
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol. , 02 September 2013
Sec. Translational and Clinical Endocrinology
Volume 4 - 2013 | https://doi.org/10.3389/fendo.2013.00111
This article is part of the Research Topic Circadian clocks and pregnancy View all 8 articles
Circadian aspect of reproduction has gained much attention in recent years. In mammals, it is very important that the timing of greatest sexual motivation is in line with the highest fertility. Peripheral clocks have been found to reside also in reproductive organs, such as the uterus and ovary. The timing signal from the suprachiasmatic nucleus is suggested to be transmitted via hormonal and neural mechanisms, and could thus mediate circadian expression of target genes in these organs. In turn, estrogens from the ovary have been found to signal back to the hypothalamus, completing the feedback loop. In this review we will focus on the interplay between clock and estrogens. Estradiol has been directly linked with expression of Per1 and Per2 in the uterus. CLOCK, on the other hand, has been shown to alter estradiol signaling. We also present the idea that cholesterol could play a vital role in the regulation of reproduction. Cholesterol synthesis itself is circadially regulated and has been found to interfere with steroidogenesis in the ovary on the molecular level. This review presents a systems view on how the interplay between circadian clock, steroidogenesis, and cholesterol synthesis affect various aspects of mammalian reproduction.
Like other organisms, mammals are adapted to the 24 h environmental day/night cycle via an internal clock. These evolutionary conserved rhythms are related to daily and yearly changes due to Earth’s rotation and translation as well as to food availability, social interactions, and increased chances of reproduction and survival. Virtually every mammalian cell contains an autonomous circadian clock. In peripheral tissues they are proposed to drive rhythms of gene transcription and in turn govern daily oscillation of many physiological processes. The mammalian timing system is organized in a hierarchical manner. The central pacemaker resides in the suprachiasmatic nucleus (SCN) in the hypothalamus. SCN neurons receive direct photic input from the retina entraining them to the environmental light and dark cycle (1). In turn, the SCN synchronizes other oscillators in the brain and peripheral tissues through endocrine or neural mechanisms, modulation of body temperature and feeding behavior (2). In mammals, it is very important that the timing of greatest sexual motivation is in line with the highest fertility. Therefore hormonal stimuli governing these aspects of reproduction must be under strict control. The circadian control of these mechanisms has been known for a long time as well as the dependency of the reproductive cycle on estradiol levels (3, 4). The SCN mediates its effects on reproduction through direct and indirect neural projections to the hypothalamic-pituitary-gonadal (HPG) axis (5). In females it is responsible for providing a stimulatory signal for the onset of the preovulatory luteinizing hormone (LH) surge (6). Aside from the SCN, sufficiently high concentrations of estradiol are necessary for the LH surge to begin (7, 8). Estradiol is synthesized mainly by the ovary in response to the stimulation by gonadotropins from the HPG axis. Two proteins have been the center of research regarding its production. The steroidogenic acute regulatory (StAR) protein is responsible for cholesterol transportation to the mitochondrial inner membrane. Aromatase is the final enzyme in estrogen synthesis, converting testosterone to estradiol and androstendione to estrone. The expression of both is induced by gonadotropin stimulation via cAMP responsive transcription factors, such as the cAMP response element binding protein (CREB) (9). Estradiol was found to influence the expression of clock genes in peripheral tissues, including the uterus, while clock proteins were found to interfere with estradiol signaling, providing an interplay between both systems (10–13).
In this review we will approach the complex relationships between estrogens and circadian rhythmicity and how this influences the female reproductive cycle. Since most research was done on model organisms, mainly rodents, the nomenclature of genes will be written accordingly, unless otherwise specified.
The basic mechanism of the mammalian circadian rhythm is a transcriptional-translational-post-translational autoregulatory feedback loop. The core of the loop consists of Clock and Bmal1. CLOCK and BMAL1 proteins form a dimer which binds to the E-box region in promoters of period (Per1, Per2, Per3) and cryptochrome (Cry1, Cry2) genes (14–17). Following transcription and translation, PER, and CRY proteins form a complex with casein kinase 1ɛ and translocate into the nucleus. Here they bind to BMAL1/CLOCK complex and inhibit their own transcription, which completes the basic autoregulatory loop (18). PER and CRY proteins are then tagged for proteasomal degradation via phosphorylation by casein kinase 1ɛ and 1δ and subsequently by ubiquitination. This cycle lasts approximately 24 h. BMAL1/CLOCK heterodimer also upregulates the transcription of Rev-erbα and Rora. Their protein products interact with ROR elements (RORE) in the promoter of Bmal1 gene, up (RORα) or downregulating (REV-ERBα) its transcription (19, 20).
As a master pacemaker and synchronizer the SCN maintains a near 24 h daily rhythm in all cells, hence the name circadian (“circa” meaning approximately and “diem” meaning day) (21). It is responsible for sensing the time of the day via outside cues and transmitting that information to oscillators throughout the body, in order to synchronize and entrain their cycles. Neurons within the SCN are organized in a coupling manner to provide a more robust and precise rhythm than individual cells (22). Light information is detected by cells within the retina. Intrinsically photosensitive retinal ganglion cells (ipRGCs), which contain the photopigment melanopsin, gather data through their intrinsic phototransduction mechanism as well as by extrinsic signals from rods and cones (23–26). This signal is then transmitted via the retinohypothalamic tract (RHT) to the core region of the SCN (1, 27). Upon stimulation, RHT neurons release glutamate and pituitary adenylate cyclase-activating polypeptide (PACAP) at synaptic contacts with the SCN (28). This in turn leads to calcium influx into the SCN cells and activation of various kinase pathways [MAPK, CaMK, protein kinase A (PKA)] (29). Kinases phosphorylate and activate CREB, which upon phosphorylation binds to CREB binding protein. This complex then binds to cAMP response elements (CRE) in the promoter regions of target genes, altering their transcription (30). Per1 and Per2 are two target genes that are influenced by light. They are activated by light signals only during the night and are involved in phase shifts and clock resetting (31–33). Additionally, it has been shown that the human PER1 gene can also be activated by extracellular stimulators acting through PKA and PKC pathways, and that this activation is distinct from BMAL1/CLOCK regulation (34, 35).
The SCN needs to transmit its timing signals to all other oscillators throughout the body. This is accomplished by utilizing humoral and neural signaling mechanisms (36). Locomotor activity is supposed to be maintained by peptides, such as vasopressin, transforming growth factor α (TGF-α), prokineticin 2, and cardiotrophin-like cytokine (37–40). There are also several proposed pathways used by the SCN to synchronize and entrain peripheral oscillators, such as signaling by hormones and autonomic neural connections, as well as more indirect ways, as is modulation of body temperature and feeding behavior (41–44).
The circadian system is important for successful reproduction, as it ensures that the period of maximal fertility is in line with highest sexual motivation (4). It influences the maturation of the follicle and ovulation as well as timely and successful mating behavior. These complex events and behavior are coordinated in part by the HPG axis and its hormones. Gonadotropin-releasing hormone (GnRH) is the first hormone in the HPG axis. It is released in a pulsatile fashion from the hypothalamus to the anterior pituitary, where it regulates the release of gonadotropins – LH and follicle-stimulating hormone (FSH). Gonadotropins than travel to the reproductive organs, where they trigger the release of sex steroids. On the day of proestrus there is a high release of GnRH with a subsequent LH surge that triggers ovulation.
But where does the SCN come into the picture? A signal from the SCN is crucial for the initiation of the LH surge and for subsequent ovulation (6, 8). When animals are kept under different light/dark cycles, the LH surge still occurs around the time of activity onset (45). In rats a high release of GnRH from the hypothalamus and a subsequent LH surge occur once every 4–5 days. The administration of barbiturates that prevent signaling within the SCN to rats prior to this event blocks the LH surge and ovulation and delays them for 24 h (3). Additionally, studies on SCN lesions in rats resulted in the absence of ovulation (46, 47). Combined, these studies provide evidence that a signal from the circadian system is crucial for successful ovulation.
The LH surge presents a strong signal by the SCN to the periphery. As previously mentioned, circadian clocks are present in peripheral organs, such as the ovary (48). Both FSH and LH have been shown to induce Per1 and Per2 expression in rat granulosa cells (49). The effect is most likely mediated by CREB since the promoters of both genes contain CRE elements (50, 51). The importance of the LH surge on the day of proestrus is that it might provide a resetting signal for clocks in the ovary and is thus involved in their synchronization (49, 52).
As for the mechanism by which the SCN coordinates these events, the SCN signals both directly and indirectly to the GnRH neurons in the medial preoptic area (MPOA) (53). Direct signals are believed to be transmitted via vasoactive intestinal peptide (VIP) synthesizing neurons (54). These neurons project from the SCN core to the GnRH neurons that contain the VIP receptor (VIPR2/VPAC2) (55). The indirect signals are transmitted by vasopressinergic (AVPergic) cells from the SCN shell to the anteroventral paraventricular nucleus (AVPV) (56, 57). The AVPV contains Kiss1 neurons that form an additional link between the SCN and the GnRH neurons. The Kiss1/Kiss1r system has recently been shown to have a big impact on mammalian reproduction. It has been implicated in the onset of puberty (58), preovulatory LH surge (59), GnRH release, including a positive (60) and a negative feedback (61) by sex steroids.
Prior to the discovery of the Kiss1 system, the gonadotropin inhibitory hormone (GnIH) had been identified in the quail brain (62). Since then it has been found in the brains of many other species, including mammals (63). GnIH acts as a negative regulator of the HPG axis suppressing gonadotropin secretion at the pituitary level (64, 65) as well as inhibiting their synthesis (66). In rodents, the GnIH neuron bodies are present in the dorsomedial hypothalamus (DMH) and project monosynaptically to the GnRH neurons (63, 65). The SCN was found to project to the GnIH cells in hamsters, providing a mechanism for the clock to inhibit the negative effect of GnIH on the HPG axis. The SCN could thus have a dual role in ovulatory control, on one hand stimulating GnRH release and on the other preventing the inhibition by the GnIH (67). The GnIH-immunoreactive cells express estrogen receptor-α (ERα), suggesting a feedback by gonadal estrogens (68). GnIH was also found to affect mice ovary, where it inhibits steroidogenesis probably via the inhibition of LH receptor (LHR) as well as StAR and 3β-hydroxysteroid dehydrogenase (3β-HSD) proteins (69).
Sex steroids present the final part of the HPG axis. They are synthesized from cholesterol in the gonads in response to gonadotropin stimulation. In females, the ovary is the primary organ for the synthesis of estrogens and progestins apart from the placenta during pregnancy. Estradiol is an important mediator of the positive and negative feedback of GnRH release. During most of the female estrous cycle, low levels of estradiol reduce the amplitude of GnRH pulses (70). Levels of LH are thus kept low while the follicle develops. As the levels of estradiol rise toward the end of the proestrus, the feedback turns to positive with an induction of a high amplitude GnRH surge, followed by the LH surge and ovulation (71). The action of estradiol is mediated through Kiss1 neurons. They are present in the AVPV and the arcuate nucleus (ARC) (72). In contrast to GnRH neurons that express estrogen receptor-β (ERβ), the Kiss1 neurons express mainly ERα, through which estradiol can transmit its signals. The expression of ERβ was also discovered, though its role seems to be less significant since the ovariectomized (OVX) ERβ knockout mice still respond to estradiol stimulation with the upregulation of the Kiss1 expression, but not the OVX ERα knockout mice. The negative feedback during most of the estrous cycle is a result of estradiol acting on Kiss1 neurons in the ARC, with a following decrease in Kiss1 mRNA (73). On the other hand, positive feedback on the day of proestrus is a result of the action of estradiol on Kiss1 neurons in the AVPV, where it increases Kiss1 expression (74). GnIH neurons also express ERα, through which estradiol can exert its influence, though the exact mechanism of this interaction remains to be elucidated. Together these studies show that apart from the signal by the circadian system, estradiol plays a vital part in the induction of the LH surge and triggering of ovulation (Figure 1).
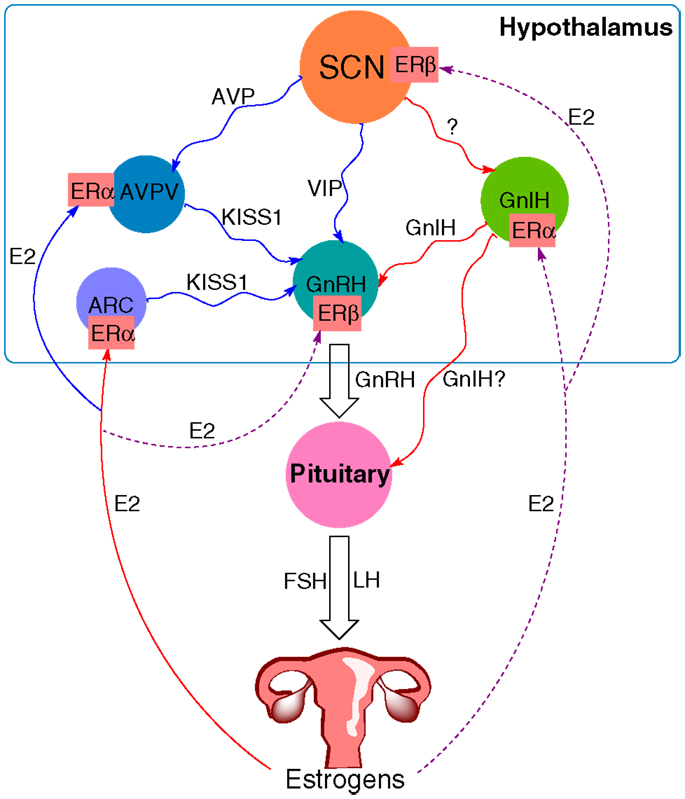
Figure 1. The proposed model of circadian influence on the HPG axis and feedback by estrogens. Blue lines depict positive interaction, red lines negative interaction, and purple (solid and dashed) lines depict a complex or not yet known interaction. The SCN signals to various neurons within the hypothalamus, including GnIH, GnRH, and Kiss1 neurons in the AVPV. GnIH neurons negatively regulate GnRH release, whereas the effect of Kiss1 neurons is stimulatory. Estradiol (E2) from the ovary transmits its signals via estrogen receptors (ER). It sends negative feedback to Kiss1 neurons in the ARC and positive to Kiss1 neurons in the AVPV. ER are also present on GnRH neurons, GnIH neurons, and the SCN, suggesting a feedback mechanism to these structures as well, although the exact mechanism for these interactions remains yet to be elucidated.
The SCN itself also contains estrogen receptors, mainly ERβ (75, 76). This enables it to detect plasma estrogen concentrations and react to them accordingly. An increase in estradiol levels during follicular development promotes the formation of synapses between the SCN and GnRH neurons (77) and increases the SCN sensitivity to light (78). To further study the role of circadian timing in reproduction and pregnancy, several mouse mutant models were developed (for summary, see Table 1). Female ClockΔ19 mutant mice (79) produce a dysfunctional CLOCK protein that can form a complex with BMAL1, but the dimer fails to initiate transcription through E-box elements in the promoters of target genes (14, 80). These mice lose central and peripheral rhythmicity in constant DD conditions (79). In view of reproduction, they are fertile with some reports of parturition difficulties, irregular estrous cycles, lack of a coordinated LH surge, and have a higher rate of pregnancy failure (81, 82). These effects are somewhat less in ClockΔ19 + MEL mice, which are able to synthesize melatonin due to a different genetic background (82). Mice lacking a functional Bmal1 gene show a complete loss of rhythmicity in total darkness (83). Despite this, ovulation does still occur even though there is no apparent LH surge. These mice also have a prolonged estrous cycle, but are infertile due to impaired steroidogenesis and low progesterone levels. There is no evident implantation of the embryo, but it can be reinstituted by progesterone supplementation (84, 85). Two other mouse models were developed, lacking either Per1 or Per2 gene (17, 86). Young adult female mutant mice (aged 2–6 months) have regular estrous cycles and the same reproductive success as wild-type females. On the other hand, middle-aged female mutant mice (aged 9–12 months) show lower incidence of estrous cyclicity and have a significantly lower reproductive rate as compared to wild-types. Together these results indicate an accelerated reproductive aging as a consequence of Per gene disruption (87). Outside the core clock genes, Vipr2 null mice were also generated. VIP signaling is important for maintaining rhythmicity and synchrony of neurons within the SCN (88). It is also involved in signaling by VIP and PACAP to the GnRH neurons (89). Mice lacking a functional VPAC2 receptor showed elongated estrous cycles exacerbated by constant darkness, however pregnancy rates were not affected (90). Taken together, these studies show that the disruption of core clock genes or SCN signaling pathways has an impact on estrous cyclicity and reproduction. The mechanism underlying these changes remains a matter of debate. Many of the studied mouse models retain rhythmicity under LD conditions, yet still exhibit disrupted reproduction. One possible explanation could be that, while being part of the core clock machinery, many of the disrupted or knocked out genes are transcription factors and could alter the expression of genes involved in reproduction. An example of this would be BMAL1, which may directly regulate StAR expression (84).
Estrogens are female sex hormones important for the development of secondary female sexual characteristics and enabling successful reproduction. They are produced from cholesterol mainly by the ovary following stimulation by FSH. An enzyme from cholesterol synthesis, lanosterol 14α-demethylase (CYP51) (93, 94) was detected in the rat oocytes on the mRNA and protein levels, suggesting the potential of oocytes to synthesize cholesterol de novo (95). This has been confirmed in mice where the CYP51 protein was detected in primary mouse oocytes in a stage- and cell type-specific manner, suggesting distinct regulatory pathways for its expression in the oocyte and the surrounding cumulus cells (96). While it has not yet been proven that the oocyte can indeed synthesize cholesterol, sperm cells retain this ability, which was a surprising discovery (97). In sperm, the major role of the cholesterol synthesis pathway might not be to synthesize cholesterol, but to produce meiosis activating sterols (MAS) (98), whose roles have been reviewed recently (99).
The plasma concentrations of cholesterol vary according to the time of the day, which is of great clinical importance for the hyperlipidemia therapy by statins (100, 101). There have been several studies performed on mouse models with disrupted genes from cholesterol synthesis pathway (102) as well as on models with defects in clock components. It has been shown that mutations in the Clock gene abolish circadian expression of Hmgcr, the regulatory gene in cholesterol synthesis (103). Signaling by cAMP presents an important mechanism for transmitting circadian information. The proximal promoter of Hmgcr reveals one CRE element, but the effect on expression of this gene was mild to none when coupled with overexpression of immediate cAMP early repressor (ICER) or CRE modulator (CREMτ). This suggests an indirect signaling mechanism that controls Hmgcr expression (104). The promoter analysis of Cyp51 from the latter part of cholesterol synthesis identified three CRE elements, and both CREMτ and ICER had a significant effect on Cyp51 expression. These results are in concordance with findings from Crem knockout mice, where in the absence of Crem, the circadian regulation of Cyp51 was abolished, while the expression of Hmgcr remained circadian (104). ICER is transcribed from the Crem gene and is part of the negative loop of cAMP signaling. It is expressed in a circadian manner and is able to repress its own transcription. ICER contributes to attenuation of cAMP signaling. The canonical signaling pathway (Gαs/cAMP/PKA) has been discovered over 20 years ago (105, 106). After the binding of FSH to its receptor (FSHR), Gαs functionally couples with FSHR and in turn activates adenylate cyclase (AC). This leads to cAMP production by AC and subsequent activation of PKA. Activated PKA’s catalytic subunits then phosphorylate various targets within the cytosol or the nucleus (107). Gene transcription regulated by FSH is controlled by translocation of the PKA catalytic subunit to the nucleus, where it phosphorylates and activates CREB. CREB binds to genes that contain CRE regions in their promoters and activates their transcription (108). ICER has also been found to bind to CRE elements in the promoter of the Per1 gene and to attenuate its transcription in the adrenal gland. It is proposed to act as a noise filter and might be a mechanism for the central clock to mediate the fine tuning of peripheral clocks (51). Together this presents an interesting aspect, whereby ICER could be involved in circadian regulation of cholesterol and sex steroid synthesis as well as in mediating the effect of the central pacemaker to the periphery. Whether this is true for the ovary still needs to be proven.
Similar to cholesterol synthesis, cAMP signaling is also important for the production of sex steroids (109). The first step in estrogen synthesis is cleavage of cholesterol by the CYP11A1 forming a C21 product pregnenolone. Pregnenolone presents a branching point in the biosynthetic pathway, as it can be converted to either glucocorticoids and mineralocorticoids or to sex steroids. The CYP17A1 and 3β-HSD then transform pregnenolone to androstenedione. In the next step, androstenedione can be converted by 17β-HSD into testosterone. The final enzyme in the pathway is CYP19A1 (aromatase) that catalyzes the conversion of androstenedione or testosterone by aromatization of the first ring to estrone or estradiol, respectively (Figure 2).
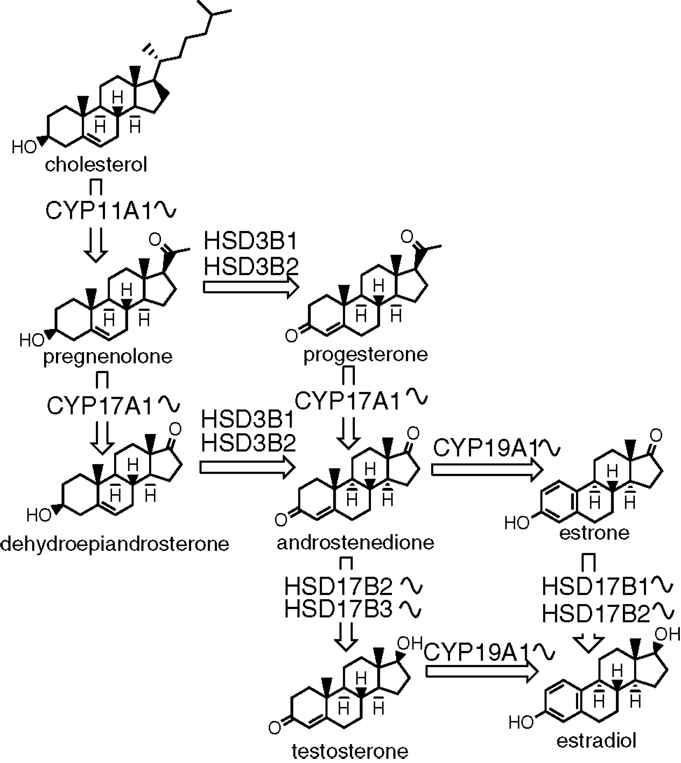
Figure 2. The biosynthesis of sex steroids. The sinusoid curve next to an enzyme depicts its known circadian expression. The synthesis starts with cholesterol and consists of several steps catalyzed by either enzymes from the cytochrome P450 or hydroxysteroid dehydrogenase families.
Aromatase encoded by the Cyp19a1 gene catalyzes the final reaction in estradiol biosynthesis. Cyp19a1 contains two promoter regions that mediate the effects of FSH through cAMP/PKA pathway: a steroidogenic factor-1 (SF-1) binding site and a CRE like element (9, 110, 111). Apart from the canonical signaling pathway, FSH utilizes various other signaling mechanisms, such as phosphatidylinositol-3 kinase (PI3K) which activates Akt (112). Akt was shown to alleviate the repressive effect of forkhead box O1 (FOXO1) on aromatase expression (113). Additionally, the LH surge induces the expression of the transcription factors CCAAT/enhancer-binding protein β (CEBPB) and ICER (114, 115), both of which are involved in the silencing of Cyp19a1 (116, 117). In the aromatase knockout mice (ArKO) (118), the females lack endogenous estrogens but have increased levels of testosterone, FSH, and LH. These mice are infertile due to the disruption of folliculogenesis and the lack of corpora lutea (119). The high basal levels of LH and the lack of an LH surge were found responsible for the lack of ovulation (120). An interesting aspect of disrupted estrogen synthesis is the altered sleep-wake rhythms of ArKO mice. They are less nocturnal and are generally less active when compared to WT mice (121, 122).
If we move away from the synthesis itself, we find another crucial protein for the production of sex steroids. The StAR protein is a rate limiting step in steroidogenesis. It is involved in the transport of cholesterol from the outer to the inner mitochondrial membrane. StAR knockout mice on steroid replacement therapy show premature ovarian failure with progressive lipid deposition and without detectable corpora lutea (92). The phenotype can in part be explained by insufficient production of progesterone, as progesterone receptor knockout mice exhibit similar defects (123). StAR expression and activity in the ovary is regulated by LH via cAMP/PKA pathway (124). While the promoter of the human STAR gene lacks a consensus CRE sequence (125), CREB, CREM, and activating transcription factor-1 (ATF-1) are still able to bind to it. This is possible by binding to three CRE half-sites identified in the promoter region (126). Several other cAMP responsive transcription factors are involved in StAR transcription, such as SF-1, CCAAT/enhancer-binding proteins (CEBPs), sterol regulatory element binding protein (SREBP), and DAX-1 (127–130). Interestingly, the core clock gene Rev-erbα induces StAR expression in mouse granulosa cells (131). It has been shown that mouse StAR promoter region contains putative RORE, which can bind Rev-erbα (132). This indicates that the StAR gene might be under the direct control of one of the core clock genes, although there are mixed findings, whether this results in the induction or repression of StAR transcription (131, 132). Further research is needed to confirm this hypothesis and to describe the mechanistic background.
The effect of steroids on the phase, amplitude, and period of circadian rhythms has been known for a long time (133). This might result from their direct action on the SCN or from one of the previously discussed pathways from the SCN to the periphery. Estradiol shortens the period of Per2 expression in the uterus of OVX mice, but not in the SCN (11). Estradiol also alters the rhythms of Per1 and Per2 in the liver, kidney, and uterus of OVX rats (10). From this we can see that estradiol differentially regulates the expression of clock genes in central and peripheral tissues. The reason for this might in part be due to the differential expression of ERs. As mentioned, the SCN expresses mainly ERβ while liver, kidney, and uterus express predominantly ERα (134, 135). An interesting study by Li et al. directly links the CLOCK protein with ERα activity. Two sumoylation sites were located on the human CLOCK protein and treatment with estradiol promotes this post-translational modification. Sumoylated but not native CLOCK protein was found to modify the transcriptional activity of ERα. This was most likely the result of the direct interaction between sumoylated CLOCK and ERα as concentrations of ERα remained unchanged (13). ERβ was also shown to be directly linked to the core clock genes. The promoter of ERβ contains an evolutionary conserved E-box, which binds core clock proteins to drive rhythmic expression of this receptor. This is in line with the fact that CLOCK/BMAL1 heterodimer has been found to bind to E-box elements in a rhythmic manner (136, 137). These studies provide evidence that there is a complex relationship between estradiol signaling and clock proteins. On one hand, estradiol influences the core clock machinery, and on the other, the core clock proteins may influence ERα transcriptional activity and the expression of ERβ. If and how this affects fertility and reproduction remains to be determined.
Cholesterol synthesis is one of the circadially regulated processes (104). In humans cholesterol levels peak toward the morning and are lowest in the afternoon (100). Also, SCN signaling via the HPG axis influences in part the production of sex steroids. Both are therefore linked to the central pacemaker, yet little is known whether there is a link connecting them directly. There is one example of the interplay between the two pathways through the regulation of LHR. LHR is expressed in ovarian cells and shows a marked downregulation following the LH surge (138, 139). When LH binds to its receptor, it activates the cAMP/PKA signaling pathway (140). This results in an increase in sex steroid hormone biosynthesis. The increased steroidogenesis leads to cholesterol depletion in the cell, triggering upregulation of genes involved in cholesterol biosynthesis, such as mevalonate kinase (MVK), via SREBP (141, 142). MVK catalyzes the conversion of mevalonate to 5-phosphomevalonate in the cholesterol synthesis pathway. Interestingly, MVK also acts as an RNA binding protein. It was found to form a complex with LHR mRNA and prevent its translation, which results in LHR mRNA degradation (143, 144). In the meantime, steroidogenesis is temporarily interrupted until MVK is able to restore its catalytic function.
With every new publication the importance of biological rhythms in various aspects of our lives becomes more evident. In recent years research has extensively focused on the implications of circadian rhythmicity in reproduction. Mouse knockout models have proven to be invaluable in determining how defects of the clock or impaired steroidogenesis influence reproduction. Interesting to note is that even the defects in functionally related genes, such as Clock and Bmal1, have different outcomes, with former showing decreased reproduction rates, while the latter proving infertile. In females estrogens present a link between circadian rhythmicity and reproduction. Signals by estradiol and the SCN together are crucial for the start of an LH surge with the resulting ovulation. The neural network within the hypothalamus presents a complex system that integrates both environmental and hormonal signals to ensure successful reproduction. But the relationship between the two doesn’t stop there. It seems that estradiol and circadian signaling pathways are more intimately connected with evidence for their interplay on the molecular level. The core clock genes were found to interfere with estradiol signaling, while estradiol was shown to alter the rhythms of Per1 and Per2 gene expression in various tissues. To add another piece to the puzzle, cholesterol synthesis itself is rhythmic, and interferes with steroidogenesis in the ovary via the downregulation of LHR.
Despite the complexity of this network of interactions, there is growing evidence that the disruption of one of these systems has an influence on reproduction. But the questions regarding the mechanistic background and the potential application in medicine both require additional research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the Slovenian Research Agency program grants P1-0104 and J7-4053. Ziga Urlep is funded as a young researcher by the Slovenian Research Agency.
1. Do MT, Yau KW. Intrinsically photosensitive retinal ganglion cells. Physiol Rev (2010) 90(4):1547–81. doi:10.1152/physrev.00013
2. Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol (2010) 72:551–77. doi:10.1146/annurev-physiol-021909-135919
3. Everett JW, Sawyer CH. A 24-hour periodicity in the “LH-release apparatus” of female rats, disclosed by barbiturate sedation. Endocrinology (1950) 47(3):198–218. doi:10.1210/endo-47-3-198
4. Sarkar DK, Chiappa SA, Fink G, Sherwood NM. Gonadotropin-releasing hormone surge in pro-oestrous rats. Nature (1976) 264(5585):461–3. doi:10.1038/264461a0
5. Boden MJ, Kennaway DJ. Circadian rhythms and reproduction. Reproduction (2006) 132(3):379–92. doi:10.1530/rep.1.00614
6. Chappell PE. Clocks and the black box: circadian influences on gonadotropin-releasing hormone secretion. J Neuroendocrinol (2005) 17(2):119–30. doi:10.1111/j.1365-2826.2005.01270.x
7. Christian CA, Mobley JL, Moenter SM. Diurnal and estradiol-dependent changes in gonadotropin-releasing hormone neuron firing activity. Proc Natl Acad Sci U S A (2005) 102(43):15682–7. doi:10.1073/pnas.0504270102
8. Legan SJ, Coon GA, Karsch FJ. Role of estrogen as initiator of daily LH surges in the ovariectomized rat. Endocrinology (1975) 96(1):50–6. doi:10.1210/endo-96-1-50
9. Fitzpatrick SL, Richards JS. Identification of a cyclic adenosine 3′,5′-monophosphate-response element in the rat aromatase promoter that is required for transcriptional activation in rat granulosa cells and R2C leydig cells. Mol Endocrinol (1994) 8(10):1309–19. doi:10.1210/me.8.10.1309
10. Nakamura TJ, Moriya T, Inoue S, Shimazoe T, Watanabe S, Ebihara S, et al. Estrogen differentially regulates expression of Per1 and Per2 genes between central and peripheral clocks and between reproductive and nonreproductive tissues in female rats. J Neurosci Res (2005) 82(5):622–30. doi:10.1002/jnr.20677
11. Nakamura TJ, Sellix MT, Menaker M, Block GD. Estrogen directly modulates circadian rhythms of PER2 expression in the uterus. Am J Physiol Endocrinol Metab (2008) 295(5):E1025–31. doi:10.1152/ajpendo.90392.2008
12. Cai W, Rambaud J, Teboul M, Masse I, Benoit G, Gustafsson JA, et al. Expression levels of estrogen receptor beta are modulated by components of the molecular clock. Mol Cell Biol (2008) 28(2):784–93. doi:10.1128/MCB.00233-07
13. Li S, Wang M, Ao X, Chang AK, Yang C, Zhao F, et al. CLOCK is a substrate of SUMO and sumoylation of CLOCK upregulates the transcriptional activity of estrogen receptor-alpha. Oncogene (2012). doi:10.1038/onc.2012.518
14. Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science (1998) 280(5369):1564–9. doi:10.1126/science.280.5369.1564
15. Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A (1998) 95(10):5474–9. doi:10.1073/pnas.95.10.5474
16. Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell (1999) 98(2):193–205. doi:10.1016/S0092-8674(00)81014-4
17. Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell (2001) 105(5):683–94. doi:10.1016/S0092-8674(01)00380-4
18. Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell (2001) 107(7):855–67. doi:10.1016/S0092-8674(01)00610-9
19. Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell (2002) 110(2):251–60. doi:10.1016/S0092-8674(02)00825-5
20. Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron (2004) 43(4):527–37. doi:10.1016/j.neuron.2004.07.018
21. Halberg F. Circadian (about twenty-four-hour) rhythms in experimental medicine. Proc R Soc Med (1963) 56:253–7.
22. Aton SJ, Herzog ED. Come together, right...now: synchronization of rhythms in a mammalian circadian clock. Neuron (2005) 48(4):531–4. doi:10.1016/j.neuron.2005.11.001
23. Berson DM, Dunn FA, Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Science (2002) 295(5557):1070–3. doi:10.1126/science.1067262
24. Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science (2002) 295(5557):1065–70. doi:10.1126/science.1069609
25. Provencio I, Jiang G, De Grip WJ, Hayes WP, Rollag MD. Melanopsin: an opsin in melanophores, brain, and eye. Proc Natl Acad Sci U S A (1998) 95(1):340–5. doi:10.1073/pnas.95.1.340
26. Schmidt TM, Taniguchi K, Kofuji P. Intrinsic and extrinsic light responses in melanopsin-expressing ganglion cells during mouse development. J Neurophysiol (2008) 100(1):371–84. doi:10.1152/jn.00062.2008
27. Hankins MW, Peirson SN, Foster RG. Melanopsin: an exciting photopigment. Trends Neurosci (2008) 31(1):27–36. doi:10.1016/j.tins.2007.11.002
28. Hannibal J. Neurotransmitters of the retino-hypothalamic tract. Cell Tissue Res (2002) 309(1):73–88. doi:10.1007/s00441-002-0574-3
29. Golombek DA, Rosenstein RE. Physiology of circadian entrainment. Physiol Rev (2010) 90(3):1063–102. doi:10.1152/physrev.00009.2009
30. Carlezon WA Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci (2005) 28(8):436–45. doi:10.1016/j.tins.2005.06.005
31. Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, et al. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci (1999) 19(3):1115–21.
32. Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell (1997) 91(7):1043–53. doi:10.1016/S0092-8674(00)80494-8
33. Wakamatsu H, Takahashi S, Moriya T, Inouye ST, Okamura H, Akiyama M, et al. Additive effect of mPer1 and mPer2 antisense oligonucleotides on light-induced phase shift. Neuroreport (2001) 12(1):127–31. doi:10.1097/00001756-200101220-00033
34. Motzkus D, Maronde E, Grunenberg U, Lee CC, Forssmann W, Albrecht U. The human PER1 gene is transcriptionally regulated by multiple signaling pathways. FEBS Lett (2000) 486(3):315–9. doi:10.1016/S0014-5793(00)02315-2
35. Motzkus D, Loumi S, Cadenas C, Vinson C, Forssmann WG, Maronde E. Activation of human period-1 by PKA or CLOCK/BMAL1 is conferred by separate signal transduction pathways. Chronobiol Int (2007) 24(5):783–92. doi:10.1080/07420520701672481
36. Kalsbeek A, Yi CX, Cailotto C, la Fleur SE, Fliers E, Buijs RM. Mammalian clock output mechanisms. Essays Biochem (2011) 49(1):137–51. doi:10.1042/BSE0480137
37. Cheng MY, Bullock CM, Li C, Lee AG, Bermak JC, Belluzzi J, et al. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature (2002) 417(6887):405–10. doi:10.1038/417405a
38. Kalsbeek A, Fliers E, Hofman MA, Swaab DF, Buijs RM. Vasopressin and the output of the hypothalamic biological clock. J Neuroendocrinol (2010) 22(5):362–72. doi:10.1111/j.1365-2826.2010.01956.x
39. Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, Davis FC, et al. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science (2001) 294(5551):2511–5. doi:10.1126/science.1067716
40. Kraves S, Weitz CJ. A role for cardiotrophin-like cytokine in the circadian control of mammalian locomotor activity. Nat Neurosci (2006) 9(2):212–9. doi:10.1038/nn1633
41. Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol (2002) 12(18):1574–83. doi:10.1016/S0960-9822(02)01145-4
42. Damiola F, Le MinhN, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev (2000) 14(23):2950–61.
43. Vujovic N, Davidson AJ, Menaker M. Sympathetic input modulates, but does not determine, phase of peripheral circadian oscillators. Am J Physiol Regul Integr Comp Physiol (2008) 295(1):R355–60. doi:10.1152/ajpregu.00498.2007
44. Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science (2000) 289(5488):2344–7. doi:10.1126/science.289.5488.2344
45. Moline ML, Albers HE. Response of circadian locomotor activity and the proestrus luteinizing hormone surge to phase shifts of the light-dark cycle in the hamster. Physiol Behav (1988) 43(4):435–40.
46. Gray GD, Söderstein P, Tallentire D, Davidson JM. Effects of lesions in various structures of the suprachiasmatic-preoptic region on LH regulation and sexual behavior in female rats. Neuroendocrinology (1978) 25(3):174–91. doi:10.1159/000122739
47. Mosko SS, Moore RY. Neonatal ablation of the suprachiasmatic nucleus. Effects on the development of the pituitary-gonadal axis in the female rat. Neuroendocrinology (1979) 29(5):350–61. doi:10.1159/000122944
48. Fahrenkrug J, Georg B, Hannibal J, Hindersson P, Gräs S. Diurnal rhythmicity of the clock genes Per1 and Per2 in the rat ovary. Endocrinology (2006) 147(8):3769–76. doi:10.1210/en.2006-0305
49. He PJ, Hirata M, Yamauchi N, Hashimoto S, Hattori MA. Gonadotropic regulation of circadian clockwork in rat granulosa cells. Mol Cell Biochem (2007) 302(1–2):111–8. doi:10.1007/s11010-007-9432-7
50. Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A (2002) 99(11):7728–33. doi:10.1073/pnas.102075599
51. Zmrzljak UP, Korencic A, Košir R, Golicnik M, Sassone-Corsi P, Rozman D. Inducible cAMP early repressor regulates the Period 1 gene of the hepatic and adrenal clocks. J Biol Chem (2013) 288(15):10318–27. doi:10.1074/jbc.M112.445692
52. Yoshikawa T, Sellix M, Pezuk P, Menaker M. Timing of the ovarian circadian clock is regulated by gonadotropins. Endocrinology (2009) 150(9):4338–47. doi:10.1210/en.2008-1280
53. Colledge WH. Kisspeptins and GnRH neuronal signalling. Trends Endocrinol Metab (2009) 20(3):115–21. doi:10.1016/j.tem.2008.10.005
54. Tanaka M, Ichitani Y, Okamura H, Tanaka Y, Ibata Y. The direct retinal projection to VIP neuronal elements in the rat SCN. Brain Res Bull (1993) 31(6):637–40. doi:10.1016/0361-9230(93)90134-W
55. Smith MJ, Jiennes L, Wise PM. Localization of the VIP2 receptor protein on GnRH neurons in the female rat. Endocrinology (2000) 141(11):4317–20. doi:10.1210/en.141.11.4317
56. Leak RK, Moore RY. Topographic organization of suprachiasmatic nucleus projection neurons. J Comp Neurol (2001) 433(3):312–34. doi:10.1002/cne.1142
57. Watson RE Jr, Langub MC Jr, Engle MG, Maley BE. Estrogen-receptive neurons in the anteroventral periventricular nucleus are synaptic targets of the suprachiasmatic nucleus and peri-suprachiasmatic region. Brain Res (1995) 689(2):254–64. doi:10.1016/0006-8993(95)00548-5
58. Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS Jr, Shagoury JK, et al. The GPR54 gene as a regulator of puberty. N Engl J Med (2003) 349(17):1614–27. doi:10.1056/NEJMoa035322
59. Clarkson J, Herbison AE. Oestrogen, kisspeptin, GPR54 and the pre-ovulatory luteinising hormone surge. J Neuroendocrinol (2009) 21(4):305–11. doi:10.1111/j.1365-2826.2009.01835.x
60. Wintermantel TM, Campbell RE, Porteous R, Bock D, Gröne HJ, Todman MG, et al. Definition of estrogen receptor pathway critical for estrogen positive feedback to gonadotropin-releasing hormone neurons and fertility. Neuron (2006) 52(2):271–80. doi:10.1016/j.neuron.2006.07.023
61. Navarro VM, Castellano JM, Fernández-Fernández R, Barreiro ML, Roa J, Sanchez-Criado JE, et al. Developmental and hormonally regulated messenger ribonucleic acid expression of KiSS-1 and its putative receptor, GPR54, in rat hypothalamus and potent luteinizing hormone-releasing activity of KiSS-1 peptide. Endocrinology (2004) 145(10):4565–74. doi:10.1210/en.2004-0413
62. Tsutsui K, Saigoh E, Ukena K, Teranishi H, Fujisawa Y, Kikuchi M, et al. A novel avian hypothalamic peptide inhibiting gonadotropin release. Biochem Biophys Res Commun (2000) 275(2):661–7. doi:10.1006/bbrc.2000.3350
63. Kriegsfeld LJ, Gibson EM, Williams WP 3rd, Zhao S, Mason AO, Bentley GE, et al. The roles of RFamide-related peptide-3 in mammalian reproductive function and behaviour. J Neuroendocrinol (2010) 22(7):692–700. doi:10.1111/j.1365-2826.2010.02031.x
64. Clarke IJ, Sari IP, Qi Y, Smith JT, Parkington HC, Ubuka T, et al. Potent action of RFamide-related peptide-3 on pituitary gonadotropes indicative of a hypophysiotropic role in the negative regulation of gonadotropin secretion. Endocrinology (2008) 149(11):5811–21. doi:10.1210/en.2008-0575
65. Johnson MA, Tsutsui K, Fraley GS. Rat RFamide-related peptide-3 stimulates GH secretion, inhibits LH secretion, and has variable effects on sex behavior in the adult male rat. Horm Behav (2007) 51(1):171–80. doi:10.1016/j.yhbeh.2006.09.009
66. Sari IP, Rao A, Smith JT, Tilbrook AJ, Clarke IJ. Effect of RF-amide-related peptide-3 on luteinizing hormone and follicle-stimulating hormone synthesis and secretion in ovine pituitary gonadotropes. Endocrinology (2009) 150(12):5549–56. doi:10.1210/en.2009-0775
67. Gibson EM, Humber SA, Jain S, Williams WP 3rd, Zhao S, Bentley GE, et al. Alterations in RFamide-related peptide expression are coordinated with the preovulatory luteinizing hormone surge. Endocrinology (2008) 149(10):4958–69. doi:10.1210/en.2008-0316
68. Kriegsfeld LJ, Mei DF, Bentley GE, Ubuka T, Mason AO, Inoue K, et al. Identification and characterization of a gonadotropin-inhibitory system in the brains of mammals. Proc Natl Acad Sci U S A (2006) 103(7):2410–5. doi:10.1073/pnas.0511003103
69. Singh P, Krishna A, Tsutsui K. Effects of gonadotropin-inhibitory hormone on folliculogenesis and steroidogenesis of cyclic mice. Fertil Steril (2011) 95(4):1397–404. doi:10.1016/j.fertnstert.2010.03.052
70. Caraty A, Locatelli A, Martin GB. Biphasic response in the secretion of gonadotrophin-releasing hormone in ovariectomized ewes injected with oestradiol. J Endocrinol (1989) 123(3):375–82. doi:10.1677/joe.0.1230375
71. Moenter SM, Caraty A, Karsch FJ. The estradiol-induced surge of gonadotropin-releasing hormone in the ewe. Endocrinology (1990) 127(3):1375–84. doi:10.1210/endo-127-3-1375
72. Gottsch ML, Cunningham MJ, Smith JT, Popa SM, Acohido BV, Crowley WF, et al. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology (2004) 145(9):4073–7. doi:10.1210/en.2004-0431
73. Smith JT, Cunningham MJ, Rissman EF, Clifton DK, Steiner RA. Regulation of Kiss1 gene expression in the brain of the female mouse. Endocrinology (2005) 146(9):3686–92. doi:10.1210/en.2005-0323
74. Herbison AE. Estrogen positive feedback to gonadotropin-releasing hormone (GnRH) neurons in the rodent: the case for the rostral periventricular area of the third ventricle (RP3V). Brain Res Rev (2008) 57(2):277–87. doi:10.1016/j.brainresrev.2007.05.006
75. Gundlah C, Kohama SG, Mirkes SJ, Garyfallou VT, Urbanski HF, Bethea CL. Distribution of estrogen receptor beta (ERbeta) mRNA in hypothalamus, midbrain and temporal lobe of spayed macaque: continued expression with hormone replacement. Brain Res Mol Brain Res (2000) 76(2):191–204. doi:10.1016/S0006-8993(99)02475-0
76. Shughrue PJ, Lane MV, Merchenthaler I. Comparative distribution of estrogen receptor-alpha and -beta mRNA in the rat central nervous system. J Comp Neurol (1997) 388(4):507–25. doi:10.1002/(SICI)1096-9861(19971201)388:4<507::AID-CNE1>3.0.CO;2-6
77. Vida B, Deli L, Hrabovszky E, Kalamatianos T, Caraty A, Coen CW, et al. Evidence for suprachiasmatic vasopressin neurones innervating kisspeptin neurones in the rostral periventricular area of the mouse brain: regulation by oestrogen. J Neuroendocrinol (2010) 22(9):1032–9. doi:10.1111/j.1365-2826.2010.02045.x
78. Abizaid A, Mezei G, Horvath TL. Estradiol enhances light-induced expression of transcription factors in the SCN. Brain Res (2004) 1010(1–2):35–44. doi:10.1016/j.brainres.2004.01.089
79. Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science (1994) 264(5159):719–25. doi:10.1126/science.8171325
80. King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, et al. Positional cloning of the mouse circadian clock gene. Cell (1997) 89(4):641–53. doi:10.1016/S0092-8674(00)80245-7
81. Miller BH, Olson SL, Turek FW, Levine JE, Horton TH, Takahashi JS. Circadian clock mutation disrupts estrous cyclicity and maintenance of pregnancy. Curr Biol (2004) 14(15):1367–73. doi:10.1016/j.cub.2004.07.055
82. Kennaway DJ, Boden MJ, Voultsios A. Reproductive performance in female Clock Delta19 mutant mice. Reprod Fertil Dev (2004) 16(8):801–10. doi:10.1071/RD04023
83. Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell (2000) 103(7):1009–17. doi:10.1016/S0092-8674(00)00205-1
84. Ratajczak CK, Boehle KL, Muglia LJ. Impaired steroidogenesis and implantation failure in Bmal1-/- mice. Endocrinology (2009) 150(4):1879–85. doi:10.1210/en.2008-1021
85. Boden MJ, Varcoe TJ, Voultsios A, Kennaway DJ. Reproductive biology of female Bmal1 null mice. Reproduction (2010) 139(6):1077–90. doi:10.1530/REP-09-0523
86. Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, et al. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature (1999) 400(6740):169–73. doi:10.1038/22659
87. Pilorz V, Steinlechner S. Low reproductive success in Per1 and Per2 mutant mouse females due to accelerated ageing? Reproduction (2008) 135(4):559–68. doi:10.1530/REP-07-0434
88. Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci (2005) 8(4):476–83.
89. Gerhold LM, Rosewell KL, Wise PM. Suppression of vasoactive intestinal polypeptide in the suprachiasmatic nucleus leads to aging-like alterations in cAMP rhythms and activation of gonadotropin-releasing hormone neurons. J Neurosci (2005) 25(1):62–7. doi:10.1523/JNEUROSCI.3598-04.2005
90. Dolatshad H, Campbell EA, O’Hara L, Maywood ES, Hastings MH, Johnson MH. Developmental and reproductive performance in circadian mutant mice. Hum Reprod (2006) 21(1):68–79. doi:10.1093/humrep/dei313
91. Britt KL, Drummond AE, Dyson M, Wreford NG, Jones ME, Simpson ER, et al. The ovarian phenotype of the aromatase knockout (ArKO) mouse. J Steroid Biochem Mol Biol (2001) 79(1–5):181–5. doi:10.1016/S0960-0760(01)00158-3
92. Hasegawa T, Zhao L, Caron KM, Majdic G, Suzuki T, Shizawa S, et al. Developmental roles of the steroidogenic acute regulatory protein (StAR) as revealed by StAR knockout mice. Mol Endocrinol (2000) 14(9):1462–71. doi:10.1210/me.14.9.1462
93. Keber R, Motaln H, Wagner KD, Debeljak N, Rassoulzadegan M, Acimovic J, et al. Mouse knockout of the cholesterogenic cytochrome P450 lanosterol 14alpha-demethylase (Cyp51) resembles Antley-Bixler syndrome. J Biol Chem (2011) 286(33):29086–97. doi:10.1074/jbc.M111.253245
94. Rozman D, Strömstedt M, Tsui LC, Scherer SW, Waterman MR. Structure and mapping of the human lanosterol 14alpha-demethylase gene (CYP51) encoding the cytochrome P450 involved in cholesterol biosynthesis; comparison of exon/intron organization with other mammalian and fungal CYP genes. Genomics (1996) 38(3):371–81. doi:10.1006/geno.1996.0640
95. Vaknin KM, Lazar S, Popliker M, Tsafriri A. Role of meiosis-activating sterols in rat oocyte maturation: effects of specific inhibitors and changes in the expression of lanosterol 14alpha-demethylase during the preovulatory period. Biol Reprod (2001) 64(1):299–309. doi:10.1095/biolreprod64.1.299
96. Rozman D, Cotman M, Frangez R. Lanosterol 14alpha-demethylase and MAS sterols in mammalian gametogenesis. Mol Cell Endocrinol (2002) 187(1–2):179–87. doi:10.1016/S0303-7207(01)00693-1
97. Cotman M, Jezek D, Fon TacerK, Frangez R, Rozman D. A functional cytochrome P450 lanosterol 14 alpha-demethylase CYP51 enzyme in the acrosome: transport through the Golgi and synthesis of meiosis-activating sterols. Endocrinology (2004) 145(3):1419–26. doi:10.1210/en.2003-1332
98. Rozman D, Waterman MR. Lanosterol 14alpha-demethylase (CYP51) and spermatogenesis. Drug Metab Dispos (1998) 26(12):1199–201.
99. Keber R, Rozman D, Horvat S. Sterols in spermatogenesis and sperm maturation. J Lipid Res (2013) 54(1):20–33. doi:10.1194/jlr.R032326
100. Jones PJ, Schoeller DA. Evidence for diurnal periodicity in human cholesterol synthesis. J Lipid Res (1990) 31(4):667–73.
101. Rozman D, Monostory K. Perspectives of the non-statin hypolipidemic agents. Pharmacol Ther (2010) 127(1):19–40. doi:10.1016/j.pharmthera.2010.03.007
102. Horvat S, McWhir J, Rozman D. Defects in cholesterol synthesis genes in mouse and in humans: lessons for drug development and safer treatments. Drug Metab Rev (2011) 43(1):69–90. doi:10.3109/03602532.2010.540580
103. Kudo T, Kawashima M, Tamagawa T, Shibata S. Clock mutation facilitates accumulation of cholesterol in the liver of mice fed a cholesterol and/or cholic acid diet. Am J Physiol Endocrinol Metab (2008) 294(1):E120–30. doi:10.1152/ajpendo.00061.2007
104. Acimovic J, Fink M, Pompon D, Bjorkhem I, Hirayama J, Sassone-Corsi P, et al. CREM modulates the circadian expression of CYP51, HMGCR and cholesterogenesis in the liver. Biochem Biophys Res Commun (2008) 376(1):206–10. doi:10.1016/j.bbrc.2008.08.126
105. Dattatreyamurty B, Figgs LW, Reichert LE Jr. Physical and functional association of follitropin receptors with cholera toxin-sensitive guanine nucleotide-binding protein. J Biol Chem (1987) 262(24):11737–45.
106. Means AR, MacDougall E, Soderling TR, Corbin JD. Testicular adenosine 3′:5′-monophosphate-dependent protein kinase. Regulation by follicle-stimulating hormone. J Biol Chem (1974) 249(4):1231–8.
107. Landmark BF, Fauske B, Eskild W, Skålhegg B, Lohmann SM, Hansson V, et al. Identification, characterization, and hormonal regulation of 3′, 5′-cyclic adenosine monophosphate-dependent protein kinases in rat Sertoli cells. Endocrinology (1991) 129(5):2345–54. doi:10.1210/endo-129-5-2345
108. Cottom J, Salvador LM, Maizels ET, Reierstad S, Park Y, Carr DW, et al. Follicle-stimulating hormone activates extracellular signal-regulated kinase but not extracellular signal-regulated kinase kinase through a 100-kDa phosphotyrosine phosphatase. J Biol Chem (2003) 278(9):7167–79. doi:10.1074/jbc.M203901200
109. Richards JS, Hedin L. Molecular aspects of hormone action in ovarian follicular development, ovulation, and luteinization. Annu Rev Physiol (1988) 50:441–63. doi:10.1146/annurev.ph.50.030188.002301
110. Fitzpatrick SL, Richards JS. cis-Acting elements of the rat aromatase promoter required for cyclic adenosine 3′,5′-monophosphate induction in ovarian granulosa cells and constitutive expression in R2C Leydig cells. Mol Endocrinol (1993) 7(3):341–54. doi:10.1210/me.7.3.341
111. Lynch JP, Lala DS, Peluso JJ, Luo W, Parker KL, White BA. Steroidogenic factor 1, an orphan nuclear receptor, regulates the expression of the rat aromatase gene in gonadal tissues. Mol Endocrinol (1993) 7(6):776–86. doi:10.1210/me.7.6.776
112. Zeleznik AJ, Saxena D, Little-Ihrig L. Protein kinase B is obligatory for follicle-stimulating hormone-induced granulosa cell differentiation. Endocrinology (2003) 144(9):3985–94. doi:10.1210/en.2003-0293
113. Park Y, Maizels ET, Feiger ZJ, Alam H, Peters CA, Woodruff TK, et al. Induction of cyclin D2 in rat granulosa cells requires FSH-dependent relief from FOXO1 repression coupled with positive signals from Smad. J Biol Chem (2005) 280(10):9135–48. doi:10.1074/jbc.M409486200
114. Mukherjee A, Urban J, Sassone-Corsi P, Mayo KE. Gonadotropins regulate inducible cyclic adenosine 3′,5′-monophosphate early repressor in the rat ovary: implications for inhibin alpha subunit gene expression. Mol Endocrinol (1998) 12(6):785–800. doi:10.1210/me.12.6.785
115. Sirois J, Richards JS. Transcriptional regulation of the rat prostaglandin endoperoxide synthase 2 gene in granulosa cells. Evidence for the role of a cis-acting C/EBP beta promoter element. J Biol Chem (1993) 268(29):21931–8.
116. Yang S, Fang Z, Suzuki T, Sasano H, Zhou J, Gurates B, et al. Regulation of aromatase P450 expression in endometriotic and endometrial stromal cells by CCAAT/enhancer binding proteins (C/EBPs): decreased C/EBPbeta in endometriosis is associated with overexpression of aromatase. J Clin Endocrinol Metab (2002) 87(5):2336–45. doi:10.1210/jc.87.5.2336
117. Morales V, Gonzalez-Robayna I, Hernandez I, Quintana J, Santana P, Ruiz deGalarreta CM, et al. The inducible isoform of CREM (inducible cAMP early repressor, ICER) is a repressor of CYP19 rat ovarian promoter. J Endocrinol (2003) 179(3):417–25. doi:10.1677/joe.0.1790417
118. Fisher CR, Graves KH, Parlow AF, Simpson ER. Characterization of mice deficient in aromatase (ArKO) because of targeted disruption of the cyp19 gene. Proc Natl Acad Sci U S A (1998) 95(12):6965–70. doi:10.1073/pnas.95.12.6965
119. Honda S, Harada N, Ito S, Takagi Y, Maeda S. Disruption of sexual behavior in male aromatase-deficient mice lacking exons 1 and 2 of the cyp19 gene. Biochem Biophys Res Commun (1998) 252(2):445–9. doi:10.1006/bbrc.1998.9672
120. Mann RJ, Keri RA, Nilson JH. Transgenic mice with chronically elevated luteinizing hormone are infertile due to anovulation, defects in uterine receptivity, and midgestation pregnancy failure. Endocrinology (1999) 140(6):2592–601. doi:10.1210/en.140.6.2592
121. Brockman R, Bunick D, Mahoney MM. Estradiol deficiency during development modulates the expression of circadian and daily rhythms in male and female aromatase knockout mice. Horm Behav (2011) 60(4):439–47. doi:10.1016/j.yhbeh.2011.07.011
122. Vyazovskiy VV, Kopp C, Wigger E, Jones ME, Simpson ER, Tobler I. Sleep and rest regulation in young and old oestrogen-deficient female mice. J Neuroendocrinol (2006) 18(8):567–76. doi:10.1111/j.1365-2826.2006.01452.x
123. Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA Jr, et al. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev (1995) 9(18):2266–78.
124. Wood JR, Strauss JF 3rd. Multiple signal transduction pathways regulate ovarian steroidogenesis. Rev Endocr Metab Disord (2002) 3(1):33–46.
125. Sugawara T, Lin D, Holt JA, Martin KO, Javitt NB, Miller WL, et al. Structure of the human steroidogenic acute regulatory protein (StAR) gene: StAR stimulates mitochondrial cholesterol 27-hydroxylase activity. Biochemistry (1995) 34(39):12506–12. doi:10.1021/bi00039a004
126. Manna PR, Dyson MT, Eubank DW, Clark BJ, Lalli E, Sassone-Corsi P, et al. Regulation of steroidogenesis and the steroidogenic acute regulatory protein by a member of the cAMP response-element binding protein family. Mol Endocrinol (2002) 16(1):184–99. doi:10.1210/me.16.1.184
127. Christenson LK, McAllister JM, Martin KO, Javitt NB, Osborne TF, Strauss JF 3rd. Oxysterol regulation of steroidogenic acute regulatory protein gene expression. Structural specificity and transcriptional and posttranscriptional actions. J Biol Chem (1998) 273(46):30729–35. doi:10.1074/jbc.273.46.30729
128. Reinhart AJ, Williams SC, Clark BJ, Stocco DM. SF-1 (steroidogenic factor-1) and C/EBP beta (CCAAT/enhancer binding protein-beta) cooperate to regulate the murine StAR (steroidogenic acute regulatory) promoter. Mol Endocrinol (1999) 13(5):729–41. doi:10.1210/me.13.5.729
129. Sandhoff TW, Hales DB, Hales KH, McLean MP. Transcriptional regulation of the rat steroidogenic acute regulatory protein gene by steroidogenic factor 1. Endocrinology (1998) 139(12):4820–31. doi:10.1210/en.139.12.4820
130. Zazopoulos E, Lalli E, Stocco DM, Sassone-Corsi P. DNA binding and transcriptional repression by DAX-1 blocks steroidogenesis. Nature (1997) 390(6657):311–5. doi:10.1038/36899
131. Chen H, Chu G, Zhao L, Yamauchi N, Shigeyoshi Y, Hashimoto S, et al. Rev-erbalpha regulates circadian rhythms and StAR expression in rat granulosa cells as identified by the agonist GSK4112. Biochem Biophys Res Commun (2012) 420(2):374–9. doi:10.1016/j.bbrc.2012.02.164
132. Kocerha J, Prucha MS, Kroll KJ, Steinhilber D, Denslow N. Regulation of steroidogenic acute regulatory protein transcription in largemouth bass by orphan nuclear receptor signaling pathways. Endocrinology (2010) 151(1):341–9. doi:10.1210/en.2009-0551
133. Albers HE. Gonadal hormones organize and modulate the circadian system of the rat. Am J Physiol (1981) 241(1):R62–6.
134. Hiroi H, Inoue S, Watanabe T, Goto W, Orimo A, Momoeda M, et al. Differential immunolocalization of estrogen receptor alpha and beta in rat ovary and uterus. J Mol Endocrinol (1999) 22(1):37–44. doi:10.1677/jme.0.0220037
135. Kuiper GG, Carlsson B, Grandien K, Enmark E, Häggblad J, Nilsson S, et al. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology (1997) 138(3):863–70. doi:10.1210/en.138.3.863
136. Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science (2012) 338(6105):349–54. doi:10.1126/science.1226339
137. Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet (2006) 38(3):369–74. doi:10.1038/ng1738
138. Hoffman YM, Peegel H, Sprock MJ, Zhang QY, Menon KM. Evidence that human chorionic gonadotropin/luteinizing hormone receptor down-regulation involves decreased levels of receptor messenger ribonucleic acid. Endocrinology (1991) 128(1):388–93. doi:10.1210/endo-128-1-388
139. LaPolt PS, Oikawa M, Jia XC, Dargan C, Hsueh AJ. Gonadotropin-induced up- and down-regulation of rat ovarian LH receptor message levels during follicular growth, ovulation and luteinization. Endocrinology (1990) 126(6):3277–9. doi:10.1210/endo-126-6-3277
140. Clark MR, Azhar S, Menon KM. Ovarian adenosine 3′:5′-cyclic monophosphate-dependent protein kinase(s). Regulation by choriogonadotropin and lutropin in rat ovarian cells. Biochem J (1976) 158(2):175–82.
141. Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell (2006) 124(1):35–46. doi:10.1016/j.cell.2005.12.022
142. Shimano H. Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Prog Lipid Res (2001) 40(6):439–52. doi:10.1016/S0163-7827(01)00010-8
143. Kash JC, Menon KM. Identification of a hormonally regulated luteinizing hormone/human chorionic gonadotropin receptor mRNA binding protein. Increased mrna binding during receptor down-regulation. J Biol Chem (1998) 273(17):10658–64. doi:10.1074/jbc.273.17.10658
Keywords: cholesterol, estrogens, signaling, circadian clocks, steroid hormones, regulation
Citation: Urlep Z and Rozman D (2013) The interplay between circadian system, cholesterol synthesis, and steroidogenesis affects various aspects of female reproduction. Front. Endocrinol. 4:111. doi: 10.3389/fendo.2013.00111
Received: 16 June 2013; Accepted: 13 August 2013;
Published online: 02 September 2013.
Edited by:
James Olcese, Florida State University College of Medicine, USAReviewed by:
Urs Albrecht, University of Fribourg, SwitzerlandCopyright: © 2013 Urlep and Rozman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Damjana Rozman, Center for Functional Genomics and Bio-Chips, Institute of Biochemistry, Faculty of Medicine, University of Ljubljana, Zaloska 4, SI-1000 Ljubljana, Slovenia e-mail:ZGFtamFuYS5yb3ptYW5AbWYudW5pLWxqLnNp
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.