
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Earth Sci. , 10 January 2023
Sec. Quaternary Science, Geomorphology and Paleoenvironment
Volume 10 - 2022 | https://doi.org/10.3389/feart.2022.1059196
This article is part of the Research Topic Bioarchaeology in East Asia View all 13 articles
Mitochondrial DNA was first successfully extracted from ancient remains approximately 4 decades ago. Research into ancient DNA has been revolutionized due to improvements in next-generation sequencing (NGS) technology in the early 21st century, as well as advances in the field of ancient DNA extraction and enhancement. In recent years, a large number of paleogenomic data has shed light on the origin and evolution of humans, and provided new insights into the migration and admixture events of populations, as well as the spread of languages and technologies. As China is located in the eastern part of Eurasia, it plays an integral role in exploration of the genetic history of Eurasians throughout the history of modern human habitation. Here we review recent progress deriving from paleogenomic analysis, which helps to reconstruct the prehistory of China.
Over the past decades, the field of ancient DNA research has witnessed a revolution. Many remarkable achievements have been made in regards to the reconstruction of the history of humans (Pickrell and Reich, 2014; Parks et al., 2015; Slatkin and Racimo, 2016; Marciniak and Perry, 2017; Skoglund and Mathieson, 2018; Yang and Fu, 2018; Racimo et al., 2020; Bergström et al., 2021; Liu et al., 2021; Liu et al., 2022). Initially, ancient DNA studies focused on highly variable and partially coding regions of mitochondrial DNA and the informative single nucleotide polymorphisms (SNPs) of the Y chromosome. After 2006, the introduction of NGS technology and the optimization of the methods on ancient DNA extraction, library construction and DNA enrichment (Meyer and Kircher, 2010; Kircher et al., 2011; Barnett and Larson, 2012; Dabney and Meyer, 2012; Carpenter et al., 2013; Gansauge and Meyer, 2013; Korlević et al., 2015; Slon et al., 2017), enabled large scale genome-wide ancient DNA investigations on prehistoric populations, which opened new windows into the past. Paleogenomic study leads to a more comprehensive understanding of patterns of human variation and mobility through time and space. Genome-wide data from archaic human, such as Neanderthals and Denisovans, provides direct insights into human evolution (Green et al., 2010; Reich et al., 2010; Fu et al., 2015; Racimo et al., 2015; Prüfer et al., 2017; Slon et al., 2018; Chen et al., 2020; Mafessoni et al., 2020; Hajdinjak et al., 2021). Large-scale studies of continental populations on the scale of deeply sampled time transects can offer direct evidence for the genetic origin of populations, and enrich the details of human migration and interaction, both, temporally and spatially (Olalde and Posth, 2020; Vicente and Schlebusch, 2020; Zhang and Fu, 2020; Choin et al., 2021; Willerslev and Meltzer, 2021).
Situated in the eastern part of Eurasia and connecting North, Central and Southeast Asia, China serves as a vital area for the reconstruction of human history of Asia. Its vast territory and diverse topography was a cradle for a long history of human settlement and thriving ancient civilizations. The origin and evolutionary history of Chinese populations has been studied extensively within different research fields, such as archaeology, history, linguistics, anthropology, genetics, and more recently, palaeogenomics. Although paleogenomic research in China was launched relatively late, genome-wide data of ancient Chinese populations has been accumulating rapidly in recent years. The ancient DNA studies have provided valuable information on demographic history in China and East Asia, and have been pivotal in the efforts to better understand the formation and evolution of Chinese civilization.
The origins of anatomically modern humans (AMH) in China and even East Asia have been debated at great length. There are mainly two hypotheses: multi-regional hypothesis and recent African origin hypothesis (Stringer and Andrews, 1988; Wu, 2006). Evidences collected from fossils of ancient humans unearthed in China support the multi-regional hypothesis, while genetic studies agree with the recent out-of-Africa hypothesis (Wu, 2006; López et al., 2015).
In 2017, Yang et al. sequenced the whole genome of a 40,000-year-old individual uncovered from Tianyuan Cave near Beijing, China (Yang et al., 2017). Yang et al.’s accomplishments marked the first genome-wide analysis of early modern humans in China, thus filling the sampling gap in time and space. Tianyuan individual’s genome, possessed genetic characteristics similar to the one of modern Asians, who carry approximately 4%–5% Neanderthal DNA which is shared by Upper Paleolithic Eurasians. Tianyuan individual has a relatively closer relationship with present-day and ancient Asians than with Europeans. However, the genetic connection between the Tianyuan individual and the ancient European individuals of the same period (GoyetQ116-1 and Bacho Kiro) was observed, which may be related to the spread of Paleolithic Aurignacian culture (Fu et al., 2016; Hajdinjak et al., 2021). The evidence from genomic analysis showed that the Tianyuan individual was not a direct ancestor of modern Chinese populations, indicating the diversity of Asian populations 40,000 years ago. In addition, the individual dating back about 33,000 years from the Amur region (AR33K) shared similar genetic components with the Tianyuan individual (Mao et al., 2021), which suggested that before the Last Glacial period the Tianyuan-related population was widespread in northern China.
Early Chinese civilizations formed in multiple regions while displaying a variety of characteristics, hence the diversity of material cultures (Wu et al., 2018). There is a plethora of topics on the genetic history of various cultural populations, their exchanges and interactions with the surrounding populations, as well as their influence on the formation of modern populations which are widely discussed. Recent paleogenomic studies have provided new insights into these issues.
The northeast region of China geographically connects North Asia and the Far East, including Liaoning, Jilin, Heilongjiang provinces, and the southeastern part of Inner Mongolia. This region has been populated since the Paleolithic Age (Pitulko, 2001). The material cultures of this area began to flourish in the Neolithic Age, and became one of the cultural and dryland agricultural centers of northern China (Liu et al., 2015). This region had a profound impact on the formation and evolution of Chinese civilization, as well as on the proliferation of agriculture in northern East Asia (Liu et al., 2015). Recently, genome-wide data from the Amur and West-Liao River regions aided researchers in the comprehension of the dynamic demographic processes of this area in China (Ning et al., 2020; Mao et al., 2021).
The Amur River (AR) region, adjacent to the Far East, is rich in natural resources. As a result, fishing, hunting and animal husbandry constitute the primary subsistence strategies in this area (Kato, 2006). The demographic history in this region was reconstructed through the genomic analysis of AR individuals dated from the Paleolithic to the Iron Age (Ning et al., 2020; Mao et al., 2021). Before the Last Glacial Maximum (LGM), the population in AR (AR33k) had similar genetic profile with the Tianyuan individual. While, at the end of the LGM, the genetic makeup (AR19k) in this area changed, and the previously widespread Tianyuan-related component may have been replaced by modern East Asian component, reflecting a genetic discontinuity from 33,000 to 19,000 years old (Mao et al., 2021). When compared to the ancient southern coastal populations of East Asia, AR19k has a closer genetic relationship with those of the ancient coastal northerners (Mao et al., 2021). Based on this observation, the genetic differences between the northern and southern East Asians had been formed about 19,000 years ago, nearly 10,000 years earlier than previously reported potentialities. After the LGM, the populations in the AR region showed a genetic continuity, and had the closest genetic relationship with the Devil’s Gate cave population (DevilsCave_N), Neolithic forager/farmers recovered from Devil’s Gate cave in Far East (Sikora et al., 2019). In addition, after 14ka, although the genetic components of the population did not change, the local population size had increased, which may be related to the advent of sedentary farming in the region (Mao et al., 2021).
The West-Liao River region (WLR), an important area for the early formation and development of Chinese civilization, is one of the dryland millet agricultural centers in northern China (Teng, 2013). The people in this region had lived primarily on agriculture and fishing and hunting (Sun, 2015). In the middle of the Neolithic Age, well-developed material culture, represented by the Hongshan culture (6,500–5,000 BP), had expanded in this and neighboring regions (Teng, 2017). Researches focusing on the population origin in this area and its contacts with other millet planting regions, such as Yellow River Basin, are crucial for enlarging our knowledge on the broadcast of agriculture in China. Unlike the demographic process in the AR region, population dynamics in this area from the Middle Neolithic Age to the Bronze Age can be characterized by a series of interactions with surrounding populations (Ning et al., 2020). The populations in this region carried a mixture of ancestries from the Neolithic AR (AR_EN) and mid-Neolithic Yellow River region (YR_MN), while the proportions of these two ancestries varied with spatial and temporal scales. The mid-Neolithic Haminmangha (HMMH) population in the northern extent of the WLR, where neighbors the AR region, had more AR_EN ancestral component than contemporaneous Hongshan population from the southern part of WLR, while the late Neolithic populations had more YR_MN ancestral component than the mid-Neolithic ones. This variation of genetic makeup reflected a northward migration of the Central Plain populations, and a contact between populations from the two major dryland agricultural centers in northern China. Genetic evidence inferred that the pattern of agricultural diffusion in northern China may be driven by population movements. As a consequence of climatic changes during the Bronze Age, the region was no longer suitable for millet cultivation, the subsistence strategy transformed to nomadic pastoralism, accompanied with a genetic shift of the incremental AR_EN ancestry component (Jia et al., 2016).
The Central Plain, with good natural conditions, is the cradle of Chinese civilization. Complex societies emerged in this region as early as 10,000 years ago, and the advent of agriculture can be dated back to 8,700BP (Pang and Gao, 2006). The Yangshao and Longshan cultures, representatives of the Neolithic cultures of the Central Plain, are well-known for painted pottery (Wu, 2001). Archaeological evidences revealed the interactions between the Central Plain and surrounding regions (Shui, 2001). The genetic profile of populations from the Neolithic Age to the Late Bronze/Iron Age in Central Plain was similar, but the genetic components from southern China and Southeast Asia (SC-SEA) increased after the Late Neolithic Age, when rice farming intensified in this region (Ning et al., 2020). The correlation between the population’s genetic change and the northward spread of rice farming technology from the Yangtze River Basin, inferred a demic diffusion pattern of agriculture in China. Compared with the ancient Central Plain populations, the modern Han has more ancestral components in common with SC-SEA, which may be a result of the continuous northward migration of rice-based agricultural populations during the formation of the Han. In addition, from the Neolithic Age to the Iron Age, the populations carried ancestral components similar with the Central Plain population were widely distributed in the surrounding areas, such as the upper Yellow River, Shaanxi, and Inner Mongolia, indicating continuous genetic exchanges between the Central Plain and the surrounding areas (Ning et al., 2020; Wang et al., 2021a).
The northwest region of China is a pathway connecting the East and the West, playing a vital role in cultural and population exchanges. The genetic history of this region has been debated for a long time.
Laying at a junction between East and Central Asia, Xinjiang has a complex demographic history. Cultural exchanges and population interactions between East and West Eurasia had occurred in this region long before the opening of the Silk Road (Shao, 2009; Damgaard et al., 2018). The peopling of Xinjiang is a highly debated topic. Mainly two hypotheses were proposed to explain the origins, the steppe hypothesis and the oasis hypothesis. The steppe hypothesis posits that the initial immigrants to Xinjiang were the Afanasievo-related populations from the Altai region north of the Tarim Basin (Mallory and Mair, 2000). In contrast, the oasis hypothesis suggests that farmers from Bactria-Margiana Archaeological Complex (BMAC) region arrived in the Tarim Basin during the Bronze Age (Chen and Hiebert, 1995). In 2021, Zhang et al. reported the findings from ancient DNA study conducted on the mummies from the Xiaohe culture in the Tarim Basin (Zhang et al., 2021). Supporting neither of the previous hypotheses, the paleogenomic analysis of the Bronze Age Tarim Basin population showed a local origin model. The genetic components of the Tarim population were neither from steppe populations nor related to BMAC populations, but rather a mixture of ancient North Eurasians (ANE) and ancient Northeastern Asians (ANA). The date of this admixture event was estimated at 10,000–6,000 years ago. The ANE-related component, currently best represented by the Tarim mummies, may have been widespread in Eurasia during the Bronze Age and prior. Although cultural exchanges occurred between Xiaohe and the surrounding populations, Xiaohe population was genetically isolated and maintained long-term genetic stability. Unlike the Xiaohe population, the early populations from the Dzungarian Basin derived a majority of their ancestral components from the Afanasievo population, and additionally, from the local ANE ancestry.
Subsequently, a genomic study of more than 200 ancient individuals in Xinjiang also proved that the ancestral components of the early Bronze Age populations in northwestern Xinjiang derived not only from the local populations, represented by the Xiaohe population, but also from the Afanasievo, the Chemurchek, and the Shamanka populations (Kumar et al., 2022). Frequent interactions and admixtures between these populations during the Bronze Age in Xinjiang and the surrounding regions, such as the western steppe, Central Asia, and Northeast Asia, reflected a complex genetic history of the Xinjiang populations. In the middle and late Bronze Age, the ancestral component from the Andronovo emerged in Xinjiang populations, and an increasing influx of ancestry from East Asia was observed. Compared to the Bronze Age populations, the Iron Age populations presented high influxes of Central Asian and East Asian ancestral components with more diverse origins, including Han and Xiongnu. The genetic profile of the Iron Age populations was still observed in historical and present-day Xinjiang populations. The presence of the Saka component in the Iron Age Xinjiang may be related to the spread of the Indo-Iranian languages in Xinjiang (Bailey, 1970).
These genomic studies of Xinjiang aid in efforts to comprehend the demographic history of the region and further contribute to the literature on the transmission of technology and language (Zhang et al., 2021; Kumar et al., 2022).
Located in the northwest, the middle and upper Yellow River region is one of the important birthplaces of Chinese civilization. Both archaeological and linguistic studies had indicated that early populations in this region had a substantial impact on the formation and development of Sino-Tibetan populations (Van, 2005). The genetic analysis of the genomes from a 5,000-year-old population in this region (Wang et al., 2021a) revealed that the Neolithic population in the Yellow River Basin once migrated to the Tibetan and the Central Plain, likely broadcasting Sino-Tibetan languages, and may be one of the common ancestors of Sino-Tibetan speakers. The Iron Age population in Taiwan derived their ancestry not only from farmers residing in the Yangtze River Basin, but also from ancient people habituated northern China (Wang et al., 2021a). The trail of findings confirmed a southward migration of the northern Chinese people. The genetic composition of modern Han can be modeled as a mixture of the ancestries from the Yellow River Basin and the Iron Age Taiwan to varying degrees, which inferred the causes of the genetic differences between the northern and southern Han.
The genetic patterns and divergent processes of the northern and southern Chinese populations are important to explore the deep population history of East Asia. To gain insights from the genomic perspective, the researchers collected Neolithic human remains spanning the coastal areas of northern China (Shandong) and Inner Mongolia (9,500–7,700 years ago), and the southern coastal areas (12,000–500 years ago) (Yang et al., 2020; Wang et al., 2021b). The genomic data indicated that the northern East Asian ancestral component, represented by the Neolithic Shandong individuals spread to the Yellow River Basin and even northward to the eastern Siberian steppes at least 9,500 years ago, whereas the populations from the southern coastal areas and Taiwan Strait island had the genetic components of the southern East Asian ancestry, represented by the Neolithic individuals from Fujian and its adjacent islands 8,400 years ago (Yang et al., 2020). These two genetic components separated into two distinct genetic lineages. Over time, the genetic differences between the northern and southern populations have gradually narrowed, suggesting frequent population migration and mixing between the north and the south since the Neolithic Age.
The southern coastal region of China, bordering Southeast Asia, has a long history of modern human occupation. It is a key area for studying the origin and evolutionary history of modern humans in eastern Eurasia and Oceania.
The paleogenomic analysis on the individuals from Fujian and the surrounding areas, dated back to 9,000 to 8,000 years ago, indicated that the southern ancestry was greatly reduced in the current southern mainland populations (Yang et al., 2020; Wang et al., 2021b). However, this ancestral component had significant influence on the present-day Austronesians, and was closely related to the Southeast Asian populations in the late Neolithic period.
The genome-wide study conducted on the human remains dating 10,686–294 years ago in Guangxi identified a previously unsampled East Asian ancestry, Longlin (10,686–10,439 cal BP, Guangxi ancestry), whose genetic profile is different from the ancient southern ancestry (Fujian ancestry) and the 8,000-year-old Hòabìnhian from Southeast Asia (Wang et al., 2021b). The interaction between populations carrying Fujian ancestry and Guangxi ancestry occurred 9,000 years ago. The Guangxi ancestry persisted until 6,400 BP in southern China. The admixture of populations between southern China and Southeast Asia occurred as early as 9,000–6,400 years ago, that is, before introducing of the agricultural technology in the region.
The observed 5 years-long rapid development of paleogenomic research conducted in China has provided a large amount of valuable information for tracing the origin and demographic history of Chinese populations, for instance, the genetic characteristics of the 40,000-year-old Tianyuan individual and the widespread of this ancestry (Yang et al., 2017; Mao et al., 2021). China has a vast territory and diverse topography. The genetic landscape and the demographic events of prehistoric populations differ regionally (Figure 1, Supplementary Table S1). The northern and southern populations diverged as early as 19,000 years ago and the genetic differences between them reduced through time, indicating continuous population interactions. Amur region experienced genetic shift at the end of the LGM, and after that, this region exhibited long-term genetic continuity. Genetic shift and population movement accompanying with the spread of agriculture were observed in West-Liao River region. Central Plain population expanded to surrounding areas and their genetic components from SC-SEA were increased through time. The dynamic pattern of Xinjiang populations can be characterized by a series of admixture between local ancestry, represented by early Xiaohe population, and surrounding populations. The southern coastal region experienced complex interaction between different genetic ancetries before the introducing of farming.
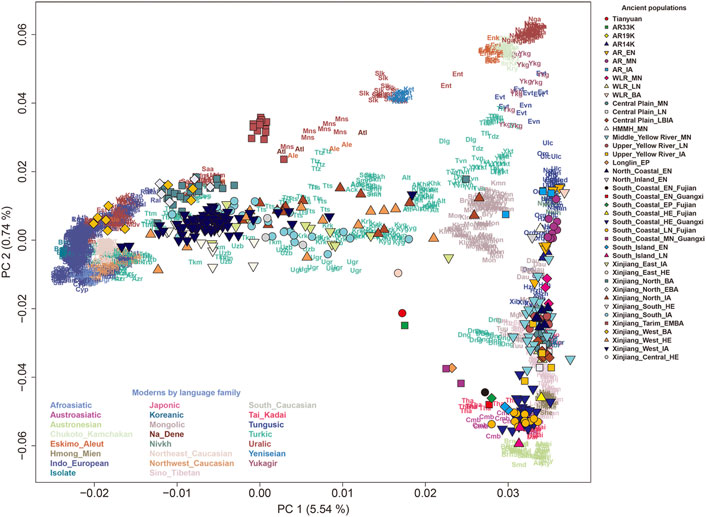
FIGURE 1. PCA Analysis for ancient Chinese populations. The PCA was constructed from present-day Eurasians in the Human Origins dataset, and the ancient individuals were projected onto the top PCs.
However, our understanding of the history of Chinese populations is far from complete. Numerous questions were put forth, such as the direct ancestry of Chinese populations, when and how the northern populations interact with the southern ones, the genetic composition and the expansion of the ancient farmers from the Yangtze River region, the origin and evolution of the Tibetans, and so on. Although there has been some accumulation of ancient genomic data, there is still a long way to reconstruct the complex genetic process of the Chinese people. In the future, with more paleogenomic data from previous unsampled regions under time and space transections, for instance, the central region of China, as well as the advances in archeometry, linguistics, bioinformatics, genomics and proteomics, multidisciplinary research can provide us with extraordinary insights into the origin of Chinese populations, the pattern of population migration and interaction, as well as the adaptation to the environment through time.
Briefly, the contributions from the individual co-authors are: YC lead the study and the writing of the MS; SG collect the published data and participate the writhing of the MS.
This work was supported by the Science and technology development project of Jilin Province (20200201138JC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer SW declared a past co-authorship with the author YC to the handling editor.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/feart.2022.1059196/full#supplementary-material
Barnett, R., and Larson, G. (2012). “A phenol–chloroform protocol for extracting DNA from ancient samples,” in Ancient DNA: Methods and protocols. Editors B. Shapiro, and M. Hofreiter (Totowa, NJ: Humana Press).
Bergström, A., Stringer, C., Hajdinjak, M., Scerri, E. M. L., and Skoglund, P. (2021). Origins of modern human ancestry. Nature 590, 229–237. doi:10.1038/s41586-021-03244-5
Carpenter, M., Buenrostro, J. D., Valdiosera, C., Schroeder, H., Allentoft, M. E., Sikora, M., et al. (2013). Pulling out the 1%: Whole-genome capture for the targeted enrichment of ancient DNA sequencing libraries. Am. J. Hum. Genet. 93, 852–864. doi:10.1016/j.ajhg.2013.10.002
Chen, K. T., and Hiebert, F. T. (1995). The late prehistory of xinjiang in relation to its neighbors. J. World Prehist. 9, 243–300. doi:10.1007/bf02221840
Chen, L., Wolf, A. B., Fu, W., Li, L., and Akey, J. M. (2020). Identifying and interpreting apparent neanderthal ancestry in african individuals. Cell. 180, 677–687.e16. e16. doi:10.1016/j.cell.2020.01.012
Choin, J., Mendoza-Revilla, J., Arauna, L. R., Cuadros-Espinoza, S., Cassar, O., Larena, M., et al. (2021). Genomic insights into population history and biological adaptation in Oceania. Nature 592, 583–589. doi:10.1038/s41586-021-03236-5
Dabney, J., and Meyer, M. (2012). Length and GC-biases during sequencing library amplification: A comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries. BioTechniques 52, 87–94. doi:10.2144/000113809
Damgaard, P. D. B., Marchi, N., Rasmussen, S., Peyrot, M., Renaud, G., Korneliussen, T., et al. (2018). 137 ancient human genomes from across the Eurasian steppes. Nature 557, 369–374. doi:10.1038/s41586-018-0094-2
Fu, Q., Hajdinjak, M., Moldovan, O. T., Constantin, S., Mallick, S., Skoglund, P., et al. (2015). An early modern human from Romania with a recent Neanderthal ancestor. Nature 524, 216–219. doi:10.1038/nature14558
Fu, Q., Posth, C., Hajdinjak, M., Petr, M., Mallick, S., Fernandes, D., et al. (2016). The genetic history of ice Age europe. Nature 534, 200–205. doi:10.1038/nature17993
Gansauge, M.-T., and Meyer, M. (2013). Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protoc. 8, 737–748. doi:10.1038/nprot.2013.038
Green, R. E., Krause, J., Briggs, A. W., Maricic, T., Stenzel, U., Kircher, M., et al. (2010). A draft sequence of the neandertal genome. Science 328, 710–722. doi:10.1126/science.1188021
Hajdinjak, M., Mafessoni, F., Skov, L., Vernot, B., HüBNER, A., Fu, Q., et al. (2021). Initial upper palaeolithic humans in europe had recent neanderthal ancestry. Nature 592, 253–257. doi:10.1038/s41586-021-03335-3
Jia, X., Sun, Y., Wang, L., Sun, W., Zhao, Z., Lee, H. F., et al. (2016). The transition of human subsistence strategies in relation to climate change during the Bronze Age in the West Liao River Basin, Northeast China. Holocene 26, 781–789. doi:10.1177/0959683615618262
Kato, H. (2006). Neolithic culture in amurland : The formation process of a prehistoric complex hunter-gatherers society. J. Graduate Sch. Lett. 1, 3–15.
Kircher, M., Sawyer, S., and Meyer, M. (2011). Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3. doi:10.1093/nar/gkr771
Korlević, P., Gerber, T., Gansauge, M. T., Hajdinjak, M., Nagel, S., Aximu-Petri, A., et al. (2015). Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. Biotechniques 59, 87–93. doi:10.2144/000114320
Kumar, V., Wang, W., Zhang, J., Wang, Y., Ruan, Q., Yu, J., et al. (2022). Bronze and Iron Age population movements underlie Xinjiang population history. Science 376, 62–69. doi:10.1126/science.abk1534
Liu, L., Duncan, N. A., Chen, X., Liu, G., and Zhao, H. (2015). Plant domestication, cultivation, and foraging by the first farmers in early Neolithic Northeast China: Evidence from microbotanical remains. Holocene 25, 1965–1978. doi:10.1177/0959683615596830
Liu, Y., Bennett, E. A., and Fu, Q. (2022). Evolving ancient DNA techniques and the future of human history. Cell. 185, 2632–2635. doi:10.1016/j.cell.2022.06.009
Liu, Y., Mao, X., Krause, J., and Fu, Q. (2021). Insights into human history from the first decade of ancient human genomics. Science 373, 1479–1484. doi:10.1126/science.abi8202
López, S., Van Dorp, L., and Hellenthal, G. (2015). Human dispersal out of Africa: A lasting debate. Evol. Bioinform. Online. 11s2, 33489. doi:10.4137/ebo.s33489
Mafessoni, F., Grote, S., De Filippo, C., Slon, V., Kolobova, K. A., Viola, B., et al. (2020). A high-coverage neandertal genome from chagyrskaya cave. Proc. Natl. Acad. Sci. U. S. A. 117, 15132–15136. doi:10.1073/pnas.2004944117
Mallory, J. P., and Mair, V. H. (2000). The Tarim mummies: Ancient China and the mystery of the earliest peoples from the west. London: Thames & Hudson.
Mao, X., Zhang, H., Qiao, S., Liu, Y., Chang, F., Xie, P., et al. (2021). The deep population history of northern East Asia from the late pleistocene to the holocene. Cell. 184, 3256–3266.e13. e13. doi:10.1016/j.cell.2021.04.040
Marciniak, S., and Perry, G. H. (2017). Harnessing ancient genomes to study the history of human adaptation. Nat. Rev. Genet. 18, 659–674. doi:10.1038/nrg.2017.65
Meyer, M., and Kircher, M. (2010). Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, 5448. doi:10.1101/pdb.prot5448
Ning, C., Li, T., Wang, K., Zhang, F., Li, T., Wu, X., et al. (2020). Ancient genomes from northern China suggest links between subsistence changes and human migration. Nat. Commun. 11, 2700. doi:10.1038/s41467-020-16557-2
Olalde, I., and Posth, C. (2020). Latest trends in archaeogenetic research of west Eurasians. Curr. Opin. Genet. Dev. 62, 36–43. doi:10.1016/j.gde.2020.05.021
Pang, X., and Gao, J. (2006). Investigation of agricultural economy in the process of civilization in the Central Plains. Agric. Archaeol. 2006, 1–13.
Parks, M., Subramanian, S., Baroni, C., Salvatore, M. C., Zhang, G., Millar, C. D., et al. (2015). Ancient population genomics and the study of evolution. Phil. Trans. R. Soc. B 370, 20130381. doi:10.1098/rstb.2013.0381
Pickrell, J. K., and Reich, D. (2014). Toward a new history and geography of human genes informed by ancient DNA. Trends Genet. 30, 377–389. doi:10.1016/j.tig.2014.07.007
Pitulko, V. (2001). “Holocene stone Age of northeastern Asia,” in Encyclopedia of prehistory: Volume 2: Arctic and subarctic. Editors P. N. PEREGRINE, and M. EMBER (Boston, MA: Springer US).
Prüfer, K., De Filippo, C., Grote, S., Mafessoni, F., Korlević, P., Hajdinjak, M., et al. (2017). A high-coverage neandertal genome from vindija cave in Croatia. Science 358, 655–658. doi:10.1126/science.aao1887
Racimo, F., Sankararaman, S., Nielsen, R., and Huerta-SáNCHEZ, E. (2015). Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 16, 359–371. doi:10.1038/nrg3936
Racimo, F., Sikora, M., Vander Linden, M., Schroeder, H., and Lalueza-Fox, C. (2020). Beyond broad strokes: Sociocultural insights from the study of ancient genomes. Nat. Rev. Genet. 21, 355–366. doi:10.1038/s41576-020-0218-z
Reich, D., Green, R. E., Kircher, M., Krause, J., Patterson, N., Durand, E. Y., et al. (2010). Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060. doi:10.1038/nature09710
Shao, H. (2009). The early collision and integration of eastern and western cultures: The cultural pattern in prehistoric xinjiang. Soc. Sci. Front 12, 146–150. doi:10.1038/s41598-022-09033-y
Shui, T. (2001). On the role and influence of the Central Plains in the process of Chinese civilization. Cultural Relics of Central China 06, 29–31.
Sikora, M., Pitulko, V. V., Sousa, V. C., Allentoft, M. E., Vinner, L., Rasmussen, S., et al. (2019). The population history of northeastern Siberia since the Pleistocene. Nature 570, 182–188. doi:10.1038/s41586-019-1279-z
Skoglund, P., and Mathieson, I. (2018). Ancient genomics of modern humans: The first decade. Annu. Rev. Genomics Hum. Genet. 19, 381–404. doi:10.1146/annurev-genom-083117-021749
Slatkin, M., and Racimo, F. (2016). Ancient DNA and human history. Proc. Natl. Acad. Sci. U. S. A. 113, 6380–6387. doi:10.1073/pnas.1524306113
Slon, V., Hopfe, C., Weiß, C. L., Mafessoni, F., De La Rasilla, M., Lalueza-Fox, C., et al. (2017). Neandertal and denisovan DNA from pleistocene sediments. Science 356, 605–608. doi:10.1126/science.aam9695
Slon, V., Mafessoni, F., Vernot, B., De Filippo, C., Grote, S., Viola, B., et al. (2018). The genome of the offspring of a Neanderthal mother and a Denisovan father. Nature 561, 113–116. doi:10.1038/s41586-018-0455-x
Stringer, C. B., and Andrews, P. (1988). Genetic and fossil evidence for the origin of modern humans. Science 239, 1263–1268. doi:10.1126/science.3125610
Sun, Y. (2015). A comparative study of prehistoric subsistence on West-Liao River region and Central Plain of China. Journal. of Liaoning Normal University (Social Science Edition) 38, 269–274. doi:10.16216/j.cnki.lsxbwk.201502269
Teng, H. (2013). Ancient environmental change impact on the origin of dryland agriculture Basin and cultural development in the West-Liao River region. Soc. Sci. J. 209, 167–170.
Teng, H. (2017). The staging and types of Hongshan culture. J. Chifeng Univ. Soc. Sci. Chin. Ed. 38, 21–24. doi:10.13398/j.cnki.issn1673-2596.2017.12.002
Van, D. G. (2005). “Tibeto-Burman vs Indo-Chinese: Implications for population geneticists, archaeologists and prehistorians,” in The Peopling of East Asia: Putting together Archaeology, Linguistics and Genetics. Editors R. Blench, L. Sagart, and A. Sanchez-Mazas (London: Routledge), 71–96.
Vicente, M., and Schlebusch, C. M. (2020). African population history: An ancient DNA perspective. Curr. Opin. Genet. Dev. 62, 8–15. doi:10.1016/j.gde.2020.05.008
Wang, C.-C., Yeh, H.-Y., Popov, A. N., Zhang, H.-Q., Matsumura, H., Sirak, K., et al. (2021a). Genomic insights into the formation of human populations in East Asia. Nature 591, 413–419. doi:10.1038/s41586-021-03336-2
Wang, T., Wang, W., Xie, G., Li, Z., Fan, X., Yang, Q., et al. (2021b). Human population history at the crossroads of East and Southeast Asia since 11, 000 years ago. Cell. 184, 3829–3841. e21. doi:10.1016/j.cell.2021.05.018
Willerslev, E., and Meltzer, D. J. (2021). Peopling of the Americas as inferred from ancient genomics. Nature 594, 356–364. doi:10.1038/s41586-021-03499-y
Wu, W., Zheng, H., Hou, M., and Ge, Q. (2018). The 5.5 cal ka BP climate event, population growth, circumscription and the emergence of the earliest complex societies in China. Sci. China Earth Sci. 61, 134–148. doi:10.1007/s11430-017-9157-1
Wu, X. (2006). Evidence of multiregional human evolution hypothesis from China. Quat. Sci. 26, 702–709.
Wu, Y. (2001). The origin and formation of the Central Plain civilization. Cultural Relics of Central China 4, 16–23.
Yang, M. A., Fan, X., Sun, B., Chen, C., Lang, J., Ko, Y.-C., et al. (2020). Ancient DNA indicates human population shifts and admixture in northern and southern China. Science 369, 282–288. doi:10.1126/science.aba0909
Yang, M. A., and Fu, Q. (2018). Insights into modern human prehistory using ancient genomes. Trends Genet. 34, 184–196. doi:10.1016/j.tig.2017.11.008
Yang, M. A., Gao, X., Theunert, C., Tong, H., Aximu-Petri, A., Nickel, B., et al. (2017). 40, 000-year-old individual from Asia provides insight into early population structure in Eurasia. Curr. Biol. 27, 3202–3208. e9. doi:10.1016/j.cub.2017.09.030
Zhang, F., Ning, C., Scott, A., Fu, Q., BjøRN, R., Li, W., et al. (2021). The genomic origins of the Bronze Age Tarim Basin mummies. Nature 599, 256–261. doi:10.1038/s41586-021-04052-7
Keywords: paleogenomics, Chinese populations, origin, evolutionary history, ancient DNA
Citation: Gao S and Cui Y (2023) Ancient genomes reveal the origin and evolutionary history of Chinese populations. Front. Earth Sci. 10:1059196. doi: 10.3389/feart.2022.1059196
Received: 01 October 2022; Accepted: 31 October 2022;
Published: 10 January 2023.
Edited by:
Yaowu Hu, Fudan University, ChinaReviewed by:
Emmanuel Paul Gilissen, Royal Museum for Central Africa, BelgiumCopyright © 2023 Gao and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinqiu Cui, Y3VpeXFAamx1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.