
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Earth Sci. , 27 September 2022
Sec. Sedimentology, Stratigraphy and Diagenesis
Volume 10 - 2022 | https://doi.org/10.3389/feart.2022.1014976
This article is part of the Research Topic Gas Hydrate Appearance Accumulation, Exploration and Exploitation in Continental Margins - Volume 2 View all 15 articles
Microbial communities and their associated metabolic activities, methanogenesis and anaerobic oxidation of methane (AOM), are the key components of carbon biocycles in continental margin sediments. The composition and diversity of microbial communities in a methanic environment have been widely investigated, but identifying direct correlations between microbial communities and their activities remains a challenge. Here, we investigated shifting microbial communities that performed methanogenesis and AOM in long-term incubations (the longest is up to 199 days). AOM, methanogenesis, sulfate reduction and iron reduction occurred during the incubation, and 16S rRNA gene sequencing showed that some bacteria were maintained or even enriched during the incubation compared to the environmental samples. In contrast, archaeal diversity was reduced, and only some uncultured archaea belonging to the phylum Bathyarchaeota were enriched after treatment with a high sulfate concentration (29.38 mM), suggesting that sulfate might promote their enrichment. Well-known anaerobic methanotrophic archaea (ANME) were not detected, and SEEP-SRB1, which is in syntrophy with ANME, decreased to approximately zero after the incubation. The abundance of known methanogens, such as genera Methanococcoides and Methanosarcina, increased slightly in some incubations but was still present at a low relative abundance (<0.15%). Incubation with a lower sulfate concentration (4 mM) and higher iron content resulted in greater δ13CO2 accumulation, indicating that iron may be the additional electron acceptor for AOM. Based on these results, other unknown or unconventional phylotypes or pathways of methanogenesis and AOM may occur during the incubation. Thus, the diversity of methanogens and anaerobic methanotrophs warrants further investigation.
Marine sediments on continental margins often contain substantial concentrations of organic carbon, which can be used by microbes for methane production (Ferry and Lessner, 2008; Zhuang et al., 2018). The produced methane may be gaseous sources of methane hydrate (Paull et al., 1994; Davie and Buffett, 2003) or even migrates out of the sedimentary column to the seawater or atmosphere (Reeburgh, 2007; Ferry and Lessner, 2008). However, most methane produced by microbe methanogenesis or seeping from gas hydrate is removed by microbial-mediated anaerobic oxidation of methane (AOM) (Reeburgh, 2007; Regnier et al., 2011; Egger et al., 2018). Therefore, AOM is considered the major biological sink of methane in marine sediments (Hinrichs and Boetius, 2003), and methanogenesis and AOM are the key components of carbon biogeocycles in continental margin sediments (Barnes and Goldberg, 1976).
AOM depends on the reduction of electron acceptors, such as SO2- 4 (Martens and Berner, 1974; Joye et al., 2004), NO- 3/NO- 2 (Ettwig et al., 2010) and Fe3+/Mn4+ (Beal et al., 2009; Joye, 2012; Egger et al., 2015). Moreover, the discovery of the artificial electron acceptor, 9,10-anthraquinone-2,6-disulfonate (AQDS), in marine sediments enabled anaerobic methanotrophic archaea (ANME) to survive alone. And that suggests potential independence of respiratory AOM (Rotaru and Thamdrup, 2016; Scheller et al., 2016). The microbes known to be responsible for sulfate-dependent AOM are ANME and sulfate-reducing bacteria (SRB) (Hinrichs et al., 1999; Boetius et al., 2000; Orphan et al., 2001). Candidatus Methylomirabilis oxyfera (M. oxyfera) and Candidatus Methanoperedens nitroreducens (M. nitroreducens) are responsible for nitrate dismutation-coupled AOM (Ettwig et al., 2010). Candidatus Methanoperedens ferrireducens can use Fe3+ as the terminal electron acceptor to mediate AOM (Cai et al., 2018) and members of the Methanoperedenaceae, Candidatus Methanoperedensmanganicus and Candidatus Methanoperedens manganireducens, can be responsible for Mn (IV)-dependent AOM (Leu et al., 2020). Moreover, studies using culture-independent biomolecular techniques have shown that the dominant microorganisms vary in the niche where AOM may occur, including subsurface sediments within the sulfate-methane transition zone (SMTZ) from gas hydrate-bearing areas and cold seep system sediments and the total prokaryotic population of ANME increases or is missing in the SMTZ. Bacterial groups, such as the JS1 group, Planctomycetes, and Chloroflexi are not typically associated with AOM and are also enriched in these environments (Inagaki et al., 2006; Reed et al., 2009; Pachiadaki and Kormas, 2013; Yanagawa et al., 2014; Gong et al., 2017; Cui et al., 2019). These findings prompted questions regarding the AOM and the microorganisms responsible.
Hence, we performed incubations with sediments from three different sites in potential gas hydrate-bearing areas in the northern South China Sea, and we treated the mixtures with 13CH4. Because the sediments from the three sites included in the incubations received a large amount of organic carbon inputs (>1%) (Supplementary Table S1), we treated the incubations with a low sulfate condition characteristic of the SMTZ to eliminate the potential utilization of other substrates (mainly organic matter) in addition to methane, as previously reported (Webster et al., 2011). However, as we reported in a previous study, AOM might be conducted above the SMTZ, where the sulfate concentration is relatively high (up to 18.76 mM) (Kong et al., 2021). We also established incubations treated with a high sulfate concentration to assess other potential microbial activities and their effects on microbial diversity and abundance. After the incubation, we used high-throughput sequencing data from microbial 16S rRNA gene amplicons in the incubation slurries and related environmental samples to analyze the shifting microbial communities and their implications.
Three sediment cores were collected during the “NORC 2018-05” cruise conducted by the “TAN KAH KEE” research vessel in 2018 in the northeastern South China Sea. The A27 and SH1 sites were sampled at sea water depths of 677 m below sea level (mbsl) and 1,451 mbsl, respectively, within the bottom stimulating reflector (BSR, one of the signs to determine the potential gas hydrate-bearing) distribution in the Shenhu area (Figure 1). This area is considered a location favorable for gas hydrate formation and conservation (Lu et al., 2011), and gas hydrate has been recovered during multiple drilling expeditions (Wu et al., 2011; Zhang et al., 2015; Zhang et al., 2017). The depth of sulfate-methane transition (SMT) in this area ranges from 7.7 m below the sea floor (mbsf) to 87.9 mbsf, and most of the SMT is shallower than 50 mbsf (Wu L. et al., 2013). The SMT at sites A27 and SH1 was predicted to be located at depths of 10.76 mbsf and 9.63 mbsf, respectively, according to the linear fitting of SO2- 4 concentrations from 4 mbsf (Kong et al., 2021). The JL1 site was sampled at a sea water depth of 3,011 mbsl near the 973-4 site, which has been reported to be influenced by gas hydrate decomposition and AOM, and the SMT was located at 9 mbsf (Liu et al., 2018; Zhang et al., 2018). We selected sediments at 4.9–5.0 mbsf from A27, 4.2–4.3 mbsf from SH1 and 5.05–5.15 mbsf from JL1, and we used samples from A27 to assess the effects of the sulfate concentration on the activities, diversity and abundance of microbial communities. According to our preliminary research, the depths of all these sediments did not reach the SMT (Liu et al., 2018; Zhang et al., 2018; Kong et al., 2021) but still they had the ability to perform AOM (Kong et al., 2021). The detailed geochemical conditions of the sampling layers are included in the Supplementary material (Supplementary Table S1).
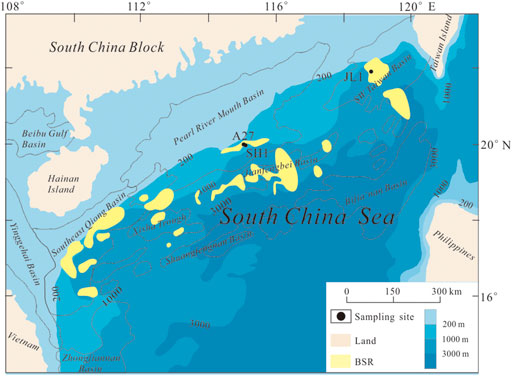
FIGURE 1. Sampling site. The bottom stimulating reflector (BSR) distribution is delineated from previous literature (Wu N. et al., 2013).
After retrieval, the sediment core was cut into two halves. One of the two halves was used to collect environmental samples for DNA extraction. Samples were cut with a sterile knife, stored in a sterile centrifuge tube, placed into a liquid nitrogen tank after the collection of 10 samples and transferred to a −80°C freezer after one sediment core was sampled. The other half was used to collect incubation and other geochemical samples. The incubation sediment samples were sliced at intervals of 10 cm with a sterile knife, placed into two sterile Zip-lock homogenous plastic bags with no headspace and preserved at 4°C in a deck refrigerator. All sampling was performed in the laboratory of the cabin during the voyage. After the field expedition, DNA samples were transported on dry ice, and the incubation samples were transported on ice to the Marine Geological Laboratory, Department of Geological Oceanography, Xiamen University where they were stored at −80°C or 4°C.
The incubation experiments were started 4 months after sampling. For the initial experimental protocol, 7 experimental and control incubation bottles were prepared for each study sediment core. We periodically extracted the headspace gas from one experimental and one control bottle and then opened it to take water and mud samples for major ion and microbial tests. However, after three tests, the headspace gas test results were unstable but showed obvious 13CO2 accumulation, indicating that this scheme was not feasible. Therefore, we changed the protocols to periodically extract the headspace gas and supernatant from one bottle and test the indicators. Therefore, we randomly selected one of the remaining culture bottles from every site as the parallel experiment for the third test culture bottle. So each culture group (two parallel experiments and one control) was pretreated as follows (calculated according to the modified experimental protocol): 51 g of wet sediments (∼45 ml) in each selected layer was homogenized in 135 ml of bottom seawater (treated with or without BaCl2), then 5 ml of this suspension were extracted each time and placed separately into 3 culture bottles, and finally 60-ml suspension was distributed in approximately 110-ml culture bottles. The approximate ratio of sediment to medium was selected according to previously reported incubation studies (Beal et al., 2009; Segarra et al., 2013; Egger et al., 2015). The bottom seawater was collected at site A27 using a conductivity-temperature-depth (CTD) instrument. The sulfate concentration of the bottom seawater was 29.38 mM. The bottom seawater treated with BaCl2 was filtered twice with membrane filters (0.2 μm) after being well mixed to remove BaSO4, resulting in approximately 4 mM sulfate. All bottom seawater samples treated with or without BaCl2 were filter-sterilized and degassed with N2 (99.999%; the same nitrogen was used throughout the study) before mixing with sediments. After the contents were mixed, the culture bottles were sealed with airtight butyl rubber stoppers and secured with open-top Al screw caps. After sealing, the contents were purged with nitrogen for 20 min. After all pretreatment procedures, 50 ml of labeled methane (99% 13CH4 and 1% 12CH4) and nitrogen were injected into the headspace of the experimental and control groups, respectively, to yield 2 bar overpressures (volume headspace ≈50 ml). Each incubation was numbered (see details in Table 1) and incubated in the dark at 20°C with shaking (300 r/min).
Headspace samples were taken periodically. The CO2 content and the δ13C-CO2 in the headspace of all samples were analyzed with a gas chromatograph (Agilent 6,820, United States) coupled to a mass spectrometer (Delta V Advantage, Thermo Fisher, Bremen, Germany). The analysis column was a Poraplot Q-type capillary column, and He was used as the carrier gas. The heating procedure was as follows: an initial temperature of 50°C, a constant temperature for 2 min, an increase in temperature 180°C at a rate of 25°C/min, and a constant temperature for 8 min. The assay was repeated three times. The test results are presented as the content of δ13CCO2 (in ‰ vs. Vienna Peedee Belemnite (VPDB); precision and accuracy were ±0.2‰) and R, which was defined as 13C/12C. The headspace CH4 concentration in the control groups was analyzed in the final sampling period with a gas chromatograph (with a pyrolysis furnace at 960°C and reducing furnace at 600°C) coupled to a mass spectrometer using a HP-PLOT Q column with helium as the carrier gas (flow rate of 1.5 ml/min). The measurement error was< 3% (obtained by repeated the test testing twice for each sample). The CH4 concentrations in the headspace were then converted to dissolved CH4 contents as previously reported (Johnson et al., 1990). All tests were performed at the Third Institute of Oceanography, Ministry of Natural Resources.
Because 13CH4 was the only stable isotope tracer added during the incubation, we defined that the 13CO2 that formed in the headspace was fully converted from 13CH4 oxidation. The fractional abundances of 13CO2 [13F=13C/(12C+13C)] were used to quantify the AOM activity as previously reported (Beal et al., 2009; Scheller et al., 2016; Bray et al., 2017). The AOM rates were calculated during the period of the linear increase in 13CO2 formation (Beal et al., 2009; Egger et al., 2015; Scheller et al., 2016; Bray et al., 2017). However, accurate quantification of the concentration of inorganic carbon formed from methane oxidation is challenging (Scheller et al., 2016). Therefore, we estimated inorganic carbon formed from the total headspace CO2 and the dissolved CO2 in the incubation liquid, which were calculated according to Henry’s law based on only the loss due to each headspace sampling procedure.
Supernatant samples were collected after every headspace sampling procedure and allowed to stand until the supernatant was clarified. Approximately 2 ml of supernatant liquid was drawn out by the disposable syringe with a long sterile needle (20G, inner, 0.6 mm, outter, 0.9 mm). The supernatant liquid was used to analyze the SO2- 4 concentration by ion chromatography (IC) in a 100-fold diluted sample (0.01 ml of supernatant liquid with 9.99 ml of de-oxygenated UHQ water) as Zhang et al. (2015) and the Fe2+ concentration was measured by the UV spectrophotometry in a 5-fold diluted sample (0.5 ml of supernatant liquid with 2 ml of ion chromatography). Utilized 0-phenanthroline reacted with Fe2+ then colorated, a wave-length of 510 nm was determined by UV-absorption spectroscopy. Linear and well reproducible calibration curves were obtained in the concentration range of 0–5 mg/L Fe2+ standard solution made by Fe3+ standard solution with 10% Hydroxylamine Hydrochloride. The standard deviation of this method was found less than 2%.
We collected 2.5 ml of slurry from every incubation after the final headspace gas sample was collected, and we centrifuged the samples at 12,000 g for 5 min and removed the supernatant. DNA was extracted from 0.5 g of sediment using the E.Z.N.A. Soil DNA Kit (OMEGA, United States) according to the manufacturer’s instructions.
The V4 regions of the 16S rRNA genes were amplified using the ArBa515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and Arch806R (5′-GGACTACVSGGGTATCTAAT-3′) primers, which target conserved sequences present in bacteria and archaea (Bates et al., 2011). Based on the preliminary experiments, we used the TransGen AP221-02: TransStart Fastpfu DNA Polymerase (20 μL) system, and the minimum number of thermal cycles was set to 27. The thermal cycling conditions were as follows: initial denaturation at 95°C for 3 min; 27 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 45 s; elongation at 72°C for 10 min; and a hold at 10°C. The correct size of the amplicons (∼250 bp) was verified by 2% agarose gel electrophoresis and purification using Diffinity RapidTips (Sigma-Aldrich, United States). Purified amplicons were sequenced using the Illumina MiSeq platform at Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China, according to standard protocols.
The methyl coenzyme reductase subunit A (mcrA) geneencodes the key enzyme in methane metabolism in all well-known methanogens (Luton et al., 2002). We amplified the functional mcrA gene after final headspace sampling using the 5′-GGTGGTGTMGGATTCACACARTAYGCWACAGC-3′ and 5′-TTCATTGCRTAGTTWGGRTAGTT-3′ primers as previously reported (Luton et al., 2002). The Double Taq Plus Master Mix (10 μL) system was used, and the minimum number of thermal cycles was set to 37. The thermal cycling conditions were as follows: initial denaturation at 95°C for 3 min; 37 cycles of denaturation at 95°C for 30 s, annealing at 53°C/55°C/58°C for 30 s and extension at 72°C for 45 s; elongation at 72°C for 10 min; and a hold at 10°C.
Raw fastq files were quality-filtered using QIIME61 (version 1.9.1 http://qiime.org/install/index.html). Reads that were unable to be assembled were rejected. Operational taxonomic units (OTUs) were clustered by UPARSE (version 11 http://drive5.com/uparse/) with 97% similarity. After quality filtering and clustering, OTUs at the 97% identity level were obtained, and phylotypes were assigned an identity based on comparisons with sequences in the SILVA (SSU111) database (http://www.arb-silva.de). After each sample was normalized to the minimum OTUs, the alpha diversity, Good’s coverage, beta diversity, composition and abundance analyses of the libraries were completed. The data preprocessing and OTU-based analysis were performed using Mothur (version v.1.30.1 http://www.mothur.org/wiki/Schloss_SOP#Alpha_diversity) and the R language toolkit at the free online Majorbio I-Sanger Cloud Platform (www.i-sanger.com).
Source sequences are available in the Sequence Read Archive (SRA) database under the accession number PRJNA627291.
Initially, we used incubations injected with N2 in the headspace as the controls for incubations injected with 13CH4. At the end of the incubation period, however, CH4 concentrations were present in the headspace of the control groups. This result indicated that methanogenesis occurred in the controls during the incubation. Compared to the controls, the experimental groups exhibited a significant accumulation of 13CO2 in the headspace gas, indicating that the AOM occurred. Meanwhile, the SO2- 4 and Fe2+ concentrations in the supernatant of each incubation fluctuated revealing that sulfate reduction and iron reduction occurred during the incubation.
Methanogenesis: At the end of the 117-days incubation, we measured the headspace methane content of all control groups. Methane production rates were calculated over the period of linear increase of the dissolved CH4 concentration. The headspace of all control groups contained a certain methane concentration (52.56–193.81 ppm, Table 2). The A27s group produced less methane than theA27, and the SH1 group produced more methane than the A27 group. The methane production rate in each group was calculated. SH1_cntl had the highest methane production (81.67 nmol/g/yr) followed by A27_cntl (62.27 nmol/g/yr), A27s_cntl (58.86 nmol/g/yr) and JL1_cntl (22.15 nmol/g/yr).
Anaerobic methane oxidation: Using the calculation method mentioned in the Methods section, the headspace gas tests and calculations are provided in the supplementary material (Supplementary Table S2). As shown in Figure 2, all the experimental groups showed much greater 13CO2 enrichment but much lower total CO2 production than the control groups. The headspace CO2 concentration did not change accordingly with 13FCO2. Moreover, the A27s group produced more CO2 than the A27 group. However, the patterns among A27, SH1 and JL were difficult to distinguish. We determined the AOM rate in the experimental group to be 4.5353–27.0239 nmol/g/yr. At each stage, the AOM rate in group A27 was higher than that in the other two site groups, and the A27s group showed a slightlly higher AOM rate than the A27 group.
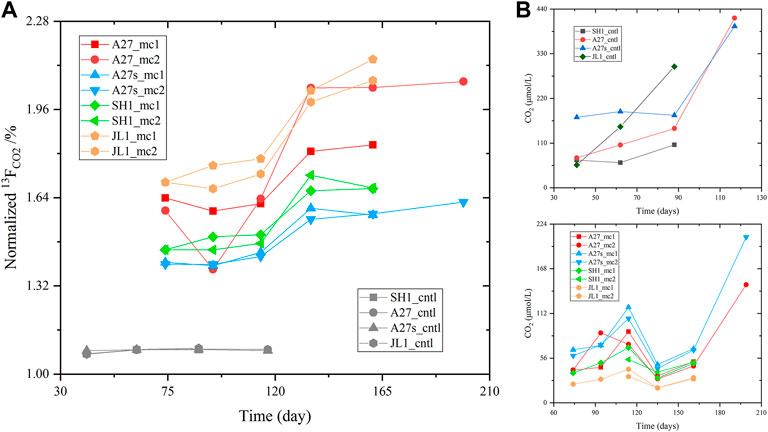
FIGURE 2. Time course of tested (A) normalized 13FCO2 (13CO2/(12CO2+13CO2), which was calculated based on the δ13C-CO2 measured during the incubation period) and (B) CO2 concentrations in the headspace. The serial number of each incubation sample and its corresponding incubation conditions are listed in Table 1.
Sulfate reduction: The sulfate concentration in the supernatant showed a fluctuating trend during the incubation (Figure 3A). A clear pattern of change was not observed in the comparison. However, a clear decreasing was detected in some experimental groups (A27, A27s, and SH) at the end of incubation.
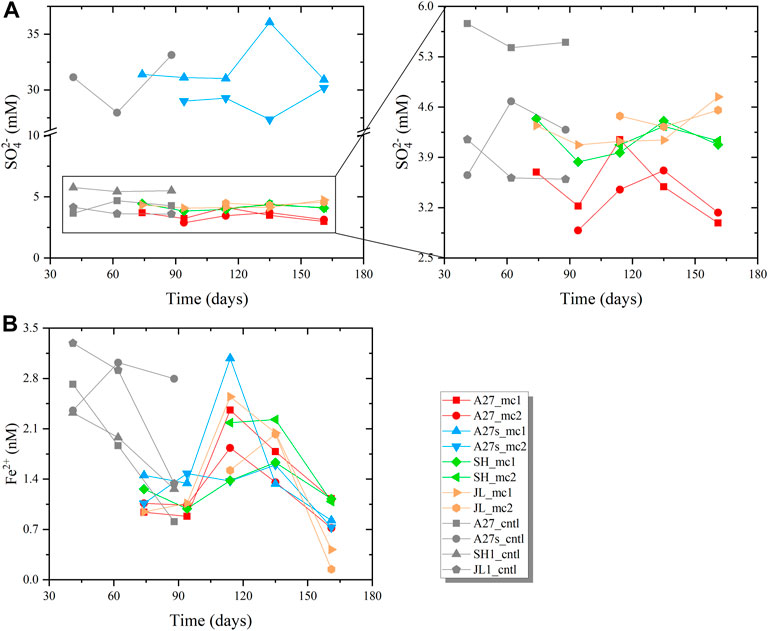
FIGURE 3. Time course of (A) the measured SO2- 4 concentrations and (B) Fe2+ concentrations in the supernatant liquid. The enlarged graph in (A) on the right shows the detailed changes observed in incubations with treated bottom seawater. Because the incubation scheme was modified, the supernatant of the control groups was only sampled three times, and the sampling times were inconsistent with the experimental group.
Iron reduction: Figure 3B shows the increasing trend for the Fe2+ concentration in the supernatant at the early stage, especially during the period from 94 to 117 days, and the Fe2+ content increased rapidly. However, the Fe2+ concentration decreased in all experimental groups. In addition, the Fe2+ concentration in the supernatant exhibited a decreasing trend in almost of the control groups (Figure 3B). The trends in all experimental groups indicated that iron reduction occurred during the first 117 days. The decreasing Fe2+ concentration in the late stage (after 117 days) may have been caused by precipitation with other elements.
To determine the differences of microbial diversity after the incubation, 16S rRNA gene libraries were constructed with the DNA extracted from the 8 slurry samples and 3 corresponding environmental sediment samples collected at the following depths: 5.0 mbsf in the A27 core, 4.2 mbsf in the SH1 core and 5.15 mbsf in the JL1 core. A total of 621,679 sequences were obtained after quality filtering. After clustering, each sample was normalized to the minimum sequences (35,449), and 389,939 sequences were included in the final OTU table (Supplementary Table S3). Estimators of alpha diversity were calculated after normalization, and Good’s coverage indexes were 0.9785–0.9961 (Supplementary Table S4), indicating an adequate sequencing depth, which was further confirmed by rarefaction analysis (Supplementary Figure S5).
The species richness (Chao 1 and sobs indexes) and evenness (Shannon and Simpson indexes) of the microbial communities showed a decreasing trend in the slurry samples compared to those in the environmental sediment samples (Figure 4). Groups amended with sufficient SO2- 4 (A27s and A27s_cntl, 29.38 mM) showed higher values than those amended with low SO2- 4 concentrations (A27 and A27_cntl, 4 mM), and the experimental groups (A27_mc, A27s_mc, SH1_mc and JL1_mc) typically showed lower values than controls (A27_cntl, A27s_cntl, SH1_cntl and JL1_cntl).
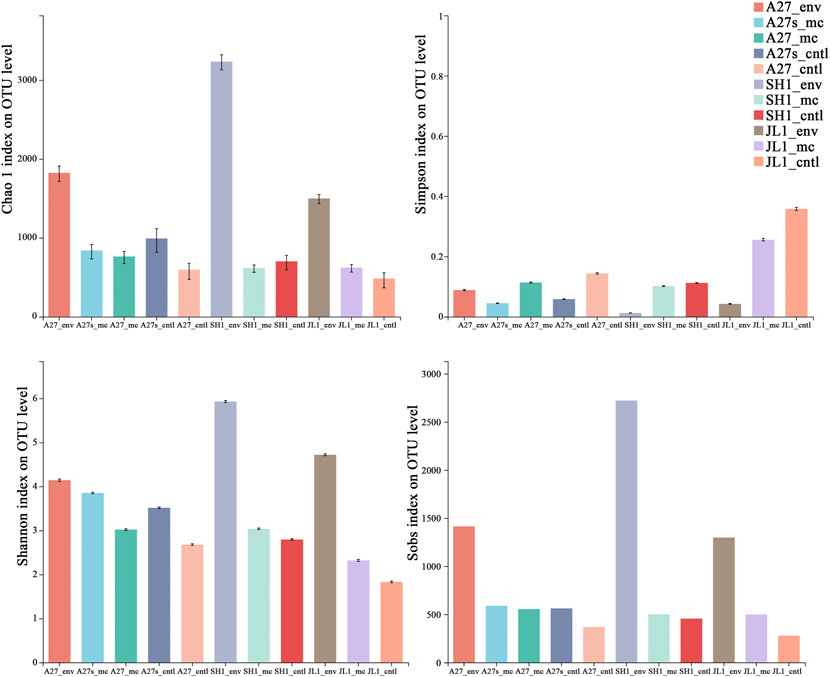
FIGURE 4. Alpha diversity estimators of microbial community 16S gene sequences at the OTU level for environmental sediment samples (sampling site_env) and the related incubation slurry samples amended with 13CH4 (sampling site_mc) and N2 (sampling site_cntl).
The cluster analysis reflected the effects of incubation conditions for the incubation slurries and environmental sediment communities. The incubation slurries amended with different SO2- 4 concentrations were distinctly separated from one another (Figure 5A).
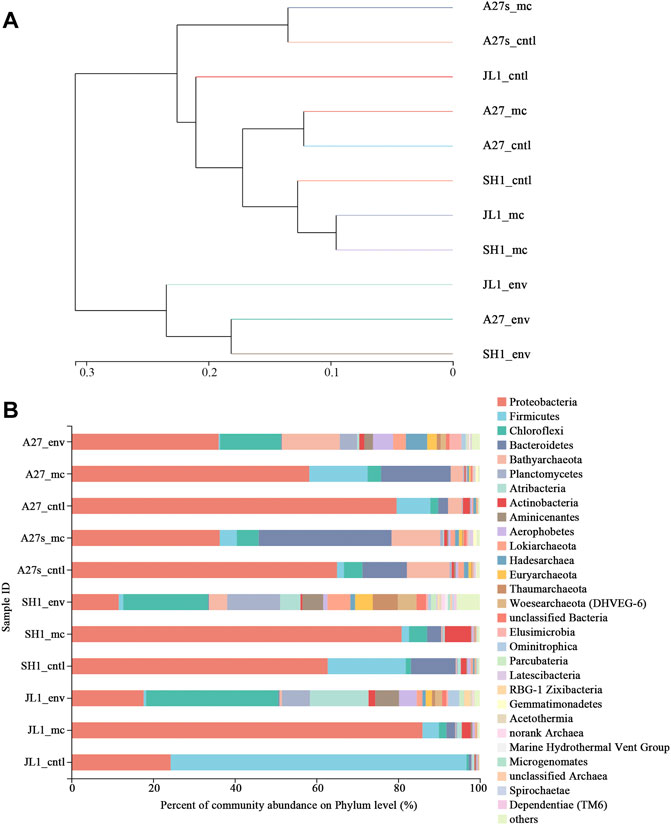
FIGURE 5. Comparison of microbial communities among all samples. (A) Hierarchical cluster analysis using pairwise weighted UniFrac distances. The scale bar between branches represents the distance between samples. (B) Community abundance of microorganisms at the phylum level for environmental sediment samples (sampling site_env) and the related incubation slurry samples amended with 13CH4 (sampling site_mc) and N2 (sampling site_cntl). Taxa with abundances <0.5% were included in “others”.
Seventy-four prokaryotic phyla were identified from the 16S rRNA gene sequences (Supplementary Table S3). Phyla with less than 0.5% abundance were classified as others, and the abundance of each phylum is shown in Figure 5B. The changes in community structure after incubation were mainly focused on the abundance of bacteria. The proportion of the phyla Proteobacteria, Bacteroidetes, Firmicutes, Actinobacteria, Latescibacteria, and Gemmatimonadetes increased after the incubation, and the proportions of other bacteria decreased but were maintained at a certain value. In contrast, the abundances of archaea were mainly reduced after incubation. Archaea sequences belonging to Bathyarchaeota, Lokiarchaeota, Hadesarchaea, Euryarchaeota, Thaumarchaeota, and Woseearchaeota (DHVEG-6) were maintained after incubation.
The 5 most dominant OTUs of bacteria in the incubation samples belonged to the genera Pseudomonas, Halomonas, Marinobacter, family Sva1033 and order Clostridiales. The 5 most dominant OTUs of archaea in the incubation samples belonged to the phyla Bathyarchaeota, Lokiarchaeota, and Hadesarchaea. Only OTU2228 and OTU 5554 were enriched after the incubation, and they belonged to uncultured or unclassified Bathyarchaeota. The lineages of OTU 2228 and OTU 5554 were similar to those of the MCG-B and MCG-C groups, respectively (Figure 6B). The proportion of OTU 2228 increased from 7.40% (A27_env) before incubation to 9.23% (A27s_mc) in the A27 incubations amended with sufficient sulfate. OTU 5554 was slightly enriched in JL1_mc (from 0.06% in the environmental sample to 0.10% in the slurry) and was maintained at a certain proportion in other groups. The proportion of other OTUs belonging to the phylum Bathyarchaeota were slightly decreased but maintained certain abundances during the incubation (Figure 6A). Enriched OTUs of phylum Bathyarchaeota were present in the experimental and control sediments amended with a sufficient sulfate concentration (29.38 mM), and a higher proportion of phylum Bathyarchaeota was present in groups treated with a sufficient sulfate concentration (29.38 mM) compared to groups treated with a low sulfate concentration (4 mM).
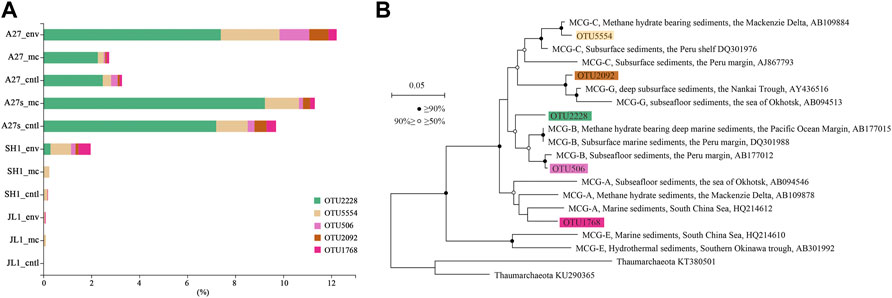
FIGURE 6. Community abundance and lineage of enriched OTUs in the phylum Bathyarchaeota. (A). The top five OTUs in the phylum Bathyarchaeota. (B). Neighbor-joining 16S rRNA gene tree showing the placement of representative members of Bathyarchaeotal relative to environmental sequences, including genes recovered from marine sediments. Thaumarchaeota 16S rRNA sequences from reference genomes were used as an outgroup. Bathyarchaeota (formerly MCG) groups are mainly based on a previous classification (Meng et al., 2014). The stability of the topology was evaluated by bootstrapping (1,000 replicates). Nonparametric support values are indicated with white (≥70%) and black (≥90%) circles. The environmental context and National Center for Biotechnology Information (NCBI) accession numbers are shown. Scale bars indicate the expected number of substitutions per site.
Well-known ANME sequences were not detected in any of the samples. Well-known methanogens in the phylum Euryarchaeota, genera Methanococcoides, Methanobrevibacter, Methanosarcina and Methanobacterium were detected in the incubation groups (Supplementary Table S3). However, the proportion of these genera were low, and the highest value was only 0.15% (A27s_mc). Compared to the environmental samples, the abundance of some methanogens belonging to uncultured or unclassified species in the genera Methanococcoides and Methanosarcina increased, but none were present across all samples from the control groups. We also attempted toamplify mcrA from the DNA samples from all incubation slurries under three different annealing conditions (50/55/58°C), but all failed. Considering the amount of sample available, we did not perform further experiments.
We divided the 11 samples into the following 3 groups to perform the one-way ANOVA: environmental group (Env), incubation group amended with 13CH4 (Exp) and incubation group amended with N2 (Cntl). In terms of the top 15 relative abundances of microorganisms at the phylum level, the phyla Proteobacteria and Chloroflexi presented significant differences (0.01 <p value <0.05)between the three groups of samples (Figure 7A). In terms of the top 15 relative abundances of microorganisms at the genus level, the genera Halomonas, Marinobacter, and norank genus in MSBL9 presented significant differences (0.001 <p value <0.05) among the three groups (Figure 7B).
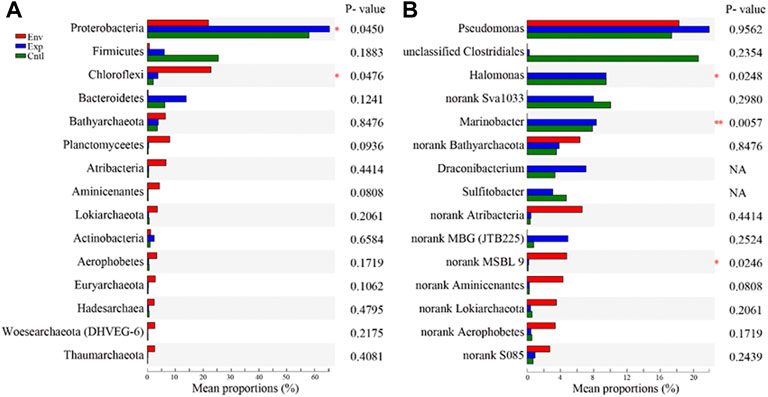
FIGURE 7. Statistical comparison of the relative abundances (A) at the phylum level and (B) at the genus level among the environmental sediment samples (Env) and the related incubation slurries amended with 13CH4 (Exp)and N2 (Cntl). One-way ANOVA was used to evaluate the significance of differences between the indicated groups. *p <0.05 and **p <0.01.
No significant differences in archaea and bacteria were observed at the phylum level between the experimental groups and control groups. At the genus level, however, the low abundance genera Alcanivorax, Streptomyces, Filomicrobium and Pelomonas presented significant differences (Supplementary Figure S6). We also compared the differences in microbial communities between the two experimental incubation slurries and their controls treated with different sulfate concentrations. A higher proportion of archaea was present in the experiment and control samples treated with high sulfate concentrations (29.38 mM) (Supplementary Figure S7).
In the present study, the enriched microorganisms differed from those previously reported in previous studies using incubations to detect AOM (Beal et al., 2009; Webster et al., 2011). The most striking feature of the enriched microorganisms was the overwhelming dominance of bacteria, including the uncultured or unclassified species in Pseudomonas, Halomonas, Marinobacter, Sva 1,033 and Desulfobulbaceae, but none were widely enriched across all incubation slurries. These phylogenetic groups are presentin various marine sediments and are regularly detected in methanic environments (Reed et al., 2002; Inagaki et al., 2006), but their abundance is relatively low. AOM, methanogenesis, sulfate reduction and iron reduction occurred during the incubation in the present study. Thus, the enrichment of these unclassified or uncultured species in our anaerobic incubation indicated that they may be directly or indirectly involved in these biogeochemical processes.
Notably, ANME sequences were not present in any of the samples. However, an abundance of labeled methane was oxidized anaerobically during the incubation. Previous studies have shown that ANME are not present in some geochemical environments where AOM dominates (Lin et al., 2014; Yanagawa et al., 2014; Katayama et al., 2016). Some researchers have speculated that the dominant phylum Bathyarchaeota might participate directly or indirectly in AOM in methane-rich environments (Lin et al., 2014; Katayama et al., 2016; Fan et al., 2017; Cui et al., 2019). Moreover, Bathyarchaeota might have the potential for methane metabolism (Evans et al., 2015). In addition, researchers have recently speculated that Bathyarchaeota express genes encoding MCR enzymes that are potentially required for alkane metabolism rather than methane metabolism (Laso-Pérez et al., 2016; Evans et al., 2019). In the present study, the phylum Bathyarchaeota was enriched in experimental groups with no ANME, and AOM still occurred during the incubation further confirming the hypothesis that Bathyarchaeota may participate in AOM in some way.
In addition, methanogens have been previously reported to perform AOM (Soo et al., 2016), and they contain the mcrA gene, which conducts AOM through “reverse methanogenesis” (Hallam et al., 2004). After incubation in the present study, the abundance of some methanogens belonging to uncultured or unclassified species in the genera Methanococcoides and Methanosarcina increased in the A27 experimental groups but was still considered relatively low (<0.15%). However, this change was not universal in all culture groups. The other control groups that produced a certain amount of methane contained fewer or no methanogen sequences, which may explain why we failed to amplify the mcrA gene. Thus, these findings suggested that other unknown or unconventional phylotypes or pathways may perform AOM and methanogenesis during the incubation. The uncultured or unclassified lineage belonging to the phylum Bathyarchaeota may be the most promising lineage and is worthy of further investigation.
Webster et al. (2011) suggested that a low sulfate concentration promotes the enrichment of representative SMTZ prokaryotes. However, we only detected some of the representative SMTZ bacteria, such as Gammaproteobacteria, Deltaproteobacteria, Firmicutes, and Bacteroidetes, and the bacteria not known to be involved in AOM, such as Actinobacteria (Harrison et al., 2009), were enriched in thelow concentration of sulfate (4 mM) group. We also found that the sulfate concentration exerted a marked effect on archaeal community enrichment/maintenance as reflected by the higher proportion of archaea sequences in the slurries amended with high concentrations of sulfate (29.38 mM) group. The enrichment of some lineages of the phylum Bathyarchaeota in slurries amended with high concentration of sulfate (29.38 mM) suggested that sulfate promotes their enrichment.
Sulfate reduction related AOM has been suggested to be the most strongly cooperative metabolic process that controls methane emissions from marine sediments (Barnes and Goldberg, 1976; Iversen and Jorgensen, 1985; Nauhaus et al., 2002; Knittel and Boetius, 2009). SR-AOM is presumed to be most active in the SMTZ where sulfate is depleted to approximately zero and the AOM rate is the highest (Iversen and Jorgensen, 1985). In our incubations, more 13CH4 was oxidized in the experimental groups with a low concentration of sulfate (A27, ∼4 mM) than in those with a high concentration of sulfate (A27s, 29.38 mM), but we did not determine whether a high sulfate concentration reduces the AOM rate or whether electron acceptors other than sulfate are used during the incubation, leading to a higher AOM rate. Because a high concentration of iron was present in the sediment, reactive iron reduction in sediments may have stimulated sulfate-driven AOM (Sivan et al., 2014) or coupled to AOM to remove large amounts of CH4 (Egger et al., 2015). Thus, a reasonable assumption is that iron may be involved in the AOM during the incubation. Iron reduction was observed in all groups in the initial period of incubation, the Fe2+ levels were clearly increased and, even showed a rapid increasing trend from 74 to 117 days in this stage. The iron reduction rate was similar to the AOM rate in the A27 and SH1 groups. Our major element test and sequential reactive iron extraction results showed that the incubation of sediments of A27 contained a higher reactive iron content (1.33%) and total iron (6.18%) than those of SH1 (0.67 and 4.82%, respectively) (Supplementary Table S1). If iron is merely a stimulant, the iron content does not exert much of an effect on the AOM rate. However, iron may be the electron acceptor, which is energetically more favorable than sulfate in AOM (Beal et al., 2009; Ettwig et al., 2010; Joye, 2012). Therefore, we suggest that the higher reactive iron content led to the anaerobic oxidation of additional methane, resulting in higher AOM rates in the two groups incubated with lower sulfate concentrations.
Numerous bacterial genera enriched in our incubations have been identified to reduce ferric iron, including the dominant and enriched genus Pseudomonas (Johnson and Mcginness, 1991; Naganuma et al., 2006), the genus Halomonas (Hajizadeh et al., 2015) and the genus Shewanella (Kim et al., 1999; Weber et al., 2006; Wang et al., 2008). The enriched family Sva1033, which is most closely related to Desulfuromonas palmitatis, has been reported to be a dissimilatory iron reducer capable of oxidizing long-chain fatty acids (Coates et al., 1995) and is hypothesized to undergo metabolic switching from metal reduction to sulfate reduction (Buongiorno et al., 2019). An investigation of microbial communities in metal reduction-coupled AOM incubations has suggested that metal-reducing bacteria play a vital role in metal-dependent AOM (Beal et al., 2009). The same conclusion has been proposed based on investigations of the microbial community in methanic sediments (Li et al., 2019). Thus, the enriched dissimilatory iron-reducing bacteria might directly or indirectly participate in AOM, further suggesting a role for iron reduction in AOM in our incubations.
In this study, we reported the incubation results and the shifting diversity and composition of microbial communities. The results of headspace gas indicated that AOM occurred in the experimental groups and that methane production occurred in the control groups during incubations. The enriched microorganisms in our incubations have been reported to degrade organic matter, reduce sulfate, reduce iron and produce methane. However, the well-known ANME, responsible organisms of AOM, were not detected. We propose that the enriched Bathyarchaeata have the potential to perform AOM during incubation. Additionally, more 13CH4 was oxidized to 13CO2 in the experimental groups treated with low concentrations of sulfate (∼4 mM) than in those treated with high concentrations of sulfate (29.38 mM), and more 13CH4 was oxidized to 13CO2 in the A27 groups, which contained more reactive iron and total iron than SH1 groups. Furthermore, bacteria with dissimilatory iron-reducing metabolism were enriched after incubation. These results suggested that additional electron acceptors may be involved in AOM and that iron is the best candidate. The known methanogens were only detected at low abundance in some samples from the control group, suggesting that methanogenesis may be more phylogenetically widespread than currently appreciated. The shifting of microbial communities before and after culture in this study provided a better understanding of AOM, methanogenesis and the responsible microorganisms.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YK; Data curation, Formal analysis, Investigation, Software, Visualisation, Writing-original draft, Writing-review and editing. HL; Project administration, Supervision, Writing-review and editing, Validation. WC; Investigation, Methodology, Data curation, BW; Investigation, Methodology. FP; Investigation, Software, Visualization. FH; Investigation, Formal analysis.
This work was funded by the National Natural Science Foundation of China (Grant Nos. 41773078 and 41276046), the Fundamental Research Funds of Xiamen University (Grant No. 20720180114) and the China Ocean Mineral Resources Research and Development Association (DY135-B2-13).
We would like to thank the National Natural Science Foundation of China (Grant Nos. 41773078 and 41276046) and the Fundamental Research Funds of Xiamen University (Grant Nos. 20720180114) for funding this study. We would also like to thank the captain, crew and scientists of the NORC 2018–05 voyage for the gravity core sampling.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/feart.2022.1014976/full#supplementary-material
Barnes, R. O., and Goldberg, E. D. (1976). Methane production and consumption in anoxic marine sediments. Geology 4 (1976), 297–300. doi:10.1130/0091-7613(1976)4<297:MPACIA>2.0.CO;2
Bates, S. T., Berg-Lyons, D., Caporaso, J. G., Walters, W. A., Knight, R., and Fierer, N. (2011). Examining the global distribution of dominant archaeal populations in soil. ISME J. 5 (5), 908–917. doi:10.1038/ismej.2010.171
Beal, E. J., House, C. H., and Orphan, V. J. (2009). manganese- and iron-dependent marine methane oxidation. Science 325 (5937), 184–187. doi:10.1126/science.1169984
Boetius, A., Ravenschlag, K., Schubert, C. J., Rickert, D., Widdel, F., Gieseke, A., et al. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407 (6804), 623–626. doi:10.1038/35036572
Bray, M. S., Wu, J., Reed, B. C., Kretz, C. B., Belli, K. M., Simister, R. L., et al. (2017). Shifting microbial communities sustain multiyear iron reduction and methanogenesis in ferruginous sediment incubations. Geobiology 15, 678–689. doi:10.1111/gbi.12239
Buongiorno, J., Herbert, L. C., Wehrmann, L. M., Michaud, A. B., Laufer, K., Røy, H., et al. (2019). Complex microbial communities drive iron and sulfur cycling in arctic fjord sediments. Appl. Environ. Microbiol. 85 (14), e00949–00919. doi:10.1128/aem.00949-19
Cai, C., Leu, A. O., Xie, G. J., Guo, J., Feng, Y., Zhao, J. X., et al. (2018). A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction. ISME J. 12 (8), 1929–1939. doi:10.1038/s41396-018-0109-x
Coates, J. D., Lonergan, D. J., Philips, E. J. P., Jenter, H., and Lovley, D. R. (1995). Desulfuromonas palmitatis sp. nov., a marine dissimilatory Fe(III) reducer that can oxidize long-chain fatty acids. Arch. Microbiol. 164 (6), 406–413. doi:10.1007/bf02529738
Cui, H., Su, X., Chen, F., Holland, M., Yang, S., Liang, J., et al. (2019). Microbial diversity of two cold seep systems in gas hydrate-bearing sediments in the South China Sea. Mar. Environ. Res. 144, 230–239. doi:10.1016/j.marenvres.2019.01.009
Davie, M. K., and Buffett, B. A. (2003). Sources of methane for marine gas hydrate: Inferences from a comparison of observations and numerical models. Earth Planet. Sci. Lett. 206 (1), 51–63. doi:10.1016/S0012-821X(02)01064-6
Egger, M., Rasigraf, O., Sapart, C. J., Jilbert, T., Jetten, M. S., Röckmann, T., et al. (2015). Iron-mediated anaerobic oxidation of methane in brackish coastal sediments. Environ. Sci. Technol. 49 (1), 277–283. doi:10.1021/es503663z
Egger, M., Riedinger, N., Mogollón, J. M., and Jørgensen, B. B. (2018). Global diffusive fluxes of methane in marine sediments. Nat. Geosci. 11 (6), 421–425. doi:10.1038/s41561-018-0122-8
Ettwig, K. F., Butler, M. K., Paslier, D. L., Pelletier, E., Mangenot, S., Kuypers, M. M. M., et al. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464 (7288), 543–548. doi:10.1038/nature08883
Evans, P. N., Boyd, J. A., Leu, A. O., Woodcroft, B. J., Parks, D. H., Hugenholtz, P., et al. (2019). An evolving view of methane metabolism in the Archaea. Nat. Rev. Microbiol. 17 (4), 219–232. doi:10.1038/s41579-018-0136-7
Evans, P. N., Parks, D. H., Chadwick, G. L., Robbins, S. J., Orphan, V. J., Golding, S. D., et al. (2015). Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350 (6259), 434–438. doi:10.1126/science.aac7745
Fan, X. B., Liang, Q. Y., Niu, M. Y., Yu, T. T., Wang, Y. S., and Wang, F. P. (2017). The diversity and richness of archaea in the northern continental slope of South China Sea. Microbiol. China 44 (7), 1589–1601. doi:10.13344/j.microbiol.china.170159
Ferry, J. G., and Lessner, D. J. (2008). Methanogenesis in marine sediments. Ann. N. Y. Acad. Sci. 1125 (1), 147–157. doi:10.1196/annals.1419.007
Gong, J., Sun, X., Xu, L., and Lu, H. (2017). Contribution of thermogenic organic matter to the formation of biogenic gas hydrate: Evidence from geochemical and microbial characteristics of hydrate-containing sediments in the Taixinan Basin, South China Sea. Mar. Petroleum Geol. 80, 432–449. doi:10.1016/j.marpetgeo.2016.12.019
Hajizadeh, N., Sefidi Heris, Y., Zununi Vahed, S., Vallipour, J., Hejazi, M., Golabi, S., et al. (2015). Fe(III) reduction by halomonas sp. TBZ9 and maribobacter sp. TBZ23, isolated from urmia lake in Iran. Adv. Environ. Biol. 8, 59–65.
Hallam, S. J., Putnam, N., Preston, C. M., Detter, J. C., Rokhsar, D., Richardson, P. M., et al. (2004). Reverse methanogenesis: Testing the hypothesis with environmental genomics. Science 305 (5689), 1457–1462. doi:10.1126/science.1100025
Harrison, B. K., Zhang, H., Berelson, W., and Orphan, V. J. (2009). Variations in archaeal and bacterial diversity associated with the sulfate-methane transition zone in continental margin sediments (santa barbara basin, California). Appl. Environ. Microbiol.and Environ. Microbiol. 75 (6), 1487–1499. doi:10.1128/aem.01812-08
Hinrichs, K.-U., and Boetius, A. (2003). “The anaerobic oxidation of methane: New insights in microbial ecology and biogeochemistry,” in Ocean margin systems. Editors G. Wefer, D. Billett, D. Hebbeln, B. B. Jørgensen, M. Schlüter, and T. C. E. van Weering (Berlin, Heidelberg: Springer Berlin Heidelberg), 457–477.
Hinrichs, K. U., Sylva, S., Brewer, P., Delong, E., and Hayes, J. (1999). Methane-consuming archaebacteria in marine sediments. Nature 398 (6730), 802–805. doi:10.1038/19751
Inagaki, F., Nunoura, T., Nakagawa, S., Teske, A., Lever, M., Lauer, A., et al. (2006). Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. U. S. A. 103 (8), 2815–2820. doi:10.1073/pnas.0511033103
Iversen, N., and Jorgensen, B. B. (1985). Anaerobic methane oxidation rates at the sulfate‐methane transition in marine sediments from Kattegat and Skagerrak (Denmark)1. Limnol. Oceanogr. 30 (5), 944–955. doi:10.4319/lo.1985.30.5.0944
Johnson, D. B., and Mcginness, S. (1991). Ferric iron reduction by acidophilic heterotrophic bacteria. Appl. Environ. Microbiol. 57 (1), 207–211. doi:10.1128/AEM.57.1.207-211.1991
Johnson, K. M., Hughes, J. E., Donaghay, P. L., and Sieburth, J. M. (1990). Bottle-calibration static head space method for the determination of methane dissolved in seawater. Anal. Chem. 62 (21), 2408–2412. doi:10.1021/ac00220a030
Joye, S. B., Boetius, A., Orcutt, B. N., Montoya, J. P., Schulz, H. N., Erickson, M. J., et al. (2004). The anaerobic oxidation of methane and sulfate reduction in sediments from Gulf of Mexico cold seeps. Chem. Geol. 205 (3), 219–238. doi:10.1016/j.chemgeo.2003.12.019
Katayama, T., Yoshioka, H., Takahashi, H. A., Amo, M., Fujii, T., and Sakata, S. (2016). Changes in microbial communities associated with gas hydrates in subseafloor sediments from the Nankai Trough. FEMS Microbiol. Ecol. 92 (8), fiw093–10. doi:10.1093/femsec/fiw093
Kim, B. H., Kim, H. J., Hyun, M. S., and Park, D. H. (1999). Direct electrode reaction of Fe(III)-reducing bacterium, Shewanella putrefaciens. J. Microbiol. Biotechnol. 9 (2), 127–131. doi:10.1002/(SICI)1099-1514(199905/06)20:33.0.CO;2-I
Knittel, K., and Boetius, A. (2009). Anaerobic oxidation of methane: Progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334. doi:10.1146/annurev.micro.61.080706.093130
Kong, Y., Lei, H. Y., Zhang, Z. L., Cheng, W. D., Wang, B., Pan, F. L., et al. (2021). Depth profiles of geochemical features, geochemical activities and biodiversity of microbial communities in marine sediments from the Shenhu area, the northern South China Sea. Sci. Total Environ. 779, 146233. doi:10.1016/j.scitotenv.2021.146233
Laso-Pérez, R., Wegener, G., Knittel, K., Widdel, F., Harding, K. J., Krukenberg, V., et al. (2016). Thermophilic archaea activate butane via alkyl-coenzyme M formation. Nature 539 (7629), 396–401. doi:10.1038/nature20152
Leu, A. O., Cai, C., McIlroy, S. J., Southam, G., Orphan, V. J., Yuan, Z., et al. (2020). Anaerobic methane oxidation coupled to manganese reduction by members of the Methanoperedenaceae. ISME J. 14 (4), 1030–1041. doi:10.1038/s41396-020-0590-x
Li, J., Li, L., Bai, S., Ta, K., Xu, H., Chen, S., et al. (2019). New insight into the biogeochemical cycling of methane, S and Fe above the sulfate-methane transition zone in methane hydrate-bearing sediments: A case study in the dongsha area, South China sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 145, 97–108. doi:10.1016/j.dsr.2019.01.011
Lin, L.-H., Wu, L.-W., Cheng, T.-W., Tu, W.-X., Lin, J.-R., Yang, T. F., et al. (2014). Distributions and assemblages of microbial communities along a sediment core retrieved from a potential hydrate-bearing region offshore southwestern Taiwan. J. Asian Earth Sci. 92, 276–292. doi:10.1016/j.jseaes.2014.02.014
Liu, J., Wang, J., Izon, G., Antler, G., Wang, Z., Zhao, J., et al. (2018). Vivianite formation in methane-rich deep-sea sediments from the South China Sea. Biogeosciences 15 (20), 6329–6348. doi:10.5194/bg-15-6329-2018
Lu, G., ShaoYong, J., Tao, Y., JingHong, Y., NengYou, W., Zhang, G., et al. (2011). Glycerol ether biomarkers and their carbon isotopic compositions in a cold seep carbonate chimney from the Shenhu area, northern South China Sea. Chin. Sci. Bull. 56 (16), 1700–1707. doi:10.1007/s11434-011-4486-z
Luton, P. E., Wayne, J. M., Sharp, R. J., and Riley, P. W. (2002). The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill b bThe GenBank accession numbers for the mcrA sequences reported in this paper are AF414034–AF414051 (see Fig. 2) and AF414007–AF414033 (environmental isolates in Fig. 3). Microbiol. Read. Engl. 148, 3521–3530. doi:10.1099/00221287-148-11-3521
Martens, C. S., and Berner, R. A. (1974). Methane production in the interstitial waters of sulfate-depleted marine sediments. Science 185 (4157), 1167–1169. doi:10.1126/science.185.4157.1167
Meng, J., Xu, J., Qin, D., He, Y., Xiao, X., and Wang, F. (2014). Genetic and functional properties of uncultivated MCG archaea assessed by metagenome and gene expression analyses. The ISME Journal 8 (3), 650–659. doi:10.1038/ismej.2013.174
Naganuma, T., Sato, M., Hoshii, D., Amanomurakami, Y., Iwatsuki, T., and Mandernack, K. W. (2006). Isolation and characterization of Pseudomonas strains capable of Fe(III) reduction with reference to redox response regulator genes. Geomicrobiol. J. 23, 145–155. doi:10.1080/01490450600596565
Nauhaus, K., Antje, B., Martin, K., and Widdel, F. (2002). In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 4 (5), 296–305. doi:10.1046/j.1462-2920.2002.00299.x
Orphan, V. J., House, C. H., Hinrichs, K. U., Mckeegan, K. D., and Delong, E. F. (2001). Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293 (5529), 484–487. doi:10.1126/science.1061338
Pachiadaki, M. G., and Kormas, K. A. (2013). Interconnectivity vs. isolation of prokaryotic communities in European deep-sea mud volcanoes. Biogeosciences 10 (5), 2821–2831. doi:10.5194/bg-10-2821-2013
Paull, C. K., Ussler, W., and Borowski, W. S. (1994). Sources of biogenic methane to form marine gas hydrates. Ann. N. Y. Acad. Sci. 715, 392–409. doi:10.1111/j.1749-6632.1994.tb38852.x
Reeburgh, W. S. (2007). Oceanic methane biogeochemistry. Chem. Rev. 107 (2), 486–513. doi:10.1021/cr050362v
Reed, A. J., Dorn, R., Van Dover, C. L., Lutz, R. A., and Vetriani, C. (2009). Phylogenetic diversity of methanogenic, sulfate-reducing and methanotrophic prokaryotes from deep-sea hydrothermal vents and cold seeps. Deep Sea Res. Part II Top. Stud. Oceanogr. 56 (19), 1665–1674. doi:10.1016/j.dsr2.2009.05.012
Reed, D. W., Fujita, Y., Delwiche, M. E., Blackwelder, D. B., Sheridan, P. P., Uchida, T., et al. (2002). Microbial communities from methane hydrate-bearing deep marine sediments in a forearc basin. Appl. Environ. Microbiol. 68 (8), 3759–3770. doi:10.1128/AEM.68.8.3759-3770.2002
Regnier, P., Dale, A. W., Arndt, S., Larowe, D. E., Mogollón, J., and Cappellen, P. V. (2011). Quantitative analysis of anaerobic oxidation of methane (AOM) in marine sediments: A modeling perspective. Earth. Sci. Rev. 106 (1–2), 105–130. doi:10.1016/j.earscirev.2011.01.002
Rotaru, A. E., and Thamdrup, B. (2016). A new diet for methane oxidizers. Science 351 (6274), 658. doi:10.1126/science.aaf0741
Scheller, S., Yu, H., Chadwick, G. L., Mcglynn, S. E., and Orphan, V. J. (2016). Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 351 (6274), 703–707. doi:10.1126/science.aad7154
Segarra, K. E. A., Comerford, C., Slaughter, J., and Joye, S. B. (2013). Impact of electron acceptor availability on the anaerobic oxidation of methane in coastal freshwater and brackish wetland sediments. Geochimica Cosmochimica Acta 115 (5), 15–30. doi:10.1016/j.gca.2013.03.029
Sivan, O., Antler, G., Turchyn, A. V., Marlow, J. J., and Orphan, V. J. (2014). Iron oxides stimulate sulfate-driven anaerobic methane oxidation in seeps. Proc. Natl. Acad. Sci. U. S. A. 111 (40), 4139–4147. doi:10.1073/pnas.1412269111
Soo, V. W. C., McAnulty, M. J., Tripathi, A., Zhu, F., Zhang, L., Hatzakis, E., et al. (2016). Reversing methanogenesis to capture methane for liquid biofuel precursors. Microb. Cell. Fact. 15 (1), 11. doi:10.1186/s12934-015-0397-z
Wang, F., Wang, J., Jian, H., Zhang, B., Li, S., Wang, F., et al. (2008). Environmental adaptation: Genomic analysis of the piezotolerant and psychrotolerant deep-sea iron reducing bacterium Shewanella piezotolerans WP3. PLOS ONE 3 (4), 1937–1949. doi:10.1371/journal.pone.0001937
Weber, K. A., Achenbach, L. A., and Coates, J. D. (2006). Microorganisms pumping iron: Anaerobic microbial iron oxidation and reduction. Nat. Rev. Microbiol. 4 (10), 752–764. doi:10.1038/nrmicro1490
Webster, G., Sass, H., Cragg, B. A., Gorra, R., Knab, N. J., Green, C. J., et al. (2011). Enrichment and cultivation of prokaryotes associated with the sulphate–methane transition zone of diffusion-controlled sediments of Aarhus Bay, Denmark, under heterotrophic conditions. FEMS Microbiol. Ecol. 77 (2), 248–263. doi:10.1111/j.1574-6941.2011.01109.x
Wu, L., Yang, S., Liang, J., Su, X., Fu, S., Sha, Z., et al. (2013). Variations of pore water sulfate gradients in sediments as indicator for underlying gas hydrate in Shenhu Area, the South China Sea. Sci. China Earth Sci. 56 (4), 530–540. doi:10.1007/s11430-012-4545-6
Wu, N., Zhang, G., Liang, J., Su, Z., Wu, D., Lu, H., et al. (2013). Progress of gas hydrate research in northern South China sea. Adv. New Renew. Energy 1 (1), 80–94. doi:10.3969/j.issn.2095-560X.2013.01.008
Wu, N., Zhang, H., Yang, S., Zhang, G., Liang, J., Lu, J. a., et al. (2011). Gas hydrate system of Shenhu area, northern South China sea: Geochemical results. J. Geol. Res. 2011, 1–10. doi:10.1155/2011/370298
Yanagawa, K., Kouduka, M., Nakamura, Y., Hachikubo, A., Tomaru, H., and Suzuki, Y. (2014). Distinct microbial communities thriving in gas hydrate-associated sediments from the eastern Japan Sea. J. Asian Earth Sci. 90, 243–249. doi:10.1016/j.jseaes.2013.10.019
Zhang, G., Liang, J., Lu, J. a., Yang, S., Zhang, M., Holland, M., et al. (2015). Geological features, controlling factors and potential prospects of the gas hydrate occurrence in the east part of the Pearl River Mouth Basin, South China Sea. Mar. Petroleum Geol. 67, 356–367. doi:10.1016/j.marpetgeo.2015.05.021
Zhang, J., Lei, H., Chen, Y., Kong, Y., Kandasamy, S., Ou, W., et al. (2018). Carbon and oxygen isotope composition of carbonate in bulk sediment in the southwest Taiwan Basin, South China Sea: Methane hydrate decomposition history and its link to mud volcano eruption. Mar. Petroleum Geol. 98, 687–696. doi:10.1016/j.marpetgeo.2018.08.031
Zhang, W., Liang, J., Lu, J. a., Wei, J., Su, P., Fang, Y., et al. (2017). Accumulation features and mechanisms of high saturation natural gas hydrate in Shenhu Area, northern South China Sea. Petroleum Explor. Dev. 44 (5), 708–719. doi:10.1016/S1876-3804(17)30082-4
Keywords: anaerobic oxidation of methane, methanogenesis, microbial communities, incubation, marine sediments
Citation: Kong Y, Lei H, Cheng W, Wang B, Pan F and Huang F (2022) Shifting microbial communities perform anaerobic oxidation of methane and methanogenesis in sediments from the Shenhu area of northern south China sea during long-term incubations. Front. Earth Sci. 10:1014976. doi: 10.3389/feart.2022.1014976
Received: 09 August 2022; Accepted: 31 August 2022;
Published: 27 September 2022.
Edited by:
Zhifeng Wan, Sun Yat-sen University, ChinaReviewed by:
Jiliang Wang, Institute of Deep-Sea Science and Engineering (CAS), ChinaCopyright © 2022 Kong, Lei, Cheng, Wang, Pan and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huaiyan Lei, bGh5QHhtdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.