- 1College of Geographical Science, Harbin Normal University, Harbin, China
- 2Institute of Nature and Ecology, Heilongjiang Academy of Sciences, Harbin, China
- 3Heilongjiang Academy of Forestry, Harbin, China
Soil bacteria are a crucial component of forest soil biodiversity and play important functions in numerous ecosystem processes. Hence, studying the variation of diversity and composition of soil bacteria between latitude gradients and the driving factors responsible for these differences is important for understanding the changes of soil bacteria. We used Illumina MiSeq sequencing of bacterial 16S rRNA to investigate the distribution pattern and driving factors of bacterial diversity and composition in temperate forest soils at three different latitudes in northeast China, with samples taken at low, middle and high latitude. Each sample area was located at a distance of 1,200 km. Our results indicate that the soil bacterial diversity decreased with increasing latitude. Members of the phyla Acidobacteria and Proteobacteria were the dominant in all investigated soils, the highest relative abundances of these phyla were: Acidobacteria and Proteobacteria in high-latitude forest, Rokubacteria and Actinobacteria in low-latitude forest. The dominant bacterial genera in the three different latitude forests were Candidatus_Solibacter, Bryobacter, Roseiarcus and Granulicella. Mean average temperature, soil pH and total nitrogen content were the key environmental factors shaping the soil bacterial diversity and composition in different latitudes of these temperate forests. The results of this study contribute to a deeper understanding and better predictions the latitudinal pattern of soil biodiversity.
1 Introduction
Soil microorganisms represent some of the most abundant and diverse biological populations on Earth, and play an important role in material conversion, energy flow, and biogeochemical cycling (Cardinale et al., 2015; Bahram et al., 2018; Deng et al., 2020). Understanding the distribution patterns of soil microorganism on a spatial scale is a central goal of ecology (Wang et al., 2015). However, compared with macrobiology, the distribution and driving mechanisms of soil microorganisms is still limitedly investigated (Fuhrman, et al., 2008; Zhou et al., 2016). It has been reported that soil microorganisms produce a random distribution pattern on a global scale (Finlay, 2002), although on a local or regional scale, regular, spatial distribution patterns of soil microbial community structures have been observed (Shen et al., 2014; Sun et al., 2021; Ji et al., 2022). However, the conclusions of previous studies, performed at different scales, are inconsistent (Tedersoo et al., 2014; Neu et al., 2021). Even under the same environmental conditions can the distribution of soil microorganisms be inconsistent (Han et al., 2015; Li et al., 2019; Tian et al., 2022). Therefore, further studies are needed to reveal the driving mechanism of changes in the spatial distribution of soil microorganisms.
Forest ecosystems represent highly important terrestrial ecosystems with a rich species diversity, which plays an important role in the material cycle and energy flow (Andrew, 2020; Hu et al., 2021; Chen et al., 2022). Soil microorganisms are highly abundant and diverse in forest soils. Understanding their community composition and diversity, predicting dynamic changes, and exploring the ecosystem’s response mechanism to habitat changes, all have great significance on the protection of forest ecosystems and their stability (Chen et al., 2020; Chen et al., 2022; Zhou et al., 2022). Tripathi et al. (2014) showed that environmental factors played a decisive role in the variation of soil microorganisms community structure and diversity, among the environmental factors, pH had the most significant effect followed by total nitrogen and altitude (Tripathi et al., 2014). Temperature also regulates the diversity of microorganisms in forest soil on a continental scale, as was demonstrated by Zhou et al. (2016) who investigated a temperature gradient showed it affected the diversity of different microbiota variably. Tian et al. (2018) studied microorganisms in forest soil at different latitudes (including tropical, subtropical, and temperate forest), and found that soil bacterial diversity was significantly higher in temperate forests than the others. In that study, climate factors were the main driving factors of forest soil bacterial community structure, together with pH and organic matter, but vegetation diversity had relatively little impact on forest soil bacterial community structure (Tian et al., 2018). Although studies have focused on the spatial and temporal distribution patterns of soil microorganisms and their main driving factors, a unified conclusion has not been reached, especially due to the lack of large-scale data and the lack of universal rules, that hinder an accurate prediction of soil bacterial diversity changes and the development of ecological theories (Zhang et al., 2020; Sun et al., 2021). Therefore, there is an urgent need to study the distribution and driving mechanism of soil microorganisms in a more systematic way on a large scale.
Along latitude gradients, the geographic variation can be associated with changes in environmental factors such as temperature, precipitation, vegetation type, and soil properties (Yang et al., 2019; Zheng et al., 2021). These provide a natural experimental platform for assessing how soil bacterial communities respond to environmental change (Tian et al., 2018). There remains controversy regarding the patterns of changes in soil bacterial diversity and community structure with increasing latitude, with available works describing patterns with declining (Zhou et al., 2016), humped (Tian et al., 2018) or stepped (Lee et al., 2018) trends of soil bacterial diversity. Furthermore, Zhang et al. (2020) studied the variation of soil bacterial community diversity in 11 latitudinal gradients in eastern China and considered the variation of bacterial diversity patterns could be explained by a local community aggregation mechanism. The inconsistent conclusions of these studies may be caused by the different response patterns of soil bacteria to latitudinal changes in different vegetation types and ecosystems. Bacterial community composition significantly corresponds to vegetation and habitat selection (Srivastava et al., 2021), and therefore the composition and diversity of bacterial communities are strongly influenced by the composition of above-ground vegetation and soil environmental conditions (Sui et al., 2021). As the most representative forest type in a certain area, typical forests are often used as the research objects to explain the spatial distribution characteristics of soil (Lee et al., 2018), plants (Liu et al., 2022) and microorganisms communities (Gaytán et al., 2022) in different latitude forests. There are various temperate forest ecosystems in northeast China, which create an important barrier to the agricultural development of the Northeast Great Plain and play an important ecological role in the maintenance and regulation of the regional climate (Zheng et al., 2018; Sang et al., 2021). The typical forests in this area are cold temperate Larix gmelinii forest, temperate Coniferous-broad Leaved Korean pine Mixed Forest and warm temperate broadleaf Quercus forest (Jia et al., 2019; Chen et al., 2022). Patterns of plants and animals decreasing with increasing latitude have been well documented (Berdugo et al., 2018; Yu et al., 2021), but it is unknown whether microorganisms exhibit similar latitudinal diversity gradients (Zhou et al., 2016). This study was performed to fill in this knowledge gap.
Three typical temperate forest areas were selected from the China Forest Biodiversity Monitoring Network to investigate the spatial distribution pattern of soil bacterial community composition and diversity by high-throughput sequencing. We aim to elaborate: (1) the distribution pattern of soil bacterial community in the three different latitudes forest ecosystems. (2) the key environmental factors driving the changes of soil bacterial community and diversity in three different latitudes forest ecosystems. The obtained results can not only promote the development of biogeography, but also provide a theoretical basis for the improvement of forest latitudinal diversity patterns. At the same time, it is conducive to the functional regulation and management of terrestrial ecosystems, so as to better cope with important environmental problems such as global warming.
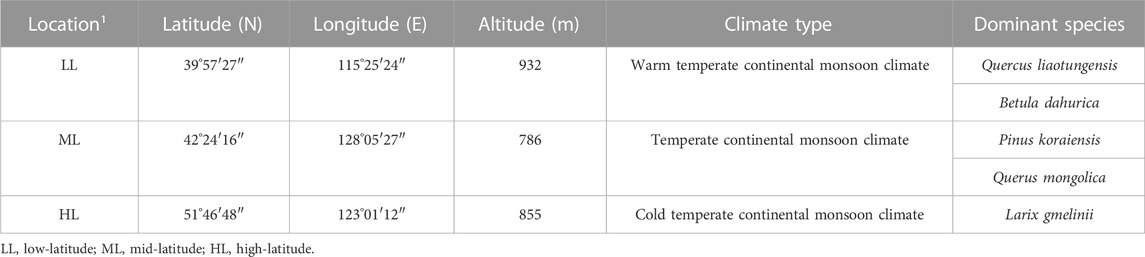
2 Materials and Method
2.1 Site description
The study areas covered a longitude range of 115°–129°E and latitude range 39°–54°N, and included a warm, a moderate, and a cold temperate zone, with altitudes ranging from 786 m to 932 m. The average annual precipitation ranges from 383.53 mm to 836.23 mm. Three sampling areas were identified in each zone of low, middle, and high latitude. The survey was mainly conducted in national nature reserves or scientific research bases, away from habitation, and mature forests with good preservation and strong regional representation were selected. The specific areas of low latitude to high latitude included: Long-term Biodiversity Monitoring Plot of Dongling Mountain in Beijing (LL): The experimental samples were located in Beijing Forest Ecosystem 20 hm2 Research Station, Chinese Academy of Sciences, five 20 m × 20 m standard quadrates were selected, and each quadrate was 100 m apart, the vegetation type of the sample plot is Quercus liaotungensis forest; Changbai Mountain National Nature Reserve of Jilin Province (ML): The experimental samples were located in the forest monitoring plots of 25 hm2 in Changbai Mountain, five 20 m × 20 m standard quadrates were selected, and each quadrate was 100 m apart, the vegetation type of the sample plot are Quercus mongolica and Pinus koraiensis mixed forest, and Huzhong National Nature Reserve in Daxing’anling, Heilongjiang Province (HL): the experimental samples were located in the forest monitoring plots of 25 hm2 in Daxing’anling, five 20 m × 20 m standard quadrates were selected, and each quadrate was 100 m apart, the vegetation type of the sample plot is Larix gmelinii forest. A summary of these three research areas is shown in Table 1.
2.2 Soil sampling
From each of the three areas, forest soil samples were acquired in July 2020. For this, standard experimental plots were set up in the core areas of the national nature reserves (see Table 1). Within each forest type, five sampling plots were established, and within each plot soil samples (0–20 cm) were collected using an 8 cm diameter soil auger from 15–20 locations along an S-shaped path. Before sampling, superficial debris (e.g., leaves and dry vegetation) was removed. The collected soil samples were transferred to sterile seal bags and stored in an icebox. Following transport to the laboratory, the soil was immediately sieved (2 mm mesh) to remove stones and plant material. One portion of the samples was air-dried to conduct physical and chemical analyses, whereas the remainder was kept at −80°C until required for microbial analysis.
2.3 Characterization of soil chemical parameters
A soil-water (1:2.5 w/v) suspension was shaken for 30 min prior to measuring the pH with a pH meter. The soil total N (TN) and organic C (SOC) were quantified using an Elemental Analyzer (Elementar, Langenselbold, Germany). Available nitrogen (AN) was sequentially digested in H2SO4–HClO4, 0.5 M NaHCO3, and 2.0 M KCl, and then examined with a continuous flow analysis system (SKALAR SAN++, the Netherlands). After wet digestion with HClO4–H2SO4, total phosphorus (TP) was determined spectrophotometrically using standard procedures.
2.4 DNA extraction, PCR amplification, and MiSeq sequencing
Total DNA of soil microorganisms was extracted using the QIAGEN DNeasy PowerSoil Pro Kit according to the manufacturer’s instructions. The V3, V4 region of the bacterial 16S rRNA region was amplified using universal primers 338 F and 806 R. Each sample was tagged with a different six bp barcode present at the 5′end of the primer. The PCR amplification protocol consisted of an initial denaturation stage at 94°C for 10 min, followed by 30 cycles of 90°C for 60 s, 55°C for 60 s, and 72°C for 60 s, and final extension at 72°C for 10 min. The product was purified using the QIAquick PCR Purification Kit (QIAGEN). The PCR products of different samples were sequenced by BMC Biotechnology Co., Ltd., using an Illumina MiSeq sequencer.
The raw sequences were processed using QIIME1. Forward and reverse reads were merged using FLASH software. ‘Trim’ was used to remove low-quality sequences shorter than 150 bp, reads with a mean quality score below 20, and sequences containing ambiguous nucleotides. Chimeric sequences were identified and removed with the uchime algorithm. A similarity of 97% was applied to divide the reads into operational taxonomic units (OTU) through UPARSE (V.7.0.1090). Operational taxonomic identities were determined using QIIME by executing the BLAST algorithm against sequences in the SILVA database (https://www.arb-silva). Diversity indices Chao1, ACE, Shannon and Simpson were calculated using ‘alpha_diversity.py’ to reflect the a diversity of the bacterial community, and ‘beta_diversity.py’ was used to calculate the Bray-Curtis distance matrix to reflect the ß diversity.
2.5 Statistical analyses
Differences in soil physicochemical properties between the three treatments were analyzed using one-way ANOVA at a 0.05 significance level coupled with Duncan’s tests, using SPSS 23.0 (IBM SPSS Statistics for Windows). OTU- level a diversity indices, such as Chao1 index, ACE index, Shannon index and Simpson index, were calculated using the OTU table in QIIME1. Venn diagrams and heatmap representations of the relative abundances of bacterial OTUs among soil samples were generated using the “vegan” package in R. Non-metric multidimensional scaling (NMDS) was conducted based on the Bray—Curtis dissimilarity at the OTU level, also using “vegan” (Frey et al., 2021). Redundancy analysis (RDA), implemented in “vegan”, was used to generate compositional profiles, agglomerating the OTUs to the phylum level and genus level. Indicator species analysis based on the OTU level was performed as outlined in Rime et al. (2015) using “vegan” and the “labdsv” (version 1.2–2) R package.
3 Results
3.1 Forest soil chemical properties at different latitudes
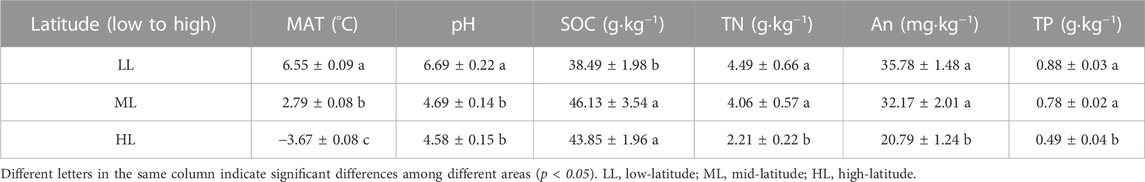
The determined chemical properties of the soil collected at three different latitudes is summarized in Table 2. The mean annual temperature (MAT) decreased with increasing latitude in the three locations. The pH and content of SOC, TN, AN and AP differed as well. The SOC content was lower and the soil pH was higher at the lowest latitude but they did not differ for the other two locations. Contents of TN, AN and AP values were lower for the highest latitude than for the other two locations.
3.2 Latitude variation pattern of forest soil bacterial a- and ß-diversities
Analysis of 16S rDNA gene sequences in the 15 samples revealed information on the structures of the bacterial communities. The vast majority of OTUs were detected at all three latitudes (n=1,267), but unique and partially shared OTUs were recognized as well, as visualized in a Venn diagram (Figure 1). There were 1,472 identified bacterial OTUs recognized in HL, 1,591 in ML and 1,507 in LL, and these soils contained 44, 11 and 34 unique OTUs, respectively (Figure 1). The fewest number of OTUs were exclusively shared by LL and HL (n=27), whose locations were most distant, and the most OTUs were exclusively shared by ML and LL (n=179).
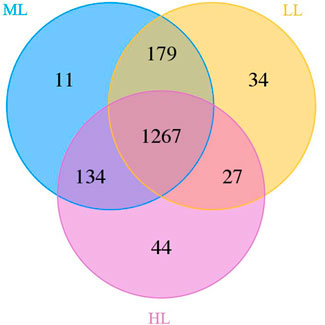
FIGURE 1. Venn diagram of soil bacterial OTUs. LL, low-latitude; ML, mid-latitude; HL, high-latitude.
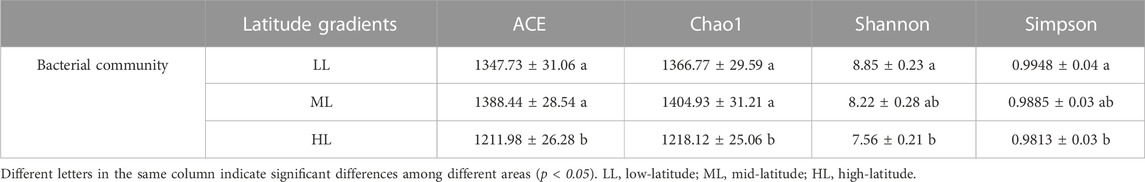
As shown in Table 3, the bacterial a diversity of the forest soils at different latitudes varied significantly. HL produced the lowest values for the ACE, Chao 1, Shannon and Simpson index and these were all significantly lower compared to LL (p<0.05). The highest values for Shannon and Simpson index were observed in LL with 8.85 and 0.9948, respectively, followed by ML and HL. The higher ACE and Chao one index of ML was not significant different compared to HL or LL (p>0.05) (Table 3).
NMDS plots based on confidence intervals (p<0.05) were produced (Figure 2A) that showed an absence of overlap between the different sites, which illustrated that the soil bacterial communities (stress = 0.0225) were clearly different in the three areas. This was confirmed by PERMANOVA analysis, whose findings are shown in a box plot (Figure 2B) (significant difference between groups, R2 = 0.743, p<0.01).
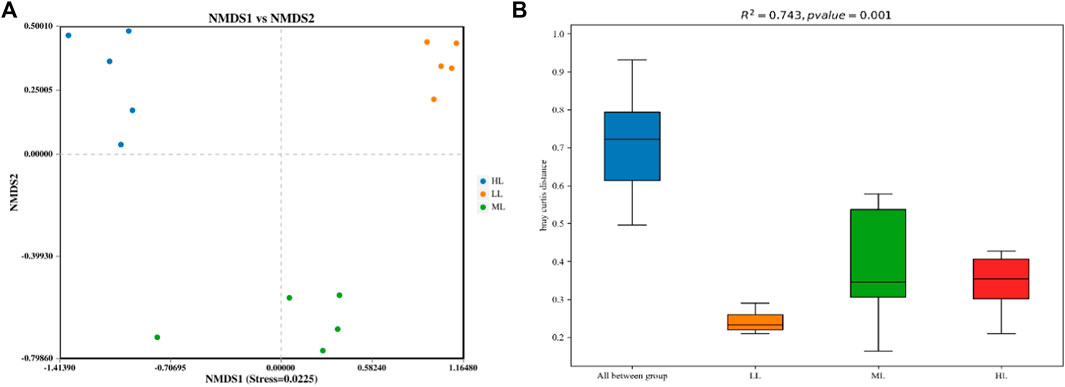
FIGURE 2. NMDS plot (A) and PERMANOVA analysis (B) of the soil bacterial OTUs based on Bray Curtis metrics among all samples. LL, low-latitude; ML, mid-latitude; HL, high-latitude.
3.3 Variation in bacterial composition at different latitudes
At the phylum level (Figure 3), the soil bacterial communities of the LL, ML, and HL were dominated by Acidobacteria (35%, 43%, and 43%, respectively) and Proteobacteria (37%, 35%, 39%). Much lower levels were reported for Rokubacteria (8.6%, 7%, 0.5%), Actinobacteria (7%, 4%, 6%), and Gemmatimonadetes (4.9%, 2.6%, 1.8%), and even lower abundance was noted for other phyla (Figure 3). There were differences between the samples taken at the different latitudes, notably for lower levels of Acidobacteria in LL (p<0.001), but also for the other phyla shown in the figure. Proteobacteria members were lowest in soil collected in ML(p<0.01). The relative abundances of Gemmatimonadetes, Rokubacteria, Chloroflexi and Nitrospirae produced decreasing trends (of variable strengths) with an increase of latitude (Figure 3).
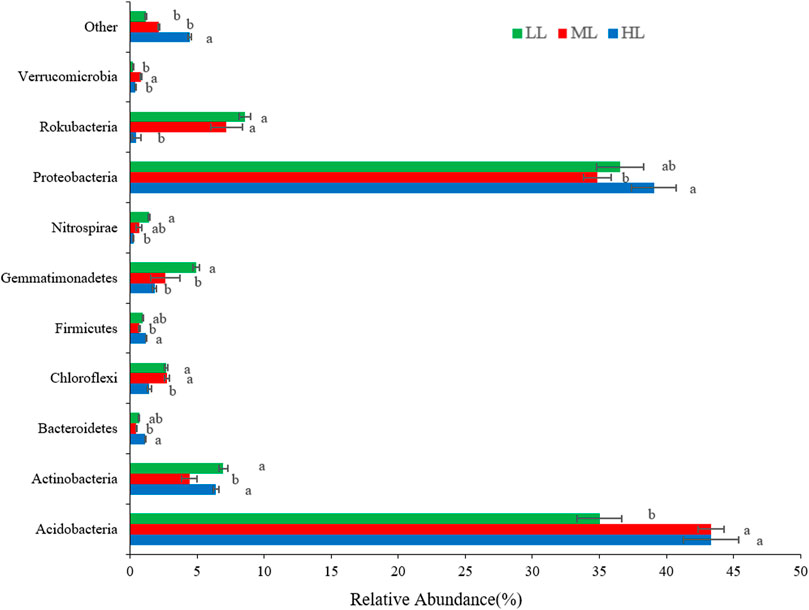
FIGURE 3. Histogram of the relative abundance of bacterial phyla detected in the forest soil at different latitudes. Different lowercase letters indicate significant differences at the p < 0.05 level among the treatments. LL, low-latitude; ML, mid-latitude; HL, high-latitude.
At the genus level, the soil of HL and LL was dominated by Candidatus_Solibacter (4.1%, and 5.0%, respectively) and Bryobacter (4.5% and 3.8%), while these genera were present at significantly lower (p<0.001) levels in ML (Figure 4). Roseiarcus (4.5%) and Granulicella (4.3%) were particularly abundant in HL but undetected in ML. Conversely, Sphingomonas and MND1 were dominant in ML, while Lactococcus was particularly abundant in LL (p<0.001). Thus, Bryobacter and Roseiarcus were the most abundant genera in the HL where Nitrospira and MND1 were the least abundant. Sphingomonas was the most abundant genus in ML, where Roseiarcus and Granulicella were least abundant. Lastly, Candidatus_Solibacter was the most abundant genus in LL while Granulicella was present at lowest abundance here (Figure 4).
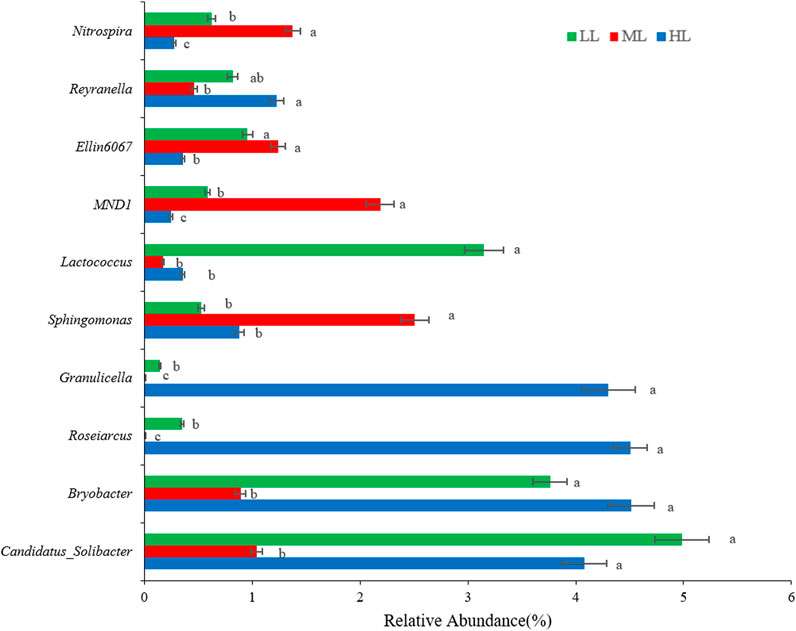
FIGURE 4. Genus-level composition of bacteria detected in the forest soil at different latitudes. Different lowercase letters indicate significant differences at the p< 0.05 level among the treatments. LL, low-latitude; ML, mid-latitude; HL, high-latitude.
A heatmap analysis was performed to compare over- and under-abundance of the 24 most abundant bacterial genera in the five replicates for each latitude (Figure 5). This revealed co-enrichment patterns, as indicated that the genera Elin6067, MND2, Sphingomonas and Dongia in LL, Candidatus_Udaeobacter, GAS113, Reyranella and Candidatus-Koribacter in ML, Burkhoderia, Rosearcus, Granulicella and Occallatibacter in HL, (Figure 5).
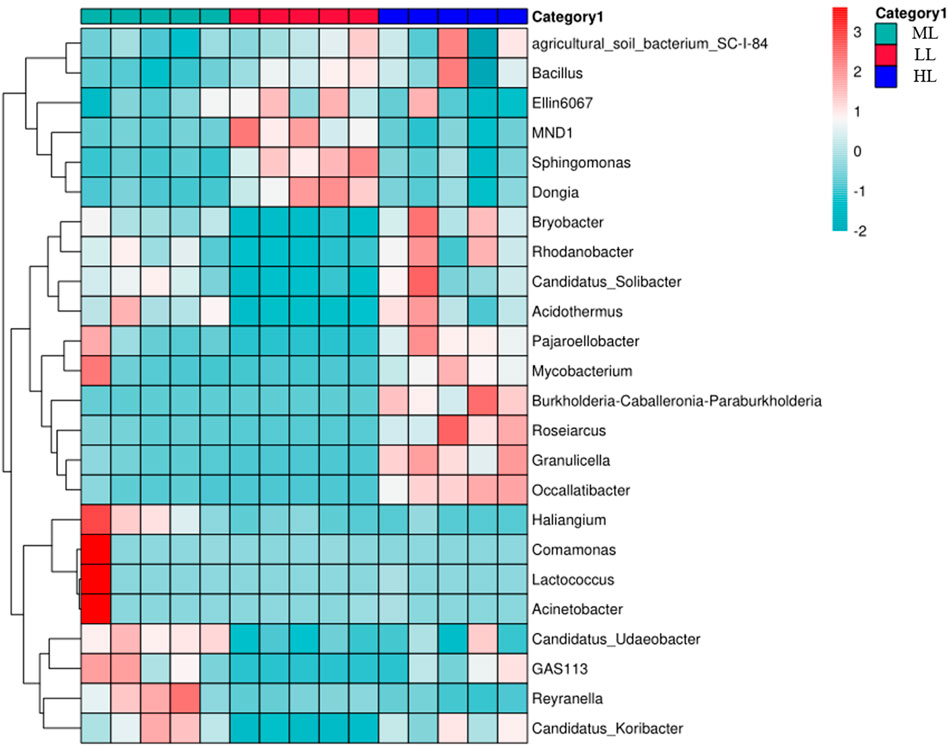
FIGURE 5. Heatmap and hierarchical clustering of the relative abundances of the top 24 genera detected in the individual soil samples. Co-enriched genera are ioutlined in red. LL, low-latitude; ML, mid-latitude; HL, high-latitude.
3.4 Indicator species
A LEfSe analysis was conducted to determine the classified bacterial taxa with significant relative abundance differences among the different soil samples. Significant differences were observed among 16 taxa in the three latitudes, as indicated by LDA effect size scores of >4.5 (Figure 6). These are shown to the right of the cladogram in the figure.
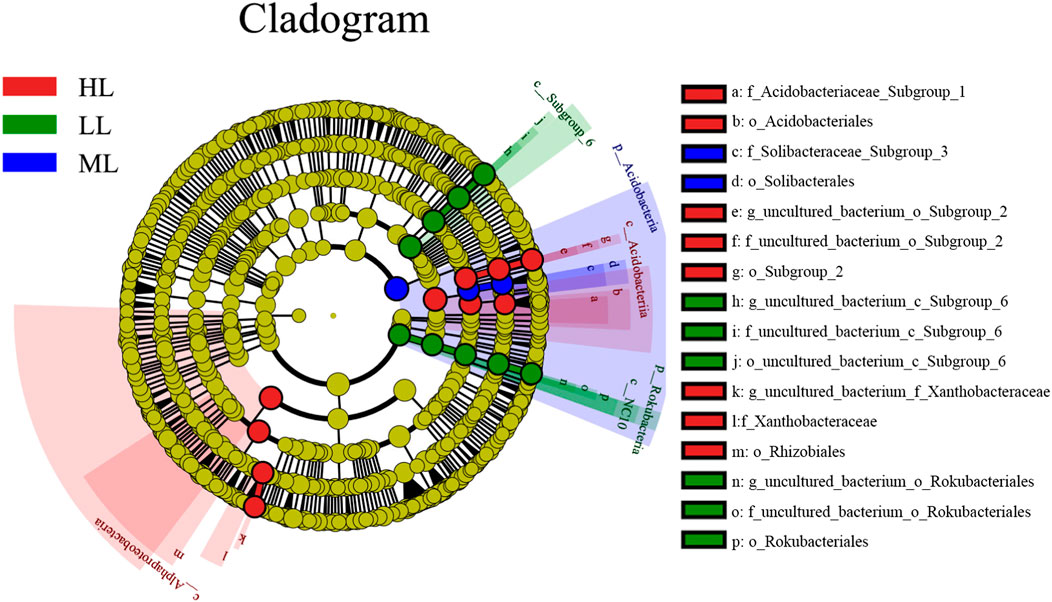
FIGURE 6. Cladogram of the bacterial communities at the three latitude soils with LDA >4.5 (A). LL, low-latitude; ML, mid-latitude; HL, high-latitude.
3.5 Relationship between soil chemical properties and bacterial communities
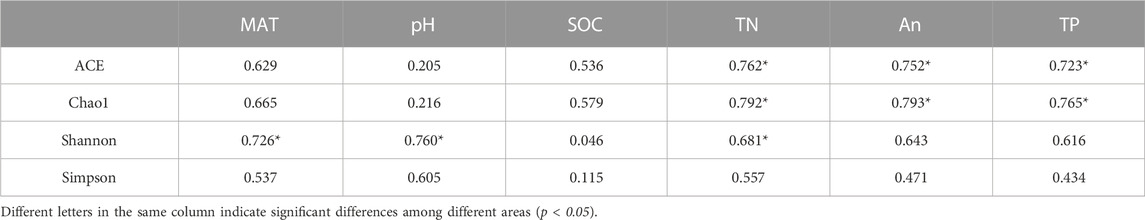
Pearson correlation analysis was used to detect correlations between soil bacteria a-diversity and soil chemical factors. The results (Table 4) indicated that the ACE and Chao1 diversity indices correlated with TN, AN and TP (p<0.05), while the Shannon diversity was correlated with MAT and pH (p<0.05). The Simpson diversity did not correlate with any of the determined soil chemical properties (Table 4).
An RDA analysis indicated that soil chemical properties were important factors driving the structural and compositional changes of the soil bacterial communities (Figure 7A). The cumulative variations in the first and second RDA axes were 42.95% and 13.40%, respectively. Soil pH, MAT, SOC and TN were the major factors explaining the composition differences of the soil bacterial community. Further, through the correlation heat map analysis found the soil pH produced significant positive correlations with Rokubacteria, Nitrospirae and Chloroflexi, and negative correlations with Acidobacteria. Members of Rokubacteria correlated positively with all soil parameters except for SOC, while that soil parameter produced significant negative correlations with Bacteroides, Proteobacteria and Actinobacteria. Lastly, Cloroflexi correlated positively with AN and Nitrospirae with MAT (Figure 7B).
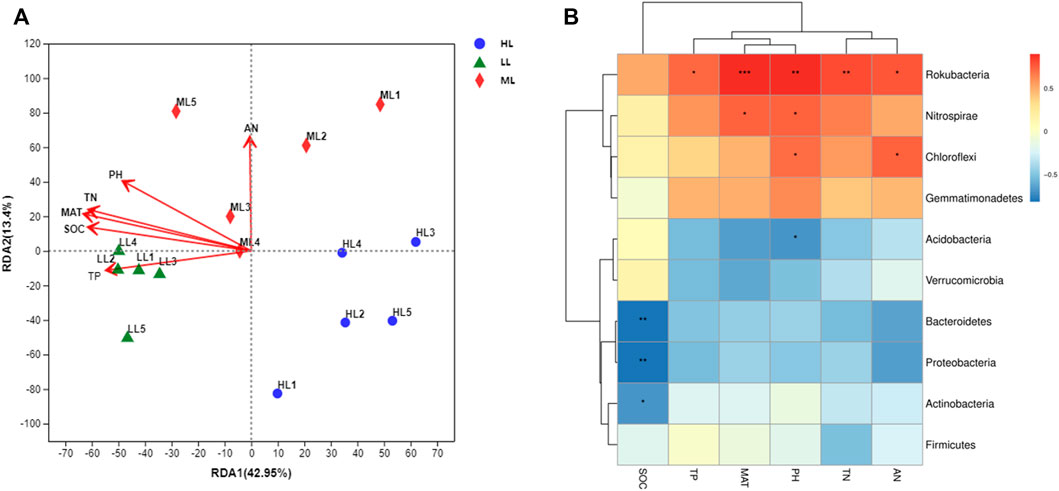
FIGURE 7. Correlations between soil chemical properties and bacterial phyla. RDA (A), Heatmap (B). LL, low-latitude; ML, mid-latitude; HL, high-latitude.
4 Discussion
4.1 Effects of latitude changes on bacterial alpha diversity
Most available studies have suggested presence of specific soil microbial communities at specific latitudes (Zhou et al., 2016; Tian et al., 2018; Zheng et al., 2021). The present study shows that the bacterial Shannon index of forest soil decreased gradually with increasing latitude, and the difference between highest and lowest latitude was significant (Table 3). There are two possible reasons for this. On the one hand, higher temperatures at lower latitudes promote the metabolism and population growth of soil bacteria, and accelerating litter decomposition and improving nutrient availability (Anderson et al., 2009; Zhou et al., 2020). On the other hand, lower pH at high latitudes acidifies forest soils, leading to an increase in acidophilus, which may reduce the number of other species and thus reduce soil bacterial diversity. The mid-latitude soil exhibited the highest bacterial ACE and Chao1 index, though this was not significantly different from low-latitude. Variations in ACE and Chao1 indices may be attributed to the effect of soil physicochemical factors, as levels of TN, AN and TP correlated significantly with soil ACE and Chao1 indices. This indicates that changes in latitudes can affect soil physicochemical factors and therefore affect the abundance of bacteria.
Explaining how biodiversity changes with spatial, temporal, and environmental gradients and its main drivers remains a central focus on biodiversity science. In recent years, research was performed to identify the factors that influence and drive the spatial distribution of microorganisms. Most studies have identified soil pH as the main factor driving soil microorganisms diversity and spatial distribution (Fiere and Jackson, 2006; Griffiths et al., 2011; Tripathi et al., 2018; Chen et al., 2022). In this study, we found that pH directly affected the diversity of soil bacterial communities in different latitudes and produced significant correlations with four phyla, suggesting that pH is a major factor driving alpha diversity in forest soil bacterial communities. This is consistent with results from the literature. Likewise, Bahram et al. (2018) found that pH caused niche differentiation of soil bacteria and fungi on a global scale and identfied certain trends of composition and diversity of soil bacterial communities with pH. However, according to a recent study, on a continental scale temperature regulates the diversity of microorganisms in forest soils (Zhou et al., 2016). Our study found that the soil bacterial alpha-diversity decreased with increasing latitude, with a significant correlation between temperature and soil bacterial Shannon diversity (Table 3), indicating that soil bacteria had strong latitudinal diversity patterns related to temperature in the temperate forests of China. Temperature can affect soil bacterial diversity through various mechanisms. First of all, the most important direct mechanism is that a higher temperature will increase the soil bacteria’s metabolic rates and growth (Wang et al., 2009; Segura et al., 2015). Secondly, higher temperatures promotes plant diversity and productivity, which in turn provides substrates and nutrients for bacterial growth, thus improving soil bacterial diversity (Lange et al., 2015; Deng et al., 2020). Thirdly, higher temperatures support more effective litter decomposition and promote nutrient release, increasing nutrient availability that allows higher bacterial diversity (Zhou et al., 2012; Gao et al., 2022). Finally, the temperature may interact with soil pH, soil nutrients and other environmental factors to regulate the change of soil bacterial diversity (Shen et al., 2013; Delgado-Baquerizo and Eldridge, 2019).
4.2 Effects of latitude changes on bacterial compositions
Our results identifiud the main bacterial phyla at different latitudes as Acidobacteria and Proteobacteria, followed by Rokubacteria, Actinobacteria and Gemmatimonadetes. Furthermore, we found that increasing latitude significantly decreased the abundance of Acidobacteria. Members of that phylum have a strong sensibility to the environment, such as vegetation type and soil pH, and these can affect the diversity and community composition of soil Acidobacteria. In both HL and ML, the soil pH was under 5.0, Acidobacteria in these soils can degrade cellulose under microaerobic and anoxic conditions, and the refractory needles in coniferous forests increased, which promoted the expansion of Acidobacteria populations. Previous studies have shown that Proteobacteria and Actinobacteria are mainly involved in the decomposition of organic matter, and the relative abundance of Proteobacteria is positively correlated with soil carbon content (Li et al., 2018; Li et al., 2022; Zhang et al., 2022). Indeed, both the SOC content and the relative abundance of Proteobacteria was highest in HL, which was consistent with previous results. Most Actinomycetes are aerobic, grow at an optimum temperature of 28°C–30 °C, and are abundant in neutral or slightly alkaline soils. The relative abundance of Actinobacteria was highest in LL, this is mainly because the low latitudes provides suitable temperature and soil pH for the growth of Actinobacteria. Increasing latitude brought about a reduction in the relative abundances of Gemmatimonadetes, Rokubacteria, Chloroflexi and Nitrospirae, which was somewhat inconsistent with published observations that described revegetation resulted in initial increases and then decreases of Gemmatimonadetes, Chloroflexi, and Nitrospirae (Zhou et al., 2016; Tian et al., 2018). Further research is needed to address the reasons for this discrepancy, which may be attributed to the different vegetation types and different latitudinal gradients.
At the genus level, Candidatus_Solibacter and Bryobacter were highly abundant in two or three of the investigated soils, while Roseiarcus, Granulicella and Lactococcus were abundant in one soil type (Figure 4), which was consistent with other studies (Challacombe et al., 2017; Zhang et al., 2021; Kim et al., 2021). Members of the most abundant Candidatus_Solibacter and Bryobacter genera are involved in decomposing organic matter and use this as a carbon source. Interestingly, their relative abundance was significantly lower in ML. This may be due to the acidity of soil in high latitude coniferous forest, which provides a suitable environment for their growth, while low latitude broad-leaved forest provides sufficient carbon sources. The high relative abundance of Roseiarcus in the high-latitude coniferous forest soil of HL was consistent with previous results (Zhang et al., 2014; Rime et al., 2015; Du et al., 2016). The effects of latitude on the diversity and abundance of bacteria have been linked to the quantity and characteristics of the organic matter derived from plant litter, as it constitutes an important nutrient source for soil microorganism growth (Wagai et al., 2011).
The main drivers of soil bacterial composition can result in inconsistent responses at different study scales. Many studies showed that difference of soil factors at local and regional scales had a major impact on microorganisms communities, while climate factors and geography plays a more important role at larger scales (Tripathi et al., 2012; Zhang et al., 2020). In the present study, the soil bacterial community structure was affected by multiple environmental factors and was not determined by a single factor, which was similar to other results (Griffithset al., 2011; Delgado-Baquerizo and Eldridge, 2019). According to Pearson correlation analyses, the 10 most abundant bacterial phyla were mainly influenced by pH, MAT, SOC and TN: Proteobacteria, Acidobacteria, Chloroflexi, Rokubacteria and Nitrospirae all produced significant, though variable correlations with one or more of these soil factors (Figure 7B). Although temperature is considered a major factor regulating soil bacterial composition at large scales, it only signficiantly affected abundance of Rokubacteria and Nitrospirae in our study, wherea pH played an important role in the distribution of soil bacterial core species in the different latitudes. This may be because pH can affect specific communities, such as increasing the relative abundance of Acidobacteria, while decreasing the relative abundance of actinomycetes, thus affecting the spatial distribution of the communities (Xia et al., 2016; Tian et al., 2021). In addition, soil nutrient factors such as SOC and TN are also closely related to bacterial composition, suggesting these too are main drivers of soil bacteria at different latitudes. Thus, there are no single climatic factors or soil physical and chemical properties, but more likely multiple factors interact with each other to regulate changes in soil bacterial composition. Therefore in the future, environmental factors shoud more comprehensively be considered in combination to explore their interactions and respective contributions, it can better reveal the driving mechanism of soil microbial distribution and stability on a large scale.
5 Conclusion
Our results indicate that the bacterial diversity of temperate forests in China decreases with increasing latitude. MAT, pH and TN were significantly correlated with bacterial diversity. At the same time, Acidobacteria, Proteobacteria and Actinobacteria were the dominant phyla in soil bacteria at different latitudes. MAT, pH and TN were significantly correlated with the relative abundance of most dominant phyla. These results indicated that they were the main driving factors affecting the latitudinal patterns of soil bacteria in temperate forests. From a microbiological point of view, this study revealed that the distribution of bacterial communities in temperate forests followed the latitudinal diversity pattern, which has important reference value for enrich the latitudinal pattern of biodiversity.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
X-YF performed this experiment, analyzed the data, and wrote this MS. Z-CC performed this experiment and help to write this MS. H-WN, and R-TZ designed this experiment and revised this manuscript. The author(s) read and approved the final MS.
Funding
This work was supported by the Science and Technology Development Special Project of Central government guides local government (ZY20B15), the Applied Technology Research and Development Program in Heilongjiang (GA19C006-6), the Research expenses of provincial research institutes in Heilongjiang (CZKYF 2022-1-C009; ZNBZ2019ZR02), and the Special Project Foundation of the Heilongjiang Academy of Sciences (KY2021ZR01).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/feart.2022.1096931/full#supplementary-material
References
Anderson, J. A., Hooper, M. J., Zak, J. C., and Cox, S. B. (2009). Characterization of the structural and functional diversity of indigenous soil microbial communities in smelter-impacted and nonimpacted soils. Enviro. Toxicol. Chem. 28, 534. doi:10.1897/08-281.1
Andrew, M. S. (2020). Land-use change and forest biodiversity. Science 368 (6497), 1324.7–1325. doi:10.1126/science.368.6497.1324-g
Bahram, M., Hildebrand, F., Forslund, S. K., Anderson, J. L., Soudzilovskaia, N. A., Bodegom, P. M., et al. (2018). Structure and function of the global topsoil microbiome. Nature 560, 233–237. doi:10.1038/s41586-018-0386-6
Berdugo, M., Quant, J. M., Wason, J. W., and Dovciak, M. (2018). Latitudinal patterns and environmental drivers of moss layer cover in extratropical forests. Glob. Ecol. Biogeogr. 27 (10), 1213–1224. doi:10.1111/geb.12778
Cardinale, M., Grube, M., Erlacher, A., Quehenberger, J., and Berg, G. (2015). Bacterial networks and co-occurrence relationships in the lettuce root microbiota. Environ. Microbiol. 17, 239–252. doi:10.1111/1462-2920.12686
Challacombe, J., Eichorst, S., Hauser, L., Land, M., Xie, G., and Kuske, C. (2017). Biological consequences of ancient gene acquisition and duplication in the large genome of Candidatus Solibacter usitatus Ellin6076. Plos One 6 (9), e24882. doi:10.1371/journal.pone.0024882
Chen, G., Ma, S., Tian, D., Xiao, W., Jiang, L., Xing, A., et al. (2020). Patterns and determinants of soil microbial residues from tropical to boreal forests. Soil. Biol. biochem. 151, 108059. doi:10.1016/j.soilbio.2020.108059
Chen, W., Su, F., Nie, Y., Zhong, B., Zheng, Y., Mo, J., et al. (2022). Divergent responses of soil microbial functional groups to long-term high nitrogen presence in the tropical forests. Sci. Total. Environ. 821, 153251. doi:10.1016/J.SCITOTENV.2022.153251
Chen, Y., Xi, J., Xiao, M., Wang, S., Chen, W., Liu, F., et al. (2022). Soil fungal communities show more specificity than bacteria for plant species composition in a temperate forest in China. BMC Microbiol. 22 (1), 208. doi:10.1186/S12866-022-02591-1
Degrune, F., Dufrêne, M., Colinet, G., Massart, S., Taminiau, B., Bodson, B., et al. (2015). A novel sub-phylum method discriminates better the impact of crop management on soil microbial community. Agron. Sustain. Dev. 35, 1157–1166. doi:10.1007/s13593-015-0291-4
Delgado-Baquerizo, M., and Eldridge, D. J. (2019). Cross-biome drivers of soil bacterial alpha diversity on a worldwide scale. Ecosystems 22, 1220–1231. doi:10.1007/s10021-018-0333-2
Deng, J., Bai, X., Zhou, Y., Zhu, W., and Yin, Y. (2020). Variations of soil microbial communities accompanied by different vegetation restoration in an open-cut iron mining area. Sci. Total. Environ. 704 (C), 135243. doi:10.1016/j.scitotenv.2019.135243
Deng, J., Zhang, Y., Yin, Y., Zhu, X., Zhu, W., and Zhou, Y. (2019b). Comparison of soil bacterial community and functional characteristics following afforestation in the semi-arid areas. PeerJ 7, e7141. doi:10.7717/peerj.7141
Du, S., Yu, M., Liu, F., Xiao, L., Zhang, H., Tao, J., et al. (2016). Effect of facility management regimes on soil bacterial diversity and community structure. Chin. Jour. Eco-Agr. 25 (11), 1615–1625. doi:10.13930/j.cnki.cjea.170291
Finlay, B. J. (2002). Global dispersal of free-living microbial eukaryote species. Science 296, 1061–1063. doi:10.1126/science.1070710
Frey, B., Walthert, L., Perez- Mon, C., Stierli, B., Koechli, R., Dharmarajah, A., et al. (2021). Deep soil layers of drought-exposed forests harbor poorly known bacterial and fungal communities. Front. Microbiol. 12, 674160. doi:10.3389/fmicb.2021.674160
Fuhrman, J. A., Steele, J. A., Hewson, I., Schwalbach, M. S., Brown, M. V., Green, J. L., et al. (2008). A latitudinal diversity gradient in planktonic marine bacteria. Proc. Natl. Acad. Sci. U. S. A. 105 (22), 7774–7778. doi:10.1073/pnas.0803070105
Gao, S., Song, Y., Song, C., Wang, X., Ma, X., Gao, J., et al. (2022). Effects of temperature increase and nitrogen addition on the early litter decomposition in permafrost peatlands. Catena 209 (P1), 105801. doi:10.1016/J.CATENA.2021.105801
Griffiths, R. I., Thomson, B. C., James, P., Bell, T., Bailey, M., and Whiteley, A. S. (2011). The bacterial biogeography of British soils. Environ. Microbiol. 13, 1642–1654. doi:10.1111/j.1462-2920.2011.02480.x
Han, S., Gao, R., Li, A., Ma, H., Yin, Y., Si, Y., et al. (2015). Soil microbial community structure of two types of forests in the mid-subtropics of China. Jour. Appl. Ecol. 26 (7), 2151–2158. doi:10.13287/j.1001-9332.20150506.011
Hu, T., Zhao, B., Li, F., Dou, X., Hu, H., and Sun, L. (2021). Effects of fire on soil respiration and its components in a Dahurian larch (Larix gmelinii) forest in northeast China: Implications for forest ecosystem carbon cycling. Geoderma 402, 115273. doi:10.1016/J.GEODERMA.2021.115273
Ji, L., Shen, F., Liu, Y., Yang, Y., Wang, J., Purahong, W., et al. (2022). Contrasting altitudinal patterns and co-occurrence networks of soil bacterial and fungal communities along soil depths in the cold-temperate montane forests of China. Catena 209 (P2), 105844. doi:10.1016/J.CATENA.2021.105844
Jia, H., Chen, Y., Wang, X., Li, P., Yuan, Z., and Ye, Y. (2019). The relationships among topographically-driven habitats, dominant species and vertical layers in temperate forest in China. Russ. J. Ecol. 50 (2), 172–186. doi:10.1134/S1067413619020061
Kim, H., Lee, S., Jo, H., Finneran, K. T., and Kwon, M. (2021). Diversity and composition of soil Acidobacteria and Proteobacteria communities as a bacterial indicator of past land-use change from forest to farmland. Sci. Total. Environ. 797, 148944–148954. doi:10.1016/J.SCITOTENV.2021.148944
Lange, M., Eisenhauer, N., Sierra, C. A., Bessler, H., Engels, C., Griffiths, R., et al. (2015). Plant diversity increases soil microbial activity and soil carbon storage. Nat. Commun. 6, 6707. doi:10.1038/ncomms7707
Lee, J., Cho, J., Cho, Y., Cho, A., Woo, J., Lee, J., et al. (2018). The latitudinal gradient in rock-inhabiting bacterial community compositions in Victoria Land, Antarctica. Sci. Total. Environ. 657, 731–738. doi:10.1016/j.scitotenv.2018.12.073
Li, D., Chen, L., Xu, J., Ma, L., Olk, D., Zhao, B., et al. (2018). Chemical nature of soil organic carbon under different long-term fertilization regimes is coupled with changes in the bacterial community composition in a Calcaric Fluvisol. Biol. Fert. Soils 54 (8), 999–1012. doi:10.1007/s00374-018-1319-0
Li, P., Shen, C., Jiang, L., Feng, Z., and Fang, J. (2019). Difference in soil bacterial community composition depends on forest type rather than nitrogen and phosphorus additions in tropical montane rainforests. Biol. Fert. Soils 55 (3), 313–323. doi:10.1007/s00374-019-01349-8
Li, Y., Wang, L., Tian, L., Zheng, H., Ou, Y., Yan, B., et al. (2022). Dissolved organic carbon, an indicator of soil bacterial succession in restored wetland under freeze-thaw cycle. Ecol. Eng. 177, 106569. doi:10.1016/J.ECOLENG.2022.106569
Neu, A. T., Allen, E. E., and Roy, K. (2021). Do host-associated microbes show a contrarian latitudinal diversity gradient? Insights from Mytilus californianus, an intertidal foundation host. J. Biogeogr. 48 (11), 2839–2852. doi:10.1111/JBI.14243
Rime, T., Hartmann, M., Brunner, I., Widmer, F., Zeyer, J., and Frey, B. (2015). Vertical distribution of the soil microbiota along a successional gradient in a glacier forefield. Mol. Ecol. 24, 1091–1108. doi:10.1111/mec.13051
Sang, C., Xia, Z., Sun, L., Sun, H., Jiang, P., Wang, C., et al. (2021). Responses of soil microbial communities to freeze–thaw cycles in a Chinese temperate forest. Ecol. Process 10 (1), 66. doi:10.1186/S13717-021-00337-X
Segura, A. M., Calliari, D., Kruk, C., Fort, H., Izaguirre, I., Saad, J. F., et al. (2015). Metabolic dependence of phytoplankton species richness. Glob. Ecol. Biogeogr. 24, 472–482. doi:10.1111/geb.12258
Shen, C., Liang, W., Shi, Y., Lin, X., Zhang, H., Wu, X., et al. (2014). Contrasting elevational diversity patterns between eukaryotic soil microbes and plants. Ecology 95, 3190–3202. doi:10.1890/14-0310.1
Shen, C., Xiong, J., Zhang, H., Feng, Y., Lin, X., Li, X., et al. (2013). Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil. Biol. biochem. 57, 204–211. doi:10.1016/j.soilbio.2012.07.013
Srivastava, A. K., Kashyap, P. L., Santoyo, G., and Newcombe, G. (2021). Editorial: Plant microbiome: Interactions, mechanisms of action, and applications. Front. Microbiol. 12, 706049. doi:10.3389/FMICB.2021.706049
Sui, X., Zhang, R., Frey, B., Yang, L., Liu, Y., Ni, H., et al. (2021). Soil physicochemical properties drive the variation in soil microbial communities along a forest successional series in a degraded wetland in northeastern China. Ecol. Evol. 11 (5), 2194–2208. doi:10.1002/ECE3.7184
Sun, X., Cao, X., Zhao, D., Zeng, J., Huang, R., Duan, M., et al. (2021). The pattern of sedimentary bacterial communities varies with latitude within a large eutrophic lake. Limnologica 87, 125860. doi:10.1016/J.LIMNO.2021.125860
Tedersoo, L., Bahram, M., Põlme, S., Kõljalg, U., Yorou, N. S., Wijesundera, R., et al. (2014). Fungal biogeography. Global diversity and geography of soil fungi. Science 346, 1256688. doi:10.1126/science.1256688
Tian, J., He, N., Hale, L., Niu, S., Yu, G., Liu, Yuan., et al. (2018). Soil organic matter availability and climate drive latitudinal patterns in bacterial diversity from tropical to cold temperate forests. Funct. Ecol. 32 (1), 61–70. doi:10.1111/1365-2435.12952
Tian, Q., Jiang, Q., Huang, L., Li, D., Lin, Q., Tang, Z., et al. (2022). Vertical distribution of soil bacterial communities in different forest types along an elevation gradient. Microb. Ecol. 1-14. doi:10.1007/S00248-021-01949-8
Tian, Q., Jiang, Y., Tang, Y., Wu, Y., Tang, Z., and Liu, F. (2021). Soil pH and organic carbon properties drive soil bacterial communities in surface and deep layers along an elevational gradient. Front. Microbiol. 12, 646124. doi:10.3389/FMICB.2021.646124
Tripathi, B. M., Kim, M., Singh, D., Lee-Cruz, L., Lai-Hoe, A., Ainuddin, A. N., et al. (2012). Tropical soil bacterial communities in Malaysia: pH dominates in the equatorial tropics too. Microb. Ecol. 64, 474–484. doi:10.1007/s00248-012-0028-8
Tripathi, B. M., Lee-Cruz, L., Kim, M., Singh, D., Go, R., Shukor, N. A., et al. (2014). Spatial scaling effects on soil bacterial communities in Malaysian tropical forests. Microb. Ecol. 68, 247–258. doi:10.1007/s00248-014-0404-7
Tripathi, B. M., Stegen, J. C., Kim, M., Dong, K., Adams, J. M., and Lee, Y. K. (2018). Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 12, 1072–1083. doi:10.1038/s41396-018-0082-4
Wagai, R., Kitayama, K., Satomura, T., Fujinuma, R., and Balser, T. (2011). Interactive influences of climate and parent material on soil microbial community structure in Bornean tropical forest ecosystems. Ecol. Res. 26, 627–636. doi:10.1007/s11284-011-0822-7
Wang, X., Van, J. D., Deng, Y., Lü, X., Wang, C., Zhou, J., et al. (2015). Scale-dependent effects of climate and geographic distance on bacterial diversity patterns across northern China's grasslands. FEMS Microbiol. Ecol. 91, fiv133. doi:10.1093/femsec/fiv133
Wang, Z., Brown, J. H., Tang, Z., and Fang, J. (2009). Temperature dependence, spatial scale, and tree species diversity in eastern Asia and North America. Proc. Natl. Acad. Sci. U. S. A. 106, 13388–13392. doi:10.1073/pnas.0905030106
Xia, Z., Bai, E., Wang, Q., Gao, D., Zhou, J., Jiang, P., et al. (2016). Biogeographic distribution patterns of bacteria in typical Chinese forest soils. Front. Microbiol. 7, 1106. doi:10.3389/fmicb.2016.01106
Yang, S., Yao, F., Ye, J., Fang, S., Wang, Z., Wang, R., et al. (2019). Latitudinal pattern of soil lignin/cellulose content and the activity of their degrading enzymes across a temperate forest ecosystem. Ecol. Indic. 102, 557–568. doi:10.1016/j.ecolind.2019.03.009
Yu, C., Fan, C., Zhang, C., Zhao, X., and Gadow, K. (2021). Decomposing spatial β-diversity in the temperate forests of northeastern China. Ecol. Evol. 11 (16), 11362–11372. doi:10.1002/ECE3.7926
Zhang, F., Huang, H., Cui, Y., Sun, Q., Zhu, J., Liu, M., et al. (2014). Isolation and diversity of bacillus species from Jiaxi tropical rain forest soil. J. Microbiol. 34 (4), 42–46. doi:10.3969/j.issn.1005-7021.2014.04.008
Zhang, R., Liu, Y., Zhong, H., Chen, X., and Sui, X. (2022). Effects of simulated nitrogen deposition on the soil microbial community diversity of a Deyeuxia angustifolia wetland in the Sanjiang Plain, Northeastern China. Ann. Microbiol. 72 (1), 11. doi:10.1186/S13213-022-01666-8
Zhang, X., Liu, S., Wang, J., Huang, Y., Freedman, Z., Fu, S., et al. (2020). Local community assembly mechanisms shape soil bacterial β diversity patterns along a latitudinal gradient. Nat. Commun. 11 (1), 5428. doi:10.1038/s41467-020-19228-4
Zhang, Y., Ding, K., Yrjälä, K., Liu, H., Tong, Z., and Zhang, J. (2021). Introduction of broadleaf species into monospecific Cunninghamia lanceolata plantations changed the soil Acidobacteria subgroups composition and nitrogen-cycling gene abundances. Plant Soil 467 (1-2), 29–46. doi:10.1007/S11104-021-05014-8
Zheng, S., Bian, H., Quan, Q., Xu, L., Chen, Z., and He, N. (2018). Effect of nitrogen and acid deposition on soil respiration in a temperate forest in China. Geoderma 329, 82–90. doi:10.1016/j.geoderma.2018.05.022
Zheng, Y., Chen, L., Ji, N., Wang, Y., Gao, C., Jin, S., et al. (2021). Assembly processes lead to divergent soil fungal communities within and among 12 forest ecosystems along a latitudinal gradient. New phytol. 231 (3), 1183–1194. doi:10.1111/NPH.17457
Zhou, B., Liao, Z., Chen, S., Jia, H., Zhu, J., and Fei, X. (2022). Net primary productivity of forest ecosystems in the southwest karst region from the perspective of carbon neutralization. Forests 13 (9), 1367. doi:10.3390/F13091367
Zhou, J., Deng, Y., Shen, L., Wen, C., Yan, Q., Ning, D., et al. (2016). Temperature mediates continental-scale diversity of microbes in forest soils. Nat. Commun. 7 (1), 12083. doi:10.1038/ncomms12083
Zhou, J., Xue, K., Xie, J., Deng, Y., Wu, L., Cheng, X., et al. (2012). Microbial mediation of carbon-cycle feedbacks to climate warming. Nat. Clim. Change 2, 106–110. doi:10.1038/nclimate1331
Keywords: temperate forest, latitude pattern, bacterial diversity, effect factors, soil nutrient
Citation: Fu X-Y, Cheng Z-C, Ni H-W and Zhang R-T (2023) Latitude variations of soil bacterial community diversity and composition in three typical forests of temperate, northeastern of China. Front. Earth Sci. 10:1096931. doi: 10.3389/feart.2022.1096931
Received: 13 November 2022; Accepted: 22 December 2022;
Published: 09 January 2023.
Edited by:
Xin Sui, Heilongjiang University, ChinaReviewed by:
Dongxing Zhou, Northeast Agricultural University, ChinaZhijie Chen, Fujian Normal University, China
Xiang-Min Fang, Jiangxi Agricultural University, China
Copyright © 2023 Fu, Cheng, Ni and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rong-Tao Zhang, emhhbmdyb25ndGFvMTRAMTYzLmNvbQ==; Hong-Wei Ni, NDY5OTA5NzYxQHFxLmNvbQ==
 Xiao-Yu Fu1,2
Xiao-Yu Fu1,2 Rong-Tao Zhang
Rong-Tao Zhang