- 1Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin, China
- 2National Technology Innovation Center of Synthetic Biology, Tianjin, China
- 3Key Laboratory of Cenozoic Geology and Environment, Institute of Geology and Geophysics, Chinese Academy of Sciences, Center for Excellence in Life and Paleoenvironment, Beijing, China
The study on sediments in the marginal basins of the Tibetan Plateau is of great significance for global climate change. The geological information of the Linxia Basin has been intensely investigated; however, the profiles of the microbial communities in this basin remain largely unknown. Here, based on the 16S rRNA high-throughput sequencing method, the bacterial community structure vertical succession is studied with different thicknesses of sedimentary samples. The bacterial community with a total of 1,729,658 paired reads distributed within 1,042 phylogenetic amplicon sequence variants (ASVs) from twenty sediments, and three surrounding soil samples were sequenced. First, high-throughput sequencing results highlight the surrounding soil sample bacterial community structures were significantly different from those recovered from the sediment samples. In addition, as observed in the PCoA and PERMANOVA, there is a dramatic change shift event of the community structure at M311. Our data suggest that shifts in relative abundances of the abundant taxa (˃1%) and the significant variations in the diversity of bacterial community implied community structure responses to changes in different sedimentary layers. Predicted community function changes demonstrate that the sediment bacterial community aerobic chemoheterotrophy has been significantly increased, and we believe that the possible influence of the lithofacies changes from the anaerobic system to the aerobic environment, possibly accompanied by the significant uplift of the plateau that has previously been associated with enhanced aridity in Central Asia at ∼8 Ma. Taken together, these results illustrate the potential for the microbial community as a biological indicator in sediment ecosystems to reconstruct paleoenvironments.
Introduction
Microorganisms play key ecological roles in the process of geological and climate change. With the rapid development of modern molecular microbiology, we are able to study the abundance, distribution, and types of microorganisms in various extreme environments (Bard et al., 1997; Hofman et al., 2015). Scientists use these new methods to study microbes in modern environments and also microbes in ancient rocks, such as microbial ancient DNA (Coolen et al., 2005; Coolen et al., 2006). According to relevant research reports, the ancient microbial community structure and paleoenvironmental conditions can be reconstructed by studying ancient microbial DNA and biomarkers retained in sediments and sedimentary rocks. Coolen and Ovemann (1998) and Coolen et at. (2006) have confirmed that a certain amount of DNA was detected in the Holocene sediments, which was well preserved and protected against further degradation (Coolen and Overmann, 1998; Coolen et al., 2006). In 2007, 217000-year-old DNA sequences of green sulfur bacteria were implicated for the reconstruction of the paleoenvironment (Coolen et al., 2007). Inagaki et al. reported an unusual archaeal community, may be microbial relicts more than 2 million years old, serves as potential geomicrobiological evidence (Inagaki et al., 2001), and bacterial ribosomal RNA genes were amplified from a continental drilled core of black shale approximately 100 million years in age, and then “Paleome” was proposed (Inagaki et al., 2005). In particular, oceanic sulfate-reducing bacteria were found predominated at the mid-Cretaceous oceanic anoxic events layer, which is very consistent with major geological events. Recent research studies showed ancient DNA preserved in Qinghai Lake sediments was used to reconstruct the temporal succession of plankton communities in the past 18,500 years (Li et al., 2016). Moreover, a time series approach to infer past taxa richness from sedimentary ancient DNA from the southeastern Tibetan Plateau revealed a threat of warming-induced alpine habitat loss to Tibetan Plateau plant diversity (Liu et al., 2021). Therefore, the characteristics of climate change, the understanding of the continuous acquisition of fine molecular-level organic components, and the recording of large-scale environmental regional changes in climate characteristics and other scientific issues could be answered with a combination of interpretation from microbiology study (Coolen et al., 2004; 2013; Cao et al., 2017; Han et al., 2017).
All along, with the severe impact of the changing global climate and temperature, the changes and environmental effects of the inland arid regions of Asia have attracted more attention from many researchers because it is closely related to the uplift of the Tibetan Plateau (Li and Fang, 1999; Zachos et al., 2001). The arid region of northwestern China, located in the inland of Asia, is one of the most significant arid regions in the northern hemisphere (Guo et al., 2008). It is characterized by temperate desert and temperate grassland landscapes with dry and humid temperatures, unlike latitude zonality in other parts of the world’s arid area. At the same time, many basins around the Tibetan Plateau are in key areas of environmental change. During the uplift of the Tibetan Plateau, sedimentary organic matter plays an important role in the aridification mechanism (Guo et al., 2004). Therefore, the study on sediments in the marginal basins of the Tibetan Plateau is of great significance. There are some large inland basins in Northwest China, which accumulate in extra-thick Cenozoic sediments. Many previous studies have already confirmed the value of these sediments, and related chronostratigraphic work has been carried out to record changes in the arid environment within Asia (Fang et al., 1999). According to previous geologists, we know that the sedimentation of the Linxia Basin began at about ∼54 Ma with almost no interruption until ∼1.7 Ma (Feng et al., 2021, GR). They have an almost continuous and complete sedimentary sequence controlled by precise ages since the Cenozoic in the Linxia Basin (Fang et al., 2003). In the natural geographical position, the Linxia Basin is located at the intersection of the three major natural belts in the eastern monsoon region. It is also at the apex of the monsoon triangle, affected by the two systems of the westerly circulation and the monsoon circulation. The Cenozoic strata of the entire basin have been almost continuously deposited from the Oligocene to the present, providing a more complete Cenozoic stratigraphic record than the South Valley West Valley, which provides a very favorable geological condition for in-depth study of environmental evolution in the region.
Many major geological or climatic events on the Earth, especially the uplift of the Qinghai–Tibet Plateau since the Cenozoic, and the aridification of the inland of Asia are even more interrelated with the lithosphere, hydrosphere, atmosphere, and biosphere. The role is closely related, so it is necessary to break through the traditional geological point of view, and it is necessary to examine and react with a new perspective to confirm the plateau uplift, and its resource environment, and climate effects. Previous scholars have successfully used physical and chemical methods to study climate change in this region, but the use of microbial methods to study the community structure of prokaryotic microbes in this region and the inference of paleoenvironmental and paleoclimate have rarely been reported.
Therefore, we make an exploratory attempt to determine the bacterial community recovered from Maogou section sediments in the Linxia Basin on the northeastern edge of the Tibetan Plateau. This study focuses on the microbial community structure and function from different stratigraphy sediment samples, in hopes of revealing the bacterial community evolution characteristics and the potential for the microbial community as a biological indicator in sediment ecosystems to reconstruct paleoenvironments.
Materials and Methods
Site Description
The Linxia Basin is located in the northeastern part of the Qinghai–Tibet Plateau (120°30′–104°E, 35°10′–35°51′N, Figure 1). The sedimentary samples are mainly mudstone and sandstone of fluvial and lacustrine origin. The Maogou section is located in the middle of the Linxia Basin and is a representative section of the Tertiary red bed, and the strata are almost vertically stacked (Li et al., 2014). It is a set of river-lake-phase clastic rocks. The previous study provides a detailed magnetostratigraphic record of subsidence in the basin. The red bed in this area is from 30.6 to 4.0 Ma, that is, from the upper Oligocene to the Pliocene, the total thickness of the section is about 443 m (Wang et al., 2012). Figure 1 shows the location of each sample in the studied section.
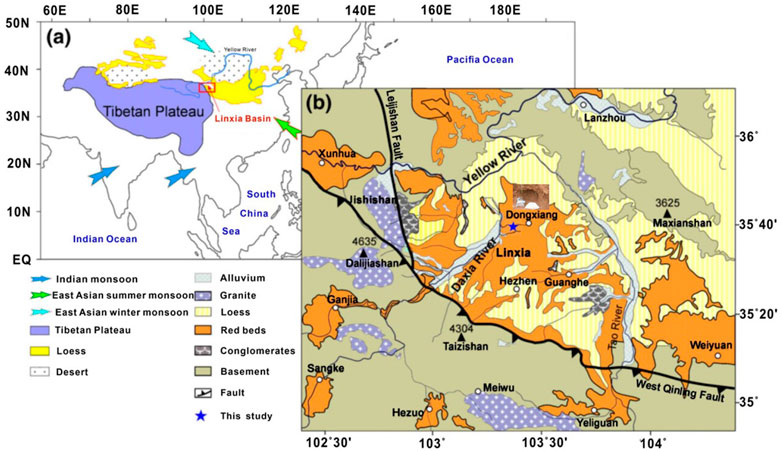
FIGURE 1. Location of the Linxia Basin (A) and the geological map of the Linxia Basin and adjacent regions in this study area (modified from Yang et al., 2016).
Sampling Method
Twenty representative sediment thicknesses were selected from 289 to 387 m, respectively, as well as three surrounding soil samples. Three lacustrine sediment samples were collected from each thickness, and then samples from each thickness were pooled and homogenized in the laboratory. Figure 2 shows the thicknesses, lithology, and stratigraphic division of each sample in the study section. In order to minimize pollution, we climbed and reached the location of the specific stratum, removed the ∼1-m exposed surface samples from the layer, and then took the sediment samples. All collected sedimentary samples were stored in transparent capsules and sealed bottles that had been completely sterilized, numbered, and sealed. Samples for microbial community analysis were transported on ice and stored at −80°C for subsequent DNA extraction. Samples for geochemical analysis were sealed and stored at room temperature.
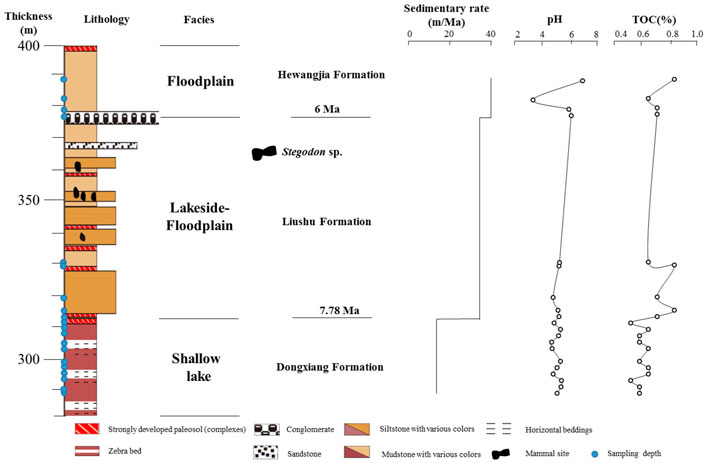
FIGURE 2. Stratigraphic division, sedimentary facies, lithology (modified from Fang et al., 2016), sampling depth (solid blue circle), sedimentary rate (He et al., 2020), pH, and total organic carbon (TOC) content in Maogou section.
Geochemical Measurements
Surface soil and lacustrine sediment samples total organic carbon (TOC) were measured with a PE 2400 series II CHNS/O analyzer (Perkin Elmer, United States). pH was determined using a pH meter (pH700, Eutech, and China).
DNA Extraction, PCR Amplicon Libraries Construction, and Sequencing
In this study, all the sediment and soil samples were extracted using a Power Max Soil DNA Isolation Kit (MO Bio Laboratories, Solana Beach, CA). According to the instructions of the kit, 0.25 g of sedimentary rock samples were taken and the subsequent steps were carried out. The extracted genome was identified by electrophoresis, and the content and quality of the genome were determined by Nanodrop 2000. The genomic DNA of the flora extracted from sediment samples was used as a template, and the V3-V4 hypervariable region of 16S rRNA was detected by the Illumina MiSeq sequencing platform. In the present study, the bacterial universal primer sequences were used as F515 5′-GTG CCA GCM GCC GCG GTA A-3′ and R806 5′-GGA CTA CHV GGG TWT CTA AT-3'.
Accession Numbers
The 16S rRNA gene sequences were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession number PRJNA732793.
Statistical and Data Analysis
Each sample yields at least 40,000 reads, and basic data analysis is performed by QIIME 2 (version v.2019-07) software (http://qiime.org/scripts/assign_taxonomy.html), according to the relevant process (Bolyen et al., 2019). The sequences were aligned against the Silva database (version: http://www.arb-silva.de/), denoised, high-quality sequences were clustered, ASVs (features) were divided, and their species classification was carried out according to the sequence composition of the feature. The principal coordinate analysis (PCoA) and alpha diversity including Chao 1, ACE, Simpson diversity, and Shannon diversity were subjected to statistical analysis using the QIIME 2. Data were analyzed using multivariate correlational and ordination methods in the R statistical environment (R Core Team 2019), using R packages vegan (version 2.0) and labdsv (version 1.6). The effect size and statistical significance between group A and group B on each of the experimental analyses were determined with permutational multivariate analysis of variance (PerMANOVA, function adonis) (Ramos-Robles et al., 2016). PICRUSt2 was used to analyze the functional characteristics of the bacterial community.
Results
Environmental Characteristics
The strata section here is divided into three formations, with their depths mentioned next to the name: Dongxiang formation at 289–313 m, Liushu formation at 313–330 m, and Hewangjia formation at 376–387 m (Figure 2). Environmental parameters varied greatly between sediment and soil samples. For instance, the contents of TOC in the soil samples were higher than 1%, while those were less than 0.1% in the sediment samples, indicating the organic matter content in the sediment samples is very low. The total organic carbon (TOC) varied slightly between different thicknesses sediment samples, and total organic carbon varies between 0.03 and 0.08%, averaging 0.05% (of total dry weight). Due to the large sampling interval and low total organic carbon, it is difficult to identify consistent trends in the data and especially for the sediment samples. The pH in the sediments is in the range of 8.29–9.81, with the highest values in the sediments from the M387, and the lowest values in the sediment from the M381. It is worth noting that all samples are alkaline, including soil samples and sediment samples, and the highest sediment sample alkaline is as high as 9.81 ± 0.02.
Bacterial Community Composition
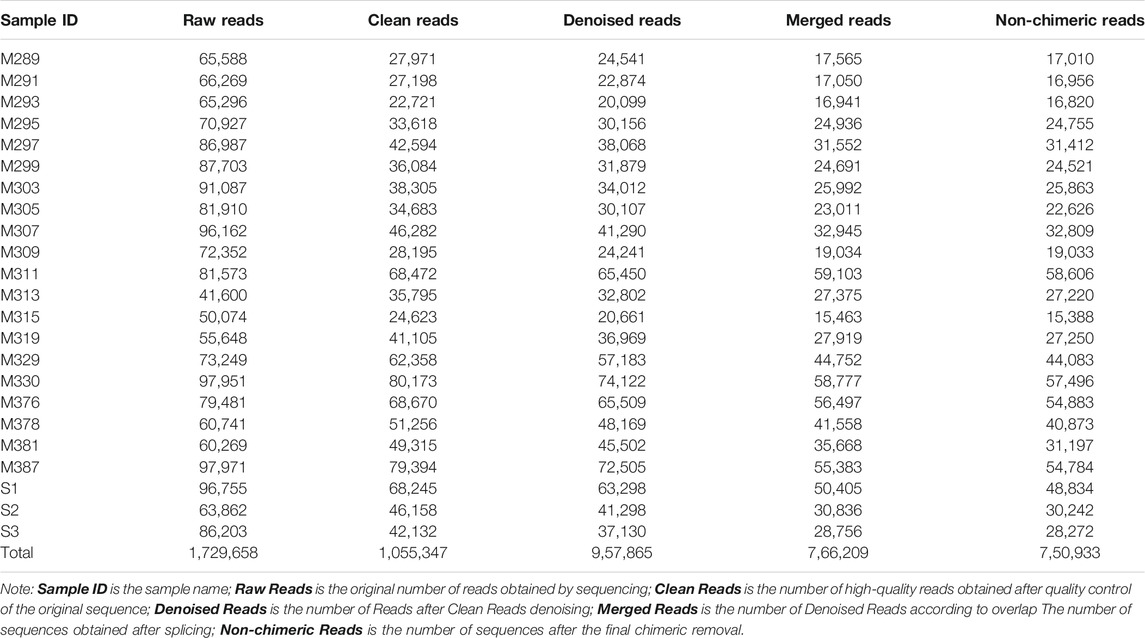
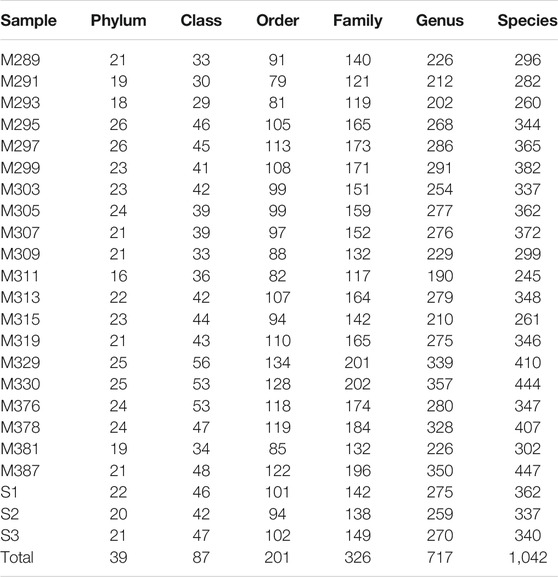
The rarefaction curve indicated variation in amplicon sequence variants (ASVs; or features) density within all the samples, while the sequence coverage sufficiently captured the bacterial community diversity (Supplementary Figure S1; Supplementary Table S1). After the denoising, normalization, and clustering, a total of 1,729,658 pairs of reads were obtained by sequencing 23 samples. After quality control and splicing of double-ended reads, a total of 1,055,347 clean reads were generated. Each sample generated at least 22,721 clean reads, with an average of 45,885 clean reads (Table 1). Overall, these sequences were assigned to 39 phyla, 87 classes, 201 orders, 326 families, 717 genera, and 1,042 species (Table 2).
The top 10 abundant classes and families in each sample were selected for a detailed understanding of the differences in the microbial community structure. The top ten classes were affiliated with Gammaproteobacteria, Clostridia, Alphaproteobacteria, Bacilli, Actinobacteria, Bacteroidia, Thermoleophilia, Gemmatimonadetes, Acidimicrobiia, and Deltaproteobacteria, accounting for 30.46, 14.46, 11.22, 10.98, 9.08, 4.29, 3.94, 1.84, 1.53, and 1.50% of the total bacterial microbial community, respectively (Figure 3A). In addition, the top ten families were Burkholderiaceae, Ruminococcaceae, Lachnospiraceae, Lactobacillaceae, Pseudomonadaceae, Sphingomonadaceae, Xanthomonadaceae, Enterobacteriaceae, Oxalobacteraceae, and Micrococcaceae, accounting for 8.31, 5.89, 5.31, 4.45, 3.60, 3.28, 2.32, 2.19, 2.07, and 2.05% of the total bacterial community, respectively (Figure 3B).
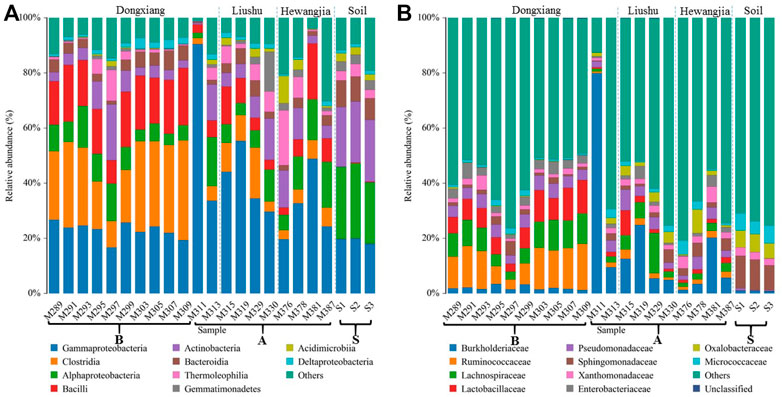
FIGURE 3. Relative abundance of the microbial 16S rRNA gene from the samples collected in different sedimentary layers and surrounding soil, respectively, at classes (A) and families (B) level (top 10).
Figure 3 shows the differences in the relative abundance of dominant bacteria at different samples. Overall, the bacterial community structure varied greatly between the sediments and soil profile. At classes, Gammaproteobacteria, Clostridia, Bacilli, and Alphaproteobacteria dominated in the sediment samples, while Alphaproteobacteria, Actinobacteria, Gammaproteobacteria, and Bacteroidia were the most dominant in the soil samples. At families, Burkholderiaceae, Ruminococcaceae, Lachnospiraceae, and Lactobacillaceae were the most dominant in the sediment samples, and Sphingomonadaceae, Oxalobacteraceae, Micrococcaceae, and Xanthomonadaceae dominated in the soil samples. The structures of the sediment bacterial communities differed among the studied stratigraphic division groups. Noteworthily, compared with their abundances in the twenty sediment samples, it is worth noting that the relative abundance of Gammaproteobacteria was significantly increased, while Clostridia was reduced in sediment M311 (Figure 3A). According to the sediment thicknesses of the samples, the bacterial community composition could be clustered into two distinctive groups: the first group occurred from M289 to M309 (group B) and the second group occurred from M311 to M387 (group A). At classes, group B contained higher relative abundances of Clostridia, Gammaproteobacteria, and Bacilli, but group A had higher relative abundances of Gammaproteobacteria, Alphaproteobacteria, and Clostridia, respectively. The Ruminococcaceae, Lachnospiraceae, Lactobacillaceae, and Burkholderiaceae were the most dominant in group B sediment samples, and Burkholderiaceae, Lachnospiraceae, Pseudomonadaceae, and Oxalobacteraceae dominated in group A sediment samples at families, respectively.
Comparison of Bacterial Communities’ Beta Diversity
PCoA can be used to evaluate the similarity or variability of sample community compositions, and the species composition of two samples is closer when their distance in a PCoA map is closer. The bacterial community composition is clustered into three groups based on the Bray–Curtis distance calculation on PCoA (Figure 4). Three groups of samples are discriminated for the location and geological stratification sequence series (Figure 4): group B, group A, and soil. As shown in Figure 4, the effect of the main factor Ⅰ was 23.80%, and the effect of the main factor Ⅱ was 16.32%. The microbial community composition is significantly different between sediment and soil samples. In comparison to the soil samples, the sediment samples had more similar bacterial communities, suggesting there are greater similarities among the bacterial communities in the same ancient geological samples, and it may reflect from the side that the microbial community structures of these sediment samples strongly differed from the community structures of the surface soil samples from the surrounding environment. Meanwhile, different sediment microbial structures were also observed between group B and group A, which might indicate that some factors affect the microbial structure more significantly than lithology or chronological sequence.
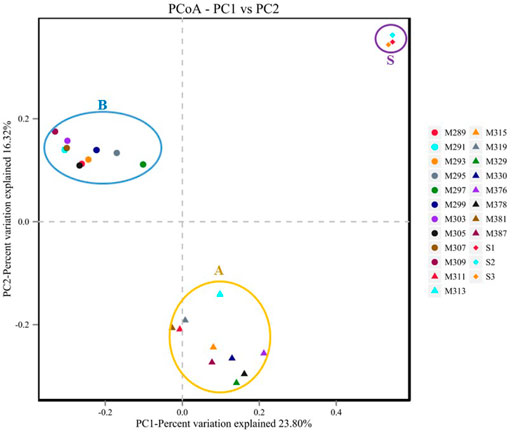
FIGURE 4. Partial correspondence analysis ordinations of the microbial community diversity as revealed by 16S rRNA gene sequencing from all samples (yellow circle A: group A, blue circle B: group B, purple circle S: soil).
Bacterial Community Alpha Diversity Change Trends
The alpha diversity can reflect the abundance and diversity of microbial communities, which includes a series of indices based on statistical analyses. Considering the focus on sediment samples, the Chao 1 and ACE indices reflect the community richness, and the Simpson and Shannon indices reflect the community evenness in both sediment samples and without soil samples. Overall, the Good’s coverage index for sequence depth ranged from 0.997 to 1; the Ace and Chao 1 indices for community richness ranged from 312.14 to 795.89 and 312.00 to 794.95, respectively, and the Shannon and Simpson indexes for community diversity ranged from 2.777 to 8.618 and 0.480 to 0.995, respectively (Supplementary Table S1). By calculating the community structure alpha diversity index, the community richness of bacterial community structure at the four thicknesses of M293, M311, M315, and M381 is relatively low (Figure 5), especially at M293 (Chao 1 and ACE indexes, 312.14 and 312.00) and M311 (Chao 1 and ACE indexes, 320.24 and 324.79). In regard to the community evenness, the Shannon index and the Simpson index also sharply decreased at M311 (Shannon and Simpson indexes, 2.777 and 0.480) and M381 (Shannon and Simpson indexes, 6.757 and 0.972).
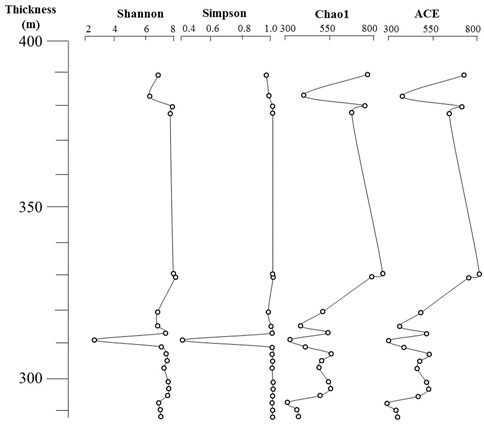
FIGURE 5. Bacterial community complexity was evaluated using the alpha components including the species richness (ACE and Chao 1 index) and diversity (Simpson and Shannon index) in all sedimentary samples.
PERMANOVA Between Sediment Samples
Microbial community structures inferred from ASV sequences were significantly dissimilar between group A and group B, with PerMANOVA R2 = 0.299, p = 0.001 for unweighted UniFrac analysis and R2 = 0.344, p = 0.001 for weighted UniFrac analysis. PCoA clearly demonstrates that bacterial communities obtained from the same group are similar to each other and distinct from communities under other environments (Figure 6), reinforcing the finding based on community diversity metrics that drastic climate or environment changes are associated with distinct microbial communities. UniFrac analysis showed that the community clustered strongly, suggesting a history of evolutionary specialization in relation to a distinctive factor change at M311.
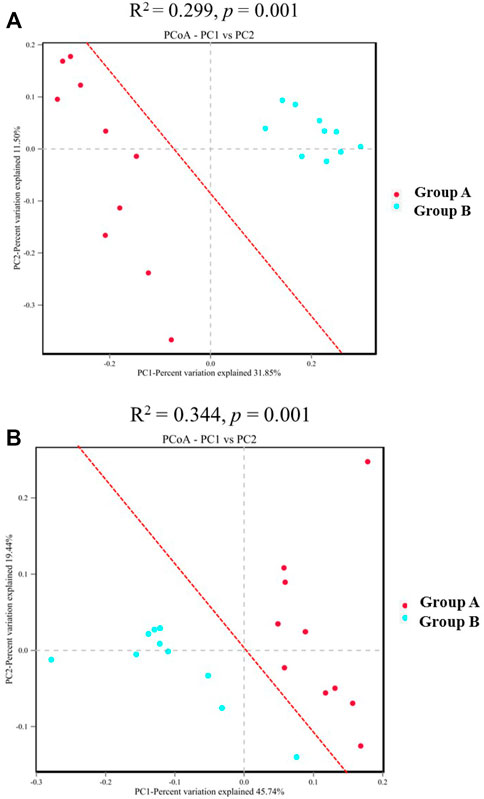
FIGURE 6. Partial correspondence analysis ordinations of the sedimentary microbial community diversity. ((A) unweighted (B) weighted). The R2 obtained by the PERMANOVA indicates the degree of interpretation of the difference between different sedimentary groups. The larger the R2, the higher is the degree of interpretation of the difference between the groups, the greater is the difference between the groups, and the higher is the reliability of the test when the p value is less than 0.05.
ANOVA Between Sediment Samples
In the present study, a clear differentiation between the bacterial communities in the group B and group A was observed in terms of taxonomic composition and community structure. Top 20 genus were selected and calculated the ANOVA analysis between these two groups, especially the genus with the average relative abundance greater than 1% (Figure 7). The average relative abundances of Romboutsia and Massilia in group A were 116.72 and 23.24 times than those in group B, while the average relative abundances of Lactobacillus, Faecalibacterium, Blautia, Romboutsia, Streptococcus, Turicibacter, and Bacteroides in group B were significantly higher than those in group A (p < 0.01).
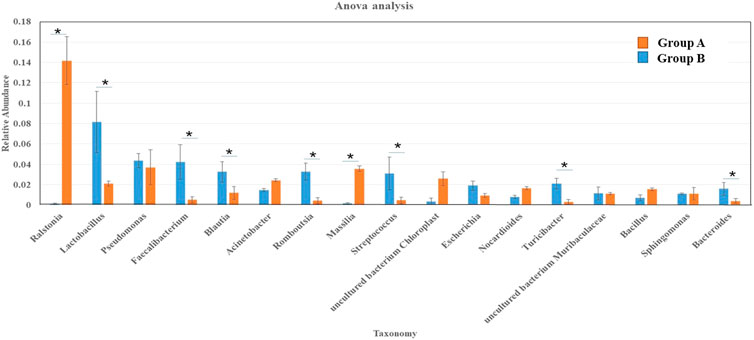
FIGURE 7. Significant analysis of differences between sedimentary groups—analysis of variance. Note: The abscissa represents species (showing the top 20 species with the average relative abundance greater than 1%), the ordinate represents the relative abundance of species, the columns with different colors represent each sample, and * on the column indicates significant difference (p < 0.05).
Community Function Prediction and Comparative Analysis
The metabolic function of microbes was predicted by PICRUSt based on the KEGG pathway (Figure 8A). The KEGG pathway results showed that the pathways were mainly involved in genetic information processing, metabolism, environmental information processing, and cellular processes and signaling, according to the relative abundances of sequences in all sediment samples. A total of 29 metabolic functions were predicted in all samples with the most enrichment in carbohydrate metabolism, amino acid metabolism, nucleotide metabolism, replication and repair, translation, and signal transduction. In the Group B samples, the relative abundances of sequences involved in replication and repair, nucleotide metabolism, translation, folding, sorting and degradation, glycan biosynthesis and degradation, carbohydrate metabolism, and drug resistance: Antimicrobial were significantly higher than those in Group A (p < 0.01), while the relative abundances of sequences involved in xenobiotics biodegradation and metabolism, aging, metabolism of other amino acids, lipid metabolism, neurodegenerative disease, signal transduction and cell motility were significantly lower than those in Group A (p < 0.01).
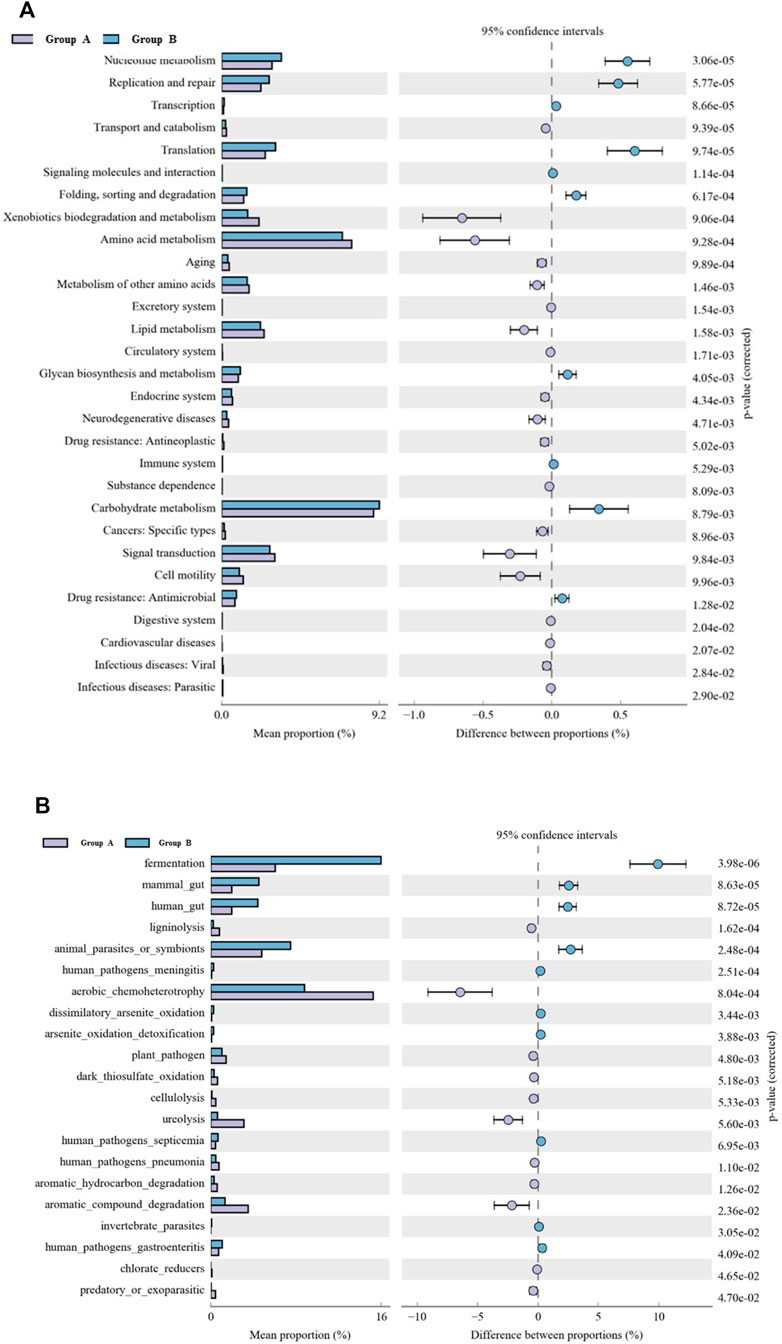
FIGURE 8. Variation in bacterial function profiles between group A and group B analyzed by PICRUSt (A) and FAPROTAX (B). Note: The level 2 KEGG ortholog function predictions about the abundance of the main metabolic functions. The different colors in the picture represent different groups. The left figure in the picture shows the abundance ratio of different functions in two samples or two groups of samples, the middle shows the difference ratio of the function abundance within the 95% confidence interval, and the rightmost value is the p value.
A total of 21 functional assignments were obtained using the Functional Annotation of Prokaryotic Taxa (FAPROTAX) tool (Figure 8B). Functional group fermentation was significantly less abundant in group A (p < 0.01), but its abundance increased in group B. In contrast, the abundant functions, such as ureolysis, and aromatic hydrocarbon degradation, which are related to compound degradation, are significantly increased in group A samples (p < 0.01). Similarly, functions associated with the carbon cycle, such as aerobic chemoheterotrophy, increased in group A. Also, the number of plant pathogens increased in group A. However, animal parasites or symbionts, mammal gut, and human gut significantly decreased (p < 0.01).
Discussion
Microorganisms play key ecological roles in geologically aquatic and terrestrial environments. Environmental and climate changes as well as their interactions may drive different microbial community formation, and microbial assemblages will affect the subsequent operation of the ecosystem (Schadt, 2010). In this study, the microbial assemblages in the sedimentary samples of the Maogou section at the Linxia Basin on the northeastern margin, TP, were selected as experimental objects as well as the surrounding soil samples.
The Reliability of Ancient Microbial Community Structure
Conventionally, DNA is easily damaged by various processes such as hydrolysis, oxidation, ultraviolet radiation, and enzymatic hydrolysis. However, more and more research studies have confirmed that permafrost, ancient fossil bones, and lacustrine sediment samples contain ancient DNA that has been preserved for thousands to even millions of years (Inagaki et al., 2005). In the process of DNA extraction, we found that the DNA content of sediment samples was much lower than modern surrounding surface soil samples. In addition, through the PCoA of the microbial community structure, it can be seen that the community structure of the surface soil was significantly different from the community structure of the sediment samples. That proved these sediment samples’ DNAs were not contaminated or exchanged by the modern microbial environment. With the research deepening, it was discovered that minerals and organic macromolecules (such as humic acid) have strong adsorption to DNA, which can protect DNA from degradation, and are influenced by diagenesis (Pietramellara et al., 2009; Schelble et al., 2010), especially in a deposition environment which consists of anoxic, low temperature, high pressure, hypersaline, and dry conditions, which can constraint the activity, and fluidity of microorganisms and enable long-term preservation of DNA (Panieri, et al., 2010; Danovaro et al., 2010). The results of this study confirmed the sediment samples of the Maogou section have the characteristics of low organic matter, anoxic, hypersaline, dry, and low biomass. Furthermore, the sedimentary community structure compositions were layer-specific and different which proved the possible barriers that prevent the exchange of DNA between the different sedimentary layers. In summary, these results illustrated that the sediment community structures in this study could represent pristine sedimentary environment, which may be used for paleoenvironment reconstruction.
Vertical Trends in Microbial Community Structure Composition
The relative abundance of the bacterial genus was also clearly impacted by the sudden environmental and climate events, which led to shifts in the relative abundance of Lactobacillus and Ralstonia. The relative abundance of Lactobacillus was greater in group B relative to group A, whereas Ralstonia abundance was greater in group A. Lactobacillus belonging to phylum Firmicutes is a kind of anaerobic, mesophilic, and protein-specific utilizing bacteria. It is easy to understand its dominance for group B (8.15%); meanwhile, its abundance was lower (2.12%) for group A. The genus Ralstonia was aerobic Gram-negative bacteria, established in 1995, and initially contained only one recognized pathogen. The function of this genus is related to the withering of plants and the degradation of organic matter. For example, Ralstonia pickettii was often found in moist environments such as soils, rivers, and lake sediment, which can survive in areas with a very low concentration of nutrients. Species diversity is the most critical level of biodiversity because the microbial diversity index can closely reflect the function of the ecosystem and the changes in the surrounding environment. In the evolution of herbaceous plants, the temperature dropped sharply, the microbial community became single, and the diversity index decreased significantly. In the past 20 years, many taxonomic diversity index–related application studies have been carried out in marine, freshwater, land, and other habitats (Leonard et al., 2006; Jiang et al., 2014). Diversity indices provide important information about the rarity or commonness of species in a community. The temporal and spatial variations of the proto-nuclear microbial community structure of sedimentary rocks at different thicknesses to speculate on the major climate events. Compared with other sediments, the diversity of M311 and M381 community structures is poor, and these two positions (historical period) may reflect the poor climatic environment, resulting in poor bacterial community diversity. The data indicate that striking changes in diversity of microbial communities are accompanied by significant climatic variations in the marginal basins of the Tibetan Plateau. The results of this study found that among the sediment samples assessed, there is a significant microbial diversity index declined in the arid-cold environment, which was generally in accordance with the previous pollen evidence, that is, the vegetation type shifted from that of a forest to that of a steppe after approximately 8.0 Ma in the Linxia Basin (Ma et al., 1998), nearby Tianshui Basin (Hui et al., 2011), and Jiuxi Basin (Ma et al., 2005).
In addition, this study shows that the impacts of stratigraphic division and lithology may be overpassed by the geological structure and climatic fluctuation impacts, as observed in the PCoA and PERMANOVA. Therefore, based on this, we can infer the changes in the environment through the changes in the relative abundance, diversity index, PCoA, and PERMANOVA. And through comprehensive analysis to compare the trends of the relative abundances of bacterial community, ACE and Chao 1, Shannon index and Simpson index, can fully confirm the aforementioned change in the environment. At the same time, Ma and colleagues obtained statistical data based on vegetation research, indicating that the Maogou section in the eastern part of the Linxia Basin can obtain a severe aridification event (Manabe and Broccoli, 1990; Jiang et al., 2014), which is consistent with our data based on microbial community analysis.
Vertical Trends in Microbial Community Function Prediction
Furthermore, previous studies have shown that changes in the microbial community function are affected by the fluctuations in the surrounding environment and climate at that time and then make corresponding metabolic reactions (Xiong et al., 2010; Comte and Giorgio, 2012; Deng et al., 2021). There were also significant differences in the functional composition between group A and group B probably because of the increased environmental pressure, amino acid metabolism, xenobiotics biodegradation and metabolism, and lipid metabolism were more enriched, especially aerobic chemoheterotrophy. In this study, our results demonstrate that the sediment microbial community aerobic chemoheterotrophy has been significantly increased since M311 to M387 from the metabolic function of microbes was predicted (p < 0.01). Based on the predicted microbial community function changes, the tectonic uplift of the NE TP may have caused the probable effects of changes in lithofacies from lacustrine systems (anaerobic, waterlogged environments) to a thick alluvial fan (aerobic environments).
Possible Aridity Enhanced Inferred From Microbial Evidences
Previous studies have shown that n-alkane distribution characteristics can characterize organic matter from different sources, reflecting the relative changes of algal organisms and higher plants and indicating the relative sources of aquatic and terrestrial plants (Zmudczynska-Skarbek et al., 2013). Moreover, through the comprehensive analysis of the geological background data information, it can be found that at the Asian inland, the event is characterized by aridification at ∼8.0 Ma. Our study shows that these climatic fluctuations induced significant modifications within bacterial community assemblages in the sediment samples, whereas marked arid-cold periods led to drastic rearrangements of community possible associated with significant uplift of the Tibetan Plateau at about 8.0 Ma. From the time window, the inland aridification in Asia is mainly caused by regional environmental changes, that is, the aridification-driving mechanism is associated with events such as regional structural uplift. This gives a more reasonable explanation for the mechanism of the inland aridification in Asia from the perspective of molecular microbiology. Microbial communities have the potential to deepen our understanding of these environmental and climate changes and provide insights into the possible consequences for the planet’s ecosystems.
Conclusion
In this study, the bacterial community succession in the different thickness lacustrine–fluvial sediments from the Linxia Basin, NE Tibetan Plateau, was compared and analyzed. Obviously, we found that there was a dramatic change shift event regarding the bacterial community alpha or beta diversity in the different sediments. Furthermore, predicted bacterial community function demonstrates environment changes from the anaerobic system to the aerobic system. This significant congruence might be primarily underpinned by aridity-enhanced processes. Thus, our findings inferred that the microbial community could also serve as a potential biological indicator in sediment ecosystems to reconstruct paleoenvironments. Future studies and technical methods are also encouraged to confirm our findings by contrasting the relationships with the microbial community under climatic changes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA732793).
Author Contributions
SX contributed to investigation, conceptualization, formal analysis, writing—original draft, and writing—review and editing. JW helped with formal analysis. XZ curated the data curation. RY: resources. WZ framed the methodology. ZH helped with conceptualization, funding acquisition, supervision, and writing—review and editing. YW acquired funding.
Funding
This work was financially supported by the State Key Program of the National Natural Science of China (Grant No. 41831176), the Science and Technology Support Program of Tianjin Municipality (Grant Nos. 18ZXSZSF00100 and 19YFZCSF00750), and the General Program from the National Natural Science Foundation of China (Grant No. 41977200).
Conflict of Interest
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer ZS declared a past co-authorship with one of the authors (YW) to the handling editor.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Wei He for her help and assistance in geology analysis and reviewers for their constructive and helpful comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/feart.2021.714352/full#supplementary-material
References
Bard, E., Rostek, F., and Sonzogni, C. (1997). Interhemispheric Synchrony of the Last Deglaciation Inferred from Alkenone Palaeothermometry. Nature 385, 707–710. doi:10.1038/385707a0
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., and Caporaso, J. G. (2018). QIIME 2: Reproducible, Interactive, Scalable, and Extensible Microbiome Data. Science 37, 852–857. doi:10.1038/s41587-019-0209-9
Cao, J., Rao, Z., Jia, G., Xu, Q., and Chen, F. (2017). A 15 Ka pH Record from an alpine lake in north China Derived from the Cyclization Ratio index of Aquatic brGDGTs and its Paleoclimatic Significance. Org. Geochem. 109, 31–46. doi:10.1016/j.orggeochem.2017.02.005
Comte, J., and del Giorgio, P. A. (2012). Composition Influences the Pathway but Not the Outcome of the Metabolic Response of Bacterioplankton to Resource Shifts. Plos One 6, e25266. doi:10.1371/journal.pone.0025266
Coolen, M. .J. L., Boere, A., Abbas, B., Muyzer, G., and Damste, J. (2005). “Fossil DNA as a Recorder of Ancient Microbial Communities and Palaeoenvironments,” in AGU Fall Meeting Abstracts, Den Burg. American Geophysical Union [abstract].
Coolen, M. J. L., Boere, A., Abbas, B., Baas, M., Wakeham, S. G., and Sinninghe Damsté, J. S. (2006). Ancient DNA Derived from Alkenone-Biosynthesizing Haptophytes and Other Algae in Holocene Sediments from the Black Sea. Paleoceanography 21 (1), a–n. doi:10.1029/2005PA001188
Coolen, M. J. L., Orsi, W. D., Balkema, C., Quince, C., Harris, K., Sylva, S. P., et al. (2013). Evolution of the Plankton Paleome in the Black Sea from the Deglacial to Anthropocene. Proc. Natl. Acad. Sci. 110, 8609–8614. doi:10.1073/pnas.1219283110
Coolen, M. J. L., and Overmann, J. (1998). Analysis of Subfossil Molecular Remains of Purple Sulfur Bacteria in a lake Sediment. Appl. Environ. Microbiol. 64 (11), 4513–4521. doi:10.1128/AEM.64.11.4513-4521.1998
Coolen, M. J. L., Volkman, J. K., Abbas, B., Muyzer, G., Schouten, S., and Damsté, J. S. S. (2007). Identification of Organic Matter Sources in Sulfidic Late Holocene Antarctic Fjord Sediments from Fossil rDNA Sequence Analysis. Paleoceanography 22, 1357–1377. doi:10.1029/2006pa001309
Coolen, M., Muyzer, G., Rijpstra, W., Schouten, S., Volkman, J. K., and Damsté, J. (2004). Combined DNA and Lipid Analyses of Sediments Reveal Changes in Holocene Haptophyte and Diatom Populations in an Antarctic lake. Earth Planet. Sci. Lett. 223, 225–239. doi:10.1016/j.epsl.2004.04.014
Danovaro, R., Corinaldesi, C., Dell'Anno, A., Fabiano, M., and Corselli, C. (2005). Viruses, Prokaryotes and DNA in the Sediments of a Deep-Hypersaline Anoxic basin (DHAB) of the Mediterranean Sea. Environ. Microbiol. 7, 586–592. doi:10.1111/j.1462-2920.2005.00727.x
Deng, Y., Mao, C., Chen, H., Wang, B., Cheng, C., Ma, H., et al. (2021). Shifts in Pond Water Bacterial Communities Are Associated with the Health Status of Sea Bass (Lateolabrax Maculatus). Ecol. Indicators 127, 107775. doi:10.1016/j.ecolind.2021.107775
Fang, X.-M., Ono, Y., Fukusawa, H., Bao-Tian, P., Li, J.-J., Dong-Hong, G., et al. (1999). Asian Summer Monsoon Instability during the Past 60,000 Years: Magnetic Susceptibility and Pedogenic Evidence from the Western Chinese Loess Plateau. Earth Planet. Sci. Lett. 168, 219–232. doi:10.1016/s0012-821x(99)00053-9
Fang, X. M., Garzione, C., and Van, der. Voo, R. (2003). Flexural Subsidence by 29 Ma on the NE Edge of Tibet from the Magnetostratigraphy of Linxia Basin, China. Earth Planet. Sci. Lett. 210 (3-4), 545–560. doi:10.1016/S0012-821X(03)00142-0
Feng, Z. T., Zhang, W. L., Fang, X. M., Zan, J. B., Zhang, T., Song, C. H., et al. (2021). Eocene Deformation of the NE Tibetan Plateau: Indications from Magnetostratigraphic Constraints on the Oldest Sedimentary Sequence in the Linxia Basin. Gondwana Res. 101, 77–93. doi:10.1016/j.gr.2021.07.027
Guo, Z., Peng, S. Z., Hao, Q. Z., Biscaye, P. E., An, Z. S., and Liu, T. S. (2004). Late Miocene?Pliocene Development of Asian Aridification as Recorded in the Red-Earth Formation in Northern China. Glob. Planet. Change 41 (3-4), 135–145. doi:10.1016/j.gloplacha.2004.01.002
Guo, Z. T., Sun, B., Zhang, Z. S., Peng, S. Z., Xiao, G. Q., Ge, J. Y., et al. (2008). A Major Reorganization of Asian Climate by the Early Miocene. Clim. Past 4 (3), 153–174. doi:10.5194/cp-4-153-2008
Han, D., Nam, S.-I., Kim, J.-H., Stein, R., Niessen, F., Joe, Y. J., et al. (2017). Inference on Paleoclimate Change Using Microbial Habitat Preference in Arctic Holocene Sediments. Sci. Rep. 7 (1), 9652. doi:10.1038/s41598-017-08757-6
He, W., Wang, G., Wang, Y., Wei, Z., Huang, Z., Zhang, T., et al. (2020). Microbial Communities and Lipid Records of the Linxia Basin, NE Tibetan Plateau: Implications for Enhanced Aridity in the Late Miocene. J. Asian Earth Sci. 193, 104290. doi:10.1016/j.jseaes.2020.104290
Hofman, C. A., Rick, T. C., Fleischer, R. C., and Maldonado, J. E. (2015). Conservation Archaeogenomics: Ancient DNA and Biodiversity in the Anthropocene. Trends Ecol. Evol. 30, 540–549. doi:10.1038/s41598-017-08757-610.1016/j.tree.2015.06.008
Hui, Z., Li, J., Xu, Q., Song, C., Zhang, J., Wu, F., et al. (2011). Miocene Vegetation and Climatic Changes Reconstructed from a Sporopollen Record of the Tianshui Basin, NE Tibetan Plateau. Palaeogeogr. Palaeoclimatol. Palaeoecol. 308, 373–382. doi:10.1016/j.palaeo.2011.05.043
Inagaki, F., Okada, H., Tsapin, A. I., and Nealson, K. H. (2005). MICROBIAL SURVIVAL: The Paleome: A Sedimentary Genetic Record of Past Microbial Communities. Astrobiology 5 (2), 141–153. doi:10.1089/ast.2005.5.141
Inagaki, F., Takai, K., Komatsu, T., Kanamatsu, T., Fujioka, K., and Horikoshi, K. (2001). Archaeology of Archaea: Geomicrobiological Record of Pleistocene thermal Events Concealed in a Deep-Sea Subseafloor Environment. Extremophiles 5 (6), 385–392. doi:10.1007/s007920100211
Jiang, X., Song, Z., Xiong, J., and Xie, Z. (2014). Can Excluding Non-insect Taxa from Stream Macroinvertebrate Surveys Enhance the Sensitivity of Taxonomic Distinctness Indices to Human Disturbance Ecol. Indicators 41 (6), 175–182. doi:10.1016/j.ecolind.2014.01.036
Leonard, D. R. P., Robert Clarke, K., Somerfield, P. J., and Warwick, R. M. (2006). The Application of an Indicator Based on Taxonomic Distinctness for UK marine Biodiversity Assessments. J. Environ. Manage. 78, 52–62. doi:10.1016/j.jenvman.2005.04.008
Li, G., Dong, H., Hou, W., Wang, S., Jiang, H., Yang, J., et al. (2016). Temporal Succession of Ancient Phytoplankton Community in Qinghai Lake and Implication for Paleo-Environmental Change. Sci. Rep. 6 (1), 19769. doi:10.1038/srep19769
Li, J., Fang, X., Song, C., Pan, B., Ma, Y., and Yan, M. (2014). Late Miocene-Quaternary Rapid Stepwise Uplift of the NE Tibetan Plateau and its Effects on Climatic and Environmental Changes. Quat. Res. 81, 400–423. doi:10.1016/j.yqres.2014.01.002
Li, J., and Fang, X. (1999). Uplift of the Tibetan Plateau and Environmental Changes. Chin. Sci. Bull. 44 (23), 2117–2124. doi:10.1007/BF03182692
Liu, S., Kruse, S., Scherler, D., Ree, R. H., Zimmermann, H. H., Stoof-Leichsenring, K. R., et al. (2021). Sedimentary Ancient DNA Reveals a Threat of Warming-Induced alpine Habitat Loss to Tibetan Plateau Plant Diversity. Nat. Commun. 12, 2995. doi:10.1038/s41467-021-22986-4
Ma, Y., Fang, X., Li, J., Wu, F., and Zhang, J. (2005). The Vegetation and Climate Change during Neocene and Early Quaternary in Jiuxi Basin, China. Sci. China Ser. D-earth Sci. 48, 676–688. doi:10.1360/03yd0110
Ma, Y. Z., Li, J. J., and Fang, X. M. (1998). Pollen Assemblage in 30.6-5.0 Ma Redbeds of Linxia Region and Climate Evolution. Chin. Sci. Bull. 43, 301–304. doi:10.1007/bf02883929
Manabe, S., and Broccoli, A. J. (1990). Mountains and Arid Climates of Middle Latitudes. Science 247, 192–195. doi:10.1126/science.247.4939.192
Panieri, G., Lugli, S., Manzi, V., Roveri, M., Schreiber, B. C., and Palinska, K. A. (2010). Ribosomal RNA Gene Fragments from Fossilized Cyanobacteria Identified in Primary gypsum from the Late Miocene, Italy. Italy. Geobiology 8 (2), 101–111. doi:10.1111/j.1472-4669.2009.00230.x
Pietramellara, G., Ascher, J., Borgogni, F., Ceccherini, M. T., Guerri, G., and Nannipieri, P. (2009). Extracellular Dna in Soil and Sediment: Fate and Ecological Relevance. Biol. Fertil. Soils 45 (3), 219–235. doi:10.1007/s00374-008-0345-8
Ramos-Robles, M., Andresen, E., and Díaz-Castelazo, C. (2016). Temporal Changes in the Structure of a Plant-Frugivore Network Are Influenced by Bird Migration and Fruit Availability. Peerj 4 (6), e2048. doi:10.7717/peerj.2048
Schadt, C. W. (2010). Soil Microbial Community Responses to Multiple Experimental Climate Change Drivers. Appl. Environ. Microbiol. 76, 999–1007. doi:10.1128/AEM.02874-09
Schelble, R. T., Hall, J. A., Nealson, K. H., and Steele, A. (2008). DNA Perseverance of Microorganisms Exposed to Silica: an Experimental Study. Geobiology 6 (5), 503–511. doi:10.1111/j.1472-4669.2008.00177.x
Wang, Y., Fang, X., Zhang, T., Li, Y., Wu, Y., He, D., et al. (2012). Distribution of Biomarkers in Lacustrine Sediments of the Linxia Basin, NE Tibetan Plateau, NW China: Significance for Climate Change. Sediment. Geology. 243-244, 108–116. doi:10.1016/j.sedgeo.2011.10.006
Xiong, J., Wu, L., Tu, S., Van Nostrand, J. D., He, Z., Zhou, J., et al. (2010). Microbial Communities and Functional Genes Associated with Soil Arsenic Contamination and the Rhizosphere of the Arsenic-Hyperaccumulating Plant Pteris Vittata L. Appl. Environ. Microbiol. 76, 7277–7284. doi:10.1128/aem.00500-10
Yang, Y., Fang, X., Galy, A., Jin, Z., Wu, F., Yang, R., et al. (2016). Plateau Uplift Forcing Climate Change Around 8.6 Ma on the Northeastern Tibetan Plateau: Evidence from an Integrated Sedimentary Sr Record. Palaeogeogr. Palaeoclimatol. Palaeoecol. 461, 418–431. doi:10.1016/j.palaeo.2016.09.002
Zachos, J., Pagani, M., Sloan, L., Thomas, E., and Billups, K. (2001). Trends, Rhythms, and Aberrations in Global Climate 65 Ma to Present. Science 292, 686–693. doi:10.1126/science.1059412
Keywords: NE Tibetan plateau, biological indicator, microbial diversity, aridification, paleoenvironments reconstructions
Citation: Xu S, Wang J, Zhang X, Yang R, Zhao W, Huang Z and Wang Y (2022) Lacustrine Sediments Bacterial Community Structure Vertical Succession of the Linxia Basin, NE Tibetan Plateau: Significance for Paleoenvironment Reconstruction. Front. Earth Sci. 9:714352. doi: 10.3389/feart.2021.714352
Received: 26 May 2021; Accepted: 17 December 2021;
Published: 11 January 2022.
Edited by:
Chunhui Song, Lanzhou University, ChinaReviewed by:
Bowen Song, China University of Geosciences Wuhan, ChinaZepeng Sun, Shanxi Agricultural University, China
Copyright © 2022 Xu, Wang, Zhang, Yang, Zhao, Huang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyong Huang, aHVhbmdfenlAdGliLmNhcy5jbg==
 Song Xu
Song Xu Jingjing Wang
Jingjing Wang Xiaoxia Zhang1,2
Xiaoxia Zhang1,2 Wei Zhao
Wei Zhao Zhiyong Huang
Zhiyong Huang Yongli Wang
Yongli Wang