaded osteocytes regulate osteoblast activity through various mechanisms including the downregulation of sclerostin (SOST) expression (22, 23), release of NO, which has an anabolic effect on osteoblast activity (9), and release of prostaglandin E2 (PGE2), which regulates osteoblast proliferation and differentiation (24). Whilst the methodology developed here focuses on osteocyte control of osteoblasts, it is clear that osteocytes regulate other cells activities in response to load (25–27).
A major problem with investigating osteocytes is the difficulty in isolation and culture of these cells in vitro. Studies using sequential digestion of bone to enrich for osteocytes have proved difficult and, so far, limited to chick (28–30), rat (31), and mouse (32). Mouse cell lines representing late osteoblast/early osteocytes (MLO-A5) (33) and osteocyte-like (MLO-Y4) (33, 34) cells have been developed to facilitate in vitro investigation of osteocytes, but these are routinely cultured in monolayer on type I collagen-coated plastic. More recently the IDG-SW3 mouse derived cells have been shown to replicate osteoblast-to-late osteocyte differentiation under both two-dimensional (2D) and three-dimensional (3D) collagen culture conditions in vitro (35).
There have been very few publications on co-culture of osteoblasts and osteocytes, despite the known physiological interactions between these cell types. Taylor et al. (36) describe a co-culture system in which the two cell types are grown in 2D, either side of a semi permeable cell culture insert membrane. Stimulation of the osteocyte layer by fluid shear enhanced alkaline phosphatase (ALP) expression by the osteoblasts, an effect at least partially dependent on cell–cell contact and gap junction communication (36). This system is useful but does not allow osteocytes to form a 3D network. The three-dimensionality of osteocyte environment is important; firstly embedding primary osteoblasts within 3D matrices induces differentiation to osteocyte-like cells in vitro (37), recapitulating the in vivo differentiation pathway, and secondly it facilitates a more realistic model of a 3D lacunocanalicular system (LCS) of cells that can be subjected to appropriate mechanical cues.
In vitro, 3D bone models where bone cells are embedded in type I collagen gels have not been used to investigate osteocyte loading or osteocyte–osteoblast interactions (38–42). 3D cultures made out of polybicarbonate membranes (37) and scaffolds (43–46) do not embed cells within a 3D matrix, but instead attach them to the scaffold surface and therefore do not accurately capture the environment of an osteocyte within bone. Whilst these systems have proven the feasibility of reproducing the synthesis of an organized matrix (44) and cell-mediated matrix degradation (47–49), there are no models that co-culture osteoblasts and osteocytes in 3D under mechanical stimulation. This highlights a major gap in the understanding of the interactions that lead to mechanically induced bone formation.
Here, we describe the methodology for a new 3D co-culture model, cultured within a custom built multi-well silicone loading plate, to investigate how mechanical loading of osteocytes regulates osteoblast function. MLO-Y4 cells were cultured within type I collagen gels, with an osteoblast-like cell line [MC3T3-E1(14) or MG63] layered on top of the gel (Figure 1). Both osteoblasts and osteocytes maintain cell viability, morphology, and phenotype when cultured in 3D co-cultures and express CX43, a component of network formation. These co-cultures resulted in anabolic responses when stimulated with bone morphogenetic protein 2 (BMP-2) or mechanically loaded. This model will be useful in elucidating osteocyte-driven mechanical mechanisms that regulate bone formation.
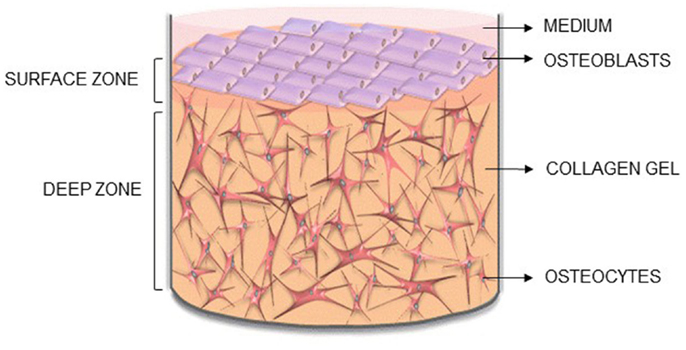
Figure 1. Novel 3D osteocyte–osteoblast co-culture model. Diagram of the 3D in vitro model indicating the surface and deep zone, and positions of the surface osteoblasts and embedded osteocytes.
Materials and Methods
Cells
MLO-Y4 cells were a kind gift from Professor Lynda Bonewald, University of Missouri-Kansas City, USA. MC3T3-E1(14) and MG63 cells were obtained from the European Collection of Cell Cultures, Salisbury, UK.
MLO-Y4 cells (34) were cultured on collagen-coated flasks (rat tail tendon type I collagen, 0.15 mg/mL in 0.02 N glacial acetic acid) in alpha minimum essential medium (αMEM, Invitrogen) supplemented with 2.5% Heat Inactivated Fetal Bovine Serum (HIFBS, Invitrogen) and 2.5% Heat Inactivated Newborn Calf Serum (HINCS, Invitrogen) (50). MC3T3-E1(14) cells were cultured in αMEM supplemented with 10% FBS (Invitrogen) (51). MG63 cells were cultured in Dulbecco’s Minimum Essential Medium (DMEM, Invitrogen) and supplemented with 5% FBS (Invitrogen). All three cell lines were supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin and grown at 37°C in 5% CO2. At 70–80% (MLO-Y4) or 80–90% [MC3T3-E1(14) and MG63] confluency, cells were sub-cultured by treating with trypsin/ethylenediaminetetraacetic acid (EDTA) (0.25% w/v of each; Invitrogen).
3D Co-Cultures
MLO-Y4 cells were incorporated within type I collagen gels and either MC3T3-E1(14) or MG63 cells layered on top. Rat tail tendon type I collagen (Sigma, in 7 mM glacial acetic acid) was mixed 4:1 with 5X MEM (Invitrogen) containing 11 g/L sodium bicarbonate on ice and neutralized [1 M tris(hydroxymethyl)aminomethane (Tris) base, pH 11.5] to give 2–2.6 mg/mL type I collagen gels. MLO-Y4 cells (1.5 × 106 cells/mL gel) diluted in αMEM (<10% of total gel volume) were added to the collagen on ice and 500 or 250 μL distributed into 24 or 48-well plastic plates, respectively for polymerization at 37°C for 1 h. MC3T3-E1(14) or MG63 cells (1.5 × 105 cells/well) in DMEM with 5% FBS (MG63) or 5% dialyzed FBS (DFBS) [MC3T3-E1(14)] were applied onto the surface of each gel after 1 h and incubated at 37°C for up to 1 week (Figure 1). Medium was changed after 24 h and every 2 days thereafter. To test cell responses, co-cultures were treated with human recombinant BMP-2 (250 ng/mL, Peprotech) for 5 days.
Cell Viability
Co-cultures grown in plastic plates were rinsed with phosphate buffered saline, pH 7.3 (PBS), incubated with 1 μM ethidium homodimer (Invitrogen) in serum free medium for 2 h at 4°C and then for a further 2.5 h at 37°C before washing overnight at 37°C in normal culture medium with gentle agitation. Positive controls co-cultures were freeze-thawed at −20°C three times, before treatment. For cell death analysis of the surface zone, confocal microscopy was performed directly on whole co-cultures. Samples were scanned using appropriate excitation and emission settings for simultaneous recording of 4′,6-diamidino-2-phenylindole (DAPI) [358 nm Excitation (Ex(max)); 461 nm Emission (Em(max))] and ethidium homodimer [590 nm Ex(max); 617 nm Em(max)]. Samples were optically sectioned, over five defined arbitrary regions per gel quarter, using a x10 objective lens with 2.32 zoom. 5 μm step size z-stack optical sections were reconstructed using Leica Confocal Software. Maximum intensity models were prepared showing detail of the surface zone. Counts were made of DAPI (blue) labeled nuclei (to give total number of cells) and ethidium homodimer and DAPI (purple) co-labeled nuclei (to give number of dead cells).
For deep zone viability, cultures were fixed with 1% paraformaldehyde (Sigma) in 0.05 M PBS for 30 min at 4°C and then washed in PBS. Some were labeled whole for filamentous actin and type I pro-collagen (see below). Cultures were infiltrated with 50% OCT compound (Tissue Tek) in PBS overnight at 4°C and then frozen in fresh OCT compound onto cryostat stubs using dry ice. Cryosections were cut at 20 μm using a Bright OTF5000 cryostat and collected on Polysine slides (VWR). Five random slides of each co-culture containing 4–6 sections each were mounted in Vectashield mounting medium with DAPI as a nuclear counterstain. One random section from each slide was observed under epi-fluorescence as above, and 10 random fields of view using the x20 objective photographed for each section under both DAPI and ethidium homodimer illumination. Counts were made as above.
Microscopy and Imaging of Cells and Cell Markers
Osteocyte and osteoblast morphology was assessed in live cultures grown in plastic plates with a Leica DMRB inverted microscope equipped with a Moticam 2000 digital camera. For labeling procedures, 3D cultures were treated, fixed, frozen, and cryosectioned as previously outlined.
Filamentous actin was labeled by incubating fixed intact cultures or sections with 5 μM Atto488 conjugated phalloidin (Sigma) in PBS containing 0.1% tween 20 (PBST; Sigma) for 40 min at 4°C. Specimens were then washed thoroughly in PBST and mounted in Vectashield containing DAPI to label nuclei (Vector Laboratories). Labels for type I pro-collagen, gap junction protein CX43 and dendritic cell marker podoplanin (E11) were performed by indirect immunohistochemical procedures. In all cases, the primary antibody was replaced in control sections with PBST alone or with non-immune immunoglobulins (IgG), to reveal any non-specific binding. In all cases both PBST and IgG controls were negative. Type I pro-collagen was labeled using monoclonal antibody M38 recognizing the C-terminus of pro-collagen I in a wide range of species except mouse [5 μg/mL; (52); Developmental Studies Hybridoma Bank], CX43 using monoclonal antibody CXN-6 (8 μg/mL; Sigma) and E11 with goat anti-mouse podoplanin (E11) primary antibody (2.5 μg/mL; R&D Systems). Sections were blocked with 5% goat serum (those stained with antibodies M38 and CXN-6; Dako) or 5% rabbit serum (E11; Dako) for 30 min and then incubated with primary antibodies overnight at 4°C (E11) or 1 h at room temperature (M38, CXN-6). After extensive washing with PBST, M38 and CXN-6 sections were incubated with AlexaFlour594 or AlexaFluor488 goat anti-mouse conjugates, for 30 min at room temperature. All sections were thoroughly washed and mounted in Vectashield/DAPI. E11 sections were incubated with a horseradish peroxidase rabbit anti-goat conjugate (Vector Laboratories; 1:800 dilution) for 30 min, washed in PBST and label visualized using a nickel-enhanced diaminobenzidine peroxidase substrate kit (Vector Laboratories). Fluorescence specimens were examined with an Olympus BX61 epi-fluorescence microscope equipped with an F-view camera and AnalySISimage capture and analysis software, or using a Leica TCS SP2 AOBS confocal scanning laser microscope and using the Leica Confocal Software, and peroxidase labels with a Leica DMRB microscope with a Moticam 2000 camera. Confocal microscopy was carried out on samples using appropriate excitation and emission settings for simultaneous recording of DAPI and ethidium homodimer as previously outlined, Phalloidin-Atto488 [495 nm Ex(max), 519 nm Em(max)], and Alexa 594 [590 nm Ex(max), 617 nm Em(max)]. Specimens were optically sectioned using a x63 objective with an arbitrary zoom (surface and deep zone actin filament stain, and CX43 immunofluorescence). 5 μm (surface zone) or 0.5 μm (deep zone) step size z-stack optical sections through the specimen were reconstructed using Leica Confocal Software. Maximum intensity models were prepared showing detail of the surface zone or deep zone.
Molecular Analysis of Osteocyte and Osteoblast Phenotype
Osteocyte and osteoblast phenotype was determined at the molecular level using reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR). Ribonucleic acid (RNA) was extracted separately from surface and embedded cells of co-cultures grown in plastic plates by dispensing 1 mL Trizol (Invitrogen) onto the surface for 10 s to extract osteoblast RNA, and subsequently dissolving the underlying gel within a separate 1 mL Trizol aliquot to extract RNA from gel embedded cells. After RNA extraction according to manufacturer’s protocol and DNAse treatment (DNA-free, Ambion) RNA was re-precipitated with 3 M sodium acetate (pH 5.5, Ambion) and dissolved in molecular biology grade water (25 μL for surface cells, 50 μL for deep cells). RNA with A260/A280 and A260/A230 ratios of ≥1.8 was deemed good quality. RNA (1.7–253.3 ng for the surface zone; 108.8–1864.9 ng for the deep zone) was primed with random hexadeoxynucleotides (Promega, Southampton, UK) and reverse transcribed according to manufacturer’s instructions (Superscript III, Invitrogen). Gene expression of E11, osteocalcin (OCN), runt-related transcription factor 2 (Runx2), type I collagen (Col1a1), ALP, and simian vacuolating virus 40 (SV40) large T-antigen were analyzed by RT-qPCR [SYBR Green I master mix (Sigma), 2.5 mM MgCl2, 200 nM each primer, Stratagene MX3000P]. All targets except OCN used intron-spanning primers (Table 1) and no template controls were included for all reactions. Thermocycling included 1 cycle of 95°C for 3 min, 40 cycles of 95°C for 20 s, 60°C for 40 s, 72°C for 20 s, and 1 cycle of 72°C for 10 min. Specificity of RT-qPCR was confirmed by melting curve analysis (Stratagene MX3000P) with complementary deoxyribonucleic acid (cDNA) standard curves of 90–110% efficiency and with an R2 value of ≥0.99 accepted as valid.
Relative quantification of messenger ribonucleic acid (mRNA) expression by RT-qPCR was calculated using the 2−ΔΔCT method (53). In all cases, RT-qPCR was carried out using three reference genes (RG), 18S rRNA, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and hypoxanthine–guanine phosphoribosyltransferase (HPRT1). The most stable RG was determined via NormFinder. Expression data for each gene in an experiment were transformed from log10 to linear scale using a standard curve and then loaded into the NormFinder Microsoft Excel Add-In1 (54). The most stable gene or combination of two genes, combined by calculating the geometric mean, was used for normalization and is clearly stated in the relevant figures. Target gene expression levels were expressed as relative expression units (REU), using the highest expressor within all samples as the calibrator sample.
Mechanical Loading
A 16-well loading plate (Figure 2A) was manufactured from solid silicone (Technovent TechSil 25)2 so that the wells of the plate were of the same dimensions (10 mm diameter) as a standard Greiner (Stonehouse, UK) 48-well tissue culture plate but with a 150 μm thick base. The spaces between the wells were filled with silicone and a series of holes were made on each side of the plate to accommodate hooks for attachment to a BOSE loading instrument. A Dantec Dynamics Digital Image Correlation (DIC) system was used to measure strain in the loading plate in order to calibrate the system. DIC compares two digital images of two different mechanical states of a particular object: a reference state and a deformed state. A previously applied speckle pattern (here applied using black face paint)3 follows the strain of the object, and so the displacement that occurs between both reference and deformed state can be measured by matching the speckle pattern in small regions of the image (55, 56). By using two cameras (Limess Messtechnik)4 and matching speckle patterns in each image, the position and displacement in 3D can be obtained, after calibrating the system using a grid of known dimensions to determine the position of the cameras. DIC validated strains of 4000–4500 με in the majority of the wells of the loading plate when a force of 2.5 N was applied.
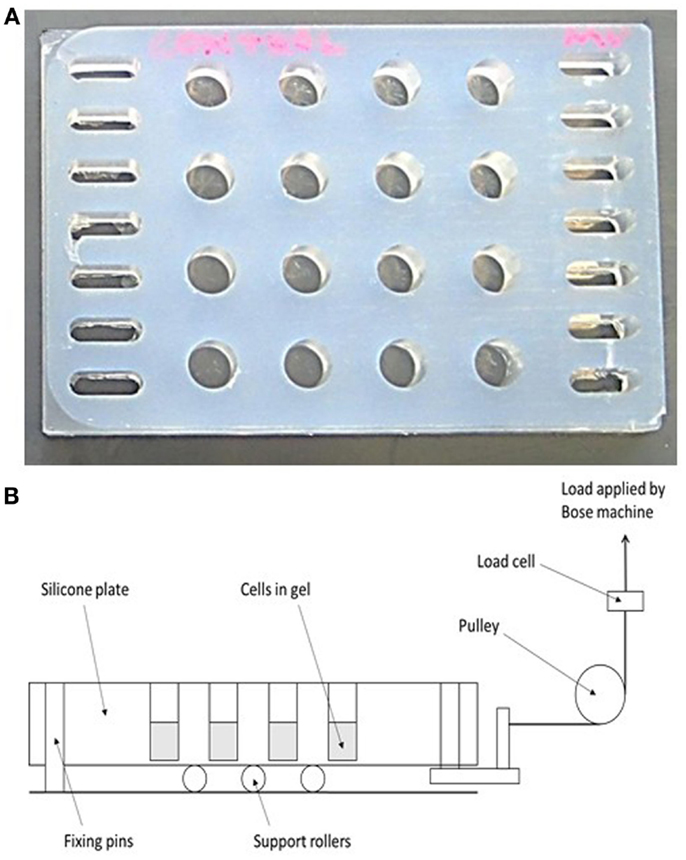
Figure 2. A novel mechanical loading device. (A) A 16-well rubber loading plate of the same dimensions (10 mm diameter) as a standard 48-well tissue culture plate but with a 150 μm thick base and with holes for its attachment to a BOSE loading instrument. (B) Schematic showing the operation of the loading device. The collagen gels are contained in the wells of the silicone plate, and the entire plate is stretched to apply a strain to the gels.
For mechanical loading the silicone plate was attached to a BOSE ElectroForce® 3200 (Kent, UK) loading instrument by a custom-made device (Figure 2B) in order to stretch the plate from one end causing cyclic compression in all wells. A 250 N load cell was used to apply a loading regime of 5 min, 10 Hz, 2.5 N to 3D osteocyte mono-cultures. Loading was controlled using WinTest® Software 4.1 with TuneIQ control optimization (BOSE).
For loading, 3D osteocyte mono-cultures were prepared and cultured in the silicone plate in 800 μl of DMEM GlutaMAX™ supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 5% DFBS incubated at 37°C in 5% CO2/95% air atmosphere for 24, 48, or 72 h without changing culture medium prior to load, or 7 days where culture medium was changed every 2–3 days and prior to loading. 3D co-cultures were prepared and cultured in the silicone plate as described previously for plastic plates and cultured for 7 days prior to load, changing culture medium every 2–3 days and immediately prior to loading.
Prostaglandin E2 release was measured at 0, 0.5, 1, 3, 6, 12, and 24 h post-load using an Enzo Life Sciences PGE2 kit (Exeter, UK) and following manufacturer’s instructions. Experimental samples were diluted 1:64 (gels set up 24 h before loading), 1:16 (gels set up 48 and 72 h before loading), or 1:40 (gels set up 7 days before loading) in order to fit within the standard curve of the assay. All samples were within the standard curve range. The sensitivity of the assay is 8.26 μg as stated by the manufacturer. Absorbance was recorded using a BMG Labtech FLUOstar Optima plate reader (Bucks, UK) and using the Optima Software for FLUOstar V2.00 R3 (BMG Labtech). PGE2 concentration of the experimental samples was determined according to the standard curve. Data was normalized to total cell number by lysing cultures and performing LDH assay (CytoTox 96® Non-Radioactive Cytotoxicity Assay, Promega, Southampton, UK).
PINP (type I pro-collagen) synthesis was measured at day 1 and 5 post-load using a Rat/Mouse PINP Enzyme Immunoassay (EIA) kit (Immunodiagnostic systems, Tyne & Wear, UK) following the manufacturer’s instructions. All samples were within the standard curve range. The sensitivity of the assay was 0.7 ng/ml as stated by the manufacturer. Data were normalized to total DNA content (extracted using TRIzol® reagent and quantified after precipitation using a Quant-iT™ dsDNA High-Sensitivity Assay Kit, both following the manufacturer’s instructions). The sensitivity of the DNA assay was 0.5 ng/μl.
Statistics
Data are expressed as the mean ± Standard Error of the Mean (SEM). Residuals were tested for normality (Anderson–Darling) and equal variance (Bartlett’s and Levene’s tests) and transformed if necessary, before applying analysis of variance (ANOVA) and post hoc Fisher’s or Tukey’s tests or General Linear Model (GLM) for crossed factors with pairwise comparisons where P < 0.05 were recorded. Data were deemed to be significantly different when P < 0.05. 3D cultures prepared in individual wells within a plate, cultured for 1–12 days and loaded/treated were considered independent replicates. Group of independent replicates (3D cultures) prepared, cultured, and loaded/treated in separate plates on separate occasions were considered independent experiments.
Results
Osteocyte and Osteoblast Viability in 3D Co-Cultures
Confocal images of the surface zone across five arbitrary fields of view were taken for all replicates of both MLO-Y4/MC3T3-E1(14) (three independent experiments of n = 3) or MLO-Y4/MG63 (two independent experiments of n = 3) 3D co-cultures grown in plastic plates. At both day 1 and day 7, the viability of MC3T3-E1(14) (Figures 3A,B, respectively) or MG63 (Figures 3C,D, respectively) osteoblast-like cells was 100%. Freeze-thaw controls showed 100% death of the surface zone of the model (Figure 3I).
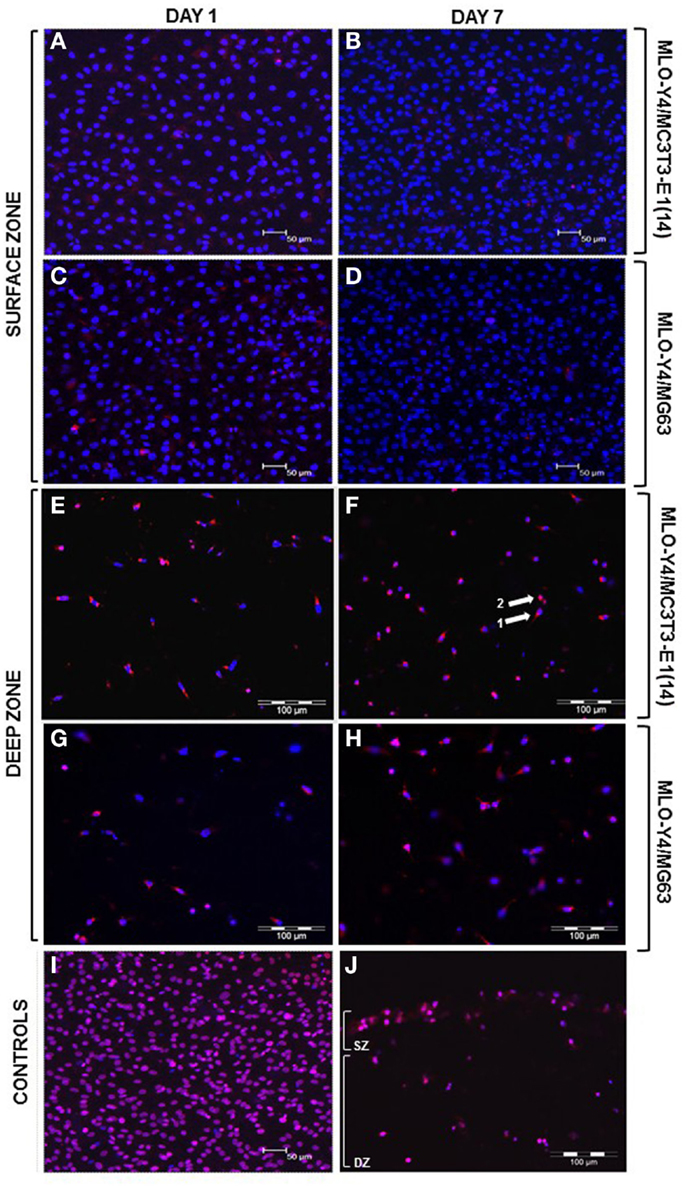
Figure 3. Viability of the surface and deep zone cells in 3D co-cultures (red: ethidium homodimer; blue: DAPI, purple: combination of both dyes). Confocal microscope images of an arbitrary field of view showing only live (blue nuclei) surface cells at day 1 (A,C) and at day 7 (B,D) of both MLO-Y4/MC3T3-E1(14) (A,B) and MLO-Y4/MG63 (C,D) co-cultures, respectively. Some live osteoblasts show red staining in their cytoplasm but not their nucleus at both time-points. Fluorescence microscope images of an arbitrary field of view showing a mixture of live and dead embedded cells at day 1 (E,G) and at day 7 (F,H) in both MLO-Y4/MC3T3-E1(14) (E,F) and MLO-Y4/MG63 (G,H) co-cultures, respectively. Some live osteocytes also show red staining [arrow 1 in (F)] in the cytoplasm but not their nucleus, whereas, dead cells are red throughout [arrow 2, (F)]. Confocal microscope image of an arbitrary field of view from a freeze-thaw control where all surface cells are dead (I). Fluorescence microscope image of an arbitrary field of view from a freeze-thaw control where all surface and embedded cells are dead (J) (SZ: surface zone, DZ: deep zone). For the osteoblasts, images were taken at the level of the surface zone and are representative of three independent experiments, n = 3 per experiment, five arbitrary fields of view per replicate. Scale bars: 50 μm (A–D, I). For the osteocytes, images were taken from 3D co-culture transverse cryosections and are representative of two or three independent experiments, n = 3 per experiment, 5 sections per replicate, 10 arbitrary fields of view per section [scale bars: 100 μm (E–H, J)].
Ten arbitrary fields of view from five random transverse cryosections from all replicates were used for quantification of MLO-Y4 cell death as a proportion of total MLO-Y4 number in both MLO-Y4/MC3T3-E1(14) (three independent experiments of n = 3) or MLO-Y4/MG63 (one independent experiment of n = 3 for day 1; two independent experiments of n = 3 for day 7) 3D co-cultures. At day 1 and day 7, a mixture of live and dead MLO-Y4 cells was observed for both MLO-Y4/MC3T3-E1(14) (Figures 3E,F, respectively) and MLO-Y4/MG63 (Figures 3G,H, respectively) co-cultures. Live osteocytes had a blue nucleus and dendritic morphology, whereas, a purple nucleus and rounded morphology was observed for dead osteocytes. Some live MLO-Y4 cells had red staining in their cytoplasm but not their nucleus (arrow 1, Figure 3F). Freeze-thaw controls showed 100% osteocyte death (Figure 3J).
In MLO-Y4/MC3T3-E1(14) co-cultures, an average of 16.13 ± 3.16% osteocyte cell death was observed at day 1 and 13.85 ± 2.35% at day 7 (Figure 4A). Mean MLO-Y4 cell death within 3D co-cultures did not differ between day 1 and day 7, however, MLO-Y4 cell death varied significantly between independent experiments (GLM, P = 0.002) with a significant interaction between day and experiment (GLM, P = 0.018). MLO-Y4 cell death at day 1 was significantly reduced in experiment 3 (10.04 ± 1.14%) compared with experiment 1 (20.62 ± 2.28%, P = 0.007) and experiment 2 (17.32 ± 1.43%, P = 0.004) whereas, MLO-Y4 cell death at day 7 differed significantly between experiment 1 (18.08 ± 1.86%) and experiment 2 (9.35 ± 1.39%) (P = 0.041).
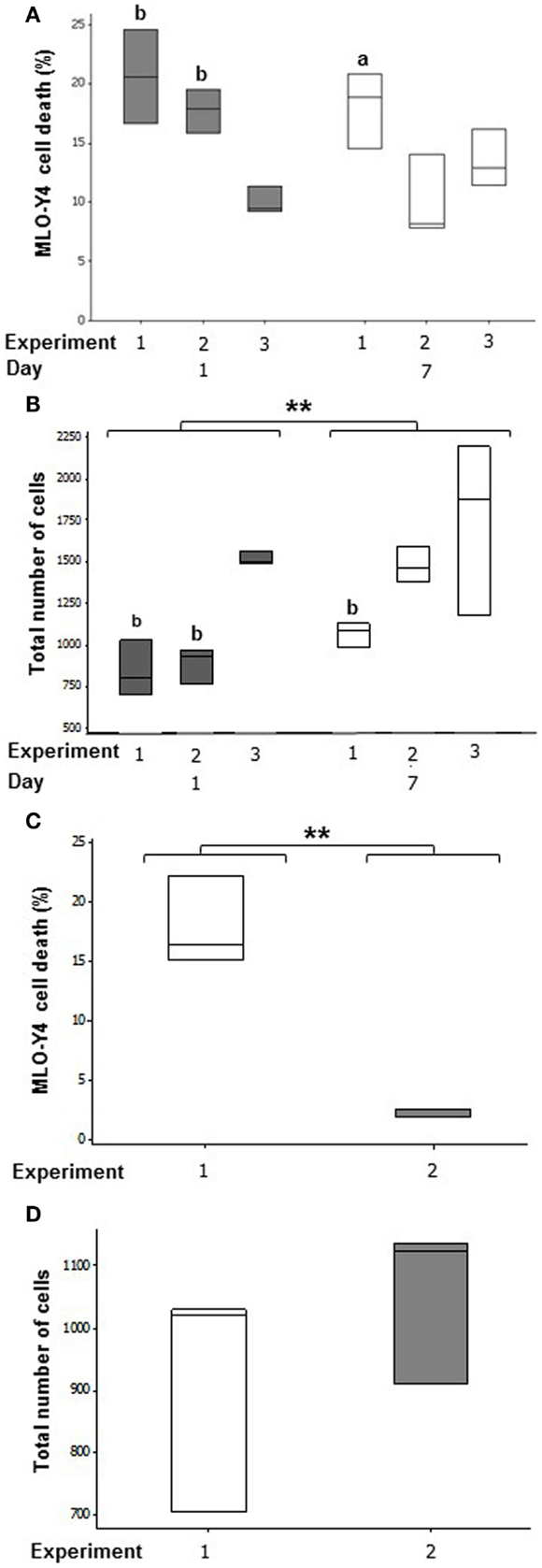
Figure 4. Quantification of cell number and cell death in the deep zone of 3D co-cultures. Boxplots of percentage cell death as a proportion of total number of cells at day 1 and day 7 in MLO-Y4/MC3T3-E1(14) (A) and MLO-Y4/MG63 (C) co- cultures. Boxplots showing total cell number counted for each replicate experiment at day 1 and day 7 in each independent experiment for MLO-Y4/MC3T3-E1(14) (B) and MLO-Y4/MG63 (D). For total cell number, significant differences obtained by GLM of log10 data between day 1 and day 7 denoted by **P < 0.01. Significant differences from pairwise comparisons, within each day, between independent experiments are shown by “a,” with respect to experiment 2; and “b,” with respect to experiment 3.
Total cell number increased between day 1 and day 7 (GLM, P = 0.003 of log10 data), and varied between replicate experiments (GLM, P < 0.001 of log10 data) in MLO-Y4/MC3T3-E1(14) co-cultures. At day 1, total cell number was 2-fold higher in experiment 3 when compared to experiment 1 (GLM, P = 0.009 of log10 data), and 1.5-fold higher when compared to experiment 2 (GLM, P = 0.019 of log10 data). At day 7, total cell number was 1.7-fold higher in experiment 3 when compared to experiment 1 (GLM, P = 0.049 of log10 data) (Figure 4B).
After 7 days of co-culture, the mean percentage of dead MLO-Y4 cells in MLO-Y4/MG63 co-cultures was 9.98 ± 3.66% across both independent experiments. However, there was a significant difference observed between mean percentage death of experiments 1 (17.88 ± 2.16%) and 2 (2.08 ± 0.22%) (One-way ANOVA, P = 0.002) (Figure 4C). There was no significant difference observed in total MLO-Y4 cell number at day 7 between experiments 1 and 2 in MLO-Y4/MG63 co-cultures (Figure 4D).
Osteocytes and Osteoblasts Assume Appropriate Morphology in Co-Cultures
MLO-Y4 osteocyte-like cells embedded within a 3D type I collagen gel overlaid with either MC3T3-E1(14) or MG63 osteoblasts grown in plastic plates revealed a single osteoblast surface cell layer and dendritic MLO-Y4 cells embedded throughout the depth of the type I collagen gel (Figure 5).
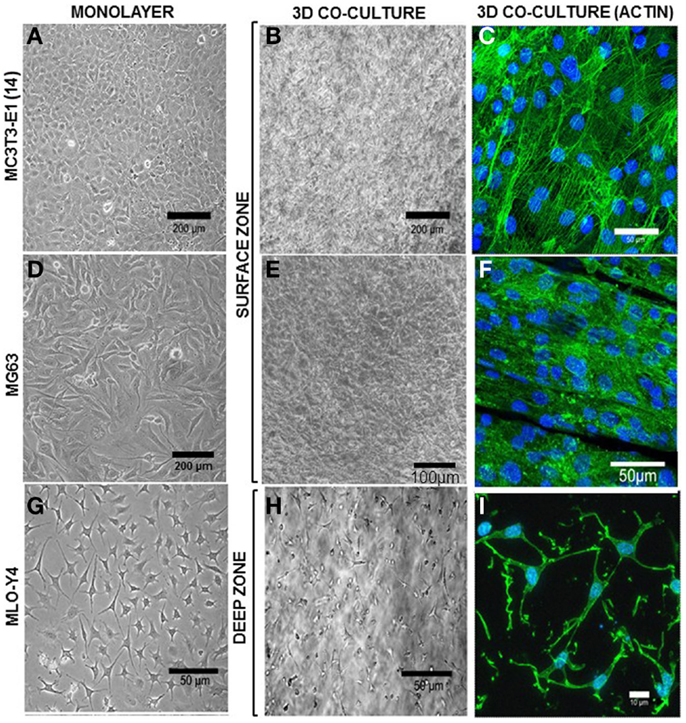
Figure 5. Morphology of surface and deep zone cells in 3D co-cultures. Inverse light microscope images of MC3T3-E1(14) (A), MG63 (D), and MLO-Y4 (G) cells in monolayer. MC3T3-E1(14) and MG63 osteoblast-like cells present typical osteoblastic morphologies, mainly ovoid or pyriform, and MLO-Y4 osteocyte-like cells have a typical dendritic morphology. Inverse light microscope images taken from the surface zone (B,E) of the 3D model where a confluent monolayer of surface cells is observed, and halfway through the depth of a 3D co-culture (H) showing embedded cells with a similar dendritic morphology to that seen in MLO-Y4 monolayer cultures. Confocal microscope focusing directly onto the surface zone of a 3D co-culture at day 7 (C,F), where surface cells were stained to reveal actin filaments (Phalloidin-Atto488) and cell nuclei (DAPI). The image shows a pavement-like monolayer, with individual cells containing well-developed stress-fibers, and maintaining an osteoblastic morphology. Confocal microscope image stack of a 3D co-culture transverse cryosection at day 7 (I) where embedded cells were stained to reveal actin filaments (Phalloidin-Atto488) and cell nuclei (DAPI), showing a dendritic morphology and connections between neighboring cells. Images are arbitrary fields of view representative of three independent experiments, n = 3 per experiment.
In MLO-Y4/MC3T3-E1(14) co-cultures, MC3T3-E1(14) cells had a similar ovoid or pyriform morphology to those in monolayer cultures (Figure 5A). The MC3T3-E1(14) osteoblasts formed a thin pavement-like single cell layer on top of the 3D co-cultures, which were difficult to image under the inverse light microscope (Figure 5B). However, actin filament labeling (Figure 5C) revealed MC3T3-E1(14) osteoblast morphology more clearly, with stress-fibers throughout their cell bodies. With similar morphology to MG63 monolayer cultures (Figure 5D), MG63 cells formed a pavement-like single cell layer in MLO-Y4/MG63 co-cultures under both light microscopy (Figure 5E) and immunofluorescence, when filamentous actin was labeled (Figure 5F). MLO-Y4 dendritic morphology observed in monolayer cultures (Figure 5G) was similar in co-cultures (Figure 5H), although projections extended in three dimensions. Contacts between neighboring osteocytes were revealed by actin filament staining (Figure 5I).
Osteocytes and Osteoblasts Display Appropriate Phenotype in 3D Co-Cultures
mRNA expression was assessed in both surface and deep zones of day 7 MLO-Y4/MC3T3-E1(14) 3D collagen co-cultures grown in plastic plates by relative RT-qPCR using primers against osteoblast and osteocyte phenotypic markers. Data were expressed in REU and normalized to Gapdh, which was ranked as the most stable reference gene (NormFinder stability value = 0.398, intergroup variation = 0.376, and intragroup variation = 0.012). Data were analyzed from three independent experiments, each with three replicates for the surface zone, and four replicates for the deep zone, for all genes except Col1a1 (two independent experiments).
In MLO-Y4/MC3T3-E1(14) co-cultures, no significant difference in expression was detected between zones of the model for E11 (surface zone, 0.264 ± 0.072 REU; deep zone, 0.361 ± 0.087 REU) (Figure 6A), OCN (surface zone, 0.212 ± 0.076 REU; deep zone, 0.269 ± 0.080 REU) (Figure 6B), and Runx2 (surface zone, 0.275 ± 0.083 REU; deep zone, 0.157 ± 0.025) (Figure 6C). However, the surface zone of the model showed 6-fold increases in expression of Col1a1 compared to the deep zone (0.168 ± 0.085 vs. 0.028 ± 0.007 REU, GLM, P < 0.001 of log10 data) (Figure 6D). In contrast, the deep zone of the 3D co-culture showed 2-fold increases in ALP expression over the surface zone (0.366 ± 0.075 vs. 0.185 ± 0.047 REU, GLM, P = 0.001 of ranked data) (Figure 6E). Whilst REU of all genes varied significantly between replicate experiments (GLM, E11, OCN, and Col1a1, P < 0.001 of log10 data; Runx2 P = 0.013 of ranked data; ALP P < 0.001 of ranked data; P < 0.05 for all pairwise comparisons) the trend in terms of surface compared with deep REUs within each experiment was consistent. Consistent with this, RT-PCR of MLO-Y4/MG63 co-cultures, revealed surface osteoblasts and embedded osteocytes expressed E11, OCN, Runx2, and COL1A1 mRNA (data not shown, three independent experiments of n = 3 for both surface and deep zones). Quantification of mRNA expression could not be compared between surface MG63 and embedded MLO-Y4 cells as the respective human and mouse cDNA sequences are not sufficiently homologous to use the same primers.
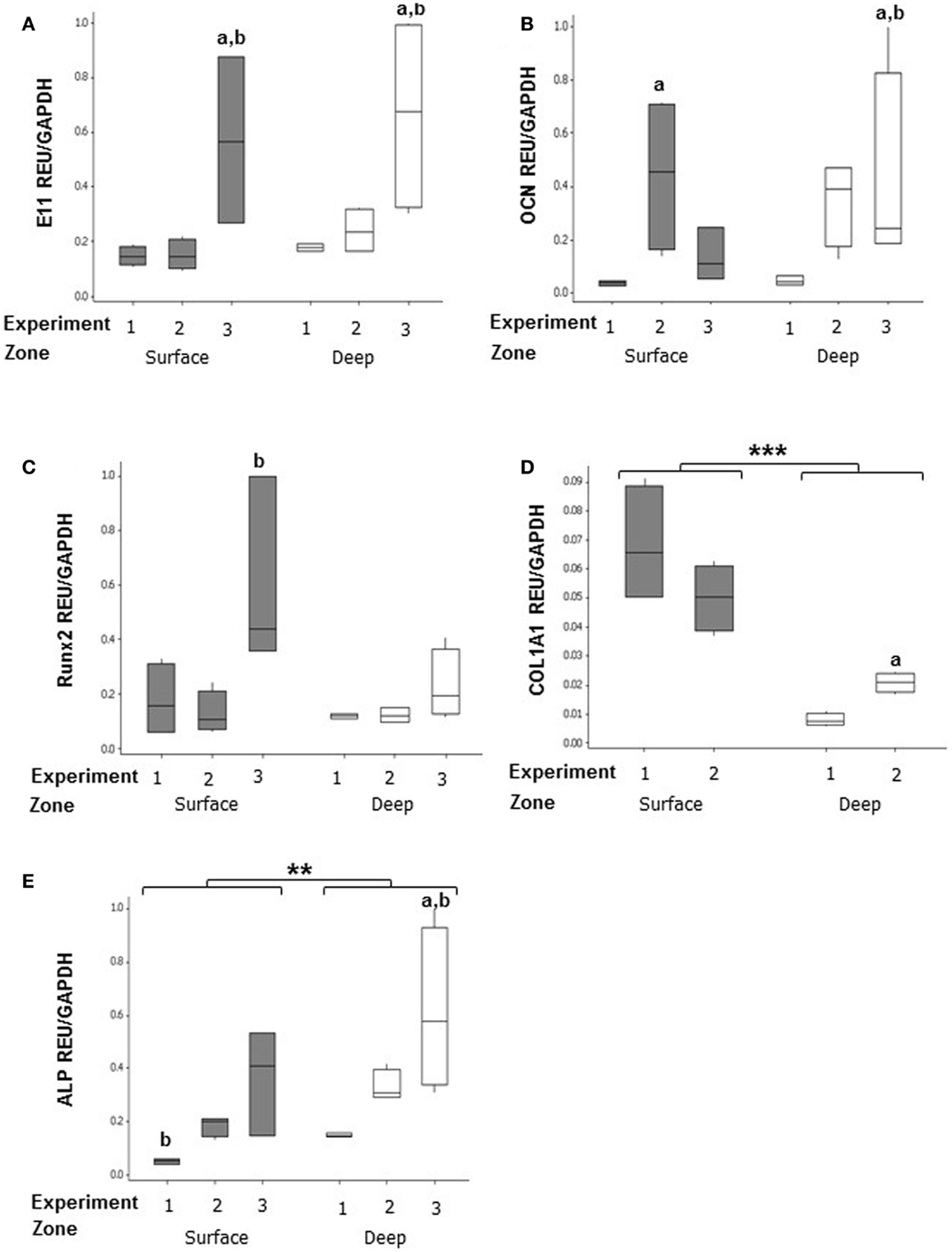
Figure 6. Gene expression of cellular markers in surface and deep zone cells in MLO-Y4/MC3T3-E1(14) 3D co-cultures. Quantification of gene expression in the 3D co-culture after 7 days by relative RT-qPCR, boxplots of E11 (A), OCN (B), RUNX2 (C), Col1a1(D), and ALP (E) expressed as REU and normalized to Gapdh expression. Significant differences obtained by GLM of log10 data (E11, Col1a1, and OCN) or ranked data (ALP and Runx2) between surface and deep zones denoted by **P < 0.01, ***P < 0.0001. Significant differences from pairwise comparisons, within each zone, between independent experiments denoted by “a,” with respect to experiment 2; and “b,” with respect to experiment 3. Values derived from two (Col1a1) or three (all others) independent experiments, n = 3 for surface and four for deep zones.
Osteoblasts and osteocytes in MLO-Y4/MC3T3-E1(14) co-cultures showed strong, uniform immunolabelling for the dendricity marker E11 (Figures 7A,B). Intense E11 immunolabelling was also observed in embedded MLO-Y4 cells within the MLO-Y4/MG63 co-cultures, but not in surface MG63 cells (Figures 7C,D). In both 3D co-culture systems abundant CX43 immunostaining was observed in the cell membrane and cytoplasm of osteoblasts and in osteocytes along their processes, as well as within the cytoplasm, around the nucleus (Figures 7E–G) and in contacts between cells (Figures 7F inset; 7H). Immunohistochemistry images are representative of day 7, 3D co-cultures from three independent experiments where n = 3 [MLO-Y4/MC3T3-E1(14)], or two independent experiments where n = 3 (MLO-Y4/MG63). Four to six cryosections from all replicates were observed. PBST and IgG controls were negative.
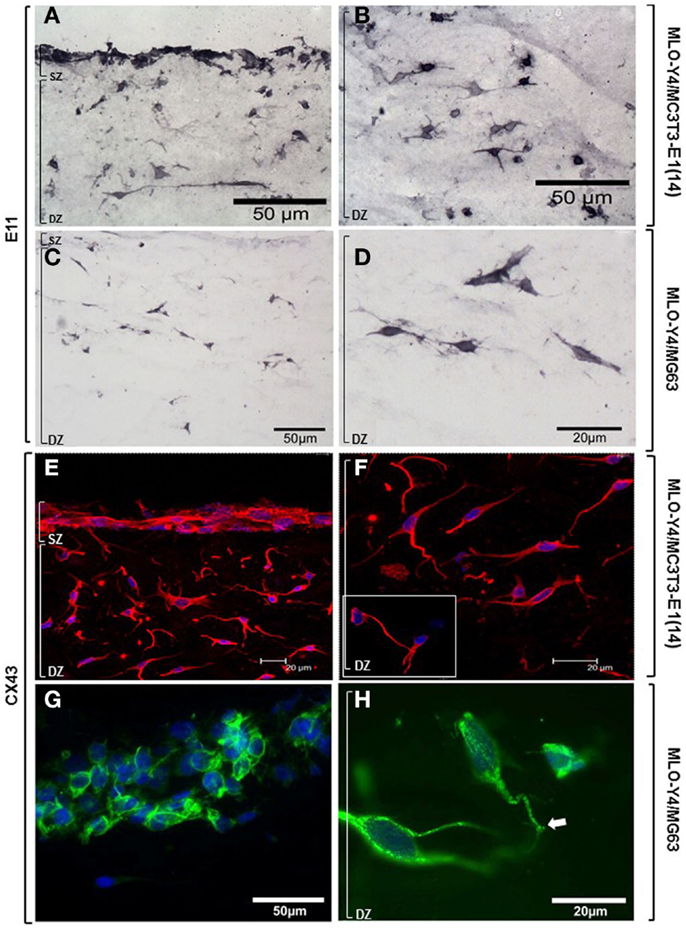
Figure 7. Protein expression of cellular markers in surface (SZ) and deep (DZ) zone cells of the 3D co-culture systems. Brightfield photomicrographs showing immunostaining for the dendricity marker E11 in both surface and embedded cells (A) and showing E11 immunostaining in the osteocytes highlighting their morphology (B), in MLO-Y4/MC3T3- E1(14) 3D co-cultures. Light microscope images revealing immunostaining for the dendricity marker E11 in embedded cells (C,D) but not in surface cells (C), in MLO-Y4/MG63 co-cultures. Confocal microscope images showing CX43 (Dylight594) immunolabelling and cell nuclei stain (DAPI) in surface and deep zone cells (E) of MLO-Y4/MC3T3-E1(14) co-cultures. Image reveals abundant quantities of CX43 present in the cytoplasm and cell membranes of both cell types, around the nucleus of the embedded cells (F), and connections between neighboring cells [(F), inset] (inset scale bar: 10 μm). Fluorescent photomicrographs of surface (G) and deep (H) zone cells of MLO-Y4/MG63 co-cultures labeled for CX43 (green) and counterstained with DAPI (blue) reveals that the surface cell layer, in this case several cells thick, intensely labels for CX43 along cell–cell interfaces (G). High magnification of embedded cells within the same co-culture gel reveals extensive punctate labeling within the cytoplasm and at the cell surface, including at interfaces between cell processes (arrow) (H). Images are arbitrary fields of view taken from 3D co-culture transverse cryosections representative of two or three independent experiments, n = 3 per experiment. In all cases, controls performed by omitting or substituting the primary antibody, showed no labeling.
Cell Migration in Co-Cultures
To detect whether MLO-Y4 cells moved to the surface zone, expression of the SV40 large T-antigen (only expressed by MLO-Y4 cells) was determined in MLO-Y4/MC3T3-E1(14) co-cultures grown in plastic plates (Figure 8). Whilst low levels of SV40 large T-antigen mRNA expression were detected in the surface zone (Figure 8A), SV40 large T-antigen immunolabelling was completely absent from the surface zone of the model (Figure 8B).
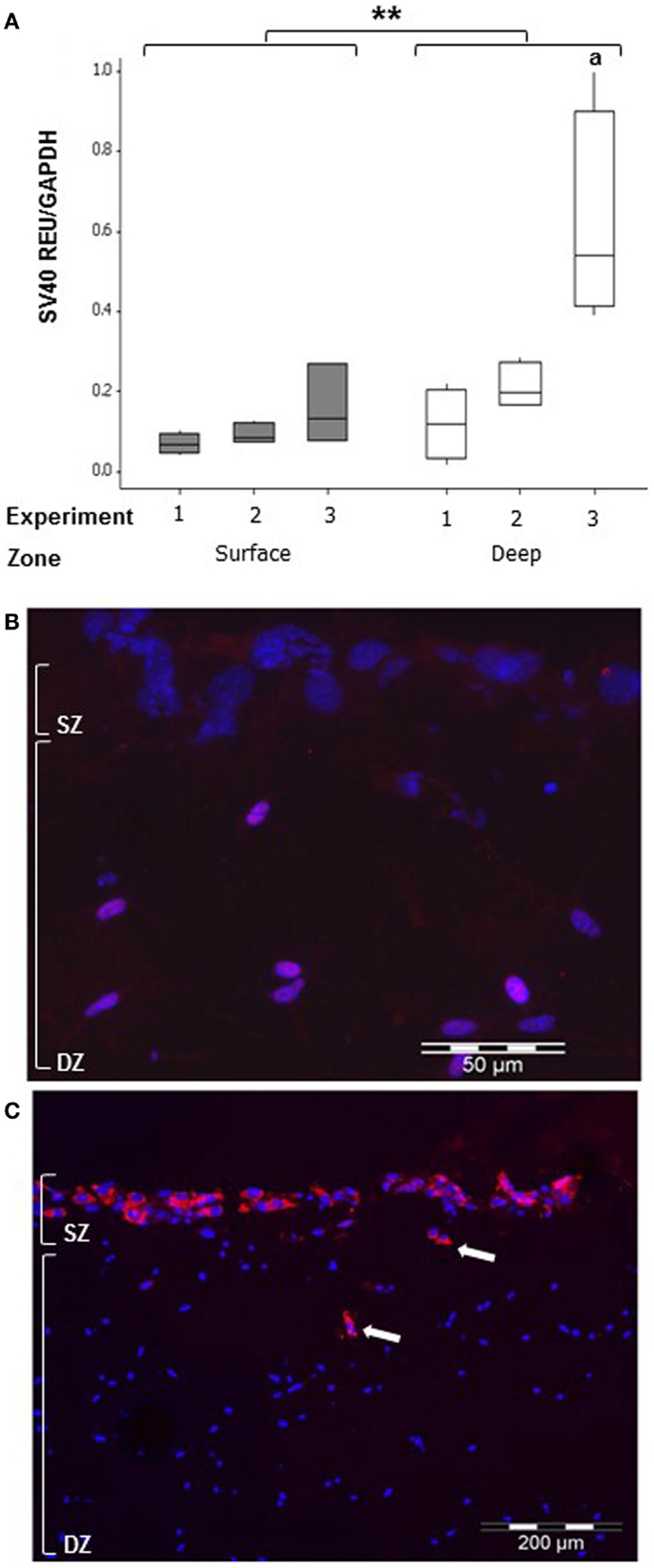
Figure 8. Co-cultures retain zone separation for up to 7 days (SZ: surface zone; DZ: deep zone). Boxplot showing quantification of SV40 large T-antigen gene expression in the MLO-Y4/MC3T3-E1(14) co-culture after 7 days by relative RT-qPCR (A) expressed as REU and normalized to Gapdh expression. Significant differences obtained by GLM of log10 data between surface and deep zones denoted by **P < 0.01. Significant differences from pairwise comparisons, within each zone, between independent experiments denoted by “a” with respect to experiment 1 (three independent experiments, n = 3 for surface and 4 for deep zones). Fluorescent photomicrograph of transverse cryosection from day 7 MLO-Y4/MC3T3-E1(14) co-culture shows immunolabelling for SV40 large T-antigen (red) and cell nuclei stain (blue) in osteocytes only, represented by the purple color (red and blue co-localization) (B). However, no SV40 large T-antigen immunostaining in the osteoblasts was present (three independent experiments, n = 3). SZ, surface zone; DZ, deep zone. Fluorescent photomicrograph of transverse cryosection from BMP-2 treated MLO-Y4/MG63 co-cultures at day 5 (C) revealed presence of type I pro-collagen in the upper layer of cells and in cells up to 100 μm beneath the surface, which are all MG63 cells since M38 antibody does not recognize mouse type I pro-collagen (two independent experiments,n = 3).
Osteoblast migration from surface to deep zone could be tracked in MLO-Y4/MG63 co-cultures, using a type I pro-collagen antibody that only detects human (i.e., MG63-derived) pro-collagen and not that expressed by mouse. Immunolocalization revealed that MG63 cells synthesizing human type I pro-collagen, whilst abundant in the upper layer of cells were also occasionally observed in cells up to 100 μm beneath the surface zone (Figure 8C).
BMP-2 Treatment Regulates MG63 Expression of Type I Collagen in Co-Cultures
In order to determine whether osteoblasts in co-cultures could respond to an osteogenic signal, we stimulated the MLO-Y4/MG63 co-cultures grown in plastic plates with BMP-2 (Figure 9). We used the mouse/human model so that we could discriminate between MLO-Y4-derived and MG63-derived type I collagen expression. BMP-2 treatment significantly increased MG63 COL1A1 mRNA expression at day 5 compared to day 1 (Figure 9A) (GLM of log10 data, P = 0.03, two independent experiments of n = 3). However, BMP-2 treatment had no effect on MLO-Y4 Col1a1 (Figure 9B), MG63 or MLO-Y4 OCN (Figures 9C,D), and E11 (Figures 9E,F) mRNA expression.
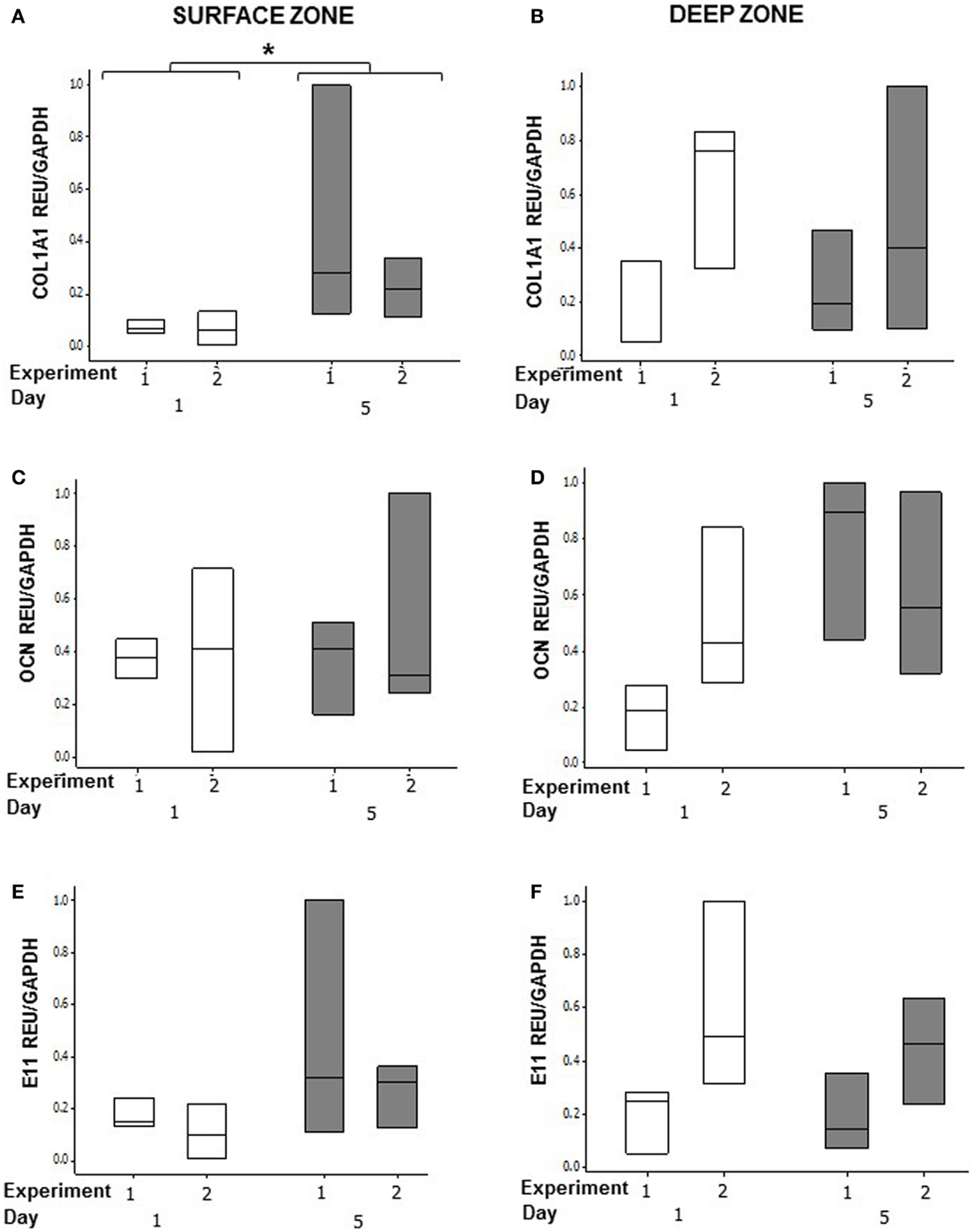
Figure 9. Effects of BMP-2 treatment on osteoblast and osteocyte phenotype in MLO-Y4/MG63 3D co-cultures. Quantification of gene expression in surface and deep zone cells of 3D co-cultures at day 1 and 5 after BMP-2 treatment by relative RT-qPCR, boxplots of COL1A1 (A,B), OCN (C,D), and E11 (E,F) expressed as REU and normalized to GAPDH expression. Significant differences obtained by GLM of log10 data denoted by *P < 0.05. Data are from two independent experiments of n = 3 for surface and deep zones.
Immunolabelling with monoclonal antibody M38 that recognizes the C-terminus of human type I pro-collagen (an epitope not present in the collagen used to make the gel) revealed that type I pro-collagen is abundant in surface MG63 cells after 5 days BMP-2 treatment (Figures 10A,B).
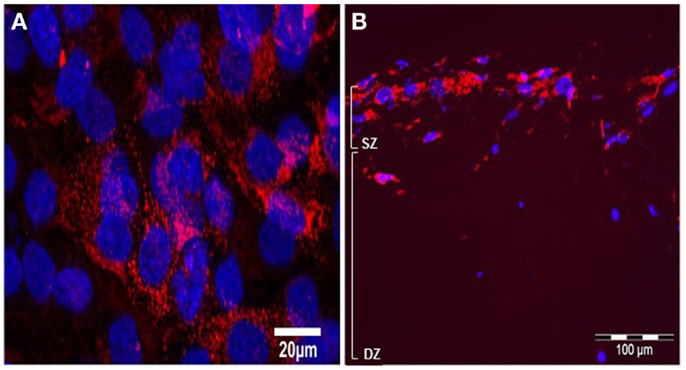
Figure 10. Effects of BMP-2 treatment on type I pro-collagen synthesis in co-cultures of MLO-Y4 and MG63 cells (type I pro-collagen: red; DAPI: blue). Confocal microscope image of the surface cells from an untreated 3D co-culture revealed presence of particulate type I pro-collagen in all surface cells (A). Fluorescence microscope images of transverse cryosections from BMP-2 treated 3D co-cultures at day 5 (B) revealed presence of type I pro-collagen in the upper layer of cells and in cells up to 100 μm beneath the surface, which are all MG63 cells since M38 antibody does not recognize mouse type I pro-collagen.
Embedded MLO-Y4 Cells Release PGE2 in Response to Mechanical Loading
A pilot experiment to determine whether mechanical loading induced PGE2 release in MLO-Y4 cells in 3D gels in the silicone plate, revealed that load (5 min, 10 Hz, 2.5 N) increased PGE2 release between 0.5 and 24 h. Mean PGE2 release was increased approximately 4-fold at 0.5 h post-load (control 1206.55 ± 37.32 pg/ml; loaded 4632.91 ± 1773.78 pg/ml; n = 2 at each time) (Figure 11A).
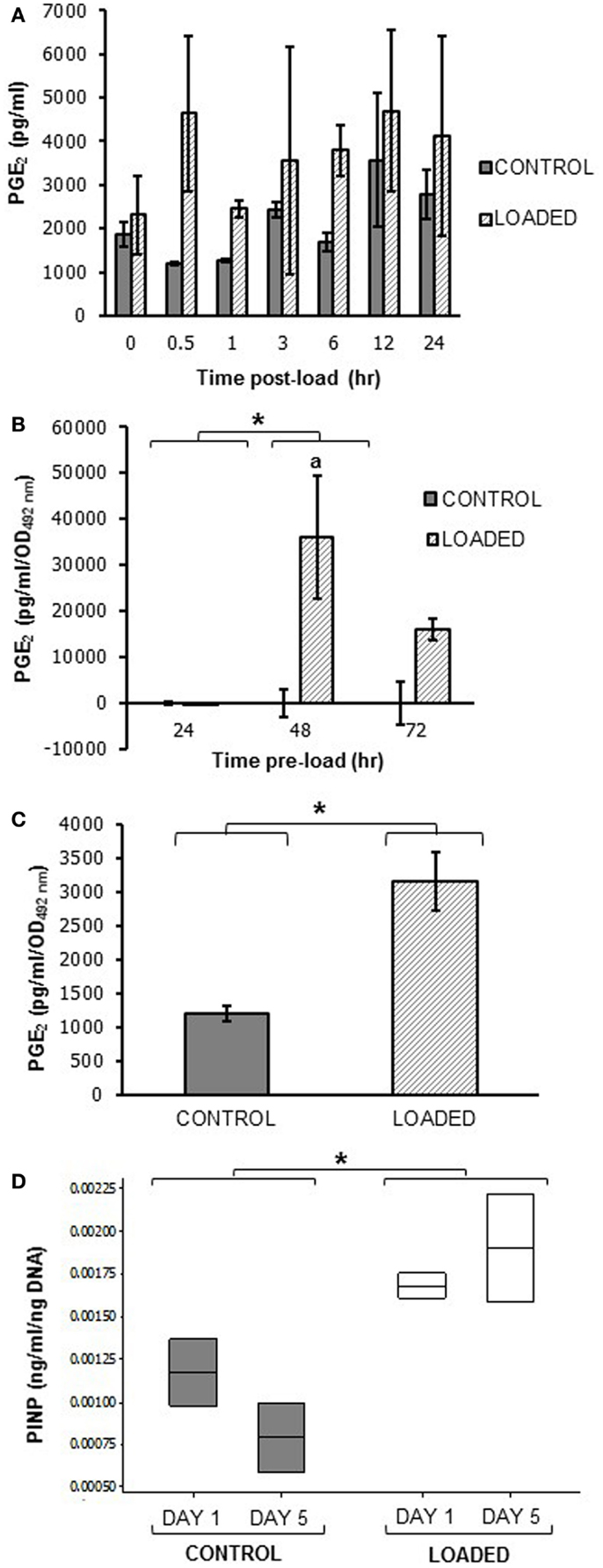
Figure 11. Prostaglandin E2 and PINP release in mechanically loaded (5 min, 10 Hz, 2.5 N) 3D cultures by ELISA. Graphs showing PGE2 release from 3D osteocyte mono-cultures in a pilot experiment of 24 h cultures (A), categorized by time of culture (B), and 7 days cultures (C) at 0.5 h post-load unless other time-points are indicated. Data were normalized to the absorbance (OD492 nm) of LDH lysates (cell number) (B,C). (D) Boxplot of PINP release from control and loaded MLO-Y4/MC3T3-E1(14) 3D co-cultures cultures at day 1 and day 5 post-load, normalized to total DNA. *P < 0.05 as obtained by GLM, GLM of ranked data (B) or one-way ANOVA (C,D). Significant differences as obtained by GLM pairwise comparisons denoted by “a” with respect to 24 h loaded cultures (B). Data presented are from (A) one independent experiment, n = 2 or 3; (B) one (48–72 h cultures) or two (24 h cultures) independent experiments, n = 3; (C,D) one independent experiment, n = 2 or 3.
To determine whether load-induced PGE2 release was affected by MLO-Y4 culture time in 3D gels prior to loading, MLO-Y4 cells were pre-cultured in gels for 24, 48, or 72 h and PGE2 measured 0.5 h after loading as before. After normalizing to cell number, PGE2 was not detectable in loaded or control 3D MLO-Y4 mono-cultures pre-cultured for 24 h, whereas mean PGE2 was increased in loaded osteocytes pre-cultured for 48 h and for 72 h compared with their respective unloaded controls (Figure 11B, n = 3 per pre-culture time). When MLO-Y4 cells were pre-cultured for 7 days prior to mechanical loading, mean PGE2 release, normalized to cell number, was also increased 0.5 h post-load [control 1195.40 ± 109.72 pg/ml/OD492 nm, loaded 3152.26 ± 435.20 pg/ml/OD492 nm; n = 2 or 3 (Figure 11C)].
To determine whether mechanical loading could induce type I pro-collagen synthesis a pilot experiment assessed PINP synthesis 1 and 5 days post-load. This was carried out in 3D MLO-Y4/MC3T3-E1(14) co-cultures grown in the silicone plate pre-cultured for 7 days prior to load. Our preliminary findings reveal mean PINP release was increased in loaded 3D co-cultures when compared to control cultures at day 1 or 5 (Figure 11D, n = 2 control and loaded).
Discussion
This paper describes methodology for a novel in vitro 3D osteocyte–osteoblast co-culture model, which can be used to assess how osteocytes regulate osteoblasts in response to mechanical load. The model has been morphologically and phenotypically characterized, and methodology optimized based on responses to mechanical load. The model is a two-phase culture system where osteocytes are embedded within collagen gels and cultured overnight before osteoblasts were added to the surface of the gel. In this model, cells were viable, expressed appropriate phenotypic markers and contacted neighboring cells. A 16-well silicone plate was developed to enable application of physiological and osteogenic forces within each gel. Our preliminary findings indicate that these 3D cultures increase PGE2 synthesis and PINP release in response to mechanical loading.
Osteocyte and Osteoblast Viability in 3D Co-Cultures
In the 3D co-cultures, both MC3T3-E1(14) and MG63 surface osteoblasts were 100% viable. It is likely that osteoblasts on the surface of the co-culture behave like a monolayer of cells, with dead osteoblasts detaching from the top of the collagen gel and being replaced by new osteoblasts to maintain the single cell layer.
In the 3D co-cultures, embedded osteocytes displayed 16% death after 1 day of culture [MLO-Y4/MC3T3-E1(14)] and 10–14% death at day 7 (both co-culture systems). Osteocyte death is common in normal human bone (57) and increases from <1% at birth up to 75% by 80 years old (58–60). If there is a linear relationship between age and osteocyte death, 20% osteocyte death would occur in humans in their early 20s, consistent with osteocyte viability observed in both 3D models. Cells undergoing cell death are usually destroyed by neighboring or phagocytic cells (61), but dead osteocytes, embedded within a mineralized matrix are inaccessible, and can be detected within their lacunae in vivo (62, 63). In the 3D co-culture a similar percentage osteocyte cell death was observed at day 1 and day 7, which may reflect dead osteocyte retention within the matrix, but this remains to be determined.
Osteocyte and Osteoblast Morphology in 3D Co-Cultures
In the 3D co-culture model, both MC3T3-E1(14) and MG63 cells displayed a range of osteoblastic, ovoid, and pyriform morphologies, when maintained for 7 days. They formed a pavement-like monolayer on top of the 3D culture with well-defined stress-fibers. Whilst both MC3T3-E1(14) (64) and MG63 (65) monolayer cultures show fibroblastic morphology during logarithmic growth in vitro, they assume a pyriform shape with prominent stress-fibers across their cell bodies when confluent (39, 64). In vivo, osteoblasts can be ovoid, rectangular, columnar, cuboidal, or pyriform (66). Osteoblasts form a pavement-like or “overlapping roof tiles” monolayer on the bone surface [Bidder, 1906 as cited in Bourne (66) and Sudo et al. (64)] overlaying osteocytes within the bone matrix [Gegenbaur, 1864 as cited in Bourne (66)]. Osteoblast position is essential for osteocyte–osteoblast interactions, which ultimately regulate bone matrix formation (36, 67–69). Osteoblast morphology in the 3D co-culture is thus consistent with in vitro and in vivo observations.
In the 3D co-culture model, MLO-Y4 cells maintain their osteocytic morphology throughout all gel depths for 7 days, with cell projections from adjacent cells in contact. In vivo, osteocytes present a dendritic morphology that allows communication with neighboring osteocytes. This forms an extensive network known as the LCS (12, 70–73), which permits metabolic traffic and exchange within the mineralized environment of the bone matrix. In vitro, monolayer cultures of MLO-Y4 cells display a 2D dendritic morphology, which becomes 3D in collagen gel cultures (34, 39). Furthermore, IDG-SW3 cells also display dendritic morphology in 3D gels (35). The osteocyte morphology in the 3D co-cultures is consistent with both in vivo and in vitro observations, with morphological characteristics indicative of a 3D network throughout the co-culture.
Osteocyte and Osteoblast Phenotype in 3D Co-Cultures
In 3D co-cultures, MC3T3-E1(14) cells expressed E11 mRNA and protein and MG63 cells expressed E11 mRNA (antibody does not recognize human E11). E11 has been detected in mature osteoblasts and osteoblasts undergoing bone matrix synthesis (74–78). The E11 expression detected in the surface zone of the model suggests that osteoblasts may be sending out projections to connect with neighboring osteoblasts and/or embedded osteocytes. Previous studies have shown that osteoblasts have cytoplasmic processes connecting them to neighboring cells [Spuler, 1899 as cited in Bourne (66), Wetterwald et al. (75), and Schulze et al. (79)], with prominent stress-fibers that stretch across their cell bodies into small cytoplasmic processes (39).
OCN, Runx2, and Col1a1 were also expressed in MC3T3-E1(14) and MG63 cells when in 3D co-cultures, consistent with in vivo and in vitro studies (51, 80–84). ALP mRNA expression was also found in MC3T3-E1(14) cells in 3D co-cultures consistent with previous reports (51, 80, 81, 84–86). CX43 protein expression was observed throughout osteoblast cytoplasm and cell membrane in both MC3T3-E1(14) and MG63 cells in 3D co-cultures, suggesting that the surface osteoblasts have the potential to connect to neighboring cells. Osteoblasts have been shown to express the gap junction CX43 protein (36, 87, 88), which is important in responses to mechanical loading (21) and skeletal function (68, 89, 90).
In 3D co-cultures MLO-Y4 cells expressed mRNA and protein for the early osteocyte marker E11 (75–77, 79) consistent with previous data (75–77). OCN, Runx2, Col1a1, and ALP mRNAs were also detected in these cells in co-cultures. Osteocytes have been previously shown to express OCN, Col1a1, and ALP in vivo and in vitro (34, 91) and Runx2 in vitro (92).
Confocal imaging of osteocytes in 3D co-cultures showed the presence of CX43 throughout the cytoplasm, osteocytic processes, and around the nucleus. Osteocytes express CX43 both in vivo and in vitro (34, 36, 93, 94), which allows the formation of the LCS within the bone matrix, and connects osteocytes to surface osteoblasts (36, 95). CX43 gap junctions in osteocytes contribute to bone remodeling and formation (96) and they are also mediate load-induced PGE2 release in osteocytes (97). The expression of CX43 indicates that osteocytes within the 3D co-culture are potentially able to form a network similar to the LCS, as well as connect to the osteoblasts on the surface.
In 3D co-cultures, MC3T3-E1(14) cells showed a significantly higher expression of Col1a1 mRNA compared to the deep zone of the model. Interestingly, deep zone cells within the 3D co-cultures expressed significantly higher levels of ALP compared to the surface zone of the model. This suggests that under appropriate conditions the MLO-Y4 cells within the 3D model may contribute to mineralization the collagen matrix within which they are embedded, as previously seen in embedded IDG-SW3 cells (35).
Testing Osteogenic Responses in the 3D Co-Culture Model
Pilot experiments were performed to see whether the co-culture methodology could reveal an osteogenic response. This was tested firstly by treatment with BMP-2 and secondly by testing responses to mechanical loading.
In the MLO-Y4/MG63 co-culture, osteoblasts were able to respond to BMP-2, by significantly increasing their COL1A1 mRNA expression and showing abundant type I pro-collagen protein expression after 5 days of BMP-2 treatment. These data are consistent with previous in vitro studies, which showed that BMP-2 stimulates collagen synthesis in MC3T3-E1 cells (85).
To determine whether the osteocytes within the co-culture model responded to loading, we cultured MLO-Y4 in 3D collagen gels, without surface osteoblasts, and measured PGE2 release in response to loading. To facilitate loading of the 3D model, a 16-well silicone plate was developed that applied uniform strain within each gel. The loading regime applied (5 min, 10 Hz, 2.5 N) was based on previous publications showing that 10 min of 10 Hz, 4000–4500 με loading is physiological and osteogenic in vivo (91, 98, 99). In 3D osteocyte mono-cultures, loading induced PGE2 release over 24 h with maximum PGE2 release occurred after 0.5 h. In osteocytes pre-cultured in 3D collagen gels for 48, 72 h, or 7 days, mechanical loading increased PGE2 release 0.5 h post-load. No PGE2 release occurred in osteocytes pre-cultured in 3D gels for 24 h. This suggests that the osteocytes may require at least 48 h in 3D collagen gels to develop an osteocytic phenotype, form dendrites and the CX43 gap junctions that are involved in the release of PGE2 from osteocytes in vitro (100, 101). Others have shown that mechanically loaded osteocytes in monolayer increase PGE2 release (24, 93, 102, 103), as early as 0.5 h post-load (93) but no previous studies have investigated osteocyte response to load in 3D.
To determine whether mechanical loading in 3D co-cultures could elicit an osteogenic response, co-cultures were mechanically loaded as before and type I collagen synthesis quantified. In 3D co-cultures, mechanical loading increased PINP release, suggesting that mechanical stimuli of 3D co-cultures elicit an osteogenic response. PINP synthesis was measured from whole 3D co-cultures, therefore, PINP synthesis may not only be from surface osteoblasts, but also from embedded osteocytes. Both osteoblasts and osteocytes produce type I collagen in vitro (34, 104) although MLO-Y4 cells express reduced Col1a1 mRNA compared to osteoblasts both in monolayer (34) and here in 3D co-cultures.
Our preliminary data showing that both BMP-2 and mechanical loading can induce type I collagen synthesis, reveals the potential for the new 3D co-culture and loading methodology described in this paper in investigating osteogenic responses regulated by osteocytes.
Limitations of the 3D Co-Culture Model
Cell migration in co-cultures
The 3D co-culture method is subject to the possibility of cross-contamination of RNA between surface osteoblasts and embedded osteocytes, due to the extraction protocol, or mixing of cell types between zones due to osteoblast and/or osteocyte migration. We used expression of the SV40 large T-antigen, exclusive to MLO-Y4 cells [derived from mice expressing the SV40 large T-antigen oncogene under the control of the OCN promoter (34)], and an antibody that detects human but not mouse type I pro-collagen, to investigate this. The expression of SV40 large T-antigen mRNA in RNA extracted from the surface zone, suggests that there is low level RNA cross-contamination from the osteocytes, or MLO-Y4 cell migration to the surface in MLO-Y4/MC3T3-E1(14) co-cultures. Since no SV40 large T-antigen immunostaining was observed in the surface zone of the model even after 7 days of co-culture, we conclude that no osteocytes migrated to the surface zone of the 3D co-culture and that the SV40 large T-antigen mRNA contamination in the surface zone is due to MLO-Y4 cells immediately underlying gel surface, lysing, and releasing RNA during extraction from surface cells. However, the human-specific type I pro-collagen antibody revealed that, although rare, some MG63 cells migrated from the surface to the deep zone of the model, in MLO-Y4/MG63 co-cultures.
Phenotype and function
Whilst the 3D co-culture model described here has shown osteoblast and osteocyte cell viabilities (58–60, 105, 106), morphologies (66, 71, 107, 108), phenotypes (75, 81, 86, 91, 109–114), and loading (104) and osteogenic responses (115, 116) consistent with those found in vivo, the use of MLO-Y4 cells means that the important mechanically regulated factor, SOST, is not expressed in the osteocytes in the model. This limitation could be solved by replacing the MLO-Y4 cells with the IDG-SW3 cell line, which are able to differentiate into mature osteocytes and express SOST (35). Furthermore, the phenotypic characterization was performed in 3D co-cultures grown in plastic plates and it is possible that aspects of the phenotype would be affected by growing cells in silicone plates.
Although the 3D model is designed to investigate mechanically induced osteogenesis in a similar in vivo physiological environment, it is not mineralized and so it would only represent interactions that occur in newly formed osteoid rather than mineralized bone. Previous studies have shown the mineralization of 3D collagen gels is possible with IDG-SW3 cells (35) during differentiation to osteocytes, and therefore the 3D co-culture could be mineralized. Mineralization of the 3D collagen gel would affect the properties of the matrix and cell–ECM interactions. Previous in vitro studies have shown that after mineralization the ECM of monolayer cultures became gradually stiffer (117). Furthermore, ECM composition has been shown to affect gene expression (118) and osteoblast differentiation and behavior (119). Therefore, mineralizing the 3D co-culture would make the collagen gels stiffer and alter phenotype, further mimicking a physiological environment. If the 3D co-culture was mineralized, further investigations should be done to test the mechanical properties of the mineralized 3D co-cultures as well as the viability and phenotype of the cells within the model and assess whether the medium nutrients can still diffuse to all areas of the 3D gel.
A technical challenge is to ensure that MLO-Y4 cells are evenly distributed within the collagen solution when gels are being set up, otherwise some 3D cultures will have more osteocytes than others. This will lead to cell number variability between experimental replicates, which could cause differences in osteocytic network and loading responses. This was further affected by a variation of up to 20% in the weight of collagen supplied by Sigma, meaning that, since the defined collagen mass was dissolved in a set volume of acid, the collagen gels varied from 2.0 to 2.6 mg/ml. These are potential explanations for differences in magnitude of responses across independent experiments and the essential 3D pre-culture time of at least 48 h for a consistent increase in PGE2 in response to mechanical loading.
Mechanical loading device
Currently, there are two devices similar to the one developed here (120, 121). Tata et al. (121) developed a silicone plate in a six-well plate format to mechanically load vascular smooth muscle cells (VSMCs) in monolayers, whereas, the device developed by Neidlinger-Wilke et al. (120) is a single-well silicone plate, which was designed to load 3D collagen cultures of intervertebral disk cells. Both devices applied cyclic mechanical stimuli by stretching in a similar fashion to the device described here, but used much higher strains at low frequency (Neidlinger-Wilke et al., 24 h, 0.1 Hz, 10,000 με; Tata et al., 6–72 h, 1 Hz, 10–20% strain) (120, 121). Neidlinger-Wilke et al. (120) did not publish how they assessed strain associated with their device. Tata et al. (121) assessed the strain field at the bottom surface of the wells using finite element (FE) modeling, but did not validate this FE model with DIC, or any other methods. Therefore, our loading device is the first where the strains have been directly measured, albeit on the plate surface rather than within the gel.
Digital image correlation showed that when 2.5 N is applied to the silicone plate, the majority of the wells experienced strains of 4000–4500 με. Peak strain values in vertebrate bone range from 2000 to 3500 με (122–125), 4000–4500 με loading is physiological and osteogenic (91, 98, 99), whereas, 6000 με is pathophysiological (126). The strain testing performed was carried out on an empty plate. Testing a silicone plate with 3D cultures within the wells would further validate the loading plate. Whilst incorporation of particles into the 3D gels (127) would enable strains to be measured directly within the gels, we were unable to achieve this by DIC given the limited well size and the pink color and reflective properties of the gels. Further work is necessary to confirm the strain experienced by the cells in the gels is similar to that on the base of the plate.
Conclusion
There is a great need for a fully characterized in vitro 3D matrix based bone model. The majority of the available 3D models involve culturing cells on scaffolds (44–46, 128), which does not represent the bone environment in vivo where osteocytes, are embedded within a matrix. Published models involving embedding osteoblasts (39, 129), MLO-Y4 (38, 39), primary osteocytes (42), or normal human bone-derived cells (NHBCs) (41) within a matrix showed maintenance of cell viability (38, 129), osteocyte cell morphology (38, 39, 41, 42), connectivity (38), and gene expression (41, 42). However, none of these models have been individually assessed in all key areas. Furthermore, none of the available 3D collagen based cultures involve co-culturing osteocytes and osteoblasts, nor they have been exposed to mechanical stimuli. Therefore, none investigate the important interactions between these cell types, which lead to mechanically induced bone formation.
This co-culture model facilitates a 3D network of osteocyte-like cells that can be subjected to appropriate anabolic and mechanical loading cues to act upon osteoblasts. Osteoblasts and osteocytes retain appropriate morphology, phenotype, and viability, and osteoblasts increase COL1A1 expression when stimulated with BMP-2 and mechanical load. Furthermore, embedded osteocytes respond to mechanical loading by releasing PGE2. Potentially, this model may be useful in elucidating osteocyte-driven mechanisms that regulate bone formation as a result of mechanical loading, something other current 3D models do not provide (38, 39, 41, 42). The 3D co-culture, combined with a multi-well loading system could provide a novel platform for drug discovery and development for the treatment of age-related bone diseases.
Author Contributions
Conception and design: Marisol Vazquez, Bronwen A. J. Evans, Daniela Riccardi, Sam L. Evans, Jim R. Ralphs, Christopher M. Dillingham, Deborah J. Mason. Collection and assembly of data: Marisol Vazquez, Sam L. Evans, Jim R. Ralphs, Christopher M. Dillingham. Analysis and interpretation of data: Marisol Vazquez, Bronwen A. J. Evans, Daniela Riccardi, Sam L. Evans, Deborah J. Mason. Drafting of the manuscript: Marisol Vazquez, Deborah J. Mason. Critical revision: Marisol Vazquez, Bronwen A. J. Evans, Daniela Riccardi, Sam L. Evans, Deborah J. Mason. Final approval of the article: Marisol Vazquez, Bronwen A. J. Evans, Daniela Riccardi, Sam L. Evans, Jim R. Ralphs, Christopher M. Dillingham, Deborah J. Mason.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Professor Lynda Bonewald for the provision of the MLO-Y4 cell line, and Mrs. Carole Elford, Dr. Emma Blain, and Dr. Karen Brakspear for their contribution. This project was funded by Cardiff University and the Arthritis Research UK Biomechanics and Bioengineering Center.
Footnotes
References
1. Franz-Odendaal TA, Hall BK, Witten PE. Buried alive: how osteoblasts become osteocytes. Dev Dyn (2006) 235:176–90. doi: 10.1002/dvdy.20603
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. Pead MJ, Suswillo R, Skerry TM, Vedi S, Lanyon LE. Increased 3H-uridine levels in osteocytes following a single short period of dynamic bone loading in vivo. Calcif Tissue Int (1988) 43:92–6. doi:10.1007/BF02555153
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
3. Skerry TM, Bitensky L, Chayen J, Lanyon LE. Early strain-related changes in enzyme activity in osteocytes following bone loading in vivo. J Bone Miner Res (1989) 4:783–8. doi:10.1002/jbmr.5650040519
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Gluhak-Heinrich J, Ye L, Bonewald LF, Feng JQ, Macdougall M, Harris SE, et al. Mechanical loading stimulates dentin matrix protein 1 (DMP1) expression in osteocytes in vivo. J Bone Miner Res (2003) 18:807–17. doi:10.1359/jbmr.2003.18.5.807
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Harris SE, Gluhak-Heinrich J, Harris MA, Yang W, Bonewald LF, Riha D, et al. DMP1 and MEPE expression are elevated in osteocytes after mechanical loading in vivo: theoretical role in controlling mineral quality in the perilacunar matrix. J Musculoskelet Neuronal Interact (2007) 7:313–5.
6. Reijnders CM, Bravenboer N, Holzmann PJ, Bhoelan F, Blankenstein MA, Lips P. In vivo mechanical loading modulates insulin-like growth factor binding protein-2 gene expression in rat osteocytes. Calcif Tissue Int (2007) 80:137–43. doi:10.1007/s00223-006-0077-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Reijnders CM, Bravenboer N, Tromp AM, Blankenstein MA, Lips P. Effect of mechanical loading on insulin-like growth factor-I gene expression in rat tibia. J Endocrinol (2007) 192:131–40. doi:10.1677/joe.1.06880
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Zaman G, Pitsillides AA, Rawlinson SC, Suswillo RF, Mosley JR, Cheng MZ, et al. Mechanical strain stimulates nitric oxide production by rapid activation of endothelial nitric oxide synthase in osteocytes. J Bone Miner Res (1999) 14:1123–31. doi:10.1359/jbmr.1999.14.7.1123
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Fox SW, Chambers TJ, Chow JW. Nitric oxide is an early mediator of the increase in bone formation by mechanical stimulation. Am J Physiol (1996) 270:E955–60.
10. Mullender MG, Huiskes R. Osteocytes and bone lining cells: which are the best candidates for mechano-sensors in cancellous bone? Bone (1997) 20:527–32. doi:10.1016/S8756-3282(97)00036-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Nomura S, Takano-Yamamoto T. Molecular events caused by mechanical stress in bone. Matrix Biol (2000) 19:91–6. doi:10.1016/S0945-053X(00)00050-0
12. Plotkin LI, Manolagas SC, Bellido T. Transduction of cell survival signals by connexin-43 hemichannels. J Biol Chem (2002) 277:8648–57. doi:10.1074/jbc.M108625200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Turner CH, Robling AG, Duncan RL, Burr DB. Do bone cells behave like a neuronal network? Calcif Tissue Int (2002) 70:435–42. doi:10.1007/s00223-001-1024-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Klein-Nulend J, Bakker AD, Bacabac RG, Vatsa A, Weinbaum S. Mechanosensation and transduction in osteocytes. Bone (2013) 54:182–90. doi:10.1016/j.bone.2012.10.013
15. You L, Cowin SC, Schaffler MB, Weinbaum S. A model for strain amplification in the actin cytoskeleton of osteocytes due to fluid drag on pericellular matrix. J Biomech (2001) 34:1375–86. doi:10.1016/S0021-9290(01)00107-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Mcgarry JG, Klein-Nulend J, Prendergast PJ. The effect of cytoskeletal disruption on pulsatile fluid flow-induced nitric oxide and prostaglandin E2 release in osteocytes and osteoblasts. Biochem Biophys Res Commun (2005) 330:341–8. doi:10.1016/j.bbrc.2005.02.175
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Hoey DA, Kelly DJ, Jacobs CR. A role for the primary cilium in paracrine signaling between mechanically stimulated osteocytes and mesenchymal stem cells. Biochem Biophys Res Commun (2011) 412:182–7. doi:10.1016/j.bbrc.2011.07.072
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Xiao Z, Dallas M, Qiu N, Nicolella D, Cao L, Johnson M, et al. Conditional deletion of Pkd1 in osteocytes disrupts skeletal mechanosensing in mice. FASEB J (2011) 25:2418–32. doi:10.1096/fj.10-180299
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Nguyen AM, Jacobs CR. Emerging role of primary cilia as mechanosensors in osteocytes. Bone (2013) 54:196–204. doi:10.1016/j.bone.2012.11.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Litzenberger JB, Kim JB, Tummala P, Jacobs CR. Beta1 integrins mediate mechanosensitive signaling pathways in osteocytes. Calcif Tissue Int (2010) 86:325–32. doi:10.1007/s00223-010-9343-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Zhang Y, Paul EM, Sathyendra V, Davison A, Sharkey N, Bronson S, et al. Enhanced osteoclastic resorption and responsiveness to mechanical load in gap junction deficient bone. PLoS One (2011) 6:e23516. doi:10.1371/journal.pone.0023516
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Robling AG, Niziolek PJ, Baldridge LA, Condon KW, Allen MR, Alam I, et al. Mechanical stimulation of bone in vivo reduces osteocyte expression of SOST/sclerostin. J Biol Chem (2008) 283:5866–75. doi:10.1074/jbc.M705092200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Tu X, Rhee Y, Condon KW, Bivi N, Allen MR, Dwyer D, et al. SOST downregulation and local Wnt signaling are required for the osteogenic response to mechanical loading. Bone (2012) 50:209–17. doi:10.1016/j.bone.2011.10.025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Li L, Yang Z, Zhang H, Chen W, Chen M, Zhu Z. Low-intensity pulsed ultrasound regulates proliferation and differentiation of osteoblasts through osteocytes. Biochem Biophys Res Commun (2012) 418:296–300. doi:10.1016/j.bbrc.2012.01.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Tan SD, Kuijpers-Jagtman AM, Semeins CM, Bronckers ALJJ, Maltha JC, Hoff JW, et al. Fluid shear stress inhibits TNFα-induced osteocyte apoptosis. J Dent Res (2006) 85:905–9. doi:10.1177/154405910608501006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Nakashima T, Hayashi M, Fukunaga T, Kurata K, Oh-Hora M, Feng JQ, et al. Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat Med (2011) 17:1231–4. doi:10.1038/nm.2452
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Xiong J, Onal M, Jilka RL, Weinstein RS, Manolagas SC, O’brien CA. Matrix-embedded cells control osteoclast formation. Nat Med (2011) 17:1235–41. doi:10.1038/nm.2448
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Nijweide PJ, Van Der Plas A, Alblas MJ, Klein-Nulend J. Osteocyte isolation and culture. Methods Mol Med (2003) 80:41–50.
29. Van Der Plas A, Nijweide PJ. JBMR anniversary classic. Isolation and purification of osteocytes. A van der Plas A, PJ Nijweide. Originally published in Volume 7, Number 4, pp 389-96 (1992). J Bone Miner Res (2005) 20(4):706–14. doi:10.1002/jbmr.5650070406
30. Semeins CM, Bakker AD, Klein-Nulend J. Isolation of primary avian osteocytes. Methods Mol Biol (2012) 816:43–53. doi:10.1007/978-1-61779-415-5_4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Gu G, Nars M, Hentunen TA, Metsikko K, Vaananen HK. Isolated primary osteocytes express functional gap junctions in vitro. Cell Tissue Res (2006) 323:263–71. doi:10.1007/s00441-005-0066-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Halleux C, Kramer I, Allard C, Kneissel M. Isolation of mouse osteocytes using cell fractionation for gene expression analysis. Methods Mol Biol (2012) 816:55–66. doi:10.1007/978-1-61779-415-5_5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. Kato Y, Boskey A, Spevak L, Dallas M, Hori M, Bonewald LF. Establishment of an osteoid preosteocyte-like cell MLO-A5 that spontaneously mineralizes in culture. J Bone Miner Res (2001) 16:1622–33. doi:10.1359/jbmr.2001.16.9.1622
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Kato Y, Windle JJ, Koop BA, Mundy GR, Bonewald LF. Establishment of an osteocyte-like cell line, MLO-Y4. J Bone Miner Res (1997) 12:2014–23. doi:10.1359/jbmr.1997.12.12.2014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. Woo SM, Rosser J, Dusevich V, Kalajzic I, Bonewald LF. Cell line IDG-SW3 replicates osteoblast-to-late-osteocyte differentiation in vitro and accelerates bone formation in vivo. J Bone Miner Res (2011) 26:2634–46. doi:10.1002/jbmr.465
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Taylor AF, Saunders MM, Shingle DL, Cimbala JM, Zhou Z, Donahue HJ. Mechanically stimulated osteocytes regulate osteoblastic activity via gap junctions. Am J Physiol Cell Physiol (2007) 292:C545–52. doi:10.1152/ajpcell.00611.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Boukhechba F, Balaguer T, Michiels JF, Ackermann K, Quincey D, Bouler JM, et al. Human primary osteocyte differentiation in a 3D culture system. J Bone Miner Res (2009) 24:1927–35. doi:10.1359/jbmr.090517
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Kurata K, Heino TJ, Higaki H, Vaananen HK. Bone marrow cell differentiation induced by mechanically damaged osteocytes in 3D gel-embedded culture. J Bone Miner Res (2006) 21:616–25. doi:10.1359/jbmr.060106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Murshid SA, Kamioka H, Ishihara Y, Ando R, Sugawara Y, Takano-Yamamoto T. Actin and microtubule cytoskeletons of the processes of 3D-cultured MC3T3-E1 cells and osteocytes. J Bone Miner Metab (2007) 25:151–8. doi:10.1007/s00774-007-0772-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. Qi J, Chi L, Faber J, Koller B, Banes AJ. ATP reduces gel compaction in osteoblast-populated collagen gels. J Appl Physiol (2007) 102:1152–60. doi:10.1152/japplphysiol.00535.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Atkins GJ, Welldon KJ, Holding CA, Haynes DR, Howie DW, Findlay DM. The induction of a catabolic phenotype in human primary osteoblasts and osteocytes by polyethylene particles. Biomaterials (2009) 30:3672–81. doi:10.1016/j.biomaterials.2009.03.035
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Honma M, Ikebuchi Y, Kariya Y, Suzuki H. Establishment of optimized in vitro assay methods for evaluating osteocyte functions. J Bone Miner Metab (2014). doi:10.1007/s00774-013-0555-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. Santos MI, Unger RE, Sousa RA, Reis RL, Kirkpatrick CJ. Crosstalk between osteoblasts and endothelial cells co-cultured on a polycaprolactone-starch scaffold and the in vitro development of vascularization. Biomaterials (2009) 30:4407–15. doi:10.1016/j.biomaterials.2009.05.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Tortelli F, Pujic N, Liu Y, Laroche N, Vico L, Cancedda R. Osteoblast and osteoclast differentiation in an in vitro three-dimensional model of bone. Tissue Eng Part A (2009) 15:2373–83. doi:10.1089/ten.tea.2008.0501
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Barron MJ, Tsai CJ, Donahue SW. Mechanical stimulation mediates gene expression in MC3T3 osteoblastic cells differently in 2D and 3D environments. J Biomech Eng (2010) 132:041005. doi:10.1115/1.4001162
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. Krishnan V, Dhurjati R, Vogler EA, Mastro AM. Osteogenesis in vitro: from pre-osteoblasts to osteocytes: a contribution from the Osteobiology Research Group, The Pennsylvania State University. In vitro Cell Dev Biol Anim (2010) 46:28–35. doi:10.1007/s11626-009-9238-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Nakagawa K, Abukawa H, Shin MY, Terai H, Troulis MJ, Vacanti JP. Osteoclastogenesis on tissue-engineered bone. Tissue Eng (2004) 10:93–100. doi:10.1089/107632704322791736
48. Domaschke H, Gelinsky M, Burmeister B, Fleig R, Hanke T, Reinstorf A, et al. In vitro ossification and remodeling of mineralized collagen I scaffolds. Tissue Eng (2006) 12:949–58. doi:10.1089/ten.2006.12.949
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Tortelli F, Cancedda R. Three-dimensional cultures of osteogenic and chondrogenic cells: a tissue engineering approach to mimic bone and cartilage in vitro. Eur Cell Mater (2009) 17:1–14.
50. Kitase Y, Barragan L, Qing H, Kondoh S, Jiang JX, Johnson ML, et al. Mechanical induction of PGE2 in osteocytes blocks glucocorticoid-induced apoptosis through both the beta-catenin and PKA pathways. J Bone Miner Res (2010) 25:2657–68. doi:10.1002/jbmr.168
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Wang D, Christensen K, Chawla K, Xiao G, Krebsbach PH, Franceschi RT. Isolation and characterization of MC3T3-E1 preosteoblast subclones with distinct in vitro and in vivo differentiation/mineralization potential. J Bone Miner Res (1999) 14:893–903. doi:10.1359/jbmr.1999.14.6.893
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
52. Mcdonald JA, Broekelmann TJ, Matheke ML, Crouch E, Koo M, Kuhn C III. A monoclonal antibody to the carboxyterminal domain of procollagen type I visualizes collagen-synthesizing fibroblasts. Detection of an altered fibroblast phenotype in lungs of patients with pulmonary fibrosis. J Clin Invest (1986) 78:1237–44. doi:10.1172/JCI112707
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods (2001) 25:402–8. doi:10.1006/meth.2001.1262
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res (2004) 64:5245–50. doi:10.1158/0008-5472.CAN-04-0496
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Peters WH, Ranson WF. Digital imaging techniques in experimental stress analysis. Opt Eng (1982) 21:213427–213427. doi:10.1117/12.7972925
56. Chu T, Ranson W, Sutton M. Applications of digital-image-correlation techniques to experimental mechanics. Exp Mech (1985) 25:232–44. doi:10.1016/j.jmbbm.2013.02.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
57. Noble BS, Stevens H, Loveridge N, Reeve J. Identification of apoptotic changes in osteocytes in normal and pathological human bone. Bone (1997) 20:273–82. doi:10.1016/S8756-3282(96)00365-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
59. Mullender MG, Van Der Meer DD, Huiskes R, Lips P. Osteocyte density changes in aging and osteoporosis. Bone (1996) 18:109–13. doi:10.1016/8756-3282(95)00444-0
60. Tomkinson A, Reeve J, Shaw RW, Noble BS. The death of osteocytes via apoptosis accompanies estrogen withdrawal in human bone. J Clin Endocrinol Metab (1997) 82:3128–35. doi:10.1210/jc.82.9.3128
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Gregory CD, Pound JD. Cell death in the neighbourhood: direct microenvironmental effects of apoptosis in normal and neoplastic tissues. J Pathol (2011) 223:178–95. doi:10.1002/path.2792
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
62. Tomkinson A, Gevers EF, Wit JM, Reeve J, Noble BS. The role of estrogen in the control of rat osteocyte apoptosis. J Bone Miner Res (1998) 13:1243–50. doi:10.1359/jbmr.1998.13.8.1243
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Weinstein RS, Nicholas RW, Manolagas SC. Apoptosis of osteocytes in glucocorticoid-induced osteonecrosis of the hip. J Clin Endocrinol Metab (2000) 85:2907–12. doi:10.1210/jc.85.8.2907
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Sudo H, Kodama HA, Amagai Y, Yamamoto S, Kasai S. In vitro differentiation and calcification in a new clonal osteogenic cell line derived from newborn mouse calvaria. J Cell Biol (1983) 96:191–8. doi:10.1083/jcb.96.1.191
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
65. Billiau A, Edy VG, Heremans H, Van Damme J, Desmyter J, Georgiades JA, et al. Human interferon: mass production in a newly established cell line, MG-63. Antimicrob Agents Chemother (1977) 12:11–5. doi:10.1128/AAC.12.1.11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Bourne GH. The Biochemistry and Physiology of Bone. 2nd ed. New York, NY: Academic Press (1972). p. 21–40.
67. Heino TJ, Hentunen TA, Vaananen HK. Conditioned medium from osteocytes stimulates the proliferation of bone marrow mesenchymal stem cells and their differentiation into osteoblasts. Exp Cell Res (2004) 294:458–68. doi:10.1016/j.yexcr.2003.11.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
68. Rhee Y, Allen MR, Condon K, Lezcano V, Ronda AC, Galli C, et al. PTH receptor signaling in osteocytes governs periosteal bone formation and intracortical remodeling. J Bone Miner Res (2011) 26:1035–46. doi:10.1002/jbmr.304
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
69. Zarrinkalam MR, Mulaibrahimovic A, Atkins GJ, Moore RJ. Changes in osteocyte density correspond with changes in osteoblast and osteoclast activity in an osteoporotic sheep model. Osteoporos Int (2012) 23:1329–36. doi:10.1007/s00198-011-1672-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
70. Doty SB. Morphological evidence of gap junctions between bone cells. Calcif Tissue Int (1981) 33:509–12. doi:10.1007/BF02409482
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
71. Menton DN, Simmons DJ, Chang SL, Orr BY. From bone lining cell to osteocyte – an SEM study. Anat Rec (1984) 209:29–39. doi:10.1002/ar.1092090105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
72. Palumbo C, Palazzini S, Marotti G. Morphological study of intercellular junctions during osteocyte differentiation. Bone (1990) 11:401–6. doi:10.1016/8756-3282(90)90134-K
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
73. Bonewald LF. Establishment and characterization of an osteocyte-like cell line, MLO-Y4. J Bone Miner Metab (1999) 17:61–5. doi:10.1007/s007740050066
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
74. Nose K, Saito H, Kuroki T. Isolation of a gene sequence induced later by tumor-promoting 12-O-tetradecanoylphorbol-13-acetate in mouse osteoblastic cells (MC3T3-E1) and expressed constitutively in ras-transformed cells. Cell Growth Differ (1990) 1:511–8.
75. Wetterwald A, Hoffstetter W, Cecchini MG, Lanske B, Wagner C, Fleisch H, et al. Characterization and cloning of the E11 antigen, a marker expressed by rat osteoblasts and osteocytes. Bone (1996) 18:125–32. doi:10.1016/8756-3282(95)00457-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
76. Hadjiargyrou M, Rightmire EP, Ando T, Lombardo FT. The E11 osteoblastic lineage marker is differentially expressed during fracture healing. Bone (2001) 29:149–54. doi:10.1016/S8756-3282(01)00489-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
77. Zhang K, Barragan-Adjemian C, Ye L, Kotha S, Dallas M, Lu Y, et al. E11/gp38 selective expression in osteocytes: regulation by mechanical strain and role in dendrite elongation. Mol Cell Biol (2006) 26:4539–52. doi:10.1128/MCB.02120-05
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
78. Jahn K, Richards RG, Archer CW, Stoddart MJ. Pellet culture model for human primary osteoblasts. Eur Cell Mater (2010) 20:149–61.
79. Schulze E, Witt M, Kasper M, Lowik CW, Funk RH. Immunohistochemical investigations on the differentiation marker protein E11 in rat calvaria, calvaria cell culture and the osteoblastic cell line ROS 17/2.8. Histochem Cell Biol (1999) 111:61–9. doi:10.1007/s004180050334
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
80. Collin P, Nefussi JR, Wetterwald A, Nicolas V, Boy-Lefevre ML, Fleisch H, et al. Expression of collagen, osteocalcin, and bone alkaline phosphatase in a mineralizing rat osteoblastic cell culture. Calcif Tissue Int (1992) 50:175–83. doi:10.1007/BF00298797
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
81. Zhou H, Choong P, Mccarthy R, Chou ST, Martin TJ, Ng KW. In situ hybridization to show sequential expression of osteoblast gene markers during bone formation in vivo. J Bone Miner Res (1994) 9:1489–99. doi:10.1002/jbmr.5650090922
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
82. Shi S, Kirk M, Kahn AJ. The role of type I collagen in the regulation of the osteoblast phenotype. J Bone Miner Res (1996) 11:1139–45. doi:10.1002/jbmr.5650110813
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
83. Katopodis H, Philippou A, Tenta R, Doillon C, Papachroni KK, Papavassiliou AG, et al. MG-63 osteoblast-like cells enhance the osteoprotegerin expression of PC-3 prostate cancer cells. Anticancer Res (2009) 29:4013–8.
84. Tsai SW, Liou HM, Lin CJ, Kuo KL, Hung YS, Weng RC, et al. MG63 osteoblast-like cells exhibit different behavior when grown on electrospun collagen matrix versus electrospun gelatin matrix. PLoS One (2012) 7:e31200. doi:10.1371/journal.pone.0031200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
85. Takuwa Y, Ohse C, Wang EA, Wozney JM, Yamashita K. Bone morphogenetic protein-2 stimulates alkaline phosphatase activity and collagen synthesis in cultured osteoblastic cells, MC3T3-E1. Biochem Biophys Res Commun (1991) 174:96–101. doi:10.1016/0006-291X(91)90490-X
86. Whyte MP. Hypophosphatasia and the role of alkaline phosphatase in skeletal mineralization. Endocr Rev (1994) 15:439–61. doi:10.1210/edrv-15-4-439
87. Lecanda F, Warlow PM, Sheikh S, Furlan F, Steinberg TH, Civitelli R. Connexin43 deficiency causes delayed ossification, craniofacial abnormalities, and osteoblast dysfunction. J Cell Biol (2000) 151:931–44. doi:10.1083/jcb.151.4.931
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
88. Yamaguchi DT, Ma D. Mechanism of pH regulation of connexin 43 expression in MC3T3-E1 cells. Biochem Biophys Res Commun (2003) 304:736–9. doi:10.1016/S0006-291X(03)00633-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
89. Stains JP, Civitelli R. Gap junctions in skeletal development and function. Biochim Biophys Acta (2005) 1719:69–81. doi:10.1016/j.bbamem.2005.10.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
90. Sharrow AC, Li Y, Micsenyi A, Griswold RD, Wells A, Monga SS, et al. Modulation of osteoblast gap junction connectivity by serum, TNFalpha, and TRAIL. Exp Cell Res (2008) 314:297–308. doi:10.1016/j.yexcr.2007.10.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
91. Mason DJ, Hillam RA, Skerry TM. Constitutive in vivo mRNA expression by osteocytes of beta-actin, osteocalcin, connexin-43, IGF-I, c-fos and c-jun, but not TNF-alpha nor tartrate-resistant acid phosphatase. J Bone Miner Res (1996) 11:350–7. doi:10.1002/jbmr.5650110308
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
92. Fujita T, Izumo N, Fukuyama R, Meguro T, Nakamuta H, Kohno T, et al. Phosphate provides an extracellular signal that drives nuclear export of Runx2/Cbfa1 in bone cells. Biochem Biophys Res Commun (2001) 280:348–52. doi:10.1006/bbrc.2000.4108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
93. Cheng B, Kato Y, Zhao S, Luo J, Sprague E, Bonewald LF, et al. PGE(2) is essential for gap junction-mediated intercellular communication between osteocyte-like MLO-Y4 cells in response to mechanical strain. Endocrinology (2001) 142:3464–73. doi:10.1210/endo.142.8.8338
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
94. Cheng B, Zhao S, Luo J, Sprague E, Bonewald LF, Jiang JX. Expression of functional gap junctions and regulation by fluid flow in osteocyte-like MLO-Y4 cells. J Bone Miner Res (2001) 16:249–59. doi:10.1359/jbmr.2001.16.2.249
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
95. Shalhoub V, Shatzen E, Henley C, Boedigheimer M, Mcninch J, Manoukian R, et al. Calcification inhibitors and Wnt signaling proteins are implicated in bovine artery smooth muscle cell calcification in the presence of phosphate and vitamin D sterols. Calcif Tissue Int (2006) 79:431–42. doi:10.1007/s00223-006-0126-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
96. Bivi N, Condon KW, Allen MR, Farlow N, Passeri G, Brun LR, et al. Cell autonomous requirement of connexin 43 for osteocyte survival: consequences for endocortical resorption and periosteal bone formation. J Bone Miner Res (2012) 27:374–89. doi:10.1002/jbmr.548
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
97. Chan ME, Lu XL, Huo B, Baik AD, Chiang V, Guldberg RE, et al. A trabecular bone explant model of osteocyte-osteoblast co-culture for bone mechanobiology. Cell Mol Bioeng (2009) 2:405–15. doi:10.1007/s12195-009-0075-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
98. Hillam RA, Skerry TM. Inhibition of bone resorption and stimulation of formation by mechanical loading of the modeling rat ulna in vivo. J Bone Miner Res (1995) 10:683–9. doi:10.1002/jbmr.5650100503
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
99. Rubin CT, Sommerfeldt DW, Judex S, Qin Y-X. Inhibition of osteopenia by low magnitude, high-frequency mechanical stimuli. Drug Discov Today (2001) 6:848–58. doi:10.1016/S1359-6446(01)01872-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
100. Jiang JX, Cherian PP. Hemichannels formed by connexin 43 play an important role in the release of prostaglandin E(2) by osteocytes in response to mechanical strain. Cell Commun Adhes (2003) 10:259–64. doi:10.1080/cac.10.4-6.259.264
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
101. Cherian PP, Siller-Jackson AJ, Gu S, Wang X, Bonewald LF, Sprague E, et al. Mechanical strain opens connexin 43 hemichannels in osteocytes: a novel mechanism for the release of prostaglandin. Mol Biol Cell (2005) 16:3100–6. doi:10.1091/mbc.E04-10-0912
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
102. Jiang JX, Cheng B. Mechanical stimulation of gap junctions in bone osteocytes is mediated by prostaglandin E2. Cell Commun Adhes (2001) 8:283–8. doi:10.3109/15419060109080738
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
103. Saini V, Yadav S, Mccormick S. Low-intensity pulsed ultrasound modulates shear stress induced PGHS-2 expression and PGE2 synthesis in MLO-Y4 osteocyte-like cells. Ann Biomed Eng (2011) 39:378–93. doi:10.1007/s10439-010-0156-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
104. Yang M, Huang L, Xiao L, Liao E. [Effects of mechanical stimulation on proliferation and differentiation in MG-63 osteoblast-like cells]. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi (2012) 29:894–7.
105. Parfitt A. Bone-forming cells in clinical conditions. In: Bk H, editor. Bone: The Osteoblast and Osteocyte. Boca Raton, FL: Telford Press and CRC Press (1990). p. 351–429.
106. Jilka RL, Weinstein RS, Bellido T, Parfitt AM, Manolagas SC. Osteoblast programmed cell death (apoptosis): modulation by growth factors and cytokines. J Bone Miner Res (1998) 13:793–802. doi:10.1359/jbmr.1998.13.5.793
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
107. Doty SB, Robinson RA, Schofield B. Morphology of bone and histochemical staining characteristics of bone cells. In: Greep RO, Astwood EB, editors. Handbook of Physiology. Washington, DC: American Physiological Society (1976). 3 p.
108. Palumbo C. A three-dimensional ultrastructural study of osteoid-osteocytes in the tibia of chick embryos. Cell Tissue Res (1986) 246:125–31. doi:10.1007/BF00219008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
109. Dodds RA, Ali N, Pead MJ, Lanyon LE. Early loading-related changes in the activity of glucose 6-phosphate dehydrogenase and alkaline phosphatase in osteocytes and periosteal osteoblasts in rat fibulae in vivo. J Bone Miner Res (1993) 8:261–7. doi:10.1002/jbmr.5650080303
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
110. Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell (1997) 89:747–54. doi:10.1016/S0092-8674(00)80257-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
111. Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell (1997) 89:755–64. doi:10.1016/S0092-8674(00)80258-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
112. Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell (1997) 89:765–71. doi:10.1016/S0092-8674(00)80259-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
113. Schwab W, Schulze E, Witt M, Funk RH, Kasper M. Immunohistochemical localization of the differentiation marker E11 in dental development of rats. Acta Histochem (1999) 101:431–6. doi:10.1016/S0065-1281(99)80043-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
114. Li X, Ominsky MS, Niu QT, Sun N, Daugherty B, D’agostin D, et al. Targeted deletion of the sclerostin gene in mice results in increased bone formation and bone strength. J Bone Miner Res (2008) 23:860–9. doi:10.1359/jbmr.080216
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
115. Chen X, Yang L, Tian W. [Heterotopic osteogenesis of autogenous marrow stromal cells with recombinant human bone morphogenetic protein 2 gene transfection and porous calcium phosphate ceramic as a scaffold]. Hua Xi Kou Qiang Yi Xue Za Zhi (2003) 21:419–21.
116. Chen G, Deng C, Li YP. TGF-beta and BMP signaling in osteoblast differentiation and bone formation. Int J Biol Sci (2012) 8:272–88. doi:10.7150/ijbs.2929
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
117. Meng Y, Qin YX, Dimasi E, Ba X, Rafailovich M, Pernodet N. Biomineralization of a self-assembled extracellular matrix for bone tissue engineering. Tissue Eng Part A (2009) 15:355–66. doi:10.1089/ten.tea.2007.0371
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
118. Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J Theor Biol (1982) 99:31–68. doi:10.1016/0022-5193(82)90388-5
119. Prideaux M, Loveridge N, Pitsillides AA, Farquharson C. Extracellular matrix mineralization promotes E11/gp38 glycoprotein expression and drives osteocytic differentiation. PLoS One (2012) 7:e36786. doi:10.1371/journal.pone.0036786
120. Neidlinger-Wilke C, Wurtz K, Liedert A, Schmidt C, Borm W, Ignatius A, et al. A three-dimensional collagen matrix as a suitable culture system for the comparison of cyclic strain and hydrostatic pressure effects on intervertebral disc cells. J Neurosurg Spine (2005) 2:457–65. doi:10.3171/spi.2005.2.4.0457
121. Tata U, Xu H, Rao SMN, Chuong C-J, Nguyen KT, Chiao JC. A novel multiwell device to study vascular smooth muscle cell responses under cyclic strain. J Nanotechnol Eng Med (2011) 2:021007–021007. doi:10.1115/1.4003928
122. Rubin CT. Skeletal strain and the functional significance of bone architecture. Calcif Tissue Int (1984) 36(Suppl 1):S11–8. doi:10.1007/BF02406128
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
123. Rubin CT, Lanyon LE. Dynamic strain similarity in vertebrates; an alternative to allometric limb bone scaling. J Theor Biol (1984) 107:321–7. doi:10.1016/S0022-5193(84)80031-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
124. Rubin CT, Lanyon LE. Regulation of bone formation by applied dynamic loads. J Bone Joint Surg Am (1984) 66:397–402.
125. Rubin CT, Mcleod KJ, Bain SD. Functional strains and cortical bone adaptation: epigenetic assurance of skeletal integrity. J Biomech (1990) 23(Suppl 1):43–54. doi:10.1016/0021-9290(90)90040-A
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
126. Bailey DA, Faulkner RA, Mckay HA. Growth, physical activity, and bone mineral acquisition. Exerc Sport Sci Rev (1996) 24:233–66.
127. Sutton MA, Orteu J, Schreier HW. Digital image correlation (DIC). Image Correlation for Shape, Motion and Deformation Measurements: Basic Concepts, Theory and Applications. New York: Springer (2009).
128. Papadimitropoulos A, Scherberich A, Guven S, Theilgaard N, Crooijmans HJ, Santini F, et al. A 3D in vitro bone organ model using human progenitor cells. Eur Cell Mater (2011) 21:445–58.
129. Maeno S, Niki Y, Matsumoto H, Morioka H, Yatabe T, Funayama A, et al. The effect of calcium ion concentration on osteoblast viability, proliferation and differentiation in monolayer and 3D culture. Biomaterials (2005) 26:4847–55. doi:10.1016/j.biomaterials.2005.01.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: osteocyte, osteoblast, 3 dimensional, co-culture, model, loading
Citation: Vazquez M, Evans BAJ, Riccardi D, Evans SL, Ralphs JR, Dillingham CM and Mason DJ (2014) A new method to investigate how mechanical loading of osteocytes controls osteoblasts. Front. Endocrinol. 5:208. doi: 10.3389/fendo.2014.00208
Received: 16 October 2014; Paper pending published: 03 November 2014;
Accepted: 18 November 2014; Published online: 09 December 2014.
Edited by:
Jonathan H. Tobias, University of Bristol, UKReviewed by:
Andy Sunters, Royal Veterinary College, UKJonathan Gooi, The University of Melbourne, Australia
Copyright: © 2014 Vazquez, Evans, Riccardi, Evans, Ralphs, Dillingham and Mason. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Deborah J. Mason, Division of Pathophysiology and Repair, School of Biosciences, Cardiff University, Museum Avenue, Cardiff, CF10 3AX, UK e-mail: masondj@cardiff.ac.uk
 Marisol Vazquez
Marisol Vazquez Bronwen A. J. Evans
Bronwen A. J. Evans Daniela Riccardi3
Daniela Riccardi3 Christopher Mark Dillingham
Christopher Mark Dillingham Deborah J. Mason
Deborah J. Mason