- 1Department of Biology, City College of New York and The Graduate Center, City University of New York, New York, NY, USA
- 2Department of Brain Science, Daegu Gyeongbuk Institute of Science and Technology (DGIST), Daegu, South Korea
Neuropeptides regulate all aspects of behavior in multicellular organisms. Because of their ability to act at long distances, neuropeptides can exert their effects beyond the conventional synaptic connections, thereby adding an intricate layer of complexity to the activity of neural networks. In the nematode Caenorhabditis elegans, a large number of neuropeptide genes that are expressed throughout the nervous system have been identified. The actions of these peptides supplement the synaptic connections of the 302 neurons, allowing for fine tuning of neural networks and increasing the ways in which behaviors can be regulated. In this review, we focus on a large family of genes encoding FMRFamide-related peptides (FaRPs). These genes, the flp genes, have been used as a starting point to identifying flp genes throughout Nematoda. Nematodes have the largest family of FaRPs described thus far. The challenges in the future are the elucidation of their functions and the identification of the receptors and signaling pathways through which they function.
Introduction
Behavior in all animals is the output of multiple neural networks, whose individual circuits are activated, inhibited, or modulated at any one time by the combinatorial action of many neurotransmitters. In recent years, there has been a push to determine the neural connectome of organisms, including the human connectome (Human Connectome Project). The first completed neural connectome was that of the nematode Caenorhabditis elegans (1). In a heroic effort and with much foresight into the usefulness of the project, John White and collaborators embarked on deciphering the entire neural circuitry and mapping all chemical synapses and gap junctions between the 302 neurons of the C. elegans adult hermaphrodite (1); more recently, Scott Emmons and David Hall have initiated work on the C. elegans male neural connectome (2). The mapping of all the synaptic connections between neurons has been highly informative and helpful to researchers. However, the neural connectome, while representing a large fraction of neural network activity, underestimates the complexity of the nervous system. Layered on top of the connectome is the action of hormones and neuropeptides, whose effects can be exerted over large distances and whose actions, therefore, are not wholly represented by the neural connectome. Hence, hormones and neuropeptides add considerable complexity to neural networks to affect behavior.
In C. elegans and other invertebrates, neuropeptides, which are short sequences of amino acids, can act as primary transmitters as well as neuromodulators. Most neurons in C. elegans express at least one neuropeptide gene and in the majority of neurons, neuropeptides are co-localized with classical small molecule transmitters, such as acetylcholine, GABA, serotonin (5-HT), and dopamine (3, 4). Based on BLAST and bioinformatic screening, two major families of neuropeptides emerged in C. elegans: insulin-like peptides (ILPs) (5–9) and FMRFamide-related peptides (FaRPs) (10–12). In addition, C. elegans also has a large complement of non-insulin, non-FaRP neuropeptides (NLPs), many of which have invertebrate and vertebrate orthologs (3, 13). In this review, we will focus on the C. elegans FaRPs, which, by definition, all share a C-terminal Arg-Phe-NH2 and which are more commonly referred to as FLPs in C. elegans. We will describe their processing and some of their functions, and compare the diversity of FLPs in other nematodes.
Identification of flp Genes
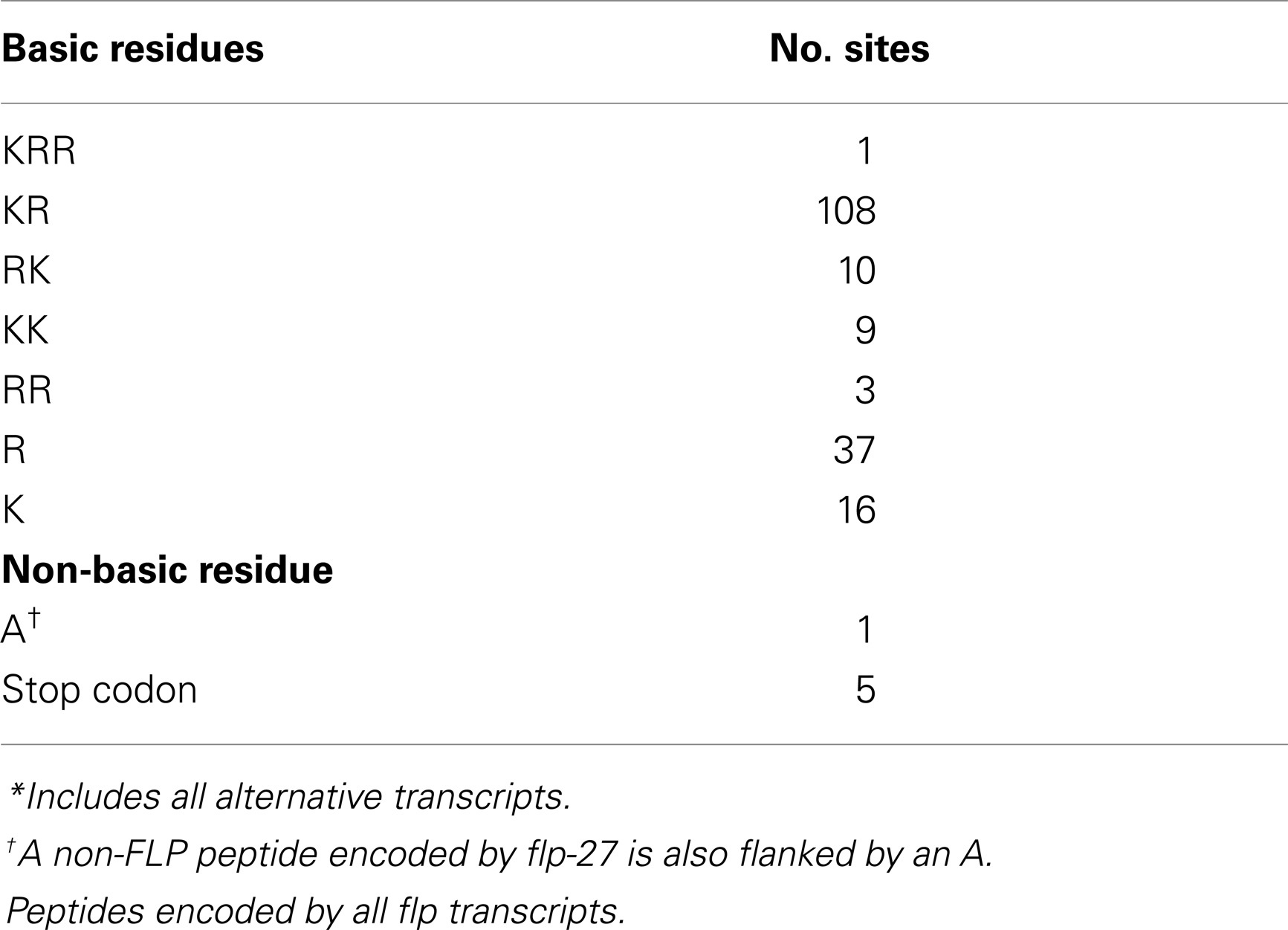
By cDNA library screening, bioinformatic searches, and genome/transcriptome data mining for proteins with Arg-Phe-Gly sequences flanked by N- and C-terminal tri-, di-, or mono-basic residues, we and others identified 31 genes encoding FLPs in C. elegans (10, 12). Each flp gene encodes a unique set of FLPs for a total of 71 possible distinct FLPs (Table 1) (10, 12). Fourteen flp genes encode a single FLP, although seven of these genes encode multiple copies of the same FLP [flp-6 (6 copies of KSAYMRF-NH2), 8 (3–4 copies of KNEFIRF-NH2), 9 (2 copies of KPSFVRF-NH2), 10 (1 copy of QPKARSGYIRF-NH2), 12 (1 copy of RNKFEFIRF-NH2), 14 (4 copies of KHEYLRF-NH2; Figure 1A), 20 (2 copies of AMMRF-NH2), 21 (1 copy of GLGRPLRF-NH2), 22 (3 copies of SPSAKWMRF-NH2), 24 (1 copy of VPSAGDMMVRF-NH2), 27 (1 copy of GLGGRMRF-NH2), 28 (1 copy of APNRVLMRF-NH2), 32 (1 copy of AMRNSLVRF-NH2), 33 (1 copy of APLEGFEDMSGFLRTIDGIQKPRF-NH2), and 34 (1 copy of ALNRDSVASLNNAERLRF-NH2)]. At least nine flp genes encode non-FLP peptides [flp-1, 3 (Figure 1A), 7, 11, 20, 23, 26, 27, and 34] and six flp genes (flp-1, 2, 8, 11, 22, and 23) are alternatively spliced (WormBase). The organization of flp genes ranges from the simplest at only two small exons to the largest at six exons. Similarly, the FLP protein precursors are small, ranging from 66 to 184 amino acids, sizes that include the signal peptide; the bioactive peptides vary from 5 to 24 amino acids in length (Table 1).
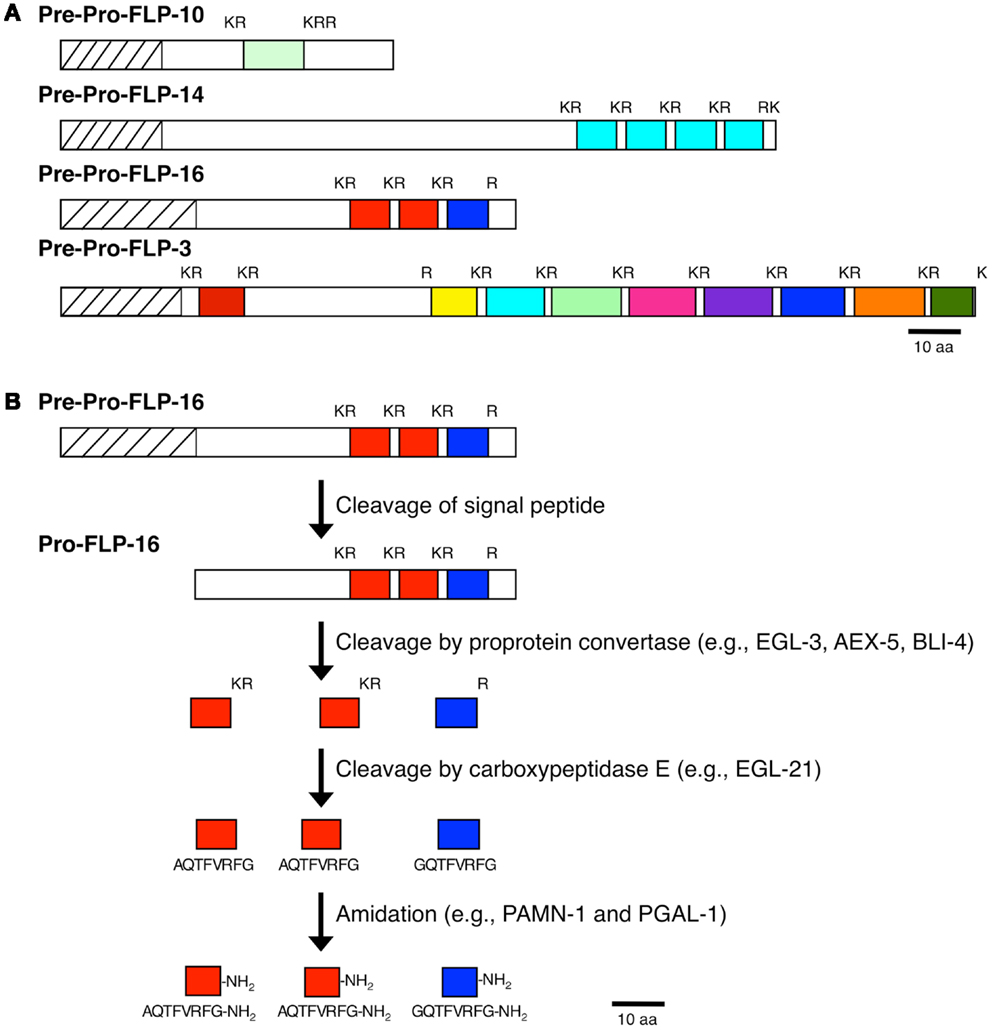
Figure 1. Processing of neuropeptide precursor molecules. (A) Representative FLP precursor molecules that contain one FLP peptide (FLP-10), multiple copies of the same FLP peptide (FLP-14), multiple distinct FLP peptides (FLP-3), and multiple distinct FLP peptides and multiple copies of the same FLP (FLP-16) are shown. (B) Proteolytic pathway of the FLP-16 precursor molecule to yield the active FLP-16 peptides.
Processing of Pre-Pro-Neuropeptide Precursors
As with other neuropeptides (25), the FLPs are derived from pre-propeptides (Figure 1), which are large precursor molecules that must be proteolytically cleaved and post-translationally modified to yield the active peptides (Figure 1B). The FLP precursor molecules all have a signal peptide, which is removed to yield the propeptide. The propeptides are then cleaved by proprotein convertases (PCs), serine endoproteases that preferentially recognize dibasic residues and cleave C-terminal to the basic residues (26). In C. elegans, PCs also cleave after tribasic and monobasic residues and very rarely, after a non-basic residue. Of the FLP precursor molecules, including different isoforms encoded by one gene, there are 184 instances of FLP sequences flanked by basic amino acids (Table 2). Seventy-one percent of the FLPs are flanked by the typical dibasic residues, of which the majority (83%) are Lys-Arg, and 29% are flanked by monobasic residues of which Arg is the most common residue (70%). Five FLP peptides are directly followed by a stop codon and one FLP peptide has an unusual non-basic flanking residue (Ala).
There are four genes encoding PCs in C. elegans: kpc-1, egl-3/kpc-2, aex-5/kpc-3, and bli-4/kpc-4; with the exception of aex-5/kpc-3, all are alternatively spliced to yield isoforms that differ at the C-terminus [(27, 28); WormBase], suggesting that different isoforms may have different substrate targets or preferences. Mutant alleles of all the PC genes have been isolated (27–31). By mass spectrometry (MS) analysis of neuropeptide profiles, the major PC that cleaves FLP propeptides is EGL-3/KPC-2 (22), whose mammalian ortholog is proprotein convertase 2 (PC2) (32). The number of isolated FLP peptides is significantly decreased in egl-3/kpc-2 mutants compared to in kpc-1, aex-5/kpc-3, and bli-4/kpc-4 mutants (22). However, the biochemical data also indicate that besides EGL-3/KPC-2 other PCs cleave propeptides (22). Furthermore, using an antibody that only recognizes mature FLPs, anti-FMRFamide immunoreactivity was detected in egl-3 mutants, suggesting that other PC2s function in neuropeptide processing in C. elegans (33). egl-3 is widely expressed in the nervous system and acts within post-Golgi secretory vesicles (32). Knockout of egl-3 PC2 causes several defects, including in egg-laying (30) and body mechanosensation (32).
The minor PCs also have the capability to cleave neuropeptide precursors. kpc-1 encodes a PC similar to mammalian furin (28) and is widely expressed in the nervous system and epithelial cells (34). kpc-1 PC mutants show slight uncoordination and slowed growth (28) and have defects in the dendritic remodeling of the IL2 neurons during dauer formation and nictation (34). While the dendritic remodeling defects may be due to lack of cleavage of semaphorins or TGFβ substrates (34), the nictation defects may be due to lack of cleavage of neuropeptide precursors to yield active peptides that regulate nictation (35). aex-5/kpc-3 mutants show a defect in the defecation motor pattern (31). Furthermore, knockdown of aex-5/kpc-3 by RNA-mediated interference (RNAi) feeding gives a low rate of embryonic lethality (36), suggesting other essential substrates for AEX-5/KPC-3. The genomic organization of aex-5/kpc-3 is slightly unusual because it is part of an operon with unc-54, which encodes a muscle myosin heavy chain (28); the regulation of the operon would predict that aex-5/kpc-3 is expressed only in muscle, similar to unc-54 myosin. However, intestinal specific expression of aex-5/kpc-3 can rescue the defection motor program in aex-5/kpc-3 mutants, suggesting that the intestine also expresses aex-5/kpc-3 (37). bli-4/kpc-4 encodes a furin-like protease that is expressed in neurons, hypodermal cells, vulval muscles, and the intestine; loss of bli-4/kpc-4 results in late embryonic lethality (27). Although DPY-5 is a procollagen that must be cleaved by BLI-4/KPC-4 for proper adult cuticle formation (38), BLI-4/KPC-4 must have other substrates, currently unidentified, during embryogenesis. The relevance of the different PC isoforms is unknown, but may relate to substrate specificity.
The PC2s themselves are also synthesized as precursor molecules that must be proteolytically cleaved to become activated and transported (39). SBT-1 is the C. elegans neuroendocrine 7B2 ortholog (40) that presumably cleaves pro-EGL-3/PC2 for its activation. In high pressure liquic chromatography (HPLC)-MS isolations, 29 FLPs were isolated from extracts of sbt-1/7B2 mutants compared to 53 FLPs isolated from wild-type extracts, suggesting that loss of sbt-1 interferes with neuropeptide maturation (23). sbt-1/7B2 mutants also showed resistance to the effects of aldicarb (40), an acetylcholinesterase inhibitor that causes paralysis in C. elegans (41).
After cleavage by the PCs, the C-terminal basic residues are removed by the action of carboxypeptidase E (CPE), which cleaves N-terminal to the basic residue(s). In C. elegans, one CPE ortholog is egl-21, which is expressed widely in the nervous system (33). Loss of egl-21 CPE results in defects in egg-laying (30), locomotion, and defecation and decreased FMRFamide-like immunoreactivity and aldicarb sensitivity (33). Because residual FMRFamide-like immunoreactivity is present in egl-21 CPE mutants (33), other carboxypeptidases must also function to process peptide intermediates. There are two other carboxypeptidases with similarity to CPE and carboxypeptidase D (33): CPD-1/F59A3.1 and CPD-2/T27A8.11. Knockdown of cpd-2/T27A8.1 had no phenotype, whereas knockdown of cpd-1 by RNAi showed variable uncoordination and larval lethality (42). Elucidation of cpd-1 and cpd-2 function in neuropeptide processing awaits future mutant analysis.
To prolong their effects, many neuropeptides are post-translationally modified to prevent their immediate degradation. In the case of the FLPs, the peptides are amidated and this amidation is required for the bioactivity of the peptide (43). The amide at the C-terminus of each mature FLP is donated by the C-terminal glycine. In mammals, amidation occurs as the result of two sequential enzymatic steps from a bifunctional protein peptidylgylcine-α-amidating monooxygenase (PAM): the peptide is subjected to the action of a peptidylgylcine-α-hydroxylating monooxygenase (PHM), followed by the action of a peptidyl- α-hydroxyglycine α-amidating lyase (PAL) (44, 45). By contrast, C. elegans has two independent enzymes: PAMN-1/T19B4.1 and PGAL-1/F21F3.1 have similarity to PHM and PAL, respectively, and could function in neuropeptide amidation [(46); WormBase].
After release, inactivation of neuropeptide signaling is regulated by their proteolytic cleavage by the class of neprilysin (NEP) zinc metallopeptidases (47). In C. elegans, NEP activity cleaves the amino group of hydrophobic amino acids; for instance, FLP-1 (SDPNFLRF-NH2) is cleaved at multiple sites (e.g., NF, FL, and RF), each including the Phe residue (48). At least 20 NEP-encoding genes are present in C. elegans; 10 of these are known to be expressed (48–50). nep-1 is expressed in pharyngeal cells and a single head neuron (51), suggesting that NEPs are active at different synapses. A nep-1 knockout showed an uncoordinated phenotype; in addition, in isolated pharyngeal preparations, nep-1 mutants showed decreased basal activity (51). Which, if any, of these NEP genes are relevant to peptide processing is still unknown.
Packaging and Release of FLPs
The processing of neuropeptide precursor molecules into bioactive peptides starts in the endoplasmic reticulum (ER), when the signal peptide is first removed. Subsequent cleavages of the propeptide and post-translational modifications of the cleaved peptides occur in the ER and Golgi network or as the vesicle, which in the case of neuropeptides is large, dense core vesicles (DCVs), is transported to the nerve terminal. In C. elegans transport of DCVs to the nerve terminals is dependent on UNC-104/KIF1A/kinesin-3 (33, 52, 53) and UNC-116/kinesin-1 (53).
Small molecule transmitters and neuropeptides are packaged into distinct vesicles, small, clear synaptic vesicles (SVs) versus large, DCVs, respectively. Calcium triggers the release of their contents with a latency of 4.5 ms after the peak of an action potential for SVs or 16.7 ms for DCVs, which show better release after repeated or prolonged stimulation (54). In addition, the contents of the vesicles are significantly different; one SV releases about 4,700 transmitter molecules with a time constant of decay of transmitter discharge of 260 μs, while one DCV releases about 18,000 molecules with a time constant of 1.6 ms (54). DCVs are marked by the presence of IDA-1/IA-2/phogrin, which is a receptor protein tyrosine phosphatase found specifically on DCVs and which regulates levels of phosphatidylinositol phospholipids (PIPs) (55, 56). Regulated release of either type of vesicle is a multi-step process: vesicle docking, priming, fusion, and release. DCVs are primarily found in C. elegans axons and not in cell somas; activity of the UNC-43 CaMKII regulates DCV exocytosis from the axon rather than the cell soma (57). The core SNARE complex, which includes vesicular SNB-1/synaptobrevin-1 (v-SNARE) and plasma membrane proteins RIC-4/SNAP-25 and UNC-64/syntaxin (t-SNAREs), is essential for this process and interacts with a number of other proteins, some specific to DCVs, to facilitate vesicular release (58). Presumably, v-SNARE SNT-1/synaptotagmin (59) has a similar function with DCVs and acts as a calcium sensor. Both types of vesicles also require the cytoplasmic protein UNC-18 for vesicular release; UNC-18/Munc-18 has multiple functions: (1) it helps in the efficient trafficking from the ER and anterograde transport of UNC-64/syntaxin (60); (2) it binds UNC-64/syntaxin and mediates the conformational switch of UNC-64/syntaxin from a closed to open state to allow SNARE complex assembly, thereby priming the vesicle (61–63); and (3) it allows vesicle and plasma membrane fusion (62). Unlike SVs that are docked and released from active zones, DCVs are released at sites distinct from active zones (64). These DCV docking sites may be determined by the cytoplasmic protein RIC-7, which is a nematode-specific protein with no known homologies (65). UNC-31/CAPS, a calcium-dependent activator protein that contains a DCV-binding domain and pleckstrin-homology domain, serves as a bridge between DCVs and the plasma membrane (66, 67) to promote DCV docking and release (64, 68, 69). UNC-13, which promotes SNARE complex assembly (69, 70), and PKC-1 both contain C1 domains that bind phorbol esters to regulate vesicular release, although PKC-1 is only involved in DCV release (69). UNC-13/Munc13 stabilizes the open conformation of syntaxin to promote vesicle priming and this activity is antagonized by TOM-1/tomosyn, which associates with UNC-64/syntaxin and RIC-4/SNAP-25 to form an inhibitory SNARE complex (71) and prevent DCV release (68). Unlike small molecule transmitters, neuropeptides are not recycled and excess neuropeptides are cleared from the synaptic regions by degradation (see above); new neuropropeptides must be synthesized de novo in the cell body, processed and packaged into DCVs, and transported to the nerve terminals.
Isolation of FLPs and Expression Patterns
Among the 31 flp genes, there are 71 predicted FLPs. The initial isolation of FLPs was through laborious extracts from grams worth of animals (14–19, 21). Fortunately, the advent of new technologies with high pressure liquid chromatography in tandem with MALDI-TOF mass spectrometry have greatly decreased the amount of starting materials necessary for peptidomic studies and increased the resolution of peptide identities. Hence, microgram amounts of animals are now sufficient to isolate even low abundance peptides. The group of Liliane Schoofs has taken on the challenge to decipher the FLP peptidome in C. elegans and other nematodes (22, 24). Fifty-four of the 71 predicted FLPs representing 25 of the 31 flp genes have now been isolated (Table 1) (22, 24). However, there are also three FLPs, APNRVLMRLVRF-NH2, HFYNFSSESRKPNFLRF-NH2, and KPXPXFIRF-NH2, that have been biochemically isolated, but the corresponding gene has not been identified (N. Marks, A. Maule, and A.O. Stretton, pers. comm.). These results suggest that most, if not all, predicted FLPs are produced and additional flp genes have yet to be identified. Given that many of the peptides encoded by one gene are highly similar, we predict that these peptides have slightly different binding affinities to receptors to allow fine tweaking of a circuit. For instance, flp-18 encodes six distinct FLPs that share a common C-terminal PGVLRF-NH2; NMR analysis of two of the peptides DGDGAMPGVLRF-NH2 and EMPGVLRF-NH2 revealed that the N-terminal aspartates of DGDGAMPGVLRF-NH2 form long-range electrostatic interactions with the C-terminal arginines, thereby forming a loop that decreases peptide binding to one of its receptors, NPR-1 (72). Similarly, other N-terminal extensions can form secondary structures that decrease the affinity of the peptide to its receptors, thereby modulating the biological effects of the peptides.
cDNAs from all flp genes have been isolated, indicating that all flp genes are expressed [(4, 10, 73, 74); WormBase]. Because FLPs are relatively small, ranging from 5 to 24 amino acids, and all share a C-terminal Arg-Phe-NH2 motif, making antibodies against specific FLPs have been difficult. A general anti-FaRP antiserum that recognizes FaRPs encoded by multiple flp genes showed that roughly 10% of the neurons were immunoreactive (75). This figure, however, appears to grossly underestimate the widespread expression of the flp genes. Using transcriptional reporters, over 50% of the nervous system was found to express one or more flp genes (4, 76–78). Transcriptional reporters, however, have many inherent caveats, such as incorrect/incomplete promoter regions used or intronic sequences with regulatory elements missing; in addition, researchers injecting similar constructs have sometimes reported different expression patterns (4, 77). Using a monoclonal antibody specific for FLP-8 (79, 80), we showed that the immunoreactivity pattern matched that of the reporter expression pattern, suggesting that transcriptional reporters can reflect protein expression patterns. Although each flp gene is expressed in a distinct set of neurons, there is considerable overlap in the expression pattern of the different flp genes and a single neuron can show a wide diversity of flp expression. For instance, the chemosensory neuron ASE and oxygen-sensor URX express five and four flp genes, respectively (4). In addition, most flp genes are expressed in neurons that also express a small molecule transmitter. Some flp genes are also expressed in non-neuronal cells, such as the intestine or gonad (4, 77). The complex flp expression pattern allows exceptionally intricate modulation of neural networks to generate behavior.
Function of FLPs in C. elegans
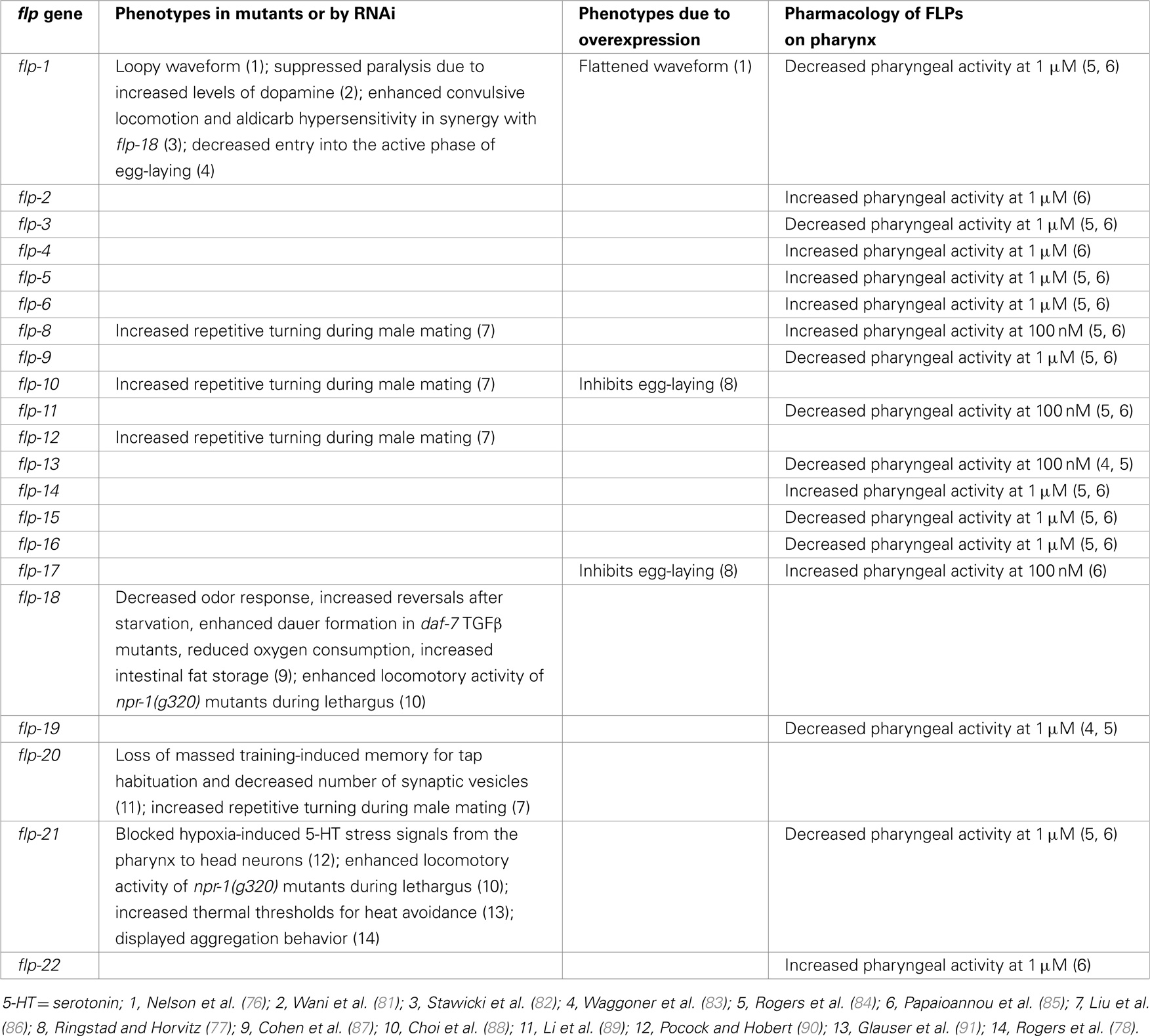
FLP neuropeptides have been shown to function in various behaviors in C. elegans (Table 3). A common functional modality among FLPs is that these neuropeptides appear to inhibit circuit activity in most, but not all, behaviors tested. Pharmacological application of FLPs, however, shows that FLPs also has excitatory effects (Table 3).
Locomotion
Wild-type animals move in a sinusoidal waveform on a solid surface and initiate swimming in liquid. flp-1 mutants displayed an exaggerated waveform on a solid surface; conversely, overexpression of flp-1 caused a flattening of the waveform and extreme sluggishness, suggesting that flp-1 modulates the locomotory circuits (76). dat-1 encodes a dopamine transporter and mutations in dat-1 caused paralysis due to increased synaptic dopamine concentration (92). Chase and coworkers showed that a flp-1 mutation fully suppressed this locomotion defect of dat-1 mutants (81). Although the exact mechanism has not been elucidated, these results further indicate a role of flp-1 in locomotory circuits (76).
ACR-2 is a nicotinic acetylcholine receptor subunit expressed in cholinergic motoneurons (93). acr-2 gain-of-function (gf) mutants exhibited excitation–inhibition imbalance due to an increase in cholinergic excitation and a decrease in GABAergic inhibition in the locomotory circuit, resulting in spontaneous convulsive behavior and increased aldicarb sensitivity (93). Loss of egl-3 PC2 decreased GABAergic inhibition and enhanced the convulsion phenotype of acr-2(gf), suggesting that neuropeptides were involved (82). After testing multiple neuropeptide mutants, flp-1; flp-18 double mutants but not single mutants, were identified as enhancing convulsive locomotion and aldicarb hypersensitivity of acr-2(gf) by further reducing GABAergic inhibition (82). In addition, acr-2(gf) mutants showed increased expression of flp-18 in the ventral cord cholinergic B-type motoneurons and overexpression of flp-18 decreased convulsion phenotype of acr-2(gf) (82). NPR-1 and NPR-5 G protein-coupled receptors (GPCRs) appeared to mediate the action of FLP-18 in acr-2 (gf) mutants (82). These results indicate that flp-1 and flp-18 play important roles in suppressing overexcitation of the locomotory circuit.
Egg-Laying
C. elegans egg-laying behavior is regulated by the actions of both small molecule neurotransmitters, such as acetylcholine and 5-HT, and neuropeptides (94). The HSN and VC4/5 serotoninergic neurons control the temporal switch between active/inactive states of egg-laying; during the active phase, release of acetylcholine triggers the egg-laying event (95). flp-1 functions in parallel with serotonin to promote entry into the active phase of egg-laying (83). A different peptidergic circuit regulating egg-laying behavior was revealed by analysis of egl-6 mutants, which were isolated on the basis of their egg-laying defects (30). EGL-6 is a GPCR; a gain-of-function mutation or overexpression of egl-6 exclusively in the HSN motoneurons inhibited egg-laying, while egl-6 deletion mutants did not exhibit egg-laying defects (77). However, egl-6 deletion mutants fully suppressed egg-laying defects caused by overexpression of flp-10 or flp-17 (77). Surprisingly, the effects of the FLP-10 peptide were conferred by expression in non-neuronal cells, including vulval cells and spermatheca, as well as the CO2-sensing BAG neurons, whereas flp-17 appeared to function primarily in the BAG neurons (77). The FLP-10/FLP-17 peptidergic circuit acted redundantly with a cholinergic circuit distinct from HSN/VCs to inhibit egg-laying (77). Using the Xenopus oocyte heterologous system, FLP-10 and FLP-17 were verified as cognate ligands for EGL-6 GPCR (77).
Metabolism
de Bono and coworkers found that flp-18 deletion mutants showed a multitude of defects, including decreased odor response, increased reversals and turns after 1 h of starvation, enhanced dauer formation in daf-7 TGFβ mutant background, reduced oxygen consumption, and increased intestinal fat storage (87). These flp-18 phenotypes were rescued by expression of flp-18 in the AIY interneurons (or additionally RIG interneurons for the fat phenotype), which are major synaptic targets of multiple chemosensory neurons, indicating that the AIY neurons may integrate and process food information to determine release of FLP-18 (87). FLP-18 activated two GPCRs, NPR-4 and NPR-5, in Xenopus oocytes; furthermore, activation of NPR-4 in the intestine and NPR-5 in a set of chemosensory neurons mediated fat storage, activation of NPR-4 in the AVA and RIV interneurons mediated reversals and turns, and activation of NPR-5 in the ASJ chemosensory neurons mediated dauer formation (87). Fat storage was also increased in two other NPY-like receptor mutants, npr-1 or npr-7, suggesting additional roles of non-FLP-18 neuropeptides in fat metabolism (87).
Stress Responses
Pocock and Hobert noticed that hypoxic stress increased levels of serotonin (5-HT) in a subset of gustatory neurons (ASG and ADF) of C. elegans via direct regulation by the hypoxia-inducible transcription factor HIF-1 and enhanced response to NaCl (90). Serotonin released from the ASG and ADF neurons activated the M4 pharyngeal motoneuron via its SER-7 5-HT receptor; activation of M4 resulted in the release of the FLP-21 peptide to relay these hypoxia-induced 5-HT signals from the pharynx to NPR-1-expressing neurons in the head (AQR, PQR, and URX), which may act upstream of the gustatory circuit (90). Thus, FLP-21 and its receptor NPR-1 play roles in transmitting stress signals.
Aggregation
The wild-type laboratory strain N2 var. Bristol displays solitary feeding behavior (96). However, under stressful conditions, such as high population density, low food, or low O2 concentration, animals will aggregate (96–98). In addition, a mutation in the Bristol npr-1 GPCR gene that corresponded to an NPR-1 isoform found in many naturally isolated wild strains caused aggregation even under non-stressful conditions, indicating that low activity of NPR-1 GPCR may represent stress-related behavioral states (96, 99, 100); expression of npr-1 in the AQR, PQR, and oxygen-sensor URX neurons regulates aggregation behavior (101). Animals lacking FLP-21, a cognate ligand of NPR-1 GPCR, also displayed aggregation behavior, providing genetic evidence for interactions between FLP-21 and NPR-1 (78, 102). Recently, Bargmann, Sengupta, and coworkers found a “hub-and-spoke” circuit motif that generates behavioral differences depending upon NPR-1 activity (103, 104). In this circuit, the RMG command interneuron/motoneuron, which also expresses npr-1 (101), serves as the hub of seven spoke neurons, including two pheromone-sensing neurons ASK and ADL and an oxygen-sensing URX, which are connected to the RMG hub via gap junctions; high RMG activity is required for aggregation behavior (103, 104). The ASK neuron is involved in male attraction and hermaphrodite repulsion at low and high concentrations of pheromone, respectively (105). High NPR-1 activity in solitary N2 hermaphrodites resulted in decreased RMG activity, leading to an avoidance response due to the enhanced ADL (repulsion) and reduced ASK (attraction) pheromone response (103, 104). Conversely, low or absent NPR-1 activity resulted in increased RMG activity, leading to an increased ASK–RMG mediated attraction and decreased ADL mediated repulsion, resulting in hermaphrodites being neutral to pheromone (103, 104). Thus, NPR-1 signaling in RMG regulates its activity level to integrate environmental signals, such as pheromones and oxygen.
Sleep-Like Behavior
During each of the four larval molts, C. elegans exhibits a sleep-like behavioral state during a period called lethargus, during which wild-type animals show reduced sensory and motor activity and feeding behaviors (106). By contrast, npr-1 GPCR mutants exhibited increased locomotive activity compared to that of quiescent wild-type animals; this activity was further enhanced in flp-18 or flp-21 single or double mutants (88). FLP-18/21 activation of NPR-1 signaling in the RMG hub-and-spoke interneuron was found to decrease sensory activity of the ASK sensory neurons, resulting in decreased PDF-1 secretion from the ASK neurons (88). Decreased PDF-1 secretion led to decreased activation of the PDF-1 receptor in mechanosensory neurons and muscles, resulting in decreased arousal (88). Thus, flp-18 and flp-21 have roles in decreasing neuronal and circuit activities underlying lethargus.
Learning and Memory
C. elegans have been shown to habituate to mechanical stimuli generated by tapping the worm plate; repeated tap stimulation caused decremented responses to taps (107). Massed training-induced memory for tap habituation lasted at least 12 h (89). This 12-h memory retention of tap habituation training was abolished in flp-20 mutants and the increased number of SVs in mechanosensory neurons after massed training was also reduced in flp-20 mutants, indicating that FLP-20 is required for the 12-h memory retention (89). Since flp-20 was expressed in and acted in the mechanosensory neurons, FLP-20 may regulate the number of DCVs in mechanosensory neurons via a feedback circuit (89).
Heat Avoidance
Goodman and coworkers developed new assays to identify genes involved in heat avoidance (91). Mutations in either flp-21 or npr-1 were found to increase thermal thresholds of heat avoidance. These effects may be due to FLP-21 activation of the RMG interneurons to decrease thermal thresholds (91).
Male Mating
During mating, males undergo a stereotypic series of movements that includes hermaphrodite contact, backing, turning, locating the vulva, spicule insertion, and sperm transfer (108, 109). The male tail contains many specialized ray neurons that function in concert with core neurons for efficient male mating. In particular, glutamatergic mechanosensory neurons are required independently of ray neurons for turning behavior (86). In addition, loss of flp-8, 10, 12, and 20, all of which are expressed in all or subsets of the mechanosensory neurons (4), increased repetitive turning behavior, suggesting that these FLPs are necessary for the accurate timing of turning (86).
Pharyngeal Pharmacology
Holden-Dye’s group has made excellent use of a dissected pharyngeal preparation that contains the pharynx and nerve ring; the pharyngeal musculature has myogenic properties that can be stimulated with 500 nM serotonin (84). By recording from the pharyngeal muscle with either an intracellular electrode or a suction pipette, the group found that numerous FLP peptides increased (FLP-17 and 8 at 100 nM; FLP-2, 4, 5, 6, 14, and 22 at 1 μM) or decreased (FLP-11 and 13 at 100 nM; FLP-1, 3, 9, 15, 16, 19, and 21 at 1 μM) pharyngeal activity (84, 85). Several of the flp genes with bioactive peptides are expressed within pharyngeal neurons, but many of the genes encoding the bioactive peptides are expressed in neurons outside the pharyngeal system (4). Whether the peptides are acting directly on pharyngeal muscle or indirectly via other neurons remains to be determined. In addition, because some of the peptides were bioactive only at high concentrations, the physiological roles of the peptides await further confirmation.
Identifying G Protein-Coupled Receptors through Which FLPs Signal
With one exception (110), neuropeptides signal through GPCRs. In C. elegans, there are an estimated 1,100 GPCRs (111) of which most have been classified as chemoreceptors and 50–125 are likely to correspond to neuropeptide GPCRs (111, 112). Knocking down activity of a subset of the identified neuropeptide receptors suggest that signaling through these receptors affect reproduction and locomotion (113).
A current initiative by several labs has been to de-orphanize the GPCRs by identifying their respective ligand(s) and determining the pathways in which they function. This task is daunting in any system, because GPCRs are promiscuous and can bind more than one ligand; similarly, neuropeptides are also promiscuous and one peptide may bind multiple GPCRs with different affinities. Whether these differing binding affinities reflect a biological function, whereby activation by a specific peptide may only occur after low or high frequency stimulation, or are an artifact of the experimental system remains to be clarified.
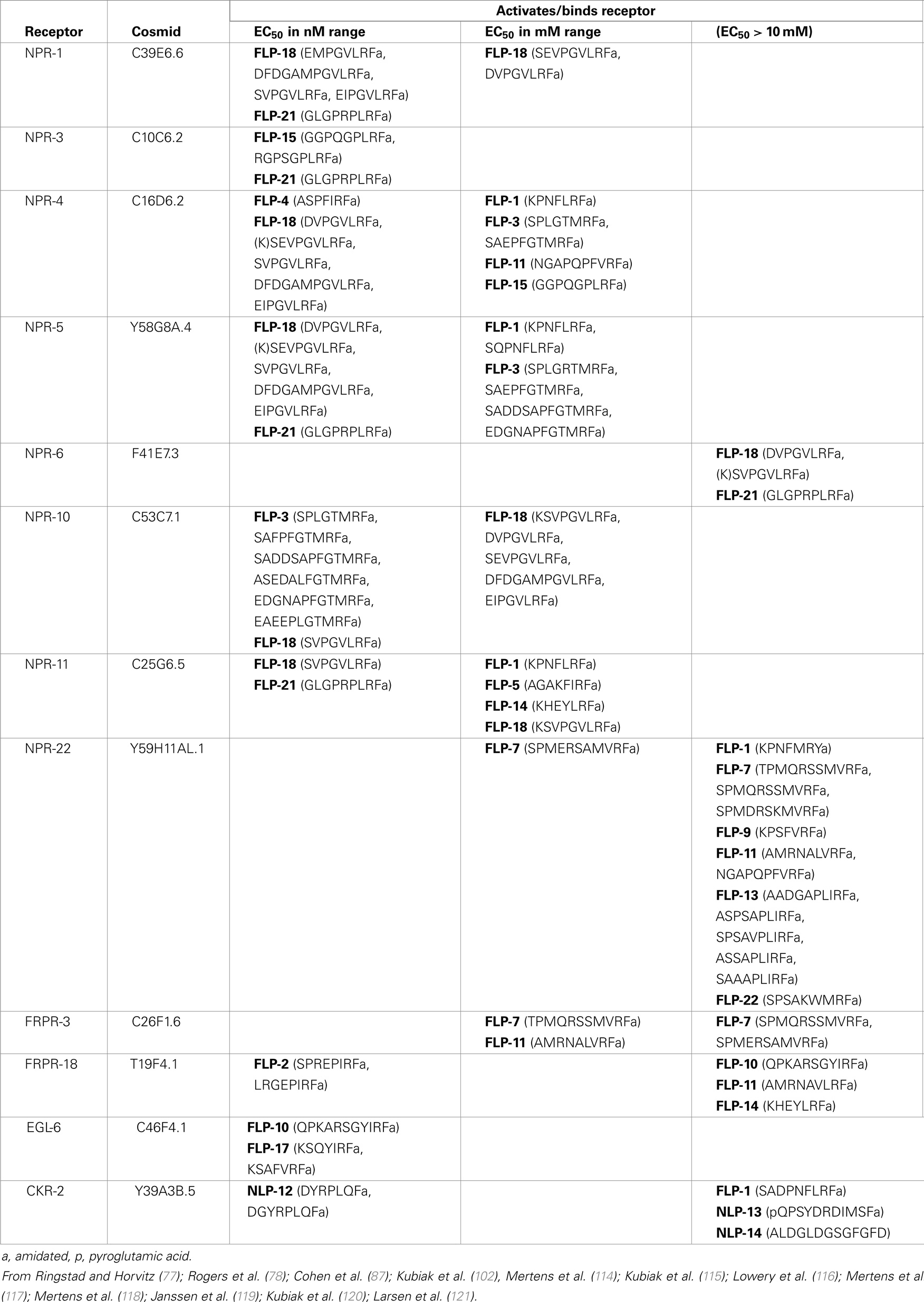
Several heterologous systems have been used to de-orphanize FLP receptors: expression of receptors in Xenopus laevis oocytes or in cell lines and use of different signaling readouts, including whole cell voltage-clamp, GTPγS binding, cAMP levels, and calcium indicators, to determine receptor activation. The EC50 (concentration which produces 50% maximal activation) varies substantially among the different readouts (nanomolar to micromolar), suggesting different sensitivity among the readout systems. These studies have revealed that as with vertebrate and other invertebrate neuropeptide receptors, C. elegans FLP receptors are generally activated by multiple FLPs, which may be encoded by the same or, more likely, distinct flp genes (Table 4). FRPR-18/T19F4.1, is the only receptor identified thus far that is activated by peptides encoded by only one flp gene, flp-2, with a physiological EC50 (114). Most receptors are activated by peptides encoded by multiple flp genes. For instance, NPR-3/C10C6.2 was activated at an EC50 in the nM range by both peptides encoded by flp-15 (115) and the single flp-21 peptide, which all share a C-terminal PLRFamide (116). FRPR-3/C26F1.6 was activated by one FLP-11 and one FLP-7 peptide, both of which share a C-terminal VRFamide, while other closely related FLP-7 and FLP-11 peptides did not activate the receptor (117). EGL-6/C46F4.1 was activated by two C-terminally related peptides (YIRFamide), FLP-10, and FLP-17 (77). Several receptors can be activated by a large number of FLP peptides that do not appear to share structural similarities except for the C-terminal RFamide: NPR-22/Y59H11AL.1 binds 15 peptides encoded by six flp genes (flp-1, 7, 9, 13, 11, and 22) with an EC50 ranging from 25 nM to 5 μM (116, 118), NPR-4/C16D6.2 binds 13 peptides encoded by seven flp genes (flp-1, 3, 4, 11, 14, 15, and 18) with an EC50 ranging from 5 nM to >10 μM (116), and NPR-11/C25G6.5 binds eight FLP peptides encoded by five genes (flp-1, 5, 14, 18, and 21) with an EC50 ranging from 1 nM to 8 μM (116). Despite being able to activate receptors at high concentrations (e.g., EC50 > 10 μM), such peptides may not be physiological ligands. For instance, FLP-1 activates multiple receptors, but all at an EC50 that is unlikely to be physiological; hence, the FLP-1 receptor has yet to be identified. One receptor, CKR-2, has been found to bind both FLP and non-FLP peptides. CKR-2 binds NLP-12 and NLP-13 at high affinity (nanomolar range), but also binds a FLP-1 peptide at low affinity (>10 μM) (119). The complexity with which the peptides binds receptors and the receptors bind peptides suggests that behaviors can be subtly refined by modulating peptide levels.
Diversity of FLPs in Nematodes
The accessibility of C. elegans, ease of manipulation and maintenance in the laboratory, and wealth of previous research have made C. elegans a useful model in the study of FLPs in other nematodes, and, in particular, parasitic nematodes. Parasitic nematodes infect over 1 billion people worldwide (University of Cambridge) and contributes to greater than $126 billion of crop damage globally (122). The recent sequencing of multiple nematode genomes and transcriptomes has greatly enhanced our understanding of the FLP complement in different nematodes. In addition to C. elegans, there are now 16 nematode genomes and/or transcriptomes that have been completed, although some are not as well annotated as others; this collection includes both parasitic and non-parasitic nematodes2. Other nematode sequences have also been deposited in the NCBI database, allowing us to use bioinformatic techniques to scan for flp genes among these genomes. By comparison to the C. elegans flp genes, there are 30 parasitic and 6 free-living nematodes that have flp orthologs [(123); unpubl. obs.].
Some striking observations can be noted among this collection of sequences. First, most nematodes, including free-living and parasitic (animal and plant), have a large complement of flp genes (Table 5). The most frequently appearing genes (>65%) among the scanned genomes are flp-1, 11, 14, and 18; other common genes (>50%) include flp-6, 12, 13, 16, 19, 21, 22, and 24. There were also some genes that appear infrequently. For instance, flp-10 has only been identified in free-living nematodes (both hermaphrodite and male/female species), whereas, flp-31 only appears in plant parasitic nematodes (123). Among the nematodes whose genomes have been completely determined, free-living and animal parasitic nematodes express flp-8, 15, 23, 24, and 33; none of the plant parasitic nematodes have been found to express these flp genes thus far. By contrast, we have not identified any flp gene that is expressed in free-living and plant parasitic nematodes, but not in animal parasitic nematodes. Some flp genes appear in free-living and parasitic nematodes, but are poorly represented, appearing in <25% of the genomes scanned (e.g., flp-9, 26, and 28). As the genomes/transcriptomes become better annotated, these numbers may change. However, the diversity and number of FLPs suggest that FLPs are widely used as neuromodulators in many, if not all, nematodes.
Function of FLPs in Non-C. elegans Nematodes
Because of the difficulty of propagating some of the parasitic nematodes in the lab, the function of peptides has been difficult to query. Furthermore, many genetic tools, such as isolating mutants or generating transgenic strains, are not available. However, the larger sizes of some nematodes allow the ability to do dissections and perform electrophysiological recordings and in situ hybridization techniques (128–130). Hence, the flp expression patterns described in many pathogenic nematodes based on in situ data may more accurately reflect the expression patterns of the genes than in C. elegans, where expression patterns are generally determined by use of transcriptional reporters (12). In addition, while the decreased expression of the SID-1 receptor in neurons hampers analysis of neuropeptides in C. elegans (131, 132), RNAi by soaking has been effective in neurons of some pathogenic nematodes (133). However, as in C. elegans, there are many caveats with the use of RNAi, particularly as there are no mutants among parasitic nematodes that can be used to confirm RNAi phenotypes (134). For simplicity in the discussion below, any C. elegans FLP peptide that is identical and conserved through different nematode species will be referred to by the C. elegans name, while peptides that are specific to a certain species will be indicated as such.
Locomotion
The neuromuscular system is highly sensitive to disruptions in FLP peptide levels. In a sand column motility assay, knockdown of Gp-flp-1 and 18 and flp-6, 12, and 14 inhibited motility of second-stage juvenile (J2) Globodera pallida animals by 70–100% after 24 h of dsRNA soaking; the effects were transitory and by 6 days post-treatment, 27.1% of the animals recovered 39.4% of their normal activity (133). By contrast, when Gp-flp-32 was knocked down, G. pallida and Meloidogyne incognita juveniles became more mobile, which translated to an enhanced ability to infect potato plant and other plant roots (135).
Chemosensation
When infecting the roots of tomato plants, RNAi knockdown of Mi-flp-18 in J2 M. incognita dramatically decreased chemotaxis to tomato plant roots, thereby decreasing rates of infection (136). The exudate of Chrysanthemum coronarium, which is often planted with tomato, contains lauric acid, which decreases Mi-flp-18 expression, hence providing a mechanism to alleviate crop damage (136).
Pharmacology of FLPs
Muscle preparations of some of the larger nematodes have allowed the pharmacological application of different FLPs at nanomolar to micromolar concentrations. For instance, Ascaris suum somatic muscle strips can be isolated, muscle tension recorded, and the effects of different FLPs examined (126). As-FLP-18 (0.1 μM) caused contraction and occasionally rhythmic activity; this action could be reversed by FLP-1 (SADPNFLRF-NH2; 0.1 μM) (137). As-FLP-1, FLP-1 (SADPNFLRF-NH2, SDPNFLRF-NH2; 1 nM), FLP-13 (APEASPFIRF-NH2;10 μM) caused relaxation of muscle strips whether innervated or denervated (16, 138). Furthermore, As-FLP-1 (1 nM) inhibited contractions of spontaneously active preparations (138) and hyperpolarized muscle membrane (139). Some of the FLPs had more complex actions and produced biphasic responses. FLP-8 (0.1–1 μM) and FLP-14 initially caused relaxation, followed by rhythmic contractions; FLP-14 (nanomolar) was 10–100 times more potent on the muscle strips than FLP-8 (138, 140). FLP-6 (0.1–10 μM) caused a rapid (within 30 s) increase in muscle tension in ventral muscle strips, but relaxation of dorsal muscle strips (126, 138). Intracellular recordings from A. suum muscle support the effects of some FLPs on muscle strips. Namely, FLP-1 (SDPNFLRF-NH2 and SADPNFLRF-NH2; EC50 ~300 nM) caused a slow hyperpolarization and decreased amplitude of the excitatory junction potentials (141).
The reproductive system of A. suum also provides a robust biological system to examine the activity of multiple FLPs. In particular, the ovijector contains circular muscle that regulates egg release and sperm influx; the muscle is innervated by a nerve plexus, which is FMRFamide-like immunoreactive (142). When the ovijector was hooked to a photo-optic transducer, the ovipositor revealed rhythmic spontaneous contractions (143). Most FLPs induced an inhibitory response, whereby there was a decrease in contraction frequency and amplitude; these peptides included FLP-3 (SPLGTMRF-NH2; 0.1–10 μM), 4 (ASPSFIRF-NH2; 0.01 μM), 7 (SPMERSAMVRF-NH2; 0.1 μM), 10 (QPKARSGYIRF-NH2; 1 μM), 11 (NGAPQPFVRF-NH2; 1 μM), 12 (1 μM), 13 (ASSAPLIRF-NH2; 1 μM), 15 (GPSGPLRF-NH2; 0.1 μM), FLP-16 (GQTFVRF-NH2; 0.1 μM), 17 (KSQYIRF-NH2; 0.1 μM), and 20 (AMMRF-NH2; 10 μM) (143). A few FLPs caused an excitatory response [FLP-2 (SPREPIRF-NH2; 10–100 nM), 18 (SVPGVLRF-NH2; 10 nM–10 μM), 19 (WANQVRF-NH2; 10 nM–10 μM; ASWASSVRF-NH2; 10 μM), and 21 (1–10 μM)] or showed a shortening of the ovipositor before an increase in contraction frequency [FLP-11 (AMRNALVRF-NH2; 100 nM)] (143). The activities of some peptides were only a transient contraction [FLP-2 (LRGEPIRF-NH2; 0.1–10 μM)] or a transient contraction followed by an extended period of inactivity [FLP-5 (AGAKFIRF-NH2, APKPKFIRF-NH2), 8, and 22 (SPSAKWMRF-NH2)] (143). Whether these FLP responses are physiological are unclear, particularly as some activities were only induced at high, micromolar concentrations.
To determine the effect of peptides in intact animals, peptides were directly injected into the Ascaris pseudocelom (injected at 100 μM; final concentration within pseudocelom estimated at 10 μM). FLP-8 and 14 inhibited locomotion, caused shallower waveforms with no propagation, and shortened body length, while injection of FLP-4 (ASPSFIRF-NH2), 6 (KSAYMRF-NH2), 7 (SPMQRSMVRF-NH2), 13 (APEASPIRF-NH2), and 16 (AQTFVRF-NH2) and AsFLP-13 abolished body waves and increased body length; As-FLP-1 produced a more severe phenotype by causing a complete paralysis (144, 145). By contrast, injection of As-FLP-18 increased the number of body waves, but the body waves did not propagate and body length was decreased (145). FLP-6 and 9 caused ventral coils, while FLP-12 caused uncoordination (145).
Physiology of FLP Peptides on A. suum Motoneurons
Several FLPs are expressed in the motoneurons of many nematodes. A. suum presents a relatively accessible system in which DE2 excitatory and DI inhibitory motoneurons can be exposed in situ and recorded while saline containing test solutions (generally 10 μM except where noted) are superfused; recorded responses can be direct actions of peptides on motoneurons or indirect effects via presynaptic neurons (128, 146). FLP-8 (1–100 nM) abolished slow oscillatory membrane potentials and reduced input resistances (147). As-FLP-4 and FLP-8 induced strong depolarizations in DE2 and weak depolarizations in DI motoneurons and generally decreased input resistances; FLP-16 elicited strong depolarizations in DE2 and DI (144). FLP-14 induced a stronger depolarization, but showed a transient increase in input resistance followed by a longer decreased input resistance in DE2 neurons; although FLP-14 caused a modest decrease in the input resistance of DI motoneurons, there was no membrane potential effect (144). The depolarizations elicited by the FLPs were in many cases the result of increased frequency of EPSPs. FLP-21 caused a complex response in which there was an initial hyperpolarization, followed by a sustained depolarization in DE2 neurons, but only a hyperpolarization in DI neurons; no change or a decrease was seen in the input resistance of DE2 and DI neurons, respectively. Application of different As-FLP-18 peptides or As-FLP-1 elicited weak depolarizations in DE2. FLP-6 elicited a weak hyperpolarization in DE2, but a strong depolarization, followed by miniature IPSPs in DI (144). As-FLP-13 elicited weak hyperpolarizations in DE2 and DI motoneurons. No effect was seen with application of As-FLP-28 in DE2 or DI neurons. As seen in other Ascaris preparations, FLPs exert a diversity of effects, but the physiological relevance has yet to be determined.
Summary
The diversity and plethora of neuropeptides that can act over long distances allows invertebrates to greatly increase the complexity of their neural networks despite the relatively small number of neurons. A single neuron may mediate response to several sensory modalities and activate different or conserved downstream pathways to ensure correct motor outputs. Despite the relatively simple nervous system of C. elegans, the clever use of neuropeptides, gap junctions, hub neurons, and conventional synaptic connections allows the animal to have a wide behavioral repertoire that can be finely tuned in response to different environmental stimuli. Understanding how these simple neural networks determine behavior remains the challenge for the future.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the NIH and NSF (Chris Li) and from the DGIST R&D Program of the Ministry of Science, ICT and Future Planning (14-BD-06), and the National Research Foundation of Korea (NRF-2012R1A1A2009385) (Kyuhyung Kim). We thank members of the Li and Kim labs for helpful discussions.
Footnotes
References
1. White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci (1986) 314:1–340. doi: 10.1098/rstb.1986.0056
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. Jarrell TA, Wang Y, Bloniarz AE, Brittin CA, Xu M, Thomson JN, et al. The connectome of a decision-making circuit. Science (2012) 337:437–44. doi:10.1126/science.1221762
3. Nathoo AN, Moeller RA, Westlund BA, Hart AC. Identification of neuropeptide-like protein gene families in Caenorhabditis elegans and other species. Proc Natl Acad Sci USA (2001) 98:14000–5. doi:10.1073/pnas.241231298
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Kim K, Li C. Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J Comp Neurol (2004) 475:540–50. doi:10.1002/cne.20189
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Duret L, Guex N, Peitsch MC, Bairoch A. New insulin-like proteins with atypical disulfide bond pattern characterized in Caenorhabditis elegans by comparative sequence analysis and homology modeling. Genome Res (1998) 8:348–53.
6. Gregoire FM, Chomiki N, Kachinskas D, Warden CH. Cloning and developmental regulation of a novel member of the insulin-like gene family in Caenorhabditis elegans. Biochem Biophys Res Commun (1998) 249:385–90. doi:10.1006/bbrc.1998.9164
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Kawano T, Ito Y, Ishiguro M, Takuwa K, Nakajima T, Kimura Y. Molecular cloning and characterization of a new insulin/IGF-like peptide of the nematode Caenorhabditis elegans. Biochem Biophys Res Commun (2000) 273:431–6. doi:10.1006/bbrc.2000.2971
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Li W, Kennedy SG, Ruvkun G. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev (2003) 17:844–58. doi:10.1101/gad.1066503
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, Buchman AR, et al. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev (2001) 15:672–86. doi:10.1101/gad.867301
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Nelson LS, Kim K, Memmott JE, Li C. FMRFamide-related gene family in the nematode, Caenorhabditis elegans. Brain Res Mol Brain Res (1998) 58:103–11. doi:10.1016/S0169-328X(98)00106-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Li C, Kim K, Nelson LS. FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. Brain Res (1999) 848:26–34. doi:10.1016/S0006-8993(99)01972-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Li C, Kim K. Neuropeptides (September 25, 2008). In: WormBook, editor. The C. elegans Research Community, WormBook (2008). Available from: http://www.wormbook.org
13. Li C, Nelson LS, Kim K, Nathoo A, Hart AC. Neuropeptide gene families in the nematode Caenorhabditis elegans. Ann N Y Acad Sci (1999) 897:239–52. doi:10.1111/j.1749-6632.1999.tb07895.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Rosoff ML, Doble KE, Price DA, Li C. The FLP-1 propeptide is processed into multiple, highly similar FMRFamide-like peptides in Caenorhabditis elegans. Peptides (1993) 14:331–8. doi:10.1016/0196-9781(93)90049-M
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Marks NJ, Shaw C, Maule AG, Davis JP, Halton DW, Verhaert P, et al. Isolation of AF2 (KHEYLRFamide) from Caenorhabditis elegans: evidence for the presence of more than one FMRFamide-related peptide-encoding gene. Biochem Biophys Res Commun (1995) 217:845–51. doi:10.1006/bbrc.1995.2849
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Marks NJ, Maule AG, Geary TG, Thompson DP, Davis JP, Halton DW, et al. APEASPFIRFamide, a novel FMRFamide-related decapeptide from Caenorhabditis elegans: structure and myoactivity. Biochem Biophys Res Commun (1997) 231:591–5. doi:10.1006/bbrc.1997.6155
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Marks NJ, Maule AG, Geary TG, Thompson DP, Li C, Halton DW, et al. KSAYMRFamide (PF3/AF8) is present in the free-living nematode, Caenorhabditis elegans. Biochem Biophys Res Commun (1998) 248:422–5. doi:10.1006/bbrc.1998.8982
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Marks NJ, Maule AG, Li C, Nelson LS, Thompson DP, Alexander-Bowman S, et al. Isolation, pharmacology and gene organization of KPSFVRFamide: a neuropeptide from Caenorhabditis elegans. Biochem Biophys Res Commun (1999) 254:222–30. doi:10.1006/bbrc.1998.9920
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Marks NJ, Shaw C, Halton DW, Thompson DP, Geary TG, Li C, et al. Isolation and preliminary biological assessment of AADGAPLIRFamide and SVPGVLRFamide from Caenorhabditis elegans. Biochem Biophys Res Commun (2001) 286:1170–6. doi:10.1006/bbrc.2001.5524
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Davis RE, Stretton AO. The motornervous system of Ascaris: electrophysiology and anatomy of the neurons and their control by neuromodulators. Parasitol (1996) 113:S97–117.
21. Husson SJ, Clynen E, Baggerman G, De Loof A, Schoofs L. Discovering neuropeptides in Caenorhabditis elegans by two dimensional liquid chromatography and mass spectrometry. Biochem Biophys Res Commun (2005) 335:76–86. doi:10.1016/j.bbrc.2005.07.044
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Husson SJ, Clynen E, Baggerman G, Janssen T, Schoofs L. Defective processing of neuropeptide precursors in Caenorhabditis elegans lacking proprotein convertase 2 (KPC-2/EGL-3): mutant analysis by mass spectrometry. J Neurochem (2006) 98:1999–2012. doi:10.1111/j.1471-4159.2006.04014.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Husson SJ, Schoofs L. Altered neuropeptide profile of Caenorhabditis elegans lacking the chaperone protein 7B2 as analyzed by mass spectrometry. FEBS Lett (2007) 581:4288–92. doi:10.1016/j.febslet.2007.08.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Husson SJ, Landuyt B, Nys T, Baggerman G, Boonen K, Clynen E, et al. Comparative peptidomics of Caenorhabditis elegans versus C. briggsae by LC-MALDI-TOF MS. Peptides (2009) 30:449–57. doi:10.1016/j.peptides.2008.07.021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Hallberg M. Neuropeptides: metabolism to bioactive fragments and the pharmacology of their receptors. Med Res Rev (2014). doi:10.1002/med.21323
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Steiner DF. The proprotein convertases. Curr Opin Chem Biol (1998) 2:31–9. doi:10.1016/S1367-5931(98)80033-1
27. Thacker C, Peters K, Srayko M, Rose AM. The bli-4 locus of Caenorhabditis elegans encodes structurally distinct kex2/subtilisin-like endoproteases essential for early development and adult morphology. Genes Dev (1995) 9:956–71. doi:10.1101/gad.9.8.956
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Thacker C, Rose AM. A look at the Caenorhabditis elegans kex2/subtilisin-like proprotein convertase family. Bioessays (2000) 22:545–53. doi:10.1002/(SICI)1521-1878(200006)22:6<545::AID-BIES7>3.0.CO;2-F
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Trent C, Tsung N, Horvitz HR. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics (1983) 104:619–47.
31. Thomas JH. Genetic analysis of defecation in Caenorhabditis elegans. Genetics (1990) 124:855–72.
32. Kass J, Jacob TC, Kim P, Kaplan JM. The EGL-3 proprotein convertase regulates mechanosensory responses of Caenorhabditis elegans. J Neurosci (2001) 21:9265–72.
33. Jacob TC, Kaplan JM. The EGL-21 carboxypeptidase E facilitates acetylcholine release at Caenorhabditis elegans neuromuscular junctions. J Neurosci (2003) 23:2122–30.
34. Schroeder NE, Androwski RJ, Rashid A, Lee H, Lee J, Barr MM. Dauer-specific dendrite arborization in C. elegans is regulated by KPC-1/Furin. Curr Biol (2013) 23:1527–35. doi:10.1016/j.cub.2013.06.058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. Lee H, Choi MK, Lee D, Kim HS, Hwang H, Kim H, et al. Nictation, a dispersal behavior of the nematode Caenorhabditis elegans, is regulated by IL2 neurons. Nat Neurosci (2011) 15:107–12. doi:10.1038/nn.2975
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Rual J-F, Ceron J, Koreth J, Hao T, Nicot A-S, Hirozane-Kishikawa T, et al. Towards improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res (2004) 14:2162–68. doi:10.1101/gr.2505604
37. Mahoney TR, Luo S, Round EK, Brauner M, Gottschalk A, Thomas JH, et al. Intestinal signaling to GABAergic neurons regulates a rhythmic behavior in Caenorhabditis elegans. Proc Natl Acad Sci U S A (2008) 105:16350–5. doi:10.1073/pnas.0803617105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Thacker C, Sheps JA, Rose AM. Caenorhabditis elegans dpy-5 is a cuticle procollagen processed by a proprotein convertase. Cell Mol Life Sci (2006) 63:1193–204. doi:10.1007/s00018-006-6012-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Seidah NG, Day R, Marcinkiewicz M, Benjannet S, Chrétien M. Mammalian neural and endocrine pro-protein and pro-hormone convertases belonging to the subtilisin family of serine proteases. Enzyme (1991) 45:271–84.
40. Sieburth D, Ch’ng Q, Dybbs M, Tavazoie M, Kennedy S, Wang D, et al. Systematic analysis of genes required for synapse structure and function. Nature (2005) 436:510–7. doi:10.1038/nature03809
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, Rand JB. A genetic selection for Caenorhabditis elegans synaptic transmission mutant. Proc Natl Acad Sci U S A (1996) 93:12593–8. doi:10.1073/pnas.93.22.12593
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol (2003) 1(1):e12. doi:10.1371/journal.pbio.0000012
43. Bowman JW, Friedman AR, Thompson DP, Ichhpurani AK, Kellman MF, Marks N, et al. Structure-activity relationships of KNEFIRFamide (AF1), a nematode FMRFamide-related peptide, on Ascaris suum muscle. Peptides (1996) 17:381–7. doi:10.1016/0196-9781(96)00007-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Eipper BA, Perkins SN, Husten EJ, Johnson RC, Keutmann HT, Mains RE. Peptidyl-alpha-hydroxyglycine alpha-amidating lyase. Purification, characterization, and expression. J Biol Chem (1991) 266:7828–33.
45. Eipper BA, Milgram SL, Husten EJ, Yun HY, Mains RE. Peptidylglycine alpha-amidating monooxygenase: a multifunctional protein with catalytic, processing, and routing domains. Protein Sci (1993) 2:489–97. doi:10.1002/pro.5560020401
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. Han M, Park D, Vanderzalm PJ, Mains RE, Eipper BA, Taghert PH. Drosophila uses two distinct neuropeptide amidating enzymes, dPAL1 and dPAL2. J Neurochem (2004) 90:129–41. doi:10.1111/j.1471-4159.2004.02464.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Roques BP, Noble F, Daugé V, Fournié-Zaluski MC, Beaumont A. Neutral endopeptidase 24.11: structure, inhibition, and experimental and clinical pharmacology. Pharmacol Rev (1993) 45:87–146.
48. Isaac RE, Siviter RJ, Stancombe P, Coates D, Shirras AD. Conserved roles for peptidases in the processing of invertebrate neuropeptides. Biochem Soc Trans (2000) 28:460–4. doi:10.1042/0300-5127:0280460
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Coates D, Siviter R, Isaac RE. Exploring the Caenorhabditis elegans and Drosophila melanogaster genomes to understand neuropeptide and peptidase function. Biochem Soc Trans (2000) 28:464–9. doi:10.1042/0300-5127:0280464
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
50. Turner AJ, Isaac RE, Coates D. The neprilysin (NEP) family of zinc metalloendopeptidases: genomics and function. Bioessays (2001) 23:261–9. doi:10.1002/1521-1878(200103)23:3<261::AID-BIES1036>3.0.CO;2-K
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Spanier B, Stürzenbaum SR, Holden-Dye LM, Baumeister R. Caenorhabditis elegans neprilysin NEP-1: an effector of locomotion and pharyngeal pumping. J Mol Biol (2005) 352:429–37. doi:10.1016/j.jmb.2005.06.063
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
52. Hall DH, Hedgecock EM. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell (1991) 65:837–47. doi:10.1016/0092-8674(91)90391-B
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Schinkmann K. FMRFamide-like peptides in the nematodes Caenorhabditis elegans and Caenorhabditis vulgaris. PhD thesis. Boston, MA: Boston University (1994).
54. Bruns D, Jahn R. Real-time measurement of transmitter release from single synaptic vesicles. Nature (1995) 377:62–5. doi:10.1038/377062a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Caromile LA, Oganesian A, Coats SA, Seifert RA, Bowen-Pope DF. The neurosecretory vesicle protein phogrin functions as a phosphatidylinositol phosphatase to regulate insulin secretion. J Biol Chem (2010) 285:10487–96. doi:10.1074/jbc.M109.066563
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Zahn TR, Macmorris MA, Dong W, Day R, Hutton JC. IDA-1, a Caenorhabditis elegans homolog of the diabetic autoantigens IA-2 and phogrin, is expressed in peptidergic neurons in the worm. J Comp Neurol (2001) 429:127–43. doi:10.1002/1096-9861(20000101)429:1<127::AID-CNE10>3.0.CO;2-H
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
57. Hoover CM, Edwards SL, Yu SC, Kittelmann M, Richmond JE, Eimer S, et al. A novel CaM kinase II pathway controls the location of neuropeptide release from Caenorhabditis elegans motor neurons. Genetics (2014) 196:745–65. doi:10.1534/genetics.113.158568
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Richmond JE, Broadie KS. The synaptic vesicle cycle: exocytosis and endocytosis in Drosophila and C. elegans. Curr Opin Neurobiol (2002) 12:499–507. doi:10.1016/S0959-4388(02)00360-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
59. Brose N, Petrenko AG, Südhof TC, Jahn R. Synaptotagmin: a Ca2+ sensor on the synaptic vesicle surface. Science (1992) 256:1021–5. doi:10.1126/science.1589771
60. McEwen JM, Kaplan JM. UNC-18 promotes both the anterograde trafficking and synaptic function of syntaxin. Mol Biol Cell (2008) 19:3836–46. doi:10.1091/mbc.E08-02-0160
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Ogawa H, Harada S, Sassa T, Yamamoto H, Hosono R. Functional properties of the unc-64 gene encoding a Caenorhabditis elegans syntaxin. J Biol Chem (1998) 273(4):2192–8.
62. Johnson JR, Ferdek P, Lian LY, Barclay JW, Burgoyne RD, Morgan A. Binding of UNC-18 to the N-terminus of syntaxin is essential for neurotransmission in Caenorhabditis elegans. Biochem J (2009) 418:73–80. doi:10.1042/BJ20081956
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Gracheva EO, Maryon EB, Berthelot-Grosjean M, Richmond JE. Differential regulation of synaptic vesicle tethering and docking by UNC-18 and TOM-1. Front Synaptic Neurosci (2010) 5(2):141. doi:10.3389/fnsyn.2010.00141
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Hammarlund M, Watanabe S, Schuske K, Jorgensen EM. CAPS and syntaxin dock dense core vesicles to the plasma membrane in neurons. J Cell Biol (2008) 180:483–91. doi:10.1083/jcb.200708018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
65. Hao Y, Hu Z, Sieburth D, Kaplan JM. RIC-7 promotes neuropeptide secretion. PLoS Genet (2012) 8(1):e1002464. doi:10.1371/journal.pgen.1002464
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Renden R, Berwin B, Davis W, Ann K, Chin CT, Kreber R, et al. Drosophila CAPS is an essential gene that regulates dense-core vesicle release and synaptic vesicle fusion. Neuron (2001) 31:421–37. doi:10.1016/S0896-6273(01)00382-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
67. Grishanin RN, Klenchin VA, Loyet KM, Kowalchyk JA, Ann K, Martin TF. Membrane association domains in Ca2+-dependent activator protein for secretion mediate plasma membrane and dense-core vesicle binding required for Ca2+-dependent exocytosis. J Biol Chem (2002) 277:22025–34. doi:10.1074/jbc.M201614200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
68. Gracheva EO, Burdina AO, Touroutine D, Berthelot-Grosjean M, Parekh H, Richmond JE. Tomosyn negatively regulates CAPS-dependent peptide release at Caenorhaditis elegans synapses. J Neurosci (2007) 27:10176–84. doi:10.1523/JNEUROSCI.2339-07.2007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
69. Sieburth D, Madison JM, Kaplan JM. PKC-1 regulates secretion of neuropeptides. Nat Neurosci (2007) 10:49–57. doi:10.1038/nn1810
70. Richmond JE, Davis WS, Jorgensen EM. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat Neurosci (1999) 2:959–64. doi:10.1038/12160
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
71. McEwen JM, Madison JM, Dybbs M, Kaplan JM. Antagonistic regulation of synaptic vesicle priming by tomosyn and UNC-13. Neuron (2006) 51:303–15. doi:10.1016/j.neuron.2006.06.025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
72. Dossey AT, Reale V, Chatwin H, Zachariah C, de Bono M, Evans PD, et al. NMR analysis of Caenorhabditis elegans FLP-18 neuropeptides: implications for NPR-1 activation. Biochemistry (2006) 45:7586–97. doi:10.1021/bi0603928
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
73. Rosoff ML, Burglin TR, Li C. Alternatively spliced transcripts of the flp-1 gene encode distinct FMRFamide-like peptides in Caenorhabditis elegans. J Neurosci (1992) 12:2356–61.
74. McVeigh P, Leech S, Mair GR, Marks NJ, Geary TG, Maule AG. Analysis of FMRFamide-like peptide (FLP) diversity in phylum Nematoda. Int J Parasitol (2005) 35:1043–60. doi:10.1016/j.ijpara.2005.05.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
75. Schinkmann K, Li C. Comparison of two Caenorhabditis genes encoding FMRFamide (Phe-Met-Arg-Phe-NH2)-like peptides. Brain Res Mol Brain Res (1994) 24:238–46. doi:10.1016/0169-328X(94)90137-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
76. Nelson LS, Rosoff M, Li C. Disruption of a neuropeptide gene, flp-1, causes multiple behavioral defects in Caenorhabditis elegans. Science (1998) 281:1686–90. doi:10.1126/science.281.5383.1686
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
77. Ringstad N, Horvitz HR. FMRFamide neuropeptides and acetylcholine synergistically inhibit egg-laying in C. elegans. Nat Neurosci (2008) 11:1168–76. doi:10.1038/nn.2186
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
78. Rogers C, Reale V, Kim K, Chatwin H, Li C, Evans P, et al. Inhibition of Caenorhabditis elegans social feeding by FMRFamide-related peptide activation of NPR-1. Nat Neurosci (2003) 6:1178–85. doi:10.1038/nn1140
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
79. Sithigorngul P, Jarecki JL, Stretton AO. A specific antibody to neuropeptide AF1 (KNEFIRFamide) recognizes a small subset of neurons in Ascaris suum. J Comp Neurol (2011) 519:1546–61. doi:10.1002/cne.22584
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
80. Kim K. Function of a FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. Ph.D. thesis. Boston, MA: Boston University (2003).
81. Wani KA, Catanese M, Normantowicz R, Herd M, Maher KN, Chase DL. D1 dopamine receptor signaling is modulated by the R7 RGS protein EAT-16 and the R7 binding protein RSBP-1 in Caenorhabditis elegans motor neurons. PLoS One (2012) 7(5):e37831. doi:10.1371/journal.pone.0037831
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
82. Stawicki TM, Takayanagi-Kiya S, Zhou K, Jin Y. Neuropeptides function in a homeostatic manner to modulate excitation-inhibition imbalance in C. elegans. PLoS Genet (2013) 9(5):e1003472. doi:10.1371/journal.pgen.1003472
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
83. Waggoner LE, Hardaker LA, Golik S, Schafer WR. Effect of a neuropeptide gene on behavioral states in Caenorhabditis elegans egg-laying. Genetics (2000) 154:1181–92.
84. Rogers CM, Franks CJ, Walker RJ, Burke JF, Holden-Dye L. Regulation of the pharynx of Caenohrabditis elegans by 5-HT, octopamine, and FMRFamide-like neuropeptides. J Neurobiol (2001) 49:235–44. doi:10.1002/neu.1078
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
85. Papaioannou S, Marsden D, Franks CJ, Walker RJ, Holden-Dye L. Role of a FMRFamide-like family of neuropeptides in the pharyngeal nervous system of Caenorhabditis elegans. J Neurobiol (2005) 65:304–19. doi:10.1002/neu.20201
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
86. Liu T, Kim K, Li C, Barr MM. FMRFamide-like neuropeptides and mechanosensory touch receptor neurons regulate male sexual turning behavior in Caenorhabditis elegans. J Neurosci (2007) 27:7174–82. doi:10.1523/JNEUROSCI.1405-07.2007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
87. Cohen M, Reale V, Olofsson B, Knights A, Evans P, de Bono M. Coordinated regulation of foraging and metabolism in C. elegans by RFamide neuropeptide signaling. Cell Metab (2009) 9:375–85. doi:10.1016/j.cmet.2009.02.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
88. Choi S, Chatzigeorgiou M, Taylor KP, Schafer WR, Kaplan JM. Analysis of NPR-1 reveals a circuit mechanism for behavioral quiescence in C. elegans. Neuron (2013) 78:869–80. doi:10.1016/j.neuron.2013.04.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
89. Li C, Timbers TA, Rose JK, Bozorgmehr T, McEwan A, Rankin CH. The FMRFamide-related neuropeptide FLP-20 is required in the mechanosensory neurons during memory for massed training in C. elegans. Learn Mem (2013) 20:103–8. doi:10.1101/lm.028993.112
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
90. Pocock R, Hobert O. Hypoxia activates a latent circuit for processing gustatory information in C. elegans. Nat Neurosci (2010) 13:610–4. doi:10.1038/nn.2537
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
91. Glauser DA, Johnson BE, Aldrich RW, Goodman MB. Intragenic alternative-splicing coordination is essential for Caenorhabditis elegans slo-1 gene function. Proc Natl Acad Sci USA (2011) 108:20790–5. doi:10.1073/pnas.1116712108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
92. McDonald PW, Hardie SL, Jessen TN, Carvelli L, Matthies DS, Blakely RD. Vigorous motor activity in Caenorhabditis elegans requires efficient clearance of dopamine mediated by synaptic localization of the dopamine transporter DAT-1. J Neurosci (2007) 27:14216–27. doi:10.1523/JNEUROSCI.2992-07.2007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
93. Jospin M, Qi YB, Stawicki TM, Boulin T, Schuske KR, Horvitz HR, et al. A neuronal acetylcholine receptor regulates the balance of muscle excitation and inhibition in Caenorhabditis elegans. PLoS Biol (2009) 7(12):e1000265. doi:10.1371/journal.pbio.1000265
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
94. Schafer WR. Deciphering the neural and molecular mechanisms of C. elegans behavior. Curr Biol (2005) 15:R723–9. doi:10.1016/j.cub.2005.08.020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
95. Waggoner LE, Zhou GT, Schafer RW, Schafer WR. Control of alternative behavioral states by serotonin in Caenorhabditis elegans. Neuron (1998) 21:203–14. doi:10.1016/S0896-6273(00)80527-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
96. de Bono M, Bargmann CI. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell (1998) 94:679–89. doi:10.1016/S0092-8674(00)81609-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
97. Cheung BH, Arellano-Carbajal F, Rybicki I, de Bono M. Soluble guanylate cyclases act in neurons exposed to the body fluid to promote C. elegans aggregation behavior. Curr Biol (2004) 14:1105–11. doi:10.1016/j.cub.2004.06.027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
98. Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature (2004) 430:317–22. doi:10.1038/nature02714
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
99. de Bono M, Tobin DM, Davis MW, Avery L, Bargmann CI. Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature (2002) 419:899–903. doi:10.1038/nature01169
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
100. Rogers C, Persson A, Cheung B, de Bono M. Behavioral motifs and neural pathways coordinating O2 responses and aggregation in C. elegans. Curr Biol (2006) 16:649–59. doi:10.1016/j.cub.2006.03.023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
101. Coates JC, de Bono M. Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans. Nature (2002) 419:925–9. doi:10.1038/nature01170
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
102. Kubiak TM, Larsen MJ, Nulf SC, Zantello MR, Burton KJ, Bowman JW, et al. Differential activation of “social” and “solitary” variants of the Caenorhabditis elegans G protein-coupled receptor NPR-1 by its cognate ligand AF9. J Biol Chem (2003) 278:33724–9. doi:10.1074/jbc.M304861200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
103. Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RA, Clardy J, et al. A hub-and-spoke circuit drives pheromone attraction and social behavior in C. elegans. Nature (2009) 458:1171–5. doi:10.1038/nature07886
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
104. Jang H, Kim K, Neal SJ, Macosko E, Kim D, Butcher RA, et al. Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. elegans. Neuron (2012) 75:585–92. doi:10.1016/j.neuron.2012.06.034
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
105. Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn HT, Teal PE, et al. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature (2008) 454:1115–8. doi:10.1038/nature07168
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
106. Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV, et al. Lethargus is a Caenorhabditis elegans sleep-like state. Nature (2008) 451:569–72. doi:10.1038/nature06535
107. Ardiel EL, Rankin CH. An elegant mind: learning and memory in Caenorhabditis elegans. Learn Mem (2010) 17:191–201. doi:10.1101/lm.960510
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
108. Loer CM, Kenyon CJ. Serotonin-deficient mutants and male mating behavior in the nematode Caenorhabditis elegans. J Neurosci (1993) 13:5407–17.
109. Liu KS, Sternberg PW. Sensory regulation of male mating behavior in Caenorhabditis elegans. Neuron (1995) 14:79–89. doi:10.1016/0896-6273(95)90242-2
110. Lingueglia E, Champigny G, Lazdunski M, Barbry P. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature (1995) 378:730–3. doi:10.1038/378730a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
111. Bargmann CI. Neurobiology of the Caenorhabditis elegans genome. Science (1998) 282:2028–33. doi:10.1126/science.282.5396.2028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
112. Janssen T, Lindemans M, Meelkop E, Temmerman L, Schoofs L. Coevolution of neuropeptidergic signaling systems: from worm to man. Ann N Y Acad Sci (2010) 1200:1–14. doi:10.1111/j.1749-6632.2010.05506.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
113. Keating CD, Kriek N, Daniels M, Ashcroft NR, Hopper NA, Siney EJ, et al. Whole-genome analysis of 60 G protein-coupled receptors in Caenorhabditis elegans by gene knockout with RNAi. Curr Biol (2003) 13:1715–20. doi:10.1016/j.cub.2003.09.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
114. Mertens I, Meeusen T, Janssen T, Nachman R, Schoofs L. Molecular characterization of two G protein-coupled receptor splice variants as FLP2 receptors in Caenorhabditis elegans. Biochem Biophys Res Commun (2005) 330:967–74. doi:10.1016/j.bbrc.2005.03.071
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
115. Kubiak TM, Larsen MJ, Zantello MR, Bowman JW, Nulf SC, Lowery DE. Functional annotation of the putative orphan Caenorhabditis elegans G-protein-coupled receptor C10C6.2 as a FLP15 peptide receptor. J Biol Chem (2003) 278:42115–20. doi:10.1074/jbc.M304056200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
116. Lowery DE, Geary TG, Kubiak TM, Larsen MJ. G protein-Coupled Receptor-Like Receptors and Modulators Thereof. Pharmacia & Upjohn Company. United States Patent No. 6632621 (2003).
117. Mertens I, Vandingenen A, Meeusen T, Janssen T, Luyten W, Nachman RJ, et al. Functional characterization of the putative orphan neuropeptide G-protein coupled receptor C26F1.6 in Caenorhabditis elegans. FEBS Lett (2004) 573:55–60. doi:10.1016/j.febslet.2004.07.058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
118. Mertens I, Clinckspoor I, Janssen T, Nachman R, Schoofs L. FMRFamide related peptide ligands activate the Caenorhabditis elegans orphan GPCR Y59H11AL.1. Peptides (2006) 27:1291–6. doi:10.1016/j.peptides.2005.11.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
119. Janssen T, Meelkop E, Lindemans M, Verstraelen K, Husson SJ, Temmerman L, et al. Discovery of a cholecystokinin-gastrin-like signaling system in nematodes. Endocrinology (2008) 149:2826–39. doi:10.1210/en.2007-1772
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
120. Kubiak TM, Larsen MJ, Bowman JW, Geary TG, Lowery DE. FMRFamide-like peptides encoded on the flp-18 precursor gene activate two isoforms of the orphan Caenorhabditis elegans G-protein-coupled receptor Y58G8A.4 heterologously expressed in mammalian cells. Biopolymers (2008) 90:339–48. doi:10.1002/bip.20850
121. Larsen MJ, Lancheros ER, Williams T, Lowery DE, Geary TG, Kubiak TM. Functinal expression and characterization of the C. elegans G-protein-coupled FLP-2 receptor (T19F4.1) in mammalian cells and yeast. Int J Parasitol Drugs Drug Resist (2009) 3:1–7. doi:10.1016/j.ijpddr.2012.10.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
122. Chitwood DJ. Research on plant-parasitic nematode biology conducted by the United States department of agriculture-agricultural research service. Pest Manag Sci (2003) 59:748–53. doi:10.1002/ps.684
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
123. McCoy CJ, Atkinson LE, Zamanian M, McVeigh P, Day TA, Kimber MJ, et al. New insights into the FLPergic complements of parasitic nematodes: informing deorphanisation approaches. EuPA Open Proteom (2014). doi:10.1016/j.euprot.2014.04.002
124. van Megen H, van den Elsen S, Holterman M, Karssen G, Mooyman P, Bongers T, et al. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology (2009) 11:927–50. doi:10.1163/156854109X456862
125. Geary TG, Klein RD, Vanover L, Bowman JW, Thompson DP. The nervous system of helmiths as targets for drugs. J Parasitol (1992) 78:215–30. doi:10.2307/3283469
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
126. Maule AG, Shaw C, Bowman JW, Halton DW, Thompson DP, Geary TG, et al. KSAYMRFamide: a novel FMRFamide-related heptapeptide from the free-living nematode, Panagrellus redivivus, which is myoactive in the parasitic nematode, Ascaris suum. Biochem Biophys Res Commun (1994) 200:973–80. doi:10.1006/bbrc.1994.1545
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
127. Thakur P, Sharma A, Rao SB, Kumar M, G NP, Tyagi N, et al. Cloning and characterization of two neuropeptide genes from cereal cyst nematode, Heterodera avenae from India. Bioinformation (2012) 8:617–21. doi:10.6026/97320630008617
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
128. Stretton AO, Fishpool RM, Southgate E, Donmoyer JE, Walrond JP, Moses JE, et al. Structure and physiological activity of the motoneurons of the nematode Ascaris. Proc Natl Acad Sci U S A (1978) 75:3493–7. doi:10.1073/pnas.75.7.3493
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
129. Tuan RS, Shepley KJ, Mulligan MM, Abraham D, Perler FB. Histochemical localization of gene expression in Onchocerca volvulus: in situ DNA histohyridization and immunocytochemistry. Mol Biochem Parasitol (1991) 49:191–203. doi:10.1016/0166-6851(91)90063-C
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
130. Nanda JC, Stretton AO. In situ hybridization of neuropeptide-encoding transcripts afp-1, afp-3, and afp-4 in neurons of the nematode Ascaris suum. J Comp Neurol (2010) 518:896–910. doi:10.1002/cne.22251
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
131. Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science (2002) 295:2456–9. doi:10.1126/science.1068836
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
132. Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol (2001) 2(1):RESEARCH0002.
133. Kimber MJ, McKinney S, McMaster S, Day TA, Fleming CC, Maule AG. flp gene disruption in a parasitic nematode reveals motor dysfunction and unusual neuronal sensitivity to RNA interference. FASEB J (2007) 21:1233–43. doi:10.1096/fj.06-7343com
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
134. Dalzell JJ, Warnock ND, McVeigh P, Marks NJ, Mousley A, Atkinson L, et al. Considering RNAi experimental design in parasitic helminthes. Parasitology (2012) 139:589–604. doi:10.1017/S0031182011001946
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
135. Atkinson LE, Stevenson M, McCoy CJ, Marks NJ, Fleming C, Zamanian M, et al. FLP-32 ligand/receptor silencing phenocopy faster plant pathogenic nematodes. PLoS Pathog (2013) 9(2):e1003169. doi:10.1371/journal.ppat.1003169
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
136. Dong L, Li X, Huang L, Gao Y, Zhong L, Zheng Y, et al. Lauric acid in crown daisy root exudate potently regulates root-knot nematode chemotaxis and disrupts Mi-flp-18 expression to block infection. J Exp Bot (2014) 65:131–41. doi:10.1093/jxb/ert356
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
137. Cowden C, Stretton AO. Eight novel FMRFamide-like neuropeptides isolated from the nematode Ascaris suum. Peptides (1995) 16:491–500. doi:10.1016/0196-9781(94)00211-N
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
138. Maule AG, Geary TG, Bowman JW, Marks NJ, Blair KL, Halton DW, et al. Inhibitory effects of nematode FMRFamide-related peptides (FaRPs) on muscle strips from Ascaris suum. Invert Neurosci (1995) 1:255–65. doi:10.1007/BF02211027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
139. Bowman JW, Winterrowd CA, Friedman AR, Thompson DP, Klein RD, Davis JP, et al. Nitric oxide mediates the inhibitory effects of SDPNFLRFamide, a nematode FMRFamide-related neuropeptide, in Ascaris suum. J Neurophysiol (1995) 74:1880–8.
140. Cowden C, Stretton AO. AF2, an Ascaris neuropeptide: isolation, sequence, and bioactivity. Peptides (1993) 14:423–30. doi:10.1016/0196-9781(93)90127-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
141. Holden-Dye L, Franks CJ, Williams RG, Walker RJ. The effect of the nematode peptides SDPNFLRFamide (PF1) and SADPNFLRFamide (PF2) on synaptic transmission in the parasitic nematode Ascaris suum. Parasitology (1995) 110:449–55. doi:10.1017/S0031182000064787
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
142. Fellowes RA, Maule AG, Marks NJ, Geary TG, Thompson DP, Shaw C, et al. Modulation of the motility of the vagina vera of Ascaris suum in vitro by FMRF amide-related peptides. Parasitology (1998) 116:277–87. doi:10.1017/S0031182097002229
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
143. Moffett CL, Beckett AM, Mousley A, Geary TG, Marks NJ, Halton DW, et al. The ovijector of Ascaris suum: multiple response types revealed by Caenorhabditis elegans FMRFamide-related peptides. Int J Parasitol (2003) 33:859–76. doi:10.1016/S0020-7519(03)00109-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
144. Davis RE, Stretton AO. Structure-activity relationships of 18 endogenous neuropeptides on the motor nervous system of the nematode Ascaris suum. Peptides (2001) 22:7–23. doi:10.1016/S0196-9781(00)00351-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
145. Reinitz CA, Herfel HG, Messinger LA, Stretton AO. Changes in locomotory behavior and cAMP produced in Ascaris suum by neuropeptides from Ascaris suum or Caenorhabditis elegans. Mol Biochem Parasitol (2000) 111:185–97. doi:10.1016/S0166-6851(00)00317-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
146. Walrond JP, Kass IS, Stretton AO, Donmoyer JE. Identification of excitatory and inhibitory motoneurons in the nematode Ascaris by electrophysiological techniques. J Neurosci (1985) 5:1–8.
147. Cowden C, Stretton AO, Davis RE. AF1, a sequenced bioactive neuropeptide isolated from the nematode Ascaris suum. Neuron (1989) 2:1465–73. doi:10.1016/0896-6273(89)90192-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: neuropeptides, neural circuits, behavior, nematodes, worms
Citation: Li C and Kim K (2014) Family of FLP peptides in Caenorhabditis elegans and related nematodes. Front. Endocrinol. 5:150. doi: 10.3389/fendo.2014.00150
Received: 04 August 2014; Paper pending published: 22 August 2014;
Accepted: 09 September 2014; Published online: 14 October 2014.
Edited by:
Karine Rousseau, Museum National d’Histoire Naturelle, FranceReviewed by:
Joao Carlos Dos Reis Cardoso, University of Algarve, PortugalGert Jansen, Erasmus MC, Netherlands
Copyright: © 2014 Li and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chris Li, 160 Convent Avenue, MR526, New York, NY 10031, USA e-mail:Y2xpQHNjaS5jY255LmN1bnkuZWR1;
Kyuhyung Kim, 333 Techno Jungang-Daero, Hyeonpung-Myeon, Dalseong-Gun, Daegu 711-873, South Korea e-mail:a2hraW1AZGdpc3QuYWMua3I=
 Chris Li
Chris Li Kyuhyung Kim
Kyuhyung Kim