
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 20 March 2012
Sec. Experimental Endocrinology
Volume 3 - 2012 | https://doi.org/10.3389/fendo.2012.00042
This article is part of the Research Topic Molecular mechanisms underlying insect behaviors: receptors, peptides, & biosynthetic pathways View all 16 articles
In most lepidopteran insects, the biosynthesis of sex pheromones is regulated by pheromone biosynthesis-activating neuropeptide (PBAN). Bombyx mori PBAN (BomPBAN) consists of 33 amino acid residues and contains a C-terminus FSPRLamide motif as the active core. Among neuropeptides containing the FXPRLamide motif, the arginine (Arg, R) residue at the second position from the C-terminus is highly conserved across several neuropeptides, which can be designated as RXamide peptides. The purpose of this study was to clarify the role of the Arg residue in the BomPBAN active core. We synthesized 10-residue peptides corresponding to the C-terminal part of BomPBAN with a series of replacements at the second position from the C-terminus, termed the C2 position, and measured their efficacy in stimulating Ca2+ influx in insect cells expressing a fluorescent PBAN receptor chimera (PBANR–EGFP) using the fluorescent Ca2+ indicator, Fura Red–AM. The PBAN analogs with the C2 position replaced with alanine (Ala, A), aspartic acid (Asp, D), serine (Ser, S), or L-2-aminooctanoic acid (Aoc) decreased PBAN-like activity. RC2A (SKTRYFSPALamide) and RC2D (SKTRYFSPDLamide) had the lowest activity and could not inhibit the activity of PBAN C10 (SKTRYFSPRLamide). We also prepared Rhodamine Red-labeled peptides of the PBAN analogs and examined their ability to bind PBANR. In contrast to Rhodamine Red-PBAN C10 at 100 nM, none of the synthetic analogs exhibited PBANR binding at the same concentration. Taken together, our results demonstrate that the C2 Arg residue in BomPBAN is essential for PBANR binding and activation.
Virgin female moths attract conspecific males using sex pheromone for species-specific mating. Pheromone biosynthesis and emission from the pheromone gland are activated during a calling period that is synchronized with the photoperiodic cycle through the function of pheromone biosynthesis-activating neuropeptide (PBAN), which is produced in the subesophageal ganglion (Kitamura et al., 1989; Raina et al., 1989; Raina, 1993; Rafaeli, 2002). In Bombyx mori, PBAN consists of 33 amino acid residues and contains as its active core a C-terminal FXPRLamide motif (X = S, T, G, or V) that is essential for PBAN activity (Kuniyoshi et al., 1992).
Several neuropeptides containing the FXPRLamide motif at the C-terminus have been identified from a number of insect orders including Lepidoptera, Diptera, and Orthoptera (Predel and Wegener, 2006). These peptides have been reported to regulate various biological activities such as initiation of B. mori embryonic diapause (Imai et al., 1991) and heliothine pupal (Xu and Denlinger, 2003), melanization of larvae in lepidopteran species (Matsumoto et al., 1990), hindgut/oviduct contraction in the cockroach and the locust (Nachman et al., 1986; Schoofs et al., 1992), and acceleration of pupariation in several flies (Zdarek et al., 1998). Moreover, PRX or RXamide (X = F, L, V, or Y) peptides have also been identified in many organisms as biologically active peptides (Predel and Wegener, 2006; Walker et al., 2009). These peptides have been widely identified from insect species (Baggerman et al., 2002; Hummon et al., 2006; Hauser et al., 2008; Weaver and Audsley, 2008). The presence of a C-terminal amide and an Arg residue at the second position from the C-terminus, termed the C2 position, in so many diverse insect neuropeptides suggests some functional importance.
The receptors for many of the peptides discussed above have been identified as G protein-coupled receptors (GPCRs). The insect GPCRs have been identified using cultured cells such as HEK293, CHO mammalian cells (Iversen et al., 2002; Yamanaka et al., 2008), and Sf9 insect cells (Choi et al., 2003; Hull et al., 2004). The PBAN receptor (PBANR) was first cloned from the corn earworm, Helicoverpa zea, as a homolog of the neuromedin U receptor (NmUR) in vertebrates that recognized FXPRNamide peptides (Fujii et al., 2000; Choi et al., 2003). Since then, PBANRs have been identified as class-A GPCR family members from B. mori, Heliothis virescens, Manduca sexta, and Plutella xylostella (Hull et al., 2004; Kim et al., 2008; Lee et al., 2011). In B. mori, PBAN binding has been shown in vitro using isolated pheromone glands to trigger an influx of extracellular Ca2+ in the pheromone producing cells (Mastumoto, 2010), as well as insect cells transiently expressing PBANR (Choi et al., 2003; Hull et al., 2004).
In the course of our study on the structure–activity relationship of PBAN and search for PBAN antagonists to control pests and to obtain a B. mori PBANR (BomPBANR)/ligand complex appropriate for crystallographic structural analysis, we established an in vitro PBAN assay system by observing Ca2+ influx into Sf9 cells, which had been prepared from immature ovaries of Spodoptera frugiperda (Vaughn et al., 1977), constitutively expressing BomPBANR–EGFP (Hull et al., 2011; Kawai et al., 2011). We first focused on the C6 residue and determined that a syntheticYC6N [SKTRNFSPRLamide] was a partial agonist (Kawai et al., 2010). Next, we prepared cyclic peptides, cycloYC6 [cyclo(TCYFSPRL)] and cycloNC6 [cyclo(TCNFSPRL)], from linear precursor peptides fused to an intein protein (Kimura et al., 2006) and determined that these partial peptides were also partial agonists (Kawai et al., 2011). These results showed that BomPBANR recognizes the amino acid residue at C6 position and type I β-turn secondary structure (Nachman et al., 1993). We also synthesized and examined a peptide substituted with D-Phe at C4 position [RYFdFPRLamide, LA-4] which had been reported as a linear antagonist (Zeltser et al., 2000; Ben-Aziz et al., 2005) and demonstrated that LA-4 was a partial agonist to BomPBANR (Kawai et al., 2009).
In this study, we focused on the Arg residue at the C2 position in the PBAN active core, because this residue is highly conserved in not only PBAN but also many other related neuropeptides. We synthesized four PBAN analogs consisting of 10 amino acid residues, in which the Arg residue was substituted with Ala, Asp, Ser, or L-2-aminooctanoic acid (Aoc), and assessed their in vitro PBAN-like activities by a Ca2+ imaging assay using stably transformed Sf9 cells expressing BomPBANR–EGFP and a confocal laser scanning microscope.
To investigate the effect of the Arg side chain on in vitro PBAN-like activity, we prepared four synthetic PBAN analogs, in which the Arg residue at the C2 position was substituted with amino acid residues exhibiting different side chain properties (Figure 1A). For this purpose, we selected Ala, Asp, Ser, and Aoc for the following reasons: Ala has a simple small side chain that lacks a functional group, Asp has a negatively charged carboxyl group, Ser has a hydroxyl group with an uncharged polarity, while the Aoc side chain is similar in length to the Arg side chain but lacks any functional groups. To determine the agonistic activities of the Ala (RC2A), Asp (RC2D), Ser (RC2S), and Aoc (RC2Aoc) analogs, we compared the Ca2+ response of the respective analogs with a corresponding PBAN analog composed of the C-terminal 10 residues (PBAN C10). Sf9 cells stably expressing BomPBANR–EGFP were loaded with Fura Red–AM, a fluorescent Ca2+ indicator, and their fluorescent profile in response to the each ligand was examined with a confocal laser scanning microscope (Kawai et al., 2011). As shown in Figure 1B, the PBAN-like activities of all of the RC2X peptides were much lower than that of PBAN C10 at 1 nM. RC2A and RC2D showed full PBAN-like activity at 100 nM, while RC2S and RC2Aoc showed full activity at 10 nM.
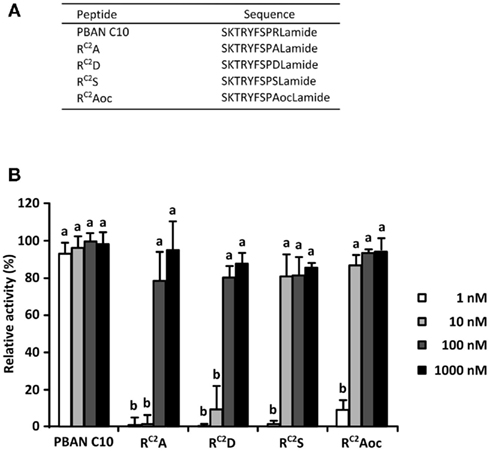
Figure 1. Pheromone biosynthesis-activating neuropeptide-like activities of RC2X peptides. (A) Amino acid sequences of the RC2X peptides. (B) Dose dependency of PBAN-like effects of the RC2X peptides measured via a Ca2+ imaging technique with Fura Red as the Ca2+ indicator. Bars represent the mean + SD of five replicates. Different letters (a and b) indicate significant difference at P < 0.05 by the Tukey–Kramer test.
Because RC2A and RC2D showed lower PBAN-like activities than the other two mutations, we also examined their antagonistic activities against PBAN C10.We found that the activity of PBAN C10 at 1 nM decreased to 87 and 85% following the addition of a 10-fold concentration of RC2A and RC2D, respectively (Figure 2), although this reduction in activity was not statistically significant.
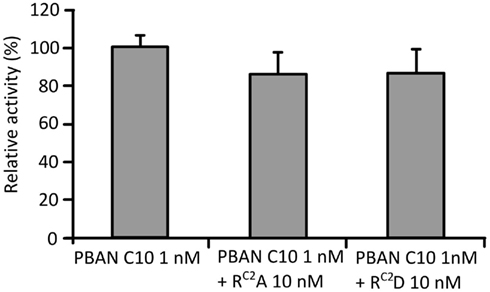
Figure 2. Antagonistic effects of RC2A and RC2D. Effects were measured using the Ca2+ imaging technique with Fura Red as the Ca2+ indicator. PBAN C10 was added after the addition of RC2A and RC2D. Bars represent the mean + SD of five replicates.
To confirm the binding of the RC2X mutations to BomPBANR–EGFP, we prepared fluorescent analogs of the synthetic peptides via conjugation with Rhodamine Red-X succinimidyl ester. As before, we used Sf9 cells stably expressing BomPBANR–EGFP to examine binding of each labeled peptide, Sf9 cells were imaged using a confocal laser scanning microscope using excitation at 488 and 560 nm for EGFP and Rhodamine Red-X, respectively. The binding intensity was assessed by the fluorescence ratio of Rhodamine Red-X to EGFP. Figure 3 shows the results. The expression of BomPBANR–EGFP on the cell surface was visualized as a ring of green fluorescence by EGFP in all cases. It was clearly demonstrated that PBAN C10 bound to BomPBANR–EGFP as observed as a red fluorescent ring on the cell surface, which was merged with green fluorescence to be yellow. However, the binding of the RC2X peptides to BomPBANR–EGFP could not be detected clearly.
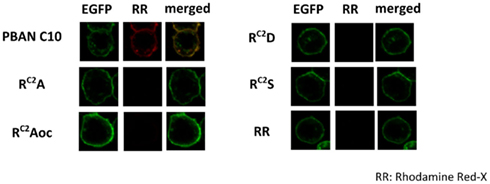
Figure 3. Confocal imaging of BomPBANR–EGFP expressing Sf9 cells after incubation with synthetic peptides labeled with Rhodamine Red-X. Each fluorescence labeled peptide was added at a final concentration of 100 nM. Co-localization is indicated by yellow in the merged images.
Neuropeptides mediate various biological events in both vertebrates and invertebrates. Among them, there is a group of neuropeptides that share chemical characteristics of C-terminal amidation and the presence of Arg at the C2 position. PBAN, an insect neuropeptide, belongs to this group. In this study, we sought to clarify the role of the Arg residue in PBAN activity. Instead of an ordinary in vivo PBAN assay using female moths, we adopted a bioassay assessing PBAN-like activity in vitro by using insect (Sf9) cells artificially expressing BomPBANR–EGFP, which we have previously established (Mastumoto, 2010; Kawai et al., 2011). We synthesized and examined four synthetic peptides, in which Arg at the C2 position was replaced by each of four representative amino acid residues with different chemical properties from Arg.
The activity of RC2A was significantly lower than PBAN C10, which is consistent with previous results demonstrating the effect of Ala substitutions of this position in a Heliothis virescens PBAN C-terminal hexapeptide (Kim et al., 2008). It has also been reported that the substitution of Arg at the C2 position of neuropeptide Y, a vertebrate neuropeptide, with Ala reduced the affinity to both human Y1 and Y2 receptors (Beck-Sickinger et al., 1994). In addition, the reduced in vivo PBAN activity was reported following Gln, Lys, or norvaline substitution of Arg at the C2 position in B. mori (Kuniyoshi et al., 1991). While these results suggest the potential importance of the Arg guanidine group, the length of the side chain may also influence the activity. We consequently prepared a peptide with Aoc at the C2 position because Aoc has a side chain of similar length to Arg but without any functional groups, thus making it hydrophobic. Unexpectedly, the RC2Aoc peptide, showed higher activity than RC2A (Figure 1B). While the guanidino functional group is important for binding, it is apparent that the length of the side chain is also important.
Concerning the hydrophilicity of the side chain, we compared the activity of RC2A with that of RC2S. RC2S was found to be more active than RC2A, indicating some contribution of the hydroxyl group. However, the increased bulkiness of the Ser hydroxymethyl group over the Ala methyl group may also contribute to some extent. RC2S was a little less active than RC2Aoc, indicating the side chain length is more important than hydrophilicity. RC2D, which carried a charge opposite to the natural peptide, showed activity stronger than RC2A, but was weaker than RC2S, indicating that a negative charge is not preferable for binding to the receptor.
Neither RC2A nor RC2D was able to inhibit the activity of 1 nM PBAN C10 even at a 10-fold higher concentration (10 nM), at which concentration that RC2A and RC2D exhibited almost no and very low agonistic activity, respectively. This result suggests that both RC2A and RC2D bind very weakly to BomPBANR; it is unlikely that they bind BomPBANR as strongly as PBAN C10 but cannot trigger the Ca2+ influx. The weak binding of the four synthetic analogs was directly demonstrated by observation of the Rhodamine Red-labeled conjugates. We therefore concluded that the Arg residue at C2 position is important for PBANR binding. Although the four mutants exhibited PBAN-like activity comparable to PBAN C10 at 100 nM, we failed to observe any binding at that concentration, perhaps because the Rhodamine Red-labeled analogs were weakly bound and removed during the final washing steps prior to imaging. It is unlikely that the Rhodamine Red-labeled analogs have lower activity than non-labeled peptides, because the possible labeling sites are the ε-amino group of Lys at the C9 position and the N-terminal amino group, which are far from the active core. Experiments at higher concentrations of labeled peptides will be needed.
Pheromone biosynthesis-activating neuropeptide analogs were synthesized with an automated peptide synthesizer Apex 369 (AAPPTec, Louisville, KY, USA) using the Fmoc solid-phase strategy according to the manufacturer’s instructions. The Arg residue at the C9 position of all PBAN analogs was replaced with Lys for labeling with Rhodamine Red-X succinimidyl ester. After deprotection and cleavage from the resin, synthetic peptides were purified by HPLC with a PEGASIL ODS column (10 i.d. × 150 mm, Senshu Kagaku, Tokyo, Japan) using two solvents, 0.05% trifluoroacetic acid (TFA; solvent A) and 90% CH3CN/0.05% TFA (solvent B). Peptides were eluted at a flow rate of 4.0 ml/min using a step-gradient program starting from 0% B in 2.5 min, then to 75% B in 15 min, and then 100% B over the next 30 min. Reagents for peptide synthesis were purchased from WATANABE CHEMICAL IND., LTD (Hiroshima, Japan). Synthetic peptides were lyophilized and stored at −20°C until use.
A 200-μl aliquot containing 250 μg Rhodamine Red succinimidyl ester (Life technologies, Tokyo, Japan) was added to a solution of 0.1 M sodium bicarbonate (pH 8.2) and 1.0 μmol synthetic peptide. The mixture was stirred overnight at room temperature and the conjugated peptide purified by HPLC under the same conditions as above. RR-C10 PBAN and RR–RC2X were stored at 4°C until use.
An expression plasmid encoding the BomPBANR fused with EGFP at its C-terminus was produced using a pIB/V5-His TOPO TA cloning kit (Life Technologies). Sf9 cells in IPL-41 insect cell medium (Life Technologies) supplemented with 10% fetal bovine serum (FBS) were transfected with the expression plasmid using Cellfectin (Life Technologies). Transfected Sf9 cells were selected using blasticidin S according to the manufacturer’s procedures.
BomPBANR–EGFP stably expressing cells were harvested and incubated in 24 well glass bottom plate (AGC, Tokyo, Japan) at 28°C for 2 days with 10% FBS in IPL-41. On the day of experiment, cells were washed three times with IPL-41 and incubated in 250 μl of IPL-41 with 0.75 μl of Pluronic F-127 and 1 mM Fura Red AM (Life Technologies) for 30 min in the dark. After incubation, the cells were washed three times with IPL-41, and 300 μl IPL-41 was added to each well. The plate was left in the dark for 20 min to allow hydrolysis of the Fura Red AM ester bond. EGFP and Fura Red fluorescence were obtained on a FV-1000D confocal laser microscope (OLYMPUS, Tokyo, Japan) using 488 and 548 nm laser lines, respectively. To measure agonistic activity, fluorescence was monitored for 40 scans (1.08 s/scan), and a 100-μl peptide solution which were prepared to the concentration of 1, 10, 100, and 1000 nM was added to the well after 10 scans. For measuring antagonistic activity of RC2A and RC2D, the fluorescence was monitored for 50 scans. RC2A and RC2D solution (100 μl) was added to the well after 10 scans, and then 100 μl PBAN C10 was added after 25 scans. The Fura Red fluorescence was analyzed using the FV-1000D software “FluoView” (OLYMPUS). The data obtained were statistically analyzed using a Tukey–Kramer test. These experiments were performed with five replicates.
The binding ability of Rhodamine Red–RC2X peptides (X = Ala, Asp, Ser, and Aoc) to BomPBANR–EGFP was analyzed using a confocal laser scanning microscope. Sf9 cells adherently cultured on 35 mm glass bottom dishes (AGC, Tokyo, Japan) were transfected with 5 μg of expression plasmid containing BomPBANR–EGFP. Sixteen hours after transfection, the Sf9 cells were washed with 10% FBS/IPL-41 containing kanamycin at the final concentration of 200 μg/ml and incubated for 1 day at 28°C. On the day of the experiment, transfected cells were washed with serum free IPL-41 medium and incubated at 4°C for 15 min. To examine the binding of the labeled peptides to BomPBANR–EGFP, transfected cells were incubated in 500 μl IPL-41 containing each RR-peptide at the concentration of 100 nM at 4°C for 60 min in the dark, and then fixed with 4% formaldehyde/PBS. The fluorescence of BomPBANR–EGFP and RR-peptides were obtained on a FV-1000D confocal laser scanning microscope with 488 and 548 nm laser lines, respectively.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was supported by Targeted Proteins Research Program (TPRP) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Baggerman, G., Cerstiaens, A., De Loof, A., and Schoofs, L. (2002). Peptidomics of the larval Drosophila melanogaster central nervous system. J. Biol. Chem. 277, 40368–40374.
Beck-Sickinger, A. G., Wieland, H. A., Willim, K.-D., Rudlf, K., and Jung, G. (1994). Complete L-alanine scan of neuropeptide Y reveals ligands binding to Y1 and Y2 receptors with distinguished conformations. Eur. J. Biochem. 225, 947–958.
Ben-Aziz, O., Zeltser, I., and Altstein, M. (2005). PBAN selective antagonists: inhibition of PBAN induced cuticular melanization and sex pheromone biosynthesis in moth. J. Insect Physiol. 51, 305–314.
Choi, M. Y., Fuerst, E. J., Rafaeli, A., and Jurenka, R. (2003). Identification of a G protein-coupled receptor for pheromone biosynthesis activating neuropeptide from pheromone glands of the moth Helicoverpa zea. Proc. Natl. Acad. Sci. U.S.A. 100, 9721–9726.
Fujii, R., Hosoya, M., Fukusumi, S., Kawamata, Y., Habata, Y., Hinuma, S., and Onda, H. (2000). Identification of neuromedin U as the cognate ligand of the orphan G protein-coupled receptor FM-3. J. Biol. Chem. 275, 21068–21074.
Hauser, F., Cazzamali, G., Williamson, M., Park, M., Li, B., Tanaka, Y., Predel, R., Neupert, S., Schachtner, J., Verleyen, P., and Grimmelikhuijzen, C. J. P. (2008). A genome-wide inventory of neurohormone GPCRs in the red flour beetle Tribolium castaneum. Front. Neuroendocrinol. 29, 142–165.
Hull, J. J., Lee, H. M., and Matsumoto, S. (2011). Identification of specific sites in the third intracellular loop and carboxyl terminus of the Bombyx mori pheromone biosynthesis activating neuropeptide receptor crucial for ligand-induced internalization. Insect Mol. Biol. 20, 801–811.
Hull, J. J., Ohnisi, A., Moto, K., Kawasaki, Y., Kurata, R., Suzuki, M. G., and Matsumoto, S. (2004). Cloning and characterization of the pheromone biosynthesis activating neuropeptide receptor from the silkmoth, Bombyx mori: significance of the carboxyl terminus in receptor internalization. J. Biol. Chem. 279, 51500–51507.
Hummon, A. B., Richmond, T. A., Verleyen, P., Baggerman, G., Huybrechts, J., Ewing, M. A., Vierstrate, E., Rodriguez-Zas, S. L., Schoofs, L., Robinson, G. E., and Sweedler, J. V. (2006). From the genome to the proteome: uncovering peptides in the Apis brain. Science 314, 647–649.
Imai, K., Konno, T., Nakazawa, T., Komiya, T., Isobe, M., Koga, K., Goto, T., Yaginuma, T., Sakakibara, K., Hasegawa, K., and Yamashita, O. (1991). Isolation and structure of diapause hormone of the silkworm, Bombyx mori. Proc. Jpn. Acad. 67B, 98–101.
Iversen, A., Cazzamali, G., Williamson, M., Hauser, F., and Grimmelikhuijzen, C. J. P. (2002). Molecular cloning of functional expression of a Drosophila receptor for the neuropeptides capa-1 and -2. Biochem. Biophys. Res. Commun. 299, 628–633.
Kawai, T., Hull, J. J., Matsumoto, S., Nagata, J., Tanokura, M., and Nagasawa, H. (2010). “Studies on the structure-activity relationship of pheromone biosynthesis-activating neuropeptide (PBAN),” in Peptide Science 2009: Proceedings of the 46th Japanese Peptide Symposium, ed. K. Okamoto (Osaka: The Japanese Peptide Society Press), 231–234.
Kawai, T., Nagata, K., Okada, K., Hayakawa, K., Hull, J. J., Lee, H. M., Matsumoto, S., Tanokura, M., and Nagasawa, H. (2011). “Structure-activity relationship studies of the pheromone biosynthesis-activating neuropeptide of the silkworm, Bombyx mori,” in Peptide science 2010: Proceedings of the Fifth International Peptide Symposium, eds N. Fujii, and Y. Kiso (Osaka: The Japanese Peptide Society Press), 126.
Kawai, T., Sugisaka, A., Hull, J. J., Matsumoto, S., Nagata, K., Tanokura, M., and Nagasawa, H. (2009). “Development of a novel bioassay system for pheromone biosynthesis-activating neuropeptide (PBAN) using the Bombyx receptor expressed in insect cell,” in Peptide Science 2008: Proceedings of the 45th Japanese Peptide Symposium, ed. M. Nomizu (Osaka: The Japanese Peptide Society Press), 287–288.
Kim, Y. J., Nachman, R. J., Aimanova, K., Gill, S., and Adams, M. E. (2008). The pheromone biosynthesis activating neuropeptide (PBAN) receptor of Heliothis virescens: identification, functional expression, and structure-activity relationships of ligand analogs. Peptides 29, 268–275.
Kimura, R. H., Tran, A.-T., and Camaero, J. A. (2006). Biosynthesis of the cyclotide Kalata B1 by using protein splicing. Angew. Chem. Int. Ed. Engl. 45, 973–976.
Kitamura, A., Nagasawa, H., Kataoka, H., Inoue, T., Ando, T., and Suzuki, A. (1989). Amino acid sequence of pheromone biosynthesis-activating neuropeptide (PBAN) of the silkworm, Bombyx mori. Biochem. Biophys. Res. Commun. 163, 520–526.
Kuniyoshi, H., Kitamura, A., Nagasawa, H., Chuman, T., Shimazaki, K., Ando, T., and Suzuki, A. (1991). “Structure-activity relationship of pheromone biosynthesis activating neuropeptide (PBAN) from the silkmoth, Bombyx mori,” in Peptide Chemistry 1990: Proceedings of the28th Symposium on Peptide Chemistry, ed. Y. Shimonishi (Osaka: The Japanese Peptide Society Press), 251–254.
Kuniyoshi, H., Nagasawa, H., Ando, T., Suzuki, A., Nachman, R. J., and Holman, G. M. (1992). Cross-activity between pheromone biosynthesis activating neuropeptide (PBAN) and myotropic pyrokinin insect peptides. Biosci. Biotechnol. Biochem. 56, 167–168.
Lee, D. W., Shrestha, S., Kim, A. Y., Park, S. J., Yang, C. Y., Kim, Y., and Koh, Y. H. (2011). RNA interference of pheromone biosynthesis-activating neuropeptide receptor suppresses mating behavior by inhibiting sex pheromone production in Plutella xylostella (L.). Insect Biochem. Mol. Biol. 41, 236–243.
Mastumoto, S. (2010). Molecular mechanisms underlying sex pheromone production in moths. Biosci. Biotechnol. Biochem. 74, 223–231.
Matsumoto, S., Kitamura, A., Nagasawa, H., Kataoka, H., Orikasa, C., Mitsui, T., and Suzuki, A. (1990). Functional diversity of a neurohormone produced by the suboesophageal ganglion: molecular identify of melanization and reddish coloration hormone and pheromone biosynthesis activating neuropeptide. J. Insect Physiol. 53, 752–759.
Nachman, R. J., Holman, G. M., and Cook, B. J. (1986). Active fragments and analogs of the insect neuropeptide leucopyrokinin; structure-function studies. Biochem. Biophys. Res. Commun. 137, 936–942.
Nachman, R. J., Kuniyoshi, H., Roberts, V. A., Holman, G. M., and Suzuki, A. (1993). Active conformation of the pyrokinin/PBAN neuropeptide family for pheromone biosynthesis in the silkworm. Biochem. Biophys. Res. Commun. 193, 661–666.
Predel, R., and Wegener, C. (2006). Biology of the CAPA peptides in insects. Cell. Mol. Life Sci. 63, 2477–2490.
Rafaeli, A. (2002). Neuroendocrine control of pheromone biosynthesis in moths. Int. Rev. Cytol. 213, 41–49.
Raina, A., Jaffe, K., Kempe, T. G., Keim, P., Blacher, R. W., Fales, H. M., Riley, C. T., Klun, J. A., Ridgway, R. L., and Hayes, D. K. (1989). Identification of neuropeptide hormone that regulates sex pheromone production in female moth. Science 244, 796–798.
Raina, A. K. (1993). Neuroendocrine control of sex pheromone biosynthesis in lepidoptera. Annu. Rev. Entomol. 38, 329–349.
Schoofs, L., Holman, G. M., Hayes, T. K., Nachman, R. J., Kochansky, J. P., and Deloof, A. (1992). Isolation identification and synthesis of locastamyotropin III and IV, two additional neuropeptides of Locusta migratoria; members of the locustamyotropin family. Insect Biochem. Mol. Biol. 22, 447–452.
Vaughn, J. L., Goodwin, R. H., Tompkins, G. J., and McCawley, P. (1977). The establishment of two cell lines from the insect Spodoptera frugiperda (lepidoptera: noctuidae). In vitro 13, 213–217.
Walker, R. J., Papaioannou, S., and Holden-Dye, L. (2009). A review of FMRFamide- and RFamide-like peptides in metazoan. Invert. Neurosci. 9, 111–153.
Weaver, R. J., and Audsley, N. (2008). Neuropeptides of the beetle, Tenebrio molitor, identified using MALDI-TOF mass spectrometry and deduced sequences from the Tribolium castaneum genome. Peptides 29, 168–178.
Xu, W.-H., and Denlinger, D. L. (2003). Molecular characterization of prothoracicotropic hormone and diapause hormone in Heliothis virescens during diapauses, and a new role for diapauses hormone. Insect Mol. Biol. 12, 509–516.
Yamanaka, N., Ymamoto, S., Zitnan, D., Watanabe, K., Kawada, T., Satake, H., Kaneko, Y., Himura, K., Tanaka, Y., Shinoda, T., and Kataoka, H. (2008). Neuropeptide receptor transcriptome reveals unidentified neuroendcrine pathway. PLoS ONE 3, e3048. doi:10.1371/journal.pone.0003048
Zdarek, J., Nachman, R. J., and Hayes, T. K. (1998). Structure-activity relationships of insect neuropeptides of the pyrokinin/PBAN family and their selective action on pupariation in fresh fly (Neobelleria bullata) larvae (diptera: sarcophagidae). Eur. J. Entomol. 95, 9–16.
Keywords: FXPRLamide peptides, GPCR, Lepidoptera, neuropeptide, PBAN, PBANR, sex pheromone silkmoth
Citation: Kawai T, Lee JM, Nagata K, Matsumoto S, Tanokura M and Nagasawa H (2012) The arginine residue within the C-terminal active core of Bombyx mori pheromone biosynthesis-activating neuropeptide is essential for receptor binding and activation. Front. Endocrin. 3:42. doi: 10.3389/fendo.2012.00042
Received: 09 November 2011; Accepted: 05 March 2012;
Published online: 20 March 2012.
Edited by:
Joe Hull, USDA Agricultural Research Service, USAReviewed by:
Adrien Fónagy, Plant Protection Institute of Hungarian Academy of Sciences, HungaryCopyright: © 2012 Kawai, Lee, Nagata, Matsumoto, Tanokura and Nagasawa. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Hiromichi Nagasawa, Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1, Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan. e-mail:YW5hZ2FoaUBtYWlsLmVjYy51LXRva3lvLmFjLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.