- Functional Genomics and Proteomics Group, Department of Biology, KU Leuven, Leuven, Belgium
In the three decades since the FMRFamide peptide was isolated from the mollusk Macrocallista nimbosa, structurally similar peptides sharing a C-terminal RFamide motif have been identified across the animal kingdom. FMRFamide-like peptides (FLPs) represent the largest known family of neuropeptides in invertebrates. In the phylum Nematoda, at least 32 flp-genes are classified, making the FLP system of nematodes unusually complex. The diversity of the nematode FLP complement is most extensively mapped in Caenorhabditis elegans, where over 70 FLPs have been predicted. FLPs have shown to be expressed in the majority of the 302 C. elegans neurons including interneurons, sensory neurons, and motor neurons. The vast expression of FLPs is reflected in the broad functional repertoire of nematode FLP signaling, including neuroendocrine and neuromodulatory effects on locomotory activity, reproduction, feeding, and behavior. In contrast to the many identified nematode FLPs, only few peptides have been assigned a receptor and there is the need to clarify the pathway components and working mechanisms of the FLP signaling network. Here, we review the diversity, distribution, and functions of FLPs in nematodes.
Introduction
FMRFamide-like peptides (FLPs) are the largest and most diverse family of neuropeptides known (1, 2). Since the identification of the founder sequence FMRFamide from the clam Macrocallista nimbosa (3), structurally similar peptides have shown to be present in animals of all major phyla (4, 5). Sequence variants of the authentic tetrapeptide have been mainly identified in lophotrochozoans (6). In most phyla and especially in nematodes, however, a diverse repertoire of extended peptides sharing the C-terminal RFamide motif is found (5, 6). Though, they are thought to have a common eumetazoan origin, the relatedness of subfamilies of FLPs remains unclear because of the large sequence diversity (7, 8). Some peptides show high sequence similarity to FMRFamide suggesting homology to the tetrapeptide, and are therefore often referred to as FMRFamide-related peptides (FaRPs). FaRPs are broadly defined as peptides containing the C-terminal sequence X1 X2 RFamide, with X1 generally representing an aromatic amino acid, whereas X2 denotes a hydrophobic residue (6, 7). As many described RFamides differ from the tetrapeptide core for which evolutionary relationships are difficult to determine, the more general term FLP will be used here to address all peptides with a C-terminal RFamide sequence.
FMRFamide-like peptides are intimately involved in a broad pattern of biological processes as diverse as feeding, cardiovascular function, and water homeostasis (4–6, 9). Despite the large sequence diversity, typified by more than 70 family members in the nematode Caenorhabditis elegans, several functions of FLPs in the control of energy balance, feeding behavior, reproduction, and neuromodulation emerge consistently throughout evolution (10, 11). Biochemical and genetic studies, exploiting mainly C. elegans, have provided insight into the FLP-coordinated regulation of these processes in nematodes. The central role of FLPs in nematode biology including reproduction and locomotory activity has also boosted research on FLP signaling as a target for parasite control in pathogenic nematodes (12–14). However, a lack of data on functional nematode FLP-receptor couples slows down the progress in understanding and exploiting the FLP signaling system. Here, we focus on the evolutionary aspect of FLPs, discussing both the sequence conservation and diversity in the phylum Nematoda, and review our knowledge of conserved FLP signaling functions in nematodes.
FLP Repertoire of Nematodes
Initial attempts to identify FLPs relied on molecular cloning of flp-genes (15, 16), and biochemical characterization of immunoreactive peptide fractions by Edman degradation or gas-phase sequencing [reviewed by Maule et al. (17) and Day and Maule (18)]. The first nematode FLP, named AF1 (KNEFIRFa), was biochemically isolated in this way from the parasite Ascaris suum (19). The completion of the C. elegans genome sequence revealed a large diversity in the nematode FLP system, boosting the prediction of neuropeptides through in silico data-mining (20–23). To date, at least 31 flp precursor genes are predicted in C. elegans that give rise to around 70 distinct FLPs [Table 1; Ref. (24)]. Expression has been confirmed for the majority of these peptides (Table 1), mainly by peptidomic strategies enabling a comprehensive analysis of the whole peptide content of organisms (25–27). In addition, these approaches allow determining the presence of posttranslational modifications and the exact processing into bioactive peptides, which may be difficult to accurately predict when multiple or non-conventional cleavage sites are present (25). Peptidomic techniques have also been successfully adopted for characterizing and localizing FLPs in other nematodes, mostly in A. suum (28–32).
Although the FLP repertoire of nematodes has been best studied in C. elegans, evidence emerges on the distribution of FLPs in parasitic and free-living species across the nematode phylum (Table 1). Complete genome sequences have been determined for few phylum members so far, but transcriptome data is available for over 60 nematode species (66, 67). In 2005, McVeigh and co-workers performed a systematic BLAST analysis of EST databases to investigate FLP sequence diversity within the phylum Nematoda identifying more than 500 FLPs across 46 species (13). The FLP complement of other nematodes seems to be generally similar to that of C. elegans (13, 30, 44). This finding is confirmed by a very recent study of McCoy and colleagues, who re-investigated the available genome and transcriptome resources for 17 pathogenic nematodes (24). Many nematode FLPs display a high degree of inter-species structural conservation that is independent of their parasitic or free-living lifestyle [Table 1; Ref. (13)], supporting a fundamental role of FLPs in nematode biology. Corroborating this, only one nematode flp gene is thought to be parasite-specific; the flp-31 gene is absent from the C. elegans genome, but occurs in several plant parasitic nematodes suggesting a function specific in phytoparasitism (13, 24, 44). Although flp-31 was previously predicted from A. suum (32), this gene is considered to be a sequelog of C. elegans flp-15 (24). Initially, flp-29 and flp-30 were also found to be parasite-specific (13), but a recent investigation of their C-terminal motif and genomic location suggests that these genes should be re-designated to respectively flp-28 and flp-2 (24).
Whereas most FLPs are likely widespread throughout the nematode phylum, variable conservation has been reported for some family members. Highly conserved nematode flp-genes include flp-1, flp-6, flp-11, flp-12, flp-14, flp-16, flp-18, flp-19, flp-21, and flp-22; other genes such as flp-2 and flp-10 have shown to be more restricted and structurally diverse (13, 24). Interestingly, parasitic nematodes appear to possess variable proportions of the C. elegans flp-gene complement and variation is highest among distinct clades (24). Genome-wide analysis of the parasite Meloidogyne incognita showed that its FLP complement is reduced to about 60% compared to that of C. elegans (44). Two other nematodes, Trichuris muris and Trichinella spiralis, were shown to display a dramatically reduced complement of only 13%, whereas A. suum possesses 84% of C. elegans flp-genes (24) The finding that fewer flp-genes are expressed in parasitic nematodes as compared to free-living species has been postulated to be an indication of the more contained repertoire of stimuli these nematodes encounter during their endoparasitic stage (68). Furthermore, more FLPs seem to be present in the animal parasitic datasets compared to plant parasitic nematodes (68). Our view on the diversity of FLPs in nematodes however strongly depends on the available sequence data. In depth analyses of the increasing number of completed genome sequences and transcriptome resources should further expand our understanding of the nematode FLP repertoire in the near future.
Recent studies estimate the presence of 32 distinct flp-genes in nematodes (24). Among them are 15 genes that code for N-terminally extended peptides carrying the classical FaRP motif, whereas most others peptides share the restricted RFamide core (Table 1). Although the relatedness of FLPs across metazoans is often unclear, sequences of the neuropeptide F (NPF) family have been identified in several invertebrate groups and predicted in the nematode phylum as well (69). NPF-like peptides are encoded by the flp-27 precursor that is highly conserved in nematodes, and contains the C-terminal RXRFamide motif characteristic of the invertebrate NPF family (70). The plethora of FLPs in nematodes is high, given the structural simplicity of their nervous system harboring around 300 neurons (2, 5, 6). This diversity of neural messengers is magnified by classical neurotransmitters and a broad range of other neuropeptides of insulin (ins) and neuropeptide-like protein (nlp) families, of which about 200 peptides are predicted in C. elegans (46, 71).
Localization of FLPs
Immunocytochemical localization of FLPs has been performed in various nematode species, mainly using antibodies raised against synthetic FMRFamide or the RFamide motif (18, 37, 55). These studies suggest that FLPs are widely expressed in the nervous system of all nematodes, supporting a general role for FLP signaling in nematode biology. The broad distribution of immunoreactive neurons, including in motor neurons, fueled the research effort to decipher nematode FLP signaling and its role in neuromotor function, which had already proven to be a successful target for parasite control (12). Although the vast patterns of FLP immunoreactivity are generally similar between nematode species, HPLC-ELISA studies have identified qualitative differences between free-living and plant parasitic nematodes, suggesting that the distinct peptides present in plant nematodes are structurally different (72, 73).
Despite immunocytochemistry being immensely useful to study gross patterns of FLP localization, most of the C-terminally directed antibodies used were incapable of reliably discriminating between structurally related FLPs. Gene-specific flp expression has been mainly investigated in C. elegans, using reporter transgenes in which LacZ or a fluorescent protein gene is placed under the control of the endogenous promoter region. Li and co-workers applied this molecular approach to map the specific expression patterns of flp-1 to flp-23 genes (22, 23). Just over 50% of the total number of neurons were found to express flp’s, a wide distribution in stark contrast to earlier immunochemical studies in which only 10% of all C. elegans neurons showed FLP reactivity (74). Expression could be detected in all neuronal cell types, including interneurons, sensory neurons, and motor neurons. Six flp-genes were also expressed in non-neuronal cells, including in head muscle (flp-2 and flp-11), pharyngeal muscle (flp-5 and flp-15), socket and/or sheath cells (flp-11 and flp-15), vulval cells (flp-10), and uterine cells (flp-11 and flp-2). Although the expression of each flp gene can be precisely delineated, there is a considerable overlap with many cells expressing more than one flp gene (23). Most flp-genes are also expressed in multiple neurons suggesting that some FLPs have overlapping functions, unlike others fulfilling unique roles.
In situ hybridization (ISH), which uses nucleotide probes complementary to specific gene transcripts, offers an attractive alternative to delineate flp gene expression in other nematodes that are less amenable to transgenesis than C. elegans (38, 42, 54, 75). Furthermore, as immunochemistry allows the determination of neurite morphology facilitating neuronal identification, several antibodies highly specific to certain A. suum FLPs were recently generated (42, 76). Another approach for FLP localization consists of direct mass spectrometric analysis on dissected neuronal tissue without the need of an extensive extraction process (29, 76). Importantly, this technique can identify previously unknown peptides as no prior sequence information is required. Yew et al. (29) subjected individually dissected nerve ganglia from A. suum to mass spectrometric analysis, producing a peptidomic map of the individual anterior ganglions. FLP distribution appeared to be much less restricted to specific cells as compared to the gene expression studies in C. elegans or G. pallida using reporter constructs and ISH, thus supporting the notion that FLP expression differs among nematode species. The authors however also stress the sampling biases inherent in the use of mass spectrometry. In large nematodes such as the foot-long A. suum, cell-specific FLP content can also be rapidly determined by precisely dissecting individual cells (31, 42).
Taken together, abovementioned studies paint a picture in which FLPs widely occur in all known nematode neuronal subtypes and even in non-neuronal tissues. Whereas the gross patterns of FLP distribution remain consistent across the phylum, more recent studies indicate that the cellular expression of homologous FLPs can substantially differ between nematode species (2, 42). This is remarkable, given that both the general FLP complement and the basic nervous architecture are conserved, with C. elegans even considered as a miniature version of A. suum at the level of neuronal morphology (2, 24, 51, 77, 78). Caution is however warranted as dissimilarities could be attributed to experimental differences, with each technique suffering from inherent caveats. Besides the limited specificity of the commonly used antibodies, transgenic reporter constructs may not contain all regulatory sequences necessary to recapitulate endogenous gene expression. Moreover, variability of cellular expression patterns of different gene products has repeatedly been observed in A. suum, partially due to genetic differences since the worms are not isogenic (31, 42, 75, 76).
FLP-Receptors in Nematodes
Most FLPs are known to act through binding of G protein-coupled receptors (GPCRs) (79–81). Although the early work on nematode FLPs primarily focused on peptide identifications, in vitro and functional studies have started to address the biology of their receptors and mode of action. In C. elegans, sequence similarity or homology to the FLP-receptor family has been postulated for several of the more than 100 peptide GPCR genes predicted in the genome (81, 82). The neuropeptide receptor NPR-1 was previously suggested as a member of the invertebrate NPF receptor (NPFR) family and related neuropeptide Y receptors (NPYRs) in mammals (83). Sequence similarity and phylogenetic clustering suggests additional NPFR/NPYR-like family members are likely to be present in C. elegans, as well as representatives related to vertebrate neuropeptide FF receptors, and Drosophila myosuppressin and FMRFamide receptors (8, 82, 84). Peptides that functionally activate these GPCRs, with exception of NPR-1, unfortunately remain unknown.
In general, few nematode FLPs have been matched to their receptor(s) and the identification of FLP-receptor couples has only been undertaken in C. elegans (2, 81). Activation by FLPs has been reported for 13 C. elegans receptors encoded by 10 genes (Table 1), all of which are members of the rhodopsin family of GPCRs [reviewed in Ref. (81)]. Deorphanization, i.e., the identification of receptor ligand(s), is typically done by expressing GPCRs in a heterologous cellular system such as Xenopus oocytes, mammalian cells or yeast. Receptor activation can then be detected by monitoring downstream steps in the GPCR signaling pathway including levels of secondary messenger molecules or GTP exchange upon G protein activation (85). When heterologous GPCRs are challenged with a peptide library, multiple FLPs are generally found to activate a single receptor. Peptide motifs essential for receptor activation are often shared by FLPs derived from the same precursor protein. For example, Kubiak and co-workers showed that all peptides from the FLP-15 precursor carrying the highly similar GPXGPLRFamide motif, recognize the neuropeptide receptor NPR-3 (60). Likewise, two structurally similar FLPs processed from the FLP-2 precursor were found to activate C. elegans receptors encoded by the frpr-18 locus (45). By monitoring intracellular calcium levels, Mertens et al. showed that both FLP-2 peptides activate two isoforms of the receptor FRPR-18 though with different potencies. Whereas SPREPIRFamide (FLP-2A) was active with nanomolar half-maximal effective concentrations (EC50 values), FRPR-18 receptors were only activated at micromolar concentrations by LRGEPIRFamide (FLP-2B). In contrast, Larsen and co-workers found FLP-2A and FLP-2B to be equipotent on the FRPR-18b isoform using a similar calcium mobilization assay in a different type of cells (14). Receptor pharmacology can thus vary dependent on the heterologous system, which may be due to differences in the available G protein signaling machinery or folding properties that affect the functional expression of a GPCR. Although in vitro expression systems may not fully reflect endogenous settings, most ligand-receptor couples identified in C. elegans are supported by functional studies on FLPs and their receptors (61, 62, 81). Functional evidence on peptide GPCRs and putative FLP ligands is also emerging in other nematodes (86, 87), which may serve as a lead in the search for FLP-receptors in these species.
In C. elegans and likely other nematodes, the FLP signaling network is highly expanded by GPCRs able of binding multiple FLPs that, can even originate from different precursor proteins (Table 1). The neuropeptide receptor NPR-1 was the first FLP-receptor to be deorphanized in C. elegans, and shown to recognize both FLP-18 and FLP-21 peptides (61, 63). Interestingly, this GPCR exists in two variants differing by a single amino acid at position 215, NPR-1.215F or NPR-1.215V that is likely implicated in G protein coupling. Substitution of this residue is sufficient for affecting ligand binding and potency resulting in the differential regulation of feeding behavior (61, 63, 83). Both receptor variants are activated by the FLP-21 peptide that is, however, 10-fold more potent in binding NPR-1.215V than NPR-1.215F (61, 63). In addition, Rogers and co-workers found that the NPR-1.215V variant expressed in Xenopus oocytes can be activated by peptides from the FLP-18 precursor, albeit with lower potencies than FLP-21 (61). A second study reported by Kubiak et al. did not identify FLP-18 peptides as the ligands of NPR-1.215V (63), but the receptor variant was expressed in mammalian cells and differences in expression system may account for the discrepancy in identified ligands in both studies. Both FLP-21 and FLP-18 peptides have been found to activate other C. elegans receptors as well, including NPR-11 and NPR-5 (40, 62, 88). In addition, FLP-18 peptides were also identified as ligands of the receptors NPR-4 and NPR-10 (40, 62). An unusual structure-activity relationship has been suggested for the C. elegans receptor FRPR-3 (47, 50). Ligands identified for this GPCR include a FLP-7 (TPMQRSSMVRFamide) and FLP-11 (AMRNALVRFamide) peptide, whereas structurally similar peptides encoded on the same precursor proteins were ineffective at activating the receptor (50). However, it should be noted that EC50 values for both peptides reside in the micromolar range (50), and other functional ligands might activate FRPR-3 with higher potency. FLP-7 and FLP-11 peptides were also shown to activate another receptor, NPR-22, together with an array of FLPs including FLP-1, FLP-9, FLP-13, and FLP-22 peptides (35). The potencies of receptor activation varied from the nanomolar to the micromolar range (Table 1). Finally, the EGL-6 receptor found to be involved in C. elegans egg-laying has been coupled to its FLP-10 and FLP-17 ligands in two ways, by making use of an in vitro assay but also by screening neuropeptide-encoding transgenes for the ability to inhibit egg-laying (53).
Although knowledge has been gathered on the receptor biology of several C. elegans FLPs, our view of nematode FLP-receptors is far from complete. Deorphanization of GPCRs has been successful in matching some FLP-receptor couples; however, often a sub-set of the predicted peptide repertoire is tested such that the array of ligands acting on a receptor remains incomplete. FLPs are thought to exert most of their effects through the activation of GPCRs, but some family members are capable of eliciting fast responses by gating ion channels (89–91). This mode of action likely also applies for several nematode FLPs (90–94). The coupling of multiple peptides to a single GPCR and vice versa greatly enhances the complexity of FLP signaling in C. elegans. However, the characterization of all functional FLP-receptor couples will be crucial to further expand our understanding of nematode FLP signaling, and will uncover whether promiscuity of FLP-receptors can be generalized in nematodes.
FLP-Mediated Modulation of Nematode Physiology and Behavior
Despite the apparent simplicity of the nematode nervous system, harboring around 300 cells, a surprisingly rich behavioral repertoire has been described (86, 87, 95, 96). The structural and spatiotemporal gene expression diversity of the nematode FLP system is reflected in the range of FLP-induced physiological responses. The role of FLPs has been extensively described in previous reviews (2, 5, 6); here, we focus on FLP signaling functions emerging consistently throughout evolution to illustrate some of the general principles of FLP signaling gleaned from the study of nematode peptides. Although C. elegans has been heavily exploited to investigate the basic biology of FLP signaling, we highlight some pharmacological and behavioral studies performed on related nematodes.
Nematode FLPs in the Control of Feeding Behavior
Although FLPs display a tremendous diversity in structure and biological activity, their involvement in the regulation of energy balance and feeding behavior has been described in both invertebrate and vertebrate lineages (11). Feeding state is a paramount environmental factor that guides C. elegans behavior, with a central role for FLP signaling in for instance the regulation of locomotory activity, foraging and food intake (5, 6, 46).
C. elegans NPR-1 signaling regulates food-dependent aggregation behavior
The best characterized example of FLP-modulated behavior in C. elegans is food-related aggregation. Certain wild-type isolates, including the standard laboratory strain N2, mainly show a “solitary feeding” phenotype in which worms disperse to feed alone. Others have a propensity to aggregate into clumps in areas of high food density, a behavior that is termed “social feeding” (83). This behavioral polymorphism can be attributed to a single amino acid difference in the npr-1 gene, which encodes a member of the NPYR/NPFR family (83). Worms expressing the partial loss-of-function isoform with a phenylalanine, NPR-1.215F, are social feeders, whereas strains bearing the npr-1 allele encoding the version with a valine, NPR-1.215V, are solitary. Since chemically generated null mutations of npr-1 convert the solitary wild-type N2 lab strain into an aggregating one, NPR-1 activity is suggested to repress aggregating behavior (83).
Both loss-of-function and gain-of-function studies confirm that FLP-21 acts as the endogenous NPR-1 ligand required for its activation and consequent suppression of food-dependent aggregation (61). Whereas transgenic overexpression of flp-21 rescues the social feeding phenotype of NPR-1.215F worms, genomic deletion of flp-21 further enhances worm clumping. However, loss of flp-21 only slightly increases aggregation in animals bearing the Val-215 allele, suggesting that another ligand most likely encoded by the flp-18 gene may functionally substitute for the loss of FLP-21 ligands (61, 95). FLP-21 furthermore does not appear to act in NPR-1 dependent acute ethanol tolerance, once again suggesting that FLP-18 may be a physiological active ligand (97).
The food-dependent aggregation of social npr-1 mutant worms relies on chemosensory responses in a number of different sensory neurons exposed to the environment and the pseudocoelomic body fluid. Due to their specific localization in C. elegans, these cells are able to detect various adverse or stressful conditions (98, 99). Despite expression of npr-1 in at least 20 neurons (99), the inter/motorneuron RMG seems to be the cellular hub of the NPR-1 mediated feeding behavior (100). Anatomical gap junctions connect RMG to five sensory neurons known to promote aggregation, including the nociceptive ASH and ADL neurons and the URX oxygen sensor (101). In a hub-and-spoke model in which RMG functions as the central hub, RMG is suggested to integrate signals from various sensory neurons to stimulate aggregation using its own chemical synapses. Furthermore, due to the bidirectionality of the gap junctions, RMG in turn modulates the responses of its associated sensory neurons having their own synaptic outputs (100). NPR-1 however inhibits the gap junction driven activation of RMG, either by downregulating the gap junctions or by altering RMG excitability (99, 100).
NPR-1 functions as a modulator of many neurons and behavioral responses, not only in response to food but also to other key environmental parameters of which ambient O2 and CO2 levels appear to have a major influence. NPR-1 modulates aerotaxis as well as the integration of sensory cues of food availability, internal metabolic state and O2 levels (100, 102–106). NPR-1 also alters the sensitivity to environmental repellents (e.g., pheromones, CO2), innate immune responses, and tolerance to ethanol (107–113). The fact that flp-18 and flp-21 mutants often display more subtle phenotypes in these studies indicates that NPR-1 might have additional ligands. Furthermore, the expression patterns of flp-18 and flp-21 have limited overlap and it is not known how the expression of both genes or release of their peptide products is regulated. Despite the wide neuronal expression pattern of npr-1, RMG can be pinpointed as the cellular locus of NPR-1 function for a number of behaviors. Besides “social feeding,” npr-1 expression in RMG acts synergistically with the primary heat sensing machinery to regulate aversive behaviors at high temperature, with npr-1 or flp-21 loss-of-function animals showing an increased threshold for heat avoidance (112). Similarly, RMG-specific rescue of npr-1 restores pheromone avoidance defects in the npr-1 mutant background (114). Furthermore, NPR-1 and its FLP-18 and FLP-21 ligands are required for locomotion quiescence during lethargus, a quiescent behavioral state occurring before each of the four molts, with increased activity in the RMG circuit promoting locomotion arousal (115). The RMG-hub-and-spoke circuit therefore appears to be a multifunctional sensory circuit integrating various stimuli that heavily depends on FLP neuropeptidergic signaling in order to coordinate behavioral output.
Other FLPs modulating food-related behaviors
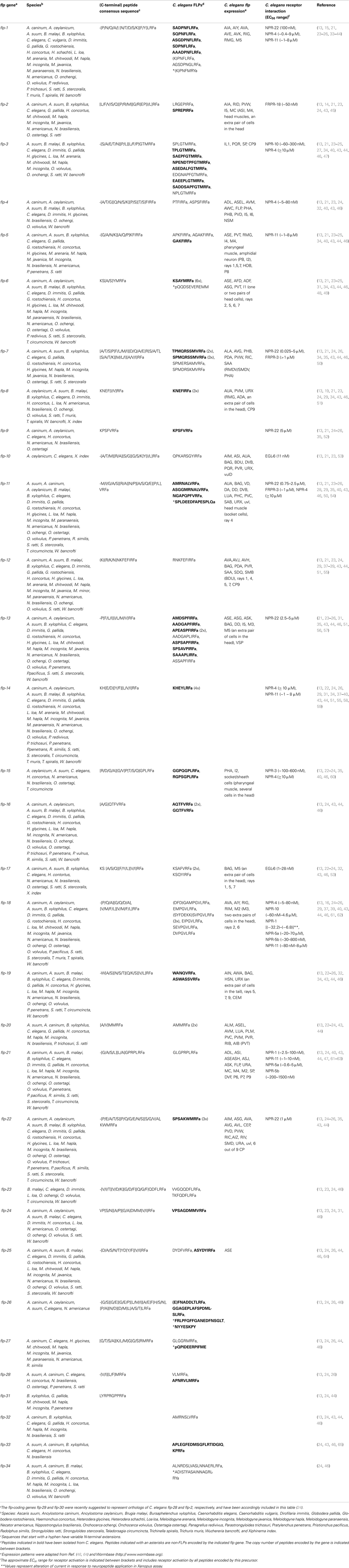
Besides aggregation, FLPs are implicated in other feeding behaviors such as the regulation of energy balance and metabolism according to perceived food availability (11). In C. elegans, loss-of-function of the flp-18 precursor gene causes defects in chemosensation, foraging, and formation of the arrested dauer developmental stage that is induced by stress conditions (62). In addition, these mutants display increased levels in intestinal fat and reduced aerobic metabolism, strongly suggesting that FLP-18 neuropeptides are involved in fat storage and metabolism (62). FLP-18 peptides activate the neuropeptide receptors NPR-4 and NPR-5, and loss-of-function of these GPCRs recapitulates some of the phenotypic effects observed in flp-18 mutants (62, 88). Cohen and co-workers found that npr-4 is expressed in a number of sites including the intestine, whereas NPR-5 is present in several sensory neurons and head, neck, and body wall muscles. FLP-18 signaling through activation of NPR-4 in intestinal muscle was shown to regulate the accumulation of intestinal fat. NPR-5 however modulates the activity of a number of amphid sensory neurons that directly sense environmental cues, of which the chemosensory ASJ neurons are critical in dauer formation (Figure 1A) (116).
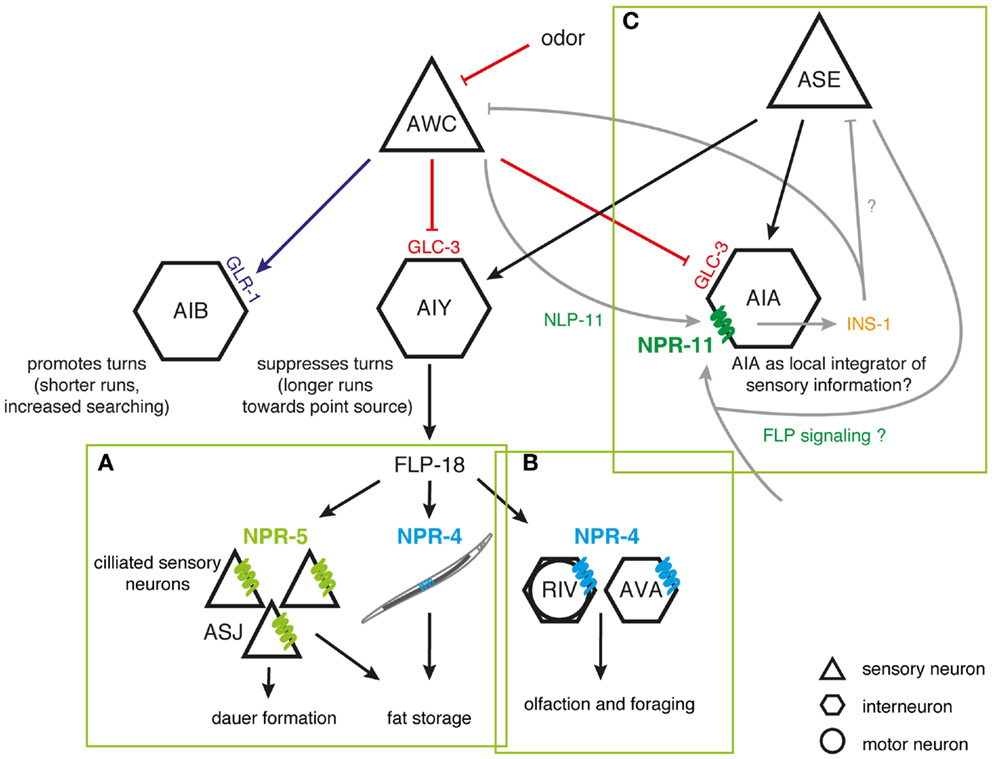
Figure 1. FLP signaling regulates C. elegans foraging and metabolism. (A) FLP-18 peptides are released from AIY in response to sensory cues relaying food availability. By acting on the receptors NPR-4 in the intestine and NPR-5 in ciliated neurons, FLP-18 peptides control fat storage; while activation of NPR-5 in ASJ neurons regulates dauer formation. (B) To regulate odor responses and foraging strategy, FLP-18 peptides signal through NPR-4 in AVA and RIV neurons that control reversal frequency and turning bias, respectively. (C) Peptidergic feedback modulates sensory responses in C. elegans. In response to odor, the AWC olfactory neuron releases NLP-1 neuropeptides, which act on the NPR-11 receptor on AIA to modulate INS-1 peptide secretion. INS-1 subsequently closes the feedback loop by modulating AWC’s responsiveness to sensory stimuli. AIA could act as a local integrator of sensory information, with FLP sensory peptides driving similar neuropeptidergic feedback loops to modulate the responsiveness to sensory stimuli [adapted from Ref. (62, 117–121)].
Environmental food availability also strongly influences C. elegans food-seeking behavior. When feeding on a bacterial lawn, C. elegans spends most of its time slowly moving within a restricted area. Upon removal from their food source, animals however initiate an intensive local search behavior characterized by repetitive bursts of reversing and turning in a restricted immediate area. After prolonged food withdrawal (≥15 min off-food), FLP-18 neuropeptides released from the primary interneuron AIY and to a lesser extent from RIG interneurons, activate a switch in behavioral state from this local search to dispersal in which turning events are suppressed (62, 117). FLP-18 peptides were found to act on the neuropeptide receptor NPR-4 in AVA interneurons and RIV motor neurons that regulate reversal frequency and turning bias, respectively (Figure 1B) (117, 122). NPR-4 signaling by FLP-18 peptides may therefore reduce local search behavior by modulating the activity of these neurons (62). Upstream in the circuit, AIY interneurons, which release FLP-18, receive synaptic input from various sensory neurons. As such, they presumably play an integrative role enabling the regulation of locomotory behaviors in response to environmental perception. Among the presynaptic partners is the AWC olfactory neuron pair that is a prominent player in local search behavior (117). Both neurons are stimulated following the removal of an attractive odorant that serves as a cue for food presence (119). Upon odorant removal, the AWC neurons provide glutamatergic input to downstream interneurons that will accordingly reorient locomotory behavior by stimulating local search behavior. Glutamate release was found to hyperpolarize AIY neurons that express FLP-18 and suppress turning, via the glutamate-gated Cl− channel GLC-3. On the other hand, AIB interneurons that promote turning are depolarized by glutamate via the AMPA/kainate-like glutamate receptor GLR-1, resulting in directed chemotaxis behavior along odor gradients (119, 120, 123). Taken together, these observations fit within a model in which sensory detection of food availability can coordinately regulate adequate responses such as foraging behavior and energy metabolism via FLP signaling (Figure 1).
Interestingly, there is evidence for a neuropeptide-mediated sensorimotor feedback loop that dampens the odor-evoked activity of the AWC neurons, hereby limiting local search behavior (120). When odor is sensed, the AWC neurons release buccalin-related NLP-1 peptides, which in turn act upon the NPR-11 receptor on AIA to modulate INS-1 peptide secretion. Closing the feedback loop, INS-1 acts on the AWC sensory neurons to modulate their responsiveness to sensory stimuli (Figure 1C). Although strong evidence from mutant and other studies demonstrate a functional NLP-1/NPR-11 relationship, other peptides have been shown to activate the receptor with EC50 values in the nanomolar range, including FLP-21 (1–10 nM) and FLP-18 [(SYFDEKK)SVPGVLRFa, 80–800 nM] (40). As mentioned above, the FLP-21 peptide modulates behavior in the context of food and other environmental parameters through activation of the receptor NPR-1, and is expressed in ADL, ASE, and ASH sensory neurons among others (61, 63, 111). ASE neurons in particular are implicated in food-dependent behavior, as they are mainly responsible for chemotaxis to water-soluble attractants (116, 124). Which neurons functionally act downstream of ASE in water-soluble chemotaxis has not been fully understood. AIA interneurons are prominent targets of ASE, hereby hinting on a functional FLP-21/NPR-11 interaction consistent with the observed in vitro data. Although this interaction has not been uncovered in previous studies, it remains interesting to investigate whether NPR-11 signaling in AIA by FLP sensory peptides can activate a neuropeptidergic feedback loop to modulate the gain or temporal properties of the sensory activation process, analogous to that for AWC olfactory neurons (118, 120). AIA could in that respect act as a local integrator of sensory information (Figure 1).
In addition to the regulation of foraging behavior and metabolism, feeding in C. elegans is closely linked to pharyngeal pumping activity (125). Pumping activity is regulated by an intrinsic pharyngeal nervous system (126), but neurohormones released from neurons extrinsic to this cellular system can also influence pumping behavior (127). Several FLPs act on pharyngeal muscle to either excite or inhibit pumping (34, 128, 129). Despite the disadvantage of its size, numerous electrophysiological studies have been performed to reveal the effect of FLPs on pharyngeal preparations in C. elegans. Surprisingly, many of the tested FLPs modulate action potential frequency, suggesting an impressive neurochemical complexity of the feeding circuit (34, 128, 129). Different FLPs have been found to exert opposite effects on action potential frequencies of pharyngeal muscles. Stimulatory peptides include FLPs derived from the flp-5, 6, 8, and 14 precursor genes, whereas others elicit inhibitory effects on serotonin-induced depolarization of pharyngeal muscles like flp-1, 3, 9, 13, and 16 encoded peptides. By using wild-type worms and mutants with deficits in synaptic signaling, it was shown that FLP-13 (APEASPFIRFa) acts directly on the pharyngeal muscle, while FLP-8 acts via the pharyngeal neuronal circuit (34). These results are consistent with the fact that the majority of excitatory and inhibitory peptides were encoded on genes shown to be expressed in the C. elegans pharyngeal nervous system (23). It therefore appears that multiple FLPs are involved in feeding behavior by modulating pharyngeal activity, as supported by findings in A. suum (54, 130–132). Using a modified pressure transducer, Brownlee and colleagues measured changes in intrapharyngeal pressure to monitor the contraction of the Ascaris radial pharynx muscle. PF3 (AF8, KSAYMRFa) causes a biphasic response in the pharynx of A. suum, with hyper-contraction following an initial relaxation. AF1 (KNEFIRFa), however, leaves the muscle in a more relaxed state (130, 132).
Effects of FLPs on locomotion
Besides locomotory activity, the control of the neuromuscular junctions that drive locomotion is certainly also to be considered in the context of nematode feeding, as it enables the worm to migrate toward food sources. Diverse inhibitory and excitatory activities have been reported in A. suum on body wall muscles upon the application of FLPs (54, 91, 133–136). Data obtained from this type of studies do, however, not always facilitate a better understanding of in vivo physiological functions. Although an array of FLP peptides clearly shows muscle-based effects, denervation of somatic muscle strips can alter or even abolish the activity of many FLPs indicating that FLP-receptors do not only reside on muscles (1, 92). Electrophysiological studies indeed demonstrate that FLPs, such as AF1, can act through modulation of neuronal conductance of motor neurons in addition to their muscle-based effects (137). Contrary to AF1, the effects of AF8 (KSAYMRFa) on somatic muscle of A. suum are uniquely differential and context-dependent. While application to dorsal muscles causes slow relaxation, AF8 has profound excitatory effects on ventral muscles (49). Remarkably, this is the only known nematode peptide to show such differential neuromusculatory activity. Stretton et al. have characterized the effects of C. elegans and Ascaris FLPs on the synaptic activity of Ascaris motor neurons (138). They identified five major neuronal response types, theoretically corresponding to at least five FLP-receptor subtypes. These differences might possibly be attributed to different receptors, second messengers, or the combination of both.
In order to understand the in vivo functions of neuropeptides, comprehensive analyses on locomotory behaviors of intact nematodes have been carried out. In A. suum, direct injection of synthetic FLPs into the body cavity elicits diverse behavioral responses including effects on body waveforms, body length, and paralysis (19, 96, 138, 139). Similarly, the normal locomotory behavior is severely disrupted when flp-coding genes are silenced by RNAi, as was shown for flp-14 and flp-32 in G. pallida (87, 140). FLPs also have profound impacts on the migrational abilities of parasitic nematodes toward their host, as illustrated by RNAi silencing of flp-14 and flp-18 in M. incognita (86, 141). Host delivered RNAi of flps as non-chemical based control strategy for parasitic nematodes is therefore gaining importance (142).
When on a solid surface, C. elegans lays on its side and moves in a sinusoidal fashion by undulating contractions and relaxations of dorsal and ventral longitudinal body wall muscles. These muscles use acetylcholine (ACh) and GABA as their primary excitatory and inhibitory neurotransmitters, respectively, and disruption of either of these transmitter biosynthetic pathways leads to severely uncoordinated locomotion (143–145). FLP-1 peptides are also required for the smooth sinusoidal movement of the animals, as inactivation of flp-1 in C. elegans causes hyperactive movement (146). FLP-1 has been found to modulate ACh signaling (147), hereby providing a possible direct link to the regulation of locomotion. FLP-1 as well as FLP-18 peptides were also recently implied in the homeostatic balance of excitation-inhibition coupling in the locomotor circuit that drives body wall muscle contractions (148). This neuropeptide modulation primarily acts on the GABAergic neural transmission at the neuromuscular junctions, where FLP-18 peptides act directly on muscles via the NPR-5 receptor to either inhibit contraction or to promote relaxation. However, the FLPs also appear to have an effect on other cell types to coordinate locomotory output. In addition, Wani and co-workers performed a large-scale RNAi screen to identify genes that mediate endogenous dopamine signaling in C. elegans, an important system controlling worm locomotion (149). The identification of FLP-1 peptides in this study suggests that FLP signaling may be required for dopamine synthesis and release from dopaminergic neurons or for modulating dopamine signaling in dopamine-receptive neurons.
FLP-Coordinated Regulation of Feeding and Nociception
One salient feature of neuropeptide modulation common to both vertebrates as invertebrates is their role in gating and controlling the gain of peripheral sensory inputs (150, 151). In vertebrates, FLP signaling has been repeatedly linked to the modulation of opioid signaling and nociception, whereas the opioid system participates in the regulation of feeding (11, 152, 153). This recurrent interplay makes it conceivable to state that FLP and opioid systems could interact to integrate feeding with stress. Such coordinated regulation would enable animals to decide whether to engage in feeding-related behaviors when presented with an attractive food source in the presence of aversive or noxious stimuli (11). Furthermore, the primary FMRFamide sequence is embedded within an endogenous mammalian opioid peptide derived from the Met-enkephalin precursor, suggesting that enkephalins and FLPs may have coevolved from a common ancestral peptide and share functional links (154). These findings imply that synergistic pathways between stress and feeding behavior might have been evolutionary conserved.
The coordinated regulation of food-dependent behavior (aggregation) and stress perception (nociception) has been thoroughly documented in C. elegans. The manifestation of aggregating behavior involves multiple pathways linking the RMG hub neuron by gap junctions to nociceptive (ASH and ADL), oxygen-sensing (URX), and chemosensory neuron spokes (98–100). Simultaneous ablation of ASH and ADL attenuates aggregation, implying that this behavior may be a response to repulsive or stressful environmental stimuli (98). Aggregation could supply a defense to the animal, with group feeding stimulating dauer formation or prompting the secretion of enzymes that inactivate bacterial toxins (98). The induction of solitary behavior by the FLP receptor NPR-1 hints that its actions may antagonize responses of ASH and ADL to stressful cues. As both neuron types synthesize FLP-21 (61), they are believed to be able to induce solitary behavior under certain conditions. Given that NPR-1 is expressed in ASH nociceptors, it may also directly modulate their sensory responses correlated to feeding state and food availability (99).
Surprisingly, NPR-1 is able to uncouple two overlapping circuits downstream of the ASH nociceptor (151). ASH utilizes glutamatergic synapses to signal to interneurons that control backward locomotion associated with the avoidance response to noxious stimuli (155, 156). In contrast, aggregation is driven by electrical gap junction signaling between ASH and the RMG hub neuron. Neuromodulation of RMG by NPR-1 uncouples the aggregation circuitry thus making it functionally silent, while sparing the function of the ASH-mediated avoidance circuit. This organization allows ASH to differentially generate behaviors depending on the neuromodulatory state, with aggregation occurring only when NPR-1 activity is low, and avoidance occurring regardless of modulation. One attractive hypothesis is that the dynamics of this circuit is differentially regulated by distinct sensory cues. A high intensity aversive cue might trigger the release of FLP-21 peptides and subsequently suppress the aggregation behavior, hereby facilitating efficient escape from highly noxious stimuli without the interference of motor programs for aggregation. The polymodal ASH nociceptor is exquisitely suited to detect various aversive stimuli, but other (FLP releasing) sensory neurons may also impinge on the RMG circuit. The RMG-hub-and-spoke circuit perfectly illustrates how information flow through worm circuits depends on neuromodulatory states defined by neuropeptides (Figure 2). The principle of circuit flexibility relying on connectivity modulation also extends to vertebrates as exemplified by stress-induced analgesia, an acute suppression of pain generated mediated by opioids (151, 157, 158).
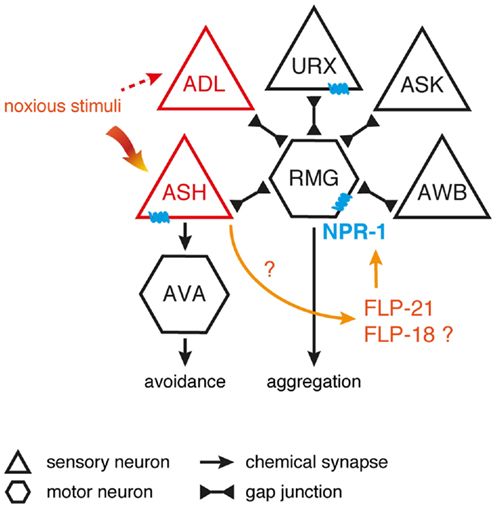
Figure 2. Inhibition of the RMG interneuron by NPR-1. Signaling from ASH and ADL neurons induces aggregation through gap junctions with RMG. RMG is the hub neuron of a gap junction network connecting various sensory neurons known to trigger aggregation. ASH and ADL also mediate acute avoidance behavior through synaptic signaling. Both types of connections are differentially regulated by the NPR-1 receptor, with FLP signaling inhibiting the gap junction driven activation of RMG and not being essential to ASH-mediated avoidance [adapted from Ref. (100,151)].
Nematode Reproduction: FLP Modulation of Egg-Laying and Sexual Behavior
FMRFamide-like peptide signaling modulates nematode reproductive behaviors such as egg-laying and copulation. Neuropeptides encoded by the C. elegans flp-1 gene are suggested to modulate egg-laying rates, since flp-1 deletion mutants show a defect in the timing of these events (159). This FLP-1-dependent regulation is furthermore dependent on food abundance (159). In a genome-wide RNAi study, Keating et al. (160) reported that knockdown of the FLP receptor FRPR-3 increases brood size and the rate of egg-laying (160). Besides genetic studies, FLPs have been directly tested for activity on muscles associated with the female reproductive system (134, 161). When applied to the ovijector of A. suum, for example, AF1 causes a biphasic effect transiently relaxing and then contracting the tissue, whereas both AF2 (KHEYLRFa) and PF3 (AF8, KSAYMRFa) have inhibitory effects (133).
Egg-laying in C. elegans is also modulated by flp-10 and flp-17 encoded peptides (53). These FLPs are able to activate the EGL-6 receptor that is present in the HSN motor neurons innervating the vulval musculature, hence regulating egg-laying behavior (53, 101, 162). In comparison to wild-type C. elegans, egl-6 overexpression and gain-of-function mutants display slower egg-laying rates, suggesting an inhibitory receptor function (53). Both peptides encoded by flp-17 are expressed in a pair of BAG sensory neurons, whereas flp-10 is expressed in several neuronal and non-neuronal tissue. Laser-ablation and overexpression experiments suggest that the vulva and spermatheca are the principal source of the endogenous FLP-10 peptide acting on EGL-6 (23, 53). This leads to a simple model in which relevant sensory cues control FLP-10/FLP-17 secretion, hereby directly modulating the activity of the egg-laying motor neurons to suppress egg-laying in unsuitable environments (Figure 3). Inhibition of the HSN motor neuron by EGL-6 seems to be synergistically to cholinergic inhibition of egg-laying upon unfavorable conditions (53).
Figure 3. FLP signaling suppresses egg-laying in unsuitable environments. BAG neurons release FLP-17 neuropeptides in response to unfavorable conditions. These peptides are able to activate the EGL-6 receptor on HSN motor neurons, hereby inhibiting egg-laying. Release of FLP-10 by the vulva and spermatheca along with subsequent EGL-6 signaling further inhibits egg-laying, with the exact triggering stimuli still uncharacterized. Under unfavorable conditions, cholinergic signals (ACh) may be independently invoked by other sensory circuits to synergistically inhibit egg-laying [adapted from Ref. (53,81)].
Although C. elegans populations almost entirely consist of self-fertilizing hermaphrodites, males arise infrequently under certain environmental conditions. Males strikingly differ from their hermaphrodite counterparts in their complex mating behavior in which males turn backwards along the hermaphrodite body until their tail contacts the vulva, after which copulation is engaged. Peptidergic signaling by FLP-8, FLP-10, FLP-12, and FLP-20 is required for the sensory transduction in male turning behavior (163). Loss-of-function mutations in corresponding genes each induce repeated turning, with males continually circling the hermaphrodite instead of initiating copulation after a single turn. Although these flp-genes are somewhat dispersedly expressed in various sensory neurons and interneurons, flp-20 expression in the mechanosensitive cells completely rescues the mutant’s turning phenotype (163). FLP-20 is therefore hypothesized to convey somatosensory information to terminate the turning program and initiate copulation. How gender-specific modifications of the shared touch circuitry of male and hermaphrodite nervous systems contribute to copulatory behaviors still remains unknown.
FLP Signaling in Learning Behavior
A growing body of evidence, including from studies on mollusks and arthropods, implicates FLPs in the regulation of learning behavior (164–166). C. elegans displays a remarkable level of behavioral plasticity similar to that observed in higher organisms (95, 167, 168), including non-associative (adaptation, habituation) and associative learning behaviors (169). For example, C. elegans can learn to approach or avoid tastes, odors, oxygen, or temperatures that predict the presence or absence of food. Both short-term and long-term forms of memory have been demonstrated in C. elegans (95).
In C. elegans, FLP-20 is involved in tap habituation, a type of non-associative learning behavior (170). C. elegans reverses its locomotion in response to a non-localized mechanical stimulus generated by tapping the culture plate containing the animal, a behavior known as the tap withdrawal response. Repeated taps result in habituation as measured in a decrement of both the amplitude and the frequency of this reversal (171). Mutants for the flp-20 gene show deficits in the relatively short-term 12-h memory following a massed training session. On the other hand, flp-20 is not required for long-term memory of tap habituation that lasts up to 48-h after temporally spaced training in which the same amount of training is presented with interval resting periods (172). This and other studies illustrate how two types of memory within the same learning paradigm are induced by distinct molecular mechanisms that are differentially initiated depending on the temporal pattern of the training regimen. The flp-20 gene is specifically required within the mechanosensory neurons that presumably release FLP-20 peptides to activate downstream neurons required for short-term memory consolidation. This type of memory correlates with a flp-20-dependent increase of synaptic vesicles in the terminals of the mechanosensory neurons (170). This and other studies suggest that the molecular changes underlying short-term memory arise and are maintained at the level of the sensory neurons. Pre-synaptic changes in particular seem indispensable, and likely entail differential release of signaling molecules to dampen the reversal response in the context of tap habituation.
Conclusion
The nematode FLP system comprises an intertwined signaling network with a broad array of neuropeptides operating within an anatomically small nervous system. FLP diversity translates into a central role of this neuropeptide family in various aspects of nematode biology. Functional studies in nematodes support the evolutionary continuity of FLPs as key regulators of energy balance, feeding behavior, reproduction, and sensory modulation. In general, the FLP complement has shown to be widely conserved throughout the phylum though some peptides show a more restricted distribution, with the latter potentially as a consequence of adaptation to a specific lifestyle such as parasitism (2, 24, 42). The particular cellular distribution of FLPs appears not to be fully conserved across nematodes, in contrast to the slow rate at which the nematode nervous system evolves at the cellular level. Rapidly evolving peptide expression could therefore reveal to be an essential factor in the generation of species-specific behavior, furthermore facilitating the radiation of nematodes into a variety of habitats including as parasites of both animals and plants (12).
In C. elegans, the flp-genes have overlapping expression patterns, with at least half of all neurons expressing one or more FLPs (173). This implies that some neurons use a repertoire of FLP peptides in addition to other messengers, which may be deployed in a context-dependent way rendering these cells multifunctional. Such multiplexing could contribute to increase the complexity of information processing in a numerally simple nervous system, hereby supporting the rich behavioral palette of nematodes. Given their broad diversity and expression, neuropeptides are exquisitely suited to actively recruit particular cellular circuits depending on the environmental and internal context. This type of neuromodulation appears to be an irreducible part of circuit flexibility in the nematode nervous system (174).
The considerable amounts of data on nematode FLP function derived from neuronal and neuromuscular bioassays demonstrate an impressive complexity in the FLP signaling system. On the other hand, the knowledge of FLP-receptor interplay remains sparse, and most of our current understanding is derived from C. elegans in which several FLP-receptors have been coupled to their peptide ligands by in vitro assays and in vivo functional studies. A common theme in these studies is that a single receptor can be activated by multiple FLPs encoded by one or more genes. However, this apparent receptor promiscuity will need to be proven physiologically relevant, as a whole layer in the control of FLP signaling may reside in the spatiotemporal expression patterns of both receptor and ligand molecules. With the increasing number of completed genome projects and transcriptome resources, putative FLP-receptors can readily be identified using bioinformatics, and C. elegans data as a scaffold, broadening our view on FLP signaling in other nematodes. In addition, further receptor deorphanization and subsequent localization of these proteins will, together with the extensive data regarding FLP distribution, shed light on specific FLP functioning within the modulation and coordination of nematode behavior and physiology.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge the Research Foundation – Flanders (G0089.12 and G0697.13) and the European Research Council (ERC-2013-ADG-340318) for financial support. J. Watteyne and I. Beets benefit from a PhD and postdoctoral fellowship from the Research Foundation – Flanders, respectively.
References
1. Maule AG, Mousley A, Marks NJ, Day TA, Thompson DP, Geary TG, et al. Neuropeptide signaling systems – potential drug targets for parasite and pest control. Curr Top Med Chem (2002) 2:733–58. doi: 10.2174/1568026023393697
2. McVeigh P, Geary TG, Marks NJ, Maule AG. The FLP-side of nematodes. Trends Parasitol (2006) 22:385–96. doi:10.1016/j.pt.2006.06.010
3. Price DA, Greenberg MJ. Structure of a molluscan cardioexcitatory neuropeptide. Science (1977) 197:670–1. doi:10.1126/science.877582
4. Dockray GJ. The expanding family of RFamide peptides and their effects on feeding behaviour. Exp Physiol (2004) 89:229–35. doi:10.1113/expphysiol.2004.027169
5. Walker R, Papaioannou S, Holden-Dye L. A review of FMRFamide- and RFamide-like peptides in metazoa. Invertebr Neurosci (2009) 9:111–53. doi:10.1007/s10158-010-0097-7
6. Krajniak KG. Invertebrate FMRFamide related peptides. Protein Pept Lett (2013) 20:647–70. doi:10.2174/0929866511320060005
7. Espinoza E, Carrigan M, Thomas S, Shaw G, Edison A. A statistical view of FMRFamide neuropeptide diversity. Mol Neurobiol (2000) 21:35–56. doi:10.1385/MN:21:1-2:035
8. Jékely G. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc Natl Acad Sci U S A (2013) 110:8702–7. doi:10.1073/pnas.1221833110
9. Salzet M, Bulet P, Wattez C, Malecha J. FMRFamide-related peptides in the sex segmental ganglia of the Pharyngobdellid leech Erpobdella octoculata. Eur J Biochem (1994) 221:269–75. doi:10.1111/j.1432-1033.1994.tb18738.x
10. Kriegsfeld LJ. Driving reproduction: RFamide peptides behind the wheel. Horm Behav (2006) 50:655–66. doi:10.1016/j.yhbeh.2006.06.004
11. Bechtold DA, Luckman SM. The role of RFamide peptides in feeding. J Endocrinol (2007) 192:3–15. doi:10.1677/JOE-06-0069
12. Stretton AOW, Cowden C, Sithigorngul P, Davis RE. Neuropeptides in the nematode Ascaris suum. Parasitology (1991) 102:S107–16. doi:10.1017/S0031182000073339
13. McVeigh P, Leech S, Mair GR, Marks NJ, Geary TG, Maule AG. Analysis of FMRFamide-like peptide (FLP) diversity in phylum Nematoda. Int J Parasitol (2005) 35:1043–60. doi:10.1016/j.ijpara.2005.05.010
14. Larsen MJ, Lancheros ER, Williams T, Lowery DE, Geary TG, Kubiak TM. Functional expression and characterization of the C. elegans G-protein-coupled FLP-2 Receptor (T19F4.1) in mammalian cells and yeast. Int J Parasitol Drugs drug Resist (2013) 3:1–7. doi:10.1016/j.ijpddr.2012.10.002
15. Rosoff ML, Burglin TR, Li C. Alternatively spliced transcripts of the flp-1 gene encode distinct FMRFamide-like peptides in Caenorhabditis elegans. J Neurosci (1992) 12:2356–61.
16. Edison AS, Messinger LA, Stretton AO. afp-1: a gene encoding multiple transcripts of a new class of FMRFamide-like neuropeptides in the nematode Ascaris suum. Peptides (1997) 18:929–35. doi:10.1016/S0196-9781(97)00047-8
17. Maule A, Geary T, Bowman J, Shaw C, Falton D, Thompson D. The pharmacology of nematode FMRFamide-related peptides. Parasitol Today (1996) 12:351–7. doi:10.1016/0169-4758(96)10051-X
18. Day TA, Maule AG. Parasitic peptides! The structure and function of neuropeptides in parasitic worms. Peptides (1999) 20:999–1019. doi:10.1016/S0196-9781(99)00093-5
19. Cowden C, Stretton AO, Davis RE. AF1, a sequenced bioactive neuropeptide isolated from the nematode Ascaris suum. Neuron (1989) 2:1465–73. doi:10.1016/0896-6273(89)90192-X
20. The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science (1998) 282:2012–8. doi:10.1126/science.282.5396.2012
21. Nelson LS, Kim K, Memmott JE, Li C. FMRFamide-related gene family in the nematode, Caenorhabditis elegans. Mol Brain Res (1998) 58:103–11. doi:10.1016/S0169-328X(98)00106-5
22. Li C, Kim K, Nelson LS. FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. Brain Res (1999) 848:26–34. doi:10.1016/S0006-8993(99)01972-1
23. Kim K, Li C. Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J Comp Neurol (2004) 475:540–50. doi:10.1002/cne.20189
24. McCoy CJ, Atkinson LE, Zamanian M, McVeigh P, Day TA, Kimber MJ, et al. New insights into the FLPergic complements of parasitic nematodes: informing deorphanisation approaches. EuPA Open Proteomics (2014) 3:262–72. doi:10.1016/j.euprot.2014.04.002
25. Husson SJ, Clynen E, Baggerman G, De Loof A, Schoofs L. Discovering neuropeptides in Caenorhabditis elegans by two dimensional liquid chromatography and mass spectrometry. Biochem Biophys Res Commun (2005) 335:76–86. doi:10.1016/j.bbrc.2005.07.044
26. Husson SJ, Clynen E, Baggerman G, Janssen T, Schoofs L. Defective processing of neuropeptide precursors in Caenorhabditis elegans lacking proprotein convertase 2 (KPC-2/EGL-3): mutant analysis by mass spectrometry. J Neurochem (2006) 98:1999–2012. doi:10.1111/j.1471-4159.2006.04014.x
27. Husson SJ, Janssen T, Baggerman G, Bogert B, Kahn-Kirby AH, Ashrafi K, et al. Impaired processing of FLP and NLP peptides in carboxypeptidase E (EGL-21)-deficient Caenorhabditis elegans as analyzed by mass spectrometry. J Neurochem (2007) 102:246–60. doi:10.1111/j.1471-4159.2007.04474.x
28. Yew JY, Dikler S, Stretton AO. De novo sequencing of novel neuropeptides directly from Ascaris suum tissue using matrix-assisted laser desorption/ionization time-of-flight/time-of-flight. Rapid Commun Mass Spectrom (2003) 17:2693–8. doi:10.1002/rcm.1240
29. Yew JY, Kutz KK, Dikler S, Messinger L, Li L, Stretton AO. Mass spectrometric map of neuropeptide expression in Ascaris suum. J Comp Neurol (2005) 488:396–413. doi:10.1002/cne.20587
30. Husson SJ, Landuyt B, Nys T, Baggerman G, Boonen K, Clynen E, et al. Comparative peptidomics of Caenorhabditis elegans versus C. briggsae by LC-MALDI-TOF MS. Peptides (2009) 30:449–57. doi:10.1016/j.peptides.2008.07.021
31. Jarecki JL, Andersen K, Konop CJ, Knickelbine JJ, Vestling MM, Stretton AO. Mapping neuropeptide expression by mass spectrometry in single dissected identified neurons from the dorsal ganglion of the nematode Ascaris suum. ACS Chem Neurosci (2010) 1:505–19. doi:10.1021/cn1000217
32. Jarecki JL, Frey BL, Smith LM, Stretton AO. Discovery of neuropeptides in the nematode Ascaris suum by database mining and tandem mass spectrometry. J Proteome Res (2011) 10:3098–106. doi:10.1021/pr2001176
33. Rosoff ML, Doble KE, Price DA, Li C. The flp-1 propeptide is processed into multiple, highly similar FMRFamide-like peptides in Caenorhabditis elegans. Peptides (1993) 14:331–8. doi:10.1016/0196-9781(93)90049-M
34. Rogers CM, Franks CJ, Walker RJ, Burke JF, Holden-Dye L. Regulation of the pharynx of Caenorhabditis elegans by 5-HT, octopamine, and FMRFamide-like neuropeptides. J Neurobiol (2001) 49:235–44. doi:10.1002/neu.1078
35. Mertens I, Clinckspoor I, Janssen T, Nachman R, Schoofs L. FMRFamide related peptide ligands activate the Caenorhabditis elegans orphan GPCR Y59H11AL.1. Peptides (2006) 27:1291–6. doi:10.1016/j.peptides.2005.11.017
36. Schinkmann K, Li C. Comparison of two Caenorhabditis genes encoding FMRFamide(Phe-Met-Arg-Phe-NH2)-like peptides. Mol Brain Res (1994) 24:238–46. doi:10.1016/0169-328X(94)90137-6
37. Kimber MJ, Fleming CC, Bjourson AJ, Halton DW, Maule AG. FMRFamide-related peptides in potato cyst nematodes. Mol Biochem Parasitol (2001) 116:199–208. doi:10.1016/S0166-6851(01)00323-1
38. Kimber MJ, Fleming CC, Prior A, Jones JT, Halton DW, Maule AG. Localisation of Globodera pallida FMRFamide-related peptide encoding genes using in situ hybridisation. Int J Parasitol (2002) 32:1095–105. doi:10.1016/S0020-7519(02)00084-X
39. Kimber MJ, Fleming CC. Neuromuscular function in plant parasitic nematodes: a target for novel control strategies? Parasitology (2005) 131:S129–42. doi:10.1017/S0031182005009157
40. Lowery DE, Geary TG, Kubiak TM, Larsen MJ. G Protein-Coupled Receptor-like Receptors and Modulators Thereof United States, Patent 6,632,621. (2003). Available at: http://www.google.com/patents/EP1238076A2?cl=nl
41. Geary TG, Price DA, Bowman JW, Winterrowd CA, Mackenzie CD, Garrison RD, et al. Two FMRFamide-like peptides from the free-living nematode Panagrellus redivivus. Peptides (1992) 13:209–14. doi:10.1016/0196-9781(92)90098-N
42. Jarecki JL, Viola IR, Andersen KM, Miller AH, Ramaker MA, Vestling MM, et al. Three independent techniques localize expression of transcript afp-11 and its bioactive peptide products to the paired AVK neurons in Ascaris suum: in situ hybridization, immunocytochemistry, and single cell mass spectrometry. ACS Chem Neurosci (2013) 4:418–34. doi:10.1021/cn3001334
43. Kikuchi T, Cotton JA, Dalzell JJ, Hasegawa K, Kanzaki N, McVeigh P, et al. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog (2011) 7:e1002219. doi:10.1371/journal.ppat.1002219
44. Abad P, Gouzy J, Aury J-M, Castagnone-Sereno P, Danchin EGJ, Deleury E, et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol (2008) 26:909–15. doi:10.1038/nbt.1482
45. Mertens I, Meeusen T, Janssen T, Nachman R, Schoofs L. Molecular characterization of two G protein-coupled receptor splice variants as FLP2 receptors in Caenorhabditis elegans. Biochem Biophys Res Commun (2005) 330:967–74. doi:10.1016/j.bbrc.2005.03.071
46. Li C, Kim K. Neuropeptide gene families in Caenorhabditis elegans. Adv Exp Med Biol (2010) 692:98–137. doi:10.1007/978-1-4419-6902-6_6
47. Greenwood K, Williams T, Geary T. Nematode neuropeptide receptors and their development as anthelmintic screens. Parasitology (2005) 131:S169–77. doi:10.1017/S003118200500819X
48. Marks NJ, Maule AG, Geary TG, Thompson DP, Li C, Halton DW, et al. KSAYMRFamide (PF3/AF8) is present in the free-living nematode, Caenorhabditis elegans. Biochem Biophys Res Commun (1998) 248:422–5. doi:10.1006/bbrc.1998.8982
49. Maule AG, Shaw C, Bowman JW, Halton DW, Thompson DP, Geary TG, et al. KSAYMRFamide: a novel FMRamide-related heptapeptide from the free-living nematode, Panagrellus redivivus, which is myoactive in the parasitic nematode, Ascaris suum. Biochem Biophys Res Commun (1994) 200:973–80. doi:10.1006/bbrc.1994.1545
50. Mertens I, Vandingenen A, Meeusen T, Janssen T, Luyten W, Nachman RJ, et al. Functional characterization of the putative orphan neuropeptide G-protein coupled receptor C26F1.6 in Caenorhabditis elegans. FEBS Lett (2004) 573:55–60. doi:10.1016/j.febslet.2004.07.058
51. Davis RE, Stretton AOW. The motornervous system of Ascaris: electrophysiology and anatomy of the neurons and their control by neuromodulators. Parasitology (1996) 113:S97–117. doi:10.1017/S0031182000077921
52. Marks NJ, Maule AG, Li C, Nelson LS, Thompson DP, Alexander-Bowman S, et al. Isolation, pharmacology and gene organization of KPSFVRFamide: a neuropeptide from Caenorhabditis elegans. Biochem Biophys Res Commun (1999) 254:222–30. doi:10.1006/bbrc.1998.9920
53. Ringstad N, Horvitz HR. FMRFamide neuropeptides and acetylcholine synergistically inhibit egg-laying by C. elegans. Nat Neurosci (2008) 11:1168–76. doi:10.1038/nn.2186
54. Yew JY, Davis R, Dikler S, Nanda J, Reinders B, Stretton AO. Peptide products of the afp-6 gene of the nematode Ascaris suum have different biological actions. J Comp Neurol (2007) 502:872–82. doi:10.1002/cne.21357
55. Johnston MJG, McVeigh P, McMaster S, Fleming CC, Maule AG. FMRFamide-like peptides in root knot nematodes and their potential role in nematode physiology. J Helminthol (2010) 84:253–65. doi:10.1017/S0022149X09990630
56. Marks NJ, Maule AG, Geary TG, Thompson DP, Davis JP, Halton DW, et al. APEASPFIRFamide, a novel FMRFamide-related decapeptide from Caenorhabditis elegans: structure and myoactivity. Biochem Biophys Res Commun (1997) 231:591–5. doi:10.1006/bbrc.1997.6155
57. Marks NJ, Shaw C, Halton DW, Thompson DP, Geary TG, Li C, et al. Isolation and preliminary biological assessment of AADGAPLIRFamide and SVPGVLRFamide from Caenorhabditis elegans. Biochem Biophys Res Commun (2001) 286:1170–6. doi:10.1006/bbrc.2001.5524
58. Cowden C, Stretton AO. AF2, an Ascaris neuropeptide: isolation, sequence, and bioactivity. Peptides (1993) 14:423–30. doi:10.1016/0196-9781(93)90127-3
59. Maule AG, Shaw C, Bowman JW, Halton DW, Thompson DP, Geary TG, et al. The FMRFamide-like neuropeptide AF2 (Ascaris suum) is present in the free-living nematode, Panagrellus redivivus (Nematoda, Rhabditida). Parasitology (1994) 109:351–6. doi:10.1017/S0031182000078380
60. Kubiak TM, Larsen MJ, Zantello MR, Bowman JW, Nulf SC, Lowery DE. Functional annotation of the putative orphan Caenorhabditis elegans G-protein-coupled receptor C10C6.2 as a FLP15 peptide receptor. J Biol Chem (2003) 278:42115–20. doi:10.1074/jbc.M304056200
61. Rogers C, Reale V, Kim K, Chatwin H, Li C, Evans P, et al. Inhibition of Caenorhabditis elegans social feeding by FMRFamide-related peptide activation of NPR-1. Nat Neurosci (2003) 6:1178–85. doi:10.1038/nn1140
62. Cohen M, Reale V, Olofsson B, Knights A, Evans P, de Bono M. Coordinated regulation of foraging and metabolism in C. elegans by RFamide neuropeptide signaling. Cell Metab (2009) 9:375–85. doi:10.1016/j.cmet.2009.02.003
63. Kubiak TM, Larsen MJ, Nulf SC, Zantello MR, Burton KJ, Bowman JW, et al. Differential activation of “social” and “solitary” variants of the Caenorhabditis elegans G protein-coupled receptor NPR-1 by its cognate ligand AF9. J Biol Chem (2003) 278:33724–9. doi:10.1074/jbc.M304861200
64. Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, Marra MA, et al. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev (2007) 21:1653–74. doi:10.1101/gad.1560107
65. Husson SJ, Mertens I, Janssen T, Lindemans M, Schoofs L. Neuropeptidergic signaling in the nematode Caenorhabditis elegans. Prog Neurobiol (2007) 82:33–55. doi:10.1016/j.pneurobio.2007.01.006
66. Elsworth B, Wasmuth J, Blaxter M. NEMBASE4: the nematode transcriptome resource. Int J Parasitol (2011) 41:881–94. doi:10.1016/j.ijpara.2011.03.009
67. Kumar S, Koutsovoulos G, Kaur G, Blaxter M. Toward 959 nematode genomes. Worm (2012) 1:42–50. doi:10.4161/worm.19046
68. Maule A, Curtis R. Parallels between plant and animal parasitic nematodes. In: Jones J, Gheysen G, Fenoll C, editors. Genomics and Molecular Genetics of Plant-Nematode Interactions SE – 11. Dordrecht: Springer (2011). p. 221–51.
69. Nässel DR, Wegener C. A comparative review of short and long neuropeptide F signaling in invertebrates: any similarities to vertebrate neuropeptide Y signaling? Peptides (2011) 32:1335–55. doi:10.1016/j.peptides.2011.03.013
70. Clynen E, Husson SJ, Schoofs L. Identification of new members of the (short) neuropeptide F family in Locusts and Caenorhabditis elegans. Ann N Y Acad Sci (2009) 1163:60–74. doi:10.1111/j.1749-6632.2008.03624.x
71. Bargmann CI, Kaplan JM. Signal transduction in the Caenorhabditis elegans nervous system. Annu Rev Neurosci (1998) 21:279–308. doi:10.1146/annurev.neuro.21.1.279
72. Atkinson HJ, Isaac RE, Harris PD, Sharpe CM. FMRFamide-like immunoreactivity within the nervous system of the nematodes Panagrellus redivius, Caenorhabditis elegans and Heterodera glycines. J Zool (1988) 216:663–71. doi:10.1111/j.1469-7998.1988.tb02464.x
73. Masler EP, Kovaleva ES, Sardanelli S. Comparison of FaRP immunoreactivity in free-living nematodes and in the plant-parasitic nematode Heterodera glycines. Ann N Y Acad Sci (1999) 897:253–63. doi:10.1111/j.1749-6632.1999.tb07896.x
74. Schinkmann K, Li C. Localization of FMRF amide-like peptides in Caenorhabditis elegans. J Comp Neurol (1992) 316:251–60. doi:10.1002/cne.903160209
75. Nanda JC, Stretton AOW. In situ hybridization of neuropeptide-encoding transcripts afp-1, afp-3, and afp-4 in neurons of the nematode Ascaris suum. J Comp Neurol (2010) 518:896–910. doi:10.1002/cne.22251
76. Sithigorngul P, Jarecki JL, Stretton AOW. A specific antibody to neuropeptide AF1 (KNEFIRFamide) recognizes a small subset of neurons in Ascaris suum: differences from Caenorhabditis elegans. J Comp Neurol (2011) 519:1546–61. doi:10.1002/cne.22584
77. Stretton AOW, Davis RE, Angstadt JD, Donmoyer JE, Johnson CD. Neural control of behaviour in Ascaris. Trends Neurosci (1985) 8:294–300. doi:10.1016/0166-2236(85)90105-5
78. Angstadt JD, Donmoyer JE, Stretton AO. Retrovesicular ganglion of the nematode Ascaris. J Comp Neurol (1989) 284:374–88. doi:10.1002/cne.902840305
79. Gilchrist A. Modulating G-protein-coupled receptors: from traditional pharmacology to allosterics. Trends Pharmacol Sci (2007) 28:431–7. doi:10.1016/j.tips.2007.06.012
80. Bridges TM, Lindsley CW. G-protein coupled receptors: from classical modes of modulation to allosteric mechanisms. ACS Chem Biol (2008) 3:530–41. doi:10.1021/cb800116f
81. Frooninckx L, Van Rompay L, Temmerman L, Van Sinay E, Beets I, Janssen T, et al. Neuropeptide GPCRs in C. elegans. Front Endocrinol (2012) 3:167. doi:10.3389/fendo.2012.00167
82. Hobert O. The Neuronal Genome of Caenorhabditis elegans. In: WormBook. The C. elegans Research Community editor. WormBook (2013). doi:10.1895/wormbook.1.75.1
83. de Bono M, Bargmann CI. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell (1998) 94:679–89. doi:10.1016/S0092-8674(00)81609-8
84. Mirabeau O, Joly J-S. Molecular evolution of peptidergic signaling systems in bilaterians. Proc Natl Acad Sci U S A (2013) 110:E2028–37. doi:10.1073/pnas.1219956110
85. Mertens I, Vandingenen A, Meeusen T, De Loof A, Schoofs L. Postgenomic characterization of G-protein-coupled receptors. Pharmacogenomics (2004) 5:657–72. doi:10.1517/14622416.5.6.657
86. Dalzell JJ, McMaster S, Fleming CC, Maule AG. Short interfering RNA-mediated gene silencing in Globodera pallida and Meloidogyne incognita infective stage juveniles. Int J Parasitol (2010) 40:91–100. doi:10.1016/j.ijpara.2009.07.003
87. Atkinson LE, Stevenson M, McCoy CJ, Marks NJ, Fleming C, Zamanian M, et al. flp-32 ligand/receptor silencing phenocopy faster plant pathogenic nematodes. PLoS Pathog (2013) 9:e1003169. doi:10.1371/journal.ppat.1003169
88. Kubiak TM, Larsen MJ, Bowman JW, Geary TG, Lowery DE. FMRFamide-like peptides encoded on the flp-18 precursor gene activate two isoforms of the orphan Caenorhabditis elegans G-protein-coupled receptor Y58G8A.4 heterologously expressed in mammalian cells. Biopolymers (2008) 90:339–48. doi:10.1002/bip.20850
89. Lingueglia E, Champigny G, Lazdunski M, Barbry P. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature (1995) 378:730–3. doi:10.1038/378730a0
90. Purcell J, Robertson AP, Thompson DP, Martin RJ. PF4, a FMRFamide-related peptide, gates low-conductance Cl- channels in Ascaris suum. Eur J Pharmacol (2002) 456:11–7. doi:10.1016/S0014-2999(02)02622-5
91. Bowman JW, Friedman AR, Thompson DP, Maule AG, Alexander-Bowman SJ, Geary TG. Structure–activity relationships of an inhibitory nematode FMRFamide-related peptide, SDPNFLRFamide (PF1), on Ascaris suum muscle. Int J Parasitol (2002) 32:1765–71. doi:10.1016/S0020-7519(02)00213-8
92. Maule AG, Geary TG, Bowman JW, Marks NJ, Blair KL, Halton DW, et al. Inhibitory effects of nematode FMRFamide-related peptides (FaRPs) on muscle strips from Ascaris suum. Invert Neurosci (1995) 1:255–65. doi:10.1007/BF02211027
93. Bowman JW, Friedman AR, Thompson DP, Ichhpurani AK, Kellman MF, Marks N, et al. Structure-activity relationships of KNEFIRFamide (AF1), a nematode FMRFamide-related peptide, on Ascaris suum muscle. Peptides (1996) 17:381–7. doi:10.1016/0196-9781(96)00007-1
94. Holden-Dye L, Brownlee DJ, Walker RJ. The effects of the peptide KPNFIRFamide (PF4) on the somatic muscle cells of the parasitic nematode Ascaris suum. Br J Pharmacol (1997) 120:379–86. doi:10.1038/sj.bjp.0700906
95. de Bono M, Maricq AV. Neuronal substrates of complex behaviors in C. elegans. Annu Rev Neurosci (2005) 28:451–501. doi:10.1146/annurev.neuro.27.070203.144259
96. Reinitz CA, Pleva AE, Stretton AOW. Changes in cyclic nucleotides, locomotory behavior, and body length produced by novel endogenous neuropeptides in the parasitic nematode Ascaris suum. Mol Biochem Parasitol (2011) 180:27–34. doi:10.1016/j.molbiopara.2011.08.001
97. Bendena WG, Campbell J, Zara L, Tobe SS, Chin-Sang ID. Select neuropeptides and their G-protein coupled receptors in Caenorhabditis elegans and Drosophila melanogaster. Front Endocrinol (2012) 3:93. doi:10.3389/fendo.2012.00093
98. de Bono M, Tobin DM, Davis MW, Avery L, Bargmann C. Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature (2002) 419:899–903. doi:10.1038/nature01169
99. Coates JC, de Bono M. Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans. Nature (2002) 419:925–9. doi:10.1038/nature01170
100. Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RA, Clardy J, et al. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature (2009) 458:1171–5. doi:10.1038/nature07886
101. White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc B Biol Sci (1986) 314:1–340. doi:10.1098/rstb.1986.0056
102. Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature (2004) 430:317–22. doi:10.1038/nature02714
103. Cheung BHH, Cohen M, Rogers C, Albayram O, de Bono M. Experience-dependent modulation of C. elegans behavior by ambient oxygen. Curr Biol (2005) 15:905–17. doi:10.1016/j.cub.2005.04.017
104. Chang AJ, Chronis N, Karow DS, Marletta MA, Bargmann CI. A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biol (2006) 4:e274. doi:10.1371/journal.pbio.0040274
105. Rogers C, Persson A, Cheung B, de Bono M. Behavioral motifs and neural pathways coordinating O2 responses and aggregation in C. elegans. Curr Biol (2006) 16:649–59. doi:10.1016/j.cub.2006.03.023
106. Chang AJ, Bargmann CI. Hypoxia and the HIF-1 transcriptional pathway reorganize a neuronal circuit for oxygen-dependent behavior in Caenorhabditis elegans. Proc Natl Acad Sci U S A (2008) 105:7321–6. doi:10.1073/pnas.0802164105
107. Davies AG, Bettinger JC, Thiele TR, Judy ME, McIntire SL. Natural variation in the npr-1 gene modifies ethanol responses of wild strains of C. elegans. Neuron (2004) 42:731–43. doi:10.1016/j.neuron.2004.05.004
108. Hallem EA, Sternberg PW. Acute carbon dioxide avoidance in Caenorhabditis elegans. Proc Natl Acad Sci U S A (2008) 105:8038–43. doi:10.1073/pnas.0707469105
109. Styer KL, Singh V, Macosko E, Steele SE, Bargmann CI, Aballay A. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science (2008) 322:460–4. doi:10.1126/science.1163673
110. Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science (2009) 323:382–4. doi:10.1126/science.1166527
111. Pocock R, Hobert O. Hypoxia activates a latent circuit for processing gustatory information in C. elegans. Nat Neurosci (2010) 13:610–4. doi:10.1038/nn.2537
112. Glauser DA, Chen WC, Agin R, Macinnis BL, Hellman AB, Garrity PA, et al. Heat avoidance is regulated by transient receptor potential (TRP) channels and a neuropeptide signaling pathway in Caenorhabditis elegans. Genetics (2011) 188:91–103. doi:10.1534/genetics.111.127100
113. Carrillo MA, Guillermin ML, Rengarajan S, Okubo RP, Hallem EA. O2-sensing neurons control CO2 response in C. elegans. J Neurosci (2013) 33:9675–83. doi:10.1523/JNEUROSCI.4541-12.2013
114. Jang H, Kim K, Neal SJ, Macosko E, Kim D, Butcher RA, et al. Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. elegans. Neuron (2012) 75:585–92. doi:10.1016/j.neuron.2012.06.034
115. Choi S, Chatzigeorgiou M, Taylor KP, Schafer WR, Kaplan JM. Analysis of NPR-1 reveals a circuit mechanism for behavioral quiescence in C. elegans. Neuron (2013) 78:869–80. doi:10.1016/j.neuron.2013.04.002
116. Bargmann CI, Horvitz HR. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science (1991) 251:1243–6. doi:10.1126/science.2006412
117. Gray JM, Hill JJ, Bargmann C. A circuit for navigation in Caenorhabditis elegans. Proc Natl Acad Sci U S A (2005) 102:3184–91. doi:10.1073/pnas.0409009101
118. Tomioka M, Adachi T, Suzuki H, Kunitomo H, Schafer WR, Iino Y. The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron (2006) 51:613–25. doi:10.1016/j.neuron.2006.07.024
119. Chalasani SH, Chronis N, Tsunozaki M, Gray JM, Ramot D, Goodman M, et al. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature (2007) 450:63–70. doi:10.1038/nature06292
120. Chalasani SH, Kato S, Albrecht DR, Nakagawa T, Abbott LF, Bargmann C. Neuropeptide feedback modifies odor-evoked dynamics in Caenorhabditis elegans olfactory neurons. Nat Neurosci (2010) 13:615–21. doi:10.1038/nn.2526
121. Hart A, Chao MY. From Odors to Behaviors in Caenorhabditis elegans. The Neurobiology of Olfaction. Boca Raton, FL: CRC Press (2010).
122. Chalfie M, Sulston J, White JG, Southgate E, Thomson JN, Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J Neurosci (1985) 5:956–64.
123. Horoszok L, Raymond V, Sattelle DB, Wolstenholme AJ. GLC-3: a novel fipronil and BIDN-sensitive, but picrotoxinin-insensitive, L-glutamate-gated chloride channel subunit from Caenorhabditis elegans. Br J Pharmacol (2001) 132:1247–54. doi:10.1038/sj.bjp.0703937
124. Kaufman A, Keinan A, Meilijson I, Kupiec M, Ruppin E. Quantitative analysis of genetic and neuronal multi-perturbation experiments. PLoS Comput Biol (2005) 1:e64. doi:10.1371/journal.pcbi.0010064
126. Albertson DG, Thomson JN. The pharynx of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci (1976) 275:299–325. doi:10.1098/rstb.1976.0085
127. Nass R, Merchant KM, Ryan T. Caenohabditis elegans in Parkinson’s disease drug discovery: addressing an unmet medical need. Mol Interv (2008) 8:284–93. doi:10.1124/mi.8.6.6
128. Papaioannou S, Marsden D, Franks CJ, Walker RJ, Holden-Dye L. Role of a FMRFamide-like family of neuropeptides in the pharyngeal nervous system of Caenorhabditis elegans. J Neurobiol (2005) 65:304–19. doi:10.1002/neu.20201
129. Papaioannou S, Holden-Dye L, Walker RJ. Evidence for a role for cyclic AMP in modulating the action of 5-HT and an excitatory neuropeptide, FLP17A, in the pharyngeal muscle of Caenorhabditis elegans. Invert Neurosci (2008) 8:91–100. doi:10.1007/s10158-008-0072-8
130. Brownlee DJA, Holden-Dye L, Fairweather I, Walker RJ. The action of serotonin and the nematode neuropeptide KSAYMRFamide on the pharyngeal muscle of the parasitic nematode, Ascaris suum. Parasitology (1995) 111:379–84. doi:10.1017/S0031182000081932
131. Brownlee DJ, Holden-Dye L, Walker RJ, Fairweather I. The pharynx of the nematode Ascaris suum: structure and function. Acta Biol Hung (1995) 46:195–204.
132. Brownlee DJ, Walker RJ. Actions of nematode FMRFamide-related peptides on the pharyngeal muscle of the parasitic nematode, Ascaris suum. Ann N Y Acad Sci (1999) 897:228–38. doi:10.1111/j.1749-6632.1999.tb07894.x
133. Fellowes RA, Maule AG, Marks NJ, Geary TG, Thompson DP, Shaw C, et al. Modulation of the motility of the vagina vera of Ascaris suum in vitro by FMRF amide-related peptides. Parasitology (1998) 116(Pt 3):277–87. doi:10.1017/S0031182097002229
134. Moffett CL, Beckett AM, Mousley A, Geary TG, Marks NJ, Halton DW, et al. The ovijector of Ascaris suum: multiple response types revealed by Caenorhabditis elegans FMRFamide-related peptides. Int J Parasitol (2003) 33:859–76. doi:10.1016/S0020-7519(03)00109-7
135. Trailovic SM, Clark CL, Robertson AP, Martin RJ. Brief application of AF2 produces long lasting potentiation of nAChR responses in Ascaris suum. Mol Biochem Parasitol (2005) 139:51–64. doi:10.1016/j.molbiopara.2004.10.001
136. Verma S, Robertson AP, Martin RJ. The nematode neuropeptide, AF2 (KHEYLRF-NH2), increases voltage-activated calcium currents in Ascaris suum muscle. Br J Pharmacol (2007) 151:888–99. doi:10.1038/sj.bjp.0707296
137. Thompson DP, Davis JP, Larsen MJ, Coscarelli EM, Zinser EW, Bowman JW, et al. Effects of KHEYLRFamide and KNEFIRFamide on cyclic adenosine monophosphate levels in Ascaris suum somatic muscle. Int J Parasitol (2003) 33:199–208. doi:10.1016/S0020-7519(02)00259-X
138. Davis RE, Stretton AO. Structure-activity relationships of 18 endogenous neuropeptides on the motor nervous system of the nematode Ascaris suum. Peptides (2001) 22:7–23. doi:10.1016/S0196-9781(00)00351-X
139. Reinitz CA, Herfel HG, Messinger LA, Stretton AOW. Changes in locomotory behavior and cAMP produced in Ascaris suum by neuropeptides from Ascaris suum or Caenorhabditis elegans. Mol Biochem Parasitol (2000) 111:185–97. doi:10.1016/S0166-6851(00)00317-0
140. Kimber MJ, McKinney S, McMaster S, Day TA, Fleming CC, Maule AG. flp gene disruption in a parasitic nematode reveals motor dysfunction and unusual neuronal sensitivity to RNA interference. FASEB J (2007) 21:1233–43. doi:10.1096/fj.06-7343com
141. Dalzell JJ, McMaster S, Johnston MJ, Kerr R, Fleming CC, Maule AG. Non-nematode-derived double-stranded RNAs induce profound phenotypic changes in Meloidogyne incognita and Globodera pallida infective juveniles. Int J Parasitol (2009) 39:1503–16. doi:10.1016/j.ijpara.2009.05.006
142. Papolu PK, Gantasala NP, Kamaraju D, Banakar P, Sreevathsa R, Rao U. Utility of host delivered RNAi of two FMRF amide like peptides, flp-14 and flp-18, for the management of root knot nematode, Meloidogyne incognita. PLoS One (2013) 8:e80603. doi:10.1371/journal.pone.0080603
144. Alfonso A, Grundahl K, Duerr JS, Han HP, Rand JB. The Caenorhabditis elegans unc-17 gene: a putative vesicular acetylcholine transporter. Science (1993) 261:617–9. doi:10.1126/science.8342028
145. McIntire SL, Jorgensen E, Kaplan J, Horvitz HR. The GABAergic nervous system of Caenorhabditis elegans. Nature (1993) 364:337–41. doi:10.1038/364337a0
146. Nelson LS, Rosoff ML, Li C. Disruption of a neuropeptide gene, flp-1, causes multiple behavioral defects in Caenorhabditis elegans. Science (1998) 281:1686–90. doi:10.1126/science.281.5383.1686
147. Sieburth D, Ch’ng Q, Dybbs M, Tavazoie M, Kennedy S, Wang D, et al. Systematic analysis of genes required for synapse structure and function. Nature (2005) 436:510–7. doi:10.1038/nature03809
148. Stawicki TM, Takayanagi-Kiya S, Zhou K, Jin Y. Neuropeptides function in a homeostatic manner to modulate excitation-inhibition imbalance in C. elegans. PLoS Genet (2013) 9:e1003472. doi:10.1371/journal.pgen.1003472
149. Wani KA, Catanese M, Normantowicz R, Herd M, Maher KN, Chase DL. D1 dopamine receptor signaling is modulated by the R7 RGS protein EAT-16 and the R7 binding protein RSBP-1 in Caenorhabditis elegans motor neurons. PLoS One (2012) 7:e37831. doi:10.1371/journal.pone.0037831
150. Taghert PH, Nitabach MN. Peptide neuromodulation in invertebrate model systems. Neuron (2012) 76:82–97. doi:10.1016/j.neuron.2012.08.035
151. Bargmann CI. Beyond the connectome: how neuromodulators shape neural circuits. Bioessays (2012) 34:458–65. doi:10.1002/bies.201100185
152. Yang H-YT, Tao T, Iadarola MJ. Modulatory role of neuropeptide FF system in nociception and opiate analgesia. Neuropeptides (2008) 42:1–18. doi:10.1016/j.npep.2007.06.004
153. Feng Y, He X, Yang Y, Chao D, Lazarus LH, Xia Y. Current research on opioid receptor function. Curr Drug Targets (2012) 13:230–46. doi:10.2174/138945012799201612
154. Greenberg MJ, Painter SD, Doble KE, Nagle GT, Price DA, Lehman HK. The molluscan neurosecretory peptide FMRFamide: comparative pharmacology and relationship to the enkephalins. Fed Proc (1983) 42:82–6.
155. Hart A, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature (1995) 378:82–5. doi:10.1038/378082a0
156. Chao MY, Komatsu H, Fukuto HS, Dionne HM, Hart A. Feeding status and serotonin rapidly and reversibly modulate a Caenorhabditis elegans chemosensory circuit. Proc Natl Acad Sci U S A (2004) 101:15512–7. doi:10.1073/pnas.0403369101
157. Akil H, Watson SJ, Young E, Lewis ME, Khachaturian H, Walker JM. Endogenous opioids: biology and function. Annu Rev Neurosci (1984) 7:223–55. doi:10.1146/annurev.ne.07.030184.001255
158. Fields H. State-dependent opioid control of pain. Nat Rev Neurosci (2004) 5:565–75. doi:10.1038/nrn1431
159. Waggoner LE, Hardaker LA, Golik S, Schafer WR. Effect of a neuropeptide gene on behavioral states in Caenorhabditis elegans egg-laying. Genetics (2000) 154:1181–92.
160. Keating CD, Kriek N, Daniels M, Ashcroft NR, Hopper NA, Siney EJ, et al. Whole-genome analysis of 60 G protein-coupled receptors in Caenorhabditis elegans by gene knockout with RNAi. Curr Biol (2003) 13:1715–20. doi:10.1016/j.cub.2003.09.003
161. Marks NJ, Maule AG. Neuropeptides in helminths: occurrence and distribution. Adv Exp Med Biol (2010) 692:49–77. doi:10.1007/978-1-4419-6902-6_4
162. Trent C, Tsuing N, Horvitz HR. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics (1983) 104:619–47.
163. Liu T, Kim K, Li C, Barr MM. FMRFamide-like neuropeptides and mechanosensory touch receptor neurons regulate male sexual turning behavior in Caenorhabditis elegans. J Neurosci (2007) 27:7174–82. doi:10.1523/JNEUROSCI.1405-07.2007
164. Guan Z, Giustetto M, Lomvardas S, Kim J-H, Miniaci MC, Schwartz JH, et al. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell (2002) 111:483–93. doi:10.1016/S0092-8674(02)01074-7
165. Guan Z, Kim J-H, Lomvardas S, Holick K, Xu S, Kandel ER, et al. p38 MAP kinase mediates both short-term and long-term synaptic depression in aplysia. J Neurosci (2003) 23:7317–25.
166. Krashes MJ, DasGupta S, Vreede A, White B, Armstrong JD, Waddell S. A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell (2009) 139:416–27. doi:10.1016/j.cell.2009.08.035
167. Ardiel EL, Rankin CH. An elegant mind: learning and memory in Caenorhabditis elegans. Learn Mem (2010) 17:191–201. doi:10.1101/lm.960510
168. Beets I, Janssen T, Meelkop E, Temmerman L, Suetens N, Rademakers S, et al. Vasopressin/oxytocin-related signaling regulates gustatory associative learning in C. elegans. Science (2012) 338:543–5. doi:10.1126/science.1226860
169. Giles AC, Rankin CH. Behavioral and genetic characterization of habituation using Caenorhabditis elegans. Neurobiol Learn Mem (2009) 92:139–46. doi:10.1016/j.nlm.2008.08.004
170. Li C, Timbers TA, Rose JK, Bozorgmehr T, McEwan A, Rankin CH. The FMRFamide-related neuropeptide FLP-20 is required in the mechanosensory neurons during memory for massed training in C. elegans. Learn Mem (2013) 20:103–8. doi:10.1101/lm.028993.112
171. Rankin CH, Beck CD, Chiba CM. Caenorhabditis elegans: a new model system for the study of learning and memory. Behav Brain Res (1990) 37:89–92. doi:10.1016/0166-4328(90)90074-O
172. Rose JK, Rankin CH. Blocking memory reconsolidation reverses memory-associated changes in glutamate receptor expression. J Neurosci (2006) 26:11582–7. doi:10.1523/JNEUROSCI.2049-06.2006
173. Li C, Kim K. Neuropeptides. In WormBook. The C. elegans Research Community, editor. WormBook (2008). Available at: http://www.wormbook.org
Keywords: FMRFamide-like peptides, nematodes, C. elegans, neuropeptide, G protein-coupled receptor, feeding behavior, reproduction
Citation: Peymen K, Watteyne J, Frooninckx L, Schoofs L and Beets I (2014) The FMRFamide-like peptide family in nematodes. Front. Endocrinol. 5:90. doi: 10.3389/fendo.2014.00090
Received: 06 May 2014; Accepted: 31 May 2014;
Published online: 16 June 2014.
Edited by:
Hubert Vaudry, University of Rouen, FranceReviewed by:
Gregoy Y. Bedecarrats, University of Guelph, CanadaPascal Favrel, University of Caen Lower Normandy, France
Copyright: © 2014 Peymen, Watteyne, Frooninckx, Schoofs and Beets. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isabel Beets, Functional Genomics and Proteomics Group, Department of Biology, KU Leuven, Naamsestraat 59, Leuven 3000, Belgium e-mail:aXNhYmVsLmJlZXRzQGJpby5rdWxldXZlbi5iZQ==
†Katleen Peymen and Jan Watteyne have contributed equally to this work.
 Katleen Peymen
Katleen Peymen Jan Watteyne
Jan Watteyne Lotte Frooninckx
Lotte Frooninckx Liliane Schoofs
Liliane Schoofs Isabel Beets
Isabel Beets